-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
Echinoderms, which are phylogenetically related to vertebrates and produce large numbers of transparent embryos that can be experimentally manipulated, offer many advantages for the analysis of the gene regulatory networks (GRN) regulating germ layer formation. During development of the sea urchin embryo, the ectoderm is the source of signals that pattern all three germ layers along the dorsal-ventral axis. How this signaling center controls patterning and morphogenesis of the embryo is not understood. Here, we report a large-scale analysis of the GRN deployed in response to the activity of this signaling center in the embryos of the Mediterranean sea urchin Paracentrotus lividus, in which studies with high spatial resolution are possible. By using a combination of in situ hybridization screening, overexpression of mRNA, recombinant ligand treatments, and morpholino-based loss-of-function studies, we identified a cohort of transcription factors and signaling molecules expressed in the ventral ectoderm, dorsal ectoderm, and interposed neurogenic (“ciliary band”) region in response to the known key signaling molecules Nodal and BMP2/4 and defined the epistatic relationships between the most important genes. The resultant GRN showed a number of striking features. First, Nodal was found to be essential for the expression of all ventral and dorsal marker genes, and BMP2/4 for all dorsal genes. Second, goosecoid was identified as a central player in a regulatory sub-circuit controlling mouth formation, while tbx2/3 emerged as a critical factor for differentiation of the dorsal ectoderm. Finally, and unexpectedly, a neurogenic ectoderm regulatory circuit characterized by expression of “ciliary band” genes was triggered in the absence of TGF beta signaling. We propose a novel model for ectoderm regionalization, in which neural ectoderm is the default fate in the absence of TGF beta signaling, and suggest that the stomodeal and neural subcircuits that we uncovered may represent ancient regulatory pathways controlling embryonic patterning.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001259
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001259Summary
Echinoderms, which are phylogenetically related to vertebrates and produce large numbers of transparent embryos that can be experimentally manipulated, offer many advantages for the analysis of the gene regulatory networks (GRN) regulating germ layer formation. During development of the sea urchin embryo, the ectoderm is the source of signals that pattern all three germ layers along the dorsal-ventral axis. How this signaling center controls patterning and morphogenesis of the embryo is not understood. Here, we report a large-scale analysis of the GRN deployed in response to the activity of this signaling center in the embryos of the Mediterranean sea urchin Paracentrotus lividus, in which studies with high spatial resolution are possible. By using a combination of in situ hybridization screening, overexpression of mRNA, recombinant ligand treatments, and morpholino-based loss-of-function studies, we identified a cohort of transcription factors and signaling molecules expressed in the ventral ectoderm, dorsal ectoderm, and interposed neurogenic (“ciliary band”) region in response to the known key signaling molecules Nodal and BMP2/4 and defined the epistatic relationships between the most important genes. The resultant GRN showed a number of striking features. First, Nodal was found to be essential for the expression of all ventral and dorsal marker genes, and BMP2/4 for all dorsal genes. Second, goosecoid was identified as a central player in a regulatory sub-circuit controlling mouth formation, while tbx2/3 emerged as a critical factor for differentiation of the dorsal ectoderm. Finally, and unexpectedly, a neurogenic ectoderm regulatory circuit characterized by expression of “ciliary band” genes was triggered in the absence of TGF beta signaling. We propose a novel model for ectoderm regionalization, in which neural ectoderm is the default fate in the absence of TGF beta signaling, and suggest that the stomodeal and neural subcircuits that we uncovered may represent ancient regulatory pathways controlling embryonic patterning.
Introduction
It is becoming increasingly apparent that most developmental processes are controlled by dozens or hundreds of regulatory genes assembled into complex gene regulatory networks (GRNs), rather than by a small number of master genes. By describing the functional relationships between these genes, GRNs allow integration of various levels of information on the activity of transcription factors and signaling pathways that regulate developmental processes. Over the last few years, a number of GRNs have been elucidated, including regulatory networks that drive specification of germ layers or organs in various organisms [1]–[7].
Sea urchin embryos offer many advantages for GRN analysis [8]. Unlike vertebrates, sea urchin embryos have a relatively small number of cells (about 800 cells in a gastrula) are fully transparent, and their embryos, available in huge number, develop rapidly as free-swimming larvae. A panoply of techniques is available for the functional analysis of developmental genes including treatments with pharmacological inhibitors and exogenous ligands, microinjection of antisense morpholino oligonucleotides for gene loss of function, and overexpression of mRNA for gain of function. Analysis of the first full sea urchin genome sequence from Strongylocentrotus purpuratus has revealed that echinoderms have a vast genetic repertoire but a low level of genetic redundancy, with almost all developmental regulatory genes being present as single copy [9]. Furthermore the sea urchin embryo has a rich history of experimental embryology and a wealth of biological knowledge is available on various aspects of its development. Finally, echinoderms occupy a basal position within the deuterostome lineage and are more related to chordates than most other invertebrate phyla. These various properties mean that echinoderms are a key phylum to study the evolution of developmental mechanisms and to understand the evolutionary origin of certain features of the chordate body plan. Axis specification has been extensively studied in the sea urchin [10]. Pioneer studies on endomesoderm patterning have shown that it is possible to dissect a complex GRN without the use of classical genetics by combining cis-regulatory and functional analysis, embryological, cell biological and genomic/computational approaches [11]. However, while considerable knowledge is available regarding the functional relationships between genes controlling specification of the territories along the animal vegetal axis, much less was known until recently on the genes that regulate ectoderm patterning and morphogenesis of the embryo along the dorsal-ventral axis. This gap started to be filled recently by the identification in Paracentrotus lividus of the TGFβ Nodal, Univin, and BMP2/4 as key regulators of ectoderm patterning [12]–[15]. Nodal is expressed zygotically, starting at the 32-cell stage. Its expression is initially very broad then it is rapidly restricted to a discrete sector of the ectoderm that corresponds to the presumptive ventral ectoderm. The restricted expression of nodal is so far the earliest known regional difference in zygotic gene expression detectable along the dorsal ventral axis. However, experiments performed at the beginning of the century have shown that as early as the 8-cell stage, respiratory gradients, visualized by mitochondrial cytochrome oxidase activity, prefigure the dorsal-ventral axis of the early embryo [16]. In addition, orientation of the dorsal-ventral axis can be biased by using respiratory inhibitors or by culturing embryos in hypoxic conditions [17]–[19]. Recent studies reported that mitochondria are asymmetrically distributed in some batches of eggs of Strongylocentrotus purpuratus with the ventral side displaying the highest concentration, and that microinjection of purified mitochondria can bias orientation of the dorsal-ventral axis [20], [21]. A possible link between the transcriptional activation of nodal and these redox gradients is suggested by the finding that the stress activated kinase p38 is required for nodal expression [22]. An attractive model therefore emerges in which an asymmetry in the distribution of mitochondria may generate a redox gradient, which would activate p38 anisotropically leading to the spatially restricted expression of nodal. However, strong experimental evidence supporting this model are presently lacking and experimental manipulations that perturb the redox gradient have very modest effects on the spatial expression of nodal [21] (Thierry Lepage unpublished results). If the role of redox gradients in the establishment of nodal expression is still unclear, in contrast, the role of a reaction diffusion mechanism, which involves a short range Nodal positive autoregulation and a long range inhibition mechanism by the Nodal antagonist Lefty, is probably essential to convert a subtle initial anisotropy into a sharply defined pattern [12].
Overexpression of nodal strongly ventralizes the embryos and largely mimics the effects of treatments with nickel chloride [23], knockdown of Nodal function using morpholinos or by overexpressing lefty, completely eliminates dorsal-ventral polarity and results in embryos with disorganized skeletal elements, no mouth and a straight archenteron. The same, strongly-radialized, phenotypes are obtained by blocking translation of the univin transcript which encodes a Vg1/GDF1 ortholog expressed maternally [14], suggesting that Univin may either act upstream of nodal expression or that it may heterodimerize with Nodal as suggested in vertebrates [24], [25]. Intriguingly, in the absence of Nodal, not only is the expression of ventral marker genes such as brachyury, goosecoid or lefty abolished, but the expression of dorsal marker genes such as tbx2/3 and of the novel transmembrane protein 29D is suppressed as well [13]. As a consequence, most of the ectoderm (except the ectoderm surrounding the animal and vegetal poles) of Nodal morphants differentiates into a thick ectoderm consisting of cuboidal ciliated cells that morphologically resembles the neurogenic ectoderm of the ciliary band. Injection of synthetic mRNA encoding either Nodal or an activated Nodal receptor into one blastomere of Nodal morphant embryos at the 8-cell stage is sufficient to rescue both the ventral and the dorsal side of these embryos, indicating that a distinct relay molecule specifies dorsal fates. This relay molecule was recently identified as BMP2/4, which is transcribed in the ventral ectoderm downstream of Nodal signaling, has a strong dorsalizing activity when overexpressed, and mediates the “rescue” of dorsal structures when Nodal signaling pathway is ectopically activated in a cell-autonomous manner in a Nodal loss of function background [26]. Furthermore, despite its ventral transcription, BMP2/4 has been shown to trigger receptor mediated signaling exclusively on the dorsal side of the embryo. Based on this series of findings, a basic model for sea urchin embryo dorso-ventral patterning emerges in which the dorsal ectoderm is induced by BMP2/4 signals emanating from the opposite side of the embryo. The ventral side produces inducing factors such as Nodal and BMP2/4 but it is also a source of inhibitors such as Lefty, which restricts Nodal signaling to the ventral side, and Chordin, which prevents BMP2/4 signaling in the ventral ectoderm. In the absence of lefty function, Nodal signaling is unrestricted and propagates throughout a large belt of cells surrounding the embryo while in the absence of chordin, ectopic BMP2/4 signaling occurs on the ventral side and causes abnormal patterning of the embryo [12], [26]. Therefore, in the sea urchin as in vertebrates patterning of the embryo critically relies on sequential inductive events mediated by Nodal and BMP2/4 and on the interplay between ligands and their antagonists. However, in the sea urchin embryo, both the ligands (Nodal and BMP2/4) and their antagonists (Chordin and Lefty) are co-expressed in the ventral ectoderm, which may represent a D/V organizer, and D/V patterning requires translocation of BMP2/4 from the ventral side where it is produced to the dorsal side where it activates its receptor.
Another pathway that plays a crucial role in ectoderm patterning is the Wnt pathway. Wnt signaling from the vegetal pole region is required to restrict formation of the animal pole domain. The animal pole domain is a small ectodermal territory made of thick ciliated ectoderm that forms in the apical region of the embryo. This six3 expressing neurogenic territory appears to be specified at mesenchyme blastula stage and is thought to be resistant to Wnt and TGF beta signaling [10], [13], [27], [28]. When the Wnt pathway is blocked by overexpression of cadherin or of a dominant negative form of TCF, the animal plate expands towards the vegetal pole and most of the ectoderm differentiates into neuroectodem, which contains scattered serotonergic neurons normally restricted to the animal plate region [27]. In contrast, inhibition of Nodal/Vg1/Activin signaling with a pharmacological inhibitor of the Nodal receptor causes formation of a thickened ciliated ectoderm, but this ciliated ectoderm does not appear to be specified as animal plate ectoderm since serotonergic neurons remain localized to the animal pole in these embryos. Instead, this ectoderm may have a ciliary band like identity as first proposed by Duboc et al. [13]. This idea is supported by the finding that the ectoderm of Nodal morphants abundantly expresses the ciliary band marker tubulinß3 [13] and by the presence of ectopic neurons as revealed by staining for the pan-neural marker synaptotagmin [27]. However, more in depth analysis of the specification state of this ectoderm in the absence of Nodal signaling is required to further test this idea.
Deciphering the gene regulatory network that controls patterning of the ectoderm is of special importance for several reasons. The first reason is that patterning of all three germ layers relies on the activity of a signaling center located in the ventral ectoderm and analyzing how this signaling center works is essential to understand how dorsal ventral polarity of the embryo is established. Another reason is that, despite a wealth of information available on establishment of D/V polarity during normal and regulative development, the GRN that controls specification of the main ectodermal territories (ventral ectoderm, dorsal ectoderm and ciliary band) remains incompletely described and the molecular mechanisms involved in regionalization of the embryo along the D/V axis in normal and perturbed embryos have just started to be investigated [29]. A third reason to study the D/V GRN comes from the basal evolutionary position of echinoderms within the deuterostome superclade, and of the notion that studying D/V axis formation in echinoderms will contribute to better understand the evolution of the patterning mechanisms that shaped the deuterostome body plan. Indeed, recent studies have shown that this GRN relies extensively on cell interactions mediated by TGF beta family members such as Nodal, Univin/Vg1 and BMP2/4, molecules that play crucial roles during vertebrate development [13], [14], [26]. Finally, since major morphogenetic processes such as mouth formation, skeleton formation and elongation of the arms and apex of the larva occur along the D/V axis, dissecting the D/V GRN offers the promise to study how morphogenetic processes are encoded in the genomic program of development. This will help to fill the gap that presently exists between our understanding of cell fate specification and our knowledge of how genes work together to regulate morphogenesis.
We previously described the core of the GRN that acts downstream of Nodal and is responsible for patterning of the ectoderm along the dorsal-ventral axis [13]. We showed that on the ventral side, Nodal acts at the top of this GRN by regulating the expression of lefty, bmp2/4, goosecoid and brachyury while on the dorsal side BMP2/4 activates the expression of tbx2/3. Although the functional relationships between these key genes was elucidated in this initial study, recent molecular screens conducted by us (Thierry Lepage unpublished) and others [30] revealed that many more downstream genes are likely involved in patterning of the ectoderm along the dorsal ventral axis. A large scale effort to dissect the ectoderm GRN in S. purpuratus was recently published by Su and colleagues who used the nanostring technology to monitor the effects of gene perturbations [29]. However, this technique, which measures RNA concentrations in whole embryos, lacks the spatial resolution that is required to analyze the changes in the complex spatial expression patterns of many developmental genes.
To understand better how the ectoderm of the sea urchin embryo is patterned by Nodal and BMP2/4 signals and to expand our provisional GRN, we conducted a large-scale study. Using a combination of gain of function and loss of function studies, and taking advantage of the amenability of Paracentrotus lividus embryos to detailed phenotypic analyses and in situ hybridization studies, we analyzed at high spatial resolution the expression and regulation by Nodal and BMP2/4 of 18 transcription factors and 8 signaling molecules that displayed a restricted expression along the D/V axis. Using an assay with recombinant proteins, we identified direct targets of Nodal and BMP2/4. Finally, by conducting a large-scale analysis of the epistatic relationships between these genes, we were able to start ordering them into a hierarchy and to identify key regulators acting downstream of Nodal and BMP2/4. Not only our results uncover novel and probably ancient regulatory circuits that drive morphogenetic processes such as mouth formation and neural induction, but they elicit a model for patterning of the ectoderm in which two successive inductive events regionalize the ectoderm into three territories: the ventral ectoderm that is specified by Nodal, the dorsal ectoderm that is specified by BMP2/4 and the neurogenic ectoderm of the ciliary band, which forms between the ventral and the dorsal ectoderm in a region protected from Nodal and BMP signaling. In addition, these findings highlight a striking parallel between the mouse embryo and the sea urchin embryo by showing that in both models a neurogenic ectoderm is the default state of ectoderm differentiation in the absence of Nodal and BMP signaling. Our analysis provides a picture of this GRN significantly different from that proposed by Su et al. in S.purpuratus and stresses the importance of the spatial resolution level in the analysis of gene regulatory networks in early embryos.
Results
Novel markers of regional differences in gene expression within the ectoderm
To elucidate the gene regulatory network that controls specification and patterning of the ectoderm in Paracentrotus, we first performed large scale in situ hybridization screens. In addition to a random screen initiated several years ago, which allowed us to characterize the expression of 4000 randomly selected cDNAs (Thierry Lepage unpublished), we screened a P. lividus EST database against S. purpuratus sequences encoding transcription factors and signaling molecules and analyzed the expression of all those that were expressed during development of the sea urchin embryo [30]–[34] (Table 1). This allowed us to assemble a list of 36 genes displaying a robust expression in either the ventral ectoderm, the dorsal ectoderm or in the ciliary band territory (Table 1) (Figure 1A, 1B). Genes expressed in the animal pole domain were largely excluded from this analysis since most of them do not display a restricted expression along the D/V axis. The expression patterns of a number of the genes presented in this study had previously been described at various degrees in S. purpuratus [30]–[34] but they had never been described in Paracentrotus. In addition, the expression of several genes analyzed here, including smad6, gfi1, id, admp2, BMP1, and oasis has not been described previously in either species.
Fig. 1. Gene expression profiles of transcription factors and signaling molecules analyzed in this study. 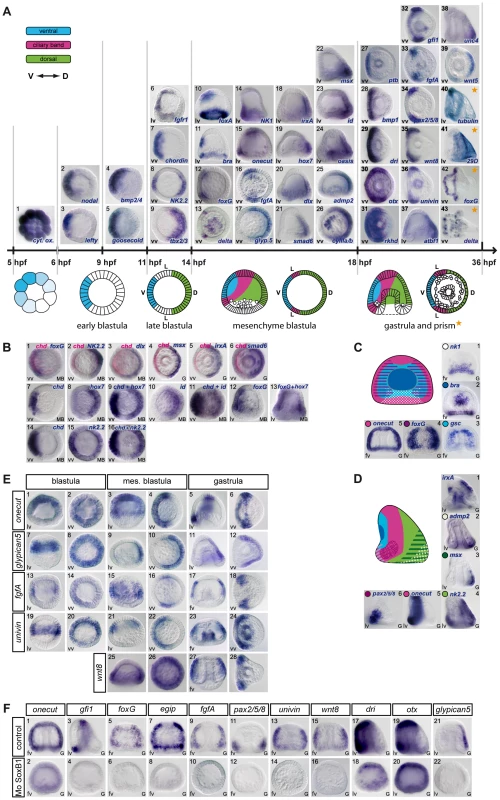
(A) Spatial and temporal expression profiles. The expression of 21 genes encoding transcription factors (goosecoid, nk2.2, tbx2.3, nk1, foxA, brachyury, foxG, onecut/hnf6, irxA, hox7, dlx, smad6, msx, id, oasis, deadringer, otx, gfi, pax2/5/8, atbf1, unc4), 13 signaling molecules (nodal, lefty, bmp2/4, chordin, fgfA, fgfr1, glypican5, admp2, bmp1, wnt8, univin, wnt5, Delta), 2 RNA binding proteins (rkhd, ptb), 3 differentiation genes (cyIIIa, 29D, tubulinß3) and a mitochondrial gene (cytochrome oxidase) is depicted above a scheme of early development of the sea urchin embryo. The genes are classified into 5 groups according to the timing of their expression. (1) Maternal cytochrome oxidase transcripts show a graded distribution in cleaving embryos. (2,3) Starting at the early blastula stage, nodal and lefty are the first zygotic genes to be expressed in a restricted pattern along the D/V axis, followed by bmp2/4 and goosecoid before hatching (4,5). (6–9) After hatching, expression of fgfr1 and chordin is initiated ventrally while nk2.2 and tbx2/3 start to be expressed dorsally. (10–26) At mesenchyme blastula stage foxA, brachyury, foxG, Delta, nk1, onecut, fgfA, glypican5, irxA, hox7, dlx, smad6, msx, id, oasis, admp2, and CyIII start to be expressed in a restricted pattern. (27–36) A restricted expression is established slightly later at the early gastrula stage for ptb, bmp1, dri, otx, rkhd, gfi1, pax2/5/8, wnt8, univin, and for atbf1, unc4 and wnt5 (37–39). (40–43) tubulinß3 and 29D are expressed in the presumptive ciliary band or dorsal ectoderm starting at prism/early pluteus stages while foxG is expressed in a ventral subdomain of the ciliary band and Delta in individual cells within the ciliary band and facial ectoderm. (B) (1) Two color double in situ hybridization showing that foxG is expressed ventrally. (2,15,16) nk2.2 is expressed in two discrete regions along the D/V axis. At the late blastula stage, the territory with the strongest expression is located on the ventral side while at mesenchyme blastula stage, the highest level of transcripts is detected on the dorsal side. (3–6) Two color double in situ hybridization with chordin shows that dlx, msx, irxA and smad6 are expressed on the dorsal side. (7–13) One color double in situs hybridizations confirm that id and hox7 are expressed on the dorsal side. (C) At late gastrula stage, differences in expression of marker genes along the A/V and D/V axes identify differently specified territories within the ventral ectoderm. (1)The NK1 homeobox gene is expressed in a trapezoidal domain that abuts the stomodeal domain and the lateral ectodermal regions that express fgfA and pax2/5/8. (2) brachyury is expressed in a group of 30 cells located in the center of the ventral ectoderm and that likely constitute the stomodeal precursors. (3–5) This group of cells is surrounded by a large belt of goosecoid and foxG expressing cells that are themselves surrounded by a thinner belt of onecut/hnf6 expressing ciliary band precursors. (D) Nested expression domains are also apparent within the dorsal ectoderm and ciliary band. (1) irxA is expressed in a medial sub domain of the dorsal ectoderm that abuts the ciliary band. Note that irxA is also expressed in the stomodeal region at this stage. (2,3) Genes like admp2 and msx are expressed in nested patterns in the dorsal most region that corresponds to the presumptive apex of the larva. (4) nk2.2, like tbx2/3, is expressed in most cells of the dorsal ectoderm. (5,6) Onecut is expressed in the whole ciliary band, while pax2/5/8 is expressed in the vegetal portion of the presumptive ciliary band. (E) A set of ectodermal genes including onecut/hnf6, glypican5, fgfA, univin and wnt8 are expressed broadly in the ectoderm at blastula stages and subsequently restricted to either the dorsal ectoderm (glypican5) or the ciliary band (onecut/hnf6, fgfA, univin, wnt8). (F) Expression of several ciliary band genes including onecut, gfi1, foxG, egip, fgfA, pax2/5/8, univin, wnt8, dri, otx and of the dorsal marker glypican5 critically relies on the activity of the transcription factor SoxB1. V, ventral, D, dorsal, L, lateral. lv, lateral view, vv, vegetal pole view, fv, frontal view. Tab. 1. Genes examined in this study. 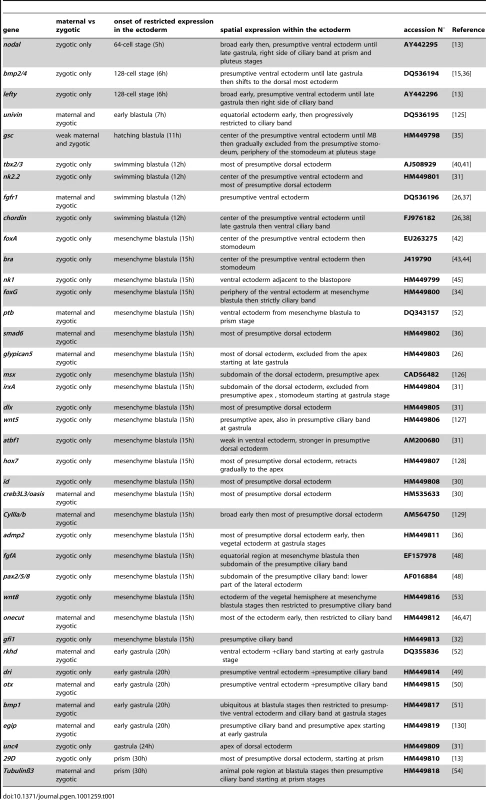
The earliest asymmetrically distributed transcript that we identified in the in situ screens is the maternal transcript encoding mitochondrial cytochrome oxidase, with cleavage stage embryos frequently displaying a graded distribution of transcripts in the presumptive ectoderm (Figure 1A1). This asymmetrical distribution of a mitochondrial transcript likely reflects the asymmetrical distribution of mitochondria previously reported by Coffman and colleagues [20], [21]. At the zygotic level, the first signs of tissue regionalization within the ectoderm are seen at 64/128 cell-stage with nodal and lefty transcripts starting to accumulate in the presumptive ventral territory (Figure 1A2,3) [12], [13]. The second wave of zygotic genes displaying a restricted expression along the D/V axis starts at the prehatching blastula with bmp2/4 and goosecoid starting to be transcribed in the ventral ectoderm rapidly followed by fgfr1, chordin and nk2.2 at the swimming blastula stage (Figure 1A4–8) [15], . In Paracentrotus, there is no known example of genes displaying a restricted expression in the dorsal ectoderm before the swimming blastula stage. The first genes to be expressed in the dorsal ectoderm are nk2.2 and tbx2/3, whose expression increases abruptly in the presumptive dorsal territory after hatching (Figure 1A8,9) [31], [40], [41]. These genes are therefore good candidates as immediate early targets of Nodal or BMP2/4 signaling and are likely to play an early role in specification of these territories. Soon after ingression of the primary mesenchyme cells, when the embryo acquires its bilateral symmetry, a third wave of zygotic genes starts to be expressed. This includes the largest number of genes such as foxA, brachyury, foxG, Delta, NK1, in the ventral ectoderm (Figure 1A10–14) [34], [42]–[45], onecut/hnf6 and fgfA (Figure 1A15–16) [46]–[48] in the lateral ectoderm and glypican5, irxA, hox7, dlx, smad6, msx, id, oasis, admp2 and cyIII in the dorsal ectoderm (Figure 1A17–26) [26], [30], [31], [36], [39]. Based on the timing of their expression, genes in this category are likely secondary targets of Nodal or BMP2/4 signaling.
Starting at the early gastrula stage, additional genes start to be expressed with a restricted pattern along the D/V axis, with ptb transcripts accumulating in the ventral ectoderm (Figure 1A27), bmp1, deadringer (dri), otx, and rkhd being expressed in a broad domain encompassing the ventral ectoderm and ciliary band territory (Figure 1A28–31) [13], [49]–[52], and gfi1, pax2/5/8, wnt8, univin transcripts starting to be expressed in the presumptive ciliary band (Figure 1A32–36) [32], [36], [48], [53]. Similarly, atbf1, unc4, wnt5 start to be expressed in the dorsal ectoderm at the early gastrula stage. (Figure 1A37–39). Finally, at prism stage, tubulinß3 transcripts accumulate in the presumptive ciliary band while transcripts encoding the sea urchin specific transmembrane protein 29D accumulate in the presumptive dorsal ectoderm (Figure 1A 40,41) [13], [54].
At the mesenchyme blastula stage, foxG (also known as Brain factor1 or Bf1) is expressed in two broad ventro-lateral stripes that largely overlap with the goosecoid expression territory (Figure 1A12), while Delta is first expressed in the ectoderm in a cluster of cells at the animal pole as well as in individual cells, possibly neurons, first on the ventral side then on the dorsal side, within the vegetal part of the foxG expression domain. At the prism/early pluteus stage, the pattern of foxG resolves into a thin belt of cells on the ventral side of the presumptive ciliary band (Figure 1A42) [34] while Delta expression now occurs in a salt and pepper pattern within the ciliary band and facial ectoderm (Figure 1A43) [55].
For simplification we divide the ectoderm into three main territories along the dorsal-ventral axis, however there are additional regional differences in gene expression that show that more than three regions can be defined (Figure 1C). For example, the homeobox gene nk1 is expressed in the ventral-vegetal ectoderm in a region fated to become the ventral supra-anal ectoderm (Figure 1C1). Similarly, several dorsally expressed genes such as msx, id, oasis, admp2 or unc4 are strongly expressed in the dorsal-vegetal region fated to become the dorsal supra-anal ectoderm (Figure 1A22–25; 38; Figure 1D2,3). Thus, the ectoderm near the vegetal pole is divided into at least two sub domains along the D/V axis. Gene expression patterns also revealed that the ventral and dorsal ectodermal regions are progressively regionalized into different domains. This is best illustrated by the dynamics of goosecoid expression. goosecoid and brachyury are initially co-expressed within the ventral ectoderm (Figure 1A5,11), but during gastrulation, the expression domain of goosecoid is progressively cleared from the center of the ventral ectoderm (Figure 1C3). While goosecoid expression is progressively shifted at the periphery of the ventral ectoderm, forming a belt of cells abutting the ciliary band, brachyury and foxA remain expressed at the center of the ventral ectoderm, where the stomodeum will form (Figure 1C2). Similarly, analysis of gene expression within the dorsal ectoderm revealed the existence of nested patterns, with genes like nk2.2, tbx2/3 and dlx (Figure 1D4, Figure 1A,9,20) being expressed in a broader domain than genes like msx, wnt5 or smad6 (Figure.1A22,29,39; 1D3,4 see also [26]) and genes like irxA being expressed in a sub domain of the dorsal ectoderm that excludes the dorsal apex (Figure 1D1). Finally, sub regions can also be recognized within the ciliary band territory starting at the early gastrula stage, with genes like fgfA, vegf, pax2/5/8 and sprouty being expressed in the ventral lateral region (Figure 1A33,34; Figure 1D6 and data not shown) [48], [56], genes like onecut/hnf6 or gfi1 being expressed in the entire presumptive ciliary band territory (Figure 1C5; Figure 1D5), and genes like foxG, which in vertebrates is expressed in and required for specification of the ventral telencephalon [57], [58], being expressed in a ventral subdomain of the ciliary band (Figure 1A42).
Interestingly, several genes whose expression is later confined to the ciliary band are initially expressed much more broadly in the ectoderm (Figure 1E). This is particularly apparent for glypican5, fgfA, univin, and wnt8, which are expressed in a large belt of ectodermal cells at blastula stage and also for the neural marker onecut/hnf6 which is first expressed ubiquitously, then in a broad ventro-lateral domain, and only later in the ciliary band (Figure 1E1–6) [26], [46]–[48]. This suggests that the expression of these ciliary band marker genes is initiated by broadly distributed transcription factors and later repressed on the ventral and/or dorsal sides by additional factors. As a first step to dissect the ectoderm gene regulatory network, we analyzed the regulation of these broadly expressed ciliary band genes. Since SoxB1 plays a critical role in ectoderm patterning in the sea urchin [59] and in the specification and maintenance of neural regions in vertebrates [60], we tested if SoxB1 is required for expression of ciliary band marker genes (Figure 1F). Injection of morpholinos against SoxB1 abrogated the expression of most markers of the neurogenic ectoderm of the ciliary band including onecut, gfi1, foxG, egip, fgfA, pax2/5/8, univin, wnt8 and strongly affected the spatial expression of dri and otx [14] (Figure 1F1–20). This result supports the idea that transcription of at least a subset of ciliary band marker genes is initiated by broadly distributed transcription factors such as SoxB1 and later restricted to the ciliary band by zygotic factors induced by Nodal and/or BMP signaling.
Nodal and BMP2/4 promote ventral and dorsal ectodermal fates and repress ciliary band gene expression
We next tested how Nodal and/or BMP2/4 regulate the expression of the 36 genes identified in the in situ screen. We focused on Nodal and BMP2/4 since previous studies showed that these two ligands are essential for specification and patterning of the ventral and dorsal territories. We first analyzed the effects of overexpressing nodal or bmp2/4 on the expression of ectodermal markers. Embryos were injected with nodal or bmp2/4 mRNA and the expression of the ventral, dorsal, or ciliary band markers was monitored at different stages. In most cases, results were confirmed by treatments with recombinant mouse Nodal or BMP4.
Overexpression of nodal mRNA or treatments with recombinant Nodal protein dramatically expanded the expression of nodal, bmp2/4, chordin, lefty, goosecoid and brachyury as reported previously (Figure 2) [13], [26]. Overexpression of Nodal also expanded the ectodermal domain of expression of foxA and fgfr1 at mesenchyme blastula stages. Similarly, the expression domain of nk1, which is normally restricted to the ventral vegetal ectoderm, became radial in nodal overexpressing embryos. Genes expressed in the ciliary band behaved differently depending on the gene. In the case of deadringer, bmp1 and univin, which are expressed in the ciliary band and in the ventral ectoderm, overexpression of nodal expanded their expression to the whole ectoderm. In the case of wnt8, which is expressed in two broad lateral stripes at gastrula stages, as well as in the case of fgfA and its downstream target pax2/5/8, which are expressed in the ventral sub domain of the ciliary band, all expression was eliminated by exogenous nodal. However, in the case of foxG, egip, onecut/hnf6, gfi1, otx, exogenous nodal suppressed expression in most of the ectoderm except in the animal and/or vegetal most domains of the ectoderm. Overexpression of nodal increased the number of ventral-vegetal cells that normally express Delta at the early gastrula stage and, at 48h, produced ventralized embryos in which most Delta expressing cells were located at the animal pole and in the vegetal most ectoderm. Largely similar phenotypes were obtained following treatments with nickel chloride (Figure S3) although we noted intriguing differences in the behavior of a few genes including wnt8, univin, fgfA and pax2/5/8, in response to these perturbations. Overall, these data are consistent with the idea that in nodal-overexpressing or nickel treated embryos, radially expressed Nodal promotes specification of ventral ectodermal fates and suppresses specification of the ciliary band in a large equatorial region but not in the animal pole region or in the ectoderm surrounding the blastopore. One likely reason that may explain why the vegetal ectoderm is refractory to Nodal overexpression or to nickel treatment is that in these embryos, Nodal signaling is restricted to the equatorial region [13]. The vegetal ectoderm may therefore be protected from Nodal activity by Lefty which is thought to diffuse farther than Nodal [12], . Consistent with this idea, in Nodal treated embryos and in nickel treated embryos, nodal expression expands to a large belt of cells in the equator and a ciliary band differentiates in the vegetal most ectoderm while in lefty morphants, which also display unrestricted Nodal signaling, ciliary band marker genes such as tubulinß3 and onecut/hnf6 are expressed in the animal pole region but not in the vegetal ectoderm (Figure 2) [12]. Taken together, these results suggest that a Lefty dependent inhibition of Nodal signaling is required for ciliary band formation in the vegetal pole region. Finally, as expected, overexpression of nodal eliminated the expression of all the dorsal marker genes we tested including, nk2.2, tbx2/3, smad6, msx, atbf1, wnt5, admp2, unc4, hox7, dlx, and 29D (Figure 2).
Fig. 2. Overexpression of nodal represses the expression of ciliary band and dorsal marker genes and expands the expression of ventral markers genes. 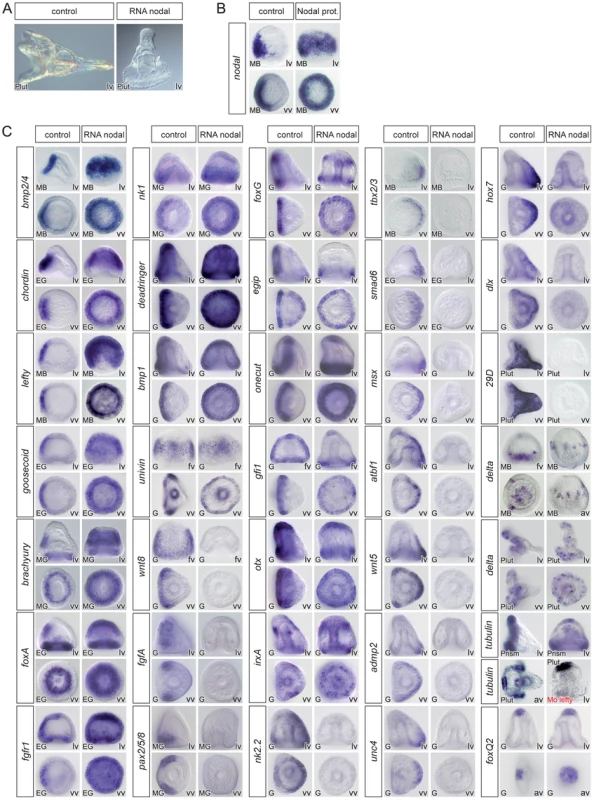
(A) morphology of nodal overexpressing embryos at 72h. (B) treatment with recombinant Nodal protein induces ectopic expression of nodal in a large belt of ectodermal cells. (C) Injection of nodal mRNA (200–400 µg/ml) caused the expression of most ventral marker genes to become radial. In most cases, however, the animal pole domain appeared to be resistant to ectopic expression of nodal. Overexpression of nodal also radialized the expression of dri, bmp1 and univin, which are expressed in a broad domain encompassing the presumptive ventral ectoderm and ciliary band. In contrast, Nodal strongly antagonized the expression of ciliary band marker genes such as wnt8, fgfA, pax2/5/8, foxG, egip, onecut, gfi1, otx, tubulinß3 or Delta in the equatorial region. Nodal overexpression efficiently repressed the expression of all the genes expressed in the dorsal ectoderm but did not affect the expression of marker genes of the animal pole (tubulinß3, foxQ2, Delta). Note that, starting at gastrula stage, irxA is expressed in a patch of animal-ventral cells that likely corresponds to the upper part of the presumptive stomodeum. Therefore in nodal overexpressing embryos, this territory becomes radial and forms a belt of cells near the animal pole. Also note that while a ciliary band does form in the vegetal region of nodal overexpressing embryos, as shown by the expression of ciliary band genes around the blastopore, no ciliary band forms in the vegetal region of lefty morphants as indicated by the absence of tubulinß3 expression. In contrast, nodal overexpression does not affect the expression of animal pole marker genes such as foxQ2. lv, lateral view, vv, vegetal pole view, av, animal pole view, fv, frontal view. Reciprocally, overexpression of bmp2/4 or treatments with recombinant BMP4 protein eliminated expression of all the ventral marker genes we tested including nodal, bmp2/4, chordin, goosecoid, foxA, lefty (not shown), brachyury, and nk1 (Figure 3). As in the case of nodal overexpression, misexpression of bmp2/4 or of the activated Alk3/6 BMP receptor (Alk3/6QD) [26] strongly suppressed the expression of the ciliary band markers such as bmp1, foxG, onecut/hnf6, otx, gfi1, tubulinß3, egip, dri, univin, wnt8, fgfA and pax2/5/8. However, unlike in the case of nodal overexpressing or nickel treated embryos, which conserved expression of ciliary band markers in the animal pole and in vegetal ectodermal regions, overexpression of bmp2/4 or of the activated type I BMP receptor (Alk3/6QD) efficiently eliminated the expression of all the ciliary band markers at the animal pole and in the vegetal most ectoderm as well as the expression of animal pole specific markers such as foxQ2 highlighting the very strong antagonism existing between high level of BMP2/4 signaling and specification of the animal pole and ciliary band cell fates. Finally, misexpression of BMP2/4 dramatically expanded the expression of all the dorsal marker genes including tbx2/3, smad6, nk2.2, wnt5, oasis, msx, irxA, dlx, atbf1, hox7, unc4, admp2, id and 29D.
Fig. 3. Overactivation of BMP signaling eliminates the expression of ventral and ciliary band marker genes and expands the dorsal territory. 
(A) Morphology of BMP2/4 overexpressing or BMP4 treated embryos at 72h. Embryos with overactivated BMP signaling are elongated and covered with a thin and squamous ectoderm and possess ectopic spicules. (B) Expression of ventral, dorsal and ciliary band marker genes in embryos misexpressing BMP2/4. The results presented were obtained using treatments with recombinant BMP4 protein except one experiment in which an activated BMP receptor (Alk3/6QD) [26] was used. For all these genes, identical results were obtained by overexpression of BMP2/4 mRNA. Overactivation of BMP2/4 signaling eliminated the expression of ventral markers and of ciliary band genes such as bmp1, foxG, onecut/hnf6, otx, gfi1, tubulinß3, egip, deadringer, univin, fgfA, and pax2/5/8. Overactivation of BMP signaling dramatically expanded the expression of all the dorsal marker genes and eliminated the expression of markers of the animal pole domain such as foxQ2. lv, lateral view, vv, vegetal pole view, av, animal pole view, fv, frontal view. Discrimination between direct versus indirect targets of Nodal and BMP2/4
We next sought to determine which genes are direct targets of Nodal and BMP2/4 signaling. Based on the timing of expression of the ventral or dorsal markers genes, it was expected that only a subset would be direct targets of Nodal or BMP2/4 signaling. For example, only lefty, bmp2/4, chordin, goosecoid, nk2.2, fgfr1 and tbx2/3 are expressed at swimming blastula stage, the expression of most of the other starting only at mesenchyme blastula stage. We therefore tested whether the ventral marker genes are transcribed in direct response to Nodal and whether the dorsal marker genes are transcribed in direct response to BMP2/4 signaling or if transcription of these genes requires protein synthesis. To achieve this, we treated embryos at the hatching blastula, mesenchyme blastula or gastrula stages with recombinant mouse Nodal or BMP2/4 proteins in the presence or absence of a protein synthesis inhibitor (Figure 4), and analyzed the expression of all the ventral and all the dorsal marker genes.
Fig. 4. nodal, lefty, bmp2/4, chordin, goosecoid, nk2.2, and fgfr1 are direct targets of Nodal while tbx2/3, nk2.2, and smad6 are direct targets of BMP2/4 signaling. 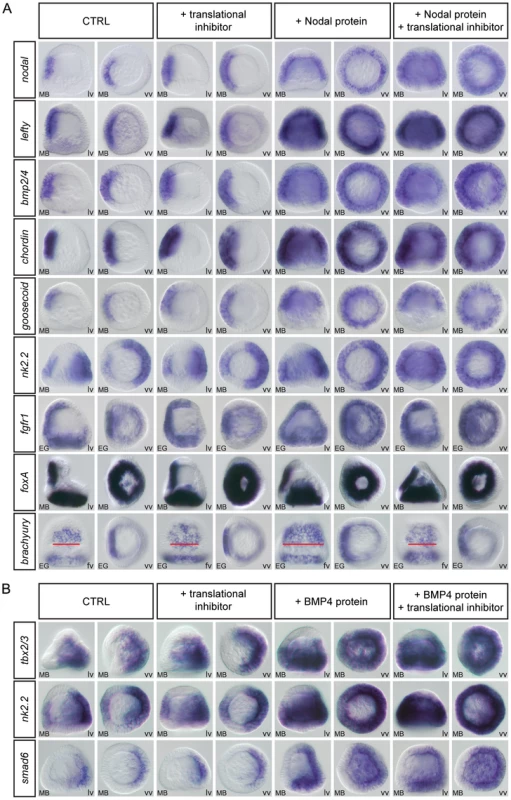
(A,B) Embryos at the late blastula stage were treated for two hours with recombinant Nodal or BMP4 protein in the presence or absence of puromycin. At the end of the treatment the embryos were fixed and the expression of the indicated genes was analyzed by in situ hybridization. In control experiments, DMSO or the translational inhibitor were added alone. (A) A short treatment with recombinant Nodal protein induced strong ectopic expression of early expressed genes such as nodal, lefty, bmp2/4, chordin, goosecoid, nk2.2 and fgfr1. This ectopic expression was still observed in the presence of protein synthesis inhibitor indicating that these genes are early targets of Nodal signaling. In contrast, this 2h treatment with Nodal did not induce ectopic expression of genes expressed later such as foxA and only induced a partial expansion of brachyury expression, an effect that disappeared in the presence of the protein synthesis inhibitor. These genes are therefore likely secondary targets of Nodal signaling. Note that in control embryos the width of the brachyury expression territory encompasses 8–9 cells (red bar) while in Nodal treated embryos the width of this stomodeal field increases to about 12–15 cells. In embryos treated with Nodal and the translation inhibitor, the width of the stomodeal field is similar to that in control embryos. (B) Similarly, BMP4 treatment induced massive ectopic expression of the early expressed genes tbx2/3, nk2.2 and smad6 even in the presence of a translational inhibitor but failed to induce ectopic expression of dorsal markers genes expressed later. tbx2/3, nk2.2 and smad6 are therefore likely direct targets of BMP2/4 signaling while the other dorsally expressed genes are likely indirect targets whose expression requires protein synthesis downstream of activation of the BMP receptors. Identical results were obtained by using emetine as translational inhibitor. lv, lateral view, vv, vegetal pole view fv, frontal view. Short treatments with recombinant Nodal protein at blastula stage strongly induced expression of nodal, lefty, bmp2/4, chordin, goosecoid, nk2.2 and fgfr1 throughout most of the ectoderm (Figure 4A). These effects were observed even in the presence of a translational inhibitor suggesting that these genes are direct targets of Nodal signaling. In contrast, short treatments with Nodal at either mesenchyme blastula or gastrula stages failed to induce any ectopic expression of the other ventral genes such as foxA (Figure 4A) foxG, nk1, or deadringer (data not shown), which are expressed in the ectoderm starting at or after mesenchyme blastula. This suggests that these genes are indirect targets of Nodal signaling that cannot be induced during the short interval of the treatment. Interestingly, in the case of brachyury, a weak but consistent broadening of the ectodermal domain of expression was observed following treatment with Nodal. However, this effect was abolished by treatment with the protein synthesis inhibitor, consistent with this gene being an indirect target of Nodal signaling. Similarly, among all the dorsal marker genes we tested, 3 genes were strongly induced by treatments with BMP2/4, even in the presence of protein synthesis inhibitors. These were tbx2/3, nk2.2 and smad6 (Figure 4B). Short treatments with high doses of BMP2/4 failed to induce expression of irxA, dlx, msx, atbf1, hox7, id, unc4, oasis, wnt5, admp2 or glypican 5 (data not shown) suggesting that these genes may be indirect targets of BMP signaling. The very good correlation between the results of this induction assay and the timing of expression of the downstream targets of Nodal and BMP2/4 indicates that this assay predicts with good confidence the direct, and probably also the indirect, target genes of these ligands at swimming blastula stage. It should be kept in mind however, that at later stages, this assay does not allow to rule-out completely the existence of a direct input from Nodal or BMP2/4 to downstream target genes. An alternative explanation for the fact that several genes appear to be refractory to induction by recombinant Nodal or BMP4 proteins is that after swimming blastula stage, the ventral and dorsal ectoderm may no longer be competent to switch their gene regulatory networks to a state that supports expression of dorsal or ventral genes respectively.
Nodal and BMP2/4 dependence of ectodermal gene expression
We next attempted to determine if the activity of Nodal and BMP2/4 accounts for the restricted expression of all of the ventral and all the dorsal genes. Embryos were injected with a nodal morpholino and the expression of ventral, dorsal or ciliary band markers analyzed at successive stages (Figure 5). Expression of all the ventral marker genes that we tested including, bmp2/4, goosecoid, fgfr1, nk1, chordin, brachyury, foxA and lefty disappeared in the Nodal morphants, consistent with previous results (Figure 5B) [13], [29], [38]. Injection of the nodal morpholino also largely prevented expression of foxG, confirming that this gene is induced downstream of Nodal signaling [29]. We also found that in Nodal morphants, the expression of all dorsal markers genes was strongly downregulated in most of the ectoderm, with responses falling into two categories: for some genes, e.g. glypican5, oasis, msx, dlx, hox7, wnt5, smad6, or unc4, expression completely disappeared in the Nodal morphants (Figure 5C). Others, e.g. tbx2/3, id, irxA, nk2.2, atbf1, admp2 and 29D displayed residual expression in the vegetal-most ectoderm and/or in the PMCs indicating Nodal-independent expression of these genes in the presumptive dorsal vegetal ectoderm.
Fig. 5. Blocking Nodal function prevents expression of ventral and dorsal marker genes in the presumptive ectoderm and causes massive ectopic expression of ciliary band genes. 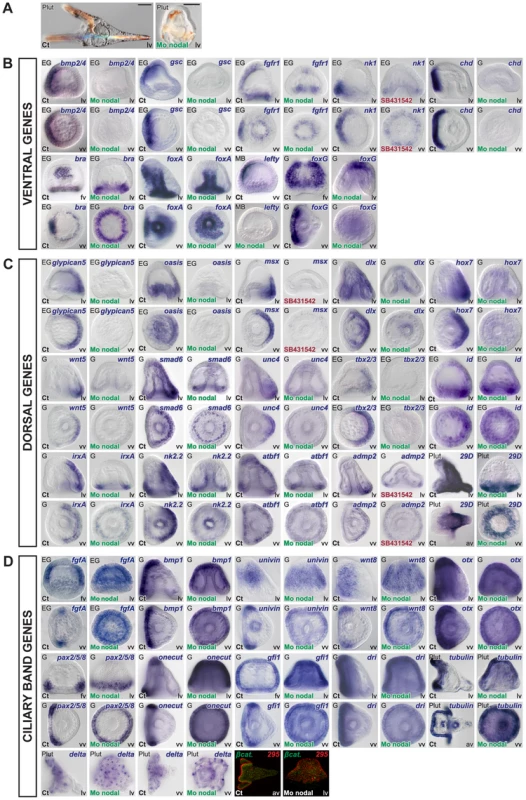
(A) morphology of nodal morphants at 72h. Note that most of the ectoderm of these embryos differentiates into a thick ciliated ectoderm that resembles the ciliary band ectoderm. (B–D) Embryos were injected with a Nodal morpholino or treated with the Nodal receptor inhibitor SB431542 and the expression of ventral, dorsal or ciliary band genes was analyzed at the relevant stages. (B,C) All the ventral and all the dorsal marker genes tested required Nodal to be expressed in the presumptive ventral and presumptive dorsal ectoderm respectively. However, in the nodal morphants, a number of genes expressed dorsally continued to be expressed in the ectoderm derived from the vegetal region and/or in the mesendoderm including tbx2/3, id, irxA, nk2.2, atbf1, admp2 and 29D (the residual expression of tbx2/3 is not visible here since it is mostly visible at gastrula stages). This indicates that in the nodal morphants there is a residual D/V polarity with the vegetal most ectoderm adopting a dorsal identity. (D) Inhibition of Nodal signaling caused a massive ectopic expression of ciliary band genes throughout the ectoderm. Note that genes expressed throughout the ciliary band territory such as bmp1, otx, onecut, gfi1, dri, or tubulinß3 are ectopically expressed throughout the ectoderm of Nodal morphants. Genes that are expressed in sub domains of the ciliary band such as pax2/5/8, fgfA, univin or wnt8 are also ectopically expressed and display a radial expression but in accordance with their normal animal-vegetal boundaries. The neural marker Delta and the ciliary band antigen 295, which in control embryos labels the ciliated cuboidal cells of the ciliary band, are also expressed ectopically throughout the thick ciliated ectoderm typical of nodal morphants. lv, lateral view, vv, vegetal pole view, fv, frontal view. Scale bar: 100µm. A striking result was obtained when we analyzed the expression of ciliary band markers in the nodal morphants (Figure 5D). The expression of most ciliary band markers dramatically expanded to most of the ectoderm following inhibition of Nodal signaling. This was the case for fgfA, bmp1, univin, wnt8, otx, pax2/5/8, onecut/hnf6, gfi1, dri, as well as of the late ciliary band marker tubulinß3 and the ciliary band antigen 295. Importantly, expression of Delta, which at pluteus stages identifies individual neurons of the facial ectoderm and ciliary band region [26], [55], was expanded to the whole ectoderm in Nodal morphants, strongly suggesting that most of the ectoderm is converted into neurogenic ectoderm in these embryos. Largely similar results were obtained using a pharmacological inhibitor of the Nodal receptor [62] (Figure S4). Taken together, these results show that Nodal signaling is essential for expression of all the ventral and of all the dorsal marker genes within the ectoderm. In the absence of Nodal, expression of all the ventral and dorsal marker genes is abolished and ciliary band genes are ectopically expressed throughout most of the ectoderm.
We also examined the effect of knocking down BMP signaling on the expression of the ventral, dorsal and ciliary band markers (Figure 6). As expected, we found that expression of all the ventral markers that we tested was independent of BMP2/4 signaling: nodal, bmp2/4, chordin, brachyury or foxA were expressed at similar levels and in similar domains in the controls and in the alk3/6 morphants (Figure 6B). Removing BMP2/4 or Alk3/6 function affected the expression of dorsal marker genes in a way very similar to that caused by removing Nodal: expression of most genes including wnt5, atbf1, hox7, msx, dlx, smad6, tbx2/3, unc4 was abolished while for irxA, nk2.2 and id, residual expression was still observed in the vegetal most ectoderm on the presumptive dorsal side (Figure 6C). These results confirm that expression of all the dorsal ectodermal genes stringently relies on BMP2/4 signaling and that in the absence of Nodal or BMP2/4 signals, no other signals compensate for the lack of these inducers. Again, a striking result was observed when we analyzed the expression of ciliary band markers in the bmp2/4 or Alk3/6 morphants. For all of them, including gfi1, onecut/hnf6, otx, deadringer, pax2/5/8, foxG, wnt8, fgfA, univin, bmp1 and tubulinß3, loss of BMP2/4 signaling caused a dramatic ectopic expression in the dorsal ectoderm (Figure 6D). This ectopic expression transformed the normally bilateral expression domains of fgfA, pax2/5/8, foxG, gfi1, univin, and wnt8 into a horseshoe shaped domain covering the lateral and dorsal regions and caused the expression domain of deadringer and otx to become radial. These results reveal that in addition to promoting specification of dorsal cell fates, an essential function of BMP2/4 signaling is to repress ciliary band gene expression within the dorsal ectoderm.
Fig. 6. Blocking BMP2/4 or Alk3/6 signaling strongly downregulates the expression of dorsal genes and causes massive ectopic expression of ciliary band marker genes on the dorsal side. 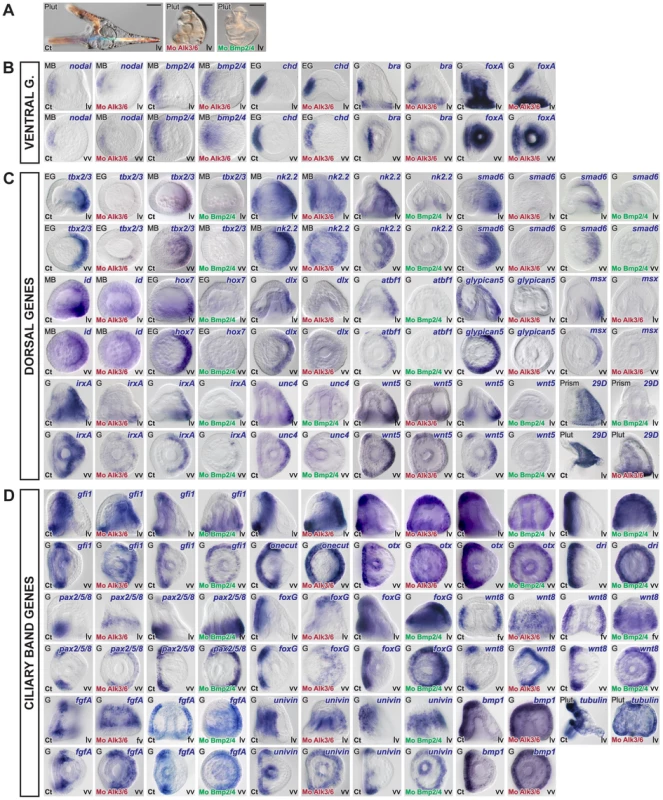
(A) Morphology of alk3/6 and BMP2/4 morphants at 72h. Ventral structures such as the stomodeum do form in embryos injected with the alk3/6 or BMP2/4 morpholinos but dorsal structures such as the apex fail to differentiate. (B–D) Embryos were injected with BMP2/4 or Alk3/6 morpholinos and the expression of ventral, dorsal or ciliary band marker genes was analyzed at the relevant stages. (B) Inhibition of BMP2/4 signaling does not interfere with the expression of ventral genes. (C) The expression of all the dorsal marker genes is strongly reduced or abolished following inhibition of BMP2/4 or Alk3/6 function. A residual expression in the vegetal most region is still observed for certain genes such as irxA, wnt5, id. (D) Inhibition of Alk3/6 or BMP2/4 causes a striking ectopic expression of ciliary band genes throughout the dorsal ectoderm. lv, lateral view, vv, vegetal pole view, av, animal pole view, fv, frontal view. Scale bar: 100µm. Construction of a provisional gene regulatory network
To establish the functional hierarchy between key ventral, dorsal and ciliary band genes, we designed morpholinos against 17 transcription factors and 8 signaling molecules expressed within the ectoderm with a restricted pattern along the dorsal-ventral axis. Among these 25 morpholinos, 19 (alk4/5/7, alk3/6, brachyury, bmp2/4, chordin, foxA, foxG, fgfA, goosecoid, irxA, lefty, tbx2/3, dlx, msx, nodal, onecut/hnf6, soxB1, univin, wnt8) gave a clearly recognizable morphological phenotype (Figure 5–9). The expression of 15 transcription factors (goosecoid, brachyury, foxA, nk1, nk2.2, tbx2/3, msx, smad6, hox7, irxA, onecut, gfi1, dri, pax2/5/8, foxG) and 8 signaling factors (nodal, bmp2/4, fgfA, chordin, wnt8, univin, wnt5, glypican5) was analyzed at different stages in the 17 morphant backgrounds while in the case of nodal and bmp2/4 morphants we analyzed the expression of an additional set of 17 marker genes (Tables S1, S2). In addition, we overexpressed a subset of genes encoding transcription factors (goosecoid, foxA, foxG, deadringer, nk2.2, tbx2/3, msx, smad6) and signaling molecules (nodal, bmp2/4, chordin) and analyzed the expression of ventral, dorsal and ciliary band genes in these embryos. Since many of the genes identified in our screens including brachyury, foxA, otx, smad6, tbx2/3, wnt5, oasis, univin, wnt8, rkhd, ptb, fgfA, Delta are expressed not only in the ectoderm but also in the mesendoderm and since many other markers such as atbf1, irxA, nk2.2 or egip, oasis, wnt5, glypican5, wnt8, Delta, otx or bmp1 are expressed in more than one region and sometimes in both the ventral and dorsal ectoderm, we used in situ hybridization rather than QPCR to monitor the consequences of the perturbations. In situ hybridization is usually not used as the primary technique in large-scale projects such as gene regulatory network analysis since it is time and effort consuming and requires large numbers of injected embryos. However, we believe it is the only technique that provides the necessary spatial resolution to accurately analyze the expression of genes with complex expression patterns in perturbed embryos. Furthermore, when used with appropriate controls, in situ hybridization can provide a good estimate of the level of expression in perturbed embryos compared to controls. To provide a temporal view of the consequences of these perturbations and avoid secondary effects, the expression of the genes analyzed in response to nodal or bmp2/4 overexpression was examined at two different stages, soon after the onset of their restricted expression, and at a later stage, most often early or late gastrula stage depending on the gene analyzed.
Fig. 7. Epistasis analysis of ventral genes: goosecoid as a key regulator of brachyury and foxA expression and a repressor of ciliary band genes. 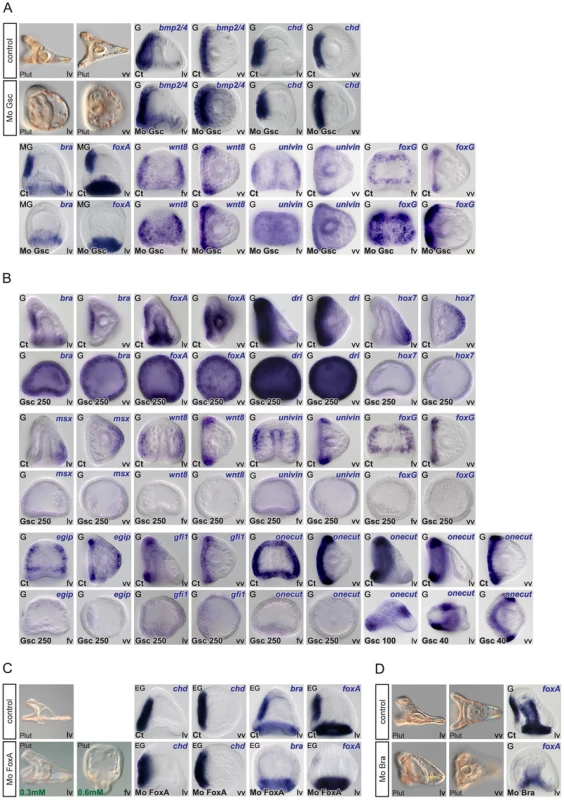
(A) Morphology of goosecoid morphants at 48h. Interfering with goosecoid function strongly delays gastrulation and produces partially radialized embryos as reported previously [35]. bmp2/4 and chordin are expressed normally in these embryos but expression of brachyury and foxA is lost while wnt8, univin and foxG and are ectopically expressed within the ventral ectoderm. (B) Injection of goosecoid mRNA at 250 µg/ml produced radialized embryos as reported previously [35]. Overexpression of goosecoid caused ectopic expression of brachyury, foxA and deadringer throughout the ectoderm. Overexpression of goosecoid also led to repression of dorsal markers genes such as hox7 and msx and of ciliary band marker gene expression as shown for wnt8, univin, foxG, egip, gfi1 and onecut/hnf6. Note the dose dependent repression of onecut/hnf6, with low doses (40–100 µg/ml) causing a dorsal shift of the expression domain of onecut/hnf6 and high doses (250 µg/ml) leading to complete repression. (C) Embryos injected with low doses (0.3 mM) of the foxA morpholino lacked a stomodeum as reported previously. At higher doses (0.6 mM), the foxA morpholino strongly interfered with gastrulation resulting in embryos with no gut or a small exogastrulated gut. foxA morphants had a normal expression of chordin but lacked expression of brachyury and foxA. (D) brachyury morphants, like foxA morphants, lacked a stomodeum and did not express foxA in the presumptive stomodeal region. lv, lateral view, vv, vegetal pole view, fv, frontal view. Fig. 8. Epistasis analysis of dorsal genes: irxA as a repressor of ciliary band gene expression downstream of tbx2/3. 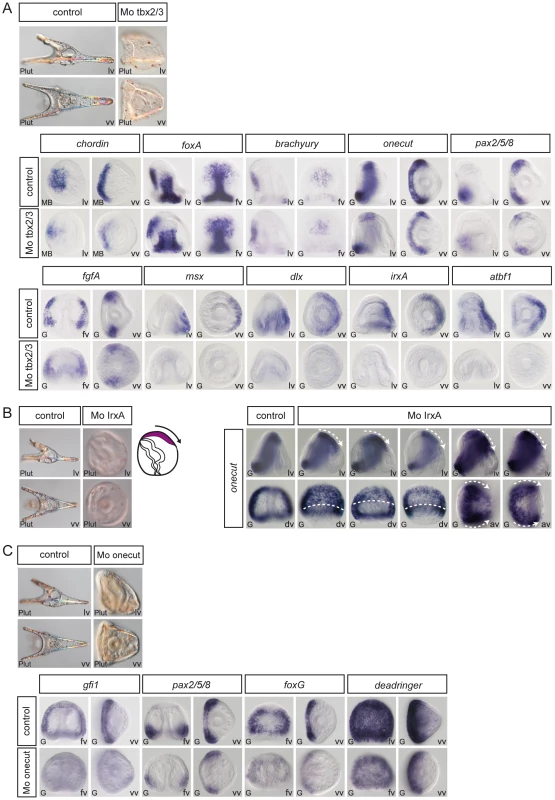
(A) tbx2/3 morphants are partially radialized and do not elongate along the D/V axis. The expression of ventral genes such as chordin, foxA, brachyury, onecut/hnf6 and of ciliary band genes such as pax2/5/8 and fgfA is largely normal in these embryos. In contrast, expression of dorsal marker genes such as msx, dlx, irxA and atbf1 is lost at gastrula stages. (B) irxA morphants are partially radialized and occasionally show an ectopic ciliary band like thickened region within the dorsal region. Expression of onecut/hnf6 is dramatically expanded towards the dorsal side of irxA morphants. (C) Onecut/hnf6 morphants are slightly radialized but do form a morphologically recognizable ciliary band. In these embryos, the expression of ciliary band genes gfi1 is absent while the expression of pax2/5/8, foxG or deadringer is strongly reduced. lv, lateral view, vv, vegetal pole view, av, animal pole view, dv, dorsal view, fv, frontal view. Fig. 9. Regulation of D/V patterning by extracellular and intracellular modulators of Nodal and BMP signaling. 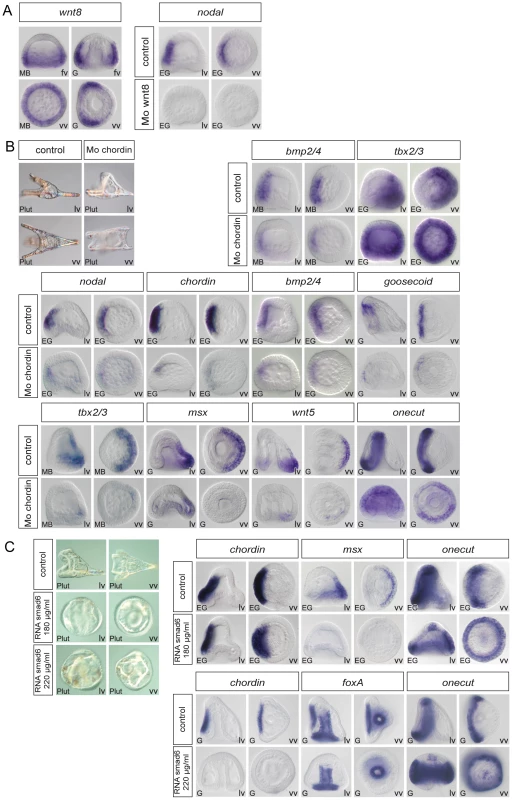
(A) Wnt8 regulates maintenance of nodal expression. At mesenchyme blastula stage, wnt8 starts to be expressed in a large belt of ectodermal cells within the vegetal hemisphere. At gastrula stage wnt8 expression is detected in the animal and vegetal hemispheres in two broad lateral stripes that flank the ventral ectoderm. In wnt8 morphants, expression of nodal is lost at mesenchyme blastula stage. (B) Chordin is essential for normal patterning along the D/V axis and plays a key role in restricting the expression of ciliary band genes. In chordin morphants, most of the ectoderm derived from the animal hemisphere differentiates into a ciliary band-like ectoderm. Expression of ventral (nodal, chordin, bmp2/4, goosecoid) and dorsal (msx, wnt5) marker genes is strongly downregulated in these embryos. A transient ectopic expression of tbx2/3 is detected at early mesenchyme blastula stage in these embryos followed by downregulation of the gene. Restriction of the expression of the ciliary band gene onecut/hnf6 is disrupted in chordin morphants and ectopic expression of this gene is detected in the ectoderm. (C) At high concentration (>220 µg/ml) smad6 mRNA suppressed the ectodermal expression of Nodal targets genes such as chordin and foxA and caused a dramatic ectopic expression of onecut/hnf6 throughout the ectoderm. At lower doses (<180 µg/ml), smad6 mRNA did not interfere with the expression of chordin but specifically antagonized with the expression of BMP target genes as msx and caused the ectopic expression of onecut/hnf6 in the dorsal ectoderm. lv, lateral view, vv, vegetal pole view, fv, frontal view. Information derived from these perturbations analyses was combined with earlier results to build a provisional gene regulatory network. The main features of this gene regulatory network are described below.
Goosecoid is required to initiate a stomodeal regulatory sub circuit and to repress ciliary band genes
Low levels of goosecoid transcripts are present maternally then their abundance increases sharply at swimming blastula stage, shortly after the peak of Nodal expression [35] (Figure S2). Expression of lefty, chordin, bmp2/4, fgfr1 and goosecoid, was unchanged in the goosecoid morphants consistent with these genes being direct targets of Nodal signaling and with previous studies [38] (Figure 7A and data not shown). Interestingly, at gastrula stages, strong ectopic expression of wnt8, univin and foxG was detected in the ventral ectoderm of goosecoid morphants indicating that one function of Goosecoid is to repress expression of these three genes in the ventral ectoderm between blastula and gastrula stages. In contrast, ectodermal expression of foxA and brachyury, two likely indirect targets of Nodal required for mouth formation, was lost in the goosecoid morphants, consistent with the lack of stomodeum in these embryos (Figure 7A) [42]. Reciprocally, overexpression of goosecoid caused a dramatic expansion of foxA and brachyury (Figure 7B). Therefore, in the sea urchin as in vertebrates, brachyury and foxA are targets of Nodal signaling but unlike in vertebrates, in the sea urchin, they are not primary targets of Nodal since their expression depends on the zygotic expression of goosecoid [63]–[65]. Overexpression of goosecoid also expanded the expression of deadringer as reported previously by Bradham et al. [22], [66]. In contrast, the two dorsal marker genes hox7 and msx failed to be expressed in the goosecoid overexpressing embryos consistent with previous studies showing that goosecoid overexpression suppresses expression of dorsal genes such as tbx2/3 and spec1 [35], [40]. Overexpression of goosecoid also abolished the expression of all the other ciliary band genes that we tested including wnt8, univin, foxG, egip, gfi1 and onecut/hnf6. Taken together these observations suggest that goosecoid plays a double function, first by allowing expression of stomodeal genes such as foxA and brachyury and second by suppressing the expression of ciliary band and dorsal genes.
Once goosecoid and foxA have been turned on, Brachyury and FoxA cross regulate each other so that brachyury maintains foxA expression while foxA promotes brachyury expression (Figure 7C, 7D). When the function of either of the two genes was blocked with a morpholino, expression of the other gene was lost and the resulting embryos developed without a stomodeum. The role of these cross regulatory interactions between brachyury and foxA may be to stabilize and lock the specification of the ventral ectoderm that has been initiated by Nodal as described in the endomesoderm GRN, for example between the transcription factors hex and tgif [7].
Tbx2/3: an early regulator of dorsal gene expression downstream of BMP2/4 signaling
Inhibition of tbx2/3 function strongly perturbed establishment of dorsal-ventral polarity resulting in embryos with a rounded shape, which lacked ventral arms and had a strongly reduced dorsal region (Figure 8A). Molecular analysis revealed that ventral markers such as chordin, foxA or brachyury were expressed in tbx2/3 morphants, albeit with reduced levels compared to controls (Figure 8A). A similar slight reduction was observed for the ciliary band markers onecut/hnf6, fgfA and pax2/5/8. In contrast, inhibition of tbx2/3 function abolished the expression of several dorsal genes encoding transcription factors including msx, dlx, irxA and atbf1 while the expression of other genes such as smad6, glypican5, oasis and wnt5 appeared unaffected. These results identify tbx2/3 as a key regulator of dorsal gene expression downstream of BMP2/4.
IrxA: a negative regulator of onecut/hnf6 downstream of tbx2/3
Since loss of BMP2/4 or Alk3/6 signaling causes ectopic expression of ciliary band genes in the dorsal ectoderm, it follows that in unperturbed embryos, a transcriptional repressor must act in the dorsal ectoderm downstream of BMP2/4 to prevent expression of ciliary band genes. Of the four transcription factors expressed in the dorsal ectoderm that we tested, only in the case of one of them did we observe robust ectopic expression of a ciliary band gene. This gene is irxA. In embryos injected with morpholinos against the irxA transcript, onecut/hnf6 expression was strikingly expanded in the dorsal ectoderm (Figure 8B). This effect was very robust and the territory in which the ectopic expression of onecut/hnf6 was observed was congruent with the expression territory of irxA. Interestingly, a small number of embryos injected with irxA morpholinos later developed with a thickened ectodermal region on the dorsal side that resembled an ectopic ciliary band (Figure 8B). This suggests that IrxA is a repressor of ciliary band genes downstream of BMP2/4.
Onecut/hnf6: an upstream positive regulator of ciliary band gene expression
onecut/hnf6 is of one of the earliest marker genes expressed in the presumptive ciliary band. onecut/hnf6 morphants developed with a slightly reduced D/V axis but they clearly displayed a D/V polarity and a well-developed ciliary band (Figure 8C). Nevertheless, we found that the expression of several marker genes of the ciliary band was affected in the onecut/hnf6 morphants. A reduced level of expression in the onecut/hnf6 morphants was observed in the case of pax2/5/8, foxG and dri while in the case of gfi1, no expression was detected. onecut/hnf6 is thus an upstream regulator of gfi1. Gfi proteins are conserved in C. elegans (Pag3), Drosophila (Senseless) and mice (Gfi1). In all three species, these zinc finger proteins play conserved roles in neural development [67]. Mice mutant for gfi1 are deaf and ataxic while flies mutant for senseless lack sensory organs indicating that Gfi proteins regulate sensory organ development [67], [68]. One can therefore anticipate that Gfi1 likely plays a role in neural development in the sea urchin embryo as it does in vertebrates and in flies. Since gfi1 is downstream of onecut, the ciliary band network therefore appears to be composed of at least two layers of zygotic factors.
Positive and negative feed back loops downstream of Nodal and BMP signaling
Wnt8 signaling is required for maintenance of nodal expression
To identify the Wnt ligands responsible for maintenance of nodal expression, we examined the expression pattern of candidate genes encoding Wnt factors and tested them for their requirement to maintain nodal expression. Of the various Wnt ligands examined, only wnt8 had a dynamic expression in the ectoderm at blastula stages (Figure 9A). Consistent with previous studies, we found that wnt8 morphants are radialized [53]. In these embryos, nodal expression is strongly reduced or absent (Figure 9A). These findings indicate that, in addition to the essential nodal auto regulatory loop that is required early to maintain the expression of nodal, Wnt8 function is required zygotically to maintain nodal expression at blastula stages through a second positive feedback loop.
Chordin and Smad6 act as negative regulators of BMP signaling on the ventral and dorsal side respectively
We showed previously that most of the ectoderm of chordin morphants differentiated into an enlarged ciliary band, a finding that was difficult to reconcile with the demonstrated activity of Chordin as an inhibitor of BMP signaling. We found that the abnormal patterning of the ectoderm in the chordin morphants is associated with a strongly reduced expression of ventral genes such as nodal, chordin, bmp2/4 and goosecoid and of dorsal genes such as msx and wnt5 (Figure 9B). We confirmed that following injection of the chordin morpholino, tbx2/3 was transiently ectopically expressed within the ventral ectoderm, consistent with the presence of ectopic BMP signaling in chordin morphants at mesenchyme blastula stage [26]. However, following this transient ectopic expression, tbx2/3 expression was rapidly downregulated like that of other dorsal marker genes such as msx and wnt5 (Figure 9B). Strikingly, expression of onecut/hnf6 was de-repressed in the ectoderm of the chordin morphants at gastrula stages consistent with the reduced expression of nodal and bmp2/4 in these embryos and with the development of an expanded ciliary band at later stages.
Overexpression of smad6 phenocopied the BMP2/4 loss of function phenotype, resulting in partially radialized embryos with ectopic spicules and expanded ciliary band on the dorsal side (Figure 9C). This phenotype was correlated with the loss of msx expression and ectopic expression of onecut/hnf6. These observations together with the finding that expression of smad6 is regulated by BMP signaling, strongly suggests that in the sea urchin as in vertebrates, Smad6 acts in a negative feedback loop required for the fine tuning of BMP signaling. High-level overexpression of smad6 abolished the expression of Nodal target genes such as chordin and foxA and caused a massive ectopic expression of onecut/hnf6 throughout the ectoderm indicating that one function of Smad6 may also be to restrict Nodal signaling to the ventral side.
Discussion
Essential roles of Nodal and BMP2/4 in patterning of the ectoderm along the D/V axis
In this study, taking advantage of the detailed phenotypic analyses and robust in situ hybridization procedures available in Paracentrotus lividus, we analyzed with a high level of spatial resolution the expression, the regulation and the function of most of the zygotic transcription factors and signaling molecules displaying restricted expression within the ectoderm of the sea urchin embryo. This analysis allowed us to assemble a gene regulatory network, the D/V GRN, which describes the regulatory interactions between these genes and provides a framework for understanding the developmental program responsible for patterning the embryo along the dorsal-ventral axis. Several interesting conclusions emerged from the resultant GRN. First, it provides a clear demonstration that the activities of Nodal and BMP2/4 account fully for the spatially restricted expression of all the known genes of this network: Nodal controls the expression of all the genes expressed specifically in the ventral ectoderm, and through BMP2/4, the expression of all the genes expressed specifically in the dorsal ectoderm. Both overexpression of these ligands and corresponding loss of function experiments produced very strong, all or none, effects consistent with the idea that Nodal and BMP2/4 are critical inputs that drive the D/V GRN. It should be noted that despite their essential roles, Nodal and BMP2/4 are certainly not the only ligands involved in D/V patterning of the ectoderm and other ligands more broadly expressed likely cooperate with Nodal and BMP2/4 to specify the ventral and dorsal regions. In particular, Nodal may bind to its receptor as a heterodimer with Univin, a GDF1/Vg1 ortholog, as shown in other models [24], [25] while BMP2/4 may heterodimerize with BMP5/8 to specify the dorsal ectoderm as shown in vertebrates and in Drosophila [69], [70]. Nevertheless, the key roles played by Nodal in this GRN together with the essential function of Nodal factors in D/V axis formation in vertebrates and basal chordates [71] reinforce the hypothesis that an ancestral function of Nodal may have been in the regulation of D/V axis formation in deuterostomes.
Goosecoid, a repressor that drives a stomodeal regulatory sub circuit and represses ciliary band and dorsal genes
A second key conclusion emerging from our D/V GRN is that in the sea urchin, Goosecoid is a key upstream element of a small regulatory circuit that controls mouth formation. In vertebrates ectopic expression of goosecoid promotes cell migration and induces incomplete secondary axes while loss of function studies implicate goosecoid in the function of the Spemann organizer and head formation [72]. The function of goosecoid during development of other deuterostome embryos has not been studied. In the sea urchin, previous studies reported that both overexpression and loss of function of goosecoid strongly perturbed establishment of the dorsal-ventral axis, however the target genes of goosecoid were not known and the role of this repressor within the ventral ectoderm remained largely unclear [35], [38], [40]. Our finding that goosecoid is a direct target of Nodal signaling strongly suggested that this gene could play a key role in specification of the ventral ectoderm downstream of Nodal. We have shown that Goosecoid likely regulates the expression of deadringer and foxG in the ventral ectoderm. Furthermore, we demonstrated that Goosecoid plays a critical role in mouth formation by regulating downstream target genes such as the stomodeal genes brachyury and foxA. This raises the possibility that an ancestral function of goosecoid may have been in the regulation of stomodeum formation. Consistent with this idea, goosecoid is expressed in the stomodeal region in both protostomes and deuterostomes and is co-expressed with brachyury and foxA in the oral region of cnidarians [73]. Since Goosecoid is a transcriptional repressor [74], this suggests that zygotic goosecoid activates foxA and brachyury by repressing the expression of a transcriptional repressor, the identity of which is presently unknown (Figure 10). Similar double repression mechanisms have been described in different GRNs. For example, in the sea urchin the skeletogenic mesoderm GRN, the repressor pMar has been proposed to repress hes-C as well as unidentified repressors to allow expression of genes specific of the PMC lineage [75], [76]. Similarly Schnurri, represses the expression of brinker to allow the expression of Dpp target genes in Drosophila imaginal discs [77]. One candidate for a repressor acting downstream of goosecoid is the transcriptional repressor ZEB1/Smad Interacting Protein 1 (Sip1) [78]. In the sea urchin embryo, Sip1 is expressed early in the presumptive ectoderm and its expression is downregulated at blastula stage, coincident with the onset of goosecoid expression [31] (see Figure S2 and S5). Experiments are currently being carried out in different labs to test this hypothesis.
Fig. 10. Representation of the gene regulatory network regulating regionalization of the ectoderm of the sea urchin embryo. 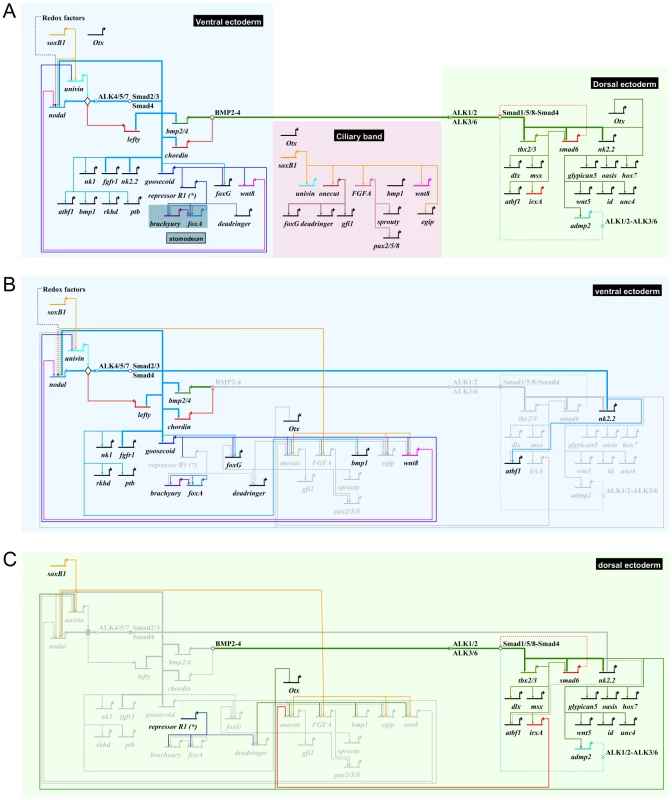
(A) Biotapestry [123], [124] diagram of the provisional gene regulatory network describing the regulatory interactions that have been identified in this study. Arrows indicate positive transcriptional activation. Flat arrows indicate repression. The colored boxes represent the spatial domains as indicated. The linkages between Nodal and its immediate direct target genes as well as the linkages between BMP2/4 and its direct target genes are shown as bold arrows. The gene regulatory linkages sustained by solid evidence are presented as solid lines. The gene regulatory linkages that are hypothetical or suspected to be indirect are represented as dotted lines. Except for the bold arrows, no assumption on whether these interactions are direct or indirect is made. The linkage between Nodal and Delta, expressed in single cells of the ciliary band, is not represented here. (B) Gene regulatory interactions within the ventral ectoderm highlighting the repressive action of Goosecoid on ciliary band genes. The genes that are inactive are represented in light grey. (C) Gene regulatory interactions within the dorsal ectoderm highlighting the repressive action of irxA on onecut/hnf6 and more generally of Nodal signaling on the expression of ciliary band genes. Another important function of Goosecoid appears to be in the repression of ciliary band and dorsal genes. Overexpression of goosecoid potently repressed expression of ciliary band markers. Furthermore, knockdown of Goosecoid function caused ectopic expression of univin, wnt8 and foxG in the ventral ectoderm. However, additional repressors likely cooperate with Goosecoid in this repression since inhibition of goosecoid function, unlike inhibition of irxA on the dorsal side, was not sufficient to derepress ciliary band markers genes such as onecut within the ventral ectoderm.
Tbx2/3 an early mediator of BMP2/4 signaling
Tbx2/3 has a special status amongst dorsal genes since it is one of the earliest zygotic genes expressed on the presumptive dorsal side [40], [41]. Previous studies had shown that tbx2/3 is expressed dynamically in a broad dorsal territory in all three germ layers and that its expression is regulated by BMP signaling [13], [26], [40], [41]. Indeed we showed that tbx2/3 is a direct target of BMP2/4 signaling in the ectoderm and that its function is required for expression of several dorsally expressed transcription factors such as msx, dlx, irxA and atbf1. Intriguingly, previous studies in Paracentrotus failed to detect any D/V polarity defect in tbx2/3 morphants [40]. In contrast, we found that tbx2/3 is essential for D/V axis formation in this species. The reasons for this discrepancy are unclear. Interestingly, in vertebrates, tbx2 is also a target of BMP4 signaling during D/V patterning of the optic cup [79]. Similarly, in hemichordates, which are positioned phylogenetically as the sister phylum of echinoderms, tbx2/3 is a target of BMP2/4 suggesting that key genes that drive the D/V GRN are conserved in these two closely related phyla [80]. In vertebrates, tbx2 and tbx3, unlike brachyury, which is a transcriptional activator, act as transcriptional repressors due to the presence of a strong repressor domain in their C-terminal region [81], [82]. It is therefore possible that the sea urchin Tbx2/3 protein also functions as a transcriptional repressor and that, like Goosecoid, it stimulates gene expression by relieving the repressive action of a transcriptional repressor. The identity of this hypothetical transcriptional repressor is presently unknown.
IrxA - and BMP2/4-dependent repression of ciliary band gene expression
One of the most important findings of this study is the identification of irxA as a gene which acts downstream of BMP signaling to repress the ciliary band gene onecut. We previously reported that inhibition of BMP2/4 or Alk3/6 function causes an expansion of the presumptive ciliary band territory towards the dorsal side, and that this expansion is accompanied by the ectopic expression of the neural gene onecut/hnf6 [26]. On the basis of this result we anticipated that one function of the BMP pathway in the dorsal ectoderm was to repress ciliary band gene expression and we postulated the existence of a BMP2/4 dependent repressor of ciliary band genes. We have now identified IrxA as one such repressor based on the following evidence. First, we showed that irxA expression is regulated by BMP2/4 signaling. Second, we showed that blocking irxA translation with morpholinos caused a robust ectopic expression of onecut in a sector of the dorsal ectoderm that coincides with the expression domain of irxA. Finally, it is established that Irx proteins can function as repressors by recruiting the Groucho Co-repressor [83], [84]. Since irxA is downstream of tbx2/3 in the GRN, we might predict that blocking tbx2/3 function should also result in ectopic expression of ciliary band genes. Surprisingly, we never observed ectopic expression of ciliary band marker genes in tbx2/3 morphants. This observation is consistent with previous GRN studies, which reported that direct target genes are more strongly affected than indirect target genes or in other words, that when a perturbation affects the driver gene, it causes stronger effects on target genes than when the perturbation affects genes further upstream in the pathway [29]. However, the simplest explanation is that our tbx2/3 morpholino may not be completely effective and that residual irxA expression may prevent ectopic expression of onecut in these embryos.
In vertebrates and in Drosophila, irx genes are involved in neural development [85]. In Xenopus for example, irx1 promotes neural development by repressing bmp4 expression in the neural plate. It was therefore surprising to find that in the sea urchin embryo, irxA acts downstream of BMP2/4 to negatively regulate neural marker genes. Nevertheless, the identification of irxA as a BMP2/4 dependent repressor of ciliary band gene expression strongly supports our proposal that the default state of the ectoderm in the absence of TGF beta signaling is the ciliary band and that the ectoderm is patterned by two successive inductive events that repress the ciliary band fate on the ventral and dorsal sides.
Extracellular and intracellular modulators of Nodal and BMP2/4 signaling
admp2 and glypican5: two components of a positive feedback loop downstream of BMP2/4?
Positive and negative feedback loops are essential components of gene regulatory networks. Indeed, in the sea urchin like in vertebrates, maintenance of nodal expression critically relies on autoregulation [12], [13], [14], [86]. Another auto regulatory loop that is highly conserved in vertebrates is found in the BMP signaling pathway with BMP signaling stimulating BMP transcription. However, in the sea urchin unlike in vertebrates, BMP2/4 signaling does not stimulate its own expression since transcription of bmp2/4 occurs on the ventral side while BMP signaling occurs on the dorsal side [26]. While BMP2/4 signaling did not induce bmp2/4 expression in dorsal cells, we found that BMP2/4 signaling instead induces expression of admp2 and promotes expression of glypican5, a positive regulator of BMP signaling [26]. The dorsal expression of admp2 and glypican5 was expanded following overexpression of bmp2/4 (Figure 3) and abolished in Nodal and/or BMP2/4 morphants (Figure 5, Figure 6) indicating that expression of these genes is regulated by BMP2/4 signaling. The positive regulation of admp2 by BMP2/4 in the sea urchin contrasts with the regulation of admp genes in vertebrates. In Xenopus or zebrafish, bmp4 and admp are expressed at opposite poles of the embryo and are under opposite transcriptional control: high levels of BMP signaling stimulate bmp4 expression but repress transcription of admp, a property that has been correlated with the ability of dorsal halves of early embryos to regulate and re-establish a D/V axis [87], [88], [89], [90]. In the sea urchin, both dorsal and ventral halves of partial embryos can regulate and regenerate a complete D/V axis [91]. However, in this embryo, bmp2/4 and admp2 are not under opposite transcriptional control since BMP signaling does not repress but stimulates admp2 expression.
Whether admp2 cooperates with BMP2/4 and contributes to specification of dorsal cells is presently not known but one can speculate on the possible role of this ligand as part of positive feed back loop downstream of BMP2/4 signaling. Therefore, while a BMP2/4 synexpression group does not exist in the sea urchin, BMP2/4 may activate a positive regulatory loop by inducing expression of admp2 to reinforce BMP signaling within the dorsal territory. Future studies are required to clarify the function of ADMP2 during D/V patterning of the ectoderm and to determine if this BMP ligand contributes to the activation of pSmad1/5/8 signaling during normal and regulative development.
Wnt signaling and maintenance of nodal expression
Previous studies had shown that when the canonical Wnt pathway is inhibited by using a dominant negative form of TCF, expression of nodal is lost at blastula stage [13]. By analyzing nodal expression at early stages in cadherin injected embryos Angerer and colleagues showed that nodal expression is initiated normally in animalized embryos but that it is not maintained [92]. They further provided evidence that the loss of nodal expression in these animalized embryos is caused by the persistence of the expression of the FoxQ2 repressor in the ectoderm. Here we have identified Wnt8 as a ligand required to maintain nodal expression. The exact period when Wnt8 signals are required to maintain nodal expression is not known. During cleavage and blastula stage, wnt8 is expressed in the vegetal pole region and starting at mesenchyme blastula stage, wnt8 expression in a large belt of ectodermal cells (Figure 9A). One possibility is that Wnt8 signals emitted by the ventral and/or lateral ectoderm are responsible for the maintenance of nodal at the beginning of gastrulation. Alternatively, Wnt8 signals may be required earlier for the restriction of foxQ2 expression at early blastula stages. Future studies are required to resolve this issue. It is remarkable that, during mouse embryogenesis, a Wnt signal is also required to maintain high levels of nodal expression in posterior cells through a TCF dependent enhancer and that in the absence of Wnt3, nodal transcription is initiated but it is not maintained [93]. Therefore, both in the sea urchin embryo and in the mouse embryo, in addition to the nodal auto regulatory loop, a second positive regulatory input from a Wnt ligand is required to maintain nodal expression.
Restriction of BMP signaling by Chordin is essential for D/V axis formation
Lastly, this study helped to resolve a paradox regarding the chordin loss of function phenotype. Chordin morphants display an expanded ciliary band covering the animal hemisphere, a reduced dorsal side and a pair of parallel spicules [26]. In vertebrates, chordin is expressed within the organizer and promotes neural differentiation by interrupting a BMP positive auto regulatory loop in the prospective neural ectoderm. In the sea urchin, chordin likely functions in a similar way by preventing BMP signaling within the ventral and dorsal ectoderm [26]. Since the function of Chordin as an inhibitor of BMP signaling is conserved, the reason why most of the ectoderm adopted a ciliary band fate instead of an epidermal fate in the chordin morphants remained unclear. We showed that in addition to ectopic BMP signaling, chordin morphants suffer from a strong downregulation of nodal expression leading ultimately to derepression of ciliary band and neural genes. One possibility is that the transient ectopic BMP signaling observed on the ventral side of chordin morphants interferes with Nodal signaling and interrupts the Nodal auto regulatory loop causing the subsequent loss of bmp2/4 expression. Taken together, these findings highlight the crucial role played by chordin in D/V patterning of the ectoderm in the sea urchin embryo by showing that chordin is required for normal patterning along the whole dorsal-ventral axis.
A new model of specification and regionalization of the ectoderm: the ciliary band as the default state of the ectoderm in the absence of Nodal and BMP signaling
The results obtained in this study largely support this idea that the default state of the ectoderm in the absence of Nodal and BMP signaling is a ciliary band-like ectoderm that expresses a number of neural genes and that Nodal and BMP2/4 restrict this ciliary band fate by specifying the ventral and dorsal ectoderm. The first hint that the default state of the ectoderm in the absence of TGF beta signaling is the ciliary band is that several genes whose expression is later restricted to the ciliary band territory are expressed throughout the ectoderm at earlier stages. This is for example apparent for fgfA, univin and wnt8, which are expressed in a belt of cells that includes most of the presumptive ectoderm at blastula stages. The expression of fgfA, univin and wnt8 is subsequently repressed on the ventral and dorsal sides during gastrulation thereby restricting the expression of these genes to the ciliary band domain. Several additional lines of evidence support the idea that the default state of the ectoderm in the absence of TGF beta signaling is a ciliary band and neural fate and that alternative ectodermal fates must be induced by active signaling. First, overexpression of both nodal and bmp2/4 strongly antagonized the expression of ciliary band and neural markers such as onecut, foxG and gfi1, with bmp2/4 leading to a very potent inhibition of ciliary band formation. Second, in the lefty morphants the ciliary band failed to form while in the absence of Nodal and BMP2/4 signaling, the ventral and dorsal ectodermal regions were not specified and most of the ectoderm differentiated instead into a thickened ciliated ectoderm that resembled the ciliary band ectoderm and expressed all tested ciliary band markers. These ciliary band markers were de-repressed throughout the ventral and dorsal ectoderm in the nodal morphants while in the absence of BMP2/4, which acts as a dorsal inducer, or of alk3/6, which is required to transduce BMP2/4 signals, only specification of the dorsal ectoderm was perturbed and ectopic expression of these ciliary band genes was detected only on the dorsal side. A third argument is that the presumptive ciliary band territory is also a region in which fgfA and vegf are expressed and where MAP kinase activity is high [48], [56], [94]. Studies in vertebrates have shown that the activity of the MAP kinase ERK inhibits both BMP signaling and neuralization by phosphorylating Smad1 in the linker region thereby preventing its nuclearization. We thus predict that during normal development of the sea urchin embryo, the high MAP kinase activity present in the lateral ectoderm promotes neural fates within the presumptive ciliary band by inhibiting the activity of pSMAD1/5/8 and pSMAD2/3. Thus, in the absence of Nodal and BMP signaling, signals such as FGFA that are normally present at the level of the lateral ectoderm are ectopically expressed in the ventral and dorsal regions where they may promote ectopic neuron formation [26], [27]. One last but crucial argument that supports our model of the ciliary band as a default state of the ectoderm in the absence of TGF beta signaling is that we identified irxA and possibly Goosecoid as repressors of a subset of ciliary band genes downstream of Nodal or BMP signaling. One read-out of Nodal and BMP2/4 signaling therefore appears to be active repression of the ciliary band fate as we had predicted [26].
Yaguchi and colleagues previously demonstrated that in the absence of Wnt signaling, most of the ectoderm differentiates as a neurogenic ectoderm that expresses markers of the animal pole [27]. Since many ciliary band genes are also expressed in the animal pole, it could be argued that the ectopic expression of ciliary band marker genes observed following inhibition of Nodal or BMP signaling also reflects an expansion of the animal pole domain. This can be ruled out for several reasons. First, we showed that the expression of animal pole markers such as foxQ2, is unaffected in Nodal morphants or in embryos treated with a pharmacological inhibitor of the Nodal receptor. Second, Yaguchi et al. showed that the number and location of serotonergic neurons of the apical organ are unaffected by inhibition of Nodal signaling. Importantly, we showed that pax2/5/8, which is expressed in the vegetal part of the ciliary band but not in the animal pole region behaved exactly like the other ciliary band marker genes and was strongly derepressed in the ventral and dorsal ectoderm of Nodal morphants. Taken together these observations indicate that the lateral ectoderm of the prospective ciliary band, not the animal pole domain, is expanded in the Nodal morphants.
Our study suggests that specification of the ciliary band is likely initiated by a combination of maternal factors such as SoxB1 and by zygotic factors such as FGFA, Otx and Onecut/Hnf6 whose expression is initiated independently of the Nodal and BMP2/4 signals (Figure 10). These zygotic genes initially show a broad expression in the ectoderm, which then becomes restricted to the presumptive ciliary band by the activity of transcriptional repressors such as Goosecoid and IrxA expressed in the ventral or dorsal ectoderm downstream of Nodal or BMP2/4. Collectively our results suggest that the neural ectoderm of the ciliary band forms in a territory that is devoid of Nodal and BMP2/4 signaling (Figure 11). On the dorsal side, inhibition of BMP signaling appears to be sufficient to trigger formation of the ciliary band as was observed in BMP2/4 or Alk3/6 morphants or in embryos injected with low doses of smad6 mRNA. Similarly, on the ventral side, inhibition of Nodal signaling is sufficient to initiate formation of a ciliary band since BMP signaling does not occur on the ventral side but on the dorsal side [26]. In this case, ectopic neural differentiation likely results from inhibition of ventral differentiation. This highlights that, in the sea urchin ectoderm, preventing ventral cells to differentiate downstream of Nodal signaling promotes neural differentiation just as efficiently as inhibiting BMP signaling on the dorsal side. Similarly, in zebrafish embryos, inhibition of Nodal signaling causes the transfating of prospective mesendodermal cells into neural cells [95], [96] and in the mouse, lack of Nodal signaling causes precocious neural differentiation [97]. Therefore, in the sea urchin embryo like in vertebrate embryo models, neural differentiation can result both from inhibition of BMP signals as well as from inhibition of other signals that regulate the fate of early blastomeres and allocate cells to embryonic territories and germ layers.
Fig. 11. Changes in identity of ectodermal territories following perturbations of Nodal or BMP signaling and novel model of ectoderm patterning. 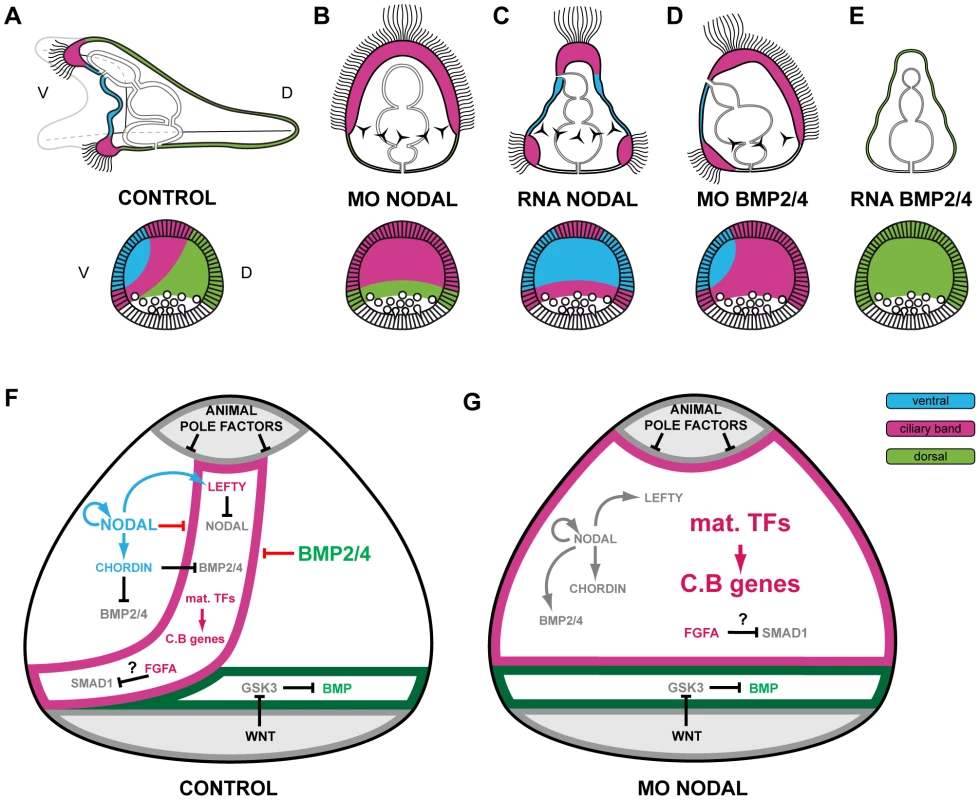
Schemes describing the morphology of control embryos and perturbed embryos. (A) control embryo. The thick ciliated epithelium of the ciliary band is restricted to a belt of cells at the interface between the ventral and dorsal ectoderm. (B) Nodal morphant. Most of the ectoderm differentiates into an expanded large ciliary band. An animal pole domain is nevertheless present in these embryos as shown by the presence of the apical tuft and at the molecular level by the expression of apical domain marker genes. In these embryos, the ectoderm surrounding the blastopore differentiates into dorsal ectoderm. (C) embryo overexpressing Nodal. Most of the ectoderm differentiates into ventral ectoderm. A ciliary band-like ectoderm forms at the animal pole and in the ectoderm surrounding the blastopore. (D) BMP2/4 morphants. An ectopic ciliary band forms in the dorsal ectoderm in addition to the normal ciliary band. (E) bmp2/4 overexpressing embryo. All the ectoderm has a dorsal identity. The animal pole domain is largely absent. The triradiated stars represent the spicule rudiments. (F) Proposed model for regionalization of the ectoderm of the sea urchin embryo through restriction of the ciliary band fate by Nodal and BMP signaling. Maternal factors such as SoxB1 promote the early expression of ciliary band genes within the ectoderm. Nodal signaling on the ventral side promotes differentiation of the ventral ectoderm and stomodeum and represses the ciliary band fate probably through the activity of Goosecoid as well as of additional repressors. Nodal induces its antagonist Lefty, which diffuses away from the ventral ectoderm up to the presumptive ciliary band territory. Within the ventral ectoderm, Nodal induces expression of bmp2/4 and of its antagonist chordin. Chordin prevents BMP signaling within the ventral ectoderm and probably within the presumptive ciliary band region. At blastula stages, protein complexes containing BMP2/4 and Chordin can diffuse towards the dorsal side to specify dorsal fates. In the dorsal ectoderm, BMP signaling strongly repress the ciliary band fate partly by inducing the expression of the irxA repressor. A high level of MAP kinase activity resulting from FGFA signaling in the lateral ectoderm likely contributes to maintain a low level of Nodal and BMP signaling within the presumptive ciliary band region by phosphorylating Smad1/5/8 and Smad2/3 in the linker region, which inhibits their activity. The presence of Chordin and Lefty in the prospective ciliary band allows expression of ciliary band genes to be maintained in this region. The ectoderm surrounding the blastopore differentiates into dorsal ectoderm likely because it receives Wnt signals that antagonize GSK3 and promote BMP signaling. (G) In the absence of Nodal signaling, both the ventral and the dorsal inducing signals are not produced, ciliary band genes are not repressed and unrestricted MAP kinase signaling promotes differentiation of the ventral and dorsal ectoderm into neural ectoderm and ciliary band. The genes or proteins that are inactive are represented in light grey. In summary, our results show that in the sea urchin embryo, the neurogenic territory of the ciliary band is not induced by an interaction between the ventral and dorsal territories as previously suggested [98], but that it represents the default state of the ectoderm in the absence of Nodal and BMP signaling. Nodal and BMP2/4 may therefore be regarded as factors that are required to prevent premature differentiation of ectodermal cells into neural cells as much as factors that are required for specification of the ventral and dorsal ectoderm.
Comparison with previous GRNs
Another recent GRN analysis of ectoderm specification in S. purpuratus was performed using nanostring technology [29]. A comparison of the architecture of the gene regulatory networks derived from this study and ours reveals the expected similarities but also some major differences. A common central element in the architecture of both networks is the critical dependence of dorsal genes on non-autonomous signaling by BMP2/4, a feature already proposed previously [13]. Another point of convergence is that both studies pointed to goosecoid and tbx2/3 as important early zygotic genes downstream of Nodal and BMP2/4: both studies identified brachyury as a downstream target of Goosecoid, and dlx and irxA as downstream targets of Tbx2/3. Finally, both studies identified foxG and deadringer as downstream targets of Nodal.
The first important difference in the architecture of the two proposed networks is that whereas our study defines the default state of the ectoderm in the absence of Nodal and BMP signals as a ciliary band-like ectoderm, the network proposed by Su et al. largely ignores formation of the ciliary band. Another important difference between the two studies concerns the dependence of ventral genes on Nodal. Su et al. argued that only part of the oral ectoderm specification system is downstream of Nodal [29]. According to the authors, a number of regionally expressed genes including onecut/hnf6, otx2, lim1, and foxA, are activated “specifically in the oral ectoderm…exactly the same with or without nodal”, leading them to speculate that hypothetical Nodal independent early oral ectoderm signals regulate these genes in the ventral ectoderm. We do not agree with this interpretation, since from our in situ analysis, it is clear that these genes cannot be considered as oral-specific markers. Furthermore, we showed that the expression of onecut/hnf6, otx2, lim1, and foxA in the presumptive ectoderm region of Nodal morphants was not regionalized, consistent with the absence of any oral territory in these embryos. The expression of onecut/hnf6 and otx2 is first initiated in a territory much larger than the ventral ectoderm, before subsequently becoming restricted to either to the ciliary band (onecut/hnf6) or to a broader territory that also includes the ventral ectoderm (otx2). We thus interpret the continued expression of onecut/hnf6 and otx2 in the ectoderm as reflecting adoption of a ciliary band character by the entire ectoderm. Concerning foxA, the nodal-independent detection of the mRNA reported by Su et al is undoubtedly due to the abundant expression of this gene in a distinct endodermal territory, which, unlike the oral ectoderm expression, is largely Nodal-independent. The foxA example highlights the importance of using methods that allow spatial resolution to analyze the expression of genes with complex expression patterns in epistasis experiments.
According to Otim and colleagues and Su and colleagues, two genes, onecut/hnf6 and deadringer, play essential roles in the DV GRN. Using an “unconventional morpholino” that targeted a sequence 660 bp downstream of the first ATG but that did not target a splice junction, Otim et al. reported that “inhibition” of hnf6/onecut function eliminated D/V polarity and caused a radialized phenotype that strikingly resembled the Nodal loss of function. Using the same reagent, Su et al. expanded this analysis and further argued that a positive regulatory input from onecut/hnf6 is required for the expression of several key regulators such as nodal, goosecoid, lefty, chordin, and bmp2/4 [29], [46]. These results are highly surprising since morpholinos are predicted to be ineffective at blocking translation when they target sequences after the first 25 bases following the initiator ATG [99], [100]. Using two different and more conventional morpholinos targeting the 5′ leader or the translation start site of the P. lividus hnf6/onecut transcript, we were unable to reproduce either the striking hnf6/onecut morphant phenotypes originally reported by Otim and colleagues or the effects on nodal, goosecoid, lefty, chordin, and bmp2/4 reported by Su and colleagues. It is therefore very unlikely that onecut/hnf6, which is expressed only transiently within the ventral ectoderm, plays the crucial role proposed by these authors in this gene regulatory network. Regarding deadringer, Su et al. found that deadringer morphants display a much reduced expression of ventral genes such as goosecoid, NK1 and hes as well as a strongly reduced expression of dorsal genes such as irx, nk2.2 and tbx2.3. Again, these results are surprising since the published cDNA sequence of deadringer used by Su et al. to design their morpholino as well as the associated predictions of the translation start site of the protein are probably incorrect and correspond to a truncated protein sequence as suggested by our sequence analysis of the genomic S. purpuratus deadringer locus and the analysis of the deadringer cDNAs in Paracentrotus (Figure S1). In addition, using two different morpholinos against the P. lividus deadringer transcript, we were unable to reproduce the published drastic effects of deadringer morpholinos on the expression of ventral and dorsal marker genes. It is therefore also unlikely that deadringer plays the role that it had been previously attributed in the S. purpuratus GRN.
Finally, it has been argued that specific aboral differentiation genes such as CyIIIa and spec1 are transcriptionally activated in the aboral ectoderm long before late blastula and that this implied the existence of an early asymmetry in the aboral ectoderm that affected transcriptional activity. Su et al. postulated that this asymmetry may be a redox gradient that would directly regulate the transcriptional activity of aboral genes such as CyIIIa and tbx2/3. Our results oppose this view. In Paracentrotus, the ectodermal expression of tbx2/3 is essentially lost following inhibition of Nodal or BMP2/4 signaling. While it is true that a residual tbx2/3 expression is observed in the Nodal morphants at gastrula stage, this expression is restricted to the vegetal most regions and therefore likely reflects the response of this gene to signals that act along the animal-vegetal axis rather than response to a redox gradient along the D/V axis. Furthermore, in Paracentrotus, expression of CyIII genes is first ubiquitous and only becomes restricted to the dorsal ectoderm at mesenchyme blastula stage (see Figure S5), coinciding with the nuclear translocation of pSmad1/5/8 in dorsal cells. In other words, we never observed any marker gene that was expressed specifically in the dorsal ectoderm before the onset of BMP signaling i.e. at late blastula stage. Our observations therefore do not support the view that the asymmetrical CyIIIa or tbx2/3 expression is driven by an early red-ox gradient, at least not in Paracentrotus, but suggest that their expression is more likely driven by differential Nodal and BMP signaling along the dorsal-ventral axis.
Nodal and BMP inhibition as an ancestral mechanism of neural induction?
A comparison of the mechanisms of neural induction in different species reveals both similarities and divergences regarding the signaling pathways involved. In Xenopus, inhibition of both Nodal and BMP signaling appears to be essential for neural induction, although FGF signaling is likely implicated in the early steps of this process [101], [102]. Similarly, in mammals, both Nodal and BMP signaling have been involved in neural differentiation, the strongest evidence being that most epibast cells of mouse embryos mutant for nodal or bmpr1 display widespread and precocious expression of anterior neural markers [97], [103]. In the chick and in zebrafish, there is strong evidence that FGF signaling regulates neural induction partly through the regulation of expression of BMP ligands and of BMP antagonists [104], [105], [106]. In contrast in ascidians, which are basally branching but divergent chordates, FGF signals are the key players in neural induction by directly regulating the expression of neural markers such as otx [107]–[109]. Inhibition of BMP signaling does not appear to play a role in this process [110] while Nodal plays a distinct, inductive role in patterning of the neural plate [111]. Similarly, in hemichordates, which together with the echinoderms form a sister group of the chordates and have a diffuse neural system, BMP signaling does not appear to play a role in the choice between neural and epidermis [80].
Our experiments in the sea urchin embryo show that inhibition of Nodal and BMP signaling is central to neural induction in echinoderms and that in the absence of Nodal or BMP signaling, most cells of the ectoderm differentiate into a neurogenic ectoderm. Since BMP signaling also regulates neural differentiation in insects [112] and annelids [113], it appears likely that inhibition of Nodal and BMP signaling may have been an ancestral mechanism to specify neural cells not only in deuterostomes but also perhaps in bilateria, and thus that the neural specification mechanisms used in ascidians and hemichordates have diverged during evolution.
Although in the sea urchin inhibition of Nodal causes the ventral ectoderm to adopt ultimately a neurogenic ectodermal fate, it should be kept in mind that our experiments also suggest that Nodal may have an early and positive role in specification and/or patterning of the neurogenic territory of the ciliary band since we showed that Nodal promotes the expression of Delta in a subpopulation of ciliary band cells and drives the early expression of the neural gene foxG. Therefore, in the sea urchin as in chordates, in addition to its general inhibitory role on neural induction, Nodal may also play a positive role in specification and/or patterning of the neural territory [111], [114], [115].
In conclusion, this large scale, systematic GRN analysis has allowed us to identify a number of key gene regulatory interactions and to build a provisional gene regulatory network describing specification of the three main ectodermal territories of the sea urchin embryo. It has not only uncovered key and probably ancient regulatory sub circuits that drive morphogenesis of the ectoderm, but has also allowed us to propose a new model of how specific regions of the ectoderm are induced over a default state, and of how the ectoderm is patterned by successive rounds of induction by TGF beta ligands. This relatively simple model captures most of the results derived from the functional analyses of Nodal and BMP2/4 in the sea urchin embryo and provides testable predictions for futures studies. Finally, our study illustrates the power of the GRN based approaches which can provide a global perspective on a set of genes regulating a biological process, explaining how this process works and what happens when it fails.
Materials and Methods
Animals, embryos, and treatments with recombinant proteins and inhibitors
Adults sea urchins (Paracentrotus lividus) were collected in the bay of Villefranche-sur-Mer. Embryos were cultured as described previously [116], [117]. When required, fertilization envelopes were removed by adding 2mM 3-amino-1,2,4 triazole 1 min before insemination to prevent hardening of this envelope followed by filtration through a 75µm nylon net. SB431542 (10 µM in sea water) was diluted from stocks solutions in DMSO, and embryos incubated in 24 well plates protected from light. In controls experiments, DMSO was added at 0.1% final concentration. NiCl2 was used at 0.5 mM. SB431542 and nickel treatments were performed continuously starting 30 min after fertilization. Continuous treatments with recombinant mouse Nodal (1µg/ml) and BMP4 proteins (0.5 µg/ml) (R&D) started at the 16-cell-stage and used embryos lacking the fertilization envelope. We verified with a set of 10 genes that RNA overexpression and recombinant proteins produced equivalent effects for both Nodal and BMP.
To determine if marker genes are direct or indirect targets of Nodal or BMP4 signaling, embryos at the swimming blastula/late blastula, early mesenchyme blastula stage or at gastrula stage from which the fertilization envelope had been removed were treated for 2h with recombinant proteins in the presence or absence of protein synthesis inhibitors. To block protein synthesis, puromycin or emetine was added at a final concentration of 360µM (200µg/ml) or 5µM (10µg/ml) respectively using stock solutions prepared in DMSO. In control experiments embryos were treated with 0.1% DMSO or with Puromycin at 200µg/ml or emetine at 10µg/ml. Development of the treated embryos was usually arrested 30 min after addition of the inhibitor, an indication of the effectiveness of the reagent and after 3–4h, all the treated embryos underwent a massive and brutal apoptosis, an effect characteristic of treatments with protein synthesis inhibitors. In the case of nodal, bmp2/4, lefty, goosecoid, fgfr1, chordin, nk2.2, tbx2.3, treatments were performed at the swimming blastula stage. In the case of nk1, foxA, brachyury, foxG, dlx, hox7, id, irxA, glypican5, cyIIIa, admp2, smad6 and msx, treatments were performed at the early mesenchyme blastula sage. In the case of deadringer, atbf1, msx, wnt5, irxA and dlx treatments were also performed at gastrula stage. Short treatments with Nodal or BMP4 failed to induce ectopic expression of any marker gene at gastrula stage suggesting that most of the genes expressed at this stage are indirect targets of Nodal and BMP2/4 or alternatively that at this stage, ectodermal territories are resistant to respecification by exogenous Nodal or BMP4.
cDNA sequences and cloning of full-length transcripts
Most of the genes analyzed in this study were discovered in the course of a random in situ hybridization screen using cDNA libraries from various stages (T. Lepage unpublished). Additional marker genes were discovered in a second in situ screen aimed at analyzing the expression profiles of all the transcription factors and signaling molecules expressed during early sea urchin development [30] using a Paracentrotus lividus EST library (http://goblet.molgen.mpg.de/cgi-bin/webapps/paracentrotus.cgi). When the isolated clones were incomplete, full-length cDNA sequences were obtained either by screening cDNA libraries with conventional methods and sequencing the corresponding clones. In certain cases, 5′RACE was performed using the Smart RACE kit (Clontech) to obtain the 5′ sequences. A list of all the Paracentrotus transcripts analyzed in this study with a summary of their temporal and spatial expression patterns is provided in Table 1 together with the corresponding accession numbers and original references describing these genes. Note that in the case of deadringer, the sequence of the Paracentrotus lividus clones diverged significantly from the published Strongylocentrotus purpuratus sequence. The published S. purpuratus deadringer transcript is predicted to encode a 490 amino acid protein. However, all the 13 independent deadringer cDNA clones that we sequenced encoded a protein 100 amino acids longer on the N-terminal side. Furthermore, translation of the S. purpuratus genomic sequence upstream of the predicted first ATG revealed the presence of a much longer open reading frame compared to the published deadringer protein sequence that encoded a protein highly similar to the deduced protein sequence from Paracentrotus (see Figure S1). This indicates that the previously published deadringer mRNA sequence was probably incorrect on the 5′ end and that the predicted deadringer protein sequence deduced from this mRNA was truncated. Since morpholinos fail to block translation when their target sequence is located after the first 25 bp following the initiator ATG [100], the conclusions derived from previous functional studies of deadringer in S. purpuratus, which relied on a truncated sequence, are probably erroneous.
Characterization of the temporal and spatial expression of regulatory genes of the network
For each gene of the network, a detailed analysis of the expression pattern was performed using digoxygenin labeled probes and in some cases, the temporal expression was analyzed by Northern blotting to verify maternal expression and to determine the exact onset of zygotic gene expression (Figure S2). In situ hybridization was performed following a protocol adapted from Harland [118] with antisense RNA probes and staged embryos. For marker genes expressed in ventral or dorsal territories at early stages, and for genes with complex expression profiles, double in situ hybridization was performed to confirm the orientations of the expression pattern. In this case, the two probes were hybridized and developed simultaneously. Probes derived from pBluescript vectors were synthesized with T7 RNA polymerase after linearization of the plasmids by NotI, while probes derived from pSport were synthesized with SP6 polymerase after linearization with SfiI. Control and experimental embryos were developed for the same time in the same experiments. Two color in situ hybridization was used following the procedure of Thisse et al. [119].
Overexpression analysis
For overexpression studies the coding sequence of the genes analyzed was amplified by PCR with the Pfx DNA polymerase (Invitrogen) using oligonucleotides containing restriction sites and cloned into pCS2 [120]. Capped mRNAs were synthesized from NotI-linearized templates using mMessage mMachine kit (Ambion). After synthesis, capped RNAs were purified on Sephadex G50 columns and quantitated by spectrophotometry. RNAs were mixed with Tetramethyl Rhodamine Dextran (10000 MW) or Texas Red Dextran (70000 MW) or Fluoresceinated Dextran (70000 MW) at 5 mg/ml and injected in the concentration range 100–800µg/ml. The nodal, bmp2/4, fgfA, univin, alk3/6QD, and chordin pCS2 constructs have been described in Duboc et al. (2004), Röttinger et al. (2008), Range et al. (2007) and Lapraz et al. (2009). The pCS2 goosecoid construct is described in [40]. RNA derived from the following additional constructs were made (the cloning sites are indicated in parenthesis): pCS2foxA (ClaI-XbaI); pCS2deadringer (EcoRI-XhoI); pCS2foxG (ClaI-XhoI); pCs2smad6 (EcoRI-XbaI); pCS2pax2/5/8 (BamHI-XhoI); pCS2tbx2/3 (BamHI-XhoI); pCS2msx (BamHI-XhoI); pCS2nk2.2 (BamHI-XhoI).
Perturbation analysis with morpholinos
Morpholino antisense oligonucleotides were obtained from GeneTools LLC (Eugene, OR). The nodal, BMP2/4, Alk4/5/7,Alk3/6, univin, lefty and soxB1 morpholinos are described in [12]–[14], [26]. Since morpholinos can have side effects or display toxicity or produce variable reductions in gene activity [121], we designed and tested several morpholinos for each gene. A pair of morpholinos that did not display toxic effects was selected for further use (a morpholino was considered toxic if it caused developmental arrest during cleavage or a massive cell death at the onset of gastrulation when injected at low doses (0.1–0.3 mM)). In the cases of nodal, bmp2/4, alk3/6, Alk4/5/7, univin and soxB1, the efficiency of the morpholino to downregulate the expression of previously characterized targets genes was systematically assessed in control experiments [13], [14], [26]. The phenotypes observed for nodal, bmp2/4, brachyury chordin, foxA, fgfA, goosecoid, irxA, lefty, tbx2/3, dlx, msx, onecut/hnf6, soxB1, univin, wnt8 morpholinos were considered specific since they were confirmed with a separate, non-overlapping morpholino. In the case of alk3/6, alk4/5/7 and nodal, a rescue experiment had previously been performed demonstrating the specificity of these reagents [13], [14], [26]. The phenotypes observed were always consistent with the zygotic expression pattern of the targeted genes and with previous well-established functional data [13], [14], [26], [35], [42], [122]. We did not observe inconsistent phenotypes among several knockdowns except in one case, in which knocking down Tbx2/3, an upstream regulatory gene of irxA, did not cause the same effect on the IrxA target gene onecut/hnf6 as knocking down irxA itself suggesting that the tbx2/3 morphant phenotype is a hypomorphic phenotype and not a null. In the case of the ventrally expressed genes nodal and bmp2/4, we observed strong non autonomous effects consistent with the demonstrated translocation of BMP2/4 from the ventral to the dorsal ectoderm and with the role of BMP2/4 as relay downstream of Nodal [26]. In contrast, we never observed strong effects on the expression of ventral markers by morpholinos targeting genes expressed dorsally. In three cases, (dlx, msx, foxG) a morphological phenotype was consistently observed but molecular analysis failed to detect significant perturbations in the expression of the genes analyzed. Other morpholinos pairs (deadringer, hox7, nk2.2, oasis, wnt5) gave very weak or not always reproducible phenotypes. Molecular analysis on embryos injected with these morpholinos failed to detect significant and reproducible changes in gene expression in any of the ventral, dorsal or ciliary band markers genes that we tested. In a few cases, (atbf1, klf2/4) all the morpholinos synthesized were highly toxic and were not studied further. The loss of function phenotypes of 29D, tubulinß3, egip, CyIIIa, admp2, fgfr1, pax2/5/8, unc4, nk1, id, rkhd and ptb and otx were not analyzed in this study and these genes were only used as markers in the following experiments. The sequences of all the morpholino oligomers used in this study are listed below. The most efficient morpholino of each pair is labeled with a star.
alk4/5/7 Mo 1: TAAGTATAGCACGTTCCAATGCCAT
alk3/6: Mo1: TAGTGTTACATCTGTCGCCATATTC
brachyury Mo1: AGCATCGGCGCTCATAGCAGGCATA
brachyury Mo2*: CTGGCAGAAGATGTACTTCGACGAT
bmp2/4 Mo1*: GACCCCAGTTTGAGGTGGTAACCAT
bmp2/4 Mo2: CATGATGGGTGGGATAACACAATGT
chordin Mo1*: GGTATAAATCACGACACGGTACATG
chordin Mo2: CGAAGATAAAAACTTCCAAGGTGTC
deadringer Mo1: TGCTCGCGGTAACAAGTGATTCCAT
deadringer Mo2: TTATATGGCAAAGGACTTCTACAGC
dlx Mo1: CCCACGTCAAATGAATACATCAACA
dlx Mo2: AAACACGTTTAGAATCCTCACGACT
fgfA Mo1: ACTTTCATCCATTTTCGCTTTCATG
fgfA Mo2*: ACACATTTTGGATACTTACAGCTCC
foxA Mo1: CATGGGTTCCTCCTTGAAATCCACG
foxA Mo2*: TGAAAGATTAAAGTAGCACAGTCAG
foxG Mo1*: TCCGATGAATGTGCATGAAAAACTG
foxG Mo2: CTTCTTGCTAAATACCAAGTTGGAG
goosecoid Mo1*: TGTCTGGAAGGTAATAGTCCATCTC
goosecoid Mo2: AGATCAGAGCTAACCACTTAGGACG
hnf6/onecut: Mo1: AGCCGCTGGACCTCAAACGCGAAGA
hnf6/onecut Mo2*: AAAATGATAATGTGGTCTCCGTCGC
hox7 Mo1: TGACGAAATACGAACTCGAACTCAT
hox7 Mo2: ACCACTTCATTAATAGCCAAAACCT
irxA Mo1: ATTGTGGATAACTGCTCGTCGTCAT
irxA Mo2: TTGTTGAAATCAACTTTGAGACGAT
Lefty Mo1: GGAGCGCCATGAGATAATTCCATAT
Lefty Mo2: GGAGATGGGCAAAATATGAAGATAC
msx Mo1: CGACTTGATGGAAGAAAATTATTCC
msx Mo2 : TTATCGCTTTAAGAATGACCAAGGA
NK1 Mo1: AAGCATTGAGAATCCCTAAAACTGC
NK1 Mo2: CATGTGCTCTGTTCAGACGGTCAAC
nk2.2 Mo1: ATCAACATTCATACGATGTCTCTAT
nk2.2 Mo2: ATAGTTAATTCCACACCACCCACTT
nodal Mo1*: ACTTTGCGACTTTAGCTAATGATGC
nodal Mo2: ATGAGAAGAGTTGCTCCGATGGTTG
tbx2/3 Mo1: TCGACGAACCACCAAATCTTGAGCA
tbx2/3 Mo2* : TCGGCAAAAGCCTCCGAGTCCAAAT
Oasis Mo1: CTCTTCACCTAAAAGCCCATCCATG
Oasis Mo2: CCAATTTGGGCCGTAGTCGAGGGAC
soxB1 Mo 1*: GACAGTCTCTTTGAAATTAGACGAC
soxB1 Mo2: GAAATAAAGCCAAAGTCTTTTGATG
univin Mo1*: ACGTCCATATTTAGCTCGTGTTTGT
univin Mo2: GTTAAACTCACCTTTCTAAACTCAC
wnt8 Mo1: GAACAACTGCCGTAAAGATATCCAT
wnt8 Mo2*: AACAGTCCAAATATGAAGTTCAAAC
As a control for defects related to injection and egg quality, we used morpholinos directed against the hatching enzyme gene: 5′-GCAATATCAAGCCAGAATTCGCCAT-3′ or against the Nemo like kinase transcript -5′-TCGGAGGCAGACCAGCAGCGAGAAA-3′. Embryos injected with either of these morpholinos at 1mM normally develop into pluteus larvae. Morpholinos oligonucleotides were dissolved in sterile water and injected at the one-cell stage together with Tetramethyl Rhodamine Dextran (10000 MW) at 5 mg/ml. For each morpholino a dose-response curve was obtained and a concentration at which the oligomer did not elicit non-specific defect was chosen. Approximately 2–4 pl of oligonucleotide solution at 0.5 mM were used in most of the experiments described here. For morphological observations, about 150–200 eggs were injected in each experiment. To analyze gene expression in the morphants a minimum of 50–75 injected embryos were hybridized with a given probe. All the experiments were repeated at least twice and only representative phenotypes observed in more than 80% of embryos are presented.
Supporting Information
Zdroje
1. ChristiaenL
DavidsonB
KawashimaT
PowellW
NollaH
2008 The transcription/migration interface in heart precursors of Ciona intestinalis. Science 320 1349 1352
2. ImaiKS
LevineM
SatohN
SatouY
2006 Regulatory blueprint for a chordate embryo. Science 312 1183 1187
3. ImaiKS
StolfiA
LevineM
SatouY
2009 Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136 285 293
4. OliveriP
DavidsonEH
2004 Gene regulatory network controlling embryonic specification in the sea urchin. Curr Opin Genet Dev 14 351 360
5. LevineM
DavidsonEH
2005 Gene regulatory networks for development. Proc Natl Acad Sci U S A 102 4936 4942
6. StathopoulosA
LevineM
2005 Genomic regulatory networks and animal development. Dev Cell 9 449 462
7. DavidsonEH
RastJP
OliveriP
RansickA
CalestaniC
2002 A genomic regulatory network for development. Science 295 1669 1678
8. OliveriP
DavidsonEH
2004 Gene regulatory network analysis in sea urchin embryos. Methods Cell Biol 74 775 794
9. SodergrenE
WeinstockGM
DavidsonEH
CameronRA
GibbsRA
2006 The genome of the sea urchin Strongylocentrotus purpuratus. Science 314 941 952
10. AngererLM
AngererRC
2003 Patterning the sea urchin embryo: gene regulatory networks, signaling pathways, and cellular interactions. Curr Top Dev Biol 53 159 198
11. DavidsonEH
RastJP
OliveriP
RansickA
CalestaniC
2002 A genomic regulatory network for development. Science 295 1669 1678
12. DubocV
LaprazF
BesnardeauL
LepageT
2008 Lefty acts as an essential modulator of Nodal activity during sea urchin oral-aboral axis formation. Dev Biol 320 49 59
13. DubocV
RottingerE
BesnardeauL
LepageT
2004 Nodal and BMP2/4 signaling organizes the oral-aboral axis of the sea urchin embryo. Dev Cell 6 397 410
14. RangeR
LaprazF
QuirinM
MarroS
BesnardeauL
2007 Cis-regulatory analysis of nodal and maternal control of dorsal-ventral axis formation by Univin, a TGF-β related to Vg1. Development 134 3649 3664
15. AngererLM
OleksynDW
LoganCY
McClayDR
DaleL
2000 A BMP pathway regulates cell fate allocation along the sea urchin animal-vegetal embryonic axis. Development 127 1105 1114
16. CzihakG
1963 Entwicklungsphysiologische Untersuchungen an Echininiden (Verteilung und bedeutung der Cytochomoxydase). Whilhem roux's Archiv EntwickMechOrg 154 272 292
17. ChildCM
1948 Exogastrulation by Sodium Azide and other inhibiting conditions in Strongylocentrotus purpuratus. J Exp Zool 107 1 38
18. PeaseDC
1941 Echinoderm bilateral determination in chemical concentration gradients I. The effects of cyanide, fericyanide, iodoacetate, picrate,dinitrophenol,urethane,iodine, malonate, etc. J Exp Zool 86 381 405
19. CoffmanJA
DavidsonEH
2001 Oral-aboral axis specification in the sea urchin embryo. I. Axis entrainment by respiratory asymmetry. Dev Biol 230 18 28
20. CoffmanJA
McCarthyJJ
Dickey-SimsC
RobertsonAJ
2004 Oral-aboral axis specification in the sea urchin embryo II. Mitochondrial distribution and redox state contribute to establishing polarity in Strongylocentrotus purpuratus. Dev Biol 273 160 171
21. CoffmanJA
ColuccioA
PlanchartA
RobertsonAJ
2009 Oral-aboral axis specification in the sea urchin embryo III. Role of mitochondrial redox signaling via H2O2. Dev Biol 330 123 130
22. BradhamCA
McClayDR
2006 p38 MAPK is essential for secondary axis specification and patterning in sea urchin embryos. Development 133 21 32
23. HardinJ
CoffmanJA
BlackSD
McClayDR
1992 Commitment along the dorsoventral axis of the sea urchin embryo is altered in response to NiCl2. Development 116 671 685
24. AnderssonO
ReissmannE
JornvallH
IbanezCF
2006 Synergistic interaction between Gdf1 and Nodal during anterior axis development. Dev Biol 293 370 381
25. TanakaC
SakumaR
NakamuraT
HamadaH
SaijohY
2007 Long-range action of Nodal requires interaction with GDF1. Genes Dev 21 3272 3282
26. LaprazF
BesnardeauL
LepageT
2009 Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. PLoS Biol 7 e1000248 doi:10.1371/journal.pbio.1000248
27. YaguchiS
YaguchiJ
BurkeRD
2006 Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development 133 2337 2346
28. WeiZ
YaguchiJ
YaguchiS
AngererRC
AngererLM
2009 The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development 136 1179 1189
29. SuYH
LiE
GeissGK
LongabaughWJ
KramerA
2009 A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev Biol 329 410 421
30. Howard-AshbyM
MaternaSC
BrownCT
ChenL
CameronRA
2006 Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol 300 90 107
31. Howard-AshbyM
MaternaSC
BrownCT
ChenL
CameronRA
2006 Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev Biol 300 74 89
32. MaternaSC
Howard-AshbyM
GrayRF
DavidsonEH
2006 The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol 300 108 120
33. RizzoF
Fernandez-SerraM
SquarzoniP
ArchimandritisA
ArnoneMI
2006 Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev Biol 300 35 48
34. TuQ
BrownCT
DavidsonEH
OliveriP
2006 Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol
35. AngererLM
OleksynDW
LevineAM
LiX
KleinWH
2001 Sea urchin goosecoid function links fate specification along the animal - vegetal and oral-aboral embryonic axes. Development 128 4393 4404
36. LaprazF
RottingerE
DubocV
RangeR
DuloquinL
2006 RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol 300 132 152
37. McCoonPE
AngererRC
AngererLM
1996 SpFGFR, a new member of the fibroblast growth factor receptor family, is developmentally regulated during early sea urchin development. J Biol Chem 271 20119 20125
38. BradhamCA
OikonomouC
KühnA
CoreAB
ModellJW
2009 Chordin is required for neural but not axial development in sea urchin embryos. Developmental Biology 328 221 233
39. PoustkaAJ
KuhnA
GrothD
WeiseV
YaguchiS
2007 A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol 8 R85
40. CroceJ
LhomondG
GacheC
2003 Coquillette, a sea urchin T-box gene of the Tbx2 subfamily, is expressed asymmetrically along the oral-aboral axis of the embryo and is involved in skeletogenesis. Mech Dev 120 561 572
41. GrossJM
PetersonRE
WuSY
McClayDR
2003 LvTbx2/3: a T-box family transcription factor involved in formation of the oral/aboral axis of the sea urchin embryo. Development 130 1989 1999
42. OliveriP
WaltonKD
DavidsonEH
McClayDR
2006 Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development 133 4173 4181
43. CroceJ
LhomondG
GacheC
2001 Expression pattern of Brachyury in the embryo of the sea urchin Paracentrotus lividus. Dev Genes Evol 211 617 619
44. GrossJM
McClayDR
2001 The Role of Brachyury (T) during Gastrulation Movements in the Sea Urchin Lytechinus variegatus. Dev Biol 239 132 147
45. MinokawaT
RastJP
Arenas-MenaC
FrancoCB
DavidsonEH
2004 Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns 4 449 456
46. OtimO
AmoreG
MinokawaT
McClayDR
DavidsonEH
2004 SpHnf6, a transcription factor that executes multiple functions in sea urchin embryogenesis. Dev Biol 273 226 243
47. PoustkaAJ
KuhnA
RadosavljevicV
WellenreutherR
LehrachH
2004 On the origin of the chordate central nervous system: expression of onecut in the sea urchin embryo. Evol Dev 6 227 236
48. RottingerE
SaudemontA
DubocV
BesnardeauL
McClayD
2008 FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development 135 353 365
49. AmoreG
YavrouianRG
PetersonKJ
RansickA
McClayDR
2003 Spdeadringer, a sea urchin embryo gene required separately in skeletogenic and oral ectoderm gene regulatory networks. Dev Biol 261 55 81
50. LiX
ChuangCK
MaoCA
AngererLM
KleinWH
1997 Two Otx proteins generated from multiple transcripts of a single gene in Strongylocentrotus purpuratus. Dev Biol 187 253 266
51. HwangSP
PartinJS
LennarzWJ
1994 Characterization of a homolog of human bone morphogenetic protein 1 in the embryo of the sea urchin, Strongylocentrotus purpuratus. Development 120 559 568
52. RottingerE
BesnardeauL
LepageT
2006 Expression pattern of three putative RNA-binding proteins during early development of the sea urchin Paracentrotus lividus. Gene Expr Patterns 6 864 872
53. WikramanayakeAH
PetersonR
ChenJ
HuangL
BinceJM
2004 Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis 39 194 205
54. Di BernardoMG
GianguzzaF
CiaccioM
PallaF
ColomboP
1989 Nucleotide sequence of a full length cDNA clone encoding for beta-tubulin of the sea urchin Paracentrotus lividus. Nucleic Acids Res 17 5851
55. RottingerE
CroceJ
LhomondG
BesnardeauL
GacheC
2006 Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development 133 4341 4353
56. DuloquinL
LhomondG
GacheC
2007 Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134 2293 2302
57. ManuelM
MartynogaB
YuT
WestJD
MasonJO
2010 The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development 137 487 497
58. MartynogaB
MorrisonH
PriceDJ
MasonJO
2005 Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev Biol 283 113 127
59. KennyAP
KozlowskiD
OleksynDW
AngererLM
AngererRC
1999 SpSoxB1, a maternally encoded transcription factor asymmetrically distributed among early sea urchin blastomeres. Development 126 5473 5483
60. MiyagiS
KatoH
OkudaA
2009 Role of SoxB1 transcription factors in development. Cell Mol Life Sci 66 3675 3684
61. SakumaR
Ohnishi YiY
MenoC
FujiiH
JuanH
2002 Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells 7 401 412
62. InmanGJ
NicolasFJ
CallahanJF
HarlingJD
GasterLM
2002 SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol 62 65 74
63. DirksenML
JamrichM
1992 A novel, activin-inducible, blastopore lip-specific gene of Xenopus laevis contains a fork head DNA-binding domain. Genes Dev 6 599 608
64. KnochelS
LefJ
ClementJ
KlockeB
HilleS
1992 Activin A induced expression of a fork head related gene in posterior chordamesoderm (notochord) of Xenopus laevis embryos. Mech Dev 38 157 165
65. Ruiz i AltabaA
JessellTM
1992 Pintallavis, a gene expressed in the organizer and midline cells of frog embryos: involvement in development of the neural axis. Development 116 81 93
66. BradhamC
McClayDR
2006 p38 MAPK in development and cancer. Cell Cycle 5 824 828
67. Jafar-NejadH
AcarM
NoloR
LacinH
PanH
2003 Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev 17 2966 2978
68. WallisD
HamblenM
ZhouY
VenkenKJ
SchumacherA
2003 The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development 130 221 232
69. SuzukiA
KanekoE
MaedaJ
UenoN
1997 Mesoderm induction by BMP-4 and -7 heterodimers. Biochem Biophys Res Commun 232 153 156
70. ShimmiO
UmulisD
OthmerH
O'ConnorMB
2005 Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell 120 873 886
71. OnaiT
YuJK
BlitzIL
ChoKW
HollandLZ
2010 Opposing Nodal/Vg1 and BMP signals mediate axial patterning in embryos of the basal chordate amphioxus. Dev Biol 344 1 377 89
72. DeRobertisEM
2004 Goosecoid and gastrulation.
SternCD
Gastrulation, from cells to embryo London Cold Spring Harbor Laboratory press 581 589
73. MartindaleMQ
HejnolA
2009 A developmental perspective: changes in the position of the blastopore during bilaterian evolution. Dev Cell 17 162 174
74. De RobertisEM
BlumM
NiehrsC
SteinbeisserH
1992 Goosecoid and the organizer. Dev Suppl 167 171
75. OliveriP
TuQ
DavidsonEH
2008 Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci U S A 105 5955 5962
76. SharmaT
EttensohnCA
2010 Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development 137 1149 1157
77. AffolterM
MartyT
ViganoMA
JazwinskaA
2001 Nuclear interpretation of Dpp signaling in Drosophila. EMBO J 20 3298 3305
78. VerschuerenK
RemacleJE
CollartC
KraftH
BakerBS
1999 SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem 274 20489 20498
79. BehestiH
HoltJK
SowdenJC
2006 The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev Biol 6 62
80. LoweCJ
TerasakiM
WuM
FreemanRMJr
RunftL
2006 Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol 4 e291 doi:10.1371/journal.pbio.0040291
81. HeM
WenL
CampbellCE
WuJY
RaoY
1999 Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proc Natl Acad Sci U S A 96 10212 10217
82. CarreiraS
DexterTJ
YavuzerU
EastyDJ
GodingCR
1998 Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol 18 5099 5108
83. Gomez-SkarmetaJ
de La Calle-MustienesE
ModolellJ
2001 The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and downregulates Bmp4. Development 128 551 560
84. ItohM
KudohT
DedekianM
KimCH
ChitnisAB
2002 A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development 129 2317 2327
85. Gomez-SkarmetaJL
ModolellJ
2002 Iroquois genes: genomic organization and function in vertebrate neural development. Curr Opin Genet Dev 12 403 408
86. NamJ
SuYH
LeePY
RobertsonAJ
CoffmanJA
2007 Cis-regulatory control of the nodal gene, initiator of the sea urchin oral ectoderm gene network. Dev Biol 306 860 869
87. LeleZ
NowakM
HammerschmidtM
2001 Zebrafish admp is required to restrict the size of the organizer and to promote posterior and ventral development. Dev Dyn 222 681 687
88. WillotV
MathieuJ
LuY
SchmidB
SidiS
2002 Cooperative action of ADMP - and BMP-mediated pathways in regulating cell fates in the zebrafish gastrula. Dev Biol 241 59 78
89. MoosMJr
WangS
KrinksM
1995 Anti-dorsalizing morphogenetic protein is a novel TGF-beta homolog expressed in the Spemann organizer. Development 121 4293 4301
90. ReversadeB
De RobertisEM
2005 Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell 123 1147 1160
91. HorstadiusS
1973 Experimental Embryology of Echinoderms Oxford Clarendon Press
92. YaguchiS
YaguchiJ
AngererRC
AngererLM
2008 A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell 14 97 107
93. Ben-HaimN
LuC
Guzman-AyalaM
PescatoreL
MesnardD
2006 The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 11 313 323
94. RottingerE
BesnardeauL
LepageT
2004 A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development 131 1075 1087
95. FeldmanB
DouganST
SchierAF
TalbotWS
2000 Nodal-related signals establish mesendodermal fate and trunk neural identity in zebrafish [In Process Citation]. Curr Biol 10 531 534
96. SchierAF
2001 Axis formation and patterning in zebrafish. Curr Opin Genet Dev 11 393 404
97. CamusA
Perea-GomezA
MoreauA
CollignonJ
2006 Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol 295 743 755
98. CameronRA
BrittenRJ
DavidsonEH
1993 The embryonic ciliated band of the sea urchin, Strongylocentrotus purpuratus derives from both oral and aboral ectoderm. Dev Biol 160 369 376
99. SummertonJ
1999 Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta 1489 141 158
100. MoultonJD
YanYL
2008 Using Morpholinos to control gene expression. Curr Protoc Mol Biol Chapter 26 Unit 26 28
101. WilsonSI
EdlundT
2001 Neural induction: toward a unifying mechanism. Nat Neurosci 4 Suppl 1161 1168
102. ChangC
HarlandRM
2007 Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development 134 3861 3872
103. Di-GregorioA
SanchoM
StuckeyDW
CromptonLA
GodwinJ
2007 BMP signalling inhibits premature neural differentiation in the mouse embryo. Development 134 3359 3369
104. AlvarezIS
AraujoM
NietoMA
1998 Neural induction in whole chick embryo cultures by FGF. Dev Biol 199 42 54
105. StreitA
BerlinerAJ
PapanayotouC
SirulnikA
SternCD
2000 Initiation of neural induction by FGF signalling before gastrulation. Nature 406 74 78
106. WilsonSI
GrazianoE
HarlandR
JessellTM
EdlundT
2000 An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr Biol 10 421 429
107. BertrandV
HudsonC
CaillolD
PopoviciC
LemaireP
2003 Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell 115 615 627
108. HudsonC
DarrasS
CaillolD
YasuoH
LemaireP
2003 A conserved role for the MEK signalling pathway in neural tissue specification and posteriorisation in the invertebrate chordate, the ascidian Ciona intestinalis. Development 130 147 159
109. HudsonC
LemaireP
2001 Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev 100 189 203
110. DarrasS
NishidaH
2001 The BMP/CHORDIN antagonism controls sensory pigment cell specification and differentiation in the ascidian embryo. Dev Biol 236 271 288
111. HudsonC
YasuoH
2005 Patterning across the ascidian neural plate by lateral Nodal signalling sources. Development 132 1199 1210
112. Mieko MizutaniC
BierE
2008 EvoD/Vo: the origins of BMP signalling in the neuroectoderm. Nat Rev Genet 9 663 677
113. DenesAS
JekelyG
SteinmetzPR
RaibleF
SnymanH
2007 Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129 277 288
114. HudsonC
YasuoH
2006 A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development 133 2855 2864
115. SampathK
RubinsteinAL
ChengAM
LiangJO
FekanyK
1998 Induction of the zebrafish ventral brain and floorplate requires cyclops/nodal signalling. Nature 395 185 189
116. LepageT
GacheC
1989 Purification and characterization of the sea urchin embryo hatching enzyme. J Biol Chem 264 4787 4793
117. LepageT
GacheC
1990 Early expression of a collagenase-like hatching enzyme gene in the sea urchin embryo. Embo J 9 3003 3012
118. HarlandRM
1991 In situ hybridization: an improved whole mount method for Xenopus embryos.
KayBK
PengHJ
Methods in Cell Biology San Diego, Calif. Academic Press Inc. 685 695
119. ThisseB
HeyerV
LuxA
AlunniV
DegraveA
2004 Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol 77 505 519
120. TurnerDL
WeintraubH
1994 Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev 8 1434 1447
121. EisenJS
SmithJC
2008 Controlling morpholino experiments: don't stop making antisense. Development 135 1735 1743
122. KennyAP
OleksynDW
NewmanLA
AngererRC
AngererLM
2003 Tight regulation of SpSoxB factors is required for patterning and morphogenesis in sea urchin embryos. Dev Biol 261 412 425
123. LongabaughWJ
DavidsonEH
BolouriH
2005 Computational representation of developmental genetic regulatory networks. Dev Biol 283 1 16
124. LongabaughWJ
DavidsonEH
BolouriH
2009 Visualization, documentation, analysis, and communication of large-scale gene regulatory networks. Biochim Biophys Acta 1789 363 374
125. StenzelP
AngererLM
SmithBJ
AngererRC
ValeWW
1994 The univin gene encodes a member of the transforming growth factor-beta superfamily with restricted expression in the sea urchin embryo. Dev Biol 166 149 158
126. DobiasSL
MaL
WuH
BellJR
MaxsonR
1997 The evolution of Msx gene function: expression and regulation of a sea urchin Msx class homeobox gene. Mech Dev 61 37 48
127. CroceJC
McClayDR
2006 The canonical Wnt pathway in embryonic axis polarity. Semin Cell Dev Biol 17 168 174
128. AngererLM
DoleckiGJ
GagnonML
LumR
WangG
1989 Progressively restricted expression of a homeo box gene within the aboral ectoderm of developing sea urchin embryos. Genes Dev 3 370 383
129. CoxKH
AngererLM
LeeJJ
DavidsonEH
AngererRC
1986 Cell lineage-specific programs of expression of multiple actin genes during sea urchin embryogenesis. J Mol Biol 188 159 172
130. YangQ
AngererLM
AngererRC
1989 Unusual pattern of accumulation of mRNA encoding EGF-related protein in sea urchin embryos. Science 246 806 808
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



