-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
Inside the nucleus of a cell, DNA is associated with proteins to form a complex three-dimensional structure referred to as chromatin. The structure of chromatin influences how accessible specific DNA sequences are to transcription factors, and therefore chromatin accessibility is an important determinant of gene expression. To better understand how patterns of chromatin accessibility change over time, we quantitatively measured levels of chromatin accessibility in two yeast species and their diploid hybrid. We show that significant differences in chromatin accessibility exist between these two species and occur upstream of genes that are enriched for specific biological functions. We also develop new statistical methods to understand the genetics of variation in chromatin accessibility. Finally, we show that the relationship between chromatin accessibility and gene expression is complex, and many of the observed differences in chromatin accessibility between these two species may not influence gene expression levels. Thus, our work highlights the need to develop additional experimental and statistical methods to distinguish between functionally significant and benign changes in chromatin accessibility.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004427
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004427Summary
Inside the nucleus of a cell, DNA is associated with proteins to form a complex three-dimensional structure referred to as chromatin. The structure of chromatin influences how accessible specific DNA sequences are to transcription factors, and therefore chromatin accessibility is an important determinant of gene expression. To better understand how patterns of chromatin accessibility change over time, we quantitatively measured levels of chromatin accessibility in two yeast species and their diploid hybrid. We show that significant differences in chromatin accessibility exist between these two species and occur upstream of genes that are enriched for specific biological functions. We also develop new statistical methods to understand the genetics of variation in chromatin accessibility. Finally, we show that the relationship between chromatin accessibility and gene expression is complex, and many of the observed differences in chromatin accessibility between these two species may not influence gene expression levels. Thus, our work highlights the need to develop additional experimental and statistical methods to distinguish between functionally significant and benign changes in chromatin accessibility.
Introduction
Changes in gene regulation have long been hypothesized to be an important mechanism of evolutionary diversification [1]–[3] and to contribute to phenotypic variation [4]–[7]. An increasing catalog of adaptive regulatory changes has been identified, such as lactase persistence [8], [9] and the effect of the Duffy blood group chemokine receptor on malaria resistance in humans [10], [11] and beak morphology in Darwin's finches [12]. Furthermore, it has also been suggested that a substantial fraction of SNPs associated with human diseases through genome-wide association studies may act through regulatory changes with genes [13], [14].
On a genome-wide scale, molecular studies have uncovered pervasive transcriptional variation within and between species [15]–[20]. A substantial amount of gene expression variation is heritable, and thousands of regulatory QTL have been mapped in numerous organisms [17], [21]–[24]. In general, regulatory variation can act in cis or trans. Cis-acting regulatory QTL influence transcript levels in an allele-specific manner, typically from variation located within or near the gene being studied. In contrast, trans-acting regulatory QTL does not result in allelic differences in expression and arises from variation that is usually located at a position distinct from the gene being studied [7]. Although both cis and trans regulatory variation make important contributions to heritable variation of transcript abundance, cis-acting variants are thought to be more numerous, have larger effect sizes, and accumulate at a faster rate between species [21], [25].
Despite the progress in mapping cis and trans-acting regulatory QTL, the mechanisms they act through are less well understood. Chromatin structure is a fundamentally important determinant of gene regulation, and changes in the position and number of nucleosomes can affect transcript abundance [26]–[29]. New technologies have enabled genome-wide maps of chromatin architecture to be constructed across different cell types [30], [31] individuals [32]–[34] and species [20], [35]. Although these studies have revealed extensive variation in chromatin structure, many outstanding issues remain, including how much of variation in chromatin accessibility is heritable, the relative contributions of cis and trans-acting regulatory variation to differences in chromatin architecture [32], and how often variation in chromatin structure results in gene expression variation [22], [36].
To address these issues, we describe a genome-wide analysis of chromatin accessibility between two closely related Saccharomyces sensu stricto yeast species, Saccharomyces cerevisiae and Saccharomyces paradoxus, and their hybrid. S. cerevisiae is the yeast model species and has been extensively studied. S. paradoxus is the most closely related species to S. cerevisiae, with an estimated divergence time of 5 million years ago [37]. Chromatin structure in S. cerevisiae has been studied previously [38], [39] and across a single genome, open chromatin regions are weakly associated with increased expression [39]. In addition, nucleosome locations have been compared across multiple yeast species, including S. cerevisiae and S. paradoxus, and cis changes, such as anti-nucleosomal sequences and binding sites for general regulatory factors, were found to contribute to differences in nucleosome location [20]. Within species, the genetic architecture of chromatin accessibility has been studied using QTL mapping [34]; however, this has not been addressed between species.
We assessed chromatin accessibility using FAIRE-Seq and found considerable divergence in chromatin structure between S. cerevisiae and S. paradoxus. Moreover, we developed a novel statistical approach to identify cis and trans-acting effects on chromatin accessibility in hybrids and found cis effects on chromatin structure are more common than trans effects, are of greater magnitude, and that the direction of cis and trans effects are often in opposite directions suggesting compensatory evolution. Finally, we show that the relationship between chromatin structure and transcript levels in S. cerevisiae and S. paradoxus is complex, and a significant proportion of differences in chromatin accessibility might be functionally benign.
Results
Differences in chromatin accessibility within and between species
We first assessed differences in chromatin structure between haploid strains of S. cerevisiae and S. paradoxus. We generated FAIRE-Seq (Formaldehyde-Assisted Isolation of Regulatory Elements) data [40] for two biological replicates for two strains of S. cerevisiae (DBVPG1373, a wine strain, and UWOPS05_217_3, a wild isolate) and one strain of the sister species S. paradoxus, CBS432 (see Methods). FAIRE isolates DNA that is not bound to proteins, resulting in increased signal in regions with increased chromatin accessibility. We sequenced FAIRE DNA samples to approximately 10× coverage using short read sequencing (see Methods). As expected, sequencing reads were enriched in intergenic regions (mean of 2.4× enrichment compared to coding regions).
We first asked which specific areas of the genome have undergone changes in chromatin accessibility between species. We focused on the nucleosome-free region (NFR) found upstream of the transcription start site of many yeast genes because this region is known to harbor important regulatory information; this was also where the dominant FAIRE signal was found in our data [41], (see Figure S1). We computationally identified the nucleosome-free region from the FAIRE data (see Methods) by identifying the peak in FAIRE signal found upstream of each gene and extended the region in either direction until a background level of signal was observed. We then merged NFR calls across the two species (see Methods). We also carried out extensive filtering to eliminate peaks whose differences might be caused by duplications between species or mapping issues (see Methods). In total, we identified 3,498 NFRs that passed our filtering and had an average size of 253 bp.
We first compared one strain of S. paradoxus, CBS432, and one strain of S. cerevisiae, UWOPS05_217_3. Overall, the locations of NFRs called were well-conserved across species, and on average the location of 42% of NFRs overlapped between the two species. As a complementary analysis, we compared levels of chromatin accessibility in the set of all 3,498 NFRs, and found them to be strongly correlated (R2 = 0.68 between species, p<2.2×10−16) suggesting that broad-scale patterns of accessibility are conserved over time.
Next, we tested each of the 3,498 NFRs for differences in chromatin accessibility between the two parental haploid species, S. cerevisiae and S. paradoxus, and used the R package DESeq to test for significant differences. We found 947 NFRs showed significant differences in FAIRE signal (FDR = 0.05, Figure 1, see Methods). Furthermore, by analyzing the distribution of p-values [42], we estimate that π0 (the proportion of NFRs with no differences in chromatin accessibility) is 0.53, suggesting that 47 percent of NFRs are differentially accessible between species. These 947 NFRs were upstream of 1,149 distinct genes and on average resulted in a 2.17-fold difference in FAIRE signal between the two species. 483 of the NFRs showed higher accessibility in UWOPS05_217_3, while 464 NFRs showed higher accessibility in CBS432. We carried out a test for GO enrichment at the genes downstream of differentially accessible peaks and found that several GO biological process terms were enriched compared to the genome as a whole (corrected p<0.05), specifically intracellular transport, protein localization, protein transport, and establishment of protein localization [43].
Fig. 1. Patterns of chromatin accessibility within and between S. cerevisiae and S. paradoxus. 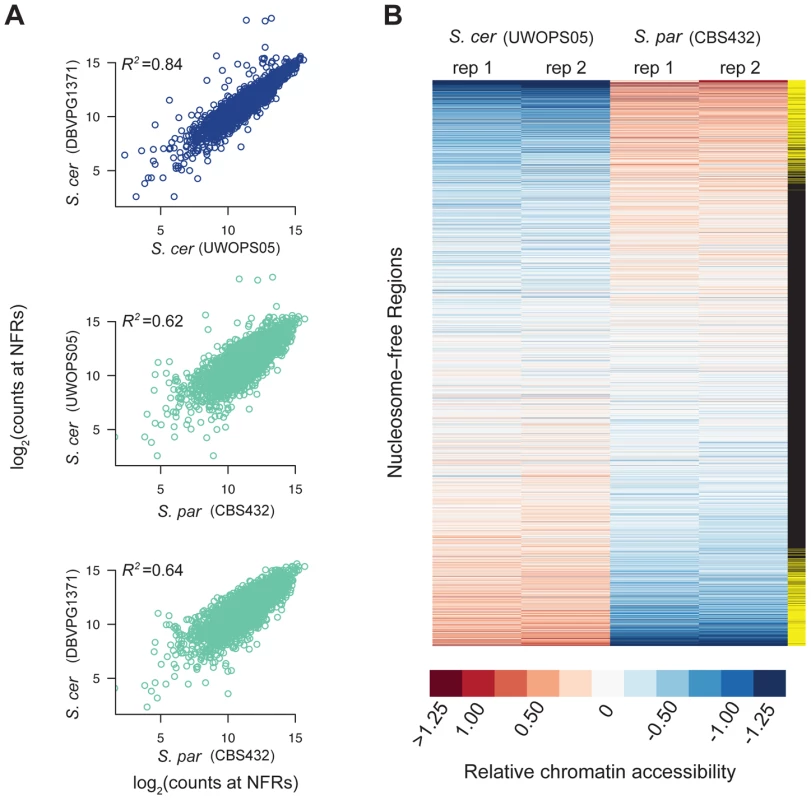
A. Scatterplots of relative chromatin accessibility between S. cerevisiae strains DBVPG1373 and UWOPS05_217_3 (top), S. cerevisiae strain UWOPS05_217_3 and S. paradoxus strain CBS432 (middle), and S. cerevisiae strain DBVPG1373 and S. paradoxus strain CBS432 (bottom). Note, comparisons within and between species are shown as blue and light green, respectively. B. Heatmap representation of chromatin accessibility at all NFRs in S. cerevisiae strain UWOPS05_217_3 versus S. paradoxus strain CBS432. Each row is a NFR, and columns are the two biological replicates of S. cerevisiae strain UWOPS05_217_3 and S. paradoxus strain CBS432. Rows are sorted by average difference in signal at NFRs between S. cerevisiae and S. paradoxus. The far right column indicates if the difference in chromatin accessibility between species is significant (yellow rectangles). To assess the robustness of these results, we also generated FAIRE-Seq data for a second strain of S. cerevisiae (DBVPG1373, a wine strain). Divergence at synonymous sites between these species is estimated to be 0.29 [37]. Levels of chromatin accessibility in NFRs were highly similar between the two S. cerevisiae strains (Figure 1; R2 = 0.84; p<2.2×10−16), and of similar magnitude between species (Figure 1; mean R2 = 0.63; p<2.2×10−16). Similarly, of the 947 NFRs that showed differential accessibility between UWOPS05_217_3 and CBS432, 515 were also significantly different between DBVPG1373 and CBS432. Thus, patterns of chromatin accessibility are highly reproducible between genetically diverse strains of S. cerevisiae and S. paradoxus.
Genetic architecture of chromatin differences
To better understand the genetic architecture of the widespread differences in chromatin accessibility observed between S. cerevisiae and S. paradoxus, we developed novel statistical tests for the presence of cis and trans effects (see Methods; Figure 2). Simulations showed that these tests had high power and maintained correct false positive rates over a range of parameters (see Methods; Table S1). Briefly, we tested for allele-specific chromatin accessibility within the hybrid to identify cis effects and tested for differences between the ratio of chromatin accessibility in the two parental species and the ratio of chromatin accessibility observed in the hybrid to identify trans effects (Figure 2). Over 99% of all NFRs identified in the parental strains contained one or more variants (median = 32) and could therefore be assessed for cis and trans effects. We identified 2,256 NFRs showing a significant cis effect (posterior probability >0.95, see Figure 3A) and 1,020 NFRs showing a significant trans effect (posterior probability >0.95, see Figure 3B). Interestingly, 782 NFRs showed both significant cis and significant trans effects. Cis effects were both more numerous as well as of greater magnitude on average compared to trans effects (1.8 and 1.6-fold difference in chromatin accessibility for cis and trans effects, respectively; Mann Whitney test, p<2.2×10−16, Figure 3C). Strikingly, we found that cis and trans effects were negatively correlated (r = −0.32, p<1×10−16), which suggests a widespread role for compensatory evolution to stabilize chromatin structure (Figure 3D).
Fig. 2. Schematic of approach to detect cis and trans effects on chromatin accessibility. 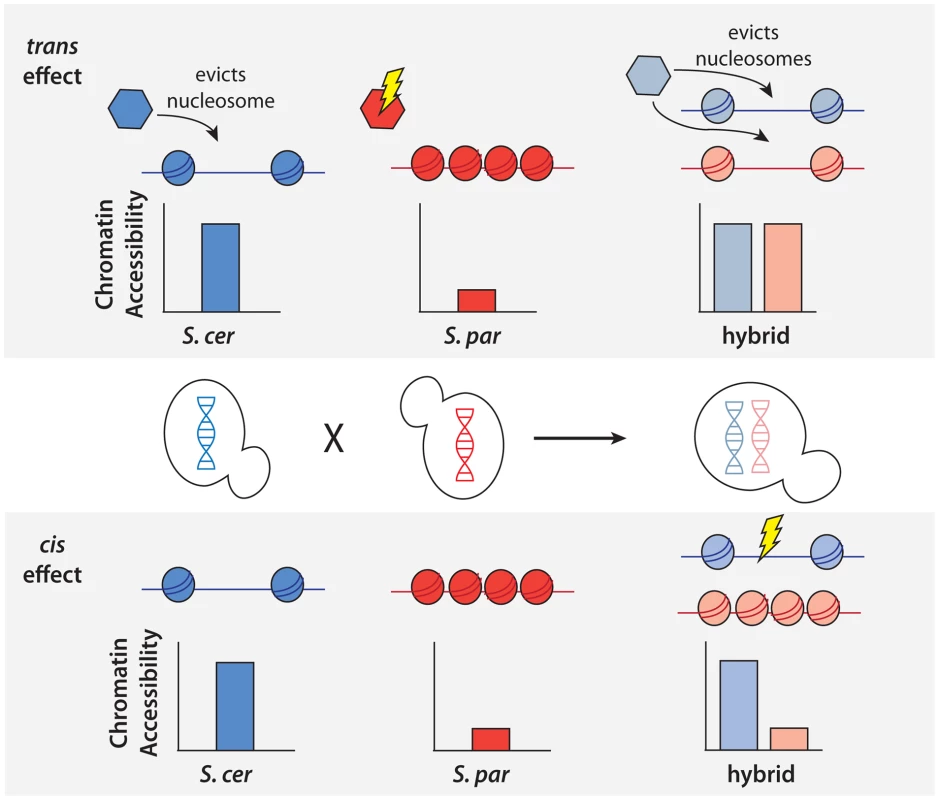
Top, an example of a NFR showing only a trans effect on chromatin accessibility. A trans effect is detected as a case where there is a difference in chromatin accessibility between the two parental haploid species, but there is no difference in chromatin accessibility between the two alleles in the hybrid. As shown above, this could be explained by a case where a nucleosome remodeler (shown as a hexagon) acts to evict nucleosomes and increase accessibility in S. cerevisiae, but a mutation in S. paradoxus has rendered it inactive and it is unable to evict the nucleosomes. In the diploid hybrid, the chromatin remodeler from S. cerevisiae is able to evict nucleosomes from both the S. cerevisiae and S. paradoxus chromosomes. Bottom, an example of a NFR showing only a cis effect on chromatin accessibility. A cis effect is detected as a difference between the accessibility detected between the two alleles in the diploid, and the lack of a trans effect is shown by the same difference being detected between the parental species. In this case, there has been a mutation at the NFR on the S. cerevisiae allele, leading to a difference in the number of nucleosomes binding in the region. Fig. 3. Cis and trans effects on chromatin accessibility. 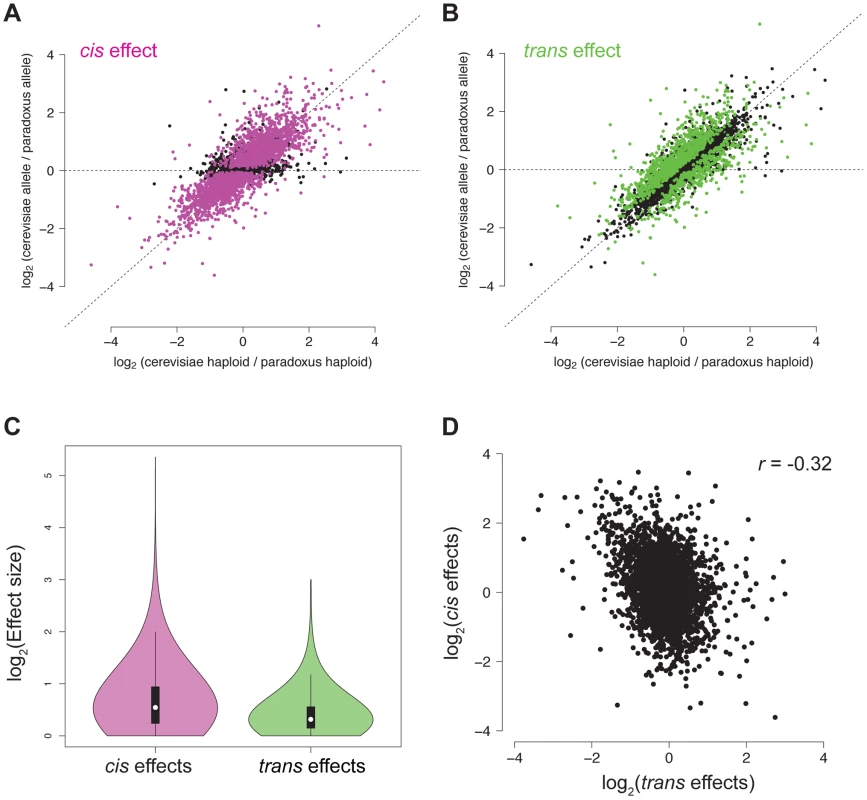
A. For each NFR, the relative chromatin accessibility in the haploid is plotted versus the relative chromatin accessibility in the diploid. NFRs with a significant cis effect are shown in pink. B. Reproduction of the plot from (A), but NFRs with a significant trans effect are shown in green. C. Violin plot showing the effect size distribution of cis and trans effects. D. Scatter plot of relative cis and trans effect sizes. Positive effects indicate higher accessibility in S. cerevisiae and negative effects indicate higher accessibility in S. paradoxus. Disrupted motifs are associated with cis effects
To test the hypothesis that cis-acting chromatin QTL result from variation in regulatory motifs, we identified motifs independently in the two species and computationally inferred whether sequence differences abrogated motif usage. Specifically, we define disrupted motifs as those that were called in only one of the two species (see Methods). Disrupted motifs were strongly enriched in NFRs with significant cis-acting chromatin QTL (p = 2.4×10−7). We also found that overall nucleotide divergence was higher at NFRs with significant cis effects compared to regions without significant cis effects (Mann Whitney test, p = 3.48×10−6). Note, this observation parallels previous findings that polymorphism is higher for genes that show significant allele-specific expression in S. cerevisiae hybrids [44].
We next asked if any of the 106 motifs were overrepresented for being disrupted in the set of significant cis-acting chromatin QTL. We found two overrepresented motifs, GCN4 and GZF3 (FDR = 0.10; Figure 4A). GCN4 is an activator of amino acid biosynthetic genes, which itself is a tightly regulated pathway [45]. GZF3 is a negative regulator of nitrogen catabolic gene expression [46]. While it is not immediately clear why disruption of these two genes is associated with changes in chromatin structure, it is interesting that both play an important role in metabolism, which is a highly regulated process.
Fig. 4. Motifs contributing to cis and trans effects. 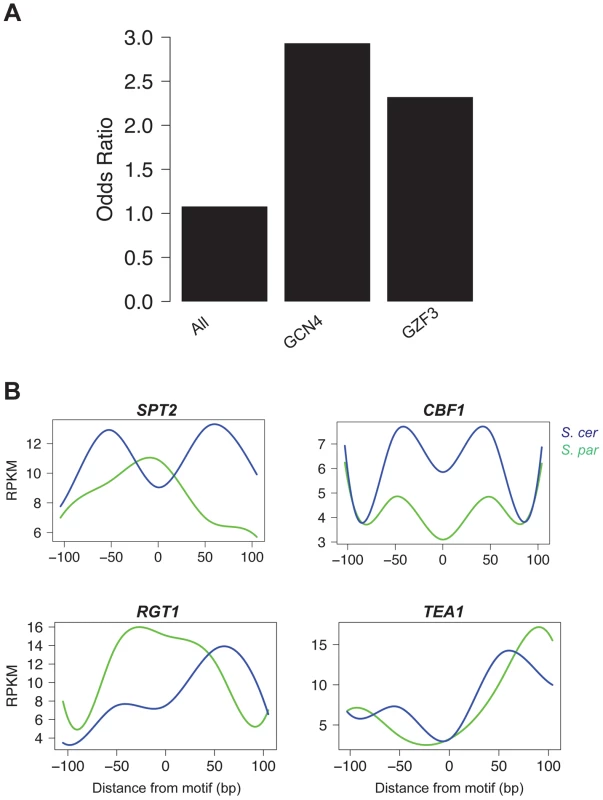
A. The odds ratio of observing a disrupted motif compared to a non-disrupted motif in NFRs with a significant cis effect. Odds ratios are shown for all motifs, as well as the two individual motifs (GCN4 and GZF3) that were found to be significant by permutations (FDR = 0.10). B. Pattern of accessibility for four motifs found within trans effect NFRs that vary between S. cerevisiae and S. paradoxus. Differential footprints for certain DNA binding factors found at trans effects loci
To identify factors contributing to trans effects, we searched for cases where there was no disruption to the motif but the occupancy of the site changed between species. Such patterns could result from mutations that either alter the binding specificity of a trans-acting regulatory protein or change its regulation. We used the FAIRE data surrounding each motif to determine occupancy, analogous to a DNase I footprint [39]. We then tested whether there was a significant difference in the pattern of occupancy between species by fitting splines to the mean occupancy across conserved sites in trans regions and testing whether the splines were significantly different in a 100 bp window surrounding the motif using bootstrapping (see Methods). We identified four motifs whose pattern of occupancy had significantly (p<0.05) changed between species (Figure 4B). SPT2, a transcription factor that interacts with histones and the SWI/SNF complex, showed a clear footprint in S. paradoxus, but nearly the opposite pattern in S. cerevisiae, implying decreased occupancy in S. cerevisiae at these trans regions. Similarly, TEA1, a Ty enhancer activator, and RGT1, a glucose-responsive transcription factor, showed increased occupancy in S. paradoxus. Conversely, CBF1, a centromere binding factor also involved in stress response, showed higher FAIRE signals in S. paradoxus than S. cerevisiae, implying increased occupancy in S. cerevisiae.
Effects on gene expression
To examine the relationship between differences in chromatin accessibility and transcriptional divergence between S. cerevisiae and S. paradoxus, we performed RNA-Seq on the haploid parents and interspecific hybrid and tested for the cis and trans effects on gene expression values. Out of the 4,899 genes that could be aligned between species, 4,181 exhibited significant cis effects and 3,117 showed significant trans effects. Overall, cis and trans effects on gene expression levels were smaller than those on chromatin accessibility, (Spearman rank-sum test, p<2.2×10−16 for both cis and trans effects, Figure 5A).
Fig. 5. Gene expression and chromatin accessibility. 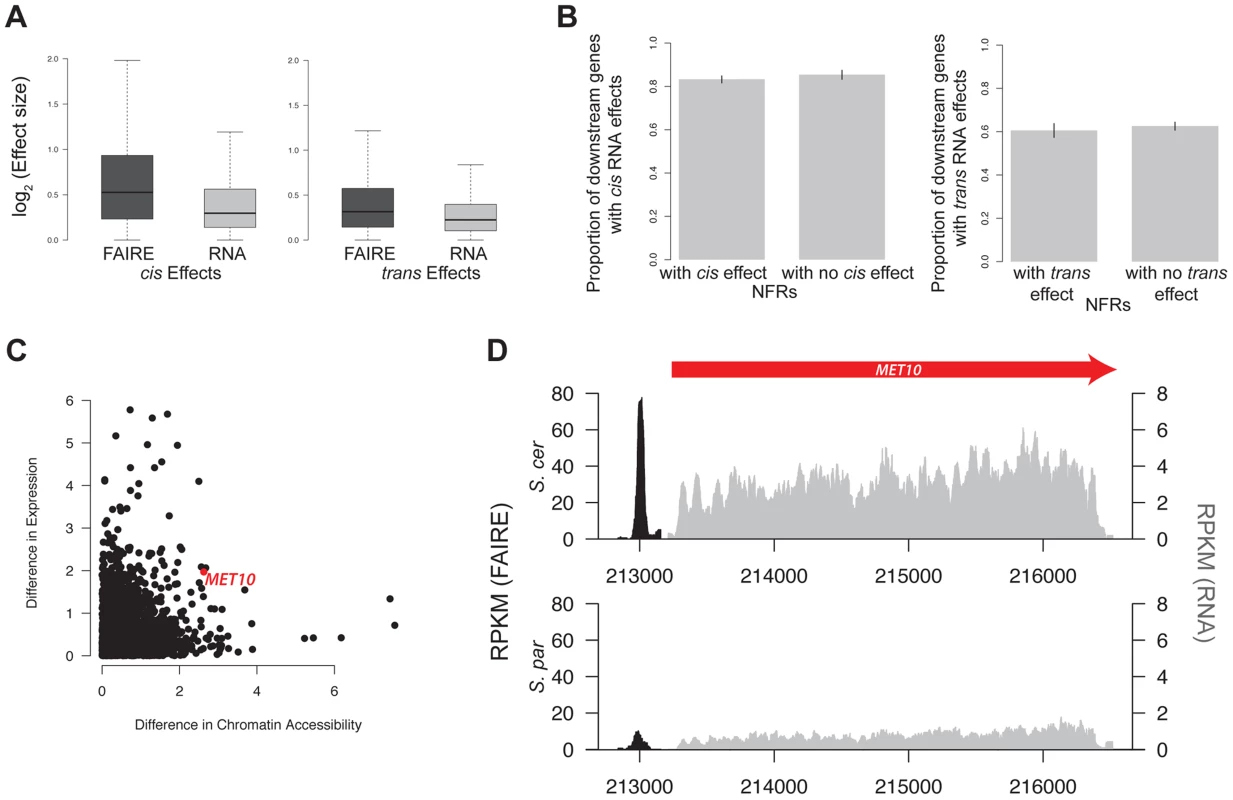
A. Boxplot of log2(effect size) of both cis and trans effects for FAIRE (dark grey) and RNA (light grey). B. Barplot of the percentage of genes with significant cis effects in RNA that are downstream of NFRs with and without cis effects (left). Barplot of the percentage of genes with significant trans effects in RNA that are downstream of NFRs with and without trans effects (right). C. Scatterplot of the log2(absolute value of the difference in chromatin accessibility between the two species) vs log2(absolute value of the difference in expression between the two species. The red dot indicates data from the MET10 gene, whose FAIRE-Seq and RNA data are shown in panel D. For clarity, the FAIRE-Seq data is only shown in a 100 bp window on either side of the NFR. FAIRE signal is shown in black, and RNA signal is shown in grey. We next tested whether genes with a significant cis or trans effect in chromatin were more likely to have a significant cis or trans effect in transcript abundance. Specifically, we divided genes into categories of those downstream of an NFR with a cis effect on chromatin versus those downstream of an NFR without a cis effect on chromatin. We then compared the percentage of genes showing cis effects on RNA in these two categories. Surprisingly, we did not find evidence that cis or trans effects in NFRs were more likely to be upstream of cis or trans effects on RNA, as would be expected if there was a simple correspondence between cis and trans effects in NFRs and RNA (see Figure 5B, Table S2). This was true even when using varying cutoffs for the cis and trans effects, including ones that took into account the magnitude of effect sizes (Table S2).
The relationship of cis and trans effects observed in gene expression and chromatin structure may be complicated by differences in statistical power. For example, 85% of all genes show significant cis effects on RNA. Thus, even if cis effects in NFRs are not more likely to be found upstream of cis effects on RNA, they could still contribute to gene expression variation between S. cerevisiae and S. paradoxus. To this end, we assessed whether expression differences between species could be modeled as a function of the cis and trans effects found upstream of each gene. Specifically, we fit the simple linear model: expression difference = Intercept + cis effect + trans effect + cis * trans effect + error, using the lm function in R. We found that both cis effects and trans effects on chromatin were significantly related to expression differences between species (p = 0.002, p = 4.18×10−5 respectively) though they explained a very small proportion of the total variance in expression between species (0.8% combined). The interaction term of cis and trans effects was not significant (p>0.05). The motif for GZF3, which is significantly overrepresented in cis NFRs, was overrepresented in cis NFRs upstream of genes with cis effects on RNA.
Finally, we found no significant correlation between the magnitude of differences in chromatin accessibility and differences in gene expression between the parental species (Spearman rank-sum test, p = 0.11, Figure 5C). However, for a subset of NFRs, differences in chromatin accessibility and gene expression do appear to be highly correlated. To identify these regions, we compared the log2(S. paradoxus/S. cerevisiae) for NFRs and gene expression at downstream genes and identified those whose absolute value of the difference between the two ratios was less than 0.25. We identified 701 such regions; one example is shown in Figure 5D.
Discussion
The ability to assay chromatin accessibility at high-resolution and on a genome-wide scale has enabled comprehensive insights into the structure and function of chromatin in many cell types, developmental stages, and organisms. Here, we were particularly interested in the evolutionary dynamics of changes in chromatin accessibility between two closely related yeast species. Broad-scale patterns of chromatin accessibility have been well conserved between S. cerevisiae and S. paradoxus (Figure 1), but superimposed on this background of conservation, we estimate that nearly 50% of NFRs exhibit differential accessibility.
To better understand the relative contributions of cis and trans effects on differences in chromatin accessibility observed between S. cerevisiae and S. paradoxus, we developed novel statistical methods to analyze FAIRE-Seq data from diploid hybrids. Similar to previous findings on RNA levels [17], [21], [22], [25], differences in chromatin accessibility are caused by changes both in cis and in trans. In our data, cis effects were of greater magnitude and were more abundant. Recently, Lee et al. performed a study similar to ours and assessed cis and trans effects on chromatin structure in a cross between two strains of S. cerevisiae [34]. In contrast to our observations, they found that trans QTL were more pervasive than cis QTL (92.1% of associations versus 7.9% of associations) [34]. We hypothesize that these disparate observations are primarily the consequence of differences in the evolutionary trajectory of chromatin accessibility QTL in within versus between species data. In particular, trans-acting chromatin QTL are likely to be subject to more intense purifying selection due to their potential pleiotropic effects, and tend to be eliminated over longer time periods [47]. This hypothesis is consistent with findings for expression QTL studies, which showed that trans-eQTL were more common within species and cis-eQTL were more common between species [22], [23]. Consistent with this hypothesis, we found that cis and trans effects were significantly negatively correlated, indicating that chromatin accessibility in each species is subject to stabilizing selection and perturbations of chromatin structure are, on average, deleterious.
We estimated cis and trans effects for both chromatin accessibility and gene expression levels. Unexpectedly, the presence of cis or trans effects on chromatin accessibility in NFRs was not significantly associated with cis or trans effects on RNA. In other words, gene expression levels with significant cis or trans effects were not more likely to have an NFR with significant cis or trans effects on chromatin accessibility. Thus, it appears that many of the changes in chromatin accessibility in NFRs between S. cerevisiae and S. paradoxus do not necessarily have transcriptional consequences. One factor that may contribute to this observation is that compensatory changes downstream of chromatin accessibility, such as mutations that influence mRNA stability, may evolve to maintain levels of gene expression. In addition, many changes in chromatin accessibility may simply be functionally benign.
Furthermore, an important caveat is that our data was obtained from a single environmental condition, and it is plausible that stronger correlations between chromatin and gene expression QTL may exist when analyzing data from either a different environment or across multiple environments. Nonetheless, the lack of a clear relationship between chromatin and gene expression QTL in our data is interesting in light of recent observations from the ENCODE Project [48] that have found a large proportion of the human genome has reproducible biochemical activity. Our results suggest caution in assuming all, or perhaps even most, of such sequences are functionally important.
Materials and Methods
Strain growth, FAIRE, and RNA-Seq
65 ml of each of 2 biological replicates of the S. paradoxus strain CBS432 and the two S. cerevisiae strains DBVPG1373 and UWOPS05_217_3 were grown to mid-log phase. 15 ml were used for RNA-seq and 50 ml were used for FAIRE. We performed FAIRE as described in Simon et al. [40], with some modification. The cells were fixed with 1% formaldehyde for 35 minutes with mixing. Cells were sonicated using the Fisher Scientific Sonic Dismembrator Model 100 for three cycles of 15 one-second bursts with 1 second rest in between, keeping the cells on ice for at least 30 seconds between cycles. The remainder of the protocol was followed as in Simon et al. [40]. RNA isolation was performed using the hot phenol protocol [49], and RNA was treated with Turbo DNAse before library construction.
Library construction and sequencing
Libraries were constructed for the FAIRE samples using the Illumina TruSeq DNA kit, starting with approximately 200 ng FAIRE DNA, following their standard kit protocol but omitting the fragmentation step. RNA libraries were prepared using the Illumina TruSeq RNA kit, following their standard protocol. Libraries were pooled into two lanes, one for the FAIRE samples and one for the RNA samples, and were sequenced on the HiSeq 2000. Raw sequence data and processed files are available at the GEO database with accession number GSE55717.
Read mapping
Reads were mapped to genomes assembled in Skelly et al. [50] for the S. cerevisiae haploid samples using bwa and samtools [51], [52]. For the S. paradoxus strain CBS432, we used the last updated reference version from the SGRP [53]. For the diploid samples, we mapped to a combined FASTA containing both genomes. We tested whether mapping to each genome separately for the diploid samples resulted in increased mapping; it did not. For the diploid samples, we generated simulated reads and mapped to the combined FASTA. For all further analyses, we restricted analysis to NFRs for which greater than 90 percent of simulated reads mapped back to the correct region. We also sequenced a genomic DNA sample. We also filtered out NFRs where the absolute value of the log2(ratio of reads between the two species) for the genomic DNA was greater than 0.3.
Identifying NFRs
We identified NFRs as follows: specifically, starting at the beginning of the coding region of the gene, we looked for the peak of chromatin accessibility within 300 bp upstream of the start codon. We then defined the edges of the NFR as the base-pair after which at least 3 bases had had a chromatin accessibility count of less than 10. We did this separately for each biological replicate and each species. For each gene separately, we then merged NFRs if they were within 200 bp.
Filtering NFRs and genes
In order to convert between the two species coordinates, we created a multiple alignment between the two species using LASTZ and TBA [54], [55]. We inferred scoring parameters using the two species of interest. Using this multiple alignment, we then converted the NFRs called in CBS432 to S. cerevisiae coordinates, and found the union of all NFRs called across the samples. We used this union of NFRs for further tests. We also filtered the NFRs based on a reciprocal alignment filter, where we required that NFRs align to only one region in the other species, based on the multiple alignment. This allowed us to filter out regions with duplications or deletions between the two species.
Identifying differentially accessible NFRs
Using samtools, we summed the count of reads mapping in each species across each NFR or gene in both biological replicates. Note that we did this in the native coordinates for each species, filtering out sites that were called as indels in the multiple alignment. We then used the R package DESeq [56] to assess differential FAIRE signal between species. This method takes into account biological replicates, and models the count distribution using a negative binomial distribution. We used the R package qvalue [42] to estimate q-values. We used a significance threshold of FDR = 0.05 unless otherwise noted.
Statistical model to detect trans effects
If differences in chromatin accessibility between S. cerevisiae and S. paradoxus are due to trans-acting factors, the relative chromatin accessibility in the haploid parents will be different than the relative chromatin accessibility in the diploid hybrid (Fig. 2). We leveraged the FAIRE-Seq data to detect differences in the relative levels of chromatin accessibility between F1 hybrids and the parental species. Specifically, let Nc and Np be the total number of reads across the genome mapping to polymorphic sites in the S. cerevisiae and S. paradoxus haploid parents, respectively. For a particular locus j, Yc and Yp denote the observed number of reads mapping to S. cerevisiae and S. paradoxus, respectively. Then assume: Yc|rc∼Binomial (Nc, rc) and Yp|rp∼Binomial(Np, rp), where rc and rp denote the probabilities of observing a read mapping to S. cerevisiae or S. paradoxus for a particular locus, respectively. Since Nc and Np are large, and rc and rp are small, we can approximate these binomials by Poissons to give: Yc|rc∼Poisson(Nc rc) and Yp|rp∼Poisson(Np rp).
We define θP = rc/rp to be the ratio of these probabilities in the parents and R = Nc/Np to be the ratio of the total numbers of reads in each parent. Then, Yc|Yc+Yp, sc∼Binomial(Yc+Yp, sc), where sc = Ncrc/(Ncrc+Nprp) = RθP/(RθP+1) is the probability of observing a read map to S. cerevisiae, without adjusting for differences in the total number of reads mapping to each species. We can thus write log(Sc/1−Sc) = log R+log θP, such that θP is the odds of observing a read map to S. cerevisiae compared to S. paradoxus for a particular locus in the haploid parents, adjusted for differences in the total number of reads mapping to each species.
For the diploid hybrid, let Zc and Zp denote the number of reads mapping to S. cerevisiae and S. paradoxus SNPs within locus j, respectively. Thus, Zc|Zc+Zp, pc∼Binomial(Zc+Zp, pc), where pc is the probability of observing a read map to the S. cerevisiae allele for a particular locus. The odds of observing a read map to S. cerevisiae in the hybrid for a particular gene is θH = pc/(1−pc). In the following, let Ycj, Ypj, Zcj, and Zpj represent the data as defined above, but with j = [1], [2] indexing biological replicate.
Thus, the locus specific models are:
Ycj|Ycj+Ypj, scj∼Binomial(Ycj+Ypj, scj),
Zcj|Zcj+Zpj, pcj∼Binomial(Zcj+Zpj, pcj)
logit scj = log Rj+log θP+δj
logit pcj = log θP+Δ+εj
where Rj = Ncj/Npj, δj∼N(0, σ2) and εj∼N(0, σ2) represent random effects that allow for excess-binomial variation. Here, Δ is the parameter of interest and provides an estimate of the difference between log(θP) and log(θH), as described above. The above framework is an example of a generalized linear mixed model (GLMM) and we used a Bayesian approach to inference with relatively flat hyperpriors. One computationally intensive method for summarizing the posterior would be Markov chain Monte Carlo (MCMC) but the integrated nested Laplace approximation (INLA) as described in [57] provides an efficient alternative for GLMMs [58]. We used the R implementation of INLA to estimate Δ. We examined a 95% posterior interval estimate for Δ and recorded whether this interval contained 0 or not. If the interval does not contain 0 it indicates that chromatin accessibility differs.
Statistical model to detect cis effects
To detect cis effects, we developed a model to test for differential accessibility between alleles within the diploid hybrid. Let Zcj, and Zpj represent the data as defined above. We can therefore write:
Zcj|Zcj+Zpj, pcj∼Binomial(Zcj+Zpj, pcj)
logit pcj = log θH+εj
with εj∼N(0,σ2) representing random effects that allow for excess-binomial variation. In this model, θH is the parameter of interest and provides an estimate of the odds of a read mapping to the S. cerevisiae allele compared to the S. paradoxus allele in the diploid hybrid for a particular gene. We again used the R program INLA to estimate the posterior for log(θH) and in particular examine whether the 95% posterior interval estimate contains 0.
Simulations
We carried out extensive simulations to evaluate the operating characteristics of our model. Specifically, for the trans model, we set the total number of reads mapping to polymorphic sites for species 1 (Nc1) equal to 5×106, and drew the total number of reads mapping to polymorphic sites for the other species and replicate from a normal distribution with mean Nc1 and standard deviation Nc1. We then drew the value for rc, the probability of a read mapping to S. cerevisiae for a particular locus from an exponential distribution with rate 10,000. For Nc1 = 5×106, this results in a mean of 500 reads mapping to a locus, with most having less than 500 reads, consistent with the observed data. We drew the value for rp, the probability of a read mapping to S. paradoxus for a particular locus, from a normal distribution with mean rc and standard deviation rc and took the absolute value to ensure rp was greater than zero. Using these values, we derived Yc and Yp, the number of reads mapping to S. cerevisiae and S. paradoxus, respectively, for a particular locus, for two biological replicates as specified by the model. For Zc and Zp, the number of reads mapping to the S. cerevisiae and S. paradoxus alleles in the hybrid summed across polymorphic sites in a particular locus, we either derived these using the same rc and rp values as above, to simulate a locus which showed no trans effect, or we set the value of log2(θP)−log2(θH) equal to 0.1, 0.5, or 0.8, to simulate a locus with a trans effect. Note, this spans the range of detected trans effects. For 100 replicates, we simulated a collection of 6000 loci, 5000 of which did not show a trans effect and 1000 or which did show a trans effect. For each of the 100 replicates, we then used the method described above to test whether the 95% posterior interval estimate for Δ for each locus contained zero.
To evaluate the cis test, we again started with the same values for the total number of reads. To simulate a locus with no cis effect, we set the value of log2(Zc/Zp) equal to zero, and to simulate a locus with a cis effect, we set the value of log2(Zc/Zp) equal to 0.1, 0.5, or 0.8. Again, for 100 simulations, we simulated a collection of 6000 loci, 5000 showing no cis effect and 1000 showing a cis effect. For each simulated set of loci, we then used the statistical method above to test whether the 95% posterior interval estimate for log(θH) for each locus contained zero to test for a significant cis effect.
We found that the false discovery rate for both cis and trans based on a test based on a 95% interval was 0.05. Moreover, we found that the trans test has reduced power compared to the cis test, as expected because there were more parameters that could vary across biological replicates. However, with an effect size = 0.5 for both the cis and trans tests, there was significant power to detect the cis or trans effects (Table S1).
Motif analysis
We called motifs separately in both species, using MEME, using their standard p value cutoff of p<10−4 [59]. This results in the same cutoffs used for both species. Motifs that were not called in both species were considered polymorphic. We filtered out motifs where the polymorphism was due to indels in order to mitigate alignment errors. The motif calls used for this analysis are available as supplementary data on our website (http://akeylab.gs.washington.edu/downloads.shtml). We compared the proportion of disrupted motifs (those that were called in only one species) in cis NFRs to non-cis NFRs using the Fisher exact test. We determined significance by permutations; we permuted the assignment of cis or not cis NFRs 1000 times and obtained p values from the permutations. We then used the positive False Discovery Rate approach to determine significance [42].
Occupancy at trans NFRs
We obtained the RPKM in a 200 bp window surrounding motifs that were conserved across species in trans NFR regions for each of the two species. We filtered out motifs that did not have at least five instances of conserved motifs. We fit a cubic smoothing spline to the mean coverage using the R function spline. We then bootstrapped the data 1000 times by resampling from the motifs for each species. At five bp intervals across the region, we then tested whether the coverage was significantly different between the species, using the confidence intervals obtained from the bootstrapping. We then manually inspected the significant motifs (p<0.05) to identify those which appeared to affect the FAIRE signal at or the near the motif.
Supporting Information
Zdroje
1. BrittenRJ, DavidsonEH (1971) Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol 46 : 111–38.
2. KingMC, WilsonAC (1975) Evolution at two levels in humans and chimpanzees. Science 188 : 107–16.
3. WrayGA (2007) The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics 8 : 206–16 doi:10.1038/nrg2063
4. ShapiroMD, MarksME, PeichelCL, BlackmanBK, NerengKS, et al. (2004) Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. 428 : 717–23 Nature 428 : 717–23.
5. AkeyJ, RuheA, AkeyD, WongA, ConnellyC, et al. (2010) Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci USA 107 : 1160–1165.
6. MouC, PitelF, GourichonD, VignolesF, TzikaA, et al. (2011) Cryptic Patterning of Avian Skin Confers a Developmental Facility for Loss of Neck Feathering. PLoS Biol 9: e1001028 doi:10.1371/journal.pbio.1001028.g007
7. SkellyDA, RonaldJ, AkeyJM (2009) Inherited variation in gene expression. Annu Rev Genomics Hum Genet 10 : 313–332 doi:10.1146/annurev-genom-082908-150121
8. EnattahNS, SahiT, SavilahtiE, TerwilligerJD, PeltonenL, et al. (2002) Identification of a variant associated with adult-type hypolactasia. Nature Genet 30 : 233–237.
9. TishkoffSA, ReedFA, RanciaroA, VoightBF, BabbittCC, et al. (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet 39 : 31–40.
10. TournamilleC, ColinY, CartronJP, Le Van KimC (1995) Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nature Genet 10 : 224–228.
11. HamblinMT, Di RienzoA (2000) Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66 : 1669–1679.
12. AbzhanovA, ProtasM, GrantBR, GrantPR, TabinCJ (2004) Bmp4 and morphological variation of beaks in Darwin's finches. Science 305 : 1462–5.
13. ViselA, RubinEM, PennacchioLA (2009) Genomic views of distant-acting enhancers. Nature 461 : 199–205 doi:10.1038/nature08451
14. MauranoMT, HumbertR, RynesE, ThurmanRE, HaugenE, et al. (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337 : 1190–5 doi:10.1126/science.1222794
15. PrimigM, WilliamsRM, WinzelerEA, TevzadzeGG, ConwayAR, et al. (2000) The core meiotic transcriptome in budding yeasts. Nat Genet 26 : 415–23 doi:10.1038/82539
16. SandbergR, YasudaR, PankratzDG, CarterTA, Del RioJA, et al. (2000) Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci USA 97 : 11038–43.
17. BremRB, YvertG, ClintonR, KruglyakL (2002) Genetic dissection of transcriptional regulation in budding yeast. Science 296 : 752–5 doi:10.1126/science.1069516
18. KhaitovichP, HellmannI, EnardW, NowickK, LeinweberM, et al. (2005) Parallel Patterns of Evolution in the Genomes and Transcriptomes of Humans and Chimpanzees. Science 309 : 1850–1854 doi:10.1126/science.1108296
19. PickrellJK, MarioniJC, PaiAA, DegnerJF, EngelhardtBE, et al. (2010) Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464 : 768–772 doi:10.1038/nature08872
20. TsankovAM, ThompsonDA, SochaA, RegevA, RandoO (2010) The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol 8: e1000414 doi:10.1371/journal.pbio.1000414
21. WittkoppPJ, HaerumBK, ClarkAG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430 : 85–8 doi:10.1038/nature02698
22. TiroshI, ReikhavS, LevyA, BarkaiN (2009) A Yeast Hybrid Provides Insight into the Evolution of Gene Expression Regulation. Science 324 : 659–662 doi:10.1126/science.1169766
23. EmersonJ, HsiehL, SungH, WangT, HuangC, et al. (2010) Natural selection on cis and trans regulation in yeasts. Genome Res 20 : 826–836 doi:10.1101/gr.101576.109
24. SkellyD, JohanssonM, MadeoyJ, WakefieldJ, AkeyJ (2011) A powerful and flexible statistical framework for testing hypotheses of allele-specific gene expression from RNA-seq data. Genome Res 21 : 1728–1737 doi:10.1101/gr.119784.110
25. WittkoppPJ, HaerumB, ClarkAG (2008) Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40 : 346–350 doi:10.1038/ng.77
26. HanM, GrunsteinM (1988) Nucleosome loss activates yeast downstream promoters in vivo. Cell 55 : 1137–45.
27. GrossDS, AdamsCC, LeeS, StentzB (1993) A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO 12 : 3931–45.
28. BirneyE, LiebJD, FureyTS, CrawfordGE, IyerVR (2010) Allele-specific and heritable chromatin signatures in humans. Hum Mol Genet 19: R204–R209 doi:10.1093/hmg/ddq404
29. GossettAJ, LiebJD (2012) In Vivo Effects of Histone H3 Depletion on Nucleosome Occupancy and Position in Saccharomyces cerevisiae. PLoS Genet 8: e1002771 doi:10.1371/journal.pgen.1002771.g005
30. ThurmanRE, RynesE, HumbertR, VierstraJ, MauranoMT, et al. (2012) The accessible chromatin landscape of the human genome. Nature 489 : 75–82 doi:10.1038/nature11232
31. StergachisAB, NephS, ReynoldsA, HumbertR, MillerB, et al. (2013) Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell 154 : 888–903 doi:10.1016/j.cell.2013.07.020
32. McDaniellR, LeeB, SongL, LiuZ, BoyleA, et al. (2010) Heritable Individual-Specific and Allele-Specific Chromatin Signatures in Humans. Science 328 : 235–239 doi:10.1126/science.1184655
33. KasowskiM, Kyriazopoulou-PanagiotopoulouS, GrubertF, ZauggJB, KundajeA, et al. (2013) Extensive Variation in Chromatin States Across Humans. Science 342 : 750–2 doi:10.1126/science.1242510
34. LeeK, KimSC, JungI, KimK, SeoJ, et al. (2013) Genetic landscape of open chromatin in yeast. PLoS Genet 9: e1003229 doi:10.1371/journal.pgen.1003229
35. ShibataY, SheffieldN, FedrigoO, BabbittC, WorthamM, et al. (2012) Extensive Evolutionary Changes in Regulatory Element Activity during Human Origins Are Associated with Altered Gene Expression and Positive Selection. PLoS Genet 8: e1002789 doi:10.1371/journal.pgen.1002789.g007
36. DegnerJF, PaiAA, Pique-RegiR, VeyrierasJ, GaffneyDJ, et al. (2012) DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482 : 390–4 doi:10.1038/nature10808
37. KellisM, PattersonN, EndrizziM, BirrenB, LanderES (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 : 241–54 doi:10.1038/nature01644
38. HoganGJ, LeeCK, LiebJD (2006) Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet 2: e158 doi:10.1371/journal.pgen.0020158
39. HesselberthJR, ChenX, ZhangZ, SaboPJ, SandstromR (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6 : 283–9 doi:10.1038/nmeth.1313
40. SimonJM, GiresiPG, DavisIJ, LiebJD (2013) A detailed protocol for formaldehyde-assisted isolation of regulatory elements (FAIRE). Curr Protoc Mol Biol Chapter 21: Unit 21.26 doi:10.1002/0471142727.mb2126s102
41. RandoO, ChangH (2009) Genome-Wide Views of Chromatin Structure. Annu Rev Biochem 78 : 245–271 doi:10.1146/annurev.biochem.78.071107.134639
42. StoreyJD, TibshiraniR (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 : 9440–5 doi:10.1073/pnas.1530509100
43. MedinaI, CarbonellJ, PulidoL, MadeiraSC, GoetzS, et al. (2010) Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res 38: W210–3 doi:10.1093/nar/gkq388
44. RonaldJ, BremRB, WhittleJ, KruglyakL (2005) Local regulatory variation in Saccharomyces cerevisiae. PLoS Genet 1: e25 doi:10.1371/journal.pgen.0010025
45. HinnebuschA, NatarajanK (2002) Gcn4p, a Master Regulator of Gene Expression, Is Controlled at Multiple Levels by Diverse Signals of Starvation and Stress. Eukaryot Cell 1 : 22–32 doi:10.1128/EC.01.1.22-32.2002
46. StanbroughM, RowenDW, MagasanikB (1995) Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA 92 : 9450–4.
47. RonaldJ, AkeyJ (2007) The Evolution of Gene Expression QTL in Saccharomyces cerevisiae. PLoS One 2: e678 doi:10.1371/journal.pone.0000678.t001
48. ENCODE Project Consortium (2012) BernsteinBE, BirneyE, DunhamI, GreenED, et al. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 447 : 799–816 doi:10.1038/nature11247
49. Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. 198 p.
50. SkellyDA, MerrihewGE, RiffleM, ConnellyCF, KerrEO, et al. (2013) Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res 23 : 1496–1504 doi:10.1101/gr.155762.113
51. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760 doi:10.1093/bioinformatics/btp324
52. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–9 doi:10.1093/bioinformatics/btp352
53. LitiG, CarterDM, MosesAM, WarringerJ, PartsL, et al. (2009) Population genomics of domestic and wild yeasts. Nature 458 : 337–341 doi:10.1038/nature07743
54. Harris RS (2007) Improved pairwise alignment of genomic DNA [PhD thesis]. [University Park (PA)]: The Pennsylvania State University.
55. BlanchetteM (2004) Aligning Multiple Genomic Sequences With the Threaded Blockset Aligner. Genome Res 14 : 708–715 doi:10.1101/gr.1933104
56. AndersS, HuberW (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106.
57. PaulM, RieblerA, BachmannLM, RueH, HeldL (2010) Bayesian bivariate meta-analysis of diagnostic test studies using integrated nested Laplace approximations. Stat Med 29 : 1325–39 doi:10.1002/sim.3858
58. FongY, RueH, WakefieldJ (2010) Bayesian inference for generalized linear mixed models. Biostatistics 11 : 397–412.
59. BaileyTL, BodenM, BuskeFA, FrithM, GrantCE, et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–8 doi:10.1093/nar/gkp335
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



