-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
Streptococcus mutans is a bacterium of the human dental plaque that contributes to caries development. It controls two important survival mechanisms via a cell-density dependent communication system (quorum sensing): The synthesis of peptide antibiotics, and of a membrane apparatus for genetic competence, i.e. the ability to take up external DNA and integrate it into its own genome. S. mutans synthesizes two different signalling peptides to this end. It has remained elusive, how exactly these signals are propagated within the cell and why only a fraction of the population becomes competent. To actually observe under the microscope which bacterium in the population is activated, and which genes are required for the activation, we constructed strains of S. mutans that reported on the transcription of a gene by starting to fluoresce green. We even constructed strains that reported on two genes simultaneously, by fluorescing either green or blue or both. With these tools, and by additionally knocking out or modifying key genes as needed, we investigated the complete signaling cascade under various conditions. Thus we discovered a central regulatory switch. S. mutans makes sure that external DNA is available when it becomes genetically competent–by killing cells in the environment.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005353
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005353Summary
Streptococcus mutans is a bacterium of the human dental plaque that contributes to caries development. It controls two important survival mechanisms via a cell-density dependent communication system (quorum sensing): The synthesis of peptide antibiotics, and of a membrane apparatus for genetic competence, i.e. the ability to take up external DNA and integrate it into its own genome. S. mutans synthesizes two different signalling peptides to this end. It has remained elusive, how exactly these signals are propagated within the cell and why only a fraction of the population becomes competent. To actually observe under the microscope which bacterium in the population is activated, and which genes are required for the activation, we constructed strains of S. mutans that reported on the transcription of a gene by starting to fluoresce green. We even constructed strains that reported on two genes simultaneously, by fluorescing either green or blue or both. With these tools, and by additionally knocking out or modifying key genes as needed, we investigated the complete signaling cascade under various conditions. Thus we discovered a central regulatory switch. S. mutans makes sure that external DNA is available when it becomes genetically competent–by killing cells in the environment.
Introduction
Horizontal gene transfer in prokaryotes is mediated via three distinct mechanisms comprising conjugation, transduction, and transformation [1,2]. Studying those mechanisms in detail is needed because they are among the reasons for the spread of antibiotic resistance and virulence determinants between bacteria [3]. Natural transformation, i.e. the uptake of extracellular DNA from the environment via genetic competence, is a powerful process able to expand and modify the gene inventory in both Proteobacteria and Firmicutes [4]. It requires a multi-protein complex localized in the cell membrane, many elements of which are highly conserved, and was studied in most detail in Bacillus subtilis and Vibrio cholerae [5,6].
In streptococci competence is a tightly controlled transient state whose activation involves detection of quorum sensing (QS) signaling peptides, which in streptococci are termed pheromones. In all streptococci studied so far the proximal master regulator of competence, and final receiver of the transduced signals, is the alternative sigma factor SigX (previously termed ComX). SigX binding to the RNA polymerase activates transcription of a core set of ~ 20 “late” competence effector genes [7]. They carry a nine bp cin-box in their promoter region and mediate the synthesis of proteins for DNA uptake and recombination [8]. The SigX regulon has been found in all streptococci sequenced to date, suggesting that genetic competence is ubiquitous in streptococci, although until now it could only be demonstrated experimentally in very few [9].
Although all streptococci use peptide pheromones for density dependent activation of sigX expression as well as bacteriocin synthesis, the details of signal transduction and integration differ widely between the different species. Two principal types of pheromones and signal detection mechanisms have been found [10]: Pheromones derived from pre-peptides carrying a double-glycine leader sequence are cleaved and exported by the membrane localized ComAB complex, and after an additional processing step the mature pheromone is detected in the extracellular environment through the transmembrane sensor histidine kinase of a two component signal transduction system that phosphorylates its corresponding response regulator. The phosphorylated response regulator induces transcription of the alternative sigma-factor sigX. The competence-stimulating peptide (CSP) of S. pneumoniae belongs into this group. The second type of pheromones was recently discovered and shown to be conserved in streptococci [7,11–13]. A pre-peptide is synthesized, exported and processed to yield a short hydrophobic mature peptide termed XIP (sigX inducing peptide), which is then re-imported by a permease. Its detection occurs by an intracellular transcription factor that interacts directly with the peptide signal [10]. The activated, dimeric regulator induces transcription of sigX by binding to its promoter region [12], much like the LuxR type QS regulators that are activated by binding of acylated homoserine lactons (AHLs), the QS signals of Proteobacteria.
In S. mutans competence is regulated via both types of QS signaling peptides and the medium composition determines which signal is active. Moreover the medium also determines whether competence is induced uni-modally [14] or only in a subpopulation of cells [7,15].
The current understanding of competence development in S. mutans is shown in Fig 1. In complex media competence can only be induced by CSP (Fig 1A). The 46 amino acid CSP precursor is encoded by the comC gene and processed and exported by the comAB encoded ABC transporter, yielding the extracellular 21 residue CSP-peptide [16]. This is cleaved to its biologically active form by the protease SepM, thereby removing the 3 C-terminal amino acids [17]. Binding of CSP-18 to the histidine kinase ComD induces autophosphorylation of the protein and results in the transfer of the phosphorylgroup to a conserved aspartic acid residue of the cognate response regulator ComE. Phosphorylated ComE binds to two direct repeats in the promoter regions of genes encoding bacteriocins and their corresponding immunity proteins resulting in transcription of the “early competence genes” [18–20]. Via an unknown link comDE activation induces comRS expression and finally competence development in a subpopulation of the cells [7,15].
Fig. 1. Previous model of competence development in S. mutans in complex (A) and defined medium (B). 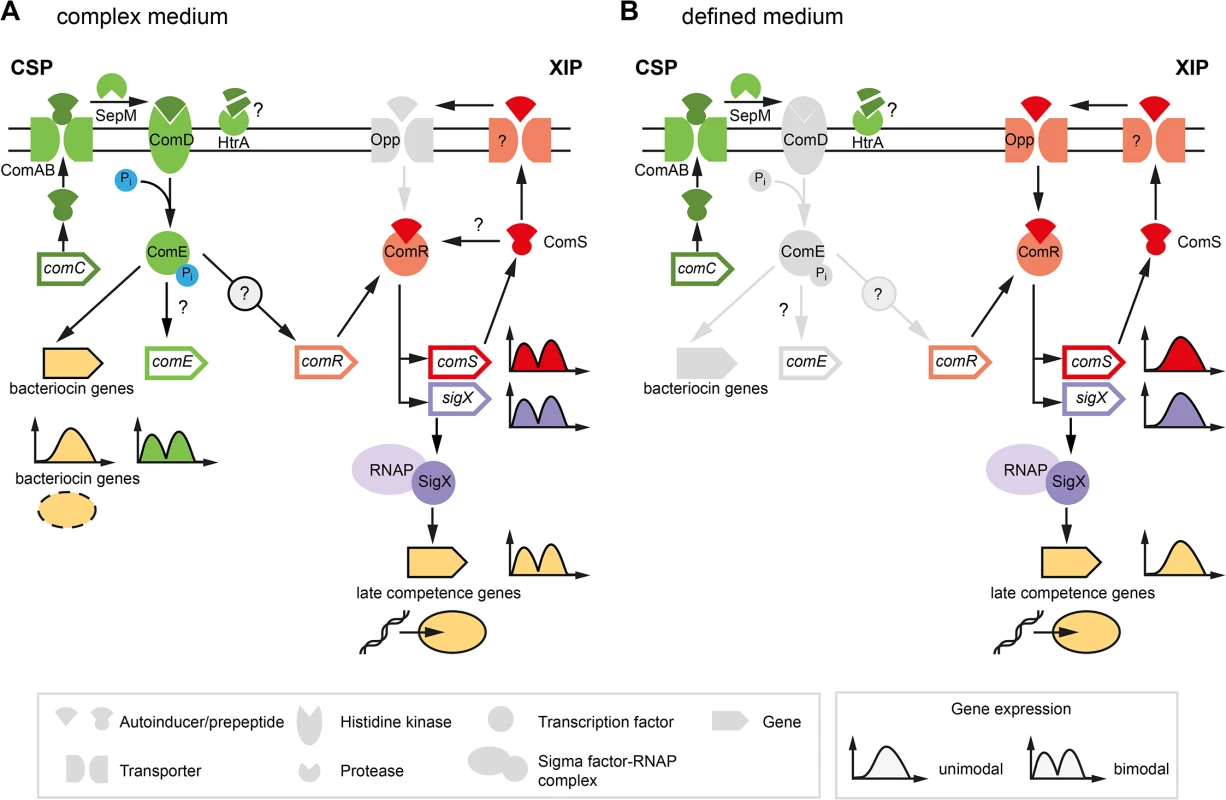
The different classes of structural components of the competence network are explained below in the legend (grey box in the left). Two quorum sensing pathways regulate competence in S. mutans. Components of the ComDE circuit with the autoinducer CSP are shown in green while components of the ComRS circuit and the autoinducer XIP are illustrated in red color. Inactive elements of the circiuts are shown in grey. The two QS pathways are operating in parallel and converge on the central comRS system. Both circuits are connected via an unknown link between ComDE and ComRS in complex medium. The medium determines which autoinducer is biologically active and whether competence is induced bimodally or unimodally across the population. This is illustrated by the gene expression shemes (grey box in the right). The modality of the expression of involved genes is illustrated in the same coloring as used for the gene symbol. The output of the systems, bacteriocin expression and competence, are symbolized below the bacteriocin encoding genes and the late competence genes in yellow. Bacteriocin expression is exclusively observed in complex medium while competence development in defined medium is not coupled to bacteriocin expression. In the peptide free chemically definde medium (CDM), competence can only be induced by XIP, the second autoinducer of S. mutans (Fig 1B), via the ComRS system, that is also present in other mutans, pyogenic and bovis group streptococci [7,21]. It consists of an Rgg type transcriptional regulator (ComR) and a small XIP peptide, encoded by the comS gene [7]. The XIP pre-peptide is processed and the active seven residue pheromone is secreted into the environment by an unknown process. After accumulation extracellular XIP is internalized, most likely via the Opp permease, and binds to ComR in a proposed stoichiometry of 2 : 1 (XIP:ComR) [7,12]. Resulting conformational changes promote dimerization of the XIP2/ComR complex and binding to DNA targets containing the ComR binding motif. In S. mutans binding motifs for ComR were found upstream of the comS and the sigX gene [7,12]. Thus XIP controls its own expression in a positive auto-regulatory feedback loop. XIP mediated competence induction is exclusively observed in chemically defined media and results in uni-modal expression of competence.
The effect of the medium is hypothesized to be caused by differences in transport and degradation, respectively, of the autoinducers. In S. pneumoniae it was shown that CSP is degraded by the membrane bound protease HtrA, and that this degradation is reduced in the presence of unfolded proteins [22]. Therefore, it was hypothesized that the homologous HtrA of S. mutans might be inhibited from CSP degradation by small peptides present in complex medium, resulting in active CSP in complex but not in minimal medium [15]. Conversely, XIP is active in defined, but not in complex media. It was suggested that the import of XIP might be inhibited in complex media due to the clogging of the Opp permease [15]. Small amounts of peptides that are added to the cultivation medium completely eliminate the activity of XIP [15]. However, it remains unclear how under CSP induced conditions ComR might be activated by XIP, if this signal cannot be imported into the cell (Fig 1A).
Bacteria often respond to environmental stimuli in a non-uniform manner; even in isogenic cultures and under homogenous conditions the appearance of multiple phenotypes (phenotypic variation) is observed [23]. Phenotypic variability has strong implications for the treatment of bacterial infections and is relevant for cellular differentiation [24]. Two distinct phenotypes occurring simultaneously are referred to as bimodal and were observed e.g. for lactose utilization, chemotaxis and persister cell occurence in E.coli [25] and for competence development and sporulation in B. subtilis [23,26,27]. It was previously shown that phenotypic heterogeneity enhances the overall fitness of the population under fluctuating conditions and helps bacteria to colonize different ecological niches within an ecosystem [23,28]. Thus phenotypic variability can be considered as a bet-hedging strategy and an evolvable trait.
Signaling in bacteria is never discrete since stochastic fluctuations of the components that determine a cellular state occur [29]. This phenomenon is called noise and it is most pronounced for processes involving a limited number of molecules such as transcription and translation [30]. Noise is one critical determinant for the establishment of phenotypic variation [23,29]. Phenotypic heterogeneity can also be a result of the architecture of regulatory networks comprising positive feedback loops or toggle switches (two regulators that negatively regulate each other) which can amplify signals and respond nonlinearly to stimuli [23,24]. Noisy gene expression in conjunction with such networks can convert a graded signal into a bimodal response which is stably maintained in the population.
An intriguing feature of the competence cascade in S. mutans is its bimodality under CSP induced conditions in THBY medium. It was hypothesized to be caused by the auto-regulatory feedbackloop of ComS [15]. A different hypothesis was developed by Lemme et al. [14] who found that the entire population expressed the bacteriocin related genes in an uni-modal way, while only a subpopulation expressed sigX and entered the competent state. However, bimodal expression of comE was observed at the same time. Since comE is supposed to act upstream of ComRS signaling, it was therefore hypothesized that comE expression might be the origin of bimodality [14].
Finally, an intriguing question is the autolysis of a fraction of the population after stimulation by XIP [31] or CSP [14,32,33]. In S. pneumoniae, it is the non-competent subpopulation which is killed by the competent siblings, a phenomenon termed fratricide [9]. The CbpD murein hydrolase which is responsible for this process has a homologue termed LytFsm in S. mutans. It has however been shown that lysis occurs in a fraction of the competent cells [14,34] and that LytFsm is a self-acting autolysin, not a fratricin [32].
The QS signaling cascade of S. mutans thus comprises two different signals, one that is detected in the external environment (CSP), and one that is detected intracellularly (XIP). They are linked with each other in different ways in different media. The system has a temporal dimension (“early” and “late” competence genes), and it can result in phenotypic heterogeneity of the population. Here we asked (1) At which level in the competence signaling cascade is bimodal gene expression triggered? (2) How is the ComE signaling cascade connected to the ComRS signaling cascade? (3) How can bacteriocin expression be unimodal, although comE expression is bimodal? (4) How is the medium effect on signaling mediated—is CSP really degraded in CDM, and how can an active XIP/ComR complex be formed in THBY?
To answer these questions, it was necessary to follow the activation of the key genes along the signaling cascade on the single cell level. We developed a tool-box of integrative reporter plasmids carrying a fusion of the promoter of interest with a fluorescent protein. Those constructs were integrated into the chromosome of S. mutans. We used the wild-type, as well as strains that had key genes of the signaling cascade deleted, over-expressed, or modified. To monitor co-expression of “early” and “late” competence genes, we developed dual-fluorescent reporter strains that clearly demonstrated which of the cells induced upstream became finally competent. Population heterogeneity was observed directly under the microscope or using flow cytometry. RNA sequencing was used to determine gene expression of all involved components during the first 30 min post induction. The data result in a new understanding of the QS regulatory circuit of S. mutans that resolves the discordant observations described above.
Results
Signal propagation through the competence cascade
The construction and experimental verification of all single and dual fluorescent reporter strains used in this study is described in detail in S1 Text and S1–S6 and S9 Figs. Using dual fluorescent reporter strains we analyzed signal propagation along the entire CSP-induced competence cascade in S. mutans, from the upstream comDE system to the central regulatory comRS module and the alternative sigma-factor sigX to the late competence gene lytFsm, a component of the transformasome. First the effect of the CSP inducer concentration was evaluated. S7 Fig (flow cytometry) and S8 Fig (microscopy) show that bimodal lytFsm expression was observed above a concentration of 20 nM CSP; the percentage of cells expressing lytFsm was not significantly changed between 200 nM and 100 μM CSP. Thus 2 μM CSP was chosen as inducer concentration in all following experiments. No significant influence on cell growth and viability was observed under these conditions while higher concentrations of CSP induce cell death and growth arrest [34]. The analysis was conducted 2.5 h post induction because the expression of the late competence genes reaches its maximum after 2 h [14] and the full maturation of the fluorescent proteins requires additional 30 minutes [35,36]. Fig 2 shows that the comE gene is co-expressed with comS, sigX and the late competence gene lytFsm. Moreover and not surprisingly comS is coxpressed with sigX (S5B Fig), since both genes share the same ComR binding sequence in their 5`UTR [7].,. All four genes are expressed in a bimodal way. The same coexpression patterns were also observed when we switched fluorophores for the two genes under study in the reporter strains (S9 Fig). To conclude, we observed an identical bimodal gene expression pattern for all genes in the signaling cascade. Two mechanistic explanations would be in accordance with the observed coexpression pattern and the bimodality of comE expression. Both models assume a basal expression of comE.
Fig. 2. Single cell co-expression analysis of comE with the late competence genes lytFsm (A), comS (B) and sigX (C) using dual fluorescent reporter strains. 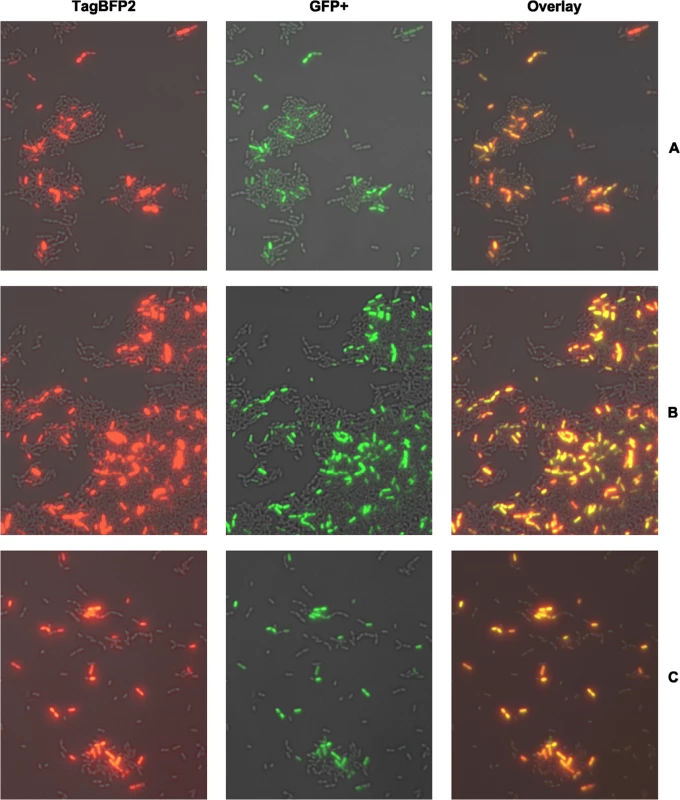
Strains were grown in complex THBY medium and gene expression induced with 2 μM CSP. 150 minutes post induction cells were collected and imaged using fluorescence microscopy. ComE expressing cells show green fluorescence (middle column), while the blue fluorescence (left column) indicates expression of co-expression partners. An overlay of the green and blue channel is shown in the right column. For better visualization the blue fluorescence is shown in red false color. ComE itself might represent the origin of bimodality and propagates the signal linearly to the downstream genes.
One of the downstream regulatory systems (e.g. ComRS or SigX) induces expression of the upstream localized comE gene in a subpopulation, leading to bimodality.
Overexpression of ComDE and ComRS
To determine whether ComD/ComE or ComR/ComS represent the origin of bimodality during the CSP-induced competence development of S. mutans, we constructed overexpression strains strongly and constitutively expressing those proteins independent from their native promoters and thus equally across the entire population. Overexpression of the cellular component(s) representing the origin of bimodality should cause unimodal lytFsm expression (indicative of unimodal competence) across the population. We utilized plasmid pIB166 [37] and cloned the genes downstream of the strong constitutive Lactococcal P23 promoter. Plasmids were transformed into the LytFsm pAE03 reporter strain. LytFsm is a late competence gene [32] and it is the most strongly expressed late competence gene of the tested reporters (S4 Fig). We used the comRS overexpression strain in defined medium to test whether unimodal lytFsm expression is observed, as expected according to Masburn-Warren et al.[7]. LytFsm expression was unimodally induced even at low cell densities and without addition of external XIP (S10 Fig), indicating that endogeneous production of XIP is sufficient to induce competence unimodally. To analyse the origin of bimodality, the experiments were conducted in THBY under CSP inducing conditions. Fig 3 shows that for all overexpression strains CSP was needed to induce lytFsm expression. In a transcriptional signaling cascade from comDE to sigX it would have been expected that overexpression of one component should be sufficient to induce competence. The strong constitutive overexpression of comE or comD in the entire population did not enhance the percentage of lytFsm induced cells (Figs 3 and S11). Thus we can exclude that ComE or ComD is the origin of bimodality. Overexpression of only comR or only comS still resulted in a biphasic population behavior, although the proportion of LytFsm expressing cells was significantly enhanced compared to the WT or the comE overexpression strain. However, overexpression of both genes together (comRS) from the lactococcal P23 promoter resulted in unimodal expression of LytFsm, proving that comRS represents the origin of bimodality (Figs 3 and S11). ComR needs to be activated by XIP which is synthesized by comS [12]. Thus overexpression of comR or comS alone is not sufficient to induce sigX and the late competence genes. Both comR and comS are needed for the positive feedback loop to occur.
Fig. 3. Overexpression analysis of comE, comR, comS and comRS in the LytFsm reporter strain background. 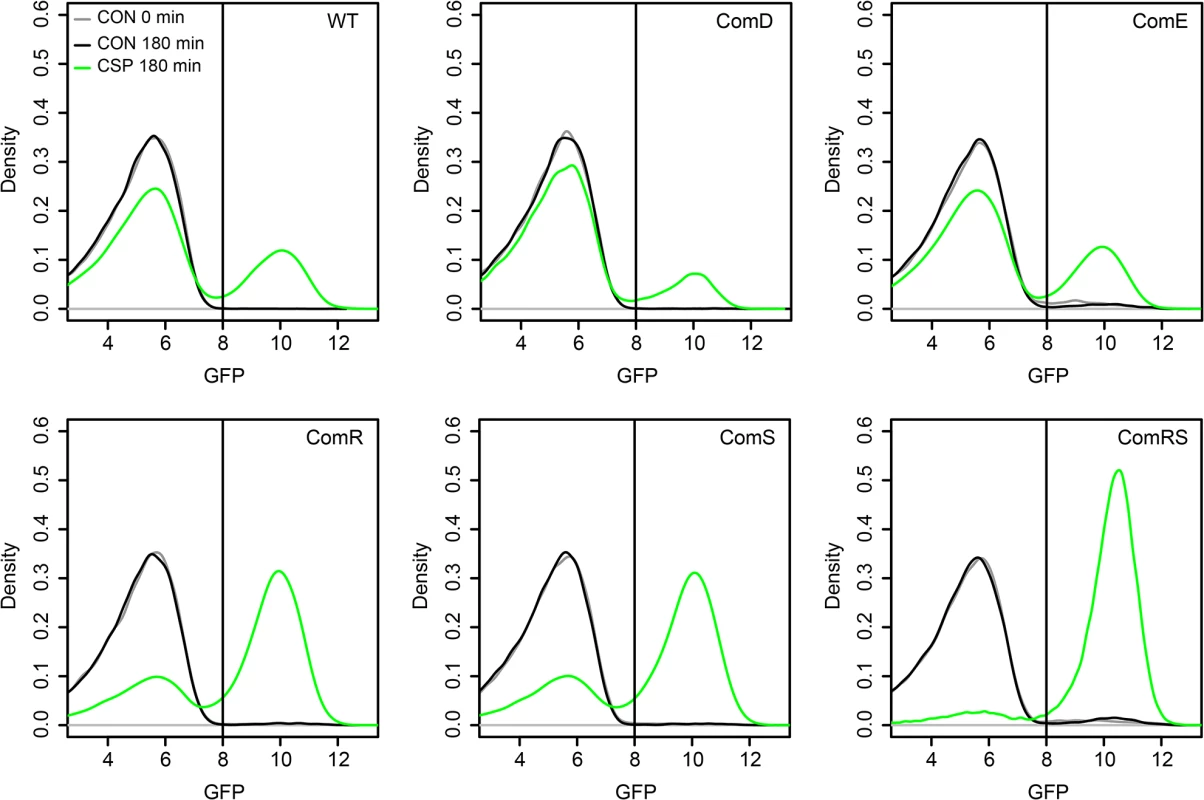
Expression of the different genes was under the control of the strong constitutive lactococcal P23 promoter on the replicative plasmid pIB166. Strains were grown in THBY under CSP induced (2μM) and uninduced conditions. 180 min post induction samples were collected and analyzed using a flow cytometer. A sample collected directly before induction (0 min) was used as negative control. The distribution of the gfp fluorescence intensity of 50.000 analysed cells as determined by flow cytometry is shown in the density plots. Density plots derived from uninduced cells at t = 180 min are shown in black while the density plots of CSP induced cells at that timepoint are shown in green. Density plots of the control at t = 0 min are shown in grey. Surprisingly, however, unimodal competence development in the comRS overexpression strain absolutely required CSP induction in THBY medium. No competence development was observed in the absence of CSP. A regulator upstream of comRS as assumed currently (Fig 1B) cannot be responsible for this observation, because overexpression of comRS would be able to overcome it. In complex medium it is thought that import of secreted XIP is impossible due to the block of the Opp peremease by small peptides (Fig 1A). The data suggest that CSP itself or induced cellular changes are able to bypass this block.
Combined effect of XIP and CSP on lytFsm expression
Thus we tested how the percentage of cells expressing the late competence gene lytFsm was influenced by XIP supplementation to a CSP induced LytFsm reporter strain grown in complex medium and how the relative amounts of those two signals affect bimodal gene expression. Accordingly different combinations of CSP and XIP with concentrations ranging from 0.2 μM to 20 μM of each inducer were tested. The results of the flow cytometric analysis of the LytFsm pAE03 reporter strain 120 minutes post XIP and/or CSP supplement are presented in Fig 4. As expected, XIP supplementation alone did not induce expression of the late competence gene lytFsm, regardless of the used concentration (Fig 4A). Conversely, and as expected, too, addition of CSP alone induced bimodal lytFsm expression. The percentage of cells expressing lytFsm was always constant, independent of the used CSP concentration in a range from 0.2 μM to 100 μM (Figs 4B and S7 and microscopic images in S8 Fig). However, addition of both XIP and CSP in a ratio of 1 : 1 resulted in expression of lytFsm (Fig 4C) while no expression had been observed with XIP alone up to a concentration of 20 μM. At the highest concentration of XIP and CSP tested here, unimodal expression of competence was observed. Lowering the CSP concentration to 0.2 μM still resulted in unimodal lytFsm expression (Fig 4D). The XIP concentration strongly determines the percentage of lytFsm expressing cells, while the CSP concentration is of minor importance.
Fig. 4. Combined effect of CSP and XIP peptides on lytFsm expression in complex medium. 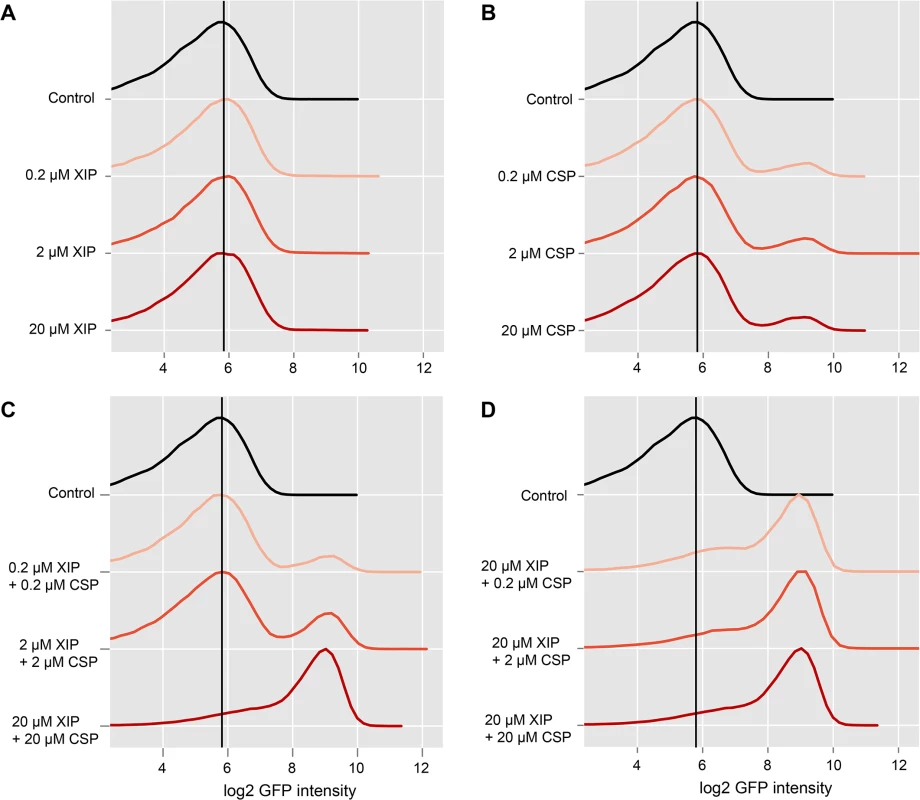
The LytFsm pAE03 reporter strain was grown in complex medium until the culture reached an OD600 of 0.2. The culture was divided and induced with either CSP or XIP alone or treated with combinations of XIP and CSP. Concentrations between 200 nM and 20 μM of each inducer were tested. 2 h post induction cells were harvested and analyzed using flow cytometry. 50.000 cells were analyzed for each condition. Density plots of the log2 gfp fluorescence intensity for the differently treated reporter cells are shown. Controls are shown in black. Increasing inducer concentrations are visualized by increased intensity of the red color. The vertical black line represents the peak fluorescence intensity of the uninduced control. A Effect of increasing concentrations of XIP on the fluorescence of the LytFsm reporter strain. B Effect of increasing concentrations of CSP on the reporter strain. C LytSsm fluorescenc of cells treated with combinations of XIP and CSP in equimolar ratios. D Effect of different molar ratios of XIP to CSP. We hypothesize that extracellular XIP is imported into the cell in a concentration dependent way, but only in the presence of CSP. How can this be achieved? The XIP uptake must be independent of Opp, since its deletion does not affect the CSP induced bimodal competence phenotype in complex medium [15]. We suggest that CSP induces import of XIP in an indirect way by inducing expression of bacteriocins, resulting in permeabilization of the cell. As relatively high XIP concentrations are required, it is likely that the import of the small XIP peptide is ineffective in THBY. To further substantiate this hypothesis we deleted the cipB gene in the LytFsm reporter strain background and tested whether addition of CSP or XIP or the combination of both petides still induces lytFsm expression. Strinkingly no induction of lytFsm expression was found in the CipB deletion strain, regardless which peptide or peptide combination was used for induction (S12 Fig). Accordingly, Perry et al.[34] observed that CipB deletion completely abolished CSP induced competence development.
Since there is no direct regulatory link between comDE and comRS (see above), and since competence induction by CSP in complex medium appears to be an indirect effect, we hypothesize that CSP controls only bacteriocin synthesis, while competence is exclusively controlled via XIP.
Biological activity of CSP in chemically defined medium
Previously only competence development has been tested in both CDM and THBYmedia, and it was shown that in CDM competence is not induced by CSP, regardless of the used concentration [15]. It was speculated that CSP might be proteolytically degraded by the HtrA protease in CDM, while in complex medium this protease was thought to be saturated by small peptides that are constituents of the medium, thus protecting CSP from degradation [15]. However, it was not yet tested if bacteriocin encoding genes are transcribed in CDM under CSP induced conditions. Using the CipB pMR1 reporter strains in the different deletion backgrounds we therefore tested whether CSP induction in CDM promotes transcriptional activation of bacteriocins. We show (Table 1 and S13 Fig) that CSP promotes a strong induction of cipB expression in CDM and thus is biologically active and not degraded. Induction of cipB expression occurs via ComDE, as deletion of either comD or comE completely abolished it. The induction of cipB expression was unchanged in the ΔcomRS, ΔcomS and ΔsigX background. An influence of those regulatory systems can thus be excluded. To summarize, CSP is active in CDM. It induces transcription of bacteriocin encoding genes via ComDE but does not induce competence via the ComRS system.
Tab. 1. Expression of cipB in different gene deletion background in CDM under CSP induced conditions. 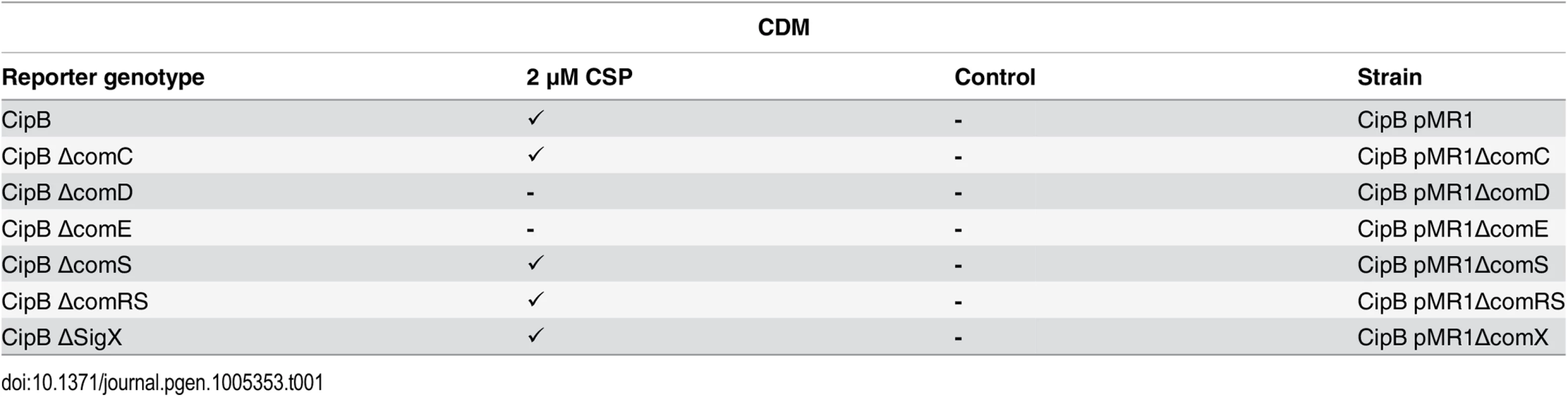
In the genetic background of the fluorescent reporter strain CipB pMR1 deletion mutants for ΔcomC, ΔcomD, ΔcomE, ΔcomS and ΔcomRS were constructed, cultivated in CDM medium and induced by CSP (2 μM). Fluorescence microscopic images were obtained 3 hours after induction. Uninduced strains were used as controls. Tickmarks indicate detected fluorescence, tickmarks in brackets indicate weak fluorescence, a minus indicates no detectable fluorescence. Transcriptome analysis of CSP and XIP signaling in CDM
To demonstrate that XIP signaling induces exclusively competence, while CSP controls only bacteriocin expression, we performed a time resolved analysis of the transcriptional response to these two autoinducers during the first 30 min after supplementation using RNA sequencing.
Differentially expressed genes are presented in Fig 5 and are grouped according to quorum sensing related genes, mutacins, competence and others. CSP supplemention in CDM induced the expression of mutacins and their corresponding immunity proteins, while competence related genes were not found to be differentially expressed. The exact opposite behavior was observed for XIP stimulation in CDM, confirming our hypothesis. For the XIP signaling cascade it can be seen that the transcription of the central XIP responsive transcriptional regulator ComR was only very slightly induced, in accordance with the general observation that essential key regulators are unlikely to be strongly transcriptionally regulated. The primary targets of ComR, comS and sigX were instantly induced by XIP, representing the strongest responding transcripts. The up-regulation of the two genes already 5 minutes after XIP addition is in accordance with the post-translational activation of the ComR protein by XIP binding. Interestingly, XIP addition resulted in the activation of comDE expression for the later time-points (15 and 30 min). The CSP signaling cascade, by contrast, did not induce sigX or comS expression as expected. comDE expression was only very slightly induced upon CSP stimulation. However, mutacins, the primary targets of ComE, were strongly transcribed already 5 min post supplementation, suggesting post-transcriptional activation of the regulator ComE.
Fig. 5. Time resolved transcriptome analysis of XIP and CSP induced S. mutans WT cultures in CDM. 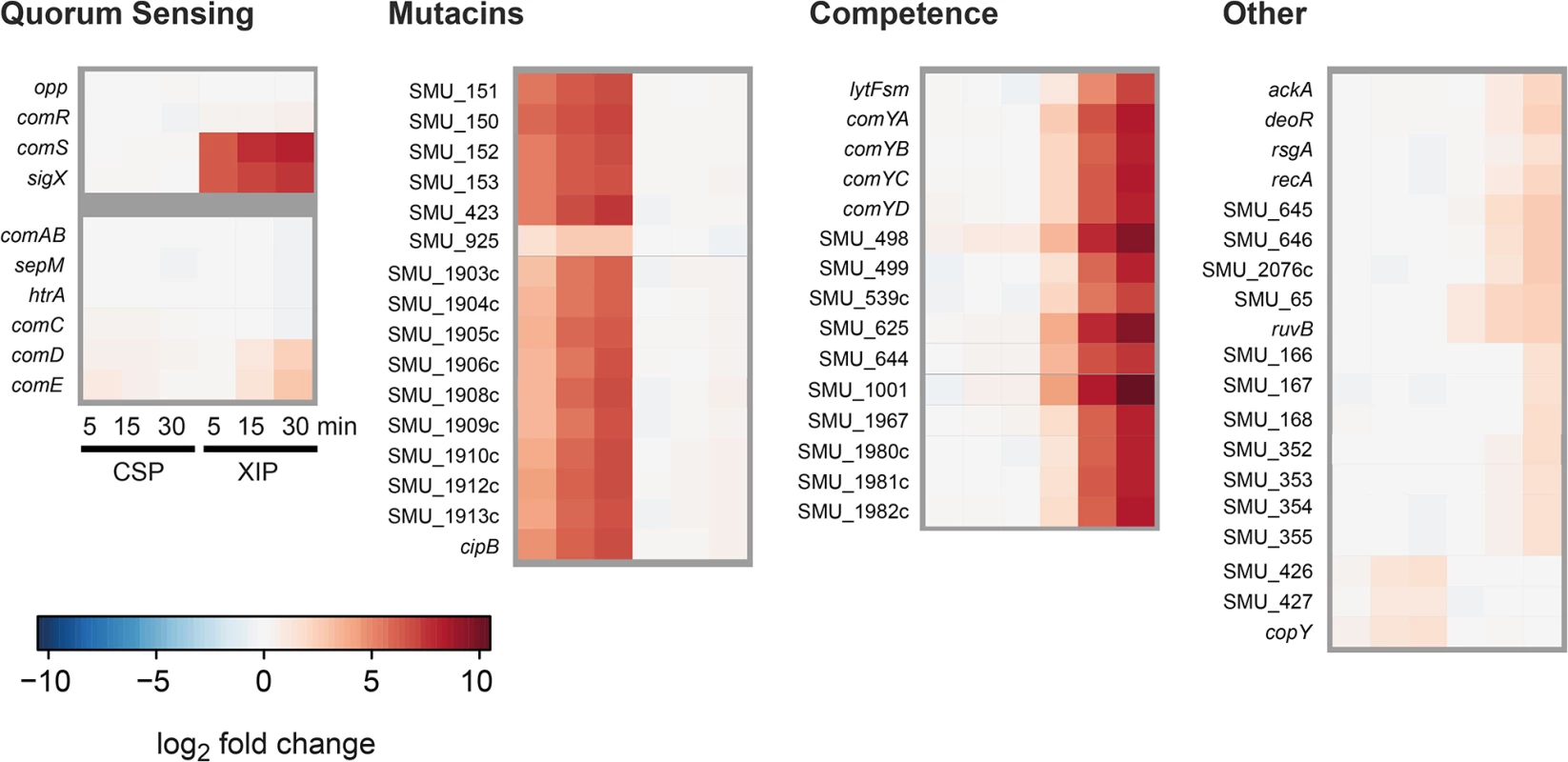
S. mutans WT cells were grown in CDM and induced either with 2 μM CSP or 2 μM XIP. Samples were taken 0, 5, 15 and 30 minutes post induction. Samples from uninduced controls were taken at corresponding timepoints. Differentially expressed genes (log2 fold > 1) were clustered into the categories competence, mutacins, quorum sensing and others. Differential expression of the respective genes is displayed in a heatmap. Genes that belong to the two quorum sensing regulons controlling competence in S. mutans but were no differentially expressed are also shown. Genes encoding transporters and proteases (ComAB, HtrA, Opp and SepM) as well as comC, encoding the CSP precursor, were not differentially expressed, regardless if XIP or CSP was used for stimulation.
Both QS systems responded very fast, within the first 5 minutes, to their stimuli. Thus the results of the transcriptome analysis indicate that QS in S. mutans is initiated by the regulators ComE and ComR on the post-transcriptional level. The transcription of the target genes mutacins, comRS and sigX can already be observed 5–15 minutes after stimulation. The response of the “late competence” genes is slightly delayed compared to that of the mutacins, since the activation of SigX is additionally required. The expression of comE and cipB was confirmed by quantitative PCR (S1 Text and S15 Fig).
To summarize our findings up to this point: The data show that comRS is the origin of bimodality. There is no direct signaling cascade from CSP to ComRS. CSP induces bacteriocins. Its role in competence stimulation in complex medium is an indirect one, most likely as a result of bacteriocin synthesis causing permeabilization of the cells and reimport of endogenously produced and secreted XIP.
The co-expression analysis described above however raised the question, why comE is coexpressed with comS, sigX and the late competence gene lytFsm. We hypothesize that transcription of comE is controlled by ComR or SigX.
XIP controls comE expression via SigX
If SigX or ComR would control comE transcription, then the XIP signal should induce comE expression independent of ComDE. We tested this hypothesis using the comE reporter strain in different gene deletion backgrounds under XIP induced conditions in CDM. Table 2 (and microscopic pictures in S16 Fig) shows that this is indeed the case. The transcriptional activation of comE is independent of internal production and external sensing of CSP as it also occurs in ∆comDE and ∆comC deletion strains. Deletion of comRS and surprisingly also sigX completely abolished transcriptional activation of comE. In the ΔcomS background, transcription of comE is slightly reduced, since the internal production of XIP is missing and cannot be completely restored by externally added XIP. In THBY, comE transcription was similarly dependend on ComRS and SigX (S1 Text and S5 Table). These data provide strong evidence for a direct regulatory role of SigX for the expression of comE. Since the targets of SigX contain a cin box in their 5`UTR [38], we analysed the promoter region of comE.
Tab. 2. Expression of comE in different gene deletion background in CDM under XIP induced conditions. 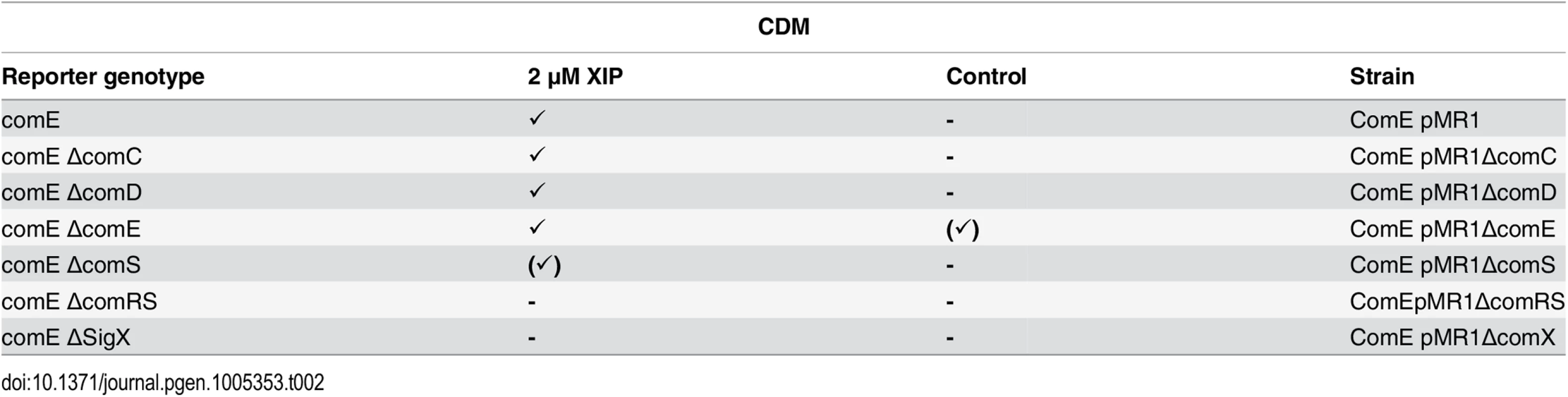
In the genetic background of the fluorescent reporter strain ComE pMR1 deletion mutants for ΔcomC, ΔcomD, ΔcomE, ΔcomS and ΔcomRS were constructed, cultivated in CDM medium and induced by XIP (2 μM). Further explanations as in Table 1. Identification of a cin-box in the promoter region of comE
We searched for the cin-box, a known binding motif for SigX [38], in the 5´ UTR of comE. Indeed we found a sequence 109 bp upstream from the start codon of comE which has only one mismatch with the consensus sequence of the cin-box (TGCGAATA, Fig 6). The late competence gene lytFsm, which is controlled by SigX [32,39] and is highly expressed (our data) also showed one mismatch at the same position (TCCGAATA), thus we conclude that the cin-box homologue identified upstream of comE is most likely functional. We then looked for a promoter sequence. Directly adjacent to the cin-box a -10 region (Pribnow box) was found (TATATT). A corresponding -35 region was not identified. The transcriptome analysis allowed to identify the transcriptional start site of the comE gene. It was localized directly downstream of the cin-box, in complete agreement with the -10 localization of the Pribnow box. Sequence analysis and RNAseq thus strongly suggest that SigX can bind to the 5`UTR of comE and induce transcription.
Fig. 6. RNAsequencing and bioinformatics analysis of the 5`UTR of the comE gene of S. mutans. 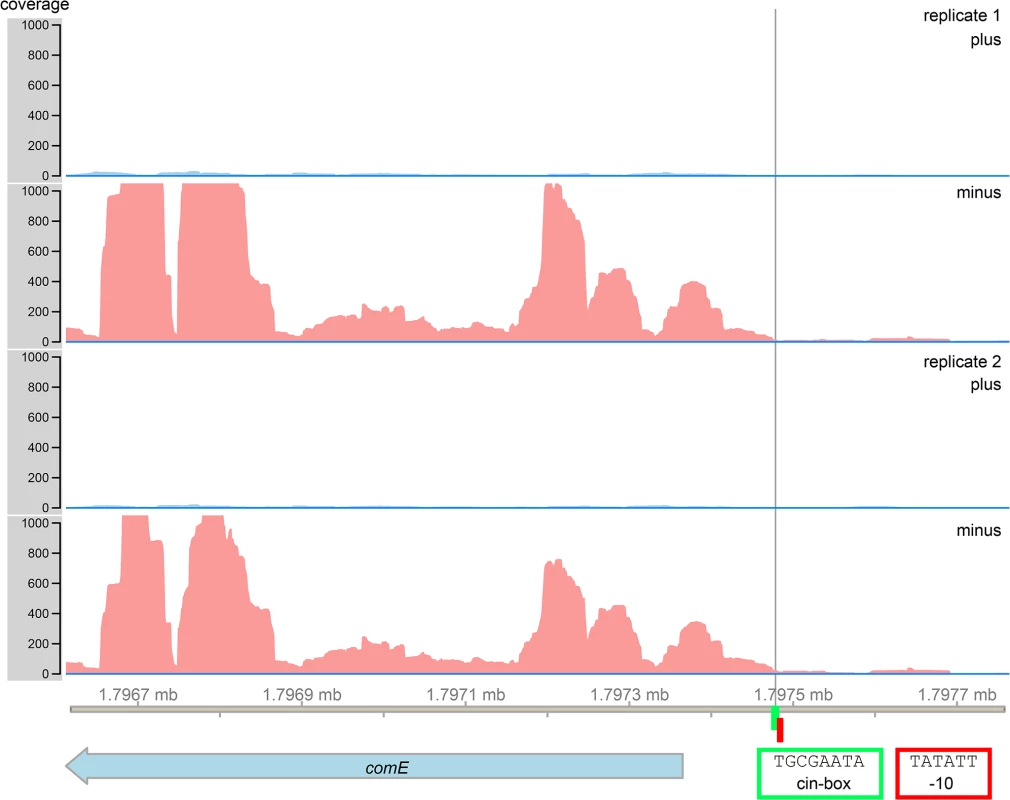
A SigX binding motif (cin-box, highlighted in green) was identified 109 bp upstream of the comE start codon. Directly adjacent to the cin-box a Pribnow box (highlighted in red) was found (red corners). RNA sequencing data revealed the transcriptional start site of the comE gene (black line). Control of bacteriocin transcription and synthesis by XIP
If XIP induces comE expression via SigX it should also induce bacteriocin expression. We analysed the transcription of the mutacin V encoding gene cipB in the cipB pMR1 reporter strain with different gene deletion backgrounds in CDM medium under XIP induced conditions. If SigX controls comE and thus cipB transcription, then fluorescence of the reporter should be induced by XIP, but not in a ΔcomRS or ΔsigX background, and independent of CSP signaling.Table 3 (and microscopic images in S13 Fig) shows that the expression of cipB required the presence of comE,comRS and sigX, and was weakened in the ΔcomD background. As the deletion of comE completely abolished cipB expression we can exclude an influence of other bacteriocin regulatory systems. The results on the CSP induced comE and cipB reporter strains in THBY are shown in S5 Table and S6 Table. Thus we conclude that SigX indeed induces transcription of bacteriocins in S. mutans via ComE. This is a novel role for SigX, therefore we asked if bacteriocins could be found in the culture supernatant.
Tab. 3. Expression of cipB in different gene deletion background in CDM under XIP induced conditions. 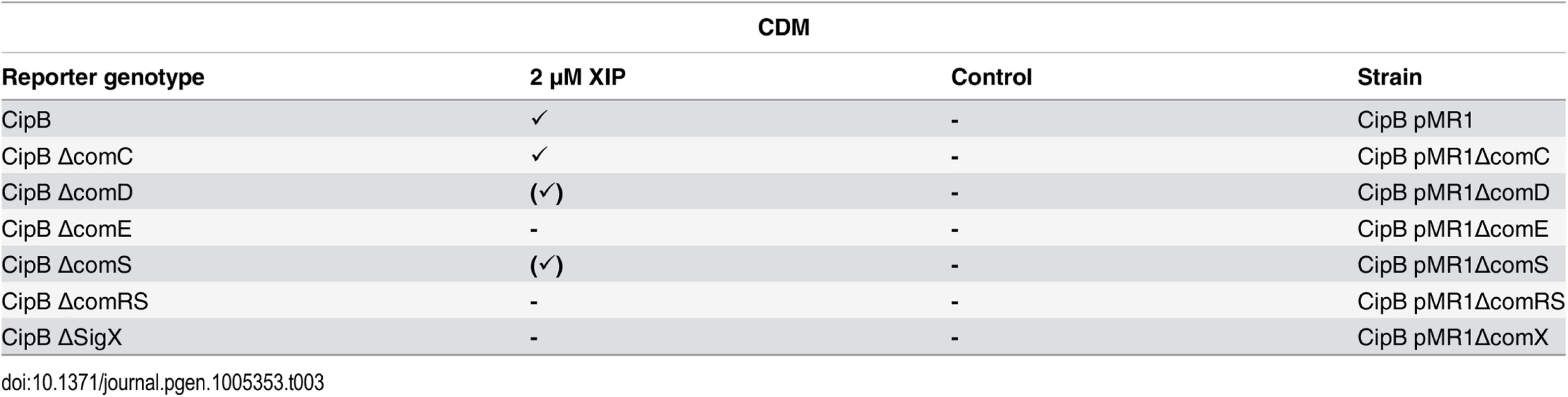
In the genetic background of the fluorescent reporter strain CipB pMR1 deletion mutants for ΔcomC, ΔcomD, ΔcomE, ΔcomS and ΔcomRS were constructed, cultivated in CDM medium and induced by XIP (2 μM). Further explanations as in Table 1. As a positive control, we analysed CSP induced bacteriocin production in THBY. CSP induction caused clear inhibition zones of the overlaid indicator strains L lactis and S. sanguinis, which are sensitive for mutacins V and IV, in the wild-type and the ΔcomC and ΔcomS mutant, respectively (S17 Fig). Deletion of comC could be complemented by externally added CSP, and deletion of comS had no effect as expected. We then tested bacteriocin production on CDM agar plates. No inhibition of the indicator strains could be observed, neither for CSP nor for XIP stimulated cultures. Bactericidal activity was also not detected in the concentrated supernatants of XIP induced and planktonically growing S. mutans strains in CDM. Since bacteriocins were clearly transcribed under these conditions, various mechanism could account for the lack of bacteriocidal activity: Bacteriocin synthesis could be regulated posttranscriptionally, they might not be secreted, or they could be degraded. Interestingly also CSP was either not produced, degraded or not secreted (S18 Fig and S1 Text).
Population heterogeneity and timing of bacteriocin expression
Our data show that bacteriocin expression in S. mutans is controlled by two different mechanisms: (1) A fast unimodal post-transcriptional activation of ComE by CSP independent from the growth medium. It is likely caused by phosphorylation of the response regulator upon CSP detection by the ComDE two-component system. (2) A delayed activation of comE transcription by XIP in CDM mediated by SigX, and thus showing the same modality as SigX.
To investigate the timing and modality of bacteriocin expression, we analyzed CSP and XIP mediated signaling using time-resolved flow cytometric analysis of the reporter strains for comE and its immediate target, cipB. The density plots of uninduced comE and cipB reporter strains (controls) can be found S19 Fig. Fig 7A shows that comE transcription was unimodal in CDM using XIP as an inducer (green density plots), but bimodal in THBY under CSP inducing conditions (red density plots). Exactly the same behavior had previously been observed for sigX [15]. Apparently the modality of sigX expression determines whether comE expression is uni - or bimodal.
Fig. 7. Time resolved analysis of comE and cipB expression using flow cytometry. 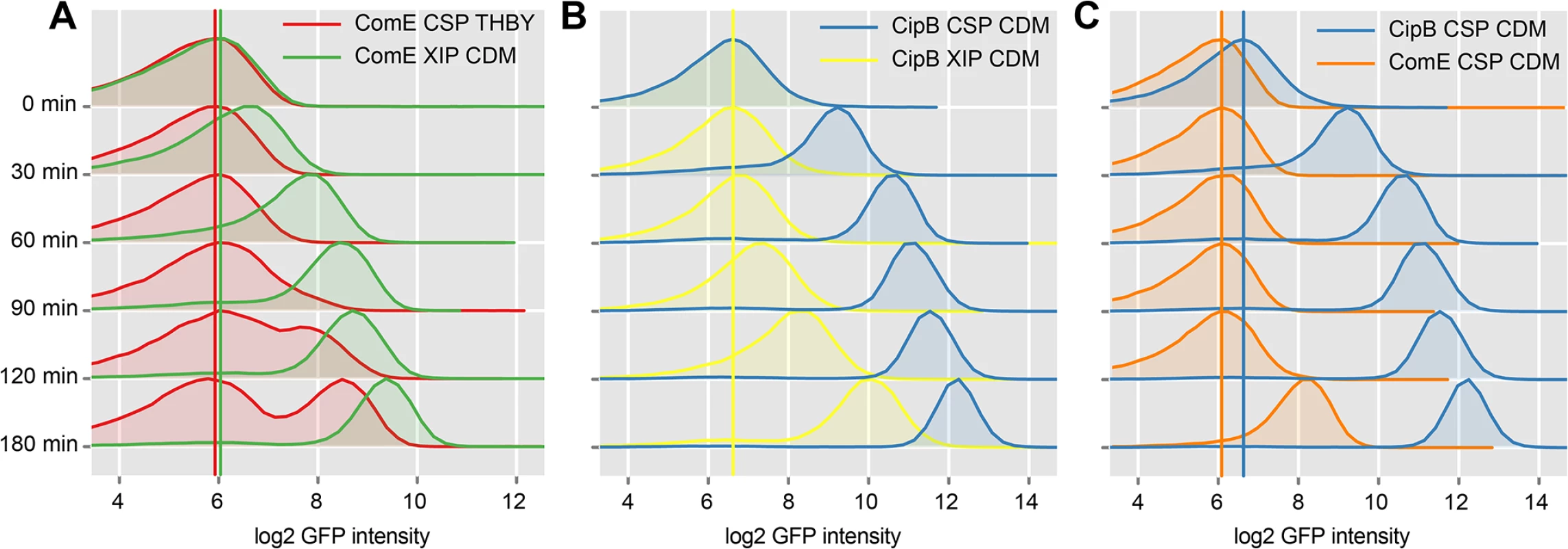
CipB pAE03 and ComE pAE03 GFP reporter strains were grown in CDM or THBY and were induced with 2 μM CSP or 2 μM XIP. 0, 30, 60, 90, 120 and 180 minutes post induction samples were taken and analyzed using a flow cytometer. Samples from uninduced controls were taken at corresponding timepoints. Density plots of GFP fluorescence (log2 units) are shown during time. In A the density plots of the XIP-induced ComE pAE03 reporter strain grown in CDM (green plots) are compared with the density plots derived from the same strain grown under CSP-induced conditions in THBY (red plots). The colored vertical lines represent the peak log2 GFP fluorescence intensity of the distribution at timepoint 0h. In part B density plots derived from the XIP induced CipB pAE03 reporter strain grown in CDM (yellow plots) are plotted against the density plots of the same strain grown in the same medium under CSP induced conditions (blue plots). C showed the comparative analysis of the ComE pAE03 reporter strain (orange density plots) and the CipB pAE03 strain (blue density plots) both grown in CDM under CSP (2μM) induced conditions. The induction of comE expression by XIP in CDM starts faster (30–60 minutes post XIP addition) than the CSP induced transcriptional activation of comE expression in THBY which begins 90–120 minutes post induction. CSP causes medium independent unimodal expression of bacteriocins, as also shown above. We have also shown above, that addition of CSP to THBY allows XIP signaling, but not because of a direct regulatory link as previously hypothesized. Instead an indirect effect is operating. We suggest that bacteriocins make the cells permeable and thus allow reimport of XIP into the producing cell. Thus in our experiment here, bacteriocin synthesis is induced by CSP, likely resulting in permeabilzation of the cell and reimport of XIP that is endogeneously produced. XIP then activates sigX, resulting in transcription of comE via binding of the SigX-RNA-Polymerase complex to the cin-Box. This explains why comE expression is delayed in THBY by about 90 min in comparison to CDM.
Fig 7B clearly shows that the bacteriocin encoding gene cipB was strongly and instantaneously induced upon CSP stimulation in CDM in an unimodal way (blue density plots). Already 30 minutes after induction, which corresponds to the maturing time of the GFP+ fluorophore, the expression of cipB was significantly enhanced. Addition of XIP in CDM medium (yellow density plots) also induced cipB expression unimodally but more weakly and significantly delayed. It starts 90 minutes after addition of XIP and reaches its maximum 180 minutes post induction. Independent from the growth medium, induction of cipB expression occurs within 30 min post CSP supplementation (S20 Fig). Thus, the data are consistent with the proposed transcriptional control of comE by SigX.
To observe post-transcriptional and transcriptional regulation in more detail, we then compared the time-course of expression of comE with that of its direct target cipB under CSP induced conditions in CDM directly (Fig 7C). CipB expression was observed instantaneously already 30 minutes post CSP supplement (blue density plots), while comE expression was unchanged and started to increase only 180 min post CSP addition (orange density plots). Microscopic analysis of the comE and cipB reporter strains confirmed that comE is not induced within the first 120 min while its target cipB is already fully induced 30 min post induction by CSP in CDM (S14 Fig and S1 Text). Thus CSP-induced activation of the bacteriocins via ComDE must be mediated via post-transcriptional regulation. The basal level of ComE transcripts is apparently sufficient for this reaction.
We then investigated the influence of the key genes in the signalling cascade on timing and modality of comE and cipB expression by analyzing the fluorescence of the comE and cipB reporter strains in different gene deletion backgrounds. Fig 8 shows that transcriptional activation of comE requires comRS, comS and sigX while the transcriptional activation of the ComE target cipB is independent of these genes. Moreover transcription of comE is bimodal while the transcription of its target cipB is unimodal. These findings confirm that indeed two different mechanisms (post-transcriptional and transcriptional regulation) are operating that activate ComE thus fully supporting our hypothesis.
Fig. 8. Modality of CSP induced comE and cipB expression in THBY in different gene deletion backgrounds. 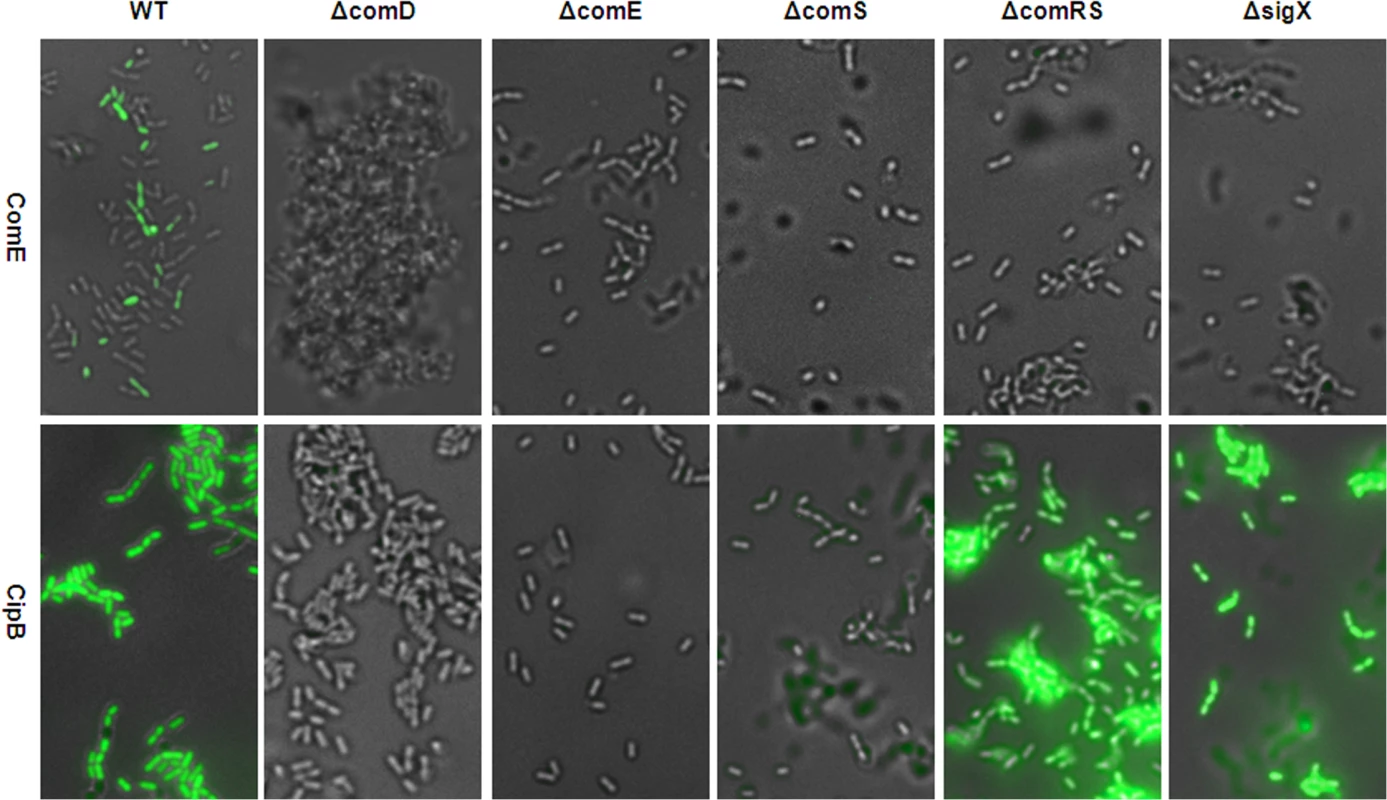
ComE pMR1 and CipB pMR1 reporter strains were grown in THBY under CSP (2 μM) induced conditions. 3 h post induction images were collected using fluorescence microscopy. GFP fluorescence images (green) of the reporters were overlaid with the phase contrast (grey) images of the corresponding strain. Images of the comE reporter strain are shown in the upper panel, while images of the cipB reporter strain are shown in the lower panel. Posttranscriptional activation of ComE by phosphorylation
To determine if phosphorylation of ComE controls transcription of the bacteriocin encoding genes we constructed mutants of comE where the aspartate residue at position 60 was replaced by a phosphomimetic, namely by glutamate (D60E) or alanine (D60A). Glutamate enhances the negative charge of the side chain and thus mimics a phosphorylated aspartate. Conversely, alanine cannot be phosphorylated and thus ComE should be inactive under all circumstances.
We utilized the CipB pMR1 reporter strain in the ΔcomE deletion background and transformed it with replicative pIB166 [37] based plasmids carrying either the native or a mutated comE gene under the control of the native comE promoter. As a control, we introduced the native comE gene into the ΔcomE reporter strain, which restored the CSP responsive phenotype. We also introduced a comE overexpression plasmid into the reporter strain and found CSP independent unimodal cipB expression. Reporter strains carrying the empty plasmid showed no detectable fluorescence during growth or CSP induction (Fig 9). As expected, strain CipB pMR1 ΔcomE D60A did not fluoresce either under CSP induced or uninduced conditions. Due to the D60A mutation ComE could not be phosphorylated and looses its ability to induce cipB expression. In the CipB pMR1 ΔcomE D60E strain, which mimicked stable phosphorylation, CipB induction was independent of the presence of CSP, in contrast to the native comE gene. The fluorescence of the D60E mutant was lower than that of the CSP-induced wild-type. Hung et al. found that ComE phosphorylation affected dimerization of S. mutans ComE in vitro and that the D60E mutation prevents that dimerization [40]. To conclude here we demonstrate that phosphorylation of ComE is the mechanism responsible for the induction of bacteriocin expression under CSP induced conditions.
Fig. 9. Phophorylmimetic study. 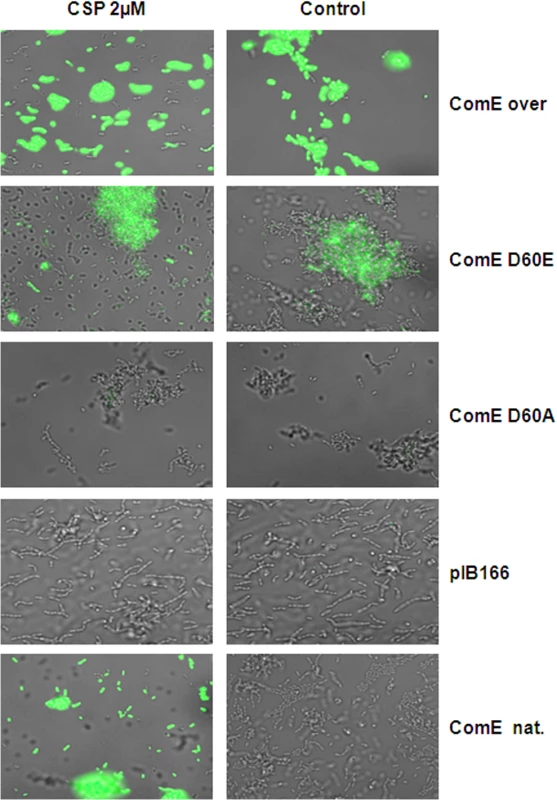
A CipB reporter strain in the ΔcomE deletion background was transformed with plasmids carrying the coding sequence of the natural comE (ComE nat.), the coding sequence of comE under the control of the strong lactococcal P23 promoter (P23-ComE), or the mutant genes ComE D60E (mimicking stable phorphorylation) and ComE D60A (no phosphorylation possible). A strain transformed with the empty vector (pIB166) was used as negative control. All strains were grown in THBY and fluorescence microscopy was performed 1 h after addition of CSP. Overlays of phase contrast and green fluorescence microscopic images are shown. The right column shows the un-induced controls. Discussion
Model of competence development in S. mutans
Our analysis results in a new understanding of the QS regulatory network in S. mutans which is depicted in Fig 10. The two signals CSP and XIP control different traits: CSP signaling (green box) mediates bacteriocin expression while XIP signaling induces competence development (red box). The model contains three different temporal layers of signaling: (I) Fast post transcriptional activation of the response regulator ComE and the transcription factor ComR induced by their respective signals within the first 5 min after detection. ComE is activated by phosphorylation, and ComR is activated by binding of XIP. (II) Early transcriptional response of the activated regulators resulting in bacteriocin synthesis (CSP signaling) and transcription of the comRS genes and the alternative sigma-factor sigX (XIP signaling). (III) Late transcriptional response mediated by the alternative sigma-factor SigX. Transcription of the gene encoding the regulator of bacteriocin synthesis, comE, is controlled by SigX. In such a way both QS systems are connected. SigX controls both competence development and—via comE–bacteriocin synthesis. In the competence cascade comE is localized downstream of comRS and sigX and not upstream as previously thought. ComE could thus be viewed as a “late competence gene”, although it is only indirectly involved in competence development. This model explains why co-expression of comS, sigX, comE and the late competence genes is observed and why the modality of sigX expression determines the modality of comE. Our model discriminates between post-transcriptional and transcriptional regulation, which has not been taken into account by previous modeling approaches.
Fig. 10. Model of competence development in S. mutans. 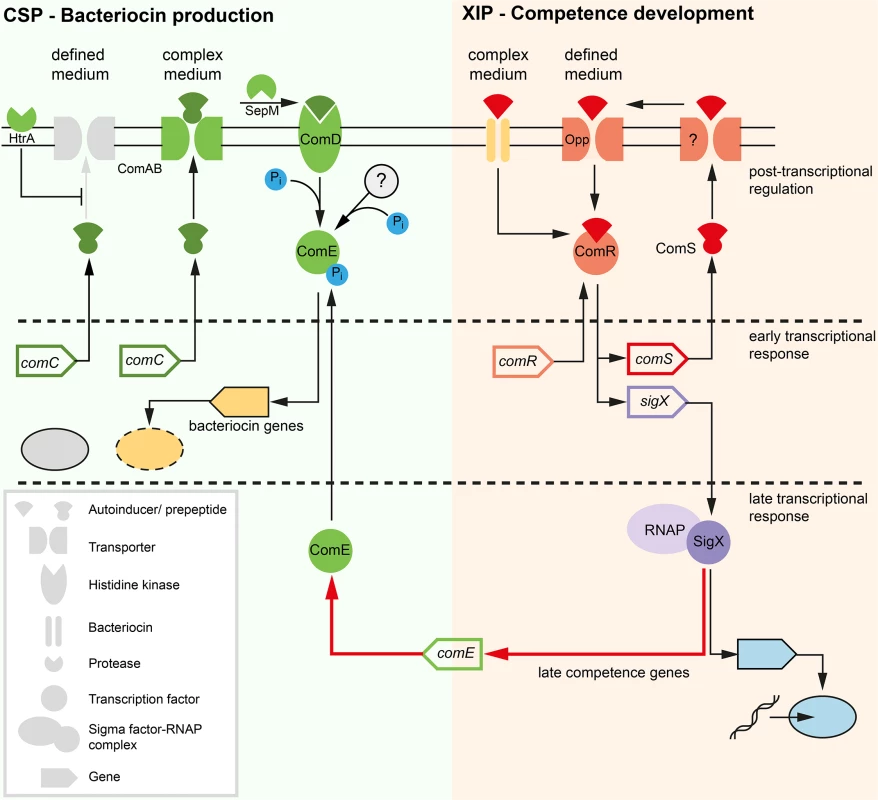
Two quorum sensing pathways are operating in S. mutans. Bacteriocin expression is regulated via CSP signaling and the ComDE two-component system (green box). CSP should be renamed MIP (mutacin inducing peptide). Competence development is regulated via XIP signaling and the ComR regulator (red box). Both systems are connected via the alternative sigma-factor SigX which controls comE transcription (red arrow). Competence regulation proceeds in three successive steps: (1) Instantaneous post-transcriptional activation of the regulator ComR. (2) Early transcriptional response: Transcription of comS, resulting in a positive feedback loop for signal synthesis, and of sigX. (3) Late transcriptional response: Transcription of the SigX regulon, including the competence genes and the comE response regulator. Upon competence development the medium determines via which mechanism XIP is imported into the cell. In defined medium XIP is specifically imported via the Opp permease, leading to unimodal competence development. XIP import in complex medium requires bacteriocin activity and might be mediated via permeabilization of the cells. In complex medium, competence development is bimodal. Active bacteriocins and CSP areexclusively observed in complex medium. Post transcriptional mechanisms must be operating in the defined medium, e.g. down-regulation of bacteriocin translation, lack of secretion, or degradation, since the cells sense externally added CSP and transcription of bacteriocin encoding genes is induced. Instead of two QS systems operating in parallel connected through a hypothetical regulator as assumed before (Fig 1), we have two QS systems operating independently, triggered by their respective signals CSP and XIP. The competence cascade, however, is linked to bacteriocin synthesis through the alternative sigma-factor SigX which controls comE. Thus, SigX induces competence, and with a time delay, also bacteriocin synthesis. Typically QS networks are organized hierarchically (e.g. in Pseudomonas aeruginosa) [41]. By contrast, in Vibrio harveyi three parallel input channels responding to three different autoinducer signals are integrated by a central response regulator [42]. The type of network architecture found here comprises two different input channels which are connected by the alternative sigma-factor SigX. SigX is stimulated directly by XIP and indirectly by CSP. This type of network architecture has not been found before.
A hierarchical cascade mediates competence. XIP is the signal inducing competence, independent from the growth medium. However the medium determines via which way XIP is imported into the cell. We hypothesize that bacteriocin expression is required to induce permeability of the cell for external XIP. CSP signaling is independent from XIP signaling and is mediated via phosphorylation of ComDE. CSP-induced unimodal activation of bacteriocin synthesis is regulated post-transcriptionally and occurs before the transcription of comE which is mediated via XIP signaling. CSP induces competence only indirectly. It is therefore not a competence inducing peptide and should be renamed MIP (mutacin inducing peptide). CSP signaling is also in principle independent from the cultivation medium, but in defined medium bacteriocin excretion is inhibited.The new model simplifies our understanding of QS in S. mutans and resolves the open questions initially formulated. We will subsequently discuss them point by point.
Bimodality of competence development
Lemme et al. [14] postulated comE being the origin of bimodality upon CSP induced competence development. Here we clearly show that this not the case since strong overexpression of comE independent of its native promoter did not enhance the percentage of competent cells. For comS a positive feedback loop was identified, which is a prerequisite for bimodality to occur [7] and thus a model assuming bistable expression of comS was developed [15]. However, here we show that neither comR nor comS alone can induce unimodal competence development when they are strongly overexpressed. Both genes or rather their gene products are required. Bistable expression of comS is not necessarily required. How then is the bimodality established in the system? Besides a positive feedback loop, noise is the second critical determinant for bimodality [29,43]. We suggest that noise in comR and comS expression accounts for bimodality since the overexpression analysis showed that both are limiting factors for unimodal competence development. Bimodality would then be a feature of the quorum sensing network, and not necessarily the result of bistable expression of one component [44].
XIP is a secreted factor, and thus it is unclear how phenotypic heterogeneity can be stably established if all cells are able to sense XIP, not only the overproducers. Previously it was found that the mature XIP signal of S. thermophilus is not released into the medium, but remains cell associated [13]. Thus cells with higher comS expression levels can re-import more XIP resulting in the observed positive feedback loop.
Link between CSP and XIP signaling
It was previously suggested that an unknown regulator transfers the CSP induced signal from ComDE to ComRS. Here we show that this regulator does not exist. If a regulator would be required for transmitting the CSP induced signal to ComRS, then it would be possible to overcome its effect by overexpression of comRS, which should result in unimodal competence development. However, here we show that overexpression of comRS does not induce competence at all in complex media.
We suggest that CSP induces competence only indirectly by allowing XIP to be imported into the cell in complex media. The following findings support this hypothesis: 1. Competence is not induced in CDM by CSP, although externally added CSP is fully active and not degraded as previously thought. 2. A small quantity of CSP is required to induce unimodal competence development in THBY in a strain overexpressing comRS. 3. An increase in the concentration of CSP does not increase the percentage of competent cells in this experiment, suggesting that CSP triggers a process required for competence to occur and is not inducing it directly. 4. The time-lag of 60–90 min between CSP addition and sigX transcription in THBY clearly supports an indirect activation mechanism. 5. Deletion of CipB completely abolished CSP and CSP/XIP induced expression of the late competence gene lytFsm. Our findings are in accordance with a recent study showing that deletion of the bacteriocin encoding gene cipB almost completely abolishes competence development through CSP stimulation [34]. Thus it was suggested that the CipB bacteriocin itself might act as a regulator of competence [39]. This scenario is highly unlikely, because a regulatory role for a bacteriocin has never been demonstrated. Moreover, bacteriocins are secreted into the environment and thus can hardly exert intracellular regulatory roles. Based on our findings we suggest that bacteriocins are required to induce permeability of the producer cell for external XIP.
It was shown that deletion of Opp has no influence on CSP induced competence development in complex medium in S. mutans [15]. Thus XIP import upon bacteriocin expression in complex medium is not mediated via Opp and its precise mechanism remains to be elucidated. Pore formation is the mode of action of many bacteriocins [45]. Via these pores the small XIP molecule might enter the cell. Alternatively some of the bacteriocin exporters might allow XIP import. Cell wall and membrane alterations which confer immunity to the bacteriocin producer cells against their own bacteriocin [45] may also account for the altered permeability of the cell for XIP. Presently it is not known how and by which system the XIP precursor is secreted and processed in S. mutans [7,46,47].
Why is comE expression biomodal, while bacteriocins are synthesized by the whole population?
Bacteriocin synthesis is induced by post-transcriptional unimodal activation of the ComE response regulator. The CSP signal causes phosphorylation of the ComE protein, resulting in instantaneous unimodal bacteriocin transcription and synthesis. This process is not affected by transport processes across the membrane and therefore it is independent of the cultivation medium. By contrast, the transcription of the comE gene is controlled by SigX, which is regulated via the XIP signaling cascade. Therefore bimodal comE expression occurs much later (90–120 post induction).
How is the medium effect on signaling mediated?
It was thought that external CSP is degraded by the HtrA protease in CDM, while this protease is inhibited by media components in THBY and thus allows CSP to remain intact [15]. Here we show that external CSP is not degraded in CDM. For the HtrA serine protease of S. pneumoniae it was demonstrated that it indeed cleaves the pneumococcal CSP. The enzyme was not inhibited by bovine serum albumin, but by denatured protein, prompting the authors to suggest that CSP signaling is a means to detect intracellular stress [22]. The CSP of S. mutans is more homologous to the BlpC peptide which induces bacteriocin synthesis than to the CSP autoinducer of S. pneumoniae. Extracellular BlpC is not degraded by HtrA [48] in accordance with the lack of degradation of S. mutans CSP in CDM. These data, too, demonstrate that CSP of S. mutans is functionally related to BlpC of S. pneumoniae.
CSP is a mutacin stimulating peptide
Activated ComR represents the only regulator mediating competence in S. mutans. In a recent review it was proposed that the ComDE two-component system (TCS) of S. mutans should be renamed to Blp since it primarily regulates bacteriocin expression [1]. However, this suggestion was based on the situation in S. pneumoniae, where the BlpRH TCS is exclusively involved in regulating bacteriocin production [48]. In S. mutans, comE is controlled by the master regulator of competence SigX. Thus we would not propose to rename the system Blp. However, since CSP does not stimulate competence directly, and in S. mutans comE is not an early competence gene as the name suggests, we propose to rename CSP to MIP (mutacin inducing peptide) and ComDE to MutDE.
How is ComE phosphorylated in the absence of a CSP signal?
Upon XIP induction SigX enhances the ComE level in the cell. To be active, ComE needs to be phosphorylated, yet the histidine kinase ComD is not receiving a signal under these conditions and its deletion still allows cipB expression [34]. Therefore non-cognate kinases must be able to phosphorylate ComE. Recently cross-talk between TCS systems was observed [49]. For the serine/threonine protein kinase PknB of S. mutans a regulatory role in bacteriocin expression was found [50], suggesting that this enzyme might also be able to phosphorylate ComE.
Why are bacteriocins not secreted in CDM medium?
Although the transcription of bacteriocin encoding genes was strongly induced by externally added CSP or XIP in CDM, no bactericidal activity was found in the culture supernatants, suggesting that bacteriocin synthesis is regulated post-transcriptionally. This is supported by a study of two phenotypic variants (transparent/opaque) of the S. pneumoniae R6 strain [48]. They showed an identical transcriptional response to the bacteriocin inducer BlpC. However only for cells of the transparent phenotype bacteriocin activity could be detected, while none was found in the opaque phenotype. The authors demonstrated that the HtrA protease abolished bacteriocin synthesis post transcriptionally in these cells [48]. This might be the reason why we detected no CSP and no bactericidal activity in the CDM supernatants. HtrA activity likely prevents CSP and bacteriocin processing and/or secretion in CDM. Multiple environmental factors additionally influence bacteriocin expression in S. mutans, including cell density and nutrient availability [51].
A novel role of SigX and its biological implications
The observation that comE transcription is under the control of SigX represents a novel regulatory role for SigX, which thus not only controls the expression of the transformasome, but also the synthesis of bacteriocins. In S. pneumoniae and B. subtilis the master regulators of competence, SigX and ComK respectively, exclusively regulate transformasome related genes [1,43]. Competence and bacteriocin production is uncoupled in these organisms. Coupling it like in S. mutans makes ecological sense, because it provides the genetic variability which makes competence an adaptive trait, as suggested previously [52]. In its natural niche S. mutans is part of the multispecies oral biofilm which consists of over 600 different species [53]. Thus S. mutans is faced with a strong competitive environment which is at the same time a rich source of genetic variability. In a dual species biofilm of S. mutans and Candida albicans the CSP and XIP triggered QS cascades were induced simultaneously [54]. The ComR regulator has a low stringency with respect to the exact sequence of the XIP peptide required for activation [12], and consequently, Streptococci respond to heterologous peptides [12,55]. Thus S. mutans might sense the presence of other Streptococci via XIP signaling and directly respond to this stimulus by producing bacteriocins and killing the competitor, while exploiting its genetic information at the same time.
To conclude, here we deciphered the complex QS signaling network of S. mutans on the single cell level. Competence is exclusively mediated via XIP signaling in a hierarchical network structure. We show that competence development is coupled to bacteriocin synthesis through the alternative sigma-factor SigX, which makes this QS network highly efficient for acquiring new genetic information in the competitive environment of dental biofilms.
Methods
Strains and media
All S. mutans strains were routinely propagated in in Todd Hewitt broth medium supplemented with 0.5% (wt/vol.) yeast extract (THBY; Becton Dickinson, Heidelberg, Germany) in an incubator (5% CO2, 37°C) without agitation. When indicated, antibiotics were added to the medium (chloramphenicol 10 μg/ml, tetracycline 12.5 μg/ml and erythromycin 10μg/ml). For experiments conducted in chemically defined medium (CDM) [56] a 10 ml overnight culture of the appropriate strain grown in THBY was centrifuged for 10 min at 5000 rpm and 4°C. The supernatant was removed and the cell pellet was gently resuspended in fresh CDM medium and again centrifuged as described above. After removal of the supernatant the pellet was finally gently resuspended in 5 ml of CDM. Strains were diluted to an OD of 0.1 and grown at 37°C and 5% CO2.
Construction and verification of single and dual fluorescent reporter strains
The plasmids used for the construction of reporter strains (S1 Fig), an analysis of maturation times of various fluorescent proteins in S. mutans (S2 Fig), a scheme showing the integration of the plasmids into the chromosome (S3 Fig), comparison of expression levels of different late competence genes (S4 Fig), comparison of two LytFsm constructs with different fluorophores (S5B Fig), spectral separation of TagBFP2 and GFP+ (S6 Fig), dye swap of comE with the late competence gene lytFsm (S9 Fig), and construction of reporters in various gene deletion backgrounds are described in full detail in S1 Text, which also provides a discussion of the reporter constructs. A list of the constructed strains (S1 Table) plasmids (S2 Table) and primers (S3 Table) can be found in the supplements.
Flow cytometric analysis
Overnight cultures of the strains grown in THBY were diluted to an OD of 0.1. For the strains intended to grow in CDM the above described washing procedure was applied to remove all traces of the complex media and finally strains were diluted to an OD of 0.1 and grown at 37°C and 5% CO2. When the bacteria had reached an OD of 0.15 the culture was divided into three equal fractions. One fraction was treated with 2 μM synthetic CSP, the second with 2 μM synthetic XIP, and the third fraction was used as an uninduced control. Samples (0.5 ml) were taken after 30, 60, 90, 120 and 180 minutes post induction. Samples derived from strains growing in either CDM or THBY were centrifuged (5 min and 7000 rpm) and washed once with PBS. Subsequently the strains were resuspended in 1 ml of ice-cold PBS and sonicated using a MS72 sonotrode with the Sonoplus HD2200 device (Bandelin, Germany) for at least 20 sec at 10% power. Settings were a 0.5 sec impulse which was followed by a 0.5 sec break. Live/Dead staining before and after sonication was performed to exclude that sonication significantly interfered with membrane integrity. For flow cytometry the LSR Fortessa Cell analyser (BD, Germany) was used. 0.22 μM filtered PBS was applied as sheath fluid. Cytometer settings were chosen as previously reported [14]. 50000 cells were analysed and the resulting data processed with a self-written R-Script.
Construction of phosphomimetic mutants of ComE
The mutation of the comE gene was accomplished using a PCR-driven overlap extension approach [57]. The triplet encoding the aspartate residue at position 60 (GAT) was changed to GCT (Ala) and GAG (Glu), respectively, using the primers listed in S3 Table. In a first PCR the comE gene and its native promoter region were amplified in two separate parts thereby introducing the desired mutation via the 5`termini of the inner primers (primer pairs PcomE_F1/D60E_R1 and PcomE/D60A_R1 for the first part and primer pairs D60E_F2/ComE_R2 and D60A_F2/ComE_R2 for the second part of the comE gene. The two PCR amplified parts of comE contained homologous flanks of approximately 22 bp to each other and were used as template for a second PCR with only the outer primers (PcomE_F1/ComE_R2), amplifying the entire comE coding sequence including the native promoter. The PCR products were purified and cloned into the vector pIB166 [37] in opposite direction to the constitutive P23 promoter to allow transcription from the native promoter. The plasmid sequence was verified by sequencing and the plasmid transformed into the S. mutans cipB reporter strain in the ΔcomE deletion background. As positive control the native comE sequence was cloned into pIB166 and the transformed empty vector was used as negative control.
Overexpression strains in the LytFsm reporter background
To construct overexpression strains for ComD, ComE, ComR, ComS and ComR+ComS, independent from their native promoter, plasmid pIB166 containing the strong constitutive P23 promoter from Lactococcus sp. was used [37]. The coding sequences including the native ribosomal binding sites were PCR-amplified (see S3 Table) and cloned blunt end via the SmaI restriction site into the vector. The correct plasmid sequence was verified by sequencing and 100 ng of the plasmid transformed into the LytFsm reporter strain.
While this cloning approach was successful for comE, comS, comRS and comR, we were not able to obtain a correct plasmid sequence for comD. Point mutations and frameshift mutations were observed in the ribosomal binding site or the coding region of the gene and strongly suggest that this protein is toxic for E.coli. As it was already successfully cloned in E. coli under the control of the native promoter we assume that the P23 promoter has a significantly higher basal transcription than the native comD promoter in E. coli. Attempts to fuse the GtfB or GtfC promoter to the coding sequence of comD were also not successful, although these constructs could be easily obtained for comE and comR, respectively. We therefore used a PCR-driven overlap extension approach ([57]) to amplify a fusion construct consisting of five parts; the upstream region of gene SMU_1342, the P23 promoter, the comD gene, the chloramphenicol resistance cassette (CAT) and downstream flanking regions of SMU_1342. Homology flanks of the different parts were introduced via the 5`terminus of the PCR primers. This construct allows the integration of the comD gene under the control of the P23 promoter via double homologous recombination at the SMU_1342 locus. In a first PCR all parts of the fusion construct were amplified separately and purified using the PCR Purification Kit (Qiagen, Germany). Subsequently equal amounts (100 ng) of all different PCR products were used as template for a second PCR with primers UP1342_F/D1342_R spanning the entire construct (outer primers). The PCR reaction was directly used as template to transform the LytFsm reporter strain. Chloramphenicol resistant clones were selected on THBY agar plates, picked and cultivated in liquid THBY medium containing 10 μg/ml chloramphenicol. Genomic DNA was isolated from 2 ml of the exponentially growing culture and correct constructs were verified by PCR using the primer pairs UP1342_F/D1342_R and P23_F/ CAT_R).
Analysis of gene expression
A detailed description of methods for RNA extraction, RNA sequencing and qRT-PCR can be found in the S1 Text. Raw and processed RNAseq data have been deposited in the gene expression omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE65982.
Bacteriocin overlay assay
O/N cultures of S. mutans WT, ΔcomS and ΔcomC deletion strains with a starting OD600 of 0.1 were grown in CDM or THBY until the cultures reached an OD600 = 0.25. Then the cultures were split and induced with either 2 μM XIP or 2 μM CSP. Uninduced cultures were used as controls. After 2 hours of further growth (37°C, 5% CO2) 2.5 μl of the culture were spotted on CDM or THBY agar plates. The plates were incubated for further 4 h until the induced cultures were overlaid with a 2.5 μl drop of a 2 μM CSP (or 2 μM XIP) solution in CDM (THBY). The drops were allowed to soak in the plate. Finally the plate was incubated for 24 h at 37°C and 5% CO2. Overnight cultures of the indicator strains S. sanguis and L. lactis were diluted 1 : 200 in fresh CDM Top Agar (0.7% Agar) placed at 37°C and 5 ml of the mixture was poured over the agar plates containing the spotted S. mutans producer strains. The overlaid plate was incubated for further 16 h in a CO2 incubator. Zones of inhibition were documented using a conventional camera.
Supporting Information
Zdroje
1. Johnston C, Martin B, Fichant G, Polard P, Claverys JP (2014) Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12 : 181–196. nrmicro3199 [pii]. doi: 10.1038/nrmicro3199 24509783
2. Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3 : 711–721. nrmicro1234 [pii]. 16138099
3. Weigel LM, Donlan RM, Shin DH, Jensen B, Clark NC, McDougal LK, et al. (2007) High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob Agents Chemother 51 : 231–238. AAC.00576-06 [pii]. 17074796
4. Dalia AB, McDonough E, Camilli A (2014) Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A 111 : 8937–8942. 1406478111 [pii]. doi: 10.1073/pnas.1406478111 24889608
5. Burton B, Dubnau D (2010) Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol 2: a000406. cshperspect.a000406 [pii]. doi: 10.1101/cshperspect.a000406 20573715
6. Seitz P, Blokesch M (2013) DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci U S A 110 : 17987–17992. 1315647110 [pii]. doi: 10.1073/pnas.1315647110 24127573
7. Mashburn-Warren L, Morrison DA, Federle MJ (2010) A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78 : 589–606. doi: 10.1111/j.1365-2958.2010.07361.x 20969646
8. Claverys JP, Martin B, Polard P (2009) The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33 : 643–656. FMR164 [pii]. doi: 10.1111/j.1574-6976.2009.00164.x 19228200
9. Berg KH, Biornstad TJ, Johnsborg O, Havarstein LS (2012) Properties and biological role of streptococcal fratricins. Appl Environ Microbiol 78 : 3515–3522. AEM.00098-12 [pii]. doi: 10.1128/AEM.00098-12 22407687
10. Cook LC, Federle MJ (2014) Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38 : 473–492. doi: 10.1111/1574-6976.12046 24118108
11. Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, et al. (2011) Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol 80 : 1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x 21435032
12. Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, et al. (2013) Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87 : 1113–1132. doi: 10.1111/mmi.12157 23323845
13. Gardan R, Besset C, Gitton C, Guillot A, Fontaine L, Hols P, et al. (2013) Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J Bacteriol 195 : 1845–1855. JB.02196-12 [pii]. doi: 10.1128/JB.02196-12 23396911
14. Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I (2011) Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193 : 1863–1877. JB.01363-10 [pii]. doi: 10.1128/JB.01363-10 21317319
15. Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ (2012) Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86 : 258–272. doi: 10.1111/j.1365-2958.2012.08187.x 22845615
16. Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG (2001) Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183 : 897–908. 11208787
17. Hossain MS, Biswas I (2012) An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol 194 : 5886–5896. JB.01381-12 [pii]. doi: 10.1128/JB.01381-12 22923597
18. Kreth J, Merritt J, Zhu L, Shi W, Qi F (2006) Cell density - and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett 265 : 11–17. FML459 [pii]. 16981904
19. Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, et al. (2007) The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153 : 1799–1807. 153/6/1799 [pii]. 17526837
20. van der Ploeg JR (2005) Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol 187 : 3980–3989. 187/12/3980 [pii]. 15937160
21. Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, et al. (2010) A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192 : 1444–1454. JB.01251-09 [pii]. doi: 10.1128/JB.01251-09 20023010
22. Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME (2012) The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem 287 : 38449–38459. M112.391482 [pii];doi: 10.1074/jbc.M112.391482 23012372
23. Veening JW, Smits WK, Kuipers OP (2008) Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62 : 193–210. doi: 10.1146/annurev.micro.62.081307.163002 18537474
24. Smits WK, Kuipers OP, Veening JW (2006) Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4 : 259–271. nrmicro1381 [pii]. 16541134
25. Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305 : 1622–1625. 15308767
26. Avery SV (2006) Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4 : 577–587. nrmicro1460 [pii]. 16845428
27. Veening JW, Smits WK, Hamoen LW, Kuipers OP (2006) Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J Appl Microbiol 101 : 531–541. JAM2911 [pii]. 16907804
28. Acar M, Mettetal JT, van OA (2008) Stochastic switching as a survival strategy in fluctuating environments. Nat Genet 40 : 471–475. ng.110 [pii]. doi: 10.1038/ng.110 18362885
29. Eldar A, Elowitz MB (2010) Functional roles for noise in genetic circuits. Nature 467 : 167–173. nature09326 [pii]. doi: 10.1038/nature09326 20829787
30. Raj A, van OA (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135 : 216–226. S0092-8674(08)01243-9 [pii]. doi: 10.1016/j.cell.2008.09.050 18957198
31. Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB (2012) A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol Lett 336 : 104–112. doi: 10.1111/j.1574-6968.2012.02660.x 22900705
32. Dufour D, Levesque CM (2013) Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin. J Bacteriol 195 : 105–114. JB.00926-12 [pii]. doi: 10.1128/JB.00926-12 23104806
33. Perry JA, Cvitkovitch DG, Levesque CM (2009) Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol Lett 299 : 261–266. FML1758 [pii]. doi: 10.1111/j.1574-6968.2009.01758.x 19735463
34. Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM (2009) Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72 : 905–917. MMI6693 [pii]. doi: 10.1111/j.1365-2958.2009.06693.x 19400789
35. Newman RH, Fosbrink MD, Zhang J (2011) Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev 111 : 3614–3666. doi: 10.1021/cr100002u 21456512
36. Scholz O, Thiel A, Hillen W, Niederweis M (2000) Quantitative analysis of gene expression with an improved green fluorescent protein. p6. Eur J Biochem 267 : 1565–1570. ejb1170 [pii]. 10712585
37. Biswas I, Jha JK, Fromm N (2008) Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154 : 2275–2282. 154/8/2275 [pii]. doi: 10.1099/mic.0.2008/019265-0 18667560
38. Campbell EA, Choi SY, Masure HR (1998) A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol 27 : 929–939. 9535083
39. Dufour D, Cordova M, Cvitkovitch DG, Levesque CM (2011) Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol 193 : 6552–6559. JB.05968-11 [pii]. doi: 10.1128/JB.05968-11 21984782
40. Hung DC, Downey JS, Kreth J, Qi F, Shi W, Cvitkovitch DG, et al. (2012) Oligomerization of the response regulator ComE from Streptococcus mutans is affected by phosphorylation. J Bacteriol 194 : 1127–1135. JB.06565-11 [pii]. doi: 10.1128/JB.06565-11 22210762
41. Lee J, Zhang L (2015) The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6 : 26–41. doi: 10.1007/s13238-014-0100-x 25249263
42. Plener L, Lorenz N, Reiger M, Ramalho T, Gerland U, Jung K (2015) The phosphorylation flow of the Vibrio harveyi quorum sensing cascade determines levels of phenotypic heterogeneity in the population. J Bacteriol. JB.02544-14 [pii].
43. Maamar H, Raj A, Dubnau D (2007) Noise in gene expression determines cell fate in Bacillus subtilis. Science 317 : 526–529. 1140818 [pii]. 17569828
44. To TL, Maheshri N (2010) Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science 327 : 1142–1145. 327/5969/1142 [pii]. doi: 10.1126/science.1178962 20185727
45. Cotter PD, Ross RP, Hill C (2013) Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11 : 95–105. nrmicro2937 [pii]. doi: 10.1038/nrmicro2937 23268227
46. Khan R, Rukke HV, Ricomini Filho AP, Fimland G, Arntzen MO, Thiede B, et al. (2012) Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J Bacteriol 194 : 3781–3788. JB.00624-12 [pii]. doi: 10.1128/JB.00624-12 22609914
47. Desai K, Mashburn-Warren L, Federle MJ, Morrison DA (2012) Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194 : 3774–3780. JB.00337-12 [pii]. doi: 10.1128/JB.00337-12 22609913
48. Kochan TJ, Dawid S (2013) The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol 195 : 1561–1572. JB.01964-12 [pii]. doi: 10.1128/JB.01964-12 23354751
49. Laub MT, Goulian M (2007) Specificity in two-component signal transduction pathways. Annu Rev Genet 41 : 121–145. 18076326
50. Banu LD, Conrads G, Rehrauer H, Hussain H, Allan E, van der Ploeg JR (2010) The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect Immun 78 : 2209–2220. IAI.01167-09 [pii]. doi: 10.1128/IAI.01167-09 20231406
51. Merritt J, Qi F (2012) The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27 : 57–69. doi: 10.1111/j.2041-1014.2011.00634.x 22394465
52. Kreth J, Merritt J, Shi W, Qi F (2005) Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol 57 : 392–404. MMI4695 [pii]. 15978073
53. Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, Mira A (2012) The oral metagenome in health and disease. ISME J 6 : 46–56. ismej201185 [pii]. doi: 10.1038/ismej.2011.85 21716308
54. Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, et al. (2014) Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J 8 : 2256–2271. ismej201473 [pii]. doi: 10.1038/ismej.2014.73 24824668
55. Fleuchot B, Guillot A, Mezange C, Besset C, Chambellon E, Monnet V, et al. (2013) Rgg-associated SHP signaling peptides mediate cross-talk in Streptococci. PLoS One 8: e66042. doi: 10.1371/journal.pone.0066042 23776602
56. van dR I, Kessler RE (1980) Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27 : 444–448. 6991416
57. Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2 : 924–932. nprot.2007.132 [pii]. 17446874
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



