-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
Transgenic crops producing insecticidal toxins from Bacillus thuringiensis (Bt) are commercially successful in reducing pest damage, yet knowledge of resistance mechanisms that threaten their sustainability is incomplete. Insect resistance to the pore-forming Cry1Ac toxin is correlated with the loss of high-affinity, irreversible binding to the mid-gut membrane, but the genetic factors responsible for this change have been elusive. Mutations in a 12-cadherin-domain protein confer some Cry1Ac resistance but do not block this toxin binding in in vitro assays. We sought to identify mutations in other genes that might be responsible for the loss of binding. We employed a map-based cloning approach using a series of backcrosses with 1,060 progeny to identify a resistance gene in the cotton pest Heliothis virescens that segregated independently from the cadherin mutation. We found an inactivating mutation of the ABC transporter ABCC2 that is genetically linked to Cry1Ac resistance and is correlated with loss of Cry1Ac binding to membrane vesicles. ABC proteins are integral membrane proteins with many functions, including export of toxic molecules from the cell, but have not been implicated in the mode of action of Bt toxins before. The reduction in toxin binding due to the inactivating mutation suggests that ABCC2 is involved in membrane integration of the toxin pore. Our findings suggest that ABC proteins may play a key role in the mode of action of Bt toxins and that ABC protein mutations can confer high levels of resistance that could threaten the continued utilization of Bt–expressing crops. However, such mutations may impose a physiological cost on resistant insects, by reducing export of other toxins such as plant secondary compounds from the cell. This weakness could be exploited to manage this mechanism of Bt resistance in the field.
Published in the journal: . PLoS Genet 6(12): e32767. doi:10.1371/journal.pgen.1001248
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001248Summary
Transgenic crops producing insecticidal toxins from Bacillus thuringiensis (Bt) are commercially successful in reducing pest damage, yet knowledge of resistance mechanisms that threaten their sustainability is incomplete. Insect resistance to the pore-forming Cry1Ac toxin is correlated with the loss of high-affinity, irreversible binding to the mid-gut membrane, but the genetic factors responsible for this change have been elusive. Mutations in a 12-cadherin-domain protein confer some Cry1Ac resistance but do not block this toxin binding in in vitro assays. We sought to identify mutations in other genes that might be responsible for the loss of binding. We employed a map-based cloning approach using a series of backcrosses with 1,060 progeny to identify a resistance gene in the cotton pest Heliothis virescens that segregated independently from the cadherin mutation. We found an inactivating mutation of the ABC transporter ABCC2 that is genetically linked to Cry1Ac resistance and is correlated with loss of Cry1Ac binding to membrane vesicles. ABC proteins are integral membrane proteins with many functions, including export of toxic molecules from the cell, but have not been implicated in the mode of action of Bt toxins before. The reduction in toxin binding due to the inactivating mutation suggests that ABCC2 is involved in membrane integration of the toxin pore. Our findings suggest that ABC proteins may play a key role in the mode of action of Bt toxins and that ABC protein mutations can confer high levels of resistance that could threaten the continued utilization of Bt–expressing crops. However, such mutations may impose a physiological cost on resistant insects, by reducing export of other toxins such as plant secondary compounds from the cell. This weakness could be exploited to manage this mechanism of Bt resistance in the field.
Introduction
Insecticidal protein toxins of the Cry1A family produced by certain strains of the gram-positive bacterium Bacillus thuringiensis (Bt) are highly active against many Lepidoptera but nontoxic to most other animal species. Transgenic cotton producing Cry1Ac and transgenic maize producing Cry1Ab have been grown commercially since 1996 and offer protection against some major pests, including species in the genera Heliothis, Helicoverpa, Ostrinia, and Pectinophora [1], [2]. After ingestion and solubilization in the alkaline midgut lumen of the caterpillar, the protoxin is cleaved by digestive proteases to yield an active 60 kDa toxin which interacts with high-affinity binding sites on the brush border epithelium, eventually oligomerizing to form a transmembrane pore, leading to lysis of epithelial cells [3], [4]. Additional mechanisms of toxicity involving an adenylyl cyclase/PKA signaling pathway have also been described [5]. High toxin concentrations are lethal, lower toxin concentrations inhibit larval growth in a dose-dependent manner. The binding targets are critical in determining the range of species on which the toxin is active [6], and reduction or loss of binding is an important mechanism of genetically based resistance in the target pest species [7].
The most common type of Bt toxin resistance (“Mode 1”) [8] which has evolved in field populations of Plutella xylostella in response to sprays of formulated Bt toxins [9] and in laboratory-selected strains of other Lepidoptera [10]–[12] is characterized by recessive inheritance, >500-fold resistance to at least one Cry1A toxin, much less resistance to Cry1C, and greatly reduced binding of Cry1A toxins to target sites in the midgut membrane. Several cases of resistance to Bt crops in field populations of insect pests have also been reported, but the genetic basis of resistance has not been identified in any of these cases [2], [13], [14]. Genetic mutations linked to Cry1A resistance have been identified in laboratory strains, but their role in Mode 1 resistance is still not fully understood. The mutations most commonly found in Cry1A-resistant strains inactivate a gene encoding a 12-cadherin-domain protein of about 1750 amino acids, expressed in the larval midgut [15]–[17]. These mutations confer resistance to Cry1A toxins including Cry1Ac, but do not block irreversible Cry1Ac binding to midgut membranes as measured by in vitro assays [12], [18], [19]. Conversely, Mode 1 resistance in the NO-QA strain of P. xylostella which includes loss of Cry1Ac binding [20] is determined by a single gene that segregates independently from the 12-cadherin-domain protein gene [21].
What could account for the apparent independence of resistance-conferring cadherin mutations and resistance-conferring loss of irreversible membrane binding? There is evidence for a multi-step mechanism that could offer an explanation. Bravo et al. [22] have proposed that activated Cry1A toxin monomers first bind to an extracellular membrane-proximal domain of the 12-cadherin-domain protein. The toxin undergoes a conformational change, facilitating proteolytic cleavage of the Domain I helix α1 from the toxin N-terminus by a yet-uncharacterized protease. The resulting “clipped” toxin monomers subsequently assemble into a oligomeric pre-pore structure in solution, which binds reversibly to several other membrane-bound proteins, and finally inserts irreversibly into the membrane [22]. Thus absence of the cadherin protein in resistant strains would slow the rate of monomer clipping and oligomerization of the active pore structure, but not directly affect the subsequent irreversible binding and insertion of the pore into the membrane. This would predict that even higher levels of resistance could be attained by interfering with the later binding steps in this sequential binding model. To test this idea, it would be useful to examine toxin binding to membranes of resistant strains, with or without the cadherin protein.
The first Bt-resistant cadherin mutation was identified [17] as the resistant allele 4r of the previously-mapped gene BtR-4 [23] in the YHD2 strain of the cotton pest Heliothis virescens. This strain had evolved >10,000 fold resistance in response to laboratory selection by diet-incorporated Cry1Ab and Cry1Ac toxin over four years [10]. Insertion of an LTR retrotransposon into the coding sequence of the 12-cadherin-domain protein defines the 4r allele, and resulted in a truncated 622-amino acid protein lacking the last 7 cadherin domains, membrane-proximal toxin binding region, transmembrane domain, and cytoplasmic domain. The absence of the 12-cadherin-domain protein from the midgut membranes of YHD2 was confirmed with antibodies [19]. The first binding measurements on homozygous resistant 4r4r YHD2 published in 1995 showed greatly reduced binding of midgut epithelial brush border membrane vesicles (BBMV) to Cry1Aa, but surprisingly no reduction in Cry1Ab or Cry1Ac binding [18]. YHD2 was subsequently selected to even higher levels of resistance, and later studies published in 2002 and 2004 showed a loss of membrane binding by Cry1Ab and Cry1Ac also, as well as a reduction in their pore-forming ability [19], [24]. This suggested the existence of a second gene (which we named BtR-6) with a mutant allele 6r in the more resistant strain responsible for its increased resistance and decreased binding affinity to Cry1Ac.
In order to test the hypothesis that a separate mechanism affecting later steps in toxin binding existed in this more resistant strain, we sought to identify BtR-6 and the nature of the 6r allele by map-based cloning. We first isolated the two resistance mechanisms into separate strains and characterized their toxin-binding properties. We then used these strains in a series of backcrosses that were assayed for resistance using a sublethal, growth-inhibition bioassay. Fine-scale linkage mapping identified a cluster of ABC transporter genes, one of which showed an inactivating mutation in the most resistant strain. This implicates the ABC transporter family for the first time in the mode of action of Bt Cry1A toxins, and offers an explanation for Mode 1 resistance that is compatible with the sequential binding model.
Results
Construction of Strains
In order to synthesize strains that were homozygous for different combinations of resistant and susceptible alleles at the BtR-4 and BtR-6 loci (4r vs 4s, 6r vs 6s), we used a combination of progeny testing on Cry1Ac-containing diet and marker-assisted selection of parents with a PCR (polymerase chain reaction) test diagnostic for 4r [25]. These strains were then maintained on artificial diet containing the highest concentration of Cry1Ac that would allow the same larval growth rate as toxin-free diet. Strain YHD3 was homozygous resistant for both genes (4r4r/6r6r), had a resistance level similar to the newer YHD2, and was reared on 200 µg/ml Cry1Ac. YFO was 4r4r/6s6s and could be reared on at most 5 µg/ml Cry1Ac. YEE was 4s4s/6r6r and was reared on 50 µg/ml Cry1Ac. Fully susceptible strains CNW and JEN were 4s4s/6s6s and were reared on toxin-free diet; their growth rate was reduced 50% by only 0.064 µg/ml Cry1Ac.
Toxin Binding Measurements
Qualitative in vitro binding studies with Cry1Aa, Cry1Ab, and Cry1Ac using BBMV (Figure 1) showed that the doubly homozygous susceptible JEN strain (4s4s/6s6s) bound all three toxins as expected. The doubly homozygous resistant YHD3 strain (4r4r/6r6r) bound to none of the three, similar to YHD2 in 2002 [24] and 2004 [19]. The two intermediately resistant strains showed a complementary pattern: YFO (4r4r/6s6s, the hypothesized genotype of the older YHD2 strain) had lost only the ability to bind Cry1Aa, similar to YHD2 in 1995 [18]. YEE (4s4s/6r6r) still bound Cry1Aa but failed to bind Cry1Ab and Cry1Ac, a pattern that has not been previously reported (Figure 1). Thus homozygosity for the 6r allele but not 4r is correlated with loss of Cry1Ac binding.
Fig. 1. Cry1A toxin binding to membrane vesicles. 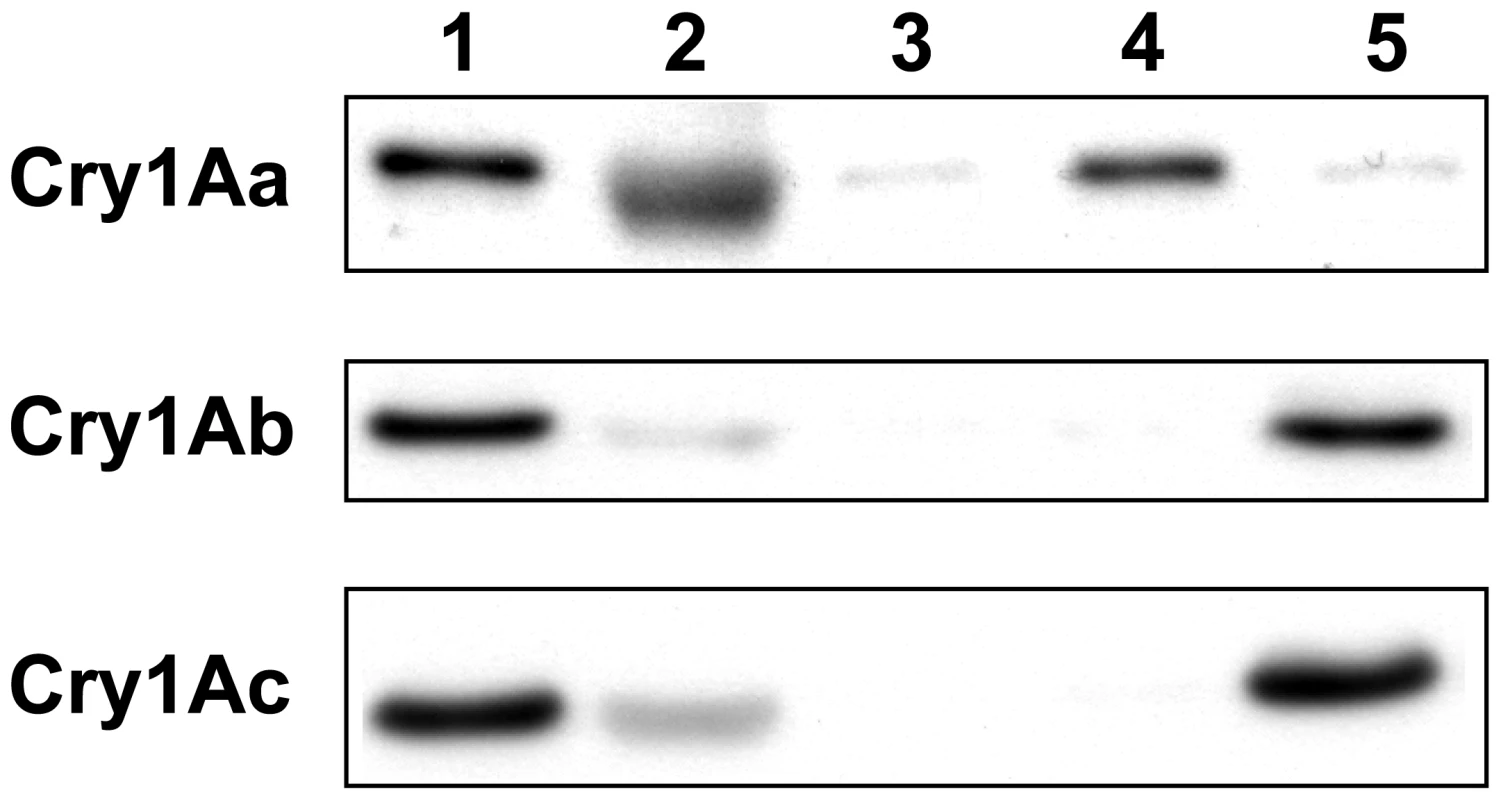
Qualitative binding of biotinylated Cry1Aa, Cry1Ab and Cry1Ac to brush border membrane vesicles (BBMV) of midguts from susceptible and resistant H. virescens strains. Biotinylated toxins (2.5 nM) were incubated with BBMV (20 µg protein) from the following strains: susceptible JEN (lane 1) and resistant YHD3 (lane 3), YEE (lane 4) and YFO (lane 5). BBMV were pelleted by centrifugation, and bound toxins were resolved by electrophoresis, blotted onto membranes and detected by a chemiluminescent-coupled streptavidin probe. Binding specificity was assessed by incubating biotinylated toxin with BBMV from the susceptible strain JEN in the presence of a 200-fold excess of unlabeled toxin (lane 2). Linkage Mapping
We explored the genetic basis of these resistance and binding differences by linkage mapping using a larval growth bioassay with Cry1Ac conducted on backcrosses as done previously [17]. Backcrosses to YHD3 using F1 (YHD3 x YFO) mothers were first screened with a panel of RFLP markers to identify the linkage group containing BtR-6, by exploiting the absence of crossing-over during meiosis in female Lepidoptera. A probe previously mapped to linkage group 2 (LG2) with similarity to a microsomal glutathione transferase (GenBank HM150720) showed a highly significant association with resistance as measured by larval weight on the Cry1Ac diet. This confirmed that BtR-6 was genetically distinct from the two previously mapped resistance genes in this species, BtR-4 (the 12-cadherin-domain protein) on LG9 [17] and BtR-5 on LG10 [26]. Neither LG9 nor LG10 had a significant association with resistance in these crosses (in the backcross to YFO, all progeny are BtR-4r4r). The significant effect of LG2 was confirmed in backcrosses to YEE using F1 (YEE x YFO) mothers, which were also segregating at BtR-6. Ribosomal protein genes RpP0, RpS5, RpL8, RpL10A, and RpL30 also mapped to LG2 in H. virescens, indicating homology with Chromosome 15 (Chr15) of the domesticated silkmoth Bombyx mori, where these same genes had been mapped by recombinational [27] and cytogenetic [28] methods.
We localized BtR-6 relative to marker genes along LG2 using recombinational mapping in backcrosses with F1 males, which do undergo crossing-over during meiosis. In the first step, markers were chosen from H. virescens and Helicoverpa armigera cDNA clones homologous to genes that had been genetically mapped to Chr15 in B. mori. The second step at a finer scale used genes physically mapped to Chr15 after the assembled B. mori genome sequence was made available to the public in April 2008. The linkage map of LG2 in H. virescens was entirely collinear with the genetic and physical maps of Chr15 of B. mori (Figure 2). BtR-6 was localized within the interval between markers b7730 and b7793, showing zero recombinants out of a total of 1060 informative progeny from 3 sets of mapping families that had been reared on Cry1Ac-containing diet. The physical map of this region in B. mori contains 10 predicted genes, nine of which showed expression in B. mori larval midgut as indicated by microarray studies, and which also had homologs in cDNA libraries constructed from midgut tissue of larval H. armigera (Table S2).
Fig. 2. Linkage map of Heliothis virescens Linkage Group 2. 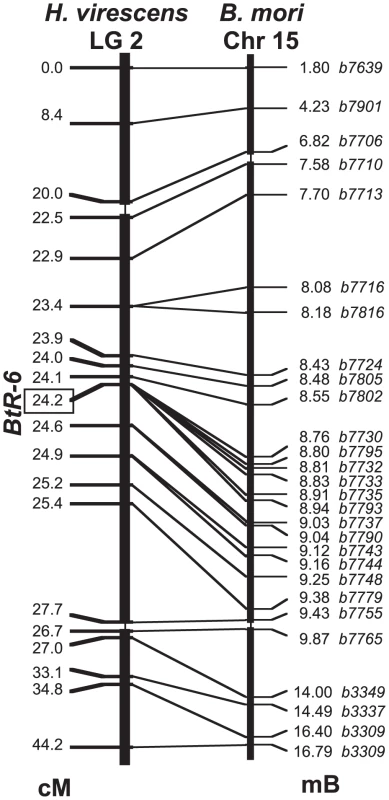
Markers were mapped in 1,060 offspring from 3 sets of backcross families (cM = Haldane centimorgans). The linkage map is compared with the physical map of homologs on Bombyx mori Chromosome 15 (Mb = megabases of DNA). The scale of the middle portion of both maps is magnified 10-fold. Sequence Analysis
A PCR product corresponding to B. mori predicted gene BGIBMGA007793 was amplified from H. virescens midgut cDNA, and used to screen BAC libraries of H. virescens and its sister species H. subflexa. The latter library yielded a positive clone which was sequenced (GenBank Accession No. GQ332573, Figure 3A), revealing a cluster of three genes with high sequence similarity to ABC transporters (ABCC1, ABCC2, and ABCC3), in the same orientation as the corresponding region in B. mori (Figure 3B). Genomic sequence comparison of the ABCC2 gene from YHD3 (GenBank GQ332572) and YFO (GenBank GQ332571) strains revealed a 22-bp deletion in exon 2 occurring only in YHD3 (Figure S2). The same deletion was found in RT-PCR products from YHD3 larval midgut cDNA. The frameshift generated by this deletion predicts a truncated 99-residue protein from YHD3 mRNA. In contrast, the full-length ABCC2 protein of 1339 amino acids predicted from the YFO or H. subflexa sequence (97% amino acid identity) has all the features of the bipartite structure of ABC transporters, with six transmembrane segments and a large cytoplasmic ATP-binding domain in each half [29] (Figure 4, Figure 5). A PCR assay using primers flanking the exon 2 deletion region was used to determine genotypes of individual backcross progeny (Figure S1). Only those larvae with two copies of the exon 2 deletion allele grew rapidly on Cry1Ac-containing diet. The deletion was present in all YHD3 and YEE individuals tested; no YFO or CNW individuals had the deletion. This 22-bp deletion is taken to define the 6r allele of BtR-6 in YHD3.
Fig. 3. ABCC2 genomic region. 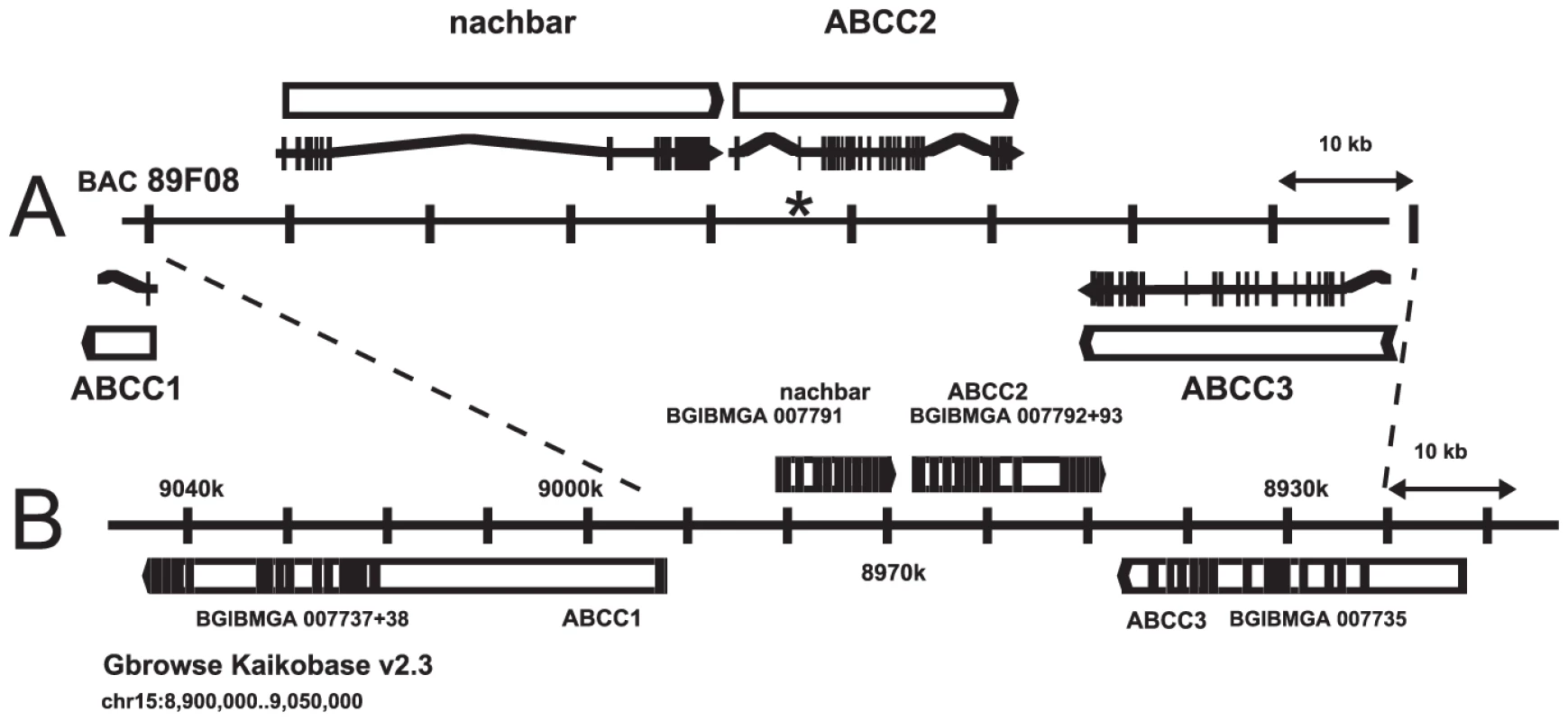
H. subflexa BAC clone 89F08 (A) and corresponding region from Chromosome 15 of B. mori with BGI protein predictions (B). The asterisk marks exon 2 of ABCC2 containing the 22 bp deletion characterizing the 6r allele of BtR-6 in H. virescens. Fig. 4. Sequence alignment of ABCC proteins from Lepidoptera, Drosophila, and mouse, Part 1. 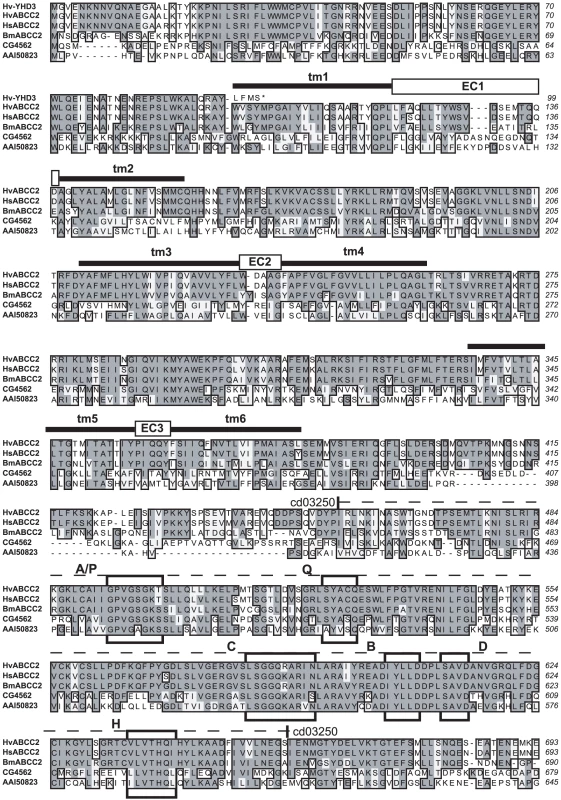
Hv-YHD3 is the predicted protein sequence of the YHD3 exon 2 frameshift mutant (GenBank GQ332572); HvABCC2 is from the YFO strain of Heliothis virescens (GQ332571); HsABCC2 is from Heliothis subflexa (GQ332573); BmABCC2 is from Bombyx mori; CG4562 is isoform A of CG4562 from Drosophila melanogaster (AAF44707); AAI50823 is ATP-binding cassette, sub-family C (CFTR/MRP) from Mus musculus (AAI50823). Sequence features determined for HvABCC2 include tm1 through tm12: transmembrane domains as predicted by Phobius [81]. EC1 through EC6: predicted extracellular loops. Dashed line, cd03250: ABCC_MRP_domain1, Domain 1 of the ABC subfamily C (E = 5e-77); dotted line, cd03244: ABCC_MRP_domain2, Domain 2 of the ABC subfamily C (E = 2e-95). Boxed regions are A/P: Walker A motif/P-loop; Q: Q-loop/lid; C: ABC transporter signature motif; B: Walker B motif; D: D-loop; H: H-loop/switch region [82]. Fig. 5. Sequence alignment of ABCC proteins from Lepidoptera, Drosophila, and mouse, Part 2. 
Sequences and features are described in the caption to Figure 4. Temporal Allele Frequency Correlations with Toxin Binding
We used PCR analysis of archival DNA samples to investigate whether an increase of the 6r allele frequency occurred concomitantly with the decrease in Cry1Ac binding affinity of YHD2 over the years. DNA from parents of YHD2 backcrosses conducted in March 1993 [23] yielded a 6r allele frequency estimate of 14% and an expected 2% frequency of 6r6r homozygotes; not high enough to appreciably reduce the binding to Cry1Ac [18]. Thus the 6r allele was present although rare in the YHD2 strain as early as 1993. When we screened for 6r in DNA that had been isolated in December 2002 from the YHD2 larvae whose BBMV showed a loss of Cry1Ab and Cry1Ac binding [19], we found that the frequency of 6r had increased to 100%. Thus a loss of Cry1Ab and Cry1Ac binding was correlated with an increase in 6r within YHD2 over approximately 100 generations while the Cry1Ac resistance level as measured by bioassay also increased. This correlation also extended to other strains. DNA samples from the Cry1Ac-resistant KCBhyb strain had a 6r frequency of 5%; membranes from these larvae retained Cry1Ab and Cry1Ac binding, and binding of Cry1Aa only was dependent on the BtR-4 genotype [19]. Both the Cry1Ac-resistant strain CxC with a 6r allele frequency of 0% and the Cry1Ac-susceptible strain YDK with 6% retained Cry1Aa, Cry1Ab and Cry1Ac binding [19] (Table S3).
Discussion
Recent research has shown that the mode of action of Bt toxins is more complex than originally envisioned. Cry toxins may induce cell death by interacting with the 12-cadherin-domain protein without forming pores [5]; responses to Cry toxins may involve intracellular signal transduction pathways that protect cells against pore forming toxins [30], [31]. Yet a major feature of Cry1A toxin action in Lepidoptera is the formation of pores in the plasma membrane leading to cell disruption by colloid-osmotic lysis [32]. At high enough concentrations, Cry toxins can eventually insert and form pores in planar lipid bilayer membranes devoid of any other protein [33], [34]. However, these toxins have evolved to interact with a series of host proteins in the midgut membrane to form pores much faster and at much lower concentrations. These interactions are toxin - and host-specific, e.g. Cry1A toxins are active against certain Lepidoptera, but not Diptera or Coleoptera. Interfering with one or more of these steps may confer resistance, such that higher concentrations of toxin are required to achieve the same mortality endpoint. Identifying the molecular changes that accompany resistance is a useful first step to posing hypotheses about the mode of toxin action. Based on the mapping results and binding correlations described here, we hypothesize that the ABCC2 protein participates in the mechanism of Cry1Ab and Cry1Ac toxicity by binding and facilitating insertion into the membrane, in an extension of the multi-step model of Bravo et al. [35].
In the first step of this model, reversible toxin binding to the 12-cadherin-domain protein accelerates the formation of clipped toxin monomers which are more competent to form the oligomeric pre-pore structure in solution. Evidence supporting this mechanism includes the enhanced toxicity of Cry1Ab or Cry1Ac toxin when fed to larvae along with a peptide fragment from the toxin-binding domain of the cadherin protein [36]. This fragment itself binds to Cry1Ab and Cry1Ac [36], and accelerates the rate of formation of a 250 kDa oligomer of Cry1Ac [37]. Additional evidence is provided by the elevated potency of “pre-clipped” Cry1Ab or Cry1Ac monomers generated by recombinant methods, which lack the α1 helix [38]. These modified Cry1AbMod and Cry1AcMod toxin monomers rapidly form oligomers in the absence of the cadherin, and are more potent than unmodified toxins against Cry1Ac-resistant Pectinophora gossypiella with cadherin mutations [38], although possessing similar properties in most other respects [39].
According to this model, absence of the 12-cadherin-domain protein confers a certain level of resistance to Cry1Ab or Cry1Ac by slowing down the formation of oligomers, not by stopping it completely. Evidently oligomerization of these two toxins can occur in the absence of cadherin binding, but at a slower rate; since higher concentrations of Cry1Ab or Cry1Ac are still capable of killing resistant insects with cadherin mutations. Moreover, even if the cadherin functions in accelerating the “clipping” of Cry1Ab and Cry1Ac toxin monomers, this does not rule out a separate role in additional binding events. The cadherin appears to be the major binding protein for Cry1Aa; as BBMV from strains lacking it have also lost their ability to bind Cry1Aa [18], [19]. Furthermore, presence of the cadherin appears to be necessary and sufficient for binding of BBMV to Cry1Aa, but not Cry1Ab or Cry1Ac (Figure 1). The 12-cadherin-domain protein from B. mori also binds to Cry1Aa [40], but experiments on the effect of the cadherin on oligomerization have not yet been conducted on Cry1Aa. Therefore, cadherin binding may play more than one role, depending on the toxin.
In the second binding step in the hypothesized mode of action [35], toxin oligomers bind to the soluble ectodomains of membrane-associated glycosylated proteins such as aminopeptidase N (APN) [41], [42], alkaline phosphatase [43], [44], P252 glycoprotein [45], or BTR-270 glycoprotein [46]. These proteins are GPI-anchored and enriched in lipid rafts, and disruption of lipid rafts by cholesterol depletion reduces pore formation by Cry1Ab [47]. Experimental cleavage of GPI anchors removes APN from the BBMV surface and reduces the amount of Cry1Ab toxin inserted into the membrane [22]. Massive shedding of GPI-anchored proteins by the action of endogenous phospholipase C has been shown to occur in response to toxin consumption [48], which might be a defense mechanism against the second binding step, but so far this has not been observed to occur in any resistant strains.
Toxin binding to these glycoproteins appears to be predominantly reversible; e.g. binding of Cry1Ac to purified APN exhibits measurable on - and off-kinetics by surface plasmon resonance [49]–[51]. No single glycoprotein appears to be essential for Cry1A toxicity; e.g. mutants of Cry1Ac which eliminate binding to a 115 kDa APN only result in a two-fold decrease in toxicity [52]. RNA interference directed against midgut APNs produces a measurable but slight decrease of toxicity [53], [54]. Therefore the main significance of Cry1A toxin binding to these glycoproteins seems to be to increase the concentration of the pre-pore oligomer at the membrane surface, increasing the probability of eventual insertion by some other mechanism.
The final binding step proposed here involves interactions of the oligomeric toxin pre-pore structure with the ABCC2 protein. ABC transporters cycle between closed and open configurations as they transport small molecules out of the cell, driven by binding and hydrolysis of ATP by the intracellular nucleotide-binding domains. A recently determined structure for the ABCB1 P-glycoprotein shows that in the closed configuration, the extracellular loops between the transmembrane domains completely cover the channel opening, resulting in a large internal cavity facing the cytoplasm [29]. In this pretransport state, the small molecule to be transported is located within the internal cavity. Binding of ATP by the two cytoplasmic nucleotide-binding domains causes their dimerization and a large conformational change resulting in the open state, in which several hydrophobic surfaces of the channel are transiently exposed to the outside of the cell while the small molecule is expelled [29]. Hydrolysis of the ATP restores the ABC protein to the closed configuration. We hypothesize that Cry1Ab and Cry1Ac toxins, as pre-formed oligomers or possibly also as monomers, bind to the open configuration of ABCC2 and that this facilitates subsequent membrane insertion. The resistance conferred by BtR-6r would thus be due to the absence of this binding site for Cry1Ab and Cry1Ac. Direct toxin binding assays with the membrane-integrated ABCC2 protein would be required for evaluation of this hypothesis.
To our knowledge, ABC transporters have not yet been suggested as binding targets for Bt toxins. Failure to detect them may be due to the under-representation or absence of integral membrane proteins in 1-D or 2-D gels used in ligand binding studies with labelled toxin [55], [56]. Failure to isolate them could be due to the general difficulty of isolating membrane proteins. The midgut proteins from Lepidoptera previously isolated on the basis of binding to Cry1A-toxin-immobilized affinity columns [42], immunoprecipitation [57], [58] or preparative gel electrophoresis [46] all have a large ectodomain projecting into the lumen available for binding, and are readily solubilized, being attached to the membrane by a GPI anchor or a single transmembrane domain. The predicted structure of ABCC2, however, presents only 6 small loops (of 19, 5, 5, 43, 5, and 5 residues respectively) projecting into the lumen, which connect the 6 α-helices of each of the two transmembrane domains buried in the lipid bilayer (Figure 4, Figure 5). Cry1A toxins are known to bind to carbohydrate residues of glycoproteins, but none of the 6 loops of ABCC2 have predicted glycosylation sites. If the toxin binds primarily to the hydrophobic interior of the channel, then methods stringent enough to solubilize the ABC protein would likely disrupt this interaction.
If confirmed, the role of an ABC transporter in Bt toxin action proposed here could have implications for the management of Cry1Ac resistance in field populations of H. virescens and other lepidopteran pests currently controlled by Bt-cotton or Bt-maize. We emphasize that as no attention has been paid to ABC transporters in Bt resistance previously, we do not know whether this or similar mutations occur in the field in H. virescens or any other species. However, the genetic basis of field-evolved resistance to Bt sprays by Plutella xylostella [21], [59] and Trichoplusia ni [60], and to Bt crops by Helicoverpa zea, Spodoptera frugiperda, and Busseola fusca [2] has not yet been identified, and these strains should be examined for ABC transporter mutations. We do not know whether H. virescens larvae homogozygous for the ABCC2 mutation can survive on cotton, with or without Cry1Ac toxin. Developmental arrest in the last larval instar of the YHD2 strain feeding on non-transgenic cotton was observed prior to 1993 [25], when BtR-4r was nearly fixed, indicating a strong fitness cost to the cadherin mutation; but at that time BtR-6r was still at a very low frequency. We do not know how ABCC2 mutations would respond to selection for Cry1A-toxin resistance in the field. In India, China, and many other countries, the predominant varieties of Bt-cotton still produce the single toxin Cry1Ac, thus selection for Cry1Ac resistance is strong. The Bt-cotton currently used in the USA and Australia produces Cry2Ab in addition to Cry1Ac; the different modes of action of these two toxins are thought to produce a “redundant killing” effect whereby selection for resistance to either single toxin is greatly weakened. However, we do not know whether ABC transporter mutations confer cross-resistance to Cry2Ab. The binding targets of Cry2Ab are unknown and ABC proteins have not yet been investigated as candidates. Moreover, Cry2Ab resistance is detectable using F2 screens in Australian populations of Helicoverpa armigera [61] and H. punctigera [62], and the molecular basis of the resistance mechanism involves binding site alterations in both species [63].
The biological function of ABCC2 is unknown, but its similarity to multidrug resistance proteins suggests that it could export small hydrophobic toxins from midgut epithelial cells for eventual elimination in the feces. Homozygous deletions of ABCC2 as seen in the YEE and YHD3 strains have no obvious effect on insects consuming artificial toxin-free diet in the laboratory. However, plant secondary compounds that deter herbivory or poison the herbivore would be encountered by larvae consuming plants in nature, affecting Bt-susceptible and resistant insects in different ways. If exported by an active ABCC2 in Bt-susceptible insects, they could potentiate the Bt-toxin by increasing the proportion of time the channel is in the open state, exposing the hydrophobic inner surfaces to toxin binding. There is evidence for an effect of different plant tissues with different amounts of secondary compounds on the potency of Cry1Ac [64]. Additionally, by imposing a fitness cost on Bt-resistant insects they could select against resistance alleles encoding defective variants of the ABCC2 protein that fail to export them. For example, Bt-resistant Pectinophora gossypiella is more sensitive to the cotton secondary compound, gossypol [65]. Even a slight fitness cost of ABCC2 mutations would be effective in delaying the increase of resistance alleles, the goal of the high-dose/refuge strategy mandated by the US Environmental Protection Agency. PCR-based DNA diagnostics for specific ABCC2 mutants shown to be present in the field could be useful in supporting the continued success of this strategy by monitoring resistance alleles in field populations of insect pests.
Materials and Methods
Marker-Assisted Selection to Develop Resistant Strains
All crosses used virgin adults of Heliothis virescens in single-pair matings. Resistant strain YHD2 was crossed to the susceptible strain CNW (July 2001) and F1 offspring were intercrossed. F2 progeny were reared on artificial diet [66] containing 0.2 µg/ml Cry1Ac toxin for 10 days, individually weighed, and transferred to toxin-free diet for rearing to adulthood. The 10-day weights were used as an indication of the ability to resist the growth-inhibiting effect of this sublethal Cry1Ac concentration, due to the presence of different combinations of resistant and susceptible alleles at the two resistance genes BtR-4 and BtR-6. F2 adults from the top third of the weight distribution were intercrossed to form the YHD3 strain, which was subjected to selection on Cry1Ac-containing diet over 25 generations, eventually attaining the same resistance level as the parent YHD2. It was maintained on artificial diet with 200 µg/ml Cry1Ac. To develop the YFO strain, F2 adults from the middle third of the weight distribution were repeatedly backcrossed in single-pair matings to the susceptible CNW strain. Parents were scored for the presence of the BtR-4r allele by PCR using the primers SF1, SR2, and RR3 (Figure S1) [25], after collection of fertile eggs. Only progeny of parents that still carried the BtR-4r allele were retained for subsequent matings. Larvae of these generations were reared on toxin-free diet to avoid any toxin-based selection of resistance alleles. After 6 generations of backcrossing and PCR screening, YFO adults were intercrossed and subsequent generations made homozygous for BtR-4r, after which the strain was raised on 5 µg/ml Cry1Ac. The YEE strain was developed by intercrossing the F2 from the lower third of the weight distribution and subsequent generations, and keeping only progeny of parents with the lowest frequency of BtR-4r alleles as detected by PCR. Larvae of this strain were reared on diet with 5 µg/ml Cry1Ac toxin to select for BtR-6r alleles. After no parents were found to carry BtR-4r alleles, the YEE strain was maintained on 50 µg/ml Cry1Ac. As YFO was homozygous BtR-4r4r and YEE was subsquently shown to be homozygous BtR-6r6r, the ABCC2 mutation permits larvae to consume 10 times as much Cry1Ac without growth retardation as does the cadherin mutation. All strains showed equivalent growth in the laboratory on artificial diet with no toxin.
Linkage Mapping
Backcross larval progeny were tested by rearing on a sublethal concentration of Cry1Ac in artificial diet [17], allowing normal growth (i. e. equivalent to susceptible individuals on non-Bt diet over the same time period) in individuals homozygous resistant for the gene segregating in the cross, but suppressing growth in heterozygotes. Larvae were weighed to the nearest 1 mg after 7 days; backcross size distributions were strongly bimodal consistent with segregation of a single major resistance gene (Figure S3). All larvae were then transferred to toxin-free diet and reared to adults for DNA extraction. Polymorphisms at genetic marker loci were scored using RFLPs (restriction fragment length polymorphisms) visualized by Southern blots of restriction-digested genomic DNA, or scored by screening for intron size polymorphisms by PCR using primers placed in adjacent exons.
Three series of interstrain crosses were used to generate backcross families (BRX) segregating at BtR-6. In BRX28 (February 2006) and BRX35 (December 2007), F1 progeny from crosses between YHD3 (4r4r 6r6r) and YFO (4r4r 6s6s) were backcrossed to YHD3. Backcross progeny were expected to be 4r4r 6r6r or 4r4r 6r6s; they were tested on 25 µg/ml Cry1Ac. In BRX36 (June 2008), F1 progeny from crosses between YFO and YEE (4s4s 6r6r) were backcrossed to YEE. Backcross progeny were expected to be 4s4s 6r6r, 4r4s 6r6r, 4s4s 6r6s, or 4r4s 6r6s. To minimize the effect of segregation of the 4r allele, which is recessive at high concentrations, backcross progeny were tested on 50 µg/ml Cry1Ac and otherwise treated as in the other two series.
The linkage analysis strategy exploited the absence of crossing-over during meiosis in female Lepidoptera [67]. Female-informative backcrosses (with F1 mothers) were examined first to verify that segregation of LG2 markers correlated with larval weight. Male-informative backcrosses (with F1 fathers in which crossing-over occurs) were then used to estimate linkage relationships among LG2 markers and resistance as measured by larval weight on Cry1Ac-containing diet.
For RFLP analysis, DNA was isolated from adults using phenol and chloroform, digested with HindIII or PstI, electrophoresed on 0.8% agarose gels, and transferred to Hybond N+ filters for probing with 32P-labelled probes. RFLP probes for LG2 markers were generated from H. virescens or Helicoverpa armigera cDNA probes previously mapped to LG2, or from genes mapped to Bombyx mori Chromosome 15. These were used to search EST databases of H. virescens and H. armigera by BLAST, or to design degenerate PCR primers for amplification and sequencing from H. virescens cDNA or gDNA. Intron size polymorphisms in some markers were scored by agarose gel electrophoresis of PCR products generated using primers positioned in adjacent exons.
Three strategies were used to screen B. mori Chromosome 15 for markers that could be used in mapping, in a sequential approach to narrow the interval containing BtR-6. First, sequence information from the RAPD-based linkage map of Yasukochi et al. [27], [68] was used in BLASTN searches of the wgs section of GenBank (http://www.ncbi.nlm.nih.gov) to identify whole-genome-shotgun contigs produced by the first [69] and second [70] genome assemblies, and these in turn were screened for conserved coding sequences present in the H. armigera and H. virescens cDNA libraries. This approach was limited by small contig size and frequent occurrence of chimeric contigs. Second, a BAC-walking strategy was employed using BAC-end sequences deposited in the gss section of GenBank [71]. BAC ends occuring in contigs were identified by BLASTN to gss, the other end was obtained by a text search using the BAC clone name, and used to identify the contig in which it occurred by BLASTN to wgs. Third, when the third genome assembly [72] was made available to the public on-line on SilkDB (http://silkworm.swu.edu.cn/silkdb/) [73] and Kaikobase (http://sgp.dna.affrc.go.jp/index.html) [74], predicted genes in the genome browser view were used. Serial numbers of BGI predicted genes are represented here by the last four digits; e.g. b7795 for BGIBMGA007795. These approaches were successful because of the high degree of evolutionary conservation of gene order among Bombyx and Heliothis for this linkage group.
Recombinants were identified by reference to parental and grandparental genotypes and tallied by hand in order to guide the direction of search for additional markers. The final linkage map was constructed using 20 markers and 1060 offspring using the program Mapmaker3 [75] with Haldane centimorgans. A Macintosh PowerBook running the MacPort implementation of the unix version was used, as the MS-DOS version of this program running under Windows crashed with our dataset.
After BtR-6 was localized within the interval between markers b7730 and b7793 showing zero recombinants, the linkage map of B. mori Chr15 was examined and found to also have zero recombinants out of 190 informative progeny in the corresponding region [71]. The physical map of this region in B. mori contains 10 predicted genes [72], nine of which showed expression in B. mori larval midgut as indicated by microarray studies [76] and also had homologs in cDNA libraries constructed from midgut tissue of larval H. armigera (Table S2).
Preparation of Brush Border Membrane Vesicles and Binding Assays
Actively feeding early fifth-instar H. virescens larvae were chilled on ice and dissected, (May 2007). Tracheae, Malpighian tubules, peritrophic matrix and food bolus were removed and the midgut tissue was rinsed briefly in ice-cold phosphate-buffered saline (PBS). Brush border membrane vesicles (BBMV) were prepared by the Mg2+ precipitation method according to Wolfersberger et al. [77]. The final BBMV pellet was resuspended at a protein concentration of 1 mg/ml in PBS (determined by the BCA protein assay with BSA as standard, Bio-Rad) and stored at −80°C until use. Brush border membrane enrichment was estimated by measuring the aminopeptidase activity using L-leucine-p-nitroanilide as a substrate. Typical enrichment of the leucyl-aminopeptidase activity in the BBMV preparation was between 5 and 6 fold compared to the initial midgut homogenate.
E. coli strains harboring individual Cry1Aa, Cry1Ab, or Cry1Ac genes cloned into pKK223-3 were obtained from the Bacillus Genetic Stock Center (Ohio State University). Cry1A protoxins were prepared according to Lee et al. [78], and were activated by trypsin at a trypsin/protoxin ratio of 1/50 (w/w) at 37°C for 1 h. Activated toxins were further purified by anion exchange chromatography using a 1 ml RESOURCE Q column (GE Healthcare). For toxin biotinylation, 0.5 mg of purified toxins was incubated (1∶30 molar ratio) with NHS-Biotin (Sigma) for 30 min at room temperature. To remove excess biotin, samples were run through a 5 ml HiTrap desalting column (GE healthcare).
Qualitative binding assays were performed by incubating 2.5 nM of each biotinylated Cry1A toxin with BBMV (containing 20 µg protein) for 1 h at room temperature. Then, BBMV were pelleted by centrifugation (13,000 g, 10 min, 4°C) and washed three times with PBS to remove unbound toxin. The final pellet was resuspended in SDS-PAGE sample buffer, boiled for 5 min, and proteins were resolved on a 10% SDS-PAGE gel. Toxin binding was revealed by western blot using streptavidin-HRP (Sigma) and ECL (GE Healthcare). The homologous competition experiment was performed as described above except that biotinylated toxin and BBMV were incubated in the presence of a 200-fold excess of the corresponding unlabeled Cry1A toxin.
BAC Library Screening and Sequencing
High-density filters for a BAC library of H. virescens [79] were obtained from the Texas A&M BAC Center (http://hbz7.tamu.edu), and high-density filters for a BAC library of H. subflexa were obtained from the Clemson University Genomics Institute (CUGI, http://www.genome.clemson.edu). These were screened by hybridization using a 32P-labelled 236-bp PCR product amplified from H. virescens larval midgut cDNA using primers Ha-ABC2-U14-F1 (5′ AACAA TCGTT ACCTG ATGGC GT) and Ha-ABC2-U14-R2 (5′ AGGAT TGGTA TCGAA AAATC TCATT AC) for the H. subflexa filters, and a 252-bp PCR product from the nachbar gene using primers Ha-bgi07733-F7 (5′ GAACT TGGGA CCTAC AGGTG GTAT) and Ha-bgi07733-R10 (5′ GCAGC ATTAC GGATA TTAAT TTCAA C). The H. virescens filters yielded two positive clones, and the H. subflexa filters 20 positive clones, which were obtained from CUGI and re-screened by PCR with primers Hs-BACscr02-F1 (5′-CACCG GCTCA ACACC ATCAT) and Hs-BACscr02-R2 (5′-GTCCT TGGCC ATGCT GTAGAA). Clone HS_Ba 89F08 was chosen and shot-gun sequenced at the Max Planck Institute for Chemical Ecology, Department of Entomology and deposited in GenBank as GQ332573. Primers designed from the H. subflexa sequence (Table S1) were used to amplify the ABCC2 gene in overlapping fragments from genomic DNA; sequence from YHD3 was deposited as GQ332572 and from YFO as GQ332571. Alignment of exon 2 of the YHD3 and YFO sequence revealed a 22-bp deletion in the former, causing a frameshift and resulting in a predicted stop codon after residue 99 (Figure S2).
Predicted Structure of ABCC2 Protein
Conceptual translations of the ABCC2 coding sequence from H. subflexa and the YFO allele of H. virescens were subjected to analysis for conserved domains by blastp to the Conserved Domain Database of NCBI (http://www.ncbi.nlm.nih.gov/cdd) and for transmembrane topology by the server (http://phobius.sbc.su.se/) for the prediction program Phobius [80]. Potential glycosylation sites were screened for using the CBS Prediction Servers (http://www.cbs.dtu.dk/services/); none were found in the sequences examined. Conserved domains, predicted transmembrane domains and extracellular loops are depicted on a sequence alignment of ABCC2 from H. virescens, H. subflexa, B. mori, and homologues from Drosophila melanogaster and Mus musculus (Figure 4, Figure 5).
Estimation of Allele Frequencies from Archival DNA Samples
PCR with primers eU02-F1 and eiT02-R10 (Table S1) flanking the region containing the 22-bp deletion in the 6r allele were used to genotype individuals (Figure S1) used in previous mapping crosses and binding studies. YHD2 strain individuals from March 1993 are the adults used in crosses to map BtR-4 from which DNA was still available; no binding data are available from that generation. No DNA was available from individuals in the binding studies of Lee et al. [18] in 1995. All other samples come from binding studies of Jurat-Fuentes et al. in 2004 [19] in which midguts were dissected from individual larvae in December 2002, the genotypes at BtR-4 were determined by PCR, and midguts from individuals with the same BtR-4 genotypes were pooled for binding analysis as shown in Figure 2 of that publication [19] (Table S3).
Supporting Information
Zdroje
1. JamesC
2008
Global Status of Commercialized Biotech/GM Crops: 2008.
Ithaca, NY
International Service for the Acquisition of Agri-biotech Applications
2. TabashnikBE
Van RensburgJBJ
CarrièreY
2009
Field-evolved insect resistance to Bt crops: Definition, theory, and data.
J Econ Entomol
102
2011
2025
3. PietrantonioPV
GillSS
1996
Bacillus thuringiensis toxins: Action on the insect midgut.
LehaneMJ
BillingsleyPF
Biology of the Insect Midgut
London
Chapman & Hall
345
372
4. SoberónM
GillSS
BravoA
2009
Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells?
Cell Mol Life Sci
66
1337
1349
5. ZhangXB
CandasM
GrikoNB
TaussigR
BullaLA
2006
A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis.
Proc Natl Acad Sci USA
103
9897
9902
6. PigottCR
EllarDJ
2007
Role of receptors in Bacillus thuringiensis crystal toxin activity.
Microbiol Mol Biol Rev
71
255
281
7. FerréJ
Van RieJ
2002
Biochemistry and genetics of insect resistance to Bacillus thuringiensis.
Annu Rev Entomol
47
501
533
8. TabashnikBE
LiuYB
MalvarT
HeckelDG
MassonL
1998
Insect resistance to Bacillus thuringiensis: uniform or diverse?
Philos Trans R Soc Lond, Ser B: Biol Sci
353
1751
1756
9. TabashnikBE
1994
Evolution of resistance to Bacillus thuringiensis.
Annu Rev Entomol
39
47
97
10. GouldF
AndersonA
ReynoldsA
BumgarnerL
MoarW
1995
Selection and genetic analysis of a Heliothis virescens (Lepidoptera: Noctuidae) strain with high levels of resistance to Bacillus thuringiensis toxins.
J Econ Entomol
88
1545
1559
11. AkhurstRJ
JamesW
BirdLJ
BeardC
2003
Resistance to the Cry1Ac delta-endotoxin of Bacillus thuringiensis in the cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae).
J Econ Entomol
96
1290
1299
12. González-CabreraJ
EscricheB
TabashnikBE
FerréJ
2003
Binding of Bacillus thuringiensis toxins in resistant and susceptible strains of pink bollworm (Pectinophora gossypiella).
Insect Biochem Mol Biol
33
929
935
13. CarrièreY
CrowderDW
TabashnikBE
2010
Evolutionary ecology of insect adaptation to Bt crops.
Evolutionary Applications
3
561
573
14. StorerNP
BabcockJM
SchlenzM
MeadeT
ThompsonGD
2010
Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico.
J Econ Entomol
103
1031
1038
15. XuXJ
YuLY
WuYD
2005
Disruption of a cadherin gene associated with resistance to Cry1Ac delta-endotoxin of Bacillus thuringiensis in Helicoverpa armigera.
Appl Environ Microbiol
71
948
954
16. MorinS
BiggsRW
SistersonMS
ShriverL
Ellers-KirkC
2003
Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm.
Proc Natl Acad Sci USA
100
5004
5009
17. GahanLJ
GouldF
HeckelDG
2001
Identification of a gene associated with Bt resistance in Heliothis virescens.
Science
293
857
860
18. LeeMK
RajamohanF
GouldF
DeanDH
1995
Resistance to Bacillus thuringiensis CryIA delta-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration.
Appl Environ Microbiol
61
3836
3842
19. Jurat-FuentesJL
GahanLJ
GouldFL
HeckelDG
AdangMJ
2004
The HevCaLP protein mediates binding specificity of the Cry1A class of Bacillus thuringiensis toxins in Heliothis virescens.
Biochemistry
43
14299
14305
20. TabashnikBE
LiuYB
MalvarT
HeckelDG
MassonL
1997
Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis.
Proc Natl Acad Sci USA
94
12780
12785
21. BaxterSW
ZhaoJZ
GahanLJ
SheltonAM
TabashnikBE
2005
Novel genetic basis of field-evolved resistance to Bt toxins in Plutella xylostella.
Insect Mol Biol
14
327
334
22. BravoA
GómezI
CondeJ
Muñoz-GarayC
SánchezJ
2004
Oligomerization triggers binding of a Bacillus thuringiensis Cry1Ab pore-forming toxin to aminopeptidase N receptor leading to insertion into membrane microdomains.
Biochim Biophys Acta Biomemb
1667
38
46
23. HeckelDG
GahanLC
GouldF
AndersonA
1997
Identification of a linkage group with a major effect on resistance to Bacillus thuringiensis Cry1Ac endotoxin in the tobacco budworm (Lepidoptera: Noctuidae).
J Econ Entomol
90
75
86
24. Jurat-FuentesJL
GouldFL
AdangMJ
2002
Altered glycosylation of 63 - and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins.
Appl Environ Microbiol
68
5711
5717
25. GahanLJ
GouldF
LópezJD
MicinskiS
HeckelDG
2007
A polymerase chain reaction screen of field populations of Heliothis virescens for a retrotransposon insertion conferring resistance to Bacillus thuringiensis toxin.
J Econ Entomol
100
187
194
26. GahanLJ
MaYT
CobleMLM
GouldF
MoarWJ
2005
Genetic basis of resistance to Cry1Ac and Cry2Aa in Heliothis virescens (Lepidoptera: Noctuidae).
J Econ Entomol
98
1357
1368
27. YasukochiY
AshakumaryLA
BabaK
YoshidoA
SaharaK
2006
A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects.
Genetics
173
1319
1328
28. YoshidoA
BandoH
YasukochiY
SaharaK
2005
The Bombyx mori karyotype and the assignment of linkage groups.
Genetics
170
675
685
29. AllerSG
YuJ
WardA
WengY
ChittaboinaS
2009
Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science
323
1718
1722
30. BellierA
ChenCS
KaoCY
CinarHN
AroianRV
2009
Hypoxia and the hypoxic response pathway protect against pore-forming toxins in C. elegans.
PLoS Pathog
5
e1000689
doi:10.1371/journal.ppat.1000689
31. ChenCS
BellierA
KaoCY
YangYL
ChenHD
2010
WWP-1 Is a novel modulator of the DAF-2 insulin-like signaling network involved in pore-forming toxin cellular defenses in Caenorhabditis elegans.
PLoS ONE
5
e9494
doi:10.1371/journal.pone.0009494
32. KnowlesBH
EllarDJ
1987
Colloid-osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis delta-endotoxins with different insect specificity.
Biochim Biophys Acta
924
509
518
33. SchwartzJL
GarneauL
SavariaD
MassonL
BrousseauR
1993
Lepidoperan-specific crystal toxins from Bacillus thuringiensis form cation-selective and anion-selective channels in planar lipid bilayers.
J Membr Biol
132
53
62
34. SlatinSL
AbramsCK
EnglishL
1990
Delta-endotoxins form cation-selective channels in planar lipid bilayers.
Biochem Biophys Res Commun
169
765
772
35. BravoA
GillSS
SoberónM
2007
Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control.
Toxicon
49
423
435
36. ChenJ
HuaG
Jurat-FuentesJL
AbdullahMA
AdangMJ
2007
Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin.
Proc Natl Acad Sci USA
104
13901
13906
37. PengDH
XuXH
YeWX
YuZN
SunM
2010
Helicoverpa armigera cadherin fragment enhances Cry1Ac insecticidal activity by facilitating toxin-oligomer formation.
Appl Microbiol Biotechnol
85
1033
1040
38. SoberónM
Pardo-LópezL
LópezI
GómezI
TabashnikBE
2007
Engineering modified Bt toxins to counter insect resistance.
Science
318
1640
1642
39. Muñoz-GarayC
PortugalL
Pardo-LópezL
Jiménez-JuárezN
ArenasI
2009
Characterization of the mechanism of action of the genetically modified Cry1AbMod toxin that is active against Cry1Ab-resistant insects.
Biochim Biophys Acta
1788
2229
2237
40. NagamatsuY
TodaS
KoikeT
MiyoshiY
ShigematsuS
1998
Cloning, sequencing, and expression of the Bombyx mori receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin.
Biosci Biotechnol Biochem
62
727
734
41. SangadalaS
WaltersFS
EnglishLH
AdangMJ
1994
A mixture of Manduca sexta aminopeptidase and phosphatase enhances Bacillus thuringiensis insecticidal CryIA(c) toxin binding and (Rb+-K+)-Rb-86 efflux in vitro.
J Biol Chem
269
10088
10092
42. KnightPJK
CrickmoreN
EllarDJ
1994
The receptor for Bacillus thuringiensis CrylA(c) delta-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N.
Mol Microbiol
11
429
436
43. Jurat-FuentesJL
AdangMJ
2004
Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae.
Eur J Biochem
271
3127
3135
44. ArenasI
BravoA
SoberónM
GómezI
2010
Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin.
J Biol Chem
285
12497
12503
45. PandianGN
IshikawaT
TogashiM
ShitomiY
HaginoyaK
2008
Bombyx mori midgut membrane protein P252, which binds to Bacillus thuringiensis Cry1A, is a chlorophyllide-binding protein, and the resulting complex has antimicrobial activity.
Appl Environ Microbiol
74
1324
1331
46. ValaitisAP
JenkinsJL
LeeMK
DeanDH
GarnerKJ
2001
Isolation and partial characterization of gypsy moth BTR-270, an anionic brush border membrane glycoconjugate that binds Bacillus thuringiensis Cry1A toxins with high affinity.
Arch Insect Biochem Physiol
46
186
200
47. ZhuangMB
OlteanDI
GómezI
PullikuthAK
SoberónM
2002
Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation.
J Biol Chem
277
13863
13872
48. ValaitisAP
2008
Bacillus thuringiensis pore-forming toxins trigger massive shedding of GPI-anchored aminopeptidase N from gypsy moth midgut epithelial cells.
Insect Biochem Mol Biol
38
611
618
49. MassonL
LunYJ
MazzaA
BrousseauR
AdangMJ
1995
The CryIA(c) receptor purified from Manduca sexta displays multiple specificities.
J Biol Chem
270
20309
20315
50. LuoK
SangadalaS
MassonL
MazzaA
BrousseauR
1997
The Heliothis virescens 170kDa aminopeptidase functions as Receptor A by mediating specific Bacillus thuringiensis Cry1a delta-endotoxin binding and pore formation.
Insect Biochem Mol Biol
27
735
743
51. LeeMK
JenkinsJL
YouTH
CurtissA
SonJJ
2001
Mutations at the arginine residues in alpha 8 loop of Bacillus thuringiensis delta-endotoxin Cry1Ac affect toxicity and binding to Manduca sexta and Lymantria dispar aminopeptidase N.
FEBS Lett
497
108
112
52. JenkinsJL
LeeMK
SangadalaS
AdangMJ
DeanDH
1999
Binding of Bacillus thuringiensis Cry1Ac toxin to Manduca sexta aminopeptidase N receptor is not directly related to toxicity.
FEBS Lett
462
373
376
53. RajagopalR
SivakumarS
AgrawalN
MalhotraP
BhatnagarRK
2002
Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor.
J Biol Chem
277
46849
46851
54. YangYL
ZhuYC
OtteaJ
HussenederC
LeonardBR
2010
Molecular characterization and RNA interference of three midgut aminopeptidase N isozymes from Bacillus thuringiensis-susceptible and -resistant strains of sugarcane borer, Diatraea saccharalis.
Insect Biochem Mol Biol
40
592
603
55. CandasM
LosevaO
OppertB
KosarajuP
BullaLA
2003
Insect resistance to Bacillus thuringiensis - Alterations in the indianmeal moth larval gut proteome.
Mol Cell Proteomics
2
19
28
56. KrishnamoorthyM
Jurat-FuentesJL
McNallRJ
AndachtT
AdangMJ
2007
Identification of novel CrylAc binding proteins in midgut membranes from Heliothis virescens using proteomic analyses.
Insect Biochem Mol Biol
37
189
201
57. VadlamudiRK
JiTH
BullaLA
1993
A specific binding protein from Manduca sexta for the insecticidal toxin of Bacillus thuringiensis subsp Berliner.
J Biol Chem
268
12334
12340
58. NagamatsuY
TodaS
YamaguchiF
OgoM
KogureM
1998
Identification of Bombyx mori midgut receptor for Bacillus thuringiensis insecticidal CryIA(a) toxin.
Biosci Biotechnol Biochem
62
718
726
59. BaxterSW
ZhaoJZ
SheltonAM
VogelH
HeckelDG
2008
Genetic mapping of Bt-toxin binding proteins in a Cry1A-toxin resistant strain of diamondback moth Plutella xylostella.
Insect Biochem Mol Biol
38
125
135
60. WangP
ZhaoJZ
Rodrigo-SimónA
KainW
JanmaatAF
2007
Mechanism of resistance to Bacillus thuringiensis toxin Cry1Ac in a greenhouse population of the cabbage looper, Trichoplusia ni.
Appl Environ Microbiol
73
1199
1207
61. MahonRJ
OlsenKM
DownesS
AddisonS
2007
Frequency of alleles conferring resistance to the Bt toxins Cry1Ac and Cry2Ab in Australian populations of Helicoverpa armigera (Lepidoptera: noctuidae).
J Econ Entomol
100
1844
1853
62. DownesS
ParkerT
MahonR
2010
Incipient resistance of Helicoverpa punctigera to the Cry2Ab Bt toxin in Bollgard II (R) cotton.
PLoS ONE
5
e12567
doi:10.1371/journal.pone.0012567
63. CacciaS
Hernández-RodríguezCS
MahonRJ
DownesS
JamesW
2010
Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species.
PLoS ONE
5
e9975
doi:10.1371/journal.pone.0009975
64. OlsenKM
DalyJC
2000
Plant-toxin interactions in transgenic Bt cotton and their effect on mortality of Helicoverpa armigera (Lepidoptera: Noctuidae).
J Econ Entomol
93
1293
1299
65. CarrièreY
Ellers-KirkC
BiggsR
HigginsonDM
DennehyTJ
2004
Effects of gossypol on fitness costs associated with resistance to Bt cotton in pink bollworm.
J Econ Entomol
97
1710
1718
66. JoynerK
GouldF
1985
Developmental consequences of cannibalism in Heliothis zea (Lepidoptera: Noctuidae).
Ann Entomol Soc Am
78
24
28
67. RobinsonR
1971
Lepidoptera Genetics.
Oxford
Pergamon Press
68. YasukochiY
1998
A dense genetic map of the silkworm, Bombyx mori, covering all chromosomes based on 1018 molecular markers.
Genetics
150
1513
1525
69. MitaK
KasaharaM
SasakiS
NagayasuY
YamadaT
2004
The genome sequence of silkworm, Bombyx mori.
DNA Res
11
27
35
70. XiaQY
ZhouZY
LuC
ChengDJ
DaiFY
2004
A draft sequence for the genome of the domesticated silkworm (Bombyx mori).
Science
306
1937
1940
71. YamamotoK
NohataJ
Kadono-OkudaK
NarukawaJ
SasanumaM
2008
A BAC-based integrated linkage m0ap of the silkworm Bombyx mori.
Genome Biology
9
R21
72. XiaQY
WangJ
ZhouZY
LiRQ
FanW
2008
The genome of a lepidopteran model insect, the silkworm Bombyx mori.
Insect Biochem Mol Biol
38
1036
1045
73. WangJ
XiaQY
HeXM
DaiMT
RuanJ
2005
SilkDB: a knowledgebase for silkworm biology and genomics.
Nucleic Acids Res
33
D399
D402
74. ShimomuraM
MinamiH
SuetsuguY
OhyanagiH
SatohC
2009
KAIKObase: An integrated silkworm genome database and data mining tool.
BMC Genomics
10
486
75. LanderES
GreenP
AbrahamsonJ
BarlowA
DalyMJ
1987
MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations.
Genomics
1
174
181
76. XiaQY
ChengDJ
DuanJ
WangGH
ChengTC
2007
Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori.
Genome Biology
8
R162
77. WolfersbergerMG
LuethyP
MaurerP
PrentiP
SacchiVF
1987
Preparation and partial characterisation of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae).
Comp Biochem Physiol
86A
301
308
78. LeeMK
MilneRE
GeAZ
DeanDH
1992
Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis delta-endotoxin.
J Biol Chem
267
3115
3121
79. WuCC
ProestouD
CarterD
NicholsonE
SantosF
2009
Construction and sequence sampling of deep-coverage, large-insert BAC libraries for three model lepidopteran species.
BMC Genomics
10
283
80. KallL
KroghA
SonnhammerELL
2007
Advantages of combined transmembrane topology and signal peptide prediction - the Phobius web server.
Nucleic Acids Res
35
W429
W432
81. KallL
KroghA
SonnhammerELL
2004
A combined transmembrane topology and signal peptide prediction method.
J Mol Biol
338
1027
1036
82. GaudetR
WileyDC
2001
Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing.
EMBO J
20
4964
4972
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome DeletionsČlánek Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable EpiallelesČlánek A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular StressČlánek Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis inČlánek The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 inČlánek Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrAČlánek Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 12- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Whole-Genome and Chromosome Evolution Associated with Host Adaptation and Speciation of the Wheat Pathogen
- Association of Variants at 1q32 and with Ankylosing Spondylitis Suggests Genetic Overlap with Crohn's Disease
- Initiator Elements Function to Determine the Activity State of BX-C Enhancers
- Identification of Genes Required for Neural-Specific Glycosylation Using Functional Genomics
- A Young Duplicate Gene Plays Essential Roles in Spermatogenesis by Regulating Several Y-Linked Male Fertility Genes
- The EpsE Flagellar Clutch Is Bifunctional and Synergizes with EPS Biosynthesis to Promote Biofilm Formation
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
- Season of Conception in Rural Gambia Affects DNA Methylation at Putative Human Metastable Epialleles
- A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress
- GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells
- A Single Enhancer Regulating the Differential Expression of Duplicated Red-Sensitive Opsin Genes in Zebrafish
- Investigation and Functional Characterization of Rare Genetic Variants in the Adipose Triglyceride Lipase in a Large Healthy Working Population
- Reduction of Protein Translation and Activation of Autophagy Protect against PINK1 Pathogenesis in
- Noisy Splicing Drives mRNA Isoform Diversity in Human Cells
- The Loss of PGAM5 Suppresses the Mitochondrial Degeneration Caused by Inactivation of PINK1 in
- Thymus-Associated Parathyroid Hormone Has Two Cellular Origins with Distinct Endocrine and Immunological Functions
- An ABC Transporter Mutation Is Correlated with Insect Resistance to Cry1Ac Toxin
- Role of Individual Subunits of the CSN Complex in Regulation of Deneddylation and Stability of Cullin Proteins
- The C-Terminal Domain of the Bacterial SSB Protein Acts as a DNA Maintenance Hub at Active Chromosome Replication Forks
- The DNA Damage Response Pathway Contributes to the Stability of Chromosome III Derivatives Lacking Efficient Replicators
- Cleavage of Phosphorothioated DNA and Methylated DNA by the Type IV Restriction Endonuclease ScoMcrA
- LaeA Control of Velvet Family Regulatory Proteins for Light-Dependent Development and Fungal Cell-Type Specificity
- Competitive Repair by Naturally Dispersed Repetitive DNA during Non-Allelic Homologous Recombination
- Distinct Functions for the piRNA Pathway in Genome Maintenance and Telomere Protection
- MOS11: A New Component in the mRNA Export Pathway
- Self-Mating in the Definitive Host Potentiates Clonal Outbreaks of the Apicomplexan Parasites and
- A Role for ATF2 in Regulating MITF and Melanoma Development
- Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm
- Cancer and Neurodegeneration: Between the Devil and the Deep Blue Sea
- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Linking Crohn's Disease and Ankylosing Spondylitis: It's All about Genes!
- Genomics Meets Glycomics—The First GWAS Study of Human N-Glycome Identifies HNF1α as a Master Regulator of Plasma Protein Fucosylation
- Continuous and Periodic Expansion of CAG Repeats in Huntington's Disease R6/1 Mice
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Endocytic Sorting and Recycling Require Membrane Phosphatidylserine Asymmetry Maintained by TAT-1/CHAT-1
- Histone Deacetylases Suppress CGG Repeat–Induced Neurodegeneration Via Transcriptional Silencing in Models of Fragile X Tremor Ataxia Syndrome
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Comparison of Innate Immune Signaling Pathways in Primates
- Expression of Linear and Novel Circular Forms of an -Associated Non-Coding RNA Correlates with Atherosclerosis Risk
- Genome-Wide Interrogation of Mammalian Stem Cell Fate Determinants by Nested Chromosome Deletions
- Histone H2A C-Terminus Regulates Chromatin Dynamics, Remodeling, and Histone H1 Binding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



