-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
Ants are a highly successful family of insects that thrive in a variety of habitats across the world. Perhaps their best-known features are complex social organization and strict division of labor, separating reproduction from the day-to-day maintenance and care of the colony, as well as strict discrimination against foreign individuals. Since these social characteristics in ants are thought to be mediated by semiochemicals, a thorough analysis of these signals, and the receptors that detect them, is critical in revealing mechanisms that lead to stereotypic behaviors. To address these questions, we have defined and characterized the major chemoreceptor families in a pair of behaviorally and evolutionarily distinct ant species, Camponotus floridanus and Harpegnathos saltator. Through comprehensive re-annotation, we show that these ant species harbor some of the largest yet known repertoires of odorant receptors (Ors) among insects, as well as a more modest number of gustatory receptors (Grs) and variant ionotropic glutamate receptors (Irs). Our phylogenetic analyses further demonstrate remarkably rapid gains and losses of ant Ors, while Grs and Irs have also experienced birth-and-death evolution to different degrees. In addition, comparisons of antennal transcriptomes between sexes identify many chemoreceptors that are differentially expressed between males and females and between species. We have also revealed an agonist for a worker-enriched OR from C. floridanus, representing the first case of a heterologously characterized ant tuning Or. Collectively, our analysis reveals a large number of ant chemoreceptors exhibiting patterns of differential expression and evolution consistent with sex/species-specific functions. These differentially expressed genes are likely associated with sex-based differences, as well as the radically different social lifestyles observed between C. floridanus and H. saltator, and thus are targets for further functional characterization. Our findings represent an important advance toward understanding the molecular basis of social interactions and the differential chemical ecologies among ant species.
Published in the journal: . PLoS Genet 8(8): e32767. doi:10.1371/journal.pgen.1002930
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002930Summary
Ants are a highly successful family of insects that thrive in a variety of habitats across the world. Perhaps their best-known features are complex social organization and strict division of labor, separating reproduction from the day-to-day maintenance and care of the colony, as well as strict discrimination against foreign individuals. Since these social characteristics in ants are thought to be mediated by semiochemicals, a thorough analysis of these signals, and the receptors that detect them, is critical in revealing mechanisms that lead to stereotypic behaviors. To address these questions, we have defined and characterized the major chemoreceptor families in a pair of behaviorally and evolutionarily distinct ant species, Camponotus floridanus and Harpegnathos saltator. Through comprehensive re-annotation, we show that these ant species harbor some of the largest yet known repertoires of odorant receptors (Ors) among insects, as well as a more modest number of gustatory receptors (Grs) and variant ionotropic glutamate receptors (Irs). Our phylogenetic analyses further demonstrate remarkably rapid gains and losses of ant Ors, while Grs and Irs have also experienced birth-and-death evolution to different degrees. In addition, comparisons of antennal transcriptomes between sexes identify many chemoreceptors that are differentially expressed between males and females and between species. We have also revealed an agonist for a worker-enriched OR from C. floridanus, representing the first case of a heterologously characterized ant tuning Or. Collectively, our analysis reveals a large number of ant chemoreceptors exhibiting patterns of differential expression and evolution consistent with sex/species-specific functions. These differentially expressed genes are likely associated with sex-based differences, as well as the radically different social lifestyles observed between C. floridanus and H. saltator, and thus are targets for further functional characterization. Our findings represent an important advance toward understanding the molecular basis of social interactions and the differential chemical ecologies among ant species.
Introduction
The family of insects commonly known as ants (family Formicidae) originated during the Cretaceous period, approximately 140 million years ago [1]. Since that time, they have established a global presence, with only the most remote locations lacking ant species [2]. Indeed, in some cases, such as lowland tropical rainforest canopies, ants have come to dominate the biomass [3], [4]. Their ecological success is reflected in the number and diversity of ants, of which there were 283 known genera [5].
There is a wide diversity in the behavior and morphology of different ant subfamilies that includes both the level and complexity of social organizations. For instance, Camponotus floridanus (the Florida Carpenter Ant), is a Formicine ant from the South-Eastern United States which belongs to one of the most globally prevalent ant genera [6]. These ants feature a rigid caste structure, with strict division of labor between the reproductive queens and the non-reproductive workers that is primarily regulated through pheromones [7], [8], [9]. Workers have a high threshold to lay eggs, and regulation of their reproduction through aggressive interactions does not occur [10]. Furthermore, the worker caste is divided into two classes: minor workers and major workers, which differ in size and morphology [2], [6]. On the other hand, Harpegnathos saltator, a predatory species of Ponerine ant endemic to India and Sri Lanka is characterized by a more flexible reproductive system. H. saltator colonies are relatively small (averaging 65 to 225 individuals, depending on season and region) [11], and queen to worker dimorphism is weak [11], [12]. When a H. saltator colony loses its queen, one or more of the workers will begin laying eggs and become functional reproductives (referred to as gamergates) [12] and this behavioral transition is initiated with strong aggressive interactions [13].
Sociality in ants is considered to be a simple model for complex behaviors in humans and other mammals [14]. The success of ants is thought to have arisen in large part from their well-developed eusociality, wherein individuals live together in colonies with one or several highly fertile female “queens” surrounded by a host of non-reproductive female “workers.” These workers then support and defend the queen and her progeny. The fact that the workers are the queen's own daughters is thought to provide the evolutionary advantage for the workers to protect and support the queen [6].
While it is generally accepted that a variety of chemical signals mediate many of the interactions between these castes, as well as interactions between individuals from competing colonies, there is great interest in determining the particular pheromones and their cognate molecular receptors that mediate these interactions [2]. It is likely that these semiochemicals are initially detected in peripheral sensory neurons by members of three major insect chemosensory receptor gene families: odorant receptors (Ors) [15], [16], [17], [18], [19], gustatory receptors (Grs) [15], [20], [21], [22], [23], and the more recently discovered variant ionotropic glutamate receptors (Irs) [24], [25], [26].
Ors and Grs belong to the same superfamily and both encode seven-transmembrane-domain proteins [17], [22]. Ors are mainly expressed in olfactory receptor neurons (ORNs) within sensory appendages such as antennae and maxillary palps, where they are responsible for the perception of volatile chemical signals [17], [19]. Conventional insect Ors (so-called “tuning” Ors) are associated with odorant specificity. They are typically highly divergent and their orthologous relationships are usually difficult to determine even within order (e.g. Drosophila vs. Anopheles [27], and Nasonia vs. Apis [28]). In contrast, one member of this gene family, which is now uniformly known as Orco, is both highly conserved across insect orders and widely expressed in a majority of ORNs [29], [30]. Orco is necessary and sufficient for the proper localization and retention of other tuning Ors at the dendritic membrane, and is required for proper function of tuning Ors [29], [31]. Rather than playing a role in odorant specificity, Orco forms an essential part of a heteromeric ion channel in cooperation with a tuning Or that is gated by its cognate odor ligand [32], [33], [34], [35], [36].
In contrast with the Ors, Grs are highly expressed in gustatory organs [20], [21], [22], and a large portion of these receptors respond to soluble tastants [37], [38], [39] and pheromones [40], [41], [42], leading to the “gustatory” designation for this group of chemoreceptors. However, there are some exceptions; for example, one unusual group of Grs respond to the volatile chemical carbon dioxide [43], [44], demonstrating that members of this receptor family are not necessarily limited to gustatory or pheromonal responses. This is further supported by the expression of some Grs in non-gustatory organs such as the arista and Johnston's organ [45].
Irs are homologous to ionotropic glutamate receptors (iGluRs) and thus are evolutionarily unrelated to Ors and Grs [24], [26]. The role of IRs as chemosensory receptors has recently been uncovered based on multiple lines of evidence, including their divergence from conventional iGluRs at sequence level and the expression of several Irs in chemosensory neurons [24]. While Irs are generally thought to mediate responses to acids and amines [25], members of this family of chemosensory receptors may also sense other classes of chemicals.
We hypothesize that the striking contrast between C. floridanus, with its strict queen-worker dimorphism and largely pheromone-regulated reproduction, and H. saltator, with its flexible reproductive system that is associated with behavioral and pheromonal regulation of reproduction, is correlated with distinctive semiochemical and chemoreceptor profiles, which in turn generate differences in their chemical ecologies. The same is likely to be true of caste - or sex-based differences in behavior within each species.
To test these hypotheses, we first developed a custom gene annotation pipeline to comprehensively describe the chemosensory receptor repertoires of C. floridanus and H. saltator. We then investigated the evolutionary patterns (e.g. gene gain-and-loss) of these chemosensory receptor genes, in order to gain insight on their functional diversification. Furthermore, we performed RNAseq analyses of caste - and sex-specific antennal transcriptomes to identity chemoreceptors that are differentially expressed between males/females and between species. We found multiple clades of chemosensory receptor genes that show differential expansion/contraction among ant species. In addition, a large number of chemosensory receptor genes exhibited sex-specific expression or male/female-enrichment. These chemosensory receptor genes exhibiting interesting evolutionary and expression patterns may have potentially contributed to the different chemical ecology between sexes/species. We also successfully identified agonists for two Or genes to further validate these annotations. The findings of this study inform us as to the genetic basis for the differences in chemical ecology between C. floridanus and H. saltator, as well as the potential role of chemosensory receptors in the biology and evolution of eusociality in ants.
Results
Annotation of C. floridanus and H. saltator chemosensory receptor genes
The automated genome annotations of C. floridanus and H. saltator revealed about 100 Or and about 10 Gr genes [46], which is substantially fewer than the number of Or and Gr genes in two other sequenced ant genomes (e.g. argentine ant: Linepithema humile [47], and harvester ant: Pogonomyrmex barbatus [48]; Figure 1). These low numbers were not surprising because the annotation of Or/Gr genes in other insect genomes has been difficult and usually requires extensive manual efforts [47], [48]. In order to address this potential discrepancy and comprehensively elucidate the genomic repertoire of chemosensory receptor genes in C. floridanus and H. saltator, we rigorously re-annotated Or, Gr, and Ir genes in these two ant species using a custom automated pipeline followed by careful manual inspection.
Fig. 1. Annotation of C. floridanus and H. saltator chemosensory receptor genes. 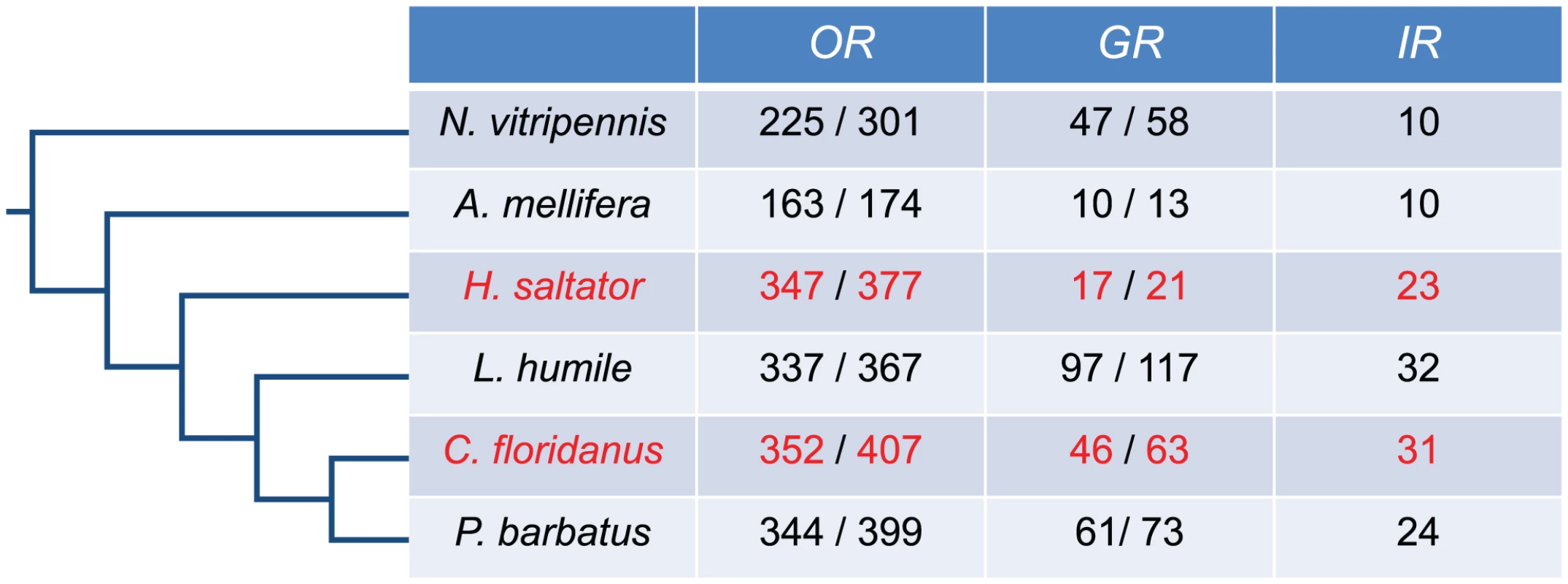
Number of Or, Gr, and Ir gene predictions in six hymenopteran species. For Or and Gr genes, the number to the right is the number of all gene models (coding for proteins longer than 300 aa in C. floridanus and H. saltator, or 200 aa in other species), while the number to the left is the number of seemingly intact gene models. To maximize the sensitivity of our re-annotation, we collected reported Or, Gr, and Ir gene sequences from other sequenced Hymenoptera and insect relatives of C. floridanus and H. saltator, including Apis mellifera, Acyrthosiphon pisum, Drosophila melanogaster, Nasonia vitripennis, L. humile, and P. barbatus. These insect chemosensory receptor genes were used to identify putative Or/Gr/Ir coding regions within the C. floridanus and H. saltator genomes and to guide homology-based gene prediction. As a result, we discovered a large number of previously unannotated chemosensory receptor genes and corrected several previously reported gene models [46]. All these annotations were manually inspected in multiple sequences alignments to identify and correct for potential errors (e.g. missing exons, unrelated sequences). This analysis indicates that C. floridanus contains 407 putative Or coding loci, of which 352 loci encode intact Or genes, which is similar to those newly annotated in H. saltator, with 377 loci in total and 347 intact loci (all chemosensory receptor genes annotated in this study are available in Dataset S1). The number of Ir predictions is also similar between the two ants, with 31 Ir genes in C. floridanus and 23 in H. saltator. On the other hand, C. floridanus contains 46 intact Gr genes, which is significantly higher than the 17 intact Gr genes found in H. saltator (Figure 1). Moreover, all three families of chemosensory receptor genes exhibited high degrees of sequence divergence among family members (Table S1).
In addition to the chemosensory receptor genes listed above, we also found a large number of incomplete gene models in these two ant genomes. For example, in C. floridanus and H. saltator, there are respectively ∼100 and ∼80 Or gene models encoding proteins shorter than 300 amino acids. In parallel to the difference in intact Gr genes, only three fragmented Gr gene models were found in H. saltator, while C. floridanus has ∼30 short Gr genes. Close examination of their genomic sequences revealed two principal mechanisms apparently leading to these fragmented Or/Gr gene models: 1) the presence of multiple frame-shift mutations and premature stop-codons, suggesting that they represent pseudogenes; and 2) their locations around undetermined genomic regions (e.g. edges of contigs/scaffolds), indicative of incomplete assembly as expected from a draft genome. The latter mechanism explains about 80% of the incomplete gene models.
Furthermore, similar to other insects [28], [47], [48], [49], [50], [51], most chemosensory receptor genes are tandemly arrayed in the C. floridanus and H. saltator genomes. In both cases, about 75% of Or genes are located in gene clusters of 4 to about 40 genes, and these occur in 24 and 20 Or gene clusters (n≥4) in C. floridanus and H. saltator, respectively (Figure S1). Although to a lesser degree than the Ors, half of the Gr and Ir genes in both ants have at least one neighboring homolog.
Phylogenetic analysis
To better understand the evolutionary history of chemosensory receptor genes in the two ant species, we performed Hymenoptera-wide phylogenetic analysis on each of the OR, GR, and IR gene families. Additional analyses including D. melanogaster and Tribolium castaneum showed that most relationships among hymenopteran and non-hymenopteran sequences were not resolved within the OR and GR families (see below). In this study, while they are generally categorized as belonging to the same receptor superfamily [22], we elected to analyze the OR and GR families separately due to their high level of divergence.
OR family
Our phylogenetic analysis of hymenopteran Or genes revealed a highly dynamic evolutionary history of this gene family featuring rapid gene birth and death (Figure 2A). Due to the rapid divergence of Or genes (average amino acid distance = 2.56; overall protein sequence identity = 19.45%), most deep relationships in the OR phylogeny lacked support (see Figure S2 and Dataset S2 for the full version of OR phylogeny with gene names and bootstrap values). In spite of this, we found 24 well-supported clades (referred to as subfamilies; A-V, Orco, and 9-exon in Figure 2), each potentially representing one Or gene copy in the common ancestor of Hymenoptera (also see Figure S3 for the OR phylogeny with D. melanogaster and T. castaneum sequences). These subfamilies exhibited vastly different patterns of expansion/contraction, which can be divided into three types (Figure 2A, 2B): 1) strict single-copy representation in each of the six analyzed hymenopterans was observed for the Orco subfamily, which is the only Or gene with clear orthologous relationships throughout insects [15], [28], [50], [51], [52], [53], [54] (Figure S3); 2) 11 subfamilies showed either gene loss only, or a limited number of gene duplication events (e.g. B and C); 3) the remaining 12 subfamilies had experienced substantial expansions within and/or shared by hymenopteran lineages (e.g. A and D).
Fig. 2. Phylogenetic relationships of Hymenoptera Or genes. 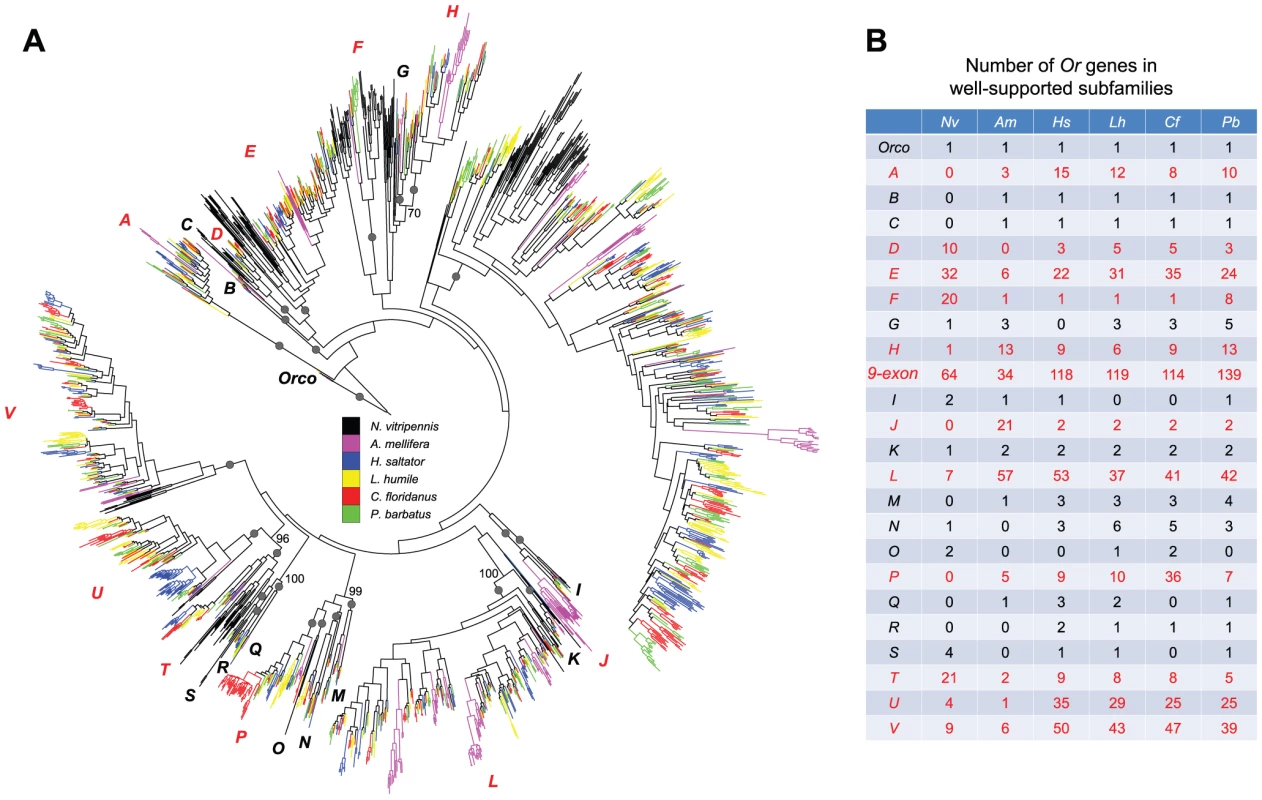
(A) A maximum-likelihood tree of hymenopteran Or genes estimated by using RAxML with Le-Gascuel (LG) model. Reliability of internal nodes was evaluated by 100 bootstrap replicates. Grey round dots indicate well-supported subfamilies (bootstrap value ≥80). Bootstrap values ≥70 are shown for relationships among subfamilies. Ors in different species are color-coded as following: N. vitripennis, black; A. mellifera, purple; H. saltator, blue; L. humile, yellow; P. barbatus, green; and C. floridanus, red. Subfamilies with rapid changes in gene copy numbers are highlighted in red. (B) Numbers of hymenopteran Or genes in well supported subfamilies. In particular, the most dramatic expansion was found in the subfamily composed of Or genes with 9 exons (Figure 2A, 2B). This 9-exon subfamily encompasses more than 30% of the entire repertoires of Or genes in the six hymenopterans, which is in agreement with previous observations in other ants [47], [48]. Furthermore, our analysis revealed highly dynamic Or evolution within these subfamilies; subclades were often differentially expanded and/or contracted in different species and rapid expansions were usually accompanied by frequent gene losses (Figure S4).
As described above, most Or genes in C. floridanus and H. saltator were found in tandem arrays in their respective genomes. Our phylogenetic results provided further evidence that these clustered Or genes were derived from tandem whole-gene duplication events. Moreover, more than 60% of all tandem duplicates in the two ants were due to lineage-specific expansions, while the others were generated during or even before the divergence of Hymenoptera. For example, the neighboring Or genes on C. floridanus scaffold538 and H. saltator scaffold105 belonged to four different subfamilies, suggesting that these two gene clusters were established before the divergence of the six hymenopterans, and underwent further expansion within each lineage (Figure S5).
Although Hymenoptera-wide orthology of Or genes may have been obscured by rapid gene gain and loss, we were still able to identify several clear 1-to-1 relationships among Formicidae. In total, we found 35 ant-specific clades that are composed of a single copy of Or gene from each of the four ants. Among these, genes in 30 clades are located in gene clusters while all others occur as singletons. A chi-square test showed that neither tandem duplicates nor singletons are significantly enriched in the 35 orthologous gene clades (p-value = 0.05).
GR family
Similar to the OR family, the GR phylogeny provided evidence for birth-and-death evolution in this family (Figure 3; see Figure S6 and Dataset S3 for the full version of GR phylogeny with gene names and bootstrap values). Within the Gr phylogeny, 13 well-supported subfamilies were found, most of which were likely generated by ancestral duplications before the divergence of Hymenoptera (although the precise relationships among them remained unresolved). Within Hymenoptera, ancestral duplications and/or lineage-specific expansions were found in most subfamilies, except for the GR1 and GR2 subfamilies. Indeed, significant lineage-specific expansions of ant Gr genes include the GR3 (L. humile and P. barbatus) and GR8/9 (C. floridanus) subfamilies. The most dramatic expansion was observed in an ant-specific subfamily which underwent multiple rounds of amplifications before and after the separation of H. saltator, especially within C. floridanus and L. humile. In contrast to the other three ants, H. saltator specific duplication was only observed once (in the GR4 subfamily), which explains the low number of Gr genes in this species. Moreover, our GR phylogeny showed that the formation of Gr gene clusters was likely due to tandem duplication, highlighting the importance of this duplication mechanism in the evolution of chemosensory receptor genes.
Fig. 3. Phylogenetic relationships of Hymenoptera Gr genes. 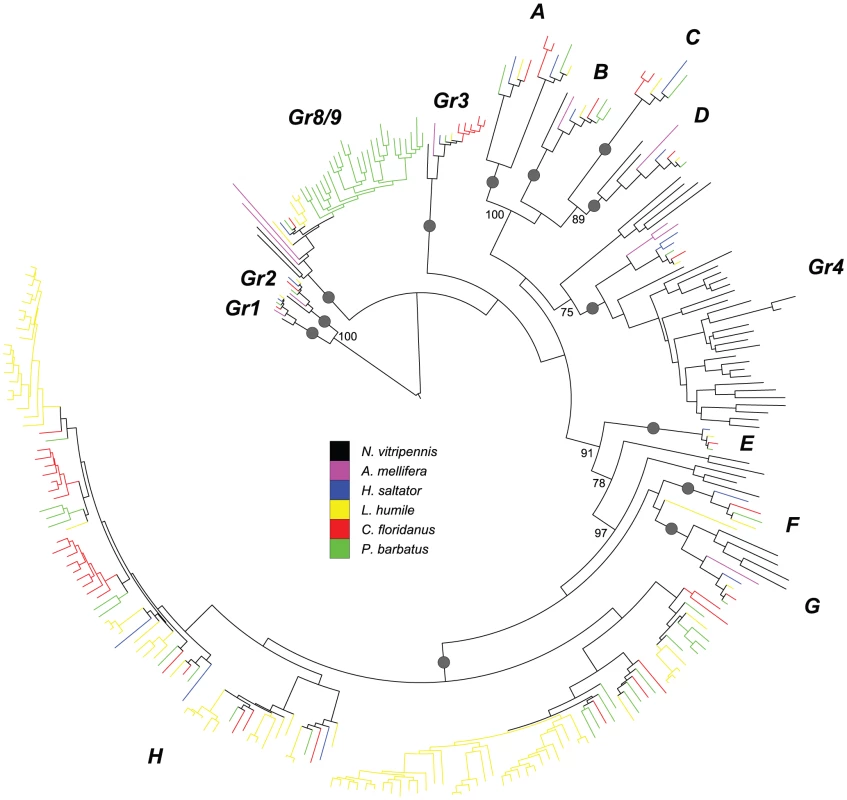
A maximum-likelihood tree of hymenopteran Gr genes estimated by using RAxML with Le-Gascuel (LG) model. Reliability of internal nodes was evaluated by 100 bootstrap replicates. Grey round dots indicate well-supported subfamilies (bootstrap value ≥80). Bootstrap values ≥70 are shown for relationships among subfamilies. Subfamilies showing interesting evolutionary patterns are named after the orthologs in N. vitripennis and A. mellifera. The other subfamilies are named as A–H. Grs in different species are color-coded as following: N. vitripennis, black; A. mellifera, purple; H. saltator, blue; L. humile, yellow; P. barbatus, green; and C. floridanus, red. Among the Grs, orthologs of the known sugar receptor genes (Gr1 and Gr2) [55], [56], [57], [58] and another insect-wide conserved Gr, D. melanogaster Gr43a (Gr3) [28], [53], [54], were observed in all of the species examined (also see Figure S7 for the GR phylogeny with D. melanogaster and T. castaneum sequences). However, no orthologs of the well-described dipteran carbon dioxide (CO2) receptor genes [43], [59] were found (Figure S7), consistent with the proposed loss of dipteran CO2 receptors in the ancestor of Hymenoptera [60]. Interestingly, it is known that the ability to perceive CO2 is present in ants [61], suggesting that different receptor genes are involved.
IR family
Unlike Ors and Grs, Ir genes have maintained relatively stable copy numbers during ant evolution (Figure 4; see Figure S8 and Dataset S4 for the full version of IR phylogeny with gene names and bootstrap values). While multiple duplications are likely to have occurred in the ancestor of Formicidae, unambiguous orthology among H. saltator, C. floridanus, L. humile, and P. barbatus genes has been maintained across most IR clades. The only lineage-specific expansion of ant Ir genes occurred in the IR317 subfamily, in which the number of C. floridanus genes increased from 1 to 7, partially due to tandem duplications. The evolutionary history of Ir genes across Protostomia (e.g. nematodes, arthropods, and molluscs) has been described, where Ir genes are classified into “antennal IRs”, which are more conserved, and “divergent IRs”; of the seven antennal IRs, one (IR21a) was only found in N. vitripennis [26]. Nevertheless, orthologs of the other 6 antennal IRs, including IR8a, IR25a, and IR76b (which are thought to code for Ir co-receptors, that may play similar roles as the Orco Or coreceptor) [24], [25], as well as IR68a, IR75u/f, and IR93a,—were found in ants (also see Figure S9 for the IR phylogeny with D. melanogaster and T. castaneum sequences). In addition, there were 13 other subfamilies of divergent IRs. Of these divergent IRs, no ortholog is present in the genome of N. vitripennis and only one is found in A. mellifera, which could be due to ant specific duplications and/or preferential retention of these divergent IRs occurred in ants.
Fig. 4. Phylogenetic relationships of Hymenoptera Ir genes. 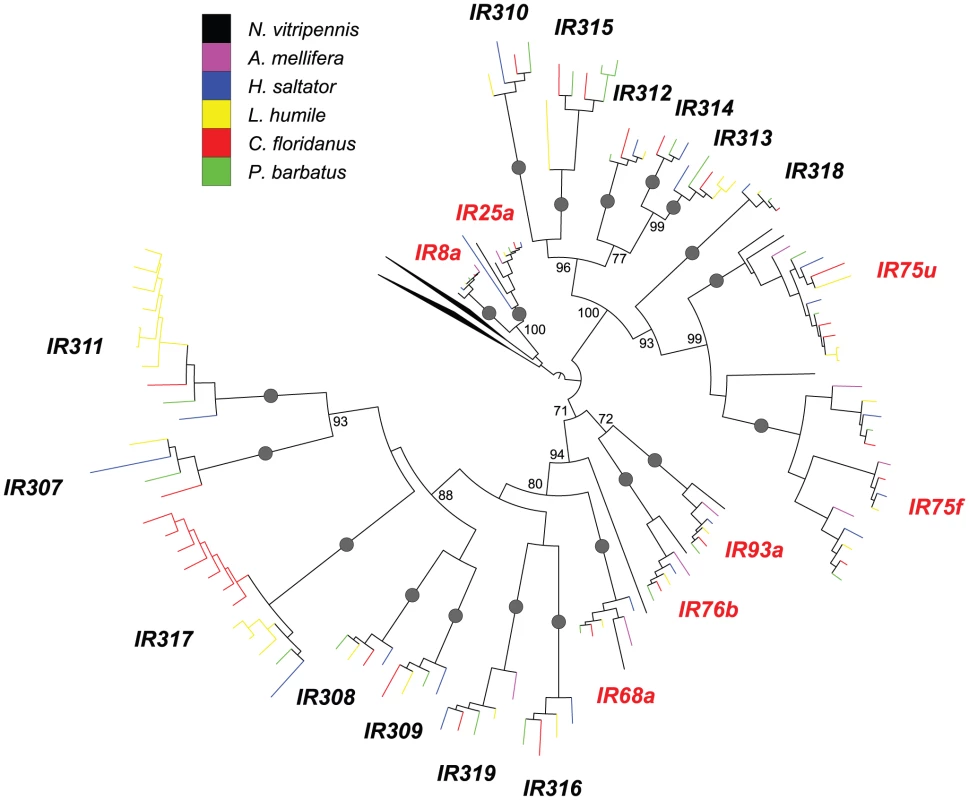
A maximum-likelihood tree of hymenopteran Ir genes estimated by using RAxML with Le-Gascuel (LG) model. Reliability of internal nodes was evaluated by 100 bootstrap replicates. Grey round dots indicate well-supported clades of “antennal” or “divergent” IRs (bootstrap value ≥80). Bootstrap values ≥70 are shown for relationships among the well supported clades. Subfamilies are named after the orthologs in L. humile and P. barbatus. Irs in different species are color-coded as following: N. vitripennis, black; A. mellifera, purple; H. saltator, blue; L. humile, yellow; P. barbatus, green; and C. floridanus, red. Clades for “antennal IRs” are highlighted in red. The clades for ionotropic glutamate receptors were collapsed. Evolutionary dynamics
To further understand the evolutionary dynamics of chemosensory receptor genes, we quantified the gene birth and death events and estimated the number of ancestral gene copies in each family using both the maximum-likelihood (ML) and the parsimony based methods implemented in CAFÉ [62] and Notung [63], respectively. For all three families, the ML method suggested relatively high copy numbers in the ancestor of Hymenoptera (Figure 5). For instance, it estimated a repertoire of 266 Or genes in the hymenopteran ancestor, which was expanded in all ant lineages, but significantly contracted in both N. vitripennis and A. mellifera. A similar pattern was also observed in both the GR and IR families. Moreover, the ML analysis suggested that the low number of Gr genes in H. saltator is due to a significant gene loss in this lineage.
Fig. 5. Estimated numbers of gene birth-and-death events and ancestral gene copies for chemosensory gene families. 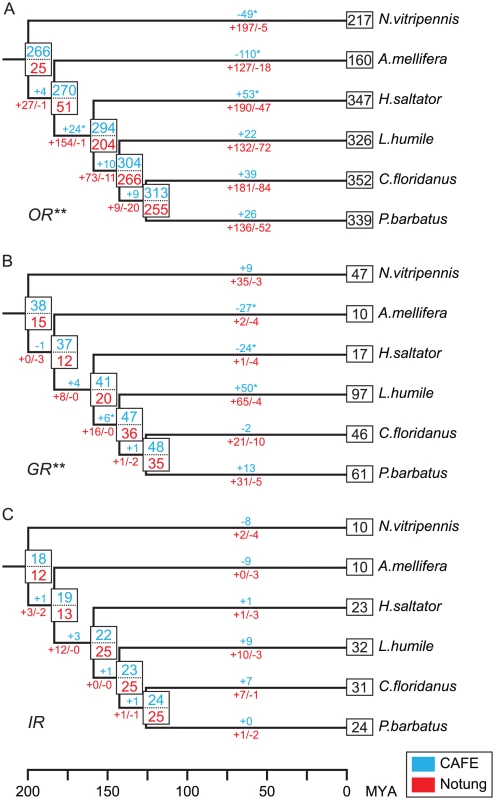
(A) OR family. (B) GR family. (C) IR family. The results of CAFÉ and Notung are highlighted in blue and red, respectively. Numbers above branches indicate net copy number changes estimated by CAFÉ. Numbers below branches with plus and negative signs indicate the number of gene gain and loss events estimated by Notung, respectively. Single asterisk indicates significant branch-specific expansions/contractions. Double asterisk indicates gene families that significantly violate the random gene birth and death assumption of CAFÉ. The phylogeny of Hymenoptera and the time scale are from [1], [102]. On the other hand, the parsimony approach gave conservative estimates of ancestral copy numbers and showed that many more gene-gain events occurred during later stages of hymenopteran evolution. According to the parsimony analysis, the number of Or genes increased from 25 in the last common ancestor of Hymenoptera to about 200 in N. vitripennis and A. mellifera, and more than 300 in all four ants (Figure 5A). Most notably, the repertoire of Or genes increased by three-fold in the ancestor of ants (from 51 to 204 copies), after the separation of A. mellifera, and continued to expand greatly along each ant lineage. Interestingly, although to a lesser degree, the ML method also identified significant expansion on the branch leading to the ant ancestor. In addition to the large number of gene gains, substantial gene losses also occurred in all ants. On the other hand, most duplications of ant Grs occurred in C. floridanus, L. humile, and P. barbatus, while there were only one gene gain and four gene loss events on the lineage to H. saltator (Figure 5B). Similar to the OR and GR families, the number of Ir genes also doubled in the ancestor of ants after its separation from other Hymenoptera (Figure 5C). Subsequent increase of Ir gene number was only observed in C. floridanus and L. humile.
Overall, the ML and parsimony analyses gave different estimates of the ancestral copy numbers and gene gain and loss events. The ML method assumes a random gene birth and death process [64], which is significantly violated by both the OR and GR families (p-values<0.01). On the other hand, the parsimony approach aims to minimize the number of gene gain and loss events, and thus might underestimate the number of ancestral copies. Nonetheless, both analyses support the hypothesis that chemosensory genes have distinct evolutionary dynamics in ant lineages in comparison to the other two hymenopterans.
Antennal expression profiles of ant chemosensory receptor genes
In insects, most Ors and some Grs/Irs are expressed in antennal ORNs [18], [24], [49], [65]. As best illustrated in studies of the Drosophila olfactory system, each ORN expresses a single tuning Or which is responsible for the odorant response profile and all the ORNs expressing that singular tuning Or send axonal connections to a single antennal lobe glomerulus thereby providing a mechanistic basis for the initial stages of odor coding [18]. Therefore, we analyzed antennal transcriptomes of workers and males for both C. floridanus and H. saltator, to identify chemosensory receptor genes that are differentially expressed between castes (minors and majors in C. floridanus) and between different sexes, and which might play salient roles in social communication (see Table S2 for information on transcriptome datasets).
We performed pairwise comparisons between males and females within C. floridanus and H. saltator (Dataset S5). At the whole transcriptome level, there was a very high similarity between major and minor worker of C. floridanus (r2 = 0.99; Figure S10A), while greater diversity was found between workers and males (r2 values around 0.85 for all comparisons), largely due to mild up-regulation of many genes in males (Figure S10B, S10C). Similar trends were also observed for chemosensory receptor genes (Figure S10D).
OR family
In both sexes of C. floridanus and H. saltator, the ortholog of Orco was consistently the most highly expressed Or gene. It accounted for ∼15%–20% of all the Or gene expression in C. floridanus and ∼6%–8% in H. saltator. For the repertoire of tuning Ors within each species, almost all of them were expressed in workers at levels above the medians of their respective antennal transcriptomes (which was used as the criterion for expression versus non-expression of chemosensory gene in the present study). In contrast, only one third of the tuning Or genes were expressed in males of both ants. These comparisons identified almost 40 Ors in C. floridanus and 120 Ors in H. saltator that displayed significant differential expression between workers and males (Table 1, Dataset S5). Interestingly, ∼95% of these genes were enriched in workers, almost all of which had below-median expression levels in males. In addition, we found 13 Or genes that were differentially expressed between major and minor workers of C. floridanus (Table 1, Dataset S5). However, the log2 fold-changes of these genes (less than 1.5) were much lower than those of the genes (greater than 3) revealed in worker vs. male comparisons.
Tab. 1. Significantly differentially expressed C. floridanus and H. saltator chemosensory receptor genes revealed by analysis of antennal transcriptomes. 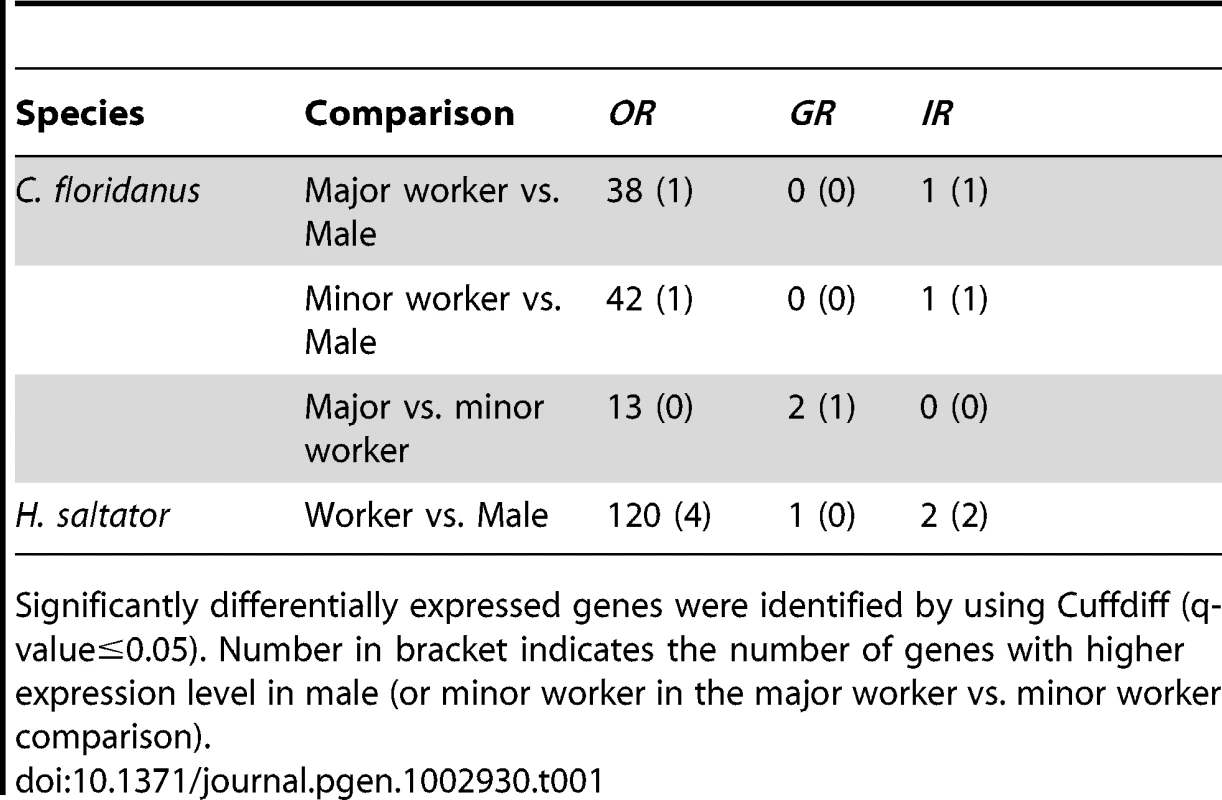
Significantly differentially expressed genes were identified by using Cuffdiff (q-value≤0.05). Number in bracket indicates the number of genes with higher expression level in male (or minor worker in the major worker vs. minor worker comparison). To investigate the relationship between evolutionary relatedness and expression regulation of Or genes, we mapped results of worker vs. male comparisons to the phylogeny of C. floridanus and H. saltator Or genes. As shown in Figure 6A, there are multiple examples where Or genes in one ant species showed sex-specific-enrichment patterns similar (or opposite) to closely related homologs in the other ant species. Notably, the 9-exon Or subfamily illustrates both situations described above (Figure 6B, 6C). In the three basal clades, C. floridanus genes were mostly enriched in male, while all but one H. saltator gene had higher expression levels in workers (Figure 6B). In contrast, all the remaining Or genes formed a well-supported monophyletic clade and almost all of them were enriched in workers for both C. floridanus and H. saltator (Figure 6C). We further examined the expression patterns of (co-)orthologous genes in the two ant species. Using bootstrap values of 70 as threshold, we delineated 98 orthologous groups of C. floridanus and H. saltator Or genes, of which 41 groups included at least one gene from each ant species being differentially expressed by at least two-fold between males and females. C. floridanus and H. saltator genes were enriched in the same sex in 29 of the 41 groups and in different sexes in other 10 groups (Table S3). The remaining 2 groups showed conflicting expression patterns within species.
Fig. 6. Diversified expressions of evolutionarily related Or genes. 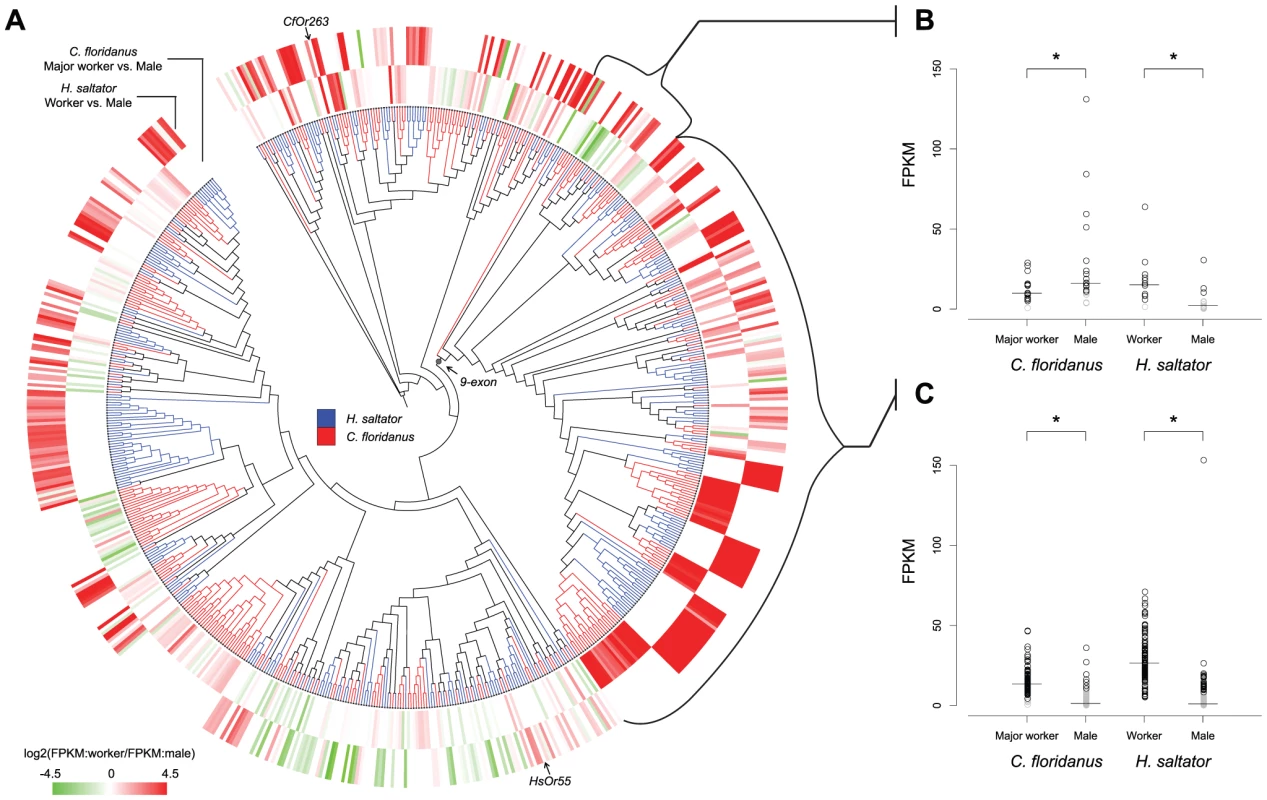
(A) Expression profiles of C. floridanus and H. saltator Ors shown along with the phylogeny of Or genes. In the phylogenetic tree, C. floridanus Ors are labeled by red and H. saltator Ors by blue. In the heat-map, red color indicates higher expression level in worker and green indicates higher expression level in male. The inner circle shows the relative expressions of C. floridanus Ors between major worker and male (comparison between minor worker and male not shown because of the highly similar expression profiles of C. floridanus major and minor workers); the outer circle shows the relative expressions of H. saltator Ors between worker and male. FPKM stands for Fragments Per Kilobase of exon per Million fragments mapped. (B) Expression levels of Ors belonging to the three basal clades in the 9-exon subfamilies. C. floridanus Ors had significantly higher expression in male (p-value<0.05; Wilcoxon ranked-sum test), while H. saltator Ors had significantly higher expressions in worker (p-value<1e-4; Wilcoxon ranked-sum test). (C) Expression levels of the remaining Ors in the 9-exon subfamilies. For both C. floridanus and H. saltator, Ors had significantly higher expressions in worker (p-value<1e-15; Wilcoxon ranked-sum test). Short lines indicate median expression levels for each gene set. For both panels (B) and (C), genes expressed below the medians of their respective transcriptomes were labeled by grey. GR family
Unlike Or genes, only a portion of Gr genes within each species were expressed in workers (less than 25% for C. floridanus and ∼35% for H. saltator) and males (less than 15% for both ants) (Figure 7A). Furthermore, worker vs. male comparisons revealed only one H. saltator Gr gene that was differentially expressed between worker and male (HsGr7), and none in C. floridanus. While two C. floridanus Gr genes were found to have differential expressions between major and minor workers (CfGr9 and CfGr54), their absolute expression values were close to or below the median of their respective transcriptomes.
Fig. 7. Expression levels of Gr and Ir genes. 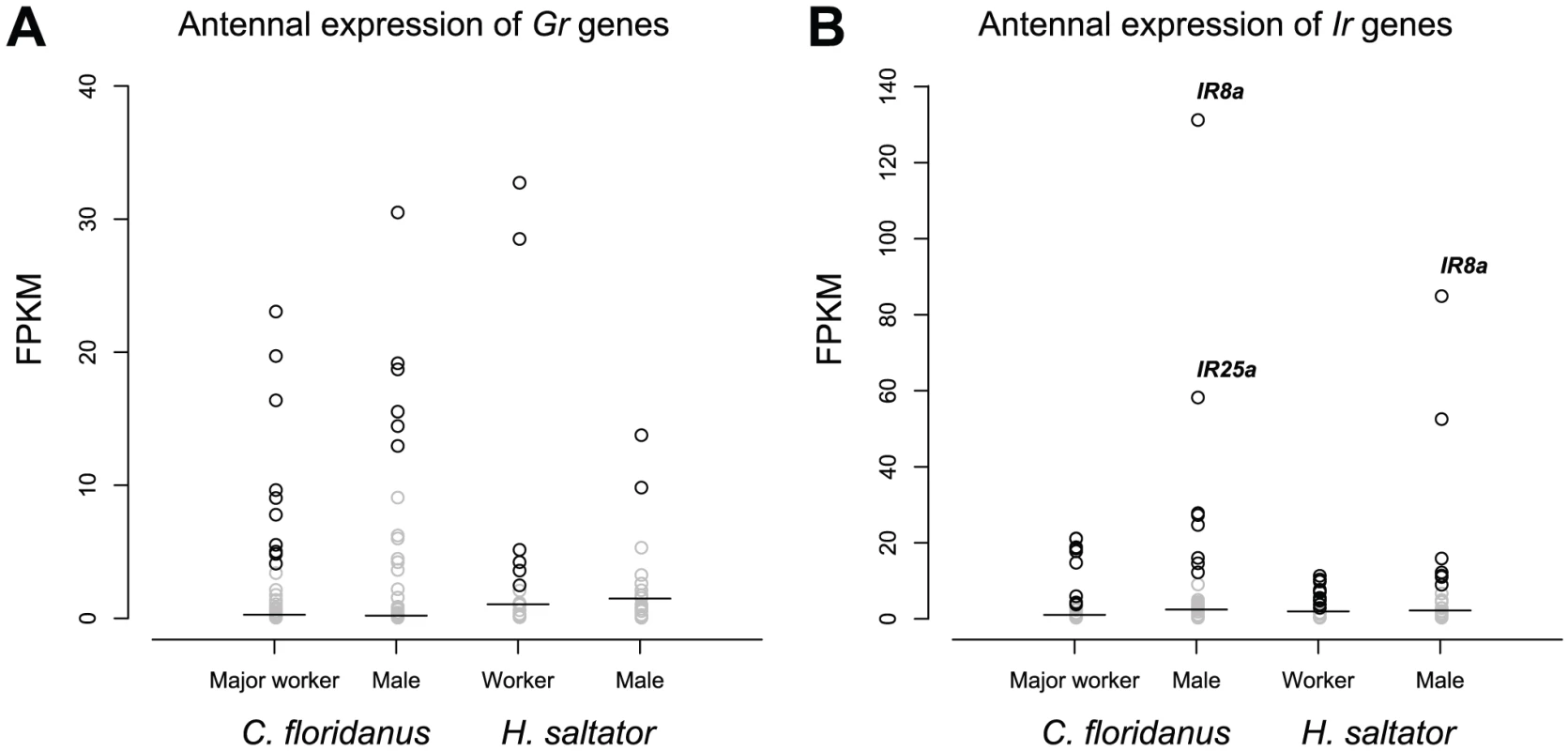
(A) The GR family. (B) The IR family. Short lines indicate median expression levels for each gene set. In both panels, genes expressed below the medians of their respective transcriptomes were labeled by grey. IR family
50% or less of Ir genes of each species were expressed in any given sex (Figure 7B), and almost all expressed Ir genes are conserved “antennal IRs”. We identified only one C. floridanus Ir gene and two H. saltator Ir genes that have differential expressions between workers and males. Interestingly, all of these Ir genes were enriched in male. The ortholog of IR8a, encoding one of the Ir co-receptors [24], [25], was differentially expressed in both C. floridanus and H. saltator, and was also the most highly expressed Ir gene in males of both ants, while another “antennal IR” (HsIR75u.2) was also found to be more highly expressed in H. saltator males than in workers.
Identification of a ligand for a differentially expressed ant odorant receptor
In order to validate our bioinformatic annotations and in an attempt to link functional data to the antennal expression data, we have cloned a small subset of 14 C. floridanus and H. saltator Or genes, drawn from 6 subfamilies in the Or phylogeny (D, E, H, L, V, and 9-exon). These include four genes (CfOr263, HsOr212, HsOr213, and HsOr279) that display significant differential expression in our transcriptome analysis (see Methods and Materials for full list). This allowed us to carry out deorphanization studies to decipher the odorant response profiles of these receptors through the use of two-electrode voltage clamp recordings in Xenopus oocytes heterologously expressing ant Ors [44], [66]. After first confirming that the C. floridanus and H. saltator Orco proteins showed coreceptor function in combination with a previously deorphanized mosquito tuning Or (Figure 8A, 8B), candidate ant tuning Ors were screened against a panel of 73 unitary and complex stimuli (Table S4). These stimuli consisted of a variety of general odorants, as well as hydrocarbons known to be produced by H. saltator or C. floridanus.
Fig. 8. Identification of ligands for C. floridanus and H. saltator Ors. 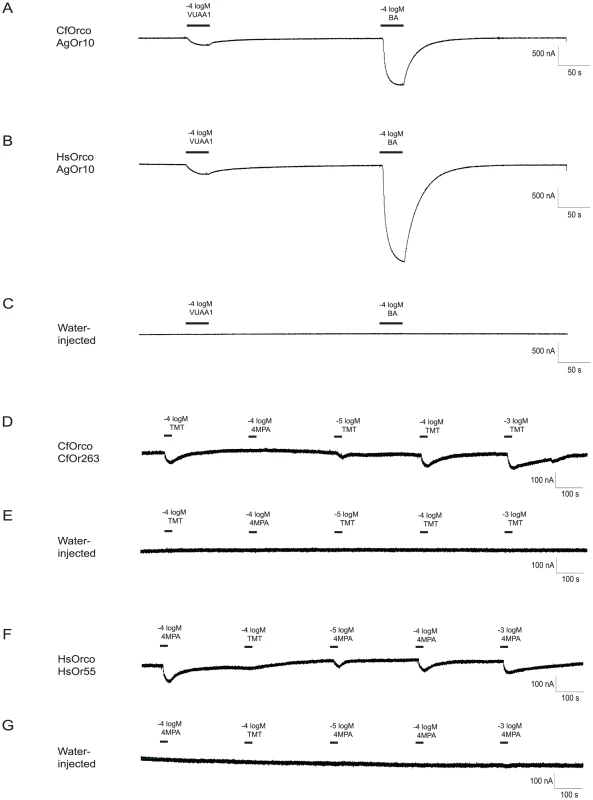
When paired with a previously characterized mosquito Or (AgOR10) [103], both CfOrco (C. floridanus Orco) (A) and HsOrco (H.saltator Orco) (B) produced responses to VUAA1 (an agonist for Orco) [35] and benzaldehyde (BA, an agonist for AgOR10) in Xenopus oocytes. These responses were not observed in water-injected control oocytes (C). The novel C. floridanus tuning Or CfOr263 also shows a specific and dose-dependent response to 2,4,5-trimethylthiazole (TMT) (D), while the novel H. saltator tuning Or HsOr55 shows a similar dose-dependent response to 4-methoxyphenylacetone (4 MPA) (F). Neither response is observed in water-injected oocytes (E,G). Out of the 14 tuning Ors initially screened, CfOr263 (from OR subfamily D; Figure 2), which is highly expressed in workers as compared to males (Figure 6A), produced specific and dose-dependent responses to 2,4,5-trimethylthiazole (Figure 8D, 8F), a naturally occurring odorant found in cooked beef and pork [67] found in the library of general odorants that we screened. An additional Or from H. saltator, HsOr55 (from OR subfamily L; Figure 2), showed a dose-dependent response to another odorant from our general odorant library, 4-methoxyphenylacetone (Figure 8E, 8G), which is a naturally occurring odorant found in anise essential oil [68]. However, this particular Or has not been shown to be differentially expressed between males and females. It should also be noted that, as is the case for most ant Ors, both receptors have multiple closely related homologs that may possess similar chemosensory functions (Figure 6A).
Discussion
Expanded ant chemosensory receptor repertoire
We have developed and used a dedicated annotation scheme to comprehensively elucidate the repertoire of chemosensory receptor genes in both C. floridanus and H. saltator. Through exhaustive homology search and careful manual curation, we significantly improved upon previous studies to identify roughly equivalent numbers of Or/Gr/Ir genes in the genomes of C. floridanus and H. saltator as compared to two other sequenced ant genomes [47], [48], providing a solid foundation for subsequent study.
It is striking that, in general, ants have the most expanded repertoire of chemosensory receptor genes in Hymenoptera (Figure 1). The numbers of ant OR and IR family members are much greater than those of the other two hymenopteran genomes currently available. Indeed, thus far, ant genomes have the largest number of Or genes among all insects [69]. Furthermore, although the number of the Gr genes varies greatly among hymenopterans and also within ants, L. humile carries the largest Gr family; it has about 2 - and 10-fold more Grs than N. vitripennis and A. mellifera, respectively. Interestingly, although ants and honey bees are both social insects, ants have much larger repertoires of all three chemosensory receptor gene families than honey bees, possibly indicative of a more sophisticated communication system relying on chemicals [70].
Our phylogenetic analyses of hymenopteran chemosensory receptor genes reveal distinct evolutionary patterns among gene families. Among chemosensory receptors, the OR family shows the most dramatic birth-and-death evolution, with many OR subfamilies displaying diversified patterns of gene gain-and-loss. For example, the 9-exon subfamily and others have experienced rapid gene duplications at almost all stages of Hymenoptera evolution, followed by numerous losses of duplicates. In contrast, there are 35 subclades that have only one ortholog in all four ants. Further, the IR family has maintained relatively stable copy numbers in ants; lineage-specific expansion only occurred in C. floridanus and L. humile for two of the 13 “divergent IRs”. In between these extremes is the GR family that has expanded moderately in N. vitripennis and three of the four ants.
Recent studies of chemosensory receptors in mammals and Drosophila, as well as other genes with important regulatory and physiological functions, have suggested a possible correlation between functional requirements and the variations of gene numbers [52], [71], [72]. Genes with conserved roles tend to have relatively stable copy numbers while those with diversified functions have higher rates of birth-and-death, although the degrees of copy number changes are somewhat random. Our results suggest that this pattern could also hold true for the evolution of the hymenopteran chemosensory receptor genes. For example, as an obligatory co-receptor for all other Ors [29], Orco is the most conserved insect Or gene and also the only one that has maintained unambiguous orthology in all insects studied to date, including ants [69]. Similarly, orthologs of most “antennal IRs” [26] have also maintained strict single-copy in Hymenoptera. It has been proposed that these conserved “antennal IRs” represent the earliest insect chemosensory receptors and perform functions important for all insects [26]. Therefore, we suggest that the chemosensory receptor genes that have constant copy numbers in ants (e.g. the 35 single-copy tuning Ors) are likely to carry out important functions common for all ants.
On the other hand, prevalent rapid expansions in chemosensory receptor gene families could allow for diversification in ligand specificity/sensitivity among duplicated receptor genes. Such functional divergences would offer tremendous opportunities for organisms to explore different chemical niches, thus facilitating the adaption to new environments and/or the evolution of novel life styles such as sociality. In all three gene families, we found either retention of the complete ancestral repertoire (according to the ML method) or dramatic increases in gene numbers (according to the parsimony method) in the ancestor of ants (Figure 5), which might have contributed to the success and subsequent diversification of this group.
In addition, there are many cases of unbalanced expansions/contractions among lineages in specific (sub-)families, suggesting that the chemosensory receptor repertoire has been differentially exploited among ants, which might shed light on the evolution of different lifestyles of ants. For example, our results indicate expansions of Grs in C. floridanus, L. humile, and P. barbatus, but not H. saltator, which are likely to reflect differences in their feeding behaviors. In this view, scavengers like C. floridanus might require a highly expanded repertoire of taste receptors to discriminate nutritious food sources from spoiled, contaminated, or poisoned substrates. In contrast, H. saltator workers likely rely more on visual cues to track down prey, as suggested by their large eyes and expanded number of ommatidia [73]. Furthermore, Grs which act as contact chemoreceptors would be far less useful for identifying and capturing prey. In fact, ponerine ants in general rarely use liquid food sources, since they normally lack the ability to exchange liquids stored in their crop [74] which further reduces the potential benefit of a large Gr repertoire.
Another intriguing possibility is that Grs are involved in the contact chemosensation of species-specific, nonvolatile CHCs (e.g. queen pheromone, nestmate recognition signals, etc.), and that C. floridanus has more Grs precisely because they utilize a greater number and variety of pheromones to support their more rigid and complex social lifestyle. Presumably, these Grs would be in addition to the large number of worker enhanced Ors that are likely to be involved in the same process. Furthermore, C. floridanus has expansions in multiple GR subfamilies, including 5 homologs of the DmGr43a/AmGr3 gene, which has been recently shown to be a fructose receptor [75]. Taken together, our results indicate a correlation between the expanded GR family and the more complex chemical ecology of C. floridanus.
Diversified expression of chemosensory receptor gene
The antenna is perhaps the most important chemosensory organ for ants, where a variety of ant species have been observed to closely inspect their environment and each other by touching their antennae in a process known as antennation [2]. This makes it likely that most of the behaviorally important chemosensory neurons (and their corresponding chemosensory receptors) are located in this organ. Our comparative analysis of antennal transcriptomes of workers and males in both C. floridanus and H. saltator reveal differential expressions of chemosensory receptor genes both within and between species, providing important clues on their functional divergence.
One major pattern revealed by our results is the substantial sexual dimorphism in chemosensory receptor gene expression in ants. For both C. floridanus and H. saltator, almost all Ors were expressed in workers, but only one third were expressed in male. Similarly, workers consistently had more expressed Grs and Irs than males. In contrast, expression of chemosensory receptor genes was highly similar between major and minor workers in C. floridanus. Previous studies have shown that the antennal lobes of males from both C. floridanus and H. saltator lack a large subset of glomeruli relative to workers [76], [77], [78], which may explain the low number of chemosensory receptor genes expressed in males. Given that the number of glomeruli in insects generally correlates with the number of functional odorant receptors [18], [65], it is likely that most of the Ors that are only expressed in C. floridanus and H. saltator workers project to these female-specific glomeruli. Furthermore, it has been shown in another Camponotus species (Camponotus japanicus) that females exclusively possess the olfactory sensilla necessary to detect non-nestmate CHCs, [79], [80]. It is therefore likely that the CHCs receptors are encoded by some of the worker-specific Ors in C. floridanus. In particular, the 9-exon subfamily represents the largest expansion of Ors in all ants and it harbors close to 100 worker-specific Ors in both C. floridanus and H. saltator. These results strongly support previous hypothesis that members of the 9-exon subfamily are likely candidates for ant CHCs receptors [47], [48]. These Ors are potentially involved in detecting CHCs involved in worker-to-worker or worker-to-queen intracolonial social communication.
Interestingly, we also noticed discrepancies between the overall number of Ors and the number of glomeruli in the adults of these two ant species. H. saltator workers and males both have far more expressed Ors than the number of glomeruli in the adult antennal lobe (approximately 78 in the adult male and 178 in the adult worker [77]). The discrepancy in H. saltator could possibly be the result of co-expression of multiple tuning Ors in the same ORN and/or the projection of ORNs expressing different, but related tuning Ors to the same glomerulus, which have both been observed for a small number of Ors/ORNs in D. melanogaster [81], [82], [83], [84].
However, given that the number of expressed Ors is about twice the number of observed glomeruli, this would mean that each glomerulus received input from, on average, two odorant receptors. Although co-expression of tuning Ors has not been observed to such a broad extent in any insect olfactory system studied to date, it should be noted that many of the receptor pairs that are co-expressed in Drosophila appear to be the result of tandem duplication events [84]. Therefore, it is possible that the extensive tandem duplication of H. saltator Or genes may also result in the co-expression of closely related odorant receptors from the same clusters. All of these are highly interesting hypotheses that may be examined in future studies.
In contrast to H. saltator, C. floridanus has approximately 80 fewer Ors than the number of adult worker glomeruli (about 454 [76]). In this instance it is possible that many of those glomeruli receive projections from Gr and Ir expressing ORNs, as there is precedence for this in Drosophila [24], [43] and the number of predicted Grs and Irs would be enough to fill the gap. Moreover, it could be that several Ors have been missed by the current analysis due to incomplete genome assembly; some of the fragmented Or gene models might represent genuine genes, and further genomic/transcriptomic data would help address this possibility.
Although chemosensory receptor genes in general had higher expression in workers, our studies have nevertheless identified a single Or (CfOr267, in subfamily 9-exon) and a single Ir (CfIR8a) in C. floridanus, as well as 4 Ors (HsOr32, HsOr35, and HsOr37, in subfamily L; and HsOr224, in subfamily E) and 2 Irs (HsIR8a and HsIR75u.2) in H. saltator that were significantly male-enriched. The male-enrichment of a receptor gene could be due to elevated expression of the gene in ORNs of males relative to workers, and/or increased number of ORNs expressing the gene in males. No matter which of the possibilities is indeed the case, our results indicate higher overall abundances of these chemosensory receptor genes in male antennae. These genes are viable candidates for receptors that are specifically tuned for male-specific social cues, including queen pheromones. In fact, at least one male-specific honeybee odorant receptor that responds to a queen-specific pheromone has already been revealed through microarray analysis and subsequent functional characterization in Xenopus oocytes [85]. It would not be surprising to see that similar results will be found with the male-enriched ant Ors.
In insects, the co-receptors IR8a and IR25a are the two most conserved Irs [26]. Although a systematic profiling of sexual dimorphic Ir expression is still lacking, a previous study has shown that the Anopheles gambiae orthologs of both IR8a and IR25a have higher expression in female than male [49]. Interestingly, IR8a was the most male-enriched Ir in both C. floridanus and H. saltator. While IR25a also displayed higher expression in C. floridanus male, it was not expressed in the male of H. saltator. These results could possibly indicate a functional divergence of IR8a and IR25a between Diptera and Hymenoptera. In addition, the high expression of IR25a in males of C. floridanus, but not H. saltator, suggests that IR25a-mediated signaling might have contributed to the more expanded roles for males within the colony of the former species. It may be that C. floridanus males are more involved in intracolonial interactions than H. saltator males, since males from other Camponotus species are known to participate in food exchange in the colony [86], which has not observed in H. saltator males.
We have also found diversified expression of closely related Ors within and between species. For example, in the basal clades of the 9-exon OR subfamily, closely related C. floridanus and H. saltator Ors showed opposite sexual dimorphism in their expression (Figure 6B). Although the well-supported monophyletic clade within the 9-exon OR subfamily mostly consists of worker-enriched genes, it also harbors a few genes that are highly enriched in male (Figure 6C). Thus, while our expression results are generally (and strongly) consistent with the idea that members of the 9-exon OR subfamily are involved in the detection of CHCs by workers [47], a subset of these receptors have apparently been adapted for use in males, possibly for detecting queen mating pheromones.
Taken together, these results indicate that ant Or genes have experienced not only extensive gain-and-loss, but also rapid changes in their expression, once again highlighting the highly dynamic nature of chemosensory receptor gene evolution. Our phylogenetic and transcriptomic analyses, in combination, have identified ant chemosensory receptor genes that exhibit evolutionary and expression patterns indicative of species/sex-specific functions. Ultimately, deorphanization of these receptors will greatly facilitate our understanding of the chemical ecology of social lifestyle in ants.
Heterologous characterization of differentially expressed C. floridanus Ors
In our heterologous studies of ant tuning Ors, we have identified chemical agonists for a single receptor from each of the two species analyzed. These data provide conclusive validations for our bioinformatic-based annotations. Although a honeybee odorant receptor has been previously shown to respond to the queen substance 9-oxo-2-decenoic acid [85], we believe that this represents the first published report of ligand activators for odorant receptors from ants.
In these studies, HsOr55 from H. saltator, display significant responses to 4-methoxyphenylacetone, a naturally occurring odorant found in anise essential oil [68]. Since anise essential oil has been shown to have a repellent and/or insecticidal effect on at least some species of insects [87], [88], 4-methoxyphenylacetone might represent a general insect repellent, with HsOr55 acting as the detector for this repellent in H. saltator. Whatever HsOr55's role may be, it is likely to be a very general one, since HsOr55 transcripts do not appear to be differentially expressed between workers and males.
The other odorant receptor characterized in this study, CfOr263 from C. floridanus, displayed sensitivity to 2,4,5-trimethylethiazole, a naturally occurring odorant found in cooked beef and pork [67] that has been previously shown to induce strong responses in the CpC neuron of the maxillary palp in the mosquito Anopheles gambiae [44]. While the relevance of this chemical to C. floridanus remains unclear, the fact that CfOr263 transcripts are enriched in workers relative to males suggests that this odorant may be an important volatile semiochemical for C. floridanus workers. Regardless, the successful identification of odors that activate CfOr263 and HsOr55 strongly validates the role of ant Ors as chemosensory receptors. Furthermore, the large differential expression of CfOr263 between workers and males indicates that it is detecting a sex - specific signal that is relevant to workers but not to males, and testing a broader panel of odorants in the future will provide a better understanding of what that signal might be.
Conclusions
We have revealed a greatly expanded repertoire of chemosensory receptor genes for a pair of divergent ant species, including about 400 Ors and an order of magnitude smaller number of Grs and Irs. Phylogenetic analysis of these newly annotated genes indicates that there are likely to be vast differences in the importance of particular chemoreceptor families and subfamilies between the four ant species examined, which is likely to reflect the variety of ecological and social demands experienced the members of each species. These analyses also reveal high rates of gene birth-and-death evolution among the olfactory and gustatory receptor genes, suggesting that some factor (such as changes in the complex CHC profiles that control ant social behavior) is driving rapid evolution in their chemical response profiles. The large repertoire of ant chemosensory genes might be either due to preferential retention of ancestral genes or rapid expansions in the ant ancestor and during later stages of ant evolution. To further complement these phylogenetic results, we have generated and analyzed antennal-specific RNAseq expression data to identify ∼40 C. floridanus and ∼120 H. saltator chemosensory receptors that exhibit significant sexual dimorphism in expression. This expression data has, in turn, informed studies towards the identification of odorant ligands for socially relevant receptors, a process that we have already successfully accomplished in a heterologous system for one of the differentially expressed C. floridanus Ors. Taken together, our evolutionary analysis, transcriptome profiling, and heterologous characterization provide new insights into the roles of the chemosensory receptors in inter-sex behavioral and social differences of ants.
Materials and Methods
Gene annotations
The assemblies of C. floridanus (version 3.5) and H. saltator (version 3.5) were downloaded from the Hymenoptera Genome Database [89]. Protein sequences of reported chemosensory gene were also collected from Apis mellifera, Acyrthosiphon pisum, Drosophila melanogaster, Nasonia vitripennis, L. humile, and P. barbatus [15], [26], [28], [47], [48], [50], [54]. An in-house bioinformatics pipeline was developed to identify candidate chemosensory genes in C. floridanus and H. saltator. First, all collected chemosensory gene sequences were searched against the two ant genomes using TBLASTN [90] with an e-value cutoff of 1e-5. Resulting High-scoring Segment Pairs (HSPs) were sorted by their blast bit-scores, and an average bit-score of the top 75% HSPs were calculated. Any HSPs with a bit-score less than 25% of the average was discarded. Chains of HSPs were than created from retained HSPs. Two HSPs were chained together if the following criteria were met: 1) they are derived from the same query; 2) they are located within 3 kb on the same strand of a scaffold/contig; and 3) the corresponding query region of the upstream HSPs must also be N-terminal to that of the downstream HSPs. The third criterion was applied to avoid artificial concatenation of neighboring chemosensory genes. Genomic regions covered by HSPs chains were considered putative chemosensory gene coding regions. For each putative gene, we then selected the query corresponding to the highest scoring HSPs at that region as reference sequence for homology-based gene prediction using GeneWise (version 2.2.0) [91]. All predictions were sorted by ORF length and the lowest 25% was filtered. This pipeline was iterated by adding results of previous run to input until no additional genes were found.
Multiple sequence alignments (MSAs) of predicted OR/GR/IRs were constructed using MUSCLE (version 3.8) [92] and manually inspected. Attempts to improve annotations were made whenever an obvious problem was identified (e.g. missing exon, incorrect exon-exon junction). In addition, in the OR and GR families, we observed many fragmented gene models, likely due to pseudogenization and incomplete genome assembly. For the convenience of subsequent analyses, a minimum size cutoff of 300 amino acids was used for the ORs and GRs. For IRs, we screened all predicted protein sequences with InterProScan (V4.8) [93] and filtered the ones without characteristic domains of IR (PF10613 and PF00060) [26].
Phylogenetic analysis
We included in our phylogenetic analysis chemosensory receptor genes in six hymenopteran species, including A. mellifera, C. floridanus, H. saltator, N. vitripennis, L. humile, and P. barbatus. For each of the OR/GR/IR families, all family members were firstly aligned at once using MUSCLE (version 3.8) and a preliminary phylogenetic tree was built using RAxML (version 7.2.8) [94]. Sequences were then divided into groups corresponding to highly supported clades in the preliminary phylogeny. Groups were aligned individually using PROBALIGN (version 1.4) [95] and then combined together using the profile alignment function of MUSCLE. The complete alignment were further manually inspected and adjusted using GeneDoc (version 2.6) [96]. In addition, poorly aligned regions in the alignment were removed using trimAl (version 1.4) [97]. The final maximum-likelihood tree was constructed using RAxML with Le-Gascuel (LG) substitution model [98] and GAMMA correction for rate variation among sites. Reliability of tree topology was evaluated by 100 bootstrap replicates. To estimate the number of gene gain and loss events, we used a maximum-likelihood based approach implemented in CAFÉ (version 2.2) [62] with default settings. As an alternative approach, we also used the parsimony based “modified reconciliation method” [99]; we first collapsed branches with bootstrap support lower than 70 in phylogenies of OR/GR/IR families and then reconciled condensed trees with known organismal relationships using Notung (version 2.6) [63].
Antenna collection, RNA extraction, and Illumina sequencing
Samples originated from C. floridanus colonies that had been founded in the Liebig lab from queens captured in southern Florida between 2002 and 2009 and from H. saltator colonies collected in Karnataka, India between 1995 and 1999. Antennae were collected from each of five groups of adult ants: H. saltator workers and males and C. floridanus major workers, minor workers, and males. Whole ants were flash-frozen in liquid nitrogen and kept on dry ice as 100 antennae from each group were removed with forceps. Antennae were placed directly into RNAlater ICE (Ambion) that had been pre-chilled on dry ice in a conical, ground-glass, tissue homogenizer. RNAlater ICE was replaced with 1 ml Trizol (Invitrogen), in which antennae were homogenized. Total RNA was isolated following Trizol manufacturer instructions; briefly, after addition of 200 µl of a chloroform∶isoamylalcohol mixture (24∶1), each sample was mixed vigorously and the RNA-containing aqueous layer was isolated with centrifugation. RNA was further purified and DNAse-treated with the RNeasy Miniprep kit (Qiagen). After ethanol-precipitation, the RNA pellet was resuspended in 30 µl nuclease-free water. Male samples were sequenced using Illumina HiSeq2000 at the NYULMC Genome Technology Center, generating ∼33 million 50 bp single-end reads for C. floridanus male and ∼164 million 51 bp single-end reads for H. saltator male. All worker samples were sequenced at Hudson Alpha, generating more than 20 million 50 bp paired-end reads for each sample (sum of two technical replicates).
Analysis of ant antennal transcriptome
Reads of C. floridanus male sample were trimmed to 34 bp (8 bp trimmed from both ends) to remove low-quality positions. In addition, for all worker datasets, we treated each paired-end read as two single-end reads. Therefore, all datasets in our subsequent analyses consist of only single-end reads. Alternative strategies for data processing led to highly similar estimations of gene expression values (Table S5). For each dataset, reads were mapped to the corresponding ant genome using TopHat (version 1.3.3) [100] with default setting. Gene annotations for C. floridanus (version 3.5) and H. saltator (version 3.5) were downloaded from the Hymenoptera Genome Database and used in combination with our annotation of chemosensory genes to guide the reads mapping. Gene expression levels (in FPKM values) and differentially expressed genes were determined using Cuffdiff v1.3.0 [101] with frag-bias-correct, multi-read-correct, and upper-quartile-norm options turned on.
Heterologous analysis of ant odorant receptors
Predicted Or coding sequences were amplified, by PCR, from H. saltator and C. floridanus worker antennal cDNA samples obtained from colonies established at Arizona State University (Tempe, AZ). The PCR-amplified sequences were then TOPO cloned into the Gateway Entry vector pENTR/D-TOPO (Life Technologies), followed by an additional cloning step into a destination vector derived from pSP64T. To obtain cRNA for each Or, the pSP64T vector containing the appropriate coding sequence was linearized by restriction digest and used as a template for cRNA synthesis using the mMessage mMachine Sp6 Kit (Ambion). Heterologous expression of ORs was accomplished as described previously [66]. Briefly, mature oocytes were surgically extracted from Xenopus leavis adult females, treated with 2 mg/mL collagenase II in 1× Ringer's solution (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, and 5 mM Hepes, pH 7.6) for 30–45 minutes at room temperature, and then injected with 27.6 nL of a 1∶1 mixture (by mass) of a given tuning Or in combination with the appropriate Orco ortholog (either HsOrco or CfOrco). After injection, oocytes were stored in Incubation Medium (10% dialyzed horse serum in 1× Ringer's solution) at 18C for 3–7 days before testing. Responses to odorants were measured by recording whole-cell currents in Clampex 10.2 (Molecular Devices) using a two-electrode voltage-clamp setup (OC-725C, Warner Instruments) maintained at a −80 mV holding potential. Odorants were first dissolved in DMSO, and then further diluted into Ringer's solution before being introduced to the oocyte recording chamber using a perfusion system. For the hydrocarbons that were tested, 0.01% Triton X-100 (Sigma) was also added to the Ringer's solution to aid in dissolving the odorant. The following odorant receptors were tested with the odorants listed in Table S4: CfOr183, CfOr215, CfOr263, HsOr19, HsOr55, HsOr132, HsOr170, HsOr175, HsOr212, HsOr213, HsOr234, HsOr239, HsOr279, HsOr287.
Chemicals
Odorant chemicals were purchased from commercial sources at the highest purity available. Henkel 100, a mixture of 100 different volatile chemicals, was obtained from Henkel (Düsseldorf, Germany), and the C7–C40 saturated alkane mixture was purchased from Supelco (Bellefonte, PA, USA).
Supporting Information
Zdroje
1. MoreauCS, BellCD, VilaR, ArchibaldSB, PierceNE (2006) Phylogeny of the ants: diversification in the age of angiosperms. Science 312 : 101–104.
2. Hölldobler B, Wilson EO (1990) The ants. Cambridge, Mass.: Belknap Press of Harvard University Press. xii, 732 p., 724 p. of plates p.
3. DavidsonDW, CookSC, SnellingRR, ChuaTH (2003) Explaining the abundance of ants in lowland tropical rainforest canopies. Science 300 : 969–972.
4. DavidsonDW (1997) The role of resource imbalances in the evolutionary ecology of tropical arboreal ants. Biol J Linn Soc 61 : 153–181.
5. BoltonB (2003) Synopsis and Classification of Formicidae. Mem Am Entomol Inst 71 : 1–370.
6. WilsonEO (1976) Which are the most prevalent ant genera? Stud Entomol 19 : 187–200.
7. MooreD, LiebigJ (2010) Mechanisms of social regulation change across colony development in an ant. BMC Evol Biol 10 : 328.
8. EndlerA, LiebigJ, HölldoblerB (2006) Queen fertility, egg marking and colony size in the ant Camponotus floridanus. Behav Ecol Sociobiol 59 : 490–499.
9. EndlerA, LiebigJ, SchmittT, ParkerJE, JonesGR, et al. (2004) Surface hydrocarbons of queen eggs regulate worker reproduction in a social insect. Proc Natl Acad Sci U S A 101 : 2945–2950.
10. EndlerA, HölldoblerB, LiebigJ (2007) Lack of physical policing and fertility cues in egg-laying workers of the ant Camponotus floridanus. Anim Behav 74 : 1171–1180.
11. PeetersC, LiebigJ, HölldoblerB (2000) Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator. Insect Soc 47 : 325–332.
12. PeetersC, HölldoblerB (1995) Reproductive cooperation between queens and their mated workers: the complex life history of an ant with a valuable nest. Proc Natl Acad Sci U S A 92 : 10977–10979.
13. LiebigJ, PeetersC, HölldoblerB (1999) Worker policing limits the number of reproductives in a ponerine ant. Proc R Soc B 266 : 1865–1870.
14. LiangZS, NguyenT, MattilaHR, Rodriguez-ZasSL, SeeleyTD, et al. (2012) Molecular determinants of scouting behavior in honey bees. Science 335 : 1225–1228.
15. RobertsonHM, WarrCG, CarlsonJR (2003) Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A 100 Suppl 2 : 14537–14542.
16. GaoQ, ChessA (1999) Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60 : 31–39.
17. ClynePJ, WarrCG, FreemanMR, LessingD, KimJ, et al. (1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22 : 327–338.
18. VosshallLB, AmreinH, MorozovPS, RzhetskyA, AxelR (1999) A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96 : 725–736.
19. FoxAN, PittsRJ, RobertsonHM, CarlsonJR, ZwiebelLJ (2001) Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci U S A 98 : 14693–14697.
20. ScottK, BradyRJr, CravchikA, MorozovP, RzhetskyA, et al. (2001) A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104 : 661–673.
21. DunipaceL, MeisterS, McNealyC, AmreinH (2001) Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol 11 : 822–835.
22. ClynePJ, WarrCG, CarlsonJR (2000) Candidate taste receptors in Drosophila. Science 287 : 1830–1834.
23. WangL, HanX, MehrenJ, HiroiM, BilleterJ-C, et al. (2011) Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci 14 : 757–U392.
24. BentonR, VanniceKS, Gomez-DiazC, VosshallLB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136 : 149–162.
25. AbuinL, BargetonB, UlbrichMH, IsacoffEY, KellenbergerS, et al. (2011) Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69 : 44–60.
26. CrosetV, RytzR, CumminsSF, BuddA, BrawandD, et al. (2010) Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6: e1001064 doi:10.1371/journal.pgen.1001064.
27. HillCA, FoxAN, PittsRJ, KentLB, TanPL, et al. (2002) G protein-coupled receptors in Anopheles gambiae. Science 298 : 176–178.
28. RobertsonHM, GadauJ, WannerKW (2010) The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol 19 Suppl 1 : 121–136.
29. LarssonMC, DomingosAI, JonesWD, ChiappeME, AmreinH, et al. (2004) Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43 : 703–714.
30. BentonR, SachseS, MichnickSW, VosshallLB (2006) Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol 4: e20 doi:10.1371/journal.pbio.0040020.
31. KriegerJ, KlinkO, MohlC, RamingK, BreerH (2003) A candidate olfactory receptor subtype highly conserved across different insect orders. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 189 : 519–526.
32. SatoK, PellegrinoM, NakagawaT, NakagawaT, VosshallLB, et al. (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452 : 1002–1006.
33. WicherD, SchaferR, BauernfeindR, StensmyrMC, HellerR, et al. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452 : 1007–1011.
34. SmartR, KielyA, BealeM, VargasE, CarraherC, et al. (2008) Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol 38 : 770–780.
35. JonesPL, PaskGM, RinkerDC, ZwiebelLJ (2011) Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci U S A 108 : 8821–8825.
36. PaskGM, JonesPL, RutzlerM, RinkerDC, ZwiebelLJ (2011) Heteromeric anopheline odorant receptors exhibit distinct channel properties. PLoS ONE 6: e28774 doi:10.1371/journal.pone.0028774.
37. DahanukarA, FosterK, van der Goes van NatersWM, CarlsonJR (2001) A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci 4 : 1182–1186.
38. UenoK, OhtaM, MoritaH, MikuniY, NakajimaS, et al. (2001) Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Curr Biol 11 : 1451–1455.
39. MoonSJ, KottgenM, JiaoY, XuH, MontellC (2006) A taste receptor required for the caffeine response in vivo. Curr Biol 16 : 1812–1817.
40. BrayS, AmreinH (2003) A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron 39 : 1019–1029.
41. WangL, HanX, MehrenJ, HiroiM, BilleterJC, et al. (2011) Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat Neurosci 14 : 757–762.
42. MiyamotoT, AmreinH (2008) Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci 11 : 874–876.
43. JonesWD, CayirliogluP, KadowIG, VosshallLB (2007) Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445 : 86–90.
44. LuT, QiuYT, WangG, KwonJY, RutzlerM, et al. (2007) Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr Biol 17 : 1533–1544.
45. ThorneN, AmreinH (2008) Atypical expression of Drosophila gustatory receptor genes in sensory and central neurons. J Comp Neurol 506 : 548–568.
46. BonasioR, ZhangG, YeC, MuttiNS, FangX, et al. (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329 : 1068–1071.
47. SmithCD, ZiminA, HoltC, AbouheifE, BentonR, et al. (2011) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci U S A 108 : 5673–5678.
48. SmithCR, SmithCD, RobertsonHM, HelmkampfM, ZiminA, et al. (2011) Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci U S A 108 : 5667–5672.
49. PittsRJ, RinkerDC, JonesPL, RokasA, ZwiebelLJ (2011) Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue - and sex-specific signatures of odor coding. BMC Genomics 12 : 271.
50. RobertsonHM, WannerKW (2006) The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res 16 : 1395–1403.
51. EngsontiaP, SandersonAP, CobbM, WaldenKK, RobertsonHM, et al. (2008) The red flour beetle's large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem Mol Biol 38 : 387–397.
52. NozawaM, NeiM (2007) Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci U S A 104 : 7122–7127.
53. Penalva-AranaDC, LynchM, RobertsonHM (2009) The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol 9 : 79.
54. SmadjaC, ShiP, ButlinRK, RobertsonHM (2009) Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol Biol Evol 26 : 2073–2086.
55. KentLB, RobertsonHM (2009) Evolution of the sugar receptors in insects. BMC Evol Biol 9 : 41.
56. SloneJ, DanielsJ, AmreinH (2007) Sugar receptors in Drosophila. Curr Biol 17 : 1809–1816.
57. DahanukarA, LeiYT, KwonJY, CarlsonJR (2007) Two Gr genes underlie sugar reception in Drosophila. Neuron 56 : 503–516.
58. JiaoY, MoonSJ, MontellC (2007) A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc Natl Acad Sci U S A 104 : 14110–14115.
59. KwonJY, DahanukarA, WeissLA, CarlsonJR (2007) The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A 104 : 3574–3578.
60. RobertsonHM, KentLB (2009) Evolution of the gene lineage encoding the carbon dioxide receptor in insects. J Insect Sci 9 : 19.
61. KleineidamC, TautzJ (1996) Perception of carbon dioxide and other “air-condition” parameters in the leaf cutting ant Atta cephalotes. Naturwissenschaften 83 : 566–568.
62. De BieT, CristianiniN, DemuthJP, HahnMW (2006) CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22 : 1269–1271.
63. ChenK, DurandD, Farach-ColtonM (2000) NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol 7 : 429–447.
64. HahnMW, De BieT, StajichJE, NguyenC, CristianiniN (2005) Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res 15 : 1153–1160.
65. VosshallLB, WongAM, AxelR (2000) An olfactory sensory map in the fly brain. Cell 102 : 147–159.
66. WangG, CareyAF, CarlsonJR, ZwiebelLJ (2010) Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 107 : 4418–4423.
67. Burdock GA (1997) Encyclopedia of food and color additives. Boca Raton: CRC Press.
68. Khan IA, Abourashed EA (2010) Leung's encyclopedia of common natural ingredients: used in food, drugs, and cosmetics. Hoboken, N.J.: John Wiley & Sons, Inc. xxix, 810 p. p.
69. Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J (2011) Comparative Genomics of the Major Chemosensory Gene Families in Arthropods. eLS. Chichester: John Wiley & Sons Ltd.
70. GadauJ, HelmkampfM, NygaardS, RouxJ, SimolaDF, et al. (2012) The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet 28 : 14–21.
71. NeiM, NiimuraY, NozawaM (2008) The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet 9 : 951–963.
72. XuG, MaH, NeiM, KongH (2009) Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc Natl Acad Sci U S A 106 : 835–840.
73. UrbaniCB, BoyanGS, BlarerA, BillenJ, AliTMM (1994) A Novel Mechanism for Jumping in the Indian Ant Harpegnathos Saltator (Jerdon) (Formicidae, Ponerinae). Experientia 50 : 63–71.
74. LiebigJ, HeinzeJ, HölldoblerB (1997) Trophallaxis and aggression in the ponerine ant, Ponera coarctata: Implications for the evolution of liquid food exchange in the Hymenoptera. Ethology 103 : 707–722.
75. SatoK, TanakaK, TouharaK (2011) Sugar-regulated cation channel formed by an insect gustatory receptor. Proc Natl Acad Sci U S A 108 : 11680–11685.
76. ZubeC, RosslerW (2008) Caste - and sex-specific adaptations within the olfactory pathway in the brain of the ant Camponotus floridanus. Arthropod Struct Dev 37 : 469–479.
77. HoyerSC, LiebigJ, RösslerW (2005) Biogenic amines in the ponerine ant Harpegnathos saltator: serotonin and dopamine immunoreactivity in the brain. Arthropod Struct Dev 34 : 429–440.
78. NakanishiA, NishinoH, WatanabeH, YokohariF, NishikawaM (2010) Sex-specific antennal sensory system in the ant Camponotus japonicus: glomerular organizations of antennal lobes. J Comp Neurol 518 : 2186–2201.
79. OzakiM, Wada-KatsumataA, FujikawaK, IwasakiM, YokohariF, et al. (2005) Ant nestmate and non-nestmate discrimination by a chemosensory sensillum. Science 309 : 311–314.
80. NakanishiA, NishinoH, WatanabeH, YokohariF, NishikawaM (2009) Sex-specific antennal sensory system in the ant Camponotus japonicus: structure and distribution of sensilla on the flagellum. Cell Tissue Res 338 : 79–97.
81. DobritsaAA, van der Goes van NatersW, WarrCG, SteinbrechtRA, CarlsonJR (2003) Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37 : 827–841.
82. GoldmanAL, Van der Goes van NatersW, LessingD, WarrCG, CarlsonJR (2005) Coexpression of two functional odor receptors in one neuron. Neuron 45 : 661–666.
83. FishilevichE, VosshallLB (2005) Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol 15 : 1548–1553.
84. CoutoA, AleniusM, DicksonBJ (2005) Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol 15 : 1535–1547.
85. WannerKW, NicholsAS, WaldenKK, BrockmannA, LuetjeCW, et al. (2007) A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. Proc Natl Acad Sci U S A 104 : 14383–14388.
86. HölldoblerB (1966) Futterverteilung durch Männchen im Ameisenstaat. Z Vergl Physiol 52 : 430–455.
87. ErlerF, UlugI, YalcinkayaB (2006) Repellent activity of five essential oils against Culex pipiens. Fitoterapia 77 : 491–494.
88. PrajapatiV, TripathiAK, AggarwalKK, KhanujaSP (2005) Insecticidal, repellent and oviposition-deterrent activity of selected essential oils against Anopheles stephensi, Aedes aegypti and Culex quinquefasciatus. Bioresour Technol 96 : 1749–1757.
89. Munoz-TorresMC, ReeseJT, ChildersCP, BennettAK, SundaramJP, et al. (2011) Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res 39: D658–662.
90. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
91. BirneyE, ClampM, DurbinR (2004) GeneWise and Genomewise. Genome Res 14 : 988–995.
92. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 : 1792–1797.
93. ZdobnovEM, ApweilerR (2001) InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17 : 847–848.
94. StamatakisA (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 : 2688–2690.
95. RoshanU, LivesayDR (2006) Probalign: multiple sequence alignment using partition function posterior probabilities. Bioinformatics 22 : 2715–2721.
96. NicholasKB, NicholasHB, DeerfieldDW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNETnews 4 : 1–4.
97. Capella-GutierrezS, Silla-MartinezJM, GabaldonT (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 : 1972–1973.
98. LeSQ, GascuelO (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25 : 1307–1320.
99. NamJ, NeiM (2005) Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol 22 : 2386–2394.
100. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
101. TrapnellC, WilliamsBA, PerteaG, MortazaviA, KwanG, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515.
102. BradySG, SchultzTR, FisherBL, WardPS (2006) Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci U S A 103 : 18172–18177.
103. BohbotJD, JonesPL, WangG, PittsRJ, PaskGM, et al. (2011) Conservation of indole responsive odorant receptors in mosquitoes reveals an ancient olfactory trait. Chem Senses 36 : 149–160.
Štítky
Genetika Reprodukční medicína
Článek Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma GenomesČlánek Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail ExpansionČlánek Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid CellsČlánek Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug ActivationČlánek The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin DynamicsČlánek Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 8- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Mutational Signatures of De-Differentiation in Functional Non-Coding Regions of Melanoma Genomes
- Rescuing Alu: Recovery of Inserts Shows LINE-1 Preserves Alu Activity through A-Tail Expansion
- Genetics and Regulatory Impact of Alternative Polyadenylation in Human B-Lymphoblastoid Cells
- Chromosome Territories Meet a Condensin
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Fine-Mapping and Initial Characterization of QT Interval Loci in African Americans
- Genome Patterns of Selection and Introgression of Haplotypes in Natural Populations of the House Mouse ()
- A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins
- Advances in Quantitative Trait Analysis in Yeast
- Experimental Evolution of a Novel Sexually Antagonistic Allele
- Variation of Contributes to Dog Breed Skull Diversity
- , a Gene Involved in Axonal Pathfinding, Is Mutated in Patients with Kallmann Syndrome
- A Single Origin for Nymphalid Butterfly Eyespots Followed by Widespread Loss of Associated Gene Expression
- Cryptocephal, the ATF4, Is a Specific Coactivator for Ecdysone Receptor Isoform B2
- Retrovolution: HIV–Driven Evolution of Cellular Genes and Improvement of Anticancer Drug Activation
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
- A Sexual Ornament in Chickens Is Affected by Pleiotropic Alleles at and , Selected during Domestication
- Use of Allele-Specific FAIRE to Determine Functional Regulatory Polymorphism Using Large-Scale Genotyping Arrays
- Novel Loci for Metabolic Networks and Multi-Tissue Expression Studies Reveal Genes for Atherosclerosis
- The Genetic Basis of Pollinator Adaptation in a Sexually Deceptive Orchid
- Uncovering the Genome-Wide Transcriptional Responses of the Filamentous Fungus to Lignocellulose Using RNA Sequencing
- Inheritance Beyond Plain Heritability: Variance-Controlling Genes in
- The Metabochip, a Custom Genotyping Array for Genetic Studies of Metabolic, Cardiovascular, and Anthropometric Traits
- Reprogramming to Pluripotency Can Conceal Somatic Cell Chromosomal Instability
- Condensin II Promotes the Formation of Chromosome Territories by Inducing Axial Compaction of Polyploid Interphase Chromosomes
- PTEN Negatively Regulates MAPK Signaling during Vulval Development
- A Dynamic Response Regulator Protein Modulates G-Protein–Dependent Polarity in the Bacterium
- Population Genomics of the Facultatively Mutualistic Bacteria and
- Components of a Fanconi-Like Pathway Control Pso2-Independent DNA Interstrand Crosslink Repair in Yeast
- Polysome Profiling in Liver Identifies Dynamic Regulation of Endoplasmic Reticulum Translatome by Obesity and Fasting
- Stromal Liver Kinase B1 [STK11] Signaling Loss Induces Oviductal Adenomas and Endometrial Cancer by Activating Mammalian Target of Rapamycin Complex 1
- Reprogramming of H3K27me3 Is Critical for Acquisition of Pluripotency from Cultured Tissues
- Transgene Induced Co-Suppression during Vegetative Growth in
- Hox and Sex-Determination Genes Control Segment Elimination through EGFR and Activity
- A Quantitative Comparison of the Similarity between Genes and Geography in Worldwide Human Populations
- Minibrain/Dyrk1a Regulates Food Intake through the Sir2-FOXO-sNPF/NPY Pathway in and Mammals
- Comparative Analysis of Regulatory Elements between and by Genome-Wide Transcription Start Site Profiling
- Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome
- S Phase–Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
- Cell-Nonautonomous Signaling of FOXO/DAF-16 to the Stem Cells of
- The Mi-2 Chromatin-Remodeling Factor Regulates Higher-Order Chromatin Structure and Cohesin Dynamics
- Comparative Analysis of the Genomes of Two Field Isolates of the Rice Blast Fungus
- Role of Mex67-Mtr2 in the Nuclear Export of 40S Pre-Ribosomes
- Genetic Modulation of Lipid Profiles following Lifestyle Modification or Metformin Treatment: The Diabetes Prevention Program
- HAL-2 Promotes Homologous Pairing during Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors
- SLX-1 Is Required for Maintaining Genomic Integrity and Promoting Meiotic Noncrossovers in the Germline
- Phylogenetic and Transcriptomic Analysis of Chemosensory Receptors in a Pair of Divergent Ant Species Reveals Sex-Specific Signatures of Odor Coding
- Reduced Prostasin (CAP1/PRSS8) Activity Eliminates HAI-1 and HAI-2 Deficiency–Associated Developmental Defects by Preventing Matriptase Activation
- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- Identification of Human Proteins That Modify Misfolding and Proteotoxicity of Pathogenic Ataxin-1
- and Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR
- CDK9 and H2B Monoubiquitination: A Well-Choreographed Dance
- Rare Copy Number Variations in Adults with Tetralogy of Fallot Implicate Novel Risk Gene Pathways
- Ccdc94 Protects Cells from Ionizing Radiation by Inhibiting the Expression of
- NOL11, Implicated in the Pathogenesis of North American Indian Childhood Cirrhosis, Is Required for Pre-rRNA Transcription and Processing
- Human Developmental Enhancers Conserved between Deuterostomes and Protostomes
- A Luminal Glycoprotein Drives Dose-Dependent Diameter Expansion of the Hindgut Tube
- Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Dynamic Distribution of Linker Histone H1.5 in Cellular Differentiation
- Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration
- Chemical Genetics Reveals a Specific Requirement for Cdk2 Activity in the DNA Damage Response and Identifies Nbs1 as a Cdk2 Substrate in Human Cells
- Experimental Relocation of the Mitochondrial Gene to the Nucleus Reveals Forces Underlying Mitochondrial Genome Evolution
- Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome
- Mutations in a P-Type ATPase Gene Cause Axonal Degeneration
- A General G1/S-Phase Cell-Cycle Control Module in the Flowering Plant
- Multiple Roles and Interactions of and in Development of the Respiratory System
- UNC-40/DCC, SAX-3/Robo, and VAB-1/Eph Polarize F-Actin during Embryonic Morphogenesis by Regulating the WAVE/SCAR Actin Nucleation Complex
- Epigenetic Remodeling of Meiotic Crossover Frequency in DNA Methyltransferase Mutants
- Modulating the Strength and Threshold of NOTCH Oncogenic Signals by
- Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer's Disease–Related Tau Phosphorylation Via PAR-1
- Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System
- ATXN2-CAG42 Sequesters PABPC1 into Insolubility and Induces FBXW8 in Cerebellum of Old Ataxic Knock-In Mice
- Cohesin Rings Devoid of Scc3 and Pds5 Maintain Their Stable Association with the DNA
- The MicroRNA Inhibits Calcium Signaling by Targeting the TIR-1/Sarm1 Adaptor Protein to Control Stochastic L/R Neuronal Asymmetry in
- Rapid-Throughput Skeletal Phenotyping of 100 Knockout Mice Identifies 9 New Genes That Determine Bone Strength
- The Genes Define Unique Classes of Two-Partner Secretion and Contact Dependent Growth Inhibition Systems
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dissecting the Gene Network of Dietary Restriction to Identify Evolutionarily Conserved Pathways and New Functional Genes
- It's All in the Timing: Too Much E2F Is a Bad Thing
- Variation of Contributes to Dog Breed Skull Diversity
- The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



