-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Cooperativity of , , and in Malignant Breast Cancer Evolution
Breast cancers that are “triple-negative” for the clinical markers ESR1, PGR, and HER2 typically belong to the Basal-like molecular subtype. Defective Rb, p53, and Brca1 pathways are each associated with triple-negative and Basal-like subtypes. Our mouse genetic studies demonstrate that the combined inactivation of Rb and p53 pathways is sufficient to suppress the physiological cell death of mammary involution. Furthermore, concomitant inactivation of all three pathways in mammary epithelium has an additive effect on tumor latency and predisposes highly penetrant, metastatic adenocarcinomas. The tumors are poorly differentiated and have histologic features that are common among human Brca1-mutated tumors, including heterogeneous morphology, metaplasia, and necrosis. Gene expression analyses demonstrate that the tumors share attributes of both Basal-like and Claudin-low signatures, two molecular subtypes encompassed by the broader, triple-negative class defined by clinical markers.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003027
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003027Summary
Breast cancers that are “triple-negative” for the clinical markers ESR1, PGR, and HER2 typically belong to the Basal-like molecular subtype. Defective Rb, p53, and Brca1 pathways are each associated with triple-negative and Basal-like subtypes. Our mouse genetic studies demonstrate that the combined inactivation of Rb and p53 pathways is sufficient to suppress the physiological cell death of mammary involution. Furthermore, concomitant inactivation of all three pathways in mammary epithelium has an additive effect on tumor latency and predisposes highly penetrant, metastatic adenocarcinomas. The tumors are poorly differentiated and have histologic features that are common among human Brca1-mutated tumors, including heterogeneous morphology, metaplasia, and necrosis. Gene expression analyses demonstrate that the tumors share attributes of both Basal-like and Claudin-low signatures, two molecular subtypes encompassed by the broader, triple-negative class defined by clinical markers.
Introduction
The dire need for more effective treatments for aggressive breast cancers has motivated intensive investigations into their cellular and molecular etiology. Breast cancers classified as “triple-negative” by clinical diagnostic markers (ESR1, PGR, and HER2 negative) are heterogeneous in their clinical behavior, morphology, and molecular biology. Triple-negative breast cancers (TNBC) typically express the Basal-like molecular signature, thus TNBC and Basal cancer classifications are frequently used interchangeably. However, they are not completely synonymous [1], [2]. TNBCs also include the Claudin-low molecular subtype [3], which is characterized by greatly reduced expression of intercellular junction components and by activation of molecular pathways associated with epithelial-to-mesenchymal transition (EMT), cancer stem cells, and the immune response [4].
Histologically, most triple-negative breast cancers are invasive ductal carcinomas, but TNBCs also include the metaplastic, medullary, and adenocystic histologic special types, distinctive morphologies that are prevalent among Claudin-low tumors [4]. TNBCs are insensitive to endocrine therapy and HER2 antagonists, but they are sensitive to chemotherapy. Nevertheless, long-term patient outcomes are poor due to high rates of relapse and acquired chemoresistance [5], [6]. Mouse models that mimic the complexity of TNBC will be invaluable tools for defining the diverse cellular biology and behavior of these tumors and for rigorously triaging new drug candidates.
Basal-like breast cancers often simultaneously inactivate three tumor suppressors that are infamous for their roles in familial cancers: Rb (RB1) [7], p53 (TP53) [8], and BRCA1 [9]. p53 is mutated in 20–30% of human breast cancers and defective pathway intermediates also increase breast cancer risk [10]. Moreover, nearly all Basal-like cancers with BRCA1 mutation have concomitant p53 mutation [11]. Germline BRCA1 mutation predisposes early-onset breast cancers that are often triple-negative and that correlate with Basal-like tumors in microarray analyses [9]. BRCA1 mutation accounts for nearly half of familial breast cancers (OMIM 604370), but BRCA1 is also down-regulated in sporadic breast tumors without germline mutation [12]. The overall effect of BRCA1 loss is likely pleiotropic. The best studied function of BRCA1 is in orchestrating DNA double-strand break repair through homologous recombination or non-homologous end-joining. The importance of defective DNA repair in breast cancer is underscored by the observation that all known genes associated with inherited forms of the disease safeguard genomic integrity [13]. BRCA1 also functions as a transcription factor [14] that appears to be required for the differentiation of stem/progenitor cells into mature luminal cells [15]. This finding is consistent with the recent characterization of BRCA1-associated tumors as aberrant luminal progenitor cells [16].
The pair-wise cooperativity of Brca1 and p53 in mammary tumorigenesis has been studied in mouse models [17]. We and others have investigated the impact of combined Rb and p53 inactivation on mammary tumorigenesis in vivo [18]–[20]. We showed that following Rb perturbation in vivo, tumor progression is limited largely by p53-dependent apoptosis, and that loss of the second p53 allele in MFT121+/p53f/+ tumors is likely a prerequisite for mammary tumor progression [19], since the vast majority of tumors lose the wild type p53 allele during tumorigenesis. In the context of a brain carcinoma model initiated by T121, apoptosis appears to be the critical function of the normal p53 allele that is the target of selective pressure [21].
Our motivation to combine Rb inactivation with Brca1 and p53 mutation derives, in part, from the observation that the Rb gene is among the most frequently deleted loci in Brca1/p53-mutated mouse tumors [22], indicating that Rb is a critical barrier to tumor progression. Rb pathway inactivation is strongly associated with human triple negative breast cancers. A cardinal feature of basal-like breast cancers is the abundant expression of the “proliferation cluster” genes [7], which include many E2F-regulated genes that are de-repressed following Rb inactivation. In human breast cancers, reduced pRb activity correlates with higher tumor grade [23], but also predicts improved chemotherapy responsiveness [24]. The Rb gene itself is mutated in breast cancer [25], and recent genomic studies have indicated an overrepresentation of mutations within pRb-binding sites of human gene regulatory domains [26].
In this study we show that mammary tumors caused by inactivation of the pRb family (pRbf) of proteins (pRb, p107, p130), together with Brca1 and p53 inactivation, mimic several aspects of the most aggressive forms of breast cancer, including rapid tumor progression, poor differentiation, distant metastasis, necrosis, metaplasia, and genomic instability. Our findings illustrate the compounding effect of acquiring multiple tumor suppressor mutations during tumor evolution and underscore the distinct requirements of each of these canonical tumor suppressor proteins.
Results
Conditional T121 expression in mammary epithelium
We constructed the MFT121 (MMTV-Floxed-eGFP-T121) transgene to conditionally inactivate the pRb family (pRbf) of pocket proteins in mammary epithelium (Figure 1A). The T121 protein is an amino-terminal fragment of the SV40 large T antigen that perturbs pRbf activity and predisposes tumorigenesis in a range of tissues [27]. While Rb inactivation alone is sufficient to induce mammary tumors [20], the shorter latency of TgWAP-T121 tumors indicates there is functional redundancy or compensation by the related pocket proteins p107 or p130 [19]. In the MFT121 model, the MMTV-LTR promotes mammary-specific transgene expression (Figure 1B). The approach of inactivating pRBf, Brca1, and p53 specifically in mammary epithelial cells via the Wap-Cre transgene [28] enabled us to avoid the appearance of lymphomas [20] or sarcomas [29]. Wap-Cre excised the LoxP-eGFP-stop-LoxP reporter cassette and initiated T121 expression in ductal and alveolar luminal epithelial cells (Figure 1C). Virgin glands of MFT121; WAP-Cre mice appeared normal. Lactating glands (WAP-Cre-induced) showed reduced alveolar density, similar to the gland atrophy phenotype in the TgWap-T121 model, which was associated with apoptosis caused by T121-induced proliferation [19] and lactation defects observed in WAP-Cre; Rbfl/fl; p107−/− mice [20].
Fig. 1. The MFT121 transgene construct. 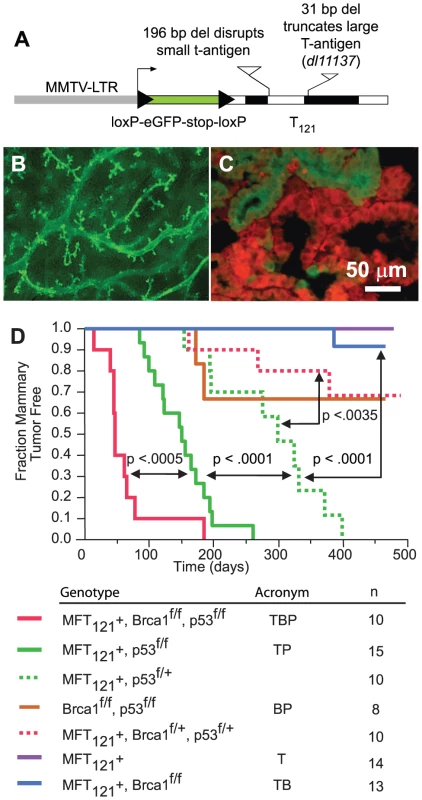
(A) The MFT121 transgene construct. The “floxed” eGFP-stop cassette was expressed throughout virgin mammary epithelium (B, original magnification 50×). Following Cre-induced excision, T121 was expressed (red) in the majority of luminal epithelial cells (C). eGFP immunolabeling (green) revealed non-recombined cells (original mag. 400×). Kaplan-Meier analysis of tumor onset (D). p53 was haploinsufficient (dashed green) for tumor suppression (p<0.0001, log-rank test). Homozygous p53 mutation (solid green) shortened tumor latency. Brca1 loss (solid red) further accelerated tumor onset (p<0.0005). Median tumor latency of TBP mice was approximately seven weeks. Parturition Day 1 = Time 0. All mice harbored the Wap-Cre transgene (not shown). Significance levels for critical comparisons are indicated. Concomitant pRbf, p53, and Brca1 inactivation significantly accelerates tumor onset
Multiparous TgMFT121; TgWap-Cre mice remained tumor-free for more than a year after Cre-induction, but mice that were either heterozygous or homozygous for a conditional p53 allele [30] (TgMFT121; TgWAP-Cre; p53f/+ or TgMFT121; TgWAP-Cre; p53f/f) developed mammary tumors with 100% penetrance and clear evidence of p53 haploinsufficiency (Figure 1D, p<0.0001, log-rank test). Heterozygous p53 mice developed tumors with a median latency of 299 days, while p53 homozygous mice had a median tumor latency of 150 days. These results are consistent with previous findings that p53 activity is rate limiting for mammary tumor progression initiated by Rb inactivation [18]–[20]. In contrast to the high penetrance we previously found in our TgWAP-T121 breast tumor model [19], the present studies indicate that the TgMFT121 transgene only partially inhibits pRbf pathways, possibly owing to the reduction in the transgene gene copy number following Cre excision. Similar dosage effects were observed in a conditional T121 transgenic model of astrocytoma [31].
Brca1 mutation dramatically accelerated tumor onset in mice with Rbf/p53 inactivation (Figure 1D, p<0.0005). Only a single mouse (n = 13), doubly defective for Rbf and Brca1 activity (TgMFT121;TgWAP-Cre;Brca1f/f), developed mammary tumors at 386 days following Cre induction by multiple pregnancies. In contrast, 100% (n = 10) of TgMFT121; Brca1f/f p53f/f; TgWAP-Cre mice (hereafter, TBP) developed mammary tumors with a median latency of only 47 days. Thus, a single pregnancy was sufficient for 100% TBP tumor penetrance. Once palpable, TBP tumors grew rapidly to 1500 cc within several days (data not shown). Therefore, inactivation of the three canonical tumor suppressors showed additive effects on tumor latency, since TBP mammary tumors developed significantly faster than did pair-wise combinations.
Enhanced survival and increased proliferation rates cause rapid tumor progression in pRbf/p53/Brca1-perturbed epithelium
To investigate the early effects of tumor suppressor inactivation, we examined mammary gland biopsies (n = 5 mice) from time points (0, 2, 6 weeks) following forced weaning at Day 1 parturition. At the earliest time point, T121 expression alone was sufficient to cause benign, hyperproliferative lesions (Figure 2A, 2B). T121-expressing cells showed a higher Ki67 index than did cells without T121, as expected (p<0.0063, Figure 3P). Combinations with Brca1 and p53 mutation caused higher grade, premalignant lesions (Figure 2C–2F). Tall columnar epithelia of darkly staining cells and papillary tufting (Figure 2C) were characteristic of the disrupted epithelial morphology. Pleomorphic, faintly staining nuclei with prominent nucleoli were common among T121 expressing cells (Figure 3F).
Fig. 2. Early lesions in lactating mammary glands. 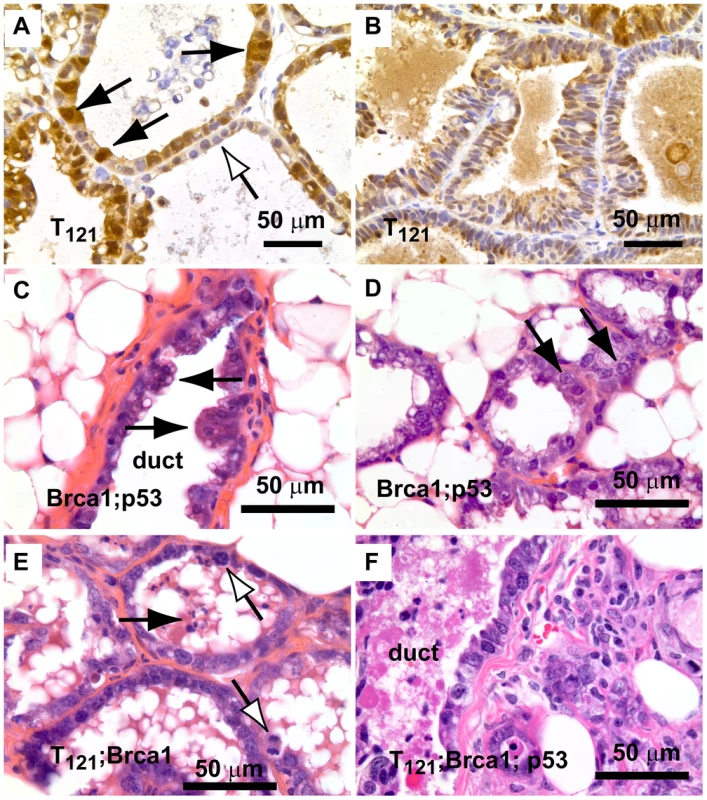
(A) Nuclear and cytoplasmic T121 expression (brown) is associated with enlarged, pleiomorphic nuclei (filled arrows) compared to low-expressing cells (open arrow). (B) Benign, multi-layered, mammary intra-epithelial neoplasia (MIN) within the same gland as A. (C) Dual inactivation of Brca1 and p53 disrupted epithelial architecture in primary ducts (C) and lactating alveoli (D), shown by papillary tufting (C, arrows) and high grade nuclei and prominent nucleoli (D, arrows). High grade nuclei and mitotic figures (E, open arrows) and dead cell debris (arrow) in MFT121; Brca1 gland. Severely pleomorphic and high grade nuclei are visible throughout MFT121; Brca1; p53 gland (F). Fig. 3. Combined inactivation of pRbf and p53 causes a durable block in involution. 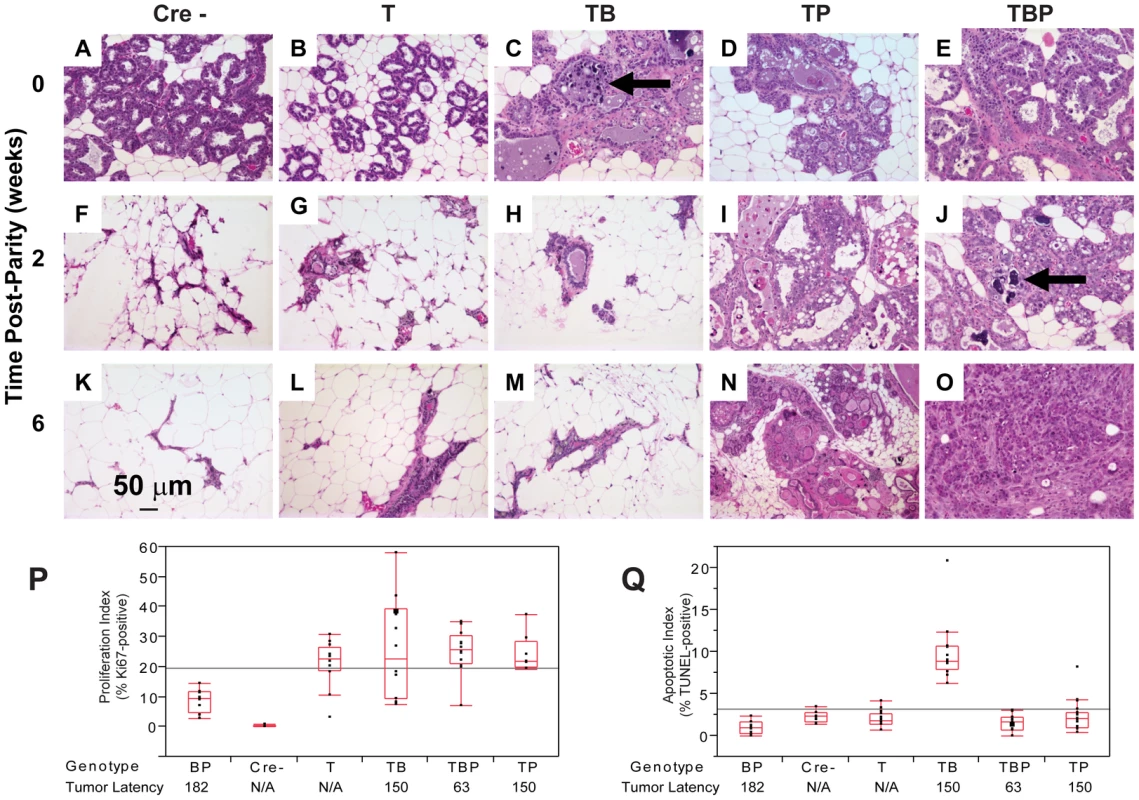
Lactating mammary epithelium of Cre-negative control mice (A, F, K) and T121-expressing mice (B, G, L) involute normally (A–E: 0 wks, F–J: 2 wks, K–O: 6 wks). T121-expressing mice have reduced alveolar density. Cell death and debris were abundant in TB glands (arrows panels C, J). TP glands failed to involute (N) and persisted as highly cystic glands. Frank tumors were present by 6 wks in TBP mice. An invasive adenocarcinoma fills the field of panel O. Measured at 0 wks, T121 increased the Ki67 index but without added effect by Brca1 and/or p53 loss (P). Combined inactivation of pRbf and Brca1 increased TUNEL-positive cells, which was suppressed by p53 loss (Q, p<0.0001). For each genotype n = 5 mice. Original magnification of each panel was 200×. T = T121, B = Brca1, P = p53. The combined inactivation of pRbf and p53 also dramatically suppressed the physiologic cell death of mammary involution (Figure 3I, 3N), although pRbf perturbation alone had little effect (Figure 3B, 3G, 3L). It was shown that p53 mutation alone delayed involution by several days [32]. Here, we observed that the combined inactivation of pRbf and p53 blocked involution through six weeks (Figure 3I, 3N). Biopsies from mice harboring Brca1 mutation (Figure 3C, 3J) showed extensive cellular debris from dead or dying cells within their lumen. The dual loss of pRbf and Brca1 activities (TgMFT121;TgWAP-Cre;Brca1f/f) triggered elevated cell death that was p53-dependent (p<0.0001, Figure 3Q). The combined loss of pRbf, Brca1, and p53 activities accelerated tumor progression, indicated by frank tumors that appeared by six weeks (Figure 3O). However, when measured at the earliest time point, neither an increased Ki67 proliferation index (Figure 3P) nor a decreased cell death index (Figure 3Q) presaged faster onset of TBP tumors compared to Rbf/p53 tumors. Thus, full transformation occurred in only a minority cell population, which likely reflects the requirement for additional collaborating oncogenic events.
Histopathologic features of tumors with conditional inactivation of pRbf, p53, and Brca1
T121/p53 (TP) and TBP mice developed high grade mammary adenocarcinomas with heterogeneous phenotypes indicative of highly perturbed differentiation, summarized in Table 1. Tumors of both genotypes showed mixed solid and glandular morphologies. More TBP mice than TP mice (13/14 vs. 8/16 cases, p = 0.0169, two-tailed Fisher's exact) developed solid tumors that were largely devoid of glandular architecture (Figure 4A). All solid tumors showed a mixture of pushing and infiltrative borders, and nearly half of the tumors were vascularized (Figure 4A). All carcinomas expressed varying levels of luminal epithelial markers, including Keratin-8 (Figure 4E, 4G, 4K) and E-Cadherin (Figure 4I, 4L). These markers were abundant in well-differentiated tumor regions but were greatly diminished or absent in poorly differentiated areas (Figure 4E, 4I, 4K, 4L). Nests of carcinoma cells variably expressed basal/myoepithelial lineage markers (Keratins-5, -14, Figure 4E, 4K).
Fig. 4. Perturbed differentiation in TBP tumors. 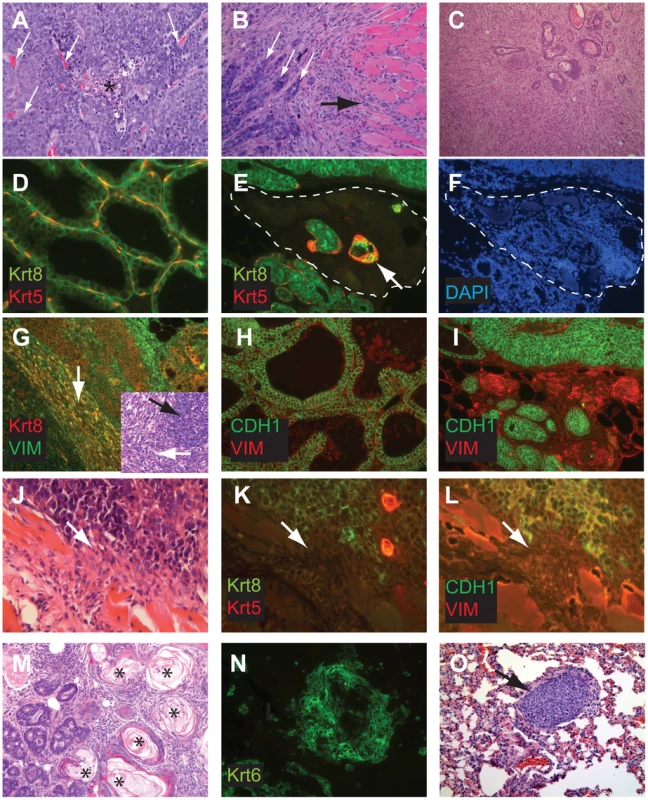
Poorly differentiated tumors with solid morphology were most common (A), with and without vascularization (white arrows) or central necrosis (asterisk). Other tumors retained remnant glandular architecture (B, white arrows). Metaplastic cells invaded adjacent muscle and stroma (B, black arrow). Homogeneous spindloid cells of a carcinosarcoma entrap carcinomatous cells (C, 100×). Keratin-8 (Krt8, green) and Keratin-5 (Krt5, red) immunolabeling of luminal and myoepithelial cells, respectively (D, E, K). Greatly reduced expression of both Krt5 and Krt8 (E dashed lines, and K arrow). DAPI staining (blue) indicates the high cellularity of the region devoid of epithelial markers in panel E (F). Metaplastic tumor cells (G) with dual staining of Krt8 (red) and the mesenchymal marker Vimentin (green), or reduced Krt staining (K). Abundant E-cadherin (CDH1, green) in normal adjacent (H) or well-differentiated tumor (I). Reduced or absent CDH1 along invasive tumor fronts (J–L). Keratinic whorls in squamous metaplastic cells (M, asterisks). Whorl-associated and disseminated Keratin 6 expression (N, green). Pulmonary metastases were observed in both T121/p53 and TBP mice (O, arrow). Tab. 1. Characteristics of TP and TBP tumors. 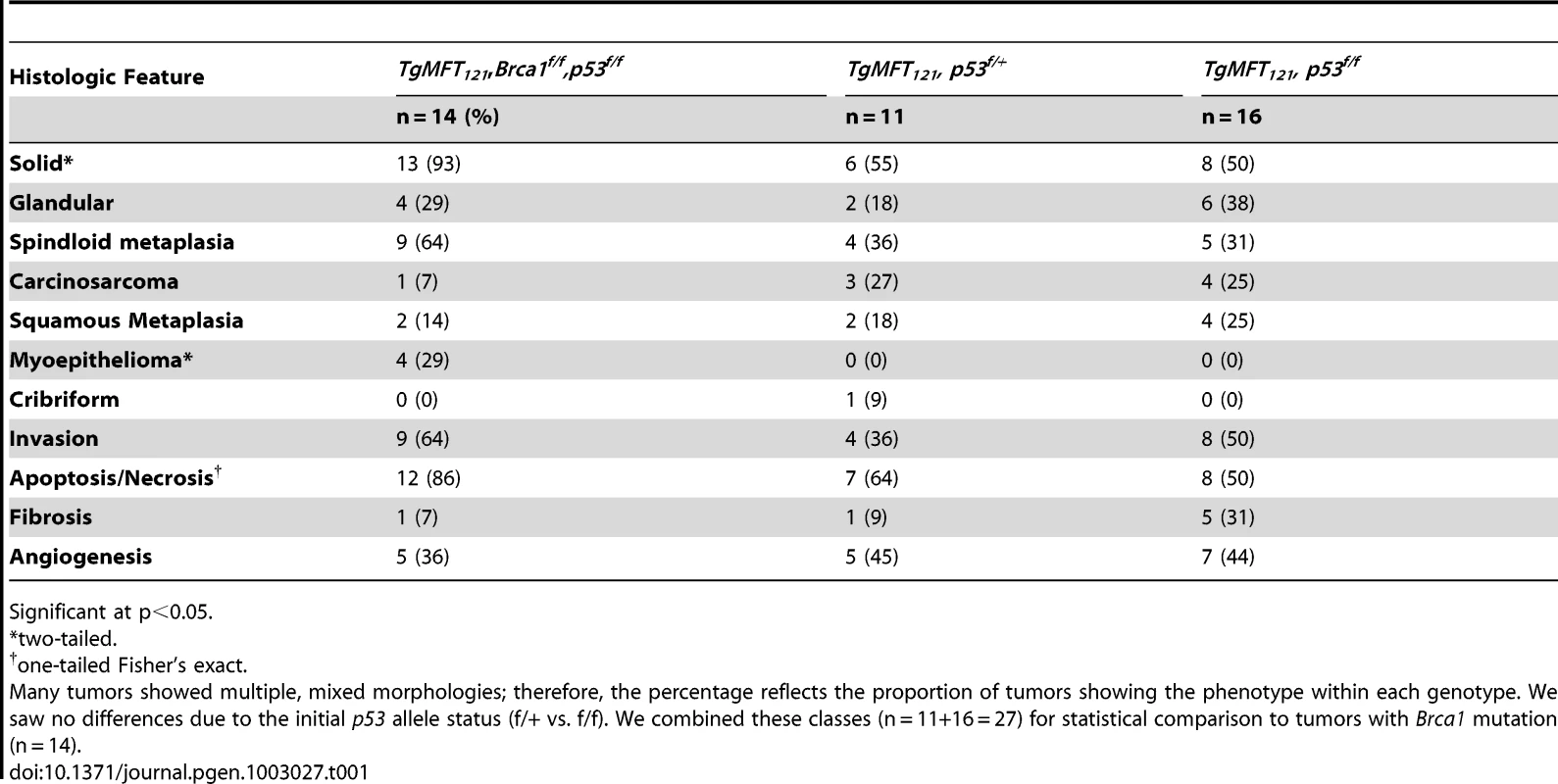
Significant at p<0.05. Both TP and TBP mice (4/16 vs. 1/14 cases, p = 0.3359) developed carcinosarcomas (Figure 4C), also known as “EMT” (Epithelial to Mesenchymal Transition) tumors [33]. These tumors are comprised of faintly-staining, fusiform, spindloid cells and are characteristic of p53 mutant mice. The histology of hematoxylin and eosin stained carcinosarcomas is relatively homogeneous and is visibly distinct from carcinomas with spindloid metaplasia that showed biphasic carcinomatous and spindloid morphologies (Figure 4B, 4G). Among both carcinosarcomas and metaplastic tumors, we observed dual expression of mesenchymal (Vimentin) and epithelial (Keratin-8) markers (Figure 4G), which are mutually exclusive lineage markers in the normal gland. Dual expression is also a feature of human Claudin-low [4] and mouse EMT tumors [33]. Poorly differentiated spindloid cells along invasive tumor fronts showed either dual expression (Figure 4G) or greatly diminished epithelial marker expression (Figure 4J–4L).
In both TP and TBP tumors, we observed squamous metaplasia that was characterized by keratin nests (Figure 4M) and high expression of Keratin 6 (Figure 4N), a marker of progenitor cells that is expanded in Wnt-1-induced mammary tumors. Four of fourteen cases (29%) of TBP tumors, but zero cases of TP tumors (p = 0.0365), included whorls of spindloid cells that resembled myoepithelial cells (not shown). Finally, central necrosis, a hallmark feature of human BRCA1-mutated tumors, was present in twelve of fourteen TBP cases, a greater proportion than the eight of sixteen T121/p53 cases (p = 0.0577), indicating that the selective pressure of cell death persists even in the absence of p53 activity.
Distant metastases were observed in the lungs of both TP (4 of 14 cases) and TBP (3 of 6 cases) mice (Figure 4P), which is noteworthy because relatively few transgenic mammary tumor models metastasize. Among mouse models that do metastasize, most are derivatives of the PyMT model, which more closely resembles human Luminal subtype tumors than Basal-like tumors [3]. Metastases were not evident in the sternum, liver, or spleen of these mice. A primary cell line established from a TBP tumor formed secondary tumors following serial transplantation into syngeneic (FVB) mammary fat pads (Patel, unpublished), confirming the malignant capacity of TBP tumors.
In summary, TBP tumors displayed heterogeneous histology, including high grade, central necrosis, metaplasia, pushing boarders, and metastasis, features that are characteristic of human BRCA1-mutated and Claudin-low breast tumors.
Global gene expression analysis
The majority of the TgMFT121 mouse tumors (76%) co-segregated with human Basal-like breast cancers by hierarchical clustering of the top ∼1000 most variable genes in a combined data set of mouse (n = 135) and human (n = 337) tumor expression profiles (Figure 5, Figure S1, File S2). Using a similar approach that we reported previously [3], we assayed global transcript levels by microarray using tumors derived from TBP (n = 8) and TP mice (n = 9), and we compared them directly to tumors and normal tissue from other mouse models [3] and patient samples [4]. Two TP tumors clustered with human tumors that showed a mixture of the PAM50 molecular subtypes that were assigned by Prat and colleagues (2010). The two remaining TP tumors clustered with Claudin-low subtype human tumors.
Fig. 5. Cross-species comparison of breast cancers. 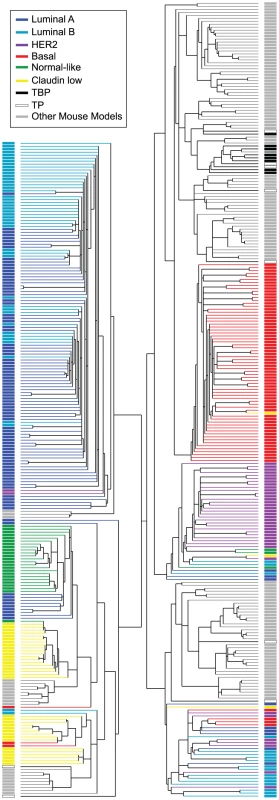
TBP (black boxes, n = 8) and TP (open black boxes, n = 9) tumors were compared to published mouse (gray boxes, n = 135) and human (n = 337) microarray expression profiles (colored according to PAM50 subtype). Most (76%) of our TgMFT121 mouse tumors cluster with human Basal-like breast cancers (red boxes). The Treeview files of the clustering analysis are available in File S2. The effect of Brca1 mutation on TP tumors was more evident when we focused our analysis on the mouse specimens alone (Figure 6, File S3). 56% of the TP tumors (5 of 9) clustered with mouse tumors that we previously showed resemble human Luminal tumors [3]. Brca1 mutation shifted the tumor phenotype (p = 0.0529, Fisher's exact). All eight TBP tumors segregated with mouse tumors, including other Brca1-mutant models that more closely resemble human Basal-like TNBC (Figure 6A). TBP tumors and the other Basal-like mouse tumors expressed low levels of luminal markers and high levels of both Proliferation and Basal cluster transcripts, including Keratins-14, -6b, -17 (Figure 6C). In contrast, the TP tumors that clustered with Luminal-B-like tumors (Figure 5A, blue box) showed higher expression of luminal marker genes that correlate with the estrogen pathway target Xbp1 (Figure 5A). Interestingly, TBP tumors were distinct from most other Basal-like mouse tumors in their elevated expression of a subset of Claudin-low signature genes [3], [4], including Snail1, Tgfbi, Dtr, and Timp1 (Figure 6C).
Fig. 6. TBP tumors share features of Basal-like and Claudin-low expression signatures. 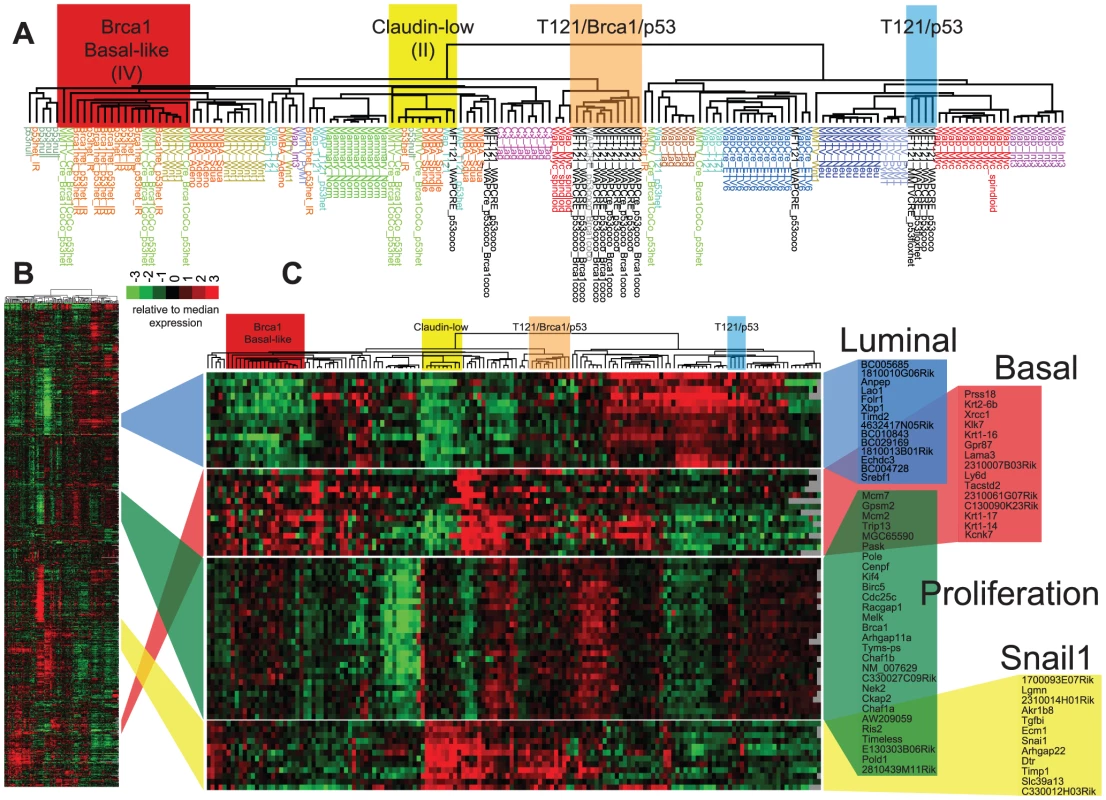
(A) Expression of 866 reference genes of TP (n = 9) and TBP (n = 8) tumors and 13 models of breast cancer. Tumor annotations of the expanded dendrogram (A) are color coded by tumor model. Selected gene clusters (C) correspond to the full data matrix (B). TBP tumors (orange box) cluster with mouse tumors that more closely resemble TNBC on the left branch. 56% (5 of 9) of TP tumors clustered with Luminal-B tumors (blue box). TBP tumors show high expression of Snai1-correlated genes in the Claudin-low cluster. The Treeview files of the clustering analysis are included in Table S1 in File S1. Four TP tumors (44%) did not segregate with Luminal-like tumors. This finding is consistent with previous reports by us and others that Rb/p53 tumors can also resemble TNBC and the Claudin-low molecular phenotype [3], [18], [20]. A single TP tumor clustered among the previously designated Group II tumors (Figure 6A, yellow box), which are the paradigm cases of the Claudin-low subtype [3]. In addition, a single TP tumor clustered with tumors with a squamous metaplastic histology. Finally, two TP tumors co-segregated with the TBP tumors (Figure 6A, orange box), which is not surprising given that Rb is one of the most frequently deleted loci among Brca1/p53-mutated mouse tumors [22].
Pathway analysis
The similarity between TBP tumors and human Claudin-low and Basal-like cancers was also evident from pathway analysis of up-regulated genes of each of the three tumor types (Figure 7A, File S1). We queried the KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) databases with lists of genes that were differentially expressed by TBP tumors (see Methods) and by human Claudin-low and Basal-like tumors [4]. Cytokine, chemokine, and MAPK signaling pathways ranked highly among both Claudin-low and TBP tumors. Pathways that are enriched in cancers of diverse origins ranked highly in both Basal-like and murine TBP tumors.
Fig. 7. Intersecting KEGG pathway terms. 
(A) The genes differentially expressed among mouse TBP tumors show enrichment of KEGG pathways associated with human Claudin-low and Basal-like tumors. (B) The pathway terms are colored according to the intersections depicted in the Venn diagram. The full KEGG pathway lists are included in Table S1 in File S1. The GO terms associated with the respective tumor types were consistent with the enriched KEGG pathways. Cell-cycle progression (GO:0007049, p = 2.43551E-59) and DNA repair (GO:0034984, p = 6.95081E-22) dominate the list of functions enriched in Basal-like tumors (File S1). Similarly, regulation of cell proliferation (GO:0042127, p = 6.01E-13) is among the top terms for TBP tumors. The three top scoring, inter-related GO terms for TBP tumors are regulation of developmental process (GO:0050793, p = 7.50E-16), organ morphogenesis (GO:0009887, p = 3.53E-14), and tissue development (GO:0009888, p = 1.36E-13). These GO terms are reflective of the enrichment of the Wnt, ErbB, TGF-β, and VEGF signaling pathways identified by KEGG pathway analysis. Claudin-low tumors are enriched for wound (GO:0009611, p = 4.29939E-66) and inflammatory responses (GO:0006954, p = 1.26817E-50), which are also among the top functions associated with TBP tumors (7.37E-13 and 6.46E-12, respectively).
CGH analysis
Given the requirement for BRCA1 in DNA damage repair and centrosome regulation, we tested the hypothesis that TBP tumors harbor more genomic copy number aberrations (CNAs) than do TP tumors with intact Brca1. We enumerated CNAs by counting “copy number transitions,” the number of changes in the CGH profile from one copy number level to another that occur within chromosomes [34]. Unexpectedly, we found no statistically significant difference (p = 0.8374) in the mean number of CNAs between TBP tumors (n = 8) and TP tumors (n = 10) using array-based comparative genomic hybridization (aCGH).
The low multiplicity of TBP and TP tumors (1–3 per mouse) and their latency indicate that combinations of pRbf, Brca1, and p53 pathway perturbations are not sufficient for malignant transformation in our models. To identify potentially collaborating oncogenic events, we manually curated loci with copy number changes (see Methods). In nine tumors (50%), we observed recurrent losses of large, variable regions spanning chr4 and chr10 (Figure 8). Both chromosomes harbor many potential tumor suppressors, including regulators of cell death, such as Tm2d1, Utp11l, Trp73, Dffa, Runx3, Lck, Dhcr24, Faf1, Pax7, and Casp9, and effectors of cell death, such as Col18a1, Gadd45b, Dapk3, and Casp14. Among all the tumors assayed (n = 18), we identified nearly five-hundred loci (Table S9 in File S1) with potential copy number gains. Approximately half of the genes are included on curated lists of cancer-associated genes, including the Cancer Gene Census (Sanger Institute) and the KEGG Pathways in Cancer. We observed focal amplification of several canonical proto-oncogenes, including c-Myc amplification (log2ratio = 3.64, p<0.0001) in a single TP tumor, H-ras amplification in two of ten TP tumors, and K-ras amplification in two of eight TBP tumors. Pathway analysis of these five-hundred putative collaborating genes revealed enrichment of several signaling pathways, including the MAP Kinase, Focal Adhesion, Wnt, and ErbB pathways (Table S10 in File S1).
Fig. 8. Comparative genomic hybridization. 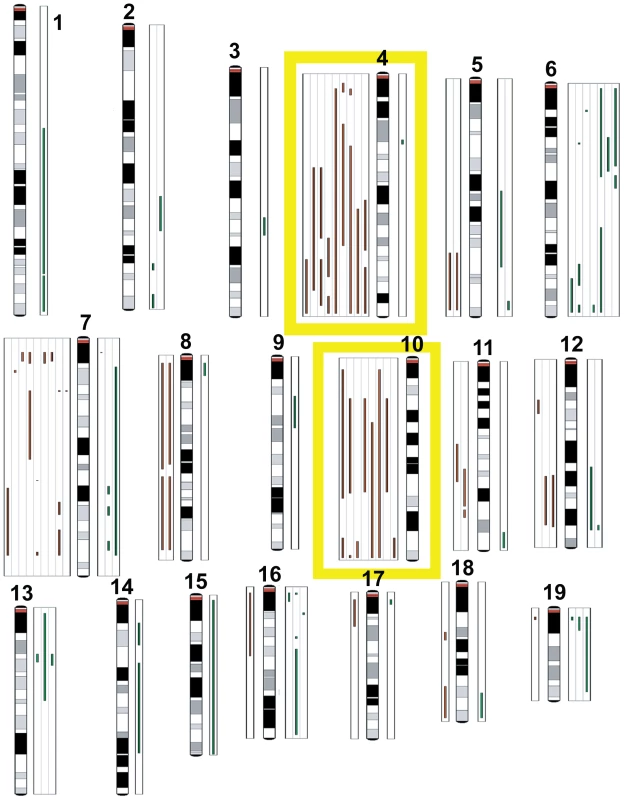
TBP tumors (n = 8) and TP tumors (n = 10) were analyzed by array CGH to identify copy number aberrations (CNAs). Green lines to the right of the chromosome ideograms indicate gains and red lines to the left indicate losses of individual tumor samples. No significant difference was found between the average number of CNAs between TBP and TP tumors (p = 0.8374). Frequent losses were seen on chromosomes 4 and 10. Other recurrent losses included chromosome 7. Frequent gains were associated with chromosome 6. Discussion
Here we report a highly penetrant engineered mouse model of TNBC. Our previous work showed that when pRbf and p53 are simultaneously perturbed in mammary epithelium, adenocarcinomas develop with long latency, suggesting a requirement for additional oncogenic events. However, these mouse tumors displayed only limited chromosomal copy number aberrations [19]. Because genomic instability is a hallmark of malignant transformation [35], especially among BRCA1 familial cancers [36] and aggressive sporadic breast cancers [37], we hypothesized that Brca1 mutation would accelerate the tumor development we observed following dual inactivation of pRbf and p53. Our results show that concomitant inactivation of all three tumor suppressor pathways in mammary epithelium has an additive effect on tumor latency and predisposes highly penetrant, malignant carcinomas. Although Brca1 inactivation accelerated tumorigenesis compared to TP tumors, we observed no statistical difference between chromosomal copy number transitions in tumors with or without Brca1, despite the extensive CNAs observed by others in Brca1/p53 tumors [22].
The pRb-regulated cell cycle network is frequently disrupted in TNBC tumors [7], [26], [37], [38], and the Rb locus is among the most frequently lost in Brca1/p53 mouse tumors [22], indicating that there is strong selective pressure for Rb pathway inactivation. We speculate that direct inactivation of pRbf by T121 may allow TBP cells to escape this rate-limiting barrier of transformation without accruing numerous chromosomal aberrations. Thus, in the context of defective pRb and p53 function, tumor progression may be unrelated to the proportion of the genome altered by copy number alterations. It will be important to determine the effect of Brca1 loss on the abundance and identities of somatic mutations that are not detectable by CGH.
The importance of p53 mutation in breast cancers is well documented and is confirmed in the present study. The dual inactivation of pRbf and Brca1 caused markedly increased cell death that was reduced by p53 mutation. p53-independent cell death likely remains a significant barrier to tumor progression among TBP tumors and may account, in part, for the observed loss of genomic regions that harbor cell death regulatory genes, most notably on chr4 where the p53 paralog p73 resides. Identifying the genomic alterations that are conserved across species will be useful in evaluating the impact of the myriad of CNAs observed in breast cancers and may help to explain the heterogeneity of TNBCs.
Brca1 mutation not only accelerated tumor development but also shifted the tumor spectrum. Whereas T121/p53 mouse tumors often resembled the Luminal-B molecular subtype breast cancers, which show relatively abundant expression of luminal epithelial cell differentiation markers, TBP tumors consistently shared features of Basal-like and Claudin-low molecular subtypes. Others have argued that Basal-like and Claudin-low gene expression signatures reflect progenitor and stem cell phenotypes, respectively [4], [16], consistent with a role for Brca1 in mediating stem/progenitor cell maturation [15]. Loss of BRCA1 activity may also alter tumor phenotype through deregulation of the EMT inducer SLUG [39].
The CGH analysis of our mouse tumors revealed CNAs consistent with mutations observed in genomic surveys of human breast cancers [40], [41]. Similar to the studies of human tumors, we saw increased copy numbers of known oncogenic driver genes, including myc, egfr, crebbp, jak1, H-ras, and K-ras, as well as enrichment of pathways implicated in tumor progression, including the WNT signaling pathway, regulation of actin cytoskeleton, focal adhesion, cell shape, and mobility proteins. Far fewer investigations have focused on genetic deletions and cancer development mechanisms. We also found decreased copy numbers of known tumor suppressors, including map2k, ppp2r, and pten. Given the strong similarities between our mouse model and aggressive human breast cancers, the TBP model provides an invaluable preclinical platform to identify and assess potential therapeutics for aggressive and chemoresistant breast cancer subtypes [42], [43].
Materials and Methods
Ethics statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Derivation of MFT121 transgenic mice
The LoxP-eGFP-Stop-LoxP cassette and T121-encoding DNA were cloned into EcoRI HindIII sites of MMTV-SV40-Bssk (Addgene plasmid 1824). The LoxP-eGFP-Stop-LoxP cassette was provided courtesy of the T. Jacks lab. Resulting and subsequent generation MFT121 transgenic mice were identified by PCR amplification of a 215-bp fragment using the oligo pair: 5′-GCATCCAGAAGCCTCCAAAG -3′ and 5′-GAATCTTTGCAGCTAATGGACC-3′ complementary to the T121 sequence. Cre transgenic mice were identified using the oligo pair: 5′-TGATGAGGTTCGCAAGAACC-3′ and 5′-CCATGAGTGAACGAACCTGG-3′. The cycling profile was 94°C for 2 min., 35 cycles of 94°C for 20 sec., 62°C for 45 sec., and 72°C for 45 sec.; the final incubation of 72°C was for 2 min. We established five TgMFT121 founder transgenic lines, though three lines failed to express the eGFP reporter. We describe here our studies of the single mouse line with higher eGFP expression in virgin mammary glands. eGFP expression was also evident in salivary glands and foot pads in this line (data not shown).
Transgenic breeding strategies
TgMFT121;TgWAP-Cre mice were mated to p53 conditional allele mice (p53f/f) [44]. p53 genotypes were determined by PCR using two reactions: neomycin primer 5′-TCCTCGTGCTTTACGGTATC-3′, p53 primer 5′-TATACTCAGAGCCGGCCT-3′, 525-bp product; the endogenous p53 allele; substituting 5′-ACAGCGTGGTGGTACCTTAT-3′ for the neo primer, 475-bp product. Cycling parameters were the same as they were for the T121 reaction. We produced female mice with the genotypes TgMFT121;TgWAP-Cre;p53f/+ and TgMFT121;TgWAP-Cre;p53f/f, and female littermates served as controls. To study the effect of Brca1 loss, TgMFT121; TgWAP-Cre mice were mated to Brca1f/f,p53f/f mice [44]. Brca1 genotypes were determined by PCR using two reactions. We generated female mice with the genotypes TgMFT121; TgWAP-Cre; Brca1f/+,p53f/+ and TgMFT121; TgWAP-Cre; Brca1 f/f,p53f/f with nontransgenic (Cre negative) littermate controls for each cohort. Pregnancy induced WAP-Cre transgene expression. Parturition of the first litter was designated as Day 1 for all aging studies. Matings with TgMMTV-Cre mice (Line F) yielded small litter sizes; therefore, experiments reported here employed TgWap-Cre unless otherwise indicated.
Histopathology and apoptosis assays
A portion of each mammary sample was fixed overnight in 10% phosphate-buffered formalin, transferred to 70% ethanol, and then embedded in paraffin. Samples were sectioned for 10 successive layers at 5-µm intervals and stained with hematoxylin and eosin for histopathologic examination, as described previously. Apoptosis levels were assessed using the terminal deoxynucleotidyl transferase–mediated dUTP-biotin nick end labeling (TUNEL) method with standard protocols. Differences in apoptosis levels between mice with different genotypes were evaluated by the t test (p<0.05 was deemed statistically significant).
Immunostaining
Immunohistochemical analysis was performed using formalin-fixed paraffin sections. Antigen retrieval for all antibodies was done by boiling the slides in citrate buffer (pH 6.0) for 15 min. Antibodies were α-cytokeratins 8/18 (Ker8/18, 1∶450 Progen, GP11), α-cytokeratin 5 (K5, 1∶8000, Covance, PRB-160P), smooth muscle actin (SMA; 1∶1,000, mouse A2537; Sigma, St. Louis, MO), anti–phosphorylated histone H3 (1∶100, rabbit 06-570; Upstate, Waltham, MA) and SV40 (monoclonal Ab2, 1∶100; Oncogene, Cambridge, MA). All immunofluorescence reactions were done using AlexaFlour-conjugated secondary antibodies (AlexaFluor 488 and 594, Molecular Probes). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) using Hardset Mounting Media (Vector Laboratories).
Microarray analysis
We compared microarray profiles of T121/p53 (n = 9) and TBP (n = 8) tumors to published microarray profiles (n = 152) using two-way hierarchical clustering (centroid linkage) of 866 “intrinsic genes” [3], [45]. Total RNA was collected from end-stage tumors. RNA was purified using the Qiagen RNeasy Mini Kit according to the manufacturer's protocol using 20–30 mg tissue. RNA integrity was assessed using the RNA 6000 Nano LabChip by Bioanalyzer (Agilent). Two micrograms of total RNA were reverse transcribed, amplified, and labeled with Cy5 using a Low RNA Input Amplification kit (Agilent). Common reference RNA consisted of total RNA harvested from equal numbers of C57Bl6/J and 129 male and female Day 1 pups (courtesy of Dr. Cam Patterson, UNC). Reference RNA was reverse transcribed, amplified, and labeled with Cy3. The amplified sample and reference were co-hybridized overnight to Agilent Mouse Oligo Microarrays (G4121A). They were then washed and scanned on an Axon GenePix 4000B scanner, analyzed using GenePix 4.1 software, and uploaded into the UMD database (https://genome.unc.edu/) where Lowess normalization is automatically performed. All data were submitted to GEO (GSE34479). The genes for all analyses were filtered by: 1) requiring intensity values in both channels to have a mean Lowess normalized intensity of >10, 2) Values being reported in >70% of the samples, and 3) the absolute value of the log2 of the ratio of Channel 2/Channel 1 for at least three arrays having to be >1.6. Hierarchical clustering was performed using Cluster v3.0 and displayed using JavaTreeview v1.0.8.
We identified 871 differentially expressed TBP transcripts using SAM implemented in BRB-ArrayTools (R. Simon and the BRB-ArrayTools Development Team, NCI; Table 1; FDR 0.0485, delta 0.92931). Gene ontology analyses were performed using the FatiGO tool (Babelomics ver. 4.2, babelomics.bioinfo.cipf.es). Mouse gene symbols were converted to human EntrezIDs using Agilent annotations and the Mouse Genome Informatics database of The Jackson Laboratory for comparisons. The Fisher's exact two-tailed test was used to determine significance of GO (biological process [levels 3–9]) and KEGG pathway terms. Terms with p<0.05 are reported.
Combined murine and human expression data sets
For the mouse tumor data set, IDs for 21,670 unfiltered probes were retrieved (GSE34479). Human EntrezIDs were assigned to orthologous genes as above. Mean values of redundant mouse probes were calculated, missing values were imputed, the columns were standardized to N(0,1), and the rows were median centered using R (ver. 2.10.0). For the human data set, we downloaded the file “UNC337arraydata_imputedCollapsed.txt” [4] from the UNC MicroArray Database (genome.unc.edu). The two data sets were corrected for systemic biases using Distance Weighted Discrimination [46]. The combined data set was used for centroid linkage hierarchical clustering analysis.
CGH analysis
We performed array CGH essentially as previously described [47]. Briefly, genomic DNA was isolated from tumors, fluorescently labeled, and competitively hybridized with wt DNA spotted BAC (Bacterial Artificial Chromosomes) arrays. All data were submitted to GEO (GSE40925). We used the HaarSeg algorithm with default parameters implemented in waviCGH (wavi.bioinfo.cnio.es) for chromosomal segmentation of mutations and for CNA calling. We manually curated BAC clones spanning putative CNAs with a conservative tumor:normal DNA threshold of log2ratio >0.5 or <−0.5. Genes mapping to the BAC clones were identified using the National Center for Biotechnology Map Viewer and the Jackson Laboratory Mouse Genome Database. Gene lists were compared to the Cancer Gene Census (Sanger Institute), the KEGG Pathways in Cancer, Atlas Genetics Oncology, and the Michigan Molecular Interactions database. For the pathway analysis, we used the SAM algorithm implemented in BRB-Array Tools to identify differentially expressed genes (TBP_SAM) among TBP tumors versus the rest of the mouse tumors in the data set. We interrogated KEGG (TBP_KEGG) and GO (TBP_GO_BP) databases using the FatiGO algorithm implemented in the Babelomics (ver. 4.2) suite of bioinformatics tools (babelomics.bioinfo.cipf.es). We compared these results to the differentially expressed genes reported by Prat et al. (2010) of human Claudin-low (CL_KEGG, CL_GO_BP) and Basal-like (Basa_KEGG, Basal-_GO_BP) tumors. We also used the FatiGO tool in the Babelomics 4.3.0 integrative platform with parameters set for the Fisher's exact two-tailed test to determine the most alternately expressed KEGG pathways among the amplified genes determined by CGH analysis.
Supporting Information
Zdroje
1. CareyL, WinerE, VialeG, CameronD, GianniL (2010) Triple-negative breast cancer: Disease entity or title of convenience? Nat Rev Clin Oncol 7 (12)
683–692.
2. RakhaEA, El-SayedME, Reis-FilhoJ, EllisIO (2009) Patho-biological aspects of basal-like breast cancer. Breast Cancer Res Treat 113 (3)
411–422.
3. HerschkowitzJI, SiminK, WeigmanVJ, MikaelianI, UsaryJ, et al. (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8 (5)
R76.
4. PratA, ParkerJS, KarginovaO, FanC, LivasyC, et al. (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12 (5)
R68.
5. LiedtkeC, MazouniC, HessKR, AndreF, TordaiA, et al. (2008) Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 26 (8)
1275–1281.
6. CareyLA, DeesEC, SawyerL, GattiL, MooreDT, et al. (2007) The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res 13 (8)
2329–2334.
7. HerschkowitzJI, HeX, FanC, PerouCM (2008) The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res 10 (5)
R75.
8. SorlieT, PerouCM, TibshiraniR, AasT, GeislerS, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98 (19)
10869–10874.
9. SorlieT, TibshiraniR, ParkerJ, HastieT, MarronJS, et al. (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A 100 (14)
8418–8423.
10. Borresen-DaleAL (2003) TP53 and breast cancer. Hum Mutat 21 (3)
292–300.
11. SchuyerM, BernsEM (1999) Is TP53 dysfunction required for BRCA1-associated carcinogenesis? Mol Cell Endocrinol 155 (1–2)
143–152.
12. EstellerM, SilvaJM, DominguezG, BonillaF, Matias-GuiuX, et al. (2000) Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92 (7)
564–569.
13. WalshT, KingMC (2007) Ten genes for inherited breast cancer. Cancer Cell 11 (2)
103–105.
14. WelcshPL, LeeMK, Gonzalez-HernandezRM, BlackDJ, MahadevappaM, et al. (2002) BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc Natl Acad Sci U S A 99 (11)
7560–7565.
15. LiuS, GinestierC, Charafe-JauffretE, FocoH, KleerCG, et al. (2008) BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A 105 (5)
1680–1685.
16. LimE, VaillantF, WuD, ForrestNC, PalB, et al. (2009) Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15 (8)
907–913.
17. DrostRM, JonkersJ (2009) Preclinical mouse models for BRCA1-associated breast cancer. Br J Cancer 101 (10)
1651–1657.
18. ChengL, ZhouZ, Flesken-NikitinA, ToshkovIA, WangW, et al. (2010) Rb inactivation accelerates neoplastic growth and substitutes for recurrent amplification of cIAP1, cIAP2 and Yap1 in sporadic mammary carcinoma associated with p53 deficiency. Oncogene 29 (42)
5700–5711.
19. SiminK, WuH, LuL, PinkelD, AlbertsonD, et al. (2004) pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol 2: e22 doi:10.1371/journal.pcbi.0010022.
20. JiangZ, DengT, JonesR, LiH, HerschkowitzJI, et al. (2010) Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest 120 (9)
3296–3309.
21. LuX, YangC, YinC, Van DykeT, SiminKJ (2011) Apoptosis is the essential tumor suppression function of p53 during carcinoma progression. Mol Cancer Res
22. HolstegeH, van BeersE, VeldsA, LiuX, JoosseSA, et al. (2010) Cross-species comparison of aCGH data from mouse and human BRCA1 - and BRCA2-mutated breast cancers. BMC Cancer 10 : 455.
23. ScambiaG, LovergineS, MasciulloV (2006) RB family members as predictive and prognostic factors in human cancer. Oncogene 25 (38)
5302–5308.
24. ErtelA, DeanJL, RuiH, LiuC, WitkiewiczAK, et al. (2010) RB-pathway disruption in breast cancer: Differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle 9 (20)
4153–4163.
25. BiecheI, LidereauR (2000) Loss of heterozygosity at 13q14 correlates with RB1 gene underexpression in human breast cancer. Mol Carcinog 29 (3)
151–158.
26. ShahSP, RothA, GoyaR, OloumiA, HaG, et al. (2012) The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature
27. SiminK, HillR, SongY, ZhangQ, BashR, et al. (2005) Deciphering cancer complexities in genetically engineered mice. Cold Spring Harb Symp Quant Biol 70 : 283–290.
28. WagnerKU, WallRJ, St-OngeL, GrussP, Wynshaw-BorisA, et al. (1997) Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res 25 (21)
4323–4330.
29. Clark-KnowlesKV, SentermanMK, CollinsO, VanderhydenBC (2009) Conditional inactivation of Brca1, p53 and rb in mouse ovaries results in the development of leiomyosarcomas. PLoS ONE 4: e8534 doi:10.1371/journal.pone.0008534.
30. JonkersJ (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature Genetics 29 (4)
418.
31. XiaoA, WuH, PandolfiPP, LouisDN, Van DykeT (2002) Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell 1 (2)
157–168.
32. JerryDJ, KuperwasserC, DowningSR, PinkasJ, HeC, et al. (1998) Delayed involution of the mammary epithelium in BALB/c-p53null mice. Oncogene 17 (18)
2305–2312.
33. CardiffRD (2010) The pathology of EMT in mouse mammary tumorigenesis. J Mammary Gland Biol Neoplasia 15 (2)
225–233.
34. SnijdersAM, FridlyandJ, MansDA, SegravesR, JainAN, et al. (2003) Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene 22 (28)
4370–4379.
35. HanahanD, WeinbergRA (2011) Hallmarks of cancer: The next generation. Cell 144 (5)
646–674.
36. JonssonG, NaylorTL, Vallon-ChristerssonJ, StaafJ, HuangJ, et al. (2005) Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res 65 (17)
7612–7621.
37. FridlyandJ, SnijdersA, YlstraB, LiH, OlshenA, et al. (2006) Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer 6 (1)
96.
38. GauthierML, BermanHK, MillerC, KozakeiwiczK, ChewK, et al. (2007) Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 12 (5)
479–491.
39. ProiaTA, KellerPJ, GuptaPB, KlebbaI, JonesAD, et al. (2011) Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8 (2)
149–163.
40. CurtisC, ShahSP, ChinSF, TurashviliG, RuedaOM, et al. (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486 (7403)
346–352.
41. ChinK, DeVriesS, FridlyandJ, SpellmanPT, RoydasguptaR, et al. (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10 (6)
529–541.
42. HennessyBT, Gonzalez-AnguloAM, Stemke-HaleK, GilcreaseMZ, KrishnamurthyS, et al. (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69 (10)
4116–4124.
43. TaubeJH, HerschkowitzJI, KomurovK, ZhouAY, GuptaS, et al. (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A 107 (35)
15449–15454.
44. LiuX, HolstegeH, van der GuldenH, Treur-MulderM, ZevenhovenJ, et al. (2007) Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A 104 (29)
12111–12116.
45. LiZ, TognonCE, GodinhoFJ, YasaitisL, HockH, et al. (2007) ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell 12 (6)
542–558.
46. BenitoM, ParkerJ, DuQ, WuJ, XiangD, et al. (2004) Adjustment of systematic microarray data biases. Bioinformatics 20 (1)
105–114.
47. SnijdersAM, NowakNJ, HueyB, FridlyandJ, LawS, et al. (2005) Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res 15 (2)
302–311.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



