-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003425
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003425Summary
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
Introduction
The Pax-Six-Eya-Dach genetic network was first identified in Drosophila as a key transcriptional regulator of compound eye development. Within this network, the Pax gene, Eyeless, is an upstream regulator of genes for the Six transcription factor sine oculis and of its co-factor Eyes absent (Eya), with feedback regulation between these genes [1], [2]. Vertebrate homologues are involved in eye development [3] but also in other developmental processes, suggesting that the mechanisms orchestrated by this genetic network are conserved and used for multiple types of organogenesis and tissue specification during embryonic development [4], [5], [6]. Indeed in Drosophila, Pox meso, dSix4 and Eya are involved in somatic myogenesis [7], [8], [9].
During vertebrate myogenesis, Pax3 and Pax7 are important upstream regulators of myogenic progenitor cell behaviour, survival and fate, as shown by genetic manipulations in the mouse embryo [5], [10]. Skeletal muscles of the trunk and limbs are derived from progenitors present in the dorsal dermomyotome domain of the somites which segment from paraxial mesoderm and mature following an anterior/posterior gradient along the axis of the vertebrate embryo. Pax3 is expressed throughout the epithelial dermomyotome and Pax7 in its central domain that will give rise to the progenitor cells of the myotome [5]. Six1/4, together with the Six co-activators, are also present in the dermomyotome together with Eya1/2, expressed at a high level in the epaxial and hypaxial domains. These Six and Eya genes have been shown to control the myogenic progenitor cell population, particularly in the hypaxial domain, where Pax3 also plays a key role in the survival and delamination/migration of myogenic progenitors. Interactions between these genes in the myogenic context, were suggested by overexpression experiments in the chick embryo, in somite explants [11] and in cell culture [12]. Analysis of compound Six1/4 and Eya1/2 mutants show that these factors regulate Pax3 in the hypaxial dermomyotome, whereas Pax3 expression is increased in the posterior dermomyotome in the absence of Six transactivation [13], [14], [15]. In the head muscles, which form from anterior unsegmented paraxial mesoderm, Pax3 is not expressed in myogenic progenitors, and Pax7 only later, whereas Six1 and Eya1 co-factors are present and active [15], [16], [17], as well as Pitx2 which acts as an upstream regulator of craniofacial myogenesis [18].
Entry into the myogenic programme, both in the head and trunk, depends on the myogenic determination factors Myf5/Mrf4 and Myod. Another member of this family of basic-helix-loop helix transcription factors, Myogenin, intervenes at the level of myogenic differentiation [19]. During the onset of myogenesis in the mouse embryo, Myf5 is expressed before Myod and in the absence of Myf5 and Mrf4 the activation of Myod is delayed [20]. In Pax3;Myf5/Mrf4 double mutants, Myod is not activated and skeletal muscle does not form in the trunk and limbs. In the absence of Pax3, the onset of myogenesis in the epaxial somite, although perturbed [21] takes place, with Myf5/Mrf4 activation through Wnt, and Shh signalling pathways [22], acting on an early epaxial enhancer of Myf5. Later activation of Myf5 in the hypaxial somite and in myogenic progenitor cells that have migrated to the limb, depends on another enhancer element which is directly regulated by Pax3 [23] and by Six1/4 [24], illustrating the synergistic action of these upstream regulators in driving the expression of myogenic determination genes. Double mutant analyses of Six1/4 and Eya1/2 show a reduction in Myod expression [15] in the somites during embryonic development and Six binding to Myod regulatory elements has been shown in cultured muscle cells [25]. This, together with the absence of Pax3 expression in the hypaxial somite observed in Six1/4 and Eya1/2 mutants, suggests that Six/Eya may intervene in the Pax3/Myod genetic cascade. Six/Eya also, unlike Pax3, are expressed in craniofacial myogenic progenitors [17], and play a role in the onset of differentiation, when they directly regulate the activation of Myogenin [26] [15], and later when they directly regulate the expression of genes coding for sarcomeric proteins [27], [28].
In this paper, we use genetic tools to further investigate how Six1/4, with the participation of Eya1/Eya2, intervene in the myogenic hierarchy. By analysis of Six/Pax and Six/Myf5 mutants, together with mutants in which a dominant negative form of the Six coding sequence has been targeted to an allele of Pax3, we show that Six1/4 are essential regulators in the Pax3/Myod genetic cascade, revealed in the absence of Myf5 at the body level. This is confirmed by the demonstration that Myod activation depends directly on Six binding to key enhancer sequences upstream of the Myod gene, which controls Myod expression in all myogenic lineages.
Results
Pax3 and Six1 act through a common genetic pathway
In order to investigate whether Pax3 and Six1 act in the same genetic pathway, we analysed Pax3/Six1 double mutants. Comparison of Six1nLacZ/nLacZ and Pax3Sp/Sp mutant embryos from the same litter shows that somite defects are similar at E11.5 and E13.5 with more cell dispersion of SixnlacZ cells, notably hypaxially, in the Pax3 mutant (Figure 1A–1I). The somitic phenotype of Six1/Pax3 double mutants is similar to that of the Pax3 mutant, but with more somite truncation at E11.5 (Figure 1J–1K), consistent with partially overlapping function of Pax3 and Six1 at this stage. At E13.5 however, the phenotype of Pax3/Six1 double mutant embryos is clearly more pronounced than either single mutant (Figure 1F, 1I, 1L), indicating that Pax3 and Six1 have separate functions at later stages. We had shown already that the expression of Eya1 and Eya2 is maintained in Six1−/− :Six4−/− mutants, and that the expression of Eya1 is preserved in the Pax3 mutant [15]. We now show that this is also the case for Eya2, which continues to be transcribed in the myogenic cells still present in somites of the Pax3 mutant (Figure 1M, 1N, 1Q, 1U). Activation of Eya1 and Eya2 is therefore independent of Six1/4 and Pax3. Furthermore, we note that the expression of Myod is only detected in Eya2 expressing cells of the somite, in the absence of Pax3 (Figure 1T, 1U, 1V), consistent with the proposed involvement of Eya co-factors acting with Six1/4, upstream of Myod during mouse embryogenesis [15].
Fig. 1. Genetic relationships between Six1 and Pax3. 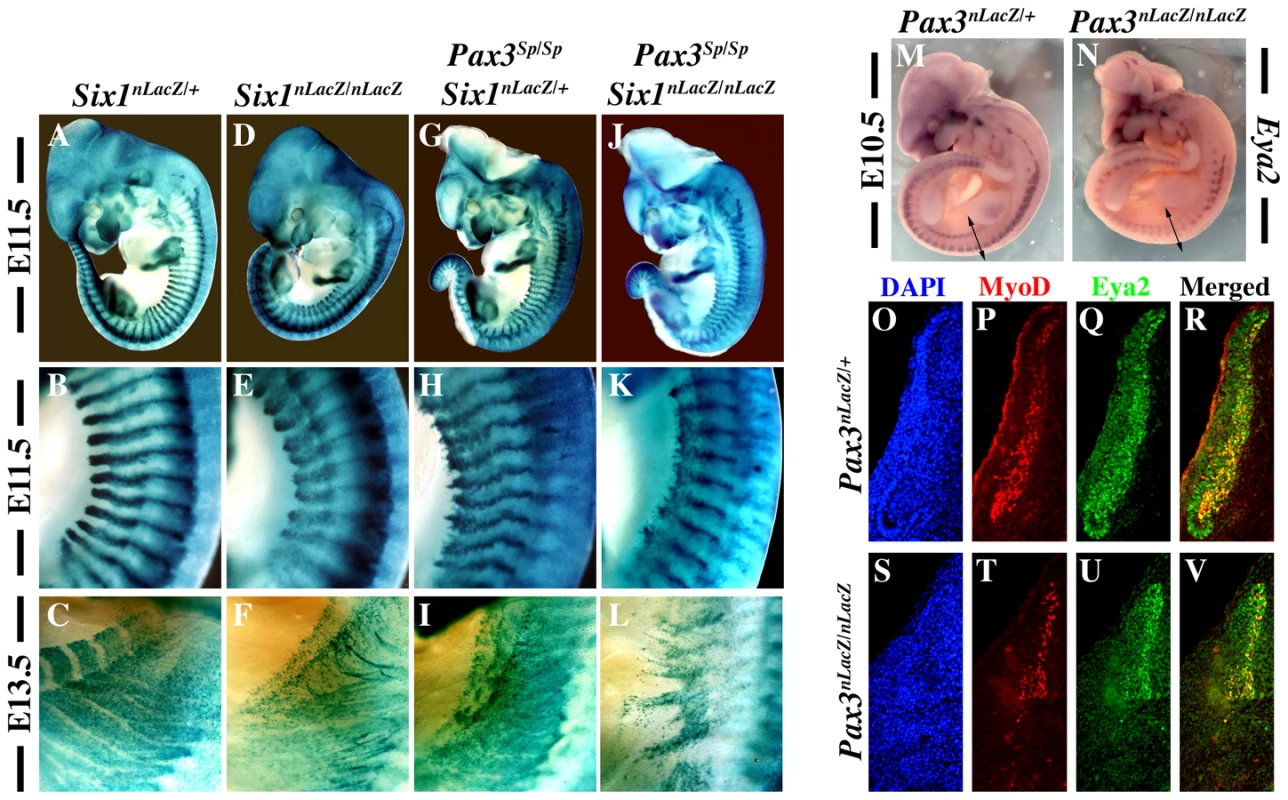
X-Gal staining of Six1nLacZ/+ heterozygous embryos on a wild type background (A–C) and on a Pax3 mutant background (Pax3Sp/Sp) (G–I) at E11.5 (A–B, G–H) and E13.5 (C, I) shows that Six1 expression, followed by the nLacZ reporter, is maintained in the absence of Pax3. Comparison of Pax3Sp/Sp : Six1nLacZ/+ embryos (G–I) with Six1nLacZ/nLacZ mutants (D–F) shows a reduction of the extent of the somite where SixnLacZ is expressed, particularly hypaxially at E11.5 (E, H). Disorganisation and loss of hypaxial muscle fibers is observed at E13.5 (F, I) in the interlimb level. These phenotypes are more severe in Pax3Sp/Sp : Six1−/− double mutants (J,K), notably at E13.5 (L). B,E,H,K and C,F,I,L show enlargements in the interlimb region. M-N, Whole mount in situ hybridization using an Eya2 probe on Pax3nLacZ/+ (M) and Pax3nLacZ/nLacZ (N) embryos at E10.5 shows that Eya2 expression is independent of Pax3. O-V, co-immunohistochemistry with Eya2 (Q,R,U,V) and Myod (P,T,R,V) antibodies on interlimb sections of Pax3nLacZ/+ and Pax3nLacZ/nLacZ embryos confirms continuing expression of Eya2 in the absence of Pax3. Reduction of Eya2 expression, notably in hypaxial lips of the somites, is due to dermomyotome reduction in the Pax3 mutant. O,S, DAPI staining. Targeting of a dominant negative form of Six4 (Six4Δ) into the Pax3 locus
To investigate the role of the Pax3-Six-Eya network in vivo while bypassing both functional compensation between genes in the same family, and potential problems of cell loss due to the function of Pax3 in cell survival, we adopted a dominant negative approach. We selected a Six coding sequence mutated in the Eya interaction domain, but nevertheless able to bind specifically to the Six (MEF3) binding site. Nuclear translocation of the co-activator Eya depends on the Six-Eya interaction which requires the N-terminal Six domain [29], [30], however this domain is also required for DNA binding specificity [31]. We therefore used a sequence encoding an alternative splice variant of Six4, Six4Δ (isolated from a mouse muscle cDNA library), which is divergent in the N-terminal-region of the conserved Six binding domain (Figure 2A). The truncated Six protein encoded by Six4Δ is still able to bind DNA, but has lost the capacity to associate with Eya2, as shown in gel mobility shift analyses (GMSA) (Figure 2B). While full length Six4 protein synergizes with Eya2 to activate MEF3 reporter activity in transient transfection assays, Six4Δ is unable to synergize with Eya2, and increasing amounts of added Six4Δ competes with the Six4-Eya2 transcription complex, leading to decreased transcriptional activation (Figure 2C).
Fig. 2. Targeting of a sequence encoding dominant negative Six4 into the Pax3 locus. 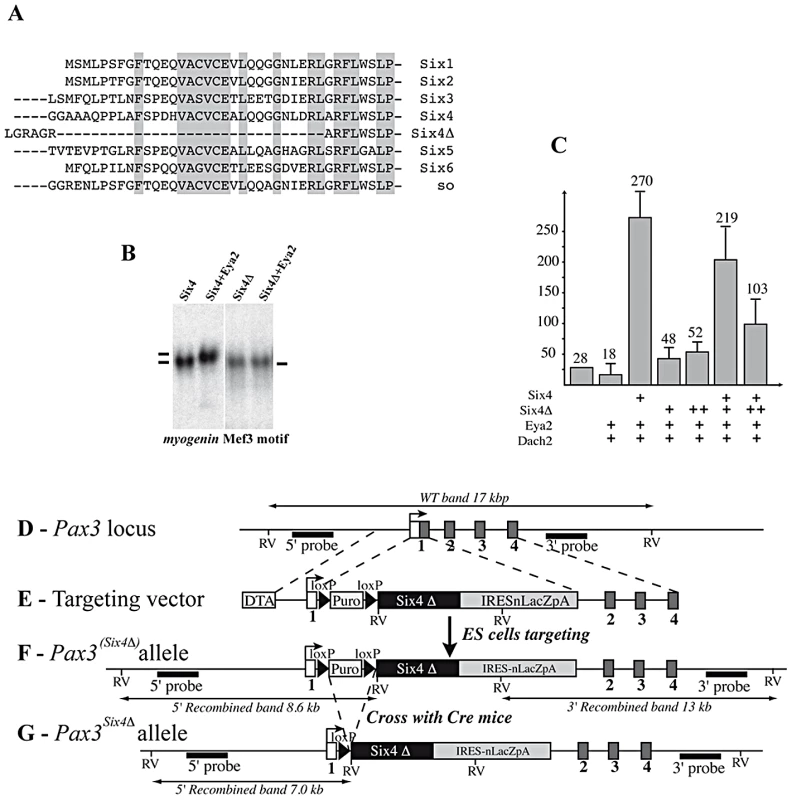
A, Alignment of Six protein sequences shows conservation of the N-terminal-most regions of the Six-domain. This region is absent in the Six4Δ mRNA splicing variant. B, Bandshift assays show that Six4 and Six4Δ bind the MEF3 site, but that only Six4 can interact with Eya2 protein to form a larger complex. C, Transfection experiments performed in primary cultures of chick myoblasts show that Six4 and Eya2 synergistically activate transcription of a luciferase reporter driven by the multimerized MEF3 sequence. In contrast, Six4Δ and Eya2 display no functional synergy, and increasing amounts of Six4Δ compete for Six4-Eya2 transcriptional activation. Y axis, ratio between Luciferase and Renilla activities in arbitrary units. D-G, Strategy for targeting the Six4Δ coding sequence into an allele of Pax3. The probes and restriction enzymes (EcoRV: RV) are indicated, with the size of the resulting wild-type and recombined restriction fragments. The targeting construct (E) contains 2.4 kb and 4 kb of 5′ and 3′ genomic flanking sequences of the mouse Pax3 gene. A floxed puromycin-pA selection marker (Puro), replaces the coding sequence in exon 1 of Pax3 (D), followed by a di-cistronic cassette containing the murine Six4Δ cDNA comprising the whole coding region, followed by an IRESnLacZ cassette and by a final pA signal. The IRESnLacZ allows easy detection of Six4Δ expression [32]. A counter-selection cassette encoding the A subunit of Diptheria Toxin (DTA) was inserted at the 5′end of the vector. After homologous recombination in embryonic stem (ES) cells, Six4Δ-IRESnLacZ expression from the Pax3(Six4Δ-IRESnLacZ) allele is blocked by the floxed puromycin-pA cassette (F) and is therefore conditional to removal by crossing with a PGK-Cre mouse [52]. This generates the Pax3Six4Δ-IRESnLacZ allele (abbreviated Pax3Six4Δ) (G). We targeted the Six4Δ sequence into an allele of Pax3, to evaluate the function of the Six-Eya interaction during myogenesis in vivo. To avoid potential problems of lethality, we used a conditional strategy (see Figure 2D–2G), similar to that previously reported [32], with an IRESnLacZ reporter following the Six4Δ sequence, to monitor expression. X-Gal staining revealed correct expression of the reporter at E10.5 when compared to embryos where an nLacZ reporter is targeted into an allele of Pax3 (Pax3IRESnLacZ/+, abbreviated Pax3ILZ/+) (Figure 3A–3B). This was also the case after Pax3 in situ hybridization, compared to wild type (WT) embryos (data not shown, and see [32]). Despite robust Six4Δ-IRESnLacZ expression, these embryos did not present any obvious defects and indeed Pax3Six4Δ-IRESnLacZ/+ (abbreviated Pax3Six4Δ/+) mice are viable and fertile.
Fig. 3. Expression of Six4Δ does not perturb normal embryonic development nor rescue Pax3 mutant deficiencies. 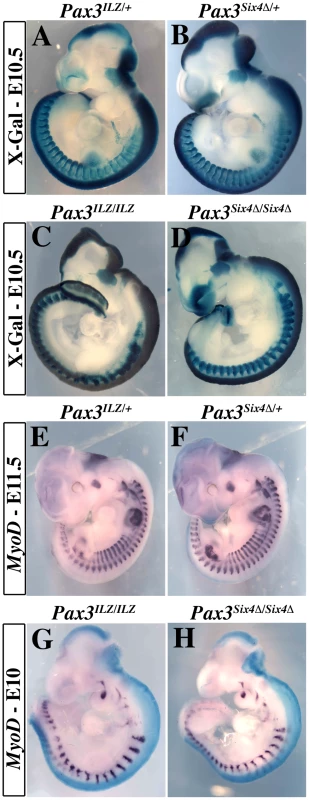
A–B, X-Gal staining of Pax3IRESnLacZ/+ (Pax3ILZ/+, A) and Pax3Six4Δ/+ (B) embryos at E10.5 demonstrates correct expression of the Six4Δ transgene. C–D, X-Gal staining of homozygotes Pax3ILZ/ILZ (C) and Pax3Six4Δ/Six4Δ (D) embryos at E10.5 demonstrates that the Six4Δ sequence does not rescue deficiencies due to the absence of Pax3 (Exencephaly, spina bifida, lack of limb muscles, somitic defects and neural crest cell deficiencies). E–F, Whole mount in situ hybridization using a Myod probe on Pax3ILZ/+ (E) and Pax3Six4Δ/+ (F) embryos at E11.5 shows that the Six4Δ sequence does not overtly perturb Myod expression. G–H, Whole mount in situ hybridization using a Myod probe on homozygote Pax3ILZ/ILZ (G) and Pax3Six4Δ/Six4Δ (H) embryos at E10 shows that the onset of Myod expression is similar to that of a Pax3 mutant, in the absence of Pax3 but in the presence of a dominant negative Six4Δ (H). We went on to test whether expression of Six4Δ driven by Pax3 was able to rescue aspects of the Pax3 mutant phenotype. X-Gal staining of Pax3ILZ/ILZ or Pax3Six4Δ/Six4Δ homozygote embryos at E10.5 (Figure 3C–3D) showed the same defects previously reported for Pax3 mutant mice (dorsal neural tube, neural crest and myogenic defects). From these data, we conclude that expression of Six4Δ under Pax3 regulation does not perturb normal embryonic development nor rescue Pax3 deficiencies.
We next examined Myod expression in Pax3Six4Δ/+ embryos using in situ hybridization. These experiments did not reveal any perturbation in Myod transcription (Figure 3E–3F). Comparison of Myod transcription in Pax3 mutant embryos and in Pax3Six4Δ/Six4Δ homozygote embryos indicated that expression of Six4Δ does not prevent Myod expression and myogenesis when the Myf5/Mrf4 myogenic pathway is active (Figure 3G–3H). The decreased expression of Myod observed in Pax3Six4Δ/Six4Δ embryos is similar to that observed in Pax3ILZ/ILZ embryos, as a result of cell death in the absence of Pax3 (data not shown).
Expression of Six4Δ in vivo specifically impairs the Pax3-mediated myogenic pathway
In order to determine if Six-Eya lies in the Pax3-Myod myogenic pathway, we crossed the Pax3Six4Δ/+ mice with Myf5nLacZ/+ (abbreviated Myf5+/−) mice [33]. In wild type embryos, Myod expression is initiated around E10 in the hypaxial domain of thoracic somites [34]. However, in Myf5 mutant embryos, Myod expression is delayed by about 24 h. Muscle formation is normal at later stages, indicating that Myod is able to rescue myotome formation in the absence of Myf5 after E11.5 [20]. In Myf5+/− and Myf5+/−: Pax3Six4Δ/+ embryos, Myod is activated normally and by E11.5 Myod expression is seen throughout the myotome (Figure 4A–4B, 4E–4F). As previously shown (Tajbakhsh et al., 1997), in Myf5 mutant embryos (Myf5−/−, Figure 4C, 4G) Myod is activated later in the muscle precursor cells which are blocked in the epaxial and hypaxial somite, and at E11.5 the hypaxial part of the myotome is partially rescued (Figure 4G, arrowheads). In contrast, in Myf5−/−: Pax3Six4Δ/+ embryos, Myod expression is significantly reduced in epaxial and, notably, in hypaxial muscle precursor cells at E11.5 (Figure 4D, 4D′, 4H, 4H′). This impaired Myod expression leads to only partial rescue of myotome development; the cells that are expressing Myod in the hypaxial domain, despite activation of myogenic differentiation genes, like Myogenin, remain restricted to this part of the somite (data not shown). At E12.5, trunk muscles still show some disorganisation in Myf5−/− embryos, but this is more pronounced in Myf5−/−: Pax3Six4Δ/+ embryos (Figure 5A–5D, 5A′–5D′). By E14–E14.5, myogenesis is rescued in Myf5−/− fetuses (Figure 5G–5G′, 5K–5K′). In contrast, Myf5−/−: Pax3Six4Δ/+ fetuses display a reduction in trunk muscles (Figure 5H–5H′) which is more severe than in Myf5−/−:Pax3+/− embryos at this stage (Figure 5L–5L′). These results indicate that Six/Eya intervene in the Pax3-dependent pathway of Myod activation.
Fig. 4. Six4Δ affects Myod expression and myogenesis in the absence of Myf5. 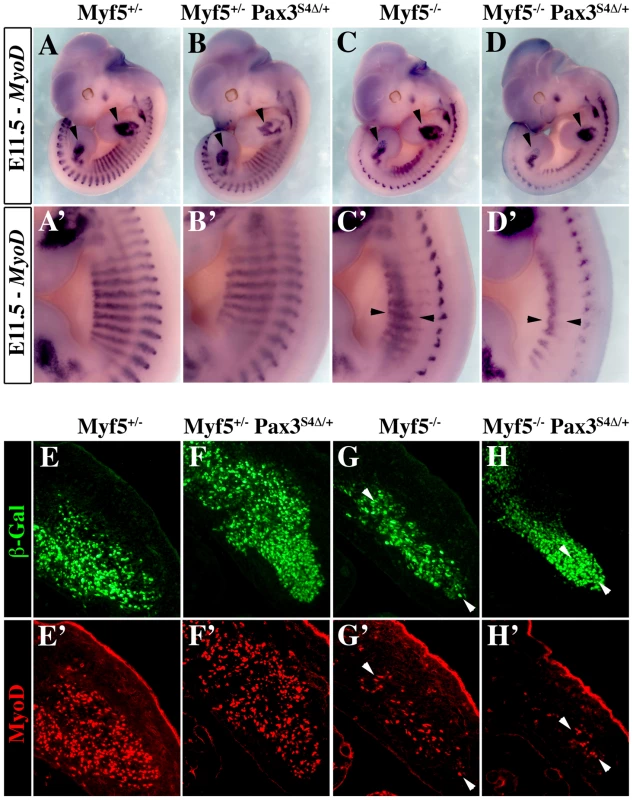
A–D′, Whole mount in situ hybridization experiments using a Myod probe on Myf5+/− (A, A′), Myf5+/− : Pax3Six4Δ/+ (B, B′), Myf5−/− (C, C′) and Myf5−/− : Pax3Six4Δ/+ (D, D′) embryos at E11.5. At this stage, in Myf5−/− embryos (C, C′), Myod is activated and begins to rescue the formation of the myotome (arrowheads in C′). However, in Myf5 deficient embryos which express Six4Δ under the control of Pax3 regulatory elements, Myf5−/− : Pax3Six4Δ/+ (D, D′), Myod expression is reduced, affecting the rescue of myotome formation (D′, arrowheads). In contrast, in thoracic somites of Myf5+/− : Pax3Six4Δ/+ (B, B′) Myod expression is not altered compared to Myf5+/− embryos (A,A′). A′–D′, show enlargements in the interlimb region of A–D. E–H′, co-immunohistochemistry on transverse sections of hypaxial somites from Myf5+/− (E, E′), Myf5+/− : Pax3Six4Δ/+ (F, F′), Myf5−/− (G, G′) and Myf5−/− : Pax3Six4Δ/+ (H, H′) embryos at E11.5 using anti-β-Galactosidase (β-Gal) (green, E–H) and anti-Myod (red, E′–H′) antibodies confirms the severe reduction of Myod expression in Myf5−/− : Pax3Six4Δ/+ (H, H′) embryos. Arrowheads indicate examples of cells in which the β-Gal reporter from the Myf5nLacZ allele is expressed and which co-express Myod. Fig. 5. Impaired myogenesis in the presence of Six4Δ, in the absence of Myf5. 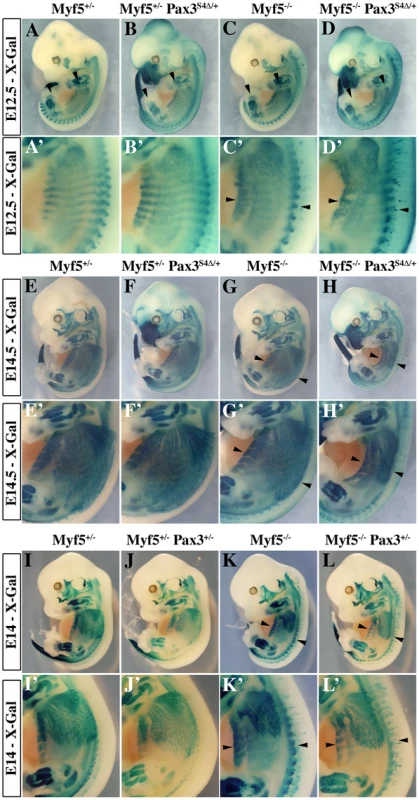
A–L′; X-Gal staining of E12.5 (A–D′), E14.5 (E–H′) or E14 (I–L′) Myf5+/− (A, A′, E, E′, I, I′), Myf5+/− : Pax3Six4Δ/+ (B, B′, F, F′), Myf5−/− (C, C′, G, G′,K, K′) and Myf5−/− : Pax3Six4Δ/+ (D, D′, H, H′), Myf5+/− : Pax3+/− (J, J′), Myf5−/− : Pax3+/− (L, L′) embryos demonstrates that in Myf5 deficient embryos which express Six4Δ under the control of Pax3 regulatory elements, the localisation of myogenic cells, marked by the Myf5nLacZ reporter is impaired, notably in trunk muscles (H′ compared with L′). A′–D′, E′–H′, and I′–L′ are enlargements in the interlimb region of A–D, E–H and I–L respectively. Myf5 is required for Myod activation in the absence of Six1 and Six4
We had previously shown that Myod expression is severely compromised in Six1−/−/Six4−/− double mutant embryos [13]. In these embryos, Pax3 expression is maintained in anterior and posterior domains of the dermomyotomes, while impaired in the epaxial and hypaxial domains. Early Myf5 expression is detectable, although decreased [13]. To test whether the remaining expression of Pax3 and/or Myf5 is responsible for the somitic activation of Myod observed in Six double mutant embryos, we examined Myf5−/−:Six1−/−/Six4−/− and Pax3sp/sp:Six1−/−/Six4−/− embryos. As shown in Figure 6, the expression of Myod is higher in Pax3 mutants (Pax3sp/sp) compared to Six1/Six4 double mutant embryos. Myod expression is still detectable, although decreased, in Pax3sp/sp:Six1−/−/Six4−/− embryos at E11.5. In contrast, Myod transcripts are not detectable in the somites of Myf5−/−:Six1−/−/Six4−/− embryos at E11.5 (Figure 6H), where Myod expression persists in the branchial arches (Figure 6H). X-Gal staining of compound Myf5−/−:Six1−/−/Six4−/− embryos at E12.5 further illustrates lack of axial myogenesis at a later stage (revealed by Myf5-LacZ and Six1-LacZ), while craniofacial musculature is still present (Figure 6L). Loss of trunk muscles is confirmed by desmin immunohistochemistry on E12.5 sections (Figure 6i′–6l′). The presence of myogenic desmin-positive cells in extra-ocular and masseter muscles (Figure 6i″–6l″, Figure S1) shows that craniofacial myogenesis is not abrogated in Myf5−/−:Six1−/−/Six4−/− embryos.
Fig. 6. Axial Myod expression is lost in Myf5−/−:Six1−/−/Six4−/− embryos. 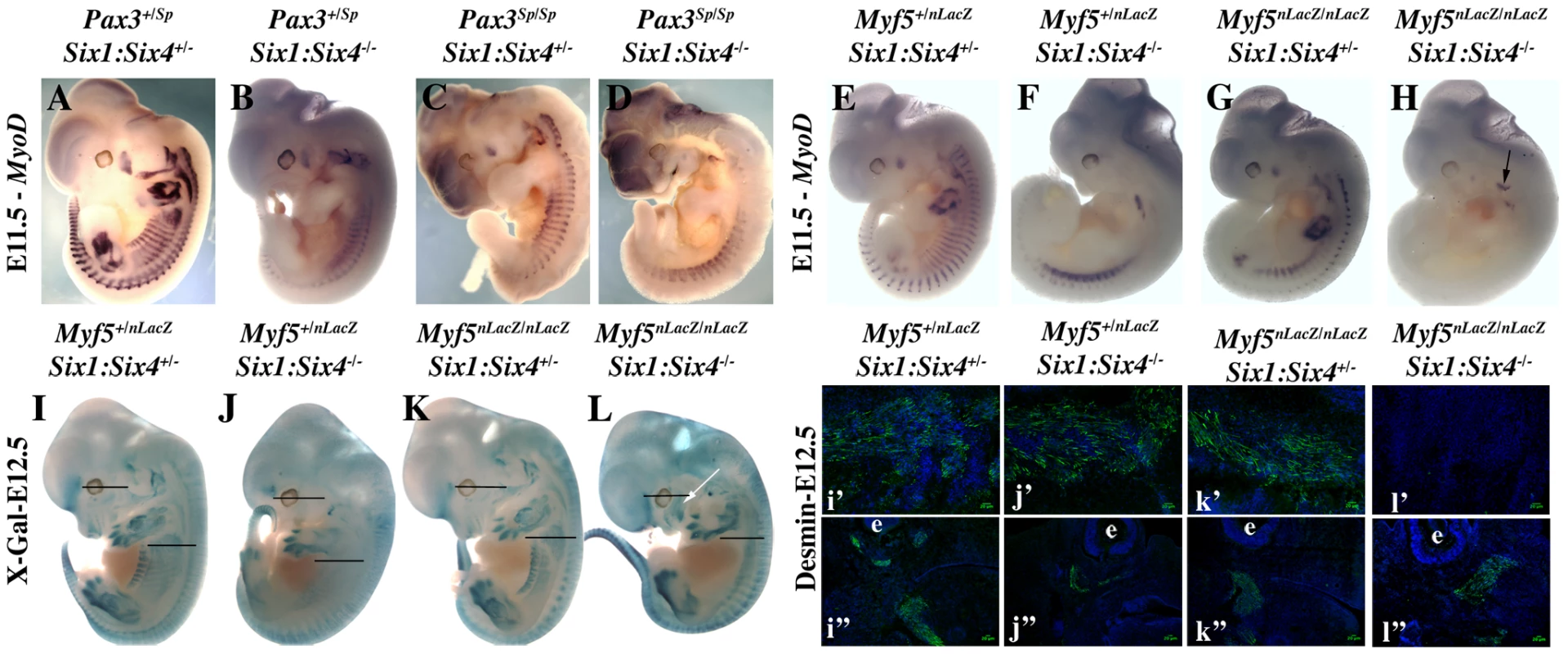
A–H, Whole mount in situ hybridization using a Myod probe, I–L, X-Gal staining, and i′–l′, i″–l″ immunohistochemistry on sagittal sections of I–L embryos at the interlimb somites or head level using Desmin antibodies, at E11.5 (A–H) or E12.5 (I–L) with Pax3+/SpSix1+/−Six4+/− (A), Pax3+/SpSix1−/−Six4−/− (B) Pax3Sp/SpSix1+/−Six4+/− (C), Pax3Sp/SpSix1−/−Six4−/− (D), Myf5+/−Six1+/−Six4+/− (E, I), Myf5−/−Six1+/−Six4+/− (F, J) Myf5+/−Six1−/−Six4−/− (G, K), Myf5−/−Six1−/−Six4−/− (H, L) embryos, showing the role of Pax3/Six proteins and Myf5 acting upstream of Myod during trunk myogenesis. Desmin expression in E12.5 compound embryos at the axial level (i′–l′) and at the head level (i″–l″) is not detected in Myf5−/−Six1−/−Six4−/− embryos at the axial level (l′) but at the head level (l″), showing that craniofacial myogenesis can take place in this compound mutant. e: eye. White arrow in L shows the presence of craniofacial muscles. Six proteins directly activate Myod regulatory elements
Myod expression has been shown to be under the control of at least three separate DNA elements, a promoter region, a distal regulatory region (DRR), 6 kb 5′ of the transcription start site [35], and a core enhancer (CE) region, located 20 kb 5′ of the transcription start site (TSS) [36]. Both the CE and the DRR drive expression of a LacZ reporter to sites of myogenesis in transgenic embryos, where the CE shows a higher and more precocious activity [35], [37]. A specific deletion of either enhancer by the CREloxP system indicates functional redundancy [38], [39]. Both CE and DRR elements have been shown to bind Six1 and Six4 homeoproteins in growing and differentiating cells in the C2 muscle cell line [25]. Furthermore, both the CE and DRR are bound by Eya proteins in vivo, as shown by ChIP experiments on Pax3-GFP positive cells purified by flow cytometry from E11.5 embryos (Figure 7A and data not shown). One MEF3 element that binds Six proteins, is present in the DRR (5′TCcGGTTTC, which is conserved in the human sequence), and two in the CE (5′TaAaaTTaC, corresponding to part of the conserved box4 of the human sequence, shown to affect activity of the human enhancer in transgenic embryos [37], and 5′TCcGGTTTC, overlapping boxes 15 and 16 of the human CE sequence) (Figure 7B). These potential MEF3 sites bind Six1 and Six4 proteins, as shown in gel mobility shift experiments (Figure 7C). We next tested these sites for function in transgenic embryos. For these experiments we constructed a transgene in which the Myod CE sequence was inserted 5′ of the −5.8 kb flanking sequence of Myod, 340 bp upstream of the DRR element [35] to give a CE-MD5.8-LacZ transgene (Figure 7D). 6 out of 10 CE-MD5.8-LacZ transgenic embryos show X-Gal staining similar to that of endogenous Myod expression at E12.5 (Figure 7E and data not shown). Mutation of the three MEF3 sites compromised transgene activity, such that only 3 out of 8 transgenic embryos carrying the mutant sequences (mut3MEF3-CE-MD5.8-LacZ) show any LacZ expression at E12.5. In two of them, very low expression is detected in myogenic territories at the thoracic and limb levels (Figure 7E, 7Fe,e′,f,f′), while in the third (Figure 7E, 7Fd,d′, Figure S2) most of the LacZ transgene expression does not overlap with endogenous Myod expression. Expression in head muscles is detected with all wild type transgenes in Myod expressing cells (Figure 7E, 7Fc″,c′″, Figure S2). This is not the case with mutant transgenes, where most Myod expressing cells at the temporalis muscle or eye level do not express the mutant transgene (Figure 7E, 7F d″–f″; d′″–f′″, Figure S2). Sections of wild type and mutant embryos are shown in Figure 7F at trunk (left panels) and head (right panels) levels. Wild type Myod transgenes drive the expression of the LacZ reporter in 64 to 95% of Myod-positive cells, while mutant transgenes drive low expression of the LacZ reporter in 3 to 10% of Myod-positive cells. Expression is never detected in the tail somites in the posterior part of the embryo with the mutant transgene. Taken together, these experiments demonstrate a direct function of the Six binding sites in the activation of Myod during myogenesis in the embryo both in the trunk and head.
Fig. 7. Six proteins are required for Myod expression in the mouse embryo. 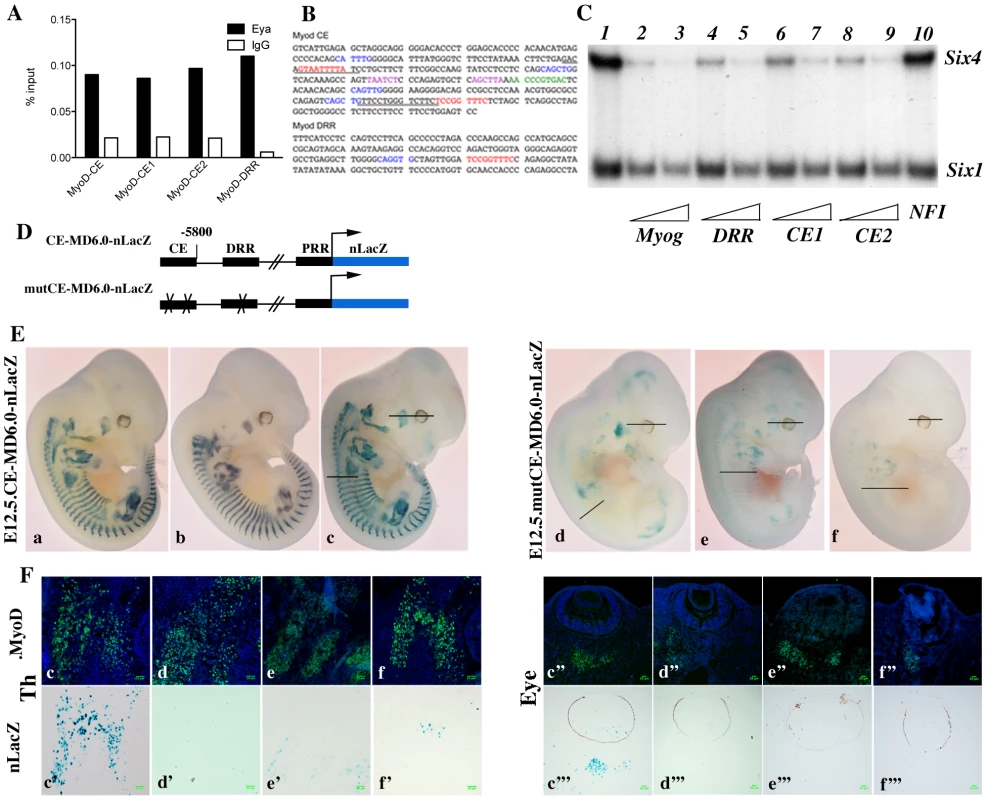
A- Chromatin Immunoprecipitation (ChIP) experiments performed with Eya antibodies or control IgG, on chromatin prepared from Pax3-GFP cells separated by flow cytometry from the trunk region of Pax3GFP/+ embryos [32] at E11.5. ChIP experiments reveal association of Eya proteins with the core enhancer (CE) and distal regulatory region (DRR) 5′ of the Myod gene. B- Sequence of mouse Myod core enhancer (CE) and DRR. MEF3 sites are in red, E boxes in blue, Pitx sites in purple and Pax3 site in green. Underlined sequences correspond to the LS4 and LS15 linker-scanner mutagenesis performed on the human core enhancer [37]. C- Electromobility shift assays showing the interaction of Six1 and Six4 proteins with three distinct MEF3 DNA elements present in the regulatory regions of Myod. Radioactively labelled oligonucleotides with the Myogenin MEF3 site (Myog) were incubated with in vitro translated Six1 and Six4 proteins as a control (1). A 60 or 300 fold excess of unlabelled oligonucleotides containing the MEF3 Myogenin site (2,3), the MEF3 DRR site (4,5), the MEF3 CE1 site (6,7), the MEF3 CE2 site (8,9) or a 300 fold excess of unrelated Myogenin NFI oligonucleotides (10) were added in the reaction mix. D- Wild type and MEF3 mutant Myod transgenes used in the study, (not to scale). PRR, proximal regulatory region corresponding to the Myod promoter. E-Transient transgenic embryos with wild type or mutant Myod sequences at E12-E12.5. X-Gal staining of transgenic embryos with wt CE-MD6.0-nLacZ (a–c) or mut3MEF3-CE-MD6.0-nLacZ (d–f) transgenes. Six out of ten wild type transgenes expressed the LacZ reporter with the same expression pattern, three of them are shown. The number of transgenes inserted varied between 3 and 34 for X-Gal-positive (X-Gal+) embryos, and from 1 to 14 for X-Gal-negative (X-Gal−) embryos. Three out of eight mutant transgenic embryos expressed the LacZ reporter, all three are shown. The number of transgenes inserted was 23 (f), 39 (d) and 40 (e) for X-Gal+ embryos, and from 1 to 51 for X-Gal− embryos. F- Sections for one wild type (c) and for the three mutant transgenic embryos expressing the LacZ transgene were analysed for Myod protein by immunohistochemistry at the thoracic (Th) (c–f), and eye (c″–f″) levels to detect myogenic cells, thus revealing the % of transgene expression (X-Gal+ cells, c′–f′ and c′″–f′″) in the myogenic cell population (Myod-positive cells). While most Myod+ cells express the wt Myod transgene (c′, c′″), very few are marked by expression of the mutant Myod transgene (d′–f′, d′″–f′″). Six2 is expressed in myogenic territories in the embryo
To explain the discrepancies observed between mut3MEF3-CE-MD5.8-LacZ expression and the expression of Myod in Six1/Six4 mutant embryos, we looked for other Six genes expressed in myogenic territories during embryogenesis [40] that could be responsible for the rescue of Myod expression observed in Six1/Six4 embryos at the epaxial and craniofacial levels. Six2 [41] and Six5 [42], [43], [44] are the two other Six genes expressed in myogenic cells. By whole mount in situ hybridization, we further show that Six2 is expressed in the first branchial arch of E9.5 embryos, and also in the dorsal regions of newly formed somites, where early epaxial Myf5 is first activated (Figure 8A). Both Six2 and Six5 bind efficiently to the three Myod MEF3 elements, as determined by gel mobility shift experiments (Figure 8B). We next isolated chromatin for ChIP experiments to check if Six2 binds in vivo on Myod regulatory elements. With wild type embryos we did not observe significant binding. However with chromatin from E12 Six1/Six4 mutant embryos we observed efficient binding of Six2 on Myod CE and DRR elements, demonstrating that Six2 can bind to Myod regulatory elements in the embryo (Figure 8C). We examined Six2 protein in the masseter muscle of Six1−/−/Six4−/− and Myf5−/−:Six1−/−/Six4−/− mutant embryos by immunocytochemistry at E12.5 and show that it co-localizes with Myod protein (Figure 8D). These results indicate that Six2 is a good candidate for the activation of Myod expression in the absence of Six1 and Six4. They also suggest that Six2 is upregulated under these conditions (Figure 8C, 8D).
Fig. 8. Six2 proteins bind Myod regulatory elements. 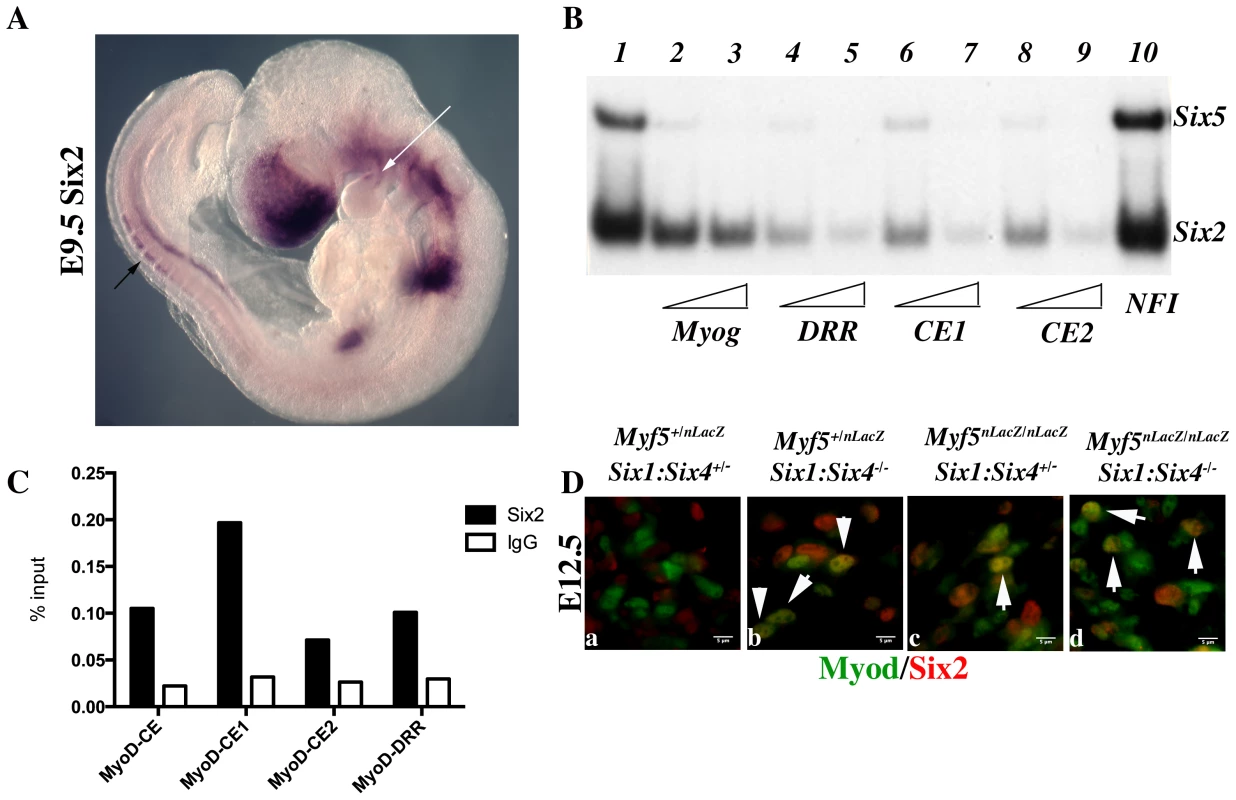
Whole-mount in situ hybridization using a Six2 probe on an E9.5 embryo. Note Six2 expression in dorsal aspects of newly formed somites (black arrow), and in the center of the first branchial arch (white arrow). B- Electromobility shift assays showing the interaction of Six2 and Six5 proteins with three distinct MEF3 DNA elements present in the regulatory regions of Myod. Radioactively labelled oligonucleotides with the Myogenin MEF3 site (Myog) were incubated with in vitro translated Six2 and Six5 proteins as a control (1). A 60 or 300 fold excess of unlabelled oligonucleotides containing the MEF3 Myogenin site (2,3), the MEF3 DRR site (4,5), the MEF3 CE1 site (6,7), the MEF3 CE2 site (8,9) or a 300 fold excess of unrelated Myogenin NFI oligonucleotides (10) were added in the reaction mix. C- Chromatin Immunoprecipitation (ChIP) experiments performed with Six2 antibodies or control IgG, on chromatin prepared from E12 Six1−/−Six4−/− embryos, and showing binding of Six2 in vivo on the regulatory elements of Myod. D- Immunocytochemistry performed with Six2 and Myod antibodies on E12.5 Myf5+/−Six1+/−Six4+/− (a), Myf5+/−Six1−/−Six4−/− (b) Myf5−/−Six1+/−Six4+/− (c), Myf5−/−Six1−/−Six4−/− embryos (d) at the masseter level, demonstrating Six2 (red) accumulation in Myod-positive (green) cells (white arrowheads). Discussion
We show that Six1/4 play an essential role in the Pax3/Myod genetic pathway that regulates the onset of myogenesis [20]. This is revealed on a Myf5 mutant background. Since the Myf5 mutation that we use also affects Mrf4, entry into the myogenic program depends entirely on the myogenic determination factor Myod in the absence of Myf5/Mrf4 [45]. We illustrate with the Six4Δ sequence that Eya co-activators are required for Six transactivation, as previously shown [15], [30]. Furthermore our results show that key enhancer sequences of the Myod gene are directly regulated by MEF3 sites that are required in vivo at all sites of myogenesis to control Myod expression through the recruitment of Six1, Six2 and Six4 transcription factors.
The Pax/Six/Eya pathway to tissue specification is therefore important for the formation of skeletal muscle in the mouse embryo. However this network appears to be more complex than the Eyeless/sine oculis/Eyes absent cascade that leads to eye formation in Drosophila [1], [5]. As we show here for Six1 and Eya2, their activation takes place in the absence of Pax3, whereas Eyeless initiates the cascade in Drosophila. In the mouse somite, Pax7 is also expressed in the central domain of the dermomyotome and may compensate. However, prior to the extensive cell death seen in the hypaxial somite in the absence of Pax3, Six1/4 genes are transcribed. Furthermore during craniofacial myogenesis, the Six1 gene and genes for Eya co-factors are expressed [41], [46] and the polyMEF3-LacZ reporter of Six transcriptional activity is high [15], in the absence of Pax3 that is not expressed during head myogenesis [5]. In Six1−/−/Six4−/− or Eya1−/−/Eya2−/− double mutants, Pax3 expression is compromised in the hypaxial domain [15] indicating that Six/Eya can also regulate Pax3. Our analyses of Six1−/− and Pax3−/− mutants shows that they have overlapping but not identical myogenic phenotypes, confirmed by the double mutant phenotype which is more severe, particularly at later stages.
The Six4Δ sequence, which we targeted to an allele of Pax3, encodes a protein that still binds DNA, but does not bind Eya and is transcriptionally inactive, thus acting as a dominant-negative factor. The effectiveness of its action will depend on competition with wild type Six factors present in Pax3 expressing cells. By diminishing the effects of Six factors (Six2 and Six5, also expressed at sites of myogenesis [41] [42], as well as Six1 and Six4), it serves as a probe, under conditions that are less radical than double mutants. This type of strategy, with a Pax3Pax3-En/+ mouse line had previously proved valuable for probing Pax3 function [23]. In the absence of Myf5/Mrf4, when Six4Δ is present, down-regulation of Myod expression is clearly observed, under conditions in which somites are less perturbed, at E11.5–12.5. Later, the failure of skeletal muscles to develop leads to severe perturbations at sites of myogenesis in the trunk. Head musculature on the other hand appears normal, as do the forming limb muscles. In Six1−/−/Six4−/− double mutants, in the absence of Myf5/Mrf4, Myod is not transcribed in the trunk and limbs and myogenesis does not occur, whereas Myod transcripts are detectable in head muscle progenitors and muscle markers are present. These observations show that head myogenic progenitors, that are not derived from the somites, activate Myod and form head muscles in the absence of Six1 and Six4 [13]. This is in contrast to a report on zebrafish where Six1a was found to be essential for craniofacial myogenesis [16], and with reduced head myogenesis observed in compound Six1;Eya1 mutant mice [17]. In addition to Six1 and Six4, Six2 and Six5 genes are also transcribed in myogenic cells in the mouse embryo [41], [42], and we now show that Six2 can regulate Myod expression. Other transcriptional regulators, such as Pitx2, play an important upstream role in head myogenesis. Pitx2 has been shown to activate Myod in the trunk [47], where Pitx2 lies genetically downstream of Pax3. However in the head, where Pax3 is not expressed at the onset of myogenesis [18], Pitx2 acts independently. In keeping with this, Pax3/Mrf4/Myf5 triple mutants do not have defects in Myod activation and myogenesis in the head. However Myod is not activated in the trunk where skeletal muscles do not form [20]. In this context, Six genes do not rescue the phenotype.
Activation of Myod relies on two enhancer elements at 5 kb (DRR) and 20 kb (CE) upstream of the gene, as well as on the proximal promoter [35], [36]. In adult myogenic cells, Pax7 activates the promoter [48] and Pax3/7 have been shown to bind the CE in myogenic cell cultures [49], but there are no data on such a role of Pax3/7 in the embryo. The CE is an important regulator of embryonic Myod expression, but the DRR is also implicated in this activity. When the CE is deleted, delayed Myod expression is still observed, notably in the branchial arches and limb buds [39]. Deletion of the DRR does not abolish embryonic Myod expression [38], in keeping with the important role of the CE. We identify three separate DNA elements in the CE and DRR of Myod that are bound by Six1 and Six4 [25], and bound in vivo by Eya. In a transgene controlled by the proximal promoter, DRR and CE, we show expected expression of the nLacZ reporter at all sites of myogenesis in E12.5 embryos. When the Six/MEF3 binding sites are mutated, this activity is mainly lost, with low level expression at sites of myogenesis, in a few Myod-positive cells. These results show that Six transactivation is required for the function of these regulatory elements. Residual activity may be due to Myf5 activation of Myod regulatory sequences, through E-boxes that are also known to play an important role [37], [50]. Indeed our genetic experiments, which show that a major effect on Myod activation in the Six1/4 double mutant is only seen in the absence of Myf5, are in keeping with this. In Myf5/Mrf4 mutant embryos, Pax3-dependent rescue of CE enhancer activity is observed [39], potentially due to Six transactivation acting in the Pax3/Six/Myod pathway. In the linker scanning experiments where human MYOD CE elements were sequentially mutated [37], box 4 was found to be essential for expression in all skeletal muscle lineages. This sequence contains the Six binding site CE1. In contrast mutation of box 16 which contains our box CE2 did not lead to loss of activity, demonstrating that CE1 is the main functional MEF3 site [37].
In our transgenic analysis, mutation of Six/MEF3 sites leads to loss of transgene expression in most embryos, at sites of myogenesis in the head, as well as in the trunk. This contrasts with our findings with Six1−/−/Six4−/− and Myf5−/−:Six1−/−/Six4−/− mutants. An explanation for these discrepancies is that other Six proteins known to be expressed in myogenic cells compensate in some embryonic territories for the lack of Six1 and Six4. We provide evidence that Six2 may play such a role since it is expressed at myogenic sites (Figure 8A and [41]). In the Six1−/−/Six4−/− double mutant, Six2 is expressed in Myod-positive cells and binds the Myod regulatory elements. We have not been able to examine Six5 in this context due to the lack of appropriate antibodies.
We conclude that during skeletal muscle formation in the trunk the Pax3 genetic cascade that leads to Myod activation functions through Six genes and that in the absence of Myf5/Mrf4, the Six transactivation complex plays a key role in the activation of myogenesis. During the onset of craniofacial myogenesis, where Pax transcription factors do not play a role, Six expression is also a key determinant for Myod activation (Figure 9). Our analysis of the Pax/Six/Eya genetic cascade in the context of myogenesis has implications for the derivation of skeletal muscle from stem cell populations [44] and also, more generally, for other examples of tissue specification and organogenesis in vertebrates that also employ this genetic network [5].
Fig. 9. Schematic representation of genetic networks that activate Myod during myogenesis in the trunk and head. 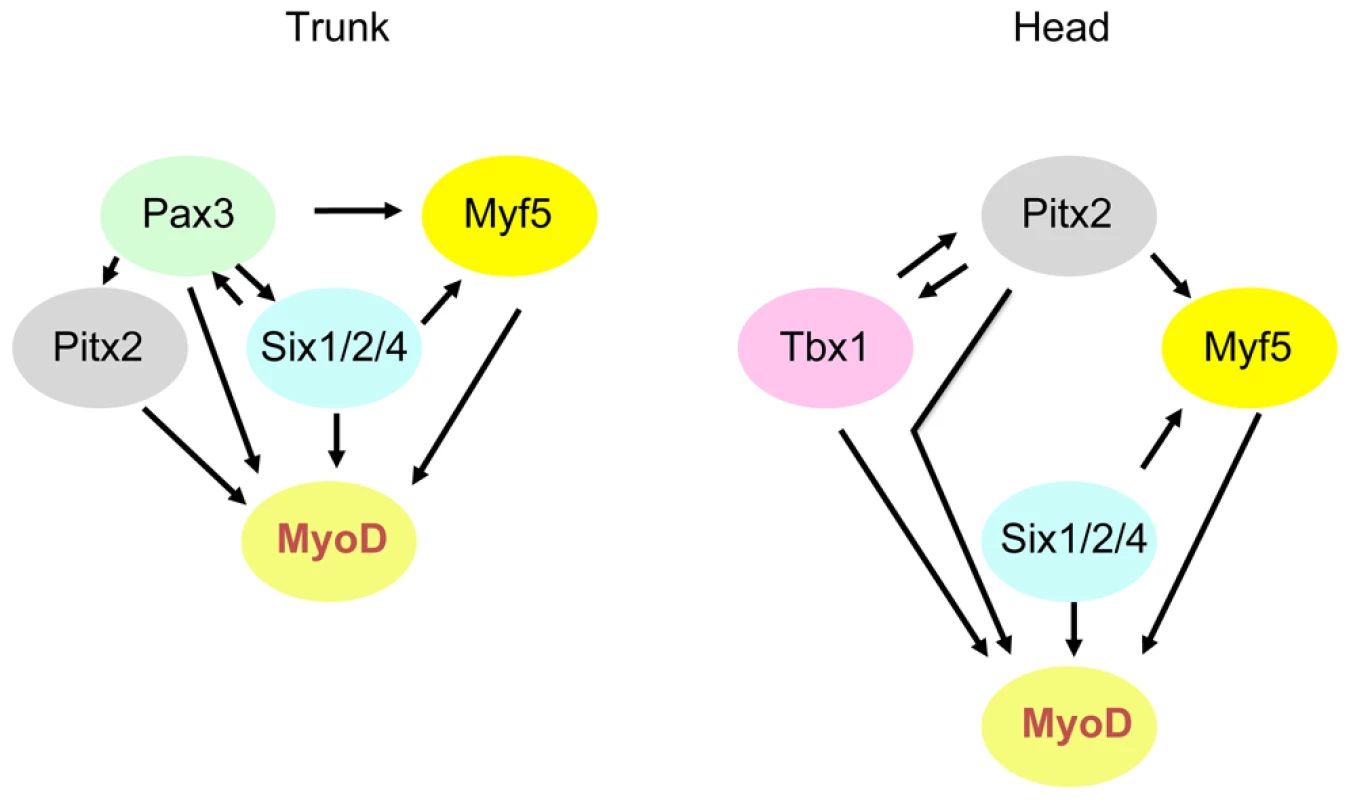
Variations in the interactions of factors in sub-domains at different developmental stages are not included here. Materials and Methods
Cloning, targeting vectors, and mice
Six4 and Six4Δ cDNAs were obtained by screening a λgt11 library from adult mouse muscle (Clontech) [26]. The Pax3Six4Δ/+ construct is derived from a construct previously reported [32]. The Pax3Six4Δ-ILZ) allele contains 2.4 kb of 5′ Pax3 genomic region, in which the coding sequence of exon 1 is replaced by targeted sequences and 4 kb of 3′ sequence containing exons 2–4. The genomic sequences surround a Floxed Puromycin (Puro) cassette followed by 2.6 kb of Six4ΔcDNA, then an IRESnLacZ cassette. In addition, a PGK-DTA cassette encoding the A subunit of the Diptheria toxin gene (DTA) was inserted at the 5′ end of the construct to allow negative selection in ES cells. The targeting vector was electroporated into CK35 ES cells [51]. ES cells were selected and screened for recombination events by Southern blot analysis using EcoRV (RV in Figure 2) digests and a 5′-flanking probe (Figure 2E). Targeted ES cells were recovered with a 0.5–1% frequency and injected into blastocysts to generate chimaeras. Germline tramsmitted alleles were identified by the classical Splotch (Pax3Sp/+) heterozygote phenotype (lack of melanocyte colonization of the belly), and by PCR or by Southern blotting. PGK-Cre transgenic mice have previously been described [52]. Six1nLacZ/+ mice, were crossed with Pax3Sp/+ mice and X-Gal staining was performed as previously described [53]. Six1−/+/Six4−/+ mice [13] were crossed with Pax3Sp/+or Myf5nLacZ/+ mice [20].
All experiments with mice were performed according to the European Community Council Directive of 11/24/1986 (86/609/EEC) and with permission from the French Veterinary Services (permit number 75-1373) and approval by the Cochin General Animal Facility Service (accreditation number A-75-14-02). All efforts were made to minimize suffering.
X-Gal staining, immunohistochemistry, and whole-mount in situ hybridization
We collected mouse embryos after natural overnight matings; for staging, embryonic day (E) 0.5 corresponded to midday assuming that fertilization had taken place at 6 a.m. Genotyping was carried out by X-Gal staining in X-Gal, with 0.2% PAF for 30 minutes following 1–2 h fixation in 4% PAF, on ice. When a light blue color had developed, embryos were rinsed in PBS and post-fixed overnight in 4% PAF. Whole mount in situ hybridization with digoxigenin-labelled riboprobes was performed as described [20]. The Myod riboprobe has also been previously described [20]. Fluoresencent co-immunohistochemistry was carried out according to [32], using the following antibodies : polyclonal anti-β-Gal (Molecular Probe, diluted 1∶200), monoclonal anti Myod (DAKO, 1∶200), monoclonal anti-Desmin (Abcam, 1/100), polyclonal anti Six2 (Proteintech, 1/200) and monoclonal anti-Myogenin (DAKO, 1∶200). Secondary antibodies were coupled to Alexa 488 1/250 and 546 1/1000 (Molecular Probes).
DNA binding assays and transactivation
Gel mobility shift assays (GMSA) were performed essentially as previously described [26], with a labeled probe corresponding to the MEF3 site of the Myogenin promoter or Myod DRR and CE MEF3 elements and with Six1, Six2, Six4, Six5, Six4Δ and Eya2 proteins produced using the TNT T7 Coupled Reticulocyte Lysate System (Promega). The DNA templates used for in vitro transcription of mouse Six1, Six2, Six4, Six4Δ, Six5 and Eya2 were cloned in the pCR3 vector (Invitrogen). Because of the high molecular weight of Six4 and Six4Δ (about 90 kDa) we used a 3% acrylamide gel, run overnight at +4°C. To ensure that proteins were appropriately translated, parallel reactions were performed in the presence of [35S]methionine, separated on SDS-PAGE gels and visualized using autoradiography. The sequences of the double stranded oligonucleotides containing MEF3 sites used for bandshift assays are: DRR: 5′ AGT TGG ATC CGG TTT CCA GAG GC, CE1 : 5′ TGA GAC AGT AAT TTT ATC CTG CT, CE2 : 5′ GGT CTT CTC CGG TTT CTC TAG CT, Myogenin MEF3 : 5′TGG GGG GGC TCA GGT TTC TGT GGC GT, Myogenin NFI: TAT CTC TGG GTT CAT GCC AGC AGG G. The TCAGGTTTC MEF3 sequence is underlined.
Chick primary myoblasts were grown and transfected as previously described [26], using RSV-Renilla as a control for transfection efficiency. Eya2, Six4 or Six4Δ expression was driven by the CMV promoter-enhancer present in pCR3, with the luciferase reporter gene under the control of a multimerized MEF3 element cloned upstream of the human Aldolase A minimal −35 to +45 bp promoter [54]. Two days after transfection, luciferase activity was measured using standard procedures.
Generation and analysis of transient transgenic embryos
For the construction of the CE-MD5.8-lacZ and mutated Mut3-CE-MD5.8-lacZ sequences, mouse DNA was first used as a template to clone the core enhancer (CE) of Myod [36] with forward Apa1/Not1 5′ GGG-CCC-GCG-GCC-GCT-GAG-CCC-CAC-AGC-ATT-TGG and reverse 5′ GAA-TTC-CCC-CAG-CCC-TAG-GCC-TGA-GCT oligonucleotides; the MEF3 sequence is underlined. This 262 bp CE fragment was subsequently inserted into an Apa1-Pml1 site (position −5792 to−5652) of the pMD6.8-lacZ linearised plasmid [35], 340 bp upstream of the distal regulatory region (DRR) lying at −5310 bp from the Myod gene. The sequence of the pCE-MD6.8-lacZ reporter vector was verified by sequencing. To obtain mutated pCE-MD6.8-lacZ, one MEF3 site in the DRR (position −5176 to −5167) and two MEF3 sites in the CE (at position 55 and position 229) were mutated by substitution with a Hind III site (TCCGGTTTC->AAGCTTTTC), a XhoI site (GTAATTTTA ->CTCGAGTTA) or a BglII site (TCCGGTTTC->TCCAGATCT), respectively. The pMutCE-MD6.8-lacZ reporter vector was verified by sequencing. The plasmid was digested with Not1. Migration on an agarose gel allowed removal of plasmid sequences. Transgenic mice were generated by microinjection of the purified construct into fertilized F2 eggs from C57BL/6JxSJL mice, at a concentration of approximately 1 ng/µl using standard techniques. Injected eggs were reimplanted the same day or the day after the injection into outbred pseudo-pregnant foster mothers. Transient transgenic embryos were dated taking the day of reimplantation into the pseudo-pregnant foster mothers as E0.5. Embryos were dissected in PBS, fixed in 4% paraformaldehyde for 15 minutes, rinsed 3 times in PBS and stained in X-gal solution [33] at 37°C overnight. DNA was prepared from the vitelline membrane from each embryo and analysed by PCR, using nlacZ primers, and Myod primers in the DRR and in the CE.
ChIP experiments
Pax3GFP/+ males were crossed with C57Bl6N females to obtain Pax3GFP/+ embryos. Somites were collected from E11.5 embryos by removing heads, neural tubes and internal organs. These samples were enzymatically digested with collagenase and dissociated cells were fixed with 1% formaldehyde at room temperature for 15 min. The GFP-positive cells were sorted by flow cytometry (BD FACs ARIA III). The gates for positive and negative GFP cells were determined using an equivalent sample isolated from wild type embryos and from Pax3GFP/+ heterozygous embryos. About 7.5×105 cells were collected for ChIP experiments from nine embryos. E12 Six1−/−/Six4−/− embryos were collected and enriched myogenic tissues were pooled after removing limbs, neural tube and internal organs. Dounce dissociated cells were fixed with 1% formaldehyde at room temperature for 15 min.
The chromatin immunoprecipitation procedure was performed according to the manufacturer's protocol (EZ-Magna ChIP G Kit; Merck Millipore) with antibodies recognizing all Eya proteins (Santa Cruz), Six2 protein (Proteintech), and Normal Mouse IgG provided in the EZ-Magna kit as a control. Input DNA and immunoprecipitated DNA were analyzed by quantitative-PCR (Roche, Light Cycler 480). Results were normalized with a negative control from an intergenic region without a MEF3 site (NC2). The sequences of primers were as follows:
Myod-CE1 : Fwd : 5′ GGG CAT TTA TGG GTC TTC CT, Rev : 5′ GCC CTA GGC CTG AGC TAG A ; Myod-CE4 : Fwd : 5′ GGG CAT TTA TGG GTC TTC CT, Rev : 5′ GCT GAG CAC TCT GGG AGA TT; Myod-CE5 : Fwd : 5′ TCA GCT GTT CCT GGG TCT TC, Rev : 5′ GAC CTC TCA TGC CTG GTG TT; Myod-CE7 : Fwd : 5′ AAC CCG TGA CTC ACA ACA CA, Rev : 5′ AGC CCT AGG CCT GAG CTA GA ; Myod-DRR : Fwd : 5′ GCC CGC AGT AGC AAA GTA AG, Rev : 5′ GCT CCC TTG GCT AGT CTT CC; NC2 : Fwd : 5′ GAG TTG GCA GGA ATC AGC TC, Rev : 5′ GCC AGC AAT TTG GTT TGA AT.
Supporting Information
Zdroje
1. GehringWJ, IkeoK (1999) Pax 6: mastering eye morphogenesis and eye evolution. Trends in genetics 15 : 371–377.
2. KumarJP (2010) Retinal determination the beginning of eye development. Current topics in developmental biology 93 : 1–28.
3. van HeyningenV, WilliamsonKA (2002) PAX6 in sensory development. Human molecular genetics 11 : 1161–1167.
4. JemcJ, RebayI (2007) The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annual review of biochemistry 76 : 513–538.
5. BuckinghamM, RelaixF (2007) The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annual review of cell and developmental biology 23 : 645–673.
6. Xu PX (2012) The EYA-SO/SIX complex in development and disease. Pediatric nephrology (Berlin, Germany).
7. DuanH, ZhangC, ChenJ, SinkH, FreiE, et al. (2007) A key role of Pox meso in somatic myogenesis of Drosophila. Development (Cambridge, England) 134 : 3985–3997.
8. KirbyRJ, HamiltonGM, FinneganDJ, JohnsonKJ, JarmanAP (2001) Drosophila homolog of the myotonic dystrophy-associated gene, SIX5, is required for muscle and gonad development. Current biology 11 : 1044–1049.
9. LiuYH, JakobsenJS, ValentinG, AmarantosI, GilmourDT, et al. (2009) A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Developmental cell 16 : 280–291.
10. LaghaM, BrunelliS, MessinaG, CumanoA, KumeT, et al. (2009) Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Developmental cell 17 : 892–899.
11. HeanueTA, ReshefR, DavisRJ, MardonG, OliverG, et al. (1999) Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes & development 13 : 3231–3243.
12. RidgewayAG, SkerjancIS (2001) Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. The Journal of biological chemistry 276 : 19033–19039.
13. GrifoneR, DemignonJ, HoubronC, SouilE, NiroC, et al. (2005) Six1 and Six4 homeoproteins are required for Pax3 and MRF expression during myogenesis in the mouse embryo. Development 132 : 2235–2249.
14. BrownCB, EnglekaKA, WenningJ, Min LuM, EpsteinJA (2005) Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis (New York, NY) 41 : 202–209.
15. GrifoneR, DemignonJ, GiordaniJ, NiroC, SouilE, et al. (2007) Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol 302 : 602–616.
16. LinCY, ChenWT, LeeHC, YangPH, YangHJ, et al. (2009) The transcription factor Six1a plays an essential role in the craniofacial myogenesis of zebrafish. Developmental biology 331 : 152–166.
17. GuoC, SunY, ZhouB, AdamRM, LiX, et al. (2011) A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. The Journal of clinical investigation 121 : 1585–1595.
18. SambasivanR, KurataniS, TajbakhshS (2011) An eye on the head: the development and evolution of craniofacial muscles. Development (Cambridge, England) 138 : 2401–2415.
19. BuckinghamM, VincentSD (2009) Distinct and dynamic myogenic populations in the vertebrate embryo. Current opinion in genetics & development 19 : 444–453.
20. TajbakhshS, RocancourtD, CossuG, BuckinghamM (1997) Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell 89 : 127–138.
21. SatoT, RocancourtD, MarquesL, ThorsteinsdottirS, BuckinghamM (2010) A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet 6: e1000897 doi:10.1371/journal.pgen.1000897.
22. BorelloU, BerarducciB, MurphyP, BajardL, BuffaV, et al. (2006) The Wnt/beta-catenin pathway regulates Gli-mediated Myf5 expression during somitogenesis. Development (Cambridge, England) 133 : 3723–3732.
23. BajardL, RelaixF, LaghaM, RocancourtD, DaubasP, et al. (2006) A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev 20 : 2450–2464.
24. GiordaniJ, BajardL, DemignonJ, DaubasP, BuckinghamM, et al. (2007) Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc Natl Acad Sci U S A 104 : 11310–11315.
25. LiuY, ChuA, ChakrounI, IslamU, BlaisA (2010) Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic acids research 38 : 6857–6871.
26. SpitzF, DemignonJ, PorteuA, KahnA, ConcordetJP, et al. (1998) Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci U S A 95 : 14220–14225.
27. NiroC, DemignonJ, VincentS, LiuY, GiordaniJ, et al. (2010) Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Developmental biology 338 : 168–182.
28. RichardAF, DemignonJ, SakakibaraI, PujolJ, FavierM, et al. (2011) Genesis of muscle fiber-type diversity during mouse embryogenesis relies on Six1 and Six4 gene expression. Developmental biology 359 : 303–320.
29. PignoniF, HuB, ZavitzKH, XiaoJ, GarrityPA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91 : 881–891.
30. OhtoH, KamadaS, TagoK, TominagaSI, OzakiH, et al. (1999) Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Molecular and cellular biology 19 : 6815–6824.
31. KawakamiK, OhtoH, IkedaK, RoederRG (1996) Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic acids research 24 : 303–310.
32. RelaixF, PolimeniM, RocancourtD, PonzettoC, SchaferBW, et al. (2003) The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes & development 17 : 2950–2965.
33. TajbakhshS, BoberE, BabinetC, PourninS, ArnoldH, et al. (1996) Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Developmental dynamics 206 : 291–300.
34. SassoonD, LyonsG, WrightWE, LinV, LassarA, et al. (1989) Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature 341 : 303–307.
35. AsakuraA, LyonsGE, TapscottSJ (1995) The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Developmental biology 171 : 386–398.
36. GoldhamerDJ, BrunkBP, FaermanA, KingA, ShaniM, et al. (1995) Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development (Cambridge, England) 121 : 637–649.
37. KucharczukKL, LoveCM, DoughertyNM, GoldhamerDJ (1999) Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development (Cambridge, England) 126 : 1957–1965.
38. ChenJC, RamachandranR, GoldhamerDJ (2002) Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Developmental biology 245 : 213–223.
39. ChenJC, GoldhamerDJ (2004) The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Developmental biology 265 : 502–512.
40. KawakamiK, SatoS, OzakiH, IkedaK (2000) Six family genes–structure and function as transcription factors and their roles in development. BioEssays 22 : 616–626.
41. OliverG, WehrR, JenkinsNA, CopelandNG, CheyetteBN, et al. (1995) Homeobox genes and connective tissue patterning. Development (Cambridge, England) 121 : 693–705.
42. KlesertTR, ChoDH, ClarkJI, MaylieJ, AdelmanJ, et al. (2000) Mice deficient in Six5 develop cataracts: implications for myotonic dystrophy. Nature genetics 25 : 105–109.
43. YajimaH, MotohashiN, OnoY, SatoS, IkedaK, et al. (2010) Six family genes control the proliferation and differentiation of muscle satellite cells. Experimental cell research 316 : 2932–2944.
44. Le GrandF, GrifoneR, MourikisP, HoubronC, GigaudC, et al. (2012) Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J Cell Biol 198 : 815–832.
45. Kassar-DuchossoyL, Gayraud-MorelB, GomesD, RocancourtD, BuckinghamM, et al. (2004) Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431 : 466–471.
46. XuPX, WooI, HerH, BeierDR, MaasRL (1997) Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development (Cambridge, England) 124 : 219–231.
47. L'HonoreA, OuimetteJF, Lavertu-JolinM, DrouinJ (2010) Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development (Cambridge, England) 137 : 3847–3856.
48. HuP, GelesKG, PaikJH, DePinhoRA, TjianR (2008) Codependent activators direct myoblast-specific MyoD transcription. Developmental cell 15 : 534–546.
49. CaoL, YuY, BilkeS, WalkerRL, MayeenuddinLH, et al. (2010) Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer research 70 : 6497–6508.
50. CaoY, YaoZ, SarkarD, LawrenceM, SanchezGJ, et al. (2010) Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Developmental cell 18 : 662–674.
51. KressC, Vandormael-PourninS, BaldacciP, Cohen-TannoudjiM, BabinetC (1998) Nonpermissiveness for mouse embryonic stem (ES) cell derivation circumvented by a single backcross to 129/Sv strain: establishment of ES cell lines bearing the Omd conditional lethal mutation. Mammalian genome 9 : 998–1001.
52. LallemandY, LuriaV, Haffner-KrauszR, LonaiP (1998) Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic research 7 : 105–112.
53. LaclefC, HamardG, DemignonJ, SouilE, HoubronC, et al. (2003) Altered myogenesis in Six1-deficient mice. Development 130 : 2239–2252.
54. GrifoneR, LaclefC, SpitzF, LopezS, DemignonJ, et al. (2004) Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol Cell Biol 24 : 6253–6267.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic LethalityČlánek DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



