-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Unveiling the Intracellular Survival Gene Kit of Trypanosomatid Parasites
Trypanosomatids are unicellular protozoans of medical and economical relevance since they are the etiologic agents of infectious diseases in humans as well as livestock. Whereas Trypanosoma cruzi and different species of Leishmania are obligate intracellular parasites, Trypanosoma brucei and other trypanosomatids develop extracellularly throughout their entire life cycle. After their genomes have been sequenced, various comparative genomic studies aimed at identifying sequences involved with host cell invasion and intracellular survival have been described. However, for only a handful of genes, most of them present exclusively in the T. cruzi or Leishmania genomes, has there been any experimental evidence associating them with intracellular parasitism. With the increasing number of published complete genome sequences of members of the trypanosomatid family, including not only different Trypanosoma and Leishmania strains and subspecies but also trypanosomatids that do not infect humans or other mammals, we may now be able to contemplate a slightly better picture regarding the specific set of parasite factors that defines each organism's mode of living and the associated disease phenotypes. Here, we review the studies concerning T. cruzi and Leishmania genes that have been implicated with cell invasion and intracellular parasitism and also summarize the wealth of new information regarding the mode of living of intracellular parasites that is resulting from comparative genome studies that are based on increasingly larger trypanosomatid genome datasets.
Published in the journal: . PLoS Pathog 10(12): e32767. doi:10.1371/journal.ppat.1004399
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1004399Summary
Trypanosomatids are unicellular protozoans of medical and economical relevance since they are the etiologic agents of infectious diseases in humans as well as livestock. Whereas Trypanosoma cruzi and different species of Leishmania are obligate intracellular parasites, Trypanosoma brucei and other trypanosomatids develop extracellularly throughout their entire life cycle. After their genomes have been sequenced, various comparative genomic studies aimed at identifying sequences involved with host cell invasion and intracellular survival have been described. However, for only a handful of genes, most of them present exclusively in the T. cruzi or Leishmania genomes, has there been any experimental evidence associating them with intracellular parasitism. With the increasing number of published complete genome sequences of members of the trypanosomatid family, including not only different Trypanosoma and Leishmania strains and subspecies but also trypanosomatids that do not infect humans or other mammals, we may now be able to contemplate a slightly better picture regarding the specific set of parasite factors that defines each organism's mode of living and the associated disease phenotypes. Here, we review the studies concerning T. cruzi and Leishmania genes that have been implicated with cell invasion and intracellular parasitism and also summarize the wealth of new information regarding the mode of living of intracellular parasites that is resulting from comparative genome studies that are based on increasingly larger trypanosomatid genome datasets.
Trypanosomatids: Distinct Life Cycles, but Not So Divergent Genomes
Trypanosomatids (order Kinetoplastida) constitute a group of early-branching unicellular eukaryotes, which includes several human parasites responsible for diseases that affect over 20 million people and cause countless infections in other mammals, primarily in developing countries. Chagas disease (American trypanosomiasis), caused by T. cruzi—sleeping sickness or Human African Trypanosomiasis (HAT)—caused by Trypanosoma brucei gambiensis, Trypanosoma brucei rhodesiensis, and different forms of leishmaniases, caused by various species of Leishmania, are categorized amongst the most important neglected diseases causing approximately 150,000 deaths annually. In addition, Trypanosoma vivax, Trypanosoma congolense, and Trypanosoma brucei are pathogenic species in livestock and responsible for considerable production losses in South American and African countries (www.who.int/topics/tropical_diseases/en/). In spite of this large burden and the increasing efforts made by a relatively small group of researchers, no suitable vaccines for these diseases are available and the treatment is limited to a few drugs that have several undesirable side effects.
Kinetoplastids are protozoans characterized by the presence of a single branched mitochondrion containing a unique mitochondrial DNA structure known as kinetoplast [1]. Being early-branching eukaryotes, these organisms possess many peculiar characteristics, some of them reminiscent of their prokaryotic ancestors. Among the unusual features are genomic organization consisting of large, unidirectional gene clusters that are polycistronically transcribed [2], RNA polymerase I-mediated transcription of protein coding genes [3], RNA trans-splicing coupled to poly(A) addition [4], [5], and extensive RNA editing of mitochondrial mRNAs [6]. Besides their medical relevance, these unusual characteristics have driven the focus of intense research which may, hopefully, result in the development of new forms of treatment and disease prevention.
The life cycles of all Trypanosoma and Leishmania species that cause human diseases depend on insect vectors (Figure 1). Leishmania spp proliferates as promastigotes in the midgut of phlebotomine sand flies and is transmitted to several species of mammals as metacyclic promastigotes when the fly takes a blood meal. In the mammalian host, Leishmania major is phagocytosed by macrophages and, once in the phagolysossome, metacyclic forms are converted into amastigotes, which multiply numerous times before being released during cell lysis [7]. T. cruzi replicates as epimastigotes in the midgut of different species of reduviid bugs and develops into infective metacyclic trypomastigotes once they reach the rectum and are excreted with the insect feces. In contrast to L. major, which multiplies only inside mammalian host macrophages, T. cruzi trypomastigotes invade essentially any nucleated cell type by a mechanism involving either lysosomal recruitment at the parasite invasion site or invagination of the plasma membrane followed by intracellular fusion with lysosomes. Also in contrast to Leishmania spp, T. cruzi trypomastigotes are able to escape from the phagolysosome into the cytosol where they differentiate into amastigotes [8]. After several rounds of cell division, amastigotes differentiate again into trypomastigotes that are released from the infected cell. T. cruzi amastigotes prematurely released from heavily infected cells can be also taken up and replicate within neighboring cells (See [9] and [10] for recent reviews on T. cruzi and Leishmania internalization processes). Different from T. cruzi and Leishmania spp, T. brucei develops extracellularly throughout its entire life cycle. It multiplies as procyclic forms in the intestinal tract of the tsetse fly before being transformed into infective metacyclic forms in the salivary glands. After being injected into the host during a blood meal, T. brucei proliferates in the bloodstream [11]. Therefore, unlike T. cruzi and Leishmania, which are able to hide inside mammalian cells, T. brucei needs to cope with the direct exposure to a strong antibody response in the host. To achieve this, it acquired a sophisticated immune evasion protocol, known as variant surface glycoprotein (VSG) switching. VSGs are encoded by a large family of T. brucei-specific genes whose monoallelic expression in bloodstream trypomastigotes results in a tightly packed surface coat of variant glycoproteins that shields other invariant surface proteins from the attack by the host immune system, allowing the parasite to multiply indefinitely in the bloodstream [12]. Thus, the lack of VSG genes in T. cruzi and Leishmania correlates with their ability to invade mammalian cells and by doing so, they do not need to cope with the continuous attack by the host humoral immune response.
Fig. 1. The distinct life cycles of tritryp parasites. 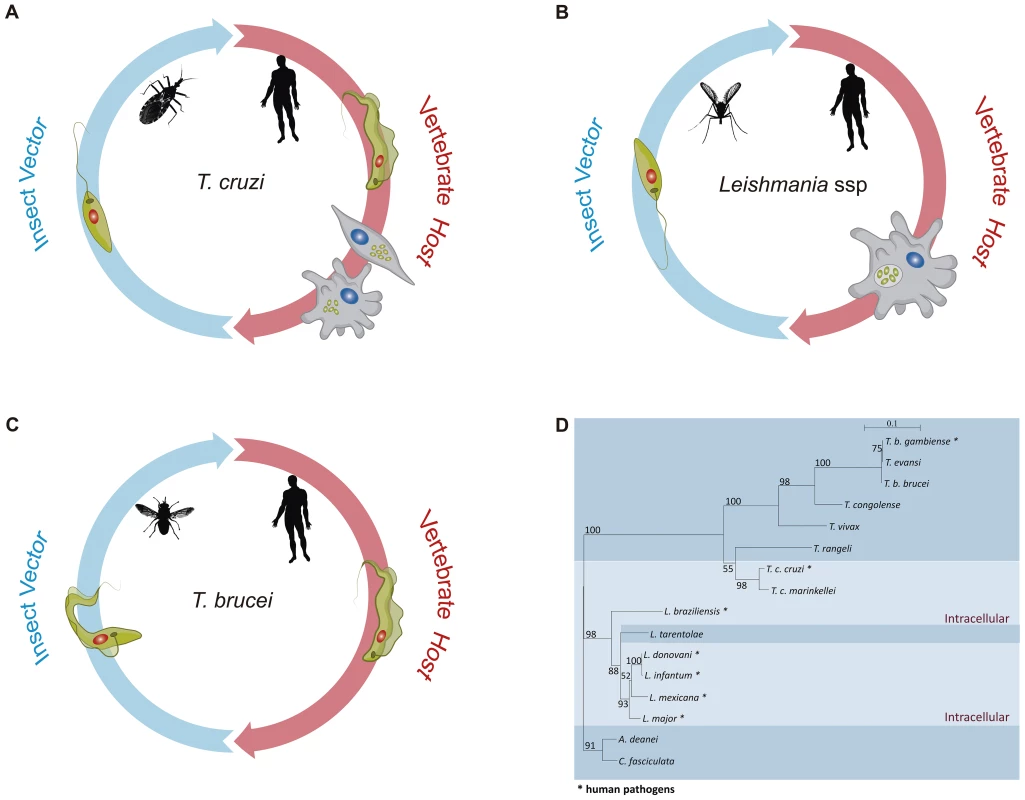
Panels A–C show the life cycles of T. cruzi, Leishmania spp, and T. brucei, respectively. In each panel, some of the parasite stages present in their insect vectors, T. cruzi epimastigotes, Leishmania promastigotes, and T. brucei procyclic forms, are shown on the left. Different sand fly species of the genera Lutzomyia and Phlebotomus are vectors for Leishmania. Triatoma infestans and Rhodnius prolixus are the most important vector species in the transmission of T. cruzi to man, whereas different species of Glossina, also known as tse-tse fly, are vectors of African trypanosomes. Leishmania and T. brucei parasites move from the fly midgut up to the mouthparts before being inoculated into the human host as metacyclic, infective forms. Although Leishmania promastigotes achieve their journey in sand flies by being regurgitated from the stomodeal valve to the mouthparts, T. brucei epimastigotes do not stay in the mouthparts, as they have to first migrate from the proventriculus to the salivary glands where they develop into metacyclic forms and are expelled with the insect saliva. In contrast, T. cruzi infective metacyclic trypomastigotes develop in the hindgut of the triatomine bug and, after being excreted with the insect feces, gain access to the mammalian host bloodstream through skin wounds or the mucous membranes. On the right side of each panel, parasite forms present in the mammalian host, T. cruzi trypomastigotes, and intracellular amastigotes, Leishmania intracellular amastigotes, and T. brucei bloodstream forms are shown. Whereas Leishmania promastigotes are internalized by host phagocytes and reside into the phagolysosome, T. cruzi trypomastigotes actively invade a variety of nonphagocytic cells and are able to escape from the phagocytic vacuole and multiply in the host cell cytoplasm. Although distinct developmental forms of T. brucei are found in the mammalian host, namely stumpy and slender trypomastigotes, they remain extracellular during the entire parasite life cycle and were represented here as bloodstream trypomastigotes. Panel D shows a phylogenetic analysis inferred from glycosomal glyceraldehyde 3-phosphate dehydrogenase (gapdh) nucleotide sequences from 16 trypanosomatid species, with the species that have an intracellular stage shown with a light blue color. The maximum likelihood tree was constructed with 849 nt (80% of gapdh coding sequences), using SeaView v.04 and rooted at the Crithidia fasciculata/A. deanei clade, with the bootstrap values for 1,000 replicates shown in the major basal nodes. The complete genome sequences of T. brucei [13], T. cruzi [14], and L. major [15], known as the tritryp genomes, represent a landmark in the study of these parasites. In contrast to the T. brucei and L. major genomes, whose 25 and 33 Mb sequences were assembled into 11 and 36 chromosomes, respectively, the much larger CL Brener T. cruzi genome (55 Mb haploid genome) has not been fully assembled, due to its repetitive and hybrid nature and the high level of allelic polymorphism. Although additional efforts resulted in the assembly of 41 pairs of chromosomes [16]. the exact chromosome number in T. cruzi is still not known. Despite a divergence period estimated between 200 to 500 million years, a conserved proteomic core derived from about 6,200 genes and a surprisingly large conservation of gene synteny were found between the tritryp genomes [17]. In the first comparative analysis published together with the description of the tritryp genomes, the authors remarked that the intracellular parasites, L. major and T. cruzi, appear to share slightly more genes than do T. brucei and T. cruzi and considerably more than do L. major and T. brucei. Whereas a total of 482 genes are shared between L. major and T. cruzi, only 74 common genes are present in the genomes of T. brucei and L. major [17]. Multigene families, retroelements, and structural RNAs often present in regions of synteny breaks were considered important elements that have shaped each parasite genome accordingly to their life cycles. Besides VSG genes, genes encoding elements of the RNA interference (RNAi) machinery were found exclusively in the T. brucei genome [17], [18]. Since a major role of RNAi silencing pathways is to down-regulate transcripts derived from transposons and repeats to maintain genome integrity, the absence of an RNAi machinery in L. major seems compatible with their lack of active transposons as well as with the mechanism of gene amplification involving the production of extrachromosomal circular DNA elements [19]. Like in other organisms, RNAi has been largely used as a tool for functional studies in T. brucei [20]. In contrast, due to the absence of functional RNAi machinery in T. cruzi and L. major [21], [19], functional genomics has evolved at a much slower pace in these two parasites. However, with the recent discovery that RNAi is functional in Leishmania (Viannia) braziliensis and other members of the Viannia subgenus [19], gene function studies in this Leishmania subgroup may start to catch up. Even more importantly, the conservation of the RNAi pathway in only a few Leishmania species may be correlated with the presence of dsRNA virus, known as Leishmania RNA Virus (LRV), with possible medical implications derived from recent data showing that infection by Leishmania isolates bearing LRVs results in metastatic forms of disease [19], [22].
T. cruzi and Leishmania Genes Involved with Host Cell Invasion and Intracellular Multiplication
Work from several groups has been dedicated to the characterization of genes known as virulence factors in T. cruzi and in different Leishmania species (Table 1) [23], [24]. Two groups of proteins present exclusively in Leishmania, the Promastigote Surface Proteins (PSAs), also known as the GP46 family, and the surface protein known as A2, both of which contain large amino acid repetitive domains, have been extensively characterized (Figure 2). PSAs are Leucine Rich Repeat (LRR)–containing surface proteins that bind to host cell macrophages and protect Leishmania infantum from complement-mediated lysis [25], [26]. The A2 genes encode a family of 42 to 100 kDa proteins made up almost entirely of 40 to 90 copies of a repetitive amino acid sequence, mainly expressed in amastigotes of Leishmania donovani and L. infantum, and that are known to be involved with parasite visceralization [27], [28]. Exogenous expression of A2 genes in L. major, which possesses only a truncated A2 pseudogene, enhanced the ability of L. major–infected cells to migrate out of the dermis and increase parasite survival in visceral organs [29]. Similarly, expression of A2 in Leishmania tarentolae, a lizard parasite which is nonpathogenic to mammals, resulted in increased L. tarentolae survival in mouse visceral organs [30].
Fig. 2. Surface proteins present in Leishmania and T. cruzi. 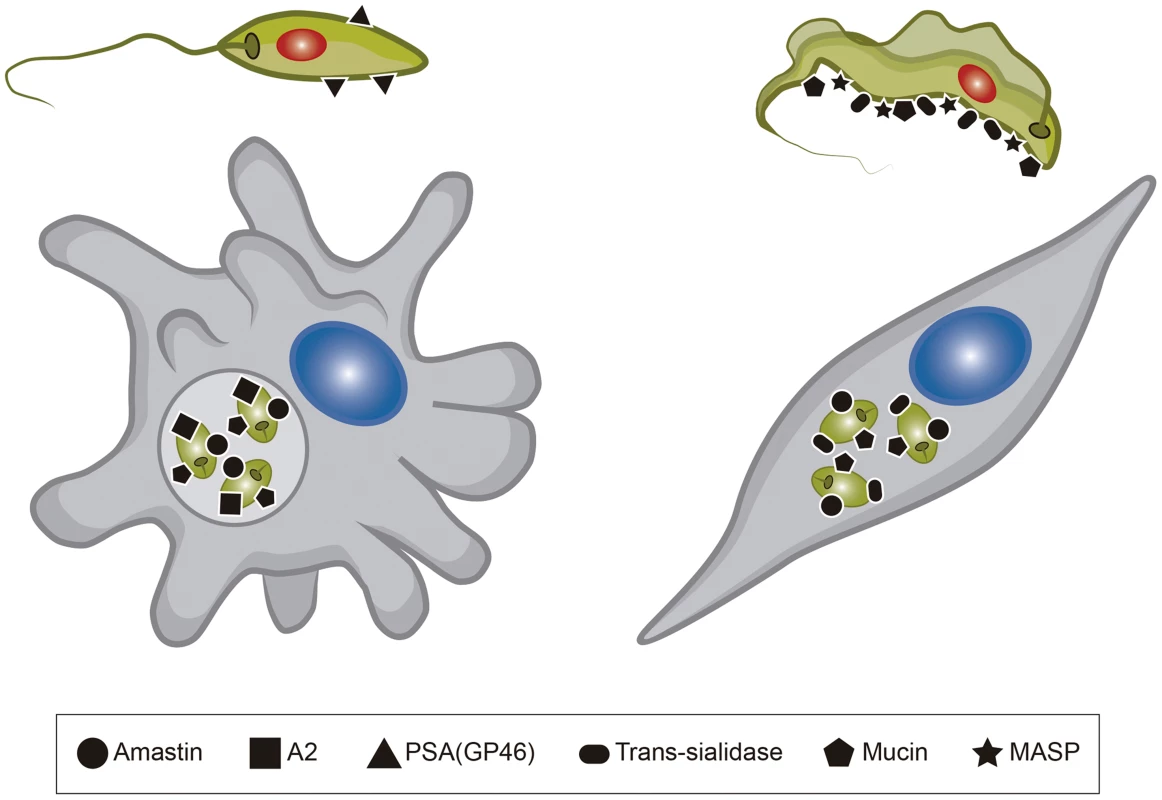
The figure shows six different surface molecules known to be present in promastigote and amastigote forms of Leishmania (left) and trypomastigote and amastigote forms of T. cruzi (right). Each protein is represented by the symbols indicated below the figure. Tab. 1. <i>T. cruzi</i> and <i>Leishmania</i> genes involved with host cell invasion and intracellular survival. 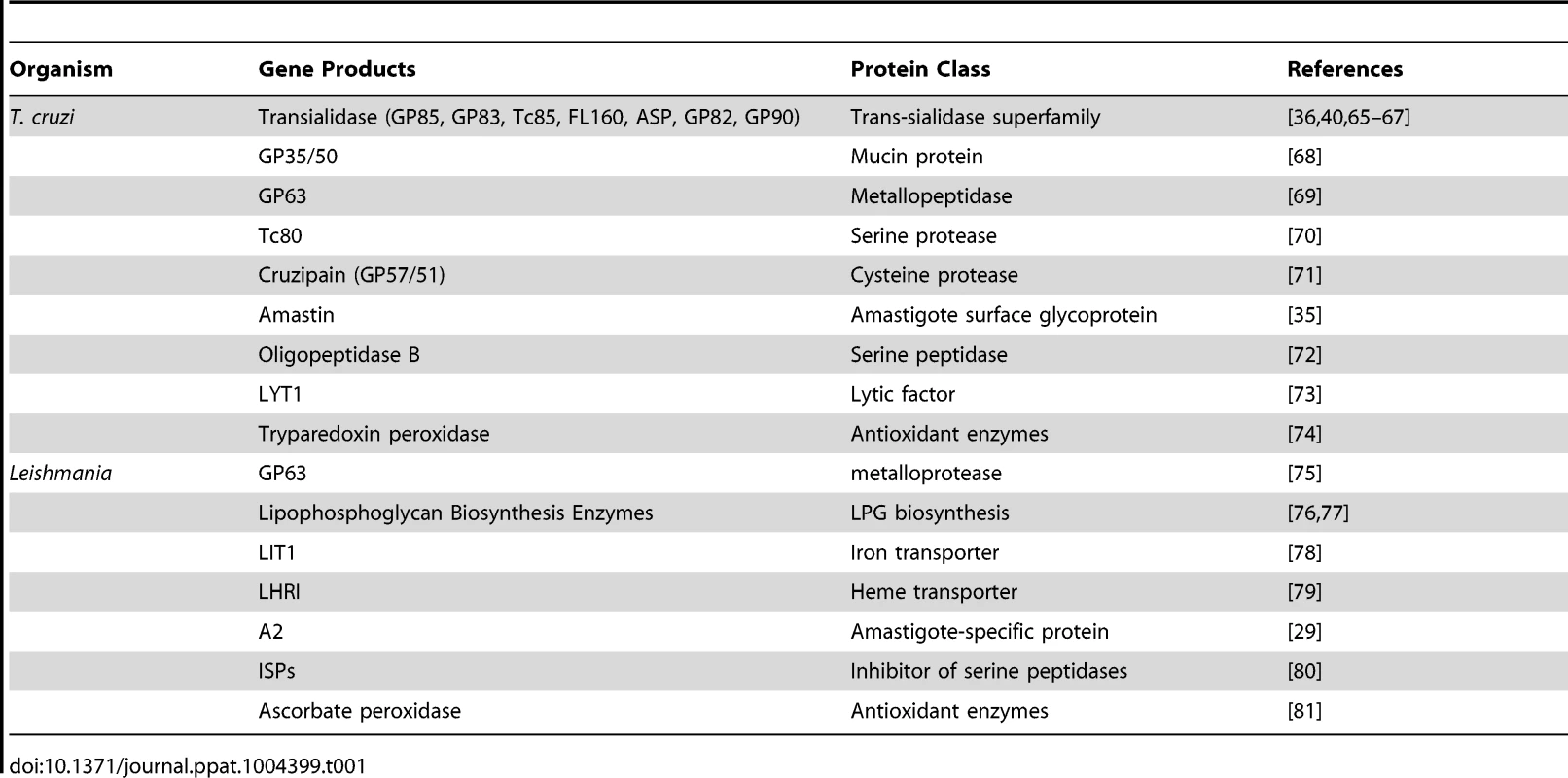
Amastin genes, which are also present in T. cruzi, are part of the group of trypanosomatid genes with up-regulated expression in amastigotes [31]. Initially characterized as T. cruzi and Leishmania amastigote specific genes [32], amastin genes have been identified in the genomes of other trypanosomatids, including the insect parasites, Leptomonas seymouri, Angomonas deanei, and Strigomonas culicis [33], [34]. Evidence indicating a role of this family of surface proteins related to intracellular survival and parasite dissemination within the mammalian host has been found recently in studies where amastin genes were overexpressed in the low infective T. cruzi G strain, which has reduced levels of amastin expression [35].
Mucins, Trans-sialidases (TSs), and Mucin Associated Surface Proteins (MASPs) are products of the three largest and highly heterogeneous gene families present in the T. cruzi genome [14]. TS catalyzes the transfer of sialic acid from sialylated donors present in the host cells to the terminal galactose residues of mucins present in the T. cruzi cell surface [36]. Whereas Mucin and MASP genes are exclusively found in T. cruzi, TS genes are also present in the genome of African trypanosomes, but not in Leishmania spp. However, in contrast to T. cruzi that has more than 1,400 copies of TS genes, the TS-like gene family has only nine members in T. brucei [37], 17 members in T. congolense [38], and about 100 members in Trypanosoma rangeli (E. Grisard, personal communication). Considerably less copies of TS are also present in T. rangeli, an insect parasite nonpathogenic for humans that does not have TS activity [39]. Evidence for the involvement of T. cruzi TS in the earlier steps of host cell invasion exit from the parasitophorous vacuole to the cytoplasm and the subsequent differentiation of trypomastigotes into amastigotes has been described by several groups [40], [41]. Thus, the massive expansion of the TS gene family in T. cruzi compared to T. brucei, its absence in Leishmania spp, and the lack of TS activity of the T. rangeli enzyme is likely to reflect not only the distinct niches these different trypanosomatids occupy within the infected host but also the distinct mechanisms of parasite internalization used by intracellular parasites. Also in agreement with the role of TS in T. cruzi is its large repertoire of genes encoding mucins, which form a thick glycocalyx barrier at the surface of different forms of the parasite [42]. Since mucins are the major acceptors of sialic acid, the absence of mucin genes in T. brucei [13] and the presence of much fewer members of mucin genes in T. rangeli [39] are also consistent with the incapability of these two Trypanosoma species to invade mammalian cells. Conversely, the ability of T. cruzi to invade and multiply in the cytoplasm of distinct host cell types has been also associated with its large repertoire of MASP genes, which, similar to mucin genes, is a highly polymorphic gene family [43], [44] that is absent in T. brucei and has a much smaller allele repertoire in T. rangeli [45].
Comparative Genomics among Trypanosomatids As a Tool to Identify a Gene Set Specific for Intracellular Parasites
With the advent of next generation sequencing technologies, full genome analyses of an increasingly large number of members of the trypanosomatid family are being published, allowing us to dig deeper into the analysis of the genetic similarities and differences behind their distinct life styles. Surprisingly, comparative genome analyses of sequences from two other Leishmania species, L. infantum and L. braziliensis, with the L. major genome revealed not only a remarkable conservation in overall gene synteny, but also no more than 200 genes presenting a differential distribution between the three species [46]. Similar analyses that include sequences from other species of the Viannia complex, also known as New World Leishmania, Leishmania mexicana and Leishmania amazonensis, confirmed that there is little variation in the overall gene content and indicated that gene amplification as well as variation in chromosome number and ploidy constitute major sources of genomic variation across Leishmania species [47], [48]. Since human leishmaniasis is characterized by a highly diverse spectrum of clinical symptoms, these studies suggest that additional factors besides differences in gene content across Leishmania species are likely to play a role in determining disease phenotype. Likewise, in spite of the fact that the lizard parasite L. tarentolae, which belongs to a third subgenera, the Sauroleishmania, is nonpathogenic to humans and does not multiply intracellularly, comparative studies identified only 95 predicted coding sequences unique to L. (S) tarentolae [49]. Furthermore, also highlighting our scarce understanding of the biology of intracellular parasites, most of the genes that are present in pathogenic Leishmania species and absent in L. tarentolae encode hypothetical proteins or proteins with unknown function.
Comparative genomic analyses between two T. cruzi strains, CL Brener and Sylvio X-10 strain [50], which belong to two phylogenetically distinct groups as well as from Trypanosoma cruzi marinkellei, a bat-associated parasite of the subgenus Schizotrypanum [51], also revealed few differences in their genome content. Again, copy number variation within the large multigene families appears to be a major determinant of subspecies variation in Trypanosoma. Similar to mammals infected by T. cruzi, bats infected by T. cruzi marinkellei contain intracellular amastigotes in cardiac, skeletal, and stomach muscle cells. Therefore, differences found in the genomes of T. cruzi, T. cruzi marinkellei, and the various T. brucei subspecies, such as the absence of MASP and mucin genes in T. brucei, can be associated with the lack of capacity of T. brucei to invade host cells. On the other hand, the observations that all Trypanosoma species, including T. cruzi, T. cruzi marinkellei, and T. brucei, have TS genes suggest that TS plays a role in the biology of trypanosomatid parasites regarding not only host cell invasion and intracellular survival but also parasite survival in the insect or in the mammalian bloodstream. Indeed, in GPI-anchored defective T. brucei, lack of surface TS strongly affects parasite survival in the insect midgut [52].
Genome studies of trypanosomatids other than Trypanosoma and Leishmania such as the recent sequence analysis of the genomes of two monoxenic insect parasites, A. deanei and S. culicis, again revealed strikingly conserved features when compared to pathogenic trypanosomatids, such as gene families encoding amastin and cysteine proteases. As expected, no sequences homologous to VSG, mucin-like glycoproteins, and TS genes were found [34]. Besides helping understand the distinct life cycles of these organisms, the relevance of this study relies on the fact that these trypanosomatids bear endosymbiotic bacteria and are considered excellent models for evolutionary studies, specifically how a host protozoan coevolved with an intracellular bacterium in a mutualistic relationship.
Another recent genome-wide study based on the complete genome sequences of 27 protozoans, 17 of them obligate intracellular parasites, six of them exclusively extracellular, and four free-living protists, showed that the predicted proteome of intracellular parasites have a higher content of repetitive sequences compared to extracellular parasites and free-living protists [53]. Therefore, this study suggests that the ability to invade host cells may have shaped the expansion and maintenance of amino acid repeats in the proteome of intracellular parasites. Indeed, tandemly repeated amino acid sequences are characteristic of many surface proteins of other intracellular protozoan parasites such as Plasmodium spp, that have been implicated with binding to host receptors and immune-evasion mechanisms [54]. Similarly, several Leishmania spp and T. cruzi proteins containing repeated amino acid motifs have been described as targets of B cell immune response, and a bias towards the expression of these proteins in the amastigote stage further suggests their involvement with intracellular parasitism [55], [56].
Genome mining of the Trypanosomatid sequence databases can still provide new sets of valuable information about T. cruzi and Leishmania genes that may be part of their intracellular survival gene kit. We analyzed the predicted protein sequences from 15 trypanosomatids for which the genome sequences are available and that were divided into two groups according to their ability to invade and survive inside mammalian host cells. As shown in Table S1, the first group is formed by species that have an intracellular stage and the second group includes trypanosomatids that are either nonpathogenic to mammals or that do not have an intracellular stage. From a total of 13,609 OrthoMCL clusters identified in the analyzed dataset, 3,340 were present only in intracellular parasites and approximately 1.0% of them (37 clusters) are shared between the two T. cruzi strains (CL Brener and Sylvio X-10), T. cruzi marinkellei, and six species of Leishmania (Figure 3). Over 60% of these clusters contain genes annotated as hypothetical proteins with no functional characterization (Table S2). Among the few proteins in this group that have been characterized as virulence factors, we identified kinases and phosphatases that are known to play important roles in host cell invasion in T. cruzi [57], [58] and differentiation and proliferation of amastigotes in L. mexicana [59], L. donovani, and L. major [60]. Also present in this group are mannosyltransferases and a putative fatty acid transporter. Mannan constitutes over 80% of the cellular carbohydrate content of Leishmania intracellular amastigotes [61], and the involvement of this sugar modification has been highlighted in studies with parasite mutants deficient in mannan metabolism which are unable to infect macrophages [62]. Increased fatty acid uptake is required in response to changes in the metabolism of intracellular parasites since a dramatic shift from carbohydrate - to lipid-dependent energy metabolism has been observed in Leishmania and Trypanosoma as an adaptation to the intracellular environment [63], [64]. Not surprisingly, the small number of common genes found between Trypanosoma and Leishmania is in agreement with the distinct mechanisms of host cell invasion adopted by these organisms as well as differences in the intracellular niches they occupy within their host cells.
Fig. 3. Common genes present exclusively in intracellular parasites. 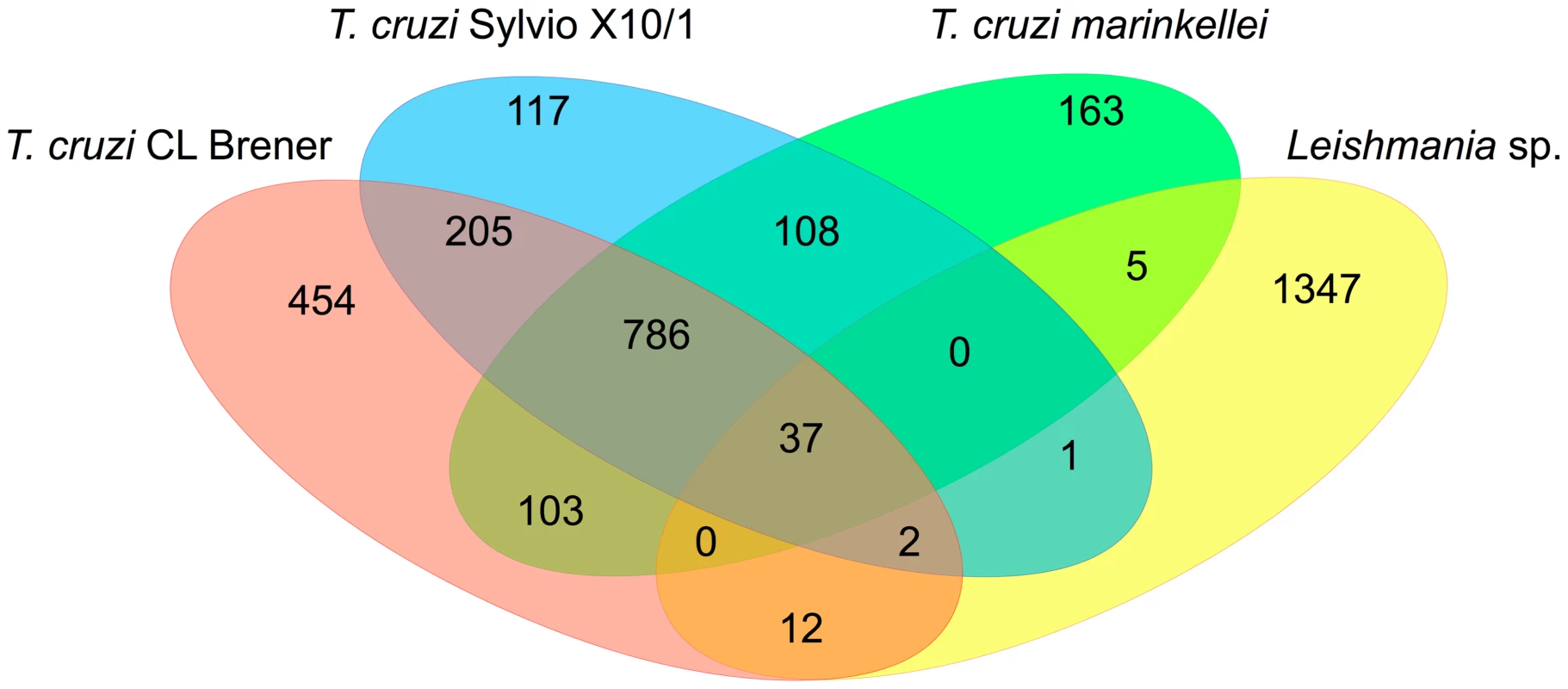
After identifying orthologous proteins, by performing an all-versus-all alignment between the amino acid sequences, the results of the pairwise alignments were used as input to the OrthoMCL software V1.4 with its default parameters. Specific OrthoMCL clusters of intracellular and extracellular/apathogenic trypanosomatids and functional enrichment analysis based in genome annotation were performed using in-house PERL scripts. By delivering more and more genomic sequence information, trypanosomatid genome databases will still provide a large portion of the foundations for our studies on the molecular basis of the intracellular life style of these parasites. Yet, experimental approaches based on genetic manipulation of these parasites are more necessary than ever to better characterize such a distinctive gene set. It is noteworthy the increasingly large difference in the pace with which these genomes are being explored, due to the faster advancement of genetic manipulation tools developed for T. brucei. For T. cruzi and Leishmania, efforts towards genome-wide experimental characterizations of sequences that can be associated with their intracellular life style are, therefore, much welcome.
Supporting Information
Zdroje
1. StevensJR (2008) Kinetoplastid phylogenetics, with special reference to the evolution of parasitic trypanosomes. Parasite 15 : 226–232.
2. TeixeiraSM, El-SayedNM, AraujoPR (2011) The genome and its implications. Adv Parasitol 75 : 209–230.
3. RudenkoG, BishopD, GottesdienerK, Van der PloegLH (1989) Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. Embo J 8 : 4259–4263.
4. MatthewsKR, TschudiC, UlluE (1994) A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev 8 : 491–501.
5. LeBowitzJH, SmithHQ, RuscheL, BeverleySM (1993) Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev 7 : 996–1007.
6. HajdukS, OchsenreiterT (2010) RNA editing in kinetoplastids. RNA Biol 7 : 229–236.
7. SmithDF, PeacockCS, CruzAK (2007) Comparative genomics: from genotype to disease phenotype in the leishmaniases. Int J Parasitol 37 : 1173–1186.
8. BrenerZ (1973) Biology of Trypanosoma cruzi. Annu Rev Microbiol 27 : 347–382.
9. FernandesMC, AndrewsNW (2012) Host cell invasion by Trypanosoma cruzi: a unique strategy that promotes persistence. FEMS Microbiol Rev 36 : 734–747.
10. UenoN, WilsonME (2012) Receptor-mediated phagocytosis of Leishmania: implications for intracellular survival. Trends Parasitol 28 : 335–344.
11. FennK, MatthewsKR (2007) The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol 10 : 539–546.
12. HornD, McCullochR (2010) Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol 13 : 700–705.
13. BerrimanM, GhedinE, Hertz-FowlerC, BlandinG, RenauldH, et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309 : 416–422.
14. El-SayedNM, MylerPJ, BartholomeuDC, NilssonD, AggarwalG, et al. (2005) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309 : 409–415.
15. IvensAC, PeacockCS, WortheyEA, MurphyL, AggarwalG, et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309 : 436–442.
16. WeatherlyDB, BoehlkeC, TarletonRL (2009) Chromosome level assembly of the hybrid Trypanosoma cruzi genome. BMC Genomics 10 : 255.
17. El-SayedNM, MylerPJ, BlandinG, BerrimanM, CrabtreeJ, et al. (2005) Comparative genomics of trypanosomatid parasitic protozoa. Science 309 : 404–409.
18. NgoH, TschudiC, GullK, UlluE (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci U S A 95 : 14687–14692.
19. LyeLF, OwensK, ShiH, MurtaSM, VieiraAC, et al. (2010) Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog 6: e1001161.
20. KolevNG, TschudiC, UlluE (2011) RNA interference in protozoan parasites: achievements and challenges. Eukaryot Cell 10 : 1156–1163.
21. DaRochaWD, OtsuK, TeixeiraSM, DonelsonJE (2004) Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol 133 : 175–186.
22. HartleyMA, KohlK, RonetC, FaselN (2013) The therapeutic potential of immune cross-talk in leishmaniasis. Clin Microbiol Infect 19 : 119–130.
23. TeixeiraSM, de PaivaRM, Kangussu-MarcolinoMM, DarochaWD (2012) Trypanosomatid comparative genomics: Contributions to the study of parasite biology and different parasitic diseases. Genet Mol Biol 35 : 1–17.
24. McCallLI, McKerrowJH (2014) Determinants of disease phenotype in trypanosomatid parasites. Trends Parasitol 30 : 342–349.
25. KedzierskiL, MontgomeryJ, BullenD, CurtisJ, GardinerE, et al. (2004) A leucine-rich repeat motif of Leishmania parasite surface antigen 2 binds to macrophages through the complement receptor 3. J Immunol 172 : 4902–4906.
26. LincolnLM, OzakiM, DonelsonJE, BeethamJK (2004) Genetic complementation of Leishmania deficient in PSA (GP46) restores their resistance to lysis by complement. Mol Biochem Parasitol 137 : 185–189.
27. MatlashewskiG (2001) Leishmania infection and virulence. Med Microbiol Immunol 190 : 37–42.
28. ZhangWW, MatlashewskiG (2000) Analysis of antisense and double stranded RNA downregulation of A2 protein expression in Leishmania donovani. Mol Biochem Parasitol 107 : 315–319.
29. ZhangWW, MendezS, GhoshA, MylerP, IvensA, et al. (2003) Comparison of the A2 gene locus in Leishmania donovani and Leishmania major and its control over cutaneous infection. J Biol Chem 278 : 35508–35515.
30. MizbaniA, TaslimiY, ZahedifardF, TaheriT, RafatiS (2011) Effect of A2 gene on infectivity of the nonpathogenic parasite Leishmania tarentolae. Parasitol Res 109 : 793–799.
31. TeixeiraSM, RussellDG, KirchhoffLV, DonelsonJE (1994) A differentially expressed gene family encoding “amastin,” a surface protein of Trypanosoma cruzi amastigotes. J Biol Chem 269 : 20509–20516.
32. RochetteA, McNicollF, GirardJ, BretonM, LeblancE, et al. (2005) Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol Biochem Parasitol 140 : 205–220.
33. JacksonAP (2010) The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol Biol Evol 27 : 33–45.
34. MottaMC, MartinsAC, de SouzaSS, Catta-PretaCM, SilvaR, et al. (2013) Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS ONE 8: e60209.
35. CruzMC, Souza-MeloN, da SilvaCV, DarochaWD, BahiaD, et al. (2012) Trypanosoma cruzi: role of delta-amastin on extracellular amastigote cell invasion and differentiation. PLoS ONE 7: e51804.
36. Dc-RubinSS, SchenkmanS (2012) T rypanosoma cruzi trans-sialidase as a multifunctional enzyme in Chagas' disease. Cell Microbiol 14 : 1522–1530.
37. MontagnaG, CremonaML, ParisG, AmayaMF, BuschiazzoA, et al. (2002) The trans-sialidase from the african trypanosome Trypanosoma brucei. Eur J Biochem 269 : 2941–2950.
38. TiralongoE, MartensenI, GrotzingerJ, TiralongoJ, SchauerR (2003) Trans-sialidase-like sequences from Trypanosoma congolense conserve most of the critical active site residues found in other trans-sialidases. Biol Chem 384 : 1203–1213.
39. WagnerG, Eiko YamanakaL, MouraH, Denardin LuckemeyerD, SchlindweinAD, et al. (2013) The Trypanosoma rangeli trypomastigote surfaceome reveals novel proteins and targets for specific diagnosis. J Proteomics 82 : 52–63.
40. Rubin-de-CelisSS, UemuraH, YoshidaN, SchenkmanS (2006) Expression of trypomastigote trans-sialidase in metacyclic forms of Trypanosoma cruzi increases parasite escape from its parasitophorous vacuole. Cell Microbiol 8 : 1888–1898.
41. YoshidaN (2006) Molecular basis of mammalian cell invasion by Trypanosoma cruzi. An Acad Bras Cienc 78 : 87–111.
42. Acosta-SerranoA, AlmeidaIC, Freitas-JuniorLH, YoshidaN, SchenkmanS (2001) The mucin-like glycoprotein super-family of Trypanosoma cruzi: structure and biological roles. Mol Biochem Parasitol 114 : 143–150.
43. BartholomeuDC, CerqueiraGC, LeaoAC, daRochaWD, PaisFS, et al. (2009) Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Res 37 : 3407–3417.
44. dos SantosSL, FreitasLM, LoboFP, Rodrigues-LuizGF, MendesTA, et al. (2012) The MASP family of Trypanosoma cruzi: changes in gene expression and antigenic profile during the acute phase of experimental infection. PLoS Negl Trop Dis 6: e1779.
45. StocoPH, WagnerG, Talavera-LopezC, GerberA, ZahaA, et al. (2014) Genome of the Avirulent Human-infective Trypanosome – Trypanosoma rangeli. PLoS Negl Trop Dis 8: e3176.
46. PeacockCS, SeegerK, HarrisD, MurphyL, RuizJC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39 : 839–847.
47. RogersMB, HilleyJD, DickensNJ, WilkesJ, BatesPA, et al. (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21 : 2129–2142.
48. RealF, VidalRO, CarazzolleMF, MondegoJM, CostaGG, et al. (2013) The Genome Sequence of Leishmania (Leishmania) amazonensis: Functional Annotation and Extended Analysis of Gene Models. DNA Res 20 : 567–581.
49. RaymondF, BoisvertS, RoyG, RittJF, LegareD, et al. (2012) Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res 40 : 1131–1147.
50. FranzenO, OchayaS, SherwoodE, LewisMD, LlewellynMS, et al. (2011) Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis 5: e984.
51. FranzenO, Talavera-LopezC, OchayaS, ButlerCE, MessengerLA, et al. (2012) Comparative genomic analysis of human infective Trypanosoma cruzi lineages with the bat-restricted subspecies T. cruzi marinkellei. BMC Genomics 13 : 531.
52. NagamuneK, Acosta-SerranoA, UemuraH, BrunR, Kunz-RenggliC, et al. (2004) Surface sialic acids taken from the host allow trypanosome survival in tsetse fly vectors. J Exp Med 199 : 1445–1450.
53. MendesTA, LoboFP, RodriguesTS, Rodrigues-LuizGF, daRochaWD, et al. (2013) Repeat-enriched proteins are related to host cell invasion and immune evasion in parasitic protozoa. Mol Biol Evol 30 : 951–963.
54. HughesAL (2004) The evolution of amino acid repeat arrays in Plasmodium and other organisms. J Mol Evol 59 : 528–535.
55. PaisFS, DaRochaWD, AlmeidaRM, LeclercqSY, PenidoML, et al. (2008) Molecular characterization of ribonucleoproteic antigens containing repeated amino acid sequences from Trypanosoma cruzi. Microbes Infect 10 : 716–725.
56. GotoY, CarterD, GuderianJ, InoueN, KawazuS, et al. (2010) Upregulated expression of B-cell antigen family tandem repeat proteins by Leishmania amastigotes. Infect Immun 78 : 2138–2145.
57. MaedaFY, CortezC, YoshidaN (2012) Cell signaling during Trypanosoma cruzi invasion. Front Immunol 3 : 361.
58. OrregoPR, OlivaresH, CorderoEM, BressanA, CortezM, et al. (2014) A cytoplasmic new catalytic subunit of Calcineurin in Trypanosoma cruzi and its molecular and functional characterization. PLoS Negl Trop Dis 8: e2676.
59. GrantKM, DunionMH, YardleyV, SkaltsounisAL, MarkoD, et al. (2004) Inhibitors of Leishmania mexicana CRK3 cyclin-dependent kinase: chemical library screen and antileishmanial activity. Antimicrob Agents Chemother 48 : 3033–3042.
60. NascimentoM, ZhangWW, GhoshA, HoustonDR, BerghuisAM, et al. (2006) Identification and characterization of a protein-tyrosine phosphatase in Leishmania: Involvement in virulence. J Biol Chem 281 : 36257–36268.
61. RaltonJE, NadererT, PirainoHL, BashtannykTA, CallaghanJM, et al. (2003) Evidence that intracellular beta1-2 mannan is a virulence factor in Leishmania parasites. J Biol Chem 278 : 40757–40763.
62. GaramiA, MehlertA, IlgT (2001) Glycosylation defects and virulence phenotypes of Leishmania mexicana phosphomannomutase and dolicholphosphate-mannose synthase gene deletion mutants. Mol Cell Biol 21 : 8168–8183.
63. AtwoodJA3rd, WeatherlyDB, MinningTA, BundyB, CavolaC, et al. (2005) The Trypanosoma cruzi proteome. Science 309 : 473–476.
64. BermanJD, GallaleeJV, BestJM, HillT (1987) Uptake, distribution, and oxidation of fatty acids by Leishmania mexicana amastigotes. J Parasitol 73 : 555–560.
65. SchenkmanS, JiangMS, HartGW, NussenzweigV (1991) A novel cell surface trans-sialidase of Trypanosoma cruzi generates a stage-specific epitope required for invasion of mammalian cells. Cell 65 : 1117–1125.
66. RamirezMI, Ruiz RdeC, ArayaJE, Da SilveiraJF, YoshidaN (1993) Involvement of the stage-specific 82-kilodalton adhesion molecule of Trypanosoma cruzi metacyclic trypomastigotes in host cell invasion. Infect Immun 61 : 3636–3641.
67. MagdesianMH, TonelliRR, FesselMR, SilveiraMS, SchumacherRI, et al. (2007) A conserved domain of the gp85/trans-sialidase family activates host cell extracellular signal-regulated kinase and facilitates Trypanosoma cruzi infection. Exp Cell Res 313 : 210–218.
68. YoshidaN, MortaraRA, AraguthMF, GonzalezJC, RussoM (1989) Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35 - and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect Immun 57 : 1663–1667.
69. KulkarniMM, OlsonCL, EngmanDM, McGwireBS (2009) Trypanosoma cruzi GP63 proteins undergo stage-specific differential posttranslational modification and are important for host cell infection. Infect Immun 77 : 2193–2200.
70. GrellierP, VendevilleS, JoyeauR, BastosIM, DrobecqH, et al. (2001) Trypanosoma cruzi prolyl oligopeptidase Tc80 is involved in nonphagocytic mammalian cell invasion by trypomastigotes. J Biol Chem 276 : 47078–47086.
71. MeirellesMN, JulianoL, CarmonaE, SilvaSG, CostaEM, et al. (1992) Inhibitors of the major cysteinyl proteinase (GP57/51) impair host cell invasion and arrest the intracellular development of Trypanosoma cruzi in vitro. Mol Biochem Parasitol 52 : 175–184.
72. CalerEV, Vaena de AvalosS, HaynesPA, AndrewsNW, BurleighBA (1998) Oligopeptidase B-dependent signaling mediates host cell invasion by Trypanosoma cruzi. Embo j 17 : 4975–4986.
73. Manning-CelaR, CortesA, Gonzalez-ReyE, Van VoorhisWC, SwindleJ, et al. (2001) LYT1 protein is required for efficient in vitro infection by Trypanosoma cruzi. Infect Immun 69 : 3916–3923.
74. AlvarezMN, PeluffoG, PiacenzaL, RadiR (2011) Intraphagosomal peroxynitrite as a macrophage-derived cytotoxin against internalized Trypanosoma cruzi: consequences for oxidative killing and role of microbial peroxiredoxins in infectivity. J Biol Chem 286 : 6627–6640.
75. BrittinghamA, MorrisonCJ, McMasterWR, McGwireBS, ChangKP, et al. (1995) Role of the Leishmania surface protease gp63 in complement fixation, cell adhesion, and resistance to complement-mediated lysis. J Immunol 155 : 3102–3111.
76. SpathGF, EpsteinL, LeaderB, SingerSM, AvilaHA, et al. (2000) Lipophosphoglycan is a virulence factor distinct from related glycoconjugates in the protozoan parasite Leishmania major. Proc Natl Acad Sci U S A 97 : 9258–9263.
77. VinetAF, JananjiS, TurcoSJ, FukudaM, DescoteauxA (2011) Exclusion of synaptotagmin V at the phagocytic cup by Leishmania donovani lipophosphoglycan results in decreased promastigote internalization. Microbiology 157 : 2619–2628.
78. HuynhC, SacksDL, AndrewsNW (2006) A Leishmania amazonensis ZIP family iron transporter is essential for parasite replication within macrophage phagolysosomes. J Exp Med 203 : 2363–2375.
79. MiguelDC, FlanneryAR, MittraB, AndrewsNW (2013) Heme uptake mediated by LHR1 is essential for Leishmania amazonensis virulence. Infect Immun 81 : 3620–3626.
80. FariaMS, ReisFC, Azevedo-PereiraRL, MorrisonLS, MottramJC, et al. (2011) Leishmania inhibitor of serine peptidase 2 prevents TLR4 activation by neutrophil elastase promoting parasite survival in murine macrophages. J Immunol 186 : 411–422.
81. DolaiS, YadavRK, PalS, AdakS (2009) Overexpression of mitochondrial Leishmania major ascorbate peroxidase enhances tolerance to oxidative stress-induced programmed cell death and protein damage. Eukaryot Cell 8 : 1721–1731.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus InfectionČlánek P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to InfectionČlánek Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 12- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Microbial Programming of Systemic Innate Immunity and Resistance to Infection
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
- Measles Immune Suppression: Functional Impairment or Numbers Game?
- Cellular Mechanisms of Alpha Herpesvirus Egress: Live Cell Fluorescence Microscopy of Pseudorabies Virus Exocytosis
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus Infection
- Loss of Dynamin-Related Protein 2B Reveals Separation of Innate Immune Signaling Pathways
- Intraspecies Competition for Niches in the Distal Gut Dictate Transmission during Persistent Infection
- Unveiling the Intracellular Survival Gene Kit of Trypanosomatid Parasites
- Extreme Divergence of Tropism for the Stem-Cell-Niche in the Testis
- HTLV-1 Tax-Mediated Inhibition of FOXO3a Activity Is Critical for the Persistence of Terminally Differentiated CD4 T Cells
- P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to Infection
- Hypercytotoxicity and Rapid Loss of NKp44 Innate Lymphoid Cells during Acute SIV Infection
- Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
- Crystal Structure of Calcium Binding Protein-5 from and Its Involvement in Initiation of Phagocytosis of Human Erythrocytes
- Chronic Parasitic Infection Maintains High Frequencies of Short-Lived Ly6CCD4 Effector T Cells That Are Required for Protection against Re-infection
- Specific Dysregulation of IFNγ Production by Natural Killer Cells Confers Susceptibility to Viral Infection
- HSV-2-Driven Increase in the Expression of αβ Correlates with Increased Susceptibility to Vaginal SHIV Infection
- Murine Anti-vaccinia Virus D8 Antibodies Target Different Epitopes and Differ in Their Ability to Block D8 Binding to CS-E
- Brothers in Arms: Th17 and Treg Responses in Immunity
- Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Typhimurium Colitis
- A Negative Feedback Modulator of Antigen Processing Evolved from a Frameshift in the Cowpox Virus Genome
- Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms
- The Non-receptor Tyrosine Kinase Tec Controls Assembly and Activity of the Noncanonical Caspase-8 Inflammasome
- Targeted Changes of the Cell Wall Proteome Influence Ability to Form Single- and Multi-strain Biofilms
- Apoplastic Venom Allergen-like Proteins of Cyst Nematodes Modulate the Activation of Basal Plant Innate Immunity by Cell Surface Receptors
- The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in
- Anti-α4 Antibody Treatment Blocks Virus Traffic to the Brain and Gut Early, and Stabilizes CNS Injury Late in Infection
- Initiation of ART during Early Acute HIV Infection Preserves Mucosal Th17 Function and Reverses HIV-Related Immune Activation
- Microbial Urease in Health and Disease
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Blocking Junctional Adhesion Molecule C Enhances Dendritic Cell Migration and Boosts the Immune Responses against
- IL-28B is a Key Regulator of B- and T-Cell Vaccine Responses against Influenza
- A Natural Genetic Variant of Granzyme B Confers Lethality to a Common Viral Infection
- Neutral Sphingomyelinase in Physiological and Measles Virus Induced T Cell Suppression
- Differential PfEMP1 Expression Is Associated with Cerebral Malaria Pathology
- The Role of the NADPH Oxidase NOX2 in Prion Pathogenesis
- Rapid Evolution of Virus Sequences in Intrinsically Disordered Protein Regions
- The Central Role of cAMP in Regulating Merozoite Invasion of Human Erythrocytes
- Expression of Suppressor of Cytokine Signaling 1 (SOCS1) Impairs Viral Clearance and Exacerbates Lung Injury during Influenza Infection
- Cellular Oxidative Stress Response Controls the Antiviral and Apoptotic Programs in Dengue Virus-Infected Dendritic Cells
- SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in
- Monocyte Recruitment to the Dermis and Differentiation to Dendritic Cells Increases the Targets for Dengue Virus Replication
- Oral Streptococci Utilize a Siglec-Like Domain of Serine-Rich Repeat Adhesins to Preferentially Target Platelet Sialoglycans in Human Blood
- SV40 Utilizes ATM Kinase Activity to Prevent Non-homologous End Joining of Broken Viral DNA Replication Products
- Amphipathic α-Helices in Apolipoproteins Are Crucial to the Formation of Infectious Hepatitis C Virus Particles
- Proteomic Analysis of the Acidocalcisome, an Organelle Conserved from Bacteria to Human Cells
- Experimental Cerebral Malaria Pathogenesis—Hemodynamics at the Blood Brain Barrier
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



