-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
[], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
The yeast prion [SWI+], formed of heritable amyloid aggregates of the Swi1 protein, results in a partial loss of function of the SWI/SNF chromatin-remodeling complex, required for the regulation of a diverse set of genes. Our genetic analysis revealed that [SWI+] propagation is highly dependent upon the action of members of the Hsp70 molecular chaperone system, specifically the Hsp70 Ssa, two of its J-protein co-chaperones, Sis1 and Ydj1, and the nucleotide exchange factors of the Hsp110 family (Sse1/2). Notably, while all yeast prions tested thus far require Sis1, [SWI+] is the only one known to require the activity of Ydj1, the most abundant J-protein in yeast. The C-terminal region of Ydj1, which contains the client protein interaction domain, is required for [SWI+] propagation. However, Ydj1 is not unique in this regard, as another, closely related J-protein, Apj1, can substitute for it when expressed at a level approaching that of Ydj1. While dependent upon Ydj1 and Sis1 for propagation, [SWI+] is also highly sensitive to overexpression of both J-proteins. However, this increased prion-loss requires only the highly conserved 70 amino acid J-domain, which serves to stimulate the ATPase activity of Hsp70 and thus to stabilize its interaction with client protein. Overexpression of the J-domain from Sis1, Ydj1, or Apj1 is sufficient to destabilize [SWI+]. In addition, [SWI+] is lost upon overexpression of Sse nucleotide exchange factors, which act to destabilize Hsp70's interaction with client proteins. Given the plethora of genes affected by the activity of the SWI/SNF chromatin-remodeling complex, it is possible that this sensitivity of [SWI+] to the activity of Hsp70 chaperone machinery may serve a regulatory role, keeping this prion in an easily-lost, meta-stable state. Such sensitivity may provide a means to reach an optimal balance of phenotypic diversity within a cell population to better adapt to stressful environments.
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1001309
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001309Summary
The yeast prion [SWI+], formed of heritable amyloid aggregates of the Swi1 protein, results in a partial loss of function of the SWI/SNF chromatin-remodeling complex, required for the regulation of a diverse set of genes. Our genetic analysis revealed that [SWI+] propagation is highly dependent upon the action of members of the Hsp70 molecular chaperone system, specifically the Hsp70 Ssa, two of its J-protein co-chaperones, Sis1 and Ydj1, and the nucleotide exchange factors of the Hsp110 family (Sse1/2). Notably, while all yeast prions tested thus far require Sis1, [SWI+] is the only one known to require the activity of Ydj1, the most abundant J-protein in yeast. The C-terminal region of Ydj1, which contains the client protein interaction domain, is required for [SWI+] propagation. However, Ydj1 is not unique in this regard, as another, closely related J-protein, Apj1, can substitute for it when expressed at a level approaching that of Ydj1. While dependent upon Ydj1 and Sis1 for propagation, [SWI+] is also highly sensitive to overexpression of both J-proteins. However, this increased prion-loss requires only the highly conserved 70 amino acid J-domain, which serves to stimulate the ATPase activity of Hsp70 and thus to stabilize its interaction with client protein. Overexpression of the J-domain from Sis1, Ydj1, or Apj1 is sufficient to destabilize [SWI+]. In addition, [SWI+] is lost upon overexpression of Sse nucleotide exchange factors, which act to destabilize Hsp70's interaction with client proteins. Given the plethora of genes affected by the activity of the SWI/SNF chromatin-remodeling complex, it is possible that this sensitivity of [SWI+] to the activity of Hsp70 chaperone machinery may serve a regulatory role, keeping this prion in an easily-lost, meta-stable state. Such sensitivity may provide a means to reach an optimal balance of phenotypic diversity within a cell population to better adapt to stressful environments.
Introduction
Yeast prions are non-Mendelian genetic elements, most of which are amyloid aggregates formed by single proteins [1]–[8]. These aggregates, often referred to as seeds or propagons, whose number varies depending on the prion, serve as templates for the conversion of newly synthesized protein to the prion conformation [8]–[10]. The presence of an amyloid prion is often associated with phenotypes that arise from the partial loss of function of the prion-forming protein due to its sequestration in the aggregates. For example, [PSI+] and [URE3], the prion forms of a translation termination factor and a transcriptional regulator, cause misreading of nonsense codons and misregulation of a set of genes involved in nitrogen catabolism, respectively [4], [11]. In addition, a single prion-forming protein can take on different conformational states, thus resulting in prion “strains” having varying levels of severity of these heritable traits; so-called “weak” and “strong” [PSI+] are an example of such variants [12]–[15]. Of particular interest in regard to prion-associated phenotypes is the recently identified prion, [SWI+], formed from Swi1, an important component of the SWI/SNF chromatin remodeling complex [16], [17]. [SWI+] cells exhibit a partial loss of SWI/SNF function, resulting in the impaired uptake of certain sugars, slow growth on synthetic media, and poor germination [16]. However, despite these mal-adaptive phenotypes, [SWI +] cells grow indistinguishably from wild-type cells on rich media and under at least one condition, the presence of microtubule-inhibiting fungicide benomyl, grow strikingly better than cells lacking the prion [18].
Propagation of yeast prions appears to be inexorably reliant on the function of molecular chaperones, proteins more generally known for their ability to facilitate protein folding and prevent misfolding [19]–[21]. In the case of amyloid prion propagation, they are required for the fragmentation of the aggregates needed to generate additional seeds [22]–[26]. Physical transmission of these aggregates/seeds to daughter cells is required for propagation of the prion in the cell population [8], [27]–[29]. It has been known for some time that the action of the molecular chaperone Hsp104, which functions as a dissaggregase by threading partially folded proteins through its central pore, is required for amyloid prion fragmentation [19], [20], [22], [26], [30]. More recent work has underscored the importance of the cytosolic Hsp70 Ssa and its co-chaperones [9], [23]–[25], [31]–[36]. Like all Hsp70 systems, Ssa functions in conjunction with two sets of co-chaperone proteins, J-proteins (also known as Hsp40s) and nucleotide exchange factors (NEFs) [21]. J-proteins and NEFs are critical because they affect the ATPase activity of Hsp70 and thus the cycle of interaction with client polypeptides such as prion proteins [21]. J-proteins act to stimulate the ATPase activity of Hsp70s, increasing the affinity for client polypeptides; NEFs stimulate the release of ADP from Hsp70, and thereby facilitate polypeptide release [21]. J-proteins are particularly diverse in structure, with some binding client proteins and “delivering” them to their partner Hsp70 [21], [37]. However, all have a highly conserved J-domain, which is responsible for ATPase stimulation [37]. Although Ssa has 12 J-protein partners [21], [38], only one, Sis1, is required for the propagation of the three best-studied prions, [PSI+], [RNQ+], and [URE3] [24], [25], [33]. Current models assert that Sis1∶Ssa and Hsp104 act sequentially to fragment prion aggregates [23]–[25].
The global effects of [SWI+] raise the intriguing possibility that it may have had an impact during evolution, as it is likely that its presence or absence affects growth differentially under a variety of environmental conditions. Therefore, we initiated a genetic analysis of [SWI+], concentrating on the effects of molecular chaperones on its maintenance. We found that, relative to other prions, [SWI+] propagation is highly sensitive to perturbations in the activity of the Hsp70 machinery. The idea that stress conditions may particularly affect the stability of [SWI+] in cell populations due to this sensitivity to chaperone activity is addressed.
Results
[SWI+] has a low number of prion seeds per cell
As our first step in the characterization of [SWI+] and its dependence on molecular chaperones for its propagation, we determined its seed number. We employed an established method, a ‘propagon counting assay’, which is based on monitoring prion-loss upon inactivation of Hsp104 activity [39]. The Hsp104 inhibitor GdnHCl was used [40], as [SWI+], like other yeast prions, requires Hsp104 activity for seed generation [16]. At various times after treatment of a culture of [SWI+] cells with GdnHCl, aliquots of cells were plated onto glucose-based medium. To determine the percentage of cells having lost the prion at each time point, cells from a minimum of 24 individual colonies were transformed with a plasmid containing the Asn - and Gln-rich segment of Swi1, which contains the prion-forming domains, fused to YFP (Swi1NQ-YFP). Resulting transformants were observed under the microscope and scored for Swi1 aggregation based on punctuate or diffuse fluorescence, indicative of presence or absence of the prion, respectively (Figure 1A) [16]. As expected, [SWI+] was lost from the population with time; after six generations, approximately 50% of the population was [swi−] (Figure 1C). 13 generations after addition of GdnHCl, none of the colonies tested positive for the prion. No prion loss was observed for the control culture to which no GdnHCl was added. To confirm that the microscopic analysis provided an accurate indicator of the presence or absence of the prion, we also tested the growth of cells on medium containing raffinose as the carbon source. Growth of [SWI+], but not [swi−], cells is greatly impaired on such medium because raffinose transport into cells is reduced when activity of the SWI/SNF complex is impaired [16]. All 24 isolates taken from a culture prior to addition of GdnHCl grew poorly on raffinose-based medium; all 24 isolates taken from a culture 13 generations after GdnHCl addition grew well, confirming loss of the prion (Figure 1B and Figure S1).
Fig. 1. Determination of [SWI+] seed number using a propagon counting assay. ![Determination of [<i>SWI</i><sup>+</sup>] seed number using a propagon counting assay.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/619aafb59b39f13f92ff96c445507f25.png)
[SWI+] cells were grown in the presence or absence of 4mM GdnHCl and plated onto glucose-based media at time intervals at the indicated number of cell generations to obtain single colonies. (A) The presence of [SWI+] in the isolated colonies (n = 24) from each time point was assessed by transformation with a plasmid carrying the Swi1NQ-YFP fusion. Resulting transformants (n≥10 for each colony) were observed under the microscope, scoring for punctuate [SWI+] or diffuse [swi−] fluorescence. The presence of [SWI+] in any transformant indicates the presence of the prion in the original colony. A representative example of cells from a colony collected before (0g) and 13 generations after GdnHCl addition (13g) is shown. (B) The presence or absence of [SWI+] was also tested by assessing growth on raffinose-based medium. 24 individual colonies were also assayed before (0g) and 13 generations after (13g) the addition of GdnHCl (Figure S1A). 10-fold serial dilutions of one representative isolate from each condition were spotted onto raffinose and glucose-based medium. (C) Data obtained from cells grown in the presence (diamonds) or absence (circles) of 4mM GdnHCl, as described in A, were plotted. Solid bold line represents the best-fit curve for [SWI+] using the model of Cox et al. 2003 [29], 45 seeds/cell. Colored dotted lines represent reference curves previously published for other prions using this same model (left to right): Magenta, [URE3], 20 seeds/cell; Orange, weak [PSI+], 60 seeds/cell; Green, [RNQ+], 100 seeds/cell; Blue, strong [PSI+], 260 seeds/cell [15], [24], [39], [41]. To estimate seed number, we fit our [SWI+] curing data to an established model in which prion seeds are assumed to be diluted two-fold upon cell division in the absence of Hsp104-mediated fragmentation [29], [39]. This model allows comparison of relative seed number values among prions. A best-fit curve was generated when ∼45 seeds/cell was used as the initial estimate (Figure 1C, bold line). Utilizing the same model, similar seed number estimates have been generated for other prions: 200–300 seeds/cell for strong [PSI+] strains [15], [24], [39], ∼100 seeds for [RNQ+] [24], 40–60 seeds/cell for the weak [PSI+] strain [PSI+]Sc37 [15], [24] and 20–25 seeds/cell for [URE3] [24], [41]. Thus, we conclude that [SWI+] has a low seed number, higher only than that observed for [URE3].
[SWI+] is cured upon depletion of the J-protein Sis1
Sis1 is required for the propagation of at least three prions, [PSI+], [URE3], and [RNQ+] [24], [25], [33]. Therefore, we next tested the dependence of [SWI+] on this J-protein. Because Sis1 is an essential protein, we utilized a system having SIS1 under the control of the tetR promoter (TETr). This system, which, upon addition of the drug doxycycline, allows repression of Sis1 synthesis to a minimal level required for cell growth (Figure 2A), was previously used to analyze the role of Sis1 in the maintenance of other prions [23]–[25]. As a control, samples of cells cultured in the absence of drug were collected at time intervals and plated onto glucose-based media. As discussed above, the status of [SWI+] was assessed after subsequent transformation of resulting individual colonies (n = 24) with a plasmid expressing Swi1NQ-YFP. sis1-Δ [TETr-Sis1] cells initially maintained [SWI+] at a high frequency, but showed gradual loss of the prion (Figure 2B,) as approximately 40% of the cells became [swi−] over 23 cell generations of cell culture in the absence of drug.
Fig. 2. Sis1 is required for [SWI+] propagation. ![Sis1 is required for [<i>SWI</i><sup>+</sup>] propagation.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/5d84691e601ac15adb23f5552e319dec.png)
(A) Western blot showing doxycycline-dependent Sis1 repression. sis1-Δ [TETr-Sis1] cells in liquid culture were harvested before (−Dox., 0g) or the indicated number of generations after the addition of the drug doxycycline (+Dox.). Cell extracts were subjected to immunoblot analysis using either antibody specific for Sis1 or, as a loading control, Ssc1 (control). (B) Time course of [SWI+] loss upon repression of Sis1 expression. [SWI+] sis1-Δ [TETr-Sis1] cells were harvested after the indicated number of generations of growth in the presence (diamonds) or absence (squares) of doxycycline and plated onto glucose-based media. Following transformation of cells from individual colonies with a plasmid bearing Swi1NQ-YFP, the fraction of cells that were [SWI+] was determined by subsequent examination of Swi1 aggregation based on fluorescence (right) and plotted (left). Points represent the average of three independent experiments. Error bars represent ± σ deviation. The solid line represents a best-fit line through the averaged data. (C) Loss of [SWI+] following Sis1 depletion was also confirmed by testing for the restoration of robust growth on raffinose-based media. Serial dilutions of one representative isolate are shown above from a culture before (0g) or 21generations after addition of doxycycline (21g). A total of 24 individual colonies from each of these two time-points were assayed (Figure S1A). Upon addition of drug to cells to repress Sis1 synthesis, [SWI+] propagation was severely affected. [SWI+] was lost from the population with sigmoidal kinetics, exhibiting ∼50% loss after only 9–10 generations and complete curing within ∼20 generations. Curing in these cultures was confirmed by testing for the restoration of robust growth on raffinose media (Figure 2C, and Figure S1B). From these data we conclude that [SWI+], like all other yeast prions thus far tested, is reliant upon Sis1 activity for continued propagation.
[SWI+] is cured upon overexpression of Sis1
A comparison of the control experiments in Figure 1C and Figure 2B indicates that [SWI+] is more stable in a wild-type background than in the sis1-Δ [TETr-Sis1] strains. To test whether this loss was due to lower than normal Sis1 expression, we asked whether the prion would be stabilized by supplemental expression from a second plasmid. However, we found that the presence of the second Sis1 plasmid exacerbated prion loss (data not shown), suggesting that the instability of [SWI+] might be due to overexpression rather than underexpression. Thus, we tested two sis1-Δ strains, one expressing Sis1 from the native SIS1 promoter and one expressing Sis1 from the stronger GPD promoter, resulting in either normal or approximately two-fold higher Sis1 expression, respectively (Figure 3A). After passage of the strains for one week on glucose-based media, the presence of [SWI+] was assessed by observing the growth of cells on medium containing raffinose as the carbon source. The cells overexpressing Sis1 from the GPD promoter grew more robustly, similar to the control [swi−] cells. Those having normal levels of Sis1 expression grew poorly, similar to the [SWI+] control (Figure 3B), indicating the prion is indeed sensitive to overexpression of Sis1. The maintenance or loss of [SWI+] in these strains was also confirmed using fluorescence analysis (Figure 3C).
Fig. 3. Overexpression of Sis1 cures [SWI+]. ![Overexpression of Sis1 cures [<i>SWI</i><sup>+</sup>].](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/ce609a7a7c5e9d1d1df306067f295dd4.png)
(A) Extracts were prepared from either wild-type (WT) or sis1-Δ cells expressing Sis1 from a centromeric plasmid, either under the control of its own (SIS1-Sis1) or the GPD (GPD-Sis1) promoter. The extracts were subjected to immunoblot analysis using either antibody specific for Sis1 or, as a loading control, Ssc1 (control). (A and B) Dashed lines in both figures indicate some sections of the blot or plate have been cropped for clarity. (B) Sis1 expressing cells described in (A) were passaged for one week and then the presence of [SWI+] was assessed by streaking onto raffinose media. Wild-type [SWI+] and [swi−] cells are shown as controls. (C) Loss of [SWI+] from cells shown in (B) was also confirmed by fluorescence analysis following transformation with a plasmid bearing the Swi1NQ-YFP construct. The loss of [SWI+] upon Sis1 overexpression was somewhat surprising, as we previously reported that overexpression of Sis1 did not affect [RNQ+], [URE3], or several strains of [PSI+] [24]. However, these previous studies were not conducted in the 74D-694 genetic background that was used here. Therefore, we tested the effect of Sis1 overexpression on the maintenance of [RNQ+] and both weak and strong variants of [PSI+] in the same strain background, using the same expression constructs described above. [PRION+] cells were transformed with SIS1-bearing plasmids and serially passaged for two weeks on selective media before assaying for the continued presence of the prion. The results were the same as those obtained in the W303 background: no loss of [RNQ+] or [PSI+] was detected (Figure S2). Thus we conclude that the sensitivity of [SWI+] to Sis1 overexpression is a property of the prion, not a characteristic of the 74D-694 genetic background.
[SWI+] is generally sensitive to J-domain levels
Since Sis1 overexpression so potently affected [SWI+], we decided to test two other J-proteins, Ydj1 and Apj1, which had previously been found to affect yeast prions when overexpressed [12], [24], [42]–[45]. To this end, wild-type [SWI+] cells were transformed with either high-copy overexpression plasmids or empty vector. Transformants were repatched once before a second transformation with plasmid bearing Swi1NQ-YFP for prion scoring. The fate of at least 30 independent transformants was determined in each case. [SWI+] was maintained in >90% of control transformants receiving empty vector but, as expected, none of the transformants overexpressing Sis1 maintained the prion (Figure 4).
Fig. 4. [SWI+] is sensitive to J-protein or J-domain overexpression. ![[<i>SWI</i><sup>+</sup>] is sensitive to J-protein or J-domain overexpression.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/2be7b3f5908618df6c5a479f89b3be39.png)
High copy plasmids expressing various J-proteins or J-domain containing fragments under the control of the constitutive GPD promoter were used to transform a wild type [SWI+] strain. Individual transformants were isolated, repatched once on media selective for the plasmid, and then retransformed with a Swi1NQ-YFP expression vector. YFP aggregation was used to score the original transformants for [SWI+] maintenance. The number of transformants remaining [SWI+] is reported as a fraction of the total examined (Fraction [SWI+]). Gene domain structures for the J-proteins Sis1, Ydj1, and Apj1 are shown using the following notation: J, J-domain; G/F, glycine/phenylalanine-rich region; G/M, glycine/methionine-rich region; peptide-binding, C-terminal peptide-binding domains I & II; Zn2+ & peptide-binding, Zinc finger-like region and peptide binding domains I & II; Gly, glycine-rich region; HA, hemegglutinin A tag. Random peptide sequence is indicated by a thin line. Overexpression of both Ydj1 and Apj1 destabilized [SWI+], with 6 out of 36 YDJ1 transformants and 4 out of 48 APJ1 transformants tested maintaining the prion (Figure 4). The expression of only a J-domain has been shown to be sufficient for carrying out some J-protein functions. For example, the severe growth defects of ydj1-Δ cells can be rescued by expression of only the J-domain of Ydj1 [38]. Given that overexpression of all three J-proteins tested thus far affected [SWI+], we decided to determine whether this curing effect could also be accomplished by only a J-domain. We used a set of constructs that express J-domains attached to a C-terminal random peptide region and the hemagglutinin A tag (HA) [38]. These constructs are all expressed at high levels and are all sufficient to rescue the slow growth phenotype of ydj1-Δ cells ([38] and unpublished observations, Sahi and Craig). As we suspected, [SWI+] was severely destabilized by all three J-domain constructs tested (Figure 4), demonstrating that overexpression of a J-domain is sufficient to destabilize the prion.
Finally, to determine if the effects of J-protein and J-domain overexpression are specific to [SWI+], and not the result of strain background, we also examined whether the vectors described above also cured [RNQ+], or strong or weak strains of [PSI+] in the 74D-694 strain background. To do this, we transformed [PRION+] strains with vectors bearing J-proteins and passaged transformants for two weeks on media selective for the plasmid. As expected from results obtained using other strain backgrounds, none of the vectors used in these experiments had any discernable effect on [RNQ+] or either variant of [PSI+] (Figure S3), suggesting that J-protein and J-domain overexpression affects [SWI+] in a specific manner.
[SWI+] is cured in the absence of Ydj1
Ydj1 is the most highly expressed J-protein in the yeast cytosol [46]. Since we found [SWI+] to be highly sensitive to J-protein levels, we decided to test the ability of cells lacking Ydj1 to propagate [SWI+]. To obtain ydj1-Δ strains, [SWI+] wild-type cells were transformed with a YDJ1 disruption cassette carrying a selectable marker. The presence or absence of [SWI+] was assayed by transformation with plasmid expressing Swi1NQ-YFP. Strikingly, all 19 isolated ydj1-Δ transformants exhibited the diffuse fluorescence indicative of [swi−] cells. 39 transformants that contained the selectable marker, but did not disrupt the YDJ1 gene, were used as controls. 38 retained the punctuate fluorescence pattern of [SWI+] cells. Thus prion loss strongly correlated with the absence of Ydj1.
As an additional test for requirement of Ydj1 in [SWI+] maintenance, we obtained cells which did not express Ydj1 by a different method, plasmid shuffling. First, we constructed a [SWI+] strain having a deletion of the YDJ1 gene on the chromosome, but carrying YDJ1, driven by its native promoter in a URA3-based plasmid (ydj1-Δ [YDJ1-Ydj1, URA3]). Cells not expressing Ydj1 were obtained by selecting for resistance to 5-fluoro-orotic acid (5-FOA), a counter-selection against the URA3-based Ydj1 expression plasmid. Microscopic evaluation of cells subsequently obtained by transformation with Swi1NQ-YFP for [SWI+] scoring revealed loss of the prion in all 20 transformants evaluated, consistent with the requirement for Ydj1 in the maintenance of [SWI+].
We wanted to ensure that strain background did not account for the apparent variation in the requirement of different prions for Ydj1. Our previous experiments that indicated that Ydj1 was not required for propagation of either [PSI+] or [RNQ+] were carried out in the W303 genetic background [24]. Therefore, we crossed two strains of the 74D-694 background: a [psi−] [rnq−] ydj1-Δ strain with a [PSI+] [RNQ+] wild-type strain. 22 wild-type and 22 ydj1-Δ haploids obtained from the cross were repatched for two weeks and then tested for the presence of the two prions. All 44 strains were [PSI+] [RNQ+], indicated that, as we previously reported for the W303 strain background, [PSI+] and [RNQ+] are stably propagated in the absence of Ydj1 (Figure S4). Thus, we conclude that [SWI+], unlike other prions, requires the expression of the abundant J-protein Ydj1 for continued propagation in yeast.
The C-terminal regions of Ydj1 are critical for [SWI+] maintenance
The plasmid-shuffling system described above also opened up an avenue to test the possibility that overexpression of other J-proteins might be able to substitute for full-length Ydj1 in [SWI+] propagation. As a control to test this system, ydj1-Δ [YDJ1-Ydj1, URA3] was transformed with a second plasmid, either one carrying a second YDJ1 gene, or one lacking an insert, thus serving as a vector control. As expected, after counter-selection against the URA3-based Ydj1-expressing plasmid, none of the 26 vector transformants tested were [SWI+] (Figure 5). However, 37 of 42 isolates having the Ydj1-expressing plasmid tested positive for [SWI+]. It is likely that the loss of the prion in a small portion of these transformants was due to the increased level of Ydj1 during the time that the cells carried two YDJ1 genes, because modest overexpression of J-domains can result in [SWI+] loss, as discussed above. Therefore, we included in our analysis a wild-type strain expressing Ydj1 from the endogenous gene, as a control for possible prion loss due to increased J-protein function during the construction of these strains, rather than a lack of Ydj1 function in the ydj1-Δ test strain. Wild-type control strains carrying either the vector or YDJ1 on the plasmid sustained some loss of [SWI+], with 4 of 46 and 3 of 24 transformants being [swi−], respectively. However, the stability was sufficient to allow use of this system to test the effectiveness of J-protein constructs to substitute for Ydj1 in [SWI+] propagation.
Fig. 5. [SWI+] requires a function of the C-terminal domains of Ydj1. ![[<i>SWI</i><sup>+</sup>] requires a function of the C-terminal domains of Ydj1.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/fffe6fcdc310c236e3095556aaa092a6.png)
Plasmids expressing J-protein constructs were used to transform a strain expressing Ydj1 from a centromeric plasmid carrying the URA3 genetic marker (ydj1-Δ [YDJ1-Ydj1, URA3] [SWI+]) for plasmid-shuffling experiments (far right column). Following counter-selection against the Ydj1 plasmid by growth on 5-fluoro-orotic acid (5-FOA), individual transformants were then passaged once on selective media, and retransformed with the Swi1NQ-YFP plasmid for [SWI+] scoring by fluorescence analysis. Results are expressed as the fraction of the original transformants that remained [SWI+] over the total number examined (Fraction [SWI+]). In experiments where [SWI+] loss was observed, J-protein-bearing plasmids were also transformed into a wild-type [SWI+] strain to control for potential prion-loss due to protein overexpression (second column from right). For completeness and clarity, results for empty vector and GPD-Apj1 shown in Figure 4 are also included here. Gene domain structure elements are described in Figure 4. The construct S/Y consists of the J-domain and Gly/Phe-rich regions of Sis1 (residues 1–112) and the C-terminal Zn2+- and peptide-binding regions of Ydj1 (residues 93–409). Ydj11–134 contains the J-domain and adjacent Gly/Phe-rich region. The construct Y/A is a chimera substituting the C-terminal Zn2+- and peptide-binding regions of Ydj1 (residues 114–409) with those of Apj1 (residues 162–529). Ydj1H34Q contains a single amino acid alteration (His34→Q) in the J-domain (asterisk) which renders it unable to stimulate Hsp70 ATPase activity [48]. A full list of plasmids used can be found in Table 1. N.T. = Not Tested. Because the requirement of Ydj1 for the maintenance of [SWI+] is unique among yeast prions analyzed to date, we wanted to know whether the requirement was for Ydj1 specifically. As a first step, we tested an N-terminal containing fragment of Ydj1, Ydj11–134, which lacks the C-terminal domains involved in client protein binding, but contains the J-domain. Expression of Ydj11–134 could not support [SWI+] propagation, as none of the 23 ydj1-Δ transformants tested were [SWI+] (Figure 5), suggesting that Ydj1 J-domain function may not be sufficient. To test more directly whether the C-terminal client protein binding domain of Ydj1 is important for [SWI+] maintenance, we made use of a chimera between Sis1 and Ydj1, S/Y, which contains the C-terminal client protein binding domain of Ydj1 and the N-terminal J-domain and glycine-rich region of Sis1. We transformed our test and experimental plasmid-shuffling strains with centromeric plasmids expressing either the S/Y chimera or full-length Sis1. Full-length Sis1 was not able to replace Ydj1 in the ydj1-Δ strain, as none of the 29 transformants analyzed were [SWI+]. It is possible that this failure to replace Ydj1 is due to loss caused by overexpression of Sis1 as discussed above. However, the prion was only mildly destabilized in our control strain, as 15 of 24 transformants maintained the prion. Interestingly, the chimeric S/Y protein was able to maintain the prion in most cases (18 out of 27 transformants) (Figure 5). We conclude that [SWI+] propagation requires a function that can be accomplished by the C-terminal segment of Ydj1, but not Sis1.
To begin to address the question of whether Ydj1 was functionally unique, we asked whether another J-protein that is more closely related to Ydj1 than Sis1 could substitute. Although Sis1 has a client protein binding domain bearing some structural similarity with Ydj1, it does not possess the Zn2+-binding region characteristic of the so-called “class I” J-proteins [47]. Therefore we tested the ability of the class I J-protein Apj1, which is normally expressed at much lower levels than Ydj1 [46], to substitute for Ydj1 when overexpressed. As discussed above, Apj1 expression driven by the strong GPD promoter resulted in [SWI+] loss in the wild-type control strain, with only 4 of 48 transformants retaining the prion (Figure 5), indicating that its overexpression in the presence of wild-type Ydj1 destabilizes the prion. However, 17 of 25 ydj1-Δ transformants expressing Apj1 retained the prion, indicating that Apj1 can at least partially substitute for Ydj1 in [SWI+] propagation. To more directly test the idea that the C-terminal region of Apj1 can functionally substitute for that of Ydj1, we constructed a Ydj1/Apj1 chimera, called Y/A, by substituting the Zn2+ and putative peptide-binding regions of Apj1 for that of Ydj1. This chimera was competent to substitute for Ydj1 when expressed from a single-copy plasmid under a constitutive promoter, as 28 of 30 ydj1-Δ transformants remained [SWI+]. Thus, we conclude that there is functional overlap between Ydj1 and Apj1 and that a function of the C-terminal regions of Ydj1 or Apj1 is required for [SWI+] propagation.
Expression of a single amino acid variant of the Hsp70 Ssa1, Ssa1–21, dominantly cures [SWI+]
The above observations implicate several J-proteins in [SWI+] biology. Because Hsp70 ATPase stimulation is the only known function of a J-domain [21], [37], the sensitivity of [SWI+] to J-domain overexpression strongly indicates that this curing effect is likely mediated through alteration of Hsp70 activity. To test this more directly, we made use of a variant of Ydj1, Ydj1H34Q, which lacks a histidine residue critical for J-domain-mediated stimulation of Hsp70 [21], [37], [48]. We subjected Ydj1H34Q to the tests described in the previous section. We found that it was unable to cure [SWI+] when overexpressed in a wild-type strain and unable to replace Ydj1 in [SWI+] maintenance (Figure 5), indicating that J-domain function, and therefore likely Ssa stimulation, is required in both [SWI+] curing and maintenance.
We next wanted to test for Hsp70's involvement more directly. Since Ssa-type Hsp70s, the partner of both Sis1 and Ydj1, are essential, we turned to a previously identified Ssa1 variant, Ssa1–21 known to affect the maintenance of other yeast prions [35]. Ssa1–21 bears a single amino acid substitution in the C-terminal domain (L483→W). Expression of Ssa1–21 can destabilize [PSI+], even in presence of the wild-type protein [35]. To test whether [SWI+] is similarly affected by Ssa1–21 expression, we transformed [SWI+] cells with a vector expressing Ssa1–21 under the constitutive TEF promoter. As controls, we also transformed cells with either vector expressing wild-type Ssa1, or empty vector. Transformants were re-patched once before individual colonies (n≥21) were transformed with Swi1NQ-YFP for [SWI+] scoring. [SWI+] was maintained in a high percentage of transformants regardless of whether cells expressed the wild-type Ssa1 (19 out of 21), or empty vector (27 out of 30). Ssa1–21, on the other hand, greatly affected [SWI+], dominantly curing the prion in all 22 transformants examined. Thus, we conclude that Ssa1, along with its J-protein co-chaperones, is involved in [SWI+] propagation.
The degree to which [SWI+] was cured by Ssa1–21 expression was surprising considering that [PSI+] is only mildly affected by Ssa1–21 [35]. However, because these observations were made in a different yeast genetic background, direct comparisons between [SWI+] and [PSI+] are not possible. To address whether strain background is a confounding factor in evaluating our data, we tested the effects of Ssa1–21 overexpression on [PSI+] in the 74D-694 background. To do this, we transformed strains bearing either weak or strong [PSI+] with the Ssa1–21 expression vector. After only one passage of the transformants on selective media, the time period used in the [SWI+] experiments, we observed no [PSI+] loss in any strain. Therefore, we continued passaging cells for two weeks to allow adequate time for prion curing. Indeed, while occasional [psi−] (red) colonies were observed in cultures of the weak [PSI+] strain (not shown), [PSI+] was maintained in the overwhelming majority of cells in all cultures (Figure S5). These results indicate that strain background is not the causative factor for the high curing rate of [SWI+], compared to [PSI+], we observed when Ssa1–21 is expressed. Thus, we conclude that [SWI+] is markedly more sensitive than either weak or strong [PSI+] to this alteration in Hsp70 activity.
[SWI+] is sensitive to under - and over-expression of Hsp110-type nucleotide exchange factors
The function of Hsp70 chaperone machinery requires the action of NEFs to stimulate ADP/ATP exchange, and subsequently, peptide release [21]. Consistent with this requirement, one particular NEF of the Hsp110 family, Sse1, has been found to be important for the continued propagation of [URE3] and some weak strains of [PSI+], but not [RNQ+] or strong [PSI+] [43], [49]. To test whether [SWI+] also requires Sse1, we created sse1-Δ strains by transformation of a wild-type [SWI+] strain with an SSE1 deletion cassette bearing the LEU2 selectable marker. The absence of Sse1 expression was verified by immunoblot analysis (Figure 6A). All 31 resulting sse1-Δ transformants became [swi−], as judged by both their ability to grow robustly on raffinose-based media and by the absence of punctate fluorescence (Figure 6B–6D). In contrast, 11 transformants, which obtained the selectable marker but preserved wild-type Sse1 expression, maintained the prion, indicating that like [URE3] and weak [PSI+] strains, [SWI+] is lost upon SSE1 deletion.
Fig. 6. [SWI+] is cured upon deletion of SSE1. ![[<i>SWI</i><sup>+</sup>] is cured upon deletion of <i>SSE1</i>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/df294a81f04b3ecc747785f39a89a658.png)
[SWI+] cells were transformed with an SSE1-deletion cassette bearing a LEU2 marker and transformants selected on media lacking leucine. (A) sse1-Δ transformants were identified by loss of Sse1 expression, as visualized by SDS-PAGE and immunoblotting using antibody specific for Sse1. Cells which received the marker but maintained Sse1 expression were classified as SSE1 transformants and used as controls in subsequent experiments. Wild-type Sse1 expression is also shown for comparison (SSE1 control) (B and C) Two representative transformants (one sse1-Δ and one SSE1) are shown along with a [SWI+] control strain. (B) sse1-Δ cells regain robust growth on raffinose indicative of [SWI+] loss. To test for [SWI+] maintenance, transformants were streaked onto raffinose- or glucose-based media and growth rates compared to control strains. (C) Loss of prion-specific fluorescent puncta in sse1-Δ cells. The presence or absence of [SWI+] in transformants was also confirmed by subsequently transforming cells with a plasmid expressing Swi1NQ-YFP and scoring for the presence of characteristic punctuate foci. (D) Summary of results for 31 sse1-Δ and 11 SSE1 transformants scored for [SWI+] maintenance using both growth on raffinose and Swi1NQ-YFP aggregation assays. The number of transformants remaining [SWI+] is reported as a fraction of the total examined (Fraction [SWI+]). The Hsp110-type Sse protein family consists of two homologous isoforms Sse1 and Sse2 [50]. Because [SWI+] has exhibited a high sensitivity to ectopic chaperone expression, we also tested whether overexpression of either isoform would affect [SWI+]. To do this, [SWI+] cells were transformed with high-copy plasmids expressing either Sse1 or Sse2 from the constitutive GPD promoter or, as a control, empty vector. Overexpression of either isoform significantly destabilized [SWI+] relative to strains transformed with empty vector (Figure 7). Greater than 92% of the transformants overexpressing an NEF became [swi−], while less than 4% of the vector control did. We conclude that stable [SWI+] propagation requires moderate expression of Sse proteins.
Fig. 7. [SWI+] is destabilized by overexpression of Sse1 or Sse2. ![[<i>SWI</i><sup>+</sup>] is destabilized by overexpression of Sse1 or Sse2.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/2d865ff62b6ea2cb214ece5f19bf0749.png)
Wild-type [SWI+] cells were transformed with either empty vector or vectors expressing Sse1 or Sse2 from high copy plasmids under the control of the constitutive GPD promoter. (A and B) One representative transformant for each vector is shown. (A) Serial dilutions of individual transformants were spotted onto raffinose- and glucose-based media to test for [SWI+] loss. [SWI+] cells receiving only empty vector and [swi−] cells were used as controls. (B) [SWI+] maintenance was also monitored by transformation of the original transformants with vector expressing Swi1NQ-YFP, and subsequent fluorescence analysis. (C) Results for each vector are expressed as the fraction of the original transformants which remained [SWI+] over the total number examined (Fraction [SWI+]). Although Sse1 has NEF activity [51]–[53], the fact that it has a domain structure similar to that of Hsp70s suggests that it might have additional functions as well. For example, Sse1 is known to interact with client proteins, though the relationship between client protein binding and NEF activity is less clear [54], [55]. To assess whether loss of [SWI+] caused by Sse1 overexpression was due to increased NEF activity we took advantage of previously characterized SSE1 mutants. After transformation of plasmids carrying the mutant genes into a wild-type [SWI+] strain, transformants were tested for prion maintenance, using both the raffinose growth assay and visualization of Swi1 distribution. First, we tested whether expression of either the N-terminal ATP-binding domain or the C-terminal putative peptide-binding domain of Sse1 is sufficient to effect prion loss by expressing either Sse1Δ394–693 or Sse1Δ1–396. In both cases, all of the 60 transformants tested positive for [SWI+], indicating a requirement of both domains for prion curing (Figure 8). We then tested two point mutations encoding single amino acid alterations within the N-terminal domain: (1) G233→D in the ATP binding site, which impairs, both in vitro and in vivo, Sse1's NEF activity; (2) K69→Q in a site predicted to be required for ATP hydrolysis, which has no measureable effect on Sse1 NEF function [51], [52]. 57 out of 60 transformants overfexpressing Sse1K69Q lost [SWI+], while the prion was stable in those expressing Sse1G233D. These data are consistent with the hypothesis that Sse1 acts as a nucleotide exchange factor for Hsp70 in [SWI+] curing, rather than performing another uncharacterized function.
Fig. 8. Alterations that impair Sse1 NEF function also impair [SWI+] curing. ![Alterations that impair Sse1 NEF function also impair [<i>SWI</i><sup>+</sup>] curing.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/265404f5bf7449af44056ad8f9737533.png)
(A) Domain structure of Sse1. Sse1 shares domain homology with Hsp70s, possessing both an N-terminal ATP-binding domain (NTD) and a C-terminal peptide-binding domain (PBD). Single amino acid alterations tested in this study are indicated by asterisks. (B and C) [SWI+] cells were transformed with either empty vector, or vectors expressing the indicated variants of Sse1 (wild-type Sse1, WT Sse1; Sse1K69Q, K69Q; Sse1G233D, G233D; Sse1Δ1–396, Δ1–396, Sse1Δ394–693, Δ394–693) from the TEF promoter. Analysis of one representative transformant of each variant is shown. (B) Transformants were streaked onto raffinose- and glucose-based media along with [SWI+] and [swi−] control strains for scoring. (C) Transformants and [SWI+] and [swi−] control strains were transformed with vector expressing Swi1NQ-YFP. [SWI+] maintenance was assessed by fluorescence analysis. A representative image of the fluorescence pattern observed in the majority of transformants from each vector and [SWI+] or [swi−] control strains is shown. (D) For each Sse1 variant tested and vector control, results are presented as the fraction of the original transformants which remained [SWI+] over the total number examined (Fraction [SWI+]). For each variant listed, the presence (+) or absence (−) of previously determined nucleotide exchange factor activity (NEF Activity) is shown [51], [52]. Prolonged but not transient heat stress destabilizes [SWI+]
Our results described above indicate that [SWI+] is highly sensitive to various perturbations of the activity of the Hsp70 chaperone machinery brought about by ectopic expression or mutation. As it is well understood that yeast naturally encounter stressful environmental conditions known to alter chaperone expression, we wanted to ask if [SWI+] is sensitive to such conditions. We subjected our wild-type [SWI+] strain to a variety of cell stresses, including heat and ethanol shock, as well as acute exposure to severe oxidative stress. No appreciable loss of the prion was found compared to untreated control cells for any of these conditions (data not shown). However, because prion curing typically requires multiple cell divisions, we next tested whether extended growth at elevated temperatures, a condition known to cause prolonged alteration in chaperone activity altered [SWI+] stability. Indeed, cells grown for 8 days at 37°C reproducibly regained the ability to grow well on raffinose-based media, relative to cultures grown at 23°C (Figure 9A). To confirm that the improved growth on raffinose was in fact due to loss of the prion, rather than some other alteration acquired during growth at 37°C, we also assayed these cultures for Swi1 aggregation using the YFP assay. The fraction of [SWI+] cells in a culture grown at 37°C was significantly less than that from a control culture grown at 23°C (Figure 9B), confirming that unlike [PSI+] [35], [SWI+] is destabilized by prolonged growth at elevated temperatures.
Fig. 9. [SWI+] is destabilized by growth at elevated temperatures. ![[<i>SWI</i><sup>+</sup>] is destabilized by growth at elevated temperatures.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/bcb88ea950baf482b3d7438958ef2359.png)
Wild-type [SWI+] cells were grown on glucose-based media at either 23°C, 30°C, or 37°C for 8 days to test the effect of elevated temperature on [SWI+] stability. Cells were allowed to recover for 1 day at 30°C on fresh media before assaying for the continued presence of the prion. (A) Presence or absence of [SWI+] was tested by assessing growth on raffinose-based medium. 10-fold serial dilutions of cells from each culture, or untreated control [SWI+] and [swi−] cultures, were spotted onto raffinose and glucose-based medium and grown at 30°C. The results of two independent experiments are shown. Dashed lines indicate some sections of the plate have been cropped for clarity. (B) The presence of [SWI+] in isolated colonies (n = 24) obtained from plating of cultures grown at various temperature was assessed by transformation with a plasmid carrying the Swi1NQ-YFP fusion. Resulting transformants were observed under the microscope, scoring for punctuate [SWI+] or diffuse [swi−] fluorescence. The number of transformants remaining [SWI+] is reported as a fraction of the total examined (Fraction [SWI+]). Discussion
[SWI+], the prion formed by Swi1, an important component of the SWI/SNF chromatin remodeling complex, is highly sensitive to alterations in the activity of the Hsp70 chaperone machinery, a characteristic that we hypothesize may have an evolutionary impact.
[SWI+] is the only prion known to require Ydj1
The J-protein Sis1 is required for the propagation of [SWI+], as this prion is lost upon repression of Sis1 expression. This is not a surprising result. The three prions previously analyzed, [PSI+], [RNQ+] and [URE3], also require Sis1 [24], [25], [33]. However, amongst these four prions, [SWI+] is unique in that it also requires Ydj1, as [SWI+] is lost when YDJ1 is deleted. Ydj1, the most abundant J-protein in the cell, is involved in many physiological processes [21], [46]. Because of these global roles, ydj1-Δ cells grow extremely poorly, thus raising the possibility that the requirement for Ydj1 is due to a general effect on cell growth, rather than a direct role in prion dynamics. However, our results point to a direct and important role of client protein binding in Ydj1 function in [SWI+] propagation.
Ydj1 is structurally complex, containing an N-terminal J-domain, critical for functional interaction with Hsp70s, and a C-terminal region, capable of binding client proteins [21]. The N-terminal fragment Ydj11–134 was not able to provide the [SWI+] propagation function. However, as we previously reported [38] the J-domain itself is sufficient to restore robust growth. Thus, the ability of Ydj11–134 to suppress the slow growth phenotype of ydj1-Δ cells, but not the prion propagation defect, indicates that the requirement for Ydj1 is not an indirect effect of poor cell growth but rather a specific requirement for Ydj1. The fact that the C-terminal region containing the client protein binding domain, when fused to the N-terminal J-domain containing region of Sis1, was able to substitute for full-length Ydj1 in [SWI+] propagation, supports the idea that Ydj1 binding to a client protein, presumably Swi1 itself, is critical.
However, the role that Ydj1 plays remains elusive. It is possible, that it, like Sis1, functions in fragmentation to generate seeds. Alternatively, Ydj1 may play a role in conversion of the soluble form of Swi1 to the prion conformation. Although, because Ydj1 inhibits [URE3] fiber formation in vitro [56], [57], it seems more likely that Ydj1 may oppose polymerization, perhaps preventing the formation of dead-end aggregates. It is possible that Ydj1 plays a role, though clearly a non-essential one, in the maintenance of other prions. Indeed, Ydj1 has also been found to associate with Sup35 and Rnq1, the proteins which form [PSI+] and [RNQ+] [32], [58], [59]. This idea is also supported by the observation that, at least for [PSI+], deletion of the YDJ1 gene exacerbates the negative phenotypes of Ssa1–21, indicating that Ydj1 may perform a beneficial function in [PSI+] maintenance under normal circumstances as well [60]. In addition, the observation that Apj1, a J-protein predicted to have a structure quite similar to that of Ydj1, could compensate for Ydj1 in [SWI+] maintenance when expressed at a sufficiently high level does support the idea that other J-proteins in the cytosol may be compensatory in the absence of Ydj1, at least for prions other than [SWI+]. It is also interesting to note that Apj1, named Anti-prion J-protein 1, was originally identified in a screen for factors capable of curing a synthetic prion when overexpressed [42].
[SWI+] is highly sensitive to the alteration in activity of Hsp70 machinery
The data presented here support the idea that [SWI+] is more sensitive to the change in balance of the Hsp70 chaperone system than other prions such as [RNQ+] and [PSI+]. The dependence on Ydj1 for [SWI+] propagation described above is one example. The destabilization of [SWI+], unlike [PSI+] and [RNQ+], by overexpression of J-domains and NEFs, is another example. In addition, the observation that [SWI+] was cured in all cells expressing the Ssa1–21 variant underscores the idea that a variety of alterations in Hsp70 chaperone activity can cause destabilization of this prion. The explanation of the mechanism(s) behind the observed sensitivity is not obvious. For example, from the general understanding of the Hsp70 machinery, overexpression of J-proteins or NEFs would be expected to bias Hsp70 toward the ADP - or ATP-bound states, respectively. But how this precisely affects the overall rate of client protein cycling and the residence time spent associated with Hsp70 is not understood. Regardless of the precise mechanism, the overall picture that emerges is one in which [SWI +] propagation is delicately balanced, requiring a steady-state level of Hsp70 machinery activity, the disruption of which causes dramatic instability. It should be noted that while “strong” and “weak” variants of several prions have been identified, only one form of the recently identified [SWI+] prion is known. It will be of interest to analyze other stronger and weaker variants when they become available.
One other prion, [URE3], stands out as being sensitive to Hsp70 machinery activity. Although it does not require Ydj1 for propagation [24], it is sensitive to overexpression of both J-domains and the NEF Sse1 [24], [43], [45]. Intriguingly, a recent analysis of the amino acid composition of known yeast prion - domains (PrDs) revealed several distinctive features placing the four prions discussed here into two groups: [PSI+] and [RNQ+] in one; [SWI+] and [URE3] in the other [61]. While all four are abundant in Q and N residues, a distinctive feature of yeast prion-forming proteins, the ratio of these residues (Q∶N) in the PrDs of Sup35 and Rnq1 is nearly 2∶1, whereas the PrDs of Swi1 and Ure2 are N-rich, having Q∶N ratios approximating 2∶3 and 1∶3, respectively. A recent study of Q/N-rich proteins in yeast revealed that those richest in N residues were more likely to form prions, supporting the idea that asparagines are more prionogenic than glutamines [18]. Congruent with this idea, the N-terminal 323 residue “N-domain” of Swi1 alone, which is N-rich, is sufficient for amyloid formation and prion induction, whereas the adjacent Q-rich region is not [62]. Additionally, PrDs of Swi1 and Ure2 are more abundant in bulky hydrophobic residues (F, I, L, M, V, W) known to promote amyloid formation [63]–[65] (19% and 15%, respectively) than the corresponding domains of Sup35 and Rnq1 (4% and 9%). On the other hand, Sup35 and Rnq1 are highly abundant in glycine residues (17% vs. 3% and 6% for Swi1 and Ure2). Taken together, the amino acid compositions of the PrDs which form prions that are particularly sensitive to chaperone function ([SWI+] and [URE3]) appear to be skewed in favor of amyloid formation relative to those that are less sensitive ([PSI+] and [RNQ+]). This correlation also extends to a fifth prion [PSI+PS]. [PSI+PS] is the prion form of a chimeric protein in which the PrD of Sup35 from Saccharomyces cerevisiae (Sup35-NSc) is replaced by the corresponding domain from Pichia methanolytica (Sup35-NPm) [66]. Like [SWI+], [PSI+PS] is also highly sensitive to ectopic chaperone expression, being destabilized by overexpression of Sis1, Ydj1, Apj1 or Sse1, or by deletion of SSE1 [42], [44]. Strikingly, like the PrD of Swi1, Sup35-NPm is N-rich (Q∶N≈1∶2) and has a lower content of glycine and a higher content of bulky hydrophobics than Sup35-NSc, consistent with the idea that PrDs which favor amyloid formation form prions with a higher degree of chaperone sensitivity. Thus, we think that the available data makes the idea that the amino acid composition of a PrD plays an important role in determining the sensitivity of a prion to chaperone activity an idea worthy of testing. Of course, it must be kept in mind that other factors, such as the character of the adjacent non-PrD, may play either a primary or secondary role.
How might propensity to form amyloid relate to chaperone sensitivity? One possibility is that prion forming proteins which are optimized for amyloid formation may form stable amyloid fibers which are more difficult to fragment than those of other prions. An alternate, but not mutually exclusive possibility is that these proteins may exhibit higher fiber extension rates in vivo. Rapidly formed fibers may laterally associate [67], occluding chaperone interaction sites, before fibers can be fragmented by the Hsp70·Hsp104 chaperone machinery. In either case, the expected result would be prions with larger and less numerous prion seeds. Indeed, as discussed above, [URE3] and [SWI+] have a lower number of prion-forming seeds/cell than [RNQ+] or [PSI+] [15], [24], [41]. However, since all the parameters that determine seed number are unknown, other factors besides amyloid propensity may also be involved. For example, prions with a low number of seeds/cell may simply be less able to withstand even small changes in chaperone activity, which would lead to a further decrease in seed number, and failure to disseminate seeds efficiently to daughter cells.
Functional implications
It is intriguing that the two prions found to be more sensitive to Hsp70 chaperone activity, [URE3] and [SWI+], are formed by proteins that regulate S. cerevisiae's use of the essential nutrients, nitrogen and carbon, respectively. The presence of [URE3] results in indiscriminate utilization of nitrogen sources due to de-repression of genes typically involved in utilization of nitrogen under starvation conditions, while [SWI+] results in altered carbon source utilization [4], [16], [68]. It is easy to imagine that the presence and/or rapid loss of these prions could profoundly affect the ability of cells to survive and thrive under particular stressful environmental conditions [68]. For example, [SWI+] is clearly disadvantageous when grown in the presence of particular carbon sources such as raffinose, but has been reported to be advantageous in the presence of a mircotubule-inhibiting drug [18]. Thus, high chaperone sensitivity may be a beneficial counterbalancing factor to allow curing of the prion under certain unfavorable environmental conditions, and provide a means to reach an optimal balance of phenotypic diversity within a cell population. Our data indicating that [SWI+] is prone to being lost upon prolonged stress points to such a possibility. It is not known if [SWI+] is unique in this regard. However, [PSI+] has been reported to be quite stable in cells grown at elevated temperatures [35]. It is also interesting to note that two very recently identified prions, [MOT3] and [OCT+], are formed by the proteins, Mot3 and Cyc8, respectively, which play roles in regulation of gene expression [18], [69]. Mot3, like Ure2, plays a rather specific role, regulating the expression of genes needed for robust growth when oxygen is limited. Cyc8, on the other hand, is a global regulator, like Swi1. It acts as a general co-repressor of RNA polymerase II, as well as playing a role in global chromatin structure [68]. It is possible that these prions might also exhibit high sensitivity towards chaperone activity, and thereby, along with [SWI+] and [URE3], be candidates for prions that may play particularly important roles in adaptation of yeast to stressful environments.
Methods
Yeast strain construction
Unless otherwise noted, the originally described [SWI+] 74D-694 strain ([SWI+] [psi−] [rnq−] MATa ade1–14 ura3–5 leu2–3, 112 trp1–289 his3–200 SNF5YFP::kanMX4) [16]was considered the wild-type strain used in all experiments to characterize the [SWI+] prion. All other strains are of the 74D-694 genetic background.
To create [SWI+] cells for Sis1 repression, homozygous diploid cells ([psi−] [RNQ+] ade1–14 ura3–52 leu2–3, 112 trp1–289 his3–200, a gift from Susan Liebman) were transformed with a Δsis1::LEU2 PCR fragment. The recombinant SIS1/sis1-Δ::LEU2 heterozygous diploid (Y1544) was transformed by a plasmid bearing SIS1 and a URA3 marker and the resulting transformants were sporulated and subjected to tetrad dissection. A resulting haploid strain ([RNQ+] MATα sis1::LEU2 [SIS1-Sis1, URA3]) was mated to the wild-type [SWI+] strain. Two haploid strains ([SWI+] [RNQ+] sis1::LEU2 [SIS1-Sis1, URA3]) were obtained from one complete tetrad and the presence of [SWI+] confirmed. Transformation of these strains with p414-TETr-SIS1 followed by passage onto 5-FOA, which counter-selects against the URA3 plasmid, resulted in the isolation of three [SWI+], sis1::LEU2, [TETr-Sis1] strains.
To test the effect of YDJ1 deletion on [SWI+], we transformed the wild-type [SWI+] strain with a PCR-generated ydj1::LEU2 integration cassette, and transformants selected on –Leu media. Transformants were identified as ydj1-Δ by slow growth at 30°C and 23°C and by the ability to rescue this phenotype by subsequent transformation with a plasmid expressing normal levels of Ydj1 protein. Original Leu+ transformants which grew normally at both temperatures were classified as YDJ1 and used as controls. To construct a strain suitable for YDJ1 gene plasmid shuffling experiments, a resulting [swi−] ydj1-Δ strain was transformed with a Ydj1-expressing plasmid [YDJ1-Ydj1, URA3] and mated to a MATα [SWI+] strain which is otherwise isogenic to our wild-type strain. Isolated diploids were screened for [SWI+] and sporulated on media selective for the YDJ1 plasmid. One haploid strain ([SWI+] ydj1::LEU2 [YDJ1-Ydj1, URA3]) was isolated from a single complete tetrad and used for plasmid-shuffling manipulations. A prion-cured version of this strain (MATa) was also mated to a [PSI+], [RNQ+] MATα strain (Y1682, described below), and subjected to tetrad dissection and 5-FOA treatment to test the ability of these prions to propagate in a ydj1-Δ strain.
Two haploid strains were used to test the effects of chaperone overexpression on other prions. One strain, Y2051 ([PSI+]Sc37 [rnq−] ade1–14 ura3–52 leu2–3, 112 trp1–289 his3–200, a gift from Jonathan Weissman), bears the well-characterized weak [PSI+] variant [PSI+]Sc37 (referred to in the text as “weak [PSI+]”) [14], [15]. The second strain, Y1682 ([PSI+] [RNQ+] ade1–14 ura3–52 leu2–3, 112 trp1–289 his3–200) carries both [RNQ+] (derived from a strain Y1505, a gift from Susan Liebman) [12]and a strong [PSI+] variant generated by transient Sup35 overexpression. This strain was used in all other investigation involving [RNQ+] or [PSI+] (referred to in text as “strong [PSI+]”). The [RNQ+] variant in this strain is a mitotically stable variant from the Liebman laboratory (derived from strain L1842) [12]. This variant is resistant to curing by Ydj1 overexpression.
To create sse1-Δ strains, a plasmid-based disruption construct sse1-Δ::LEU2 (a gift from Kevin Morano) was digested with Sac II and Pst I [52]. The resulting digestion mixture was used to transform the wild-type [SWI+] strain and transformants selected on –Leu media. SSE1 disruption was confirmed by immunoblotting.
Plasmids
A complete list of plasmids used in this study is shown in Table 1, and unless otherwise indicated, are based on the pRS plasmid series [70]. The URA3-marked plasmid carrying the Swi1NQ-YFP construct was described elsewhere [62]. A HIS3-marked version of this vector was constructed by digestion with the enzymes Spe I and Xho I, and ligation into the pRS vector p413TEF. The SSA1 gene was cloned by polymerase chain reaction (PCR) using genomic DNA from a wild-type yeast strain from the W303 genetic background. A double-stranded product was then digested with BamH I and Xho I and ligated into predigested p416TEF to created the plasmid p416TEF-SSA1. The plasmid p416TEF-SSA1-21 was then constructed by site-directed mutagenesis PCR (Quikchange) to introduce a single Trp residue in place of Leu483. The plasmid p414GPD-YDJ1H34Q was created by first subcloning the YDJ1 open reading frame into p414GPD and subsequently a single Gln was introduced in place of His34 by Quikchange. The plasmid p414ADH1-Y/A was constructed by “PCR sewing” to combine PCR amplified fragments encoding residues 1–113 of Ydj1 and 162–529 of Apj1, flanked by restriction sites for the enzymes XbaI and SalI at the 5′ and 3′ ends, respectively. The amplified linear insert was subsequently double digested and ligated into the vector p414ADH1.
Tab. 1. Plasmids used in this study. 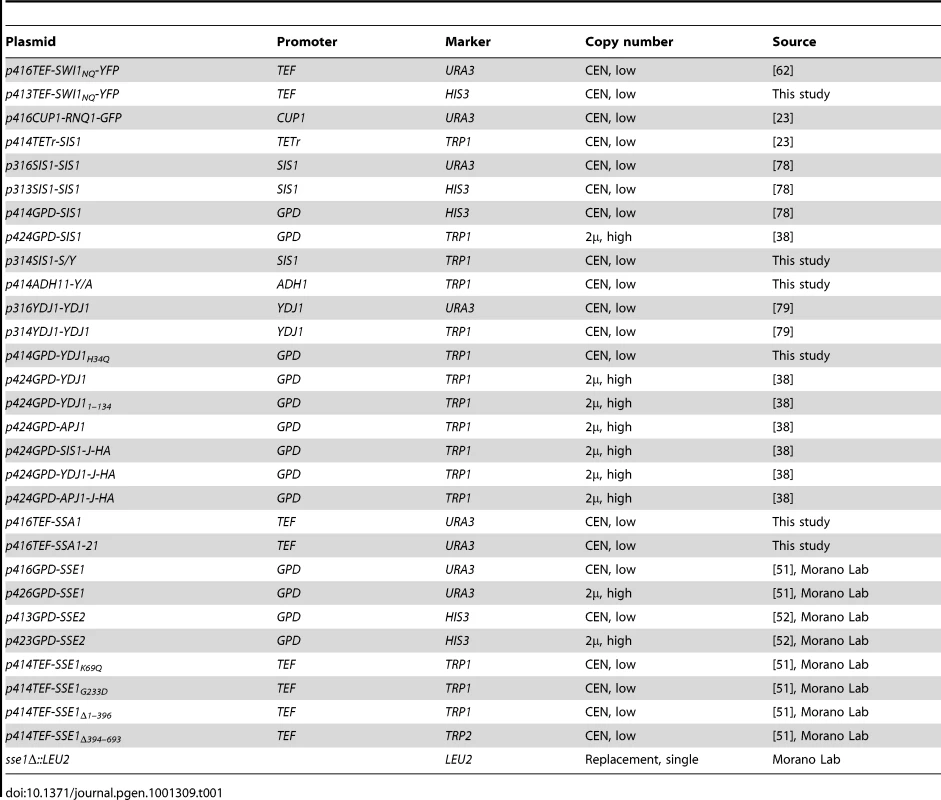
Assays for prion loss
To test SWI/SNF-dependent phenotypes, cells were plated onto synthetic media supplemented with 1 µg/ml antimycin A from Streptomyces sp. (Sigma) and 2% raffinose as the carbon source (defined as raffinose-based media throughout). The glucose-based rich media YEPD (Teknova) is used as a control. Cells were grown at 30°C for 2–3 days prior to imaging. Prion-mediated Swi1 aggregation was observed directly by transforming cells with a plasmid expressing the Asn - and Gln-rich regions of Swi1 (residues 1–554), which contains the prion-forming domains, fused to YFP (Swi1NQ-YFP). [SWI+] cells exhibit a characteristic punctuate fluorescence in the cytoplasm against a dark cytosolic background, whereas [swi−] cells exhibit a diffuse cytosolic fluorescence in combination with occasional increased nuclear fluorescence due to Swi1 accumulation in the nucleus and/or a single extremely large non-prion aggregate in less than 5% of cells [16], [62]. Transformation of [SWI+] cultures routinely results in >85% of cells presenting punctuate fluorescence [16], [62]. To allow time for prion curing, cells were serially passaged by repatching onto solid media every 2 days for the duration specified. ydj1-Δ cultures were grown at 23°C; all other yeast cultures were grown at 30°C unless otherwise noted.
The presence or absence of [PSI+] was confirmed by observation of colony color on rich media where [PSI+]-mediated aggregation of Sup35, a translation termination factor, causes read-through of the premature nonsense codon in the ade1–14 mutant allele [71], [72]. Strains which are otherwise wild-type for adenine production appear pink or white in the presence of [PSI+] or dark red in the absence of [PSI+] due to the accumulation of a red intermediate when adenine production is blocked [73]. [RNQ+] aggregates in cells were observed directly following transformation with a vector expressing Rnq1 fused to GFP (Rnq1-GFP). [RNQ+] cells can be easily distinguished from [rnq−] cells when examined under a microscope by characteristic punctuate or diffuse fluorescence patterns, respectively [23]. Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) was also used to resolve [RNQ+]-dependent detergent resistant aggregates and was performed as described elsewhere [24], [74], [75]. To visualize aggregates, protein was transferred to a nitrocellulose membrane at 1A for 1 hr at 25°C in a tris-glycine/methanol buffer and probed with antibodies specific for Rnq1.
Time course experiments for [SWI+] curing were performed as previously reported for [PSI+] and [URE3] [24]. Cell cultures were maintained in exponential growth phase by continual subculturing in YEPD in the presence of either 5 µg/ml doxycycline (Sigma) or 4 mM GdnHCl when indicated. Sis1 depleted cells remained viable for the duration of the experiment with a typical growth rate of 2.0–2.5 hrs/generation. GdnHCl curing experiments used to estimate relative seed numbers utilize the ‘propagon counting assay’ model of Cox et al. 2003, which may underestimate actual propagon number [76], but enables direct and unbiased comparisons between estimates generated for different prions [29].
Cell stress assays
Cells grown at either 23°C or 37°C were subjected to heat shock (2 min. at 51°C), ethanol shock (2 min. with 12% EtOH), or exposure to severe oxidative stress (3 min. with 4mM H2O2) and then allowed to recover for in fresh media without additional additives at 30°C for 2 days before assaying for the presence of the prion. To test the effects of prolonged heat stress, cells were patched onto glucose-based media and grown at either 23°C or 37°C. Cells were grown for 4 days before repatching to fresh media and allowed to grow an additional 4 days. Cells were then repatched to fresh media and grown overnight to allow time to recover before being assayed for the presence of [SWI+] by raffinose growth assay or Swi1NQ-YFP transformation.
SDS-PAGE and immunoblot analysis
Total protein extracts were prepared by harvested yeast cells in mid-log phase followed by lysis in NaOH.
The resulting protein extracts were analyzed by SDS-PAGE and immunoblot analysis. Sse1 antibody used in this study was kindly provided by Jeff Brodsky. Antibodies specific to Sis1 and Rnq1 have been described elsewhere [23]. Densitometry measurements used to estimate relative protein expression levels were made using the program ImageJ [77].
Supporting Information
Zdroje
1. UptainSM
LindquistS
2002 Prions as protein-based genetic elements. Annu Rev Microbiol 56 703 741
2. WicknerRB
1994 [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264 566 569
3. WicknerRB
MasisonDC
EdskesHK
1995 [PSI] and [URE3] as yeast prions. Yeast 11 1671 1685
4. WicknerRB
EdskesHK
ShewmakerF
NakayashikiT
2007 Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol 5 611 618
5. PatinoMM
LiuJJ
GloverJR
LindquistS
1996 Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273 622 626
6. SerioTR
CashikarAG
KowalAS
SawickiGJ
LindquistSL
2001 Self-perpetuating changes in Sup35 protein conformation as a mechanism of heredity in yeast. Biochem Soc Symp 35 43
7. DerkatchIL
BradleyME
MasseSV
ZadorskySP
PolozkovGV
2000 Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? Embo J 19 1942 1952
8. SindiSS
SerioTR
2009 Prion dynamics and the quest for the genetic determinant in protein-only inheritance. Curr Opin Microbiol 12 623 630
9. JonesGW
TuiteMF
2005 Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays 27 823 832
10. Satpute-KrishnanP
SerioTR
2005 Prion protein remodelling confers an immediate phenotypic switch. Nature 437 262 265
11. TuiteMF
CoxBS
2006 The [PSI+] prion of yeast: a problem of inheritance. Methods 39 9 22
12. BradleyME
EdskesHK
HongJY
WicknerRB
LiebmanSW
2002 Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A 99 Suppl 4 16392 16399
13. DerkatchIL
ChernoffYO
KushnirovVV
Inge-VechtomovSG
LiebmanSW
1996 Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144 1375 1386
14. TanakaM
ChienP
NaberN
CookeR
WeissmanJS
2004 Conformational variations in an infectious protein determine prion strain differences. Nature 428 323 328
15. TanakaM
CollinsSR
ToyamaBH
WeissmanJS
2006 The physical basis of how prion conformations determine strain phenotypes. Nature 442 585 589
16. DuZ
ParkKW
YuH
FanQ
LiL
2008 Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40 460 465
17. SahaA
WittmeyerJ
CairnsBR
2006 Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7 437 447
18. AlbertiS
HalfmannR
KingO
KapilaA
LindquistS
2009 A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137 146 158
19. ChernoffYO
LindquistSL
OnoB
Inge-VechtomovSG
LiebmanSW
1995 Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268 880 884
20. GloverJR
LindquistS
1998 Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94 73 82
21. KampingaHH
CraigEA
2010 The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11 579 592
22. Satpute-KrishnanP
LangsethSX
SerioTR
2007 Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol 5 e24 doi:10.1371/journal.pbio.0050024
23. AronR
HigurashiT
SahiC
CraigEA
2007 J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. Embo J 26 3794 3803
24. HigurashiT
HinesJK
SahiC
AronR
CraigEA
2008 Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A 105 16596 16601
25. TiptonKA
VergesKJ
WeissmanJS
2008 In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell 32 584 591
26. PaushkinSV
KushnirovVV
SmirnovVN
Ter-AvanesyanMD
1996 Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. Embo J 15 3127 3134
27. ByrneLJ
CoxBS
ColeDJ
RidoutMS
MorganBJ
2007 Cell division is essential for elimination of the yeast [PSI+] prion by guanidine hydrochloride. Proc Natl Acad Sci U S A 104 11688 11693
28. TuiteMF
CoxBS
2003 Propagation of yeast prions. Nat Rev Mol Cell Biol 4 878 890
29. CoxB
NessF
TuiteM
2003 Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165 23 33
30. MogkA
HaslbergerT
TessarzP
BukauB
2008 Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans 36 120 125
31. AllenKD
WegrzynRD
ChernovaTA
MullerS
NewnamGP
2005 Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169 1227 1242
32. BagriantsevSN
GrachevaEO
RichmondJE
LiebmanSW
2008 Variant-specific [PSI+] Infection Is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol Biol Cell 19 2433 2443
33. SondheimerN
LopezN
CraigEA
LindquistS
2001 The role of Sis1 in the maintenance of the [RNQ+] prion. Embo J 20 2435 2442
34. SongY
WuYX
JungG
TutarY
EisenbergE
2005 Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell 4 289 297
35. JungG
JonesG
WegrzynRD
MasisonDC
2000 A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics 156 559 570
36. ChernoffYO
2007 Stress and prions: lessons from the yeast model. FEBS Lett 581 3695 3701
37. CheethamME
CaplanAJ
1998 Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3 28 36
38. SahiC
CraigEA
2007 Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A 104 7163 7168
39. EaglestoneSS
RuddockLW
CoxBS
TuiteMF
2000 Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 97 240 244
40. GrimmingerV
RichterK
ImhofA
BuchnerJ
WalterS
2004 The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem 279 7378 7383
41. RipaudL
MailletL
CullinC
2003 The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. Embo J 22 5251 5259
42. KryndushkinDS
SmirnovVN
Ter-AvanesyanMD
KushnirovVV
2002 Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem 277 23702 23708
43. KryndushkinD
WicknerRB
2007 Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell 18 2149 2154
44. KushnirovVV
KryndushkinDS
BogutaM
SmirnovVN
Ter-AvanesyanMD
2000 Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 10 1443 1446
45. SharmaD
StanleyRF
MasisonDC
2009 Curing of yeast [URE3] prion by the Hsp40 cochaperone Ydj1p is mediated by Hsp70. Genetics 181 129 137
46. GhaemmaghamiS
HuhWK
BowerK
HowsonRW
BelleA
2003 Global analysis of protein expression in yeast. Nature 425 737 741
47. CraigEA
HuangP
AronR
AndrewA
2006 The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 156 1 21
48. TsaiJ
DouglasMG
1996 A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem 271 9347 9354
49. FanQ
ParkKW
DuZ
MoranoKA
LiL
2007 The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177 1583 1593
50. MukaiH
KunoT
TanakaH
HirataD
MiyakawaT
1993 Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132 57 66
51. ShanerL
TrottA
GoeckelerJL
BrodskyJL
MoranoKA
2004 The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem 279 21992 22001
52. ShanerL
SousaR
MoranoKA
2006 Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry 45 15075 15084
53. DragovicZ
BroadleySA
ShomuraY
BracherA
HartlFU
2006 Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. Embo J 25 2519 2528
54. GoeckelerJL
PetrusoAP
AguirreJ
ClementCC
ChiosisG
2008 The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett 582 2393 2396
55. PolierS
HartlFU
BracherA
2010 Interaction of the Hsp110 Molecular Chaperones from S. cerevisiae with Substrate Protein. J Mol Biol
56. LianHY
ZhangH
ZhangZR
LooversHM
JonesGW
2007 Hsp40 interacts directly with the native state of the yeast prion protein Ure2 and inhibits formation of amyloid-like fibrils. J Biol Chem 282 11931 11940
57. SavistchenkoJ
KrzewskaJ
FayN
MelkiR
2008 Molecular chaperones and the assembly of the prion ure2p in vitro. J Biol Chem 283 15732 15739
58. KrzewskaJ
MelkiR
2006 Molecular chaperones and the assembly of the prion Sup35p, an in vitro study. Embo J 25 822 833
59. LopezN
AronR
CraigEA
2003 Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol Biol Cell 14 1172 1181
60. JonesGW
MasisonDC
2003 Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163 495 506
61. CrowE
DuZ
LiL
2008 New insights into prion biology from the novel [SWI+] system. Prion 2 141 144
62. DuZ
CrowET
KangHS
LiL
2010 Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol Cell Biol 30 4644 4655
63. ToombsJA
McCartyBR
RossED
2010 Compositional determinants of prion formation in yeast. Mol Cell Biol 30 319 332
64. PetkovaAT
IshiiY
BalbachJJ
AntzutkinON
LeapmanRD
2002 A structural model for Alzheimer's beta -amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A 99 16742 16747
65. RossED
ToombsJA
2010 The effects of amino acid composition on yeast prion formation and prion domain interactions. Prion 4
66. KushnirovVV
Kochneva-PervukhovaNV
ChechenovaMB
FrolovaNS
Ter-AvanesyanMD
2000 Prion properties of the Sup35 protein of yeast Pichia methanolica. Embo J 19 324 331
67. Kawai-NomaS
PackCG
KojidaniT
AsakawaH
HiraokaY
2010 In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J Cell Biol 190 223 231
68. HalfmannR
AlbertiS
LindquistS
2010 Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol 20 125 133
69. PatelBK
Gavin-SmythJ
LiebmanSW
2009 The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 11 344 349
70. MumbergD
MullerR
FunkM
1995 Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119 122
71. Inge-VechtomovSG
TikhodeevON
KarpovaTS
1988 [Selective systems for obtaining recessive ribosomal suppressors in saccharomycete yeasts]. Genetika 24 1159 1165
72. CoxBS
1965 [PSI], a cytoplasmic suppressor of super-suppression in yeast. Heredity 20 505 521
73. BoussetL
SavistchenkoJ
MelkiR
2008 Assembly of the asparagine - and glutamine-rich yeast prions into protein fibrils. Curr Alzheimer Res 5 251 259
74. BagriantsevSN
KushnirovVV
LiebmanSW
2006 Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol 412 33 48
75. KryndushkinDS
AlexandrovIM
Ter-AvanesyanMD
KushnirovVV
2003 Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278 49636 49643
76. ByrneLJ
ColeDJ
CoxBS
RidoutMS
MorganBJ
2009 The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae. PLoS ONE 4 e4670 doi:10.1371/journal.pone.0004670
77. AbramoffMD
MagelhaesPJ
RamSJ
2004 Image Processing with ImageJ. Biophotonics International 11 36 42
78. YanW
CraigEA
1999 The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol 19 7751 7758
79. JohnsonJL
CraigEA
2000 A role for the Hsp40 Ydj1 in repression of basal steroid receptor activity in yeast. Mol Cell Biol 20 3027 3036
Štítky
Genetika Reprodukční medicína
Článek Break to Make a ConnectionČlánek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



