-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
Transcription of the switch (S) regions of immunoglobulin genes in B cells generates stable R-loops that are targeted by Activation Induced Cytidine Deaminase (AID), triggering class switch recombination (CSR), as well as translocations with c-MYC responsible for Burkitt's lymphomas. In Saccharomyces cerevisiae, stable R-loops are formed co-transcriptionally in mutants of THO, a conserved nuclear complex involved in mRNP biogenesis. Such R-loops trigger genome instability and facilitate deamination by human AID. To understand the mechanisms that generate genome instability mediated by mRNP biogenesis impairment and by AID, we devised a yeast chromosomal system based on different segments of mammalian S regions and c-MYC for the analysis of chromosomal rearrangements in both wild-type and THO mutants. We demonstrate that AID acts in yeast at heterologous S and c-MYC transcribed sequences leading to double-strand breaks (DSBs) which in turn cause chromosomal translocations via Non-Homologous End Joining (NHEJ). AID–induced translocations were strongly enhanced in yeast THO null mutants, consistent with the idea that AID–mediated DSBs depend on R-loop formation. Our study not only provides new clues to understand the role of mRNP biogenesis in preventing genome rearrangements and the mechanism of AID-mediated genome instability, but also shows that, once uracil residues are produced by AID–mediated deamination, these are processed into DSBs and chromosomal rearrangements by the general and conserved DNA repair functions present from yeast to human cells.
Published in the journal: . PLoS Genet 7(2): e32767. doi:10.1371/journal.pgen.1002009
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002009Summary
Transcription of the switch (S) regions of immunoglobulin genes in B cells generates stable R-loops that are targeted by Activation Induced Cytidine Deaminase (AID), triggering class switch recombination (CSR), as well as translocations with c-MYC responsible for Burkitt's lymphomas. In Saccharomyces cerevisiae, stable R-loops are formed co-transcriptionally in mutants of THO, a conserved nuclear complex involved in mRNP biogenesis. Such R-loops trigger genome instability and facilitate deamination by human AID. To understand the mechanisms that generate genome instability mediated by mRNP biogenesis impairment and by AID, we devised a yeast chromosomal system based on different segments of mammalian S regions and c-MYC for the analysis of chromosomal rearrangements in both wild-type and THO mutants. We demonstrate that AID acts in yeast at heterologous S and c-MYC transcribed sequences leading to double-strand breaks (DSBs) which in turn cause chromosomal translocations via Non-Homologous End Joining (NHEJ). AID–induced translocations were strongly enhanced in yeast THO null mutants, consistent with the idea that AID–mediated DSBs depend on R-loop formation. Our study not only provides new clues to understand the role of mRNP biogenesis in preventing genome rearrangements and the mechanism of AID-mediated genome instability, but also shows that, once uracil residues are produced by AID–mediated deamination, these are processed into DSBs and chromosomal rearrangements by the general and conserved DNA repair functions present from yeast to human cells.
Introduction
Transcription is a source of genetic instability in eukaryotic cells (reviewed in [1]). High transcription rates can result in increased recombination and mutagenesis [2], [3], and a variety of factors have an influence on this relationship, including the collision between replication forks and the transcription machinery [4], and the occurrence of RNA–DNA hybrids (R-loops) [5], [6]. In mammalian B cells, class switch recombination (CSR) and somatic hypermutation (SHM) are strongly dependent on transcription. Transcription of the switch (S) regions generates DNA intermediates in which the C-rich template strand forms stable R-loops and the non-template G-rich strand can form secondary structures (reviewed in [7]). Activation Induced Deaminase (AID), an enzyme specifically expressed in mature B cells [8] that is essential in both CSR and SHM, appears to be associated with the transcription machinery [9]–[11], and AID-mediated mutations increase when RNAPII is stalled [12]. Current models of CSR suggest that AID triggers the process by deaminating cytosines in the non-template DNA strand displaced by the R-loop [13]–[15]. In addition, during transcription the DNA sequences upstream of the elongating RNA polymerase are negatively supercoiled, and this transient change in DNA topology may allow AID to access both DNA strands [16]. Cytosine residues deaminated by AID at S regions are converted into uracils, which are processed by base excision repair (BER) and/or mismatch repair (MMR) pathways into DSBs responsible for CSR [17]–[19]. However, the reason why in B cells uracils lead to DSBs instead of being repaired remains unclear, although a breakdown of the cellular protective error-free repair has been suggested [20]. An open question is whether B cells have specialized functions or DNA modifying enzymes not present in other eukaryotic cells to process uracils into DSBs.
The same mechanism leading to CSR seems to be involved in the formation of chromosomal translocations (CTs) between the immunoglobulin S regions and c-MYC (Ig/myc) that are associated with B cell-derived Burkitt's lymphomas, also mediated by AID [21]–[23]. CTs are indeed a hallmark of cancer cells [24], [25], and are mediated by DSBs [26]. Repair of AID-mediated DSBs occurs via Non-Homologous End Joining (NHEJ) [27], but in the presence of sequence homology, CTs may also occur by homologous recombination [28]–[32].
R-loops have been shown to accumulate in yeast THO mutants. The THO complex is functionally involved in transcription elongation and mRNP biogenesis [33], and depletion of some of its components generates strong transcription impairment and hyper-recombination phenotypes, which are linked to the formation of R-loops [5]. These phenotypes depend on gene length, GC content and transcription levels [34], [35]. Importantly, THO depletion enhances AID-dependent mutations as a consequence of its ability to accumulate R-loops [36]. Taking advantage of this ability, we developed a yeast genetic system to study the mechanisms of AID-mediated chromosomal rearrangements. With this system we observed that transcription of S and c-MYC sequences in yeast generates AID-mediated DSBs that can be joined with an ectopic HO-induced DSB to yield a reciprocal CT. AID-induced CTs were strongly enhanced in yeast THO null mutants but not in hpr1-101 mutants, which do not form R-loops, consistent with the idea that AID-mediated DSBs depend on R-loop formation. CTs analyzed took place through NHEJ, confirming that the initial event was a DSB. This study not only provides new clues to understanding the role of mRNP biogenesis in preventing genome rearrangements and the mechanism of AID-mediated genome instability, but it shows that yeast cells, as mammalian B cells, have the factors and pathways required to channel AID-mediated DNA lesions into DSBs responsible for NHEJ-dependent genome rearrangements and CSR.
Results
Murine S sequences are hotspots of recombination in yeast THO null mutants overexpressing human AID
We developed different genetic systems to analyze how R-loops initiate genome instability, whether or not via DSBs. As S regions are known to form R-loops in B cells both in vivo and in vitro [6], [37]–[39], which in turn lead to DSBs responsible for CSR, we assayed the ability of these regions to induce recombination between repeats in yeast. We used a direct-repeat recombination assay based on a plasmid with two truncated GFP direct repeats sharing 200-bp of homology under control of the GAL1 promoter [36] (Figure 1). Different S region fragments, which are characterized by a specific repetitive motif and length, were inserted between the repeats, and recombinants were scored as GFP+ cells by FACS among total cells. We analyzed sequences derived from murine Sµ and Sγ3 regions, which were 350-bp and 600-bp long, respectively (Sµ350 and Sγ600) [39], a 1050-bp longer version of Sµ carrying three consecutive repeats of Sµ350 (Sµ1050), and the Sµ350 in reversal orientation (Sµ350rev). None of these sequences yielded a significant increase of recombination compared with the control carrying no S sequences in wild-type cells (Figure 1).
Fig. 1. AID–induced recombinogenic effect of different murine S sequences in wild-type and THO mutants. 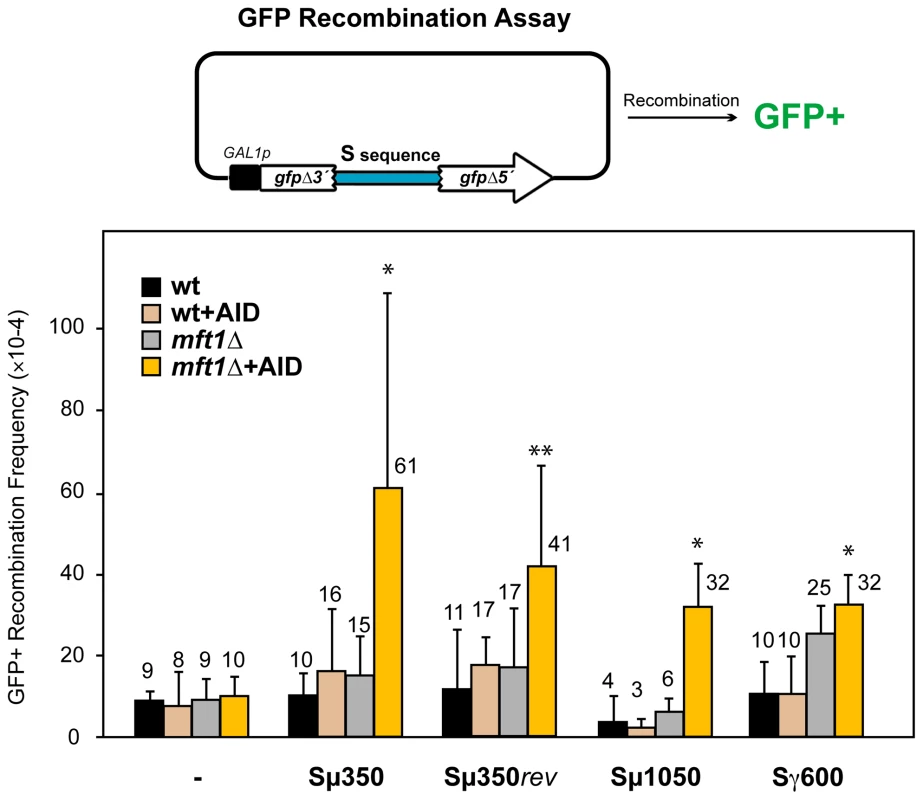
A diagram of the direct-repeat recombination system is shown on top. GFP-based direct-repeat recombination was detected by FACS in wild-type and mft1Δ strains, with or without AID overexpression. Recombination frequencies were obtained as the median value of at least six independent colonies, and are indicated on top of each bar. Median values and the corresponding standard deviations are shown. Statistical analyses were performed with a Mann-Whitney test compared with the control carrying no S sequence cells. * p<0.001, ** p<0.01. We next tested whether AID had the ability to stimulate recombination at S regions, given its preference to act on ssDNA within R-loops [13], [14]. AID expression did not significantly increase the recombination frequency of the different sequences analyzed in wild-type cells (Figure 1). We then analyzed the effect of the mft1Δ THO mutation, which induces R-loop formation but do not compromise viability [35]. Whereas the frequency of GFP+ recombinants in mft1Δ cells not expressing AID was the same as in the wild-type for the different S regions tested, AID overexpression significantly increased it 3-6-fold above wild-type levels (Figure 1). These results suggest that the S fragments used here are too short to form R-loops capable of causing detectable hyper-recombination. However, they are able to facilitate the action of AID. As the highest recombination frequency was obtained with the Sµ350 sequence (6.1×10−3), with little differences to those of Sµ1050 and Sµ350rev (Figure 1), we selected Sµ350 for further analyses as the better candidate for the analysis of AID action in vivo.
High levels of reciprocal CTs in yeast THO null mutants overexpressing AID
In order to unequivocally assay the capacity of R-loops to form DSBs, we developed a yeast system for the detection of reciprocal CTs that should occur via DSBs in the absence of DNA homology. The system is based on two non-homologous halves of the LEU2 gene (leu2Δ5′ and leu2Δ3′) integrated at chromosomes XV and III, respectively (Figure 2A). Upstream to the leu2Δ5′ at chromosome XV, an HO endonuclease cut site fused to the hygromycin gene and the 246-bp 3′-end of the ACT1 intron were integrated (HO-ACT1iΔ5’-leu2Δ5’). The leu2Δ3′ sequence, under control of the GAL1 promoter, followed by the remaining 62-bp of the ACT1 intron and the Sµ350 sequence (leu2Δ3′-ACT1iΔ3′-Sµ350) was integrated at the right arm of chromosome III together with the URA3 gene (Figure 2A). As a control we used a similar system carrying no Sµ350 sequence (leu2Δ3′-ACT1iΔ3′). These genetically engineered chromosomes III and XV were made in haploid MATa-inc cells containing the HO endonuclease gene under the GAL1 promoter, and the endogenous LEU2 gene and the ACT1 intron deleted from their chromosomal loci. In galactose, the HO endonuclease is activated causing a DSB at the HO-ACT1iΔ5’-leu2Δ5’ construct on chromosome XV, whereas the leu2Δ3′-ACT1iΔ3′-Sµ350 on chromosome III is heavily transcribed from the GAL1 promoter. Therefore, the HO-induced DSBs on chromosome XV could be sealed with a transcription-mediated DSB on chromosome III to lead a Leu+ translocations (Figure 2A). These translocations would be expected to contain the breakpoints embedded in a functional ACT1 intron located between the two LEU2 gene halves (Figure 2A).
Fig. 2. Intron-based assay for chromosomal translocations in yeast. 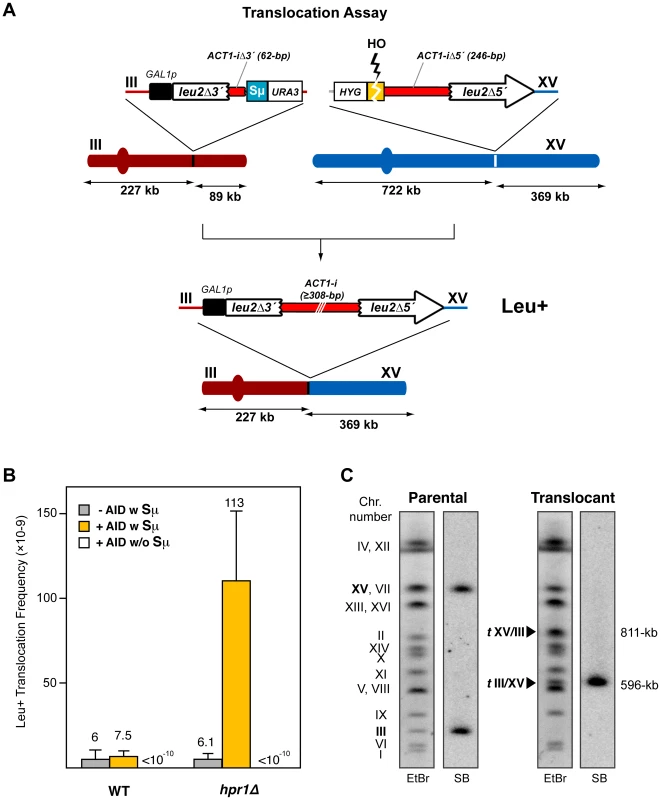
(A) Scheme of the translocation system used. There is no sequence homology between the two DNA constructs integrated in chromosomes III and XV. Chromosomal translocations detected generate a galactose-inducible full-length LEU2 gene harboring a yeast ACT1 intron sequence inside. Size of chromosomal fragments is indicated. (B) Leu+ translocation frequencies after HO-induced DSBs in both wild-type and hpr1Δ cells. The presence (w) or absence (w/o) of the Sµ350 sequence and AID overexpression are indicated. Translocation frequencies were obtained as the median value of at least ten independent colonies for each strain. Median values and the corresponding standard deviations are shown. (C) Molecular karyotype of Leu+ translocants analyzed by PFGE. Agarose gels were stained with ethidium bromide (left) and analyzed by Southern using a radiolabeled LEU2 probe (right). The electrophoretic mobility of natural yeast chromosomes is indicated. The LEU2 signal in parental cells corresponds to the two non-homologous halves of LEU2 integrated in chromosomes III and XV (bold). Chromosomes XV and VII have the same electrophoretic mobility in the experimental conditions used here. The translocated chromosomes containing LEU2 (tIII/XV) or not containing it (tXV/III), are marked with black triangles, and their sizes are indicated. After 24h of growth in galactose, Leu+ translocants were obtained at a frequency of 6×10-9 in wild-type cells (Figure 2B), which indicated that transcription through leu2Δ3′-ACT1iΔ3′-Sµ350 was indeed able to generate DSBs that were joined to the HO-induced DSB on chromosome XV. This Leu+ frequency was unaffected by overexpression of AID (Figure 2B). We then analyzed the effect of a THO null mutation, in this case hpr1Δ for its known strong effect in transcription [40]. Whereas the frequency of translocations in hpr1Δ was the same as in wild-type cells not expressing AID (6.1×10−9), it increased 19-fold when AID was overexpressed (Figure 2B). This effect was specific for the system containing the S sequence (leu2Δ3′-ACT1iΔ3′-Sµ350), as in the absence of S sequences translocations were not detectable (<10−10; Figure 2B). As it has been reported that secondary structures do not form in vitro when the sequence is inverted and the G-rich strand is transcribed [39], we also analyzed the effect of the inverted S sequence (Sµ350rev) in our translocation assay, for which we constructed the corresponding leu2Δ3′-ACT1iΔ3′-Sµ350rev on chromosome III. Interestingly, Sµ350rev also increased the frequency of translocations in hpr1Δ cells overexpressing AID respect to wild-type levels, although it reached lower levels than the forward Sµ350 sequence (Figure S1).
Consistent with previous analyses of direct-repeats recombination (Figure 1), the S sequences used here seem to be too short as to form sufficient R-loops to give a significant increase of CTs in wild-type cells with AID or in hpr1Δ cells without AID, but they stimulated recombination in hpr1Δ cells expressing AID. Altogether, these results indicate that transcription through S regions in yeast can lead to DSBs able to generate CTs. The specific increase caused by AID overexpression in THO mutants suggests that at least a proportion of such DSBs are mediated by R-loops.
AID–mediated translocation breakpoints occur at the S regions
To assess whether Leu+ events corresponded to the expected translocations, chromosomes from twenty independent Leu+ recombinants obtained from hpr1Δ yeast overexpressing AID were analyzed by pulsed-field gel electrophoresis (PFGE). Ethidium bromide staining and Southern analysis with a LEU2 specific probe revealed that all translocated chromosomes showed the pattern expected from the reciprocal joining of the DSBs generated at the S region and HO site in chromosomes III and XV, respectively (Figure 2C and Figure S2). This reciprocal translocation resulted in two new chromosomes of 596 - and 811-kb long, respectively, with the LEU2 signal specifically detected in the smaller one, the chromosome in which the two halves of the LEU2 gene are joined (Figure 2C and Figure S2). Concomitantly, parental chromosomes III and XV disappeared. This indicates that translocations in hpr1Δ yeast overexpressing AID show a unique and recognizable structure originated by a DSB at the S region.
To map the CT breakpoints we amplified the ACT1 intron reconstituted within the LEU2 gene by PCR using specific primers that anneal at the corresponding sequences involved in the translocations on chromosomes III and XV (Figure 3A). This amplification generated a variety of DNA fragments longer than the 308-bp of the natural ACT1 intron (Figure 3A), suggesting a non site-specific distribution of the transcription-mediated DSBs along the Sµ350 region. DSBs appeared mainly in the 5′-half of the Sµ350 region, resulting in 13 to 122-nt long S sequence stretches within the reconstituted ACT1 intron of the translocated chromosomes (Figure 3B), which was similar regardless of AID expression (Figure 3B and 3C). Analysis of the junctions by sequencing of the PCR products revealed frequent short microhomologies (1-2-nt) between the Sµ and HO breakpoints (Figure 3C), and the lost of a few nucleotides (0-3-nt) from the HO cut site in the end-joining process (Figure 3C). These data suggest that DSBs are generated by AID deamination at hpr1Δ-mediated R-loops in the transcribed Sµ350 regions.
Fig. 3. PCR analysis of CT breakpoints in hpr1Δ yeast with the Sµ350 sequence integrated at chromosome III. 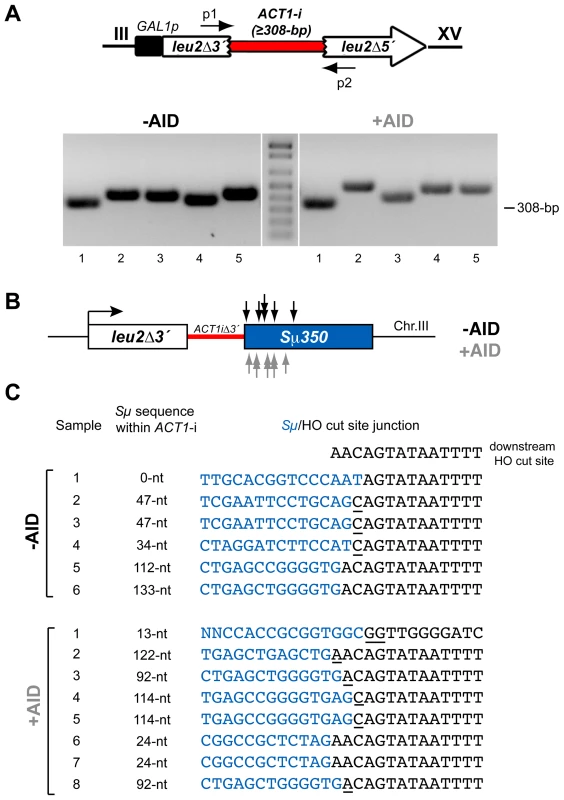
(A) Genomic DNA extracted from Leu+ translocants obtained from hpr1Δ yeast with and without AID-overexpression were used to amplify the reconstituted ACT1 intron by PCR using specific p1 and p2 primers, which anneal at the sequences involved in the translocations on chromosomes III and XV. The PCR reaction yielded fragments with heterogeneous sizes longer than the 308-bp of the full-length ACT1 intron. (B) Identification of CT breakpoints. PCR products were sequenced to identify the breakpoints at the Sµ350 sequence. Breakpoints from cells with (grey) and without (black) AID-overexpression are indicated by arrows. (C) Sequence analysis of breakpoint junctions. The Sµ350/HO site junctions from six AID- and eight AID+ independent Leu+ translocant strains are shown. The length of the S sequence within the reconstituted ACT1 intron after the translocation is indicated. Underlined nucleotide residues indicate microhomologies between the Sµ350 and HO cut site sequences. The sequence downstream of the HO cleavage site is shown on top. AID does not stimulate DSBs in hpr1-101 mutants not forming R-loops
Depletion of specific mRNP biogenesis factors, such as THO, generates a transcription elongation impairment that is linked to the accumulation and/or stabilization of R-loops [5]. Accordingly, we found that transcription of the Sµ350 sequence in hpr1Δ yeast reduced the efficiency of the transcription process in our system (Figure S3). In order to determine whether R-loops formed in hpr1Δ cells are required for AID-dependent DSBs and CTs, we analyzed CTs in the hpr1-101 mutant strain, which has been shown not to accumulate stable R-loops during transcription [41]. Interestingly, hpr1-101 cells overexpressing AID showed a strong decrease in CT formation after galactose induction (Figure 4), suggesting that the formation of R-loops during transcription of the Sµ region contributes to the generation of the DSBs responsible for CTs in this system.
Fig. 4. Analysis of Leu+ translocations in mutant strains overexpressing AID. 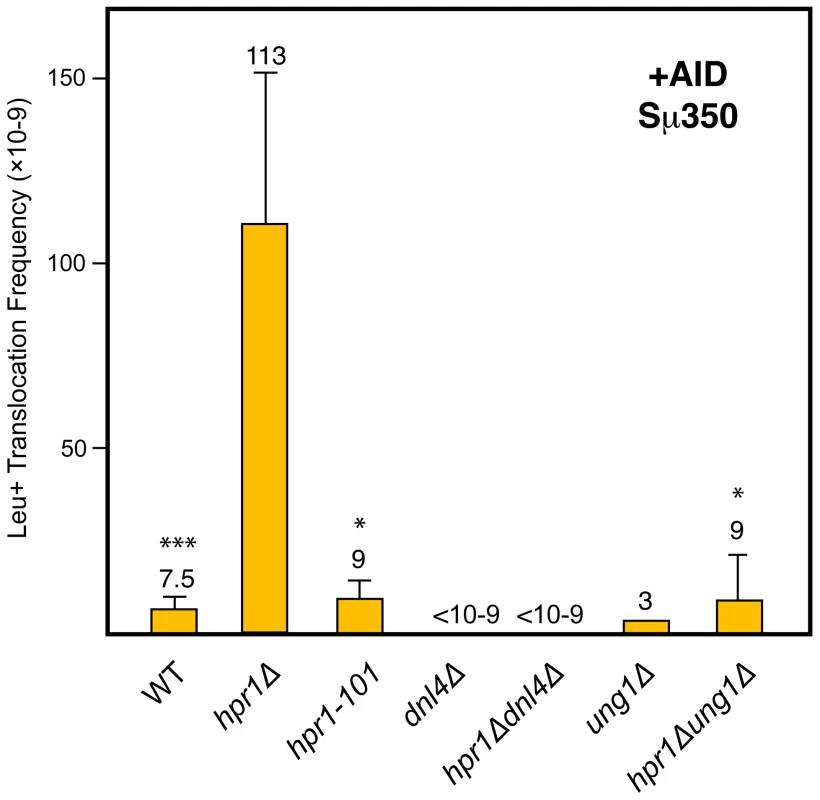
Details as in Figure 2B. Statistical analyses were performed with a Mann-Whitney test compared with hpr1Δ cells with the Sµ350 sequence overexpressing AID. * p<0.001, *** p<0.05. AID–induced translocations in yeast THO null mutants are mediated by Ung1 and NHEJ
The current model proposed for the formation of AID-dependent DSBs suggests that uracils generated after AID-mediated deamination of cytosines are primarily processed by the uracil glycosylase Ung1 leading to abasic sites [19]. Subsequent processing of these abasic sites generates the DSBs that are repaired by NHEJ during CSR in mammalian B cells. To assay whether AID-mediated DSBs generated in our yeast system were processed as in B cells, we determined the effect of both UNG1 and DNL4 deletions on CT formation. As can be seen in Figure 4, both ung1Δ and dnl4Δ abolished the capacity of AID to induce DSB-mediated translocations in hpr1Δ cells. Therefore, as occurs in mammalian B cells, AID-dependent DSBs in yeast are also mediated by Ung1 and repaired by NHEJ. These results validate this yeast chromosomal translocation assay as a model system to study the mechanism by which AID can induce CTs in vivo. It reveals that once cytosine deamination is produced, further steps leading to DSBs and rearrangements in B cells may occur by standard and conserved DNA repair functions present from yeast to human cells, and not by B cell specific DNA modifying enzymes.
The c-MYC oncogene is a recombination hotspot in yeast THO null mutants
The observation that the S regions used in this study become a hotspot for DSBs only in R-loop-forming hpr1Δ cells, and by the action of AID, suggests that the sequences used are not long enough to harbor the same structural features as full natural S regions in B cells. In order to test this hypothesis, one possibility was to enlarge the size of the S sequence cloned into our yeast systems. The use of such sequences with high abundance of internal repeats, however, makes them particularly unstable, as deletions between the repeats by single-strand annealing occur at high frequency [42], [43], a problem accentuated in hpr1Δ cells. Consequently, we used as an equivalent control the c-MYC oncogene. As the S regions of Ig genes, c-MYC is characterized by its GC-richness, especially in the 5′ region that includes the first exon and first intron (up to 60% of GC residues). This region forms co-transcriptional secondary structures during transcription in vitro [44], and is frequently hypermutated and involved in reciprocal translocations with the S regions [22]. The recent identification of c-MYC as a main target of AID-dependent translocations and the demonstration that nuclease-mediated DSBs at c-MYC causes similar translocations [23] suggest that c-MYC carries the same structural features that make S regions highly recombinogenic during transcription in B cells.
Therefore, we cloned a 3.8-kb fragment of the human c-MYC gene that included the first exon and the first intron in our GFP direct-repeat recombination assay, as a functional and structural equivalent of a long S region. As can be seen in Figure 5A, whereas recombination was not affected by the presence of the MYC sequence in wild-type cells, regardless of AID expression, a strong increase was seen in hpr1Δ cells both without and with AID expression (24-fold and 32-fold increase with respect to the control wild-type, respectively; Figure 5A). To test whether these high recombination levels, notably higher than those obtained with the Sµ350 sequence (compare Figure 1 and Figure 5A), were not just due to the presence of a longer DNA sequence between the GFP repeats, we tested the effect of the long (4.1 kb) and low GC-rich (40%) sequence of the LYS2 gene. Recombination levels in this system containing the LYS2 sequence dropped markedly (Figure 5A), indicating that the high recombinogenic behavior of c-MYC in this system in hpr1Δ was due to both its high GC-content (as compared with LYS2) and length (as compared with Sµ350). Therefore, c-MYC behaves as a hotspot of recombination in hpr1Δ cells that is slightly increased by the action of AID.
Fig. 5. Analysis of recombinogenic potential of the c-MYC sequence. 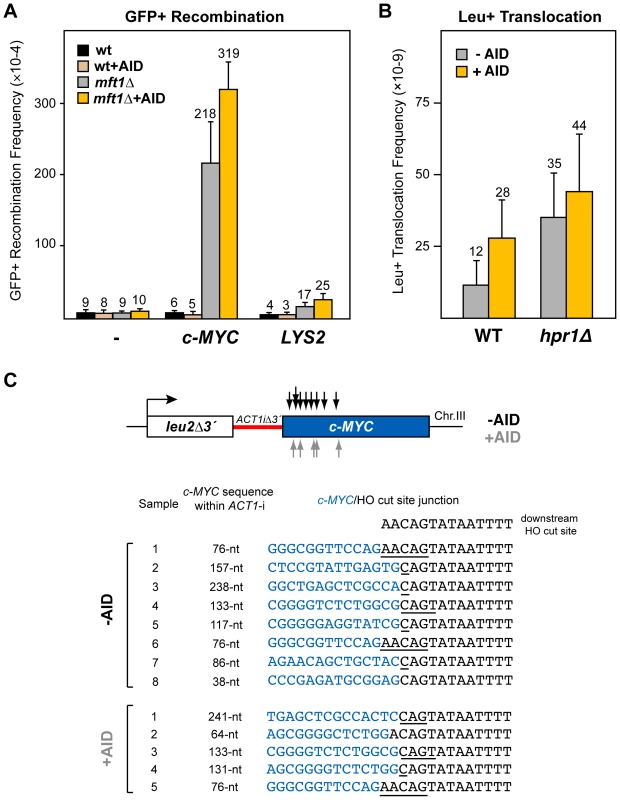
(A) AID-independent recombinogenic behavior of c-MYC in THO mutants. A 3.8-kb fragment of the human c-MYC, as well as the control 4.1-kb long LYS2 sequence, were cloned into the GFP-based direct-repeat recombination system. Details as in Figure 1. (B) Leu+ translocation frequencies in c-MYC-containing wild-type and hpr1Δ cells after HO-induced DSBs. Details as in Figure 2. (C) Analysis by PCR of CT breakpoints in hpr1Δ yeast at the c-MYC sequence. Breakpoints from cells with (grey) and without (black) AID-overexpression are indicated by arrows (top). The analysis of the breakpoint junctions is shown at the bottom. The c-MYC/HO cut site junctions from eight AID- and five AID+ independent Leu+ translocant strains are shown. The length of the c-MYC sequence within the reconstituted ACT1 intron in the translocant chromosome is indicated. Underlined nucleotide residues indicate microhomologies between the MYC and HO cut site sequences. The sequence downstream of the HO cleavage site is shown on top. To determine whether the recombinogenic behavior of the c-MYC sequence in hpr1Δ was due to an accumulation of DSBs, we next tested its effect on the chromosomal translocation assay (Figure 5B). For this test, a 2.5-kb c-MYC 5′-end fragment, including the first exon and the first intron, was integrated under the inducible GAL1 promoter at chromosome III, instead of the Sµ350 sequence (leu2Δ3′-ACT1iΔ3′-cMYC). Interestingly, after galactose induction, Leu+ translocants were recovered in wild-type cells more efficiently in the system with c-MYC than with Sµ350 (2-fold and 4-fold increase without and with AID expression, respectively; compare Figure 2B and Figure 5B), suggesting that DSBs are spontaneously induced during transcription of the c-MYC sequences, and slightly enhanced by AID. Slightly higher translocation frequencies were obtained in hpr1Δ cells (3-fold and 2-fold increase with respect to wild-type cells without or with AID expression, respectively; Figure 5B). This translocation frequency, however, was 3-fold lower than that induced by the Sµ350 in hpr1Δ cells expressing AID (compare Figure 2B and Figure 5B). We mapped the translocation breakpoints by PCR and sequencing as previously described. PCR of the breakpoint fragments rendered heterogeneous sizes that were longer than the 308-bp full-length ACT1 intron (not shown). Breakpoints occurred mainly in the 5′-region of c-MYC, resulting in a reconstituted ACT1 intron slightly longer than that obtained with S sequences (from 38 to 241-nt long; Figure 5C). The length of c-MYC stretches within the reconstituted ACT1 intron was the same regardless of whether translocations were obtained with or without AID overexpression (Figure 5C). Sequencing of the translocation junctions revealed also frequent microhomologies between the c-MYC and the HO cut site ends (Figure 5C), which were longer (1-5 nt) than those obtained with Sµ350, regardless of AID expression. The number of nucleotides lost from the HO cut site in the end-joining event was similar to that of Sµ350 (Figure 5C).
These data suggest that c-MYC behaves as an intrinsic recombination hotspot in yeast THO mutants forming R-loops. Even though AID induce more recombination and DSBs at c-MYC, it only causes a slight enhancement of c-MYC recombination hotspot activity. This suggests that c-MYC can be targeted by recombinogenic DNA breaks in AID-dependent and independent manners in THO mutants, suggesting that a suboptimal mRNP structure is sufficient for c-MYC to behave as a fragile site.
Discussion
We show here that AID induces DSBs in the immunoglobulin S regions and in the c-MYC sequences in yeast THO mutants, which are impaired in mRNP biogenesis. Our data suggest that AID-mediated deamination of cytosines at S sequences in yeast are channeled by the Ung1 activity into DSBs as occurs in mammalian B cells, which are then repaired by NHEJ. The c-MYC oncogene, which shares features of S sequences, also acts as a hotspot of recombination in yeast THO mutants. Using a novel translocation assay we show that translocations involving S or c-MYC sequences occur efficiently in yeast THO mutants, but poorly in wild-type cells. This study demonstrates that the mechanisms of AID-mediated CSR and AID-dependent translocations are mediated by standard DNA repair functions existing in yeast and human cells. The only requirement for these events to occur is the formation of the appropriate substrates for AID action, as they are the R-loops known to be accumulated in THO null mutants.
It is widely accepted that transcription of S regions plays a primary role in CSR in B cells by generating R-loops that are substrates for efficient AID action [7]. However, once cytosines at the single-stranded DNA sequence of the R-loop are converted into uracils by AID deamination, it is unclear whether B cells have specific DNA modification enzymes or pathways to channel such uracils into DSBs responsible for CSR or Ig/myc translocations instead of being efficiently repaired by either BER or MMR, although a breakdown of the protective accurate repair pathways during the process of B cell maturation has been suggested [20]. Our yeast assay for the detection of translocations generated by the NHEJ-mediated processing of DSBs, reveals that yeast cells also have the capability of processing AID-mediated lesions into DSBs. We found that transcription through mammalian Sµ regions in yeast generates AID-mediated DSBs that can lead to CTs, similarly to B cells. The breakpoints of these translocations are located at the 5' end of Sµ sequences, where most CSR junctions occur in vivo [45], and show short nucleotide microhomologies, consistent with data from mammalian B cells [46]. Notably, AID-dependent CTs in our assay occur at a low frequency in wild-type cells, but are synergistically enhanced in yeast THO null mutants, which are affected in mRNP biogenesis. This is in agreement with previous results showing the mutagenic and recombinogenic phenotype of yeast overexpressing AID [47], and that the transcription impairment in yeast THO null mutants enhances AID-dependent mutation of the non-template DNA strand, consistent with its exposure as a single stranded DNA during the transcription elongation in the form of R-loops [36]. These results, and the fact that AID-dependent CTs are poorly observed in hpr1-101 mutants, which do not form R-loops [41], suggest that the formation of a suboptimal structure of the ribonucleoparticle around the S region RNA fragment may be a major contributor to the formation and/or stabilization of a R-loop as an optimal substrate for AID action. Therefore, our data open the possibility that mRNP biogenesis may have an impact on the generation of genetic variability in mammalian B cells by controlling the formation of R-loops as an efficient substrate for the action of AID.
It has been reported that CSR is orientation-dependent and that inversion of the S sequences decreases the efficiency of CSR [38], [45], as it happens with the formation of sequence-dependent secondary structures in DNA during transcription in vitro [39]. We found in our assays a lesser, although substantial, recombinogenic effect of the Sµ sequence in the reversal orientation, which suggests that in our yeast system the presence of G-rich non-transcribed sequences is not an unique requisite for recombinogenicity. The size of the S region seems important for the recombination hotspot activity in B cells, as longer S regions could generate more extensive R-loops [48]. A slight increase of the size of the Sµ sequence in our yeast assay did not enhance the translocation frequency, because it was still below a minimum size. Notably, the long GC-rich c-MYC oncogene, used in this study as a structural equivalent of the longer S region, was a hotspot for recombination between direct-repeats and NHEJ-mediated CTs in wild-type yeast in our assays. Genetic instability in c-MYC and other oncogenes has been correlated with the GC-richness [44], [49], and the identification of c-MYC as a target of AID [23] also suggests that c-MYC shares structural features with S regions that make them highly recombinogenic during transcription in B cells. Interestingly, the recombinogenic potential generated by the transcription of the c-MYC sequence in our assays in hpr1Δ cells was also observed in the absence of AID expression. This is explained by the direct effect of hpr1Δ mutation observed in stability of long and GC-rich sequences such as c-MYC, as previously shown for bacterial DNA sequences [40], and by the lower density of AID-recognition motifs along this sequence. Altogether, our results confirm that c-MYC behaves as a natural hotspot of recombination, but this is more pronounced in THO depleted cells. The results suggest that the lower efficiency of mRNP biogenesis caused by THO mutation predispose c-MYC to breakage, leading to translocations. The similarity of phenotypes observed for mutations in the THSC/TREX-2 complex [50], which is located at the nuclear periphery in close contact with the nuclear pore complex and also causes transcription and RNA-dependent hyper-recombination, suggests that AID action would enhance DSB formation at S and c-MYC sequences provoking NHEJ-mediated translocations also in other mRNP biogenesis mutants, THO mutants being a paradigmatic example.
In summary, our work points to the idea that the structure of the nascent mRNA molecule may be a major determinant in the formation of R-loops at the S regions in B cells in vivo. Accordingly, previous data demonstrate that the RNA transcript formed during transcription of S sequences in vitro can thread back onto the template DNA strand, leading to an R-loop [51], [52]. Our model yeast system provides a validated assay to study the molecular mechanisms mediated by AID leading to DSBs at S regions and the c-MYC oncogene, such as those occurring in B cells responsible for class switching or the Ig/myc translocations that are associated with Burkitt's lymphomas, thus opening new perspectives in understanding AID-mediated rearrangements.
Materials and Methods
Yeast strains and plasmids
Yeast strains used were isogenic to W303, and are listed in Table S1. GFP direct-repeat system constructs and translocation systems were done as described [32], [36]. The hpr1Δ, dnl4Δ and ung1Δ alleles were generated by PCR-based gene replacement and confirmed by PCR and Southern analysis. hpr1-101 and double mutants (hpr1Δ dnl4Δ, hpr1Δ ung1Δ) with the translocation system were obtained by genetic crosses.
Determination of recombination frequencies
Yeast strains were grown on rich YEPD plates. Recombination tests using the GFP direct-repeat system and translocation assays were done as previously described [32], [36].
Physical analysis of translocations
Translocation analysis was performed by Pulsed Field Gel Electrophoresis (PFGE) and Southern analysis as described [32].
Chromosomal breakpoint analysis
Breakpoint regions were obtained by PCR amplification of genomic DNA extracted from yeast translocants using primers p1 (5′-GTATGTTCTAGCGCTTGC) and p2 (5′-CTAAACATATAATATAGCAACA), purified and sequenced.
Supporting Information
Zdroje
1. AguileraA 2002 The connection between transcription and genomic instability. EMBO J 21 195 201
2. DattaAJinks-RobertsonS 1995 Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science 268 1616 1619
3. SaxeDDattaAJinks-RobertsonS 2000 Stimulation of mitotic recombination events by high levels of RNA polymerase II transcription in yeast. Mol Cell Biol 20 5404 5414
4. MirkinEVMirkinSM 2007 Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71 13 35
5. HuertasPAguileraA 2003 Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12 711 721
6. YuKChedinFHsiehCLWilsonTELieberMR 2003 R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol 4 442 451
7. ChaudhuriJAltFW 2004 Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol 4 541 552
8. MuramatsuMSankaranandVSAnantSSugaiMKinoshitaK 1999 Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274 18470 18476
9. NambuYSugaiMGondaHLeeCGKatakaiT 2003 Transcription-coupled events associating with immunoglobulin switch region chromatin. Science 302 2137 2140
10. BesmerEMarketEPapavasiliouFN 2006 The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol Cell Biol 26 4378 4385
11. PavriRGazumyanAJankovicMDi VirgilioMKleinI Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell 143 122 133
12. CanugoviCSamaranayakeMBhagwatAS 2009 Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J 23 34 44
13. ChaudhuriJTianMKhuongCChuaKPinaudE 2003 Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422 726 730
14. RamiroARStavropoulosPJankovicMNussenzweigMC 2003 Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol 4 452 456
15. YuKRoyDBayramyanMHaworthISLieberMR 2005 Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol 25 1730 1736
16. ShenHMStorbU 2004 Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A 101 12997 13002
17. RadaCDi NoiaJMNeubergerMS 2004 Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16 163 171
18. SchraderCELinehanEKMochegovaSNWoodlandRTStavnezerJ 2005 Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med 202 561 568
19. Di NoiaJMWilliamsGTChanDTBuersteddeJMBaldwinGS 2007 Dependence of antibody gene diversification on uracil excision. J Exp Med 204 3209 3219
20. LiuMDukeJLRichterDJVinuesaCGGoodnowCC 2008 Two levels of protection for the B cell genome during somatic hypermutation. Nature 451 841 845
21. KuppersRDalla-FaveraR 2001 Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 20 5580 5594
22. RamiroARJankovicMEisenreichTDifilippantonioSChen-KiangS 2004 AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118 431 438
23. RobbianiDFBothmerACallenEReina-San-MartinBDorsettY 2008 AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135 1028 1038
24. RabbittsTH 1994 Chromosomal translocations in human cancer. Nature 372 143 149
25. NussenzweigANussenzweigMC Origin of chromosomal translocations in lymphoid cancer. Cell 141 27 38
26. RichardsonCJasinM 2000 Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 405 697 700
27. LieberMR The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79 181 211
28. ElliottBRichardsonCJasinM 2005 Chromosomal translocation mechanisms at intronic alu elements in mammalian cells. Mol Cell 17 885 894
29. PutnamCDPennaneachVKolodnerRD 2005 Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype. Mol Cell Biol 25 7226 7238
30. MieczkowskiPALemoineFJPetesTD 2006 Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair (Amst) 5 1010 1020
31. LeeJACarvalhoCMLupskiJR 2007 A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131 1235 1247
32. RuizJFGomez-GonzalezBAguileraA 2009 Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol Cell Biol 29 5441 5454
33. AguileraA 2005 Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol 17 242 250
34. ChavezSGarcia-RubioMPradoFAguileraA 2001 Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 21 7054 7064
35. Garcia-RubioMChavezSHuertasPTousCJimenoS 2008 Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics 279 123 132
36. Gomez-GonzalezBAguileraA 2007 Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci U S A 104 8409 8414
37. DanielsGALieberMR 1995 RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucleic Acids Res 23 5006 5011
38. TianMAltFW 2000 Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J Biol Chem 275 24163 24172
39. DuquetteMLHandaPVincentJATaylorAFMaizelsN 2004 Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18 1618 1629
40. ChavezSAguileraA 1997 The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11 3459 3470
41. Gomez-GonzalezBAguileraA 2009 R-loops do not accumulate in transcription-defective hpr1-101 mutants: implications for the functional role of THO/TREX. Nucleic Acids Res 37 4315 4321
42. HuangFTYuKBalterBBSelsingEOrucZ 2007 Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol 27 5921 5932
43. RajagopalDMaulRWGhoshAChakrabortyTKhamlichiAA 2009 Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med 206 1237 1244
44. DuquetteMLPhamPGoodmanMFMaizelsN 2005 AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene 24 5791 5798
45. ShinkuraRTianMSmithMChuaKFujiwaraY 2003 The influence of transcriptional orientation on endogenous switch region function. Nat Immunol 4 435 441
46. DunnickWHertzGZScappinoLGritzmacherC 1993 DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res 21 365 372
47. PoltoratskyVPWilsonSHKunkelTAPavlovYI 2004 Recombinogenic phenotype of human activation-induced cytosine deaminase. J Immunol 172 4308 4313
48. ZarrinAATianMWangJBorjesonTAltFW 2005 Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci U S A 102 2466 2470
49. DuquetteMLHuberMDMaizelsN 2007 G-rich proto-oncogenes are targeted for genomic instability in B-cell lymphomas. Cancer Res 67 2586 2594
50. Gonzalez-AguileraCTousCGomez-GonzalezBHuertasPLunaR 2008 The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell 19 4310 4318
51. RoyDYuKLieberMR 2008 Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 28 50 60
52. RoyDZhangZLuZHsiehCLLieberMR 2010 Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol 30 146 159
Štítky
Genetika Reprodukční medicína
Článek Break to Make a ConnectionČlánek A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on DiseaseČlánek The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Break to Make a Connection
- A New Testing Strategy to Identify Rare Variants with Either Risk or Protective Effect on Disease
- Single-Tissue and Cross-Tissue Heritability of Gene Expression Via Identity-by-Descent in Related or Unrelated Individuals
- Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in
- The Architecture of Gene Regulatory Variation across Multiple Human Tissues: The MuTHER Study
- MiRNA Control of Vegetative Phase Change in Trees
- New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion
- Genome-Wide Association Studies of the PR Interval in African Americans
- Mapping of the Disease Locus and Identification of As a Candidate Gene in a Canine Model of Primary Open Angle Glaucoma
- Mapping a New Spontaneous Preterm Birth Susceptibility Gene, , Using Linkage, Haplotype Sharing, and Association Analysis
- A Population Genetic Approach to Mapping Neurological Disorder Genes Using Deep Resequencing
- and Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust
- Targeted Sister Chromatid Cohesion by Sir2
- Correlated Evolution of Nearby Residues in Drosophilid Proteins
- Parallel Evolution of a Type IV Secretion System in Radiating Lineages of the Host-Restricted Bacterial Pathogen
- Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism
- Genome-Wide Association Study of Coronary Heart Disease and Its Risk Factors in 8,090 African Americans: The NHLBI CARe Project
- The Evolution of Host Specialization in the Vertebrate Gut Symbiont
- Genome-Wide Association of Familial Late-Onset Alzheimer's Disease Replicates and and Nominates in Interaction with
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- Association between Common Variation at the Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development
- AID Induces Double-Strand Breaks at Immunoglobulin Switch Regions and Causing Chromosomal Translocations in Yeast THO Mutants
- A Study of CNVs As Trait-Associated Polymorphisms and As Expression Quantitative Trait Loci
- Whole-Genome Comparison Reveals Novel Genetic Elements That Characterize the Genome of Industrial Strains of
- Prevalence of Epistasis in the Evolution of Influenza A Surface Proteins
- Srf1 Is a Novel Regulator of Phospholipase D Activity and Is Essential to Buffer the Toxic Effects of C16:0 Platelet Activating Factor
- Two Frizzled Planar Cell Polarity Signals in the Wing Are Differentially Organized by the Fat/Dachsous Pathway
- Phosphoinositide Regulation of Integrin Trafficking Required for Muscle Attachment and Maintenance
- Pathogenic VCP/TER94 Alleles Are Dominant Actives and Contribute to Neurodegeneration by Altering Cellular ATP Level in a IBMPFD Model
- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues
- Nucleosomes Containing Methylated DNA Stabilize DNA Methyltransferases 3A/3B and Ensure Faithful Epigenetic Inheritance
- Mutations in Zebrafish Result in Adult-Onset Ocular Pathogenesis That Models Myopia and Other Risk Factors for Glaucoma
- [], the Prion Formed by the Chromatin Remodeling Factor Swi1, Is Highly Sensitive to Alterations in Hsp70 Chaperone System Activity
- Characterization of Transcriptome Remodeling during Cambium Formation Identifies and As Opposing Regulators of Secondary Growth
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
- Epistatic Interaction Maps Relative to Multiple Metabolic Phenotypes
- Quantitative Models of the Mechanisms That Control Genome-Wide Patterns of Transcription Factor Binding during Early Development
- Genome-Wide Transcript Profiling of Endosperm without Paternal Contribution Identifies Parent-of-Origin–Dependent Regulation of
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in Celiac Disease and Rheumatoid Arthritis Identifies Fourteen Non-HLA Shared Loci
- MiRNA Control of Vegetative Phase Change in Trees
- Risk Alleles for Systemic Lupus Erythematosus in a Large Case-Control Collection and Associations with Clinical Subphenotypes
- The Cardiac Transcription Network Modulated by Gata4, Mef2a, Nkx2.5, Srf, Histone Modifications, and MicroRNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



