-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
Fanconi anemia (FA) is a devastating genetic disease, associated with genomic instability and defects in DNA interstrand cross-link (ICL) repair. The FA repair pathway is not thought to be conserved in budding yeast, and although the yeast Mph1 helicase is a putative homolog of human FANCM, yeast cells disrupted for MPH1 are not sensitive to ICLs. Here, we reveal a key role for Mph1 in ICL repair when the Pso2 exonuclease is inactivated. We find that the yeast FANCM ortholog Mph1 physically and functionally interacts with Mgm101, a protein previously implicated in mitochondrial DNA repair, and the MutSα mismatch repair factor (Msh2-Msh6). Co-disruption of MPH1, MGM101, MSH6, or MSH2 with PSO2 produces a lesion-specific increase in ICL sensitivity, the elevation of ICL-induced chromosomal rearrangements, and persistence of ICL-associated DNA double-strand breaks. We find that Mph1-Mgm101-MutSα directs the ICL-induced recruitment of Exo1 to chromatin, and we propose that Exo1 is an alternative 5′-3′ exonuclease utilised for ICL repair in the absence of Pso2. Moreover, ICL-induced Rad51 chromatin loading is delayed when both Pso2 and components of the Mph1-Mgm101-MutSα and Exo1 pathway are inactivated, demonstrating that the homologous recombination stages of ICL repair are inhibited. Finally, the FANCJ - and FANCP-related factors Chl1 and Slx4, respectively, are also components of the genetic pathway controlled by Mph1-Mgm101-MutSα. Together this suggests that a prototypical FA–related ICL repair pathway operates in budding yeast, which acts redundantly with the pathway controlled by Pso2, and is required for the targeting of Exo1 to chromatin to execute ICL repair.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002884
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002884Summary
Fanconi anemia (FA) is a devastating genetic disease, associated with genomic instability and defects in DNA interstrand cross-link (ICL) repair. The FA repair pathway is not thought to be conserved in budding yeast, and although the yeast Mph1 helicase is a putative homolog of human FANCM, yeast cells disrupted for MPH1 are not sensitive to ICLs. Here, we reveal a key role for Mph1 in ICL repair when the Pso2 exonuclease is inactivated. We find that the yeast FANCM ortholog Mph1 physically and functionally interacts with Mgm101, a protein previously implicated in mitochondrial DNA repair, and the MutSα mismatch repair factor (Msh2-Msh6). Co-disruption of MPH1, MGM101, MSH6, or MSH2 with PSO2 produces a lesion-specific increase in ICL sensitivity, the elevation of ICL-induced chromosomal rearrangements, and persistence of ICL-associated DNA double-strand breaks. We find that Mph1-Mgm101-MutSα directs the ICL-induced recruitment of Exo1 to chromatin, and we propose that Exo1 is an alternative 5′-3′ exonuclease utilised for ICL repair in the absence of Pso2. Moreover, ICL-induced Rad51 chromatin loading is delayed when both Pso2 and components of the Mph1-Mgm101-MutSα and Exo1 pathway are inactivated, demonstrating that the homologous recombination stages of ICL repair are inhibited. Finally, the FANCJ - and FANCP-related factors Chl1 and Slx4, respectively, are also components of the genetic pathway controlled by Mph1-Mgm101-MutSα. Together this suggests that a prototypical FA–related ICL repair pathway operates in budding yeast, which acts redundantly with the pathway controlled by Pso2, and is required for the targeting of Exo1 to chromatin to execute ICL repair.
Introduction
DNA interstrand cross-links (ICLs) represent an extremely toxic form of genomic damage; a consequence of their ability to inhibit basic cellular processes such as transcription and replication [1]–[3]. Eukaryotic ICL repair remains relatively poorly understood, although an initial ICL incision step followed by homologous recombination repair (HRR) and translesion DNA synthesis (TLS) steps have been implicated by genetic studies [4]–[7]. Molecular evidence for some of these events has been provided, although many mechanistic details remain obscure [8]–[11]. Patients suffering from the inherited condition Fanconi anemia (FA) are defective in ICL repair, where ICL incision and the subsequent TLS and HRR stages are all affected [12]–[14]. This repair defect is associated with bone marrow failure and a predisposition to haematological and solid malignancies [15]. Despite some differences in ICL processing between yeast and human cells, most notably the absence of readily identifiable homologs of many FA factors, yeast does possess a putative ortholog (Mph1) of the protein mutated in FA complementation group M (FANCM) [16], [17], indicating that yeast could be a powerful model for understanding some aspects of the repair defect in FA. Strikingly, however, and in contrast to higher eukaryotic fancm mutants, yeast mph1 cells are not significantly ICL sensitive.
MPH1 was identified following genetic screens for strains with an elevated polymerase ζ-dependent spontaneous mutator phenotype [18], suggesting that Mph1 contributes to error-free damage tolerance pathways [19], [20]. Cells lacking Mph1 are mildly sensitive to a range of DNA alkylating agents [19], [20]. Both budding yeast Mph1 and its fission yeast homolog Fml1 have been shown to directly regulate the outcome of HRR events [21]–[23], limiting crossovers. Biochemically, Mph1 is a member of the superfamily-2 (SF2) helicases, and purified Mph1 is an ATP-dependent 3′-5′ helicase, whose activity is augmented in the presence of replication protein A [24]. FANCM is found in the FA core complex, and, like other members of the FA family, it is required for normal cellular resistance to ICLs [16], [17]. Similar to Mph1, FANCM demonstrates ATP-dependent helicase activity, and is able to unwind a number of substrates including model replication forks, D-loops and Holliday junctions [21], [25]. Recent studies demonstrated that the entire helicase domain of human FANCM and ATP binding by the Walker A motif is required for normal resistance to ICLs [26], although mutation of the Walker B motif suggests that ICL repair is not dependent upon functional helicase activity [27]. Finally, FANCM has a role in the ATR-mediated replication checkpoint in vertebrate cells [28]–[30].
Like human FA cells, budding yeast pso2 mutants (originally also known as snm1) are highly sensitive to ICLs, but not to other forms of DNA damage [31]–[33]. Several groups have concluded that the initial unhooking incisions at ICLs are normal in pso2 mutants [34]–[36]. However, ICL-associated DNA double-strand breaks (DSBs) accumulate in pso2 cells [35]–[38]. It was subsequently demonstrated that such DSBs are the result of replication fork cleavage when an ICL stalls replication [6], [11], [39]–[43]. Recent studies confirm that a human homolog of Pso2, hSNM1A, also plays a key role in ICL repair during S-phase [43]. Finally, several lines of evidence also point to a role for mismatch repair (MMR) factors in ICL repair [44]–[46]. In budding yeast, Msh2 and Exo1 appear to play a role that is redundant with Pso2 in ICL repair during S-phase [38], although the basis for this remains unknown.
The identification of novel Pso2 - and Mph1-interacting factors will be critical to an improved understanding of ICL repair. Here, we demonstrate that Mgm101, a factor hitherto thought to only repair mitochondrial DNA, interacts with both Mph1 and MutSα (Msh2-Msh6). Our subsequent analysis reveals a critical role for Mph1-MutSα-Mgm101 and Exo1, as well as the FANCJ - and FANCP-related factors Chl1 and Slx4, respectively, in the Pso2-independent repair of ICLs, suggestive of an FA-like repair pathway in budding yeast.
Results
Mgm101 interacts with Mph1 and Msh2-Msh6
Large-scale protein-interactome studies suggested interactions between Pso2 and the Mgm101 protein [47]. Mgm101 was originally identified as a factor required for the maintenance of the mitochondrial genome [48], and is a component of the mitochondrial nucleoid, where it co-localises and interacts with the mitochondrial genome maintenance factor, Mmm1 [49], [50]. In addition to genome transmission, Mgm101 might be required for the repair of oxidative DNA lesions in mitochondria [49], as well as HR in mitochondria [51]. Outside of fungi and marine invertebrates Mgm101 is not clearly conserved in higher eukaryotes, and it has recently been noticed that Mgm101 has some similarity to Rad52 [51], [52]. Our extensive yeast two-hybrid and co-immunoprecipitation studies failed to produce evidence of an interaction between Pso2 and Mgm101 (data not shown). However, when we employed FLAG-tagged Mgm101, separated Mgm101-FLAG associated proteins by liquid chromatography (LC) and determined their identity by MALDI-TOF mass-spectrometry (Figure 1A), three nuclear DNA repair proteins, Mph1, Msh2, Msh6 as well as Fkh1, the nucleo-cytoplasmic factor Lsm7 and a mitochondrial factor Mrp51 were identified.
Fig. 1. Mgm101 interacts with Mph1 and MutSα. 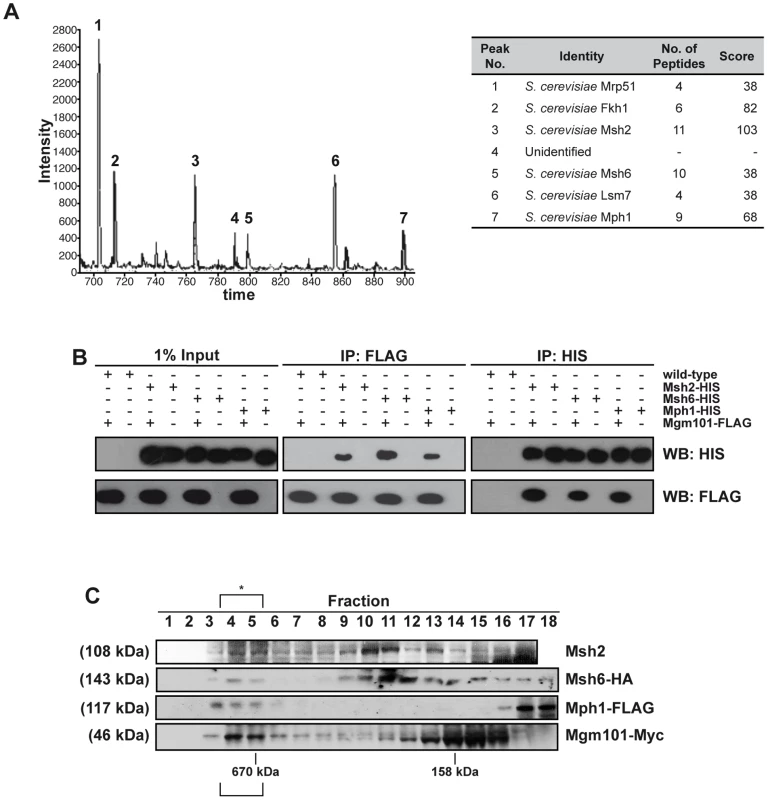
(A) LC-MS. Prominent peaks from LC analysis with intensity above 400 were subjected for MALDI-TOF analysis. (B) Co-IPs. Reciprocal immunoprecipitations using FLAG-tagged Mgm101 expressed in cells with chromosomally 6xHis-tagged Msh2, Msh6 or Mph1. The empty vector transformants (wild type) were used as controls. A portion of the lysates (1%) was immunoblotted to show input proteins (left-hand panel). (C) The elution profiles of Mgm101-Myc, Mph1-FLAG and Msh6-HA as well as Msh2 following Superdex 200 chromatography reveal coincident elution in a high molecular weight complex at around 670 kDa, suggesting that they associate in a high molecular-weight complex, marked with an asterisk. We confirmed this pattern of interaction with the three DNA repair factors by performing reciprocal immunoprecipitations using C-terminally FLAG-tagged Mgm101 introduced into strains with C-terminal chromosomally 6xHis-tagged Msh2, Msh6 or Mph1. By incubating cell extracts with anti-FLAG coated agarose or nickel resin to immunoprecipitate the FLAG - and 6xHIS-tagged proteins, respectively, reciprocal interactions between Mgm101 and Msh2, Msh6 and Mph1 were all confirmed (Figure 1B). Note that in all immunoprecipitation experiments the cell extracts were treated with a nuclease mix, excluding the possibility that the interactions observed are mediated via DNA. To further explore the possibility of a complex containing the identified Mgm101-interacting factors, we tagged each factor with a unique epitope in a single strain and performed Superdex 200 gel filtration analysis, followed by immunoblotting to establish the elution profile of each factor. Consistent with recently published data suggesting that Mgm101 multimerises efficiently [51], much of the Mgm101 was found in fractions peaking at around 160 kDa (fractions 12–16). The major elution peaks for Msh2 and Msh6 were around 250 kDa (fractions 10–11), consistent with the majority of the cellular pool of these factors existing together in the MutSα complex [53]. The presence of all these factors in a co-eluting high molecular weight fraction (>670 kDa, fractions 4–5) was also observed, consistent with a sub-population of each of these proteins residing in a high molecular weight complex with the same elution characteristics.
Since Msh2-Msh6 and Mph1 are primarily nuclear factors, our interaction results suggested that Mgm101 could, therefore, reside in the nucleus as well as the mitochondrial nucleoid [49]. Cellular fractionation of the nuclear and mitochondrial compartments confirmed that Mgm101-FLAG was present in both fractions, with enrichment in both the nucleus and mitochondria compared to levels in whole cell extracts (Figure S1A).
Mph1, MutSα, and Mgm101 are required for nuclear ICL repair in the absence of Pso2
When strains disrupted for PSO2, MSH2, MPH1 and MGM101 were tested for sensitivity to a range of DNA damaging agents, including hydrogen peroxide (H2O2), ionising radiation (IR), methyl methanesulfonate (MMS) and ultraviolet light (UV), no significant sensitivity was apparent (Figure S2, panels A–F and J–O). Moreover, none of the mph1, msh2, msh6 or mgm101 single mutants demonstrated any sensitivity to the cross-linking drug nitrogen mustard (HN2) (Figure 2A, B and D). Since Mgm101 interacts with both Mph1 and Msh2-Msh6, and our previous studies demonstrated a functional overlap between Pso2 and Msh2 in ICL repair [38], we deleted MGM101, MSH6 and MPH1 in a pso2 mutant background. This revealed an approximately ten-fold increase in HN2 sensitivity for the pso2 mph1, pso2 mgm101 and pso2 msh6 strains over the pso2 single mutant at doses over 10 mM (Figure 2A, B and D). This sensitisation to ICLs is highly specific, since it was also observed for another cross-linking drug cisplatin (CDDP, Figure S2G–I), but not other forms of DNA damage including those induced by H2O2, IR, UV or MMS treatment (Figure S2A–F and J–O). Since Mgm101 and Mph1 physically interact, and show an indistinguishable phenotype upon co-deletion with PSO2, we asked whether they were epistatic for ICL repair. Creation of a pso2 mgm101 mph1 triple mutant confirmed this to be the case (Figure 2C).
Fig. 2. Pso2 and Mph1-Mgm101-MutSα control redundant ICL repair pathways. 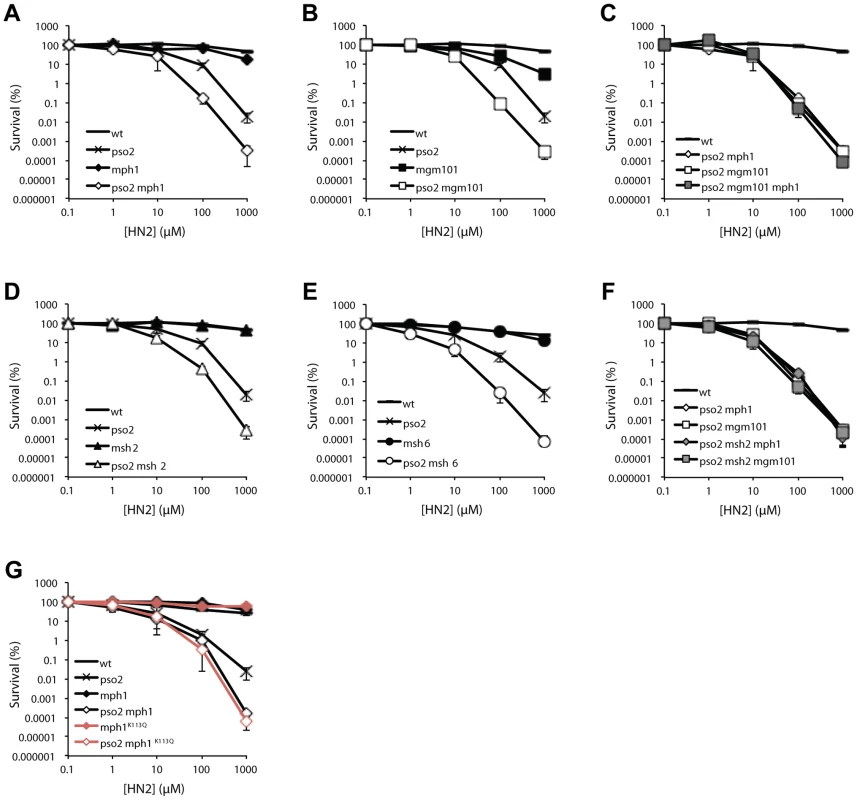
(A to G) Analysis of the ICL sensitivity of combinations of PSO2 with MPH1, MGM101, MSH6 and MSH2 gene disruptions, and an mph1K113Q mutant. The ICL-inducing agent was HN2. All results are the mean of at least three independent experiments and the error bars show the standard error of the mean. The genetic relationship between pso2 and mph1/mgm101 is strongly reminiscent of our previous observations, in a different strain background (W303), of the relationship between pso2 and msh2 double disruptant (recapitulated in the genetic background used in these studies, RDKY3615, Figure 2D and Figure S2G) [38]. It is clear (Figure 2E) that the pso2 msh2 mgm101 and pso2 msh2 mph1 triple disruptants show equivalent sensitivity to both their double disruptant parents, confirming that Msh2, Mph1 and Mgm101 act in a common pathway during ICL repair, which is redundant with the pathway controlled by Pso2. Consistent with this, co-deletion of MSH2 and MGM101 or MSH2 and MPH1, in strains where PSO2 remains functional, does not result in increased HN2 sensitivity (data not shown).
As studies of human FANCM have revealed a critical role for the helicase/translocase domain for normal resistance to ICLs, we also determined whether the helicase domain of Mph1 is required for its ICL repair role. Comparing the HN2 sensitivity of pso2 mph1 strain to that deleted for PSO2 and harbouring an amino acid substitution at a critical lysine (K113Q) predicted to prevent ATP binding in the Walker type A box of the helicase, revealed that an intact helicase domain is required for Mph1 to protect against ICLs in a pso2 background (Figure 2F).
No role for Pso2, Mph1, or Msh2 in mitochondrial genome maintenance
We also wished to explore the possibility that Mph1, Msh2 or Pso2 play some role in mitochondrial genome maintenance, like Mgm101. We examined the rate of loss of mitochondrial function in msh2, mph1 and pso2 mutants, measured as spontaneous petite mutant formation [54]. This was determined by scoring ability to form colonies on non-fermentable (glycerol-containing) media after multiple generations of growth under non-selective conditions (Figure S1B). We found that pso2 and mph1 cells did not exhibit any increased spontaneous loss of functional mitochondria, nor did msh2 mutants, as previously reported [55]. Moreover, the loss of mitochondria in pso2, mph1 and msh2 cells was not increased by treatment with HN2 (Figure S1C) or the oxidative agent H2O2 (Figure S1D). Together our data indicate that Pso2, Mph1 and Msh2 are dispensable for mitochondrial genome stability, even under conditions of stress.
Gross chromosomal rearrangements are increased in pso2 msh2, pso2 mph1, and pso2 mgm101 mutants following ICL induction
Many genes that are required for the maintenance of genome stability act to suppress the induction of gross chromosomal rearrangements (GCRs) [56], and the FA factors play such a role in mammalian cells. We therefore examined the rate of GCRs in our pso2, msh2, mph1 and mgm101 single - and double-mutants, using an assay that detects large interstitial deletions, translocations, chromosome fusions and loss of an entire chromosome arm [56], [57] (Figure 3A). Compared to wild type parent, the pso2 mutant showed an increase of 6-fold in the rate of GCRs (Figure 3A). This was not increased by treatment with 10 mM HN2, which induces negligible lethality in these strains. Consistent with previously published data [58], an msh2 single mutant demonstrated an 10-fold increase in spontaneous GCRs, but this, again, was not increased by ICL induction. However, in a pso2 msh2 double disruptant, while the rate of spontaneous GCRs was similar to that of the msh2 single mutant, the rate of GCRs was enhanced by treatment with HN2. For cells disrupted for MGM101, no increase in the rate of GCR over wild type was observed for either spontaneous or ICL-induced conditions. Deletion of PSO2 in mgm101 cells increased the level of spontaneous GCRs to a level similar to that of the pso2 single mutant. Strikingly, like the pso2 msh2 double mutant, the pso2 mgm101 double disruptant exhibited an increase in GCRs following ICL induction. As previously noted, deletion of MPH1 alone produces a modest elevation in spontaneous GCRs [59]. Once again, while co-deletion of PSO2 and MPH1 did not further affect the levels of spontaneous GCRs, HN2-induced GCRs were significantly elevated in pso2 mph1 cells. Notably, this increase was greater than that seen in pso2 msh2 and pso2 mgm101 double mutants, suggesting that Mph1 might further suppress ICL-induced GCRs by pathway(s) additional to that involving Mgm101 and Msh2. Together, our results suggest that Pso2, Msh2 and Mph1 all suppress spontaneous GCRs, but that Mgm101 is not required. In contrast, the GCRs induced by ICLs are suppressed by two redundant pathways, controlled by Pso2 and Mph1-Mgm101-Msh2, respectively.
Fig. 3. Pso2 and Mph1-Mgm101-MutSα protect against ICL-induced GCRs. 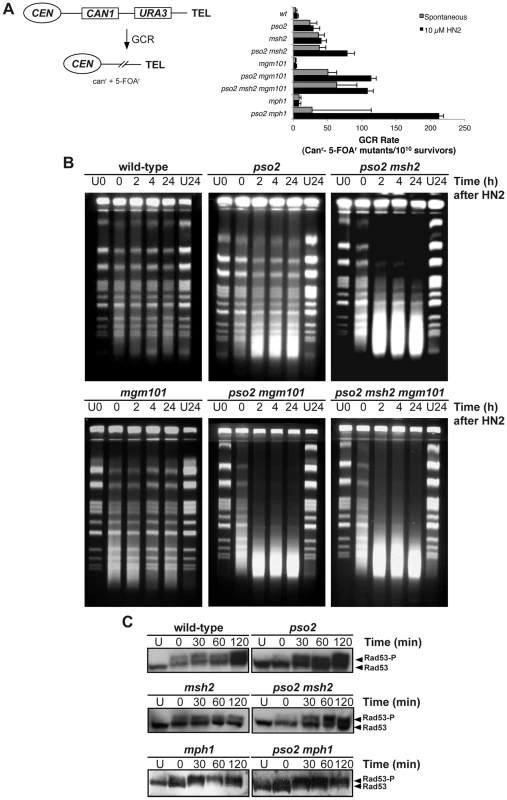
(A) Left panel - schematic of the genomically-integrated (at the HXT13 locus on chromosome V) substrate used to measure GCRs. Simultaneous loss of both the CAN1 and URA3 genes can only occur via GCRs, producing cells able to form colonies able to grow on media containing canavanine and 5-FOA. See text for further details. Right panel - GCRs rates in wild type, pso2, mgm101, mph1 and their respective double mutants, both spontaneously occurring and induced by a sub-lethal dose of HN2 (10 mM). Rates were determined as described in the Materials and Methods section. (B) Accumulation of DSBs in HN2-treated wild type, pso2, pso2 msh2, mgm101, pso2 mgm101 and pso2 mgm101 msh2 cells. Exponentially growing cells were treated with 100 mM HN2 for 2 hours at 28°C, or mock-treated (U) with water, and subsequently allowed to repair in minimal medium for 2, 4 and 24 hours. The U24 sample was mock-treated allowed to repair for 24 hours. Samples were analysed on PFGE gels. (C) Rad53 phosphorylation following treatment with 100 mM HN2 and up to 2 hours recovery in wild type, pso2, msh2, mph1, pso2 msh2 and pso2 mph1 mutants. Mgm101, Mph1, and Msh2 are required for the efficient repair of ICL-associated DSBs in the absence of Pso2
A consequence of PSO2 disruption is the accumulation of ICL-associated DSBs [38], and it is likely that the HN2-induced GCRs observed in pso2 mph1/msh2/mgm101 double mutant cells are the result of aberrant repair of ICL-associated DSBs. We therefore determined whether loss of MGM101 and MSH2 in pso2 cells also affects ICL-associated DSB induction and repair. Pulsed-field gel electrophoresis (PFGE) analysis of whole chromosomes prepared from cells treated with 100 mM HN2 showed that, as previously demonstrated [38], few DSBs accumulate in a wild type strain at this dose (Figure 3B). In pso2 mutants there is a modest accumulation of DSBs at this dose, persisting throughout 24 hours of repair incubation. Furthermore, as previously reported [38], co-deletion of PSO2 and MSH2 causes a dramatic increase in the accumulation of DSBs over that observed in the pso2 single mutant. Consistent with Mgm101 acting in the same pathway of ICL repair as Msh2, deletion of MGM101 in a pso2 strain also caused a dramatic increase in DSB accumulation, and a pso2 msh2 mgm101 triple mutant behaves identically to the pso2 msh2 and pso2 mgm101 double disruptants. Interestingly, although it exhibits no major increase in sensitivity to HN2, the mgm101 single mutant does appear to accumulate slightly more DSBs than its isogenic wild type parent.
Consistent with the accumulation of replication-associated DSBs, phosphorylation of Rad53, a key effector of the yeast S-phase checkpoint, occurs rapidly following ICL treatment in the wild type strain (Figure 3C). Since FANCM has been implicated in efficient S-phase checkpoint activation in human cells [29], we determined whether checkpoint activation by ICLs is affected by loss of Pso2 and the Mph1-Mgm101-Msh2 pathway. Two lines of evidence indicate the major checkpoints are largely intact in these mutants. First, FACS analysis indicates that wild type cells accumulate briefly in G2/M phase 4 hours following HN2 treatment, whereas msh2 cells show a slightly greater accumulation in G2/M at this time, but both strains recover by 6 hours (Figure S3A). Second, Rad53 phosphorylation is induced and sustained in pso2, msh2, pso2 msh2 as well mph1 and pso2 mph1 cells (Figure 3C and Figure S3B). By contrast, pso2 single and pso2 msh2 double mutants arrest in S-phase, and this persists at least until 8 hours following treatment, consistent with the accumulation of the increased levels of broken chromosomes as detected by PFGE.
Pso2 and Mph1-Mgm101-MutSα are required for the repair of ICL-associated DSBs by HR and NHEJ
ICL-associated DSBs form as a result of replication fork collapse and cleavage, and are primarily dependent upon HRR apparatus (RAD52 pathway) for their repair, with NHEJ acting as a back-up [6]. While disruption of RAD52 in a pso2 single mutant leads to an increase in HN2 sensitivity (Figure 4A) inactivation of RAD52 in a pso2 msh2 strain did not further increase its ICL sensitivity (Figure 4D). Similarly, when RAD52 is deleted in a pso2 mph1 double and pso2 msh2 mgm101 triple disruptants, the resulting strains demonstrate no further increase in sensitivity to HN2 (Figure S4A and B). This means either that HRR is not functional in these strains, or that an intermediate accumulates at the site of ICLs that cannot be processed efficiently for downstream repair by HRR. Since, in contrast to a rad52 disruptant, none of the pso2 msh2, pso2 mgm101 or pso2 mph1 double disruptant cells are markedly sensitive to IR-induced DSBs, which require HRR for their repair, the latter explanation appears very likely (Figure S2A–C). Interestingly, although yku70 single mutant displays no sensitivity to ICLs, a pso2 yku70 double mutant also exhibited an increase in HN2 sensitivity (Figure 4B), suggesting that NHEJ plays a role in the repair of ICL-associated DSBs persisting in the absence of Pso2. This effect is not limited to Yku70-defective cells, as cells disrupted for another member of the NHEJ pathway, DNA ligase IV (Dnl4), behave indistinguishably (Figure 4C). Moreover, disruption of YKU70 or DNL4 in a pso2 msh2 background did not lead to an increase in ICL sensitivity (Figure 4E and F). Together, this suggests that neither HRR nor NHEJ can be utilised for the repair of ICL-associated DSBs in strains lacking both the Pso2 and Mph1-Mgm101-MutSα controlled pathways, consistent with the accumulation of high levels of DSBs observed by PFGE (Figure 3).
Fig. 4. HRR and NHEJ mediated repair of ICL-associated DSBs is precluded in cells lacking Pso2 and components of the Mph1-Mgm101-MutSα repair pathways. 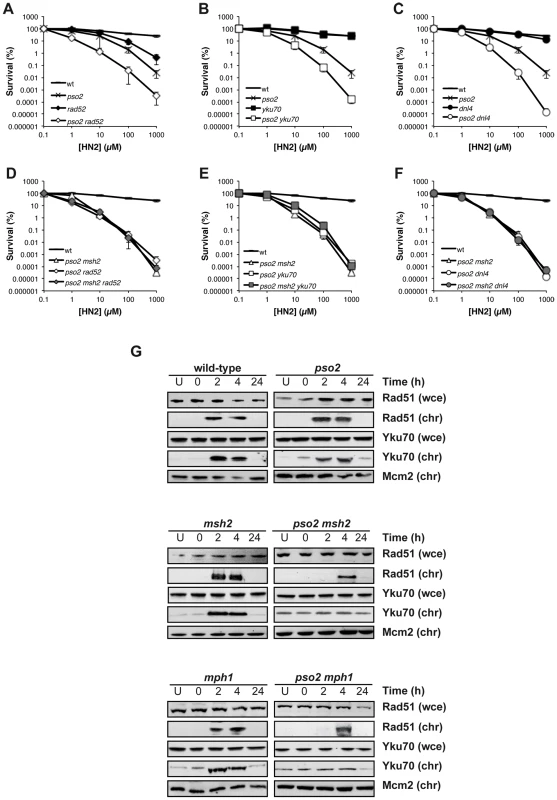
(A–F) HN2 sensitivity of combinations of pso2, msh2, rad52 and yku70 disruptants treated with HN2. All results are the mean of at least three independent experiments and the error bars show the standard error of the mean. (G) HN2-induced chromatin recruitment of Rad51 and Yku70 in wild type, pso2, msh2 and mph1 single and double disruptants. Mcm2 is shown as a loading control for chromatin associated protein. Chromatin bound material is labelled chr, and that from whole cell extract labelled wce. To explore the relationship of Pso2 and Mph1-Mgm101-Msh2 with HRR and NHEJ more directly, we followed the chromatin recruitment of Rad51, required for the early strand invasion step of HRR, and Yku70 that binds to broken DSB ends early in NHEJ, following ICL induction. In wild type cells, Rad51 is recruited to chromatin, within 2 hours following ICL induction (Figure 4G), concomitant with DSB induction (Figure 3), and persists for 4 hours. This is consistent with ongoing repair by HRR, and indeed wild type cells escape from the S/G2 checkpoint block at between 4 and 6 hours following HN2 treatment (Figure S3A). In pso2 cells, Rad51 recruitment is still apparent, and occurs with normal kinetics. The msh2 and mph1 single mutants also behave indistinguishably from wild type cells. Co-deletion of PSO2 with MSH2 or MPH1 led to a delay and reduction in Rad51 recruitment, which could only be observed 4 hours following ICL induction (Figure 4G), as did co-deletion of MGM101 (Figure S4C). Together our genetic and molecular data suggest that processing of ICL repair intermediates by Pso2 and Mph1-Mgm101-MutSα occurs prior to, or early during the recombination steps of repair, since pso2 rad52 is epistatic with pso2 msh2/mph1 double disruption, and Rad51 chromatin loading is delayed in such cells. The recruitment of Yku70 to chromatin followed similar kinetics to Rad51 recruitment in wild type, pso2, msh2 and mph1 disruptants (Figure 4G), suggesting that NHEJ is also precluded at ICL-associated DSBs in these cells.
Exo1 is targeted to chromatin in a Pso2 - and Msh2-dependent fashion
Exonuclease1 (Exo1) interacts with components of the MMR apparatus, including Msh2 [60], and like MutSα factors plays a redundant role with Pso2 during ICL repair [38]. To further explore this, we first asked whether the nuclease activity of Exo1 is required for its role in ICL repair. An exo1 disruptant exhibits no ICL sensitivity, but when combined with pso2 disruption we observed increased sensitivity as previously reported in a different genetic background [38] (Figure 5A). Mutation of aspartic acid 173 to alanine (D173A) in Exo1, that abolishes its exonuclease activity in vitro [61], produces a phenocopy of the exo1 disruptant, where the single mutant has no sensitivity to HN2, but the pso2 exo1-D173A strain exhibits increased sensitivity, confirming that the nuclease activity of Exo1 is required for its role in ICL repair (Figure 5B). We next asked whether the Mph1-Mgm101-MutSα complex is involved in the targeting of Exo1 to chromatin during ICL repair. We followed the recruitment of Exo1-FLAG to chromatin after HN2 treatment for up to 24 hours. Exo1 was recruited to chromatin in wild type, pso2 and mph1 cells within 2 hours of ICL induction (Figure 5C). In contrast, in a pso2 mph1 double disruptant, Exo1-FLAG recruitment was much reduced, suggesting that both pathways, involving Pso2 and Mph1-Mgm101-MutSα, respectively, are required for the efficient chromatin targeting of Exo1-FLAG to effect its role in ICL repair. Since, like Pso2, Exo1 is a 5′-3′ exonuclease it is possible that these two factors are functionally redundant during ICL repair (see Discussion). As with pso2 mph1 and pso2 msh2 cells, Rad51 was recruited to chromatin with delayed kinetics in pso2 exo1 double disruptant compared to the exo1 single mutant, indicating that the initiation of recombination was delayed (Figure 5D). It is clear that the absence of Pso2 alone does impact on the early phase of ICL repair, but the defect is compensated for to a significant extent by the activity of Exo1, since a delay in Rad51 recruitment at the whole chromatin level in pso2 cells is not observed, at least at the whole chromatin level. It is, of course, possible that a loading defect would be observed if it was technically possible to examine individual ICL repair events in pso2 cells. This absence of a delay in Rad51-chromatin association in the exo1 single disruptant indicates that the delay in Rad51 recruitment in pso2 exo1 cells is not a result of the (very mild) DSB resection defects incurred by Exo1 loss [62], but is most likely to be the result of defective ICL processing.
Fig. 5. Exo 1 is involved in the pathways controlled by Pso2 and Mph1-Mgm101-MutSα. 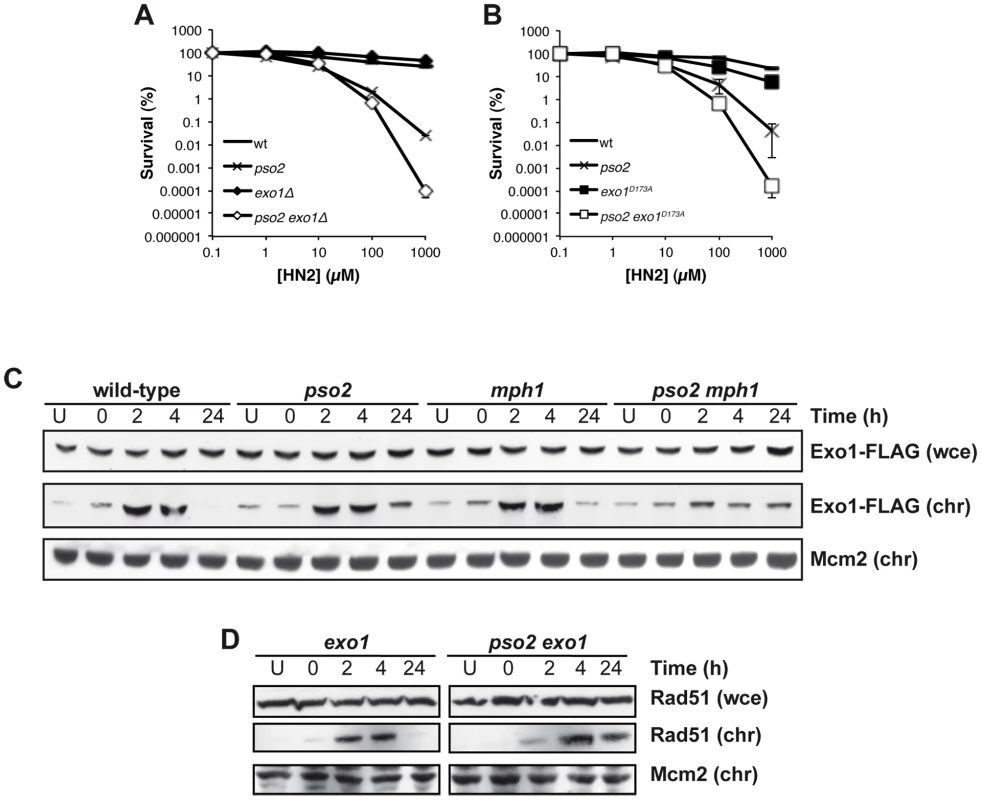
(A and B) HN2 sensitivity of combinations of pso2 and exo1 disruptants, and exo1-D173A mutants treated with HN2. All results are the mean of at least three independent experiments and the error bars show the standard error of the mean. (C) HN2-induced chromatin recruitment of Exo1-FLAG in wt, pso2, mph1 and pso2 mph1 disruptants. Mcm2 is shown as a loading control for chromatin associated protein. Chromatin bound material is labelled chr, and that from whole cell extract labelled wce. (D) HN2-induced chromatin recruitment of Rad51 in wild type cells, pso2 and exo1 single and double disruptants. Mcm2 is shown as a loading control for chromatin associated protein. Chromatin bound material is labelled chr, and that from whole cell extract labelled wce. The ICL repair pathway controlled by Mph1-Mgm101-MutSα also involves the putative FANCJ - and FANCP-related factors Chl1 and Slx4
The existence of an ICL repair pathway involving Mph1 that is only revealed in the absence of Pso2, suggests that a simplified FA-related pathway might be operational in yeast. To this end, analysis of the yeast genome and proteome reveals two further factors that are putative FA homologs, notably the Chl1 helicase that has similarity with FANCJ [63] and Slx4/FANCP [64]–[66]. We therefore inactivated CHL1 and SLX4, alone and in combination with PSO2 and MSH2 (Figure 6). This revealed that both chl1 and slx4 shows the same pattern of genetic interaction with pso2 as msh2, mph1 and mgm101 (Figure 6A and C). Moreover, disruption of MSH2 in a pso2 chl1 and pso2 slx4 double disruptants does not further increase the strains sensitivity to HN2 (Figure 6B and D), indicating that Chl1 and Slx4 operate within the ICL repair pathway controlled by Mph1-Mgm101-MutSα. Together, our data suggest that a prototypical FA-related repair pathway may operate in budding yeast.
Fig. 6. The FANCJ- and FANCP-related factors Chl1 and Slx4, respectively, are both involved in the ICL repair pathway controlled by Mph1-Mgm101-MutSα. 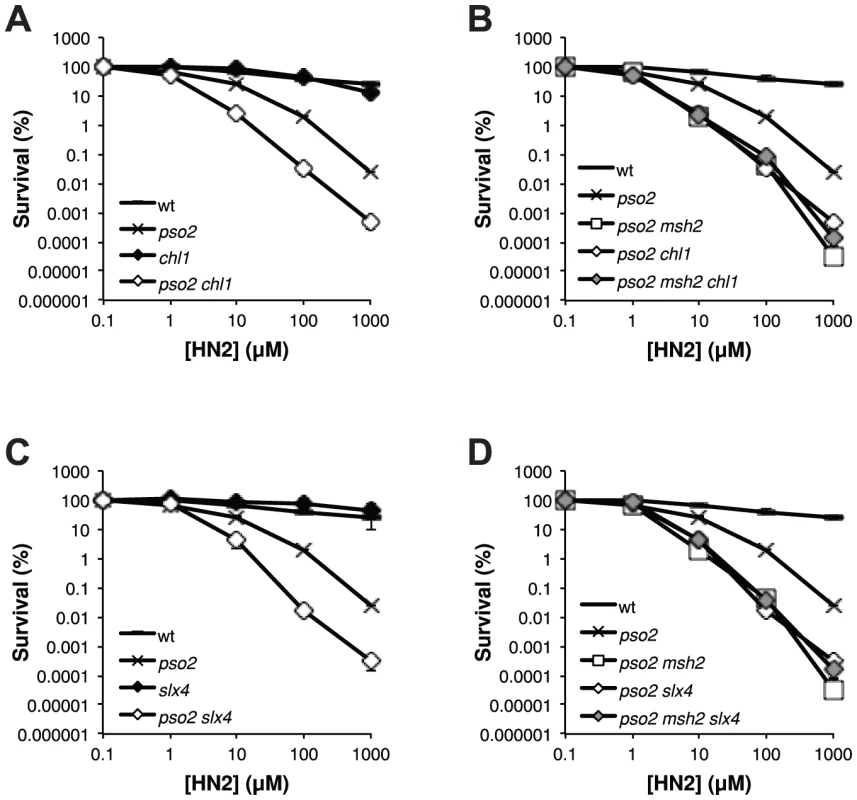
(A–D) HN2 sensitivity analysis of cells disrupted for PSO2 and MSH2 in combination with the FANCJ-related factor CHL1 and FANCP related factor SLX4. All results are the mean of at least three independent experiments and the error bars show the standard error of the mean. Discussion
Here we show that Mgm101 interacts with MutSα and Mph1, and collectively these factors control a key ICL repair pathway. Although Mph1 has significant similarity to FANCM, the disruption of MPH1 alone does not cause any increase in ICL sensitivity, in contrast to vertebrate fancm mutants [16], [17], [26]. Co-disruption of PSO2 revealed an important role for Mph1 in ICL repair. Mgm101 has a well-established role in the transmission and repair of the mitochondrial genome [48], [49], [67], and we now show that Mgm101 also contributes to nuclear DNA repair. Cells disrupted for MGM101 exhibit no increase in spontaneous forward mutation frequency (Figure S1E) or GCR rates (Figure 3A), suggesting that they undertake nuclear DNA replication with close to normal fidelity and efficiency. However, the interactions of Mgm101 with the nuclear repair factors Mph1 and MutSα promoted us to examine a role for Mgm101 in nuclear DNA metabolism. From our fractionation studies we were able to demonstrate that Mgm101 resides in both the nucleus and mitochondria, consistent with a role in DNA repair in both compartments.
It is well-established that several pathways contribute to the repair of ICLs, and these are, at least partly, utilised in a cell cycle phase-dependent manner [9], [10], [38], [42]. Indeed, genetic and molecular evidence has been presented for a pathway involving the sequential action of NER followed by TLS, involving DNA polymerase ζ [6], [10]. This pathway is thought to predominate in the G1-phase and early S-phase of the cell cycle, where a favoured HRR substrate (sister chromatid) is absent. Moreover, the sensitivity profile of rad52 deficient yeast cells though the cell cycle suggests that Rad52 is indeed dispensable in the primary ICL repair pathway operating in G1-phase and in quiescent (stationary phase) cells [6], [38]. Further, it is well-established that ICLs induce replication-associated DSBs [42], [43], and that the repair of these DSBs is mainly dependent upon HRR, and hence Rad52 for their repair, in both yeast and mammalian cells [6], [7], [68]. Therefore, the fact that pso2 msh2 and pso2 mgm101 deficient cells accumulate ICL-associated DSBs argues that their repair by HR is blocked when both these pathways are deleted. This is corroborated by our observation that deleting RAD52 in pso2 mgm101, pso2 mph1 or pso2 msh2 strains does not lead to a further increase in sensitivity to ICL-inducing agents, and also by the delay in Rad51 chromatin recruitment we observed following ICL induction. Notably, a rad52 single mutant is less sensitive to ICLs than pso2 mph1, pso2 msh2 and pso2 mgm101 double mutants, indicating that together these factors control ICL repair events in addition to HRR. In this regard, NHEJ factors appear to be utilised to repair ICL-associated DSBs in the absence of Pso2. Indeed, we have previously observed that rad52 yku70 double disruptant has a more severe DSB repair defective phenotype than a rad52 single mutant following HN2 treatment, consistent with a role for both HRR and NHEJ in the repair of ICL-associated DSBs [6].
We therefore suggest that the primary defect in cells lacking Pso2 and components of the Mph1-Mgm101-MutSα (and Exo1) pathway lie in processing an ICL repair intermediate that must be dealt with prior to the initiation of the recombinational/DSB repair phase of ICL repair (see Figure 7 for a model). Compelling data showing that pso2 mutants incise ICLs has been presented by several groups [34]–[36]. This suggests that a tethered cross-linked oligonucleotide is a possible substrate for degradation by exonuclease activity of Pso2, by analogy with the postulated role for hSNM1A [43]. We speculate that in the absence of Pso2 activity the same intermediate is recognised by MutSα, Mph1 and Mgm101. This leads to recruitment of Exo1, which operates with the same polarity (5′-to-3′) as Pso2 [69], [70], to degrade the tethered oligonucleotide. In this regard, it is possible that the D-loop dissociation activity of Mph1 [23] controls commitment to the recombinational phase of repair until the ICL has been processed to provide a qualitatively useful substrate for HRR. Alternatively, or in addition, the ICL processing pathway that depends upon Mph1-Mgm101-MutSα might rely on the stable formation of a regressed fork for the recruitment of Exo1, another potential function ascribed to Mph1 [21]. Given the increase in GCRs induced by the presence of ICLs in pso2 mgm101/msh2/mph1 cells, the inability to process this ICL repair intermediate is not only highly toxic, but also leads to drastic rearrangements of the genome, presumably through the use of low fidelity pathways to heal broken chromosomes [57]. Clearly the biochemical analysis of ICL intermediates as substrates for Pso2, Mph1, MutSα, Mgm101 and Exo1 factors is an important future area of study.
Fig. 7. A model for the two major ICL processing pathways in budding yeast, one controlled by Pso2 and the other by Mph1-Mgm101-MutSα in collaboration with Exo1. 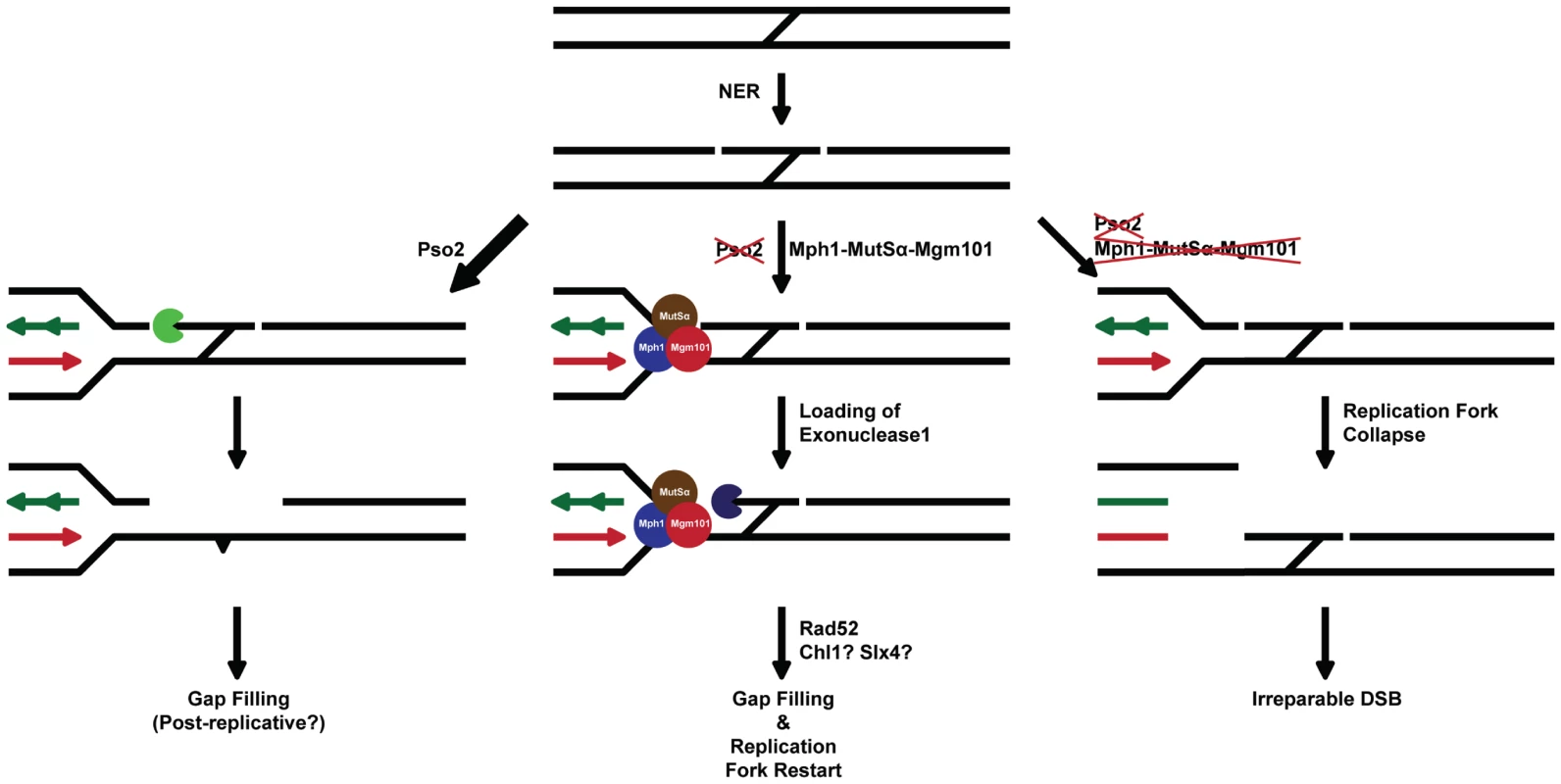
See associated text for details. Recent reports suggest that human MMR factors might be important effectors of the FA repair pathway [71]–[73], reminiscent of a role for MutSα collaborating with Mph1 during ICL repair in yeast. Our studies in yeast suggest that Pso2 is dominant over Mph1-Mgm101-MutSα and Exo1, as well as the FANCJ and FANCP homologs/orthologs Chl1 and Slx4, during ICL repair. However, it is possible that mammalian cells prioritise differently, sharing the burden between the FA pathway and SNM1A, since depletion of FA factors or SNM1A both produces pronounced ICL sensitisation in human cells. Indeed, a recent study indicated that breeding FANCD2−/− mice with SNM1−/− produced enhanced perinatal mortality, consistent with redundancy between these pathways in mammals [74]. Moreover, evidence that hSNM1A recruitment to ICLs is controlled by Rad18, in an FA pathway-independent manner, has recently been presented [75]. Finally, it appears that a significant number of tumours somatically inactivate the FA pathway [76]–[78] or the MMR apparatus [79], which might help drive initial genomic instability. In such cases the inhibition of human SNM1 factors might be a means for selective sensitisation to ICL-inducing anticancer drugs.
Materials and Methods
Chemicals and enzymes
Analytical grade nitrogen mustard, L-canavanine, methyl methanesulfonate, cisplatin and hydrogen peroxide were purchased from Sigma-Aldrich (Poole, UK). Arthrobacter Luteus Zymolyase-20T was obtained from MP Biomedicals (Cambridge, UK). 5-fluoroorotic acid (5-FOA) was from Zymo Reseach (Irvine, CA, USA).
Yeast strains
Strains used in this study are listed in Table S1. Gene deletion and C-terminal tagging were carried out by PCR-based micro-homology targeted gene disruption and targeting strategies, using the pFA6a vector series and its derivatives [80]–[82]. Some deletions were also generated by synthetic genetic analysis methodology [83]. Deletions and tagging were confirmed by PCR analysis and restriction enzyme digestion.
Antibodies
For detection of FLAG-tagged Mgm101 protein, an affinity-purified rabbit antibody to the FLAG epitope tag was obtained from Sigma-Aldrich. Polyclonal anti-6xHIS antibodies (Abcam) was used to detect 6xHIS-tagged proteins. An anti-GFP polyclonal antibody from Roche was used to detect Mph1-GFP. A rabbit polyclonal, ChIP-grade histone H3 (Abcam) and mouse monoclonal antibody to porin (Invitrogen) were used as controls for nuclear and mitochondrial localisation, respectively. Anti-Rad51 polyclonal antibodies were a kind gift of Patrick Sung. Immuno-detection of Rad53 was performed using a goat polyclonal antibody (Santa Cruz Biotechnology). Mouse anti-c-Myc (9E11, Abcam), mouse anti-HA (ab9110, Abcam), monoclonal mouse anti-FLAG (Sigma F1804) and goat polyclonal Msh2 (Santa Cruz Biotechnologies Inc, SC-26230) antibodies were employed in the gel filtration analysis.
Plasmids
MGM101 was amplified from yeast genomic DNA using primers containing cloning restriction sites and the nucleotide sequence for the FLAG epitope tag. Following amplification, the MGM101-FLAG construct was ligated into appropriately digested pYES2.0 yeast expression vector (Invitrogen, UK). Expression was induced by growth in glucose-free synthetic complete media containing 2% galactose and 1% raffinose as a carbon source.
Immunoprecipitation and co-immunoprecipitation
Galactose-induced (5 hours) cell cultures were split into two separate aliquots, spun down, washed once in ddH2O, and resuspended in lysis buffer (50 mM sodium phosphate, pH 7.4, 1 mM EDTA, 5% glycerol, supplemented with protease inhibitor cocktail and nuclease mix (both from Amersham Biosciences, Piscataway, NJ, USA), EDTA in lysis buffer was omitted in case of TALON Metal Affinity resin binding assay. Cells were disrupted by sonication and whole cell extracts were cleared by centrifugation. Six milligrams of whole cell extracts were incubated with Anti-FLAG M2 Affinity Gel (Sigma-Aldrich, St. Louis, MO, USA) or TALON Metal Affinity Resin (Clontech, Palo Alta, CA, USA), and affinity purification of FLAG - and 6xHIS-tagged proteins was performed according to the protocols outlined in the manufacturer's manuals.
Liquid chromatography and MALDI-TOF/MS-based protein identification
Anti-FLAG M2 Affinity gel elute was subjected to the liquid chromatography (EASY -NLC, Bruker, Germany). A total of nine potential peaks (Cutoff >400 intensity) were selected for further protein analysis. Proteins were trypsin digested automatically using a Proteineer DP protein digestion station (Bruker-Daltonics, Bremen, Germany). An aliquot of digestion solution was mixed with an aliquot of α-cyano-4-hydroxycinnamic acid (Bruker-Daltonics) in 33% aqueous acetonitrile and 0.25% trifluoroacetic acid. This mixture was deposited onto a 600 µm AnchorChip prestructured MALDI probe (Bruker-Daltonics) and allowed to dry at room temperature. MALDI-MS data were obtained in an automated analysis loop using an Ultraflex time-of-flight (TOF) mass spectrometer (Bruker-Daltonics) equipped with a LIFT-MS/MS device [84]. Spectra were acquired in the positive-ion mode at 50 Hz laser frequency, and 100 to 1000 individual spectra were averaged. Subsequently, selected precursor ions were subject to fragment ion analysis in the tandem time-of-flight (TOF/TOF) mode to obtain the corresponding MALDI-MS/MS spectra. Automated analysis of mass data was performed using the flexAnalysis software (Bruker-Daltonics). MALDI-MS and MALDI-MS/MS data were combined through the BioTools program (Bruker-Daltonics) to search a non-redundant protein database (NCBInr) using Mascot software (Matrix Science, London, UK). MALDI-MS and MALDI-MS/MS spectra as well as database search results were inspected in detail using flexAnalysis as well as in-house software.
Yeast cellular fractionation
Fractionation of yeast to isolate nuclear and mitochondrial fractions were performed as described [85], [86], with minor modifications.
Gel filtration analysis
Approximately 60 mg of yeast whole cell extract prepared from a strain harbouring Msh6-HA, Mph1-FLAG and Mgm101-Myc constructs was loaded in a 200 ml volume onto a Superdex 200 column on an AKTA FPLC (Amersham Pharmacia Biotech) equilibrated with (25 mM HEPES pH 7.9, 10% glycerol, 150 mM NaCl, 1 mM EDTA and 1 mM DTT). The flow rate was set at 0.5 ml/min and fractions of 2 ml were collected following a void volume of 40 ml. The proteins from aliquots were subjected to SDS-PAGE on a 4–12% Bis-Tris gel (Invitrogen) and blotted onto Hybond-C (Amersham) membranes.
Chromatin binding assays
These were performed as described [86].
Nitrogen mustard, cisplatin, methyl methanesulfonate, ultraviolet light, and hydrogen peroxide sensitivity assays
Exponentially growing cells were diluted in serial in PBSA to a concentration of 2×107 cells/ml and incubated with HN2, CDDP, MMS or H2O2 at the stated doses for 1 hour at 30°C with shaking, or exposed to 254 nm UV light at the doses shown. Samples were subsequently plated onto YPD plates and incubated at 30°C for 3 days before scoring survival.
Canavanine resistance forward mutation assays
These were performed as previously described [87].
Pulsed-field gel electrophoresis analysis
This was performed as previously described [6], except plugs were analysed on a Beckmann TAFE machine.
Mitochondrial petite induction
Yeast cells were cultivated in YPD media until the stationary phase of growth was reached. The cells were then collected by centrifugation and washed with, and resuspended in, sterile water to the final concentration of 2×107 cells/ml. The resulting cell suspension was subsequently treated with 10 µM HN2 or 100 µM H2O2 for 1 hour at 30°C. After treatment, cells were harvested by centrifugation, and washed twice with, and resuspended in, potassium phosphate buffer. Appropriately diluted cell suspension was plated in triplicate onto YPD, YPG or YPGE plates. Plates were incubated at 30°C for 2–3 days (YPD), 5 days (YPG, YPGE) before being scored.
Measurement of Gross Chromosomal Rearrangements
Yeast cultures grown in SC media overnight were inoculated into fresh YPD media, where incubation continued until the cell suspension reached a density of 2×107 cells/ml. 2×1010 of cells were collected by centrifugation, washed with, and resuspended in, potassium phosphate buffer to the final concentration of 2×108 cells/ml. The cell suspension was treated with 10 µM HN2 for 1 hour at 30°C. After treatment, cells were harvested, washed twice with, and resuspended in, potassium phosphate buffer. Cell suspension was diluted and plated in triplicate onto YPD plates to determine cell viability. For GCR measurement, the undiluted cell suspension (approximately 1010 of cells) was plated on 5-FOA plates supplemented with canavanine.
Supporting Information
Zdroje
1. McHughPJ, SpanswickVJ, HartleyJA (2001) Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol 2 : 483–490.
2. DronkertML, KanaarR (2001) Repair of DNA interstrand cross-links. Mutat Res 486 : 217–247.
3. LehoczkyP, McHughPJ, ChovanecM (2007) DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol Rev 31 : 109–133.
4. RuhlandA, BrendelM (1979) Mutagenesis by cytostatic alkylating agents in yeast strains of differing repair capacities. Genetics 92 : 83–97.
5. MillerRD, PrakashL, PrakashS (1982) Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol Cell Biol 2 : 939–948.
6. McHughPJ, SonesWR, HartleyJA (2000) Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol 20 : 3425–3433.
7. JachymczykWJ, von BorstelRC, MowatMR, HastingsPJ (1981) Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol Gen Genet 182 : 196–205.
8. ZhengH, WangX, WarrenAJ, LegerskiRJ, NairnRS, et al. (2003) Nucleotide excision repair - and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol Cell Biol 23 : 754–761.
9. NiedernhoferLJ, OdijkH, BudzowskaM, van DrunenE, MaasA, et al. (2004) The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol 24 : 5776–5787.
10. SarkarS, DaviesAA, UlrichHD, McHughPJ (2006) DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. Embo J 25 : 1285–1294.
11. RaschleM, KnipscheerP, EnoiuM, AngelovT, SunJ, et al. (2008) Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134 : 969–980.
12. NiedzwiedzW, MosedaleG, JohnsonM, OngCY, PaceP, et al. (2004) The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 15 : 607–620.
13. KnipscheerP, RaschleM, SmogorzewskaA, EnoiuM, HoTV, et al. (2009) The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326 : 1698–1701.
14. LongDT, RaschleM, JoukovV, WalterJC (2011) Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333 : 84–87.
15. JoenjeH, PatelKJ (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet 2 : 446–457.
16. MeeteiAR, MedhurstAL, LingC, XueY, SinghTR, et al. (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37 : 958–963.
17. MosedaleG, NiedzwiedzW, AlpiA, PerrinaF, Pereira-LealJB, et al. (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol 12 : 763–771.
18. EntianKD, SchusterT, HegemannJH, BecherD, FeldmannH, et al. (1999) Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet 262 : 683–702.
19. SchellerJ, SchurerA, RudolphC, HettwerS, KramerW (2000) MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155 : 1069–1081.
20. SchurerKA, RudolphC, UlrichHD, KramerW (2004) Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics 166 : 1673–1686.
21. SunW, NandiS, OsmanF, AhnJS, JakovleskaJ, et al. (2008) The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32 : 118–128.
22. BanerjeeS, SmithS, OumJH, LiawHJ, HwangJY, et al. (2008) Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J Cell Biol 181 : 1083–1093.
23. PrakashR, SatoryD, DrayE, PapushaA, SchellerJ, et al. (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23 : 67–79.
24. PrakashR, KrejciL, Van KomenS, Anke SchurerK, KramerW, et al. (2005) Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem 280 : 7854–7860.
25. GariK, DecailletC, StasiakAZ, StasiakA, ConstantinouA (2008) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29 : 141–148.
26. XueY, LiY, GuoR, LingC, WangW (2008) FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet
27. RosadoIV, NiedzwiedzW, AlpiAF, PatelKJ (2009) The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res 37 : 4360–4370.
28. SchwabRA, BlackfordAN, NiedzwiedzW (2010) ATR activation and replication fork restart are defective in FANCM-deficient cells. Embo J 29 : 806–818.
29. CollisSJ, CicciaA, DeansAJ, HorejsiZ, MartinJS, et al. (2008) FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32 : 313–324.
30. Luke-GlaserS, LukeB, GrossiS, ConstantinouA (2010) FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. Embo J 29 : 795–805.
31. HenriquesJA, MoustacchiE (1980) Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95 : 273–288.
32. RuhlandA, HaaseE, SiedeW, BrendelM (1981) Isolation of yeast mutants sensitive to the bifunctional alkylating agent nitrogen mustard. Mol Gen Genet 181 : 346–351.
33. RuhlandA, KircherM, WilbornF, BrendelM (1981) A yeast mutant specifically sensitive to bifunctional alkylation. Mutat Res 91 : 457–462.
34. GrossmannKF, WardAM, MosesRE (2000) Saccharomyces cerevisiae lacking Snm1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat Res 461 : 1–13.
35. Magana-SchwenckeN, HenriquesJA, ChanetR, MoustacchiE (1982) The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci U S A 79 : 1722–1726.
36. WilbornF, BrendelM (1989) Formation and stability of interstrand cross-links induced by cis - and trans-diamminedichloroplatinum (II) in the DNA of Saccharomyces cerevisiae strains differing in repair capacity. Curr Genet 16 : 331–338.
37. LiX, MosesRE (2003) The beta-lactamase motif in Snm1 is required for repair of DNA double-strand breaks caused by interstrand crosslinks in S. cerevisiae. DNA Repair (Amst) 2 : 121–129.
38. BarberLJ, WardTA, HartleyJA, McHughPJ (2005) DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol 25 : 2297–2309.
39. De SilvaIU, McHughPJ, ClingenPH, HartleyJA (2000) Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol 20 : 7980–7990.
40. AkkariYM, BatemanRL, ReifsteckCA, OlsonSB, GrompeM (2000) DNA replication is required To elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol 20 : 8283–8289.
41. BesshoT (2003) Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J Biol Chem 278 : 5250–5254.
42. HanadaK, BudzowskaM, ModestiM, MaasA, WymanC, et al. (2006) The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J 25 : 4921–4932.
43. WangAT, SengerovaB, CattellE, InagawaT, HartleyJM, et al. (2011) Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 25 : 1859–1870.
44. ZhangN, LuX, ZhangX, PetersonCA, LegerskiRJ (2002) hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol Cell Biol 22 : 2388–2397.
45. WuQ, ChristensenLA, LegerskiRJ, VasquezKM (2005) Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep 6 : 551–557.
46. LanL, HayashiT, RabeyaRM, NakajimaS, KannoS, et al. (2004) Functional and physical interactions between ERCC1 and MSH2 complexes for resistance to cis-diamminedichloroplatinum(II) in mammalian cells. DNA Repair (Amst) 3 : 135–143.
47. HoY, GruhlerA, HeilbutA, BaderGD, MooreL, et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415 : 180–183.
48. ChenXJ, GuanMX, Clark-WalkerGD (1993) MGM101, a nuclear gene involved in maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Nucleic Acids Res 21 : 3473–3477.
49. MeeusenS, TieuQ, WongE, WeissE, SchieltzD, et al. (1999) Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J Cell Biol 145 : 291–304.
50. MeeusenS, NunnariJ (2003) Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol 163 : 503–510.
51. MbantenkhuM, WangX, NardozziJD, WilkensS, HoffmanE, et al. (2011) Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J Biol Chem
52. ZuoX, XueD, LiN, Clark-WalkerGD (2007) A functional core of the mitochondrial genome maintenance protein Mgm101p in Saccharomyces cerevisiae determined with a temperature-conditional allele. FEMS Yeast Res 7 : 131–140.
53. DrummondJT, GenschelJ, WolfE, ModrichP (1997) DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci U S A 94 : 10144–10149.
54. ContamineV, PicardM (2000) Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev 64 : 281–315.
55. ReenanRA, KolodnerRD (1992) Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics 132 : 963–973.
56. ChenC, KolodnerRD (1999) Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23 : 81–85.
57. KolodnerRD, PutnamCD, MyungK (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297 : 552–557.
58. MyungK, DattaA, ChenC, KolodnerRD (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet 27 : 113–116.
59. SmithS, HwangJY, BanerjeeS, MajeedA, GuptaA, et al. (2004) Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 101 : 9039–9044.
60. TishkoffDX, BoergerAL, BertrandP, FilosiN, GaidaGM, et al. (1997) Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A 94 : 7487–7492.
61. TranPT, ErdenizN, DudleyS, LiskayRM (2002) Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 1 : 895–912.
62. SymingtonLS, GautierJ (2010) Double-Strand Break End Resection and Repair Pathway Choice. Annu Rev Genet
63. WhiteMF (2009) Structure, function and evolution of the XPD family of iron-sulfur-containing 5′→3′ DNA helicases. Biochem Soc Trans 37 : 547–551.
64. StoepkerC, HainK, SchusterB, Hilhorst-HofsteeY, RooimansMA, et al. (2011) SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet 43 : 138–141.
65. CrossanGP, van der WeydenL, RosadoIV, LangevinF, GaillardPH, et al. (2011) Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet 43 : 147–152.
66. KimY, LachFP, DesettyR, HanenbergH, AuerbachAD, et al. (2011) Mutations of the SLX4 gene in Fanconi anemia. Nat Genet 43 : 142–146.
67. ZuoXM, Clark-WalkerGD, ChenXJ (2002) The mitochondrial nucleoid protein, Mgm101p, of Saccharomyces cerevisiae is involved in the maintenance of rho(+) and ori/rep-devoid petite genomes but is not required for hypersuppressive rho(−) mtDNA. Genetics 160 : 1389–1400.
68. De SilvaIU, McHughPJ, ClingenPH, HartleyJA (2002) Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res 30 : 3848–3856.
69. LiX, HejnaJ, MosesRE (2005) The yeast Snm1 protein is a DNA 5′-exonuclease. DNA Repair (Amst) 4 : 163–170.
70. HazratiA, Ramis-CastelltortM, SarkarS, BarberLJ, SchofieldCJ, et al. (2007) Human SNM1A rescues the DNA repair defects of yeast pso2 mutants. DNA Repair (Amst) in press.
71. PengM, LitmanR, XieJ, SharmaS, BroshRMJr, et al. (2007) The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. Embo J 26 : 3238–3249.
72. WilliamsSA, WilsonJB, ClarkAP, Mitson-SalazarA, TomashevskiA, et al. (2011) Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet
73. HuangM, KennedyR, AliAM, MoreauLA, MeeteiAR, et al. (2011) Human MutS and FANCM complexes function as redundant DNA damage sensors in the Fanconi Anemia pathway. DNA Repair (Amst)
74. HemphillAW, BruunD, ThrunL, AkkariY, TorimaruY, et al. (2008) Mammalian SNM1 is required for genome stability. Mol Genet Metab 94 : 38–45.
75. YangK, MoldovanGL, D'AndreaAD (2010) RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J Biol Chem 285 : 19085–19091.
76. MarsitCJ, LiuM, NelsonHH, PosnerM, SuzukiM, et al. (2004) Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene 23 : 1000–1004.
77. WreesmannVB, EstiloC, EiseleDW, SinghB, WangSJ (2007) Downregulation of Fanconi anemia genes in sporadic head and neck squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec 69 : 218–225.
78. TaniguchiT, TischkowitzM, AmezianeN, HodgsonSV, MathewCG, et al. (2003) Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med 9 : 568–574.
79. JiricnyJ (1998) Eukaryotic mismatch repair: an update. Mutat Res 409 : 107–121.
80. WachA, BrachatA, PohlmannR, PhilippsenP (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 : 1793–1808.
81. LongtineMS, McKenzieA3rd, DemariniDJ, ShahNG, WachA, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 : 953–961.
82. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
83. TongAH, EvangelistaM, ParsonsAB, XuH, BaderGD, et al. (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294 : 2364–2368.
84. SuckauD, ResemannA, SchuerenbergM, HufnagelP, FranzenJ, et al. (2003) A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem 376 : 952–965.
85. ZinserE, DaumG (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11 : 493–536.
86. LiangC, StillmanB (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev 11 : 3375–3386.
87. McHughPJ, GillRD, WatersR, HartleyJA (1999) Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res 27 : 3259–3266.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



