-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
Certain proteins can misfold into β-sheet-rich, self-seeding aggregates. Such proteins appear to be associated with neurodegenerative diseases such as prion, Alzheimer's and Parkinson's. Yeast prions also misfold into self-seeding aggregates and provide a good model to study how these rogue polymers first appear. De novo prion appearance can be made very frequent in yeast by transient overexpression of the prion protein in the presence of heterologous prions or prion-like aggregates. Here, we show that the aggregates of one such newly induced prion are initially formed in a dot-like structure near the vacuole. These dots then grow into rings at the periphery of the cell prior to becoming smaller rings surrounding the vacuole and maturing into the characteristic heritable prion tiny dots found throughout the cytoplasm. We found considerable colocalization of two heterologous prion/prion-like aggregates with the newly appearing prion protein aggregates, which is consistent with the prevalent model that existing prion aggregates can cross-seed the de novo aggregation of a heterologous prion protein. However, we failed to find any physical interaction between another heterologous aggregating protein and the newly appearing prion aggregates it stimulated to appear, which is inconsistent with cross-seeding.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004814
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004814Summary
Certain proteins can misfold into β-sheet-rich, self-seeding aggregates. Such proteins appear to be associated with neurodegenerative diseases such as prion, Alzheimer's and Parkinson's. Yeast prions also misfold into self-seeding aggregates and provide a good model to study how these rogue polymers first appear. De novo prion appearance can be made very frequent in yeast by transient overexpression of the prion protein in the presence of heterologous prions or prion-like aggregates. Here, we show that the aggregates of one such newly induced prion are initially formed in a dot-like structure near the vacuole. These dots then grow into rings at the periphery of the cell prior to becoming smaller rings surrounding the vacuole and maturing into the characteristic heritable prion tiny dots found throughout the cytoplasm. We found considerable colocalization of two heterologous prion/prion-like aggregates with the newly appearing prion protein aggregates, which is consistent with the prevalent model that existing prion aggregates can cross-seed the de novo aggregation of a heterologous prion protein. However, we failed to find any physical interaction between another heterologous aggregating protein and the newly appearing prion aggregates it stimulated to appear, which is inconsistent with cross-seeding.
Introduction
Prions were first described as self-perpetuating infectious agents devoid of nucleic acids that cause several fatal neurodegenerative diseases. Prion diseases, also known as transmissible spongiform encephalopathies (TSEs), were shown to infect a variety of mammals [1]. All known mammalian prion diseases are caused by conversion of largely α-helical cellular prion protein PrPC into fibrous β-sheet-rich ordered aggregates (amyloids) called PrPSc (associated with scrapie) [2]. Curiously, PrPSc can exist in different heritable forms, called strains, which cause neurodegenerative diseases with different characteristics and pathologies [3]–[5].
A number of other neurodegenerative diseases are also associated with conversion of a soluble protein to amyloid. For example, amyloid-like forms of Aβ and Tau, α-synuclein, huntingtin, FUS/TLS, TDP-43 or SOD1 are linked respectively to Alzheimer's (AD) [6], Parkinson's (PD) [7], [8], Huntington's (Htt) [9] and Amyotrophic Lateral Sclerosis (ALS) diseases [10]–[15]. Factors that influence the spontaneous conversion to amyloid are of considerable interest as possible disease risk factors. One important finding is that heterologous amyloid can promote the de novo conversion of a protein to amyloid. For example, Aβ accelerated the in vivo aggregation of tau [16], and Aβ and α-synuclein seeded each other's aggregation in vitro [17]. Indeed, recently distinct conformational variants of α-synuclein aggregates were shown to differentially promote the aggregation of tau in neurons [18].
Several proteins in the simple eukaryote yeast have been shown to convert from soluble to amyloid. The amyloid forms of these proteins are self-propagating prions associated with transmissible phenotypes [19]–[28]. These proteins provide good models for the amyloid conversion of human disease proteins.
For both human and yeast proteins, only a portion of the protein, called the prion domain (PrD), converts to amyloid. This portion of the protein is required for prion induction and propagation [29], [30]. The PrD of most known yeast prions is glutamine (Q) and asparagine (N) rich. Likewise, several human aggregating disease proteins e.g. huntingtin, TDP-43 and FUS contain Q/N-rich regions [31], [32]. In contrast, the recently discovered yeast [MOD+] prion, composed of Mod5, as well as PrP, Aβ and α-synuclein do not contain Q/N rich domains [27]. Similar to the mammalian PrP strains, yeast prions can also fold into numerous heritable conformations, called variants, leading to different degrees of altered phenotypes [33]–[36].
The most well-studied yeast prion is [PSI+], the prion form of Sup35. In its native form, Sup35 (release factor 3) works with Sup45 (release factor 1) to promote translational termination at stop codons [37]. The Sup35 protein consists of three major domains: N-proximal (N domain) required for prion induction and propagation; a highly charged middle (M) domain conferring solubility to the molecule and containing Hsp104 binding sites [38] and a C-terminal (C) domain essential for translational termination and viability [33], [39]–[41]. [PSI+] forms when Sup35 molecules assemble into amyloid-like aggregates, causing loss-of-function in translation termination, which leads to read-through of stop codons [20], [42], [43].
The spontaneous appearance of prions in yeast is extremely rare. Indeed, the conversion of prion-free cells, [psi-] to [PSI+] was determined to be ∼5.8×10−7 [44]–[47]. On the other hand, overproduction of full-length Sup35 or its prion containing domain (Sup35NM) can increase the de novo appearance of [PSI+] dramatically, presumably by increasing the chance of Sup35 prion domains to misfold and interact [48]. This enhanced formation of [PSI+] requires either the presence of another prion [22], [49], [50] or the simultaneous overexpression of heterologous Q/N-rich domains [22], [51]. The best studied example of this stimulation by a prion involves [PIN+], the prion form of the Rnq1 protein. Although [PIN+] dramatically enhances the appearance of [PSI+], it is not required for [PSI+] propagation [49].
Understanding how [PIN+] enhances de novo induction of [PSI+] will help us understand analogous interactions between heterologous human disease proteins. Several models have been proposed (reviewed in [52]). The cross-seeding model suggests that [PIN+] initially acts as a seed for the conversion of the Sup35 prion domain into the [PSI+] conformation. Once [PSI+] is established, it is proposed to create its own seeds independent of [PIN+], allowing it to continue to propagate efficiently [53]. Several in vitro studies provide evidence in favor of induction of [PSI+] via cross-seeding [52], [54], and for the enhanced rate of polymerization of other proteins in the presence of heterologous aggregates [55]–[61]. Most notably, mCherry:FUS fibers were extended in length heterotypically when exposed to monomeric GFP:hnRNPA1 [55]. Definitive evidence for cross-seeding in vivo is much more difficult to obtain. Still, a fusion of the prion domain of Sup35 (NM) and Rnq1 lead to the efficient induction of [PSI+] in the presence of [PIN+], even without Sup35 overexpression [62], presumably because the fusion efficiently brings Sup35NM to the Rnq1 aggregates, thereby increasing the chance of physical association and resulting cross-seeding. Also, different [PIN+] variants preferentially cause the genesis of different variants of [PSI+] [63], [64], which can be easily explained by cross-seeding but not by chaperone titration.
The titration model postulates that cellular factors responsible for the disassembly of aggregates and the refolding of misfolded proteins are so busy working on the existing [PIN+] prion that they are not available to prevent the appearance of the new prion, [PSI+] [22], [51], [65]. In support of this model, prion-like aggregates have been shown to colocalize with chaperones, reducing the cytosolic level of chaperones and thereby affecting the stability of heterologous prion aggregates in the cell [66]–[70].
Molecular chaperones, which are normally involved in proper protein folding play a critical role in the maintenance of yeast prions (reviewed in [71]). Particularly, the Hsp104 chaperone in conjunction with chaperones of the Hsp70 (Ssa/Ssb) and Hsp40 (Sis1) families was shown to shear prion aggregates into smaller fragments that promote fiber growth and transmission to daughter cells [72]–[77]. The shearing activity of Hsp104 is antagonized by millimolar concentrations of guanidine hydrochloride (GuHCl), leading to the loss of [PSI+] [78], [79], and other yeast prions [19], [50]. Hsp104 is required for the propagation of almost all known yeast prions [72], [80]–[82].
Stimulation of de novo generation of prions in yeast is achieved by inducing overexpression of the corresponding prion protein. The resulting aggregates have been monitored with fluorescent derivatives. The de novo induction of [PSI+] promoted by [PIN+] was shown to display various Sup35 aggregates and go through several stages. Overexpression of Sup35NM-GFP, gave rise to fluorescent dot, line and ring-like assemblies [83]–[85]. The fluorescent rings induced by Sup35NM-GFP overexpression is a hallmark of [PSI+] induction. Indeed, most viable ring/dot-bearing cells gave rise to [PSI+] progeny [83]–[86]. Sup35 dots appeared earlier than rings and lines [84]. Ring-like aggregates were shown to be first peripheral along the cell membrane, and later internal surrounding the vacuole [83], [85]. When cells with such rings were followed in media that turned down Sup35 overexpression, Sup35NM-GFP appeared as dots in daughter cells, a typical feature of [PSI+] [83]–[85]. Once [PSI+] is established, Sup35NM-GFP overexpression results in one or a few large mature dots, or clumps but rings do not appear at all [29], [43]. These large dots replace the numerous small Sup35-GFP aggregates seen in [PSI+] cells with endogenous Sup35 tagged with GFP prior to overexpression [75]. When Sup35NM-GFP was constitutively overproduced in [PIN+] cells with a deletion of the Sup35 prion domain, only internal rings were observed prior to the transition to mature dots [87].
In this study, we report that the de novo appearance of [PSI+] aggregates begins with dots that co-localize with the main Rnq1 aggregate near the vacuole, that grow into peripheral rings and lines prior to the appearance of internal rings. Our studies also reveal preferential colocalization of Rnq1 and Pin4C aggregates with newly appearing vs. established [PSI+] aggregates, which is consistent with the cross-seeding model for [PSI+] induction. However, the failure of Mod5 to physically interact with Sup35 during Mod5-promoted [PSI+] induction suggests that cross-seeding is not involved. Finally, we provide evidence for the [PIN+]-independent requirement of Hsp104 during [PSI+] induction in vivo.
Results
Newly induced Sup35 aggregates arise near the vacuole and then grow into peripheral rings
We used GFP-tagged SUP35 constructs to visualize the initial conversion of Sup35 from soluble to aggregated protein when [psi-] cells were induced to become [PSI+] by overexpressing Sup35(NM). Sup35NM-GFP overexpressed in [PIN+][psi-] cells (Fig. 1A) progressed over time from diffuse cytoplasmic fluorescence in all cells to some cells with one to three fluorescent foci one of which was always near the vacuole, to more cells with dots. Later, peripheral rings and lines started to replace dots in some cells (see Table 1 for details). In contrast, [pin-] cells showed no aggregates of Sup35NM-GFP at any time point. Even when we dramatically reduced the level of Sup35NM-GFP overexpression by growing cells in 0.2 rather than 2% Gal, the type and order of appearance of these aggregates did not change (S1 Table).
Fig. 1. Sup35 forms vacuole-associated dots first, then rings during [PSI+] induction. ![Sup35 forms vacuole-associated dots first, then rings during [<i>PSI<sup>+</sup></i>] induction.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/a8d746eeb57809627630e94b3eb95cf8.png)
A. Visualization of Sup35 aggregates induced by Sup35NM-GFP in [PIN+] cells. 74D-694 [PIN+][psi-] and [pin-][psi-] cells were grown in inducing media (2% Gal) to overexpress Sup35NM-GFP (p1951, see Table 5 for plasmid nomenclature). From the time of induction, Sup35NM-GFP signals were visualized with fluorescent microscope over a time course (detailed in Table 1). Percentages are based on n≈500-800. B. Visualization of Sup35 aggregates induced by untagged Sup35NM in [PIN+] cells with endogenous SUP35-GFP. [PIN+] cells with endogenous SUP35 labeled with GFP (L3107) were grown in 2% Gal to overexpress untagged Sup35NM (p2036). High levels of Sup35NM induced endogenous Sup35-GFP to form dots and rings at the indicated times (n≈600). C. Visualization of Sup35 aggregates induced by Sup35NM BiFC constructs in [PIN+] cells. Sup35NM-VC (p1892) and Sup35NM-VN (p1893) in 74D-694 [PIN+][psi-] cells were co-overexpressed by growth in 0.2% Gal. The picture on the right (magnified ∼3X) at 8 h represents FM4-64 stained (red) cells with Sup35 BiFC dot (yellow). BiFC fluorescence was detected with a YFP filter. (n≈600). Tab. 1. Quantitative data for the aggregation of fluorescently-tagged Sup35 shown in Fig. 1. 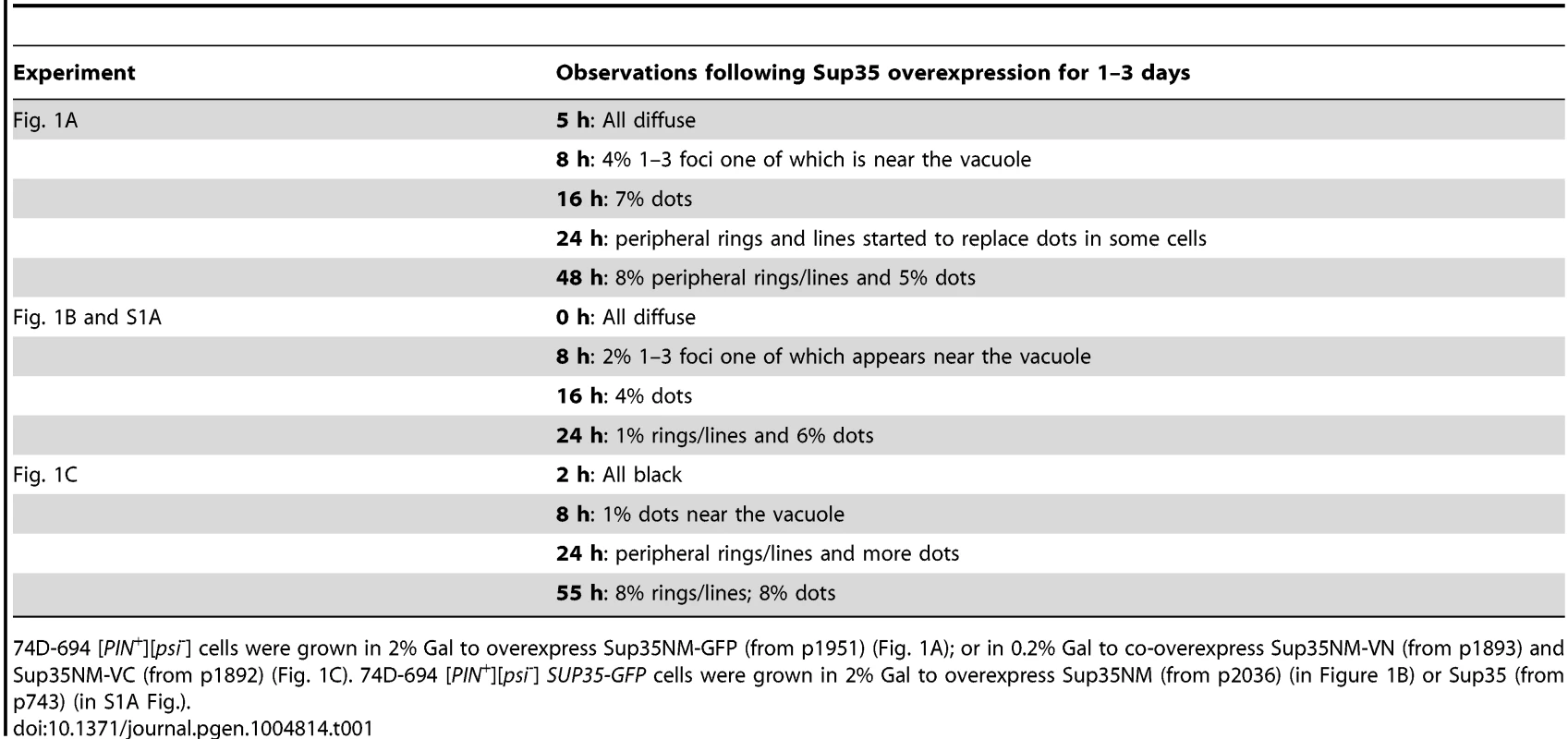
74D-694 [PIN+][psi-] cells were grown in 2% Gal to overexpress Sup35NM-GFP (from p1951) (Fig. 1A); or in 0.2% Gal to co-overexpress Sup35NM-VN (from p1893) and Sup35NM-VC (from p1892) (Fig. 1C). 74D-694 [PIN+][psi-] SUP35-GFP cells were grown in 2% Gal to overexpress Sup35NM (from p2036) (in Figure 1B) or Sup35 (from p743) (in S1A Fig.). In another approach, we examined endogenous Sup35 tagged with GFP when untagged Sup35NM was overexpressed in [PIN+][psi-] cells (Fig. 1B, Table 1). Upon induction of Sup35NM, all cells initially showed diffuse Sup35-GFP fluorescence, which was later seen as cytoplasmic dots one of which was near the vacuole and then, lines and rings in some cells. Similar results were observed when untagged full-length Sup35 was overexpressed (S1A Fig.). As expected, [pin-] cells always displayed diffuse Sup35-GFP molecules in the presence of Sup35NM overexpression (S1B Fig.).
Because the diffuse fluorescence of Sup35NM-GFP observed in the experiments above might have masked the visualization of initial Sup35 aggregates during [PSI+] induction, we overexpressed Sup35NM from Bimolecular Fluorescence Complementation [88], [89] (BiFC) constructs, Sup35NM-VN and Sup35NM-VC, simultaneously in [PIN+][psi-] cells (Fig. 1C, Table 1). Prior to 8 h of induction, no fluorescence was detected. The lack of diffuse fluorescence suggests that Sup35 aggregation does not begin all over the cell. At 8 h, a few cells showed fluorescent dots near the vacuole revealed by FM4-64 staining [90], but no lines/rings were visible. By 24-55 h, peripheral rings and lines appeared and more cells displayed dots. In control [pin-] cells, no fluorescence was detected.
To determine if the dots and rings/lines that appeared within 24 h of induction of Sup35NM-YFP overexpression show amyloid-like properties, we stained the newly appearing Sup35NM-YFP aggregates in [PIN+][psi-] cells with Thioflavin T (ThT) (Fig. 2): 30% of the dots and 60% of the rings were ThT-positive. As expected, Sup35NM-YFP mature dots in [pin-][PSI+] were all stained with ThT. In control [pin-][psi-] cells however, diffuse Sup35NM-YFP fluorescence did not show any ThT fluorescence. These data suggested that Sup35NM-YFP does not always form amyloidogenic aggregates during [PSI+] induction, but eventually becomes amyloid in mature [PSI+].
Fig. 2. Sup35 newly induced aggregates are not always amyloid-like, but mature Sup35 aggregates are. 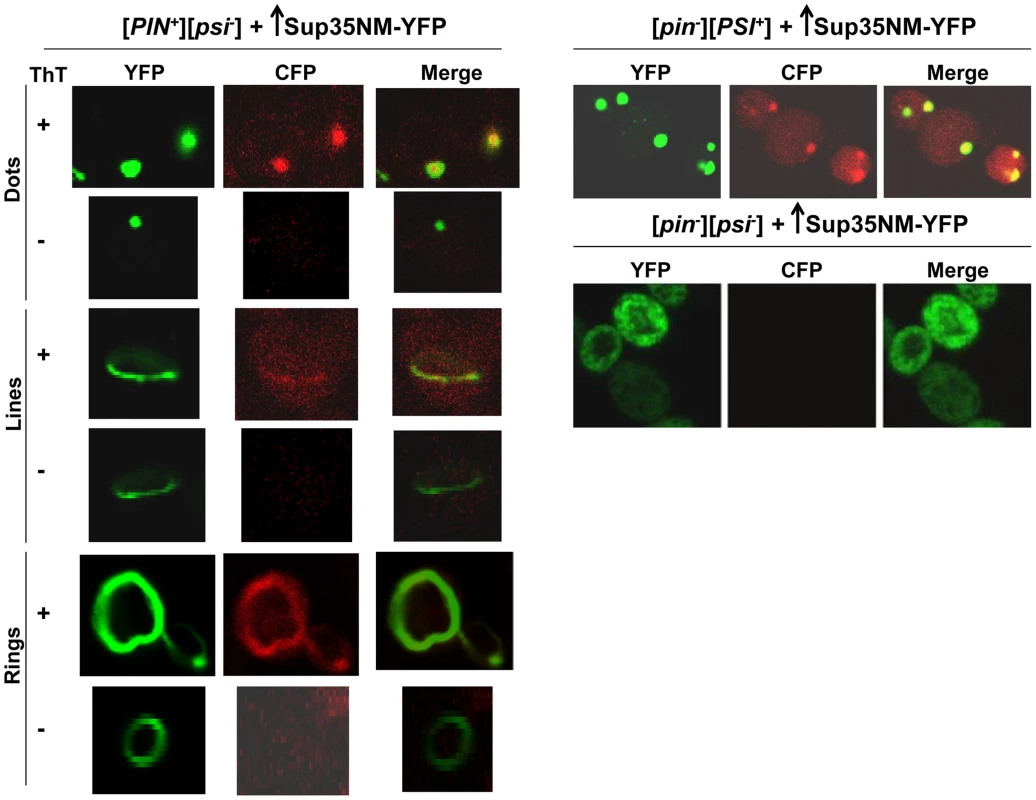
[PIN+][psi-], [pin-][PSI+], [pin-][psi-] yeast cells overexpressing YFP-fusions of Sup35NM (p1753) for 24 h in 2% Gal were stained with Thioflavin T to assess amyloid formation. Sup35NM-YFP inclusions in [pin-][PSI+] contained amyloid (ThT fluorescence signal in CFP channel), however in [pin-][psi-], there were no inclusions formed, and these cells were negative in ThT fluorescence. In [PIN+][psi-], overexpression of Sup35NM-YFP induced the formation of dots, lines and rings, which were sometimes but not always ThT-positive. From top to bottom, pictures represent ThT positive and negative, respectively, for dots, lines and rings. To further investigate the subcellular localization of the initial Sup35 dots, we used BY4741 cells with genomic HSP42 tagged with GFP [91] (see Table 2 for details). Hsp42 is a small heat shock protein that appears as one big dot near the vacuole, sometimes referred to as the IPOD for the site(s) of deposit of insoluble protein aggregates [92]–[94]. Overexpression of Sup35NM-RFP in [PIN+][psi-] HSP42-GFP cells first caused the occasional appearance of cells with 1-6 dots, one of which always colocalized with the Hsp42-GFP dot (Fig. 3). Later, in some cells, Sup35NM-RFP fluorescence extended from a bright dot that colocalized with the Hsp42-GFP dot as short lines tangent to the vacuole or as lines extending to the cell periphery. Interestingly, the multiple Sup35NM-RFP dots observed initially were never seen later once lines appeared, suggesting that Sup35NM-RFP aggregates that did not colocalize with Hsp42-GFP were solubilized, or may have joined the lines. Eventually, in some cells, Sup35 formed internal rings surrounding the vacuole as seen previously [83], [84], intersecting the Hsp42-GFP dot, and in a very few cells, lines were seen to extend from the Hsp42-GFP dot peripherally and around the vacuole simultaneously.
Fig. 3. Sup35 aggregates initially appear near the vacuole, from which short lines extend to the periphery to form rings. 
Sup35NM-RFP was overexpressed from p2017 by growing [PIN+] BY4741 cells with endogenous HSP42-GFP in 2% Gal. Time shown is after the addition of 2% Gal (see details in Table 2). Tab. 2. Data for the aggregation of Sup35NM-RFP in HSP42-GFP cells shown in Fig. 3. 
BY4741 [PIN+][psi-] HSP42-GFP cells were grown in 2% Gal to overexpress Sup35NM-RFP (from p2017). Data for the florescence of only Sup35NM-RFP is tabulated. Hsp42-GFP was always seen as 1 big spot near the vacuole in all cells. To determine the localization of Sup35 newly induced aggregates with respect to the vacuole, we overexpressed Sup35NM-RFP in [PIN+] cells with genomic VPH1 tagged with GFP (S2 Fig.). Vph1 is a subunit of the vacuolar-ATPase protein and marks the vacuolar membrane [95]. We found that Sup35 early dots (after 24 h of Sup35NM-RFP overexpression) were localized near the vacuole, and later, short lines extended outward from the vacuole to the periphery of the cell. Then, as expected, Sup35 formed peripheral rings, and eventually perivacuolar rings.
In summary, the various experiments above showed that during the de novo aggregation of Sup35 induced by its overexpression, Sup35 initially formed dots, one of which perfectly colocalized with the Hsp42-GFP dot near the vacuole. Then, Sup35 lines extended from this dot to form peripheral and eventually perivacuolar rings, while the other initial Sup35 dots disappeared.
Colocalization of Rnq1 with Sup35 dots
To visualize the relationship of Sup35 and Rnq1 during the de novo induction of [PSI+], we expressed Rnq1-GFP under its own promoter, and overproduced Sup35-RFP in [PIN+][psi-] cells (Fig. 4A, top). Sup35-RFP initially formed fluorescent dots but no lines or rings (Details in S2 Table). All the Rnq1-GFP dots perfectly colocalized with Sup35-RFP dots, but only 60% of the Sup35-RFP dots colocalized with Rnq1-GFP foci. Interestingly, the colocalized Rnq1-Sup35 dots were always associated with the vacuole revealed by FM4-64 staining (Fig. 4A, middle). Additional Sup35 dots that did not overlap Rnq1 were away from the vacuole (Fig. 4A, gray arrows).
Fig. 4. Rnq1 aggregates colocalize with Sup35 aggregates when [PSI+] is induced de novo. ![Rnq1 aggregates colocalize with Sup35 aggregates when [<i>PSI<sup>+</sup></i>] is induced <i>de novo</i>.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/4ddaf437c1bbc727f79d1ee6f26421f7.png)
A. Colocalization of Sup35 newly appearing vs. mature [PSI+] dots with normal levels of Rnq1. Sup35-RFP was overexpressed (p1678) in [PIN+][psi-] cells with the plasmid p1730 expressing Rnq1-GFP from its own promoter by growth in 2% Gal (top). Gray arrows show additional Sup35 dots that did not overlap Rnq1 and that are away from the vacuole. The middle panel (magnified ∼2X) shows an Rnq1-GFP dot colocalized with Sup35-RFP dot (white arrow) located near the vacuole stained with FM4-64 (red circle). [PIN+] cells with established [PSI+] were grown in 0.05% Gal for 3–4 h to stain mature [PSI+] dots marked by Sup35NM-YFP (bottom). These dots showed partial colocalization (enlarged box) with [PIN+] dots marked by Rnq1-CFP. Blue arrows indicate partially colocalized Rnq1 dots; yellow arrows indicate non-colocalized additional Rnq1 dots. B. Colocalization of Sup35 rings with normal levels of Rnq1 expressed from a plasmid. Sup35-RFP was overexpressed from p1678 in [PIN+][psi-] cells with the CEN plasmid p1730 expressing Rnq1-GFP from its own promoter by growth in 2% Gal. Sup35-RFP rings colocalized with Rnq1-GFP rings after 24 h. C. Colocalization of Sup35 rings with normal levels of Rnq1 expressed from an integrated construct. Rnq1 tagged with CFP, integrated into genomic TRP1 was expressed from its own promoter in [PIN+] cells overexpressing Sup35NM-YFP from p1753, by growth in 2% Gal for the indicated times. D. Visualization of [PIN+] aggregates during [PSI+] induction with overexpressed Rnq1 and Sup35. Rnq1-YFP was co-overexpressed with Sup35NM respectively from p1728 and p1893, by growth in 2% Gal for 24 h. Rnq1-YFP formed lines and mesh-like aggregates in the presence of [PIN+] and Sup35NM overexpression, but remained diffuse in [pin-]. In the absence of Sup35NM overexpression, [PIN+] contained only multiple dots of Rnq1-YFP. E. Visualization of [PIN+] aggregates during Sup35 overexpression with normal levels of Rnq1. Untagged Sup35NM (p2036) was overexpressed in [PIN+] RNQ1-CFP integrants in 2% Gal for 48 h. Rnq1-CFP displayed ring/line-like structures in the presence of Sup35NM overexpression in 10% of [PIN+] cells (4 trials, each with n≈350). These Sup35 dots have different characteristics from Sup35 dots seen in mature [PSI+] cells. For example, while the results above showed that newly appearing vacuole-associated Sup35 aggregates perfectly colocalize with Rnq1, mature Sup35 aggregates in established [PSI+] cells did not entirely overlap Rnq1 (seen as in two sets intersecting in a Venn diagram) (Fig. 4A, bottom). Also, additional Rnq1-CFP dots existed in [PIN+][PSI+] cells that did not show any colocalization with Sup35 (Fig. 4A bottom, arrows, enlarged box).
In summary, after 6 h of [PIN+]-promoted Sup35-RFP aggregation, all Rnq1 dots perfectly overlapped newly induced Sup35 dots around the vacuole, but additional Sup35 dots (away from the vacuole) without overlapping Rnq1 sometimes existed in those cells. In contrast, in established [PSI+] cells, all Sup35 dots partially overlapped Rnq1 dots, but additional Rnq1 dots without overlapping Sup35 also existed.
Colocalization of Rnq1 with Sup35 rings
To look for colocalization of Rnq1 with Sup35 rings, we induced overexpression of Sup35-RFP for 24 h in [PIN+] cells also expressing Rnq1-GFP from its own promoter. Sup35-RFP formed rings 70% of which colocalized with Rnq1-GFP (Fig. 4B). In the remaining 30% of cells with Sup35-RFP rings that did not colocalize, Rnq1-GFP fluorescence was instead diffuse or in the form of dots (S3A Fig., S3 Table). In contrast, essentially all Rnq1-GFP rings colocalized with Sup35-RFP rings (S4 Table). As expected, Rnq1-GFP and Sup35-RFP always remained diffuse in [pin-] cells (S3B Fig.).
In another version of this experiment, Sup35NM-YFP was overexpressed in [PIN+][psi-] cells expressing Rnq1-CFP under its own promoter. Sup35NM-YFP overexpressed for 24 h formed fluorescent dots or rings respectively, in 7 and 0.8% of the cells. In these cells, 90% of Sup35NM-YFP dots colocalized with Rnq1-CFP dots, and all Sup35NM-YFP rings colocalized with Rnq1-CFP rings (Fig. 4C top, S5 Table). Curiously, these Rnq1-CFP rings looked like beads on a string, rather than an uninterrupted full ring.
After 48 h of Sup35NM-YFP overexpression, Sup35NM-YFP formed rings in 11% of the cells, and 75% of these Sup35NM-YFP rings colocalized with Rnq1-CFP rings (Fig. 4C bottom). In the remaining 25% of cells with Sup35NM-YFP rings, Rnq1-CFP showed diffuse or dot fluorescence (S6 Table). On the other hand, essentially all Rnq1-CFP rings colocalized with Sup35NM-YFP rings (S7 Table). In control experiments, [pin-] cells never formed any aggregates when Sup35NM-YFP was overexpressed (S4A Fig.). [PIN+] cells showed only Rnq1-CFP dots, but no lines/rings when Sup35NM-YFP expression remained repressed in 2% Glucose (S4B Fig.) and when cells with empty vector expressing YFP were grown in 2% Gal (S4C Fig.). These colocalization data were based on visually checking different planes of the cells by moving the focal plane up and down, and were also confirmed by collecting z-stacks from representative cells (S5 Fig.).
Next, we co-overexpressed Rnq1-YFP and Sup35NM-VN in [PIN+][psi-] cells (Fig. 4D). In 24 h, 7% of the cells had one Rnq1-YFP dot with lines extending from it in all directions, referred to as mesh-like aggregates. In controls, when Rnq1-YFP was overexpressed in [PIN+] without overexpressing Sup35NM, 90% of the cells showed multiple fluorescent dots, 10% had diffuse fluorescence, and none had mesh-like aggregates. Also, overexpressing Rnq1-YFP and Sup35NM-VN simultaneously in [pin-] control cells did not result in any aggregate formation. When cells from cultures with Rnq1-YFP mesh-like aggregates (7%) were scored for [PSI+], 6.5% of these cells formed pink or white colonies and were able to grow on media lacking adenine (SD-Ade), indicative of [PSI+] (See Methods).
To visualize if Sup35 and Rnq1 have a close physical interaction during [PSI+] induction, we co-overexpressed Sup35NM-VN and Rnq1-VC in [PIN+][psi-] cells (S6 Fig., S8 Table). Initially (16 h post induction) 1.8% of the cells showed dots, but no lines/rings; but later (40 h post induction) peripheral rings, lines and mesh-like aggregates appeared and more cells displayed dots. We also co-overexpressed Sup35NM-VN and Rnq1-VC in established [PSI+] as well as in [pin-][psi-] cells and did not observe any fluorescence. These data indicate that Rnq1 and Sup35 interact in a close proximity during [PSI+] induction, but not in established [PSI+].
As expected, overexpression of untagged Sup35NM caused Rnq1-CFP expressed from its own promoter to align in ring/line-like aggregates in 10% of the [PIN+] cells (Fig. 4E). The Rnq1-CFP lines looked like beads on a string as seen previously in the presence of overexpressed YFP-tagged Sup35NM (Fig. 4C top). Such beads on a string never appeared in [PIN+] cells without overexpressed Sup35NM, where Rnq1-CFP fluorescent dots remained dispersed, or in [pin-], where Rnq1-CFP remained diffuse (S7 Fig.). When cells from the culture with Rnq1-CFP beads on a string (10%) were scored for [PSI+], 11% of these cells formed pink or white colonies, indicative of [PSI+].
The above experiments show that during the de novo aggregation of Sup35 in [PIN+] cells, overexpression of Sup35NM induced Rnq1 to form mesh-like or line/ring-like aggregates. Essentially all Rnq1 line/rings perfectly overlapped Sup35 line/rings, while some Sup35 line/rings did not overlap Rnq1. BiFC between Rnq1 and Sup35 confirmed that they form a close physical interaction during the induction of [PSI+], and initially form dots, and then lines/meshes; while they do not form such an interaction in established [PSI+].
Localization of other proteins during [PSI+] induction
In order to determine what other proteins (particularly chaperones) colocalize with Sup35 during its aggregation in the presence of [PIN+], we overexpressed Sup35NM-RFP in [PIN+] cells with endogenously tagged GFP proteins [91]. As seen previously [96], we found that the molecular chaperones Hsp104, Sis1, and Ssa1 involved in [PSI+] propagation via their involvement in prion shearing [72], [97]–[104], colocalized with Sup35 rings and dots perfectly during de novo induction of [PSI+] (Fig. 5A, S8 Fig.). However, Ydj1, a Hsp40 co-chaperone shown to co-immunoprecipitate with [PSI+] aggregates as a minor component along with Hsp104, Ssb, Sis1, Sse1 [104], [105] did not colocalize with Sup35 rings (Fig. 5A).
Fig. 5. Induced Sup35 rings do not colocalize with Ydj1, but do colocalize with Hsp104 and Sup45. 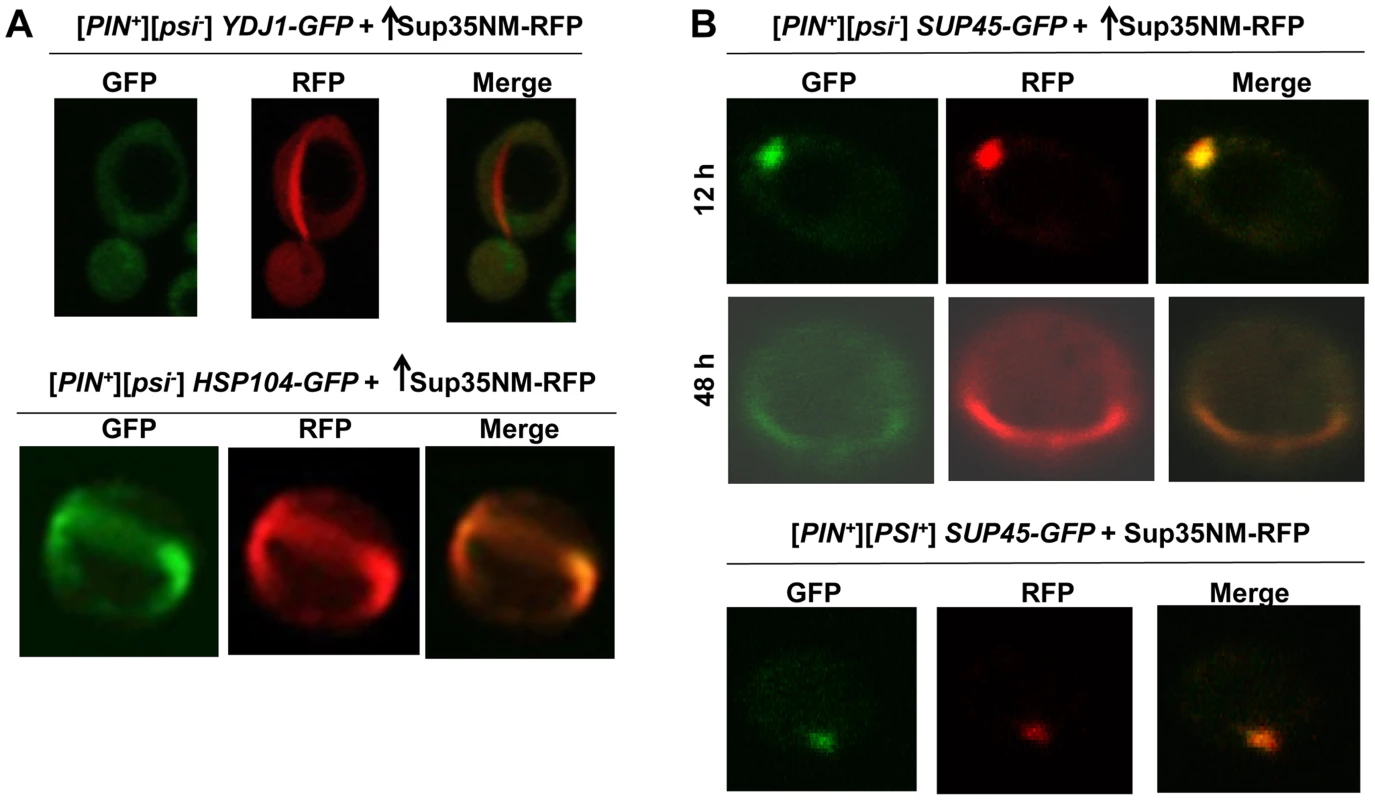
A. Colocalization of newly induced Sup35 aggregates with chaperones. Sup35NM-RFP was overexpressed (p2018) in BY4741 [PIN+] cells with GFP-tagged chaperones in 2% Gal for 48 h. While 9.3% of the cells contained Sup35NM-RFP peripheral rings and dots, Ydj1-GFP always remained cytoplasmically diffuse (n≈450) (top). In HSP104-GFP cells, by 48 h, Hsp104-GFP formed rings in 5.6% of the [PIN+] cells all of which colocalized with Sup35 rings during [PSI+] induction. (n≈350) (bottom). B. Colocalization of newly induced vs. established Sup35 aggregates with Sup45. Sup35NM-RFP was overexpressed from p2018 in [PIN+] cells with endogenous SUP45 tagged with GFP by growth in 2% Gal for the indicated times. Although overexpression of Sup45 inhibits [PSI+] induction [107], Sup45-GFP perfectly colocalized with Sup35NM-RFP aggregates in [PIN+][psi-] cells (top, n≈400). After these cells were allowed to propagate [PSI+], Sup45-GFP also perfectly overlapped Sup35 mature dot (bottom). We also asked if proteins other than chaperones, previously found to influence the maintenance or induction of [PSI+] [22], [106], [107], would colocalize with newly appearing Sup35 aggregates. We found that Sup45-GFP perfectly colocalized with newly appearing Sup35NM-RFP aggregates, as well as with established [PSI+] aggregates (Fig. 5B). Upon testing other candidate proteins (Cyc8, New1, Pin3, Pin4, Tup1, Mod5, Sgt2), we found that none of them displayed any colocalization with Sup35NM-RFP aggregates (S9 Fig.).
Colocalization of Q/N-rich Pin4C vs. non-Q/N rich Mod5 with newly appearing Sup35 aggregates during [PSI+] induction in the absence of Rnq1
Next, we investigated if high levels of Pin4C (120–668 a.a.), which were previously shown to substitute for [PIN+] in promoting de novo induction of [PSI+] [22], [66], would overlap Sup35 aggregates during [PSI+] induction in the absence of Rnq1. We simultaneously overexpressed Pin4C-RFP and Sup35NM-GFP in 74D-694 rnq1Δ [psi-] cells (Fig. 6, Table 3). Both proteins initially remained diffuse, but by 8 h, tiny Pin4C-RFP fluorescent dots appeared in some of the cells, all of which still had diffuse fluorescence of Sup35NM-GFP. At 16 h, Sup35NM-GFP fluorescent dots appeared and essentially all of these colocalized with Pin4C-RFP near the vacuole. At 24 h, Sup35NM-GFP started to appear as fluorescent rings in addition to dots. In these cells, essentially all Sup35NM-GFP aggregates overlapped Pin4C-RFP. At 48 h, the number of cells with Sup35NM-GFP dots decreased, while cells with rings increased, and all Sup35 aggregates still overlapped Pin4C-RFP. At 72 h, the Sup35NM-GFP dots still colocalized with Pin4C-RFP. However, almost all of the cells that had Sup35NM-GFP rings failed to show Pin4C-RFP rings. Instead, they contained large fluorescence spots of Pin4C-RFP that did not colocalize with Sup35NM-GFP. In control rnq1Δ cells that separately overexpressed either Sup35NM-GFP or Pin4C-RFP for 72 h, respectively, Sup35NM-GFP always remained diffuse, while in 50% of the cells (n≈450) Pin4C-RFP formed large fluorescent dots (S10A Fig.). As expected, [PSI+] appeared de novo in rnq1Δ cultures with overexpressed Sup35NM-GFP and Pin4C-RFP (S9 Table).
Fig. 6. Excess Pin4C temporarily colocalize with Sup35 aggregates during [PSI+] induction in the absence of Rnq1. ![Excess Pin4C temporarily colocalize with Sup35 aggregates during [<i>PSI<sup>+</sup></i>] induction in the absence of Rnq1.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/a42b868cb9b9bde50901607184b6e4a3.png)
Pin4C-RFP (p1708) and Sup35NM-GFP (p1951) were simultaneously overexpressed in 74D-694 rnq1Δ cells with 2% Gal for the indicated times. While Sup35NM-GFP early dots and rings (up to 48 h) colocalized with Pin4C-RFP, Pin4C-RFP lost colocalization with Sup35 rings later (72 h) (data in Table 3). Tab. 3. The data for the fluorescence patterns of Pin4C-RFP and Sup35NM-RFP in rnq1Δ cells (refer to Fig. 6). 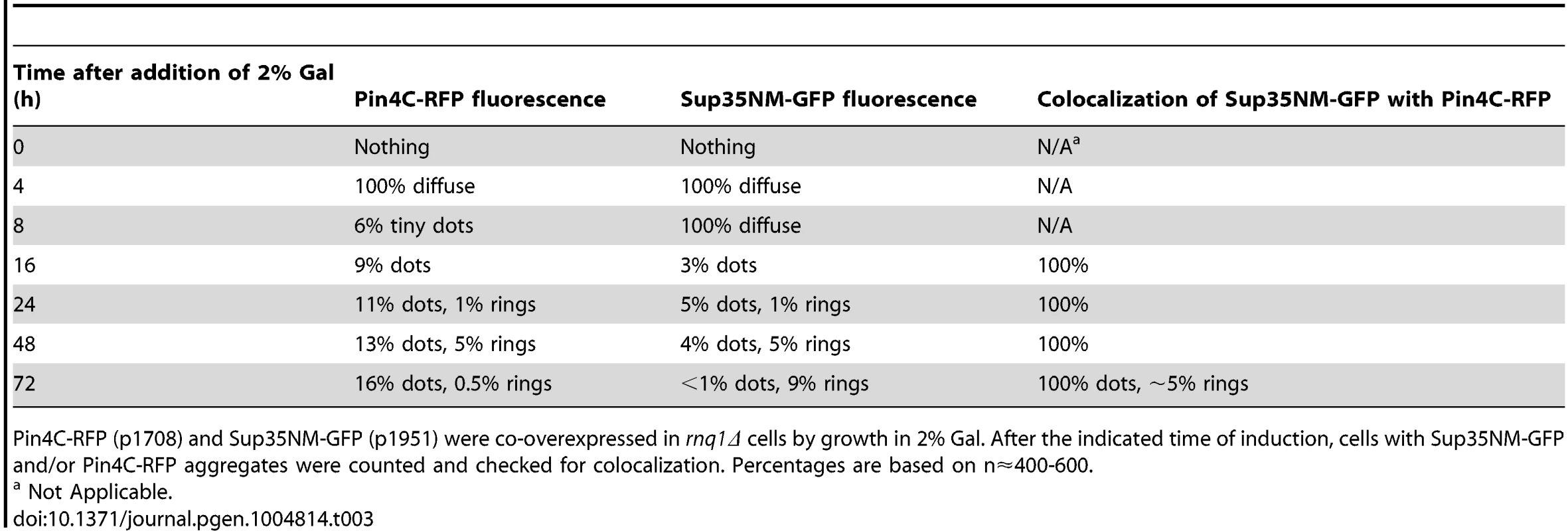
Pin4C-RFP (p1708) and Sup35NM-GFP (p1951) were co-overexpressed in rnq1Δ cells by growth in 2% Gal. After the indicated time of induction, cells with Sup35NM-GFP and/or Pin4C-RFP aggregates were counted and checked for colocalization. Percentages are based on n≈400-600. To determine the location of Pin4C aggregates relative to the vacuole during Sup35 de novo aggregation, we overexpressed Pin4C-RFP in [pin-] HSP42-GFP cells in the presence vs. absence of Sup35NM overexpression (S10B Fig.). In the presence of Sup35NM overexpression, Pin4C-RFP formed one to a few foci, one of which perfectly (in 6.5% of the cells; n∼800) or partially (in 93.5% of the cells) overlapped the Hsp42-GFP dot. In the absence of Sup35NM overexpression, Pin4C-RFP dots never perfectly overlapped the Hsp42-GFP dot; rather one of the Pin4C-RFP aggregates was juxtaposed to, or partially colocalized with the Hsp42-GFP dot. This suggests that Sup35NM overexpression promotes a more frequent closer association of the Pin4C-RFP aggregate and the vacuole-associated Hsp42-GFP protein deposit.
When we simultaneously overexpressed the non-Q/N rich prion protein Mod5 tagged with GFP and Sup35NM-RFP in 74D-694 rnq1Δ cells, Sup35NM-RFP initially formed dots, and then rings/lines, while Mod5-GFP always remained diffuse (Fig. 7A, Table 4). In control rnq1Δ cells that separately overexpressed either Sup35NM-RFP or Mod5-GFP, both proteins remained diffuse (S11A Fig.). Since we could not see fluorescent aggregates of Mod5-GFP, we turned to BiFC to look for an interaction between Mod5 and Sup35NM (Fig. 7B, S11B Fig.). Overexpression of Mod5-VN and Sup35NM-VC did not result in any fluorescence in [pin-] cells, although [PSI+] was induced in this culture with a frequency of 0.9% (n∼1000) suggesting that cross-seeding may not be universal for [PSI+] induction. Curiously, in [PIN+] cells, overexpression of Mod5-VN and Sup35NM-VC resulted in first diffuse fluorescence and then the formation of a single (near the vacuole) to multiple dots over time in 15% of cells (Fig. 7B). Also, Mod5-VN and Mod5-VC overexpression in [pin-] cells did not show any fluorescence, but in [PIN+] cells they showed diffuse fluorescence (S11C Fig.). Possibly, newly appearing Sup35NM aggregates in [PIN+] cells are attracted to the Mod5 aggregates seen as diffuse fluorescence.
Fig. 7. Overexpressed Mod5 does not interact with Sup35 during [PSI+] induction in the absence of Rnq1. ![Overexpressed Mod5 does not interact with Sup35 during [<i>PSI<sup>+</sup></i>] induction in the absence of Rnq1.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/085436da488db2447e172ec7eb2e6791.png)
A. Mod5 did not form visible aggregates although Sup35 newly appearing aggregates formed. Mod5-GFP (p2061) and Sup35NM-RFP (p2018) were simultaneously overexpressed in 74D-694 rnq1Δ cells with 2% Gal for the indicated times. Mod5-GFP remained diffuse although Sup35NM–RFP formed dots and rings (data in Table 4). B. Mod5 did not form a close physical interaction with Sup35 during [PSI+] induction in the absence of [PIN+]. Mod5-VN (p2170) and Sup35NM-VC (p1892) were simultaneously overexpressed in 74D-694 [pin-] (top) and [PIN+] (bottom) cells with 2% Gal for the indicated times. No fluorescence was detected in [pin-], but Sup35-Mod5 dots appeared in [PIN+] cells mostly near the vacuole (arrows) following the diffuse fluorescence. (see also S11B Fig.) Tab. 4. The data for the fluorescence patterns of Sup35NM-RFP and Mod5-GFP in rnq1Δ cells (refer to Fig. 7A). 
Sup35NM-RFP (p2018) and Mod5-GFP (p2061) were co-overexpressed in rnq1Δ cells by growth in 2% Gal. After the indicated time of induction, cells with Sup35NM-RFP and Mod5-GFP fluorescence were counted (n≈600). These findings indicate that during the de novo aggregation of overexpressed Sup35 promoted by overexpression of Q/N-rich Pin4C in the absence of Rnq1 [22], [66], Sup35 aggregates initially colocalize with Pin4C aggregates (near the vacuole), but Pin4C falls off the Sup35 rings later. Furthermore, Pin4C-RFP perfectly overlapped Hsp42-GFP only in the presence of Sup35 overexpression. The data is consistent with the cross-seeding of Sup35 aggregation by Pin4C aggregates in the absence of Rnq1. However, although the overexpression of the non-Q/N rich protein Mod5 promotes [PSI+] induction [27], we could not visualize Mod5-Sup35 direct interaction in the absence of [PIN+] suggesting that Mod5-promoted de novo Sup35 aggregation occurs via a different mechanism.
The role of Hsp104 in de novo [PSI+] induction
The Hsp104 chaperone requirement for the maintenance of [PSI+] [72] and the colocalization of Hsp104 with Sup35 dots and rings during the induction of [PSI+] ([96], Fig. 5A) led us to ask if Hsp104 is also required for the de novo aggregation of Sup35 during [PSI+] induction. Since Hsp104 is required for the maintenance of [PIN+], and the requirement for [PIN+] in the de novo induction of [PSI+] can be overcome by overexpressing certain Sup35NM-containing fragments, e.g. with a short extension of hydrophobic residues [108], we overexpressed Sup35NM with a short extension of hydrophobic residues (magic tail), previously shown to induce [PSI+] even in [pin-] cells [49], [108]. Overexpression of Sup35NM with this magic tail (Sup35NM-mt) in [pin-] HSP104 cells, caused endogenous Sup35-GFP molecules to form dots and short lines in 6% of the cells (n≈1200) (Fig. 8A) and this increased to 15% (n≈1400) of the cells in the presence of [PIN+]. Unlike the Sup35 dots induced in [PIN+] cells by 24 h of overexpression of Sup35NM without magic tail, Sup35 dots induced by Sup35NM-mt in [pin-] HSP104 or [PIN+] HSP104 cells did not always appear near the vacuole. In [pin-] hsp104Δ cells however, no dots were seen and only 0.4% of the cells (n≈1700) formed Sup35-GFP lines. Furthermore, all these lines appeared to be at the cell membrane as opposed to those seen in the cytoplasm in HSP104 cells.
Fig. 8. Hsp104 is required for the de novo [PSI+] induction. ![Hsp104 is required for the <i>de novo</i> [<i>PSI<sup>+</sup></i>] induction.](https://www.prolekare.cz/media/cache/resolve/media_object_image_small/media/image/ad90eb00fab056d727cd199c8eca6b51.png)
A. Absence of Hsp104 dramatically reduces the frequency of de novo Sup35 aggregates. Sup35NM with a short tail of hydrophobic residues (Sup35NM-mt) was overexpressed from p1984 by growing hsp104Δ (GF844) or [pin-] HSP104 (GF658) cells with endogenous SUP35-GFP in 2% Gal. Sup35-GFP remained mostly diffuse in hsp104Δ cells with a few cells with short lines located at the cell membrane, but formed dots and short lines throughout the cytoplasm in [pin-] HSP104 cells after 48 h of induction. B. Hsp104 is necessary to induce Sup35 newly appearing rings and lines during Pin4C-promoted [PSI+] induction. Sup35NM-GFP and Pin4C-RFP were respectively co-overexpressed from p1181 and p1708, by growing hsp104Δ (L1802, or L1803), HSP104 [pin-] (L2910) or HSP104 [PIN+] (L1749) cells in 50 µM CuSO4 and 2% Gal. In the absence of Hsp104, Sup35NM-GFP failed to form any aggregates in contrast to HSP104 cells. C. Inhibition of Hsp104 does not induce [PSI+] de novo. Sup35NM-GFP and Pin4C-RFP were co-overexpressed respectively from p1181 and p1708, by growing HSP104 [pin-] (L2910) cells in 50 µM CuSO4, 2% Gal and with or without 10 mM GuHCl. In comparison to dots and ring formation by Sup35NM-GFP in the absence of GuHCl (Fig. 8B), Sup35NM-GFP never formed rings/lines in the presence of GuHCl. 2.8% of cells had dots and 97.2% of cells were diffuse. None of cells grown in the presence of GuHCl induced [PSI+]. The pictures were taken 72 h after the addition of copper, galactose and GuHCl to the growth media. In an alternative approach to test the role of Hsp104 in [PSI+] induction, we used Pin4C overexpression to substitute for [PIN+]. We co-overexpressed Sup35NM-GFP and Pin4C-RFP in [pin-] hsp104Δ cells (Fig. 8B), and Sup35NM-GFP remained diffuse, while in control [pin-] HSP104 or [PIN+] HSP104 cells, as expected, Sup35NM-GFP formed dots and rings/lines respectively in 7% and 9% of the cells. Also, Pin4C-RFP aggregates were not affected by the presence vs. absence of Hsp104 in [pin-] cells, but were larger in the presence of [PIN+] (S12 Fig.). Co-overexpression of Sup35NM-GFP and Pin4C-RFP in [pin-] HSP104 cells in the presence of GuHCl, which inhibits Hsp104's ATPase activity [79] resulted in the formation of only Sup35NM-GFP dots in 2.8% of cells, but no rings/lines and failed to induce any [PSI+] cells (Fig. 8C). Taken together, the striking differences in the level and types of Sup35 aggregate formation in the presence vs. absence of Hsp104 and in the presence vs. absence of GuHCl suggest that Hsp104 is required for the formation of de novo Sup35 aggregates, and induction of [PSI+] de novo.
Discussion
Protein aggregates have been implicated in a wide variety of diseases including Amyotrophic Lateral Sclerosis, Alzheimer's, Parkinson's and prion disease [109], [110]. Interactions between proteins associated with protein misfolding diseases (PMD) are of great interest since molecular cross-talk between disease aggregates of one protein can accelerate the de novo appearance of heterologous disease protein aggregates [16], [60], [111]–[115]. Several reports implicate cross-seeding as a mechanism to explain this [17], [18]. Here, our data provide insight into the mechanism of prion induction, which is a model for such heterologous interactions in human diseases. Our data suggest that cross-seeding is not the only mechanism for this cross-talk phenomenon. While our findings that [PIN+] or Pin4C aggregates physically interact with Sup35 de novo aggregates are consistent with the cross-seeding model, the failure of Mod5 to interact with Sup35 suggests the existence of another mechanism, possibly via hindering the chaperone network.
We stimulated the de novo generation of a prion in yeast by overexpressing fluorescent derivatives of the prion protein and then monitored its aggregation status. Based on our findings (Fig. 1–3, 6, S1–2 Fig.), we propose a model to explain the pathway followed by Sup35 during its de novo conversion into mature [PSI+] (Fig. 9A). Bimolecular complementation [88], [89] using BiFC-tagged Sup35NM (Fig. 1C) showed that Sup35NM molecules do not associate throughout the cytoplasm, but first interact at specific sites in the cell. This means that the de novo aggregation of Sup35 first occurs at these sites rather than being brought to them as pre-existing aggregates. This may be true of other prion-like aggregating proteins as well.
Fig. 9. Models we propose to explain de novo Sup35 aggregation. 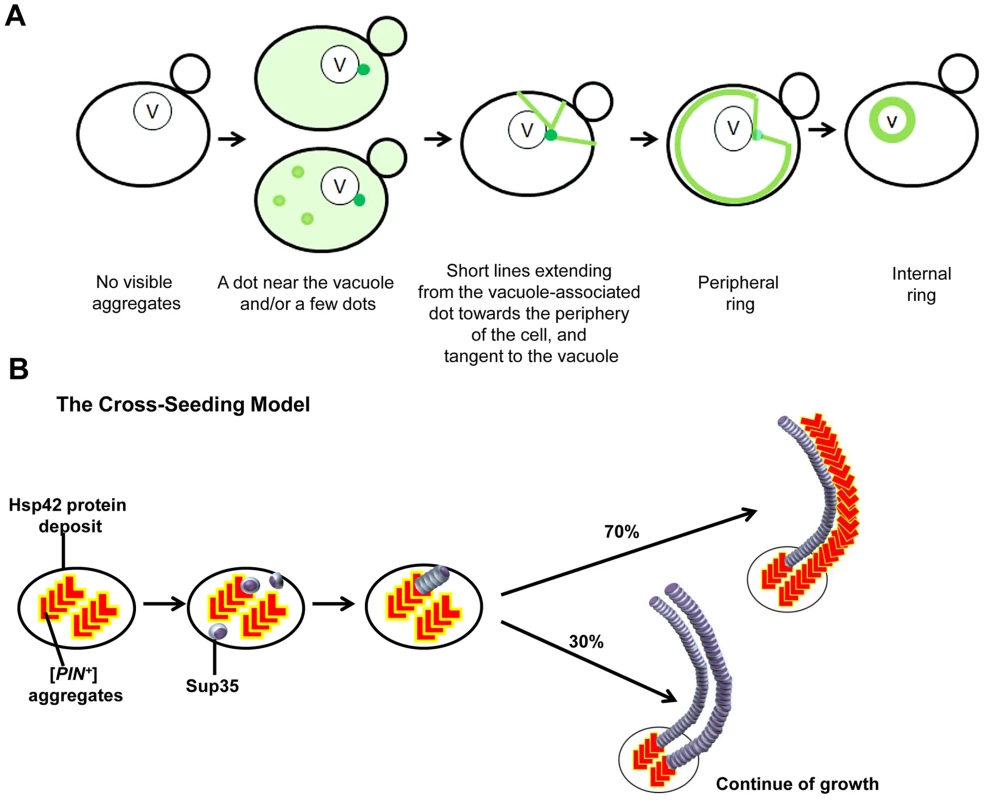
A. The stages of the induced appearance of [PSI+] de novo. Induction of [PSI+] by overexpression of fluorescently-labeled Sup35 in [PIN+][psi-] cells displays early Sup35 fluorescent foci ranging from one to a few dots, one of which is located near the vacuole (shown as darker green). At this perivacuolar Hsp42 protein deposit site, preexisting heterologous aggregates such as [PIN+] or Pin4C cross-seed the de novo aggregation of Sup35 (see panel B). Later, short lines appear to emanate from this dot towards the periphery of the cell while dots away from the vacuole disappear. Peripheral rings appear next, followed by perivacuolar internal rings. B. The cross-seeding model to explain how heterologous aggregates facilitate de novo Sup35 aggregation. Preexisting [PIN+]/Pin4C aggregates (shown in red ‘L’ shape) located at the Hsp42 protein deposit near the vacuole physically interact with soluble misfolded Sup35 proteins (shown as purple marbles). This interaction causes Sup35 to form oligomers. Then, Sup35 grows in length in a homotypic manner. Based on our data, we propose that [PIN+]/Pin4C aggregates can also grow in length laterally guided by Sup35 fibrils. Over time, laterally growing [PIN+]/Pin4C aggregates are less frequent. For simplicity, only two fibrils are depicted. How could prion-like proteins first interact within discrete inclusions? We speculate that upon initial expression, Sup35 is soluble, but upon overexpression, some molecules of the intrinsically unstructured Sup35NM misfold. We suggest that this misfolded protein could be captured by quality control compartments (QCCs) as inclusions, i.e. recently discovered Q-bodies [116], where misfolded proteins accumulate en route to degradation. The high local concentration of Sup35 at these sites increases the likelihood of prion induction. Furthermore, [PIN+] prion aggregates, which are required for [PSI+] induction [21], [22], [51], are also located at these sites, where they could facilitate nucleation for Sup35 [53], [87], [117], [118] to polymerize Sup35 into amyloid. In Pin4C-promoted [PSI+] induction, Pin4C appears to take over the role of [PIN+] aggregates. This suggests that the co-existence of misfolded protein with heterologous amyloid in inclusions accelerates de novo conversion of the misfolded protein into amyloid.
However, not all of these inclusions give rise to larger Sup35 fibrils. Rather, Sup35 aggregates away from the perivacuolar site disappear, and Sup35 lines and rings emanate only from the single inclusion in the cell near the vacuole (Fig. 1–3,6). This perivacuolar inclusion also differs from the other inclusions because it alone colocalizes with Hsp42. Furthermore, the finding of only one fluorescent dot in [PIN+] cells with overexpressed BiFC-tagged Sup35NM and Rnq1 (S6 Fig.) suggests that Sup35 and Rnq1 interact only at the perivacuolar site. It is unclear what happens to the other inclusions, they could be solubilized, degraded or join the remaining perivacuolar aggregate. This suggests that the fibrillar growth of de novo aggregates requires site-specific chaperones.
Curiously, heterologous aggregates are not only involved in the initial cross-seeding, but continue to be associated with some newly seeded heterologous fibrils. This is surprising since once nucleated by [PIN+]/Pin4C amyloid, Sup35 polymerization should continue without the need for Rnq1/Pin4C seed. Indeed, due to the higher efficiency of homotypic polymerization [55], [118], [PIN+]/Pin4C aggregates are not expected to incorporate into the growing Sup35 fibrils. Paradoxically, Rnq1 frequently (Pin4C always) are found to overlap newly appearing Sup35 rings (Fig. 4, 6). We propose that although the initial step in nucleation is heterotypical, [PIN+]/Pin4C aggregates might also grow in length homotypically in close proximity to the Sup35 aggregates via a lateral interaction, possibly with the help of chaperones that are associated with the aggregates. Indeed, our findings that all Rnq1/Pin4C rings colocalized with Sup35 rings (S4 Table); and that Rnq1/Pin4C formed mesh/ring-like aggregates only in the presence of Sup35 overexpression (Fig. 4D–E, S6 Fig.) support this hypothesis, which predicts that Sup35 rings template the continued growth of Rnq1/Pin4C into rings and not vice versa (see Fig. 9B).
Mature [PSI+] dots do not associate with [PIN+] in the same manner as newly induced [PSI+] aggregates do. Indeed, although Rnq1 was found to co-immunoprecipitate with newly appearing Sup35 aggregates [63], [119], it was not detected in purified [PSI+] aggregates [104]. Here, we show the formation of fluorescent dots and mesh-like aggregates by co-overexpressed Rnq1-VC and Sup35NM-VN during [PSI+] induction, and the inability of Rnq1-VC and Sup35NM-VN to cause fluorescence in established [PSI+], suggesting that Rnq1 and Sup35 are in close proximity during [PSI+] induction, but not so close in established [PSI+] (S6 Fig., S8 Table).
Mature and newly induced Sup35 aggregates also differ in their amyloid characteristics as our data showed that all of the mature [PSI+] aggregates were stained in situ with amyloid-binding dye, thioflavin T, while only 30% and 60% of respectively, newly appearing Sup35 dots and rings were stained with ThT (Fig. 2). This could be either because cross-talk between heterologous amyloid aggregates may not always convert the prion-like protein into amyloid vs. amorphous aggregates, or interference of other proteins attracted to aggregates in situ may cause false negative results. We favor the latter possibility as most viable Sup35 ring-bearing cells give rise to [PSI+] progeny [84], [85]. However, it is possible that dead cells have non-amyloid Sup35 rings. Alternatively, it is also possible that newly appearing Sup35 aggregates that are stained with Thioflavin T harbor Rnq1 amyloid, leading to the ThT staining, while those Sup35 aggregates that are not stained do not harbor Rnq1 amyloid, leading to the failure of ThT staining. This would imply that the Sup35 molecules in the dot and ring structures are not amyloid.
While colocalization of a protein with heterologous aggregates is consistent with cross-seeding, it is not proof of cross-seeding. Indeed, the strict and permanent colocalization of Sup45 with Sup35 newly appearing aggregates and with mature [PSI+] aggregates (Fig. 5B) suggests that Sup45-GFP simply decorates all Sup35 aggregates. However, the considerable and transient colocalization of respectively, Rnq1 and Pin4C with Sup35 newly appearing aggregates, but only partial and no colocalization of respectively, Rnq1 and Pin4C [66] with mature [PSI+] aggregates supports the idea that as opposed to Sup45, Rnq1 and Pin4C actually cross-seed Sup35 de novo aggregates. It is noteworthy that Sup45 is not required for [PSI+] induction [86], but [PIN+], or one of its substitutes, Pin4C is.
Also, the colocalization of the molecular chaperone Hsp104 with all Sup35 aggregates (Fig. 5A, [120]) suggests that Hsp104 decorates rather than cross-seeds Sup35. Such decoration could enable Hsp104 to perform its known shearing activity of [PSI+] aggregates that is required for [PSI+] propagation [72], [80]–[82]. We showed that Hsp104 itself is also required for de novo induction of [PSI+] by finding inhibition of the de novo aggregation of Sup35 in [pin-] cells lacking Hsp104 (Fig. 8A,B) and inhibition of de novo induction of [PSI+] in [PIN+] cells in the presence of GuHCl (Fig. 8C), which inhibits Hsp104's activity [79]. This is consistent with a previous report that overexpression of Hsp104 enhances prion appearance [121].
Although cross-seeding is generally thought to explain the cross-talk that enables amyloid aggregates to promote conversion of heterologous prion-like protein to amyloid, our data suggest that another mechanism is also involved. Indeed, we showed that in the absence of [PIN+], overexpression of the non-Q/N rich prion protein Mod5 enhances [PSI+] formation without direct physical interaction with Sup35 (Fig. 7B, S11B Fig.). Possibly, instead of cross-seeding which requires a physical interaction, excessive amounts of misfolded Mod5 proteins in [pin-] cells sequester chaperones away from newly forming Sup35 aggregates in the cell, and thus allow them to mature into a prion. Since Mod5-promoted [PSI+] induction is rare compared to [PIN+]-promoted induction, cross-seeding appears to be more efficient than the other mechanism in promoting de novo prion formation.
[PIN+] also appears to influence de novo aggregation of Mod5. Bimolecular complementation using BiFC-tagged Mod5 (S11C Fig.) showed that Mod5 prion protein interacts with itself, resulting in diffuse fluorescence only in [PIN+], but not in [pin-] cells. Curiously, an interaction between Sup35NM-VC and Mod5-VN is also seen as diffuse fluorescence, and again only in [PIN+] cells (Fig. 7B). Possibly, the Mod5 diffuse aggregates present in [PIN+] cells attract and interact with Sup35NM aggregates.
The data presented here aid our understanding of how prion formation occurs in yeast, and provide clues to the molecular mechanisms underlying many human aggregating neurodegenerative diseases, particularly these arising more frequently in people with preexisting neurodegenerative disease.
Methods
Strains and plasmids
Yeast plasmids and strains used in this study are listed in Tables 5 and 6, respectively. All [PIN+] cells used in the study were high [PIN+] [34]. GF657, GF658 and GF844 are, respectively, [PSI+][PIN+] HSP104, [psi-][pin-] HSP104 and [psi-][pin-] hsp104Δ versions of 74-D694 with endogenous SUP35 replaced with SUP35-GFP (kindly supplied as SY80, SY84, and SY97 by T. R. Serio, U. Arizona) [75]. L3107 and L2903 were obtained independently by curing GF657 of [PSI+] by overexpressing Pin4C [66]. GF852 and GF855, respectively, are [psi-][PIN+] and [psi-][pin-] versions of 74-D694 with RNQ1-CFP under its own promoter integrated into the genomic TRP1 (kindly supplied as 645 and 651 by L. Li, Northwestern U.). GF647 was constructed by replacing chromosomal RNQ1 with KanMX4 in 74D-694 [122]. All other yeast strains used in the studies of colocalization with Sup35 are from the BY4741 GFP library strain (Life Technologies, CA) harboring the gene of interest tagged endogenously with GFP [91]. To obtain [pin-] versions of these cells, they were grown on YPD plates with 5 mM GuHCl for three passes [50].
Tab. 5. Plasmids used in this study. 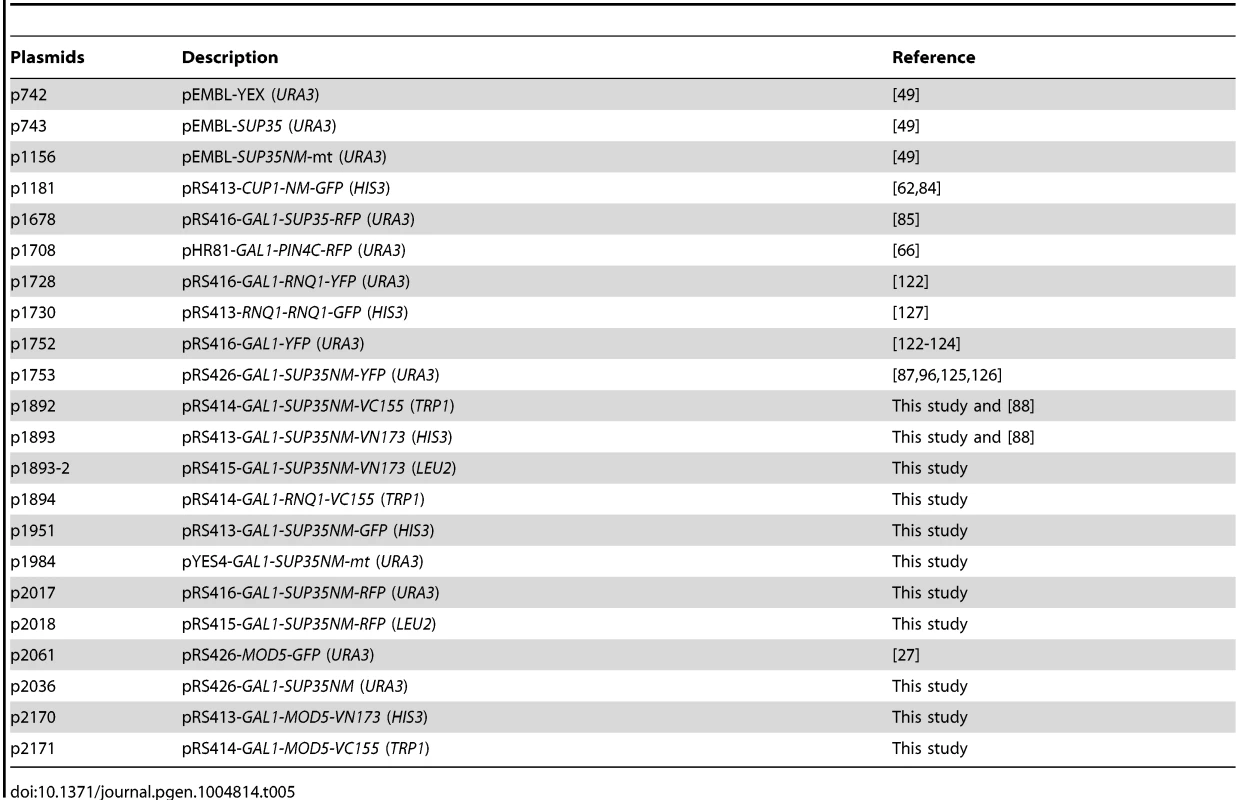
Tab. 6. Yeast strains used in this study. 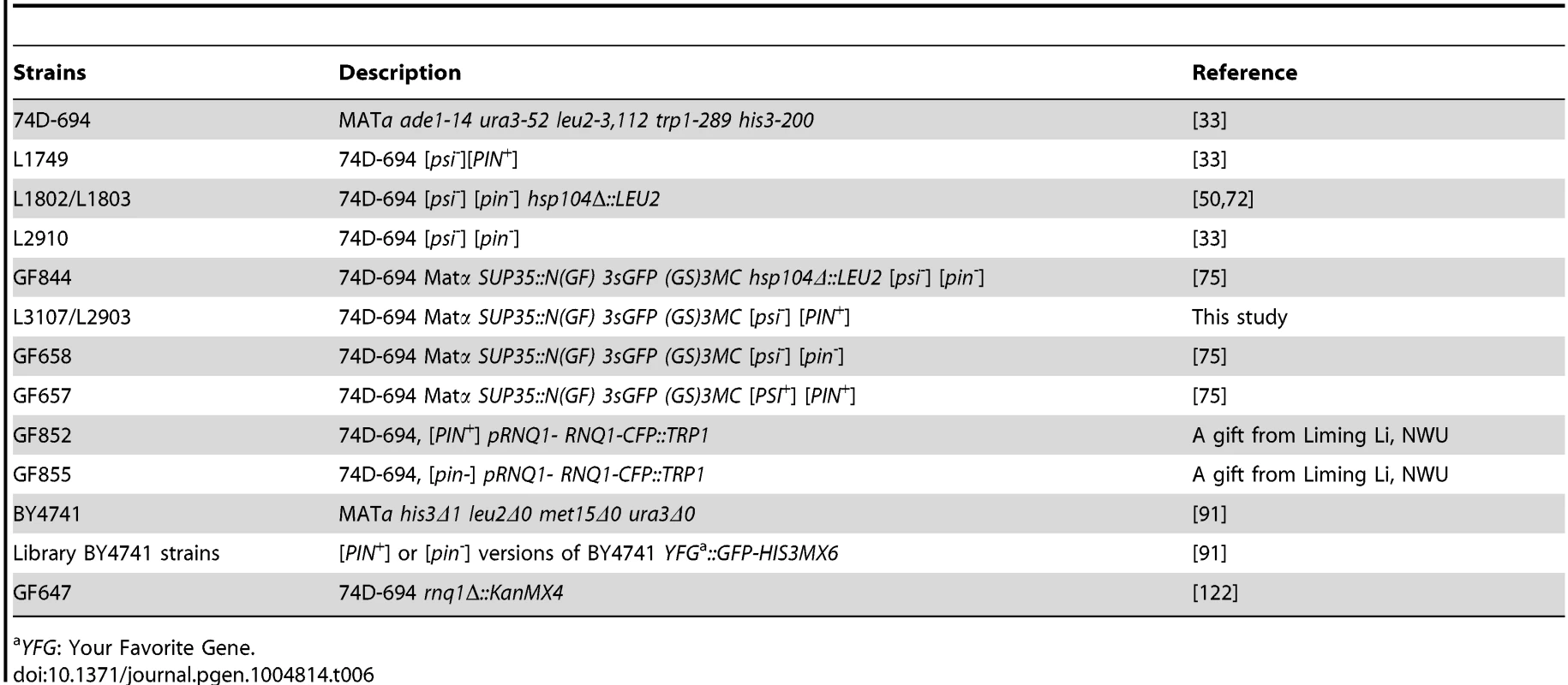
YFG: Your Favorite Gene. All overexpression plasmids in this study were driven by the GAL1 promoter unless otherwise stated. p1951 (Sup35NM-GFP) was constructed by inserting the SUP35NM BamHI-NotI fragment, and the GFP NotI-SacI fragment in-frame into the pRS413GAL1 plasmid backbone. p1893 (Sup35NM-VN) was constructed by replacing the GFP NotI-SacI fragment in p1951 with the VN173 NotI-SacI fragment PCR-amplified from pFA6a-HIS3MX6-pGAL1-VN173 [88]. p1893-2 was constructed by moving the GAL1-SUP35NM-VN XhoI-SacI fragment in p1893 to the pRS415 backbone. p2170 was constructed by replacing the SUP35NM BamHI-NotI fragment in p1893 with the MOD5 BamHI-NotI fragment PCR-amplified from the genome. p2171 was constructed by replacing the SUP35NM BamHI-NotI fragment in p1892 with the MOD5 BamHI-NotI fragment PCR-amplified from the genome. p1892 (Sup35NM-VC) was constructed by (1) replacing the GFP NotI-SacI fragment in p1951 with the VC155 NotI-SacI PCR-amplified from pFA6a-HIS3MX6-pGAL1-VC155 [88], and (2) moving this GAL1-SUP35NM-VC155 XhoI-SacI fragment into the XhoI-SacI sites of pRS414. p1894 was constructed by replacing the SUP35NM BamHI-NotI fragment in p1892 with the RNQ1 BamHI-NotI fragment PCR-amplified from the genome. p1984 was constructed by cloning the XmaI-Sal1 fragment of p1156 into p742. p2036 was constructed by replacing the YFP SpeI-XhoI fragment in p1752 with the NM(TAA) SpeI-XhoI fragment PCR-amplified from the genome [122]–[124]. p1753 was described previously [87], [96], [125], [126]. p2017 and p2018 vectors (Sup35NM-RFP) with URA3 or LEU2 markers, respectively were constructed in a two-step cloning: (1) The GFP NotI-SacI fragment in p1951 was replaced with the RFP NotI-SacI fragment amplified from p1708. (2) Then, this GAL1-NM-RFP XhoI-SacI fragment was cloned into pRS416 (URA3) and pRS415 (LEU2) backbones, respectively. All other plasmids listed in Table 5 were described previously [49], [62], [66], [85], [122], [127].
Cultivation procedures
Yeast strains were cultivated using standard media and growth conditions [128]. Rich media contained 2% dextrose (YPD). Synthetic complete media contained all amino acids except for those used for selection and 2% dextrose (SD, 2% Dex) or 2% galactose (2% Gal). Synthetic liquid media contained amino acids lacking the selective ones and 2% raffinose plus (SRGal) or minus 2% Gal (SRaf).
Induction of [PSI+] de novo
Yeast cells with Sup35 overexpression plasmids were grown in synthetic liquid selection media (SRaf) overnight. Unless otherwise stated, 2% Gal was added to the culture (OD∼0.5) to induce de novo Sup35 aggregation. In time course experiments where time exceeded 48 h, cultures were diluted back to OD∼0.5 in a fresh growth media to keep cells in exponential phase.
Staining of mature [PSI+] dots
After de novo [PSI+] was induced in [psi-][PIN+] cells, they were grown on synthetic dropout media with 2% dextrose (SD) for many generations to maintain [PSI+] (∼8 days). Then, they were grown in 0.05% Gal for 3-4 h to allow Sup35NM-YFP to decorate existing [PSI+] aggregates.
Color and plate assay for [PSI+] induction
[PSI+] induced de novo was scored in yeast as described previously [33], [49], [72], [129], [130]. [PSI+] but not [psi-] causes read-through of the nonsense mutation, ade1-14. The ade1-14 mutation causes the accumulation of a red by-product in the adenine biosynthesis pathway, so [psi-] ade1-14 cells are red. In [PSI+] cells, when nonsense mutations are suppressed, ade1-14 cells become pink or white in rich media (YPD). In addition, the read-through of the ade1-14 allele in [PSI+] allows them to grow in media lacking adenine (SD-Ade), while [psi-] cells cannot grow on this medium [49]. To confirm that Ade+ cells are of [PSI+] rather than suppressor mutants, they were mated with a tester strain ([psi-] SUP35-GFP, GF658) to look for multiple fluorescent dots formed by endogenous Sup35-GFP in only [PSI+] cells.
FM4-64 and Thioflavin T staining
To locate the vacuole, cells were stained with FM4-64 as described previously [90]. Yeast cells were stained with Thioflavin T according to a protocol adapted from ref. [131] with the addition of two extra washes in PMST [0.1M KPO4 (pH 7.5), 1 mM MgCl2, 1 M Sorbitol, 0.1% Tween 20].
Visualization of aggregates and colocalization studies
Aggregates formed in cells by fluorescently labeled proteins were examined with a Nikon Eclipse E600 fluorescent microscope (100X oil immersion) and/or an Olympus FV1000 confocal microscope (60X oil immersion, with 1.6 magnifier). Colocalization was visualized by the confocal microscope using the channels of interest and by moving the focal plane up and down. Z-stacks were analyzed to confirm colocalization of proteins with 8-12 layers, with 0.5-1 µm increments. In dual color RFP/GFP studies, the RFP channel was always examined first to prevent visualization of activated GFP in the RFP channel.
Detection of protein levels
Upon the induction of overexpression of proteins tagged with VN or VC with galactose, cells were lysed as described previously [132]. Equal amounts of total proteins in precleared lysates were analyzed by Western blot using previously described antibodies [27], [63], [66], [104].
Supporting Information
Zdroje
1. PrusinerSB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216 : 136–144.
2. PrusinerSB (1998) Prions. Proc Natl Acad Sci U S A 95 : 13363–13383.
3. FraserH, DickinsonAG (1973) Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol 83 : 29–40.
4. CollingeJ, SidleKC, MeadsJ, IronsideJ, HillAF (1996) Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383 : 685–690.
5. BessenRA, MarshRF (1994) Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 68 : 7859–7868.
6. GoedertM, WischikCM, CrowtherRA, WalkerJE, KlugA (1988) Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A 85 : 4051–4055.
7. SpillantiniMG, SchmidtML, LeeVM, TrojanowskiJQ, JakesR, et al. (1997) Alpha-synuclein in Lewy bodies. Nature 388 : 839–840.
8. MezeyE, DehejiaA, HartaG, PappMI, PolymeropoulosMH, et al. (1998) Alpha synuclein in neurodegenerative disorders: murderer or accomplice? Nat Med 4 : 755–757.
9. MacDonald MEAC, DuyaoMP, MyersRH, LinC, et al. (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 72 : 971–983.
10. Al-ChalabiA, LeighPN (2000) Recent advances in amyotrophic lateral sclerosis. Curr Opin Neurol 13 : 397–405.
11. KwiatkowskiTJJr, BoscoDA, LeclercAL, TamrazianE, VanderburgCR, et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323 : 1205–1208.
12. VanceC, RogeljB, HortobagyiT, De VosKJ, NishimuraAL, et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323 : 1208–1211.
13. NeumannM, SampathuDM, KwongLK, TruaxAC, MicsenyiMC, et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314 : 130–133.
14. SreedharanJ, BlairIP, TripathiVB, HuX, VanceC, et al. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319 : 1668–1672.
15. ConwitRA (2006) Preventing familial ALS: a clinical trial may be feasible but is an efficacy trial warranted? J Neurol Sci 251 : 1–2.
16. GotzJ, ChenF, van DorpeJ, NitschRM (2001) Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science 293 : 1491–1495.
17. OnoK, TakahashiR, IkedaT, YamadaM (2012) Cross-seeding effects of amyloid beta-protein and alpha-synuclein. J Neurochem 122 : 883–890.
18. GuoJL, CovellDJ, DanielsJP, IbaM, StieberA, et al. (2013) Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell 154 : 103–117.
19. WicknerRB (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 264 : 566–569.
20. WicknerRB, MasisonDC, EdskesHK (1995) [PSI] and [URE3] as yeast prions. Yeast 11 : 1671–1685.
21. SondheimerN, LindquistS (2000) Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5 : 163–172.
22. DerkatchIL, BradleyME, HongJY, LiebmanSW (2001) Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106 : 171–182.
23. DuZ, ParkKW, YuH, FanQ, LiL (2008) Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet 40 : 460–465.
24. AlbertiS, HalfmannR, KingO, KapilaA, LindquistS (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137 : 146–158.
25. PatelBK, Gavin-SmythJ, LiebmanSW (2009) The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol 11 : 344–349.
26. CrowET, LiL (2011) Newly identified prions in budding yeast, and their possible functions. Semin Cell Dev Biol 22 : 452–459.
27. SuzukiG, ShimazuN, TanakaM (2012) A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336 : 355–359.
28. GloverJR, KowalAS, SchirmerEC, PatinoMM, LiuJJ, et al. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89 : 811–819.
29. DePaceAH, SantosoA, HillnerP, WeissmanJS (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93 : 1241–1252.
30. Wickner RB LS, Saupe SJ (2004) Prions of Yeast and Filamentous Fungi: [URE3], [PSI+], [PIN+], and [Het-s]. In: SB P, editor. Prion Biology and Diseases. 2 ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. pp.305–377.
31. FuentealbaRA, UdanM, BellS, WegorzewskaI, ShaoJ, et al. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J Biol Chem 285 : 26304–26314.
32. CushmanM, JohnsonBS, KingOD, GitlerAD, ShorterJ (2010) Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci 123 : 1191–1201.
33. DerkatchIL, ChernoffYO, KushnirovVV, Inge-VechtomovSG, LiebmanSW (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144 : 1375–1386.
34. BradleyME, EdskesHK, HongJY, WicknerRB, LiebmanSW (2002) Interactions among prions and prion "strains" in yeast. Proc Natl Acad Sci U S A 99 Suppl 416392–16399.
35. SchlumpbergerM, PrusinerSB, HerskowitzI (2001) Induction of distinct [URE3] yeast prion strains. Mol Cell Biol 21 : 7035–7046.
36. KrishnanR, LindquistSL (2005) Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435 : 765–772.
37. StansfieldI, JonesKM, KushnirovVV, DagkesamanskayaAR, PoznyakovskiAI, et al. (1995) The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14 : 4365–4373.
38. HelsenCW, GloverJR (2012) Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J Biol Chem 287 : 542–556.
39. Ter-AvanesyanMD, KushnirovVV, DagkesamanskayaAR, DidichenkoSA, ChernoffYO, et al. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 7 : 683–692.
40. Ter-AvanesyanMD, DagkesamanskayaAR, KushnirovVV, SmirnovVN (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137 : 671–676.
41. LiuJJ, SondheimerN, LindquistSL (2002) Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proc Natl Acad Sci U S A 99 Suppl 416446–16453.
42. PaushkinSV, KushnirovVV, SmirnovVN, Ter-AvanesyanMD (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15 : 3127–3134.
43. PatinoMM, LiuJJ, GloverJR, LindquistS (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273 : 622–626.
44. LundPM, CoxBS (1981) Reversion analysis of [psi-] mutations in Saccharomyces cerevisiae. Genet Res 37 : 173–182.
45. LancasterAK, BardillJP, TrueHL, MaselJ (2010) The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 184 : 393–400.
46. ChernoffYO, NewnamGP, KumarJ, AllenK, ZinkAD (1999) Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol 19 : 8103–8112.
47. AllenKD, ChernovaTA, TennantEP, WilkinsonKD, ChernoffYO (2007) Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 282 : 3004–3013.
48. ChernoffYO, DerkachIL, Inge-VechtomovSG (1993) Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet 24 : 268–270.
49. DerkatchIL, BradleyME, MasseSV, ZadorskySP, PolozkovGV, et al. (2000) Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J 19 : 1942–1952.
50. DerkatchIL, BradleyME, ZhouP, ChernoffYO, LiebmanSW (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147 : 507–519.
51. OsherovichLZ, WeissmanJS (2001) Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 106 : 183–194.
52. DerkatchIL, LiebmanSW (2007) Prion-prion interactions. Prion 1 : 161–169.
53. DerkatchIL, UptainSM, OuteiroTF, KrishnanR, LindquistSL, et al. (2004) Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 101 : 12934–12939.
54. VitrenkoYA, GrachevaEO, RichmondJE, LiebmanSW (2007) Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem 282 : 1779–1787.
55. KatoM, HanTW, XieS, ShiK, DuX, et al. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149 : 753–767.
56. WasmerC, ZimmerA, SabateR, SoragniA, SaupeSJ, et al. (2010) Structural similarity between the prion domain of HET-s and a homologue can explain amyloid cross-seeding in spite of limited sequence identity. J Mol Biol 402 : 311–325.
57. FurukawaY, KanekoK, MatsumotoG, KurosawaM, NukinaN (2009) Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J Neurosci 29 : 5153–5162.
58. O'NuallainB, WilliamsAD, WestermarkP, WetzelR (2004) Seeding specificity in amyloid growth induced by heterologous fibrils. J Biol Chem 279 : 17490–17499.
59. KrebsMR, Morozova-RocheLA, DanielK, RobinsonCV, DobsonCM (2004) Observation of sequence specificity in the seeding of protein amyloid fibrils. Protein Sci 13 : 1933–1938.
60. BudkaH, AguzziA, BrownP, BrucherJM, BugianiO, et al. (1996) [Consensus report: tissue handling in suspected Creutzfeldt-Jakob disease and other spongiform encephalopathies (prion diseases) in the human. European Union Biomed-1 Concerted Action]. Pathologe 17 : 171–175.
61. GuoJP, AraiT, MiklossyJ, McGeerPL (2006) Abeta and tau form soluble complexes that may promote self aggregation of both into the insoluble forms observed in Alzheimer's disease. Proc Natl Acad Sci U S A 103 : 1953–1958.
62. ChoeYJ, RyuY, KimHJ, SeokYJ (2009) Increased [PSI+] appearance by fusion of Rnq1 with the prion domain of Sup35 in Saccharomyces cerevisiae. Eukaryot Cell 8 : 968–976.
63. SharmaJ, LiebmanSW (2013) Exploring the basis of [PIN(+)] variant differences in [PSI(+)] induction. J Mol Biol 425 : 3046–3059.
64. HuangVJ, SteinKC, TrueHL (2013) Spontaneous variants of the [RNQ+] prion in yeast demonstrate the extensive conformational diversity possible with prion proteins. PLoS One 8: e79582.
65. OsherovichLZ, WeissmanJS (2002) The utility of prions. Dev Cell 2 : 143–151.
66. YangZ, HongJY, DerkatchIL, LiebmanSW (2013) Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS Genet 9: e1003236.
67. WinklerJ, TyedmersJ, BukauB, MogkA (2012) Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 198 : 387–404.
68. WinklerJ, TyedmersJ, BukauB, MogkA (2012) Chaperone networks in protein disaggregation and prion propagation. J Struct Biol 179 : 152–160.
69. RomanovaNV, ChernoffYO (2009) Hsp104 and prion propagation. Protein Pept Lett 16 : 598–605.
70. DerkatchIL, LiebmanSW (2013) The story of stolen chaperones: how overexpression of Q/N proteins cures yeast prions. Prion 7 : 294–300.
71. LiebmanSW, ChernoffYO (2012) Prions in yeast. Genetics 191 : 1041–1072.
72. ChernoffYO, LindquistSL, OnoB, Inge-VechtomovSG, LiebmanSW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268 : 880–884.
73. CoxB, NessF, TuiteM (2003) Analysis of the generation and segregation of propagons: entities that propagate the [PSI+] prion in yeast. Genetics 165 : 23–33.
74. KryndushkinDS, AlexandrovIM, Ter-AvanesyanMD, KushnirovVV (2003) Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278 : 49636–49643.
75. Satpute-KrishnanP, LangsethSX, SerioTR (2007) Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol 5: e24.
76. ShorterJ, LindquistS (2004) Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304 : 1793–1797.
77. TessarzP, MogkA, BukauB (2008) Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol 68 : 87–97.
78. CoxBS, TuiteMF, McLaughlinCS (1988) The psi factor of yeast: a problem in inheritance. Yeast 4 : 159–178.
79. EaglestoneSS, RuddockLW, CoxBS, TuiteMF (2000) Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 97 : 240–244.
80. HaslbergerT, BukauB, MogkA (2010) Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem Cell Biol 88 : 63–75.
81. RogozaT, GoginashviliA, RodionovaS, IvanovM, ViktorovskayaO, et al. (2010) Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci U S A 107 : 10573–10577.
82. VolkovKV, AksenovaAY, SoomMJ, OsipovKV, SvitinAV, et al. (2002) Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics 160 : 25–36.
83. MathurV, TanejaV, SunY, LiebmanSW (2010) Analyzing the birth and propagation of two distinct prions, [PSI+] and [Het-s](y), in yeast. Mol Biol Cell 21 : 1449–1461.
84. ZhouP, DerkatchIL, LiebmanSW (2001) The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol Microbiol 39 : 37–46.
85. GanusovaEE, OzolinsLN, BhagatS, NewnamGP, WegrzynRD, et al. (2006) Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol 26 : 617–629.
86. ManogaranAL, HongJY, HufanaJ, TyedmersJ, LindquistS, et al. (2011) Prion formation and polyglutamine aggregation are controlled by two classes of genes. PLoS Genet 7: e1001386.
87. TyedmersJ, TreuschS, DongJ, McCafferyJM, BevisB, et al. (2010) Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci U S A 107 : 8633–8638.
88. SungMK, HuhWK (2007) Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 24 : 767–775.
89. KerppolaTK (2006) Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc 1 : 1278–1286.
90. VidaTA, EmrSD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128 : 779–792.
91. HuhWK, FalvoJV, GerkeLC, CarrollAS, HowsonRW, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425 : 686–691.
92. WangY, MeriinAB, CostelloCE, ShermanMY (2007) Characterization of proteins associated with polyglutamine aggregates: a novel approach towards isolation of aggregates from protein conformation disorders. Prion 1 : 128–135.
93. SpechtS, MillerSB, MogkA, BukauB (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195 : 617–629.
94. CollingeJ (1996) New diagnostic tests for prion diseases. N Engl J Med 335 : 963–965.
95. ManolsonMF, WuBG, ProteauD, TaillonBE, RobertsBT, et al. (1994) Stv1 Gene Encodes Functional Homolog of 95-Kda Yeast Vacuolar H+-Atpase Subunit Vph1p. Journal of Biological Chemistry 269 : 14064–14074.
96. SaibilHR, SeybertA, HabermannA, WinklerJ, EltsovM, et al. (2012) Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc Natl Acad Sci U S A 109 : 14906–14911.
97. NewnamGP, WegrzynRD, LindquistSL, ChernoffYO (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol 19 : 1325–1333.
98. AllenKD, WegrzynRD, ChernovaTA, MullerS, NewnamGP, et al. (2005) Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169 : 1227–1242.
99. JungG, JonesG, WegrzynRD, MasisonDC (2000) A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics 156 : 559–570.
100. JonesGW, MasisonDC (2003) Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163 : 495–506.
101. JonesG, SongY, ChungS, MasisonDC (2004) Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol 24 : 3928–3937.
102. SongY, WuYX, JungG, TutarY, EisenbergE, et al. (2005) Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot Cell 4 : 289–297.
103. SchwimmerC, MasisonDC (2002) Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol 22 : 3590–3598.
104. BagriantsevSN, GrachevaEO, RichmondJE, LiebmanSW (2008) Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell 19 : 2433–2443.
105. KushnirovVV, KryndushkinDS, BogutaM, SmirnovVN, Ter-AvanesyanMD (2000) Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol 10 : 1443–1446.
106. KiktevDA, PattersonJC, MullerS, BariarB, PanT, et al. (2012) Regulation of chaperone effects on a yeast prion by cochaperone Sgt2. Mol Cell Biol 32 : 4960–4970.
107. DerkatchIL, BradleyME, LiebmanSW (1998) Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc Natl Acad Sci U S A 95 : 2400–2405.
108. Derkatch ILBM, MasseSVL, ZadorskySP, PolozkovGV, et al. (2000) Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? The EMBO Journal 19 : 1942–1952.
109. WindlO, DempsterM, EstibeiroJP, LatheR, de SilvaR, et al. (1996) Genetic basis of Creutzfeldt-Jakob disease in the United Kingdom: a systematic analysis of predisposing mutations and allelic variation in the PRNP gene. Hum Genet 98 : 259–264.
110. NemotoT, MatsusakaT, OtaM, TakagiT, CollingeDB, et al. (1996) Dimerization characteristics of the 94-kDa glucose-regulated protein. J Biochem 120 : 249–256.
111. BrownP, SalazarAM, GibbsCJJr, GajdusekDC (1982) Alzheimer's disease and transmissible virus dementia (Creutzfeldt-Jakob disease). Ann N Y Acad Sci 396 : 131–143.
112. GajdusekDC (1994) Spontaneous generation of infectious nucleating amyloids in the transmissible and nontransmissible cerebral amyloidoses. Mol Neurobiol 8 : 1–13.
113. PrusinerSB (1984) Some speculations about prions, amyloid, and Alzheimer's disease. N Engl J Med 310 : 661–663.
114. LangerF, EiseleYS, FritschiSK, StaufenbielM, WalkerLC, et al. (2011) Soluble Abeta seeds are potent inducers of cerebral beta-amyloid deposition. J Neurosci 31 : 14488–14495.
115. Meyer-LuehmannM, CoomaraswamyJ, BolmontT, KaeserS, SchaeferC, et al. (2006) Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science 313 : 1781–1784.
116. Escusa-ToretS, VonkWI, FrydmanJ (2013) Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15 : 1231–1243.
117. KaganovichD, KopitoR, FrydmanJ (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454 : 1088–1095.
118. SerioTR, CashikarAG, KowalAS, SawickiGJ, MoslehiJJ, et al. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289 : 1317–1321.
119. SalnikovaAB, KryndushkinDS, SmirnovVN, KushnirovVV, Ter-AvanesyanMD (2005) Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem 280 : 8808–8812.
120. OhtaS, Kawai-NomaS, KitamuraA, PackCG, KinjoM, et al. (2013) The interaction of Hsp104 with yeast prion Sup35 as analyzed by fluorescence cross-correlation spectroscopy. Biochem Biophys Res Commun 442 : 28–32.
121. KryndushkinDS, EngelA, EdskesH, WicknerRB (2011) Molecular chaperone Hsp104 can promote yeast prion generation. Genetics 188 : 339–348.
122. DouglasPM, TreuschS, RenHY, HalfmannR, DuennwaldML, et al. (2008) Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci U S A 105 : 7206–7211.
123. MumbergD, MullerR, FunkM (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22 : 5767–5768.
124. RonickeV, GraulichW, MumbergD, MullerR, FunkM (1997) Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol 283 : 313–322.
125. TyedmersJ, MadariagaML, LindquistS (2008) Prion switching in response to environmental stress. PLoS Biol 6: e294.
126. BeckerJ, WalterW, YanW, CraigEA (1996) Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol 16 : 4378–4386.
127. AronR, HigurashiT, SahiC, CraigEA (2007) J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J 26 : 3794–3803.
128. Sherman F FG, Hicks JB (1986) Methods in Yeast Genetics; Sherman F FG, Hicks JB, editor. Plainview, New York: Cold Spring Harbor Press.
129. LiebmanSW, DerkatchIL (1999) The yeast [PSI+] prion: making sense of nonsense. J Biol Chem 274 : 1181–1184.
130. CoxBS (1965) Psi, A cytoplasmic suppressor of super-suppressor in yeast. Heredity 20 : 505–521.
131. JohnsonBS, McCafferyJM, LindquistS, GitlerAD (2008) A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A 105 : 6439–6444.
132. MathurV, HongJY, LiebmanSW (2009) Ssa1 overexpression and [PIN(+)] variants cure [PSI(+)] by dilution of aggregates. J Mol Biol 390 : 155–167.
133. ChernovaTA, RomanyukAV, KarpovaTS, ShanksJR, AliM, et al. (2011) Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell 43 : 242–252.
134. Yang Z, Stone DE, Liebman SW (2014) Prion promoted phosphorylation of heterologous amyloid is coupled with ubiquitin-proteasome system inhibition and toxicity. Mol Microbiol.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



