-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
Methylation of DNA is used by numerous organisms to regulate a wide variety of cellular processes, but specific roles for most DNA methyltransferases have not been defined. We studied one such enzyme in Vibrio cholerae, the cholera pathogen, using genome-wide approaches to compare DNA methylation, gene expression, and the sets of genes required or dispensable for growth in bacterial strains that produced or lacked this enzyme. These studies allowed us to identify numerous cellular processes regulated, either directly or indirectly, by this cytosine methyltransferase. In particular, we found that an absence of enzyme activity was associated with reduced levels of a bacterial stress response; consequently, a stress response pathway that is essential in wild type bacteria is not needed for survival of the mutant lacking the methyltransferase. Similar genome-wide analyses can likely to be used to define the cellular roles of many additional uncharacterized DNA methyltransferases.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005666
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005666Summary
Methylation of DNA is used by numerous organisms to regulate a wide variety of cellular processes, but specific roles for most DNA methyltransferases have not been defined. We studied one such enzyme in Vibrio cholerae, the cholera pathogen, using genome-wide approaches to compare DNA methylation, gene expression, and the sets of genes required or dispensable for growth in bacterial strains that produced or lacked this enzyme. These studies allowed us to identify numerous cellular processes regulated, either directly or indirectly, by this cytosine methyltransferase. In particular, we found that an absence of enzyme activity was associated with reduced levels of a bacterial stress response; consequently, a stress response pathway that is essential in wild type bacteria is not needed for survival of the mutant lacking the methyltransferase. Similar genome-wide analyses can likely to be used to define the cellular roles of many additional uncharacterized DNA methyltransferases.
Introduction
DNA methylation—the covalent attachment of methyl moieties to specific nucleotides in the genome by DNA methyltransferases (MTases)—is a fundamental mechanism for epigenetic regulation in all domains of life (reviewed in [1,2]). Bacterial DNA MTases principally generate three modified DNA bases [3,4]: 6-methyladenine (6mA), 4-methylcytosine (4mC) and 5-methylcytosine (5mC). Most bacterial MTases are components of restriction-modification (R-M) systems; these MTases modify target DNA sequences in order to protect them from digestion by a cognate restriction enzyme, which is typically co-transcribed. R-M systems enable digestion of horizontally acquired DNA sequences that lack appropriate methylation marks, and thus protect bacteria from selfish elements and phage predation [5]. However, a subset of MTase genes are not accompanied by a cognate restriction enzyme, and a few of these so-called ‘orphan’ MTases are known to regulate diverse host cell processes (reviewed in [2,6,7]). For example, the 6mA MTase Dam, which is found in E. coli and many other gamma proteobacteria, regulates DNA replication [8], mismatch repair [9], transposition [10] and pilus biogenesis [11], while the adenine MTase CcrM is a critical regulator of the Caulobacter crescentus cell cycle [12]. Recently, Dcm, an E. coli orphan MTase that produces 5mC, was found to modulate antibiotic resistance [13], translation [14], and stationary phase gene expression [15]. Additionally, MTases of Type III R-M systems have also been found to mediate phase variation [16,17]. Nonetheless, roles for the majority of bacterial MTases—which are predicted in over 90% of genomes [18]—have not been defined.
Here, we investigated the importance of DNA methylation in the cholera pathogen, Vibrio cholerae. The canonical El Tor O1 V. cholerae strain N16961 [19] is predicted to encode 4 MTases [18] with distinct catalytic activities. The V. cholerae Dam homologue has been shown to be critical for replication of one of the organism’s two chromosomes, and is consequently essential for survival [20,21]. In contrast, vc1769 (hsdM), vca0198 (vchM) and vca0447 have been found to be not essential, either through targeted mutagenesis [22] or in transposon insertion sequencing screens [23–25]. VC1769 is a homologue of the E. coli 6mA-generating HsdM, which is part of a type I R-M system [26], and vca0447, a putative orphan adenine MTase, remains uncharacterized to date. VchM is present with almost 100% identity in 91% (10/11) of complete V. cholerae genome sequences at NCBI, but is absent from more than 90% (20/22) of non-cholerae Vibrios (S1 Fig), and thus appears to have been acquired by horizontal gene transfer. VchM was previously characterized as an orphan 5mC MTase that targets the consensus sequence RCCGGY [22,27], but the importance of this enzyme to host gene expression has not been defined.
Here, we demonstrate that VchM is required for optimal V. cholerae growth, both in vitro and during infection. Bisulfite sequencing defined the V. cholerae 5-methylcytosine methylome, and RNA sequencing analyses revealed that VchM regulates expression of genes important in a variety of cellular processes, potentially through direct intragenic methylation. Unexpectedly, transposon insertion sequencing-based analyses of vchM genetic interactions revealed that deletion of vchM suppresses the essentiality of the σE envelope stress response pathway. Additional transposon mutagenesis studies identified host genes that are required for envelope stability, in whose absence σE is induced. Many of these genes, especially those involved in the modification of the lipopolysaccharide inner core, contain VchM recognition sites. Mutational analyses suggest that VchM cytosine methylation directly downregulates the expression of some of these LPS modification genes. Thus, our findings show that V. cholerae has co-opted the horizontally acquired VchM DNA methyltransferase to regulate a diverse array of critical cellular processes.
Results
vchM is required for optimal V. cholerae growth in vitro and during infection
We created in-frame deletion mutants of V. cholerae’s non-essential MTases—vc1769 (hsdM), vca0198 (vchM), and vca0447—and compared the growth of each mutant to an isogenic wild type (wt) El Tor biotype strain during infection of suckling mice, using competition assays. The ∆vchM mutant displayed significantly attenuated (~10 fold) growth in vivo, while the recovery of the ∆hsdM and ∆vca0447 mutants was equivalent to that of WT cells (Fig 1A). When vchM was deleted in the prototypical classical biotype V. cholerae strain, O395, ten-fold attenuation in vivo was also observed (Fig 1A). Similarly, infant mice inoculated solely with ∆vchM V. cholerae accumulated 30-fold fewer intestinal bacteria than did animals infected with WT V. cholerae (Fig 1B). The ∆vchM mutant was also outcompeted by wt cells when grown in vitro (Fig 1C) and displayed a reduced doubling rate when grown in monoculture (Fig 1D). Together, these data indicate that ∆vchM cells, unlike other mutants lacking an orphan 5mC MTase [15], have an intrinsic growth defect that manifests both during in vitro and in vivo growth.
Fig. 1. VchM methyltransferase activity promotes V. cholerae growth and pathogenicity. 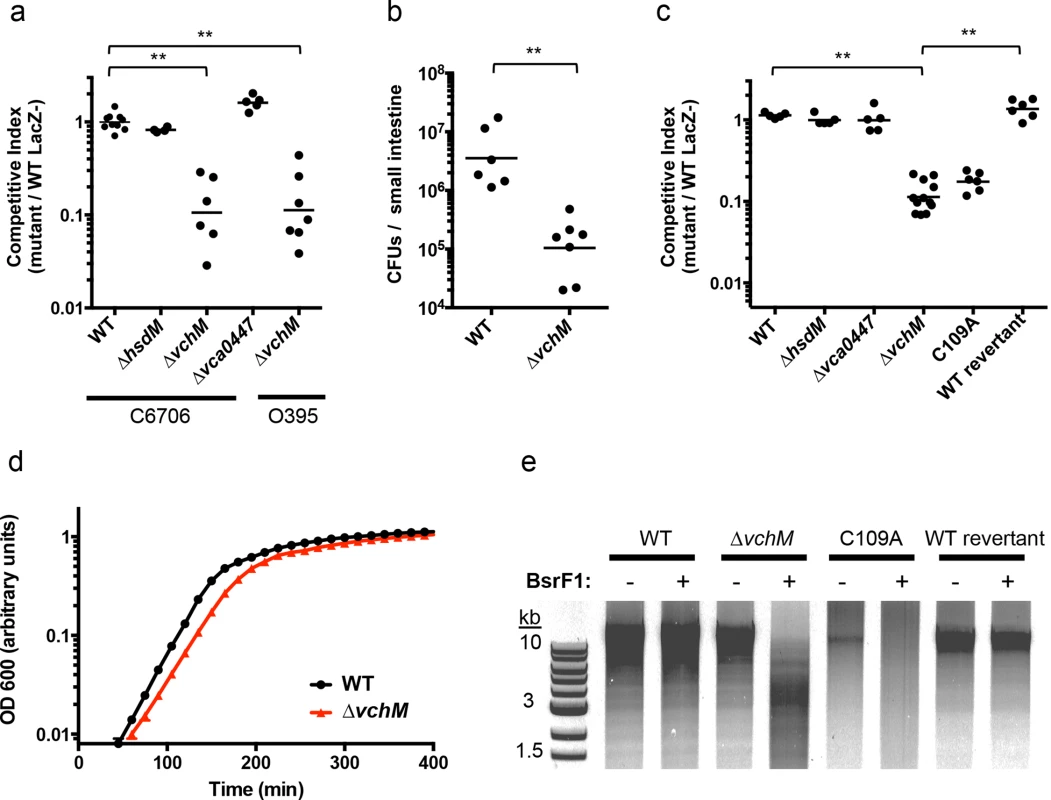
(A) Mice were co-infected with wildtype V. cholerae (either C6706 or O395) and a differentially marked methyltransferase mutant, which could be distinguished by blue/white screening. The competitive index was calculated from the ratio of mutant (LacZ+) to WT (LacZ-) cells recovered after infection divided by the corresponding ratio of the inoculum. WT represents a control competition between LacZ+ and LacZ- V. cholerae C6706. **p-value <0.01. (B) Mice were singly infected either with wt or ∆vchM V. cholerae C6706 and the total number of CFUs recovered per small intestine is shown. **p-value <0.01. (C) Differentially marked strains (LacZ-, ∆hsdM, ∆vca0447, and either ∆vchM, a vchM active site mutant (C109A), or a deletion mutant into which the wt gene had been reintroduced (wt revertant)) were co-cultured in LB with wt cells and the competitive indices were calculated as in (A). **p-value <0.01. (D) The growth rate of wt and ∆vchM cells in LB was monitored over time using OD 600 measurements. (E) Genomic DNA from the indicated strains was digested with SalI and the methylation-sensitive enzyme BsrFI, which does not cleave methylated RCCGGY motifs. To confirm that the growth defect of the ∆vchM mutant is due to the absence of 5mC methylation, rather than to a second site mutation or a non-enzymatic role of VchM, we re-introduced a wildtype or catalytically inactive vchM (C109A) allele into the ∆vchM mutant at the endogenous locus. Restriction digests confirmed that gDNA from the strain into which the wt sequence was reintroduced—like that of wt V. cholerae—was resistant to cleavage by BsrFI, which cannot cleave methylated RCCGGY motifs. In contrast, gDNA from ∆vchM and C109A V. cholerae was sensitive as expected (Fig 1E). The WT replacement, but not the C109A replacement, also fully complemented the in vitro growth defect of the ∆vchM parent (Fig 1A and 1C). Thus, the catalytic activity of VchM is required for optimal bacterial growth, suggesting 5mC DNA methylation controls processes necessary for optimal V. cholerae growth.
The VchM methylome is stable across different growth states
Previous work revealed that VchM recognizes and methylates a consensus sequence of RCCGGY (methylated residue underlined) [22]. Interestingly, the distribution of RCCGGY motifs is not uniform across the genome (S2 Fig). A previous study evaluated the methylation status of these sites and found that three VchM sites in V. cholerae are undermethylated in late exponential phase, compared to the rest of the genome [27]. However, it remained unknown whether V. cholerae’s pattern of 5mC methylation could vary between different growth states, as was observed for the E. coli orphan 5mC DNA MTase, dcm [15]. Thus, we used bisulfite sequencing, in which non-methylated cytosines are converted into uracils and detected as C to T transitions, to assess the methylation status of all cytosines in the genomes of bacteria in exponential and stationary growth phases, as well as V. cholerae that had been isolated from infected rabbit intestines. This approach was highly specific and sensitive, revealing a clear distinction between methylated cytosines within VchM’s RCCGGY target sites and non-methylated cytosines in other sequence contexts (Fig 2A, S3A Fig).
Fig. 2. V. cholerae cytosine methylation and its associations with gene expression. 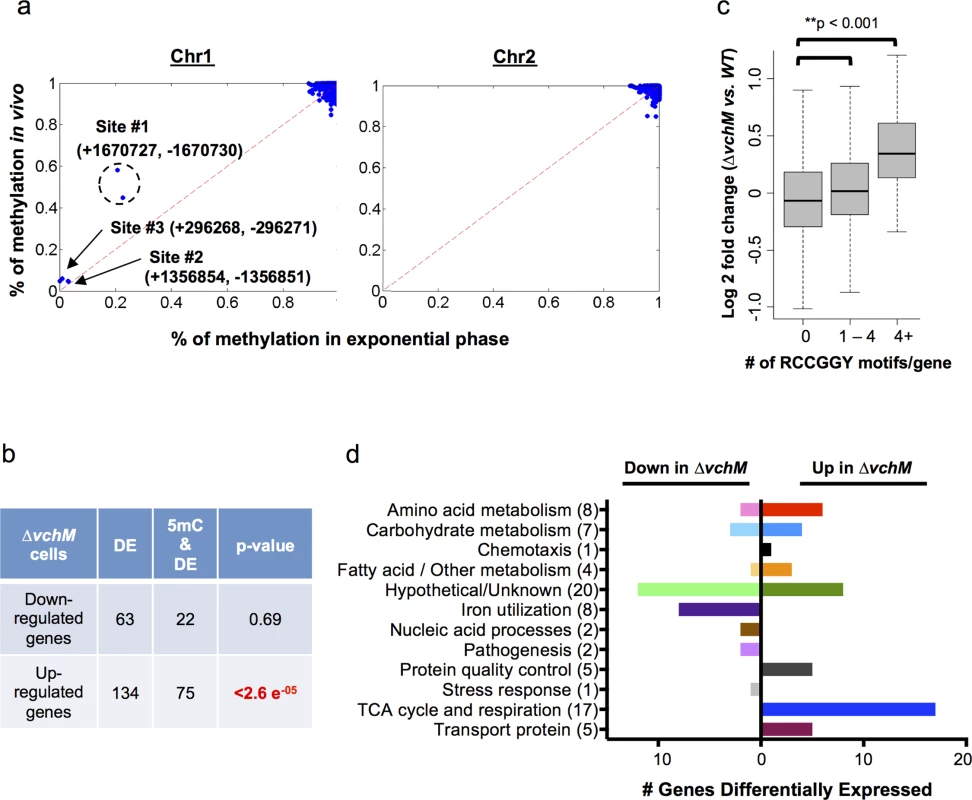
(A) The extent of VchM target site (RCCGGY) methylation on chromosomes I and II, as determined by bisulfite sequencing of DNA from exponential phase cultures, is plotted against the extent of methylation for these sites during infection. Three sites (#1-#3) were found to be persistently non-methylated; numbers designate the genomic position of each site on the forward (+) or reverse (-) strand. (B) Genes that were significantly (p-val <0.01) differentially expressed by wt and ∆vchM cells were correlated with the presence of intragenic RCCGGY motifs. Significance was assessed using a Fisher’s exact test by the comparing the fraction of differentially expressed genes containing RCCGGY motifs (e.g., 22 out of 63) to the fraction of all genes in V. cholerae containing RCCGGY motifs (1460 out of 3842 genes). Significant enrichment of motifs was found within only the genes that were upregulated in ∆vchM cells. DE = number of differentially expressed genes; 5mC = number of genes containing methylated RCCGGY motifs. (C) Differences in transcript abundance between exponential phase cultures of ∆vchM and WT cells are plotted relative to the number of RCCGGY motifs present within the coding region for each gene. The boxes represent the fold change of genes in the 25%-75% quartile with the median fold change shown as a line. The whiskers represent 1.5 fold of the interquartile range (the third quartile minus the first quartile) away from the box. (D) The numbers of genes that are differentially expressed (p-value <0.05) by ∆vchM strains of C6706 and O395 V. cholerae compared to their parental strains are shown for a variety of biological categories. The bisulfite sequencing results (GEO Accession number GSE73975) revealed that virtually all VchM motifs in the genome are methylated with high frequency in exponential phase, i.e., RCCGGY sites on >90% of the DNA molecules were fully methylated (Fig 2A). Of the ~2100 VchM motifs in the genome, only 3 were persistently non-methylated (methylated <20% of the time on both strands). These sites, which were previously described, are all located within intergenic regions between two divergently oriented genes, and are thought to be non-methylated in exponential phase due to binding by transcription factors [27]. Most VchM sites were also methylated with high frequency in stationary and in vivo grown cells (Fig 2A, S3B Fig). The non-methylated sites #2 and #3 were similarly non-methylated during infection, but site #1 (Fig 2A), which is located in between VC1558 and VC1559, showed increased methylation in vivo (~51% of cells) compared to in vitro exponential grown cells (~20% of cells). While the functional consequence of this difference remains to be defined, our data show that the methylation of VchM sites is highly saturated across the genome (i.e., virtually all RCCGGY motifs are fully methylated) and is not drastically altered during V. cholerae growth in the intestine.
VchM methylation correlates with altered gene expression
To investigate the impact of cytosine methylation on V. cholerae gene expression, we compared the transcriptomes of WT and ∆vchM cells in the C6706 strain background (GEO Accession number GSE73974). We identified 134 genes with significantly (p-value <0.05) elevated transcript abundance in the ∆vchM mutant, and 63 genes with reduced transcript abundance (S1 Table). While there was no significant enrichment of VchM motifs within the genes with reduced transcript levels, seventy-five of the genes with elevated transcript levels contained a RCCGGY motif, which is significantly more than would be predicted by chance alone (Fig 2B). The correlation between the presence of VchM target sites and increased transcript abundance in the ∆vchM mutant was also observed when analysis was not restricted to genes with significantly altered transcript levels. Genome wide, relative transcript levels were significantly higher in ∆vchM cells versus wt for genes that contained 1 or more VchM targets within 200 bp of their transcriptional start sites [28] (S4A Fig). We also observed a significant association between the number of RCCGGY target sites within a gene’s coding region and its expression change in ∆vchM cells, especially for genes containing more than 4 target sites (Fig 2C). This result suggests that VchM methylation reduces gene expression of some genes. This significant association was independent of the genes’ GC contents (S4B Fig) and was not observed for several other similar motifs (S4C Fig). Additionally, no strong association between the precise location of RCCGGY sites within genes and differential expression was observed; the motifs are similarly distributed throughout the coding region of all genes as well as those that are differentially expressed (S4D Fig).
To identify pathways that are consistently regulated (directly or indirectly) by VchM, we compared transcriptomic analyses for C6706 and O395 ∆vchM mutants and the corresponding wt strains. We identified 79 genes that were significantly and differentially (p-value < 0.01) expressed in the absence of VchM in both biotypes (Fig 2D, S2 Table). Approximately 25% of these are hypothetical genes, while the remainders are predicted to participate in a variety of critical processes, including energy production (22%), amino acid metabolism (10%) and iron utilization (10%). Expression of the iron utilization genes was reduced in ∆vchM cells compared to wt cells, and thus is unlikely to be directly controlled by VchM-dependent methylation. Likewise, while the genes involved in the TCA cycle and respiratory chain are upregulated in ∆vchM cells, most do not contain RCCGGY motifs, suggesting that their elevated expression may also be an indirect response to the loss of methylation.
vchM is required to maintain rpoE essentiality
To gain more insight into the physiology of the vchM mutant, in particular the processes that it utilizes for growth, we used a transposon-insertion sequencing (TIS) approach to screen for loci that are differentially required for survival in ∆vchM or wt V. cholerae. High-density transposon libraries were created in WT and ∆vchM cells grown in rich media, and all the transposon insertion sites in each library were sequenced. No genes specifically required by the ∆vchM mutant (i.e., genes lacking insertions only in the ∆vchM background) were identified in this screen other than the positive control, vchM itself. However, we unexpectedly identified 5 genes (Table 1, Fig 3A) in which there was a significantly higher frequency of transposon insertion in ∆vchM cells compared to the wt strain (Fig 3B, S5 Fig), suggesting such genes are potentially more important to the survival or optimal in vitro growth of the wt strain than that of the ∆vchM mutant. Remarkably, four of the five genes—rseP, degS, rpoE and rep—are known to participate in the σE envelope stress response pathway (reviewed in [29]).
Fig. 3. TIS-based identification of loci that interact genetically with vchM or rpoE. 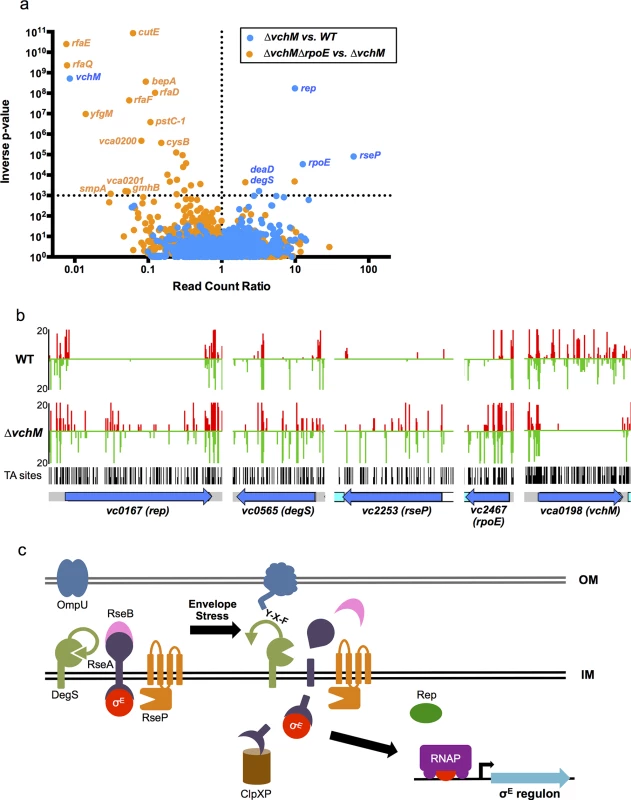
(A) For two comparative analyses (∆vchM vs. wt and ∆vchM ∆rpoE vs. ∆vchM), the read count ratio of transposons inserted into each genetic locus was determined, and is plotted against the inverse of the associated p-value as determined by Mann-Whitney U analysis of read counts. Genes plotted above the horizontal line are considered significantly different in each analysis (p-value <0.001). (B) The raw number of reads originating from insertions on the forward (red) or reverse strand (green) in wt and ∆vchM insertion libraries are shown for selected loci from both libraries. All potential insertion sites (TA dinucleotides) are marked by black bars. (C) The predicted σE stress response pathway in V. cholerae. Tab. 1. <i>vchM</i> genetic interactions identified by transposon-insertion sequencing. 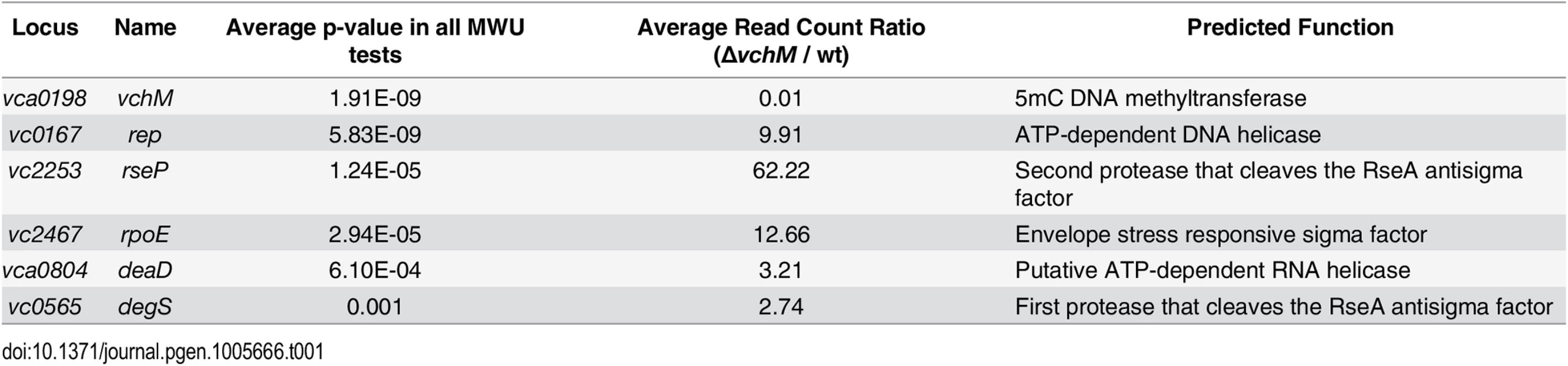
In the σE stress response pathway, which has been best described in E. coli but has also been characterized in V. cholerae (Fig 3C), σE is sequestered at the membrane by an integral membrane anti-sigma factor, RseA [30–32]. Upon envelope stress, specific outer membrane proteins become unfolded and expose a terminal YXF motif, which interacts with and activates the intramembrane protease DegS [33,34]. Envelope stress may also dysregulate outer membrane biosynthesis, leading to increased periplasmic LPS, which binds and inactivates an additional DegS inhibitory factor, RseB [35,36\. The presence of LPS and OMP stimuli permits DegS to cleave within the periplasmic domain of RseA [34,37,38], allowing a second intramembrane protease, RseP, to cleave the cytosolic domain of RseA [38–40]. These proteolytic cleavages lead to the release of σE into the cytosol, where it can engage RNA polymerase to direct transcription of the σE regulon, which reduces de novo expression of outer membrane proteins while facilitating the refolding of existing ones [41–44]. An ATP-dependent helicase, rep, was also recently found to be required for induction of some σE regulon genes [45], though the exact mechanism by which this occurs is unknown.
Both in E. coli and V. cholerae, rpoE and related factors have been found to be essential [46–49], although suppressor mutations that permit survival of rpoE-deficient cells have also been identified [47,50–52]. In V. cholerae, the outer membrane protein OmpU is the predominant determinant of rpoE activation, both under normal growth conditions and in response to envelope stress [53], and ompU mutations are the most frequently identified suppressors of rpoE essentiality [47]. Suppressors directly associated with LPS synthesis or processing have not been identified in V. cholerae.
Our TIS analysis revealed that rseP, degS, rpoE and rep, which are all required for activation of the rpoE pathway, appeared dispensable in ∆vchM cells, suggesting that disruption of vchM may be a previously unrecognized suppressor of rpoE essentiality in V. cholerae. To validate this hypothesis, we attempted to delete rpoE in both WT and ∆vchM cells, using a targeting construct designed to replace rpoE with a kanamycin-resistance cassette. As in previous analyses [47], a very high percentage (here, 100%) of the putative ∆rpoE::aphR mutants generated in the wt background were false positives that retained an additional antibiotic resistance marker, reflecting retention of the targeting vector and failure to disrupt rpoE (Fig 4A). In contrast, 99% of ∆vchM ∆rpoE candidates did not retain the allelic exchange vector, suggesting that deletion of rpoE in the ∆vchM strain can be readily achieved. Furthermore, a putative ∆vchM ∆rpoE colony was randomly selected and the absence of σE was confirmed by western blot analysis (Fig 4B). Thus, V. cholerae does not require the presence of rpoE in the absence of vchM.
Fig. 4. Deletion of vchM suppresses the essentiality of rpoE but ∆vchM ∆rpoE mutants show impaired growth under stress conditions. 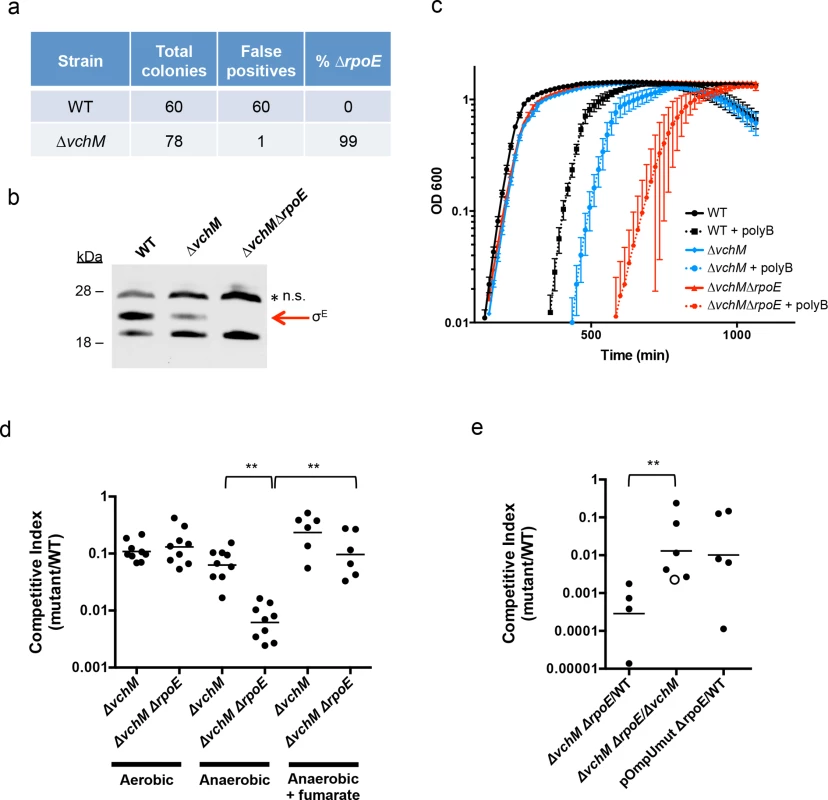
(A) Allelic exchange to replace rpoE with a kanamycin resistance cassette was performed in WT and ∆vchM cells. After patching colonies to determine if candidate colonies still retained the knockout vector, the false positive and rpoE deletion frequencies were calculated. (B) A representative western blot of lysates from cultures of WT, ∆vchM and ∆vchM ∆rpoE cells probed with polyclonal antisera against σE. n.s. = non-specific signal. (C) Culture density (OD 600) of the indicated strains was monitored for cells grown in LB and in LB supplemented with polymyxin B (poly B). (D) In vitro competition assays were performed between LacZ- V. cholerae and the indicated mutants. Cultures were grown in LB under aerobic or anaerobic conditions and supplemented with fumarate as noted. Competitive indices were calculated as in Fig 1. (E) Competitive infections were performed using differentially marked versions (LacZ+/-) of the indicated strains. Competitive indices were calculated for bacteria in intestinal homogenates as in Fig 1A. The open circle represents the limit of detection where no ∆vchM ∆rpoE mutants were recovered. Interestingly, our western blots also revealed that basal levels of σE were only ~30% of WT levels in the ∆vchM mutant (Fig 4B). Since σE expression and activity is elevated in response to envelope stress, the lower basal level σE in ∆vchM cells suggests that 5mC methylation by VchM may contribute to envelope stress, and thereby modulate production of σE. Reduced activation of σE in cells lacking VchM could allow rpoE to be dispensable in these cells, at least under the conditions at which the mutants were selected.
rpoE is required for growth under stress conditions and during infection
In addition to confirming the viability of V. cholerae lacking vchM and rpoE under relatively favorable growth conditions (rich media), we assessed whether these mutations altered bacterial growth in a variety of more challenging environments. We found that growth of the ∆vchM ∆rpoE mutant was equivalent to that of the ∆vchM parent strain in LB monocultures (Fig 4C), and that it exhibited an equivalent ~10-fold growth impairment as ∆vchM cells in competition assays with WT cells in aerobic LB culture (Fig 4D). However, in the presence of the outer membrane targeting antimicrobial peptide, polymyxin B, which is known to induce σE above baseline levels in V. cholerae [53], growth of the ∆vchM ∆rpoE mutant was reduced far more than that of the ∆vchM strain (Fig 4C). Thus, the strain that has no capacity to activate the σE stress response pathway (e.g., ∆rpoE) is at a marked growth disadvantage in envelope stress-inducing environments, and the ∆vchM mutation does not suppress V. cholerae’s need for rpoE under these conditions.
Similarly, deletion of vchM did not suppress V. cholerae’s need for σE during growth in the infant mouse intestine. In in vivo competition experiments, the ∆vchM ∆rpoE mutant was recovered at more than a 1000-fold reduced frequency (CI ~0.0007) than the wt strain from the infant mouse intestine (Fig 4E), and with 100-fold reduced frequency when competed against the ∆vchM parent strain (CI ~0.01). These results suggest that deletion of rpoE exacerbates the ~10-fold in vivo proliferation deficiency of the ∆vchM mutant, which has a CI vs the wt strain of ~0.1, by a factor of 100. A similar ~100-fold effect of rpoE disruption on V. cholerae growth in vivo was observed using an rpoE-deficient strain containing an ompU promoter mutation (pOmpU mut) that reduces porin expression. Since previous studies showed that deletion of ompU does not impair V. cholerae intestinal colonization [54], the in vivo attenuation of this double mutant can be fully explained by the absence of rpoE.
Finally, since the RNA-seq results revealed that ∆vchM cells had elevated expression of TCA cycle and respiration genes, we assessed the growth of the ∆vchM and ∆vchM ∆rpoE strains in anaerobic conditions, which would also be encountered during host infection. Relative to the WT strain, the ∆vchM mutant exhibited a similar growth defect in both aerobic and anaerobic conditions (Fig 4D). In contrast, the ∆vchM ∆rpoE mutant was significantly more attenuated than the ∆vchM parent during anaerobic growth, raising the possibility that the in vivo attenuation of ∆rpoE strains is in part explainable by the anaerobic conditions in the intestine (Fig 4D). Interestingly, the deleterious effect of the anaerobic environment on the ∆vchM ∆rpoE mutant was eliminated by the addition of fumarate, which acts as a terminal electron acceptor for V. cholerae respiration during anaerobic growth [55]. Thus, non-respiratory growth (e.g., fermentation) is specifically deleterious for V. cholerae lacking rpoE. It is possible that a fermentative byproduct is selectively toxic to ∆rpoE cells, or that physiological changes linked to fermentative growth lead to increased cell envelope stress.
TIS identifies genes required for envelope stability
We used transposon-insertion sequencing to begin to explain why the presence of VchM correlates with higher basal σE levels. We hypothesized that VchM regulates genes whose products are necessary for optimal envelope stability and conducted a screen to identify genes whose absence leads to increased envelope stress. In cells with a functional envelope stress pathway, disruption of genes that are required for envelope stability should induce envelope stress that can be ameliorated by the σE pathway. However, in cells lacking rpoE, such mutations (and the resulting stress) may be lethal. Using TIS, we identified 13 candidate genes that could tolerate transposon insertion in the ∆vchM background but not in ∆vchM ∆rpoE cells (Fig 3A, Table 2). Many of these genes are known to be involved in envelope biogenesis (Table 2), including several genes required for modifying the inner core component of LPS (orange shaded genes). Consistent with our hypothesis, disruption of many of these LPS synthesis genes (including waaC and rfaF) is known to cause induction of the σE stress response in E. coli [36,56]. We also identified non-LPS outer membrane-related genes, such as smpA, a known σE-regulating gene that is required for assembly of outer membrane beta-barrel proteins (OMPs) and for correct outer membrane biogenesis, and lnt, which acylates lipoproteins transported by the Lol complex [57,58,59]. It is known that overexpression of the major outer membrane protein, Lpp, in E. coli, induces the σE stress response, potentially by dysregulating outer membrane lipoprotein insertion through the LolA/B complex [44]. Additionally, the screen yielded genes that are not clearly tied to the rpoE pathway or outer membrane biogenesis, including cysB, a transcriptional activator that regulates sulfur transport [60], and vca0200 and vca0201, two hypothetical genes that are specific to V. cholerae and lie downstream of vchM. Using mutants from an ordered V. cholerae transposon library [24], we validated our hypothesis that these mutants contain more σE than wt cells (Fig 5A), suggesting envelope stress is increased in the absence of these genes.
Fig. 5. Other cell envelope-related loci and VchM control of LPS modification contribute to basal levels of σE. 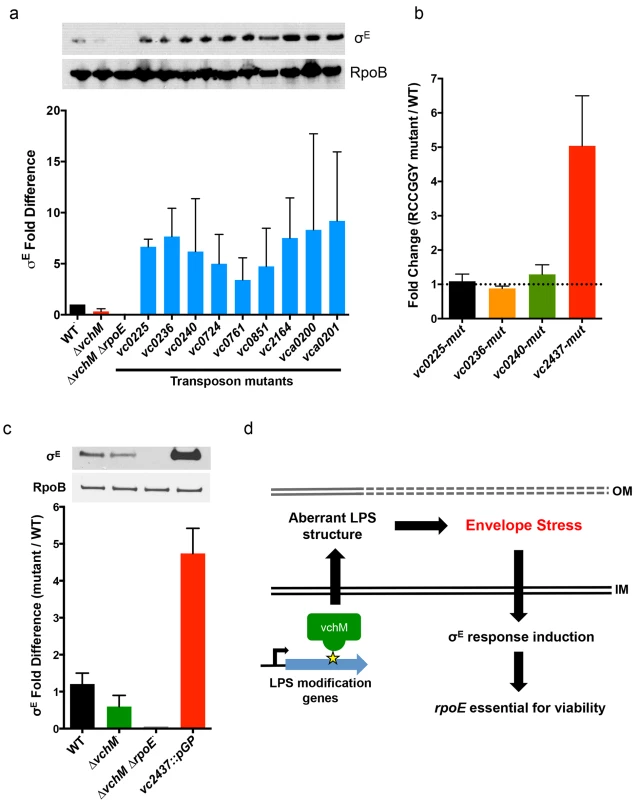
(A) Western blotting and relative abundance (compared to wt) of σE in mutants lacking genes that could not be disrupted in the ∆vchM ∆rpoE mutant. Mutants contain transposon insertions in the genes of interest, and are derived from a wt strain, not ∆vchM. RpoB was used as a loading control to normalize for total protein. Results are the average of three independent experiments. (B) Relative abundance of transcripts for LPS synthesis genes in mutants containing synonymous mutations that abolished RCCGGY motifs in these genes. Abundance is relative to transcript levels in wt cells, as determined by qRT-PCR in three independent biological replicates. (C) The relative abundance of σE in wt, ∆vchM and ∆vchM ∆rpoE cells and in a vc2437 insertion mutant (vc2437::pGP). Error bars represent data from two independent replicates normalized to wt basal σE levels. (D) Model for VchM-mediated modulation of the σE envelope stress response in V. cholerae. In wt cells, VchM methylation of some LPS modification genes restricts their expression, leading to aberrant LPS structure, generation of envelope stress and the induction of the σE response, which renders rpoE essential. Tab. 2. TIS identification of genes necessary for envelope stability. 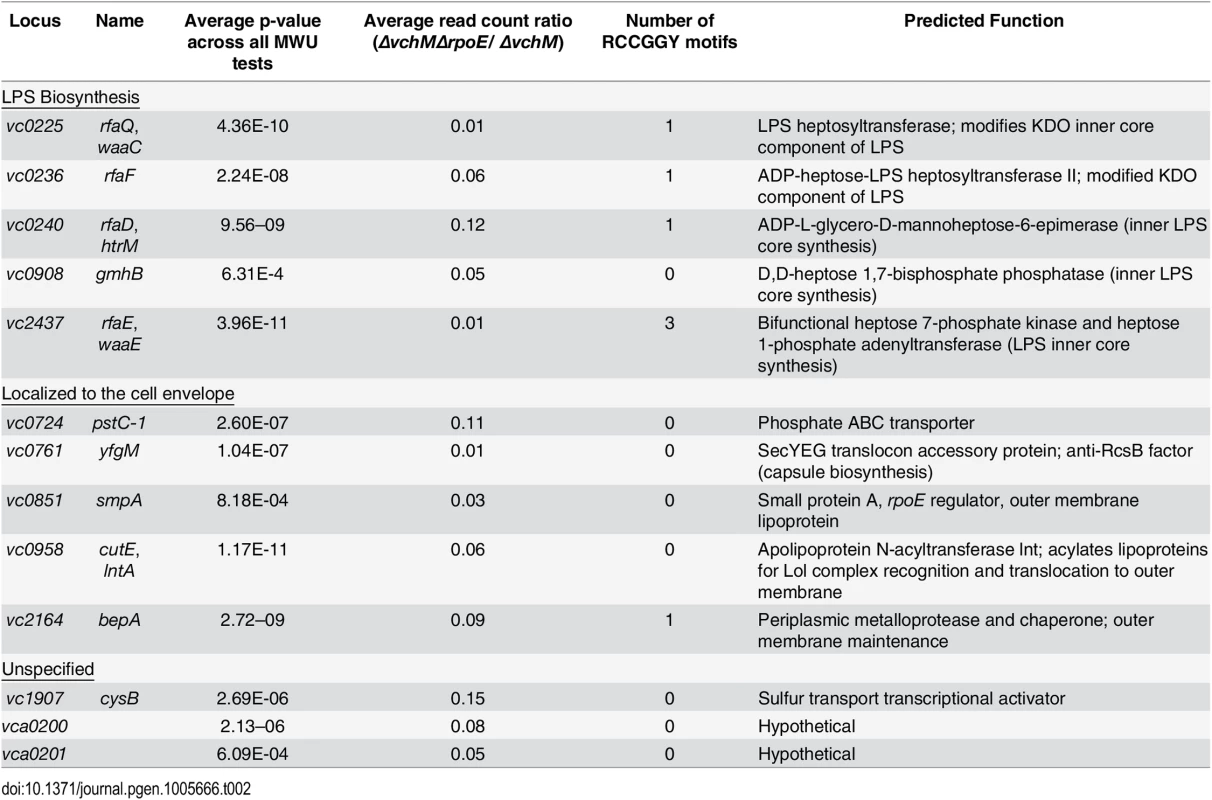
Genes are grouped based on functioning in LPS biosynthesis, localization to the cell envelope, or an unspecified contribution towards cell envelope processes. We speculated that expression of some of these loci might be directly regulated by VchM-dependent methylation, which could then allow them to modulate cellular levels of σE. Interestingly, the LPS-related candidate genes are enriched for RCCGGY motifs compared to candidate genes of the other classes (p-value < 0.05 by Fisher’s exact test), so we focused on this subset. We used site directed mutagenesis to replace the RCCGGY motifs in vc0225, vc0236, vc0240 and vc2437 with non-consensus sites that do not alter the protein sequence. While there were no significant differences in mRNA expression of vc0225, 0236 and 0240 after RCCGGY ablation, replacing all three RCCGGY motifs in vc2437 caused an approximately 5-fold increase in transcript abundance compared to WT levels (Fig 5B), suggesting that methylation of vc2437 by VchM reduces its expression. Consistent with the idea that regulation is methylation-dependent, there was a less than two-fold difference between expression of the wt and mutated locus in the vchM background. Furthermore, a mutant harboring a disruption of vc2437 (vc2437::pGP) had significantly elevated basal σE levels (Fig 5C), suggesting that downregulation of this gene’s expression can induce V. cholerae envelope stress.
Discussion
We carried out methylomic, transcriptomic, and genetic analyses of the role of the orphan cytosine MTase, VchM, and uncovered an unexpected connection between cytosine modification and outer membrane stress in V. cholerae. To our knowledge, VchM is only the second orphan 5mC-catalyzing bacterial MTase to be characterized in depth and genetic interactions comprehensively identified. Strains lacking VchM exhibit impaired growth in vivo and during both aerobic and anaerobic growth in vitro, and transcriptomic analysis indicates that many important metabolic pathways are altered in ∆vchM cells. Comparative transposon insertion sequencing analyses of wt and ∆vchM V. cholerae revealed that deletion of vchM, suppresses V. cholerae’s requirement for the σE envelope stress response pathway, which likely reflects reduced basal activity of this pathway in the ∆vchM strain. Notably, deletion of vchM does not mitigate V. cholerae’s need for rpoE under challenging growth conditions, and by studying the ∆vchM ∆rpoE mutant we were able to quantify the contribution of σE during host infection and during anaerobic growth as well as screen for genes that cannot be disrupted in the absence of the σE stress pathway. These genes, which are likely required for optimal envelope stability, include loci that catalyze the heptose modification of LPS and are enriched for VchM recognition motifs. Our data indicate that methylation directly regulates expression of at least some of these genes, and thus reveals one of the mechanisms by which vchM influences V. cholerae physiology.
Unexpectedly, we found that VchM is required for optimal V. cholerae growth in standard laboratory media (LB). As deletion of rpoE does not further impair growth of the ∆vchM mutant under this condition, it is unlikely that the mutant’s dysregulation of the σE envelope stress response pathway alone accounts for its slower proliferation. Instead, additional processes, such as those shown by transcriptomic analyses to be altered by the absence of VchM, (e.g., the amino acid and carbohydrate metabolism pathways and those mediating aerobic respiration), may account for the growth phenotype. However, it remains unclear whether the growth deficiency is due to altered expression of a single pathway or to the cumulative effect of simultaneously dysregulating numerous genetic loci. Nonetheless, our data suggests that the horizontally acquired VchM has become an integral component of V. cholerae’s regulatory networks.
In wt V. cholerae, rpoE is an essential gene [47]; thus, it is noteworthy that rpoE can be deleted in the ∆vchM strain, as relatively few suppressors of rpoE essentiality have been identified. Additionally, as noted above, our analyses indicate that growth of the ∆vchM and ∆vchM ∆rpoE strains can be equivalent. Deletion of vchM appears to reduce basal expression/activation of σE, likely reflecting a reduced need for this factor under conditions that apparently do not produce significant envelope stress. A similar reduction has been observed in strains in which OmpU levels are reduced and rpoE is not essential [47]. However, we do not observe reduced expression of OmpU in the ∆vchM cells (S6 Fig), suggesting ∆vchM suppresses σE essentiality through a novel mechanism. Furthermore, deletion of vchM does not render rpoE dispensable under all growth conditions. We observed marked differences (e.g., 10–100 fold) between the growth of ∆vchM and ∆vchM ∆rpoE strains in the presence of an antimicrobial compound that activates the σE pathway, in an animal model of infection, and during anaerobic growth. These data suggest that the vchM mutant retains the ability to activate the σE regulon in response to envelope (or potentially other) stress, despite its lower basal level of σE.
We performed a TIS screen to identify mutations that are detrimental specifically in the absence of rpoE, as such mutations are likely to activate σE, and thus might also be regulated through VchM methylation. This screen identified a variety of loci whose products are associated with the cell envelope, including several factors required for heptose modification of the LPS inner core. As anticipated, mutations in these loci were associated with increased σE abundance, suggesting these mutants have defects in cell envelope integrity. The deletion of genes involved in LPS modification, especially those catalyzing inner core and Lipid A synthesis, have been previously linked to abnormal LPS structure and σE activation in E. coli [36,56,61]. Intriguingly, many genes encoding the LPS modifying enzymes identified by our TIS screen contained RCCGGY motifs, raising the possibility that some of these genes might be directly regulated by VchM methylation. Indeed, synonymous disruption of three VchM targets sites in one locus, vc2437, led to enhanced expression of this gene, suggesting that VchM-dependent methylation may limit its expression. Since disruption of vc2437 leads to increased accumulation of σE, we hypothesize that increased levels of Vc2473 in the vchM mutant will reduce stimuli activating the σE stress response, as outlined in our model (Fig 5D). In E. coli, two signals are required for RseA cleavage and thus σE induction: activation of DegS through interaction with unfolded outer membrane proteins [33,34] and displacement of RseB from RseA through titration by periplasmic LPS [36]. Lowered expression of vc2437 could induce and sustain both of these signals. Reduced heptose modification may produce aberrant periplasmic LPS molecules that bind and compete RseB away from RseA, lowering the threshold for DegS activation [35], while simultaneously, aberrant LPS may alter outer membrane structure, facilitating the misincorporation and unfolding of outer membrane proteins.
Additional VchM targets may also exhibit methylation-regulated expression, since transcriptomic and genomic analyses showed that there is a significant correlation between the presence of intragenic RCCGGY motifs and increased expression of these genes in ∆vchM cells. However, it is likely that cytosine methylation does not universally modulate gene expression in V. cholerae, since mutation of the single RCCGGY motif in each of three LPS modification genes did not detectably alter gene expression. Genes containing 5 or more motifs showed the greatest elevation in expression in the vchM mutant, raising the possibilities that the effects of methylation may be additive within a single gene and/or that a threshold level of methylation must be present for regulation to occur. It is likely that additional regulatory factors also constrain the influence of methylated bases on gene expression.
VchM is the second bacterial orphan 5mC-catalyzing MTase to be characterized extensively at the genomic and transcriptomic levels, and the first for which genetic interactions have been comprehensively defined. Interestingly, our findings reveal striking differences between the functional consequences of methylation by VchM and the previously characterized E. coli 5mC-catalyzing orphan MTase, Dcm [15]. While Dcm regulates stationary phase expression of ribosomal proteins and mediates resistance to antibiotics in E. coli [13–15], VchM is required for optimal bacterial growth and modulates cell envelope stress responses in V. cholerae. At the genomic level, the E. coli genome is enriched in Dcm targets (CCWGG) [15], while the VchM target, RCCGGY, is not overrepresented in the V. cholerae genome. Furthermore, Dcm sites are undermethylated during exponential phase and rise during entry into stationary phase, while VchM target sites are nearly fully methylated under all growth states tested, including during infection, suggesting that the two enzymes differ in their expression and/or activity. While increases in gene expression were observed in both ∆dcm and ∆vchM cells, differentially expressed genes in ∆dcm strains are not enriched for intragenic CCWGG motifs and are instead thought to be controlled indirectly through rpoS-dependent regulation [14,15]. In contrast, there is a significant association between the presence of intragenic VchM targets and elevated gene expression in ∆vchM cells, and mutational analyses suggest that the methylation of these motifs can dampen gene expression. Thus, despite sharing similar catalytic activities, different orphan 5mC MTases can regulate diverse processes through different mechanisms.
The mechanism(s) by which methylation alters gene expression have not been characterized, but many possibilities can be envisioned. For example, methylation within transcribed sequences may influence transcriptional attenuation or transcript half life, perhaps due to effects upon transcript or template structure. Methylation might also influence transcription initiation, e.g. by influencing binding of regulatory factors that control gene expression. It should also be noted that many genes whose expression differs between wt and vchM V. cholerae are likely indirectly controlled by methylation, e.g., are governed by factors influenced by methylation, but are not themselves methylated. Furthermore, methylation may globally alter chromatin structure in ways that modulate gene expression. Investigation of the precise means by which cytosine methylation in V. cholerae influences gene expression will be the focus of future studies.
Banerjee et al. [22] found that the closest VchM homologues lie in non-Vibrio species, suggesting VchM was horizontally acquired. The introduction of a MTase such as VchM, which modifies thousands of sites and potentially alters gene expression at numerous loci, would exert selective pressure on MTase recognition sites. Consistent with this theory, we found that RCCGGY motifs in V. cholerae are not randomly distributed; instead, the genome includes regions that are enriched or depleted for VchM recognition sites (S2B Fig). Interestingly, many of the σE-related genes that become dispensable in V. cholerae lacking vchM (as well as vchM itself) are located in regions containing a disproportionately low number of RCCGGY motifs (binomial test p-value <4.8e-6; S2B and S2C Fig). It is possible that target sites in these loci have been selected against, as methylation might interfere with beneficial regulatory processes that promote σE expression.
In conclusion, we found that VchM, a 5mC-catalyzing DNA methyltransferase, serves critical roles in V. cholerae growth and envelope stress signaling. The processes and mechanisms through which VchM exerts control are strikingly different from E. coli Dcm, the other well-characterized bacterial 5mC DNA MTase. Thus, future investigations of the regulatory roles of additional bacterial DNA MTases will likely reveal new regulatory schemes for the control of diverse bacterial processes.
Materials and Methods
Ethics statement
All animal infections were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal protocols were reviewed and approved by the Harvard Medical Area Standing Committee on Animals (protocol 04316). Isofluorane was used for anesthesia.
Strains, media and culture conditions
All strains were grown on LB Miller (1% NaCl) unless otherwise noted. Antibiotic concentrations used were 200 μg/mL streptomycin (Sm), 50 μg/mL kanamycin (Km), 100 μg/mL ampicillin (Amp), and 50 μg/mL polymyxin B. Wildtype V. cholerae C6706, V. cholerae O395 and E. coli SM10 (lambda pir) carrying the Himar1 suicide transposon vector pSC189 [62] were grown at 37°C in LB + Sm and LB + Amp, respectively. Individual transposon mutants from the ordered V. cholerae transposon library [63] (which contain intact vchM) were grown overnight in LB + Sm + Km, or plated as lawns on LB + Sm + Km at 37°C.
Construction of V. cholerae deletion, revertant and RCCGGY mutant strains
All primers used for mutant generation can be found in S6 Table. Deletion plasmids for hsdM (vc1769), vchM (vca0198) and vca0447 were derived from the allelic exchange vector pCVD442 [64] using isothermal assembly [65]. Each deletion construct encodes the first five and last four amino acids of the gene of interest, with intervening sequences removed. Suicide plasmids were conjugated into V. cholerae and sucrose-based counter-selection performed as previously described [66] to create in-frame deletions. Allelic exchange of rpoE for a kanamycin resistance gene was carried out using a previously generated suicide vector [47] and a similar protocol as above. However, cells were plated on both 10% sucrose and kanamycin to select for the ∆rpoE mutant. For the introduction of a vchM C109A mutation into its endogenous locus, an allelic exchange vector containing the entire vchM gene with a C109A mutation flanked by 500 bp of upstream and downstream sequence was created, conjugated into ∆vchM cells, and counter-selected on sucrose plates. Subsequently, a second exchange vector was constructed containing wildtype vchM sequence that included 200 bp flanking sequence on each side of the C109 codon, which was used to revert the vchM C109A allele back to wildtype.
Similar constructs and methodology were used to replace the single RCCGGY motifs of vc0225, vc0236, vc0240 with ACAGGT, ACAGGC, and GCAGGT, respectively. For vc2437, a 850 bp fragment encompassing mutations in the three RCCGGY motifs (converted to GCCTGC, GCAGGT and GCAGGC) were synthesized (Integrated DNA Technologies) and used to carry out allelic exchange. For vc2437 disruption, an internal 850 bp fragment of vc2437 (derived from the PCR product using primers vc2437-mut1F and vc2437-mut3R) was blunt ligated into the suicide vector pGP704 [67] and conjugated into V. cholerae. All deletions, reversions and mutations were confirmed by Sanger sequencing (Genewiz).
Restriction digests
1 ug of purified genomic DNA from the indicated strains was subjected to digestion with SalI in the presence or absence of BsrFI at 37°C for 30 minutes. The reactions were separated by gel electrophoresis, stained with ethidium bromide and imaged on a FujiFilm FLA-5100 fluorescent imager.
Growth curves, in vitro and in vivo competition experiments
For growth curves, overnight V. cholerae cultures were diluted 1 : 1000 in LB + Sm and any additional chemicals as indicated. These cultures were grown at 37°C in a Bioscreen C optical density reader (Growth Curves USA) with OD600 measurements taken at 15 minute intervals for 12 hours. For competitive growth experiments, overnight stationary cultures of wildtype and mutant V. cholerae, one of which was lacZ-, were independently diluted 1 : 1000 in LB + Sm and then mixed in a 1 : 1 ratio. 20 μl of the diluted mixture was inoculated into 2 mL of LB + Sm and grown at 30°C for 24 hours. At the start and end of the experiment, cells were diluted and plated onto LB + 60 μg/mL X-gal to enumerate the ratio between wildtype and mutant cells. For anaerobic experiments, overnight cultures of V. cholerae were diluted into pre-warmed LB + Sm that was pre-depleted for oxygen by sitting overnight anaerobically at 37°C. Diluted cells were mixed and inoculated into 2 mL of oxygen-depleted LB + Sm (+ 50 μg/mL fumarate when indicated) and after 24 hours of growth at 37°C, cultures were removed from anaerobic conditions and the competitive index determined on X-gal as above.
For in vivo competitions, overnight stationary cultures of LacZ - wildtype or mutant V. cholerae were diluted and mixed 1 : 1 as above. 50 uL of the diluted culture was orally inoculated into 5-day-old suckling mice (Charles River). After 24 hrs, the mice were sacrificed, the small intestine homogenized using a mini-beadbeater-16 and two 3.2mm stainless steel beads (BioSpec Products Inc., Bartlesville, OK, USA) for 2 minutes, and dilutions of the homogenate were plated on LB + Sm + 60 ug/mL X-gal plates to enumerate the ratio of wildtype and mutant bacteria.
Bisulfite sequencing
Genomic DNA was extracted from two biological replicates of exponential phase, overnight stationary phase and frozen rabbit cecal fluid V. cholerae using the Wizard Genomic DNA purification kit (Promega). Bisulfite conversion of the DNA was carried using the EZ DNA Methylation Kit (Zymo) twice to ensure high conversion efficiency. The converted DNA was then amplified by PCR using the Kapa HiFi Uracil+ polymerase for 12–15 cycles (Kapa biosystems) and sequenced on the MiSeq platform (Illumina). Bismark [68] was used to call 5mC sites from the bisulfite sequencing data. Each cytosine site was assigned with two counts, representing the numbers of reads that had a C->T conversion (non-methylated) and those that did not have a C->T conversion (methylated). The fraction of methylation was then calculated for each cytosine site and a minimum total coverage of 10x was used to filter out cytosine sites with too few read counts for estimating the methylation frequency.
RNA sequencing
Purified mRNA was extracted from two biological replicates of exponential phase V. cholerae and converted to cDNA as previously described [69]. RNA sequencing was performed on a HiSeq 2500 with 100bp single-end reads. To call differentially expressed genes from RNAseq data, we first mapped raw RNA reads for each sample to the Genbank V. cholerae El Tor N16961 reference (Accession number: NC_002505 for chromosome I and NC_002506 for chromosome II), which is highly similar (>99.6% of VchM target sites are conserved). Reads that mapping to rRNA and tRNAs were excluded. A gene was included for differential expression analysis if it had more than one count per million reads (CPM = 1) in at least two samples. Differentially expressed genes (>2-fold differences, p-value <0.01, false discovery rates <0.2) were identified by the software program edgeR [70]. Expression differences for all genes in ∆vchM C6706 and O395 cells are located in S7 Table and S8 Table.
Correlation analysis between motif counts and gene expression fold changes
For motif counts within gene coding regions, we first use linear regression (regress fold change on gene length) to get residuals after removing gene length effects (Fig 2B). Furthermore, to confirm that the correlation between RCCGGY count in gene body and gene expression fold change is not due to GC bias, or more generally less-specific motifs of RCCGGY, we use a linear regression to remove the effects of all less-specific motifs. For example, for RCCGGY, the less-specific motifs are RCCGG, CCGGY, RCCG, CCGG, CGGY, RCC, CCG, CGG, GGY, RC, CC, CG, GG, GY, R, C, G, Y. We observed partial correlation between motif count and gene expression fold change after removing effects of all less-specific motifs, as well as length of the gene. As shown in S4C Fig, only the RCCGGY motif has significant correlation with fold change.
The RCCGGY motif distribution within genes was determined using a custom Python script. Briefly, RCCGGY motifs in every gene were localized to windows corresponding to 5% of the coding length of the gene and the sites in each window enumerated.
Transposon mutant library construction and sequencing
Transposon libraries were created in wildtype, ∆vchM, or ∆vchM∆rpoE V. cholerae, and genomic DNA was purified and sequenced as previously described [23], with the exception that 10 ug of purified genomic DNA was sheared to ~350 bp fragments through acoustic disruption (Covaris, Woburn, MA, USA) for each DNA library. After sequencing and mapping, the read counts for every TA site were tallied and assigned to annotated genes or intergenic regions using custom scripts [23]. The raw read count data for all libraries can be found in S1 Table. Reads in the WT and mutant TIS libraries were normalized for differences in library saturation and read depth through simulation-based resampling and then subjected to Mann-Whitney U statistical tests as previously described in the ARTIST pipeline [66]. Candidates with significant p-values (<0.001) and > 5 fold differences in normalized read counts were considered as candidates for follow-up. In total, the WT, ∆vchM and ∆vchM ∆rpoE libraries contained ~650000 colonies with 118683, 103029, and 115845 unique transposon insertions detected from 3125378, 656980, 2709183 total mapped reads, representing insertions at 62%, 53% and 60% of all TA dinucleotides, respectively. The full ARTIST analyses for the ∆vchM and ∆vchM ∆rpoE experiments as well as the raw read counts data are found in S3, S4 and S5 Tables, respectively.
Immunoblotting
Strains of interest were harvested at mid-exponential phase (OD 0.5), lysed directly in 1X NuPAGE LDS buffer (Novex) containing 6uM DTT, separated by NuPAGE Bis-Tris gel electrophoresis and transferred onto nitrocellulose using the iBlot system (Life Technologies). Blots were incubated with rabbit polyclonal antisera against σE or monoclonal antibody against RpoB (sc56766, Santa Cruz Biotechnology) in 5% milk in TBST. Horseradish-peroxidase conjugated secondary antibodies (Pierce) and Supersignal West Pico chemilumeniscent substrate (Pierce) were used to detect primary antibody signal. Blots were visualized on X-ray film, which was subsequently digitized on a FujiFilm FLA-5100 imager, and bands quantitated using MultiGuage V3.1 image analysis software.
Quantitative RT-PCR
Overnight stationary cells were inoculated into 3 ml LB + Sm medium, grown at 37°C until mid-late exponential phase (OD 600 0.5–0.8), harvested and total RNA extracted with TRIzol reagent (Life Technologies). RNA was treated with Turbo DNase I for 30 min (Life Technologies) and subjected to qRT-PCR as previously described [71]. Briefly, 1 μg total RNA was used for the reverse transcription reaction with Superscript III first strand synthesis system with random hexamers (Life Technologies). The synthesized cDNA was subjected to real time-PCR amplification using the Fast SYBR Green Master Mix kit (Life Technologies) on the StepOnePlus platform (Life Technologies) using primers shown in S6 Table. The amplification data was analyzed by ΔΔCT method utilizing rpoC mRNA as internal control.
Supporting Information
Zdroje
1. Schubeler D. Function and information content of DNA methylation. Nature 2015;517 : 321–326, doi: 10.1038/nature14192 25592537
2. Casadesús J. & Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 2006;70 : 830–856, doi: 10.1128/MMBR.00016-06 16959970
3. Pósfai J., Bhagwat A. S. & Roberts R. J. Sequence motifs specific for cytosine methyltransferases. Gene 1988;74 : 261–265. 3248729
4. Timinskas A., Butkus V. & Janulaitis A. Sequence motifs characteristic for DNA [cytosine-N4] and DNA [adenine-N6] methyltransferases. Classification of all DNA methyltransferases. Gene 1995;157 : 3–11. 7607512
5. Murray N. E. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol Mol Biol Rev 2000;64, 412–434. 10839821
6. Wion D. & Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol 2006;4 : 183–192, doi: 10.1038/nrmicro1350 16489347
7. Marinus M. G. & Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev 2009;33; 488–503, doi: 10.1111/j.1574-6976.2008.00159.x 19175412
8. Lu M., Campbell J. L., Boye E. & Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell 1994;77 : 413–426. 8011018
9. Schlagman S. L., Hattman S. & Marinus M. G. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J Bacteriol 1986;165 : 896–900. 3512529
10. Roberts D., Hoopes B. C., McClure W. R. & Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell 1985;43 : 117–130, doi:0092-8674(85)90017-0 [pii]. 3000598
11. Peterson S. N. & Reich N. O. Competitive Lrp and Dam assembly at the pap regulatory region: implications for mechanisms of epigenetic regulation. J Mol Biol 2008;383 : 92–105, doi: 10.1016/j.jmb.2008.07.086 18706913
12. Collier J., McAdams H. H. & Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA 2007;104; 17111–17116, doi: 10.1073/pnas.0708112104 17942674
13. Militello K. T., Mandarano A. H., Varechtchouk O. & Simon R. D. Cytosine DNA methylation influences drug resistance in Escherichia coli through increased sugE expression. FEMS Microbiol Lett 2014;350 : 100–106, doi: 10.1111/1574-6968.12299 24164619
14. Militello K. T. et al. Conservation of Dcm-mediated cytosine DNA methylation in Escherichia coli. FEMS Microbiol Lett 2012;328 : 78–85, doi: 10.1111/j.1574-6968.2011.02482.x 22150247
15. Kahramanoglou C. et al. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat Commun 2012;3 : 886, doi: 10.1038/ncomms1878 22673913
16. Srikhanta Y. N. et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog 2009;5: e1000400, doi: 10.1371/journal.ppat.1000400 19390608
17. Srikhanta Y. N. et al. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS ONE 2011;6: e27569, doi: 10.1371/journal.pone.0027569 22162751
18. Roberts R. J., Vincze T., Posfai J. & Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 2010;38: D234–236, doi: 10.1093/nar/gkp874 doi:gkp874 [pii] 19846593
19. Heidelberg J. F. et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000;406 : 477–483, doi: 10.1038/35020000 10952301
20. Demarre G. & Chattoraj D. K. DNA adenine methylation is required to replicate both Vibrio cholerae chromosomes once per cell cycle. PLoS Genet 2010;6: e1000939, doi: 10.1371/journal.pgen.1000939 20463886
21. Gerding MA, Chao MC, Davis BM, & Waldor MK. Molecular dissection of the essential features of the origin of replication of the second Vibrio cholerae chromosome. mBio 2015;6: e00973–15. doi: 10.1128/mBio.00973-15 26220967
22. Banerjee S. & Chowdhury R. An orphan DNA (cytosine-5-)-methyltransferase in Vibrio cholerae. Microbiology (Reading, Engl) 2006;152 : 1055–1062, doi: 10.1099/mic.0.28624–0
23. Chao M. C. et al. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res 2013;41 : 9033–9048, doi: 10.1093/nar/gkt654 23901011
24. Cameron D. E., Urbach J. M. & Mekalanos J. J. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci USA 2008;105 : 8736–8741, doi: 10.1073/pnas.0803281105 18574146
25. Kamp H. D., Patimalla-Dipali B., Lazinski D. W., Wallace-Gadsden F. & Camilli A. Gene fitness landscapes of Vibrio cholerae at important stages of its life cycle. PLoS Pathog 2013;9: e1003800, doi: 10.1371/journal.ppat.1003800 24385900
26. Haberman A., Heywood J. & Meselson M. DNA modification methylase activity of Escherichia coli restriction endonucleases K and P. Proc Natl Acad Sci U S A 1972;69 : 3138–3141. 4564204
27. Dalia A. B., Lazinski D. W. & Camilli A. Characterization of undermethylated sites in Vibrio cholerae. J Bacteriol 2013;195 : 2389–2399, doi: 10.1128/JB.02112-12 23504020
28. Papenfort K., Forstner K. U., Cong J. P., Sharma C. M. & Bassler B. L. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci U S A 2015;112: E766–775, doi: 10.1073/pnas.1500203112 25646441
29. Ades S. E. Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol 2008;11 : 535–540, doi: 10.1016/j.mib.2008.10.004 18983936
30. De Las Penas A., Connolly L. & Gross C. A. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol Microbiol 1997;24 : 373–385. 9159523
31. Missiakas D., Mayer M. P., Lemaire M., Georgopoulos C. & Raina S. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol 1997;24 : 355–371. 9159522
32. Campbell E. A. et al. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol Cell 2003;11 : 1067–1078. 12718891
33. Wilken C., Kitzing K., Kurzbauer R., Ehrmann M. & Clausen T. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell 2004;117 : 483–494. 15137941
34. Walsh N. P., Alba B. M., Bose B., Gross C. A. & Sauer R. T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003;113 : 61–71. 12679035
35. Chaba R. et al. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli. Proc Natl Acad Sci U S A 2011;108 : 2106–2111, doi: 10.1073/pnas.1019277108 21245315
36. Lima S., Guo M. S., Chaba R., Gross C. A. & Sauer R. T. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013;340 : 837–841, doi: 10.1126/science.1235358 23687042
37. Ades S. E., Connolly L. E., Alba B. M. & Gross C. A. The Escherichia coli sigma(E)-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 1999;13 : 2449–2461. 10500101
38. Alba B. M., Leeds J. A., Onufryk C., Lu C. Z. & Gross C. A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev 2002;16 : 2156–2168, doi: 10.1101/gad.1008902 12183369
39. Kanehara K., Ito K. & Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev 2002;16 : 2147–2155, doi: 10.1101/gad.1002302 12183368
40. Akiyama Y., Kanehara K. & Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J 2004;23 : 4434–4442, doi: 10.1038/sj.emboj.7600449 15496982
41. Dartigalongue C., Missiakas D. & Raina S. Characterization of the Escherichia coli sigma E regulon. J Biol Chem 2001;276 : 20866–20875, doi: 10.1074/jbc.M100464200 11274153
42. Rezuchova B., Miticka H., Homerova D., Roberts M. & Kormanec J. New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol Lett 2003;225 : 1–7. 12900013
43. Rhodius V. A., Suh W. C., Nonaka G., West J. & Gross C. A. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS biology 2006;4: e2, doi: 10.1371/journal.pbio.0040002 16336047
44. Guo M. S. et al. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 2014;28 : 1620–1634, doi: 10.1101/gad.243485.114 25030700
45. Thompson K. M., Rhodius V. A. & Gottesman S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol 2007;189 : 4243–4256, doi: 10.1128/JB.00020-07 17416652
46. De Las Penas A., Connolly L. & Gross C. A. SigmaE is an essential sigma factor in Escherichia coli. J Bacteriol 1997;179 : 6862–6864. 9352942
47. Davis B. M. & Waldor M. K. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res 2009;37 : 5757–5767, doi: 10.1093/nar/gkp568 19620211
48. Dartigalongue C., Loferer H. & Raina S. EcfE, a new essential inner membrane protease: its role in the regulation of heat shock response in Escherichia coli. EMBO J 2001;20 : 5908–5918, doi: 10.1093/emboj/20.21.5908 11689431
49. Alba B. M., Zhong H. J., Pelayo J. C. & Gross C. A. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide sigma (E) activity. Mol Microbiol 2001;40 : 1323–1333. 11442831
50. Button J. E., Silhavy T. J. & Ruiz N. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol 2007;189 : 1523–1530, doi: 10.1128/JB.01534-06 17172327
51. Daimon Y., Narita S. & Akiyama Y. Activation of Toxin-Antitoxin System Toxins Suppresses Lethality Caused by the Loss of sigmaE in Escherichia coli. J Bacteriol 2015;197 : 2316–2324, doi: 10.1128/JB.00079-15 25917909
52. Douchin V., Bohn C. & Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J Biol Chem 2006;281 : 12253–12259, doi: 10.1074/jbc.M600819200 16513633
53. Mathur J., Davis B. M. & Waldor M. K. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 2007;63 : 848–858, doi: 10.1111/j.1365-2958.2006.05544.x 17181782
54. Provenzano D., Lauriano C. M. & Klose K. E. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol 2001;183 : 3652–3662, doi: 10.1128/JB.183.12.3652–3662.2001 11371530
55. Lee K. M. et al. Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J Biol Chem 2012;287 : 39742–39752, doi: 10.1074/jbc.M112.394932 23019319
56. Klein G., Lindner B., Brabetz W., Brade H. & Raina S. Escherichia coli K-12 Suppressor-free Mutants Lacking Early Glycosyltransferases and Late Acyltransferases: minimal lipopolysaccharide structure and induction of envelope stress response. J Biol Chem 2009;284 : 15369–15389, doi: 10.1074/jbc.M900490200 19346244
57. Fukuda A. et al. Aminoacylation of the N-terminal cysteine is essential for Lol-dependent release of lipoproteins from membranes but does not depend on lipoprotein sorting signals. J Biol Chem 2002;277 : 43512–43518, doi: 10.1074/jbc.M206816200 12198129
58. Robichon C., Vidal-Ingigliardi D. & Pugsley A. P. Depletion of apolipoprotein N-acyltransferase causes mislocalization of outer membrane lipoproteins in Escherichia coli. J Biol Chem 2005;280 : 974–983, doi: 10.1074/jbc.M411059200 15513925
59. Sklar J. G. et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 2007;104 : 6400–6405, doi: 10.1073/pnas.0701579104 17404237
60. Kredich N. M. Regulation of L-cysteine biosynthesis in Salmonella typhimurium. I. Effects of growth of varying sulfur sources and O-acetyl-L-serine on gene expression. J Biol Chem 1971;246 : 3474–3484. 4931306
61. Tam C. & Missiakas D. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol Microbiol 2005;55 : 1403–1412, doi: 10.1111/j.1365-2958.2005.04497.x 15720549
62. Chiang S. L. & Rubin E. J. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 2002;296 : 179–185, doi:S0378111902008569 [pii]. 12383515
63. Cameron D. E., Urbach J. M. & Mekalanos J. J. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 2008;105 : 8736–8741, 0803281105 [pii]. doi: 10.1073/pnas.0803281105 18574146
64. Donnenberg M. S. & Kaper J. B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 1991;59 : 4310–4317. 1937792
65. Gibson D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 2009;6 : 343–345, doi: 10.1038/nmeth.1318 19363495
66. Pritchard J. R. et al. ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing. PLoS Genet 2014;10: e1004782, doi: 10.1371/journal.pgen.1004782 25375795
67. Miller V. L. & Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 1988;170 : 2575–2583. 2836362
68. Krueger F. & Andrews S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27 : 1571–1572, doi: 10.1093/bioinformatics/btr167 21493656
69. Mandlik A. et al. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 2011;10 : 165–174, S1931-3128(11)00224-1 [pii]. doi: 10.1016/j.chom.2011.07.007 21843873
70. Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26 : 139–140, doi: 10.1093/bioinformatics/btp616 19910308
71. Wang Q. et al. A genome wide screen reveals that Vibrio cholerae phosphoenolpyruvate phosphotransferase system (PTS) modulates virulence gene expression. Infect Immun 2015; doi: 10.1128/IAI.00411-15 26056384
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



