-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
In bacterial cells, bidirectional replication of the circular chromosome is initiated from a single origin (oriC) and terminates in an antipodal terminus region such that movement of the pair of replication forks is largely codirectional with transcription. The terminus region is flanked by discrete Ter sequences that act as polar, or direction-dependent, arrest sites for fork progression. Alternative oriC-independent modes of replication initiation are possible, one of which is constitutive stable DNA replication (cSDR) from transcription-associated RNA–DNA hybrids or R-loops. Here, I discuss the distinctive attributes of fork progression and termination associated with different modes of bacterial replication initiation. Two hypothetical models are proposed: that head-on collisions between pairs of replication forks, which are a feature of replication termination in all kingdoms of life, provoke bilateral fork reversal reactions; and that cSDR is characterized by existence of distinct subpopulations in bacterial cultures and a widespread distribution of origins in the genome, each with a small firing potential. Since R-loops are known to exist in eukaryotic cells and to inflict genome damage in G1 phase, it is possible that cSDR-like events promote aberrant replication initiation even in eukaryotes.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004909
Category: Review
doi: https://doi.org/10.1371/journal.pgen.1004909Summary
In bacterial cells, bidirectional replication of the circular chromosome is initiated from a single origin (oriC) and terminates in an antipodal terminus region such that movement of the pair of replication forks is largely codirectional with transcription. The terminus region is flanked by discrete Ter sequences that act as polar, or direction-dependent, arrest sites for fork progression. Alternative oriC-independent modes of replication initiation are possible, one of which is constitutive stable DNA replication (cSDR) from transcription-associated RNA–DNA hybrids or R-loops. Here, I discuss the distinctive attributes of fork progression and termination associated with different modes of bacterial replication initiation. Two hypothetical models are proposed: that head-on collisions between pairs of replication forks, which are a feature of replication termination in all kingdoms of life, provoke bilateral fork reversal reactions; and that cSDR is characterized by existence of distinct subpopulations in bacterial cultures and a widespread distribution of origins in the genome, each with a small firing potential. Since R-loops are known to exist in eukaryotic cells and to inflict genome damage in G1 phase, it is possible that cSDR-like events promote aberrant replication initiation even in eukaryotes.
Introduction
Many features of chromosomal DNA replication are shared across the three kingdoms of life, including initiation from discrete origins, bidirectional fork progression, and termination by merger of opposing replication forks [1]. Whereas replication in eukaryotes is initiated from multiple origins on linear chromosomes, in bacteria most often there is a single circular chromosome whose replication is initiated from an oriC locus and proceeds bidirectionally for forks to meet in an antipodal terminus region. With this arrangement, replication and transcription of highly transcribed genes are rendered majorly codirectional in bacterial genomes. oriC-like sequences have been identified in more than 1,500 bacteria [2].
Alternative (oriC-independent) means of bacterial chromosomal replication have been characterized. These include (i) “integrative suppression” with replicons of phage or plasmids, and (ii) replication initiated from RNA–DNA hybrids or R-loops. The latter is called constitutive stable DNA replication (cSDR), whose mechanism is poorly understood. Significant perturbations, both of codirectionality between replication and transcription and of the arrangement for opposing replication forks to meet in the terminus region, are expected when bidirectional replication is not oriC-initiated.
This review explores the dynamics of fork progression and termination in Escherichia coli (gram-negative) bacterial cells exhibiting oriC-dependent and oriC-independent replication initiation to support two new concepts: (i) that when pairs of forks collide, bilateral fork reversal reactions take place; and (ii) that cSDR is characterized by stochastic replication initiation events distributed genome-wide. Table 1 summarizes relevant features and functions in E. coli that are shared in Bacillus subtilis (a gram-positive bacterium) and in eukaryotes, as is further elaborated in the text.
Tab. 1. Counterparts in B. subtilis and eukaryotes of E. coli functions related to chromosomal DNA replication and repair. 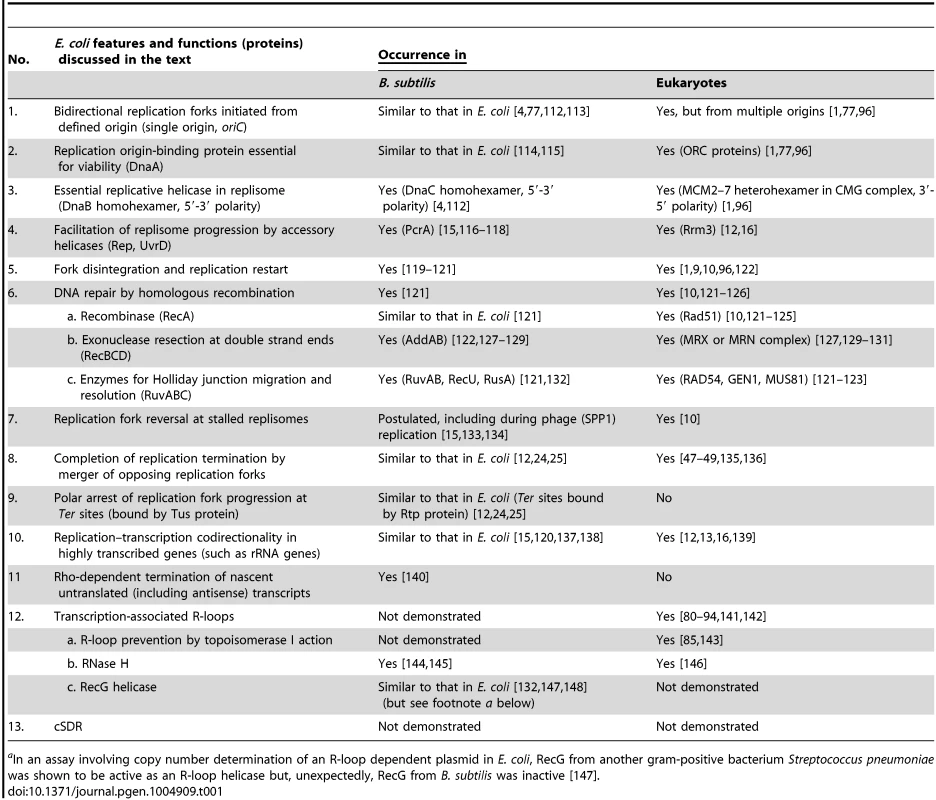
aIn an assay involving copy number determination of an R-loop dependent plasmid in E. coli, RecG from another gram-positive bacterium Streptococcus pneumoniae was shown to be active as an R-loop helicase but, unexpectedly, RecG from B. subtilis was inactive [147]. Replication Initiated from oriC and Its Termination
Bidirectional replication initiation at oriC is dependent on the protein DnaA, and included within the replisome complex at each fork is the 5′-3′ replicative helicase DnaB [3]–[6]. The forks move divergently around the circular chromosome to meet in the terminus region, and their traversed paths represent the (clockwise and counterclockwise) replichores (Fig. 1A). Both oriC and DnaA are essential for viability.
Fig. 1. Features of oriC-initiated replication in E. coli. 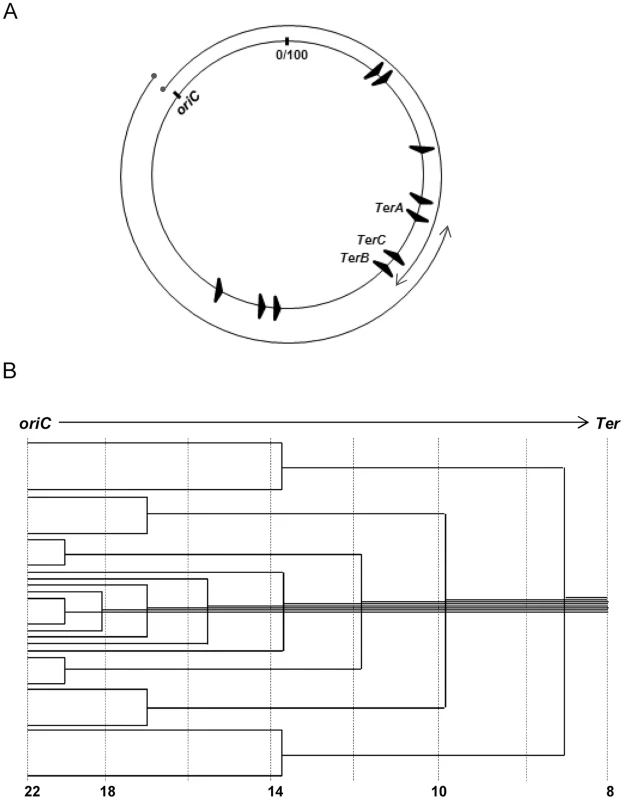
(A) Depiction of oriC, TerA, TerB and TerC loci on the 100 minute long circular E. coli chromosome, and of the clockwise and counterclockwise replichores; locations of the seven other Ter sites are also shown. (B) Schematic depiction of the copy number gradient, from oriC to Ter, created by the different extents to which replication forks have progressed on a single replichore in individual cells of an asynchronously dividing population. Aggregate copy numbers at the indicated positions are given at the bottom, but these are only illustrative and not to scale. Advancing forks may suffer disintegration [7], [8], whose frequency is increased with DNA damage [7]–[10] or by transcription–replication conflicts [11]–[16]. For example, all seven ribosomal RNA operons are codirectional with the replichores, and inversion of any of them leads to slowing or disintegration of replication forks [17]–[19]. Accessory helicases Rep and UvrD with 3′-5′ polarity facilitate replisome progression across DNA–protein barriers, including at sites of transcription–replication conflict [15], [18]–[20]. Fork disintegration also occurs when one fork runs into a preceding one stalled on the same replichore [21], [22]. Reassembly of disintegrated forks is mediated by replication restart proteins acting together with the proteins for homologous recombination RecA, RecBCD, and RuvABC (Table 1) [7]–[10].
At the terminus region, the Tus protein binds to discrete Ter sequences and mediates polar, or direction-specific, arrest of replisome progression (by inactivating DnaB helicase) [12], [23]–[25]. Thus, this region contains at (i) its clockwise end, TerA where counterclockwise forks are terminated, and (ii) its counterclockwise end, TerC and TerB where clockwise forks are arrested (Fig. 1A). Hence, most often chromosomal replication is completed when opposing forks meet at, or in, the interval between TerA and TerC or TerB. However, replisome arrest at Tus-bound Ter sites is not absolute [26], [27]. In addition to TerA, -B and -C, seven other Ter sequences are present on the E. coli replichores (Fig. 1A), oriented such as to cause arrest only of the oppositely directed replisomes [12], [24], [28].
Copy Number Analysis in Chromosome Replication Studies
When replication forks advance from origin to terminus in cells of an asynchronously dividing cell population, a decreasing gradient of gene copy numbers is expected from the former to the latter (Fig. 1B) [29], [30]. Analysis of copy number distributions has therefore enabled identification of origins and termini of replication [26], [27], [31]–[36] as well as of chromosome rearrangements [37]. Two caveats are (i) that copy numbers can change on account not only of fork progression but also of recombination (for example, tandem amplification [38]) or DNA degradation [39], [40]; and (ii) that the values represent an average of all cells in a population, which may comprise distinct subpopulations including inviable cells [38], [41].
When Replisomes Collide: Evidence for Bilateral Replication Fork Reversals
Replication fork reversal is the process by which nascent leading and lagging daughter strands at a fork are extruded to anneal to one another, thus forming a cruciform or “chicken-foot” structure. Such extrusion could occur when replisome progression is halted for any reason, and would be promoted by accumulation of positive supercoils ahead of the fork. Fork reversals can competitively be either limited by “end-resection” activity of the RecBCD complex, or exacerbated by the RuvABC proteins that catalyze branch migration and cleavage at Holliday junctions [19], [42]–[45], reviewed in [10], [46]. In RecBC-deficient strains, fork reversal is also accompanied by excessive chromosome degradation (which may indeed seem paradoxical given that RecBCD is itself a potent DNA exonuclease), that is mediated by RecJ nuclease [19].
Kuong et al. [40] and Rudolph et al. [41] have shown that chromosomal terminus region copy numbers are severely reduced in RecBC-deficient strains (the former studies were done with thymine starvation). This suggests that irreparable chromosome breakage and degradation occurs in the terminus region of some proportion of recBC mutant cells which, for example, may result from bilateral fork reversals provoked by head-on collisions between two replisomes, as depicted in Fig. 2. Additional experiments are required to confirm this hypothesis. Since collisions between pairs of replisomes are a common feature of replication termination in all organisms [25], [47]–[49], it is also possible that consequential fork reversals may be universal.
Fig. 2. Model of bilateral fork reversal reaction at a site where oncoming replisomes meet during replication termination. 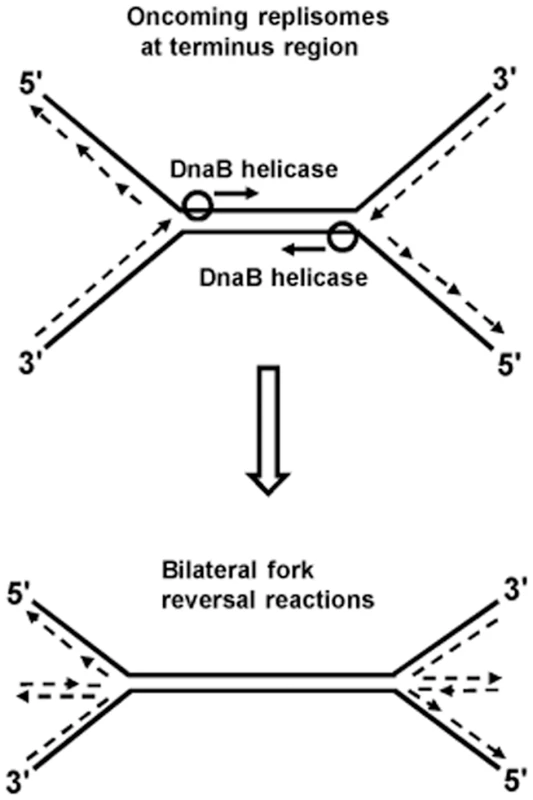
Chromosome Replication in Absence of oriC or DnaA: Integrative Suppression
In integrative suppression, the replication origin of a plasmid or phage is integrated into the chromosome of a strain defective in oriC or DnaA [26], [27], [33]–[36], [50]–[53]. In general, a strain's health is more compromised when the exogenous origin has integrated further from oriC, and when replication is unidirectional rather than bidirectional. Retrograde replication fork progression (towards oriC) in strains with ectopic origins is slow [27], [54], presumably because codirectionality between replication and transcription is lost, providing support to the model of impedance of fork movement by head-on transcription [17]–[19].
For strains where the exogenous origin is integrated at oriC-distal sites, the terminus region is replicated (as expected) by the fork which traverses the shorter arc between it and the integration site, but additionally there is a sharp decrease in copy numbers immediately before the Ter site that arrests its passage [26], [27]. A similar decrease in copy numbers proximal to the Ter arrest site of a prematurely arriving fork is evident in a strain possessing two chromosomal replication origins [41]. It is possible that these decreases are related to changes in fork architecture at the arrest sites, leading to DNA degradation by endo - and exonucleolytic enzymes.
Chromosome Replication in Absence of oriC or DnaA: cSDR
Another means to confer viability to cells lacking oriC or DnaA is cSDR, wherein transcription-associated R-loops serve as primers for initial DNA synthesis following which replication forks are established by the mechanisms of replication restart [55], [56]. Enzymes RNase HI (rnhA-encoded) and RecG (recG-encoded) act to eliminate R-loops by hydrolysis and by unwinding, respectively, and DNA synthesis by cSDR has been demonstrated in both rnhA and recG single mutants (while the double-deficiency is lethal) [55]. R-loops similarly initiate replication in ColE1 plasmids [57], [58]. cSDR has also been implicated in stress-induced mutagenesis and genome instability [59].
cSDR Origin Sites in RNase HI-Deficient Mutants
By examining the copy number gradient in rnhA mutants lacking oriC-initiated replication, the late Kogoma's group reported several putative replication initiation sites (termed oriKs), at least two of which were in the chromosomal terminus region [55]. Madiuke et al. [60] have revisited this question through a deep sequencing approach, and their results have again demonstrated a prominent copy number peak in the terminus region of rnhA mutants. However, this peak was abolished in a Tus-deficient derivative, leading the authors to suggest that it may not represent an oriK site but instead could be a consequence of trapping by polar Ter sites of replication forks that were initiated outside, and had then progressed into, the terminus region [60]. This idea is further developed in my model proposed below. Furthermore, no oriK locus was detected in a chromosome-wide search for sequences that could confer autonomous replication ability in an RNase HI-deficient strain [38], [61]. Thus, a definitive identification of the so-called oriK sites for cSDR has remained elusive.
Where Do R-loops Occur in the E. coli Genome?
One way to identify replication initiation sites in cSDR would be to determine the locations of R-loops in the genome, even while it is recognized that their occurrence is necessary but may not be sufficient for establishing origin activity [62]. R-loop mapping studies have not been reported for rnhA or recG mutants, but they have been done [63] for a mutant defective in Rho-dependent transcription termination (RDTT) as explained below.
RDTT is a process by which nascent non-rRNA transcripts that are not being simultaneously translated are prematurely terminated. RDTT-deficient mutants exhibit an increased prevalence of R-loops [63]–[65], which is assumed to arise from the reannealing of nascent untranslated transcripts with upstream DNA [66], [67]. R-loop formation in these situations is facilitated also by backtracking of RNA polymerase leading to stalled or arrested transcription elongation complexes [68], [69], but why this is so is unclear.
In an RDTT-deficient mutant, R-loops are distributed genome-wide, being generated from both sense and antisense transcripts [63]; Peters et al. [70] have also shown that antisense transcription is increased when RDTT is compromised (reviewed in [71]). Therefore, it is likely that oriK sites for cSDR are also widespread, and that they may indeed be stochastically different amongst individual cells in a population. This would explain the earlier findings [60], [61] that no distinct oriK sites were unambiguously identified in RNase HI-deficient mutants.
cSDR in RecG-Deficient Mutants
cSDR with RNase HI - or RecG-deficiency: Similar findings, different models
Copy number studies in both rnhA [60] and recG [41] mutants have demonstrated the similar occurrence in them of Tus-dependent (i) peak in the chromosomal terminus region, and (ii) inversion of the classical oriC-peaked curve when replication initiation from oriC is abolished. However, cSDR in the recG mutant has been explained to be the consequence of aberrant replication reinitiation events following fork collisions [41], [72].
According to this model [41], when opposing forks meet in the terminus region, DnaB helicase acts to unwind and extrude the oncoming fork's leading strand at its 3′ end, which then serves as a substrate for aberrant replication restart unless RecG and at least one of three single-strand DNA 3′ exonucleases are available. Combined deficiency of RecG together with the three 3′ exonucleases is lethal [73], which has been attributed to excessive occurrence of such over-replication in these cells. However, the source of origin of forks which are postulated to collide in the terminus region to mediate cSDR in recG mutants has not been explained, since these strains were also DnaA-deficient [41].
This raises the question of replication initiation in cSDR occurring by completely different mechanisms in rnhA and recG mutants, the former from R-loops [55], [60] and the latter from fork collisions in the terminus region [41]. However, the similarities cited above would suggest that cSDR in both indeed operates by a common mechanism, as is further explored below. An additional similarity is that, just as with RecG deficiency, RNase HI deficiency is also lethal in the combined absence of the three 3′ exonucleases [73].
A model invoking subpopulations with distinct replication dynamics during cSDR
The sharpness of the observed copy number peak in the terminus region of recG mutants is inconsistent with the excessive reinitiation model [41], which would predict that copy numbers exhibit a plateau (with no peaks) across this entire interval between TerA and TerB or TerC (since replication reinitiation anywhere within this region will duplicate all markers between the Tus-bound Ter boundaries). An alternative way to explain the observed peak in the mid-terminus region of a recG or rnhA mutant is to assume that its copy number curve is a composite of distributions from subpopulations with one or more of three different kinds of replication initiation events, as represented in Fig. 3.
Fig. 3. Predicted copy number distribution patterns for different categories of replication events in recG or rnhA mutants. 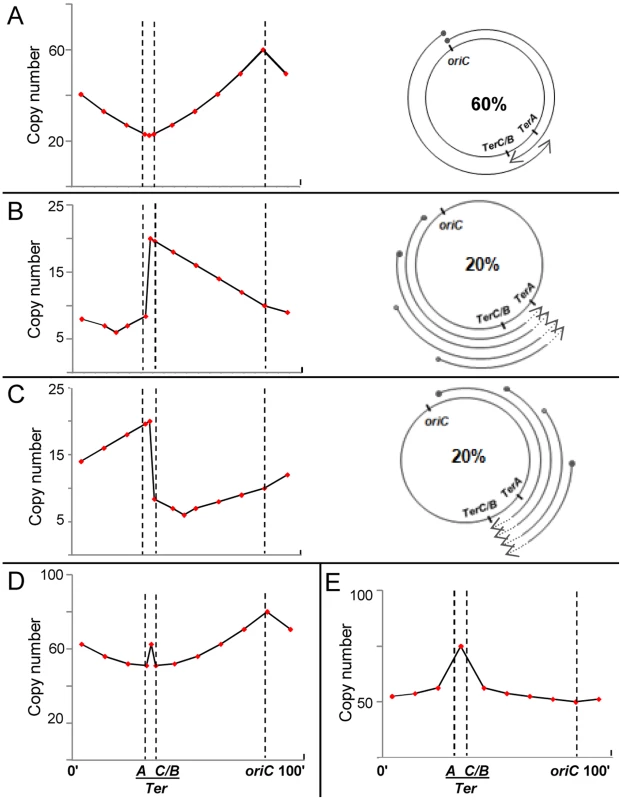
For all curves, positions of oriC, TerA, and TerC or TerB (TerC/B), are marked by the interrupted vertical lines; and copy number values are plotted on a linear instead of log scale to enable comparison with curves shown in Rudolph et al. [41]. (A–C) Three categories of replication events are shown, comprising those with forks initiated, respectively, (i) at oriC, DnaA-mediated (60%); (ii) on the counterclockwise replichore at various locations, R-loop mediated (20%); and (iii) on the clockwise replichore at various locations, R-loop mediated (20%). An individual cell in the population may harbor more than one category of event (see text). On the right in each of the three panels is a schematic depiction of progression of forks, each beginning at a solid circle and progressing to the position of arrowhead; in panels B and C, terminus region chromosomal DNA degradation (proximal to the sites of fork arrest at Ter) is shown as interrupted lines on the arcs, but retrograde fork advancements towards oriC (which are expected to occur at low efficiency [27], [54]) are not marked. On the left in each of the three panels is shown the expected copy number distribution for that category. (D) Expected copy number distribution for the entire cell population, obtained by summation of the distributions shown in panels A–C. (E) Expected copy number distribution for recG or rnhA mutant lacking oriC-initiated replication. For the purpose of this depiction, 60% of replication initiations are envisaged to have occurred from oriC (Fig. 3A), and 20% each from R-loops in the counterclockwise and clockwise replichore arms (Figs. 3B and 3C, respectively). Nevertheless, a single cell may harbor more than one category of replication event: for example, a simple representation of the percentages above would have it that for every three cells in the culture per generation, one suffers a (supernumerary) cSDR initiation event on the clockwise replichore, another a similar event on the counterclockwise replichore, whereas all exhibit oriC-initiated replication.
For cSDR events initiated from sites on the counterclockwise replichore (Fig. 3B), retrograde progression of forks towards oriC would be slow (as noted in other cases earlier [27], [54]), whereas they would progress smoothly towards and beyond TerC/B into the terminus region to be arrested at TerA; the small proportion of forks that overcome arrest would then progress in retrograde direction beyond TerA. Since R-loops are evenly distributed [63], cSDR origins are also likely to occur at a uniform, but low, probability across the genome, such that the copy number for an arbitrary locus on the counterclockwise arm is higher the further its distance from oriC (which is opposite to that with DnaA-mediated initiations from oriC; compare Figs. 3A and 3B).
The earlier studies [26], [27], [41] have also indicated that prolonged arrest of replication forks at a Ter site, in the absence of arrival of forks of the opposite replichore, is associated with a sharp copy number drop in the region preceding the fork arrest site (which is depicted in Figure 3B adjacent to TerA, in the interval between TerA and TerC or TerB). The mirror symmetrically reverse situation would apply for cSDR initiation events on the clockwise replichore, as shown in Figure 3C.
The composite pattern for the entire population, derived by summation of the three distributions above, is shown in Figure 3D. Two features of the data reported for mutants lacking RecG [41] or RNase HI [60] are recapitulated here, namely, a peak in the mid-terminus region and a smaller enrichment of oriC-proximal to oriC-distal markers (compare Figs. 3A and 3D). As has also been suggested earlier [41], many cSDR events may likely contribute only to copy number values without concomitantly increasing viable cell numbers, since every event would not necessarily lead to duplication of the entire chromosome.
With the same assumptions, the copy number distribution in an rnhA or recG mutant that is additionally defective for DnaA can be derived as the approximate composite of Figs. 3B and 3C, as depicted in Fig. 3E. The derived curve broadly recapitulates the inversion in these mutants of the “classical” curve so that the peak and trough are now at the terminus and oriC, respectively [41], [60].
In strains exhibiting cSDR, the frequency of replication fork disintegration events is expected to be elevated when replisomes advance towards oriC; this would explain their dependence for viability on proteins of replication restart and homologous recombination [55], [56]. Since R-loop prevalence is less upon loss of RecG than of RNase HI [55], cSDR-mediated viability of a recG dnaA mutant requires the presence of additional mutations in Tus and RNA polymerase (rpoB*35) [41]. While absence of Tus permits passage of counterdirectional forks across Ter sites, rpoB*35 mitigates the adversity associated with transcription–replication conflicts [44], [68], [74]; in cSDR, rpoB*35 would promote retrograde fork progression both from cSDR initiation sites and in regions beyond the Ter sites.
Comparisons in Other Organisms
The similarities listed in Table 1 between E. coli and B. subtilis would suggest that the models proposed here for the former may apply to the latter, although cSDR has so far not been demonstrated in B. subtilis. Archaeal and eukaryotic DNA replication mechanisms are very similar [1], [75]–[77], and in the archaeon Haloferax volcanii, the circular chromosome possesses four replication origins, but derivatives in which all were deleted unexpectedly exhibited greater fitness than the parental strains [78]; a cSDR-like mechanism has been proposed in the originless mutant [78], although an alternative possibility is that dormant replication origins were activated under these conditions [79].
Transcription-associated R-loops exist in eukaryotes [80]–[84], and their prevalence is increased when either elongation or cotranscriptional processing of mRNA is impeded [85]–[94]. DNA double-strand breaks occur in the G1 phase following R-loop formation [81], but the mechanism is not known, and one could thus speculate whether cSDR-like events may be a contributory factor. Furthermore, replication stress in eukaryotes triggers new initiation sites that are generally thought to arise by activation of dormant origins [1], [10], [95]–[98]; once again, it is possible that some of them arise from R-loops, given that they are located predominantly in transcribed gene regions [99], [100]. That R-loops in eukaryotic cells may confer genome instability by priming new DNA synthesis has been suggested earlier [59], [86]–[88], [101]; the BRCA2 protein, which functions as “chromosome custodian” and cancer suppressor [102], has also recently been suggested to exert its oncoprotective role by preventing R-loop accumulation [84].
Conclusions and the Future Perspective
The major ideas proposed in this review, which need to be tested in future studies, are that fork reversal reactions occur when opposing replisomes meet; that replication origins for bacterial cSDR are widespread in the genome (although the firing potential of any single origin is small); that replication fork progression in cSDR faces two separate obstacles, of conflicts with transcription and of arrest at Tus-bound Ter sites; and that cSDR-like events may contribute to R-loop mediated genome damage in eukaryotes.
The additional questions to be addressed in the bacterial systems include the following [67]: What are the determinants of R-loop propensity? What regulates conversion of R-loops to replication origins? Would cSDR occur in other instances of increased R-loop prevalence, such as in mutants deficient in Rho (discussed above) or topoisomerase I [103]–[105]? When forks undergo polar arrest at Tus-bound Ter sites in absence of oncoming forks, how does the postulated DNA degradation occur proximal to Ter? And what are the roles for single-strand DNA exonucleases in replication?
The bacterial chromosome is organized into macrodomains [106]–[109], one of which is the Terminus macrodomain. Whether such organization influences (or is influenced by) replication fork dynamics near the terminus is unclear. Chromosome replication is also linked to downstream events of chromosome segregation, nucleoid condensation and cytokinesis [5], [110], [111], and the repercussions thereon of perturbations in replisome progression remain to be characterized.
Zdroje
1. O'DonnellM, LangstonL, StillmanB (2013) Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol 5: a010108.
2. GaoF, LuoH, ZhangCT (2013) DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res 41: D90–93.
3. Kornberg A, Baker TA (2005) DNA Replication, 2nd Edition: University Science Books.
4. MottML, BergerJM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5 : 343–354.
5. Reyes-LamotheR, NicolasE, SherrattDJ (2012) Chromosome replication and segregation in bacteria. Annu Rev Genet 46 : 121–143.
6. SkarstadK, KatayamaT (2013) Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol 5: a012922.
7. KuzminovA (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev 63 : 751–813.
8. CoxMM, GoodmanMF, KreuzerKN, SherrattDJ, SandlerSJ, et al. (2000) The importance of repairing stalled replication forks. Nature 404 : 37–41.
9. HellerRC, MariansKJ (2006) Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7 : 932–943.
10. YeelesJT, PoliJ, MariansKJ, PaseroP (2013) Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 5: a012815.
11. MirkinEV, MirkinSM (2005) Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol 25 : 888–895.
12. MirkinEV, MirkinSM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71 : 13–35.
13. RudolphCJ, DhillonP, MooreT, LloydRG (2007) Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 6 : 981–993.
14. McGlynnP, SaveryNJ, DillinghamMS (2012) The conflict between DNA replication and transcription. Mol Microbiol 85 : 12–20.
15. MerrikhH, ZhangY, GrossmanAD, WangJD (2012) Replication-transcription conflicts in bacteria. Nat Rev Microbiol 10 : 449–458.
16. HelmrichA, BallarinoM, NudlerE, ToraL (2013) Transcription-replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20 : 412–418.
17. FrenchS (1992) Consequences of replication fork movement through transcription units in vivo. Science 258 : 1362–1365.
18. BoubakriH, de SeptenvilleAL, VigueraE, MichelB (2010) The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29 : 145–157.
19. De SeptenvilleAL, DuigouS, BoubakriH, MichelB (2012) Replication fork reversal after replication-transcription collision. PLoS Genet 8: e1002622.
20. BidnenkoV, LestiniR, MichelB (2006) The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol Microbiol 62 : 382–396.
21. BidnenkoV, EhrlichSD, MichelB (2002) Replication fork collapse at replication terminator sequences. EMBO J 21 : 3898–3907.
22. SimmonsLA, BreierAM, CozzarelliNR, KaguniJM (2004) Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol Microbiol 51 : 349–358.
23. NeylonC, KralicekAV, HillTM, DixonNE (2005) Replication termination in Escherichia coli: structure and antihelicase activity of the Tus-Ter complex. Microbiol Mol Biol Rev 69 : 501–526.
24. DugginIG, WakeRG, BellSD, HillTM (2008) The replication fork trap and termination of chromosome replication. Mol Microbiol 70 : 1323–1333.
25. KaplanDL, BastiaD (2009) Mechanisms of polar arrest of a replication fork. Mol Microbiol 72 : 279–285.
26. HillTM, HensonJM, KuempelPL (1987) The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci U S A 84 : 1754–1758.
27. de MassyB, BejarS, LouarnJ, LouarnJM, BoucheJP (1987) Inhibition of replication forks exiting the terminus region of the Escherichia coli chromosome occurs at two loci separated by 5 min. Proc Natl Acad Sci U S A 84 : 1759–1763.
28. EsnaultE, ValensM, EspeliO, BoccardF (2007) Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3: e226.
29. YoshikawaH, SueokaN (1963) Sequential replication of Bacillus subtilis chromosome. I. Comparison of marker frequencies in exponential and stationary growth phases. Proc Natl Acad Sci U S A 49 : 559–566.
30. SueokaN, YoshikawaH (1965) The chromosome of Bacillus subtilis. I. Theory of marker frequency analysis. Genetics 52 : 747–757.
31. MastersM, BrodaP (1971) Evidence for the bidirectional replication of the Escherichia coli chromosome. Nat New Biol 232 : 137–140.
32. BirdRE, LouarnJ, MartuscelliJ, CaroL (1972) Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol 70 : 549–566.
33. LouarnJ, PatteJ, LouarnJM (1977) Evidence for a fixed termination site of chromosome replication in Escherichia coli K12. J Mol Biol 115 : 295–314.
34. ChandlerM, SilverL, CaroL (1977) Suppression of an Escherichia coli dnaA mutation by the integrated R factor R100.1: origin of chromosome replication during exponential growth. J Bacteriol 131 : 421–430.
35. KuempelPL, DuerrSA, MaglothinPD (1978) Chromosome replication in an Escherichia coli dnaA mutant integratively suppressed by prophage P2. J Bacteriol 134 : 902–912.
36. Maisnier-PatinS, DasguptaS, KrabbeM, NordstromK (1998) Conversion to bidirectional replication after unidirectional initiation from R1 plasmid origin integrated at oriC in Escherichia coli. Mol Microbiol 30 : 1067–1079.
37. SkovgaardO, BakM, Lobner-OlesenA, TommerupN (2011) Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res 21 : 1388–1393.
38. KodamaK, KobayashiT, NikiH, HiragaS, OshimaT, et al. (2002) Amplification of Hot DNA segments in Escherichia coli. Mol Microbiol 45 : 1575–1588.
39. SangurdekarDP, HamannBL, SmirnovD, SriencF, HanawaltPC, et al. (2010) Thymineless death is associated with loss of essential genetic information from the replication origin. Mol Microbiol 75 : 1455–1467.
40. KuongKJ, KuzminovA (2012) Disintegration of nascent replication bubbles during thymine starvation triggers RecA - and RecBCD-dependent replication origin destruction. J Biol Chem 287 : 23958–23970.
41. RudolphCJ, UptonAL, StockumA, NieduszynskiCA, LloydRG (2013) Avoiding chromosome pathology when replication forks collide. Nature 500 : 608–611.
42. SeigneurM, BidnenkoV, EhrlichSD, MichelB (1998) RuvAB acts at arrested replication forks. Cell 95 : 419–430.
43. SeigneurM, EhrlichSD, MichelB (2000) RuvABC-dependent double-strand breaks in dnaBts mutants require RecA. Mol Microbiol 38 : 565–574.
44. McGlynnP, LloydRG (2000) Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101 : 35–45.
45. KhanSR, KuzminovA (2012) Replication forks stalled at ultraviolet lesions are rescued via RecA and RuvABC protein-catalyzed disintegration in Escherichia coli. J Biol Chem 287 : 6250–6265.
46. MichelB, BoubakriH, BaharogluZ, LeMassonM, LestiniR (2007) Recombination proteins and rescue of arrested replication forks. DNA Repair 6 : 967–980.
47. AlverRC, BielinskyAK (2010) Termination at sTop2. Mol Cell 39 : 487–489.
48. FachinettiD, BermejoR, CocitoA, MinardiS, KatouY, et al. (2010) Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39 : 595–605.
49. SteinacherR, OsmanF, DalgaardJZ, LorenzA, WhitbyMC (2012) The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev 26 : 594–602.
50. NishimuraY, CaroL, BergCM, HirotaY (1971) Chromosome replication in Escherichia coli. IV. Control of chromosome replication and cell division by an integrated episome. J Mol Biol 55 : 441–456.
51. LindahlG, HirotaY, JacobF (1971) On the process of cellular division in Escherichia coli: replication of the bacterial chromosome under control of prophage P2. Proc Natl Acad Sci U S A 68 : 2407–2411.
52. LouarnJ, PatteJ, LouarnJM (1982) Suppression of Escherichia coli dnaA46 mutations by integration of plasmid R100.1. derivatives: constraints imposed by the replication terminus. J Bacteriol 151 : 657–667.
53. Maisnier-PatinS, NordstromK, DasguptaS (2001) RecA-mediated rescue of Escherichia coli strains with replication forks arrested at the terminus. J Bacteriol 183 : 6065–6073.
54. KouzminovaEA, KuzminovA (2008) Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Mol Microbiol 68 : 202–215.
55. KogomaT (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61 : 212–238.
56. SandlerSJ (2005) Requirements for replication restart proteins during constitutive stable DNA replication in Escherichia coli K-12. Genetics 169 : 1799–1806.
57. ItohT, TomizawaJ (1980) Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A 77 : 2450–2454.
58. KuesU, StahlU (1989) Replication of plasmids in Gram-negative bacteria. Microbiol Rev 53 : 491–516.
59. WimberlyH, SheeC, ThorntonPC, SivaramakrishnanP, RosenbergSM, et al. (2013) R-loops and nicks initiate DNA breakage and genome instability in non-growing Escherichia coli. Nat Commun 4 : 2115.
60. MaduikeNZ, TehranchiAK, WangJD, KreuzerKN (2014) Replication of the Escherichia coli chromosome in RNase HI-deficient cells: multiple initiation regions and fork dynamics. Mol Microbiol 91 : 39–56.
61. NishitaniH, HidakaM, HoriuchiT (1993) Specific chromosomal sites enhancing homologous recombination in Escherichia coli mutants defective in RNase H. Mol Gen Genet 240 : 307–314.
62. InoueN, UchidaH (1991) Transcription and initiation of ColE1 DNA replication in Escherichia coli K-12. J Bacteriol 173 : 1208–1214.
63. LeelaJK, SyedaAH, AnupamaK, GowrishankarJ (2013) Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A 110 : 258–263.
64. HarinarayananR, GowrishankarJ (2003) Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol 332 : 31–46.
65. AnupamaK, LeelaJK, GowrishankarJ (2011) Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol 82 : 1330–1348.
66. GowrishankarJ, HarinarayananR (2004) Why is transcription coupled to translation in bacteria? Mol Microbiol 54 : 598–603.
67. GowrishankarJ, LeelaJK, AnupamaK (2013) R-loops in bacterial transcription: their causes and consequences. Transcription 4 : 153–157.
68. DuttaD, ShatalinK, EpshteinV, GottesmanME, NudlerE (2011) Linking RNA polymerase backtracking to genome instability in E. coli. Cell 146 : 533–543.
69. NudlerE (2012) RNA polymerase backtracking in gene regulation and genome instability. Cell 149 : 1438–1445.
70. PetersJM, MooneyRA, GrassJA, JessenED, TranF, et al. (2012) Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev 26 : 2621–2633.
71. WadeJT, GraingerDC (2014) Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12 : 647–653.
72. RudolphCJ, UptonAL, BriggsGS, LloydRG (2010) Is RecG a general guardian of the bacterial genome? DNA Repair 9 : 210–223.
73. RudolphCJ, MahdiAA, UptonAL, LloydRG (2010) RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics 186 : 473–492.
74. TrautingerBW, JaktajiRP, RusakovaE, LloydRG (2005) RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19 : 247–258.
75. BarryER, BellSD (2006) DNA replication in the archaea. Microbiol Mol Biol Rev 70 : 876–887.
76. MakarovaKS, KooninEV (2013) Archaeology of eukaryotic DNA replication. Cold Spring Harb Perspect Biol 5: a012963.
77. LeonardAC, MechaliM (2013) DNA replication origins. Cold Spring Harb Perspect Biol 5: a010116.
78. HawkinsM, MallaS, BlytheMJ, NieduszynskiCA, AllersT (2013) Accelerated growth in the absence of DNA replication origins. Nature 503 : 544–547.
79. MichelB, BernanderR (2014) Chromosome replication origins: do we really need them? BioEssays 36 : 585–590.
80. HelmrichA, BallarinoM, ToraL (2011) Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44 : 966–977.
81. WahbaL, AmonJD, KoshlandD, Vuica-RossM (2011) RNase H and multiple RNA biogenesis factors cooperate to prevent RNA: DNA hybrids from generating genome instability. Mol Cell 44 : 978–988.
82. MischoHE, Gomez-GonzalezB, GrzechnikP, RondonAG, WeiW, et al. (2011) Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41 : 21–32.
83. GinnoPA, LottPL, ChristensenHC, KorfI, ChedinF (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45 : 814–825.
84. BhatiaV, BarrosoSI, Garcia-RubioML, TuminiE, Herrera-MoyanoE, et al. (2014) BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511 : 362–365.
85. TuduriS, CrabbeL, TourriereH, CoquelleA, PaseroP (2010) Does interference between replication and transcription contribute to genomic instability in cancer cells? Cell Cycle 9 : 1886–1892.
86. BermejoR, LaiMS, FoianiM (2012) Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell 45 : 710–718.
87. AguileraA, Garcia-MuseT (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46 : 115–124.
88. FongYW, CattoglioC, TjianR (2013) The intertwined roles of transcription and repair proteins. Mol Cell 52 : 291–302.
89. MontecuccoA, BiamontiG (2013) Pre-mRNA processing factors meet the DNA damage response. Front Genet 4 : 102.
90. ChanYA, HieterP, StirlingPC (2014) Mechanisms of genome instability induced by RNA-processing defects. Trends Genet 30 : 245–253.
91. HamperlS, CimprichKA (2014) The contribution of co-transcriptional RNA: DNA hybrid structures to DNA damage and genome instability. DNA Repair 19 : 84–94.
92. Skourti-StathakiK, ProudfootNJ (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28 : 1384–1396.
93. AguileraA, GaillardH (2014) Transcription and recombination: when RNA meets DNA. Cold Spring Harb Perspect Biol 6: a016543.
94. GrohM, GromakN (2014) Out of balance: R-loops in human disease. PLoS Genet 10: e1004630.
95. MechaliM (2010) Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11 : 728–738.
96. MasaiH, MatsumotoS, YouZ, Yoshizawa-SugataN, OdaM (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79 : 89–130.
97. TuduriS, TourriereH, PaseroP (2010) Defining replication origin efficiency using DNA fiber assays. Chromosome Res 18 : 91–102.
98. BlowJJ, GeXQ, JacksonDA (2011) How dormant origins promote complete genome replication. Trends Biochem Sci 36 : 405–414.
99. KarnaniN, DuttaA (2011) The effect of the intra-S-phase checkpoint on origins of replication in human cells. Genes Dev 25 : 621–633.
100. ImJS, KeatonM, LeeKY, KumarP, ParkJ, et al. (2014) ATR checkpoint kinase and CRL1βTRCP collaborate to degrade ASF1a and thus repress genes overlapping with clusters of stalled replication forks. Genes Dev 28 : 875–887.
101. KimN, Jinks-RobertsonS (2012) Transcription as a source of genome instability. Nat Rev Genet 13 : 204–214.
102. VenkitaramanAR (2014) Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 343 : 1470–1475.
103. MasseE, DroletM (1999) R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol 294 : 321–332.
104. MasseE, DroletM (1999) Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem 274 : 16659–16664.
105. UsongoV, DroletM (2014) Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet 10: e1004543.
106. BoccardF, EsnaultE, ValensM (2005) Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol Microbiol 57 : 9–16.
107. RochaEP (2008) The organization of the bacterial genome. Annu Rev Genet 42 : 211–233.
108. MercierR, PetitMA, SchbathS, RobinS, El KarouiM, et al. (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 : 475–485.
109. DameRT, KalmykowaOJ, GraingerDC (2011) Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in Gram negative bacteria. PLoS Genet 7: e1002123.
110. KuzminovA (2013) The chromosome cycle of prokaryotes. Mol Microbiol 90 : 214–227.
111. YoungrenB, NielsenHJ, JunS, AustinS (2014) The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev 28 : 71–84.
112. Zakrzewska-CzerwinskaJ, JakimowiczD, Zawilak-PawlikA, MesserW (2007) Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev 31 : 378–387.
113. KatayamaT, OzakiS, KeyamuraK, FujimitsuK (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8 : 163–170.
114. HassanAK, MoriyaS, OguraM, TanakaT, KawamuraF, et al. (1997) Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J Bacteriol 179 : 2494–2502.
115. MoriyaS, HassanAK, KadoyaR, OgasawaraN (1997) Mechanism of anucleate cell production in the oriC-deleted mutants of Bacillus subtilis. DNA Res 4 : 115–126.
116. PetitMA, DervynE, RoseM, EntianKD, McGovernS, et al. (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29 : 261–273.
117. PetitMA, EhrlichD (2002) Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J 21 : 3137–3147.
118. MerrikhC, MerrikhH (2014) The B. subtilis accessory helicase PcrA facilitates replication through transcription units genome-wide. FASEB J 28 Supp. LB126
119. GabbaiCB, MariansKJ (2010) Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair 9 : 202–209.
120. MerrikhH, MachonC, GraingerWH, GrossmanAD, SoultanasP (2011) Co-directional replication-transcription conflicts lead to replication restart. Nature 470 : 554–557.
121. AyoraS, CarrascoB, CardenasPP, CesarCE, CanasC, et al. (2011) Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol Rev 35 : 1055–1081.
122. CarrAM, LambertS (2013) Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol 425 : 4733–4744.
123. JasinM, RothsteinR (2013) Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol 5: a012740.
124. DaleyJM, KwonY, NiuH, SungP (2013) Investigations of homologous recombination pathways and their regulation. Yale J Biol Med 86 : 453–461.
125. AzeA, ZhouJC, CostaA, CostanzoV (2013) DNA replication and homologous recombination factors: acting together to maintain genome stability. Chromosoma 122 : 401–413.
126. WillisNA, ChandramoulyG, HuangB, KwokA, FollonierC, et al. (2014) BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature 510 : 556–559.
127. YeelesJT, DillinghamMS (2010) The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair 9 : 276–285.
128. WigleyDB (2013) Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol 11 : 9–13.
129. BlackwoodJK, RzechorzekNJ, BraySM, MamanJD, PellegriniL, et al. (2013) End-resection at DNA double-strand breaks in the three domains of life. Biochem Soc Trans 41 : 314–320.
130. MimitouEP, SymingtonLS (2011) DNA end resection–unraveling the tail. DNA Repair 10 : 344–348.
131. SymingtonLS (2014) End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol 6: a016436.
132. SharplesGJ, InglestonSM, LloydRG (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J Bacteriol 181 : 5543–5550.
133. Lo PianoA, Martinez-JimenezMI, ZecchiL, AyoraS (2011) Recombination-dependent concatemeric viral DNA replication. Virus Res 160 : 1–14.
134. ZecchiL, Lo PianoA, SuzukiY, CanasC, TakeyasuK, et al. (2012) Characterization of the Holliday junction resolving enzyme encoded by the Bacillus subtilis bacteriophage SPP1. PLoS One 7: e48440.
135. MaricM, MaculinsT, De PiccoliG, LabibK (2014) Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346 : 440.
136. MorenoSP, BaileyR, CampionN, HerronS, GambusA (2014) Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346 : 477–481.
137. WangJD, BerkmenMB, GrossmanAD (2007) Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci U S A 104 : 5608–5613.
138. SrivatsanA, TehranchiA, MacAlpineDM, WangJD (2010) Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6: e1000810.
139. HuvetM, NicolayS, TouchonM, AuditB, d'Aubenton-CarafaY, et al. (2007) Human gene organization driven by the coordination of replication and transcription. Genome Res 17 : 1278–1285.
140. NicolasP, MaderU, DervynE, RochatT, LeducA, et al. (2012) Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335 : 1103–1106.
141. ChanYA, AristizabalMJ, LuPYT, LuoZ, HamzaA, et al. (2014) Genome-wide profiling of yeast DNA: RNA hybrid prone sites with DRIP-Chip. PLoS Genet 10: e1004288.
142. El HageA, WebbS, KerrA, TollerveyD (2014) Genome-wide distribution of RNA-DNA hybrids identifies RNase H targets in tRNA genes, retrotransposons and mitochondria. PLoS Genet 10: e1004716.
143. El HageA, FrenchSL, BeyerAL, TollerveyD (2010) Loss of topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24 : 1546–1558.
144. FukushimaS, ItayaM, KatoH, OgasawaraN, YoshikawaH (2007) Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J Bacteriol 189 : 8575–8583.
145. TadokoroT, KanayaS (2009) Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J 276 : 1482–1493.
146. CerritelliSM, CrouchRJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J 276 : 1494–1505.
147. WenQ, MahdiAA, BriggsGS, SharplesGJ, LloydRG (2005) Conservation of RecG activity from pathogens to hyperthermophiles. DNA Repair 4 : 23–31.
148. SanchezH, CarrascoB, CozarMC, AlonsoJC (2007) Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation. Mol Microbiol 65 : 920–935.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



