-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
Multicellular organisms deploy various strategies to fight microbial infections. Invading pathogens may be eradicated directly by antimicrobial effectors of the immune system. Another strategy consists of increasing the tolerance of the host to infection, for example, by limiting the adverse effects of the immune response. The molecular mechanisms underlying this novel concept remain largely uncharacterized. Here, we demonstrate that the epigenetic regulator G9a mediates tolerance to virus infection in Drosophila. We found that G9a-deficient flies succumb faster than control flies to infection with RNA viruses, but that the viral burden did not significantly differ. Unexpectedly, mutant flies express higher levels of genes that are regulated by the Jak-Stat signaling pathway, which in other studies was found to be important for antiviral defense. Exploiting the genetic toolbox in Drosophila, we demonstrate that Jak-Stat hyperactivation induces early mortality after virus infection. Precise control of immune pathways is essential to ensure efficient immunity, while preventing damage due to excessive immune responses. Our results indicate that G9a, an epigenetic modifier, dampens Jak-Stat responses to prevent immunopathology. Therefore, we propose epigenetic regulation of immunity as a new paradigm for disease tolerance.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004692
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004692Summary
Multicellular organisms deploy various strategies to fight microbial infections. Invading pathogens may be eradicated directly by antimicrobial effectors of the immune system. Another strategy consists of increasing the tolerance of the host to infection, for example, by limiting the adverse effects of the immune response. The molecular mechanisms underlying this novel concept remain largely uncharacterized. Here, we demonstrate that the epigenetic regulator G9a mediates tolerance to virus infection in Drosophila. We found that G9a-deficient flies succumb faster than control flies to infection with RNA viruses, but that the viral burden did not significantly differ. Unexpectedly, mutant flies express higher levels of genes that are regulated by the Jak-Stat signaling pathway, which in other studies was found to be important for antiviral defense. Exploiting the genetic toolbox in Drosophila, we demonstrate that Jak-Stat hyperactivation induces early mortality after virus infection. Precise control of immune pathways is essential to ensure efficient immunity, while preventing damage due to excessive immune responses. Our results indicate that G9a, an epigenetic modifier, dampens Jak-Stat responses to prevent immunopathology. Therefore, we propose epigenetic regulation of immunity as a new paradigm for disease tolerance.
Introduction
Efficient immunity against pathogens requires the coordinated activation and repression of genes within multiple signaling networks. Insufficient immune activation results in high microbial burden, severe pathogenesis, and high mortality from infection; overly strong immune responses may lead to tissue damage, immunopathology, and auto-inflammatory diseases. The inevitable tradeoff between immunity and immunopathology necessitates tightly regulated induction and resolution of immune responses. This is achieved by negative regulatory circuits within and among immune signaling cascades and by complex cellular and molecular programs that terminate inflammation [1,2].
It was recently proposed that host defense depends on a combination of resistance mechanisms, which lower or eliminate pathogen burden, and tolerance mechanisms [3,4]. Tolerance reduces the negative effects of an infection on host fitness, which could be either direct damage inflicted by the pathogen itself or adverse effects of the immune response on host tissues. Little is known about the molecular basis for tolerance, but it likely involves regulatory mechanisms that control the magnitude of the immune response [3,4].
The fruit fly Drosophila melanogaster is a powerful model to genetically and functionally dissect innate immunity. Past studies found that the evolutionarily conserved NF-κB pathways Toll and Immune Deficiency (Imd) mediate the humoral response against bacteria and fungi [5]. Defense against viruses, in contrast, requires the constitutively expressed RNA interference (RNAi) pathway [6]. In addition, the RNA viruses Drosophila C virus (DCV), Cricket paralysis virus (CrPV), and Drosophila X virus (DXV) activate the Janus Kinase-Signal transducers and activators of transcription (Jak-Stat) pathway that orchestrates a transcriptional response to fight the infection [7,8].
The evolutionarily conserved Jak-Stat pathway controls important developmental and homeostatic processes, including hematopoiesis and immunity [9,10]. Deficiencies in Jak-Stat pathway genes cause serious immune disorders and increase susceptibility to infections [11–13], whereas hyperactivated Jak-Stat responses are associated with autoimmune diseases and carcinogenesis in humans [13,14]. Also in insects, the Jak-Stat pathway needs to be tightly controlled. The Jak-Stat pathway is required for efficient antiviral immunity in fruit flies and Aedes aegypti mosquitoes, [15]. For example, loss-of-function fly mutants for the Jak kinase hopscotch (hop) support high levels of virus replication and show increased mortality rates upon infection with the RNA viruses DCV and CrPV [7,8]. Yet, hyperactivation of the Jak-Stat pathway in Drosophila can have serious consequences, such as the formation of lethal hematopoietic melanotic tumors in hop gain-of-function mutants [16].
Spatiotemporal regulation of immune responses occurs via a variety of mechanisms. At the transcriptional level, chromatin structure is a major determinant of gene expression. Histone-modifying enzymes deposit covalent modifications on specific amino acid residues of histone tails that alter the structure of chromatin and its accessibility to the transcriptional machinery. Histone H3 lysine 9 dimethylation (H3K9me2) is commonly regarded as a marker for heterochromatic genomic regions and transcriptional repression. Yet, G9a, one of the three H3K9 methyltransferases in Drosophila, mediates H3K9 dimethylation in vivo, but is associated with euchromatic regions [17]. Loss of G9a does not affect heterochromatin formation or global heterochromatic H3K9me2 levels in flies and mice [18,19], but G9a fly mutants show loss of H3K9 dimethylation at about 5% of the euchromatic genome [20]. Moreover, H3K9me2 can be associated with actively transcribed genes [21] and its presence does not globally correlate well with gene repression, unlike other repressive marks such as H3K27me2 and H3K27me3 [20,22]. These observations suggest that H3K9me2 is not solely associated with stably repressed genes, and that G9a might regulate defined sets of euchromatic genes.
A previous study revealed that G9a controls genes that are involved in processes that require tight and dynamic regulation and high transcriptional plasticity, including neuronal processes, stress responses, and immunity [20]. These observations prompted us to study the role of G9a in antiviral defense. Here, we report that G9a mutant flies are hypersensitive to RNA virus infection and that their inducible immune responses are highly dysregulated. We show that G9a and the Jak-Stat pathway epigenetically and genetically interact to modulate immune defense. Genetic hyperactivation of Jak-Stat signaling causes early lethality after viral infection, thus phenocopying loss of G9a. Together, our results uncover an epigenetic mechanism for tolerance that shapes Jak-Stat pathway activity in response to virus infection.
Results
Reduced survival of G9a mutant flies after RNA virus infection
To investigate the role of G9a in Drosophila antiviral defense, we used the loss-of-function allele G9aDD2 and its wild-type genetic background control (hereafter referred to as G9a-/- and G9a+/+)[20]. Since the H3K9me2 mark is essential for the establishment of heterochromatin and proper gene regulation, we first assessed the overall fitness of G9a-deficient flies. G9a mutants were viable, fertile, and showed no obvious defects in development, confirming previous observations [20,23]. Moreover, the average life span of G9a-/- flies was slightly longer than that of wild-type controls (mean survival = 105.8 days and 87,8 days, respectively; P < 0.001) (Fig 1A).
Fig. 1. G9a mutants have a normal life span but are hypersensitive to RNA virus infection. 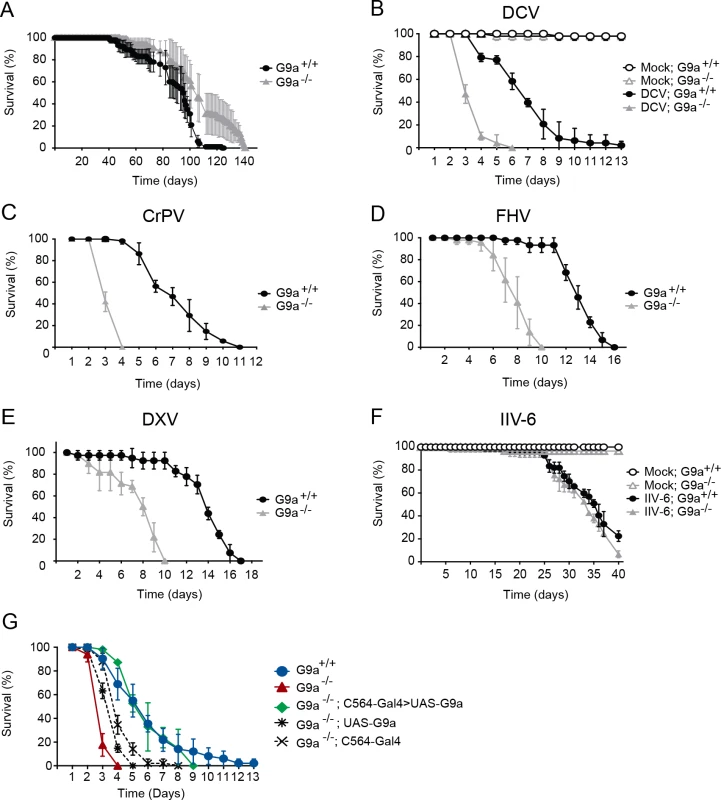
(A) Life span of non-infected female wild-type (G9a+/+) or G9a mutant (G9a-/-) flies at 20°C. (B-F) Survival of wild-type or G9a mutants infected with (B) DCV, (D) CrPV, (D) FHV, (E) DXV, (F) IIV-6, and Tris buffer as a control (mock). (G) Survival of wild-type or G9a mutant flies expressing a G9a transgene in the fat body using the UAS/Gal4 system upon DCV infection (1,000 TCID50 units). The fat body-specific C564 driver line (C564-Gal4) was used to drive expression of the transcription factor Gal4, which binds to the Upstream Activating Sequence to induce expression of a G9a transgene (UAS-G9a). Flies expressing only the C564-Gal4 driver or the UAS-G9a responder construct were included as controls. Mock infections were performed in all experiments (B-G), and no difference in survival was observed between wild-type or G9a mutant flies, as shown in panel B and F. All survival data are available in S1 Dataset. Data represent means and s.d. of five (A) or three (B-G) biological replicates of at least 15 female flies (A-F), or 15 male flies (G) per replicate for each genotype. Data are from one experiment representative of at least 3 (B,C,F,G), or 2 (D,E) independent experiments. We challenged wild-type and mutant flies with DCV, a positive-sense RNA virus from the Dicistroviridae family, by intra-thoraxic injection. G9a mutants were more sensitive to infection than their wild-type controls, with a mean survival of 3.6 and 6.9 days for G9a-/- and G9a+/+ female flies, respectively (P < 0.001; Fig 1B). Male G9a-deficient flies were also more sensitive to DCV infection than control flies, indicating that hypersensitivity to virus infection was not sex-dependent (S1A Fig).
To analyze whether G9a-/- flies were also more sensitive to other virus infections, we challenged flies with a panel of viruses with different genome organization and genetic makeup. Upon challenge with another Dicistrovirus, Cricket paralysis virus (CrPV), the mean survival of G9a mutants was 3.4 days, compared to 7.3 days for wild-type flies (P < 0.001; Fig 1C). Similarly, when infected with Flock House Virus (FHV), a positive-sense virus of the Nodaviridae family, G9a mutants succumbed faster than wild-type flies to infection (mean survival = 7.9 days and 13.1 days, respectively; P < 0.001; Fig 1D). Also upon challenge with the dsRNA virus Drosophila X Virus (DXV, member of the Birnaviridae), G9a mutant flies displayed higher lethality rates compared to wild-type controls (mean survival 7.6 days and 13.6 days, respectively, P < 0.001; Fig 1E). To analyze whether G9a mutants are also more sensitive to DNA virus infection, we challenged flies with Invertebrate iridescent virus 6 (IIV-6). As we observed before, IIV-6 infected wild-type flies survived for prolonged periods of time and mortality only became apparent in the later stages of the infection (>25 days post infection) [24]. In contrast to their hypersensitivity to RNA virus infection, survival rates of G9a mutants after IIV-6 infection were similar to wild-type levels (mean survival = 33.6 days and 34.4 days, respectively; P = 0.2; Fig 1F). Of note, mock infection with Tris buffer did not affect the survival rate of G9a mutants for up to 40 days (Fig 1F). Flies carrying another loss-of-function allele, G9aDD3, exhibited the same phenotype and succumbed more rapidly than their wild-type controls to DCV, but not to IIV-6 infection (S1B–S1D Fig). As G9a mutants displayed increased sensitivity against all RNA viruses tested, we used the model RNA virus DCV for follow-up studies.
Fat body specific expression of G9a rescues hypersensitivity to virus infection
To confirm the role of G9a in antiviral defense, we performed genetic rescue experiments by expression of a G9a transgene in the mutant background using the UAS/Gal4 system. We were unable to recover adult flies expressing the G9a transgene under control of the drivers actin-Gal4, daughterless-Gal4 and tubulin-Gal4, suggesting that ubiquitous overexpression of G9a is detrimental to fly development. The fat body, an organ that is involved in metabolism and immunity [5], is a major target organ of DCV [25]. We therefore used a fat body driver (C564-Gal4) to induce tissue-specific expression of the G9a transgene. Early lethality of infected G9a mutants (mean survival = 3.1 days) was rescued to control levels by fat body-specific G9a expression in the G9a-deficient background (mean survival = 5.6 days, compared to 6.3 days for G9a+/+; P = 0.482; Fig 1G). Survival of genetic control flies that only express the C564-Gal4 driver or the UAS-responder in the G9a-/- background remained significantly different from wild-type flies (mean survival = 4.1 and 3.8 days, respectively, P < 0.001 for both), indicating that the observed rescue was dependent on functional expression of the G9a transgene (Fig 1G). The rescue was tissue specific, since expression of G9a in other tissues, such as hemocytes (using the hemolectin-Gal4 driver), or glia (using the repo-Gal4 driver) did not rescue the phenotype of G9a mutants (S2A and S2B Fig). These experiments indicate that G9a is required specifically in the fat body during virus infection. Moreover, these experiments genetically segregate the role of G9a in antiviral defense from its function in other organs [20].
Reduced tolerance of G9a mutants to RNA virus infection
To analyze whether the reduced survival of G9a mutants is due to a defect in resistance or to reduced tolerance to infection, we analyzed viral load over time. No differences in infectious viral titers were observed between G9a-/- and G9a+/+ flies during the first 3 days post-infection (dpi) (Fig 2A). Since G9a was specifically required in the fat body during DCV infection (Fig 1G), we analyzed viral titers in dissected fat bodies of virus-challenged flies. Virus titers in G9a-/- flies were slightly higher than in wild-type flies, but no significant difference was observed at any time point (Fig 2B). To confirm these data, we measured viral RNA levels in whole flies and fat bodies by quantitative RT-PCR (RT-qPCR). Consistent with the results from the titration, we did not detect significant differences in DCV RNA levels in wild-type and mutant flies over three days post-infection. In the fat body, we observed a modest 3-fold increase in viral RNA at 1 dpi (P = 0.014), but not at the other time points (Fig 2C and 2D).
Fig. 2. Loss of G9a does not affect viral loads upon DCV infection. 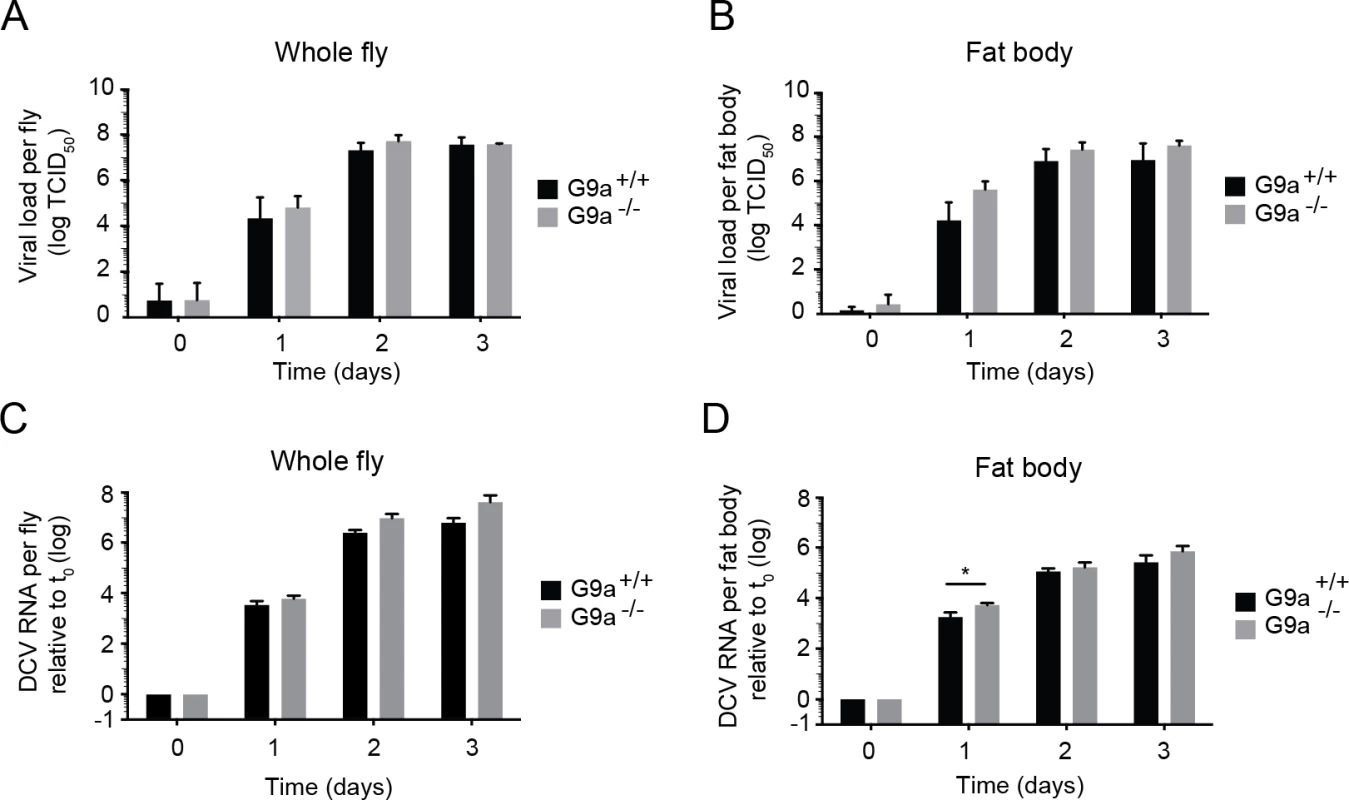
(A,B) Wild-type or G9a mutant flies were inoculated with DCV and viral titers were determined over time in (A) whole flies, and (B) dissected fat bodies. Data represent means and s.d of three independent experiments. Each experiment contained three biological replicates of 5 female flies (A), or 10 fat bodies (B) per replicate for each genotype. (C,D) DCV RNA levels over the course of 3 days post-infection analyzed by RT-qPCR in (C) whole flies or (D) fat bodies of wild-type and G9a mutant flies. DCV RNA levels were normalized to transcript levels of the housekeeping gene Ribosomal Protein 49 and are calculated relative to the viral RNA levels in flies harvested immediately after inoculation (t0). Data represent means and s.d. of three biological replicates of 5 female flies (C) or 10 fat bodies (D) per replicate for each genotype. Data in panel C and D are from one experiment representative of 2 independent experiments. *P < 0.05 (Student’s t-test). Together, our results demonstrate that G9a mutants are more sensitive to DCV infection, but that this is not associated with a major and generalized increase in viral titers. Moreover, the modest increase in viral load at 1 dpi in the fat body seems insufficient to explain the strongly reduced survival upon virus infection. We conclude that G9a mutant flies exhibit defects in tolerance to RNA virus infection.
The antiviral RNAi pathway is functional in G9a mutants
RNA interference (RNAi) is a major antiviral pathway in Drosophila [6]. Given the hypersensitivity of G9a-/- flies to virus infection, we analyzed whether this pathway is functional in mutant flies. To this end, we first monitored RNAi activity using an in vivo sensor assay, in which the inhibitor of apoptosis thread (th) is silenced by expression of an RNAi-inducing hairpin RNA (thRNAi) [26,27]. Expression of thRNAi using the eye-specific driver (GMR-Gal4) leads to severe apoptosis in the developing eye. Consequently, adult thRNAi flies display a reduced eye size, roughening of the eye surface, and loss of pigmentation (S3A Fig). This phenotype is fully dependent on the RNAi pathway, since the phenotype is lost in mutants lacking the central catalytic component of the pathway, Argonaute 2 (AGO2) [26,27]. We expressed the thRNAi hairpin in the eye of G9a-/- and G9a+/+ flies and analyzed the phenotype. Both in G9a-/- and G9a+/+ flies, expression of thRNAi resulted in strong RNAi-induced eye phenotypes (S3A Fig). These results suggest that G9a-/- mutant flies have no major defect in RNAi.
To further evaluate the efficiency of the RNAi response of G9a mutants, we adapted a luciferase-based RNAi sensor assay that we routinely use in Drosophila S2 cells [27,28], to adult flies. Flies were subjected to in vivo transfection with firefly and Renilla luciferase reporter plasmids along with either firefly luciferase-specific dsRNA or control dsRNA, and three days later, efficiency of silencing was assessed in whole fly lysates. As controls, we included Ago2 null mutants and their wild-type controls (w1118). As expected, silencing was abolished in Ago2-/- flies, confirming that loss of FLuc expression was RNAi dependent (S3B Fig, left panel). Efficiency of silencing was similar in G9a-/- and G9a+/+ flies (S3B Fig, right panel), indicating the RNAi pathway is fully proficient in G9a mutant flies.
Hyperactivation of the Jak-Stat pathway in G9a mutants upon virus infection
Since the RNAi pathway was fully functional in G9a-/- flies, we next analyzed whether inducible immune responses were intact in these flies. Virus infection of Drosophila activates the Jak-Stat pathway to induce expression of downstream genes, such as virus induced RNA-1 (vir-1) [7,8]. In addition, the NF-κB pathways Toll and IMD have been implicated in the response to virus infections in some studies [29–31]. We measured expression of vir-1, the stress-induced genes Turandot A and M (TotA and TotM), and the antimicrobial-like peptide Listericin as markers for Jak-Stat activation. To monitor activation of the Toll and IMD pathways, we measured expression of genes encoding the antimicrobial peptides Drosomycin (Drs), Metchnikowin (Mtk), Diptericin (Dpt). In addition, we measured expression of Vago, which is induced in DCV infection via an unknown signaling pathway [25].
We monitored expression of these genes by RT-qPCR at 24 hours after DCV infection (hpi) in whole flies (Fig 3A) and isolated fat bodies (Fig 3B). As observed before [7,8], DCV infection induced expression of the Jak-Stat dependent genes vir-1, TotA, and TotM, but not of NF-κB dependent Drs, Mtk, and Dpt genes. Strikingly, in G9a-/- flies we noted a much higher induction of Jak-Stat dependent genes than in wild-type flies, but no induction of NF-κB dependent genes (Fig 3A). In the fat body, even stronger overactivation of Jak-Stat dependent pathway genes was observed in G9a mutants (Fig 3B). However, basal expression levels of these genes did not differ between non-challenged G9a+/+ and G9a-/- flies (Fig 3C and 3D), suggesting that G9a is not required for steady-state repression of these genes, but that it mitigates their inducibility in response to viral infection.
Fig. 3. Hyperactivation of the Jak-Stat pathway by virus infection of G9a mutants. 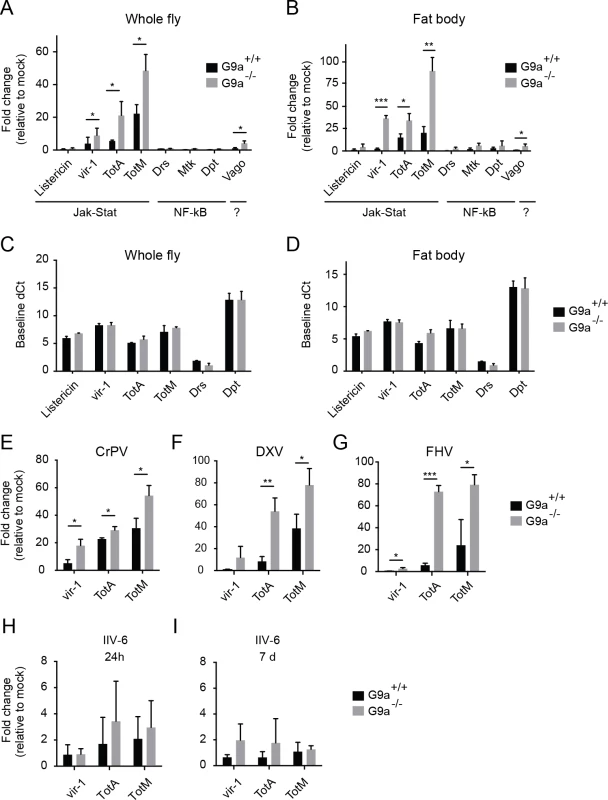
(A,B) Expression of inducible immune genes at 24 hours after DCV infection (TCID50 = 10,000) determined by RT-qPCR in (A) whole flies, and (B) fat bodies of wild-type or G9a mutant flies. Expression of the gene of interest was normalized to transcript levels of the housekeeping gene Ribosomal Protein 49 and expressed as fold change relative to mock infection (Tris buffer). Data are means and s.d. of three independent pools of (A) 30 female flies and (B) 30 fat bodies for each genotype. (C,D) Basal expression levels of the indicated genes measured by RT-qPCR on 3 to 5-day-old unchallenged wild-type and G9a mutant female flies (C) or fat bodies (D). Basal expression levels are expressed as dCt values (difference between Ct of the gene of interest and the Ct of Ribosomal Protein 49). (E-I) Expression of inducible Jak-Stat dependent immune genes at (E-H) 24 hpi or (I) 7 dpi with 10,000 TCID50 units of (E) CrPV, (F) DXV, (G) FHV or (H,I) 14,000 TCID50 units of IIV-6. Data are means and s.d. of three independent pools of at least 15 female flies (C,E-I) or at least 10 fat bodies (D) per genotype. Data are from one experiment representative of 3 (A,B,E), and 2 (C,D) independent experiments. *P < 0.05; ** P < 0.01; *** P < 0.001 (Student’s t-test). We also monitored expression of the Jak-Stat dependent genes upon infection with 3 other RNA viruses: CrPV, DXV and FHV (Fig 3E–3G). As observed upon DCV infection, a strong upregulation of vir-1, TotA, and TotM was found in G9a mutants compared to wild-type flies. Upon infection with the DNA virus IIV-6, we detected only slight expression of these genes (1 to 4-fold, at 24 hpi and 7 dpi, when the replication plateau is reached), and expression levels were not significantly different between wild-type and G9a mutant flies (Fig 3H and 3I). We note that those viruses that induce higher Jak-Stat activation also induce higher mortality rates in G9a mutants (Fig 1B–1F). Our results are in line with a previous report showing that DCV, CrPV, DXV, and FHV, but not IIV-6, induce expression of the Jak-Stat dependent genes vir-1 or TotM [7]. In that study, DXV induces strong TotM expression, and DCV, CrPV and FHV induce mainly vir-1 expression, whereas under our experimental conditions, TotA and TotM are induced at higher levels than vir-1 for all viruses.
Jak-Stat deficient flies were reported to display higher viral load and increased mortality upon DCV and CrPV infection [7,8], suggesting that the Jak-Stat pathway controls expression of antiviral effectors. Our data suggest that robust induction of Jak-Stat dependent genes is not sufficient for efficient host defense, which is in line with previous observations [8]. Moreover, the G9a phenotype seems counter-intuitive, since the antiviral Jak-Stat pathway is strongly activated in G9a mutant flies, yet they are hypersensitive to virus infection.
G9a mutants display an altered transcriptional response to virus infection
To analyze the transcriptional response to viral infection at a genome-wide scale, we performed transcriptome analyses by next-generation sequencing (RNA-seq). We infected wild-type or G9a mutant flies with DCV, and collected whole flies or dissected fat bodies at 24 hpi (Fig 4A). At this time point, flies do not yet exhibit pathological symptoms, such as reduced locomotion and abdominal swelling.
Fig. 4. RNA sequencing and transcriptome analysis of wild-type and G9a mutant flies following DCV infection. 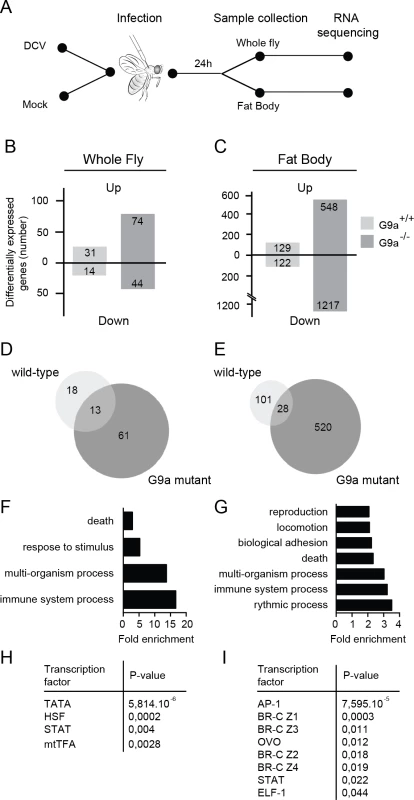
(A) Experimental workflow. Three to five-day-old female flies were infected with DCV or mock-infected with Tris buffer as a control and total RNA was extracted for next-generation sequencing from whole flies or dissected fat bodies at 24 hpi. (B,C) Number of differentially expressed genes in (B) whole flies or (C) fat bodies of wild-type (G9a+/+) or mutant (G9a-/-) flies upon virus infection normalized to their respective mock control. Numbers of genes with ≥2-fold change are shown. (D,E) Venn diagrams representing the overlap of DCV-induced genes (relative to mock) between wild-type and G9a mutant flies in (D) whole flies or (E) fat bodies. (F-I) Gene ontology (GO) and predicted transcription factor binding sites of genes that are expressed at ≥2-fold higher levels in DCV infected G9a mutants than in infected wild-type flies. (F,G) All significantly enriched GO terms of level 2 are shown (P < 0.05 in a hypergeometric test with Benjamini & Hochberg correction). (H,I) Pscan was used to predict transcription factor binding sites in the 500-bp region upstream of the transcription start site using the TRANSFAC database. Significantly enriched transcription factors compared to the genome-wide mean are shown (P < 0.05 in a z-test). Data are from whole flies (F,H) or dissected fat bodies (G,I). We first determined the number of differentially expressed genes (≥ 2-fold) upon DCV infection in whole fly (Fig 4B) or fat body (Fig 4C) relative to mock-infected flies. We noted that only a limited number of genes were induced upon DCV infection in whole wild-type flies (n = 31), whereas many more genes were induced in the fat body (n = 129), possibly because the fat body is a major immune organ and a target organ for DCV [25]. In G9a mutants, significantly more genes were induced upon DCV infection than in wild-type flies, both in whole flies and in dissected fat bodies (n = 74, P < 0.0001 and n = 548, P < 0.0001, respectively, Pearson’s chi-squared test). We also observed a large number of genes that were downregulated upon DCV infection. These genes followed the same trends as the virus-induced genes, with greater number of genes affected in G9a mutants both in whole fly and fat body. These observations are in agreement with the results from Fig 3 and suggest that the transcriptional response to virus infection is dysregulated in G9a mutants.
Only a limited number of genes were induced by DCV in both wild-type and G9a mutant flies (13 and 28 genes in whole fly and fat body, respectively; Fig 4D and 4E). This core set of virus-induced genes consisted of genes involved in stress responses such as heat shock proteins (Hsp70 family, Hsp68) and the Jak-Stat dependent Turandot proteins (TotM, TotX, TotC), as well as other Jak-Stat dependent genes, Diedel [32] and Suppressor of Cytokine Signaling 36E (Socs36E) [8], and genes of unknown function (S4A and S4B Fig).
Jak-Stat dependent genes are enriched in the G9a transcriptome
We focused our subsequent analyses on the genes that were differentially expressed (≥2-fold) in G9a mutants, based on the prediction that if G9a represses genes by depositing H3K9me2 marks, direct target genes are most likely de-repressed in G9a mutants. To analyze whether specific biological processes are dysregulated in G9a mutants, we analyzed Gene Ontology (GO) terms of genes that were expressed at least 2-fold higher in DCV-infected G9a mutant flies over infected wild-type flies. In the whole fly dataset, we observed significant enrichment for GO terms, such as “response to abiotic stimulus" and "response to stress" (within the ancestral GO term "response to stimulus") and "immune response" (ancestral GO term "immune system process") (Fig 4F and S4C Fig). GO term analysis on the fat body dataset identified several additional processes, including "reproduction" and “locomotion” (Fig 4G and S4D Fig). Using Pscan [33] to predict transcription factor binding sites in the promoter regions of the differentially expressed genes, we observed, in addition to the TATA-box binding motif, strong enrichment of Stat binding sites, and target sites of the JNK cascade transcription factor, AP-1 (Fig 4H and 4I). In accordance, we noted among the categories “response to stress” and “immune system processes” genes of the Jak-Stat and c-Jun N-terminal Kinase (JNK) signaling pathways, which included pathways components (dPIAS, Socs36E for Jak-Stat; Hemipterous, Gadd45, Jra, Kay for JNK) as well as some of their downstream targets (Socs36E, vir-1, CG13559, CG1572 for Jak-Stat; Puckered and Rab-30 for JNK) [34].
G9a targets genes of the Jak-Stat pathway
Our results indicate that the transcriptional response to infection is highly dysregulated in the absence of G9a and that Jak-Stat pathway components and downstream targets are among the genes that are derepressed in G9a mutants. We then asked whether these derepressed Jak-Stat genes are direct targets of G9a, or whether they are affected indirectly.
A previous study identified putative G9a target sites by comparing genome-wide H3K9me2 profiles obtained by chromatin immunoprecipation (ChIP) followed by next generation sequencing in wild-type and G9a mutant larvae [20]. Interestingly, these predicted targets are enriched for the GO term "Jak-Stat cascade" (P = 0.0011, 2.3-fold enrichment). We therefore selected Jak-Stat genes that fulfilled three criteria for further analysis: i) harboring a reported loss-of-methylation site in G9a mutants, ii) previously shown to be involved in defense responses, iii) being upregulated in the transcriptome sets of challenged G9a mutants. This set of five genes consisted of pathway components and regulators (domeless, dPIAS, Socs36E), as well as the downstream targets vir-1 and TotM [10,35–38]. Using RT-qPCR, we confirmed that all five predicted G9a target genes show over-induction in response to virus infection in G9a mutant fat bodies (domeless, dPIAS, Socs36E, Fig 5A; vir-1, TotM, Fig 3B). For none of these genes, a difference in basal expression was observed in the absence of viral infection, indicating that these genes are only derepressed upon viral infection in G9a mutants (Fig 5B and Fig 3D).
Fig. 5. G9a targets genes of the Jak-Stat pathway. 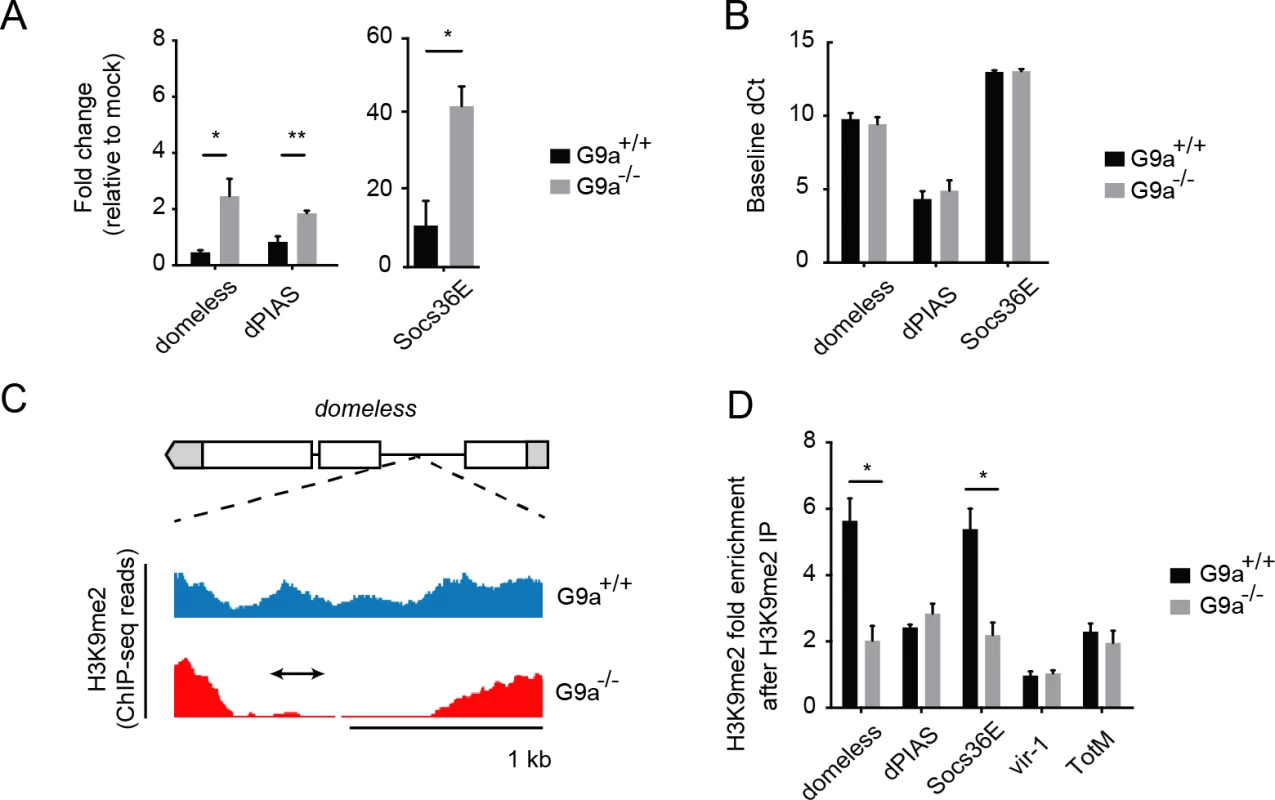
(A) Expression levels of domeless, dPIAS, and Socs36E at 24 hpi in fat bodies of 3 to 5-day-old female wild-type or G9a mutant flies challenged with DCV (10,000 TCID50 units). Data are expressed as fold change relative to mock infection (Tris buffer). (B) Basal expression levels of Jak-Stat genes measured by RT-qPCR on fat bodies of 3 to 5-day-old unchallenged female wild-type and G9a mutant flies. Basal expression is presented as dCt (difference between Ct of the gene of interest and the Ct of Ribosomal Protein 49). (C) Representative example of a G9a target locus within the domeless gene, defined as a genomic region in which the H3K9me2 mark is present in wild-type flies, but not in G9a mutants, in a previous study [20]. Blue and red plots represent sequence reads in H3K9me2 ChIP-seq analyses of wild-type and G9a mutants, respectively [20]. Gene structure is indicated with boxes for exons, lines for introns, and gray boxes for untranslated regions. The arrow represents the position of the amplicon generated by qPCR after Chromatin-Immunoprecipitation (ChIP-qPCR). (D) H3K9me2 ChIP-qPCR on fat bodies of wild-type or G9a mutant flies. Fold enrichment is the percentage of input of the gene of interest normalized to that of a reference gene with very low H3K9me2 marks (moca). Specificity control experiments for ChIP-qPCR experiments are shown in S5E–S5J Fig. Data are means and s.d. of (A,B) three independent pools of at least 10 fat bodies, or (D) three independent pools of 80 female fat bodies, for each genotype. Data are from one experiment representative of 2 (A,B) or 6 (D) independent experiments. *P < 0.05 (Student’s t-test). We next analyzed G9a-dependent targeting of these Jak-Stat genes by H3K9me2 ChIP followed by qPCR (ChIP-qPCR) in dissected fat bodies of wild-type and G9a mutants. We designed qPCR primers in the loss-of-methylation regions observed in ChIP-seq, as shown (domeless in Fig 5C; dPIAS, Socs36E, vir-1, TotM in S5A–S5J Fig). We found that Socs36E and domeless were significantly depleted of H3K9me2 in the fat body of G9a mutants at previously predicted G9a target sites [20] (Fig 5D). Not all G9a targets sites could be confirmed, possibly because ChIP-seq and ChIP-qPCR have been performed at different developmental stages and tissues (whole larvae versus adult fat body, respectively). Although we could not confirm direct targeting by G9a of dPIAS, vir-1 and TotM using ChIP-PCR, we did observe higher expression of these genes in infected G9a mutants. Upregulation of these genes could be a secondary effect resulting from the dysregulation of pathway components, such as domeless and Socs36E, in G9a mutants, rather than from direct epigenetic regulation by G9a. Taken together, these observations suggest that G9a epigenetically regulates a subset of Jak-Stat genes in the adult fat body to shape their transcriptional response to virus infection.
G9a regulates tolerance through modulation of Jak-Stat pathway activity
Our data suggest that G9a regulates Jak-Stat responses to prevent excessive expression of downstream target genes. We performed genetic epistasis tests to analyze the relationship between G9a and the Jak-Stat pathway. Epistasis is defined as a genetic interaction in which a mutation in one gene masks the phenotype of a mutation in another gene. We hypothesized that if G9a mediates viral tolerance through dampening Jak-Stat-induced transcription, inactivation of the Jak-Stat pathway would mask the hypersensitivity of G9a mutants to virus infection. Alternatively, if G9a confers tolerance to DCV infection in a Jak-Stat independent manner, simultaneous loss of G9a and Jak-Stat function would result in more dramatic hypersensitivity to virus infection.
To test our hypothesis, we combined the mutant G9a allele with a dominant negative allele of the Jak-Stat pathway receptor domeless (domeΔCyt) under control of a UAS enhancer [39]. We drove expression of domeΔCyt in the background of G9a mutants and wild-type controls using the ubiquitous actin-Gal4 driver and challenged flies with DCV. As expected [8], overexpression of domeΔCyt increased mortality rates in a G9a+/+ background. Remarkably, the difference in survival between G9a-/- and G9a+/+ flies was masked in the Jak-Stat impaired genetic background (Fig 6A). Moreover, mortality rates of double mutant flies (G9a-/- and Jak-Stat deficient) were similar to those of flies in which either G9a or Jak-Stat was inactivated. Therefore, our data suggests a genetic interaction between G9a and the Jak-Stat pathway receptor, domeless. Additionally, we found that domeΔCyt negated the over-induction of TotA and vir-1 in DCV-infected G9a mutants, demonstrating that G9a regulates these genes in a Jak-Stat dependent manner (Fig 6C).
Fig. 6. Genetic interaction between G9a and the Jak-Stat pathway. 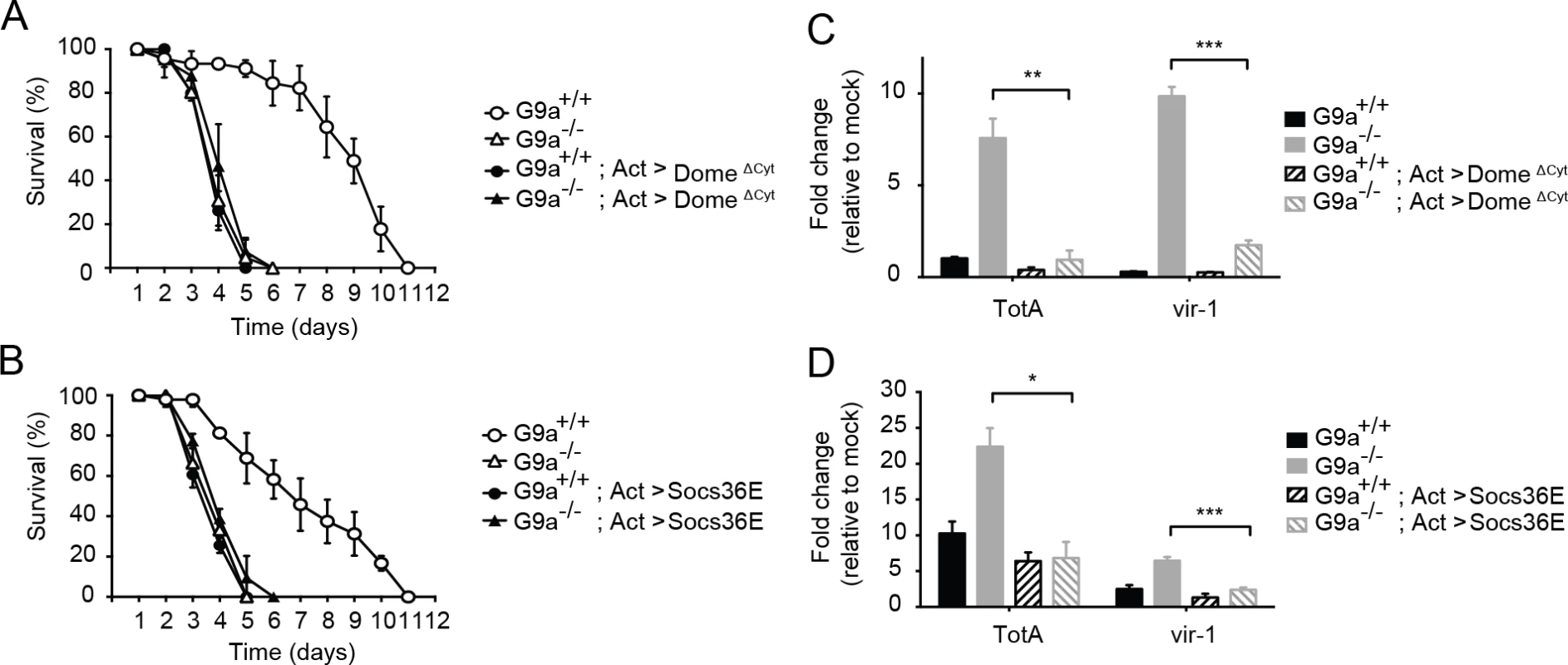
(A,B) Survival upon DCV infection (1,000 TCID50 units) of wild-type or G9a mutant and wild-type flies overexpressing (A) a dominant negative version of the domeless receptor (domeΔCyt), or (B) the negative regulator of Jak-Stat signaling Socs36E. The UAS/Gal4 system was used to drive transgene expression. Gal4 is expressed under control of the actin promoter (Act-Gal4) to drive ubiquitous expression of the UAS-domeΔCyt and UAS-Socs36E transgenes. Control flies expressing only the Act-Gal4, the UAS-domeΔCyt, or the UAS-Socs36E transgenes were included as controls (see S5A and S5B Dataset). Mock infections where performed along the experiments and are shown in S6A and S6B Fig. (C,D) Expression of TotA and vir-1 upon DCV infection of wild-type or G9a mutant flies, expressing (C) domeΔCyt, or (D) Socs36E. Expression of the gene of interest (by RT-qPCR) was normalized to transcript levels of the housekeeping gene Ribosomal Protein 49 and expressed as fold change relative to mock infection (Tris buffer). Data are means and s.d. of three independent pools of at least 15 male flies for each genotype. (A,B) A representative experiment of two independent experiments is shown. Differences in expression of TotA and vir-1 were evaluated with a Student’s t-test (*P < 0.05; ** P < 0.01; *** P < 0.001). To confirm these results with another Jak-Stat loss-of-function allele, we performed a second epistasis experiment using a fly strain overexpressing the negative regulator of the Jak-Stat pathway Socs36E under control of the UAS sequence [40]. Similar to the experiment with domeΔCyt, overexpression of Socs36E masked the hypersensitivity phenotype of G9a mutants to virus infection, suggesting a genetic interaction between G9a and Socs36E (Fig 6B). Again, as expected, Socs36E overexpression significantly reduced expression of TotA and vir-1 (Fig 6D). In both assays, mock infections were performed in parallel, confirming that differences in survival cannot be attributed to the injury caused by the injection itself (S6A and S6B Fig). As the DCV inoculum of 1,000 TCID50 induced high mortality rates in G9a mutants, as well as in Jak-Stat deficient flies, it remained possible that we may have missed higher mortality rates in flies carrying both mutations. Therefore, we repeated the epistasis experiments using a lower inoculum of 100 TCID50, and confirmed that combining Jak-Stat inactivation with G9a loss-of-function did not yield higher mortality rates than in single mutants (S6C and S6D Fig).
In both cases, inhibition of Jak-Stat signaling in wild-type flies masked the effect of a G9a null mutation upon viral challenge, indicating a genetic interaction between G9a and the Jak-Stat components. Taken together, these results suggest that G9a regulates viral tolerance through modulation of Jak-Stat pathway activity.
Jak-Stat hyperactivation induces early mortality after virus infection
Our results suggest that G9a buffers Jak-Stat dependent responses to prevent excessive expression of Jak-Stat dependent genes. We hypothesize that hyperactivation of the Jak-Stat response induces immunopathology that causes increased mortality of G9a mutants upon virus infection. This hypothesis predicts that ectopic activation of the Jak-Stat pathway results in increased rates of mortality upon virus infection.
To test this prediction, we activated the Jak-Stat pathway in adult flies by ubiquitous expression of Unpaired (Upd), a ligand for the domeless receptor, and subsequently infected flies with DCV. Since the Jak-Stat pathway has important functions in development, we used the temperature sensitive Gal80ts allele [41] to induce ubiquitous Upd expression in adult flies by transferring them from 18–20°C (non-permissive temperature) to 29°C (permissive temperature) (Fig 7A). We confirmed by RT-qPCR that Upd as well as the Jak-Stat target gene TotA were strongly induced at 3 days after the shift to 29°C (Fig 7B). We next challenged Upd-overexpressing adult flies with virus. Strikingly, flies with a hyperactivated Jak-Stat pathway succumbed earlier to DCV infection (mean survival = 3.3 days) than genetic control flies expressing only the UAS-Upd transgene or the tubulin-Gal4, tubulin-Gal80ts drivers (mean survival = 5.5 days for both, P < 0.001) (Fig 7C). Moreover, irrespective of the genotype, mock infection did not induce mortality, excluding the possibility that incubation at 29°C is a stressor that triggers early lethality. We conclude that ectopic Jak-Stat activation phenocopies loss of G9a, indicating that immune hyperactivation may underlie the hypersensitivity of G9a mutant flies to DCV infection.
Fig. 7. Hyperactivation of the Jak-Stat pathway renders flies hypersensitive to virus infection. 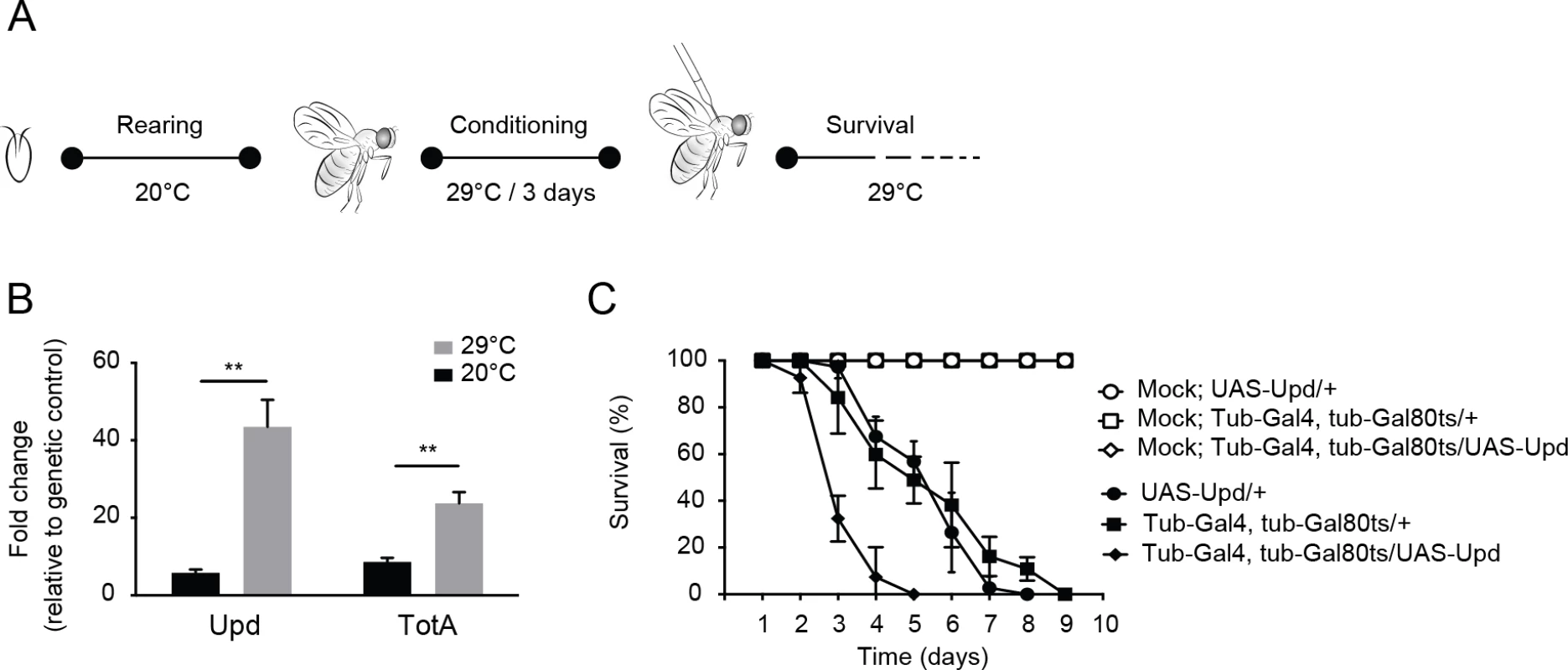
(A) Experimental set-up. Expression of the Upd transgene was induced specifically in adult flies using the Gal4/Gal80ts system. Gal80ts is a temperature-sensitive allele of the Gal80 inhibitor that binds Gal4 to prevent activation of gene expression at 20°C. At 29°C, Gal80ts is degraded, allowing Gal4 to bind to the Upstream Activating Sequences (UAS) to induce gene expression. Flies were reared at 20°C, and 0 to 3-day-old adults were conditioned at 29°C for 3 days prior to viral challenge. (B) Expression levels by RT-qPCR of Upd and TotA in flies carrying the temperature-dependent Upd overexpression system (UAS-Upd; tubulin-Gal4/Gal80ts) after 3 days conditioning at 29°C. The Gal4 and Gal80ts transgenes were combined with the UAS-Upd by standard genetic crosses at 20°C and 0 to 3-day-old adult offspring was cultured for 3 days at 20°C or at 29°C before RNA levels were analyzed by RT-qPCR. Transcript levels of Upd and TotA were normalized to RNA levels of the housekeeping gene Ribosomal Protein 49, and expressed as fold change relative to control flies carrying only the UAS-Upd transgene. (C) Survival of flies carrying the temperature-dependent Upd overexpression system (UAS-Upd; tubulin-Gal4/Gal80ts) and genetic control flies upon DCV infection (1,000 TCID50 = units) at 29°C. Data are means and s.d. of three independent pools of at least 10 male flies for each genotype. Data in (C) are from one experiment representative of 2 independent experiments. Differences in expression of Upd and TotA were evaluated with a Student’s t-test (*P < 0.05; ** P < 0.01; *** P < 0.001). Discussion
Disease tolerance was recently defined as a defense strategy that reduces the negative impact of infection on host fitness, without a concomitant reduction of pathogen burden [3,4]. The concept of tolerance (also termed resilience) provides an exciting, novel perspective on pathogen-host interactions in metazoans. A few examples of tolerance to bacterial or viral infections have been described in flies [42–48], but the mechanisms of tolerance remain largely unknown. In this study, we elucidate a novel epigenetics-based mechanism for tolerance. We provide evidence that the histone methyltransferase G9a contributes to tolerance by regulating the antiviral Jak-Stat signaling pathway.
G9a mutant flies are hypersensitive to RNA virus infection. Transcriptome analyses indicate that Jak-Stat pathway genes are highly upregulated upon DCV challenge in G9a mutants, whereas their basal levels prior to viral infection are normal. This phenotype, like others reported previously [49,50], seems paradoxical: the antiviral Jak-Stat pathway is strongly activated, yet G9a flies are hypersensitive to infection, showing that immune induction per se is not sufficient for efficient host defense. We propose that increased expression of Jak-Stat dependent genes causes immunopathology, eventually resulting in earlier mortality upon virus infection. In support of this hypothesis, we demonstrated that G9a limits the strength of the immune response through Jak-Stat and that ectopic hyperactivation of Jak-Stat signaling triggered early lethality after DCV infection, thus phenocopying the G9a phenotype. Therefore, we propose that epigenetic regulation by G9a dampens Jak-Stat signaling to avoid immune hyperactivation and subsequent mortality.
G9a seems to be required for tolerance to RNA viruses, but not to DNA viruses. G9a mutants induce higher expression of the Jak-Stat dependent genes vir-1, TotA, and TotM and show increased lethality rates upon infection with four RNA viruses (DCV, CrPV, FHV, and DXV). A previous study found that these viruses all induce either vir-1 or TotM to some extent, but that a resistance phenotype for Jak-Stat mutants (higher lethality rates in combination with increased viral load) was only observed after DCV and CrPV infection [7]. Thus, whereas Jak-Stat is only required for resistance to DCV and CrPV infection, our results suggest that all RNA viruses activate the Jak-Stat pathway and that precise epigenetic control of the pathway is required to prevent immunopathology.
Like in mammals, hyperactivation of immune pathways in Drosophila is detrimental for fitness and survival. For instance, overexpression of antimicrobial peptides or loss of negative regulators such as Caudal or the catalytic peptidoglycan receptor proteins (PGRP-LB and PGRP-SCs) triggers severe tissue pathology in the gut that are reminiscent of chronic inflammatory syndromes in mammals [49,51]. The mechanism by which Jak-Stat overactivation triggers lethality remains to be determined, but may involve expression of potentially toxic gene products that require tight regulation. Moreover, we cannot exclude that additional derepressed genes upon loss of G9a contribute to increased mortality of mutant flies. Alternatively, the G9a phenotype might be caused by defects in cell growth, differentiation, tissue homeostasis or apoptosis, which are also under control of the Jak-Stat pathway. We do note, however, that an external infectious stimulus, i.e. virus infection, was required to cause increased mortality upon genetic hyperactivation of the Jak-Stat pathway, and that G9a mutants appear to develop normally, thus excluding more generalized defects.
Our transcriptome analysis uncovered that, in addition to the Jak-Stat pathway, a multitude of pathways are activated by virus infection, many of which are of interest for follow-up studies. We observed a strong activation of the JNK pathway upon DCV infection. In accordance, predicted binding sites for the AP-1 complex, the transcriptional module of the JNK pathway, were highly enriched in promoters of genes upregulated upon DCV infection in G9a mutant fat bodies. Whether Stat and AP-1 associate upon virus infection to regulate immune genes cooperatively, as previously described in lipopolysaccharide stimulated Drosophila cells [52,53], is an interesting question for future investigation.
Our study makes an important contribution to understanding tolerance mechanisms beyond Drosophila. Two EHMT/G9a paralogs exist in mammals, EHMT1/GLP and EHMT2/G9a [20]. They form a heterodimeric complex, and loss of either protein results in nearly identical phenotypes [54]. We analyzed published microarray data of mice in which the G9a and GLP genes were inactivated in forebrain neurons and observed enrichment for the GO term "immune response", and over-representation of NF-κB binding sites in differentially regulated genes, suggesting that G9a also regulates immune signaling cascades in mammals (S7 Fig). Indeed, a previous study suggested that the G9a-dependent H3K9me2 mark is an epigenetic determinant of the interferon response in murine and human cells [55]. In that study, the abundance of H3K9me2 at the promoters of the Interferon-β (Ifnβ) gene and Interferon stimulated genes (ISG) correlates with expression levels of these genes in different cell types, but deficiency in G9a did not affect basal gene expression. Pharmacological inhibition or genetic ablation of G9a increased Ifnβ and ISG expression in mouse fibroblasts and rendered these cells resistant to viral infection.
Our results demonstrate that the role of G9a in controlling the responsiveness to immune challenge is evolutionarily conserved. Moreover, while the in vitro cell culture model suggested that loss of G9a would be beneficial to the antiviral response of the host [55], our data show that loss of G9a disrupts tolerance mechanisms at the organismal level, and is therefore detrimental for survival. This seems to better match the observations in humans. Heterozygous loss of EHMT1/GLP causes Kleefstra syndrome (OMIM number 610253). This rare disorder is characterized by developmental delay and severe intellectual disability. Interestingly, up to 60% of Kleefstra syndrome patients suffer from recurrent infections; yet, these patients do not suffer from primary immune deficiencies [56]. Whether defects in tolerance explain this aspect of the clinical presentation of Kleefstra syndrome remains an interesting hypothesis.
Materials and Methods
Fly strains and husbandry
Flies were reared on standard cornmeal-agar media at 25°C on a light/dark cycle of 12h/12h. G9aDD2 mutants were generated previously by mobilization of the P-element KG01242 located in the 5’ UTR of the gene [20]. G9aDD2 has been used throughout the main text and is referred to as G9a-/-. A precise transposon excision line, referred to as G9a+/+, has been generated in the same genetic background and serves as a control in all experiments. An independent null allele, G9aDD3, has been generated by mobilization of the same P element and contains a deletion of 1850 bp that spans the translation start site [20] (S1B Fig). The following fly stocks and alleles have been described before: UAS-G9a (ref. [20]), C564-Gal4 fat body driver (ref. [57]), Hml-Gal4 hemocyte driver (ref. [58]), UAS-domeΔCYT (ref. [8,37]), UAS-Socs36E (ref. [59]), UAS-Upd (ref. [59]), tubulin-Gal4, tubulin-Gal80ts (ref. [60]), and Argonaute 2414 (ref. [61]). The driver lines armadillo-Gal4 and repo-Gal4 were obtained from the Bloomington Stock Center. In vivo RNAi experiments were performed by crossing GMR-Gal4, UAS-thRNAi/CyO male flies [26] with G9a+/+or G9a-/- virgins. The eye phenotype was monitored in two to four-day-old male F1 offspring lacking the CyO balancer. Upd was conditionally overexpressed by crossing tubulin-Gal4, tubulin-Gal80ts with UAS-Upd flies. Flies were reared at 20°C until hatching. Zero to three-day-old F1 offspring were then incubated at 29°C for 3 days prior to viral challenge, and cultured at 29°C throughout the remainder of the experiment.
Virus infection
Fly stocks were raised for two generations on standard fly flood containing 0.05 mg/ml tetracycline hypochloride (Sigma) to clear Wolbachia infection. Absence of Wolbachia was verified by PCR on DNA of whole flies using Wolbachia-specific primers, as described previously [24]. Persistent virus infections were cleared by bleaching embryos, and absence of DCV, DAV and Nora virus was verified by RT-PCR, as previously described [24].
Virus stocks were prepared as described [24]. Three to five-day-old flies were anesthetized with CO2 and injected with virus suspension using a Nanoject II injector (Drummond) in the thorax, between the mesopleura and the pteropleura. Virus suspensions in 10 mM Tris-HCl, pH 7.3 contained 1,000 median tissue culture infectious dose (TCID50) of DCV and CrPV; 14,000 TCID50 of IIV-6; 3,000 TCID50 of FHV and 2,000 TCID50 of DXV for all survival experiments. 10,000 TCID50 of DCV was used in experiments in which transcriptional responses were analyzed. Flies were cultured at 25°C and transferred to fresh food every 3 days. Survival was monitored daily; lethality at day 1 was attributed to the injection procedure and subtracted from the survival analysis. Unless noted otherwise, three pools of 10 to 15 flies were injected per condition with independent dilutions of virus stock. Fat body tissues were isolated by careful dissection of the abdominal carcasses of adult flies and removal of the gut and reproductive system. This procedure recovers cuticle-associated fat body with minor contamination by muscular and epidermal tissues [62].
Virus titration
Drosophila S2 cells (Invitrogen) were cultured at 25°C in Schneider’s Drosophila Media (Gibco) supplemented with 10% heat-inactivated Fetal Calf Serum (PAA), 50 U/mL Penicillin and 50 μg/mL Streptomycin (Gibco). DCV titers were determined by end-point dilution, as described previously [24]. Briefly, 2.104 cells were seeded in 96-well plates and ten-fold dilutions of fly homogenate were inoculated in quadruplicate. Cells were transferred to fresh medium at day 5, and cytopathic effect (CPE) was monitored until day 14. Viral titers were calculated according to the method of Reed and Muench [63].
RNA analysis
RNA was isolated from flies using Isol-RNA lysis Agent (5-Prime), treated with DNase I (Ambion), and cDNA synthesis was performed on 1 μg RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer’s instructions. qPCR was performed on a LightCycler 480 using SYBR Green I Master Mix (Roche). The qPCR program was the following: 95°C for 5 min, and 45 cycles of 95°C for 5s, 60°C for 10s, 72°C for 20s. Expression of the gene of interest was normalized to transcript levels of the housekeeping gene Ribosomal Protein 49 (Rp49). The following primers were used for qPCR:
Rp49 forward, 5’ - ATGACCATCCGCCCAGCATAC-3’;
Rp49 reverse, 5’-CTGCATGAGCAGGACCTCCA-3’;
Vago forward, 5’ - CAGCCAAGCGATTCCTTATC-3’;
Vago reverse, 5’ - CTCATACAGTGGGCAGCATC-3’;
vir-1 forward, 5’-ATTACTCCGAATTCGAAGCTTCC-3’;
vir-1 reverse, 5’ - CGAATTCTTCACGCTCCTTC-3’;
Listericin forward, 5’-TTGCGGCCATTCTGGCCATG-3’,
Listericin reverse, 5’ - TTTACGTCCCCAACTGGAAC-3’;
TotA forward, 5’ - CCCTGAGGAACGGGAGAGTA-3’;
TotA reverse, 5’ - CTTTCCAACGATCCTCGCCT-3’;
TotM forward, 5’ - ACCGGAACATCGACAGCC-3’;
TotM reverse, 5’ - CCAGAATCCGCCTTGTGC-3’;
Drosomycin forward 5’-GTACTTGTTCGCCCTCTTCG-3’;
Drosomycin reverse, 5’ - ACAGGTCTCGTTGTCCCAGA-3’;
Metchnikowin forward 5’ - TACATCAGTGCTGGCAGAGC-3’;
Metchnikowin reverse, 5’ - AATAAATTGGACCCGGTCTTG-3’;
Diptericin forward, 5’ - TGTGAATCTGCAGCCTGAAC-3’;
Diptericin reverse, 5’ - GCTCAGATCGAATCCTTGCT-3’;
DCV forward, 5’ - TTGCCATTGCACCACTAAAA -3’;
DCV reverse, 5’ - AAAATTTCGTTTTAGCCCAGAA -3’;
Domeless forward, 5’ - AGCTCTGATCCGGATTGTTG-3’;
Domeless reverse, 5’-ATCTCACCGCATTCACCAAG-3’;
dPIAS forward, 5’-AACTGCCCTGTATGCGACAA-3’;
dPIAS reverse, 5’-ACACCTCCTGGAAGTAGCCA-3’;
Socs36E forward, 5’-GTTGCTGCTCCCATTGAAAG-3’;
Socs36E reverse, 5’-GCAAAAGTCGGAGTGTGAGAG-3’;
In vivo RNAi reporter assay
RNAi competency of adult flies was analyzed using a reporter assay that was adapted from a previously published method in S2 cells [27,28]. In vivo plasmid transfection was based on a method described for Aedes aegypti mosquitoes [64,65]. Three to five-day-old female flies were injected in the abdomen with a 100 nl suspension containing a 1 : 1 mixture of Schneider’s Drosophila Media (Gibco) and Lipofectamine 2000 (Invitrogen) complexed with 80 ng pMT-GL3 (encoding firefly luciferase, FLuc), 50 ng pMT-Ren (encoding Renilla luciferase, RLuc) and 1 ng FLuc-specific or non-specific control dsRNA. After incubation for 3 days at 25°C, flies were homogenized with a Douncer in passive lysis buffer (Promega). Supernatant was collected after 10 min centrifugation at 16,000 × g and transferred to a new tube, followed by centrifugation for 5 min at 16,000 × g. 25μL of fly lysate was used to measure FLuc and RLuc activity using the Dual Luciferase assay reporter system (Promega). Ratios of FLuc/Rluc were calculated for each sample, and data are presented as fold silencing relative to the non-specific dsRNA control (GFP).
Chromatin immunoprecipitation followed by qPCR
Eighty dissected fat bodies were homogenized in PBS with a douncer and crosslinked with 3.7% formaldehyde for 30 minutes at room temperature. The cross-linking reaction was quenched by addition of 1.25 mM glycine, and the samples were washed with 1 mL PBS and resuspended in a buffer containing 150 mM Tris-HCl (pH 7.5), 600 mM KCl, 150 mM NaCl, 10 mM EDTA, 1 mM EGTA, 1.5 mM spermine (Sigma) and 5 mM spermidine (Sigma). Tissue was further homogenized using a QiaShredder column, and cells were lysed by adding the same buffer supplemented with 2% Triton-X100. Nuclei were pelleted by centrifugation at 6,000 rpm for 10 min, and resuspended in 250 μL incubation buffer (0.75% SDS, 5% Triton-X100, 750 mM NaCl, 5mM EDTA, 2.5 mM EGTA, 50 mM Tris pH 8.0, 0.4% BSA, 1x protease inhibitor cocktail complete (Roche)). After nuclei purification, chromatin was sonicated at 4°C using a Bioruptor sonicator (Diagenode) for 30 minutes at high power with cycles of 30 seconds ON, and 30 s OFF. Anti-H3K9me2 (ab1220, Abcam), anti - H3 (ab1791, Abcam), anti-V5 (R960-20, Invitrogen) antibodies, and Prot A/G beads (Santa Cruz) were used to capture antibody-bound chromatin overnight at 4°C on a rotating wheel. Chromatin was eluted and de-crosslinked for 4 hours at 65°C in 416 μL elution buffer containing 0.2 M NaCl, 1% SDS and 0.1 M NaHCO3. DNA was then isolated using phenol/chloroform, precipitated overnight at -20°C with 1 mL 100% ethanol, 5 μg linear acrylamide, 0.1 M NaAc, pH 5.2. The pellet was washed with 70% ethanol and resuspended in water. Non-immunoprecipitated DNA was isolated in parallel from purified nuclei and used as an input control in qPCR.
qPCR was performed on a LightCycler 480 using SYBR Green I Master Mix (Roche) using the following qPCR program: 95°C for 10 min, and 40 cycles of 95°C for 15s, 60°C for 1 min. The percentage of immunoprecipitated DNA relative to the input was calculated after qPCR. Fold enrichment in H3K9me2 positive DNA was calculated by normalizing the percentage of input of the gene of interest to the euchromatic control gene previously shown to lack H3K9me2, moca [66]. We confirmed in our conditions that H3K9me2 marks are indeed nearly absent on moca in both wild-type flies and G9a mutants. Also, we show that histone H3 levels are identical between G9a mutant and wild-type flies, both on moca and domeless. Using an aspecific IgG isotypic control antibody, we verified very low aspecific background binding to chromatin (S5E–S5J Fig).
Primers for qPCR were designed in regions previously shown to be depleted of H3K9me2 in G9a mutants by ChIP-sequencing [20]. Sequences are as follows:
Socs36E forward, 5’-GAAATCCGATGTGCTGAAG-3’;
Socs36E reverse, 5’-ACATGGGGGTGTTTTACAGG-3’;
Domeless forward: 5’-CACGTGGATCCAAAATACCC-3’;
Domeless reverse, 5’-GATTGCGATTCCGAGAACTG-3’;
dPIAS forward, 5’ - CACTGACTCAACCACGCTTC-3’;
dPIAS reverse, 5’-CCGTAAAAGGTGAACCGAAA-3’;
vir-1 forward, 5’ - TTGTTCTGGGGCAGAGAAAG-3’;
vir-1 reverse, 5’ - ATCGCTTCATGTCAGTGTCC-3’;
TotM forward, 5’-TTCGGGACGGTCACAGATAG-3’;
TotM reverse, 5’-TCTCGAAAAACCCCTGTAGC-3’;
RNA sequencing
Thirty whole flies or 100 fat bodies of three to five-day-old flies were collected at 24 hours after infection with 10,000 TCID50 of DCV. Samples were frozen on dry ice and stored at -80°C before RNA was isolated using Isol-RNA Lysis reagent as described above. The cDNA library was prepared with the Illumina TruSeq mRNA kit and single-end sequencing was performed on an Illumina HiSeq 2000 (Baseclear BV, Leiden, the Netherlands). RNAseq was performed on a single biological replicate, and should be considered an exploratory analysis.
The FastQ sequence reads were generated in the Illumina Casava pipeline version 1.8.0. Initial quality assessment was based on data passing the Illumina Chastity filter. Reads containing only adapters or PhiX control sequences were removed by filtering protocols developed by Baseclear BV. The second quality control on the remaining reads was performed with FastQC quality control tool 0.10.0. Reads were mapped to the reference genome (Drosophila melanogaster R5/dm3, released in April 2006, UCSC Bioinformatics) using TopHat version 1.4.0. Differential expression between two datasets was analyzed with the Genomatix analysis suite (using DESeq 1.0.6). Gene Ontology enrichment was analyzed using GoToolBox [67], with a hypergeometric test with Benjamini & Hochberg correction. Level 2 GO terms are shown in Fig 4, and level 3 GO terms in S4 Fig Fold enrichment is the ratio of the GO term frequency in the G9a datasets to the genome-wide GO term frequency. Promoter binding-sites for transcription factors were predicted with Pscan [33] on the 500-bp region upstream of the transcriptional start site using the TRANSFAC database. Venn diagrams were generated using Biovenn [68]. The RNA-Seq data are available at the NCBI Gene Expression Omnibus under series accession number GSE56013.
Statistical analysis
Kaplan-Meier analyses and log-rank tests, as implemented in SPSS Statistics (version 20, IBM), were used to evaluate whether differences in survival were statistically significant. For all other experiments, unpaired two-tailed Student’s t-tests and Pearson’s chi-squared test, as implemented in Graphpad Prism version 6, were used to determine statistical significance. P-values below 0.05 were considered statistically significant.
Supporting Information
Zdroje
1. Schneider DS (2007) How and Why Does a Fly Turn Its Immune System Off? PLoS Biol 5: e247. 17880266
2. Han J, Ulevitch RJ (2005) Limiting inflammatory responses during activation of innate immunity. Nat Immunol 6 : 1198–1205. 16369559
3. Medzhitov R, Schneider DS, Soares MP (2012) Disease Tolerance as a Defense Strategy. Science 335 : 936–941. doi: 10.1126/science.1214935 22363001
4. Ayres JS, Schneider DS (2012) Tolerance of infections. Annu Rev Immunol 30 : 271–294. doi: 10.1146/annurev-immunol-020711-075030 22224770
5. Lemaitre B, Hoffmann J (2007) The host defense of Drosophila melanogaster. Annu Rev Immunol 25 : 697–743. 17201680
6. van Mierlo JT, van Cleef KW, van Rij RP (2011) Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol Biol 721 : 3–22. doi: 10.1007/978-1-61779-037-9_1 21431676
7. Kemp C, Mueller S, Goto A, Barbier V, Paro S, et al. (2013) Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol 190 : 650–658. doi: 10.4049/jimmunol.1102486 23255357
8. Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, et al. (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol 6 : 946–953. 16086017
9. Rawlings JS, Rosler KM, Harrison DA (2004) The JAK/STAT signaling pathway. J Cell Sci 117 : 1281–1283. 15020666
10. Zeidler MP, Bach EA, Perrimon N (2000) The roles of the Drosophila JAK/STAT pathway. Oncogene 19 : 2598–2606. 10851058
11. Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, et al. (2003) Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 33 : 388–391. 12590259
12. van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, et al. (2011) STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med 365 : 54–61. doi: 10.1056/NEJMoa1100102 21714643
13. O'Shea JJ, Holland SM, Staudt LM (2013) JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med 368 : 161–170. doi: 10.1056/NEJMra1202117 23301733
14. Theofilopoulos AN, Baccala R, Beutler B, Kono DH (2005) Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 23 : 307–336. 15771573
15. Merkling SH, van Rij RP (2013) Beyond RNAi: antiviral defense strategies in Drosophila and mosquito. J Insect Physiol 59 : 159–170. doi: 10.1016/j.jinsphys.2012.07.004 22824741
16. Arbouzova NI, Zeidler MP (2006) JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133 : 2605–2616. 16794031
17. Stabell M, Eskeland R, Bjorkmo M, Larsson J, Aalen RB, et al. (2006) The Drosophila G9a gene encodes a multi-catalytic histone methyltransferase required for normal development. Nucleic Acids Res 34 : 4609–4621. 16963494
18. Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, et al. (2009) Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 64 : 678–691. doi: 10.1016/j.neuron.2009.11.019 20005824
19. Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC (2009) Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181 : 1303–1319. doi: 10.1534/genetics.108.100271 19189944
20. Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, et al. (2011) Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol 9: e1000569. doi: 10.1371/journal.pbio.1000569 21245904
21. Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19 : 381–391. 16061184
22. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129 : 823–837. 17512414
23. Seum C, Bontron S, Reo E, Delattre M, Spierer P (2007) Drosophila G9a is a nonessential gene. Genetics 177 : 1955–1957. 18039887
24. Bronkhorst AW, van Cleef KW, Vodovar N, Ince IA, Blanc H, et al. (2012) The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc Natl Acad Sci U S A 109: E3604–3613. doi: 10.1073/pnas.1207213109 23151511
25. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, et al. (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol 9 : 1425–1432. doi: 10.1038/ni.1664 18953338
26. Meyer WJ, Schreiber S, Guo Y, Volkmann T, Welte MA, et al. (2006) Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet 2: e134. 16934003
27. van Mierlo JT, Bronkhorst AW, Overheul GJ, Sadanandan SA, Ekstrom JO, et al. (2012) Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog 8: e1002872. doi: 10.1371/journal.ppat.1002872 22916019
28. van Cleef KW, van Mierlo JT, van den Beek M, van Rij RP (2011) Identification of viral suppressors of RNAi by a reporter assay in Drosophila S2 cell culture. Methods Mol Biol 721 : 201–213. doi: 10.1007/978-1-61779-037-9_12 21431687
29. Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, et al. (2014) The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog 10: e1004507. doi: 10.1371/journal.ppat.1004507 25473839
30. Zambon Ra, Nandakumar M, Vakharia VN, Wu LP (2005) The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 102 : 7257–7262. 15878994
31. Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW (2009) A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog 5: e1000582. doi: 10.1371/journal.ppat.1000582 19763182
32. Coste F, Kemp C, Bobezeau V, Hetru C, Kellenberger C, et al. (2012) Crystal structure of Diedel, a marker of the immune response of Drosophila melanogaster. PLoS One 7: e33416. doi: 10.1371/journal.pone.0033416 22442689
33. Zambelli F, Pesole G, Pavesi G (2009) Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 37: W247–252. doi: 10.1093/nar/gkp464 19487240
34. Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP (2010) Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep 11 : 201–207. doi: 10.1038/embor.2010.1 20168330
35. Karsten P, Hader S, Zeidler MP (2002) Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev 117 : 343–346. 12204282
36. Rivas ML, Cobreros L, Zeidler MP, Hombria JC (2008) Plasticity of Drosophila Stat DNA binding shows an evolutionary basis for Stat transcription factor preferences. EMBO Rep 9 : 1114–1120. doi: 10.1038/embor.2008.170 18802449
37. Agaisse H, Perrimon N (2004) The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev 198 : 72–82. 15199955
38. Hombria JC, Brown S (2002) The fertile field of Drosophila Jak/STAT signalling. Curr Biol 12: R569–575. 12194841
39. Brown S, Hu N, Hombria JC (2001) Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol 11 : 1700–1705. 11696329
40. Croker BA, Kiu H, Nicholson SE (2008) SOCS regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol 19 : 414–422. doi: 10.1016/j.semcdb.2008.07.010 18708154
41. McGuire SE, Roman G, Davis RL (2004) Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet 20 : 384–391. 15262411
42. Ferrandon D (2013) The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr Opin Immunol 25 : 59–70. doi: 10.1016/j.coi.2012.11.008 23228366
43. Ayres JS, Schneider DS (2009) The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS biology 7: e1000150–e1000150. doi: 10.1371/journal.pbio.1000150 19597539
44. Ayres JS, Freitag N, Schneider DS (2008) Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178 : 1807–1815. doi: 10.1534/genetics.107.083782 18245331
45. Ayres JS, Schneider DS (2008) A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS biology 6 : 2764–2773. doi: 10.1371/journal.pbio.0060305 19071960
46. Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6: e2. doi: 10.1371/journal.pbio.1000002 19222304
47. Osborne SE, Leong YS, O'Neill SL, Johnson KN (2009) Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS pathogens 5: e1000656–e1000656. doi: 10.1371/journal.ppat.1000656 19911047
48. Taillebourg E, Schneider DS, Fauvarque MO (2014) The Drosophila deubiquitinating enzyme dUSP36 acts in the hemocytes for tolerance to Listeria monocytogenes infections. J Innate Immun 6 : 632–638. doi: 10.1159/000360293 24777180
49. Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, et al. (2008) Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319 : 777–782. doi: 10.1126/science.1149357 18218863
50. Gordon MD, Dionne MS, Schneider DS, Nusse R (2005) WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature 437 : 746–749. 16107793
51. Paredes JC, Welchman DP, Poidevin M, Lemaitre B (2011) Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35 : 770–779. doi: 10.1016/j.immuni.2011.09.018 22118526
52. Kim LK, Choi UY, Cho HS, Lee JS, Lee WB, et al. (2007) Down-regulation of NF-kappaB target genes by the AP-1 and STAT complex during the innate immune response in Drosophila. PLoS Biol 5: e238. 17803358
53. Kim T, Yoon J, Cho H, Lee WB, Kim J, et al. (2005) Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat Immunol 6 : 211–218. 15640802
54. Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, et al. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16 : 1779–1791. 12130538
55. Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, et al. (2012) Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J Exp Med 209 : 661–669. doi: 10.1084/jem.20112343 22412156
56. Willemsen MH, Vulto-van Silfhout AT, Nillesen WM, Wissink-Lindhout WM, van Bokhoven H, et al. (2012) Update on Kleefstra Syndrome. Mol Syndromol 2 : 202–212. 22670141
57. Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, et al. (2002) Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis 34 : 51–57. 12324947
58. Goto A, Kadowaki T, Kitagawa Y (2003) Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Developmental Biology 264 : 582–591. 14651939
59. Bach EA, Vincent S, Zeidler MP, Perrimon N (2003) A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics 165 : 1149–1166. 14668372
60. McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302 : 1765–1768. 14657498
61. Okamura K, Ishizuka A, Siomi H, Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18 : 1655–1666. 15231716
62. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann Ja (1996) The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86 : 973–983. 8808632
63. Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints Am J Epidemiol 27 : 493–497.
64. Isoe J, Kunz S, Manhart C, Wells MA, Miesfeld RL (2007) Regulated expression of microinjected DNA in adult Aedes aegypti mosquitoes. Insect Mol Biol 16 : 83–92. 17257211
65. Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, et al. (2011) Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog 7: e1002189. doi: 10.1371/journal.ppat.1002189 21909258
66. Yasuhara JC, Wakimoto BT (2008) Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet 4: e16. doi: 10.1371/journal.pgen.0040016 18208336
67. Martin D, Brun C, Remy E, Mouren P, Thieffry D, et al. (2004) GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol 5: R101. 15575967
68. Hulsen T, de Vlieg J, Alkema W (2008) BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9 : 488. doi: 10.1186/1471-2164-9-488 18925949
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



