-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
An Intronic microRNA Links Rb/E2F and EGFR Signaling
Animal genomes encode hundreds of microRNA genes that impact all areas of biology by limiting the expression of their targets. What remains largely unappreciated is that a significant proportion of microRNA genes are embedded within protein-coding genes, and are often co-expressed with their hosts, which raises the possibility of a functional interaction between them. The mir-998 gene is located within an intron of the gene encoding Drosophila E2F1 transcription factor. E2F1 can induce the expression of cell death genes, and its activity is negatively regulated by the pRB tumour suppressor protein. In certain settings, unrestrained E2F1 activity is sufficient to induce cell death in cells lacking functional pRB. Here, we show that miR-998 limits cell death in Rb-deficient cells by repressing dCbl, a negative regulator of Epidermal Growth Factor Receptor signaling (EGFR). miR-998 also augments EGFR signaling in differentiating photoreceptor cells. Furthermore, we show that the interaction between miR-998 and Cbl is conserved: in human cells, miR-29, a mir-29/998 seed family member, enhances EGFR signaling by targeting c-Cbl. Therefore, by examining the role of an intronic microRNA in the context of its host's function, we identified an important microRNA target and uncovered a biological function of the microRNA.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004493
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004493Summary
Animal genomes encode hundreds of microRNA genes that impact all areas of biology by limiting the expression of their targets. What remains largely unappreciated is that a significant proportion of microRNA genes are embedded within protein-coding genes, and are often co-expressed with their hosts, which raises the possibility of a functional interaction between them. The mir-998 gene is located within an intron of the gene encoding Drosophila E2F1 transcription factor. E2F1 can induce the expression of cell death genes, and its activity is negatively regulated by the pRB tumour suppressor protein. In certain settings, unrestrained E2F1 activity is sufficient to induce cell death in cells lacking functional pRB. Here, we show that miR-998 limits cell death in Rb-deficient cells by repressing dCbl, a negative regulator of Epidermal Growth Factor Receptor signaling (EGFR). miR-998 also augments EGFR signaling in differentiating photoreceptor cells. Furthermore, we show that the interaction between miR-998 and Cbl is conserved: in human cells, miR-29, a mir-29/998 seed family member, enhances EGFR signaling by targeting c-Cbl. Therefore, by examining the role of an intronic microRNA in the context of its host's function, we identified an important microRNA target and uncovered a biological function of the microRNA.
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs that regulate the expression of mRNA targets, thereby modulating biological processes including development, proliferation, metabolism, homeostasis and tumorigenesis. While some miRNAs elicit strong effects, many miRNAs operate more subtly to buffer a system or response to a signal. There is significant redundancy among miRNAs of the same family in regulating their target genes, making it difficult to identify the physiological role of an individual miRNA. The absence of strong loss-of-function phenotypes of a significant proportion of miRNAs has significantly hampered the characterization of their functions in vivo [1]. A number of approaches have been used to reveal miRNA functions, including combining mutations to generate synthetic phenotypes [2]–[5]. What remains largely unappreciated is that approximately 40% of miRNA genes are embedded within, and frequently co-expressed with protein-coding genes [6], [7]. There is a growing number of examples of intronic miRNAs directly impacting the function of the genes in which they reside [8]–[12]. Therefore, investigating a miRNA in the context of its host gene function could potentially provide insight into the biological roles of a large number of miRNAs. The value of such an approach is illustrated by recent studies of the Drosophila melanogaster dE2f1 gene.
The dE2f1 transcription factor, and its mammalian homologs coordinate the expression of genes involved in cell proliferation and cell death. In a variety of systems, E2F is rate-limiting for S phase entry while it triggers apoptosis in specific contexts. The last intron of the Drosophila E2F gene dE2f1 harbors a miRNA, mir-11, which is co-expressed with dE2f1 (Figure 1A and Figure S1). The loss of mir-11 was shown to strongly enhance dE2F1-dependent DNA damage-induced apoptosis even though it was insufficient to cause cell death in unprovoked settings. Therefore, the physiological role of mir-11 was revealed only when examined in the sensitized background of its host gene. This function of miR-11 is explained by its ability to directly regulate the expression of dE2F1-regulated cell death genes, thus highlighting a complex interaction between an intronic miRNA and its host gene [1], [8]. In addition to mir-11, the last intron of the dE2f1 gene contains another miRNA, mir-998. The sequence of mature miR-998 is different than that of miR-11, particularly at the 5′ end in the seed sequence (Figure 1A), which is the primary determinant of miRNA target selection. Therefore the two miRNAs are likely to regulate distinct sets of genes and, consequently, may have different functions. However since miR-998 was only recently identified, nothing was known about its biological function. Here we show that miR-998 limits dE2F-dependent cell death, but it does so in a different context and by a different mechanism than miR-11. While miR-11 repressed components of the core cell death machinery, including rpr and hid, miR-998 limited E2F-dependent cell death by elevating prosurvival signaling downstream of the Epidermal Growth Factor Receptor (EGFR) through regulation of dCbl, a negative regulator of EGFR. Thus, our data reveal a novel layer of intrinsic regulation at the dE2f1 genomic locus involving intronic miRNAs.
Fig. 1. miR-998 limits dE2F1-dependent cell death in rbf1120a eye discs. 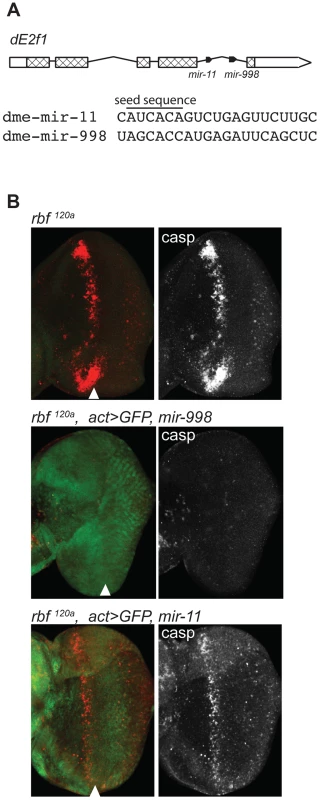
(A) Diagram of the dE2f1 transcript. Two miRNAs are in the last intron: mir-11 and mir-998, and are co-transcribed with dE2f1. See also Figure S1. The aligned sequences of mature miR-11 and miR-998 are shown, with the seed sequence highlighted. (B) Third instar larval eye discs were immunostained with an antibody that recognizes active caspases in dying cells (C3 antibody). GFP and miR-998 or miR-11 were expressed in the entire eye disc of rbf1120a hemizygous males using a flip-out technique induced by ey-FLP. Full genotypes are: rbf1120a, ey-FLP; +/+ (top), rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP/UAS-mir-998 (middle), and rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP/UAS-mir-11 (bottom). Results
miR-11 and miR-998 have different functions
Rbf is a negative regulator of dE2F1. The loss of rbf sensitizes cells to dE2F1-dependent apoptosis. As has been previously shown, there is a high level of apoptosis in a band running along the anterior edge of the morphogenetic furrow of rbf mutant eye discs [2]–[5], [13]. This stripe of apoptotic cells can be revealed by staining with the C3 antibody, which specifically recognizes activated caspases (Figure 1B). Importantly, apoptosis is dependent on dE2F1 since it was suppressed in rbf, dE2f1 double mutant animals [6], [7], [13]. To examine the effect of miR-998, we expressed a UAS-mir-998 transgene in the developing eyes of rbf mutant animals using the ey-FLP; Act≫Gal4 (Flip-out) system. Remarkably, no apoptotic cells were found in rbf120a, act>mir-998 eye discs, indicating that miR-998 strongly suppressed E2F-dependent cell death in this context (Figure 1B). Interestingly, unlike miR-998, miR-11 failed to block apoptosis in rbf mutants as the number of C3 positive cells was similar between rbf120a, act>mir-11 and rbf120a eye discs.
Differences in suppression of cell death in rbf mutant cells by miR-11 and miR-998 prompted us to investigate the impact of the two miRNAs on dE2F1-dependent apoptosis in other settings. When dE2F1 expression is driven by the Act88F-Gal4 driver, high levels of apoptosis in the wings of newly eclosed adults give rise to gnarled, blistered wings that have a downward curvature [8]–[12], [14]. This cell death phenotype is strongly rescued by miR-11 (Figure S2A and [8]). However, the wings of Act88F>dE2f1, mir-998 animals were indistinguishable from the wings of Act88F>dE2f1 adults, suggesting that expression of miR-998 was insufficient to suppress apoptosis (Figure S2A). Next, we performed genetic interaction tests in the eye imaginal disc. Ectopic expression of dE2f1 in the posterior compartment of the eye imaginal disc potently induces apoptosis [15]. This apoptosis is strongly suppressed by co-expression of miR-11 (Figure S2B and [8]). In contrast, miR-998 had no effect on E2F1-induced cell death in the posterior compartment, as the level of C3 staining was indistinguishable between GMR>dE2f1/dDP/mir-998 and GMR>dE2f1/dDP eye discs (Figure S2B). In addition to apoptosis, GMR>dE2f1/dDP had been shown to induce unscheduled proliferation that can be visualized by BrdU labeling [15]. However, neither miR-11 nor miR-998 modulated dE2F1-induced proliferation, as the level of E2F1-induced ectopic BrdU incorporation was largely unchanged by co-expression of miR-11, as was previously shown [8], or miR-998 (Figure S2B).
Therefore we concluded that overexpression of miR-998 suppressed dE2F1-dependent apoptosis in rbf mutants but not when dE2F1 was ectopically expressed in the eye or in the wing. In contrast, miR-11 suppressed dE2F1-induced phenotypes but failed to block apoptosis in rbf mutants. Thus, miR-998 and miR-11 both suppressed E2F-dependent cell death, but did so in mutually exclusive contexts.
Loss of mir-998 enhances apoptosis in rbf mutants
The results described above using a miR-998 transgene raise the question of whether endogenous miR-998 operates in a similar manner and blocks apoptosis in rbf mutants. Therefore we examined the consequence of the loss of mir-998 in the background of the rbf120a mutation. Since there were no pre-existing mir-998 mutants, we generated a mir-998 null allele (for details see Materials and Methods and Figure S3). It was essential that the mir-998 loss-of-function allele did not disrupt the expression of mir-11 or dE2f1, such that any observed phenotype could be attributed specifically to the function of miR-998. We used one piggyBac element and two P elements inserted near the dE2f1 gene and screened for local transpositions of these transposons into intron 5, which harbors miR-11 and miR-998. Out of 4,254 transposition events a single P element insertion into intron 5 was recovered and then used to screen for imprecise excisions that specifically disrupted mir-998. Among 400 excision events only two imprecise events were isolated and both of them retained a small piece of the P-element. The mir-998exc222 allele contained an 87 bp insertion within the mir-998 hairpin that is expected to disrupt the correct folding and processing of miR-998. Indeed, no mature miR-998 was detected in whole 3rd instar larvae, larval eye imaginal discs, or adult heads from mir-998exc222 mutant animals (Figure 2A). Importantly, the expression of dE2f1 and miR-11 were not affected in the mir-998exc222 mutant animals (Figure 2B). We concluded that mir-998exc222 is a null allele of mir-998.
Fig. 2. mir-998exc222 mutant allele enhances apoptosis in rbf mutant eye discs. 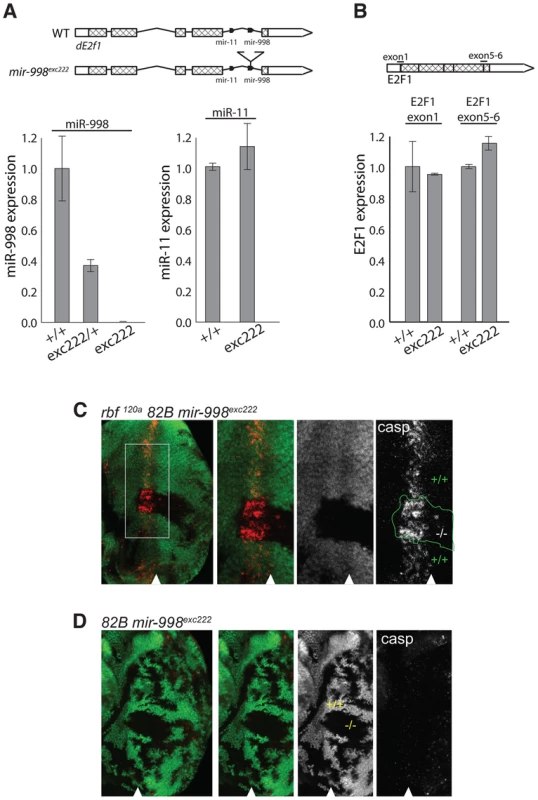
(A) cDNA was prepared from RNA extracted from +/+ (Canton S, wild-type), mir-998exc222/+, or mir-998exc222 adult fly heads. The expression of mir-998 and mir-11 were measured by Taqman assay, and normalized to β-tubulin levels. Expression in +/+ was designated as 1.0 and the expression in mir-998exc222was compared. (B) cDNA was prepared from RNA extracted from +/+ (Canton S, wild-type) or mir-998exc222 third instar larvae. The expression of de2f1 mRNA was measured using qPCR, and normalized to β-tubulin levels. Expression in +/+ was designated as 1.0, and the expression in mir-998exc222was compared. E2F1 exon1 primers span the translation start site and E2F1 5–6 primers span the exon 5-exon 6 junction. (C and D) Clones of mir-998 mutant tissue were generated using ey-FLP in rbf 120a hemizygous (top), or rbf wild-type (bottom) animals. The full genotypes are rbf1120a, ey-FLP/Y; FRT 82B mir-998exc222/82B GFP (C) and ey-FLP/+; FRT 82B mir-998exc222/82B GFP (D). Third instar larval eye discs were dissected, fixed, and stained with an antibody recognizing active caspase (casp). The full disc is shown in the left panel, while a higher magnification in panels on the right. Mutant clones were outlined and wild-type mir-998 was indicated by +/+, and −/− for mir-998exc222. A minimum of 10 larvae were analyzed for each genotype, and representative results are shown. Clones of mir-998exc222 homozygous mutant cells in rbf mutant eye discs were generated using the ey-Flp/FRT system and apoptotic cells were visualized with the C3 antibody. The mir-998 mutant tissue was marked by the absence of GFP, while tissue that contained a wild-type mir-998 allele expressed GFP. Since the entire disc was mutant for rbf, cell death occurred in the distinctive pattern in the morphogenetic furrow in both mir-998 mutant and mir-998 wild type tissue. However, significantly elevated C3 staining was consistently observed in clones of mir-998exc222 mutant cells compared to adjacent mir-998 wild-type tissue (Figure 2C). In contrast, no apoptosis was detected in the furrow when clones of mir-998 mutant cells were induced in wild type eye discs (Figure 2D). Thus, the loss of mir-998 specifically sensitized rbf mutant cells to apoptosis, while overexpression of miR-998 blocked cell death in rbf mutants.
miR-998 modulates EGFR signaling
How does miR-998 suppress apoptosis of rbf mutant cells? The specific pattern of apoptosis in rbf mutant eye discs is due to the coincident transient reduction of EGFR signaling in the morphogenetic furrow that, in turn, lowers prosurvival cues. As a result, the level of unrestrained dE2F1 becomes sufficient to trigger cell death in this region of rbf mutant but not in wild type eye discs [13]. In addition, the cell death in rbf mutants was shown to be dependent on the expression of the pro-apoptotic genes reaper (rpr) and hid. Therefore we examined impact of miR-998 on EGFR signaling and on hid and rpr.
We began by determining whether miR-998 regulates expression of rpr and hid in luciferase sensor assays. The 3′UTRs of rpr and hid were cloned downstream of the constitutively expressed luciferase gene. Increasing amounts of miR-998 were co-transfected with rpr or hid 3′UTR sensors, and luciferase activity was measured. As shown in Figure 3A, expression of miR-998 did not modulate rpr or hid 3′ UTR reporters. To corroborate this result we compiled a list of predicted miR-998 targets and performed Gene Ontology of Biological Processes (GOBP) enrichment analysis. Interestingly, none of the GOBP terms associated with apoptosis were statistically enriched among miR-998 targets (Figure 3B, Table S2). In contrast, as it has been shown previously [8], GOBP terms that relate to the induction and positive regulation of cell death were significantly enriched among miR-11 targets (Figure 3B, Table S2). Furthermore, although miR-11 and miR-998 share 170 common targets, cell death GOBP terms were not enriched among them but were overrepresented among genes that are exclusively miR-11 targets. Thus, the sensor assays and bioinformatics analyses do not support the explanation that suppression of apoptosis in rbf mutants by miR-998 occurs through the direct regulation of cell death genes.
Fig. 3. miR-11 and miR-998 limit dE2F-dependent cell death through different targets. 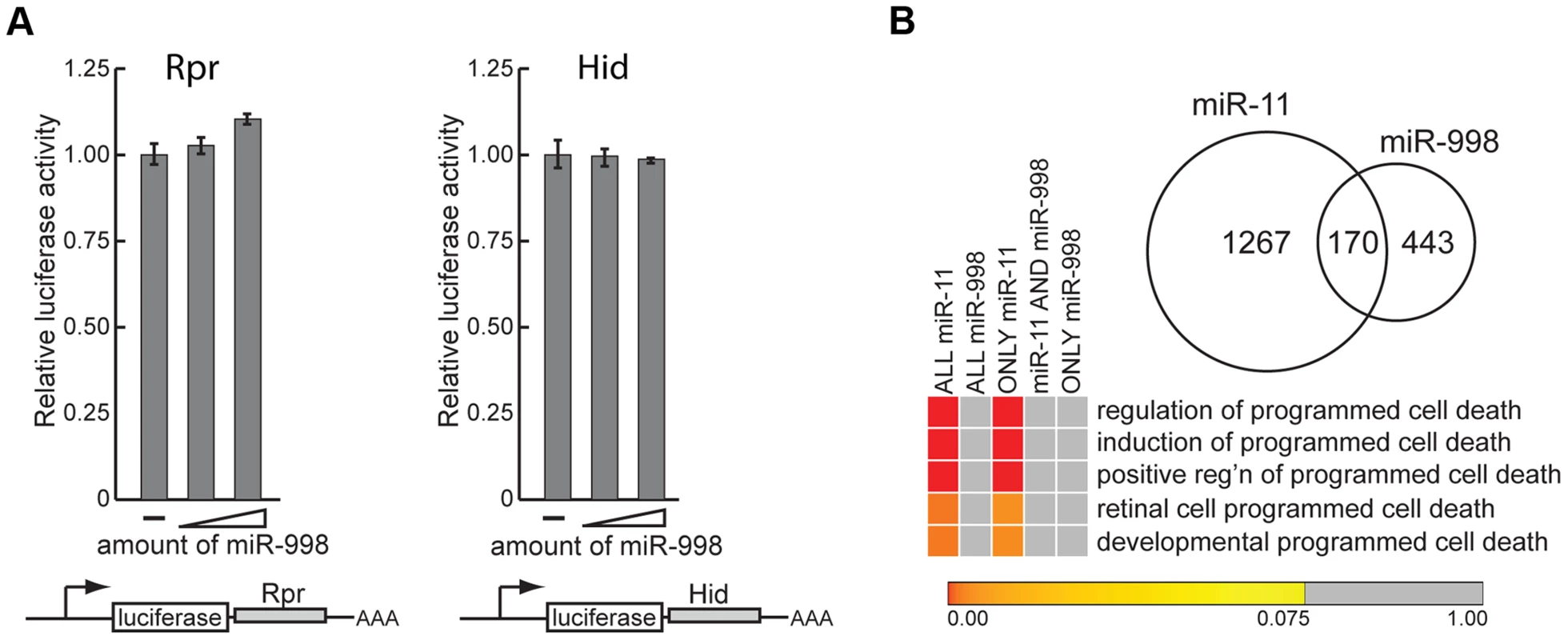
(A) 3′ UTR sensor assays were performed in HeLa cells using rpr and hid 3′ UTR sensors. The indicated 3′ UTR sensor plasmid was transfected with increasing amounts of pcDNA3/mir-998 plasmids. Cells were harvested 40–48 h post-transfection, and Renilla and Firefly luciferase activities were measured. (B) Comparison of predicted miR-11 and miR-998 targets (see Table S1 for lists of predicted miR-11 and miR-998 targets). Heat map of GOBP enrichment analysis of predicted miR-11 and miR-998 targets (FDR <or = 0.075). Next, we asked whether EGFR activity is altered in mir-998 mutants. The level of EGFR activity is accurately reflected by di-phosphorylated, activated ERK (dpERK) [16]. During eye development, EGFR signaling is transiently reduced within the morphogenetic furrow, while EGFR activity is high in groups of cells that form the ommatidial preclusters in a column immediately posterior to the morphogenetic furrow, which is revealed by the dpERK antibody. Within a column, clusters are specified sequentially in short intervals beginning at the midline. This gives rise to a gradual rise and fall of dpERK staining within a column (Figure 4A, left panel). To examine the level of EGFR signaling in mir-998 mutants, clones of mir-998exc222 mutant cells were generated in rbf120a mutant eye discs and stained with the dpERK antibody. While the pattern of natural variation of dpERK staining was not altered in mir-998 mutant tissue, the intensity of dpERK staining was reduced within clones of mir-998 mutant cells, as well as in wild type tissue immediately adjacent to the clonal boundary (Figure 4A, see yellow arrowheads). Since the level of dpERK expression reflects the level of EGFR signaling, this indicated that the loss of mir-998 reduces EGFR activity and this may explain the enhancement of apoptosis in rbf mutants.
Fig. 4. mir-998exc222 suppresses EGFR signaling. 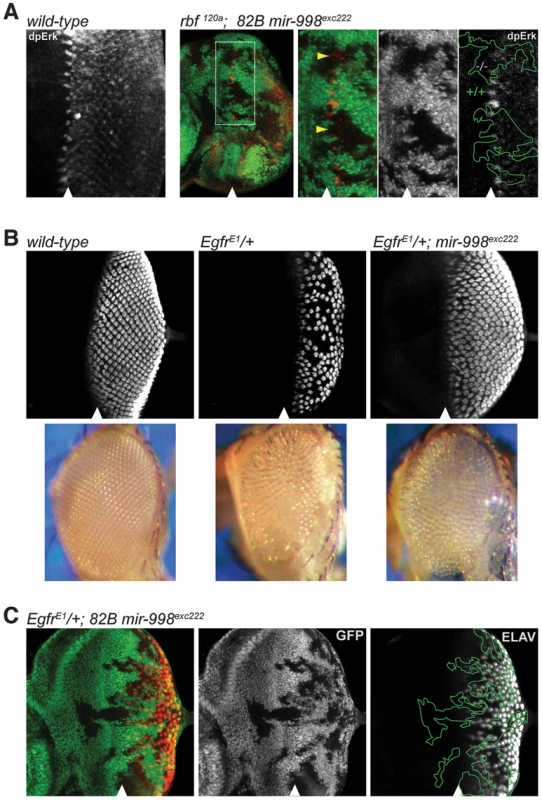
(A) Third instar larval eye discs were dissected, fixed, and stained with an antibody recognizing active Erk (dpErk). A wild-type larval eye disc is shown in the left-most panel. To the right: clones of mir-998exc222 mutant tissue were generated using ey-FLP in rbf 120a hemizygous animals as in Fig. 2C. The full disc as well as a higher magnification are shown. Mutant clones were outlined and wild-type mir-998 was indicated by +/+, and −/− for mir-998exc222. Yellow arrowheads show regions in mir-998 mutant tissue with significantly less dpERK staining than expected. (B) Third instar larval eye discs were harvested from wild-type control, EgfrElp/+, and EgfrElp/+; 82B mir-998exc222 animals. Dissected tissue was fixed and stained with an antibody recognizing ELAV (red), which is expressed in photoreceptor clusters. Below are images of adult eyes from wild-type control, EgfrElp/+, and EgfrElp/+; 82B mir-998exc222 animals. Images were taken at the same magnification. (C) Third instar larval eye discs were harvested from ey-FLP; EgfrElp/+; 82B mir-998exc222/82B GFP animals. Tissue that is wild-type for mir-998 is marked by GFP (green), while clones of mir-998 mutant tissue are marked by the absence of GFP. Mutant clones were outlined and wild-type mir-998 was indicated by +/+, and −/− for mir-998exc222. Analysis was performed on a minimum of 10 larvae of each genotype. The position of the morphogenetic furrow is marked with an arrowhead. EGFR signaling is used reiteratively throughout development, including in the recruitment of photoreceptor cells into the ommatidial clusters of the developing larval eye [17]–[19]. To confirm that EGFR signaling is reduced in mir-998 mutants, we performed genetic interaction tests between the mir-998 mutant allele and the dominant gain-of-function Ellipse (Elp) allele of the Egfr gene. The number of ommatidial clusters is significantly reduced in EgfrElp/+ larval eye discs as revealed by staining with an ELAV antibody that marks differentiated photoreceptors (Figure 4B and [18], [20]). Strikingly, the mir-998exc222 mutation strongly suppressed this phenotype: there was a dramatic increase in the number of ommatidial clusters in EgfrElp/+, mir-998exc222 double mutant eye discs compared to EgfrElp/+ single mutant eye discs (Figure 4B). Consistently, the small, rough eye phenotype of EgfrElp/+ adult flies was suppressed by the loss of mir-998 (Figure 4B). To determine whether the effect of mir-998 is cell autonomous, we generated clones of mir-998exc222 mutant cells in heterozygous EgfrElp/+ eye discs. As shown in Figure 4C, the EgfrElp mutant phenotype was partially suppressed in cells that were immediately adjacent to the mir-998exc222 mutant tissue suggesting some non-cell autonomous effects. Therefore, the loss of mir-998 suppresses the EGFR gain-of-function phenotype, which is consistent with the reduction of dpERK activity (Figure 4A) and therefore EGFR signaling in mir-998 mutant tissue.
dCbl is downregulated in mir-998 mutant eye discs, phenocopies the response of the mir-998 mutant to E2F-dependent apoptosis, and can be directly repressed by miR-998 in vitro
To gain insight into the molecular basis of the genetic interaction between miR-998 and EGFR signaling, we asked whether the loss of mir-998 results in misexpression of gene(s) that are known to be connected to the EGFR signaling pathway. To identify such genes in an unbiased manner we performed gene expression microarrays using RNA isolated from mir-998exc222 homozygous mutant and wild type eye discs. This analysis led to identification of a set of 382 genes that were differentially expressed (DE) in the mir-998 mutant. The list of DE genes was then mined for genes with terms related to the EGFR signaling pathway by referring to AmiGO and KEGG pathway databases, as well as the literature [21]–[23] (Table S3).
Of 80 genes associated with the EGFR pathway, only one was significantly differentially expressed in the mir-998exc222 microarray: dCbl-S. dCbl negatively regulates signaling from EGFR by binding to the activated, phosphorylated receptor, and inducing its ubiquitination and endocytosis [24]–[27]. According to the microarray data, the expression of dCbl-S was increased 3.6-fold in the absence of mir-998, which made it a good candidate as a target of repression by miR-998, the prevailing mechanism of miRNA function. To confirm the results of the gene expression microarray, dCbl expression was measured in mir-998 homozygous mutant eye discs by quantitative RT-PCR (qRT-PCR). The dCbl gene encodes two dCbl proteins generated from alternatively spliced transcripts which, like their mammalian Cbl homologs, both negatively regulate EGFR signaling in Drosophila [28], [29]. As shown in Figure 5A, both dCbl transcripts were significantly upregulated in mir-998exc222 eye discs (dCbl-S: 4.4-fold, dCbl-L: 2.3-fold). Together with the results of genetic interaction tests described above, this suggested a model where miR-998 represses dCbl, a negative regulator of EGFR signaling, to enhance signaling downstream of the EGF receptor.
Fig. 5. miR-998 enhances EGFR signaling by repressing dCbl. 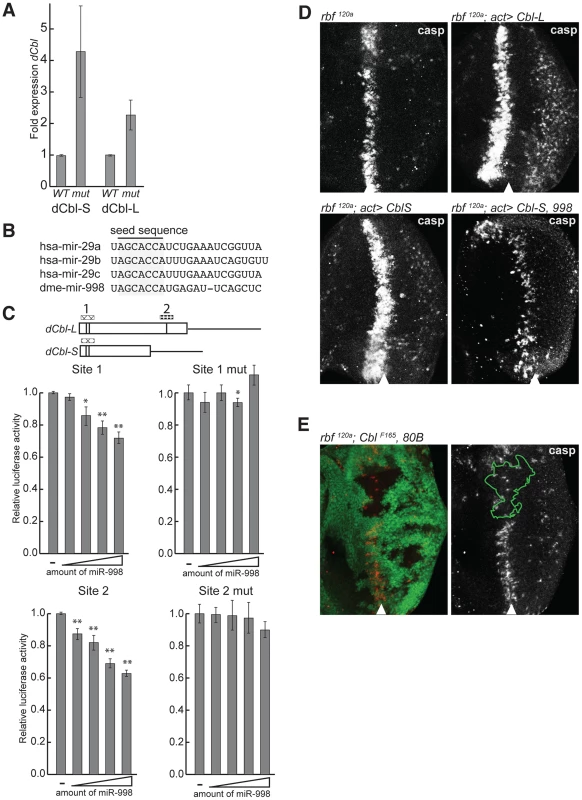
(A) cDNA was prepared from RNA extracted from +/+ (Canton S, wild-type) or mir-998exc222 third instar larval eye discs. The expression of dCbl-S, and dCbl-L were measured using qPCR, and normalized to rp49 or β-tubulin levels. Expression in +/+ was designated as 1.0 and the expression in mir-998 exc222 was compared. (B) Alignment of mature miRNA sequences of mir-29 seed family homologs. Seed sequences are highlighted. (C) 3′ UTR sensor assays were performed in HeLa cells using sequences predicted to be regulated by miR-29 or miR-998, or sequences carrying mutations in miR-998 binding sites. The indicated 3′ UTR sensor plasmid was transfected with pcDNA3/empty or increasing amounts of pcDNA3/mir-998 plasmid. Cells were harvested 24–48 h post-transfection, and luciferase activities were measured. A minimum of three independent transfections was performed for each sensor construct. Error bars represent standard error. Asterisks are shown for differences between vector and miR-998 that are statistically significant by t-test (* is p<0.05, ** is p<0.01). (D) Third instar larval eye discs were immunostained with an antibody that recognizes active caspases in dying cells. GFP and dCblS, dCblL, or miR-998 were expressed in the entire eye disc of of males hemizygous for rbf1120a using a flip-out technique induced by ey-FLP. Full genotypes are: rbf1120a, ey-FLP; +/+ (top left), rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP/UAS-dCblL (top right), and rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP; UAS-dCblS (bottom left), and rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP/UAS-miR-998; UAS-dCblS (bottom right). (E) Third instar larval eye discs from rbf1120a, ey-FLP/Y; dCblF165, FRT 80B/GFP, FRT 80B animals were dissected, fixed, and stained with an antibody recognizing active caspase (casp). This model was tested in three sets of experiments. First, we asked whether dCbl is a miR-998 target. Since dCbl was not identified as a target of miR-998 by bioinformatic prediction, we asked whether the mammalian ortholog, c-Cbl, was a predicted target of the mammalian miRNA members of the mir-29/998 miRNA seed family: mir-29a-c [30] (Figure 5B and Table S1). Analysis revealed that c-Cbl contains five predicted miR-29 target sites including two sites in tandem within a highly conserved region that encodes the tyrosine kinase-binding (TKB) domain. Further analysis of dCbl using the RNA22 prediction algorithm with low stringency parameters [31] revealed a putative target site for miR-998 in dCbl-L (Figure 5C). The functionality of these sites was then tested in luciferase sensor assays. The identified predicted target sites for miR-998 in dCbl were cloned downstream of the Renilla luciferase gene, and were transfected with increasing amounts of a miR-998 expression plasmid. Significantly, miR-998 exhibited dose-dependent repression of luciferase 3′UTR sensor constructs carrying either the double miR-29 target site within the highly conserved TKB domain (site 1), or the miR-998 site near the 3′ end of the dCbl-L coding sequence (site 2) (Figure 5C). Moreover, the repression of site 1 and site 2 was completely blocked when the target sequences were mutated. Thus, miR-998 directly repressed luciferase sensor constructs carrying dCbl target sequences. While our sensor assay results are consistent with the notion that miR-998 directly represses dCbl in eye discs, we acknowledge the possibility that miR-998 represses dCbl indirectly through a different mechanism in vivo.
Since dCbl is a negative regulator of EGFR signaling, its elevated expression in mir-998 mutants may explain the increased apoptosis in the morphogenetic furrow of rbf, mir-998 double mutants. To test this idea, we examined the effect of dCbl overexpression on cell death in rbf mutant eye discs. When dCbl-S or dCbl-L were expressed in rbf120a mutant eye discs using the Flip-out system, a wider band of C3 antibody staining was detected than in the control rbf120a mutant eye disc. Importantly, co-expression of miR-998 blocked the increase in cell death induced by dCbl, which is consistent with the notion that miR-998 can repress dCbl expression in vivo (Figure 5D). In the converse experiment, clones of dCbl mutant cells were generated in rbf120a mutant background and mosaic eye discs were stained with the C3 antibody. Consistent with results of dCbl overexpression described above, the loss of dCbl strongly suppressed apoptosis in rbf mutants as the number of C3 positive cells was dramatically reduced in the dCbl mutant tissue (Figure 5E). Therefore miR-998 represses dCbl, a negative regulator of EGFR signaling that is functionally important for triggering cell death, in the morphogenetic furrow of rbf mutant eye discs. Furthermore, dCbl is a critical target of miR-998 in modulating apoptosis in rbf-deficient cells since overexpression of dCbl in eye discs mimics the mir-998 mutant phenotype, while the dCbl mutant mimics the miR-998 overexpression phenotype.
miRNA-dependent repression of dCbl is conserved in mammalian cells
miR-998 is part of the mir-29 seed family of miRNAs, which is defined by having identical seed sequences ([30] and Figure 5B). The presence of multiple miR-29 target sites in mammalian c-Cbl raises the question of whether miRNA-dependent regulation of dCbl is conserved in mammals. To address this question we generated and expressed three different luciferase sensors carrying single, or paired predicted miR-29 target sites from the human c-Cbl gene, along with increasing amounts of a miR-29a expression plasmid (Figure 6A). While some miRNAs exert strong repression of their targets, others including miR-29 have been shown to elicit more modest effects in sensor assays and in vivo [32]–[36]. Indeed, miR-29a exerted modest but clearly dose-dependent repression of a sensor carrying two sites in tandem in the TKB domain near the 5′ end of the CDS (site 1). miR-29a also repressed a luciferase sensor carrying a predicted miR-29 site in the 3′UTR (site 2), while a sensor carrying tandem sites near the 3′ end of the 3′UTR failed to respond to miR-29 (site 3) (Figure 6A). To address the specificity of repression, we introduced mutations into the mir-29 target sequences in the c-Cbl site 1 and site 2 luciferase sensors and found that miR-29 did not repress these mutant sensors, regardless of the amount of transfected miR-29. Therefore, even though miR-29 modestly repressed sensors containing site 1 and site 2, this repression was specific since the response to miR-29 was dose-dependent, while mutating the sites completely blocked the effect of miR-29. We concluded that miR-29 seed family members can directly target c-Cbl through corresponding paired sites that are present within the highly conserved region in both Drosophila and human genes encoding the TKB domain. In addition, Drosophila and human miR-29 seed family members can regulate Cbl sensors through distinct target sites that are not conserved between the two species (Figure 5C and Figure 6A).
Fig. 6. c-Cbl is a target of hsa-miR-29 in mammalian cells. 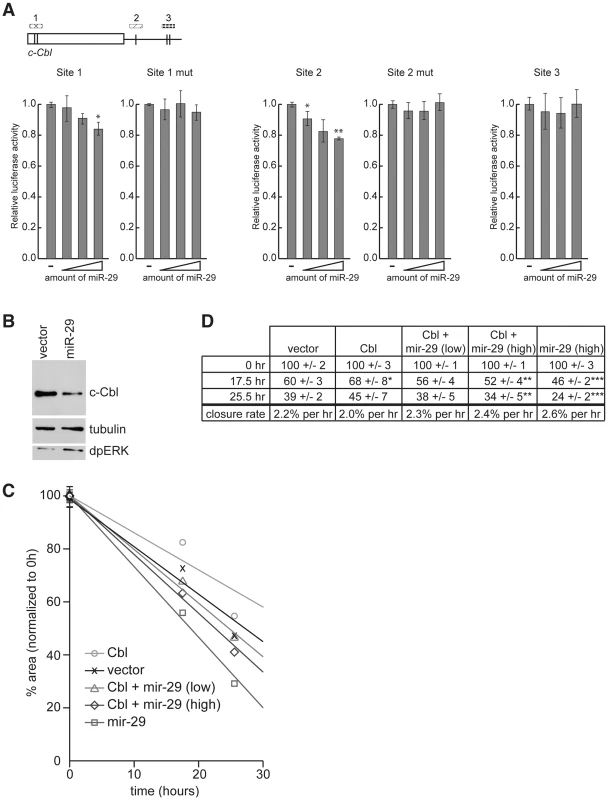
(A) 3′ UTR sensor assays were performed in HeLa cells using sequences predicted to be regulated by miR-29. The indicated wild-type or mutant 3′ UTR sensor plasmids were transfected with pcDNA3/empty alone, or increasing amounts of pcDNA3/mir-29 plasmids. Cells were harvested 24–48 h post-transfection, and luciferase activities were measured. A minimum of three independent transfections was performed for each sensor construct. Error bars represent standard error. Asterisks are shown for differences between vector and miR-29 that are statistically significant by t-test (* is p<0.05, ** is p<0.01). (B) Cell lysates were prepared from HeLa cells transiently transfected with pcDNA3/empty or pcDNA3/mir-29 plasmids. The expression of c-Cbl, dpERK, and tubulin were detected by Western blot. (C and D) Wound healing assays were performed on HeLa cells transfected with c-Cbl, miR-29a, or an empty vector control. The area of the wound was measured at each time point and normalized to 0 h. Eight wound areas were measured for each sample at each time point. (C) Average wound areas relative to the initial scratch were plotted for each transfection condition. (D) Summary of scratch assay data. Shown are average wound area and standard error for each transfection condition, and the rate of wound closure (change in % scratch area/time) represented in (C) as trendlines. Asterisks are shown for differences between sample and vector control that are statistically significant by t-test (* is p<0.25, ** is p<0.15, and *** is p<0.005). Having established the functionality of putative miR-29 target sites in the c-Cbl sequence using sensor assays, we asked whether the endogenous c-Cbl could be repressed by miR-29. HeLa cells were transfected with a miR-29 expression plasmid or an empty vector and the level of c-Cbl was analyzed by Western blot analysis 48 hr after transfection. As shown in Figure 6B, the expression of endogenous c-Cbl protein was significantly reduced in cells expressing miR-29 compared to the vector control. Importantly, downregulation of c-Cbl was accompanied by an elevated level of di-phosphorylated ERK (dpERK), which reflects an increase in EGFR/MAPK activity, in response to miR-29a expression.
Previous work showed that in the absence of functional Cbl, the sensitivity of cells to signals from the extracellular milieu is enhanced, and cells exhibit increased growth factor-induced motility in wound-healing scratch assays. The cell migration is mediated in part by ERK-MAPK signaling [37]–[39]. Therefore we tested whether reduction of c-Cbl by miR-29a expression alters the rate of wound-healing in scratch assays. Scratch assays were performed on HeLa cells transfected with constructs expressing miR-29a, c-Cbl, c-Cbl and miR-29a, or an empty vector control. The area of the wound was measured 0, 17.5, and 25.5 hours after the scratch was introduced (Figure 6C and 6D). As expected, cells expressing c-Cbl exhibited decreased motility and delayed wound closure compared to the control. In contrast, expression of miR-29a significantly increased the rate of wound healing compared to both cells expressing c-Cbl, and cells transfected with empty vector. miR-29a also increased the rate of wound healing in cells transfected with c-Cbl, which was consistent with the notion that miR-29 directly represses c-Cbl expression and, as a consequence, its function in vivo. 25.5 hours after the scratches were introduced, cells expressing miR-29a had closed all but 24% of the original area of the scratch wound, while the scratch wound of controls cells occupied 39% of the original area, and the scratch in cells expressing c-Cbl was 55% of the original area (Figure 6C and 6D). Co-expression of lower and higher amounts of miR-29 with c-Cbl led to scratches that were 38% and 34% the original area after 25.5 hours, suggesting that miR-29 can suppress the function of its target in vivo.
From these experiments we concluded that c-Cbl expression is modulated by miR-29a in mammalian cells and that miR-29a represses endogenous c-Cbl expression. Importantly, repression of c-Cbl expression is accompanied by increased ERK-MAPK signaling and an elevated rate of growth factor-regulated cell migration.
Discussion
The potential for complex interactions between intronic miRNAs and their host is illustrated by the Drosophila dE2f1 gene, and the two miRNAs embedded in its last intron: mir-11 and mir-998. We previously showed that mir-11 directly represses a subset of apoptotic genes that are transcriptional targets of the host gene dE2f1, and in doing so, miR-11 limits E2F-dependent cell death induced by DNA damage. Here, we identified a novel layer of regulation in the dE2f1 locus as miR-998 enhances EGFR cell survival signaling, thereby suppressing E2F-dependent cell death in rbf mutant animals. Therefore, the proapoptotic function of dE2F1 is under intrinsic control by two distinct and complementary mechanisms.
Many miRNAs elicit relatively weak effects and therefore their mutant phenotypes are rather subtle. Therefore it is not surprising that both the mir-11 and mir-998 mutant alleles were viable, and exhibited no phenotypes on their own. The lack of a mutant phenotype represents a major hurdle in identifying the physiological function of a miRNA [2], [40]. A number of approaches have been taken to reveal miRNA functions such as generating compound mutants, and analyzing mutant phenotypes in the context of disruptions to core regulatory pathways [3]. In our work we have used a different strategy and investigated miRNAs in the context of the function of their host gene. This novel approach turned out to be highly informative and allowed us to identify the elusive functions of two intronic miRNAs embedded within the dE2f1 gene. Notably, both miRNAs exhibited phenotypes only in E2F-sensitized backgrounds but lacked phenotypes on their own. One implication of our work is that the functions of intronic miRNAs can be linked to their host gene, and where known, the host gene function can be exploited to uncover the physiological roles of embedded miRNAs. This idea is particularly relevant given that approximately 40% of all miRNAs are embedded in protein-coding genes and therefore such approach can be applicable beyond the dE2f1 locus.
In various systems, inactivation of Rb provides a cellular context to investigate E2F-dependent apoptosis. Interestingly, animal models revealed that not every Rb mutant cell is equally sensitive to apoptosis. In the Drosophila rbf mutant eye disc, apoptosis occurs in a highly reproducible pattern that is determined by the level of prosurvival signaling from the EGF receptor [13]. Our data show that the loss of mir-998 enhanced cell death in rbf mutant eye discs but did not alter the overall pattern of apoptosis. Notably, we did not find evidence that miR-998 directly repressed cell death genes. Therefore, miR-998 is unlikely to function by altering the expression of apoptosis genes. Rather, our results support a scenario where miR-998 represses cell death in rbf mutants by enhancing pro-survival EGFR signaling. Using genome-wide approaches we identified dCbl as a highly upregulated gene in mir-998 mutant eye discs. Genetic interaction tests demonstrated that dCbl phenocopies the effect of mir-998 on dE2f1-dependent cell death in rbf mutants. Therefore dCbl behaves as a critical player that mediates the effect of mir-998 on apoptosis. We acknowledge that our data do not rule out the possibility that the effect of miR-998 on dCbl is indirect. For example, miR-998 may function by limiting the level of a positive regulator of dCbl expression. However, miR-998 can directly repress dCbl in sensor assays and this repression occurs in a sequence-dependent manner mediated by three different miR-998 target sites in dCbl. Similarly, a miR-998 seed family homolog, mir-29 repressed mammalian c-Cbl sensors in HeLa cells. Thus, while the mechanism of regulation of Cbl by the mir-998/mir-29 seed family in vivo may involve indirect or direct regulation, we favor a model where miR-998 modulates EGFR signaling by directly regulating dCbl. Further testing of this model would require introducing mutations in the miR-998 binding sites in the endogenous dCbl gene in order to generate dCbl alleles that are insensitive to direct targeting by miR-998.
Cbl is a negative regulator of EGFR signaling that binds the activated receptor and induces its ubiquitination and subsequent endocytosis, after which either further downregulation occurs through receptor degradation, or the receptor is retained in endosomes, or recycled back to the plasma membrane [24]. In the eye disc, the transient decrease in EGFR signaling prior to ommatidial specification occurs in part through the sequestration of the EGFR ligand, Spitz, which limits communication from neighbouring cells [13], [41], [42]. Moreover, expression of an EGFR isoform that cannot transmit ligand-initiated signals also stimulated EGFR-dependent cell death in rbf mutants [13]. Consistent with this mode of regulation, we showed that changes in the levels of dCbl, which limits EGFR signaling from the plasma membrane, modulated E2F-dependent cell death in rbf mutants. We suggest that through repression of dCbl expression, miR-998 supported ligand-dependent EGFR signaling in this context, which limited the proapoptotic activity of dE2F1.
miR-998 belongs to the miR-29 seed family of miRNAs, which also includes miR-285 and miR-998 in Drosophila [30]. While no mutant phenotypes have been reported, a recent investigation of gain-of-function phenotypes showed similar wing defects caused by overexpression of miR-285 and miR-995, although their molecular basis unknown [43]. Similarly, the functions of miR-29 seed family members cel-miR-49 and cel-miR-83 have not been reported. Seed family members in humans include the less abundant mature miRNA generated from the mir-21 oncomir, mir-21* (mir-21-3p), and mir-593* (mir-593-5p), and miR-29a, miR-29b and miR-29c. Unlike for other seed family members, a number of functions and targets of miR-29 have been reported. While it is not clear whether these functions and targets are common to other seed family members, this possibility warrants further investigation.
In humans, two intergenic mir-29 clusters give rise to three different mature miR-29 miRNAs that share an identical seed sequence, but differ slightly in their 3′ sequences. miR-29 was shown to disrupt epithelial polarity, and cooperate with oncogenic Ras in inducing epithelial-to-mesenchymal transition (EMT) and metastasis through the repression of the tristetraprolin nuclease [44]. Here, we identify a previously unknown miR-29 target, c-Cbl, which functions upstream of Ras in many signaling pathways. This novel link raises the question of whether miR-29 modulates receptor tyrosine kinase-induced EMT. Additional evidence for an oncogenic function of miR-29 came from the identification of PTEN as a miR-29 target [45]. Moreover, the oncogenic viral miRNA BLV-miR-B4 shares the same seed sequence as miR-29 and directly regulates miR-29 targets peroxidasin and HBP1 [46]. However, in different contexts, miR-29 has been shown to function as a tumor suppressor [47]–[49], and miR-29c repressed cancer cell proliferation, and limited E2F activity through the indirect activation of Rb [50]. It is not yet clear whether either of the two mir-29 clusters are transcriptionally regulated by E2F, although the expression of miR-29a is induced in the G1 phase of the cell cycle [51], which is coincident with increased E2F activity.
c-Myc bound both mir-29 cluster gene promoters and repressed miR-29 expression, suggesting that miR-29 is directly regulated by c-Myc [52]. Furthermore, expression of miR-29 lead to a decrease in the expression of E2F targets Cyclin A2, MCM2, and PCNA [52]. It is currently unclear whether the expression of E2F and miR-29 are linked directly, or through c-Myc, which is well-known to induce the expression of E2F1. We note that while miR-998 and miR-29 share at least one common target, their mechanisms of interaction with E2F function may differ. Interestingly, overexpression of dMyc in Drosophila embryos lead to both cell death, and decreased expression of miR-998 [53]. It is not known whether miR-998 could block dMyc-induced cell death in Drosophila embryos, or whether this would be accomplished through repression of dCbl, and a consequent increase of EGFR signaling.
EGFR/MAPK and Rb/E2F pathways intersect in the regulation of proliferation and cell death and are frequently disrupted in cancer. Our results reveal a novel connection between Rb/E2F and EGFR signaling: miR-998 interacts with both pathways and integrates their activities to effect an overall cellular response. This link may represent an attractive target to intentionally perturb cellular homeostasis thereby sensitizing cells to therapeutic reagents in the treatment of malignancy.
Materials and Methods
Fly stocks
All fly crosses were done at 25°C. The following stocks were obtained from Bloomington Drosophila Stock Center at Indiana University: GMR-Gal4, w; Dr/Δ2-3 99B, Sb, dE2f1EY05005 (FBal0160590), dE2f17172 and; egfrE1. The following stocks were previously published: UAS-miR-11 (Brennecke et al. 2005), Act88F-Gal4 and Act88F-Gal4, UAS-dE2f1 (from Erick Morris and Teiichi Tanimura), CblF165, FRT 80B, UAS-Cbl-L (A18) and UAS-Cbl-S (A1) (from Trudi Schupbach), rbf1120a, ey-FLP; act5c>CD2>GAL4, UAS-GFP/CyO, GFPAct, and rbf1120a, ey-FLP/FM7, GFPAct;; FRT 82B, GFPUbi, and rbf1120a, ey-FLP/FM7, GFPAct;; GFPUbi, FRT 80B (from Nam Sung Moon). dE2F1Δ1 is a deletion which lacks the genomic region between P element insertion P[XP]E2fd01508 and piggyBac insertion PBac[RB]InRe01952 and generated according to [54]. E2f1d01508 (FBal0183912), and InRd03668 (FBti0055281) were obtained from the Exelixis collection at Harvard Medical School. Generation of mir-998exc222 mutant allele is described in Protocol S1.
Construction of UAS-mir-998 transgene
For in vivo expression of miR-998, an insert encoding miR-998 was cloned into the pUAST plasmid using standard molecular cloning techniques. This insert contained the de2f1 intron 5′ sequence flanking two mir-1 chimeras: mir1/mCherry shmiR [55] replaced the mir-11 gene, and mir-1/998 replaced the mir-998 gene, which was designed as in Haley et al. (2008) and was synthesized by GenScript USA. Flies carrying UAS-miR-998 were generated by P-element transformation at The Best Gene, Inc.
Wound healing assay
HeLa cells were seeded in 6-well plates, transfected as described, and cultured until 100% confluent. Straight scratches were made across the cell layer using a 0.2 ml pipette tip. The cells were then gently washed three times with PBS to remove cellular debris and the media was replaced at 0 and 17.5 hours. Photographs of the wound region were taken using a Zeiss AxioObserver A1 microscope and AxioCam IC camera. The wound area was calculated using Image J software.
3′ UTR sensor plasmid construction
Sequences were cloned downstream of the Renilla luciferase coding sequence in the psiCheck2 (Promega) plasmid using standard cloning techniques. See Table S6 for sensor sequences.
Cell culture, transfection and Luciferase assay
HeLa cells were cultured in DMEM+10% FBS, and were transfected with the X-treme Gene HP transfection reagent (Roche) according to the manufacturer's protocol. Cells were harvested 24–48 hours post-transfection. pcDNA3/mir-998 generated by PCR amplification of the mir-998 gene and standard molecular cloning techniques. pcDNA3/hsa-mir-29a was generated by insertion of a GeneString (Invitrogen) with the mir-29a gene sequence in pcDNA3. Insert sequences were verified by sequencing analysis. Sequences are in Table S4. Firefly and Renilla luciferase activity were measured using the Dual Luciferase Assay protocol (Promega).
Western blot
Antibodies: c-Cbl 1∶200 (sc-170), from Santa Cruz; di-phosphorylated p42/p44 ERK 1∶5000 (M8159) from Sigma-Aldrich; mouse anti-tubulin 1∶10000 (cat# T9026) from Sigma-Aldrich; goat anti-rabbit-HRP (#31460) and goat anti-mouse-HRP (#31430) from Thermo Fisher.
Immunohistochemistry
Antibodies used were as follows: rabbit anti-C3 (Cleaved Caspase3), lot 26, 1∶75 (Cell Signaling), mouse anti-BrdU 1∶50 (Beckton Dickinson), rat anti-ELAV 1∶50, (Developmental Studies Hybridoma Bank), phosphorylated p42/p44 ERK 1∶200 (M8159) from Sigma-Aldrich, and Cy3-, and Cy5 - conjugated anti-mouse, and anti-rabbit secondary antibodies (Jackson Immunoresearch Laboratories). Larval tissues were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 30 minutes, permeabilized in 0.3% Triton X-100 in PBS twice for 10 minutes each, blocked in PBS with 0.1% Triton X-100 for 30 minutes at 4°C, and then incubated with antibodies overnight at 4°C in10% normal goat serum, and 0.3% Triton X-100 in PBS. After washing three times for 10 minutes each at room temperature in 0.1% Triton X-100 (in PBS), samples were incubated with appropriate conjugated secondary antibodies for 45 minutes at room temperature in 10% normal goat serum, and 0.3% Triton X-100 (in PBS). After washing with 0.1% Triton X-100 (in PBS), tissues were stored in glycerol+antifade reagents and then mounted on glass slides. To detect S phases, dissected larval eye discs were labeled with BrdU for 2 hrs at room temperature and then fixed overnight in 1.5% formaldehyde, 0.2% Tween 20 in PBS at 4°C. Samples were then digested with DNase (Promega) for 30 minutes at 37°C. Samples were then probed with primary and secondary antibodies as described above. All immunofluorescence was done on a Zeiss Confocal microscope and images were prepared using Adobe Photoshop CS4. All images are confocal single plane images unless otherwise stated as projection images. A minimum of 10 larvae were used for each analysis.
miRNA target prediction
Comprehensive lists of predicted miR-998 targets and miR-11 targets were compiled from TargetScan [56], MinoTar [57], PITA [58], miRanda [59], and RNAhybrid [60]. Target predictions use for hsa-miR-29 were from TargetScan [61].
qRT-PCR
Total RNA was isolated from 10 adult heads, 10 larvae, or 30–50 eye discs, with TRIzol (Invitrogen). Reverse transcription to measure standard mRNAs was performed using the iScript kit (BioRad) according to manufacturer's specifications. Quantitative PCR was performed with the SYBR Green I Master (Roche) on a Light Cycler 480 (Roche). miR-11 and miR-998 were measured by Taqman assay (Applied Biosystems). Primer sequences are in Table S5.
Isolation of mir-998exc222 by P-element excision mutagenesis
See Protocol S1.
Microarray data analysis and enrichment analysis
Microarray gene expression data were analyzed using “Affy” package [62] and differential expression analysis by “Limma” package [63]. Functional and pathway enrichment analysis of differentially expressed genes were counducted using Gitools [64]. See Protocol S1 for detail.
Supporting Information
Zdroje
1. MiskaEA, Alvarez-SaavedraE, AbbottAL, LauNC, HellmanAB, et al. (2007) Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3: e215 doi:10.1371/journal.pgen.0030215
2. Alvarez-SaavedraE, HorvitzHR (2010) Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20 : 367–373 doi:10.1016/j.cub.2009.12.051
3. BrennerJL, JasiewiczKL, FahleyAF, KempBJ, AbbottAL (2010) Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol 20 : 1321–1325 doi:10.1016/j.cub.2010.05.062
4. IzumiyaM, OkamotoK, TsuchiyaN, NakagamaH (2010) Functional screening using a microRNA virus library and microarrays: a new high-throughput assay to identify tumor-suppressive microRNAs. Carcinogenesis 31 : 1354–1359 doi:10.1093/carcin/bgq112
5. HerranzH, HongX, HungNT, VoorhoevePM, CohenSM (2012) Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes & Development 26 : 1602–1611 doi:10.1101/gad.192021.112
6. KimVN, HanJ, SiomiMC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10 : 126–139 doi:10.1038/nrm2632
7. BaskervilleS, BartelDP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11 : 241–247 doi:10.1261/rna.7240905
8. TruscottM, IslamABMMK, López-BigasN, FrolovMV (2011) mir-11 limits the proapoptotic function of its host gene, dE2f1. Genes & Development 25 : 1820–1834 doi:10.1101/gad.16947411
9. Najafi-ShoushtariSH, KristoF, LiY, ShiodaT, CohenDE, et al. (2010) MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science 328 : 1566–1569 doi:10.1126/science.1189123
10. DillH, LinderB, FehrA, FischerU (2012) Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes & Development 26 : 25–30 doi:10.1101/gad.177774.111
11. HinskeLCG, GalantePAF, KuoWP, Ohno-MachadoL (2010) A potential role for intragenic miRNAs on their hosts' interactome. BMC Genomics 11 : 533 doi:10.1186/1471-2164-11-533
12. van RooijE, SutherlandLB, QiX, RichardsonJA, HillJ, et al. (2007) Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316 : 575–579 doi:10.1126/science.1139089
13. MoonN-S, Di StefanoL, DysonN (2006) A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Molecular and Cellular Biology 26 : 7601–7615 doi:10.1128/MCB.00836-06
14. MorrisEJ, MichaudWA, JiJ-Y, MoonN-S, RoccoJW, et al. (2006) Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet 2: e196 doi:10.1371/journal.pgen.0020196
15. DuW, XieJE, DysonN (1996) Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J 15 : 3684–3692.
16. GabayL, SegerR, ShiloBZ (1997) In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277 : 1103–1106.
17. KumarJP, MosesK (2001) EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell 104 : 687–697 doi:10.1016/S0092-8674(01)00265-3
18. LesokhinAM, YuSY, KatzJ, BakerNE (1999) Several levels of EGF receptor signaling during photoreceptor specification in wild-type, Ellipse, and null mutant Drosophila. Dev Biol 205 : 129–144 doi:10.1006/dbio.1998.9121
19. FreemanM (1996) Reiterative Use of the EGF Receptor Triggers Differentiation of All Cell Types in the Drosophila Eye. Cell 87 : 651–660.
20. ZakNB, ShiloBZ (1992) Localization of DER and the pattern of cell divisions in wild-type and Ellipse eye imaginal discs. Dev Biol 149 : 448–456.
21. KanehisaM, GotoS (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28 : 27–30.
22. CarbonS, IrelandA, MungallCJ, ShuS, MarshallB, et al. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25 : 288–289 doi:10.1093/bioinformatics/btn615
23. Moses K1 (2002) Drosophila eye development. Berlin ; New York : Springer.
24. AbellaJV, ParkM (2009) Breakdown of endocytosis in the oncogenic activation of receptor tyrosine kinases. Am J Physiol Endocrinol Metab 296: E973–E984 doi:10.1152/ajpendo.90857.2008
25. PaiLM, BarceloG, SchüpbachT (2000) D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103 : 51–61 doi:10.1016/S0092-8674(00)00104-5
26. JékelyG, SungH-H, LuqueCM, RørthP (2005) Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Developmental Cell 9 : 197–207 doi:10.1016/j.devcel.2005.06.004
27. ChangW-L, LiouW, PenH-C, ChouH-Y, ChangY-W, et al. (2008) The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development 135 : 1923–1933 doi:10.1242/dev.017103
28. PaiL-M, WangP-Y, ChenS-R, BarceloG, ChangW-L, et al. (2006) Differential effects of Cbl isoforms on Egfr signaling in Drosophila. Mech Dev 123 : 450–462 doi:10.1016/j.mod.2006.04.001
29. WangP-Y, PaiL-M (2011) D-Cbl Binding to Drk Leads to Dose-Dependent Down-Regulation of EGFR Signaling and Increases Receptor-Ligand Endocytosis. PLoS ONE 6: e17097 Available: http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0017097.
30. Ibáñez-VentosoC, VoraM, DriscollM (2008) Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 3: e2818 doi:10.1371/journal.pone.0002818
31. MirandaKC, HuynhT, TayY, AngY-S, TamW-L, et al. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126 : 1203–1217 doi:10.1016/j.cell.2006.07.031
32. HyunS, LeeJH, JinH, NamJ, NamkoongB, et al. (2009) Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139 : 1096–1108.
33. BellareP, GanemD (2009) Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6 : 570–575 doi:10.1016/j.chom.2009.11.008
34. MuniyappaMK, DowlingP, HenryM, MeleadyP, DoolanP, et al. (2009) MiRNA-29a regulates the expression of numerous proteins and reduces the invasiveness and proliferation of human carcinoma cell lines. Eur J Cancer 45 : 3104–3118 doi:10.1016/j.ejca.2009.09.014
35. BaekD, VillénJ, ShinC, CamargoFD, GygiSP, et al. (2008) The impact of microRNAs on protein output. Nature 455 : 64–71 doi:10.1038/nature07242
36. MukherjiS, EbertMS, ZhengGXY, TsangJS, SharpPA, et al. (2011) MicroRNAs can generate thresholds in target gene expression. Nat Genet 43 : 854–859 doi:10.1038/ng.905
37. HirschDS, ShenY, WuWJ (2006) Growth and motility inhibition of breast cancer cells by epidermal growth factor receptor degradation is correlated with inactivation of Cdc42. Cancer Research 66 : 3523–3530 doi:10.1158/0008-5472.CAN-05-1547
38. TanY-HC, KrishnaswamyS, NandiS, KantetiR, VoraS, et al. (2010) CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS ONE 5: e8972 doi:10.1371/journal.pone.0008972
39. VialE, SahaiE, MarshallCJ (2003) ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 4 : 67–79 doi:10.1016/S1535-6108(03)00162-4
40. AmbrosV (2010) MicroRNAs: genetically sensitized worms reveal new secrets. Curr Biol 20: R598–R600 doi:10.1016/j.cub.2010.05.054
41. KleinDE, NappiVM, ReevesGT, ShvartsmanSY, LemmonMA (2004) Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature 430 : 1040–1044 doi:10.1038/nature02840
42. FreemanM (1997) Cell determination strategies in the Drosophila eye. Development 124 : 261–270.
43. BejaranoF, Bortolamiol-BecetD, DaiQ, SunK, SajA, et al. (2012) A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139 : 2821–2831 doi:10.1242/dev.079939
44. GebeshuberCA, ZatloukalK, MartinezJ (2009) miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 10 : 400–405 doi:10.1038/embor.2009.9
45. TumanengK, SchlegelmilchK, RussellRC, YimlamaiD, BasnetH, et al. (2012) YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol 14 : 1322–1329 doi:10.1038/ncb2615
46. KincaidRP, BurkeJM, SullivanCS (2012) RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci USA 109 : 3077–3082 doi:10.1073/pnas.1116107109
47. MottJL, KobayashiS, BronkSF, GoresGJ (2007) mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene 26 : 6133–6140 doi:10.1038/sj.onc.1210436
48. AnastasiadouE, BoccellatoF, VincentiS, RosatoP, BozzoniI, et al. (2010) Epstein-Barr virus encoded LMP1 downregulates TCL1 oncogene through miR-29b. Oncogene 29 : 1316–1328 doi:10.1038/onc.2009.439
49. ParkS-Y, LeeJH, HaM, NamJ-W, KimVN (2009) miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16 : 23–29 doi:10.1038/nsmb.1533
50. BaeHJ, NohJH, KimJK, EunJW, JungKH, et al. (2013) MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene 33 : 2557–67 doi:10.1038/onc.2013.216
51. BuenoMJ, Gómez de CedrónM, LaresgoitiU, Fernández-PiquerasJ, ZubiagaAM, et al. (2010) Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Molecular and Cellular Biology 30 : 2983–2995 doi:10.1128/MCB.01372-09
52. MarziMJ, PuggioniEMR, Dall'OlioV, BucciG, BernardL, et al. (2012) Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. J Cell Biol 199 : 77–95 doi:10.1083/jcb.201206033
53. DaneshvarK, NathS, KhanA, ShoverW, RichardsonC, et al. (2013) MicroRNA miR-308 regulates dMyc through a negative feedback loop in Drosophila. Biol Open 2 : 1–9 doi:10.1242/bio.20122725
54. ParksAL, CookKR, BelvinM, DompeNA, FawcettR, et al. (2004) Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36 : 288–292 doi:10.1038/ng1312
55. HaleyB, HendrixD, TrangV, LevineM (2008) A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev Biol 321 : 482–490 doi:10.1016/j.ydbio.2008.06.015
56. RubyJG, StarkA, JohnstonWK, KellisM, BartelDP, et al. (2007) Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Research 17 : 1850–1864 doi:10.1101/gr.6597907
57. Schnall-LevinM, ZhaoY, PerrimonN, BergerB (2010) Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci USA 107 : 15751–15756 doi:10.1073/pnas.1006172107
58. KerteszM, IovinoN, UnnerstallU, GaulU, SegalE (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39 : 1278–1284 doi:10.1038/ng2135
59. EnrightAJ, JohnB, GaulU, TuschlT, SanderC, et al. (2003) MicroRNA targets in Drosophila. Genome Biol 5: R1 doi:10.1186/gb-2003-5-1-r1
60. RehmsmeierM, SteffenP, HÖchsmannM, GiegerichR (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10 : 1507–1517 doi:10.1261/rna.5248604
61. LewisBP, BurgeCB, BartelDP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 : 15–20 doi:10.1016/j.cell.2004.12.035
62. GautierL, CopeL, BolstadBM, IrizarryRA (2004) affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20 : 307–315 doi:10.1093/bioinformatics/btg405
63. SmythGK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3 doi:10.2202/1544-6115.1027
64. Perez-LlamasC, López-BigasN (2011) Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE 6: e19541 doi:10.1371/journal.pone.0019541
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



