-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
There are three members of the α1,2fucosyltransferases gene family in mammalian genomes, Fut1, Fut2 and Sec1. The encoded fucosyltransferases are key enzymes for the synthesis of glycans that can be used as ligands by pathogens. However, the polymorphism of expression of these fucosylated glycans on epithelial cell types contributes to protection at the species level. In most mammalian species Sec1 is a pseudogene and in humans, genetic variation of α1,2fucosylated glycans is provided by FUT2 polymorphisms. Rabbit haemorrhagic disease virus (RHDV) uses α1,2fucosylated glycans as attachment factors. It induces an acute disease with very high mortalities in rabbit populations. We now confirm an association between genetic markers in the rabbit Sec1-Fut2 genomic region and survival to RHDV. We show that the Fut1 gene is the main contributor to the synthesis of RHDV binding sites although individual variation is not achieved by Fut1 polymorphisms but by variation in levels of Sec1 transcription. The Sec1 protein acting as a dominant-negative of Fut1, high Sec1 expression leads to a decreased number of RHDV binding sites. Thus, unlike in other mammals, in rabbits Sec1 underwent neofunctionalization. It contributes to generate diversity of fucosylated glycans, a key mechanism for escaping pathogens such as RHDV.
Published in the journal: . PLoS Pathog 11(4): e32767. doi:10.1371/journal.ppat.1004759
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004759Summary
There are three members of the α1,2fucosyltransferases gene family in mammalian genomes, Fut1, Fut2 and Sec1. The encoded fucosyltransferases are key enzymes for the synthesis of glycans that can be used as ligands by pathogens. However, the polymorphism of expression of these fucosylated glycans on epithelial cell types contributes to protection at the species level. In most mammalian species Sec1 is a pseudogene and in humans, genetic variation of α1,2fucosylated glycans is provided by FUT2 polymorphisms. Rabbit haemorrhagic disease virus (RHDV) uses α1,2fucosylated glycans as attachment factors. It induces an acute disease with very high mortalities in rabbit populations. We now confirm an association between genetic markers in the rabbit Sec1-Fut2 genomic region and survival to RHDV. We show that the Fut1 gene is the main contributor to the synthesis of RHDV binding sites although individual variation is not achieved by Fut1 polymorphisms but by variation in levels of Sec1 transcription. The Sec1 protein acting as a dominant-negative of Fut1, high Sec1 expression leads to a decreased number of RHDV binding sites. Thus, unlike in other mammals, in rabbits Sec1 underwent neofunctionalization. It contributes to generate diversity of fucosylated glycans, a key mechanism for escaping pathogens such as RHDV.
Introduction
Following gene duplication the fate of paralogues can be quite variable. Both duplicate copies may be maintained due to a beneficial gene dosage effect or because of functional divergence from the ancestral locus, generating novel gene functions. Alternatively, one of the two duplicates might be driven to pseudogenization because of redundancy with the paralogue [1,2]. In this context, previous analyses of the α1,2fucosyltransferase gene family in mammals indicated that the three members of the family called Fut1, Fut2 and Sec1 arose by two successive rounds of duplication. The first duplication gave rise to Fut1 and to the ancestor of Fut2 and Sec1. The second one generated Fut2 and Sec1, these two genes being arranged in tandem in the genome [3,4,5]. Yet, their biological functions are not fully understood. In several mammalian species Sec1 has become a pseudogene, suggesting functional redundancy with either Fut1 or Fut2 [6,7,8,9]. In humans and mice, Fut1 and Fut2 are generally not expressed in the same cell types, strongly suggesting functional differentiation [9,10,11]. In accordance, recent data indicate that Fut1 would be involved in cellular functions such as the development of the olfactory bulb and possibly angiogenesis or adhesion of leukocytes to the vascular endothelium [12,13,14,15], whilst in human, FUT2 would be involved in a relationship with environmental pathogens including bacteria and enteric viruses [16,17,18,19,20,21]. However, in European rabbit (Oryctolagus cuniculus), Sec1 is not as clearly a pseudogene as in other mammalian species. The coding region of these fucosyltransferases genes is entirely comprised within one exon and it has been observed that in rabbits the Sec1 exon can encode a full protein with all the characteristics of a fucosyltransferase, albeit with a reduced enzymatic activity compared to those of Fut1 and Fut2 [22,23]. It was later observed that 5 out of 7 Sec1 alleles encoded proteins devoid of detectable enzymatic activity [23]. This indicated that in European rabbit Sec1 might still be in the process of pseudogenization, confirming its redundancy as in other mammalian species. Nevertheless, we observed extensive gene conversion involving Sec1 and both Fut1 and Fut2 in rabbits, unlike in several other mammalian species [4]. In those species, including primates, gene conversion could be documented between Sec1 and Fut2 only and was limited to the region coding the N-terminal part of the catalytic domain. Since gene conversion leads to homogenization [24], these observations did not fit with the idea that Sec1 was undergoing pseudogenization in rabbits, but suggested that this gene family was undergoing a specific path of evolution in that species. Interestingly, it was later observed that genetic polymorphisms at the Sec1 locus are associated with resistance to rabbit haemorrhagic disease virus (RHDV) [23].
RHDV is a member of the genus Lagovirus of the Caliciviridae family responsible for the rabbit haemorrhagic disease (RHD) that was first reported in China in 1984 [25]. RHD is extremely lethal and highly contagious in domestic and wild adult European rabbits (Oryctolagus cuniculus) of both subspecies O. c. cuniculus and O. c. algirus [26]. Resistance to RHD in rabbits below two months of age was readily noticed in the first outbreaks. It was later hypothesized that this could be explained by the weak expression in young rabbits of histo-blood group antigen H type 2 to which RHDV was shown to attach on the epithelial cells of the upper respiratory and digestive tracts, the most likely doors of entry of the virus [27]. Histo-blood group antigens (HBGAs) are carbohydrate structures of glycoproteins and glycolipids that show diversity of expression between individuals. Their function is not completely clear but their location on the most outer part of epithelia suggests that they may be involved in establishing the first contact between hosts and pathogens [16]. In support of this concept various bacterial adhesins, as well as viral capsid proteins, show specificity for these polymorphic glycans and associations between HBGA phenotypes and risk of infection have been described [16,28,29,30]. Human and animal caliciviruses of the Norovirus genus also use HBGAs as attachment factors to infect their host [28]. It appeared that distinct strains of virus bind to different HBGAs structures and that due to the human HBGA polymorphisms, none of these strains can infect the entire population [19]. Because of its high virulence and its documented ability to recognize HBGA structures, RHDV provides a unique model to study the impact of a pathogen on HBGA diversity and reciprocally on the effect of this diversity on the pathogen’s transmission and virulence. However, if the genetic basis of HBGAs diversity in humans is well described, it has been very poorly studied in other mammals, including rabbits, particularly in wild animals.
HBGAs result from the sequential addition of monosaccharides to a precursor glycan chain through specific glycosyltransferases. In particular, synthesis of H type 2 is performed by addition of a fucose residue onto the galactose of the precursor by an α1,2fucosyltransferase. Synthesis can then proceed by addition of either an N-acetylgalactosamine or a galactose in α1,3 linkage to the galactose residue of the precursor to generate the A and B blood group antigens of the ABO system [16]. As mentioned above, there are three α1,2fucosyltransferase genes in mammals: Fut1, Fut2 and Sec1 and in humans and several other species Sec1 is a pseudogene. In humans FUT2 is a highly polymorphic gene with several null or weak alleles undergoing balancing selection [31,32,33]. Homozygous individuals for these alleles possess the nonsecretor phenotype characterized by the absence of blood group A, B, and H antigens in secretions and on various types of epithelial cells. So-called nonsecretor individuals are resistant to various strains of human calicivirus belonging to the Norovirus genus that use α1,2fucosylated ligands for attachment [34]. In this context, it was anticipated that polymorphisms of the rabbit α1,2fucosyltransferases could similarly contribute to control the resistance/susceptibility to RHDV. However, a study on the polymorphisms in the coding sequence (CDS) of rabbit Fut2 and Sec1 revealed no effect on Fut2 enzyme activity, whereas for Sec1 either weakly active or inactive enzyme variants were found [23]. Most importantly, Guillon and co-workers found an association between a Sec1 variant and survival to a devastating RHDV outbreak in a wild rabbit population. This variant encoded a weakly functional enzyme and was always associated with functional Fut2 variants. It was thus suggested that this Sec1 allele was in linkage disequilibrium with an unknown polymorphism affecting Fut2 expression since the two genes are located in tandem in the genome [23].
To progress in our understanding of the genetics of HBGAs expression in rabbits and the relationship with resistance/susceptibility to RHDV, we analysed the variation in Fut1, Fut2 and Sec1 RNA expression in rabbit duodenum tissue, the impact of rabbit Fut1 CDS polymorphisms on enzyme activity and the relationship between the expression of these three fucosyltransferases genes and virus binding to the duodenum. We unexpectedly uncovered a new mechanism of intra-species production of glycan diversity whereby a paralogue gene (Sec1) encodes a protein with a dominant-negative effect on its active isoform (Fut1), corresponding to a neofunctionalization of Sec1 in rabbits. Further analysis of gene conversion between the three members of the α1,2fucosyltransferase family in mammals suggests that this mechanism is unique to leporids and that the fate of the three members of this gene family diverged between leporids and other mammals.
Results
Genetic polymorphisms of the rFut2 5’ genomic region and lack of association with mRNA expression
It was hypothesized earlier that, in European rabbit, differences in expression of H type 2, a main RHDV ligand or precursor to the A type 2 and B type 2 RHDV ligands, may result from variation in Fut2 gene transcription rather than enzyme activity [23]. Since the rabbit genome is only partially sequenced, in order to characterize polymorphisms in the promoter region of the rabbit Fut2 gene (rFut2), the genomic region harboring the three α1,2fucosyltransferase genes was first reconstructed from sequences acquired from a genome walking approach and from sequences available in databases. The general organization of this region was found similar to the orthologous human genomic region located on chromosome 19 (Fig 1). Sequencing of 930 bp 5’ of the untranslated first exon of rFut2 in 65 wild rabbits from Chèvreloup, France, 23 wild rabbits from the Pyrénées Orientales, France, and 29 domestic rabbits, yielded 27 polymorphic positions that segregated into 13 haplotypes, called variants 1–13 (Table 1). Animals from Chèvreloup had been sampled prior to the 1995 and 1996 outbreaks and their survival had been monitored. Variants 1–7 were found in this population. As shown on Fig 2, an association between the presence of variant 3 and survival was observed, confirming the existence of an association between polymorphisms in this genetic region and survival to RHD in wild animals. Indeed, the proportion of rabbits that died during the RHD outbreak differs significantly between variant 3-rabbits (2/25) and other rabbits (42/105) (χ2 = 9.23, df = 1, p = 0.002). In order to test if rFut2 promoter region variant 3 was responsible for a low transcription of the rFut2 gene, we looked for the relationship between promoter region variants and expression of all three α1,2fucosyltransferase genes by quantitative RT-PCR in the duodenum from a group of 23 wild and 6 domestic rabbits. The RT-PCR data were normalized against two house-keeping genes, GAPDH and β-actin, giving similar results. There was no relationship between rFut2 expression and the presence of the promoter variant 3 (v3) or any other variant. Indeed, relative expression of rFut2 mRNA normalized to GAPDH in v3 positive rabbits and v3 negative rabbits were 944 +/ - 240 and 612 +/ - 90, respectively and normalized to β-actin 143 +/ - 33 and 157 +/ - 49, respectively. These data indicate that the v3 promoter variant did not affect rFut2 gene transcription, which invalidated the hypothesis.
Fig. 1. Comparative scheme of the organization of the genome region encompassing α1,2fucosyltransferases genes in European rabbit and human. 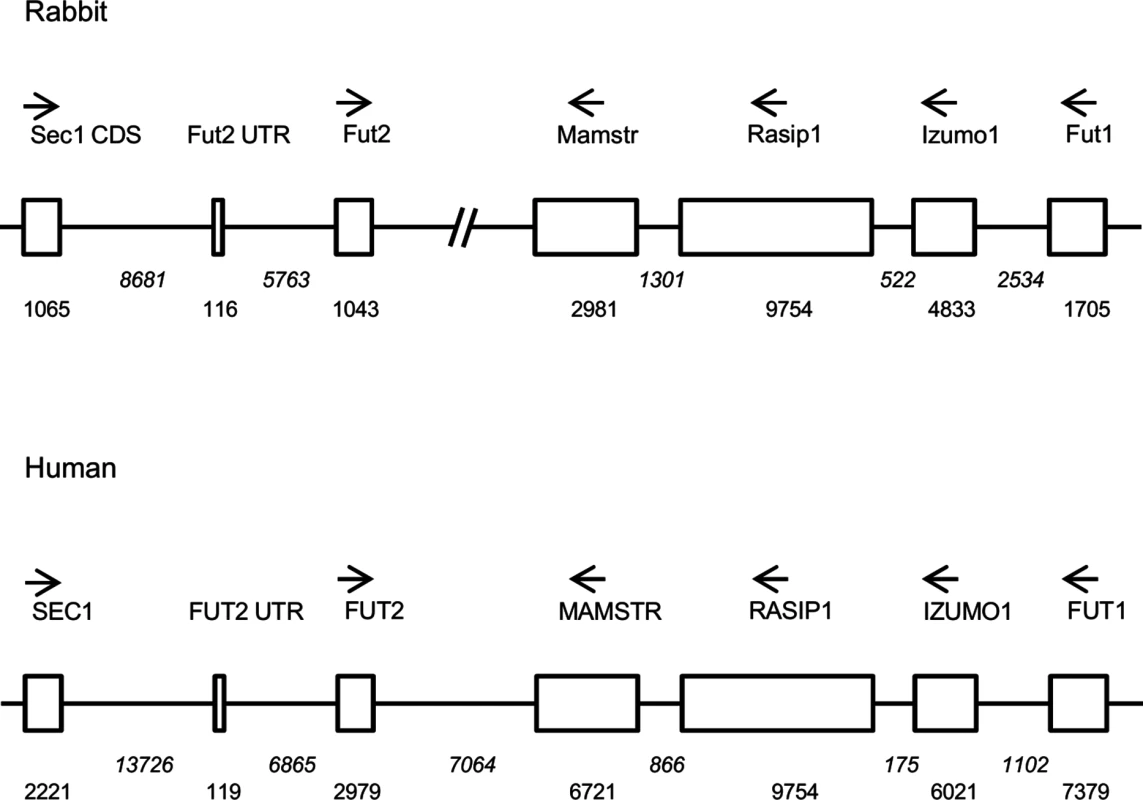
Genes including coding regions are boxed. Orientation of the genes is given by arrows and distances in nucleotides are given below boxes (in italics for introns and intergenic regions). In the case of Fut2, location of the non coding first exon (Fut2 UTR) is given in order to show the distance between its 5’ putative regulatory region and the Sec1 gene coding determining region (Sec1 CDS). Fig. 2. Allele frequencies of the rFut2 promoter region of dead and survival animals in the 1995–1996 RHDV outbreaks at Chèvreloup, France. 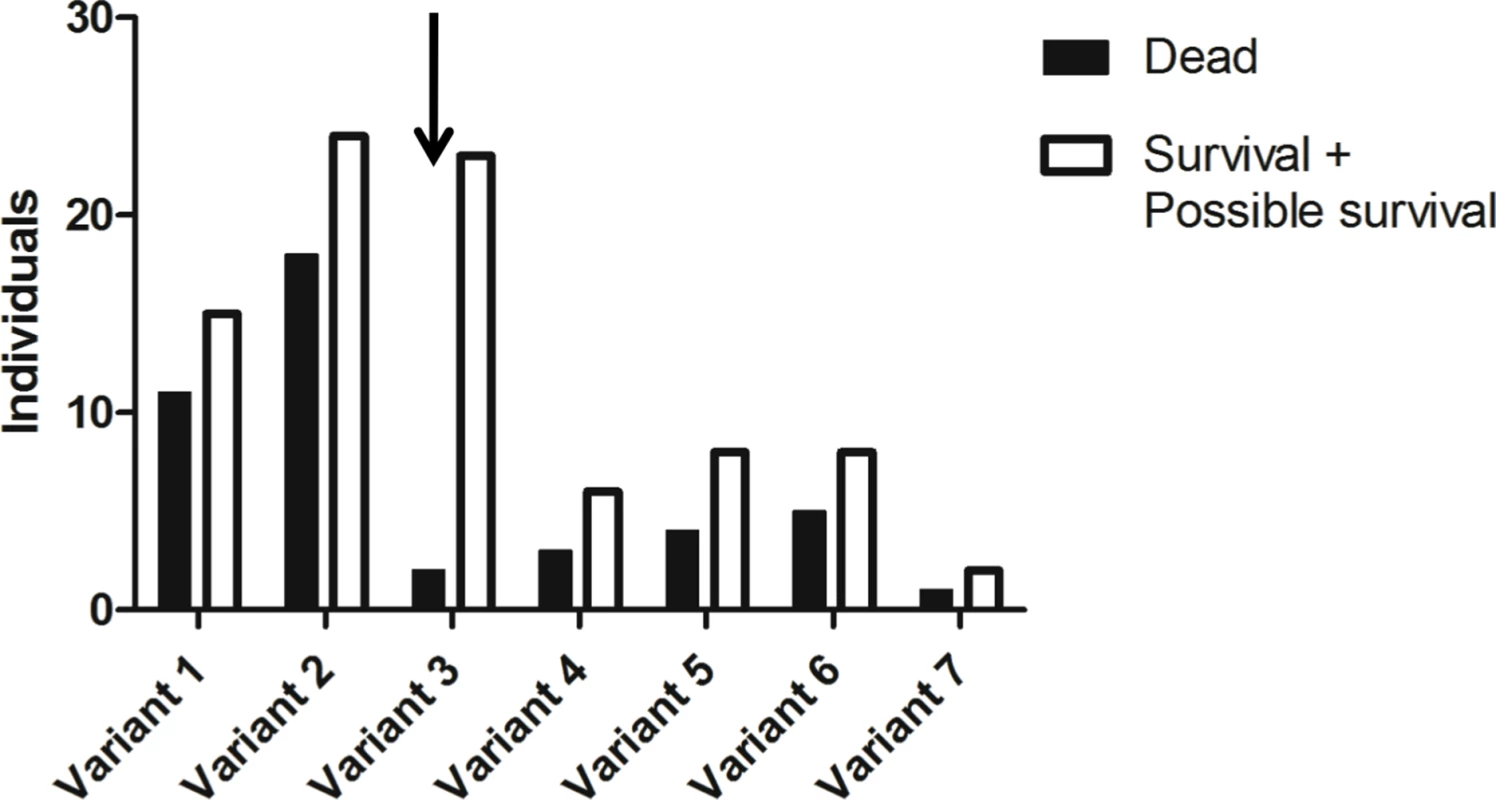
The group of survivors may contain a few animals that are adults of the first descendants of survivors rather than direct survivors due to sampling uncertainties (probable survivors). Frequency of the variant 3 allele (arrow) is significantly increased among survivors compared to nonsurvivors (P = 0.002). Tab. 1. Nucleotide variation found in French wild and laboratory animals in the putative promoter region of rFut2 and inferred haplotypes. 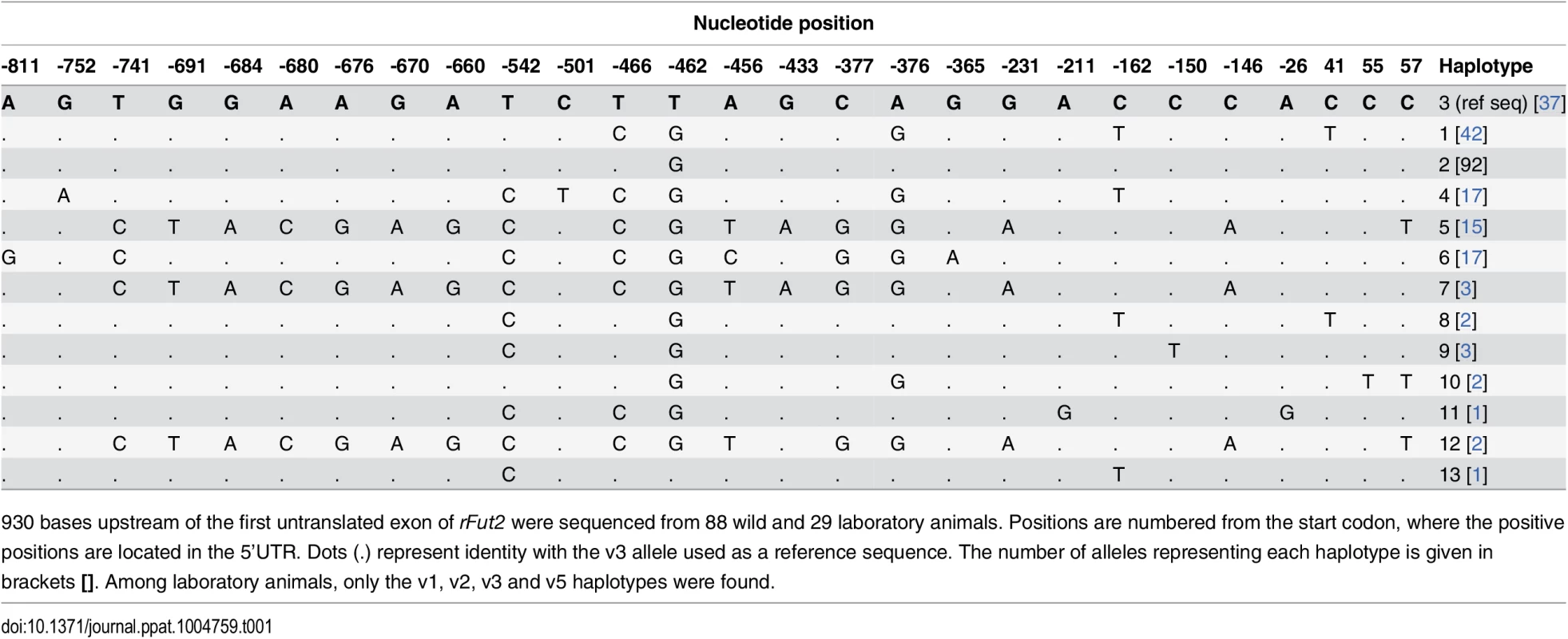
930 bases upstream of the first untranslated exon of rFut2 were sequenced from 88 wild and 29 laboratory animals. Positions are numbered from the start codon, where the positive positions are located in the 5’UTR. Dots (.) represent identity with the v3 allele used as a reference sequence. The number of alleles representing each haplotype is given in brackets []. Among laboratory animals, only the v1, v2, v3 and v5 haplotypes were found. rFut1 is the main contributor to RHDV binding sites in the rabbit gut
Quantitative RT-PCR analysis of rFut1 and rFut2 expression indicated that the two genes were expressed at similar levels with variation between individuals in the 10-fold range (Fig 3). This was unexpected since it had been reported earlier that rFut1 was not expressed in the gut [35]. In order to determine which gene, of rFut1 and rFut2, contributes the most to RHDV binding sites, we first tested the binding of six distinct RHDV strains, representative of the diversity of RHDV, to a set of immobilized synthetic oligosaccharides of the HBGA family. As shown on Fig 4, the six strains bound to various degrees to either H, A or B type 2 structures. However, no binding was observed to either type 1 or type 3-based A, B or H structures as previously observed [36]. Therefore H type 2, but not H type 1 or H type 3, is either a preferred ligand or the precursor to a preferred RHDV ligand (A type 2 or B type 2). Since it has been previously shown that the human FUT1 enzyme shows some preference for the type 2 precursor over the types 1 or 3 precursors whereas the opposite is true for the human FUT2 enzyme [37,38,39], we sought to determine whether similar preferences for the acceptor substrates would also exist for the rabbit Fut1 and Fut2 enzymes and whether some rFut1 polymorphisms could contribute to a low expression of RHDV ligands.
Fig. 3. RNA expression of the α1,2fucosyltransferases genes in the rabbit duodenum. 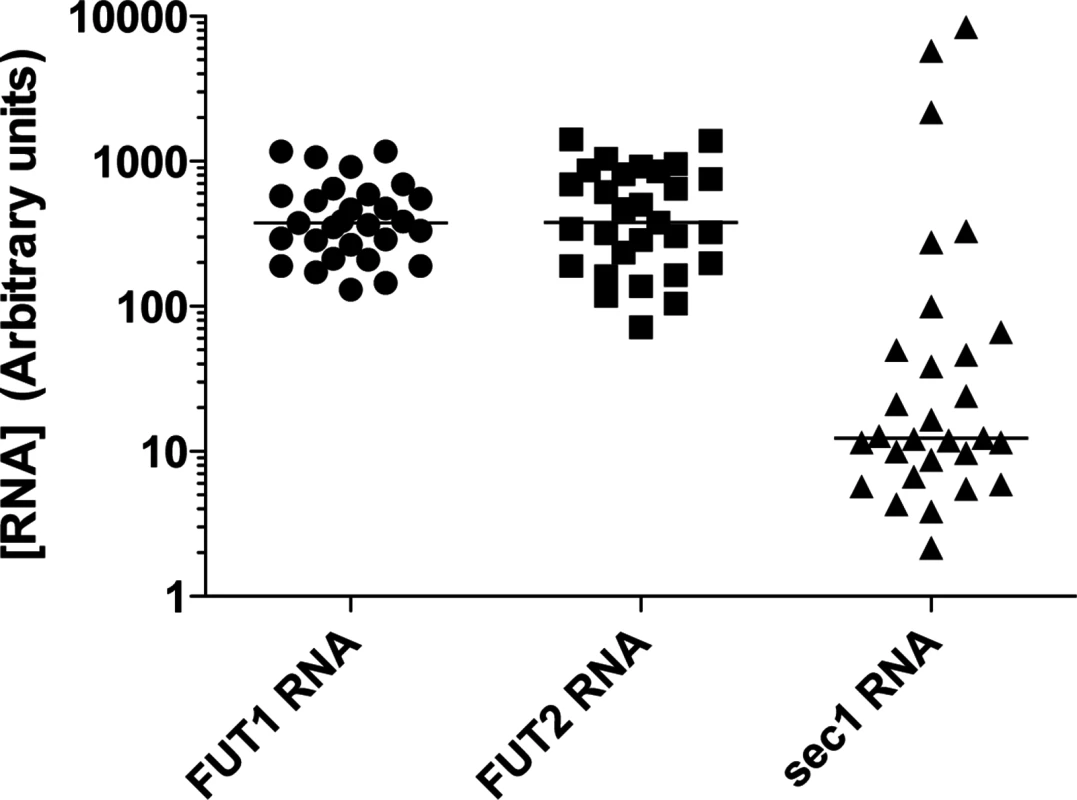
Total RNA from duodenum scrapings of individual animals was analyzed using quantitative real-time RT-PCR. Each circle, square, or triangle represents RNA concentration of Fut2, Fut1 and Sec1 of individual animals normalized to GAPDH. The bars indicate median values for each gene. Fig. 4. Binding of RHDV strains to immobilized synthetic oligosaccharides. 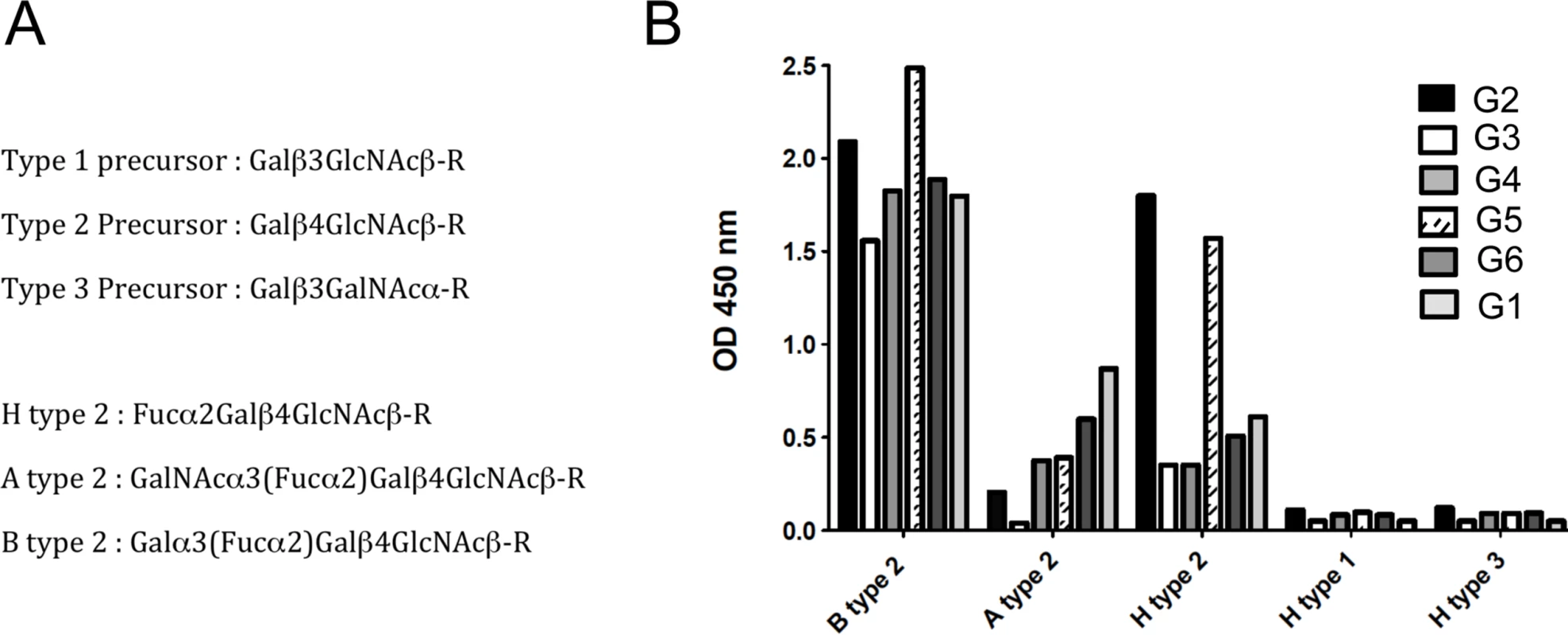
RHDV liver extracts containing 2x109 viral RNA copies, determined by quantitative real-time RT-PCR as previously described, were incubated on immobilized neoglycoconjugates. Binding was detected using primary antibodies against the RHDV capsid protein and HRP-conjugated secondary reagents as described in the materials and methods section. Structures of the types 1, 2 and 3 precursors from which A, B and H structures are built upon addition of monosaccharides. Structures of the H type 2, A type 2 and B type 2 epitopes based on the type 2 precursor are also shown (A). Binding to B type 2, A type 2 and H type 2 of the different RHDV strains is shown as OD 450nm values. No binding was observed to H type 1 or H type 3 (B). The rFut1 coding sequence was thus obtained from 41 animals from France (Chèvreloup n = 19, Cerisay n = 22) and 27 animals from Portugal (Pancas n = 15, Mértola n = 5, Santarém n = 7), yielding a total of 33 polymorphic positions when compared to the rabbit Fut1 reference sequence (GenBank accession number X80226) (Fig 5A). All 68 individuals presented a silent mutation compared to the reference sequence, which for that reason is not displayed on the figure. Three of the polymorphic positions, with one being non-synonymous, were found in the French populations. In the Portuguese populations, 10 mutations out of 30 were non-synonymous. The mutations were distributed almost uniformly along the coding sequence, although the non-synonymous substitutions were mainly located toward the 5’ end. Five polymorphisms were shared with those previously described for rFut2 and rSec1, while two where shared only with rFut2 [23]. According to the amino acid variation, two and eleven haplotypes (or variants) were inferred for the French and Portuguese populations, respectively (Table 2). No shared variants were detected between these two groups of populations. In Portugal variants 4, 8, 9, 12 and 14 were rare (frequency<0.02), variants 3 and 10 had a frequency of 0.037, variant 11 a frequency of 0.055, variant 7 a frequency of 0.11, variant 6 a frequency of 0.17 and variant 5 a frequency of 0.5. Variant 2 was the most common in France with a frequency of 0.9. For amino acid position 140, three different amino acids were detected: S, P and Q. The low genetic diversity observed for the French populations when comparing with the Portuguese populations is in accordance with what has been shown for other genetic markers [40,41,42].
Fig. 5. (A) Distribution of rFut1 polymorphisms along the coding region. 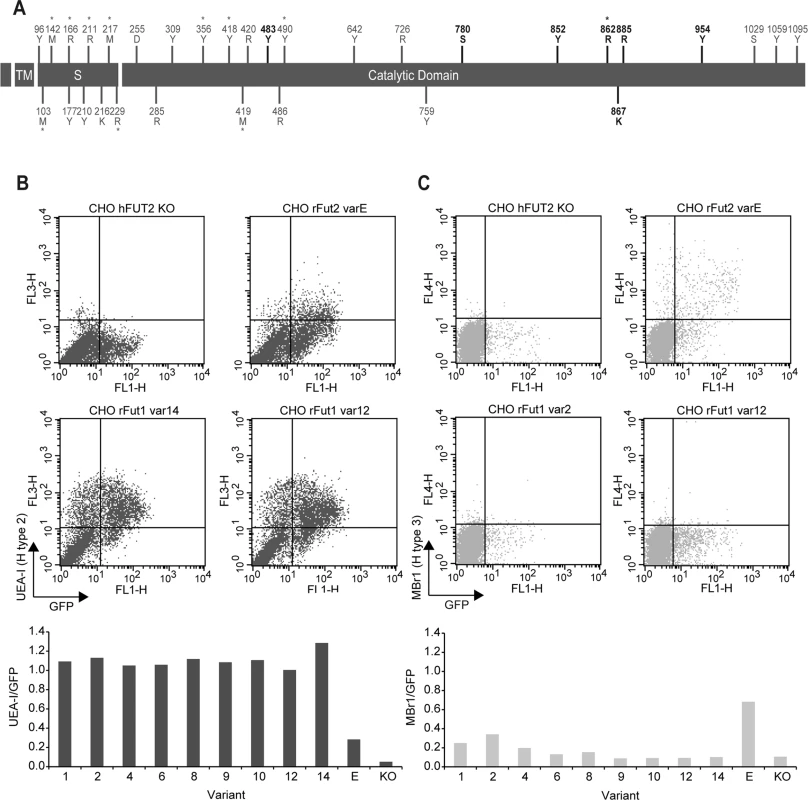
The protein domains are represented by black boxes (tm, transmembrane domain; S, stem region). Nucleotide variations are shown above boxes (R = A or G; Y = C or T; K = G or T; S = G or C; M = A or C; D = A, G or T). Amino acid variations are shown below boxes. Positions numbering begins at the start codon and first methionine respectively. The positions in bold refer to the polymorphic positions shared by Sec1, Fut2 and Fut1 (positions 780, 852, 862, 867 and 885) and to the polymorphic positions shared by Fut2 and Fut1 (positions 483 and 954). Asterisks represent non-synonymous substitutions. (B; C) Flow cytometry analysis of wild rabbit rFut1 variants. Enzymes were produced by transfection in CHO cells as N-term GFP fusion proteins. Synthesis of H type 2 and H type 3 was determined using the UEA-I lectin (B) and the Mbr1 mAb (C), respectively. Transfection of a human null allele (hFUT2 KO) fused to GFP provided a negative control and transfection of the rFut2 allele variant E corresponding to the rFut2 reference sequence provided a positive control. The upper panels show examples of results obtained with two selected rFut1 variants. The lower panels show the ratios between the percentages of UEA-I (H type 2) or Mbr1 (H type 3) positive cells and the percentage of GFP positive cells. In each case, the activity of rFut1 variants is compared to that of the reference rFut2 (variant E). Tab. 2. Fut1 amino acid variation found in the Portuguese and French rabbit populations and the inferred haplotypes. 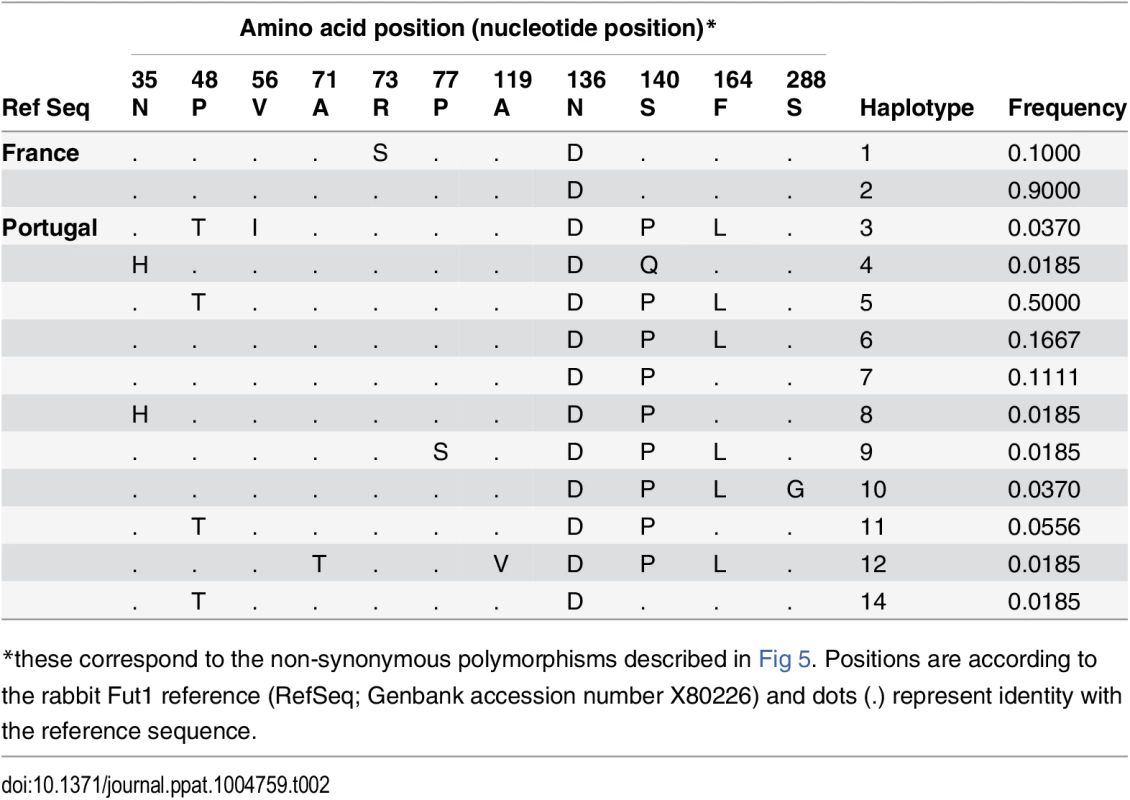
*these correspond to the non-synonymous polymorphisms described in Fig 5. Positions are according to the rabbit Fut1 reference (RefSeq; Genbank accession number X80226) and dots (.) represent identity with the reference sequence. After cloning in an expression vector, rFut1 variants 1, 2, 4, 6, 8, 9, 10, 12 and 14 were tested for their catalytic activity. Variants 5 and 7 were not tested since the amino acid variation described above was almost completely covered by the other variants. Only variation at amino acid position 56 (V/I) was not included as it is located in the stem region and thought to be highly conservative. Variant 11 was not tested since cloning of the full coding sequence failed despite several attempts. Following transfection into CHO cells, the presence of fucose in α1,2 linkage was measured by flow cytometry using the UEA-I lectin that detects H type 2 and a monoclonal antibody (MBr1) that detects H type 3. CHO cells do not express type 1 precursor and therefore synthesis of H type 1 was not measured. The presence of a GFP tail in the recombinant enzymes allowed for control and normalization of transfection and protein expression efficiencies. Synthesis of H type 2 was similar for all variants tested and much higher than for the rFut2 enzyme (rFut2 variant E) used as a positive control (Fig 5B). By contrast, synthesis of H type 3 was either low or undetectable, whereas it was clearly visible for the rFut2 control (Fig 5C). Since, as described above, rFut1 and rFut2 mRNA are expressed at similar levels in the duodenum, these results indicate that rFut1 should be the main contributor to H type 2 synthesis and therefore to RHDV binding sites.
rSec1 gene expression is associated with the level of RHDV binding to the duodenum
Quantitative RT-PCR indicated that on average Sec1 mRNA levels were much lower than those of rFut1 and rFut2 in individual rabbits (p<0.0001). However, the 10-fold variation in rFut1 and rFut2 mRNA expression appeared quite limited compared to the extreme individual variation found in rSec1 mRNA levels that varied over 1000-fold (Fig 3).
In order to determine whether transcription of the α1,2fucosyltransferase genes could be related to the presence of RHDV binding sites, attachment of the virus to the duodenum extracts was determined in the same group of animals. To this aim, six RHDV strains representative of each virus genotype were used [36]. No relationship was observed between either rFut1 or rFut2 expression. However, there was a clear relationship between rSec1 mRNA expression and RHDV binding (Fig 6). Thus, all animals classified as high rSec1 mRNA expressers were among the poor binders of most RHDV strains.
Fig. 6. Relationship between expression of rSec1 RNA and recognition of individual rabbits by RHDV strains. 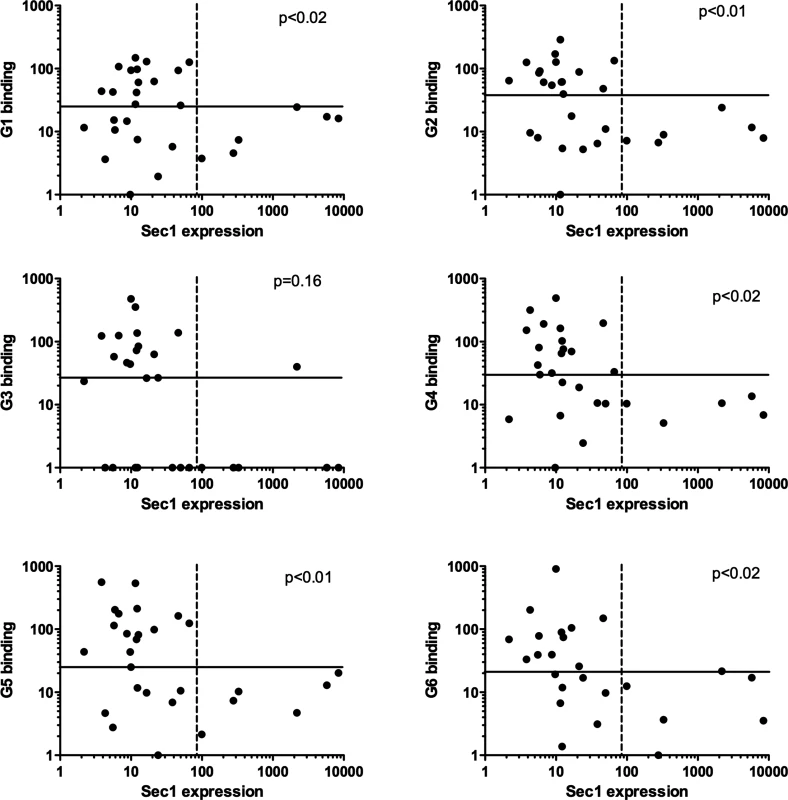
Binding to individual duodenum extracts of six strains of RHDV representative of the virus diversity was determined as described in the materials and methods section. rSec1 RNA expression in the duodenum of the same rabbits corresponds to the values shown in Fig 3. Plain lines separate high and low RHDV binders defined as above or below median values. Dashed lines separate high and low expressors of rSec1 mRNA defined as animals being above or below the Fut1 and Fut2 range of expression. For each RHDV strain P values were obtained from analysis of these categories by Fisher’s exact test. Treating values as continuous variable and testing correlations between rSec1 RNA expression and RHDV binding using the Spearman correlation test also yielded significant relationships (p<0.05) for G2, G3, G4 and G6. rSec1 inactive protein acts as a dominant-negative of rFut1
Since high expression of rSec1 mRNA was related to alleles encoding an inactive enzyme and to low binding of RHDV, we reasoned that the catalytically inactive rSec1 protein might impair synthesis of RHDV binding sites (A, B or H type 2). As described above, rFut1 appears to be the main enzyme controlling for synthesis of A, B and H type 2 in the rabbit duodenum, thus we analyzed the expression of RHDV binding sites at the surface of CHO cells co-transfected with rFut1 and an inactive rSec1 variant. In absence of rFut1 transfection no RHDV binding sites were detected on CHO cells, consistent with their lack of endogenous α1,2fucosyltransferase activity (Fig 7). rFut1 and rSec1 were fused to a GFP at their N-terminus in order to control for the efficacy of transfection and of protein synthesis. In addition, transfections were performed using the same total amount of plasmid DNA encoding for either rFut1-GFP, rSec1-GFP or a truncated inactive rFUT2-GFP fusion protein in order to maintain a constant amount of translated GFP-fusion protein. Under all conditions, GFP was expressed at similar levels and transfection of Sec1 alone did not generate RHDV binding sites. By contrast, transfection of Fut1 alone allowed synthesis of RHDV binding sites since strong RHDV binding was observed in over 10% of the total population of transiently transfected cells. Interestingly, we observed that in the presence of rSec1, synthesis of RHDV binding sites by transfected rFut1 was significantly decreased. The effect was amplified when a higher amount of rSec1 than of rFut1 encoding plasmid was used. Indeed, a 50% decrease in percentage of RHDV binding cells as well as a decrease in their intensity of fluorescence was observed when a ratio of rFut1/rSec1 plasmids of 1 : 9 was used in the transfection experiment (Fig 7A). To confirm that the decreased RHDV binding was due to a decreased synthesis of H type 2 ligands, the same experiment was performed using an anti-H type 2 antibody. In the presence of rSec1, both the anti-H type 2 and RHDV cell labeling were decreased compared to those of cells transfected with rFut1 alone (Fig 7B). To further confirm these results, we then generated stable transfectants of rFut1. These cells express H type 2 epitopes and upon transfection of rSec1, the percentage of cells expressing the H epitope and attaching RHDV were clearly decreased (Fig 7C). The loss of H type 2 RHDV-binding sites was only partial, likely because only a fraction of the cells was actually transfected by rSec1. These results indicate that the presence of an inactive rSec1 protein blocks synthesis of H type 2 ligands by rFut1. In order to determine how rSec1 blocks the activity of rFut1, we performed confocal microscopy experiments to observe the intracellular localization of these enzymes. This type of glycosyltransferases is known to be expressed in the Golgi apparatus [43]. We observed that cells transfected with rFut1 containing a DSRed tag in N-terminal position showed a typical Golgi-like perinuclear labeling. Cells transfected with a GFP-tagged rSec1 were labeled in a less well-defined cellular compartment that also may be compatible with the Golgi. Strikingly, all cells doubly expressing rFut1 and rSec1 showed a cytoplasmic labeling with both the red (rFut1) and the green (rSec1) tags, clearly indicating that the presence of rSec1 displaced rFut1 out of the Golgi to the cytoplasm, explaining the impairment of its function (Fig 8).
Fig. 7. Assay of rSec1 expression on rFut1 activity. 
CHO cells were co-transfected with a total amount of 10 μg of DNA from an expression plasmid encoding either the Sec1-GFP fusion protein (9 μg) and the truncated hFUT2-GFP (1 μg): Fut1/Sec1 0:9; the truncated rFut2-GFP alone (10 μg): Fut1/Sec1 0:0; the rFut1-GFP fusion protein (1 μg) and the truncated rFut2-GFP (9 μg): Fut1/Sec1 1:0; the rFut1-GFP plasmid (1 μg), the rSec1-GFP plasmid (1 μg) and the truncated rFut2-GFP (8 μg): Fut1/Sec1 1:1; the rFut1-GFP plasmid (1 μg) and the rSec1-GFP plasmid (9 μg). Forty eight hours after transfection, binding of a G2 RHDV strain was determined by flow cytometry. Percentage of positive cells recorded on the FL1 channel for GFP, the FL4 channel for RHDV binding (among total cells) and mean fluorescent intensities of RHDV positive cells are shown. Values represent means and S.D. from 3 independent experiments. * P<0.05; **P<0.01, ***P<0.002 (A). Transfection of CHO cells with the truncated rFut2-GFP alone (10 μg): Fut1/Sec1 0:0; the rFut1-GFP fusion protein (1 μg) and the truncated rFut2-GFP (9 μg): Fut1/Sec1 1:0; the rFut1-GFP plasmid (1 μg), the rSec1-GFP plasmid (1 μg) and the truncated rFut2-GFP (8 μg): Fut1/Sec1 1:1; the rFut1-GFP plasmid (1 μg) and the rSec1-GFP plasmid (9 μg): Fut1/Sec1 1:9. Forty-eight hours after transfection, binding of either an anti-H antibody (12.4-LE) or a G2 RHDV strain were determined by flow cytometry. Data represent mean % positive cells from an experiment performed in duplicates (B). Flow cytometry plot of the anti-H antibody staining of stably rFut1-transfected CHO cells (negative control in grey). Transfection of rSec1 into stably rFut1-transfected cells. Cells were transfected with either truncated rFut2-GFP (10 μg): Sec1:0; truncated rFut2-GFP (9 μg) and Sec1-GFP (1 μg): Sec1:1; or Sec1-GFP(10 μg): Sec1:10. Forty-eight hours following transfection binding of the anti-H and of RHDV were assessed as above. Results are shown as % of positive cells relative to the negative controls performed in absence of primary antibody or of virus from an experiment performed in duplicates (C). Fig. 8. Intracellular localization of the rFut1 and rSec1 proteins. 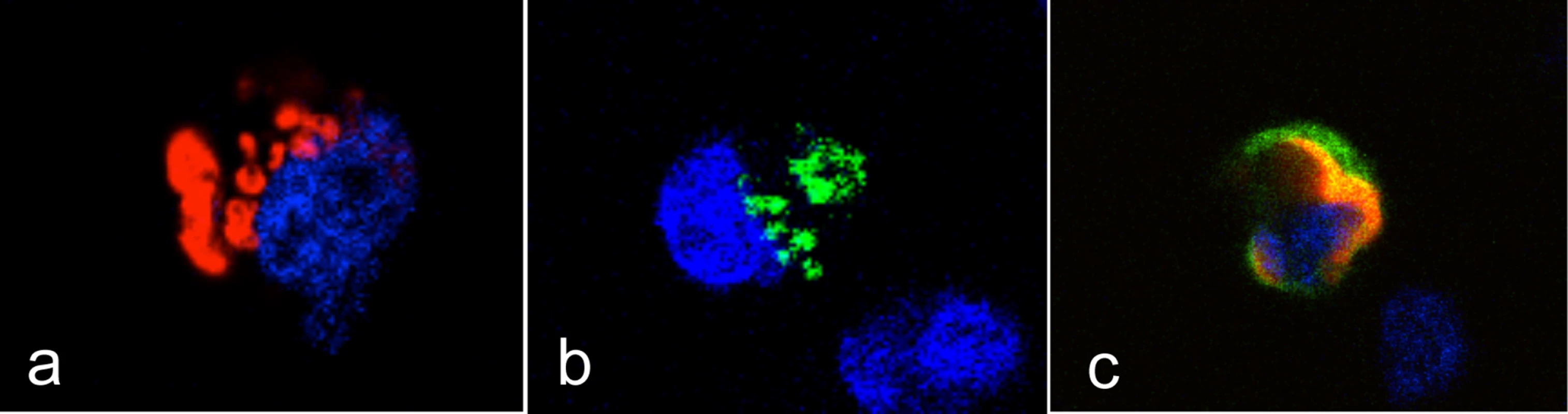
CHO cells were transfected with either rFut1 with a DSred tag (a), rSec1 with a GFP tag (b), or both (c). Images of representative cells expressing either protein alone or co-expressing both proteins are shown. Nuclei are stained in blue. Evolution of the α1,2fucosyltransferases genes in leporids: A particular gene conversion pattern
A limited and most likely ancient gene conversion between Sec1 and Fut2 in primates and several other mammalian species has been described earlier [4]. In these species gene conversion was restricted to the gene fragment coding the N-terminal domain of the catalytic region. Since in the same species Sec1 is a pseudogene presenting clear coding-sequence alterations we concluded that gene conversion with either Fut1 or Fut2 would likely no longer be observed because of deleterious effects on the Fut1 and Fut2 enzymes. However, we additionally observed that in European rabbit, the three α1,2fucosyltransferases genes are involved in gene conversion all along the catalytic domain coding sequences. This observation led us to hypothesize that in rabbits, Sec1 might have acquired a function distinct from those of the two other α1,2fucosyltransferases genes. The results presented above indicating that rSec1 acts as a dominant-negative of rFut1 are fully consistent with its neofunctionalization in rabbits. In order to determine if gene conversion involving the three members of the α1,2fucosyltransferase gene family could also be observed in other species, which would be consistent with the acquisition of a new function in Sec1, we analyzed the sequences of the three genes (Fut1, Fut2, Sec1) in 23 mammalian species including six genera of leporids.
Visual inspection of the alignment of the catalytic domain of the three enzymes in the different mammalian species highlighted major differences in the gene conversion extent, particularly between leporids and the remaining mammals (Fig 9 and S1 Fig). In leporids, amino acid motifs shared between the Fut2 and Sec1 encoded enzymes are visible from the beginning of the alignment and extend up to amino acid position 217. In contrast, for the remaining mammals, motifs are only shared between amino acids 133 and 179. Other Fut2-Sec1 shared motifs are observed, but they extend for a few amino-acids only; in pigs, however, these motifs are larger although Sec1 is a pseudogene. Regarding Fut1-Fut2-Sec1 shared motifs, and although they can be identified for most species, in leporids they extend from amino acid position 262 to 305. In addition, in the 3’ region (amino acid positions 341 to 351), a short fragment of gene conversion can also be observed for leporids (Fig 9). Interestingly, in primates, this region is located immediately downstream of the Sec1 premature stop codon in gorillas, humans and chimpanzee (S1 Fig).
Fig. 9. Alignment of the catalytic domain of α1,2fucosyltransferases for 6 leporid species illustrating the gene conversion events. 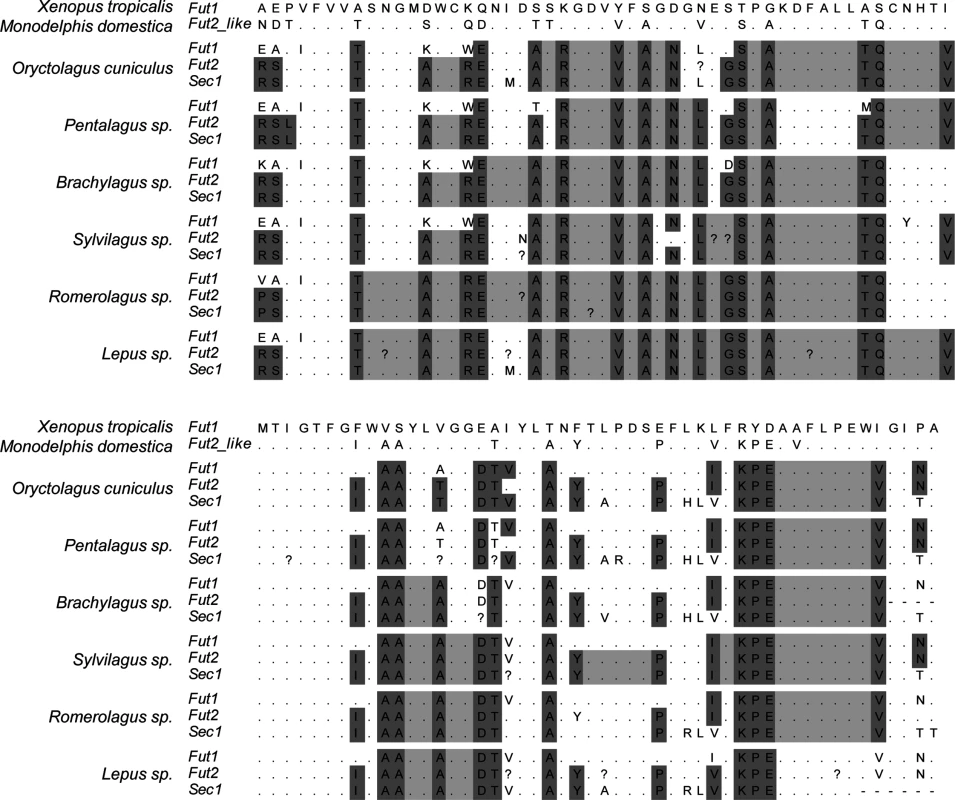
For each species the 3 enzymes are shown. Dark grey: positions that differ in more than one gene relatively to Xenopus tropicalis. Light grey: positions that are identical in more than one gene between the positions identified by dark grey. Black: premature stop codons. Dots mean identity with the X. tropicalis sequence.? indicate positions that were ambiguous upon sequencing. The upper part of the alignment comprises amino acid positions 254 to 304 of Homo sapiens FUT1, the lower part comprises amino acids positions 305 to 354 of Homo sapiens FUT1. Full names of each species and accession numbers of sequences are given in Supplementary Table 1. In order to determine recombination breakpoints, a GARD analysis was performed considering three distinct groups: all mammals, all mammals except leporids and only leporids. This partition was according to the different extent of gene conversion as observed from the amino acid alignment. One recombination breakpoint was consistently detected when analyzing mammals at nucleotide 375 of the alignment. When analyzing leporids only, the recombination breakpoint was detected at nucleotide 501 (Table 3).
Tab. 3. List of the breakpoints detected by GARD and their statistical significance. 
1 Akaike Information Criteria (AICc) for b = 0, b = 1 and improvement relative to the model with one fewer breakpoint (ΔAICc). The phylogenetic trees (S2 Fig) constructed for the three genes considering the different dataset partitions and the recombination breakpoints identified by GARD (Table 3) showed a wider extent of gene conversion between Fut2 and Sec1 in leporids than in the other groups of mammals analyzed. Although gene conversion events involving Fut1 are apparent from visual inspection of Fig 9 and S1 Fig, which suggested its involvement in the gene conversion in leporids, they are not clear from the phylogenetic trees.
Discussion
A major role of α1,2fucosyltransferases, particularly documented for FUT2 in humans, is their contribution to the synthesis of glycan motifs that present common polymorphisms [16]. These carbohydrates expressed on epithelial surfaces in contact with the environment, such as the gut, the upper airways and the lower genito-urinary tract, constitute attachment factors for various pathogens. By restricting transmission of the pathogen their polymorphism provides what may be called “herd innate protection” by reference to “herd immunity” or indirect immunity whereby the presence of immunized individuals in a population confers protection to immunologically naïve individuals [28]. The earlier observation that a rabbit Sec1 allele was associated with survival to a devastating RHDV outbreak led to the hypothesis that this allele was in linkage disequilibrium with a rFut2 promoter allele responsible for low expression of the corresponding Fut2 enzyme. This was justified by the observation that although rabbits express highly variable levels of A, B or H antigens that require an α1,2fucosyltransferase for synthesis, all allelic variants of rFut2 presented equal enzymatic activities [23]. Thus individual differences in rFut2 transcription rather than in enzyme catalytic properties could have accounted for large differences in the expression of the A, B or H glycan motifs. Sequencing of the rFut2 promoter region revealed an association between a genetic variant and survival to RHDV in a wild rabbit population. However, this promoter variant could not be associated with an altered transcription level of rFut2 and therefore it is unlikely that a low transcription of rFut2 could be responsible for resistance to RHDV. Unexpectedly, we observed that in the small intestine mucosa of rabbits, unlike in humans, rFut1 transcription was as high as that of rFut2. The six RHDV strains tested, spanning the genetic diversity of RHDV, recognized carbohydrate motifs based exclusively on the type 2 precursor. We observed that rFut1 enzyme variants were all equally active, generating H type 2 motifs but not H type 3 motifs. By contrast, the rFut2 enzyme preferentially generated H type 3, indicating that rFut1 is the main contributor to the synthesis of RHDV ligands. This is at variance with the synthesis of HBGAs in the human small intestine that is essentially dependent upon FUT2 and HBGAs based on type 1 precursor [11].
The mRNA levels of rFut1 and rFut2 were relatively homogeneous and were not related to RHDV binding. Therefore the wide individual variation in α1,2fucosylated motifs used as attachment factors by RHDV cannot be accounted for by either rFut1 or rFut2 allelic enzyme variants of catalytic activity, or by allelic variants of transcription. How then is the individual variation generated?
Compared to the expression of rFut1 and rFut2 transcripts, that of rSec1 was much lower for most animals, apparently consistent with the lack of enzyme activity and evolution toward a complete loss of function. Nevertheless, in a small fraction of animals, rSec1 mRNA levels were in the same range or even higher than the rFut1 and rFut2 mRNA levels and this elevated rSec1 expression was associated with low RHDV binding. This suggested that the rSec1 protein could function as a dominant-negative of the rFut1 enzyme and in support of this hypothesis we observed that co-transfection of rSec1 decreased the level of RHDV binding sites synthesized by rFut1. Although it is not clear at present how rSec1 expression represses rFut1 activity, it has been shown that various glycosyltransferases, including fucosyltransferases function as dimers or oligomers [44,45,46]. Furthermore, a report indicated that human FUT1 transfected in CHO cells is exclusively present as dimers [47]. Dominant-negative effects of artificially introduced mutations that inactivate the enzyme activity have been observed for several glycosyltransferases and a dominant-negative function of an inactive splice variant isoform of a glucuronyltransferase was demonstrated [48,49,50,51]. It is therefore reasonable to hypothesize that enzymatically deficient rSec1 could form dimers with rFut1, leading to a decrease of rFut1 catalytic ability. Further studies will be required to understand the molecular mechanism underlying the rSec1 dominant-negative function.
Overall our data indicate that in rabbits there are no functionally significant individual variations in rFut1 or rFut2. Instead, variations in levels of rSec1 transcription contribute to generate individual differences in the degree of α1,2fucosylation of the gut mucosa, leading to differences in recognition by at least one dreadful pathogen. Nevertheless, since not all animals classified as low binders of RHDV expressed high levels of rSec1 RNA, additional factors must contribute to the variation in RHDV attachment. We previously reported that A and B antigen expression were contributing to modulate binding in a strain-specific manner [36]. Indeed, the ability of individual RHDV strains to recognize A, B and H type 2 motifs is highly variable and the expression of the A or B epitopes are differentially expressed between rabbits. Moreover, some strains such as G1 strains preferentially recognize the difucosylated motif Lewis y. Synthesis of this motif requires an α1,3fucosyltransferase in addition to an α1,2fucosyltransferase. Individual variation in the levels of α1,3fucosyltransferase in the rabbit gut may also exist. Understanding the genetic determinisms of these additional glycan modifications of rabbit gut mucosa will require further investigation. Moreover, other unknown factors may contribute to generate individual variation in the repertoire of rabbit gut glycans recognized by RHDV.
The genomic region where the three α1,2fucosyltransferases genes lie presents the same organization in rabbits and humans with the Sec1 and Fut2 genes in tandem and the Fut1 gene localized further apart and separated from its paralogues by three other genes. Since gene conversion is facilitated by genomic proximity [24] the differences observed between primates and other mammals on the one hand and leporids on the other hand cannot be explained by differences in genomic organization. In leporids extensive conversion between the three members of the gene family appears to take place. This is expected to homogenize these sequences, suggesting that each of the family members retains some functionality. By contrast, in other mammalian species, including primates, gene conversion is limited to a small part of the Fut2 and Sec1 coding regions and never involves Fut1, consistent with the pseudogenization of Sec1 and functional divergence between the other two members of the family in these species. Indeed, in species where Sec1 is a pseudogene, either due to frameshifts, premature stop codons or mutations that impair the enzyme’s activity, the extent of gene conversion is limited to regions where Sec1 has no deleterious mutations or where it retains its characteristics as α1,2fucosyltransferase. For example, in pig, Sec1 is a pseudogene due to the lack of the start codon (Meijerink et al 1997), but the remaining putative coding sequence presents no further deleterious mutations allowing to a larger extent of Fut2-Sec1 gene conversion when comparing to the other species (except leporids).
These observations show that the α1,2fucosyltransferases paralogues have undergone different evolutionary pathways in leporids than in other mammals. The following scenario can be proposed. The first duplication event generated subfunctionalization of the two paralogues Fut1 and the ancestor of Fut2 and Sec1 in all mammals, through expression in different cell types. In humans and likely in most other mammalian species, FUT1 is involved in cellular functions such as the development of sensory neurons, angiogenesis or leukocytes adhesion [12,13,14,15,52]. Consistent with these roles, the frequency of FUT1 null alleles is well below 0.01, although not associated with any obvious defects in the homozygous state [53]. As discussed above, with null alleles at a near 0.5 frequency, FUT2 is involved in herd innate protection due to its common polymorphisms and its expression on epithelial cells in direct contact with microbes [16,28]. For this reason it is strongly under balancing selection [32]. The second round of duplication generated Fut2 and Sec1. Following this duplication, Sec1 has been redundant in most species and evolved toward pseudogenization and since then it has no longer been involved in gene conversion with its paralogues [4]. This is clear from S1 Fig where clustering of Fut2 and Sec1 together rather than with their orthologues can be observed, although with distinct patterns: in (A), where the phylogenetic relationships were inferred for the entire catalytic domain, for most species, Fut2 and Sec1 cluster apart; in pig, rat and mouse, Fut2 and Sec1 sequences cluster together, as in these species the homogenization due to gene conversion is quite extended when compared to the remaining species; in (B), Fut2 and Sec1 cluster together for more species as the segment used corresponds to the region where gene conversion is most evident; in (C) the pattern resembles the tree in (A) as a consequence of the more extended gene conversion in pig, rat and mouse and the absence of gene conversion in the other mammals. Thus, in mammals other than leporids, gene conversion also occurs, but it is quite limited in extent and most likely has no impact on the enzymes’ activity. Yet, in leporids where gene conversion between Sec1 and both Fut1 and Fut2 has been maintained (Fig 9 and S2D, S2E, and S2F Fig), the Sec1 protein follows a convergent evolution with the other two members of the gene family. It has essentially lost its enzyme activity but has acquired a dominant-negative function of its Fut1 and possibly Fut2 isoforms in tissues where they are co-expressed. In European rabbit, there is no functional polymorphism of either Fut1 or Fut2, but the levels of Sec1 expression contribute to produce glycan diversity required for herd innate protection. Thus, rather than becoming a pseudogene as in other mammals, in leporids, Sec1 acquired a new function through regulation of the activity of the other α1,2fucosyltransferases.
Materials and Methods
Ethics statement
Work involving the acquisition and sampling of French wild rabbits in Chèvreloup and Cerisay was carried out with permission from the bylaw N° 2009–014 issued by the Paris Prefecture to the Office National de la Chasse et de la Faune Sauvage (ONCFS) and the specific permit N° 11/04 to Dr. Stéphane Marchandeau issued by the ONCFS. Rabbits were caught alive in cage traps or with ferrets. The use of laboratory animals was carried out in a group V animal facility (agreement N° 44267), under specific agreement N° 006933 from the National Committee of Ethics on Animal Experiments of the Ministry of Higher Education and Research. Animal care and handling were performed in strict accordance with the recommendations of the French National Guide for the Ethics of Animal Experiments and euthanasia was performed under xylacine and ketamine anesthesia.
Rabbit samples
For the characterisation of the CDS of rFut1 and rSec1, total genomic DNA was extracted from ear samples collected upon ear tagging of surveyed wild populations in France (Chèvreloup, n = 19 and Cerisay, n = 22) and Portugal (Santarém, n = 7, Mértola, n = 5 and Pancas, n = 15) with the QIAamp DNA Mini kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s protocol. The French population from Chèvreloup experienced two consecutive outbreaks in 1995 and 1996 and samples had been collected prior to the outbreaks in 1994 and 1996 [54,55]. For the characterization of rFut2 5’UTR, 65 DNA samples from the Chèvreloup population were used in addition to DNA obtained from duodenum samples.
Rabbit duodenums were collected from domestic New Zealand White rabbits (29 animals) and wild rabbits from the Pyrénnées Orientales, France that had been freshly killed by hunters (23 animals). The first 5 cm posterior to the gastroduodenal junction was removed after clearing the section from intestinal contents, the sample was vigorously rinsed in phosphate buffered saline (PBS) and stored in RNAlater (Ambion, Life Technologies, Paisley, UK) at -20°C. Sections of the duodenum were then rinsed in PBS, opened and scraped into RLT lysis buffer (Qiagen, Hilden, Germany) containing β-mercaptoethanol. The tissue scrapings were homogenized and split into three parts for ELISA assays, RNA and DNA extraction. The ELISA scrapings were boiled for 10 minutes.
Reconstruction of the rabbit α1,2fucosyltransferases locus
To locate the 5’UTR of Fut2, a RACE was performed on rabbit RNA extracted from duodenum tissue. Ten μg of RNA were used and the RACE was performed according to the manufacturer’s instructions (FirstChoice RLM-RACE kit, Life Technologies, Paisley UK), giving the 118 bp 5’ UTR. Primers were designed in this UTR for a genome walk in the 5’ and 3’ direction. This was performed using the GenomeWalker Universal Kit (Clontech, Mountain View CA) according to the manufacturer’s instructions. These acquired sequences allowed to perform BLAST searches of the trace archives where long matches of at least 400bp were added on in the 5’ and 3’ direction from the UTR as well as in the 5’ and 3’ direction of Fut2 and Sec1, respectively. The 930 bp 5’ of the 5’UTR of Fut2 were then sequenced for the promoter region of Fut2.
RT-PCR quantification of Fut1, Fut2 and Sec1 RNA expression
The RNeasy Mini kit (Qiagen) was used for purification of total RNA according to the manufacturer’s instruction. Reverse transcription was performed with 400 ng of RNA, using the Superscript II Reverse Transcriptase (Life Technologies, Paisley, UK) according to the manufacturer’s instructions. Forty ng of cDNA were then used for real-time PCR (Universal Mastermix, Applied Biosystems, Life Technologies, Paisley, UK), also according to manufacturer’s instructions, on a Mx3005P (Stratagene Agilent Technologies, Massy, France). All primers and probes were designed using Primer Express (Applied Biosystems, Life Technologies, Paisley, UK) and plasmids containing the amplicon were used as a standard for each run. Relative concentrations of transcripts from the different α1,2fucosyltransferase genes were determined using the ΔCT method according to manufacturer’s instruction regarding TaqMan chemistry. The rabbit house-keeping genes GAPDH and β-actin were both separately used as normalization standards, linearising the real-time PCR data against house-keeping gene, giving very similar results.
RHDV binding assay
RHDV binding to rabbit duodenum scrapings or synthetic sugars was analyzed as previously described [36]. Briefly, rabbit duodenum scrapings were coated diluted in a range of dilutions in 0.1M sodium carbonate buffer or 1μg of synthetic sugars was coated in the same buffer. Plates were blocked with 5% non-fat dry milk diluted in PBS or distilled water. RHDV from six strains belonging to genetic groups G1 to G6 as defined in [56] was then added to the plates in similar amounts. To this aim, RHDV strains were prepared from infected liver PBS extracts and quantified by real time RT-PCR using a plasmid containing an RHDV inserted sequence as an internal standard. Binding of RHDV strains was detected using a high-titered rabbit serum Lp4 or an RHDV monoclonal antibody 2G3 for G1–G5 and G6 detection, respectively. Secondary antibodies anti-rabbit conjugated with HRP were used against Lp4 and anti-mouse biotin followed by avidin-HRP was used for RHDV detection with 2G3 anti-RHDVa (G6) monoclonal antibody. TMB (BD Bioscience, San Jose CA) was used as a substrate for all assays and O.D. values were measured at 450nm. To compare binding data between individuals, a threshold was set at 3 times the background. Dilution values of each sample for crossing the threshold were analyzed. All values were normalized against values obtained with the mannose-binding lectin Concavalin A to control for differences in the amount of material scraped, as protein quantification proved to be difficult due to the addition of β-mercaptoethanol to remove any potential anti-RHDV antibodies in the duodenum that would interfere with the analysis.
Amplification and cloning of the open reading frame of the rFut1 and rSec1 genes
Amplification of the coding sequences was performed using the AmpliTaq gold kit with GenAmp 10x PCR Gold Buffer and MgCl2 Solution (Applied Biosystems, Foster city, USA) with the primers rFut1 ATG 5’ ATGTGGCCTCCGAGCC 3’ and rFut1 STOP 5’ CTATCCCAGAAATCTCCAGGGC 3’ for rFut1 or rSec1.3 5’ ATGAGATTCGCCCCTGACTA TGTCC 3’ and rSec1.2 inv 5’ CTAGAGGCCACTCCACAAGGC 3’ for Sec1. The amplified PCR products were 1122 bp and 1044 bp long, respectively. Cloning was performed either in the pcDNA 3.1 vector using the NT-GFP Fusion TOPO TA Expression kit (Invitrogen, Paisley, UK), or in the pDSred vector (Clontech, Rockville, MD), according to the manufacturers’ instructions. The bacteria used for transformation were E. coli One Shot TOP10F’ (Invitrogen, Paisley, UK). After controlling for correct orientation of the inserted sequences several clones were sequenced and the resulting correctly oriented sequences generated GFP or DSred fusion proteins with the GFP or DSred tags located at the N-terminus.
Assay of the α1,2fucosyltransferase activity of rFut1
CHO cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM L-glutamine, free nucleotides (10 μg/mL), 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco, Paisley, UK). Cells were cultured to confluence after dispersal with 0.025% trypsin and 0.02% EDTA. Following amplification and purification with the Qiagen Miniprep kit, the recombinant pcDNA3.1 constructs were transfected into the CHO cells using ICAFECTIN®448 (In-Cell-Art, Nantes, France) following the manufacturer’s instructions. A stable rFut1 expressing cell line was obtained by transfection of the GFP-rFut1 plasmid followed by selection of the cells using G418 (Promega France, Charbonnières, France). Stable expression of the H epitopes on the selected cells was controlled by flow cytometry using either the UEA-I lectin or the 12-4LE anti-H type mAb. For transient transfections, cells were collected 24 hours after transfection and the presence of α1,2-linked fucose residues was tested by flow cytometry using the UEA-I lectin or the 12-4LE mAb that detect the H type 2 (Fucα 2Galβ4GlcNAcβ-R) motif and the MBr1 mAb that detects the H type 3 (Fucα 2Galβ3GalNAcα -R) motif. To this aim, 2.5 x 105 viable transfected cells were incubated in the presence of either biotin-labeled lectin at 5 μg/mL (Vector labs, Peterborough, UK) or the MBr1 mAb (Alexis Biochemicals, San Diego, CA) at 5 μg/mL for 20 min at 4°C. After three washings with PBS, cells were incubated under the same conditions in the presence of either PerCP-conjugated streptavidin (BD Biosciences, San José, CA) at 0.5 μg/mL or a Cy5-conjugated anti-mouse IgG (BD Biosciences) at a 1/500 dilution. After three more washings with PBS, cell fluorescence was measured on a FACScalibur flow cytometer (Becton-Dickinson) and analysed using the CellQuest program (Becton-Dickinson). The transfected protein expression was quantified by the GFP fluorescence recorded on the FL1 channel. The presence of the H type 2 and H type 3 motifs was detected on the FL3 and FL4 channels, respectively.
Co-transfection experiments were conducted to determine the effect of rSec1 on rFut1 enzyme activity. They were performed in 6-well plates using 10 μg plasmid DNA and 20 μL Lipofectamin 2000 (Life Technologies) according to the manufacturer’s instructions. Positive control of rFut1 activity was obtained by transfecting CHO cells with 1 μg of the pcDNA3.1 plasmid containing an rFut1-GFP construct and 9 μg of the pcDNA3.1-truncated hFUT2-GFP vector. This last construct was designed to allow expression of an inactive enzyme fused to GFP (GFP plasmid). To obtain a 1 : 1 rFut1/rSec1 ratio, co-transfection was performed with 1 μg rFut1-GFP plasmid, 1 μg rSec1-GFP plasmid and 8 μg truncated rFut1-FP plasmid; to obtain a 1 : 9 rFut1/rSec1 ratio, 1 μg rFut1-GFP plasmid and 9 μg rSec1-GFP plasmid were used. Forty-eight hours following transfection, expression of RHDV binding sites was determined by flow cytometry. To this aim the transfected cells were incubated for 30 min at 4°C with a 10 μg/mL suspension of virus-like particles from a G2 RHDV strain prepared as described previously [27]. After washings, their binding was detected using the monoclonal 13B5 anti-RHDV diluted at 1.2 μg/mL followed by a Cy5-conjugated anti-mouse IgG (BD Biosciences) at a 1/500 dilution as described above.
Confocal microscopy analysis
CHO cells were transiently transfected with either 1 μg rFut1-DSred plasmid and 1 μg empty vector, or 1 μg rSec1-GFP plasmid and 1 μg empty vector, or with both rFut1-DSred plasmid and rSec1-GFP plasmid, 1 μg each. Forty-eight hours later, cells were detached and plated on Lab-Tek chamber slides (NUNC) overnight. Following washes with PBS, cells were fixed with 4% paraformaldehyde for 15 min at room temperature, washed with PBS and nuclei were stained using Hoescht 33422 from Molecular Probes (Life Technologies). After PBS washes, slides were mounted with Prolong Gold antifading from Molecular Probes. Fluorescence was observed using a Nikon A1 confocal microscope and image processing performed with ImageJ.
Gene conversion analyses
In order to explore and fully determine the extent of gene conversion in mammals, the available sequences of the three α1,2fucosyltransferase genes Fut1, Fut2 and Sec1 from mammals were retrieved from public databases (see S1 Table for accession numbers and gene scaffolds). Only mammals for which sequences of the three genes were available were used in this study. Given the ancestry relatively to other mammals, and thus the closer relation to the ancestral forms of the α1,2fucosyltransferases, sequences from Monodelphis domestica and Xenopus tropicalis were also included. The family Leporidae started to radiate around 14 million years ago [57]. This family is composed by 11 genera. In this study, gDNA samples from five genera were amplified and sequenced for the three genes: Romerolagus diazi, Lepus spp., Brachylagus idahoensis, Sylvilagus spp. and Pentalagus furnessi. These genera diverged from Oryctolagus at 14, 12, 10, 10 and 9 million years, respectively [58,59]. For amplification of Fut1, a combination of two pairs of primers was used. For the first pair, Fut1ATG (5' ATGTGGCCTCCGAGCC 3') and Fut1conservR (5’AAGCCGAAGGTGCCGATGGTCAT 3’), PCR conditions consisted in 5’ at 94°C, 35 cycles of 45” at 94°C, 30” at 52°C and 1’15” at 72°C, and a final extension of 10’ at 72°C. For the second pair, IntrFut1 (5’ ACTGGATGTCGGAGGAGTA 3') and Fut1STOP (5' CTATCCCAGAAATCTCCAGGGC 3'), the conditions were: 3’ at 98°C, 40 cycles of 30” at 98°C, 20” at 52°C and 40” at 72°C, and a final extension of 5’ at 72°C. For Fut2 the PCR consisted in 3’ at 98°C, followed by 40 cycles of 30” at 98°C, 20” at 58°C and 1’ at 72°C and a final extension of 5’ at 72°C with primers rFut2.3 (5’ATGAGCACCGCCCAGGTCCCCTTC 3’) and rFut2.2inv (5’ TCAGTGCTTGAGCAATGGGGACAG 3’). PCR conditions for amplification of Sec1 were the same as for Fut2, except the annealing temperature (56°C); primers used were Sec1.3 (5' ATGAGATTCGCCCCTGACTATGTCC 3') and Sec1.2inv (5' CTAGAGGCCACTCCACAAGGC 3'). PCR products were sequenced on an automatic sequencer ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems) using the amplification primers. Sequences were submitted to GenBank (see S1 Table for accession numbers).
Sequences were aligned using BioEdit software version 7.0.9 [60]. Since these genes are highly divergent in the transmembrane domain and in the stem region, only their catalytic domain was considered and tested for gene conversion by using GARD, as implemented in the Datamonkey Web Server [61,62]. All the analyses were performed considering three distinct groups (all mammals, all mammals except leporids and leporids only) and a general reversible process model (REV) with beta-gamma variation and two rate classes. GARD infers the number of recombination breakpoints (b) by starting with b = 0 and increasing b in increments of one until the corrected Akaike Information Criteria (AICc) score does not improve further [63]. For each dataset, neighbor-joining (NJ) trees were constructed for each segment identified by GARD. Trees were estimated with software MEGA version 5.1 [64] under the Tamura 3-parameter model with the following parameters: 10,000 replicates, bootstrap method, gamma distributed rates and partial deletion.
Statistical analyses
Analysis of the frequencies of Fut2 promoter variant alleles between the survivors and dead animals in the Chèvreloup population was performed by a likelihood ratio Chi-square test. Comparisons of RNA levels between the Sec1 and/or Fut1 and Fut2 genes in the duodenum of rabbits was performed using a Mann-Whitney test. The two-tailed Fisher’s exact test was used to compare groups of rabbits with Sec1 mRNA expression within/above or below the Fut1 and Fut2 range of expression (high or low Sec1 mRNA expressors, respectively) to groups above or below median values of virus binding to duodenum extracts (high or low virus binders, respectively). Mean values of positive cells obtained by flow cytometry from independent experiments were compared by a Student’s t-test.
Supporting Information
Zdroje
1. Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11 : 97–108. doi: 10.1038/nrg2689 20051986
2. Katju V (2012) In with the Old, in with the New: The Promiscuity of the Duplication Process Engenders Diverse Pathways for Novel Gene Creation. International Journal of Evolutionary Biology 2012 : 341932. 23008799
3. Saunier K, Barreaud JP, Eggen J, Oriol R, Levéziel H, et al. (2001) Organization of the bovine alpha2-fucosyltransferase gene cluster suggests that the Sec1 gene might have been shaped through a nonautonomous L1-retrotransposition event within the same locus. Mol Biol Evol 18 : 2083–2091. 11606704
4. Abrantes J, Posada D, Guillon P, Esteves PJ, Le Pendu J (2009) Widespread gene conversion of alpha-2-fucosyltransferase genes in mammals. J Mol Evol 69 : 22–31. doi: 10.1007/s00239-009-9239-0 19533213
5. Oriol R, Candelier JJ, Mollicone R (2000) Molecular genetics of H. Vox Sang 78 : 105–118. 10938937
6. Apoil PA, Roubinet F, Despiau S, Mollicone R, Oriol R, et al. (2000) Evolution of a2-fucosyltransferase genes in primates: relation between an intronic Alu-Y element and red cell expression of ABH antigen. Mol Biol Evol 17 : 337–351. 10723735
7. Bureau V, Marionneau S, Cailleau-Thomas A, Le Moullac-Vaydie B, Liehr T, et al. (2001) Comparison of the three rat GDP-L-fucose:b-D-galactoside 2-a-L-fucosyltransferases FTA, FTB and FTC. Eur J Biochem 268 : 1006–1019. 11179967
8. Borges BN, Paiva TS, Harada ML (2008) Evolution of the SEC1 gene in New World monkey lineages (Primates, Platyrrhini). Genet Mol Res 7 : 663–678. 18752194
9. Iwamori M, Domino SE (2004) Tissue-specific loss of fucosylated glycolipids in mice targeted deletion of alpha(1,2)fucosyltransferase genes. Biochem J 380 : 75–81. 14967068
10. Oriol R, Le Pendu J, Mollicone R (1986) Genetics of ABO, H, Lewis, X and related antigens. Vox Sang 51 : 161–171. 2433836
11. Ravn V, Dabelsteen E (2000) Tissue distribution of histo-blood group antigens. APMIS 108 : 1–28. 10698081
12. Amin MA, Ruth JH, Haas CS, Pakozdi A, Mansfield PJ, et al. (2008) H-2g, a glucose analog of blood group H antigen, mediates mononuclear cell recruitment via Src and phosphatidylinositol 3-kinase pathways. Arthritis Rheum 58 : 689–695. doi: 10.1002/art.23296 18311817
13. Garcia-Vallejo JJ, van Liempt E, da Costa Martins P, Beckers C, van het Hof B, et al. (2008) DC-SIGN mediates adhesion and rolling of dendritic cells on primary human umbilical vein endothelial cells through Lewis Y antigen expressed on ICAM-2. Mol Immunol 45 : 2359–2369. 18155766
14. Moehler TM, Sauer S, Witzel M, Andrulis M, Garcia-Vallejo JJ, et al. (2008) Involvement of alpha 1-2-fucosyltransferase (FUT1) and surface-expressed Lewis(y) (CD174) in first endothelial cell-cell contacts during angiogenesis. J Cell Physiol 215 : 27–36. doi: 10.1002/jcp.21285 18205178
15. St John JA, Claxton C, Robinson MW, Yamamoto F, Domino SE, et al. (2006) Genetic manipulation of blood group carbohydrates alters development and pathfinding of primary sensory axons of the olfactory systems. Dev Biol 298 : 470–484. 16884711
16. Marionneau S, Cailleau-Thomas A, Rocher J, Le Moullac-Vaidye B, Ruvoën-clouet N, et al. (2001) ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83 : 565–573. 11522384
17. Stapleton A, Nudelman E, Clausen E, Hakomori S, I, Stamm W, E (1992) Binding of uropathogenic Escherichia coli R45 to glycolipids extracted from vaginal epithelial cells is dependent on histo-blood group secretor status. J Clin Invest 90 : 965–972. 1522244
18. Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS (2003) Campylobacter jejuni binds intestinal H(O) antigen and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 278 : 14112–14120. 12562767
19. Tan M, Jiang X (2011) Norovirus-host interaction: Multi-selections by human histo-blood group antigens. Trends Microbiol 19 : 382–388. doi: 10.1016/j.tim.2011.05.007 21705222
20. Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, et al. (2012) Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485 : 256–259. doi: 10.1038/nature10996 22504179
21. Liu Y, Huang P, Tan M, Biesiada J, Meller J, et al. (2012) Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol 86 : 9899–9910. doi: 10.1128/JVI.00979-12 22761376
22. Hitoshi S, Kusunoki S, Kanazawa I, Tsuji S (1995) Molecular cloning and expression of two types of rabbit b-galactoside a1,2fucosyltransferase. J Biol Chem 270 : 8844–8850. 7721792
23. Guillon P, Ruvöen-Clouet N, Le Moullac-Vaidye B, Marchandeau S, Le Pendu J (2009) Association between expression of the H histo-blood group antigen, α1,2 fucosyltransferases polymorphism of wild rabbits, and sensitivity to rabbit hemorrhagic disease virus. Glycobiology 19 : 21–28. doi: 10.1093/glycob/cwn098 18842963
24. Teshima KM, Innan H (2004) The effect of conversion on the divergence between duplicated genes. Genetics 166 : 1553–1560. 15082568
25. Liu SJ, Xue HP, Pu BQ, Quian NH (1984) A new viral disease in rabbit. Anim Husb Vet Med 16 : 235–255.
26. Abrantes J, van der Loo W, Le Pendu J, Esteves PJ (2011) Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Veterinary Research in press. doi: 10.1186/1297-9716-43-12 22325049
27. Ruvoën-Clouet N, Ganière JP, André-Fontaine G, Blanchard D, Le Pendu J (2000) Binding of Rabbit Hemorrhagic Disease Virus to antigens of the ABH histo-blood group family. J Virol 74 : 11950–11954. 11090195
28. Ruvöen-Clouet N, Belliot G, Le Pendu J (2013) Noroviruses and histo-blood groups: the impact of common host genetic polymorphisms on virus transmission and evolution. Rev Med Virol 23 : 355–366. doi: 10.1002/rmv.1757 23959967
29. Ilver D, Arnqvist A, Ögren J, Frick IM, Kersulyte D, et al. (1998) Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279 : 373–377. 9430586
30. Azevedo M, Eriksson S, Mendes N, Serpa J, Figueiredo C, et al. (2008) Infection by Helicobacter pylori expressing the BabA adhesin is influenced by the secretor phenotype. J Pathol 215 : 308–316. doi: 10.1002/path.2363 18498114
31. Koda Y, Soejima M, Kimura H (2001) The polymorphisms of fucosyltransferases. Leg Med (Tokyo) 3 : 2–14.
32. Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, et al. (2009) A natural history of FUT2 polymorphism in humans. Mol Biol Evol 26 : 1993–2003. doi: 10.1093/molbev/msp108 19487333
33. Silva LM, Carvalho AS, Guillon P, Seixas S, Azevedo M, et al. (2010) Infection-associated FUT2 (fucosyltransferase 2) genetic variation and impact on functionality assessed by in vivo studies. Glycoconj J 27 : 61–68. doi: 10.1007/s10719-009-9255-8 19757028
34. Le Pendu J, Ruvöen-Clouet N, Kindberg E, Svensson L (2006) Mendelian resistance to human norovirus infections. Sem Immunol 18 : 375–386.
35. Hitoshi S, Kusunoki S, Kanazawa I, Tsuji S (1996) Molecular cloning and expression of a third type of rabbit GDP-L-fucose: b-D-galactoside 2-a-L-fucosyltransferase. J Biol Chem 271 : 16975–16981. 8663168
36. Nyström K, Le Gall-Recule G, Grassi P, Abrantes J, Ruvoën-Clouet N, et al. (2011) Histo-Blood Group Antigens act as attachment factors of Rabbit Hemorrhagic Disease Virus infection in a virus strain-dependent manner. PLoS Pathogens 7: e1002188. doi: 10.1371/journal.ppat.1002188 21901093
37. Le Pendu J, Cartron JP, Lemieux R, U, Oriol R (1985) The presence of at least two different H-blood group-related b-D-gal a-2-L-fucosyltransferases in human serum and the genetics of blood group H substances. Am J Hum Genet 37 : 749–760. 9556663
38. Sarnesto A, Köhlin T, Hindsgaul O, Thurin J, Blaszczyk-Thurin M (1992) Purification of the secretor-type b-galactoside a1-2fucosyltransferase from human serum. J Biol Chem 267 : 2737–2744. 1733969
39. Masutani H, Kimura H (1995) Purification and characterization of secretory-type GDP-L-fucose: b-D-galactoside 2-a-L-fucosyltransferase from human gastric mucosa. J Biochem 118 : 541–545. 8690714
40. Esteves PJ, Lanning D, Ferrand N, Knight KL, Zhai SK, et al. (2004) Allelic variation at the VHa locus in natural populations of rabbit (Oryctolagus cuniculus, L.). J Immunol 172 : 1044–1053. 14707078
41. Surridge AK, van der Loo W, Abrantes J, Carneiro M, Hewitt GM, et al. (2008) Diversity and evolutionary history of the MHC DQA gene in leporids. Immunogenetics 60 : 515–525. doi: 10.1007/s00251-008-0309-z 18584169
42. Abrantes J, Areal H, Esteves PJ (2013) Insights into the European rabbit (Oryctolagus cuniculus) innate immune system: genetic diversity of the toll-like receptor 3 (TLR3) in wild populations and domestic breeds BMC Genetics 14 : 73. doi: 10.1186/1471-2156-14-73 23964588
43. Varki A, Esko JD, Colley KJ (2009) Cellular organization of localization. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P et al., editors. Essentials of Glycobiology. 2dn Edition ed. New York: Cold Spring Harbor.
44. Hashimoto K, Madej T, Bryant SH, Panchenko AR (2010) Functional states of homooligomers: insights from the evolutionof glycosyltransferases. J Mol Biol 399 : 196–206. doi: 10.1016/j.jmb.2010.03.059 20381499
45. Faik A, Bar-Peled M, DeRocher AE, Zeng WQ, Perrin RM, et al. (2000) Biochemical characterization of an alpha-1,2fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosyntesis in pea. J Biol Chem 275 : 15082–15089. 10747946
46. Sousa VL, Costa MT, Palma AS, Enguita F, Costa J (2001) Localization, purification and specificity of the full-length membrane-bound form of human recombinant alpha 1,3/4-fucosyltransferase from BHK-21B cells. Biochem J 357 : 803–810. 11463351
47. El-Battari A, Prorok M, Angata K, Mathieu S, Zerfaoui M, et al. (2003) Different glycosyltransferases are differentially processed for secretion, dimerization, and autoglycosylation. Glycobiology 13 : 941–953. 14514709
48. Ihara H, Ikeda Y, Koyota S, Endo T, Honke H, et al. (2002) A catalytically inactive beta 1,4-N-acetylglucosaminyltransferase III (GnT-III) behaves as a dominant nege-ative GnT-III inhibitor. Eur J Biochem 269 : 193–201. 11784313
49. Matsunami K, Miyagawa S, Nakagawa K, Hideaki O, Shirakura R (2006) Molecular cloning of pigGnT-I and I.2: an application to xenotransplantation. Biochemical and Biophysical Resarch Communication 343 : 677–683. 16563346
50. Olson FJ, Bäckström M, Karlsson H, Burchell J, Hansson GC (2005) A MUC1 tandem repeat reporter protein produced in CHO-K1 cells has sialylated core 1 O-glycans and becomes more densely glycosylated if coexpressed with polypeptide-GalNAc-T4 transferase. Glycobiology 15 : 177–191. 15456735
51. Bellemare J, Rouleau M, Harvey M, Têtu B, Guillemette C (2010) Alternative-splicing forms of major phase II conjugating UGT1A gene negatively regulate glucuronidation in human carcinoma cell lines. Pharmacogenomics J 10 : 431–441. doi: 10.1038/tpj.2009.64 19997083
52. Hitoshi S, Koijima N, Kusunoki S, Inokuchi J, Kanazawa I, et al. (1996) Expression of the beta-galactoside alpha 1,2-fucosyltransferase gene suppresses axonal outgrowth of neuro2a neuroblastoma cells. J Neurochem 66 : 1633–1640. 8627320
53. Wagner FF, Flegel WA (1997) Polymorphism of the h allele and the population frequency of sporadic nonfunctional alleles. Transfusion 37 : 284–290. 9122901
54. Queney G, Ferrand N, Marchandeau S, Azevodo M, Mougel F, et al. (2000) Absence of a genetic bottleneck in a wild rabbit (Oryctolagus cuniculus) population exposed to a severe viral epizootic. Molecular Ecology 9 : 1253–1264. 10972766
55. Queney G, Ferrand N, Weiss S, Mougel F, Monnerot M (2001) Stationary distributions of microsatellite loci between divergent population groups of the European rabbit (Oryctolagus cuniculus). Mol Biol and Evol 18 : 2169–2178. 11719566
56. Le Gall-Recule G, Zwingelstein F, Laurent S, De Boisseron C, Portejoie Y, et al. (2003) Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000 and the characterization of RHDV antigenic variants. Arch Virol 148 : 65–81. 12536296
57. Matthee CA, van Vuuren BJ, Bell D, Robinson TJ (2004) A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst Biol 53 : 443–447.
58. Esteves PJ, Lanning D, Ferrand N, Knight KL, Zhai SK, et al. (2005) The evolution of the immunoglobulin heavy chain variable region (IgVH) in Leporids: an unusual case of transspecies polymorphism. Immunogenetics 57 : 874–882. 16247606
59. van der Loo W, Abrantes J, Esteves PJ (2009) Sharing of endogenous lentiviral gene fragments among leporid lineages separated for more than 12 million years. J Virol 83 : 2386–2388. doi: 10.1128/JVI.01116-08 19109386
60. Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41 : 95–98.
61. Kosakovsky-Pond SL, Frost SD (2005b) Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21 : 2531–2533. 15713735
62. Delport W, Poon AF, Frost SDW, Kosakovsky-Pond SL (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26 : 2455–2457. doi: 10.1093/bioinformatics/btq429 20671151
63. Kosakovsky-Pond SL, Posada D, Gravenor MB, Woelk CH, Frost S (2006) GARD: A Genetic Algorithm for Recombination Detection. Bioinformatics 22 : 3096–3098. 17110367
64. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol and Evol 28 : 2731–2739. doi: 10.1093/molbev/msr121 21546353
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human SkinČlánek Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?Článek The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious DiseasesČlánek Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting PolypeptideČlánek A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 4- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Pathogens as Biological Weapons of Invasive Species
- Selection and Spread of Artemisinin-Resistant Alleles in Thailand Prior to the Global Artemisinin Resistance Containment Campaign
- Endopeptidase-Mediated Beta Lactam Tolerance
- Prospective Large-Scale Field Study Generates Predictive Model Identifying Major Contributors to Colony Losses
- Relay of Herpes Simplex Virus between Langerhans Cells and Dermal Dendritic Cells in Human Skin
- Structural Determinants of Phenotypic Diversity and Replication Rate of Human Prions
- Sigma Factor SigB Is Crucial to Mediate Adaptation during Chronic Infections
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Heterologous Expression in Remodeled . : A Platform for Monoaminergic Agonist Identification and Anthelmintic Screening
- Novel Disease Susceptibility Factors for Fungal Necrotrophic Pathogens in Arabidopsis
- Interleukin 21 Signaling in B Cells Is Required for Efficient Establishment of Murine Gammaherpesvirus Latency
- Phosphorylation at the Homotypic Interface Regulates Nucleoprotein Oligomerization and Assembly of the Influenza Virus Replication Machinery
- Human Papillomaviruses Activate and Recruit SMC1 Cohesin Proteins for the Differentiation-Dependent Life Cycle through Association with CTCF Insulators
- Ubiquitous Promoter-Localization of Essential Virulence Regulators in
- TGF-β Suppression of HBV RNA through AID-Dependent Recruitment of an RNA Exosome Complex
- The Immune Adaptor ADAP Regulates Reciprocal TGF-β1-Integrin Crosstalk to Protect from Influenza Virus Infection
- Antagonism of miR-328 Increases the Antimicrobial Function of Macrophages and Neutrophils and Rapid Clearance of Non-typeable (NTHi) from Infected Lung
- The Epigenetic Regulator G9a Mediates Tolerance to RNA Virus Infection in
- Does the Arthropod Microbiota Impact the Establishment of Vector-Borne Diseases in Mammalian Hosts?
- Hantaan Virus Infection Induces Both Th1 and ThGranzyme B+ Cell Immune Responses That Associated with Viral Control and Clinical Outcome in Humans
- Viral Inhibition of the Transporter Associated with Antigen Processing (TAP): A Striking Example of Functional Convergent Evolution
- Plasma Membrane Profiling Defines an Expanded Class of Cell Surface Proteins Selectively Targeted for Degradation by HCMV US2 in Cooperation with UL141
- Optineurin Regulates the Interferon Response in a Cell Cycle-Dependent Manner
- IFIT1 Differentially Interferes with Translation and Replication of Alphavirus Genomes and Promotes Induction of Type I Interferon
- The EBNA3 Family of Epstein-Barr Virus Nuclear Proteins Associates with the USP46/USP12 Deubiquitination Complexes to Regulate Lymphoblastoid Cell Line Growth
- Hepatitis C Virus RNA Replication Depends on Specific and -Acting Activities of Viral Nonstructural Proteins
- A Neuron-Specific Antiviral Mechanism Prevents Lethal Flaviviral Infection of Mosquitoes
- The Aspartate-Less Receiver (ALR) Domains: Distribution, Structure and Function
- Global Genome and Transcriptome Analyses of Epidemic Isolate 98-06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution
- The Ebola Epidemic Crystallizes the Potential of Passive Antibody Therapy for Infectious Diseases
- Ebola Virus Entry: A Curious and Complex Series of Events
- Conserved Spirosomes Suggest a Single Type of Transformation Pilus in Competence
- Spatial Structure, Transmission Modes and the Evolution of Viral Exploitation Strategies
- Bacterial Cooperation Causes Systematic Errors in Pathogen Risk Assessment due to the Failure of the Independent Action Hypothesis
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Cerebrospinal Fluid Cytokine Profiles Predict Risk of Early Mortality and Immune Reconstitution Inflammatory Syndrome in HIV-Associated Cryptococcal Meningitis
- Utilize Host Actin for Efficient Maternal Transmission in
- Borna Disease Virus Phosphoprotein Impairs the Developmental Program Controlling Neurogenesis and Reduces Human GABAergic Neurogenesis
- An Effector Peptide Family Required for Toll-Mediated Immunity
- Hepatitis D Virus Infection of Mice Expressing Human Sodium Taurocholate Co-transporting Polypeptide
- A Redox Regulatory System Critical for Mycobacterial Survival in Macrophages and Biofilm Development
- Quadruple Quorum-Sensing Inputs Control Virulence and Maintain System Robustness
- Leukocyte-Derived IFN-α/β and Epithelial IFN-λ Constitute a Compartmentalized Mucosal Defense System that Restricts Enteric Virus Infections
- A Strategy for O-Glycoproteomics of Enveloped Viruses—the O-Glycoproteome of Herpes Simplex Virus Type 1
- Macrocyclic Lactones Differ in Interaction with Recombinant P-Glycoprotein 9 of the Parasitic Nematode and Ketoconazole in a Yeast Growth Assay
- Neofunctionalization of the α1,2fucosyltransferase Paralogue in Leporids Contributes to Glycan Polymorphism and Resistance to Rabbit Hemorrhagic Disease Virus
- The Extracytoplasmic Linker Peptide of the Sensor Protein SaeS Tunes the Kinase Activity Required for Staphylococcal Virulence in Response to Host Signals
- Murine CMV-Induced Hearing Loss Is Associated with Inner Ear Inflammation and Loss of Spiral Ganglia Neurons
- Dual miRNA Targeting Restricts Host Range and Attenuates Neurovirulence of Flaviviruses
- GATA-Dependent Glutaminolysis Drives Appressorium Formation in by Suppressing TOR Inhibition of cAMP/PKA Signaling
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- Genetic Analysis Using an Isogenic Mating Pair of Identifies Azole Resistance Genes and Lack of Locus’s Role in Virulence
- A Temporal Gate for Viral Enhancers to Co-opt Toll-Like-Receptor Transcriptional Activation Pathways upon Acute Infection
- Neutrophil Recruitment to Lymph Nodes Limits Local Humoral Response to
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Toxin-Induced Necroptosis Is a Major Mechanism of Lung Damage
- Transgenic Fatal Familial Insomnia Mice Indicate Prion Infectivity-Independent Mechanisms of Pathogenesis and Phenotypic Expression of Disease
- Role of Hypoxia Inducible Factor-1α (HIF-1α) in Innate Defense against Uropathogenic Infection
- EphrinA2 Receptor (EphA2) Is an Invasion and Intracellular Signaling Receptor for
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



