-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
A key challenge in the production of second generation biofuels is the conversion of lignocellulosic substrates into fermentable sugars. Enzymes, particularly those from fungi, are a central part of this process, and many have been isolated and characterised. However, relatively little is known of how fungi respond to lignocellulose and produce the enzymes necessary for dis-assembly of plant biomass. We studied the physiological response of the fungus Aspergillus niger when exposed to wheat straw as a model lignocellulosic substrate. Using RNA sequencing we showed that, 24 hours after exposure to straw, gene expression of known and presumptive plant cell wall–degrading enzymes represents a huge investment for the cells (about 20% of the total mRNA). Our results also uncovered new esterases and surface interacting proteins that might form part of the fungal arsenal of enzymes for the degradation of plant biomass. Using transcription factor deletion mutants (xlnR and creA) to study the response to both lignocellulosic substrates and low carbon source concentrations, we showed that a subset of genes coding for degradative enzymes is induced by starvation. Our data support a model whereby this subset of enzymes plays a scouting role under starvation conditions, testing for available complex polysaccharides and liberating inducing sugars, that triggers the subsequent induction of the majority of hydrolases. We also showed that antisense transcripts are abundant and that their expression can be regulated by growth conditions.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002875
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002875Summary
A key challenge in the production of second generation biofuels is the conversion of lignocellulosic substrates into fermentable sugars. Enzymes, particularly those from fungi, are a central part of this process, and many have been isolated and characterised. However, relatively little is known of how fungi respond to lignocellulose and produce the enzymes necessary for dis-assembly of plant biomass. We studied the physiological response of the fungus Aspergillus niger when exposed to wheat straw as a model lignocellulosic substrate. Using RNA sequencing we showed that, 24 hours after exposure to straw, gene expression of known and presumptive plant cell wall–degrading enzymes represents a huge investment for the cells (about 20% of the total mRNA). Our results also uncovered new esterases and surface interacting proteins that might form part of the fungal arsenal of enzymes for the degradation of plant biomass. Using transcription factor deletion mutants (xlnR and creA) to study the response to both lignocellulosic substrates and low carbon source concentrations, we showed that a subset of genes coding for degradative enzymes is induced by starvation. Our data support a model whereby this subset of enzymes plays a scouting role under starvation conditions, testing for available complex polysaccharides and liberating inducing sugars, that triggers the subsequent induction of the majority of hydrolases. We also showed that antisense transcripts are abundant and that their expression can be regulated by growth conditions.
Introduction
The conversion of cellulose and hemicellulose, from non-food crop sources into fermentable sugars is one of the key challenges in the production of second generation biofuels. Fungi are the predominant source of enzymes currently being used on an industrial scale for this purpose [1], [2]. Many relevant enzymes have been isolated and characterised for functionality [3], [4]. The overall aim of our study was to look beyond the simple array of hydrolytic enzymes produced by fungi and to understand the strategies that fungi employ to degrade complex polysaccharides. This approach may provide novel insights into the development of strategies for the production of second generation biofuels.
Aspergillus niger is a filamentous, black-spored fungus that has been used in many industrial processes, including the production of enzymes, food products and pharmaceuticals [5]. This historical importance has led to the development of a wide array of genetic tools [6]. These include a variety of mutagenesis systems, both targeted [7] and random [8], highly tuneable expression systems [9], and complete genome sequences for both the enzyme-producing industrial strain CBS 513.88 [10] and the citric acid-producing strain, ATCC 1015 [11]. The Carbohydrate-Active Enzymes Database (http://www.cazy.org/ [12]) identifies the CBS 513.88 genome as encoding 281 putative polysaccharide degrading enzymes, that represent 61 different enzyme families. Thus, the genome of A. niger encodes one of the most diverse CAZy enzyme arsenals among currently sequenced fungal genomes. The availability of DNA microarrays for A. niger has led to discoveries in the areas of protein secretion [13] and of global transcriptional responses to simple sugars such as glucose, xylose and glycerol [14]–[16]. This genomic information is complemented by studies that have elucidated some of the basic molecular pathways by which hydrolytic enzyme production and sugar metabolism are regulated in A. niger [17], [18]. Furthermore, genome-wide approaches provide a basis for comparability with other fungal species [19], [20]. This wealth of background knowledge of A. niger and closely-related species, and their responses to monomeric sugars and simple polysaccharides, provides an excellent foundation on which to build more complete studies of growth on more complex, industrially relevant substrates. Measuring gene expression of Neurospora crassa during growth on Miscanthus [21], and transcriptional changes when A. niger is exposed to sugar cane bagasse [13] using microarray technology have previously provided important insights into degradation of those substrates.
Wheat straw is one of the most attractive potential feed stocks for biofuel production. It is a co-product of cereal grain production and is available in significant quantities; for example, in the order of 10 million tonnes are produced in the UK each year [22]. This study aims to gain a thorough understanding of the mechanisms employed by A. niger to degrade and grow upon this complex lignocellulosic substrate, beginning with the transcriptional changes associated with exposure to wheat straw compared to simple sugars. Our data were acquired using Next Generation RNA-sequencing (RNA-seq) technology which provides a wealth of information for the confirmation of gene predictions from both published genomes, as well as in identifying alternative splicing patterns, novel genes/exons, transcription start/end points and antisense (AS) transcripts.
Results/Discussion
Saccharification of wheat straw by A. niger
The wheat straw used was composed of 37±1.69% cellulose and 32±1.2% hemicelluloses, 22±0.1% lignin and, after ball milling, the substrate retained 25±0.76% crystallinity (data are the mean of three replicates and are shown ± standard deviation). In order to determine the time at which degradation of the wheat straw by A. niger had begun to take place, the monomeric sugar content of the culture supernatant was analysed by HPLC. Figure 1A shows that, prior to inoculation, in minimal media containing 1% straw, the concentration of free monomeric sugar present in the liquid fraction was 76±0.9 µM. After 12 h of incubation, control samples, which had not been inoculated with A. niger, contained similar levels of each sugar. In the A. niger cultures, the total concentration of free monomeric sugar present in the liquid fraction increased to 166±26.9 µM, showing that degradation of wheat straw polysaccharides had begun to take place. There were also changes detected in the proportions of individual sugars (Figure 1A). Xylose, arabinose and galactose levels were increased by a statistically significant level, whilst glucose levels were not. Two non-exclusive hypotheses could explain this observation; i) the hemicellulose fraction of the lignocelluloses substrate is degraded first, and/or ii) glucose is preferentially imported by the fungus. Indeed a requirement for glucose depletion prior to xylose utilisation, when both sugars are present, has been observed in Aspergillus nidulans [23]. After 24 h of incubation, levels of free sugar had not increased any further suggesting that the balance of degradation and sugar uptake had reached a steady state. RT-PCR showed that transcription of several genes encoding glycoside hydrolases, (endoglucanase; eglA, cellobiohydrolase; cbhA and the endoxylanases; xynA and xynB) that are transcriptionally activated in response to xylose by the xylanolytic regulator XlnR, were highly induced at this point, when compared to expression in the Glucose 48 h cultures (data not shown). These two time-points were therefore chosen for RNA-seq analysis (Glucose 48 h and Straw 24 h). After 24 h incubation in the straw media, the particles of straw were in intimate association with A.niger mycelia (Figure 1B). It is possible therefore that the responses seen are due not only to the presence of inducing molecules, but also due to the physical interaction between the fungal mycelia and the straw.
Fig. 1. Free sugars in the straw media. 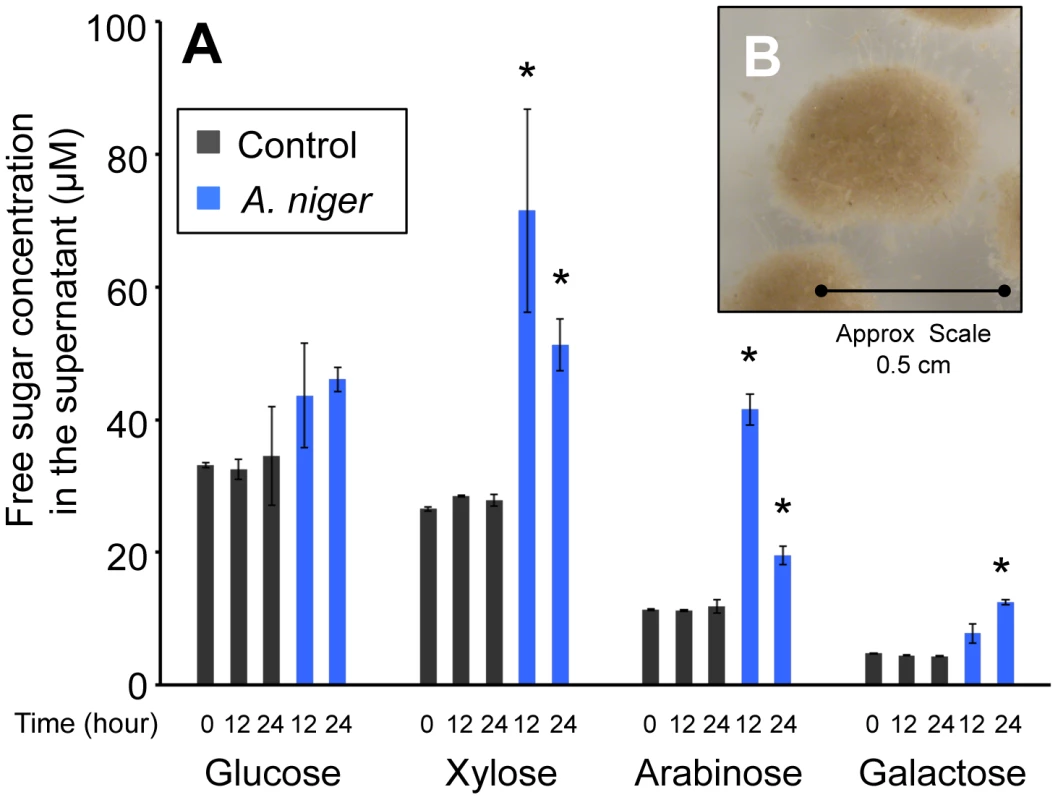
(A) The monomeric sugar content of culture supernatants was analysed by HPLC at 0, 12 and 24 h after the transfer of the mycelia to the straw media. Each bar represents the mean +/− the standard deviation of values from three independent experiments where black represents control cultures and blue represents cultures containing A. niger. The asterisk indicates p-values<0.05 relative to the control culture at the corresponding time by unpaired t-test. (B) A. niger mycelial clump from a culture grown for 24 hours in minimum media containing straw as sole carbon source. To investigate the repressive effect of glucose on expression of degradative enzymes, glucose was added exogenously to the wheat straw-incubated 24 h cultures to a final concentration of 1% (w/v). Samples were taken for RT-PCR analysis after 30 min, 1, 2 and 5 h. For all genes tested the levels of hydrolase expression decreased over this time course, reaching for most of the genes a similar level to that seen in the Glucose 48 h cultures after 5 hours of exposure to the exogenously added glucose (data not shown). Therefore, 5 hours after the addition of glucose to the straw cultures was selected as the final time point for RNA-seq analysis (Straw+Glucose 5 h). These represent three physiologically distinct conditions; long term growth under glucose repressing conditions, growth in the presence of an inducing lignocellulosic substrate and growth in the presence of the inducing substrate and glucose simultaneously. RNA was extracted from triplicate independent cultures in each condition for RNA-seq analysis.
The wheat straw-induced transcriptome of A. niger
Reads were mapped to the A. niger ATCC 1015 genome sequence [11] as it is phylogenetically very close (based on the β-tubulin sequence) to the N402 strain used in this study, and RPKMs (Reads Per Kilobase of exon model per Million mapped reads) were calculated for each annotated gene. The ATCC 1015 gene model is thought to under-predict the true number of genes present in the A. niger genome [11] so, in order to extract the maximum amount of data from our transcriptome sequencing, reads were also mapped to the CBS 513.88 sequence [10], which has a greater number of predicted genes. Approximately 2.5% more reads were successfully mapped to the ATCC 1015 genome than the CBS 513.88, reflecting the closer relationship between this strain and N402 used in this study. The CBS 513.88 genome model contains 4213 genes not included within the ATCC 1015 genome model and 939 of these genes were found to have an RPKM of 1 or more in at least one of the conditions tested in our transcriptome and are therefore very likely to be present within the ATCC 1015 genome also. A full list of the gene expression for the 4213 genes is included in supplementary material (Table S1). Eight of these genes encoded potential polysaccharide degrading enzymes or demonstrated a transcriptional pattern in the CBS 513.8 of interest to this study and so were added to the ATCC 1015 gene model and their RPKM values from the ATCC 1015 mapping were calculated.
RPKM values for all genes were calculated for each of the separate biological replicates, as well as for the combined mapping of all three, (Table S2). Table S3 shows that inter-replicate reproducibility was extremely high (R-squared values of >0.9). The results shown within the main text are from the combined mapping scores. Three statistical significance tests were applied to changes in gene expression, the Likelihood Ratio Test [24], Fisher's Exact Test [25], and an MA-plot-based method with Random Sampling model [26]. The results of these tests for each gene are listed in Table S2. All gene inductions discussed within the text had a p-value of <0.001 for all three statistical tests.
Expression of CAZy genes
Deconstruction of plant cell wall polysaccharides is mediated by enzymes of three major classes: Glycoside Hydrolases (GH), Carbohydrate Esterases (CE) and Polysaccharide Lyases (PL). The Carbohydrate Active Enzyme database (CAZy - http://www.cazy.org) subdivides enzymes within these three classes into families based on their related activity and sequence. CAZy defines 281 of these enzymes within the CBS 513.88 genome, all but two of which are also predicted in the ATCC 1015 genome model (CBS 513.88 annotation: An14g07390 and An14g07420). The ATCC 1015 genome encodes 246 predicted GHs, representing 51 families; 25 CEs representing 9 families and 8 PLs representing 2 families.
After 48 hours of growth in minimal media with 1% glucose, CAZy genes represent approximately 3 percent of total mRNA, with the glucoamylase glaA (GH15 family) accounting for the majority (over 65 percent) of this (Figure 2A). SDS-PAGE of the culture supernatant revealed the presence of a highly predominant protein band, which was identified by tandem MS as GlaA (data not shown). Twenty-four hours after the mycelia were transferred to straw, expression of CAZy genes made up more than 19 percent of total mRNA. This is a strong over-representation of the CAZy group of genes, as they represent only ∼2.5 percent of the coding genome. Thirty of the induced CAZy genes reached an expression level above 50 RPKM. They represent 14 families of GH, 2 of CE and 1 of PL (Table 1).
Fig. 2. CAZy gene expression. 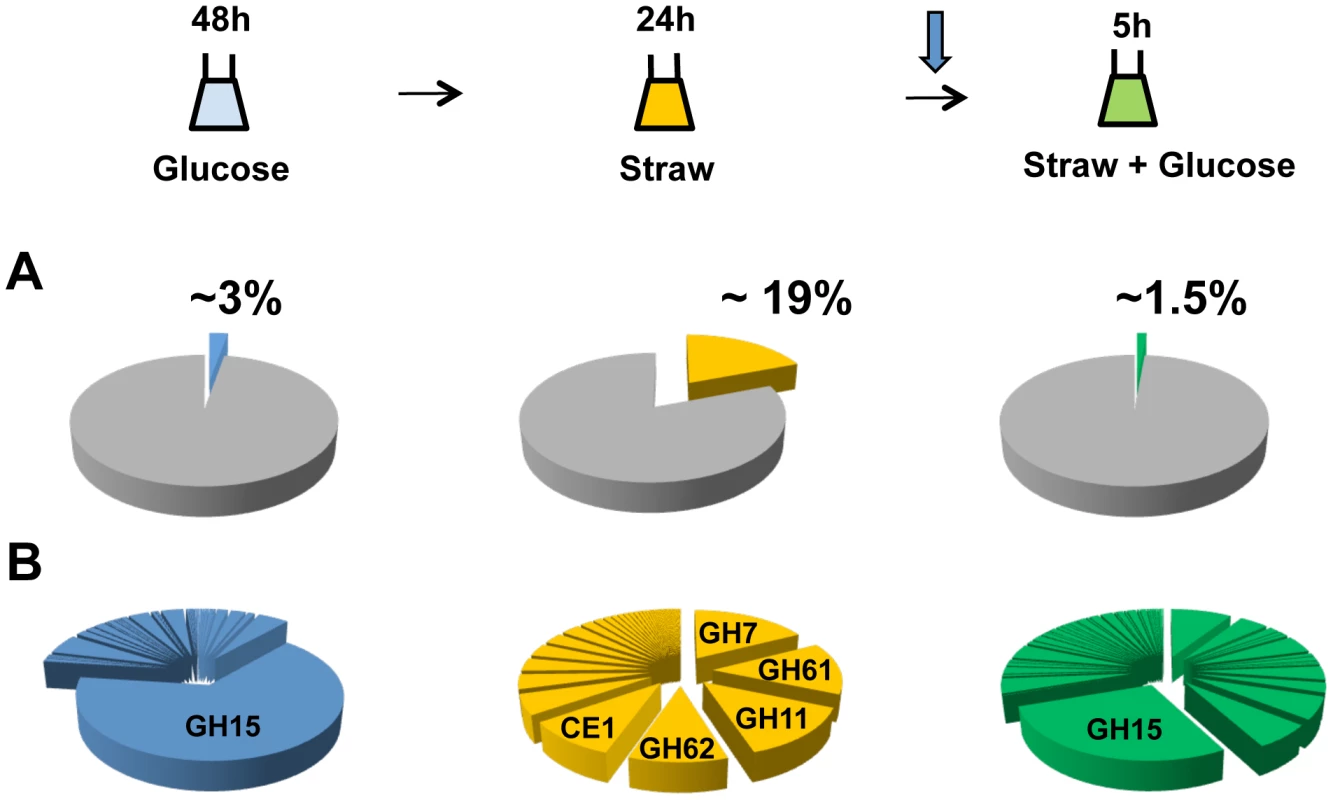
The sampling conditions are shown for the RNA-seq study. Cells were grown in glucose (48 h), washed and transferred to straw (24 h) and then glucose was added (downward arrow) followed by a further 5 h incubation. (A) Percentage of total mRNA (calculated from RPKM values) represented by CAZy family genes from each condition of the transcriptome study. (B) Proportions of total of CAZY gene mRNA from each enzyme family. The families are listed in decreasing order of expression in the Straw 24 h condition. Tab. 1. Straw-induced CAZy genes. 
CAZy genes strongly induced (≥20 x) and expressed (≥50 RPKM) after 24 h in Straw. An02g02540 has not been annotated in the ATCC 1015 genome but was found on chromosome 4_2 (5′-669026 - 667767-3′). Annotation are from CADRE [66] (http://www.cadre-genomes.org.uk/index.html). RPKM values are from the combined mapping of three biologically-independent samples under each condition, values for each individual sample are listed in Table S2. All genes listed showed a statistically significant induction when switched from glucose to straw (p>0.001). The diverse categories of CAZy genes expressed during exposure to straw reflect the complexity of the carbohydrates present within the substrate. However, it is interesting to note that around 65 percent of the mRNA from the CAZy group at this time-point is from genes encoding just 5 families of enzyme, GH7, 11, 61 and 62 (cellobiohydrolases, xylanases, polysaccharide monooxygenases and arabinofuranosidases, respectively) and CE1 (acetyl xylan esterases) (Figure 2B and Table 1). Proteins from each of these categories, except GH61, were also identified within culture supernatants by SDS-PAGE and tandem MS (data not shown). The fact we did not identify any GH61 proteins, that play a role in the oxidative cleavage of recalcitrant plant biomass [27]–[29], amongst the major bands could be due to a discrepancy between transcript and protein levels, or simply a technical issue in detection, such as the protein not staining well, or the protein being attached to the substrate or the fungal membrane, whilst only the supernatant was analysed. Based on transcript abundance, the 5 categories of encoded enzyme might provide the bulk of activities required for the degradation of straw. Catabolite repression, by addition of glucose to the straw cultures, exerts strong repression upon CAZy gene expression; after 5 hours, CAZy gene mRNA is reduced to only 1% of total mRNA. The glucoamylase glaA (GH15) becoming again the most expressed CAZy gene under this condition (Figure 2B).
Expression of non-CAZy genes
Twenty-eight genes that were transcriptionally induced more than 20-fold, and reached an RPKM of greater than 50 after the switch to straw, do not encode hydrolases as classified by CAZy. These genes are listed in Table 2 and 23 of them fall into four broad functional categories: lipases/esterases, surface interacting proteins, enzymes of carbon and nitrogen metabolism and transporters.
Tab. 2. Straw-induced non-CAZy genes. 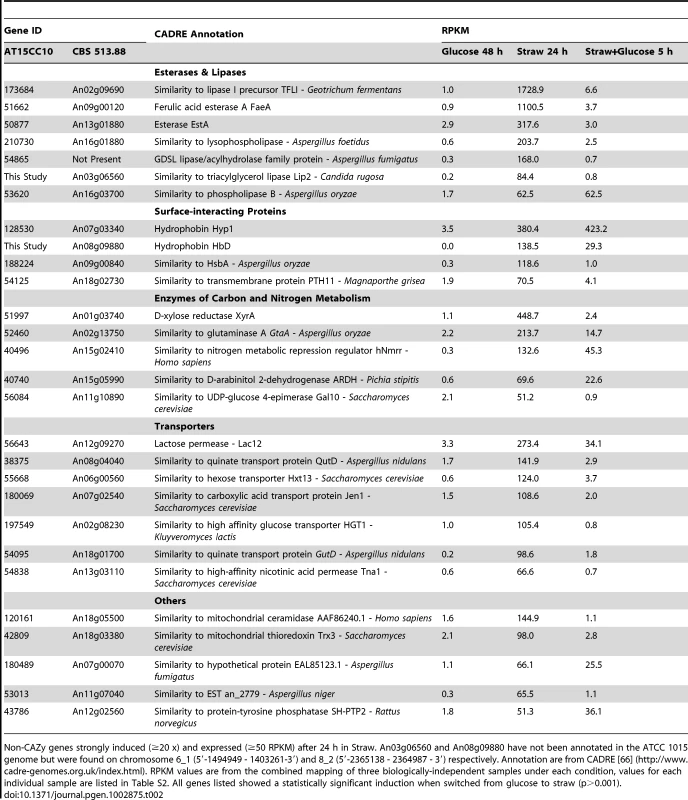
Non-CAZy genes strongly induced (≥20 x) and expressed (≥50 RPKM) after 24 h in Straw. An03g06560 and An08g09880 have not been annotated in the ATCC 1015 genome but were found on chromosome 6_1 (5′-1494949 - 1403261-3′) and 8_2 (5′-2365138 - 2364987 - 3′) respectively. Annotation are from CADRE [66] (http://www.cadre-genomes.org.uk/index.html). RPKM values are from the combined mapping of three biologically-independent samples under each condition, values for each individual sample are listed in Table S2. All genes listed showed a statistically significant induction when switched from glucose to straw (p>0.001). Lipases and esterases
Seven putative lipases or esterases not classified as CEs by CAZy were strongly induced after the transfer to straw, and 6 of these were repressed by addition of glucose (Table 2). The esterase EstA (TID_50877) was shown to be regulated by XlnR [30], and we identified it by SDS-PAGE and tandem MS as being one of the major proteins present in the supernatant after 24 hours in straw, while the ferulic acid esterase (FaeA) has been shown to have activity against wheat arabinoxylan [31]. Whilst the others have not previously been associated with polysaccharide degradation, their co-expression alongside the well characterised estA and faeA, raises the possibility that these enzymes may be involved in the saccharification of wheat straw lignocellulose.
Surface-interacting proteins
Two genes encoding hydrophobin family proteins and one hydrophobic surface binding protein (HsbA) were strongly induced by the switch from glucose to straw (Table 2). The gene hyp1 is not repressed by the re-addition of glucose to the culture, but hfbD and and hsbA were strongly repressed; their transcriptional profile is therefore similar to many of the genes of the CAZy group. Hydrophobins are amphipathic, surface-active proteins produced by filamentous fungi, with numerous biological functions, often relating to mycelial interactions with solid surfaces [32]. RolA (homologous to Hyp1) and HsbA of Aspergillus oryzae have been shown to associate with the synthetic polyester polybutylene succinate-coadipate and promote its degradation through the recruitment of a specific polyesterase [33], [34]. It is striking therefore that A. niger genes that encode proteins bearing homology to each, are highly induced by the presence of straw, suggesting that these proteins could have a role in recruiting degradative enzymes to the straw surface. The last gene in this group TID_54125 encodes a G-protein couple receptor homologous to Pth11p from the rice pathogen Magnaporthe grisea. Pth11p is required for signalling in appressorium formation [35] through the sensing of the plant host surface via both hydrophobicity and the presence of cutin monomers (which are also a component of the wheat straw cuticula [36]). The induction of the A. niger homologue of pth11 in the presence of straw suggests that a similar strategy could perhaps be employed by A. niger in the sensing of solid substrate surfaces.
Carbon metabolism
The increased expression of the xylose reductase xyrA and other genes of the xylose utilisation pathway, such as xylitol dehydrogenase (TID_203198) and D-xylulokinase (TID_209771) (both induced approximately 10-fold, Table S2), along with decreased expression of glycolytic pathway enzymes (e.g. phosphofructokinase, TID_54093, repressed 5-fold), suggests that 24 hours after the transfer to straw, pentose sugars are the prominent carbon source available. These results, and the fact that A. niger is an efficient xylan-degrading organism [37], support the first of the hypotheses discussed earlier, that the reason xylose is the predominant free sugar observed after 24 hours incubation of wheat straw with A. niger (Figure 1A) is that the hemicellulose fraction of the straw is the first to be degraded, rather than being due to a greater rate of glucose uptake by the fungus.
The degradative response is sequential and triggered by carbon starvation
To establish the order of induction of the genes that were highly expressed by 24 h, a time-course experiment was performed. RNA was extracted 0.5, 1, 2, 3, 6, 9 and 12 h after the switch from glucose to straw, and the expression of genes of interest was measured using RT-PCR (Figure S1). The results show the cellobiohydrolase cbhB to be induced after 6 hours of exposure to straw, whilst cbhA and GH61a do not appear to be induced until the 9 hour time point. These timings were verified, and shown statistically significant, using quantitative RT-PCR (Figure S2). To investigate the regulatory basis for the differential response of these genes, expression under the glucose and straw conditions was examined in strains deleted for either the gene encoding the xylanolytic activator XlnR, that mediates xylose-induction of some GHs and esterases, or CreA, which mediates wide-domain carbon catabolite repression [38]. All three of the glycosyl hydrolases showed a statistically significant (p-value of >0.01 in an equal variance, one-tailed T-test) dependence upon XlnR for maximal induction at 24 h in straw (Figure S3), which is typical of hydrolases in A. niger [39], [40]. Interestingly, a smaller scale induction of cbhB in the straw media was still observed in the ΔxlnR strain. This induction is mediated by alleviation of CreA repression, since in the ΔcreA strain, cbhB was expressed at a significantly higher level than the wild-type during growth on glucose, as assessed by qRT-PCR, whilst cbhA and GH61a were not (Figure S4). This observation led to the hypothesis that the earlier induction seen for cbhB is triggered, not by the presence of an inducing sugar such as xylose, but instead by the absence of an available carbon source. Transferring mycelia from glucose (48 h) to media completely devoid of carbon source for 24 h had a significant inductive effect upon cbhB expression, whereas it did not induce cbhA or GH61a (Figure S5). As only a very low concentration of free xylose is present in the straw media until several hours after incubation with A. niger (Figure 1A), this might explain why the induction of cbhB occurs earlier than the induction of cbhA or GH61a.
CbhA and CbhB are the only two GH7-family cellobiohydrolases in A. niger. CbhB contains a family 1 Carbohydrate-Binding-Module (CBM), whilst CbhA has no CBM [41]. CBMs aid the attachment of the enzymes containing them to the complex polysaccharide surfaces of intact cell walls [42], [43]. It could be speculated that the CBM-containing enzyme is induced early because it plays an important role in targeting the relatively intact plant cell wall. Whilst the enzyme lacking the CBM is induced later, once soluble oligosaccharides have been released. Six other genes that were induced after 24 h in straw also encoded proteins containing CBM domains. Two of these genes, TID_205580 (encoding a member of the GH family 5, containing a CBM 1) and abfB (an arabinofuranosidase containing a CBM 42), showed the same pattern of expression and regulation as cbhB; i.e. de-repression in a ΔcreA strain, induction 6 h after transfer to straw, and induction by carbon source starvation (Figures S3, S5 and S6). This CreA de-repression-dependent/XlnR activation-independent mechanism of induction may allow a subset of hydrolases that are targeted more specifically to the intact plant cell wall to play a “scouting” role under carbon starvation conditions, testing for available complex polysaccharides and liberating small quantities of sugar and oligosaccharides which trigger the subsequent larger scale induction of the genes themselves and the remaining majority of hydrolases by XlnR. Such a system would allow the organism to probe the surrounding environment for complex substrates when undergoing carbon starvation, without over-committing resources, until the presence of a degradable substrate is sensed from the release of inducing sugars. The expression of the “scouting” enzymes themselves, as well as the majority of hydrolases, which are completely dependent upon an active form of XlnR for expression, are then induced to extremely high levels (around one-fifth of the total mRNA inside the cell 24 hour after the transfer to straw, Figure 2A). None of the genes identified here as being induced early are predicted to encode enzymatic activities capable of releasing xylose from wheat straw, which would lead to XlnR activation. However, our preliminary data (not shown) indicates that the genes shown here are only part of a larger subset, which may encode such activity. Defining the full subset of hydrolase genes that are induced concurrently with cbhB is the subject of ongoing study.
Of the non-CAZy genes that are responsive to straw, both the putative lipase gene (TID_173684) and esterase, estA (TID_51662), are induced by the 6 h time point (Figure S1); they therefore form part of the early response, along with cbhB, abfB and the GH5 family member TID_205580. The lipase-encoding gene shares the same CreA-dependent expression pattern as cbhB (Figure S7B). The genes encoding the Pth11 homologue and HsbA are also induced by a lack of carbon source but their induction shows dependence upon neither XlnR nor CreA (data not shown). The extremely early expression of the homologue of pth11, after 3 h, may indicate a role in the early stages of the signalling pathway and is the subject of current investigation. The concurrent timing of expression of hsbA with the majority of hydrolases, at 9 h, raises the possibility of a role in recruiting hydrolases to the surface of the straw, i.e. analogous to the recruitment of degradative enzymes to the surface of certain plastics [33], [34]. The gene encoding the hydrophobin HfbD was induced later, at 12 h. The possible functionality of surface binding proteins in the recognition of surfaces, and the possibility that those proteins recruit hydrolytic enzymes to the surface, is intriguing and may provide new avenues to enhance the action of hydrolases in the saccharification of biomass in the generation of biofuels and the synthesis of other chemicals within a bio-based economy.
The mechanism by which the presence of a complex substrate, which cannot be imported into the cell, is signalled and triggers expression of the requisite hydrolytic enzyme mix, is widely debated and may vary with fungal species. The predominant induction model in fungal systems [21], [44] proposes that general basal expression of small quantities of hydrolase begins the degradation of complex polysaccharides, thereby producing inducing compounds that elicit the full transcriptional response. It has alternatively been suggested that some relevant genes are induced by carbon source depletion, and that the derived enzymes might play a foraging role, under starvation conditions [45]. Temporally differential expression of complex polysaccharide degrading enzymes and the presence of “scouting” enzymes that do not require the presence of a substrate-derived inducer for expression, but work to release inducing molecules and trigger a larger degradative response, has also been observed for the A. niger response to pectin [46], [47] and starch [48] perhaps indicating a conserved response pattern to insoluble substrates. Our data support this model for lignocellulose and we take it further by proposing a succession of events that not only includes the timing of expression of genes encoding hydrolases but also involves sensing proteins and the possibility that surface-binding proteins serve as a scaffold for recruitment of hydrolases. In summary the overall strategy appears to be an induction of a specific, small scale, sensory response by carbon source starvation, mediated at least partially by alleviation of CreA-dependent catabolite repression. This leads, in the event of successful liberation of free sugars from the wheat straw substrate, to the full scale degradative response, which is activated by XlnR (Figure 3).
Fig. 3. Induction model based on the sequential expression of responsive genes. 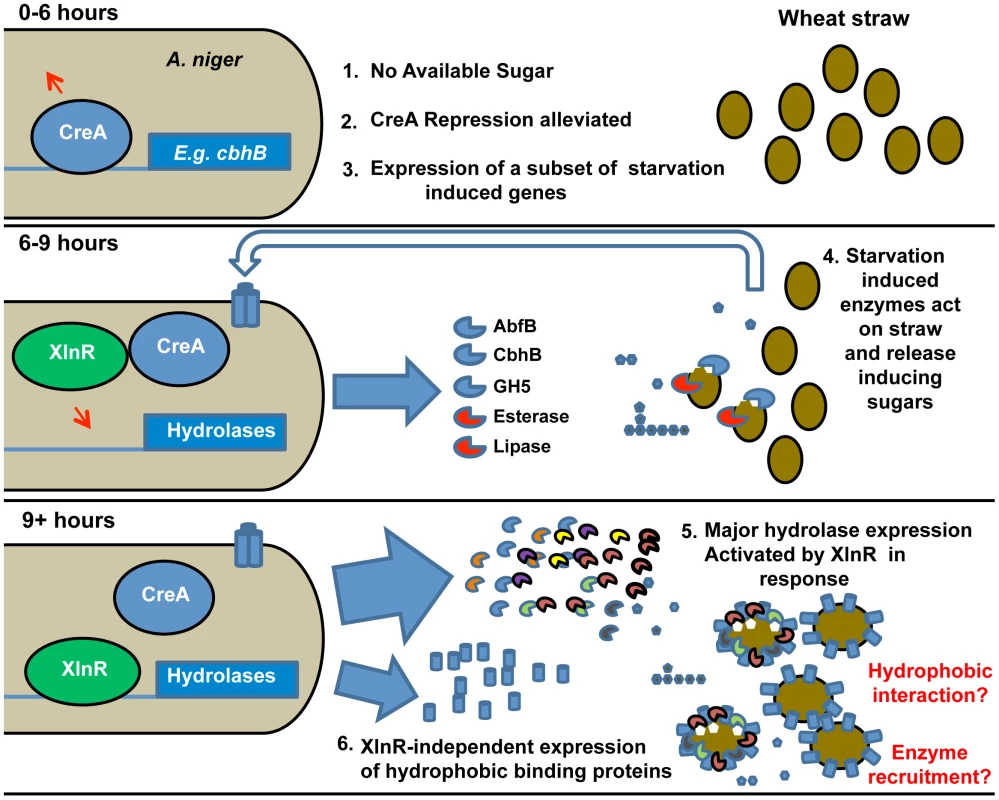
The sequence of events is illustrated and key events are numbered. The upper panel represents the transcriptional events in A. niger upon exposure (0–6 h) to straw represented by filled ovals. Lack of easily-available carbon source leads to the alleviation of CreA repression (represented by the arrow above CreA) and induction of a subset of starvation-induced genes represented by cbhB. At 6–9 h exposure to straw (middle panel), the expressed hydrolases and other enzymes (examples named in the Figure.) act upon the wheat straw, releasing small quantities of inducing sugars such as xylose (filled pentamer) as well as glucose (filled hexamer). Transporters for the sugars are induced (indicated by the trans-membrane cylinders and un-filled large arrow). By 9 hours (lower panel) the presence of xylose has caused activation of XlnR and, thereby, large scale expression of hydrolases genes. Also induced by 9 hours, in an XlnR-independent manner, is the hydrophobic binding protein HsbA. The hydrophobin HfbD is induced by 12 hours. A physical association of hydrophobic binding proteins with straw and degradative enzymes is hypothesised and represented. Note that the functionality of XlnR and CreA is indicated by attachment to recognition sequences in target promoters and is meant only to indicate the functional control of those promoters by the transcription factors. Modifications to those transcription factors (e.g. phosphorylation of XlnR in A. oryzae [65]) may occur without necessarily implying that their location changes. Antisense transcription
Natural antisense transcripts (NATs) are RNAs transcribed from a region of the genome that lies antisense (AS) to a gene. They have been found in a number of organisms, including several fungi [49]–[51], and play various regulatory roles [52]. The number of reads that fell upon the AS strand was counted for each gene in our study and AS RPKM values were calculated in each condition. AS reads accounted for approximately 2% of total reads under the conditions tested (2.41, 1.94 and 2.04% in Glucose 48 h, Straw 24 h and Straw+Glucose 5 h, respectively). Although the vast majority of genes had just a few associated AS transcripts, 521 genes had an AS RPKM of greater than 1 and up to about 120 (Figure S6 and Table S4).
In order to confirm that the NATs observed were biologically significant, and not simply an artefact of RNA-sequencing, a specific example, TID_53176, encoding a predicted membrane protein belonging to the GPR1/FUN34/YaaH family, which is required for acetate uptake in A. nidulans [53], was chosen for further analysis. The TID_53176 sense transcript is expressed in the straw media, whilst the AS transcript is expressed in the presence of glucose (Figure 4A).
Fig. 4. Sense and antisense transcription from TID_53176. 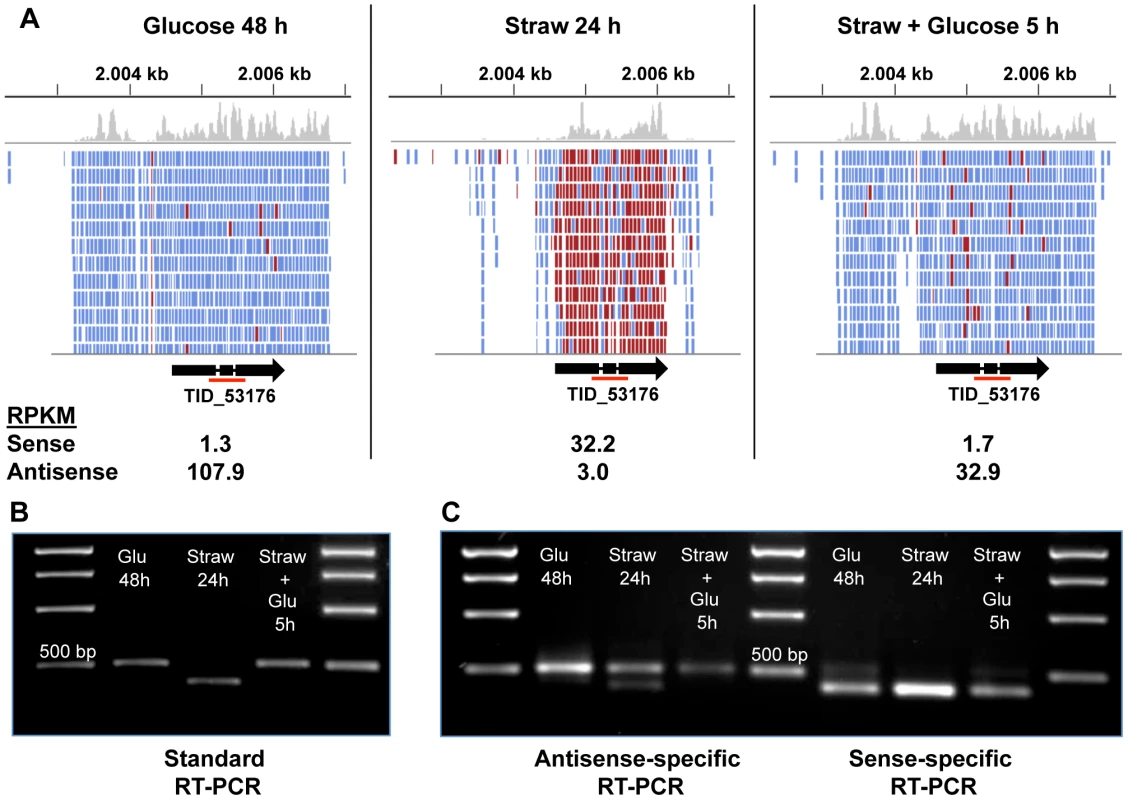
(A) Alignment of RNA-seq reads to the TID_53176 genome region under each condition. Reads represented in blue are antisense, those in red are sense. The Figure was constructed using the Integrative Genomics Viewer [62]. (B) Oligo(dT) primed RT-PCR using TID_53176 specific primers. The expected band size from the spliced sense transcript is 411 bp and the size of the non-spliced antisense transcript is 524 bp. The red line under the gene model in panel A indicates the amplified region. (C) Strand-specific RT-PCR. One of the standard PCR primers, with an added sequence tag (Table S5), was used to synthesise cDNA from one strand only and then the PCR step was performed by using the tagged sequencing primer together with the opposing gene-specific primer. The larger band is only seen in the antisense-specific reaction, confirming it does represent an antisense transcript. The smaller band is the only band present in the sense-specific reaction. The AS coverage level is high in both glucose conditions, and extends over the full length of the predicted gene including the two introns and extends both upstream and downstream, but does not overlap any neighbouring genes (Figure 4A). The sense coverage, seen in the straw condition, is shorter in length and there is almost zero coverage of the introns, indicating that the vast majority of sense transcripts are fully spliced. Figure 4B shows that RT-PCR, using primers upstream of the first intron and downstream of the second, can distinguish between the larger AS product and the shorter, fully spliced, sense transcript. Since oligo(dT) was used as the primer for cDNA synthesis, the AS transcript must be polyadenylated. To verify the strandedness of the two products, strand-specific RT-PCR was performed using a tagged primer approach and the results confirmed that all of the larger product is generated from antisense transcripts, whilst the smaller product is the only band seen in a sense-specific reaction (Figure 4C). A trace amount of a smaller antisense product can be seen in the antisense-specific assay under straw 24 h conditions. This may represent a true RNA intermediate, or it could be due to the high level of sense transcript self-priming and slight carryover of primer from the cDNA synthesis priming its amplification.
To identify AS transcripts that responded to the change in carbon source we calculated the ratio of antisense∶sense expression under Glucose 48 h and Straw 24 h conditions for the 521 genes with an AS RPKM of >1 (Table S4). Genes where sense transcription is induced on straw but AS predominates on glucose, include examples of transporters and permeases, CAZy enzymes and the putative lipase TID_173684.
This putative lipase TID_173684, one of the most highly induced genes upon exposure to straw, is also one of the genes showing the most marked antisense∶sense ratio switch (Table S4). Under glucose 48 h conditions there is significant expression of both sense and AS transcripts with a 60% greater level of AS (RPKMs of 1 and 1.6 respectively). After the switch to straw, at 24 h there is a large induction of the sense transcript (∼1700-fold) and AS transcription is cut to less than a third of the initial level (Figure S7A). Standard and strand-specific RT-PCR reactions under the same conditions give a similar pattern of bands to that seen above for TID_53176 (Figure 4). Interestingly, in the ΔcreA strain sense transcription is seen in both glucose and straw conditions, suggesting that the AS/S ratio switch is regulated either directly or indirectly by CreA. This was confirmed by strand-specific RT-PCR (Figure S7B). The timeline of induction experiment detailed earlier (Figure S1) shows that the AS/S switch in the expression of the putative lipase can be seen to occur between 3 and 6 h after the transfer to straw, which is concurrent with the expression of the carbon starvation induced subset of genes (of which the lipase gene is part). This suggests a possible relationship between carbon source responsive regulation by CreA and antisense transcription, providing an interesting area for further study.
Methods
Wheat straw analysis
Ball milling
The wheat straw (Cordiale variety) was milled using a Laboratory Mill (Laboratory Mill 3600, Perten, Sweden) and passed through a sieve with a mesh size of 700 µm for size reduction prior to ball milling. 5 g of pre-milled wheat straw was ball-milled in 80 mL stainless steel grinding bowls with 10-mm-diameter steel balls in a Planetary Mill (Pulverisette 5 classic line, Fritsch, Germany), at 400 rpm for a grinding time of 20 min, resulting in an average particle size of ∼75 µm.
Sugar analysis
The total sugar in processed ball-milled wheat straw was quantified in the hydrosylate after acid hydrolysis. 30 mg of dried wheat straw was weighed and subjected to a two stage acid hydrolysis initially with 12 M sulphuric acid for 1 hour at 37°C followed by 1 M sulphuric acid for 2 hours at 100°C. The monosaccharide analysis was performed on the supernatant. Sugar monomers in culture supernatants, collected at appropriate times points and separated from mycelia and insoluble straw by filtration through Miracloth (CalBioChem), and fully acid hydrolysed residues were determined by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (Dionex, UK) using a CarboPac PA20 column with 50 mM NaOH isocratic system at working flow rate of 0.5 ml/min at 30°C. Glucose, xylose, arabinose and galactose were used as standards with mannitol as an internal standard.
Lignin analysis
The acetyl bromide method [54], [55] was performed to quantify lignin in wheat straw samples. 100 mg of dried sample was digested with 4 ml of 25% v/v acetyl bromide in glacial acetic acid (HAc) in a Teflon capped tube at 50°C for 2 h with occasional mixing. After cooling, the digested sample volume was made up to 16 ml with HAc. After centrifugation at 3000 g for 15 min, 0.5 ml of the supernatant was mixed with 2.5 mL of HAc and 1.5 mL of 0.3M NaOH. 0.5 mL of 0.5M hydroxylamine hydrochloride solution was then added and the volume was made up to 10 mL with HAc. An analytical sample of low sulphate lignin (Sigma) was used as the standard for quantitative analysis by the acetyl bromide method. 10 mg of the lignin was dissolved in 5 ml of dioxane and 0.2, 0.3, 0.4, 0.5 and 0.6 mL was added in Teflon capped tubes to determine a standard curve. A reagent blank was also included. 0.5 mL of 25% acetyl bromide in glacial acetic acid (HAcBr) was added to each tube and incubated at 50°C for 30 min. The samples were cooled and 2.5 mL of HAc, 1.5 mL of 0.3 M NaOH, and 0.5 mL of 0.5 M hydroxylamine hydrochloride solution were added. The final volume was made up to 10 mL with HAc. Optical density was measured by scanning from 250 to 400 nm in a UV-Visible spectrophotometer (Varian, UK). The concentration of acetyl bromide soluble lignin for the samples was determined from the standard curve measured at 280 nm.
X-ray diffraction
The percentage cystallinity in the milled straw samples was estimated using X-ray diffraction, according to a variation of the method of Ruland and Vonk [56]. Cellulose is presumed to be the only crystallisable polymer in the cell-wall matrix so non-cellulose components such as lignin and hemicelluloses also contribute to the non-crystalline proportion. Powder measurements were carried out on a Siemens D5000 system, with copper Kα X-ray source, with scans performed from 5 to 50° 2è, A linear baseline was applied between the scan limits and a diffractogram of a ball-milled (fully amorphous) straw sample was then subtracted from the experimental profiles.
Strains and growth conditions
The A. niger strains used were N402 [57] and AB4.1 pyrG [7] or as specified otherwise. Strains were maintained on potato dextrose agar (Oxoid). All AB4.1 cultures were supplemented by 10 mM uridine (Sigma). Cultures were incubated at 28°C until they had conidiated. Spores were harvested into 0.1% (v/v) Tween 20 (Sigma). ΔxlnR and ΔcreA strains are A. niger AB4.1 pyrG containing a deletion of the respective open reading frame. Strains were constructed using the method developed by Scherer and Davis. [58] based on recombination between a plasmid containing the flanking region of the gene of interest and the chromosome. As a selection/counter-selection marker we used the gene coding for the orotidine-5-phosphate decarboxylase [59] (pyrG, from Aspergillus oryzae). After transformation of A. niger, cells were selected for uridine prototrophy, confirming integration of the plasmid into the chromosome. After purification of the transformants, release of the selective pressure for the integrated plasmid was achieved by propagating the clones twice on potato dextrose agar containing 10 mM uridine. Selection for cells that had excised the plasmid from the chromosome was done by plating them on media containing 4 mM of 5-fluoro-orotic acid (Melford) and 1.6 mM uridine. Deletion of creA or xlnR was confirmed by PCR using internal and external oligonucleotide primers and by sequencing around the respective loci.
Liquid batch cultures were inoculated with spores to a final concentration of 106 spores/ml. A. niger was grown in 100 ml of minimal media [all l−1: NaNO3, 6 g; KCl, 0.52 g; MgSO4.7H2O, 0.52 g; KH2PO4, 1.52 g; Na2B4O7.10H2O, 0.008 mg; CuSO4.5H2O, 0.16 mg; FePO4.H2O, 0.16 mg; MnSO4.4H2O, 0.16 mg; NaMoO4.2H2O, 0.16 mg; ZnSO4, 1.6 mg] with the appropriate carbon source added to a final concentration of 1% (w/v) in 250 ml Erlenmeyer flasks at 28°C, shaken at 150 rpm. The standard time-course consisted of growth for 48 h in 1% (w/v) glucose media, after which mycelia were removed by filtration through Miracloth (Merck), washed thoroughly with media devoid of carbon source, and transferred to fresh media containing 1% (w/v) ball-milled wheat straw as sole carbon source. Incubation was continued for 24 h. Glucose was then added exogenously to a final concentration of 1% (w/v) and incubation continued for 5 hours. Figure 1 shows an image of a mycelial clump that was magnified using a Nikon SMZ1000 stereomicroscope and the picture taken using a Nikon 4500 camera.
RNA extraction
Mycelia from each condition were frozen and ground under liquid nitrogen using a mortar and pestle. Total RNA was extracted from the ground material using the TRIzol reagent protocol (Invitrogen). An additional clean-up was performed using the RNEasy Mini Kit (Qiagen), following the manufacturer's RNA Clean-up protocol, including the additional on-column DNAse digest.
RT-PCR
SuperScript III Reverse Transcriptase (Invitrogen) was used to synthesise cDNA from total RNA according to manufacturer's instructions, using oligo(dT) as primer or tagged gene specific primers for strand-specific experiments. 0.5 µg of total RNA was used for each reverse transcription. PCR reactions were performed using Phusion (New England Biolabs) using 1 µl of cDNA in a 25 µl reaction. PCR conditions were 30 cycles of denaturation at 98°C for 30 s, annealing at 60°C for 30 s and extension at 72°C for 30 s. For PCR amplification of cDNA synthesised strand-specifically, a primer identical to the added tag sequence was paired with the opposing standard primer. In this way only cDNA synthesised from the tagged primer, and not non-specifically self-primed transcripts, was amplified.
qPCR amplifications were carried out using the Applied Biosystems 7500 Fast Real-Time PCR system. The PCR reaction mixture (10 µl) contained 1 µl of cDNA, specific primer sets (175 nM final concentration), and FAST SYBR-Green Master Mix (Applied Biosystems). PCRs were carried out for 40 cycles; denaturation at 95°C for 15 seconds, annealing at 67°C for 30 seconds, and extension at 60°C for 60 seconds. All measurements were independently conducted 3 times on 2 separate biological isolates. The specificity of primer sets used for qRT-PCR amplification was evaluated by melting curve analysis. The Standard Curve Method was used for quantification against a known concentration of genomic DNA (Li et al 2009). All primers used, and sequences are listed in Table S5.
RNA-seq
10 µg of Total RNA was depleted of ribosomal RNA using the Ribominus Eukaryotic kit (Invitrogen). SOLiD whole transcriptome libraries were made as outlined in the SOLiD Whole transcriptome kit protocol (Applied Biosystems). The Quant-it HS dsDNA assay kit (Invitrogen) was used to measure the concentration of libraries in order to pool equimolar amounts. Pooled libraries were gel-purified using 2% size-select E-gels to 200–300 bp (Invitrogen). Emulsion PCR (0.5 pM final concentration of pooled libraries) and bead-based enrichment was carried out according to the SOLiD 4 Templated bead preparation guide containing library. Sequencing was performed on a SOLiD 4 ABi sequencer according to the manufacturer's instructions to generate 50 bp reads in colour space.
Data analysis
SOLiD 4 RNA-seq reads from each experimental sample were mapped to the two published genome sequences of A. niger (JGI A. niger version 3.0 unmasked assembly scaffolds and gene annotation and CADRE A. niger assembly) using the BioScope 1.3.1 Whole Transcriptome Pipeline (LifeTechnologies). This included the initial filtering of reads against a collection of published A. niger rRNA sequences prior to mapping (Genbank [60] sequence IDs: 197115286, 300675560, 222105557, 312434471, 34304202, 241017633, 157267321, 213866863). BioScope provided the primary read alignment position of each read mapped against the complete genome sequence and exon spanning junctions using available gene coordinate information. Read alignment results were recorded in BAM format for further downstream analysis. Read counts per gene were calculated for each sample with Htseq-count (http://www-huber.embl.de/users/anders/HTSeq) using the BAM file and genome annotation as the input. Strand-specific RNA-seq reads, as generated by SOLiD, can be specified for when executing Htseq-count in determining accurate read counts per gene. These counts were then used to calculate normalized expression values (RPKM) for each gene [61]. Additionally, antisense transcription could be detected by comparing gene counts generated by Htseq-count when opting to ignore or include strand-specificity in the calculations.. Data were further visualised using Integrative Genomic Viewer (IGV) [62]. Raw and processed data files have been submitted to the Gene Expression Omnibus, under accession number GSE33852. Reads were mapped against an EST assembly of Genbank EST records [60] for the wheat Triticum aestivum (taxon id 4565; 1,073,845 EST's March 2012) a publicly available wheat genome [63]. The number of reads that mapped was no higher in the wheat-grown A. niger cultures than it was in the glucose-grown cultures. This suggests wheat transcript levels are insignificant, and any reads mapping to the wheat genome are simply due to a basic level of homology between genomes.
Identification of secreted proteins
Proteins in culture supernatant from relevant times were separated on 4–20% Tris-glycine Novex SDS-PAGE gels (Invitrogen). 15 µl Tris-glycine-SDS loading buffer was added to 12 µl supernatant plus 3 µl 1 M DTT. The sample mixture was heated to 100°C for 5 minutes before being loaded onto the gel with running conditions of 100 V for 3 hours. The gel was subsequently silver stained [64]. Bands resolved by size were excised from the gel, diced into cubes (∼1 mm3) and placed into individual wells of a microtitre plate, processed (destained, reduced, alkylated) and digested with trypsin using standard procedures on a MassPREP station (Waters). Resulting peptides were delivered via nanoflow reversed phase-HPLC to a Q-ToF2 (Waters) for tandem MS analysis. An automated experiment (DDA = data dependent acquisition) was run where selected peptides automatically enter MSMS for fragmentation. The data was searched against the Swissprot and NCBInr databases using the MS/MSIONS search on the MASCOT web site (http://www.matrixscience.com) using the variable modifications of carbamidomethylation of cysteine and oxidation of methionine.
Supporting Information
Zdroje
1. SimsRE, MabeeW, SaddlerJN, TaylorM (2010) An overview of second generation biofuel technologies. Bioresour Technol 101 : 1570–1580.
2. GusakovAV (2011) Alternatives to Trichoderma reesei in biofuel production. Trends Biotechnol 29 : 419–425.
3. StephanopoulosG (2007) Challenges in engineering microbes for biofuels production. Science 315 : 801–804.
4. WeberC, FarwickA, BenischF, BratD, DietzH, et al. (2010) Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol 87 : 1303–1315.
5. ArcherDB (2000) Filamentous fungi as microbial cell factories for food use. Curr Opin Biotechnol 11 : 478–483.
6. MeyerV, WuB, RamAF (2011) Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol Lett 33 : 469–476.
7. van HartingsveldtW, MatternIE, van ZeijlCM, PouwelsPH, van den HondelCA (1987) Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet 206 : 71–75.
8. HihlalE, BraumannI, van den BergM, KempkenF (2011) Suitability of Vader for transposon-mediated mutagenesis in Aspergillus niger. Appl Environ Microbiol 77 : 2332–2336.
9. MeyerV, WankaF, van GentJ, ArentshorstM, van den HondelCA, et al. (2011) Fungal Gene Expression on Demand: an Inducible, Tunable, and Metabolism-Independent Expression System for Aspergillus niger. Appl Environ Microbiol 77 : 2975–2983.
10. PelHJ, de WindeJH, ArcherDB, DyerPS, HofmannG, et al. (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25 : 221–231.
11. AndersenMR, SalazarMP, SchaapPJ, van de VondervoortPJI, CulleyD, et al. (2011) Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Research 21 : 885–897.
12. CantarelBL, CoutinhoPM, RancurelC, BernardT, LombardV, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37: D233–238.
13. GuillemetteT, van PeijNN, GoosenT, LanthalerK, RobsonGD, et al. (2007) Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8 : 158.
14. AndersenMR, VongsangnakW, PanagiotouG, SalazarMP, LehmannL, et al. (2008) A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc Natl Acad Sci U S A 105 : 4387–4392.
15. SalazarM, VongsangnakW, PanagiotouG, AndersenMR, NielsenJ (2009) Uncovering transcriptional regulation of glycerol metabolism in Aspergilli through genome-wide gene expression data analysis. Mol Genet Genomics 282 : 571–586.
16. JorgensenTR, GoosenT, HondelCA, RamAF, IversenJJ (2009) Transcriptomic comparison of Aspergillus niger growing on two different sugars reveals coordinated regulation of the secretory pathway. BMC Genomics. England. 44.
17. de VriesRP (2003) Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl Microbiol Biotechnol 61 : 10–20.
18. de SouzaWR, de GouveaPF, SavoldiM, MalavaziI, de Souza BernardesLA, et al. (2011) Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels 4 : 40.
19. Le CromS, SchackwitzW, PennacchioL, MagnusonJK, CulleyDE, et al. (2009) Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci U S A. United States 16151–16156.
20. CoutinhoPM, AndersenMR, KolenovaK, vanKuykPA, BenoitI, et al. (2009) Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol 46 Suppl 1: S161–S169.
21. TianC, BeesonWT, IavaroneAT, SunJ, MarlettaMA, et al. (2009) Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci U S A 106 : 22157–22162.
22. BranderM, HutchisonC, SherringtonC, BallingerA, BeswickC, et al. (2009) Methodology and Evidence Base on the Indirect Greenhouse Gas Effects of Using Wastes, Residues, and By-products for Biofuels and Bioenergy : Report to the Renewable Fuels Agency and the Department for Energy and Climate Change. Ecometrica, Eunomia, Imperial College London.
23. PrathumpaiW, McIntyreM, NielsenJ (2004) The effect of CreA in glucose and xylose catabolism in Aspergillus nidulans. Appl Microbiol Biotechnol 63 : 748–753.
24. MarioniJC, MasonCE, ManeSM, StephensM, GiladY (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18 : 1509–1517.
25. BloomJS, KhanZ, KruglyakL, SinghM, CaudyAA (2009) Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 10 : 221.
26. WangL, FengZ, WangX, ZhangX (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010 26 : 136–138.
27. HarrisPV, WelnerD, McFarlandKC, ReE, Navarro PoulsenJC, et al. (2010) Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry 49 : 3305–3316.
28. QuinlanRJ, SweeneyMD, Lo LeggioL, OttenH, PoulsenJC, et al. (2011) Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci U S A 108 : 15079–15084.
29. BeesonWT, PhillipsCM, CateJH, MarlettaMA (2012) Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc 134 : 890–892.
30. BourneY, HasperAA, ChahinianH, JuinM, De GraaffLH, et al. (2004) Aspergillus niger protein EstA defines a new class of fungal esterases within the alpha/beta hydrolase fold superfamily of proteins. Structure 12 : 677–687.
31. de VriesRP, MichelsenB, PoulsenCH, KroonPA, van den HeuvelRH, et al. (1997) The faeA genes from Aspergillus niger and Aspergillus tubingensis encode ferulic acid esterases involved in degradation of complex cell wall polysaccharides. Appl Environ Microbiol 63 : 4638–4644.
32. LinderMB, SzilvayGR, Nakari-SetäläT, PenttiläME (2005) Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev 29 : 877–896.
33. TakahashiT, MaedaH, YonedaS, OhtakiS, YamagataY, et al. (2005) The fungal hydrophobin RolA recruits polyesterase and laterally moves on hydrophobic surfaces. Mol Microbiol 57 : 1780–1796.
34. OhtakiS, MaedaH, TakahashiT, YamagataY, HasegawaF, et al. (2006) Novel hydrophobic surface binding protein, HsbA, produced by Aspergillus oryzae. Appl Environ Microbiol 72 : 2407–2413.
35. DeZwaanTM, CarrollAM, ValentB, SweigardJA (1999) Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11 : 2013–2030.
36. StelteW, ClemonsC, HolmJ, AhrenfeldtJ, HenriksenU, et al. (2012) Fuel pellets from wheat straw: The effect of lignin glass transition and surface waxes on pelletizing properties. Bioenerg. Res. (2012) 5 : 450–458.
37. KingBC, WaxmanKD, NenniNV, WalkerLP, BergstromGC, et al. (2011) Arsenal of plant cell wall degrading enzymes reflects host preference among plant pathogenic fungi. Biotechnol Biofuels 4 : 4.
38. Kobayashi T, Kato M (2012) Transcriptional Regulation in Aspergillus. In: Machida M, Gomi K, editors. Aspergillus Molecular Biology and Genomics. UK: Caister Academic Press. 85–114.
39. de VriesRP, VisserJ, de GraaffLH (1999) CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol 150 : 281–285.
40. van PeijNN, GielkensMM, de VriesRP, VisserJ, de GraaffLH (1998) The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol 64 : 3615–3619.
41. GielkensMM, DekkersE, VisserJ, de GraaffLH (1999) Two cellobiohydrolase-encoding genes from Aspergillus niger require D-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol 65 : 4340–4345.
42. BorastonAB, BolamDN, GilbertHJ, DaviesGJ (2004) Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382 : 769–781.
43. HerveC, RogowskiA, BlakeAW, MarcusSE, GilbertHJ, et al. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A 107 : 15293–15298.
44. Carle-UriosteJC, Escobar-VeraJ, El-GogaryS, Henrique-SilvaF, TorigoiE, et al. (1997) Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J Biol Chem 272 : 10169–10174.
45. ForemanPK, BrownD, DankmeyerL, DeanR, DienerS, et al. (2003) Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem 278 : 31988–31997.
46. ParenicovaL, BenenJA, KesterHC, VisserJ (2000) pgaA and pgaB encode two constitutively expressed endopolygalacturonases of Aspergillus niger. Biochem J 345 : 637–644.
47. de VriesRP, JansenJ, AguilarG, ParenicovaL, JoostenV, et al. (2002) Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett 530 : 41–47.
48. YuanXL, van der KaaijRM, van den HondelCA, PuntPJ, van der MaarelMJ, et al. (2008) Aspergillus niger genome-wide analysis reveals a large number of novel alpha-glucan acting enzymes with unexpected expression profiles. Mol Genet Genomics 279 : 545–561.
49. GowdaM, VenuRC, RaghupathyMB, NobutaK, LiH, et al. (2006) Deep and comparative analysis of the mycelium and appressorium transcriptomes of Magnaporthe grisea using MPSS, RL-SAGE, and oligoarray methods. BMC Genomics 7 : 310.
50. NiT, TuK, WangZ, SongS, WuH, et al. (2010) The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One 5: e15271.
51. OhmRA, de JongJF, LugonesLG, AertsA, KotheE, et al. (2010) Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 28 : 957–963.
52. LapidotM, PilpelY (2006) Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep 7 : 1216–1222.
53. RobelletX, FlipphiM, PegotS, MaccabeAP, VelotC (2008) AcpA, a member of the GPR1/FUN34/YaaH membrane protein family, is essential for acetate permease activity in the hyphal fungus Aspergillus nidulans. Biochem J 412 : 485–493.
54. FukushimaRS, HatfieldRD (2001) Extraction and isolation of lignin for utilization as a standard to determine lignin concentration using the acetyl bromide spectrophotometric method. J Agric Food Chem 49 : 3133–3139.
55. HatfieldR, FukushimaRS (2005) Can lignin be accurately measured? Crop Sci 45 : 832–839.
56. MaierG, ZipperP, StubičarM, SchurzJ (2005) Amorphization of different cellulose samples by ballmilling. Cellulose Chem Technol 39 : 167–177.
57. BosCJ, DebetsAJ, SwartK, HuybersA, KobusG, et al. (1988) Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet 14 : 437–443.
58. SchererS, DavisRW (1979) Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A 76 : 4951–4955.
59. BoekeJD, LaCrouteF, FinkGR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197 : 345–346.
60. BensonDA, Karsch-MizrachiI, LipmanDJ, OstellJ, SayersEW (2011) GenBank. Nucleic Acids Res 39: D32–37.
61. MortazaviA, WilliamsBA, McCueK, SchaefferL, WoldB (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5 : 621–628.
62. RobinsonJT, ThorvaldsdóttirH, WincklerW, GuttmanM, LanderES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26.
63. LaiK, BerkmanPJ, LorencMT, DuranC, SmitsL, et al. (2012) WheatGenome.info: an integrated database and portal for wheat genome information. Plant Cell Physiol 53: e2.
64. YanJX, WaitR, BerkelmanT, HarryRA, WestbrookJA, et al. (2000) A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 21 : 3666–3672.
65. NoguchiY, TanakaH, KanamaruK, KatoM, KobayashiT (2011) Xylose triggers reversible phosphorylation of XlnR, the fungal transcriptional activator of xylanolytic and cellulolytic genes in Aspergillus oryzae. Biosci Biotechnol Biochem 75 : 953–959.
66. Mabey GilsenanJ, CooleyJ, BowyerP (2012) CADRE: the Central Aspergillus Data REpository 2012. Nucleic Acids Res 40: D660–666.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



