-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Microbial Urease in Health and Disease
article has not abstract
Published in the journal: . PLoS Pathog 10(12): e32767. doi:10.1371/journal.ppat.1004472
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004472Summary
article has not abstract
Overview
Since the discovery of Helicobacter pylori, the urease activity of this bacterial pathogen has been identified as the key factor in infection and acid acclimation in the human stomach. Ureolytic activity plays a key role in the pathogenesis of several bacteria, and urease has also been described as an emerging pathogenic factor during fungal infection. However, urease produced by the oral bacteria community has been shown to counteract caries, and caries-free subjects have high levels of urease activity in plaque samples. Some lactic acid bacteria with documented probiotic behavior are urease-positive. Likewise, other lactic acid bacterial species that are widely used in yogurt production and other fermented dairy products use urease activity to counteract acid stress and to feed several biosynthetic pathways with carbon dioxide and ammonia derived from urea hydrolysis. Urease is also diffused in several species belonging to the human gut microbiota, and it is estimated that this complex microbial community is able to hydrolyze 15%–30% of the urea synthesized in normal subjects. In this context, urease was proposed to serve as a microbial biomarker to distinguish microbiomes based on age and geography, thus highlighting the crucial involvement of this enzymatic activity in nitrogen recycling when dietary nitrogen is limiting. In light of these considerations, the designation of urease as a microbial virulence factor would be misleading, and the proposed use of urease as a therapeutic target to counteract microbial infections should be carefully evaluated.
Urease: Multifunctional Roles in Microbial Physiology
Urease and its substrate urea represent historically important milestones in early scientific investigation. Urea was the first organic molecule synthesized, and urease from jack bean was the first enzyme crystallized, in addition to being the first enzyme shown to contain nickel [1]–[4]. The scientific interest in microbial urease is largely related to the relevance of this enzymatic activity in infection. This interest has been strongly stimulated since the discovery of the association of H. pylori with gastritis and stomach cancer [5]. Moreover, urease has served as a paradigm for understanding the activation mechanisms of many metalloenzymes that require accessory proteins for their catalytic activity [3], [4]. Urease is a urea amidohydrolase (EC 3.5.1.5) that catalyzes the hydrolysis of urea to yield ammonia and carbamate, which spontaneously decomposes to yield a second molecule of ammonia and carbonic acid. The released carbonic acid and the two molecules of ammonia are in equilibrium with their deprotonated and protonated forms, respectively, and the net effect of these reactions is an increase in the pH of the environment that surrounds the urease-positive microorganisms (Fig. 1). For this reason, urease is considered a stress response that was developed by several bacteria to counteract a low environmental pH [6]. In Streptococcus thermophilus, urease is metabolically related to the biosynthetic pathways involved in aspartate, glutamine, arginine, and carbon dioxide metabolism [7], [8]. Notably, urea hydrolysis increases the catabolic efficiency of S. thermophilus by modulating the intracellular pH and increasing the activity of β-galactosidase, glycolytic enzymes, and lactate dehydrogenase [9]. Urea hydrolysis results in increases in both the pHin and the pHout due to the rapid diffusion of ammonia outside of the cell. Consequently, in the presence of urea and a urease-positive microorganism, urease-negative microorganisms share the environmental benefit derived from the transient local pH increase [9].
Fig. 1. Schematic representation of the reaction catalyzed by microbial urease and the involvement of these enzymes in microbial physiology, human health, and disease. 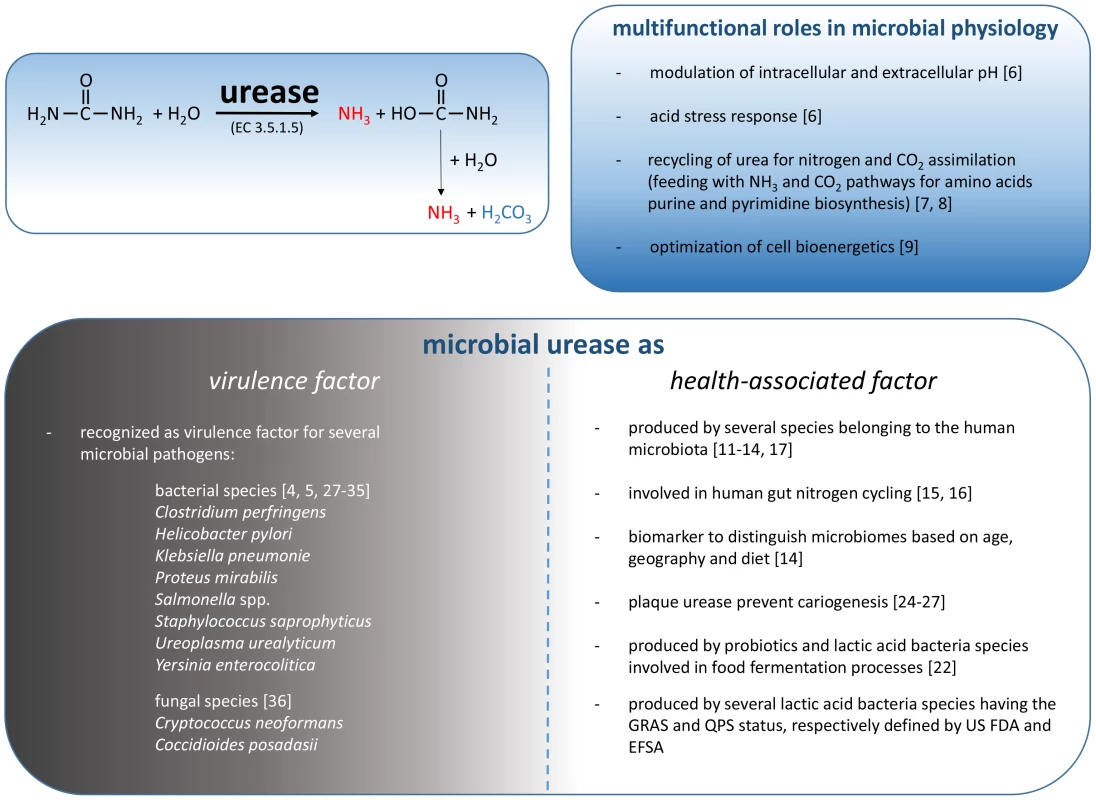
Beyond that, the role of urease in most of the microorganisms that show this enzymatic activity is primarily linked to the recycling of nitrogenous wastes and nitrogen assimilation.
Human Microbiota Urease As a “Health-Associated Factor”
Urea is the major nitrogenous waste product of most terrestrial animals. Ammonium, released from the urea present in the secretions of major and minor exocrine glands, provides a nitrogen source for bacteria that colonize the human body. It was estimated that 15%–30% of the urea synthesized in healthy subjects is continually hydrolyzed by microbial ureases [10]. Several microbial species belonging to the human microbiota produce active urease, and these species take advantage of urea hydrolysis, as has been demonstrated for the oral bacteria Streptococcus salivarius [11] and Actinomyces naeslundii [12] and hypothesized for other species of the gut microbiota [13], [14]. Urea is also present in human milk. Human milk contains only approximately 15% of its nitrogen in the form of urea. It is therefore believed that the total nitrogen in the infant lower gastrointestinal tract (GIT) may be present at suboptimal levels [15]. An increase in postnatal nitrogen levels is likely necessary to satisfy the growth and metabolism requirements of the infant and the GIT microbiota. In human infants, it has been determined that the amino acids in plasma can be derived from urea after hydrolysis and utilization of nitrogen by the intestinal microbiota [16]. It is therefore not surprising that an early colonizer of the GIT of humans, the Bifidobacterium longum subsp. Infantis, produced an active urease [17]. By contrast, in subjects with acute liver failure, the major clinical problem is the development of hepatic encephalopathy (HE) that is associated with high level of gut-derived ammonia. In this scenario, the microbial gut community, especially urease-positive species such as Klebsiella spp. and Proteus spp., is an important source of ammonia in humans in the pathogenesis of HE [18]. Several clinical cases have suggested that the severity of HE can be reduced by modulating the microbial gut community using agents that lead to a normalization of gut microbiota, such as rifaximin, lactulose, prebiotics, and probiotics. However, even probiotics can be urease-positive. Interestingly, a recent study performed using a murine model reported that the administration of the probiotic urease-positive Lactobacillus reuteri reduced the amount of urease activity in the murine gut, presumably due to the suppression of fecal bacteria [19]. Another bacterial species that is currently used as a probiotic for the oropharyngeal tract is the urease-positive S. salivarius strain K12 [20]. Following oral administration, strain K12 can colonize the oral mucosae of infants and adults and down-regulate the innate immune responses of human epithelial cells. It is also active against S. pyogenes and safe and well tolerated by the human host [21]. In a more general food context, it is worth mentioning that yogurt consumption is commonly associated with a health benefit by the consumers [21], and one of the two species of the yogurt consortium, S. thermophilus, is urease-positive [22]. S. thermophilus is widely used in the manufacturing of dairy products, yogurt, fermented milk, and cheeses, and as a consequence, over 1021 living cells carrying active urease molecules are ingested annually by the human population.
A recent study [14] focused on the characterization of gut microbial communities in two human populations revealed that urease gene frequency was significantly higher in Malawian and Amerindian infant microbiomes and that it decreased with age in these two populations, unlike in the United States, where it remains low from infancy to adulthood. Considering that urease has a crucial involvement in nitrogen recycling, particularly when diets are deficient in protein, the ability of the microbiome to use urea would presumably be advantageous to both microbes and host.
Urea is secreted into all parts of the digestive tract starting from the oral cavity. In saliva, urea is present at a concentration of 3–10 mM, and it represents a relevant nitrogen source for several species belonging to the oral microbiota, including S. salivarius, S. vestibularis and Actinomyces naeslundii. A substantial body of evidence is beginning to accumulate that indicates a direct contribution of alkali generation in dental biofilms to the inhibition of dental caries [23]. The development of dental caries is favored by tooth demineralization that happens as a consequence of the frequent acidification of dental biofilms and the subsequent emergence of acidogenic and acid-tolerant microorganisms, including mutans streptococci and Lactobacillus spp., which ferment dietary carbohydrates rapidly and lower the pH. The increasing number of acid-tolerant microorganisms results in a simultaneous decrease in the less acid-tolerant species that are often associated with dental health [24]. Notably, bacteria associated with dental health are able to use urea and/or arginine to generate ammonia. Alkali production by these microorganisms positively affects the balance between the remineralization and demineralization of the tooth and may help prevent the emergence of cariogenic microorganisms [25]. The real scenario is actually more complex than it might appear. In fact, while the urease activity associated with plaque seems to correlate with a decrease in the incidence of caries, the urease activity associated with the saliva had a significant effect on the risk of developing caries, and this effect was not protective but instead promoted the development of caries [26]. It therefore appears that the oral localization of urease activity is fundamental in preventing caries. Interestingly, in mice, the carcinogenicity of the plaque bacterium S. mutans (naturally urease-negative) was dramatically reduced in a derivative recombinant strain of S. mutans that was able to produce an active urease, thus suggesting that recombinant ureolytic bacteria may be useful in promoting dental health [27].
Microbial Urease As a General “Virulence Factor”
In addition to the positive aspects of microbial ureases in human health, a consistent body of evidence has identified urease as a virulence factor for several microbial pathogens (Fig. 1). In fact, ureolytic activity has a key role in the pathogenesis of bacteria such as Clostridium perfringens, Helicobacter pylori, Klebsiella pneumoniae, Proteus mirabilis, Salmonella spp., Staphylococcus saprophyticus, Ureoplasma urealyticum, and Yersinia enterocolitica, and such activity has been reported in diseases such as urolithiasis, pyelonephritis, ammonia encephalopathy, HE, hepatic coma, and gastroduodenal infections [4], [28]. The role of urease in microbial infection has been well established in H. pylori. Hydrolysis of urea in the human stomach provides NH3 that is essential for acid neutralization, enabling H. pylori to raise the pH in its microenvironment and periplasm, thus maintaining the proton motive force [28]. Moreover, the urea-dependent ammonia production appears to be partially responsible for the gastric mucosal injury found in association with H. pylori infection [29]. A proton-gated channel, UreI, which regulates the uptake of urea [4], is only active at acidic pH and therefore does not allow for the transport of urea into the bacterial cell at neutral pH, thus preventing lethal alkalinization of the cytoplasm [4]. Without this mechanism, H. pylori is unable to develop the infection process in the stomach [30], [31]. Similarly, the urease activity allows the survival to the gastric transit of Y. enterocolitica [32].
The role of urease activity in urinary tract infections and struvite and carbonate apatite stones formation was described for P. mirabilis and Sta. saprophyticus. The urease-dependent invasive property of P. mirabilis was supported by in vitro observation and by the use of urease-negative mutants. P. mirabilis defective in urease exhibited in a mouse model an ID50 more than 1,000-fold higher than the wild-type strain, and only the wild-type strain was able to persist significantly [33]. Likewise, the contribution of urease to the cytopathogenicity of Sta. saprophyticus has been demonstrated in a rat model using a chemically mutagenized urease-deficient strain, and by the heterologous expression of an active urease in the nonureolytic Staphylococcus carnosus strain [34], [35].
More recently [36], the role of urease as a general microbial virulence factor was proposed, highlighting the emerging pathogenic roles of urease during infection of the fungal species Cryptococcus neoformans (a basidiomycete) and Coccidioides posadasii (an ascomycete). During fungal lung infection, the urea present in the epithelial lining fluid of the lungs is hydrolyzed by fungal urease, and the generated ammonia inhibits immune function and contributes to lung tissue damage [36].
Other human pathogens are urease-positive, and in many cases, urea hydrolysis is thought to have a role in the infectivity or persistence of the microorganisms. In this context, although largely unexplored, the positive role of urease in microbial physiology (Fig. 1) can be an advantage for a pathogen during the various stages of the infection process in terms of competition with commensal microorganisms associated with the human body.
Perspectives
Because of the facts that the human genome does not contain urease-encoding genes and that no human nickel-containing enzymes are known, urease was proposed as a potential therapeutic target [36] without taking into consideration all the positive aspects linked to the microbial ureases of the human microbiota. In this context, the use of the term “virulence factor” for microbial ureases should be carefully evaluated. Microbiologists working on infectious organisms routinely define any gene product that contribute to the virulence potential of a pathogen as a “virulence factor.” Recently, the increasing interest in the human microbiota raises questions about the terminology we use to describe the molecular and metabolic strategies that pathogenic microbes use to compete in these complex biological systems [37]. In the GIT, many pathogens and commensals use similar strategies to overcome the challenges associated with this particular environment. It would therefore be misleading to describe the same strategies and structures found in harmless or beneficial commensals as “virulence factors” simply because they were acquired or evolved to survive in the GIT. The term “niche factors” was therefore proposed [37] to describe the molecular and metabolic strategies evolved by beneficial gut microbes to colonize this complex environment.
Zdroje
1. SumnerJB (1926) The isolation and crystallization of the enzyme urease. J Biol Chem 69 : 435–441.
2. DixonNE, GazzolaC, BlakeleyRL, ZernerB (1975) Jack bean urease (EC 3.5.1.5). A metalloenzyme. A simple biological role for nickel? J American Chem Society 97 : 4131–4133.
3. AndrewsRK, BlakeleyRL, ZernerB (1984) Urea and urease. Adv Inorg Biochem 6 : 245–283.
4. WeeksDl, EskandariS, ScottDR, SachsG (2000) A H+ - gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287 : 482–485.
5. MobleyHLT, IslandMD, HausingerRP (1995) Molecular biology of microbial ureases. Microbiol Rev 59 : 451–480.
6. CotterPD, HillC (2003) Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol Biol Rev 67 : 429–453.
7. ArioliS, MonnetC, GuglielmettiS, PariniC, DeNoni, et al. (2007) Aspartate biosynthesis is essential for the growth of Streptococcus thermophilus in milk, and aspartate availability modulates the level of urease activity. Appl Environ Microbiol 73 : 5789–5796.
8. ArioliS, RoncadaP, SalzanoAM, DeriuF, CoronaS, et al. (2009) The relevance of carbon dioxide metabolism in Streptococcus thermophilus. Microbiol 155 : 1953–1965.
9. ArioliS, RaggEM, ScaglioniL, FessasD, SignorelliM, et al. (2010) Alkalizing reactions streamline cellular metabolism in acidogenic microorganisms. PLoS ONE 5: e15520.
10. WalserM, BodenlosL (1959) Urea metabolism in man. J Clin Invest 38 : 1617–1626.
11. ChenYM, BurneRA (2003) Identification and characterization of the nickel transport system for urease biogenesis in Streptococcus salivaius 57.1. J Bacteriol 185 : 6773–6779.
12. Morou-BermudezE, BurneRA (2000) Genetic and physiologic characterization of urease of Actinomyces naeslundii. Infect Immun 67 : 504–512.
13. WegmannU, LouisP, GoesmannA, HenrissatB, DuncanSH, et al. (2013) Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ Microbiol 16 : 2879–2890 doi:10.1111/1462-2920.12217
14. YatsunenkoT, ReyFE, ManaryM, TrehanI, Dominguez-BelloMG, et al. (2012) Human gut microbiome viewed across age and geography. Nature 486 : 222–227.
15. Fuller MF, Reeds PJ 1998. Nitrogen cycling in the gut. Annu Rev Nutr 18 : 385–411.
16. MillwardDJ, ForresterT, Ah-SingE, YeboahN, GibsonN, et al. (2000) The transfer of 15N from urea to lysine in the human infant. British J Nut 83 : 505–512.
17. LoCascioRG, DesaiP, SelaDA, WeimerB, MillsDA (2010) Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol 76 : 7373–7381.
18. RiordanSM, WilliamsR (2010) Gut flora and hepatic encephalopathy in patients with cirrhosis. N Engl J Med 362 : 1140–1142.
19. WilsonCM, LoachD, LawleyB, BellT, SimsI, et al. (2014) Lactobacillus reuteri 100-23 modulates urea hydrolysis in the murine stomach. Appl Environ Microbiol 80 : 6104–6113 doi:10.1128/AEM.01876-14
20. PowerDA, BurtonJP, ChilcottCN, DawesPJ, TaggJR (2008) Preliminary investigations of the colonization of upper respiratory tract tissues of infants using a pediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur J Clin Microbiol Infect Dis 27 : 1261–1263.
21. BurtonJP, CowleyS, SimonRR, McKinneyJ, WescombePA, et al. (2011) Evaluation of safety and human tolerance of the oral probiotic Streptococcus salivarius K12: A randomized, placebo-controlled, double-blind study. Food Chem Toxicol 49 : 2356–2364.
22. MoraD, MaguinE, MasieroM, RicciG, PariniC, et al. (2004) Characterization of urease genes cluster of Streptococcus thermophilus. J Appl Microbiol 96 : 209–219.
23. Zhu Y, Wang H, Hollis JH, Jacques PF (2014) The association between yogurt consumption, diet quality, and metabolic profiles in children in the USA. Eur J Nutr. E-pub ahead of print. doi:10.1007/s00394-014-0735-7
24. LiuY-L, NascimentoM, BurneR (2012) Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int J Oral Sci 4 : 135–140.
25. BeckerMR, PasterBJ, LeysEJ, MoeschbergerML, KenyonSG, et al. (2002) Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40 : 1001–1009.
26. Morou-BermudezE, Elias-BonetaA, BillingsRJ, BurneRA, Garcia-RivasV, et al. (2011) Urease activity as a risk factor for caries development in children during a three-year study period: A survival analysis approach. Archives Oral Microbiol 56 : 1560–1568.
27. ClancyKA, PearsonS, BowenWH, BurneRA (2000) Characterization of recombinant, ureolytic Streptococcus mutans demonstrates an inverse relationship between dental plaque ureolytic capacity and cariogenicity. Infect Immun 68 : 2621–2629.
28. BurneRA, ChenYM (2000) Bacterial ureases in infectious diseases. Microb Infect 2 : 533–542.
29. SmootDT, MobleyHL, ChippendaleGR, LewisonJF, ResauJH (1990) Helicobacter pylori urease activity is toxic to human gastric epithelial cells. Infect Immun 58 : 1992–1994.
30. Mollenhauer-RektorschekM, HanauerG, SacksG, MelchersK (2002) Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res Microbiol 153 : 659–666.
31. SkouloubrisS, ThibergeJM, LabigneA, De ReuseH (1998) The helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect Immun 66 : 4517–4521.
32. De Koning-WardTF, Robins-BrowneRM (1995) Contribution of urease to acid tolerance in Yersinia enterocolitica.. Infect Immun 63 : 3790–3795.
33. JohnsonDE, RusselRG, LockatellCV, ZultyJC, WarrenJW, et al. (1993) Contribution of Proteus mirabilis urease to persistence, urolithiasis, and acute pyelonephritis in a mouse model of ascending urinary tract infection. Infect Immun 61 : 2748–2754.
34. GatermannS, JohnJ, MarreR (1989) Staphylococcus saprophyticus urease: characterization and contribution to uropathogenicity in unobstructed urinary tract infection of rats. Infect Immun 57 : 110–116.
35. GatermannS, MarreR (1989) Cloning and expression of Staphylococcus saprophyticus urease gene sequences in Staphylococcus carnosus and contribution of the enzyme to virulence. Infect Immun 57 : 2998–3002.
36. RutherfordJC (2014) The emerging role of urease as a general microbial virulence factor. PLoS Pathog 10: e1004062.
37. HillC (2012) Virulence or niche factors: what's in a name? J Bacteriol 194 : 5725–5727.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus InfectionČlánek P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to InfectionČlánek Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 12- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Microbial Programming of Systemic Innate Immunity and Resistance to Infection
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
- Measles Immune Suppression: Functional Impairment or Numbers Game?
- Cellular Mechanisms of Alpha Herpesvirus Egress: Live Cell Fluorescence Microscopy of Pseudorabies Virus Exocytosis
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus Infection
- Loss of Dynamin-Related Protein 2B Reveals Separation of Innate Immune Signaling Pathways
- Intraspecies Competition for Niches in the Distal Gut Dictate Transmission during Persistent Infection
- Unveiling the Intracellular Survival Gene Kit of Trypanosomatid Parasites
- Extreme Divergence of Tropism for the Stem-Cell-Niche in the Testis
- HTLV-1 Tax-Mediated Inhibition of FOXO3a Activity Is Critical for the Persistence of Terminally Differentiated CD4 T Cells
- P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to Infection
- Hypercytotoxicity and Rapid Loss of NKp44 Innate Lymphoid Cells during Acute SIV Infection
- Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
- Crystal Structure of Calcium Binding Protein-5 from and Its Involvement in Initiation of Phagocytosis of Human Erythrocytes
- Chronic Parasitic Infection Maintains High Frequencies of Short-Lived Ly6CCD4 Effector T Cells That Are Required for Protection against Re-infection
- Specific Dysregulation of IFNγ Production by Natural Killer Cells Confers Susceptibility to Viral Infection
- HSV-2-Driven Increase in the Expression of αβ Correlates with Increased Susceptibility to Vaginal SHIV Infection
- Murine Anti-vaccinia Virus D8 Antibodies Target Different Epitopes and Differ in Their Ability to Block D8 Binding to CS-E
- Brothers in Arms: Th17 and Treg Responses in Immunity
- Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Typhimurium Colitis
- A Negative Feedback Modulator of Antigen Processing Evolved from a Frameshift in the Cowpox Virus Genome
- Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms
- The Non-receptor Tyrosine Kinase Tec Controls Assembly and Activity of the Noncanonical Caspase-8 Inflammasome
- Targeted Changes of the Cell Wall Proteome Influence Ability to Form Single- and Multi-strain Biofilms
- Apoplastic Venom Allergen-like Proteins of Cyst Nematodes Modulate the Activation of Basal Plant Innate Immunity by Cell Surface Receptors
- The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in
- Anti-α4 Antibody Treatment Blocks Virus Traffic to the Brain and Gut Early, and Stabilizes CNS Injury Late in Infection
- Initiation of ART during Early Acute HIV Infection Preserves Mucosal Th17 Function and Reverses HIV-Related Immune Activation
- Microbial Urease in Health and Disease
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Blocking Junctional Adhesion Molecule C Enhances Dendritic Cell Migration and Boosts the Immune Responses against
- IL-28B is a Key Regulator of B- and T-Cell Vaccine Responses against Influenza
- A Natural Genetic Variant of Granzyme B Confers Lethality to a Common Viral Infection
- Neutral Sphingomyelinase in Physiological and Measles Virus Induced T Cell Suppression
- Differential PfEMP1 Expression Is Associated with Cerebral Malaria Pathology
- The Role of the NADPH Oxidase NOX2 in Prion Pathogenesis
- Rapid Evolution of Virus Sequences in Intrinsically Disordered Protein Regions
- The Central Role of cAMP in Regulating Merozoite Invasion of Human Erythrocytes
- Expression of Suppressor of Cytokine Signaling 1 (SOCS1) Impairs Viral Clearance and Exacerbates Lung Injury during Influenza Infection
- Cellular Oxidative Stress Response Controls the Antiviral and Apoptotic Programs in Dengue Virus-Infected Dendritic Cells
- SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in
- Monocyte Recruitment to the Dermis and Differentiation to Dendritic Cells Increases the Targets for Dengue Virus Replication
- Oral Streptococci Utilize a Siglec-Like Domain of Serine-Rich Repeat Adhesins to Preferentially Target Platelet Sialoglycans in Human Blood
- SV40 Utilizes ATM Kinase Activity to Prevent Non-homologous End Joining of Broken Viral DNA Replication Products
- Amphipathic α-Helices in Apolipoproteins Are Crucial to the Formation of Infectious Hepatitis C Virus Particles
- Proteomic Analysis of the Acidocalcisome, an Organelle Conserved from Bacteria to Human Cells
- Experimental Cerebral Malaria Pathogenesis—Hemodynamics at the Blood Brain Barrier
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



