-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRegulating Maf1 Expression and Its Expanding Biological Functions
article has not abstract
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004896
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1004896Summary
article has not abstract
Maf1 is a nutrient - and stress-sensitive global repressor of transcription by RNA polymerase (pol) III [1], [2]. Its primary function in this context is to limit the synthesis of highly abundant 5S rRNA and tRNAs in response to nutrient availability and cellular stress [3]. Thus, Maf1 ensures the efficient use of metabolic resources while balancing the need for protein synthetic components during cell growth, proliferation, differentiation, and quiescence. Less abundant products of pol III transcription (such as the spliceosomal U6 snRNA and the 7SL RNA component of signal recognition particle) are also repressed by Maf1, since its mechanism of action involves direct inhibitory interactions with proteins required for transcription by all pol III genes (i.e., the TFIIB-related initiation factors Brf1 or Brf2 and the polymerase) [4]–[7]. In the previous issue of PLOS Genetics, Palian et al. describe new insights into the regulation of Maf1 and its function in mammalian systems [8].
Studies on Maf1 regulation up until now have focused on posttranslational mechanisms, notably phosphorylation, which controls Maf1 localization (in yeast) and its interaction with the polymerase (in yeast and humans) [2], [3]. The new work from Palian and colleagues shows that the steady-state level of the Maf1 protein is also regulated. This is achieved through PI3K/AKT/FoxO1 signaling (Fig. 1). To reach this conclusion, tissue-specific PTEN knockout mice and a human PTEN null mutant cell line with inducible PTEN expression were used to perturb PI3K/AKT/FoxO1 signaling and show that Maf1 expression can be varied in both directions. Other manipulations of signaling through the pathway yielded consistent results. Importantly, mouse embryo fibroblasts in which the AKT substrate FoxO1 was knocked down or constitutively active showed reduced and elevated Maf1 protein levels, respectively, with corresponding reciprocal effects on the levels of precursor tRNAs (reflecting pol III transcription). Finally, the physiological relevance of the regulation was demonstrated by feeding mice a diet high in carbohydrates, which activates the pathway, and finding that Maf1 expression was decreased. One intriguing aspect of the work is that changes in PI3K/AKT/FoxO1 signaling affected Maf1 protein levels but had little influence on Maf1 mRNA. Additional studies are needed to determine how FoxO1, an insulin-sensitive DNA-binding transcription factor, alters the synthesis or stability of the Maf1 protein.
Fig. 1. PI3K signaling via FoxO1 regulates Maf1 abundance and downstream processes. 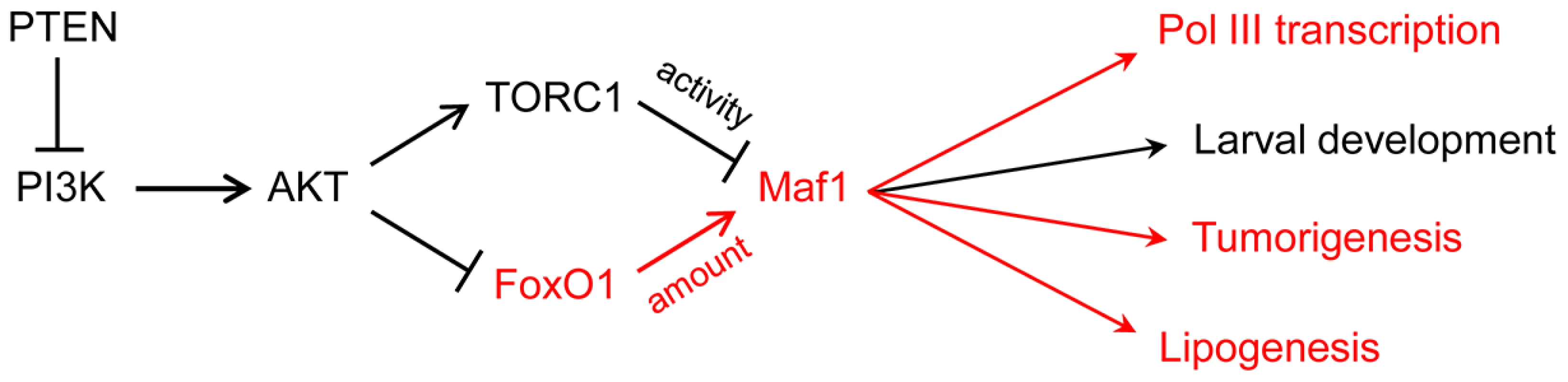
FoxO1 signaling to Maf1 and the biological processes that are sensitive to this regulation are shown in red. The larval development phenotype in Drosophila is primarily due to increased pol III transcription and elevated initiator tRNAMet. Maf1-regulation of tumorigenesis, previously thought to result from changes in pol III transcription, may also include direct effects of Maf1 on pol II transcription. In addition to its regulation by nutrients and stress, pol III transcription is inhibited by tumor suppressors, increased by oncogenic activation and cell transformation, regulated during the cell cycle, and targeted by viruses and other pathogens [9]–[13]. The extent to which Maf1 is involved in these transcriptional changes is either not well understood or has not been examined. However, the potential for Maf1 to impact cancer-related phenotypes is suggested by several observations including (i) its control by conserved oncogenic signaling pathways, e.g., the Ras/PKA pathway as demonstrated in yeast and the TOR pathway as shown in yeast, flies and mammalian cells [3], [14]–[16]; (ii) the requirement for elevated levels of pol III transcripts for Myc-driven cell transformation and tumorigenesis [17]; (iii) the increased growth of cells with elevated levels of initiator methionine tRNA [16], [18], [19]; and (iv) the ability of Maf1 overexpression to suppress anchorage-independent growth of PTEN-deficient human glioblastoma cells [20]. Expanding on this, Palian et al., report that Maf1 levels are reduced in PTEN-negative human prostate and liver cancers compared to matched normal tissue. Moreover, they show that hepatoma cells engineered to overexpress Maf1 exhibit less anchorage-independent growth and delayed onset of tumorigenesis when the cells are injected into mice (Fig. 1). These new experiments add to the growing importance of the pol III system in cancer biology and highlight its potential as a target for cancer therapeutics. One surprising aspect of these experiments is the low level of Maf1 overexpression that was apparently needed to affect a change in function. In future studies, it will be interesting to benchmark the phenotypic changes against specific cellular quantities of Maf1.
Beyond its role in the pol III system, previous work by Johnson et al. [20] found that knockdown and overexpression of Maf1 in human cell lines affected pol II transcription of the TBP and Egr1 genes. Maf1 is not known to be a DNA-binding protein. However, its effect on transcription at the human TBP promoter is thought to be direct since it was delimited to a promoter proximal region that crosslinked to Maf1 in chromatin immunoprecipitations (ChIPs). Since this initial report, few details have emerged on the scope of Maf1 in pol II gene regulation. One recent exception is the finding that deletion of MAF1 in yeast affects the expression of gluconeogenic genes [21]. The work from Palian et al., provides new knowledge in this regard, showing that genes encoding key lipogenic enzymes, acetyl CoA carboxylase (ACC1) and fatty acid synthase (FASN), are repressed by Maf1 (Fig. 1). As with the effect on pol III transcription, increasing or decreasing Maf1 expression had reciprocal effects on the expression of both enzymes. Moreover, these changes impacted the accumulation of lipid droplets in mammalian cell lines and triglyceride levels in mouse liver. Given the effect of Maf1 on pol III transcription and TBP expression, it is possible for Maf1 to affect the expression of additional genes by indirect mechanisms. A striking example of this capacity is the cell non-autonomous phenotype of a Maf1 knockdown in the fat body of Drosophila. In this case, increased organismal growth and accelerated larval development resulted from a diffusible signal, generated in the fat body, that affected systemic signaling by insulin-like peptides synthesized in the brain [16]. Arguing against an indirect effect of Maf1 in lipogenesis in mammalian cells is the fact that the protein ChIPs to the Fasn promoter. Clearly, genome-wide transcriptional profiling in cells with altered Maf1 expression is necessary to better appreciate the function of this global regulator.
Altogether, the new study identifies an important mechanism of Maf1 regulation along with new Maf1-regulated protein-coding genes that impact a novel biological function. Since research on Maf1 has only just scratched the surface for a limited number of model organisms, it seems certain that additional regulatory targets and biological functions have yet to be discovered in your favorite eukaryote.
Zdroje
1. UpadhyaR, LeeJ, WillisIM (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10 : 1489–1494.
2. BogutaM (2013) Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829 : 376–384.
3. MoirRD, WillisIM (2013) Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta 1829 : 361–375.
4. PlutaK, LefebvreO, MartinNC, SmagowiczWJ, StanfordDR, et al. (2001) Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21 : 5031–5040.
5. DesaiN, LeeJ, UpadhyaR, ChuY, MoirRD, et al. (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J Biol Chem 280 : 6455–6462.
6. ReinaJH, AzzouzTN, HernandezN (2006) Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS ONE 1: e134.
7. RollinsJ, VerasI, CabarcasS, WillisI, SchrammL (2007) Human Maf1 negatively regulates RNA polymerase III transcription via the TFIIB family members Brf1 and Brf2. Int J Biol Sci 3 : 292–302.
8. PalianBM, RohiraAD, JohnsonSAS, HeL, ZhengN (2014) Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet 10 (12) e1004789 doi:10.1371/journal.pgen.1004789
9. MarshallL, WhiteRJ (2008) Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer 8 : 911–914.
10. Dumay-OdelotH, Durrieu-GaillardS, Da SilvaD, RoederRG, TeichmannM (2010) Cell growth - and differentiation-dependent regulation of RNA polymerase III transcription. Cell Cycle 9 : 3687–3699.
11. RanaT, MisraS, MittalMK, FarrowAL, WilsonKT, et al. (2011) Mechanism of down-regulation of RNA polymerase III-transcribed non-coding RNA genes in macrophages by Leishmania. J Biol Chem 286 : 6614–6626.
12. FairleyJA, MitchellLE, BergT, KennethNS, von SchubertC, et al. (2012) Direct regulation of tRNA and 5S rRNA gene transcription by Polo-like kinase 1. Mol Cell 45 : 541–552.
13. GjidodaA, HenryRW (2013) RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim Biophys Acta 1829 : 385–392.
14. MichelsAA, RobitailleAM, Buczynski-RuchonnetD, HodrojW, ReinaJH, et al. (2010) mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 30 : 3749–3757.
15. KantidakisT, RamsbottomBA, BirchJL, DowdingSN, WhiteRJ (2010) mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A 107 : 11823–11828.
16. RideoutEJ, MarshallL, GrewalSS (2012) Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci U S A 109 : 1139–1144.
17. JohnsonSA, DubeauL, JohnsonDL (2008) Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem 283 : 19184–19191.
18. WhiteRJ (2008) RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet 24 : 622–629.
19. Pavon-EternodM, GomesS, RosnerMR, PanT (2013) Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19 : 461–466.
20. JohnsonSS, ZhangC, FrommJ, WillisIM, JohnsonDL (2007) Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell 26 : 367–379.
21. MorawiecE, WichtowskaD, GraczykD, ConesaC, LefebvreO, BogutaM (2013) Maf1, repressor of tRNA transcription, is involved in the control of gluconeogenetic genes in Saccharomyces cerevisiae. Gene 526 : 16–22.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



