-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
Hybridization between species is an important mechanism for the origin of novel lineages and adaptation to new environments. Increased allelic variation and modification of the transcriptional network are the two recognized forces currently deemed to be responsible for the phenotypic properties seen in hybrids. However, since the majority of the biological functions in a cell are carried out by protein complexes, inter-specific protein assemblies therefore represent another important source of natural variation upon which evolutionary forces can act. Here we studied the composition of six protein complexes in two different Saccharomyces “sensu stricto” hybrids, to understand whether chimeric interactions can be freely formed in the cell in spite of species-specific co-evolutionary forces, and whether the different types of complexes cause a change in hybrid fitness. The protein assemblies were isolated from the hybrids via affinity chromatography and identified via mass spectrometry. We found evidence of spontaneous chimericity for four of the six protein assemblies tested and we showed that different types of complexes can cause a variety of phenotypes in selected environments. In the case of TRP2/TRP3 complex, the effect of such chimeric formation resulted in the fitness advantage of the hybrid in an environment lacking tryptophan, while only one type of parental combination of the MBF complex allowed the hybrid to grow under respiratory conditions. These phenotypes were dependent on both genetic and environmental backgrounds. This study provides empirical evidence that chimeric protein complexes can freely assemble in cells and reveals a new mechanism to generate phenotypic novelty and plasticity in hybrids to complement the genomic innovation resulting from gene duplication. The ability to exchange orthologous members has also important implications for the adaptation and subsequent genome evolution of the hybrids in terms of pattern of gene loss.
Published in the journal: . PLoS Genet 9(10): e32767. doi:10.1371/journal.pgen.1003836
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003836Summary
Hybridization between species is an important mechanism for the origin of novel lineages and adaptation to new environments. Increased allelic variation and modification of the transcriptional network are the two recognized forces currently deemed to be responsible for the phenotypic properties seen in hybrids. However, since the majority of the biological functions in a cell are carried out by protein complexes, inter-specific protein assemblies therefore represent another important source of natural variation upon which evolutionary forces can act. Here we studied the composition of six protein complexes in two different Saccharomyces “sensu stricto” hybrids, to understand whether chimeric interactions can be freely formed in the cell in spite of species-specific co-evolutionary forces, and whether the different types of complexes cause a change in hybrid fitness. The protein assemblies were isolated from the hybrids via affinity chromatography and identified via mass spectrometry. We found evidence of spontaneous chimericity for four of the six protein assemblies tested and we showed that different types of complexes can cause a variety of phenotypes in selected environments. In the case of TRP2/TRP3 complex, the effect of such chimeric formation resulted in the fitness advantage of the hybrid in an environment lacking tryptophan, while only one type of parental combination of the MBF complex allowed the hybrid to grow under respiratory conditions. These phenotypes were dependent on both genetic and environmental backgrounds. This study provides empirical evidence that chimeric protein complexes can freely assemble in cells and reveals a new mechanism to generate phenotypic novelty and plasticity in hybrids to complement the genomic innovation resulting from gene duplication. The ability to exchange orthologous members has also important implications for the adaptation and subsequent genome evolution of the hybrids in terms of pattern of gene loss.
Introduction
The Saccharomyces sensu stricto yeasts represent a diverse, monophyletic group of species that have the ability to produce viable and stable hybrids that can propagate mitotically. Hybrids among yeast species and strains seem to be common, especially amongst wine, and beer brewing yeasts [1], [2], but also within natural ecological niches [3]. When two parental genomes merge in yeast hybrids there is a potential for genetic novelty but also for a genetic conflict to occur. Dominant genetic incompatibilities do not seem to occur in the S. cerevisiae sensu stricto group [4], however evidence of recessive allelic incompatibilities between nuclear and mitochondrial genomes have recently been uncovered [5].
Hybridization can play an important role in evolution since hybrids could occupy a different niche from both parental species and eventually establish a new lineage. The presence of naturally occurring yeast hybrids isolated from specific environments seem to confirms this hypothesis [6], [7]. So far, many unique characteristics of the Saccharomyces “sensu stricto” species and hybrids have been attributed to changes in gene expression, including novel cis-trans interactions [8] and to divergence in regulatory regions [9]. Nevertheless, in the hybrid cellular environment, where two sets of homologous proteomes coexist, there is also the potential for the cell to form chimeric assemblies between homologus protein complexes. Analysis of large-scale proteomics data has shown that the majority of cellular processes are carried out by protein assemblies rather than single proteins and that over 60% of yeast proteins form obligate complexes [10]. Since the correct formation of a complex is essential to carry out the biological function, we would expect that any sub-optimal protein interaction would be detrimental to the cell and therefore discouraged by the cell. On the other hand, spontaneous chimeric assemblies may widen the adaptation potential of the cell, since several different combinations of the same protein complex can be used. Therefore, such situation can lead to new phenotypic variants that are beneficial to the hybrid in novel contexts. The primary aim of this work is to establish proof of principle that chimeric protein complexes can form freely in hybrids of Saccharomyces species despite the intra-specific co-evolutionary forces and to quantify the impact that such complexes can have on the overall fitness of the hybrids. In fact, chimericity in protein-protein interaction represents a potentially important mechanism for generating phenotypic diversity upon which evolutionary forces can act, and may constitute a molecular explanation of hybrid vigour.
Results and Discussion
Experimental strategy for the analysis of chimeric complexes in yeast hybrids
To test for the existence of natural chimeric complexes in yeast hybrids, we analysed six physically stable ‘obligatory’ protein complexes (Table S1) each of which have constitutively expressed members that were previously recovered by large-scale protein interaction studies and also by independent small-scale biochemical studies [11], [12].
We created S cerevisiae/S. mikatae (Sc/Sm) and S. cerevisiae/S. uvarum (Sc/Su) hybrids by crossing either S. mikatae or S. uvarum with S. cerevisiae strains carrying a molecular tag (TAP-tag) at the C-terminus of a selected member of the protein complex (Figure S1). Tagged proteins, along with their interacting partners, were isolated via affinity chromatography and all the members of the protein complex were identified via mass spectrometry. If only species-specific parental complexes are established in the hybrid, just proteins from the species carrying the TAP-tag (S. cerevisiae) will be identified. However, if chimeric protein complexes are formed, proteins from the other parental species (S. mikatae or S. uvarum) will also be isolated and identified (Figure 1). The protein fractions were analyzed by mass spectrometry to identify tryptic peptides in a custom protein database of six Saccharomyces sensu stricto yeast proteomes. Species-specific peptides were distinguished from the shared peptides that are identical between the two parental species. As control experiment to test whether in vitro chimeric interactions were generated artefactually during the protein extraction procedure (as opposed to in vivo within the hybrid cellular environment), a mixture of parental cells (i.e. S. cerevisiae and S. mikatae or S. uvarum) were grown separately and mixed together just prior to cell lysis. To establish that both parental genomes were present, all hybrids were screened for chromosomal content via PCR using species-specific primers (Figure S2). To check for genomic alterations after hybridisation, meiosis was induced and spore viability was assessed. Hybrids between yeast species are sterile (<1% survival rate) but they can present a higher rate of spore viability if the cells undergo aneuploidy incrementing their chromosomes number. After dissecting 128 tetrads per hybrid background, no viable cells were detected (Figure S3), suggesting that the hybrids were 2n.
Fig. 1. TAP-strategy for recovery and identification of hybrid protein complexes. 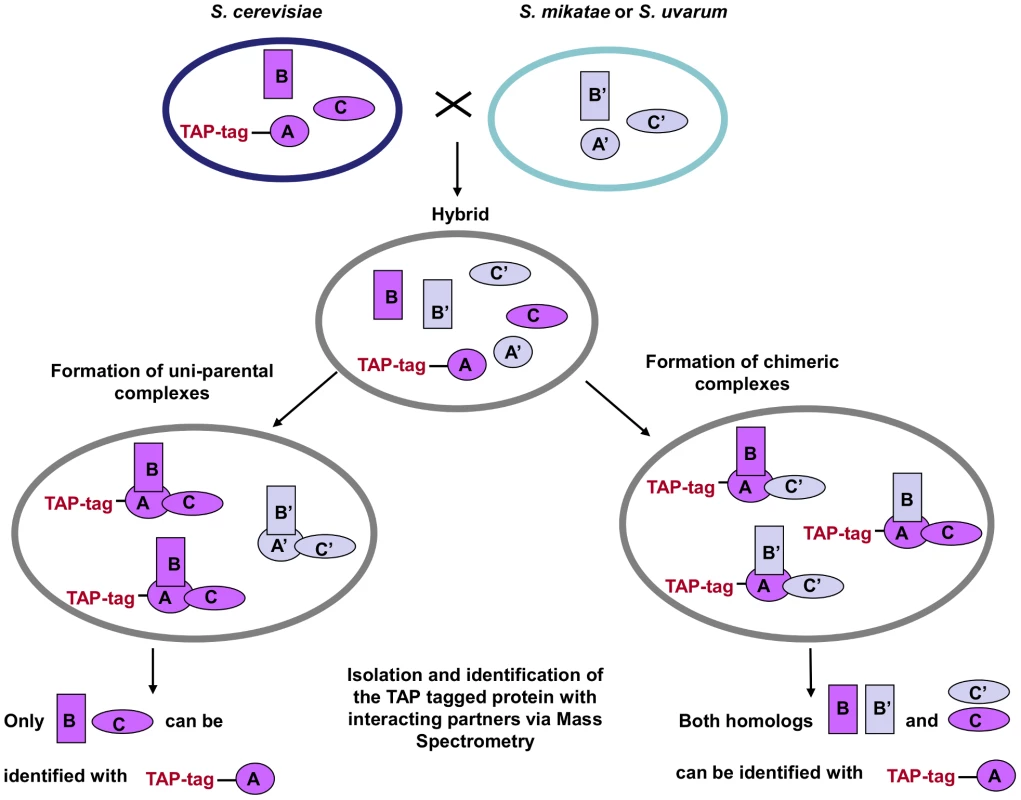
S. cerevisiae strains with the TAP cassette inserted into the C-terminal of one member of the complex (TAP-tag A) were crossed with S. mikatae and S. uvarum species. The complexes that freely formed in the hybrids were then isolated and the interacting members identified via MS analysis. A', B' and C' represent the orthologs of the S. cerevisiae A, B, C proteins, respectively. Transcription of the homologous members of the protein complexes in the hybrids was also confirmed via RT-PCR (Figures S4, S5, S6, S7, S8, S9).
Analysis of the nature of the protein complexes in yeast hybrids
The first complex we considered was the Sec 62/63 complex, a tetramer that is involved in the transport of proteins across the ER membrane, composed of two essential proteins, Sec62p and Sec63p and two non-essential proteins, Sec66p and Sec72p [13]. In both hybrids Sc/Sm and Sc/Su, the mass spectrometry analysis identified Sec63p and Sec72p from either S. mikatae or S. uvarum, respectively, demonstrating that in yeast hybrids the assembly of the Sec62/63 complex can be spontaneously chimeric (Figure 2, Figure S10, S11, S12, S13, S14, S15, S16, S17, Table S2 and S3).
Fig. 2. Peptide map of the S. uvarum Sec63p. 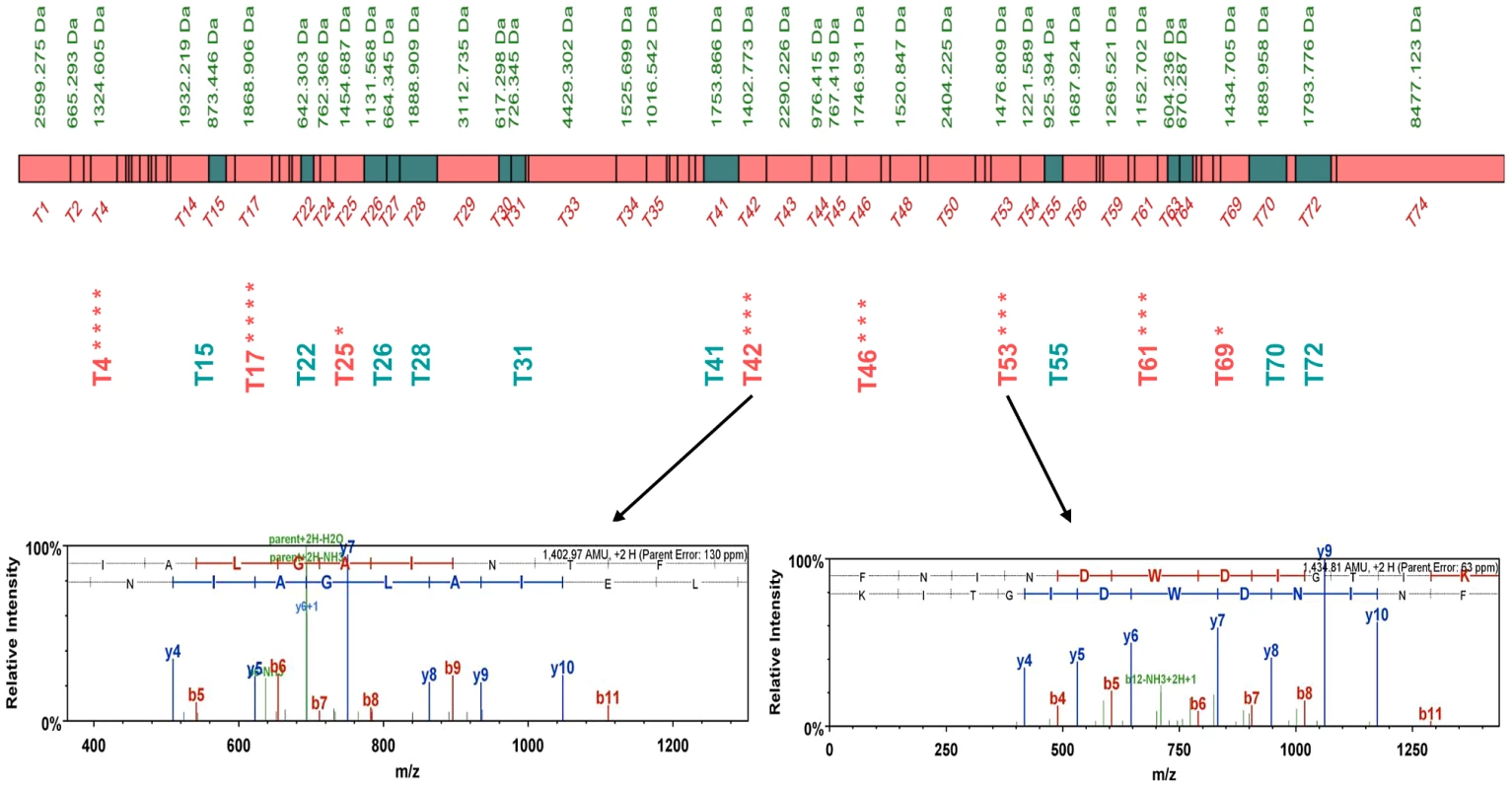
The peptides in common for both S. cerevisiae and S. uvarum species are shown as green boxes, while S. uvarum specific peptides are shown as pink boxes. Unique peptides detected independently in different biological repeats are marked with asterisks. The MS spectra of unique S. uvarum ion peptides T42 and T53 are shown below. Evidence of chimeric interactions were also detected between members of the TRP2/TRP3 complex, involved in the tryptophan biosynthesis [14] (Figures S18 and S19, Tables S4 and S5) and the CTK complex, involved in transcription and translation regulation [15] (Ctk1p, Ctk2p, Ctk3p; see Figures S20 and S21, Table S6 and S7), in both Sc/Sm and Sc/Su hybrids.
In the case of the MBF complex, a dimer composed of two proteins, Mbp1 (a transcription factor responsible for DNA synthesis at the G1/S phase of the cell cycle) and Swi6p (a trans-activating component) [16], chimeric complexes were only identified in hybrids Sc/Su, while, surprisingly, no free interaction was detected in the hybrids of the more closely related species S. cerevisiae and S. mikatae (Figures S22 and S23, Tables S8 and S9). Targeted mass-spectrometry was also performed on Sc/Sm hybrid to seek specifically S. mikatae Swi6 peptides, which constituted the majority of the tryptic digest (ca 76% of all peptides). However, no specific Sm peptides were detected, indicating that this protein was not present in the complex at significant levels (Table S10). The level of expression of Sm Swi6p is higher than that that one of Sc Swi6p in Sc/Sm background, and is also higher than that one of Su Swi6p in Sc/Su hybrids, as showed by Real time PCR experiments (Figure S24), ruling out the lack of detection due to the insufficient expression of Swi6p in the Sc/Sm hybrid. This results indicates that, given the choice, Mbp1p from Sc prefer to form uni-specific complexes with Swi6p from Sc in Sc/Sm background. When considering protein-protein interactions the sequence identity of the binding interfaces is likely to be more important than the phylogenetic relationship. In fact, Swi6p shows greater gene sequence similarity between S. cerevisiae and S. uvarum than between S. cerevisiae and S. mikatae, despite their phylogeny (Figure S25).
The remaining two complexes tested, the RAM (Ram1p and Ram2p, farnesyltransferase complex involved in the prenylation of Ras proteins) [17] and KU (Yku70p and Yku80p), involved in double strand breaks repair and non-homologous end joining) [18], appeared unable to form chimeric complexes in any hybrid background (Tables S11, S12, S13, S14). In fact, using Yku70p as TAP-bait, no specific Yku80p peptides from S. uvarum and S. mikatae parental species were ever found in any biological replica tested, while numerous S. cerevisiae specific Yku80p peptides were consistently isolated. Although the failure to detect such interactions in mass spectrometry is not a definite proof that chimeric complexes are not at all assembled, this data suggests that chimericity within RAM and Ku complexes may at least occur rarely, and that the proteins forming such complexes tend to assemble in uni-specific manner if given the option. Interestingly, an independent study of the KU complex in hybrids of two diverged strains of S. paradoxus showed that negative epistatic interactions occur between the different homologues of Yku70p and Yku80p, suggesting either lack of assembly or functionality of the heterodimer [19]. The inability to detect spontaneous chimeric complex formation in both Sc/Sm and Sc/Su hybrids observed in this work support the idea that the prevention of complex formation could be the possible mechanism for the negative epistasis identified between Yku70p and Yku80p in the S. paradoxus strains.
Phenotypic variations caused by different types of protein assemblies
We evaluated the impact that chimeric interactions have on fitness by forcing the hybrids to use only one specific type of complex to carry out the biological function. We chose to investigate the TRP2/TRP3 ad the MBF complex, since the relationship between the functional complexes and the resulting output fitness could be clearly measured under tryptophan starvation and respiratory growth condition, respectively. In fact, the TRP2/TRP3 complex is involved in the first step of the tryptophan biosynthesis [14], and null mutants of Mbp1p and Swi6p display a range of fitness defects including decrease rate of respiratory growth and abnormal mitochondrial morphology [20].
We created different combinations of the TRP2/TRP3 and MBF complexes by deleting different protein members in both Sc/Sm and Sc/Su hybrid backgrounds (Figures 3A and 4A), and then scored the growth rates of the hybrids carrying either uni-specific or chimeric complexes.
Fig. 3. Fitness assays of the engineered Sc/Su hybrids carrying different type of TRP2/TRP3 chimeric complexes. 
Sc/Su hybrids were genetically modified to carry either the two different chimeric complexes, Trp2pSu/Trp3pSc and Trp2pSc/Trp3pSu, or the two parental hemizygous controls, Trp2pSu/Trp3pSu and Trp2pSc/Trp3pSc (panel A). The growth curves of S. cerevisiae, S. uvarum, the hybrid Sc/Su and the engineered hybrids shows that Trp2pSu/Trp3pSc grows better than the other combinations in SD media lacking tryptophan (panel B). The fitness competition assay between Sc/Su hybrids with different combination of the TRP2/TRP3 complex and the GFP reference strain shows again that Trp2pSu/Trp3pSc grows faster (panel C). The competitive fitness coefficient Sg represents the difference between the ln of the ratio of hybrid and reference strain between final and initial time points, normalized for the number of generations. An equal fitness between hybrid and reference strains would be indicated by a value of zero (see Method section). Fig. 4. Growth assays of Sc/Su hybrids carrying different types of MBF chimeric complexes. 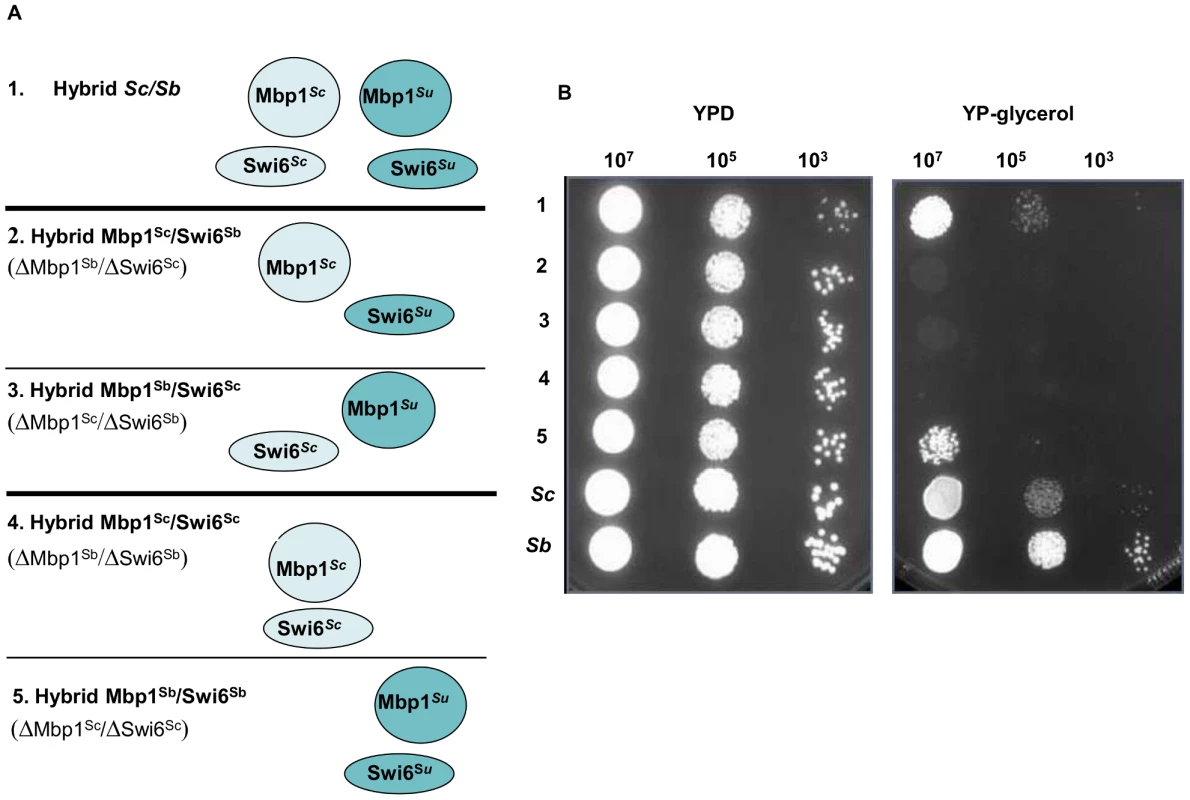
Sc/Su hybrids were genetically modified either to carry the two different chimeric complexes, Mbp1Su/Swi6Sc and Mbp1Sc/Swi6Su, or the two uni-parental controls, Mbp1Su/Swi6Su and Mbp1Sc/Swi6Sc (Panel A). The growth spot assay of the engineered hybrids in rich YPD and YP-glycerol media are shown in Panel B. The strain carrying the S. uvarum homologous Mbp1Su and Swi6Su is the only one that performs respiratory growth and grows normally in the presence of glycerol a sole carbon source. For the TRP2/TRP3 complex in the Sc/Su background, a large range of fitness levels was detected for the different types of assemblies (Figure 3B). The S. uvarum parent grows poorly compared to the S. cerevisiae parent, while the hybrid shows an intermediate fitness (Figure 3B). When comparing the growth of the four strains bearing different combinations of TRP2/TRP3 protein complexes (i.e. possessing the same TRP2/TRP3 copy number in the same hybrid genetic background), we found that the strain with the Trp2pSu/Trp3pSc chimeric complex grew much better than all the other strains in a medium lacking tryptophan (Figure 3B). The uni-parental hemizygous controls Trp2pSu/Trp3pSu showed the lowest fitness, while the chimeric Trp2pSc/Trp3pSu and the hemizygote Trp2pSc/Trp3pSc showed an intermediate growth (Figure 3B). When tryptophan was added to the SD medium the phenotypic difference between the hybrids carrying different protein complexes was minimised (Figure S26).
The strain with the chimeric combination Trp2pSu/Trp3pSc seems to grow similarly to the S. cerevisiae parent and better than the original hybrid. It is possible that, in the parent hybrid, a higher percentage of uni-specific S. uvarum complexes are formed, which are the most unfit of all four combination (Trp2pSu/Trp3pSu, Figure 3B and C), and could therefore partially compromise the fitness of the hybrid. In fact, although the quantitative expression of the two TRP2 orthologs is similar in the hybrid, the S. uvarum TRP3 copy is more expressed than the S. cerevisiae counterpart (Figure S27).
To confirm the increased fitness of the strain expressing a Trp2pSu/Trp3pSc chimeric complex, competition experiments between the chimeric hybrids and a GFP reference strain was carried out using FACS analysis [21]. The results showed that strains with the chimeric Trp2pSu/Trp3pSc complex were more fit than those with the other chimeric complex (Trp2pSc/Trp3pSu) and those with both uni-specific protein-protein interaction combinations (Figure 3C). Moreover, a competitive growth essay between the hybrid carrying the fittest chimeric complex Trp2pSu/Trp3pSc and the reference strain was carried out in SD medium with and without tryptophan. The fitness gain of the strain carrying Trp2pSu/Trp3pSc complex was lessened in the medium containing tryptophan (Figure S28).
For the MBF complex in the Sc/Su background all the engineered hybrids carrying the different type of complexes were able to grow on glucose medium, however only the hybrid carrying the uni-specific combination Mbp1pSu and Swi6pSu derived from S. uvarum was able to grow in media containing glycerol, a carbon source that can only be respired (Figure 4). The other parental combination of Mbp1pSc/Swi6pSc could not be rescued by adding either Mbp1pSu or Swi6pSu to its genotype, showing that the presence of both S. uvarum members of the MBF complex is required for hybrid growth on glycerol (Figure S29). Interestingly, the restriction analysis of the mitochondrial genes COX2 and COX3 indicated that the Sc/Su hybrids harbour the Su mitochondrial DNA (data not shown). Recently, incompatibilities between nuclear and mitochondrial genes have been proposed as general mechanism causing reproductive isolation between species.
This is a type of Dobzhansky-Muller incompatibility involving lack of interaction or malfunctioning of interacting alleles derived from two different species. For example, the S. uvarum nuclear encoded mitochondrial protein Aep2p is unable to regulate the translation of the S. cerevisiae mitochondrial gene OLI [5], and the S. cerevisiae Mrs1p is not able to splice either the S. paradoxus or the S. uvarum COX1 gene [22].
In the case of MBF complex, we have shown an example of phenotypic plasticity of different chimeric assemblies, and found a novel case of hybrid incompatibility between S. cerevisiae and S. uvarum when cells are grown on a non–fermentable medium and the mitochondria function become essential for cell viability.
Fitness variation between the different types of protein assemblies was not otherwise observed in Sc/Sm hybrids either for the TRP2/TRP3 or for the MBF complex (Figure S30), underlying the dependency of these phenotypes on their genetic background (manifesting in Sc/Su but not in Sc/Sm hybrids). This background dependency is not entirely surprising given the fact that, even between two strains belonging to the same S. cerevisiae species (i.e. BY4743 and Sigma 1278b) several conditional essential genes have been discovered [23].
Conclusions
Here we have shown that protein complexes in hybrids of S. cerevisiae/S. mikatae and S. cerevisiae/S. uvarum are able to spontaneously exchange components for inter-specific orthologs, and, while this manuscript was under review, a study on protein-protein interactions among members of the nuclear pore complex and the RNA polymerase II complex in other S. cerevisiae “sensu stricto” hybrids (i.e. S. cerevisiae/S. kudriazvevii) also concluded that chimeric protein complexes could assemble [24].
Out of the six complexes studied four were convincingly found to form natural chimeric protein assemblies in either one or both genetic hybrid background (i.e. Sec62–63, TRP2/TRP3, MBF, and CTK complex). These results provide evidence that chimeric protein interactions in hybrids can arise to generate evolutionary novelty in protein-protein interaction networks, providing a new evolutionary mechanism to complement innovation by gene duplication [25].
We also found that some complexes prefer to form species-specific configurations in the natural hybrid cell environment (i.e. Ku and RAM complex). The lack of spontaneous chimeric assembly in these cases could be due to less favourable changes in the binding interfaces of the proteins, or to stoichiometry imbalance between homologous proteins in the hybrid [26]. The inability to create chimeric interaction can be responsible for some negative epistatic effect seen in hybrids [19].
We showed that different type of complexes can cause a variety of phenotypes in selected environments. In the case of TRP2/TRP3, we find that chimeric complex formation can lead to hybrid vigour, reinforcing the idea that the ability to form different types of protein assemblies could be advantageous to the hybrid in specific nutritional contexts. We can speculate that the advantage of the chimeric combination can be due to a more harmonious expression of some alleles leading to a better stoichiometry of that specific type of complex. Alternatively, the chimeric complex may be more efficient in its biological function in the hybrid background.
In the case of MBF complex only one parental combination of protein-protein interaction was compatible with cell viability under respiratory condition, highlighting a new case of allelic incompatibilities in yeast hybrids. These phenotypes were proved to be dependent on both genetic and environmental backgrounds since we did not observe any fitness change in Sc/Sm hybrids and the advantages could be lost or gained in different media, such as in the case of the strains carrying different combination of the MBF complex grown in YPD or YP-glycerol (Figure 4B).
Ultimately, this study proposes a novel molecular mechanism for creating phenotypic variation within a hybrid cell, with important implications for understanding the evolutionary forces that govern the reshaping of hybrid genomes. The genomic fate of the homolog genes will in fact be influenced by the ability or not of the hybrid to create inter-specific protein assemblies (Figure S31). Moreover, chimeric complexes may be able to recruit new proteins and evolve new functions in the cell [27]. In the future, the genomic information of naturally occurring hybrids (like S. pastorianus strains) will provide insight into the nature of how the formation of chimeric interactions influences selective gene retention of members of protein complexes and networks.
Materials and Methods
Generation of yeast hybrids
All the TAP-tagged constructs, based on S. cerevisiae MGD353-13D strain, were obtained from the EUROSCARF strains collection (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/cellzome.html). Hybrids between S. cerevisiae strains (bearing the TAP-tag in selected members of different protein complexes) and wild-type S. mikatae 1815 and S. uvarum NCYC2669 species were generated using a Singer Instruments MSM micromanipulator as previously described [28]. To enable selection of hybrid colonies, we made the S. cerevisiae TAP strains geneticin-resistant by inserting a kanMX in the neutral AAD3 locus. Hybrid colonies were then selected on minimal media containing geneticin G418 (see Figure S1). The nature of the chromosomes were verified by chromosomal PCR using genomic DNA from the hybrid as template and species-specific primers designed to distinguish between S. cerevisiae, S. mikatae and S. uvarum alleles (see Figure S2 and Table S15, S16, S17).
After the hybrid was created it took ca. 24 generations (growing in two different selective plates) to select the hybrids before the PCR was made to check the chromosomes, and another 16 generations before the TAP tagging experiment (total of about 40 generations since the production of the hybrid). The hybrid was then maintained in glycerol stock at −80 C.
Hybrid genomic DNA and RNA was isolated using the DNasy Blood & Tissue kit and the RNeasy mini kit (Qiagen, Crawley, UK), respectively.
Expression analysis by real-time quantitative PCR
The expression levels of S. cerevisiae, S. uvarum and S. mikatae SWI6, TRP2 and TRP3 alleles in Sc/Su and Sc/Sm background were performed on the cDNA samples amplified using the Quantitect real time PCR kit from Qiagen. Optimized reactions were carried out using 10 ng/µl of cDNA, 5 pmoles of each primer and syber green according to the manufacturer instructions (Table S18). Actin (ACT1) was used as a housekeeping reference gene. The expression of each gene was estimated using the Ct Values.
Purification of protein complexes from yeast hybrids and mass spectrometry analysis
Purification of the protein complexes was carried out using the standard TAP protocol [29] optimized for these specific classes of proteins. In particular, two affinity binding steps, the IgG Sepharose and Calmodulin Binding Protein (CBP) binding and TEV protease cleavage were carried out for 2 hours at 4°C instead of 16°C. The protein mixtures were resolved using 1D gel electrophoresis, stained with Coomassie Bio Safe (Bio-Rad) and digested with trypsin (Promega). The trypsin digest was carried out overnight at 37°C according to Shevchenko, A. et al. [30]. The digested protein mixture was separated by the high performance liquid chromatography (HPLC) and analyzed by tandem mass spectrometry (ESI MS/MS) (Micromass CapLC-Q-ToF, Waters, Manchester, UK). The system was either used in a discovery manner with the system selecting peptides automatically or in a targeted manner with the system selecting peptides directed from a list of peptides of interest. Spectra acquired for every protein complex member were compared against a custom database containing all proteins from S. cerevisiae “sensu stricto” species, using Mascot version 2.2.06 (Matrix Science Inc., Boston, MA). Scaffold (Scaffold_2_01_00, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide identification. A peptide match was acknowledged if it could be established at greater than 50.0% probability as specified by the Peptide Prophet algorithm [31]. The peptide criteria were set to 50% as we were looking specifically at homologous proteins and shared peptides are generally given lower confidence scores because it cannot be determined which protein the peptides originate from. Significant peptides were checked manually to ensure all the major fragments were matched and a contiguous series of at least 4 y or b ions were present. Protein identifications were accepted if they could be established at greater than 95.0% probability by Protein Prophet and contained at least 2 identified peptides. The Liverpool Peptide Mapping Tool (http://www.liv.ac.uk/pfg/Tools/Pmap/pmap.html) was used to generate proteolytic peptide maps of protein complex members. The peptide maps were generated with one trypsin miscleavage per site after lysine and arginine (K-X, R-X) but not at lysine-proline and arginine-proline (K-P, R-P) sites.
Generation of chimeric protein complexes in Sc/Sm and Sc/Su hybrids and fitness assays
Chimeric and unispecific versions of the TRP2/TRP3 and MBF complexes in both Sc/Sm and Sc/Su hybrids were generated by PCR-mediated gene deletion strategy using hygromycin (HPH) and nourseothricin (NAT) as selectable markers [32]. The S. cerevisiae TRP2 and TRP3 copies were replaced with HPH while the S. uvarum ones were deleted using NAT (see Figure 3). Similarly for the MBF complex, the S. cerevisiae orthologs of Mbp1 and Swi6 were disrupted using HPH, while the S. uvarum copies of Mbp1 and Swi6 were deleted using NAT (see Figure 4). Yeast hybrids were grown in YPD, SD and minimal F1 media [33] at 30°C for 40 hours with continuous shaking. Growth rates were measured by absorbance at OD595 at 5 minutes intervals using Fluostar Optima bioscreen workstation (BMG Labtech).
Fitness competition assays were carried out by FACS analysis according to Lang et al. [21]. As reference strain we used the FY3 strains bearing the GFP tag at the C-terminus of CDC33p (generated for the purpose of this experiment), and the competition was carried out in SD media lacking tryptophan. The hybrids strains were mixed with the reference strain in 4∶1 ratio, and a total of 1×105 cells, counted on a cellometer (Auto M10, Nexcelom), were inoculated into a 1 ml of fresh medium. The strains were allowed to grow for 12 hours and then the ratio of the number of hybrid cells over the fluorescent reference was determined using the Dako CyAn flow cytometer, with a total counting total 50,000 cells for each time point. Three biological and three technical replicates were performed for each fitness measurement. The sg fitness coefficient was calculated using the following equation:
where, H and R are the cell number of the hybrid and reference strain and g0 and gf are the number of generations at the beginning and after a time interval (12 hours).Supporting Information
Zdroje
1. MasneufI, HansenJ, GrothC, PiskurJ, DubourdieuD (1998) New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol 64 : 3887–3892.
2. GanglH, BatusicM, TscheikG, TiefenbrunnerW, HackC, et al. (2009) Exceptional fermentation characteristics of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Nat Biotechnol 25 : 244–251.
3. LitiG, CarterDM, MosesAM, WarringerJ, PartsL, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458 : 337–341.
4. GreigD, BortsRH, LouisEJ, TravisanoM (2002) Epistasis and hybrid sterility in Saccharomyces. Proc Biol Sci 269 : 1167–1171.
5. LeeHY, ChouJY, CheongL, ChangNH, YangSY, et al. (2008) Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135 : 1065–1073.
6. BlieckL, ToyeG, DumortierF, VerstrepenKJ, DelvauxFR, et al. (2007) Isolation and characterization of brewer's yeast variants with improved fermentation performance under high-gravity conditions. Appl Environ Microbiol 73 : 815–824.
7. GonzálezSS, GalloL, ClimentMA, BarrioE, QuerolA, et al. (2007) Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol 116 : 11–17.
8. TiroshI, ReikhavS, LevyAA, BarkaiN (2009) A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324 : 659–662.
9. BornemanAR, GianoulisTA, ZhangZD, YuH, RozowskyJ, et al. (2007) Divergence of transcription factor binding sites across related yeast species. Science 317 : 815–819.
10. PuS, WongJ, TurnerB, ChoE, WodakSJ (2009) Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res 37 : 825–831.
11. GavinAC, BöscheM, KrauseR, GrandiP, MarziochM, et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415 : 141–147.
12. TarassovK, MessierV, LandryCR, RadinovicS, Serna MolinaMM, et al. (2008) An in vivo map of the yeast protein interactome. Science 320 : 1465–1470.
13. SteelGJ, BrownswordJ, StirlingCJ (2002) Tail-anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry 41 : 11914–11920.
14. PrasadR, NiederbergerP, HütterR (1987) Tryptophan accumulation in Saccharomyces cerevisiae under the influence of an artificial yeast TRP gene cluster. Yeast 3 : 95–105.
15. ChoEJ, KoborMS, KimM, GreenblattJ, BuratowskiS (2001) Opposing effects of Ctk1 kinase and Fep1 phosphatase at Ser 2 of the RNA polymerase II C - terminal domain. Genes Dev 15 : 3319–3329.
16. BeanJM, SiggiaED, CrossFR (2005) High functional overlap between Mlu I cell - cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 171 : 49–61.
17. HeB, ChenP, ChenSY, VancuraKL, MichaelisS, et al. (1991) RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci U S A 88 : 11373–11377.
18. TamATY, PikeBL, HammetA, HeierhorstJ (2007) Telomere-related function of yeast KU in the repair of bleomycin-induced DNA damage. Biochem Biophys Res Commun 357 : 800–803.
19. LitiG, HaricharanS, CubillosFA, TierneyAL, SharpS, et al. (2009) Segregating YKU80 and TLC1 alleles underlying natural variation in telomere properties in wild yeast. PLoS Genet 5: e1000659.
20. SteinmetzLM, ScharfeC, DeutschbauerAM, MokranjacD, HermanZS, et al. (2002) Systematic screen for human disease genes in yeast. Nat Genet 31 : 400–404.
21. LangGI, MurrayAW, BotsteinD (2009) The cost of gene expression underlies a fitness trade-off in yeast. Proc Natl Acad Sci U S A 106 : 5755–5760.
22. ChouJY, HungYS, LinKH, LeeHY, LeuJY (2010) Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol 8: e1000432.
23. DowellRD, RyanO, JansenA, CheungD, AgarwalaS, et al. (2010) Genotype to phenotype: a complex problem. Science 328 : 469.
24. LeducqJB, CharronG, DissG, Gagnon-ArsenaultI, DubéAK, et al. (2012) Evidence for the robustness of protein complexes to inter-species hybridization. PLoS Genetics 8 (12) e1003161.
25. Ohno S. (1970) Evolution by gene duplication. New York Springer-Verlag.
26. PappB, PálC, HurstLD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424 : 194–197.
27. IsalanM, LemerleC, MichalodimitrakisK, HornC, BeltraoP, et al. (2008) Evolvability and hierarchy in rewired bacterial gene networks. Nature 452 : 840–845.
28. DelneriD, ColsonI, GrammenoudiS, RobertsIN, LouisEJ, et al. (2003) Engineering evolution to study speciation in yeasts. Nature 422 : 68–72.
29. RigautG, ShevchenkoA, RutzB, WilmM, MannM, et al. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17 : 1030–1032.
30. ShevchenkoA, TomasH, HavlisJ, OlsenJV, MannM (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1 : 2856–2860.
31. NesvizhskiiAI, KellerA, KolkerE, AebersoldRA (2003) Statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75 : 4646–4658.
32. CarterZ, DelneriD (2010) New generation of loxP-mutated deletion cassettes for the genetic manipulation of yeast natural isolates. Yeast 27 : 765–775.
33. DelneriD (2011) Competition experiments coupled with high-throughput analyses for functional genomics studies in yeast. Methods Mol Biol 759 : 271–282.
Štítky
Genetika Reprodukční medicína
Článek Defending Sperm FunctionČlánek How to Choose the Right MateČlánek Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental TransitionČlánek Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in SpermatogenesisČlánek The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate EmbryogenesisČlánek Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time inČlánek Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral MyelinationČlánek A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase inČlánek Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Defending Sperm Function
- How to Choose the Right Mate
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
- Conserved Translatome Remodeling in Nematode Species Executing a Shared Developmental Transition
- A Novel Actin mRNA Splice Variant Regulates ACTG1 Expression
- Tracking Proliferative History in Lymphocyte Development with Cre-Mediated Sister Chromatid Recombination
- Correlated Occurrence and Bypass of Frame-Shifting Insertion-Deletions (InDels) to Give Functional Proteins
- Chimeric Protein Complexes in Hybrid Species Generate Novel Phenotypes
- Loss of miR-10a Activates and Collaborates with Activated Wnt Signaling in Inducing Intestinal Neoplasia in Female Mice
- Both Rare and Copy Number Variants Are Prevalent in Agenesis of the Corpus Callosum but Not in Cerebellar Hypoplasia or Polymicrogyria
- Reverse PCA, a Systematic Approach for Identifying Genes Important for the Physical Interaction between Protein Pairs
- Partial Deletion of Chromosome 8 β-defensin Cluster Confers Sperm Dysfunction and Infertility in Male Mice
- Genome-Wide and Cell-Specific Epigenetic Analysis Challenges the Role of Polycomb in Spermatogenesis
- Coordinate Regulation of Mature Dopaminergic Axon Morphology by Macroautophagy and the PTEN Signaling Pathway
- Cooperation between RUNX1-ETO9a and Novel Transcriptional Partner KLF6 in Upregulation of in Acute Myeloid Leukemia
- Mobility of the Native Conjugative Plasmid pLS20 Is Regulated by Intercellular Signaling
- FliZ Is a Global Regulatory Protein Affecting the Expression of Flagellar and Virulence Genes in Individual Bacterial Cells
- Specific Tandem Repeats Are Sufficient for Paramutation-Induced Trans-Generational Silencing
- Condensin II Subunit dCAP-D3 Restricts Retrotransposon Mobilization in Somatic Cells
- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- The Insulator Homie Promotes Expression and Protects the Adjacent Gene from Repression by Polycomb Spreading
- Human Intellectual Disability Genes Form Conserved Functional Modules in
- Coordination of Cell Proliferation and Cell Fate Determination by CES-1 Snail
- ORFs in Drosophila Are Important to Organismal Fitness and Evolved Rapidly from Previously Non-coding Sequences
- Different Roles of Eukaryotic MutS and MutL Complexes in Repair of Small Insertion and Deletion Loops in Yeast
- The Spore Differentiation Pathway in the Enteric Pathogen
- Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization
- The Human Nuclear Poly(A)-Binding Protein Promotes RNA Hyperadenylation and Decay
- Genome Wide Analysis Reveals Zic3 Interaction with Distal Regulatory Elements of Stage Specific Developmental Genes in Zebrafish
- Xbp1 Directs Global Repression of Budding Yeast Transcription during the Transition to Quiescence and Is Important for the Longevity and Reversibility of the Quiescent State
- The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
- Incorporating Motif Analysis into Gene Co-expression Networks Reveals Novel Modular Expression Pattern and New Signaling Pathways
- The Bacterial Response Regulator ArcA Uses a Diverse Binding Site Architecture to Regulate Carbon Oxidation Globally
- Direct Monitoring of the Strand Passage Reaction of DNA Topoisomerase II Triggers Checkpoint Activation
- Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in
- A Reversible Histone H3 Acetylation Cooperates with Mismatch Repair and Replicative Polymerases in Maintaining Genome Stability
- ALS-Associated Mutations Result in Compromised Alternative Splicing and Autoregulation
- Robust Demographic Inference from Genomic and SNP Data
- Preferential Binding to Elk-1 by SLE-Associated Risk Allele Upregulates Expression
- Rad52 Sumoylation Prevents the Toxicity of Unproductive Rad51 Filaments Independently of the Anti-Recombinase Srs2
- The Serum Resistome of a Globally Disseminated Multidrug Resistant Uropathogenic Clone
- Identification of 526 Conserved Metazoan Genetic Innovations Exposes a New Role for Cofactor E-like in Neuronal Microtubule Homeostasis
- SUMO Localizes to the Central Element of Synaptonemal Complex and Is Required for the Full Synapsis of Meiotic Chromosomes in Budding Yeast
- Integrated Enrichment Analysis of Variants and Pathways in Genome-Wide Association Studies Indicates Central Role for IL-2 Signaling Genes in Type 1 Diabetes, and Cytokine Signaling Genes in Crohn's Disease
- Genome-Wide High-Resolution Mapping of UV-Induced Mitotic Recombination Events in
- Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in
- Playing the Field: Sox10 Recruits Different Partners to Drive Central and Peripheral Myelination
- Two Portable Recombination Enhancers Direct Donor Choice in Fission Yeast Heterochromatin
- Mining the Human Phenome Using Allelic Scores That Index Biological Intermediates
- Yeast Tdh3 (Glyceraldehyde 3-Phosphate Dehydrogenase) Is a Sir2-Interacting Factor That Regulates Transcriptional Silencing and rDNA Recombination
- A Minimal Nitrogen Fixation Gene Cluster from sp. WLY78 Enables Expression of Active Nitrogenase in
- A Review of Bacteria-Animal Lateral Gene Transfer May Inform Our Understanding of Diseases like Cancer
- High Throughput Sequencing Reveals Alterations in the Recombination Signatures with Diminishing Spo11 Activity
- Partitioning the Heritability of Tourette Syndrome and Obsessive Compulsive Disorder Reveals Differences in Genetic Architecture
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- A GDF5 Point Mutation Strikes Twice - Causing BDA1 and SYNS2
- Systematic Unraveling of the Unsolved Pathway of Nicotine Degradation in
- Natural Genetic Variation of Integrin Alpha L () Modulates Ischemic Brain Injury in Stroke
- Evolutionary Tuning of Protein Expression Levels of a Positively Autoregulated Two-Component System
- Evolutionary Change within a Bipotential Switch Shaped the Sperm/Oocyte Decision in Hermaphroditic Nematodes
- Limiting of the Innate Immune Response by SF3A-Dependent Control of MyD88 Alternative mRNA Splicing
- Multiple Signaling Pathways Coordinate to Induce a Threshold Response in a Chordate Embryo
- Distinct Regulatory Mechanisms Act to Establish and Maintain Pax3 Expression in the Developing Neural Tube
- Genome Wide Analysis of Narcolepsy in China Implicates Novel Immune Loci and Reveals Changes in Association Prior to Versus After the 2009 H1N1 Influenza Pandemic
- Mismatch Repair Genes and Modify CAG Instability in Huntington's Disease Mice: Genome-Wide and Candidate Approaches
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- Hsp70-Hsp40 Chaperone Complex Functions in Controlling Polarized Growth by Repressing Hsf1-Driven Heat Stress-Associated Transcription
- Function and Evolution of DNA Methylation in
- Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome
- Transcription Termination and Chimeric RNA Formation Controlled by FPA
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dominant Mutations in Identify the Mlh1-Pms1 Endonuclease Active Site and an Exonuclease 1-Independent Mismatch Repair Pathway
- Eleven Candidate Susceptibility Genes for Common Familial Colorectal Cancer
- The Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters
- A Mutation in the Gene in Labrador Retrievers with Hereditary Nasal Parakeratosis (HNPK) Provides Insights into the Epigenetics of Keratinocyte Differentiation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




