-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
Malassezia are the dominant eukaryotic residents of human skin and are associated with the most common skin disorders, including dandruff, atopic dermatitis, eczema, and others. Despite significant effort, the role of Malassezia in skin disease and homeostasis remains unclear. Malassezia are also unique among fungi by requiring lipids for growth, but the breadth and genetic basis of their lipophilic lifestyle has not been comprehensively studied. Here we report the complete genomes of all 14 Malassezia species (including multiple strains of the most common species found on humans) and systematically identify features that define the genus and its sub-lineages, including horizontally transferred genes likely to represent key gain-of-function events and which may have enabled evolution of the genus from plant to animal inhabitants. Genus wide expansion of lipid hydrolases and loss of carbohydrate metabolism genes underscore the entire genus’ gradual evolution to lipid-dependency, which was confirmed even in the previously thought to be lipophilic M. pachydermatis, via genomics with experimental confirmation. Finally, these reference genomes will serve as a valuable resource for future metagenomic investigations into the role of Malassezia species in normal healthy skin and diseases.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005614
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005614Summary
Malassezia are the dominant eukaryotic residents of human skin and are associated with the most common skin disorders, including dandruff, atopic dermatitis, eczema, and others. Despite significant effort, the role of Malassezia in skin disease and homeostasis remains unclear. Malassezia are also unique among fungi by requiring lipids for growth, but the breadth and genetic basis of their lipophilic lifestyle has not been comprehensively studied. Here we report the complete genomes of all 14 Malassezia species (including multiple strains of the most common species found on humans) and systematically identify features that define the genus and its sub-lineages, including horizontally transferred genes likely to represent key gain-of-function events and which may have enabled evolution of the genus from plant to animal inhabitants. Genus wide expansion of lipid hydrolases and loss of carbohydrate metabolism genes underscore the entire genus’ gradual evolution to lipid-dependency, which was confirmed even in the previously thought to be lipophilic M. pachydermatis, via genomics with experimental confirmation. Finally, these reference genomes will serve as a valuable resource for future metagenomic investigations into the role of Malassezia species in normal healthy skin and diseases.
Introduction
Over 100 years ago Malassezia was recognized as an inhabitant of human skin and implicated in a common skin disorder i.e. seborrheic dermatitis [1]. Since then, Malassezia has been found on the skin of all tested warm blooded animals [2,3], including dogs, horses, pigs, goats, cats and lambs [4–8], and associated with other common skin disorders including dandruff [9], atopic eczema/dermatitis, pityriasis versicolor, seborrheic dermatitis, and in systemic disease [10]. Recent investigations of the skin microbiome using culture-free approaches have highlighted the overwhelming dominance of Malassezia among eukaryotes on all human surface body sites, with only the exception of three foot sites [11,12]. Other studies have suggested that they are abundant in body sites beyond skin, including the human oral microbiome [13], but a systematic characterization of Malassezia species and their functional repertoires represented in metagenomic datasets has been hampered by the lack of reference genomes (only 2 out of 14 known species have reference genomes i.e. M. globosa [2] and M. sympodialis [14]). In addition, several reports have suggested that Malassezia-like organisms are found in a wide range of environmental habitats, from deep sea sediments, hydrothermal vents and arctic soils, to marine sponges, stony corals, eels, lobster larvae, and nematodes [15]. These studies have relied on high-identity DNA sequence matches to short amplified barcode regions, but concerns about amplification bias or laboratory contamination raise doubts about the results and the lack of a comprehensive genus-wide genomic resource for known species has made it challenging to investigate this question further.
Malassezia belong to the class Malasseziomycetes in the subphylum of Ustilaginomycotina, (phylum of Basidiomycota, Kingdom of Fungi) [16], which are otherwise comprised exclusively of more than 1,500 species of plant pathogens [17]. Other known fungal residents on human skin, such as Candida albicans and the dermatophytes are in distant branches of the fungal tree of life and are likely to have evolved independently to adapt to life on animal skin [18,19]. The genetic basis of the unique lipophilic nature of Malassezia and its adaptation to animal skin (putatively starting from an ancestral state as a plant or soil resident) is thus an intriguing and open question. Answers to this question could also serve as the basis for developing new anti-fungals and therapeutics for associated skin disorders. Analysis of the two existing Malassezia genomes [2,14] highlights that their small genomes (among the smallest for free living organisms in the fungal kingdom) likely contain only the minimal complement of information necessary for existence in their specific ecological niche. In this context, the expansion of several gene families as noted before (e.g. lipases, phospholipases, and aspartyl proteases) may point to their functional importance [2,14]. However, it has not been clear if these observations are indeed genus-wide features. In addition, the limited availability of reference genomes has precluded the systematic characterization of genomic features unique to Malassezia (such as gene gain or loss, horizontal gene transfers, linkage between mating type loci, and regions undergoing positive or negative selection) that could serve as the basis of understanding its unique physiology and niche adaptation.
To address this limitation, we sequenced and assembled high-quality, annotated genomes of all known Malassezia species and multiple strains of the species most common on humans (including a re-annotation of existing references), representing a 7-fold increase in available reference genomes (from 2 to 14), and providing a comprehensive genomic resource for the investigation of Malassezia biology and its ecological distribution (24 Malassezia strains in total). Showcasing this, we established the abundance and surprising diversity of Malassezia species on various human skin sites and their scarcity in other environments. We then used comparative genomic analysis to systematically compare Malassezia genomes with a broad panel of fungal genomes to reveal genomic features unique to Malassezia, including hundreds of gene gain and loss events, gene family expansions, and positive selection events. Our analysis revealed several hallmarks of Malassezia genomes, including key horizontally transferred genes (a few of bacterial origin) that we characterized functionally and which may be prime candidates to explain the emergence of host and niche-adaptation in Malassezia. A larger set of genes (>700) were found to be lost in all Malassezia compared to other Basidiomycota, with an enrichment for glycosyl hydrolases and genes involved in carbohydrate metabolism, concordant with adaptation to a carbohydrate-deficient environment. Combining genomic analysis with an experimental re-evaluation of culture characteristics, we revert previous assumptions and established the likely lipid dependency of all Malassezia species. Finally, our analysis of lineage-specific gene family expansions revealed extensive turnover in the gene repertoire of Malassezia and underlined the importance of secretory lipases, phospholipases, aspartyl proteases and other peptidases in the experimentally observed lipid specificity of this genus.
Results
Establishing a comprehensive genomic resource for the Malassezia genus
Genome sequences for all 14 known Malassezia species, including multiple strains of the more widely studies species (24 in total) were obtained by high-throughput sequencing and de novo assembly (Table 1; see Methods). The high coverage data (median coverage of 322X) was systematically assembled with an assembly pipeline incorporating parameter optimization, contig construction, scaffolding and gap closure steps to produce assemblies with a median N50 of 54 kbp and a maximum N50 of 1.4 Mbp (Table 1). In particular, we noted that the N50s of the four new M. globosa assemblies were comparable to that of a gold-standard reference M. globosa genome [2] obtained previously using Sanger sequencing with significant directed finishing (Table 1). Assembly sizes typically varied from 7.2 Mbp (for M. restricta) to 9.0 Mbp (for M. globosa) as expected but we noted that 4 out of the 6 M. furfur assemblies were twice this size, suggesting that they might have undergone whole genome duplication or hybridization events (see S1 Text and S1 Fig for further details).
Tab. 1. Assembly and annotation statistics for Malassezia genomes in this study. 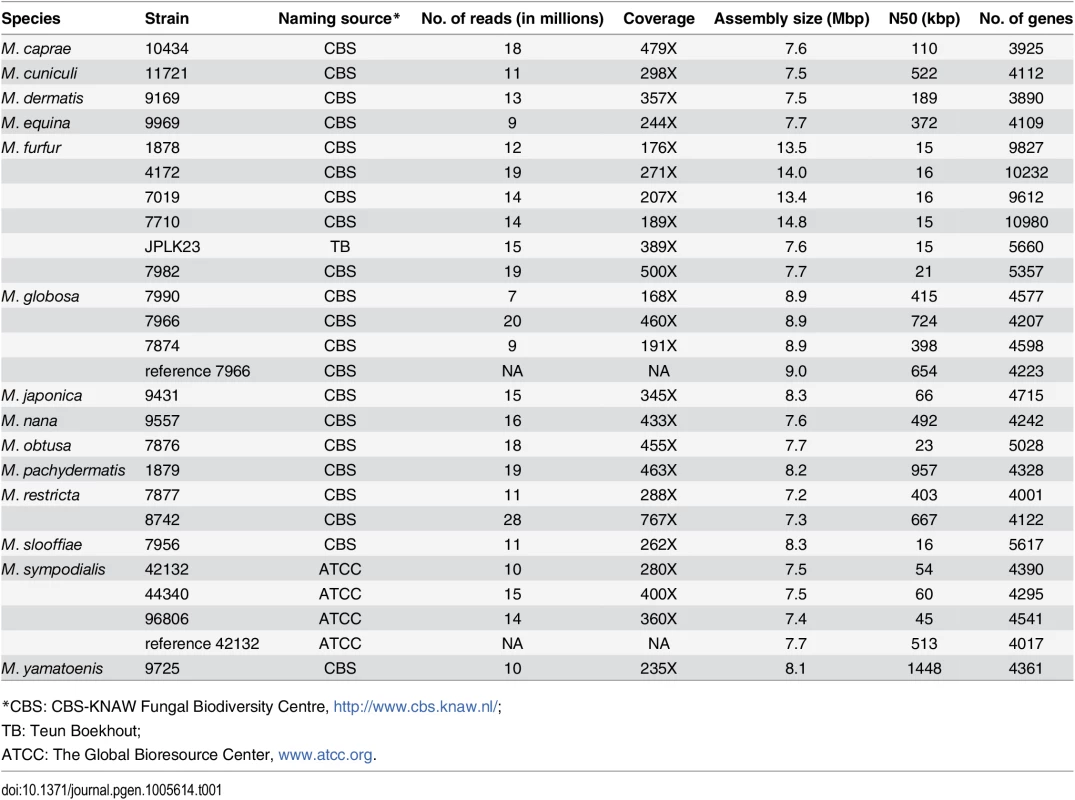
Note the statistics for the previously reported M. globosa [2] and M. sympodialis [14] assemblies are provided for reference. To assess the completeness of our assemblies we evaluated them using matches to a well-established set of core eukaryotic genes (CEGs) [20]. As can be seen in Fig 1A, our de novo assemblies are comparable to the reference genomes of M. globosa and M. sympodialis [2,14] in terms of the number of complete and partial CEGs identified. In addition, comparison to the gold standard Saccharomyces cerevisiae genome suggests that our assemblies are more than 95% complete (Fig 1A). We assessed the correctness of our assemblies by comparing them to the published reference genomes of the same strains (M. globosa 7966 and M. sympodialis 42132). Overall, we found that our assemblies agreed very well with that of the reference genomes (Fig 1B, S1 Table), showing a high degree of colinearity (<5 breakpoints per 100 kbp in our assemblies) and identity (>99.92%) as expected from the comparison of two high-quality assemblies of the same strain (Fig 1B, S1 Table). Assuming that the reference genome is correct, we noted that the observed differences in our assembly affected <0.5% of all genes in M. globosa, indicating that our assembly is of particularly high quality in genic regions.
Fig. 1. Correctness and completeness of Malassezia assembly and annotation. 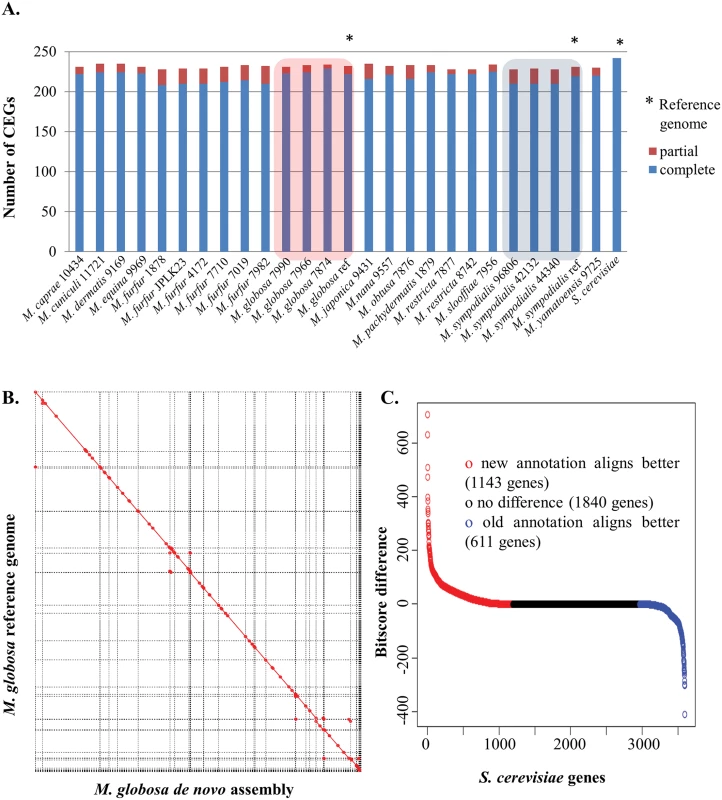
A) Assembly completeness in terms of partial and complete core eukaryotic genes that can be detected in each genome. As shown here, the assemblies from this study are comparable to published references for M. globosa and M. sympodialis and are very similar to the gold-standard S. cerevisiae genome. B) Whole-genome alignment of the assembly of M. globosa 7966 in this study as compared to the published reference, highlighting the robust assembly and the lack of clear misassemblies. C) Comparison of an annotation of M. globosa 7966 in this study with the reference annotation, using alignments to S. cerevisiae as a gold-standard. Y-axis indicates the BLAST bitscore difference between the top matches from the new and old annotations to the same S. cerevisiae protein. X-axis indicates the number of S. cerevisiae proteins. Red circles indicate S. cerevisiae proteins with a better match to the new annotation. Blue circles indicate S. cerevisiae proteins with a better match to the reference annotation. To systematically annotate the protein-coding complement of the genomes, we used an iterative and automated pipeline that combines transcriptome data (where available), ab initio predictions, and protein evidence from related species (see Methods). We evaluated results from this pipeline by comparison to the manually curated annotations for the M. globosa 7966 reference genome and using the S. cerevisiae annotations as gold standard. As shown in Fig 1C, as a whole, annotations from our pipeline match the S. cerevisiae proteome better (>1,100 S. cerevisiae proteins are better aligned to the new annotation versus ~600 proteins for the reference annotation) indicating that we have a comparable or better annotation. In addition, the new annotation has more matches to known domain families than the original annotation (unique PFam domains and total PFam domains, pfam.xfam.org/ Table 2) as well as improved identification of intron-exon boundaries, highlighting the value of the iterative approach employed here (the utility of transcriptome data is highlighted in S2 Table and the lack of alternative isoforms is noted in Methods). As observed before, we found that Malassezia species code for a compact proteome of ~4,000 genes with the exception of M. slooffiae and M. furfur (after excluding those with doubled genome sizes) which appear to have a somewhat larger set of genes (Table 1). It is of note that the lower N50 of the M. furfur and M. slooffiae assemblies may cause a spurious increase in gene count due to coding regions being split.
Tab. 2. Comparison of annotation quality for the iterative annotation pipeline in this study with a reference annotation. 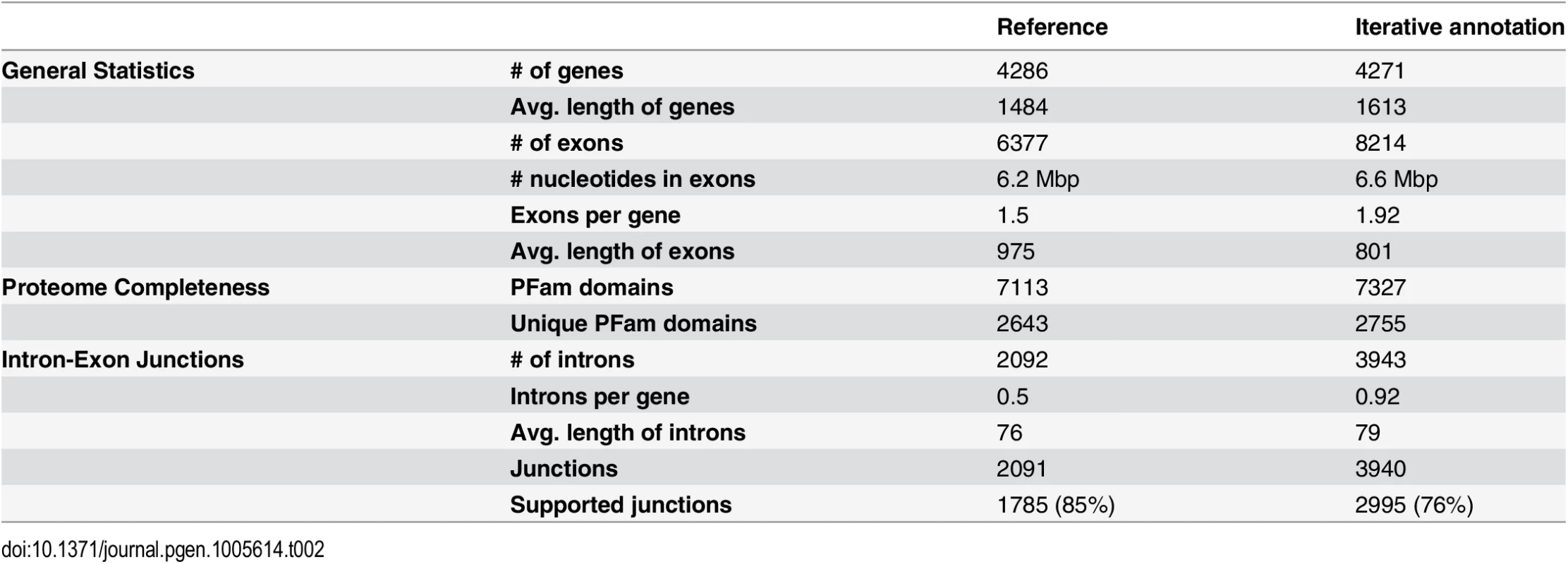
Results shown are for the M. globosa genome. To showcase the utility of this genomic resource, we studied the distribution and diversity of Malassezia species in the environment and in human microbiomes by extensive reanalysis of publicly available metagenomic datasets. We first used in silico benchmarks to confirm our analysis pipeline is highly sensitive and specific in identifying Malassezia species from short, shotgun metagenomic reads (S2 Fig). We then applied this approach to a wide spectrum of environmental metagenomic datasets including ocean (www.microb3.eu/osd), marine sediments [21], soil [22], and rhizosphere samples [23]. Despite recent reports of Malassezia-like organisms being widely distributed in the environment [15], we were unable to detect evidence for this, suggesting they are present in abundances below our detection limit or are sufficiently diverged from known Malassezia species to elude our detection based on known genomes. Similarly, we re-analyzed oral microbiome samples from six different oral sites (five samples from each site) and found no evidence for the presence of Malassezia, in contrast to a recent report [13]. These results are consistent with either contamination artefacts or Malassezia being presented at lower abundance in the oral mycobiome and thus being detectable only using more sensitive 18S rRNA sequencing approaches which utilize an amplification step [13]. In contrast, analysis of metagenomic datasets from different sites on healthy human skin [12] readily revealed the abundance and diversity of Malassezia (detected in 247 out of 280 samples analyzed, from 18 sites on 15 adults and two children; Fig 2A). In general, our analysis reconfirmed that M. globosa and M. restricta are the two most abundant species on human skin, found in 199 samples on 16 individuals and 247 samples on 17 individuals, respectively. M. sympodialis is a distant third, detectable in 69 samples on 12 individuals, though it is the most abundant species in several samples (Fig 2A). In addition, nine other species were also found either less frequently or in lower abundance. For example, M. slooffiae, which has previously not been detected on human skin via ribosomal RNA sequencing [11], was found in high abundance in several samples, mostly from one individual (Fig 2A). It is also accompanied in three samples by M. obtusa (in one individual) and apparently excluding M. sympodialis (in two individuals) (Fig 2A).
Fig. 2. Characterizing the diversity of Malassezia in skin samples. 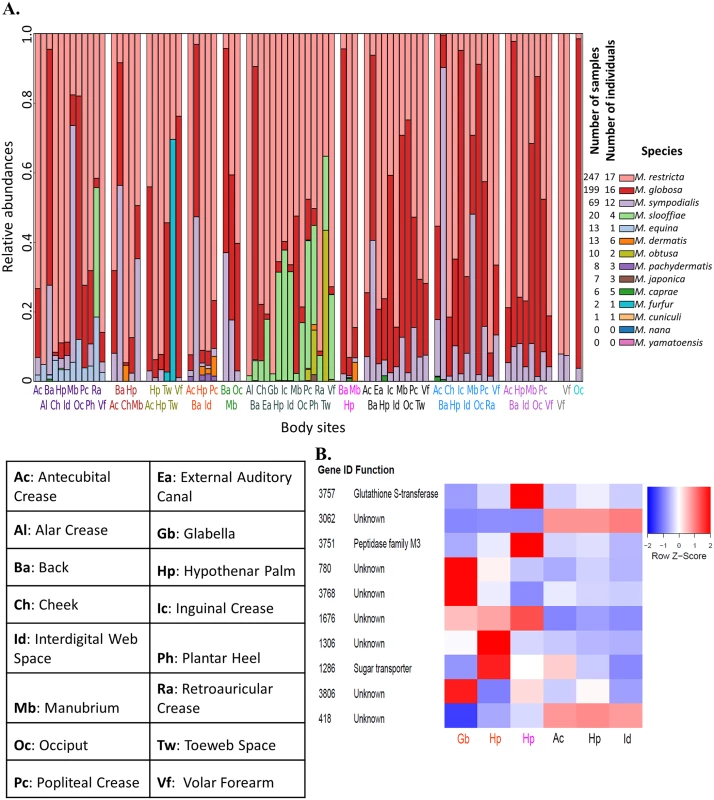
A) The relative abundance of various Malassezia species (y-axis) in skin samples from different body sites (labels on the x-axis) and individuals (separated by white columns) is depicted. Samples where >99% of reads came from M. globosa and M restricta are not shown here. The numbers of samples and the numbers of individuals in which each species was found is indicated in the legend on the right. B) Z-score transformed normalized read counts for the top 10 copy number variable genes in M. restricta 7877 (measured in terms of coefficient of variation of normalized counts) across six skin samples. We further probed the relative abundance of various genes from the Malassezia pan-genome in skin metagenomic samples [12] to identify those that are highly variable, likely reflecting strain-level variations in the commensal population [24]. As M. restricta is the most abundant species on human skin, we were readily able to find samples with sufficient read coverage of the genome (>5X) for robust analysis (see Methods). Genome-wide we found significant copy number variations in >100 genes across 6 skin samples and 4 body sites (Fig 2B, S3 Table), though analysis of more samples is likely to reveal even more variable genes. Our proof-of-concept analysis revealed several highly variable genes including genes of unknown function, a glutathione S-transferase (known to be involved in detoxification of xenobiotic substrates), a peptidase and a sugar transporter (Fig 2B). As changes in carbohydrate and lipid metabolism are key features of Malassezia genomes (see Results below), this analysis suggests our reference genomes will serve as an important resource for characterizing strain variations contributing to different phenotypes on human skin.
Identifying genetic features that define the Malassezia genus
Leveraging the comprehensiveness of our genomic resource for Malassezia, we set out to compare it against a broad panel of 16 fungal genomes (including all sequenced species in Ustilaginomycotina, a few other Basidiomycetes and several Ascomycetes as outgroups). Using a genome-wide multi-gene approach we first established a robust phylogenetic view of Malassezia’s relationship with other fungi and each other (Fig 3A and 3B; see Methods). In contrast to earlier reports placing Malassezia among the Exobasidiomycetes [17] or Ustilaginomycetes [25], our analysis suggests that it may be an isolated group (namely the class Malasseziomycetes) in the subphylum of Ustilaginomycotina, in agreement with Wang et al [16]. However, Wang et al placed Ustilaginomycetes close to Malassezia, and placed Exobasidiomycetes as the basal group [16]; our tree placed Malassezia as the basal group, indicating early divergence from its plant-pathogenic relatives. Within Malassezia, our phylogeny supports three main clusters (Fig 3B): Cluster A consists of fungemia-causing species M. furfur [26] and three other species (M. japonica, M. obtusa, and M. yamatoensis), rarely found on healthy human skin (Fig 2A); Cluster B includes a sub-cluster of the most common human skin residents M. globosa and M. restricta [11], the slightly less common M. sympodialis [14]) as well as related species in another sub-cluster; Cluster C consists of two outliers, M. cuniculi and M. slooffiae (Fig 3B), both of which are rare on human skin (Fig 2A). Notably, while broadly in agreement, our phylogeny disagrees with the placement of several species compared to a four-gene tree [27] and an earlier AFLP based tree [28], though its concordance with the mitochondrial phylogeny as well as alternative approaches to reconstruct phylogeny (S3 Fig) suggest that it is likely to be more reliable. Note that, as expected, our phylogeny also confirms the definition and molecular distinctness of various Malassezia species as well as the entire genus.
Fig. 3. Phylogenetic relationships and lineage specific events in the Malassezia genus. 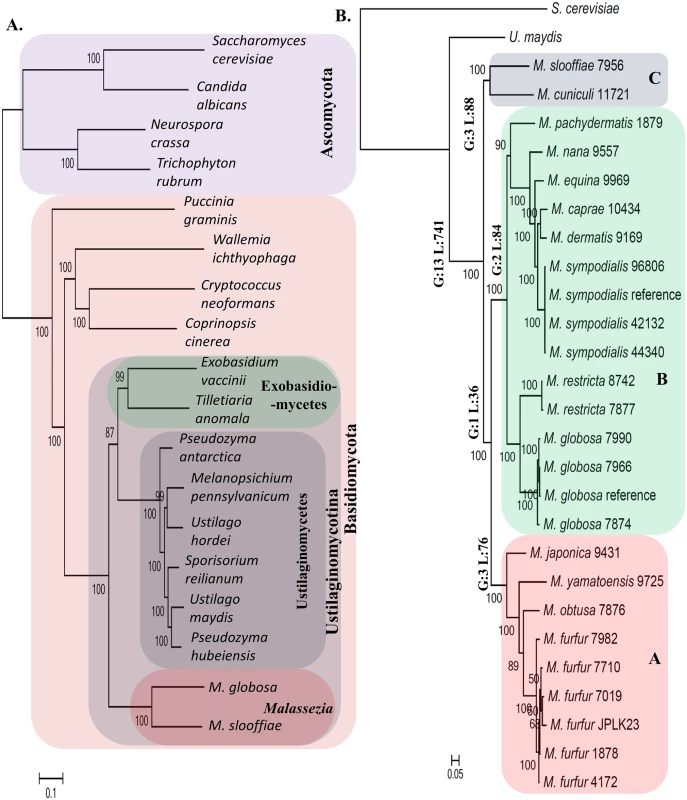
A) The relationship of the Malassezia genus with respect to other fungi with sequenced genomes. Malassezia seem to form a distinct group in the subphylum of Ustilaginomycotina contrary to earlier reports. B) An expanded phylogeny of Malassezia that includes all known species in the genus. Major lineages of the genus are annotated with the number of lineage specific events that were identified in this study (G: gene family gain; L: gene family loss based on PFam analysis; see S3 Table for details). Horizontal numbers on each branch are bootstrap values. We then used comparative genomics to reveal genomic elements unique to Malassezia, identifying a small set of 13 functional domains (PFam families [29]) to be Malassezia-specific, compared to a much larger set of 741 domains likely lost in the common ancestor to all Malassezia (Fig 3B, S3 Table; see Methods), in addition to gene family expansions and signatures of selection (S3 Table). The set of Malassezia-specific genes contains mainly genes of unknown function and is not enriched for a specific functional category (S3 Table). On the other hand, the set of genes lost in all Malassezia varies widely in function, from genes encoding enzymes to transcriptional regulators to known accessory genes (S3 Table). However, we did detect significant enrichment for two lost functional categories, specifically, enzymes involved in carbohydrate metabolic process (q-value < 4.5×10−4) and in hydrolysis activity (hydrolyzing O-glycosyl compounds; q-value < 4.5×10−4), as expected for a genus of skin-adapted fungi that use lipids as their main carbon source. In addition, we also noted that the gene encoding the fatty acid synthase (FAS) was missing in all Malassezia, indicating that the genus is lipid-dependent and not just lipophilic as suggested earlier [30]. The idea that a subset of Malassezia is not lipid-dependent is based on the observation that some M. pachydermatis isolates can grow in media (Sabouraud-dextrose agar) without added lipids, though it does require fatty acids to grow in simple defined media [31]. We experimentally re-investigated the contents of Sabouraud-dextrose agar media and noted that the added peptone contains 0.6% lipid, with 6 μg of palmitic acid per gram of peptone and lesser amounts of other fatty acids. Furthermore, in 2 X YNB defined media, M. pachydermatis strains (1879 and 7550) were able to grow only in the presence of added lipids confirming the unique lipid-dependent nature of all Malassezia species (S2 Text).
At the structural level we confirmed linkage between the two mating loci (MAT) in three Malassezia species (belonging to clusters A and B, S4 Fig; i.e. likely a pseudo-bipolar configuration), a feature that is hypothesized to contribute to pathogenesis [32], but is unique to Malassezia among Basidiomycetes (S3 Text, S4 Fig and S4 Table). We also noted a loss of the RNAi pathway and a concomitant reduction in transposon element density in all Malassezia genomes (S4 Text). Finally our selection analysis revealed a diverse set of noteworthy genes undergoing positive selection (S5 Text and S3 Table), with the strongest signal being observed in a protein (with match to the PFam domain PF12481) known to be induced by aluminum, a common component of deodorant, shaving cream and gel [33].
Acquisition and function of horizontally transferred genes in the Malassezia genus
The Malassezia-specific gene families identified using known domain families (PFam) contain many interesting candidates for horizontally transferred genes (HTGs). We also used a clustering based approach to expand this analysis to gene families with or without PFam domains, obtaining an additional set of 44 Malassezia-specific gene clusters, most of which have unknown function (S3 Table). Finally, we used two additional approaches based on similarity searches and phylogenetic analysis to catalog genes with more subtle evidence of horizontal transfer from bacteria into Malassezia [34,35] to identify 6 additional genes, many of which appear to be associated to oxidative stress response (including two oxidoreductases and one catalase; S3 Table and S6 Text).
We further investigated the role and function of three specific gene families that likely represent key gain-of-function events in Malassezia. The first of these is unique as it is the only one found to be conserved in all Malassezia. This gene family is defined by matches to the PFam domain PF06742 (a domain of unknown function) and is present in a single gene copy in all Malassezia, except for the M. furfur hybrids and M. slooffiae which have two gene copies. Its universal presence in all Malassezia and absence in all other Basidiomycetes suggests that a lateral gene transfer event in the ancestor of all Malassezia is the most parsimonious explanation. In addition, while the likely source of this gene could not be determined due to its ancient origin, we noted that it is seen in diverse and often pathogenic bacteria (e.g. Mycobacterium tuberculosis, Listeria monocytogenes and Salmonella enterica) and fungi (e.g. Aspergillus flavus) and is surprisingly well conserved (http://pfam.xfam.org/). Furthermore, we noted that the gene in M. globosa is significantly up-regulated in nutrient deficient conditions (Fig 4A) while its ortholog in Chlamydomonas reinhardtii is dramatically up-regulated under sulfur depletion conditions (http://tinyurl.com/nyjd3md), suggesting that they might serve an essential biological role. Proteomics evidence from M. sympodialis [14] indicates that this gene is likely translated (Fig 4C) and secreted (based on a signal peptide match). Homology modeling predicted its likely function to be a glycosyl hydrolase (EC 3.2.1.x, www.genome.jp/kegg/) (Fig 4D, S7 Text). The exact substrate remains to be determined but based on structural considerations there is slightly higher similarity to beta-galactosidases or mannosidases near the predicted substrate binding site (S7 Text). In addition, hydrolyzing activity on fungal cell wall glucans, which have been determined to be mainly (1->6) beta-D-glucans in M. sympodialis [36], cannot be excluded based on profile sequence searches (S7 Text). Adding to the functional context, co-expression analysis in M. globosa revealed that this putative hydrolase gene’s expression is highly correlated with that of an aspartyl protease (mgl_641, Pearson Correlation = 0.955, FDR = 2.16×10−20). Interestingly, in another fungus, Candida glabrata, an aspartyl protease is required for pH-change-induced reduction in total beta-glucan levels in the cell wall [37] which could be achieved by coordinating with a beta-glucan hydrolase. Further experimental work should help clarify this hypothesis and the gene’s impact on Malassezia biology.
Fig. 4. Functional characterization of novel, putative horizontally transferred genes in Malassezia. 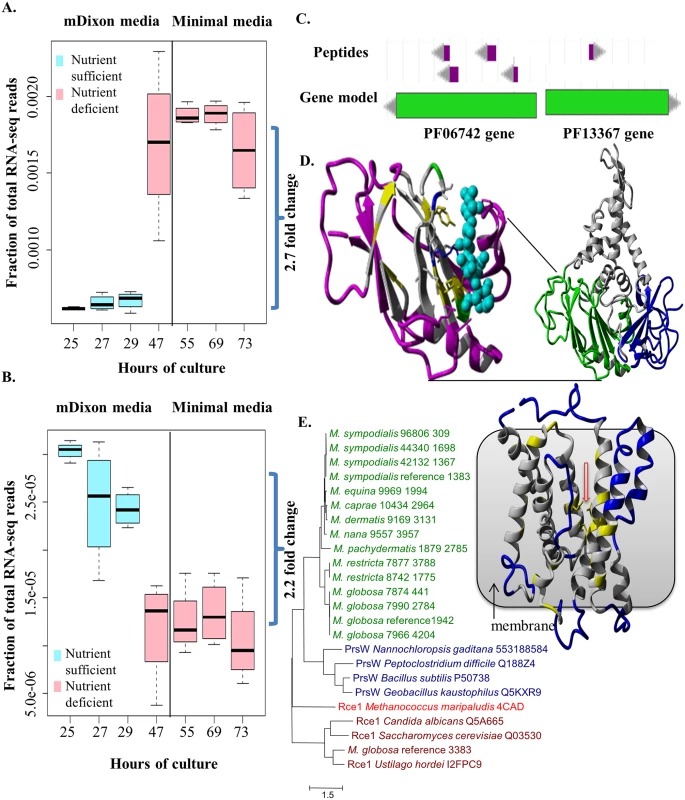
A) Upregulation of the gene containing the PFam domain PF06742 in nutrient deprived conditions in M. globosa. B) Downregulation of the gene containing PF13367 in nutrient deprived conditions in M. globosa. C) Peptide evidence for the genes containing PF06742 and PF13367 in M. sympodialis. D) Structural model of representative gene containing PF06742 (MGL_833 from M. globosa 7966 reference). Right side: full model with domains in different colors. Left side: zoom to Jelly Roll domain with predicted glycosyl hydrolase function (Coloring shows relative similarity to known hydrolase enzymes. Purple: structurally different; Gray: structurally same, amino acid different; Yellow: structurally same, amino acid identical, hydrophobic; Blue, Red, Green: structurally same, amino acid identical, non-hydrophobic [blue: positive charge; red: negative charge; green: polar]; Cyan: substrate sugar). E) Left side: Maximum likelihood phylogenetic tree of the Malassezia PrsW-like family (green) with representatives from PrsW (blue) and Rce1 (brown) families including one resolved structure (red). Gene IDs are specified behind species names and strain IDs. Right side: homology model of a PrsW-like protease (MG7966_4204 from M. globosa 7966; contains the PFam domain PF13367) with red arrow indicating conserved glutamates and histidines coming together to form the active site. Coloring: Blue: structurally different; Gray: structurally same, amino acid different; Yellow: structurally same, amino acid identical (all types). The second gene family, with a match to the PFam domain PF00199, likely represents a case of inter-kingdom gene transfer (from bacteria) of a catalase gene whose product carries out the key function of removing the reactive oxygen species H2O2 [38]. Intriguingly our phylogenetic analysis suggests that while, in general, all Malassezia have one catalase that is more closely related to bacterial catalases (from Blastomonas and Sphingomonas), M. slooffiae has an additional, presumably ancestral, catalase that is more closely related to fungal catalases (S5 Fig). The acquisition of a bacterial catalase in Malassezia could have provided a selective advantage in adapting to life on a new host, especially considering the numerous secreted proteins (for example, GMC oxidoreductases) that could generate hydrogen peroxide [2]. Within the genus, catalase genes are missing in two species, M. restricta and M. pachydermatis, and this was confirmed by BLAST search [39] to both genomes and proteomes. For M. restricta, absence of catalase enzyme activity has been confirmed by enzyme test [40]; given the fact that Malassezia live in an aerobic environment on skin [1] alternative metabolic pathways might exist to detoxify oxygen in M. restricta. For M. pachydermatis, catalase activity has been observed [40] and alternative catalases might exist which are sufficiently diverged from catalases in other Malassezia species.
The third gene family, defined by matches to the PFam domain PF13367 (a family of putative PrsW proteases), was found to be present in all genomes of Malassezia cluster B (containing species commonly found on human skin) while being absent in other Malassezia and Basidiomycetes (S3 Table), suggesting that it may have been horizontally acquired in the lineage leading to cluster B. Genes belonging to this gene family were readily found in skin resident bacteria (e.g. Propionibacterium, Streptococcus and Staphylococcus) as well as a few parasitic protists (e.g. Toxoplasma gondii, Neospora caninum, Cryptosporidium and Plasmodium) (http://pfam.xfam.org/). In Bacillus subtilis, PrsWs sense antimicrobial peptides and then cleave the anti-ϬW factor to activate the ϬW factor [41]. However, in the absence of the anti-ϬW factor or the ϬW factor in Malassezia, these genes are likely to have a different role. We confirmed that this gene is expressed and translated (Fig 4B and 4C) and significantly down-regulated in nutrient deficient conditions (Fig 4B). PrsW-like proteases belong to the endopeptidase family M82 that is related to the family M79 (that includes Rce1 peptidases) (Fig 4E) [41,42] with a recently resolved crystal structure [43]. Homology modeling confirmed the known catalytically important residues [41,43] to be conserved between these two families (S6 Fig, S7 Text) and located in the center of the transmembrane bundle forming the active site (Fig 4E). The Rce1 peptidases typically cleave C-terminal tripeptides from isoprenylated proteins (e.g. fungal mating factor a) [43]. However, this is not likely the function of Malassezia PrsW-like family due to the presence of direct Rce1 homologs in Malassezia (e.g. MGL_3383, Fig 4E).
Gene family expansion and extensive turnover underlie niche specificity in Malassezia
Malassezia are known to have varying host tropism and highly specific preferences for environmental niches and food sources [3,44]. For example, some highly sebaceous sites such as scalp (including occiput) and back are typically dominated by M. globosa [11]. To further understand niche-specificity in Malassezia, we evaluated their preference for growth in various lipid media using “Lipid Assimilation Assays” (see Methods). These experiments highlight a strong specificity in Malassezia’s preference for lipids (S4 Table) that is not well correlated with their phylogenetic relatedness. For example, M. furfur and M. sympodialis are functionally similar as the most robust of the lipid-dependent species in culture, sharing the broadest range of lipids that support growth (Fig 5A, S5 Table). However, they are not closely related and are placed in different sub-clusters of the Malassezia phylogeny (Fig 3B). Also, the closely related species M. globosa and M. restricta have different lipid assimilation profiles (Fig 5A, S5 Table). To understand the genetic basis of these phenotypes, we reexamined the list of gene family expansions and selection in Malassezia. Strikingly, the most expanded gene family in Malassezia was found to be a phospholipase family, and a secretory lipase family was also among the list of 13 families with a 2-fold increase in median copy number in Malassezia compared to other fungi (S3 Table). Lipase and phospholipase activities have been detected in multiple Malassezia species [45,46]. Their genes are highly expressed in vivo on human scalp [2,45,47], and are thought to play an essential role in supporting their growth. Therefore, we hypothesized that expansion of these gene families might explain Malassezia niche-specificity. Other than lipases, many peptidases in multiple families are found in the most expanded gene families in Malassezia, underlining their importance in Malassezia biology (S3 Table, S8 Text).
Fig. 5. Lipid specificity and extensive turnover in the lipase gene family. 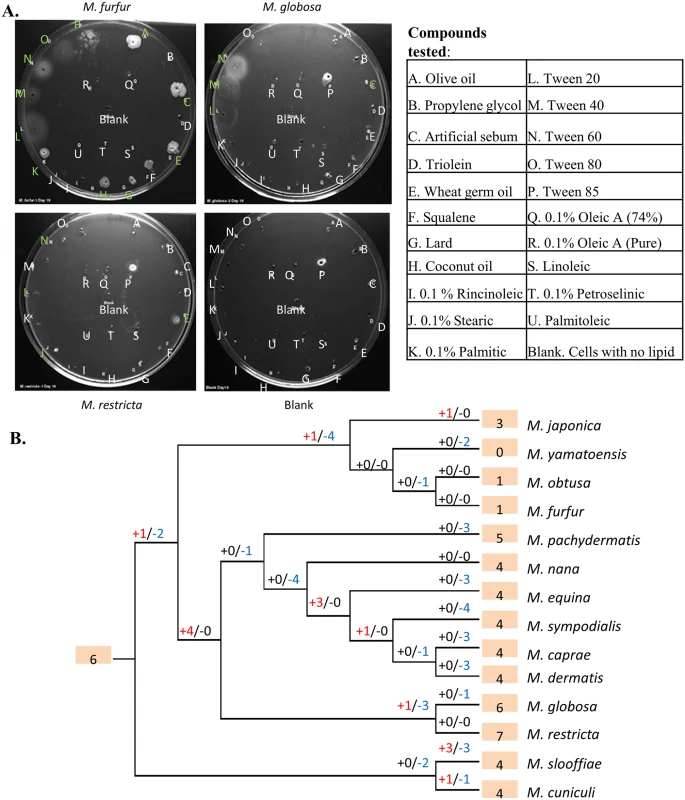
A) Representative lipid assimilation assay images. Letters correspond to lipid wells (S5 Table). White letters indicate no growth, while green letters indicate growth on visual scale. B) Gene gains and losses in phosphoesterase family PF04185, where “+” indicates the number of gene gain events while “-” indicates the number of gene loss events. Shaded numbers indicate the estimated gene copy number in the most recent common ancestor and gene copy numbers in the observed species. To understand the evolution of these families, we inferred parsimonious reconstructions of gain and loss events (see Methods). In particular, the most expanded gene family, a family of phospholipases (phosphoesterases, PF04185) showed a striking pattern of extensive turnover, with a dramatic expansion of the family in the ancestor of cluster B species followed by lineage specific losses (Fig 5B). Recent duplications were also observed in M. japonica, M. slooffiae and M. cuniculi while cluster A species seem to have experienced a significant contraction in this gene family that is thought to be relevant to fungal pathogenesis [48]. Extensive turnover in the lipase gene repertoire of Malassezia was also seen in the secretory lipases (PF03583) (S7 Fig). Species-specific duplication events were found in seven species and, in particular, there has been rapid recent expansion of the family in M. slooffiae and M. pachydermatis (S7 Fig). We observed frequent lineage-specific duplication and loss of genes in other lipase families as well (e.g. PF01764) and together these could explain the complex patterns of lipid-specificity observed in Malassezia. Further experiments should help establish the exact roles specific lipase genes play in the process of human colonization and pathogenesis.
Discussion
Malassezia, while found on all humans and associated with many common human skin diseases, are poorly understood in large part due to a lack of genomic tools. Here, we report generation and analysis of the genomes of all 14 accepted Malassezia species, including multiple strains of those most commonly found on human skin (for a total of 24 strains). Malassezia are unique in several ways, including their adaptation to life on animal skin, their dominance as eukaryotic residents on human skin (in contrast to the diversity seen among prokaryotic commensals), and their lipid-dependent lifestyle. Even within Malassezia, we noted there is substantial variability in preference for food sources and thus environmental niches. As a first step, the analysis in this study serves to systematically catalog and characterize genomic features unique to Malassezia and its lineages, which could then be associated with the observed phenotypes. This was aided by the characterization of all known species in the genus as well as multiple strains for key species, allowing robust conclusions to be drawn despite potential analysis pitfalls. Correspondingly, several of the genes identified in this study are prime candidates for further experimental study. It is tempting to speculate, for example, that the gene containing the PFam family PF06742 serves an essential function in Malassezia such that loss of the gene could be lethal. As this gene was likely horizontally acquired by the ancestor of all Malassezia, its function could also be tied to the origin of the genus, particularly if it relates to utilizing energy sources from the host. Similarly, the role of PF13367 could be linked to the ability of cluster B Malassezia to thrive on human skin. In general, Malassezia are not facile experimental systems as they are challenging to cultivate and typically recalcitrant to genetic manipulation. In this context, recent success in performing gene deletion in M. furfur is encouraging (Giuseppe Ianiri and Alexander Idnurm, personal communication) and could enable in vivo functional characterization.
Among other gene families of interest, particularly due to their association with niche-specificity, are several lipase families. Interestingly, there are a total of 25 lipases found in the two major lipase families (PF03583 and PF01764) in M. slooffiae, a species found on both animals and humans [9,49] with little known about its involvement in diseases. This is the most in any haploid Malassezia strain (S3 Table), with the closely related M. cuniculi having only 16 lipases and M. globosa 14 (S3 Table). Many lipases in M. slooffiae are derived from unique species-specific duplication events (S7 Fig). However, it remains an open question if M. slooffiae is indeed able to leverage this large arsenal of lipases to utilize a wider range of lipids and hence live in more diverse ecosystems. Intriguingly, we observed in our skin metagenomic datasets that the only three samples with relatively high abundance of M. obtusa are in co-occurrence with M. slooffiae (Fig 2A), suggesting the possibility that M. slooffiae breaks down lipids for utilization by M. obtusa, a rare and hard-to-culture species. Further studies are needed to establish this relationship but it is clear that the availability of genomes for all Malassezia species will be critical to understanding their distribution and role in human diseases.
The question of whether Malassezia or Malassezia-like species are abundant in habitats other than on the skin of warm-blooded animals is an intriguing one. Our analysis of samples from varying habitats suggests that they are either not common or not similar enough to known Malassezia species. With the availability of a catalog of Malassezia-specific genes, sensitive models can be built to detect remote homologies to these sequences [29] as a way to search for distant Malassezia-like species in the environment. This in turn should help clarify the emergence and role of Malassezia as a skin-adapted fungus.
Fungal mating is hypothesized to play an important role in pathogenesis by increasing genetic diversity [32,50]. The observation that mating loci are linked in three species in cluster A and B, suggests that bipolar or pseudo-bipolar mating systems may be present in all Malassezia. The former is observed in most human pathogenic fungi [32] and in ascomycota, and the latter is observed in M. sympodialis [14], despite the fact that the tetrapolar mating system is more common in basidiomycota [32]. Further studies using sequencing technology which enables longer read lengths and hence analysis of larger genomic structural elements are needed to define the presence and impact of pseudo-bipolar mating systems in Malassezia and any role in their pathogenesis.
Methods
Malassezia culture, extraction of DNA and RNA, and library preparation
Malassezia was grown on mDixon media for DNA extraction, and M. globosa was grown on mDixon or minimal media for RNA extraction. Sequencing was done using Illumina HiSeq 2000. Please see S9 Text for details.
De novo genome assembly
Genomes were assembled using an in-house pipeline. Specifically, we used a conservative trimming approach (removing Q2 bases and all following bases and discarding paired-end reads with one or both ends shorter than 50 bp) as recommended by Illumina. After read-trimming, de novo assemblies (contiging and scaffolding) were constructed using SOAPdenovo [51] (version 1.05, maximum insert length = 300bp). A range of assembly options were explored including testing various k-mers (41, 51, 61, 71, 81) and down-sampling of coverage on an exponential scale down to 50X to identify an optimal assembly in terms of contiguity statistics (N50). An example of this can be seen in S6 Table. The resulting assembly was then re-scaffolded using the program Opera version 1.4 [52] with default parameters and we attempted to close remaining gaps in the assembly in silico using FinIS [53]. Details of assembly statistics can be found in S7 Table.
RNA-seq assembly for M. globosa
Illumina paired-end reads were first trimmed using fastq_quality_trimmer (-t 20 -l 10) in FASTX (http://hannonlab.cshl.edu/fastx_toolkit/). Reads that could not be paired were discarded. Then, reads were assembled into transcripts using Trinity [54] (—jaccard_clip—SS_lib_type FR). The jaccard_clip option was turned on to avoid fusion of transcripts. Assembly was done individually for each sample and all assemblies were combined to generate a complete set of 727,354 transcripts. After removing redundant transcripts that were identical to another transcript, a total of 716,864 transcripts were left. Then, transcripts that were fully contained in another transcript were also removed to generate a set of 402,757 transcripts.
Gene annotation
The M. globosa 7966 reference genome was originally annotated by combining in silico prediction and a limited number of EST sequences [2]. Therefore, we re-annotated this genome to improve quality with the aid of RNA-seq datasets from two growth conditions and the application of newer ab initio gene prediction programs [55,56]. We used the MAKER pipeline [57] to integrate ab initio gene prediction (SNAP [55] and AUGUSTUS [56]), transcript evidence (from our RNA-seq dataset), and protein evidence to predict genes in an iterative manner. For protein sequences, we downloaded several fungal genomes from Genbank, including Saccharomyces cerevisiae [58], Ustilago maydis [59], Candida albicans [60], M. globosa [2], and M. sympodialis [14]. The protein sequences of M. globosa and M. sympodialis were not used in their own annotation process, respectively. Four sets of annotation were generated using the MAKER pipeline [57] in an iterative manner to improve ab initio gene prediction. First, MAKER was run with protein evidence and ab initio predictors trained with core eukaryotic genes predicted by CEGMA [20]. Then, MAKER was re-run twice additionally. For each time, ab initio predictors were retrained with gene models predicted by the previous run.
The first annotation was generated without transcripts. Compared to the reference annotation, this annotation retrieved longer genes and identified more exons and introns. The number of PFam domains identified in this annotation was slightly higher than that of the reference annotation, suggesting that it annotation captured more sequences with coding potential. The number of supported intron-exon junctions also increased, indicating that a large portion of the newly identified junctions were likely real (S2 Table, 1st set). These results suggest that even without transcript evidence, our annotation captured more coding sequences and more complete genes. Upon addition of transcript evidence in the second set, the number of introns increased, while the number of genes decreased, likely due to the false merging of genes by transcripts spanning two genes (S2 Table, 2nd set). Since the M. globosa genome is compact, with 4,223 genes (Table 1) in a 9.0 Mbp genome, false merging of genes during transcriptome assembly is likely. The number of PFam domains in the protein sequences also decreased. Therefore, we removed any transcript that overlapped with two or more gene models in order to generate the third annotation set (S2 Table, 3rd set). The full set of transcripts was aligned to the M. globosa reference genome using BLAT with minimum identity of 99% [61] and any transcript that overlapped with more than one gene model were removed to generate the reduced set of transcripts. This reduced the transcript set from 402,757 transcripts to 322,251 transcripts. We also noted that very few genes exhibited evidence for alternatives isoforms (10 with two isoforms and one with four isoforms). Then, gene models in the second set that do not overlap with any gene in the third set were added to generate the fourth annotation set (S2 Table, 4th set). Adding these transcripts led to an increased number of supported junctions, but the number of unique PFam domains and total PFam domains did not increase further (S2 Table), suggesting that ab initio prediction is sufficient to capture sequences with coding potential. For genomes other than M. globosa 7966, we used the same iterative approach for annotation without transcript evidence.
Phylogenetic analysis
Species trees were constructed using the concatenated sequences of 164 core eukaryotic genes (CEGs) predicted by CEGMA [20] that are present in all Malassezia genomes, U. maydis genome, and the S. cerevisiae genome, and the Malassezia sequences are at least 90% the length of their S. cerevisiae orthologs. Sequences were aligned using MUSCLE [62] and the phylogeny was constructed using maximum likelihood (ML) approaches as implemented by RAxML [63]. RAxML was run using “–f a –m PROTGAMMAJTT” with 400 bootstraps. (Ustilaginomycotina tree was built using the same approach.) To test the robustness of our Malassezia phylogeny beyond bootstrap values, we applied a Bayesian approach on the same concatenated sequences using MRBAYES [64]. MRBAYES was run using “prset aamodelpr = mixed” and “mcmc nchains = 1 ngen = 300000”. We also merged individual ML gene trees for CEGs into a supertree [65]. Individual gene trees were constructed using the same approach and they were merged into a supertree using Clann [65]. Using this approach, more genes could be incorporated into the final tree, as missing a strain in a gene tree could be tolerated. Both approaches yielded the same phylogeny as the concatenated ML tree (Bayesian tree in S3A Fig). We also generated a species tree derived from the mitochondrial genomes (Jack Kennell, personal communications, S3B Fig). Despite differences observed on an intra-species level within the M. furfur, M. globosa, and M. sympodialis lineages, we found that the two trees correspond perfectly on an inter-species level.
For M. furfur-specific tree (S1 Fig), MCL clusters (described later) with two genes in each of the hybrids (M. furfur 7710, M. furfur 1878, M. furfur 4172, and M. furfur 7019) and one gene in each of the haploids (M. furfur 7982 and M. furfur JPLK23) were used. Nucleotide sequences were used instead of amino acid sequences. The genes from hybrid strains were separated into two groups based on their similarity to the genes of the two haploid strains as measured by BLAST bitscore [39]. Only clusters with less than 5% of total alignment as indels were used to minimize the effect of assembly and annotation errors. A total of 1,306 clusters were concatenated and aligned using MUSCLE [62]. To generate a maximum likelihood tree, RAxML was used with “-f a -# 400 -m GTRGAMMA” [63].
Gene family analysis
PFam domains [29] were identified in all Malassezia strains and other fungal protein sequences using hmmscan (HMMER 3.1b1) with trusted cutoffs (http://hmmer.org). Malassezia-specific PFam domains are defined as present in at least seven Malassezia species and in at most two other fungi. This approach is limited to the study of protein families having at least one PFam domain. MCL clustering [66] was used to cluster Malassezia and other fungal genes based on their pairwise sequence similarity to construct gene families. This approach does not rely on PFam and can include all genes regardless whether they have PFam domains or not. Yet, unrelated genes with no orthology can be clustered together if they both have a good match to a third gene. BLAST [39] was used to align Malassezia and other fungal protein sequences in an all-against-all fashion with an e-value cutoff of 10−5. MCL was used to cluster protein sequences based on their E-values with–I 2.0. Sequences shorter than 90% of the median length in the cluster were excluded. Malassezia-specific clusters were defined as present in at least seven Malassezia species and not present in other fungi). To infer gain and loss in PFam families, PFam family phylogenies were constructed using MUSCLE and RAxML as described earlier. Then, species phylogeny and gene family phylogenies were reconciled using NOTUNG [67] to infer gain and loss events that took place along the Malassezia phylogeny.
For selection tests, we choose singleton PFam families in Malassezia species. Protein sequences were first aligned using MUSCLE (described earlier) and nucleotides were then substituted back in the sequences. PAML [68] was used for selection tests (CODEML, M7 M8 mode), and genes with bonferroni-corrected p-value < 0.05 were tabulated (χ2 test, two degrees of freedom).
Malassezia and lipid dependence
For Malassezia growth in 2X YNB media, cultures were incubated in 2X YNB at 31°C for 24, 48, 72, and 144 hours. Four strains, M. furfur 7982, M. sympodialis 42132, M. pachydermatis 1879 and 7550 (also named by CBS, http://www.cbs.knaw.nl/), were used. For each strain, two cultures, one with and one without Tween 40 at 1%, were included in this experiment.
Two separate methods were used to extract lipids from the Bactopeptone media. Please refer to S9 Text for details.
Lipid assimilation assays
Briefly, Sabouraud broth was melted with 3% Sea Plaque GTG (low-melt) agarose and equilibrated to 45°C. Malassezia cells were counted and diluted to 1x105 cells/ml in Sabouraud broth with chloramphenicol, and equilibrated to 31°C. Cell suspension (30ml) and melted agar broth (30ml) were quickly mixed and poured into 150 mm dishes. Once solidified, 18 holes were made with a 2 mm punch biopsy. To each hole, 5 μl of test compound was added and the plates were incubated for 17 days at 34°C. Test compounds are listed in S5 and S6 Tables. Please refer to [69,70] for artificial sebum.
MAT loci analysis
We examined the two mating type loci (P/R and HD) for all Malassezia species. We used the gene sequences from the reference strains for M. sympodialis and M. globosa, as queries to BLAST [39] against the genome assemblies of all Malassezia species to identify the locations of the MAT loci in these genomes. The alignments of MAT loci between Malassezia species were generated through WebACT (http://www.webact.org/WebACT/generate) using blastn with an E-value cutoff of 0.0001.
Horizontal gene transfers in Malassezia
M. sympodialis 42132 protein sequences were compared to UniProtKB v2015_02 [71] and the NCBI Nucleotide Collection database using NCBI Blast [72]. Proteins with the most significant BLAST hit against bacteria in both databases were analyzed further. Bacterial and fungal homologs of each HGT candidate were retrieved from GenBank [73]. Multiple sequence alignments were generated using MUSCLE v3.8.31 [62]. Poorly aligned or divergent regions in the alignments were identified and excluded from further analysis using Gblocks v0.91 (with options –u = y –t = p) [74]. Phylogenetic trees and 1000 bootstraps for each alignment were generated using PhyML v20120412 (with options –m JTT –d aa) [75]. Phylogenetic trees were manually inspected and candidate horizontally transferred genes were chosen, if they were closer relatives to their bacterial orthologs than their fungal orthologs.
Malassezia profiling from shotgun metagenomics datasets
We used PathoScope 2.0 [76] to estimate the abundances of the 14 Malassezia species in different shotgun metagenomics datasets (strain 7982 used for M. furfur, strain 7877 for M. restricta, strain 7966 for M. globosa, and strain 42132 for M. sympodialis). Before mapping to the reference genomes, the reads were filtered against a set of non-Malassezia fungal genomes (all fungal genomes from ftp://ftp.ncbi.nlm.nih.gov/genomes/Fungi/ plus Ustilago maydis [59]) using PathoScope 2.0's MAP module. To further account for potential false positives from PathoScope analysis, Malassezia genomes were divided into 1 kb bins and a species was considered present only if >10% of the bins were covered by at least one read. Abundances of Malassezia species were renormalized after removing false positives using this filter. The performance of PathoScope 2.0 was benchmarked by mapping the original reads of each Malassezia strain to the genomes of all Malassezia species using the protocol described here.
Accession numbers for the datasets studied in this paper are summarized in S9 Text.
To identify genes with variable copy number in Malassezia, we focused on M. restricta (using M. restricta 7877 genome), the most abundant species in skin samples [12]. We selected six samples with highest M. restricta genome coverage (MET0202, MET0207, MET0259, MET0270, MET0276, MET0278) [12]. Read counts for each gene were obtained from PathoScope [76] and normalized across all genes in each sample. Normalized read counts were used to compute the coefficient of variation (= sample standard deviation divided by sample mean). The top 10 genes with highest coefficient of variation are shown in Fig 2B and the top 15 are listed in S3 Table.
Supporting Information
Zdroje
1. Malassez L. Note sur le champignon du pityriasis simple. Arch Physiol. 1874;1 : 451–459.
2. Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci. 2007;104 : 18730–18735. 18000048
3. Kurtzman C, Fell JW, Boekhout T. The Yeasts: A Taxonomic Study. Elsevier; 2011.
4. Castellá G, Hernández JJ, Cabañes FJ. Genetic typing of Malassezia pachydermatis from different domestic animals. Vet Microbiol. 2005;108 : 291–296. 15922521
5. Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance M-A, et al. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. Int J Syst Evol Microbiol. 2004;54 : 623–627. 15023986
6. Cabañes FJ, Theelen B, Castellá G, Boekhout T. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Res. 2007;7 : 1064–1076. 17367513
7. Cabañes FJ, Hernández JJ, Castellá G. Molecular Analysis of Malassezia sympodialis-Related Strains from Domestic Animals. J Clin Microbiol. 2005;43 : 277–283. 15634983
8. Ra K, Jr RE, Nb O, Rw W. Quantity and distribution of Malassezia organisms on the skin of clinically normal dogs. J Am Vet Med Assoc. 1996;208 : 1048–1051. 8621316
9. Gemmer CM, DeAngelis YM, Theelen B, Boekhout T, Thomas L. Dawson J. Fast, Noninvasive Method for Molecular Detection and Differentiation of Malassezia Yeast Species on Human Skin and Application of the Method to Dandruff Microbiology. J Clin Microbiol. 2002;40 : 3350–3357. 12202578
10. Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia Genus in Skin and Systemic Diseases. Clin Microbiol Rev. 2012;25 : 106–141. doi: 10.1128/CMR.00021-11 22232373
11. Kong HH, Segre JA with Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al., NIH Intramural Sequencing Center Comparative Sequencing Program. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498 : 367–370. doi: 10.1038/nature12171 23698366
12. Kong HH, Segre JA with Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514 : 59–64. doi: 10.1038/nature13786 25279917
13. Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, et al. Redefining the Human Oral Mycobiome with Improved Practices in Amplicon-based Taxonomy: Discovery of Malassezia as a Prominent Commensal. PLoS ONE. 2014;9: e90899. doi: 10.1371/journal.pone.0090899 24614173
14. Gioti A, Nystedt B, Li W, Xu J, Andersson A, Averette AF, et al. Genomic Insights into the Atopic Eczema-Associated Skin Commensal Yeast Malassezia sympodialis. mBio. 2013;4: e00572–12. doi: 10.1128/mBio.00572-12 23341551
15. Amend A. From Dandruff to Deep-Sea Vents: Malassezia-like Fungi Are Ecologically Hyper-diverse. PLoS Pathog. 2014;10: e1004277. doi: 10.1371/journal.ppat.1004277 25144294
16. Wang Q-M, Theelen B, Groenewald M, Bai F-Y, Boekhout T. Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia—Mol Phylogeny Evol Fungi. 2014;33 : 41–47.
17. Begerow D, Stoll M, Bauer R. A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia. 2006;98 : 906–916. 17486967
18. Saunders CW, Scheynius A, Heitman J. Malassezia Fungi Are Specialized to Live on Skin and Associated with Dandruff, Eczema, and Other Skin Diseases. PLoS Pathog. 2012;8: e1002701. doi: 10.1371/journal.ppat.1002701 22737067
19. White TC, Findley K, Dawson TL, Scheynius A, Boekhout T, Cuomo CA, et al. Fungi on the skin: dermatophytes and Malassezia. Cold Spring Harb Perspect Med. 2014;4.
20. Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23 : 1061–1067. 17332020
21. Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Bælum J, Kimbrel J, et al. Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 2014;8 : 1464–1475. doi: 10.1038/ismej.2013.254 24451203
22. Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci. 2012;109 : 21390–21395. doi: 10.1073/pnas.1215210110 23236140
23. Rascovan N, Carbonetto B, Revale S, Reinert MD, Alvarez R, Godeas AM, et al. The PAMPA datasets: a metagenomic survey of microbial communities in Argentinean pampean soils. Microbiome. 2013;1 : 21. doi: 10.1186/2049-2618-1-21 24450949
24. Greenblum S, Carr R, Borenstein E. Extensive strain-level copy-number variation across human gut microbiome species. Cell. 2015;160 : 583–594. doi: 10.1016/j.cell.2014.12.038 25640238
25. Matheny PB, Gossmann JA, Zalar P, Kumar TKA, Hibbett DS. Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Can J Bot. 2006;84 : 1794–1805.
26. Dankner WM, Spector SA, Fierer J, Davis CE. Malassezia Fungemia in Neonates and Adults: Complication of Hyperalimentation. Rev Infect Dis. 1987;9 : 743–753. 3125578
27. Castellá G, Coutinho SDA, Cabañes FJ. Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med Mycol. 2014;52 : 99–105. doi: 10.3109/13693786.2013.815372 23902157
28. Theelen B, Silvestri M, Guého E, van Belkum A, Boekhout T. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLPTm), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 2001;1 : 79–86. 12702352
29. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32: D138–D141. 14681378
30. Guillot J, Bond R. Malassezia pachydermatis: a review. Med Mycol. 1999;37 : 295–306. 10520154
31. Gueho E, Simmons RB, Pruitt WR, Meyer SA, Ahearn DG. Association of Malassezia pachydermatis with systemic infections of humans. J Clin Microbiol. 1987;25 : 1789–1790. 3654952
32. Nielsen K, Heitman J. Sex and Virulence of Human Pathogenic Fungi. In: Dunlap JC, editor. Advances in Genetics. Academic Press; 2007. pp. 143–173. Available: http://www.sciencedirect.com/science/article/pii/S006526600657004X 17352904
33. Alzomor AK, Moharram AS, Absi NMA. Formulation and evaluation of potash alum as deodorant lotion and after shaving astringent as cream and gel. Int Curr Pharm J. 2014;3 : 228–233.
34. Fitzpatrick DA, Logue ME, Butler G. Evidence of recent interkingdom horizontal gene transfer between bacteria and Candida parapsilosis. BMC Evol Biol. 2008;8 : 181. doi: 10.1186/1471-2148-8-181 18577206
35. Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet TIG. 2010;26 : 5–8. doi: 10.1016/j.tig.2009.11.007 19969385
36. Kruppa MD, Lowman DW, Chen Y-H, Selander C, Scheynius A, Monteiro MA, et al. Identification of (1—>6)-beta-D-glucan as the major carbohydrate component of the Malassezia sympodialis cell wall. Carbohydr Res. 2009;344 : 2474–2479. doi: 10.1016/j.carres.2009.09.029 19853245
37. Bairwa G, Kaur R. A novel role for a glycosylphosphatidylinositol-anchored aspartyl protease, CgYps1, in the regulation of pH homeostasis in Candida glabrata. Mol Microbiol. 2011;79 : 900–913. doi: 10.1111/j.1365-2958.2010.07496.x 21299646
38. Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci CMLS. 2004;61 : 192–208. 14745498
39. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215 : 403–410. 2231712
40. Kaneko T, Makimura K, Abe M, Shiota R, Nakamura Y, Kano R, et al. Revised Culture-Based System for Identification of Malassezia Species. J Clin Microbiol. 2007;45 : 3737–3742. 17881545
41. Ellermeier CD, Losick R. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 2006;20 : 1911–1922. 16816000
42. Pei J, Grishin NV. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem Sci. 2001;26 : 275–277. 11343912
43. Manolaridis I, Kulkarni K, Dodd RB, Ogasawara S, Zhang Z, Bineva G, et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature. 2013;504 : 301–305. doi: 10.1038/nature12754 24291792
44. Batra R, Boekhout T, Guého E, Cabañes FJ, Dawson TL, Gupta AK. Malassezia Baillon, emerging clinical yeasts. FEMS Yeast Res. 2005;5 : 1101–1113. 16084129
45. DeAngelis YM, Saunders CW, Johnstone KR, Reeder NL, Coleman CG, Kaczvinsky JR, et al. Isolation and Expression of a Malassezia globosa Lipase Gene, LIP1. J Invest Dermatol. 2007;127 : 2138–2146. 17460728
46. Cafarchia C, Otranto D. Association between Phospholipase Production by Malassezia pachydermatis and Skin Lesions. J Clin Microbiol. 2004;42 : 4868–4869. 15472366
47. Lee YW, Lee SY, Lee Y, Jung WH. Evaluation of Expression of Lipases and Phospholipases of Malassezia restricta in Patients with Seborrheic Dermatitis. Ann Dermatol. 2013;25 : 310. doi: 10.5021/ad.2013.25.3.310 24003273
48. Ghannoum MA. Potential Role of Phospholipases in Virulence and Fungal Pathogenesis. Clin Microbiol Rev. 2000;13 : 122–143. 10627494
49. Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996;69 : 337–355. 8836432
50. Heitman J, Carter DA, Dyer PS, Soll DR. Sexual Reproduction of Human Fungal Pathogens. Cold Spring Harb Perspect Med. 2014;4: a019281. doi: 10.1101/cshperspect.a019281 25085958
51. Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1 : 18. doi: 10.1186/2047-217X-1-18 23587118
52. Gao S, Sung W-K, Nagarajan N. Opera: Reconstructing Optimal Genomic Scaffolds with High-Throughput Paired-End Sequences. J Comput Biol. 2011;18 : 1681–1691. doi: 10.1089/cmb.2011.0170 21929371
53. Gao S, Bertrand D, Nagarajan N. FinIS: Improved in silico Finishing Using an Exact Quadratic Programming Formulation. In: Raphael B, Tang J, editors. Algorithms in Bioinformatics. Springer Berlin Heidelberg; 2012. pp. 314–325. Available: http://link.springer.com.ejproxy.a-star.edu.sg/chapter/10.1007/978-3-642-33122-0_25
54. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29 : 644–652. doi: 10.1038/nbt.1883 21572440
55. Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5 : 59. 15144565
56. Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7 : 62. 16469098
57. Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12 : 491. doi: 10.1186/1471-2105-12-491 22192575
58. Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, et al. The Reference Genome Sequence of Saccharomyces cerevisiae: Then and Now. G3 GenesGenomesGenetics. 2014;4 : 389–398.
59. Kämper J, Kahmann R, Bölker M, Ma L-J, Brefort T, Saville BJ, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444 : 97–101. 17080091
60. Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101 : 7329–7334. 15123810
61. Kent WJ. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;12 : 656–664. 11932250
62. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32 : 1792–1797. 15034147
63. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22 : 2688–2690. 16928733
64. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17 : 754–755. 11524383
65. Creevey CJ, McInerney JO. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21 : 390–392. 15374874
66. Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30 : 1575–1584. 11917018
67. Chen K, Durand D, Farach-Colton M. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J Comput Biol J Comput Mol Cell Biol. 2000;7 : 429–447.
68. Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24 : 1586–1591. 17483113
69. Ro BI, Dawson TL. The role of sebaceous gland activity and scalp microfloral metabolism in the etiology of seborrheic dermatitis and dandruff. J Investig Dermatol Symp Proc Soc Investig Dermatol Inc Eur Soc Dermatol Res. 2005;10 : 194–197.
70. Troller JA. Model System for the Investigation of Dandruff. J Soc Cosmet Chem. 1971;22 : 187–198.
71. Consortium UniProt. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014;42: D191–198. doi: 10.1093/nar/gkt1140 24253303
72. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10 : 421. doi: 10.1186/1471-2105-10-421 20003500
73. Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36: D25–30. 18073190
74. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56 : 564–577. 17654362
75. Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59 : 307–321. doi: 10.1093/sysbio/syq010 20525638
76. Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2 : 1–15.
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



