-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
Human metabolic individuality is under strict control of genetic and environmental factors. In our study, we aimed to find the genetic determinants of circulating molecules in sera of large set of individuals representing the general population. First, we performed a hypothesis-free genome wide screen in this population to identify genetic regions of interest. Our study confirmed four known gene metabolite connections, but also pointed to four novel ones. Genome-wide screens enriched for common intergenic variants may miss causal genetic variations directly changing the protein sequence. To investigate this further, we zoomed into regions of interest and tested whether the association signals obtained in the first stage were direct, or whether they represent causal variations, which were not captured in the initial panel. These subsequent tests showed that protein coding and regulatory variations are involved in metabolite levels. For two genomic regions we also found that genes harbour more than one causal variant influencing metabolite levels independent of each other. We also observed strong connection between markers of cardio-metabolic health and metabolites. Taken together, our novel loci are of interest for further research to investigate the causal relation to for instance type 2 diabetes and cardiovascular disease.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004835
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004835Summary
Human metabolic individuality is under strict control of genetic and environmental factors. In our study, we aimed to find the genetic determinants of circulating molecules in sera of large set of individuals representing the general population. First, we performed a hypothesis-free genome wide screen in this population to identify genetic regions of interest. Our study confirmed four known gene metabolite connections, but also pointed to four novel ones. Genome-wide screens enriched for common intergenic variants may miss causal genetic variations directly changing the protein sequence. To investigate this further, we zoomed into regions of interest and tested whether the association signals obtained in the first stage were direct, or whether they represent causal variations, which were not captured in the initial panel. These subsequent tests showed that protein coding and regulatory variations are involved in metabolite levels. For two genomic regions we also found that genes harbour more than one causal variant influencing metabolite levels independent of each other. We also observed strong connection between markers of cardio-metabolic health and metabolites. Taken together, our novel loci are of interest for further research to investigate the causal relation to for instance type 2 diabetes and cardiovascular disease.
Introduction
Intermediary metabolites in bodily fluids seem a direct reflection of our genetic constituency in interaction with the environment, which includes eating habits, life style and other external factors. Thus, the use of metabolomic phenotypes in genetic epidemiological studies may provide specific insight in pathways underlying complex metabolic diseases, such as type 2 diabetes mellitus (T2D), stroke or cardiovascular disease (CVD) but also other complex diseases such as rheumatoid arthritis, migraine and depression [1]–[3]. The sample sizes in the first genome-wide association studies (GWAS) of metabolite quantitative traits were in general relatively small compared to GWAS on traditional phenotypes, yet revealed strong signals for association of common variants with specific metabolites. Single-proton Nuclear Magnetic Resonance (1H-NMR) spectroscopy is a metabolomics technique that requires relatively little sample preparation, yet has the capacity to reproducibly quantify dozens to more than 100 metabolite signals per measurement. Several studies have reported genetic loci that influence the metabolites quantified by 1H-NMR in plasma and urine [4]-[7]. Here, we present the results of 42 plasma metabolites quantified by 1H-NMR spectroscopy in 2,482 individuals of the family-based Erasmus Rucphen Family (ERF) study, a Dutch genetic isolate. We estimated the heritability and the effect of shared environment (household effect) for these metabolites. The GWA was followed by high-resolution analysis of coding variants in the candidate genes that were identified by physical proximity and pathway analysis. To provide further insight into the pathogenesis of cardio-metabolic diseases, we also investigated the association between the NMR metabolites and the classical risk factors for CVD and T2D.
Results
Heritability estimates and GWAS results
The study was conducted in the ERF population (see S1 Table) using fasting serum samples. After quality filtering, we resolved 42 metabolites, for which the identity was confirmed by the typical chemical shifts of the related peaks, their high correlation with other peaks and spiking of pure compounds in serum (S2 Table). Heritability estimates of the metabolites were moderate to high ranging from 10% to 52% whereas estimates for the shared environmental effect ranged from 0% to 8% (Fig. 1). The highest heritability is observed for citrate (52%), followed by phenylalanine (51%), ornithine (47%) and methanol (45%) whereas the lowest heritability estimate was 10% for 3-hydroxybutyrate. We performed genome-wide association (GWA) analysis for all metabolite SNP pairs, including 2.5 M SNPs from the HapMap2 reference panel, see S1 Fig. for the Q-Q plots of the 42 metabolites. In total, we found eight unique genomic loci that associated with NMR metabolites below the genome-wide significant P-value threshold (P-value <5.0×10−8) as shown in the Manhattan plot (Fig. 2). Regional plots of the 8 loci are shown in S2 Fig.. Four of these loci were also significant after correction for the number of metabolites analyzed (P-value <1.10×10−9) and three of these were previously shown to associate with the same metabolites: rs715 located in the 3′UTR of the carbamoyl-phosphate synthase 1 (CPS1) gene associated with glycine level (P-value = 1.27×10−32) [8], rs2540641 35 Kb distant from proline dehydrogenase (oxidase) 1 (PRODH) gene (P-value = 1.11×10−19) associated with proline levels [9] and rs1171614 in the 5’UTR of SLC16A9 (solute carrier family 16, member 9) associated with carnitine level (P-value = 4.81×10−14) [9]–[11]. The association between intronic SNP rs248386 within DMGDH (dimethyl-glycine dehydrogenase) and dimethyl-glycine level is a novel finding (P-value = 1.65×10−19). This locus has also been associated with betaine, which is a closely related metabolite [8].
Fig. 1. Heritability and sibship effects on the NMR metabolites. 
Figure shows the magnitude of heritability (H2) and sibship (household) effect estimates for each metabolic trait included in the ERF population. Fig. 2. GWAS results of the NMR metabolites. 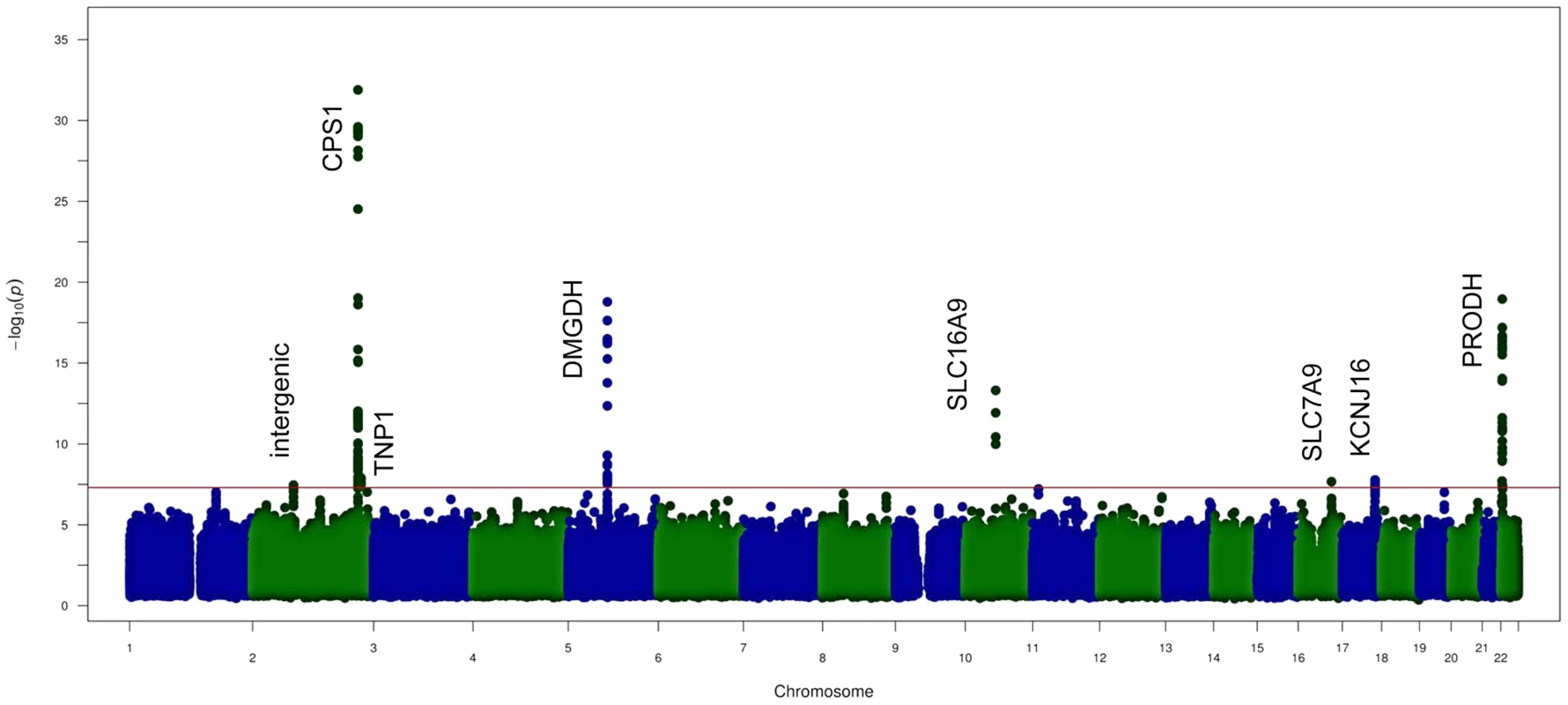
Figure shows the aggregated Manhattan plot for the 42 metabolites studied. Red line shows the suggestive genome-wide significance level with a P-value of 5×10−8.Loci harbouring DMGDH, SLC16A9, PRODH and CPS1 are reported as metabolome wide significance. Four other suggestively significant loci were uncovered by our analyses (5.0×10−8>P-value>1.10×10−9). One of these has previously been identified in urine: the association between rs8056893 within the SLC7A9 (solute carrier family 7 member 9) and lysine (P-value = 1.26×10−8) [12]. Three novel associations were found (1) rs1922005 located inside the TNP1 (transition protein 1) gene and pyruvate level (P-value = 1.26×10−8), (2) rs9896573 located nearby KCNJ16 (potassium inwardly-rectifying channel, subfamily J, member 16) and 3-hydroxybutyrate level (P-value = 1.65×10−8) (3) rs11687765 located in a non-protein coding region on chromosome 2 and valine level, (P-value = 3.49×10−8). For the 8 top loci, we also investigated the mode of inheritance. The model supervised analysis in those regions of interest shows that the recessive genetic model applies successfully for six of the effect alleles: rs715 (CPS1) on glycine, rs1922005 (TNP1) on pyruvate rs248386 (DMGDH) on dimethyl-glycine, rs1171614 (SLC16A9) on carnitine, rs8056893 (SLC7A9) on lysine and rs2540641 (PRODH) on proline. For rs11687765 (intergenic on chromosome 2) affecting valine, the mode of inheritance seems to be dominant for the effect allele. For rs9896573 (KCNJ16) affecting 3-hydroxybutyrate, the over-dominant model resulted in the strongest association among the models tested.
Fine mapping within the candidate genes
In the same study population exome sequences of 921 individuals from the ERF pedigree were analyzed for potentially causal SNPs in biologically plausible genes which were extracted using an automated workflow, within the top eight GWAS loci in Table 1. The outputs of the automated workflow are given in S1 Text. In addition to coding region variation, a number of intronic variants that were captured around the intron-exon connections, as well as nearby 5’UTR and 3′UTR variants were captured by sequencing and those were also included in analysis. This approach revealed in total seven independent SNPs with potentially causal effects located inside CPS1, KCNJ2 (potassium inwardly-rectifying channel, subfamily J, member 2), PRODH and SLC25A1 (solute carrier family 25 member 1) (Table 2). More precisely, for glycine we found evidence for two independent effects within the CPS1 gene. First, the missense mutation Thr1412Asn (rs1047891) within CPS1 is the most likely causal variant tagged by the GWAS SNP rs715, due to the high LD (R2 = 0.92) and large drop in P-values after conditioning the SNP-metabolite associations for each other. Second, we found three intronic variants in strong LD with each other (R2>0.89) in CPS1 that independently associated with glycine (lowest P-value = 2.55×10−5 for rs182548513, Table 2) when conditioned on the leading GWAS SNP. For 3-hydroxybutyrate, we found that rs173135 located 3′UTR of KCNJ2 gene is most likely the causal SNP (P-value = 1.01×10−07) influencing the circulating level of this metabolite. Rs173135 is in strong LD with the leading GWAS SNP (R2 = 0.72) showed a large drop in P-value, yet remained significant in the conditional analysis (P-value = 0.002). For proline, in total, we observed four independent effects within the PRODH locus including one missense mutation Thr116Asn (rs5747933, P-value = 1.82×10−9), two intronic SNPS (rs1076466, P-value = 6.34×10−4 and rs3213491, P-value = 7.48×10−4) and one (semi-)independent SNP rs13058335 (R2 = 0.66 with the leading GWAS SNP), explaining the GWAS finding with a conditional P-value = 1.20×10−5. We also found significant coding variations associated with dimethyl-glycine, carnitine, pyruvate and lysine however all those signals vanished after adjustment by the leading GWAS SNP, indicating that these associations so far are best explained by the leading GWAS hits in these regions (S3 Table).
Tab. 1. Unique loci that influence the NMR-metabolome. 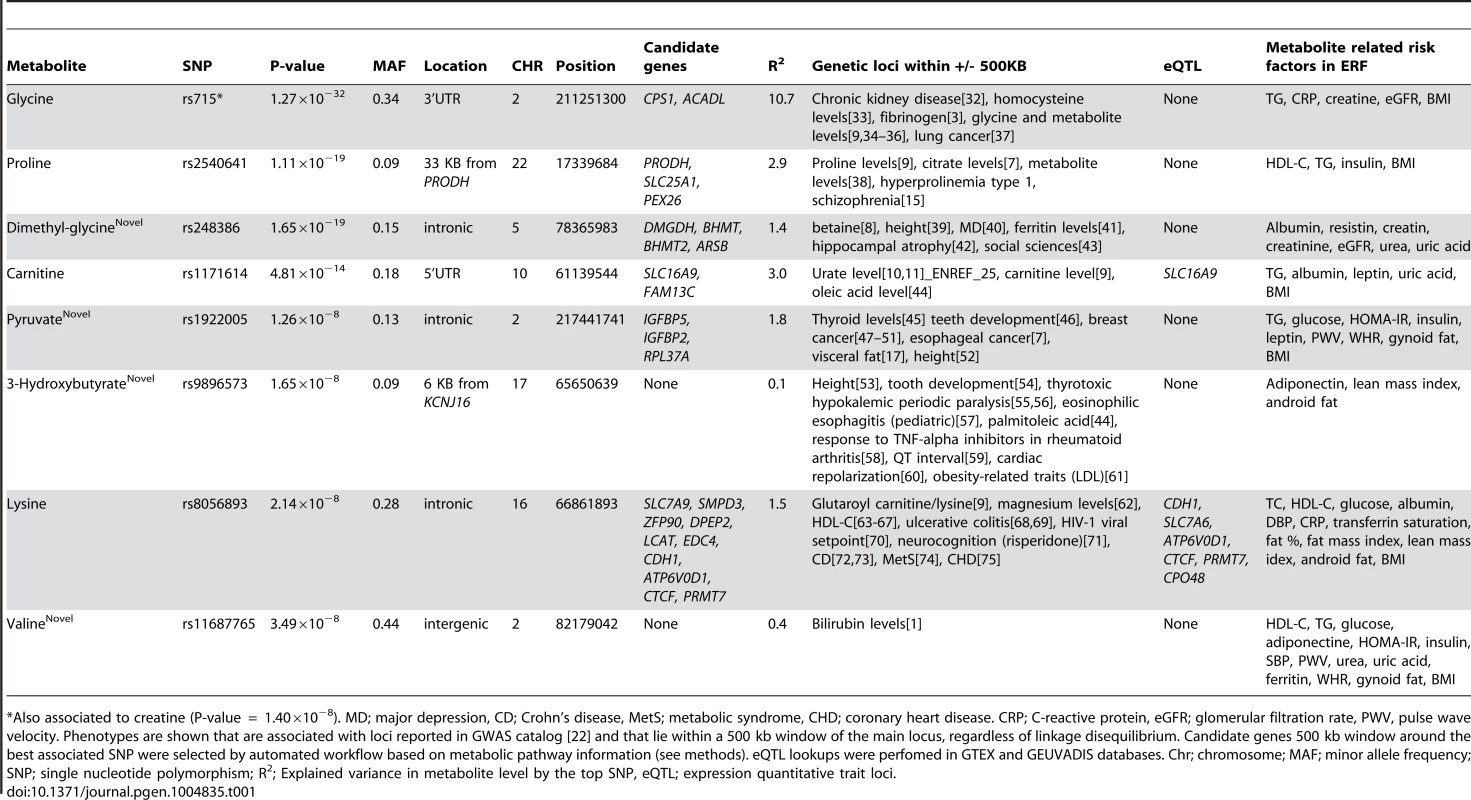
*Also associated to creatine (P-value = 1.40×10−8). MD; major depression, CD; Crohn's disease, MetS; metabolic syndrome, CHD; coronary heart disease. CRP; C-reactive protein, eGFR; glomerular filtration rate, PWV, pulse wave velocity. Phenotypes are shown that are associated with loci reported in GWAS catalog [22] and that lie within a 500 kb window of the main locus, regardless of linkage disequilibrium. Candidate genes 500 kb window around the best associated SNP were selected by automated workflow based on metabolic pathway information (see methods). eQTL lookups were perfomed in GTEX and GEUVADIS databases. Chr; chromosome; MAF; minor allele frequency; SNP; single nucleotide polymorphism; R2; Explained variance in metabolite level by the top SNP, eQTL; expression quantitative trait loci. Tab. 2. Sequence variants within the coding regions of candidate genes that influence the metabolomic levels independent of the GWAS hits. 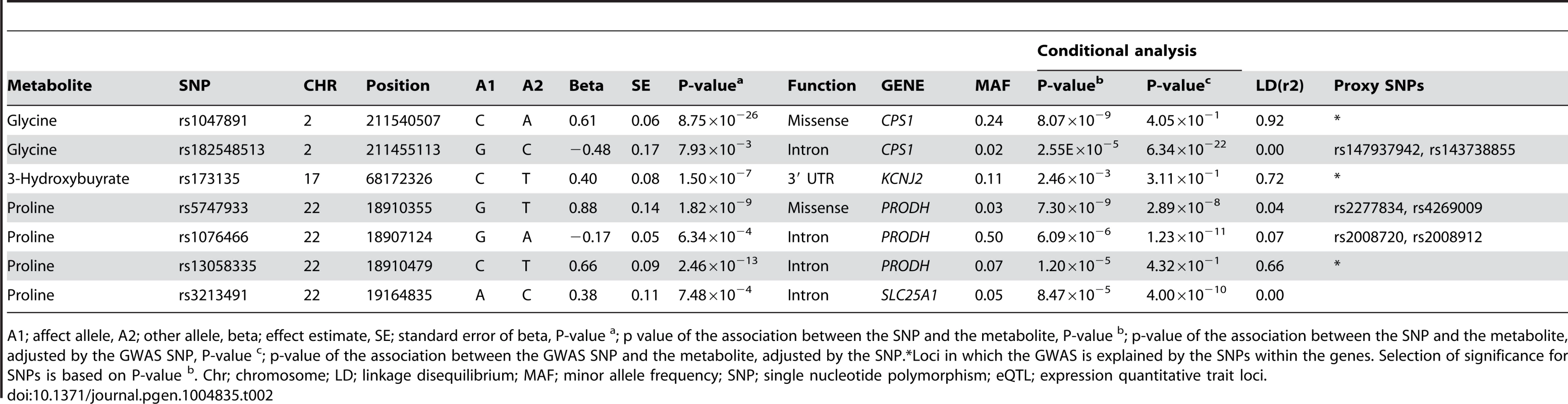
A1; affect allele, A2; other allele, beta; effect estimate, SE; standard error of beta, P-value a; p value of the association between the SNP and the metabolite, P-value b; p-value of the association between the SNP and the metabolite, adjusted by the GWAS SNP, P-value c; p-value of the association between the GWAS SNP and the metabolite, adjusted by the SNP.*Loci in which the GWAS is explained by the SNPs within the genes. Selection of significance for SNPs is based on P-value b. Chr; chromosome; LD; linkage disequilibrium; MAF; minor allele frequency; SNP; single nucleotide polymorphism; eQTL; expression quantitative trait loci. eQTL and functional effects
We used the GTEX and GEUVADIS [13] databases to check if the significantly associated SNPs affect cis gene expression. We obtained evidence that the leading GWAS SNP for carnitine (rs1171614) influenced the expression of SLC16A9 in lymphoblasts (P-value = 8.91×10−6) and rs8056893 (associated with lysine) influenced the expression of ZPF90 in lymphoblasts (P-value = 4.01×10−6) and SLC7A9 in thyroid cells (P-value = 0.00008). Rs248386 (associated with dimethyl-glycine) associated with the expression of BHMT (betaine—homocysteine S-methyltransferase) in the tibial nerve (P-value = 0.000066). One of the missense variants; Thr1412Asn (rs1047891) in CPS1 predicted to be “tolerated” by SIFT and “benign” by Polyphen functional predictions. The other missense variant Thr116Asn (rs5747933) on PRODH predicted to be “tolerated” by SIFT and “possibly damaging” by Polyphen.
Correlation with classical risk factors
Within the ERF population, we found that BMI correlated positively with carnitine (r = 0.136, P-value = 4.40×10−11), proline (r = 0.123, P-value = 2.80×10−9), pyruvate (r = 0.240, P-value = 5.40×10−32), lysine (r = 0.132, P-value = 1.45×10−10), and valine (r = 0.383, P-value = 2.05×10−82) (S4-A Table), whereas BMI correlated negatively with glycine (r = −0.178, P-value = 4.19×10−18). After additional adjustment for BMI, we observed that pyruvate, lysine and valine correlated positively with risk factors of T2DM, whereas glycine correlated negatively with triglycerides and C-reactive protein (CRP) (S4-B Table). Dimethyl-glycine particularly correlated with measures of kidney function; uric acid (r = 0.21, P-value = 2.42×10−9), glomerular filtration rate (eGFR) (r = −0.14, P-value = 2.53×10−10), urea (r = 0.18, P-value = 1.20×10−7), and creatinine (r = 0.22, P-value = 1.35×10−22).
We also explored possible relationships between the eight mQTL and the classical risk factors. Among the metabolites which associate with BMI, none of the mQTLs were associated with BMI itself in the ERF population. In addition, the association of the mQTLs with the metabolites glycine, carnitine, proline, pyruvate, lysine and valine did not change after adjustment for BMI (S5 Table). Interestingly, only for rs11687765 (valine-QTL) association with risk factors reached nominally significant P-values: specifically glucose (P-value = 0.013), HOMA, insulin resistance (P-value = 0.049) and gynoid fat mass (P-value = 0.003). Association of rs11687765 with HOMA-insulin resistance dropped when adjusted by the valine level itself (P-value = 0.122).
Discussion
In this study, we report on the heritability, GWAS, candidate genes and fine genetic mapping of 42 metabolites identified and quantified using 1H-NMR spectroscopy in the Erasmus Rucphen Family (ERF) study. In 2009, the first GWAS of metabolites identified by 1H-NMR spectroscopy measured in human plasma was reported by Chasman et al. [4]. This study focused primarily on lipoprotein particle size and content, and did not measure other metabolites such as organic acids and amino acids, yet reported 43 significant metabolite mQTL. This was followed by three reports on blood and urine samples [5], [6] the largest of which by Kettunnen et al. involving both small metabolites and lipoprotein particle sizes, reporting 31 novel mQTL [7]. Recently, Rueedi et al. reported one novel locus using an untargeted approach [12]. Here, we used 1H-NMR J-Resolved 2D spectrometry followed by spiking experiments, yielding a reliable certain metabolite identification. Traditional CVD traits in ERF and other cohorts in general show a heritability ranging from 20% to 30% [14]. In the present study, we observed a similar distribution of heritability for NMR detected metabolites, ranging from 10% to 52%. These heritability estimates seem somewhat lower than those found in the NMR GWAS by Kettunen et al. [7]. However, in that report a significant proportion of the reported NMR traits and heritability estimates concern lipoprotein particle characteristics. Since, in general, heritability for lipoproteins is high [7], ranging from 30% to 50%, this could explain the apparent discrepancy with our reported heritability data.
Using verified metabolites, we replicated three known loci and uncovered a novel association for dimethyl-glycine in the vicinity of the biologically plausible genes DMGDH and BHMT. This was expected since our study had 62 to 100% power to detect genetic variants with 0.2 to 0.5 effect size with metabolome-wide significant P-value (1.1×10−9) for a bi-allelic marker with 0.3 MAF (for instance rs715 in CPS1) based on the assumption of complete LD with the causal genetic variant. For more rare variants with larger effect size such as rs248386 in DMDGH with 0.15 MAF and 0.4 effect size the power on metabolome wide significance was 100%. Furthermore, we report suggestive common genetic variants; first in an intergenic region on chromosome 2 for valine, second in TNP1 for pyruvate and lastly in KCNJ16 for 3-hydroxybutyrate levels. Analysis of the coding sequence in the candidate genes uncovered potentially causal signals within CPS1, KCNJ2 and PRODH that explain the GWAS hits, as well as additional independent signals located in CPS1 and PRODH indicating allelic heterogeneity within these genes. Among the eight mQTL, rs715 in CPS1 explained the highest (10%)of the total phenotypic variance in circulating glycine levels (Table 1).This was higher than the total explained variance in for glycine level by age and sex. (S6 Table).
The CPS1 locus has been previously found associated with kidney disease, homocysteine, and several metabolite levels including glycine. CPS1 mutations are known to cause carbamoylphosphate synthetase I deficiency, an autosomal recessive inborn error of metabolism of the urea cycle which causes hyperammonemia. The disease may also have a delayed onset in adulthood and is associated with chronic kidney disease. Gene-network predictions for this gene included functions such as triglycerides (TG) and lipoprotein homeostasis. In our study, we also found association of the same SNP with creatine level and also observed a significant correlation between creatine and glycine (r = 0.08, P-value = 1.46×10−4), glomerular filtration rate (r = −0.09, P-value = 7.07×10−5) and TG (r = −0.08, P-value = 1.15×10−4). We identified Thr1412Asn in CPS1 as a potential variant that may alter the protein function. The second independent signal within CPS1 was located intronic (rs182548513). The neighbouring SNP, rs147937942, (Table 2) in LD with rs182548513 is located on 5′UTR of a CPS1 transcript variant (CPS1-001), and identified as transcription factor binding site according to the ENCODE database however, so far we did not find any evidence that the SNP affects expression which may be tissue specific.
The second locus, PRODH, a gene highly expressed in cerebral cortex, cerebrum and other brain tissues is known to be involved in proline metabolism, but also in central nervous system myelination. The locus was previously shown to associate with schizophrenia [15] and autism [16]. We show in total 4 independent SNPs that associate with circulating proline level; including (1) the GWAS hit, (2) one very common SNP (tagged by rs2008720), (3) a possibly damaging missense mutation with low frequency (MAF = 0.03, Thr167Asn) and (4) another with MAF = 0.05 (rs3213491). It is important to mention that rs2008720 maps to first exon of PRODH (PRODH-001 isoform) resulting the amino-acid change Pro19Gln, whereas it also maps to the promoter regulatory region of another PRODH isoform (PRODH-004). Neither for these variants did we find experimental evidence from eQTL database.
DMGDH codes for the enzyme dimethyl-glycine dehydrogenase which is involved in catabolism of choline, catalyzing the oxidative demethylation of dimethyl-glycine to form sarcosine. The gene is highly expressed in liver, followed by kidney. Mutations in this gene cause an inborn error of metabolism characterized by unusual fish-like body odour. Functional predictions for this gene by KEGG database include several functions in amino-acid metabolism and bile acid synthesis. Conditional analysis in this region showed that the GWAS hit located intronic in DMGDH (rs248386) is most likely the causal variant. Interestingly we found this SNP associated with the expression of the neighbouring gene, BHMT that is also involved in dimethyl-glycine and betaine metabolism.
SLC16A9 is involved in drug transport, bile salt and organic anion transport and has been previously shown to be associated with carnitine, uric acid levels. In the ERF population carnitine and uric acid are highly correlated (r = 0.25, P -value = 3.93×10−13). For this locus, we did not find any potentially causal coding variants. However, the GWAS hit (rs1171614) located 5′UTR of SLC16A9 influences the expression of SLC16A9 in both GTEX and GEUVADIS databases, indicating that the effect on carnitine level is possibly through expression, rather than the change in protein function.
The metabolite pyruvate is the product of anaerobic glycolysis. Pyruvate levels correlate with gynoid adipose tissue mass, BMI, waist hip ratio, TG, glucose, HOMA-IR and leptin in the ERF population (S4B Table). Genes in the TNP1 locus, particularly IGFBP5 have been previously associated with visceral adipose tissue mass in men [17]. Within these genes, we did not find any causal variants, neither for the GWAS hits were we unable to uncover downstream eQTL. For 3-hydroxybutyrate, rs173135 located in the 3′UTR of KCNJ2 is the most likely causal variant tagged by the GWAS hit for 3-hydroxybutyrate. The gene is predominantly expressed in heart muscles but also in brain and the locus has been previously associated with QT interval and cardiac repolarization. Currently, it is not known how this gene may be affecting 3-hydroxybutyrate levels. The association between SLC7A9 and valine has previously been shown [9]. Within the candidate genes in this locus, we were not able to detect any causal variants. However, the leading GWAS SNP is associated with expression of SLC7A9 and ZPF90. Finally, valine has been suggestively associated with an intergenic region with no eQTL association. This region has been previously shown to associate with bilirubin level, which is a determinant of hepatic health. The strong correlation between valine and pyruvate levels and the risk factors of T2DM suggests these loci are candidates for T2DM research. Using the data from the ERF population, for 7 out of 8 loci, we found no evidence that the mQTL discovered directly or indirectly influenced the risk factors for common diseases. Our data indicate that the association between these mQTLs and the metabolites were independent of disease risk factors. For BMI, our results support an additive effect of BMI and mQTL, both influencing the metabolite levels. We did find evidence for an association between HOMA insulin resistance, valine and rs11687765. However, this finding asks for replication in independent larger sized studies.
Altogether, our study provides strong evidence for associations of metabolic traits with a range of novel and previously detected genetic loci. These loci are potentially of biomedical and pharmaceutical interest, and may provide insight into human metabolic and disease pathways.
Methods
Study cohort
The Erasmus Rucphen Family (ERF) study is a cross-sectional cohort including 3000 living descendants of 22 couples who had at least 6 children baptized in the community church around 1850-1900. The participants are not selected on any disease or other outcome (S1 Table). Details about the genealogy of the population have been described elsewhere[18]. The study protocol was approved by the medical ethics board of the Erasmus MC Rotterdam, the Netherlands.
1H-NMR JRES measurements
2,640 sera of ERF participants were submitted for 1H-NMR experiments. All NMR experiments were acquired on a 600 MHz Bruker Avance II spectrometer (Bruker BioSpin, Karlsruhe, Germany). For this study the 2D J-resolved (JRES) and CPMG (Carr-Purcell-Meiboom-Gill) methods were used. Data processing was performed in Topspin and Matlab (R2009a, The Mathworks Inc., Natick, MA, USA). After eliminating low-quality spectra after a QC procedure, metabolite intensities were obtained from the serum CPMG spectra by applying a linear model. The model was constructed by identifying well-resolved peaks in the 2D JRES spectrum, and relating the intensity of the peak representing the metabolite with the intensity profile of the much more convoluted CPMG spectrum. This way, the higher resolution of the JRES 2D spectrum is combined with the better signal-to-noise of the CPMG spectrum. After quality control peaks in the JRES projection were automatically deconvoluted by fitting the spectra with mixed Gauss-Lorentz line-shapes using the Simplex method yielding 256 deconvoluted peaks, 42 metabolites could be reliably assigned using a combination of chemical shift interpretation, cross-correlation between peaks and spiking of pure compounds in a mixed serum sample of them were annotated to unique metabolites (S2 Table). Further selection procedure and QC and the list of unique metabolites studied are given in the supplement.
Heritability analysis
Heritability estimations for all metabolite concentrations were obtained using SOLAR version 6.6.2 software using a polygenic model and sex and age as covariates.
Genome-wide association analyses
Data points below or above 4 standard deviations from the mean were removed and non-missing data points of all variables were rank transformed using the “rank” function in R, this function takes the missing values into account. No samples were detected as outliers. DNA samples were genotyped according to the manufacturer's instructions on Illumina Infinium HumanHap300v2, HumanHap300v1 or HumanCNV370v1 SNP bead microarrays. Genotype data were imputed using MACH 1.0 (v1.0.18.c) using the HapMap CEU population (release 22, build 36). As the ERF study included related individuals, testing for association between lipid and allele dosage was performed using a mixed model approach as implemented with the ‘mmscore’ option in the GenABEL software. 1.7–4 (R 2.15.3) [19]. This option combines the Family Based Score Test for Association (FASTA) method of Abecasis et al. [20] and kinship matrix estimated from genotyped SNPs [21]. The total genotype set after imputation involved dosage information of approximately 2.5 million SNPs. Among the 2,640 samples, 2,416 were genotyped, following the exclusion of people on lipid lowering (N = 298), in total 2,118 samples were included in the final analysis. To correct for multiple testing, we used the number of unique metabolites (N = 42) which yielded a suggestive significance zone that lies between 5×10−8 and 1.2×10−9. Details are described in the S2 Text.
Automated annotation of GWAS results
In order to facilitate the manual process of assigning genes to a locus, we used an automated workflow developed in-house to generate reports containing the associated protein, enzyme, metabolic reaction, pathway, and disease phenotypes of every gene within a window of 1 MB of the locus. In addition, SNPs published in the GWAS catalog [22] and eQTLs from the GTEx-eQTL database. (http://www.ncbi.nlm.nih.gov/gtex/GTEX2) were given. In detail, the reports created by our workflow were based on the dbSNP [23], NCBI-Gene (http://www.ncbi.nlm.nih.gov/gene), GTEx-eQTL, GWAS catalog, ConsensusPathDB [24], UniProtKB [25], OMIM [26], TCDB [27], ExPASy [28] and KEGG database [29]. The databases had been downloaded earlier from the respective ftp servers and have been integrated offline in Matlab. For the KEGG database the last freely available version was used (30-6-2011).
Exome sequencing
Coding variant analysis were performed within the 3rd data freeze (N = 1309) from the ERF pedigree which were sequenced “in-house” at the Center for Biomics of the Cell Biology department of the Erasmus MC, The Netherlands, using the Agilent version V4 capture kit on an Illumina Hiseq2000 sequencer using the TruSeq Version 3 protocol. The sequence reads were aligned to the human genome build 19 (hg19) using BWA and the NARWHAL pipelines [30], [31]. After processing, genetic variants were called using the Unified Genotyper tool from the GATK. The effects of the called variants on the respective protein sequences were determined with a custom variant annotation script. For each sample, at least 4 Gigabases of sequence was aligned to the genome. All variants in the vicinity of the genes of interest were selected for further analysis. Variants with less than 5 observations were removed. Of the 1,309 individuals with exome sequencing data, 921 had eligible NMR metabolite measurements. Single variant analyses were performed using and additive model as implemented in the “mmscore” function in GenABEL v.1.7–4, adjusting for relatedness.
Ethics statement
The study protocol was approved by the medical ethics board of the Erasmus MC Rotterdam, the Netherlands. The study included only adults and written informed consents were provided by all the subjects participated in the study.
Supporting Information
Zdroje
1. ChenG, RamosE, AdeyemoA, ShrinerD, ZhouJ, et al. (2012) UGT1A1 is a major locus influencing bilirubin levels in African Americans. Eur J Hum Genet 20 : 463–468.
2. McCarthyMI, AbecasisGR, CardonLR, GoldsteinDB, LittleJ, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9 : 356–369.
3. DanikJS, PareG, ChasmanDI, ZeeRY, KwiatkowskiDJ, et al. (2009) Novel loci, including those related to Crohn disease, psoriasis, and inflammation, identified in a genome-wide association study of fibrinogen in 17 686 women: the Women's Genome Health Study. Circ Cardiovasc Genet 2 : 134–141.
4. ChasmanDI, PareG, MoraS, HopewellJC, PelosoG, et al. (2009) Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet 5: e1000730.
5. SuhreK, WallaschofskiH, RafflerJ, FriedrichN, HaringR, et al. (2011) A genome-wide association study of metabolic traits in human urine. Nat Genet 43 : 565–569.
6. NicholsonG, RantalainenM, LiJV, MaherAD, MalmodinD, et al. (2011) A genome-wide metabolic QTL analysis in Europeans implicates two loci shaped by recent positive selection. PLoS Genet 7: e1002270.
7. KettunenJ, TukiainenT, SarinAP, Ortega-AlonsoA, TikkanenE, et al. (2012) Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet 44 : 269–276.
8. Xie W, Wood AR, Lyssenko V, Weedon MN, Knowles JW, et al. (2013) Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes.
9. SuhreK, ShinSY, PetersenAK, MohneyRP, MeredithD, et al. (2011) Human metabolic individuality in biomedical and pharmaceutical research. Nature 477 : 54–60.
10. KottgenA, AlbrechtE, TeumerA, VitartV, KrumsiekJ, et al. (2013) Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45 : 145–154.
11. KolzM, JohnsonT, SannaS, TeumerA, VitartV, et al. (2009) Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet 5: e1000504.
12. Rueedi R LM, Nicholls AW., Reza M Salek, Pedro Marques-Vidal, Edgard Morya, Koichi Sameshima, Ivan Montoliu, Laeticia Da Silva, Sebastiano Collino, François-Pierre Martin, Serge Rezzi, Christoph Steinbeck, Dawn M Waterworth, Gérard Waeber, Peter Vollenweider, Jacques S Beckmann, Johannes Le Coutre, Vincent Mooser, Sven Bergmann, Ulrich K Genick, Zoltán Kutalik (2014) Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links PLoS Genet.
13. LappalainenT, SammethM, FriedlanderMR, t HoenPA, MonlongJ, et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501 : 506–511.
14. MailmanMD, FeoloM, JinY, KimuraM, TrykaK, et al. (2007) The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 39 : 1181–1186.
15. OtaVK, BelluccoFT, GadelhaA, SantoroML, NotoC, et al. (2014) PRODH Polymorphisms, Cortical Volumes and Thickness in Schizophrenia. PLoS One 9: e87686.
16. VorstmanJA, MorcusME, DuijffSN, KlaassenPW, Heineman-de BoerJA, et al. (2006) The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry 45 : 1104–1113.
17. FoxCS, LiuY, WhiteCC, FeitosaM, SmithAV, et al. (2012) Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet 8: e1002695.
18. HennemanP, AulchenkoYS, FrantsRR, van DijkKW, OostraBA, et al. (2008) Prevalence and heritability of the metabolic syndrome and its individual components in a Dutch isolate: the Erasmus Rucphen Family study. J Med Genet 45 : 572–577.
19. AulchenkoYS, RipkeS, IsaacsA, van DuijnCM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23 : 1294–1296.
20. AbecasisGR, ChernySS, CooksonWO, CardonLR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30 : 97–101.
21. AminN, van DuijnCM, AulchenkoYS (2007) A genomic background based method for association analysis in related individuals. PLoS One 2: e1274.
22. HindorffLA, SethupathyP, JunkinsHA, RamosEM, MehtaJP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106 : 9362–9367.
23. SherryST, WardMH, KholodovM, BakerJ, PhanL, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29 : 308–311.
24. KamburovA, PentchevK, GalickaH, WierlingC, LehrachH, et al. (2011) ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res 39: D712–D717.
25. Magrane M, UniProt Consortium (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011.
26. McKusick VA (1998) Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. Baltimore: Johns Hopkins University Press.
27. SaierJ, M.H., TranCV, BaraboteRD (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34: D181–D186.
28. GasteigerE, GattikerA, HooglandC, IvanyiI, AppelRD, et al. (2003) ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31 : 3784–3788.
29. KanehisaM, GotoS (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28 : 27–30.
30. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
31. Brouwer RW, van den Hout MC, Grosveld FG, van Ijcken WF NARWHAL, a primary analysis pipeline for NGS data. Bioinformatics 28 : 284–285.
32. KottgenA, PattaroC, BogerCA, FuchsbergerC, OldenM, et al. (2010) New loci associated with kidney function and chronic kidney disease. Nat Genet 42 : 376–384.
33. LangeLA, Croteau-ChonkaDC, MarvelleAF, QinL, GaultonKJ, et al. (2010) Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum Mol Genet 19 : 2050–2058.
34. XieW, WoodAR, LyssenkoV, WeedonMN, KnowlesJW, et al. (2013) Genetic variants associated with glycine metabolism and their role in insulin sensitivity and type 2 diabetes. Diabetes 62 : 2141–2150.
35. HongMG, KarlssonR, MagnussonPK, LewisMR, IsaacsW, et al. (2013) A genome-wide assessment of variability in human serum metabolism. Hum Mutat 34 : 515–524.
36. IlligT, GiegerC, ZhaiG, Romisch-MarglW, Wang-SattlerR, et al. (2010) A genome-wide perspective of genetic variation in human metabolism. Nat Genet 42 : 137–141.
37. LeeY, YoonKA, JooJ, LeeD, BaeK, et al. (2013) Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis 34 : 307–313.
38. InouyeM, RipattiS, KettunenJ, LyytikainenLP, OksalaN, et al. (2012) Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet 8: e1002907.
39. SannaS, JacksonAU, NagarajaR, WillerCJ, ChenWM, et al. (2008) Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet 40 : 198–203.
40. RietschelM, MattheisenM, FrankJ, TreutleinJ, DegenhardtF, et al. (2010) Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry 68 : 578–585.
41. BenyaminB, McRaeAF, ZhuG, GordonS, HendersAK, et al. (2009) Variants in TF and HFE explain approximately 40% of genetic variation in serum-transferrin levels. Am J Hum Genet 84 : 60–65.
42. PotkinSG, GuffantiG, LakatosA, TurnerJA, KruggelF, et al. (2009) Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One 4: e6501.
43. BenjaminDJ, CesariniD, van der LoosMJ, DawesCT, KoellingerPD, et al. (2012) The genetic architecture of economic and political preferences. Proc Natl Acad Sci U S A 109 : 8026–8031.
44. WuJH, LemaitreRN, ManichaikulA, GuanW, TanakaT, et al. (2013) Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Genet 6 : 171–183.
45. PorcuE, MediciM, PistisG, VolpatoCB, WilsonSG, et al. (2013) A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 9: e1003266.
46. GellerF, FeenstraB, ZhangH, ShafferJR, HansenT, et al. (2011) Genome-wide association study identifies four loci associated with eruption of permanent teeth. PLoS Genet 7: e1002275.
47. Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, et al. (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet 45 : 353-361, 361e351–352.
48. FletcherO, JohnsonN, OrrN, HoskingFJ, GibsonLJ, et al. (2011) Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst 103 : 425–435.
49. LiJ, HumphreysK, HeikkinenT, AittomakiK, BlomqvistC, et al. (2011) A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat 126 : 717–727.
50. ThomasG, JacobsKB, KraftP, YeagerM, WacholderS, et al. (2009) A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet 41 : 579–584.
51. StaceySN, ManolescuA, SulemP, RafnarT, GudmundssonJ, et al. (2007) Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 39 : 865–869.
52. N'DiayeA, ChenGK, PalmerCD, GeB, TayoB, et al. (2011) Identification, replication, and fine-mapping of Loci associated with adult height in individuals of african ancestry. PLoS Genet 7: e1002298.
53. Lango AllenH, EstradaK, LettreG, BerndtSI, WeedonMN, et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467 : 832–838.
54. PillasD, HoggartCJ, EvansDM, O'ReillyPF, SipilaK, et al. (2010) Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet 6: e1000856.
55. JongjaroenprasertW, PhusantisampanT, MahasirimongkolS, MushirodaT, HirankarnN, et al. (2012) A genome-wide association study identifies novel susceptibility genetic variation for thyrotoxic hypokalemic periodic paralysis. J Hum Genet 57 : 301–304.
56. CheungCL, LauKS, HoAY, LeeKK, TiuSC, et al. (2012) Genome-wide association study identifies a susceptibility locus for thyrotoxic periodic paralysis at 17q24.3. Nat Genet 44 : 1026–1029.
57. RothenbergME, SpergelJM, SherrillJD, AnnaiahK, MartinLJ, et al. (2010) Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet 42 : 289–291.
58. KrintelSB, PalermoG, JohansenJS, GermerS, EssiouxL, et al. (2012) Investigation of single nucleotide polymorphisms and biological pathways associated with response to TNFalpha inhibitors in patients with rheumatoid arthritis. Pharmacogenet Genomics 22 : 577–589.
59. PfeuferA, SannaS, ArkingDE, MullerM, GatevaV, et al. (2009) Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet 41 : 407–414.
60. MarjamaaA, OikarinenL, PorthanK, RipattiS, PelosoG, et al. (2012) A common variant near the KCNJ2 gene is associated with T-peak to T-end interval. Heart Rhythm 9 : 1099–1103.
61. ComuzzieAG, ColeSA, LastonSL, VorugantiVS, HaackK, et al. (2012) Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 7: e51954.
62. Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, et al. (2010) Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet 6.
63. SabattiC (2009) Service SK, Hartikainen AL, Pouta A, Ripatti S, et al (2009) Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet 41 : 35–46.
64. TeslovichTM, MusunuruK, SmithAV, EdmondsonAC, StylianouIM, et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 : 707–713.
65. WillerCJ, SannaS, JacksonAU, ScuteriA, BonnycastleLL, et al. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40 : 161–169.
66. AulchenkoYS, RipattiS, LindqvistI, BoomsmaD, HeidIM, et al. (2009) Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 41 : 47–55.
67. KathiresanS, WillerCJ, PelosoGM, DemissieS, MusunuruK, et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet 41 : 56–65.
68. ConsortiumUIG, BarrettJC, LeeJC, LeesCW, PrescottNJ, et al. (2009) Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 41 : 1330–1334.
69. AndersonCA, BoucherG, LeesCW, FrankeA, D'AmatoM, et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43 : 246–252.
70. LingappaJR, PetrovskiS, KahleE, FellayJ, ShiannaK, et al. (2011) Genomewide association study for determinants of HIV-1 acquisition and viral set point in HIV-1 serodiscordant couples with quantified virus exposure. PLoS One 6: e28632.
71. McClayJL, AdkinsDE, AbergK, BukszarJ, KhachaneAN, et al. (2011) Genome-wide pharmacogenomic study of neurocognition as an indicator of antipsychotic treatment response in schizophrenia. Neuropsychopharmacology 36 : 616–626.
72. JostinsL, RipkeS, WeersmaRK, DuerrRH, McGovernDP, et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491 : 119–124.
73. KennyEE, Pe'erI, KarbanA, OzeliusL, MitchellAA, et al. (2012) A genome-wide scan of Ashkenazi Jewish Crohn's disease suggests novel susceptibility loci. PLoS Genet 8: e1002559.
74. KristianssonK, PerolaM, TikkanenE, KettunenJ, SurakkaI, et al. (2012) Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet 5 : 242–249.
75. LettreG, PalmerCD, YoungT, EjebeKG, AllayeeH, et al. (2011) Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet 7: e1001300.
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



