-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAn Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
All living organisms use protein chaperones to prevent proteins from becoming insoluble either spontaneously or during cellular stress that can damage proteins. The HSP70 chaperone DnaK has been well characterized in E. coli and is important for that bacterium to resist protein denaturation from heat, but is dispensable for cell growth in the absence of stress due to redundancy with other chaperone systems. However, the function of chaperones in bacterial pathogens, which are exposed to protein stress within the host, has received less attention. Here we examine the function of DnaK in mycobacteria, a genus that includes multiple human pathogens, and find that DnaK is required for cell growth. This essential function is due to a lack of redundancy with other chaperone systems for the folding of proteins, even in the absence of stress. These findings expand the paradigm of DnaK function and identify DnaK as a promising target for antibiotic development for mycobacteria.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004516
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004516Summary
All living organisms use protein chaperones to prevent proteins from becoming insoluble either spontaneously or during cellular stress that can damage proteins. The HSP70 chaperone DnaK has been well characterized in E. coli and is important for that bacterium to resist protein denaturation from heat, but is dispensable for cell growth in the absence of stress due to redundancy with other chaperone systems. However, the function of chaperones in bacterial pathogens, which are exposed to protein stress within the host, has received less attention. Here we examine the function of DnaK in mycobacteria, a genus that includes multiple human pathogens, and find that DnaK is required for cell growth. This essential function is due to a lack of redundancy with other chaperone systems for the folding of proteins, even in the absence of stress. These findings expand the paradigm of DnaK function and identify DnaK as a promising target for antibiotic development for mycobacteria.
Introduction
Proper protein folding is essential for all organisms and assures that the primary sequence of the polypeptide forms its functional tertiary and quaternary structures. Protein chaperones are present in all domains of life and serve multiple functions in protein homeostasis. During translation, chaperones are required to assure proper protein folding and prevent protein aggregation, which can occur as hydrophobic segments of the protein emerge from the ribosome. After synthesis, protein denaturation is a common event due to exogenous proteotoxic stresses such as heat and oxidation, correction of which requires chaperone systems to refold denatured proteins when possible, and facilitate disaggregation and degradation when refolding is not possible. The importance of chaperone function for cellular viability is reflected in the frequent redundancy between chaperones for protein folding and aggregate processing [1]–[4].
The Hsp70 family of chaperones is widely distributed in both prokaryotic and eukaryotic cells [5]. The best studied Hsp70 chaperone of bacteria is E. coli DnaK. DnaK is a central hub for protein folding, shuttling misfolded peptides to other chaperones and proteases for resolution, a function that is essential during the protein denaturation that occurs during heat shock [6]–[9]. In addition to its effector function in the heat shock response, DnaK also regulates this response by destabilizing the alternative sigma factor, σ32, preventing aberrant induction of the heat shock response during non-stress conditions and turning off the response after heat shock [10]. However, In E. coli, DnaK is nonessential for native protein folding because of redundancy with Trigger Factor, which associates with proteins soon after emergence from the ribosome [11]. Although dnaK/tf double mutants are nonviable, overexpression of GroEL, SecB, or Hsp33 can suppress the synthetic lethality of dnaK/tig double mutants [10], [12]–[16]. Furthermore, examination of proteins that interact with DnaK indicates that most client proteins that require DnaK for proper folding and/or stability are largely non-essential, suggesting that loss of function of these proteins in the absence of DnaK does not impact viability [7]. However, in the absence of both DnaK and TF, the E. coli cell suffers proteostasis collapse characterized by global insolubility of nascent proteins [7].
In bacteria other than E. coli, the function of DnaK has not been studied extensively. In mycobacteria, DnaK regulates the heat shock response through its interaction with the HspR C terminal tail, which becomes insoluble upon heat shock, thereby relieving repression of chaperone genes [17]. The mycobacterial heat shock response is negatively regulated by the repressors HspR and HscA; and positively through σH [18], [19]. Deletion of hspR, which derepresses several chaperones involved in the heat shock response, including ClpB, alpha-crystalin, and DnaK-GrpE-DnaJ1, led to decreased persistence after the initial phase of infection in a mouse model suggesting that dnaK and other HspR regulated genes must be controlled during infection for optimal growth and persistence [19]. Host inflicted proteotoxic stress is likely a significant in vivo stress for M. tuberculosis during infection, yet the function of the mycobacterial chaperone network in native and stress induced proteostasis is incompletely understood. Additionally, Mtb DnaK is found in culture filtrates [20], [21] and on the bacterial surface [22], and has a role in pathogenesis by modulating host immune responses [22]–[24]. Despite substantial progress in targeting chaperone function in malignant human cells [25], [26], inhibition of chaperone function as an antimicrobial strategy is relatively unexplored, in part because of the redundancy of the chaperone network. Using a model mycobacterial species, M. smegmatis, we characterized the function of DnaK in mycobacteria and find an unanticipated lack of redundancy that places mycobacterial DnaK as a central chaperone in both native and stress induced protein folding.
Results
DnaK is essential for growth of M. smegmatis
To study the function of mycobacterial DnaK, we attempted to delete dnaK from the M. smegmatis chromosome. Initial attempts yielded no allelic replacements, suggesting that DnaK may be essential for viability. Provision of a second copy of dnaK at the attB phage integration site allowed replacement of the chromosomal dnaK with an unmarked ΔdnaK allele lacking the first 1765 bp of the 1869 bp dnaK ORF (Figure S1A). We then attempted to remove the second copy of dnaK from attB by marker exchange with either a vector, pMV306kan, or a plasmid encoding DnaK and conferring kanamycin resistance, pAJF447. Only transformation with pAJF447 yielded transformants that were kanamycin resistant at 30°C or 37°C (Figure S2A). We observed small numbers of kanamycin resistant transformants in vector transformed cells, but these cells continued to express DnaK, indicating that the second copy of dnaK was not lost in these transformants (Figure S2A). This failure to remove the copy of dnaK from attB in our ΔdnaK strain suggested that dnaK was required for growth at 30°C (low) and 37°C (high) temperatures. We observed a similar essentiality for the DnaK cofactor GrpE. After constructing a strain with a grpE deletion and a second copy of grpE at attB (MGM6023; Figure S1B) we were unable to remove this second copy of grpE from attB by marker exchange at either 30°C and 37°C (Figure S2B), demonstrating that both DnaK and GrpE are required for growth of M. smegmatis. Similar results were obtained when we attempted to delete the entire dnaK operon suggesting that all three components of the operon are essential (data not shown).
To further study the function of DnaK, we generated a depletion strain, MGM6005, which encodes an anhydrotetracycline (ATc) inducible allele of DnaK with a C terminal StrepTagII (STII). By 9 hours after withdrawal of ATc, DnaK-STII was undetectable by immunoblot with anti-STII antibodies (Figure 1A). Cells lacking DnaK continued to grow for an additional 12 to 15 hours without detectable DnaK, at which point optical density stabilized, in contrast to the continued replication of DnaK replete cells (Figure 1B). The viability of growth arrested cells lacking DnaK was determined by culturing dilutions onto selective media with ATc for both depletion and control cultures. The number of viable cells remained constant in the growth arrested, DnaK depleted population, indicating that loss of DnaK is bacteriostatic rather than bactericidal over the time course of the experiment (Figure 1B). The DnaK chaperone system has been previously implicated in protection and recovery of bacterial cells from heat shock [10], [27]–[29]. In M. tuberculosis, loss of σH has been shown to result in decreased survival at 53°C, a phenotype that was attributed to attenuation of the mycobacterial heat shock response, including the induction of DnaK [18]. To directly test the contribution of DnaK to heat shock response, we depleted DnaK for 12 hours prior to measuring cell viability at 53°C. Cells lacking DnaK were 100 fold more sensitive to killing by heat compared to DnaK replete cells (Figure S3).
Fig. 1. Depletion of DnaK is bacteriostatic in M. smegmatis and disrupts membrane structure. 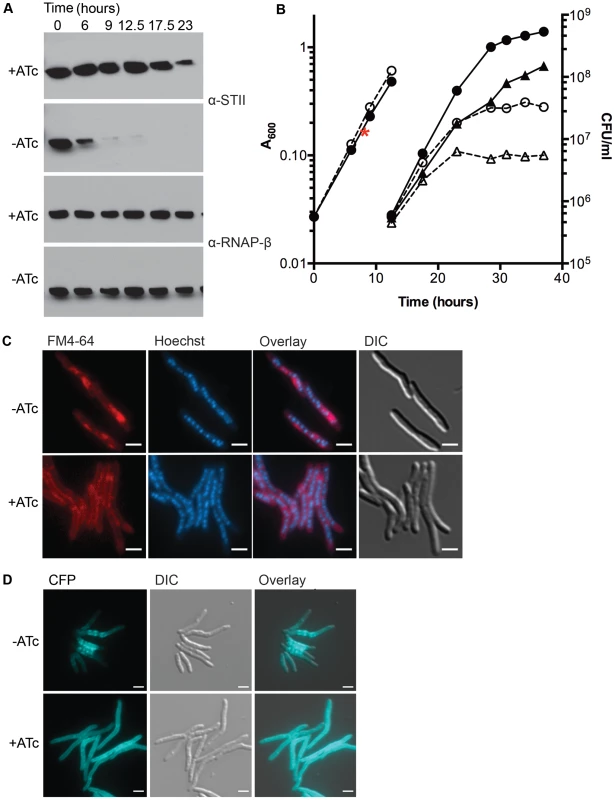
(A) Depletion of DnaK. Immunoblots of lysates prepared from DnaK-STII depletion cultures after withdrawal of the ATc inducer. Top panel is blotted for StrepTagII, bottom for RNAP-β. (B) Growth and survival of DnaK replete and depleted cells. Shown are optical density (left axis, circles) and CFU (right axis, triangles) of Tet-DnaK (MGM6005) grown with (solid lines, filled symbols) or without (dashed lines, open symbols) ATc. At 12.5 hours, cultures were diluted to maintain logarithmic growth. CFU/ml was determined for timepoints post dilution. The red asterisk marks the timepoint at which DnaK-STII was undetectable by immunoblot. (C) Loss of DnaK perturbs membrane structure but not nucleoid or cell morphology. MGM6005 grown with or without ATc for 16 hours. Cells stained with FM 4-64 and Hoechst prior to imaging. (D) DnaK is required for proper membrane protein localization. A Tet-DnaK strain expressing a membrane localized 3X-mCerulean (MGM6015) observed with (+ATc) or without (−ATc) DnaK for 16 hours. White bars indicate 2 µm. Exposure times were FM 4-64 500 ms, DAPI 500 ms and CFP 500 ms. To examine the morphologic correlates of the growth arrest that accompany DnaK depletion, we examined DnaK depleted and control cells by microscopy. Nucleoid morphology (Figure 1C, Hoechst panel) and cellular morphology (Figure 1C, DIC panel) of DnaK depleted cells were indistinguishable from that observed in wildtype cells, suggesting that DnaK depletion did not alter cell gross morphology. However, membrane staining by the lipophilic dye FM 4-64 was altered in cells lacking DnaK. In contrast to the homogenous distribution of the FM 4-64 membrane staining pattern seen in wild type cells, FM 4-64 in DnaK depleted cells was no longer evenly distributed along the entire periphery of the cell, but rather accumulated in a patchy pattern at midcell (Figure 1C). To examine whether this change in membrane staining pattern was accompanied by changes in membrane protein localization, we fused E. coli MalF transmembrane domains 1 and 2 to three copies of mCerulean. This fusion was evenly distributed in the membrane of cells expressing DnaK (Figure 1D, bottom). In contrast, upon DnaK depletion, MalF-mCerulean accumulated at midcell (Figure 1D, top) in a pattern that colocalized with FM 4-64 in the absence of DnaK (Figure S4). Taken together, these results indicate that loss of DnaK affects membrane structure and/or membrane protein localization with relative preservation of overall cell dimensions and nucleoid morphology.
To assess whether the membrane alterations seen with loss of DnaK affected cell permeability, we utilized a previously described assay for measuring ethidium bromide (EtBr) permeability [30]–[32]. DnaK depleted cells showed increased accumulation of EtBr as compared to replete cells (Figure S5). The addition of carbonyl cyanide 3-chlorophenylhydrazone (CCCP), an efflux inhibitor shown to increase EtBr accumulation [31], led to an increase of EtBr accumulation in both DnaK depleted and replete cells, indicating that DnaK depleted cells were still capable of EtBr efflux. Cells lacking DnaK still had increased accumulation relative to replete cells in the presence of CCCP (Figure S5) indicating that the increase in EtBr in DnaK depleted cells was due to an increase in cell permeability. To examine whether mycolic acid synthesis is altered with DnaK depletion, we analyzed total mycolic acid methyl esters and fatty acid methyl esters made in DnaK replete and depleted cultures. We observed no difference in the amount of total mycolic acid methyl esters or fatty acid methyl esters synthesized within 1 hour in between DnaK replete or depleted cells (Figure S6). Taken together with the FM 4-64 staining and MalF-mCerulean localization, these results indicate that loss of DnaK affects membrane structure and permeability.
DnaK is required for native firefly luciferase folding in M. smegmatis
The requirement for DnaK to sustain mycobacterial growth and membrane integrity suggested that DnaK may have a critical role in the absence of exogenous proteotoxic stress. To test the function of DnaK in native protein folding, we expressed firefly luciferase, which has been used as a model protein to study the activity of DnaK in E. coli cells after heat shock [33], [34], in our M. smegmatis dnaK depletion strain. In cells depleted of DnaK, we observed a rapid loss of luciferase activity (Figure 2A), which occurred prior to growth arrest but coincident with loss of DnaK-STII protein (Figure 2A and B). 14 hours after withdrawal of ATc, >80% of luciferase activity was lost. Although luciferase activity dropped in DnaK depleted cultures, luciferase protein levels remained stable (Figure 2B, Top panel), indicating that the loss of activity was not due to a change in steady state protein levels. By utilizing centrifugation to separate insoluble aggregate proteins, as previously described for E. coli [35], we observed that upon DnaK depletion, luciferase was depleted from the soluble fraction and accumulated in the pellet fraction (Figure 2C). Taken together, these results indicate that DnaK is required for folding of luciferase in the absence of heat shock, suggesting a nonredunant role in native protein folding.
Fig. 2. Ms DnaK is required for nascent Firefly luciferase folding. 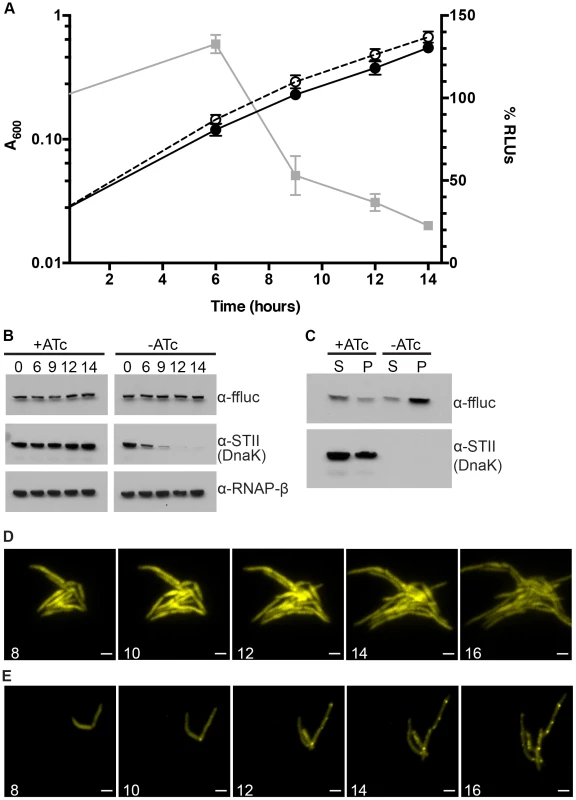
(A) A600 and relative Luciferase activity of a Tet-DnaK strain constitutively expressing firefly luciferase (MGM6006). A600 (plotted on left Y axis) for +DnaK indicated by closed circles/solid line and for −DnaK as open circles/dashed line. %RLUs (plotted on right Y axis), calculated as Counts per second (CPS) of −DnaK cultures divided by CPS of +DnaK cultures multiplied by 100, indicated by grey squares/solid line. Time indicated on X axis in hours. Each point is the mean of 3 independent cultures. Error bars indicate standard deviation of replicates. (B) Luciferase and Dnak-STII protein levels during DnaK depletion. Immunoblot of lysates prepared from one representative culture of Tet-DnaK Luciferase strain (MGM6006) shown in A. Top panel is probed for firefly luciferase, middle for StrepTagII, bottom for RNAPβ as a loading control. (C) Solubility of luciferase from DnaK depleted cultures. Soluble and pellet protein fractions of lysates prepared from DnaK replete (+ATc) and DnaK depleted (−ATc for 15 hours). Top panel is probed for firefly luciferase, bottom for StrepTagII. (D) Luciferase-mCitrine is diffuse in DnaK replete cells. Time-lapse microscopy of a Tet-DnaK strain constitutively expressing firefly luciferase fused to mCitrine (MGM6010) grown in the presence of ATc. (E) Luciferase-mCitrine forms foci in DnaK depleted cells. Time-lapse microscopy of a Tet-DnaK strain constitutively expressing firefly luciferase fused to mCitrine (MGM6010) grown in the absence of ATc. For (D) and (E), YFP images shown. White numbers indicate time in hours with respect to presence or absence of ATc and white bars indicate 2 µm. Exposure times were YFP 250 ms. To visualize the kinetics and localization of protein aggregate formation without DnaK, we generated a strain carrying a Luciferase-mCitrine fusion protein, which allowed us to simultaneously track luciferase activity and localization during DnaK depletion. Upon depletion of DnaK, the kinetics of loss of luciferase activity from the Luciferase-mCitrine fusion protein were similar to that observed with luciferase (Figure 2A). In contrast, mCitrine fluorescence was maintained in DnaK depleted cells, suggesting that whereas luciferase requires DnaK for folding, mCitrine does not [36]–[38]. Live cell time-lapse imaging in DnaK replete and depleted cells showed two patterns of localization. In the presence of DnaK, Luciferase-mCitrine was cytoplasmic and evenly distributed throughout the cell (Figure 2D). However, upon depletion of DnaK, Luciferase-mCitrine formed polar fluorescent foci which eventually coalesced into large cytoplasmic aggregates (Figure 2E). Immunoblots detecting both Luciferase and Luciferase-mCitrine during DnaK depletion did not detect any proteolytic cleavage of the protein (Figure S7), even after hours of DnaK depletion.
In E. Coli, DnaK and Trigger Factor cooperate to support the folding of nascent peptides. Their activities are largely redundant such that, at permissive temperatures, loss of either chaperone is tolerated [13], [14]. To examine potential redundancy between mycobacterial TF and DnaK, we deleted MSMEG_4674, the gene encoding TF (Figure S1C). In contrast to DnaK, mycobacterial TF was nonessential. Furthermore, overexpression of Trigger Factor did not rescue the loss of luciferase activity observed in DnaK depleted cultures (MGM6073) or allow for the loss of the dnaK gene (MGM6072) (data not shown). Loss of TF did not affect luciferase activity in a wild type background (Figure S8A) or bacterial growth (Figure S8B). DnaK depletion in the absence of Tigger Factor led to a modest acceleration in the kinetics of luciferase activity loss (Figure S8A), and in the time to growth arrest (Figure S8B). These data indicate that Trigger Factor makes a minor contribution to nascent luciferase folding and stability in M. smegmatis that is only evident when DnaK is absent, and demonstrate that DnaK is the dominant chaperone for native folding.
DnaK depletion leads to an increase in endogenous aggregate proteins
The insolubility of luciferase in DnaK depleted cells suggests a generalized role for DnaK in maintaining native protein folding and solubility. To test this idea, we examined the solubility of endogenous M. smegmatis proteins in the absence of DnaK. We depleted DnaK for 16 hours, a time point at which cells are still replicating and maintain full viability (Figure 1A), yet have no detectable DnaK (Figure 1B). We fractionated lysates from DnaK depleted and replete cells and compared fractions by SDS-PAGE. The total, soluble, and membrane (1% Triton X-100 soluble) fractions were similar from depleted and replete cells (Figure 3A). In contrast, the protein content of the pelleted (Triton X-100 insoluble) fraction was substantially increased in lysates from DnaK depleted cells (Figure 3A). To determine whether protein insolubility was the result of nascent protein misfolding versus aggregation of existing protein pools, we performed short term labeling of newly synthesized proteins with 35S-Methionine and analyzed the relative incorporation of the label into soluble and insoluble fractions with and without DnaK. In the absence of DnaK, nascent peptides accumulated in the insoluble fraction, accounting for approximately a 45% (±4.8%) increase in insoluble proteins, indicating that DnaK is required for nascent peptide folding (Figure 3B). To exclude an effect of DnaK depletion on translation rates that might account for these findings, we quantitated the rate of nascent chain synthesis using puromycin labeling and detection with an anti-Puromycin antibody [14], [39]. Both DnaK replete and depleted cells produced puromycilated chains at equal rates, as determined by immunoblotting (Figure S9), indicating that DnaK loss does not affect rate of translation.
Fig. 3. Depletion of Ms DnaK leads to an increase in insoluble protein, including large, multimodular proteins. 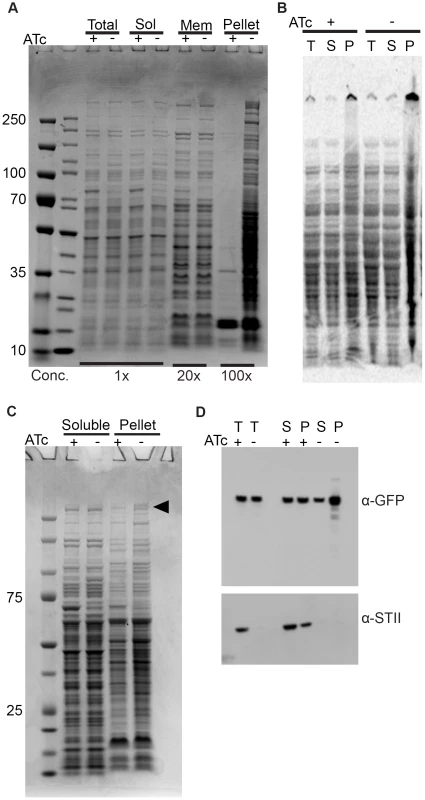
(A) DnaK depletion leads to an increase in pelleted protein. Coomassie stained SDS-PAGE gel of DnaK replete (+ATc) and DnaK depleted (−ATc) cultures of Tet-DnaK strain (MGM6005) at 16 hours of DnaK depletion. Total, Soluble, Membrane (1% Triton X-100 soluble), and Pelleted (1% Triton X-100 insoluble) fractions shown. (B) DnaK depletion leads to an increase in newly synthesized pellet protein. S35-labeled protein fractions from Tet-DnaK strain (MGM6005) cells depleted for DnaK (−ATc) or expressing DnaK (+ATc). Total (T) and soluble (S) fractions at a dilution of 1∶20 as compared to pelleted (P) fraction lanes. (C) DnaK is required for solubility of large multimodular polyketide synthases. Coomassie stained SDS-PAGE gel of DnaK depleted and control cultures of Tet-DnaK strain (MGM6005) at 15 hours post start of depletion. The black arrow indicates high molecular weight proteins that appeared in the pellet fraction in the absence of DnaK. (D) Fatty Acid Synthase I requires DnaK for solubility. Total (T), Soluble (S) and pellet (P) protein fractions of lysates prepared from a Tet-DnaK strain expressing FASI fused at the C-terminus to mCitrine (MGM6014) depleted for 15 hours. Top panel is probed for FASI-mCitrine, bottom for DnaK-STII. DnaK is required for solubility of modular polyketide synthases
Inspection of the proteins that become insoluble without DnaK revealed several high molecular weight proteins in the insoluble fraction (Figure 3C). Mass spectroscopic identification of tryptic peptides derived from these two high molecular weight proteins identified two polyketide synthetases, MSMEG_0408 (type 1 modular polyketide synthase) and MSMEG_0400 (MtbH, peptide synthase) (Figure 3C). Mycobacteria are unusual in that they encode many multimodular polyketide sythases for lipid synthesis, which are very large proteins greater than 300 kDa. The M. smegmatis chromosome encodes 8 proteins larger than 2000 amino acids, whereas the E. coli K12 chromosome encodes only 1. To extend the finding that DnaK is required for solubility of large multimodular enzymes in mycobacteria, we examined one additional essential large multimodular protein, fatty acid synthase I (FASI, 3089AA). We generated a strain expressing a single full-length copy of FASI fused to mCitrine the C terminus, expressed from its endogenous locus in our DnaK depletion strain background (strain MGM6014). Cell fractionation in DnaK replete or depleted cells revealed that, although some FASI is present in the insoluble fraction with DnaK, FASI accumulated in the insoluble fraction after DnaK depletion (Figure 3D). This demonstrates that FASI, a large, multimodular, essential protein in mycobacteria, requires DnaK for optimal folding and solubility in the absence of proteotoxic stress, suggesting that FASI may be a direct client of DnaK. Taken together our data indicates that DnaK is required for the solubility of at least 3 of the 8 large multimodular proteins in the mycobacterial proteome, all of which are lipid synthases, potentially explaining the disruption of membrane integrity observed in DnaK depleted cells.
ClpB relocalizes and is up-regulated after DnaK depletion
In addition to the large polyketide synthases that become insoluble in DnaK depleted cells, examination of soluble proteins in SDS-PAGE fractionated lysates from DnaK depleted cells revealed a protein species of approximately 90 kDa that was overrespresented in DnaK depleted cells (Figure 4A, black arrow). Mass spectrometry identified this protein as ClpB. To track both ClpB levels and localization during DnaK depletion, we fused mCitrine to the 3′ end of clpB in the DnaK depletion strain (strain MGM6008). ClpB levels were stable in DnaK replete cells (Figure 4B), and during the first 12 hours of DnaK depletion. Beginning at 21 hours after DnaK depletion, ClpB accumulated (Figure 4B). By microscopy, ClpB-mCitrine was expressed at low levels and was near the limit of detection (Figure 4C). At early time points of DnaK depletion, when native folding is lost, but before ClpB protein accumulation by immunoblot (9 to 12 hours), ClpB-mCitrine re-localized to form cytoplasmic foci, suggesting that ClpB relocalizes to protein aggregates that accumulate after loss of DnaK (Figure 4C). In several bacterial species other chaperones have been shown to be upregulated after loss of DnaK to compensate for the defect in chaperone function [10], [40]–[42]. To assess the potential compensatory upregulation of the chaperone network that may respond to DnaK loss in mycobacterial cells, we performed RT-qPCR on RNA collected from control and DnaK depleted cells. We detected upregulation of the mRNAs encoding ClpB, HspR, and Hsp20 (Figure 4D), all most likely the result of destabilization of HspR in the absence of DnaK [17]. No other chaperones or proteases tested were upregulated in the absence of DnaK, indicating that there is a lack of broad compensatory upregulation of alternative chaperone systems to handle the insoluble proteins that accumulate without DnaK function.
Fig. 4. ClpB relocalizes to polar foci and is upregulated after DnaK depletion. 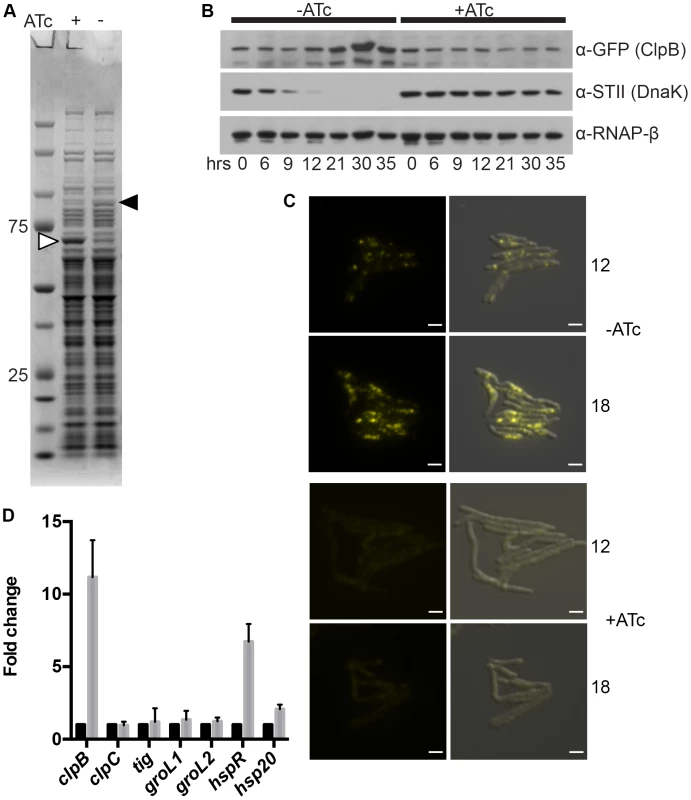
(A) DnaK depletion leads to an increase in soluble ClpB. Soluble protein fractions of lysates prepared from a Tet-DnaK strain (MGM6005) replete or depleted for 15 hours. White arrow indicates DnaK-STII, black arrow indicates band identified as ClpB. (B). DnaK depletion leads to an increase in ClpB-mCitrine after 12 hours of depletion. Immunoblot of lysates prepared from DnaK replete or depleted cells of a Tet-DnaK strain expressing ClpB fused at the C-terminus to mCitrine (MGM6008). Top panel is probed for ClpB-mCitrine, middle for StrepTagII, bottom for RNAPβ as a loading control. (C) Images of Tet-DnaK strain expressing ClpB-mCitrine (MGM6008) at 12 and 18 hours into DnaK depletion. Top panels are without DnaK (−ATc), bottom panels are with DnaK (+ATc). White arrow indicates early ClpB-mCitrine foci formed by 12 hours of depletion. Overlay is of YFP and DIC images. Exposure times were YFP 500 ms. White bars indicate 2 µm. (D) clpB, but not several other genes encoding chaperones, is upregulated after DnaK depletion. RT-qPCR analysis of the indicated chaperone genes using RNA collected from 3 independent cultures of a Tet-DnaK strain (MGM6005) grown in the presence of absence of ATc for 15 hours. sigA was used as a normalization control and all gene pairs are plotted as fold change in comparison to replete cultures. Relocalization of Dnak during proteotoxic stress
Based on our findings that mycobacterial DnaK plays a crucial role in native protein folding and in maintaining membrane protein and/or lipid composition, we hypothesized that it might localize in a peri-membrane pattern. In E. coli, DnaK localizes in a diffuse cytoplasmic pattern at 37°C and relocalizes to foci at 42°C and above [43], [44]. To assess DnaK localization in M. smegmatis, we generated a fully functional DnaK-mCitrine fusion at the native chromosomal locus such that DnaK-mCitrine was expressed as a stable fusion at the estimated full-length size (Figure S10). DnaK-mCitrine appeared to be functional for essential DnaK functions as well as for heat resistance at 53°C (Figure S11) as the DnaK-mCitrine fusion is the only copy of DnaK in the cell. DnaK-mCitrine was visible in multiple membrane peripheral foci distributed along the entire length of the cell during logarithmic growth at 30°C and 37°C (Figure 5A). DnaK foci were dynamic, changing in both number and localization within minutes (Figure 5B, Figure S12A,and Movie S1). The number of foci per micron of cell length varied among cells and within the same cell at different timepoints, however the number of foci per micron had a slightly negative correlation with the length of the cell (Pearson r −0.148, p-value 0.0013)(Figure S12B). So while longer cells had more total foci than shorter cells, they had slightly fewer foci per micron. A similar pattern of DnaK localization was observed in M. bovis BCG, indicating that this pattern is conserved across saprophytic and pathogenic mycobacteria (Figure 5C). In stationary phase cells, DnaK-mCitrine dramatically relocalized to form 1 or 2 foci in cells (Figure 5D). This pattern of localization suggested that DnaK re-localized to aggregates formed during stationary phase. To test the role of DnaK function in this localization pattern, we generated an ATPase mutant of DnaK, K70A. Ms DnaK(K70A)-mCitrine failed to complement the DnaK deletion strain and also did not form foci in log or stationary phases (Figure 5E) despite expression as a stable full-length protein at similar levels as wildtype (Figure S10).
Fig. 5. DnaK has growth phase dependent patterns of subcellular localization. 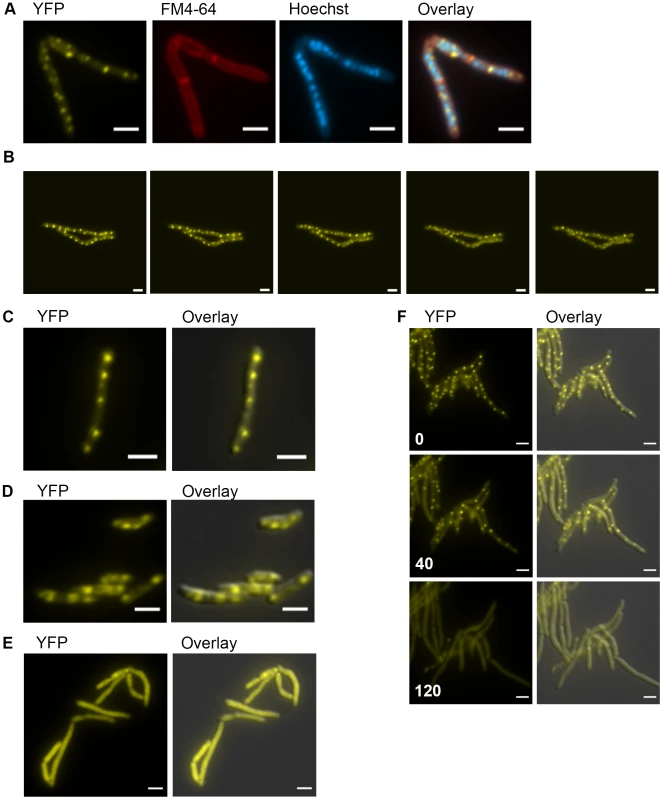
(A) DnaK-mCitrine localizes to peripheral foci during growth. Cells expressing DnaK fused at the C-terminus to mCitrine (MGM6003) at A600 0.2 stained with FM 4-64 and Hoechst prior to imaging. (B). DnaK-mCitrine foci are dynamic. Time-lapse microscopy of DnaK-mCitrine expressing strain (MGM6003) at 37°C. YFP images were acquired every 15 minutes as indicated in bottom left corner (Also see Movie S1 and Figure S12). (C). DnaK-mCitrine localizes to peripheral foci in Mycobacterium bovis BCG. BCG expressing DnaK fused at the C-terminus to mCitrine (MGM6023) at A600 0.2. Overlay is of YFP and DIC images. (D) DnaK-mCitrine localizes to 1 or 2 large foci in stationary phase cells. Cells of DnaK-mCitrine strain (MGM6003) from late stationary phase culture. Overlay is of YFP and DIC images. (E) Catalytically inactive DnaK fails to localize to peripheral foci. Cells expressing DnaK(K70A)-mCitrine (MGM6024) at A600 0.2. Overlay is of YFP and DIC images. (F) GrpE overexpression disperses DnaK-mCitrine foci. Time-lapse microscopy of Tet-GrpE DnaK-mCitrine expressing strain (MGM6016). Time at lower left of panel indicates minutes after addition of ATc to induce GrpE overexpression. Overlay is of YFP and DIC images. For all images: White bars indicate 2 µm. Exposure times were YFP 250 ms, FM 4-64 500 ms, Hoechst 500 ms. Several components of the DnaK chaperone system have been shown to alter the oligomeric state of DnaK in E. coli including levels of ATP and GrpE, a DnaK co-factor, [45]. Elevated levels of GrpE inhibit DnaK chaperone activity [46]. We tested the effect of GrpE overexpression on DnaK function and localization. Overexpression of GrpE altered the localization pattern of DnaK such that the dynamic, peripheral foci were lost (Figure 5F and Movie S2). The dispersal of DnaK by GrpE was not due to an effect on DnaK protein levels as the abundance of the DnaK-mCitrine fusion appeared unchanged by immunoblot (Figure S13A). Although prolonged overexpression of GrpE inhibited growth and inhibited luciferase activity, DnaK-mCitrine failed to localize to foci (Figure S13B), indicating that overexpression of GrpE inhibits both the log phase and stationary phase functions of DnaK.
Relocalization of DnaK-mCitrine and ClpB-mCitrine to protein aggregates
The focal relocalization of DnaK in stationary phase cells suggested that DnaK may relocalize to protein aggregates. To test this hypothesis, we induced the formation of protein aggregates by expressing the aggregating protein sequence ELK16 fused to mCerulean and confirmed that mCerulean-ELK16 accumulated in the insoluble fraction (data not shown). We observed mCerulean aggregates accumulating after the induction of mCerulean-ELK16 expression (Figure 6A). With low levels of mCerulean-ELK16 (3 hours after addition of inducer), aggregates colocalized with DnaK in the peripheral foci characteristic of the pattern of DnaK during log phase growth (Figure 6A, 3 hour panel). However, with accumulation of larger mCerulean-ELK16 aggregates, we observed relocalization of DnaK to these larger central aggregates (Figure 6A, 20 hour panel and Movie S3), a pattern that resembles that of DnaK in stationary phase cells. When we observed mCerulean-ELK16 aggregates in a strain expressing ClpB-mCitrine, we observed colocalization of ClpB to the cytoplasmic, but not peripheral, aggregates at late time points (Figure 6B, 20 hour panel). Taken together, these data indicate that DnaK has two modes of chaperone function, one in native protein folding in which it is localized in mobile peripheral foci, and one in aggregate processing in which DnaK relocalizes to central immobile foci of protein aggregates, which also contain ClpB.
Fig. 6. DnaK-mCitrine and ClpB-mCitrine co-localize with protein aggregates. 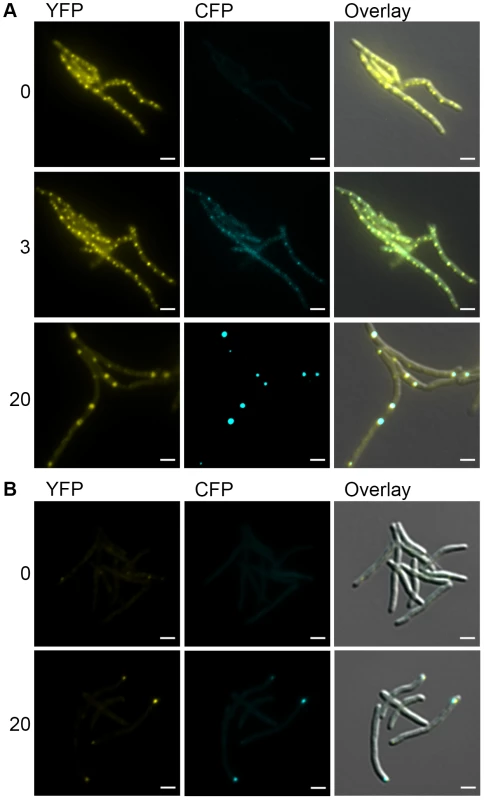
(A) DnaK-mCitrine co-localizes with mCerulean-ELK16. Time-lapse microscopy of DnaK-mCitrine Tet-mCerulean-ELK16 (MGM6011). Time shown after induction of mCerulean-ELK16 in hours to right of panels. Cells shown at 0 and 3 hours are same. Different cells from same experiment shown at 20 hours. (B) ClpB-mCitrine co-localizes with mCerulean-ELK16. Microscopy of ClpB-mCitrine Tet-mCerulean-ELK16 expression strain (MGM6012). Time shown after induction of mCerulean-ELK16 in hours to right of panels. For all images: White bars indicate 2 µm. Exposure times were YFP 250 ms and CFP 500 ms. Mycobacterial cells tolerate protein aggregates during cell growth
We observed that DnaK relocalized to aggregate proteins and that DnaK-mCitrine relocalized to similar patterns during late stationary phase. We next asked whether this aggregate relocalization was reversible by observing the pattern of DnaK localization during outgrowth from stationary phase. We observed that DnaK foci were largely immobile and persisted through several rounds of outgrowth (Figure 7A, white arrow and Movie S4). By 3 hours, larger foci were still visible, but peripheral dynamic foci had reformed. By 12 hours, the original aggregate containing cell still had some immobile DnaK-mCitrine, but all daughter cells contained only dynamic peripheral foci (Figure 7A). This suggested that protein aggregates formed during stationary phase are not dissolved prior to re-growth, but rather are tolerated by the mycobacterial cell for several rounds of division. This experiment also reveals that the two modes of DnaK function can coexist in the cell with DnaK dynamically shuttling between a function in aggregate processing and native folding.
Fig. 7. Protein aggregates persist during outgrowth of cells and are partitioned away from growing cells. 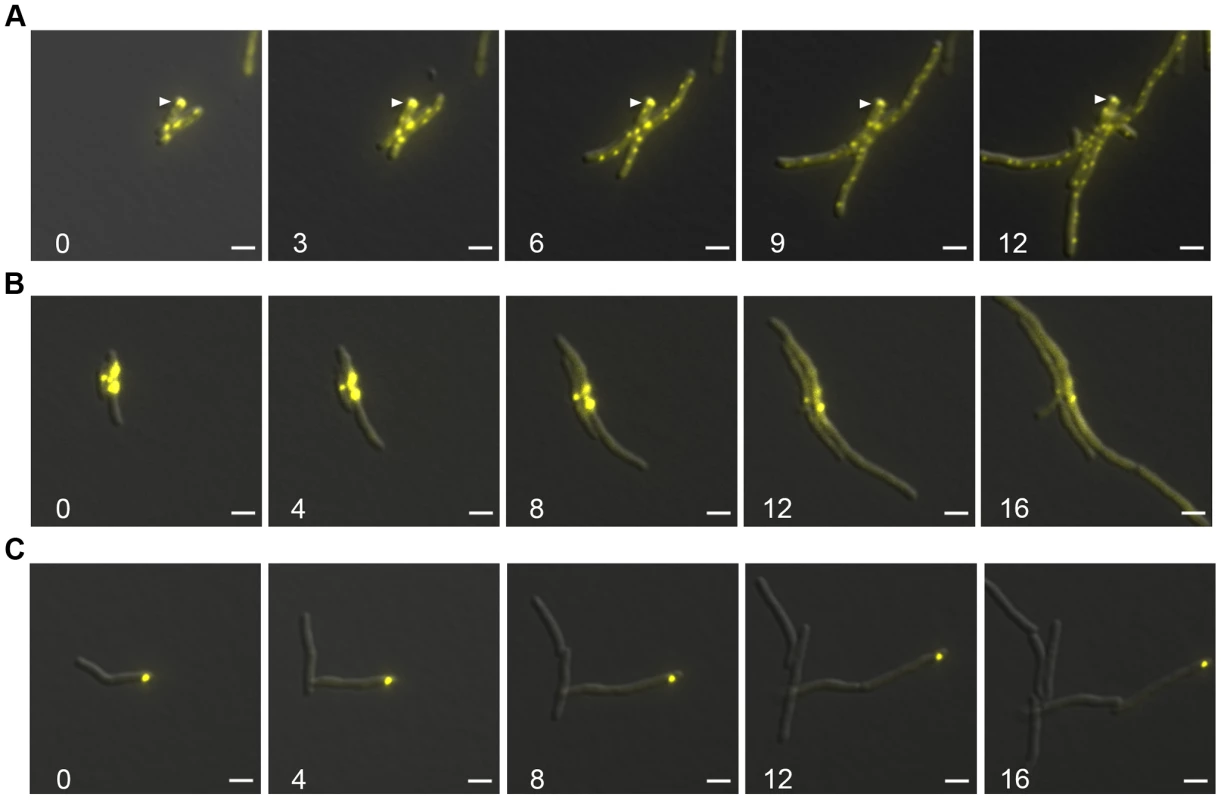
(A) Large static foci of DnaK-mCitrine persist during outgrowth of cells from stationary phase. Time-lapse microscopy of the DnaK-mCitrine strain (MGM6003). Time in white indicates hours after start of outgrowth. White arrow indicates large static foci that persists for over 12 hours. (B) Luciferase-mCitrine aggregates persist after growth is restored by DnaK expression. Outgrowth Tet-DnaK Luciferase-mCitrine strain (MGM6010) in the presence of ATc after DnaK depletion-induced bacteriostasis. Numbers indicate the time in hours after start of outgrowth. (C) Citrine-ELK16 aggregates persist during outgrowth. Time-lapse microscopy of aggregates during outgrowth after withdrawl of the ATc inducer of Tet-mCitrine-ELK16 expressing strain (MGM6022). For all images: Overlay of YFP and DIC images shown. White bars indicate 2 µm. Exposure times were YFP 250 ms. We utilized live cell time-lapse microscopy to directly observe the fate of protein aggregates during cell outgrowth. We first formed luciferase protein aggregates by transiently depleting DnaK, followed by outgrowth with DnaK reexpression. Luciferase-mCitrine aggregates persisted through several cycles of cell division, but cell growth initiated rapidly from the cell pole opposite the aggregate, eventually producing aggregate free cells. (Figure 7B and Movie S5). mCitrine-ELK16 aggregates behaved similarly: aggregates of mCitrine-ELK16 were very stable and persisted in cells after several rounds of division (Figure 7C) with a similar pattern of growth away from the aggregate, eventually forming aggregate free cells, despite persistence of the aggregate in the original cell. Thus, in three different models of protein aggregate formation (stationary phase, DnaK depletion, and heterologous protein expression) protein aggregates are stable during outgrowth and dissolution of aggregates is not required for reinitiation of cell growth. Aggregates were eventually lost in the population by dilution as they remained in the original parent cells but were not divided amongst daughters, suggesting that their loss was passive rather than by an active mechanism such as proteolysis.
Discussion
We have characterized the function of DnaK in mycobacteria and find that its cellular function differs substantially from what is known about other bacterial HSP70 chaperone systems. Our findings indicate that Mycobacterial DnaK is a central hub of the mycobacterial chaperone network with distinct nonredundant functions in both native and stress induced protein folding. These two states of DnaK function are accompanied by rapid shuttling of DnaK between two cytologic states of protein localization, one that reflects its native folding function and one at cytoplasmic aggregates.
A nonredundant role for mycobacterial DnaK in native protein folding
Our results indicate that mycobacterial DnaK is the dominant chaperone responsible for folding of native peptides in the absence of exogenous stress such as heat shock. This native folding function is evident both with model protein substrates (luciferase) and endogenous mycobacterial proteins. Trigger Factor in mycobacteria is nonessential and cannot compensate for DnaK loss, even when overexpressed. This contrasts with the function of E. coli, in which TF and DnaK have redundant functions in native protein folding and are essential in combination [13], [14].
Although loss of DnaK is accompanied by broad loss of protein solubility and formation of cytoplasmic protein aggregates, we also identified large multimodular lipid synthases as a specific class of proteins that require DnaK for solubility. We show that 3 of the 8 proteins greater than 2000 amino acids in the M. smegmatis proteome become insoluble in the absence of DnaK. One of these proteins, fatty acid synthase I (FASI), is a eukaryotic type FAS protein that is not found in bacterial taxa except for the Mycolic acid producing Actinomycetales [47] and is essential for viability [48]. The abundance of very large lipid synthases in Actinomycetales may mandate distinct chaperone functions to assure proper folding of these large multidomain proteins, as has been shown for eukaryotic proteomes that are enriched for multidomain large proteins in comparison to E. coli [49], [50]. Aggregation of large multidomain proteins is a feature of the proteostasis collapse that accompanies combined deletion of DnaK and TF in E. coli [7]. This requirement for DnaK in maintaining the solubility of large multimodular lipid synthases is consistent with our finding that the major morphologic and functional perturbation of mycobacterial cells upon growth arrest during DnaK depletion is perturbed membrane structure, rather than the filamentation phenotype seen in Caulobacter crescentus [51] and E. coli lacking DnaK [6], [52]. This requirement for DnaK to maintain the solubility of the lipid biosynthetic machinery also fits with prior literature demonstrating that GroEL1 in mycobacteria is a specialized chaperone of the FASII enzymes that elongate FASI fatty acid products to Mycolic acids [53], indicating that mycobacteria use a series of chaperones to maintain a functional lipid biosynthetic machinery.
The essential function of DnaK in mycobacteria is also apparently distinct from the mechanism of essentiality recently reported in C. crescentus. Protein aggregation that accompanies loss of DnaK activates degradation of DnaA through the Lon protease with consequent cell cycle arrest and filamentation [51]. Overexpression of DnaA restores cell growth in the DnaK mutant strain, indicating that this is the sole determinant of growth arrest in DnaK depleted cells. In our experiments, DnaK depletion in mycobacteria is not accompanied by filamentation, although DnaA depletion in mycobacteria does cause a filamentation phenotype [54]. It is also unlikely that the essentiality of DnaK is due to constitutive activation of the heat shock regulon through loss of HspR repression as previous work in mycobacteria has shown that hspR is not essential for growth [55].
Mycobacterial DnaK in stress induced protein aggregate processing
In addition to its role in native protein folding in unstressed cells, our data also indicate that mycobacterial DnaK also has a second function as a chaperone during states of protein aggregation, akin to the canonical role documented for DnaK in other bacteria. We observed rapid relocalization of DnaK from the dynamic mobile structures characteristic of rapid cell growth to focal protein aggregates that form during stationary phase or by expression of aggregating proteins. These foci remained fixed in both number and intensity and also contain ClpB, suggesting that DnaK and ClpB cooperate in protein aggregate processing, as has been shown in E. coli [56].
Once formed, we find that DnaK/ClpB containing protein aggregates are quite stable upon resumption of cell growth and that their dissolution or degradation was not necessary to restart growth after DnaK depletion-mediated growth arrest. This observation suggests that protein aggregates per se are well tolerated by the mycobacterial cell and that cytoplasmic aggregates are not per se toxic if the DnaK system is operative. In this regard, it is relevant that recent work has shown that ClpP1/P2 are essential in mycobacteria [57], [58] and form a mixed heterodimer that constitutes the function ClpP protease [59]. Loss of ClpP leaves cells susceptible to proteotoxic stress and the accumulation of misfolded proteins leading to cell death. Cells depleted for ClpP function have increased susceptibility to streptomycin, an antibiotic that caused mistranslation [58]. The findings presented here identify a second crucial susceptibility point in the mycobacterial chaperone/protein quality control network that could be targeted for antimicrobial development. The nonredunant essential role of Mycobacterial DnaK suggests that small molecule inhibition of this chaperone would be lethal for mycobacteria while sensitizing them to proteotoxic stress induced by the host. Inhibition of chaperone function is an emerging therapeutic strategy in malignant cells that depend on chaperone function and could by similarly targeted as a mechanism to sensitize pathogens to the proteotoxic stress inflicted by the host during infection.
Materials and Methods
Bacterial and DNA manipulations
Standard procedures were used to manipulate recombinant DNA and to transform E. coli. M. smegmatis strains were derivatives of mc2155 [60]. Gene deletions were made by homologous recombination and double negative selection [61]. All strains used in this study are listed in Table S1. Plasmids including relevant features, and primers are listed in Table S2 and S3. M. smegmatis was transformed by electroporation (2500 V, 2.5 µF, 1000Ω). All M. smegmatis strains were cultured in LB with 0.5% glycerol, 0.5% dextrose (LBsmeg) or 7H9 media for labeling experiments. 0.05% Tween80 was added to all liquid media. Heat sensitivity was assayed by incubating cultures at OD600 0.4 at 53°C. Aliquots were taken at indicated time points and serial dilutions were plated on selective media containing ATc. Antibiotic concentrations used for selection of M. smegmatis strains were as follows: kanamycin 20 µg/ml, hygromycin 50 µg/ml, streptomycin 20 µg/ml, zeocin 12.5 µg/ml.
Immunoblotting
For protein and epitope tag detection, the following antibodies were used: StrepTagII (Genescript, Rabbit Anti-NWSHPQFEK polyclonal antibody, 0.5 mg/ml, 1∶40, 000), YFP (Rockland Immunochemicals, Rabbit Anti-GFP polyclonal antibody, 1 mg/ml, 1∶20,000), Luciferase (Millipore, Goat Anti-Luciferase (Firefly) polyclonal antibody, 10 mg/ml 1∶20,000), Puromycin (KeraFast, Mouse anti-Puromycin (3RH11) monoclonal antibody, 1 mg/ml, 1∶2,000), and RNAP-β (Neoclone, 8RB13 Mouse Anti-E. coli RNAPβ monoclonal, 1∶20,000).
DnaK depletion using ATc withdrawal
Cultures were grown in the presence of 25 ng/ml anhydrotetracycline (ATc) to an OD600 of 0.4. Cultures were washed in equal volume of LBsmeg without ATc, then diluted back to indicated OD600. This dilution culture was then split and 25 ng/ml ATc was added to one culture, the other was grown in the absence of ATc for depletion. All depletions were carried out in the absence of antibiotic selection.
Ethidium Bromide uptake assays
A modified previously described method for detecting ethidium bromide uptake was used [31], [32], [62]. Depleted and replete cultures were collected 18 hours after ATc withdrawal, washed in an equal volume of PBS with 0.05% Tween80, then resuspended at an OD600 0.4 in PBS with 0.05% Tween80 and 0.5% glucose. 95 µl of culture was added to 0.2 ml pcr tubes. For efflux inhibition a final concentration of 5 µM CCCP was added just prior to the start of the assay. The assay was initiated by addition of ethidium bromide (0.25 µg/ml final) in PBS with 0.05% Tween80 and 0.5% glucose. Tubes were incubated at 37°C and fluorescence was determined every minute for 60 minutes using an Opticon2 instrument (MJ Research). Experiments were preformed twice in triplicate.
Mycolate analysis
DnaK depletion was carried out as described above for 16 hours. For labeling, 20 µl of 1-14C-Acetic acid, sodium salt (Perkin Elmer, 55.2 mCi/mol) was added to 20 ml of culture incubated at 37°C for 1 hour. Cells were harvested by centrifugation and washed once with 10 ml water. A final suspension of washed cells in 3 ml water was then added to 3 ml 40% tetrabutylammonium hydroxide and incubated at 100°C for 4 hours. An equal volume of dichloromethane was added then 300 µl of methyl iodide and the tubes were then rotated for 1 hour at room temperature. After the liquid layers were allowed to separate the bottom layer was removed and dried overnight. To the dried material, 2 ml ethyl ether was added and the supernatant was moved to a new tube and dried. 100 ul of ethyl ether was added just prior to spotting 5 ul samples on an HPTLC plate (Analtech, 58077). The plate was developed for 4 cycles in a 95∶5 mixture of Hexanes∶ethyl acetate. Plate imaging was preformed using a Phosphor storage cassette and Typhoon Trio (pixel size 200 microns at best sensitivity).
Luciferase assays
Culture volumes were normalized by OD600. A final concentration 5 mM D-luciferin (Gold Biotechnology) was added to each well and Counts per second (CPS) was counted using a Victor2 microplate reader (Perkin Elmer) set to read each well in triplicates, the mean of which was calculated to yield a CPS value for each well. For DnaK depletions %RLUs was calculated by the following equation on biologic triplicate cultures: 100* (CPSdepletion culture/CPScontrol culture).
Protein fractionation
3 Step Fractionation (used in Figure 3A)
DnaK depletion was carried out as described above and 50 ml of each culture cooled on ice for 5 min. Cell pellets were collected by centrifugation at 3700 g for 15 minutes at 4°C. Cells were washed with 10 ml buffer A (50 mM Tris, 100 mM NaCl, 10% glycerol, 2 mM EDTA, pH 8) and collected by centrifugation at 3700 g for 15 minutes at 4°C. Glass beads and 1 ml buffer A were added to washed cell pellets and cells were lysed using a Fastprep120 for 3 rounds at 5.0 m/sec for 25 seconds each round with cooling between rounds for 5 minutes on ice and subjected to centrifugation at 3000 g for 5 minutes at 4°C to remove beads and unlysed cells. Supernatants were collected and subjected to a second round of low speed centrifugation at 5000 g for 10 minutes at 4°C. Supernatants from this centrifugation were collected and labeled as “total” and brought up to a volume of 10 ml using buffer A. This total fraction was then centrifuged at 200,000 g for 2 hours at 4°C. Supernatants from this centrifugation were collected as the “soluble” fraction. Pellets were resuspended in 0.5 ml buffer A plus 1% Triton-X 100 by vortexing and allowed to solubilize for 1 hour at 4°C with gentle shaking. This suspension was then centrifuged at 21,130 g for 2 hours at 4°C. The supernatant of this centrifugation was collected as the “membrane” fraction. Pellets were resuspended in 0.1 ml buffer A plus 1% SDS by vortexing and sonication and labeled the “pelleted” fraction.
2 Step Fractionation (used in protein fractionation experiments except for Figure 3A)
DnaK depletion was carried out as described above and 50 ml of each culture cooled on ice for 5 min. Cell pellets were collected by centrifugation at 3700 g for 15 minutes at 4°C. Cells were washed with 10 ml buffer A (50 mM Tris, 100 mM NaCl, 10% glycerol, 2 mM EDTA, pH 8) and collected by centrifugation at 3700 g for 15 minutes at 4°C. Glass beads and 1 ml buffer A were added to washed cell pellets and cells were lysed using a Fastprep120 for 3 rounds at 5.0 m/sec for 25 seconds each round with cooling between rounds for 5 minutes on ice. A final concentration of 0.5% Triton X-100 added to disrupted cells and lysates were left to solubilize for 10 minutes on ice then subjected to centrifugation at 3000 g for 5 minutes at 4°C to remove beads and unlysed cells. Supernatants were collected and subjected to a second round of low speed centrifugation at 5000 g for 10 minutes at 4°C. Supernatants from this centrifugation were collected and labeled as “total”. This total fraction was then centrifuged at 21,130 g for 2 hours at 4°C. Supernatants from this centrifugation were collected as the “soluble” fraction. Pellets were resuspended in buffer A plus 0.5% Triton-X 100 by vortexing and sonication and labeled the “pelleted” fraction.
Puromycin labeling
DnaK depletion was carried out as described above for 16 hours during which the cultures reached an OD600 0.4. Puromycin was added at 50 µg/ml and the culture was incubated at 37°C. At indicated timepoints cells were harvested by centrifugation and immediately stored at −20°C until completion of the timecourse. For chloramphenicol inhibition controls, 10 µg/ml chloramphenicol was added immediately before the addition of puromycin. Experiments were preformed twice in triplicate.
35S protein labeling
DnaK depletion was carried out in 7H9 media for 15 hours at 37°C until cultures reached OD600 of approximately 0.4. Cultures were normalized to OD600 0.4 and 25 mls of culture was used for labeling. To label, 12.5 µl of Trans35S-LABEL (MP Biomedicals, 10.4 mCi/ml) was added and cultures were incubated with shaking for 30 minutes at 37°C. After 30 minutes 1 mM methionine was added and cultures were cooled to 4°C prior to harvesting by centrifugation. Collected cultures were fractionated for Soluble/Pelleting protein analysis as described above for the 2 Step Fractionation. After separation by SDS-PAGE, gels were dried and radioactivity quantitated using Phosphor storage cassette and Typhoon Trio (pixel size 200 microns at best sensitivity). ImageJ was used to quantitate the total radioactive signal per lane.
Microscopy
All images were acquired using a Zeiss Axio Observer Z1 microscope equipped with Definite focus, Stage top incubator (Insert P Lab-Tek S1, TempModule S1), Colibri.2 and Illuminator HXP 120 C light sources, a Hamamatsu ORCA-Flash4.0 CMOS Camera and a Plan-Apochromat 100×/1.4 oil DIC objective. Zeiss Zen software was used for acquisition and image export. The following filter sets and light sources were used for imaging: YFP (46 HE, Colibri2.0 505 LED), CFP (47 HE, HXP 120 C), Hoechst 33342 (49 HE, HXP 120 C), FM 4-64 (20, HXP 120 C). For cell staining 100 µl of culture was used. A final concentration of 1 µg/ml FM 4-64 (Invitrogen) and/or 10 µg/ml Hoechst 33342 (Invitrogen) was added. Cells were pelleted by centrifugation at 5000 g for 1 minute and resuspended in 50 µl of media. For single time point live cell imaging, 7 µl of culture was spotted onto a No. 1.5 coverslip and pressed to a slide. For time-lapse microscopy, cells were added to a 1.5% Low melting point agarose LBsmeg pad with or without 25 ng/ml ATc. For pad preparation, LBsmeg agarose was heated to 65°C and poured into a 17×28 mm geneframe (Thermoscientific, AB-0578) adhered to a 25×75 mm glass slide. A second slide was pressed down on top and the set-up was allowed to cool at room temperature for 10 minutes. The top slide was removed and the pad was cut and removed so that a 3–4 mm strip remained near the center. 2–3 µl of M. smegmatis culture was added to the pad and a No. 1.5 24×40 mm coverglass was sealed to the geneframe. Slides were incubated in stage top incubator at 37°C. For Luciferase-mCitrine aggregate imaging, DnaK depletion was carried out for 6 hours prior to transferring DnaK depleted cells to the pad. For Luciferase-mCitrine aggregate outgrowth imaging, DnaK depletion was performed for 24 hours prior to transferring DnaK depleted cells to a pad containing ATc to reinduce DnaK expression.
RT-qPCR
50 ml of cultures normalized to an OD600 of 0.4 were cooled to 4°C and harvested by centrifugation. Pellets were washed in 1 ml 10 mM Tris, pH 8.0 Pellets were then resuspended in 100 µl TE80 with 1 mg/ml lysozyme and disrupted by bead beating with a FastPrep120 2 times at 5.0 m/sec for 25 seconds. This lysate was used for RNA purification with a GeneJet RNA purification kit (Thermoscientific) following the manufacturer's protocol. RNA was eluted in 85 µl elution buffer and then treated with DNase I (Thermoscientific) for 30 minutes at 37°C. GeneJet purification columns were used to clean RNA from DNaseI reactions. First strand cDNA synthesis was carried out using Maxima Universal First Strand cDNA synthesis kit (Thermoscientific) with random hexamers and 500 ng RNA. For each RNA sample a no RT control was used to assess DNA contamination. qPCR was performed using DyNamo SYBR green qPCR kit (Thermoscientific) and an Opticon2 instrument (MJ Research). For each gene, normalized cycle threshold, C(t), was calculated using housekeeping gene sigA, and relative expression level was calculated using the equation 2−(C(t) gene – C(t) sigA). All RT-qPCR experiments were performed 2 times in triplicate.
Protein identification by nano-Liquid Chromatography coupled to tandem Mass Spectrometry (LC-MS/MS) analysis
Proteins were resolved using SDS-polyacrylamide gel electrophoresis, followed by staining with Coomassie Blue and excision of the separated protein bands; In situ trypsin digestion of polypeptides in each gel slice was performed as described [63]. The tryptic peptides were purified using a 2-µl bed volume of Poros 50 R2 (Applied Biosystems, CA) reversed-phase beads packed in Eppendorf gel-loading tips [64]. The purified peptides were diluted to 0.1% formic acid and then subjected to nano-liquid chromatography coupled to tandem mass spectrometry (nanoLC-MS/MS) analysis as detailed [65].
Initial protein/peptide identifications from the LC-MS/MS data were performed using the Mascot search engine (Matrix Science, version 2.3.02; www.matrixscience.com) with the Eubacteria segment of Uniprot protein database (12,115,765 sequences; European Bioinformatics Institute, Swiss Institute of Bioinformatics and Protein Information Resource). The search parameters were as follows: (i) two missed cleavage tryptic sites were allowed; (ii) precursor ion mass tolerance = 10 ppm; (iii) fragment ion mass tolerance = 0.8 Da; and (iv) variable protein modifications were allowed for methionine oxidation, cysteine acrylamide derivatization and deamidation of asparagines. MudPit scoring was typically applied using significance threshold score p<0.01. Decoy database search was always activated and, in general, for merged LS-MS/MS analysis of a gel lane with p<0.01, false discovery rate averaged around 1%. Scaffold (Proteome Software Inc., Portland, OR), version 4_1_1 was used to further validate and cross-tabulate the tandem mass spectrometry (MS/MS) based peptide and protein identifications. Protein and peptide probability was set at 99% with a minimum peptide requirement of 1.
Supporting Information
Zdroje
1. DeuerlingE, PatzeltH, VorderwulbeckeS, RauchT, KramerG, et al. (2003) Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol 47 : 1317–1328.
2. KernerMJ, NaylorDJ, IshihamaY, MaierT, ChangHC, et al. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122 : 209–220.
3. MogkA, DeuerlingE, VorderwulbeckeS, VierlingE, BukauB (2003) Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol 50 : 585–595.
4. TomoyasuT, MogkA, LangenH, GoloubinoffP, BukauB (2001) Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol 40 : 397–413.
5. SaibilH (2013) Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol 14 : 630–642.
6. BukauB, WalkerGC (1989) Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol 171 : 2337–2346.
7. CalloniG, ChenT, SchermannSM, ChangHC, GenevauxP, et al. (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep 1 : 251–264.
8. ItikawaH, FujitaH, WadaM (1986) High temperature induction of a stringent response in the dnaK(Ts) and dnaJ(Ts) mutants of Escherichia coli. J Biochem 99 : 1719–1724.
9. PaekKH, WalkerGC (1987) Escherichia coli dnaK null mutants are inviable at high temperature. J Bacteriol 169 : 283–290.
10. StrausD, WalterW, GrossCA (1990) DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev 4 : 2202–2209.
11. OhE, BeckerAH, SandikciA, HuberD, ChabaR, et al. (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147 : 1295–1308.
12. BruelN, Castanie-CornetMP, CirinesiAM, KoningsteinG, GeorgopoulosC, et al. (2012) Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J Biol Chem 287 : 44435–44446.
13. DeuerlingE, Schulze-SpeckingA, TomoyasuT, MogkA, BukauB (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400 : 693–696.
14. TeterSA, HouryWA, AngD, TradlerT, RockabrandD, et al. (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97 : 755–765.
15. UllersRS, LuirinkJ, HarmsN, SchwagerF, GeorgopoulosC, et al. (2004) SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci U S A 101 : 7583–7588.
16. VorderwulbeckeS, KramerG, MerzF, KurzTA, RauchT, et al. (2005) Low temperature of GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor DnaK. FEBS Lett 579 : 181–187.
17. BandyopadhyayB, Das GuptaT, RoyD, Das GuptaSK (2012) DnaK dependence of the mycobacterial stress-responsive regulator HspR is mediated through its hydrophobic C-terminal tail. J Bacteriol 194 : 4688–4697.
18. RamanS, SongT, PuyangX, BardarovS, JacobsWRJr, et al. (2001) The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183 : 6119–6125.
19. StewartGR, SnewinVA, WalzlG, HussellT, TormayP, et al. (2001) Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med 7 : 732–737.
20. MalenH, BervenFS, FladmarkKE, WikerHG (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7 : 1702–1718.
21. Berredo-PinhoM, KalumeDE, CorreaPR, GomesLH, PereiraMP, et al. (2011) Proteomic profile of culture filtrate from the Brazilian vaccine strain Mycobacterium bovis BCG Moreau compared to M. bovis BCG Pasteur. BMC Microbiol 11 : 80.
22. HickeyTB, ThorsonLM, SpeertDP, DaffeM, StokesRW (2009) Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect Immun 77 : 3389–3401.
23. FlotoRA, MacAryPA, BonameJM, MienTS, KampmannB, et al. (2006) Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science 314 : 454–458.
24. SpieringR, van der ZeeR, WagenaarJ, van EdenW, BroereF (2013) Mycobacterial and mouse HSP70 have immuno-modulatory effects on dendritic cells. Cell Stress Chaperones 18 : 439–446.
25. RodinaA, PatelPD, KangY, PatelY, BaakliniI, et al. (2013) Identification of an allosteric pocket on human hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem Biol 20 : 1469–1480.
26. PatelPD, YanP, SeidlerPM, PatelHJ, SunW, et al. (2013) Paralog-selective Hsp90 inhibitors define tumor-specific regulation of HER2. Nat Chem Biol 9 : 677–684.
27. SaitoH, UchidaH (1977) Initiation of the DNA replication of bacteriophage lambda in Escherichia coli K12. J Mol Biol 113 : 1–25.
28. SaitoH, UchidaH (1978) Organization and expression of the dnaJ and dnaK genes of Escherichia coli K12. Mol Gen Genet 164 : 1–8.
29. ItikawaH, RyuJ (1979) Isolation and characterization of a temperature-sensitive dnaK mutant of Escherichia coli B. J Bacteriol. 138 : 339–344.
30. RodriguesL, RamosJ, CoutoI, AmaralL, ViveirosM (2011) Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol 11 : 35.
31. RodriguesL, WagnerD, ViveirosM, SampaioD, CoutoI, et al. (2008) Thioridazine and chlorpromazine inhibition of ethidium bromide efflux in Mycobacterium avium and Mycobacterium smegmatis. J Antimicrob Chemother 61 : 1076–1082.
32. ViveirosM, MartinsA, PaixaoL, RodriguesL, MartinsM, et al. (2008) Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int J Antimicrob Agents 31 : 458–462.
33. SchroderH, LangerT, HartlFU, BukauB (1993) DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J 12 : 4137–4144.
34. SzaboA, LangerT, SchroderH, FlanaganJ, BukauB, et al. (1994) The ATP hydrolysis-dependent reaction cycle of the Escherichia coli Hsp70 system DnaK, DnaJ, and GrpE. Proc Natl Acad Sci U S A 91 : 10345–10349.
35. HesterkampT, BukauB (1998) Role of the DnaK and HscA homologs of Hsp70 chaperones in protein folding in E.coli. EMBO J 17 : 4818–4828.
36. EvansMS, SanderIM, ClarkPL (2008) Cotranslational folding promotes beta-helix formation and avoids aggregation in vivo. J Mol Biol 383 : 683–692.
37. ShimizuY, InoueA, TomariY, SuzukiT, YokogawaT, et al. (2001) Cell-free translation reconstituted with purified components. Nat Biotechnol 19 : 751–755.
38. UgrinovKG, ClarkPL (2010) Cotranslational folding increases GFP folding yield. Biophys J 98 : 1312–1320.
39. KelleherAR, KimballSR, DennisMD, SchilderRJ, JeffersonLS (2013) The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab 304: E229–236.
40. LemosJA, LuzardoY, BurneRA (2007) Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol 189 : 1582–1588.
41. da SilvaAC, SimaoRC, SusinMF, BaldiniRL, AvedissianM, et al. (2003) Downregulation of the heat shock response is independent of DnaK and sigma32 levels in Caulobacter crescentus. Mol Microbiol 49 : 541–553.
42. TillyK, SpenceJ, GeorgopoulosC (1989) Modulation of stability of the Escherichia coli heat shock regulatory factor sigma. J Bacteriol 171 : 1585–1589.
43. KumarM, SourjikV (2012) Physical map and dynamics of the chaperone network in Escherichia coli. Mol Microbiol 84 : 736–747.
44. WinklerJ, SeybertA, KonigL, PruggnallerS, HaselmannU, et al. (2010) Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J 29 : 910–923.
45. ThompsonAD, BernardSM, SkiniotisG, GestwickiJE (2012) Visualization and functional analysis of the oligomeric states of Escherichia coli heat shock protein 70 (Hsp70/DnaK). Cell Stress Chaperones 17 : 313–327.
46. SugimotoS, SaruwatariK, HigashiC, SonomotoK (2008) The proper ratio of GrpE to DnaK is important for protein quality control by the DnaK-DnaJ-GrpE chaperone system and for cell division. Microbiology 154 : 1876–1885.
47. SchweizerE, HofmannJ (2004) Microbial type I fatty acid synthases (FAS): major players in a network of cellular FAS systems. Microbiol Mol Biol Rev 68 : 501–517 table of contents.
48. ZimhonyO, VilchezeC, JacobsWRJr (2004) Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J Bacteriol 186 : 4051–4055.
49. RussmannF, StempMJ, MonkemeyerL, EtchellsSA, BracherA, et al. (2012) Folding of large multidomain proteins by partial encapsulation in the chaperonin TRiC/CCT. Proc Natl Acad Sci U S A 109 : 21208–21215.
50. NetzerWJ, HartlFU (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388 : 343–349.
51. JonasK, LiuJ, ChienP, LaubMT (2013) Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell 154 : 623–636.
52. KangPJ, CraigEA (1990) Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol 172 : 2055–2064.
53. OjhaA, AnandM, BhattA, KremerL, JacobsWRJr, et al. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123 : 861–873.
54. GreendykeR, RajagopalanM, ParishT, MadirajuMV (2002) Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology 148 : 3887–3900.
55. StewartGR, WernischL, StablerR, ManganJA, HindsJ, et al. (2002) Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148 : 3129–3138.
56. RosenzweigR, MoradiS, Zarrine-AfsarA, GloverJR, KayLE (2013) Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339 : 1080–1083.
57. OllingerJ, O'MalleyT, KesickiEA, OdingoJ, ParishT (2012) Validation of the essential ClpP protease in Mycobacterium tuberculosis as a novel drug target. J Bacteriol 194 : 663–668.
58. RajuRM, UnnikrishnanM, RubinDH, KrishnamoorthyV, KandrorO, et al. (2012) Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog 8: e1002511.
59. AkopianT, KandrorO, RajuRM, UnnikrishnanM, RubinEJ, et al. (2012) The active ClpP protease from M. tuberculosis is a complex composed of a heptameric ClpP1 and a ClpP2 ring. EMBO J 31 : 1529–1541.
60. SnapperSB, MeltonRE, MustafaS, KieserT, JacobsWRJr (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4 : 1911–1919.
61. BarkanD, StallingsCL, GlickmanMS (2011) An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene 470 : 31–36.
62. ViveirosM, RodriguesL, MartinsM, CoutoI, SpenglerG, et al. (2010) Evaluation of efflux activity of bacteria by a semi-automated fluorometric system. Methods Mol Biol 642 : 159–172.
63. Sebastiaan WinklerG, LacomisL, PhilipJ, Erdjument-BromageH, SvejstrupJQ, et al. (2002) Isolation and mass spectrometry of transcription factor complexes. Methods 26 : 260–269.
64. Erdjument-BromageH, LuiM, LacomisL, GrewalA, AnnanRS, et al. (1998) Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A 826 : 167–181.
65. BeverlyLJ, LockwoodWW, ShahPP, Erdjument-BromageH, VarmusH (2012) Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1. Proc Natl Acad Sci U S A 109: E119–126.
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



