-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
DJ-1, a Parkinson's disease (PD)–associated gene, has been shown to protect against oxidative stress in Drosophila. However, the molecular mechanism underlying oxidative stress-induced phenotypes, including apoptosis, locomotive defects, and lethality, in DJ-1-deficient flies is not fully understood. Here we showed that Daxx-like protein (DLP), a Drosophila homologue of the mammalian Death domain-associated protein (Daxx), was upregulated under oxidative stress conditions in the loss-of-function mutants of Drosophila DJ-1β, a Drosophila homologue of DJ-1. DLP overexpression induced apoptosis via the c-Jun N-terminal kinase (JNK)/Drosophila forkhead box subgroup O (dFOXO) pathway, whereas loss of DLP increased resistance to oxidative stress and UV irradiation. Moreover, the oxidative stress-induced phenotypes of DJ-1β mutants were dramatically rescued by DLP deficiency, suggesting that enhanced expression of DLP contributes to the DJ-1β mutant phenotypes. Interestingly, we found that dFOXO was required for the increase in DLP expression in DJ-1β mutants and that dFOXO activity was increased in the heads of DJ-1β mutants. In addition, subcellular localization of DLP appeared to be influenced by DJ-1 expression so that cytosolic DLP was increased in DJ-1β mutants. Similarly, in mammalian cells, Daxx translocation from the nucleus to the cytosol was suppressed by overexpressed DJ-1β under oxidative stress conditions; and, furthermore, targeted expression of DJ-1β to mitochondria efficiently inhibited the Daxx translocation. Taken together, our findings demonstrate that DJ-1β protects flies against oxidative stress - and UV-induced apoptosis by regulating the subcellular localization and gene expression of DLP, thus implying that Daxx-induced apoptosis is involved in the pathogenesis of DJ-1-associated PD.
Published in the journal: . PLoS Genet 9(4): e32767. doi:10.1371/journal.pgen.1003412
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003412Summary
DJ-1, a Parkinson's disease (PD)–associated gene, has been shown to protect against oxidative stress in Drosophila. However, the molecular mechanism underlying oxidative stress-induced phenotypes, including apoptosis, locomotive defects, and lethality, in DJ-1-deficient flies is not fully understood. Here we showed that Daxx-like protein (DLP), a Drosophila homologue of the mammalian Death domain-associated protein (Daxx), was upregulated under oxidative stress conditions in the loss-of-function mutants of Drosophila DJ-1β, a Drosophila homologue of DJ-1. DLP overexpression induced apoptosis via the c-Jun N-terminal kinase (JNK)/Drosophila forkhead box subgroup O (dFOXO) pathway, whereas loss of DLP increased resistance to oxidative stress and UV irradiation. Moreover, the oxidative stress-induced phenotypes of DJ-1β mutants were dramatically rescued by DLP deficiency, suggesting that enhanced expression of DLP contributes to the DJ-1β mutant phenotypes. Interestingly, we found that dFOXO was required for the increase in DLP expression in DJ-1β mutants and that dFOXO activity was increased in the heads of DJ-1β mutants. In addition, subcellular localization of DLP appeared to be influenced by DJ-1 expression so that cytosolic DLP was increased in DJ-1β mutants. Similarly, in mammalian cells, Daxx translocation from the nucleus to the cytosol was suppressed by overexpressed DJ-1β under oxidative stress conditions; and, furthermore, targeted expression of DJ-1β to mitochondria efficiently inhibited the Daxx translocation. Taken together, our findings demonstrate that DJ-1β protects flies against oxidative stress - and UV-induced apoptosis by regulating the subcellular localization and gene expression of DLP, thus implying that Daxx-induced apoptosis is involved in the pathogenesis of DJ-1-associated PD.
Introduction
Oxidative stress, a state of imbalance between the generation and elimination of reactive oxygen and nitrogen species, has been implicated in a variety of neurodegenerative diseases [1]–[3]. The central nervous system is presumed to be particularly vulnerable to oxidative stress, as it consumes abundant quantities of oxygen and employs nitric oxide as a biological messenger, both of which create reactive species as by-products [1]. Oxidative stress provokes various cytotoxic processes, such as overstimulation of glutamate receptors (excitotoxicity), ER stress, and mitochondrial dysfunction, which lead to apoptosis, the predominant form of cell death in aging-related neurodegenerative diseases [1], [2].
Parkinson's disease (PD) is characterized by typical motor dysfunction and is thought to be caused by the loss of nigrostriatal dopaminergic (DA) neurons that connect the substantia nigra pars compacta (SNpc) to other brain regions [3]–. The death of these neurons has been closely linked to oxidative stress [3]–[5]. Markers of oxidative damage to lipids, proteins and DNA, as well as mitochondrial DNA deletions, which can be caused by oxidative stress, are significantly elevated in postmortem samples of the SNpc of PD patients [4], [5]. The nigrostriatal pathway is sensitive to 6-hydroxydopamine and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)/MPP+, which destroy DA neurons via induction of oxidative stress [6]. Moreover, oxidative stress plays an important role in the function of the familial PD-related genes, α-synuclein and parkin [3]. Oxidative damaged α-synuclein is aggregated into Lewy bodies [7], and the function of Parkin is impaired by oxidative modifications [8], [9]. Additionally, various animal models of familial PD show greater damage in response to oxidative stress [10]–[16]. Although these data demonstrate a correlation between PD and oxidative stress, the molecular mechanisms underlying oxidative stress-induced DA neuronal death in PD are not well understood.
Among the genes related to PD, DJ-1 is the most closely associated with oxidative stress [17]. DJ-1 was originally identified as an oncogene that transforms mouse NIH3T3 cells in cooperation with ras [18], and its gene expression is increased in various types of cancer [19]–[21]. Later, DJ-1 was linked to an autosomal-recessive early-onset type of familial PD [22], [23]. The impairment of DJ-1 function sensitizes animal models to oxidative stress [11], [12], [14], [24]–[26]. DJ-1 performs several critical functions in response to oxidative stress via diverse cellular mechanisms [5], [17], [23]. First, DJ-1 functions as an atypical peroxiredoxin-like peroxidase that scavenges peroxides by oxidizing Cys106 [14]. Second, DJ-1 regulates expression of several antioxidant genes [27]–[30] and stabilizes the antioxidant transcriptional master regulator, Nrf2 [29], [31]. Third, DJ-1 inhibits UV - and oxidative stress-induced cell death by suppressing pro-apoptotic factors [32]–[34]. In Drosophila, there are 2 homologues of human DJ-1: DJ-1α and β [11], [12], [24]. DJ-1α is predominantly expressed in the testes, whereas DJ-1β is expressed in most tissues [11], [24], similar to the expression pattern of mammalian DJ-1 [18]. Several previous studies have demonstrated that DJ-1β loss-of-function mutants are acutely sensitive to oxidative stress and prone to locomotive dysfunction, resembling the phenotypes seen in PD [11], [12], [14], [24].
In this study, we identified a Drosophila homologue of death domain-associated protein (Daxx), Daxx-like protein (DLP), as a mediator of Drosophila DJ-1β mutant phenotypes. Daxx, originally identified as a binding partner of the pro-apoptotic receptor Fas (also called CD95) [35], performs a pivotal function in apoptosis [36], [37]. Daxx activates apoptosis signal-regulating kinase 1 (ASK1), which in turn increases c-Jun N-terminal kinase (JNK) activity leading to apoptosis [38], [39]. Daxx is increased in cells upon exposure to hydrogen peroxide and functions as a mediator of oxidative stress-induced apoptosis [33], [40], [41]. In neuronal cells, dominant negative-Daxx blocks Fas-induced cell death [42]. In addition, FADD/caspase-8 cascade-triggered cell death requires the transcriptional activation of Daxx in normal embryonic motor neurons [43]. Furthermore, Daxx has been identified as a potential component of the pathogenesis of neurodegenerative diseases, including PD [33], [44]. For example, Daxx interacts with DJ-1 [33], and MPTP induces translocation of Daxx from the nucleus to the cytoplasm and activates the ASK1 signaling pathway in mouse SNpc [44].
Although previous studies have demonstrated that Drosophila DJ-1β mutants are acutely sensitive to oxidative stress [11], [12], [14], [24], the cellular consequence of DJ-1β deficiency in the oxidative stress response remains unclear. Furthermore, the link between DJ-1 and PD, especially in the context of oxidative stress, has not been thoroughly explored. In this study, we used Drosophila and mammalian cells to investigate the functional interaction between DJ-1 and its downstream target, Daxx/DLP. We also characterized the relationship between loss of DJ-1 and PD-related phenotypes, such as DA neuronal degeneration and locomotive dysfunction, and examined the molecular mechanism of oxidative stress sensitivity in Drosophila DJ-1 mutants.
Results
Oxidative stress-sensitive neuronal death in DJ-1β mutants
Drosophila DJ-1β mutants do not have gross morphological defects or loss of DA neurons when raised under standard laboratory conditions (Figure S1) [24]. However, expression level of tyrosine hydroxylase and number of DA neurons were significantly reduced in DJ-1β mutant flies after hydrogen peroxide treatment compared to those in wild-type animals (Figure 1A). The numbers of DA neurons in 3 major clusters of the posterior brain, dorsomedial clusters, dorsolateral clusters 1, and posteriomedial clusters, were significantly decreased by oxidative stress (Figure 1B). Next, we tested the effect of similar oxidative stress conditions on the neurons of larvae. As shown in Figure 1C, oxidative stress-induced cell death was dramatically increased in the brains of DJ-1β mutant larvae compared to that in wild-type controls. This suggests that DJ-1β mediates developmentally universal protection of DA neurons from oxidative stress-induced cell death.
Fig. 1. Decreased DA neurons and increased apoptosis in DJ-1β mutant under oxidative stress conditions. 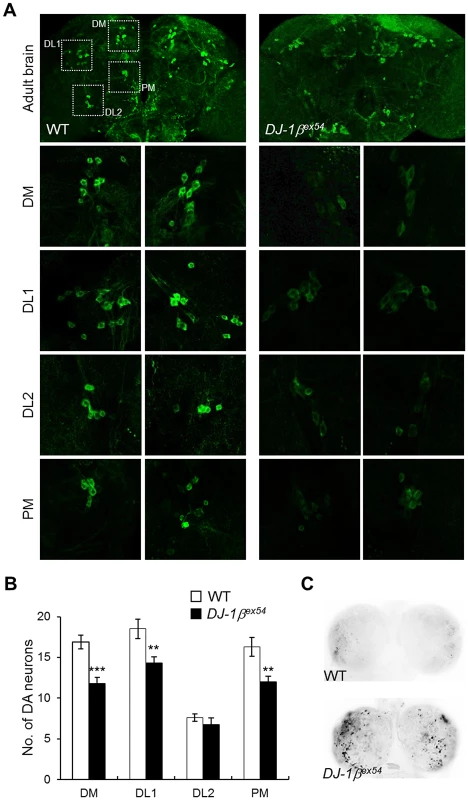
(A) DA neurons visualized by immunohistochemical analysis with anti-tyrosine hydroxylase antibody in the brains of wild-type (WT) and DJ-1β mutant (DJ-1βex54) flies fed with 1% H2O2 for 3 days. Dotted boxed areas indicate DA neuron clusters. The lower pictures, including DM, DL1, DL2, and PM, are the magnified 4 dotted boxed areas of the upper pictures. Magnification of the upper pictures, 100×; Magnification of the lower pictures, 400×. (B) Graphs showing the number of DA neurons in each cluster of WT and DJ-1βex54 flies after feeding with H2O2 for 3 days (n = 10, Student's t-test: DM, *** p<0.001; DL1 and PM, ** p<0.01). The data are expressed as means ± s.e. values. (C) Acridine orange staining of 0.1% hydrogen peroxide-treated larval brains showed that increased oxidative stress-induced apoptosis in DJ-1βex54 compared to the WT controls. DM, dorsomedial clusters; DL, dorsolateral clusters; PM, posteriomedial clusters. DLP is a downstream gene of DJ-1β
To characterize the protective mechanism of DJ-1β, we used microarrays to compare the gene expression profiles of DJ-1β mutants and wild-type controls under oxidative stress conditions. We identified 143 upregulated and 134 downregulated genes (>1.5-fold changes in expression DJ-1β mutants) in the mRNA extracted from fly heads (Table S1). The role of DLP in the DJ-1-dependent oxidative stress response was first examined among the upregulated genes because Daxx, the mammalian homologue of DLP, has been implicated in oxidative stress-induced apoptosis [40] and identified as a potential component of PD pathogenesis [33], [44]. DLP is a 183.9-kDa protein with approximately 46% similarity to human Daxx in the Daxx-homology region; it is the only Daxx homologue in the Drosophila genome [45]. Interestingly, DLP protein levels were significantly higher in the fly head than in the body (Figure 2A and Figure S2), suggesting that it performs an important function in the brain. Furthermore, DLP mRNA and protein levels were increased by both UV irradiation and oxidative stress (Figure 2B and 2C), implying that DLP is involved in these stress responses, similar to its mammalian counterpart.
Fig. 2. Gene expression of DLP by oxidative stress, UV, and DJ-1β. 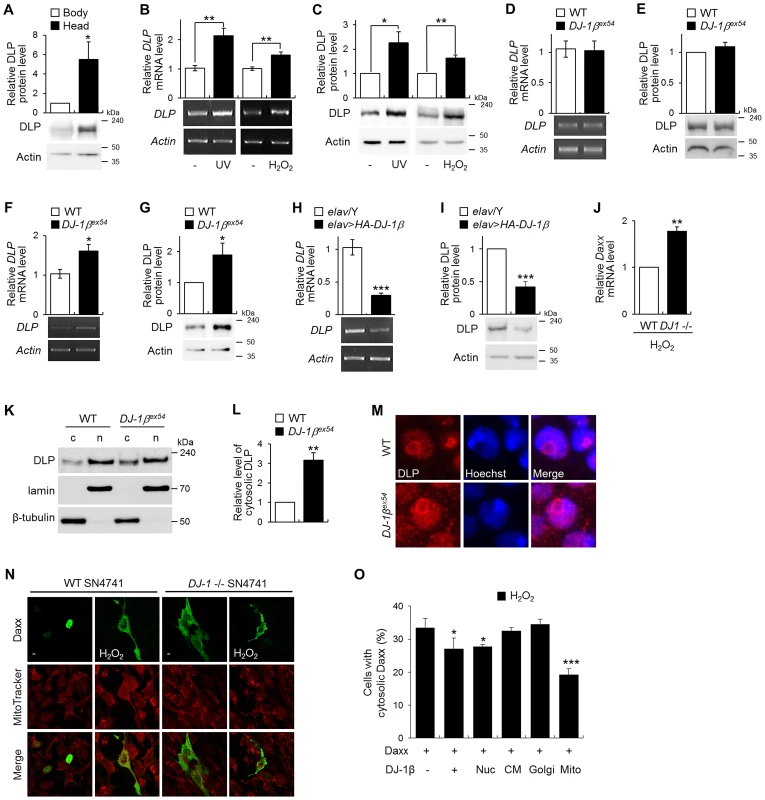
(A) DLP protein levels in the wild-type (WT) fly body and head (Student's t-test, n = 5, * p<0.05). (B–C) DLP mRNA (B) and protein (C) levels in WT embryos exposed to UV (50 mJ/cm2) and the heads of WT flies fed with 1% H2O2 (B, Student's t-test: UV, n = 5, ** p<0.01; H2O2, n = 5, ** p<0.01; C, Student's t-test: UV, n = 5, * p<0.05; H2O2, n = 4, ** p<0.01). (D–G) DLP mRNA (D, F) and protein (E, G) levels in the heads of WT and DJ-1βex54 flies grown with cornmeal-soybean standard fly food (D–E) and fed with 1% H2O2 for 3 days (F–G). (D, Student's t-test, n = 8; E, Student's t-test, n = 7; F, Student's t-test, n = 6, * p<0.05; G, Student's t-test, n = 7, * p<0.05). (H–I) DLP mRNA (H) and protein (I) levels in control (elav/Y) and pan-neuronally DJ-1β-overexpressing (elav>HA-DJ-1β) fly head (H, Student's t-test, n = 5, *** p<0.001; I, Student's t-test, n = 4, *** p<0.001). (J) Daxx mRNA level in the WT and DJ-1 null SN4741 cells treated with 1 mM H2O2 (Student's t-test, n = 3, ** p<0.01). (K–L) Western blot (K) and statistical (L) analysis of cytosolic (c) and nuclear (n) fractions of WT and DJ-1βex54 fly head extracts showed that increased translocation of DLP to the cytosol in DJ-1βex54 compared to WT controls (Student's t-test, n = 4, ** p<0.01). Relative cytosolic DLP levels were calculated by dividing the normalized cytosolic DLP level by the normalized nuclear DLP level. Lamin and β-tubulin were used as loading controls for nuclear and cytosolic fractions, respectively. (M) Confocal images of DLP immunohistochemistry in the larval brains of WT and DJ-1βex54. Hoechst-stained regions represent nuclei. Magnification, 8,000×. (N) Confocal images showing subcellular localization of Daxx in WT and DJ-1 null SN4741 cells. MitoTracker-stained spots represent mitochondria. The cells were treated with 0.4 mM H2O2 for 1 h. (O) The ratio of the cells with cytosolic localized Daxx in DJ-1 null SN4741 cells transfected with wild-type DJ-1β or nucleus (Nuc)-, cytoplasmic membrane (CM)-, Golgi (Golgi)-, or mitochondria (Mito)-targeted DJ-1β. The cells were treated with 0.4 mM H2O2 for 1 h. More than 70 cells per each samples were counted to calculate the ratio of the cells with cytosolic Daxx (Student's t-test, n = 4, * p<0.05, *** p<0.001). All data are expressed as means ± s.e. values. Actin was employed as an internal control of total extract. To confirm our microarray data, we evaluated DLP expression in the heads of wild-type and DJ-1β mutant flies following treatment with H2O2 using real-time quantitative PCR and western blot analysis. The levels of DLP mRNA and protein did not differ between the DJ-1β mutants and wild-type controls under standard laboratory conditions (Figure 2D and 2E). However, as anticipated, the levels of DLP mRNA and protein were significantly elevated in DJ-1β mutants in comparison to wild-type controls under oxidative stress (Figure 2F and 2G), suggesting that the increasing rate of DLP expression by oxidative stress in DJ-1β mutants is higher than that in wild type. When DJ-1β was overexpressed with a pan-neuronal elav-GAL4 driver, the levels of DLP transcript and protein were significantly reduced (Figure 2H and 2I). Interestingly, mammalian Daxx gene expression was also higher in DJ-1 null cells than in wild-type controls under oxidative stress condition (Figure 2J). These results indicate that DJ-1 functions as a negative regulator of Daxx/DLP gene expression under oxidative stress conditions.
Altered subcellular localization of DLP in DJ-1β mutants
As mammalian DJ-1 inhibits translocation of the nuclear Daxx to the cytosol [33], [46], we examined whether Drosophila DJ-1β also regulates subcellular localization of DLP. First, we fractionated DLP protein from the cytosol and nucleus of wild-type and DJ-1β mutant fly heads (Figure 2K). The proportion of cytosolic DLP relative to nucleic DLP was increased more than 3-fold in DJ-1β fly heads (Figure 2K and 2L). Consistently, immunohistochemical analysis with anti-DLP antibody showed that the cytosolic DLP level was significantly increased in the brain and eye imaginal disc of DJ-1β mutant flies compared to that in wild-type flies (Figure 2M and Figure S3, respectively), which was highly similar in DJ-1 null mouse DA neuroblastoma cells (Figure 2N).
Since we previously showed that DJ-1 is partially localized in mitochondria [24], we examined whether mitochondrial translocation of DJ-1 is important for the cytosolic localization of Daxx by comparing the effect of several forms of DJ-1 that are targeted to various subcellular regions including mitochondria, Golgi, nucleus, and cytoplasmic membrane. Interestingly, the mitochondrial DJ-1β efficiently inhibited translocation of Daxx from the nucleus to the cytosol under oxidative stress conditions, like wild-type or nucleus targeted DJ-1β (Figure 2O and Figure S4). These results suggest that DJ-1 regulates the translocation of Daxx/DLP as well as their gene expression in response to oxidative stress.
DLP mediates oxidative stress - and UV-induced apoptosis
To evaluate the role of DLP in the hypersensitivity of DJ-1β mutants to oxidative stress and UV irradiation, we generated and characterized DLP mutants. The mutant EY09290, which harbors a P-element inserted in the 5′ region of the DLP gene, was acquired from the Bloomington Drosophila Stock Center, and deletion mutants were generated via P-element mobilization (Figure 3A). Two deletion alleles were generated and designated DLP1 and DLP2. The genomic deletions were confirmed by PCR (Figure 3B) and DNA sequencing. These alleles have deletions of 1,311 and 1,076 bp, respectively, which removes the first 2 exons, including the translational start site of DLP (Figure 3A). Western blot analysis and RNA in situ hybridization results confirmed that DLP protein and mRNA levels were markedly reduced in DLP1 and DLP2 compared to the levels in wild-type controls (Figure 3C and 3D). In order to exclude the genetic background effect, we generated two other DLP deletion mutants, DLP3 and DLP4, using another P-element line, KG01694 (Figure 3A), and we obtained another DLP mutant, DLPU42, with a different genetic lineage [45]. Therefore, we used five mutant alleles in three different genetic backgrounds to characterize the DLP mutants. Furthermore, Upstream Activation Sequence (UAS)-DLP-RNAi was used for DLP knockdown to confirm the DLP mutant phenotypes.
Fig. 3. Generation and characterization of DLP mutants. 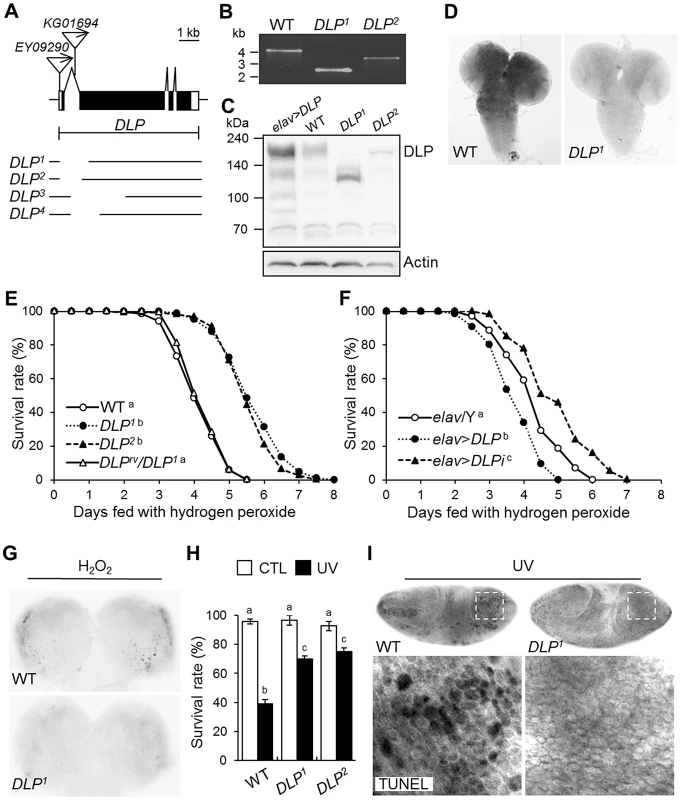
(A) Genomic structure of the DLP gene. Exons of the DLP gene are shown in black (coding region) and white (non-coding region) boxes. The inverted triangles indicate the P-elements, EY09290 and KG01694. The deletion sites of DLP1, DLP2, DLP3, and DLP4 are illustrated under the genomic structures. (B) Determination of the deleted size in DLP mutants by genomic DNA PCR. (C) Western blotting of DLP in wild type (WT), DLP loss-of-function (DLP1 and DLP2) and gain-of-function (elav>DLP) mutants. Intact DLP protein is not detected in the DLP mutants. (D) Comparison of DLP gene expression in the third-instar larval brains of WT and a DLP mutant via RNA in situ hybridization. (E–F) Survival rates of DLP loss-of-function (DLP1, DLP2, and elav>DLPi) and gain-of-function (elav>DLP) mutants under oxidative stress conditions. (E) WT and DLPrv/DLP1 were used as controls (log-rank test: WT, n = 300; DLP1, n = 250; DLP2, n = 250; DLPrv/DLP1, n = 300, p<0.01, groups with the same letter do not differ significantly). (F) elav/Y was used as a control (log-rank test: elav/Y, n = 350; elav>DLP, n = 300; elav>DLPi, n = 300, p<0.01, groups with the different letter differ significantly). The genotypes of the samples were elav/Y (elav-GAL4/Y), elav>DLP (elav-GAL4/Y; EY09290/+), and elav>DLPi (elav-GAL4/Y; UAS-DLP-RNAi/+). (G) Acridine orange staining of larval brains of DLP1 and WT treated with 0.1% H2O2 for 24 h. (H) Survival rates of WT and DLP mutant (DLP1 and DLP2) pupae after exposure to UV irradiation (10 mJ/cm2; black bars) as described in the Materials and Methods (Kruskal-Wallis test: CTL, n≥6, p<0.1; UV, n = 6, p<0.01, groups with the same letter do not differ significantly). CTL, UV-untreated control pupae; UV, UV-treated pupae. All data are expressed as means ± s.e. values. (I) TUNEL-stained images of UV-exposed 0–3 h embryos of WT and DLP1. The lower panels are higher-magnification images of the boxes indicated with dotted lines in the upper panels. CTL, control; rv, revertant; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling. Because Daxx has been implicated in oxidative stress and UV responses, the role of DLP in oxidative stress-induced lethality was assessed using these DLP mutants. As expected, DLP mutants proved more resistant to H2O2 treatment than wild-type or trans-heterozygotes between DLP revertant (DLPrv) and DLP1 (Figure 3E and Figure S5). Moreover, DLP knockdown in all neurons using UAS-DLP-RNAi conferred increased resistance to oxidative stress, whereas DLP overexpression in the same neurons increased their sensitivity to oxidative stress (Figure 3F). Oxidative stress-induced cell death was also attenuated in the DLP mutant brain (Figure 3G). UV irradiation-induced pupal lethality and apoptosis were also significantly reduced in DLP mutants (Figure 3H and 3I), indicating that DLP is involved in UV-induced stress responses. These results indicate that the neuronal function of DLP is important to the oxidative and UV stress responses at both cellular and organism level.
DLP functions as a pro-apoptotic gene by activating the JNK/dFOXO signaling pathway
Daxx was originally identified as a pro-apoptotic gene that induced cell death [35]. However, the role of Daxx and DLP in apoptosis is somewhat controversial [38], [45]. In order to confirm the function of DLP in non-stress-induced apoptosis, we utilized the UAS-GAL4 system to evaluate the effects of DLP overexpression on developing tissues. As EY09290 harbors a P-element with UAS in the 5′ region of DLP and the direction of the P-element is oriented to induce DLP gene expression (Figure 3A), the mutant was employed to study DLP overexpression. The induction of DLP expression was confirmed by RNA in situ hybridization in the wing imaginal discs of the EY09290 line harboring MS1096-GAL4 driver (Figure S6A). Interestingly, DLP overexpression in the developing wing under the control of MS1096-GAL4 reduced organ size in a dose-dependent manner (Figure 4A). When DLP was overexpressed in neurons, reduced survival and defects in locomotive behavior were observed (Figure S6B and S6C), suggesting that increased DLP expression has an adverse effect on neuronal development and function in Drosophila. Indeed, DLP overexpression strongly induced cell death in the imaginal disc (Figure 4B; compared with MS1096/Y and MS1096>DLP×2) without affecting the cell cycle or differentiation (Figure S6D and S6E). Furthermore, the DLP-induced reduction in wing size was suppressed almost completely by co-expression of Drosophila inhibitor of apoptosis protein 1 (DIAP1; Figure 4A), a caspase inhibitor. These results demonstrate that DLP activity induces apoptosis through caspase activation and reduces overall survival, as demonstrated by the reduced survival of DLP-overexpressing flies (Figure S6B).
Fig. 4. DLP activates apoptosis and the JNK/dFOXO signaling pathway. 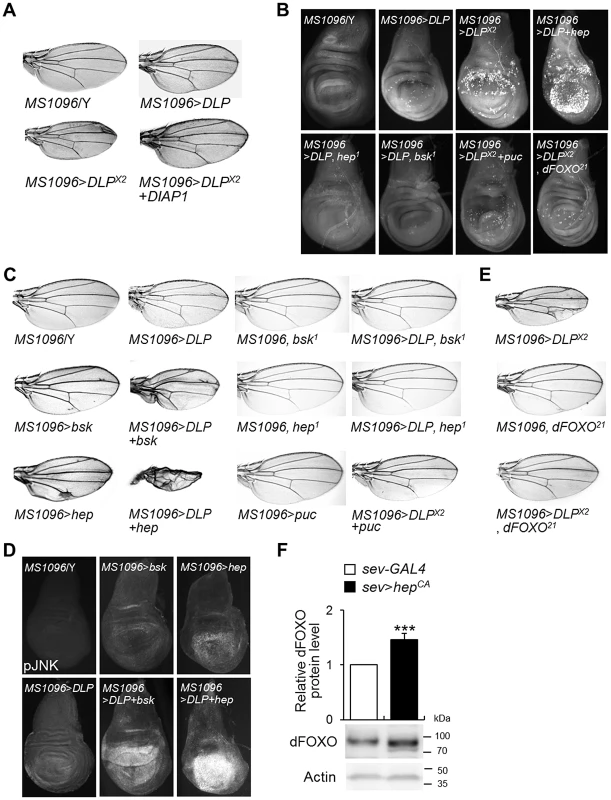
(A) Comparison of tissue sizes of control (MS1096/Y), DLP-overexpressing (MS1096>DLP and MS1096>DLP×2), and DLP- and DIAP1-coexpressing (MS1096>DLP×2+DIAP1) fly wings. Two copies of the DLP gene were overexpressed in MS1096>DLP×2. (B) Acridine orange-stained images of control (MS1096/Y), DLP-overexpressing (MS1096>DLP and MS1096>DLP×2), DLP- and hep-coexpressing (MS1096>DLP+hep), DLP-overexpressing and hep deficient (MS1096>DLP, hep1), DLP-overexpressing and bsk deficient (MS1096>DLP, bsk1), DLP- and puc-coexpressing (MS1096>DLP×2+puc), and DLP-overexpressing and dFOXO deficient (MS1096>DLP×2, dFOXO21) wing imaginal discs. (C) Genetic interactions of DLP with bsk, hep, and puc in the developing wing. The reduced wing phenotype induced by DLP overexpression (MS1096>DLP) was strongly exacerbated by bsk (MS1096>DLP+bsk) or hep (MS1096>DLP+hep) overexpression, and suppressed by bsk (MS1096>DLP, bsk1) or hep (MS1096>DLP, hep1) deficiency or co-expression of puc (MS1096>DLP×2+puc). (D) Comparison of JNK activity in the DLP- and bsk-coexpressing or DLP- and hep-coexpressing wing imaginal discs (MS1096>DLP+bsk or MS1096>DLP+hep) with DLP-, bsk- or hep-overexpressing wings (MS1096>bsk or MS1096>hep) by anti-phospho-JNK antibody staining. (E) Genetic interactions of DLP with dFOXO in the developing wing. The wing phenotype of DLP overexpression (MS1096>DLP×2) was strongly suppressed by dFOXO deficiency (MS1096>DLP×2, dFOXO21). MS1096 with dFOXO deficiency (MS1096, dFOXO21) was used as controls. (F) DLP protein levels in the control (sev-GAL4) and constitutive active hep-overexpressing (sev>hepCA) fly heads (Student's t-test, n = 9, *** p<0.001). sev, sevenless-GAL4. The data are expressed as means ± s.e. values. The genotypes of the samples were MS1096/Y (MS1096-GAL4/Y), MS1096>bsk (MS1096-GAL4/Y; UAS-bsk/+), MS1096>hep (MS1096-GAL4/Y; UAS-hep/+), MS1096, bsk1 (MS1096-GAL4/Y; bsk1/+), MS1096, hep1 (MS1096-GAL4/hep1), MS1096>DLP (MS1096-GAL4/Y; EY09290/+), MS1096>DLP+bsk (MS1096-GAL4/Y; EY09290/UAS-bsk), MS1096>DLP+hep (MS1096-GAL4/Y; EY09290/UAS-hep), MS1096>DLP, bsk1 (MS1096-GAL4/Y; EY09290/bsk1), MS1096>DLP, hep1 (MS1096-GAL4/hep1; EY09290/+), MS1096>DLP×2 (MS1096-GAL4/Y; EY09290/EY09290), MS1096>puc (MS1096-GAL4/Y; UAS-puc/+), MS1096>DLP×2+puc (MS1096-GAL4/Y; EY09290/EY09290; UAS-puc/+), MS1096, dFOXO21 (MS1096-GAL4/Y;; dFOXO21/+), MS1096>DLP×2, dFOXO21 (MS1096-GAL4/Y; EY09290/EY09290; dFOXO21/+), MS1096>DLP×2+DIAP1 (MS1096-GAL4/Y; EY09290/EY09290; UAS-DIAP1/+), sev-GAL4 (sev-GAL4/+), and sev>hepCA (sev-GAL4/+; UAS-hepCA/+). bsk, basket; DIAP1, Drosophila inhibitor of apoptosis protein 1; hep, hemipterous; pJNK, phospho-JNK; puc, puckered. In mammals, Daxx induces apoptosis by activating the JNK signaling pathway [35], [39]. We attempted to determine whether the JNK signaling pathway is activated by DLP in Drosophila. We initially examined the genetic interaction of DLP with basket (bsk), a Drosophila JNK, and hemipterous (hep), a Drosophila JNK kinase (JNKK). Although overexpression of DLP, bsk, or hep in the wing produced only a slight reduction in wing size, overexpression of either bsk or hep in conjunction with DLP resulted in severely rumpled and shrunken wings (Figure 4C). Consistently, co-expression of hep with DLP strongly induced cell death in the wing imaginal disc (Figure 4B). On the other hand, bsk or hep loss-of-function mutation or co-expression of puckered (puc), a negative regulator of JNK (a JNK phosphatase), suppressed the cell death and wing deformities induced by DLP overexpression (Figure 4B and 4C). Furthermore, co-expression of bsk or hep with DLP resulted in a strong increase of JNK phosphorylation (Figure 4D). These results demonstrate that DLP activates the JNK signaling pathway, similar to mammalian Daxx.
As FOXO is a target of JNK in mammalian systems [47] and acts downstream of JNK signaling in the control of apoptosis [48], we investigated the role of FOXO in DLP-induced apoptosis. We examined the effect of FOXO deficiency on the DLP-induced gain-of-function phenotype and apoptosis. FOXO deficiency (dFOXO21) suppressed the DLP-induced wing (Figure 4E) and cell death (Figure 4B) phenotypes, suggesting that the JNK/dFOXO pathway is downstream of DLP.
It has been shown that JNK activates FOXO4 activity through phosphorylation at Thr447 and Thr451 [47]. Therefore, we assessed whether dFOXO is also phosphorylated by JNK. Amino acid sequence analysis between mammalian FOXOs and dFOXO did not reveal the conserved Thr447 and Thr451 phosphorylation sites in dFOXO. Furthermore, we could not see any evidence to support direct phosphorylation of dFOXO by JNK in in vitro phosphorylation experiments (data not shown). However, interestingly, expression of constitutively active JNKK (hepCA) significantly increased the protein level of dFOXO (Figure 4F), implicating that JNK regulates dFOXO activity by increasing its protein level or stability in Drosophila.
DLP contributes to the oxidative stress-related phenotypes of DJ-1β mutants
Because DLP expression is regulated by DJ-1β under oxidative stress conditions (Figure 2) and DLP is important for oxidative stress-induced apoptosis and lethality (Figure 3 and Figure S5), we assessed the role of DLP in the oxidative stress-related phenotypes of DJ-1β mutants, specifically their acute sensitivity to oxidative stress and locomotive dysfunction. To accomplish this, we generated DLP and DJ-1β double mutants and asked whether DLP deficiency could rescue various DJ-1β mutant phenotypes. We first evaluated the H2O2 sensitivity of these lines. As shown in Figure 5A, DLP and DJ-1β double mutants displayed survival rates similar to those of wild-type controls, whereas DJ-1β mutants were acutely sensitive to H2O2. However, the survival rates of DLP and DJ-1β double mutants was still lower than those of DLP mutants (compare Figure 5A with Figure 3E), which suggests that the H2O2 sensitivity of DJ-1β is not fully dependent on DLP. Supporting this, gene expression of 18 oxidative stress-related genes (11 up-regulated and 7 down-regulated) was altered in DJ-1β mutants versus wild-type controls (Table S1). Although DLP is not expected to mediate the whole effect of the oxidative stress responses in DJ-1β mutants, DLP deficiency strongly suppressed the oxidative stress-induced cell death observed in DJ-1β mutant brains (Figure 5B). Consistently, oxidative stress-induced DA neuronal death in DJ-1β mutants was almost completely inhibited by DLP deficiency (Figure 5C and Figure S7). These findings indicate that DLP is a key mediator in the oxidative stress-induced neuronal cell death of DJ-1β mutants. Furthermore, DLP deficiency rescued the UV sensitivity (Figure 5D) and locomotive dysfunction (Figure 5E) of DJ-1β mutants. These results strongly suggest that DLP mediates the H2O2-induced oxidative stress responses, UV sensitivity, and locomotive dysfunction of DJ-1β mutants.
Fig. 5. DLP deficiency reduces acute sensitivity to oxidative stress and UV, and improves locomotive dysfunction in DJ-1β mutant. 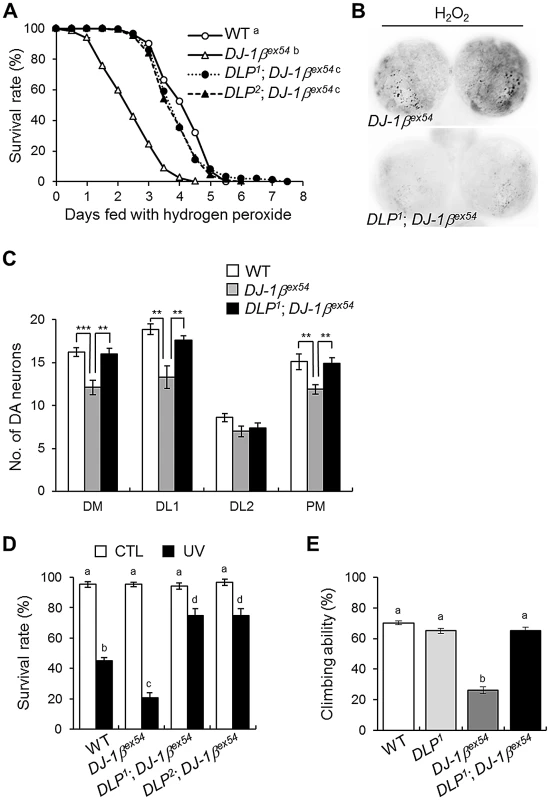
(A) Comparison of the survival rates of DLP and DJ-1β double mutants (DLP1; DJ-1βex54 and DLP2; DJ-1βex54) with wild-type (WT) and DJ-1βex54 flies under oxidative stress conditions (log-rank test: WT, n = 250; DJ-1βex54, n = 250; DLP1; DJ-1βex54, n = 250; DLP2; DJ-1βex54, n = 250, p<0.01, groups with the same letter do not differ significantly). (B) Reduced oxidative stress-induced apoptosis was noted in the larval brain of the DLP and DJ-1β double mutant (DLP1; DJ-1βex54) compared to DJ-1βex54. The larval brains were treated with 0.1% H2O2 for 24 h and cell death was detected via acridine orange staining. (C) Sensitized DA neuronal death of DJ-1βex54 under oxidative stress conditions was rescued by DLP deficiency. The flies were fed with 1% H2O2 for 3 days. (n = 10, Student's t-test: DM, ** p<0.01, *** p<0.001; DL1, ** p<0.01; PM, ** p<0.01). (D) Survival rates of WT, DJ-1βex54, and double mutant of DLP and DJ-1β (DLP1; DJ-1βex54 and DLP2; DJ-1βex54) pupae after exposure to UV irradiation (10 mJ/cm2; black bars) as described in the Materials and Methods (Kruskal-Wallis test: CTL, n≥6, p>0.1; UV, n≥5, p<0.01, groups with the same letter do not differ significantly). CTL, UV-untreated control pupae; UV, UV-treated pupae. (E) Comparison of climbing abilities of WT, DLP1, DJ-1βex54, and double mutants of DLP and DJ-1β (DLP1; DJ-1βex54). The climbing abilities of 5-day-old flies for each group were tested as described in the Materials and Methods (ANOVA and Tukey's HSD analysis: n≥12, p<0.01, groups with the same letter do not differ significantly). All data are expressed as means ± s.e. values. DJ-1β negatively regulates DLP expression through dFOXO
Next, we investigated the mechanism by which DJ-1β regulates DLP expression. Previous studies with mammalian DJ-1 suggest that DJ-1 affects oxidative stress-related gene expression by stabilizing Nrf2 [29] or by suppressing p53 transcriptional activity [34]. Alternatively, DJ-1 regulates phosphatidylinositol 3-kinase (PI3K)/Akt signaling [19], [49], [50], which inhibits FOXO. Therefore, we tested whether cap'n'collar C (cncC, Drosophila Nrf2), p53, or dFOXO is involved in the regulation of DLP gene expression by DJ-1β under oxidative stress conditions. As demonstrated in Figure 6A and Figure S8, neither cncC nor p53 affected the DLP levels in wild-type or DJ-1β mutant flies. However, the increased DLP mRNA (Figure 6B) and protein (Figure 6C) levels in DJ-1β mutants were restored to normal levels by dFOXO deficiency, suggesting dFOXO is required for elevation of DLP expression in DJ-1β mutants. Thus, when dFOXO was overexpressed in neurons, DLP expression was elevated more than 2-fold for both mRNA (Figure 6D) and protein (Figure 6E). Since a putative FOXO recognition element (FRE, AAAAACA) is located at 1,041 bp upstream of the transcription start site of the DLP gene (Figure 6F) [51], to examine the positive effect of dFOXO on the DLP gene expression at the transcriptional level, we cloned two different sizes of the DLP promoter region into a firefly reporter plasmid; 0.5 kb and 1.3 kb, respectively. Upon transient co-transfection with a construct expressing the constitutively active form of dFOXO (pMT-dFOXO A3), the construct containing the 1.3-kb fragment of the promoter region, but not the 0.5-kb fragment, exhibited the dFOXO-dependant promoter activity in a dose dependent manner (Figure 6F). To confirm whether this putative FRE site is critical, a mutation that disrupts the dFOXO binding was introduced into the 1.3-kb promoter construct by site-directed mutagenesis. Upon transfection, the construct containing the mutant 1.3-kb promoter region no longer showed dFOXO-dependent promoter activity (Figure 6F), indicating that dFOXO regulates DLP expression through this FRE site.
Fig. 6. The role of dFOXO in the regulation of DLP by DJ-1β. 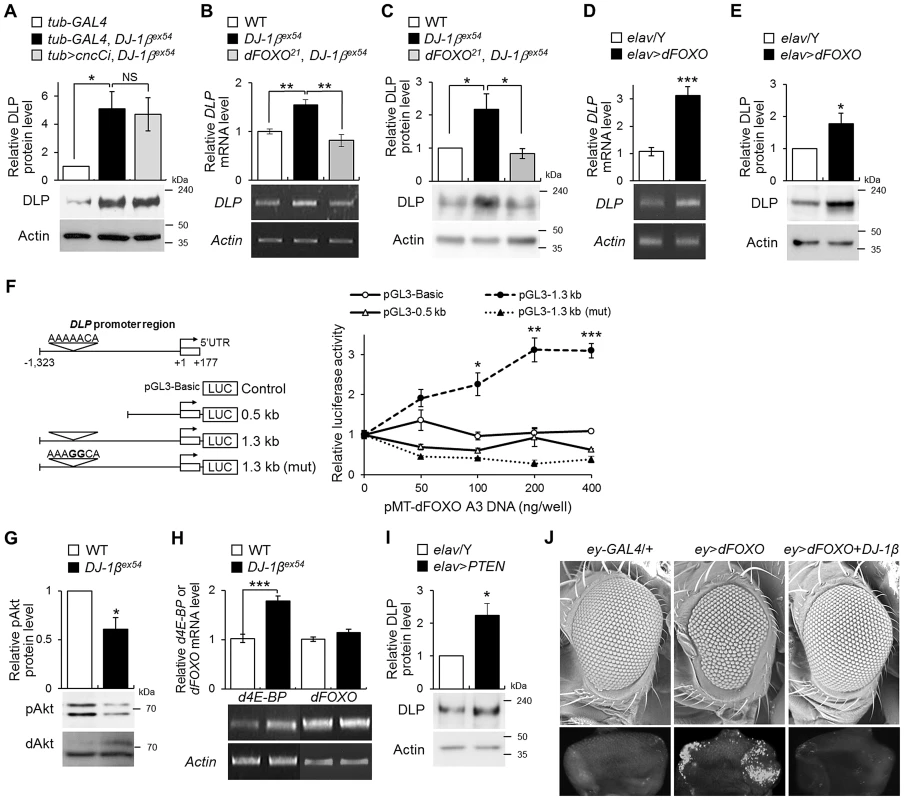
(A) DLP protein levels in the heads of control (tub-GAL4), DJ-1β mutant (tub-GAL4, DJ-1βex54) and the double mutant of cncC and DJ-1β (tub>cncCi, DJ-1βex54) flies fed with 1% H2O2 (Student's t-test, n = 4, * p<0.05). NS, not significant. (B–C) DLP mRNA (B) and protein (C) levels in the head of WT, DJ-1βex54, and dFOXO and DJ-1β double mutant (dFOXO21, DJ-1βex54) flies fed with 1% H2O2 (B, Student's t-test, n = 5, ** p<0.01; C, Student's t-test, n = 4, * p<0.05). (D–E) DLP mRNA (D) and protein (E) levels in control (elav/Y) and pan-neuronally dFOXO-overexpressing (elav>dFOXO) fly heads (D, Student's t-test, n = 6, *** p<0.001; E, Student's t-test, n = 5, * p<0.05). (F) Luciferase assays showed activation of DLP promoters in S2 cells after cotransfection with dFOXO-A3. (Open circle) Empty vector. (Open triangle) 0.5-kb fragment of DLP promoter. (Filled circle) 1.3-kb fragment of DLP promoter. (Filled triangle) 1.3-kb fragment of DLP promoter with mutation in the putative FRE site: pGL3–1.3 kb (mut). Bold characters in the putative FRE site represent the mutated nucleotides. (Student's t-test, n = 3, * p<0.05; ** p<0.01; *** p<0.001). (G) The levels of phospho-Akt in the head of WT and DJ-1βex54 flies fed with 1% H2O2 (Student's t-test, n = 3, * p<0.05). dAkt was used as an internal control. (H) d4E-BP, a target of dFOXO, and dFOXO mRNA levels in the head of WT and DJ-1βex54 flies fed with 1% H2O2 (Student's t-test: d4E-BP, n = 7, *** p<0.001; dFOXO, n = 7). (I) DLP protein levels in the control (elav/Y) and pan-neuronally PTEN-overexpressing (elav>PTEN) fly heads (Student's t-test, n = 4, * p<0.05). (J) Genetic interactions of dFOXO with DJ-1β in the developing eye. The upper pictures are scanning electron micrographs of the fly eyes. The lower pictures are acridine orange-stained images of the eye imaginal discs. The genotypes of the samples were tub-GAL4 (tub-GAL4/+), tub-GAL4, DJ-1βex54 (tub-GAL4, DJ-1βex54/DJ-1βex54), tub>cncCi, DJ-1βex54 (UAS-cncC-RNAi/+; tub-GAL4, DJ-1βex54/DJ-1βex54), dFOXO21, DJ-1βex54 (dFOXO21, DJ-1βex54/DJ-1βex54), elav/Y (elav-GAL4/Y), elav>dFOXO (elav-GAL4/Y; UAS-dFOXO/+), elav>PTEN (elav-GAL4/Y; UAS-PTEN/+), ey-GAL4 (ey-GAL4/+), ey>dFOXO (UAS-dFOXO/+; ey-GAL4/+), and ey>dFOXO+DJ-1β (UAS-dFOXO/UAS-HA-DJ-1β; ey-GAL4/+). pAkt, phospho-Akt; ey, eyeless. All data are expressed as means ± s.e. values. Actin was used as an internal control. Additionally, we revealed that phospho-Akt (an activated form of Drosophila Akt (dAkt), a negative regulator of dFOXO) was significantly reduced in DJ-1β mutants (Figure 6G). We also found that gene expression of Drosophila 4E-BP, a target of dFOXO, increased in DJ-1β mutants, although dFOXO gene expression remained unaltered (Figure 6H). Moreover, overexpression of PTEN, a negative regulator of the Akt signaling pathway and, therefore, an activator of dFOXO, elevated the DLP level (Figure 6I). These results consistently indicate that DJ-1β regulates DLP gene expression through the PI3K-Akt-dFOXO pathway.
Finally, we investigated whether DJ-1β could suppress dFOXO-induced apoptosis. As shown in Figure 6J, DJ-1β overexpression strongly suppressed dFOXO-induced eye degeneration and apoptosis (compare ey>dFOXO with ey>dFOXO+DJ-1β).
Discussion
Recently, several Drosophila PD models, each of which represents a different PD-associated gene mutant, have been developed and characterized [10]–[13], [16], [24], [49], [52]–[54]. Although each exhibits a distinct phenotype, a common feature of all these models is sensitization to oxidative stress. This, along with pathology data from PD patients [55]–[57], strongly indicates a significant role for oxidative stress in the development and progression of PD. Since the DJ-1 mutations have been linked to familial PD and hypersensitivity to toxins that induce oxidative stress [23], we examined a signaling pathway that controls oxidative stress responses by DJ-1 in Drosophila. We demonstrated that DJ-1β inhibits oxidative stress-induced neuronal apoptosis by regulating DLP gene expression and protein subcellular localization, suggesting a causal relationship between DJ-1β mutation and oxidative stress-induced DA neuronal loss in PD.
Our genetic and cellular analyses indicate DLP functions as a pro-apoptotic gene and as a JNK activator in Drosophila, like its mammalian homologue Daxx. Previous studies have shown that Daxx is upregulated in response to oxidative stress and UV irradiation; it also mediates apoptosis in these contexts [40], [41]. Consistent with these reports, our results demonstrate that DLP expression is elevated by H2O2 and UV exposure. Moreover, the apoptosis induced by these insults is reduced dramatically in DLP mutants. In contrast to oxidative stress or UV irradiation, γ-ray irradiation (40 gray)-induced apoptosis was unaffected by DLP deficiency (data not shown). This is consistent with a previous report, which demonstrated that DLP is not associated with radiosensitivity [45]. These findings suggest DLP does not function as a general pro-apoptotic factor, but rather exerts a pro-apoptotic function in response to specific insults, including oxidative stress and UV irradiation. Moreover, the level of DLP in neurons was associated with fly survival rates under oxidative stress conditions. The pan-neuronal overexpression of DLP rendered flies more sensitive to oxidative stress than controls, while knockdown or loss of DLP resulted in resistance to oxidative stress. Therefore, we believe DLP functions as a stress response mediator that generates appropriate cellular responses to oxidative stress. The similarities between the functions of DLP and Daxx suggest that this oxidative stress response pathway is highly conserved from insects to mammals.
Due to the pronounced increase in oxidative damage within DJ-1β mutants [52], we hypothesized that DLP functions as an important mediator of hypersensitivity to oxidative stress in DJ-1β mutants. Indeed, DLP expression and translocation from the nucleus to the cytoplasm increased in DJ-1β mutants, and DLP deficiency almost completely rescued the phenotypes of DJ-1β mutants, including oxidative stress-induced DA neuronal loss. Moreover, overexpression of DJ-1β reduced the level of endogenous DLP. These findings suggest DLP plays an important function in the oxidative stress-related phenotypes of DJ-1β mutant flies and that DJ-1β protects flies against oxidative stress, at least in part, by suppression of DLP expression and cytosolic localization.
These observations raised the question of how DJ-1β negatively regulates DLP expression at the transcriptional level under oxidative stress conditions. Our data indicate that DJ-1β controls DLP gene expression by regulating the activity of dFOXO. Furthermore, DLP harbors a consensus FRE in its promoter region and dFOXO overexpression increased DLP expression in neurons. Previous studies as well as this work identified DJ-1 as a positive regulator of the PI3K/Akt pathway [19], [49], [50], which suppresses the activity of FOXO by phosphorylation [58]. Therefore, it was not surprising to see that loss of DJ-1β function reduced dAkt activity and increased the transcriptional activity of dFOXO (Figure 6G–6H). FOXO activity may be crucial for setting the sensitivity threshold for oxidative stress and determining the appropriate level of stress responses, which ultimately determines whether the cells live or die. From this perspective, the elevated level of dFOXO activity in DJ-1β mutants may render their neurons more sensitive to stress, and thus neurons in mutant animals die more readily than their wild-type counterparts. We also found that dFOXO performs a dual role in DJ-1β mutant flies, as it is required for the upregulation of DLP and is an effector of DLP. Both of these roles increase the DLP-mediated apoptosis in response to oxidative stress in DJ-1β mutants. This suggests that dFOXO is involved in the loss of neurons due to oxidative stress, and possibly, DJ-1 mutation-associated familial PD cases.
In addition to transcriptional regulation, DJ-1 controls DLP translocation from the nucleus to the cytosol. Daxx is translocated to the cytosol under oxidative stress conditions [59] and this translocation is important for its pro-apoptotic function [60]. These studies and our results showed that both mammalian and Drosophila DJ-1 strongly suppress the cytosolic translocation of Daxx/DLP. The molecular mechanism by which DJ-1 suppresses the DLP translocation is elusive. It has been proposed that mammalian DJ-1 directly binds to Daxx and inhibits its translocation. However, we did not observe prominent binding between Drosophila DJ-1 and DLP (data not shown), suggesting Drosophila DJ-1 may regulate the DLP translocation by an alternative mechanism. Interestingly, the mitochondrial targeted DJ-1β efficiently inhibited the Daxx translocation under oxidative stress conditions, suggesting the function of DJ-1 in mitochondria is important for the translocation. Further studies are necessary to understand how DJ-1 inhibits the cytosolic localization of Daxx/DLP under oxidative stress.
Our work with DJ-1β and its downstream effector, DLP, has led us to propose the models illustrated in Figure 7. In a wild-type animal, DJ-1 protects the cells from oxidative stress-induced apoptosis. This protection is a result of activation of the PI3K/Akt pathway that inhibits dFOXO. dFOXO, among its many functions, induces DLP transcription. DLP expression levels have a direct positive correlation with the likelihood of a cell to undergo apoptosis in response to oxidative stress. It is important to note that not only does DJ-1β suppress DLP expression, but DJ-1β also prevents DLP translocation to the cytosol, which may be critical for the pro-apoptotic function of DLP. However, once cells are damaged by oxidative stress and UV irradiation, the DLP protein acts through the JNK pathway to initiate apoptosis. Since the JNK pathway can increase dFOXO activity, DLP expression can be further increased by a hypothetical feed-forward loop of DLP-JNK-dFOXO. In wild-type animals, this pro-apoptotic loop can be negatively regulated by DJ-1β, while in DJ-1β mutant animals, the inability to control this process leads to increased DLP levels and apoptosis. This increased chance of apoptosis may be an important factor in the development of PD.
Fig. 7. Schematic representation of the role of Drosophila DJ-1 in the cellular response to oxidative stress or UV. 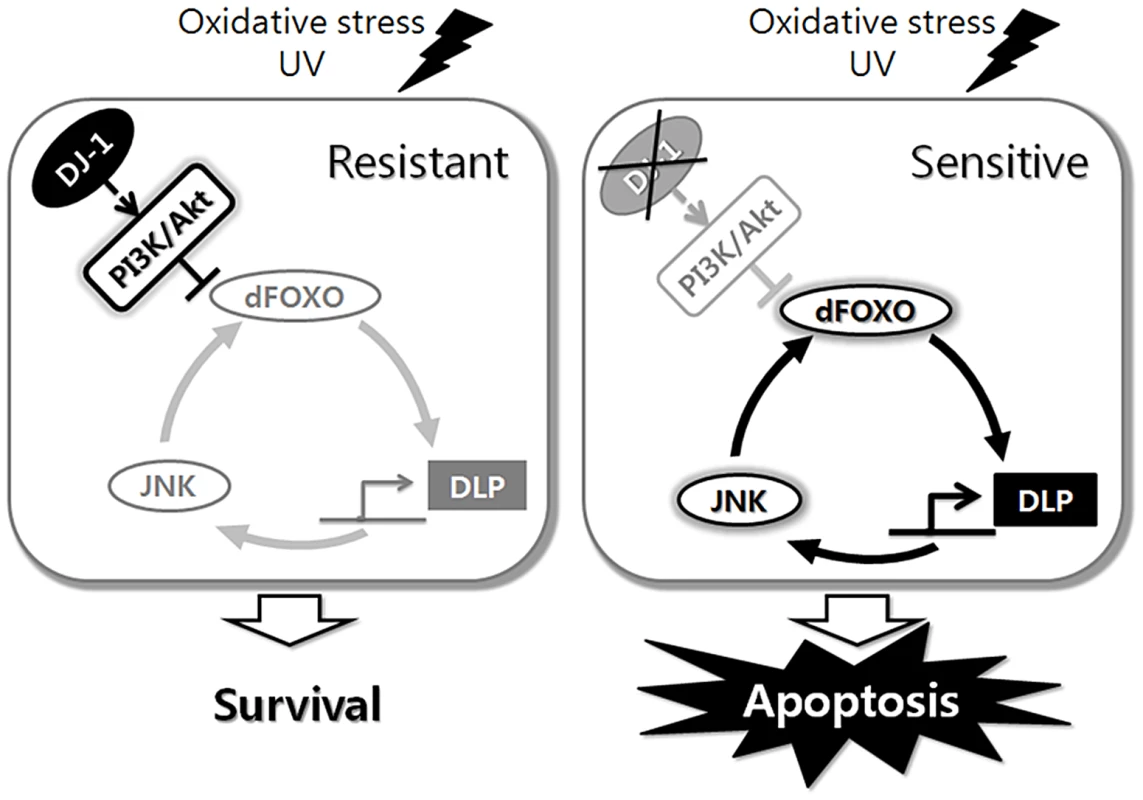
dFOXO, DLP, and JNK form a circuit that controls cellular responses to stress, and DJ-1 sets neural sensitivity to stress by regulating this circuit. Materials and Methods
Drosophila strains
DJ-1βex54 and p53E4 were previously described [24], [61], and UAS-HA-DJ-1β was generated via microinjection of the corresponding plasmid into w1118 embryos. EY09290, KG01694, basket1 (bsk1), UAS-DIAP1, UAS-dFOXO, UAS-PTEN, elav-GAL4, Glass multimer reporter (GMR)-GAL4, tubulin (tub)-GAL4, eyeless (ey)-GAL4, sevenless (sev)-GAL4, wingless (wg)-lacZ, and engrailed (en)-lacZ were acquired from the Bloomington Drosophila Stock Center (Bloomington, IN, USA). UAS-DLP-RNAi was obtained from the Vienna Drosophila RNAi Center (Vienna, Austria). UAS-basket (bsk) and UAS-hemipterous (hep) were gifts from Dr. M. Mlodzik (EMBL, Germany). UAS-puckerd (puc) and MS1096-GAL4 were generously provided by Dr. M. Peifer (University of North Carolina, Pembroke, NC) and Dr. M. Freeman (MRC Laboratory of Molecular Biology, Cambridge, UK), respectively. UAS-cncC and UAS-cncC-RNAi were gifts from Dr. D. Bohmann (University of Rochester Medical Center). DLPU42 and dFOXO21 were gifts from Dr. I. M. Boros (University of Szeged, Hungary) and Dr. E. Hafen (University of Zurich, Switzerland), respectively. hemipterous1 (hep1) was gift from Dr. S. Noselli (CNRS, France). UAS-hemipterousCA (hepCA, the constitutively active form of Drosophila JNKK) was gift from Dr. K. Mastsumoto (Nagoya University, Japan). All fly strains were maintained at 25°C.
Microarray
Total RNA was extracted from the heads of hydrogen peroxide-treated wild-type and DJ-1β mutant flies using an RNeasy Mini kit (Qiagen) in accordance with the manufacturer's instructions. Total RNA was used as a probe for microarray analyses. GeneChip Drosophila Genome 2.0 Arrays for Drosophila melanogaster were probed, hybridized, stained, and washed in accordance with the manufacturer's recommendations. Hybridized arrays were scanned using an Affymetrix Command Console, and normalization was conducted using an Affymetrix Expression Console 1.1 (MAS5). These experiments were repeated three times for each sample. We required that the fold change difference between the average of three independent wild-type samples and the average of three independent DJ-1β mutant samples exceed 1.5 (p≤0.005).
Generation of DLP mutants
To generate DLP mutant flies, we used the EY09290 and KG01694 lines (Bloomington, USA) containing the UAS in the first exon and first intron of the DLP gene, respectively. Two DLP mutants, DLP1 and DLP2, were generated via the imprecise excision of a P-element in the EY09290 line, and DLP3 and DLP4 were obtained from the KG01694 line. Additionally, the revertant line, DLPrv, was also generated via precise excision of the P-element in the EY09290 line. Among the excision lines, the deletion lines were selected via genomic DNA PCR. Genomic DNA was prepared from each independent line, and PCR was conducted with primer pairs to amplify sequences lying upstream and downstream of the P-element insertion site. The deletion sites of the selected lines were determined by sequencing the PCR products. To isogenize the genetic background, DLP1 and DLP2 were backcrossed with w1118 ten times.
Immunoblotting
A polyclonal antibody against the C-terminus of Drosophila DLP (amino acids 1300–1559) was generated in rabbits via injection of pGEX-fused DLP. The specificity of the DLP antibody was verified by immunoblotting and immunohistochemistry using wild-type and DLP mutant fly tissues (Figure 3C and Figure S2). Cell and fly lysates were prepared in lysis buffer A (150 ml NaCl, 25 mM Tris, 10% Glycerol, 0.1% NP-40, and 1 mM EDTA). For western blotting, the membranes were probed with anti-DLP (1∶1,000 in Tris-buffered saline with Tween 20 (TBST)), anti-lamin (1∶2,000 in TBST; Developmental Studies Hybridoma Bank (DSHB)), anti-β-tubulin (1∶2,000 in TBST; DSHB), anti-phospho-Drosophila Akt (Ser505) (1∶1,000 in TBST; Cell Signaling Technology), anti-Akt (1∶1,000 in TBST; Cell Signaling Technology), anti-dFOXO (1∶2,000 in TBST; Cosmo Bio, Japan), or anti-Actin (1∶2,000 in TBST; DSHB at the University of Iowa) antibodies. Western blot analyses were conducted with standard procedures using horseradish peroxidase-conjugated secondary antibodies (1∶2,000 in TBST; Cell Signaling Technology).
Nuclear/cytosolic fractionation
To separate nuclear and cytosolic fractions, 15 fly heads were collected and homogenized. Nuclear and cytosolic fractions from the fly heads were isolated with Nuclear Extract Kit (Active Motif) in accordance with the manufacturer's instructions.
RNA in situ hybridization
In situ hybridization experiments were conducted using a digoxigenin-labeled RNA probe (Roche Applied Science) in accordance with the manufacturer's instructions. The probe was prepared using the PCR product of DLP. Hybridization was conducted at 55°C, and the RNA hybrids were detected with alkaline phosphatase (AP)-conjugated anti-digoxigenin antibody followed by nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl-phosphate (BCIP) staining.
Analysis of Drosophila development
One hundred embryos of each genotype were placed on grape juice agar plates. After incubation for 2 days at 25°C, the number of larvae hatched was counted to determine embryonic lethality. These larvae were then transferred to standard media (cornmeal, yeast, molasses, agar) and aged at 25°C in upright standard plastic shell vials. Larvae were maintained under non-crowded conditions with 20 individuals per vial. The numbers of pupae and enclosed adult flies were counted. Experiments were repeated 3 times with 100 flies per genotype.
UV irradiation
UV irradiation experiments were conducted as previously described with some modifications [48]. In brief, 10 mid-aged pupae were collected, and the pupal shells surrounding the heads were surgically removed. The samples were UV irradiated at 10 mJ/cm2 using a UV crosslinker (CL-1000, UV Products). Following irradiation, the pupae were kept in darkness until processing, and the survival rate was determined. Each experiment was repeated more than 5 times. To analyze the effects of UV irradiation on cell death or DLP gene expression in the embryos, 0–3 h embryos were UV-irradiated at a dose of 50 mJ/cm2.
Oxidative stress test
The effects of oxidative stress on the survival of the indicated lines were evaluated by feeding with hydrogen peroxide. Fifty 3-day-old male flies of the indicated lines were starved for 6 h and then transferred to vials containing 1% hydrogen peroxide in 5% sucrose solution. Surviving flies were counted semi-diurnally. We carried out each survival experiment at least 5 times with 50 flies per genotype (n≥250). In order to evaluate gene expression and protein levels under oxidative stress conditions, the flies were fed with 1% hydrogen peroxide for 3 days.
Preparation of RNA and real-time quantitative PCR
For real-time quantitative PCR, total RNA from the 20 fly heads was isolated with an RNeasy Protect Mini kit (Qiagen). Then, cDNA was synthesized with a Maxime kit (iNtRON Biotechnology), and real-time quantitative PCR was undertaken using SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's recommended protocols. Real-time quantitative PCR was performed using StepOne Real-time PCR system (Applied Biosystems). Quantification was performed using the ‘delta-delta Ct’ method to normalize to Actin transcript levels and to control. Each experiment was repeated at least 5 times (n≥5). The relative level of DLP, d4E-BP or dFOXO mRNA to Actin mRNA was statistically analyzed by Student's t-test.
Primers
To determine the deletion sites of the DLP mutant lines, PCR was conducted using the following primer pairs: 5′-ACTGCAAATAGTGAATTAAGGCAAC-3′ and 5′ - TGCAACATGGGAAGTCTCTG-3′. To quantify the level of gene expression, real-time quantitative PCR was conducted using the following primer pairs: DLP, 5′-CACATCCCCAGTGGAATCAC-3′ and 5′-TGCCAACATTGATCTGCTTC-3′; Actin, 5′-CACCGGTATCGTTCTGGACT-3′ and 5′-GCGGTGGTGGTGAAAGAGTA-3′; dFOXO, 5′-GCCTGGAGGTGCTCAATAAC-3′ and 5′-GTGGCCAGCGGTATATTGAT-3′; and d4E-BP, 5′-CCATGATCACCAGGAAGGTT-3′ and 5′-GAAAGCCCGCTCGTAGATAA-3′.
Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end-labeling (TUNEL) assay
For the TUNEL assay, embryos treated with UV were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature. The samples were then washed with PBS and permeabilized by a 2-min incubation in PBS containing proteinase K (10 µg/mL) and 0.1% Triton X-100 on ice. After extensive washing, the samples were incubated for an additional 3 h in TUNEL reaction solution (Roche Applied Science) at 37°C in accordance with the manufacturer's recommendations. After three rinses with PBS, the embryos were incubated with anti-digoxigenin-AP antibody and stained with NBT/BCIP.
Construction of subcellular organelle-targeted DJ-1β
Full-length DJ-1β cDNA was cloned into dsRed2-Mito vector (Clontech) to target DJ-1β to mitochondria. dsRed2-Mito vector contains the mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase at the N-terminus. For the nucleus-targeted DJ-1β, pEF/3×NLS/Myc vector (Invitrogen) was used. To construct the cytoplasmic membrane-targeted DJ-1β, 20-amino acid farnesylation signal from c-Ha-Ras was fused to the C-terminus of DJ-1β resulting in pcDNA3 3×HA DJ-1β-F. For Golgi-targeted DJ-1β, Golgi targeting sequence from β 1,4-galactosyltransferase was fused to the N-terminus of DJ-1β resulting in pcDNA3 Golgi DJ-1β.
Mammalian cell culture and DNA transfection
HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2. WT and DJ-1 null SN4741 cells were established from the substantia nigra region of E13.5 wild-type and DJ-1 knockout mouse embryos, respectively [62]. SN4741 cells were grown in RF medium containing DMEM supplemented with 10% fetal bovine serum, 1% glucose, and L-glutamine (2 mM) at 33°C with 5% CO2. The transfection of expression plasmids was performed using Lipofectamine plus reagent (Invitrogen), or PEI (Polyethylenimine, Sigma) according to the manufacturer's instruction.
Immunocytochemistry
For immunocytochemistry, SN4741 or HeLa cells were sub-cultured on 12-well culture plates coated with poly-L-lysine (Sigma). Appropriately treated cells were washed once with PBS and fixed in 2% paraformaldehyde for 15 min, followed by permeabilization with 0.5% Triton X-100 in PBS for 5 min. Then, the cells were washed with 0.1% Triton X-100 in PBS (PBS-T) and incubated in blocking solution (4% BSA and 1% normal goat serum in PBS-T) for 1 h. Primary antibodies were added to the blocking solution and the cells were incubated overnight at 4°C. After washing with PBS-T 3 times, the cells were incubated with appropriate secondary antibodies in blocking solution for 45 min at room temperature. The antibody-labeled cells were washed with PBS-T 6 times and mounted with mounting solution [100 mg/mL 1,4-diazabicyclo[2.2.2]octane (DABCO) in 90% glycerol]. The slides were observed with LSM710 laser-scanning confocal microscope (Carl Zeiss). All immunostaining experiments with HeLa cells were conducted at least 3 times (n = 300). Anti-mouse DJ-1β [24] and anti-rabbit Daxx (Cell Signaling Technology) were used as primary antibodies. MitoTracker Red CMXRos (Invitrogen) was used to visualize mitochondria.
Immunohistochemistry
For immunohistochemistry, the wing or eye imaginal discs or adult brains were fixed in 4% paraformaldehyde in PBS at room temperature. The tissues were then washed in PBT (PBS+0.5% Triton X-100) and blocked in PBT with 2% normal goat serum (NGS). The samples were incubated first with rabbit anti-phospho-JNK antibody (1∶200 in PBT containing 2% NGS; Promega) or rabbit anti-phospho-histone H3 antibody (1∶200 in PBT containing 2% NGS; Upstate Biotechnology) or rabbit anti-DLP antibody (1∶200 in PBT containing 2% NGS) or rabbit anti-tyrosine hydroxylase antibody (1∶50 in PBT containing 2% NGS; Pel-Freez Biologicals) and then subsequently incubated with rhodamine-labeled goat anti-rabbit immunoglobulin G secondary antibody (1∶200 in PBT; Sigma-Aldrich).
Ectopic gene expression with the UAS-GAL4 system
The UAS-GAL4 system was used to evaluate the phenotypes induced by the overexpression of several target genes, including DLP. The GAL4 gene was placed near a tissue-specific enhancer, allowing for the ectopic expression of the target gene in the desired tissue. GMR-GAL4, MS1096-GAL4, elav-GAL4, tub-GAL4, and ey-GAL4 were used to induce target gene expression in the eye, the whole wing, the nervous system, the whole body, and the eye, respectively.
Acridine orange staining
Acridine orange staining was conducted as previously described [63] with some modifications. The wing or eye imaginal discs of stage L3 larvae were dissected in PBS. In order to characterize the effects of oxidative stress on cell death, we incubated the larval brains for 24 h in Schneider's Drosophila media with 0.1% hydrogen peroxide. The discs or brains were then incubated for 5 min in 1.6×10−6 M acridine orange (Sigma-Aldrich) and briefly rinsed in PBS. The samples were subsequently observed under an Axiophot2 fluorescence microscope (Carl Zeiss).
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) staining
For X-gal staining, the wing discs were fixed for 4 min in 4% formaldehyde in PBS, washed, and incubated in standard X-gal staining solution (4.9 mM X-gal, 3.1 mM K4Fe(CN)6, 3.1 mM K3Fe(CN)6, 1 mM MgCl2, 150 mM NaCl, 10 mM Na2HPO4, 10 mM NaH2PO4, 0.3% Triton X-100) for 30 min at 37°C before observation.
S2 cell culture and luciferase assay
Drosophila S2 cells were transiently transfected using the Effectene transfection reagent (Qiagen Inc., Valencia, CA) using a standard protocol. After 24 h, CuSO4 (Sigma) was added to a final concentration of 0.6 mM for the optimal expression of the dFOXO A3 construct (a gift from Dr. Oscar Puig, Roche). Dual luciferase assays were performed using the dual luciferase assay system (Promega Corp., Madison, WI). Harvested cells were lysed with luciferase cell lysis buffer. Cell lysates (20 µl out of 30 µl total lysate per sample) were analyzed for firefly luciferase activity by adding 20 µl of firefly reaction buffer. Furthermore, Renilla luciferase activity was measured by adding 20 µl of Renilla reaction buffer. Luminescence was measured from a 96-well plate by using VictorX5 multilabel plate reader (Perkin-Elmer). The data are presented as fold changes relative to the negative control (transfection with the pMT empty vector as an effecter plasmid) normalized to 1.
Climbing assay
The climbing assay was conducted as previously described [64], [65] with some modifications. Ten male flies of the indicated lines were transferred into the climbing ability test vial and incubated for 1 h at room temperature for environmental acclimation. After tapping the flies down to the bottom, we counted the number of flies that climbed to the top of the vial within 4 sec. Ten trials were conducted for each group. The experiment was repeated at least 10 times with independently derived transgenic lines. Climbing scores (ratio of the number of flies that climbed to the top to the total number of flies, expressed as a percentage) were obtained for each test group, and the mean climbing score for 10 repeated tests was compared to the scores of the wild-type flies. All climbing assay experiments were conducted at 25°C.
Statistics
Western blotting data were measured using the Multi gauge V3.1 (Fuji, Japan) software program and converted into ratios of band intensity relative to the controls. Using the non-parametric Wilcoxon signed-rank test or the Kruskal-Wallis test, the data were analyzed to detect any statistical differences between treatments. In particular, when the data analyzed with the Kruskal-Wallis test revealed a statistical difference, the data were arcsine-transformed and subsequently analyzed by ANOVA followed by Tukey's HSD post-hoc analysis. The climbing assay data were arcsine-transformed, and then ANOVA with Tukey's HSD post hoc analyses were conducted to detect any differences in climbing ability between treatments. The Kaplan-Meier estimator and the log-rank test were conducted on the pooled cumulative survival data to determine whether each treatment had any effect on the longevity of individuals using Online Application Survival Analysis Lifespan Assays (http://sbi.postech.ac.kr/oasis) [66].
Supporting Information
Zdroje
1. EmeritJ, EdeasM, BricaireF (2004) Neurodegenerative diseases and oxidative stress. Biomed Pharmacother 58 : 39–46.
2. LinMT, BealMF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443 : 787–795.
3. TsangAHK, ChungKKK (2009) Oxidative and nitrosative stress in Parkinson's disease. Biochim Biophys Acta 1792 : 643–650.
4. JennerP (2007) Oxidative stress and Parkinson's disease. Handb Clin Neurol 83 : 507–520.
5. ZhouC, HuangY, PrzedborskiS (2008) Oxidative Stress in Parkinson's Disease. Ann N Y Acad Sci 1147 : 93–104.
6. JennerP (2003) Oxidative stress in Parkinson's disease. Ann Neurol 53: S26–S38.
7. GiassonBI, LeeVMY (2000) A new link between pesticides and Parkinson's disease. Nat Neurosci 3 : 1227–1228.
8. ChungKKK, ThomasB, LiX, PletnikovaO, TroncosoJC, et al. (2004) S-nitrosylation of parkin regulates ubiquitination and compromises Parkin's protective function. Science 304 : 1328–1331.
9. LaVoieMJ, OstaszewskiBL, WeihofenA, SchlossmacherMG, SelkoeDJ (2005) Dopamine covalently modifies and functionally inactivates parkin. Nat Med 11 : 1214–1221.
10. PesahY, PhamT, BurgessH, MiddlebrooksB, VerstrekenP, et al. (2004) Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131 : 2183–2194.
11. MenziesFM, YenisettiSC, MinK-T (2005) Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol 15 : 1578–1582.
12. MeulenerM, WhitworthAJ, Armstrong-GoldCE, RizzuP, HeutinkP, et al. (2005) Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson's disease. Curr Biol 15 : 1572–1577.
13. ClarkIE, DodsonMW, JiangC, CaoJH, HuhJR, et al. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441 : 1162–1166.
14. Andres-MateosE, PerierC, ZhangL, Blanchard-FillionB, GrecoTM, et al. (2007) DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc Natl Acad Sci U S A 104 : 14807–14812.
15. GautierCA, KitadaT, ShenJ (2008) Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A 105 : 11364–11369.
16. WangD, TangB, ZhaoG, PanQ, XiaK, et al. (2008) Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Mol Neurodegener 3 : 1–7.
17. LevyO, MalageladaC, GreeneL (2009) Cell death pathways in Parkinson's disease: proximal triggers, distal effectors, and final steps. Apoptosis 14 : 478–500.
18. NagakuboD, TairaT, KitauraH, IkedaM, TamaiK, et al. (1997) DJ-1, a novel oncogene which transforms Mouse NIH3T3 cells in cooperation withras. Biochem Biophys Res Commun 231 : 509–513.
19. KimRH, PetersM, JangY, ShiW, PintilieM, et al. (2005) DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 7 : 263–273.
20. PardoM, GarcíaÁ, ThomasB, PiñeiroA, AkoulitchevA, et al. (2006) The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int J Cancer 119 : 1014–1022.
21. TianM, CuiY-Z, SongG-H, ZongM-J, ZhouX-Y, et al. (2008) Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer 8 : 1–11.
22. BonifatiV, RizzuP, SquitieriF, KriegerE, VanacoreN, et al. (2003) DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci 24 : 159–160.
23. KahlePJ, WaakJ, GasserT (2009) DJ-1 and prevention of oxidative stress in Parkinson's disease and other age-related disorders. Free Radic Biol Med 47 : 1354–1361.
24. ParkJ, KimSY, ChaG-H, LeeSB, KimS, et al. (2005) Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene 361 : 133–139.
25. KimRH, SmithPD, AleyasinH, HayleyS, MountMP, et al. (2005) Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A 102 : 5215–5220.
26. BretaudS, AllenC, InghamPW, BandmannO (2007) p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson's disease. J Neurochem 00 : 1626–1635.
27. LiuF, NguyenJL, HullemanJD, LiL, RochetJ-C (2008) Mechanisms of DJ-1 neuroprotection in a cellular model of Parkinson's disease. J Neurochem 105 : 2435–2453.
28. ZhouW, FreedCR (2005) DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T α-synuclein toxicity. J Biol Chem 280 : 43150–43158.
29. ClementsCM, McNallyRS, ContiBJ, MakTW, TingJP-Y (2006) DJ-1, a cancer - and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A 103 : 15091–15096.
30. ZhongN, XuJ (2008) Synergistic activation of the human MnSOD promoter by DJ-1 and PGC-1α: regulation by SUMOylation and oxidation. Hum Mol Genet 17 : 3357–3367.
31. ImJ-Y, LeeK-W, WooJ-M, JunnE, MouradianMM (2012) DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum Mol Genet 21 : 3013–3024.
32. MoJS, KimMY, AnnEJ, HongJA, ParkHS (2008) DJ-1 modulates UV-induced oxidative stress signaling through the suppression of MEKK1 and cell death. Cell Death Differ 15 : 1030–1041.
33. JunnE, TaniguchiH, JeongBS, ZhaoX, IchijoH, et al. (2005) Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci U S A 102 : 9691–9696.
34. FanJ, RenH, JiaN, FeiE, ZhouT, et al. (2008) DJ-1 decreases Bax expression through repressing p53 transcriptional acctivity. J Biol Chem 283 : 4022–4030.
35. YangX, Khosravi-FarR, ChangHY, BaltimoreD (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89 : 1067–1076.
36. ToriiS, EganDA, EvansRA, ReedJC (1999) Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J 18 : 6037–6049.
37. WajantH (2002) The Fas Signaling Pathway: More Than a Paradigm. Science 296 : 1635–1636.
38. SalomoniP, KhelifiAF (2006) Daxx: death or survival protein? Trends Cell Biol 16 : 97–104.
39. ChangHY, NishitohH, YangX, IchijoH, BaltimoreD (1998) Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science 281 : 1860–1863.
40. KhelifiAF, D'AlcontresMS, SalomoniP (2005) Daxx is required for stress-induced cell death and JNK activation. Cell Death Differ 12 : 724–733.
41. KimKS, HwangH-A, ChaeS-K, HaH, KwonK-S (2005) Upregulation of Daxx mediates apoptosis in response to oxidative stress. J Cell Biochem 96 : 330–338.
42. RaoulC, BarthelemyC, CouzinetA, HancockD, PettmannB, et al. (2005) Expression of a dominant negative form of Daxx in vivo rescues motoneurons from Fas (CD95)-induced cell death. J Neurobiol 62 : 178–188.
43. RaoulC, EstévezAG, NishimuneH, ClevelandDW, deLapeyrièreO, et al. (2002) Motoneuron death triggered by a specific pathway downstream of Fas: Potentiation by ALS-linked SOD1 mutations. Neuron 35 : 1067–1083.
44. KarunakaranS, DiwakarL, SaeedU, AgarwalV, RamakrishnanS, et al. (2007) Activation of apoptosis signal regulating kinase 1 (ASK1) and translocation of death-associated protein, Daxx, in substantia nigra pars compacta in a mouse model of Parkinson's disease: protection by α-lipoic acid. FASEB J 21 : 2226–2236.
45. BodaiL, PardiN, ÚjfaludiZ, BereczkiO, KomonyiO, et al. (2007) Daxx-like protein of Drosophila interacts with Dmp53 and affects longevity and Ark mRNA level. J Biol Chem 282 : 36386–36393.
46. WaakJ, WeberSS, GörnerK, SchallC, IchijoH, et al. (2009) Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J Biol Chem 284 : 14245–14257.
47. EssersMA, WeijzenS, de Vries-SmitsAM, SaarloosI, de RuiterND, et al. (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 23 : 4802–4812.
48. LuoX, PuigO, HyunJ, BohmannD, JasperH (2007) Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. EMBO J 26 : 380–390.
49. YangY, GehrkeS, HaqueME, ImaiY, KosekJ, et al. (2005) Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci U S A 102 : 13670–13675.
50. AleyasinH, RousseauxMWC, MarcogliesePC, HewittSJ, IrrcherI, et al. (2010) DJ-1 protects the nigrostriatal axis from the neurotoxin MPTP by modulation of the AKT pathway. Proc Natl Acad Sci U S A 107 : 3186–3191.
51. BiggsWH3rd, CaveneeWK, ArdenKC (2001) Identification and characterization of members of the FKHR (FOXO) subclass of winged-helix transcription factors in the mouse. Mamm Genome 12 : 416–425.
52. Lavara-CulebrasE, Muñoz-SorianoV, Gómez-PastorR, MatallanaE, ParicioN (2010) Effects of pharmacological agents on the lifespan phenotype of Drosophila DJ-1β mutants. Gene 462 : 26–33.
53. ParkJ, LeeSB, LeeS, KimY, SongS, et al. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441 : 1157–1161.
54. KohH, ChungJ (2012) PINK1 as a molecular checkpoint in the maintenance of mitochondrial function and integrity. Mol Cells 34 : 7–13.
55. FahnS, CohenG (1992) The oxidant stress hypothesis in Parkinson's disease: Evidence supporting it. Ann Neurol 32 : 804–812.
56. JennerP, OlanowCW (1996) Oxidative stress and the pathogenesis of Parkinson's disease. Neurology 47 : 161S–170S.
57. HaldA, LothariusJ (2005) Oxidative stress and inflammation in Parkinson's disease: is there a causal link? Exp Neurol 193 : 279–290.
58. BrunetA, BonniA, ZigmondMJ, LinMZ, JuoP, et al. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96 : 857–868.
59. SongJJ, LeeYJ (2003) Catalase, but not MnSOD, inhibits glucose deprivation-activated ASK1-MEK-MAPK signal transduction pathway and prevents relocalization of Daxx: Hydrogen peroxide as a major second messenger of metabolic oxidative stress. J Cell Biochem 90 : 304–314.
60. CharetteSJ, LavoieJN, LambertH, LandryJ (2000) Inhibition of Daxx-mediated apoptosis by Heat Shock Protein 27. Mol Cell Biol 20 : 7602–7612.
61. LeeJH, LeeE, ParkJ, KimE, KimJ, et al. (2003) In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett 550 : 5–10.
62. SonJH, ChunHS, JohTH, ChoS, ContiB, et al. (1999) Neuroprotection and neuronal differentiation studies using substantia nigra dopaminergic cells derived from transgenic Mouse embryos. J Neurosci 19 : 10–20.
63. LeeNG, HongYK, YuSY, HanSY, GeumD, et al. (2007) dXNP, a Drosophila homolog of XNP/ATRX, induces apoptosis via Jun-N-terminal kinase activation. FEBS Lett 581 : 2625–2632.
64. FeanyMB, BenderWW (2000) A Drosophila model of Parkinson's disease. Nature 404 : 394–398.
65. HongYK, LeeNG, LeeMJ, ParkMS, ChoiG, et al. (2009) dXNP/DATRX increases apoptosis via the JNK and dFOXO pathway in Drosophila neurons. Biochem Biophys Res Commun 384 : 160–166.
66. YangJS, NamHJ, SeoM, HanSK, ChoiY, et al. (2011) OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 6: e23525 doi:10.1371/journal.pone.0023525.
Štítky
Genetika Reprodukční medicína
Článek The G4 GenomeČlánek Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance inČlánek RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations inČlánek Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during DevelopmentČlánek Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Epigenetic Upregulation of lncRNAs at 13q14.3 in Leukemia Is Linked to the Downregulation of a Gene Cluster That Targets NF-kB
- A Big Catch for Germ Cell Tumour Research
- The Quest for the Identification of Genetic Variants in Unexplained Cardiac Arrest and Idiopathic Ventricular Fibrillation
- A Nonsynonymous Polymorphism in as a Risk Factor for Human Unexplained Cardiac Arrest with Documented Ventricular Fibrillation
- The Hourglass and the Early Conservation Models—Co-Existing Patterns of Developmental Constraints in Vertebrates
- Smaug/SAMD4A Restores Translational Activity of CUGBP1 and Suppresses CUG-Induced Myopathy
- Balancing Selection on a Regulatory Region Exhibiting Ancient Variation That Predates Human–Neandertal Divergence
- The G4 Genome
- Extensive Natural Epigenetic Variation at a Originated Gene
- Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases
- The Environment Affects Epistatic Interactions to Alter the Topology of an Empirical Fitness Landscape
- TIP48/Reptin and H2A.Z Requirement for Initiating Chromatin Remodeling in Estrogen-Activated Transcription
- Aconitase Causes Iron Toxicity in Mutants
- Tbx2 Terminates Shh/Fgf Signaling in the Developing Mouse Limb Bud by Direct Repression of
- Mondo/ChREBP-Mlx-Regulated Transcriptional Network Is Essential for Dietary Sugar Tolerance in
- Sex-Differential Selection and the Evolution of X Inactivation Strategies
- Identification of a Tissue-Selective Heat Shock Response Regulatory Network
- Phosphorylation-Coupled Proteolysis of the Transcription Factor MYC2 Is Important for Jasmonate-Signaled Plant Immunity
- RpoS Plays a Central Role in the SOS Induction by Sub-Lethal Aminoglycoside Concentrations in
- Six Homeoproteins Directly Activate Expression in the Gene Regulatory Networks That Control Early Myogenesis
- Rtt109 Prevents Hyper-Amplification of Ribosomal RNA Genes through Histone Modification in Budding Yeast
- ATP-Dependent Chromatin Remodeling by Cockayne Syndrome Protein B and NAP1-Like Histone Chaperones Is Required for Efficient Transcription-Coupled DNA Repair
- Iron-Responsive miR-485-3p Regulates Cellular Iron Homeostasis by Targeting Ferroportin
- Mutations in Predispose Zebrafish and Humans to Seminomas
- Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands
- Tissue Homeostasis in the Wing Disc of : Immediate Response to Massive Damage during Development
- All SNPs Are Not Created Equal: Genome-Wide Association Studies Reveal a Consistent Pattern of Enrichment among Functionally Annotated SNPs
- Functional 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- Genetic Requirements for Signaling from an Autoactive Plant NB-LRR Intracellular Innate Immune Receptor
- SNF5 Is an Essential Executor of Epigenetic Regulation during Differentiation
- Dialects of the DNA Uptake Sequence in
- Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate–Invertebrate Gap
- Senataxin Plays an Essential Role with DNA Damage Response Proteins in Meiotic Recombination and Gene Silencing
- High-Resolution Mapping of Spontaneous Mitotic Recombination Hotspots on the 1.1 Mb Arm of Yeast Chromosome IV
- Rod Monochromacy and the Coevolution of Cetacean Retinal Opsins
- Evolution after Introduction of a Novel Metabolic Pathway Consistently Leads to Restoration of Wild-Type Physiology
- Disruption of TTDA Results in Complete Nucleotide Excision Repair Deficiency and Embryonic Lethality
- Insulators Target Active Genes to Transcription Factories and Polycomb-Repressed Genes to Polycomb Bodies
- Signatures of Diversifying Selection in European Pig Breeds
- The Chromosomal Passenger Protein Birc5b Organizes Microfilaments and Germ Plasm in the Zebrafish Embryo
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- Regulates Synaptic Development and Endocytosis by Suppressing Filamentous Actin Assembly
- Sensory Neuron-Derived Eph Regulates Glomerular Arbors and Modulatory Function of a Central Serotonergic Neuron
- Analysis of Rare, Exonic Variation amongst Subjects with Autism Spectrum Disorders and Population Controls
- Scavenger Receptors Mediate the Role of SUMO and Ftz-f1 in Steroidogenesis
- DNA Double-Strand Breaks Coupled with PARP1 and HNRNPA2B1 Binding Sites Flank Coordinately Expressed Domains in Human Chromosomes
- High-Resolution Mapping of H1 Linker Histone Variants in Embryonic Stem Cells
- Comparative Genomics of and the Bacterial Species Concept
- Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming
- Genome-Wide Association Studies Identify Two Novel Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep
- The Genetic Correlation between Height and IQ: Shared Genes or Assortative Mating?
- Comprehensive Assignment of Roles for Typhimurium Genes in Intestinal Colonization of Food-Producing Animals
- An Essential Role for Zygotic Expression in the Pre-Cellular Drosophila Embryo
- The Genome Organization of Reflects Its Lifestyle
- Coordinated Cell Type–Specific Epigenetic Remodeling in Prefrontal Cortex Begins before Birth and Continues into Early Adulthood
- Improved Detection of Common Variants Associated with Schizophrenia and Bipolar Disorder Using Pleiotropy-Informed Conditional False Discovery Rate
- Site-Specific Phosphorylation of the DNA Damage Response Mediator Rad9 by Cyclin-Dependent Kinases Regulates Activation of Checkpoint Kinase 1
- Npc1 Acting in Neurons and Glia Is Essential for the Formation and Maintenance of CNS Myelin
- Identification of , a Retrotransposon-Derived Imprinted Gene, as a Novel Driver of Hepatocarcinogenesis
- Aag DNA Glycosylase Promotes Alkylation-Induced Tissue Damage Mediated by Parp1
- DJ-1 Decreases Neural Sensitivity to Stress by Negatively Regulating Daxx-Like Protein through dFOXO
- Asynchronous Replication, Mono-Allelic Expression, and Long Range -Effects of
- Differential Association of the Conserved SUMO Ligase Zip3 with Meiotic Double-Strand Break Sites Reveals Regional Variations in the Outcome of Meiotic Recombination
- Focusing In on the Complex Genetics of Myopia
- Continent-Wide Decoupling of Y-Chromosomal Genetic Variation from Language and Geography in Native South Americans
- Breakpoint Analysis of Transcriptional and Genomic Profiles Uncovers Novel Gene Fusions Spanning Multiple Human Cancer Types
- Intrinsic Epigenetic Regulation of the D4Z4 Macrosatellite Repeat in a Transgenic Mouse Model for FSHD
- Bisphenol A Exposure Disrupts Genomic Imprinting in the Mouse
- Genetic and Genomic Architecture of the Evolution of Resistance to Antifungal Drug Combinations
- Transposable Elements Are Major Contributors to the Origin, Diversification, and Regulation of Vertebrate Long Noncoding RNAs
- Functional Dissection of the Condensin Subunit Cap-G Reveals Its Exclusive Association with Condensin I
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The G4 Genome
- Neutral Genomic Microevolution of a Recently Emerged Pathogen, Serovar Agona
- The Histone Demethylase Jarid1b Ensures Faithful Mouse Development by Protecting Developmental Genes from Aberrant H3K4me3
- The Tissue-Specific RNA Binding Protein T-STAR Controls Regional Splicing Patterns of Pre-mRNAs in the Brain
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



