-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
The rapidly proliferating hematopoietic stem/progenitor cells (HSPCs) require well-established DNA damage response/repair pathways to resolve the DNA replication stress-induced DNA damage, which is deleterious for the genome stability and cell survival. Impairment of these pathways could lead to the progressive bone marrow failure (BMF) and hematopoietic malignancies. Here we reported a novel function of topoisomerase II β binding protein 1 (TopBP1) in definitive hematopoiesis through characterizing zebrafish mutantcas003 with a nonsense mutation in topbp1 gene encoding TopBP1. The homozygous topbp1 mutants manifested decreased HSPCs during their pool expansion in the caudal hematopoietic tissue (CHT, an equivalent of the fetal liver in mammals) due to the p53-dependent apoptosis. Further investigation revealed that the deficient TopBP1-ATR-Chk1 pathway upon DNA replication stress in topbp1 mutants led to accumulated DNA damage and further affected HSPCs survival. These studies therefore emphasized the importance of topbp1 function as well as DNA damage response pathways during the fetal HSPC rapid proliferation.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005346
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005346Summary
The rapidly proliferating hematopoietic stem/progenitor cells (HSPCs) require well-established DNA damage response/repair pathways to resolve the DNA replication stress-induced DNA damage, which is deleterious for the genome stability and cell survival. Impairment of these pathways could lead to the progressive bone marrow failure (BMF) and hematopoietic malignancies. Here we reported a novel function of topoisomerase II β binding protein 1 (TopBP1) in definitive hematopoiesis through characterizing zebrafish mutantcas003 with a nonsense mutation in topbp1 gene encoding TopBP1. The homozygous topbp1 mutants manifested decreased HSPCs during their pool expansion in the caudal hematopoietic tissue (CHT, an equivalent of the fetal liver in mammals) due to the p53-dependent apoptosis. Further investigation revealed that the deficient TopBP1-ATR-Chk1 pathway upon DNA replication stress in topbp1 mutants led to accumulated DNA damage and further affected HSPCs survival. These studies therefore emphasized the importance of topbp1 function as well as DNA damage response pathways during the fetal HSPC rapid proliferation.
Introduction
Hematopoietic stem/progenitor cells (HSPCs) possess the capabilities of self-renewal and differentiation into all lineages of mature blood cells [1]. Dysregulated self-renewal of HSPCs is tightly associated with the human blood diseases including leukemia and bone marrow failure (BMF) syndrome [2–4]. Previous studies have illustrated that the genes causative for adult hematopoietic diseases virtually play critical roles in the early hematopoiesis [5,6]. Therefore, exploring the unknown genetic regulators of HSPCs in the hematopoiesis would give us better understanding of the sophisticated mechanisms of hematopoietic diseases in adults.
Recently, zebrafish has emerged as an excellent animal model to study the development of hematopoiesis [7–9]. With multiple unique advantages including external fertilization and development, optically transparent embryos, small size and high fecundity, zebrafish is extraordinarily suitable for the unbiased large scale forward genetics screening to identify novel genes regulating HSPCs self-renewal in the embryonic development [10]. More importantly, the hematopoietic anatomy and the critical transcriptional factors involved in the development of hematopoiesis are highly conserved between zebrafish and mammals [1,11]. Similar to mammals, zebrafish hematopoiesis consists of two waves of hematopoiesis, i.e. primitive hematopoiesis and definitive hematopoiesis. The primitive hematopoiesis takes place in the anterior lateral plate mesoderm (ALPM) and intermediate cell mass (ICM) at ~12–14 somites stage, producing primitive macrophages and erythrocytes, respectively [12]. In zebrafish definitive hematopoiesis, HSPCs originate in the ventral wall of dorsal aorta (an equivalent of the aorta-gonad-mesonephros [AGM] in mammals) through endothelium to hematopoietic transition (EHT) from 26 hours post fertilization (hpf) [13,14], and then colonize in caudal hematopoietic tissue (CHT, an equivalent to the fetal liver [FL] in mammal) (at 2 days post fertilization [dpf]), thymus (at 3dpf) and ultimately kidney marrow to support adult hematopoiesis (equivalent to bone marrow (BM) in mammal) (after 5dpf) [15,16]. During fetal hematopoiesis in CHT, the nascent HSPCs undergo extensive proliferation for the pool expansion to support the embryo development [15]. It has been reported that 95–100% of HSPCs are actively cycling in the mouse fetal liver, whereas most of adult HSPCs are in a quiescent state [17].
During DNA replication, the slowed or stalled DNA replication fork, which is termed as DNA replication stress, occurs frequently due to intracellular and extracellular sources including the by-products of cellular metabolism (e.g. dNTP misincorporation, reactive oxygen species etc.), ultraviolet light and chemical mutagens [18,19]. Because the stalled replication forks are vulnerable and the collapse of the forks can result in DNA double strand breaks (DSBs) that are deleterious for the genome stability and cell survival, the DNA replication stress-induced DNA damage needs to be efficiently resolved by DNA damage response (DDR) pathways [18]. The phosphoinositide kinase-related kinase ataxia telangiectasia mutated (ATM) and ATM and Rad3-related (ATR) are two important kinases involved in DDR. ATM mainly participates in the DSBs response, whereas ATR is activated by the single-stranded DNA (ssDNA) damage and DNA replication stress [20]. Recent studies have shed the light on the association between hematopoietic homeostasis and DDR. DDR impairment can lead to progressive BMF and hematopoietic malignancies [21–23]. Fanconi anemia (FA) pathway, which consists of 15 FA genes, mainly participates in repairing the DNA interstrand crosslinks (ICL). Most of the FA genes are associated with the replication fork protection and ATR activation pathway [24,25], and they are causally mutated in BMF or acute myelogenous leukemia [26].
Topoisomerase II β binding protein 1 (TopBP1) is a structurally and functionally conserved protein from yeast to human, which is essential as a scaffold protein in DNA replication initiation and DNA damage checkpoint activation [27–30]. TopBP1 plays a vital role in the DDR, it mainly protects against the ssDNA damage and DNA replication stress through the TopBP1-ATR-Chk1 axis [31–33]. In this process, the stalled replication forks will generate a typical double-stranded DNA-single-stranded DNA (dsDNA-ssDNA) structure. Following the replication protein A (RPA) coating, TopBP1-associated proteins including Rad9-Rad1-Hus1 (9-1-1 complex), ATR interaction protein (ATRIP) and ATR are recruited to the damage locus, then TopBP1 largely activates the ATR kinase activity through its ATR activation domain (AAD), which triggers the phosphorylation of Chk1 and stabilization of replication forks until the stress is resolved [34–38]. Other TopBP1 interacting components also facilitate the establishment of the TopBP1-ATR-Chk1 axis, including the mediator of DNA-damage checkpoint 1 (MDC1) and BRCA1 interacting protein C-terminal helicase (BRIP1, aka, FANCJ) [39–42].
Although the cellular function of TopBP1 has been established, its physiological role, especially the tissue specific requirement, is still largely unknown. TopBP1 null mice are embryonic lethal due to accumulated DNA damage and reduced cell proliferation, which is phenocopied by TopBP1 W1147R knock-in mice with abrogated AAD domain of TopBP1 [43,44]. Moreover, neuronal specific deletion of TopBP1 in mice demonstrates that TopBP1 is essential for neural progenitor cells to survive from the DNA replication stress [45]. Specific disruption of TopBP1 in the lymphoid cells blocks lymphocyte development due to aberrant V(D)J rearrangement [46]. However, whether TopBP1 participates in the HSPCs development is still unknown.
Here we report a novel zebrafish mutantcas003, in which HSPCs can be generated normally, but fail thereafter in definitive hematopoiesis. Positional cloning and functional validation indicated that a nonsense mutation-caused C-terminal truncation of TopBP1 was responsible for its subcellular mislocalization and hematopoietic deficits. Disrupted TopBP1-ATR-Chk1 pathway and the accumulation of DNA damage were associated with the HSPCs defect and triggered apoptosis via a p53-dependent pathway. Our findings demonstrate that topbp1 is essential for the HSPCs survival under extensive DNA replication stress during the highly proliferative fetal definitive hematopoiesis.
Results
Mutantscas003 display defective definitive hematopoiesis
To explore new genes and regulatory mechanisms in vertebrate definitive hematopoiesis, we carried out a large-scale forward genetics screen on ENU-mutagenized F2 families in zebrafish by whole mount in situ hybridization (WISH) using c-myb probe (a key transcription factor and marker of HSPCs) [15,47]. In 5dpf wild-type zebrafish embryos, c-myb was expressed in all hematopoietic tissues including caudal hematopoietic tissue (CHT), thymus, and kidney (Fig 1); whereas homozygous mutantscas003 displayed normal morphogenesis (Fig 1A–A’), but dramatically decreased c-myb expression in CHT, kidney and thymus (Fig 1B–B’), suggesting the expansion of HSPCs was defective. To confirm the defective definitive hematopoiesis in mutantscas003, we further examined the expression of downstream hematopoietic lineage cell markers including ae1-globin (erythrocyte marker), mpx (granulocyte marker), lyz (macrophage marker) and rag1 (lymphocyte marker). The expression of all these markers was substantially decreased in the homozygous mutantcas003 embryos at 5dpf (Fig 1C–G’), which suggested hematopoiesis failure.
Fig. 1. The definitive hematopoiesis is defective in zebrafish mutantcas003 embryos. 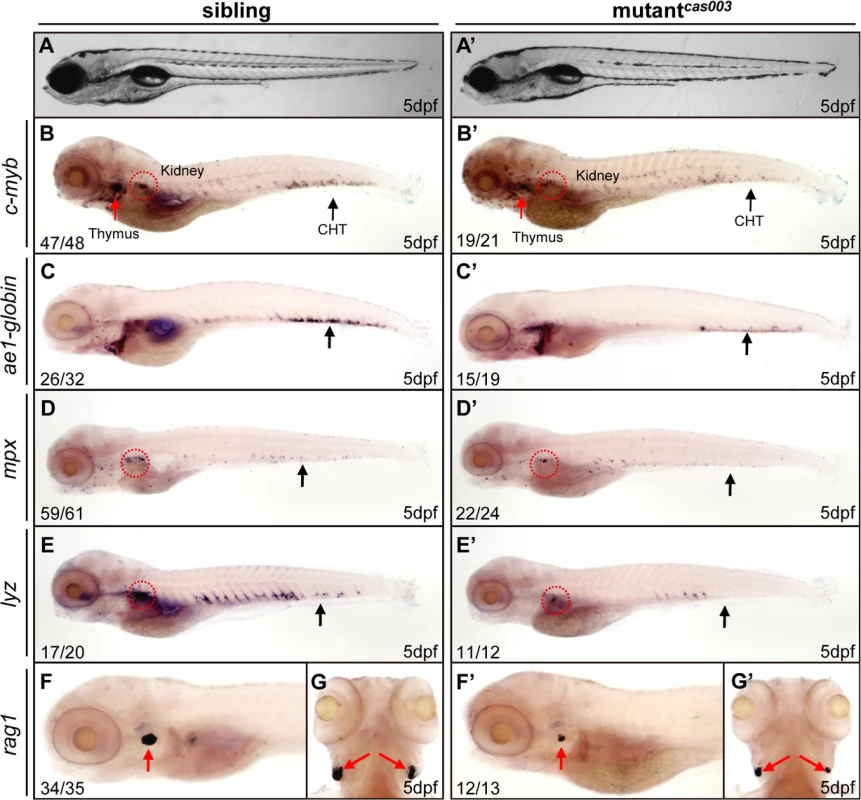
(A, A’) The bright field images of zebrafish wild-type sibling (A) and mutantcas003 embryos (A’) showing no obvious difference at 5dpf. (B-G’) Whole-mount in situ hybridization (WISH) results of c-myb, ae1-globin, mpx, lyz and rag1 showing defective definitive hematopoiesis in mutantcas003 embryos (B’-G’) but not in sibling embryos (B-G) at 5dpf. The penetrance of the indicated phenotype is shown in the bottom left of each panel. (B-F, B’-F’) Lateral views; (G, G’) dorsal views. Black arrows indicate the position of caudal hematopoietic tissue (CHT); red arrows and circles show the position of thymus and kidney, respectively. Recent studies have demonstrated that vasculogenesis and blood flow are essential for HSPCs initiation and maintenance [48,49]. We examined the expression pattern of a pan-endothelial cell marker flk1 at 36hpf and an artery vessel marker ephrinB2 at 26hpf respectively, our results revealed that both of them were intact in mutantcas003 (S1A–S1D Fig). Consistently, heart beating rate and blood circulation were comparable between mutantcas003 and sibling control (S1 and S2 movie). In addition, live observation on mutantcas003, within Tg(fli1: EGFP) transgenic background [50], indicated that the vascular plexus in the CHT region was normal from 2dpf to 5dpf (S1E–S1L Fig). We further investigated the primitive hematopoiesis in mutantcas003. The WISH analysis data demonstrated that the expression of primitive hematopoietic cell markers were identical between siblings and mutantcas003 at 22hpf, including scl (hematopoietic progenitor marker), gata1 (erythrocyte progenitor marker), pu.1 (myeloid progenitor marker), lyz, l-plastin (myeloid cell marker) and mpx (S2A–S2L Fig, quantified in M). Taken together, we concluded that mutantcas003 displayed specific deficiency in definitive hematopoiesis during zebrafish circulation system development.
HSPCs defects initiate in the CHT of mutantcas003
HSPCs are generated from the ventral wall of dorsal aorta through the endothelia to hematopoietic transition (EHT) from 26hpf [13,14], and then migrate to the CHT, a proliferative hematopoietic microenvironment, for pool expansion at 2dpf [15,16]. To figure out when the HSPCs defect initiated in mutantcas003, we performed a time course analysis of c-myb expression from 36hpf to 5dpf. The WISH results demonstrated that the generation of HSC was intact in mutantcas003 as both c-myb and runx1 [51] expression were undisturbed at 36hpf (Fig 2A–B’ and 2F–G’), and the c-myb expression was still intact in the CHT at 2dpf in mutantcas003 (Fig 2C–C’ and 2H–H’). However, mutantcas003 displayed reduced c-myb expression in the CHT at 3dpf (Fig 2D–D’ and 2I–I’), and such defect was more profound at 4dpf (Fig 2E–E’ and 2J–J’), indicating that the HSPCs proliferation or maintenance was impaired in the CHT of mutantcas003. To consolidate this discovery, we carried out quantitative RT-PCR analysis on the c-myb mRNA level in zebrafish tails region including CHT from 2dpf to 5dpf. As expected, the c-myb expression level was attenuated from 3dpf to 5dpf (Fig 2K), which was consistent with the results of WISH analysis. To further confirm these findings, we crossed mutantcas003 with Tg(c-myb: EGFP), in which HSPCs could be visualized by EGFP [52]. Statistically significant reduction of EGPF+ cells was observed at 4dpf (Fig 2L, 2N and 2Q) and was more severe at 5dpf in mutantcas003 (Fig 2L, 2O and 2R) (Due to the long half-life of EGFP protein, the dynamics of c-myb expression indicated via Tg(c-myb: EGFP) was delayed, compared to WISH analysis via c-myb probe [5]). Collectively, our data revealed that, in mutantcas003, neither HSPCs specification in AGM nor their migration to CHT was affected, but their transitory expansion in the CHT was compromised.
Fig. 2. The number of HSPCs is reduced in the mutantcas003 CHT. 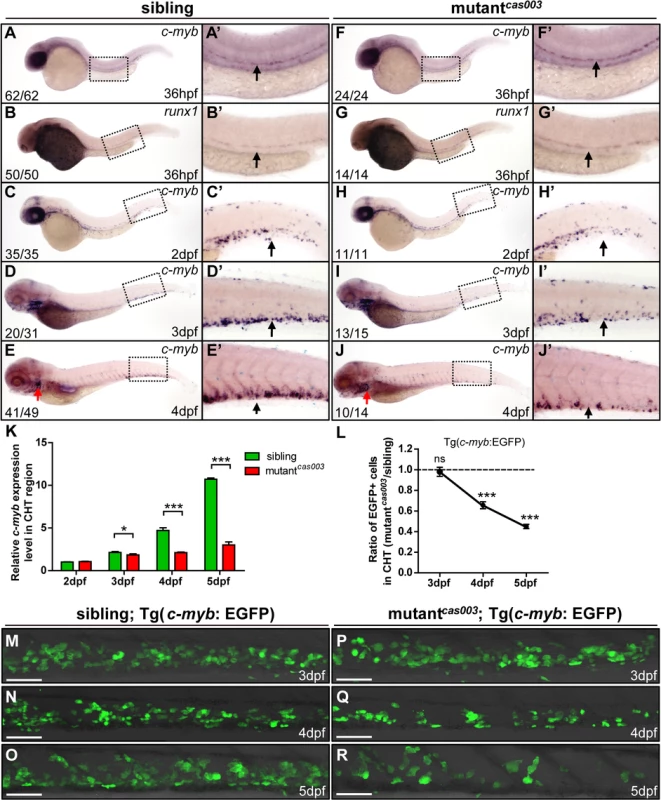
(A-J’) The time-course analysis of definitive HSPCs from 36hpf to 4dpf by WISH with c-myb (A, A’, C-E’, F, F’, H-J’) or runx1 (B, B’, G, G’) indicating the HSPCs are decreased from 3dpf in mutantcas003 embryos comparing to siblings. The penetrance of the indicated phenotype is shown in the bottom left of each panel. (A’-J’) Enlarged views of AGM or CHT regions indicated by the dotted rectangles in the left columns. Black arrows represent the AGM (A’, B’, F’, G’) or CHT (C’-E’, H’-J’); red arrows indicate the thymus. (K) Quantitative PCR analysis of c-myb expression level in the tail region from 2dpf to 5dpf showing the attenuated c-myb expression after 3dpf in mutantcas003 embryos comparing to siblings. Error bars represent SEM. *, p<0.05; ***, p<0.001. (L-R) Immunofluorescence stain of EGFP in the CHT region in mutantcas003; Tg(c-myb: EGFP) embryos (P-R) and siblings (M-O) from 3dpf to 5dpf. The statistics result of the ratio of EGFP+ cell number in the CHT region of mutantcas003 embryos to that in the siblings is indicated in L (n>6 embryos are counted in each panel). These results indicate that EGFP+ HSPCs begin to be decreased obviously in CHT region in mutantcas003 embryos comparing to siblings from 4dpf. Scale bars represent 50μm. Error bars represent SEM; ns, no significance; *** represents p<0.001. The topbp1 gene is disrupted in mutantcas003
In order to elucidate the mechanism of hematopoietic failure in mutantcas003, we carried out positional cloning of the mutant. The mutation was first mapped to chromosome 24 by bulk segregation analysis (BSA). With a high resolution mapping approach, the mutation was revealed to be flanked by two closely linked SSLP markers, L0310_5 and R0310_4. The flanked region contained four candidate genes: topbp1 (topoisomerase II β binding protein 1), tmem108, cdv3 and vps41 (Fig 3A). After sequencing cDNA of all 4 genes, we identified a C to T nonsense mutation in topbp1 gene in mutantcas003 (Fig 3B), and confirmed this result through genomic sequencing. This mutation caused an earlier stop codon before the eighth BRCT (BRCA1 C-terminus) domain and a putative C-terminus nuclear localization signal (NLS) of TopBP1 protein (Fig 3C). This truncated form of endogenous TopBP1 (TopBP1cas003) protein was further confirmed by immunoblotting analysis of the CHT of heterozygote (Het cas003) and mutantcas003 embryos at 3dpf (Fig 3D).
Fig. 3. The topbp1 gene is disrupted in mutantcas003. 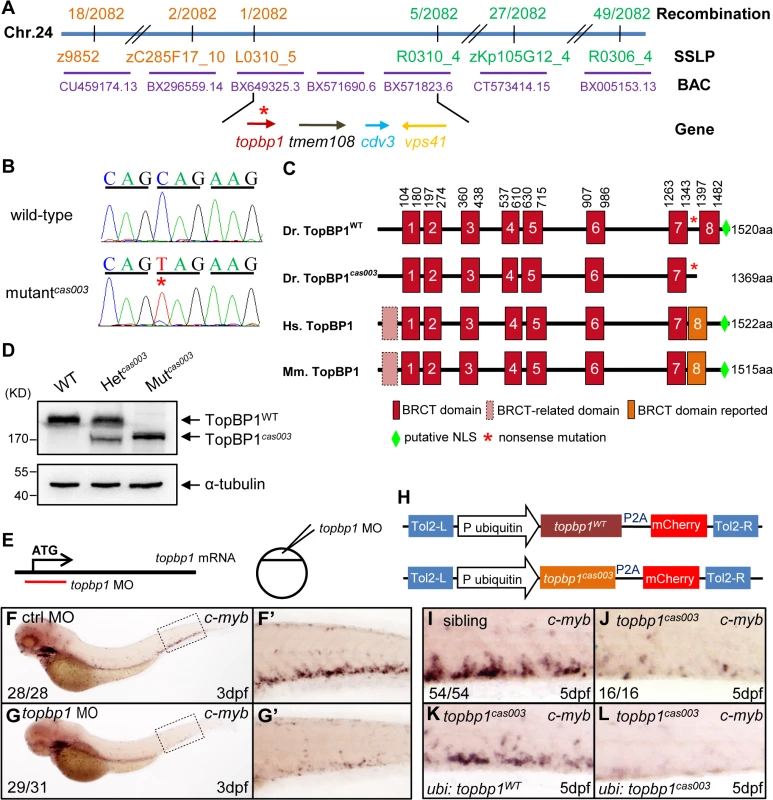
(A) Positional cloning of mutantcas003. Bulk segregation analysis (BSA) revealed that the mutation occurred on chromosome 24 (Chr. 24). After high-resolution mapping with SSLPs, the point mutation was flanked by SSLP markers L0310_5 (1 recombinant out of 2082 meiosis) and R0310_4 (5 recombinants out of 2082 meiosis). This region contains four genes including topbp1, tmem108, cdv3 and vps41. The blue line represents Chr. 24; the positions and the recombinations of the SSLP markers on Chr. 24 or BACs are indicated. The SSLP markers which are on the same side of the mutation site are shown in the same color. (B) The coding region of topbp1 was sequenced in the wild-type sibling (top) and mutantcas003 (bottom). There is a C to T mutation in the mutantcas003 embryos, which is a nonsense mutation. (C) Comparison of vertebrate TopBP1 and zebrafish TopBP1cas003. As human TopBP1 (Hs. TopBP1) and mice TopBP1 (Mm. TopBP1), zebrafish TopBP1WT (Dr. TopBP1) contains eight BRCT domains, while the eighth BRCT domain and putative nuclear localization signal (NLS) is missing in TopBP1cas003 due to the nonsense mutation (*). The red and pink BRCT domains were predicted by SMART software. The orange BRCT domains were reported previously. The molecular sizes of the protein are indicated in the right side. (D) Western blotting analysis showing reduced protein size of TopBP1cas003. (E-G’) Topbp1 morphants can phenocopy topbp1cas003. (E) Topbp1 MO can block the translation of topbp1 mRNA. The topbp1 MO, as validated in S3 Fig, was injected into one-cell stage wild-type embryos to produce topbp1 morphants. (F-G’) WISH results of c-myb in the control morphants (F, F’) and topbp1 morphants (G, G’) at 3dpf. topbp1 morphants show decreased c-myb expression. (F’, G’) The enlarged views of the dotted rectangle region in the left columns. The penetrance of the indicated phenotype is shown in the bottom left of each panel. (H) Construction of the plasmids used in Tol2-transposease-mediated rescue assays. topbp1WT and topbp1cas003 driven by the ubiquitin promoter, followed by the P2A peptide and the mCherry coding sequence, are cloned into Tol2 transposon vector. These constructs are abbreviated as ubi: topbp1WT and ubi: topbp1cas003 respectively. (I-L) WISH analysis with c-myb probe in the CHT region (at 5dpf) of sibling, topbp1cas003 mutant and topbp1cas003 mutant with transient transgenesis of ubi: topbp1WT or ubi: topbp1cas003. 28 embryos out of 45 topbp1cas003 mutants were rescued by topbp1WT, but none by topbp1cas003 (n = 14). In order to examine whether the disruption of topbp1 was causative for phenotype of mutantcas003, we injected a validated topbp1 ATG morpholino oligo (MO) (S3A–S3B Fig) into one-cell stage wild-type embryos to block the translation of endogenous topbp1 mRNA (Fig 3E). Since topbp1 MO acted in a dose-dependent manner (S3C Fig), we applied morpholino microinjection causing no morphologic phenotype in the following studies. Topbp1 morphants manifested severe defective definitive hematopoiesis as that in mutantscas003 from 36hpf to 5dpf, while the primitive hematopoiesis at 22hpf, HSPC generation in AGM at 36hpf and vascular system in CHT at 3dpf were all intact in the morphants (Fig 3F–G’ and S4A–S4R Fig). To further consolidate our findings, we performed rescue assay by ectopic expression of wild-type topbp1 in mutantcas003. Consistent with previous report on the instability of topbp1 mRNA [53], ectopic expression of TopBP1 was barely detected at 3dpf after injection of in vitro synthesized topbp1 mRNA into 1-cell stage embryos. In order to overcome this obstacle, we employed a Tol2 transposase-mediated transgenic rescue approach [54]. The ubiquitin promoter (driving ubiquitous expression) and the coding sequence of topbp1WT or topbp1cas003 followed by P2A peptide-mCherry fusion protein (P2A peptide allows self-cleavage of transgenesis efficacy indicator-mCherry without affecting TopBP1 protein) were constructed into the plasmid containing Tol2 arms (hereinafter referred to as ubi: topbp1WT and ubi: topbp1cas003, Fig 3H) [55,56]. After co-injection with Tol2 transposase mRNA and ubi: topbp1WT or ubi: topbp1cas003 constructs into one cell stage mutantcas003 embryos, ubi: topbp1WT driven ectopic expression of wild-type topbp1 could rescue mutantcas003 phenotype at 5dpf (Fig 3I–3K), but not the ubi: topbp1cas003 construct (Fig 3I–3J and 3L). Taken together, the MO phenocopy assays and the wild-type topbp1 rescue assays revealed that the nonsense mutation in topbp1 was the causative mutation in mutantcas003. Meanwhile, we changed the name of mutantcas003 into topbp1cas003.
Apoptotic HSPCs are enriched in the CHT of topbp1cas003 mutants
To explore how topbp1 affected maintenance of HSPCs in CHT region, we first investigated the expression pattern of topbp1 during embryonic development. WISH analysis data indicated that topbp1 was a maternal mRNA, and was ubiquitously expressed during embryogenesis (S5A–S5J Fig). Previous reports had showed that topbp1 knock-out or knock-down could result in either cell proliferation blockage or cell apoptosis activation [44,45]. To investigate the cause of HSPCs abrogation, we conducted cell biology assessment of HSPCs in topbp1cas003 mutants in Tg(c-myb: EGFP) transgenic background. Double staining of c-myb and phospho-histone 3 (pH3) showed no significant difference in topbp1cas003 mutants, compared with siblings at 3.5dpf (Fig 4A–D’, quantified in Q), suggesting that the cell cycle of HSPCs was not affected in topbp1cas003 mutants. Furthermore, we performed 5-bromo-2-deoxyuridine (BrdU) incorporation assay on HSPCs, BrdU and EGFP double immunostaining results indicated that there was no significant difference in the percentage of BrdU+ HSPCs between siblings and topbp1cas003 mutants at 3.5dpf (Fig 4E–H’, quantified in R). However, TUNEL assay showed a significant increase of apoptotic EGFP+ HSPCs in CHT region of topbp1cas003 mutants, compared with that in wild-type siblings at 3.5dpf (Fig 4I–L’, quantified in S). At 4dpf, the percentage of apoptotic EGFP+ HSPCs was even more significantly increased in topbp1cas003 mutants in comparison with siblings (Fig 4M–P’, quantified in S), while the number of EGFP+ HSPCs were dramatically decreased (Fig 2N and 2Q). Notably, we could also detect the increased apoptosis in the cranial region and the neural tube in the topbp1cas003 mutants at 3.5dpf and 4dpf. Collectively, we concluded that the increased apoptosis in HSPCs was linked to the defective hematopoiesis in topbp1cas003 mutants.
Fig. 4. The defect of definitive hematopoiesis is due to the increased apoptosis in the HSPCs of topbp1cas003 mutants. 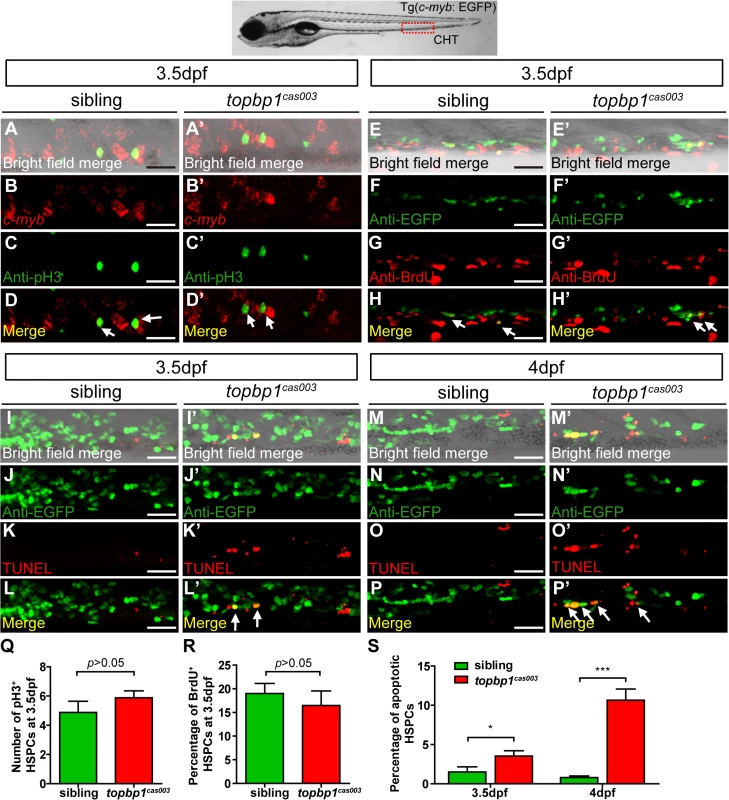
(A-D’) The c-myb in situ hybridization and phospho-histone H3 (pH3) immunostaining at 3.5dpf in sibling and topbp1cas003 mutant embryos, indicating no significant difference in the number of pH3+ HSPCs (pH3 and c-myb double positive cells, white arrows) between wild-type sibling and topbp1cas003 mutant embryos. The images show the stain in the enlarged CHT region. Scale bars represent 25μm. (E-H’) Double immunostaining of c-myb: EGFP and BrdU at 3.5dpf in the sibling and topbp1cas003 mutant embryos. The number of BrdU+ HSPCs (BrdU and EGFP double positive cells, white arrows) in topbp1cas003 mutant embryos is comparable to that in wild-type siblings. Scale bars represent 25μm. (I-P’) Double immunostaining of c-myb: EGFP and TUNEL at 3.5dpf (I-L’) and 4dpf (M-P’) in the sibling and topbp1cas003 mutant embryos. The apoptotic HSPCs (TUNEL and EGFP double positive cells, white arrows) are increased in the topbp1cas003 mutants comparing to siblings, which is more profound at 4dpf. Scale bars represent 25μm. (Q) Quantification of the number of the pH3+ HSPCs in sibling and topbp1cas003 mutant embryos at 3.5dpf. (R) Statistics result of the percentage of BrdU+ HSPCs at 3.5dpf. (Q) and (R) indicate no significant difference in the number of proliferating HSPCs between topbp1cas003 mutant embryos and siblings at 3.5dpf. (S) Quantitative analysis of apoptotic HSPCs in the CHT region in sibling and topbp1cas003 mutant embryos at 3.5dpf and 4dpf, showing the increased apoptotic HSPCs in the topbp1cas003 mutant embryos. For the Quantitative analysis, at least 6 embryos were analysis for each experimental group. Error bars represent SEM. *, p<0.05; ***, p<0.001. Apoptosis in the HSPCs of topbp1cas003 mutants is p53-dependent
To determine how TopBP1 deficiency triggered apoptosis, we firstly checked the expression of several apoptosis-related genes in the CHT regions of topbp1cas003 mutants at 3dpf. The quantitative PCR results showed that the expression of p53, p21, cyclin G1 and mdm2 were upregulated in the CHT region of topbp1cas003 mutants, indicating the p53 signaling pathway was activated (Fig 5A). Furthermore, we employed ectopic expression of Bcl2 into topbp1cas003 mutants, which was known to inhibit p53 dependent apoptosis pathway [57]. WISH analysis on c-myb expression showed that bcl2 mRNA could partially restore the c-myb expression in the CHT regions of topbp1cas003 mutants (25 out of 43 embryos were partially rescued, S6B–S6D’ Fig, quantified in S6A Fig). To confirm the apoptosis in topbp1cas003 HSPCs mainly depended on the p53 pathway, we crossed topbp1cas003 mutant with the tp53M214K mutant (abbreviated as p53-/- below), which had been reported to abrogate p53 function in apoptosis [58]. Further investigation showed that the expression of c-myb in topbp1cas003 mutants was partially rescued in p53+/- heterozygous background at 4dpf (3/12 embryos were well rescued, 3/12 embryos were partially rescued, Fig 5B–F’), and the rescue effect was more obviously in p53-/- background at 4dpf (7/13 embryos were well rescued, 4/13 embryos were partially rescued, Fig 5B–F’). Taken together, we concluded that the apoptosis of HSPCs in topbp1cas003 mutants was p53-dependent.
Fig. 5. The apoptosis of HSPCs is p53 dependent in topbp1cas003 mutants. 
(A) Quantitative PCR results of c-myb, p53, p21, cyclin G1 and mdm2 in the CHT region of wild-type sibling and topbp1cas003 mutant embryos at 3dpf. All these p53 dependent apoptosis-related genes are upregulated. Error bars represent SEM. *, p<0.05; ***, p<0.001. (B) Quantitative analysis of HSPCs phenotype, monitored by c-myb WISH analysis, showing inactivation of p53 can partially rescue c-myb expression in topbp1cas003 mutants. The 3 kinds of standards of c-myb WISH results in the CHT at 4dpf are shown in the right bottom. The numbers of embryos are shown above the columns. (C-F’) c-myb WISH analysis of siblings, topbp1cas003, topbp1cas003; p53+/- and topbp1cas003; p53-/- embryos at 4dpf, which are quantified in B. E and E’ represent partially rescued embryos; F and F’ represent well rescued embryos. (C-D’, C-D’) 23/25 siblings and 8/9 topbp1cas003 mutants show indicated phenotype. (C’-F’) Enlarged views of CHT region indicated by arrows in the left column. Mislocalized TopBP1cas003 causes hematopoiesis defects
To further understand the molecular mechanism of HSPC apoptosis which was induced by this particular defective TopBP1 without its 8th BRCT domain and the putative NLS domain, we analyzed the subcellular localization of TopBP1cas003. Confocal imaging showed that flag-tagged TopBP1WT was predominantly localized in the nucleus of transfected HeLa cells (Fig 6A, left column). However, TopBP1cas003 was mistakenly localized in cytoplasm (Fig 6A, middle column), which was consistent with our previous sequence analysis on the lack of putative NLS in TopBP1cas003 (Fig 3C) and immunoblotting analysis on TopBP1WT/TopBP1cas003 protein in cytoplasmic and nucleus fractions of pooled embryos from heterozygote incrossing (S5L Fig). Moreover, addition of SV40 NLS at C terminus of TopBP1cas003 was sufficient to correct TopBP1cas003 subcellular localization defect (Fig 6A, right column). To test whether the hematopoietic deficiency in topbp1cas003 mutants could also be rescued by the nuclear localized TopBP1cas003, we carried out transient transgenesis of topbp1cas003-NLS or topbp1WT (as the positive control) in the topbp1cas003 mutants. WISH results of c-myb at 4dpf indicated that ectopic expression of topbp1cas003-NLS could rescue c-myb expression in topbp1cas003 mutants, as efficient as transgenesis with topbp1WT (Fig 6B–E’, quantified in F). Collectively, we concluded that the loss of NLS in TopBP1cas003 and the failure of nuclear localization directly caused HSPCs deficiency in topbp1cas003 mutants.
Fig. 6. Subcellular mislocalization and defective ATR/Chk1 activation link to defects in HSPCs in the topbp1cas003 mutants. 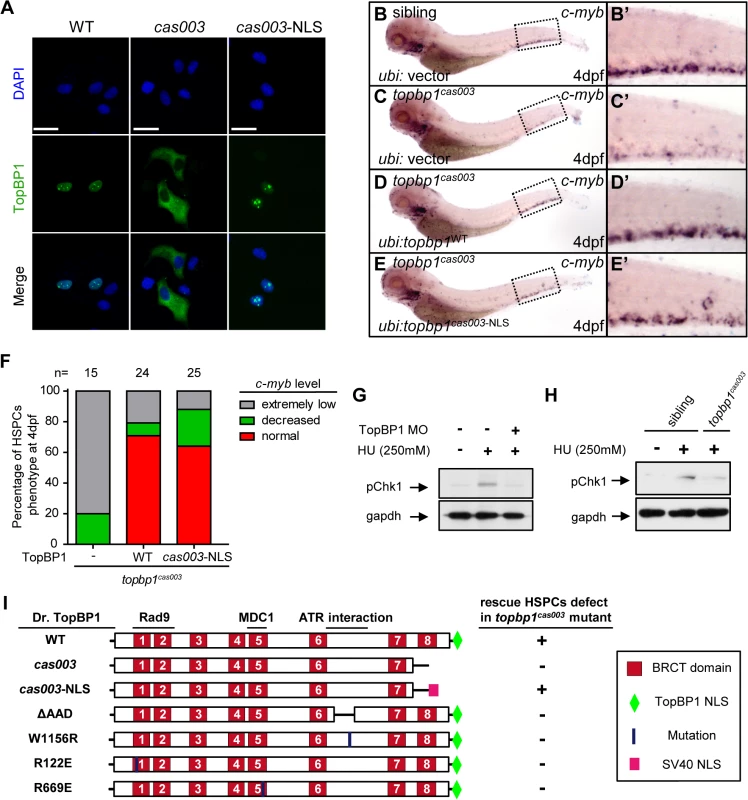
(A) Confocal imaging analysis of anti-FLAG immunostaining (green) and DAPI (blue) in HeLa cells transfected with FLAG tagged TopBP1WT (WT), TopBP1cas003 (cas003) and TopBP1cas003-NLS (cas003-NLS). TopBP1WT is mainly localized in the nucleus (left column), cytosol mislocalization of TopBP1cas003 (middle column) can be corrected by the additional SV40 NLS on its C-terminus (TopBP1cas003-NLS, right column). Scale bars represent 25 μm. (B-F) WISH analysis of hematopoiesis in siblings and topbp1cas003 mutant embryos with Tol2-transposase mediated topbp1WT and topbp1cas003-NLS transgenesis at 4dpf, which are quantified in F (the numbers of embryos are shown above). Defective c-myb expression in topbp1cas003 mutants can be rescued by topbp1cas003–NLS as well as topbp1WT. (B-C, B’-C’) 28/32 siblings and 12/15 topbp1cas003 mutants show indicated WISH results. (D-E, D’-E’) WISH results of well rescued mutants. (B’-E’) Enlarged views of CHT region in the left column. (G) Western blot with pChk1 antibody in control morphants and hydroxyurea (HU) treated control or topbp1 morphants. The morphants were treated with 250mM HU or mock from 60hpf to 76hpf. topbp1 knockdown could abrogate the Chk1 phosphorylation in the tail region upon HU treatment. (H) Immunoblotting analysis showing reduced phospho-Chk1 level in tail region of topbp1cas003 mutants upon HU treatment, comparing to that in wild-type siblings. The embryos were treated with 250mM HU or mock from 60hpf to 76hpf. (I) Schematic diagram of variant forms of TopBP1, including wild-type (WT), cas003, cas003-NLS, ΔAAD, W1156R, R122E and R669E mutation. The regions associated with ATR activation and Rad9 or MDC1 interactions are indicated. After Tol2-mediated transient transgenesis of variant forms of TopBP1 into topbp1cas003 mutant embryos, quantitative analysis of the c-myb expression in the CHT region at 4dpf was performed for the evaluation of rescue capability (n>20). “+” (rescue); “-” (not rescue effect). Except WT and cas003-NLS, all TopBP1 mutation forms are unable to rescue hematopoietic defects in topbp1cas003 mutants. Chk1 activation is reduced in topbp1cas003 mutants
Previous studies have demonstrated that TopBP1 plays conserved roles as a scaffold protein that is important for DNA replication and DNA damage response (DDR) [27,29,37]. Since the proliferation of HSPCs was not disrupted in topbp1cas003 mutants (Fig 4A–H’), it seemed that the function of TopBP1 in DDR instead of DNA replication was responsible for the HSPCs defect in the mutants. Firstly, we checked the activation of TopBP1-ATR-Chk1 pathway in topbp1cas003 mutants and siblings under the hydroxyurea (HU) treatment, which was extensively applied to mimic DNA replication stress and could activate ATR-Chk1 axis in mammalian cells and zebrafish embryos [30,59,60]. The phospho-Chk1 (pChk1) level in CHT region was significantly increased after 250mM HU treatment from 60hpf to 76hpf (Fig 6G, lane1 and 2). However, the activation of pChk1 was abrogated in topbp1 morphants (Fig 6G, lane 3). Consistently, we also observed dramatic ablation of pChk1 elevation in the CHT of topbp1cas003 mutants compared with wild-type siblings (Fig 6H).
Furthermore, we analyzed protein-protein interaction sites in TopBP1 on the basis of previous biochemical and structural investigations [31,41–43,61,62]. The R122, R669 and W1156 sites in TopBP1 are involved in Rad9 or MDC1 interaction and ATR activation, respectively. All these sites are highly conserved among zebrafish, human and mouse (S7 Fig), and they are critical for TopBP1-ATR pathway [31,41,61,63]. Transient transgenesis of TopBP1ΔAAD, TopBP1W1156R, TopBP1R122E, TopBP1R669E and TopBP1WT (as positive control) in topbp1cas003 mutants was analyzed for hematopoiesis monitored by c-myb WISH. None of these mutated TopBP1 could rescue the hematopoietic failure in topbp1cas003 mutants, compared with TopBP1WT (Fig 6I), indicating that ATR activation function of TopBP1 was essential for HSPCs survival in topbp1cas003 mutants. Taken together, these data implied that the blockage of TopBP1-ATR-Chk1 pathway was correlated to the defective HSPCs in topbp1cas003 mutants.
DNA damage response is impaired in topbp1cas003 mutants
Since TopBP1-ATR-Chk1 axis was disrupted in topbp1cas003 mutants, the unresolved DNA replication stress would result in collapse of replication forks, which could introduce DNA double-stranded breaks ultimately [18]. To check whether the apoptosis of HSPCs was due to the accumulation of DNA damage in CHT region, we carried out fluorescent c-myb WISH analysis and immunostaining with phosphorylated histone H2AX (γH2AX) antibody, which was a typical DNA damage marker [64], from 39hpf to 3.5dpf. Interestingly, we couldn’t detect any γH2AX+ cells in AGM region at 39hpf in both topbp1cas003 mutants and siblings, but γH2AX+ HSPCs emerged in CHT region in topbp1cas003 mutants at the same stage (S8A–S8B Fig). Moreover, γH2AX+ HSPCs were accumulated in CHT region of topbp1cas003 mutants afterward (S8C Fig), and they were obviously increased at 3.5dpf, (Fig 7A–H’, S8C Fig) indicating the DNA damage was indeed accumulated in HSPCs in topbp1cas003 mutant. In addition, we could also observed several γH2AX+ cells in neuronal tissue (Fig 7G), which was consistent with previous investigation [45]. Furthermore, the immunoblotting of γH2AX within CHT regions of topbp1cas003 mutants at 3dpf also showed an increase of DNA damage (Fig 7I). Collectively, we found that DNA damage was accumulated in HSPCs in topbp1cas003 mutants.
Fig. 7. topbp1cas003 mutants manifest deficient DNA damage repair capability. 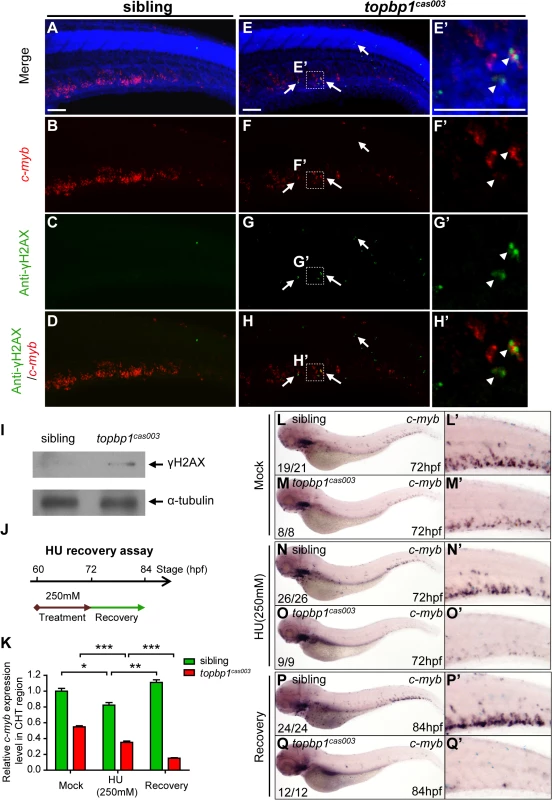
(A-H’) Immuno-staining of γH2AX (green), c-myb fluorescent in situ hybridization (red) and DAPI stain (blue) in sibling and topbp1cas003 mutant embryos at 3.5dpf. (E’-H’) Magnified views of the regions in the dashed boxes showing γH2AX+; c-myb+; DAPI+ cell in the CHT region. Arrows indicate the γH2AX+; DAPI+ cells. The γH2AX+; DAPI+ cells are increased in topbp1cas003 mutants comparing to siblings. Moreover, γH2AX+; c-myb+; DAPI+ cells can only be detected in topbp1cas003 mutants (H’). Scale bars represent 50μm. (I) Immunoblotting showing increased γH2AX level in the CHT of topbp1cas003 mutant embryos at 3dpf. (J) Procedure of the hydroxyurea (HU) recovery assay. Brown arrow line represents 250mM HU or mock treatment from 60hpf to 72hpf; green arrow line indicates removal of HU or mock treatment from 72hpf to 84hpf for recovery. (K-Q) The c-myb expression is further decreased in topbp1cas003 mutant embryos under HU treatment. Wild-type siblings or topbp1cas003 mutant embryos, after 12-hours mock treatment or after 12-hours 250mM HU treatment and sequential 12-hours recovery, were analyzed by either quantitative PCR (K) or c-myb WISH analysis (L-Q). c-myb level can be recovered in wild-type siblings, but not in topbp1cas003 mutant embryos. The penetrance of the indicated phenotype is shown in the bottom left of each panel. Error bars represent SEM. * p<0.05; ** p<0.01; *** p<0.001. (L’-Q’) Enlarged views of the CHT region of embryos in the left columns. To further examine whether the hematopoietic failure was due to the defective DDR upon DNA replication stress in topbp1cas003 mutants, we challenged the embryos with HU. Indeed, high concentration treatment from 52hpf to 76hpf directly caused embryonic lethality in topbp1cas003 mutants (over 65%), however the effect on wild-type siblings was much milder (<15%) (S9A and S9B Fig) [60]. Furthermore, we carried out a recovery assay in the HU-treated zebrafish embryos (Fig 7J). Interestingly, despite of a suppression by HU treatment, the c-myb expression was recovered in wild-type sibling embryos after challenge removal (Fig 7K, 7L–L’, 7N–N’ and 7P–P’). In contrast, the c-myb expression level was not recovered, but decreased further in the HU-treated topbp1cas003 mutant embryos (Fig 7K, 7M–M’, 7O–O’ and 7Q–Q’). Taken together, all these observations suggested that the HSPCs in CHT of topbp1cas003 mutants were defective in replicative DNA damage response and they eventually underwent apoptosis through a p53-dependent signaling pathway.
Discussion
In this study, we reported a novel zebrafish mutant topbp1cas003, which manifested severe defect in definitive hematopoiesis. The reduction of HSPCs started from 3dpf, which was mainly due to the increased p53-dependent apoptosis, rather than proliferation deficiency. Genetic assessment revealed that a nonsense mutation in topbp1 gene was causative for the hematopoiesis failure. Further investigation revealed that the mutated TopBP1cas003 protein was decreased and mislocalized from nucleus to cytoplasm which compromised the DNA damage response. As a result, it led to accumulated DNA damage that triggered sequential apoptosis of HSPCs in topbp1cas003 mutants.
In zebrafish definitive hematopoiesis, HSPCs undergo extensive proliferation in the CHT region around 3dpf, during which the replication stress, characterized by the stalled replication forks, can be induced by various endogenous and exogenous factors [18]. The stalled replication forks will generate typical dsDNA-ssDNA structure, followed with proper loading of RPA, ATR-ATRIP and 9-1-1 complex [37]. Sequential recruitment of TopBP1 can largely activate ATR kinase activity, and the latter will phosphorylate downstream molecules including Chk1. Activated Chk1 stabilizes the replication forks and arrests cell cycle in order to leave enough time for DNA damage repair machinery to work and to restart the replication fork, so that HSPCs can survive the stress and finish their pool expansion (Fig 8).
Fig. 8. TopBP1 governs HSPCs survival during pool expansion in CHT. 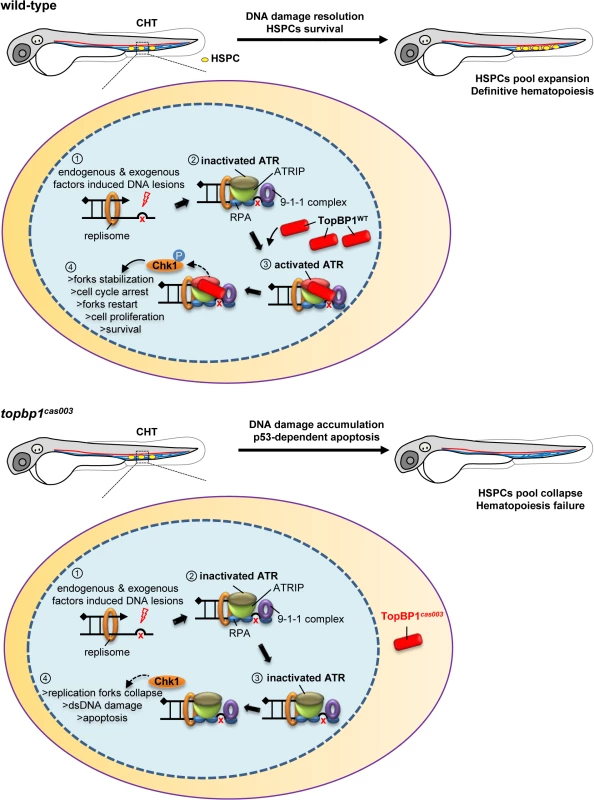
In zebrafish embryogenesis, the nascent HSPCs undergo extensive pool expansion in CHT after 2dpf, while the replication stress causes DNA damage. In normal HSPCs, the stalled replication forks will generate the typical dsDNA-ssDNA structure. After the loading of replication protein A (RPA), ATR/ATRIP, and 9-1-1 complex, sequential participation of TopBP1 can largely activate ATR kinase activity, and the latter phosphorylate downstream targets such as Chk1. The pChk1 can stabilize replication forks and arrest the cell cycle in order to give more time for DNA damage repair for the replication fork restart. As a result, the HSPCs can survive and go through the pool expansion continuously. In topbp1cas003 HSPCs, TopBP1cas003 decreases and mislocalizes in cytosol. ATR and Chk1 are prevented from activation in the circumstance of replication stress. Moreover, the stalled replication forks may collapse and trigger p53-dependent apoptosis, which finally results in hematopoiesis failure. The quantitative analysis and WISH results demonstrated that nonsense mutation in topbp1 might lead to nonsense mediated mRNA decay. The expression level of topbp1 was decreased over 80% in the whole embryo and about 50% in CHT of topbp1cas003 mutants (S5M–S5Q Fig). Although around 50% TopBP1cas003 protein remains in CHT, it was mistakenly localized in cytosol, while TopBP1WT was mainly in nucleus to play its role in DDR (S5L Fig). Our results suggest that TopBP1cas003 is decreased and fails in its nucleus entry due to the loss of its C-terminal NLS, abrogating the later ATR/Chk1 activation. In topbp1cas003 HSPCs, the unresolved stalled replication forks would collapse and generate multiple DNA fragile sites, which can induce dsDNA break [18]. As a result, p53-dependent apoptosis is elevated in topbp1cas003 HSPCs, impairing the HSPCs pool severely (Fig 8).
Recently an improved clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated proteins (Cas9) system with custom guide RNAs (gRNAs) and a zebrafish codon-optimized Cas9 protein showed high mutagenesis rate in zebrafish, which could even generate biallelic mutations in the F0 generation [65,66]. In order to confirm that the deficiency of TopBP1 could disrupt the development of HSPCs, we adapted this optimized CRISPR/Cas9 system to obtain other topbp1 zebrafish mutants (S10A–S10B Fig). Some of the topbp1 Cas9 injected wild-type embryos displayed dramatically decreased c-myb expression as same as topbp1cas003 mutant at 4dpf (S10C–S10D’ Fig). And this phenotype could be reached in higher efficiency when the injected embryos were generated from the outcross between topbp1cas003 heterozygote and wild-type fish (S10E–S10F’ Fig). Conclusively, these data provided additional evidence that definitive HSPCs were defective in the TopBP1 loss-of-function embryos.
It is an intriguing finding that topbp1 plays an essential role in proliferative tissues, especially in the definitive hematopoiesis without affecting the morphogenesis at the early stage, whereas its transcripts were ubiquitously distributed in the embryogenesis (S5 Fig), and TopBP1 knockout mice were reported to be lethal at the peri-implantation stage [44]. The WISH analysis showed maternal expression of topbp1 (S5A Fig), suggesting that homozygote topbp1 mutant embryos can inherit wild-type topbp1 mRNA from the female parents to support its early development until zygotic topbp1 expresses latter in the development. Nevertheless, we attempted to figure out whether topbp1 was expressed and functional in the HSPCs. Quantitative PCR analysis on the CD41+ cell population in the tail region of Tg(CD41: EGFP) embryos, which was reported to be an enriched population of HSPCs at 5dpf [67,68], showed that the level of topbp1 mRNA was 3-fold enriched in CD41+ cells, compared to cells in the whole tails, demonstrating its expression in HSPCs (S5K Fig) [5]. Furthermore, due to the lack of definitive hematopoiesis-specific promoter, we used hemangiogenic promoter lmo2, which was also expressed in definitive HSPCs, to drive the ectopic expression of wild-type topbp1 into topbp1cas003 mutants [52,69], we could indeed observe the expression of mCherry driven by lmo2 promoter in CHT region at 5dpf, and this construct could partially rescue the HSPCs deficits at 5dpf (S11 Fig). In addition, the vascular plexus in CHT region was normal in topbp1cas003 mutants or morphants from 2dpf to 5dpf (S1E–S1L Fig and S4O–S4R Fig), and low dose microinjection of topbp1 morpholino was sufficient to induce definitive hematopoiesis deficits in CHT without affecting the primitive hematopoiesis and vascular system in wild-type embryos (S3C–S3D Fig, S4 Fig). Taken all these data together, we concluded that TopBP1 played an essential and HSPC-intrinsic mechanism during definitive hematopoiesis.
It is intriguing that whether the truncated TopBP1 can potentially function as a dominant negative protein. Ectopic expression of cas003 mutant form of TopBP1 (TopBP1cas003) driven by ubiquitin promoter was performed in wild-type fish, and it did not cause defective definitive hematopoiesis (S12 Fig). The possible reason for this phenomenon was that the mutated TopBP1 could not enter nucleus to compete with wild-type TopBP1. Meanwhile, the hematopoietic phenotype of topbp1cas003 heterozygotes was checked, and no HSPCs defect was observed. Taking these results together, we concluded that TopBP1cas003 could not function as a dominant negative form.
In definitive hematopoiesis, nascent HSPCs seldom proliferate in AGM region, but they become active in cell cycle and undergo extensive proliferation in CHT region supported by niche cells, meanwhile, they have to overcome DNA replicative stress [13,15,18]. BrdU incorporation assays within Tg(c-myb: EGFP) embryos confirmed that HSPCs underwent high proliferation at a constant rate from 2dpf to 5dpf, although the expansion of neural tube cells was gradually attenuated (S13 Fig). As a result, the defect in HSPCs was more profound after 3dpf in the topbp1cas003 mutant. Consistently, we indeed found obvious accumulation of γH2AX positive cells (2.5dpf) and increased apoptotic cells (3.5dpf) in cranial and neuron tube tissues of topbp1cas003 mutant, which was in agreement with previous observations in neuron-specific TopBP1 knock-out mice [45]. Besides, some of homozygote topbp1 mutant embryos developed smaller head and eyes after 6dpf, and all of them eventually died around 10–20 dpf.
Previous works within zebrafish mutants revealed several genes and pathways which were critical for the HSPCs development in CHT region, including genes involved in mitotic spindle assembly, maintenance of centrosome integrity and mitotic progression; pre-mRNA processing; sumoylation of genes participating in DNA replication or cell cycle regulation [5,70,71]. All these genes were indispensable for cell to complete proliferation or division. Because the HSPCs were highly proliferative in CHT, these data depict a picture that the HSPCs in fetal stage are extremely sensitive to the disruption of genes participating in various processing to complete cell division successfully and faithfully. As the DDR pathway is essential for genomic fidelity and stability during DNA replication, our work revealed that DDR pathway is also critical for HSPCs development in fetal stage.
It has been reported that Fanconi anemia pathway is critical for the repair of DNA cross-link damage [26]. Biallelic mutations in any of 15 FANC genes will result in Fanconi anemia (FA), which can most frequently develop into inherited bone marrow failure (BMF) syndrome [72]. The work of Raphael Ceccaldi et al. revealed that the FA patients showed profound HSPCs defect before the onset of BMF [73]. The p53-p21 axis, triggered by replicative stress, was highly elevated in FA HSPCs, and the p53 silence can rescue hematopoietic deficits [73]. They also pointed out that p53 activation, caused by unresolved cellular abnormality, may be the signaling mechanism for inherited BMF, and the p53 activation was commonly found in other types of inherited BMF syndromes, such as Diamond Blackfan anemia (DBA) and dyskeratosis congenital (DC) [73]. HSPCs in topbp1cas003 mutants manifested similar features as that in FA (Fig 5), whether topbp1 could be a putative pathogenic gene in human BMF syndrome needs further investigation.
Zebrafish fancd2 morphant exhibited developmental abnormalities and p53-dependent apoptosis, however its hematopoietic phenotype had not been extensively investigated [57]. The emi1 homozygous mutants showed disrupted genomic integrity and hematopoiesis failure [74]. Studies on topbp1cas003 mutants revealed that DNA damage and apoptosis signaling was accumulated in the HSPCs of topbp1cas003 homozygous embryos, which linked to the hyper-activated p53-p21 axis (Fig 5) and failed ATR/Chk1 activation (Fig 6). Furthermore, TopBP1-involved c-myb regulated DDR pathway was proposed by recent studies on castration-resistant prostate cancer [75]. HU treatment of the developing zebrafish further emphasized the importance of DNA damage response and repair pathway for HSPCs survival during high proliferation stage.
Collectively, we demonstrated a novel and essential role of TopBP1 in HSPCs during their rapid proliferation in fetal hematopoiesis. Due to the dramatic definitive hematopoiesis phenotype in embryogenesis, topbp1cas003 mutants provide a unique model for the mechanism study and small molecular chemical screen on BMF-like hematopoiesis failure, which is caused by defective replicative DNA damage response.
Materials and Methods
Ethics statement
The zebrafish facility and study were approved by the Institutional Review Board of the Institute of Health Sciences, Shanghai Institutes of Biological Sciences, Chinese Academy of Sciences (Shanghai, China), and zebrafish were maintained according to the guidelines of the Institutional Animal Care and Use Committee.
Zebrafish maintenance and manipulation
Wild-type (WT) zebrafish strains Tubingen (TU) and WIK, the transgenic zebrafish line Tg(c-myb: EGFP) [52], Tg(fli1: EGFP) [50], Tg(CD41: EGFP) [76], the mutant zebrafish line tp53M214K/M214K [58] were maintained as previously described [77]. For the forward genetics screen, WT TU zebrafish line was treated with ethylnitrosourea (ENU, Sigma) to generate mutants, the screen approach was performed as previously described [78,79]. The desired mutants within F3 generation were identified by the whole-mount in situ hybridization (WISH) using c-myb probe at 5dpf. For the chemical treatment, the hydroxyurea (HU, Sigma) was dissolved with distilled water into 1M and stored at -20℃. The embryos were treated with 250mM HU as the indicated procedures in the egg water at 28.5℃ [59,60]. To prevent the formation of melanin pigment, the embryos were incubated in egg water containing 0.045% 1-phenyl-2-thiourea (PTU, Sigma) after gastrulation stage. The embryos were collected at the desired stages [80].
Positional cloning and genotyping of mutantcas003
Positional cloning was carried out with WIK line as previously described [81]. Firstly, the mutation was mapped to chromosome 24 by bulk segregation analysis (BSA) with simple sequence length polymorphism (SSLP) markers. Through high resolution mapping analysis on 1041 mutants, the mutation was finally flanked by two SSLP markers, L0310_5 and R0310_4. The candidate genes in this range were sequenced with wild type sibling and mutant cDNA, and the putative mutation was confirmed by genomic DNA sequencing. The primers used in the positional cloning were provided in supplemental S1 Table. Most experiments in this study were carried out with the embryos generated by the incross of mutantcas003 heterozygote pairs (TU/WIK background) used in the positional cloning if possible. The mutants can be identified by flanked SSLP markers, such as Z9852 and R0306_4. Alternatively, the mutants can be distinguished by restriction fragment length polymorphism (RFLP) using EcoP15I (NEB), the RFLP primers were provided in supplemental data (S1 Table).
Plasmid construction
To construct Tol2 transgenesis vectors, the ubiquitin promoter [55] or lmo2 promoter [69] followed by P2A [56] and in-frame mCherry was cloned into modified Tol2 backbone [82]. The vectors were referred as pUbi-Tol2 or pLmo2-Tol2 below. The genes of interest can be inserted between the promoter and P2A. Zebrafish topbp1WT or topbp1cas003 were amplified and inserted into pUbi-Tol2 or pLmo2-Tol2 vectors. To generate the mutated forms of topbp1, the mutagenesis was carried out following QuikChange mutagenesis kit instruction using pUbi-topbp1WT-Tol2 plasmid as the template. The region of TopBP1 (984–1206) are the putative ATR activation domain (AAD) between BRCT6 and BRCT7. In TopBP1ΔAAD, the coding sequence of TopBP1 (1083–1159) containing conserved RQLQ and WDDP sequences are deleted [31]. The fragment of topbp1 (-9–692) was amplified and inserted into the pCS2+ vector for in situ probe preparation. To construct topbp1 MO effect evaluation plasmid, a DNA fragment containing topbp1 ATG MO targeting site was inserted into the upstream of EGFP coding region in pCS2+. Zebrafish topbp1WTand topbp1cas003 were cloned into pCMV4-FLAG-4 vector (Sigma). The SV40 NLS (nuclear localization signal) sequence (5’-CCAAAAAAGAAGAGAAAGGTA-3’) [83] was firstly cloned into pCMV4-FLAG-4 vector in the 3’ end of FLAG tag, and then the topbp1cas003 sequence was inserted into the pCMV4-FLAG-NLS plasmid. All of the primers used were listed in S1 Table.
Microinjection and Cas9 mutagenesis
The mRNA was synthesized in vitro by SP6 mMessage mMachine Transcription Kit (Ambion). The topbp1 gRNA was synthesized as described [66]. The information of the topbp1 gRNA target site was shown in S1 Table. The zebrafish optimized Cas9 mRNA was synthesized in vitro from the pCS2-nCas9n plasmid (addgene, #47929) as described [65]. bcl2-egfp mRNA (~100pg) was injected into 1-cell stage embryos [54]. For the ectopic-expression, Tol2 transposon-mediated transient transgenesis was applied and performed as previously described [84]. A series of topbp1 transgene constructs within Tol2 vectors (~40 ng/μl) were mixed with transposase mRNA (~60 ng/μl) and 0.2 M KCl, and then injected into 1-cell stage embryos, respectively [85]. The volume of the mixture injected was about 0.5nL. The topbp1 ATG morpholino oligo (MO) (5’-CCTTGCTGGCTTTCGACATGGTGAC-3’) and control morpholino (5’ - CCTCTTACCTCAGTTACAATTTATA-3’) were synthesized by Gene Tool company and was injected into 1-cell stage embryos. For Cas9 assay, topbp1 gRNA (50pg) and Cas9 mRNA (150pg) were co-injected into one-cell stage embryos. The T7EI assay was performed as described [65].
WISH and immuno-fluorescence double staining
c-myb, runx1, ae1-globin, mpx, lyz, rag1 and topbp1 probes were transcribed in vitro by T3 or T7 polymerase (Ambion) with Digoxigenin RNA Labeling Mix (Roche). One color WISH was performed as described previously [54]. Images were photographed by the Nikon SMZ1500 microscope with Nikon DXM 1200F CCD or Olympus SZX16 microscope with Olympus DP80 CCD. c-myb RNA and immuno-fluorescence double staining was carried out as described previously [70]. For the immunostaining, rabbit anti-pH3 antibody (1 : 500, Santa Cruz) and rabbit anti-γH2AX antibody (1 : 500, gift from Dr. James Amatruda, University of Texas Southwestern) were used. The embryos were stained with goat-Alexa Fluore488-conjugated anti-rabbit secondary antibody (1 : 500, Invitrogen). DAPI (1 : 500, Beyotime) staining was carried out along with the secondary antibody incubation if necessary.
BrdU incorporation and TUNEL immunostaining
The 3.5dpf topbp1cas003 mutant/Tg(c-myb:EGFP) or sibling embryos were soaked in egg water containing 10mM BrdU (Sigma)/15% DMSO for 30 minutes at 28.5℃ or injected with 1nL 30mM BrdU into the yolk sac. Then they were transferred into fresh egg water and incubated for 2 hours. After fixation in 4% paraformaldehyde (PFA, Sigma), the embryos were dehydrated with methanol and stored at -20℃ overnight. For BrdU immunostaining, the rehydrated embryos were digested with Proteinase K(12 μg/ml, Roche) at 30℃ for 28 minutes and treated with acetone at -20℃ for 30 minutes. After re-fixation with 4% PFA, the embryos were blocked with the block solution (PBS + 0.3% Triton-X -100 +1% DMSO+ 10 mg/ml BSA+10% normal goat serum) for 2 hours at RT. The embryos were then incubated with anti-GFP Rabbit Serum (1 : 500, Invitrogen) followed by goat-Alexa Fluore488-conjugated anti-rabbit secondary antibody (1 : 500, Invitrogen) incubation. 2N HCl was used to treat the embryos for 1 hour at room temperature (RT). After that, the embryos were stained with mouse anti-BrdU primary antibody (1 : 50, Roche) and goat-Alexa Fluore546-conjugated anti-mouse secondary antibody (1 : 500, Invitrogen). TUNEL assay was performed with the In Situ Cell Death Detection Kit TMR red (Roche). Similar to the BrdU immunostaining, 3.5dpf and 4dpf topbp1cas003 mutant/Tg(c-myb:EGFP) or sibling embryos were fixed with 4% PFA. After methanol dehydration, rehydration, Proteinase K digestion and acetone treatment, the embryos were permeated with permeabilisation solution (0.5% Triton X–100, 0.1% sodium citrate in PBS) at RT for 4 hours. Then the embryos were stained with the TUNEL Kit (100ul, enzyme: labeling solution = 1 : 9) at 37℃ for 2 hours. Finally, the EGFP immunostaining was carried out as described above.
Cell sorting, RNA extraction and quantitative PCR
The CD41+ cells were sorted from the tails of Tg(CD41: EGFP) embryos including the CHT region at 5dpf as previously described [86,87]. The total RNA was extracted from TRIzol (invitrogen) dissolved zebrafish whole embryos or the tails including CHT region or the sorted cells, and then transcribed into cDNA by PrimerScript RT Master Mix (TaKaRa). The quantitative PCR was carried out with SYBR Green Real-time PCR Master Mix (TOYOBO) with ABI 7900HT real-time PCR machine, and analyzed with Graphpad 5.1 software. The primers used were listed in S1 Table.
Cell culture, plasmid transfection and immunostaining
HeLa and HEK293T cells were maintained in DMEM with 10% Fetal Bovine Serum (FBS) and penicillin-streptomycin antibiotics (1 : 100). Plasmid transfection was carried out with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instruction. The immunostaining was carried out in HeLa cells as previously described [70]. FLAG-topbp1WT, FLAG-topbp1cas003 and FLAG-topbp1cas003-NLS plasmids were transfected into HeLa cells. Mouse anti-FLAG primary antibody (1 : 1000; Genomics Technology) and goat-Alexa Fluore488-conjugated anti-mouse secondary antibody (1 : 500) were used for immunostaining. DAPI (1 : 500, Beyotime) was applied for nucleus staining.
Protein extraction and immunoblotting analysis
To extract the protein from the cell line, the cells were homogenized directly with 2 X SDS sample buffer and boiled for 5 minutes at 95℃. To obtain fish protein from the CHT region, the tails of embryos including the CHT region were cut down, then ultrasonicated in RIPA lysis buffer (50mM Tris(pH7.4), 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). After centrifugation at 12000rpm for 15 minutes, the supernatant was mixed with 2XSDS sample buffer and boiled for 10 minutes. Cytoplasmic and nuclear extracts were prepared from the 3dpf embryos with Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime) according to the manufacturer’s instruction. The immunoblotting was carried out as previously described [85], with rabbit anti-phospho-Chk1 (Ser345) (133D) antibody (Cell Signaling Technology), rabbit anti-γH2AX antibody, rabbit anti-zebrafish TopBP1 antibody (generated by 840–940 amino acid of zebrafish TopBP1 protein as antigen), mouse anti-GAPDH antibody (1D4) (Santa Cruz), mouse anti-alpha-tubulin antibody (Sigma) or rabbit anti-Histon3 (H3) antibody (Abcam).
Imaging
Images of zebrafish immunofluorescence staining or live transgenic embryos were taken by Olympus FV1000 scanning confocal microscope. The embryos were mounted in 1% low-melt agarose in a self-made 35mm coverslip-bottom dish. The confocal images were captured with an UPLSAPO 20X or 60X objective. To obtain images of HeLa cells immunostaining, the slides were directly immersed in the PBS solution in a 10cm dish. The images were captured with an UPLSAPO 40X objective. The transient transgenesis embryos and embryos for bright field imaging were anesthetized with 0.03% Tricaine (Sigma-Aldrich), mounted in 3% methylcellulose and imaged using a Zeiss Axio Zoom. V16 microscope equipped with a Zeiss AxioCam MRm digital camera.
Statistics analysis
Data were analyzed with the Graphpad Prism 5 software using the two-tailed Student’s t-test. The plot error values were calculated by standard error of the mean (SEM). All data in this study were repeated for at least twice.
Supporting Information
Zdroje
1. Orkin SH, Zon LI (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132 : 631–644. doi: 10.1016/j.cell.2008.01.025 18295580
2. Chen J (2005) Senescence of hematopoietic stem cells and bone marrow failure. Int J Hematol 82 : 190–195. 16207589
3. Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414 : 105–111. 11689955
4. Bereshchenko O, Mancini E, Moore S, Bilbao D, Mansson R, et al. (2009) Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell 16 : 390–400. doi: 10.1016/j.ccr.2009.09.036 19878871
5. Bolli N, Payne EM, Rhodes J, Gjini E, Johnston AB, et al. (2011) cpsf1 is required for definitive hematopoietic stem cell survival in zebrafish. Blood 117 : 3996–4007. doi: 10.1182/blood-2010-08-304030 21330472
6. Komeno Y, Kitaura J, Kitamura T (2009) Molecular bases of myelodysplastic syndromes: lessons from animal models. J Cell Physiol 219 : 529–534. doi: 10.1002/jcp.21739 19259975
7. Jing LL, Zon LI (2011) Zebrafish as a model for normal and malignant hematopoiesis. Disease Models and Mechanisms 4 : 433–438. doi: 10.1242/dmm.006791 21708900
8. Jagannathan-Bogdan M, Zon LI (2013) Hematopoiesis. Development 140 : 2463–2467. doi: 10.1242/dev.083147 23715539
9. Xu J, Du L, Wen Z (2012) Myelopoiesis during zebrafish early development. J Genet Genomics 39 : 435–442. doi: 10.1016/j.jgg.2012.06.005 23021543
10. Wang K, Huang Z, Zhao L, Liu W, Chen X, et al. (2012) Large-scale forward genetic screening analysis of development of hematopoiesis in zebrafish. J Genet Genomics 39 : 473–480. doi: 10.1016/j.jgg.2012.07.008 23021547
11. Song HD, Sun XJ, Deng M, Zhang GW, Zhou Y, et al. (2004) Hematopoietic gene expression profile in zebrafish kidney marrow. Proc Natl Acad Sci U S A 101 : 16240–16245. 15520368
12. Chen AT, Zon LI (2009) Zebrafish blood stem cells. J Cell Biochem 108 : 35–42. doi: 10.1002/jcb.22251 19565566
13. Kissa K, Herbomel P (2010) Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464 : 112–U125. doi: 10.1038/nature08761 20154732
14. Bertrand JY, Chi NC, Santoso B, Teng ST, Stainier DYR, et al. (2010) Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464 : 108–U120. doi: 10.1038/nature08738 20154733
15. Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, et al. (2006) Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25 : 963–975. 17157041
16. Jin H, Xu J, Wen Z (2007) Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood 109 : 5208–5214. 17327398
17. Pietras EM, Warr MR, Passegue E (2011) Cell cycle regulation in hematopoietic stem cells. J Cell Biol 195 : 709–720. doi: 10.1083/jcb.201102131 22123859
18. Zeman MK, Cimprich KA (2014) Causes and consequences of replication stress. Nature cell biology 16 : 2–9. doi: 10.1038/ncb2897 24366029
19. Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40 : 179–204. doi: 10.1016/j.molcel.2010.09.019 20965415
20. Marechal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5.
21. Zhang S, Yajima H, Huynh H, Zheng J, Callen E, et al. (2011) Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol 193 : 295–305. doi: 10.1083/jcb.201009074 21482716
22. Niedernhofer LJ (2008) DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair (Amst) 7 : 523–529.
23. Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, et al. (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447 : 725–729. 17554309
24. Gari K, Constantinou A (2009) The role of the Fanconi anemia network in the response to DNA replication stress. Critical reviews in biochemistry and molecular biology 44 : 292–325. doi: 10.1080/10409230903154150 19728769
25. Schwab RA, Blackford AN, Niedzwiedz W (2010) ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J 29 : 806–818. doi: 10.1038/emboj.2009.385 20057355
26. Deans AJ, West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11 : 467–480. doi: 10.1038/nrc3088 21701511
27. Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, et al. (2001) BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem 276 : 30399–30406. 11395493
28. Garcia V, Furuya K, Carr AM (2005) Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4 : 1227–1239.
29. Mueller AC, Keaton MA, Dutta A (2011) DNA replication: mammalian Treslin-TopBP1 interaction mirrors yeast Sld3-Dpb11. Current biology: CB 21: R638–640. doi: 10.1016/j.cub.2011.07.004 21855008
30. Kim JE, McAvoy SA, Smith DI, Chen J (2005) Human TopBP1 ensures genome integrity during normal S phase. Mol Cell Biol 25 : 10907–10915. 16314514
31. Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124 : 943–955. 16530042
32. Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9 : 616–627. doi: 10.1038/nrm2450 18594563
33. Flynn RL, Zou L (2011) ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem Sci 36 : 133–140. doi: 10.1016/j.tibs.2010.09.005 20947357
34. Zhao H, Piwnica-Worms H (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 21 : 4129–4139. 11390642
35. Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, et al. (2006) Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26 : 6056–6064. 16880517
36. Burrows AE, Elledge SJ (2008) How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev 22 : 1416–1421. doi: 10.1101/gad.1685108 18519633
37. Sokka M, Parkkinen S, Pospiech H, Syvaoja JE (2010) Function of TopBP1 in genome stability. Subcell Biochem 50 : 119–141. doi: 10.1007/978-90-481-3471-7_7 20012580
38. Lee J, Kumagai A, Dunphy WG (2007) The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282 : 28036–28044. 17636252
39. Gong Z, Kim JE, Leung CC, Glover JN, Chen J (2010) BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell 37 : 438–446. doi: 10.1016/j.molcel.2010.01.002 20159562
40. Leung CC, Gong Z, Chen J, Glover JN (2011) Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J Biol Chem 286 : 4292–4301. doi: 10.1074/jbc.M110.189555 21127055
41. Wang J, Gong Z, Chen J (2011) MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 193 : 267–273. doi: 10.1083/jcb.201010026 21482717
42. Leung CC, Sun L, Gong Z, Burkat M, Edwards R, et al. (2013) Structural insights into recognition of MDC1 by TopBP1 in DNA replication checkpoint control. Structure 21 : 1450–1459. doi: 10.1016/j.str.2013.06.015 23891287
43. Zhou ZW, Liu C, Li TL, Bruhn C, Krueger A, et al. (2013) An essential function for the ATR-activation-domain (AAD) of TopBP1 in mouse development and cellular senescence. PLoS Genet 9: e1003702. doi: 10.1371/journal.pgen.1003702 23950734
44. Jeon Y, Ko E, Lee KY, Ko MJ, Park SY, et al. (2011) TopBP1 deficiency causes an early embryonic lethality and induces cellular senescence in primary cells. J Biol Chem 286 : 5414–5422. doi: 10.1074/jbc.M110.189704 21149450
45. Lee Y, Katyal S, Downing SM, Zhao J, Russell HR, et al. (2012) Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat Neurosci 15 : 819–826. doi: 10.1038/nn.3097 22522401
46. Kim J, Lee SK, Jeon Y, Kim Y, Lee C, et al. (2014) TopBP1 deficiency impairs V(D)J recombination during lymphocyte development. EMBO J 33 : 217–228. doi: 10.1002/embj.201284316 24442639
47. Greig KT, Carotta S, Nutt SL (2008) Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol 20 : 247–256. doi: 10.1016/j.smim.2008.05.003 18585056
48. Burns CE, Galloway JL, Smith AC, Keefe MD, Cashman TJ, et al. (2009) A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood 113 : 5776–5782. doi: 10.1182/blood-2008-12-193607 19332767
49. North TE, Goessling W, Peeters M, Li P, Ceol C, et al. (2009) Hematopoietic stem cell development is dependent on blood flow. Cell 137 : 736–748. doi: 10.1016/j.cell.2009.04.023 19450519
50. Lawson ND, Weinstein BM (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248 : 307–318. 12167406
51. Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI (2005) Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev 19 : 2331–2342. 16166372
52. North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, et al. (2007) Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447 : 1007–1011. 17581586
53. Yamane K, Kawabata M, Tsuruo T (1997) A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem 250 : 794–799. 9461304
54. Jing CB, Chen Y, Dong M, Peng XL, Jia XE, et al. (2013) Phospholipase C gamma-1 is required for granulocyte maturation in zebrafish. Dev Biol 374 : 24–31. doi: 10.1016/j.ydbio.2012.11.032 23220656
55. Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, et al. (2011) Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138 : 169–177. doi: 10.1242/dev.059345 21138979
56. Kim JH, Lee SR, Li LH, Park HJ, Park JH, et al. (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. Plos One 6: e18556. doi: 10.1371/journal.pone.0018556 21602908
57. Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, et al. (2003) Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell 5 : 903–914. 14667412
58. Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, et al. (2005) tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A 102 : 407–412. 15630097
59. Stern HM, Murphey RD, Shepard JL, Amatruda JF, Straub CT, et al. (2005) Small molecules that delay S phase suppress a zebrafish bmyb mutant. Nat Chem Biol 1 : 366–370. 16372403
60. Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, et al. (2008) Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet 4: e1000240. doi: 10.1371/journal.pgen.1000240 18974873
61. Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21 : 1472–1477. 17575048
62. Rappas M, Oliver AW, Pearl LH (2011) Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 39 : 313–324. doi: 10.1093/nar/gkq743 20724438
63. Duursma AM, Driscoll R, Elias JE, Cimprich KA (2013) A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 50 : 116–122. doi: 10.1016/j.molcel.2013.03.006 23582259
64. Ward IM, Chen J (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem 276 : 47759–47762. 11673449
65. Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A 110 : 13904–13909. doi: 10.1073/pnas.1308335110 23918387
66. Liu D, Wang Z, Xiao A, Zhang Y, Li W, et al. (2014) Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J Genet Genomics 41 : 43–46. doi: 10.1016/j.jgg.2013.11.004 24480746
67. Zhang Y, Duc AC, Rao S, Sun XL, Bilbee AN, et al. (2013) Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev Cell 24 : 411–425. doi: 10.1016/j.devcel.2013.01.018 23449473
68. Ma D, Zhang J, Lin HF, Italiano J, Handin RI (2011) The identification and characterization of zebrafish hematopoietic stem cells. Blood 118 : 289–297. doi: 10.1182/blood-2010-12-327403 21586750
69. Wang L, Zhang Y, Zhou T, Fu YF, Du TT, et al. (2008) Functional characterization of Lmo2-Cre transgenic zebrafish. Dev Dyn 237 : 2139–2146. doi: 10.1002/dvdy.21630 18627109
70. Du L, Xu J, Li X, Ma N, Liu Y, et al. (2011) Rumba and Haus3 are essential factors for the maintenance of hematopoietic stem/progenitor cells during zebrafish hematopoiesis. Development 138 : 619–629. doi: 10.1242/dev.054536 21228005
71. Li X, Lan Y, Xu J, Zhang W, Wen Z (2012) SUMO1-activating enzyme subunit 1 is essential for the survival of hematopoietic stem/progenitor cells in zebrafish. Development 139 : 4321–4329. doi: 10.1242/dev.081869 23132242
72. Dokal I (2012) Heightened DNA damage response impairs hematopoiesis in Fanconi anemia. Haematologica 97 : 1117. doi: 10.3324/haematol.2012.073643 22855843
73. Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, et al. (2012) Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11 : 36–49. doi: 10.1016/j.stem.2012.05.013 22683204
74. Rhodes J, Amsterdam A, Sanda T, Moreau LA, McKenna K, et al. (2009) Emi1 maintains genomic integrity during zebrafish embryogenesis and cooperates with p53 in tumor suppression. Mol Cell Biol 29 : 5911–5922. doi: 10.1128/MCB.00558-09 19704007
75. Li L, Chang W, Yang G, Ren C, Park S, et al. (2014) Targeting poly(ADP-ribose) polymerase and the c-Myb-regulated DNA damage response pathway in castration-resistant prostate cancer. Science signaling 7: ra47.
76. Lin HF, Traver D, Zhu H, Dooley K, Paw BH, et al. (2005) Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood 106 : 3803–3810. 16099879
77. Westerfield M (2000) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene: Univ. of Oregon Press.
78. Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, et al. (1996) The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123 : 1–36. 9007226
79. Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, et al. (1996) A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123 : 37–46. 9007227
80. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203 : 253–310. 8589427
81. Bahary N, Davidson A, Ransom D, Shepard J, Stern H, et al. (2004) The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol 77 : 305–329. 15602919
82. Borovina A, Superina S, Voskas D, Ciruna B (2010) Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nature cell biology 12 : 407–412. doi: 10.1038/ncb2042 20305649
83. Lanford RE, Kanda P, Kennedy RC (1986) Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell 46 : 575–582. 3015419
84. Suster ML, Kikuta H, Urasaki A, Asakawa K, Kawakami K (2009) Transgenesis in zebrafish with the tol2 transposon system. Methods Mol Biol 561 : 41–63. doi: 10.1007/978-1-60327-019-9_3 19504063
85. Ren CG, Wang L, Jia XE, Liu YJ, Dong ZW, et al. (2013) Activated N-Ras signaling regulates arterial-venous specification in zebrafish. J Hematol Oncol 6 : 34. doi: 10.1186/1756-8722-6-34 23663822
86. Wang L, Fu C, Fan H, Du T, Dong M, et al. (2013) miR-34b regulates multiciliogenesis during organ formation in zebrafish. Development 140 : 2755–2764. doi: 10.1242/dev.092825 23698347
87. Julien Y. Bertrand ADK, Shutian Teng and David Traver (2008) CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development 135 : 1853–1862. doi: 10.1242/dev.015297 18417622
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



