-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
MOS11: A New Component in the mRNA Export Pathway
Nucleocytoplasmic trafficking is emerging as an important aspect of plant immunity. The three related pathways affecting plant immunity include Nuclear Localization Signal (NLS)–mediated nuclear protein import, Nuclear Export Signal (NES)–dependent nuclear protein export, and mRNA export relying on MOS3, a nucleoporin belonging to the Nup107–160 complex. Here we report the characterization, identification, and detailed analysis of Arabidopsis modifier of snc1, 11 (mos11). Mutations in MOS11 can partially suppress the dwarfism and enhanced disease resistance phenotypes of snc1, which carries a gain-of-function mutation in a TIR-NB-LRR type Resistance gene. MOS11 encodes a conserved eukaryotic protein with homology to the human RNA binding protein CIP29. Further functional analysis shows that MOS11 localizes to the nucleus and that the mos11 mutants accumulate more poly(A) mRNAs in the nucleus, likely resulting from reduced mRNA export activity. Epistasis analysis between mos3-1 and mos11-1 revealed that MOS11 probably functions in the same mRNA export pathway as MOS3, in a partially overlapping fashion, before the mRNA molecules pass through the nuclear pores. Taken together, MOS11 is identified as a new protein contributing to the transfer of mature mRNA from the nucleus to the cytosol.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001250
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001250Summary
Nucleocytoplasmic trafficking is emerging as an important aspect of plant immunity. The three related pathways affecting plant immunity include Nuclear Localization Signal (NLS)–mediated nuclear protein import, Nuclear Export Signal (NES)–dependent nuclear protein export, and mRNA export relying on MOS3, a nucleoporin belonging to the Nup107–160 complex. Here we report the characterization, identification, and detailed analysis of Arabidopsis modifier of snc1, 11 (mos11). Mutations in MOS11 can partially suppress the dwarfism and enhanced disease resistance phenotypes of snc1, which carries a gain-of-function mutation in a TIR-NB-LRR type Resistance gene. MOS11 encodes a conserved eukaryotic protein with homology to the human RNA binding protein CIP29. Further functional analysis shows that MOS11 localizes to the nucleus and that the mos11 mutants accumulate more poly(A) mRNAs in the nucleus, likely resulting from reduced mRNA export activity. Epistasis analysis between mos3-1 and mos11-1 revealed that MOS11 probably functions in the same mRNA export pathway as MOS3, in a partially overlapping fashion, before the mRNA molecules pass through the nuclear pores. Taken together, MOS11 is identified as a new protein contributing to the transfer of mature mRNA from the nucleus to the cytosol.
Introduction
Plants utilize a two-layered immune system to recognize and combat pathogens. Conserved pathogen-associated molecular patterns (PAMPs), which are detected by plasma membrane localized plant pattern recognition receptors (PRRs), trigger a low amplitude defense response termed PAMP-triggered immunity (PTI) [1]. To suppress PTI, pathogens have evolved a suite of effectors (also called Avirulence or Avr proteins) [2]–[5]. The second layer of the plant immune response is initiated by the recognition of effectors or their effects, by cognate Resistance (R) proteins, leading to a strong defense response termed effector-triggered immunity (ETI) that culminates with hypersensitive response (HR) [1]: a programmed cell death event believed to restrict pathogen growth. Most cloned R genes encode Nucleotide Binding Leucine Rich-Repeat (NB-LRR) proteins with either a Toll/Interleukin1 receptor (TIR) or a Coiled-Coil (CC)-domain at their N terminus [6].
Mutant snc1 (suppressor of npr1-1, constitutive 1) plants carry a gain-of-function mutation in a TIR-NB-LRR R gene. This mutation renders the snc1 protein auto-active without the presence of pathogens. As a consequence, snc1 plants constitutively express Pathogenesis Related (PR) defense marker genes, accumulate high levels of defense phytohormone salicylic acid (SA), and are more resistant to the virulent bacterial pathogen Pseudomonas syringae maculicola (P.s.m.) ES4326 and the oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2. Like most TIR-type R proteins, snc1 is fully dependent on EDS1 (Enhanced Disease Susceptibility 1) and PAD4 (PhytoAlexin Deficient 4) for its function [7], [8].
The snc1 mutant results from a point mutation in the linker region between the NB and LRR domains. This gain-of-function mutation causes activation of the ETI pathways mediated by TIR-NB-LRR R proteins. The extreme dwarf morphology of snc1 also makes it an ideal candidate to carry out genetic screens to investigate ETI components downstream of SNC1. Utilizing the unique autoimmune phenotypes of snc1, we conducted genetic screens to search for components contributing to R protein mediated immune responses. From the MOS (modifier of snc1) genetic screens, three nucleocytoplasmic pathways have been shown to affect plant immunity: Nuclear Localization Signal (NLS)-mediated nuclear protein import, Nuclear Export Signal (NES)-dependent nuclear protein export and mRNA export. MOS6, an importin α homolog involved in NLS-dependent protein import was shown to contribute to plant immunity [9]. More recently we identified MOS7, an integral nuclear pore component homologous to Nucleoporin88 (Nup88). Partial loss-of-function mos7-1 mutant plants showed increased NES-dependent protein export and exhibited reduced nuclear accumulation of the important defense regulators EDS1, NPR1 (Non-expressor of Pathogenesis Related 1) and snc1 [10]. The contribution of mRNA export to plant immunity was demonstrated in the mos3 mutant which exhibits defects in basal and R protein mediated immunity [10], [11]. MOS3/SAR3/AtNup96 is an integral nuclear pore component in the conserved Nup107–160 complex and was shown to be required for mRNA export [12].
Here we report the identification of modifier of snc1, 11 (mos11), a T-DNA insertional mutant. Mutation in MOS11 partially suppresses all the autoimmune phenotypes of snc1. MOS11 encodes a protein with unknown function. However, it is evolutionarily conserved and shares homology with a human protein CIP29. Our data suggest that MOS11 localizes to the nucleus and functions in the same mRNA export pathway as MOS3.
Results
mos11 partially suppresses the autoimmune phenotypes of snc1
The mos11-1 mutant was identified by T-DNA tagging in the snc1 background [13]. Normally snc1 plants are dwarf, have twisted dark green leaves, accumulate high levels of the defense phytohormone SA and are resistant to the virulent oomycete H.a. Noco2 and P.s.m. ES4326 bacteria. mos11-1 partially suppressed the snc1 morphology (Figure 1A). The mos11-1 snc1 plants never grew to wild type size and their leaves remained curly (Figure 1A). In addition, semi-quantitative RT-PCR confirmed that endogenous PR1 and PR2, defense markers acting downstream of R gene activation that are constitutively expressed in snc1, are greatly reduced in mos11-1 snc1 (Figure 1B). In order to investigate how the mos11 mutation suppresses snc1 morphology and PR gene expression, we assessed whether the snc1 protein levels were affected in mos11-1 snc1 plants. We observed a slight but consistent reduction in snc1 levels in mos11-1 snc1 compared with snc1 plants (Figure 1C). Total SA levels in mos11-1 snc1 were drastically reduced compared with snc1 plants, but were still significantly higher than in wild type plants (Figure 1D).
Fig. 1. Suppression of snc1-associated dwarfism and enhanced resistance phenotypes by mos11-1. 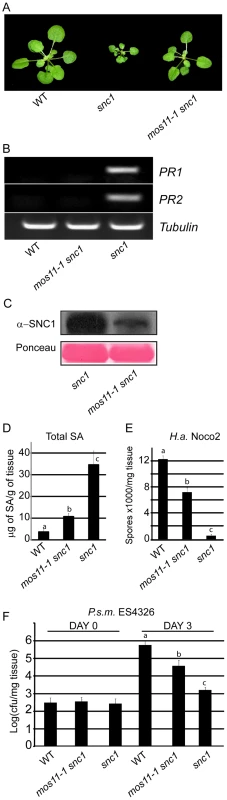
(A) Morphology of wild type (Col-0), snc1 and mos11-1 snc1. The picture was taken with 4-week-old soil grown plants. (B) PR1 and PR2 gene expression as measured by RT-PCR in WT, mos11-1 snc1 and snc1 with Tubulin as loading control. RNA was extracted from 2-week-old plants grown on 1/2 MS medium and reverse transcribed to yield cDNA. PR1, PR2, and Tubulin were amplified by 27 cycles of PCR with equal amounts of total cDNA. PCR products were stained with ethidium bromide. (C) SNC1 protein level was evaluated by Western blot using an anti-SNC1 antibody [42]. Total protein was extracted from 4-week-old soil-grown plants. As a loading control, the Rubisco band is shown from a Ponceau stain. (D) Total SA was extracted from 4-week-old plants. The values presented are averages of three replicates with error bars showing standard deviations. Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.0001). (E) Growth of H.a. Noco2 on WT, mos11-1 snc1 and snc1. Two-week-old seedlings were sprayed with a conidiospore suspension of 5×104 spores per ml of water. Spores were counted with a hemocytometer 7 days after infection. Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.000001). (F) Growth of P.s.m. ES4326 in 5-week-old plants. Leaves were infiltrated with a bacterial suspension (OD600 = 0.0001). Leaf discs were collected immediately after infiltration 0 days post inoculation (DPI) and 3 DPI. Log-transformed values are averaged over four replicates with standard deviations (cfu = colony forming units). Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.00001). To assess whether the mos11-1 mutation affects snc1-mediated resistance to virulent pathogens, we performed infection assays with the obligate biotrophic oomycete H.a. Noco2 and P.s.m. ES4326 bacteria. While snc1 plants were resistant to H.a. Noco2 and showed very low sporulation, mos11-1 snc1 displayed intermediate susceptibility to H.a. Noco2 (Figure 1E). mos11-1 snc1 supported 13-fold more sporulation than snc1, however it was still significantly more resistant than the wild type (Figure 1E). This trend was also observed when we investigated susceptibility to P.s.m. ES4326 (Figure 1F). Taken together, these results show that mos11-1 partially suppresses all the phenotypes of snc1.
Identification of the molecular lesion in mos11-1
The T-DNA insertion causing the mos11-1 snc1 phenotype was identified in the 5′ UTR of At5g02770 using inverse PCR (Figure 2A). To confirm that the mutant phenotypes in mos11-1 snc1 were caused by the T-DNA insertion, we cloned the wild type DNA of At5g02770 with 1992 base pairs (bp) upstream of the start codon and transformed it into mos11-1 snc1 plants. At5g02770 fully complemented the mos11-1 snc1 morphology (Figure 2B). When the T2 transgenic plants were challenged with H.a. Noco2, they were as resistant as snc1 plants (Figure 2C). These data confirmed that MOS11 is At5g02770.
Fig. 2. Complementation of mos11-1 snc1 by At5g02770. 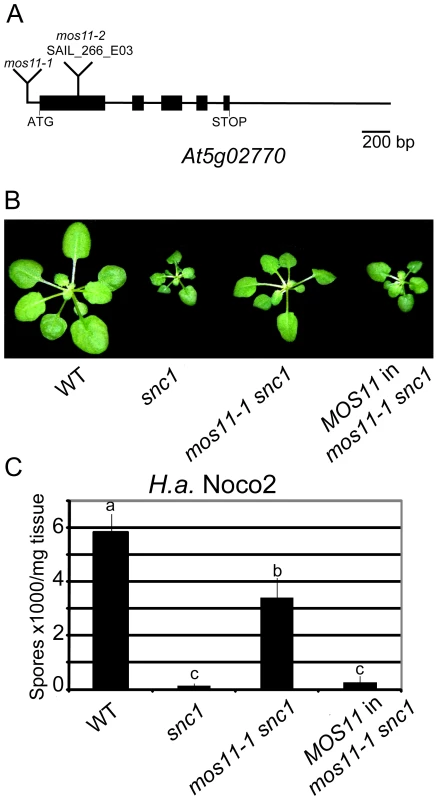
(A) Gene structure of At5g02770. Exons are represented by black boxes and introns and UTRs are shown as solid lines. Locations of mos11-1 and mos11-2 (SAIL_266_E03) insertions are as indicated. (B) Morphology of WT, snc1, mos11-1 snc1 and a representative transgenic line with MOS11 (At5g02770) driven by its own promoter transformed into mos11-1 snc1. (C) Growth of H.a. Noco2 on WT, snc1, mos11-1 snc1 and MOS11 in mos11-1 snc1. The infection was carried out as in Figure 1E. Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.001). Another T-DNA insertion allele (SAIL_266_E03) in the first exon of At5g02770 was available from the Arabidopsis Stock Center (ABRC), which we named mos11-2 (Figure 2A). The mos11-1 single mutant was obtained from a backcross between mos11-1 snc1 and the wild type Columbia ecotype (Col-0). The mos11-1 and mos11-2 single mutants had identical morphology but were slightly different than Col-0. When mos11-1 was crossed with mos11-2, the two alleles failed to complement each other in all 29 F1 plants (data not shown), confirming that mos11-2 and mos11-1 carry lesions in the same gene. When mos11-2 was crossed with snc1, the mos11-2 snc1 double mutant exhibited a similar level of snc1 suppression as mos11-1 (data not shown), further supporting that At5g02770 is MOS11.
Subcellular localization of MOS11
MOS11 is a single copy gene that encodes a 214 amino-acid protein with unknown function. Using BlastP, the human protein CIP29 (cytokine induced protein 29 kDa) was found to share 38% identity and 50% similarity over the entire length of the MOS11 protein. It is striking that islands of positively charged residues, arginine and lysine, are highly conserved (see alignment in Figure S1). Although MOS11 does not have predicted NLS (Nuclear Localization Signal) motifs, it is predicted by WoLF PSORT (http://wolfpsort.org/) to be a nuclear protein.
To determine the biochemical function of MOS11, we first investigated its subcellular localization. When MOS11-GFP with its native promoter was transformed into mos11-1 snc1, among the 9 T1 progeny, all showed snc1-like morphology (Figure 3A). The T2 transgenic plants also displayed snc1-like enhanced resistance to H.a. Noco2 (Figure 3B). These data indicate that MOS11-GFP largely reflects endogenous MOS11 in the transgenic line, and therefore, the subcellular localization of MOS11-GFP should be identical to MOS11. As shown in Figure 3C, in contrast to the MOS3-GFP control, which localizes to the nuclear envelope, strong MOS11-GFP signal was observed in the nuclei of leaf epidermal and root cells, however it appears to be excluded from the nucleolus. Nuclear localization of the MOS11-GFP signal was observed in guard cells as well. Thus we conclude that MOS11 is a nuclear protein.
Fig. 3. MOS11-GFP is localized to the nucleus. 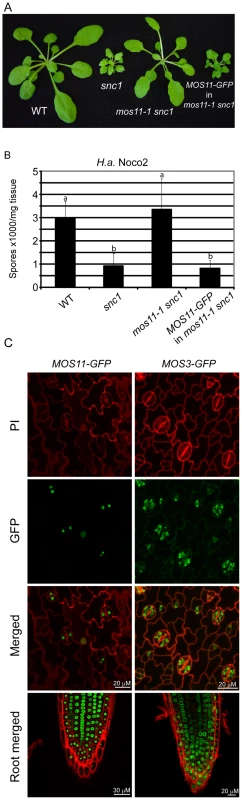
(A) Morphology of WT, snc1, mos11-1 snc1 and a transgenic line with MOS11-GFP driven by its own promoter transformed into mos11-1 snc1. (B) Growth of H.a. Noco2 on WT, snc1, mos11-1 snc1 and MOS11-GFP in mos11-1 snc1. The infection was carried out as in Figure 1E. Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.00001). (C) MOS11-GFP and MOS3-GFP fluorescence as observed by confocal microscopy in leaf cells and root cells. Cell walls were stained with propidium iodine (PI). MOS11 is required for mRNA export
In humans, CIP29 was shown to bind RNA (23). The two known interacting protein partners of CIP29 are the RNA helicase DDX39 and FUS/TLS. DDX39 shows 91% identity with UAP56, a Drosophila RNA helicase involved in mRNA export [14]. DDX39 also interacts with ALY and FUS/TLS, which were both shown to contribute to mRNA export [15]–[17]. These associations prompted us to investigate whether MOS11 also contributes to mRNA export. We first used dot blot hybridization to check whether the total mRNA level was affected in mos11-1 and mos11-2 plants. mos11-1 and mos11-2 are morphologically indistinguishable (Figure 4A). Decreasing RNA concentrations were used to avoid saturation. No visible differences in total RNA levels were observed (Figure 4B). To assess whether mRNA export from the nucleus to the cytoplasm was affected in mos11-1 and mos11-2 plants, we carried out poly(A) in situ hybridization with a 48-mer oligo d(T) 5′-end labeled with Alexa 488. Poly(A) signals were observed in the nucleus of Col-0 and the cytosol of all labeled cells (Figure 4C). When mos11-1 and mos11-2 were analyzed, most poly(A) signal was found in the nucleus, indicating that mRNA export is diminished in mos11 plants, resulting in mRNA accumulation in the nucleus (Figure 4C and Figure S2). Thus MOS11 is likely a contributing factor in the mRNA export pathway. Note that nucleolar oligo d(T) signal is weak in all in situ experiments (Figure 4C and Figure 5E), a region where MOS11-GFP was also absent (Figure 3C).
Fig. 4. mRNA export is impaired in mos11 plants. 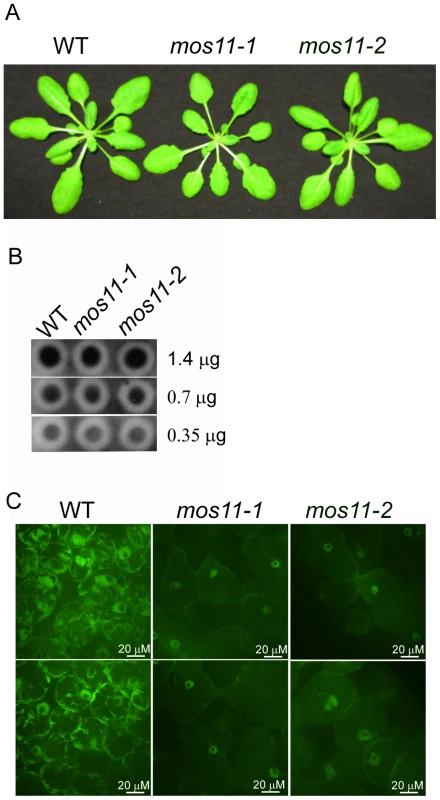
(A) Morphology of 4-week-old soil-grown WT, mos11-1 and mos11-2 plants. (B) Dot blot of WT, mos11-1 and mos11-2. The same amount of WT, mos11-1, and mos11-2 total RNA were applied to the Hybond-N+ membrane and hybridized by 32P-ATP labeled 18-mer oligo dT. The amount of total RNA loaded to each dot was indicated on the right of the blot. The radioactive signal was detected by a phosphor-imager. (C) Whole mount in situ mRNA localization of 7-day-old seedlings probed with 48-mer oligo d(T) labeled with Alexa 488. Observations were done at 63× magnification and microscope settings were identical for all samples. Fig. 5. mos11-1 mos3-1 double mutant is impaired in flowering time, basal defense, and mRNA export. 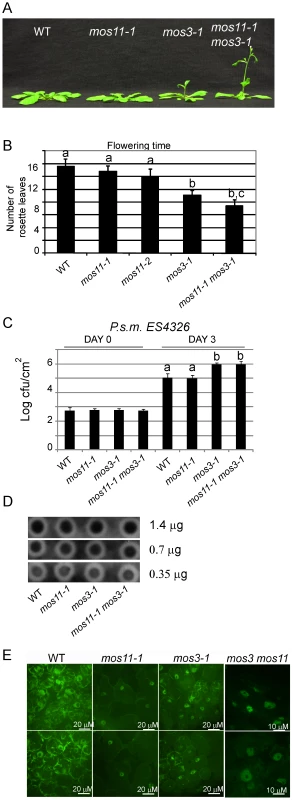
(A) Morphology of 4-week-old soil-grown WT, mos11-1, mos3-1 and mos11-1 mos3-1 plants. (B) Quantitative flowering time analysis assessed by counting the number of rosette leaves on 4-week-old soil-grown plants. (C) Growth of P.s.m. ES4326 in 5-week-old plants. The infection was carried out as in Figure 1F. Data were analyzed using one-way ANOVA. Different letters indicate statistically significant differences (p-value <0.00001). (D) Dot blot of WT, mos11-1, mos3-1 and mos3-1 mos11-1. The same amount of WT, mos11-1, mos3-1 and mos3-1 mos11-1 total RNA were applied to the Hybond-N+ membrane and hybridized by 32P-ATP labeled 18-mer oligo dT. The amount of total RNA loaded to each dot is indicated to the right of the blot. The radioactive signal was detected by the phosphor-imager. (E) Whole mount in situ mRNA localization of 7-day-old seedlings probed with oligo d(T) labeled with Alexa 488. Observations were done at 63× magnification and microscope settings were identical for all samples. MOS11 functions in the same mRNA export pathway as MOS3 in a partially overlapping manner
Since mos3 was previously shown to affect mRNA export [12], we investigated the relationship between mos3-1 and mos11-1 using epistasis analysis. The morphological phenotypes associated with both single mutants are quite different; mos3-1 shows long and narrow leaves and flowers early, whereas mos11-1 has mostly wild type-like morphology. The mos3-1 mos11-1 double mutant displayed a mos3-like morphology with leaves that are slightly narrower and longer than those of the mos3 single mutant, but flowered slightly earlier than mos3-1 or mos11-1 (Figure 5A and 5B). These data suggest that in regard to flowering time regulation, MOS3 and MOS11 seem to play partially overlapping roles.
In order to assess the relationship between mos3 and mos11 in immunity, we evaluated the response of mos3-1, mos11-1 and mos11-1 mos3-1 to the virulent bacteria P.s.m. ES4326. Virulent pathogens, ones not carrying Avirulence genes recognized by the plant R proteins, were used to evaluate the contribution of basal resistance to immunity. Both mos3-1 and mos11-1 mos3-1 plants exhibited enhanced susceptibility when infiltrated with virulent P.s.m. ES4326 and supported higher bacterial growth, whereas the response of the single mos11-1 mutant was similar to that of the wild type (Figure 5C), suggesting that MOS11 is not involved in basal resistance. No further enhanced susceptibility was observed for mos11-1 mos3-1, indicating that these two genes function in the same pathway to regulate immunity. It should be noted that due to the quantitative limit of infection assays, possible minor additive effects like those observed for the flowering time phenotype might have been missed.
To test whether mos11 affects R protein mediated resistance, we inoculated Col-0, mos11-1 and mos11-2 with Pseudomonas syringae pv tomato (P.s.t.) DC3000 strains expressing Avirulence genes avrRps4, avrRpm1, avrRpt2 or avrPphB. Avirulent pathogens carry Avirulence genes whose products or the effects of the products in the plant cells are recognized by plant R proteins. These pathogens are used to evaluate the integrity of ETI in plants. Bacterial growth was not affected in the single mutants for any of the R protein mediated pathways (Figure S3). Thus we conclude that MOS11 is not involved in the ETI response mediated by these R proteins.
We then analyzed the mRNA export status in the mos11-1 mos3-1 double mutant compared with the single mutants. Similar to the mos3-1 and mos11-1 plants, the poly(A) signal in the double mutant was mostly observed in the nucleus. However, further poly(A) accumulation was not detected in the nuclei of the double mutant (Figure 5E and Figure S2). It should be noted that in all the mRNA export deficient mutants tested, some signal, albeit much weaker than the wild type, was always observed in the cytosol, which explains why these mutations are not lethal and the plants can successfully complete their life cycle. Together, these data suggest that MOS3 and MOS11 function in the same mRNA export pathway, possibly in a partially overlapping manner. Since MOS3 is part of the nuclear pore while MOS11 localizes inside the nucleus, MOS3 probably functions downstream of MOS11 in mRNA export (Figure 6).
Fig. 6. A proposed plant mRNA export pathway. 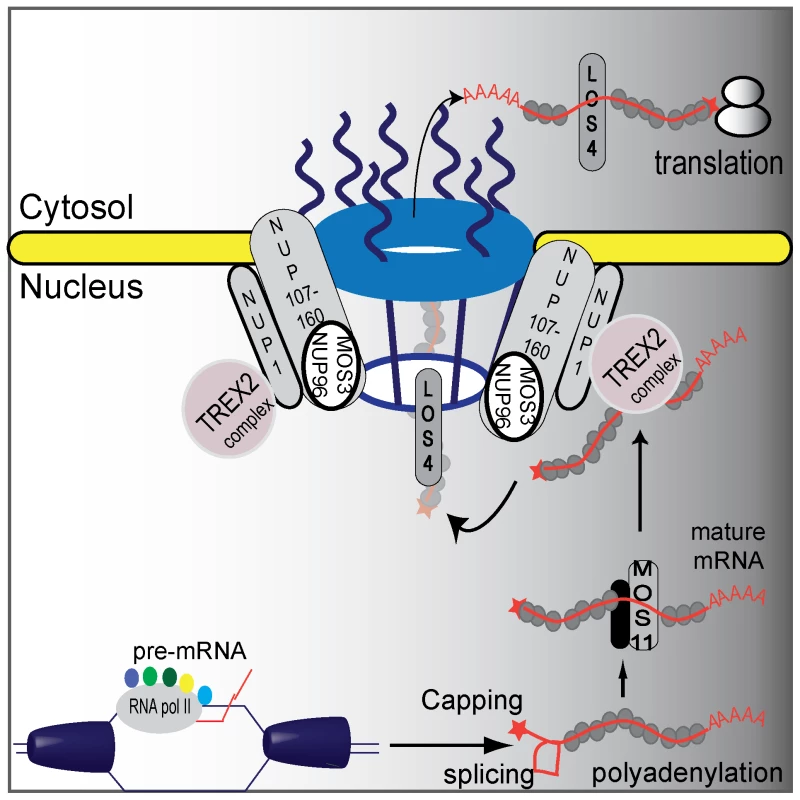
DNA helicase unwinds DNA and RNA polymerase II and co-activators proceed with transcription and mRNA maturation. Pre-mRNA is capped, polyadenylated, and spliced. Mature mRNA is then carried by MOS11 and its partners to TREX2, which associates with NUP1. LOS4 then binds the mRNA and passes the mature mRNA through the nuclear pores with the help of the NUP107–160 complex to the cytosol where it can be translated. Discussion
To search for components in R protein mediated immunity, we performed genetic screens for suppressors of the autoimmune mutant snc1. mos11-1 was identified from a T-DNA mutagenized snc1 population. Mutation in MOS11 partially abolishes the dwarf stature, elevated SA levels, and enhanced resistance to H.a. Noco2 and P.s.m. ES4326 of snc1. MOS11 encodes a protein with unknown function in the plant kingdom, however, it shares 50% similarity with human CIP29. Similarities and identities are more striking in islands of conserved positively charged residues; namely lysine and arginine (Figure S1). These basic islands are known to be present in nucleic acid binding proteins directly involved in DNA/RNA binding [18]–[20].
There are a limited number of reports on human CIP29, which was initially named HCC-1 after being isolated from the hemofiltrate of patients with chronic renal failure [21] and later renamed CCL14a [22]. Overexpression of CIP29 in human embryonic kidney cell line results in slower growth. Biochemical characterization demonstrated that CIP29 could bind RNA on its own [23]. In yeast two-hybrid assays, CIP29 interacted with two DEAD-box RNA helicases, BAT1 and DDX39 [24].
Hints on the potential biological functions of CIP29 came from the study by Sugiura et al. (2007). They showed that the ATP-dependent DEAD-box RNA helicases DDX39 can co-immunoprecipitate CIP29, confirming the earlier yeast two-hybrid interaction reported by Leaw et al. (2004). Additionally they demonstrated that CIP29 can bind RNA on its own [23], and more importantly, it enhances the RNA unwinding activity of DDX39. The same group also identified FUS/TLS, a nucleic acid binding protein participating in both transcription and splicing as a physical interactor of CIP29 [23]. In addition, they detected ALY, a well-characterized mRNA export factor, in the DDX39 immunoprecipitate. CIP29 localized mainly to the nucleus although small amounts were also detected in the cytosol [25]. Very recently, Dufu et al. (2010) reported that CIP29 physically associates with ALY via UAP56 in an ATP-dependent manner to form a trimeric complex. In addition they demonstrated that efficient recruitment of CIP29 to the mRNA is splicing - and cap-dependent [26]. The functions of the CIP29 interactors, its affinity for RNA and the conserved island of positively charged residues suggest a potential role of CIP29 in RNA processing and/or RNA export, although mRNA export defects have not been shown for a cip29 mutant. Through knockout analysis with mos11 mutants, we demonstrate that MOS11 is indeed required for mRNA export. We speculate that its human counterpart CIP29 probably has a similar function.
In eukaryotic cells, transcription and translation take place in two separate compartments, which allows fine-tuned regulation of biological processes. For proteins to be translated in the cytosol, the mRNA molecules must first be properly processed, assembled into an export competent mRNA ribonucleoprotein (mRNP) particle and transferred through the nuclear pore. This assembly starts at the onset of transcription. Several proteins bind to the nascent pre-mRNA. These proteins can be involved in transcription [27], capping, splicing [28], polyadenylation [29] or binding to nuclear pore proteins [30]. Most of the knowledge on mRNA export has been gained from studies of human and yeast cells. Little is known about the mechanisms of plant mRNA export (for reviews see [31], [32]), although it is believed to share high similarity with yeast and human pathways.
In plants, several nuclear pore complex mutants have been shown to exhibit defects in mRNA export [12], [31]–[37]. In Arabidopsis, previously reported components shown to participate in mRNA export all localize to the nuclear envelope; these include MOS3/AtNup96, AtNup160, LOS4, AtNup1 and AtTHP1. LOS4 encodes a DEAD-box RNA helicase enriched at the nuclear rim and it is also present in the cytosol. Two point mutation alleles of los4/cryophyte were shown to affect mRNA export [38]. More recently orthologs of the TREX-2 complex, which is necessary for mRNA export in yeast and humans, have been identified in Arabidopsis. The TREX-2 complex, which is tethered to the nuclear pore complex via Nup1 in yeast, is composed of Thp1, Sac3, Cdc31 and Sus1. AtNUP1 and AtTHP1 were shown to be necessary for mRNA export, but the other three proteins do not appear to be involved in mRNA export despite evidence of a physical interaction with other TREX-2 subunits [36]. Another protein, TEX1, which is not part of the THO/TREX core complex but part of a complex associated with the TREX complex is also found in Arabidopsis [37]. In Drosophila and human, TEX associates with the helicase UAP56 and the mRNA export factor ALY [39], [40]. However, co-IP experiments using HA-tagged AtTEX did not pull-down ALY or UAP56 in Arabidopsis, but they did retrieve THO1, THO2, THO5, THO6 and THO7 [37].
Subcellular localization of MOS11-GFP revealed that MOS11 is a nuclear protein. In mos11-1, mos11-2 and mos3-1, mRNA export was similarly affected. Epistasis analysis between mos11-1 and mos3-1 suggested that these two genes function in the same mRNA export pathway. Thus we drew a simplified model of the mRNA export pathway in plants (Figure 6) where MOS11 likely functions in the early steps of the mRNA export before mRNA exits the nucleus through the nuclear pore. This is consistent with their respective subcellular localization where MOS11 is nuclear and MOS3 is limited to the nuclear rim. This indicates that MOS11 has access to the mRNA prior to MOS3. Since MOS11 is conserved in eukaryotes, its human homolog CIP29 probably also functions in a similar step in mRNA export as suggested by Dufu et al. [26].
The reason why mos11 suppresses snc1 phenotypes is not clear. mos11 single mutant plants, although slightly different from the wild type, grow to full stature, produce seeds and complete their life cycle successfully. This shows that the observed impairments in mRNA export in mos11 are not sufficient to significantly affect ontology. The snc1 mutation involves an extensive transcriptional reprogramming that results in the dramatic phenotypes displayed by snc1 plants. Among the changes observed following the auto-activation of snc1 are the induction of defence gene markers and a strong stimulation of the SA biosynthetic pathway. These changes are associated with dwarf stature and increased resistance to pathogens. It is conceivable that MOS11 could promote mRNA export of genes that are up-regulated following defence activation, possibly including R genes. In support of this hypothesis, dot blot analysis demonstrated that mos11-1 does not affect the overall level of transcription since all lines analyzed had similar levels of total mRNA. However mos11-1 does have an effect on snc1 protein level in mos11-1 snc1, possibly resulting from impaired SNC1 mRNA export leading to less mRNA for translation of the protein (Figure 1C).
The detailed mechanism of MOS11 in mRNA export is not clear. Since CIP29 was recently shown to interact with ALY and UAP56 in an ATP-dependent manner [26], and it was also shown to enhance the RNA unwinding activity of DDX39 [23], we speculate that the function of MOS11 is likely to act as a co-chaperone to plant homologs of UAP56 and ALY to co-catalyze mRNA unwinding prior to its nuclear export. The specific targets of MOS11 await further investigation.
Materials and Methods
Plant growth, SA measurement, RT-PCR analysis, pathogen infections, and T-DNA screens
All plants were grown under a 16 h light/8 h dark regime. SA extractions and measurements were carried out as previously described [41]. RNA was extracted from 2-week-old plate-grown seedlings on half Murashige and Skoog (½ MS) media using the Totally RNA kit (Ambion). 200 ng of RNA was used for reverse transcription using Superscript II (Invitrogen). RT-PCR analysis of PR1 and PR2 expression was carried out as previously described [8]. Tubulin was used as an uninduced loading control with primers 5′ACGTATCGATGTCTATTTCAACG3′ and 5′ATATCGTAGAGAGCCTCATTGTCC3′. Pathogen assays with H.a. Noco2 and P.s.m. ES4326 were carried out as previously described [7]. T-DNA screen in snc1 was described previously [13].
Total protein extraction and Western blot
Total protein was extracted from 4-week-old soil-grown plants using the extraction buffer containing 100 mM Tris-HCl (pH 8), 0.2% SDS and 2% β-mercaptoethanol. 30 µl of total protein was separated on 8% SDS-PAGE and transferred onto nitrocellulose membrane. For SNC1 total protein level, the membrane was probed with purified anti-SNC1 antibody, which was generated against a SNC1-specific peptide in rabbit [42].
MOS11-GFP subcellular localization
The full-length genomic MOS11 gene plus 1992 bp upstream of its start codon was cloned in frame to the N-terminus of GFP in a modified pCambia1305 vector in which the GUS gene had been replaced by GFP. Plant transformation was carried out by Agrobacterium in planta dipping [43] and transformants were selected on ½ MS supplemented with 30 µg/ml hygromycin. T1 and T2 transgenic plants were then observed with a confocal microscope with propidium iodine as a cell wall/dying nuclei marker.
Dot blot hybridization
The total RNA was extracted by RNAiso Plus (Takara, Cat No. D9108A). The RNA samples were quantified by spectrophotometry and confirmed by agarose gel electrophoresis. The RNA samples were prepared and applied to the Hybond-N+ membrane (Amersham Cat. No. RPN303B). The membrane was cross-linked for 2 minutes at 70000 micro-joules/cm2 and baked for 2 hours at 80°C. The membrane was pre-hybridized for 1 hour at 37°C in Hyb-50 (Mylab Corporation, Beijing, P. R. C.). The 18-mer oligo-dT was labeled with 32P by the T4 poly-nucleotide kinase (NEB, Cat. No. M0201) and added to the pre-hybridization buffer. After 16 hours of hybridization, the membrane was washed following Amersham Hybond-N+ manual. The signals were detected by phospho-imager using a Typhoon scanner.
Whole mount in situ total mRNA localization
We adopted a protocol similar to that described in Parry et al. (2006). Briefly, 4–7 day-old plate-grown seedlings were immersed in 1 ml fixation cocktail (50% fixation buffer consisting of 120 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 2.7 mM KCl, 0.1% Tween 20, 80 mM EGTA, 5% formaldehyde, 10% DMSO and 50% heptane) in a glass container and gently agitated for 30 minutes at room temperature. The sample was dehydrated twice for 5 minutes each in 100% methanol, three times for 5 minutes each in 100% ethanol and incubated for 30 minutes in ethanol:xylene (50∶50) with gentle agitation. The samples were washed twice in ethanol for 5 minutes each and twice in methanol for 5 minutes each before the samples were incubated 5 minutes in methanol: fixation buffer without formaldehyde (50∶50) and fixed in fixation buffer with 5% formaldehyde for 30 minutes. Samples were rinsed twice for 5 minutes each with fixation buffer without formaldehyde and incubated for 5 minutes with PerfectHyb Plus (Sigma). 1 ml of new hybridization buffer was added and pre-hybridized at 50°C for 1 hour. Finally, 5 pmol of 5′ end-labeled Alexa-488 48-mer oligo d(T) (Invitrogen) was added and incubated overnight in the dark. The final samples were mounted in water and fluorescence was observed with a spinning disc microscope at a magnification of 63X using the following settings: Volocity 5.1 software, Hamamatsu C9100-50 camera, the exposure time was set to 100 ms and the sensitivity was adjusted to 176. Settings were the same for all samples.
Supporting Information
Zdroje
1. JonesJDG
DanglJL
2006 The plant immune system. Nature 444 323 329
2. Gimenez-IbanezS
HannDR
NtoukakisV
PetutschnigE
LipkaV
2009 AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Current Biology 19 423 429
3. GöhreV
SpallekT
HäwekerH
MersmannS
MentzelT
2008 Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Current Biology 18 1824 1832
4. ShanL
HeP
LiJ
HeeseA
PeckSC
2008 Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host & Microbe 4 17 27
5. XiangT
ZongN
ZouY
WuY
ZhangJ
2008 Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Current Biology 18 74 80
6. ChisholmST
CoakerG
DayB
StaskawiczBJ
2006 Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803 814
7. LiX
ClarkeJD
ZhangY
DongX
2001 Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Molecular Plant-Microbe Interactions 14 1131 1139
8. ZhangY
GoritschnigS
DongX
LiX
2003 A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 2636 2646
9. PalmaK
ZhangY
LiX
2005 An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Current Biology 15 1129 1135
10. ChengYT
GermainH
WiermerM
BiD
XuF
2009 Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21 2503 2516
11. ZhangY
LiX
2005 A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17 1306 1316
12. ParryG
WardS
CernacA
DharmasiriS
EstelleM
2006 The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18 1590 1603
13. ZhuZ
XuF
ZhangY
ChengYT
WiermerM
2010 Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proceeding of the National Academy of Science, USA 107 13960 13965
14. GatfieldD
Le HirH
SchmittC
BraunIC
KocherT
2001 The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Current Biology 11 1716 1721
15. MeissnerM
LopatoS
GotzmannJ
SauermannG
BartaA
2003 Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Experimental Cell Research 283 184 195
16. ZhouL
LuoMJ
StraesserK
KatahiraJ
HurtE
2000 The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407 401 405
17. ZinsznerH
SokJ
ImmanuelD
YinY
RonD
1997 TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. Journal of Cell Science 110 1741 1750
18. CherstvyAG
2009 Positively charged residues in DNA-binding domains of structural proteins follow sequence-specific positions of DNA phosphate groups. Journal of Physical Chemistry and Biology 113 4242 4247
19. HoribeK
MitakuS
2003 Positive charge cores in DNA-binding proteins. Genome Informatics 14 508 509
20. ShazmanS
Mandel-GutfreundY
2008 Classifying RNA-binding proteins based on electrostatic properties. PLoS Comput Biol 4 e1000146 doi:10.1371/journal.pcbi.1000146
21. Schulz-KnappeP
MägertHJ
DewaldB
MeyerM
CetinY
1996 HCC-1, a novel chemokine from human plasma. The Journal of Experimental Medicine 183 295 299
22. ForssmannU
MägertHJ
AdermannK
EscherSE
ForssmannWG
2001 Hemofiltrate CC chemokines with unique biochemical properties: HCC-1/CCL14a and HCC-2/CCL15. Journal of Leukocyte Biology 70 357 366
23. SugiuraT
SakuraiK
NaganoY
2007 Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Experimental Cell Research 313 782 790
24. LeawCL
RenEC
ChoongML
2004 Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cellular and Molecular Life Sciences 61 2264 2273
25. FukudaS
WuDW
StarkK
PelusLM
2002 Cloning and characterization of a proliferation-associated cytokine-inducible protein, CIP29. Biochemical and Biophysical Research Communications 292 593 600
26. DufuK
LivingstoneMJ
SeebacherJ
GygiSP
WilsonSA
2010 ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes and Development 24 2043 2053
27. Mosrin-HuamanC
HonorineR
RahmouniAR
2009 Expression of bacterial Rho factor in yeast identifies new factors involved in the functional interplay between transcription and mRNP biogenesis. Molecular and Cellular Biology 29 4033 4044
28. LuoMJ
ReedR
1999 Splicing is required for rapid and efficient mRNA export in metazoans. Proceedings of the National Academy of Sciences, USA 96 14937 14942
29. LemieuxC
BachandF
2009 Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Research 37 3418 3430
30. NapetschnigJ
KassubeSA
DeblerEW
WongRW
BlobelG
2009 Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proceedings of the National Academy of Sciences, USA 106 3089 3094
31. IglesiasN
StutzF
2008 Regulation of mRNP dynamics along the export pathway. FEBS Letters 582 1987 1996
32. VinciguerraP
StutzF
2004 mRNA export: an assembly line from genes to nuclear pores. Current Opinion in Cell Biology 16 285 292
33. DongCH
HuX
TangW
ZhengX
KimYS
2006 A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Molecular Cell Biology 26 9533 9543
34. JacobY
MongkolsiriwatanaC
VeleyKM
KimSY
MichaelsSD
2007 The nuclear pore protein AtTPR is required for RNA homeostasis, flowering time, and auxin signaling. Plant Physiology 144 1383 1390
35. JauvionV
ElmayanT
VaucheretH
2010 The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22 2697 2709
36. LuQ
TangX
TianG
WangF
LiuK
2010 Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant Journal 61 259 270
37. YelinaNE
SmithLM
JonesAM
PatelK
KellyKA
2010 Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proceeding of the National Academy of Science, USA 107 13948 13953
38. GongZ
DongC-H
LeeHS
ZhuJK
XiongL
2005 A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17 256 267
39. MasudaS
DasR
ChengH
HurtE
DormanN
2005 Recruitment of the human TREX complex to mRNA during splicing. Genes and Development 19 1512 1517
40. RehwinkelJ
HeroldA
GariK
KocherT
RodeM
2004 Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nature Structural Molecular Biology 11 558 566
41. LiX
ZhangY
ClarkeJD
LiY
DongX
1999 Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329 339
42. LiY
LiS
BiD
ChengYT
LiX
2010 SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6 e1001111 doi:10.1371/journal.ppat.1001111
43. CloughSJ
BentAF
1998 Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16 735 743
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



