-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
The organization of the Escherichia coli chromosome into a ring composed of four macrodomains and two less-structured regions influences the segregation of sister chromatids and the mobility of chromosomal DNA. The structuring of the terminus region (Ter) into a macrodomain relies on the interaction of the protein MatP with a 13-bp target called matS repeated 23 times in the 800-kb-long domain. Here, by using a new method that allows the transposition of any chromosomal segment at a defined position on the genetic map, we reveal a site-specific system that restricts to the Ter region a constraining process that reduces DNA mobility and delays loci segregation. Remarkably, the constraining process is regulated during the cell cycle and occurs only when the Ter MD is associated with the division machinery at mid-cell. The change of DNA properties does not rely on the presence of a trans-acting mechanism but rather involves a cis-effect acting at a long distance from the Ter region. Two specific 12-bp sequences located in the flanking Left and Right macrodomains and a newly identified protein designated YfbV conserved with MatP through evolution are required to impede the spreading of the constraining process to the rest of the chromosome. Our results unravel a site-specific system required to restrict to the Ter region the consequences of anchoring the Ter MD to the division machinery.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002672
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002672Summary
The organization of the Escherichia coli chromosome into a ring composed of four macrodomains and two less-structured regions influences the segregation of sister chromatids and the mobility of chromosomal DNA. The structuring of the terminus region (Ter) into a macrodomain relies on the interaction of the protein MatP with a 13-bp target called matS repeated 23 times in the 800-kb-long domain. Here, by using a new method that allows the transposition of any chromosomal segment at a defined position on the genetic map, we reveal a site-specific system that restricts to the Ter region a constraining process that reduces DNA mobility and delays loci segregation. Remarkably, the constraining process is regulated during the cell cycle and occurs only when the Ter MD is associated with the division machinery at mid-cell. The change of DNA properties does not rely on the presence of a trans-acting mechanism but rather involves a cis-effect acting at a long distance from the Ter region. Two specific 12-bp sequences located in the flanking Left and Right macrodomains and a newly identified protein designated YfbV conserved with MatP through evolution are required to impede the spreading of the constraining process to the rest of the chromosome. Our results unravel a site-specific system required to restrict to the Ter region the consequences of anchoring the Ter MD to the division machinery.
Introduction
The large size of genomes compared to cell dimensions imposes an extensive compaction of chromosomes compatible with various processes of DNA metabolism such as gene expression, replication and segregation of the genetic information. The organized bacterial chromosome, named the nucleoid, is a compact structure that presents a particular orientation inside the cell preserving the linear order of genes on DNA [1]. Although our understanding of the global organization of the bacterial chromosome is still limited, different levels of organization have been identified. At the molecular level, binding of a number of proteins including nucleoid-associated proteins (NAPs), Structural Maintenance of Chromosomes (SMC) proteins, transcriptional regulators, chromosome organizers or RNA polymerase to the DNA molecule results in the formation of a bacterial chromatin with different properties according to the identity of proteins bound [2]–[4]. The excess of negative supercoils generates the formation of plectonemes that impose a secondary structure to the DNA molecule [5]. The size of such structures has been estimated to range in size from 10 kb to 100 kb depending on the studies [6]–[8].
At a higher level, there are several levels of organization required to fully condense the E. coli chromosome. Recent studies have implicated the H-NS protein in the formation of two clusters per chromosome in which H-NS regulated genes are sequestered [9]. Using FISH and genetic approaches, a long range organization based on the existence of four insulated macrodomains (MD) and two less constrained regions called non-structured (NS) regions has been uncovered [10], [11]. MDs have been defined as large regions in which DNA interactions occurred preferentially and DNA interactions between the different MDs are highly restricted. In NS regions, DNA sites can interact with both flanking MDs [11]. The Ori MD contains oriC while the opposite Ter MD contains the replication terminus and the chromosome dimer resolution dif site. The Ter MD is flanked by the Left and Right MDs whereas the Ori MD is flanked by the two NS (NSRight and NSLeft) regions [11]. This organization influences the segregation of sister chromatids and the mobility of chromosomal DNA [12].
Structuring of the Ter MD relies on the binding of the MatP protein to a 13 bp motif called matS repeated 23 times in the 800 kb long domain. This protein accumulates in the cell as a discrete focus that colocalizes with the Ter MD. In the absence of MatP, DNA is less compacted, the mobility of markers is increased and segregation of the Ter MD occurs early in the cell cycle [13]. Moreover, recent studies showed that these two last aspects are also affected in a zapB mutant. ZapB, which is associated to the division machinery, stabilizes the Ter MD at mid-cell through a direct interaction with MatP (Espéli et al., submitted). Interestingly, bioinformatic analyses showed that matS sequences are present in different enterobacteria and Vibrio species [13] while MatP has been shown to belong to a group of proteins (including SeqA and MukBEF) exclusively identified in bacteria carrying Dam methyltransferase activity [14].
The molecular basis for structuring the other MDs remains to be characterized. However, repeated sequences analogous to matS have not been found in the other MDs and other models should be considered. In order to investigate the effect of the relative position of the MD on its properties, we designed a new method reminiscent of a cut-and-paste transposition event that allows the relocation of any chromosomal fragment at a defined position of the genetic map. Using this “transposition” technique, we generated a number of new chromosomal configurations. Very interestingly, we noticed that transposing the NS regions close to the Ter MD leads to a dramatic decrease in the mobility of fluorescent markers. We further identified two palindromic sequences called tidR and tidL (found in the Right and Left MDs, respectively) that are required in order for the NS regions to keep their specific properties. In the absence of these sequences, a constraining effect dependent upon the association of the Ter MD to the division machinery and acting in cis is detected at a very large distance, up to 750 kb of the Ter MD. Strikingly, we identified a protein, called YfbV, also exclusively found in bacteria possessing the Dam methylase which is required to prevent this constraining process of the NS regions and flanking Right and Left MDs.
Overall, this study shows that specific determinants (two palindromic sequences and a protein) are required in order to prevent a MatP dependent constraining effect to affect chromosome arms flanking the Ter MD, therefore “insulating” the Ter MD from other parts of the chromosome.
Results
The “Transposition” method: Displacement of chromosomal fragments without affecting gene orientation
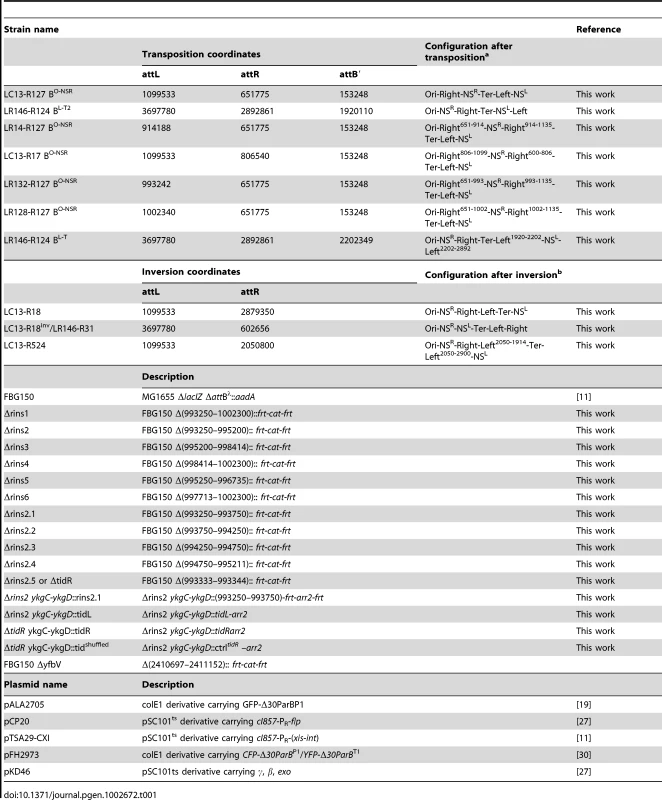
In several studies, genomic rearrangements have been used to unveil the principles of chromosome organization [15]–[18]. In these cases, recombination between inverted recombining sites was used to promote genomic rearrangement. Unfortunately, this method perturbs several parameters at the same time: for example, inversion of the segment encompassing the Ori and Right MDs changes the relative position of the involved regions in Ori and Right MDs but also affects the orientation of more than one thousand genes including those in the intervening NS region. To limit the number of parameters affected by genetic rearrangements, we devised a new strategy reminiscent of a cut-and-paste transposition event that allows relocating any chromosomal fragment at a defined position of the genetic map without changing gene orientation. It relies upon the site-specific recombination Int system from bacteriophage λ. The Int integrase mediates phage integration in the chromosome by recombination between phage attP and bacterial attB attachment sites (integrative recombination) generating two hybrid sites, attL and attR. Recombination between attL and attR (excisive recombination) requires the additional presence of the Xis excisionase. We used directly repeated attL and attR sites flanked by the 5′and 3′ parts of lacZ, respectively [11], to delimitate the genomic fragments to relocate. Recombination between these two sites leads to the formation of two circles, one carrying attB in frame within a functional lacZ gene, and one carrying attP. An additional attB site (called attB′) was integrated at the target site. The int and xis genes were cloned under the control of a thermosensitive promoter [11].
The transposition takes place in two steps (Figure 1). First, excisive recombination between attL and attR sites promoted by Int and Xis generates two circles: one carrying attP (the excised segment) and one carrying attB (the chromosome with a deletion of the excised segment). As the excised segment carries essential genes, its loss is fatal to the cell. Therefore, the second step of the transposition process consists in its reinsertion into the chromosome, either in the reconstituted attB fused to lacZ or in attB′ (giving rise to attL′ and attR′ sites) (Figure 1). The two events can be discriminated by the absence or presence of β-galactosidase activity. Conditions that provide a large amount of Xis and Int recombinases were applied to ensure that the excision of the segment between attR and attL occurs at a high frequency, greater than 95%. The level of lethality was greater than 90%, indicating that reinsertion of the excised segment occurred at low frequency. This is probably due to the presence of Xis that inhibits integrative recombination. However the amount of recombination was sufficient to obtain every transposition we tried. Insertion in the attB′ site and formation of the attL′ and attR′ sites can be detected by the appearance of blue clones on plates containing X-Gal and can be demonstrated at the DNA level by PCR analysis (Figure S1).
Fig. 1. Chromosomal rearrangement by transposition. 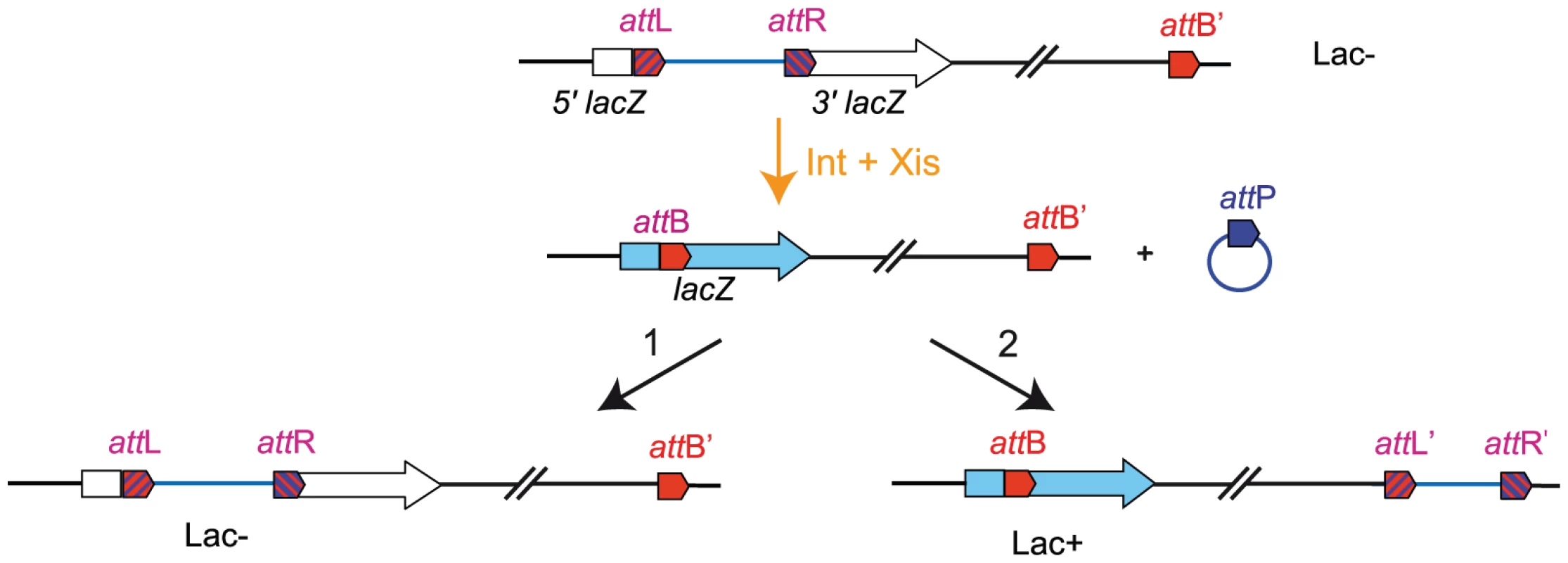
The three att sites are integrated in the chromosome in the same orientation. attL and attR sites are flanked by the 5′ and 3′ parts of lacZ, respectively [11]. An additional attB site (attB′) is inserted at a defined position. Excisive recombination promoted by Int+Xis results in the excision and circularization of the intervening segment carrying attP. This recombination event reconstitutes a functional lacZ. The excised molecule is not replicated and should be reintegrated for the viability of cells. Integration can occur in reconstituted attB (1) or in attB′ (2). Insertion in attB′ gives rise to Lac+ recombinants. Constraining of the NS regions is observed upon relocation next to the Ter MD
Using fluorescent microscopy, we showed in a previous study that the dynamic behaviour of loci was different according to their belonging to a MD or a NS region. The system used derived from the bacteriophage P1 partition module and involved the ParB-GFP fusion protein interacting with parS sites inserted in the chromosome [19]. By time-lapse imaging during a 5 min period with a 10 sec interval, tracking of parS tags inserted in different regions of the chromosome and decorated by a fluorescent ParB-GFP fusion protein allowed to measure the travelled distance by each tag and its diffusion coefficient; it revealed that constraints on mobility are higher in MDs than in NS regions [12]. In order to investigate the effect of position on these different behaviours, travelled distances (Figure 2) and diffusion coefficients (Table S1) were measured in different strains (Table 1) for a number of loci (NSR-1, NSR-2, NSR-5, Right-2, Right-5, Ter-3, Left-2, Left-1, NSL-3, NSL-4, Ori-3; for their exact location, see Table S2) in different chromosomal configurations. Results are shown in Figure 2.
Fig. 2. Travelled distance of different markers in strains with a wt or rearranged configurations. 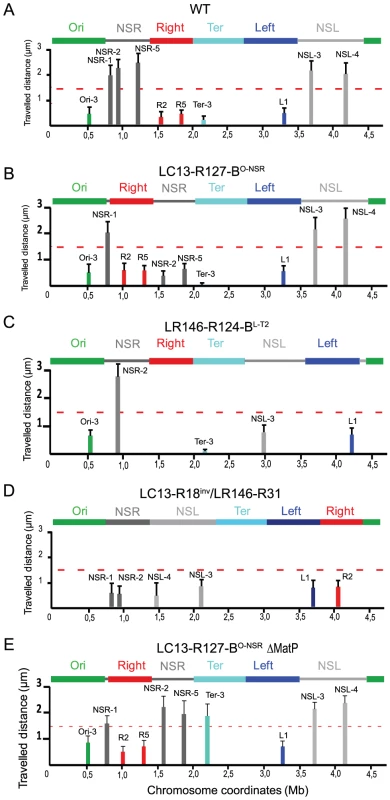
The mean value and the standard deviation calculated for 30 independent foci are indicated by columns. The x axis represents the chromosome genetic map; the positions of the loci are in Mb from oriC. The MDs and NS regions are indicated above the graph. (A) wt strain (markers Ori-3, NSR-1, NSR-2, NSR-5, Right-2 (R2), Right-5 (R5), Ter-3, Left-1 (L1), NSL-3 and NSL-4 are indicated from left to right). (B) Strain LC13-R127-BO-NSR with a transposition of the Right MD between the Ori MD and the NSRight region (markers Ori-3, NSR-1, Right-2 (R2), Right-5 (R5), NSR-2, NSR-5, Ter-3, Left-1 (L1), NSL-3, and NSL-4 are indicated from left to right). (C) Strain LR146-R124-BL-T2 with the NSLeft region transposed between the Ter and left MDs (markers Ori-3, NSR-2, Ter-3, NSL-3, and Left-1(L1) are indicated from left to right). (D) strain LC13-R18inv/LR146-R31 with the two NS regions located on the right replication arm (markers NSR-1, NSR-2, NSL-3, NSL-4, Left-1, Right-2) (E) strain LC13-R127-BO-NSR ΔmatP with a transposition of the Right MD between the Ori MD and the NSRight region with a deletion of matP (markers Ori-3, NSR-1, Right-2 (R2), Right-5 (R5), NSR-2, NSR-5, Ter-3, Left-1 (L1), NSL-3 and NSL-4 are indicated from left to right). We first transposed most of the NSRight region between the Ter and Right MDs (strain LC13-R127-BO-NSR). Such rearrangement had no dramatic effect on cell growth, cell and nucleoid aspects; a mild effect was detected in competition experiments, resulting in a 1 to 10 ratio in competition assays after 80 generations (Figure S2). Remarkably, mobility was specifically affected in the transposed NSRight region (markers NSR-2 and NSR-5) whereas it was unchanged in other regions of the chromosome, including the NSLeft region (Figure 2B).
We next transposed most of the NSLeft region between the Ter and Left MDs (strain LR146-R124 BL-T2). Strikingly, the marker NSL-3 in the NSLeft region specifically showed a decreased mobility, whereas markers in other regions (including NSR-2 in the NSRight region) were not affected (Figure 2C).
We also constructed a strain in which the two NS regions are positioned on the same replication arm of the chromosome (strain LC13-R18inv/LR146-R31 in Table 1). Strikingly, markers located in the two NS regions showed a decreased mobility (Figure 2D). This indicates that the constraining of the NS regions is not linked to their position relative to the Right or the Left MDs. Rather it may depend on proximity of the Ter MD.
Structuring of the Ter MD relies on the binding of the MatP protein to its target sequence matS. To determine whether the constraining process which originates from the Ter region requires its structuring into a MD, we analysed the changes in mobility of markers in cells devoid of MatP. We deleted the matP gene in the LC13-R127-BO-NSR strain, and assayed the mobility of markers located in different regions of the chromosome (Figure 2E). Remarkably, markers located in the NSRight region next to the Ter were mobile, in contrast to what is observed when MatP was present (see Figure 2B for comparison). The mobility of a Ter marker was also increased in a matP mutant, as expected, whereas the mobility of markers located in the Right and Left MD was not affected (Figure 2E). These results indicated that the constraining process associated to the Ter region required the MatP protein.
Identification of insulation determinants tidR and tidL
Experiments described above suggest that the Right and Left MDs may insulate the NS regions from a constraint linked to the Ter MD proximity. In order to test this hypothesis and investigate what regions of the Right MD was required to protect the NSRight region, we moved various parts of the Right MD by transposition and tested the effect of remaining parts of the Right MD on the mobility of markers in the NSRight region. Results are presented in Figure 3A. They showed that this mobility was not affected when the 221 kb region closest to the Ter MD (coordinates 914 kb to 1135 kb) remained between the Ter MD and the NS region (strain R127-LR14-BO-NSR) whereas the 206 kb region nearest from the NSRight region (coordinates 600 kb to 806 kb) was not sufficient to prevent a decrease in mobility of the NS region (strain LC13-R17-BO-NSR). In this strain, the mobility of a marker located in this 206 kb segment of the Right MD was the same as in a wild type chromosomal configuration (data not shown). This indicated that the structuring of the Right MD by itself was not affected, and that it was not sufficient to protect the NS region. This also revealed that specific insulation determinants were present in the 221 kb segment adjacent to the Ter MD.
Fig. 3. Identification of insulation determinants tidR and tidL. 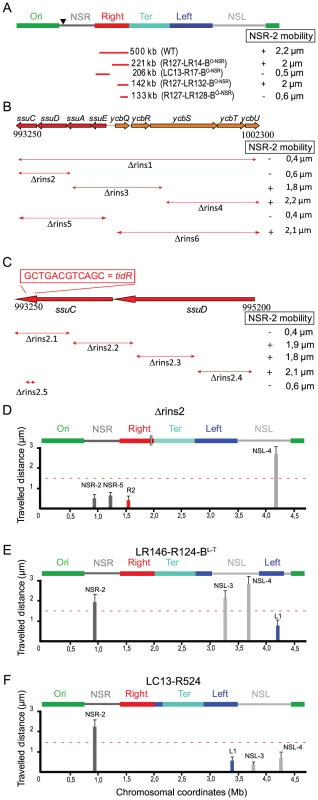
(A) Transposition of different fragments of Right MD between the Ori MD and the NSRight region (indicated by an arrowhead). The remaining part of the Right MD separating the NSRight region from the Ter MD is indicated by the red lines. The size of the remaining part is indicated in kb beside the name of the strain. Mobility of the marker NSR-2 is given and the presence of the insulation determinants is indicated by “+”. (B) Effect of deletions spanning the ssuEADC-ycbQRSTU region on NSR-2 mobility. The extent of the deletion is indicated below the map of the region (coordinates are indicated in bp). Mobility of the marker NSR-2 is given and the presence of the insulation determinants is indicated by “+”. (C) Effect of deletions spanning the ssuCD region on NSR-2 mobility. The extent of the deletion is indicated below the map of the region (coordinates are indicated in bp). Mobility of the marker NSR-2 is given and the presence of the insulation determinants is indicated by “+”. The position and sequence of tidR is indicated above the map. (D) Travelled distance of different markers in strain Δrins2 as indicated in Figure 2. The deletion of segment rins2 is schematized by a double line inside the Right MD (markers NSR-2, NSR-5, Right-2 (R2), NSL-4 are indicated from left to right). (E) Travelled distance of different markers in strain LR146-R124-BL-T with a transposition of the NSLeft region transposed inside the Left MD (markers NSR-2, NSL-3, NSL-4, and Left-1 (L1) are indicated from left to right). The insulation determinants were present in the 290 kb of the Left MD proximal to the Ter MD. (F) Travelled distance of different markers in strain LC13-R524 with an inversion between sites located in the Right and Left MD (markers NSR-2, Left-1 (L1), NSL-3, and NSL-4 are indicated from left to right). No insulation determinants were present in the 830 kb of the Left MD distal to the Ter MD; they are predicted to be present in the 135 kb segment proximal to the Ter MD and that has been positioned between the Ter and Right MDs by the inversion event. We further dissected what part of this 221 kb segment carried these determinants, using the same strategy. We showed that they were present in the 142 kb segment flanking the Ter MD (coordinates 993 kb from 1135 kb; strain R127-LR132-BO-NSR) but not in the 133 kb segment flanking the Ter MD (coordinates 1002 kb to 1135 kb; strain R127-LR128-BO-NSR in Figure 3A). This demonstrated that insulation determinants were present in the 9 kb region located between coordinate 993 kb to 1002 kb, which encompasses 10 genes (ssuCDAE-ycbQRSTU) predicted as not essential for the viability of E. coli. Deletion of this 9 kb region was engineered (strain Δrins1) and we measured the mobility of different markers. A specific reduction of mobility was observed for the marker NSR-2 in the NSRight region, confirming that insulation determinants are present in that 9 kb region.
Interestingly, these results showed that the constraining effect promoted by the Ter MD was also detectable in the native chromosome configuration provided that the insulating determinants were removed indicating that immediate proximity of the Ter MD was not required.
Deletions spanning the ssuCDAE-ycbQRSTU region were generated to map more precisely the insulation determinants and the effect of these deletions on the mobility of markers located in the NSRight region was assayed. The first series of deletions indicated that determinants were present in the ssuCD region (Figure 3B). More precise deletions were subsequently generated and the results showed that only a 12 bp palindromic sequence (GCTGACGTCAGC) located in the 3′ end region of ssuC was required for the insulation phenomenon (Figure 3C). Indeed, deletion of these 12 bp induced a specific decrease in mobility of markers NSR-2 and NSR-5, whereas other markers (such as NSL-4 in the NSLeft region) were not affected (strain ΔtidR in Figure 3D). This sequence was called tidR for ter MD insulation determinant Right. Its deletion had no important consequences for cell morphology and nucleoid distribution (Figure S3).
As described above, the constraining process seemed to be at work on both sides of the Ter MD. In order to map insulation determinants responsible for preventing a change of properties in the NSLeft region, a number of chromosomal rearrangements involving the Left MD and the NSLeft region were generated (strains LR146-R124-BL-T and LC13-R524). Results showed that insulation determinants were present in the 282 kb segment flanking the Ter MD (strains LR146-R124-BL-T in Figure 3E) and presumably present in the 135 kb segment flanking the Ter MD (strain LC13-R524 in Figure 3F and data not shown). A bioinformatic analysis of this 135 kb fragment allowed the identification of a 12 bp sequence that differed from tidR by a single nucleotide (GTTGACGTCAGC) that was called tidL for ter MD insulation determinant Left. This sequence was located in the tar gene, encoding a methyl-accepting chemotaxis protein. The effect of tidL deletion on marker mobility could not be assessed as disruption of the tar-tsp-cheRBYZ operon perturbed the number and disposition of fluorescent tags; tidL insulation properties were demonstrated by inserting it at an ectopic position (see below).
tidR and tidL insulate the NS regions from the Ter macrodomain
To determine whether tidR/L could act at another position, the 500 bp fragment carrying tidR deleted in strain ΔRins2.1 was inserted in the middle of the NSRight region, between markers NSR-2 and NSR-5, in a Δrins2 strain (Figure 4A). The NSR-5 marker which is closest to the Ter MD showed a decrease in mobility compared to the wild type situation, similar to what was observed in a ΔtidR strain. Remarkably, the NSR-2 marker, farthest from the Ter MD, displayed a mobility similar to the wild type situation, in contrast to what was observed in a ΔtidR strain. These results indicated that the fragment containing tidR protected from the constraining effect originating from the Ter MD at various positions, in the Right MD or in the NSRight region. They suggest that tidR works in cis by impeding the progression along the chromosome of a process that changes the dynamic properties of the DNA molecule.
Fig. 4. tidR and tidL insulate the Ter MD. 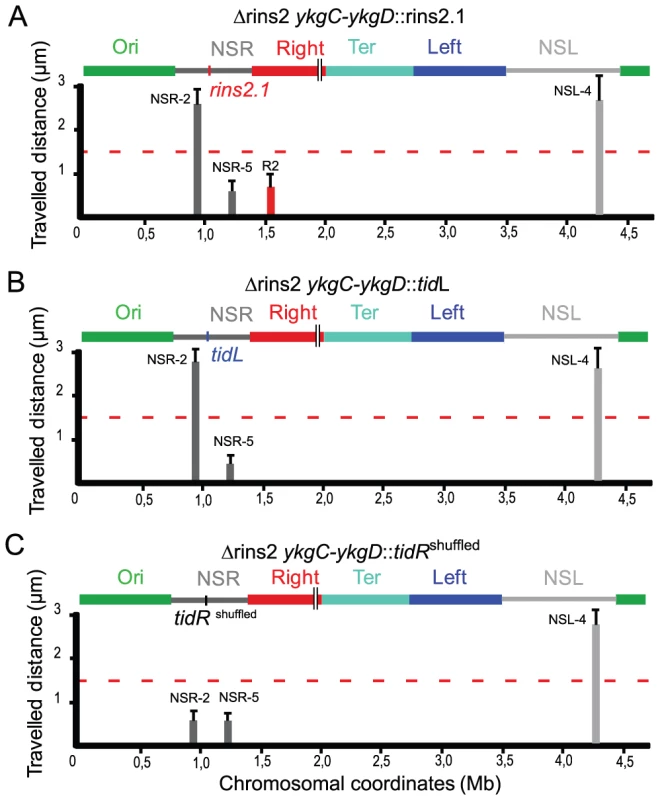
Travelled distance of different markers in strains with a Δrins2 deletion, and carrying segment rins2.1, tidL or a control sequence in the middle of the NSRight region. The representation is the same as in Figure 2. (A) strain Δrins2 ykgC-ykgD::rins2.1 with a deletion of the segment rins2 and an insertion of segment rins2.1 between ykgC and ykgD (markers NSR-2, NSR-5, Right-2 (R2), NSL-4 are indicated from left to right). (B) strain Δrins2 ykgC-ykgD::tidL with a deletion of the segment rins2 and insertion of tidL between ykgC and ykgD (markers NSR-2, NSR-5 and NSL-4 are indicated from left to right). (C) strain Δrins2 ykgC-ykgD::tidRshuffled with a deletion of the segment rins2 and an insertion of shuffled sequence with a similar palindromic organization as tidR between ykgC and ykgD (markers NSR-2, NSR-5, NSL-4 are indicated from left to right). To test whether the 12 bp tidR/L sequence was sufficient to protect from the constraining process, tidL was inserted at the same location between markers NSR-2 and NSR-5 (Figure 4B). Insertion of the tidL sequence protected the marker NSR-2 from the constraining effect indicating that the 12 bp sequence was sufficient to impede the constraining process (Figure 4B); similar results were observed with the 12 bp tidR sequence (data not shown). The effect was specific for tidR and tidL as a 12 bp control shuffled palindromic sequence GACGCTAGCGTC had no effect on impeding the change of mobility of NSR-2 (Figure 4C). These results allowed the definition of the 12-mer GYTGACGTCAGC consensus sequence that insulated the NS regions from a long range cis-acting constraining effect promoted by the Ter MD. tidR and tidL were the only occurrences of this sequence found in the E. coli genome. By bioinformatic analyses, the presence of tidRL flanking the Ter MD was found conserved only in Shigella species; other enterobacterial genomes lack tidRL sequences (e.g. Yersinia pestis) or carry multiple copies (e.g. in Salmonella typhimurium).
Insulation of the NS regions from the Ter MD involves a protein co-occurring with the Dam methylase and MatP
MatP belongs to a group of 18 proteins exclusively identified in bacteria with Dam methyltransferase activity [14]. We wondered whether one of these proteins might be involved in the insulation mechanism preventing the spreading in cis of the constraining process. In order to test this hypothesis, we analyzed the mobility of markers in mutants of each of the genes encoding proteins of unknown function. Strikingly, the mobility of markers located in the NSRight region and in the NSleft region was reduced in a yfbV mutant, whereas mobility of markers located in the different MDs was not affected (Figure 5A, see Figure 2A for comparison). This protein might thus be implicated in the insulation process promoted by both tidR and tidL. Interestingly, the yfbV gene encodes a predicted protein of 151 amino acids with two internal trans-membrane segments imposing the following topology: a 45 aa N-terminus cytoplasmic domain, a trans-membrane domain composed of two trans-membrane segments separated by a small periplasmic loop of 3 aa, and a 60 aa C-terminal cytoplasmic domain.
Fig. 5. Insulation requires the YfbV protein. 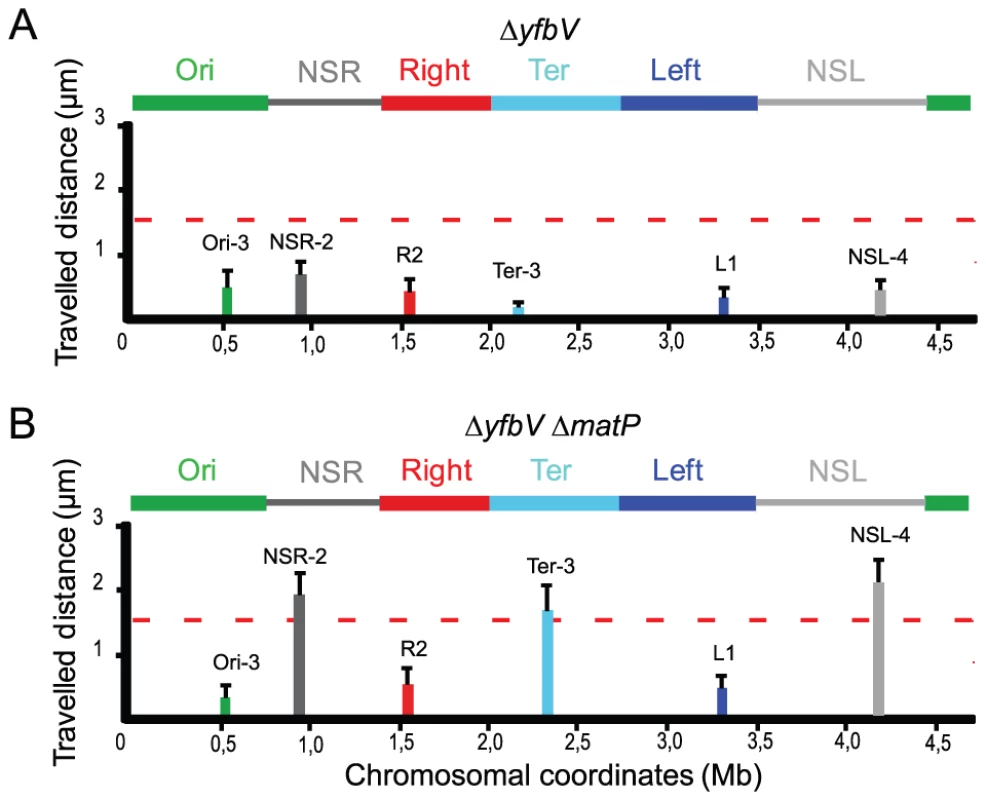
Travelled distance of different markers in strains deleted for yfbV or yfbV and matP. The representation is the same as in Figure 2. (A) strain with a deletion of yfbV (markers Ori-3, NSR-2, Right-2 (R2), Ter-3, Left-1 (L1) and NSL-4 are indicated from left to Right). (B) strain with a deletion of yfbV and matP (markers Ori-3, NSR-2, Right-2 (R2), Ter-3, Left-1 (L1) and NSL-4 are indicated from left to right). The reduction in mobility of markers located in the NSRight region and in the NSleft region observed in a yfbV mutant was not seen in a yfbV matP double mutant (Figure 5B), confirming the involvement of yfbV in impeding the spreading of the constraining process originating from the Ter MD.
The constraining process relies upon the association of the Ter MD with the division machinery
It was recently shown that the Ter MD is associated with components of the divisome during part of the cell cycle. This association relies on the interaction of MatP with ZapB (Espeli et al., submitted). In order to probe the role of this association in the constraining process, we analysed the mobility of markers in cells devoid of ZapB (Figure 6A). In a zapB mutant, the mobility of a Ter marker was increased, whereas mobility of markers located in the other MDs or in the NS regions remained the same (See Figure 2A for comparison). Remarkably, deleting the tidR palindrome in this zapB mutant context did not lead to a decreased in mobility of makers located in the NSRight region, in contrast to what was observed in a wild type background (Figure 6B, see Figure 3D for comparison).
Fig. 6. The constraining process depends on MatP and ZapB. 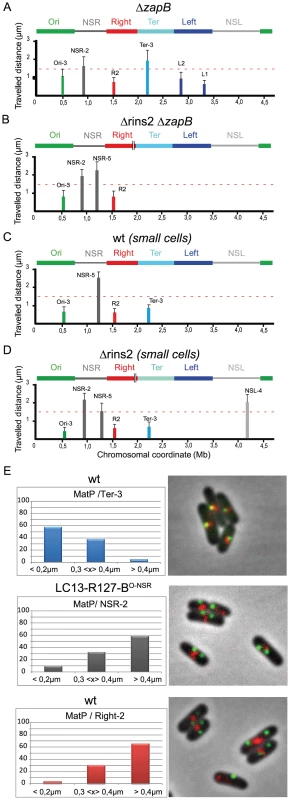
(A–D) Travelled distance of different markers in strains deleted for zapB, deleted for segment rins2 and zapB, and in wt strain. The representation is the same as in Figure 2. (A) strain with a deletion of zapB (markers Ori-3, NSR-2, Right-2 (R2), Ter-3, Left-2 (L2), Left-1 (L1) are indicated from left to right). (B) strain with a deletion of zapB and rins2 (markers Ori-3, NSR-2, NSR-5, Right-2 (R2) are indicated from left to right). (C) small cells of wt strain before Ter MD segregation at mid-cell (markers Ori-3, NSR-5, Right-2 (R2), Ter-3 are indicated from left to right). (D) small cells of strain Δrins2 before Ter MD segregation at mid-cell (markers Ori-3, NSR-2, NSR-5, Right-2 (R2), Ter-3, NSL-4 are indicated from left to right). (E) Quantification of the co-localisation of the MatP-mCherry focus (red) with a chromosomal parS tag bound by ParB-GFP (green). The percentage of cells presenting different interfocal distances is given (left column). In the top lane, the MatP-mCherry focus co-localizes with the Ter-3 marker in the wt strain. The interfocal distance is smaller than 0.4 µm in more than 90% of the cells. In the central lane, the MatP-mCherry focus does not co-localize with the NSR-2 marker in the strain LC13-R127-BO-NSR with a transposition of the Right MD between the Ori MD and the NSRight region (see chromosome configuration in Figure 2). The interfocal distance is greater than 0.3 µm in almost 90% of the cells. In bottom lane, the MatP-mCherry focus does not co-localize with the Right-2 marker in the wt strain. The interfocal distance is greater than 0.3 µm in 90% of the cells. This suggests that the constraining process affecting the NS regions in the absence of tidR and tidL may depend upon the association of the Ter MD with the divisome. This interaction lasts for about half of the cell cycle, and mobility of markers was routinely measured during that period. In order to confirm that the constraining process really required this association, we measured the mobility of markers during the part of the cell cycle when the Ter MD did not interact with the divisome, i.e. in small cells in which the Ter MD has not yet been segregated (Figure 6C). In these cells, the mobility of a Ter marker was slightly increased, which was consistent with the fact that association with the divisome added a level of constraint onto the Ter MD (Espéli et al., submitted). Strikingly, a marker in the NSRight region was as mobile in small ΔtidR cells as in small wild type cells (Figure 6D).
Altogether, these results demonstrated that the constraining process observed in the NSRight region when tidR is absent depends on the association of the Ter MD with the division machinery.
Spatial separation of the Ter MD territory and of the constrained region
It was previously shown that MatP associates with the entire 800 kb Ter region (Mercier et al., 2008). We wondered whether the constraining process originating from the Ter MD required the recruitment of the constrained NS regions within the Ter MD territory. To test this hypothesis, we analysed the co-localization of different markers with a MatP-mCherry fusion protein (Figure 6E). In the wild type chromosomal configuration, the Ter-2 marker was co-localized with the MatP focus, whereas the Right-2 marker was not (Figure 6E). In a strain where the NSRight region has been transposed next to the Ter region (strain LC13-R127-BO-NSR), the NSR-2 marker did not co-localize with the MatP focus as observed for marker Right-2 in the wild type configuration (Figure 6E). These results indicated that constraining of the NS region did not result from a recruitment of the constrained region in the Ter MD territory.
Long-range DNA interaction of the NSRight region is not affected by the MatP-associated constraining effect
Long distance DNA collisions revealed by the frequency of λ site-specific recombination between λ attR and attL sites inserted at different chromosomal locations can be used to reveal chromosome conformation [11]. Inactivation of MatP leads to increased collisions between markers in the Right MD and the Ter MD [13]. To assess whether the constraining of the NSRight region upon deletion of tidR could affect long range DNA collisions inside the NSRight region or between the NSRight region and the flanking Right MD, interactions between attL inserted in the NSRight region and attR sites located at different positions in wt or Δrins2 cells. Remarkably, interactions were as frequent in both genetic contexts, either within the NSRight region or between the NSRight region and the Right MD (Table S3).
Constraining of the NS regions affect their segregation following replication
The organization of the chromosome into macrodomains influences the segregation of sister chromatids and the mobility of chromosomal DNA [12]. Above is described the effect of the absence of the insulation determinants upon DNA mobility. We then examined its impact on the extent of co-localization following replication, by measuring the co-localization index (see Materials and Methods). In the wild type strain, this co-localisation index was high for markers located in MDs (>0.1) in contrast to markers in NS regions (<0.02). Remarkably, only the index of co-localization of the NSRight region marker increased in a ΔtidR mutant, whereas it increased for markers present in both the NSRight and NSLeft regions in the yfbV mutant (Table 2). The increase of co-localization index for markers in NS regions was correlated with a higher number of cells carrying only one focus, localized close to mid-cell.
Tab. 2. Co-localisation index of chromosomal markers. 
Discussion
A new method to engineer the bacterial chromosome
In various studies, inversion of chromosomal segments has been used to change gene positioning or chromosome configurations. Although these approaches have been very useful to reveal various features of gene and genome organization, genetic inversions affect several parameters: gene orientation relative to replication, gene dosage through the change of position relative to oriC, disruption of genetic organization at the two sites of recombination. We have devised a new method to rearrange the bacterial chromosome. The experimental design involved two steps, excision of a segment followed by the reinsertion at another location, and the desired chromosomal configuration can be directly detected on plates with a coloured indicator. This method allows the transposition of fragments ranging in size from a few kilobases to hundreds of kilobases. Because the orientation of genes and sequences are conserved, it allows the transposition of any segment on the chromosome; we succeeded in making all the rearrangements we wanted to perform. The only limitation of the method is the requirement for an essential gene in the transposed segment. In the absence of such a gene, all clones that sustained the deletion will grow making difficult the detection of clones that recombined the excised molecule.
Association of the Ter MD to the division machinery constrains DNA
Markers in the MD and NS regions can be distinguished by their mobility and by the extent of colocalization after replication. The juxtaposition of NS regions to the Ter MD, the deletion of the insulators or the inactivation of YfbV provoked a change of these two properties in the NS regions. Remarkably, the constraining process appeared to spread in cis as insertions of the insulators in the middle of the NSRight region insulated the distal part but not the part proximal to the Ter MD. This effect was regulated during the cell cycle as it was promoted by the interaction of the Ter MD with the division machinery at mid-cell.
The spreading of the constraining process can not be visualized in Right and Left MDs of wt cells because of the reduced mobility of markers in these regions. However, in yihI mutant cells in which the mobility of markers in the Right and Left MDs is released to a level similar to that of NS regions (Valens et al., in prep), mobility of markers of the Right MD located between the Ter MD and tidR were constrained by the MatP-associated constraining process (data not shown). These results indicate that, in the absence of insulation barrier tidR, the constraining process affects not only the NSRight region but also the Right MD in agreement with a process spreading in cis.
It is not yet known how MatP organizes the Ter MD at the molecular level. The association of the Ter MD with the division machinery modifies its properties and probably affects the overall structure of the Ter region; targeting of the Ter MD to the division machinery could promote the formation of loops between matS sites via the interaction of MatP with ZapB assembled in the FtsZ ring (Espéli et al., submitted). In the absence of the insulator, the macromolecular complexes assembled at mid-cell provoked a change of DNA properties detectable at several hundreds of kilobases (Figure 7). The change of properties at a long distance might either result from a change at the chromatin level by the binding/tracking of protein(s) or from modifications of physical properties of the DNA molecule. We do not favour the first hypothesis as it is hard to conceive how the effect would be related to the targeting of the Ter region to the division machinery and how binding or tracking of a protein could act in cis over a region of several hundreds of kilobases. A plausible hypothesis implies modifications of physical properties of DNA. Using two different topological reporter systems that reveal changes in local DNA supercoiling (expression of a lacZ gene under the control of the supercoiling sensitive promoter PgyrA and site-specific resolution reaction between two γδ res sites catalyzed by the γδ resolvase [6], [20]), we failed to detect modifications in the topological properties of the DNA molecule in the absence of insulation (data not shown). We must therefore consider a hypothesis involving an affect on other mechanical properties of the DNA molecule. Although the molecular mechanism is not yet identified, we can imagine that it requires two “anchorages” between which DNA is constrained. While the association of the Ter MD to the division apparatus could constitute the first anchorage, the second one would implicate the tethering of other chromosomal regions in the cell. Because the process is effective up to the NS regions, the second anchorage might involve the Ori region. By acting as a mechanical relief, tidRL would restrain the constraining process and would restrict it to the Ter region.
Fig. 7. Model for the insulation of the Ter MD by tidRL. 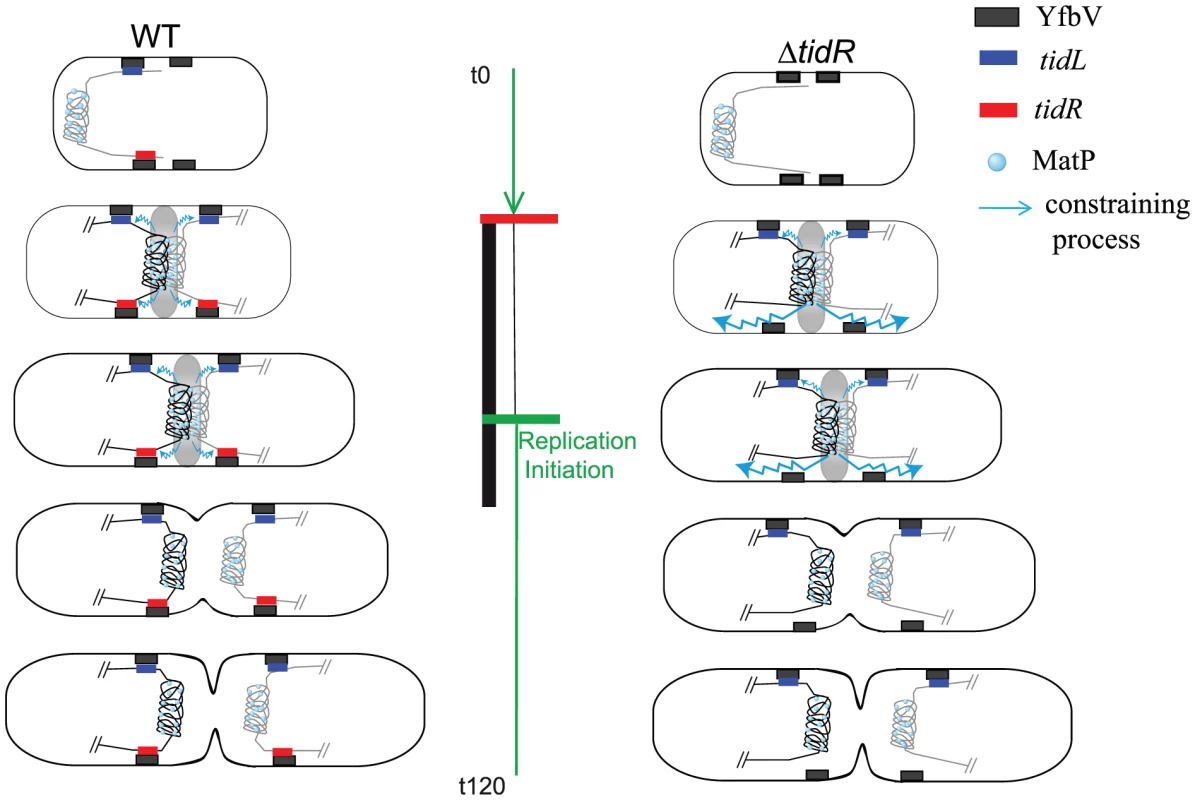
In wt cells (left panel), the Ter MD (only the Ter MD and the flanking DNA is represented) is found near the new pole. As the cell cycle progresses (see [13] for details), the Ter MD is segregated at mid-cell. Interaction of MatP with ZapB promotes the tight association of the Ter MD with the divisome (represented by a grey oval), period indicated by the vertical black bar. At this stage, the constraining process (schematized by a wavy blue line) is apparent but does not propagate behind tidR and tidL. The constraining process would abrogate when the Ter MD dissociates from the divisome. The insulation mechanism requires a direct or indirect association of tidR with YfbV. It is not known if this association lasts during the entire cell cycle. In ΔtidR cells (right panel), insulation is efficient only on the left arm of the chromosome and the constraining process spreads to the right arm of the chromosome. The replication period of wt cells is represented by the green vertical bar; replication is initiated in the mother cell (indicated by a green horizontal bar) and terminates soon after birth (indicated by a horizontal red bar). A number of processes are known to occur in the terminus of the chromosome; they include replication termination [21], decatenation of entangled chromosomes [22], chromosome dimer resolution [23] and coupling of replication termination with cell cycle progression [15]. It is tempting to speculate that the changes in DNA properties resulting from the association with the division machinery of the Ter region facilitate certain DNA metabolic processes required for the proper segregation of the chromosome. A challenge now is to characterize the molecular architecture of the Ter MD associated to the division machinery, to identify the process(es) occurring at this stage and reveal the contribution of the change of DNA properties for the efficiency of the process(es) involved.
An insulation system that controls the formation of MDs and NS regions
By genetic [11] and cytological [12] approaches, we have revealed a chromosome organization into structured MDs and less structured NS regions. Our subsequent analyses have revealed the molecular nature of the Ter MD structuring [13]. Here we have identified a new system that is related to the process involved in the structuring of the Ter MD. This system allowed restricting the effects associated to the interaction of the Ter region with the division machinery. Two 12 bp motifs that flank the Ter MD and one protein co-occurent with MatP have been shown to be required for the insulation process.
The two sequences tidR and tidL were found in the MD flanking the Ter MD, at 130 kb on the right side and at 50 kb on the left side, respectively. This positioning probably reflects the potential of MatP to act at a distance greater than 50 kb [13] that likely precludes the positioning of the insulators closer to the matS sites located at the borders of the Ter MD. Remarkably, tidR and tidL conserved their activity when moved several hundred kilobases away from their original position; their presence close to the Ter MD might thus not rely on specific positional requirements for the insulation activity but rather serve to prevent the spreading of the constraining process to the flanking regions.
tidR and tidL are the only occurrences of the 12-mer GYTGACGTCAGC consensus sequence in the E. coli genome. Allowing one nucleotide difference in the tidRL consensus sequence revealed the presence of 34 sites in the entire genome, dispersed in the diverse MD and NS regions (Figure S4). Based on their position on the genetic map and the properties of markers in various strains, 28 were predicted to be non functional for the insulation property. For 6 variants (2 in the Ter MD and 4 in the Ori MD), it was not possible to predict the functionality.
At least two types of insulation mechanisms could be envisioned at this stage. In both cases, the insulator would block the unidirectional progression of the constraining process since it acts in cis. In the first type, a nucleoprotein complex formed at the insulator would act as a barrier and block the progression of a factor tracking along the DNA molecule. This process would be reminiscent of that selected in boundary elements that prevent the spreading of heterochromatin via the binding of proteins to specific sequences in yeast or vertebrate cells [24], [25] or of that at work with the Tus/ter system that block replication fork progression in bacteria [26]. The second type may result in barrier function by interfering with the spreading of changes affecting physical properties resulting from the MatP-matS complexes. In the last case, anchoring of the DNA molecule to a cellular structure may be required to act as a point of physical relief.
We showed that the gene yfbV is required for the insulation properties of tidR and tidL. It is not yet known how YfbV could promote insulation at tidRL sequences; however it is tempting to speculate that the anchorage of YfbV in the membrane might be used to tether the insulators to a fixed structure, e.g. the membrane, within the cell. Further analyses will be required to characterize the role played by YfbV in the insulation process.
Materials and Methods
Strains and media
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown at 30°C in Lennox broth, or in minimal medium A supplemented with 0.12% of casaminoacids and 0.5% of glucose. Antibiotics were added when necessary. The different deletions targeted in the chromosome were performed by the one-step insertion-deletion technique [27]. These deletions were done in strains carrying the plasmid pKD46 or in the strain DY330 [28]. Two cassettes used to perform deletion constructions carried either a chloramphenicol or a rifampicin resistance gene. Both resistance genes are flanked by frt sites allowing their subsequent deletion [27]. Deletion coordinates are indicated in Table 1. The deletions were verified by PCR.
Induction of transposition
The strains used for transposition carried three attL, attR and attB′ sites derived from the λ site-specific integration module. The three att sites were inserted in the same orientation. Each site was flanked by a cassette carrying an antibiotic resistance gene (chloramphenicol, kanamycin and apramycin, respectively). In addition, the attL site was flanked by the 5′ part of lacZ and the attR site was flanked by the 3′ part of lacZ. The strains are transformed by the plasmid pTSA-CXI that expressed the λ recombinase [11] to promote the transposition. The strains were grown at 30°C and shifted to 39°C to induce the production of the recombinases. After the recombination step, the strains were plated on LB medium supplemented with Xgal and blue colonies were selected. The transposition reactions were verified by PCR verification with appropriate primers (Figure S1).
Fluorescence microscopy and mobility of DNA markers
Cultures were grown in minimal A medium in the presence of glucose and casaminoacids without IPTG to maintain at a minimal level expression of gfp-parB present on plasmid pALA2705 [19]. Movies were recorded automatically on a Leica microscope. Autofocus was performed at every time point on the phase contrast image and GFP fluorescence was recorded on the plane with the best phase contrast focus. Image analysis was performed with ImageJ software using the manual tracking plugin (http://rsb.info.nih.gov/ij/index.html). The XY co-ordinates of the two poles and of the foci were recorded manually and processed automatically with Excel (Microsoft) software. The travelled distance was estimated over a period of 5 minutes by adding up the absolute values of the distances for all 10 sec interval as described before [12]. The (x,y) coordinates of the foci at every time point were recorded and the distance travelled in the 10 s interval calculated. For the mobility measurements at the home position, 30 foci were analyzed (30 cells with one focus for markers in the Ter MD and 15 cells with two foci for markers in other regions of the chromosome). To measure the mobility of markers, the distance travelled by various foci was recorded over a period of 5 minutes with 10 sec intervals when markers were at home position. In the growth conditions used, at home position, markers of Ori, Right and Left MDs as well as markers from NS regions were segregated in the two halves of the cells whereas markers of the Ter MD are found at mid-cell [12].
Index of colocalisation
This index is calculated by comparing the theoretical number of genes (nbth) [29] to the experimental number of genes, i.e. the number of foci detected by microscopy (nbobs) [12]. The normalization of the index is obtained by the formula: [(nbth−nbobs)/nbth].
Supporting Information
Zdroje
1. ThanbichlerMShapiroL 2006 Chromosome organization and segregation in bacteria. J Struct Biol 156 292 303
2. BrowningDFGraingerDCBusbySJ 2010 Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr Opin Microbiol 13 773 780
3. VoraTHottesAKTavazoieS 2009 Protein occupancy landscape of a bacterial genome. Mol Cell 35 247 253
4. DameRTKalmykowaOJGraingerDC 2011 Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet 7 e1002123 doi:10.1371/journal.pgen.1002123
5. BliskaJBCozzarelliNR 1987 Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol 194 205 218
6. HigginsNPYangXFuQRothJR 1996 Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J Bacteriol 178 2825 2835
7. PostowLHardyCDArsuagaJCozzarelliNR 2004 Topological domain structure of the Escherichia coli chromosome. Genes Dev 18 1766 1779
8. SteinRADengSHigginsNP 2005 Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol Microbiol 56 1049 1061
9. WangWLiGWChenCXieXSZhuangX 2011 Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333 1445 1449
10. NikiHYamaichiYHiragaS 2000 Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14 212 223
11. ValensMPenaudSRossignolMCornetFBoccardF 2004 Macrodomain organization of the Escherichia coli chromosome. Embo J 23 4330 4341
12. EspeliOMercierRBoccardF 2008 DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68 1418 1427
13. MercierRPetitMASchbathSRobinSEl KarouiM 2008 The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 475 485
14. BrezellecPHoebekeMHietMSPasekSFeratJL 2006 DomainSieve: a protein domain-based screen that led to the identification of dam-associated genes with potential link to DNA maintenance. Bioinformatics 22 1935 1941
15. EsnaultEValensMEspeliOBoccardF 2007 Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3 e226 doi:10.1371/journal.pgen.0030226
16. RebolloJEFrancoisVLouarnJM 1988 Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc Natl Acad Sci U S A 85 9391 9395
17. SchmidMBRothJR 1983 Genetic methods for analysis and manipulation of inversion mutations in bacteria. Genetics 105 517 537
18. SegallAMahanMJRothJR 1988 Rearrangement of the bacterial chromosome: forbidden inversions. Science 241 1314 1318
19. LiYYoungrenBSergueevKAustinS 2003 Segregation of the Escherichia coli chromosome terminus. Mol Microbiol 50 825 834
20. MoulinLRahmouniARBoccardF 2005 Topological insulators inhibit diffusion of transcription-induced positive supercoils in the chromosome of Escherichia coli. Mol Microbiol 55 601 610
21. Coskun-AriFFHillTM 1997 Sequence-specific interactions in the Tus-Ter complex and the effect of base pair substitutions on arrest of DNA replication in Escherichia coli. J Biol Chem 272 26448 26456
22. HojgaardASzerlongHTaborCKuempelP 1999 Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol Microbiol 33 1027 1036
23. BlakelyGCollomsSMayGBurkeMSherrattD 1991 Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol 3 789 798
24. DormanERBusheyAMCorcesVG 2007 The role of insulator elements in large-scale chromatin structure in interphase. Semin Cell Dev Biol 18 682 690
25. GasznerMFelsenfeldG 2006 Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7 703 713
26. MulcairMDSchaefferPMOakleyAJCrossHFNeylonC 2006 A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 125 1309 1319
27. DatsenkoKAWannerBL 2000 One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 6640 6645
28. YuDEllisHMLeeECJenkinsNACopelandNG 2000 An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97 5978 5983
29. CooperSHelmstetterCE 1968 Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31 519 540
30. NielsenHJOttesenJRYoungrenBAustinSJHansenFG 2006 The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62 331 338
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání