-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEvolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
It is generally believed that the last eukaryotic common ancestor (LECA) was a unicellular organism with motile cilia. In the vertebrates, the winged-helix transcription factor FoxJ1 functions as the master regulator of motile cilia biogenesis. Despite the antiquity of cilia, their highly conserved structure, and their mechanism of motility, the evolution of the transcriptional program controlling ciliogenesis has remained incompletely understood. In particular, it is presently not known how the generation of motile cilia is programmed outside of the vertebrates, and whether and to what extent the FoxJ1-dependent regulation is conserved. We have performed a survey of numerous eukaryotic genomes and discovered that genes homologous to foxJ1 are restricted only to organisms belonging to the unikont lineage. Using a mis-expression assay, we then obtained evidence of a conserved ability of FoxJ1 proteins from a number of diverse phyletic groups to activate the expression of a host of motile ciliary genes in zebrafish embryos. Conversely, we found that inactivation of a foxJ1 gene in Schmidtea mediterranea, a platyhelminth (flatworm) that utilizes motile cilia for locomotion, led to a profound disruption in the differentiation of motile cilia. Together, all of these findings provide the first evolutionary perspective into the transcriptional control of motile ciliogenesis and allow us to propose a conserved FoxJ1-regulated mechanism for motile cilia biogenesis back to the origin of the metazoans.
Published in the journal: . PLoS Genet 8(11): e32767. doi:10.1371/journal.pgen.1003019
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003019Summary
It is generally believed that the last eukaryotic common ancestor (LECA) was a unicellular organism with motile cilia. In the vertebrates, the winged-helix transcription factor FoxJ1 functions as the master regulator of motile cilia biogenesis. Despite the antiquity of cilia, their highly conserved structure, and their mechanism of motility, the evolution of the transcriptional program controlling ciliogenesis has remained incompletely understood. In particular, it is presently not known how the generation of motile cilia is programmed outside of the vertebrates, and whether and to what extent the FoxJ1-dependent regulation is conserved. We have performed a survey of numerous eukaryotic genomes and discovered that genes homologous to foxJ1 are restricted only to organisms belonging to the unikont lineage. Using a mis-expression assay, we then obtained evidence of a conserved ability of FoxJ1 proteins from a number of diverse phyletic groups to activate the expression of a host of motile ciliary genes in zebrafish embryos. Conversely, we found that inactivation of a foxJ1 gene in Schmidtea mediterranea, a platyhelminth (flatworm) that utilizes motile cilia for locomotion, led to a profound disruption in the differentiation of motile cilia. Together, all of these findings provide the first evolutionary perspective into the transcriptional control of motile ciliogenesis and allow us to propose a conserved FoxJ1-regulated mechanism for motile cilia biogenesis back to the origin of the metazoans.
Introduction
Cilia are primitive, cell surface-associated filamentous organelles with wide-spread distribution among the protozoans and most metazoan phyla. The hair-like extension of the cilium is called the axoneme - a structure that typically comprises nine peripheral microtubule doublets arranged around a central pair of singlet microtubules (the 9+2 arrangement) as in the motile cilia, or lacking the central pair (the 9+0 arrangement) as in the immotile primary or sensory cilia [1]. Besides these differences in microtubular organization, axonemes of motile cilia are decorated with dynein arms which confer motility through ATP-dependent sliding of the microtubule doublets relative to each other. The axoneme is enveloped by the ciliary membrane, an extension of the cell membrane, and at its base are the nine triplet microtubules of the basal body. Assembly and maintenance of the cilium from the complex set of constituting proteins depends upon a unique transport mechanism termed intraflagellar transport (IFT) [2]. This process involves the continual ferry of cargo proteins by IFT particles from the base to the tip of the axoneme, and back to the base. Cilia have varied functions, which seem to have evolved in synchrony with the strategic location of the organelle at the cell surface. These range from cellular locomotion, fluid transport and as platforms for signal transduction [3]. Given these diverse roles, defective cilia have been implicated in several human pathologies [3]–[5]. Consequently, the biogenesis as well as the functions of cilia in animal development and physiology is currently under intense scrutiny.
An outstanding question in the field of ciliary biology is how the expression of the complex ciliary proteome, likely comprising several hundreds of proteins, is regulated at the transcriptional level [6]. Some insight into this problem has come from the analyses of the regulatory factor X (Rfx) family of transcription factors. Rfx proteins of Caenorhabditis elegans and Drosophila melanogaster are essential for primary cilia formation [7]–[11]. In mice, the Rfx homologs Rfx3 and Rfx4 regulate genes required for the formation and function of primary as well as motile cilia [12]–[14]. Motile cilia and motile cilia-specific genes are completely absent from C. elegans. In D. melanogaster, only sperm cells elaborate motile cilia (flagella); however, a role for the Rfx factors in the regulation of flagellar synthesis has not been defined. Yet another transcription factor, FoxJ1, plays a critical role in the regulation of ciliary gene expression in several species of vertebrates [15]–[17]. Unlike the Rfx proteins, FoxJ1 seems to have a function exclusively in the control of motile, but not primary cilia formation. Moreover, evidence from the zebrafish as well as mice suggests that FoxJ1 functions upstream of the Rfx factors in the ciliogenic pathway [17], [18].
Although the cilium has a highly conserved structure and an ancient origin, the evolutionary history of the transcriptional regulation of ciliary genes has remained an unsolved problem. Recently, two studies, using an entirely in silico approach, have garnered evidence that the Rfx family of transcription factors and the ciliary genes evolved independently [19], [20]. The authors proposed that the ciliary genes were gradually “re-wired” to come under the regulation of the Rfx proteins. It is presently not known whether any aspect of the FoxJ1-dependent control of ciliogenesis is conserved outside of the vertebrates. This is a pertinent issue, because in contrast to Rfx, FoxJ1 activity is not just necessary, but strikingly, is also sufficient for programming cells to assemble functional motile cilia [15]–[17]. In addition, it is generally agreed that the origin of the motile cilium predated the immotile primary cilium, and that the latter derived from the former through the progressive loss of the motility machinery [21]. Furthermore, whereas FoxJ1 seems to have a dedicated role in the generation of cilia [16], [17], [22], [23], the Rfx proteins have been implicated in the regulation of genes linked to many other processes, besides ciliogenesis [12], [24]. Here, we report the identification of FoxJ1 homologs from many diverse phylogenetic groups. Using transient transgenesis, we found that mis-expression of the FoxJ1 proteins from three representative invertebrate phyla – Placozoa, Platyhelminthes and Echinodermata - in zebrafish embryos, led to the ectopic induction of a number of motile ciliary genes that we have previously established to be canonical targets of vertebrate FoxJ1. To complement this sufficiency function, we explored whether these invertebrate FoxJ1 proteins in their native species are involved in the ciliogenic pathway. Indeed, RNAi-mediated abolition of FoxJ1 activity in the flatworm S. mediterranea strongly impaired the differentiation of motile cilia. These data underscore a functional conservation in motile ciliary gene regulation by FoxJ1 transcription factors across diverse groups of metazoans.
Results
FoxJ1 proteins are present only in the fungi/metazoa group within the unikonts
In order to first gain insights into the phylogenetic distribution of FoxJ1 proteins, we searched a total of 215 organisms representing all of the major phylogenetic groups within the eukaryotes for the presence of FoxJ1. FoxJ1 is part of the large Fox family of transcription factors which are characterized by a distinct DNA-binding domain (the forkhead domain or FKH) spanning ∼100 amino acids. In the mammals (mice), Fox proteins have been classified into several subfamilies ranging from FoxA-FoxS [25]. We performed BLASTP and TBLASTN searches against chosen proteomes/genomes with the FKH domain of human FOXJ1, and the proteins retrieved with an E-value less than E-2 were considered to be potential orthologs. However, the FKH family members exhibit a high degree of conservation within the FKH domain, and a large number of proteins which belonged to FKH families other than FoxJ1 were obtained using this method. In order to filter this information and identify the true FoxJ1 orthologs, a two-pronged approach was employed. First, all the identified proteins were subjected to phylogenetic analyses by aligning their FKH domain sequences with those of the FKH domains for 42 Fox family members of the mouse [25]. Independently, a reverse BLAST was also done against the human (nr) protein database using the FKH domains of the proteins identified in the initial search. A total of 60 FoxJ1 proteins could be identified using a combination of the two approaches; of these, only 43 can be considered definite orthologs as they reliably grouped with mouse FoxJ1 (bootstrap (BS)>95) (Table 1).
Tab. 1. List of organisms with an identifiable FoxJ1 based on reverse BLAST and/or phylogenetic analyses. 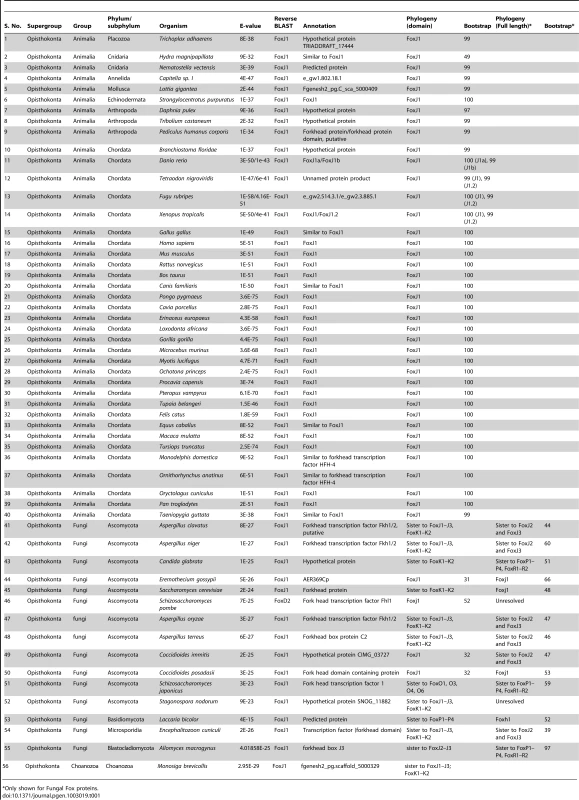
Only shown for Fungal Fox proteins. FoxJ1 homologs could not be identified from any of the bikonts (Archaeplastida, Excavata, Rhizaria and Chromalveolata); intriguingly, this includes Chlamydomonas reinhardtii, an organism where the biology of the prototypical 9+2 flagellum has been best studied. Rfx factors are also absent from this algal protist, implying that despite the high degree of similarity between its flagella and the motile cilia of metazoans, the transcriptional regulation of the biogenesis of these organelles is fundamentally different. Within the unikonts, FoxJ1 was not recovered from any organism of the amoebozoan lineage, including Acanthopodia, Archamoebae and Mycetozoa. Within the opisthokonts, true FoxJ1 orthologs are absent from Choanozoa and Nematoda (Table 1, Table S1), whereas FoxJ1 is present in many phyla such as Placozoa, Cnidaria, Annelida, Mollusca, Arthropoda, Echinodermata and Chordata (Table 1). Among the arthropods that we sampled, we found FoxJ1 in Tribolium castaneum, Pediculus humanus and Daphnia pulex, but not among species of the model genus Drosophila. As discussed earlier, like C. elegans, fruit flies are devoid of motile cilia except for the flagella that differentiate on their sperm cells. These flagella are peculiar in that they are synthesized in the cytoplasm without the involvement of IFT, and as our data show, or FoxJ1. In addition to the FKH domain, we also used the full length sequence of human FOXJ1 to search for potential homologs. The difference in the results between these two strategies centered on 13 organisms which are representatives from the Fungi and few metazoan phyla (Chordata and Arthropoda); in these, a homolog could be identified only using full-length human FOXJ1. Conversely, in 6 organisms representing the Fungi, a homolog was identifiable using the FKH domain but not with the full-length sequence. All of the additional FoxJ1 sequences that were recovered by the full-length search belonged to the Fungi/Metazoa, and as with the domain-based analysis, no homolog was identified outside this group (Table 1, Table S2).
In contrast to the metazoan FoxJ1 proteins, those identified from few fungal species, along with FoxJ1 from Monosiga brevicollis (Choanozoa), yielded inconsistent data on phylogenetic analyses as well as reverse BLAST (Table 1) – in fact, only 3 fungal species, all belonging to Ascomycota - Eremothecium gossypii, Coccidioides immitis and Coccidioides posadasii had a FoxJ1 which was identifiable using both reverse BLAST and phylogeny. However, the bootstrap for all three was less than 50% (Table 1). A previous study identified few fungal proteins as FoxJ1 orthologs since those particular sequences grouped together in the same clade as the Nematostella, Amphimedon (Porifera) as well as the bilaterian FoxJ1 proteins [26]. It is worth noting though that the particular clade had a weak bootstrap support in all three trees used (NJ<50; ML<50; PP: 0.56). In addition, although the relationship between the Fox family members are well established, a number of studies have shown discrepant data in the grouping together of certain subfamilies such as FoxR1–FoxR2, FoxL1–FoxL2, FoxJ1 with FoxJ2–FoxJ3 and FoxN1–FoxN4 with FoxN2–FoxN3 [25]. Therefore, in order to resolve the correct identity of the fungal Fox proteins identified in our search, the full length sequences of the putative FoxJ1 homologs from the 15 fungal species which showed the presence of FoxJ1 either through reverse BLAST or phylogeny (low BS support) were each aligned with full-length mouse Fox protein family members. This strategy, together with reverse BLAST and phylogeny based solely on the FKH domain yielded four species - E. gossypii, Saccharomyces cerevisiae, C. immitis and C. posadasii with identifiable FoxJ1 using at least two of the methods (although BS values were less even in the analyses based on the full length sequence (Table 1)). These findings establish the presence of FoxJ1 homologs only in the unikonts, similar to the other family of ciliogenic transcription factors, Rfx. FoxJ1, however, has a more restricted distribution, and unlike Rfx is absent from the amoebozoan lineage, as well as from the Choanozoa and the Nematoda within the opisthokonts.
FoxJ1 proteins from Placozoa sp. H4 and Strongylocentrotus purpuratus can activate motile ciliary genes in the zebrafish embryo
Studies with vertebrate FoxJ1 have established its role as a master regulator of motile ciliogenesis, meaning that the activity of the protein is both necessary as well as adequate for the generation of motile cilia [15]–[17]. Nothing is currently known about the biology of the non-vertebrate FoxJ1 proteins. To begin to investigate whether the role of FoxJ1 in regulating motile ciliogenesis is generally conserved, we first performed a mis-expression assay. For this, we selected two FoxJ1 proteins, from the placozoan species, Placozoa sp. H4 and the echinoderm S. purpuratus, as representatives, and then evaluated their transcriptional activity through transgenic expression in zebrafish embryos. Using this strategy, we have earlier shown that mis-expression of zebrafish FoxJ1 can ectopically activate a battery of motile ciliary genes [17]. The placozoans are an interesting model from an evolutionary standpoint since they are thought to represent the basal state of the metazoans [27]–[29]. However, other genome-level and phylogenomic analyses have instead placed the sponges as the most basal metazoan group [30], [31]. Regardless of the lack of a consensus view on this issue, anatomically the Placozoa are the simplest extant metazoans, with an elementary body plan and presence of only four cell-types, one of which bears motile cilia [28]. On the other hand, the echinoderms (and their sister phylum, the hemichordates), are the closest known relatives of the chordates. They typically reproduce through larval forms that have motile cilia, and hence also are an interesting group to incorporate in this study. Both the placozoan and S. purpuratus FoxJ1 proteins are 74% identical to human FoxJ1 in the FKH domain. Heat-inducible myc epitope tagged transgenic constructs for the two genes, i.e., hs::myc-Pl-foxJ1 (Placozoa) and hs::myc-Sp-foxJ1 (S. purpuratus), were made and expressed in zebrafish embryos using transient transgenesis as described previously for zebrafish FoxJ1 [17]. FoxJ1 proteins from both species localized to the nuclei of zebrafish cells (Figure 1A–1B).
Fig. 1. FoxJ1 from T. adhaerens and S. purpuratus are nuclear localized and can regulate the expression of ciliary genes. 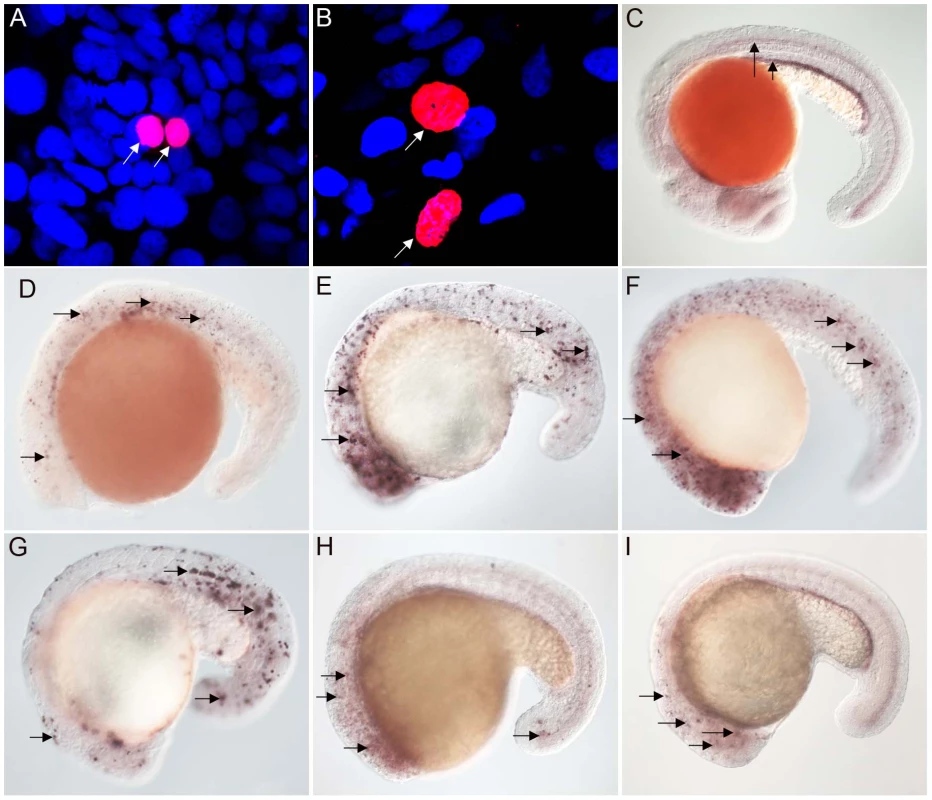
Anti-myc antibodies were used to detect Placozoa (A) and sea urchin (B) FoxJ1 (red, white arrow). Nuclei were stained with DAPI (blue). (C) Expression of dynein intermediate chain in the spinal cord (long arrow) and pronephric (kidney) duct (short arrow) of a wild-type zebrafish embryo. The wdr78 and efhc1 genes are expressed in a similar pattern in wild-type embryos (see Figure 2A and data not shown). Ectopic expression of dynein intermediate chain in embryos ectopically expressing placozoan (D) and sea urchin (E) FoxJ1, respectively. Ectopic expression of wdr78 in embryos ectopically expressing placozoan (F) and sea urchin (G) FoxJ1, respectively. Ectopic expression of efhc1 in embryos ectopically expressing placozoan (H) and sea urchin (I) FoxJ1, respectively. Mis-expression of the different ciliary genes in D–I is indicated by the arrows. Embryos depicted are at 20 hpf, oriented anterior to the left, dorsal to the top. For exploring the transcriptional activity of the placozoan and echinoderm FoxJ1, we selected five well-established zebrafish FoxJ1 targets: efhc1, spag6, wdr78, tektin1 and dynein intermediate chain that encode motile cilia-specific components and are hyper-induced in response to ectopic expression of zebrafish FoxJ1 [17]. Remarkably, both the placozoan and the S. purpuratus FoxJ1 proteins robustly induced all 5 ciliary genes (Figure 1C–1I, data not shown). Induction of these genes was lineage-independent, and could be observed in cells which under normal circumstances do not form motile cilia, indicating the sufficiency of the placozoan and echinoderm FoxJ1 proteins to activate a battery of motile ciliary genes in a wide diversity of zebrafish cell types, just like zebrafish FoxJ1 [17]. The Fox family members share a high degree of conservation in their FKH domain. For instance, mouse FoxJ1 is 56% identical in the FKH domain to the closely related FoxJ2, and 47% identical to the distantly related FoxN3 proteins. Although the different Fox family members are known to control distinct developmental and physiological processes through the regulation of discrete sets of target genes, the high degree of conservation in the DNA binding FKH domain raises the possibility that over-expression of the proteins could inappropriately lead to cross-activation of targets owing to the commonality in their DNA recognition motif. Hence, induction of the zebrafish motile ciliary genes by the placozoan and echinoderm FoxJ1 proteins could merely be reflective of such an effect. To negate this possibility, we cloned zebrafish foxJ2 and foxJ3, fox genes that are most closely related to foxJ1 [25], and mis-expressed them in zebrafish embryos using the heat-inducible transient transgenesis method described above. Unlike FoxJ1, neither FoxJ2 nor FoxJ3 was able to ectopically induce the expression of any of the ciliary genes (Figure 2A–2F). Besides the DNA binding FKH domain, a number of transcriptional activating motifs have been reported in FoxJ1. These include four regions rich in acidic amino acids (A1, A2, A3 and A4), a winged helix transcriptional activation region II motif and a proline-rich region [32], [33]. Alignment of mouse FoxJ1 and FoxJ2 sequences showed limited similarity in the acidic region A3, proline-rich region and the region II motif, but no conservation in the remaining three acidic regions (A1, A2, A4). A similar comparison with the FoxJ3 protein revealed limited homology only in the acidic region A4 and the proline-rich region, but no conservation among the remaining motifs important for transcriptional activation. Thus, a significant degree of divergence in the transcriptional regulatory domains of closely related Fox family members could explain their abilities to regulate distinct sets of target genes.
Fig. 2. Zebrafish FoxJ2 and FoxJ3 are unable to induce the expression of ciliary genes. 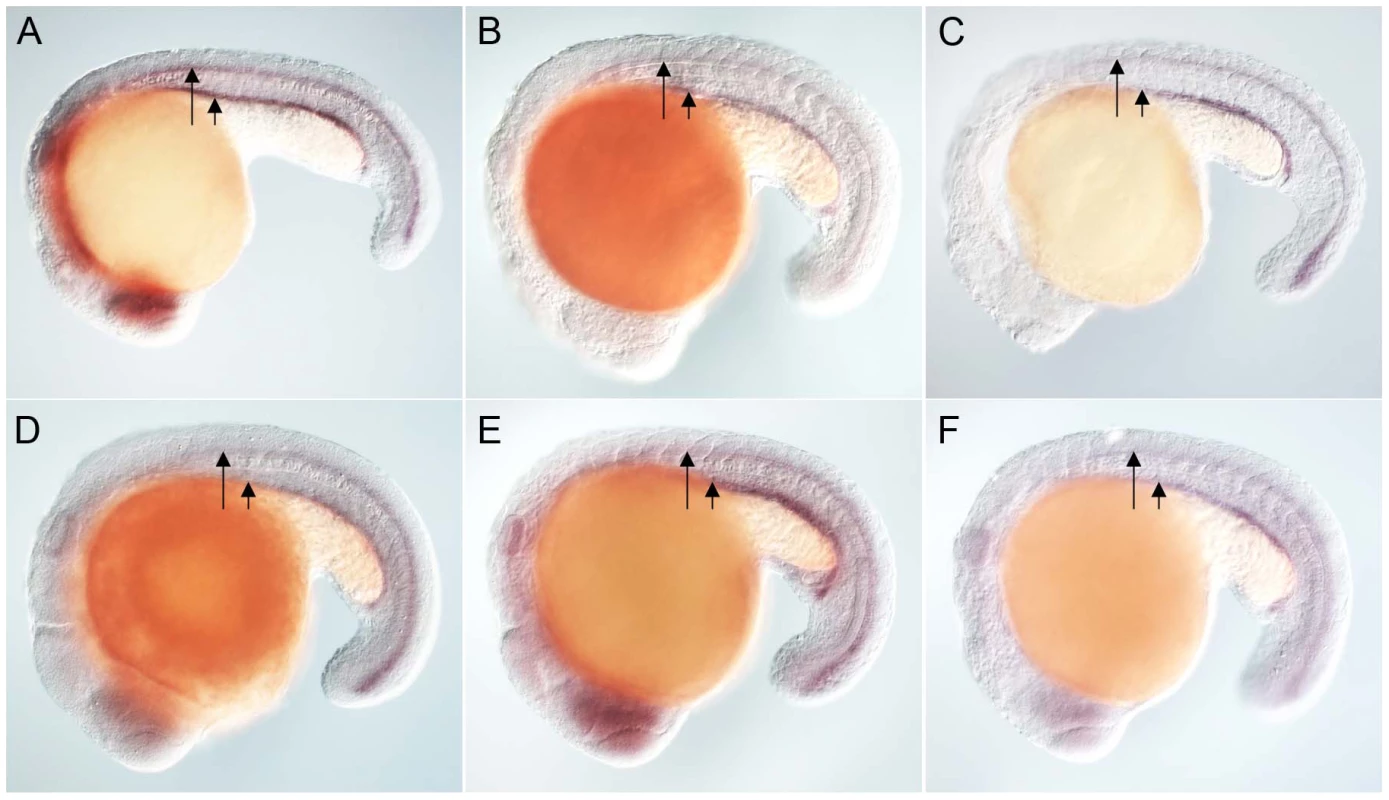
(A) Expression of efhc1 in the spinal cord (long arrow) and pronephric (kidney) duct (short arrow) of a wild-type zebrafish embryo, and in embryos ectopically expressing zebrafish FoxJ2 (B) and FoxJ3 (C), respectively. (D) Expression of spag6 in the spinal cord (long arrow) and pronephric (kidney) duct (short arrow) of a wild-type zebrafish embryo, and in embryos ectopically expressing zebrafish FoxJ2 (E) and FoxJ3 (F), respectively. Embryos depicted are at 20 hpf, oriented anterior to the left, dorsal to the top. Inactivation of a FoxJ1 homolog impairs motile cilia differentiation in S. mediterranea
Encouraged by the mis-expression studies in the zebrafish embryo, we wished to gather evidence that the non-vertebrate FoxJ1 proteins indeed control motile cilia differentiation in the context of their native species, just like their vertebrate counterparts. Among the non-vertebrate foxJ1 genes, spatio-temporal expression pattern of only the sea urchin homolog has been described previously. The gene is transcribed in larval cells that bear tufts of motile cilia, implicating a role in ciliogenesis [34]. However, the lack of well-established methods of gene manipulation in echinoderm larvae or placozoans, and the absence of FoxJ1 from the traditional genetically amenable invertebrate models, C. elegans and D. melanogaster, made us resort to the planarian S. mediterranea. S. mediterranea is a representative of the basal worm phylum Platyhelminthes (flatworms), characterized by unsegmented and acoelomic morphology. S. mediterranea is popular in regeneration research, and RNAi mediated loss of gene activity can be efficiently achieved in this organism. Much less appreciated is the fact that the ventral epithelium of the worms consists of multiciliated cells very similar to the mammalian respiratory epithelium [35]. Metachronal waves of ciliary beating propel the worm around on a film of mucus, making the system an ideal model for mammalian pulmonary mucociliary clearance. To determine whether these morphological similarities extended even to the transcriptional regulation of ciliogenesis, we first searched the genome of S. mediterranea for foxJ1 homologs. Many genes occur in multiple copies in this species, and BLAST searches with human FoxJ1 yielded four separate sequences that phylogenetic analysis confirmed to be paralogous S. mediterranea FoxJ1 proteins (SMED-FOXJ1-1 through -FOXJ1-4; BS support for FOXJ1-4 was the highest (99.9%) while that for FOXJ1-1, FOXJ1-2, FOXJ1-3 was 87.4%, 58.1% and 96.4%, respectively). In situ hybridization showed that Smed-foxJ1-4 is widely expressed in the ventral motile ciliated cells, and dorsally in the head and in a stripe along the midline (Figure 3B, 3E). The dorsal expression regions also correspond to ciliated cells, but the function of these cilia is presently unclear (motile or sensory). Overall, foxJ1-4 is expressed in a manner highly reminiscent of core ciliary genes like Smed-ift72, which encodes a highly conserved component of the IFT machinery (Figure 3D, 3F). By contrast, the expression of foxJ1-1 and foxJ1-2 is much more limited, and is confined to the mid-dorsal stripe of presumptive ciliated sensory cells (Figure 3G, 3H). We failed to observe a distinct expression pattern for foxJ1-3. In order to determine whether any of the 4 foxJ1 orthologs might function in planarian ciliogenesis, we targeted the genes individually and in combination by feeding worms with dsRNA. ift172(RNAi) served as positive control for cilia phenotypes [36]. Animals that received ift172 RNAi gradually lost their gliding ability - instead, they moved by peristaltic waves of whole body contractions (“inchworming”, Video S1). Like the ift172(RNAi) animals, those with foxJ1-4 (but not foxJ1-1, -2 or -3) RNAi also lost their gliding ability and moved around by inchworming (Video S1). Furthermore, ift172(RNAi) and some foxJ1-4(RNAi) worms developed posterior edema (Figure 4A–4C). Both inchworming and edema formation have previously been described as hallmarks of cilia defects in planarians [36], [37], and thus, strongly indicated a function of FOXJ1-4 in planarian ciliogenesis. Direct visualization of ciliary morphology confirmed this notion. Anti-α-tubulin staining of the ciliary axonemes revealed a dense lawn of cilia on the ventral epithelium of control worms (Figure 4D). ift172(RNAi) worms showed drastically shortened cilia remnants (Figure 4E) [36], whereas the foxJ1-4(RNAi) animals were almost entirely devoid of cilia (Figure 4F). To ensure that the RNAi procedure specifically knocked down the targeted genes, we performed in situ hybridization on RNAi fed worms. Control RNAi with a sequence not present in the planarian genome (DsRed) had no effect on foxJ1-4 expression (Figure 4G), nor did foxJ1-4(RNAi) affect foxJ1-1 expression (Figure 4H). foxJ1-4(RNAi), however, substantially reduced the expression levels of foxJ1-4 (Figure 4I). Using the mis-expression assay described for the placozoan and echinoderm FoxJ1 proteins in the foregoing section, we also found that SMED-FOXJ1-4 could robustly activate motile ciliary genes in zebrafish embryos (data not shown). These results demonstrate that just like the vertebrates, FoxJ1 is a critical rate-limiting component of motile cilia differentiation even in the phylogenetically distant planarians.
Fig. 3. S. mediterranea foxJ1 genes are expressed in ciliated tissues. 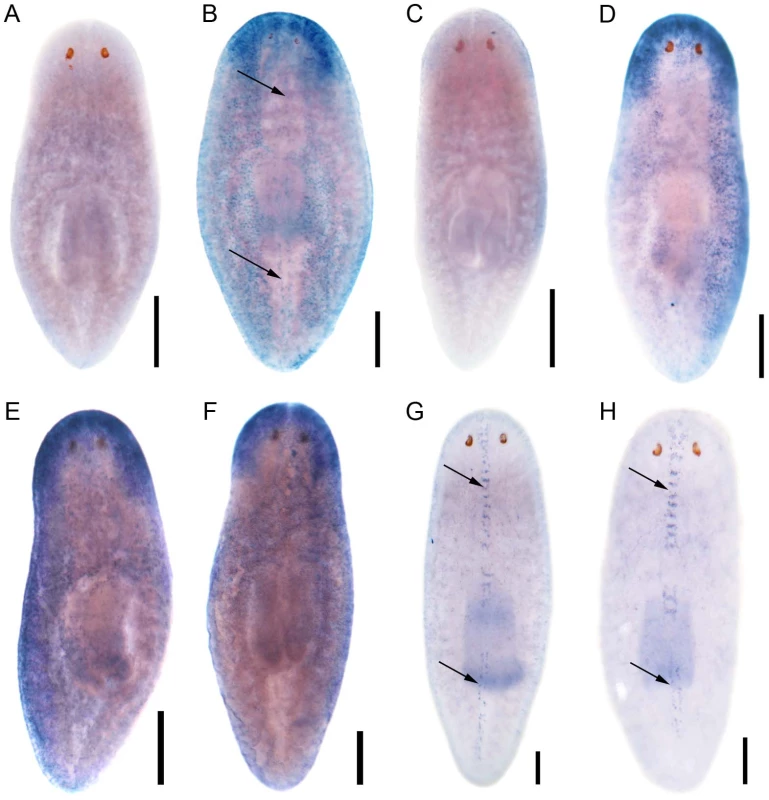
Sense control (A) and expression pattern of Smed-foxJ1-4 depicted in dorsal (B) and ventral view (E). Sense control (C) and expression pattern of Smed-ift172 shown in dorsal (D) and ventral view (F). Expression pattern of Smed-foxJ1-1 (G) and expression pattern of Smed-foxJ1-2 (H). Arrows in B, G and H denote expression in the dorsal stripe of presumptive ciliated sensory cells. Scale bars: 300 µm for A–F and 200 µm for G–H. Fig. 4. S. mediterranea foxJ1-4 is required for the differentiation of motile cilia. 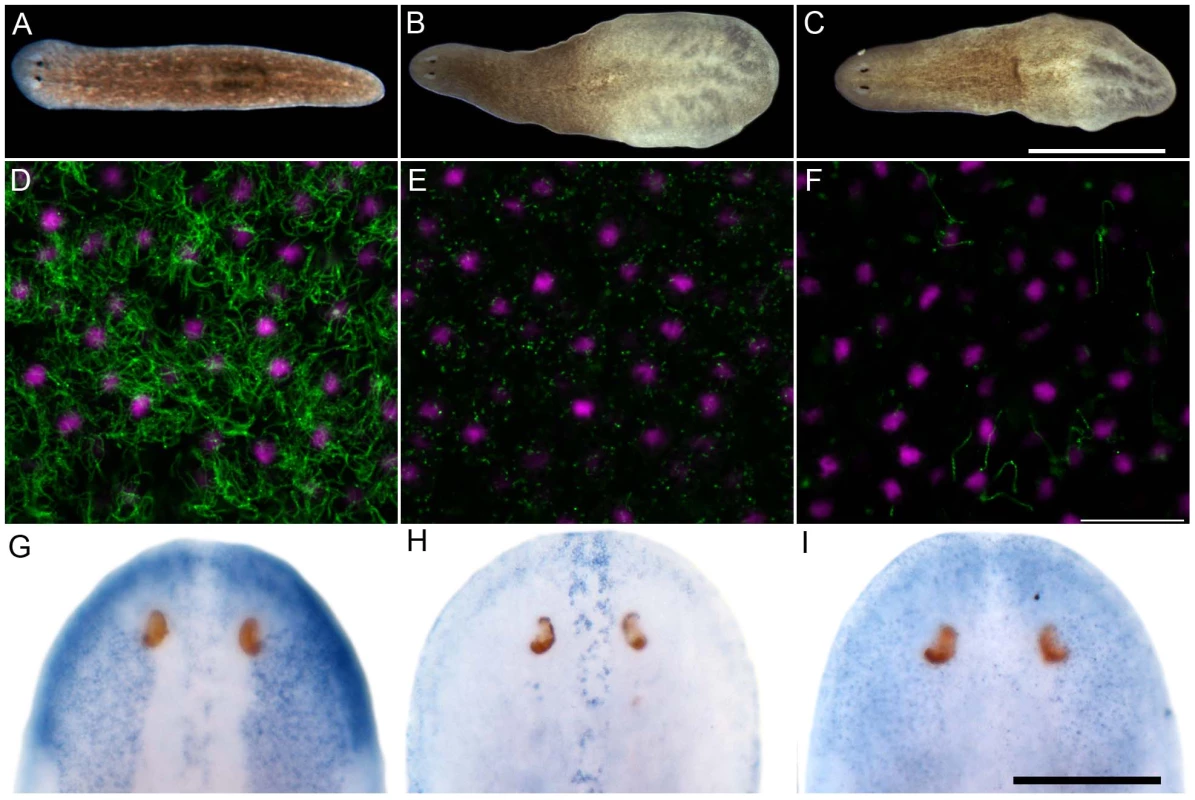
(A–C) Control (A), ift172(RNAi) (B) and foxJ1-4(RNAi) worms (C) 14 days after the last RNAi feeding. Worms shown in panels B and C display tissue edema. Scale bar: 1 mm. (D–F) Anti-α-tubulin staining (green) of the multi-ciliated ventral epithelium in control (D), ift172(RNAi) (E) and foxJ1-4(RNAi) worms (F), respectively. The even spacing of nuclei (magenta) characteristic of the ventral epithelium demonstrates epithelial integrity in E and F. Images are single optical sections. Scale bar: 20 µm. (G) foxJ1-4 expression is unaffected by control RNAi (dsred). (H) foxJ1-1 expression is not altered in a foxJ1-4(RNAi) worm. (I) foxJ1-4 expression is substantially reduced in a foxJ1-4(RNAi) worm. Scale bar: 200 µm. Discussion
A large number of transcription factors important for bilaterian development seem to have evolved even before the bilaterians and cnidarians diverged, as the two groups show considerable similarity in transcription factor family size and diversity. Similarly, the placozoans also have a comparable number of transcription factor families; for example, 16 out of the 22 bilaterian Fox subfamilies could be identified in T. adhaerens. In comparison, sponges have a more restricted representation, thus, indicating that the expansion and diversification in the suite of transcription factors was an early event in animal evolution [26], [38]. The FKH domain containing proteins have been identified only among the opisthokonts [38]. Within the Bilateria, there are FoxA-FoxS subfamilies, of which FoxL, J, N and Q have been further subdivided. Amongst these, FoxR and FoxS are specific to the vertebrates, while another subfamily, FoxAB, appears to be invertebrate-specific. These together comprise the 22 bilaterian subfamilies of Fox proteins. Most species have lost one or more subfamily members except Branchiostoma floridae, the only organism as yet sequenced whose genome encodes all of the Fox family representatives [38]. In our study too, we were unable to identify suitable FoxJ1 homologs in Archaeplastida (plants and their relatives), Excavata, Rhizaria, and Chromalveolata. Amongst the unikonts, FoxJ1 could be identified only within the opisthokont lineage. All ciliated organisms outside the unikont lineage, and even within the unikonts some species of ciliated fungi such as Batrachochytrium dendrobatidis lack FoxJ1. Conversely, FoxJ1 is present in several fungal species that are known to lack cilia.
A survey of the Rfx family of transcription factors have shown that, similar to our findings with FoxJ1, they are absent from the bikonts, but are present in Acanthamoeba castellani, a basal group within the unikont lineage, although two domains characteristic of the Rfx proteins – the dimerization domain and domain C are missing. Furthermore, compared to FoxJ1, the Rfx proteins are more wide-spread within the opisthokonts [19], [20]. Two Rfx genes have been identified in the choanoflagellate M. brevicollis [20], but we were unable to find a genuine FoxJ1 homolog. This is in contrast to an earlier study that reported FoxJ1 in M. brevicollis along with several other Fox proteins (FoxJ2/3, FoxN1/N4), and classified all these Fox family members as primitive since these were the only Fox proteins identified in M. brevicollis [38]. Another analysis identified seven Fox genes in the M. brevicollis genome, of which only FoxN1/4 and FoxJ2 were described to be true orthologs. The remaining five genes show similarity to FoxJ1/FoxJ2 on BLAST search, but their sequences are highly divergent, and hence they could not be classified into specific Fox subfamilies based on phylogeny [26]. In our present search, a M. brevicollis Fox protein was identified on BLAST using the human FOXJ1 FKH domain sequence, but it could not be classified as FoxJ1 based on phylogenetic analysis (Table 1). This concurs with a recent work that also failed to identify FoxJ1 from M. brevicollis [39]. On the other hand, reverse BLAST classifies the M. brevicollis protein as FoxJ1. The same results were obtained using the full-length human FOXJ1 sequence for identification of orthologs. In view of these conflicting lines of evidence, it seems parsimonious to state that the presence of a bona fide FoxJ1 protein in M. brevicollis remains controversial. It also emerges from the previous surveys of the Rfx factors [19], [20] and our current study of FoxJ1, that although these transcription factors have a checkered distribution, the core ciliary gene repertoire that they regulate is quite conserved across distant phyla, ranging from the protists all the way to humans. This implies that like the Rfx family members, FoxJ1 arose within the unikont lineage only after the evolution of cilia.
Using a mis-expression assay in zebrafish embryos, we have established that FoxJ1 proteins from three divergent taxonomic groups, the placozoans, platyhelminths and the echinoderms, have a conserved role in the regulation of motile ciliary genes. Despite their distant phyletic relationships, all three proteins could robustly activate the five canonical vertebrate FoxJ1 targets that we tested, whereas zebrafish FoxJ2 and FoxJ3, the two Fox proteins most closely related to zebrafish FoxJ1, failed to do so. Mis-expression of zebrafish, Xenopus and chicken FoxJ1 proteins in zebrafish, Xenopus and chicken embryos, respectively, not only activates motile ciliary genes, but also induces ectopic motile cilia biogenesis [15]–[17]. However, none of the invertebrate FoxJ1 proteins that we tested were sufficient for ectopic motile cilia formation in zebrafish embryos, indicating their incapability to fully institute the zebrafish motile ciliogenic pathway (data not shown). To rule out differences in the amounts of over-expressed FoxJ1 proteins as a cause of this discrepancy, we quantified their levels using Western blot. Although the placozoan FoxJ1 consistently showed lower levels of expression compared to the zebrafish and the sea urchin proteins (possibly due to disparity in codon usage between the placozoans and vertebrates or instability of the protein when expressed in a heterologous system), the latter two were expressed at more or less equivalent levels (Figure S1). In light of this finding, we argue that species-specific differences in the FoxJ1 DNA binding domains and their corresponding DNA recognition sequence, together with species-specific diversity in the repertoire of target genes are more plausible reasons for the inability of the invertebrate FoxJ1 proteins to induce ciliogenesis in zebrafish.
To uncover a requirement of FoxJ1 function in motile cilia differentiation outside of the vertebrates, we have complemented the mis-expression analysis with a loss-of-function study of the planarian homologs of FoxJ1. Out of four paralogous foxJ1 genes in S. mediterranea, we found that one of them, foxJ1-4, is absolutely necessary for ciliogenesis. Similar to the loss of motile cilia in FoxJ1 deficient zebrafish, frogs and mice, foxJ1-4(RNAi) planarians lost the ciliation of their ventral epithelium, where foxJ1-4 is expressed. The fact that some foxJ1-4(RNAi) animals additionally developed edema indicates that foxJ1-4 is required for ciliogenesis also in the planarian excretory system, which consists of heavily ciliated protonephridial tubules [37], just like the pronephric ducts of zebrafish embryos [40]. Although two distinct foxJ1 paralogs have been reported from Xenopus and zebrafish [17], [41], [42], [43], interestingly, planarians are unique amongst the species examined here, where we detected four foxJ1-homologues. foxJ1 is not the first example of gene amplification in planarians. noggins and noggin-like genes have been amplified to a total of ten family members in planarians [44]; other examples include six wnt11 homologues [45], and two β-catenin genes [46] apparently segregating signaling and cell adhesion function. In the zebrafish and frogs, the two foxJ1 paralogs have distinct as well as overlapping expression patterns, and functional analysis of the zebrafish genes have underscored their unique as well as redundant roles in regulating ciliogenesis in the different motile cilia bearing tissues that express them [17], [41], [42], [43]. The expression patterns of 3 of the 4 different planarian foxJ1 genes suggests that like the duplicated foxJ1 genes of fishes and amphibians, some degree of sub-functionalization within the ancestral ciliogenic role has also occurred here, with the different paralogs being delegated to the regulation of ciliogenesis in distinct tissues. Whereas foxJ1-4 is critically required for the differentiation of the motile multiple cilia on ventral epidermal cells, the foxJ1-1 and -2 genes are not. Instead, they could have a role in the formation of cilia in the dorsal midline.
In conclusion, our findings have uncovered an ancient link between FoxJ1 and the motile ciliogenic program, and provide evidence that this regulatory mechanism is an ancestral feature of metazoan evolution. The distribution of FoxJ1 across various phyletic groups seems to suggest that the transcription factor evolved in the context of multicellularity, at a time when sophisticated regulatory controls were required for the differentiation of cilia. In the zebrafish and mammals, FoxJ1 and Rfx have been shown to cross-regulate each other's expression in the ciliogenic pathway [17], [47]. It is now apparent from the earlier studies with Rfx and our current work with FoxJ1 that the two ciliogenic transcription factors coexist in several other species; to what extent they collaborate to control ciliogenesis in each of these instances remains to be determined. Finally, in organisms that lack FoxJ1 but bear motile cilia, ciliary differentiation could be programmed entirely by Rfx, or by yet undiscovered transcriptional mechanisms.
Materials and Methods
Taxa sampling to identify FoxJ1 proteins
The FKH domain of human FoxJ1 was used to search for potential orthologs in 215 species representing all the major eukaryotic lineages by conducting BLASTP and TBLASTN searches in various databases including the NCBI non-redundant protein sequences database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), Joint Genomes Institute, Department of Energy (genome.jgi-psf.org), Human Genome Sequencing Center, Baylor College of Medicine (http://www.hgsc.bcm.tmc.edu), Broad Institute (www.broadinstitute.org), Ensembl Genome Browser (http://www.ensembl.org) and Saccharomyces Genome Database (www.yeastgenome.org). Only those organisms whose proteins gave an E-value less than E-2 were considered to have a potential FoxJ1 ortholog. The FKH domain of each protein identified in BLAST was extracted using SMART (http://smart.embl-heidelberg.de) database and aligned (ClustalX multiple sequence alignment program) [48] with the 42 mouse FKH domain sequences. These data were used for making a neighbor joining tree, which was viewed using Treeview [49] to ascertain the identity of each protein. Alternatively, and independent of the phylogenetic analyses, the FKH domain of each identified protein was also used for reverse BLAST search against the Homo sapiens non-redundant protein database (NCBI) in order to assign the identified proteins to a particular subclass. The above described BLAST as well as phylogenetic analyses was also done using the full-length human FoxJ1 sequence. In instances where the full-length FoxJ1 sequence could not be retrieved for a particular species, the longest available sequence was used for the analyses.
Zebrafish strains
Wild-type zebrafish were maintained under standard conditions of fish husbandry. All experiments with zebrafish embryos were approved by the Singapore National Advisory Committee on Laboratory Animal Research.
foxJ1, foxJ2, and foxJ3 full-length cDNAs
Incomplete sequence and annotation details were available for the Trichoplax adhaerens foxJ1 gene in the public repositories (genome.jgi.doe.gov). Therefore, a full-length clone (1578 bp) (JX569795) was derived from sequencing of a Placozoa sp. H4 cDNA library [50], [51]. The full-length S. purpuratus foxJ1 (1407 bp) was amplified from a sea urchin larval cDNA pool based on the annotation available in NCBI. The full length zebrafish foxJ2 (1551 bp) and foxJ3 (1779 bp) were amplified from 1 day old zebrafish embryonic cDNA pool. Cloning of zebrafish foxJ1a was reported previously [17]. Planarian foxJ1 homologs were cloned from cDNA obtained from an 8-day regeneration series as described previously [52].
RNAi in S. mediterranea via dsRNA feeding
Gene silencing in S. mediterranea was performed as previously described [52]. Bacterial pellet resulting from 70 ml of culture was mixed with 150 µl liver paste (3 parts liver∶1 part water). For double RNAi experiments, 35 ml of each IPTG-induced culture was mixed prior to pelleting. Worms received three feedings of RNAi food (2 days in between) and were fixed for analysis 14 days after the last feed.
Ectopic foxJ1, foxJ2, and foxJ3 expression in zebrafish embryos
For generating the heat inducible placozoan, platyhelminth, sea urchin and zebrafish foxJ1 as well as zebrafish foxJ2 and foxJ3 constructs, six myc epitope tags were amplified from the pCS2+MT vector and cloned into the XbaI-SpeI sites of the pHspIG heat-shock vector. Coding sequences of the different fox genes were then cloned downstream of the myc tag to generate the hs::myc-Pl-foxJ1, hs::myc-Sm-foxJ1-4, hs::myc-Sp-foxJ1, hs::myc-Dr-foxJ1, hs::myc-Dr-foxJ2 and hs::myc-Dr-foxJ3 transgenes, respectively. For assessing the ability of the different Fox proteins to activate the expression of zebrafish motile ciliary genes, zebrafish embryos injected with the different heat inducible fox gene constructs were heat shocked at 37°C for 1 h at 14 hours post-fertilization (hpf). Following this, the embryos were allowed to grow until 20 hpf at 28°C before fixation for in situ hybridization.
Microinjections into zebrafish eggs
The different heat inducible transgene containing plamids were linearized and injected at a concentration of ∼25 ng/µl into freshly fertilized zebrafish eggs at the one-cell stage. The typical volume for injections was ∼1 nl.
RNA in situ hybridization and antibody staining
For mRNA in situ hybridization, worms were euthanized, fixed, hybridized, and developed as previously described [53]. For antibody labeling of cilia, animals were euthanized in 5% weight/volume N-acetyl cysteine in PBS under gentle rocking for 5 min and fixed for 2 h at 4°C in Carnoy's fixative (6 parts ethanol, 3 parts chloroform, 1 part glacial acetic acid). Following intensive rinsing with methanol, animals were bleached overnight in 6% H2O2 in 80% methanol, rehydrated and stained with anti-α-tubulin antibodies (Sigma), and visualized with Alexa-fluor-555 conjugated anti-mouse secondary antibodies (Invitrogen). Whole mount in situ hybridization and antibody staining of zebrafish embryos were done according to standard protocols. Antisense mRNA probes were used for the following genes: spag6, wdr78, efhc1, tektin1 and dynein intermediate chain. The following antibodies were used: mAb anti-acetylated tubulin (Sigma) and anti-c-Myc (A-14 (sc-789); Santa Cruz Biotechnology). Alexa-fluor-conjugated secondary antibodies (Molecular Probes) were used for detection of signals. Nuclei were stained with DAPI.
Microscopy
Stained S. mediterranea preparations were mounted in 80% glycerol and imaged on a Zeiss LSM700 confocal microscope equipped with multi-emersion objectives. Stained zebrafish embryos were mounted in 70% glycerol. A Zeiss compound microscope (Axio Imager Z1) fitted with a Zeiss digital camera (AxioCam HRc Rev 2) and an Olympus Fluoview laser scanning confocal microscope were used for imaging.
Supporting Information
Zdroje
1. SatirP, ChristensenST (2007) Overview of structure and function of mammalian cilia. Annu Rev Physiol 69 : 377–400.
2. IshikawaH, MarshallWF (2011) Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 12 : 222–234.
3. FliegaufM, BenzingT, OmranH (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8 : 880–893.
4. BakerK, BealesPL (2009) Making sense of cilia in disease: the human ciliopathies. Am J Med Genet C Semin Med Genet 151C: 281–295.
5. RoyS (2009) The motile cilium in development and disease: emerging new insights. Bioessays 31 : 694–699.
6. ThomasJ, MorleL, SoulavieF, LaurenconA, SagnolS, et al. (2010) Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell 102 : 499–513.
7. BlacqueOE, PerensEA, BoroevichKA, InglisPN, LiC, et al. (2005) Functional genomics of the cilium, a sensory organelle. Curr Biol 15 : 935–941.
8. ChenN, MahA, BlacqueOE, ChuJ, PhgoraK, et al. (2006) Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol 7: R126.
9. EfimenkoE, BubbK, MakHY, HolzmanT, LerouxMR, et al. (2005) Analysis of xbx genes in C. elegans. Development 132 : 1923–1934.
10. LaurenconA, DubruilleR, EfimenkoE, GrenierG, BissettR, et al. (2007) Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol 8: R195.
11. SwobodaP, AdlerHT, ThomasJH (2000) The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell 5 : 411–421.
12. Ait-LounisA, BaasD, BarrasE, BenadibaC, CharollaisA, et al. (2007) Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes 56 : 950–959.
13. BonnafeE, ToukaM, AitLounisA, BaasD, BarrasE, et al. (2004) The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol 24 : 4417–4427.
14. DornA, DurandB, MarfingC, Le MeurM, BenoistC, et al. (1987) Conserved major histocompatibility complex class II boxes–X and Y–are transcriptional control elements and specifically bind nuclear proteins. Proc Natl Acad Sci U S A 84 : 6249–6253.
15. CruzC, RibesV, KutejovaE, CayusoJ, LawsonV, et al. (2010) FoxJ1 regulates floor plate cilia architecture and modifies the response of cells to sonic hedgehog signalling. Development 137 : 4271–4282.
16. StubbsJL, OishiI, Izpisua BelmonteJC, KintnerC (2008) The forkhead protein FoxJ1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40 : 1454–1460.
17. YuX, NgCP, HabacherH, RoyS (2008) FoxJ1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40 : 1445–1453.
18. AltenL, Schuster-GosslerK, BeckersA, GroosS, UlmerB, et al. (2012) Differential regulation of node formation, nodal ciliogenesis and cilia positioning by Noto and FoxJ1. Development 139 : 1276–1284.
19. ChuJS, BaillieDL, ChenN (2010) Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol Biol 10 : 130.
20. PiaseckiBP, BurghoornJ, SwobodaP (2010) Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci U S A 107 : 12969–12974.
21. Mitchell DR (2006) The evolution of eukaryotic cilia and flagella as motile and sensory organelles. In: Jékely G, editors. Origins and Evolution of Eukaryotic Endomembranes and Cytoskeleton. Eurekah.com. pp 1–11.
22. BrodySL, YanXH, WuerffelMK, SongSK, ShapiroSD (2000) Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol 23 : 45–51.
23. ChenJ, KnowlesHJ, HebertJL, HackettBP (1998) Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest 102 : 1077–1082.
24. Ait-LounisA, BonalC, Seguin-EstevezQ, SchmidCD, BucherP, et al. (2010) The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes 59 : 1674–1685.
25. HannenhalliS, KaestnerKH (2009) The evolution of Fox genes and their role in development and disease. Nat Rev Genet 10 : 233–240.
26. LarrouxC, LukeGN, KoopmanP, RokhsarDS, ShimeldSM, et al. (2008) Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 25 : 980–996.
27. DellaportaSL, XuA, SagasserS, JakobW, MorenoMA, et al. (2006) Mitochondrial genome of Trichoplax adhaerens supports placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci U S A 103 : 8751–8756.
28. SchierwaterB (2005) My favorite animal, Trichoplax adhaerens. Bioessays 27 : 1294–1302.
29. SchierwaterB, EitelM, JakobW, OsigusHJ, HadrysH, et al. (2009) Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol 7: e20 doi:10.1371/journal.pbio.1000020.
30. PhilippeH, DerelleR, LopezP, PickK, BorchielliniC, et al. (2009) Phylogenomics revives traditional views on deep animal relationships. Curr Biol 19 : 706–712.
31. SrivastavaM, BegovicE, ChapmanJ, PutnamNH, HellstenU, et al. (2008) The Trichoplax genome and the nature of placozoans. Nature 454 : 955–960.
32. ClevidenceDE, OverdierDG, PetersonRS, PorcellaA, YeH, et al. (1994) Members of the HNF-3/forkhead family of transcription factors exhibit distinct cellular expression patterns in lung and regulate the surfactant protein B promoter. Dev Biol 166 : 195–209.
33. LimL, ZhouH, CostaRH (1997) The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc Natl Acad Sci USA 94 : 3094–3099.
34. TuQ, BrownCT, DavidsonEH, OliveriP (2006) Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol 300 : 49–62.
35. RompolasP, Patel-KingRS, KingSM (2009) Schmidtea mediterranea: a model system for analysis of motile cilia. Methods Cell Biol 93 : 81–98.
36. RinkJC, GurleyKA, ElliottSA, Sanchez AlvaradoA (2009) Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326 : 1406–1410.
37. RinkJC, VuHT, AlvaradoAS (2011) The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138 : 3769–3780.
38. ShimeldSM, DegnanB, LukeGN (2010) Evolutionary genomics of the Fox genes: origin of gene families and the ancestry of gene clusters. Genomics 95 : 256–260.
39. Sebe-PedrosA, de MendozaA, LangBF, DegnanBM, Ruiz-TrilloI (2011) Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol 28 : 1241–1254.
40. Kramer-ZuckerAG, OlaleF, HaycraftCJ, YoderBK, SchierAF, et al. (2005) Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development 132 : 1907–1921.
41. ChoiVM, HarlandRM, KhokhaMK (2006) Developmental expression of FoxJ1.2, FoxJ2 and FoxQ1 in Xenopus tropicalis. Gene Expr Patterns 6 : 443–447.
42. PohlBS, KnöchelW (2004) Isolation and developmental expression of Xenopus FoxJ1 and FoxK1. Dev Genes Evol 214 : 200–205.
43. YuX, LauD, NgCP, RoyS (2011) Cilia-driven fluid flow as an epigenetic cue for otolith biomineralization on sensory hair cells of the inner ear. Development 138 : 487–494.
44. MolinaMD, SaloE, CebriaF (2009) Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns 9 : 246–253.
45. GurleyKA, ElliottSA, SimakovO, SchmidtHA, HolsteinTW, et al. (2010) Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347 : 24–39.
46. ChaiG, MaC, BaoK, ZhengL, WangX, et al. (2010) Complete functional segregation of planarian beta-catenin-1 and -2 in mediating Wnt signaling and cell adhesion. J Biol Chem 285 : 24120–24130.
47. El ZeinL, Ait-LounisA, MorleL, ThomasJ, ChhinB, et al. (2009) RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J Cell Sci 122 : 3180–3189.
48. ThompsonJD, HigginsDG, GibsonTJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 : 4673–4680.
49. PageRD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12 : 357–358.
50. EitelM, SchierwaterB (2010) The phylogeography of the Placozoa suggests a taxon rich phylum in tropical and subtropical waters. Molecular Ecology 19 : 2315–2327.
51. EitelM, GuidiL, HadrysH, BalsamoM, SchierwaterB (2011) New insights into placozoan sexual reproduction and development. PLoS ONE 6: e19639 doi:10.1371/journal.pone.0019639.
52. GurleyKA, RinkJC, Sanchez AlvaradoA (2008) Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319 : 323–327.
53. PearsonBJ, EisenhofferGT, GurleyKA, RinkJC, MillerDE, et al. (2009) Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 238 : 443–450.
Štítky
Genetika Reprodukční medicína
Článek The Covariate's DilemmaČlánek Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene SignallingČlánek Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- The Covariate's Dilemma
- Plant Vascular Cell Division Is Maintained by an Interaction between PXY and Ethylene Signalling
- Plan B for Stimulating Stem Cell Division
- Discovering Thiamine Transporters as Targets of Chloroquine Using a Novel Functional Genomics Strategy
- Is a Modifier of Mutations in Retinitis Pigmentosa with Incomplete Penetrance
- Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program
- Genome Instability Caused by a Germline Mutation in the Human DNA Repair Gene
- Transcription Factor Oct1 Is a Somatic and Cancer Stem Cell Determinant
- Controls of Nucleosome Positioning in the Human Genome
- Disruption of Causes Defective Meiotic Recombination in Male Mice
- A Novel Human-Infection-Derived Bacterium Provides Insights into the Evolutionary Origins of Mutualistic Insect–Bacterial Symbioses
- Trps1 and Its Target Gene Regulate Epithelial Proliferation in the Developing Hair Follicle and Are Associated with Hypertrichosis
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Population-Based Resequencing of in 10,330 Individuals: Spectrum of Genetic Variation, Phenotype, and Comparison with Extreme Phenotype Approach
- HP1a Recruitment to Promoters Is Independent of H3K9 Methylation in
- Transcription Elongation and Tissue-Specific Somatic CAG Instability
- A Germline Polymorphism of DNA Polymerase Beta Induces Genomic Instability and Cellular Transformation
- Interallelic and Intergenic Incompatibilities of the () Gene in Mouse Hybrid Sterility
- Comparison of Mitochondrial Mutation Spectra in Ageing Human Colonic Epithelium and Disease: Absence of Evidence for Purifying Selection in Somatic Mitochondrial DNA Point Mutations
- Mutations in the Transcription Elongation Factor SPT5 Disrupt a Reporter for Dosage Compensation in Drosophila
- Evolution of Minimal Specificity and Promiscuity in Steroid Hormone Receptors
- Blockade of Pachytene piRNA Biogenesis Reveals a Novel Requirement for Maintaining Post-Meiotic Germline Genome Integrity
- RHOA Is a Modulator of the Cholesterol-Lowering Effects of Statin
- MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways
- Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in
- Genome-Wide Association Studies Reveal a Simple Genetic Basis of Resistance to Naturally Coevolving Viruses in
- The Principal Genetic Determinants for Nasopharyngeal Carcinoma in China Involve the Class I Antigen Recognition Groove
- Molecular, Physiological, and Motor Performance Defects in DMSXL Mice Carrying >1,000 CTG Repeats from the Human DM1 Locus
- Genomic Study of RNA Polymerase II and III SNAP-Bound Promoters Reveals a Gene Transcribed by Both Enzymes and a Broad Use of Common Activators
- Long Telomeres Produced by Telomerase-Resistant Recombination Are Established from a Single Source and Are Subject to Extreme Sequence Scrambling
- The Yeast SR-Like Protein Npl3 Links Chromatin Modification to mRNA Processing
- Deubiquitylation Machinery Is Required for Embryonic Polarity in
- dJun and Vri/dNFIL3 Are Major Regulators of Cardiac Aging in Drosophila
- CtIP Is Required to Initiate Replication-Dependent Interstrand Crosslink Repair
- Notch-Mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Intestinal Stem Cell Lineage
- A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation
- Network Analysis of a -Mouse Model of Autosomal Dominant Polycystic Kidney Disease Identifies HNF4α as a Disease Modifier
- Mitosis in Neurons: Roughex and APC/C Maintain Cell Cycle Exit to Prevent Cytokinetic and Axonal Defects in Photoreceptor Neurons
- CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function
- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- The Genomes of the Fungal Plant Pathogens and Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry
- A Genome-Scale RNA–Interference Screen Identifies RRAS Signaling as a Pathologic Feature of Huntington's Disease
- Lessons from Model Organisms: Phenotypic Robustness and Missing Heritability in Complex Disease
- Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites
- Tissue-Specific Regulation of Chromatin Insulator Function
- Disruption of Mouse Cenpj, a Regulator of Centriole Biogenesis, Phenocopies Seckel Syndrome
- Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga CCMP1779
- Antagonistic Gene Activities Determine the Formation of Pattern Elements along the Mediolateral Axis of the Fruit
- Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma
- Identification of the First ATRIP–Deficient Patient and Novel Mutations in ATR Define a Clinical Spectrum for ATR–ATRIP Seckel Syndrome
- Cooperativity of , , and in Malignant Breast Cancer Evolution
- Loss of Prohibitin Membrane Scaffolds Impairs Mitochondrial Architecture and Leads to Tau Hyperphosphorylation and Neurodegeneration
- Microhomology Directs Diverse DNA Break Repair Pathways and Chromosomal Translocations
- MicroRNA–Mediated Repression of the Seed Maturation Program during Vegetative Development in
- Selective Pressure Causes an RNA Virus to Trade Reproductive Fitness for Increased Structural and Thermal Stability of a Viral Enzyme
- The Tumor Suppressor Gene Retinoblastoma-1 Is Required for Retinotectal Development and Visual Function in Zebrafish
- Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
- Polyadenylation-Dependent Control of Long Noncoding RNA Expression by the Poly(A)-Binding Protein Nuclear 1
- A Unified Method for Detecting Secondary Trait Associations with Rare Variants: Application to Sequence Data
- Genetic and Biochemical Dissection of a HisKA Domain Identifies Residues Required Exclusively for Kinase and Phosphatase Activities
- Informed Conditioning on Clinical Covariates Increases Power in Case-Control Association Studies
- Biochemical Diversification through Foreign Gene Expression in Bdelloid Rotifers
- Genomic Variation and Its Impact on Gene Expression in
- Spastic Paraplegia Mutation N256S in the Neuronal Microtubule Motor KIF5A Disrupts Axonal Transport in a HSP Model
- Lamin B1 Polymorphism Influences Morphology of the Nuclear Envelope, Cell Cycle Progression, and Risk of Neural Tube Defects in Mice
- A Targeted Glycan-Related Gene Screen Reveals Heparan Sulfate Proteoglycan Sulfation Regulates WNT and BMP Trans-Synaptic Signaling
- Dopaminergic D2-Like Receptors Delimit Recurrent Cholinergic-Mediated Motor Programs during a Goal-Oriented Behavior
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mechanisms Employed by to Prevent Ribonucleotide Incorporation into Genomic DNA by Pol V
- Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data
- Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival
- Histone Methyltransferases MES-4 and MET-1 Promote Meiotic Checkpoint Activation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



