-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSPE-44 Implements Sperm Cell Fate
The sperm/oocyte decision in the hermaphrodite germline of Caenorhabditis elegans provides a powerful model for the characterization of stem cell fate specification and differentiation. The germline sex determination program that governs gamete fate has been well studied, but direct mediators of cell-type-specific transcription are largely unknown. We report the identification of spe-44 as a critical regulator of sperm gene expression. Deletion of spe-44 causes sperm-specific defects in cytokinesis, cell cycle progression, and organelle assembly resulting in sterility. Expression of spe-44 correlates precisely with spermatogenesis and is regulated by the germline sex determination pathway. spe-44 is required for the appropriate expression of several hundred sperm-enriched genes. The SPE-44 protein is restricted to the sperm-producing germline, where it localizes to the autosomes (which contain sperm genes) but is excluded from the transcriptionally silent X chromosome (which does not). The orthologous gene in other Caenorhabditis species is similarly expressed in a sex-biased manner, and the protein likewise exhibits autosome-specific localization in developing sperm, strongly suggestive of an evolutionarily conserved role in sperm gene expression. Our analysis represents the first identification of a transcriptional regulator whose primary function is the control of gamete-type-specific transcription in this system.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002678
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002678Summary
The sperm/oocyte decision in the hermaphrodite germline of Caenorhabditis elegans provides a powerful model for the characterization of stem cell fate specification and differentiation. The germline sex determination program that governs gamete fate has been well studied, but direct mediators of cell-type-specific transcription are largely unknown. We report the identification of spe-44 as a critical regulator of sperm gene expression. Deletion of spe-44 causes sperm-specific defects in cytokinesis, cell cycle progression, and organelle assembly resulting in sterility. Expression of spe-44 correlates precisely with spermatogenesis and is regulated by the germline sex determination pathway. spe-44 is required for the appropriate expression of several hundred sperm-enriched genes. The SPE-44 protein is restricted to the sperm-producing germline, where it localizes to the autosomes (which contain sperm genes) but is excluded from the transcriptionally silent X chromosome (which does not). The orthologous gene in other Caenorhabditis species is similarly expressed in a sex-biased manner, and the protein likewise exhibits autosome-specific localization in developing sperm, strongly suggestive of an evolutionarily conserved role in sperm gene expression. Our analysis represents the first identification of a transcriptional regulator whose primary function is the control of gamete-type-specific transcription in this system.
Introduction
Stem cells have provoked tremendous interest because of their unique ability to differentiate into multiple cell types. The specification of a particular cell fate ultimately results in a program of cell-type-specific gene expression, and the identification and characterization of the regulators that mediate these transcriptional programs are a focus of intense research. Of particular note is the class of transcription factors that act as master switches; their activities are sufficient to dictate a particular cell fate by promoting, both directly and indirectly (via the regulation of additional transcription factors), the expression of the suite of cell-type-specific target genes. The canonical example is MyoD; heterologous expression is sufficient to convert a variety of cell types into myoblasts [1]. Master switch genes therefore specify as well as implement cell fate decisions.
The hermaphrodite germline of Caenorhabditis elegans offers an attractive model for investigating the regulation of stem cell fate specification and differentiation. Cell fate is restricted to a binary choice, sperm or oocyte, which greatly simplifies the analysis. The identical cellular milieu fosters the development of both types of gametes. The switch from spermatogenesis to oogenesis is genetically determined, but can be experimentally controlled using various temperature-sensitive mutations (reviewed in [2]) and chemical reagents [3]. Alternatively, germline stem cells can be manipulated to further expand their repertoire of potential fates, as recently demonstrated by their directed transdifferentiation into neurons [4].
The sexual fate of individual germ cells is specified by an elaboration of the same sex determination program that dictates male or hermaphrodite somatic development (reviewed in [5]). In the soma, that program culminates in the terminal regulator TRA-1, a homolog of cubitus interruptus and GLI transcription factors [6], [7]. TRA-1 promotes the hermaphrodite fate and inhibits male fate, and does so by direct repression of a number of transcription factors that, in turn, regulate sex-specific gene expression in a variety of somatic tissues including the intestine [8], the nervous system [9]–[11], the vulva [12], and the tail [13]. TRA-1 thereby acts as a classic master switch in specifying somatic sexual fate.
Within the germline of C. elegans, additional regulators modify this somatic sex determination program, in part to permit the production of male gametes in an otherwise female animal. In uncommitted germ cells, the FEM and FOG proteins function as the ultimate regulators of sexual fate. Sequence homology alone reveals little about the mode of FEM activity: fem-1 encodes a protein with ankyrin repeats, fem-2 a putative serine/threonine phosphatase, and fem-3 a novel protein [14]–. Proteomic analysis has been more enlightening and shown that the FEM proteins are components of a CUL-2-dependent E3 ubiquitin ligase complex that targets TRA-1 for degradation [17]. FOG-1 is homologous to cytoplasmic polyadenylation element binding proteins, and presumably regulates translation of transcripts that govern gamete cell fate [18]. FOG-3 shares homology with the Tob/BTG family of antiproliferation proteins, and functions in both the initiation and maintenance of spermatogenesis [19]–[21].
The output of this germline sex determination program is gamete-type-specific gene expression. Microarray screening has identified thousands of genes that are differentially expressed in the germline during sperm or oocyte development [22], [23]. The functional significance of the observed transcription regulation is validated by the detection of genes known to be required for gamete development. For example, a large number of Spe (spermatogenesis-defective) genes have been isolated in mutational screens for sperm-specific sterility (reviewed in [24]), and essentially all of those genes were classified as sperm-enriched by microarray data. Transgenic studies indicate that transcriptional control is the primary mode of regulation for sperm genes [25].
Although germline sex determination ultimately governs sperm and oocyte-specific transcription, the precise mechanism of that regulation remains enigmatic. The TRA-1 transcription factor is an attractive candidate for the job, but, despite its demonstrated role in the soma, it does not appear to directly mediate sex-specific gene expression in the germline. First, TRA-1 loss-of-function mutants produce sperm and oocytes that are competent for fertilization and embryonic development [26], [27]; the only logical interpretation is that all of the genes essential for sperm and oocyte function continue to be expressed in the absence of TRA-1. Second, epistasis analysis demonstrates that the FOG proteins regulate gamete cell fate subsequent to TRA-1 [28], [29]. That observation is supported by the identification of the fog-3 gene (and, most likely, fog-1) as a direct target of TRA-1 transcriptional regulation [30]. Finally, epistasis experiments likewise indicate that the FEM proteins govern germline sex determination in the absence of TRA-1 [31], [32]; as this function is obviously independent of their role in TRA-1 degradation, it is probable that additional targets exist. Therein lies the dilemma: the known transcription factor TRA-1 cannot directly regulate gamete-specific transcription, but the known downstream regulators do not encode transcription factors.
To date, the only identified regulator of C. elegans germline transcription is the GATA factor elt-1, which targets sperm genes [33]. However, that role does not appear to be its primary function. elt-1 expression is not limited to sperm, and the gene is required for embryonic and larval development in a variety of tissues [34], [35]. The ELT-1 binding element is present in less than 5% of sperm genes, the majority of which comprise the MSP multigene family, and elt-1 expression does not appear to be directly regulated by the sex determination program. Thus, the bulk of gamete-specific transcription is governed by as-yet-unidentified regulators.
This study provides a critical piece of the puzzle by identifying spe-44 as a key regulator of sperm-specific transcription. Deletion of spe-44 results in sperm-specific sterility and is associated with multiple defects in both the cell cycle and developmental programs of sperm differentiation. The spe-44 gene is expressed in the germline at the onset of spermatogenesis, and is regulated by the terminal germline sex determination factors FEM-1, FEM-3, and FOG-1. SPE-44 promotes expression of a large fraction of sperm-enriched genes, and that role is likely conserved among nematodes. Our work highlights the mechanistic differences between sex determination in the soma, where TRA-1 specifies and directly regulates sex-specific transcription, and the germline, in which SPE-44 implements the transcriptional program specified by the FEM and FOG proteins.
Results
Identification of spe-44
A candidate gene approach was used to identify potential regulators of sperm gene expression. Previous microarray data characterized sex-specific transcriptional profiles for both the germline and soma [22], [23]. Eleven homologs of transcriptional regulators were among the 1,343 genes enriched in the sperm-producing germline (Table S1). We further restricted our list of candidates to those for which pre-existing mutations were available for functional characterization, since sperm genes as a group are generally refractory to inactivation by RNA interference [for example, see 33]. We selected C25G4.4 for further study; the gene product is most similar to the mammalian glucocorticoid modulatory element-binding proteins GMEB-1 and -2 and encodes a SAND domain (for Sp100, Aire, NucP41/75, DEAF-1) [36]–[38]. SAND-containing proteins were first identified as transcriptional activators that bind to functional regulatory elements within the promoters of target genes [39], [40]. Solution and crystal structures of SAND domains reveal a novel DNA-binding fold centered on the highly conserved KDWK motif [41], [42]. The C25G4.4 gene product contains all of the conserved residues of the SAND domain, including KDWK, but otherwise possesses no identifiable domains.
The deletion allele ok1400 of C25G4.4 is almost certainly a null allele as it lacks 1577 of the 1797 base-pair coding region, including the conserved SAND domain (Figure 1A). The original strain isolated by the C. elegans Gene Knockout Consortium [43] produced both fertile and sterile progeny in roughly 3∶1 ratio, suggestive of heterozygosity for a locus conferring recessive sterility. When PCR was used to amplify the C25G4.4 interval from individual sterile and fertile hermaphrodites, the sterile phenotype proved to be tightly linked to the deletion allele; all sterile animals were homozygous for ok1400, whereas all fertile animals contained at least one copy of the wild-type gene (Figure 1B). No additional defects in development or morphology were observed in the ok1400 mutant, suggesting that C25G4.4 might function exclusively in the context of reproduction.
Fig. 1. spe-44(ok1400) mutation causes sperm-specific sterility. 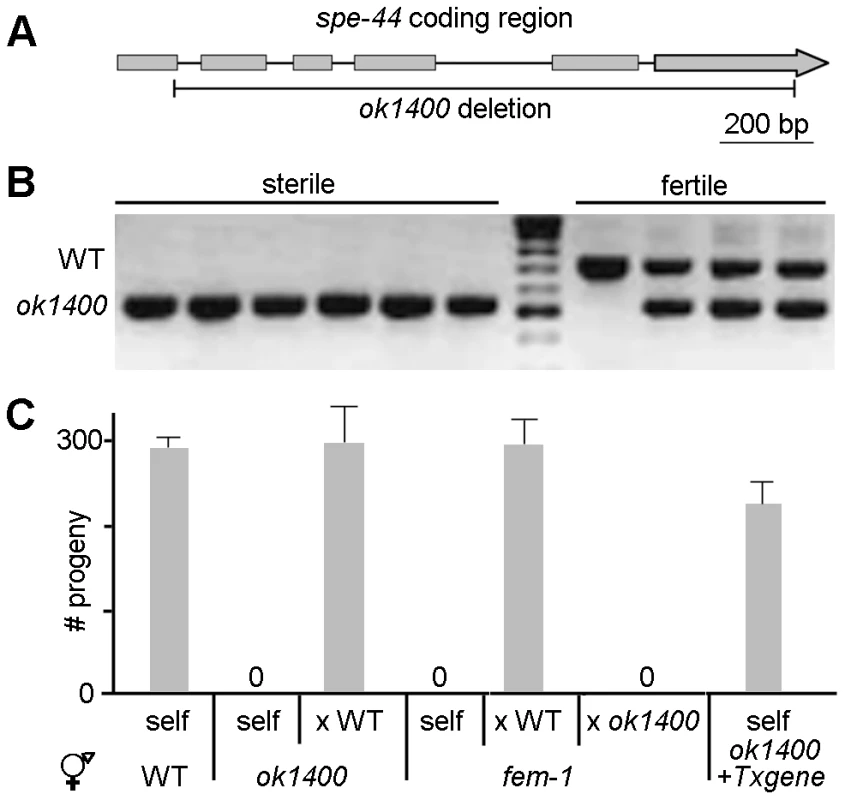
A. Schematic diagram of spe-44. Genomic interval of C25G4.4; gray boxes indicate exons. The extent of the ok1400 deletion is shown below. B. Linkage to sterile phenotype. Examples of individual sterile (N = 50) and fertile (N = 25) hermaphrodites screened by single-worm PCR for the wild-type (upper) and ok1400 (lower) alleles. C. Fertility assays. Mean values and standard deviations of total progeny counts from individual hermaphrodites (N = 20). Hermaphrodite genotype is indicated at bottom. All male strains contained dpy-20(e1282) in addition to the indicated genotype. Self, unmated; x WT, crossed with wild-type males; x ok1400, crossed with spe-44(ok1400) males; + Txgene, unmated spe-44(ok1400) hermaphrodites containing the spe-44 transgene. In self-fertilizing hermaphrodites, sterility can arise from defects in sperm and/or oocyte function. To discriminate among those possibilities, we set up reciprocal matings between ok1400 mutants and wild-type animals of the opposite sex (Figure 1C). Crosses between self-sterile ok1400 homozygous hermaphrodites and wild-type males yielded viable progeny, indicating that ok1400 hermaphrodites produce functional oocytes. By contrast, crosses between ok1400 homozygous males [marked with the tightly linked dpy-20(e1282) allele] and fem-1(hc17) hermaphrodites (which lack sperm) produced no progeny, whereas fem-1 hermaphrodites mated with homozygous dpy-20(e1282) males produced abundant outcross progeny. Microinjection of the C25G4.4 transgene rescued the sterility of unmated ok1400 homozygous hermaphrodites (Figure 1C, +Txgene), thereby confirming that the deletion allele is responsible for the observed sterility. Together these results indicate that the sterility of ok1400 results from a defect in sperm function. Based on the sperm-specificity of its sterile phenotype, the C25G4.4 gene was designated spe-44 (for spermatogenesis-defective).
Defects in spermatogenesis
Sperm cell development in C. elegans has been well characterized in both sexes [44]–[46]. Throughout the larval and adult stages, Notch signaling in the distal end of the gonad maintains a population of mitotically dividing germline stem cells with the potential to generate either oocytes or sperm [2]. During the third larval (L3) stage, the most proximal of these mitotic germ cells enter the meiotic cell cycle, an event that roughly coincides with the commitment of these cells to spermatogenesis [28]. By the fourth larval (L4) stage in both hermaphrodites and males, the various stages of spermatogenesis can be observed in a distal to proximal array within the gonad. Transcription of sperm genes typically initiates during the pachytene phase of the cell cycle [e.g., see 33]. Following the disassembly of the synaptonemal complex, the spermatocytes enter a karyosome stage during which global transcription ceases [46]. The primary spermatocytes then detach from the gonadal syncytium and undergo the two meiotic divisions. Cytokinesis during meiosis I can be complete or incomplete, producing secondary spermatocytes that contain one or two nuclei, respectively. Immediately following anaphase of meiosis II, cellular components that are not required for subsequent sperm function are partitioned away from the spermatids in a budding division and deposited in a residual body. In males, sperm production continues through adulthood, and immotile, spherical spermatids accumulate in the seminal vesicle. Upon insemination of hermaphrodites, the process of activation converts spermatids into mature, crawling spermatozoa that migrate from the uterus to the spermatheca. In hermaphrodites, gametogenesis switches abruptly from sperm to oocyte production at the L4/adult transition. Spermatids undergo activation into motile spermatozoa as they are propelled into the spermatheca ahead of the newly formed oocytes.
To characterize potential spermatogenesis defects, we examined adult male gonads, in which all stages of sperm development are readily visible, under differential interference contrast (DIC) optics. In wild-type gonads, spermatogenesis progressed in a distal-to-proximal array through the pachytene and karyosome stages of meiosis, followed by a meiotic division zone and then a region of packed spermatids (Figure 2A). The spe-44 gonads (Figure 2B) were similar in overall size, suggesting that germline proliferation occured normally. Developing spe-44 spermatocytes progressed appropriately through the pachytene and karyosome stages, although the mutant gametes were less refractile and thus smoother in appearance. However, the division zone was expanded, the cells misshapen, and normal spermatids failed to accumulate.
Fig. 2. Spermatogenesis defects in spe-44 males. 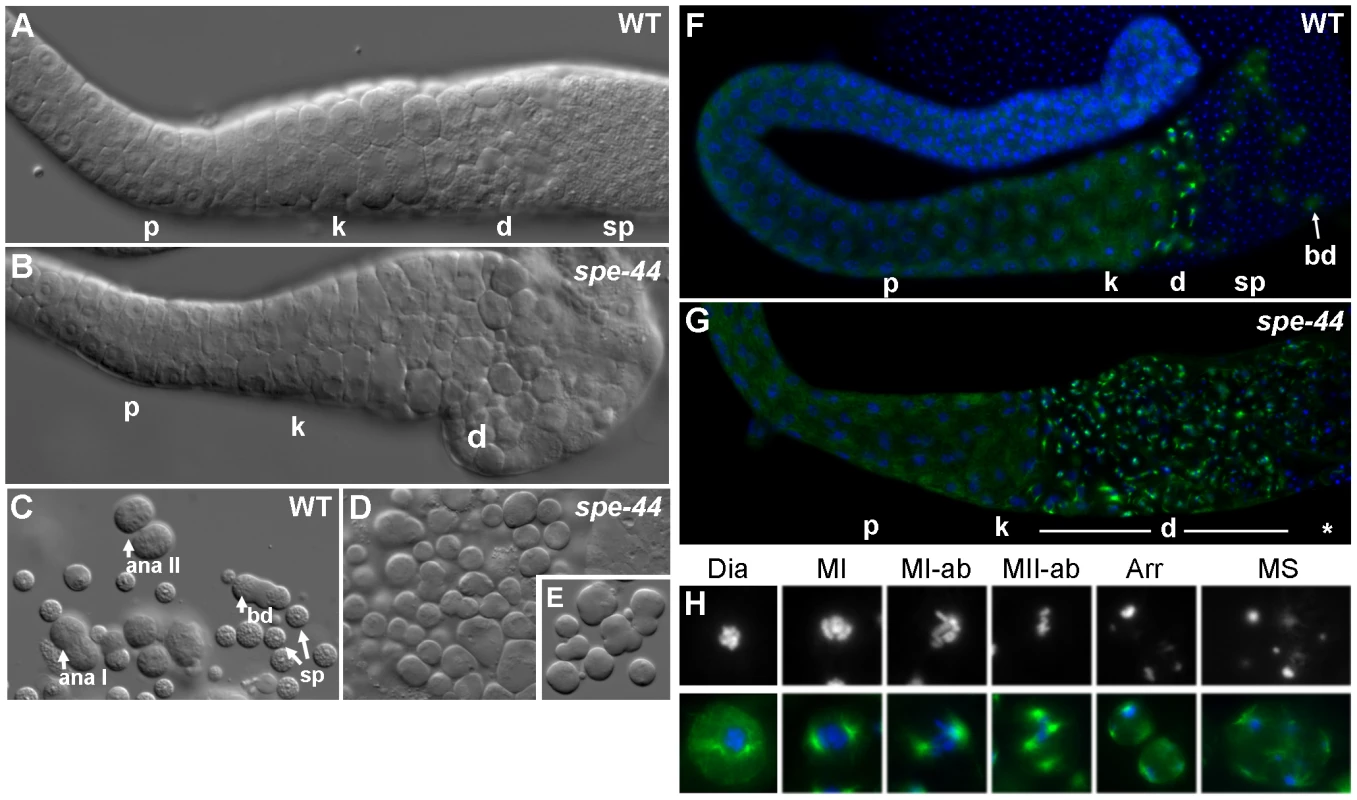
A–B. Isolated gonads from wild-type (A) and spe-44 (B) males visualized by DIC. Indicated are pachytene (p), karyosome (k), and meiotically dividing (d) spermatocytes, and spermatids (sp). C–E. Sperm spreads from wild-type (C) and spe-44 (D–E) males. C. Wild-type spermatogenesis with spermatocytes in anaphase I (ana I), anaphase II (ana II), post-meiotic budding division (bd), and mature spermatids (sp). D. spe-44 spermatogenesis with aberrantly dividing spermatocytes. E. Enlarged image of spe-44. F–G. Isolated gonads from wild-type (F) and spe-44 (G) males co-stained with anti-tubulin (green) and DAPI (blue). Labeling as in panels A–B. Additionally indicated are residual bodies from the budding division (bd), and a mix of terminally arrested and dying spermatocytes (*). H. Higher magnification of individual sperm from spe-44 males. Top row, DAPI staining to highlight nuclear morphologies; bottom row, merge of DAPI (blue) and tubulin (green). See text for labeling. These differences were even more apparent in sperm spreads, which distinguish individual spermatocytes. In wild-type samples, the meiotic division zone could be further discriminated into spermatocytes undergoing anaphase I, anaphase II, and the post-meiotic budding division of spermatids from a residual body; numerous spermatids are also visible (Figure 2C). In the spe-44 samples, dividing spermatocytes often exhibited a variety of cleavage defects, including unequal divisions and the formation of multiple partially budded structures, and no budding divisions of residual bodies were observed (Figure 2D–2E). Aberrant spermatogenesis was also observed in hermaphrodites (Figure S1). These morphological defects prompted a more detailed characterization of the spe-44 phenotype.
To that end, we examined the chromatin and microtubule dynamics in male gonads (Figure 2F–2G). Similarly to DIC, no differences were observed between the germlines of wild-type and spe-44 males through the karyosome stage. The meiotic division zone in wild-type gonads contained a small number of spermatocytes with visible asters and a few cells undergoing the budding division (in which tubulin segregates to residual bodies), followed by a large number of spermatids (Figure 2F). By contrast, spe-44 gonads exhibit a greatly expanded meiotic division zone, suggesting a cell cycle arrest (Figure 2G).
Sperm spreads revealed a myriad of defects in the spe-44 spermatocytes within the division zone (Figure 2H). Although the nucleation of asters initiated normally during diakinesis (Dia), the asters were often larger and broader than their wild-type counterparts. The asters migrated correctly to set up the metaphase I spindle (MI), but chromosome segregation was aberrant with most or all chromosomes segregating to one spindle pole (MI-ab), particularly during the second meiotic division (MII-ab). Examples of unequal chromosome segregation and extensive aneuploidy were also evident in arrested spermatocytes (Arr), and some of the smallest cells had microtubules but no chromatin (data not shown). As in wild-type spermatogenesis, both complete and incomplete cytokinesis occurred during the meiotic divisions; however, we frequently observed multiple spindles (MS), as many as eight per spermatocyte, instead of the typical two or four. The presence of these supernumerary asters is suggestive of spindle overduplication and/or fusion of terminally arrested spermatocytes. The asters in these terminal spermatocytes were almost always found adjacent to the plasma membrane, and the microtubules in these spermatocytes were often unusually long. Ultimately, the mutant spermatocytes appeared to lyse; the number of arrested spermatocytes did not increase dramatically with age, and cellular debris accumulated within the gonad (data not shown).
Cell cycle defects
The striking persistence of microtubule asters in spe-44 spermatocytes suggests an arrest of cell cycle progression. To test this hypothesis, we analyzed the pattern of histone H3 (serine 10) phosphorylation (anti-pHisH3), an indicator of Aurora kinase activity (Figure 3A–3B) [47]. In both spe-44 and wild-type gonads, anti-pHisH3 labeling was first detectable in developing spermatocytes as the synaptonemal complex was beginning to break down, became most intense during the karyosome stage, and continued to label the chromosomes through the meiotic divisions. As wild-type spermatocytes completed anaphase II and initiated the budding division, anti-pHisH3 labeling of chromatin was no longer detectable (Figure 3A). By contrast, the chromatin of terminally arrested spe-44 spermatocytes exhibited persistent anti-pHisH3 labeling (Figure 3B). Similar results were obtained with a marker of CDK-cyclin B phosphorylation. Staining of wild-type sperm spreads with monoclonal antibody MPM-2, which binds to a mitotic phospho-epitope of Cdk-CyclinB substrates [48], revealed labeling of the cytoplasm and metaphase I chromosomes during meiosis that was largely absent in spermatids (Figure 3C). In spe-44, labeling persisted in the terminally arrested spermatocytes (Figure 3D). These observations are indicative of an M-phase arrest of the cell cycle.
Fig. 3. Cell cycle defects in spe-44 sperm. 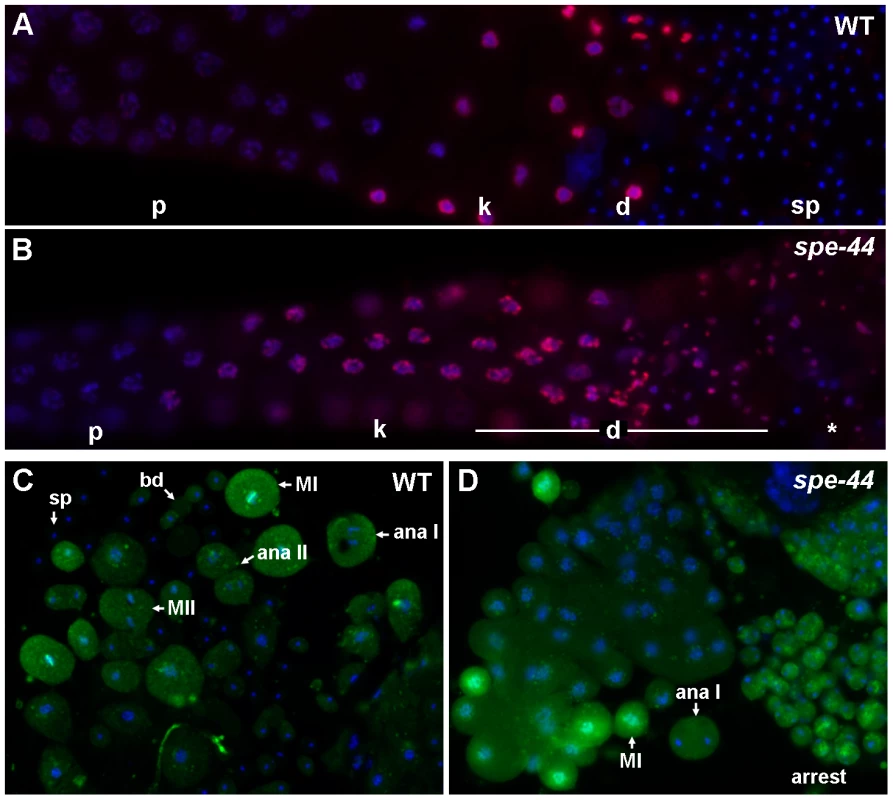
A–B. Wild-type (A) and spe-44 (B) male gonads co-stained with DAPI (blue) and anti-phosphorylated histone H3 Ser10 (red). C–D. Sperm spreads from wild-type (C) and spe-44 (D) males stained with antibody MPM-2 (green) and DAPI (blue). Labeling as in Figure 2C. Additionally indicated are metaphase I (MI), metaphase II (MII), and arrested spermatocytes (arrest). Organelle assembly defects
If SPE-44 serves as a key regulator of the spermatogenesis program, spe-44 spermatocytes might be expected to exhibit additional defects in the assembly of sperm-specific structures. Therefore, we examined the distribution and morphology of two sperm-specific components: assembly of the major sperm protein (MSP) into fibrous bodies, and the localization of Golgi-derived membranous organelles (MOs). Anti-MSP staining in wild-type sperm spreads (Figure 4A) revealed the formation of fibrous bodies, which appear as distinct puncta, early in meiosis. The fibrous bodies segregate into the spermatids during the budding division before becoming cytoplasmic in mature spermatids. In spe-44 mutants, MSP failed to assemble into fibrous bodies but instead remained diffusely distributed in the cytoplasm throughout spermatogenesis (Figure 4B).
Fig. 4. Organelle assembly defects in spe-44 sperm. 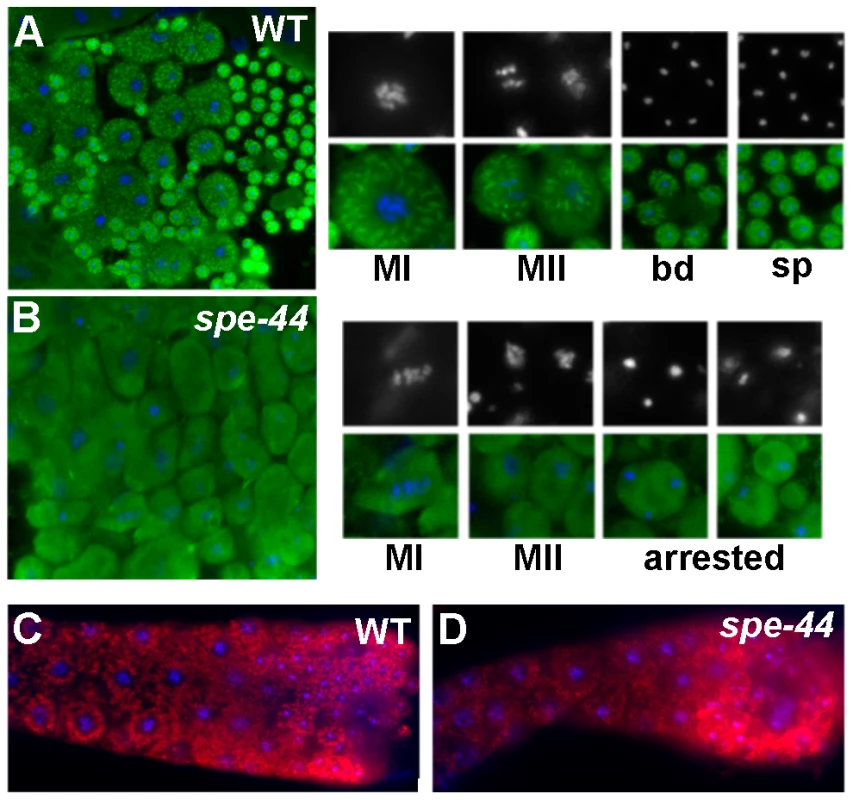
A–B. Wild-type (A) and spe-44 (B) spermatocytes co-stained with anti-MSP (green) and DAPI (blue, white). Shown at right are individual sperm at higher magnification in metaphase I (MI), metaphase II (MII), the budding division (bd), mature spermatids (sp), and arrested spermatocytes (arrested). C–D. Wild-type (C) and spe-44 (D) gonads co-stained with MO-specific 1CB4 antibody (red) and DAPI (blue). To investigate the formation and dynamics of the membranous organelles, we analyzed the immunostaining pattern of the monoclonal antibody 1CB4 [49]. In wild-type gonads (Figure 4C), MOs can first be detected as discrete structures in the late pachytene portion of the syncytial gonad; in spermatids, the MOs localized to the periphery adjacent to the cell membrane. In spe-44 gonads (Figure 4D), the structures recognized by 1CB4 labeling appeared more disorganized, and distinct localization to the cell periphery was not observed. Thus, although the key components of the fibrous bodies and membranous organelles are present in the spe-44 mutant, the assembly of these sperm-specific structures is aberrant.
Sperm-specific expression of spe-44
Previous microarray data indicated that transcription of spe-44 is elevated during spermatogenesis [22], [23], so we utilized quantitative RT-PCR to assess the dynamics of spe-44 transcription during development. Expression of spe-44 was first detected in age-synchronized populations of wild-type hermaphrodites at the L3 larval stage, when gametes become committed to the sperm cell fate (Figure 5A). Transcript levels of spe-44 declined through the L4 and adult stages, when gametogenesis switches from sperm to oocyte production. In wild-type L4 and adult males, which produce sperm continuously, spe-44 expression levels were higher than in hermaphrodites at the same stages (hermaphrodites and males are morphologically similar prior to L4, so males were not assessed at earlier larval stages).
Fig. 5. Expression and localization of spe-44 mRNA. 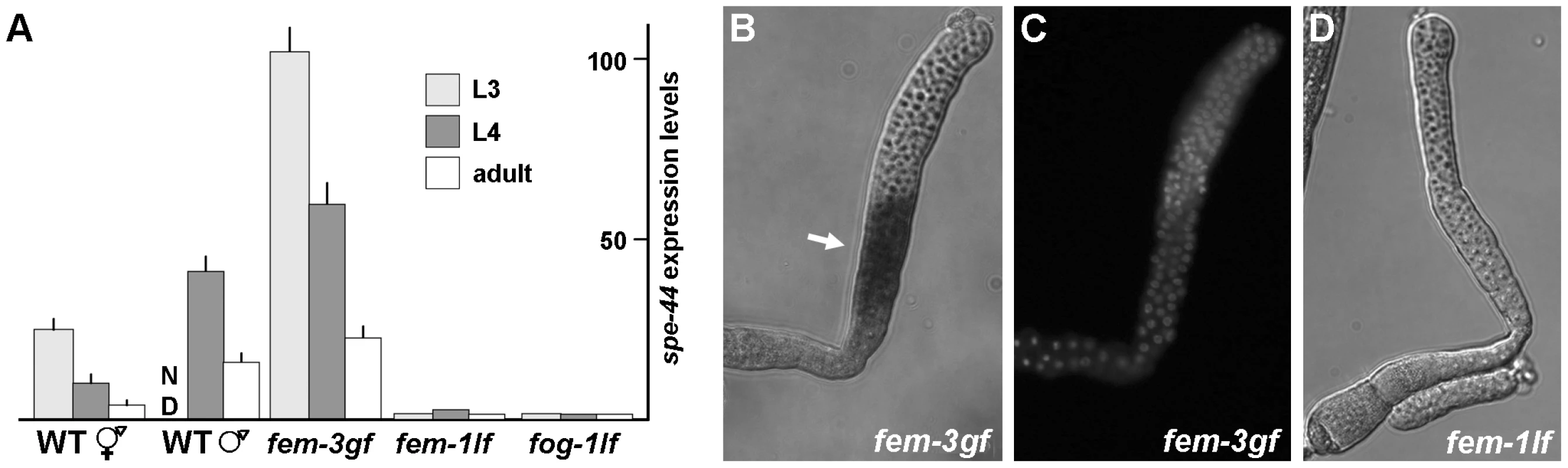
A. Quantification of spe-44 expression by qRT-PCR. Transcript levels in each sample were normalized against actin as an internal control; the scale was arbitrarily set at one for the fem-1 L3 value. Mean values and standard deviations are from triplicate samples of the indicated stage and genotype. ND, not determined. Genotypes from left to right are wild-type hermaphrodites, wild-type males, fem-3(gf) hermaphrodites, fem-1(lf) hermaphrodites, and fog-1(lf) hermaphrodites. B. In situ hybridization for spe-44 transcript (arrow) in dissected fem-3(gf) L3 gonad. C. Same gonad stained with DAPI. D. In situ hybridization for spe-44 in dissected fem-1(lf) L4 gonad. To determine the role of the germline sex determination program in spe-44 expression, we employed hermaphrodites that contained temperature-sensitive mutations in the terminal FEM and FOG regulators. When reared at the restrictive temperature, fem-3(gf) hermaphrodites produce only sperm. Expression of spe-44 was observed beginning at the L3 stage and, although the levels declined somewhat, remained elevated into adulthood (Figure 5A). Conversely, the fem-1(lf) and fog-1(lf) mutations, which cause hermaphrodites to make only oocytes, resulted in low spe-44 expression levels at all stages of development. Thus, spe-44 transcript accumulation correlates strongly with the production of sperm. Furthermore, peak expression is observed during L3 at the onset of sperm fate specification. This pattern stands in marked contrast to that observed for other sperm genes. For example, MSP, which is abundantly expressed, is not detectable until L4 [50]. Likewise, a developmental time course of global transcription demonstrates that essentially all sperm genes exhibit a sharp increase at the onset of L4 and corresponding decline at the transition to adulthood [22], [23]. Thus, the peak expression of spe-44 during L3 precedes that of other sperm genes, and is consistent with a role in the regulation of sperm gene expression. Finally, spe-44 expression is governed by terminal regulators of the germline sex determination pathway (fem-1, fem-3, and fog-1) that specify sperm or oocyte fate.
In situ hybridizations of dissected gonads were performed to ascertain the precise spatial pattern of spe-44 expression. The fem-3(gf) and fem-1(lf) mutations were used to restrict hermaphrodite gametogenesis to sperm or oocyte production, respectively. Abundant expression was first detected in fem-3(gf) animals during the L3 stage in the early meiotic germline (Figure 5B). DAPI staining of the chromatin indicated that expression coincides with early pachytene of meiosis I (Figure 5C). By contrast, spe-44 expression was undetectable in fem-1(lf) germlines at any stage of development (Figure 5D). Thus, spe-44 expression is restricted to the sperm-producing germline at a time and place consistent with the regulation of sperm gene expression.
Regulation of sperm gene transcription by SPE-44
Since SPE-44 shares homology with known transcriptional regulators, we performed DNA microarray screening to assess its role in governing gene expression. Comparison between wild-type and spe-44 L4 males at the onset of sperm production revealed statistically significant differences (p<0.05) in gene expression between the two samples. A total of 813 genes exhibited greater than two-fold changes in expression. The levels of 535 genes were reduced in the spe-44 males and 278 were elevated (Table S2).
We compared our spe-44 microarray data to previous transcriptional profiles that classified genes as sperm-enriched, oocyte-enriched, or germline-intrinsic (i.e., expressed in both sperm and oocyte) [22], [23]. Of the 535 genes that were down-regulated in the spe-44 strain, nearly two-thirds (343) were also classified as sperm-enriched (Figure 6). Note that the degree of overlap between the data sets is likely under-represented, in part due to technical differences between the microarray platforms that were employed. By contrast, very little concordance was observed with either the oocyte-enriched (four) or germline-intrinsic (25) classes. Therefore, loss of spe-44 results in a substantial defect in sperm-enriched gene expression. Those genes are hereafter referred to as spe-44 sperm targets.
Fig. 6. Summary of microarray results and transcriptional activation by SPE-44. 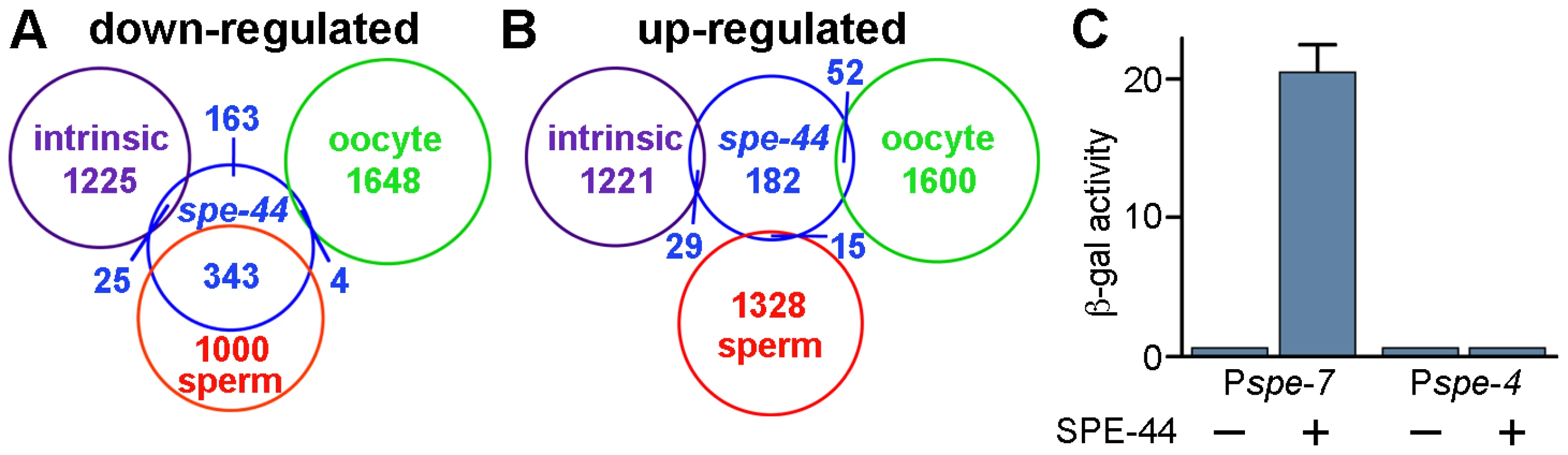
A–B. Summary of microarray results. Venn diagrams of the genes that were down-regulated (A) or up-regulated (B) in spe-44 mutant males compared to the categorization (sperm-enriched, oocyte-enriched, or germline intrinsic) defined in [23]. C. Transcriptional activation by SPE-44. Expression of lacZ reporters containing the indicating promoter plus spe-44 under repression (−) or induction (+). ß-galactosidase activity quantified by ONPG assay for triplicate samples under each condition. We performed the same comparison with the 278 genes that exhibited increased expression in the spe-44 strain. No significant overlap was observed with any of the three germline categories (Figure 6). These results indicated that germ cell fate in spe-44 mutants is not shifted from sperm to oocyte; such a fate switch would be accompanied by an increase in oocyte-enriched transcription. Therefore, spe-44 is not a component of the sex determination pathway that specifies gamete cell fate; however, it is required to implement that fate by promoting the transcription of sperm genes.
We also examined the frequency of GO terms associated with the sets of genes that were down or up-regulated in the spe-44 mutant strain. Among the genes with reduced expression levels, three categories were over-represented: protein kinases (3.2-fold higher than expected) and associated terms (e.g., protein amino acid phosphorylation); protein phosphatases (4.3-fold) and associated terms; and structural molecules (8.6-fold) (Table S3). All of these categories are consistent with a defect in sperm gene expression. The overabundance of protein kinases and phosphatases among the sperm transcriptome has been reported previously [22]; the structural molecules largely consist of the MSP and SSP gene families, which are structural components of the sperm motility apparatus (see below). Similar characterization of the genes that were up-regulated in the spe-44 strain was not as informative. The only two over-represented terms were ATP-binding (2.3-fold higher than expected) and intracellular (2.2-fold) (Table S4).
The list of spe-44 down-regulated sperm targets contains a significant number of genes with demonstrable roles in sperm (shown in Table 1). The genes fall into four categories: 1) MSP (major sperm protein) is the structural polymer responsible for nematode sperm motility, and also serves as a signaling molecule to promote ovulation and oocyte maturation [51], [52]. A subset of the multigene MSP family (17 of 28 members) exhibits greater than two-fold reduction in transcript levels in the spe-44 mutant. 2) Members of the small sperm-specific protein family (formerly SSP; designated ssp/ssq/ssr/sss) are structurally similar to MSP and play a role in MSP polymerization [53], [54]; five are found among the spe-44 targets. 3) Spe (spermatogenesis-defective) and Fer (fertilization-defective) genes have been identified in genetic screens for sperm-specific sterility [55]–[57]; four Spe genes are targets of spe-44. 4) The elt-1 gene, which encodes a GATA transcription factor that regulates the transcription of a subset of sperm genes [33], is down-regulated in the spe-44 mutant strain. Because all of these genes have known functions within sperm, the reduced transcript levels observed in the spe-44 mutant may contribute to the variety of defects that occur during spermatogenesis (see Discussion).
Tab. 1. Known sperm genes among SPE-44 targets. 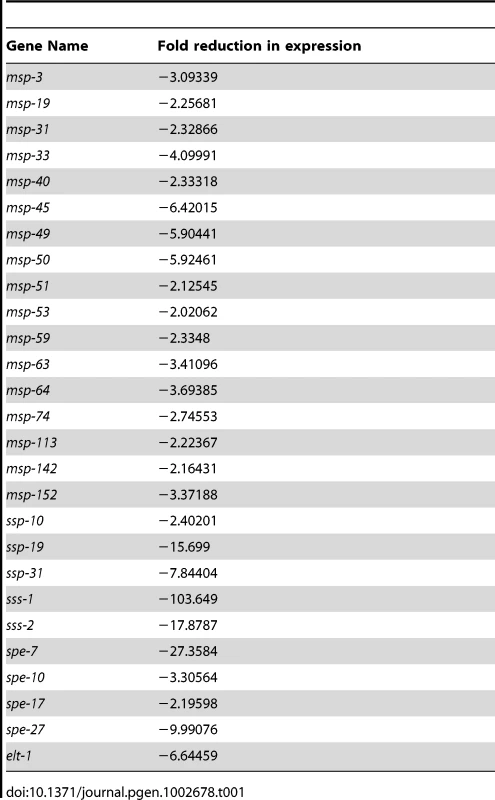
The target genes identified by microarray might be directly or indirectly regulated by SPE-44. The multigene MSP family is likely to fall within the latter category. Prior work identified the GATA transcription factor ELT-1 as a direct regulator of MSP expression [33]. Our microarray data indicate that both elt-1 and the MSP genes are expressed at lower levels in the spe-44 mutant. The simplest model is a transcriptional cascade in which SPE-44 promotes elt-1 expression, whose product in turn promotes MSP transcription. Alternatively, SPE-44 might work in conjunction with ELT-1 to promote maximal levels of MSP transcription and/or to restrict expression to sperm.
We tested the ability of SPE-44 to directly promote transcription in a heterologous system, using a variant of the yeast one-hybrid assay [58]. We constructed a yeast lacZ reporter gene that contained the putative promoter region of spe-7 (Pspe-7::lacZ), a sperm target gene that is strongly down-regulated in the spe-44 mutant strain. Expression of spe-44 was controlled by the galactose-inducible GAL1 promoter. Induction of spe-44 resulted in Pspe-7::lacZ expression as measured by ß-galactosidase activity (Figure 6B). Expression of Pspe-7::lacZ was dependent upon spe-44, as ß-galactosidase activity was not detectable under non-inducing conditions. We also examined the specificity of SPE-44 transcriptional activation for its target promoter. No expression was observed from a lacZ reporter that contained the promoter region of spe-4, a sperm gene that is not a target of spe-44. Therefore, SPE-44 can function as a transcription factor to directly activate gene expression from its cognate promoter.
Association of SPE-44 with chromatin
To determine if the SPE-44 protein localized to chromatin in vivo, we generated a SPE-44 antibody and stained dissected gonads. The timing and distribution agreed with our previous transcriptional analysis, and correlated perfectly with sperm production. Temporally, SPE-44 labeling was first detectable in L3 male germlines and persisted through adulthood (Figure S2 and Figure 7A). Spatially, SPE-44 labeling was absent in the distal stem cell niche and mitotic zone, initiated in the early meiotic zone, co-localized with chromosomes in the pachytene region, and became non-chromosomal before disappearing altogether in karyosome stage nuclei (Figure 7A; see inset for details). The observed staining pattern is dependent upon the SPE-44 protein, as it is absent in the spe-44 mutant strain (Figure S3).
Fig. 7. Pattern of SPE-44 protein expression and localization. 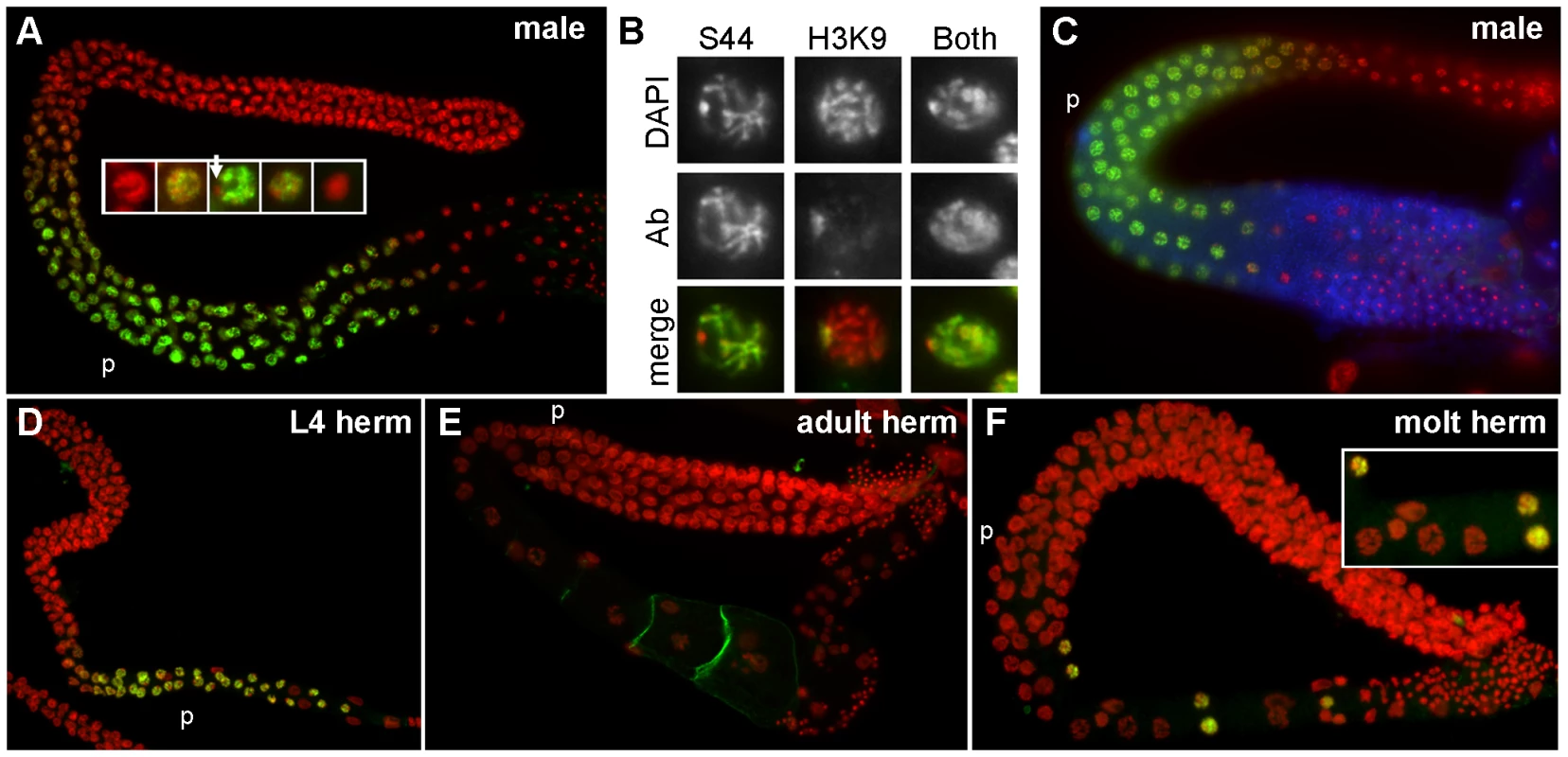
A. Wild-type adult male gonad co-stained with anti-SPE-44 (green) and DAPI (red). Insert at higher magnification shows details of nuclear morphologies during the appearance/disappearance of SPE-44 labeling. Arrow indicates unlabeled DNA. B. Individual nuclei from DAPI-stained wild-type male gonads that were co-stained with anti-SPE-44 only (S44), anti-dimethylated histone H3 (Lys9) antibody only (H3K9), or both antibodies. Top row, DAPI; middle row, antibody (Ab); bottom row, merged image. C. Wild-type male gonad stained with anti-SPE-44 (green), anti-MSP (blue), and DAPI (red). D–F. Wild-type hermaphrodite gonads stained with anti-SPE-44 (green) and DAPI (red). D. L4 larval stage. E. Adult stage. F. Spermatogenesis to oogenesis transition at the L4/adult molting stage. Insert shows details of the first oocyte nuclei interspersed with a few last SPE-44-staining nuclei. The pachytene region (p) is indicated for orientation. Closer examination revealed that SPE-44 localized along the length of most but not all of the pachytene chromosomes (Figure 7A, inset, and 7B). The unpaired X chromosome was a likely candidate for SPE-44 exclusion: X is singularly devoid of sperm genes, and contains chromatin modifications consistent with transcriptional silencing [22], [59]. To test this hypothesis, we compared the distribution of SPE-44 and histone H3(lysine 9) dimethylation, a chromatin modification that specifically labels the X chromosome in the male gonad (Figure 7B). Co-incubation with both antibodies labeled all chromosomes, indicating that the chromosome that is unlabeled by SPE-44 is indeed the X (Figure 7B). The localization of SPE-44 to autosomes (which contain sperm genes) but not X (which does not) is consistent with a role as an early acting and positive regulator of sperm gene transcription.
For sperm targets of SPE-44, we predict that their distribution would be coincident with or subsequent to the appearance of SPE-44. Therefore, we examined the localization of SPE-44 in comparison to MSP, an abundant sperm-specific marker that is a target of SPE-44 (Table 1). Co-staining clearly demonstrated that SPE-44 is detectable prior to the accumulation of MSP (Figure 7C), which is first observed near the bend of the gonad at mid to late pachytene. Thus, SPE-44 exhibits the properties predicted for a regulator of sperm gene expression: the protein appears at the onset of sperm fate specification prior to the production of sperm proteins, is bound to the chromatin of committed and developing spermatocytes, and disappears in the karyosome stage coincident with the global cessation of transcription.
Labeling of SPE-44 in hermaphrodite gonads likewise correlates with sperm production. The protein is first detectable in L3 at the time of sperm fate specification (Figure S4) and in young L4 hermaphrodites undergoing spermatogenesis (Figure 7D), but is absent in adult hermaphrodites that are undergoing oogenesis (Figure 7E). We were particularly interested in the distribution of SPE-44 during the switch in gamete fate. An intriguing property of the C. elegans hermaphrodite gonad is its ability to cleanly transition from the production of spermatocytes to the production of oocytes without making sexually ambiguous gametes. To test whether SPE-44 might be functioning in this switch, we examined the pattern of SPE-44 localization in hermaphrodite gonads during this developmental transition. Remarkably, in the proximal gonads of hermaphrodites that contained spermatocytes directly adjacent to the first enlarging oocytes, we always observed a few SPE-44 staining nuclei interspersed among these initial oocytes (Figure 7F). Although we have not ruled out the possibility that these are residual SPE-44-positive nuclei displaced from the syncytial germline by enlarging oocytes, we favor the interpretation that these are developing spermatocytes, and that SPE-44 is a marker of gamete sexual fate that is regulated at the level of individual cells. As an aside, we note that anti-SPE-44 labeling of the hermaphrodite nuclei co-localizes with most but not all of the chromatin, and (as demonstrated in males) the unlabelled region presumably represents the two X chromosomes.
Conservation of spe-44
All nematode species share an unusual mode of sperm motility (amoeboid crawling) and a novel protein (MSP) that underlies that motility. Similarly, the regulation of sperm gene expression might be conserved among nematodes. Complete genomes are available for five species of the Caenorhabditis group, so those sequences were screened for spe-44-related genes. C. elegans itself contains a paralagous gene, gmeb-3, whereas each of the remaining species contains a single spe-44 homolog. These homologs encode proteins that are more similar to SPE-44 than GMEB-3 and likely orthologous, and the tree agrees well with the current phylogeny of these species (Figure 8A). Sequences related to spe-44 were also identified in more distant nematode species (data not shown); however, conservation was limited largely to the SAND domain, which is represented in several C. elegans genes, and the degree of similarity was insufficient to distinguish the relationship unambiguously. Although the extent of spe-44 conservation across the phylum is unknown, the gene is clearly conserved within the Caenorhabditis group.
Fig. 8. spe-44 conservation in Caenorhabditis species. 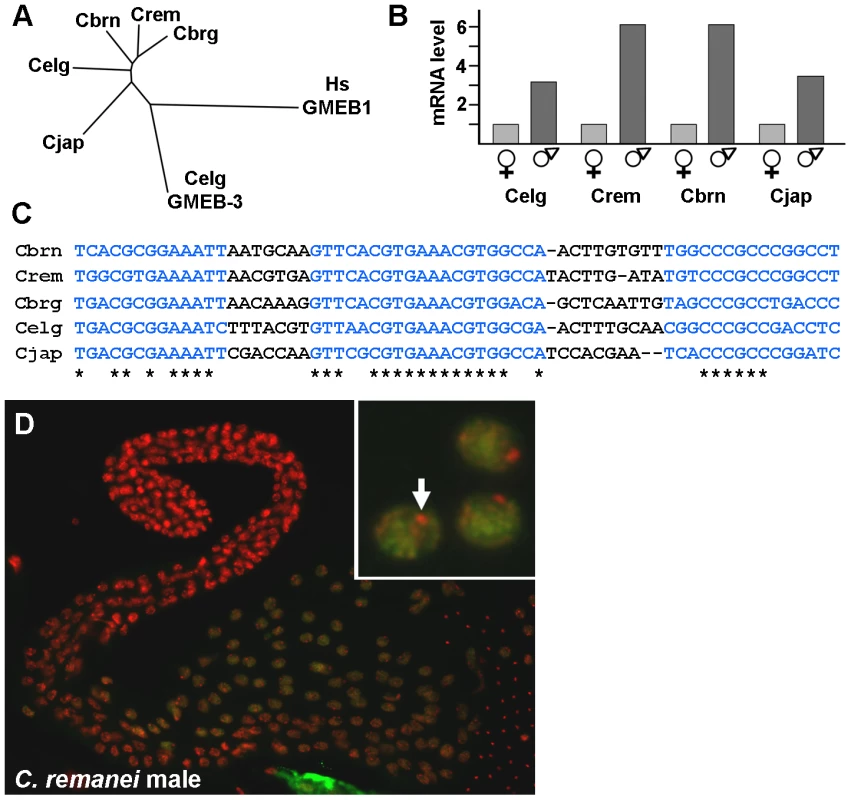
A. Tree indicating the relationship of the SPE-44 orthologs from the indicated species. CLUSTALW2 [81] was used to perform multiple sequence alignment of complete amino acid sequences for the following proteins: Wormbase accession numbers CN01523 (C. brenneri; abbreviated Cbrn), RP47564 (C. remanei; Crem), CBP08202 (C. briggsae; Cbrg), CE05305 (C. elegans; Celg), and JA05534 (C. japonica; Cjap). Also included were the paralogous GMEB-3 from C. elegans (accession number CE31443; Celg GMEB-3), and human GMEB1 as an outgroup (NCBI accession NP_077808; Hs GMEB1). Branch length reflects the degree of similarity. B. Sex-biased expression. Relative read number from RNA-Seq data; female/hermaphrodite values were normalized to one for each species. Celg, C. elegans; Crem, C. remanei; Cbrn, C. brenneri, Cjap, C. japonica. C. Conservation of spe-44 upstream sequences. Blue indicates conserved sequence elements; asterisks indicate identical nucleotides. D. SPE-44 protein localization in C. remanei. Dissected male gonad co-stained with anti-SPE-44 antibody (green) and DAPI (red). Higher magnification (inset) reveals exclusion from a single DNA focus, presumably X (arrow). An examination of sex-biased transcription in species of the Caenorhabditis group is currently underway (C. Thomas, manuscript in preparation), so those RNA-Seq data were examined for differences in spe-44 expression. In all four species under study (C. elegans, C. remanei, C. brenneri, and C. japonica), the spe-44 ortholog exhibited significant male-biased expression (Figure 8B). Note that the degree of male-biased expression may be greater than indicated by these data, which are derived from adult animals; in C. elegans, peak levels of spe-44 mRNA are observed in L3 larvae and decline through the L4 and adult stages (Figure 5A). Although the developmental profiles of spe-44 transcription in the other species are currently unknown, the data from adults clearly demonstrate that male-biased expression is a conserved property of spe-44.
The conservation of gene expression patterns among species can be reflected in the conservation of promoter elements that regulate transcription. Therefore, we performed multiple sequence alignment of the upstream regions from the five spe-44 orthologs. We discovered three highly conserved sequence elements (Figure 8C, in blue) within 150 nucleotides of the initiation codon, which could potentially represent binding sites for unknown regulatory factors. Thus, the sequence conservation of spe-44 is not limited to the coding sequences but extends to putative promoter elements.
Another distinctive property of SPE-44 in C. elegans is its restriction to meiotic autosomes in developing sperm. To determine if this distribution pattern is likewise conserved, isolated gonads from C. remanei males were immunostained with the anti-SPE-44 antibody (which recognizes a carboxy-terminal epitope whose sequence is conserved among the spe-44 orthologs). As in C. elegans, we observed SPE-44 labeling of the chromatin of pachytene nuclei, with the notable exception of the presumed X chromosome (Figure 8D, arrow in inset). The conservation of gene expression and protein distribution in both male/hermaphrodite (C. elegans) and male/female (C. remanei, C. brenneri, and C. japonica) species strongly suggests that the observed pattern represents the ancestral state among this group of nematodes. Although we have yet to directly demonstrate that spe-44 is required for spermatogenesis and sperm gene regulation in the other species, the degree of sequence conservation within the putative promoter and coding sequence, coupled with male-enriched expression and the pattern of protein distribution in the germline, makes a compelling case for an evolutionarily conserved role.
Discussion
This work identifies spe-44 as a critical regulator of sperm-specific transcription in the C. elegans germline. Mutation of spe-44 results in sperm-specific sterility, a consequence of aberrant sperm development that includes cell cycle arrest and defects in sperm organelle assembly. The spe-44 gene is expressed in the sperm-producing germline at the onset of sperm fate specification, and its product is required for the appropriate expression of several hundred sperm-enriched transcripts. Targets of spe-44 include a number of genes known to be required for sperm development. The spe-44 gene and promoter sequence, male-biased expression pattern, and germline protein distribution are conserved among nematode species, consistent with an evolutionarily conserved role in the regulation of sperm gene expression. spe-44 lies at the terminus of the germline sex determination pathway, and its primary role is to implement cell fate specification by promoting sperm-specific transcription.
The spe-44 deletion phenotype is complex and multifaceted, as might be expected for an early acting factor that regulates a large number of sperm-specific genes. Sperm cell fate is specified appropriately in spe-44 mutants, but these gametes are unable to properly implement the programs of meiotic cell division and differentiation. Initiation of meiosis appears normal, but the ensuing defects in chromosome segregation suggest that spe-44 spermatocytes either lack, or are unable to properly regulate, key spindle and kinetochore components necessary for chromosome attachment. Ultimately, spe-44 spermatocytes arrest at M-phase and fail to undergo the budding division. The relationship between these two events is unclear. Cell cycle arrest might block progression of the budding division (e.g., via a checkpoint mechanism); alternatively, the segregation of cell cycle regulatory factors (perhaps in association with the meiotic spindles) to the residual body might be required for the completion of meiosis. In either case, the defects in fibrous body and membranous organelle assembly likely contribute to the failure of the budding division. FBs are composed of MSP assembled in a paracrystalline array, and MOs contain a variety of secretory and membrane proteins. FB-MO complexes are thought to facilitate the segregation of these critical components into the budding spermatid and away from the residual body, so the failure of this partitioning event in spe-44 mutants is not surprising.
The observed phenotypes presumably reflect reduced levels of multiple components necessary for specific aspects of spermatogenesis. The spe-44 mutation causes a defect in the expression of several hundred sperm genes, which are obvious candidates for those functions. For example, mutations in the spe-44 target spe-7 are associated with defects in fibrous body assembly similar to those observed in spe-44 mutants [M. Presler, K. Messina, and D. Shakes, pers. comm.]. The reduction in MSP expression might also contribute to this phenotype. However, any effect may be modest, as the spe-44 mutation targets only a subset of the MSP genes and their expression is reduced but not abrogated. Additional targets of spe-44 that might contribute to the membranous organelle assembly defects include spe-10 and spe-17, which are required for both the structural integrity of the membranous organelles and the proper segregation of components into the residual body during spermatid formation [60], [61]. Similarly, M-phase arrest might reflect reduced expression of the spe-44 target CDC14, a phosphatase that normally functions to inactivate cyclin-dependent kinase and promote M-phase exit [62].
SPE-44 was selected as a candidate transcriptional regulator on the basis of its SAND domain. SAND domain proteins exhibit intrinsic transcriptional activation ability in a variety of reporter assays, a property shared by SPE-44 (Figure 6C). However, these proteins appear to function primarily through interactions with other transcriptional regulators. In some cases, the binding partners are canonical transcription factors. For example, GMEB-1 and GMEB-2 function with the glucocorticoid receptor to modulate target gene expression [63], while DEAF-1 cooperates with the Hox gene Deformed [40]. SAND domain proteins have also been implicated in chromatin-mediated regulation of transcription. AIRE is part of a large protein complex that includes various chromatin components [64], SP100 protein interacts with HMG and HP-1 heterochromatin proteins [65], [66], and GMEB binds the histone acetylase CBP [63]. In plants, ULTRAPETALA1 has been shown to function in the Trithorax group chromatin remodeling complex to antagonize the repressive effects of histone methylation by the Polycomb complex [67]. The observed distribution pattern of SPE-44 suggests that it, too, interacts closely with chromatin markers; the protein localizes broadly along the length of the autosomes while being specifically excluded from the X chromosome, which is known to contain repressive chromatin modifications. Identification of SPE-44-interacting factors will be crucial in determining the mechanism(s) by which SPE-44 and other SAND-domain proteins regulate gene expression.
SPE-44 is not the sole transcriptional regulator that promotes sperm-specific gene expression. The list of spe-44 targets comprises only a subset of the 1,343 sperm-enriched genes identified previously [22], [23], and a majority of Spe genes with demonstrated roles in spermatogenesis are expressed appropriately in spe-44 animals. Ten additional transcriptional regulator homologs exhibit sperm-enriched expression, and those are potential mediators of sperm gene transcription. Although elt-1 expression is governed by SPE-44, the other transcription factors are not and could promote sperm gene transcription independently of SPE-44. In addition to those candidates, transcription factors that also function in somatic tissues might not have been identified as sperm-enriched but could nonetheless promote sperm gene expression in the germline. Alternatively, the observed sperm-enriched expression of some genes could reflect repression during oogenesis, and negative regulators might be predicted to exhibit oocyte-enriched, rather than sperm-enriched, expression.
The conservation of spe-44, and its presumed role in sperm gene transcription, may be somewhat surprising given the rapid evolution of reproduction within the Caenorhabditis group of nematodes. Hermaphroditism has evolved independently at least twice from the ancestral male/female species [68], [69]. Differences in the germline sex determination programs between C. elegans and C. briggsae include gene loss, gene gain, and reordering of the pathway components [70]–[72]. Nonetheless, the data strongly suggest that spe-44 function has been retained. The orthologous gene exhibits male-biased expression among the male/female species of the group. That expression likely reflects the conservation of sequence elements within the putative promoters of these orthologs. Most remarkable is the conserved pattern of SPE-44 protein localization in the male germline of C. remanei, including binding to the autosomes and exclusion from the presumptive X chromosome. Sperm genes are largely absent from the X chromosome in C. elegans, and chromatin markers indicate that X is transcriptionally silent at the time of sperm gene expression [22], [59]. Although the chromosomal distribution of sperm genes in nematodes other than C. elegans is currently unknown, germline silencing of the X chromosome is evolutionarily conserved among this group [59]. This observation strongly predicts a similar restriction of sperm genes to the autosomes, the site of SPE-44 binding, among these Caenorhabditis species. Taken together, the data are consistent with a conserved role for spe-44 in sperm gene expression.
One unanswered question is how the expression of spe-44 is itself regulated by the germline sex determination pathway. Our quantitative RT-PCR results clearly indicate that, in C. elegans, spe-44 is governed by the terminal components FEM-1, FEM-3, and FOG-1 at the level of transcription and/or mRNA stability. The FEM proteins are part of a CUL-2-dependent E3 ubiquitin ligase complex, so one potential mechanism would be via degradation of a repressor of spe-44 expression. That repressor is unlikely to be TRA-1, despite its demonstrated role as a target of FEM-dependent degradation and a repressor of male sexual fate in the soma. The FEM proteins specify gamete fate in the absence of TRA-1, and the spe-44 promoter region does not contain sequences that match the known TRA-1 consensus binding site [73]. However, there are highly conserved sequence elements upstream of spe-44 that might serve as binding sites for as-yet-unidentified regulatory proteins, which could be targets of FEM-dependent degradation. Similarly, FOG-1 is predicted to govern the translation of unknown factors that specify gamete fate, one or more of which might control transcription of spe-44. Alternatively, the putative polyadenylation-binding activity of FOG-1 might play a more direct role in regulating spe-44 transcript stability.
The timing and specificity of spe-44 expression indicate its critical role in implementing germline sexual fate. SPE-44 is one of the earliest known markers of sperm fate in C. elegans. Both temporally and spatially, SPE-44 precedes the production of other sperm-specific components. The precise, cell-specific pattern of SPE-44 localization during the spermatogenesis-to-oogenesis transition of hermaphrodites predicts that the transcription, translation, and protein stability of SPE-44 must be tightly regulated by a variety of feedback mechanisms. This exquisite restriction of SPE-44 to developing spermatocytes provides a powerful tool for the analysis of mutants that, due to defects in the germline sex determination program, produce gametes of sexually indeterminate character.
Materials and Methods
C. elegans strains were obtained from the Caenorhabditis Genetics Center, and are derived from the wild-type isolate N2 (Bristol). The spe-44(ok1400) deletion allele was isolated by the Gene Knockout Consortium [43] and backcrossed six times to N2 prior to analysis. Additional mutations include: dpy-20(e1282)IV, fem-1(hc17)IV, fem-3(q20gf)IV, fog-1(q253)I, let-92(s677)IV, unc-22(s7)IV. A linked spe-44(ok1400) dpy-20(e1282) mutant strain, balanced by let-92(s677) unc-22(s7), was generated to facilitate discrimination of homozygous lines. Homozygous spe-44 males were obtained by mating balanced heterozygous males to homozygous Dpy Spe hermaphrodites and picking Dpy male progeny. Sodium hypochlorite treatment of gravid adults was used to obtain embryos for age-synchronized analyses. Strains were propagated on OP50-seeded NGM plates and maintained at 15°C unless otherwise indicated. Genetic manipulations were carried out by standard methods [74]. Self-fertility assays were performed by total progeny counts of individual hermaphrodites; cross-fertility was assessed by mating individual hermaphrodites with four males for 24 hours, then counting total progeny.
Single-worm PCR detection of the spe-44(ok1400) deletion utilized primers HES-343, HES-364, and HES-502 (all primers listed in Table S5). Microinjection rescue was performed with plasmid pHS584, a 6.5 kbp fragment of the spe-44 genomic interval; included in the microinjection mix were plasmid pRF4, which contains the dominant rol-6(su1006) as a morphological marker, and genomic DNA, which enhances germline expression by formation of complex arrays [75], [76]. Rescue was determined by injecting balanced spe-44(ok1400) dpy-20(e1282)/let-92(s677) unc-22(s7) hermaphrodites, picking individual dumpy rollers from the F2 generation grown at 25°C, and counting total progeny. For qPCR, RNA was isolated via Trizol (Invitrogen) from triplicate samples (each ∼500 animals) grown at 25°C at the indicated developmental stage, reverse-transcribed with Superscript II (Invitrogen), and amplified with primers HES-502 and HES-539 (for spe-44) or AKA-70 and AKA-71 (for act-1). For yeast expression assays, the spe-44 cDNA was amplified with primers HES-531 and HES-532 and inserted into a derivative of pJG4-5 [77] that lacks the transcriptional activation domain. Putative promoter fragments were amplified from genomic DNA with primers HES-612 and HES-613 (spe-7) or HES-583 and HES-584 (spe-4) and inserted into a derivative of pLacZi (Clontech); the same spe-4 upstream sequence is sufficient to confer in vivo transgene rescue of spe-4 sterility [78]. Yeast media and manipulations followed standard protocols [79].
Intact gonads were obtained by dissection of individual worms. Sperm spreads were obtained by further applying slight pressure to the coverslip. Antibody staining of dissected gonads followed established protocols [e.g., see 46]. Specimens were incubated with primary antibodies for 2–3 hours at room temperature unless otherwise indicated. Primary antibodies included: FITC-labeled anti-alpha-tubulin (DMIA, Sigma) (used at 1∶100 dilution), anti-MSP (4D5, gift from D. Greenstein) (1∶200), anti-phospho-histone H3 (serine10) (Upstate Biotechnology) (1∶150), anti-MPM2 (DAKO Corp.) (1∶100 overnight at 4°C), anti-histone H3 dimethylated Lys 9 (Upstate Biotechnology) (1∶50), 1CB4 [49]. The SPE-44 antibody (1∶100) was generated in rabbits against a C-terminal peptide epitope and affinity-purified (Open Biosystems). Affinity-purified secondary antibodies (Jackson Immunoresearch Laboratories) (1∶100) included goat anti-rabbit TRITC-labeled IgG and FITC - or DyLight-labeled goat anti-mouse IgG. Images were acquired under differential inference contrast or epifluorescence using an Olympus BX60 microscope equipped with a Cooke Sensicam cooled CCD camera and IPLab software. In some cases, images were minimally processed to enhance contrast either with IPLab software or Photoshop.
In situ hybridization for spe-44 germ line expression was performed on dissected gonads following fixation [80]. Digoxigenin-labeled, single-stranded sense and antisense probes were generated from a 1.3 kb spe-44 cDNA fragment by linear amplification according to the manufacturer's protocol (Roche). Following hybridization, probe detection was by colorimetric assay with alkaline phosphatase-conjugated anti-digoxigenin antibodies and NBT/BCIP substrate.
Worms for microarray screening were obtained by hand-picking samples of 50 L4 males of the indicated genotype. RNA was isolated by Trizol treatment and ethanol precipitation using linear polyacrylamide (GeneElute LPA; Sigma-Aldrich) as carrier. RNA was amplified and labeled according to the manufacturer's protocol (Nugen). Microarray screening was performed in triplicate using GeneChip C. elegans genome arrays (Affymetrix). Microarray data were analyzed by Microarray Suite 5.0 (Affymetrix) and Genomics Suite (Partek) software, using a p-value threshold of 0.05 for differential expression. Sequences for comparative analysis of Caenorhabditis species were obtained from WormBase release WS220 [www.wormbase.org]. Multiple sequence alignments of spe-44 homologs and promoters were performed with CLUSTALW2 [81].
Supporting Information
Zdroje
1. TapscottSJDavisRLThayerMJChengPFWeintraubH 1988 MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242 405 411
2. KimbleJCrittendenSL 2007 Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Ann Rev Cell Dev Biol 23 405 433
3. MorganCTLeeMHKimbleJ 2010 Chemical reprogramming of Caenorhabditis elegans germ cell fate. Nat Chem Biol 6 102 104
4. TursunBPatelTKratsiosPHobertO 2011 Direct conversion of C. elegans germ cells into specific neuron types. Science 331 304 308
5. ZarkowerD 2006 Somatic sex determination. WormBook Feb 10 1 12
6. HodgkinJ 1993 Molecular cloning and duplication of the nematode sex-determining gene tra-1. Genetics 133 543 560
7. ZarkowerDHodgkinJ 1992 Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70 237 249
8. YiWRossJMZarkowerD 2000 Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127 4469 4480
9. ConradtBHorvitzHR 1999 The TRA-1 sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98 317 327
10. PedenEKimberlyEGengyo-AndoKMitaniSXueD 2007 Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev 21 3195 3207
11. SchwartzHTHorvitzHR 2007 The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev 21 181 194
12. SzaboEHargitaiBRegosATihanyiBBarnaJ 2009 TRA-1/GLI controls the expression of the Hox gene lin-39 during C. elegans vulval development. Dev Biol 330 339 348
13. MasonDARabinowitzJSPortmanDS 2008 dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development 135 2373 2382
14. SpenceAMCoulsonAHodgkinJ 1990 The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell 60 981 990
15. PilgrimDMcGregorAJacklePJohnsonTHansenD 1995 The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol Biol Cell 6 1159 1171
16. RosenquistTAKimbleJ 1988 Molecular cloning and transcript analysis of fem-3, a sex-determination gene in Caenorhabditis elegans. Genes Dev 2 606 616
17. StarostinaNGLimJMSchvarzsteinMWellsLSpenceAM 2007 A CUL-2 ubiquitin ligase containing the three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell 13 127 139
18. LuitjensCGallegosMKraemerBKimbleJWickensM 2000 CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev 14 2596 2609
19. ChenPJSingalAKimbleJEllisRE 2000 A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev Biol 217 77 90
20. WinklerGS 2010 The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol 222 66 72
21. LeeMHWon KimKMorganCTMorganDEKimbleH 2011 Phosphorylation state of a Tob/BTG protein, FOG-3, regulates initiation and maintenance of the Caenorhabditis elegans sperm fate program. Proc Natl Acad Sci USA 108 9125 2130
22. ReinkeVSmithHENanceJWangJVan DorenC 2000 A global profile of germline gene expression in C. elegans. Mol Cell 6 605 616
23. ReinkeVGilISWardSKazmerK 2004 Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131 311 323
24. NishimuraHL'HernaultSW 2010 Spermatogenesis-defective (spe) mutants of the nematode Caenorhabditis elegans provide clues to solve the puzzle of male germline functions during reproduction. Dev Dyn 239 1502 1514
25. MerrittCRasolosonDKoDSeydouxG 2008 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18 1476 1482
26. HodgkinJ 1987 A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev 1 731 745
27. SchedlTGrahamPLBartonMKKimbleJ 1989 Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics 123 755 769
28. BartonMKKimbleJ 1990 fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125 29 39
29. EllisREKimbleJ 1995 The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139 561 577
30. ChenPEllisRE 2000 TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127 3119 3129
31. DoniachTHodgkinJ 1984 A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol 106 223 235
32. HodgkinJ 1986 Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114 15 52
33. del Castillo-OlivaresAKulkarniMSmithHE 2009 Regulation of sperm gene expression by the GATA factor ELT-1. Dev Biol 333 397 408
34. PageBDZhangWStewardKBlumenthalTPriessJR 1997 ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans. Genes Dev 11 1651 1661
35. SmithJAMcGarrPGilleardJS 2005 The Caenorhabditis elegans GATA factor elt-1 is essential for differentiation and maintenance of hypodermal seam cells and for normal location. J Cell Sci 118 5709 5719
36. GibsonTJRamuCGemundCAaslandR 1998 The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci 23 242 244
37. ZengHJacksonDAOshimaHSimonsSSJr 1998 Cloning and characterization of a novel binding factor (GMEB-2) of the glucocorticoid modulatory element. J Biol Chem 273 17756 17762
38. TheriaultJRCharetteSJLambertHLandryJ 1999 Cloning and characterization of hGMEB1, a novel glucocorticoid modulatory element binding protein. FEBS Lett 452 170 176
39. OshimaHSzaparyDSimonsSSJr 1995 The factor binding to the glucococorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J Biol Chem 270 21893 21901
40. GrossCTMcGinnisW 1996 DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J 15 1961 1970
41. BottomleyMJCollardMWHuggenvikJILiuZGibsonTJ 2001 The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol 8 626 633
42. SurdoPLBottomleyMJSattlerMScheffzekK 2003 Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucococorticoid modulatory element binding protein-1 reveal deoxyribonucleic acid and zinc binding regions. Mol Endocrinol 17 1283 1295
43. MoermanDGBarsteadRJ 2008 Towards a mutation in every gene in Caenorhabditis elegans. Brief Funct Genomic Proteomic 7 195 204
44. WolfNHirshDMcIntoshJR 1978 Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastruct Res 63 155 169
45. WardSArgonYNelsonGA 1981 Sperm morphogenesis in wild-type and fertilization-defective mutants of Caenorhabditis elegans. J Cell Biol 91 26 44
46. ShakesDCWuJCSadlerPLLapradeKMooreLL 2009 Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet 5 e1000611 doi:10.1371/journal.pgen.1000611
47. CrosioCFimiaGMLouryRKimuraMOkanoY 2002 Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol 22 874 885
48. DavisFMTsaoTYFowlerSKRaoPN 1983 Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA 80 2926 2930
49. OkamotoHThomsonJN 1985 Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode Caenorhabditis elegans. J Neurosci 5 643 653
50. BurkeDJWardS 1983 Identification of a large multigene family encoding the major sperm protein of Caenorhabditis elegans. J Mol Biol 171 1 29
51. SepsenwolSRisHRobertsTM 1989 A unique cytoskeleton associated with crawling in the amoeboid sperm of the nematode, Ascaris suum. J Cell Biol 108 55 66
52. MillerMANguyenVQLeeMHKosinskiMSchedlT 2001 A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291 2144 2147
53. SchormannNSymerskyJLuoM 2004 Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans. Acta Crystallogr D Biol Crystallogr 60 1840 1845
54. YiKWangXEmmettMRMarshallAGStewartM 2009 Dephosphorylation of major sperm protein (MSP) fiber protein 3 by protein phosphatase 2A during cell body retraction in the MSP-based amoeboid motility of Ascaris sperm. Mol Biol Cell 20 3200 3208
55. WardSMiwaJ 1978 Characterization of temperature-sensitive, fertilization-defective mutants of the nematode Caenorhabditis elegans. Genetics 88 285 303
56. ArgonYWardS 1980 Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics 96 413 433
57. L'HernaultSWShakesDCWardS 1988 Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Geletics 120 435 452
58. LiJJHerskowitzI 1993 Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262 1970 1874
59. KellyWGSchanerCEDernbergAFLeeMHKimSK 2002 X-chromosome silencing in the germline of C. elegans. Development 129 479 492
60. ShakesDCWardS 1989 Mutations that disrupt the morphogenesis and localization of a sperm-specific organelle in Caenorhabditis elegans. Dev Biol 134 307 316
61. L'HernaultSWBenianGMEmmonsRB 1993 Genetic and molecular characterization of the Caenorhabditis elegans spermatogenesis-defective gene spe-17. Genetics 134 769 780
62. VisintinRCraigKHwangESPrinzSTyersM 1998 The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2 709 718
63. KaulSBlackfordJAJrChenJOgryzkoVVSimonsSSJr 2000 Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol Endocrinol 14 1010 1027
64. AbramsonJGiraudMBenoistCMathisD 2010 Aire's partners in the molecular control of immunological tolerance. Cell 140 123 145
65. LehmingNLe SauxASchullerPtashneM 1998 Chromatin components as part of a putative transcriptional repressing complex. Proc Natl Acad Sci USA 95 7322 7326
66. SeelerJSMarchioASitterlinDTransyCDejeanA 1998 Interaction of SP100 with PH1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA 95 7316 7321
67. CarlesCCFletcherJC 2009 The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev 23 2723 2728
68. ChoSJinSWCohenAEllisRE 2004 A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res 14 1207 1220
69. KiontkeKGavinNPRaynesYRoehrigCPianoF 2004 Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc Natl Acad Sci USA 101 9003 9008
70. NayakSGoreeJSchedlT 2005 fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol 3 e6 doi:10.1371/journal.pbio.0030006
71. GuoYLangSEllisRE 2009 Independent recruitment of F box genes to regulate hermaphrodite development during nematode evolution. Curr Biol 19 1853 1860
72. HillRCHaagES 2009 A sensitized genetic background reveals evolution near the terminus of the Caenorhabditis germline sex determination pathway. Evol Dev 11 333 342
73. ZarkowerDHodgkinJ 1993 Zinc fingers in sex determination: only one of the two C. elegans TRA-1 proteins binds DNA in vitro. Nucleic Acids Res 21 3691 3698
74. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 71 94
75. MelloCCKramerJMStinchcombDAmbrosV 1991 Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10 3959 3970
76. KellyWGXuSMontgomeryMKFireA 1997 Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146 227 238
77. GyurisJGolemisEChertkovHBrentR 1993 Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75 791 803
78. L'HernaultSWArduengoPM 1992 Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J Cell Biol 119 55 68
79. RoseMDWinstonFHieterP 1990 Methods in Yeast Genetics: A Laboratory Course Manual CSHL Press, Cold Spring Harbor, NY
80. LeeMHSchedlT WormBook, The C. elegans Research Community, editors 2006 RNA in situ hybridization of dissected gonads.
81. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and Clustal X version 2.0. Bioinformatics 23 2947 2948
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



