-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
Primer-dependent transcription initiation, PDI, refers to an alternative mechanism of transcription initiation whereby the first phosphodiester bond within the nascent RNA is formed between a 2 - to ~4-nt RNA primer and an incoming nucleoside triphosphate. Although PDI has been shown to occur in E. coli, the impact of PDI on E. coli physiology, and the extent to which PDI occurs in other bacteria is unknown. Here we establish that PDI modulates the ability of E. coli to form biofilms, a surface attached community of bacteria encased in a polymeric matrix. We further describe a significantly improved RNA-seq based method for the detection of PDI in cells. Using this method we document the occurrence of PDI in the pathogenic bacterium Vibrio cholerae. We further show that the pattern of PDI in V. cholerae is identical to that observed in E. coli, suggesting that PDI in these two organisms may occur through a conserved process that produces identical populations of 2 - to ~4-nt RNA primers. Our findings suggest PDI may be widespread in Gammaproteobacteria.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005348
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005348Summary
Primer-dependent transcription initiation, PDI, refers to an alternative mechanism of transcription initiation whereby the first phosphodiester bond within the nascent RNA is formed between a 2 - to ~4-nt RNA primer and an incoming nucleoside triphosphate. Although PDI has been shown to occur in E. coli, the impact of PDI on E. coli physiology, and the extent to which PDI occurs in other bacteria is unknown. Here we establish that PDI modulates the ability of E. coli to form biofilms, a surface attached community of bacteria encased in a polymeric matrix. We further describe a significantly improved RNA-seq based method for the detection of PDI in cells. Using this method we document the occurrence of PDI in the pathogenic bacterium Vibrio cholerae. We further show that the pattern of PDI in V. cholerae is identical to that observed in E. coli, suggesting that PDI in these two organisms may occur through a conserved process that produces identical populations of 2 - to ~4-nt RNA primers. Our findings suggest PDI may be widespread in Gammaproteobacteria.
Introduction
Transcription in all cells is carried out by multi-subunit RNA polymerases (RNAPs) that are conserved in sequence, structure, and function from bacteria to humans. The first step in transcription, initiation, consists of a number of discrete steps that culminate in the RNAP-mediated catalysis of the first phosphodiester bond formed within the nascent RNA [1, 2]. The first phosphodiester bond within the nascent RNA can be formed between two nucleoside triphosphate (NTP) substrates, “de novo initiation,” or between a 2 - to ~4-nt oligoribonucleotide primer and an incoming NTP, “primer-dependent initiation,” PDI. Although PDI had been long known to occur during transcription reactions performed in vitro (reviewed in [3]), PDI has only recently been shown to occur during the stationary phase of growth in Escherichia coli [4, 5]. In addition, the extent of PDI relative to de novo initiation at a given promoter in vivo can influence the overall abundance of transcripts produced from the promoter as well as the sequence and phosphorylation state of the 5′ ends of transcripts produced from the promoter [3–6].
To detect PDI in vivo we developed the experimental pipeline shown in Fig 1 [5] that is based on two experimental considerations. First, studies of RNA metabolism in bacteria indicate that 2 - to ~4-nt oligoribonucleotides (species that are sometimes referred to as “nanoRNAs”) are degraded in cells by specialized ribonucleases termed “oligoribonucleases (oligoRNase)” or “nanoRNases” [7–9]. Thus, by increasing the concentration of an oligoRNase in vivo we decrease the concentrations of 2 - to ~4-nt oligoribonucleotides in vivo (Fig 1A). Second, in vitro analyses indicate that 2 - to 4-nt oligoribonucleotides effectively compete with NTPs for use as transcription primers provided the 5′ end of the RNA is complementary to sequences between positions −3 and +1 (where +1 is the position of de novo initiation) and the 3′ end is complementary to positions +1, +2 or +3 [6, 10–15]. Thus, PDI with 2 - to 4-nt oligoribonucleotides in vitro leads to the generation of transcripts emanating from template position +1 or template positions upstream of +1 (−3, −2, or −1). To unambiguously distinguish transcripts generated by PDI from those generated by de novo initiation we use high-throughput sequencing of RNA 5′ ends (5′ RNA-seq) [16] to first, identify the primary de novo start sites associated with promoters genome-wide, and second, identify transcripts that emanate from template positions upstream of these primary de novo start sites whose abundance decreases upon ectopic expression of an oligoRNase (Fig 1B and 1C).
Fig. 1. Detection of PDI in bacteria by ectopic expression of an oligoRNase coupled with 5′ RNA-seq. 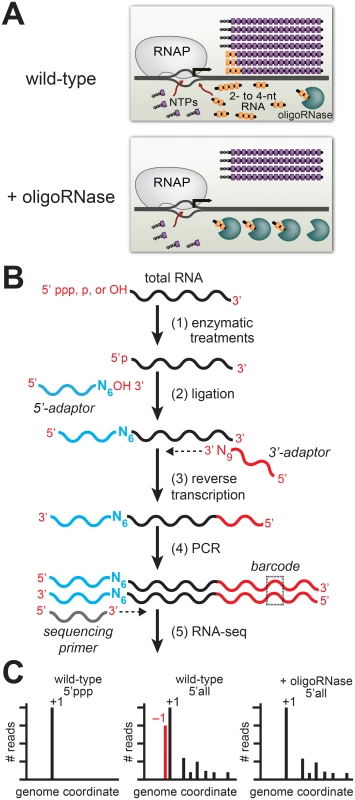
A. Inhibition of PDI through ectopic expression of an oligoRNase. Depicted is the extent of de novo initiation and PDI from a representative promoter in wild-type cells (top) and cells in which an oligoRNase is ectopically produced (bottom). 2- to 4-nt oligoribonucleotides, are depicted in yellow while NTPs are shown in purple. In the example shown here, PDI leads to generation of full-length transcripts that do not carry a 5′ triphosphate and are one base longer than the products of de novo initiation. B. Steps in 5′ RNA-seq [16]. Step 1: selective enzymatic treatments of total RNA that allow cDNA libraries to be constructed from RNAs on the basis of the phosphorylation state of the 5′ end. Step 2: ligation of 5′ adaptor (in blue) carrying six random bases (N6) at the 3′ end. Step 3: Reverse transcription using a primer with sequence of the Illumina 3′ adaptor on the 5′ end and nine random bases at the 3′ end (N9). Step 4: PCR step includes a primer that introduces a barcode (dashed box) enabling several libraries to be analyzed, in parallel, on the Illumina HiSeq. C. Histograms generated from 5′ RNA-seq. Analysis of transcripts with a 5′ triphosphate (5′ ppp) is used to identify primary start sites (+1). Comparison of results obtained from wild-type cells or cells in which an oligoRNase is ectopically expressed using the analysis of the 5′ ends of all transcripts (i.e. those carrying a 5′-triphosphate, 5′-monophosphate, or 5′-hydroxyl) identifies transcripts generated by PDI that emanate from position −1 (red). Our 5′ RNA-seq procedure facilitates the analysis of both the sequence and phosphorylation state of the portion of the transcriptome comprising the 5′ ends of RNAs. Because transcripts generated by de novo initiation carry a 5′ triphosphate the analysis of transcripts that carry a triphosphate group can be used to identify primary de novo start sites (designated position +1), each with its associated “start site region” (i.e. positions −3 to +4) (Fig 1C, left histogram). Next, we use the analysis of the 5′ ends of all transcripts (i.e. those carrying a 5′-triphosphate, 5′-monophosphate, or 5′-hydroxyl) to determine the effect of ectopic expression of an oligoRNase on the fraction of transcripts initiated from positions upstream of +1 within each start site region (Fig 1C, middle and right histograms). The inclusion of 5′-monophosphate - and 5′-hydroxyl-containing transcripts in the analysis allows us to identify PDI events that involve a primer carrying either a 5′-monophosphate or a 5′-hydroxyl. Using this experimental pipeline we established that PDI occurs in E. coli and is growth phase-dependent [4, 5]. Specifically, we found that PDI is detected during stationary phase but is not detected during exponential phase. In addition, we found that the growth phase-dependent PDI detected in E. coli leads to a significant increase in the stationary phase expression of at least two genes, bhsA and tomB [5].
Although PDI has been shown to occur in E. coli, the impact of PDI on E. coli physiology, and the extent to which PDI occurs in other bacteria is unknown. Thus, determining the full scope of PDI in E. coli and other bacteria, the mechanisms allowing or restricting PDI in E. coli and other bacteria, the specificity with which PDI is targeted to specific promoters in E. coli and other bacteria, and the role that PDI plays in cell growth in E. coli and other bacteria are significant open questions.
Here we present evidence that, in E. coli, the PDI-dependent increase in bhsA expression that occurs during stationary phase contributes to biofilm formation. Thus, our findings illuminate a previously undocumented physiological role for PDI in E. coli as a regulatory mechanism involved in biofilm growth. In addition, using a modified version of our experimental pipeline that employs a more sensitive 5′ RNA-seq protocol, we document the occurrence of PDI in the pathogenic Gammaproteobacteria Vibrio cholerae. We further find that the pattern of PDI observed in V. cholerae is strikingly similar to that observed in E. coli. Specifically, PDI in both V. cholerae and E. coli is detected during stationary phase, is not detected during exponential phase, and is preferentially targeted to promoters carrying start site regions with the sequence T−1A+1 or G−1G+1 (where position +1 corresponds to the position of de novo initiation). PDI from promoters carrying T−1A+1 and G−1G+1 start site regions produces full-length transcripts emanating from position −1 that carry a 5′ hydroxyl and begin with the sequence 5′-UA and 5′-GG, respectively. Thus, the primers used to generate these transcripts must themselves begin with the sequence 5′-UA and 5′-GG, respectively. We propose that a growth phase-dependent process that preferentially generates 5′-UA and 5′-GG oligoribonucleotides occurs in both E. coli and V. cholerae.
Results
PDI contributes to biofilm formation in E. coli
In prior work [5], we showed that PDI in E. coli leads to an increase in the stationary phase expression of bhsA, a gene that encodes a small outer membrane protein. In particular, we found that ~80% of transcripts emanating from a promoter associated with bhsA during stationary phase are produced as a consequence of PDI. Furthermore, ectopic expression of an oligoRNase reduces the expression of bhsA ~4-fold during the stationary phase of growth but does not alter bhsA expression during exponential phase.
Work from others has shown that bhsA influences the ability of E. coli cells to form biofilms [17], a mode of growth that results in the formation of a surface attached community of bacteria encased in a polymeric matrix. We therefore sought to address whether or not the changes in bhsA expression that occur as a consequence of PDI contribute to the ability of E. coli cells to form biofilms. To do this, we determined the effect of ectopic expression of a heterologous oligoRNase, Bacillus subtilis NrnB [9], or a catalytically inactive mutant, NrnBDHH [9], on biofilm formation in wild-type E. coli, or a mutant strain in which bhsA has been deleted (ΔbhsA). We placed cell suspensions into wells of a microtiter plate, allowed biofilms to form by incubating for 32 hours at 25°C without agitation, and quantified the extent of biofilm formation using a standard crystal violet staining assay [18] (Fig 2). Ectopic expression of NrnB, but not NrnBDHH, caused a ~2-fold reduction in biofilm formation in wild-type E. coli (Fig 2). Furthermore, deletion of bhsA also caused a ~2-fold reduction in biofilm formation (Fig 2). However, in contrast to what we found in wild-type E. coli, ectopic expression of NrnB in ΔbhsA cells had no effect on the ability of these cells to form biofilms (Fig 2). Thus, the reduction in biofilm formation observed upon ectopic expression of NrnB in wild-type cells requires the presence of BhsA. We conclude that the increase in bhsA expression that occurs as a consequence of PDI contributes to biofilm formation in wild-type E. coli. These findings reveal a specific role for PDI as a regulatory mechanism that modulates biofilm formation in E. coli.
Fig. 2. The PDI-dependent increase in bhsA expression contributes to biofilm formation in E. coli. 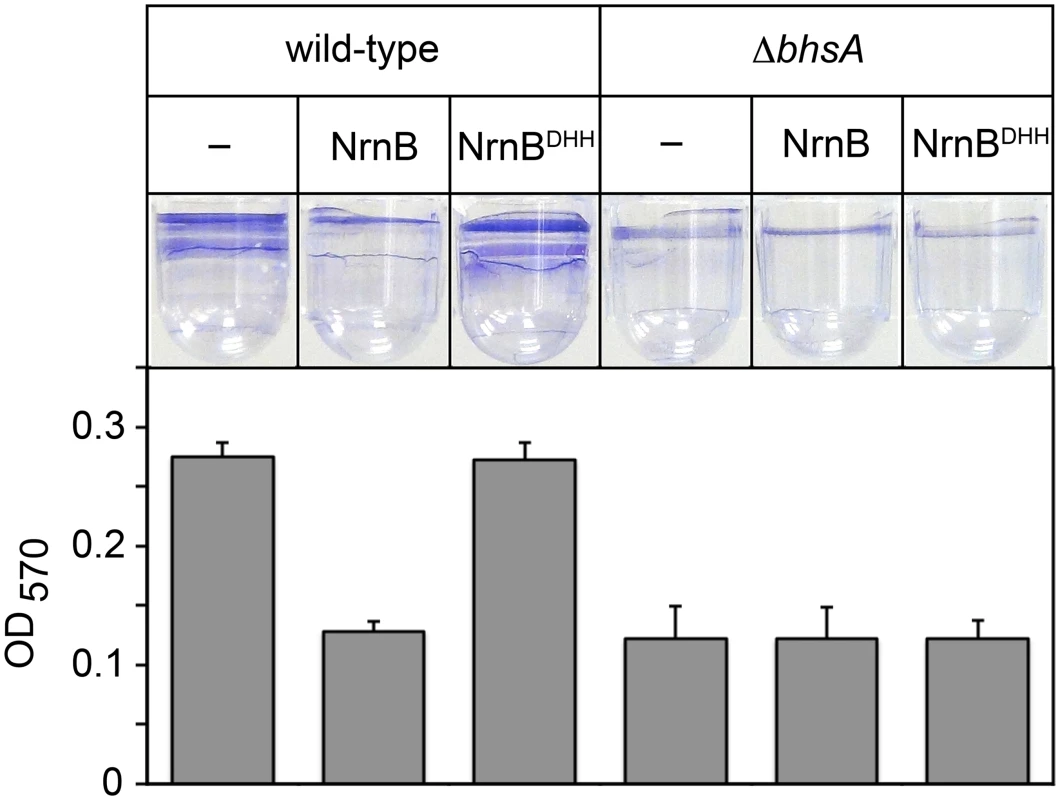
Top shows images of representative wells containing biofilms stained with crystal violet dye. Graph shows averages +SEM of the amount of crystal violet staining observed from at least four independent measurements. Assays were done using wild-type MG1655 cells or MG1655 cells carrying a deletion of bhsA (ΔbhsA). Cells carried an empty plasmid (–) or a plasmid that directs the synthesis of NrnB or NrnBDHH. Analysis of PDI in Vibrio cholerae by ectopic expression of an oligoRNase coupled with 5′ RNA-seq
Having established that PDI in E. coli can impact stationary phase gene expression [5] and cell physiology (Fig 2) we wished to investigate the occurrence of PDI in other bacteria. We therefore sought to define the extent of PDI in the pathogenic Gammaproteobacteria V. cholerae by employing the experimental pipeline used to detect PDI in E. coli (Fig 1). Thus we determined the effect of ectopically producing NrnB in V. cholerae on the distribution of transcription start sites as detected by 5′ RNA-seq.
For our analysis of PDI in V. cholerae we employed a revised 5′ RNA-seq protocol [16] that differed from that employed in our prior analysis of PDI in E. coli [5]. In particular, the 5′ and 3′ adaptors used in the revised 5′ RNA-seq protocol enable construction of cDNA compatible with analysis on the Illumina HiSeq system, whereas our prior E. coli analysis was done using 5′ and 3′ adaptors that enable construction of cDNA libraries compatible with the Applied Biosystems SOLiD system. In addition, the 5′ adaptor used in the revised protocol carries six randomized bases at the 3′ end (see Fig 1B), whereas our prior E. coli analysis was done using a 5′ adaptor that did not carry randomized bases at the 3′ end.
Having previously found that PDI in E. coli occurs during the stationary phase of growth we determined the effect of ectopically producing NrnB on the distribution of transcription start sites in V. cholerae during stationary phase. We identified several start site regions where the percentage of transcripts emanating from template positions upstream of the primary de novo start site was reduced by ectopic expression of NrnB (S1 and S2 Tables). Among these start site regions, we identified 10 where ectopic expression of NrnB reduced the percentage of transcripts emanating from upstream template positions by >25% (S1 Table).
To determine whether or not NrnB-sensitive transcripts in V. cholerae were preferentially generated from promoters carrying particular start site region sequences, we selected 1226 start site regions containing an unambiguous primary de novo start site (unambiguous Transcription Start site Regions, uTSRs). Specifically, uTSRs were identified as those where >75% of the total transcripts within the start site region emanate from position +1 in the analysis of triphosphate 5′ ends in cells containing wild-type concentrations of 2 - to ~4-nt RNAs (S2 Table). As expected, given the bias for use of a purine NTP during de novo initiation observed in E. coli [19–21], we found that 1125 of the 1226 uTSRs (~92%) carried either an A or G at position +1. On average, in cells containing wild-type concentrations of 2 - to ~4-nt RNAs, ~4% of the transcripts associated with these uTSRs emanated from position −1 in the analysis of all 5′ ends (Fig 3A). Furthermore, the proportion of transcripts initiating from position −1 was slightly reduced (to ~3%) in cells in which an NrnB was ectopically expressed (Fig 3A). We next separated the 1125 uTSRs where position +1 was an A or G into eight distinct classes on the basis of the identity of the bases at positions −1 and +1 and calculated the change in the percentage of transcripts emanating from position −1 upon ectopic expression of NrnB (Nrn effect) of each class (Fig 3A). Parsing the start site regions in this manner revealed that NrnB-sensitive transcripts initiating from position −1 are preferentially generated from uTSRs carrying T−1A+1 (an average Nrn effect of ~5% of the total transcripts for T−1A+1 start site regions versus an average Nrn effect of ~1% for all start sites regions) and G−1G+1 (an average Nrn effect of ~4% for G−1G+1 start site regions). Analysis of RNA transcripts isolated from cells carrying wild-type concentrations of 2 - to ~4-nt RNAs during exponential phase indicates that the proportion of transcripts emanating from position −1 of T−1A+1 and G−1G+1 start site regions is significantly higher during stationary phase (9.6% for T−1A+1 and 6.8% for G−1G+1) compared with exponential phase (0.8% for T−1A+1 and 2.7% for G−1G+1) (Fig 3B and S3 Table), suggesting that the generation of NrnB-sensitive transcripts in V. cholerae is growth phase-dependent.
Fig. 3. Detection of PDI in V. cholera. 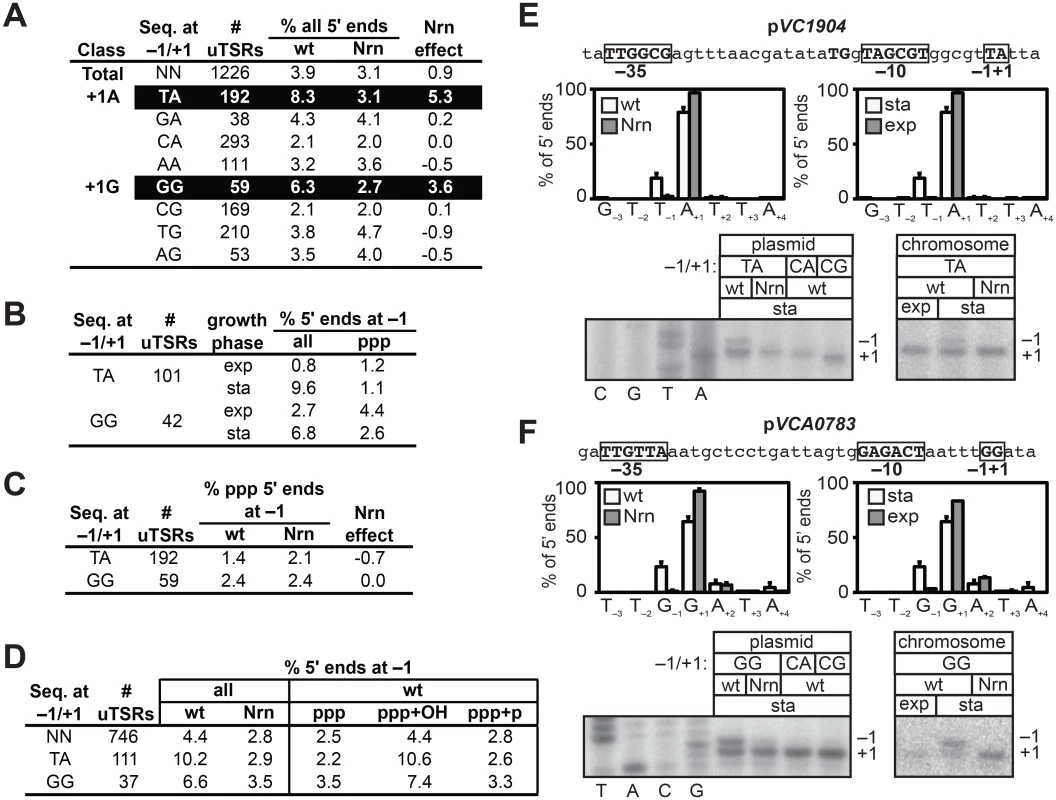
A-D. Analysis of PDI by 5′ RNA-seq. Percentage of transcripts emanating from position −1 of uTSRs (S2 and S3 and S4 Tables) with the indicated sequence at −1 and +1 in cells carrying wild-type concentrations of 2- to ~4-nt RNAs (wild-type) or cells in which the oligoRNase NrnB was ectopically expressed (Nrn). The Nrn effect reported in panels A and C represents the difference in these values. uTSRs with above average Nrn effect are highlighted in black. The total number of uTSRs used to calculate the percentages is indicated. Values in panels A and B are calculated from biological replicates listed in S2 Table, values in panel C is calculated from replicates listed in S3 Table and values in panel D are calculated from replicates listed in S4 Table. exp, exponential phase; sta, stationary phase; all, analysis of all 5′ ends; ppp, analysis of 5′ triphosphate ends; ppp+OH, analysis of 5′ triphosphate ends and 5′ hydroxyl ends; ppp+p, analysis of 5′ triphosphate ends and 5′ monophosphate ends. E. and F. Analysis of PDI at the promoters associated with VC1904 (pVC1904) and VCA0783 (pVCA0783). Top of each panel shows promoter sequence. Indicated are positions +1, −1 and the promoter −10 and −35 elements. Middle of each panel shows results of 5′ RNA-seq. Graphs on left show average distribution of all 5′ ends between positions −3 and +4 in cells carrying wild-type concentrations of 2- to ~4-nt RNAs (wt) or cells in which the oligoRNase NrnB was ectopically expressed (Nrn) as detected by 5′ RNA-seq during stationary phase. Graphs on the right show average distribution of all 5′ ends between positions −3 and +4 for pVCA0783 in wild-type cells during stationary phase (sta) or exponential phase (exp). Bottom of each panel shows primer extension analysis of plasmid-borne promoter variants carrying the indicated sequences at positions −1/+1 (left) or primer extension analysis of chromosomally encoded promoters during exponential phase (exp) or stationary phase (sta). The NrnB-sensitive transcripts initiating from position −1 were detected in the analysis of all transcripts but were not detected in the analysis of transcripts carrying only 5′ triphosphate ends (Fig 3C). These findings indicated that the NrnB-sensitive transcripts identified in V. cholerae carried a 5′ hydroxyl or a 5′ monophosphate. We therefore repeated the 5′ RNA-seq analysis in a manner that would enable us to distinguish between transcripts carrying a 5′ hydroxyl and those carrying a 5′ monophosphate. Specifically, we analyzed transcripts carrying either a 5′ triphosphate or a 5′ monophosphate (ppp + p) or transcripts carrying either a 5′ triphosphate or a 5′ hydroxyl (ppp + OH). The NrnB-sensitive transcripts emanating from position −1 of T−1A+1 and G−1G+1 start site regions were only detected when 5′ hydroxyl transcripts were included in the analysis (Fig 3D and S4 Table). Thus, we conclude that the NrnB-sensitive transcripts emanating from position −1 of T−1A+1 and G−1G+1 start site regions carry a 5′ hydroxyl group.
To validate the 5′ RNA-seq analysis we performed primer extension to analyze transcripts generated from a promoter carrying a T−1A+1 start site region, pVC1904 (Fig 3E, top), and a promoter carrying a G−1G+1 start site region, pVCA0783 (Fig 3F, top). Consistent with the results of 5′ RNA-seq (Fig 3E and 3F, middle), growth phase-dependent NrnB-sensitive transcripts emanating from position −1 of pVC1904 and position −1 of pVCA0783 were detected by primer extension (Fig 3E and 3F, bottom). In addition, transcripts emanating from position −1 were not detected in the analysis of mutant derivatives of each promoter in which the wild-type sequence at positions −1 and +1 was changed to either C−1A+1 or C−1G+1 (Fig 3E and 3F, bottom). Thus, on the basis of both 5′ RNA-seq and primer extension analysis we conclude that growth phase-dependent PDI occurs in V. cholerae, that PDI in V. cholerae is preferentially apparent at promoters carrying T−1A+1 and G−1G+1 start site regions, and that PDI in V. cholerae leads to the generation of transcripts that carry a 5′ hydroxyl group.
Reanalysis of PDI in E. coli reveals preferential targeting of both T−1A+1 and G−1G+1 start site regions
The preferential generation of oligoRNase-sensitive transcripts from T−1A+1 start site regions was observed in our prior analysis of PDI in E. coli [5], while the preferential generation of oligoRNase-sensitive transcripts from G−1G+1 start site regions was not (S5 and S6 Tables). Because our V. cholerae analysis used a 5′ RNA-seq protocol [16] that had been modified subsequent to our analysis of PDI in E. coli, we sought to establish whether or not the preferential targeting of G−1G+1 start site regions was a unique feature of PDI in V. cholerae or, alternatively, was not previously detected in E. coli due to technical limitations of our prior 5′ RNA-seq protocol [5]. We therefore reanalyzed the extent of PDI in E. coli during stationary phase using our modified 5′ RNA-seq protocol. As in our prior analysis [5], we identified several start site regions that contained NrnB-sensitive transcripts. Among these start site regions, we identified 50 where the proportion of transcripts emanating from template positions upstream of the primary start site (i.e. positions −3, −2, or −1) was reduced by >25% upon ectopic expression of NrnB (S7 Table).
To facilitate a direct comparison of the results obtained in E. coli with those obtained in V. cholerae (Fig 3A), we identified 401 uTSRs (S8 Table). On average, in cells carrying wild-type concentrations of 2 - to ~4-nt RNAs, ~11% of the transcripts associated with these 401 uTSRs emanated from position −1 in the analysis of all 5′ ends (Fig 4A). In contrast, the proportion of transcripts initiating from position −1 was reduced to ~4% in cells in which NrnB was ectopically expressed (Fig 4A). Thus, on average, ~7% of the total transcripts associated with a given uTSR are NrnB-sensitive in E. coli. Analysis of the 393 uTSRs where position +1 was an A or G revealed that, as in V. cholerae, NrnB-sensitive transcripts initiating from position −1 are preferentially generated from uTSRs carrying T−1A+1 (an average Nrn-effect of ~20% of the total transcripts for T−1A+1 start site regions versus an average Nrn-effect of ~7% for all start sites regions) and G−1G+1 (an average Nrn-effect of ~18% for G−1G+1 start site regions) (Fig 4A).
Fig. 4. Detection of PDI in E. coli. 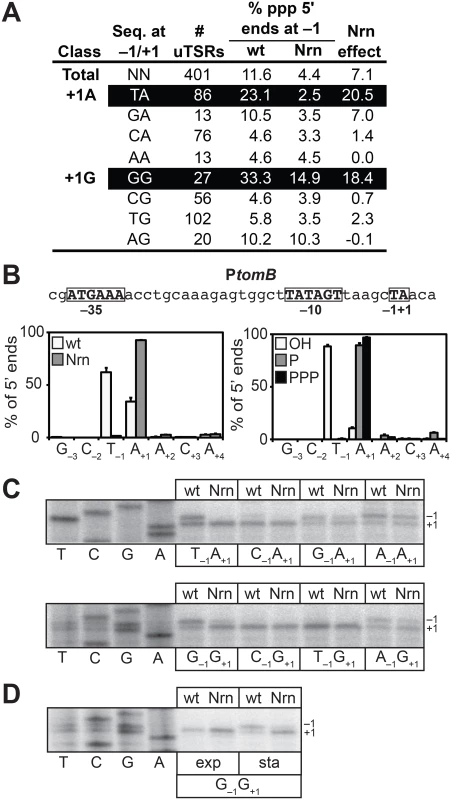
A. Analysis of PDI by 5′ RNA-seq. Percentage of transcripts emanating from position −1 of uTSRs with the indicated sequence at −1 and +1 in cells carrying wild-type concentrations of 2- to ~4-nt RNAs (wild-type) or cells in which the oligoRNase NrnB was ectopically expressed (Nrn). The Nrn effect represents the difference in these values. uTSRs with above average Nrn effect are highlighted in black. The total number of uTSRs used to calculate the percentages is indicated. Values are calculated from biological replicates listed in S8 Table. Data is derived from the analysis of all 5′ ends during stationary phase. B. Analysis of PDI at the promoter associated with tomB, ptomB, by 5′ RNA-seq. Sequence of ptomB is shown. Indicated are positions +1, −1 and the promoter −10 and −35 elements. Graph on the left shows average distribution of 5′ ends between positions −3 and +4 for ptomB in cells carrying wild-type concentrations of 2- to ~4-nt RNAs or cells in which the oligoRNase NrnB was ectopically expressed (Nrn) as detected by 5′ RNA-seq analysis of all 5′ ends during stationary phase. Graph on the right shows the average distribution of 5′ ends between positions −3 and +4 for ptomB in cells carrying wild-type concentrations of 2- to ~4-nt RNAs as detected by 5′ RNA-seq analysis of hydroxyl 5′ ends (OH), monophosphate 5′ ends (P), or triphosphate 5′ ends (PPP) during stationary phase. C. Primer extension analysis of plasmid-borne ptomB variants carrying the indicated sequence at −1 and +1 in cells carrying wild-type concentrations of 2- to ~4-nt RNAs (wt) or cells in which the oligoRNase NrnB was ectopically expressed (Nrn). D. Primer extension analysis of the plasmid-borne ptomB variant carrying the sequence G−1G+1 during exponential phase (exp) or stationary phase (sta). To validate the 5′ RNA-seq results obtained using our modified 5′ RNA-seq protocol we performed a systematic primer extension analysis of wild-type and mutant derivatives of the start site region associated with the tomB promoter (ptomB). We selected ptomB for this systematic analysis because it was among the promoters for which we detected NrnB-sensitive transcripts emanating from position −1 using both the prior version of the 5′ RNA-seq protocol [5] as well as the modified version (Fig 4B). The results show that the pattern of 5′ ends for ptomB derivatives observed using primer extension (Fig 4C) conforms to the rules inferred from the analysis of the 393 uTSRs using our modified 5′ RNA-seq protocol (Fig 4A). In particular, ~50% of the transcripts generated from the wild-type start site region (T−1A+1) or a mutant start site region carrying the sequence G−1G+1 emanate from position −1 and are significantly reduced upon ectopic expression of NrnB. In contrast, much lower levels of NrnB-sensitive transcripts emanating from position −1 are observed from derivatives with the sequence C−1A+1, C−1G+1, or T−1G+1. In addition, although a significant portion of transcripts generated from mutant derivatives carrying the sequence G−1A+1, A−1A+1, or A−1G+1 also emanate from position −1, only a small fraction of these transcripts appear NrnB-sensitive. Thus, on the basis of our 5′ RNA-seq and primer-extension analysis we conclude that PDI in E. coli, like in V. cholerae, is preferentially targeted to promoters carrying T−1A+1 and G−1G+1 start site regions. Because our prior 5′ RNA-seq protocol did not detect the preferential generation of oligoRNase-sensitive transcripts from G−1G+1 start site regions in E. coli [5] (S5 and S6 Tables), we further conclude that the modified 5′ RNA-seq protocol [16] is more sensitive and more accurate for the analysis of PDI in vivo. We suspect that the improved sensitivity of our modified 5′ RNA-seq protocol is a consequence of the use of a 5′ adaptor that carries six randomized bases at the 3′ end during Step 2 of the cDNA library construction (Fig 1B). Use of a 5′ adaptor carrying randomized bases at the 3′ end was included in our revised protocol to minimize the potential for sequence-dependent effects on ligation efficiency [22–25].
Our prior 5′ RNA-seq analysis in E. coli indicated that transcripts emanating from position −1 of T−1A+1 start site regions carry a 5′ hydroxyl [5]. To determine the phosphorylation state of the 5′ ends of transcripts emanating from position −1 of G−1G+1 start site regions we analyzed cDNA libraries generated from transcripts carrying only a 5′-hydroxyl or transcripts carrying only a 5′-monophosphate. As previously observed [5], transcripts emanating from position −1 of T−1A+1 start site regions were detected only in the analysis of transcripts carrying a 5′ hydroxyl (Fig 4B and S7 Table). Transcripts emanating from position −1 of G−1G+1 start site regions were also detected only in the analysis of transcripts carrying a 5′ hydroxyl (S1 Fig and S7 Table). We conclude that transcripts emanating from position −1 of G−1G+1 start site regions carry a 5′-hydroxyl, indicating that the primers used to generate these transcripts also carry a 5′-hydroxyl group in E. coli.
We have previously shown that oligoRNase-sensitive transcripts emanating from position −1 of T−1A+1 start site regions are not detected during exponential phase [5]. Primer extension analysis of a tomB promoter derivative carrying a G−1G+1 start site region indicates that transcripts emanating from position −1 are also not detected during exponential phase (Fig 4D). Thus, the appearance of transcripts produced by PDI from both G−1G+1 and T−1A+1 start site regions is growth phase-dependent in E. coli.
Discussion
Here, we establish that PDI in E. coli serves as a regulatory mechanism important for biofilm formation (Fig 2). We further establish that PDI also occurs in V. cholerae (Fig 3 and S1, S2, S3, and S4 Tables) and that the pattern of PDI observed in V. cholerae is strikingly similar to that observed in E. coli. Specifically, PDI in both E. coli and V. cholerae is growth phase-dependent and preferentially targeted to promoters carrying start site regions with the sequence T−1A+1 or G−1G+1 (Figs 3 and 4). Our findings support an emerging model that PDI may be widespread in Gammaproteobacteria. To account for the preferential targeting of PDI to start site regions with the sequence T−1A+1 or G−1G+1, we propose that PDI in both E. coli and V. cholerae is a consequence of a growth phase-dependent process that leads to the preferential generation of the linear dinucleotides 5′-UA-3′ and 5′-GG-3′. It is well established that cyclic di-nucleotides can influence biofilm formation [26–30]. Our finding that PDI serves as a regulatory mechanism important for biofilm formation in E. coli (Fig 2) suggests that linear dinucleotides might also influence biofilm formation by serving as primers for transcription initiation.
PDI as a mechanism for transcription initiation in vivo
The results described here, coupled with our prior analyses [5, 6], support an emerging model that PDI may be widespread in bacteria and that the extent of PDI relative to de novo initiation fluctuates as a function of growth state. During the growth conditions used in these studies we find that ~1% of transcripts generated from a given start site region in V. cholerae and ~7% of transcripts generated from a given start site region in E. coli are produced by PDI in late stationary phase (Figs 3A and 4A). However, the strict criteria we have used to measure PDI, which does not consider transcripts emanating from position +1 of a given start site region, and which considers only RNA transcripts emanating from start site regions that also produce products of de novo initiation, likely underestimates the full extent of PDI. In addition, the physiological triggers for PDI are unknown (see below). Thus, the particular growth conditions we have selected for our studies may not represent conditions where PDI is most prevalent. In this regard, over 50% of transcripts generated from a given start site region are produced by PDI in P. aeruginosa cells artificially depleted of the endogenous oligoRNase [6].
A model for growth phase-dependent PDI
The PDI we have observed in E. coli and V. cholerae is detected in late stationary phase, is not detected in exponential phase, and is preferentially apparent at promoters carrying T−1A+1 and G−1G+1 start site regions. 5′ RNA-seq and primer extension analyses indicate that PDI from T−1A+1 and G−1G+1 start site regions produces transcripts emanating from position −1. Furthermore, the 5′ RNA-seq analysis indicates that transcripts emanating from position −1 of T−1A+1 and G−1G+1 start site regions begin with the sequences 5′-UA and 5′-GG, respectively. It follows that the primers used to generate these transcripts also begin with the sequence 5′-UA and 5′-GG, respectively. We therefore propose that the growth phase-dependent PDI observed in both E. coli and V. cholerae is the result of a process that leads to the generation of RNAs beginning with the sequence 5′-UA and 5′-GG at some stage during the transition from exponential phase to late stationary phase. Among all possible 2 - to 4-nt RNAs beginning with 5′-UA and 5′-GG, only the linear dinucleotides 5′-UA-3′ and 5′-GG-3′ have the requisite template specificity to function as a primer during initiation at all T−1A+1 and G−1G+1 start site regions, respectively. Thus, the simplest model is that the linear dinucleotides 5′-UA-3′ and 5′-GG-3′ account for most, if not all, of the PDI observed from T−1A+1 and G−1G+1 start site regions.
Our 5′ RNA-seq analyses of sequence determinants that favor PDI considered only start site regions with an A or G at position +1. This was done to ensure that our conclusions were based upon the composite behavior of start site region sequences within the context of a significant number of distinct promoters. The finding that T−1A+1 and G−1G+1 start site regions are preferentially targeted among those that contain an A or G at +1 suggests that 2 - to ~4-nt RNAs beginning with 5′-UA and 5′-GG are preferentially generated in cells relative to 2 - to ~4-nt RNAs beginning with the sequence 5′-AA, 5′-CA, 5′-GA, 5′-AG, 5′-CG, or 5′-UG. However, our 5′ RNA-seq analysis of individual promoters in V. cholerae also detected oligoRNase-sensitive transcripts that began with the sequence 5′-UC (a promoter associated with VCA0982) (S1 Table). Furthermore, our 5′ RNA-seq analysis of individual promoters in E. coli detected oligoRNase-sensitive transcripts that began with the sequence 5′-CC (promoters associated with ycjX, rplU, and iclR) and 5′-UU (a promoter associated with rpoS) (S7 Table). Thus, in addition to RNAs beginning with the sequence 5′-UA and 5′-GG, other 2 - to 4-nt RNA species may be present at concentrations sufficient to enable PDI in V. cholerae and E. coli.
We do not know the source of the oligoribonucleotides used as primers in V. cholerae and E. coli. In addition, we do not know whether or not the oligoribonucleotide primers are generated by a common mechanism in both organisms. One possibility is that oligoribonucleotides beginning with the sequence 5′-UA and 5′-GG are generated during RNA degradation. Another possibility is that the linear dinucleotides 5′-UA-3′ and 5′-GG-3′ are generated through a specific enzymatic process that awaits identification. In this regard, one intriguing speculation is that enzymes involved in the synthesis and/or breakdown of cyclic dinucleotides [27, 28, 30–34] may also have the ability to generate the linear dinucleotides 5′-UA-3′ and 5′-GG-3′, and this activity may become prevalent during stationary phase. Therefore, the cellular response to fluctuations in cyclic dinucleotide concentrations may also involve alterations in the extent of PDI. Furthermore, in principle, PDI may serve to control the intracellular concentrations of linear dinucleotides such as 5′-GG-3′ and 5′-UA-3′ by acting as a “sink” that removes such species from cells. Accordingly, PDI’s impact on cell physiology may extend beyond its effect on the composition of the transcriptome.
Materials and Methods
Strains
Experiments were performed using E. coli strain MG1655 and V. cholerae strain C6706lacZ [35]. E. coli strain MG1655 ΔbhsA (also called SG107) was constructed by using P1 transduction to move the ΔbhsA::kan locus from JW1098 ΔbhsA::kan [36] into MG1655.
Plasmids
V. cholerae experiments
Plasmid pBAD-lacZ is used as an empty vector control for experiments performed in V. cholerae and has been described previously [37]. Plasmid pBADTOPO-NrnB-VSVG was generated by PCR amplification of B. subtilis nrnB from pNrnB-VSVG [5] using primers NrnBOVERFW (5′ - TAAGAGGAATAATAAATGTATCATTTATATTCACATAACG-3′), NrnBOVERRV (5′-GTCGACCTATTTTCCTAATCTATTCATTTC-3′), and cloning of this PCR product into pBADTOPO according to the manufacturer’s instructions (Life Technologies). Plasmid pNT01 carries sequences extending from −100 to +15 of the promoter associated with VCA0783 (pVCA0783) fused to the tR' terminator cloned into the HindIII and SalI sites of plasmid pACYC184 (New England Biolabs). Plasmids pNT02 and pNT03 are identical to pNT01 with the exception that the start site region associated with pVCA0783 carries the sequence C−1A+1 or C−1G+1, respectively. Plasmid pNT04 carries sequences extending from −100 to +15 of the promoter associated with VC1904 fused to the tR' terminator cloned into the HindIII and SalI sites of plasmid pACYC184. Plasmids pNT05 and pNT06 are identical to pNT04 with the exception that the start site region associated with pVC1904 carries the sequence C−1A+1 or C−1G+1, respectively.
E. coli experiments
Plasmid pPSV38 is used as an empty vector control for experiments in E. coli and has been described previously [5]. Plasmid pNrnB-VSVG expresses the wild-type nrnB gene from B. subtilis strain PY73 fused to a VSV-G epitope tag under the control of an IPTG-inducible lacUV5 promoter flanked by two lac operators and has been described previously [5]. Plasmid pNrnBDHH-VSVG is the same as pNrnB-VSVG with the exception that the nrnB gene carries three amino acid substitutions, D86A, H87A and H88A [5]. pPSV38, pNrnB-VSVG and pNrnBDHH-VSVG confer resistance to gentamicin. Plasmid pBEN 504 has been described previously [5] and carries sequences extending from −100 to +15 of the promoter associated with tomB fused to the tR' terminator cloned into the HindIII and SalI sites of plasmid pACYC184. Seven derivatives of pBEN 504 were constructed that carry the indicated base-pairs at positions −1 and +1: pBEN 512 (C−1A+1), pBEN 556 (T−1G+1), pKS 415 (G−1G+1), pKS 416 (C−1G+1), pKS 417 (G−1A+1), pKS 421 (A−1A+1), and pKS 422 (A−1G+1). Plasmid pKS 385 carries sequences extending from −100 to +15 of the promoter associated with ydiH fused to the tR' terminator cloned into the HindIII and SalI sites of plasmid pACYC184. Two derivatives of pKS 385 were constructed that carry the indicated base pairs at positions −1 and +1: pKS 386 (C−1G+1) and pKS 387 (T−1G+1).
Biofilm assays
Assays were performed using a procedure similar to that described in [18]. Plasmids pPSV38, pNrnB-VSVG or pNrnBDHH-VSVG were introduced into MG1655 or MG1655 ΔbhsA cells by transformation followed by plating on LB agar (LB-agar, Miller; EMD Millipore) containing gentamicin (10ug/mL), or gentamicin and kanamycin (50ug/mL). 5 ml overnights were grown in LB (Miller formulation: 10 g tryptone, 5 g yeast extract, and 10 g NaCl were mixed in deionized water, brought to pH 7 with 5 M NaOH and filter sterilized) containing 10 ug/ml gentamicin and 1 mM IPTG for 13.5 hours. Overnights were back-diluted 1 : 100 in 25 ml of LB broth containing 10 ug/ml gentamicin and 1 mM IPTG in a 125 ml DeLong flask (Bellco). Cultures were shaken at 220 RPM at 37°C to an OD600 of ~1.0 and 0.2 ml of the cell suspensions were placed in a 96-well microtiter plate (Greiner bio-one, cell culture treated U-bottom sterile microplate). After seeding, plates were sealed with breathable tape (World Wide Medical Products, bioexcell film for tissue culture) and incubated at 22.5°C for 32 hours. The growth medium was removed by aspiration and biofilms were washed twice with 0.15 ml of distilled water. The plate was inverted and then dried by incubating at 60°C for 2 hours. After the incubation, plates were cooled to 22.5°C and stained by addition of 0.2 ml of 0.1% crystal violet (w/v in water) for 12 minutes. The stain was removed by aspiration and the stained biofilms were washed once with 0.2 ml distilled water. The water was removed by aspiration and the plate was dried at 55°C for 5 minutes. The retained crystal violet stain was solubilized by addition of 220 μl of 33% acetic acid (v/v in water), incubation at 22.5°C for 1 hour, followed by vigorous mixing for 10 minutes. Afterwards, 100 μl aliquots of the released stain were transferred to a flat-bottom microplate and the absorbance was read at 570 nm using a microplate reader (Model 680, Biorad).
Cell growth for 5′ RNA-seq
V. cholerae experiments
Plasmids pBAD-lacZ and pBADTOPO-NrnB-VSVG were introduced separately into C6706lacZ by transformation followed by plating on LB agar [1% (w/v) Tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl, 1.5% (w/v) agar] containing streptomycin (100 μg/ml) and carbenicillin (100 μg/ml). Experiments were performed in duplicate. Each transformant was used to inoculate 5 ml of LB broth [1% (w/v) Tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl] containing streptomycin (100 μg/ml) and carbenicillin (100 μg/ml) for ~18 hr at 37°C with shaking at 200 rpm. Overnight cultures were diluted to an optical density at 655 nm (OD655) of ~0.03 in 25 ml of LB broth containing streptomycin (100 μg/ml), carbenicillin (100 μg/ml) and L-arabinose [0.2% (w/v)] in a 125 ml flask (Kimax). Cultures were shaken at 220 RPM at 37°C and harvested after ~23 hr of growth (OD655 ~3.0). 10 ml of cell cultures were placed in 50 ml Falcon tubes (VWR) and centrifuged (9 min, 10,000 x g at 4°C) to collect cell pellets.
E. coli experiments
Plasmids pPSV38 and pNrnB-VSVG were each separately introduced into MG1655 cells by transformation followed by plating onto LB-agar (LB-agar, Miller; EMD Millipore) containing gentamycin (10 ug/mL). Experiments were performed using two independent transformants of each strain. Each transformant was used to inoculate 5 ml of LB (LB-broth, Miller; EMD Millipore) containing gentamycin (10 ug/mL) in an 18 mm x 150 mm glass culture tube. Tubes were placed in a tissue culture roller and cells were rotated for ~18 hr at 37°C. 250 ul of each overnight culture was added to 25 ml of LB containing gentamycin (10 ug/mL) and IPTG (1 mM) in a 125 ml DeLong flask (Bellco). Flasks were shaken at 220 RPM at 37°C and harvested after ~23 hr of growth (OD600 ~3.5). 5 ml of the cell suspensions were placed in 50 ml Oakridge tubes (Nalgene) and centrifuged (5 min, 10,000 x g at 4°C) to collect pellets. After removal of the supernatants, the cell pellets were stored at −80°C prior to RNA isolation.
RNA isolation and purification
V. cholerae experiments
Cell pellets were resuspended in 2 ml of TRI Reagent solution (Molecular Research Center) and the suspension was split into two 1 ml aliquots that were placed into individual 2 mL microcentrifuge tubes. The suspensions were incubated at 60°C for 10 min, and centrifuged (10 min, 16,000 x g at 4°C). Each supernatant was transferred to a 2 ml tube, 200 μl of chloroform (Sigma) was added, the solution was mixed for 15 sec, incubated for 5 min at 25°C, and centrifuged (15 min, 16,000 x g at 4°C). 500 μl of each aqueous phase was added to a 2 ml tube containing 500 μl of 90% ethanol. Samples were mixed and transferred to RNeasy mini columns (Qiagen). RNA isolation was carried out according to the manufacturer’s instructions (Qiagen). RNA samples were eluted in 170 μl of nuclease-free water and treated with 20 units of Turbo DNase (Ambion) for 1 h at 37°C. 1 ml of TRI Reagent solution (Molecular Research Center) and 200 μl of chloroform (Sigma) were added, samples were mixed, incubated for 5 min at room temperature, and centrifuged (10 min, 16,000 x g at 4°C). 600 μl of the aqueous phase was transferred to a 2 ml tube containing 1.4 ml of 100% ethanol. Samples were incubated for ~12 h at −20°C, precipitated, washed twice with 1 ml of 75% ethanol, and resuspended in 50 μl of nuclease-free water.
E. coli experiments
Cell pellets were resuspended in 1 ml of TRI Reagent solution (Molecular Research Center), transferred to 1.7 ml low-binding microfuge tubes, incubated at 70°C for 10 min, and centrifuged (10 min, 21,000 x g at 4°C). The supernatant was transferred to a 1.7 ml tube, 200 μl of chloroform was added, the solution was mixed for 15 sec, incubated for 5 min at 25°C, and centrifuged (10 min, 21,000 at 4°C). 540 μl of the aqueous phase was transferred to a 1.7 ml tube, 1080 μl of 100% ethanol was added, samples were incubated at −80°C for a minimum of 2 hr, centrifuged (30 min, 21,000 at 4°C), washed with 1 ml of 75% ethanol, and resuspended in 25 μl of nuclease-free water. On-column DNase I (Qiagen) treatment was performed for 15 min at 25°C. RNA was eluted from the column into 30 μl nuclease-free water and stored at −80°C.
5′ RNA-seq: cDNA library construction
Prior to cDNA library construction the total RNA was passed through an RNeasy Mini Kit (Qiagen) to remove transcripts less than ~200 nt, and through a MICROBExpress Kit (Ambion) to remove some of the rRNAs. cDNA libraries suitable for sequencing on the Illumina HiSeq were constructed as described [16].
Ligation of the 5′ adaptor to the 5′ ends of input RNA (see Fig 1) requires the input RNA to carry a 5′ monophosphate. Thus, preparation of libraries derived from only those RNAs carrying a 5′ monophosphate requires no selective enzymatic treatments prior to ligation of the 5′ adaptor. In contrast, the exclusion of RNAs carrying a 5′ monophosphate in the analysis, the inclusion of 5′ triphosphate ends in the analysis, or the inclusion of 5′ hydroxyl ends in the analysis each requires selective enzymatic treatments to be performed prior to ligation of the 5′ adaptor. To prepare cDNA libraries derived from “all 5′ ends” rRNA-depleted RNA is treated with T4 Polynucleotide Kinase (NEB) to convert 5′ hydroxyl ends to 5′ monophosphate ends and RNA 5′-Polyphosphatase to convert 5′ triphosphate ends to 5′ monophosphate ends. To prepare cDNA libraries derived from 5′ hydroxyl ends and 5′ triphosphate ends rRNA-depleted RNA is treated with 5′-Terminator Exonuclease (Epicentre) to remove 5′ monophosphate ends, followed by T4 Polynucleotide Kinase and RNA 5`-Polyphosphatase. To prepare cDNA libraries derived from 5′ monophosphate ends and 5′ triphosphate ends rRNA-depleted RNA is treated RNA 5`-Polyphosphatase. To prepare cDNA libraries derived from 5′ triphosphate ends rRNA-depleted RNA is treated with 5′-Terminator Exonuclease followed by RNA 5`-Polyphosphatase. To prepare cDNA libraries derived from 5′ hydroxyl ends rRNA-depleted RNA is treated with 5′-Terminator Exonuclease followed by T4 Polynucleotide Kinase. Enzymatic treatments were done as described in [16].
5′ RNA-seq: Data analysis
The first six bases of each read was removed and Bowtie version 1.0.0 was used to align the raw reads to the E. coli reference genome (NC_000913.3) or the V. cholerae reference genome (NC_002505 and NC_002506). Alignment statistics are provided in S9 Table. Reads that aligned to a unique position in the genome with no mismatches were used to perform all analyses. This results in omission of non-unique sequences and excludes from analysis rRNA genes and some tRNA genes. Analysis of cDNA libraries derived from 5′ triphosphate transcripts isolated from cells carrying wild-type concentrations of 2 - to ~4-nt RNAs were used to identify primary transcription start sites. In the case of the V. cholerae analysis, primary start sites were identified on the basis of the data obtained only from RNAs isolated during stationary phase. The read counts in biological replicates were combined and genomic coordinates that met the following two criteria were identified as primary start sites: First, the combined read count was at or above a threshold value of 50 reads. Second, the read count at the genomic coordinate represented a local maximum in an 11-bp window centered on the coordinate.
The read counts at each position within the start site regions (i.e. positions −3 to +4) associated with the primary start sites were used for subsequent analyses. The start site regions shown in S1 and S7 Tables represent those where the total read count was at or above a threshold value of 50 reads in each library derived from triphosphate 5′ ends or all 5′ ends and the proportion of 5′ ends at positions −3, −2, or −1 was reduced by >25% upon ectopic expression of NrnB. The “uTSRs” shown in S2, S4, S5 and S8 Tables meet the following two criteria. First, the proportion of transcripts initiating from position +1 is >75% in the analysis of triphosphate 5′ ends in cells carrying wild-type concentrations of 2 - to ~4-nt RNAs. Second, the total read count was at or above a threshold value of 50 reads in each sample listed.
5′ RNA-seq: Data deposition
Raw reads have been deposited in the NIH/NCBI Sequence Read Archive under the study accession number SRP044667 (alias: PRJNA255655).
Cell growth for primer extension
For the experiments shown in Fig 3, V. cholerae C6706lacZ cells carrying pBAD-lacZ or pBADTOPO-NrnB-VSVG were transformed with plasmid pNT01 (pVCA0783-G−1G+1), pNT02 (pVCA0783-C−1A+1), pNT03 (pVCA0783-C−1G+1), pNT04 (pVC1904-T−1A+1), pNT05 (pVC1904-C−1A+1), or pNT06 (pVC1904-C−1G+1). Cells were grown as described above with the exception that chloramphenicol (25 μg/ml) was also present in the growth media. RNAs for primer extension were isolated using Direct-zol RNA MiniPrep according to the manufacturer’s instructions (Zymo Research).
For the experiments shown in Fig 4 and S1 Fig, E. coli MG1655 cells carrying plasmids pPSV38 or pNrnB-VSVG were transformed with plasmids pBEN 504 (ptomB-T−1A+1), pBEN 512 (ptomB-C−1A+1), pBEN 556 (ptomB-T−1G+1), pKS 415 (ptomB-G−1G+1), pKS 416 (ptomB-C−1G+1), pKS 417 (ptomB-G−1A+1), pKS 421 (ptomB-A−1A+1), pKS 422 (ptomB-A−1G+1), pKS 385 (pydiH-G−1G+1), pKS 386 (pydiH-C−1G+1), or pKS 387 (pydiH-T−1G+1). Cells were grown as described above with the exception that chloramphenicol (25 μg/ml) was also present in the growth media. RNAs were isolated from cells as described above.
Primer extension
Primer extension analyses were performed using 10–20 μg of total RNA as previously described [6] with the following modifications. First, reverse transcription was performed at 55°C to minimize the addition of nontemplated nucleotides to the 3′ ends of the cDNA products [38]. Second, reactions performed using RNAs isolated from V. cholerae contained a final concentration of 1M Betaine (Affymetrix). Sequencing ladders were prepared using a Sequenase Version 2.0 DNA sequencing Kit (Affymetrix).
The following primers were used for the indicated loci:
Chromosomally-encoded pVCA0783 (Fig 3): 5′-GGAAAGTAGTCGAGTCATGC-3′Chromosomally-encoded pVC1904 (Fig 3): 5′-CACATCGGTTATACGGGCCGC-3′ Plasmid-encoded pVCA0783 and pVC1904 derivatives (Fig 3D and 3E): 5′-ATTTTGCAGACCTCTCTGCC-3′
Plasmid-encoded ptomB (Fig 4) and pydiH (S1 Fig) derivatives: 5′-CCTCTCTGCCGGATCC-3′
Supporting Information
Zdroje
1. Feklistov A. RNA polymerase: in search of promoters. Annals of the New York Academy of Sciences. 2013;1293 : 25–32. doi: 10.1111/nyas.12197 23855603
2. Grunberg S, Hahn S. Structural insights into transcription initiation by RNA polymerase II. Trends Biochem Sci. 2013;38(12):603–11. doi: 10.1016/j.tibs.2013.09.002 24120742
3. Nickels BE, Dove SL. NanoRNAs: a class of small RNAs that can prime transcription initiation in bacteria. J Mol Biol. 2011;412(5):772–81. Epub 2011/06/28. doi: 10.1016/j.jmb.2011.06.015 21704045
4. Nickels BE. A new way to start: nanoRNA-mediated priming of transcription initiation. Transcription. 2012;3(6):300–4. doi: 10.4161/trns.21903 23117822
5. Vvedenskaya IO, Sharp JS, Goldman SR, Kanabar PN, Livny J, Dove SL, et al. Growth phase-dependent control of transcription start site selection and gene expression by nanoRNAs. Genes Dev. 2012;26(13):1498–507. doi: 10.1101/gad.192732.112 22751503
6. Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime transcription initiation in vivo. Mol Cell. 2011;42(6):817–25. Epub 2011/06/28. doi: 10.1016/j.molcel.2011.06.005 21700226
7. Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A. 1999;96(8):4372–7. 10200269
8. Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35(13):4552–61. 17586819
9. Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37(15):5114–25. Epub 2009/06/26. doi: 10.1093/nar/gkp527 19553197
10. Hoffman DJ, Niyogi SK. RNA initiation with dinucleoside monophosphates during transcription of bacteriophage T4 DNA with RNA polymerase of Escherichia coli. Proc Natl Acad Sci U S A. 1973;70(2):574–8. Epub 1973/02/01. 4568732
11. Minkley EG, Pribnow D. Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J Mol Biol. 1973;77(2):255–77. Epub 1973/06/25. 4587752
12. Di Nocera PP, Avitabile A, Blasi F. In vitro transcription of the Escherichia coli histidine operon primed by dinucleotides. Effect of the first histidine biosynthetic enzyme. J Biol Chem. 1975;250(21):8376–81. Epub 1975/11/10. 1104602
13. Smagowicz WJ, Scheit KH. Primed abortive initiation of RNA synthesis by E. coli RNA polymerase on T7 DNA. Steady state kinetic studies. Nucleic Acids Res. 1978;5(6):1919–32. Epub 1978/06/01. 353734
14. Grachev MA, Zaychikov EF, Ivanova EM, Komarova NI, Kutyavin IV, Sidelnikova NP, et al. Oligonudeotides complementary to a promoter over the region −8… + 2 as transcription primers for E. coli RNA polymerase. Nucleic Acids Res. 1984;12(22):8509–24. 6390344
15. Ruetsch N, Dennis D. RNA polymerase. Limit cognate primer for initiation and stable ternary complex formation. J Biol Chem. 1987;262(4):1674–9. 3543009
16. Vvedenskaya IO, Goldman SR, Nickels BE. Preparation of cDNA libraries for high-throughput RNA sequencing analysis of RNA 5' ends. Methods Mol Biol. 2015;1276 : 211–28. doi: 10.1007/978-1-4939-2392-2_12 25665566
17. Zhang XS, Garcia-Contreras R, Wood TK. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J Bacteriol. 2007;189(8):3051–62. 17293424
18. O'Toole GA. Microtiter dish biofilm formation assay. Journal of visualized experiments: JoVE. 2011;(47).
19. Maitra U, Hurwitz H. The role of DNA in RNA synthesis, IX. Nucleoside triphosphate termini in RNA polymerase products. Proc Natl Acad Sci U S A. 1965;54(3):815–22. 5324397
20. Jorgensen SE, Buch LB, Nierlich DP. Nucleoside triphosphate termini from RNA synthesized in vivo by Escherichia coli. Science. 1969;164(3883):1067–70. 4890175
21. Hawley DK, McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11(8):2237–55. 6344016
22. Munafo DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010;16(12):2537–52. doi: 10.1261/rna.2242610 20921270
23. Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17(9):1697–712. doi: 10.1261/rna.2799511 21775473
24. Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011;39(21):e141. doi: 10.1093/nar/gkr693 21890899
25. Zhuang F, Fuchs RT, Sun Z, Zheng Y, Robb GB. Structural bias in T4 RNA ligase-mediated 3'-adapter ligation. Nucleic Acids Res. 2012;40(7):e54. doi: 10.1093/nar/gkr1263 22241775
26. Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311(5764):1113–6. 16497924
27. Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–73. Epub 2009/03/17. doi: 10.1038/nrmicro2109 19287449
28. Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7(10):724–35. Epub 2009/09/17. doi: 10.1038/nrmicro2203 19756011
29. Boyd CD, O'Toole GA. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu Rev Cell Dev Biol. 2012;28 : 439–62. doi: 10.1146/annurev-cellbio-101011-155705 23057745
30. Sondermann H, Shikuma NJ, Yildiz FH. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol. 2012;15(2):140–6. doi: 10.1016/j.mib.2011.12.008 22226607
31. Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9(2):218–28. Epub 2006/03/15. 16530465
32. Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–70. doi: 10.1016/j.cell.2012.01.053 22500802
33. Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154(5):962–70. doi: 10.1016/j.cell.2013.08.014 23993090
34. Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11(8):513–24. doi: 10.1038/nrmicro3069 23812326
35. Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105(25):8736–41. doi: 10.1073/pnas.0803281105 18574146
36. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 : 2006 0008.
37. Pickering BS, Lopilato JE, Smith DR, Watnick PI. The Transcription Factor Mlc Promotes Vibrio cholerae Biofilm Formation through Repression of Phosphotransferase System Components. J Bacteriol. 2014;196(13):2423–30. doi: 10.1128/JB.01639-14 24769694
38. Chen D, Patton JT. Reverse transcriptase adds nontemplated nucleotides to cDNAs during 5'-RACE and primer extension. BioTechniques. 2001;30(3):574–80, 82. 11252793
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



