-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
The SUPPRESSOR OF rps4-RLD1 (SRFR1) gene was identified based on enhanced AvrRps4-triggered resistance in the naturally susceptible Arabidopsis accession RLD. No other phenotypic effects were recorded, and the extent of SRFR1 involvement in regulating effector-triggered immunity was unknown. Here we show that mutations in SRFR1 in the accession Columbia-0 (Col-0) lead to severe stunting and constitutive expression of the defense gene PR1. These phenotypes were temperature-dependent. A cross between srfr1-1 (RLD background) and srfr1-4 (Col-0) showed that stunting was caused by a recessive locus in Col-0. Mapping and targeted crosses identified the Col-0-specific resistance gene SNC1 as the locus that causes stunting. SRFR1 was proposed to function as a transcriptional repressor, and SNC1 is indeed overexpressed in srfr1-4. Interestingly, co-regulated genes in the SNC1 cluster are also upregulated in the srfr1-4 snc1-11 double mutant, indicating that the overexpression of SNC1 is not a secondary effect of constitutive defense activation. In addition, a Col-0 RPS4 mutant showed full susceptibility to bacteria expressing avrRps4 at 24°C but not at 22°C, while RLD susceptibility was not temperature-dependent. The rps4-2 snc1-11 double mutant showed increased, but not full, susceptibility at 22°C, indicating that additional cross-talk between resistance pathways may exist. Intriguingly, when transiently expressed in Nicotiana benthamiana, SRFR1, RPS4 and SNC1 are in a common protein complex in a cytoplasmic microsomal compartment. Our results highlight SRFR1 as a convergence point in at least a subset of TIR-NBS-LRR protein-mediated immunity in Arabidopsis. Based on the cross-talk evident from our results, they also suggest that reports of constitutive resistance phenotypes in Col-0 need to consider the possible involvement of SNC1.
Published in the journal: . PLoS Pathog 6(11): e32767. doi:10.1371/journal.ppat.1001172
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001172Summary
The SUPPRESSOR OF rps4-RLD1 (SRFR1) gene was identified based on enhanced AvrRps4-triggered resistance in the naturally susceptible Arabidopsis accession RLD. No other phenotypic effects were recorded, and the extent of SRFR1 involvement in regulating effector-triggered immunity was unknown. Here we show that mutations in SRFR1 in the accession Columbia-0 (Col-0) lead to severe stunting and constitutive expression of the defense gene PR1. These phenotypes were temperature-dependent. A cross between srfr1-1 (RLD background) and srfr1-4 (Col-0) showed that stunting was caused by a recessive locus in Col-0. Mapping and targeted crosses identified the Col-0-specific resistance gene SNC1 as the locus that causes stunting. SRFR1 was proposed to function as a transcriptional repressor, and SNC1 is indeed overexpressed in srfr1-4. Interestingly, co-regulated genes in the SNC1 cluster are also upregulated in the srfr1-4 snc1-11 double mutant, indicating that the overexpression of SNC1 is not a secondary effect of constitutive defense activation. In addition, a Col-0 RPS4 mutant showed full susceptibility to bacteria expressing avrRps4 at 24°C but not at 22°C, while RLD susceptibility was not temperature-dependent. The rps4-2 snc1-11 double mutant showed increased, but not full, susceptibility at 22°C, indicating that additional cross-talk between resistance pathways may exist. Intriguingly, when transiently expressed in Nicotiana benthamiana, SRFR1, RPS4 and SNC1 are in a common protein complex in a cytoplasmic microsomal compartment. Our results highlight SRFR1 as a convergence point in at least a subset of TIR-NBS-LRR protein-mediated immunity in Arabidopsis. Based on the cross-talk evident from our results, they also suggest that reports of constitutive resistance phenotypes in Col-0 need to consider the possible involvement of SNC1.
Introduction
Plants possess a highly effective immune system that responds to conserved non-self molecular patterns, or to specific pathogen-derived molecules deployed to alter host defenses [1]–[3]. The latter response, called effector-triggered immunity (ETI), is largely mediated by resistance (R) proteins that directly or indirectly detect the presence of pathogen effectors [3], [4], although mechanistically overlap between ETI and the response to molecular patterns can be observed [5], [6]. ETI can lead to programmed cell death termed the hypersensitive response (HR) [7], [8]. In the case of resistance to some viral and hemi-biotrophic bacterial pathogens, it has been shown that the HR is not causally related to resistance [9]–[13]. Nevertheless, the plant immune response is deleterious to plant growth, normal development, and seed set even in the absence of HR, and therefore needs to be tightly controlled [14].
In order to explore the molecular mechanisms that negatively regulate ETI, we performed a suppressor screen for reactivated AvrRps4-triggered resistance in the naturally susceptible Arabidopsis (Arabidopsis thaliana) accession RLD [15]. This screen yielded two mutant alleles in SUPPRESSOR OF rps4-RLD1 (SRFR1). Mutations in srfr1 enhanced resistance of RLD specifically to Pseudomonas syringae pv. tomato strain DC3000 (DC3000) expressing avrRps4, while susceptibility to the virulent strain DC3000 was unchanged [15]. Apart from re-establishing a certain level of resistance to avrRps4, no other marked phenotype was noted.
RPS4 encodes an R protein of the Toll/Interleukin-1 receptor (TIR) - nucleotide binding site (NBS) - leucine-rich repeat (LRR) (TNL) class [16], and was found to require the defense regulator EDS1 to trigger immunity [17]. This is in contrast to the coiled-coil (CC) -NBS-LRR (CNL) R proteins RPS2, RPM1 and RPS5, which require the defense gene NDR1 [17]. Combining mutations in SRFR1 and the CNL pathway genes RPM1, RPS2 or NDR1 did not measurably alter the susceptibility to the cognate effector genes. The partial resistance to avrRps4 in srfr1 mutants required EDS1 [15], [18]. In addition, mutations in RPS6, another TNL gene that requires EDS1 [12], led to susceptibility to DC3000(hopA1) that was diminished in srfr1-1 rps6-1 double mutants [19].
Taken together, these data indicated that SRFR1 function is closely associated with the EDS1 resistance pathway. Here we show that a mutation in SRFR1 in the accession Columbia-0 (Col-0), srfr1-4, activates the Col-0 specific and EDS1-dependent R-like gene SNC1, consistent with the genetic function of SRFR1 as a negative regulator of R gene-mediated resistance. Activation of constitutive defenses in srfr1-4 was temperature-dependent. In addition, RPS4 and SNC1 contributed redundantly to susceptibility to DC3000(avrRps4) in Col-0 at 22°C, whereas at 24°C RPS4 activity was the sole determinant of resistance. Interestingly, SRFR1 interacted with both RPS4 and SNC1. Our data thus provide evidence for cross-talk between these TNL pathways that converge on SRFR1, suggesting that SRFR1 may have a general function in regulating TNL protein signal output.
Results
A mutation in SRFR1 in Col-0 causes abnormal growth
We previously had isolated the mutant alleles srfr1-1 and srfr1-2 from the Arabidopsis accession RLD [15]. Apart from enhanced resistance to DC3000(avrRps4), they did not display marked phenotypes. To further investigate the function of SRFR1, we aimed at isolating T-DNA tagged lines of SRFR1 in the accession Col-0 [20], [21]. Out of four lines, one did not germinate (SALK_106212), and one was untagged (SALK_095440). We could verify a T-DNA insertion far upstream of the open reading frame in SALK_039199, without causing an apparent phenotype. Interestingly, the fourth line, SAIL_412_E08 with a T-DNA insertion in the second intron of SRFR1 (Figure 1A), showed pronounced stunting (Figure 1B) in one-fourth of plants (22 out of 97 plants; χ2 = 0.28). Genotyping showed that the T-DNA insertion in SRFR1 segregated in the original seed stock, and that stunted plants were invariably homozygous for the T-DNA insertion. Reverse transcription (RT) PCR showed that no srfr1 mRNA was detected with primers on either side of the insertion (Figure S1). A low level of srfr1 mRNA could be detected with primers located 3′ of the T-DNA insertion, but this mRNA contained the T-DNA (Figure S1), indicating that srfr1-4 mRNA does not encode functional protein. Consistent with this, Li and co-workers recently showed that no SRFR1 protein can be detected in this knock-out line [22]. We named this line srfr1-4.
Fig. 1. A mutation in SRFR1 causes severe stunting in Col-0. 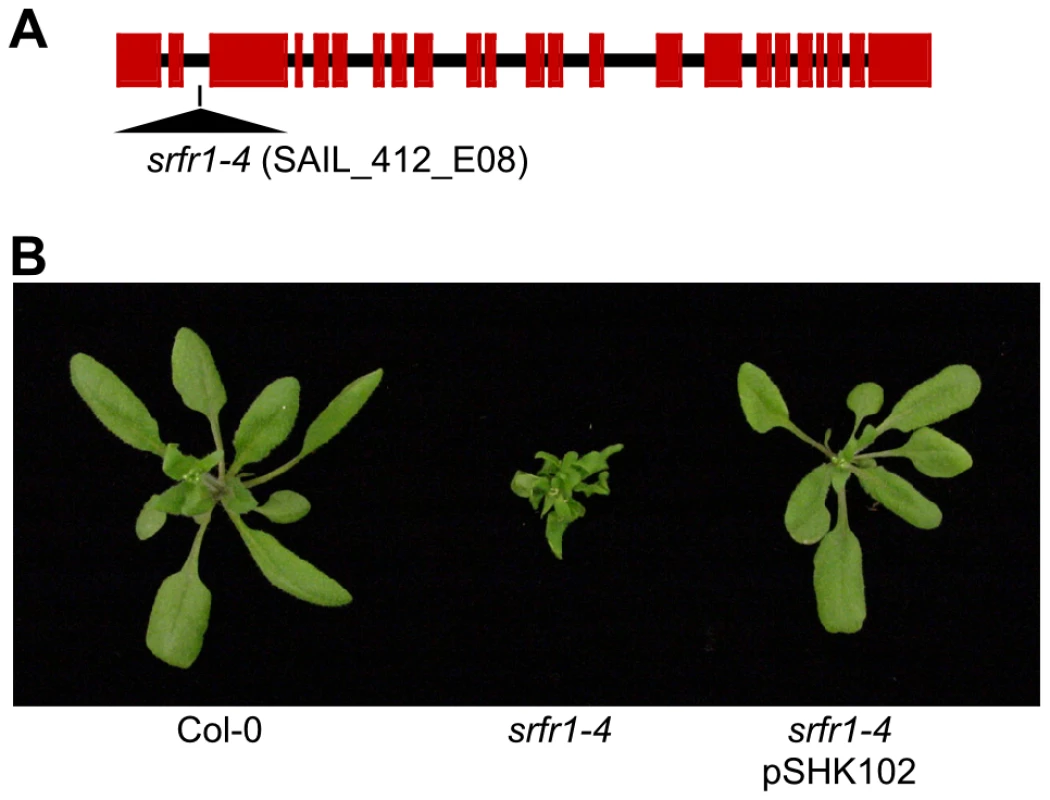
(A) Schematic gene structure of SRFR1 (At4g37460), with exons shown as boxes and introns as lines. The T-DNA insertion site in the second intron in srfr1-4 (SAIL_412_E08) is indicated. (B) Growth phenotype of srfr1-4 and complementation with a genomic copy of SRFR1 in transgenic plants. Subsequently, we back-crossed srfr1-4 to Col-0. The stunted phenotype co-segregated with homozygosity of the srfr1-4 T-DNA tagged allele in F2 plants (Table 1). To prove that the phenotype originated from the srfr1-4 allele, we transformed healthy heterozygous srfr1-4 plants with pSHK102 containing a genomic clone of SRFR1 [18], and by scoring for antibiotic resistance selected 5 single-locus homozygous transgenic SRFR1 T3 lines that contained at least one copy of the srfr1-4 T-DNA allele based on genotyping. Because the transgenic copy of SRFR1 prevented us from determining whether these T3 lines were homozygous or heterozygous for the srfr1-4 allele, we tested whether srfr1-4 segregated in the next generation by genotyping 15 progeny for each line. Three of the 5 lines were shown in this way to be homozygous for the srfr1-4 allele, and the transgenic copy of SRFR1 reversed the stunted phenotype in each case (Figure 1B). We concluded that the stunted growth phenotype is caused by the T-DNA insertion in SRFR1.
Tab. 1. The stunted phenotype co-segregates with the srfr1-4 allele in a backcross to Col-0. 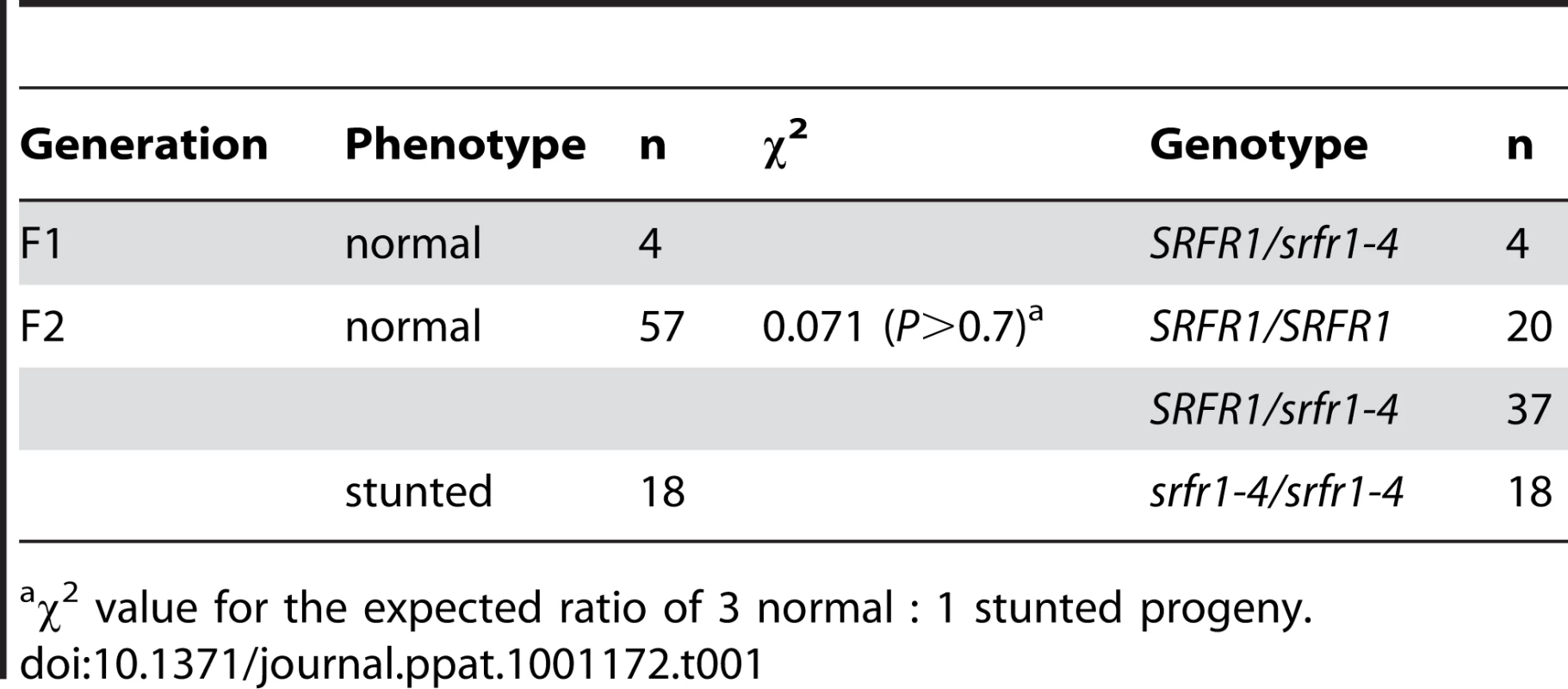
χ2 value for the expected ratio of 3 normal : 1 stunted progeny. Genetics of srfr1-mediated stunting
The stunted srfr1-4 phenotype was in marked contrast to the normal phenotype of srfr1-1 and srfr1-2 plants. To determine whether the specific allele of SRFR1 or the Col-0 genetic background causes the severe phenotype of srfr1-4, we first reexamined more closely F3 families of important break-point plants retained from the SRFR1 mapping populations. Plants in these F3 families were generated by crossing srfr1-1 or srfr1-2 (RLD background) to the SAIL RPS4 T-DNA knockout line rps4-1 (Col-0 background) [15], [18] and were progeny of F2 plants selected for resistance to DC3000(avrRps4). They were therefore homozygous for srfr1-1 or srfr1-2, with varying degrees of Col-0 background. Two out of 4 srfr1-1 and 2 out of 6 srfr1-2 F3 families contained no individuals with abnormal growth phenotypes. However, the remaining F3 families gave rise to plants with phenotypes similar to srfr1-4. The combined total number of stunted plants in these families was 20 out of 107 plants, consistent with the segregation of a single recessive gene in these populations (χ2 = 2.43, P>0.1). We concluded that most likely the mutant alleles srfr1-1 and srfr1-2 also induce stunting in the Col-0 background and that Col-0 possesses a recessive genetic modifier that alters the srfr1 phenotype.
We tested these predictions directly by out-crossing srfr1-4 to RLD and srfr1-1. In the cross to srfr1-1, 14 out of 46 plants were stunted, consistent with both srfr1-1 and srfr1-4 causing stunting and the segregation of a recessive gene (χ2 = 0.45, P>0.5). In the cross to RLD, segregation of the stunted phenotype in the F2 generation was explained by two recessive genes, and genotyping showed that while all stunted plants were homozygous srfr1-4, not all srfr1-4/srfr1-4 plants were automatically stunted (Table 2). In this cross, stunted F2 plants were also selected to determine a rough map position for the presumptive Col-0 modifier gene. This mapping placed the Col-0 modifier gene onto chromosome 4 (Table 3). Interestingly, in addition to the bottom of chromosome 4 where SRFR1 is located, individual break-point plants identified a map position towards the top of chromosome 4 between markers ciw6 and CH42 for the Col-0 modifier gene.
Tab. 2. A second recessive Col-0 allele is required for stunting in the cross RLD×srfr1-4. 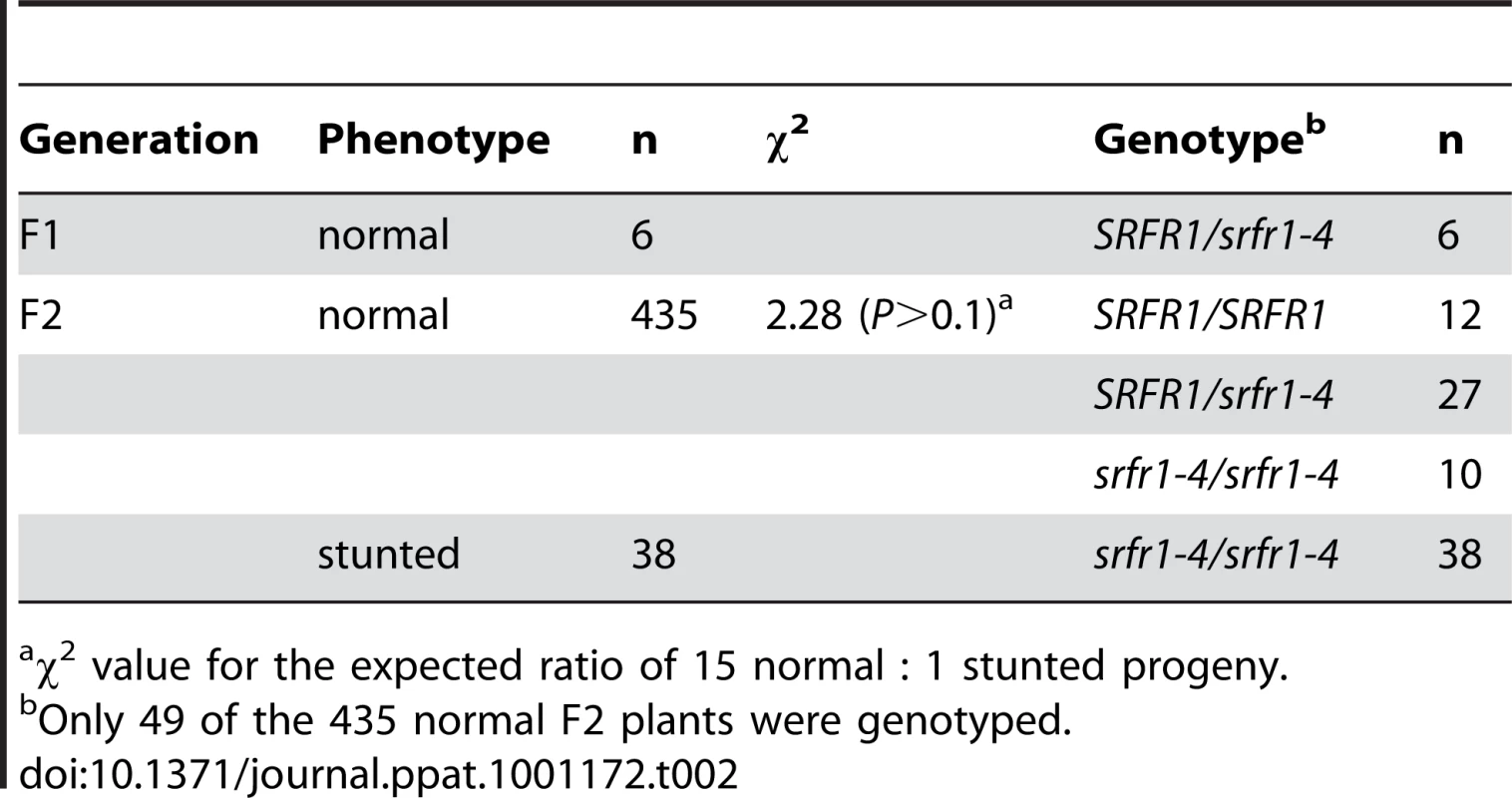
χ2 value for the expected ratio of 15 normal : 1 stunted progeny. Tab. 3. Mapping of the Col-0 modifier gene in stunted F2 plants from the cross RLD×srfr1-4. 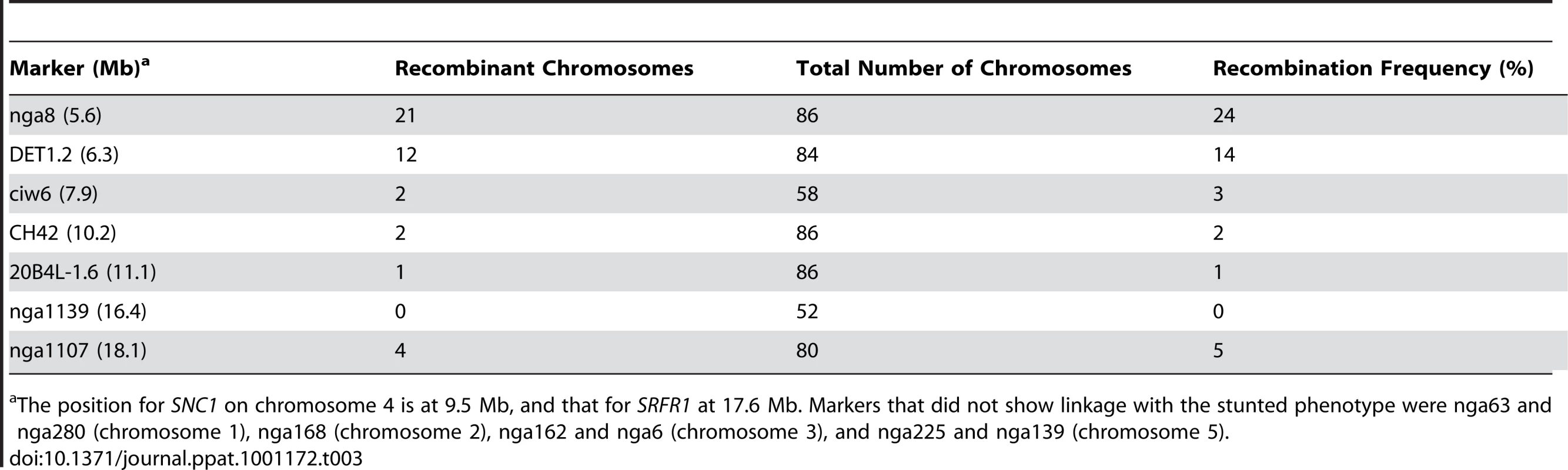
The position for SNC1 on chromosome 4 is at 9.5 Mb, and that for SRFR1 at 17.6 Mb. Markers that did not show linkage with the stunted phenotype were nga63 and nga280 (chromosome 1), nga168 (chromosome 2), nga162 and nga6 (chromosome 3), and nga225 and nga139 (chromosome 5). srfr1-4 has constitutively activated defenses caused by SNC1
The map position for the modifier gene contained the Col-0-specific TNL R gene homolog SNC1, which was originally identified through a point mutation that autoactivates the SNC1 protein and constitutively induces PR genes even in the npr1 mutant line [23]. Additional work showed that wild-type SNC1 is easily autoactivated when expression of SNC1 is misregulated [24]. For example, mutations in BON1, a member of the copine gene family encoding a plasma membrane-localized putative calcium-dependent phospholipid-binding protein [25], [26], lead to higher SNC1 expression levels, constitutive defense responses and reduced plant growth [27]. When the Col bon1-1 mutant was outcrossed to other Arabidopsis accessions, it was found that the wild-type SNC1 gene from Col-0 behaved as a recessive locus that causes stunting [27]. Our segregation data also indicated that the Col-0 modifier was recessive (Table 2). We therefore tested additional phenotypes displayed by bon1-1 plants, such as temperature dependence of constitutive defense activation and growth phenotypes. The stunted phenotype in srfr1-4 was severe at 22°C, but was intermediate at 24°C and absent at 28°C (Figure 2A), reminiscent of the Arabidopsis bon1-1 mutant phenotype.
Fig. 2. The growth phenotype of srfr1-4 is temperature-dependent and accompanied by constitutive activation of defenses. 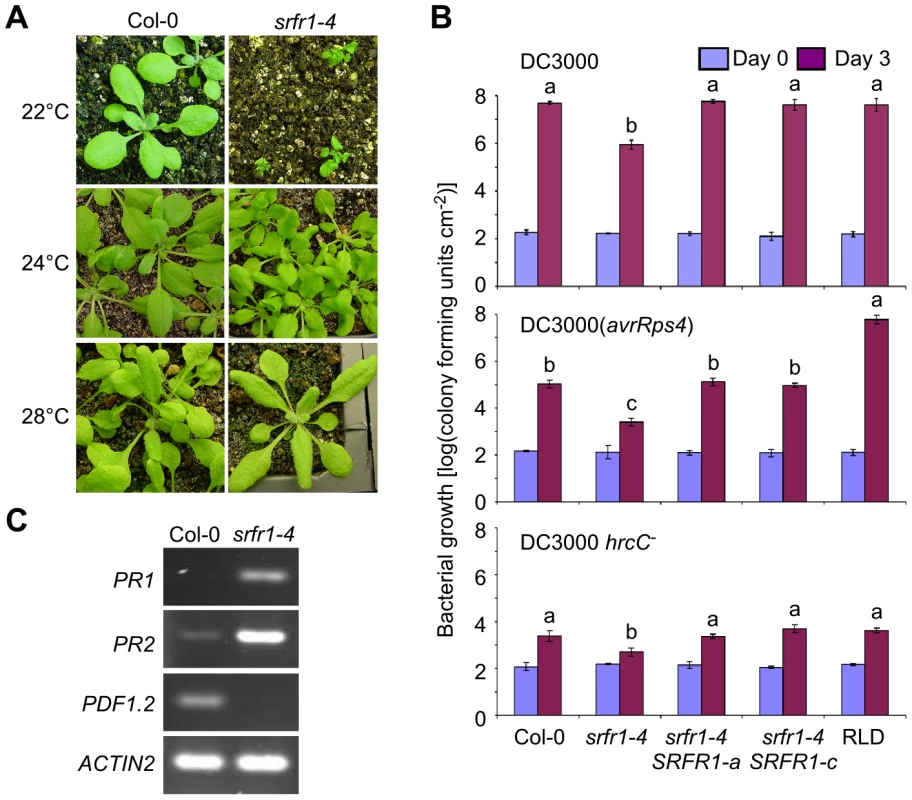
(A) Growth phenotype of wild type Col-0 (left column) and srfr1-4 (right column) at 22°C (top row), 24°C (middle) and 28°C (bottom). (B) The srfr1-4 mutation enhances both basal defenses and AvrRps4-triggered immunity in Col-0. In planta bacterial growth at 24°C of DC3000 (top), DC3000(avrRps4) (middle) and DC3000 hrcC− on day 0 (blue bars) and day 3 (purple bars) after inoculation of the indicated plant lines with bacteria at 5×104 colony-forming units (cfu) per ml. Two independent transgenic srfr1-4 lines complemented with a genomic copy of SRFR1 are shown. Values represent averages of cfu/cm2 leaf tissue from triplicate samples, and error bars denote standard deviation. Values labeled with different letters show significant differences as determined by Student's t-test (P<0.05, n = 3) on day 3. This experiment was repeated twice with similar results. (C) Altered defense gene mRNA levels in srfr1-4 at 22°C. Analysis of indicated transcripts in Col-0 and srfr1-4 by RT-PCR using 27 PCR cycles. ACTIN2 was used as an internal control. In srfr1-1 and srfr1-2 plants, resistance to DC3000(avrRps4) was enhanced, but remained unchanged to virulent DC3000, and plant growth was normal [15], [18]. Interestingly, the srfr1-4 mutants were resistant not only to avirulent DC3000(avrRps4), but also to virulent DC3000 and non-pathogenic DC3000 hrcC− (Figure 2B). The srfr1-4 line showed approximately 50-fold lower DC3000 and DC3000(avrRps4) growth than wild type Col-0, whereas the growth of DC3000 hrcC− in srfr1-4 was about 10-fold less than in Col-0, suggesting that mutations in SRFR1 in Col-0 increased basal defenses at 24°C that were additive to AvrRps4-triggered immunity (Figure 2B). Complemented srfr1-4 lines did not show either enhanced resistance phenotype (Figure 2B). We could not test bacterial growth at 22°C because srfr1-4 plants were severely stunted at this temperature. However, consistent with an upregulation of salicylic acid (SA)-based defenses, PR1 and PR2 mRNA levels were upregulated and PDF1.2 levels down-regulated in srfr1-4 at 22°C (Figure 2C).
Characterization of the srfr1-4 phenotype and mapping therefore strongly suggested that the Col-0 modifier is SNC1. To test this directly, we crossed srfr1-4 to snc1-11, a T-DNA insertion allele in the first exon of SNC1 [27]. In the F2 population, the number of stunted plants was consistent with the segregation of two recessive loci (srfr1-4 and wild-type SNC1) (Table 4). All of the stunted plants were homozygous for the srfr1-4 allele and the wild-type SNC1 allele. In contrast, all plants of normal stature that were homozygous for the srfr1-4 T-DNA allele possessed at least one copy of the snc1-11 T-DNA allele (Table 4). Therefore, the stunted phenotype of srfr1-4 plants requires two copies of SNC1 in Col-0, analogous to the phenotype of bon1-1 plants [27]. We quantified the effect of mutations in SRFR1 on plant growth by measuring the shoot weight of srfr1 mutants in Col-0 and RLD (Figure 3). Shoot weights were close to normal in the original srfr1-1 and srfr1-2 plants compared to wild-type RLD. Mutations in srfr1 caused severe reductions in shoot weight in the Col-0 background that were completely reversed by introgressing snc1-11. Interestingly, the shoot weight of srfr1 SNC1 plants was more strongly reduced than in bon1-1 plants (Figure 3), indicating that perhaps SRFR1 functions downstream of additional R genes apart from regulating SNC1. Together with the negative regulation in AvrRps4 - and HopA1-triggered immunity, these results show that SRFR1 is a negative regulator of plant immune responses of broader specificity than originally described.
Fig. 3. Stunting of srfr1-4 as measured by shoot weight is reversed by snc1-11. 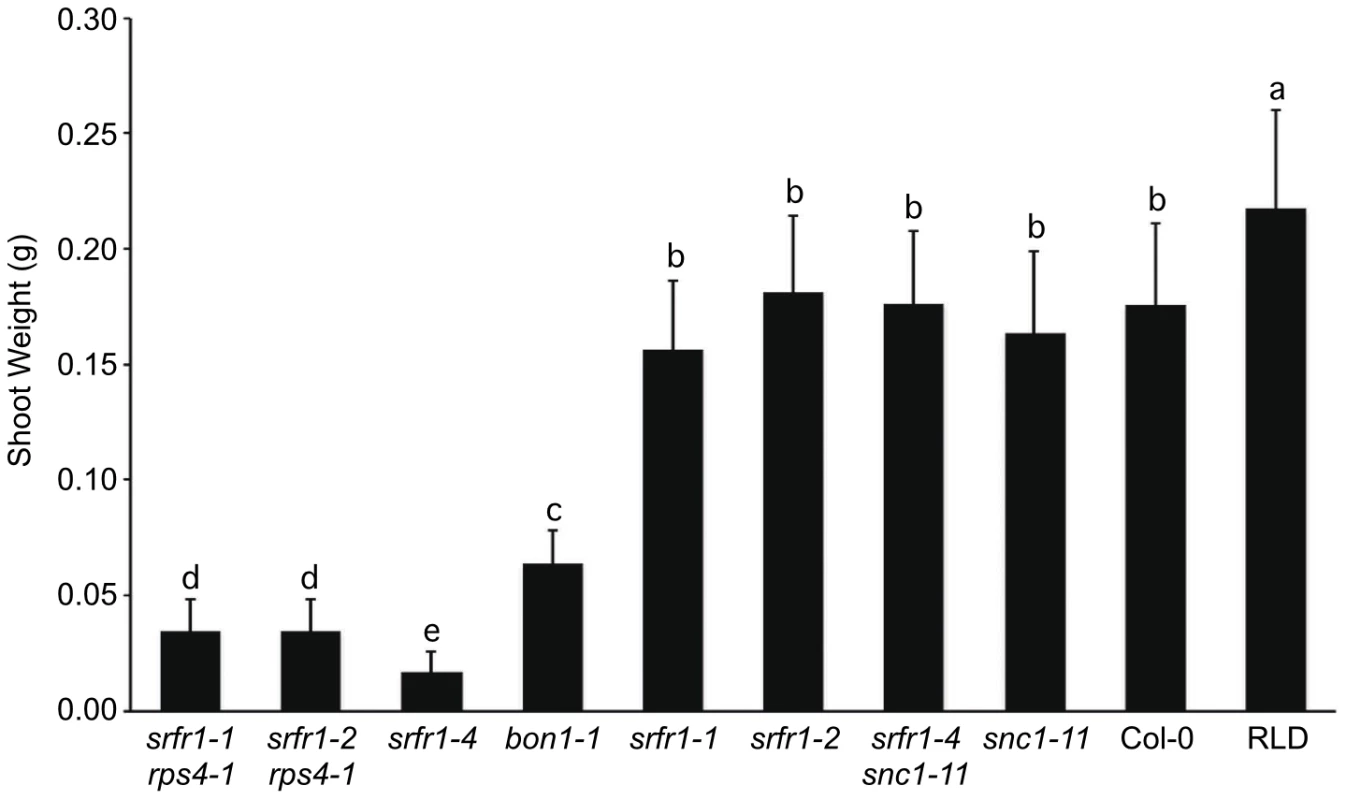
The indicated plant lines were grown for three weeks in a greenhouse at 22°C with a 16 h light/8 h dark cycle. Values represent average shoot weights of 40 to 70 plants for each line, and error bars denote standard deviation. Values labeled with different letters show significant differences as determined by Student's t-test (P<0.05). This experiment was repeated once with similar results. Tab. 4. Two copies of SNC1 are required for stunting in F2 plants from the cross snc1-11×srfr1-4. 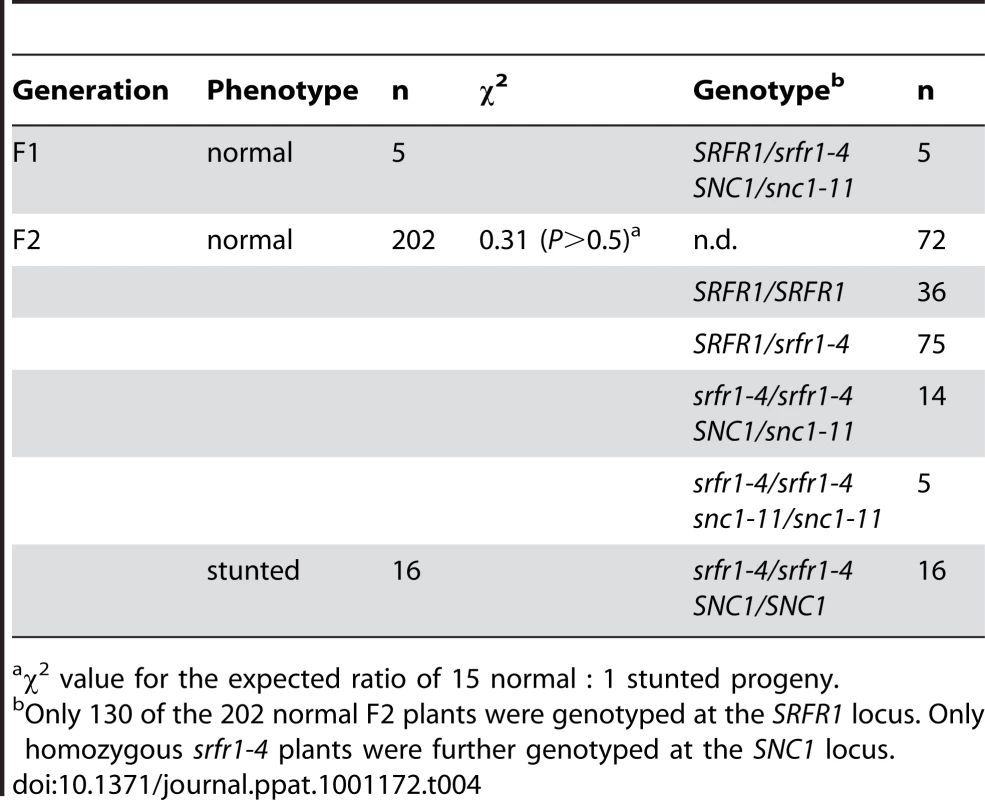
χ2 value for the expected ratio of 15 normal : 1 stunted progeny. The LRR domain is deleted in SNC1-RLD
Previous studies had suggested that the readily autoactivatable SNC1 is limited to the Col-0 accession, but these studies had not included RLD [27]. We therefore sequenced the likely RLD ortholog of SNC1 in RLD to determine the molecular basis for the very different phenotypes of Col-0 and RLD srfr1 mutants. At the 5′-end, SNC1-specific primers consistently amplified a sequence with high overall similarity to SNC1-Col (Figure 4A and 4B). SNC1-specific primers designed to amplify the complete SNC1 gene or the 3′-half of SNC1 failed to result in a unique RLD product. This reflected the very duplicated nature of the 3′-half of SNC1 in Col-0. Whole sections of the gene are not only duplicated within SNC1 with 100% sequence identity, but are also found in linked family members [28]. We were not able to experimentally determine unequivocally which genomic PCR product from the 3′-end was physically linked to the 5′-end of SNC1-RLD.
Fig. 4. SNC1-RLD encodes a truncated TNL protein. 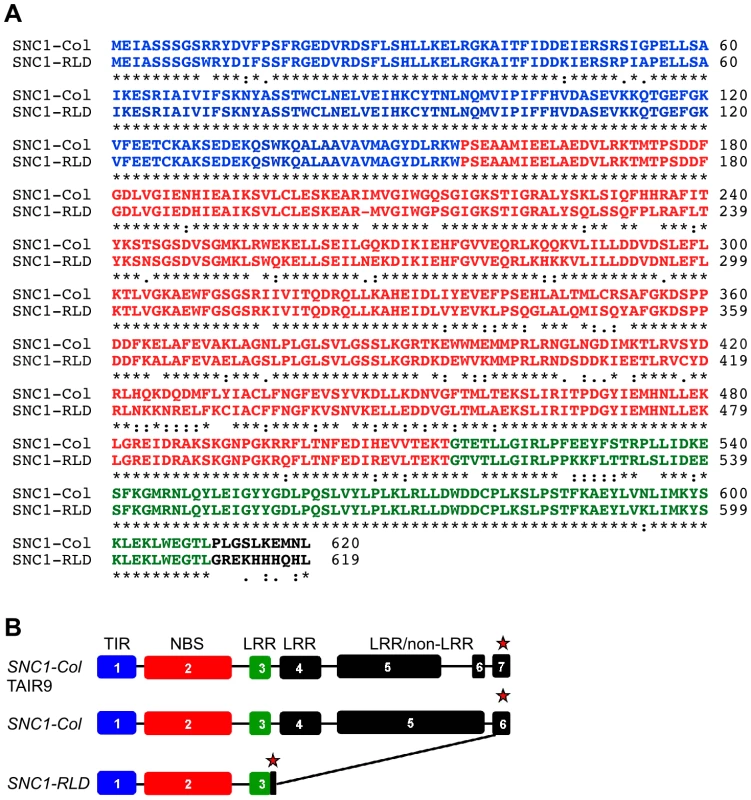
(A) Alignment of deduced amino acid sequences of SNC1-Col (top) and SNC1-RLD (bottom) using the EBI-ClustalW tool (http://www.ebi.ac.uk/Tools/clustalw/) [54]. Identical amino acids are indicated by asterisks. Colons and semi-colons show conserved substitutions and semi-conserved substitutions, respectively. Characters in blue, red and green show the amino acids corresponding to exon 1, exon 2 and exon 3, respectively. (B) SNC1 gene model as experimentally verified by reverse transcription PCR and 3′-RACE from Col-0 (middle) and RLD (bottom) compared with the TAIR9 gene model (top). Exons are indicated by boxes, introns by lines, and stop codons by red asterisks. We therefore determined the SNC1 mRNA sequence from RLD using a combination of 3′-Rapid Amplification of cDNA Ends (3′-RACE) and RT-PCR. As shown in Figure 4A, the open reading frame of SNC1-RLD predicted a protein of 619 amino acids, including a TIR and NBS domain but only a partial LRR domain. The predicted amino acid sequence identity between SNC1-Col and SNC1-RLD within the first three exons was 87%. However, our SNC1-RLD cDNA sequence was missing the fourth and fifth exons, leading to an in-frame stop codon at position 620 (Figure 4B). Interestingly, in the SNC1-RLD cDNA the very 3′-end of the open-reading frame and the 3′-untranslated region showed high nucleotide sequence identity with the corresponding region in SNC1-Col. Because we only obtained cDNA sequence of SNC1-RLD at the 3′-end, we could not determine whether the 3′-end of the SNC1-RLD coding sequence is interrupted by introns. We also obtained RT-PCR products from Col-0. These indicated that in contrast to the annotation of SNC1 in TAIR, we did not find evidence for the splicing of intron 5, which does not contain in-frame stop codons (Figure 4B). This alternative SNC1 transcript encoded a SNC1 protein of 1404 amino acids rather than the annotated 1301 amino acids. Taken together, sequencing of the RLD SNC1 ortholog provided evidence for polymorphisms at the 5′-end and major alterations in the 3′-half of the gene compared to Col-0, consistent with the fact that RLD does not have a SNC1 ortholog that triggers stunted growth in the absence of SRFR1.
Morphologically normal srfr1-4 snc-11 double mutants possess primed defenses
Activation of SNC1, either by intragenic autoactivating mutations [23] or by mutations in negative regulators of SNC1 such as BON1 [27], leads to constitutively enhanced resistance. Consistent with this and the constitutive expression of PR genes in srfr1-4 (Figure 2C), we observed with in planta bacterial growth assays increased resistance of srfr1-4 to DC3000(avrRps4) and to virulent DC3000 (Figure 2B). The latter shows that srfr1-4 plants possess elevated basal resistance that is independent of particular avirulence genes. To test if enhanced basal resistance in srfr1-4, like stunted growth, is fully dependent on SNC1, we performed in planta bacterial growth assays at varying temperatures. As noted before, we were not able to infiltrate srfr1-4 plants at 22°C because of the severe growth phenotype.
At both 22°C and 24°C, the growth of DC3000 and DC3000(avrRps4) was reduced in srfr1-4 snc1-11 compared to growth in wild type Col-0, even though the growth of DC3000(avrRps4) in srfr1-4 snc1-11 was slightly higher than that in srfr1-4 at 24°C (Figure 5A and 5B). This remnant enhanced basal resistance in srfr1-4 snc1-11 plants may be related to the induced defense gene mRNA levels observed in RLD srfr1-1 and srfr1-2 plants, although the latter plants do not show enhanced basal resistance [18], [29]. These results demonstrate that although the stunted phenotype of srfr1-4 at 22°C and 24°C is fully mediated by SNC1, enhanced basal resistance at these temperatures in srfr1-4 is not entirely mediated by SNC1. At 28°C, both basal and AvrRps4-triggered resistance were abolished in srfr1-4 and srfr1-4 snc1-11 plants (Figure S2A). In addition, AvrRps4-triggered resistance was also abolished in wild-type Col-0, confirming previous results [30], and in snc1-11 plants (Figure S2A). Consistent with normal growth and absence of resistance at 28°C, SNC1 and PR1 expression were not elevated in srfr1-4 or srfr1-4 snc1-11 plants (Figure S2B).
Fig. 5. Phenotypically normal srfr1-4 snc1-11 double mutants show enhanced basal defense and AvrRps4-triggered immunity. 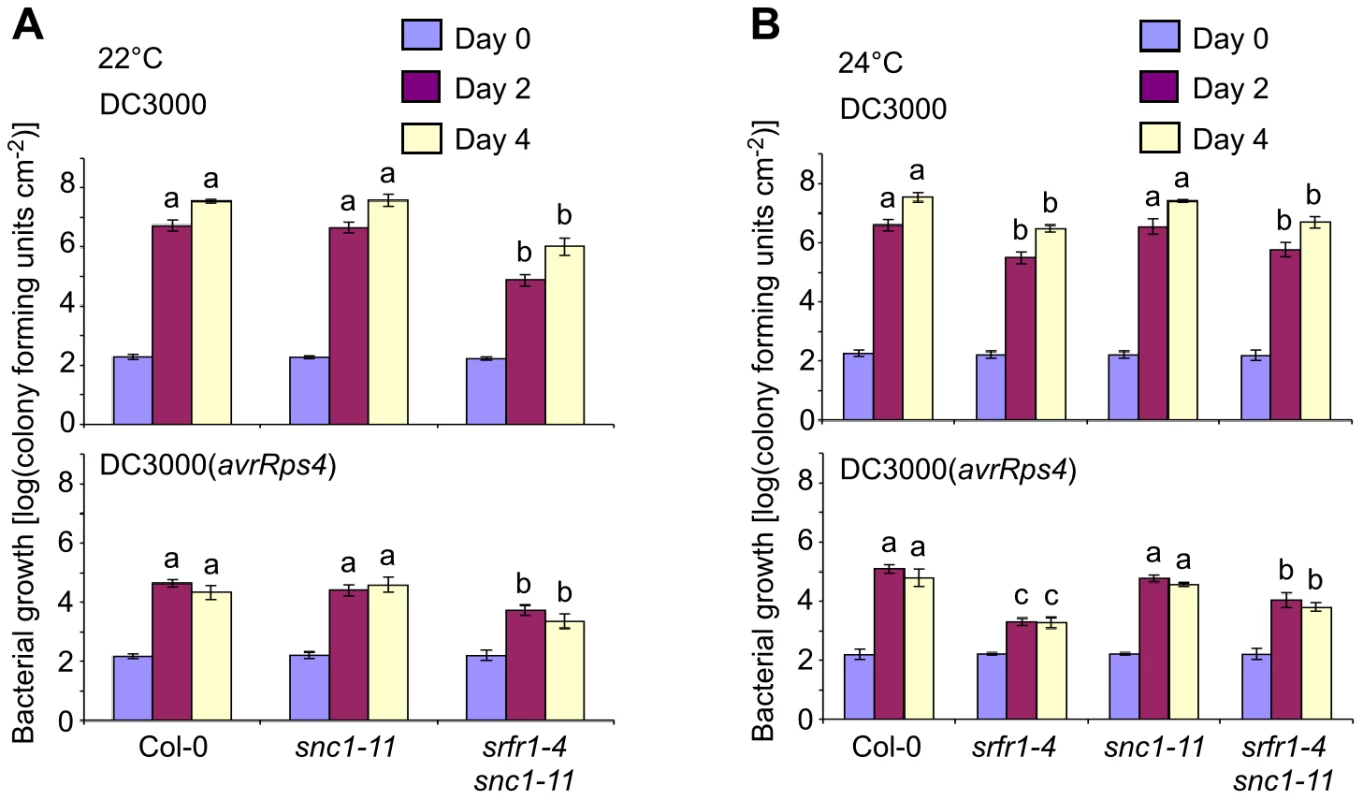
In planta bacterial growth was measured in the indicated plant lines on day 0 (blue bars), day 2 (purple) and day 4 (yellow) after inoculation with DC3000 (top) and DC3000(avrRps4) (bottom) at 5×104 cfu/ml at 22°C (A) and 24°C (B). Values represent averages of cfu/cm2 leaf tissue from triplicate samples, and error bars denote standard deviation. Values labeled with different letters show significant differences at the indicated days as determined by Student's t-test (P<0.05, n = 3). This experiment was repeated twice with similar results. Altered expression levels of defense-related genes in srfr1-4 and srfr1-4 snc1-11
Previously, we showed that several defense-related genes were up-regulated in RLD srfr1 mutants, supporting our hypothesis that SRFR1 may function as a repressor in plant innate immunity by negatively regulating defense gene expression levels [29]. The growth and constitutive defense phenotypes of srfr1-4 at 22°C and 24°C prompted us to quantify defense-related gene mRNA levels in srfr1-4 at these temperatures using quantitative reverse transcription real-time PCR (qPCR), and to determine whether all changes in expression in srfr1-4 can be attributed to SNC1. As expected, SNC1 transcript levels were higher in srfr1-4 than in Col-0 at 22°C and 24°C, as were those of RPP4 and At4g16950 (Figure 6A), two TNL genes in the SNC1 cluster that are co-regulated with SNC1 [31]. Interestingly, RPP4 and At4g16950 expression levels were higher also in the srfr1-4 snc1-11 double mutant (Figure 6A), showing that higher mRNA levels of these genes is not an indirect effect of SNC1 activation. Similarly, we observed increased mRNA levels of the CNL R gene RPS2, and to a lesser extent of RPM1, in srfr1-4 and srfr1-4 snc1-11 plants at both 22°C (Figure S3A) and 24°C (Figure S3B), indicating that upregulation of R genes by mutations in SRFR1 is not limited to TNL genes in Col-0. In contrast to SNC1-RLD, upregulation of RPM1 and RPS2 was not observed in the RLD mutant srfr1-1 (Figure S3C), possibly reflecting the presence of additional accession-specific SNC1-like genes in Col-0 [32] that may lead to enhanced expression of CNL genes.
Fig. 6. Transcript levels of defense-related genes are altered in srfr1-4 and srfr1-4 snc1-11. 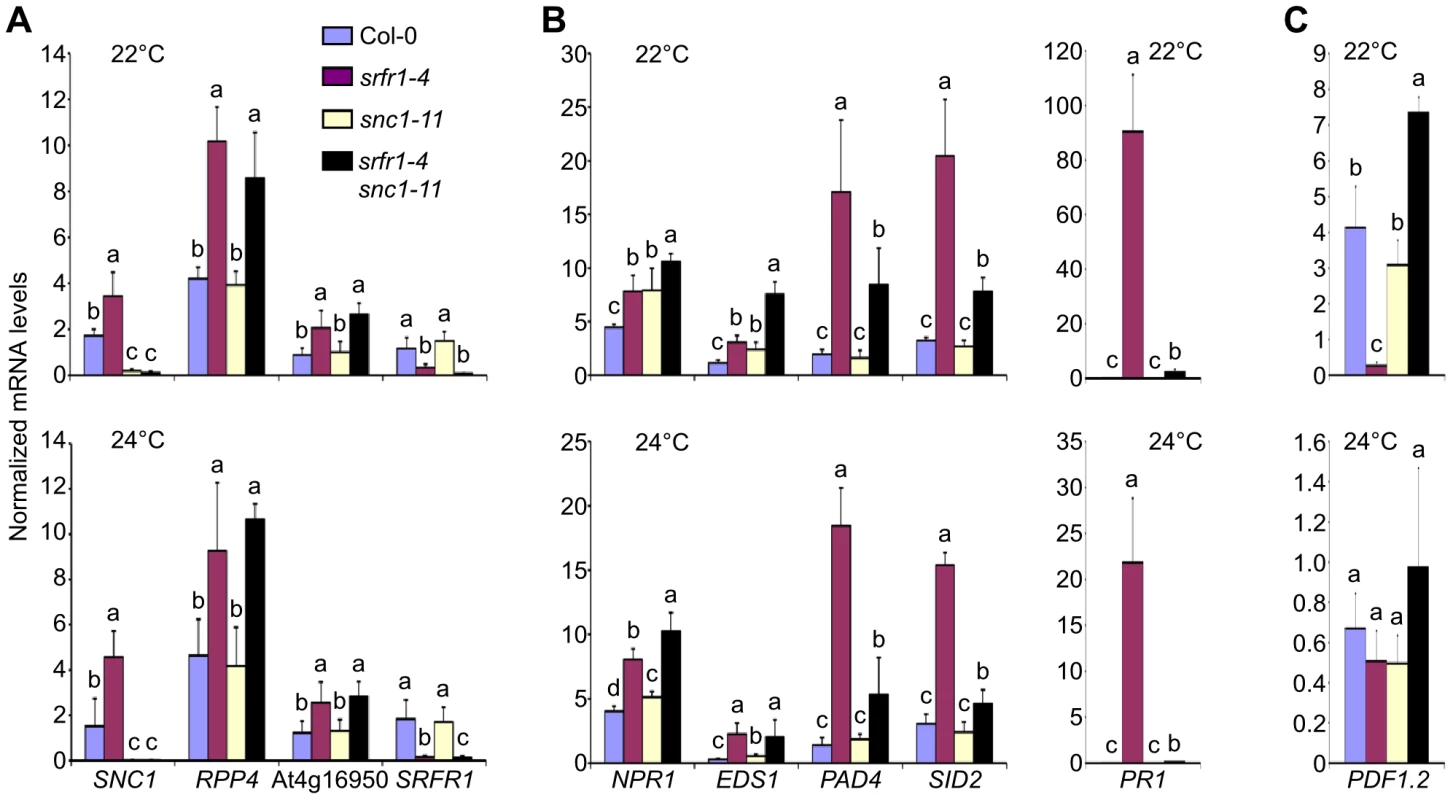
(A) Transcript levels of the co-regulated R genes SNC1, RPP4 and At4g16950. (B) Transcript levels of NPR1, EDS1, PAD4 and SID2 (left) and PR1 (right). (C) Transcript levels of PDF1.2. Transcript levels were measured by qPCR in Col-0 (blue bars), srfr1-4 (purple), snc1-11 (yellow) and srfr1-4 snc1-11 (black) at 22°C (top) and 24°C (bottom), and were normalized using SAND gene (At2g28390) mRNA levels as an internal control. Values represent averages from four biological replicates, and error bars denote standard deviation. Different letters denote significant differences between values calculated by Student's t-test (P<0.05, n = 4). This experiment was repeated once with similar results. SA-dependent defense related gene mRNA levels were also higher in srfr1-4 than in wild-type at 22°C and 24°C (Figure 6B). Unlike for TNL and CNL genes, these expression levels were reduced in srfr1-4 snc1-11 compared to srfr1-4 to varying degrees, although they were still higher than in wild-type (Figure 6B). Interestingly, NPR1 and EDS1 mRNA levels in the double srfr1-4 snc1-11 mutant showed additive increases compared to the wild-type and single mutants at 22°C (Figure 6B). In contrast, mRNA levels of PDF1.2, a defensin gene whose expression is under negative regulation by the JA-responsive transcription factor JIN1 [33], was strongly repressed at 22°C in srfr1-4 but induced in srfr1-4 snc1-11 plants compared to wild-type. PDF1.2 expression levels were not significantly different among the genotypes at 24°C (Figure 6C). These results point towards complex modular control of defense gene expression that is influenced by a combination of SRFR1, SNC1 and temperature to varying proportions.
Cross-talk between RPS4 and SNC1 in AvrRps4-triggered immunity
The Arabidopsis accession RLD carries a natural mutation in RPS4 and is fully susceptible to DC3000(avrRps4) [34], [35]. In addition, introduction of RPS4 from Col-0 or Ler into RLD is sufficient to provide full resistance to DC3000(avrRps4) when compared to Col-0 and Ler [16], [35]. We also observed susceptibility of rps4-1, an RPS4 T-DNA allele in the Col-0 background, under our conditions that were used to map SRFR1 [15]. However, it was reported that rps4-2, a second RPS4 T-DNA allele in the Col-0 background, was only slightly more susceptible to DC3000(avrRps4) [36]. Based on the accession-specific presence of SNC1 in Col-0, the temperature-dependent srfr1-4 phenotype and the fact that SRFR1 was identified in a screen for enhanced DC3000(avrRps4) resistance in RLD, we speculated that the rps4-2 phenotype might be temperature-dependent. Indeed, when directly comparing plants grown in identical growth chambers at 22°C or 24°C, we observed a strong temperature dependence: rps4-2 plants were as resistant to DC3000(avrRps4) as Col-0 at 22°C, while at 24°C they were as susceptible as Col-0 treated with virulent DC3000 and as susceptible as RLD treated with either strain (Figure 7).
Fig. 7. RPS4 and SNC1 contribute redundantly to AvrRps4-triggered resistance. 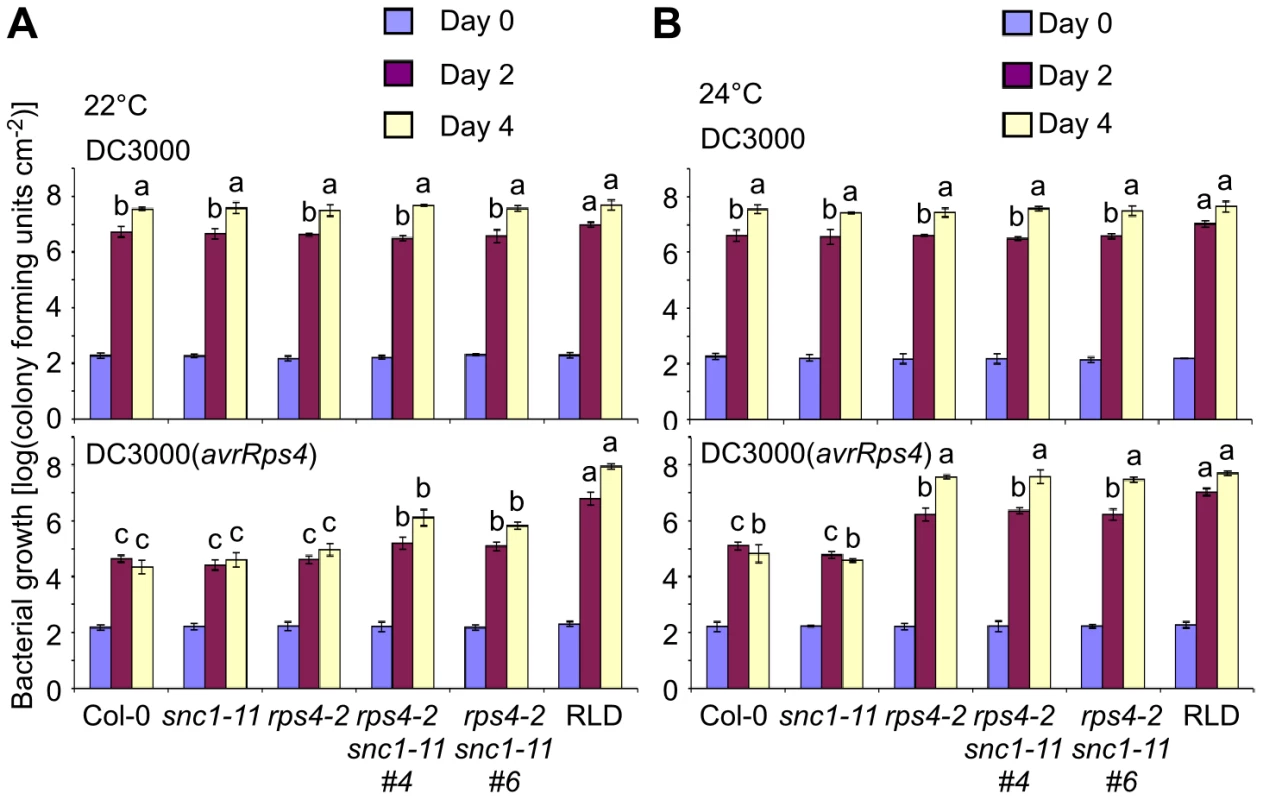
In planta bacterial growth was measured in Col-0, snc1-11, rps4-2, two independent snc1-11 rps4-2 lines and RLD on day 0 (blue bars), day 2 (purple) and day 4 (yellow) after inoculation with DC3000 (top) and DC3000(avrRps4) (bottom) at 5×104 cfu/ml at 22°C (A) and 24°C (B). Values represent averages of cfu/cm2 leaf tissue from triplicate samples, and error bars denote standard deviation. Values labeled with different letters show significant differences at the indicated days as determined by Student's t-test (P<0.05, n = 3). This experiment was repeated three times with similar results. Given the effect of temperature, we next tested whether SNC1 interferes with the susceptible phenotype at 22°C. Interestingly, rps4-2 snc1-11 double mutants displayed approximately 30-fold increased bacterial growth of DC3000(avrRps4) compared to Col-0 or rps4-2 at 22°C (Figure 7A), suggesting that SNC1 in the absence of RPS4 contributes to AvrRps4-triggered immunity at 22°C in Col-0. However, susceptibility of rps4-2 snc1-11 to DC3000(avrRps4) was not complete compared to Col-0 treated with virulent DC3000 or to RLD treated with either strain, indicating that additional factors interfere with rps4-caused susceptibility (Figure 7A). No significant difference of DC3000(avrRps4) growth in rps4-2 and rps4-2 snc1-11 was observed at 24°C, reflecting full susceptibility of rps4-2 to DC3000(avrRps4) at this temperature (Figure 7B). Recently, RRS1 was shown to be involved in DC3000(avrRps4)-mediated resistance [37], [38]. However, we observed no temperature-dependent resistance to DC3000(avrRps4) in the Ws-0 mutants rps4-21 and rrs1-1 (Figure S4). As was observed before, mutations in either RPS4 or RRS1 had equal effects on DC3000(avrRps4) susceptibility, which was qualitatively different from the redundancy between SNC1 and RPS4 (Figure 7). Interestingly, as reported before [38], we reproducibly observed approximately 10-fold higher growth of DC3000 compared to DC3000(avrRps4) in the single rps4-21 and rrs1-1 mutants and the double mutant, indicating that additional layers of resistance exist.
SNC1 and RPS4 interact with SRFR1
The redundancy between RPS4 and SNC1 suggests that they function in parallel to provide resistance to DC3000(avrRps4) at 22°C. We speculated that this cross-talk between two R proteins might occur if both interact with proteins in a common complex. Perturbation of this complex by an effector could trigger one or the other R protein, and both need to be absent to observe susceptibility. Based on the results presented here, we reasoned that SRFR1 might be a common interaction partner of RPS4 and SNC1. In the past, transient expression of SRFR1 in Nicotiana benthamiana led to variable protein expression levels and required a silencing inhibitor for detectable expression [18]. We therefore generated stable transgenic N. benthamiana plants expressing HA-SRFR1 encoded by a genomic clone driven by the native Arabidopsis SRFR1 promoter. We first determined the functionality of this genomic HA-SRFR1 construct in Arabidopsis by testing for complementation of the stunted srfr1-4 phenotype. Transgenic plants expressing HA-SRFR1 in the srfr1-4 background showed normal growth and development (Figure S5A). Immunoblot analysis detected the expression of the transgene product in these transgenic plants (Figure S5A). HA-SRFR1 in these plants localized to microsomal and nuclear fractions (Figure S5B). This localization was consistent with the nuclear and punctate cytoplasmic localization of GFP-SRFR1 transiently expressed in N. benthamiana [18].
We observed improved and reproducible HA-SRFR1 expression in the stable transgenic N. benthamiana lines. As in Arabidopsis, HA-SRFR1 localized to the microsomal and nuclear fractions in N. benthamiana (Figure 8A). A previous study showed that RPS4 was predominantly localized to microsomes [36]. Immunoblot assays of Myc-SNC1 transiently expressed in N. benthamiana suggested that SNC1 was mainly a soluble cytoplasmic protein, although a sizeable portion accumulated in the microsomal fractions (Figure 8B). We also detected some SNC1 in the nuclear fraction (Figure 8B). We tested for SRFR1 interaction with SNC1 and RPS4 by transiently expressing Myc-SNC1, Myc-RPS4 or Myc-eGFP as a negative control in transgenic HA-SRFR1 N. benthamiana plants. Co-immunoprecipitation analysis on protein isolated 48 h after infiltration of Agrobacterium tumefaciens strains showed that SRFR1 interacted with both SNC1 and RPS4 in the microsomal fraction (Figure 9). No significant interaction between SRFR1 and SNC1 was detected in the soluble fraction, even though SNC1 was detected in this fraction. No interaction with eGFP was detected in either fraction (Figure 9). As an additional control, we probed SRFR1 co-immunoprecipitated samples for the presence of GAPDH and V-ATPase. Neither protein was co-immunoprecipitated with SRFR1 (Figure S6A and S6B), indicating that the interactions of SRFR1 with SNC1 and RPS4 are specific.
Fig. 8. Localization of SRFR1 and SNC1 expressed in N. benthamiana. 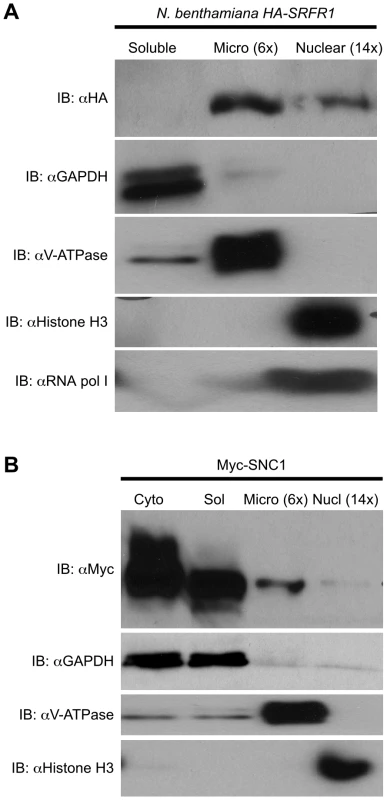
(A) HA-SRFR1 stably expressed in transgenic N. benthamiana localizes to the cytoplasmic microsomal and nuclear fractions. (B) Transiently expressed Myc-SNC1 localizes predominantly to the soluble cytoplasmic fraction, with detectable amounts in the microsomes and nuclei. In (A) and (B), the microsomal and nuclear fractions are 6 and 14 times concentrated, respectively, compared to the soluble fraction. The degree of fraction enrichment was determined using antibodies against marker proteins (anti-histone H3 and anti-RNA polymerase I subunit, nucleus; anti-GAPDH, cytoplasmic soluble; and anti-V-ATPase, microsomes). Each assay was repeated at least three times with similar results. Fig. 9. SRFR1 interacts with RPS4 and SNC1 in the microsomal fraction. 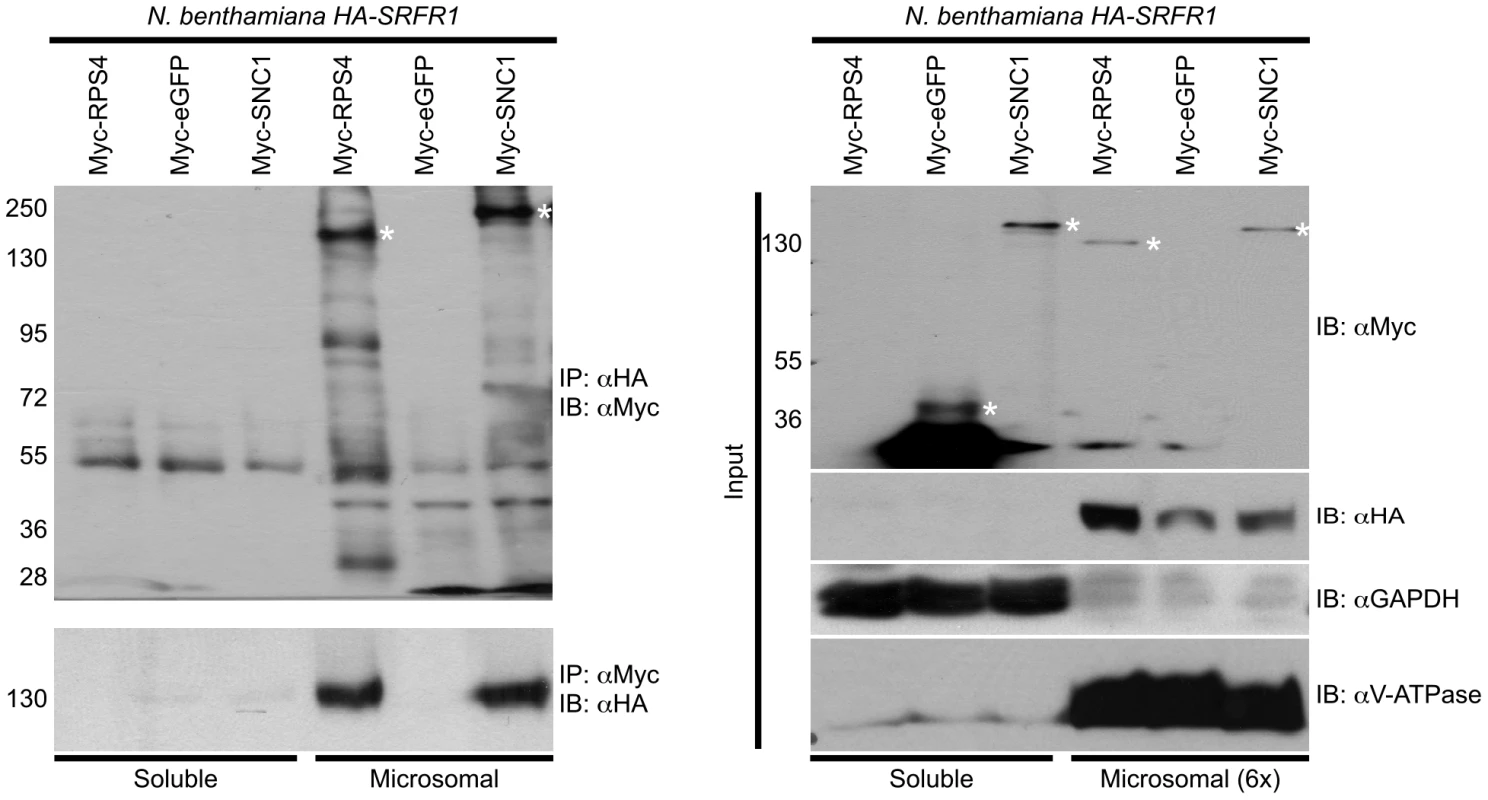
Myc-RPS4, Myc-eGFP or Myc-SNC1 were transiently expressed via Agrobacterium-mediated transient expression in N. benthamiana transgenic lines 6-4 and/or 7-1 expressing HA-SRFR1. Immunoprecipitation (IP) analysis was performed on soluble and microsomal fractions with the indicated antibodies. The immunoprecipitates were immunoblotted (IB) with the indicated antibodies (left panel). The right panel shows the corresponding protein expression levels in the input fractions for immunoprecipitation analyses. In the right panel, the microsomal fraction is 6-fold enriched compared to the soluble fraction. Molecular weight of protein standards (in kD) are shown on the left of the panels. Asterisks denote the expected sizes of the Myc-tagged proteins. The degree of soluble and microsomal fraction enrichment are shown by IB analyses with anti-GAPDH and anti-V-ATPase antibodies. The assay was repeated three times with similar results. Discussion
SRFR1 encodes a novel tetratricopeptide repeat (TPR)-containing protein that is conserved between plants and other eukaryotes [18]. Based on limited sequence similarity with the Saccharomyces cerevisiae Ssn6 and the animal OGT proteins, SRFR1 was originally proposed to function as a transcriptional repressor, perhaps with defense genes as its target. Consistent with this hypothesis, resting mRNA levels of several defense genes were slightly higher in srfr1-1 and srfr1-2 than in wild-type RLD [29]. This general priming of the defense system made it unlikely that SRFR1 function is limited to defenses triggered by AvrRps4. However, no alteration of resistance to DC3000 strains that trigger resistance via the R genes RPM1 or RPS2 were detected. The original analysis did not include a second TNL gene, which may be significant since TNL genes like RPS4 signal through EDS1, while coiled-coil (CC)-NBS-LRR genes like RPS2 and RPM1 require NDR1 [17]. With the cloning of RPS6, a second Arabidopsis TNL bacterial R gene that mediates resistance via EDS1 to the P. syringae pv. syringae effector HopA1 [12], [39], we found that rps6-1 srfr1-1 double mutants were more resistant to DC3000(hopA1) than the rps6-1 single mutant [39].
Mutations in SRFR1 activate SNC1
Here we extend our analysis to the Col-0 specific TNL R-like gene SNC1 and show that mutations in SRFR1 activate SNC1. SNC1 was originally identified based on an autoactivated allele that led to constitutive expression of PR1 [23]. Subsequently, it was shown that perturbation of wild-type SNC1 expression readily leads to autoactivation [24], [27], [40]. Our finding that SNC1 is activated in srfr1 mutants is reminiscent of the bon1/cpn1 phenotype [25]–[27]. How the absence of BON1 leads to SNC1 activation is not known. In particular, it is not known if sub-pools of BON1 and SNC1 reside in the same protein complex.
Together, our data show that mutations in SRFR1 impact three resistance specificities, namely AvrRps4-, HopA1 - and SNC1-triggered immunity. The impact of srfr1 mutations on SNC1 is novel, given that previously we observed effects of SRFR1 mutations only in the absence of the R genes RPS4 or RPS6. SNC1 is therefore the first TNL gene for which a genetically direct negative regulation by SRFR1 could be shown. Whether this is also mechanistically direct remains to be determined. Consistent with the proposed function of SRFR1 as a transcriptional repressor, we found increased mRNA levels for SNC1, RPP4 and At4g16950 in srfr1-4 plants. This altered expression level was not an indirect effect of SNC1 activation, since RPP4 and At4g18950 were also upregulated in the srfr1-4 snc1-11 double mutant. Because these members of the SNC1 locus were previously shown to be co-regulated with SNC1 [31] and because changes in SNC1 expression levels have been shown to cause autoactivation of SNC1 [24], we propose that mutations in SRFR1 lead to misregulated expression of SNC1, which in turn activates constitutive expression of an enhanced defense phenotype.
Cross-talk between RPS4 - and SNC1-mediated resistance and interactions with SRFR1
The genetic connection of SNC1 and RPS4 via SRFR1 was measurable as cross-talk between these resistance pathways in disease assays under specific environmental conditions. Because it had been convincingly shown that the Col-0 rps4-2 mutant was not fully susceptible to DC3000(avrRps4) [36], while we observed complete susceptibility, we tested whether environmental conditions had an influence on the Col-0 phenotypic response to DC3000(avrRps4). Surprisingly, we found that a mere 2°C difference in temperature changed the phenotype of rps4-2 from almost completely resistant to DC3000(avrRps4) to fully susceptible. Other environmental factors that are likely to impact this response are humidity [26], with drier conditions favoring resistance, and light intensity. Because cis or second-site mutants with activated SNC1 have a well-described conditional phenotype influenced by temperature and humidity, we tested whether the partial phenotype of rps4-2 is influenced by SNC1.
Indeed, we were able to measure a synergistic effect of mutations in RPS4 and SNC1 on susceptibility to DC3000(avrRps4) at 22°C. In addition, in the accessions RLD and Ws-0 that do not have SNC1, mutations in RPS4 result in susceptibility to DC3000(avrRps4) that is not influenced by changes in temperature in the range investigated here. SNC1 was originally identified in a screen for mutants with constitutively activated defenses, and to date no cognate avirulence gene has been identified. Nevertheless, some suppressor mutants of the constitutive snc1-1 phenotype such as mos7 also impact effector-triggered immunity [41]. Our finding that SNC1 contributes to AvrRps4-triggered immunity further indicates that SNC1 can be considered a bona fide R gene.
Conceptually, cross-talk between resistance pathways can occur if an effector protein has more than one target, or if R proteins guard a common target. The former seems to be the case for RPM1 and TAO1, which additively contribute to full resistance to DC3000(avrB) [42]. In contrast, AvrRpm1 induced measurable defenses in rpm1 plants that were dependent on RPS2, presumably because both RPM1 and RPS2 guard RIN4, a protein that is the target for both AvrRpm1 and AvrRpt2 [19]. As a first step to distinguish between these models, we tested whether SNC1 and RPS4 co-localize with a common protein. Given the regulatory function of SRFR1 on SNC1 and on AvrRps4-triggered resistance, we speculated that SRFR1 might be such a common protein. Interestingly, the microsomal pool of SRFR1 was found to be in a complex with SNC1.
Transiently expressed GFP-SRFR1 in N. benthamiana localized to the nucleus and cytoplasm [18]. The cytoplasmic localization was punctate. Here, further analysis of the cytoplasmic pool showed that most SRFR1 localized to the microsomal cytoplasmic fraction, and very little was soluble. Because the majority of SNC1 was in the soluble cytoplasmic pool, it was not possible to determine whether the microsomal pool of SNC1 diminishes in the absence of SRFR1. In addition, the native N. benthamiana pool of SRFR1 may be sufficient to localize some proportion of SNC1 to microsomes. Most likely, SNC1 is in a higher-order complex with SRFR1 in a microsomal compartment of unknown identity. Interestingly, we found that RPS4 also interacted with SRFR1 in the same cell fraction. This suggests that perhaps additional R proteins localize to a common complex. The localization of SRFR1 and interactions with RPS4 and SNC1 are reminiscent of CRT1 [43]. However, the functions of CRT1 and SRFR1 likely differ, because mutations in CRT1 compromise, not enhance, effector-triggered immunity.
Because mutations in SRFR1 lead to increased, not decreased resistance, we do not propose that SRFR1 is analogous to RIN4 as the guardee of RPS4 or SNC1, since deletion of a guardee should prevent recognition of the specific effector that targets the guardee. The function of guardee for SNC1 may be fulfilled by BON1 [44], although BON1 is localized to the plasma membrane [25] and to our knowledge it has not been determined whether BON1 interacts with SNC1. Also, because no cognate effector is known for SNC1 and because deletion of BON1 leads to autoactivation of SNC1, it is difficult to quantify the effects of BON1 mutations on disease resistance and susceptibility. Interestingly, we consistently observed a more severe growth phenotype of srfr1-4 plants compared to bon1-1 plants, yet the srfr1-4 growth phenotype is completely reversed by snc1-11. Apart from negatively regulating the activation of SNC1, SRFR1 most likely regulates additional R proteins. Because of positive feed-back, all these pathways may be turned on once SNC1 is activated. While in bon1-1 plants SRFR1 is still present to downregulate these other R proteins, this is not the case in srfr1-4 plants. Therefore, this observation is suggestive of a broad and central function of SRFR1 in downregulating R protein output.
It is currently unknown where in the cell the recognition of AvrRps4 by RPS4 occurs. Several plant R proteins, including RPS4, have been shown to function in the nucleus to trigger immunity [36], [45]. Because the cytoplasmic pool of these R proteins predominates over the nuclear pool, it is difficult to establish whether R proteins translocate to the nucleus upon effector perception, or continuously cycle between the cytoplasmic and nuclear compartment. We also detected a low amount of SNC1 in the nucleus, whereas the autoactivated mutant snc1-1 protein appears to accumulate to higher levels in the nucleus [41]. It was also found that snc1-1 needed to be in the nucleus to cause a stunted phenotype [41], and that temperature modulated the localization of snc1-1 [46]. Interestingly, a balanced partitioning of EDS1 between the cytoplasm and nucleus was recently shown to be required for full EDS1-mediated resistance [47], indicating that immune regulatory proteins may have coordinated cytoplasmic and nuclear functions during the immune response.
Here we found that SRFR1 interacts with RPS4 and SNC1 in the cytoplasm, and also that mutations in SRFR1 alter the expression of defense genes independent of a snc1 phenotype. Because of the low amount of RPS4 [36], SRFR1 and SNC1 protein in the nucleus, so far we have not been able to ascertain whether they also interact in the nucleus. However, our results seem to suggest that at resting state, the majority of SRFR1, RPS4 and SNC1 protein is extra-nuclear localized and forms a complex in the microsomal fraction. SRFR1 may therefore negatively regulate RPS4 and SNC1 translocation to the nucleus. We propose that a second point of regulation is in the nucleus, where SRFR1 may negatively regulate the transcriptional reprogramming upon pathogen perception. More detailed analyses before and during a defense response are required to substantiate these hypotheses.
Genetics of srfr1-mediated resistance revisited
The genetics of enhanced resistance in RLD srfr1 mutants were originally interpreted to signify that an additional specific R gene is required for resistance [15]. In the mapping crosses rps4-1×srfr1-1 and rps4-1×srfr1-2, resistant F2 plants were identified in the ratio 13 susceptible to 3 resistant, consistent with segregation of a recessive locus (srfr1) and a dominant locus that was proposed to be a second specific R gene with weak recognition of AvrRps4 [15]. In light of the results presented here, we needed to reinterpret these results. Retesting our mapping population provided evidence for severely stunted plants at the expected ratio of one in 16 stunted plants. These would be double recessives (srfr1 and wild-type SNC1) and would have been lost from our usual phenotypic analysis because of preferential retention of vigorously growing seedlings after planting for disease assays. Upon reinspection, the segregation of resistant plants in the two mapping populations was indeed statistically consistent with the segregation of a single recessive locus (srfr1) in a population where 1/16th of the population (genotype srfr1/srfr1 SNC1/SNC1) that would have been expected to be resistant was eliminated from consideration. In addition, in both mapping populations we had noticed an apparent suppression of recombination along chromosome 4 in retained plants [15], which is consistent with the fact that both SRFR1 and SNC1 are located on chromosome 4. At the same time, we show here that the original model for resistance in srfr1 mutants mediated by other R genes with weaker recognition of AvrRps4 is still valid because cross-talk between R genes exists in response to AvrRps4. However, we now consider it unlikely that one single additional R gene is responsible for resistance in srfr1 mutants.
In conclusion, our data contribute to evidence for extensive cross-talk between at least three TNL pathways that converge on SRFR1, indicating that SRFR1 perhaps has a central function in regulating the output of additional TNL proteins. The present data also allow us to propose more directly that SRFR1 negatively regulates R proteins or R gene expression. While models for SRFR1 so far have focused on a nuclear-localized transcriptional repressor function [18], the data here suggest that SRFR1 also has a function in the cytoplasm. Consistent with this, Li and co-workers recently showed that SRFR1 interacts with SGT1 in the cytoplasm [22]. Whether SRFR1 is merely an accessory protein in a cytoplasmic “resistasome” or has regulatory functions and migrates to the nucleus remains to be established. Nevertheless, our data highlight molecular architecture aspects of a subset of TNL-mediated resistance pathways that will allow further mechanistic insight into the function of TNL R proteins. The cross-talk evident from our results also means that any reports of constitutive resistance phenotypes in Col-0 need to consider the possible involvement of SNC1.
Materials and Methods
Plant lines
The srfr1-4 line (SAIL_412_E08) from the Syngenta Arabidopsis Insertion Library [21] was obtained from the Arabidopsis Biological Resource Center. The T-DNA insertion site in srfr1-4 in the second intron was determined by sequencing and was found to be upstream of the insertion site suggested by raw flanking sequence from the T-DNA Express website (http://signal.salk.edu/cgi-bin/tdnaexpress). rps4-2 (SALK_057697) was isolated from the Salk T-DNA knockout lines [20]. snc1-11 (SALK_047058) and bon1-1 were a kind gift from Jian Hua (Cornell University). Using snc1-11 as a recipient, srfr1-4 snc1-11 and rps4-2 snc1-11 double homozygous mutants were generated. The mutant lines rps4-21, rrs1-1 and rps4-2 rrs1-1 in the Ws-0 background were kindly provided by Yoshihiro Narusaka (Research Institute for Biological Sciences, Japan). The mapping populations generated by crossing srfr1-1 or srfr1-2 to rps4-1 (SAIL_519_B09) were described previously [15].
Complemented srfr1-4 transgenic lines were generated by transforming srfr1-4 with pSHK102, a genomic SRFR1 clone in vector pCAMBIA2300 [18], using the floral dip method [48]. Single locus transgenic lines homozygous for the transgenic copy of wild-type SRFR1 were selected by scoring for kanamycin resistance, the selectable marker of pCAMBIA2300 (the selectable marker for SAIL lines is BASTA). Among these homozygous lines, those with at least one copy of the srfr1-4 allele were selected by genotyping and propagated to the next generation. Lines homozygous for both the SRFR1 transgene and the srfr1-4 allele were identified as those where srfr1-4 did not segregate in the next generation. SNC1 was mapped by genotyping stunted plants in the F2 generation from the cross RLD×srfr1-4 using SSLP and CAPS markers [49], [50].
Plant growth and in planta bacterial growth curve assays
Unless otherwise noted, Arabidopsis plants used in this study were grown in E-7/2 reach-in growth chambers (Controlled Environments Ltd., Winnipeg, Manitoba, Canada) under an 8 h light/16 h dark cycle at 24°C and 22°C, with 70% relative humidity and a light intensity of 90–140 µmol photons m−2 s−1. Virulent Pseudomonas syringae pv. tomato strain DC3000 containing the empty vector (ev) pVSP61 or DC3000 expressing avrRps4 from plasmid pVSP61 was grown as described previously [16]. To generate DC3000 hrcC−(ev), pVSP61 was mobilized into the recipient DC3000 hrcC− mutant by triparental mating using the helper plasmid pRK2013. In planta bacterial growth assays were performed by syringe infiltration. Leaves of 4-week old plants were infiltrated with bacterial suspensions of 5×104 cfu/mL. Leaf discs with a total area of 0.5 cm2 per sample were ground in 10 mM MgCl2, and solutions were plated in serial dilutions on selective medium in triplicate at the indicated time points. Statistical comparison of bacterial growth was tested using a two-tailed Student's t-test.
Transcript profiling, reverse transcription PCR and RACE
Quantitative reverse transcription PCR was performed as described previously [18]. Briefly, total RNA was extracted from the indicated plant lines using TRIZOL (Invitrogen, Carlsbad, CA, USA). For RT-PCR experiments, cDNA was synthesized from 2 µg of total RNA using an oligo(dT)15 primer and Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI, USA) following the manufacturer's protocol. Quantitative real-time reverse transcription PCR (qPCR) was performed with SYBR GREEN PCR Master Mix and an ABI 7500 system (Applied Biosystems, Warrington, UK) according to the manufacturer's instructions. The levels of transcripts were normalized using SAND gene (At2g28390) mRNA levels as an internal standard. These experiments were performed at least twice with similar results. Semi-quantitative RT-PCR was performed from total RNA extracted from Col-0 and srfr1-4. Equivalent amounts of cDNA from both samples were used to detect PR1, PR2 and PDF1.2. ACTIN2 (At3g18780) was used as an internal control. Table S1 lists the oligonucleotide primer sequences used in qPCR and semi-quantitative RT-PCR.
To determine the SNC1 cDNA sequence from RLD and Col-0, the 3′-RACE procedure (Invitrogen, Carlsbad, CA, USA) and RT-PCR (see above) were performed as described previously [19]. PCR products were ligated into the pGEM-T Easy vector (Promega) for sequencing. See Table S1 for oligonucleotide primer sequences used in these experiments.
Molecular cloning and generation of transgenic N. benthamiana plants
All clones were verified by sequencing. To generate epitope-tagged SNC1 constructs, genomic SNC1 DNA including introns was amplified by PCR from Col-0 using SNC1 GATE primers listed in Table S1. In vitro BP Clonase recombination reactions were carried out to insert the PCR product into the pDONR201 entry vector according to the manufacturer's instructions (Invitrogen). LR reactions were performed to recombine the entry clones into GATEWAY-compatible destination vectors. Using BP and LR reactions, we constructed Myc-gSNC1 with six Myc tags under the control of the cauliflower mosaic virus 35S promoter. Similarly, Myc-gRPS4 was generated by amplifying the genomic fragment of RPS4 from the FLAG-gRPS4 construct [51] using the primers RPS4 FOR and RPS4 REV (Table S1).
To construct the binary vector expressing genomic HA-tagged SRFR1 from its native promoter (HA-gSRFR1), independent PCR reactions were performed with the primer combinations HA-SRFR1 FOR/gSRFR1 XbaI REV and pCAMBIA PmeI FOR/HA-SRFR1 REV using the template pSHK102 [18]. The PCR products were mixed and used for overlap PCR with the pCAMBIA Pme I FOR/gSRFR1 XbaI REV primers. The 2.2 kb PCR product was digested with PmeI and XbaI and used for replacing the PmeI-XbaI fragment of pSHK102. The resulting binary vector was electroporated into Agrobacterium tumefaciens strain C58C1. Transgenic N. benthamiana plants expressing HA-gSRFR1 from the Arabidopsis native promoter were generated by stable Agrobacterium-mediated transformation as previously described [52]. Transgenic plants were selected on media containing 100 µg/ml kanamycin.
Protein fractionation and immunoblot analysis
Microsomal and soluble fractions were prepared according to published procedures [53]. Briefly, plant materials were ground in buffer H (50 mM HEPES, pH 7.5, 250 mM sucrose, 15 mM EDTA, 5% glycerol, 0.5% polyvinylpyrrolidone) containing 3 mM DTT and 1×protease cocktail inhibitors (Sigma, St. Louis, MO). The extracts were filtered through two layers of miracloth pre-wetted with buffer H and centrifuged at 2000×g for 15 min at 4°C. The supernatant consisting of the cytoplasmic fraction was further subjected to ultracentrifugation at 100,000×g to separate the soluble and microsomal (pellet) fractions. The pellet was resuspended in buffer H. Nuclear extracts were prepared using the CelLytic™ PN Isolation/Extraction Kit (Sigma) following the manufacturer's instructions. Total protein concentrations of fractions were determined by Bradford assays with BSA as standard. Extracts were normalized to 1 µg/ml with buffer H. For co-immunoprecipitation assays, the nonionic detergent Igepal CA-630 (Sigma) was added to 0.2% and 1% final concentration to the soluble and microsomal fractions, respectively. The extracts were incubated overnight with 20 µl of anti-HA or anti-Myc agarose beads (Sigma). The beads were washed three times with buffer H containing 0.2% Igepal CA-630. The immunoprecipitates were analyzed by immunoblot assays with anti-Myc-HRP (Santa Cruz Biotechnology) or anti-HA-HRP (Roche) antibodies. The degree of enrichment in cellular fractionation was determined by immunoblot analyses with anti-GAPDH (Genscript, Piscataway, NJ), anti-V-ATPase (Agrisera, Vännäs, Sweden), anti-histone H3 (Abcam, Cambridge, MA) and anti-RNA pol I (Agrisera) antibodies.
TAIR accession numbers
SNC1: At4g16890; SRFR1: At4g37460; RPS4: At5g45250; RPP4: At4g16860; NPR1: At1g64280; EDS1: At3g48090; PAD4: At3g52430; SID2: At1g74710; PR1: At2g14610; PR2: At3g57260; PDF1.2: At5g44420; SAND: At2g28390; ACTIN2: At3g18780.
Supporting Information
Zdroje
1. BentAF
MackeyD
2007
Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions.
Annu Rev Phytopathol
45
399
436
2. ChisholmST
CoakerG
DayB
StaskawiczBJ
2006
Host-microbe interactions: shaping the evolution of the plant immune response.
Cell
124
803
814
3. JonesJDG
DanglJL
2006
The plant immune system.
Nature
444
323
329
4. DoddsPN
LawrenceGJ
CatanzaritiAM
TehT
WangCIA
2006
Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes.
Proc Natl Acad Sci USA
103
8888
8893
5. LeeSW
HanSW
SririyanumM
ParkCJ
SeoYS
2009
A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity.
Science
326
850
853
6. SunW
DunningFM
PfundC
WeingartenR
BentAF
2006
Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses.
Plant Cell
18
764
779
7. GoodmanRN
NovackyAJ
1994
The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon
St. Paul, MN
APS Press
8. GreenbergJT
YaoN
2004
The role and regulation of programmed cell death in plant-pathogen interactions.
Cell Microbiol
6
201
211
9. BhattacharjeeS
ZamoraA
AzharMT
SaccoMA
LambertLH
2009
Virus resistance induced by NB-LRR proteins involves Argonaute4-dependent translational control.
Plant J
58
940
951
10. CawlyJ
ColeAB
KirályL
QiuW
SchoelzJE
2005
The plant gene CCD1 selectively blocks cell death during the hypersensitive response to Cauliflower mosaic virus infection.
Mol Plant-Microbe Interact
18
212
219
11. ColeAB
KirályL
RossK
SchoelzJE
2001
Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to Cauliflower mosaic virus infection.
Mol Plant-Microbe Interact
14
31
41
12. GassmannW
2005
Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance.
Mol Plant-Microbe Interact
18
1054
1060
13. YuI-C
ParkerJ
BentAF
1998
Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant.
Proc Natl Acad Sci USA
95
7819
7824
14. McDowellJM
SimonSA
2006
Recent insights into R gene evolution.
Mol Plant Pathol
7
437
448
15. KwonSI
KoczanJM
GassmannW
2004
Two Arabidopsis srfr (suppressor of rps4-RLD) mutants exhibit avrRps4-specific disease resistance independent of RPS4.
Plant J
40
366
375
16. GassmannW
HinschME
StaskawiczBJ
1999
The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes.
Plant J
20
265
277
17. AartsN
MetzM
HolubE
StaskawiczBJ
DanielsMJ
1998
Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis.
Proc Natl Acad Sci USA
95
10306
10311
18. KwonSI
KimSH
BhattacharjeeS
NohJJ
GassmannW
2009
SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors.
Plant J
57
109
119
19. KimMG
GengX
LeeSY
MackeyD
2009
The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the Arabidopsis nucleotide-binding leucine-rich repeat protein RPS2.
Plant J
57
645
653
20. AlonsoJM
StepanovaAN
LeisseTJ
KimCJ
ChenHM
2003
Genome-wide insertional mutagenesis of Arabidopsis thaliana.
Science
301
653
657
21. SessionsA
BurkeE
PrestingG
AuxG
McElverJ
2002
A high-throughput Arabidopsis reverse genetics system.
Plant Cell
14
2985
2994
22. LiY
LiS
BiD
ChengY-T
LiX
2010
SRFR1 negatively regulates plant NB-LRR Resistance protein accumulation to prevent autoimmunity.
PLoS Pathog
6
e1001111
23. ZhangYL
GoritschnigS
DongXN
LiX
2003
A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1.
Plant Cell
15
2636
2646
24. LiYQ
YangSH
YangHJ
HuaJ
2007
The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors.
Mol Plant-Microbe Interact
20
1449
1456
25. HuaJ
GrisafiP
ChengSH
FinkGR
2001
Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes.
Genes Dev
15
2263
2272
26. JambunathanN
SianiJM
McNellisTW
2001
A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance.
Plant Cell
13
2225
2240
27. YangSH
HuaJ
2004
A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis.
Plant Cell
16
1060
1071
28. NoëlL
MooresTL
van der BiezenEA
ParniskeM
DanielsMJ
1999
Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis.
Plant Cell
11
2099
2111
29. KimSH
KwonSI
BhattacharjeeS
GassmannW
2009
Regulation of defense gene expression by Arabidopsis SRFR1.
Plant Signal Behav
4
149
150
30. WangY
BaoZ
ZhuY
HuaJ
2009
Analysis of temperature modulation of plant defense against biotrophic microbes.
Mol Plant-Microbe Interact
22
498
506
31. YiH
RichardsEJ
2007
A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing.
Plant Cell
19
2929
2939
32. LiY
PenningtonBO
HuaJ
2009
Multiple R-like genes are negatively regulated by BON1 and BON3 in Arabidopsis.
Mol Plant-Microbe Interact
22
840
848
33. Laurie-BerryN
JoardarV
StreetIH
KunkelBN
2006
The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae.
Mol Plant Microbe Interact
19
789
800
34. HinschM
StaskawiczBJ
1996
Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi.
Mol Plant-Microbe Interact
9
55
61
35. ZhangXC
GassmannW
2003
RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames.
Plant Cell
15
2333
2342
36. WirthmuellerL
ZhangY
JonesJDG
ParkerJE
2007
Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense.
Curr Biol
17
2023
2029
37. BirkerD
HeidrichK
TakaharaH
NarusakaM
DeslandesL
2009
A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions.
Plant J
60
602
613
38. NarusakaM
ShirasuK
NoutoshiY
KuboY
ShiraishiT
2009
RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens.
Plant J
60
218
226
39. KimSH
KwonSI
SahaD
AnyanwuNC
GassmannW
2009
Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1.
Plant Physiol
150
1723
1732
40. YiH
RichardsEJ
2009
Gene duplication and hypermutation of the pathogen Resistance gene SNC1 in the Arabidopsis bal variant.
Genetics
183
1227
1234
41. ChengYT
GermainH
WiermerM
BiD
XuF
2009
Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis.
Plant Cell
21
2503
2516
42. EitasTK
NimchukZL
DanglJL
2008
Arabidopsis TAO1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the Pseudomonas syringae effector AvrB.
Proc Natl Acad Sci USA
105
6475
6480
43. KangH-G
OhC-S
SatoM
KatagiriF
GlazebrookJ
2010
Endosome-associated CRT1 functions early in Resistance gene-mediated defense signaling in Arabidopsis and tobacco.
Plant Cell
22
918
936
44. BelkhadirY
SubramaniamR
DanglJL
2004
Plant disease resistance protein signaling: NBS-LRR proteins and their partners.
Curr Opin Plant Biol
7
391
399
45. ShenQH
Schulze-LefertP
2007
Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors.
EMBO J
26
4293
4301
46. ZhuY
QianW
HuaJ
2010
Temperature modulates plant defense responses through NB-LRR proteins.
PLoS Pathog
6
e1000844
47. GarcíaAV
Blanvillain-BaufuméS
HuibersRP
WiermerM
LiG
2010
Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response.
PLoS Pathog
6
e1000970
48. CloughSJ
BentAF
1998
Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana.
Plant J
16
735
743
49. BellCJ
EckerJR
1994
Assignment of 30 microsatellite loci to the linkage map of Arabidopsis.
Genomics
19
137
144
50. KoniecznyA
AusubelFM
1993
A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers.
Plant J
4
403
410
51. ZhangX-C
GassmannW
2007
Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses.
Plant Physiol
145
1577
1587
52. HorschRB
FryJE
HoffmannNL
EichholtzD
RogersSG
1985
A simple and general method of transferring genes into plants.
Science
227
1229
1231
53. HeeseA
HannDR
Gimenez-IbanezS
JonesAME
HeK
2007
The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants.
Proc Natl Acad Sci USA
104
12217
12222
54. LarkinMA
BlackshieldsG
BrownNP
ChennaR
McGettiganPA
2007
ClustalW and ClustalX version 2.0.
Bioinformatics
23
2947
2948
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease AgentČlánek TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease SusceptibilityČlánek The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive ParasiteČlánek Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Patients with Discordant Responses to Antiretroviral Therapy Have Impaired Killing of HIV-Infected T Cells
- A Molecular Mechanism for Eflornithine Resistance in African Trypanosomes
- Tyrosine Sulfation of the Amino Terminus of PSGL-1 Is Critical for Enterovirus 71 Infection
- Autoimmunity as a Predisposition for Infectious Diseases
- The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
- Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography
- Tick Histamine Release Factor Is Critical for Engorgement and Transmission of the Lyme Disease Agent
- Interferon-Inducible CXC Chemokines Directly Contribute to Host Defense against Inhalational Anthrax in a Murine Model of Infection
- TGF-b2 Induction Regulates Invasiveness of -Transformed Leukocytes and Disease Susceptibility
- The Origin of Intraspecific Variation of Virulence in an Eukaryotic Immune Suppressive Parasite
- CO Acts as a Signalling Molecule in Populations of the Fungal Pathogen
- SV2 Mediates Entry of Tetanus Neurotoxin into Central Neurons
- MAP Kinase Phosphatase-2 Plays a Critical Role in Response to Infection by
- Glycosylation Focuses Sequence Variation in the Influenza A Virus H1 Hemagglutinin Globular Domain
- Potentiation of Epithelial Innate Host Responses by Intercellular Communication
- Fcγ Receptor I Alpha Chain (CD64) Expression in Macrophages Is Critical for the Onset of Meningitis by K1
- ANK, a Host Cytoplasmic Receptor for the Cell-to-Cell Movement Protein, Facilitates Intercellular Transport through Plasmodesmata
- Analysis of the Initiating Events in HIV-1 Particle Assembly and Genome Packaging
- Evolution of Linked Avirulence Effectors in Is Affected by Genomic Environment and Exposure to Resistance Genes in Host Plants
- Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41
- Hepatitis C Virus (HCV) Evades NKG2D-Dependent NK Cell Responses through NS5A-Mediated Imbalance of Inflammatory Cytokines
- Host Cell Invasion and Virulence Mediated by Ssa1
- Global Gene Expression in Urine from Women with Urinary Tract Infection
- Should the Human Microbiome Be Considered When Developing Vaccines?
- HapX Positively and Negatively Regulates the Transcriptional Response to Iron Deprivation in
- Enhancing Oral Vaccine Potency by Targeting Intestinal M Cells
- Herpes Simplex Virus Reorganizes the Cellular DNA Repair and Protein Quality Control Machinery
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- Cryo Electron Tomography of Native HIV-1 Budding Sites
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- Modelling the Evolution and Spread of HIV Immune Escape Mutants
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
- Platelet-Activating Factor Receptor Plays a Role in Lung Injury and Death Caused by Influenza A in Mice
- Genetic and Structural Basis for Selection of a Ubiquitous T Cell Receptor Deployed in Epstein-Barr Virus Infection
- Ubiquitin-Regulated Nuclear-Cytoplasmic Trafficking of the Nipah Virus Matrix Protein Is Important for Viral Budding
- Pneumolysin Activates the NLRP3 Inflammasome and Promotes Proinflammatory Cytokines Independently of TLR4
- Immune Evasion by : Differential Targeting of Dendritic Cell Subpopulations
- Survival in Selective Sand Fly Vector Requires a Specific -Encoded Lipophosphoglycan Galactosylation Pattern
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Zn Inhibits Coronavirus and Arterivirus RNA Polymerase Activity and Zinc Ionophores Block the Replication of These Viruses in Cell Culture
- The Female Lower Genital Tract Is a Privileged Compartment with IL-10 Producing Dendritic Cells and Poor Th1 Immunity following Infection
- Crystal Structure and Size-Dependent Neutralization Properties of HK20, a Human Monoclonal Antibody Binding to the Highly Conserved Heptad Repeat 1 of gp41
- The Arabidopsis Resistance-Like Gene Is Activated by Mutations in and Contributes to Resistance to the Bacterial Effector AvrRps4
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



