-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
Small RNAs regulate diverse biological processes by directing effector proteins called Argonautes to silence complementary mRNAs. Maturation of some classes of small RNAs involves terminal 2′-O-methylation to prevent degradation. This modification is catalyzed by members of the conserved HEN1 RNA methyltransferase family. In animals, Piwi-interacting RNAs (piRNAs) and some endogenous and exogenous small interfering RNAs (siRNAs) are methylated, whereas microRNAs are not. However, the mechanisms that determine animal HEN1 substrate specificity have yet to be fully resolved. In Caenorhabditis elegans, a HEN1 ortholog has not been studied, but there is evidence for methylation of piRNAs and some endogenous siRNAs. Here, we report that the worm HEN1 ortholog, HENN-1 (HEN of Nematode), is required for methylation of C. elegans small RNAs. Our results indicate that piRNAs are universally methylated by HENN-1. In contrast, 26G RNAs, a class of primary endogenous siRNAs, are methylated in female germline and embryo, but not in male germline. Intriguingly, the methylation pattern of 26G RNAs correlates with the expression of distinct male and female germline Argonautes. Moreover, loss of the female germline Argonaute results in loss of 26G RNA methylation altogether. These findings support a model wherein methylation status of a metazoan small RNA is dictated by the Argonaute to which it binds. Loss of henn-1 results in phenotypes that reflect destabilization of substrate small RNAs: dysregulation of target mRNAs, impaired fertility, and enhanced somatic RNAi. Additionally, the henn-1 mutant shows a weakened response to RNAi knockdown of germline genes, suggesting that HENN-1 may also function in canonical RNAi. Together, our results indicate a broad role for HENN-1 in both endogenous and exogenous gene silencing pathways and provide further insight into the mechanisms of HEN1 substrate discrimination and the diversity within the Argonaute family.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002617
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002617Summary
Small RNAs regulate diverse biological processes by directing effector proteins called Argonautes to silence complementary mRNAs. Maturation of some classes of small RNAs involves terminal 2′-O-methylation to prevent degradation. This modification is catalyzed by members of the conserved HEN1 RNA methyltransferase family. In animals, Piwi-interacting RNAs (piRNAs) and some endogenous and exogenous small interfering RNAs (siRNAs) are methylated, whereas microRNAs are not. However, the mechanisms that determine animal HEN1 substrate specificity have yet to be fully resolved. In Caenorhabditis elegans, a HEN1 ortholog has not been studied, but there is evidence for methylation of piRNAs and some endogenous siRNAs. Here, we report that the worm HEN1 ortholog, HENN-1 (HEN of Nematode), is required for methylation of C. elegans small RNAs. Our results indicate that piRNAs are universally methylated by HENN-1. In contrast, 26G RNAs, a class of primary endogenous siRNAs, are methylated in female germline and embryo, but not in male germline. Intriguingly, the methylation pattern of 26G RNAs correlates with the expression of distinct male and female germline Argonautes. Moreover, loss of the female germline Argonaute results in loss of 26G RNA methylation altogether. These findings support a model wherein methylation status of a metazoan small RNA is dictated by the Argonaute to which it binds. Loss of henn-1 results in phenotypes that reflect destabilization of substrate small RNAs: dysregulation of target mRNAs, impaired fertility, and enhanced somatic RNAi. Additionally, the henn-1 mutant shows a weakened response to RNAi knockdown of germline genes, suggesting that HENN-1 may also function in canonical RNAi. Together, our results indicate a broad role for HENN-1 in both endogenous and exogenous gene silencing pathways and provide further insight into the mechanisms of HEN1 substrate discrimination and the diversity within the Argonaute family.
Introduction
Argonautes are an evolutionarily conserved family of proteins implicated in diverse cellular processes. They function as effector proteins in the RNA-induced silencing complex (RISC), a gene regulatory complex that binds small, non-coding RNAs to target its silencing effects. Small RNAs are broadly segregated into groups that differ in their mechanisms of biogenesis and silencing, as well as in the subsets of Argonaute effectors that bind them. The microRNAs (miRNAs) are highly conserved small RNAs processed from endogenous hairpin precursors that regulate networks of mRNAs primarily through post-transcriptional repression [1], [2]. The piRNAs, so named for the Piwi Argonautes that bind them, function predominantly in maintenance of germline integrity, often through repression of repetitive transposable elements. The small interfering RNAs comprise a more heterogeneous group that includes small RNAs derived from cleavage of exogenous double-stranded RNA (exo-siRNAs) or generated endogenously (endo-siRNAs).
Chemical modification has emerged as an important theme in regulation of small RNA function (for a review, see Kim et al., 2010 [3]). Internal editing has been found to occur in select miRNA precursors through the action of ADAR (adenosine deaminase acting on RNA) enzymes, with consequences for miRNA processing efficiency, stability, and targeting [4]–[8]. Some siRNAs generated in fly and mouse also show evidence of editing by ADARs [9], [10], but the significance of such internal editing among siRNAs is not yet known. In contrast, terminal editing through 2′-O-methylation, addition of untemplated nucleotides, or exonucleolytic trimming plays a more general role in small RNA metabolism. These terminal modifications are not unrelated. Evidence in plants and animals suggests that methylation of the 3′ terminal nucleotide protects small RNAs from polyuridylation and polyadenylation, signals that direct exonucleolytic degradation [11]–[16]. Thus, terminal methylation plays an important role in regulating small RNA turnover. Formation of the 2′-O-methyl group is catalyzed by HEN1, a methyltransferase discovered in Arabidopsis thaliana that is conserved across metazoa, fungi, viridiplantae, and bacteria [17]. Although plant and animal HEN1 orthologs exhibit 40–50% amino acid similarity in the conserved methyltransferase domain [18], the proteins differ in their substrate specificity. Plant HEN1 acts on small RNAs in duplex and methylates both siRNAs and miRNAs [19]–[21]. In contrast, animal HEN1 orthologs modify only single-stranded small RNAs [22]–[24], enabling methylation of small RNAs such as piRNAs, which are not derived from double-stranded RNA intermediates [25]–[29]. While animal piRNAs appear to be universally methylated [24], [26], [27], [30]–[32], animal miRNAs are generally not methylated [19], [26], [31], and the mechanisms by which animal HEN1 orthologs discriminate between substrates are not entirely clear. HEN1 orthologs that catalyze terminal methylation of small RNAs have been characterized in mouse, fish, and fly, among other organisms [15], [22]–[24], [33], yet the orthologous methyltransferase in worm [18] has yet to be investigated. With its expanded Argonaute family and diverse small RNA classes, Caenorhabditis elegans provides an advantage for studying HEN1 substrate specificity.
Since the discovery of the founding members of the microRNA family in C. elegans [1], [2], [34], many additional classes of small RNAs have been characterized. A large-scale small RNA sequencing effort revealed a class of terminally methylated 21-nucleotide RNAs with 5′ uridines [27]. These 21U RNAs were subsequently determined to represent the piRNAs of C. elegans based on their germline-specific expression, association with worm Piwi Argonautes PRG-1 and PRG-2, and function in transposon silencing and maintenance of temperature-dependent fertility [35]–[38]. Also found through small RNA cloning and deep sequencing were populations of 26 - and 22-nucleotide RNAs with a 5′ preference for guanosine (the 26G RNAs and 22G RNAs, respectively) that constitute the endo-siRNAs of C. elegans [27], [39]. The 26G RNAs are primary endo-siRNAs generated in the germline to regulate spermatogenic and zygotic gene expression. They are divided into two non-overlapping subclasses named for the Argonautes that bind them: the ERGO-1 class 26G RNAs, which are generated in the maternal germline and distributed into the embryo, and the ALG-3/ALG-4 class 26G RNAs, which are specific to the male germline and required for sperm function [40]–[42]. The 22G RNAs are composed of many small RNA classes, all of which are bound by worm-specific Argonautes (Wagos). A large population of 22G RNAs are secondary endo-siRNAs whose production by RNA-dependent RNA polymerases is triggered by the activity of 21U RNAs and 26G RNAs [36], [41]–[43]; however, many other 22G RNAs are independent of these primary small RNAs [44], [45]. Secondary siRNAs serve to amplify the signal of primary small RNAs to effect robust silencing. Production of 22G secondary siRNAs is also triggered by exogenously introduced dsRNAs [43], [45]–[47], suggesting convergence of endogenous and exogenous RNAi pathways at the level of the secondary siRNA response.
Among C. elegans small RNAs, only 21U RNAs and 26G RNAs are known to be methylated [27], [42]; 22G RNAs triggered by either primary endo - or exo-siRNAs appear to be unmethylated [45], [46]. Although the significance of worm small RNA methylation is unknown, loss of terminal methylation has been shown to decrease stability of piRNAs in many animal models [15], [22], [24] and both endo - and exo-siRNAs in fly [22], [48]. Methylation may therefore represent an essential step in stabilization of some classes of worm small RNAs.
In this study, we characterize the C. elegans hen1 ortholog, which has been named henn-1 (hen of nematode), as the name hen-1 has already been assigned to an unrelated C. elegans gene. We demonstrate that HENN-1 methylates small RNAs bound by Piwi clade Argonautes: the 21U RNAs and the ERGO-1 class 26G RNAs. However, we show that 26G RNAs bound by Ago clade Argonautes ALG-3 and ALG-4 are not methylated and are therefore henn-1-independent. Differential methylation of 26G RNAs provides evidence for an existing model [13], [22], [23], [49], [50] wherein evolutionarily divergent Argonautes either direct or prohibit HEN1-mediated methylation of associated small RNAs. In further support of this Argonaute-dictated methylation model, we find that small RNAs are likely methylated after associating with an Argonaute: the Argonaute ERGO-1 is required for 26G RNA methylation, but methylation is not required for ERGO-1 to bind a 26G RNA.
In the henn-1 mutant, levels of both 21U RNAs and ERGO-1 class 26G RNAs drop precipitously after their deposition into embryo, suggesting that HENN-1-mediated methylation is essential for perdurance of the maternal small RNA load during filial development. Accordingly, the henn-1 mutant shows enhanced somatic sensitivity to exogenous RNAi, a phenotype associated with loss of ERGO-1 class 26G RNAs. Surprisingly, however, the henn-1 mutant germline exhibits an attenuated response to RNAi, suggesting that HENN-1 may also function in the exogenous RNAi pathway. Altogether, our study supports a role for HENN-1 in diverse small RNA pathways in C. elegans and offers further insight into the mechanisms governing substrate discrimination for animal HEN1 orthologs.
Results
C02F5.6 Encodes the C. elegans HEN1 Ortholog
To examine small RNA methylation in C. elegans, we began by characterizing C02F5.6, the gene previously predicted to encode the HEN1 ortholog in worm [18]. This gene, subsequently named henn-1, encodes a protein that exhibits significant amino acid similarity across the conserved HEN1 methyltransferase domain relative to established members of the HEN1 family (Figure S1). Although two henn-1 gene models with differing 3′ ends have been proposed, 3′RACE and protein studies using a rabbit polyclonal antibody generated against a common N-terminal HENN-1 epitope detected only the longer isoform (Figure S2A, S2B).
To facilitate our studies of the function of HENN-1, we isolated and characterized the henn-1(tm4477) allele. This allele carries a deletion that encompasses henn-1 exon four, which encodes 65% of the conserved methyltransferase domain as annotated by Kamminga et al. [15]. Sequencing of the henn-1(tm4477) mRNA indicates that loss of exon four activates a cryptic splice donor site in the third intron, resulting in an extended third exon that encodes a premature termination codon (Figure S2B). The henn-1(tm4477) mRNA is readily detected by RT-PCR but does not produce a detectable protein product (Figure S2A) or exhibit methyltransferase activity (see below), suggesting that henn-1(tm4477) (hereafter, henn-1) represents a functional null allele.
HENN-1 Terminally Methylates and Stabilizes C. elegans piRNAs
Like piRNAs in fly [22], [23], [32], mouse [30], [31], and zebrafish [26], the C. elegans 21U RNAs are terminally methylated [27], but the factor catalyzing this modification has not yet been identified. To determine if 21U RNA methylation depends on henn-1, we assessed methylation status using the β-elimination assay [51]. A small RNA molecule whose terminal nucleotide has been 2′-O-methylated is resistant to this treatment, whereas the cis-diols of an unmodified 3′ terminal nucleotide are oxidized by sodium periodate, rendering the nucleotide susceptible to β-elimination under basic conditions. The resulting size difference can be resolved on a polyacrylamide gel to determine methylation status. All 21U RNAs examined were found to be terminally methylated in a henn-1-dependent manner (Figure 1A, Figure S3A), whereas a control miRNA was not methylated in either wild-type or henn-1 mutant animals (Figure 1B). Although 21U RNAs are still detectable in the henn-1 mutant, the abundance of the full-length species is visibly decreased for some 21U RNAs; this correlates with the appearance of putative degradation products of unmethylated, unprotected 21U RNAs. To demonstrate that loss of 21U RNA methylation in the henn-1 mutant is specifically due to the absence of henn-1, we used the Mos1-mediated single copy insertion technique [52] to introduce a henn-1::gfp transgene driven by the promoter of the polycistronic mRNA that encodes henn-1 (xkSi1) or by the germline-specific pie-1 promoter (xkSi2) into the henn-1 mutant (Figure S2C). Both endogenous and germline-specific expression of henn-1::gfp restore 21U RNA methylation in the henn-1 mutant (Figure 1A).
Fig. 1. Methylation of 21U RNAs Requires C. elegans HEN1 Ortholog HENN-1. 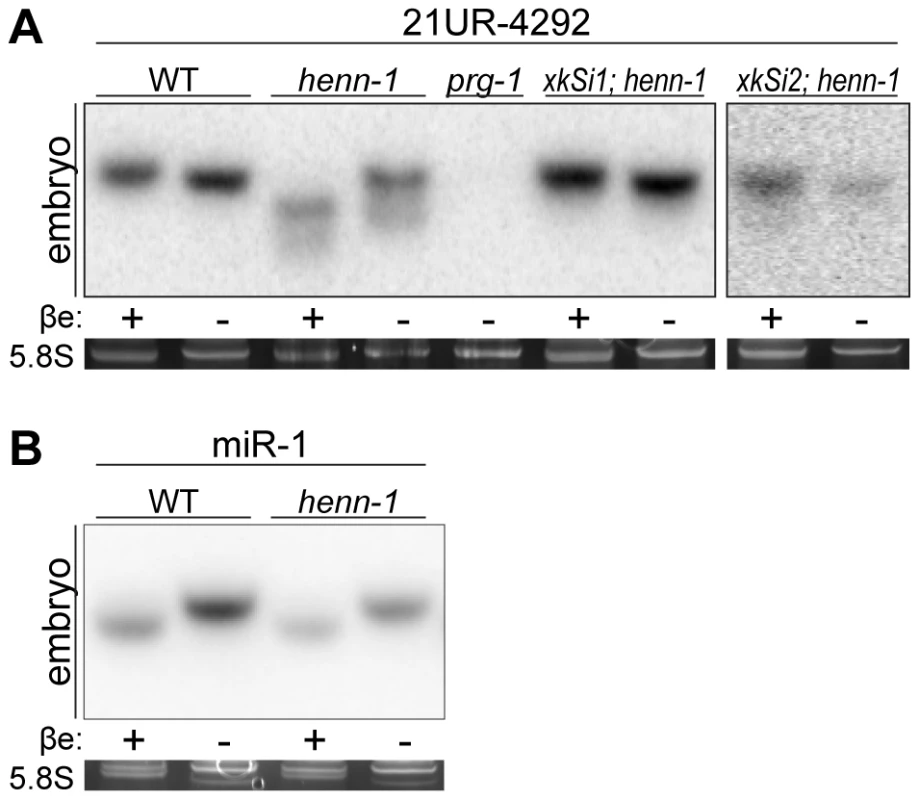
A) HENN-1 is required for 21U RNA methylation. Endogenous (xkSi1) and germline-specific (xkSi2) expression of henn-1::gfp rescue 21U RNA methylation in henn-1(tm4477) mutant embryo. Total embryo RNA of the indicated genotypes was β-eliminated (βe +) or control treated (βe −) and probed for piRNA 21UR-4292. prg-1(tm872) lacks 21U RNAs and is included as a negative control. Below, ethidium bromide staining of 5.8S rRNA is shown. Additional 21U RNA northern blots are shown in Figure S3A. B) C. elegans miRNAs are unmethylated. Total embryo RNA was probed for miR-1. Variable intensity of 5.8S rRNA bands in embryo indicates unequal loading. To investigate the relationship between terminal methylation and piRNA accumulation, we used Taqman RT-qPCR to assess 21U RNA levels in wild-type and henn-1 mutant animals across development at 25°C. Importantly, the Taqman stem-loop RT primer is capable of distinguishing between full-length and terminally degraded small RNAs [53]. For example, the let-7e miRNA differs from let-7a only in the absence of the final nucleotide and U>G substitution at the ninth nucleotide, a position likely not represented in the stem-loop Taqman primer. Absence of this final nucleotide decreases detection of let-7e by the let-7a Taqman assay by more than a thousandfold [53]. henn-1 mutant embryo and early larva show dramatically reduced detection of female germline-enriched piRNA 21UR-1848 (Figure 2A), consistent with decreased embryonic detection for some 21U RNAs observed by northern blot (Figure 1A, Figure S3A). 21U RNA levels recover to wild-type in late larval stages, coincident with the onset of germline proliferation and de novo 21U RNA biosynthesis; however, in gravid animals at 56 hours, 21UR-1848 levels in the henn-1 mutant have declined to less than 50% of those observed in wild-type (P = 0.0005; two-tailed t-test). Eight additional 21U RNAs examined show a similar pattern (Figure S4). These data suggest that henn-1 is dispensable for piRNA biogenesis but essential for robust inheritance of piRNAs. Parallel analysis of miR-1 and several additional miRNAs across development shows that effects of loss of henn-1 are specific to its substrates and not due to generalized small RNA dysregulation in the henn-1 mutant (Figure 2B, Figure S5).
Fig. 2. HENN-1 Stabilizes 21U RNAs. 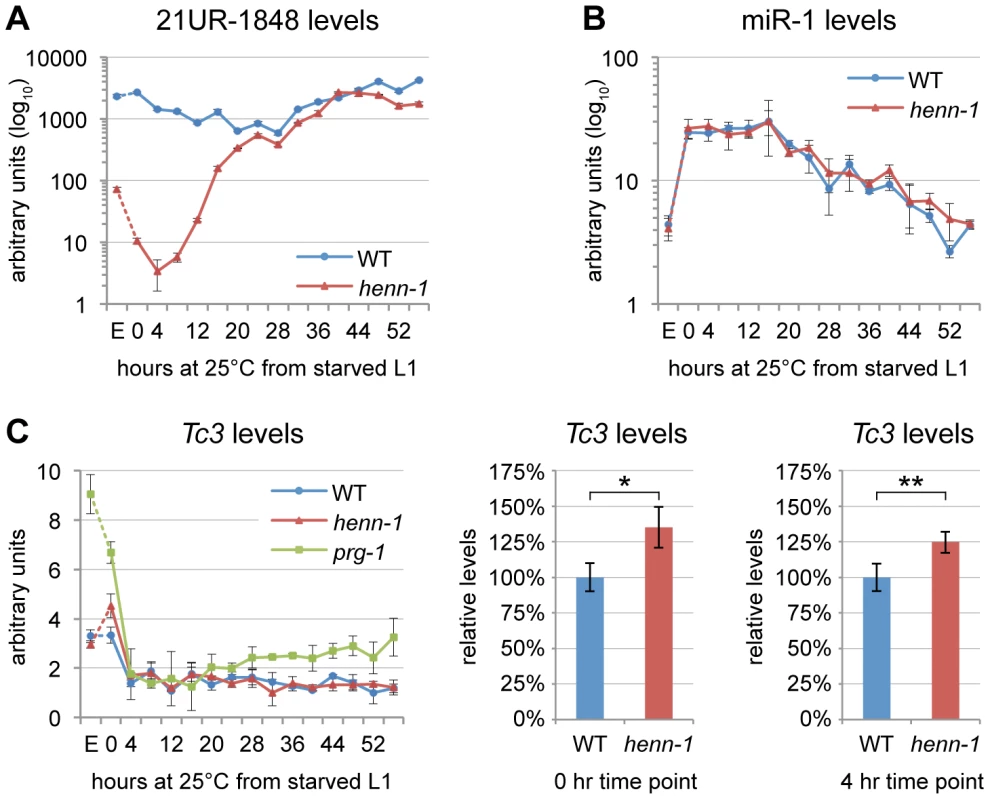
A) Loss of henn-1 impairs 21U RNA accumulation in adult, embryo, and early larva. Levels of 21UR-1848 were assayed by Taqman qPCR in embryo and every four hours across development of wild-type and henn-1(tm4477) mutant animals at 25°C. Standard deviation is shown for biological triplicates. Taqman qPCR data for eight additional 21U RNAs are shown in Figure S4. B) Effects of loss of henn-1 are restricted to its small RNA substrates. Levels of miR-1 across development were assayed by Taqman qPCR. Standard deviation is shown for biological triplicates. Additional Taqman qPCR data for miRNAs are shown in Figure S5. C) Loss of henn-1 impairs Tc3 transposase silencing primarily in early L1 larva. Tc3 transposase mRNA levels were assayed by qPCR across development and normalized to mRNA levels of eft-2, an abundantly expressed housekeeping gene. prg-1(tm872) lacks 21U RNAs and is included as a positive control for Tc3 upregulation. Significant zero and four hour time points are expanded at right (*: P = 0.0251; **: P = 0.0250, two-tailed t-test). Standard deviation is shown for biological triplicates. E, embryo; hr, hour. HENN-1 Plays a Minor Role in piRNA–Mediated Germline Regulation
We next sought to determine the extent to which decreased abundance of piRNAs in the henn-1 mutant compromises activity of the piRNA pathway. Unlike in fly, where many selfish genetic elements are desilenced in the absence of piRNAs [32], C. elegans at present has only a single established molecular readout for piRNA pathway function: increased expression of transposase mRNA from Tc3, a Tc1/mariner family transposon [35], [36]. Two 21U RNAs have been found to map to Tc3, but both map in the sense direction and thus are unlikely to act directly in Tc3 repression via canonical RNAi mechanisms [35], [36]. Rather, 21U RNAs likely mediate their repressive effects through triggering production of secondary siRNAs, 22G RNAs, that engage worm-specific Argonautes (Wagos) to effect Tc3 gene silencing [36], [45]. We therefore identified a 22G RNA that shows complete antisense complementarity to Tc3 and can be classified as a Wago-dependent, 21U RNA-dependent secondary siRNA based on its total depletion both in the MAGO12 mutant, which lacks all Wagos, and in the prg-1(n4357); prg-2(n4358) double mutant, which lacks piRNAs [36], [45]. Levels of this 22G RNA in the henn-1 mutant are reduced by 44% in embryo but not significantly altered in hatched L1 larva (Figure S6A). This suggests that the low embryonic and early larval levels of 21U RNAs in the henn-1 mutant are still sufficient to trigger production of secondary siRNAs, although to a lesser degree than in wild-type.
Consistent with the modest effect of loss of henn-1 on accumulation of piRNA-triggered secondary siRNAs, henn-1 mutant animals exhibit only a small increase (35% in starved L1 larva, 25% in L1 larva fed for 4 hours at 25°C) in Tc3 transposase mRNA levels relative to wild-type (Figure 2C). This is not unexpected due to the poor coincidence of the time intervals corresponding to piRNA dysregulation in the henn-1 mutant and Tc3 sensitivity to 21U RNAs; the henn-1 mutant shows the greatest disparity in piRNA levels in early larval development, whereas Tc3 levels are most sensitive to piRNAs in germline and embryo (Figure 2A, 2C). These findings suggest that HENN-1 is not strictly required for piRNA target repression, but contributes to robust silencing of Tc3.
In addition to Tc3 dysregulation, loss of prg-1 also results in a temperature-sensitive sterility phenotype [38], [43]. To determine if the henn-1 mutant also exhibits a fertility defect, we assessed fertility at 20°C and 25°C. At 20°C, brood size of the henn-1 mutant does not differ significantly from that of wild-type. In contrast, henn-1 mutant animals maintained at 25°C exhibit a 25% decrease in brood size relative to wild-type (P = 0.0059; two-tailed t-test) that can be rescued by germline expression of henn-1::gfp from the xkSi2 transgene (Figure S7). The impaired fertility of the henn-1 mutant is consistent with abnormal fertility phenotypes associated with loss of HEN1 methyltransferase activity in other animals. Loss of HEN1 in Tetrahymena thermophila depletes Piwi-interacting RNAs called scan RNAs, impairing DNA elimination and, consequently, the viability of progeny [24]. The zebrafish hen1 mutant fails to maintain a female germline, resulting in an exclusively male population [15]. Nevertheless, we cannot conclude that the temperature-sensitive fertility defect of the henn-1 mutant is due exclusively to compromise of the 21U RNA pathway.
ERGO-1 and ALG-3/ALG-4 Class 26G RNAs Are Differentially Methylated by HENN-1
26G RNAs were reported to be methylated in the first C. elegans small RNA deep sequencing study [27]. Subsequent studies concluded that the species assessed was an ERGO-1 class 26G RNA [40]. Consistent with these data, we found that ERGO-1 class 26G RNAs, found in female germline and embryo, are methylated. As was the case for piRNAs, this methylation occurs in a henn-1-dependent manner (Figure 3A, Figure S3B). Surprisingly, however, ALG-3/ALG-4 class 26G RNAs, specific to the male germline, showed no evidence of methylation even in wild-type animals (Figure 3B, Figure S3C). One potential explanation for this observation would be that female germline small RNAs are universally methylated, whereas male germline small RNAs are not. To explore this possibility, we assessed 21U RNAs in male and female germlines. Both were methylated (Figure 3C), indicating that differential 26G RNA methylation cannot be explained simply by a lack of methyltransferase functionality in the male germline.
Fig. 3. HENN-1 Selectively Methylates ERGO-1 Class 26G RNAs in an ERGO-1–Dependent Manner. 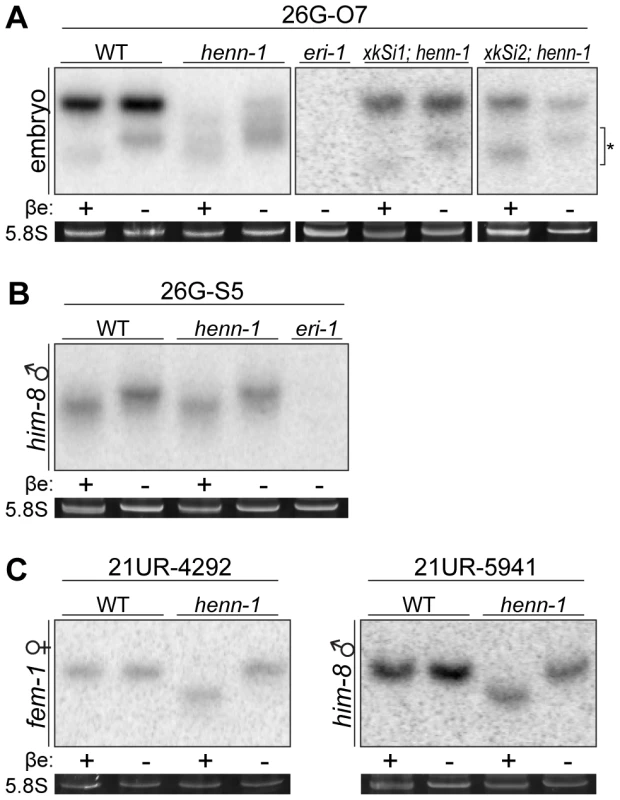
A) HENN-1 is required for ERGO-1 class 26G RNA methylation and stability. Total β-eliminated (βe +) or control treated (βe −) embryo RNA of the indicated genotypes was probed for ERGO-1 class 26G RNA 26G-O7. eri-1(mg366) lacks 26G RNAs and is included as a negative control. Asterisk indicates signal corresponding to cross-hybridization with unmethylated 22G RNAs. Below, ethidium bromide staining of 5.8S rRNA. Additional ERGO-1 class 26G RNA northern blots are shown in Figure S3B. B) ALG-3/ALG-4 class 26G RNAs are unmethylated. Total him-8(e1489) male RNA was probed for ALG-3/ALG-4 class 26G RNA 26G-S5. An additional ALG-3/ALG-4 class 26G RNA northern blot is shown in Figure S3C. C) 21U RNAs are methylated in a HENN-1-dependent manner in both female and male germlines. Total RNA of the indicated genotypes from fem-1(hc17) female or him-8(e1489) male was probed for female germline-enriched piRNA 21UR-4292 or male germline-enriched piRNA 21UR-5941, respectively. Because the two classes of 26G RNAs bind unique Argonautes in male and female germlines, we hypothesized that the Argonaute ERGO-1 might direct methylation of 26G RNAs. To address this question, we sought to assess methylation of an ERGO-1 class 26G RNA in the absence of ERGO-1. As 26G RNAs are dramatically depleted in the absence of their respective Argonautes [40], we queried published wild-type and ergo-1(tm1860) gravid adult deep sequencing libraries [42] to identify an ERGO-1 class 26G RNA that still accumulates to levels sufficient for visualization by northern blotting in the ergo-1(tm1860) mutant. 26G-O1, an extremely abundant ERGO-1 class 26G RNA, is present at roughly 0.5% wild-type levels in the ergo-1(tm1860) mutant, but still abundant enough to detect by northern blotting. Consistent with our hypothesis that ERGO-1 is required for 26G RNA methylation, we found that 26G-O1 is unmethylated in the ergo-1(tm1860) mutant embryo (Figure 4A). We next asked the converse question: Is 26G RNA methylation required for association with ERGO-1? We immunopurified ERGO-1 complexes from wild-type and henn-1 mutant embryo lysates (Figure 4B) and extracted RNA. In both wild-type and henn-1 mutant samples, ERGO-1 class 26G RNAs are readily detected (Figure 4C), indicating that ERGO-1 effectively binds both methylated and unmethylated 26G RNAs. Taken together, these data suggest that 26G RNAs bind ERGO-1 and are subsequently methylated by HENN-1.
Fig. 4. ERGO-1 Is Required for Methylation of 26G RNAs. 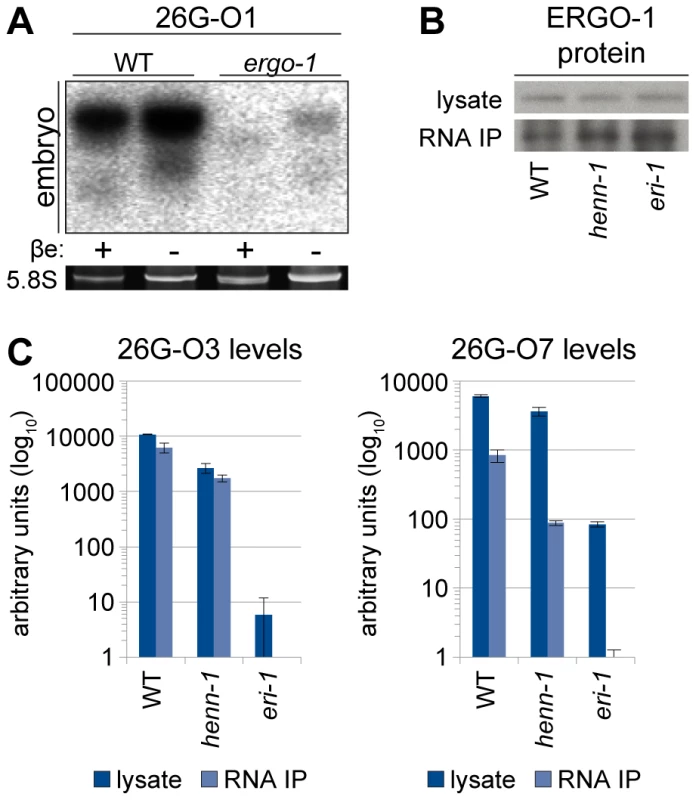
A) ERGO-1 class 26G RNA 26G-O1 is unmethylated in the absence of ERGO-1. Total embryo wild-type (5 µg) or ergo-1(tm1860) (10 µg) β-eliminated (βe +) or control treated (βe −) RNA was probed for 26G-O1. B) Anti-ERGO-1 rabbit polyclonal antibody immunoprecipitates ERGO-1 complexes. ERGO-1 complexes were immunopurified from lysates of equalized protein concentration extracted from wild-type, henn-1(tm4477) mutant, or eri-1(mg366) mutant embryo. Aliquots of lysates and immunoprecipitates (RNA IP) were probed with anti-ERGO-1 antibody. ergo-1(tm1860) mutant lysate was run in parallel to ensure specificity of ERGO-1 detection (data not shown). C) ERGO-1 binds methylated and unmethylated 26G RNAs. Taqman RT-qPCR for the indicated ERGO-1 class 26G RNAs was performed on samples described in B. The eri-1(mg366) mutant lacks 26G RNAs and serves as a negative control to demonstrate specificity of 26G RNA detection by Taqman assay. Standard deviation is shown for technical duplicates. Results are representative of two independent RNA immunoprecipitation experiments. To test whether HENN-1-mediated methylation is required to maintain levels of all substrate small RNAs, we assessed ERGO-1 class 26G RNAs for defects in accumulation in the henn-1 mutant. Loss of henn-1 has more severe consequences for this class of small RNAs than are observed for 21U RNAs: ERGO-1 class 26G RNA 26G-O3 fails to accumulate to wild-type levels at any stage of development, although the disparity is less pronounced in adulthood, during peak 26G RNA biogenesis (Figure 5A). For comparison, we assayed levels of ALG-3/ALG-4 class 26G RNA 26G-S5 across the developmental window during which it is readily detected by Taqman RT-qPCR. Levels of 26G-S5 are similar in the henn-1 mutant relative to wild-type (Figure 5B), consistent with the idea that HENN-1 is required for accumulation of ERGO-1 class 26G RNAs but dispensable for that of ALG-3/ALG-4 class 26G RNAs. Analysis of seven additional ERGO-1 class 26G RNAs and two additional ALG-3/ALG-4 class 26G RNAs corroborated these observations (Figures S8, S9).
Fig. 5. HEN1 Stabilizes ERGO-1 Class, but Not ALG-3/ALG-4 Class, 26G RNAs. 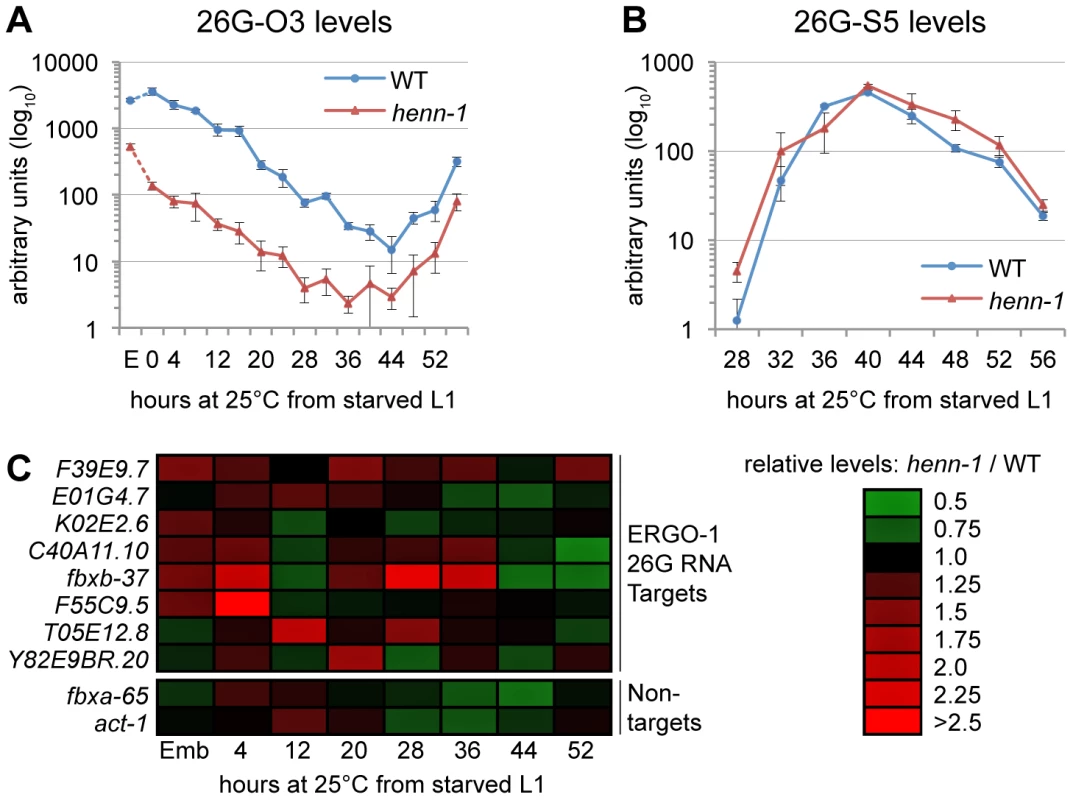
A) Loss of henn-1 impairs ERGO-1 class 26G RNA accumulation at all stages. Levels of ERGO-1 class 26G RNA 26G-O3 were assayed by Taqman qPCR across development of wild-type and henn-1(tm4477) mutant animals at 25°C. Standard deviation is shown for biological triplicates. Taqman qPCR data for seven additional ERGO-1 class 26G RNAs are shown in Figure S8. B) ALG-3/ALG-4 class 26G RNAs are henn-1-independent. Levels of ALG-3/ALG-4 class 26G RNA 26G-S5 were assayed across the period of development in which ALG-3/ALG-4 class 26G RNAs are readily detectable. Standard deviation is shown for biological triplicates. Taqman qPCR data for two additional ALG-3/ALG-4 class 26G RNAs are shown in Figure S9. C) Loss of henn-1 may result in modest, sporadic defects in ERGO-1 class 26G RNA target silencing. Levels of eight target and two non-target mRNAs were assayed across development of wild-type and henn-1(tm4477) mutant animals at 25°C and normalized to eft-2. Expression in the henn-1(tm4477) mutant relative to wild-type is represented according to the red-green color scheme indicated in the right panel. Raw data is shown in Figure S10. E, embryo. HENN-1 Contributes Minimally to ERGO-1 Class 26G RNA Target Silencing
To determine the effect of loss of henn-1 on the silencing of ERGO-1 class 26G RNA targets, we assayed levels of a panel of mRNAs targeted by ERGO-1 class 26G RNAs for desilencing in henn-1 mutant animals. During time points at which ERGO-1 class 26G RNAs are abundant, only modest upregulation of some, but not all, targets was detected; furthermore, no single target shows consistent desilencing in the henn-1 mutant (Figure 5C, Figure S10A). This is not unexpected, however, as the targets themselves vary in both expression and sensitivity to small RNA-mediated silencing across development [40]. To determine the specificity of this effect, two non-targets were examined in parallel. The maximal upregulation for either non-target does not exceed the maximal upregulation observed for any target, suggesting that the upregulation of ERGO-1 class 26G RNA targets in the henn-1 mutant may be a consequence of 26G RNA depletion (Figure 5C, Figure S10B). This connection is supported by our observation that a Wago-dependent and ERGO-1 class 26G RNA-dependent secondary siRNA that presumably enhances target silencing also shows defects in accumulation in embryo (Figure S6B). The effect is modest, indicating that, as observed for the piRNA pathway, the depleted pool of ERGO-1 class 26G RNAs in the henn-1 mutant is still sufficient for triggering fairly robust production of secondary siRNAs. Nevertheless, in an accompanying manuscript, Montgomery et al. observe that HENN-1 is required for silencing activity of a similar secondary siRNA upon a sensor transgene [54], suggesting that this pathway may indeed be compromised by loss of henn-1.
The Soma of the henn-1 Mutant Exhibits Enhanced Sensitivity to Exogenous RNAi
ALG-3/ALG-4 class 26G RNAs are restricted to the male germline, and their mRNA targets are enriched for genes involved in spermatogenesis [40]. Accordingly, loss of ALG-3/ALG-4 class 26G RNAs results in male-associated sterility at non-permissive temperatures due to defects in sperm activation that are thought to arise from target dysregulation [41]. ERGO-1 class 26G RNAs, in contrast, are dispensable for fertility and target mostly poorly conserved and incompletely annotated genes, many of which reside in duplicated regions of the genome [42]. It is therefore not unexpected that the ergo-1(tm1860) mutant, which lacks ERGO-1 class 26G RNAs, exhibits no overt phenotypes that can be traced to target dysregulation. Rather, the ergo-1(tm1860) mutant exhibits an enhanced RNAi sensitivity (Eri) phenotype that is attributed to effects of loss of the ERGO-1-dependent small RNAs themselves; presumably, depletion of ERGO-1 class 26G RNAs and dependent secondary siRNAs liberates limiting RNAi factors shared between the endogenous and exogenous RNAi pathways [43], [55], [56].
To determine whether loss of henn-1 depletes ERGO-1 class 26G RNAs sufficiently to produce an Eri phenotype, as observed in the ergo-1 mutant, we subjected L1 larvae from a panel of strains to feeding RNAi targeting various genes in the soma or germline. In order to expose subtle differences in RNAi sensitivity, we modulated the degree of knockdown, attenuating the dose of dsRNA trigger by diluting the bacterial RNAi clone with a bacterial clone harboring empty vector. RNAi of the somatic gene lir-1 causes larval arrest and lethality in wild-type animals at full strength, but dilution 1∶1 with empty vector largely eliminates the effect. In contrast, the eri-1(mg366) mutant, which lacks 26G RNAs, is affected severely by even dilute lir-1 RNAi. The henn-1 mutant also shows dramatically increased sensitivity to lir-1 feeding RNAi relative to wild-type (Figure 6A, 6B). A henn-1; eri-1 double mutant, however, shows RNAi sensitivity that is virtually identical to that of the single eri-1 mutant, suggesting that the Eri phenotype of each allele likely stems from the same defect, namely, loss of ERGO-1 class 26G RNAs. While the somatic Eri phenotype of the henn-1 mutant shows partial rescue by the germline-specific henn-1::gfp transgene xkSi2, henn-1::gfp expression under the native promoter from transgene xkSi1 rescues wild type RNAi sensitivity completely in the henn-1 mutant (Figure 6B). These findings suggest that loss of henn-1 in both germline and soma contributes to the Eri phenotype of the henn-1 mutant. The henn-1 mutant exhibits a similar somatic Eri response to RNAi of dpy-13 and lin-29 (Figure S11).
Fig. 6. The henn-1 Mutant Exhibits Opposite RNAi Sensitivity Phenotypes in Soma and Germline. 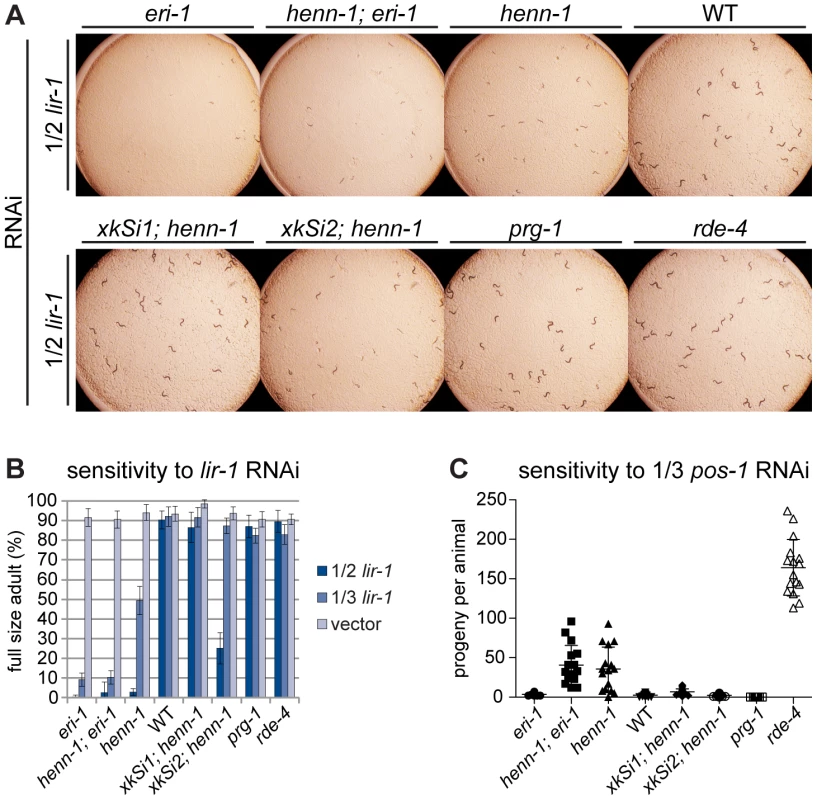
A) henn-1(tm4477) mutant animals exhibit mildly enhanced somatic RNAi. Animals of the indicated genotype were plated as L1 larvae on lir-1 feeding RNAi diluted 1∶1 with empty vector (1/2 strength) and grown for 70 hours at 20°C. Data is quantified in part B. RNAi sensitivity data for knockdown of two additional somatic transcripts are shown in Figure S11. B) Endogenous expression of henn-1::gfp from xkSi1 rescues somatic RNAi sensitivity. Percent of animals reaching full size on lir-1 feeding RNAi of the indicated strength at 70 hours is plotted. N = 8 plates of >50 animals per strain. Standard deviation is shown. C) henn-1(tm4477) mutant animals exhibit defective germline RNAi. Brood size of animals plated at 20°C as L1 larvae on pos-1 feeding RNAi diluted 1∶2 with empty vector is plotted. N≥13 animals per strain. Mean and standard deviation are shown. RNAi sensitivity data for knockdown of four additional germline transcripts are shown in Figure S12. Alleles used in this figure: eri-1(mg366), prg-1(tm872), rde-4(ne301). The Germline of the henn-1 Mutant Exhibits Decreased Sensitivity to Exogenous RNAi
While the somatic Eri phenotype of the henn-1 mutant was expected, knockdown of genes required for germline development or embryogenesis revealed that, incongruously, the henn-1 mutant maternal germline exhibits an RNAi defective (Rde) phenotype. Animals subjected to pos-1 RNAi lay dead embryos because maternally loaded pos-1 mRNA is required for specifying cell fate of many tissues during embryonic development [57]. On pos-1 RNAi diluted 1∶2 with empty vector (1/3 strength), knockdown in wild-type animals is still sufficiently robust to reduce average brood size to fewer than five offspring per animal. henn-1 mutant animals at this dilution, however, produce an average brood greater than tenfold that of wild-type, suggesting that loss of henn-1 confers resistance to RNAi-mediated knockdown of this maternally deposited mRNA (Figure 6C). A lesser but statistically significant effect was observed for RNAi of the germline-expressed transcripts par-1, par-2, pie-1, and glp-1 (Figure S12). Sensitivity to pos-1 RNAi is effectively rescued by either endogenous or germline-specific expression of henn-1::gfp, likely due to the fact that both transgenes are expressed in germline.
HENN-1 Is Expressed in Both Germline and Soma
HEN1 orthologs appear to be restricted to the germline in vertebrates [15], [33]; however, we observe phenotypes in both the germline and soma of the henn-1 mutant that suggest broader activity. To investigate expression of HENN-1 in C. elegans, we assessed henn-1 mRNA and protein levels throughout development. henn-1 mRNA levels are lowest in young larva and increase as the germline proliferates, peaking in gravid adult (Figure 7A, Figure S13A). Germline-deficient glp-4(bn2) adult hermaphrodites show approximately a 50% reduction in henn-1 mRNA levels relative to wild-type (Figure S13B), indicating that henn-1 mRNA is expressed in both germline and soma. Embryonic levels of henn-1 are high but decrease rapidly; this pattern suggests that, unlike in zebrafish [15], henn-1 mRNA may be maternally deposited into the embryo. HENN-1 protein is detectable throughout development and in both hermaphrodite and male adults (Figure 7B).
Fig. 7. HENN-1 Is Broadly Expressed in C. elegans Germline. 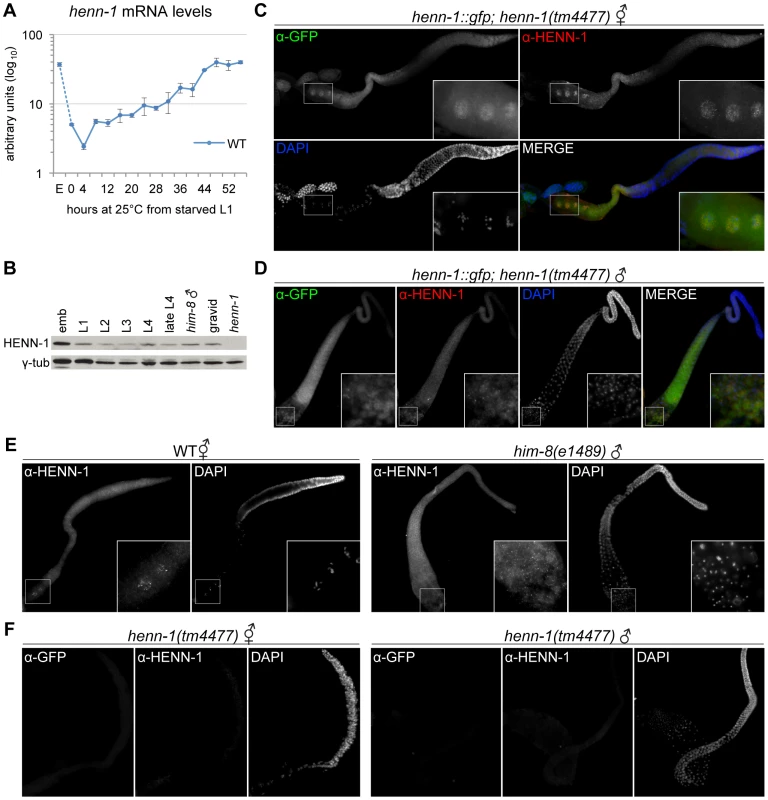
A) The henn-1 mRNA expression profile is consistent with germline enrichment. Levels of henn-1 mRNA were assayed throughout development and normalized to eft-2 mRNA. Standard deviation is shown for biological triplicates. Non-normalized levels are shown in Figure S13A. B) HENN-1 is detected at all stages of development and in male. Lysates from animals of the indicated stages were probed with anti-HENN-1 rabbit polyclonal antibody. C) HENN-1 is abundant in hermaphrodite proximal germline and enriched in proximal oocyte nucleoplasm (inset). Extruded gonads of xkSi1; henn-1(tm4477) adult hermaphrodites were stained with anti-GFP mouse monoclonal and anti-HENN-1 rabbit polyclonal antibodies. D) HENN-1 is detectable in male proximal and distal gonad, with enrichment in residual bodies during spermatid maturation (inset). Extruded gonads of xkSi1; henn-1(tm4477) adult males were stained with anti-GFP and anti-HENN-1 antibodies. E) Expression of endogenous HENN-1 mirrors expression of HENN-1::GFP from transgene xkSi1. Extruded gonads of wild-type animals were stained with anti-HENN-1 antibody. F) Detection of HENN-1 proteins by immunostaining is specific. Extruded gonads of henn-1(tm4477) mutant animals were stained with anti-GFP and anti-HENN-1 antibodies. E, embryo. We next assessed the distribution of HENN-1::GFP fusion protein expressed from xkSi1, the rescuing henn-1::gfp transgene driven by the endogenous promoter, in the henn-1 mutant background. Although single copy transgene expression levels are too low for direct visualization by fluorescence microscopy, HENN-1::GFP is readily detected using a mouse monoclonal anti-GFP antibody. Whole-mount immunostaining of transgenic L4 larvae reveals that HENN-1::GFP is expressed broadly in diverse somatic tissues and germline (Figure S13C). Non-transgenic larvae show no signal, indicating that detection of HENN-1::GFP is specific. In extruded gonads of xkSi1; henn-1 hermaphrodites, HENN-1::GFP is detected throughout the germline. Notably, the proximal oocytes show cytoplasmic and intense nucleoplasmic HENN-1::GFP expression (Figure 7C). Although nucleoplasmic enrichment is lost following fertilization, HENN-1::GFP is also abundant in embryo, with ubiquitous expression prior to gastrulation (Figure S13D). HENN-1::GFP is also expressed throughout the germline of xkSi1; henn-1 males (Figure 7D). During sperm maturation, we detect enrichment of HENN-1::GFP in residual bodies, but we cannot definitively conclude that it is excluded from sperm (Figure 7D, inset). In wild-type animals, studies of endogenous HENN-1 using the rabbit polyclonal antibody generated against an N-terminal HENN-1 epitope corroborate the above findings, although the signal is more difficult to detect (Figure 7E). Staining in the henn-1 mutant yields no signal for anti-GFP and anti-HENN-1 antibodies (Figure 7F); this demonstrates that detection of transgenic and endogenous HENN-1 proteins is specific. Together, these data define an expression pattern consistent with a role for HENN-1 in modifying small RNAs in both male and female germlines as well as in soma.
The 21U RNAs and 26G RNAs appear to be significantly stable only in the presence of their respective Argonaute proteins [35], [36], [40]; accordingly, the localization patterns of the Argonaute proteins reflect the distribution of the different classes of small RNAs. We therefore wanted to compare the expression patterns of HENN-1 and the 26G RNA-binding Argonautes to determine whether the small RNA substrate specificity of HENN-1 could be explained by differential access to Argonaute-bound small RNAs. ERGO-1, which binds methylated 26G RNAs, is abundant in embryo [42], and its transcript is enriched during oogenesis [58], but its localization has not yet been reported. We assessed the staining pattern of ERGO-1 in hermaphrodite gonad and embryo using a polyclonal antibody generated against a C-terminal ERGO-1 epitope. ERGO-1 expression in the hermaphrodite germline begins at pachytene exit and persists in embryo (Figure S13D, S13E). ERGO-1 shows cytoplasmic enrichment both in germline and embryo, suggesting that the cytoplasmic pool of HENN-1 may act in methylating 26G RNAs bound by ERGO-1. This interaction may, however, be transient, as we were unable to identify HENN-1 by mass spectrometry of immunopurified ERGO-1 complexes, nor could we detect ERGO-1 in immunopurified HENN-1::GFP complexes by western blot (data not shown). Notably, both HENN-1 and ERGO-1 remain abundant in early embryo (Figure S13D). This is consistent with the proposed existence of a somatic endo-siRNA pathway that promotes continued biosynthesis of ERGO-1 class 26G RNAs after fertilization [59].
We next assessed co-localization of HENN-1 and ALG-3. ALG-3 and its close paralog, ALG-4, bind unmethylated 26G RNAs, and their transcripts are enriched during spermatogenesis [58]. In the male gonad, a rescuing gfp::alg-3 transgene was reported to express in the proximal male germline, with localization to P granules beginning at late pachytene [41]. During sperm maturation, GFP::ALG-3 is relegated to residual bodies. Dual immunostaining of GFP::ALG-3 and endogenous HENN-1 demonstrates a large region of overlap (Figure S13F), but HENN-1 does not appear to localize to P granules. This does not explain why ALG-3/ALG-4 class 26G RNAs are not methylated, because it is likely that HENN-1 can access P granules transiently: PRG-1 localizes predominantly to P granules [35], [37], and the PRG-1-bound piRNAs are methylated. This is in contrast to zebrafish Hen1, which carries a poorly conserved C-terminal domain (Figure S1) that directs localization of Hen1 to nuage, perinuclear granules similar to C. elegans P granules, to methylate piRNAs [15].
Discussion
Differential 26G RNA Methylation Supports an Argonaute-Dictated Methylation Model
We have shown that HENN-1 is essential for methylating select classes of C. elegans small RNAs, namely, 21U RNAs and ERGO-1 class 26G RNAs. As is the case in other animals, small RNAs in C. elegans that associate with Piwi clade Argonautes require HENN-1 for maintenance of wild-type levels. Ago clade Argonaute-associated microRNAs and ALG-3/ALG-4 class 26G RNAs, in contrast, are HENN-1-independent (Figure S14A). It has been proposed that spatial and temporal regulation of HEN1 ortholog expression may contribute to small RNA substrate specificity in metazoans [24]. However, our immunostaining studies indicate that HENN-1 is coexpressed in the same tissues and subcellular compartments as Argonautes ERGO-1, PRG-1, and ALG-3 and their respective small RNAs (Figure 7, Figure S13). Therefore, differences in gross sub-cellular localization cannot explain the failure of ALG-3/ALG-4 class 26G RNAs to be methylated. Furthermore, although the two subclasses of 26G RNAs are generated in different germlines from non-overlapping targets, their sequences exhibit no obvious distinguishing characteristics that might account for their non-uniform methylation status.
One model of small RNA methylation posits that animal HEN1 orthologs only methylate small RNAs bound by Argonautes [15], [22]–[24], [49]. In support of this, work in fly shows that siRNA methylation requires assembly of DmAgo2 RISC [22], [50], and in vitro studies using lysate from a silkworm ovary-derived cell line show that methylation of synthetic RNA only occurs after the longer substrate is bound by a Piwi protein and trimmed to piRNA size [60]. This model predicts that all 26G RNAs are bound as unmethylated species by either ERGO-1 in the female germline or ALG-3/ALG-4 in the male germline and subsequently methylated or not, respectively. This is consistent with our findings in vivo that ERGO-1 is required for methylation of 26G RNAs (Figure 4A) and associates with 26G RNAs of either methylation status (Figure 4C). It has been further proposed that the identity of the Argonaute determines whether bound small RNAs are methylated [22], [23], [49], [50]. An elegant illustration of this is provided by fly miR-277, which associates with both Ago1, the canonical fly miRNA Argonaute, and Ago2, which binds methylated siRNAs [61]. The miR-277 pool contains both methylated and unmethylated species. Depletion of Ago2 in cell culture results in loss of methylated miR-277, whereas Ago1 depletion results in a completely methylated miR-277 population [22]. Similarly, fly hairpin derived hp-esiRNAs sort into Ago1 and Ago2, but accumulate mainly in Ago2 because only hp-esiRNAs bound by Ago2 are methylated and therefore protected against degradation triggered by their extensive target complementarity [50]. In C. elegans, the model of Argonaute-dictated methylation can be invoked to explain the disparate methylation of the 26G RNAs: in the male germline, only ALG-3/ALG-4 are expressed, resulting in an unmethylated male 26G RNA population, whereas exclusive expression of ERGO-1 in the female germline and embryo directs methylation of female and zygotic 26G RNAs. This raises the intriguing possibility that selective expression of Argonautes that permit or prevent methylation could represent a new mechanism for differentially regulating small RNA turnover.
It is important to note that our results do not definitively exclude an alternative model wherein 26G RNAs are methylated prior to association with Argonautes and subsequently bound by ALG-3/ALG-4 only if unmethylated or by ERGO-1 only if methylated. In this model, HEN1 would methylate 26G RNAs in both germlines, but degradation of labile unbound siRNAs would result in a purely unmethylated or methylated population of 26G RNAs in male and female germlines, respectively. Because 26G RNAs assessed in embryo are fully methylated (Figure 3A, Figure S3B), such a mechanism would require that ERGO-1 exhibit very unfavorable kinetics for association with unmethylated small RNAs. We do not find this to be the case, as ERGO-1 binds some 26G RNAs with similar efficiency when methylated and unmethylated (Figure 4C). Our data therefore provide stronger evidence for a model of Argonaute-dictated methylation of small RNAs.
Possible Advantages for Selective Methylation of Small RNAs
Differential germline expression of Argonautes could have evolved in C. elegans because of advantages conferred by selective stabilization of female germline 26G RNAs. Unlike ALG-3/ALG-4 class 26G RNAs, which appear to function exclusively during sperm development [40], [41], ERGO-1 class 26G RNAs exert much of their influence during embryonic and larval development, well beyond initiation of their biogenesis in the hermaphrodite germline [40]. Accordingly, their targets are depleted of germline-enriched genes [40], [59]. The oocyte contributes the vast majority of the initial zygotic cellular contents; therefore, methylation of 26G RNAs originating in the female germline may ensure robust inheritance and perdurance of primary small RNAs. Methylation of 26G RNAs in the male germline would likely not significantly increase their representation in sperm or zygote, as ALG-3/ALG-4 are relegated to residual bodies during spermatogenesis and exert effects in mature sperm only indirectly through dependent secondary 22G RNAs [41]. Nonetheless, it would be interesting to express ERGO-1 ectopically in sperm and determine whether ALG-3/ALG-4 class small RNAs are methylated. Such a strategy may reveal unexpected consequences related to inappropriate methylation and stabilization of ALG-3/ALG-4 class 26G RNAs.
Role of HENN-1 in the Balance between Endo – and Exo–RNAi
In the absence of henn-1, we show that response to RNAi-mediated knockdown is enhanced for somatic genes (Figure 6A and 6B, Figure S11). This is likely due to destabilization of ERGO-1 class 26G RNAs in the henn-1 mutant, which reduces competition with primary exo-siRNAs for stimulating secondary siRNA activity mediated by somatic Argonautes such as SAGO-1 and SAGO-2 [43], [55]. While germline-specific expression of henn-1::gfp only partially rescues this somatic Eri phenotype, henn-1 mutant animals rescued with an endogenous henn-1::gfp transgene, which drives both somatic and germline expression, show wild-type RNAi sensitivity. Under the model of competing endo - and exo-RNAi pathways, this suggests that HENN-1-mediated methylation of ERGO-1 class 26G RNAs in the germline alone cannot maintain small RNA levels sufficient to sequester an appropriate proportion of the limiting RNAi factors. It is possible that ERGO-1 class 26G RNA biogenesis continues in embryo and larva, as previously suggested [59], and that high concentrations of HENN-1 are necessary for continued stabilization of these small RNAs. Such a model would be consistent with our characterization of the distributions of HENN-1 and ERGO-1, both of which are still detected in abundance in developing embryo (Figure S13D, S13E).
While the majority of the phenotypes observed in the henn-1 mutant can be attributed to destabilization of endogenous small RNA substrates, the germline Rde phenotype suggests a role for HENN-1 in exogenous RNAi. It is unclear why HENN-1 is dispensable for robust exogenous RNAi in the soma but required in the germline. While this may be an indirect effect, as suggested in concurrent work by Kamminga et al. [62], one possible explanation is that HENN-1 stabilizes primary exo-siRNAs or dependent 22G secondary siRNAs. There is support in fly for methylation of exo-siRNAs and transgenic hairpin-derived siRNAs [22], [63], but this has not yet been demonstrated in C. elegans. 22G RNAs triggered by primary exo-siRNAs appear not to be methylated [47], consistent with our and others' observations that Wago-dependent 22G RNAs from diverse endogenous sources are unmethylated (Figure 3A, Figure S3B, and [45]). The methylation status of worm primary exo-siRNAs has not been definitively established, although a 22-nucleotide siRNA generated from a transgene encoding a perfect hairpin was not found to be methylated [46].
Structural Differences in Ago and Piwi Clade Argonautes May Dictate HEN1 Substrate Specificity
All Argonautes contain two signature domains, PAZ and Piwi [64]. The Piwi domain, unique to Argonautes, adopts an RNase H-like configuration and serves as the catalytic core of RISC [65], [66]. The PAZ domain recognizes and anchors the 3′ end of the small RNA [67], [68]. Comparison of Piwi and Ago clade Argonautes reveals that Piwi proteins contain a small insertion in their PAZ domains in a loop connecting two β strands [69]. Crystal structures of a human Piwi Argonaute PAZ domain suggest that this insertion results in the formation of a more spacious binding pocket capable of accommodating the 2′-O-methyl group of a piRNA. Interactions between the methyl group and hydrophobic residues lining the pocket confer a threefold to sixfold higher binding affinity for 2′-O-methyl than 2′-OH [69]. In C. elegans, only PRG-1/PRG-2 and ERGO-1 show evidence of a PAZ domain insertion (Figure S14B), consistent with their designation as Piwi clade Argonautes and association with methylated small RNAs.
In spite of their shared classification, ERGO-1 exhibits far less homology than PRG-1/PRG-2 to mammalian and insect Piwi proteins (Figure S14A) [43]. Similarly, among worm, fly, and human Argonautes, DmAgo2 and C. elegans Argonaute RDE-1 are among the most divergent members of their clades [43]. In fact, so divergent is RDE-1 that its cladistics are ambiguous, with our and other published alignments variably assigning it to each of the three clades (Figure S14A and [43], [70]). Both DmAgo2 and RDE-1 bind exo-siRNAs, although only the former has been shown to permit methylation [22]. Interestingly, both lack the insertion found in Piwi Argonaute PAZ domains (Figure S14B). The absence of this insertion in DmAgo2 suggests that it is not required for association with methylated small RNAs, raising the possibility that RDE-1 too may permit methylation of associated small RNAs. If HENN-1 does not methylate RDE-1-bound small RNAs, it is unclear what specific role HENN-1 plays in exo-RNAi in the germline. Nevertheless, its dual functions in endogenous and exogenous RNAi place HENN-1 in the company of DCR-1 and the Wago proteins at the intersection between these two RNAi pathways.
Materials and Methods
C. elegans Strains
C. elegans were maintained according to standard procedures. The Bristol strain N2 was used as the standard wild-type strain. The alleles used in this study, listed by chromosome, are: unmapped: neIs23[unc-119(+) GFP::ALG-3]; LGI: glp-4(bn2), prg-1(tm872); LGII: xkSi1[PC30A5.3::henn-1::gfp::henn-1 3′UTR cb-unc-119(+)] II, xkSi2[Ppie-1::henn-1::gfp::tbb-2 3′UTR cb-unc-119(+)] II; LGIII: rde-4(ne301), henn-1(tm4477); LGIV: eri-1(mg366), fem-1(hc17), him-8(e1489); LGV: ergo-1(tm1860). The neIs23[unc-119(+) GFP::ALG-3] strain was generously provided by Craig Mello (University of Massachusetts, Worcester, MA).
RNA Sample Preparation
For embryo samples, L1 larvae were grown at 20°C until gravid. Embryos were isolated using sodium hypochlorite solution; an aliquot of embryos was allowed to hatch overnight at room temperature to determine viability. For male samples, synchronized him-8(e1489) L1 larvae were grown at 20°C for 72–75 hours. Males were isolated by filtering through 35 µm mesh [71]. For female samples, synchronized fem-1(hc17) L1 larvae were plated and grown at 25°C for 52 hours. For time course samples, synchronized wild-type (N2) and henn-1(tm4477) L1 larvae were grown at 25°C until gravid; embryos were extracted and harvested for RNA or hatched overnight at room temperature and then grown at 25°C for the specified number of hours before harvest. The prg-1(tm872) time course samples were prepared in the same way, except that animals were grown for the first generation at 20°C to evade temperature-sensitive sterility. Samples were processed by either three rounds of freeze/thaw lysis or two rounds of homogenization for 15 sec using the Tissue Master-125 Watt Lab Homogenizer (Omni International) and the RNA was extracted in TriReagent (Ambion) following the vendor's protocol, with the following alterations: RNA was precipitated in isopropanol for one hour at −80°C; RNA was pelleted by centrifugation at 4°C for 30 min at 20,000× g; the pellet was washed three times in 75% ethanol; the pellet was resuspended in water.
β-elimination Assay for Small RNAs and Northern Blot Analysis
For detection of small RNAs, 10 or 40 µg of total RNA were β-eliminated as described [51]; control samples were processed in parallel without sodium periodate. Northern blot analysis was performed as described [72]. In brief, 5 or 10 µg of β-eliminated total RNA were resolved on 17.5% or 20% denaturing Urea-PAGE gels (SequaGel, National Diagnostics) and transferred to Hybond-NX membrane (Amersham). 21 and 26 nt synthetic RNAs were run as size markers and visualized in tandem with rRNA by ethidium bromide staining. Pre-hybridization/hybridization and washes were performed at 48°C or 50°C. Oligonucleotides corresponding to the antisense sequences of the small RNAs (Table S1) were synthesized and end-labeled with [α-32P]-dATP using the miRNA StarFire kit (Integrated DNA Technologies).
RNAi Sensitivity Assay
To test the response to exogenous RNAi, bacterial clones from the Ahringer RNAi library [73] were diluted with bacteria harboring the empty vector L4440 to achieve a level of RNAi sensitivity that allowed us to differentiate the RNAi responses in the strains examined. To determine lir-1 RNAi sensitivity, the lir-1 RNAi bacterial clone diluted with L4440 bacterial clone at a 1∶1 or 1∶2 ratio (1/2 or 1/3 strength) was used; >50 L1 larvae were plated per plate and the number of total animals assayed per plate was determined at day two after plating; the percent of animals exhibiting the larval arrest phenotype was determined at 70 hours at 20°C. Sensitivity to RNAi of dpy-13 and lin-29 was also assessed using this method, where animals subjected to dpy-13 RNAi were imaged at 70 hours and those subjected to lin-29 RNAi were evaluated for the absence of protruding vulva or bursting phenotype. For pos-1 RNAi, synchronized L1 larvae were singled onto plates with pos-1 RNAi diluted with empty vector at a 1∶2 ratio (1/3 strength) that had been induced overnight at 25°C. Animals were grown at 20°C for six days and progeny were counted. Sensitivity to RNAi of pie-1, par-1, and par-2 was assessed similarly at the indicated dilutions with 4 plates of 4 P0 animals per strain. Sensitivity to glp-1 RNAi was determined at the indicated dilutions by plating 4 plates of >50 L1 larvae per strain per gene and scoring for the absence of oocytes and embryos in both arms of the germline at 70 hours at 20°C. For all RNAi sensitivity assays, data are representative of at least two independent experiments.
Fertility Assay
To determine brood size, synchronized L1 larvae from gravid adults grown at 20°C or shifted to 25°C for two generations were singled onto plates with OP50 and grown to adulthood at their respective temperatures. Once egg-laying began, animals (N≥13 per strain) were transferred to fresh plates daily until the supply of fertilized eggs was exhausted. Progeny of the singled parents were counted as late larvae/adults. Results are representative of two independent experiments.
Quantitative RT–PCR
Taqman small RNA probes were synthesized by Applied Biosystems (Table S2) [74]. For each reaction, 50 ng of total RNA were converted into cDNA using Multiscribe Reverse Transcriptase (Applied Biosystems). The resulting cDNAs were analyzed by a Realplex thermocycler (Eppendorf) with TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems). We could not identify a small RNA whose levels were consistent across development for use in normalization. Therefore, to preserve the developmental profile of each of the small RNA assessed, back transformation was used to calculate relative small RNA levels from qRT-PCR cycle numbers. As a control for RNA quality, miR-1 Taqman assays were run in parallel for all samples excluding the ERGO-1 RNA immunoprecipitation samples, in which miRNAs are absent. For quantification of mRNA levels, 100 ng of total RNA were converted into cDNA with Multiscribe Reverse Transcriptase (Applied Biosystems) following the vendor's protocol with the following changes: 25 units of RT and 7.6 units of RNAse OUT (Invitrogen) were used per reaction. cDNAs were analyzed using Power Sybr Green PCR Master Mix (Applied Biosystems) (primers, Table S3). Relative mRNA levels were calculated based on the ΔΔ2Ct method [75] using eft-2 for normalization. For all qPCR, 40 cycles of amplification were performed; reactions whose signals were not detected were therefore assigned a cycle number of 40. All results presented are the average values of independent calculations from biological triplicates unless indicated. To determine average upregulation of ERGO-1 26G RNA targets in henn-1 relative to wild-type (Figure 5C), the mean was calculated for all of the ratios generated by dividing each henn-1 biological replicate by each wild-type biological replicate.
3′ RACE
3′ RACE was performed using the 3′ RACE System for Rapid Amplification of cDNA ends (Invitrogen) according to the manufacturer's protocol. henn-1 gene-specific primer (5′ GCAGTATGTCGCCTCCAAGTAGAT 3′) was used to amplify henn-1 3′ ends from cDNA generated from embryo. Product corresponding to only the seven-exon gene model of henn-1 was observed, consistent with detection of a single protein isoform corresponding to this model on western blot analysis.
Plasmids and Transgenic Strains
The endogenous henn-1::gfp reporter construct (xkSi1) was generated by introducing the following fragments into pCFJ151: endogenous promoter of the henn-1-containing operon CEOP3488 [76] (3.9 kb PCR fragment immediately upstream of the C30A5.3 start codon), henn-1 genomic coding region (1.8 kb PCR fragment with mutated termination codon), gfp coding region (0.9 kb fragment with multiple synthetic introns and termination codon), and henn-1 endogenous 3′UTR (1.1 kb PCR fragment immediately downstream of henn-1 termination codon). The germline-only henn-1::gfp reporter construct (xkSi2) was generated as above with the following substitutions: CEOP3488 operon promoter was replaced with the pie-1 promoter (2.4 kb PCR fragment immediately upstream of pie-1 start codon) and henn-1 endogenous 3′UTR was replaced with the C36E8.4 3′UTR (0.3 kb PCR fragment downstream of C36E8.4). Constructs were cloned into the pCFJ151 vector, confirmed by sequencing, and used to generate single-copy integrated transgenes via the MosSCI technique [52]. Gene fusion products of the expected size were specifically detected by western blot with both anti-HENN-1 and anti-GFP antibodies.
Generation of Antibodies
Synthetic antigenic peptides were conjugated to KLH and each was used to immunize two rabbits (Proteintech). Antisera were subsequently affinity purified using Affi-Gel 15 gel (Bio-Rad). Antigenic peptide sequences are as follows: N-terminal HENN-1 peptide with N-terminal added cysteine (CTYVEAYEQLEIALLEPLDR), C-terminal ERGO-1 peptide (CEVNKDMNVNEKLEGMTFV).
Western Blot Analysis
Proteins immobilized on Immobilon-FL transfer membrane (Millipore) were probed with anti-HENN-1 rabbit polyclonal antibody (1∶2000), anti-γ-tubulin rabbit polyclonal antibody (LL-17) (Sigma) (1∶2000), or anti-ERGO-1 rabbit polyclonal antibody (1∶1000). Peroxidase-AffiniPure goat anti-rabbit IgG secondary antibody was used at 1∶10000 (Jackson ImmunoResearch Laboratories) for detection using Pierce ECL Western Blotting Substrate (Thermo Scientific).
Isolation of ERGO-1–Associated RNAs
Wild-type, henn-1, or eri-1(mg366) embryos isolated from gravid adults grown at 20°C were frozen in liquid nitrogen and homogenized with a Mixer Mill MM 400 ball mill homogenizer (Retsch) Homogenates were suspended in lysis buffer (50 mM HEPES (pH 7.4), 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% glycerol, 0.05% NP-40 treated with a Complete, Mini, EDTA-free Protease Inhibitor Cocktail tablet (Roche Applied Sciences)) and clarified by centrifugation at 12,000× g for 12 minutes at 4°C. Aliquots of homogenate were reserved as crude lysate for western blot to confirm that immunoprecipitations were performed in lysates of equivalent protein concentration (2 mg/mL). For immunoprecipitations, embryo homogenates were incubated at 4°C for one hour with 75 µg anti-ERGO-1 rabbit polyclonal antibody conjugated to Dynabeads Protein A (Invitrogen), after which the beads were washed (500 mM Tris-HCl (pH 7.5), 200 mM KCl, 0.05% NP-40) and associated proteins were eluted with 200 µL glycine. Three quarters of each eluate were precipitated overnight at 4°C in trichloroacetic acid, pelleted, washed with acetone, and resuspended for western blot analysis. The remaining eluate was treated with 2 mg/ml Proteinase K (Roche) and incubated at 37°C for 30 minutes. RNA was isolated from the eluate by incubation with TriReagent and processed as described above. RNA pellets were resuspended in 10 µL water and 5 µL were used for each Taqman RT reaction.
Immunostaining
Primary antibodies were applied according to the following specifications: anti-GFP mouse monoclonal antibody 3E6 (Invitrogen) was diluted 1∶1500 to detect HENN-1::GFP and 1∶200 to detect ALG-3::GFP; anti-ERGO-1 rabbit polyclonal was diluted 1∶200; and anti-HENN-1 rabbit polyclonal antibody was preabsorbed as described [77] with henn-1(tm4477) mutant extract and diluted 1∶200. Alexa Fluor 555 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes) secondary antibodies were diluted 1∶500. All antibodies were diluted in 0.5% bovine serum albumin (Sigma). For immunostaining of gonads and embryos, synchronized gravid hermaphrodites or adult males grown at 20°C were dissected on Superfrost Plus positively charged slides (Fisherbrand) with 27 G×1/2 inch BD PrecisionGlide needles (Becton, Dickinson and Company) as described by Chan and Meyer in WormBook [78] Protocol 21 with 1.5% paraformaldehyde (Sigma). Slides were incubated with primary antibodies overnight at 4°C and with secondary antibodies for three hours at room temperature. Slides were mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). For whole-worm immunostaining, synchronized late L4 larvae grown at 20°C were transferred to subbed slides [77] in M9, fixed for six minutes in 1.5% paraformaldehyde, freeze-cracked, and incubated for 15 minutes in ice cold methanol. After fixation, slides were processed as above. Images were captured on an Olympus BX61 epifluorescence compound microscope with a Hamamatsu ORCA ER camera using Slidebook 4.0.1 digital microscopy software (Intelligent Imaging Innovations) and processed using ImageJ.
Supporting Information
Zdroje
1. LeeRCFeinbaumRLAmbrosV 1993 The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843 854
2. WightmanBHaIRuvkunG 1993 Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 855 862
3. KimY-KHeoIKimVN 2010 Modifications of small RNAs and their associated proteins. Cell 143 703 709
4. KawaharaYZinshteynBSethupathyPIizasaHHatzigeorgiouAG 2007 Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315 1137 1140
5. KawaharaYMegrawMKreiderEIizasaHValenteL 2008 Frequency and fate of microRNA editing in human brain. Nucleic Acids Research 36 5270 5280
6. KawaharaYZinshteynBChendrimadaTPShiekhattarRNishikuraK 2007 RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8 763 769
7. YangWChendrimadaTPWangQHiguchiMSeeburgPH 2006 Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13 13 21
8. HundleyHABassBL 2010 ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends in biochemical sciences 35 377 383
9. KawamuraYSaitoKKinTOnoYAsaiK 2008 Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453 793 797
10. NejepinskaJMalikRFilkowskiJFlemrMFilipowiczW 2012 dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Research 40 399 413
11. ShenBGoodmanHM 2004 Uridine addition after microRNA-directed cleavage. Science 306 997
12. van WolfswinkelJCClaycombJMBatistaPJMelloCCBerezikovE 2009 CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139 135 148
13. AmeresSLHorwichMDHungJ-HXuJGhildiyalM 2010 Target RNA-directed trimming and tailing of small silencing RNAs. Science 328 1534 1539
14. IbrahimFRymarquisLAKimEJBeckerJBalassaE 2010 Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A 107 3906 3911
15. KammingaLMLuteijnMJden BroederMJRedlSKaaijLJ 2010 Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29 3688 3700
16. LiJYangZYuBLiuJChenX 2005 Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15 1501 1507
17. TkaczukKLObarskaABujnickiJM 2006 Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol 6 6
18. ParkWLiJSongRMessingJChenX 2002 CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484 1495
19. YuBYangZLiJMinakhinaSYangM 2005 Methylation as a crucial step in plant microRNA biogenesis. Science 307 932 935
20. YangZEbrightYWYuBChenX 2006 HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34 667 675
21. VilkaitisGPlotnikovaAKlimasauskasS 2010 Kinetic and functional analysis of the small RNA methyltransferase HEN1: the catalytic domain is essential for preferential modification of duplex RNA. RNA 16 1935 1942
22. HorwichMDLiCMatrangaCVaginVFarleyG 2007 The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17 1265 1272
23. SaitoKSakaguchiYSuzukiTSiomiHSiomiMC 2007 Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi - interacting RNAs at their 3′ ends. Genes Dev 21 1603 1608
24. KurthHMMochizukiK 2009 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15 675 685
25. BrenneckeJAravinAAStarkADusMKellisM 2007 Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128 1089 1103
26. HouwingSKammingaLMBerezikovECronemboldDGirardA 2007 A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129 69 82
27. RubyJGJanCPlayerCAxtellMJLeeW 2006 Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127 1193 1207
28. GirardASachidanandamRHannonGJCarmellMA 2006 A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442 199 202
29. LauNCSetoAGKimJKuramochi-MiyagawaSNakanoT 2006 Characterization of the piRNA complex from rat testes. Science 313 363 367
30. KirinoYMourelatosZ 2007 Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14 347 348
31. OharaTSakaguchiYSuzukiTUedaHMiyauchiK 2007 The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14 349 350
32. VaginVVSigovaALiCSeitzHGvozdevV 2006 A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 320 324
33. KirinoYMourelatosZ 2007 The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13 1397 1401
34. ReinhartBJSlackFJBassonMPasquinelliAEBettingerJC 2000 The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 901 906
35. BatistaPJRubyJGClaycombJMChiangRFahlgrenN 2008 PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31 67 78
36. DasPPBagijnMPGoldsteinLDWoolfordJRLehrbachNJ 2008 Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31 79 90
37. WangGReinkeV 2008 A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Current biology : CB 18 861 867
38. CoxDNChaoABakerJChangLQiaoD 1998 A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & development 12 3715 3727
39. AmbrosVLeeRCLavanwayAWilliamsPTJewellD 2003 MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13 807 818
40. HanTManoharanAPHarkinsTTBouffardPFitzpatrickC 2009 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A 106 18674 18679
41. ConineCCBatistaPJGuWClaycombJMChavesDA 2010 Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A 107 3588 3593
42. VasaleJJGuWThiviergeCBatistaPJClaycombJM 2010 Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A 107 3582 3587
43. YigitEBatistaPJBeiYPangKMChenCC 2006 Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747 757
44. ZhangCMontgomeryTAGabelHWFischerSEPhillipsCM 2011 mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 108 1201 1208
45. GuWShirayamaMConteDJrVasaleJBatistaPJ 2009 Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36 231 244
46. SijenTSteinerFAThijssenKLPlasterkRHA 2007 Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315 244 247
47. PakJFireA 2007 Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315 241 244
48. OkamuraKChungWJRubyJGGuoHBartelDP 2008 The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453 803 806
49. HuangYJiLHuangQVassylyevDGChenX 2009 Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461 823 827
50. AmeresSLHungJ-HXuJWengZZamorePD 2011 Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA 17 54 63
51. YangZVilkaitisGYuBKlimasauskasSChenX 2007 Approaches for studying microRNA and small interfering RNA methylation in vitro and in vivo. Meth Enzymol 427 139 154
52. Frokjaer-JensenCDavisMWHopkinsCENewmanBJThummelJM 2008 Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40 1375 1383
53. ChenCRidzonDABroomerAJZhouZLeeDH 2005 Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33 e179
54. MontgomeryTARimY-SZhangCDowenRHPhillipsCM 2012 PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet 8 e1002616 doi:10.1371/journal.pgen.1002616
55. DuchaineTFWohlschlegelJAKennedySBeiYConteDJr 2006 Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124 343 354
56. LeeRCHammellCMAmbrosV 2006 Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12 589 597
57. TabaraHHillRJMelloCCPriessJRKoharaY 1999 pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126 1 11
58. ReinkeVGilISWardSKazmerK 2004 Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311 323
59. GentJILammATPavelecDMManiarJMParameswaranP 2010 Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Molecular Cell 37 679 689
60. KawaokaSIzumiNKatsumaSTomariY 2011 3′ end formation of PIWI-interacting RNAs in vitro. Molecular Cell 43 1015 1022
61. ForstemannKHorwichMDWeeLTomariYZamorePD 2007 Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130 287 297
62. KammingaLMvan WolfswinkelJLuteijnMJKaaijLJBagjinMP 2012 Differential impact of the Hen1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet In press
63. PelissonASarotEPayen-GroscheneGBuchetonA 2007 A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. Journal of Virology 81 1951 1960
64. CeruttiLMianNBatemanA 2000 Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci 25 481
65. LiuJCarmellMARivasFVMarsdenCGThomsonJM 2004 Argonaute2 is the catalytic engine of mammalian RNAi. Science 305 1437 1441
66. SongJJSmithSKHannonGJJoshua-TorL 2004 Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305 1434 1437
67. LingelASimonBIzaurraldeESattlerM 2004 Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11 576 577
68. MaJBYeKPatelDJ 2004 Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429 318 322
69. TianYSimanshuDKMaJBPatelDJ 2011 Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proceedings of the National Academy of Sciences 108 903
70. BolandAHuntzingerESchmidtSIzaurraldeEWeichenriederO 2011 Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proceedings of the National Academy of Sciences of the United States of America 108 10466 10471
71. L'HernaultSWRobertsTM 1995 Cell biology of nematode sperm. Methods in cell biology 48 273 301
72. PallGSHamiltonAJ 2008 Improved northern blot method for enhanced detection of small RNA. Nature protocols 3 1077 1084
73. KamathRSFraserAGDongYPoulinGDurbinR 2003 Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231 237
74. ChenCRidzonDABroomerAJZhouZLeeDH 2005 Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research 33 e179
75. NolanTHandsREBustinSA 2006 Quantification of mRNA using real-time RT-PCR. Nature protocols 1 1559 1582
76. AllenMAHillierLWWaterstonRHBlumenthalT 2011 A global analysis of C. elegans trans-splicing. Genome research 21 255 264
77. CrittendenSKimbleJ 2009 Preparation and immunolabeling of Caenorhabditis elegans. Cold Spring Harbor protocols 2009 pdb prot5216
78. GirardLRFiedlerTJHarrisTWCarvalhoFAntoshechkinI 2007 WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Research 35 D472 475
79. NotredameCHigginsDGHeringaJ 2000 T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology 302 205 217
80. PoirotOO'TooleENotredameC 2003 Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Research 31 3503 3506
81. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
82. GoujonMMcWilliamHLiWValentinFSquizzatoS 2010 A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Research 38 W695 699
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



