-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
Dietary restriction (DR), limiting nutrient intake from diet without causing malnutrition, delays the aging process and extends lifespan in multiple organisms. The conserved life-extending effect of DR suggests the involvement of fundamental mechanisms, although these remain a subject of debate. To help decipher the life-extending mechanisms of DR, we first compiled a list of genes that if genetically altered disrupt or prevent the life-extending effects of DR. We called these DR–essential genes and identified more than 100 in model organisms such as yeast, worms, flies, and mice. In order for other researchers to benefit from this first curated list of genes essential for DR, we established an online database called GenDR (http://genomics.senescence.info/diet/). To dissect the interactions of DR–essential genes and discover the underlying lifespan-extending mechanisms, we then used a variety of network and systems biology approaches to analyze the gene network of DR. We show that DR–essential genes are more conserved at the molecular level and have more molecular interactions than expected by chance. Furthermore, we employed a guilt-by-association method to predict novel DR–essential genes. In budding yeast, we predicted nine genes related to vacuolar functions; we show experimentally that mutations deleting eight of those genes prevent the life-extending effects of DR. Three of these mutants (OPT2, FRE6, and RCR2) had extended lifespan under ad libitum, indicating that the lack of further longevity under DR is not caused by a general compromise of fitness. These results demonstrate how network analyses of DR using GenDR can be used to make phenotypically relevant predictions. Moreover, gene-regulatory circuits reveal that the DR–induced transcriptional signature in yeast involves nutrient-sensing, stress responses and meiotic transcription factors. Finally, comparing the influence of gene expression changes during DR on the interactomes of multiple organisms led us to suggest that DR commonly suppresses translation, while stimulating an ancient reproduction-related process.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002834
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002834Summary
Dietary restriction (DR), limiting nutrient intake from diet without causing malnutrition, delays the aging process and extends lifespan in multiple organisms. The conserved life-extending effect of DR suggests the involvement of fundamental mechanisms, although these remain a subject of debate. To help decipher the life-extending mechanisms of DR, we first compiled a list of genes that if genetically altered disrupt or prevent the life-extending effects of DR. We called these DR–essential genes and identified more than 100 in model organisms such as yeast, worms, flies, and mice. In order for other researchers to benefit from this first curated list of genes essential for DR, we established an online database called GenDR (http://genomics.senescence.info/diet/). To dissect the interactions of DR–essential genes and discover the underlying lifespan-extending mechanisms, we then used a variety of network and systems biology approaches to analyze the gene network of DR. We show that DR–essential genes are more conserved at the molecular level and have more molecular interactions than expected by chance. Furthermore, we employed a guilt-by-association method to predict novel DR–essential genes. In budding yeast, we predicted nine genes related to vacuolar functions; we show experimentally that mutations deleting eight of those genes prevent the life-extending effects of DR. Three of these mutants (OPT2, FRE6, and RCR2) had extended lifespan under ad libitum, indicating that the lack of further longevity under DR is not caused by a general compromise of fitness. These results demonstrate how network analyses of DR using GenDR can be used to make phenotypically relevant predictions. Moreover, gene-regulatory circuits reveal that the DR–induced transcriptional signature in yeast involves nutrient-sensing, stress responses and meiotic transcription factors. Finally, comparing the influence of gene expression changes during DR on the interactomes of multiple organisms led us to suggest that DR commonly suppresses translation, while stimulating an ancient reproduction-related process.
Introduction
Dietary restriction (DR) in defined factors (such as calories or specific amino acids) without causing malnutrition delays the aging process, protects against age-related diseases (e.g. metabolic and cardiovascular disease, neurodegeneration and cancer) and extends lifespan in evolutionarily distant species, from unicellular yeast to rodents [1], [2]. Furthermore, there is evidence that DR might delay the aging process in non-human primates [3] and perhaps in humans as well [4]. While multiple studies in model organisms have shown that DR extends lifespan, the underlying molecular mechanisms remain largely unknown [2], [5], [6].
The mechanisms by which DR retards aging have been shown in model organisms to be mediated by genetic pathways, many of which are evolutionarily conserved and operate in humans. Considerable evidence indicates that DR is mediated by discrete signaling events that elicit specific genetic programs [1], [2], [7], [8]. Mutations in a number of genes, in fact, have been shown to prevent, shift or disrupt the lifespan-extending effect of DR, in many cases without altering the lifespan under ad libitum (AL) conditions. Such genes, which we call DR-essential, might specifically interfere with these signaling cascades and the program which leads to lifespan extension under DR. Understanding the interactions of such genes in a systematic way may provide important new clues regarding DR-mediated life-extension mechanisms.
In the post-genomic era, the vast amount of -omics data provides an opportunity to understand biological processes in a systematic fashion via data integration, network construction and analyses. Networks in biology exist on various levels, derived from the interactions between genes, transcripts, proteins and metabolites, and extending to the interactions between cells, tissues, organs and even between organisms. Examples at the molecular level are protein-protein interactions, gene-regulatory and metabolic networks. Biological networks tend to have certain discrete topological properties: They are usually scale-free (i.e. only a few nodes, termed “hubs”, have many connections, while the majority of nodes have relatively few connections), modularly composed and hierarchically structured. Scale-free networks are robust against random perturbation, but at the same time are sensitive to targeted disruption of the hubs [9]. The modular composition is due to the presence of groups of highly interconnected nodes (i.e. clusters) which perform certain biological activities (for instance protein complexes or pathways) with relatively sparse external connectivity. Lastly, these network modules associate with one another in a hierarchical manner. The challenge, however, is to integrate and combine the various networks in order to decipher biological function and gain new insights into the process under study.
Aging-associated genes are more conserved at the molecular level [10] and have a significantly higher node degree (i.e. number of interactions in a network) than expected by chance. Human orthologs of aging-associated genes found in model organisms form a continuous network and almost all of the hubs are also implicated in several age-related diseases [11]. The networks of age-related disease genes and aging-associated genes significantly overlap and approximately half of the common genes participate in signal transduction. Additionally, general disease genes have more connections to aging genes than expected by chance [12]. Numerous network studies on aging have been conducted with fruitful results [13]–[15]. To date, however, network studies on DR have been limited, in part because of a lack of adequate datasets. Therefore, we constructed GenDR, a database of DR-related genes.
Since GenDR is the first database of DR-related genes, we wished to assess its utility by performing the first network-based dissection of DR using DR-essential genes. DR can induce numerous changes in organisms, and a variety of different DR regimens have been employed. Our rationale is that by concentrating only on genes essential for DR-induced life-extension we are able to narrow down the various DR-related processes to those affecting aging. We used our list of DR-essential genes to investigate their evolutionary and network properties and identify common regulators of DR-induced lifespan extension. Novel DR-essential genes were predicted from the network and tested experimentally in yeast, which revealed new genes crucial for DR. We then integrated diverse types of data and performed a variety of network and systems biology analyses to reveal common pathways and mediators of DR effects, including putative transcription factors. Our work demonstrates the use of network approaches to study DR mechanisms, and we make our list of DR-essential genes available online for other researchers to use (http://genomics.senescence.info/diet/).
Results
Construction of GenDR, a Database of DR–Related Genes
A list of DR-essential genes was compiled from the literature. A DR-essential gene was defined as one which when genetically manipulated in a given organism blocks or disrupts the life-extending effect of DR. This criterion applies even if it was shown for only a single DR regimen. Over 100 genes were identified, mostly from the traditional biomedical model organisms (Table 1). In mice, for example, the only DR-essential gene found so far is the growth hormone receptor (Ghr) gene of which homozygous knockout mutants were long-lived and DR failed to further extend their lifespan [16]. In the case of yeast, DR-essential genes were annotated separately for replicative and chronological lifespan. However, both were combined for the analyses presented below, in order to focus on mechanisms universal to DR. For each gene, its orthologs in other model organisms and in humans (if any) were retrieved from public databases (see Materials and Methods).
Tab. 1. Number of DR–essential genes and orthologs in GenDR. 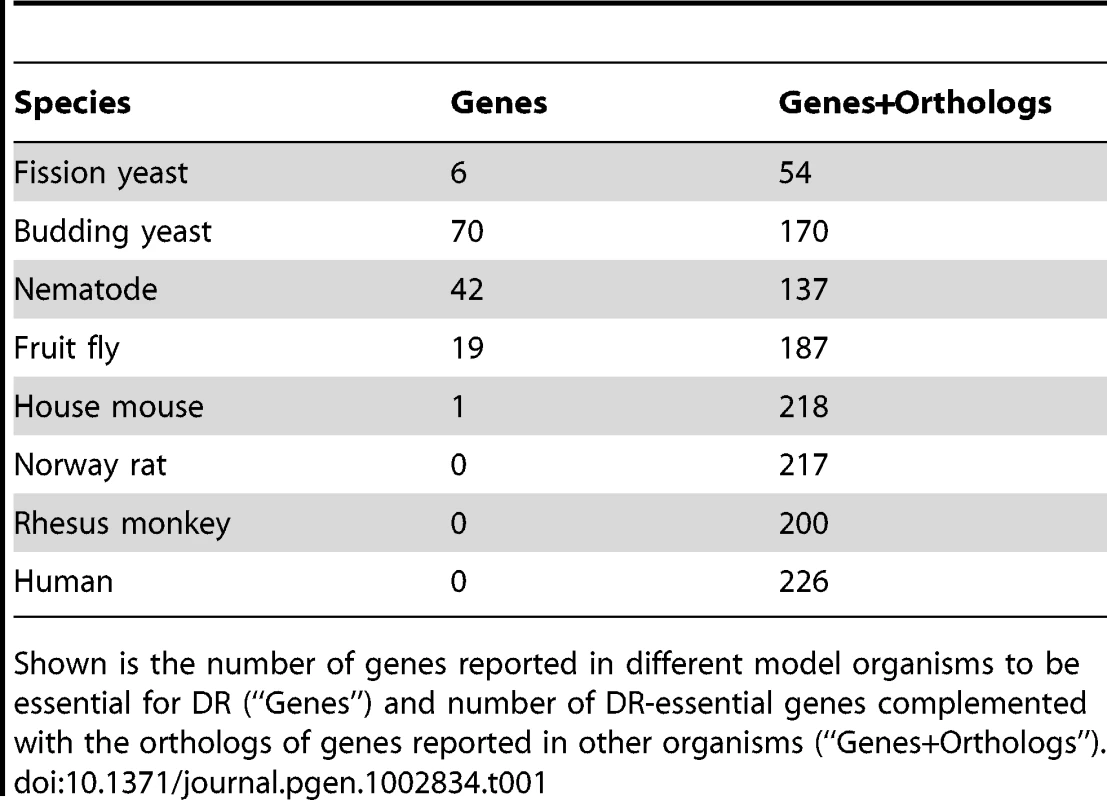
Shown is the number of genes reported in different model organisms to be essential for DR (“Genes”) and number of DR-essential genes complemented with the orthologs of genes reported in other organisms (“Genes+Orthologs”). We created a publically accessible database of DR-essential genes and their orthologs, called GenDR, which offers an important new tool to researchers working on the genetics of DR and of aging. Although other studies have emphasized the importance of genes that disrupt life-extending effects of DR [1], [2], [17], to our knowledge this is the first compilation of a list of such genes. The GenDR database employs the same system and interface as our widely-used GenAge database of aging-related genes [18]. Our inclusive selection criteria allow GenDR to incorporate a broad range of genes. Because information on each gene and reason(s) for its selection are included, however, researchers are able to focus on subsets of the database that are most relevant to their work. GenDR will be useful both as an informational website and as a research tool, and it is freely available online for the research community to use (http://genomics.senescence.info/diet/).
GenDR Is Enriched in Conserved Longevity Genes and Pathways
Given that DR works across diverse model organisms, we expected that GenDR can guide the search for evolutionarily conserved longevity mechanisms. Therefore, we analyzed GenDR at three levels: the sequences of genes/proteins, the interactome, and enriched pathways.
To test whether GenDR reflects the conserved effects of DR on longevity, we investigated the molecular evolution of genes essential for DR. In agreement with our hypothesis, the percentage of orthologs of DR-essential genes across all tested species is higher than expected by chance (Figure 1A). Likewise, the average dN/dS ratio, a measure of molecular evolution rate, as well as dN and dS rates, for DR-essential gene orthologs in mammalian species pairs were lower than expected by chance (Table S1 and Figure S1). As such, it appears that indeed DR is evolutionarily conserved at the genetic level.
Fig. 1. DR–essential genes are molecularly conserved and form a tight interaction network. 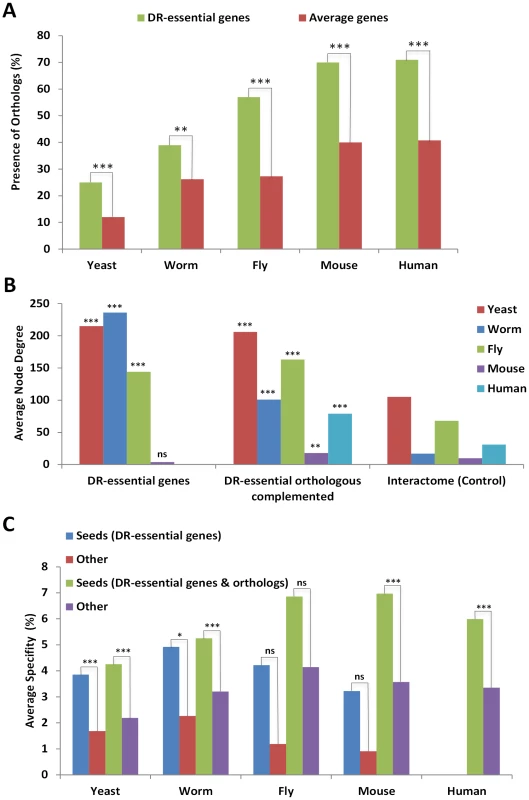
A, The proportion of genes (shown as percentages) with orthologs from multiple species in the specified organisms is in all cases higher (by 10–30%) for DR-essential genes than expected by chance. B, The average degree (number of interactions) for DR-essential genes and DR-essential gene orthologs (the ortholog-complemented set) is higher than the interactome-wide average which serves as control. C, The average percentage of specificity in interconnectivity (100× specific interactions with seed genes/all interactions) of DR-essential genes to each other is higher than for other genes in interactomes, and complementation with orthologs of DR-essential genes from other species further increases the specificity of interconnectivity. ns, P>0.05; *, P<0.05; **, P<0.01; ***, P<0.001. Interaction networks of DR-essential genes plus their orthologs were created by integrating molecular interaction information from various sources: physical, genetic and other interactions such as regulatory relationships were retrieved from public databases (see Materials and Methods). The human orthologs of DR-essential genes form an interaction network in which the total interactions appear to follow approximately a power law distribution (Figure S2). This is consistent with a scale-free network in which only a few nodes (often called “hubs”) are highly connected. In other words, a few hubs contribute highly to the DR network connectivity, as has been observed in other biological networks [11], [12].
DR-essential genes as well as their orthologs have a higher node degree than expected by chance when compared to the whole interactome (Figure 1B). Thus, DR-essential genes tend to be located in the center of the interactome rather than in the periphery. As anticipated, DR-essential genes interact with each other more than expected by chance (Figure 1C). Adding orthologs to the DR-essential gene seed lists (orthology complementation) increased the specificity (interaction with seed genes/total interactions) of the connections between seed genes and enabled the generation of interaction networks for species with few or no known DR-essential genes, such as mammals. DR-essential as well as aging-related genes are significantly enriched for signaling genes (Table S2). Nonetheless, DR-essential genes are even more evolutionarily conserved than aging-related genes (Figure S3) and have a higher average degree than signaling genes (Figure S4), indicating that these properties (evolutionary conservation and high degree) are not secondary effects expected for subsets of aging-related genes and signaling genes, respectively.
One issue, however, is that the finding of higher conservation and interconnectivity for DR-essential genes compared to other genes might reflect a selection bias of researchers, who tend to study genes that have orthologs in multiple species and/or those in pathways associated with DR. Because node degree, presence of orthologs and sequence conservation are thought to be related, we investigated the correlations among these variables for human genes. We found that log(degree) and number of species with orthologs are positively correlated (Pearson correlation coefficient, r = 0.36; p<10−200), which implies that the higher the number of connections (i.e. interactions) of a human gene the higher the number of species which have orthologs of that gene. Similarly, log(degree) was weakly negatively correlated with log(dN/dS) (r = −0.08; p<10−11), indicating that genes with more connections have slightly lower dN/dS ratios and hence more conserved sequences. Finally, the log(ortholog number) was also found to be negatively correlated with log(dN/dS) (r = −0.24; p<10−100), suggesting that genes which have orthologs across many species also tend to have lower dN/dS ratios, as expected. An interpretation of these results is that genes with high degree (more connectivity) tend to also have high numbers of orthologs and low dN/dS ratios. This makes sense, since important hubs in the network have more constraints limiting their evolutionary divergence. Next, we performed a multiple linear regression analysis (with log-transformed data) to derive equations relating each of these three variables as a function of the other two. We used the equations to determine whether DR-essential gene orthologs have higher or lower values than predicted from the other two variables. The results reveal that DR-essential gene orthologs have an average degree much higher than expected (73 vs. 12), a slightly higher number of species with orthologs (10 vs. 9) and dN/dS ratios higher than the expected value (0.08 vs. 0.04). Thus, a high node degree appears to be the most critical feature of DR-essential gene orthologs.
To ascertain which processes and functions are common to the DR gene network, DR-essential genes plus their significant interactors (p<0.05 according to specificity of interaction with DR-essential genes, as described in the Materials and Methods) for each species were subjected to functional enrichment analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery; Table S3A). Common to all DR-essential gene interaction networks in yeast, worm and fly were the terms aging (p<10−27) and mitochondrion (p<10−10). Terms common to ortholog-complemented networks in yeast, worms, flies, mice and humans were categories related primarily to phosphorylation signaling, among others (Table S3B). Kinase signaling and protein phosphorylation are also dramatically suppressed in very long-lived PI3K-null mutant worms [19], [20]. We then asked which genes among the significant interaction partners were common to multiple organisms. Although no homologous group was significant at False Discovery Rate (FDR)<0.05 (which appears to be too strict for comparisons across multiple species), by the more relaxed criterion of having a binomial p-value<0.05, four homologous groups were nominally significant: CAB39, the genes encoding 14-3-3 proteins, sirtuins, and AHCY encoding S-adenosylhomocysteinase.
Guilt-by-Association Discovers Novel Vacuolar DR–Essential Gene Functions
To predict novel DR-essential candidate genes we used a guilt-by-association strategy [21]. Essentially, the logic behind this approach is that a gene with more interactions than expected by chance with genes associated with a given process (i.e. DR-mediated lifespan extension) is likely to also play a role in that process (see Materials and Methods). The candidate genes with highest significance in their connectivity to DR-essential genes were YGR272C (EFG1; Exit From G1) in Saccharomyces cerevisiae, B0280.10 (pot-1; Protection of Telomeres 1 (Pot1) homolog) in Caenorhabditis elegans, and Akt1 (serine-threonine kinase involved in insulin-like signaling) in Drosophila melanogaster. After orthologous complementation, among the highly significant candidate genes were, for example, PAI3 (essential Proteinase A inhibitor) in S. cerevisiae, hcf-1 (human host cell factor (HCF-1) homolog) in C. elegans, CG7333 (an organic cation transmembrane transporter) in D. melanogaster, Raf1 (MAP kinase kinase kinase) in Mus musculus, and ACD (involved in telomere maintenance and meiosis) in humans. In yeast, worm, and fruit fly, respectively, 372, 1317, and 202 genes were significant at a p-value<0.05. After orthologous complementation there were 264 genes significant in yeast, 228 in worm, 743 in fruit fly, 2576 in mouse, and 996 in humans. The top candidates for each species are given in Table S4.
In order to assess the sensitivity and specificity of this guilt-by-association concept, we applied a leave-one-out test [22]. By omitting one of the DR-essential genes from the seed list and checking whether the removed gene was among the significant candidate genes (i.e. predicted correctly), we obtained a measure for sensitivity of the guilt-by-association method in the form of the percentage of genes which were recovered (i.e. rediscovered). This method achieved a recovery rate of 40% in yeast, 12% in worm and 50% in fruit fly. The lower recovery rate for worms compared to the other species (and the higher p-values) is possibly because C. elegans had the lowest number of known interactions at the time of analysis.
To test the new candidate genes, and thus to validate the usefulness of GenDR and DR network analyses as tools for aging research, we experimentally assessed the role in DR of some of our yeast candidates. Vacuole-related terms were among the most significant discrete clusters of terms in the DR-essential network of yeast (Enrichment Score: 5.2; Benjamini p-value, pB<2·10−5). Our guilt-by-association method predicted nine yeast genes whose protein products are on the vacuolar membrane and are non-essential for viability. Therefore, we decided to focus on the mutants of these genes, and measured replicative lifespan and vacuolar morphology of these nine mutants on AL and DR (Table 2 and Figure 2). Eight of the nine mutants we analyzed exhibited an impaired lifespan extension by DR (i.e. are DR-essential genes). Five of the nine mutations altered the extent of DR-induced vacuole fragmentation. While the vacuolar morphology of wild-type and rcr2Δ are indistinguishable in AL, their morphologies in DR media are different; wild-type had some cells with a few vacuolar vesicles (equal or less than 5/cell) but rcr2Δ cells had 6 or more vacuolar vesicles/cell (Figure 2). Similar to mutants (erg6Δ, nyv1Δ, etc.) with highly fragmented vacuoles under DR [23], rcr2Δ had a shortened lifespan on DR (Figure 2). Deletion of OPT2 or FRE6 showed results similar to rcr2Δ and all had an extended lifespan on AL (Table 2).
Fig. 2. DR–essential vacuole-associated genes are required for DR lifespan extension and normal vacuolar morphology. 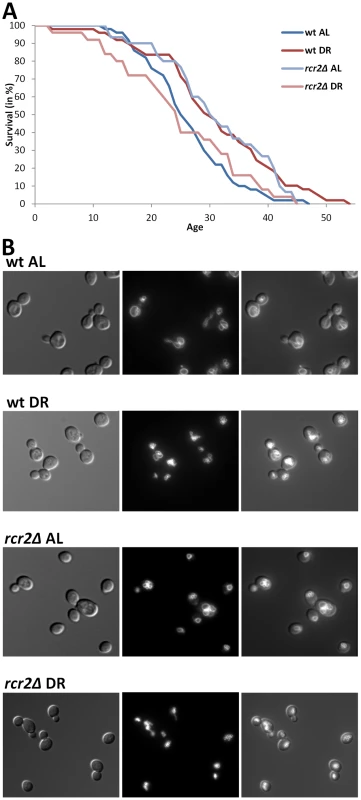
A, Survival curves of wild-type and rcr2Δ mutant strains measured on AL/YEPD and DR media. The units of age (X axis) are generations. Mean and maximum lifespans as well as sample sizes are listed in Table 2. B, Vacuolar morphology on AL and DR media of wild-type and rcr2Δ. Left panel: DIC images showing the cells; middle panel: TRITC-fluorescent images showing the vacuoles; right panel: overlay of cell and vacuole images. Tab. 2. Lifespan and vacuolar changes of novel DR–essential genes. 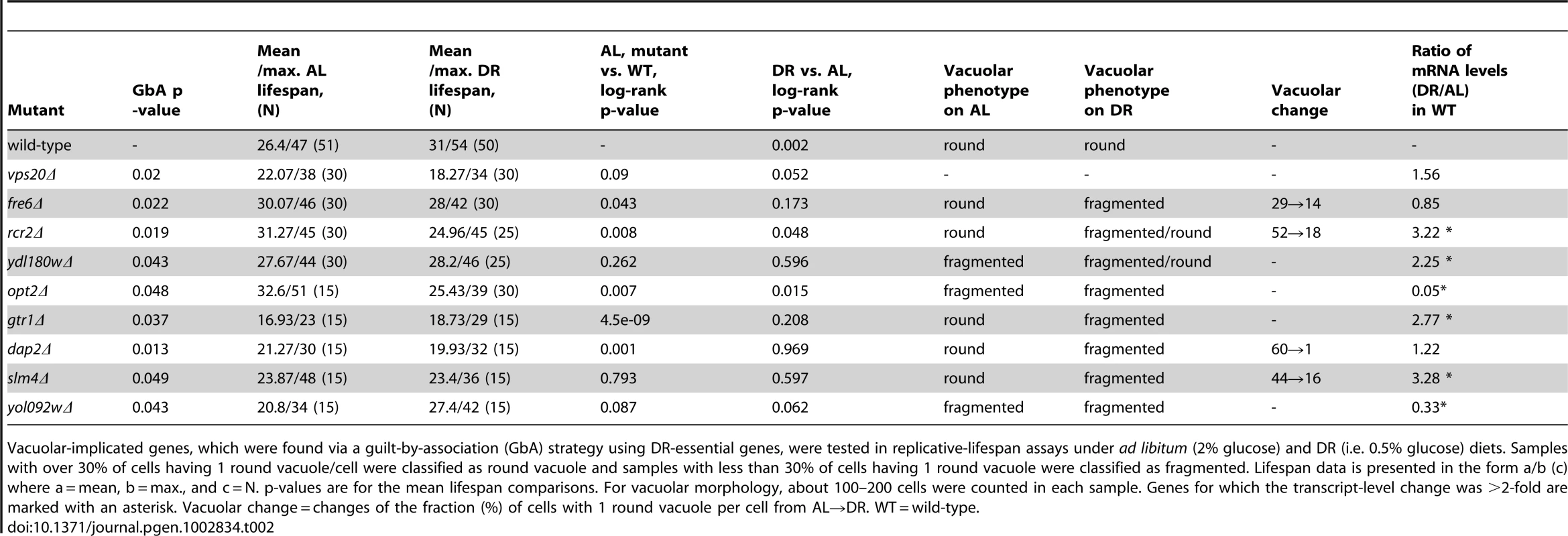
Vacuolar-implicated genes, which were found via a guilt-by-association (GbA) strategy using DR-essential genes, were tested in replicative-lifespan assays under ad libitum (2% glucose) and DR (i.e. 0.5% glucose) diets. Samples with over 30% of cells having 1 round vacuole/cell were classified as round vacuole and samples with less than 30% of cells having 1 round vacuole were classified as fragmented. Lifespan data is presented in the form a/b (c) where a = mean, b = max., and c = N. p-values are for the mean lifespan comparisons. For vacuolar morphology, about 100–200 cells were counted in each sample. Genes for which the transcript-level change was >2-fold are marked with an asterisk. Vacuolar change = changes of the fraction (%) of cells with 1 round vacuole per cell from AL→DR. WT = wild-type. Differential Expression of DR–Essential Genes upon DR
For the purpose of gaining insights into which processes on the global scale are affected by DR, we generated microarray data in yeast under DR (see Materials and Methods) and those genes more than two-fold differentially expressed were examined using DAVID by retrieving terms and clusters with FDR<5%. DR up-regulated genes were associated with heat response, mitochondria, peroxisome, transcription, mRNA processing, zinc binding, carbohydrate metabolism, sporulation, vacuole, and mitochondrial ribosomes, while DR down-regulated genes were related to ribosome/translation, nitrogen, sterol and one-carbon-source metabolism as well as DNA-replication (Table S5).
This comparison of mRNA levels between yeast cells cultured in AL and DR media, as ascertained on microarrays, also furnished further support for the involvement of the predicted vacuolar DR-essential genes. Of the 9 predicted vacuolar DR-essential genes, 6 were differentially regulated by more than two-fold either up or down (Table 2). Strikingly, OPT2 which had the strongest effect on lifespan when deleted (around 30% mean and maximum lifespan extension), was also among the genes most strongly (20-fold) down-regulated by DR ([24] and our DNA-microarray data; see Table 2).
As changes in expression of DR-essential genes via genetic manipulations mimic and/or abolish the lifespan extension conferred by DR, one interpretation is that DR-essential genes normally mediate DR effects by changing their expression or activity level in response to DR. As an approximation to investigate this, we employed large-scale microarray-expression profiles of yeast, worm [25] and fly [26], [27]. Numerous DR-essential genes change in their activity either at the transcriptional or translational level in response to DR (Tables S6, S7, S8, S9, S10). Of note among genes differentially expressed during DR, only two DR-essential homologous groups are shared in common by yeast, worm and fly: homologs of a fatty-acid elongase, and S-adenosylmethionine synthetase activity (Table S11). Experimental data support the importance of these two categories for which GenDR and DR-induced differential expression agree: (1.) worms with RNAi-suppressed fatty-acid elongase activity have increased lifespan, and long-lived C. elegans mutants tend to have shorter fatty-acid chains in proportion to their longevities [28]; and (2.) knockdown of the nematode SAM synthetase gene, sams-1, extends adult lifespan [29], [30], and also in yeast SAM1 deletion extends lifespan [31]. However, DR-essential genes in yeast are no more likely to be differentially expressed than expected by chance (30 of 70 DR-essential genes among the DR-differentially expressed genes; hypergeometric p-value = 0.6) at a threshold of two-fold change.
Transcription Factors Governing the DR Signatures in Yeast
To unravel the relationship between DR-essential genes and differential expression upon DR we utilized a gene regulatory network (transcription factor – target gene interactions [32], [33]; a subcategory of our integrated network) in yeast to identify candidate transcription factors responsible for the differential expression upon DR. Our criteria were factors which, firstly, regulate DR-differentially expressed genes with a high specificity, and secondly, interact with DR-essential genes either at the physical or genetic level. Transcription factors which had very high specificity (i.e. specific/total interactions, in %) for controlling DR-induced genes included not only factors involved in nutrient-sensing (e.g. Msn2/4 (26%), Gis1 (53%), Mig1 (40%)), and stress response (e.g., Hsf1 (34%)), both of which are known to be important for DR-lifespan extension, but also meiotic transcription factors such as Ime1 (41%), Ume1 (75%), Ume6 (38%) and Ndt80 (26%) (Table S12).
Yeast sequences upstream (within 500 bp) of the promoter regions of genes displaying >2-fold differential expression under DR were examined for motif enrichment (by hypergeometric test) and indeed, upregulated genes are significantly enriched for a number of upstream regulatory elements: STRE (stress-response element), PDS (post-diauxic shift) element, URS1 (upstream regulatory sequence 1), and MSE (middle-sporulation element) including the motifs of Ume6, Ime1 and Ndt80 (Tables S13, S14, S15). Interestingly, URS1 as well as motifs of Ume6, Sum1, Ndt80, and Ime1 are even enriched in the upstream sequences of the full set of DR-essential genes (Table S16). Also, in another signature of DR [24], meiotic transcription factor binding motifs were significantly enriched in the 500 bp upstream regions of DR-induced genes. For instance, YGNCACAAAW (NDT80) was present upstream of 14 genes, of which 11 are DR-induced genes (p-value = 0.0025, q-value = 0.029). NDT80 transcript levels increase as a function of the strength of DR [34] and at 0.5% glucose concentration in our own DNA-microarray data, were found to be 2.6-fold elevated.
DR–Essential Genes Are Triggered by Lifespan-Extending Spermidine
Comparing different longevity interventions for their commonly regulated processes may allow us to pinpoint the gene expression changes most essential for lifespan extension. Similar to DR, treatment with the polyamine spermidine extends lifespan in multiple model organisms such as yeast, worm, fruit fly, as well as in human cells in vitro [35].
We found that, in yeast [36], spermidine treatment causes differential expression of DR-essential genes more often than expected by chance: at a threshold of two-fold change, 15 of the 70 DR-essential genes were among the 727 spermidine-differentially expressed genes (hypergeometric p-value = 0.003). This effect appears to be very specific as there was no enrichment observed among genes induced by spermine, another polyamine, which has not to our knowledge been shown to affect lifespan. Moreover, DR and spermidine signatures regulate an overlapping set of genes (far more likely than expected by chance): at two-fold change, 323 of 727 spermidine-differentially expressed genes were among the 2560 DR-differentially expressed genes (hypergeometric p-value = 4·10−4), whereas genes regulated by DR and spermine did not overlap more than expected by chance at any cut-off (e.g. hypergeometric p-value = 0.69 for two-fold changes).
Comparison of the transcriptional signatures of DR (derived from our own DNA-microarray data) and spermidine treatment by looking for common functional enrichment terms among genes differentially expressed with DR and spermidine, indicates that both DR - and spermidine-upregulated genes are highly enriched in sexual sporulation (pB<10−3), peroxisome (pB = 10−3), mitochondrion (pB<0.02) and ubiquitin conjugation pathway (pB<10−3) and downregulated genes are enriched in sterol metabolic process (pB<0.05) as well as pentose transmembrane transporter activity (pB = 0.014). The set of common functional terms enriched by both spermidine and DR treatments (at a 1.5-fold-change threshold) revealed that both induce autophagy, heat response and sporulation (Tables 3 and 4). Spermidine treatment leads to both a significant induction and suppression of sporulation/meiosis genes (Table S17) and strikingly similar to DR, induces NDT80 expression.
Tab. 3. Terms associated with DR and spermidine upregulated genes. 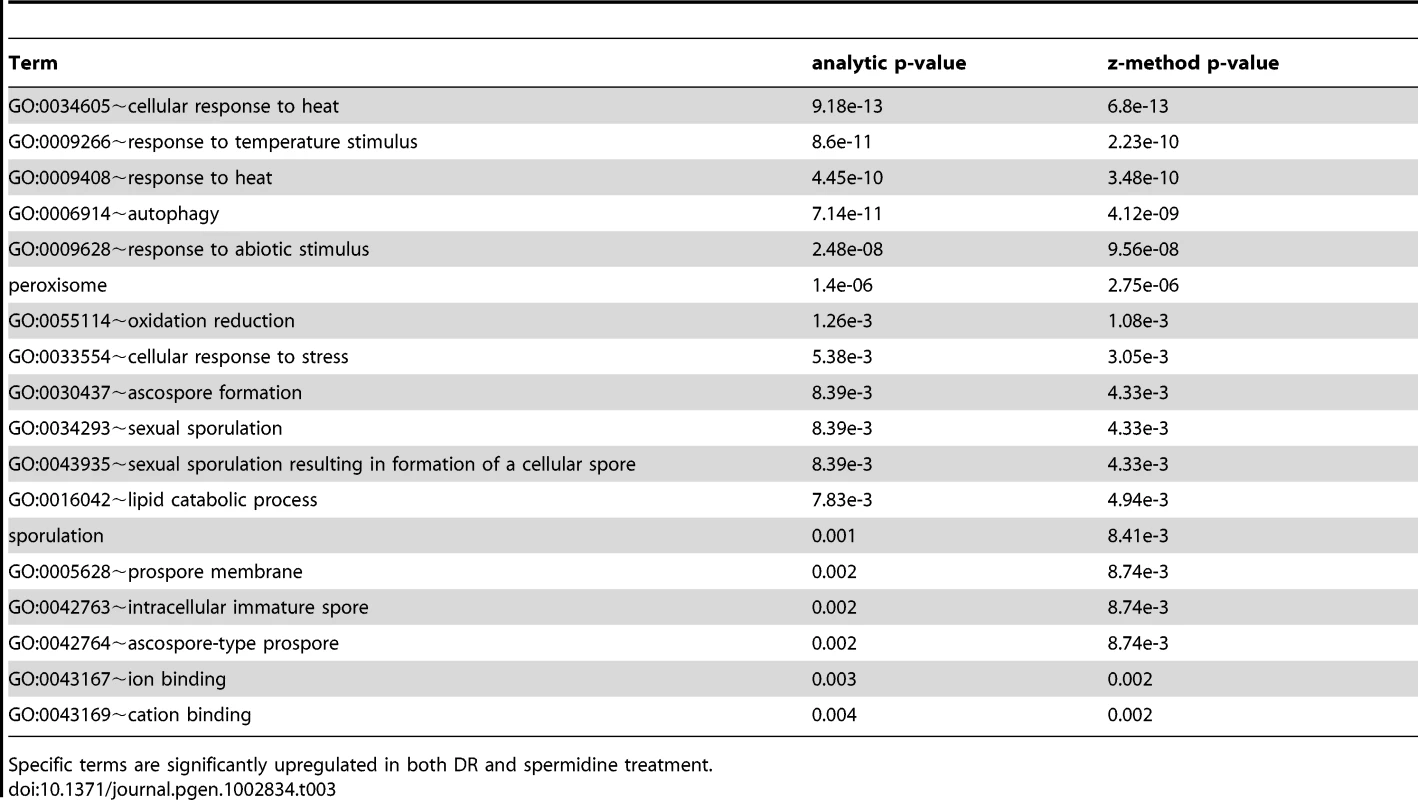
Specific terms are significantly upregulated in both DR and spermidine treatment. Tab. 4. Terms associated with DR and spermidine downregulated genes. 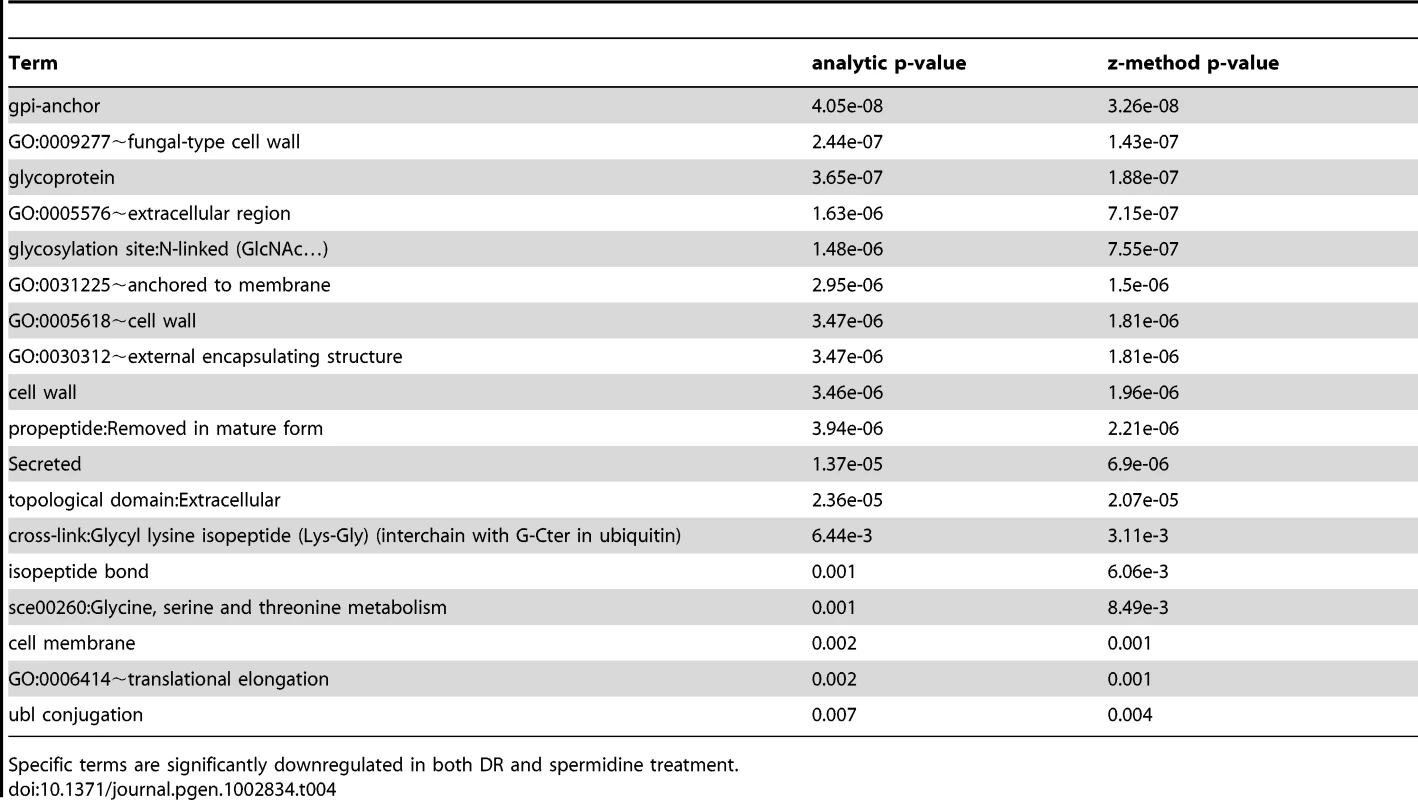
Specific terms are significantly downregulated in both DR and spermidine treatment. Comparing DR Interactomes
The validation of vacuole mutants and the responses of DR-essential genes to longevity manipulations support the biological relevance of predictions based on the DR-interactome. To further study conserved mechanisms of DR-induced life-extension, we next integrated and analyzed interactome and transcriptome data (Figure 3). The interactomes of yeast, worm and fly were integrated with their respective DR-triggered gene expression changes and condensed interaction networks were generated via the ExprEssence algorithm [37]. These networks were further condensed until the representative graph contained approximately 1,000 genes. Two graphs for each species were created, one containing only startups (induced interactions), while the other contained only shutdowns (suppressed interactions), thus partitioning the interactions of up - and down-regulated genes, respectively. Next, DAVID was used to retrieve the functionally enriched terms for each graph, retaining only the terms common to either all startups or all shutdown networks which surpass a pB<0.05. Subsequently, p-values were combined across all three species (Tables 5 and 6; see Materials and Methods). Suppressed interactions were enriched for terms associated with translation such as ribonucleoprotein, structural molecule activity and ribosome. This is consistent with previous observations that down-regulation of translation or ribosomes extends lifespan in diverse organisms from yeast to mammals [38]–[41]. Stimulated interactions mostly occurred between genes involved in modifying chromatin structure such as chromatin regulator, chromatin modification, and chromosome organization, genes associated with reproductive developmental/cellular processes as well as cell cycle and stress response. Strikingly, the majority of these cell-cycle-associated genes are involved in meiosis with p-values of <10−19, <10−46, and <10−5 for yeast, worm, and fly, respectively (Table S18).
Fig. 3. Comparing the influence of gene-expression changes upon DR, in multiple interactomes. 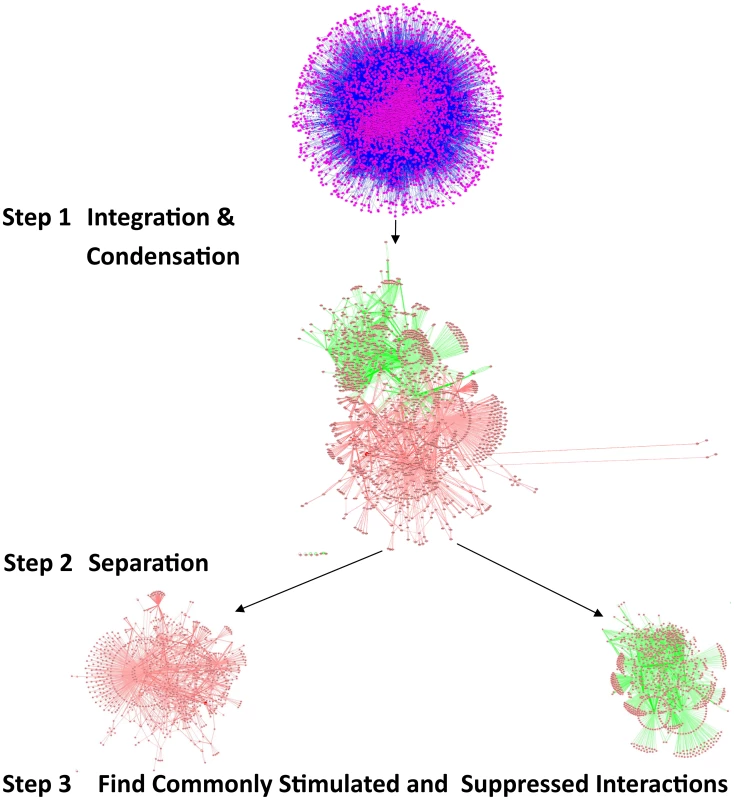
The interactomes of yeast, worm, and fly were integrated with gene expression information upon DR and condensed interaction networks generated via the ExprEssence algorithm (Step 1). Interaction networks restricted to either suppressed or stimulated interactions for each species were created (Step 2). The suppressed and stimulated networks were then separately compared for common significant functional enrichments (Step 3). Functional terms which were common to suppressed and induced interactions upon DR in yeast, worm, and fly are listed with their respective p-values (Tables 5 and 6). Tab. 5. Terms associated with DR-stimulated interactions in the comparative interactomic changes upon DR. 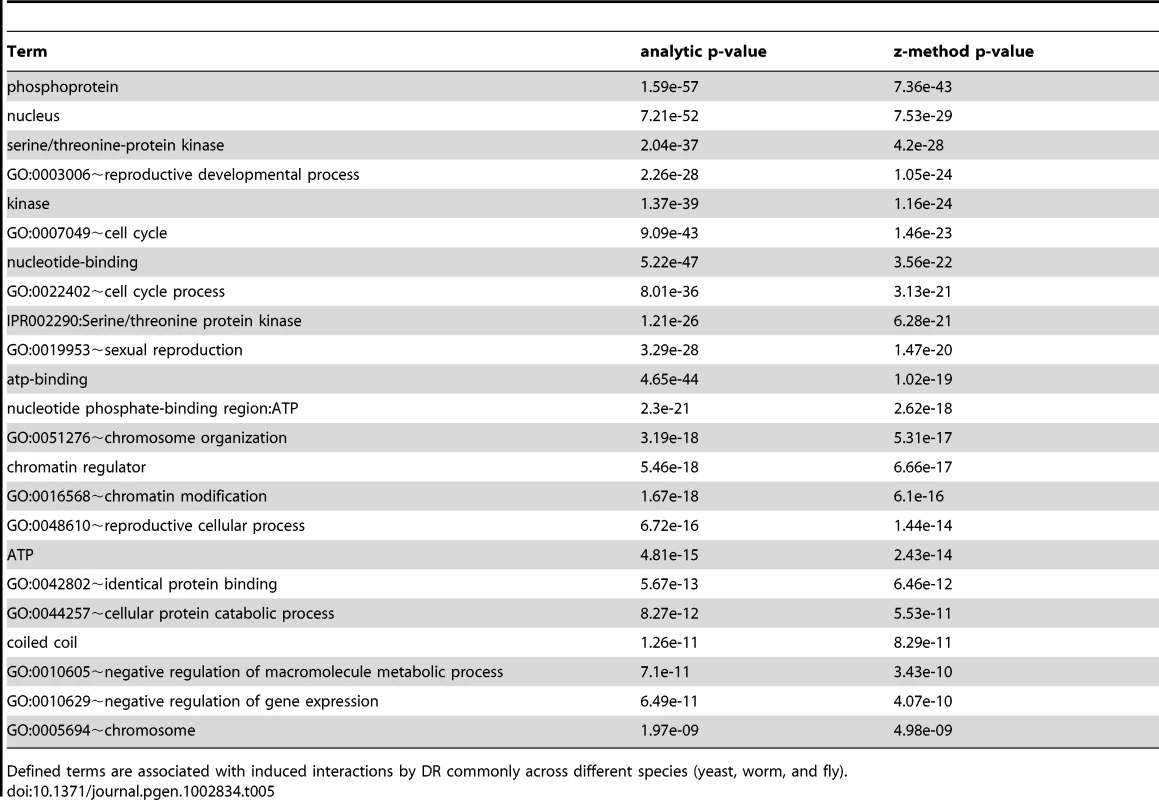
Defined terms are associated with induced interactions by DR commonly across different species (yeast, worm, and fly). Tab. 6. Terms associated to DR-suppressed interactions in the comparative interactomic changes upon DR. 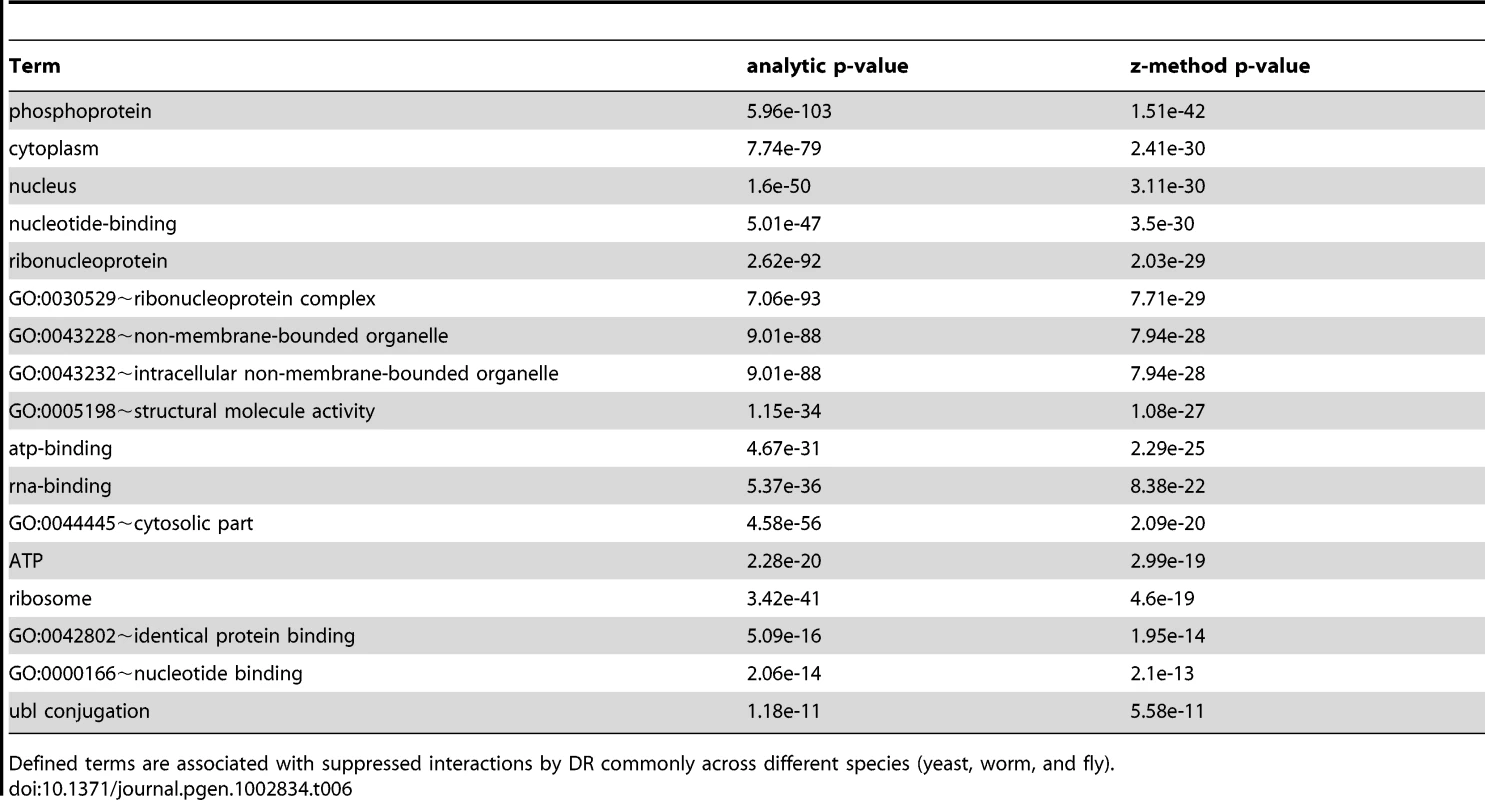
Defined terms are associated with suppressed interactions by DR commonly across different species (yeast, worm, and fly). In accordance with the functional enrichment analysis of DR-essential networks (Table S3), the comparative interactomics changes suggest that DR both stimulates and suppresses interactions associated with phosphoproteins and nucleus (Tables 5 and 6). To unravel the relationship between these two broad terms we identified all kinases and phosphoproteins in yeast known to be localized in the nucleus (using Gene Ontology annotation), that are either up - or down-regulated upon DR (Table S19). The induced and suppressed nuclear kinases (Table S20) are significantly enriched for meiosis (each pB<10−4; Table S21). While most of the induced nuclear kinases are positive regulators, the suppressed nuclear kinases are mainly negative regulators of meiosis, indicating that DR in yeast promotes the initiation of meiosis. Interestingly, yeast genes associated to meiosis, according to Gene Ontology (161 genes), had an overall average gene expression increase of 2.7 upon DR (higher than expected by chance; Mann Whitney U test p-value = 0.002) and together with all their interaction partners were on average 2-fold upregulated (Mann Whitney U-test p = 0.003). Induced nuclear phosphoproteins (i.e. substrates of kinases) are enriched for protein phosphatases (pB<10−23), ER-nuclear signaling (pB<10−4), MAPK signaling, Mn (pB<10−4), sexual reproduction/reproductive cellular process (pB<6·10−3/0.03) (Table S21), while suppressed nuclear phosphoproteins were only enriched for protein phosphatases (pB<10−10) (Table S22).
An interaction network of these kinases in yeast and their direct interaction partners restricted only to direct physical interactions in the nucleus was generated (Figure 4). IME2, which is the second most-upregulated nuclear kinase upon DR (Table S19), acts on the induced transcription factors IME1 and NDT80 as well as on the suppressed SUM1. In C. elegans, induced kinases were also enriched among other terms for female meiosis (pB<0.03; Table S23). In line with the enrichment of reproduction-related terms in the DR-essential network (Table S3) in Drosophila, induced kinases restricted to the nucleus were enriched for female gamete generation/sexual reproduction/regulation of cell cycle/multicellular organism reproduction (pB<5·10−3/0.02/0.02/0.02; Table S24). Finding upregulation of reproduction-related genes is surprising given the known suppressive effect of DR on reproduction [42].
Fig. 4. Nuclear phosphorylation network of yeast. 
DR–upregulated genes are shown in red and downregulated genes in green. Kinases are marked by a golden halo. Discussion
A Database and Analysis of Genes Essential for DR Life-Extending Effects
Understanding the genetic basis of DR is of great importance not only to help elucidate mechanisms of aging but also to understand how diet can influence aging, longevity, health, and age-related diseases, which may have major human therapeutic applications [1], [6]. DR-essential genes are no doubt crucial pieces of the puzzle to solve the mechanism of DR [1], [2], [17], but to date they have not been studied as a unified system. To address the need for a more systematic study of the mechanisms of DR-conferred lifespan extension, we created GenDR, the first database of DR-essential genes and indeed the first database of DR-related genes. GenDR provides manually-curated information on genetic mutations which interfere with the pro-longevity effect of DR as well as their respective homologous genes in the major model organisms and humans. Because different genes appear to be important in different regimens and there are conflicting findings regarding some of them, different DR regimens across multiple model organisms are covered, allowing researchers to focus on their preferred system. With the growing importance of network biology, functional genomics and systems biology to study aging and DR [6], GenDR will be a valuable tool for researchers to study the genetic and molecular mechanism of DR as evidenced by our validation of vacuole mutants (Figure 2) and gene regulatory inference (Table S16).
Our study of the molecular evolution of DR-essential genes (Figure 1) showed conservation at the molecular level reflecting the observation that DR extends lifespan in various evolutionarily distant organisms. Interestingly, DR-essential genes had lower dN/dS ratios than expected purely by chance, as well as lower dN and dS values. This could indicate that natural selection has constrained not only the amino acid sequences, but also the nucleotide sequences which may include regulatory elements. The molecular interaction data support the observation that DR-essential genes are conserved across vast evolutionary distances, as genes with relatively high degree, i.e. hubs, are more ancient and evolve more slowly than genes that encode non-hub proteins [43]–[45]. One caveat of this analysis is that these results may also reflect a bias in the selection of genes, as researchers tend to study genes which have orthologs and have been shown to affect DR in other organisms. Similarly, the result that DR-essential genes interact with each other more than expected by chance might also be affected by selection bias, as genes studied in the context of DR are often part of similar or closely interacting biological pathways (e.g. insulin/IGF1 signaling and TOR). Also, both DR-mechanism and aging-effect genes tend to be enriched for signal-transduction genes, especially those which are well known to be highly conserved [46].
Network Analyses Reveal Novel DR–Essential Genes and Candidate Mechanisms
Our network analyses successfully predicted novel DR-essential genes in yeast. This illustrates that it is possible to associate in silico genes that are of interest in a DR context, with other, little-known genes, and derive testable new hypotheses. Network analyses based on GenDR have therefore the capacity to implicate new genes related to lifespan extension by DR. In particular, we identified eight novel vacuole-related DR-essential yeast genes that impair DR-mediated longevity when deleted, of which three (OPT2, FRE6, and RCR2) extended lifespan in AL and prevented any further lifespan-extension by DR. Although the genome-wide frequency of DR-essential genes is unknown, finding 8/9 of such genes with 3/9 extending lifespan in AL is likely much more than expected by chance and illustrates the utility of our method. These genes had been previously implicated in drug detoxification (OPT2), transition metal ion homeostasis (FRE6) and endosomal-vacuolar trafficking of plasma membrane proteins (RCR2, VPS20, GTR1, SLM4) as well as a vacuolar membrane protein DAP2, plus a protein of unknown function (YDL180W). Comparing localizations of the proteins encoded by these genes with the known DR-essential genes in our database revealed endocytosis as one target of DR (Figure 5). Moreover, the different effects on lifespan of mutants missing these newly identified DR-essential genes (Table 2) prompted us to speculate on the function of each endocytic step in longevity.
Fig. 5. Localization of vacuole-related DR–essential proteins and lipids in yeast cells. 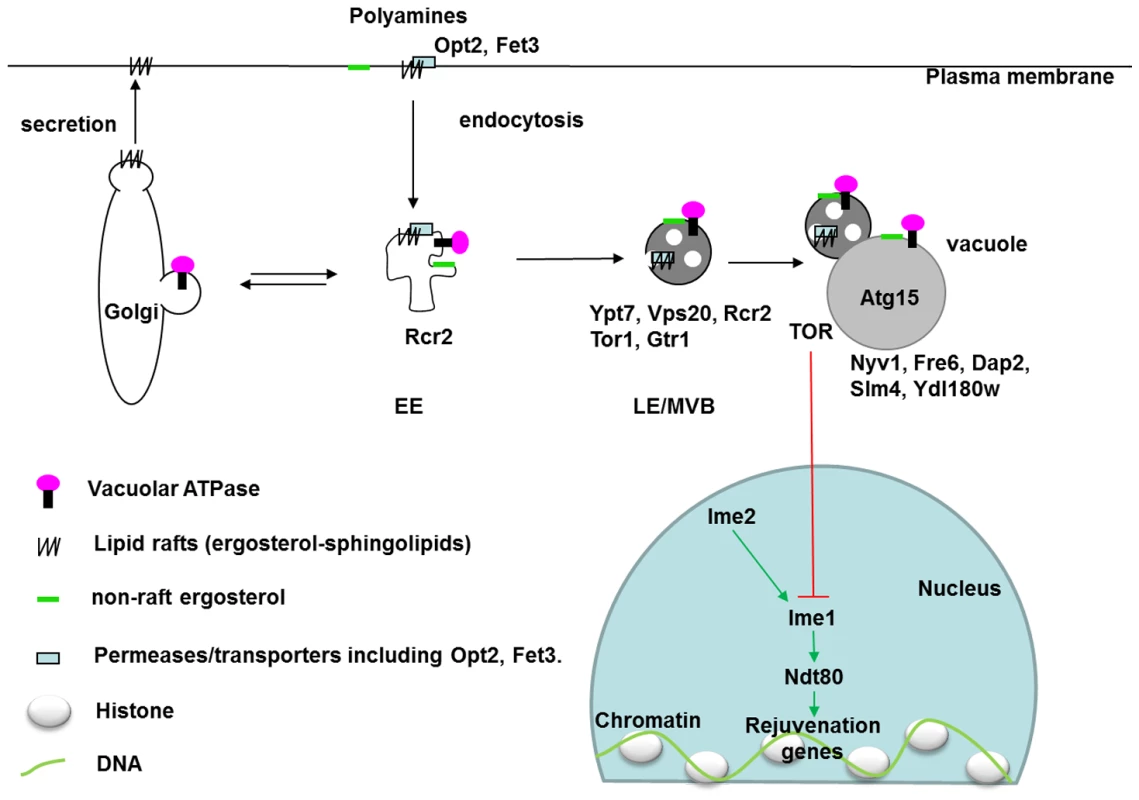
In response to environmental triggers such as DR, membrane transporters (Opt2 and others) on the plasma membrane are endocytosed through lipid rafts (ergosterol-sphingolipids) after ubiquitinylation. On early endosomes, proteins and lipids are sorted to late endosomes, Golgi, or other organelles. In late endosomes, different endosomal sorting complexes required for transport (ESCRT) push the target proteins with their membranes into the lumen of late-endosome, or multivesicular bodies (MVB). Matured MVB can fuse with vacuoles, within which proteases and lipases (Atg15) degrade target proteins and lipids. In mammalian cells, maturation of late endosomes is essential for activation of the master regulator for growth, the mammalian target of rapamycin 1 (mTORC1). In yeast cells, TORC1 suppresses meiotic transcription factors which are normally only active during gametogenesis [110]. The inhibitory effects of DR on TORC1 thus activate these rejuvenation genes. EE: early endosome. LE: late endosome. MVB: multivesicular body. Black arrows indicate movement of vesicles. DR-essential proteins are labelled in their localized organelles. Localization data were extracted from the SGD database. Ypt7 localization on late endosomes was reported by [111]. Green arrows indicate activation and a red line indicates an inhibitory effect. Cells take up nutrients through plasma-membrane transporters including oligopeptidase transporter Opt2 and ferrous transporter Fet3. Secretion and endocytosis determine the fate of these transporters depending on environmental cues (Figure 5). Both secretion and endocytosis depend on the lipid composition of membranes. Blocking ergosterol synthesis (erg6Δ) halts the degradation of Tat2, the high-affinity transporter of tryptophan, and traps it in late endosomes (or multivesicular bodies) [47]. Nutrient transporters destined for degradation in vacuoles are ubiquitinated by Rsp5 (a DR–essential ortholog of C. elegans WWP-1 [48]) on the plasma membrane or endosomes [49]. Ubiquitinated cargoes are sorted to late endosomes, where endosomal sorting complexes required for transport (ESCRT) work sequentially to encapsulate the ubiquitinated transporters within the endosomal lumen. Vps20 is a component of ESCRT III, which seals the membranes [50]. Our identification of VPS20 as a DR-essential gene may suggest the involvement of late endosomes in longevity. In support of this possibility, a screen for mutants that shorten yeast chronological lifespan also revealed a critical role of the late endosome [51], [52].
For several of the mutants tested the lifespan was shorter than wild-type, which indicates that these genes are required for normal longevity and especially for the response to DR (perhaps due to their involvement in endocytosis). There are also four mutants (opt2Δ, rcr2Δ, vps20Δ, fre6Δ) with a shorter lifespan on DR than on AL (Table 2), comparable to the “inverse DR” effect reported for erg6Δ, ypt7Δ, and nyv1Δ mutants [23]. These mutants are all likely to be blocked in the maturation of late endosomes. Deletion of Rcr2 or blockage of ergosterol synthesis (erg6Δ) led to accumulation of endocytic vesicles (or a special subset of apparent vacuoles as visualized by the dye FM4-64) that are unable to fuse with other vacuoles, and to the apparent formation of highly fragmented vacuoles (Figure 2B; also see Figure 4B of [23]). The fact that deletion of the plasma membrane transporter OPT2 also results in fragmented vacuoles [53] may indicate improper fusion between late endosome and vacuoles. The maturation of late endosomes also depends on the proton gradient across the membrane, which is maintained by a membrane H+-ATPase. FRE6 deletion could impair metal-ion efflux and consequently decrease cytosolic iron and copper levels [54]. Copper ions control the activity of membrane H+-ATPase activity [55]. As such, endocytosis appears to be essential for normal lifespan under AL and especially for the lifespan-extension response under DR conditions. Endocytosis may for instance remove damaged plasma membrane proteins, and in higher organisms even intercellular debris.
In addition to implicating the plasma-membrane-endosome-vacuole pathway as one target of DR, the longevity of three mutants (opt2Δ, fre6Δ, and rcr2Δ) under AL (Table 2) reveals a putative novel controlling mechanism of the target of rapamycin complex 1 (TORC1). OPT2 and FRE6 exhibit negative genetic interactions with TOR1 [56] and TCO89 [57], respectively, both of which encode components of TORC1. A negative genetic interaction usually suggests a cooperative relationship [57]. Rcr2 interacts with Ypt7 in a yeast 2-hybrid assay [58]. While it is not known to be true in yeast, the mammalian ortholog of Ypt7, Rab7, is essential for mammalian TORC1 activation [59]. It is thus tantalizing to speculate that TORC1 may be partially inactivated in opt2Δ, fre6Δ, or rcr2Δ strains. Considering that part of TORC1 is on the vacuolar and endosomal membranes [60], it is not surprising to observe that some vacuolar membrane mutants affect the activity of TORC1. Another beneficial effect of opt2Δ in longevity might be the uptake of life-extending polyamines (see also [61]).
Transcriptional Changes upon DR and Their Regulators
There was only a slight enrichment of DR-essential genes among those differentially expressed during DR. This is not unexpected, since DR-essential genes are likely to lie upstream in the DR-longevity pathway, whereas most genes expressed differentially upon DR would tend to lie downstream in an effector cascade. It is also possible that many DR-essential genes mediate their effects on lifespan via transient expression, changes in protein levels, or in biochemical activities that are not reflected in analyses of mRNA levels. The fact that we did not differentiate between the various DR regimens would also dilute the enrichment of essential genes shared by multiple pathways. Nonetheless, numerous DR-essential genes show strong changes at the transcriptional and/or translational level, which may correspond to a respective change in the activity level, or might represent a compensatory response to changes in regulation at other levels. For instance, the only validated DR-essential gene in mammals Ghr, extends lifespan if deleted and is also found to be downregulated upon DR (Table S10) [62]. Among the DR-essential homologs commonly differentially expressed in all examined species were fatty acid elongases, which extend lifespan when downregulated in C. elegans [28]. In addition, DR regulates S-adenosylmethionine synthetase activity at the transcriptional/translational level in multiple species; its product, S-adenosylmethionine (SAM), is the methyl donor for most methylation reactions. A common significant interactor of DR-essential genes was S-adenosylhomocysteinase catalyzing the reversible hydrolysis of S-adenosylhomocysteine (SAH), which in turn is a competitive inhibitor of methyltransferases. SAM and SAH levels, acting via DR-essential NNT1/NNMT encoding nicotinamide-N-methyltransferase [63], would impact NAD+ metabolism and consequently NAD+-dependent protein deacetylases (sirtuins) which were also common interactors of DR-essential genes. SAM and SAH levels might also exert pervasive effects via the control of DR-essential ergosterol biosynthesis in yeast, sterol modifications in C. elegans (influencing dauer formation), DNA methylation in mammals, as well as histone methylation and polyamine synthesis in all species.
It is clear that diet alters epigenetic marks, resulting in an altered chromatin state that affects gene expression [64], [65]. Our study provides evidence pointing to a fundamental role of chromatin in DR. Chromatin organization was among the most enriched categories of DR-essential orthologous networks in mammals, and most strikingly several terms related to chromatin were enriched among DR-stimulated interactions in the interactomes in multiple species (Table 5). Modified chromatin structure has been proposed as an underlying mediator of aging from yeast to humans [66], and is known to be influenced by diet [67]. Taken together, our evidence implies that an altered chromatin state in DR is a major component of its lifespan-extending mechanism, conserved across diverse species.
Clearly DR works via a conserved mechanism, but how does it interfere with the aging process? A phenomenon that is at least equally ancient and conserved is the intergenerational/meiotic reversal of all age-related changes that had accumulated, and the restoration of lifespan for progeny [68]. Yeast rejuvenation occurs during sporulation (the equivalent of the mammalian gametogenesis) [68], when transient activation of Ime1 or Ndt80 rejuvenates cells and resets their age to “zero” [68]. We found that both Ime1 (1.4-fold) and Ndt80 (2.7-fold) are induced upon DR, leading us to speculate that DR may act on aging via a mild, continuous form of “rejuvenation”. This would explain why switching the feeding regimen leads to a change in the state, rather than the rate, of aging – i.e. it lowers the mortality level almost immediately [69].
It was proposed that polyamine biosynthesis may be essential for meiosis and sporulation [70]. Spermidine and/or spermine are, in fact, required for sporulation [71] and transition into meiosis [72]. Polyamine concentration declines with age in yeast and human cells. Supplementation with spermidine increased lifespan in yeast, worms, flies, and human cells, and a polyamine-rich diet (as well as probiotics that induce polyamine production) decreased mortality in mice [73], [74]. Spermidine treatment results in hypoacetylation of H3K9, H3K14, and H3K18, probably via inactivation of histone acetyltransferases. Inactivation of the responsible histone acetyltransferases extended replicative lifespan and suppressed lifespan extension by spermidine. While spermidine treatment leads to global histone hypoacetylation, histones on specific promoters such as those of autophagy-related genes were actually hyperacetylated and induced in expression [35]. Increasing acetylation levels of H4K5 and H4K12 via deletion of DR-essential protein deacetylase RPD3 also increased lifespan in yeast and Drosophila. Glucose and nitrogen regulate the switch from histone deacetylation to acetylation of early meiosis genes [75]. It is interesting to note that spermidine could only effectively extend replicative lifespan in yeast when treatment was initiated at old ages [35], indicating that it works via a single round of light rejuvenation. Strikingly, genes differentially expressed by DR and spermidine treatment have certain chromatin marks in common, including histone acetylations, while on the global scale they lead to opposing effects (data not shown). As such, it appears that spermidine might be a downstream component of the DR-mediated lifespan extension and chromatin remodeling induced by DR awakens meiotic rejuvenation programs. Intriguingly, induction of interactions between genes annotated with reproduction-related terms was the key shared feature of the comparison of DR-interactomes from yeast, worms and flies.
Concluding Remarks
Given the multitude of changes induced by DR, the study of DR-essential genes and their networks is an important new tool to distinguish those genes and processes that are the “drivers” of the life-extending effects of DR from those that are mere “passengers”. Our GenDR database will therefore be valuable for numerous researchers and we demonstrate that DR is suited for a network-guided approach. As anticipated, DR-essential genes are conserved at the molecular level and interact with each other more than expected by chance. Crucially, our work demonstrates how gene network analyses of DR allow the identification of novel candidates that can be tested experimentally. Our results also indicate important roles for genes involved in endosomal/vacuolar trafficking in DR-lifespan extension and provide new intriguing candidates for further studies in yeast such as OPT2, FRE6, and RCR2. Genome-wide expression data provide further clues of which genes and processes are regulated by DR, and their systematic integration in interaction networks was initiated in this study. Moreover, gene-regulatory relationships and sequence-based inference both point to an important role of meiotic transcription factors in DR. Finally, by comparing the effects of DR on the interactomes of multiple species, we discovered that DR suppresses translation while stimulating chromatin reorganization and a defined meiosis-related rejuvenation process, across phyla separated by a billion years of evolution. Many questions still remain. What is the nature of the rejuvenation processes utilized by DR to extend lifespan? What is the exact role of polyamines in DR-induced rejuvenation? Are intracellular polyamine levels increased upon DR? And what factors correspond to the mammalian counterpart(s) of NDT80? We hope that our efforts in establishing GenDR will help to unravel the secrets of DR-conferred lifespan extension.
Materials and Methods
Database Creation
A list of genes, termed DR-essential genes, which if genetically manipulated (knockout by deletion, or transposition, knockdown by RNA interference, or overexpression of transgenes) interfere with the ability of DR to extend lifespan in model organisms (budding and fission yeast, nematode, fruit fly, and mouse) was compiled from the literature. The focus on genes from genetic manipulations experiments means that the selection procedure for selecting genes related to DR will be more objective and unbiased. Genes were included if they interfere with at least one kind of DR regimen which also includes by definition a shift in the response to food concentration at which lifespan is extended (e.g. chico gene). A database (GenDR) was implemented in the relational database management system MySQL and a webpage interface was designed (http://genomics.senescence.info/diet/), as for our other aging-related databases [76]. Each entry in the database contains manually curated comments about literature-based evidence and the reason for inferring an association with DR. If there are conflicting reports for a given gene, our policy is to still include the gene in our database but mention all the conflicting reports and then let visitors make their own mind on how to interpret them. This neutral stance policy is similar to the one we already employ for our GenAge database of aging-related genes. Literature citations and links to PubMed are also given. Further, conserved gene expression changes upon DR in mammals are included, although these have been described in another work [62].
Molecular Evolution
Assembling orthologs
Homologs of genes in the GenDR database were retrieved from the NCBI HomoloGene Database, Ensembl/Biomart, OrthoMCL [77], and InParanoid [78] by merging. Genes are either homologous or not, it is a binary assignment, therefore the union was taken by merging them. For the molecular evolution part specifically only Biomart (Ensembl Genes 57) was used to retrieve tables of orthologs (homologs between two different species) for Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Rattus norvegicus, Macca mulatta and Homo sapiens. Information about Saccharomyces pombe is not available in Ensembl and this species was therefore excluded from the following analyses. For calculating the presence of orthologs, Ensembl protein identifiers were employed to filter for only protein-coding genes. For a given set of genes, here DR-essential genes, it was counted how many of these genes in one species have also at least one protein-coding homologous gene in all the other species under study. This was also done for all protein-coding genes in each genome and given these two values the percentage of presence of homologs were estimated. All pairwise comparisons for each seed species were averaged and the Poisson p-values calculated.
Calculating average dN, dS, and dN/dS ratios
Non-synonymous (dN) and synonymous (dS) nucleotide substitution rates [10], [79] were retrieved from the Ensembl Compara built 65 for the orthologous pairs between a rodent (M. musculus), a non-human primate (M. mulatta) and humans (H. sapiens). Orthologs relationships were restricted to the top-hits of InParanoid. In this sense, we corrected for the number of paralogs which might be a source of bias as big gene families tend to have higher dN/dS ratios. The average values for dN and dS as well as dN/dS ratios for the orthologous relationships of DR-essential genes were compared against those without orthologs of DR-essential genes. In order to avoid biases due to overrepresentation of ancient genes, only genes which had also orthologs in lower model organisms (S. cerevisiae, C. elegans, or D. melanogaster) were selected. Thus, the comparison was between ancient genes which are presumably essential for DR versus ancient genes with no known essential role in DR. The corresponding p-values for differences in dN, dS, and dN/dS were calculated with a Mann Whitney U test.
Molecular Interaction Information
Integration
Interactions datasets were retrieved from IntAct (Download 23.01.2011; http://www.ebi.ac.uk/intact/) [80], DIP (Update 10.10.2010) [81], MINT (Version 2010-12-15 updated on 21/12/2010 11 : 07 : 00) [82], BIND (http://baderlab.org/BINDTranslation) [83], BioGRID (Release 3.1.72) [84], MPACT [85], DroID (Version 2010_08), Reactome [86], HPRD (Version 9) [87], PDZBase [88], CORUM [89], iRefIndex (05182010) [90], PhosphoSitePlus (Fri Dec 10 13 : 43 : 52 EST 2010) [91], PhosphoGRID [92], I2D (Version 1.8) [93], InteroPORC [94], InterologFinder [95], MiMI [96], [97], PINA (Update March 4, 2010) [98], YeastNet [99], WormNet [100], MouseNet [101], and TF-Atlas [102] preferentially as PSI-MAT or simple tab-delimited flat file format. Interactions were aggregated by using a unified schema consisting of synonyms for each gene/protein, their taxonomy identifiers, interaction type and detection system, PubMed identifiers for references, as well as source databases. For simplification, interactions between molecules other than genes, transcripts, or proteins were omitted and only interactions between genes/transcripts/proteins of the same species were considered.
Mapping and merging
Symbols, names, aliases and various identifiers of genes, transcripts and proteins were in the first instance mapped to unique Entrez Gene IDs, Ensembl Gene IDs, UniProt IDs and species-specific IDs (from WormBase, MGI, etc.). Ensembl Gene, UniProt, or species-specific database IDs were if possible converted into Entrez Gene IDs. In a second iteration collected synonyms from Ensembl, UniProt, and species-specific database of genes/proteins with missing Entrez Gene IDs were used as queries to map them to synonymous tables of Entrez. In each mapping the Identifier used had always the highest level of matches. For instance, Entrez Gene IDs were chosen with the highest consensus of all other database-derived synonymous tables.
The mapped interactions were merged to a directional merged interactome if the source and target interactors are the same as well as to an undirectional merged interactome in which all interactions describing the same interacting entities are fused regardless of source or target interactors.
For the integrated, mapped and merged interactions a suitable scoring system was developed. Interactions gain scores for each experimental system type, different interaction detection method that was used, and additionally for each different publication which observed an interaction. Furthermore, if an interaction involves a post-translational modification event it receives a further score.
Network Analyses
The integrated interaction data was transferred into a rational database, queried with gene lists and networks visualized via custom Python scripts or with the use of Cytoscape [103].
Each gene (candidate) was assessed for the number of interactions (degree) with genes in the query lists (seeds), as well as their total interaction number. As a measure of specificity, we calculated the ratio of specific interaction number with the seeds divided by the total number of interactions for each gene (seeds and candidates), as a percentage, and determined its corresponding p-value (based on the binomial distribution). Genes with a binomial p-value<0.05 were classified as significant interactors. Whether a set of genes has a higher average degree than expected by chance was determined using a Mann Whitney U test. We applied an exhaustively leave-one-out test by repeatedly omitting single genes from the seed list and tested whether the genes were among the significant candidate genes inferred from the remainder. From this procedure the percentage of retrieval was estimated for each of the seed genes by successively omitting each gene, one at a time, from the seed list.
Correlation of Variables and Multiple Linear Regression
We performed a multiple linear regression analysis focused on human genes which considers three variables: the degree of each gene in the interaction network, the number of species with orthologs, and the mean of its dN/dS ratios with three other mammalian species (M. musculus, R. norvegicus and M. mulatta). Genes with a degree of zero or with no given dN/dS ratios were removed, in order to log transform the data. A Pearson correlation was applied to first check whether any of those variables are correlated with each other. Then we made a regression analysis, by the method of least squares, of each variable against other variables first singly and then for a given variable as a function of the other two. The derived linear equations were used to predict the variables of greatest relevance for the set of DR-essential genes.
Functional Enrichment Analyses
By a FDR of <5% gene lists were examined using the DAVID bioinformatics resource (http://niaid.abcc.ncifcrf.gov/), under default parameters, to identify overrepresented categories [104]. Gene ontologies were retrieved from Entrez and gene descriptions for yeast genes from Saccharomyces Genome Database (SGD; http://yeastgenome.org/) [105].
Combining p-Values
To judge the overall significance of a series of experiments, the p-values were combined via two different approaches.
The first method is an analytic solution of the Fisher's test [106]. Briefly, the set point whose probability is equal to that of a set of p-values is the hyperbola:
k is the product of a set of p-values k = (x1 · x2 · x3 · … · xn), provided that the events described by these p-values are independent of one another. The volume under the surface gives the probability of obtaining a set of p-values as extreme or more extreme than the given set:
The alternative approach is the z-method as previously described [107], which gives similar, but somewhat more stringent results.
Lifespan and Vacuolar Morphology Analyses
The replicative lifespan of yeast mutants in the BY4742 (Mat α; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) background were measured on different media as described before [23]. The ad libitum (AL) medium was YEPD: yeast extract (1%), peptone (2%), agar (2%), and D-glucose (2%). The dietary restricted (DR) medium was yeast extract (1%), peptone (2%), agar (2%), and D-glucose (0.5%). For bud-counting, cells were grown at 30°C and dissected every 100 min. Five to six rounds of bud-counting were performed each day. The aging assay plate was saved at 4°C for overnight and the bud-counting was continued the next day till all cells died, which was diagnosed by either cell lysis or no budding in 2 days. The vacuolar morphology of yeast mutants under AL and DR were measured by FM4-64 labeling and chasing [108]. Briefly, cells were inoculated into AL or DR liquid media (AL: yeast extract (1%) peptone plus (2%) D-glucose; DR: yeast extract (1%) peptone plus (0.5%) D-glucose) and incubated on a shaker (150 rpm) at 30°C overnight. An aliquot of 250 µl of cells were labeled with 6 µl of 2 mg/ml FM4-64 for 1 hour. After washing off the dye, cells were chased in the corresponding medium for 3 hours and then photographed.
Significance of mean lifespan changes were assessed with the log-rank test (a.k.a. Mantel–Cox test) between mutants and wild-type as well as between dietary regimes for each genotype.
RNA Isolation and Microarray Analyses
Wild-type yeast cells (BY4742) were inoculated into AL or DR liquid media and incubated overnight on a shaker. These seed cultures were then inoculated into fresh media to an initial OD600 of 0.01. Cells were grown at 30°C for 12 hours. The final OD600 of cultures were 0.85–0.99. Three OD600 units of cells were collected and lysed with zymolyase. Total RNA was isolated using the Qiagen RNeasy mini kit as described [109]. Total RNA obtained was between 60 to 100 µg. RNA samples were processed and hybridized with GeneChip Yeast genome 2.0 arrays (Affymetrix Inc., Santa Clara, CA) by Expression Analysis (Durham, NC). The hybridization signals were normalized and analyzed with the Affymetrix statistical algorithms by Expression Analysis. By a two-fold cut-off, out of 5716 probed genes 2587 (45.26%) were differentially expressed, with 1413 genes (24.72%) upregulated and 1174 genes (20.54%) downregulated. Full results and raw microarray data are available online at GenDR (http://genomics.senescence.info/diet/Yeast_array.zip), GEO (GSE38635), and ArrayExpress (E-MTAB-1165).
For additional microarray data from yeast (GSE9217), worms (GSE9682), and flies (GSE26726 and GSE16738), GSE files were retrieved from the GEO database, replicates were averaged and fold-changes calculated via custom Python scripts.
Transcription Factor Identification
Transcription factor–target gene interactions were retrieved from YEASTRACT (http://yeastract.com/). Transcription factors interacting with DR-essential genes were identified and ordered by their specificity or binominal p-value of the ratio of regulated genes with more than two-fold differentially expression upon DR relative to the total number of genes regulated by the same transcription factor. The whole genome (sacCer3) was retrieved from UCSC and regulatory regions (+500 bp sequences from the transcription start site) annotated. Transcription factor binding motifs were integrated from SGD, YEASTRACT, and YFTD (http://biochemie.web.med.uni-muenchen.de/YTFD/YTF_alpha_2.htm), as well as from the literature. Via regular expression it was tested (hypergeometric p-value and FDR q-values) whether a defined motif is significant enriched in the regulatory regions of differential expressed (either up - or down-regulated) genes or set of genes.
Supporting Information
Zdroje
1. BishopNA, GuarenteL (2007) Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 8 : 835–844.
2. FontanaL, PartridgeL, LongoVD (2010) Extending healthy life span–from yeast to humans. Science 328 : 321–326.
3. KemnitzJW (2011) Calorie restriction and aging in nonhuman primates. ILAR J 52 : 66–77.
4. WillcoxBJ, WillcoxDC, TodorikiH, FujiyoshiA, YanoK, et al. (2007) Caloric restriction, the traditional Okinawan diet, and healthy aging: the diet of the world's longest-lived people and its potential impact on morbidity and life span. Ann N Y Acad Sci 1114 : 434–455.
5. SpindlerSR (2010) Caloric restriction: from soup to nuts. Ageing Res Rev 9 : 324–353.
6. de MagalhaesJP, WuttkeD, WoodSH, PlankM, VoraC (2012) Genome-environment interactions that modulate aging: powerful targets for drug discovery. Pharmacol Rev 64 : 88–101.
7. LakowskiB, HekimiS (1998) The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 95 : 13091–13096.
8. PartridgeL, PiperMD, MairW (2005) Dietary restriction in Drosophila. Mech Ageing Dev 126 : 938–950.
9. BarabasiAL, GulbahceN, LoscalzoJ (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12 : 56–68.
10. de MagalhaesJP, ChurchGM (2007) Analyses of human-chimpanzee orthologous gene pairs to explore evolutionary hypotheses of aging. Mech Ageing Dev 128 : 355–364.
11. BudovskyA, AbramovichA, CohenR, Chalifa-CaspiV, FraifeldV (2007) Longevity network: construction and implications. Mech Ageing Dev 128 : 117–124.
12. WangJ, ZhangS, WangY, ChenL, ZhangXS (2009) Disease-aging network reveals significant roles of aging genes in connecting genetic diseases. PLoS Comput Biol 5: e1000521 doi:10.1371/journal.pcbi.1000521.
13. PromislowDE (2004) Protein networks, pleiotropy and the evolution of senescence. Proc Biol Sci 271 : 1225–1234.
14. WittenTM, BonchevD (2007) Predicting aging/longevity-related genes in the nematode Caenorhabditis elegans. Chem Biodivers 4 : 2639–2655.
15. FortneyK, KotlyarM, JurisicaI (2010) Inferring the functions of longevity genes with modular subnetwork biomarkers of Caenorhabditis elegans aging. Genome Biol 11: R13.
16. BonkowskiMS, RochaJS, MasternakMM, Al RegaieyKA, BartkeA (2006) Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci U S A 103 : 7901–7905.
17. GemsD, PletcherS, PartridgeL (2002) Interpreting interactions between treatments that slow aging. Aging Cell 1 : 1–9.
18. de MagalhaesJP, BudovskyA, LehmannG, CostaJ, LiY, et al. (2009) The Human Ageing Genomic Resources: online databases and tools for biogerontologists. Aging Cell 8 : 65–72.
19. Shmookler ReisRJ, AyyadevaraS, CrowWA, LeeT, DelongchampRR (2011) Gene Categories Differentially Expressed in C. elegans Age-1 Mutants of Extraordinary Longevity: New Insights From Novel Data-Mining Procedures. J Gerontol A Biol Sci Med Sci 67 : 366–75.
20. TazearslanC, AyyadevaraS, BharillP, Shmookler ReisRJ (2009) Positive feedback between transcriptional and kinase suppression in nematodes with extraordinary longevity and stress resistance. PLoS Genet 5: e1000452 doi:10.1371/journal.pgen.1000452.
21. de MagalhaesJP, ToussaintO (2004) GenAge: a genomic and proteomic network map of human ageing. FEBS Lett 571 : 243–247 doi:10.1371/journal.pgen.1000452.
22. OwenAB, StuartJ, MachK, VilleneuveAM, KimS (2003) A gene recommender algorithm to identify coexpressed genes in C. elegans. Genome Res 13 : 1828–1837.
23. TangF, WatkinsJW, BermudezM, GrayR, GabanA, et al. (2008) A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy 4 : 874–886.
24. LinSJ, KaeberleinM, AndalisAA, SturtzLA, DefossezPA, et al. (2002) Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418 : 344–348.
25. HonjohS, YamamotoT, UnoM, NishidaE (2009) Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457 : 726–730.
26. BauerJ, AntochM, ChangC, SchorlC, KolliS, et al. (2010) Comparative transcriptional profiling identifies takeout as a gene that regulates life span. Aging (Albany NY) 2 : 298–310.
27. ZidBM, RogersAN, KatewaSD, VargasMA, KolipinskiMC, et al. (2009) 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139 : 149–160.
28. Shmookler ReisRJ, XuL, LeeH, ChaeM, ThadenJJ, et al. (2011) Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 3 : 125–147.
29. HansenM, HsuAL, DillinA, KenyonC (2005) New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 1: e17. doi:10.1371/journal.pgen.0010017.
30. SteinkrausKA, SmithED, DavisC, CarrD, PendergrassWR, et al. (2008) Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 7 : 394–404.
31. SmithED, TsuchiyaM, FoxLA, DangN, HuD, et al. (2008) Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res 18 : 564–570.
32. BalajiS, BabuMM, IyerLM, LuscombeNM, AravindL (2006) Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J Mol Biol 360 : 213–227.
33. AbdulrehmanD, MonteiroPT, TeixeiraMC, MiraNP, LourencoAB, et al. (2011) YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res 39: D136–140.
34. LeeYL, LeeCK (2008) Transcriptional response according to strength of calorie restriction in Saccharomyces cerevisiae. Mol Cells 26 : 299–307.
35. EisenbergT, KnauerH, SchauerA, ButtnerS, RuckenstuhlC, et al. (2009) Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11 : 1305–1314.
36. ChattopadhyayMK, ChenW, PoyG, CamM, StilesD, et al. (2009) Microarray studies on the genes responsive to the addition of spermidine or spermine to a Saccharomyces cerevisiae spermidine synthase mutant. Yeast 26 : 531–544.
37. WarsowG, GreberB, FalkSS, HarderC, SiatkowskiM, et al. (2010) ExprEssence - Revealing the essence of differential experimental data in the context of an interaction/regulation network. BMC Syst Biol 4 : 164.
38. SteffenKK, MacKayVL, KerrEO, TsuchiyaM, HuD, et al. (2008) Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133 : 292–302.
39. HansenM, TaubertS, CrawfordD, LibinaN, LeeSJ, et al. (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6 : 95–110.
40. KapahiP, ZidBM, HarperT, KosloverD, SapinV, et al. (2004) Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14 : 885–890.
41. SelmanC, TulletJM, WieserD, IrvineE, LingardSJ, et al. (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326 : 140–144.
42. ShanleyDP, KirkwoodTB (2000) Calorie restriction and aging: a life-history analysis. Evolution 54 : 740–750.
43. FraserHB, HirshAE, SteinmetzLM, ScharfeC, FeldmanMW (2002) Evolutionary rate in the protein interaction network. Science 296 : 750–752.
44. EisenbergE, LevanonEY (2003) Preferential attachment in the protein network evolution. Phys Rev Lett 91 : 138701.
45. SaeedR, DeaneCM (2006) Protein protein interactions, evolutionary rate, abundance and age. BMC Bioinformatics 7 : 128.
46. KimSK (2007) Common aging pathways in worms, flies, mice and humans. J Exp Biol 210 : 1607–1612.
47. UmebayashiK, NakanoA (2003) Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J Cell Biol 161 : 1117–1131.
48. CarranoAC, LiuZ, DillinA, HunterT (2009) A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature 460 : 396–399.
49. NikkoE, PelhamHR (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10 : 1856–1867.
50. BabstM (2011) MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol 23 : 452–457.
51. FabrizioP, HoonS, ShamalnasabM, GalbaniA, WeiM, et al. (2010) Genome-Wide Screen in Saccharomyces cerevisiae Identifies Vacuolar Protein Sorting, Autophagy, Biosynthetic, and tRNA Methylation Genes Involved in Life Span Regulation. PLoS Genet 6: e1001024 doi:10.1371/journal.pgen.1001024.
52. LongoVD, NislowC, FabrizioP (2010) Endosomal protein sorting and autophagy genes contribute to the regulation of yeast life span. Autophagy 6 : 1227–1228.
53. AouidaM, Khodami-PourA, RamotarD (2009) Novel role for the Saccharomyces cerevisiae oligopeptide transporter Opt2 in drug detoxification. Biochem Cell Biol 87 : 653–661.
54. SharmaPK, MittalN, DeswalS, RoyN (2011) Calorie restriction up-regulates iron and copper transport genes in Saccharomyces cerevisiae. Mol Biosyst 7 : 394–402.
55. KakinumaY, OhsumiY, AnrakuY (1981) Properties of H+-translocating adenosine triphosphatase in vacuolar membranes of SAccharomyces cerevisiae. J Biol Chem 256 : 10859–10863.
56. ChanTF, CarvalhoJ, RilesL, ZhengXF (2000) A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc Natl Acad Sci U S A 97 : 13227–13232.
57. CostanzoM, BaryshnikovaA, BellayJ, KimY, SpearED, et al. (2010) The genetic landscape of a cell. Science 327 : 425–431.
58. ItoT, ChibaT, OzawaR, YoshidaM, HattoriM, et al. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A 98 : 4569–4574.
59. FlinnRJ, YanY, GoswamiS, ParkerPJ, BackerJM (2010) The late endosome is essential for mTORC1 signaling. Mol Biol Cell 21 : 833–841.
60. SturgillTW, CohenA, DiefenbacherM, TrautweinM, MartinDE, et al. (2008) TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell 7 : 1819–1830.
61. EldakakA, RancatiG, RubinsteinB, PaulP, ConawayV, et al. (2010) Asymmetrically inherited multidrug resistance transporters are recessive determinants in cellular replicative ageing. Nat Cell Biol 12 : 799–805.
62. PlankM, WuttkeD, van DamS, ClarkeSA, de MagalhaesJP (2012) A meta-analysis of caloric restriction gene expression profiles to infer common signatures and regulatory mechanisms. Mol Biosyst 8 : 1339–1349.
63. AndersonRM, BittermanKJ, WoodJG, MedvedikO, SinclairDA (2003) Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423 : 181–185.
64. JaenischR, BirdA (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33 Suppl: 245–254.
65. BurdgeGC, HansonMA, Slater-JefferiesJL, LillycropKA (2007) Epigenetic regulation of transcription: a mechanism for inducing variations in phenotype (fetal programming) by differences in nutrition during early life? Br J Nutr 97 : 1036–1046.
66. FeserJ, TylerJ (2011) Chromatin structure as a mediator of aging. FEBS Lett 585 : 2041–2048.
67. VaqueroA, ReinbergD (2009) Calorie restriction and the exercise of chromatin. Genes Dev 23 : 1849–1869.
68. UnalE, KindeB, AmonA (2011) Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 332 : 1554–1557.
69. MairW, GoymerP, PletcherSD, PartridgeL (2003) Demography of dietary restriction and death in Drosophila. Science 301 : 1731–1733.
70. BrawleyJV, FerroAJ (1979) Polyamine biosynthesis during germination of yeast ascospores. J Bacteriol 140 : 649–654.
71. TaborCW (1981) Mutants of Saccharomyces cerevisiae deficient in polyamine biosynthesis: studies on the regulation of ornithine decarboxylase. Med Biol 59 : 272–278.
72. JinY, BokJW, Guzman-de-PenaD, KellerNP (2002) Requirement of spermidine for developmental transitions in Aspergillus nidulans. Mol Microbiol 46 : 801–812.
73. MatsumotoM, KuriharaS, KibeR, AshidaH, BennoY (2011) Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS ONE 6: e23652 doi:10.1371/journal.pone.0023652.
74. SodaK, DobashiY, KanoY, TsujinakaS, KonishiF (2009) Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp Gerontol 44 : 727–732.
75. PnueliL, EdryI, CohenM, KassirY (2004) Glucose and nitrogen regulate the switch from histone deacetylation to acetylation for expression of early meiosis-specific genes in budding yeast. Mol Cell Biol 24 : 5197–5208.
76. de MagalhaesJP, CostaJ, ToussaintO (2005) HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res 33: D537–543.
77. FischerS, BrunkBP, ChenF, GaoX, HarbOS, et al. (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics Chapter 6 Unit 6 12 11–19.
78. OstlundG, SchmittT, ForslundK, KostlerT, MessinaDN, et al. (2010) InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res 38: D196–203.
79. KooninEV, WolfYI (2010) Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet 11 : 487–498.
80. ArandaB, AchuthanP, Alam-FaruqueY, ArmeanI, BridgeA, et al. (2010) The IntAct molecular interaction database in 2010. Nucleic Acids Res 38: D525–531.
81. XenariosI, SalwinskiL, DuanXJ, HigneyP, KimSM, et al. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30 : 303–305.
82. CeolA, Chatr AryamontriA, LicataL, PelusoD, BrigantiL, et al. (2010) MINT, the molecular interaction database: 2009 update. Nucleic Acids Res 38: D532–539.
83. IsserlinR, El-BadrawiRA, BaderGD (2011) The Biomolecular Interaction Network Database in PSI-MI 2.5. Database (Oxford) 2011: baq037.
84. BreitkreutzBJ, StarkC, RegulyT, BoucherL, BreitkreutzA, et al. (2008) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res 36: D637–640.
85. GuldenerU, MunsterkotterM, OesterheldM, PagelP, RueppA, et al. (2006) MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res 34: D436–441.
86. SteinLD (2004) Using the Reactome database. Curr Protoc Bioinformatics Chapter 8 Unit 8 7.
87. Keshava PrasadTS, GoelR, KandasamyK, KeerthikumarS, KumarS, et al. (2009) Human Protein Reference Database–2009 update. Nucleic Acids Res 37: D767–772.
88. BeumingT, SkrabanekL, NivMY, MukherjeeP, WeinsteinH (2005) PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 21 : 827–828.
89. RueppA, WaegeleB, LechnerM, BraunerB, Dunger-KaltenbachI, et al. (2010) CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res 38: D497–501.
90. RazickS, MagklarasG, DonaldsonIM (2008) iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9 : 405.
91. HornbeckPV, ChabraI, KornhauserJM, SkrzypekE, ZhangB (2004) PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4 : 1551–1561.
92. StarkC, SuTC, BreitkreutzA, LourencoP, DahabiehM, et al. (2010) PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford) 2010: bap026.
93. BrownKR, JurisicaI (2007) Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol 8: R95.
94. MichautM, KerrienS, Montecchi-PalazziL, ChauvatF, Cassier-ChauvatC, et al. (2008) InteroPORC: automated inference of highly conserved protein interaction networks. Bioinformatics 24 : 1625–1631.
95. WilesAM, DodererM, RuanJ, GuTT, RaviD, et al. (2010) Building and analyzing protein interactome networks by cross-species comparisons. BMC Syst Biol 4 : 36.
96. JayapandianM, ChapmanA, TarceaVG, YuC, ElkissA, et al. (2007) Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res 35: D566–571.
97. TarceaVG, WeymouthT, AdeA, BookvichA, GaoJ, et al. (2009) Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res 37: D642–646.
98. WuJ, ValleniusT, OvaskaK, WestermarckJ, MakelaTP, et al. (2009) Integrated network analysis platform for protein-protein interactions. Nat Methods 6 : 75–77.
99. LeeI, LiZ, MarcotteEM (2007) An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE 2: e988 doi:10.1371/journal.pone.0000988.
100. LeeI, LehnerB, VavouriT, ShinJ, FraserAG, et al. (2010) Predicting genetic modifier loci using functional gene networks. Genome Res 20 : 1143–1153.
101. KimWK, KrumpelmanC, MarcotteEM (2008) Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol 9 Suppl 1: S5.
102. RavasiT, SuzukiH, CannistraciCV, KatayamaS, BajicVB, et al. (2010) An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140 : 744–752.
103. ClineMS, SmootM, CeramiE, KuchinskyA, LandysN, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protoc 2 : 2366–2382.
104. Huang daW, ShermanBT, TanQ, KirJ, LiuD, et al. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35: W169–175.
105. CherryJM, HongEL, AmundsenC, BalakrishnanR, BinkleyG, et al. (2012) Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40: D700–705.
106. Fisher RA (1932) Statistical methods for research workers. Edinburgh etc.: Oliver and Boyd. xiii p., 1 l. p.
107. WhitlockMC (2005) Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol 18 : 1368–1373.
108. WangYX, CatlettNL, WeismanLS (1998) Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol 140 : 1063–1074.
109. GebreS, ConnorR, XiaY, JawedS, BushJM, et al. (2012) Osh6 overexpression extends the lifespan of yeast by increasing vacuole fusion. Cell Cycle 11 : 2176–2188.
110. ColominaN, LiuY, AldeaM, GariE (2003) TOR regulates the subcellular localization of Ime1, a transcriptional activator of meiotic development in budding yeast. Mol Cell Biol 23 : 7415–7424.
111. BalderhaarHJ, ArltH, OstrowiczC, BrockerC, SundermannF, et al. (2010) The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci 123 : 4085–4094.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání





