-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
Contactins and Contactin-Associated Proteins, and Contactin-Associated Protein-Like 2 (CNTNAP2) in particular, have been widely cited as autism risk genes based on findings from homozygosity mapping, molecular cytogenetics, copy number variation analyses, and both common and rare single nucleotide association studies. However, data specifically with regard to the contribution of heterozygous single nucleotide variants (SNVs) have been inconsistent. In an effort to clarify the role of rare point mutations in CNTNAP2 and related gene families, we have conducted targeted next-generation sequencing and evaluated existing sequence data in cohorts totaling 2704 cases and 2747 controls. We find no evidence for statistically significant association of rare heterozygous mutations in any of the CNTN or CNTNAP genes, including CNTNAP2, placing marked limits on the scale of their plausible contribution to risk.
Published in the journal: . PLoS Genet 11(1): e32767. doi:10.1371/journal.pgen.1004852
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004852Summary
Contactins and Contactin-Associated Proteins, and Contactin-Associated Protein-Like 2 (CNTNAP2) in particular, have been widely cited as autism risk genes based on findings from homozygosity mapping, molecular cytogenetics, copy number variation analyses, and both common and rare single nucleotide association studies. However, data specifically with regard to the contribution of heterozygous single nucleotide variants (SNVs) have been inconsistent. In an effort to clarify the role of rare point mutations in CNTNAP2 and related gene families, we have conducted targeted next-generation sequencing and evaluated existing sequence data in cohorts totaling 2704 cases and 2747 controls. We find no evidence for statistically significant association of rare heterozygous mutations in any of the CNTN or CNTNAP genes, including CNTNAP2, placing marked limits on the scale of their plausible contribution to risk.
Introduction
Contactin-associated protein-like 2 (CNTNAP2; a.k.a CASPR2, OMIM 604569, Swiss-Prot Q9UHC6), is widely considered an established autism spectrum disorder (ASD, OMIM 209850) risk gene based on a series of findings including homozygosity mapping of rare recessive mutations[1], cytogenetics[2, 3], deep re-sequencing[2], common variant candidate gene association studies[3, 4], and behavioral phenotyping of mouse models[5]. Moreover, several of the reported human genetic studies have extended the linked or associated phenotype to include language delay[6, 7], intractable epilepsy[1], cortical malformation[1] and intellectual disability[8, 9] (OMIM 610042) and in one instance, hepatomegaly and periventricular leukomalacia[10]. These findings, in aggregate, point to an important role for CNTNAP2 in the development of the human central nervous system and provide strong evidence that the complete loss of function of this protein leads to autism and epilepsy. However, replication of the genetic association studies with regard to heterozygous variants in ASD2–4 has proven elusive. With regard to common variation, three genome wide association studies have not found further significant evidence for previously identified single nucleotide polymorphisms (SNPs)[11–13] at CNTNAP2 while a fourth found suggestive evidence falling short of genome-wide significance for a nearby SNP.[14] Moreover, a recent targeted study of common alleles at the CNTNAP2 locus did not replicate prior findings or provide new evidence for significant associations in this interval, after correction for multiple comparisons and the inclusion of a larger follow-up cohort.[15]. In addition, to our knowledge, there have been no further published investigations of rare variant mutation burden in CNTNAP2 beyond the initial report[2] from our laboratory.
In an effort to further explore the evidence for a role for rare heterozygous mutations in CNTNAP2 in ASD, we set out to re-sequence a new discovery cohort including 1030 individuals affected with ASD as well as 942 psychiatrically unscreened controls, using a combination of micro-emulsion PCR and next generation sequencing. Moreover, we extended our investigation to include the additional genes Contactins 1–6 (OMIM 600016, 190197, 601325, 607280, 607219, 607220, respectively; Swiss-Prot Q12860, Q02246, Q9P232, Q8IWV2, O94779, Q9UQ52, respectively) and Contactin Associated Proteins 1, 3, 4 and 5 (OMIM 602346, 610517, 610518, 610519, respectively; Swiss-Prot P78357, Q9UHC6, Q9BZ76, Q9C0A0, Q8WYK1, respectively), based on a range of evidence for several of these neuronal adhesion molecules in neurodevelopmental phenotypes[16–18].
In our prior analysis of CNTNAP2, sequencing of a separate cohort of 635 cases and 942 controls led to the identification of one nominally significant rare variant, I869T, found in three apparently unrelated cases and not in controls, though at the time of publication we were not able to rule out a role for population stratification in this finding[2]. In addition, we observed an approximately 2 fold increase in rare highly conserved (across 10 vertebrate lineages) mutations in cases versus controls, but this fell just short of statistical significance. In the current study, after correction for multiple comparisons, we find no additional support for an association of the I869T variant, which in contrast to our prior study, is no longer predicted to be deleterious to protein function [19, 20]. Morevoer, we find no evidence for association of rare heterozygous mutations in general in CNTNAP2 with the ASD phenotype, either in our new discovery cohort (N = 1030 cases and 942 controls), among an independent ASD sample that had previously undergone exome sequencing (N = 1039 ASD cases and 863 controls), or in a combined mutation burden analysis including these two data sets along with the cohort previously sequenced in our laboratory[2] (N = 2704 cases and 2747 controls). Our ability to assess the impact of rare loss of function mutations was limited by very few observations. However, our findings with regard to rare missense variants was not altered by predicting putative functional versus neutral variants using a range of informatics tools. Finally, we found no significant support for association of rare heterozygous mutations in other Contactins or Contactin-Associated Proteins via targeted re-sequencing of our discovery cohort (1030 cases and 942 controls).
Given the strong evidence for a link between ASD, epilepsy and homozygous loss of CNTNAP2, our data do not rule out a role for this gene and associated molecules in the risk for ASD, but place further bounds on the plausible size of the contribution from rare heterozygous missense mutations in these genes.
Results
Burden of rare mutations in cases and controls
In prior work[2] we had noted a non-significant overrepresentation of all rare (<1 observation in 4000 combined [sequenced or TaqMan-genotyped] controls) variants in affected individuals. Consequently, our primary hypothesis was that in an expanded analysis, the burden of rare mutations would be greater in cases than in well-matched controls. After QC steps and exclusion of ancestral outliers, we had a total of 1030 cases and 942 controls and conducted a gene by gene analysis, including variants that were missense, nonsense, or splice site-disrupting, fell below 2% in the high-throughput data, and were filtered against population data as described in Methods below. Variants that were present only once in cases or controls are denoted as “singleton” and are estimated to have an allele frequency approximate to that in our prior analysis (<1 observation/4000)[2]. For CNTNAP2, we found no increase in the overall burden of rare mutations (Table 1, Fig. 1) even before correcting for multiple comparisons and in fact more rare mutations in controls than cases (19 in cases, 26 in controls, Fisher exact one-tailed p = 0.113). We then evaluated all CNTN and CNTNAP genes and nominal association for rare heterozygous mutations in CNTN1 (14 in cases, 5 in controls, Fisher exact one-tailed p = 0.048) and CNTN3 (13 in cases, 3 in controls, Fisher exact one-tailed p = 0.016), however, neither results survived experiment-wide correction for multiple comparisons.
Tab. 1. Overall rare* variant mutation burden: all genes. 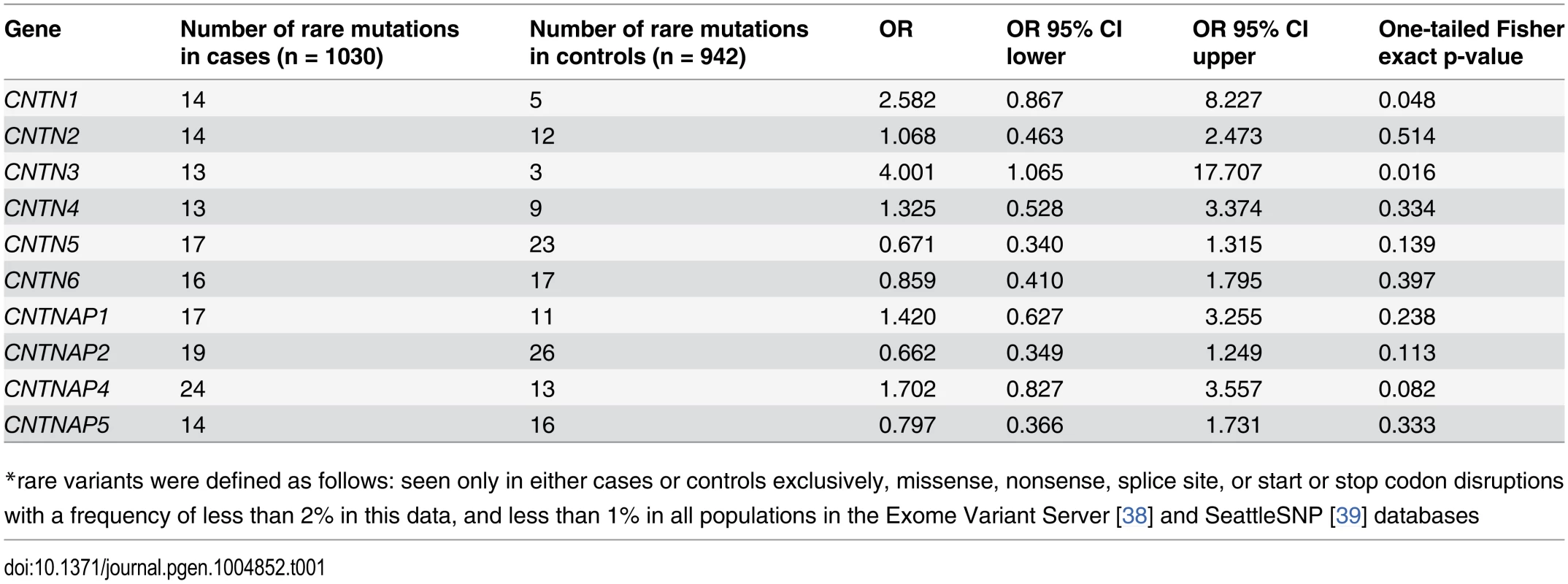
*rare variants were defined as follows: seen only in either cases or controls exclusively, missense, nonsense, splice site, or start or stop codon disruptions with a frequency of less than 2% in this data, and less than 1% in all populations in the Exome Variant Server[38] and SeattleSNP[39] databases Fig. 1. Location of all mutations of interest, i.e. rare and exclusive to cases or controls. 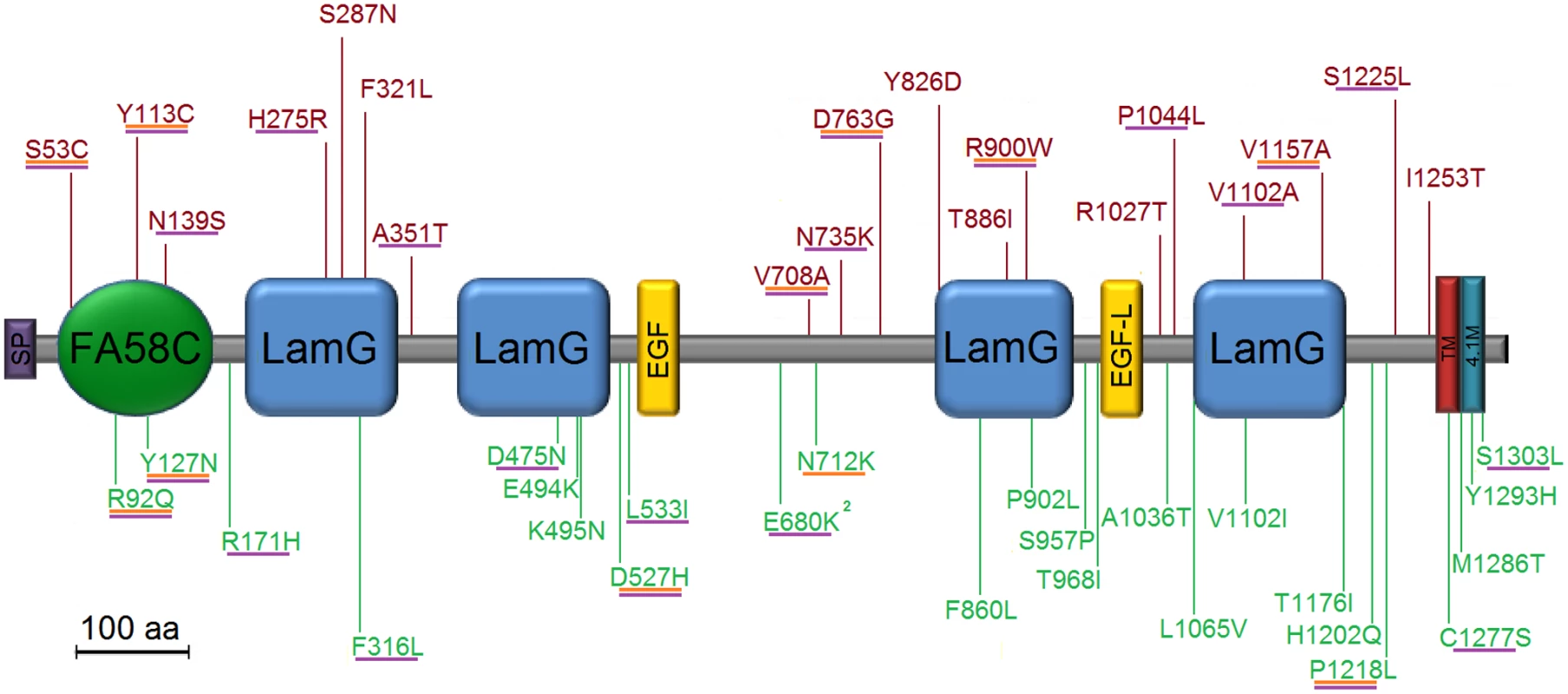
Mutations were counted if missense, nonsense, splice site, or frameshift in CNTNAP2 (in point of fact, all were missense). Variants in red are exclusive to cases; those in green, controls. Those predicted to be deleterious in SIFT are underlined in orange; in PolyPhen2, purple. Domain names and approximate locations from Bakkaloglu et al., We reasoned that the background rate of neutral rare variation in the genome might be obscuring a true difference in functional alleles in cases versus controls. Consequently, we used a variety of defining criteria to try to distinguish the subset of potentially disease-relevant mutations: First, we evaluated “singleton” mutations, i.e. those seen only once either in cases or in controls. This strategy was intended to focus on the rarest alleles in our sample, those most likely to be subject to purifying selection. A comparison of rates of singleton mutations in CNTNAP2 and all other sequenced CNTN and CNTNAP genes again found no evidence of association, prior to correction for the 10 genes evaluated in this study (Table 2). We then used two well-established informatics tools designed to predict deleterious amino acid substitutions. PolyPhen2[21] and SIFT [19], [20]. We defined mutations as deleterious if they were either ‘possibly damaging’ or ‘probably damaging’ in PolyPhen2 or ‘damaging’ in SIFT. While neither metric evaluates nonsense mutations and splice site disruptions, these were included as deleterious as well.
Tab. 2. Rates of singleton* mutations: all genes. 
*singleton mutations met the following criteria: seen only once in either cases or controls exclusively, missense, nonsense, splice site, or start or stop codon disruptions with a frequency of less than 2% in this data, and less than 1% in all populations in the Exome Variant Server[38] and SeattleSNP[39] databases Using all of these approaches separately, we compared the rates of putatively deleterious/functional alleles in the same 1030 cases versus 942 controls. We found no significant differences using any of these metrics with regard to CNTNAP2 (13 in cases, 13 in controls, Fisher exact one-tailed p = 0.486) (Table 3). CNTN1 was found to show nominally significant association (12 in cases, 2 in controls, Fisher exact one-tailed p = 0.01) but this was not significant after correcting for the 10 genes studied. No other genes were significant prior to correction. Additionally, in an analysis restricted to singletons predicted to be deleterious by SIFT or PolyPhen2 (S1 Table), none of the genes reached significance prior to correction.
Tab. 3. Rates of mutation predicted deleterious* by SIFT-or-PolyPhen2: all genes. 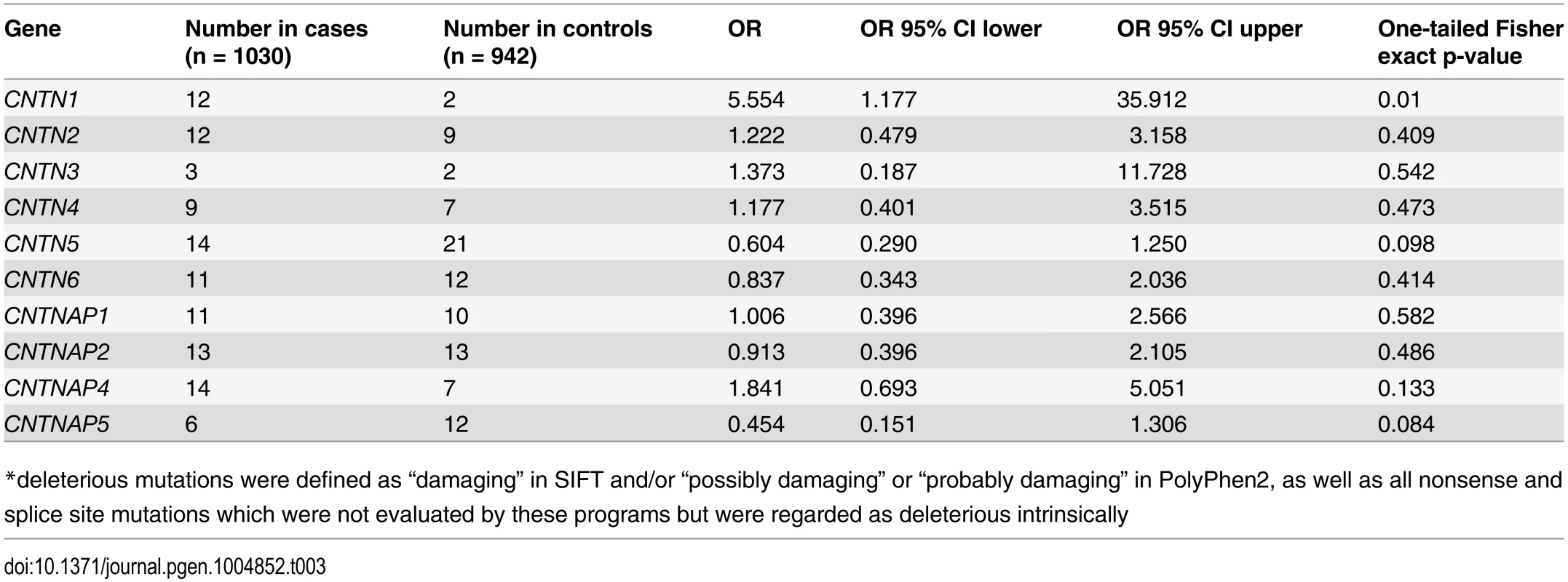
*deleterious mutations were defined as “damaging” in SIFT and/or “possibly damaging” or “probably damaging” in PolyPhen2, as well as all nonsense and splice site mutations which were not evaluated by these programs but were regarded as deleterious intrinsically Finally, we restricted our analysis to focus solely on nonsense, splice site or frameshift mutations. We found none present in CNTNAP2 in the current cohort. Moreover, this category of variation was so infrequent in both cases and controls in all other genes investigated that gene-by-gene analysis was not statistically meaningful, this limiting our conclustions about the role for true haploinsufficiency in any of the genes under study. Overall, we found 2 case individuals with a stop codon in one of the 10 genes (CNTN2 and CNTNAP1), 1 case and no controls with a splice site mutation (CNTN3), and 3 control individuals displaying a stop codon (CNTNAP1 and two in CNTN6). Of note, indel mutations were not detectable using the Raindance instrument given the short read length and the nature of the alignment process.
Of note, our primary analysis was restricted to only rare variants seen in cases or controls and not both. To be confident that this approach did not obscure a true difference in mutation burden, we re-ran all of the aforementioned analyses while including all point mutations defined as rare, regardless of whether they were present in both cases and controls. This had no impact on the interpretation of our results (S2‐S3 Tables).
Power of burden test
For the primary analysis of CNTNAP2, we compute good power (>80%) under the following parameters: observed rare variant frequency of 0.023 (Table 1), odds ratio for dominant effect of ≥ 2.0, size of the test set to 0.005 (corrected for 10 tests), and for this sample of cases (N = 1030) and controls (N = 942). The empirically derived rare variant frequency for the average gene tested is lower (across all CNTNs and CNTNAPs including CNTNAP2) at 0.015. For such a gene, good power accrues for odds ratio ≥ 2.5. Finally, it we consider the burden over all tested genes combined, for which the average frequency of rare variants is roughly 0.15, good power accrues if the odds ratio is ≥ 1.35[22].
Analysis of I869T
In our 2008 report, we regarded I869T as a significant finding given its occurrence 3 times in cases and not at all in controls (Fisher exact one-tailed p = 0.014), its deleterious rating in SIFT, and its amino acid conservation [leucine in amphibians and fish, isoleucine in mammals]. Subsequent revisions to SIFT, however, have reclassified the mutation as benign, and its PolyPhen2 rating has remained benign. Furthermore, the expanded omnibus data set of 2704 cases and 2747 controls (S4 Table) revealed just one further occurrence in cases (in the Broad-Baylor cohort), none in controls; this was no longer statistically significant in the expanded sample size (Fisher exact p = 0.06) without correction for multiple comparisons.
De Novo mutation burden
In keeping with our recent findings[23] that the identification of recurrent de novo loss-of-function point mutations is a viable strategy to identify ASD genes, we determined the transmission status of every rare/singleton mutation seen in probands. We found 2 confirmed de novo mutations using whole blood DNA for sequencing reactions. No confirmed de novo mutations were observed in CNTNAP2. CNTN6 and CNTNAP4 each showed one de novo missense variant each. Based on recent analyses of exome data, a single missense de novo mutation, regardless of other characteristics such as degree of evolutionary conservation, does not provide significant evidence of association.[23–26]
Compound heterozygous mutations
We further examined individuals for the presence of multiple rare variants in the same gene: We found 2 probands with rare mutations, one in CNTN6 and the other in CNTNAP4. A total of four controls were also identified with two rare mutations each: three were found in CNTN5, and one had two rare variants in CNTN6. We determined chromosome phase for the two cases and both were found to be compound heterozygotes. A similar analysis was not possible for controls given the absence of parental data. Again, based on recent studies of whole-exome trio data in autism, the observation of a lone missense compound heterozygous mutation in a gene carries minimal statistical evidence for association[27].
Parental origin of mutation
With the family analysis data, we were able to determine inheritance status (S5 Table) for putatively deleterious mutations of interest in each gene. There was no significant difference in the proportion of deleterious (based on ratings in SIFT (damaging) and/or PolyPhen2 (possibly or probably damaging) variants maternally inherited and paternally inherited in any gene (Table 4). We also observed no significant difference between maternal and paternal variants when this gene-by-gene analysis was restricted to singletons (S6 Table).
Tab. 4. Inheritance of mutations predicted deleterious* by SIFT-or-PolyPhen2. 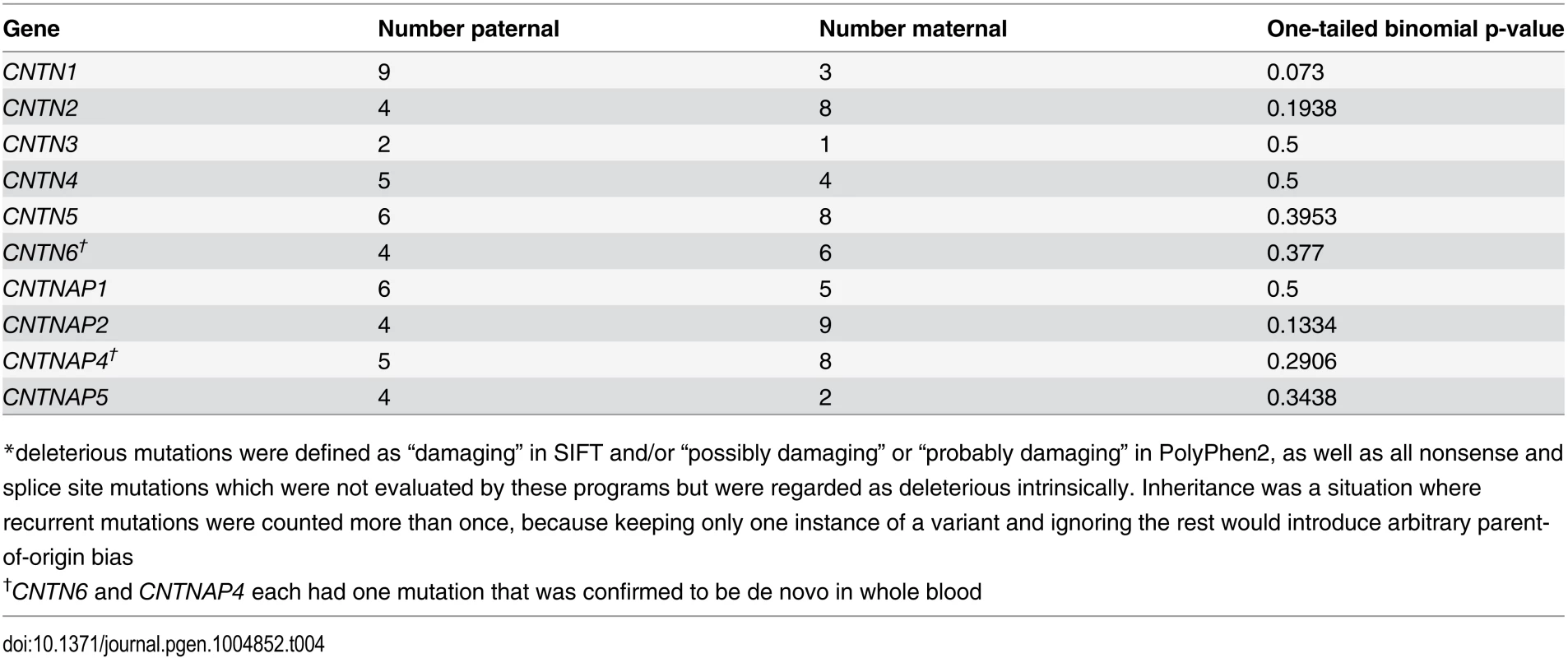
*deleterious mutations were defined as “damaging” in SIFT and/or “possibly damaging” or “probably damaging” in PolyPhen2, as well as all nonsense and splice site mutations which were not evaluated by these programs but were regarded as deleterious intrinsically. Inheritance was a situation where recurrent mutations were counted more than once, because keeping only one instance of a variant and ignoring the rest would introduce arbitrary parent-of-origin bias Combined analysis of multiple cohorts for mutations in CNTNAP2
Given the divergent findings in our initial study[2] versus the new discovery cohort with regard to CNTNAP2, we expanded our dataset. Using exome sequencing data in both cases and controls, we were able to increase the total sample for this gene to 2704 cases and 2747 controls. Consistent with the findings above, an omnibus analysis in these 2704 cases and 2747 controls showed no significant results either in a primary analysis of rare mutation burden or in any of the exploratory analyses of variant subsets, as described above (rare mutation burden 45 in cases, 49 in controls, one-tailed Fisher exact p = 0.407, deleterious mutation burden 26 in cases, 28 in controls, one-tailed Fisher exact p = 0.469, deleterious singleton mutation burden 22 in cases, 23 in controls, one-tailed Fisher exact p = 0.521) (Tables 5, S7 and S8). Furthermore, the variants from our 2008 paper show an even weaker trend for overrepresentation of deleterious mutations in cases, due to updates to the SIFT and PolyPhen2 databases (see Tables 5, S7 and S8).
Tab. 5. Combined CNTNAP2 deleterious variants**. 
*reported in Bakkaloglu et al.[2] Discussion
Strauss et al.[1] first demonstrated a role for CNTNAP2 in neurodevelopmental disorders based on a consanguineous pedigree in which probands presented with autism, cortical dysplasia and focal epilepsy. A single-base homozygous deletion resulting in a premature stop codon was identified in CNTNAP2[1]. In 2008, four studies provided varying degrees of evidence for a role for heterozygous mutations in CNTNAP2 in non-syndromic autism and language delay. Bakkaloglu et al.[2] mapped a de novo chromosomal inversion disrupting CNTNAP2 and then re-sequenced the gene in 635 cases and 942 controls, finding nominally significant association of a single rare variant and a trend toward significance for the overall burden of rare deleterious and conserved mutations in cases versus controls. Alarcon et al.[3] utilized family-based association with age at first word in 172 autism trios to identify CNTNAP2 along with three other genes; in a second-stage follow-up in 304 independent trios, only CNTNAP2 continued to show association with age at first word. Furthermore, microarray expression analysis identified 12 genes significantly enriched in human language-related association cortex, one of which was CNTNAP2. Arking et al.[4] used a linkage approach and identified a common variant in CNTNAP2; a second-stage independent cohort of 1295 autism trios showed significant overtransmission of the T allele. Finally, Vernes et al.[6], investigating the transcription factor FOXP2, previously implicated in language disorders[28–30], determined that FOXP2 binds to and downregulates CNTNAP2. Additional case reports of speech delay and/or ASD patients demonstrating disrupted CNTNAP2 have subsequently been identified[7, 31]. Most recently, Peñagarikano[5] reported a CNTNAP2 KO mouse that showed deficiencies in all three ASD core behavioral domains, seizures, and hyperactivity, with demonstrable neuronal migration aberrations, abnormal cellular morphology and a deceased number of interneurons.
Additionally, disruptions in other members of the contactin family and contactin-associated protein-like family have also been associated with autistic phenotypes. Multiple case reports have implicated contactin 4 (CNTN4) in ASD and in developmental delay[17, 32], as did a CNV study[33] described above. Contactin-associated protein-like 5 has also been recently implicated in ASD by a case study [18].
However, subsequent to these reports, the evidence for association of heterozygous variants in CNTNAP2 has been variable. As noted, multiple GWAS analyses[11–13] have not identified evidence of genome-wide significant association of SNPs mapping near CNTNAP2. In addition, to date, neither rare de novo [23–26] or compound heterozygous mutations[27, 34, 35] have implicated CNTNAP2 or other Contactin or Contactin Associated Proteins in autism or related phenotypes. Finally, other CNTN genes have so far not been subjected to large-scale case control rare variant analyses.
Against this backdrop, we undertook a follow-up of our earlier rare variant case control analysis[2] focusing on CNTNAP2, and expanded the study to include other CNTN and CNTNAP genes. As described above, our results do not convincingly replicate association of the previously nominally significant variant, I869T, nor support the association of heterozygous rare mutations, either transmitted or de novo, in CNTNAP2 with the ASD phenotype. Nor is there evidence for rare recessive risk alleles in this outbred cohort. We found similar results for all CNTN and CNTNAP genes tested here. As noted above, in addition to investigating overall mutation burden, we conducted exploratory analyses to look for any potential differences between cases and controls in subsets of patients; these included evaluating predicted deleterious mutation burden, looking for multiple mutations within a single gene or across the genes sequenced in this study (see Protocol S1), investigating parental origin of the variation and evaluating for de novo mutations. While the primary analyses were well-powered to identify odds ratios in the range of 1.35 to 2.5, the secondary analyses are truly exploratory and cannot be interpreted as excluding a role for CNTN or CNTNAP genes in ASD.
There are several potential explanations for our inability to extend our previous findings: first, the prior results with regard to overall mutation burdent showed only a trend toward signigficance and the larger, better powered sample studied here may simply be clarifying the true relationship (or absence of risk). Alternatively, the paucity of loss of function mutations in the expanded sample, coupled with the difficulty in interpreting the functional consequences of transmitted missense variants without a biologically validated assay could now be leading to a false negative result with regard to haploinsufficiency at this locus—though our results do strongly suggest that the overall contribution to population risk of heterozygous mutations of all types in CNTN and CNTNAP2 must be quite limited. With regard to the prior finding of association of the I869T variant in cases, we did not find evidence for population stratification playing a role, but neither did we add substantial evidence for association, again limiting the plausible effect size of this risk.
As noted above, the lack of a reliable functional assay allowing us to differentiate biologically meaningful from neutral variants is an important limitation of the study. It is possible that such an assay could reveal a distribution of variation consistent with an effect for heterozygous mutations in CNTNAP2—or any of the other genes we have studied. Similarly, it was notable that we did not observe any putative loss of function mutations (nonsense, frame-shift, splice site) in either cases or controls for CNTNAP2. Given the finding[1] of a severe neurodevelopmental syndrome resulting from homozygous loss of function mutations, the observation of deletion CNVs at this locus in affected individuals, and the absence of clear LOF mutations in our sample, it remains possible that very rare LOF heterozygous mutations in CNTNAP2 carry some risk for ASD. Nonetheless, based on the current study, our results place limits on the degree to which, overall, rare heterozygous missense mutations in Contactin and Contactin Associated Proteins may play a role in the population risk for ASD.
They similarly suggest that it is not yet possible to determine the clinical significance of individual rare heterozygous mutations in CNTNAP2, identified either in a research cohort or in diagnostic assays (http://www.athenadiagnostics.com/content/test-catalog/find-test/service-detail/q/id/1487).
Materials and Methods
Subjects and experimental structure
For the targeted re-sequencing discovery cohort, 1332 ASD probands were obtained from the Simons Simplex Collection (SSC), a cohort focused on simplex autism families[36]. Only probands from the SSC were sequenced. We elected to compare this cohort to 1015 unrelated controls, determined to be neurologically unaffected, drawn from the NINDS Caucasian controls repository (Coriell, Camden, NJ). As described above and in detail in Protocol S1, the final case-control cohort was filtered to 1030 ASD probands and 942 controls. For the follow-up examination of CNTNAP2, we obtained exome sequencing data from the ARRA Autism Sequencing Collaboration (AASC). AASC cases and controls were genotyped on the Illumina 550K Single BeadChip platform and filtered to exclude any individual with a genotyping call rate <95%, discrepant genotype data with regard to reported sex, Mendelian inconsistencies, or cryptic relatedness, resulting in a final sample of 1039 autism cases and 863 controls, all of whom were determined to be of European descent through ancestry matching[37] via PCA of genotyping data. Controls were screened for psychiatric conditions via questionnaire. Although AASC/ARRA variants were not confirmed via Sanger sequencing as the Yale cohort of 1030 cases and 942 controls were, we concluded that given the identical sequencing methods for cases and controls as well as the approximately equal sample sizes, any errors would be reasonably randomly distributed among cases and controls. Sample characteristics and diagnostic methodology for the AASC have been described previously.5 This study was approved by the Institutional Review Boards of both Yale University and the Broad Institute.
Library preparation
Primer emulsions representing 1152 separate amplicons were designed and synthesized by Raindance Technologies (Lexington, MA). Synthesis was infeasible for much of CNTNAP3 due to highly repetitive regions; because 16 exons out of 24 were excluded, this gene was not sequenced and therefore does not contribute to any further analyses. A single exon was omitted from each of the genes CNTN2 (out of 22 exons total) and CNTNAP4 (25 total) each due to difficulty in primer synthesis. We reasoned that any loss of data should be equally distributed across cases and controls.
In the initial sequencing effort, all DNA used was derived from human lymphoblastoid cell line DNA. All confirmations, however, were conducted in DNA derived from blood (see below). Samples were quantified on a Biotek Synergy HT fluorometer (BioTek US, Winooski, VT). After quantification, DNA was pooled, 8 individuals at a time, by case/control status (Protocol S1).
Pooled DNA samples were sheared on a Covaris S2 (Covaris, Inc., Woburn, MA) to an approximate size of 3 kb and combined with RainDance microemulsion PCR master mix prepared according to the protocol, then run with a standard protocol on the RDT1000 machine (Raindance Technologies, Lexington, MA) and subjected to conventional PCR on a thermal cycler.
After successful droplet merging and PCR, the samples were cleaned according to the RainDance protocol; aqueous PCR product phase was purified on Qiagen MinElute columns (Qiagen GmbH, Hilden, Germany) and run on an Agilent Bioanalyzer according to DNA1000 protocol. Product was then blunted and concatenated into longer DNA fragments, because the range of amplicon sizes in the microemulsion library makes sequencing uniform fragment lengths impossible without first concatenation and subsequent shearing.
Concatenated DNA was sheared on a Covaris S2 to a mean size of 200 bp. Products were processed for end repair, adenylation and adapter ligation according to modified Illumina protocol (S1 Protocol). These fragments were size selected on Invitrogen agarose e-gels (Invitrogen, Life Technologies, Carlsbad, CA); size-selected DNA fragments were enriched using Illumina primers InPE 1.0, InPE 2.0, and a selected Illumina barcode index out of 12 available. Barcode indices were selected as described in Protocol S1. PCR was performed according to standard Illumina multiplexing protocol and samples were cleaned up on MinElute columns; this product was then re-purified on an e-gel to remove primer dimer and all non-product fragments from the amplified product. 1 µL of this product was run on an Agilent 2100 Bioanalyzer according to DNA1000 protocol to quantitate final concentration.
Sequencing of the discovery cohort
Separate pools of 8 case and 8 control samples were mixed in a 1 : 1 equimolar ratio and sequenced on the Illumina Genome Analyzer IIx. Sequence reads were run through a data analysis pipeline on a high performance cluster (HPC) and aligned to the human genome (NCBI build 36, hg18) using the aligner BWA, followed by conversion to SAM format and generation of a Pileup file using SAMtools. The Pileup file was then analyzed and annotated to identify variants with a haploid coverage greater than 80X, minor allele frequency (MAF) greater than 3.5% and a PHRED-like score (generated from the quality scores in the initial reads) greater than 20. The MAF and Phred-like score cutoffs were determined empirically. Annotation was performed for all possible isoforms of each gene to ensure that any deleterious variants were detected.
Variant confirmations
Pools in which putative rare mutations of interest were identified underwent confirmation and deconvolution using Sanger sequencing. Mutations of interest were defined as missense, nonsense, splice site, or start or stop codon disruptions with a frequency of less than 2%, and less than 1% in all populations in the Exome Variant Server[38] and SeattleSNP[39] databases. Only a single region showed repeated PCR failures (chr11 : 99437155, CNTN5). This variant was excluded from any further analyses. The confirmation rate of the high-throughput sequence variant predictions of interest, as determined from all variants which successfully underwent PCR and Sanger sequencing, was 92%.
After successful Sanger confirmations, all variants identified only in cases were tested for de novo status. An initial round of re-sequencing of the proband, parents and any siblings was completed using lymphoblastoid DNA; if the mutation appeared to be de novo, a second round of sequencing in all family members was completed using whole blood DNA to rule out false positives resulting from EBV transformation. Family member DNA was not available for inheritance evaluation in the Coriell controls.
As part of our analytic strategy we planned to evaluate de novo mutations, and consequently we required a high level of certainty regarding the identity of any proband carrying a putative mutation. Consequently, we developed an in-house script to verify pool composition by comparing returned pooled next-generation sequence data to previously obtained genotyping data available on all SSC probands reported here[40]. 15 pools (9 cases and 6 control) demonstrated insoluble identity issues (ambiguity of constituent individuals) and were excluded from all analyses.
Sample quality control and adjustments for comparable ancestry
SNP Genotyping was performed on Illumina 1M arrays at the Yale Center for Genome Analysis using whole blood DNA (SSC samples) or lymphoblastoid cell line DNA (NINDS controls). Redundant samples were removed from both the case and control cohorts. After sample removal, 1266 cases and 1015 controls genotyped concurrent with being run on Raindance. Of these, PLINK[41] was used to remove those with missing genotype data or call rates <95%, to remove inconsistencies in reported sex, to detect and remove Mendelian inconsistencies, and to identify and remove cryptically related samples using an assessment of inheritance by descent up to and including 2° relatives—after all other filtering steps, one proband was excluded due to relatedness.
Principal components analysis was performed with the genotype data from these samples using Golden Helix SNP and Variation Suite v7.5.4 (Bozeman, MT, USA), evaluating 8210 tagging SNPs SNPs not found to be in high linkage disequilibrium, used in the same way as described in a recent paper[42]. Based on a scree plot, we plotted the eigenvalues of the first 3 principal components which accounted for 32%, 23% and 11% of variation. We defined the final NINDS control set of 942 as a Caucasian cohort and observed that a threshold of 5 interquartile ranges from the third quartile contained all NINDS controls; we used this criterion as a cutoff. 50 SSC probands lay outside this threshold at 6 IQR or greater and consequently were excluded from further analysis.
As noted, we removed from the analysis any variants with a greater than 1% frequency in either the Exome Variant Server[38] or SeattleSNP[39] databases.
Exome cohort
The Baylor-Broad ARRA set consisting of 1039 cases and 863 controls was used to follow-up on our targeted sequencing data given its comparable size and ancestry-matched selection of probands[37].
Statistical analyses
All statistical tests were one-tailed Fisher exact tests, with the exception of parent-of-origin analysis (see below). We used a Bonferroni correction by a factor of 10, based on the 10 genes investigated. Any variant that was seen in both cases and controls was excluded from the analysis. Because parent of origin was a binomial outcome, per-gene one-tailed binomial tests were performed. Finally, in the combined CNTNAP2 dataset of our 2008 data, the current data, and the Baylor-Broad ARRA data, any variants seen in cases and controls were removed from the set (resulting in the loss of one variant from the cumulative set.)
Supporting Information
Zdroje
1. Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, et al. (2006) Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med 354 : 1370–1377. doi: 10.1056/NEJMoa052773 16571880
2. Bakkaloglu B, O’Roak BJ, Louvi A, Gupta AR, Abelson JF, et al. (2008) Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet 82 : 165–173. doi: 10.1016/j.ajhg.2007.09.017 18179895
3. Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, et al. (2008) Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 82 : 150–159. doi: 10.1016/j.ajhg.2007.09.005 18179893
4. Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, et al. (2008) A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet 82 : 160–164. doi: 10.1016/j.ajhg.2007.09.015 18179894
5. Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, et al. (2011) Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147 : 235–246. doi: 10.1016/j.cell.2011.08.040 21962519
6. Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, et al. (2008) A functional genetic link between distinct developmental language disorders. N Engl J Med 359 : 2337–2345. doi: 10.1056/NEJMoa0802828 18987363
7. Poot M, Beyer V, Schwaab I, Damatova N, Van’t Slot R, et al. (2010) Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics 11 : 81–89. doi: 10.1007/s10048-009-0205-1 19582487
8. Zweier C, de Jong EK, Zweier M, Orrico A, Ousager LB, et al. (2009) CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet 85 : 655–666. doi: 10.1016/j.ajhg.2009.10.004 19896112
9. Zweier C (2012) Severe Intellectual Disability Associated with Recessive Defects in CNTNAP2 and NRXN1. Mol Syndromol 2 : 181–185. doi: 10.1159/000331270 22670139
10. Jackman C, Horn ND, Molleston JP, Sokol DK (2009) Gene associated with seizures, autism, and hepatomegaly in an Amish girl. Pediatr Neurol 40 : 310–313. doi: 10.1016/j.pediatrneurol.2008.10.013 19302947
11. Anney R, Klei L, Pinto D, Regan R, Conroy J, et al. (2010) A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 19 : 4072–4082. doi: 10.1093/hmg/ddq307 20663923
12. Wang K, Zhang H, Ma D, Bucan M, Glessner JT, et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459 : 528–533. doi: 10.1038/nature07999 19404256
13. Weiss LA, Arking DE, Daly MJ, Chakravarti A (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461 : 802–808. doi: 10.1038/nature08490 19812673
14. Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, et al. (2012) Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet 21 : 4781–4792. doi: 10.1093/hmg/dds301 22843504
15. Sampath S, Bhat S, Gupta S, O’Connor A, West AB, et al. (2013) Defining the contribution of CNTNAP2 to autism susceptibility. PLoS One 8: e77906. doi: 10.1371/journal.pone.0077906 24147096
16. Compton AG, Albrecht DE, Seto JT, Cooper ST, Ilkovski B, et al. (2008) Mutations in contactin-1, a neural adhesion and neuromuscular junction protein, cause a familial form of lethal congenital myopathy. Am J Hum Genet 83 : 714–724. doi: 10.1016/j.ajhg.2008.10.022 19026398
17. Fernandez T, Morgan T, Davis N, Klin A, Morris A, et al. (2004) Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet 74 : 1286–1293. doi: 10.1086/421474 15106122
18. Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TS, et al. (2010) Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry 68 : 320–328. doi: 10.1016/j.biopsych.2010.02.002 20346443
19. Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11 : 863–874. doi: 10.1101/gr.176601 11337480
20. Ng PC, Henikoff S (2003) SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 31 : 3812–3814. doi: 10.1093/nar/gkg509 12824425
21. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7 : 248–249. doi: 10.1038/nmeth0410-248 20354512
22. Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19 : 149–150. doi: 10.1093/bioinformatics/19.1.149 12499305
23. Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485 : 237–241. doi: 10.1038/nature10945 22495306
24. Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, et al. (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485 : 242–245. doi: 10.1038/nature11011 22495311
25. O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485 : 246–250. doi: 10.1038/nature10989 22495309
26. Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74 : 285–299. doi: 10.1016/j.neuron.2012.04.009 22542183
27. Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, et al. (2013) Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron 77 : 235–242. doi: 10.1016/j.neuron.2012.12.029 23352160
28. Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413 : 519–523. doi: 10.1038/35097076 11586359
29. MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, et al. (2005) Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet 76 : 1074–1080. doi: 10.1086/430841 15877281
30. Zeesman S, Nowaczyk MJ, Teshima I, Roberts W, Cardy JO, et al. (2006) Speech and language impairment and oromotor dyspraxia due to deletion of 7q31 that involves FOXP2. Am J Med Genet A 140 : 509–514. doi: 10.1002/ajmg.a.31110 16470794
31. Petrin AL, Giacheti CM, Maximino LP, Abramides DV, Zanchetta S, et al. (2010) Identification of a microdeletion at the 7q33-q35 disrupting the CNTNAP2 gene in a Brazilian stuttering case. Am J Med Genet A 152A: 3164–3172. doi: 10.1002/ajmg.a.33749 21108403
32. Roohi J, Montagna C, Tegay DH, Palmer LE, DeVincent C, et al. (2009) Disruption of contactin 4 in three subjects with autism spectrum disorder. J Med Genet 46 : 176–182. doi: 10.1136/jmg.2008.057505 18349135
33. Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. (2009) Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459 : 569–573. doi: 10.1038/nature07953 19404257
34. Chahrour MH, Yu TW, Lim ET, Ataman B, Coulter ME, et al. (2012) Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet 8: e1002635. doi: 10.1371/journal.pgen.1002635 22511880
35. Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, et al. (2013) Using whole-exome sequencing to identify inherited causes of autism. Neuron 77 : 259–273. doi: 10.1016/j.neuron.2012.11.002 23352163
36. Fischbach GD, Lord C (2010) The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68 : 192–195. doi: 10.1016/j.neuron.2010.10.006 20955926
37. Liu L, Sabo A, Neale BM, Nagaswamy U, Stevens C, et al. (2013) Analysis of rare, exonic variation amongst subjects with autism spectrum disorders and population controls. PLoS Genet 9: e1003443. doi: 10.1371/journal.pgen.1003443 23593035
38. Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/), [December 2012].
39. SeattleSNPs, NHLBI Program for Genomic Applications, SeattleSNPs, Seattle, WA (URL: http://pga.gs.washington.edu) [August 2012].
40. Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, et al. (2011) Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70 : 863–885. doi: 10.1016/j.neuron.2011.05.002 21658581
41. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575. doi: 10.1086/519795 17701901
42. Fernandez TV, Sanders SJ, Yurkiewicz IR, Ercan-Sencicek AG, Kim YS, et al. (2012) Rare copy number variants in tourette syndrome disrupt genes in histaminergic pathways and overlap with autism. Biol Psychiatry 71 : 392–402. doi: 10.1016/j.biopsych.2011.09.034 22169095
Štítky
Genetika Reprodukční medicína
Článek Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in YeastČlánek Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of HybridsČlánek Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence AnalysesČlánek ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell DivisionČlánek Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 1- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Intrauterinní inseminace a její úspěšnost
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
-
Všechny články tohoto čísla
- The Combination of Random Mutagenesis and Sequencing Highlight the Role of Unexpected Genes in an Intractable Organism
- Ataxin-3, DNA Damage Repair, and SCA3 Cerebellar Degeneration: On the Path to Parsimony?
- α-Actinin-3: Why Gene Loss Is an Evolutionary Gain
- Origins of Context-Dependent Gene Repression by Capicua
- Transposable Elements Contribute to Activation of Maize Genes in Response to Abiotic Stress
- No Evidence for Association of Autism with Rare Heterozygous Point Mutations in Contactin-Associated Protein-Like 2 (), or in Other Contactin-Associated Proteins or Contactins
- Nur1 Dephosphorylation Confers Positive Feedback to Mitotic Exit Phosphatase Activation in Budding Yeast
- A Regulatory Hierarchy Controls the Dynamic Transcriptional Response to Extreme Oxidative Stress in Archaea
- Genetic Variants Modulating CRIPTO Serum Levels Identified by Genome-Wide Association Study in Cilento Isolates
- Small RNA Sequences Support a Host Genome Origin of Satellite RNA
- Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast
- Genetic Mapping of MAPK-Mediated Complex Traits Across
- An AP Endonuclease Functions in Active DNA Demethylation and Gene Imprinting in
- Developmental Regulation of the Origin Recognition Complex
- End of the Beginning: Elongation and Termination Features of Alternative Modes of Chromosomal Replication Initiation in Bacteria
- Naturally Occurring Differences in CENH3 Affect Chromosome Segregation in Zygotic Mitosis of Hybrids
- Imputation of the Rare G84E Mutation and Cancer Risk in a Large Population-Based Cohort
- Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis
- A Genetic Strategy for Probing the Functional Diversity of Magnetosome Formation
- Interactions of Chromatin Context, Binding Site Sequence Content, and Sequence Evolution in Stress-Induced p53 Occupancy and Transactivation
- The Yeast La Related Protein Slf1p Is a Key Activator of Translation during the Oxidative Stress Response
- Integrative Analysis of DNA Methylation and Gene Expression Data Identifies as a Key Regulator of COPD
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- Functional Interplay between the 53BP1-Ortholog Rad9 and the Mre11 Complex Regulates Resection, End-Tethering and Repair of a Double-Strand Break
- Estrogenic Exposure Alters the Spermatogonial Stem Cells in the Developing Testis, Permanently Reducing Crossover Levels in the Adult
- Protein Poly(ADP-ribosyl)ation Regulates Immune Gene Expression and Defense Responses
- Sumoylation Influences DNA Break Repair Partly by Increasing the Solubility of a Conserved End Resection Protein
- A Discrete Transition Zone Organizes the Topological and Regulatory Autonomy of the Adjacent and Genes
- Elevated Mutation Rate during Meiosis in
- The Intersection of the Extrinsic Hedgehog and WNT/Wingless Signals with the Intrinsic Hox Code Underpins Branching Pattern and Tube Shape Diversity in the Airways
- MiR-24 Is Required for Hematopoietic Differentiation of Mouse Embryonic Stem Cells
- Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing
- Heterologous Aggregates Promote Prion Appearance via More than One Mechanism
- The Tumor Suppressor BCL7B Functions in the Wnt Signaling Pathway
- , A -Acting Locus that Controls Chromosome-Wide Replication Timing and Stability of Human Chromosome 15
- Regulating Maf1 Expression and Its Expanding Biological Functions
- A Polyubiquitin Chain Reaction: Parkin Recruitment to Damaged Mitochondria
- RecFOR Is Not Required for Pneumococcal Transformation but Together with XerS for Resolution of Chromosome Dimers Frequently Formed in the Process
- An Intracellular Transcriptomic Atlas of the Giant Coenocyte
- Insight in Genome-Wide Association of Metabolite Quantitative Traits by Exome Sequence Analyses
- The Role of the Mammalian DNA End-processing Enzyme Polynucleotide Kinase 3’-Phosphatase in Spinocerebellar Ataxia Type 3 Pathogenesis
- The Global Regulatory Architecture of Transcription during the Cell Cycle
- Identification and Functional Characterization of Coding Variants Influencing Glycemic Traits Define an Effector Transcript at the Locus
- Altered Ca Kinetics Associated with α-Actinin-3 Deficiency May Explain Positive Selection for Null Allele in Human Evolution
- Genetic Variation in the Nuclear and Organellar Genomes Modulates Stochastic Variation in the Metabolome, Growth, and Defense
- PRDM9 Drives Evolutionary Erosion of Hotspots in through Haplotype-Specific Initiation of Meiotic Recombination
- Transcriptional Control of an Essential Ribozyme in Reveals an Ancient Evolutionary Divide in Animals
- ALIX and ESCRT-III Coordinately Control Cytokinetic Abscission during Germline Stem Cell Division
- Century-scale Methylome Stability in a Recently Diverged Lineage
- A Re-examination of the Selection of the Sensory Organ Precursor of the Bristle Sensilla of
- Antagonistic Cross-Regulation between Sox9 and Sox10 Controls an Anti-tumorigenic Program in Melanoma
- A Dependent Pool of Phosphatidylinositol 4,5 Bisphosphate (PIP) Is Required for G-Protein Coupled Signal Transduction in Photoreceptors
- Deciphering the Genetic Programme Triggering Timely and Spatially-Regulated Chitin Deposition
- Aberrant Gene Expression in Humans
- Fascin1-Dependent Filopodia are Required for Directional Migration of a Subset of Neural Crest Cells
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
- Evolutionary Constraint and Disease Associations of Post-Translational Modification Sites in Human Genomes
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- The Genetic and Mechanistic Basis for Variation in Gene Regulation
- Inactivation of PNKP by Mutant ATXN3 Triggers Apoptosis by Activating the DNA Damage-Response Pathway in SCA3
- DNA Damage Response Factors from Diverse Pathways, Including DNA Crosslink Repair, Mediate Alternative End Joining
- hnRNP K Coordinates Transcriptional Silencing by SETDB1 in Embryonic Stem Cells
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Global Regulatory Architecture of Transcription during the Cell Cycle
- A Truncated NLR Protein, TIR-NBS2, Is Required for Activated Defense Responses in the Mutant
- Proteasomes, Sir2, and Hxk2 Form an Interconnected Aging Network That Impinges on the AMPK/Snf1-Regulated Transcriptional Repressor Mig1
- The SWI2/SNF2 Chromatin Remodeler BRAHMA Regulates Polycomb Function during Vegetative Development and Directly Activates the Flowering Repressor Gene
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



