-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Chromosomes that fail to synapse during meiosis become enriched for chromatin marks associated with heterochromatin assembly. This response, called meiotic silencing of unsynapsed or unpaired chromatin (MSUC), is conserved from fungi to mammals. In Caenorhabditis elegans, unsynapsed chromosomes also activate a meiotic checkpoint that monitors synapsis. The synapsis checkpoint signal is dependent on cis-acting loci called Pairing Centers (PCs). How PCs signal to activate the synapsis checkpoint is currently unknown. We show that a chromosomal duplication with PC activity is sufficient to activate the synapsis checkpoint and that it undergoes heterochromatin assembly less readily than a duplication of a non-PC region, suggesting that the chromatin state of these loci is important for checkpoint function. Consistent with this hypothesis, MES-4 and MET-1, chromatin-modifying enzymes associated with transcriptional activity, are required for the synapsis checkpoint. In addition, a duplication with PC activity undergoes heterochromatin assembly when mes-4 activity is reduced. MES-4 function is required specifically for the X chromosome, while MES-4 and MET-1 act redundantly to monitor autosomal synapsis. We propose that MES-4 and MET-1 antagonize heterochromatin assembly at PCs of unsynapsed chromosomes by promoting a transcriptionally permissive chromatin environment that is required for meiotic checkpoint function. Moreover, we suggest that different genetic requirements to monitor the behavior of sex chromosomes and autosomes allow for the lone unsynapsed X present in male germlines to be shielded from inappropriate checkpoint activation.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003089
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003089Summary
Chromosomes that fail to synapse during meiosis become enriched for chromatin marks associated with heterochromatin assembly. This response, called meiotic silencing of unsynapsed or unpaired chromatin (MSUC), is conserved from fungi to mammals. In Caenorhabditis elegans, unsynapsed chromosomes also activate a meiotic checkpoint that monitors synapsis. The synapsis checkpoint signal is dependent on cis-acting loci called Pairing Centers (PCs). How PCs signal to activate the synapsis checkpoint is currently unknown. We show that a chromosomal duplication with PC activity is sufficient to activate the synapsis checkpoint and that it undergoes heterochromatin assembly less readily than a duplication of a non-PC region, suggesting that the chromatin state of these loci is important for checkpoint function. Consistent with this hypothesis, MES-4 and MET-1, chromatin-modifying enzymes associated with transcriptional activity, are required for the synapsis checkpoint. In addition, a duplication with PC activity undergoes heterochromatin assembly when mes-4 activity is reduced. MES-4 function is required specifically for the X chromosome, while MES-4 and MET-1 act redundantly to monitor autosomal synapsis. We propose that MES-4 and MET-1 antagonize heterochromatin assembly at PCs of unsynapsed chromosomes by promoting a transcriptionally permissive chromatin environment that is required for meiotic checkpoint function. Moreover, we suggest that different genetic requirements to monitor the behavior of sex chromosomes and autosomes allow for the lone unsynapsed X present in male germlines to be shielded from inappropriate checkpoint activation.
Introduction
Meiosis is the specialized cell division in which a diploid cell gives rise to haploid gametes, such as eggs and sperm. To halve the chromosome complement, meiosis is composed of two divisions without an intervening S phase: meiosis I, in which homologous chromosomes are segregated, and meiosis II, in which sister chromatids are segregated. To ensure that chromosomes segregate properly in meiosis and produce gametes with the correct number of chromosomes, homologous chromosomes undergo meiosis-specific events to generate a linkage, or chiasma, that enables proper biorientation on the meiotic spindle. Homologous chromosomes identify their unique partner to pair, stabilize this pairing by assembling a proteinaceous structure called the synaptonemal complex (SC) between homologs and undergo homologous recombination in the context of the SC. Defects in SC formation prevent or severely reduce homologous recombination [1] and therefore can produce gametes with an incorrect chromosome complement. Fertilization of these defective gametes can result in embryos that are inviable or have serious developmental disorders, such as Down and Klinefelter's syndrome in humans. It is estimated that 30% of miscarriages are the product of meiotic chromosome missegregation [2].
Meiotic checkpoints maintain genomic integrity by monitoring events, such as synapsis and recombination, to make sure that they occur properly, in a timely manner and in the appropriate order. If meiotic events are disrupted, these surveillance mechanisms delay cell cycle progression to enable cells to correct errors or activate apoptosis to remove defective cells [3]. During oogenesis in Caenorhabditis elegans, a checkpoint monitors synapsis independent of recombination and activates apoptosis to remove nuclei with unsynapsed chromosomes to prevent the production of aneuploid oocytes [4]. The checkpoint signal requires the presence of unsynapsed Pairing Centers (PCs) [4], cis–acting sites near one end of each chromosome that promote pairing and synapsis [5]–[7]. How PCs activate the synapsis checkpoint is currently unknown.
The presence of unpaired or unsynapsed chromosomes results in meiotic silencing of unsynapsed chromatin (MSUC), a response conserved from fungi to mammals [8]–[10]. During MSUC, unpaired or unsynapsed chromosomes become decorated with chromatin modifications associated with transcriptional silencing or heterochromatin formation [11]. In mammals, MSUC shares features with meiotic sex chromosome inactivation (MSCI), the process by which the partially synapsed sex chromosomes (the X and Y) are transcriptionally silenced during spermatogenesis [11]. Indeed, the presence of unsynapsed autosomes can deplete factors required for MSCI [12] and induce apoptosis as a result of inappropriate gene expression from the Y chromosome [13], raising the possibility that competition for these resources may act as a reporter for defects in synapsis, at least during spermatogenesis. During oogenesis, MSUC-induced transcriptional silencing of chromosomal loci essential for meiosis has been proposed to induce apoptosis [14].
In C. elegans, both MSUC and MSCI result in decoration of unsynapsed chromosomes with dimethylated lysine 9 on histone H3 (H3K9me2), a chromatin modification consistent with transcriptional silencing [8]. However, MSCI and MSUC occur by distinct mechanisms in this organism [15]. During spermatogenesis in XO males, X chromosomes remain unsynapsed and undergo MSCI [8], which prevents meiotic checkpoint activation [16]. MSCI is dependent on a conserved SET domain histone methyltransferase, MET-2, as loss of this protein reduces H3K9me2 accumulation on the single X [17], increases the transcriptional activity of the single X and activates a DNA damage checkpoint in response to defects in recombination [15]. Loss of MET-2 during oogenesis in XX hermaphrodites also affects the chromatin state of unsynapsed chromosomes, in that they are no longer enriched with H3K9me2 [17], but there is no corresponding increase in transcriptional activity or checkpoint activation [15], indicating that MSUC and MSCI are not equivalent processes. Furthermore, MSUC appears to be the consequence of several pathways [17]–[19], one of which is not involved in MSCI [15].
In C. elegans, mes-4 and met-1 encode well-characterized histone methyltransferases associated with active transcription, specifically the catalysis of H3K36me [20]. MES-4 is a critical regulator of germline development and immortality [21] and primarily binds transcriptionally active autosomes [22]; in mes-2, mes-3 and mes-6 mutants, MES-4 mislocalizes along the X chromosome [22], possibly as a result of the inappropriate upregulation of X-linked genes in these mutants [20]. MET-1 is responsible for RNA Polymerase II dependent H3K36me3 deposition [20], most likely by the mechanism described for its ortholog, budding yeast Set2. Yeast Set2 associates with Pol II during its elongation phase and catalyzes H3K36me in the body of genes to recruit transcriptional repressors that prevent cryptic transcription initiation [23]–[25]. MET-1 also contributes to C. elegans vulval development [26].
Given the requirement for cis-acting loci in synapsis checkpoint activation, we wondered if the chromatin state of PCs was important for checkpoint function. Specifically, if unsynapsed chromosomes appear to undergo heterochromatin assembly, do PCs behave differently to contribute to checkpoint activation? To answer this question, we investigated the importance of chromatin modifying enzymes during synapsis checkpoint activation. We show that unsynapsed chromosomes are decorated with H3K9me2 whether they activate the DNA damage checkpoint or the synapsis checkpoint, indicating that heterochromatin assembly is a general response to asynapsis. Indeed, these checkpoints appear to be dispensable for H3K9me2 enrichment on unsynapsed chromosomes. However, a chromosomal duplication that harbors PC activity and is sufficient for synapsis checkpoint activation undergoes heterochromatin assembly less readily than a chromosomal duplication of a non-PC region. We also report that MES-4 and MET-1 are required for the synapsis checkpoint. Consistent with these enzymes acting at PCs, a duplication with PC activity becomes enriched with H3K9me2 when mes-4 activity is reduced. Taken together, our data suggest that these chromatin-modifying enzymes antagonize heterochromatin assembly at PCs to promote checkpoint activation. Therefore, chromatin state, and potentially transcriptional activity, at these cis-acting sites correlate with checkpoint signaling. Moreover, C. elegans sex chromosomes exhibit different genetic requirements than autosomes to activate the synapsis checkpoint: MES-4 is specifically required to monitor synapsis of X chromosomes while MES-4 and MET-1 are redundant for synapsis checkpoint activation when autosomes are unsynapsed. These results may explain why the single X chromosome in males does not activate the synapsis checkpoint despite being unsynapsed.
Results
Heterochromatin assembly is a general response to unsynapsed chromosomes
We wanted to determine whether chromosomes that activate the synapsis checkpoint also become enriched for H3K9me2. Most observations of heterochromatin assembly on unpaired or unsynapsed chromosomes have been in situations in which chromosomal duplications are present in meiotic nuclei or unpaired chromosomes activate a meiotic checkpoint that monitors recombination defects (also known as the DNA damage checkpoint) [8], [15]–[19]. We tested whether unsynapsed chromosomes became enriched for H3K9me2 in a genotype in which only the synapsis checkpoint is activated.
We have shown that a single pair of unsynapsed chromosomes can robustly activate either the synapsis checkpoint or the DNA damage checkpoint, depending on whether the unsynapsed chromosomes include active PCs [4]. meDf2 is a deficiency that removes up to 2 Mb of the left end of the X chromosome and the X chromosome Pairing Center (PC) [27]. Animals homozygous for meDf2 exhibit unsynapsed X chromosomes in almost all meiotic nuclei [5]. Unsynapsed X chromosomes in meDf2 homozygotes do not have an active PC and therefore activate the DNA damage checkpoint and not the synapsis checkpoint [4]. Animals heterozygous for meDf2 exhibit unsynapsed X chromosomes in 60% of meiotic nuclei [5]. Since the synapsis checkpoint requires an active Pairing Center (PC), meiotic nuclei with unsynapsed chromosomes in meDf2 heterozygotes activate the synapsis checkpoint [4]. However, for reasons that are not known, the DNA damage checkpoint is not activated in meDf2 heterozygotes.
We directly assessed synapsis by performing indirect immunofluorescence against the synaptonemal complex components, HTP-3 [5] and SYP-1 [28], in meiotic nuclei. HTP-3 is an axial element component that loads onto chromosomes axes prior to and independent of synapsis [5]. SYP-1 is a central element component that polymerizes between homologous chromosomes as they synapse [28]. In wildtype mid-pachytene nuclei, we observed colocalization of HTP-3 and SYP-1 (Figure 1A, 1B and 1D), consistent with complete synapsis, and chromosomal regions where H3K9me2 is depleted or enriched (Figure 1C and 1D, carets indicate regions of enrichment), as previously reported [17]. In meiotic nuclei in meDf2 homozygotes, we observed stretches of HTP-3 devoid of SYP-1 (arrows in Figure 1E and 1H): these are unsynapsed X chromosomes [5] and they are the primary source of H3K9me2 signal in these nuclei (Figure 1G). In meDf2 heterozygotes, some nuclei completed synapsis (nucleus on left in Figure 1I–1L) while other nuclei exhibited unsynapsed X chromosomes (arrows in Figure 1I, 1K and 1L). Nuclei with synapsed X chromosomes exhibited wildtype localization of H3K9me2, while nuclei with unsynapsed X chromosomes had H3K9me2 limited to regions of asynapsis (arrows in Figure 1K and 1L). Therefore, enrichment of H3K9me2 on unsynapsed chromosomes occurs whether the DNA damage or synapsis checkpoint is activated. Furthermore, heterochromatin assembly on unsynapsed chromosomes is an event each meiotic nucleus undertakes independently; neighboring nuclei can exhibit different H3K9me2 patterns depending on their state of synapsis (Figure 1I–1L).
Fig. 1. H3K9me2 is enriched on unsynapsed chromosomes when either the DNA damage checkpoint or the synapsis checkpoint is activated. 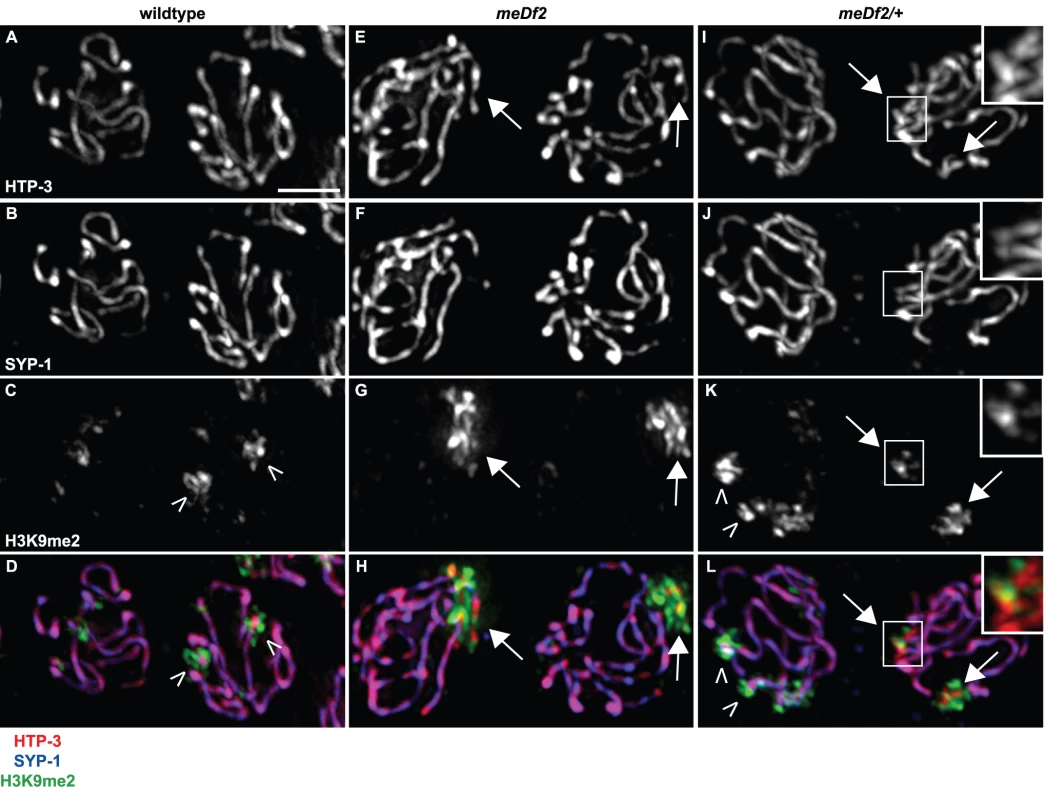
Indirect immunofluorescence was performed on meiotic nuclei in wildtype, meDf2 homozygotes and meDf2 heterozygotes using antibodies against the axial element component HTP-3, the central element component SYP-1 and H3K9me2. Wildtype meiotic nuclei exhibit colocalization of HTP-3 and SYP-1 (A, B and D), and have areas of H3K9me2 enrichment (carets) and depletion (C and D). Meiotic nuclei in both meDf2 homozygotes and meDf2 heterozygotes have chromosomes with HTP-3 but not SYP-1 (arrows in E, F, H, I, J and L, magnified insets in I, J and L). These are unsynapsed X chromosomes and are highly decorated with H3K9me2 (G, H, K and L, magnified insets in K and L). Other regions of H3K9me2 enrichment are also observed in nuclei with synapsed chromosomes in meDf2 heterozygotes (carets in K and L). Scale bar represents 4 microns. A duplication that contains PC activity and activates the synapsis checkpoint often fails to undergo heterochromatin assembly
We wanted to more closely examine the relationship between H3K9me2 and PCs. We analyzed the localization of H3K9me2 and the X chromosome PC protein HIM-8 [7] in meiotic nuclei with unsynapsed X chromosomes in meDf2 heterozygotes. In nuclei with unsynapsed X chromosomes, as identified by the enrichment of H3K9me2 (Figure 2A) and absence of SYP-1 (Figure 2B), we sometimes observed a reduction in H3K9me2 in the vicinity of HIM-8 staining (caret in Figure 2A) but we also observed robust H3K9me2 adjacent to HIM-8 (arrow in Figure 2A and Figure S1). We quantified the frequency of H3K9me2 enrichment at the PC end and non-PC end of unsynapsed chromosomes in meDf2 heterozygotes (Figure 2C). Of meiotic nuclei with unsynapsed X chromosomes (n = 320), 43% did not exhibit enriched H3K9me2 at the PC end (Figure 2C). However, this was only slightly higher than the percentage of unsynapsed X chromosomes that did not exhibit H3K9me2 enrichment at a non-PC region (35%) (Figure 2C), making it difficult for us to draw any firm conclusions about the chromatin state of unsynapsed PCs in synapsis checkpoint activating strains.
Fig. 2. A chromosomal duplication with Pairing Center activity often does not undergo heterochromatin assembly. 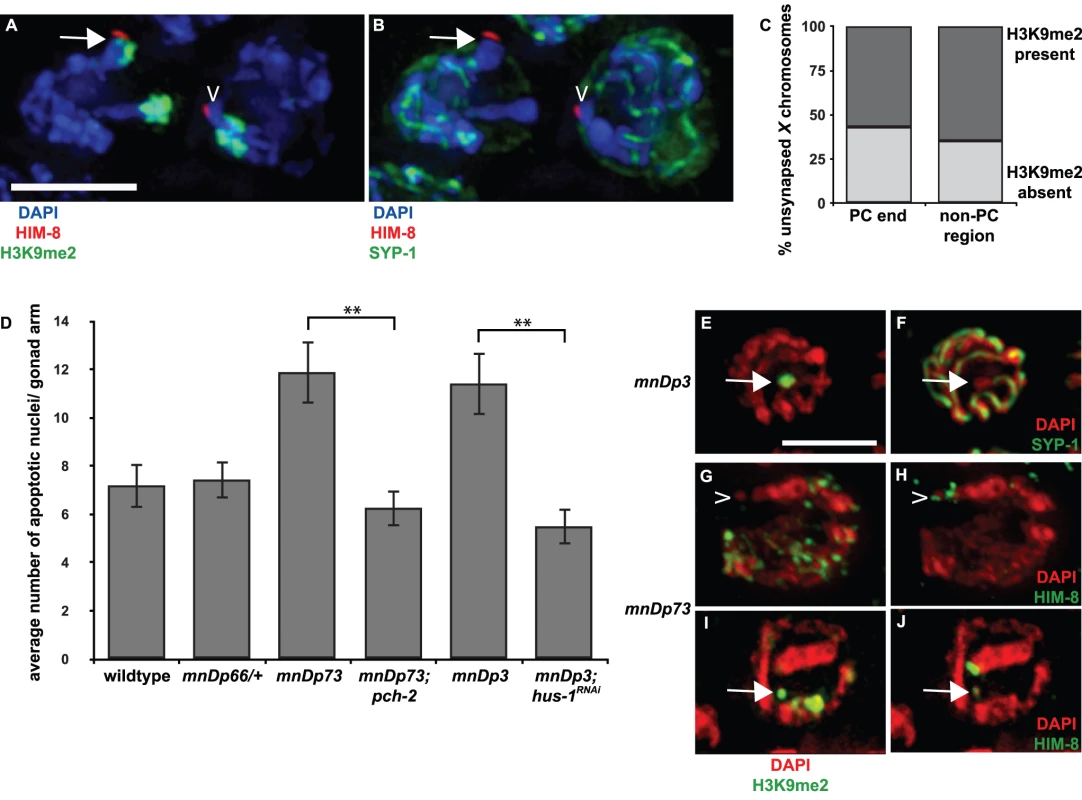
A and B. In meDf2/+ hermaphrodites, it is difficult to determine if PCs of unsynapsed X chromosomes become enriched with H3K9me2. Indirect immunofluorescence was performed on meiotic nuclei in meDf2 heterozygotes using antibodies against the X chromosome PC protein HIM-8, the central element component SYP-1 and H3K9me2. The nucleus on the left contains an unsynapsed X chromosome, as identified by HIM-8, an enrichment of H3K9me2 (A) and the absence of SYP-1 (B); the PC appears enriched with H3K9me2 (arrow in A). The nucleus on the right also contains an unsynapsed X chromosome, as indicated by the absence of SYP-1 in B, and the PC of this chromosome does not exhibit robust H3k9me2 (caret in A). C. Quantification of the percentage of unsynapsed X chromosomes in meDf2 heterozygotes that were enriched for H3K9me2 at their PC and non-PC regions. D. A chromosomal duplication with Pairing Center activity (mnDp73) activates the synapsis checkpoint. A duplication that does not have Pairing Center activity (mnDp66/+) fails to activate the synapsis checkpoint and a duplication of the non-PC portion of the X chromosome (mnDp3) activates the DNA damage checkpoint. A ** indicates a p value of <0.0001. Error bars indicate 2XSEM. E–J. Indirect immunofluorescence was performed on meiotic nuclei from hermaphrodites carrying mnDp3 (non-PC end of the X chromosome) and mnDp73 (PC end of the X chromosome) with antibodies against H3K9me2, SYP-1 or HIM-8. mnDp3 is unsynapsed (arrow in F) and highly decorated with H3K9me2 (arrow in E). This was observed in 100% of meiotic nuclei (23 of 23) in which mnDp3 could unambiguously be identified as unsynapsed. mnDp73 is identified as a free DAPI staining body that recruits HIM-8 (caret in H and arrow in J) but that is often not enriched with H3K9me2 (caret in G). This was observed in 13 out of 17 in which mnDp73 could unambiguously be identified by HIM-8 recruitment. In the remaining four nuclei in which mnDp73 was identified by HIM-8 binding (arrow in J), the duplication was enriched for H3K9me2 (arrow in I) but was not the primary signal of H3K9me2 in the nucleus. Scale bar represents 4 microns. We decided to take advantage of chromosomal duplications, such as mnDp73, that have demonstrated PC activity: they are able to recombine with full-length X chromosomes and disrupt their proper segregation in hermaphrodites [29]. To determine if mnDp73, which is a free duplication, could recapitulate all functions of PCs, we addressed whether it activated the synapsis checkpoint. When we assayed germline apoptosis in hermaphrodites carrying mnDp73, we observed an increase in apoptosis above the physiological levels of apoptosis observed in wildtype. This increase in apoptosis is dependent on pch-2 (Figure 2D and Table 1), a component of the synapsis checkpoint [4]. This is in contrast to a duplication of the PC end of the X chromosome that lacks PC activity (mnDp66, which is attached to Chromosome I) [29]. Despite being unsynapsed (data not shown), animals that are heterozygous for mnDp66 did not exhibit elevated apoptosis (Figure 2D and Table 1), indicating that synapsis checkpoint activation correlates with other measures of PC activity. Animals carrying a free duplication of the non-PC end of the X chromosome, mnDp3 [30], also exhibited elevated germline apoptosis but this elevation was dependent on the DNA damage checkpoint, as knock down of the DNA damage checkpoint component hus-1 [31] by RNA interference in animals carrying mnDp3 reduced apoptosis to wildtype levels (Figure 2D and Table 1). We have previously shown that mutation or inactivation of pch-2 or hus-1 by RNAi in an otherwise wildtype genetic background has no effect on physiological apoptosis [4].
Tab. 1. Duplications used in this study. 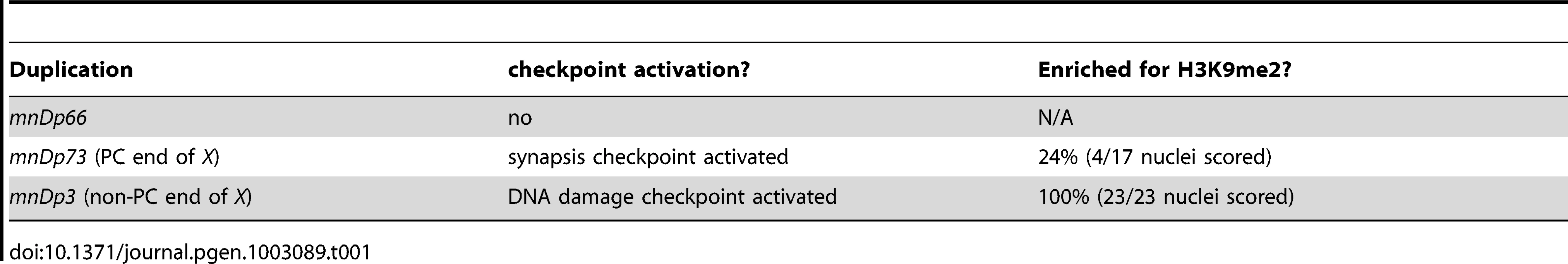
Having verified that the presence of mnDp73 was sufficient for synapsis checkpoint activation, we assessed if it, like other duplications characterized, underwent heterochromatin assembly. We performed indirect immunofluorescence on animals that contained either mnDp73 (Figure 2G and 2I) or mnDp3 (Figure 2E) [30] with an antibody against H3K9me2. In germlines of hermaphrodites carrying mnDp3, we identified free DAPI staining bodies in mid-pachytene nuclei that did not load SYP-1 (Figure 2F) and observed what proportion of these stained with H3K9me2. Of 23 nuclei in which we could unambiguously identify unsynapsed DAPI staining bodies, all 23 exhibited robust H3K9me2; the unsynapsed duplication was the primary source of H3K9me2 signal in these nuclei (Figure 2E and Table 1). We noticed that unambiguously unsynapsed DAPI staining bodies in the meiotic nuclei of mnDp3 bearing animals were rare. Often we could not observe an unsynapsed DAPI-staining body in most meiotic nuclei consisting of a single H3K9me2 body characteristic of nuclei containing a chromosomal duplication (arrows in Figure S2). We determined the percentage of nuclei that included a single H3K9me2 body that was also unsynapsed and found that only 38% (23 of 60) of H3K9me2 enriched duplications were unambiguously unsynapsed, suggesting that heterochromatin assembly is a response to the unpaired status of chromosomes as opposed to the unsynapsed status of chromosomes.
Because of the substantially smaller size of mnDp73 (see Figure 2G–2J, [29]), we identified the duplication as bound by HIM-8 (caret in Figure 2H and arrow in 2J) and observed what proportion of these were enriched with H3K9me2; this accounts for the smaller sample size we report for this duplication. In contrast to mnDp3, mnDp73 appeared as a free DAPI staining body without H3K9me2 in a majority (13 out of 17) of meiotic nuclei in which the duplication could be identified (caret in Figure 2G and Figure 3E). Four of the seventeen meiotic nuclei in which mnDp73 could be identified were enriched for H3K9me2 (Figure 2I, Figure 3E and Table 1) but this was not the dominant H3K9me2 signal in these meiotic nuclei, as observed when whole chromosomes are unsynapsed (Figure 1G and 1H) or a larger duplication is present (Figure 2E). Thus, our data support a model in which a duplication that contains PC activity and activates the synapsis checkpoint often fails to undergo heterochromatin assembly.
Fig. 3. mes-4 is required for the synapsis checkpoint and prevents heterochromatin assembly on a chromosomal duplication with PC activity. 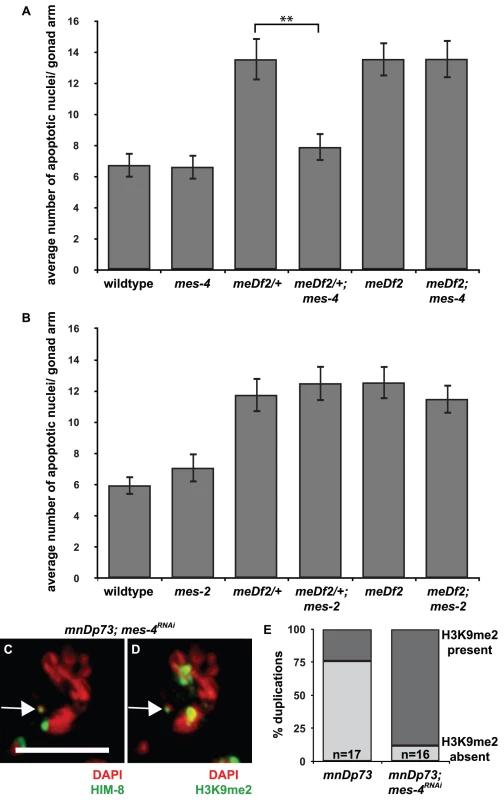
A. Mutation of mes-4 reduces apoptosis in meDf2/+ but not in meDf2 homozygotes. A ** indicates a p value of <0.0001. B. Mutation of mes-2 does not reduce apoptosis in meDf2/+ or meDf2 homozygotes. Error bars indicate 2XSEM. C and D. Indirect immunofluorescence was performed on meiotic nuclei in hermaphrodites carrying mnDp73 in which mes-4 was knocked down by RNAi with antibodies against H3K9me2 and HIM-8. mnDp73 is identified as a free duplication bound by HIM-8 (arrow in C). In 14 out of 16 in which the duplication could be identified, mnDp73 is enriched with H3K9me2 (arrow in D). Scale bar represents 4 microns. E. Quantification of the percentage of duplications that were enriched with H3K9me2 in mnDp73 bearing hermaphrodites and mnDp73 bearing hermaphrodites in which mes-4 was inactivated by RNAi. MES-4 is required for the synapsis checkpoint when X chromosomes are unsynapsed and prevents heterochromatin assembly on a duplication with PC activity
To further investigate the potential link between the exclusion of heterochromatin from PCs that we observed and synapsis checkpoint activation, we tested the role of mes-4 in the synapsis checkpoint. MES-4 is a histone methyltransferase that is required for germline viability and development [32] and methylates lysine 36 of histone H3 (H3K36me), a methyl mark associated with active chromatin [20], [33]. In embryos, it associates specifically with transcriptionally active autosomes [22] and the leftmost tip of the X, where the PC is located [33]. We introduced a mes-4 mutation into meDf2 heterozygote hermaphrodites (meDf2/+). As a result of synapsis checkpoint activation in meDf2 heterozygotes [4], germline apoptosis is elevated (Figure 3A). When we assayed apoptosis in mes-4; meDf2/+ double mutants, we observed a reduction in apoptosis compared to meDf2/+ single mutants (p value <0.0001) (Figure 3A), indicating that mes-4 is required for the synapsis checkpoint in meDf2 heterozygotes. We also assayed whether mes-4 was required for the DNA damage checkpoint by investigating levels of germline apoptosis in mes-4; meDf2 animals. meDf2 hermaphrodites activate the DNA damage checkpoint, which also produces increased levels of germline apoptosis [4]. We did not observe a decrease in apoptosis in mes-4; meDf2 double mutants, when compared to meDf2 single mutants (Figure 3A), indicating that mes-4 is not required for the DNA damage checkpoint.
In addition to mes-4, mes-2, mes-3 and mes-6 are required maternally for normal development of the germline [32] and evidence indicates that these four genes collaborate to promote germline development [22], [34]. MES-2 is a SET domain containing protein orthologous to the Drosophila Polycomb group protein Enhancer of Zeste [35]. MES-2, in a complex with MES-3 and MES-6, catalyzes repressive H3K27me to maintain X chromosome silencing in the germline [34], [36], [37]. We tested mes-2 for a role in the synapsis checkpoint by introducing a mes-2 mutation into meDf2 heterozygotes. We did not observe a reduction in apoptosis in the double mutant when compared to the meDf2 heterozygote (p value = 0.35) (Figure 3B). We also did not observe a decrease in apoptosis in meDf2 homozygotes when mes-2 function was absent, indicating that mes-2 is also not required for the DNA damage checkpoint (Figure 3B). Similar results were obtained with mes-3 mutants (data not shown). Therefore, the role of MES-4 in the synapsis checkpoint is independent of the MES-2, MES-3 and MES-6 complex.
mes-4 is not required for MSCI in male germlines [22]. We determined whether it was required for heterochromatin assembly on unsynapsed chromosomes in hermaphrodite germlines and observed no reduction in H3K9me2 on unsynapsed chromosomes in either meDf2 homozygotes or meDf2 heterozygotes in which mes-4 had been mutated (data not shown), indicating that deposition of H3K9me2 on unsynapsed chromosomes is not dependent on activating the synapsis checkpoint. We also investigated whether knock down of mes-4 by RNAi affected heterochromatin assembly on mnDp73; efficient knockdown of mes-4 in hermaphrodite germlines was verified by performing immunofluorescence against MES-4 (data not shown). We once again identified mnDp73 in mid-pachytene meiotic nuclei as bound by HIM-8 (Figure 3C) and determined its H3K9me2 state. We found that 14 out of 16 meiotic nuclei in which the duplication could be unambiguously identified as being bound by HIM-8 were also enriched for H3K9me2 (Figure 3D and 3E). Thus in the absence of MES-4, the synapsis checkpoint is inactivated and a duplication with PC activity undergoes heterochromatin assembly, suggesting a link between these events.
It has recently been shown that plk-2 regulates both a delay in meiotic progression in response to asynapsis and the synapsis checkpoint [38]. We observed that mes-4 abrogated the synapsis checkpoint in meDf2/+ hermaphrodites (Figure 3A) without affecting the accompanying delay in meiotic progression or any molecular markers that correlate with this delay, i.e. phosphorylation of SUN-1 at residues serine 8 (Figure S3) and serine 12 (Figure S4) [39], leading us to speculate that MES-4 either works downstream or independent of PLK-2 in the synapsis checkpoint pathway [38].
MES-4 and MET-1 are redundant for the synapsis checkpoint when all chromosomes are unsynapsed
We monitored synapsis checkpoint activation in mes-4 syp-1 double mutants. In syp-1 mutant animals, the SC fails to assemble between all six pairs of homologs [28] and both the DNA damage and synapsis checkpoints are activated, resulting in very high levels of apoptosis (Figure 4A and [4]). When we assayed apoptosis in mes-4 syp-1 double mutants, we did not see a decrease in the levels of apoptosis (Figure 4A), indicating that when all six pairs of homologous chromosomes are unsynapsed, mes-4 is not required for the checkpoint.
Fig. 4. mes-4 and met-1 are redundant for synapsis checkpoint activation when all chromosomes are unsynapsed. 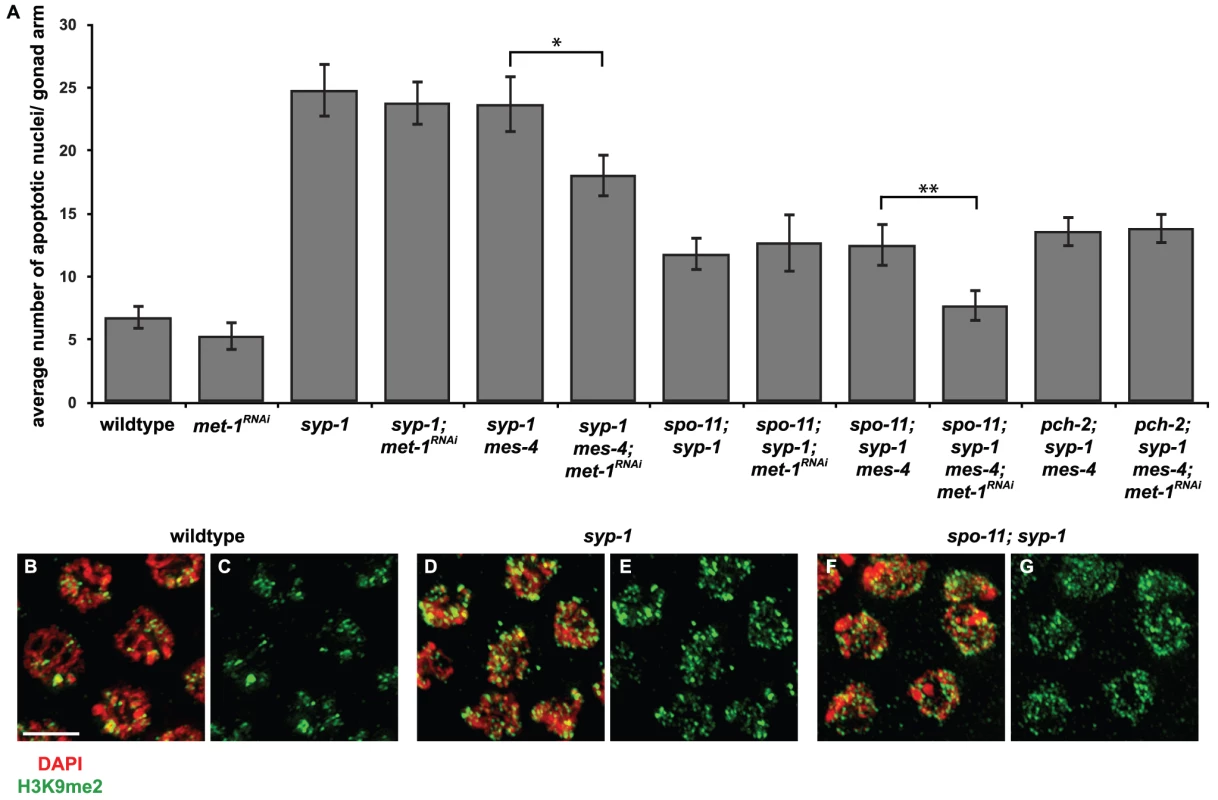
A. Inactivation of both mes-4 (by mutation) and met-1 (by RNA interference) is required to reduce apoptosis in syp-1 and spo-11; syp-1 mutants but does not affect apoptosis in pch-2; syp-1 mutants. A * indicates a p value of <0.05 and a ** indicates a p value of <0.0001. Error bars indicate 2XSEM. B–G. Heterochromatin assembly occurs on unsynapsed chromosomes in syp-1 and spo-11; syp-1 mutants. Indirect immunofluorescence was performed on meiotic nuclei in wildtype, syp-1 and spo-11; syp-1 double mutants with an antibody against H3K9me2. Wildtype nuclei exhibit regions of enrichment or depletion while both syp-1 and spo-11; syp-1 mutants exhibit dispersed H3K9me2. Scale bar represents 4 microns. Most reported examples of heterochromatin assembly on unsynapsed chromosomes in C. elegans involve situations in which there is partial asynapsis [8], [15]–[19]. If complete asynapsis does not produce robust heterochromatin assembly on all unsynapsed chromosomes, potentially leaving some chromosomal loci, including PCs, euchromatic by default, this might explain why mes-4 is dispensable for checkpoint activation in syp-1 mutants. To address this concern, we visualized H3K9me2 staining in syp-1 mutants. In contrast to wildtype animals, in which meiotic chromosomes exhibited areas of enrichment or depletion of H3K9me2 (Figure 1C and 1D, Figure 4B and 4C), syp-1 mutants exhibited dispersed H3K9me2 that colocalized with all meiotic chromosomes (Figure 4D and 4E), indicating that when all chromosomes are unsynapsed, they become enriched for H3K9me2. This has been reported in syp-2 mutants [40], which are also defective in SC formation [41]. Since unsynapsed chromosomes undergo heterochromatin assembly in syp-1 mutants (Figure 4D and 4E), we cannot attribute our finding that mes-4 function is dispensable for synapsis checkpoint activation in this mutant background to differences in H3K9me2 enrichment when all chromosomes are unsynapsed.
We reasoned that mes-4's role in the synapsis checkpoint may be redundant with another methyltransferase when all six pairs of chromosomes are unsynapsed. In both germlines and embryos, both mes-4 and met-1 are required to maintain wildtype levels of H3K36me3 (T. Takasaki and S. Strome, personal communication and [20]). To test whether met-1 is required for the synapsis checkpoint in syp-1 mutants, we knocked down met-1 in mes-4 syp-1 double mutants by RNA interference (met-1; mes-4 double mutants exhibit defects in germline organization making analysis of apoptosis problematic [data not shown]). mes-4 mutants can be resistant to RNAi but this resistance is dosage dependent [42]. RNAi (Figure 4A) or mutation (data not shown) of met-1 in wildtype and syp-1 single mutants did not reduce germline apoptosis in either genotype, indicating that loss of met-1 alone behaves like loss of mes-4 alone and does not affect physiological apoptosis or abrogate the synapsis checkpoint (Figure 4A). When we assayed germline apoptosis in mes-4 syp-1 mutant hermaphrodites in which met-1 had been knocked down by RNAi, we observed a decrease in apoptosis (Figure 4A). When compared to apoptosis in mes-4 syp-1 double mutants, this decrease was statistically significant (p value = 0.0002) and similar to the levels of apoptosis observed when other components of the synapsis checkpoint, such as pch-2 [4], are knocked down by RNAi (data not shown).
spo-11; syp-1 double mutants only activate the synapsis checkpoint since the absence of SPO-11 induced double strand breaks in this mutant background prevents DNA damage checkpoint activation [4]. The absence of double strand breaks has no effect on heterochromatin assembly on unsynapsed chromosomes in spo-11; syp-1 mutants (Figure 4F and 4G), indicating that H3K9me2 deposition occurs on unsynapsed chromosomes when the DNA damage checkpoint is not activated. When we assayed germline apoptosis in both spo-11; mes-4 syp-1 triple mutants and spo-11; syp-1 double mutants in which met-1 had been knocked down by RNAi, we observed the same levels of apoptosis as observed in spo-11; syp-1 double mutants (Figure 4A). However, when we knocked down met-1 by RNAi in spo-11; mes-4 syp-1 triple mutants, we saw a decrease in apoptosis to wildtype levels when compared to spo-11; mes-4 syp-1 (p<0.0001) (Figure 4), indicating a loss of the synapsis checkpoint. To verify that the decrease in apoptosis we observed in spo-11; mes-4 syp-1 triple mutants in which met-1 had been knocked down by RNAi was due to abrogation of the synapsis checkpoint, we also assayed apoptosis in pch-2; mes-4 syp-1 triple mutants in which met-1 had been knocked down by RNAi and did not observe any additional decrease in the level of germline apoptosis (Figure 4A). These experiments indicate that mes-4 and met-1 act redundantly in the synapsis checkpoint when all chromosomes are unsynapsed.
MET-1 is not required for the synapsis checkpoint when only X chromosomes are unsynapsed
We hypothesized that the requirement for mes-4 and met-1 activity for synapsis checkpoint activation in syp-1 mutants might be explained by a model in which MES-4's role in the checkpoint is specific to X chromosomes and MET-1's role is specific for autosomes. To test this model, we performed two experiments. First, we monitored apoptosis in meDf2 heterozygotes in which met-1 had been mutated (met-1; meDf2/+). We found that apoptosis remained as high in met-1; meDf2/+ double mutants as in meDf2/+ single mutants (Figure 5A) (p value = 0.125). Thus, met-1 is not required for the synapsis checkpoint when only X chromosomes are unsynapsed. We also determined that met-1 is not required for the DNA damage checkpoint since apoptosis in both met-1; meDf2 double mutants and meDf2 single mutants was similar (Figure 5A).
Fig. 5. met-1 is not required for the synapsis checkpoint when only X chromosomes are unsynapsed. 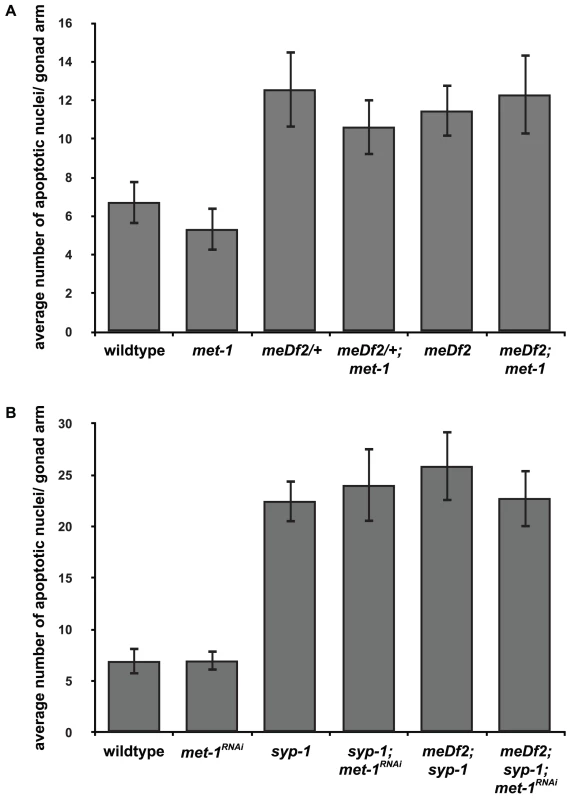
A. Mutation of met-1 does not reduce apoptosis in meDf2/+ or meDf2 homozygotes. B. Inactivation of met-1 by RNAi in meDf2; syp-1 double mutants does not reduce apoptosis. Error bars indicate 2XSEM. Having demonstrated that met-1 is not required to monitor synapsis of X chromosomes, we also tested whether met-1 is uniquely required to monitor synapsis of autosomes. We previously showed that autosomes activate the synapsis checkpoint by introducing the meDf2 deficiency into syp-1 mutants [4]. In this double mutant background, only autosomes are capable of activating the synapsis checkpoint, since the absence of an active PC prevents X chromosomes from activating the synapsis checkpoint. We knocked down met-1 gene function by RNAi in syp-1; meDf2 double mutants and observed no effect on the levels of apoptosis (Figure 5B), similar to our results in syp-1 single mutants (Figure 4A), indicating that met-1 activity is not required to monitor synapsis of autosomes, likely because of the presence of mes-4 (Figure 4A).
MES-4's role in the synapsis checkpoint does not depend on heterochromatin assembly on unsynapsed X chromosomes
The requirement for mes-4 function in the synapsis checkpoint combined with our observation that duplications with PC activity undergo heterochromatin assembly when mes-4 is inactivated raises the possibility that MES-4's primary role in the checkpoint is to antagonize H3K9me2 enrichment at X chromosome PCs when they are unsynapsed. A similar role has recently been attributed to MES-4 in the context of MES-2/MES-3/MES-6 mediated H3K27me [34]. To test this model, we monitored synapsis checkpoint activation in hermaphrodites in which both mes-4 and met-2 gene functions were knocked down. If mes-4's role in the checkpoint is dependent on met-2 function, this result would support a model in which the primary role of MES-4 (and by extension MET-1) in the synapsis checkpoint is to prevent heterochromatin assembly at PCs. If mes-4 is required for the synapsis checkpoint when met-2 function and heterochromatin assembly on unsynapsed chromosomes is reduced, this might suggest that MES-4 (and by extension MET-1) is performing some other function at PCs and this indirectly prevents H3K9me2 enrichment at PCs.
We reduced H3K9me2 by RNAi of met-2 in meDf2/+; mes-4 mutants [17]. We verified that met-2 had been sufficiently knocked down by monitoring the loss of H3K9me2 by immunofluorescence (data not shown). When met-2 is knocked down by RNAi in wildtype and meDf2/+ hermaphrodites, we did not observe a decrease in apoptosis, indicating that met-2 is not required for physiological apoptosis or activation of the synapsis checkpoint (Figure 6). Similar results were obtained when met-2 was inactivated by RNAi in meDf2 homozygotes (data not shown), consistent with other reports that met-2 is not required for the DNA damage checkpoint [15]. When met-2 was knocked down by RNAi in mes-4; meDf2/+ double mutants, the level of apoptosis was similar to what was observed in mes-4; meDf2/+ double mutants (p value = 0.20) and significantly lower than what is observed in meDf2/+ mutants (p value = 0.0005) and meDf2/+ mutants in which met-2 had been inactivated by RNAi (p value = 0.005) (Figure 6). These data argue that MES-4's role in the synapsis checkpoint does not necessarily depend on heterochromatin assembly on unsynapsed chromosomes. In the absence of met-2 and detectable H3K9me2 on unsynapsed chromosomes, the synapsis checkpoint still requires MES-4 activity, presumably at PCs.
Fig. 6. mes-4's role in the synapsis checkpoint is independent of met-2 mediated heterochromatin assembly on unsynapsed X chromosomes. 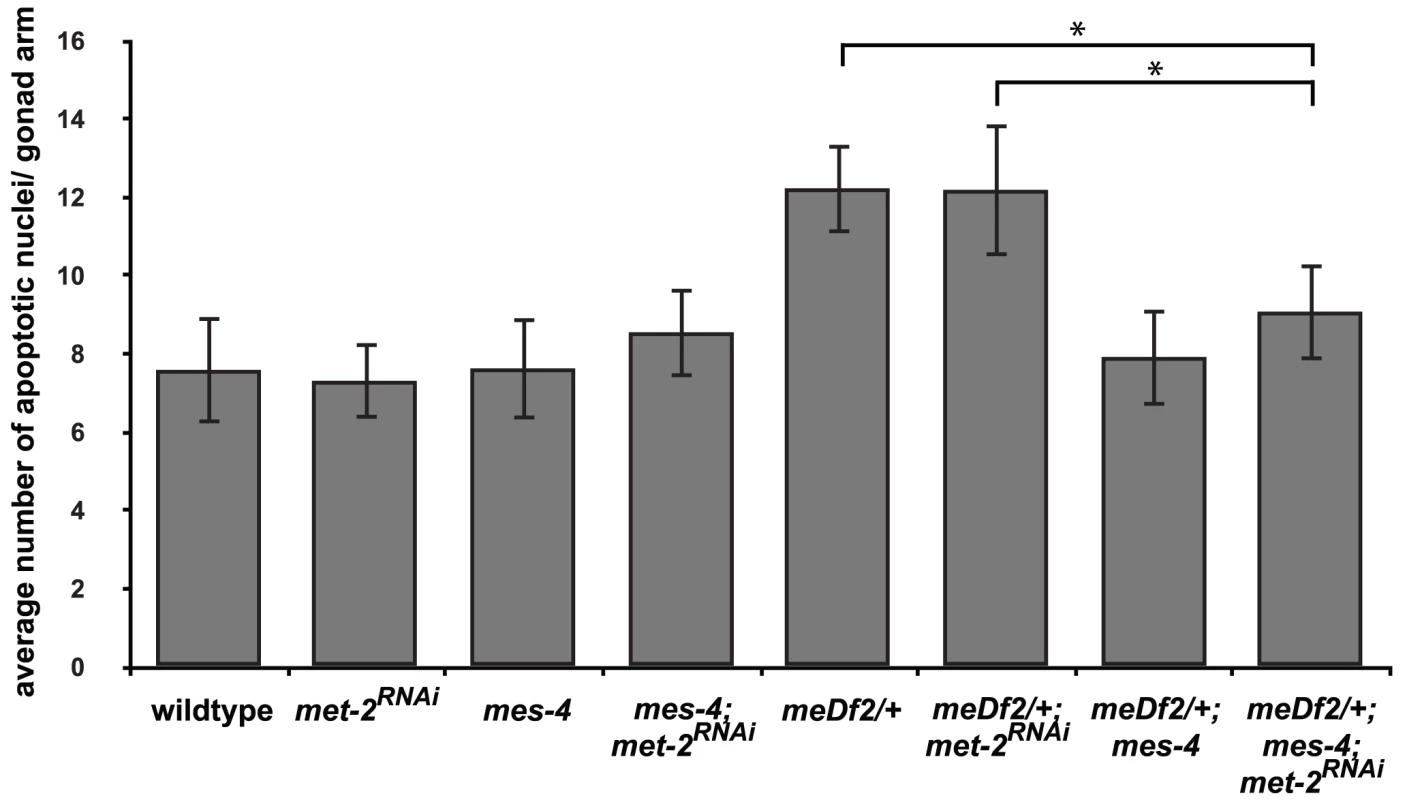
meDf2/+; mes-4 double mutants have nearly wild-type levels of apoptosis when met-2 is inactivated by RNA interference. A * indicates a p value of <0.05. Error bars indicate 2XSEM. Heterochromatin assembly in the hermaphrodite germline appears to be the consequence of several pathways [17]–[19]. For example, sin-3 activity has been shown to be required for heterochromatin assembly in the hermaphrodite germline but not for MSCI in the male germline [15]. sin-3; met-2 double mutants are sterile [15], preventing us from using the double mutant to downregulate both pathways. We tested whether mes-4 activity was required for the synapsis checkpoint when heterochromatin assembly on unsynapsed chromosomes was compromised by a reduction in sin-3 gene function but observed that mes-4 mutant hermaphrodites exposed to sin-3 RNAi were sterile a generation earlier than mes-4 single mutants (data not shown), precluding our ability to perform this experiment as well. This genetic interaction has also been observed when other chromatin modifiers, such as set-2, are depleted in mes-4 hermaphrodites [43]. In light of these results, we cannot completely rule out the possibility that the requirement for mes-4 in synapsis checkpoint activation in the absence of met-2 reveals that other pathways that regulate heterochromatin assembly on unsynapsed chromosomes are still active. However, the absence of detectable H3K9me2 in mes-4; meDf2/+ animals in which met-2 had been inactivated by RNAi leads us to favor the model that mes-4's role in the checkpoint does not necessarily depend on heterochromatin assembly on unsynapsed chromosomes.
Is H3K36me3 the relevant chromatin modification for synapsis checkpoint activation?
Mutation of mes-4 eliminates detectable H3K36me2 [33] and reduces H3K36me3 in the germline; mutation of both mes-4 and met-1 is required to lose detectable H3K36me3 in the germline (T. Takasaki and S. Strome, personal communication). Since mes-4 and met-1 are redundant for the synapsis checkpoint when autosomes are unsynapsed, our data suggest that H3K36me, in particular H3K36me3, may be an important histone modification for signaling events at the PCs of unsynapsed chromosomes. To test this possibility, we performed indirect immunofluorescence against H3K36me3 in wildtype and meDf2 heterozygotes (Figure 7A–7L). We observed visible enrichment of H3K36me3 at PC ends of X chromosomes in mid-pachytene meiotic nuclei in wildtype hermaphrodites, even as the rest of the chromosome appears depleted of this methyl mark (Figure 7D–7F), as has been reported [44]. When we quantified the percentage of mid-pachytene nuclei that exhibited H3K36me3 at the left end of the X chromosome, we determined that 55% of X chromosomes exhibited this chromatin mark in wildtype meiotic nuclei (Figure 7Q). We performed the same experiment in meDf2 heterozygotes and observed that a very similar percentage of X chromosomes exhibited H3K36me3 at the left end of X chromosomes (Figure 7Q), whether synapsed (53%) or unsynapsed (53%) (Figure 7J–7L). The slight difference in the size of this mark presented in the wildtype and meDf2 heterozygote images is not typical (data not shown). Furthermore, despite its localization to the left, or PC, end of X chromosomes [44], H3K36me3 did not colocalize with HIM-8 (Figure 7E and 7K) and is not present on duplications that contain PC activity (data not shown). We also found that enrichment of H3K36me3 at the left end of X chromosomes [44] is not dependent on HIM-8 function (data not shown), which is required for the ability of X chromosome PCs to activate the synapsis checkpoint [4].
Fig. 7. Is H3K36me3 the relevant chromatin modification for synapsis checkpoint activation? 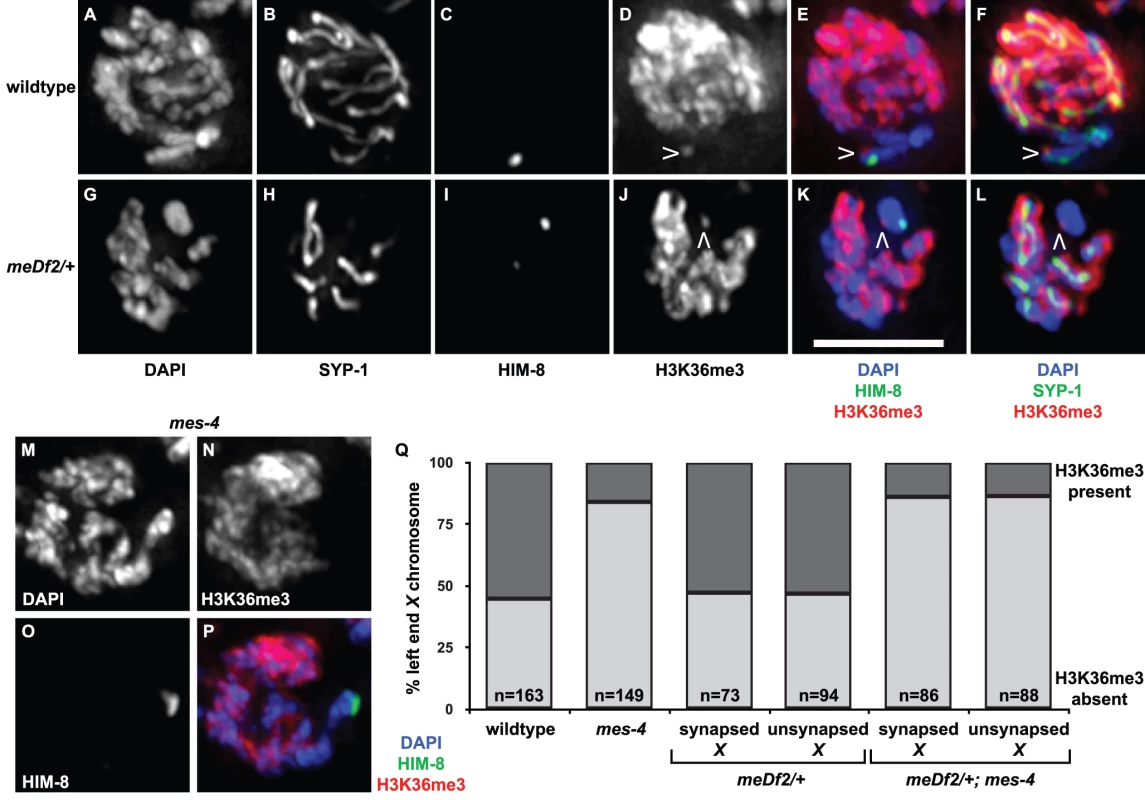
A–L. Indirect immunofluorescence was performed on meiotic nuclei in wildtype and meDf2 heterozygotes against SYP-1, HIM-8 and H3K36me3. H3K36me3 is depleted from X chromosomes in both of these genetic backgrounds except for a single dot near the end (carets in D, E, F, J, K and L) that does not colocalize with HIM-8 (E and K). Scale bar represents 4 microns. M–P. Mutation of mes-4 results in a reduction of H3K36me3 at the left end of X chromosomes. Indirect immunofluorescence was performed on meiotic nuclei in mes-4 single mutants against HIM-8 and H3K36me3. Q. Quantification of the percentage of meiotic nuclei in wildtype, mes-4 mutants, meDf2 heterozygotes and meDf2/+; mes-4 double mutants with H3K36me3 at the left end of X chromosomes. Mutation of mes-4 reduces but does not eliminate the percentage of X chromosomes that exhibit this chromatin modification. X chromosomes in meDf2 heterozygotes, whether synapsed or unsynapsed, exhibit this chromatin modification with a frequency similar to that observed in wildtype animals. We assessed whether the appearance of H3K36me3 at the left end of the X chromosome was dependent on mes-4 (Figure 7M–7P). The majority of meiotic nuclei in mid-pachytene did not exhibit H3K36me3 at the left end of the X chromosome in the absence of mes-4 (86%) (Figure 7M–7Q). A small fraction of meiotic nuclei did exhibit H3K36me3 (14%) in mes-4 mutant animals (Figure 7Q), suggesting that met-1 contributes to H3K36me3 at this locus. We observe a similar reduction in the percentage of meiotic nuclei that exhibit H3K36me3 at the left end of X chromosomes in met-1 mutants (data not shown). The percentage of X chromosomes with H3K36me3 at the left end in mes-4; meDf2/+ double mutants was similar to mes-4 single mutants, whether X chromosomes were synapsed or unsynapsed (Figure 7Q). Taken all together, these results raise the intriguing possibility that H3K36me3 may not be the relevant MES-4/MET-1 catalyzed chromatin modification for synapsis checkpoint activation (see Discussion).
We also monitored H3K36me3 at the PC end of X chromosomes in mes-4 mutant males and met-1 mutant males to determine the genetic requirements for this specific methyl mark in males. Unlike hermaphrodites (Figure 7N, 7P and data not shown), we observed that H3K36me3 at the PC end of X chromosomes was completely absent in both mes-4 mutant males and met-1 mutant males (data not shown).
Discussion
Our data suggest that chromatin state is an important aspect of the ability of PCs to activate the synapsis checkpoint. A chromosomal duplication that is sufficient to activate the synapsis checkpoint is less likely to undergo heterochromatin assembly, specifically enrichment of H3K9me2 (Figure 2D and 2G, Figure 3E). This distinguishes this duplication from duplications without PC activity, which consistently become enriched for H3K9me2 (Figure 2E and [8], [9], [19]). Furthermore, inactivation of a histone-modifying enzyme associated with active transcription and required for the synapsis checkpoint, MES-4 (Figure 3A), eliminates this distinction, producing a duplication with PC activity that undergoes heterochromatin assembly (Figure 3D and 3E). Our results lead us to propose that MES-4 and MET-1 generate a transcriptionally permissive chromatin environment at PCs that is required for synapsis checkpoint activation. We interpret the absence of H3K9me2 at PCs as an indirect result of MES-4 or MET-1 activity at PCs.
Given the difference in size between mnDp3 (∼5 Mb) and mnDp73 (∼2 Mb) [29], [30, Wormbase], our inability to observe enrichment of H3K9me2 on mnDp73 may reflect a significantly weaker fluorescence signal of the antibody against H3K9me2 on mnDp73, which could be below the detection limit. However, when mes-4 is inactivated by RNAi, mnDp73 is visibly enriched with H3K9me2 (Figure 3D and 3E) in 88% of meiotic nuclei in which the duplication can be identified by HIM-8 recruitment, indicating that we can reproducibly observe heterochromatin assembly on this smaller duplication with cytological techniques.
It is unlikely that our results could be explained by mutation of mes-4 and/or met-1 disrupting transcription of another locus (or loci) important for checkpoint function: if mes-4 and/or met-1 were regulating the transcription of some other checkpoint component(s), we would not expect that asynapsis of the X chromosome would exhibit different genetic requirements for checkpoint activation than asynapsis of autosomes. Of the multiple checkpoint components identified thus far, both published [4], [38] and unpublished (data not shown), mes-4 and met-1 are the only components that exhibit chromosome specific effects.
A role for transcription at PCs during synapsis checkpoint activation?
Both MES-4 and MET-1 are associated with active transcription: in embryos, MES-4 binds genes that were expressed in the maternal germline, potentially as a mechanism to transmit the epigenetic memory of maternal germline gene expression, while MET-1 is responsible for the deposition of transcription coupled H3K36me [20]. In the adult germline, loss of mes-4 leads to an altered distribution of MES-2/MES-3/MES-6 repressive activity on chromatin, resulting in upregulation of genes on X chromosomes and autosomal genes whose expression is normally restricted to somatic tissue [33], [34]. mes-4 mutant germ cells also exhibit downregulation of autosomal genes whose expression is normally enriched in or restricted to the germline, consistent with a role for MES-4 in promoting germline transcription. This role in promoting proper germline gene expression may be independent of the MES-2/MES-3/MES-6 complex, since mes-2; mes-4 double mutants do not alleviate the downregulation of autosomal genes, as might be expected if MES-4's only role is to antagonize the repressive activity of the MES-2/MES-3/MES-6 complex [34]. Since we observe that mes-4's role in the synapsis checkpoint is independent of the MES-2/MES-3/MES-6 complex (Figure 3B), we favor a scenario in which MES-4 (and MET-1) activity directly contributes to transcriptional activity at PCs to promote checkpoint activation.
This hypothesis is supported by our finding that mes-4 is still required for synapsis checkpoint activation in the absence of met-2 mediated heterochromatin assembly on unsynapsed chromosomes (Figure 6). Since apoptosis in response to synapsis checkpoint activation is transcriptionally regulated (J.M. Ragle and N. Bhalla, unpublished observations), we cannot directly test whether transcription is required for synapsis checkpoint activation, for example, by RNAi depletion of AMA-1, the large subunit of RNA Polymerase II [33]. We addressed this issue cytologically by using an antibody against the serine 5 phospho-epitope of RNA Polymerase II that is associated with transcriptional competence [45] and did not observe any enrichment of this specifically modified version of RNA Polymerase II on mnDp73 (data not shown). However, the observed inconsistency between cytological (reviewed in [46]) and more refined assays of transcriptional activity in the germline [20], [47], [48] raises the possibility that cytological approaches may not be adequately informative.
It is possible that MES-4 and MET-1 may not be mediating transcriptional activity at PCs. This would be consistent with our inability to localize modified RNA Polymerase II on mnDp73 (data not shown). While MES-4 and MET-1 mediated catalysis of H3K36me is associated with active transcription [20], studies in budding yeast have shown that Set2, the enzyme that methylates H3K36 in response to transcription, is responsible for creating a repressive transcriptional environment within the body of an actively transcribed gene to prevent inappropriate, or cryptic, transcriptional activation [23], [24], [25]. While we favor the interpretation that the requirement for MES-4 and MET-1 in synapsis checkpoint activation suggests a role for active transcription at PCs, we cannot rule out that MES-4 and MET-1 may contribute to a repressive chromatin environment at PCs that is distinct from the enrichment of H3K9me2 that is observed on the bulk of unsynapsed chromosomes. Additional studies will dissect the precise roles of MES-4 and MET-1 in the synapsis checkpoint.
Identifying the loci that may be transcribed to promote checkpoint activation is an obvious focus of our future investigations. Each PC is enriched with repetitive sequence elements [49] and it is tempting to speculate that transcription of these repetitive elements contributes to checkpoint activation. We tested whether extrachromosomal arrays composed of these repeats could activate the checkpoint but did not observe synapsis checkpoint activation in array-bearing hermaphrodites (data not shown). Although these arrays are competent for PC protein recruitment, pairing and synapsis and do not appear to undergo H3K9 methylation (data not shown), a chromosomal context or other sequence elements, such as promoters and enhancers, may be necessary for transcription and synapsis checkpoint activation.
To our knowledge, no data currently suggests that transcription is required for the other characterized role that PCs play in mediating pairing and synapsis [5]–[7], [38], [50]–[52]. mes-4 mutant homozygotes from heterozygote mothers are fertile (as a result of the maternal contribution of MES-4 that supports germline development) and don't exhibit a Him (high incidence of male) phenotype [21], indicating that mes-4 is not required for X chromosome segregation during meiosis. met-1; mes-4 double mutants exhibit low brood size, high embryonic lethality and some degree of larval arrest but no obvious Him phenotype among the sterile survivors (T. Takasaki and S. Strome, personal communication), suggesting that meiotic defects are not the cause of the embryonic lethality. One possible model is that transcription of loci at PCs is only required for the checkpoint. The ability to uncouple pairing and synapsis from checkpoint activation with arrays that recruit PC proteins might suggest that this is true. Alternatively, factors in addition to MES-4 and MET-1 may regulate transcription at PCs and be required for pairing and synapsis.
Four paralogous zinc-finger proteins (HIM-8, ZIM-1, ZIM-2, and ZIM-3) are required for PC function [6], [7]. PC proteins have been implicated in regulating transcription in somatic cells, independent of their roles in meiosis [53], [54]. In a variety of somatic contexts, mutations in him-8 and the zim genes suppress mutant phenotypes that arise from hypomorphic mutations in transcription factors [54]. Interestingly, this role in transcriptional regulation is strictly dependent on the ability of PC proteins to bind DNA since an allele of him-8 (me4) that supports DNA binding but not X chromosome pairing or synapsis doesn't exhibit this genetic interaction [53]. In the context of the synapsis checkpoint, the him-8(me4) allele is as defective as other alleles of him-8 (mn253 and e1489) tested for a role in the checkpoint (data not shown and [4]), indicating that any contribution to transcription at the PCs during checkpoint activation is likely to require all functions of HIM-8, ZIM-1, -2 and -3.
PCs have been compared to centromeres in monocentric organisms [50], [55]. In particular, it has been suggested that investing a single chromosomal locus with the ability to promote pairing and synapsis in the meiotic germline may contribute to genomic integrity in an organism, like C. elegans, in which chromosomes are holocentric and any deleterious changes to genome organization (i.e. the creation of translocations, rearrangements or chromosome fragments) will be faithfully transmitted during mitosis [55]. The use of PCs as sites of checkpoint activation extends this analogy since the centromere also monitors the event it engages in, namely spindle attachment. In a variety of monocentric organisms, transcription is required at centromeres for full activity and the faithful transmission of chromosomes [56]–[59]. A well-characterized contribution of transcription to centromere function involves the use of the RNAi machinery to establish and maintain pericentric heterochromatin [58]–[60]. However, these findings do not provide a satisfying framework to imagine how transcription may be contributing to PC function since the end product is silenced chromatin.
More recently, however, euchromatic domains, as defined by histone modifications associated with transcriptionally permissive chromatin, have been observed in core centromeric regions in rice [61], fruit flies, humans [62] and on human artificial chromosomes [63], suggesting that transcriptional activity aside from the formation of pericentric heterochromatin by the RNAi machinery may be required for aspects of centromere function. In some organisms, such as fission yeast, maize and humans, transcription of core centromeric regions has even been observed [64], [65], [66] and associated with centromere activity [57], [64], [67]. In budding yeast, a model organism that does not contain the RNA interference machinery, loss of transcription across centromeres results in chromosomal instability and sensitivity to microtubule depolymerizing drugs [56], phenotypic hallmarks of spindle assembly checkpoint mutants that fail to arrest mitosis in response to defects in spindle attachment [68].
What chromatin modification is critical for synapsis checkpoint activation?
Both MES-4 and MET-1 are required for all detectable H3K36me3 in germline nuclei (T. Takasaki and S. Strome, personal communication), suggesting that this histone modification is potentially relevant for synapsis checkpoint activation. Indeed, we observe a reduction in H3K36me3 at the left end of X chromosomes in mes-4 mutants (Figure 7N, 7P and 7Q). However, some of our data is inconsistent with H3K36me3 being the relevant MES-4/MET-1 catalyzed chromatin modification for synapsis checkpoint activation. Unsynapsed X chromosomes in meDf2 heterozygotes exhibit similar levels of H3K36me3 enrichment at their PC end as synapsed chromosomes in meDf2 heterozygotes or wildtype animals (Figure 7Q). Furthermore, loss of met-1 function also results in a reduction of this signal at the PC end of X chromosomes (data not shown), even though met-1 gene function is not required to monitor synapsis of X chromosomes (Figure 5A).
Other reports support the possibility that H3K36me3 is not critical for synapsis checkpoint activation. Knockdown of a characterized H3K36 demethylase (JMJD2) results in ectopic accumulation of H3K36me3 on the PC end of X chromosomes and DNA damage checkpoint activation in the germline but has no effect on synapsis checkpoint activation [44], indicating that enrichment of H3K36me3 at the left end of the X chromosome is not sufficient to activate the synapsis checkpoint. Moreover, the chromodomain protein identified as the putative H3K36me3 reader in C. elegans, MRG-1 [69], exhibits defects in synapsis [70] and activates the synapsis checkpoint when mutated [40], indicating that it is not required for the synapsis checkpoint.
Mutation of mes-4 also results in loss of detectable H3K36me2 in germline nuclei, suggesting it is the sole histone methyltransferase responsible for this methyl mark. However, given the potential inconsistencies between cytological and more refined assays to monitor chromatin modifications (see above), this does not necessarily mean that MET-1 cannot contribute to this mark. Therefore, H3K36me2 remains a potential candidate chromatin modification critical for synapsis checkpoint activation.
Another possibility is that MES-4 and MET-1 activity may be required for the catalysis of a histone modification aside from H3K36me to promote synapsis checkpoint signaling. For example, MES-2 was known to be required for H3K27me2 and H3K27me3 in the germline [36] but recent studies have also revealed that it is responsible for most of the detectable H3K9me3 as well [17]. One candidate histone modification that may be linked to synapsis checkpoint activation is H3K4me. This histone mark, which is associated with active transcription [71], has been observed on sDp1 [8], a large autosomal duplication that has PC activity [72]. Given that we suspect that PCs need to maintain transcriptional activity to monitor synapsis, this histone modification may maintain an active transcriptional state by recruiting transcriptional activators or it may specifically recruit proteins that recognize this methyl mark (e.g. a chromodomain protein) in checkpoint activating strains to promote checkpoint signaling. Future experiments will distinguish between these two models. An alternate hypothesis is that MES-4 and MET-1 are methylating something instead of histones at PCs to promote checkpoint signaling [73].
Why are there different mechanisms to monitor synapsis of the X chromosome and autosomes?
We have shown that X chromosomes require mes-4 to monitor synapsis while mes-4 and met-1 are redundant to monitor autosomal asynapsis. The X chromosome in C. elegans is distinct from the autosomes in several respects: it exhibits a more uniform distribution of gene density, recombination rates and repeat content [74]; it expresses genes in the germline [47], [48] at a reduced level [48], potentially as a result of histone modifications associated with transcriptional silencing (reviewed in [46]); it responds differently to hypomorphic mutation of certain genes required for meiosis [75]–[77]; it is the target of dosage compensation during sex determination and development [78]; and during male meiosis the single X chromosome segregates without a pairing and recombination partner.
We speculate that the unique requirement of the X for mes-4 activity in the synapsis checkpoint is a consequence of the male genotype (XO). When all germline nuclei have an unsynapsed sex chromosome, as they do in males, having distinct requirements for checkpoint activation for sex chromosomes and autosomes may allow the checkpoint to be unresponsive to the presence of an unsynapsed X chromosome while still maintaining the ability to monitor the synapsed state of the autosomes. Unsynapsed X chromosomes in males are specifically shielded from activating the DNA damage checkpoint in the germline by met-2 mediated MSCI [15]. However, loss of MSCI does not produce synapsis checkpoint activation [15] despite the proper localization of HIM-8 to the unsynapsed X chromosomes [7], indicating that some other mechanism prevents synapsis checkpoint activation in male germlines. To address whether male germlines are competent to activate the DNA damage checkpoint despite the absence of apoptosis [79], Engebrecht and colleagues have taken advantage of cytological markers of DNA damage checkpoint activation to elegantly illustrate that this checkpoint can be fully activated in males and contributes to genomic integrity [80]. Unfortunately, we currently have no comparable cytological markers of synapsis checkpoint activation and therefore can only assess checkpoint activation by monitoring apoptosis. The development of reagents that allow us to assess synapsis checkpoint activation independent of apoptosis will allow us to directly address how the unsynapsed male X is shielded from synapsis checkpoint activation.
Our model that mes-4's unique role in monitoring synapsis of X chromosomes provides a mechanism for the observed sexual dimorphism in checkpoint activation predicts that mes-4 activity is either inhibited or unnecessary in male germlines. It is currently unclear whether mes-4 is required in males for germline development (Gaydos, L. and S. Strome, personal communication). It is possible that the difference we observe between male and hermaphrodite germlines in the genetic requirements for H3K6me3 at the PC end of X chromosomes (data not shown and Figure 7N and 7P) indicates that these genes play different roles depending on the sex of the germline. This interpretation would be consistent with a recent study that found that knockdown of met-1 and mes-4 by RNAi also resulted in an increase in germline apoptosis in genotypically male germlines [15].
Materials and Methods
Genetics
The wildtype C. elegans strain background was Bristol N2. All experiments were performed at 20° under standard conditions. Mutations and rearrangements used were as follows:
LG I: mnDp66, met-1(n4337), mes-3(bn35)
LG II: mes-2(bn11), pch-2(tm1458)
LG III: met-2(n4256)
LG IV: spo-11(ok79), nT1[unc-?(n754) let-?(m435)] (IV, V)
LG V: dpy-11(e224), mes-4(bn23), syp-1(me17), bcIs39(Plin-15::ced-1::GFP)
LG X: unc-1(e1598n1201), dpy-3(e27), meDf2, mnDp73, unc-3(e151), mnDp3
Experiments with mes mutants were performed in the M+Z - generation, in which maternal load of the relevant gene function allows the generation of a fertile homozygote adult with a developed germline.
Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Scoring of apoptotic nuclei
Scoring of apoptotic nuclei was performed as in [4]. To assess significance, two-tailed unpaired t tests were performed to compare average number of apoptotic nuclei between genotypes.
RNA interference
All RNAi was performed by culturing relevant worm strains on HT115 bacteria transformed with vectors that allowed for IPTG inducible expression of dsRNA. Bacteria containing RNAi vectors were grown overnight at 37°, centrifuged and resuspended in 1/20 the original volume. 50 ul of this concentrated culture was spotted onto NGM plates containing 1 mM IPTG and 50 ug/ul carbenicillin. After incubation at 37° overnight, late L4 stage worms were picked into M9, transferred to these plates and allowed to crawl in the bacteria for two hours to clear OP50 from their gut. They were then transferred to fresh RNAi plates, allowed to produce progeny and L4s from this population were picked to score apoptosis or perform cytology.
To reproducibly knock down met-2 activity by feeding RNAi, we generated a new met-2 RNAi vector using a Gateway-compatible (Invitrogen) RNAi vector that included termination sites downstream of the T7 promoters (pDONRT7) [81]. met-2 was amplified by PCR from N2 genomic DNA using primers met-2_FOR (5′ ggg/gac/aag/ttt/tgt/aca/aaa/aag/cag/gct/gca/cca/atc/aga/atg/tcg 3′) and met-2_REV (5′ ggg/gac/cac/ttt/gta/caa/gaa/agc/tgg/gtt/ttt/cca/gct/cca/cgg/tat/c 3′). The resulting PCR product was cloned into pDONRT7 using a BP reaction (Invitrogen) and transformed into DH5α. A clone in which the met-2 PCR product was properly inserted into pDONRT7 was identified, isolated and retransformed into HT115.
Immunostaining and microscopy
Immunostaining was performed as in [4]. Primary antibodies were as follows (dilutions are indicated in parentheses): rabbit anti-SYP-1 (1∶250) [28], guinea pig anti-HTP-3 (1∶250) [7], guinea pig anti-HIM-8 (1∶250), guinea pig anti-SUN-1 pSer8 (1∶700), guinea pig anti-SUN-1 pSer12 (1∶17,000), mouse anti-H3K9me2 (1∶50,000) [82], mouse anti-H3K36me3 (1∶50,000) [20] and rabbit anti-H3K36me3 (Abcam). Secondary antibodies were Cy3 anti-mouse and anti-rabbit pig (Jackson Immunochemicals), Alexa-Fluor 555 anti-rabbit (Invitrogen) and Cy5 anti-guinea pig (Jackson Immunochemicals). We thank Anne Villeneuve, Abby Dernburg, Verena Jantsch and Hiroshi Kimura for reagents.
All images were acquired using a DeltaVision Personal DV system (Applied Precision) equipped with a 100× N.A. 1.40 oil-immersion objective (Olympus), resulting in an effective XY pixel spacing of 0.064 or 0.040 µm. Three-dimensional image stacks were collected at 0.2-µm Z-spacing and processed by constrained, iterative deconvolution. Image scaling and analysis were performed using functions in the softWoRx software package. Projections of complete meiotic nuclei were calculated by a maximum intensity algorithm. Composite images were assembled and some false coloring was performed with Adobe Photoshop. In some cases, projections of partial Z-stacks of meiotic nuclei were generated (for example, Figure 2E–2J, Figure 3C and 3D, and Figure 7G–7L) to facilitate the visualization of duplications or unsynapsed chromosomes. In Figure 2C, the non-PC region was identified as a region not adjacent to the HIM-8 signal and of similar size to observed instances when the PC region was devoid of H3K9me2.
Supporting Information
Zdroje
1. BhallaN, DernburgAF (2008) Prelude to a division. Annu Rev Cell Dev Biol 24 : 397–424.
2. HassoldT, HuntP (2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2 : 280–291.
3. MacQueenAJ, HochwagenA (2011) Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends in cell biology 21 : 393–400.
4. BhallaN, DernburgAF (2005) A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310 : 1683–1686.
5. MacQueenAJ, PhillipsCM, BhallaN, WeiserP, VilleneuveAM, et al. (2005) Chromosome Sites Play Dual Roles to Establish Homologous Synapsis during Meiosis in C. elegans. Cell 123 : 1037–1050.
6. PhillipsCM, DernburgAF (2006) A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell 11 : 817–829.
7. PhillipsCM, WongC, BhallaN, CarltonPM, WeiserP, et al. (2005) HIM-8 Binds to the X Chromosome Pairing Center and Mediates Chromosome-Specific Meiotic Synapsis. Cell 123 : 1051–1063.
8. BeanCJ, SchanerCE, KellyWG (2004) Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet 36 : 100–105.
9. ShiuPK, MetzenbergRL (2002) Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161 : 1483–1495.
10. TurnerJM, MahadevaiahSK, Fernandez-CapetilloO, NussenzweigA, XuX, et al. (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37 : 41–47.
11. TurnerJM (2007) Meiotic sex chromosome inactivation. Development 134 : 1823–1831.
12. MahadevaiahSK, Bourc'hisD, de RooijDG, BestorTH, TurnerJM, et al. (2008) Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 182 : 263–276.
13. RoyoH, PolikiewiczG, MahadevaiahSK, ProsserH, MitchellM, et al. (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Current biology: CB 20 : 2117–2123.
14. BurgoynePS, MahadevaiahSK, TurnerJM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10 : 207–216.
15. ChecchiPM, EngebrechtJ (2011) Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genet 7: e1002267 doi:10.1371/journal.pgen.1002267.
16. Jaramillo-LambertA, EngebrechtJ (2010) A single unpaired and transcriptionally silenced X chromosome locally precludes checkpoint signaling in the Caenorhabditis elegans germ line. Genetics 184 : 613–628.
17. BesslerJB, AndersenEC, VilleneuveAM (2010) Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet 6: e1000830 doi:10.1371/journal.pgen.1000830.
18. SheX, XuX, FedotovA, KellyWG, MaineEM (2009) Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet 5: e1000624 doi:10.1371/journal.pgen.1000624.
19. MaineEM, HauthJ, RatliffT, VoughtVE, SheX, et al. (2005) EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Current biology: CB 15 : 1972–1978.
20. RechtsteinerA, ErcanS, TakasakiT, PhippenTM, EgelhoferTA, et al. (2010) The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet 6: e1001091 doi:10.1371/journal.pgen.1001091.
21. CapowskiEE, MartinP, GarvinC, StromeS (1991) Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics 129 : 1061–1072.
22. FongY, BenderL, WangW, StromeS (2002) Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296 : 2235–2238.
23. CarrozzaMJ, LiB, FlorensL, SuganumaT, SwansonSK, et al. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123 : 581–592.
24. JoshiAA, StruhlK (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Molecular cell 20 : 971–978.
25. KeoghMC, KurdistaniSK, MorrisSA, AhnSH, PodolnyV, et al. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123 : 593–605.
26. AndersenEC, HorvitzHR (2007) Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134 : 2991–2999.
27. VilleneuveAM (1994) A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136 : 887–902.
28. MacQueenAJ, VilleneuveAM (2001) Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev 15 : 1674–1687.
29. HermanRK, KariCK (1989) Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics 121 : 723–737.
30. HermanRK, AlbertsonDG, BrennerS (1976) Chromosome rearrangements in Caenorhabditis elegans. Genetics 83 : 91–105.
31. HofmannER, MilsteinS, BoultonSJ, YeM, HofmannJJ, et al. (2002) Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr Biol 12 : 1908–1918.
32. GarvinC, HoldemanR, StromeS (1998) The phenotype of mes-2, mes-3, mes-4 and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics 148 : 167–185.
33. BenderLB, SuhJ, CarrollCR, FongY, FingermanIM, et al. (2006) MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development 133 : 3907–3917.
34. GaydosL, RechtsteinerA, EgelhoferTA, CarrollC, StromeS (2012) Antagonism between MES-4 and the C. elegans Polycomb Repressive Complex 2 Promotes Germ Cell-Appropriate Gene Expression. Cell Rep in press.
35. HoldemanR, NehrtS, StromeS (1998) MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development 125 : 2457–2467.
36. BenderLB, CaoR, ZhangY, StromeS (2004) The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Current biology: CB 14 : 1639–1643.
37. KetelCS, AndersenEF, VargasML, SuhJ, StromeS, et al. (2005) Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Molecular and cellular biology 25 : 6857–6868.
38. HarperNC, RilloR, Jover-GilS, AssafZJ, BhallaN, et al. (2011) Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Developmental cell 21 : 934–947.
39. PenknerAM, FridkinA, GloggnitzerJ, BaudrimontA, MachacekT, et al. (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139 : 920–933.
40. XuJ, SunX, JingY, WangM, LiuK, et al. (2012) MRG-1 is required for genomic integrity in Caenorhabditis elegans germ cells. Cell research
41. ColaiacovoMP, MacQueenAJ, Martinez-PerezE, McDonaldK, AdamoA, et al. (2003) Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Developmental cell 5 : 463–474.
42. DudleyNR, LabbeJC, GoldsteinB (2002) Using RNA interference to identify genes required for RNA interference. Proc Natl Acad Sci U S A 99 : 4191–4196.
43. XuL, StromeS (2001) Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of Caenorhabditis elegans. Genetics 159 : 1019–1029.
44. WhetstineJR, NottkeA, LanF, HuarteM, SmolikovS, et al. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125 : 467–481.
45. KellyWG, SchanerCE, DernburgAF, LeeMH, KimSK, et al. (2002) X-chromosome silencing in the germline of C. elegans. Development 129 : 479–492.
46. SchanerCE, KellyWG (2006) Germline chromatin. WormBook: the online review of C elegans biology 1–14.
47. TabuchiTM, DeplanckeB, OsatoN, ZhuLJ, BarrasaMI, et al. (2011) Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet 7: e1002074 doi:10.1371/journal.pgen.1002074.
48. WangX, ZhaoY, WongK, EhlersP, KoharaY, et al. (2009) Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10 : 213.
49. PhillipsCM, MengX, ZhangL, ChretienJH, UrnovFD, et al. (2009) Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol 11 : 934–942.
50. LabellaS, WoglarA, JantschV, ZetkaM (2011) Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Developmental cell 21 : 948–958.
51. PenknerA, TangL, NovatchkovaM, LadurnerM, FridkinA, et al. (2007) The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 12 : 873–885.
52. SatoA, IsaacB, PhillipsCM, RilloR, CarltonPM, et al. (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139 : 907–919.
53. NelmsBL, Hanna-RoseW (2006) C. elegans HIM-8 functions outside of meiosis to antagonize EGL-13 Sox protein function. Developmental biology 293 : 392–402.
54. SunH, NelmsBL, SleimanSF, ChamberlinHM, Hanna-RoseW (2007) Modulation of Caenorhabditis elegans transcription factor activity by HIM-8 and the related Zinc-Finger ZIM proteins. Genetics 177 : 1221–1226.
55. DernburgAF (2001) Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol 153: F33–38.
56. OhkuniK, KitagawaK (2011) Endogenous transcription at the centromere facilitates centromere activity in budding yeast. Current biology: CB 21 : 1695–1703.
57. DuY, ToppCN, DaweRK (2010) DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet 6: e1000835 doi:10.1371/journal.pgen.1000835.
58. FukagawaT, NogamiM, YoshikawaM, IkenoM, OkazakiT, et al. (2004) Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature cell biology 6 : 784–791.
59. VolpeTA, KidnerC, HallIM, TengG, GrewalSI, et al. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 : 1833–1837.
60. FolcoHD, PidouxAL, UranoT, AllshireRC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319 : 94–97.
61. WuY, KikuchiS, YanH, ZhangW, RosenbaumH, et al. (2011) Euchromatic subdomains in rice centromeres are associated with genes and transcription. The Plant cell 23 : 4054–4064.
62. SullivanBA, KarpenGH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature structural & molecular biology 11 : 1076–1083.
63. BergmannJH, RodriguezMG, MartinsNM, KimuraH, KellyDA, et al. (2011) Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. The EMBO journal 30 : 328–340.
64. ChanFL, MarshallOJ, SafferyR, KimBW, EarleE, et al. (2012) Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences of the United States of America 109 : 1979–1984.
65. ChoiES, StralforsA, CastilloAG, Durand-DubiefM, EkwallK, et al. (2011) Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. The Journal of biological chemistry 286 : 23600–23607.
66. SafferyR, SumerH, HassanS, WongLH, CraigJM, et al. (2003) Transcription within a functional human centromere. Molecular cell 12 : 509–516.
67. ChuehAC, NorthropEL, Brettingham-MooreKH, ChooKH, WongLH (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5: e1000354 doi:10.1371/journal.pgen.1000354.
68. LiR, MurrayAW (1991) Feedback control of mitosis in budding yeast. Cell 66 : 519–531.
69. TakasakiT, LiuZ, HabaraY, NishiwakiK, NakayamaJ, et al. (2007) MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development 134 : 757–767.
70. DombeckiCR, ChiangAC, KangHJ, BilgirC, StefanskiNA, et al. (2011) The chromodomain protein MRG-1 facilitates SC-independent homologous pairing during meiosis in Caenorhabditis elegans. Developmental cell 21 : 1092–1103.
71. KouzaridesT (2002) Histone methylation in transcriptional control. Current opinion in genetics & development 12 : 198–209.
72. RoseAM, BaillieDL, CurranJ (1984) Meiotic pairing behavior of two free duplications of linkage group I in Caenorhabditis elegans. Molecular & general genetics: MGG 195 : 52–56.
73. RathertP, DhayalanA, MaH, JeltschA (2008) Specificity of protein lysine methyltransferases and methods for detection of lysine methylation of non-histone proteins. Mol Biosyst 4 : 1186–1190.
74. Consortium CeS (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282 : 2012–2018.
75. CouteauF, NabeshimaK, VilleneuveA, ZetkaM (2004) A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol 14 : 585–592.
76. ReddyKC, VilleneuveAM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118 : 439–452.
77. Martinez-PerezE, VilleneuveAM (2005) HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 19 : 2727–2743.
78. MeyerBJ (2000) Sex in the wormcounting and compensating X-chromosome dose. Trends in genetics: TIG 16 : 247–253.
79. GumiennyTL, LambieE, HartwiegE, HorvitzHR, HengartnerMO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126 : 1011–1022.
80. Jaramillo-LambertA, HarigayaY, VittJ, VilleneuveA, EngebrechtJ (2010) Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Current biology: CB 20 : 2078–2089.
81. ReddienPW, BermangeAL, MurfittKJ, JenningsJR, Sanchez AlvaradoA (2005) Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Developmental cell 8 : 635–649.
82. EgelhoferTA, MinodaA, KlugmanS, LeeK, Kolasinska-ZwierzP, et al. (2011) An assessment of histone-modification antibody quality. Nature structural & molecular biology 18 : 91–93.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



