-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genomic Variation and Its Impact on Gene Expression in
Understanding the relationship between genetic and phenotypic variation is one of the great outstanding challenges in biology. To meet this challenge, comprehensive genomic variation maps of human as well as of model organism populations are required. Here, we present a nucleotide resolution catalog of single-nucleotide, multi-nucleotide, and structural variants in 39 Drosophila melanogaster Genetic Reference Panel inbred lines. Using an integrative, local assembly-based approach for variant discovery, we identify more than 3.6 million distinct variants, among which were more than 800,000 unique insertions, deletions (indels), and complex variants (1 to 6,000 bp). While the SNP density is higher near other variants, we find that variants themselves are not mutagenic, nor are regions with high variant density particularly mutation-prone. Rather, our data suggest that the elevated SNP density around variants is mainly due to population-level processes. We also provide insights into the regulatory architecture of gene expression variation in adult flies by mapping cis-expression quantitative trait loci (cis-eQTLs) for more than 2,000 genes. Indels comprise around 10% of all cis-eQTLs and show larger effects than SNP cis-eQTLs. In addition, we identified two-fold more gene associations in males as compared to females and found that most cis-eQTLs are sex-specific, revealing a partial decoupling of the genomic architecture between the sexes as well as the importance of genetic factors in mediating sex-biased gene expression. Finally, we performed RNA-seq-based allelic expression imbalance analyses in the offspring of crosses between sequenced lines, which revealed that the majority of strong cis-eQTLs can be validated in heterozygous individuals.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003055
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003055Summary
Understanding the relationship between genetic and phenotypic variation is one of the great outstanding challenges in biology. To meet this challenge, comprehensive genomic variation maps of human as well as of model organism populations are required. Here, we present a nucleotide resolution catalog of single-nucleotide, multi-nucleotide, and structural variants in 39 Drosophila melanogaster Genetic Reference Panel inbred lines. Using an integrative, local assembly-based approach for variant discovery, we identify more than 3.6 million distinct variants, among which were more than 800,000 unique insertions, deletions (indels), and complex variants (1 to 6,000 bp). While the SNP density is higher near other variants, we find that variants themselves are not mutagenic, nor are regions with high variant density particularly mutation-prone. Rather, our data suggest that the elevated SNP density around variants is mainly due to population-level processes. We also provide insights into the regulatory architecture of gene expression variation in adult flies by mapping cis-expression quantitative trait loci (cis-eQTLs) for more than 2,000 genes. Indels comprise around 10% of all cis-eQTLs and show larger effects than SNP cis-eQTLs. In addition, we identified two-fold more gene associations in males as compared to females and found that most cis-eQTLs are sex-specific, revealing a partial decoupling of the genomic architecture between the sexes as well as the importance of genetic factors in mediating sex-biased gene expression. Finally, we performed RNA-seq-based allelic expression imbalance analyses in the offspring of crosses between sequenced lines, which revealed that the majority of strong cis-eQTLs can be validated in heterozygous individuals.
Introduction
An important challenge in biology is to elucidate the relationship between genetic and phenotypic variation [1]. The increasing availability of comprehensive genome sequences of both human [2], [3] and model organism populations [4], [5] constitutes an important step towards meeting this challenge. The Drosophila Genetic Reference Panel (DGRP) is an example of such a recently emerging population resource, consisting of 192 sequenced wild-derived inbred Drosophila melanogaster lines [6], [7]. Drosophila is a premier model organism to understand genome function given the availability of powerful and cost-effective genetic tools and resources [8]–[10]. In addition, its genome is small, but highly polymorphic [7], [11]–[15], which has already proven helpful in studying the molecular basis of morphological evolution [16], [17]. Moreover, linkage disequilibrium (LD) decays quickly across the genome [7], [18], which is favorable to elucidating the relationship between genotypic and phenotypic variation at high resolution. This requires that we genotype all classes of segregating variants (i.e., insertions, deletions, and structural variants) and not only the commonly studied single nucleotide polymorphisms (SNPs) to investigate their effect on phenotypes. Here, we use an integrative approach to derive and characterize a genome-wide, nucleotide-resolution catalog of variants including SNPs, insertions, deletions, complex substitutions, and structural variants in 39 DGRP lines. We then investigate the impact of naturally occurring genetic variation on adult gene expression, revealing novel insights into the regulatory architecture of gene expression variation in Drosophila.
Results
Variant discovery
We generated a catalog of sequence variants using whole-genome Illumina next-generation sequencing data from 39 inbred lines from the Drosophila Genetic Reference Panel (DGRP) for which gene expression data are available for young adult flies [6]. First, we used PrInSeS-G [19], which uses de novo local assembly to generate a preliminary list of variant calls for each DGRP line; this tool works by first detecting regions with a fluctuation in sequencing coverage, then using a short fragment from the reference sequence as a seed to build a contig by extending it using the reads; the contig extending stops when another short fragment from the reference sequence is encountered. We then re-aligned each resulting genome against the reference genome to present variants in a coherent way among lines and to reduce variant fragmentation. Finally, we developed and used a genotyping algorithm, which uses the combined variant call set for all DGRP lines to improve variant discovery. This is particularly useful for genomes with low read coverage (Figure S1). This strategy allowed us to identify more than 3.6 million unique sequence variants across all 39 DGRP lines including 2.8 million SNPs, 0.6 million indels, and 0.2 million complex sequence variants (Figure 1A, Table 1). To validate our variant calls, we used five distinct approaches. First, we compared our variant catalogue to indel data from the FLYSNPdb database [20]. We validated 713 deletions and 923 insertions on the breakpoint and complete sequence level, thus covering in total 45% of the FLYSNPdb content. Second, we compared our SNP calls with those published by Mackay et al. [7], and found that 94% of calls made in that study match (Table S1). Third, we used whole genome Roche-454 reads available for the same lines to validate our SNP calls, resulting in confirmation of 98.7% of SNPs compared (Table S2). Fourth, we sequenced mRNA from three DGRP lines (see also below) and differentially aligned the reads to their respective transcriptomes and the reference transcriptome in order to validate variants in coding sequence. Thus, we examined, on average, 147,453 SNPs and 12,084 non-SNPs per line that were covered by mRNA sequencing reads, confirming on average 98.9% of all variant types within coding sequence for these lines (Table S3). The false discovery rate (FDR) is 0.5% for SNPs and 1.5% for indels and complex variants. Moreover, the FDR remains stable for indels up to 30 bp in size but increases to 8.9% for larger indels. Note that some variants could not be reliably assigned to the true or false positive categories due to low coverage. Finally, we validated 92 large indels (200 bp to 5.6 kb) in five DGRP lines using PCR (true positives 77.9%, false positives 7.2%, variant size different from predicted 14.9%, Table S4). Moreover, Sanger sequencing of 10 randomly selected large indels validated by PCR revealed a high breakpoint accuracy, whereby breakpoints of 9 out of 10 indels were perfectly reconstructed consistent with results based on query of smaller indels in the FLYSNPdb database. Together, these results indicate that our variant catalogue is of similar quality as variant data produced for other organisms using high-throughput sequencing since an overall false discovery rate of less than 10% and an overall accuracy of ∼95% or higher for all variant types (i.e., SNPs, indels and complex variants) is in line with numbers reported by these other studies [4], [21]–[23].
Fig. 1. Overview of variants. 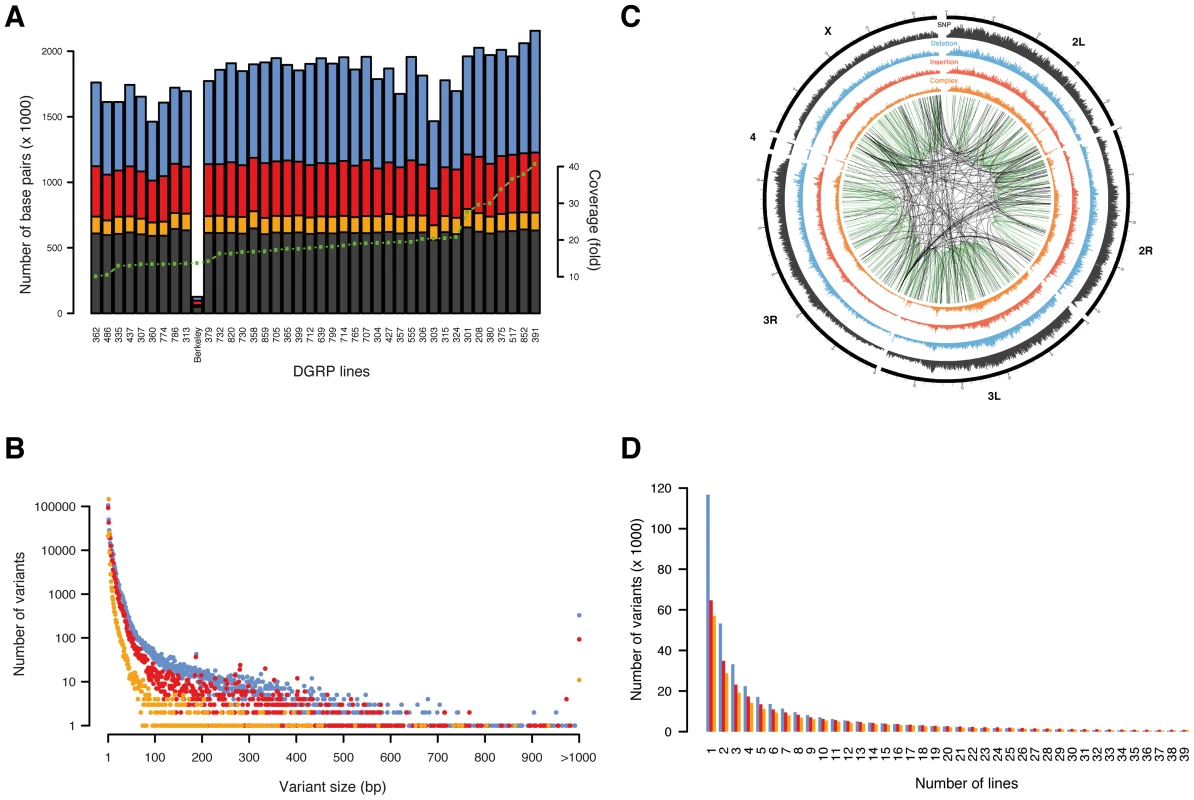
SNPs are shown in black/grey, insertions in red, deletions in blue, and complex variants in orange. (A) Number of base pairs affected by variants discovered per line, with lines ordered by depth of coverage (green dotted line). The line “Berkeley” is the reference line. (B) Number of unique variants by size (note that variants longer than 1,000 bp are grouped in a single x-coordinate). (C) Representation of variant density (0–10 SNPs/kb, 0–5 indels/kb, 0–5 complex variants/kb) across the euchromatic genome (concentric circles) in 50 kb bins. Large variants (>100 bp) mapping against a close homologous sequence (>90% sequence identify) are linked in the center with green lines representing intra-chromosomal- and black lines inter-chromosomal duplications. (D) Number of unique variants by number of lines. Tab. 1. Type classification of variants. 
Indels and complex variants
Among all lines, we identified a SNP every 43 bp and an indel or complex variant every 144 bp, together contributing a genetic marker every 33 bp on average. These findings illustrate a remarkably high density of molecular polymorphisms in Drosophila consistently greater than in humans [3], [24] and mice [4]. Deletions and complex variants affect in total 4.2% (5.0 Mb) of the reference euchromatic genome while insertions add 2.1 Mb (of which 0.5 Mb are in insertions ≥100 bp not found with similarity of 90% or above elsewhere in the reference genome). Non-SNP variants are thus a substantial source of genomic variation in Drosophila. Indel size ranges from 1 to 6,082 bp (Table 1, Figure 1B). Single base pair indels are abundant (32%) and small indels (2–10 bp) represent 50% of the indel repertoire. 6,419 structural variants (≥100 bp) represent 1% of all indels with a median size of 208 bp and encompass ∼2.5 Mb of sequence in total. 850 structural variants are homologous (≥90% sequence similarity) to another part of the assembled reference genome, representing either “young” variants or polymorphic forms of segmental duplications (i.e., copy number variants) [25]. Most (70%) of those structural variants correspond to a duplicon on the same chromosome, indicating that intra-chromosomal duplication events occur more frequently (Figure 1C). These observations support a recently proposed theoretical model governing the formation of segmental duplications in D. melanogaster, whereby, after a double strand break, a search for an ectopic homologous region which is preferentially located within the same chromosomal region triggers the repair mechanism [26].
The majority (73%) of complex variants are balanced multi-nucleotide substitutions and most (61%) constitute di-nucleotide substitutions. Balanced multi-nucleotide substitutions may arise as single nucleotide substitutions that happen to occur in adjacent positions or as multi-nucleotide mutational events [27]. Using simulation within all 1,000 bp genomic windows in all 39 lines, we find that there are on average 3.3 times more di - and 16.3 times more tri-nucleotide substitutions than the number of single nucleotide substitutions expected to be adjacent by chance, indicating that complex mutational events constribute substantially to balanced multi-nucleotide substitutions.
The allele frequency spectrum of indels and complex variants decays steeply, with 28.5% of all indels and complex variants present in only one line (Figure 1D). Comparison to neutral spectra (Figure S2) revealed an excess of low frequency alleles for deletions, consistent with previous results suggesting that more of these variants are under purifying selection [13]. We also observed a depletion of high frequency variants among indels and complex variants in protein-coding regions, splice junctions, and UTR sequences (Figure S3), again suggesting the action of pervasive purifying selection. In contrast, we found 452 deletions, 873 insertions, and 824 complex variants present among all 39 lines (Table S5). To evaluate whether these variants represent rare alleles in the reference genome, population-specific variants, or artifacts in our variant calling workflow, we focused on all 30 such insertions and deletions that were present in protein-coding regions. We found that 77% of these variants exactly recapitulate an ancestral allele (D. simulans, D. yakuba, or D. ananassae) and 90% when allowing at most one mismatch between the indel and the ancestral allele (Table S6). Therefore, these variants predominantly constitute rare alleles in the reference genome. The photoreceptor gene Rh6 is an interesting example: evolutionary conservation analysis around the two observed 17 bp and 2 bp insertions in its coding sequence revealed that both insertions perfectly match the ancestral allele of seven out of 11 Drosophila species (Figure S4). Moreover, the resulting gene model supports an Rh6 cDNA clone of the OregonR/white strain, thus revealing that the reference genome harbors a rare 19 bp deletion, which truncates the gene.
Indels and SNPs are not uniformly distributed across the genome, with autosomal centromeric regions as well as the X chromosome containing fewer variants compared to other genomic regions (Figure 1C). Several models have been proposed to explain this pattern, ranging from purifying selection to recurrent hitchhiking to demography [7], [28], [29], although we cannot rule out that low coverage or read mapping quality issues in those regions has affected variant discovery. Moreover, we found that genome-wide SNP, indel, and complex variant densities are strongly correlated on a 1 Mb scale (Spearman's ρSNP-del = 0.927, ρSNP-ins = 0.90, ρSNP-complex = 0.946; Figure S5), suggesting that similar higher-order constraints such as local selection intensities, recombination, or mutation rates may act on all types of variants. To test the impact of recombination, we tested SNP, indel, and complex variant densities against recombination rates (1 cM/Mb), and found that recombination is strongly positively correlated with variant densities (Spearman's ρSNPs = 0.54, ρinsertions = 0.55, ρdeletions = 0.62, ρcomplex = 0.46), which is line with long-held observations regarding the actions of both pervasive positive selection and background selection in Drosophila [30], [31]. We further examined the SNP distribution around detected indels in more detail, as it has been proposed that indels may act as “mutators” of surrounding sequences [32], [33]. Consistent with this hypothesis, we observed that the SNP density is elevated in close proximity to indels independent of indel type, dropping quickly to background levels (Figure 2A, 2B, and 2C). On average 26% (40%) of all SNPs per line are within 40 bp (100 bp) of a non-SNP variant. An increase in SNP density around indels has previously been observed between species [32], [33]; here, we present genome-wide evidence of this effect within a single species. Thus, the accuracy of a traditional SNP-calling method based on simple read alignment may suffer materially since a considerable portion of the reads that would contribute to SNP calling may not be mapped correctly as they also contain indels; this problem is circumvented by the integrative variant calling approach employed here.
Fig. 2. Variants in genomic context. 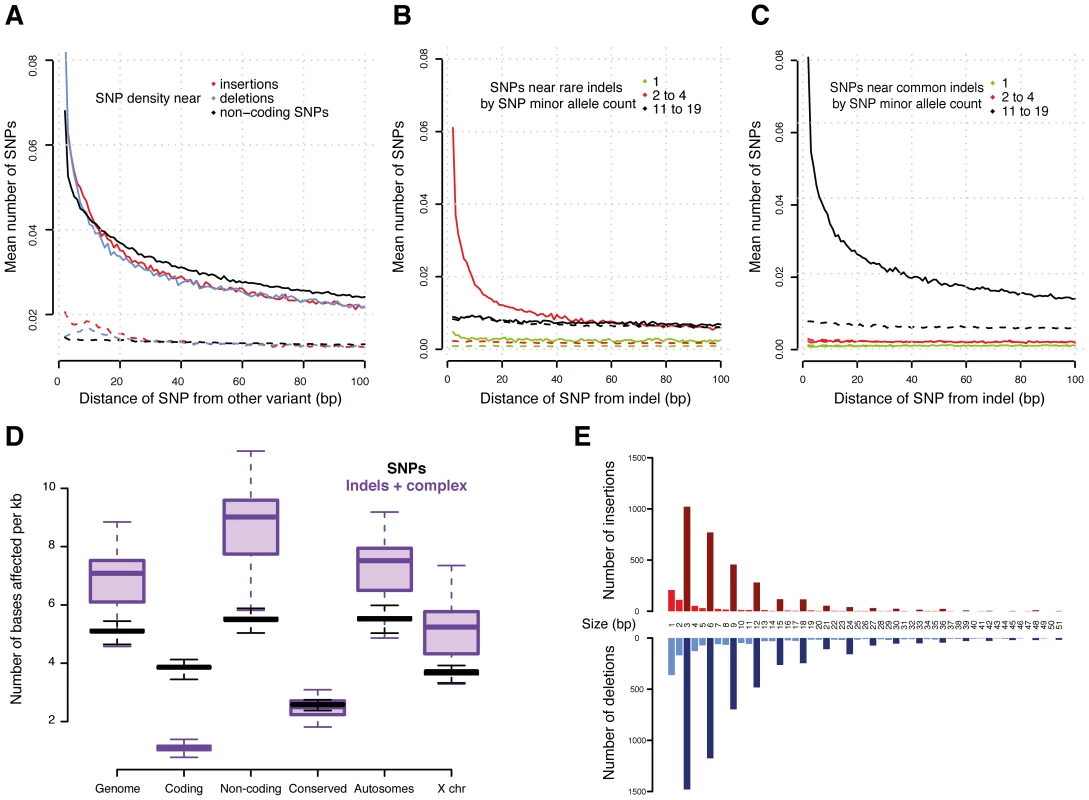
(A) Density of SNPs around variant breakpoints by variant type. The dashed lines show the SNP density at the same loci but in DGRP lines that do not have the variant. (B) and (C) Density of SNPs near indels with minor allele count 2 to 4 (B) and 11 to 19 (C). The dashed lines show the SNP density at the same locus for DGRP lines without the indel. If indels were mutagenic, one would expect enrichment for low allele count SNPs near the high allele count indels; instead, the allele count of the neighboring SNPs closely matches that of the indel. (D) Density of variants (reference bases affected per Mb) in selected genomic regions. (E) Number of indels in coding regions by indel size. Insertions are in red and deletions in blue. Bars representing indel sizes that are a multiple of three are coloured dark red and blue, respectively. Interestingly, we observed a similar SNP density increase around other SNPs (Figure 2A), suggesting that either SNPs are also mutagenic or, more likely, that alternative explanations need to be considered for the increased SNP density around variants. We plotted the allele frequencies of SNPs around variants and found that high allele frequency SNPs cluster around other high allele frequency variants (including other SNPs) and, correspondingly, low frequency variants cluster together (Figure 2B and 2C). These findings provide compelling evidence that variants in general are likely not mutagenic, since otherwise we would expect to observe a greater number of rare, thus mostly recent variants, around high frequency alleles. Alternatively, this phenomenon could occur if some genomic regions are more susceptible to mutation than others [32], [34]. We therefore examined the SNP density in the same DNA regions, but in those of the 39 lines which do not contain the focal variants. We found no material enrichment in SNP density in these regions (Figure 2A, 2B, and 2C), providing little evidence of locally increased mutation rate.
Functional impact of indels and complex variants
Non-coding regions that are conserved between species contain significantly fewer variants than other non-coding regions (Figure 2D, Mann-Whitney U test P<2.2e-16), confirming that these regions experience intra-species purifying selection as well. We obtained similar results when integrating histone modification data from the modENCODE Project [9]. We observe fewer indels affecting regulatory genomic regions - especially promoters (marked by tri-methylation of histone H3 at lysine 4, H3K4me3) - than unmarked non-coding regions, (Mann-Whitney U test P = 3.1e-14) (Figure S6).
While the majority (>90%) of indels and complex variants fall into intronic or intergenic regions, 80% of all protein-coding genes are affected by indels or complex variants (Table 2). However, only 13% of non-SNP variants within coding regions result in exon disruptions or full gene deletions. More than 50% of indels leading to frame shifts are singletons versus 35% for non-frame shifting indels, indicating that purifying selection is acting against these likely deleterious mutational events. This is illustrated by our observation that the length of indels affecting coding regions is highly biased for sizes that are multiples of three, a pattern that is still visible even up to 51 bp (Figure 2E). Consistent with previous inter-specific comparisons [35], we found that genes affected by disruptive indels or complex variants, are significantly enriched for the functional categories of sensory perception of taste and smell, proteolysis, and innate immune response as well as pathways related to food and drug metabolism (Tables S7 and S8).
Tab. 2. Potential functional indels and complex variants in genes. 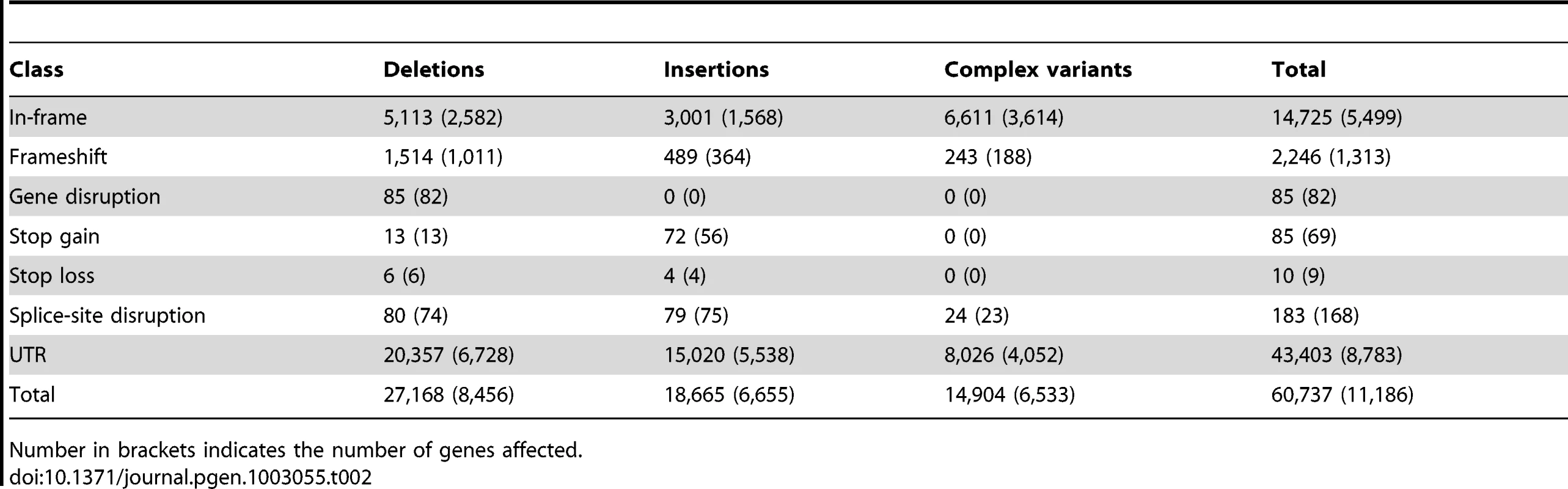
Number in brackets indicates the number of genes affected. Although the density of variants around genes is lower than the overall background, we observed an increase in variant density almost to background levels directly upstream of the transcription start site (TSS), followed by a dramatic drop across the TSS (Figure 3A). This is consistent with strong selective constraint at the TSS but not the region immediately upstream, possibly because this region has high AT content and it is typically depleted of nucleosomes [36], [37]. Indeed, we observed a strong correlation between the local AT content and indel density around the TSS (Figure S7A), strengthening earlier observations that sequence context or chromatin structure may affect the indel rate [38], [39]. We observed a similar, albeit less striking effect at the transcription end site (TES) (Figure 3B and Figure S7B).
Fig. 3. Cis-associations of variants with gene expression. 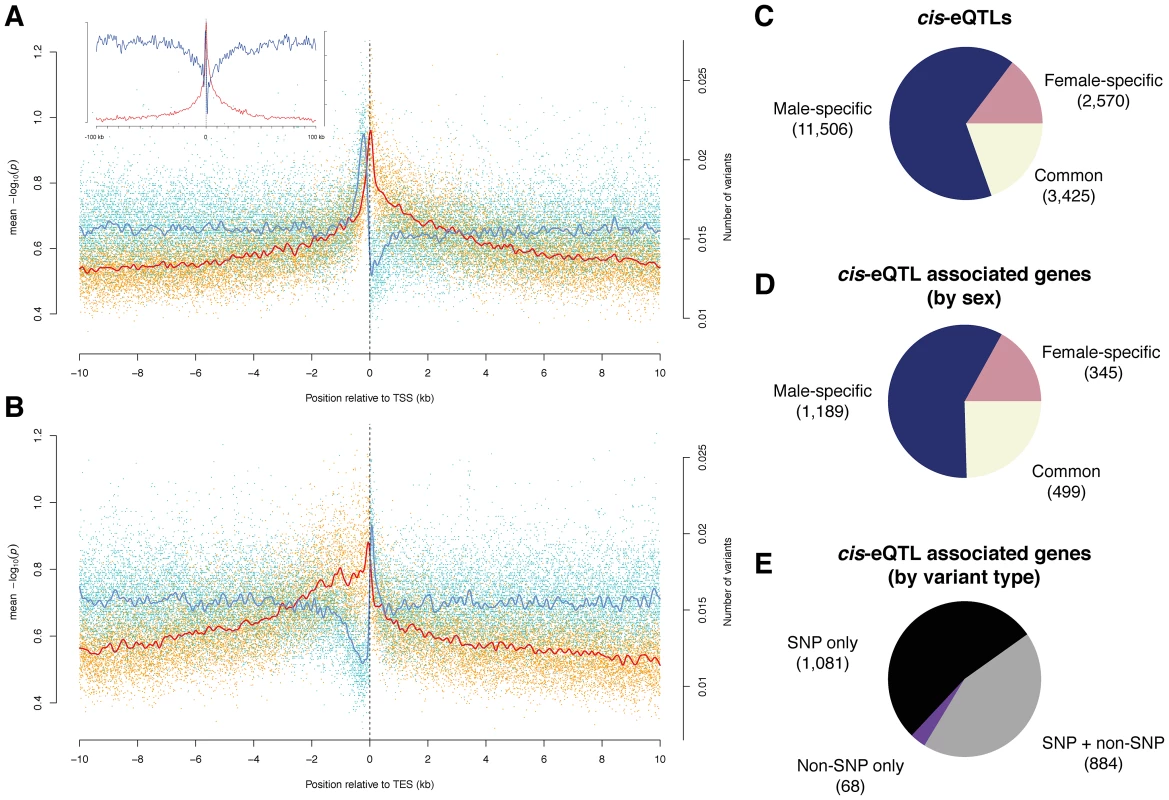
(A) and (B) Variant density (blue) and significance of allele associations (red), in males around (A) the transcription start site (TSS) and (B) the transcription end site (TES) averaged out over all transcripts in a 10 kb window. The solid lines are cubic smoothing splines, fit to the data. Transcripts on both strands are orientated such that transcription takes place in the positive direction of the x-axis. The inlet in (A) corresponds to a 100 kb window length. (C) cis-eQTLs discovered in males, females, or both sexes (FDR<10%). (D) Breakdown of cis-eQTL-associated genes by sex. (E) Breakdown of cis-eQTL associated genes, discovered in males or females, by type of variant (i.e., SNP and non-SNP). Identification and characterization of cis-eQTLs
We used published whole adult microarray gene expression data [6] to perform association analysis between the expression and variants within 10 kb of each gene. We initially identified 9,789 (9,434) genetically variable transcripts (FDR<0.001) in males (females) after removing probes from the microarray analysis that contain genomic variation. We then used the Kruskal-Wallis test followed by permutation-based multiple test correction to find significantly associated variants, termed cis-expression QTLs (cis-eQTLs), which may point to underlying functional regulatory elements. QTL studies have so far mostly associated bi-allelic variants with gene expression levels [40], [41]; given the availability of a high-resolution, comprehensive catalog of sequence variants, many of which (9.3%) are multi-allelic within the DGRP population, we grouped expression levels according to the corresponding allele as an input to each test. We conducted ∼3.8 million tests for each sex, and restricted our association analysis to variants present in at least three lines. We found 17,501 cis-eQTLs in 2,033 genes (26% of the 7,889 genes tested) at a false discovery rate of ∼10% (Figure 3C, 3D, and 3E; Tables S9 and S10; and results for 1% and 20% FDR thresholds are listed in Table S11), generating to our knowledge the first cis-eQTL map in Drosophila.
Surprisingly, the majority of cis-eQTLs were found to be sex-specific (58% specific to males and 17% to females; Figure 3C and 3D, and an example can be found in Figure 4A) with males having more than two-fold more cis-eQTL-associated genes compared to females. This result is not an artifact of the significance threshold, as a plot of the underlying association P-values in both sexes clearly reveals that the majority of cis-eQTLs are sex-specific (Figure S8). Further, the variance in gene expression among females is comparable to that among males as 56% of genetically variable transcripts without any cis-eQTL-associations display higher expression variance in females, making it unlikely that such bias affects our findings. In addition, only 38% of transcripts with male-specific cis-eQTLs exhibit greater gene expression variance in females than in males, refuting the hypothesis that the more than two-fold greater number of male - compared to female-specific cis-eQTL-associated genes may be due to a greater variability in gene expression in females compared to males. Intriguingly, we found that male-specific cis-eQTL-associated genes are significantly depleted from the X chromosome (P = 3.6e-11, χ2 test), whereas female-specific cis-eQTL (P = 0.17) and unbiased (P = 0.02) genes show a more uniform chromosomal distribution, perhaps consistent with the observed depletion of male-biased genes on the X chromosome [42], [43]. Sex-unbiased cis-eQTL-associated genes were found to have more cis-eQTLs than sex-biased ones (Figure S9) and the effect of cis-eQTLs found in both sexes is larger (Mann-Whitney U test, P<2.2e-16; Figure S10). Sex-unbiased cis-eQTL-associated genes have also a greater tendency to be expressed only in somatic tissues (Figures S11 and S12), possibly explaining why the underlying variants affect gene expression in both sexes. Nevertheless, more than 20% of sex-specific cis-eQTL-associated genes are also only expressed in somatic tissues, indicating that sex-biased changes in gene expression are pervasive [44]. Furthermore, we found that 20% of male-specific cis-eQTL-associated genes are exclusively expressed in the testes or the accessory gland in males, whereas the expression of only few (5%) female-specific counterparts are restricted to the ovaries or spermathecae in females. Approximately 60% of male-specific cis-eQTL-associated genes are expressed in female reproductive tissues (whereby 28% having greater expression than in any somatic tissue) (Figure S13), yet these genes do not yield any cis-eQTLs in females. In other words, despite the fact that these genes are expressed both in males and females, genetic mutations only produce cis-eQTLs in males, indicating that these mutations do not significantly affect gene expression in females. Together with the lower number of female-specific cis-eQTL-associated genes in general, this suggests that the underlying regulatory architecture may be more constrained in females as compared to males.
Fig. 4. Examples of cis-eQTLs and their associated genes. 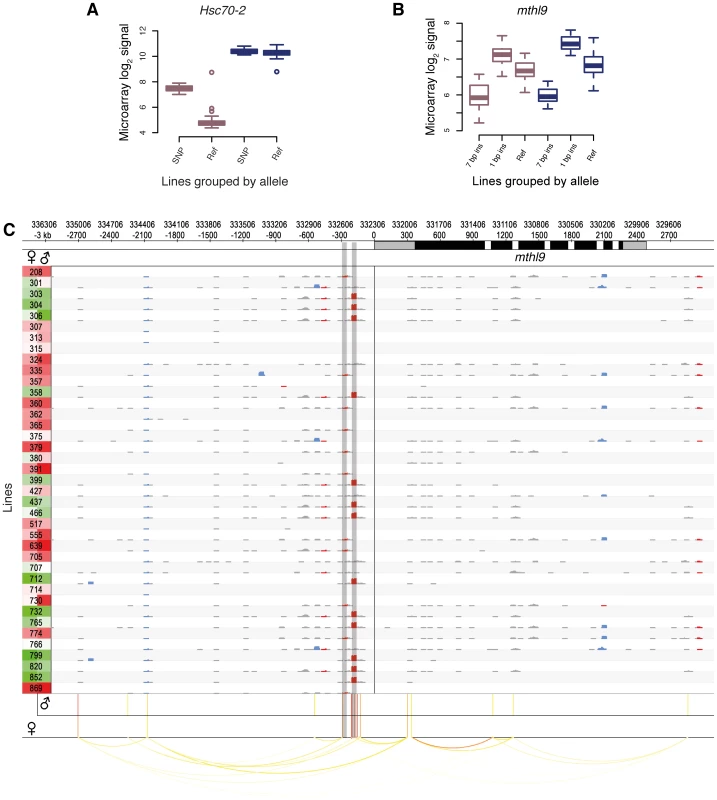
DGRP lines in (A) and (B) are grouped by their allele. Male and female expression levels are depicted in blue and dark pink, respectively. (A) Sex-biased cis-eQTL. A SNP (3R:8,875,391) is associated with higher gene expression levels in females only. (B) Indel-based cis-eQTLs associated with gene expression. Two insertions (7 bp, 3L:332,512; 1 bp, 3L:332,594, r2 = 0.20) are associated with markedly different expression levels in males and females. (C) cis-association overview. Plot illustrating the variant and association data for a single gene (mthl9) on a rolling window basis. The gene is shown on the top track, with UTRs in grey and coding regions in black. Significant cis-eQTLs are drawn below and color-coded by significance for each sex separately (red most significant). Linkage (r2>0.5) is shown by arcs, color-coded according to r2, with higher values in red. Rows represent all 39 DGRP lines and the left column shows gene expression levels for each line and sex separately (red indicates the highest expression level and green the lowest). The grid contains a representation of variants in rolling 50 bp windows (successive windows overlapping by 45 bp) with net insertions in red, net deletions in blue, and variants not affecting the sequence size (mostly SNPs) in black. The height of each variant indicates the net size of variants with the window, up to 20 bp. The two shaded vertical bars mark the cis-eQTLs shown in (B). We found that 5% of all cis-eQTL-associated genes do not exhibit detectable expression in any tissue of the corresponding sex (as represented in FlyAtlas [45]), indicating that the underlying genetic variation induces previously unreported gene expression in young adult flies. These cis-eQTLs show an enhanced effect compared to genes previously known to be expressed in adult fly tissues (Figure S14), thus strengthening the finding that these genes become strongly expressed given a specific genetic background. An example of this phenomenon involves Hsc70-2. This gene, ranking second among all female-specific associations, is known to be highly expressed in testes of adult males only [45], [46]. However, we find that females from several DGRP lines also exhibit high Hsc70-2 expression (Figure 4A), an observation which was validated using RNA-seq-based allele imbalance analyses (see below).
Of the cis-eQTL-associated genes, 53% are associated with SNPs only, 44% with SNPs and non-SNP variants, and 3% with non-SNP variants only. The latter is expected considering the increased density of SNPs near indels and complex variants (Figure 2A). In total, indels comprise 10% of all cis-eQTLs and complex variants 4%, and they have larger effect sizes than SNP cis-eQTLs (Mann-Whitney U test, P = 1.6e-10; Figure S15). One gene associated with indel cis-eQTLs is mthl9, where we identified two insertions with markedly different effects on gene expression in both males and females (Figure 4B).
Genomic distribution of cis-eQTLs
Although we considered only cis-associations, we identified 1,162 cis-eQTLs (6.6%) associated with two genes, and 58 with three genes. We found on average seven (median three) significant cis-associations per transcript (males and females combined), with the physical distance between two consecutive cis-eQTLs being 689 bp (median 121 bp), thus narrowing putatively causal variants down to a few hundred base pairs. However, cis-eQTLs associated with the expression of the same gene were found to be often in strong linkage disequilibrium (average r2 = 0.88).
We examined the genomic location of significant associations relative to TSSs and TESs (Figure 3A and 3B). Both the TSS and TES show the most significant associations independent of the variant type. We observed a quasi-symmetric distribution around TSSs in marked contrast to TESs, where upstream associations are, on average, more significant than downstream associations. These findings are consistent with previous studies in other metazoans including humans [40], [47], [48]. The high density of genetic markers in this study affords however greater resolution, clearly positioning the most significant associations within a window of less than 500 bp around the TSS and 1 kb upstream of the TES, consistent with findings in yeast [49]. The results are also consistent with enrichment for significant associations in H3K4me3 regions (2.8-fold for females and 2.6-fold for males) as compared to the rest of the genome (Table S12). Non-coding conserved regions are not enriched for cis-eQTLs, in agreement with findings in humans, where eQTLs are underrepresented in such regions [50]. One explanation could be that variants in conserved non-coding regions that dramatically affected gene expression were purged from the genome. Regions marked by tri-methylated histone H3 at lysine 27 (H3K27me3) are underrepresented for cis-eQTLs, consistent with the modification being a heterochromatin mark [51]. However, while we observe an enrichment of cis-eQTLs in functionally annotated regions, the majority of cis-eQTLs are outside of previously annotated regulatory regions. High-resolution maps of these cis-eQTL-derived putative regulatory elements (Figure 4C) may in this regard constitute a powerful resource for dissecting the regulatory architecture of gene expression in further detail.
Inheritance of gene expression
To validate cis-eQTLs genome-wide, we performed mRNA sequencing (RNA-seq) of reciprocal F1 female hybrids from two crosses involving three DGRP lines. Specifically, we aimed to identify whether transcripts predicted to be regulated by cis-eQTLs exhibit a significant allele-specific bias in gene expression. Since both alleles act in the cross in the same trans environment, differential expression in the F1 is a direct measure of cis-regulatory activity [52], [53]. As a quality control step, we first analyzed the correlation of the expression data between the reciprocal crosses, obtaining a Spearman correlation coefficient of >99% for both F1362–765-initial/F1362–765-reciprocal and F1517–765-initial/F1517–765-reciprocal, respectively. We then tested on average ∼7,100 transcripts for allelic imbalance in the two crosses. Using a 10% FDR, we found that 6% (443) of the tested transcripts show significant allele-specific gene expression (Figure 5A) with a median allelic expression difference of 1.5-fold. Of the cis-eQTLs for which the parental lines have different alleles, and whose population mean effect on microarray expression was two-, three - and four-fold among all 39 lines, we found that on average 39%, 60% and 75% of the associated transcripts respectively exhibited a significant allelic imbalance in the crosses (Figure 5B). These results are in line with similar, albeit smaller-scale analyses in yeast [49] and mouse [48], and thus provide support for our cis-eQTL map, revealing that the majority of strong cis-eQTLs can be validated in heterozygous individuals.
Fig. 5. Validation of cis-associations in F1. 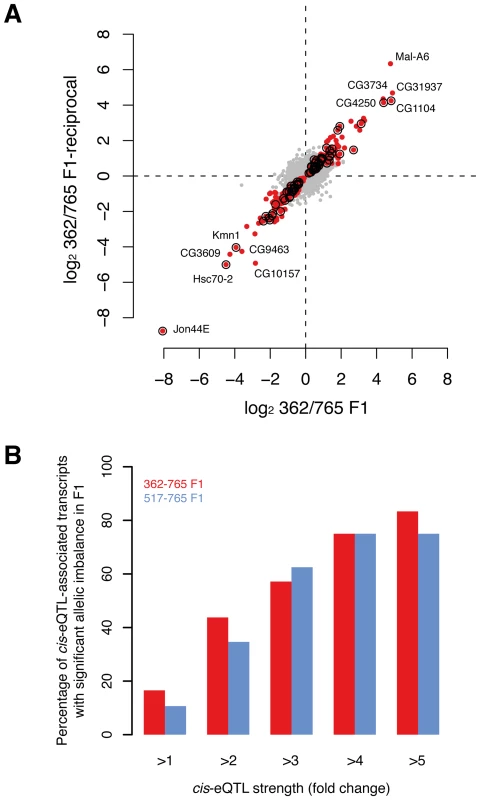
(A) Allelic imbalance measured for ∼7,100 transcripts in F1 (362/765 and reciprocal) with RNA-seq. Dots represent the fold-change (log2) between allele-specific reads counts. Red dots indicate transcripts with significant allelic imbalance in both crosses at a false discovery rate of 10%. Circles mark transcripts that demonstrate significant allelic imbalance and that were found to be associated with cis-eQTL in females (note that only cis-eQTLs were considered when the allele between both parental lines was not the same). (B) The proportion of cis-eQTL-associated transcripts that show allelic expression imbalance in F1s scales with the strength of the cis-eQTL (P<0.001, permutation-based, see Methods for details). Discussion
We present a comprehensive, genome-wide list of variants of all major types (i.e., single-, multi-nucleotide-, and structural variation) for a Drosophila wild-derived population, significantly expanding the catalog of variants that has been compiled to date for this model organism (e.g., [7], [13]–[15], [20]). Our analyses reveal extensive non-SNP variation among lines, with a high indel frequency (roughly every 150 bp) and indels in 80% of the protein-coding genes. The majority of indels do not alter the reading frame, but nevertheless reveal surprising tolerance for indels in coding regions. Our data are relevant to the inference of underlying mechanisms of varying mutation rates across genomes [32]. We show elevated SNP density around other SNPs as well as indels; that neighboring variants have similar allele frequencies; and that regions with indels are not particularly mutation-prone. These data suggest that this phenomenon reflects the existence of small haploblocks. While these may arise because of segregating mutational hotspots, a more likely explanation invokes the demographic history of the population. For example, admixture may generate such a haplotype structure to an extent largely dictated by the local genomic recombination environment [54] – potentially consistent with our observed correlation between recombination rate and variant density. Thus, while our data suggest that these population level processes may lead to incorrect inference regarding the mutagenic nature of indels, this will be a topic of further investigation in future studies during which alternative explanations (e.g., duplication hotspots) will also be explored.
The presented variant collection constitutes a useful resource for several scientific disciplines, including phylogenetics and population genetics, which are beyond the scope of this study. Here, we applied this resource to study the impact of natural polymorphisms on gene expression, identifying cis-eQTLs for >2,000 genes (10% FDR). Although further work is necessary to identify the causal variants underlying cis-eQTLs, the combined results provide general insights into the complexity and evolution of the cis-regulatory architecture of gene expression in Drosophila. More specifically, the data suggest that gene expression variation is governed by multiple cis-eQTLs of different variant types, that the most significant cis-eQTLs are tightly clustered around the TSS and immediately upstream of the TES, but that many cis-eQTLs are also located in regions currently devoid of regulatory annotations. These regions likely play an important role in gene regulation as their alteration has an expression impact that can be observed in whole adult fly expression profiles. In addition, the data suggest that the regulatory architecture is partly decoupled between males and females, at least within the tested settings, as underlying changes predominantly affect gene expression in males. However, cis-eQTLs common to both sexes typically have larger effects. This supports the hypothesis that intersexual ontogenetic conflict in D. melanogaster may stem from many sexually antagonistic alleles with low effect sizes [55]. Finally, the detection of many sex-specific cis-eQTLs indicates that genetic factors play an important role in sex-biased gene expression as changes in cis-regulatory sequences appear to significantly contribute to this phenomenon. While the relative significance of these changes and cis-regulatory variation in general in governing morphological evolution remains an important topic of debate [56], [57], we believe that the presented catalog of cis-regulatory changes constitutes a valuable resource to further illuminate this discussion.
Materials and Methods
Variant prediction
We used Release 5 of the Drosophila melanogaster reference genome for all analyses.
Stage 1: Variant discovery
We used PrInSeS-G [19], in conjunction with BWA [58] to perform the initial variant discovery using whole-genome Illumina sequencing reads.
Stage 2: Variant refinement
We computed whole-genome alignments between the chromosomes of each line and the reference genome in order to reduce the amount of variant fragmentation and to optimize the representation of variants among lines (see Text S1).
Stage 3: Variant genotyping
The purpose of genotyping is to improve variant discovery for DGRP lines or genomic regions whose sequencing coverage was originally too low for effective de novo assembly by PrInSeS-G. The algorithm is described in detail in Text S1, and Figure S16.
Gene expression analysis
Whole-adult gene expression microarray raw data files (i.e., .CEL) for 39 inbred lines were downloaded from the EBI ArrayExpress Archive (accession number E-MEXP-1594). Briefly, Ayroles et al. [6] derived inbred lines from the Raleigh (USA) population by 20 generation full-sib mating and hybridized RNA from two independent pools (25 flies/line/sex) on Affymetrix Drosophila 2.0 microarrays. The raw data set consists of gene expression measurements for males and female in two replicates. We used a four-step pipeline to analyse the microarray data:
Step 1
We obtained a custom probe set definition file (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/13.0.0/refseq.asp) for the Affymetrix Drosophila 2.0 platform, which is based on an updated set of 20,666 transcripts (UCSC RefSeq; June 22, 2010). We excluded probes that overlap with any type of sequence variants and probe sets that were covered by less than three probes after probe removal. In addition, we verified whether polymorphic duplicate regions may have contributed to gene expression variation. We aligned all insertions (>100 bp) to the annotated D. melanogaster transcriptome with the alignment program blat, retaining only high sequence similarity hits (>90%), since those would be the most likely candidates to cause problems with cross-hybridization on microarrays. We found only two insertions that could be mapped against annotated transcripts with both insertions targeting the same exon, demonstrating that duplication does not constitute an important confounding factor.
Step 2
Raw microarray data were normalized across all sexes and lines with the Robust Multichip Average (RMA) algorithm as implemented in the R affy package (default settings).
Step 3
We removed transcripts that were not or lowly expressed among lines using the Wilcoxon signed rank-based gene expression presence/absence detection algorithm (affy package, R). A transcript was classified as expressed in a single line if it was detected in either one or both sexes, respectively, and the requirement to be detected in both replicates. Finally, we only kept 16,985 probe sets (or 10,347 genes) that were classified as expressed in at least 4 out of 39 lines.
Step 4
We used ANOVA on each transcript and sex separately to test whether it is genetically variable, i.e., has a significant line term, under a conservative FDR of 0.001 [6]. In total, we found 9,789 (6,239) and 9,434 (5,797) genetically variable transcripts (genes) in males and females, respectively, of which 6,745 (4,147) were variable in both sexes.
Tissue-specific gene expression
Tissue-specific gene expression data for young adult flies was obtained from FlyBase (ftp://flybase.org/flybase/associated_files/Gelbart.2010.10.13.tar.gz). A gene was considered to be expressed in a tissue if the expression level was above 50.
Cis-associations
We grouped overlapping variants for all 39 lines as alternative alleles. For each such group of variants, we calculated associations with any transcript whose either end is within 10 kb, and for which microarray expression data was available. We employed two statistical methods: the first one, Alignment Score Association, is linear regression between the rank of alignment scores of all alleles and the rank of expression; in this context we defined alignment score to be the maximum of the number of bases inserted and that removed by the variant. Note that, since we used a non-parametric model, there was little advantage of using a more complex alignment score. This method thus associates the size of the variants rather than the exact genotype, which is important for the cases where more than two alleles have variants of different lengths. For the second method, Allele Association, we grouped the rank of expression of each line by its allele at each variant locus; we then performed a Kruskal-Wallis test. For both methods we corrected for multiple tests by repeating each test for 10,000 random permutations of gene expression, in a similar fashion to [40]. We then took for each permutation the lowest P-value for all tests in the same transcript, thus obtaining a vector of 10,000 such P-values. This vector was then sorted, and we obtained the adjusted P-value threshold from the value indexed by our unadjusted threshold. For example, the adjusted (multiple-test corrected) P-value threshold for a nominal threshold of 0.05 is the 500'th element of the sorted vector (10,000×0.05 = 500). The FDR is the ratio of the number of transcripts expected to pass the adjusted P-value threshold by chance over the number that actually passes. This, of course, implies that a different adjusted threshold is used for each transcript for the same nominal threshold. Further, since the permutations are random, each run of the above workflow will produce a slightly different list of eQTLs, since the P-value thresholds will be slightly different each time. We note that the overlap between the two association methods was more than 95% for all metrics, as both give approximately the same significance measure for bi-allelic variants, hence we only presented the results of the Allele Association method in the main manuscript.
Variant validation
We used five complementary methods to validate our variant calls. First, we compared our variant catalogue against publicly available indel data from the FLYSNPdb database, which features indels (1 bp to 360 bp) from five previously established D. melanogaster lines (but none of the DGRP lines) [20]. Second, we compared our calls with those published by Mackay et al. [7]. We used two methods (comparing all variants and comparing SNPs not within 30 bps of non-SNPs, Table S1). In many cases, SNP calls by the other study overlapped or were close to indel calls, which indicates a possible false positive since methods based on direct read alignment are confounded by indels. Third, we aligned Roche-454 whole genome sequencing reads from Mackay et al. available for all 39 lines [7] to validate our SNP calls (Massouras et al., in preparation), including those that (a) were originally supported by two thirds or more of the Illumina reads and (b) were covered by at least two Roche-454 reads (Table S2). Fourth, we sequenced mRNA from three DGRP lines at the young adult stage (see below and main manuscript for results) (Table S3). Finally, we examined 92 large indels (200 bp to 5.6 kb) in five DGRP lines using PCR after which we randomly selected 10 for subsequent Sanger sequencing to validate the predicted variant breakpoints (Table S4).
Variant validation using RNA–seq and allelic expression imbalance in F1
All fly stocks were grown at 25°C and a 12 h light-dark cycle on corn-meal fly medium. We collected virgin flies from three of the DGRP lines (lines 362, 517, and 765) during three days and we set up the following crosses: ♀ 362×♂ 517 (and reciprocal) and ♀ 517×♂ 765 (and reciprocal). We performed RNA-seq to assess transcript expression profiles of 3–5 day old F1 females. 25 flies per sample were frozen between 1–3 pm. Total RNA was extracted using the combined Trizol/RNeasy (Qiagen, http://www.qiagen.com/) protocol. RNA quality was measured using RNA Labchips and Bioanalyzer from Agilent Technologies (http://www.chem.agilent.com/). RNA-seq libraries were prepared using the RNA-True seq kit (Illumina). Prepared libraries were sequenced with an Illumina Genome Analyzer 2 DNA Sequencing Platform (GTF, Lausanne). We used the UCSC transcript annotation to derive the sequences of the annotated transcriptome from the reference genome. We mapped the reads to both the genomes and the annotated transcriptomes; any reads aligning to more than one genomic mapping coordinate were discarded. The tool we built specifically for this purpose takes as input the sequencing reads, two genome haplotypes, and the UCSC-annotated transcript coordinates in order to derive two transcriptomes. For every read in turn it attempts to align it to both transcriptome haplotypes. Reads that align to the same position in a transcript of the two haplotypes are then checked to see if the alignment overlaps with any variants. If it does, and the variants are different between the two haplotypes, the read is deemed to “support” the allele that produces the lowest number of mismatches. If the read aligns to only one transcriptome haplotype, it is also deemed to support the corresponding allele. In this way the tool measures allele-specific differential gene expression in the same line.
For the parental lines, we used the aforementioned tool, supplying it with the Stage 3 genome (see Variant prediction paragraph for more details) and the reference genome. For the crosses we gave it the two parental Stage 3 genomes. In each case, for each position with a variant we counted the number of reads best aligning to each allele. As a result, for the parental lines, this method provides a measurement of the true positive and false positive rate with regard to our variant discovery, since it measures the differential alignment of reads to the reference sequence and the target haplotype of the line. For the F1s, this method provides a measurement for differential expression for each transcript.
For variant validation, we considered a variant true positive (TP) when either all aligned reads support this variant, as opposed to the reference or when the number of reads that support it compared to that supporting the reference results in P<0.05 in a two-tailed binomial test. We considered a variant a false positive (FP) when either all aligned reads supported the reference at this position, or the number supporting the reference was significant using the same binomial test.
Allelic expression imbalance analysis
We tested for significant allelic expression imbalance between alleles in F1 using binomial exact tests (two-sided). About half of all tested transcripts were covered by at least 200 discriminative reads, thus providing a reasonable power to detect even small allelic imbalances (≥1.5-fold allelic expression differences). P-values were corrected for multiple hypothesis testing with the Benjamini & Hochberg procedure (implemented in the function p.adjust, R). Only transcripts that were tested for cis-associations in females and that were covered by at least 20 informative reads (i.e., allele1+allele2≥20) were tested for significant allelic imbalance. Transcripts that passed with 10% FDR in both reciprocal crosses were considered to have a significant allelic imbalance in gene expression.
The increase in percent-validated (Figure 5B) was tested by permutation analysis. For each “cis-eQTL strength cutoff” we obtained a random set of transcripts (from all tested cis-eQTL transcripts) and calculated how many of those where validated in F1. We repeated this sampling procedure 1,000 times and obtained an empirical P-value for each “cis-eQTL strength cutoff” by counting how many random transcript sets scored higher than the real set of transcripts and divided by the number of permutations (i.e., 1,000).
Recombination estimates
The Fiston-Lavier [59] recombination rate calculator was used to estimate the recombination rate (cM/Mb) in 1 Mb windows along each chromosome (only chromosome 4 excluded). The rate at the center of each interval was used.
Genomic distribution of cis-eQTL–associated genes
Genes were grouped into four different categories (i.e., genes with no-, male-specific-, female-specific-, and sex-unbiased cis-eQTLs) and within each category the percentage of genes located on the X chromosome was calculated. For cis-eQTL associated genes, we calculated whether the fraction of X-linked genes deviates significantly from non-cis-eQTL associated genes using the Chi-square test with Yates' correction.
Data availability
The full catalog of variants can be obtained from dgrp.epfl.ch/downloads. The RNA-seq data are available in the ArrayExpress Database using accession number E-MTAB-1266.
Supporting Information
Zdroje
1. MackayTFC, StoneEA, AyrolesJF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10 : 565–577.
2. ConsortiumH (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467 : 52–58.
3. MillsRE, WalterK, StewartC, HandsakerRE, ChenK, et al. (2011) Mapping copy number variation by population-scale genome sequencing. Nature 470 : 59–65.
4. KeaneTM, GoodstadtL, DanecekP, WhiteMA, WongK, et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477 : 289–294.
5. GanX, StegleO, BehrJ, SteffenJG, DreweP, et al. (2011) Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 477 : 419–423.
6. AyrolesJF, CarboneMA, StoneEA, JordanKW, LymanRF, et al. (2009) Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41 : 299–307.
7. MackayTFC, RichardsS, StoneEA, BarbadillaA, AyrolesJF, et al. (2012) The Drosophila melanogaster Genetic Reference Panel. Nature 482 : 173–178.
8. RubinGM, LewisEB (2000) A Brief History of Drosophila's Contributions to Genome Research. Science 287 : 2216–2218.
9. ConsortiumTm, RoyS, ErnstJ, KharchenkoPV, KheradpourP, et al. (2010) Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science 330 : 1787–1797.
10. HensK, FeuzJ-D, IsakovaA, IagovitinaA, MassourasA, et al. (2011) Automated protein-DNA interaction screening of Drosophila regulatory elements. Nat Meth 8 : 1065–1070.
11. AdamsMD, CelnikerSE, HoltRA, EvansCA, GocayneJD, et al. (2000) The Genome Sequence of Drosophila melanogaster. Science 287 : 2185–2195.
12. MoriyamaEN, PowellJR (1996) Intraspecific nuclear DNA variation in Drosophila. Mol Biol Evol 13 : 261–277.
13. EmersonJJ, Cardoso-MoreiraM, BorevitzJO, LongM (2008) Natural Selection Shapes Genome-Wide Patterns of Copy-Number Polymorphism in Drosophila melanogaster. Science 320 : 1629–1631.
14. CridlandJM, ThorntonKR (2010) Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol 2 : 83–101.
15. LangleyCH, StevensK, CardenoC, LeeYCG, SchriderDR, et al. (2012) Genomic Variation in Natural Populations of Drosophila melanogaster. Genetics
16. RebeizM, PoolJE, KassnerVA, AquadroCF, CarrollSB (2009) Stepwise Modification of a Modular Enhancer Underlies Adaptation in a Drosophila Population. Science 326 : 1663–1667.
17. WittkoppPJ, KalayG (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13 : 59–69.
18. CarboneMA, JordanKW, LymanRF, HarbisonST, LeipsJ, et al. (2006) Phenotypic variation and natural selection at Catsup, a pleiotropic quantitative trait gene in Drosophila. Current Biology 16 : 912–919.
19. MassourasA, HensK, GubelmannC, UplekarS, DecouttereF, et al. (2010) Primer-initiated sequence synthesis to detect and assemble structural variants. Nat Meth 7 : 485–486.
20. ChenD, AhlfordA, SchnorrerF, KalchhauserI, FellnerM, et al. (2008) High-resolution, high-throughput SNP mapping in Drosophila melanogaster. Nat Meth 5 : 323–329.
21. YalcinB, WongK, AgamA, GoodsonM, KeaneTM, et al. (2011) Sequence-based characterization of structural variation in the mouse genome. Nature 477 : 326–329.
22. MillsRE, PittardWS, MullaneyJM, FarooqU, CreasyTH, et al. (2011) Natural genetic variation caused by small insertions and deletions in the human genome. Genome Research 21 : 830–839.
23. CaoJ, SchneebergerK, OssowskiS, GuntherT, BenderS, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43 : 956–963.
24. A map of human genome variation from population-scale sequencing. Nature 467 : 1061–1073.
25. KimPM, LamHYK, UrbanAE, KorbelJO, AffourtitJ, et al. (2008) Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history. Genome Research 18 : 1865–1874.
26. Fiston-LavierA-S, AnxolabehereD, QuesnevilleH (2007) A model of segmental duplication formation in Drosophila melanogaster. Genome Research 17 : 000.
27. Schrider DanielR, Hourmozdi JonathanN, Hahn MatthewW (2011) Pervasive Multinucleotide Mutational Events in Eukaryotes. Current Biology 21 : 1051–1054.
28. BetancourtAJ, KimY, OrrHA (2004) A pseudohitchhiking model of X vs. autosomal diversity. Genetics 168 : 2261–2269.
29. AndolfattoP (2001) Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol 18 : 279–290.
30. BegunDJ, AquadroCF (1992) Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356 : 519–520.
31. CharlesworthB, MorganMT, CharlesworthD (1993) The effect of deleterious mutations on neutral molecular variation. Genetics 134 : 1289–1303.
32. HodgkinsonA, Eyre-WalkerA (2011) Variation in the mutation rate across mammalian genomes. Nat Rev Genet 12 : 756–766.
33. TianD, WangQ, ZhangP, ArakiH, YangS, et al. (2008) Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455 : 105–108.
34. McDonaldMJ, WangW-C, HuangH-D, LeuJ-Y (2011) Clusters of Nucleotide Substitutions and Insertion/Deletion Mutations Are Associated with Repeat Sequences. PLoS Biol 9: e1000622 doi:10.1371/journal.pbio.1000622..
35. ClarkAG, EisenMB, SmithDR, BergmanCM, OliverB, et al. (2007) Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 : 203–218.
36. MavrichTN, JiangC, IoshikhesIP, LiX, VentersBJ, et al. (2008) Nucleosome organization in the Drosophila genome. Nature 453 : 358–362.
37. PeckhamHE, ThurmanRE, FuY, StamatoyannopoulosJA, NobleWS, et al. (2007) Nucleosome positioning signals in genomic DNA. Genome Research 17 : 1170–1177.
38. TolstorukovMY, VolfovskyN, StephensRM, ParkPJ (2011) Impact of chromatin structure on sequence variability in the human genome. Nat Struct Mol Biol 18 : 510–515.
39. TanayA, SiggiaED (2008) Sequence context affects the rate of short insertions and deletions in flies and primates. Genome Biol 9: R37.
40. StrangerBE, ForrestMS, DunningM, IngleCE, BeazleyC, et al. (2007) Relative Impact of Nucleotide and Copy Number Variation on Gene Expression Phenotypes. Science 315 : 848–853.
41. CheungVG, NayakRR, WangIX, ElwynS, CousinsSM, et al. (2010) Polymorphic Cis - and Trans-Regulation of Human Gene Expression. PLoS Biol 8: e1000480 doi:10.1371/journal.pbio.1000480..
42. ParisiM, NuttallR, NaimanD, BouffardG, MalleyJ, et al. (2003) Paucity of Genes on the Drosophila X Chromosome Showing Male-Biased Expression. Science 299 : 697–700.
43. BachtrogD, TodaNRT, LocktonS (2010) Dosage Compensation and Demasculinization of X Chromosomes in Drosophila. Current Biology 20 : 1476–1481.
44. ZhangY, SturgillD, ParisiM, KumarS, OliverB (2007) Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450 : 233–237.
45. ChintapalliVR, WangJ, DowJAT (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature Genetics 39 : 715–720.
46. GraveleyBR, BrooksAN, CarlsonJW, DuffMO, LandolinJM, et al. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471 : 473–479.
47. VeyrierasJ-B, KudaravalliS, KimSY, DermitzakisET, GiladY, et al. (2008) High-Resolution Mapping of Expression-QTLs Yields Insight into Human Gene Regulation. PLoS Genet 4: e1000214 doi:10.1371/journal.pgen.1000214..
48. DossS, SchadtEE, DrakeTA, LusisAJ (2005) Cis-acting expression quantitative trait loci in mice. Genome Res 15 : 681–691.
49. RonaldJ, BremRB, WhittleJ, KruglyakL (2005) Local Regulatory Variation in Saccharomyces cerevisiae. PLoS Genet 1: e25 doi:10.1371/journal.pgen.0010025.
50. StrangerBE, NicaAC, ForrestMS, DimasA, BirdCP, et al. (2007) Population genomics of human gene expression. Nat Genet 39 : 1217–1224.
51. MikkelsenT, KuM, JaffeD, IssacB, LiebermanE, et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 : 553–560.
52. McManusCJ, CoolonJD, DuffMO, Eipper-MainsJ, GraveleyBR, et al. (2010) Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20 : 816–825.
53. WittkoppP, HaerumB, ClarkA (2004) Evolutionary changes in cis and trans gene regulation. Nature 430 : 85–88.
54. PfaffCL, ParraEJ, BonillaC, HiesterK, McKeiguePM, et al. (2001) Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet 68 : 198–207.
55. ChippindaleAK, GibsonJR, RiceWR (2001) Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proceedings of the National Academy of Sciences 98 : 1671–1675.
56. HoekstraHE, CoyneJA (2007) The locus of evolution: evo devo and the genetics of adaptation. Evolution 61 : 995–1016.
57. CarrollSB (2008) Evo-Devo and an Expanding Evolutionary Synthesis: A Genetic Theory of Morphological Evolution. Cell 134 : 25–36.
58. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25 : 1754–1760.
59. Fiston-LavierA-S, SinghND, LipatovM, PetrovDA (2010) Drosophila melanogaster recombination rate calculator. Gene 463 : 18–20.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
- Prof. Petr Urbánek: Potřebujeme najít pacienty s nediagnostikovanou akutní intermitentní porfyrií
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



