-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
In Drosophila, the nervous systems undergo extensive neuronal remodeling during metamorphosis, as many larval neurons die and adult neurons are generated while some larval neurons survive and prune their branches. Pruning that removes specific parts of neuronal branches without causing cell death is a self-destruct process, thus requiring precise regulation to prevent undesired damage to the nervous systems. Certain Drosophila sensory neurons that undergo dendrite pruning, specifically eliminating the dendrites but leaving the axons intact, provide us an opportunity to study the mechanism of how pruning activity is regulated in the dendrites. We reasoned that the distinctive microtubule polarity in dendrites and axons might be involved and factors are required to regulate the pruning activity in the dendrites through their interaction with microtubules. Here, we identified Spindle-F that mediates Ik2-dependent pruning activity in the dendrites by linking Ik2 to the microtubule motor dynein complex. We showed that elevation of Ik2 activity during dendrite pruning promotes Ik2/Spindle-F/dynein complex moving along the microtubules. We also showed that the formation and redistribution of Ik2/Spindle-F/dynein complex are essential for dendrite pruning. Our study reveals a connection between the polarized microtubules of dendrites and the pruning activity through Spindle-F for dendrite pruning.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005642
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005642Summary
In Drosophila, the nervous systems undergo extensive neuronal remodeling during metamorphosis, as many larval neurons die and adult neurons are generated while some larval neurons survive and prune their branches. Pruning that removes specific parts of neuronal branches without causing cell death is a self-destruct process, thus requiring precise regulation to prevent undesired damage to the nervous systems. Certain Drosophila sensory neurons that undergo dendrite pruning, specifically eliminating the dendrites but leaving the axons intact, provide us an opportunity to study the mechanism of how pruning activity is regulated in the dendrites. We reasoned that the distinctive microtubule polarity in dendrites and axons might be involved and factors are required to regulate the pruning activity in the dendrites through their interaction with microtubules. Here, we identified Spindle-F that mediates Ik2-dependent pruning activity in the dendrites by linking Ik2 to the microtubule motor dynein complex. We showed that elevation of Ik2 activity during dendrite pruning promotes Ik2/Spindle-F/dynein complex moving along the microtubules. We also showed that the formation and redistribution of Ik2/Spindle-F/dynein complex are essential for dendrite pruning. Our study reveals a connection between the polarized microtubules of dendrites and the pruning activity through Spindle-F for dendrite pruning.
Introduction
The precise assembly of neural circuits is crucial for the nervous system to function properly. The developing nervous systems often start with a primitive prototype, characterized by exuberant branches and excessive connections. Thus, further remodeling is required to refine the developing nervous systems to maturity. Neuronal pruning, one such remodeling mechanism, is a highly regulated self-destruct process that eliminates excessive neuronal branches in the absence of cell death. Pruning is widely observed in the nervous systems of both vertebrates and invertebrates [1,2], that not only ensures precise wiring during development, but also allows for adjustment of neuronal connections in response to injury and disease. Various studies have shown that defects in developmental pruning affect the function of the nervous systems in C. elegans [3] and Drosophila [4]. Moreover, a progressive loss of neurites far ahead of cell death is commonly observed in many neurodegenerative disorders [5,6]. Thus, any dysregulation of pruning activity even at the level of individual neurons would bring catastrophic consequences to the nervous systems. Although the primary triggers for developmental pruning and pruning that ensues upon neuronal injury and disease are diverse, the downstream machinery that eliminates neuronal processes shared some common features. For example, microtubule disruption is the earliest cellular event observed in all types of pruning [2,7,8], and the ubiquitin-proteasome system is required in all circumstances [7–10].
During Drosophila metamorphosis, substantial neuronal remodeling takes place in both the central and peripheral nervous systems [11–14]. Most of the larval peripheral neurons die during metamorphosis, whereas few, including some class IV dendritic arborization (C4da) neurons, survive and undergo large-scale dendrite pruning [13,14]. Dendrite pruning of the dorsal C4da neuron ddaC starts with severing of the proximal dendrites at 4–6 h APF (after puparium formation) [15]. Subsequently these disconnected dendrites become fragmented and eventually eliminated by the surrounding epidermal cells [16] by 16–18 h APF. In contrast to the central brain mushroom body (MB) γ neurons where both larval dendrites and axons are pruned during development, the peripheral C4da neurons specifically prune their dendrites keeping the axons intact [14]. The molecular basis for how the pruning activity is confined to the dendrites of C4da neurons remains unknown. We reasoned that molecular differences between dendrites and axons should be considered for such differential pruning activity in C4da neurons. It is known that microtubule polarity is different in the dendrites and axons of neurons [17], including in the Drosophila sensory neurons [18]. For example, C4da neurons have polarized microtubules in their proximal dendrites predominantly with microtubule minus end pointing away from the cell body, but have an opposite polarity in their axons [18,19]. This difference in microtubule polarity is essential for maintaining the proper function and compartmental identities of dendrites and axons, and might be an important determinant for spatially restricting pruning activity in the dendritic compartments of C4da neurons. Based on this assumption, some molecules are required to connect the pruning activity with the distinctive microtubule polarity of the dendrites in C4da neurons during dendrite pruning.
Previous studies have shown that dendrite pruning in C4da neurons is initiated by the steroid hormone ecdysone and its heterodimeric receptors, ecdysone receptor B1 (EcR-B1) and Ultraspiricle (Usp) [13,14]. Through transcriptional regulation of sox14, ecdysone signaling activates the Sox14 target gene mical, which encodes a cytoskeletal regulator, to regulate dendrite pruning [20]. A few other molecules mediating specific cellular activities have been shown to participate in dendrite pruning of C4da neurons, such as the ubiquitin-proteasome system [14], caspases [21,22], matrix metalloproteases [14], microtubule severing proteins [15] and mediators of dendritic calcium transients [23]. Our previous studies identified Ik2 kinase, a homologue of vertebrate IKK - ε ιν Drosophila, that plays an essential role in dendrite pruning of pupal neurons, and further demonstrated that Ik2 is sufficient to induce precocious dendrite severing in larval neurons [15]. To our knowledge, Ik2 is the only known molecule sufficient to induce premature dendrite severing in larvae, reflecting a central role of Ik2 kinase in dendrite pruning. Therefore, in this study we aimed to elucidate the mechanism by which Ik2 kinase signaling is transduced and regulated in Drosophila sensory neurons during dendrite pruning.
As Ik2 is essential for dendrite pruning, to elucidate the mechanism of Ik2 kinase signaling, we searched for candidate molecules that mediate Ik2 signals during dendrite pruning. Several lines of evidence suggested that Spn-F, a coil-coiled protein, is a good candidate. Firstly, spn-F mutant flies showed defects in developing oocytes and bristles [24], similar to the phenotypes observed in ik2 mutants [25]. Secondly, Spn-F physically interacts with Ik2 [26]. It implied that ik2 and spn-F may act in the same pathway during oogenesis and bristle morphogenesis, and raised the possibility that a similar pathway might also be involved in dendrite pruning of C4da neurons. Here, we demonstrate Spn-F playing a key role in linking Ik2 kinase to microtubule motor dynein complex for dendrite pruning. Spn-F acts downstream of Ik2 kinase in the same pathway for dendrite pruning. We show that Spn-F displays a punctate pattern in larval neurons and these Spn-F puncta become dispersed in pupal cells. The redistribution of Spn-F from puncta is essential for dendrite pruning, and depends on the activity of Ik2 kinase and the function of microtubule motor dynein complex. Our data also demonstrate that Spn-F not only links Ik2 to dynein motor complex, but also mediates the formation of Ik2/Spn-F/dynein complex, that is critical for Spn-F punctum disassembly and dendrite pruning.
Results
spn-F is required for dendrite pruning of Drosophila sensory neurons
To examine the role of spn-F in dendrite pruning, we expressed spn-F double-strand RNAs (dsRNAs) under the control of class IV-specific ppk-GAL4 [27] to reduce endogenous Spn-F expression. By 18 h APF wild-type neurons have pruned their dendrites (Fig 1A), however the primary dendrites remained connected to the cell body of C4da neurons with spn-F RNAi (RNA interference) (Fig 1B). We observed a similar phenotype in spn-F loss-of-function mutants (spn-F2, [24]) (Fig 1C). Dendrite severing was likewise suppressed in neurons of spn-F2/Df mutants (Fig 1D), which carry the spn-F2 allele and a deficiency uncovering the entire spn-F gene locus. To confirm that these pruning defects were due to loss of spn-F from C4da neurons, we expressed the full-length spn-F-GFP directed by ppk-GAL4 in spn-F2 mutants and found that the impaired dendrite pruning was rescued (Fig 1E and 1F). Moreover, we also examined the dendritic morphology of larval C4da neurons of spn-F and ik2 mutants, and found that dendrites develop normally in both mutant neurons (S1 Fig) [15], suggesting that the pruning defects observed in both mutants are not secondary to abnormal dendrite development. Taken together, these results indicated that spn-F is required for dendrite pruning in C4da neurons.
Fig. 1. Spn-F is required for dendrite pruning. 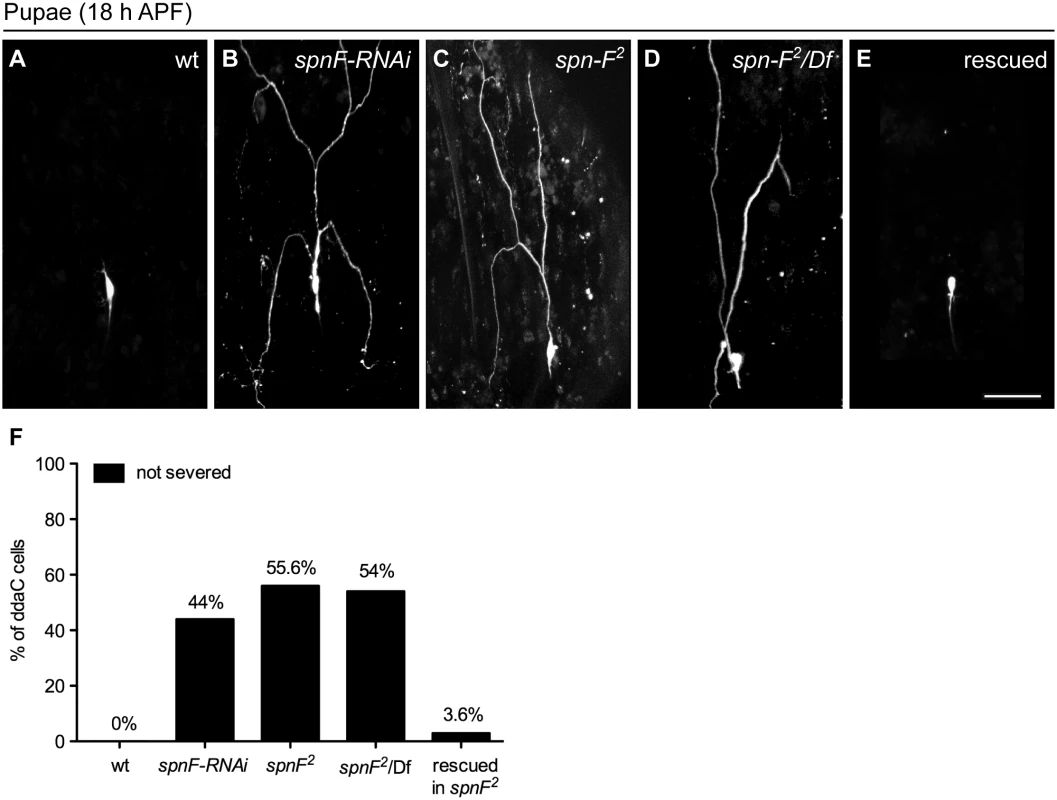
(A-E) The ddaC neurons are labeled with eGFP driven by ppk promoter (ppk-eGFP). At 18 h APF, the dendrites were severed and eliminated in wild-type ddaC neurons (A), but remained attached to the neurons of spn-F-RNAi mutants (B), of homozygous spn-F mutants (spn-F2) (C), and of spn-F/Df mutants, which carry spn-F2 on one chromosome and a deficiency Df(3R)tll-e (deletion includes the spn-F gene) on the other chromosome (D). (E) Dendrite pruning was observed normal in ddaC neurons of spn-F2 mutants rescued with wild-type spn-F-GFP expression directed by ppk-GAL4. (F) Quantitative analysis of dendrite pruning phenotypes in ddaC cells at 18 h APF. The percentage of cells was determined by dividing the number of ddaC neurons with defective pruning by the total number of cells examined for each genotype; for wild type (wt), n = 50; for spn-F-RNAi, n = 100; for spn-F2 mutants, n = 115; for spn-F2/Df, n = 90; for spn-F2 mutants rescued with wild-type spn-F-GFP, n = 88. Scale bar, 50 μm. spn-F acts downstream of ik2 in dendrite pruning
Since ik2 and spn-F mutant neurons displayed the same phenotype, we hypothesized that both genes function in the same pathway during dendrite pruning. To test this hypothesis, we performed genetic analyses between ik2 and spn-F. It was known that ik2 over-expression in larval C4da neurons causes cell death [15]. To avoid excessive apoptosis, we employed ppk-GAL4 coupled with its temperature-sensitive inhibitor GAL80ts to achieve spatial and temporal ik2 expression in larval neurons by shifting temperature. Consistent with previous studies [15], no abnormality was detected in larval neurons under permissive temperature 25°C (Fig 2A). After shifting to non-permissive temperature 29°C, ik2 overexpression not only triggered precocious dendrite severing (Fig 2B), but also caused apoptosis (Fig 2C) in wild-type larval neurons. Interestingly, we found that both precocious dendrite severing and apoptosis caused by ik2 overexpression were significantly suppressed in neurons of spn-F mutants (Fig 2D), suggesting that spn-F functions downstream (or in parallel) of ik2 in dendrite pruning. Additionally, we found no significant difference between the dendrite pruning defects observed in spn-F2 mutants and that in spn-F2 mutants with ik2 RNAi (Fig 2E), indicating that both ik2 and spn-F act in the same pathway of dendrite pruning. Together, our findings suggested that spn-F acts downstream of ik2 in the same pathway during dendrite pruning.
Fig. 2. Spn-F acts downstream of Ik2 in dendrite pruning. 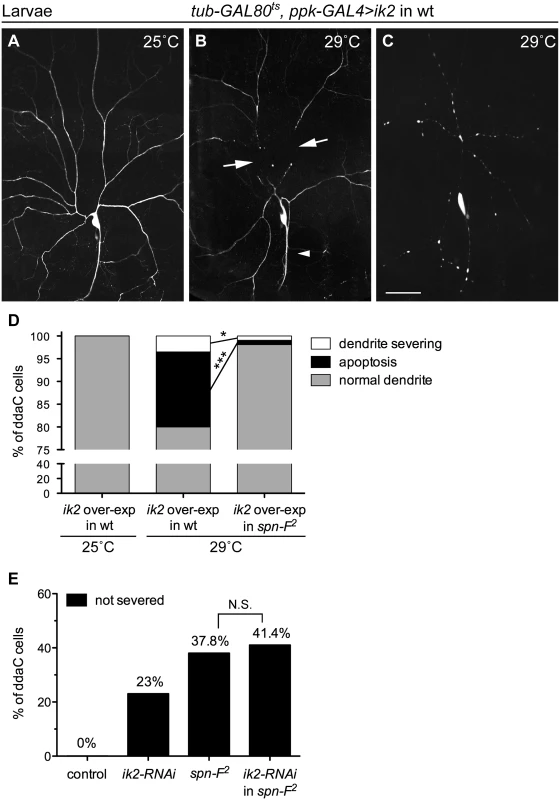
The temporally increasing Ik2 activity in ddaC neurons of wild-type larvae (A-C) was controlled by the temperature-sensitive GAL80 (GAL80ts). Each animal contained the following transgenes: tub-GAL80ts, ppk-eGFP, ppk-GAL4, and UAS-ik2. The dendrites of larval ddaC neurons displayed normal morphology at 25°C (A), but showed precocious dendrite-severing (arrows) at non-permissive temperature 29°C (B). Cell deaths were also observed in neurons at 29°C, as indicated by blebbing dendrites and axons (C). (D) Quantitative analysis of precocious dendrite-severing phenotypes and cell deaths observed in ddaC cells of wild-type and of spn-F2 mutant larvae, which have ik2 overexpression under the control of GAL80ts. In the wild-type larvae, the dendrites of all ddaC neurons showed normal morphology at 25°C (n = 100), but showed precocious dendrite-severing and apoptosis at 29°C (n = 200). Both precocious dendrite severing and apoptosis were suppressed in spn-F2 mutant larvae at 29°C (n = 420). *, p = 0.0158; ***, p <0.0001. Statistical analysis was performed with Fisher’s exact test. (E) Quantitative analysis of dendrite pruning phenotypes in ddaC cells at 16 h APF. The percentage of cells was determined by dividing the number of ddaC neurons with defective pruning by the total number of cells examined for each genotype; for wild type (wt), n = 50; for ik2-RNAi, n = 100; for spn-F2 mutants, n = 90; for ik2-RNAi in spn-F2 mutants, n = 70. Statistical analysis was performed with Chi-square test. N.S., not significant. Scale bar, 50 μm. The distribution of Spn-F proteins depends on Ik2 kinase activity
Given that the spn-F-GFP transgene could rescue observed defects in spn-F mutants, we concluded that this transgene could functionally substitute for the endogenous spn-F gene. Therefore, studying the role of Spn-F-GFP in C4da neurons should help us to uncover the molecular function of endogenous Spn-F in dendrite pruning. First, we examined the distribution of Spn-F-GFP in larval and pupal neurons. The Spn-F-GFP proteins displayed a punctate pattern in the soma, dendrites and axons of larval C4da neurons (Fig 3A and S2A Fig). However, those punctate Spn-F-GFP proteins were redistributed in the soma of pupal neurons at 5 h APF (Fig 3B). Since both Ik2 kinase and ecdysone are required for dendrite pruning [15], we asked whether Ik2 kinase activity and ecdysone signaling regulate the dispersion of Spn-F-GFP puncta in pupal neurons during pruning. To test this possibility, we examined the Spn-F-GFP distribution in mutant pupal neurons with expression of ik2-RNAi, kinase-dead mutant ik2-G250D [28] or dominantly negative ecdysone receptor (EcR-DN), and found that the Spn-F-GFP puncta remained intact in all three mutant neurons at 5 h APF (Fig 3C–3E). These results indicated that there is a negative correlation between Spn-F puncta and dendrite pruning, and both Ik2 kinase and ecdysone signaling are required to redistribute Spn-F from puncta in neurons during dendrite pruning. Moreover, we also found that redistribution of Spn-F from puncta was also observed in the larval C4da neurons with Ik2, but not with Ik2-G250D, overexpression (S2F–S2I Fig).
Fig. 3. Ik2 kinase activity regulates the distribution of Spn-F in neurons. 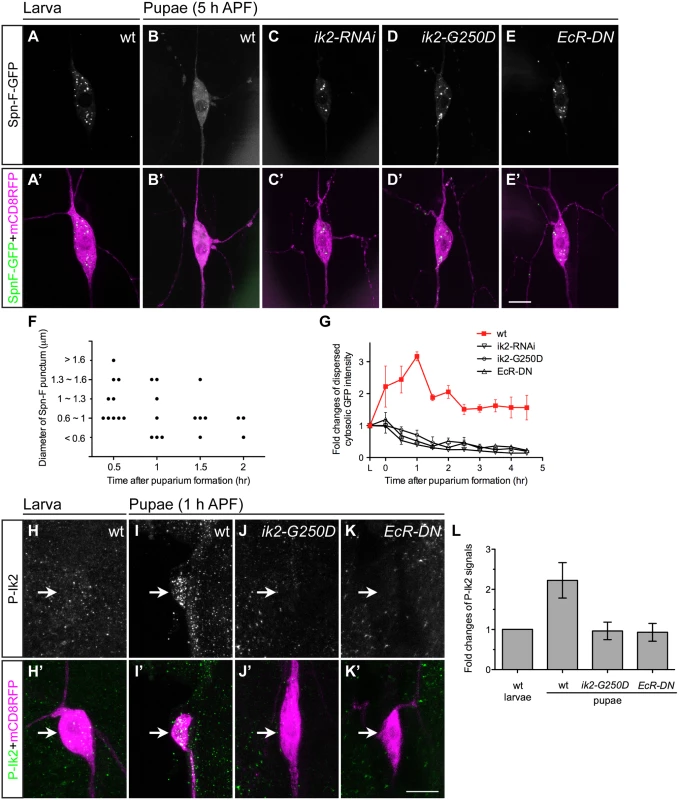
(A'-E', H'-K') The ddaC neurons were identified by ppk-GAL4 and UAS-mCD8RFP. (A-E) The expression of UAS-Spn-F-GFP was controlled by ppk-GAL4 in C4da neurons. The Spn-F-GFP showed punctate patterns in the cytosol of ddaC neurons of wild-type larvae (A). At 5 h APF, Spn-F-GFP became dispersed in wild-type pupal neurons (B), but remained punctate in mutant pupal neurons with ik2-RNAi (C), ik2-G250D (D) and EcR-DN (E) expression. (F) Quantitative analysis of the diameters and numbers of each Spn-F puncta in a single live ddaC neuron (the same neuron as in S1 Movie) at different time points. Each dot represents single Spn-F punctum in neuron at specific time point. (G) The average cytosolic fluorescent intensity of dispersed cytosolic GFP signals (an example indicated by asterisk in S2E Fig) was measured in the same ddaC neurons (n = 4 for each genotype) of larvae and at various time points of pupae in wild type and mutants with ik2-RNAi, ik2-G250D and EcR-DN expression. The fold changes were determined by dividing the average cytosolic fluorescent intensity of GFP signals in pupal ddaC neurons by that in the same larval cells, which is assigned as 1. (H-K) The activated Ik2 signals were detected by antibodies against phosphorylated Ik2 (P-Ik2) in larvae and pupae at 1 h APF. The strong P-Ik2 signals were observed in wild-type pupal C4da neurons at 1 h APF (I), but not in wild-type larval neurons (H), and not in mutant pupal neurons with either kinase-dead Ik2-G250D expression (J) or with dominantly-negative ecdysone receptor (EcR-DN) expression (K). (L) The average P-Ik2 signal intensity was measured in the cell body of ddaC neurons of wild-type larvae, and wild-type and mutant pupae at 1 h APF (n = 4 for each genotype). The fold changes were determined by dividing the average cytosolic fluorescent intensity of P-Ik2 signals in pupal ddaC neurons by that in larval cells, which is assigned as 1. Error bars show SD. Scale bars, 10 μm. To gain mechanistic insight into the redistribution of Spn-F from puncta, we performed live-cell imaging to monitor Spn-F-GFP distribution in the same C4da neurons from larvae to pupae (S1 and S2 Movies). The live imaging and signal profiling of Spn-F-GFP showed high punctate and low dispersed signals in the cytosol of larval neurons (S2B and S2E Fig). We quantified the numbers and the sizes of Spn-F puncta in single live ddaC neurons, and found both numbers and sizes of Spn-F puncta decrease along the pupation time (Fig 3F). In contrast, the dispersed Spn-F-GFP signals in the cytosol of pupal neurons increased as the punctate signals decreased (S2C–S2E Fig). We quantified the averaged fluorescent intensity of dispersed cytosolic Spn-F-GFP signals in the soma, and measured the fold changes of these signals in the same neurons at various time points. In the cytosol of wild-type C4da neurons, the dispersed cytosolic GFP signals started to increase soon after pupation (Fig 3G), peaked at 1 h APF, and decreased to a steady state after 2 h APF. We noticed that these dispersed cytosolic signals measured in pupal neurons were all higher than those in larval cells (Fig 3G). Furthermore, this prompt increase of dispersed Spn-F-GFP signals in wild-type neurons was not observed in the mutant neurons with ik2-RNAi, ik2-G250D, or EcR-DN expression (Fig 3G), indicating that Ik2 kinase and ecdysone signaling promote the increase of dispersed Spn-F in early pupal neurons.
Next, we assessed Ik2 kinase activities in C4da neurons by staining with antibodies against phosphorylated Ik2 (P-Ik2) on serine 175, whose phosphorylation is essential for Ik2 activation [29]. Consistent with the observation of maximum dispersed Spn-F-GFP signals in the cell body of pupal neurons at 1 h APF (Fig 3G), strong P-Ik2 signals were detected in C4da neurons at the same stage (Fig 3I and 3L), whereas no specific signals were found in larval cells (Fig 3H and 3L) or in mutant neurons expressing Ik2-G250D (Fig 3J and 3L). These results indicated that Ik2 kinase activity is upregulated in early pupal neurons, but downregulated in larval neurons. Since transiently increasing Ik2 expression caused precocious dendrite severing of larval neurons (Fig 2B) [15], we concluded that elevation of Ik2 activity in pupal neurons initiates dendrite severing, and suppression of Ik2 activity in larval neurons prevents premature severing. We also examined P-Ik2 signals in mutant neurons with disrupted ecdysone signaling and failed to detect activated Ik2 signals (Fig 3K and 3L), indicating that Ik2 kinase acts downstream of ecdysone signaling in dendrite pruning. Taken together, these data demonstrate that Ik2 kinase is activated promptly in early pupal neurons and regulates Spn-F distribution in C4da neurons during dendrite pruning.
Ik2-dependent phosphorylation of Spn-F is required for Spn-F distribution and dendrite pruning
The inverse correlation between Spn-F puncta and dendrite pruning suggests that redistribution of Spn-F from puncta in C4da neurons might be a critical event for dendrite pruning. If this is true, a mutant Spn-F that remains punctate and disperse-resistant in pupal neurons should have pruning defects. To generate such a mutant Spn-F, understanding the regulatory mechanism of Spn-F distribution is prerequisite. The detection of a band shifting of Spn-F-GFP, which is only expressed in C4da neurons, specifically in the larval lysates with Ik2 expression, but not with kinase dead Ik2-G250D expression or without Ik2 expression (S3 Fig), indicated that Spn-F could be phosphorylated by Ik2 kinase in C4da neurons. Since our study revealed that Ik2 kinase activity regulates Spn-F distribution, and Dubin-Bar et al. also shown that Ik2 phosphorylates Spn-F in kinase assays [26], we further characterized how the phosphorylation of Spn-F by Ik2 kinase regulates Spn-F distribution and dendrite pruning. First, to identify the residues of Spn-F phosphorylated by Ik2 kinase, we carried out mass spectrometry analyses. Given that Ik2-dependent redistribution of Spn-F-GFP in neurons could also be observed in Drosophila S2 cells, we reasoned that the mechanism underlying the reduction of Spn-F puncta is conserved in both cell types. Thus, we purified 1) Spn-F proteins from S2 cells with Ik2 expression referred to as the group of high Ik2 activity, and 2) Spn-F from S2 cells alone or 3) S2 cells with kinase-dead Ik2-K41A expression [30] referred as the group of low kinase activity for mass spectrometry analyses. The results revealed five serine residues (S53, S85, S264, S270 and S349) of Spn-F that are phosphorylated by Ik2 kinase specifically (S1 and S2 Tables and Fig 4A), and three other serine residues (S172, S202 and S325) showing increased level of phosphorylation in the group of high Ik2 activity (Fig 4A and S4 Fig). To determine the function of phospho-Spn-F, we substituted all eight serine residues with alanine to generate the phospho-deficient Spn-F-8A, and with aspartic acid to make the phospho-mimetic Spn-F-8D (Fig 4A). We observed evident gel mobility shift of Spn-F in the cell lysates of S2 cells co-expressing Ik2, but not Ik2-G250D (Fig 4B). In contrast, we detected no mobility shift of Spn-F-8A in any case (Fig 4B). This confirmed that the eight serine residues identified by mass spectrometry are the main sites of Spn-F phosphorylated by Ik2 kinase. Both Spn-F-8A and Spn-F-8D retained the interaction with Ik2 as the control Spn-F did (S5 Fig), consistent with previous reports that Ik2-dependent phosphorylation of Spn-F does not affect the interaction between Ik2 and Spn-F [26].
Fig. 4. The phosphorylation of Spn-F by Ik2 kinase is required for Spn-F distribution and dendrite pruning. 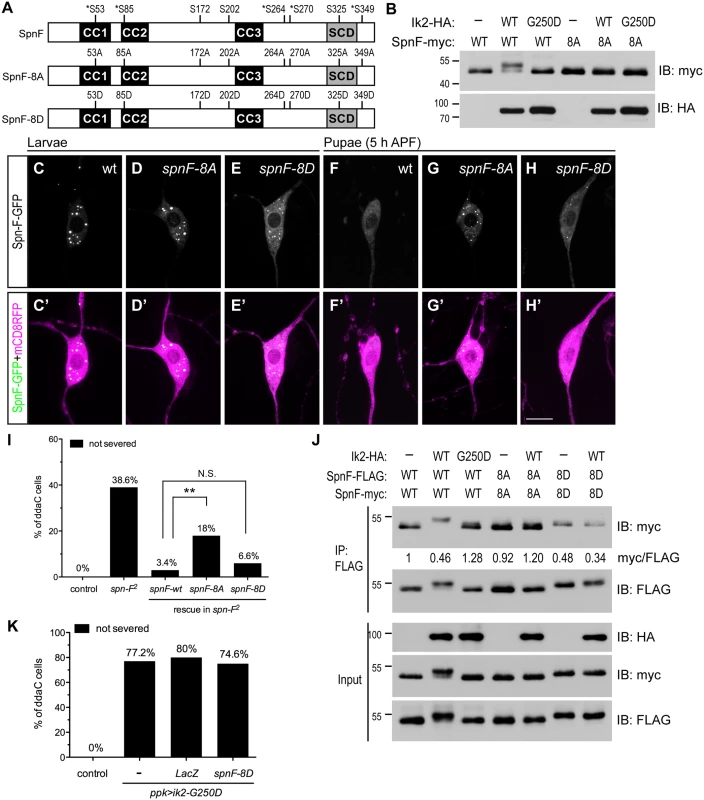
(A) Schematic shows the amino-acid residues of Spn-F protein phosphorylated by Ik2 kinase as identified by mass spectrometry. Five serine residues phosphorylated by Ik2 specifically are indicated by asterisks, and three serine residues with increased phosphorylation level in S2 cells with Ik2 overexpression are shown. These eight serine residues of Spn-F were mutated to alanines to generate phospho-resistant SpnF-8A mutant, or to aspartic acids to make phospho-mimetic SpnF-8D mutant. (B) Wild-type Spn-F or SpnF-8A was expressed in S2 cells alone, or together with wild-type Ik2 or kinase-dead Ik2-G250D mutant, and lysates were used for immunoblot analyses. (C'-H') The C4da neurons labeled by ppk-GAL4 and UAS-mCD8RFP are shown. In ddaC neurons of wild-type larvae, SpnF-GFP (C), SpnF-8A-GFP (D) and SpnF-8D-GFP (E) showed punctate pattern. At 5 h APF, both SpnF-GFP (F) and SpnF-8D-GFP (H) puncta were dispersed, whereas SpnF-8A-GFP (G) remained as puncta. (I) Quantitative analysis of dendrite pruning phenotypes in ddaC cells at 16 h APF. SpnF-8D, not SpnF-8A, rescued pruning defects in ddaC neurons of spn-F2 mutants. The percentage of ddaC neurons shows dendrite pruning defects among the total number of neurons examined. For wild-type control, n = 50; for spn-F2 mutants, n = 70; for spn-F2 mutants rescued with wild-type spn-F, n = 88; for spn-F2 mutants rescued with spnF-8A, n = 100; for spn-F2 mutants rescued with spnF-8D, n = 106. N.S., not significant. **, p = 0.0019. Statistical analysis was performed with Fisher’s exact test. (J) Co-IP experiments with lysates from S2 cells cotransfected with wild-type spn-F, spnF-8A or spnF-8D alone, and together with wild-type ik2 or mutant ik2-G250D show that phosphorylation of Spn-F by Ik2 kinase decreases the self-association of Spn-F proteins. The myc/FLAG ratio (by ImageJ) indicates the relative amounts of Spn-F-myc protein associated with Spn-F-FLAG protein in co-IP experiments with anti-FLAG antibody. (K) Quantitative analysis of dendrite pruning phenotypes in ddaC cells at 16 h APF. The percentage of ddaC neurons shows dendrite pruning defects among the total number of neurons examined. For wild-type control, n = 100; for ppk>Ik2-G250D mutants, n = 92; for ppk>Ik2-G250D, LacZ mutants, n = 50; for ppk>Ik2-G250D mutants rescued with spnF-8D, n = 122.Scale bar, 10 μm. The presence of Spn-F puncta in cells suggested that Spn-F might form oligomers through self-association. To test this possibility, we performed co-immunoprecipitation (co-IP) in S2 cells and found that Spn-F proteins could interact with themselves (Fig 4J). Moreover, we observed reduced self-association of Spn-F in the S2 cells with Ik2 co-expression, but not in either the control S2 cells alone or the S2 cells with Ik2-G250D expression (Fig 4J). These results indicated that the phosphorylation of Spn-F by Ik2 kinase decreases Spn-F self-association. As expected, the self-association of Spn-F-8D, but not Spn-F-8A, was strongly reduced in both the control and the Ik2-expressing cells (Fig 4J). These data suggested a mechanism that Ik2 phosphorylates Spn-F to decrease Spn-F self-association and promotes the redistribution of Spn-F from punctate to dispersed in cytosol. Next, we examined the distribution of Spn-F-8A-GFP and -8D-GFP proteins in larval and pupal neurons respectively. The puncta formed by SpnF-8A-GFP were comparable to those formed by Spn-F-GFP in larval neurons (Fig 4C and 4D). However, the puncta formed by SpnF-8D-GFP were generally smaller than those by Spn-F-GFP in larval cells (Fig 4E). Moreover, the dispersed GFP signals in the cytosol of larval neurons with SpnF-8D-GFP expression were higher than those with Spn-F-GFP expression (Fig 4E). Notably, SpnF-8A-GFP puncta were resistant to disperse in neurons at 5 h APF (Fig 4G), while both Spn-F-GFP and SpnF-8D-GFP puncta were redistributed (Fig 4F and 4H). These findings confirmed that phosphorylation of Spn-F by Ik2 kinase promotes the dissociation of Spn-F oligomers and facilitates Spn-F redistribution in early pupal neurons.
Finally, to determine the role of Spn-F redistribution in dendrite pruning, we examined whether SpnF-8A, whose puncta are resistant to redistribution in pupal neurons, could rescue dendrite-pruning phenotypes in spn-F mutant neurons. Compared to the pruning defects rescued by wild-type Spn-F, we found Spn-F-8A is incapable of fully rescue the pruning defects in spn-F mutants (Fig 4I, p<0.005). This result confirmed that Spn-F redistribution in pupal C4da neurons is required for dendrite pruning. Moreover, SpnF-8D rescued the pruning phenotypes in neurons of spn-F mutants as efficiently as wild-type Spn-F did (Fig 4I). However, SpnF-8D cannot rescue the pruning defects in mutant neurons with ik2-G250D overexpression (Fig 4K), suggesting that other unidentified factors are required for dendrite pruning. Together, these results demonstrated that phosphorylation of Spn-F by Ik2 kinase, Spn-F redistribution and unidentified factors are critical for dendrite pruning in C4da neurons.
Cytoplasmic dynein motor complex is required for Spn-F redistribution and dendrite pruning
Since Spn-F redistribution from puncta is critical for dendrite pruning in C4da neurons, to further study the mechanism underlying Ik2-dependent Spn-F redistribution, we searched for other candidates that involve in Spn-F redistribution during dendrite pruning. Given that Spn-F interacts with a subunit of cytoplasmic dynein complex [24], Cut up (Ctp), the Drosophila homologue of dynein light chain 1 [31,32], and dynein is a microtubule-based motor protein, we questioned whether cytoplasmic dynein is involved in dendrite pruning. To address this question, we began to examine the roles of microtubules and cytoplasmic dyneins in Ik2-dependent redistribution of Spn-F in S2 cells with disrupted microtubule cytoskeletons or impaired dynein function. The redistribution of Spn-F from punctate to dispersed in the cytosol of S2 cells is also Ik2-dependent (Fig 5A and 5B). We found that Ik2-dependent Spn-F redistribution was suppressed in cells treated with microtubule-disrupting chemical, colchicine (Fig 5C). The similar observation was also obtained from S2 cells treated with cytoplasmic dynein specific inhibitor, ciliobrevin D [33] (Fig 5D). Notably, Spn-F puncta formed normally in S2 cells treated with either colchicine or ciliobrevin D (S6 Fig), suggesting that the intact microtubule cytoskeletons and functional dynein complexes are not required for the formation of Spn-F puncta. These results indicated that Ik2-dependent redistribution of Spn-F requires both functional dynein and intact microtubule cytoskeletons, and further suggested that Spn-F redistribution is caused by the movement of cytoplasmic dynein on microtubules in cells.
Fig. 5. The function of cytoplasmic dynein complex is required for Spn-F redistribution and dendrite pruning. 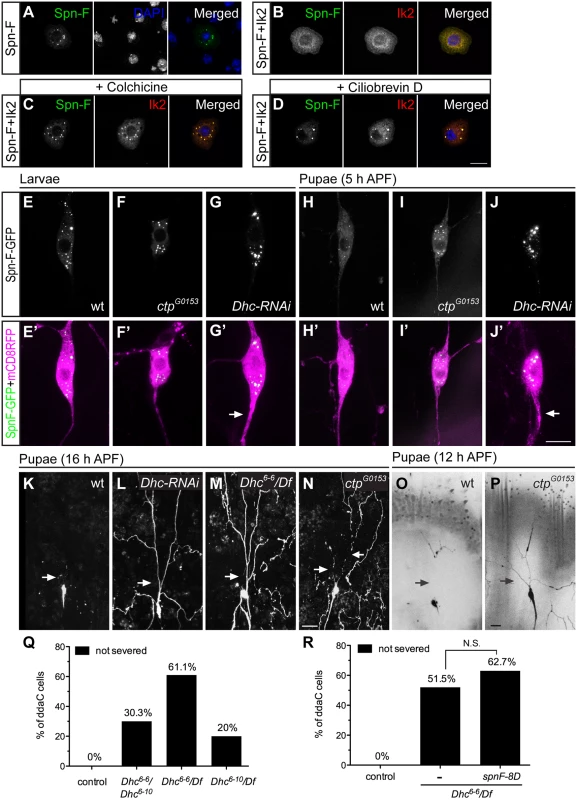
(A-D) Immunofluorescence of Spn-F-GFP, Ik2-HA and nuclear DAPI staining in S2 cells (A, B), in S2 cells treated with colchicine (C), and in S2 cells treated with ciliobrevin D (D). (E'-J') The ddaC neurons were visualized with ppk-GAL4 and UAS-mCD8RFP. The Spn-F-GFP displayed as puncta in the soma of ddaC neurons of wild type (E), ctpG0153 (F), and Dhc-RNAi (G) larvae. At 5 h APF, the Spn-F-GFP puncta were dispersed in ddaC neurons of wild-type pupae (H), but remained as puncta in both ctpG0153 (I) and Dhc-RNAi (J) pupae. The thicken axons (arrows in G’ and J’) were caused by reduction of functional dynein complexes in Dhc-RNAi neurons. (K-P) The ddaC neurons were marked with ppk-GAL4 and UAS-mCD8GFP. At 16 h APF, the dendrites of ddaC neurons were pruned in wild-type (K), but not in Dhc-RNAi (L), or Dhc6-6 /Df mutant (M) pupae. Df: deficiency line Df(3L)BSC436, which deletes Dhc gene. (N) The dendrites of ddaC cells were severed in the proximal dendrites (arrows), but not cleaned up in ctpG0153 mutant pupae at 16 h APF. At 12 h APF, the dendrites of ddaC neurons are all severed (arrows) in wild-type pupae (O), but remained attached to the soma of ddaC cells in ctpG0153 pupae (P). (Q) Quantitative analysis of dendrite pruning phenotypes in ddaC cells of Dhc mutants at 16 h APF. The percentage of ddaC neurons shows dendrite pruning defects among the total number of neurons examined. For wild-type control, n = 100; for Dhc6-6/Dhc6-10 mutants, n = 89; for Dhc6-6/Df mutants, n = 90; for Dhc6-10/Df mutants, n = 100. (R) Quantitative analysis of dendrite pruning phenotypes in ddaC cells of Dhc mutants with SpnF-8D expression at 16 h APF. The percentage of ddaC neurons shows dendrite pruning defects among the total number of neurons examined. For wild-type control, n = 100; for Dhc6-6/Df mutants, n = 130; for Dhc6-6/Df mutants rescued with spnF-8D, n = 110. Statistical analysis was performed with Chi-square test. N.S., not significant. Scale bars, 10 μm (A-J), 20 μm (K-P). Next, to verify whether the function of cytoplasmic dynein complexes is required to redistribute Spn-F in neurons for dendrite pruning, we examined the distribution of Spn-F-GFP in mutant C4da neurons with impaired dynein function. Considering the interaction between Spn-F and Ctp proteins, we first examined the Spn-F-GFP distribution in ctp mutant neurons. Since ctp is an essential gene for embryonic development [32], we used a hypomorphic ctp mutant allele ctpG0153, which had few mutant larvae developing to pupae, for analyses. The Spn-F-GFP puncta formed normally in larval neurons of ctpG0153 mutants (Fig 5F, compared to Fig 5E), but failed to disperse in mutant pupal neurons at 5 h APF (Fig 5I, compared to Fig 5H). We next examined the role of dynein heavy chain (Dhc), which encodes the only motor subunit in dynein complex, in the redistribution of Spn-F in C4da neurons. Among seven Dhc genes in fly genomes [34], Dhc64C (thereafter referred to as Dhc) is the only one exhibiting ubiquitous expression, including the nervous systems, throughout development [35]. We employed Dhc RNAi lines to knockdown endogenous Dhc. The thick axons found in larval neurons with Dhc dsRNAs expression (Fig 5G’, compared to Fig 5E’) were consistent with reported phenotypes in mutant neurons with impaired dynein function [36], and validated the knockdown efficiency. The Spn-F-GFP puncta formed normally in larval neurons of Dhc-RNAi mutants (Fig 5G), but remained intact in mutant neurons at 5 h APF (Fig 5J). These findings demonstrated the requirement of functional dynein complex to redistribute Spn-F in C4da neurons during dendrite pruning.
Finally, to verify the roles of dynein complexes in dendrite pruning, we examined whether mutant C4da neurons with impaired dynein function show pruning defects. At 16 h APF, the primary dendrites remained connected to the soma of C4da neurons in Dhc RNAi mutants (Fig 5L, compared to Fig 5K), suggesting a critical role of dynein motor in dendrite pruning. We also verified the dendrite pruning defects in neurons of trans-heterozygous Dhc mutants with hypomorphic alleles, Dhc6-6 and Dhc6-10 (Fig 5Q) [37]. In order to confirm that the phenotypes observed in hypomorphic Dhc mutants were not due to other unidentified mutations on the same chromosome, we examined C4da neurons of Dhc/Df mutants (deficiency uncovers the entire Dhc64C gene). The defective dendrite pruning was evident in C4da neurons of Dhc/Df mutants; 61% (n = 90) of ddaC neurons in Dhc6-6/Df mutants (Fig 5M and 5Q) and 20% (n = 100) of cells in Dhc6-10/Df mutants (Fig 5Q) showed phenotypes at 16 h APF. Since dynein has been shown to play an essential role in larval dendrite development [36], to verify the pruning defects in Dhc mutants were not secondary to the early dendrite development defects, we examined the dendritic morphology of C4da neurons in Dhc mutant larvae, and observed normal dendrites of most larval C4da neurons in Dhc mutants (S7 Fig). These findings demonstrated that Dhc plays a critical role in dendrite pruning of C4da neurons. Furthermore, at 16 h APF, about 22.5% of C4da neurons (n = 80) in hypomorphic ctpG0153 mutants exhibited severe dendrite clearance phenotypes, which is characterized by long disconnected dendrites surrounding the soma (Fig 5N). We hypothesized that ctpG0153 mutant neurons might have delayed dendrite severing, which resulted in deferred clearance. To test this hypothesis, we examined the dendrites of C4da neurons in wild-type and ctpG0153 mutant pupae at 12 h APF. All dendrites of wild-type neurons (n = 38) were severed at 12 h APF (Fig 5O); however, about 26.8% of neurons (n = 56) in ctpG0153 mutants still retained their primary dendrites attached to the soma at the same stage (Fig 5P). These results indicated that dendrite severing was delayed in hypomorphic ctp mutant neurons, and confirmed that ctp plays a role in dendrite pruning. Finally, we asked whether SpnF-8D could rescue the dendrite pruning defects observed in Dhc mutants. We found no significant difference between the pruning defects of C4da neurons in Dhc6-6/Df mutants and that in Dhc mutants with SpnF-8D overexpression at 16 h APF (Fig 5R), indicating that SpnF-8D cannot rescue the dendrite pruning defects in Dhc mutants. Taken together, our data demonstrates that cytoplasmic dynein motor complex is required for Spn-F redistribution in C4da neurons and for dendrite pruning.
Spn-F links Ik2 kinase to cytoplasmic dynein motor complex
Since Spn-F interacts with both Ik2 and Ctp [24,26], we hypothesized that Ik2, Spn-F and Ctp might form ternary complexes, which could be transported by cytoplasmic dynein. To test this hypothesis, we performed co-IP and antibody staining in S2 cells. The formation of Ik2/Spn-F/Ctp ternary complexes could be detected by co-IP only from the lysates of S2 cells with Spn-F co-expression, but not from that without Spn-F co-expression (Fig 6A), suggesting the central role of Spn-F in the ternary complex formation. As Ik2 kinase causes Spn-F redistribution, the even distribution of all three molecules observed in S2 cells (S8 Fig) did not provide convincing evidence to support the colocalization. To visualize the colocalization of these three molecules in S2 cells, we used Ik2-G250D for antibody staining based on the following reasons. First, the dependence of Spn-F to form ternary complex was also observed between Ik2-G250D and Ctp proteins (Fig 6A). Second, since Ik2 kinase activity does not affect Ik2/Spn-F interaction [26], kinase-dead Ik2-G250D still possesses normal interaction with Spn-F and remains as puncta in S2 cells. The immunofluorescence staining (Fig 6B and 6C) and quantification (Fig 6D) showed that the colocalization of Ik2-G250D, Spn-F and Ctp molecules were observed only in S2 cells with Spn-F co-expression, but not in cells without Spn-F co-expression, supporting the formation of Ik2/Spn-F/Ctp ternary complex in cells. To test whether Spn-F phosphorylation by Ik2 kinase may affect the formation of Ik2/Spn-F/Ctp complexes, we performed co-IP and colocalization experiments with SpnF-8A and SpnF-8D in S2 cells. Our results showed that SpnF-8A and SpnF-8D have similar abilities as wild-type Spn-F in mediating the formation of Ik2/Spn-F/Ctp complexes in S2 cells (S9 Fig), suggesting that Spn-F phosphorylation have no or minor effects on Ik2/Spn-F/Ctp complex formation. These data demonstrated that Spn-F acts as a central mediator to link Ik2 kinase to dynein motor complex via Spn-F/Ctp interaction in cells.
Fig. 6. Spn-F links Ik2 kinase to dynein complexes via SpnF/Ctp interaction. 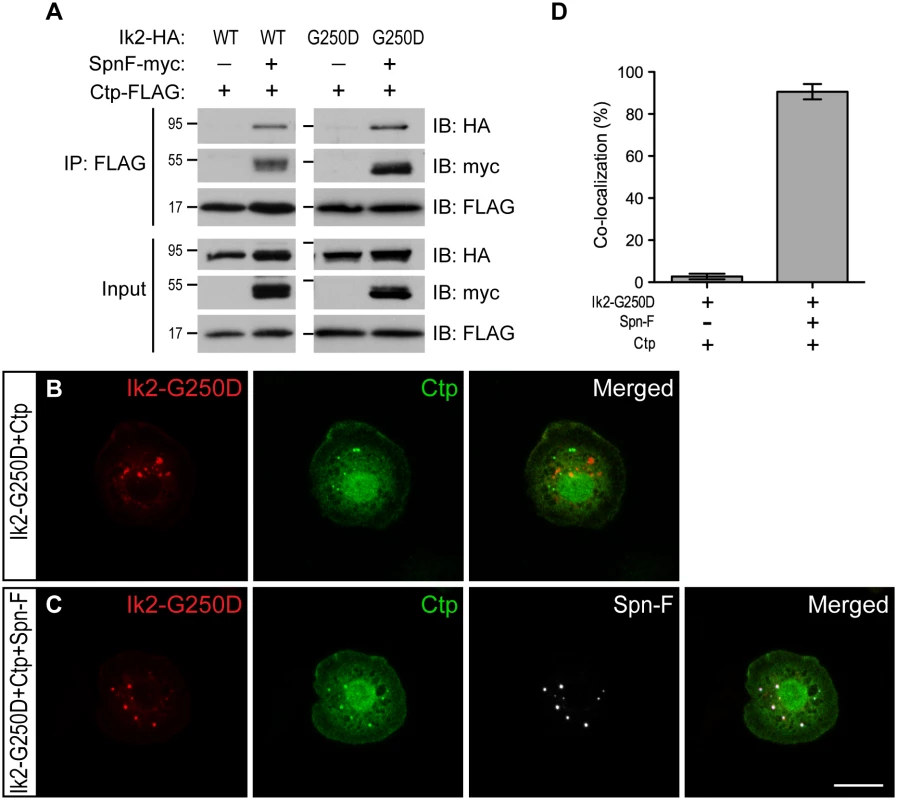
(A) Co-IP assays were performed with lysates from S2 cells transfected with Spn-F-myc, Ctp-FLAG and Ik2-HA or Ik2-G250D-HA to show that the formation of Ik2 (or Ik2-G250D)/Spn-F/Ctp tertiary complexes depends on the presence of Spn-F proteins. (B) Immunofluorescent staining in S2 cells transfected with Ik2-G250D-HA and Ctp-FLAG showed no colocalization of Ik2-G250D and Ctp. (C) Immunofluorescent staining in S2 cells transfected with Ik2-G250D-HA, Spn-F-GFP and Ctp-FLAG showed colocalization of Ik2-G250D, Spn-F and Ctp. (D) Quantitative analysis of colocalization of Ik2-G250D/Ctp and Ik2-G250D/Spn-F/Ctp in transfected S2 cells. The percentage of S2 cells shows colocalization of staining signals among the total number of examined cells that expressed all transfected proteins. The percentages of colocalization were calculated from three independent experiments: for Ik2-G250D/Ctp transfected cells, n = 5/116, 2/102 and 2/105; for Ik2-G250D/Spn-F/Ctp transfected cells, n = 102/111, 100/107 and 96/111. Error bars show SD. Scale bar, 10 μm. The Ik2/Spn-F/Ctp complex is critical for Spn-F redistribution and dendrite pruning
As Ik2, Spn-F and Ctp are crucial for dendrite pruning, and Spn-F plays a central role in the formation of Ik2/Spn-F/Ctp ternary complex in cells, we asked whether this ternary complex is required for the Spn-F redistribution and for dendrite pruning in C4da neurons. To approach this question, we set out to map the Ik2 - and Ctp-interacting domains of Spn-F, and determined the role of Ik2/Spn-F/Ctp ternary complex in neurons for dendrite pruning. Spn-F protein contains three coiled-coil domains [38], namely CC1, CC2 and CC3, and an Spn-F conserved domain (SCD) at its C terminus (Fig 4A), which is evolutionarily conserved among different insect species (S10A Fig). To identify the Ik2 - and Ctp-interacting domains of Spn-F, we generated a series of Spn-F deletion mutants (S10B Fig) and examined their interactions with Ik2 and Ctp in S2 cells by co-IP. The results showed that removal of Spn-F CC3 domain completely abolished the interaction between Ik2 and Spn-F (Fig 7A), but remained weak interaction between Spn-F and Ctp (Fig 7B), indicating that both Ik2/Spn-F and Spn-F/Ctp interactions are disrupted in SpnF-ΔCC3. Moreover, the co-IP experiments revealed normal interaction between Ik2 and SpnF-ΔSCD (Fig 7A), but weak interaction between SpnF-ΔSCD and Ctp (Fig 7B), demonstrating that SpnF-ΔSCD keeps a normal interaction with Ik2, but a defective interaction with Ctp. Both the SpnF-ΔCC3 and -ΔSCD puncta formed normally in larval neurons (Fig 7D and 7E), comparable to the wild-type Spn-F puncta (Fig 7C); however, both Spn-F deletion mutants remained punctate in neurons at 5 h APF (Fig 7G and 7H, compared to Fig 7F), indicating that both Spn-F CC3 and SCD domains are crucial for Spn-F redistribution in pupal neurons. Finally, to determine the role of Ik2/Spn-F/Ctp ternary complex in C4da neurons for dendrite pruning, we performed rescue experiments with SpnF-ΔCC3-GFP and -ΔSCD-GFP in spn-F mutant neurons. SpnF-ΔCC3-GFP failed to rescue the dendrite pruning defects of ddaC neurons in spn-F mutants at 16 h APF (Fig 7I), demonstrating that the Spn-F CC3 domain, which mediates both Ik2/Spn-F and Spn-F/Ctp interactions, is essential for dendrite pruning in C4da neurons. In contrast to SpnF-ΔCC3-GFP, SpnF-ΔSCD-GFP only partially rescued the dendrite pruning defects in spn-F mutants at 16 h APF (Fig 7I), suggesting the role of Spn-F/Ctp interaction in facilitating dendrite pruning. Collectively, our data demonstrated that the Ik2/Spn-F/Ctp complex is critical for Spn-F redistribution and for dendrite pruning in C4da neurons.
Fig. 7. The formation of Ik2/Spn-F/Ctp complex is critical for Spn-F redistribution and dendrite pruning. 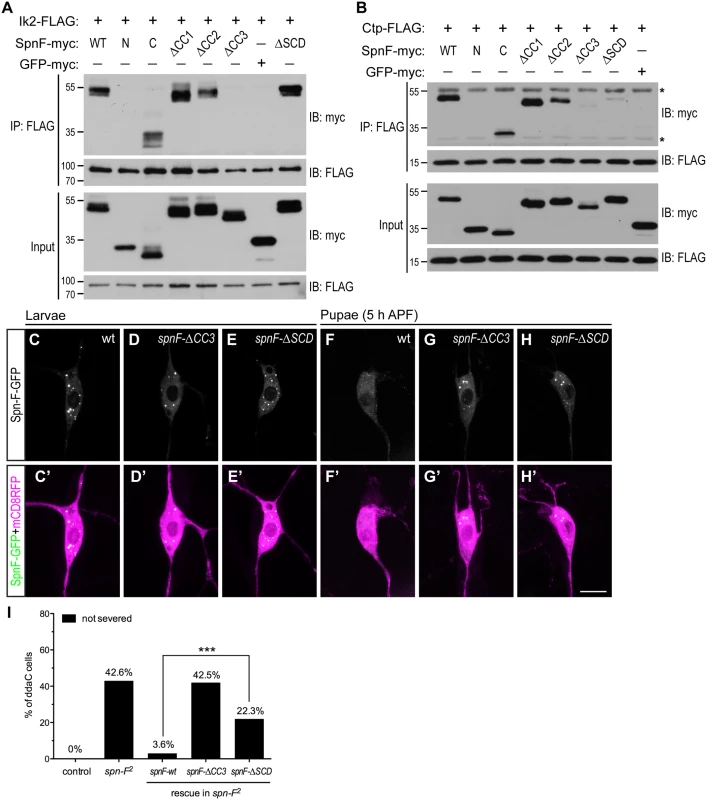
(A) The CC3 domain of Spn-F is the Ik2-interacting domain. Ik2-FLAG and full-length or Spn-F mutants lacking designated domain were expressed in S2 cells and used to make lysates for co-IP assays. GFP-myc was used as a negative control. (B) Both CC3 and SCD domains of Spn-F are required for Ctp binding. Ctp-FLAG and full-length or truncated Spn-F were expressed in S2 cells and lysates were prepared for co-IP with anti-FLAG antibodies. Asterisks indicate the position of heavy and light chains of antibodies. (C'-H') The ddaC neurons were visualized with ppk-GAL4 and UAS-mCD8RFP. The Spn-F-GFP exhibited puncta in the soma of ddaC neurons at larval stages (C), but dispersed in the pupal neurons at 5 h APF (F). The mutant SpnF-ΔCC3-GFP showed puncta in larval ddaC neurons (D), but remained as puncta in pupal neurons at 5 h APF (G). The mutant SpnF-ΔSCD-GFP formed puncta in larval ddaC neurons (E), and remained as puncta in pupal neurons at 5 h APF (H). (I) Quantitative analysis of pruning defects in ddaC neurons at 16 h APF. The percentage of ddaC neurons shows pruning defects among the total number of neurons examined. For wild-type control, n = 50; for spn-F2 mutants, n = 110; for spn-F2 mutants rescued with wild-type spn-F, n = 88; for spn-F2 mutants rescued with spnF-ΔCC3, n = 73; for spn-F2 mutants rescued with spnF-ΔSCD, n = 121. ***, p<0.0001. Statistical analysis was performed with Fisher’s exact test. Scale bar, 10 μm. Discussion
In addition to apoptosis, neurons have a second self-destruct program in their axons for axonal pruning during development and in response to neuronal injury and disorders [39]. Here, we propose a third self-destruct program, which is mediated by Ik2 kinase activity in Drosophila sensory neurons, specific for dendrite pruning. Ik2 is essential for dendrite severing in pupal C4da neurons [15], and currently is the only known molecule sufficient to cause precocious dendrite severing in larval cells [15], indicating that Ik2 activation must be regulated temporally. For temporal regulation, ecdysone signaling plays a key role in dendrite pruning [13,14]. Our studies showing that no Ik2 activation was detected in pupal C4da neurons with impaired ecdysone signaling and thus placed Ik2 kinase downstream of ecdysone signaling. Microarray studies have identified ik2 as one of the ecdysone/EcR up-regulated genes in brain MB γ neurons during axon pruning [40]. This suggests one possible mechanism where ecdysone/EcR regulates Ik2 activation through increasing ik2 expression in C4da neurons. Although Ik2 kinase activity is crucial for oogenesis and bristle morphogenesis [25,29], the activation mechanisms of Ik2 kinase in both processes remain unknown. Since pruning activity is considered as a self-destruct program, how to regulate this activity spatially in subcellular compartments within individual neurons is an intriguing issue to investigate. In this study, we identified Spn-F and cytoplasmic dynein complex as critical regulators of Ik2-mediated dendrite pruning activity in C4da neurons.
It is known that endogenous Spn-F exhibits a punctate pattern in nurse cells [24], consistent with our observation of punctate Spn-F-GFP in larval C4da neurons. The formation of Spn-F puncta in cells is through self-association, and does not depend on the integrity of microtubule network or the function of cytoplasmic dynein (S6 Fig). Since Ik2 could form oligomers in cells [29], the interaction between Ik2 and Spn-F might also play a role in Spn-F puncta formation. Indeed, we observed that SpnF-ΔCC3-GFP has normal interaction with either SpnF-ΔCC3 or full-length Spn-F (S11 Fig), but formed fewer puncta than the wild type Spn-F-GFP did in larval neurons (Fig 7D). Therefore, the Spn-F puncta formation could be attributed not only to Spn-F self-association, but also to Ik2/Spn-F interaction and Ik2 oligomerization.
In larval C4da neurons, Ik2 kinase is inactive and associates with Spn-F as puncta in the cytosol. After puparium formation, Ik2 kinase becomes activated promptly and phosphorylates Spn-F in C4da neurons. This Ik2-dependent phosphorylation on Spn-F decreases Spn-F self-association, and subsequently the numbers and sizes of Spn-F puncta were reduced (Fig 3F). One may question that protein degradation might contribute to decrease the numbers and sizes of Spn-F puncta in C4da neurons during dendrite pruning. It was known that Ik2 promotes caspase inhibitor DIAP1 degradation via proteasomes during the development of sensory organ precursors [28]; therefore, Ik2 might promote Spn-F degradation in C4da neurons during dendrite pruning. However, Ik2 overexpression does not alter the protein level of Spn-F in either S2 or germline cells [26]. Thus, protein degradation by proteasomes is unlikely the mechanism leading to decreased Spn-F puncta after Ik2 activation. Since P-Ik2 signals were indistinguishable between wild-type and Dhc64C RNAi neurons (S12 Fig), it is reasonable to presume that both Ik2 activation and Spn-F phosphorylation occur normally in dynein mutant neurons. We also found no significant differences between the pruning defects of C4da neurons in spn-F mutants and that in spn-F mutants with Dhc-RNAi (S12D Fig), and between the pruning phenotypes observed in Dhc mutants and that in Dhc mutants with ik2-RNAi (S12E Fig). These findings further support that Ik2, Spn-F and dynein complex function together in the same pathway in dendrite pruning of C4da neurons. However, our finding of Spn-F puncta in mutant pupal neurons with impaired dynein function indicated that dynein is required for Spn-F redistribution after Ik2 activation. Furthermore, Spn-F remains punctate in S2 cells with Ik2 overexpression even after microtubule depolymerization and inhibition of dynein function, suggesting that dynein might redistribute Ik2/Spn-F complexes via transporting complexes toward the minus ends of microtubules in C4da neurons during dendrite pruning. Based on our results in this study and studies in germline cells that more Spn-F puncta accumulated in nurse cells with colchicine treatment and with Dhc mutation [24], we favored the mechanism of protein redistribution for Spn-F punctum reduction in dendrite pruning of C4da neurons.
It has been shown that during Drosophila bristle elongation, directional transport of activated Ik2 and of Spn-F to the bristle tips, where the microtubule minus ends are concentrated [41], requires the function of cytoplasmic dynein, and Spn-F acts as an adaptor to link Ik2 to dynein complexes [42]. These are similar to our findings that both Ik2 activation and dynein complex are essential for Spn-F redistribution, and Spn-F plays a central role in the formation of Ik2/Spn-F/dynein complex, which is crucial for Spn-F redistribution and for dendrite pruning in C4da neurons. However, the studies in bristle elongation indicating that spn-F acts upstream of ik2 [42] disagree with our finding that ik2 acts upstream of spn-F in dendrite pruning. The discrepancy between the epistasis analyses of ik2 and spn-F in these two different processes might be due to different cell-type specific factors in these two types of cells that affect the morphological readouts in genetic studies. Moreover, we demonstrated that Ik2-dependent phosphorylation of Spn-F decreases Spn-F self-association, promotes Spn-F redistribution, and finally leads to dendrite pruning in C4da neurons.
The activated Ik2 signals are found accumulated at the microtubule minus ends in cells with polarized microtubule distribution, such as oocytes, follicle cells and bristles [29,43]. This is consistent with our conclusion that dynein transports activated Ik2 toward microtubule minus ends in C4da neurons. Since Drosophila sensory neurons have polarized microtubules in their proximal dendrites predominantly with microtubule minus end pointing away from the cell body [18], our studies revealed a possible mechanism that Spn-F and minus-end directed motor dynein complex confine Ik2-dependent pruning activity to the somatodendritic compartments of C4da neurons. During Drosophila bristle elongation, the accumulation of endogenous Spn-F observed at the bristle tip, where the microtubule minus ends are enriched [41], led us to examine Spn-F-GFP signals along the dendrites of C4da neurons during dendrite pruning. However, we did not observe enriched Spn-F-GFP signals in the proximal dendrites, where dendrite severing is expected to occur, by live imaging during pruning. Previous studies [15] showed that microtubules are first disassembled in the proximal dendrites of C4da neurons during dendrite severing, and this local disassembly of microtubules was suppressed in ik2 mutant neurons. Since our genetic studies indicated that both ik2 and spn-F acts in the same pathway of dendrite pruning, we tested whether local microtubule disassembly happens normally in spn-F mutants. We found that local breakage of microtubules in the proximal dendrites of C4da neurons was also suppressed in spn-F RNAi mutants (S13 Fig), suggesting that Spn-F, like Ik2, plays a role in dendrite severing that involves local microtubule disassembly. However, the molecular mechanisms by which activated Ik2 and Spn-F lead to dendrite severing in the proximal dendrites of C4da neurons will be an important question for future studies.
It is known that there was no decrease in cell death in wing discs with ik2 knockdown and in ik2 mutant embryos [28], indicating that the primary function of Ik2 is not involved in apoptotic pathway during development. However, ectopic Ik2 activation by overexpression leads to cell death in fly compound eyes [28] and in C4da neurons [15], suggesting that excessive Ik2 kinase signaling could trigger a crosstalk with signaling molecules in apoptotic pathway and result in apoptosis. It is known that Ik2 kinase regulates the nonapoptotic function of caspase through promoting DIAP1 degradation [28]. In a similar manner, the confinement of activated Ik2 kinase in the dendritic compartments might restrict the detected caspase activity in the degenerating dendrites after separating from the soma of C4da neurons during dendrite pruning [21]. Therefore, this raises a possibility that de-regulation of pruning activity in neurons may trigger a crosstalk with molecules in apoptotic pathway and lead to undesired cell death during neuronal injury and disorders. Recently, a caspase cascade, including caspase 3 and 6, was identified in mice to play a role in developmental axon pruning and in sensory axon pruning after trophic factor withdrawal [44,45]. Moreover, activated caspase 6 was detected in human patient brains of Alzheimer and Huntington diseases long before cell death [46], highlighting a critical role in regulating caspase activity in both diseases. Understanding the regulatory mechanisms that confine pruning activity into proper subcellular compartments of the neuron might provide molecular insights into the pathogenesis of neural disorders.
Materials and Methods
Fly stocks
spn-F2 [24], Dhc6-6, Dhc6-10 [37], ppk-eGFP [47], ppk-GAL4 [27], UAS-ik2 (UAS-DmIKK ε), and UAS-ik2-G250D [28] have been described previously. Df(3R)tll-e, Df(3L)BSC436, tub-GAL80ts, and UAS-EcR-F645A were obtained from Bloomington Stock Center. ctpG0153 was obtained from Drosophila Genetic Resource Center (DGRC). RNAi lines were obtained from Vienna Drosophila RNAi Center (VDRC), and Fly Stocks of National Institute of Genetics (NIG-FLY). Flies were raised at 25°C. For the experiments with tub-GAL80ts, flies were transferred and kept at 29°C for 40–42 h before analysis and imaging. Transgenic flies were generated by standard P-element mediated transformation.
Evaluation and quantification of the pruning phenotype
Our previous study with ddaC neurons labeled with ppk-eGFP or ppk-GAL4::UAS-mCD8GFP indicated normal severing occurs around 4–6 h APF, and complete debris clearance around 16–18 APF [15]. Therefore, we analyzed the pruning phenotype of ddaC neurons at or after 16 h APF to exclude individual temporal differences. At time of analysis, ddaC neurons having any single dendrite branch with continuous GFP signals extended from the center of soma more than 100 μm was considered pruning defective.
Molecular cloning
The spn-F full-length and deletion DNA fragments were generated by using PCR to amplify the DNA fragments from the cDNA clone LD01470 (Drosophila Genomics Resource Center) as the template. The spnF-8A and spnF-8D DNA fragments were generated by PCR-based site-directed mutagenesis. All spn-F DNA fragments were inserted together with EGFP coding sequence or the coding sequences containing HA, myc, or FLAG tags into pUAST vector to make various UAS-spn-F constructs. To generate UAS-ctp-FLAG construct, a DNA fragment encoding Ctp protein was amplified by PCR from the genomic DNA of w1118 flies and inserted together with FLAG coding sequence into pUAST vector. The DNA sequences of all spn-F and ctp inserted fragments were verified by DNA sequencing.
Cell culture, immunoblot and co-immunoprecipitation assays
Drosophila Schneider cells (S2 cells) were cultured in Shields and Sang M3 insect medium (Sigma) supplemented with 10% FCS and antibiotics at 25°C. Plasmids were transfected using Effectene (QIAGEN). To disrupt microtubule cytoskeletons, S2 cells were treated with 10 μM cholchicine. To suppress dynein function, S2 cells were treated with 10 μM ciliobrevin D. Cells were transfected with tubulin-GAL4 to drive various UAS constructs: UAS-ik2-HA, UAS-ik2-FLAG, UAS-ik2-G250D-HA, UAS-spn-F-myc, UAS-spn-F-FLAG, UAS-spnF-8A-myc, UAS-spnF-8D-myc, UAS-spnF-N-myc, UAS-spnF-C-myc, UAS-spnF-ΔCC1-myc, UAS-spnF-ΔCC2-myc, UAS-spnF-ΔCC3-myc, UAS-spnF-ΔSCD-myc, UAS-GFP-myc, and UAS-ctp-FLAG. Cells were lysed in lysis buffer (20 mM Tris pH 7.0, 150 mM NaCl, 2 mM EDTA, 1% TritonX-100, 1 mM DTT and protease inhibitors (Roche)). The cell lysates were incubated with anti-FLAG M2 agarose beads (Sigma) on ice and washed thoroughly with lysis buffer. Proteins were eluted and detected using immunoblot assays with rat antibody to HA (Roche), rabbit antibody to Myc (Santa Cruz Biotechnology), mouse antibody to FLAG (Sigma), and appropriate HRP-conjugated secondary antibodies (Jackson Immuno Research).
Immunohistochemistry and live imaging
Primary antibodies used in this study were rabbit anti-P-Ik2 antibodies (1 : 50, a gift of Dr. S. Hayashi) [29], rat anti-HA 3F10 (1 : 500, Roche), mouse anti-FLAG (1 : 500, Sigma). The secondary antibodies conjugated with DyLight dyes (Jackson Immuno Research) were used at 1 : 500 dilution. The larvae and pupae were dissected in cold PBS, fixed with 4% formaldehyde for 20 min at room temperature, and stained with proper primary antibodies and subsequent secondary antibodies. For antibody staining in S2 cells, transfected S2 cells were transferred to coverslips coated with Concanavalin A (Sigma), fixed with 4% formaldehyde, and stained with proper primary antibodies and appropriate secondary antibodies. For live imaging, pupae were placed on a double-sided tape (3M) with dorsal side up, and pupal cases were removed by forceps. The pupae were overlaid with distilled water and coverslip for imaging using laser scanning confocal microscope. Images of fluorescent signals were acquired in live animals or fixed samples on confocal microscopes of Bio-Rad MRC-600, Leica TCS SP5, or Zeiss LSM 710. The z-stack images were collected and the maximum intensity projection was used for further analysis. The images were processed using ImageJ (National Institutes of Health), and brightness and contrast were adjusted by Photoshop (Adobe).
Timelapse imaging and quantification
For timelapse imaging of Spn-F-GFP distribution in ddaC neurons from larvae to early pupae, larvae and pupae were placed with dorsal side up, overlaid with halocarbon oil and coverslip, and imaged with Zeiss LSM 510 laser scanning confocal microscope. To analyze the cytosolic fluorescence of Spn-F-GFP in neurons, we first drew a line across the center of nucleus of cells, and applied the profile analysis module of the Zen software (Zeiss) to reveal the signal intensity along the line (an example in S1B and S1E Fig). The peaks of fluorescence intensity were considered as Spn-F-GFP puncta, and the rest of cytosolic fluorescence, which excludes the peaks, was considered as the dispersed cytosolic Spn-F-GFP in cells (an example as a region marked by an asterisk in S1E Fig). The cumulative fluorescence intensities of dispersed cytosolic Spn-F-GFP (excluding the signals of any Spn-F-GFP puncta) were calculated and divided by the pixels numbers to obtain the average dispersed fluorescence intensity in the cytosol of each neuron. The average cytosolic dispersed fluorescence intensity of larval neurons was considered as one to calculate the fold changes of average cytosolic dispersed signals in the same neuron at various time points after pupation. Statistical analysis was conducted and graphs were generated in GraphPad Prism.
NanoLC-MS/MS analysis of phosphorylated Spn-F
Spn-F proteins were isolated by anti-FLAG M2 beads (Sigma) from S2 cells transfected with tubulin-GAL4 and UAS-spn-F-FLAG alone, or together with either UAS-ik2 or UAS-ik2-K41A. The bound proteins were eluted with FLAG peptide (Sigma), separated by SDS-PAGE, and stained with Coomassie brilliant blue R-250 (Sigma). The Spn-F-FLAG protein bands were manually excised from the gel and performed in-gel digestion with modified trypsin (Promega). NanoLC-nanoESI-MS/MS analysis was performed on a nanoAcquity system (Waters) connected to the Orbitrap Elite hybrid mass spectrometer (Thermo Electron) equipped with a PicoView nanospray interface (New Objective) (Core Facilities for Protein Structural Analysis at the Institute of Biological Chemistry, Academia Sinica, Taiwan). The tryptic peptide mixtures were loaded onto a 75-μm x 250-mm nanoACQUITY UPLC BEH130 column packed with C18 resin (Waters, Milford USA) and separated at a flow rate of 300 nl/min using a gradient from 5% to 80% acetonitrile in 0.1% formic acid. The effluent from the HPLC column was directly electrosprayed into the mass spectrometer. The LTQ Orbitrap Velos instrument was operated in data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. All data generated were searched against the NCBI database by the Mascot daemon software. Search criteria used were set as following: NCBI database; trypsin digestion; up to two missed cleavages allowed; variable modification set as carbamidomethyl (Cys), oxidation (Met), phosphorylation (Ser, Thr) and phosphorylation (Tyr); the mass accuracy is set as 10 ppm for precursor ion; peptide charge state set as 2+, 3+ and 4+. Phosphorylation sites and peptide sequence assignments contained in Mascot search results were validated by manual confirmation from raw MS/MS data.
Supporting Information
Zdroje
1. Kage E, Hayashi Y, Takeuchi H, Hirotsu T, Kunitomo H, et al. (2005) MBR-1, a novel helix-turn-helix transcription factor, is required for pruning excessive neurites in Caenorhabditis elegans. Curr Biol 15 : 1554–1559. 16139210
2. Luo L, O'Leary DD (2005) Axon retraction and degeneration in development and disease. Annu Rev Neurosci 28 : 127–156. 16022592
3. Hayashi Y, Hirotsu T, Iwata R, Kage-Nakadai E, Kunitomo H, et al. (2009) A trophic role for Wnt-Ror kinase signaling during developmental pruning in Caenorhabditis elegans. Nat Neurosci 12 : 981–987. doi: 10.1038/nn.2347 19561603
4. Redt-Clouet C, Trannoy S, Boulanger A, Tokmatcheva E, Savvateeva-Popova E, et al. (2012) Mushroom body neuronal remodelling is necessary for short-term but not for long-term courtship memory in Drosophila. Eur J Neurosci 35 : 1684–1691. doi: 10.1111/j.1460-9568.2012.08103.x 22571719
5. Li H, Li SH, Yu ZX, Shelbourne P, Li XJ (2001) Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J Neurosci 21 : 8473–8481. 11606636
6. Tsai J, Grutzendler J, Duff K, Gan WB (2004) Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci 7 : 1181–1183. 15475950
7. Watts RJ, Hoopfer ED, Luo L (2003) Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38 : 871–885. 12818174
8. Zhai Q, Wang J, Kim A, Liu Q, Watts R, et al. (2003) Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron 39 : 217–225. 12873380
9. Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, et al. (2006) Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron 50 : 883–895. 16772170
10. Ciechanover A, Brundin P (2003) The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40 : 427–446. 14556719
11. Lee T, Lee A, Luo L (1999) Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126 : 4065–4076. 10457015
12. Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L (2005) Developmentally programmed remodeling of the Drosophila olfactory circuit. Development 132 : 725–737. 15659487
13. Williams DW, Truman JW (2005) Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132 : 3631–3642. 16033801
14. Kuo CT, Jan LY, Jan YN (2005) Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci U S A 102 : 15230–15235. 16210248
15. Lee HH, Jan LY, Jan YN (2009) Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci U S A 106 : 6363–6368. doi: 10.1073/pnas.0902051106 19329489
16. Han C, Song Y, Xiao H, Wang D, Franc NC, et al. (2014) Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 81 : 544–560. doi: 10.1016/j.neuron.2013.11.021 24412417
17. Black MM, Baas PW (1989) The basis of polarity in neurons. Trends Neurosci 12 : 211–214. 2473556
18. Stone MC, Roegiers F, Rolls MM (2008) Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell 19 : 4122–4129. doi: 10.1091/mbc.E07-10-1079 18667536
19. Ori-McKenney KM, Jan LY, Jan YN (2012) Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron 76 : 921–930. doi: 10.1016/j.neuron.2012.10.008 23217741
20. Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, et al. (2009) A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci 12 : 1497–1505. doi: 10.1038/nn.2415 19881505
21. Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW (2006) Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 9 : 1234–1236. 16980964
22. Kuo CT, Zhu S, Younger S, Jan LY, Jan YN (2006) Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51 : 283–290. 16880123
23. Kanamori T, Kanai MI, Dairyo Y, Yasunaga K, Morikawa RK, et al. (2013) Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science 340 : 1475–1478. doi: 10.1126/science.1234879 23722427
24. Abdu U, Bar D, Schupbach T (2006) spn-F encodes a novel protein that affects oocyte patterning and bristle morphology in Drosophila. Development 133 : 1477–1484. 16540510
25. Shapiro RS, Anderson KV (2006) Drosophila Ik2, a member of the I kappa B kinase family, is required for mRNA localization during oogenesis. Development 133 : 1467–1475. 16540511
26. Dubin-Bar D, Bitan A, Bakhrat A, Kaiden-Hasson R, Etzion S, et al. (2008) The Drosophila IKK-related kinase (Ik2) and Spindle-F proteins are part of a complex that regulates cytoskeleton organization during oogenesis. BMC Cell Biol 9 : 51. doi: 10.1186/1471-2121-9-51 18796167
27. Grueber WB, Ye B, Yang CH, Younger S, Borden K, et al. (2007) Projections of Drosophila multidendritic neurons in the central nervous system: links with peripheral dendrite morphology. Development 134 : 55–64. 17164414
28. Kuranaga E, Kanuka H, Tonoki A, Takemoto K, Tomioka T, et al. (2006) Drosophila IKK-related kinase regulates nonapoptotic function of caspases via degradation of IAPs. Cell 126 : 583–596. 16887178
29. Otani T, Oshima K, Onishi S, Takeda M, Shinmyozu K, et al. (2011) IKKepsilon regulates cell elongation through recycling endosome shuttling. Dev Cell 20 : 219–232. doi: 10.1016/j.devcel.2011.02.001 21316589
30. Oshima K, Takeda M, Kuranaga E, Ueda R, Aigaki T, et al. (2006) IKK epsilon regulates F actin assembly and interacts with Drosophila IAP1 in cellular morphogenesis. Curr Biol 16 : 1531–1537. 16887350
31. Phillis R, Statton D, Caruccio P, Murphey RK (1996) Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development 122 : 2955–2963. 8898210
32. Dick T, Ray K, Salz HK, Chia W (1996) Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol Cell Biol 16 : 1966–1977. 8628263
33. Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, et al. (2012) Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 484 : 125–129. doi: 10.1038/nature10936 22425997
34. Rasmusson K, Serr M, Gepner J, Gibbons I, Hays TS (1994) A family of dynein genes in Drosophila melanogaster. Mol Biol Cell 5 : 45–55. 8186464
35. Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, et al. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471 : 473–479. doi: 10.1038/nature09715 21179090
36. Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, et al. (2008) Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol 10 : 1172–1180. doi: 10.1038/ncb1777 18758451
37. Gepner J, Li M, Ludmann S, Kortas C, Boylan K, et al. (1996) Cytoplasmic dynein function is essential in Drosophila melanogaster. Genetics 142 : 865–878. 8849893
38. Lupas A, Van Dyke M, Stock J (1991) Predicting coiled coils from protein sequences. Science 252 : 1162–1164. 2031185
39. Raff MC, Whitmore AV, Finn JT (2002) Axonal self-destruction and neurodegeneration. Science 296 : 868–871. 11988563
40. Hoopfer ED, Penton A, Watts RJ, Luo L (2008) Genomic analysis of Drosophila neuronal remodeling: a role for the RNA-binding protein Boule as a negative regulator of axon pruning. J Neurosci 28 : 6092–6103. doi: 10.1523/JNEUROSCI.0677-08.2008 18550751
41. Bitan A, Guild GM, Bar-Dubin D, Abdu U (2010) Asymmetric microtubule function is an essential requirement for polarized organization of the Drosophila bristle. Mol Cell Biol 30 : 496–507. doi: 10.1128/MCB.00861-09 19917727
42. Otani T, Oshima K, Kimpara A, Takeda M, Abdu U, Hayashi S (2015) A transport and retention mechanism for the sustained distal localization of Spn-F–IKKε during Drosophila bristle elongation. Development 142 : 2338–2351. doi: 10.1242/dev.121863 26092846
43. Amsalem S, Bakrhat A, Otani T, Hayashi S, Goldstein B, et al. (2013) Drosophila oocyte polarity and cytoskeleton organization require regulation of Ik2 activity by Spn-F and Javelin-like. Mol Cell Biol 33 : 4371–4380. doi: 10.1128/MCB.00713-13 24019068
44. Nikolaev A, McLaughlin T, O'Leary DD, Tessier-Lavigne M (2009) APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457 : 981–989. doi: 10.1038/nature07767 19225519
45. Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, et al. (2012) A caspase cascade regulating developmental axon degeneration. J Neurosci 32 : 17540–17553. doi: 10.1523/JNEUROSCI.3012-12.2012 23223278
46. Graham RK, Ehrnhoefer DE, Hayden MR (2011) Caspase-6 and neurodegeneration. Trends Neurosci 34 : 646–656. doi: 10.1016/j.tins.2011.09.001 22018804
47. Grueber WB, Ye B, Moore AW, Jan LY, Jan YN (2003) Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13 : 618–626. 12699617
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



