-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
Human genetic studies have demonstrated that quantitative human anthropometric and metabolic traits, including body mass index, waist-hip ratio, and plasma concentrations of glucose and insulin, are highly heritable, and are established risk factors for type 2 diabetes and cardiovascular diseases. Although many regions of the genome have been associated with these traits, the specific genes responsible have not yet been identified. By making use of advanced statistical “imputation” techniques applied to more than 87,000 individuals of European ancestry, and publicly available “reference panels” of more than 37 million genetic variants, we have been able to identify novel regions of the genome associated with these glycaemic and obesity-related traits and localise genes within these regions that are most likely to be causal. This improved understanding of the biological mechanisms underlying glycaemic and obesity-related traits is extremely important because it may advance drug development for downstream disease endpoints, ultimately leading to public health benefits.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005230
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005230Summary
Human genetic studies have demonstrated that quantitative human anthropometric and metabolic traits, including body mass index, waist-hip ratio, and plasma concentrations of glucose and insulin, are highly heritable, and are established risk factors for type 2 diabetes and cardiovascular diseases. Although many regions of the genome have been associated with these traits, the specific genes responsible have not yet been identified. By making use of advanced statistical “imputation” techniques applied to more than 87,000 individuals of European ancestry, and publicly available “reference panels” of more than 37 million genetic variants, we have been able to identify novel regions of the genome associated with these glycaemic and obesity-related traits and localise genes within these regions that are most likely to be causal. This improved understanding of the biological mechanisms underlying glycaemic and obesity-related traits is extremely important because it may advance drug development for downstream disease endpoints, ultimately leading to public health benefits.
Introduction
Quantitative human glycaemic and obesity-related traits, including fasting plasma glucose and insulin (FG and FI), body mass index (BMI), and waist-hip ratio (WHR) are highly heritable [1–5], and are well established risk factors for type 2 diabetes (T2D) and cardiovascular disease [6–10]. Large-scale genome-wide association studies (GWAS) have proved to be extremely successful in the identification of loci harbouring genetic variants contributing to these traits in multiple ethnic groups [11–27]. This process has been facilitated by technical advances in the development of imputation methods [28] that allow evaluation of association with genetic variants not directly assayed on genotyping arrays, but present instead in more dense phased reference panels, such as those made available through the International HapMap Consortium [29,30]. However, the detected loci are typically characterised by common variant association signals, defined by lead SNPs with minor allele frequency (MAF) of at least 5%, which extend over large genomic intervals because of linkage disequilibrium (LD). They also often map to non-coding sequence, making direct biological interpretation of their effect more difficult than for non-synonymous variants. The lead SNPs at GWAS loci are overwhelmingly of modest effect, and together account for only a small proportion (generally less than 5%) of the overall trait variance [17–19,26,27]. As a consequence, there has been limited progress in identifying the genes through which GWAS association signals are mediated, and characterisation of the downstream molecular mechanisms influencing glycaemic and obesity-related traits remains a considerable challenge.
There has been much recent debate as to the role that low frequency and rare variation (MAF<5%) might play in explaining the “missing heritability” of complex human traits [31–33]. It has been hypothesized that some of these variants will have larger effects on traits than common SNPs because they are likely to have arisen as a result of relatively recent mutation events, and thus will have been less subject to purifying selection [34]. Unfortunately, such variation is not well captured by traditional GWAS genotyping arrays, by design, even when supplemented by HapMap imputation [35–37]. However, more recent, higher density reference panels released by the 1000 Genomes (1000G) Project Consortium [38], constructed on the basis of low-pass whole-genome re-sequencing, provide haplotypes at more than 37 million variants for 1,094 individuals from multiple ethnic groups, and facilitate imputation of genetic variation with MAF as low as 0.5% across diverse populations [39–41].
Within the European Network for Genetic and Genomic Epidemiology (ENGAGE) Consortium, we sought to assess the advantages and limitations of high-density imputation for the discovery and fine-mapping of loci for glycaemic and obesity-related traits. We considered 22 European ancestry GWAS (S1 Table), each imputed up to the 1000G “all ancestries” reference panel (Phase 1 interim release, June 2011), in up to (after quality control): 87,048 individuals for BMI; 54,572 individuals for WHR; 46,694 individuals for FG; and 24,245 individuals for FI (S2 and S3 Tables). To account for the impact of overall obesity on central adiposity [18,27] and insulin sensitivity [19], we considered WHR and FI after adjustment for BMI (denoted WHRadjBMI and FIadjBMI, respectively). With these high-density imputed data, we aimed to: (i) discover novel signals of association for glycaemic and obesity-related traits, including within established GWAS loci; (ii) evaluate the impact of low-frequency variation to common SNP GWAS signals; (iii) consider the contribution of genetic variants at GWAS loci in explaining trait variance; and (iv) refine the localisation of potential causal variants underlying GWAS association signals and assess the mechanisms through which they impact glycaemic and obesity-related traits.
Results
Imputation quality
Within each study, we performed stringent quality control of the genotype scaffold before imputation, minimally including sample and variant call rate and deviation from Hardy-Weinberg equilibrium (S1 Table). Each scaffold was imputed up to the 1000G multi-ethnic reference panel (Phase 1 interim release, June 2011), which includes 762 European ancestry haplotypes, using IMPUTEv2 [42], minimac [39] or specialist in-house software (S1 Table). Making use of the multi-ethnic reference panel, including haplotypes from all ancestry groups, has been demonstrated to reduce error rates and to improve imputation quality, particularly of lower frequency variants [28]. Imputed variants were retained for downstream evaluation and association testing if they passed traditional GWAS quality control thresholds (IMPUTEv2 info score ≥ 0.4; minimac r2 ≥ 0.3) [43].
We considered the quality of imputation (as measured by the IMPUTEv2 info score) of variants from the 1000G reference panel in two contributing studies (S4 Table): the 1958 British Birth Cohort from the Wellcome Trust Case Control Consortium (58BC-WTCCC, 2,802 individuals from Great Britain); and the 1966 Northern Finnish Birth Cohort (NFBC1966, 5,276 individuals from Lapland and the Province of Oulu in Northern Finland). In 58BC-WTCCC, 98.8% of common SNPs (MAF≥5%, 6.3 million) and 97.0% of low-frequency variants (0.5%≤MAF<5%, 3.8 million) passed imputation quality control filters, of which 72.9% are not present in HapMap reference panels. However, imputation of rarer variants (0.1%≤MAF<0.5%, 3.4 million) proved less successful in 58BC-WTCCC, with only 80.5% passing quality control filters. The quality of imputation in NFBC1966 was comparable to that observed in 58BC-WTCCC: 99.7% of common SNPs (5.9 million) and 94.4% of low-frequency variants (3.7 million). However, amongst rarer variants, the quality of imputation was noticeably poorer in NFBC1966 (62.8%) than 58BC-WTCCC, presumably reflecting less representation of low-frequency haplotypes from the isolated Northern Finnish population in the 1000G reference panel.
We have demonstrated that high-density imputation provides >90% coverage of low-frequency variants present in the 1000G reference panel in two diverse European ancestry populations. Our study thus enables association testing with more than three million high-quality variants with 0.5%≤MAF<5% that would not have been directly interrogated in previous GWAS of glycaemic and obesity-related traits that have been supplemented by HapMap imputation alone. With the sample sizes available in this study, we have estimated that for any of these variants explaining at least 0.2% of the overall trait variance (i.e. effect size of 0.32 SD units for 1% MAF, and effect size of 0.15 SD units for 5% MAF), we have >99.9% power to detect their association with BMI, WHR, and FG, and >93.9% power to detect their association with FI.
Discovery of novel loci and new lead SNPs
Within each study, we tested for association of each directly typed and well imputed variant with BMI, WHRadjBMI, FG and FIadjBMI, separately in males and females, in a linear regression modelling framework (Methods, S2 and S3 Tables). Association summary statistics were then combined across studies in sex-specific and sex-combined fixed-effects meta-analyses for each trait. Variants passing quality control in fewer than 50% of the contributing studies for each trait were excluded from the meta-analysis. Association signals at genome-wide significance (p<5x10-8) and with lead SNPs independent (r2<0.05) and mapping more than 2Mb from those previously reported for the traits were considered novel. By convention, loci were labelled with the name(s) of the gene(s) located closest to the lead SNP, unless more compelling biological candidates mapped nearby (Table 1, S1, S2, S3 and S4 Figs).
Tab. 1. Novel loci for glycaemic and obesity-related traits achieving genome-wide significance (<i>p</i><5x10<sup>-8</sup>). 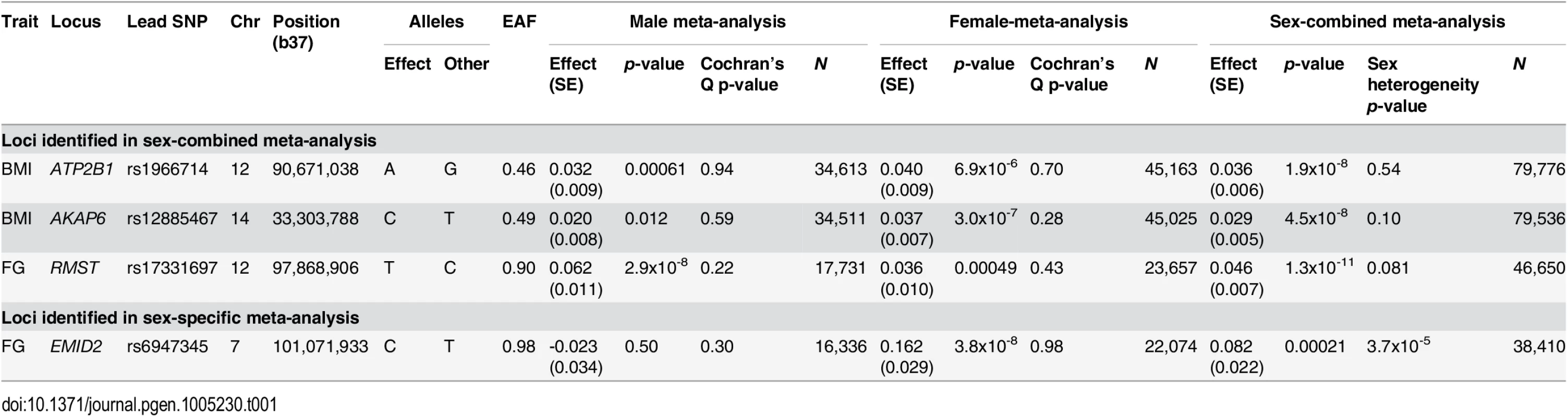
We identified two novel loci achieving genome-wide significance for BMI in the sex-combined meta-analysis: ATP2B1 (rs1966714, MAF = 0.46, p = 1.9x10-8); and AKAP6 (rs12885467, MAF = 0.49, p = 4.5x10-8). For FG, we detected one novel locus in the sex-combined meta-analysis at RMST (rs17331697, MAF = 0.10, p = 1.3x10-11) and a female-specific association at EMID2 (rs6947345, MAF = 0.017, pMALE = 0.50, pFEMALE = 3.8x10-8). We did not identify any novel loci at genome-wide significance, in either sex-combined or sex-specific analyses, for WHRadjBMI or FIadjBMI. We observed no evidence of heterogeneity in sex-specific allelic effects across studies at the lead SNPs at the novel loci (Table 1). With the exception of the sex-specific association signal at EMID2, the lead SNPs at all other novel loci were common.
At AKAP6 and RMST, the common lead SNPs were present in HapMap (S5 Fig) but did not achieve genome-wide significance in large-scale European ancestry HapMap imputed meta-analyses conducted by the GIANT Consortium [17] (for BMI in up to 123,865 individuals) and the MAGIC Investigators [16] (for FG in up to 46,186 individuals), despite substantial overlap with cohorts contributing to our study. We have estimated that, amongst individuals contributing to our 1000G imputed meta-analyses for BMI/FG, a maximum of 59%/37% also participated in the previous GIANT and MAGIC studies (S5 Table). At RMST, our lead FG SNP approaches genome-wide significance in the MAGIC meta-analysis (p = 6.5x10-6), and this likely reflects stochastic variation. However, at AKAP6, our lead BMI SNP demonstrates only nominal evidence of association (p = 0.012) in the GIANT meta-analysis, suggesting that 1000G reference panels have enabled higher quality imputation at this locus. To investigate this assertion further, we compared the quality of imputation of the lead BMI SNP using HapMap and 1000G reference panels in two contributing studies of diverse European ancestry. In 58BC-WTCCC/NFBC1966, there was a marginal improvement in the IMPUTEv2 info score from 0.972/0.939 using reference haplotypes from CEU HapMap to 0.996/0.971 using those from 1000G.
At ATP2B1, the common lead SNP was not present in HapMap (S5 Fig). The lead SNP for BMI from the GIANT HapMap imputed meta-analysis [17] was rs2579106, achieving nominal evidence for association (p = 6.4x10-5) in a reported sample size of 123,864 individuals. This SNP reached near genome-wide significance in our 1000G imputed meta-analysis, despite the smaller sample size (p = 3.3x10-7, in 86,955 individuals). Furthermore, the HapMap and 1000G lead SNPs are in only modest LD with each other (EUR r2 = 0.22). Taken together, these data suggest that the discovery of this novel locus has been due to improved coverage through 1000G imputation, despite the lead SNP being common.
We observed genome-wide significant evidence of association at 34 established loci for glycaemic and obesity-related traits, including GCKR with the same lead SNP for both FG and FI (S6 Table). At 29 of these loci, our meta-analysis identified lead SNPs that were different from previous reports in which they were first discovered, of which 23 were not present in HapMap (S7 Table). At 18 of these 29 loci, the new lead SNP was in strong LD (r2≥0.8) with that previously reported, and consequently both variants had similar MAF and allelic effect size (S6 Fig). At a further nine of the 29 loci, the new and previously reported lead SNPs were in moderate LD (0.2≤r2<0.8) with each other. For these, there was greater difference in MAF and allelic effect size for each pair of variants, but the new lead SNP was common and not consistently less frequent (S6 Fig). At the remaining two loci, the new lead SNPs were not present in HapMap and were in only weak LD with those previously reported (S7 Fig), mapping near BDNF for BMI (r2 = 0.10) and RSPO3 for WHRadjBMI (r2 = 0.04). At both loci, multiple distinct signals of association have been recently reported by the GIANT Consortium in the largest meta-analyses of BMI and WHRadjBMI in European ancestry individuals genotyped with GWAS arrays, supplemented by imputation up to reference panels from the International HapMap Consortium [29,30], and the Metabochip, in up to 339,224 and 224,459 individuals, respectively [26,27]. At BDNF, our new lead SNP (rs4517468) was in moderate LD (r2 = 0.31) with the index variant (rs10835210) for the GIANT secondary signal of association for BMI at this locus, suggesting that they represent the same underlying effect on obesity.
At established loci, amongst the 29 lead SNPs identified in our 1000G imputed meta-analysis that were different from the previous reports in which they were discovered, five of them are present on the Metabochip: NRXN3 (BMI, rs7141420), SH2B1 (BMI, rs2008514), MC4R (BMI, rs663129), LY86 (WHRadjBMI, rs1294437), and GCKR (FG/FIadjBMI, rs1260326). These variants were thus directly interrogated in the largest European ancestry meta-analyses, to date, of glycaemic and obesity related traits from the GIANT Consortium [26,27] and MAGIC Investigators [19] that made use of this array. At all five of these loci, our new lead SNP is either the same or is in strong LD (EUR r2>0.75) with that reported in the trait-equivalent Metabochip effort. Four of these loci (all except NRXN3) were densely typed as “fine-mapping” intervals on the array, providing evidence that 1000G imputation has been successful at predicting genotypes at untyped variants in these regions, even though the GWAS scaffolds used in our investigation were comparatively sparse.
Multiple distinct association signals
We investigated the evidence for multiple distinct association signals in the glycaemic and obesity-related trait loci achieving genome-wide significance in our study (four novel and 34 established) (Table 1 and S6 Table). We undertook approximate conditional analyses, implemented in GCTA [44], to select index SNPs for distinct association signals achieving “locus-wide” significance (pCOND<10−5) to reflect the number of uncorrelated variants in a 2Mb window flanking the lead SNP (Methods). We made use of summary statistics from the meta-analysis and genotypes from 58BC-WTCCC and NFBC1966 to approximate the LD between genetic variants (directly typed and well imputed) and hence the correlation in parameter estimates in the joint association model. Reassuringly, the index SNPs and association summary statistics (effect sizes and p-values) from the joint model were highly concordant for both reference studies (S8 Table). Finally, we confirmed these GCTA association signals through exact reciprocal conditional analyses by adjustment for genotypes at each index SNP as a covariate in the linear regression model (Methods, Fig 1, Table 2).
Fig. 1. Regional plots of multiple distinct signals at WHRadjBMI locus RSPO3 (A), FG loci G6PC2 (B) and GCK (C). 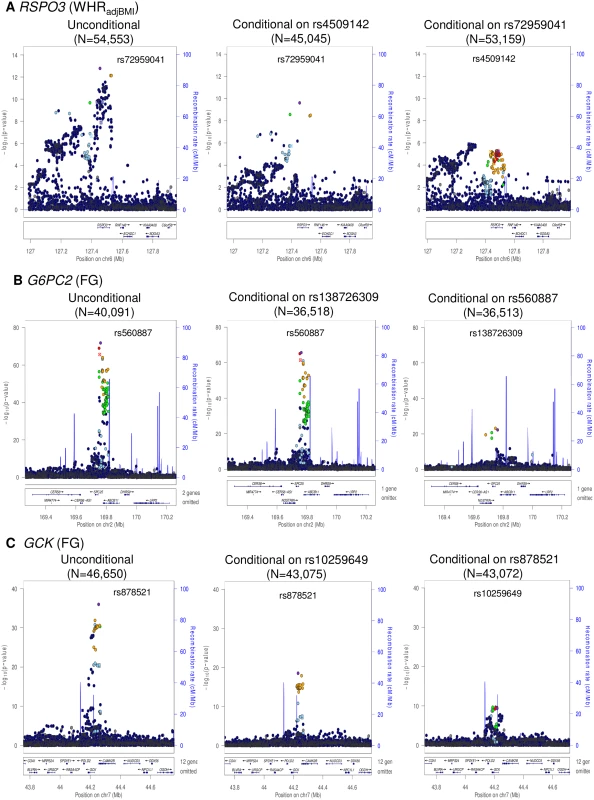
Regional plots for each locus are displayed from: the unconditional meta-analysis (left); the exact conditional meta-analysis for the primary signal after adjustment for the index variant for the secondary signal (middle); and the exact conditional meta-analysis for the secondary signal after adjustment for the index variant for the primary signal (right). The sample sizes vary due to the availability of the well imputed index SNPs of the primary and secondary signals. Directly genotyped or imputed SNPs are plotted with their association P values (on a -log10 scale) as a function of genomic position (NCBI Build 37). Estimated recombination rates are plotted to reflect the local LD structure around the associated SNPs and their correlated proxies (according to a blue to red scale from r2 = 0 to 1, based on pairwise EUR r2 values from the 1000 Genomes June 2011 release). SNP annotations are as follows: circles, no annotation; downward triangles, nonsynonymous; squares, coding or 3′ UTR; asterisks, TFBScons (in a conserved region predicted to be a transcription factor binding site); squares with an X, MCS44 placental (in a region highly conserved in placental mammals). Tab. 2. Loci with multiple distinct signals of association with glycaemic and obesity-related traits achieving “locus-wide” significance in conditional analysis (<i>p</i><sub>COND</sub><10<sup>−5</sup>). 
We identified two distinct signals of association for WHRadjBMI mapping to the RSPO3 locus, indexed by rs72959041 (MAF = 0.079, pCOND = 2.5x10-10) and rs4509142 (MAF = 0.49, pCOND = 5.8x10-6), corresponding to our new lead SNP and that previously reported [18], respectively. More recently, both signals have also been reported by large-scale meta-analyses undertaken by the GIANT Consortium [27]. Our new lead SNP (rs72959041) was reported as the index variant for their secondary association signal at this locus, whilst the index variant for our secondary signal of association (rs4509142) was in strong LD with their lead SNP (rs1936805, r2 = 0.67). The GIANT Consortium also identified a third distinct signal of association at this locus, stronger in females than in males, which was not detected in our conditional analyses, and presumably reflects reduced power due to our smaller sample size. We also identified two distinct signals of association for FG each mapping to GCK (rs878521, MAF = 0.21, pCOND = 1.3x10-18; rs10259649, MAF = 0.27, pCOND = 4.6x10-10) and G6PC2 (rs560887, MAF = 0.31, pCOND = 2.2x10-66; rs138726309, MAF = 0.015, pCOND = 5.7x10-23). None of the index variants for these distinct association signals was present in HapMap (S8 Fig), and only rs10259649 in GCK was well represented by a tag in that reference panel (rs2908292, r2 = 1.00).
Trait variance explained by novel loci and new lead SNPs
We evaluated the additional heritability of glycaemic and obesity-related traits explained by lead SNPs at novel and established loci after 1000G imputation in 5,276 individuals from NFBC1966 (Methods). For each trait, we calculated the phenotypic variance accounted for by: (i) previously reported lead SNPs at established loci; and (ii) new lead SNPs and index variants for distinct association signals at novel and established loci from the present study. The greatest increment in variance explained was observed for FG, where the novel loci and new lead SNPs after 1000G imputation together account for an increase from 1.9% to 2.3%. We also observed noticeable increments in variance explained after 1000G imputation for WHRadjBMI (from 1.1% to 1.3%) and BMI (3.2% to 3.5%). However, for FIadjBMI, only one new lead SNP at an established locus was identified after 1000G imputation, providing a negligible improvement in variance explained (from 0.46% to 0.47%).
Fine-mapping of novel and established GWAS loci
We sought to take advantage of the improved coverage of common and low-frequency variation offered by 1000G imputation to localise potential causal variants (MAF≥0.5%) for the 42 distinct association signals achieving locus-wide significance in our conditional meta-analyses (two distinct signals of association each at RSPO3, GCK, and G6PC2, one signal of association for both FG and FIadjBMI at the GCKR locus, and one signal of association at each of the other 34 novel and established loci). For each distinct signal, we constructed 99% credible sets of variants [45] that together account for 99% probability of driving the association on the basis of the (conditional) meta-analysis (Methods, S9 Table). At the 29 established loci where we identified a new lead SNP after 1000G imputation, the posterior probability of driving the association signal was consistently higher than that for the variant previously reported (S9 Fig). The greatest increases in posterior probability were observed at: GCKR (FG/FIadjBMI, increase from 2.6%/1.8% to 93.5%/89.6%); RSPO3 (WHRadjBMI, increase from 0.4% to 78.6%); PROX1 (FG, increase from 13.2% to 76.9%); and NRXN3 (BMI, increase from 2.5% to 62.2%).
Credible sets are well calibrated for common and low-frequency variants provided that imputation and meta-analysis provides complete coverage of variation with MAF≥0.5% at each locus. Smaller credible sets, in terms of the number of variants they contain, thus correspond to fine-mapping at higher resolution. We considered 99% credible sets containing fewer than 20 variants to be “tractable”, and amenable to follow-up through additional analyses of functional and regulatory annotation (Table 3, S10 Table). The most precise localisation was observed for FG loci including: MTNR1B (rs10830963 accounts for more than 99.9% of the probability of driving the association); both distinct signals at G6PC2 (two variants each, mapping to <15kb interval); and one signal at GCK (indexed by rs878521, mapping to <25kb interval). Of the 127 variants reported in these tractable credible sets, 74 (58.3%) were not present in HapMap, and accounted for 42.4% of the probability of driving the association signals. None of the HapMap variants in the tractable credible sets was of low-frequency, compared to 20.8% of those present only in 1000G (S11 Table).
Tab. 3. Association signals for glycaemic and obesity-related traits for which the 99% credible sets contain no more than 20 variants. 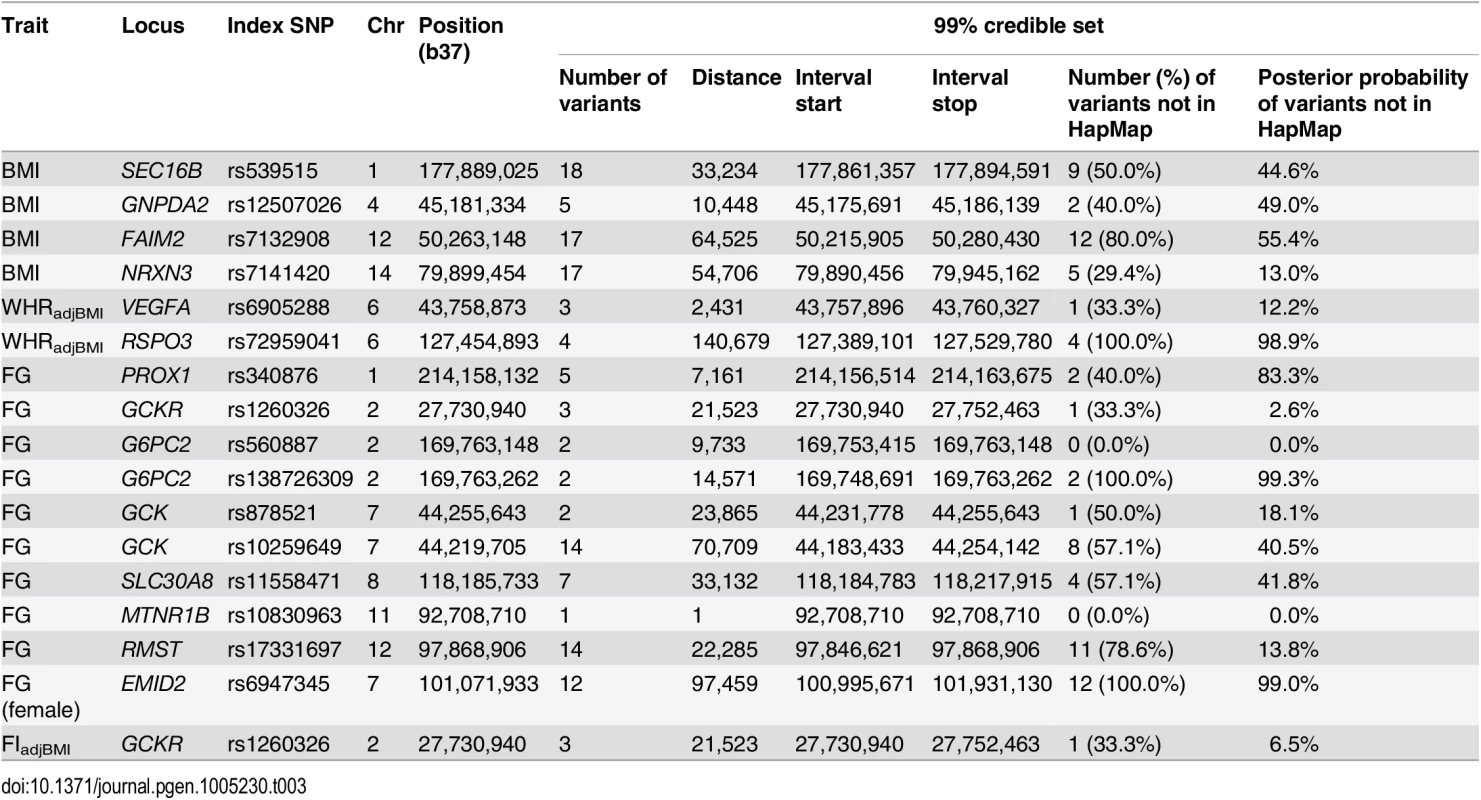
The tractable credible sets included coding variants at just three loci implicated in FG: GCKR, SLC30A8, and the low-frequency association signal at G6PC2. The lead SNP mapping to GCKR (rs1260326) was the common coding variant L446P, which accounts for 93.5% of the probability of driving the FG association signal, and was present in HapMap. At the SLC30A8 locus, the probability of driving the association for FG was shared between 7 SNPs, in strong LD with each other, and including the coding variant R325W. This variant was present in HapMap, and was sufficient to explain the association signal of the lead non-coding SNP for FG in conditional analysis (rs11558471, p = 3.2x10-10, pCOND = 0.052) at the locus. SLC30A8 R325W is also the lead SNP for T2D susceptibility at this locus in published European ancestry meta-analyses from the DIAGRAM Consortium [46]. Finally, the low-frequency index SNP for the secondary association signal mapping to G6PC2 (rs138726309, MAF = 0.015) was the coding variant H177Y, which accounts for 11.2% of the posterior probability of causality at this locus. For this association signal, none of the variants in the 99% credible set was present in HapMap, and thus would have been overlooked without 1000G imputation. This coding variant has recently been implicated in FG homeostasis in a meta-analysis of 33,407 non-diabetic individuals of European ancestry, genotyped with the Illumina exome array, and in agreement with our study, demonstrates a stronger signal of association in conditional analysis after accounting for the lead SNP at the G6PC2 locus [47].
The remaining variants in the tractable credible sets mapped to non-coding sequence. To gain insight into potential regulatory mechanisms through which these variants might impact glycaemic and obesity-related traits, we overlaid each of these credible sets, in turn, with chromatin state calls from eleven cell lines and tissues (Methods). Across all traits, 99% credible set variants were enriched for overlap with enhancer elements (Fig 2). Focussing on FG, variants within the 99% credible set showed significant enrichment (p<2.4x10-3) for active promoter and transcription factor binding site annotations compared to all others (respectively: 3.8-fold, Fisher's combined p = 9.4x10-5; and 7.2-fold, Fisher’s combined p = 2.1x10-13). Over cell types, this enrichment was most prominent in pancreatic islets (Fig 2). More than half of islet-annotated variants are not present in HapMap, and this would not have been observed without 1000G imputation. For example, at the novel FG RMST locus, 11 of the 14 variants in the 99% credible set are not present in HapMap, but all overlap active islet chromatin marks (S10 Fig).
Fig. 2. Broad category functional annotation (A) and cell-type specific annotation (B) of credible set variants. 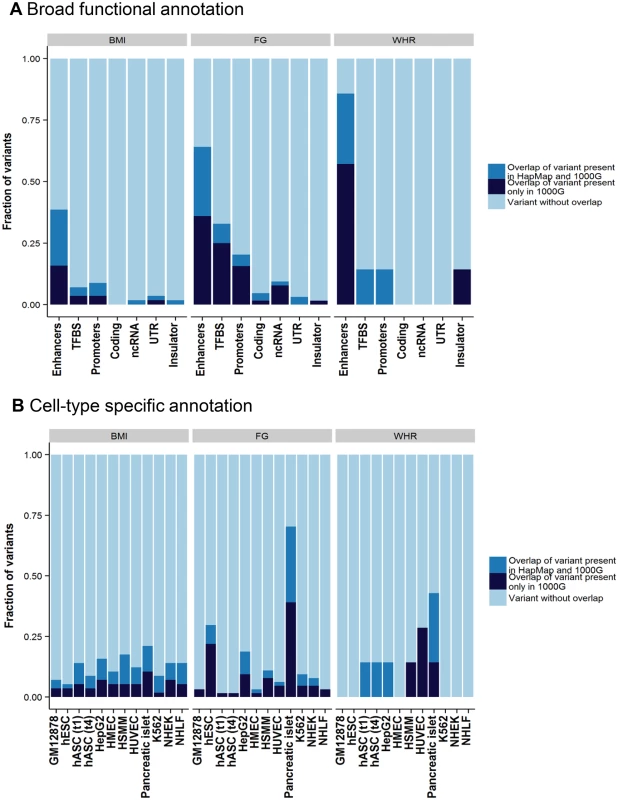
On the x-axis is each category of broad functional annotation (A) or cell-type specific annotation (B). The fraction of credible set variants that overlap with each category is shown on y-axis. The overlapping variants are further broken down into either variants that exist in both the 1000 Genomes and HapMap reference panel (green) or those that exist only in the 1000 Genomes reference panel (red). TFBS, transcription factor binding site; ncRNA, non-coding RNA; UTR, untranslated regions; GM12878, lymphoblastoid cell line from European ancestry female; hESC, H1 human embryonic stem cells; hASC(t1), human pre-adipocytes; hASC(t4), mature human adipocytes; HepG2, liver carcinoma cell-line; HMEC, human mammary epithelial cells; HSMM, human skeletal muscle myoblasts; HUVEC, human umbilical vein endothelial cells; K562, human myelogenous leukemia cell-line; NHEK, normal human epidermal keratinocytes; NHLF, normal human lung fibroblasts. Discussion
Through meta-analysis of 1000G imputed GWAS of glycaemic and obesity-related traits, we have identified two novel loci for BMI at genome-wide significance, and two for FG (including one low-frequency variant association signal that is specific to females). These loci were not reported in larger meta-analysis efforts of European ancestry undertaken by the GIANT Consortium (for BMI) and the MAGIC Investigators (for FG), despite the partial overlap of contributing studies [16–19,26,27]. Improved coverage and quality of imputation for common and low-frequency variation using 1000G reference panels has increased power. We also reported new lead SNPs at 29 established glycaemic and obesity-related trait loci achieving genome-wide significance in our meta-analyses, of which 23 were not present in HapMap, and identified multiple distinct signals of association for WHRadjBMI at RSPO3 and for FG at GCK and G6PC2. Taken together, these novel loci, distinct association signals, and new lead SNPs have increased the trait variance explained for glycaemic and obesity-related traits, although the majority of the heritability remains unaccounted for.
Despite more than 90% coverage of low-frequency variation after 1000G imputation, in diverse European ancestry populations, and equivalent power to detect association across the allele frequency spectrum for a fixed proportion of trait variance explained, the new lead SNPs at established and novel GWAS loci are predominantly common. These data argue strongly against the “synthetic association” hypothesis, which posits that common lead SNPs at GWAS loci will often reflect unobserved causal variants of lower frequency and greater effect size [32]. We recognise that our study has insufficient power to detect common or low-frequency association signals of more modest effect (S12 Table). For example, we estimated that the power to detect association in this study, at genome-wide significance, of a variant of 1% MAF, explaining 0.05% of the overall trait variance (effect size of 0.16 SD units), was 88.0% for BMI, but just 42.1% for WHRadjBMI, 27.7% for FG, and only 2.6% for FIadjBMI. Furthermore, the contribution of rare variants to glycaemic and obesity-related traits cannot be directly investigated with these data because of the low quality imputation for MAF<0.5%, but will require interrogation through deep whole-genome re-sequencing studies in large sample sizes.
We have demonstrated that integration of 1000G imputation, genetic fine-mapping, and genomic annotation, facilitates fine-mapping of GWAS loci for glycaemic and obesity-related traits, and has provided insight into potential functional and regulatory mechanisms through which the effects of these association signals are mediated. In particular, variants in the 99% credible set for the low-frequency association signal mapping to G6PC2 are completely absent from HapMap, but include H177Y. The glucose lowering allele at this variant has been demonstrated to result in a significant decrease in protein expression mediated through proteasomal degradation, leading to a loss of G6PC2 function [47]. We also demonstrated enrichment for overlap of functional elements with variants in the tractable credible sets mapping to non-coding sequence, in particular enhancers. For FG, additional enrichment was observed across credible set variants mapping to promoter and transcription factor binding sites in pancreatic islets, in particular. Uncovering these types of enrichment is essential for prioritisation of variants for functional follow-up, and can be incorporated in statistical models to elucidate causal alleles. Also, at the level of an individual locus, functional annotation can help point to the underlying molecular mechanism through which the GWAS signal is mediated. At G6PC2, for example, the lead SNP, rs560887, in the 99% credible set for the second distinct (non-coding) association signal at this locus (79.5% posterior probability) maps to an enhancer region that is active in pancreatic islets and embryonic stem cells, but repressed in most other cell types. These observations are in agreement with recent reports of clustering of T2D-associated risk variants in islet enhancers [48] and highlights a potential mechanism through which GWAS loci impact glucose homeostasis and disease risk.
Despite the success of traditional GWAS genotyping arrays for the discovery of common variant association signals for complex human traits, because of the structure of LD for variation with MAF>5%, the gold standard approach to directly interrogating lower frequency variation is through re-sequencing studies. However, in agreement with recently published investigations of the contribution of low-frequency variants to a range of phenotypes [47,49–51], our study highlights that effect sizes are modest, and require sample sizes for detection that are financially infeasible through re-sequencing on the scale of the whole genome (or exome). We have demonstrated, in this study, that imputation of existing GWAS scaffolds up to reference panels from the 1000 Genomes Project Consortium [38] enables imputation of more than 90% of low-frequency variants in diverse European populations, at no additional cost other than computation and analyst time. Future GWAS of complex traits in European ancestry populations will be further enhanced by the Haplotype Reference Consortium (www.haplotype-reference-consortium.org). This effort will create a reference panel of more than 60,000 haplotypes from re-sequencing of multiple cohorts, predominantly of European ancestry, enabling high-quality imputation to lower allele frequencies. Phase 3 of the 1000 Genomes Project includes haplotypes from diverse populations from each the five major global ethnicities, and thus would be expected to improve imputation quality over Phase 1 for low-frequency variants in East Asian, South Asian, African and American ancestry groups. The viability of imputation as an approach to recover genotypes at low-frequency variants in GWAS undertaken in populations that are not well represented by the 1000 Genomes Project might require whole-genome re-sequencing of some individuals from the study, in combination with haplotypes from the existing reference panel.
Irrespective of the population under investigation, our study suggests that imputation is unlikely to provide sufficient coverage of variation with MAF<0.5% to enable gene-based testing of rare variants [52]. Imputation is restricted to those rare variants that are present in the reference panel, which are much more likely to be population specific. Furthermore, imputation of rare variants that are present in the reference panel is generally poor, although it is not clear how well calibrated the traditional metrics of quality (such as IMPUTEv2 info score) will be. Thorough investigation of the impact of rare variation on phenotype will thus require re-sequencing, although some success in discovering rare coding variants associated with complex human traits has been achieved through exome array genotyping [47,53–55]. For the time being, arrays that combine an imputation scaffold with direct interrogation of rare coding variation likely offer the most cost-effective approach to assaying variants across the frequency spectrum.
In conclusion, our study has enabled discovery and fine-mapping of novel and established association signals for glycaemic and obesity-related traits, and through integration with genomic data from relevant tissues, has highlighted functional and regulatory processes through which these effects are mediated. Improved understanding of the biological basis of the quantitative human anthropometric and metabolic traits may advance our appreciation of the mechanisms underlying downstream disease endpoints, including T2D and cardiovascular diseases, ultimately leading to personalised treatment approaches, therapeutic development and public health benefits.
Methods
Ethics statement
All human research was approved by the relevant institutional review boards, and conducted according to the Declaration of Helsinki. All participants provided written informed consent.
Studies and samples
We considered 22 population-based and case-control GWAS of European ancestry in up to (after quality control): 87,048 individuals for BMI; 54,572 individuals for WHRadjBMI; 46,694 individuals for FG; and 24,245 individuals for FIadjBMI. Samples were limited to individuals of at least 18 years of age. Case-control studies were stratified by disease status, with each stratum analysed separately. Full details of study and sample characteristics are provided in S1 Table.
Genotyping and quality control
Samples were genotyped with a variety of GWAS arrays. Sample and SNP quality control was undertaken within each study. Sample quality control included exclusions on the basis of genome-wide call rate, extreme heterozygosity, sex discordance, cryptic relatedness, and outlying ethnicity. SNP quality control included exclusions on the basis of call rate across samples and extreme deviation from Hardy-Weinberg equilibrium. Non-autosomal SNPs were excluded from imputation and association analysis. SNPs with MAF<1% were also excluded from the genotype scaffold prior to imputation. Full details of the genotyping arrays and quality control protocols employed by each study are summarised in S1 Table.
Imputation
Within each study, the autosomal GWAS genotype scaffold was imputed up to the 1000 Genomes Project multi-ethnic reference panel (Phase I interim release, June 2011), which was the most up to date available at the time analyses were undertaken. Imputation was performed using IMPUTEv2 [42], minimac [39] or specialist in-house software. Poorly imputed variants (IMPUTE info<0.4; minimac r^2<0.3) [43], and those with minor allele count of less than three (under a dosage model) were excluded from downstream association analyses.
Trait transformations and study-level association analyses
We utilised protocols for obesity-related and glycaemic trait transformations developed by the GIANT Consortium [17,18] and MAGIC Investigators [19]. Full details of trait transformations, trait summary statistics and study-specific covariates are presented in S2 and S3 Tables.
BMI was calculated as the ratio of weight (kg) to squared height (m2). BMI was inverse normal transformed separately in males and females. Association of the transformed trait with each variant passing quality control was tested in a linear regression framework under an additive model in the dosage of the minor allele after adjustment for age, age2 and study-specific covariates, separately in males and females.
WHR was calculated as the ratio of waist circumference (m) to hip circumference (m). Residuals were obtained after adjustment for age, age2, BMI, and study-specific covariates, separately in males and females, and were subsequently inverse-rank normalised. Association of the transformed trait with each variant passing quality control was tested in a linear regression framework under an additive model in the dosage of the minor allele, separately in males and females.
FG was measured in mmol/L. Individuals with a diagnosis of diabetes (type 1 or type 2), diabetes treatment, and/or FG≥7mmol/L, non-fasting state, or pregnancy were excluded. Individuals from case cohorts (with diseases such as stroke and cardiovascular disease) were also excluded if they had undergone hospitalization or blood transfusion in the 2–3 months before measurements were taken. Association of the untransformed trait with each variant passing quality control was tested in a linear regression framework under an additive model in the dosage of the minor allele after adjustment for age, age2 and study-specific covariates, separately in males and females.
FI was measured in pmol/L with subsequent natural log transformation. Individuals with a diagnosis of diabetes (type 1 or type 2), diabetes treatment, and/or FG≥7mmol/L, non-fasting state, or pregnancy were excluded. Individuals from case cohorts (with diseases such as stroke and cardiovascular disease) were also excluded if they had undergone hospitalization or blood transfusion in the 2–3 months before measurements were taken. Association of the transformed trait with each variant passing quality control was tested in a linear regression framework under an additive model in the dosage of the minor allele after adjustment for age, age2, BMI and study-specific covariates, separately in males and females.
Meta-analysis
Summary statistics from association testing of variants passing quality control, separately in males and females, were corrected in each study for residual population structure through genomic control [56] where necessary (S2 and S3 Tables). Subsequently, association summary statistics were combined across studies in sex-specific and sex-combined fixed-effects meta-analyses (inverse-variance weighting) for each trait, as implemented in GWAMA [57]. Heterogeneity in allelic effects between males and females for each trait at each variant was assessed by means of an implementation of Cochran’s Q-statistic [58] in GWAMA [57]. Variants passing quality control in fewer than 50% of the contributing studies for each trait were excluded from the meta-analysis. After filtering, the total numbers of variants reported for each trait were: 9,953,165 for BMI; 9,954,794 for WHRadjBMI; 9,967,162 for FG; and 9,837,044 for FIadjBMI. Sex-specific or sex-combined p<5x10-8 was considered genome-wide significant for each trait. Associated loci are referred to by the name(s) of the nearest gene(s) to lead SNP, unless there are more biologically plausible candidates mapping nearby.
Approximate conditional analysis
We performed approximate conditioning in established and novel glycaemic and obesity-related trait loci in GCTA [44] on the basis of association summary statistics from the sex-combined meta-analyses after variant filtering. We utilised genotype data from two reference studies to approximate LD between variants in diverse European populations, and hence correlation between parameter estimates in the GCTA-COJO joint regression model: 58BC-WTCCC (2,802 individuals from Great Britain); and NFBC1966 (5,276 individuals from Lapland and the Province of Oulu in Northern Finland). We identified “index” variants to represent each distinct association signal achieving genome-wide significance (p<5x10-8) in the GCTA-COJO joint regression model for further validation.
Exact conditional analysis
We performed exact conditional analysis for each locus identified with multiple distinct association signals in GCTA using imputed data from all contributing studies except Rotterdam Study 1 (5,745 individuals). Within each study, we tested for association in the same linear regression framework utilised for unconditional analysis, separately in males and females, but included genotypes at each GCTA index SNP identified at the locus, in turn, as an additional covariate in the model. At each established glycaemic and obesity-related trait locus, we also performed conditioning on the previously reported lead SNP if it differed from that reported in our unconditional meta-analysis. Subsequently, association summary statistics for each signal were combined across studies in sex-specific and sex-combined fixed-effects meta-analyses (inverse-variance weighting) for each trait, as implemented in GWAMA [57].
Trait variance explained
We estimated the variance explained for each trait using genotype data from NFBC1966 (5,276 individuals) in a multiple linear regression framework. For each trait, we considered two sets of variants: (i) previously reported lead SNPs for established loci; and (ii) new lead SNPs and index variants for multiple distinct association signals in established and novel loci. We tested for association of the trait: (i) with covariates only; and (ii) with covariates and the dosage of the minor allele at each variant. For each set of variants, the trait variance explained was given by the difference in the coefficient of determination (r2) between these two regression models.
Credible set construction
For each distinct signal for each trait, we calculated the posterior probability of driving the association for the jth variant, πCj, given by
where the summation is over all variants reported in the (conditional) meta-analysis across the locus. In this expression, Λj is the approximate Bayes’ factor [59] for the jth variant, given by where βj and Vj denote the allelic effect and corresponding variance from the (conditional) meta-analysis for the association signal. The parameter ω denotes the prior variance in allelic effects, taken here to be 0.04 [59]. A 99% credible set was then constructed by: (i) ranking all variants in the locus according to their Bayes’ factor, Λj; and (ii) including ranked variants until their cumulative posterior probability exceeds 0.99.Functional and regulatory annotation
We interrogated coding variants in the 99% credible set for each association signal using Ensembl and HaploReg [60]. Their likely functional consequences were predicted by SIFT [61], PROVEAN [62] and PolyPhen2 [63].
We collected genomic annotation data from several sources. For regulatory state information, we collected sequence reads generated for six assays (H3K4me1, H3K4me3, H3K27ac, H3K27me3, H3K36me3, and CTCF) from 9 ENCODE cell types (GM12878, K562, HepG2, HSMM, HUVEC, NHEK, NHLF, hESC, HMEC) [64], pancreatic islets [65], and adipose stem cells (hASC t1, t4) [66]. Reads were mapped to the human genome reference sequence (hg19) using BWA [67]. Regulatory states for all cell types were called from the aligned reads using ChromHMM [68], assuming 10 states. We then assigned names to the resulting state definitions as follows: active promoter (High H3K4me3, H3K27ac); strong enhancer 1 (H3K4me3, H3K27ac, H3K4me1); strong enhancer 2 (H3K27ac, H3K4me1); weak enhancer (H3K4me1); poised promoter (H3K27me3, H3K4me3, H3K4me1); repressed (H3K27me3); low/no signal; insulator (CTCF); low/no signal; and transcription (H3K36me3). We also obtained transcription factor binding sites (TFBS) established using chromatin immunoprecipitation sequencing. This consisted of data on 147 proteins [64–66].
Finally, we used transcript information from GENCODEv14 [69] to define protein-coding genes, 5’ and 3’ UTR regions, and non-coding genes. For transcripts to be classified as protein-coding, the ‘protein-coding’ tag needed to be set and further filtering for either presence in the conserved coding DNA sequence (CCDS) database or experimentally confirmed mRNA start and end was applied. From this set of transcripts, 5’ UTR, exon, and 3’ UTR regions were defined. For non-coding genes, transcripts labelled as ‘lncRNA‘, ‘miRNA’, ‘snoRNA’ or ‘snRNA’ were used as non-coding genes.
Overlap between the annotations described above and variants in tractable credible sets was determined using bedtools v2.17.0. We defined seven broad functional classes from these annotation data: coding (protein-coding transcripts); ncRNA (non-coding RNA transcripts); UTR (3’ and 5’ UTR regions of coding transcripts); enhancers (strong and weak enhancer elements); promoters (active and poised promoter elements); insulators; and TFBS (sites pooled across all factors). We further used each of the cell line annotations as a distinct category. Each variant was allowed to overlap multiple annotation categories.
For each broad functional class, Fisher’s exact test as implemented in R v3.0.1 (with alternative = “greater”) was used to compare whether the set of credible variants showed a higher fold overlap of this annotation versus all of the others independently. The six resulting p-values for each class were then combined using Fisher’s method. With 21 different functional class and trait combinations, a Bonferroni adjusted significance threshold (p<2.4x10-3) was used.
Supporting Information
Zdroje
1. Rose KM, Newman B, Mayer-Davis EJ, Selby JV (1998) Genetic and behavioural determinants of waist-hip ratio and waist circumference in women twins. Obes Res 6 : 383–392. 9845227
2. Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H (1999) Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia 42 : 139–145. 10064092
3. Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, et al. (2005) Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes 54 : 275–283. 15616039
4. Silventoinen K, Rokholm B, Kaprio J, Sørensen TI (2010) The genetic and environmental influences on childhood obesity: a systematic review of twin and adoption studies. Int J Obes 34 : 29–40.
5. Van Dongen J, Willemsen G, Chen WW, de Geus EJ, Boomsma DI (2013) Heritability of metabolic syndrome traits in a large population-based sample. J Lipid Res 54 : 2914–2923. doi: 10.1194/jlr.P041673 23918046
6. American Diabetes Association (2003) The expert committee on the diagnosis and classification of diabetes mellitus: follow-up report on the diagnosis of diabetes mellitus. Diabetes Care 26 : 3160–3167. 14578255
7. Weyer C, Bogardus C, Mott DM, Pratley RE (1999) The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Invest 104 : 787–794. 10491414
8. DeFronzo RA, Ferrannini E (1991) Insulin resistance: a multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidemia, and atherosclerotic cardiovascular disease. Diabetes Care 14 : 173–194. 2044434
9. Lewis CE, McTigue KM, Burke LE, Poirier P, Eckel RH, et al. (2009) Mortality, health outcomes, and body mass index in the overweight range: a science advisory from the American Heart Association. Circulation 119 : 3263–3271. doi: 10.1161/CIRCULATIONAHA.109.192574 19506107
10. Pischon T, Boeing H, Hoffmann K, Bergmann M, Schulze MB, et al (2008) General and abdominal adiposity and risk of death in Europe. N Engl J Med 359 : 2105–2120. doi: 10.1056/NEJMoa0801891 19005195
11. Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, et al. (2008) Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet 40 : 716–718. doi: 10.1038/ng.156 18454146
12. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. (2009) Variants in MTNR1B influence fasting glucose levels. Nat Genet 41 : 77–81. doi: 10.1038/ng.290 19060907
13. Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, et al. (2009) Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 41 : 25–34. doi: 10.1038/ng.287 19079261
14. Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, et al. (2009) Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet 5: e1000508. doi: 10.1371/journal.pgen.1000508 19557161
15. Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, et al. (2009) A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet 41 : 527–534. doi: 10.1038/ng.357 19396169
16. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42 : 105–116. doi: 10.1038/ng.520 20081858
17. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. (2010) Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 42 : 937–948. doi: 10.1038/ng.686 20935630
18. Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, et al. (2010) Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet 42 : 949–960. doi: 10.1038/ng.685 20935629
19. Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, et al. (2012) Large-scale association analyses identify new loci influencing glycaemic traits and provide insight into the underlying biological pathways. Nat Genet 44 : 991–1005. doi: 10.1038/ng.2385 22885924
20. Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, et al. (2012) A genome-wide approach accounting for body-mass index identifies genetic variants influencing fasting glycaemic traits and insulin resistance. Nat Genet 44 : 659–669. doi: 10.1038/ng.2274 22581228
21. Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, et al. (2012) Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet 44 : 302–306. doi: 10.1038/ng.1086 22344221
22. Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, et al. (2012) Meta-analysis identifies common variants associated with body-mass index in east Asians. Nat Genet 44 : 307–311. doi: 10.1038/ng.1087 22344219
23. Ng MC, Hester JM, Wing MR, Li J, Xu J, et al. (2012) Genome-wide association of BMI in African Americans. Obesity 20 : 622–627. doi: 10.1038/oby.2011.154 21701570
24. Berndt SI, Gustafsson S, Mägi R, Ganna A, Wheeler E, et al. (2013) Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet 45 : 501–512. doi: 10.1038/ng.2606 23563607
25. Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, et al. (2013) A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet 45 : 690–696. doi: 10.1038/ng.2608 23583978
26. Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, et al. (2014) Genetic studies of body mass index yield new insights for obesity biology. Nature (in press).
27. Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, et al. (2014) New genetic loci link adipose and insulin biology to body fat distribution. Nature (in press).
28. Marchini J, Howie B (2010) Genotype imputation for genome-wide association studies. Nat Rev Genet 11 499–511. doi: 10.1038/nrg2796 20517342
29. The International HapMap Consortium (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449 : 851–861. 17943122
30. The International HapMap Consortium (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467 : 52–58. doi: 10.1038/nature09298 20811451
31. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. (2009) Finding the missing heritability of complex diseases. Nature 461 : 747–753. doi: 10.1038/nature08494 19812666
32. Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB (2010) Rare variants create synthetic genome-wide associations. PLoS Biol 26: e1000294.
33. Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42 : 565–569. doi: 10.1038/ng.608 20562875
34. Pritchard JK (2001) Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet 69 : 124–137. 11404818
35. Barrett JC, Cardon LR (2006) Evaluating coverage of genome-wide association studies. Nat Genet 38 : 659–662. 16715099
36. Anderson CA, Pettersson FH, Barrett JC, Zhuang JJ, Ragoussis J, et al. (2008) Evaluating the effects of imputation on the power, coverage and cost-efficiency of genome-wide SNP platforms. Am J Hum Genet 83 : 112–119.
37. Jostins L, Morley KI, Barrett JC (2011) Imputation of low-frequency variants using the HapMap3 benefits from large, diverse reference sets. Eur J Hum Genet 19 : 662–666. doi: 10.1038/ejhg.2011.10 21364697
38. The 1000 Genomes Project Consortium (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491 : 56–65. doi: 10.1038/nature11632 23128226
39. Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44 : 955–959. doi: 10.1038/ng.2354 22820512
40. Porcu E, Sanna S, Fuchsberger C, Fritsche LG (2013) Genotype imputation in genome-wide association studies. Curr Protoc Hum Genet: Chapter 1, Unit 1.25.
41. Duan Q, Liu EY, Croteau-Chonka DC, Mohlke KL, Li Y (2013) A comprehensive SNP and indel imputability database. Bioinformatics 29 : 528–531.
42. Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529. doi: 10.1371/journal.pgen.1000529 19543373
43. Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, et al (2014) Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9 : 1192–1212. doi: 10.1038/nprot.2014.071 24762786
44. Yang J, Ferreira T, Morris AP, Medland SE; Genetic Investigation of ANthropometric Traits (GIANT) Consortium, et al. (2012) Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 44 : 369–375. doi: 10.1038/ng.2213 22426310
45. Maller JB, McVean G, Byrnes J, Vukcevic D, Palin K, et al. (2012) Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet 44 : 1294–1301. doi: 10.1038/ng.2435 23104008
46. Morris AP, Voight BF, Teslovich TM, Ferreira T, Segrè AV, et al. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 44 : 981–990. doi: 10.1038/ng.2383 22885922
47. Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, et al. (2014) Identification and functional characterization of G6PC2 coding variants influencing glycaemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet 11: e1004876.
48. Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, et al. (2014) Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46 : 136–143. doi: 10.1038/ng.2870 24413736
49. Huyghe JR, Jackson AU, Fogarty MP, Buchkovich ML, Stancakova A, et al. (2013) Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet 45 : 197–201. doi: 10.1038/ng.2507 23263489
50. Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, et al. (2014) Association of low-frequency and rare coding-sequence variants with blood lipids and coronary artery disease in 56,000 whites and blacks. Am J Hum Genet 94 : 223–232. doi: 10.1016/j.ajhg.2014.01.009 24507774
51. Holmen OL, Zhang H, Zhou W, Schmidt E, Hovelson DH, et al. (2014) No large-effect low-frequency coding variation found for myocardial infarction. Hum Mol Genet 23 : 4721–4728. doi: 10.1093/hmg/ddu175 24728188
52. Moutsianas L, Morris AP (2014) Methodology for the analysis of rare genetic variation in genome-wide association and re-sequencing studies of complex human traits. Brief Funct Genomics 13 : 362–370. doi: 10.1093/bfgp/elu012 24916163
53. Chen F, Klein AP, Klein BE, Lee KE, Truitt B, et al. (2014) Exome array analysis identifies CAV1/CAV2 as a susceptibility locus for intraocular pressure. Invest Opthalmol Vis Sci 56 : 544–551.
54. Wessel J, Chu AY, Willems SM, Wang S, Yaghootkar H, et al. (2015) Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat Comms 6 : 5897.
55. Chen JA, Wang Q, Davis-Turak J, Li Y, Karydas AM, et al. (2015) A multiancestral genome-wide exome array study of Alzheimer disease, frontotemporal dementia, and progressive supranuclear palsy. JAMA Meurol (in press).
56. Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55 : 997–1004. 11315092
57. Magi R, Morris AP (2010) GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 11 : 288. doi: 10.1186/1471-2105-11-288 20509871
58. Ioannidis JP, Patsopoulos NA, Evangelou E (2007) Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE 2: e841. 17786212
59. Wakefield JA (2007) Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet 81 : 208–227. 17668372
60. Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucl Acids Res 40: D930–934. doi: 10.1093/nar/gkr917 22064851
61. Kumar P, Henikoff S, Ng P (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4 : 1073–1081. doi: 10.1038/nprot.2009.86 19561590
62. Choi Y, Sims GE, Murphy S, Miller JR, Chan AP (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7: e46688. doi: 10.1371/journal.pone.0046688 23056405
63. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7 : 248–249. doi: 10.1038/nmeth0410-248 20354512
64. The ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489 : 57–74. doi: 10.1038/nature11247 22955616
65. Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, et al. (2014) Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46 : 136–143. doi: 10.1038/ng.2870 24413736
66. Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, et al. (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143 : 156–169. doi: 10.1016/j.cell.2010.09.006 20887899
67. Li H, Durbin R. (2009) Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 25 : 1754–1760. doi: 10.1093/bioinformatics/btp324 19451168
68. Ernst J, Kellis M (2010) Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28 : 817–825. doi: 10.1038/nbt.1662 20657582
69. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, et al. (2012) GENCODE: The reference human genome annotation for the ENCODE project. Genome Res 22 : 1760–1774. doi: 10.1101/gr.135350.111 22955987
Štítky
Genetika Reprodukční medicína
Článek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání





