-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
Cornelia de Lange Syndrome (CdLS) alters many aspects of growth and development. CdLS is caused by mutations in genes encoding proteins that ensure that chromosomes are distributed equally when a cell divides. These include genes that encode components of the cohesin complex, and Nipped-B-Like (NIPBL) that puts cohesin onto chromosomes. Individuals with CdLS have only modest reductions in the activities of these genes and do not show changes in chromosome distribution. Instead, they show differences in the expression many genes that control development. Animal models of CdLS will be useful for studies aimed at understanding how development is altered, and testing methods for treating CdLS. We find that Drosophila with one mutant copy of the Nipped-B gene, which is equivalent to the NIPBL gene, show characteristics similar to individuals with CdLS. These include reduced growth, learning, memory, and altered circadian rhythms. These studies thus indicate that Drosophila Nipped-B mutants are a valuable system for investigating the causes of the CdLS birth defects, and developing potential treatments. They also reveal that the slow growth in Drosophila Nipped-B mutants is not caused by disruption of systemic hormonal growth controls, and that the learning and memory deficits may reflect changes in brain structure.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005655
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005655Summary
Cornelia de Lange Syndrome (CdLS) alters many aspects of growth and development. CdLS is caused by mutations in genes encoding proteins that ensure that chromosomes are distributed equally when a cell divides. These include genes that encode components of the cohesin complex, and Nipped-B-Like (NIPBL) that puts cohesin onto chromosomes. Individuals with CdLS have only modest reductions in the activities of these genes and do not show changes in chromosome distribution. Instead, they show differences in the expression many genes that control development. Animal models of CdLS will be useful for studies aimed at understanding how development is altered, and testing methods for treating CdLS. We find that Drosophila with one mutant copy of the Nipped-B gene, which is equivalent to the NIPBL gene, show characteristics similar to individuals with CdLS. These include reduced growth, learning, memory, and altered circadian rhythms. These studies thus indicate that Drosophila Nipped-B mutants are a valuable system for investigating the causes of the CdLS birth defects, and developing potential treatments. They also reveal that the slow growth in Drosophila Nipped-B mutants is not caused by disruption of systemic hormonal growth controls, and that the learning and memory deficits may reflect changes in brain structure.
Introduction
The ring-like cohesin complex consisting of the Smc1, Smc3, Stromalin (SA) and Rad21 proteins is required for sister chromatid cohesion and accurate chromosome segregation. Cohesin is loaded on to chromosomes by the kollerin complex, consisting of the NIPBL adherin (Nipped-B in Drosophila) and Mau-2 proteins, and is removed from chromosomes by the releasin complex containing the Wapl and Pds5 proteins [1–4]. Adherin, cohesin and releasin also function in gene transcription in a dosage-sensitive manner. Reductions in their activities alter the expression of hundreds of genes that control growth and development in metazoan organisms. These changes in gene expression occur in the absence of detectable faults in sister chromatid cohesion or chromosome segregation, and are accompanied by defects in development in Drosophila, zebrafish, mice and humans [5–20].
Adherin and cohesin regulate transcription via multiple mechanisms. They bind primarily to active genes and transcriptional enhancers, with high levels near the transcription start sites of the active genes [15,21–24]. Substantial genetic and molecular evidence supports the idea that cohesin facilitates enhancer-promoter communication. In Drosophila, the genes that bind cohesin have high levels of transcriptional pausing, in which RNA polymerase II (Pol II) stops after transcribing some 30 to 40 nucleotides, but remains transcriptionally engaged [24,25]. Adherin and cohesin can both facilitate and inhibit transition of the paused Pol II into elongation, depending on the specific gene and cell type [24,25]. Although the association of cohesin with transcriptional pausing has not been systematically explored in mammalian cells, many NIPBL - and cohesin-binding genes, such as c-myc, display high transcriptional pausing. In addition to direct control of active gene transcription, evidence from Drosophila shows that cohesin indirectly controls epigenetic silencing of homeotic genes by Polycomb complexes via sequestration of the PRC1 Polycomb complex at active genes and limiting the amount available for silencing [4]. It is currently unknown if cohesin plays a similar role in mammalian Polycomb silencing.
In humans, dominant loss-of-function mutations in NIPBL and missense or in-frame deletion mutations in the SMC1A and SMC3 cohesin subunit genes cause a striking constellation of structural and intellectual birth defects called Cornelia de Lange syndrome (CdLS, reviewed in [26,27]). Mutations in the RAD21 cohesin subunit gene, and in the HDAC8 gene encoding a deacetylase that recycles SMC3 cause a very similar set of developmental deficiencies [20,28]. These closely-related birth defect syndromes are known collectively as cohesinopathies. Individuals with classical CdLS exhibit clinical phenotypes affecting multiple organs and systems, such as characteristic craniofacial dysmorphia, pre - and postnatal growth and developmental delay, microcephaly, intellectual disability, musculoskeletal defects and diverse organ system involvement. However, mildly affected individuals may present with only mild cognitive and growth retardation and lack overt structural abnormalities [27,29].
A clinical diagnosis of CdLS is usually made by recognition of the characteristic facial features, which include arched eyebrows, long philtrum, thin lips, and low set back ears [30]. While these craniofacial features are present in 95% of classic CdLS, they can be quite subtle in milder cases. Importantly, growth reduction is the most common finding in CdLS, and may be the main clinical presentations in mild cases [11,20]. Intellectual disability and learning differences are also universally seen in individuals with CdLS, with the average IQ score being 53, (range of 30 to 100) [27]. NIPBL heterozygous mutations are present in nearly 60% of CdLS individuals and mutations in cohesin subunits and HDAC8 account for an additional 10–15% of cases overall [27,31]. Individuals with NIPBL mutations tend to be more severely affected, with slow growth, upper limb defects and organ defects and severe intellectual disabilities, while those with cohesin subunit mutations tend to have less severe growth and structural birth defects. Genotype-phenotype correlations among a large number of NIPBL probands indicate that NIPBL haploinsufficiency, resulting from nonsense mutations, splice site mutations, or frame-shift deletions/insertions, generally leads to more severe cognitive and structural phenotypes than missense mutations [32].
Understanding the origins of the developmental deficits in the cohesinopathies is complicated by their diversity, and the findings that many genes encoding components of all major signaling pathways, and transcription factors that control growth and development, such as c-Myc and HOX proteins, are bound and transcriptionally-influenced by adherin and cohesin. Much of the current understanding of how adherin and cohesin modulate transcription comes from Drosophila, due to its less complex genome and availability of substantial genetic tools. The evidence that many of the same genes and signaling pathways are regulated by adherin and cohesin in Drosophila and vertebrates raises the possibility that Drosophila could also provide fundamental insights into the developmental origins of many of the birth defects observed in human cohesinopathies. Heterozygous null mutant Nipped-B Drosophila do not appear to be as strongly affected as humans, and require weak or cryptic mutations in other genes to display overt structural alterations [5,17,33–35]. Due to the almost universal presence of cognitive, learning and growth deficits in humans with CdLS we decided to more closely examine Nipped-B mutant Drosophila for these specific phenotypes. These studies reveal that Nipped-B mutant Drosophila also display substantial growth and neurological phenotypes. Strikingly, the data indicate that there are no appreciable changes in systemically controlled developmental timing or growth regulation. The Nipped-B mutant growth and intellectual phenotypes largely correlate with changes in cell number, size and morphology.
Results
Heterozygous Nipped-B mutations reduce growth to a similar extent as mutations in the dm (myc) and Tor growth regulators
Individuals with CdLS are usually small for their age, but the reasons for their reduced size are largely unknown, and could potentially reflect altered systemic growth control, delayed developmental timing, reduced cell division, reduced utilization of nutrition, increased cell death, or any combination of these factors. We find that heterozygous Nipped-B loss-of-function mutant (Nipped-B407 / +) adult male and female Drosophila weigh 5 to 8% less than matched wild-type flies, which represent significant decreases, similar in magnitude to those caused by heterozygous null mutations in the diminutive (dm, myc) and Tor (Target of rapamycin) growth regulators (Table 1). For all experiments, the Nipped-B407 and P57B wild-type parental control chromosomes were compared after outcrossing to a wild-type Oregon R background. We chose outcrossing to a wild-type strain to minimize potential genetic background effects on growth. Outcrossing ensures that the background is heteroallelic for wild-type alleles across the genome, in contrast to inbred isogenic backgrounds, which tend to harbor recessive genetic modifiers of growth, viability and fertility.
Tab. 1. Effects of heterozygous loss-of-function Nipped-B, dm, and Tor mutations on adult body weight. 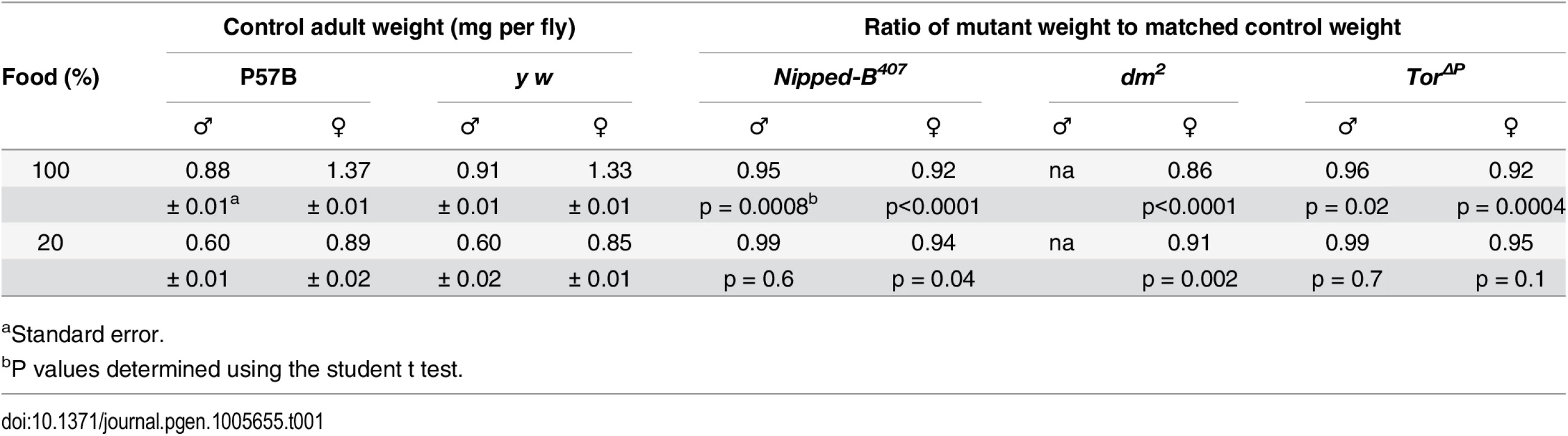
aStandard error. The myc gene (Drosophila diminutive, dm) encodes a transcription factor that promotes protein synthesis, cell growth and division. It is positively regulated by cohesin in Drosophila, zebrafish, mouse and human [14,15,17,37]. Female heterozygous mutant for a strong loss-of-function dm allele (dm2) showed a weight reduction of 14%, greater than that observed in Nipped-B mutants (Table 1). We could not measure the effects in males because dm is X-linked, and the dm2 allele is lethal. dm2 is a point mutation that causes a nonsense mutation and behaves similar to null alleles [38].
The Target of rapamycin (Tor) kinase integrates insulin and growth factor signaling to promote protein synthesis, growth and cell proliferation, and influences the expression of many of the same genes as dm [39,40]. Heterozygous Tor null mutant (TorΔP) male and female adults showed 4 to 8% reduced weight, similar to Nipped-B mutant flies (Table 1). TorΔP is a 3.5 kb deletion created by a P element transposon excision that removes the translation start site [41]. Because we do not have the parental chromosomes for the dm and Tor mutations, and both were generated in a y w background [38,41] we used a y w stock as the wild-type control for both dm2 and TorΔP. As with the Nipped-B407 and matched P57B control, the y w control and the dm and Tor mutants were compared after outcrossing into a wild-type Oregon R background to reduce the likelihood of recessive growth modifiers.
We carefully chose the Nipped-B407 allele as optimal for close examination of growth phenotypes because in contrast to all other available mutant alleles, there is substantial evidence that it is functionally null and lacks other linked mutations. Nipped-B is a large gene (~40 kb) located in centromere-proximal heterochromatin on chromosome 2R in a region refractory to meiotic recombination (S1 Fig). The introns and flanking sequences consist primarily of natural transposons, making it challenging to molecularly characterize Nipped-B mutations, and to ensure that there are no other linked mutations. When it was isolated, Nipped-B407 was backcrossed multiple times to the parental P57B wild-type chromosome and extensively tested for lack of allelism to all known essential genes in the surrounding heterochromatin [5]. Genomic sequencing and comparison to RNA-seq data indicates that Nipped-B407 is a regulatory mutation that produces little or no RNA (S1 and S2 Figs) in contrast to the sequenced chemically-induced alleles that produce missense or truncated proteins [33]. As described in detail below, Nipped-B407 dominantly reduces wing growth to a similar extent as two other mutant Nipped-B alleles, Nipped-B292.1 and T(2;3)Nipped-B359.1. Compared to other mutant alleles, Nipped-B407 usually has strong dominant genetic interactions with mutations in other genes [5,33]. Crucially, Nipped-B407 is the only Nipped-B allele shown to be the only recessive lethal mutation on its chromosome by rescuing survival beyond the normal 2nd instar lethal phase to adults or pharate adults with cDNA transgenes [36,42]. It is likely that cDNA transgenes do not fully rescue viability because of the need to use a foreign promoter, and the inability to produce the several N terminal splice variants [33] (S2 Fig).
Drosophila grown under limited nutrition (20% of the normal food concentration) showed 30 to 40% reductions in weight, which is substantially greater than the weight loss caused by the heterozygous Nipped-B, dm, or Tor mutations (Table 1). We reasoned that if Nipped-B, dm or Tor mutations decrease the ability of the flies to utilize the available nutrients to support growth, then these mutants might show a greater sensitivity to reduced nutrition than wild-type. Opposite to this expectation, Nipped-B and Tor mutant males showed the same sensitivity as wild-type to reduced nutrition, and weighed essentially the same as wild-type flies grown under the limiting conditions (Table 1). Similarly, Nipped-B, dm, and Tor mutant females showed a less significant reduction in weight relative to wild-type under limiting nutrition. In other words, restricting nutrition partially suppressed and did not enhance the relative growth deficits in Nipped-B, dm, and Tor mutants. Thus the primary growth deficit in Nipped-B mutants does not stem from decreased utilization of available food.
Systemic growth control and developmental timing are not detectably altered in heterozygous Nipped-B mutants
We tested if the reduced growth of Nipped-B, dm and Tor mutant flies correlates with altered developmental timing, or altered systemic growth controls that normally delay hormonally-driven developmental transitions until the organism reaches a critical size [43]. We collected newly-hatched larvae and measured the times from hatching to pupariation and to eclosion of adults when grown on normal and 20% food. With normal systemic growth controls, reducing nutrition delays the developmental transitions.
All genotypes showed a nearly 2-fold increase in the number of days to reach pupariation and eclosion and small insignificant decreases in viability when grown on 20% food (Fig 1). Under all conditions, heterozygous Nipped-B407 mutant flies reached these stages at the same time as the matched wild-type parental control (P57B), with a similar number of surviving pupae and adults. In some experiments, female Nipped-B407 mutants eclosed a few hours earlier than the P57B controls. Thus Nipped-B mutant flies do not show an altered rate of development or reduced viability, and the systemic growth controls are not detectably altered relative to wild-type when all tissues have the heterozygous Nipped-B407 mutation.
Fig. 1. Effects of heterozygous Nipped-B, dm, and Tor mutations on timing of pupariation and eclosion. 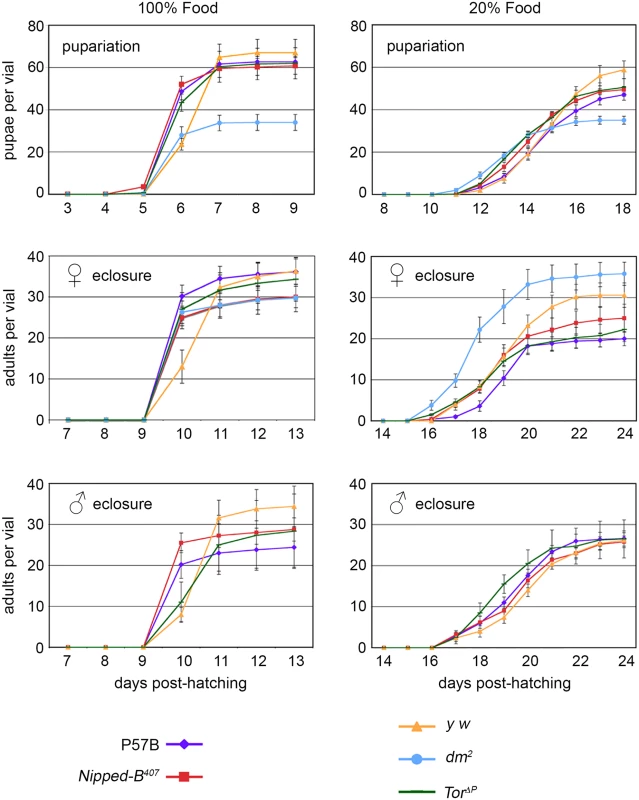
Newly hatched 1st instar larvae of the indicated genotypes were placed in vials at 100 per vial, containing either normal (100%) or diluted food (20%). The number of pupae and eclosing male and female adults were counted every day for up to 24 days. P57B (P57B/+) is the matched wild-type parental control for Nipped-B407 (Nipped-B407/+). y w (y w/+ females and y w/Y males) were the controls for the dm2 (dm2/+ females) and TorΔP (TorΔP/+) mutants. The data shown is from a single experiment and two additional independent repeats with Nipped-B407 and P57B gave very similar results. Error bars are standard errors. As expected, dm mutants gave half the number of pupae as the matched y w control flies because hemizygous males do not survive. dm mutant heterozygous females, however, eclosed earlier than the y w control females on normal food, and almost two days earlier when grown under limited nutrition, demonstrating that heterozygosity for a null dm allele alters systemic growth control, permitting premature developmental transitions under restricted nutrition (Fig 1). Tor mutant females also eclosed slightly earlier than the y w controls under normal nutrition, but at the same time as controls under limited nutrition (Fig 1). Thus, Nipped-B, dm, and Tor mutants all behave differently with regards to developmental timing, depending on nutritional conditions. Critically, Nipped-B mutants did not show significant changes in the timing of transitions between developmental stages under either normal and limited nutrition, indicating that the reduced growth of Nipped-B mutants is not caused by inefficient utilization of nutrition, or changes in systemic controls that produce hormonally-induced transitions when a critical size has been reached.
Nipped-B mutant wings have fewer and smaller cells
The three Nipped-B mutations tested (Nipped-B407, Nipped-B292.1, T(2;3)Nipped-B359.1) dominantly reduce the size of adult wings to an extent similar to the reduction of body weight in Nipped-B407 mutants (Table 2; S3 Fig). Examination of adult female wings indicates that Nipped-B407 mutants are smaller because they have fewer and smaller cells. We measured the total wing area, and counted the number of cells per unit area using the hairs produced by each cell (Fig 2A and 2B; Table 2). From these data we calculated the total number of cells per wing, and the relative cell size (Table 2). Close analysis of the Nipped-B407 mutant wings shows that the reduced size reflects a 4% decrease in cell number combined with a 3% decrease in cell size (Table 2). In Table 2, the increase in cells per mm2 from 5971 to 6163 measures the decrease in cell size. dm heterozygous mutant wings showed an approximately 9% reduction in size that reflects both a 4% decrease in cell number, and a 6% decrease in cell size (Fig 2B; Table 2). In contrast, Tor heterozygous mutant wings show only 3% decrease in wing area, with a decrease in cell size, but not cell number (Fig 2B; Table 2).
Tab. 2. Effects of heterozygous loss-of-function Nipped-B, dm, and Tor mutations on adult female wing size, cell size and cell number. 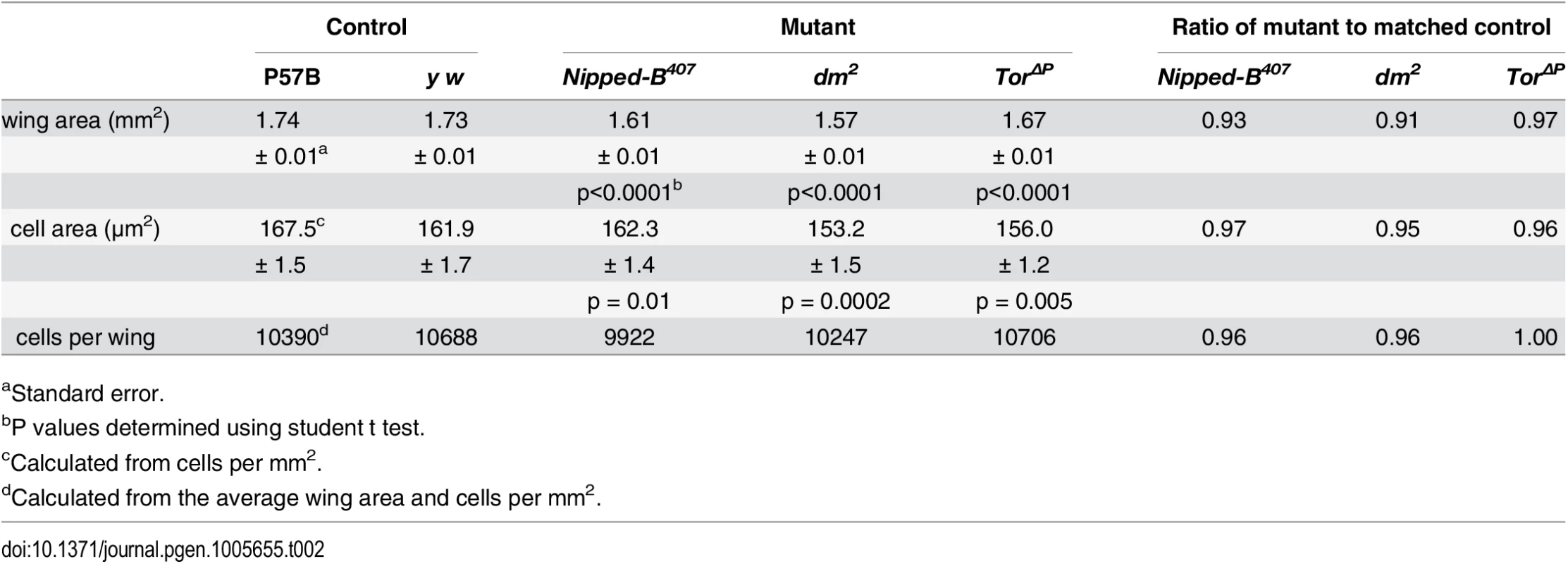
aStandard error. Fig. 2. Nipped-B, dm, and Tor mutations reduce adult wing size. 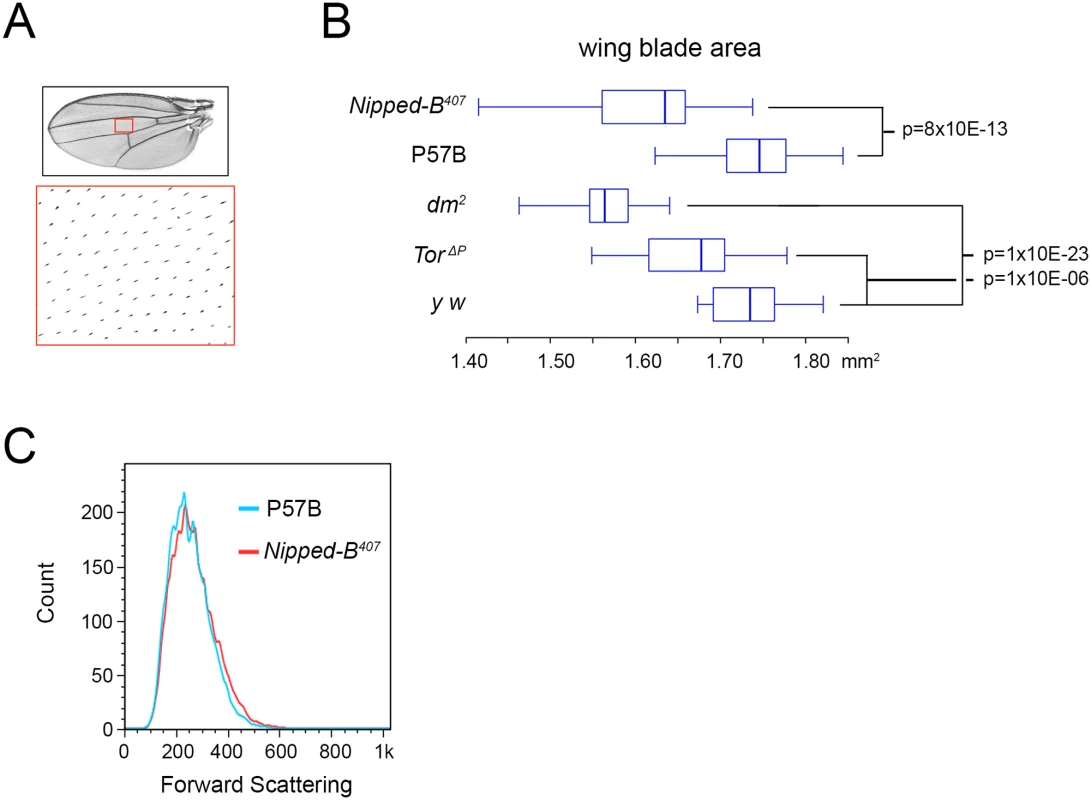
(A) Full adult female wing areas were measured, and areas of wings (red boxes) were sampled for the number of cells by counting the hairs produced by each cell at higher magnification (large red box) to determine cell size and number (Table 2). (B) Box plots showing the distributions of wing blade areas of the indicated genotypes: Nipped-B407 (Nipped-B407/+), P57B (P57B/+), dm2 (dm2/+), TorΔP (TorΔP/+), y w (y w/+). The p values of various pairwise comparisons determined by the student t test are indicated. (C) Late 3rd instar wing discs were dissected from larvae with the indicated genotypes, dissociated with trypsin and collagenase, and sorted by forward scattering in cell sorter to measure the distributions of cell sizes. We examined cell size in late 3rd instar wing imaginal discs that give rise to the adult wings by forward scattering in a cell sorter after tissue dissociation, but did not detect a significant decrease in cell size at this earlier stage, when cells are still dividing (Fig 2C). It could be that the size difference is too small to detect by forward scattering, or that a significant difference in cell size occurs only after cells have stopped dividing.
Ectopic dm expression restores wing size and increases the weight of Nipped-B mutants
Additional dm (c-myc ortholog) expression partially rescues the growth deficits of Nipped-B heterozygous null mutants (Fig 3A; Table 3). The Dm transcription factor increases protein synthesis, cell growth and cell proliferation [44]. Based on the knowledge that cohesin regulates dm expression and that Nipped-B mutant flies are small because of reduced cell number and size, we hypothesized that adding additional dm expression could rescue the growth of Nipped-B mutants. We used a tubulin-dm transgene (tub-myc, [45]) and observed an increase in the size of both Nipped-B mutant and parental wild-type control wings to a size greater than the normal wild-type (Fig 3A; Table 3). These results are consistent with the idea that the reduced growth of Nipped-B mutant flies may stem in part from reduced dm expression.
Tab. 3. Effects of additional myc (dm) expression on female wings and body weight. 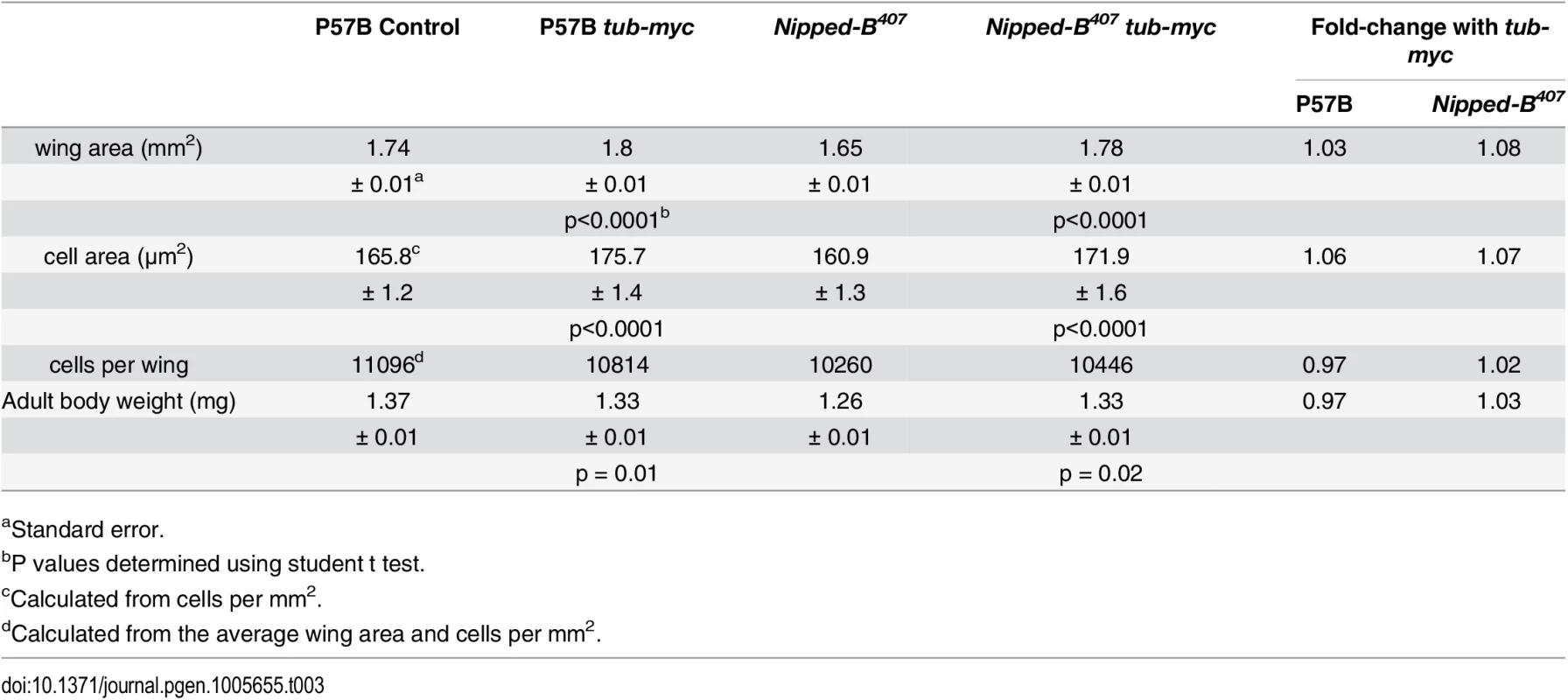
aStandard error. Fig. 3. Additional myc (dm) expression rescues adult wing size of Nipped-B407 heterozygous mutants and increases apoptosis in wild-type 3rd instar wing imaginal discs. 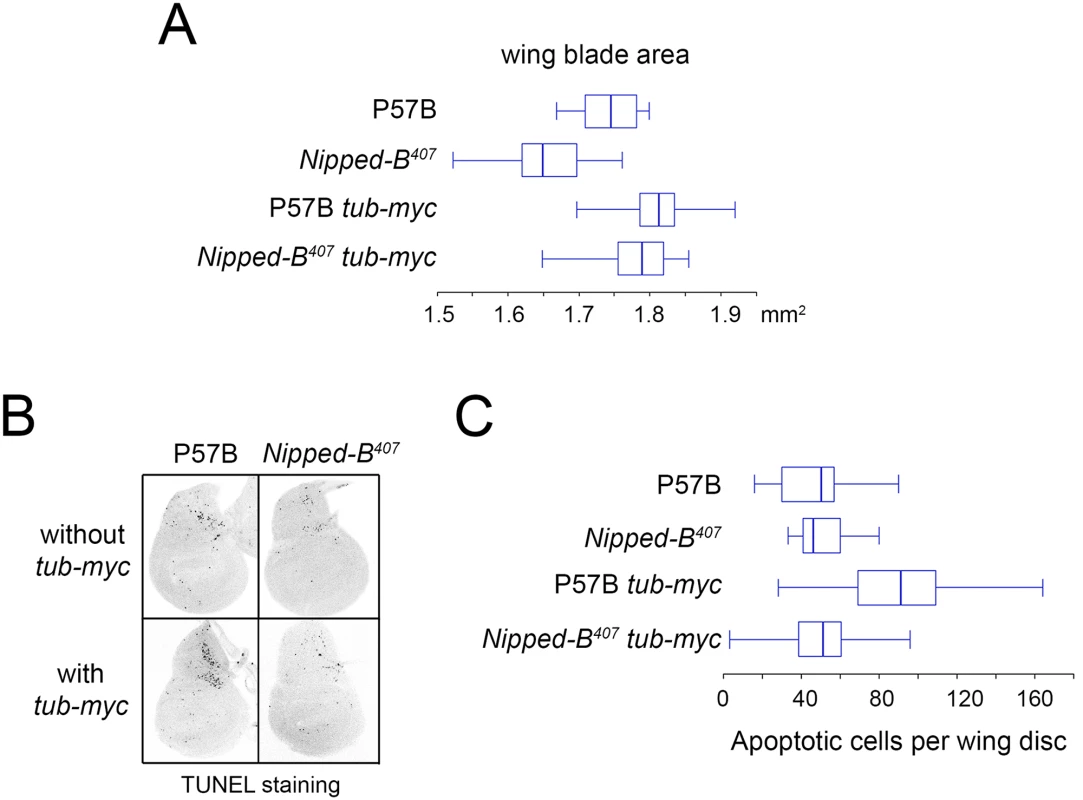
(A) The box plots indicate the distribution of the wing blade areas of the indicated genotypes, where tub-myc indicates a single copy of the tub-myc transgene that expresses Drosophila dm from a minimal tubulin gene promoter. The increases in wing area induced by tub-myc are significant for both the P57B / + and Nipped-B407 / + genotypes (p<0.0001, t test). Cell numbers and sizes were also determined for each genotype and are given in Table 3. (B) Examples of TUNEL staining of late 3rd instar wing imaginal discs of the indicated genotypes to visualize apoptotic cells. (C) Boxplot distributions of the number of apoptotic cells in 3rd instar wing discs. The number of apoptotic cells in P57B / + discs with the tub-myc transgene is significantly higher than the other genotypes (p<0.0001, student t test). Close examination of the wings showed that the increase in Nipped-B mutant wings caused by ectopic dm expression stems from both an increase in cell number and size, with an increase in the number of total cells and a decrease in the number of cells per mm2 (Table 3). In contrast, the wild-type control wings actually had fewer cells in the presence of the tub-myc transgene, and the increase in wing size thus results only from an increase in cell size (Table 3). Both mammalian c-myc and Drosophila dm promote apoptosis [46,47], and we thus tested if the tub-myc transgene reduces cell number in wild-type wings by increasing apoptosis. We quantified apoptotic cells in 3rd instar wing imaginal discs by TUNEL staining and found that tub-myc increased the number of apoptotic cells some 1.7-fold in wild-type wing discs, but had no measurable effect in Nipped-B mutant discs (Fig 3B and 3C). In other words, the Nipped-B mutation suppressed apoptotic induction by excess dm expression. Importantly, these data indicate that Nipped-B mutants do not have increased apoptosis, even in the presence of excess dm expression, providing further evidence that they have fewer cells because of reduced cell proliferation, not increased cell death.
Heterozygous Nipped-B mutant wing discs show broad modest decreases in expression of genes supporting development and growth
Beyond the decreased cell number and size, heterozygous Nipped-B mutant wing discs do not show obvious morphological defects in the absence of mutations in other genes. However, studies in cultured cells using acute RNAi depletion of Nipped-B or cohesin levels down to 10 to 20% of normal levels show that Nipped-B and cohesin directly regulate the transcription of many genes that control growth and development, such as dm, transcription factors and signaling proteins [24,35]. We thus performed RNA-seq analysis of Nipped-B407 / Oregon R mutant and P57B / Oregon R 3rd instar wing discs to see if we could identify changes in gene expression responsible for the reduced growth, and the suppression of dm-induced apoptosis. This revealed modest decreases in the expression of a large number of genes, including many that participate in the control of growth and development, but do not implicate a small set of candidate genes as the central cause of the growth deficit (Fig 4; S1 Table).
Fig. 4. Nipped-B mutant wing discs show broadly reduced gene expression by RNA-seq analysis. 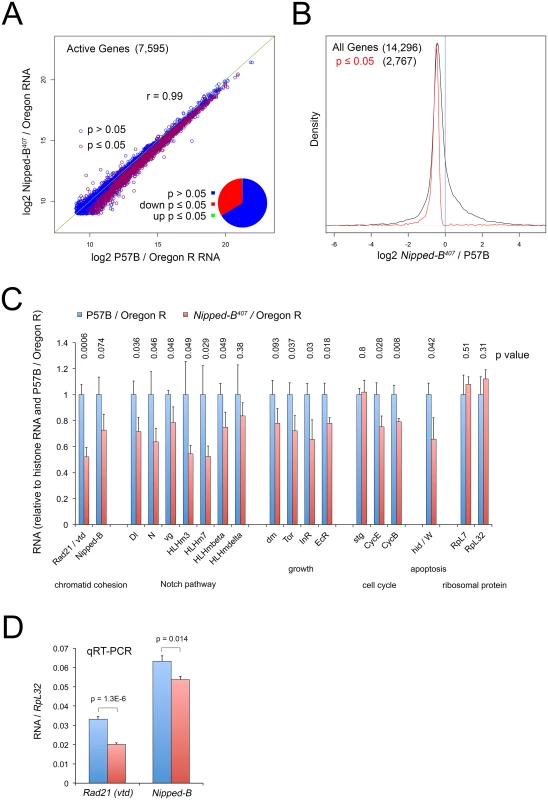
(A) Plot of expression level of all active genes in matched wild-type controls (P57B / Oregon R) versus Nipped-B407 / Oregon R 3rd instar wing discs. RNA-seq was performed using ribosomal RNA depletion and normalization to histone RNA to compensate for the broad effects of Nipped-B on gene transcription [24]. Active genes (7,595) were defined as those averaging more than 500 nucleotides of sequence coverage between replicates. Pearson correlation coefficients between replicate samples for all genes quantified (14,296) ranged from 0.96 to 1.00, indicating high reproducibility (S1 Table). The green line indicates a slope of 1, revealing that the Nipped-B mutants show diminished expression of many genes. Red circles indicate genes in which the change in RNA levels have a p value ≤ 0.05, which represent roughly a third of active genes as shown by the pie chart. (B) Density distribution plots of the fold-changes in gene expression between Nipped-B mutants and the wild-type control (log2 Nipped-B407 / P57B ratio) for all genes (black) and those genes for which the change has a p value ≤ 0.05. The number of genes in each group is indicated in parentheses. This shows that relative to histone RNA levels, most genes show lower RNA levels in the Nipped-B mutant wing discs, and that most statistically significant changes are decreases in expression. (C) Examples of expression changes for selected genes. The blue bars are set to 1 in the P57B / Oregon R control after normalization to histone RNA to allow all genes to be shown on the same scale. The error bars are the standard errors calculated from the four replicates, and p values are given above each gene. The Rad21 (vtd) cohesin subunit gene and Nipped-B both show reductions. Several genes in the Notch signaling pathway show reductions, including the Delta (Dl) ligand, Notch (N), and direct Notch targets, including vestigial (vg) and genes in the Enhancer of split complex (HLHm3, HLHm7, HLHmbeta, HLHmdelta). Several genes that regulate growth also show reductions including dm (myc), Tor, the insulin receptor (InR) and the ecdysone hormone receptor (EcR). The string (stg) cdc25 cell cycle control gene does not show a reduction, in contrast to the cyclin E (CycE) gene that drives S phase and the cyclin B (CycB) G2 cyclin gene. The Wrinkled (W, hid, head involution defective) pro-apoptosis gene shows a significant decrease. The RpL32 and RpL7 ribosomal protein genes, which are often used as normalization controls for qRT-PCR experiments and Northern blots, do not show significant changes in expression. The expression values for all genes in the RNA-seq analysis are in S1 Table, along with the ontology analysis indicating that pathways involved in development, morphogenesis and growth are broadly affected, although the only significant phenotype in adult Nipped-B mutant wings is the reduced cell number and size. (D) qRT-PCR validation of the effects on Rad21 (vtd) and Nipped-B RNA levels in 3rd instar wing discs, normalizing to RpL32 RNA levels. Ten P57B / Oregon R and eight Nipped-B407 / Oregon R replicates were assayed. The error bars are standard error calculated from all PCR replicates, and the p values for the comparisons to the controls are indicated. We used an alternative method for normalizing the RNA-seq data because prior studies using Nipped-B and cohesin depletion in cultured cells showed that standard normalization methods for genome-wide RNA analysis, which assume that most genes do not change in expression, obscure many small changes in gene expression. This is because Nipped-B and cohesin depletion alters the transcription of a majority of active genes via multiple direct and indirect mechanisms [24,35]. For instance, direct measurement of transcription by precision run-on sequencing (PRO-seq) showed that many genes with modestly reduced transcription showed increased RNA levels by standard normalization methods because their RNA decreases were smaller than the average decrease. Similar normalization artifacts have been encountered in genome-wide studies of the effects of the mammalian Myc protein on transcription because it amplifies transcription of most active genes [48]. We thus normalized to total histone RNA levels, reasoning that the tight regulation of histone mRNAs [49] will more accurately reflect DNA content and gene copy number. Histone mRNAs are not polyadenylated, and we therefore used ribosomal depletion instead of poly(A) selection to prepare the RNA-seq libraries.
With this approach, we find that roughly a third of active genes in Nipped-B mutant wing discs show modest but significant (p ≤ 0.05) decreases in RNA levels, with less than two dozen showing significant increases (Fig 4A and 4B; S1 Table). Active genes were defined as those that have at least 500 nucleotides of sequence coverage in the average of the replicate samples, which represents roughly half of all genes quantified, and captures genes that are expressed in only a few cells in the wing disc, such as cut and the Enhancer of split complex. The correlation in active gene expression levels between the averages of the P57B control and Nipped-B mutant samples is high (r = 0.99), and the correlation in expression for all genes between individual replicates ranged from 0.96 to 1.00, indicating the data is highly reproducible (Fig 4A; S1 Table).
Nipped-B RNA showed the expected decrease of 27%, which is only slightly greater than the median decrease in gene expression, and was not statistically significant (p = 0.074; Fig 4B and 4C; S1 Table). We confirmed that the decrease is significant using qRT-PCR of total RNA isolates with ten P57B and eight Nipped-B mutant replicates and RpL32 as the normalization standard (p = 0.014; Fig 4D). We used RpL32 as the reference because it did not show a significant change in the RNA-seq data (Fig 4C; S1 Table), and was also used as the standard in Northern blots showing a 25 to 30% decrease in Nipped-B mRNA in heterozygous Nipped-B407 mutant larvae [6]. Combined with the sequence analysis showing that the bulk of Nipped-B RNA in Nipped-B407 / Oregon R wing discs arises from the wild-type allele (S1 and S2 Figs) and the high resistance of Nipped-B RNA levels to in vivo RNAi [6], these results provide additional evidence for a homeostatic feedback mechanism that maintains Nipped-B mRNA levels.
Intriguingly, vtd (Rad21) cohesin subunit RNA levels were significantly reduced by some 50%, which is a larger decrease than the 27% reduction in Nipped-B (Fig 4C), which was also confirmed by qRT-PCR (Fig 4D). The reduced expression of both Nipped-B and this cohesin subunit may explain why heterozygous Nipped-B mutations generally have stronger phenotypic effects in Drosophila and humans than heterozygous cohesin subunit mutations.
Gene ontology analysis revealed enriched down-regulation of genes involved in development, morphogenesis and growth (S1 Table). These include known Nipped-B and cohesin targets in the Notch signaling pathway, including Notch (N) and genes in the Enhancer of split complex (HLHm3, HLHm7, HLHmbeta, HLHmdelta) (Fig 4, S1 Table). Although dm transcription in Nipped-B depleted cultured cells is substantially reduced as measured by PRO-seq [24], dm RNA is only modestly reduced by 22% in the heterozygous mutant wing discs, which did not achieve statistical significance (p = 0.09) (Fig 4C; S1 Table). Other growth regulators such as Tor, the insulin receptor (InR) and the ecdysone hormone receptor showed similar decreases with significant p values (Fig 4C, S1 Table). Consistent with the resistance of Nipped-B mutant wing discs to dm-induced apoptosis, the Wrinkled (W, hid) pro-apoptotic gene is also significantly reduced in expression (Fig 4C, S1 Table). The CycE and CycB genes controlling the cell cycle are also reduced, although the string (Cdc25) gene is not (Fig 4C; S1 Table).
Given the large number of modest reductions in genes involved in growth and development, the evidence for homeostatic mechanisms, and the inability to dissect direct from indirect effects, the RNA-seq data do not definitively identify a specific gene or set of genes responsible for the growth deficit of heterozygous Nipped-B mutant wing discs, and it is likely that many direct and indirect expression changes contribute to the reduced growth. We suspect that dm overexpression can partially rescue the growth deficit not only because it compensates for a modest direct decrease in dm transcription, but also because it broadly amplifies expression of many genes [48] and thus can counteract the broad down-regulation in gene expression caused by reduced Nipped-B.
Heterozygous Nipped-B mutants show learning and memory deficits
Although heterozygous Nipped-B null mutants show no overt external structural abnormalities, we investigated whether or not they have deficits in neurological and behavioral functioning. Overall, intellectual disability is present in 95% of individuals with CdLS, and is often present even in the absence of significant structural abnormalities. Homozygous deficiencies of SMC1, Stromalin, and Rad21 cause aberrant axon pruning of the Drosophila mushroom body γ-neurons, raising the possibility that there may be modest nervous system deficits in heterozygous mutants [50,51]. Mushroom bodies (MBs) are the primary brain structures involved in associative learning and memory, functions that can be tested through conditioning courtship paradigms and odor-shock classical conditioning paradigms [52–54].
We investigated the learning and memory capability of Nipped-B heterozygous males through courtship conditioning assays. Courtship is an innate behavior that involves a series of complex sensory and behavioral interactions between male and female Drosophila. Courtship rituals performed by the male fly is characterized by a sequence of stereotypic behaviors including: 1) orienting toward a female, 2) tapping the females abdomen with the males’ forelegs, 3) extending and vibrating one wing, 4) licking the female’s genitalia and 5) attempting to copulate [55,56]. Overall the courtship robustness of male flies is measured by the courtship index (CI), namely the proportion of time a male engages in active courtship behaviors during a given amount of observation time. Virgin females are receptive to copulation. Recently mated females, however, are non-receptive and reject advances to mate. The courtship activity of the male is modified by their prior sexual experience. Unsuccessful courtship attempts with a non-receptive female lead to progressive decrement of a male tester’s courtship behaviors. Such courtship suppression is retained even after the male is removed from the non-receptive female and later paired with a receptive virgin female [57]. Courtship plasticity observed in males is thus employed in conditioning courtship paradigms to measure learning and memory. During a training session, a naïve male is paired with pre-mated female “trainer” in a mating chamber and observed for 60 minutes. The amount of time the male engages in courting the trainer female is recorded for the first 10 minutes and the last 10 minutes and used to calculate the courtship indexes (CI), CIinitial (CI of the first 10 minutes) and CIfinal (CI of the last 10 minutes), respectively. The discrepancy between CIinitial and CIfinal thus reflects the extent of courtship reduction and a tester male’s capacity to learn. After the training session, the male tester is transferred into a new chamber and given an hour to rest. A receptive virgin female is subsequently introduced into the chamber. The courtship attempts of the tester male towards the virgin female are recorded for 10 minutes, which is used to deduce the courtship index CItest. A naïve fly of the same genotype is sham-trained in a courtship chamber for an hour in isolation and then tested for courtship activity towards a virgin female; the courtship of the naive fly is referred as CIsham. The difference between CItest and CIsham reflects the retention of courtship suppression resulting from the earlier unrewarding courtship experience and is used as a measure of short-term memory [58,59].
Two Nipped-B mutant alleles were used for the conditional courtship assays, Nipped-B407 and Nipped-B292.1. Like Nipped-B407, Nipped-B292.1 was one of the founder alleles defining the Nipped-B complementation group, and thus has also been extensively tested for a lack of mutations in the surrounding essential genes [5]. In the initial genetic characterization, Nipped-B407 and Nipped-B292.1 showed similar genetic interactions with other mutations on cut, Ultrabithorax, and mastermind mutations [5]. Nipped-B292.1 also has a similar effect on adult female wing size as Nipped-B407 (S3 Fig). To minimize potential influence of background variations on behavioral outputs, both Nipped-B mutants were backcrossed to an isogenized w1118 strain, wiso31, which are isogenic for chromosomes X, 2 and 3, and were rigorously tested to ensure that they have wild-type memory and circadian rhythm [60]. wiso31 flies were used as controls in all Drosophila behavioral analyses in this study and heterozygous Nipped-B mutants were generated by crossing the isogenized Nipped-B407 or Nipped-B292.1 chromosomes into a wiso31 background.
We observed that similar to wiso31 controls, heterozygous Nipped-B mutants of either allele demonstrated a substantial reduction in their courtship activities toward non-receptive female trainers, when their courtship behaviors during the first 10 minutes were compared to those seen in the final 10 minutes of a training session (Fig 5A). However, the reduction in courtship attempts seen in Nipped-B males was not as remarkable as that of wiso31 controls. We compared the amount of courtship reduction in all three groups and noticed that heterozygous mutants of either Nipped-B allele show less decrease than wiso31 controls. These results indicate that although heterozygous Nipped-B males (Nipped-B407 / + and Nipped-B292.1 / +) are capable of learning, their capacity is not as high as that of controls (Fig 5A). In addition, when tested with a receptive virgin female after an hour break, the conditioned heterozygous Nipped-B males (Nipped-B407 / + and Nipped-B292.1 / +) do not show significantly reduced courting attempts compared to the naïve males of the corresponding genotypes (Fig 5B, CIsham vs. CItest). In contrast, age-matched wiso31 controls did show significantly reduced courting attempts between the trained and naïve groups, which suggests that Nipped-B mutants have deficiencies in short term memory (Fig 5B).
Fig. 5. Nipped-B heterozygotes are deficient in learning and short-term memory. 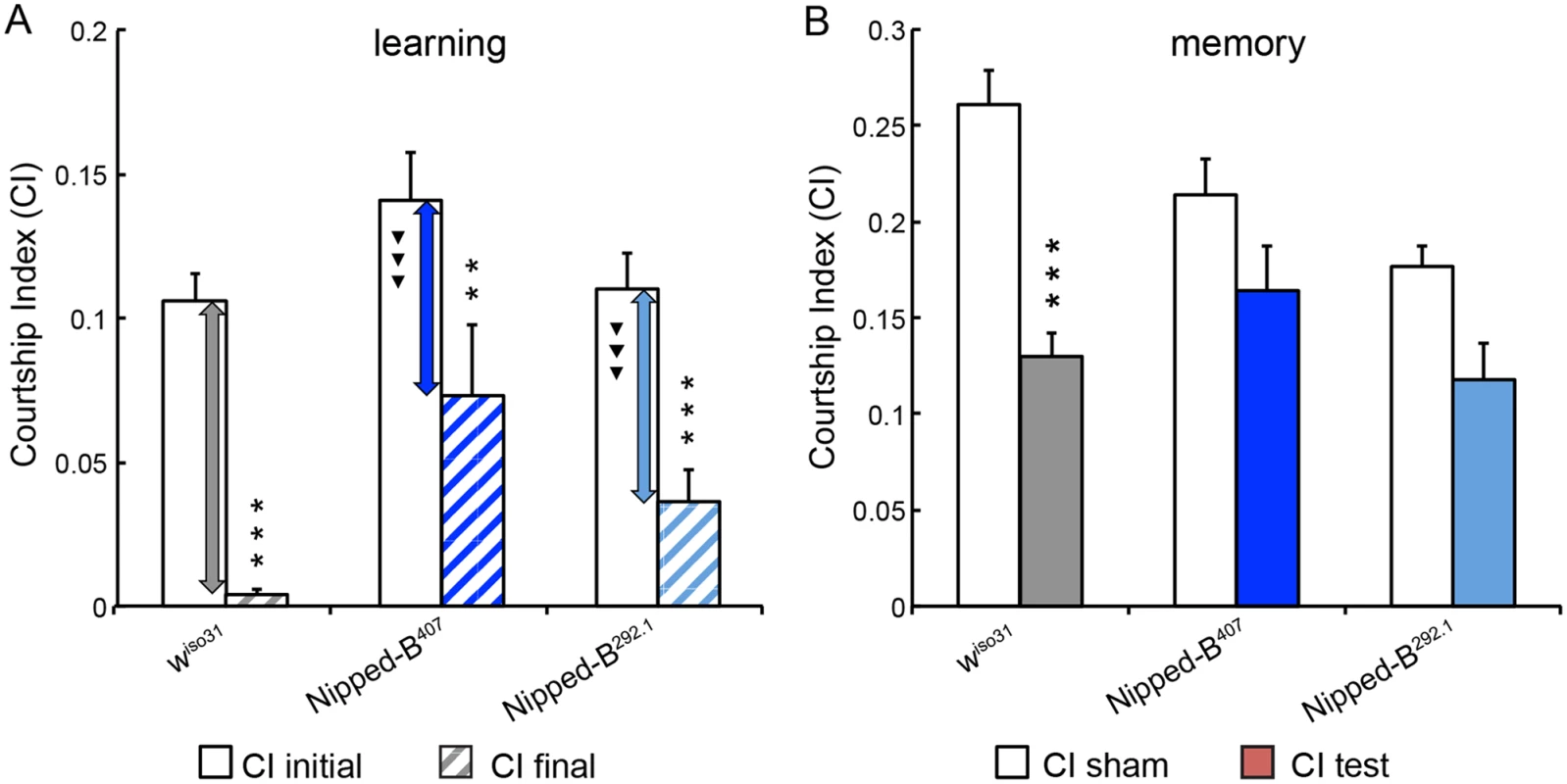
(A) Similar to wiso31 controls (p<0.0001) heterozygous Nipped-B males of both alleles (Nipped-B407 (p = 0.007) and Nipped-B292.1 (p<0.0001)) show significant decrease in their courtship activities through training sessions with non-receptive pre-mated females, suggesting that these flies display learning in experience-based conditioning. However, when the amount of decline between CI initial and CI final is compared between each Nipped-B heterozygote and the wiso31 controls, the decrements in the mutants are significantly smaller. (B) When tested for their courtship behaviors towards receptive virgin females after a short break (1 hour), trained wiso31 controls show a near 50% reduction in their courtship behaviors (CI test) compared the naïve wiso31 males (CI sham) (p<0.0001). However, heterozygotes of neither Nipped-B mutant show a significant difference to their naïve peers, for Nipped-B407, p = 0.10 and for Nipped-B292.1, p = 0.059. Over 18 flies were tested for each genotype in a given assay. Student’s t-tests were used for analyses with the levels of significance marked by number of *: *, p<0.05; **, p<0.01; ***, p<0.001. One-way ANOVA post hoc Tukey tests were used to decide statistical difference in learning capacity between Nipped-B407 or Nipped-B292.1 heterozygous males with wiso31 controls; ▼, p<0.05; ▼▼, p<0.01; ▼▼▼, p<0.001. Heterozygous Nipped-B mutants have abnormal brain structures
Mushroom body morphology in Nipped-B heterozygous males was examined to see if underlying structural abnormalities might accompany the behavioral deficits. Mushroom bodies are not required for learning in courtship conditioning, but are essential for consolidation of short-term and long-term associative memories [52]. The adult Drosophila mushroom body is composed of three specific classes of neurons, α/β, α’/β’, and γ neurons. Their axon projections give rise to five terminal lobes, α, β, α’, β’, and γ lobes [57]. Although the exact roles of the specific neurons making up the mushroom bodies are still unclear, these discrete axon clusters can be distinguished by their distinctive projection patterns, differential expression of GFP driven by various Gal4 lines, and labeling with anti-Fasciclin II (anti-FasII) [51,61].
We observed that 48% of Nipped-B407 heterozygotes (n = 29) and 27% in Nipped-B292.1 heterozygotes (n = 32) display abnormal mushroom body morphology, compared to only 4% of controls (n = 25) (Fig 6). The mushroom body lobes (α, β, γ) in control and heterozygous Nipped-B mutant adult male flies were visualized by expressing an OK201Y-Gal4>UAS-GFP fluorescent reporter gene and immunostaining with anti-FasII antibody. The OK201Y>UAS-GFP reporter is strongly expressed in the γ lobe and weakly in the α and β lobes in adults, whereas anti-FasII strongly labels the α and β lobes and the γ lobes weakly. The increase in abnormal brain structures in both Nipped-B mutants is statistically significant relative to the control, although the difference between the two mutants is not.
Fig. 6. Mushroom body abnormalities seen in Nipped-B heterozygotes. 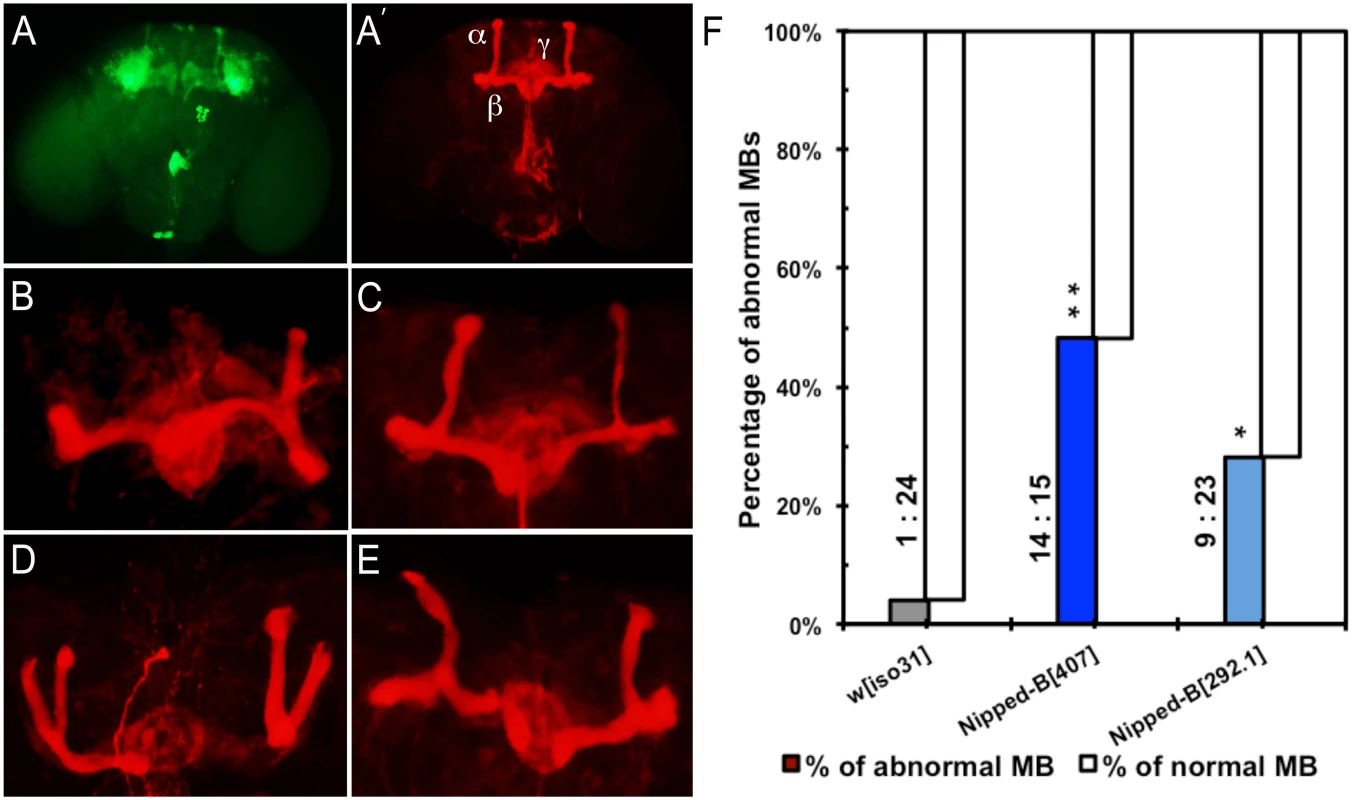
Immunofluorescence images of 2 to 4 day old adult brains are shown. Mushroom bodies (MBs) are labeled by anti-FasII (shown in red) and OK201Y>UAS-GFP (shown in green). (A, A’) Wild-type mushroom bodies are shown. α, β and γ lobes can be distinguished by their distinct projection patterns and fluorescent labeling. OK201Y >UAS-GFP is expressed mainly in the γ lobes and weakly in the α and β lobes; anti-FasII labels the α and β lobes strongly and γ lobes weakly. (B-E) Abnormal MB morphology seen in Nipped-B heterozygotes. (B) short α lobe and β lobe missing. (C) thinner α lobe and β lobes. (D) β lobe missing. (E) kinky α lobe. (F) A higher percentage of Nipped-B heterozygous MBs are morphologically abnormal compared to controls. The differences are statistically significant. Numbers of abnormal MBs versus numbers of normal MBs counted in each genotype are labeled next to the columns. Statistical significance was determined through Fisher’s exact tests. *, p<0.05; **, p<0.01. The morphological changes in mushroom bodies of Nipped-B mutants are pleiotropic and generally affect only one side of the brain. Short or kinky α lobes, fused, detached, or missing β lobes (Fig 6B–6E) are the most common defects. We observed that among 29 Nipped-B407 heterozygotes scored, 24% display missing or detached β lobe (n = 7), 10% show display short, kinky or detached α lobe (n = 3), and 7% show β lobe fusion with short or missing α lobe (n = 2). Among 32 Nipped-B292.1 heterozygotes scored, the portion of flies of each above-mentioned category is 6% (n = 2), 9% (n = 3), and 3% (ne = 1), respectively. In addition, one of the 32 Nipped-B292.1 heterozygotes displayed an α/β thinning phenotype. Although most of the structural abnormalities affect the α/β lobes, we also noted γ lobe defects in two Nipped-B407 heterozygotes and one Nipped-B292.1 heterozygote. In contrast to either Nipped-B mutant, only one out of 25 control flies showed a missing β lobe, with no other abnormalities detected. These morphological abnormalities demonstrate that heterozygous reduction of Nipped-B gene function interferes with brain development, and it is conceivable that these defects underlie the learning and memory deficits. Further studies will be required to substantiate this hypothesis and test the potential mechanistic connections.
Nipped-B mutants are arrhythmic and sleep less than controls
Sleep problems are a common clinical finding in CdLS and other neurodevelopmental disorders. Disruptive sleep and/or circadian patterns have been observed in Drosophila models for Fragile X syndrome, Neurofibromatosis Type 1 and Angelman syndrome [62]. While the molecular and physiological function of sleep is still elusive, it has long been known to have a direct correlation to learning and memory. In fact, learning and memory deficits have been observed in all the aforementioned fly models. We investigated if Nipped-B heterozygotes, like the other fly models for neurodevelopmental disorders, have aberrant sleep and/or circadian rhythm patterns.
We measured the sleep patterns of Nipped-B heterozygous mutant flies by monitoring their locomotor activities under 12 hr light/12 hr dark cycles, which revealed significant sex-dependent alterations in sleep (Fig 7). One-way ANOVA Tukey post hoc tests were used to determine if the sleep parameters of the Nipped-B mutants differ significantly from those of wiso31 controls. We found that both Nipped-B407 and Nipped-B292.1 heterozygous females sleep less than wiso31 controls, in terms of both total sleep and daytime sleep (Fig 7A and 7B). In addition, the total amount of nighttime sleep of Nipped-B407 heterozygous females was significantly less than wiso31 controls (Fig 7C). Unexpectedly, there was no significant change in sleep totals in Nipped-B mutant males, suggesting that there are sexually dimorphic effects on the expression of genes that determine sleep patterns. While Nipped-B407 mutant females showed reduced nighttime sleep, Nipped-B292.1 females did not, indicating that daytime sleep is more sensitive to Nipped-B dosage (Fig 7D and 7E).
Fig. 7. Nipped-B heterozygotes have disrupted sleep patterns. 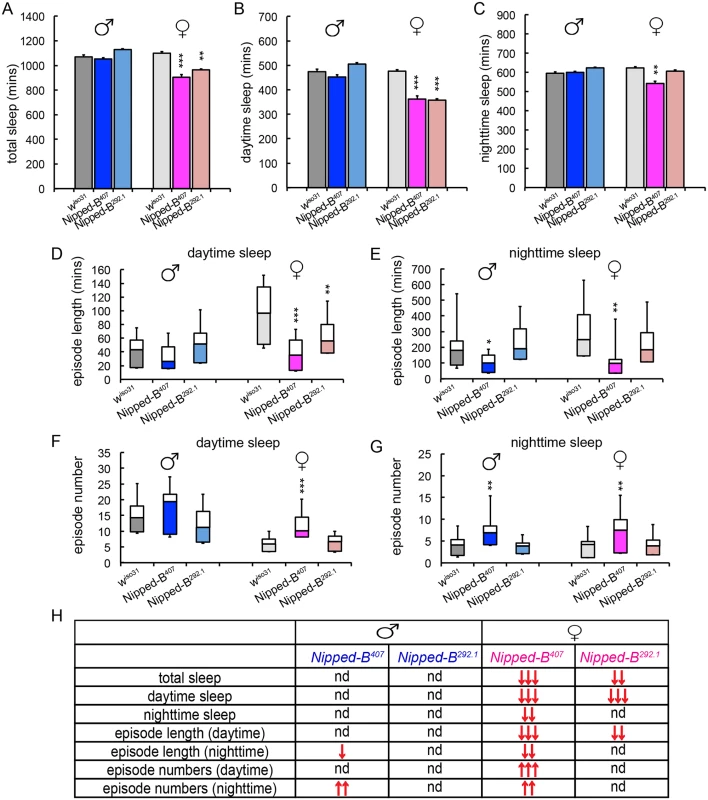
(A-C) Nipped-B heterozygous females, but not males, sleep less than wiso31 flies as shown in amounts of total sleep (A), daytime sleep (B), and nighttime sleep (C). (D-G) In general, shortened sleep episodes seen in Nipped-B407 male and female mutants are coupled with increased numbers of sleep episodes, both during daytime sleep and nighttime sleep. The lengths of sleep episodes of Nipped-B292.1 females are not significantly different from wiso31 flies. (H) Summary of statistical differences seen in Nipped-B mutants and wiso31 controls. One-way ANOVA post hoc Tukey tests were used to determine statistical difference in the sleep parameters; *, p<0.05; **, p<0.01; ***, p<0.001. ↑ and ↓ indicate more or less/fewer than controls. Compared to wiso31 flies, the average lengths of sleep episodes were significantly shortened in Nipped-B407 heterozygous females during both daytime and nighttime sleep (Fig 7D and 7E). Such reductions were partially compensated by an increased number of sleep episodes during the day and at night (Fig 7F and 7G). Nipped-B292.1 females showed a significant reduction in sleep episode length during daytime sleep (Fig 7D), which was not accompanied by an increased number of sleep bouts (Fig 7F). The nighttime sleep episode length and numbers of Nipped-B292.1 females were comparable to wiso31 control females.
Nipped-B407 and Nipped-B292.1 heterozygous males sleep for similar amounts of time as wiso31. However, a reduced sleep episode length during nighttime sleep coupled with a compensating increase in the number of episodes was also observed in Nipped-B407 males (Fig 7E and 7G). All sleep parameters of Nipped-B292.1 heterozygous males were not significantly different from wiso31 controls. Fig 7H summarizes the complex patterns of sex - and allele-specific changes in sleep patterns.
We investigated whether the aberrant sleep outputs seen in Nipped-B heterozygotes could arise from disruption of the circadian rhythm. Circadian rhythm is an intrinsic behavior independent of environmental cues. Four to five day old adult flies were loaded into a DAMS monitoring system to record their locomotor activity in constant darkness for over 14 days to measure their free-running circadian rhythms. The activity patterns of each fly were analyzed using the Clock program (Clocklab) to investigate periodogram of each individual fly to determine the period length and consolidation of the rhythm. In contrast to wiso31 controls, a significant fraction of Nipped-B407 males (p = 0.001, Fisher’s exact test) and all of the females (p = 0.0001) were arrhythmic (Fig 8). While the percentage of arrhythmic Nipped-B292.1 heterozygous females is slightly higher than wiso31 controls (p = 0.04, Fisher's exact test), there is no statistical difference between heterozygous males and the controls (p = 0.48) (Fig 8B). Periods calculated from rhythmic Nipped-B407 males, Nipped-B292.1 males and females, and wiso31 controls were all within wild-type range (~23.5 hours) (Fig 8B). There is a clear correlation between circadian rhythm and sleep disturbances, with females being more strongly affected than males. However, Nipped-B407 males show a strong circadian deficit, and only a modest change in sleep pattern, suggesting that circadian rhythm changes alone are insufficient to substantially alter sleep patterns.
Fig. 8. Nipped-B heterozygous mutants are arrhythmic. 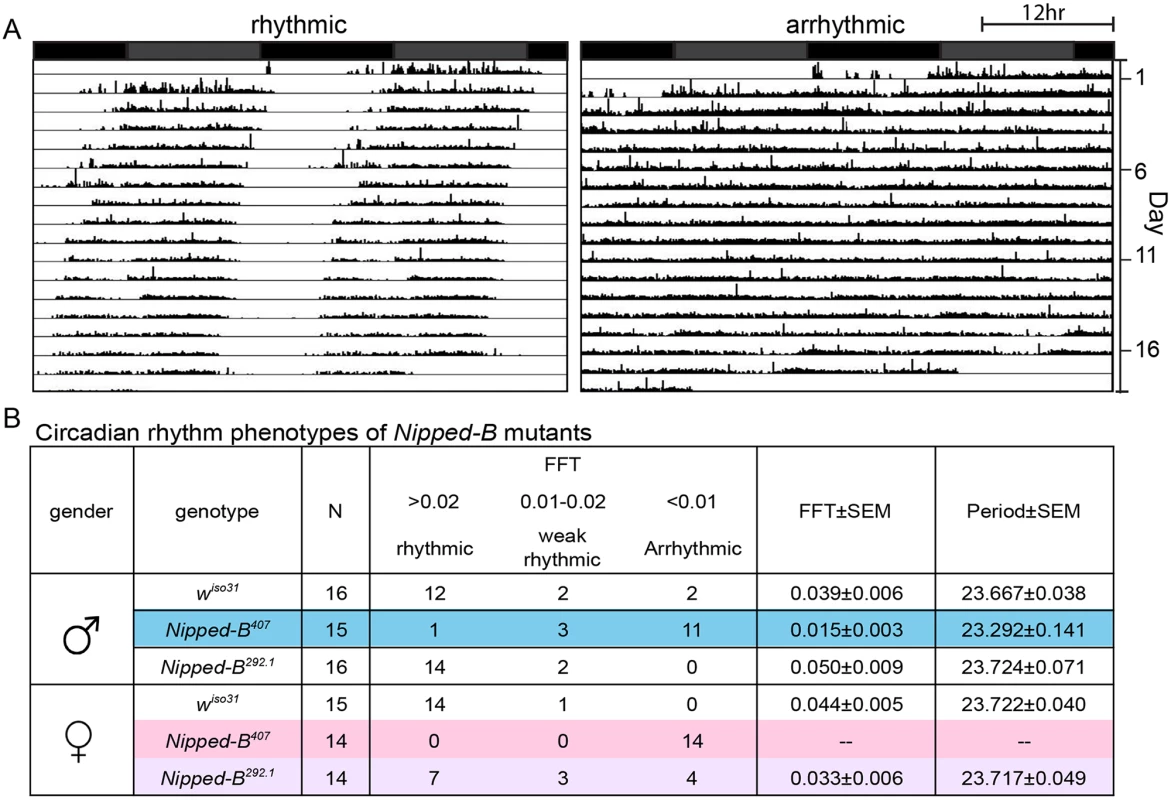
Flies were placed in constant darkness (DD) for over 14 days to record their locomotor activities, which were used to determine free-running rhythms. (A) Representative double plotted actograms are shown. Rhythmic flies show a robust rest/activity dichotomy, which is absent in arrhythmic flies. (B) Summary of circadian rhythm behaviors of Nipped-B mutants. Numbers of flies in each rhythmicity group are shown and N refers to the total numbers of flies tested. Rhythmicity was decided upon analysis of actogram, periodogram and FFT value. Significantly different from wiso31 controls, most Nipped-B407 males and all Nipped-B407 females are arrhythmic (p = 0.001 for mutant versus control males shaded in blue, p = 0.001 for mutant versus control females shaded in pink, using Fisher’s exact test, low average FFT value). Nipped-B292.1 females also show a slightly weaker rhythmicity than wiso31 females (p = 0.042, Fisher's exact test, low average FFT value, shaded purple). FFT average and Periods were calculated from flies of the rhythmic and weak rhythmic groups. Periods of all genotypes were in the wild-type range (~23.5hr). SEM: standard error. FFT: fast Fourier transform value. Actograms averaging the activities of wiso31, Nipped-B407, and Nipped-B292.1 males and females, and examples of additional individuals are presented in S4 Fig. Although the arrhythmic fly in Fig 8 appears to show more movement than the control, Nipped-B mutants are not hyperactive, as shown by the activity indices in S5 Fig.
Heterozygous Nipped-B mutants show no signs of seizures
Seizure is another common neurological manifestation seen in CdLS individuals. We tested Nipped-B mutant flies to see if they are susceptible to seizures when subject to mechanical or thermal stress. Seizure-like behaviors in Drosophila induced by intensive vortexing are characterized by a sequence of initial seizure, temporary paralysis and recovery seizure [63]. We did not detect vortexing-induced seizures in Nipped-B mutant flies with either allele. We also tested these mutants in a heat-induced seizure assay [64] and Nipped-B mutants subject to brief heat-shock at 40° showed no sign of paralysis.
Discussion
The studies reported here indicate that despite the large evolutionary divergence between Drosophila and humans, heterozygous Nipped-B mutant flies share both growth and neurological deficits with Cornelia de Lange syndrome. Although the mechanisms that cause these deficits in CdLS remain to be determined, the Drosophila studies suggest that at least some of the contributing mechanisms are highly conserved. The findings that Drosophila size is reduced primarily through mechanisms that do not involve systemic controls that sense body size in Nipped-B mutants, and that behavioral deficits are accompanied by morphological changes in brain structure are likely to have key implications for understanding the etiologies of the related deficits in CdLS.
Role of Nipped-B in growth control
Prior studies revealed that Nipped-B heterozygous mutant Drosophila display subtle and latent external morphological phenotypes that become overt in adults only when combined with mutations in key developmental regulatory genes such as cut, Ultrabithorax, Notch, mastermind, hedgehog, and genes encoding cohesin or Polycomb silencing complex subunits [5,6,9,17,33–35]. This contrasts with mice and humans, where similar deficiencies in NIPBL cause multiple specific and obvious morphological changes [30]. There are also differences between mice and humans. For instance, limb abnormalities are largely absent in Nipbl(+/-) mice, but heart defects are significantly more frequent than in individuals with CdLS [14]. Extrapolating from Drosophila genetic interaction data, we posit that the individual physical birth defects in vertebrates stem from altered expression of specific sets of developmental genes, and that the variability between individuals with CdLS reflects differences in genetic background.
We show here that although the external morphological changes in Nipped-B mutant Drosophila are minimal in an otherwise wild-type background, they share the reduced size with Nipbl(+/-) mice and CdLS. The data argue that the reduced size reflects decreases in both total cell number and size, and not changes in the systemic control of growth that sense critical body mass, deficits in the utilization of nutrition, or increased cell death. The decrease in size likely stems in part from modestly reduced expression of the myc (dm), Tor, InR and other genes that promote cell proliferation, division and growth, and not increased apoptosis. Indeed, one of the most intriguing findings is the reduced ability of excess dm expression to induce apoptosis in Nipped-B heterozygous mutants. We speculate that this may stem in part from reduced function of a Dm-dependent enhancer that drives expression of the grim and reaper pro-apoptosis genes [65]. This region binds Nipped-B and cohesin in wing discs [35], and deletion of this region permits excess dm expression using the tub-myc driver to increase wing size more than in wild-type flies because it reduced apoptosis [65].
It remains to be seen to what extent these findings in Drosophila might explain the reduced growth in CdLS and in Nipbl(+/-) mice. It seems likely that similar mechanisms at least make a significant contribution to the reduced growth in mammals because Nipbl(+/-) mice and cells from individuals with CdLS also show reduced c-myc expression [14,15]. Organisms use a variety of mechanisms to sense body size and regulate growth and developmental transitions. Drosophila transitions are timed by pulses of the ecdysone steroid hormone produced by the prothoracic gland located between the two lobes of the brain [43]. Specific neurons in the brain secrete a peptide hormone, prothoracicotropic hormone (PTTH) that stimulates ecdysone production. Insulin signaling, nutrition and many other factors, which are not all well understood, control the hormonal pathways and timing of the ecdysone pulses that determine absolute body size. For example, nutrient deprivation substantially delays the ecdysone pulses, but not enough to fully restore maximal body size. What is particularly striking is how close Nipped-B mutants are to wild type in their developmental timing and in how their developmental staging responds to nutrient deprivation. This argues that the systemic hormonal pathways that regulate body size and hormonal pulses that induce developmental stages are largely unaffected, and that the reduced size of Nipped-B mutants stems primarily from a small but significant reduction in the number of cell divisions, and in the mechanisms that determine final cell size.
It is more difficult to precisely time the developmental staging in mice and humans than in Drosophila, and thus to determine whether or not developmental timing or systemic growth pathways are significantly altered with decreased NIPBL function. Although some individuals with CdLS show slightly delayed puberty, puberty occurs at the normal age in many [30]. The slight delays, and incomplete pubertal changes might all be attributed to causes other than a general developmental delay, such as structural abnormalities or changes in hormone levels. The relatively normal timing of puberty, therefore seems to indicates that the more extreme reductions in overall size observed in CdLS are also more likely to stem from changes in cell number and size than changes in systemic body size controls.
Role of Nipped-B in neurological function
We also find that Drosophila Nipped-B heterozygotes display many behavioral and neurological features resembling those seen in CdLS patients: they are deficient in learning and short-term memory, and display disruptive sleep patterns and abnormal circadian rhythms. Intellectual disability is the most common clinical phenotype seen in individuals with CdLS. The average IQ score of typical CdLS cases, mostly with NIPBL mutations, is 53 (range 30–86) [32]. We find that although Nipped-B heterozygous mutant flies are capable of learning, their learning capacity is significantly lower than that of the wiso31 controls. Furthermore, these Nipped-B mutants are accompanied by pleiotropic structural abnormalities in mushroom bodies, the major brain structures controlling learning and memory. While it is conceivable that the morphological defects in the mushroom bodies could contribute to the learning and memory deficits observed in the Nipped-B mutants, how these structural and functional deficiencies correlate with each other warrant further detailed studies.
We observed a striking similarity in the sleep patterns of fly Nipped-B heterozygotes and those seen in CdLS patients. Sleep disturbances are common in CdLS and seen in up to 55% of CdLS individuals. Insomnia (difficulty in initiating sleep), difficulty staying asleep, frequent night wakenings, and sleepiness during the daytime are the most common sleep problems reported in CdLS [66,67]. Frequent but dramatically shortened sleep episodes was the characteristic sleep pattern observed in Nipped-B heterozygote flies. In fact, the reduced length of sleep episodes lead to a dramatic loss in daytime and nighttime sleep, which was unable to be compensated for by significant increases in the number of sleep bouts.
While it has been speculated that sleep disturbances in CdLS individuals may be in part attributable to a circadian rhythm disorder, strict and objective studies to confirm this suggestion have not been undertaken. The locomotor activity-based circadian rhythm assay is a well-established measure of circadian rhythm in Drosophila. We demonstrated that an aberrant circadian rhythm exists in a large fraction of Nipped-B haploinsufficient male and female flies. For Nipped-B mutants that still display rhythmic circadian patterns, their free-running activity rhythms are maintained at around 24 hours, similar to the periodicity of wiso31 controls. A remarkable consistency between the circadian defects and sleep aberrance was observed in these Nipped-B heterozygous mutants; flies that were profoundly arrhythmic were the ones that showed the most disturbed sleep patterns. Nevertheless, the circadian rhythm alterations are more apparent than the sleep disturbances in males, suggesting that additional sex-specific factors are involved in determining the sleep patterns. Taken together, these data suggested that at least in fly Nipped-B mutants, intrinsic circadian rhythm defects likely contribute to their aberrant sleep patterns.
Overall, our studies on fly Nipped-B mutants demonstrate a strikingly analogous growth and neurocognitive/behavioral phenotype between heterozygous Nipped-B mutants and human CdLS individuals, including small body size, learning and memory deficits, disruptive sleep patterns and circadian rhythm defects. Drosophila Nipped-B heterozygotes are thus a valuable resource, with multiple objective and readily measureable metrics, for modeling human CdLS. Studies on the function of Nipped-B and cohesin components in CdLS patient cell lines, Nipbl(+/-) mouse and zebrafish models, and Drosophila have contributed substantially to our understanding of the roles of Nipped-B and cohesin components in development and gene regulation. The presence of a sophisticated genetic tool kit and economical availability of fruit flies will make it possible to explore developmental deficits in a tissue - and stage-specific manner, as well as to test their relevance to human development and the pathogenesis of CdLS. Drosophila is an ideal model organism to address these issues, given its short life cycle, lower degree of genomic redundancy and the available genetic tools. Nipped-B mutants can also be utilized to search for and test new pharmacologic therapeutic modalities, towards amelioration of the growth and neurodevelopmental functioning of individuals with CdLS.
Materials and Methods
Drosophila culture
For growth and nutrition studies, flies were cultured on cornmeal, yeast and molasses media in agar [68] at 25°. To limit nutrition (20% food), the normal food recipe was diluted 5 fold in 0.9% agar in phosphate buffered saline (PBS). For all experiments, the mutant and controls were grown at the same time with the same batch of food preparation. To minimize differences in the genetic backgrounds, females with the balanced mutations or matched controls [(y w; Nipped-B407 P{w+}57B/CyO, Kr-GFP), (y w; P{mw+}57B), (y w, y w dm2/FM7), (y w; TorΔP P{neoFRT}40A/CyO, Kr-GFP)] were outcrossed to wild-type Oregon R males for all experiments, generating GFP negative progeny that are heteroallelic across the genome. Control and mutant crosses were always performed at the same time, and used the same batch of food.
Nipped-B407 was chosen for detailed analysis in growth experiments because it has been shown to be rescued by Nipped-B cDNA transgenes, providing evidence that Nipped-B407 is the only essential gene mutation in the chromosome [36,42]. The different fusion cDNA transgenes (FLAG-Nipped-B, Nipped-B-EGFP) used a full length cDNA of one alternatively-spliced Nipped-B transcript and were expressed using the constitutive Chi gene promoter. Depending on the transgene, they rescued Nipped-B407 homozygous mutants far beyond the normal 2nd instar lethal phase to pharate adults, or viable fertile adults. Even with the most complete rescue, however, the flies showed reduced viability and were difficult to maintain, indicating incomplete rescue. The incomplete rescue likely reflects the lack of normal Nipped-B promoter regulation and splicing patterns.
To minimize potential influence of background variations on behavioral outputs, both Nipped-B alleles were backcrossed to an isogenized w1118 strain (wiso31), which was used as the control in all Drosophila behavioral analyses. For all the behavioral assays, heterozygous Nipped-B mutants were generated by crossing the isogenized Nipped-B407 or Nipped-B292.1 chromosomes into wiso31 background and compared to the wiso31 controls.
Genomic DNA sequencing
Genomic DNA was purified from ~100 homozygous Nipped-B407 mutant 2nd instar larvae using the Zymo Research Fungal/Bacterial DNA MicroPrep kit according the manufacturer’s instructions and 50 nanograms of purified DNA was used to prepare a sequencing library with the Ion Xpress Plus Fragment Library Kit (Life Technologies) following the supplier’s methods with the exception that final library size selection was performed using Beckman Agencourt AMPure beads. The library was sequenced with an Ion Torrent Proton (Life Technologies) with a mean read length of 175 bp and aligned genome coverage of ~25-fold. Alignment to the Drosophila release 5 genome sequence (April 2006, modified by removal of chrU and Uextra sequences) was performed using the TMAP aligner map4 algorithm [69] with no soft-clipping and a minimum seed length of 20. The resulting bam files were sorted and indexed using SAMtools [70]. The sequence of the Nipped-B gene and surrounding region was inspected for indels and nucleotide changes in the aligned sequence files with the Affymetrix Integrated Genome Browser (IGB) [71].
Determination of body weight
Adult flies were aged for two to three days after eclosion at weighed in batches of 20. Four to fourteen batches (80 to 280 total flies) were weighed for each genotype.
Wing size, cell number and cell size measurements
Wings were dissected and mounted on glass slides under coverslips as previously described [72]. Photographs were taken at low magnification (4X objective) to quantify the wing area using ImageJ, and higher magnification (60X objective) of the same smaller region in each wing to count the number of cells per unit area. Cell number was counted by the number of wing hairs present in the region using a semi-automated method. The contrast of the digital high magnification photograph was increased to permit automated counting with the colony counting feature of AlphaEase software (AlphaInnotech). At least forty wings were measured per genotype. To measure cell size in developing wings, twenty wing disks each from male late 3rd instar y w; Nipped-B407 P{w+}57B/+ and y w; P{w+}57B/+ were incubated with collagenase (2 mg per ml) for 25 min in Ringer’s solution, transferred into 0.3 ml of Schneider’s medium containing 0.5% trypsin for 2.5 hours with periodic pipetting to dissociate the cells, and analyzed by forward scattering in a FACS.
Developmental timing
Forty to fifty mutant or control virgin females were crossed to a similar number of Oregon R males in small beakers with apple juice plates covered with fresh yeast paste. Embryos were collected for 3 to 4 hours, and newly hatched larvae lacking the GFP balancers were collected 23 to 26 hours after the start of egg-laying and distributed into food vials (100 or 20% food) at 100 larvae per vial. Five to ten vials (500–1000 total larvae) were scored daily for pupariation and eclosion for each genotype. The experiments for Nipped-B407 and the matched control were repeated three times. To examine the timing of the 2nd to 3rd instar molt, egg laying was restricted to a two hour period, and newly-hatched larvae collected was restricted to a one hour window.
TUNEL staining
Late third instar wing discs from y w; tub-myc/P57B, y w; tub-myc/Nipped-B407 P57B, y w; +/P57B, and y w; +/Nipped-B407 P57B were fixed in 4% paraformaldehyde in PBS and stained using the Click-iT TUNEL Alexa Fluor 488 Imaging Assay (Molecular Probes) according the manufacturer’s protocol. The stained discs were imaged using a Leica SP5 laser scanning confocal microscope to count the number of apoptotic cells. Twenty discs were scored for each genotype, and the experiment was repeated three times.
RNA-seq expression analysis
Total RNA was isolated from late 3rd instar wing discs of heterozygous P57B control and Nipped-B407 mutant female larvae after crossing to Oregon R wild type, in three separate collections from independent crosses using Zymo Research Quick-RNA MicroPrep Kits with DNaseI treatment following the manufacturer’s protocol. Ribosomal RNA depletion and sequencing library construction for each wing disc RNA isolate were performed using Eukaryotic RiboMinus and Ion Total RNA-seq v2 kits (Life Technologies) according to the manufacturer’s directions. Sequencing was performed on an Ion Torrent Proton with a mean read length of 135 nucleotides, and reads were aligned to the release 5 Drosophila genome (April 2006) using the TMAP aligner map4 algorithm. Soft-clipping at both 5’ and 3’ ends of the reads was permitted during alignment to accommodate spliced reads, with a minimum seed length of 20 nt. One P57B and one Nipped-B407 library were sequenced twice to provide additional replication, and were used as fourth replicates for statistical calculations. The combined coverage of sequencing for each genotype was >80-fold genome equivalents, with roughly half the reads corresponding to ribosomal RNA that was incompletely removed by depletion.
RNA-seq analysis methods typically normalize to total reads, which assumes that most genes do not change in expression under experimental conditions, but this assumption is false when Nipped-B or cohesin levels are reduced [24]. We thus normalized to total histone RNA, which is tightly controlled to reflect genomic DNA content [49]. To accommodate this normalization, the TMAP aligner parameters were set such that each histone RNA read was assigned randomly to only one of the top matches, and the U and Uextra sequences were removed from the genome sequence to prevent assignment of reads to unannotated histone gene copies. Genome-wide strand-specific nucleotide coverages were calculated from the aligned bam files for each sample using the genomecoveragebed program in BEDTools [73] and the nucleotide coverage for all non-redundant exons for each gene were summed using custom R [74] scripts. Most RNA-seq methods calculate expression using total read numbers, which assumes that the reads are the same length, but calculating total nucleotide coverage is more accurate for Ion Torrent sequencing, which produces reads of differing lengths. After removal of the contaminating ribosomal RNA coverage, and the coverage of small non-coding RNAs (snoRNAs, tRNAs, miRs, etc) which are not quantitatively removed during the library size selection step, the total histone RNA coverage for each sample was calculated and used to normalize the nucleotide coverage for all other genes in each sample. Normalization factors were calculated by averaging the total histone coverage for all replicates and dividing this average by the histone coverage for each sample. The total coverage for each gene in each replicate was then multiplied by these factors after adding an offset of 1 to each gene to preclude division by 0 in subsequent calculations. The averages, standard errors and p values of the coverage values for all genes in the four P57B and Nipped-B replicates (three independent RNA isolations and one resequencing for each genotype) were calculated using Microsoft Office Excel, and additional analysis including calculating correlation coefficients was performed using R. The data is presented in S1 Table and deposited in the GEO database, accession no. GSE74192. The expression values in S1 Table are the normalized strand-specific nucleotide coverage for each gene.
Courtship training and testing
Male and female flies used for courtship testing were prepared as described in [58] and [75]. Briefly, males of the appropriate genotype were collected within 4 hours of eclosion and kept individually in small food-containing vials. Virgin X^X, y f females were collected in the same manner and kept in groups of 10 to 15 in standard food vials. All flies were aged at 25° in a 12 : 12 LD cycle and courtship testing was performed during the relative light phase. Trainer females were 5 days old on the day of testing and were copulated the day before testing. Virgin females used on the day of testing were 5 days old. Male tester flies were 5 days old and were randomly assigned into training group and naive groups. The experimenter was blinded to the genotypes of the males used in courtship testing. The total amount of time a tester male engaged in active courting (orienting, taping, singing, licking and attempting copulation) was recorded for a period of 10 minutes, or upon successful copulation. The courtship index (CI) is defined as the percentage of time spent in courting during a 10 minute period or before successful copulation. For each genotype, at least 18 males were tested in the naive group and the training group. Statistical analyses were performed as described in [58].
Sleep and circadian rhythm assays
Sleep and circadian rhythm tests were performed and analyzed as described [76,77]. Adult flies were kept as groups of males and females and entrained to a 12hr light: 12hr dark (L/D) cycle at 25°C for 3 days prior to testing. They were then individually transferred into glass tubes and monitored through the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA). A beam of infrared light passes through the middle of the tube and fly activities were deduced from beam-breaking events. Behavioral quiescence longer than 5 minutes is defined as sleep. Sixteen flies of each sex were tested for each genotype. For sleep assays, the flies were loaded into the monitors at 9 am and then monitored for 6 days in L/D cycle. Sleep data collected were processed with Pysolo software [78] to calculate the amount of sleep, the number of sleep bouts, and sleep duration. For circadian assays, the flies were loaded into the monitors at 9 pm and data were collected from flies monitored in constant darkness for at least 14 days. Data collected were processed with ClockLab (Actimetrics, Wilmette, IL) to generate periodograms and actograms. Period length and FFT value (Fast Fourier Transformation, the relative power of rhythmicity) were collected from periodogram. Rhythmicity is decided by reviewing both actogram and periodogram. Statistical analyses were performed as described in [77,79].
Mushroom body labeling and morphological analysis
Brains from 2–4 day old adults were dissected in PBS, fixed, and stained as described [59,76]. Anti-Fasciclin II antibody (1 : 10 dilution, ID4, Developmental Studies Hybridoma Bank, IA) and Alexa-568 goat anti-mouse secondary antibody (1 : 400 dilution, Life Technologies, Carlsbad, CA) were used. Tissues were mounted in VectaShield (Vector Laboratories, Burlingame, CA). Images were acquired with an Olympus 1X71 fluorescence microscope. For each genotype, at least 18 males were tested in the naive group and in the training group. The experimenter was blinded to the genotypes of the brains using in morphology analyses.
Seizure tests
Mechanical stress tests (bang sensitivity) and heat shock assays were performed as described [63,80]. Over 50 flies were tested fro each stress test.
Supporting Information
Zdroje
1. Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013; 25 : 327–333. doi: 10.1016/j.ceb.2013.02.003 23465542
2. Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140 : 3715–3718. doi: 10.1242/dev.090605 23981654
3. Ball AR Jr, Chen YY, Yokomori K. Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies. Biochim Biophys Acta. 2014;1839 : 191–202. doi: 10.1016/j.bbagrm.2013.11.002 24269489
4. Dorsett D, Kassis JA. Checks and balances between cohesin and Polycomb in gene silencing and transcription. Curr Biol. 2014;24: R535–R539. doi: 10.1016/j.cub.2014.04.037 24892918
5. Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152 : 577–593. 10353901
6. Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24 : 3100–3111. 15060134
7. Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36 : 631–635. 15146186
8. Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36 : 636–641. 15146185
9. Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132 : 4743–4753. 16207752
10. Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38 : 528–530. 16604071
11. Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80 : 485–494. 17273969
12. Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, et al. Cohesin-dependent regulation of Runx genes. Development. 2007;134 : 2639–2649. 17567667
13. Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, et al. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134 : 3191–3201. 17652350
14. Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5: e1000650. doi: 10.1371/journal.pgen.1000650 19763162
15. Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7: e1000119. doi: 10.1371/journal.pbio.1000119 19468298
16. Liu J, Feldman R, Zhang Z, Deardorff MA, Haverfield EV, Kaur M, et al. SMC1A expression and mechanism of pathogenicity in probands with X-Linked Cornelia de Lange syndrome. Hum Mutat. 2009;30 : 1535–1542. doi: 10.1002/humu.21095 19701948
17. Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, et al. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4: e6202. doi: 10.1371/journal.pone.0006202 19587787
18. Zhang B, Chang J, Fu M, Huang J, Kashyap R, Salavaggione E, et al. Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One. 2009;4: e5232. doi: 10.1371/journal.pone.0005232 19412548
19. Cunningham MD, Gause M, Cheng Y, Noyes A, Dorsett D, Kennison JA, et al. Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development. 2012;139 : 4172–4179. doi: 10.1242/dev.084566 23034634
20. Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, et al. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet 90 : 2012;1014–1027. doi: 10.1016/j.ajhg.2012.04.019 22633399
21. Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117 : 89–102. 17965872
22. Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467 : 430–435. doi: 10.1038/nature09380 20720539
23. Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, et al. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J Biol Chem. 2011;286 : 17870–17878. doi: 10.1074/jbc.M110.207365 21454523
24. Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, et al. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9: e1003382. doi: 10.1371/journal.pgen.1003382 23555293
25. Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, et al. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol. 2011;21 : 1624–1634. doi: 10.1016/j.cub.2011.08.036 21962715
26. Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151 : 22–37. doi: 10.1111/j.1749-6632.2008.03450.x 19154515
27. Liu J, Krantz ID. Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet. 2009;76 : 303–314. doi: 10.1111/j.1399-0004.2009.01271.x 19793304
28. Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489 : 313–317. doi: 10.1038/nature11316 22885700
29. Mannini L, Cucco F, Quarantotti V, Krantz ID, Musio A. Mutation spectrum and genotype-phenotype correlation in Cornelia de Lange syndrome. Human Mut. 2013;34 : 1589–1596.
30. Kline AD, Krantz ID, Sommer A, Kliewer M, Jackson LG, FitzPatrick DR, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143A: 1287–1296. 17508425
31. Kaiser FJ, Ansari M, Braunholz D, Concepción Gil-Rodríguez M, Decroos C, Wilde JJ, et al. Loss-of-function HDAC8 mutations cause a phenotypic spectrum of Cornelia de Lange syndrome-like features, ocular hypertelorism, large fontanelle and X-linked inheritance. Hum Mol Genet. 2014;23 : 2888–2900. doi: 10.1093/hmg/ddu002 24403048
32. Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75 : 610–623. 15318302
33. Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, et al. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117 : 51–66. 17909832
34. Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, et al. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci U S A. 2008;105 : 12405–12410. doi: 10.1073/pnas.0801698105 18713858
35. Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, et al. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9: e1003560. doi: 10.1371/journal.pgen.1003560 23818863
36. Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4: e242. 16802858
37. Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, et al. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344 : 637–649. doi: 10.1016/j.ydbio.2010.05.493 20553708
38. Maines JZ, Stevens LM, Tong X, Stein D. Drosophila dMyc is required for ovary cell growth and endoreplication. Development. 2004;131 : 775–786. 14724122
39. Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7 : 21–32. doi: 10.1016/j.cmet.2007.11.010 18177722
40. Li L, Edgar BA, Grewal SS. Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cis-regulatory element. BMC Cell Biol. 2010;11 : 7. doi: 10.1186/1471-2121-11-7 20089194
41. Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14 : 2712–2724. 11069888
42. Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol. Cell. Biol. 2010;30 : 4940–4951. doi: 10.1128/MCB.00642-10 20696838
43. Callier V, Nijhout HF. Body size determination in insects: a review and synthesis of size - and brain-dependent and independent mechanisms. Biol Rev Camb Philos Soc. 2013;88 : 944–954. doi: 10.1111/brv.12033 23521745
44. Gallant P. Myc function in Drosophila. Cold Spring Harb Perspect Med. 2013;3: a014324. doi: 10.1101/cshperspect.a014324 24086064
45. de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117 : 107–116. 15066286
46. Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16 : 275–287. 16945552
47. Montero L, Müller N, Gallant P. Induction of apoptosis by Drosophila Myc. Genesis. 2008;46 : 104–111. doi: 10.1002/dvg.20373 18257071
48. Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, et al. Revisiting global gene expression analysis. Cell 2012;151 : 476–482. doi: 10.1016/j.cell.2012.10.012 23101621
49. Rattray AM, Müller B. The control of histone gene expression. Biochem Soc Trans. 2012;40 : 880–885. doi: 10.1042/BST20120065 22817752
50. Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, et al. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14 : 239–251. doi: 10.1016/j.devcel.2007.12.009 18267092
51. Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14 : 227–238. doi: 10.1016/j.devcel.2007.11.001 18267091
52. McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, et al. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. 1999;Neuron. 24 : 967–977. 10624959
53. Pascual A, Préat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294 : 1115–1117. 11691997
54. Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53 : 103–115. 17196534
55. Hall JC. The mating of a fly. Science. 1994;264 : 1702–1714. 8209251
56. Sokolowski MB. Drosophila: genetics meets behaviour. Nat Rev Genet. 2001;2 : 879–890. 11715043
57. Siegel RW, Hall JC. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc Natl Acad Sci U S A. 1979;76 : 3430–3434. 16592682
58. McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45 : 753–764. 15748850
59. Ejima A, Griffith LC. Assay for courtship suppression in Drosophila. Cold Spring Harb Protoc. 2011;2011:pdb.prot5575.
60. Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167 : 797–813. 15238529
61. Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126 : 4065–4076. 10457015
62. Gatto CL, Broadie K. Drosophila modeling of heritable neurodevelopmental disorders. Curr Opin Neurobiol. 2011;21 : 834–841. doi: 10.1016/j.conb.2011.04.009 21596554
63. Song J, Tanouye MA. From bench to drug: human seizure modeling using Drosophila. Prog Neurobiol. 2008;84 : 182–191. 18063465
64. Sun L, Gilligan J, Staber C, Schutte RJ, Nguyen V, O'Dowd DK, et al. A knock-in model of human epilepsy in Drosophila reveals a novel cellular mechanism associated with heat-induced seizure. J Neurosci. 2012;32 : 14145–14155. doi: 10.1523/JNEUROSCI.2932-12.2012 23055484
65. Zhang C, Casas-Tintó S, Li G, Lin N, Chung M, Moreno E, et al. An intergenic regulatory region mediates Drosophila Myc-induced apoptosis and blocks tissue hyperplasia. Oncogene 2015;34 : 2385–2397. doi: 10.1038/onc.2014.160 24931167
66. Stavinoha RC, Kline AD, Levy HP, Kimball A, Mettel TL, Ishman SL. Characterization of sleep disturbance in Cornelia de Lange Syndrome. Int J Pediatr Otorhinolaryngol. 2011;75 : 215–218. doi: 10.1016/j.ijporl.2010.11.003 21146878
67. Rajan R, Benke JR, Kline AD, Levy HP, Kimball A, Mettel TL, et al. Insomnia in Cornelia de Lange syndrome. Int J Pediatr Otorhinolaryngol. 2012;76 : 972–975. doi: 10.1016/j.ijporl.2012.03.008 22503448
68. Wirtz RA, Semey HG. The Drosophila kitchen—equipment, media preparation, and supplies. Drosophila Inform Serv. 1982;58 : 176–180.
69. Homer N. TMAP: the torrent mapping program. 2011. https://github.com/iontorrent/TMAP/blob/master/doc/tmap-book.pdf
70. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25 : 2078–2079.
71. Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009;25 : 2730–2731. doi: 10.1093/bioinformatics/btp472 19654113
72. Dorsett D. Distance-independent inactivation of an enhancer by the suppressor of Hairy-wing DNA-binding protein of Drosophila. Genetics. 1993;134 : 1135–1144. 8375652
73. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26 : 841–842. doi: 10.1093/bioinformatics/btq033 20110278
74. R Core Team. R: A Language and Environment for Statistical Computing. 2015; R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
75. Schoenfeld BP, Choi RJ, Choi CH, Terlizzi AM, Hinchey P, Kollaros M, et al. The Drosophila DmGluRA is required for social interaction and memory. Front Pharmacol. 2013;4 : 64. doi: 10.3389/fphar.2013.00064 23720628
76. Wu Y, Bolduc FV, Bell K, Tully T, Fang Y, Sehgal A, et al. A Drosophila model for Angelman syndrome. Proc Natl Acad Sci U S A. 2008;105 : 12399–12404. doi: 10.1073/pnas.0805291105 18701717
77. Chen WF, Maguire S, Sowcik M, Luo W, Koh K, Sehgal A. A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry. 2014; doi: 10.1038/mp.2014.11
78. Gilestro GF, Cirelli C. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics. 2009;25 : 1466–1467. doi: 10.1093/bioinformatics/btp237 19369499
79. Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife 2014; doi: 10.7554/eLife.01473
80. Zhang H, Tan J, Reynolds E, Kuebler D, Faulhaber S, Tanouye M. The Drosophila slamdance gene: a mutation in an aminopeptidase can cause seizure, paralysis and neuronal failure. Genetics. 2002;162 : 1283–1299. 12454073
81. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4 : 44–57.
82. Huang DW, Sherman BT, Lempicki RA. Bioinformatic enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37 : 1–13. doi: 10.1093/nar/gkn923 19033363
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



