-
Články
- Vzdělávání
- Časopisy
Top články
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
DNA topoisomerase is an enzyme that releases the torsional stress in DNA generated during DNA replication or transcription. Here, we uncovered an unexpected role of DNA topoisomerase 1α (TOP1α) in the maintenance of genome stability. Eukaryotic genomes are usually littered with transposable elements (TEs) and repeats, which pose threats to genome stability due to their tendency to move or recombine. Mechanisms are in place to silence these elements, such as RNA-directed DNA methylation (RdDM) and histone H3 lysine 9 dimethylation (H3K9me2) in plants. Two plant-specific RNA polymerases, Pol IV and Pol V, generate small and long noncoding RNAs, respectively, from TEs and repeats. These RNAs then recruit protein factors to deposit DNA methylation or H3K9me2 to silence the loci. In this study, we found that treatment of plants with camptothecin, a TOP1α inhibitor, or loss of function in TOP1α, led to the de-repression of RdDM target loci, which was accompanied by loss of H3K9me2 or DNA methylation. The role of TOP1α in RdDM could be attributed to its promotion of Pol V, but not Pol IV, transcription to generate long noncoding RNAs.
Published in the journal: . PLoS Genet 10(7): e32767. doi:10.1371/journal.pgen.1004446
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004446Summary
DNA topoisomerase is an enzyme that releases the torsional stress in DNA generated during DNA replication or transcription. Here, we uncovered an unexpected role of DNA topoisomerase 1α (TOP1α) in the maintenance of genome stability. Eukaryotic genomes are usually littered with transposable elements (TEs) and repeats, which pose threats to genome stability due to their tendency to move or recombine. Mechanisms are in place to silence these elements, such as RNA-directed DNA methylation (RdDM) and histone H3 lysine 9 dimethylation (H3K9me2) in plants. Two plant-specific RNA polymerases, Pol IV and Pol V, generate small and long noncoding RNAs, respectively, from TEs and repeats. These RNAs then recruit protein factors to deposit DNA methylation or H3K9me2 to silence the loci. In this study, we found that treatment of plants with camptothecin, a TOP1α inhibitor, or loss of function in TOP1α, led to the de-repression of RdDM target loci, which was accompanied by loss of H3K9me2 or DNA methylation. The role of TOP1α in RdDM could be attributed to its promotion of Pol V, but not Pol IV, transcription to generate long noncoding RNAs.
Introduction
DNA methylation and histone H3 lysine 9 (H3K9) methylation are two chromatin modifications widely employed by eukaryotes to maintain genome stability [1], [2]. H3K9 methylation and DNA methylation are targeted via small interfering RNAs (siRNAs) to repeats and transposable elements (TEs) and are required for their transcriptional silencing [1], [2].
In plants, cytosine methylation is established through a process known as RNA-directed DNA methylation (RdDM), which involves small and long noncoding RNAs produced by plant-specific RNA polymerases, Pol IV and Pol V, respectively [2]. Pol IV is thought to transcribe RdDM target loci and generate long precursor RNAs. These are eventually processed into 24-nucleotide (nt) siRNAs that are loaded into the Argonaute protein AGO4 [3], [4], [5], [6], [7]. In parallel, Pol V generates long non-coding RNA transcripts from RdDM target loci, and these transcripts recruit siRNA-AGO4 to chromatin [8], [9]. Through the concerted action of these two polymerases, siRNA-AGO4 becomes localized to target loci, and this ultimately recruits the methyltransferase DRM2, which effects de novo DNA methylation. In plants, DNA methylation occurs in three sequence contexts, CG, CHG, and CHH. In contrast to CG and CHG methylation, which can be maintained through the DNA methyltransferases MET1 and CMT3, respectively, CHH methylation is propagated by constant de novo methylation through RdDM [2], [10].
In plants, H3K9 dimethylation (H3K9me2) is another repressive chromatin mark associated with TE and repeat silencing [11], [12], [13]. H3K9me2 and CHG methylation act in a self-reinforcing loop to promote the maintenance of these marks by histone methyltransferases KRYPTONITE (KYP or SUVH4), SUVH5 and SUVH6 and the DNA methyltransferase CMT3 [14]. How H3K9me2 is initially deposited is less well understood, but the RdDM pathway plays a role, as mutations in RdDM pathway genes cause marked reductions in H3K9me2 levels at RdDM target loci [7], [8], [15]. In fact, a recent study revealed a strong genome-wide inter-dependence between non-CG (CHG and CHH) DNA methylation and H3K9 dimethylation [16].
DNA topoisomerases are enzymes that maintain proper DNA topology [17]. During replication or transcription, the DNA helical structure opens to form the replication or transcription fork, and the DNA in front of the fork becomes positively supercoiled, while the DNA behind the fork becomes negatively supercoiled. Topoisomerases bind these regions, nick the DNA to relieve the torsional stress, and re-ligate the DNA. Topoisomerases are divided into two major types, I and II, and further subtypes depending on their mode of action and structure [17], [18].
In Arabidopsis, there are two genes encoding type IB topoisomerases, TOP1α and TOP1β, which are tandemly arrayed in the genome. top1α mutants exhibit gross morphological defects, while top1β mutants are phenotypically normal [19]. RNAi-mediated knockdown of TOP1β in a top1α background is lethal [19]; thus these two genes are functionally redundant.
Here, we uncover a role of TOP1α in transcriptional silencing of TEs. We exploited a luciferase-based reporter (LUCL) that undergoes transcriptional silencing by DNA methylation [20] to perform a chemical genetics screen. We found that camptothecin (CPT) released the DNA methylation of LUCL and de-repressed its expression. CPT is a well-studied natural quinoline alkaloid that targets type 1B topoisomerases [21], [22]. Both the addition of CPT and loss-of-function in TOP1α led to the de-repression of RdDM target loci accompanied by a release of DNA methylation and/or a decrease in H3K9me2 levels. TOP1α is dispensable for Pol IV-mediated siRNA biogenesis but is required for the production of Pol V-dependent, long non-coding RNA transcripts. Consistent with the current model that these transcripts recruit siRNA-AGO4 to chromatin, inactivation of TOP1α resulted in reduced AGO4 occupancy at these loci. Taken together, through the identification of TOP1α as a player in RdDM, we have assigned new roles to a protein affecting DNA topology.
Results
Camptothecin releases the silencing of LUCL
To identify genes involved in DNA methylation, we performed a chemical genetics screen with LUCL, a transcriptionally-silenced luciferase (LUC)-based reporter line [20]. In LUCL, LUC is driven by a dual 35S promoter and both the 35S promoter and the LUC coding region harbor DNA methylation [20]. The DNA methylation at LUCL, and consequently its transcriptional silencing, is controlled by MET1, and to a lesser extent, by the RdDM pathway [20].
Over 3,000 compounds were screened against LUCL seedlings for their effects on LUC expression. A hit compound, camptothecin (CPT) (Figure 1A), was found to release LUC silencing in a concentration - and time-dependent manner (Figure 1B and C). Interestingly, CPT released LUC silencing in a bi-phasic manner, with optimal levels at 10 µM. Further, the release of LUC activity was not observed until one day of chemical addition in a time course assay (Figure 1B). Consistently, continuous live imaging revealed that an increase in LUC activity occurred at about 15 hr after the addition of the chemical (Figure 1C). The slow kinetics suggested that cell division is likely necessary for the de-repression of the reporter. The effects of CPT on LUC protein activity reflected a release of LUCL silencing, as the addition of CPT led to an increase in LUC transcript levels (Figure 1D). Consistent with the dose-dependent effects of CPT on LUC activity, LUC transcript levels were most de-repressed at 10 µM of CPT (Figure 1D).
Fig. 1. CPT releases LUC activity of LUCL. 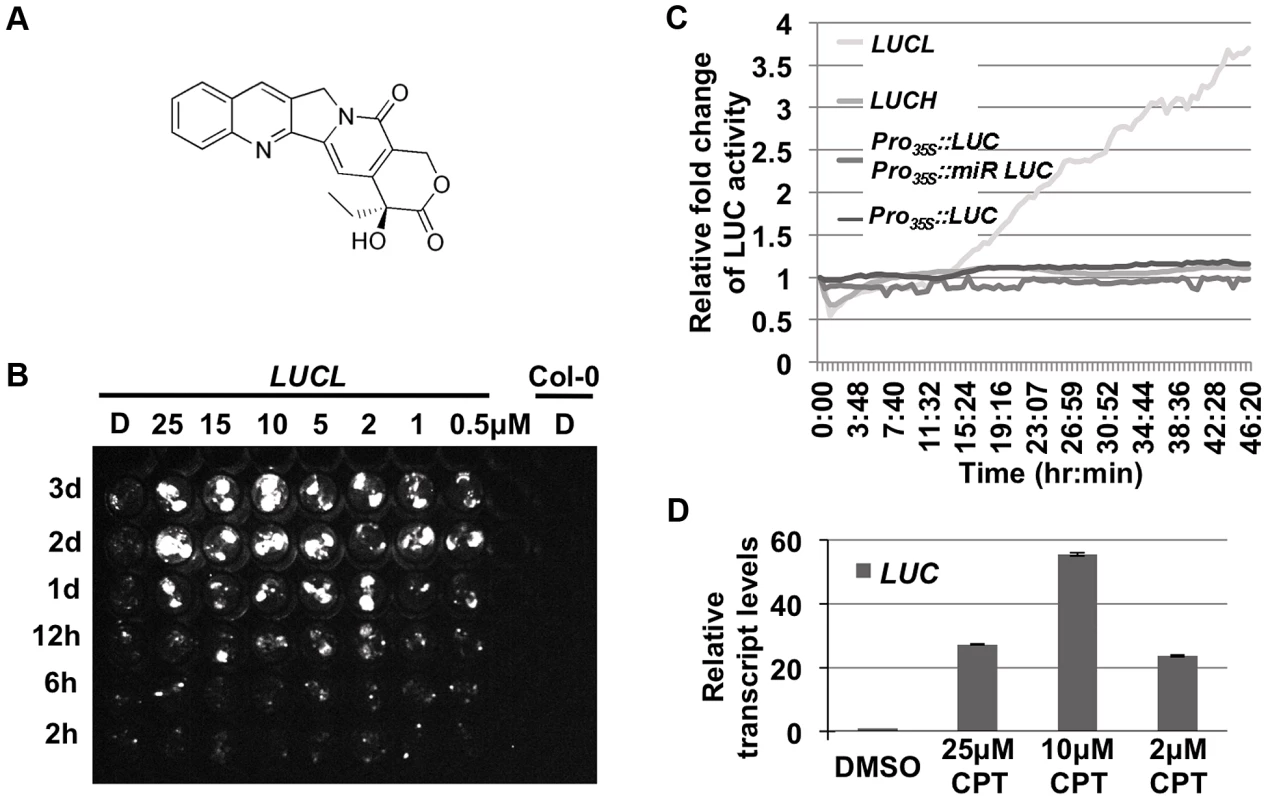
(A) Chemical structure of CPT. (B) Time- and concentration-dependent de-repression of LUCL by CPT. Seedlings were subjected to CPT treatment in 96-well plates and imaged for LUC activity at different time points (shown to the left of the image) after CPT addition. D = DMSO-treated seedlings. Col-0 wild type (without the LUCL transgene) was included as a negative control. (C) Effects of CPT treatment on various transgenes. LUC activity was measured over time using an automated luciferase reader. The x-axis indicates the time of incubation in terms of hours:minutes after addition of 10 µM CPT or DMSO. The y-axis indicates the relative fold change of LUC activity upon treatment of CPT as compared to DMSO. CPT affects the activity of LUCL but not Pro35S:LUC Pro35S:mir-LUC, a reporter line in which the LUC transgene reports miRNA activity [23], Pro35S:LUC, [23], or LUCH, a reporter line in which the LUC transgene is silenced specifically by CHH methylation [36]. (D) qRT-PCR measuring LUC transcript levels upon addition of different concentrations of CPT. Three biological replicates, each with three technical replicates, were performed. Error bars represent standard deviation from the three biological replicates. Previous experiments with LUCL ruled out that it reports miRNA activity, even though it contains the miR172 binding sequence [20]. Consistently, we found that the addition of CPT did not release the LUC activity of a miRNA reporter line, Pro35S::LUC Pro35S::miR-LUC (Figure 1C; [23]). Thus, CPT released the LUC activity of LUCL through a miRNA-independent mechanism.
To determine whether CPT increased LUC transcript levels by reducing DNA methylation, we performed McrBC-PCR to examine the methylation status of LUCL. After digestion of genomic DNA with McrBC, an enzyme that only cuts methylated DNA [24], 35S promoter sequences were amplified by PCR. In the DMSO-treated control sample, little product was observed, indicating that this region was highly methylated in LUCL. However, after CPT treatment, the amount of PCR products increased (Figure 2A), suggesting that CPT treatment led to a reduction in 35S promoter methylation. In addition, the DNA methylation status of the 35S promoter and the LUC coding region was examined by bisulfite sequencing (Figure 2B). The addition of 10 µM CPT resulted in a drastic reduction of CHH methylation, and to some extent CHG methylation, in region #1 (Figure 2C). CG methylation was largely unaffected upon CPT treatment, with the exception of region #4 (Figure 2C).
Fig. 2. Addition of CPT or loss of TOP1α releases DNA methylation. 
(A) McrBC-PCR-based methylation analysis of the 35S promoter in LUCL seedlings treated with CPT. At2g19920 is unmethylated and serves as an internal loading control. The number listed above indicates the concentration (in µM) of CPT added. D = DMSO. (B) A schematic diagram of the LUCL transgene. The numbered black lines indicate the regions for which bisulfite sequencing was performed (Figure 2C). (C) Levels of DNA methylation of LUCL in DMSO- and CPT-treated seedlings as determined by bisulfite sequencing. The regions correspond to the numbered lines in Figure 2B. (D) Loss of TOP1α results in reduced 5S rDNA methylation. Genomic DNA was digested with HpaII followed by Southern blotting. Less methylated DNA is expected to yield a higher intensity of bands lower down the gel as in nrpe1-11 (a Pol V mutant). In the CPT-treated sample, 25 µM of CPT was used. top1α-7, top1β-1, and nrpe1-11 are to be compared to Col-0 (wild type), and top1α-2 is to be compared to Ler (wild type). The CPT target, TOP1α, promotes CG methylation at 5S repeats
Due to their potent anti-cancer properties, CPT and its analogs have been intensely studied. The cellular target of CPT is topoisomerase I and the mechanism by which CPT inhibits topoisomerase I is well understood [25]. Given this knowledge, our finding that CPT de-represses LUCL implicated TOP1α in transcriptional gene silencing. A top1α mutant allele, top1α-2, had been found in an unrelated project (Xigang Liu and Xuemei Chen, unpublished results). The top1α-2 mutant carried a C→T point mutation in the second exon, which generates a premature stop codon (Figure S1A). top1α-2, which had been isolated in the Landsberg erecta background, was introgressed into Col-0 through five backcrosses to derive top1α-2Col. top1α-2Col was then crossed to LUCL in the Col-0 background. Unlike CPT, which released LUC activity, the top1α-2Col mutation was not able to release LUC activity (Figure S1B), probably due to activity of the partially redundant TOP1β gene.
We next asked whether TOP1α inactivation or CPT treatment affected DNA methylation of endogenous RdDM loci. 5S rDNA is present with thousands of copies in the genome and is under RdDM regulation [5]. We digested genomic DNA with HpaII, an enzyme that cuts unmethylated DNA in a CG context, to determine the status of 5S rDNA methylation. We found that, like nrpe1-11, a Pol V mutant, top1α-2 and CPT-treated seedlings had less methylated DNA, as indicated by the increase in intensity of the lower molecular weight restriction fragments (Figure 2D). top1α-7 (also known as mgo1-7 [26]; Figure S1A) has weaker developmental defects than top1α-2Col. The top1β-1 loss-of-function mutant in the Col-0 background (Figure S1A) has no obvious morphological defects (Xigang Liu and Xuemei Chen, unpublished results). CG methylation at 5S repeats was only weakly reduced in top1α-7 mutants and unaffected in top1β-1 mutants. Similarly, DNA blot analyses were conducted to examine CHG methylation at MEA-ISR and 180 bp repeats (Figure S1C and D), and CHH methylation at 5S rDNA repeats. Only a slight reduction in CHG methylation at the 180 bp repeats was detected in top1α-2 (Figure S1D). top1α-2 was indistinguishable from the isogenic Ler parental line in terms of CHG methylation at MEA-ISR (Figure S1C) or CHH methylation at 5S repeats (Figure S1D).
TOP1α has a limited role in DNA methylation in the genome
The studies above on a small number of loci revealed a limited role of TOP1α in DNA methylation. In order to obtain a global view of the function of TOP1α in DNA methylation, we performed whole genome bisulfite sequencing (MethylC-seq) on Ler, top1α-2, Col-0, top1α-7, nrpd1-3 (a Pol IV mutant) and nrpe1-11 seedlings. A total of 10 libraries representing one to three biological replicates of the genotypes (Table S1) were sequenced. Acceptable bisulfite conversion efficiency (Table S1) and read coverage (Table S2) were achieved for each library.
We identified differentially methylated regions (DMRs) using established procedures in the literature (see Material and Methods and Text S1). We compared each mutant to its wild-type control in the same biological replicate. We also called DMRs among the three Col-0 replicates to establish the background of spontaneous DMRs in wild type. Despite the high degree of reproducibility of the biological replicates (Table S3), when the three Col-0 replicates were subjected to the DMR analysis, we found thousands of CHH DMRs, but very few CG and CHG DMRs, between any two Col-0 replicates (Table S4A). In MethylC-seq data of three Col-0 replicates from a published study [27], we also identified thousands of CHH DMRs between any two replicates (Table S4B). This suggested that CHH methylation is considerably variable. In light of such variability, we took a conservative approach towards the identification of robust DMRs by considering only the overlap between two biological replicates or mutant alleles. For example, to derive DMRs between wild type and nrpd1-3 or nrpe1-11, we first compared the mutant to wild type within each biological replicate and then retained only DMRs that overlapped in both biological replicates (Table S5B and C). To derive DMRs between wild type and top1α, we first compared top1α-7 to Col-0 and top1α-2 to Ler, and then obtained the overlapped DMRs between the two alleles (Table S5A). In addition, hypervariability (HV) regions that are prone to changes in DNA methylation over generations [28], [29] were subtracted from the overlapped DMRs. The final set of CHH DMRs between wild type and nrpd1-3 (or nrpe1-11) consisted of over 7,500 loci showing reduced DNA methylation in the mutants (Table S5B and C), consistent with the known roles of Pol IV and Pol V in CHH methylation [27], [30]. The final set of DMRs between wild type and top1α consisted of the following: reduced in methylation in top1α — 97 (CHH), 35 (CG), and 0 (CHG); and increased in methylation in top1α — 10 (CHH), 9 (CG), and 1 (CHG) (Figure 3A, Table S5A and Table S6). The overall change in CHH methylation in top1α was very limited in comparison to that in nrpd1-3 or nrpe1-11 (Table S5). Most of the 97 WT-top1α CHH DMRs are in TEs or intergenic regions (Figure 3B). 91% of the WT-top1α CHH DMRs require Pol IV or Pol V for their CHH methylation (Figure 3C). This suggested that TOP1α promotes DNA methylation at a small number of RdDM loci.
Fig. 3. TOP1α does not globally impact DNA methylation but promotes CHH methylation at a small number of loci. 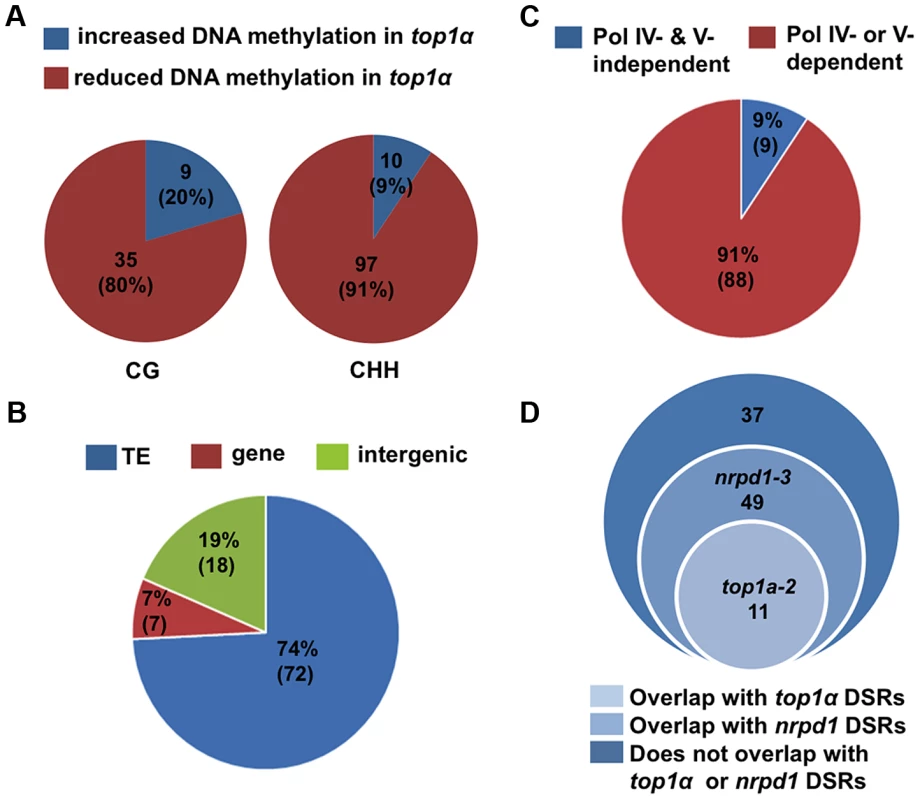
(A) Pie charts showing that the great majority of WT- top1α DMRs show reduced DNA methylation in top1α. Each circle represents total WT- top1α DMRs in a methylation context (CG or CHH). The red and blue areas represent DMRs with reduced and increased DNA methylation in top1α, respectively. The numbers indicate the numbers of DMRs in each category. The numbers in the parentheses represent the percentage of the DMR category in total DMRs. (B) The majority of WT- top1α CHH DMRs showing reduced DNA methylation in top1α overlap with TEs (74%, blue). Those that overlap with genes and intergenic regions are shown in red (7%) and green (19%), respectively. (C) The majority of WT- top1α CHH DMRs overlap with CHH DMRs between WT and nrpd1 or WT and nrpe1 (91%, red), suggesting that these regions require Pol IV or Pol V for CHH methylation. The portion of WT- top1α CHH DMRs not overlapping with WT-nrpd1 or WT-nrpe1 CHH DMRs are shown in blue (9%). (D) Overlap between WT- top1α CHH DMRs with DSRs (differential small RNA regions) between WT and nrpd1 or WT and top1α. There is little overlap between WT-top1α CHH DMRs and WT-top1α DSRs. TOP1α silences transposons through DNA methylation and H3K9 dimethylation
Since the methylation-sensitive DNA blot analyses only revealed an effect of top1α alleles on DNA methylation at the 5S and 180 bp repeats and the methylome profiling studies did not support a global role of TOP1α in DNA methylation, we sought to evaluate whether TOP1α is required for the transcriptional silencing of endogenous RdDM loci. qRT-PCR was performed to determine transcript levels from seven well-known RdDM loci. In both wild-type seedlings treated with CPT as well as top1α (both top1α-2 and top1α-7) seedlings, these endogenous siRNA target loci were de-repressed (Figure 4A). This confirmed a role of TOP1α in silencing the RdDM target loci.
Fig. 4. TOP1α promotes transposon silencing at endogenous RdDM target loci through H3K9me2 deposition. 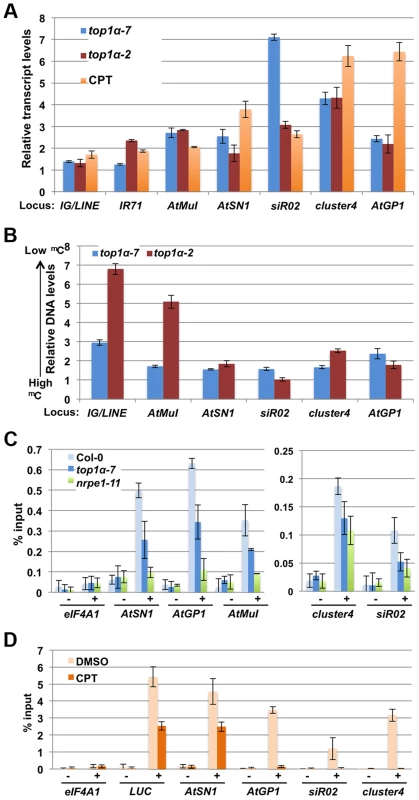
In (A) to (D), error bars represent standard deviation calculated from three biological replicates, each with three technical replicates. In (A) and (B), top1α-7, top1α-2 and CPT-treated LUCL were compared to Col-0, Ler, and DMSO-treated LUCL respectively. The relative levels to these controls (set to 1.0) are shown. The loci tested are labeled on the x axis. (A) Loss of TOP1α or addition of CPT results in RdDM target loci de-repression. (B) Loss of TOP1α or addition of CPT results in a release of DNA methylation at some loci. McrBC-qPCR analysis was performed to quantify DNA methylation levels in top1α or CPT-treated plants. Higher DNA levels in this assay correlate with lower levels of DNA methylation. At1g40129 served as an internal unmethylated control. (C) Loss of TOP1α results in reduced H3K9me2 levels at endogenous RdDM loci. ChIP-qPCR was performed to measure H3K9me2 levels at five RdDM target loci. eIF4A1, which does not harbor H3K9me2, was used as an internal control. − = samples processed without antibody. + = samples processed with anti-H3K9me2 antibodies. (D) CPT treatment results in reduced H3K9me2 levels at the LUCL transgene and four endogenous RdDM loci. LUCL plants were treated with either DMSO or 10 µM CPT and subjected to ChIP. qPCR was performed with the immunoprecipitated DNA for the LUCL transgene and four endogenous RdDM loci. eIF4A1 was used as an internal negative control. − = samples processed without antibody. + = samples processed with anti-H3K9me2 antibodies. We asked whether the release of transcriptional silencing of endogenous RdDM target loci (Figure 4A) in top1α or CPT-treated seedlings was accompanied by a loss of DNA methylation. We performed McrBC-qPCR assays to quantify the levels of DNA methylation amongst different genotypes/treatments at six endogenous RdDM loci. At most of the loci, DNA methylation was reduced in the two top1α mutants, but the reductions were small in top1α-7 (Figure 4B). Treatment of wild-type (Ler) plants with CPT resulted in reductions in DNA methylation at four of the six tested loci (Figure S1E). Although the overall trend of reduced DNA methylation in the two top1α mutants and CPT treated plants agreed with the observed de-repression of these loci, there were also inconsistencies whereby de-repression was not accompanied by reductions in DNA methylation, such as at siR02 in top1α-2 and CPT-treated plants.
This incomplete correlation between TE de-repression and a reduction in DNA methylation prompted us to ask whether TOP1α silences TEs through another mechanism. Previous studies have shown that H3K9me2 is a major repressive mark for transposon silencing and that H3K9me2-dependent silencing acts in concert or in parallel with RdDM [31], [32], [33]. Like DNA methylation, H3K9me2 is targeted to specific TEs through siRNA-AGO4 [7]. Thus, we investigated whether loss of TOP1α function or CPT treatment altered H3K9me2 levels at TEs. Chromatin immunoprecipitation (ChIP)-qPCR showed that H3K9me2 levels at AtSN1, sir02, cluster4, AtGP1, and AtMuI were reduced in both top1α-7 and nrpe1-11 (Figure 4C). We also performed ChIP-qPCR on LUCL seedlings treated with DMSO or CPT. CPT treatment was found to cause a strong reduction in H3K9me2 levels at four TE loci (Figure 4D). As CPT was initially isolated through a chemical genetics screen with LUCL, we asked whether the LUC transgene in LUCL also harbored H3K9me2 and, if so, whether CPT treatment reduced its H3K9me2 levels. Indeed, ChIP-qPCR showed that the d35S of the LUC transgene (region #1 in Figure 2B) harbored H3K9me2, with CPT treatment reducing H3K9me2 levels (Figure 4D).
As H3K9me2, which is introduced by KYP and its paralogs, and CHG methylation, which is deposited by CMT3, act in a self-reinforcing loop, and both H3K9me2 and CMT3 contribute to CHH methylation [14], [16], we asked whether the role of TOP1α in DNA methylation depends on KYP or CMT3. To address this question, we treated Ler (wild-type), kyp-2 and cmt3-7 plants with CPT to inhibit topoisomerase I activity and then assayed DNA methylation at six TE loci. CPT treatment of wild-type plants resulted in reduced DNA methylation at four of the six loci (Figure S1E). The reduction in DNA methylation caused by CPT treatment was minimal at these four loci in either cmt3-7 or kyp-2 (Figure S1E). This suggested that the effects of TOP1α in DNA methylation require CMT3 - and KYP-mediated H3K9 dimethylation.
TOP1α does not affect small RNA levels
The promotion of DNA methylation and/or H3K9me2 deposition at TEs implicates a role of TOP1α in RdDM, a process that involves Pol IV and Pol V. As topoisomerases are required to release DNA topological tension generated by transcription [17], it would be reasonable to expect that TOP1α is required for the activities of either Pol IV or Pol V. We first tested whether TOP1α is required for the activities of Pol IV, the output of which is the accumulation of 24-nt siRNAs from RdDM target loci. RNA blot analysis showed that siRNA accumulation at several loci was similar in Ler and top1α-2 (Figure S3A). To gain a global view on the potential relationship between TOP1α and Pol IV, we compared deep sequencing profiles of small RNAs from Ler, top1α-2, Col-0, nrpd1-3, and nrpe1-11. The size distributions of all small RNA reads in Ler and top1α-2 were almost identical (Figure S3B). To determine whether TOP1α affects siRNA accumulation at specific regions of the genome, we identified differential small RNA regions (DSRs). While large numbers of DSRs were found in nrpd1-3 or nrpe1-11 relative to the wild-type control, consistent with the essential role of Pol IV and the auxiliary role of Pol V in siRNA biogenesis [3], [5], [6], very few were found in top1α-2 (Table S7). Furthermore, analysis of small RNA abundance throughout the genome did not support a global role of TOP1α in small RNA accumulation (Figure S3C). Therefore, Pol IV activity does not appear to require TOP1α.
Given that we had found 71 WT-top1α DSRs (Table S7), we asked whether the reduced CHH methylation at the 97 WT-top1α DMRs was associated with reduced siRNA levels. We found that only 11 of the 97 DMRs overlapped with WT-top1α DSRs (Figure 3D). A representative of such a locus is shown in Figure S2A. Most of the 97 DMRs did not overlap with the 71 WT - top1α DSRs; two such loci are shown in Figure S2B and C. Therefore, the reduced CHH methylation in top1α could not be explained by reduced siRNA levels. On the other hand, more than 60% of the 97 WT-top1α DMRs overlapped with WT-nrpd1 DSRs (Figure 3D; Figure S2A and B), suggesting that these regions, which require TOP1α for CHH methylation, undergo Pol IV-dependent siRNA production. Therefore, TOP1α must promote CHH methylation at these RdDM loci independently of siRNA biogenesis.
TOP1α promotes the production of Pol V-dependent transcripts and AGO4 occupancy at TEs
We next tested whether TOP1α promotes the production of Pol V-dependent transcripts. We performed qRT-PCR and RT-PCR to detect Pol V-dependent transcripts from eight loci, MEA-ISR, AtSN1, and six IGN loci that produce such transcripts [9], [30]. At all eight loci, the levels of the Pol V-dependent transcripts were reduced in top1α-2 as compared to Ler (Figure 5A and B). We previously showed that Pol II generates long noncoding transcripts at the soloLTR locus [34]. The accumulation of these transcripts at soloLTR was also reduced in top1α-2 (Figure 5B). Therefore, TOP1α contributes to the production of Pol V-dependent or Pol II-dependent long noncoding transcripts.
Fig. 5. TOP1α promotes the production of Pol V-dependent transcripts and AGO4 occupancy at RdDM loci. 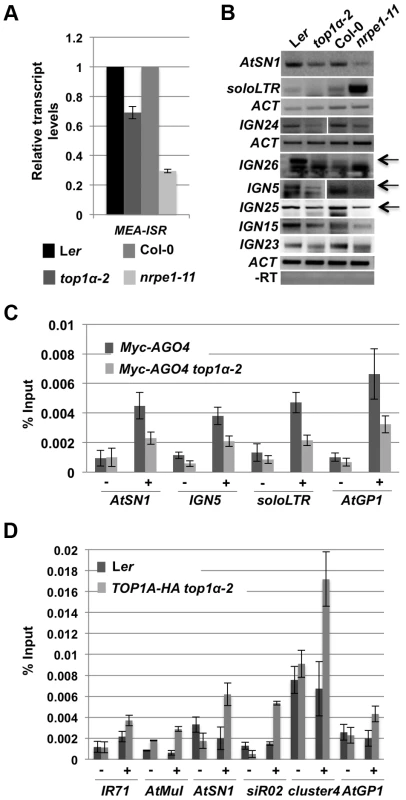
(A) TOP1α contributes to the production of Pol V-dependent transcripts at MEA-ISR. qRT-PCR was performed to quantify Pol V-dependent transcripts at MEA-ISR. The Pol V mutant nrpe1-11 was included as a control. nrpe1-11 and top1α-2 should be compared to the wild-type strains Col-0 and Ler, respectively. Error bars represent standard deviation calculated from three technical replicates. (B) TOP1α contributes to the production of Pol II- or Pol V-dependent transcripts at many loci (noted on the left of the gel images). RT-PCR was performed to detect Pol II-dependent transcripts at soloLTR and Pol V-dependent transcripts at the other seven loci (AtSN1 and six IGN loci). When multiple bands are present, the band representing the Pol V-dependent transcript is indicated by an arrow. Actin (ACT) served as an internal loading control for all the gels above. nrpe1-11 was included as a control. nrpe1-11 and top1α-2 should be compared to the wild-type strains Col-0 and Ler, respectively. –RT is the negative control in which reverse transcription was conducted in the absence of reverse transcriptase. Three biological replicates were performed and one representative image is shown. (C) Loss of TOP1α results in a decrease in AGO4 occupancy. Ten-day old wild-type and top1α-2 seedlings, both of which contain a Myc-AGO4 transgene, were subjected to ChIP with anti-MYC antibodies. qPCR was then performed on the immunoprecipitated DNA for four endogenous RdDM loci. “−” and “+” signs represent “no antibody” or “anti-Myc antibodies”, respectively. Error bars represent standard deviation calculated from three biological replicates. (D) TOP1α is present at six RdDM loci. Ten-day old Ler (used as a negative control) and TOP1A-HA top1α-2 seedlings were subjected to ChIP with anti-HA antibodies. qPCR was then performed on the immunoprecipitated DNA for six endogenous RdDM loci. “−” and “+” signs represent “no antibody” or “anti-HA antibodies”, respectively. Error bars represent standard deviation calculated from three technical replicates. Two biological replicates were performed and showed the same trend. One biological replicate is shown. As the Pol V - or Pol II-dependent long noncoding transcripts facilitate the recruitment of siRNA-AGO4 to chromatin to ultimately result in RdDM or H3K9me2 deposition, we asked whether TOP1α promotes AGO4 occupancy at these RdDM target loci. ChIP-qPCR was conducted with anti-Myc antibodies in Myc-AGO4 [35] and Myc-AGO4 top1α-2 plants. At four well-known RdDM target loci, AGO4 occupancy was reduced in top1α-2 (Figure 5C).
To determine whether TOP1α might act directly at these RdDM loci, we examined TOP1α occupancy at these loci. We first generated a TOP1α-HA fusion driven by the TOP1α promoter (TOP1α-HA) and introduced it into top1α-2. The morphological phenotypes of top1α-2 plants were completely rescued by TOP1α-HA, indicating that the transgene was functional. We then performed ChIP-qPCR using anti-HA antibodies. TOP1α was found at all six loci examined (Figure 5D).
Discussion
Beginning with a forward chemical genetics screen with a transcriptionally silenced reporter, LUCL, we have discovered that the well-studied anti-cancer compound CPT can de-repress loci undergoing transcriptional silencing by releasing H3K9 methylation and/or DNA methylation. As topoisomerase I is the cellular target of CPT, this implicates topoisomerase I in transcriptional silencing. Indeed, two top1α alleles, top1α-2 and top1α-7, mimic CPT treatment in de-repressing the expression of endogenous RdDM target loci and reducing H3K9me2 or DNA methylation levels at these loci.
Here, we first consider whether TOP1α acts through RdDM or independently of RdDM to silence TEs. RdDM requires Pol IV and Pol V, which generate siRNAs and long noncoding RNAs, respectively. We show that TOP1α is dispensable for siRNA accumulation, but is required for the production of Pol V-dependent long noncoding RNAs, which are known to recruit siRNA-AGO4 to chromatin. Consistently, TOP1α promotes the recruitment of AGO4 to RdDM target loci. Moreover, 88 out of 97 WT-top1α CHH DMRs with reduced methylation in top1α also require Pol IV or Pol V for CHH methylation (Figure 3C). 5S rDNA loci lose CG methylation in top1α-2 and nrpe1-11 mutants, and provide an example of a genomic region where CG methylation requires TOP1α, Pol IV, and Pol V. These data suggest that TOP1α acts at least in part through RdDM to silence TEs and repeats. However, MethylC-seq analyses revealed that TOP1α has a limited role in DNA methylation. We envision two possibilities for the limited role in DNA methylation observed for TOP1α. First, TOP1α may have a much broader role in DNA methylation in the genome, and the limited effects of top1α mutants on DNA methylation could be due to the redundant functions of TOP1β. So far, our efforts to knock down TOP1β in the top1α-2 background have been unsuccessful. Second, TOP1α's primary functions may lie in the promotion of H3K9 dimethylation, with DNA methylation being a secondary effect of H3K9 dimethylation. From our studies of a limited number of RdDM loci, we found that reduced H3K9me2 levels, but not necessarily reduced DNA methylation, always accompany the de-repression of these loci by CPT treatment or by mutations in TOP1α. Therefore, it is likely that the primary function of TOP1α lies in facilitating H3K9me2 deposition. Consistent with this model, the observed effects of CPT treatment on DNA methylation at four loci require CMT3 and KYP, both of which promote H3K9 dimethylation. Another observation consistent with this hypothesis is that CPT treatment had no effect on LUCH (Figure 1C), a reporter gene that is strictly repressed by CHH methylation and is insensitive to loss of function in CMT3 [36]. As CMT3-mediated DNA methylation requires H3K9me2 [14], we presume that LUCH is not repressed by H3K9me2. The lack of an effect of CPT treatment on LUCH would be consistent with TOP1α acting in TGS through H3K9me2 deposition.
Our finding that TOP1α promotes the production of Pol V-dependent transcripts is consistent with what is known about the function of topoisomerases in bacteria and yeast. Topoisomerases are thought to facilitate transcription elongation by relaxing supercoils [37]. Consistent with this model, loss of Top1 in Schizosaccharomyces pombe results in the accumulation of Pol II in gene bodies [38], [39]. The parallels of Pol V - and Pol II-mediated transcription have recently been highlighted [40], and we propose that TOP1α promotes transcription elongation by Pol V as it does for Pol II.
Although we prefer a model in which TOP1α acts in RdDM by facilitating the production of long noncoding RNAs by Pol V or Pol II, an alternative model cannot be overlooked. Studies in other systems have shown that topoisomerases interact with SMC-containing proteins acting in chromosome compaction [41], [42]. DMS3, a player of the RdDM machinery, contains an SMC domain [43]; therefore, there is a possibility that TOP1α may facilitate RdDM through DMS3.
In summary, we have discovered a role for DNA topoisomerase I in H3K9 methylation and DNA methylation in Arabidopsis. Another study showed that chemical inhibitors of topoisomerases I and II release the epigenetic silencing of an imprinted gene in mouse [44]. Together, these studies point to a role of topoisomerases in epigenetic silencing. Given that CPT is a canonical anti-cancer compound and several of its derivatives are presently used in cancer therapy [25], the emerging role of topoisomerase I in epigenetic gene silencing allude to the mode of carcinogenesis.
Materials and Methods
MethylC-seq analysis: Identification of Differentially Methylated Regions (DMRs)
Raw data from Illumina sequencing were filtered to remove reads that failed to pass the Illumina quality control and to condense multi-copy reads to a single copy. Hereafter, the reads were mapped to TAIR 10 Arabidopsis genome as well as a C-to-T converted genome using BS_Seeker [45] with default settings. Only perfectly and uniquely mapped reads were retained. For Ler and top1α, which are in the Landsberg ecotype, the reads were mapped to a pseudo-Ler genome generated by incorporating the Ler polymorphisms into the Tair10 Columbia genome (ftp://ftp.arabidopsis.org/Polymorphisms/Ecker_ler.homozygous_snp.txt). This enables the direct comparison of DMR regions between the Columbia and Landsberg samples.
DMRs were identified following a published method [27] with some modifications. In brief, the genome was split into continuous 100 bp windows. The Cs or Ts were counted in each window in the three different contexts (CG, CHG or CHH) separately. Only windows with least 4 Cs each sequenced at least 4 times in the wild-type sample were kept for the DMR analysis. The methylation level for a window was determined as:
in which ai denotes the number of read “C”s and bi denotes the number of read “T”s mapping to the ith cytosine site. The methylation level in each window in wild type is then compared to the corresponding window in a mutant. A methylation difference of 0.4, 0.2, and 0.1 for CG, CHG, and CHH, and an adjusted p-value (FDR)<0.01 (Fisher's exact test) were used as the cutoff for defining DMRs.Additional measures were taken to reduce experimental noise. First, two or three biological replicates/alleles were examined. In deriving initial DMRs, we compared each wild type/mutant pair from the same biological replicate (Table S5). Then, DMRs located within 200 bp of each other were merged. Next, the overlap in DMRs from the two biological replicates/alleles was identified (Table S5). Finally, we removed the DMRs that overlapped with the hypervariability (HV) regions found to be prone to changes in DNA methylation [28], [29] (Table S5).
See Text S1 for Supplemental Methods and Table S8 for oligonucleotides used in this study.
Accession numbers and data deposition
The gene accession numbers used in this study are At5g55310 (TOP1α), At5g55300 (TOP1β), At1g05460 (NRPD1), and At2g40030 (NRPE1). MethylC-seq and small RNA-seq read data have been deposited into NCBI GEO under the identification numbers GSE50691 and GSE50720, respectively.
Supporting Information
Zdroje
1. BirdA (2007) Perceptions of epigenetics. Nature 447 : 396–398.
2. LawJA, JacobsenSE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev Genet 11 : 204–220.
3. HerrAJ, JensenMB, DalmayT, BaulcombeDC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308 : 118–120.
4. KannoT, HuettelB, MetteMF, AufsatzW, JaligotE, et al. (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nature Genetics 37 : 761–765.
5. OnoderaY, HaagJR, ReamT, Costa NunesP, PontesO, et al. (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 : 613–622.
6. PontierD, YahubyanG, VegaD, BulskiA, Saez-VasquezJ, et al. (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Development 19 : 2030–2040.
7. ZilbermanD, CaoX, JacobsenSE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 : 716–719.
8. WierzbickiAT, HaagJR, PikaardCS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135 : 635–648.
9. WierzbickiAT, ReamTS, HaagJR, PikaardCS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nature Genetics 41 : 630–634.
10. ZemachA, KimMY, HsiehPH, Coleman-DerrD, Eshed-WilliamsL, et al. (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153 : 193–205.
11. EbbsML, BarteeL, BenderJ (2005) H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Molecular and cellular biology 25 : 10507–10515.
12. EbbsML, BenderJ (2006) Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. The Plant cell 18 : 1166–1176.
13. JacksonJP, LindrothAM, CaoX, JacobsenSE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 : 556–560.
14. DuJ, ZhongX, BernatavichuteYV, StroudH, FengS, et al. (2012) Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151 : 167–180.
15. EnkeRA, DongZ, BenderJ (2011) Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana. PLoS genetics 7: e1002350.
16. StroudH, DoT, DuJ, ZhongX, FengS, et al. (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nature structural & molecular biology 21 : 64–72.
17. ChampouxJJ (2001) DNA topoisomerases: structure, function, and mechanism. Annual Review of Biochemistry 70 : 369–413.
18. VosSM, TretterEM, SchmidtBH, BergerJM (2011) All tangled up: how cells direct, manage and exploit topoisomerase function. Nature Rev Mol Cell Biol 12 : 827–841.
19. TakahashiT, MatsuharaS, AbeM, KomedaY (2002) Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. The Plant Cell 14 : 2085–2093.
20. DinhTT, O'LearyM, WonSY, LiS, ArroyoL, et al. (2013) Generation of a luciferase-based reporter for CHH and CG DNA methylation in Arabidopsis thaliana. Silence 4 : 1.
21. JaxelC, CapranicoG, KerriganD, KohnKW, PommierY (1991) Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J Biol Chem 266 : 20418–20423.
22. StakerBL, HjerrildK, FeeseMD, BehnkeCA, BurginABJr, et al. (2002) The mechanism of topoisomerase I poisoning by a camptothecin analog. PNAS 99 : 15387–15392.
23. ManavellaPA, HagmannJ, OttF, LaubingerS, FranzM, et al. (2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151 : 859–870.
24. IrizarryRA, Ladd-AcostaC, CarvalhoB, WuH, BrandenburgSA, et al. (2008) Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 18 : 780–790.
25. LiQY, ZuYG, ShiRZ, YaoLP (2006) Review camptothecin: current perspectives. Current Medicinal Chemistry 13 : 2021–2039.
26. GrafP, DolzblaszA, WurschumT, LenhardM, PfreundtU, et al. (2010) MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. The Plant cell 22 : 716–728.
27. StroudH, GreenbergMV, FengS, BernatavichuteYV, JacobsenSE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152 : 352–364.
28. BeckerC, HagmannJ, MullerJ, KoenigD, StegleO, et al. (2011) Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480 : 245–249.
29. SchmitzRJ, SchultzMD, LewseyMG, O'MalleyRC, UrichMA, et al. (2011) Transgenerational epigenetic instability is a source of novel methylation variants. Science 334 : 369–373.
30. WierzbickiAT, CocklinR, MayampurathA, ListerR, RowleyMJ, et al. (2012) Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes & development 26 : 1825–1836.
31. BernatavichuteYV, ZhangX, CokusS, PellegriniM, JacobsenSE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PloS One 3: e3156.
32. LippmanZ, GendrelAV, BlackM, VaughnMW, DedhiaN, et al. (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430 : 471–476.
33. TranRK, ZilbermanD, de BustosC, DittRF, HenikoffJG, et al. (2005) Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biology 6: R90.
34. ZhengB, WangZ, LiS, YuB, LiuJY, et al. (2009) Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes & Development 23 : 2850–2860.
35. LiCF, PontesO, El-ShamiM, HendersonIR, BernatavichuteYV, et al. (2006) An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126 : 93–106.
36. WonSY, LiS, ZhengB, ZhaoY, LiD, et al. (2012) Development of a luciferase-based reporter of transcriptional gene silencing that enables bidirectional mutant screening in Arabidopsis thaliana. Silence 3 : 6.
37. LiuLF, WangJC (1987) Supercoiling of the DNA template during transcription. PNAS 84 : 7024–7027.
38. Durand-DubiefM, PerssonJ, NormanU, HartsuikerE, EkwallK (2010) Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J 29 : 2126–2134.
39. Durand-DubiefM, SvenssonJP, PerssonJ, EkwallK (2011) Topoisomerases, chromatin and transcription termination. Transcription 2 : 66–70.
40. PikaardCS, HaagJR, PontesOM, BlevinsT, CocklinR (2013) A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harbor Symposia on Quantitative Biology 77 : 205–212.
41. MaeshimaK, LaemmliUK (2003) A two-step scaffolding model for mitotic chromosome assembly. Developmental Cell 4 : 467–480.
42. TadesseS, MascarenhasJ, KostersB, HasilikA, GraumannPL (2005) Genetic interaction of the SMC complex with topoisomerase IV in Bacillus subtilis. Microbiology 151 : 3729–3737.
43. KannoT, BucherE, DaxingerL, HuettelB, BohmdorferG, et al. (2008) A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nature Genetics 40 : 670–675.
44. HuangHS, AllenJA, MabbAM, KingIF, MiriyalaJ, et al. (2012) Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481 : 185–189.
45. ChenPY, CokusSJ, PellegriniM (2010) BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 11 : 203.
Štítky
Genetika Reprodukční medicína
Článek Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome EvolutionČlánek Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder PopulationČlánek Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among PancrustaceansČlánek Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery DiseaseČlánek An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation inČlánek Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 7- Akutní intermitentní porfyrie
- Souvislost haplotypu M2 genu pro annexin A5 s opakovanými reprodukčními ztrátami
- Vztah užívání alkoholu a mužské fertility
- Růst a vývoj dětí narozených pomocí IVF
- Srdeční frekvence embrya může být faktorem užitečným v předpovídání výsledku IVF
-
Všechny články tohoto čísla
- Cuba: Exploring the History of Admixture and the Genetic Basis of Pigmentation Using Autosomal and Uniparental Markers
- Clonal Architecture of Secondary Acute Myeloid Leukemia Defined by Single-Cell Sequencing
- Mechanisms of Functional Variants That Impair Regulated Bicarbonate Permeation and Increase Risk for Pancreatitis but Not for Cystic Fibrosis
- Nucleosomes Shape DNA Polymorphism and Divergence
- Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection
- Comparative Phylogenomics Uncovers the Impact of Symbiotic Associations on Host Genome Evolution
- Activation of the Immune System by Combinations of Common Alleles
- Age-Associated Sperm DNA Methylation Alterations: Possible Implications in Offspring Disease Susceptibility
- Muscle-Specific SIRT1 Gain-of-Function Increases Slow-Twitch Fibers and Ameliorates Pathophysiology in a Mouse Model of Duchenne Muscular Dystrophy
- MDRL lncRNA Regulates the Processing of miR-484 Primary Transcript by Targeting miR-361
- Hypersensitivity of Primordial Germ Cells to Compromised Replication-Associated DNA Repair Involves ATM-p53-p21 Signaling
- Intrapopulation Genome Size Variation in Reflects Life History Variation and Plasticity
- SlmA Antagonism of FtsZ Assembly Employs a Two-pronged Mechanism like MinCD
- Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population
- Determinative Developmental Cell Lineages Are Robust to Cell Deaths
- DELLA Protein Degradation Is Controlled by a Type-One Protein Phosphatase, TOPP4
- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Common Transcriptional Mechanisms for Visual Photoreceptor Cell Differentiation among Pancrustaceans
- UVB Induces a Genome-Wide Acting Negative Regulatory Mechanism That Operates at the Level of Transcription Initiation in Human Cells
- The Nesprin Family Member ANC-1 Regulates Synapse Formation and Axon Termination by Functioning in a Pathway with RPM-1 and β-Catenin
- Combinatorial Interactions Are Required for the Efficient Recruitment of Pho Repressive Complex (PhoRC) to Polycomb Response Elements
- Recombination in the Human Pseudoautosomal Region PAR1
- Microsatellite Interruptions Stabilize Primate Genomes and Exist as Population-Specific Single Nucleotide Polymorphisms within Individual Human Genomes
- An Intronic microRNA Links Rb/E2F and EGFR Signaling
- An Essential Nonredundant Role for Mycobacterial DnaK in Native Protein Folding
- Integrative Genomics Reveals Novel Molecular Pathways and Gene Networks for Coronary Artery Disease
- The Genomic Landscape of the Ewing Sarcoma Family of Tumors Reveals Recurrent Mutation
- Evolution and Genetic Architecture of Chromatin Accessibility and Function in Yeast
- An ARID Domain-Containing Protein within Nuclear Bodies Is Required for Sperm Cell Formation in
- Stage-Dependent and Locus-Specific Role of Histone Demethylase Jumonji D3 (JMJD3) in the Embryonic Stages of Lung Development
- Genome Wide Association Identifies Common Variants at the Locus Influencing Plasma Cortisol and Corticosteroid Binding Globulin
- Regulation of Feto-Maternal Barrier by Matriptase- and PAR-2-Mediated Signaling Is Required for Placental Morphogenesis and Mouse Embryonic Survival
- Apomictic and Sexual Germline Development Differ with Respect to Cell Cycle, Transcriptional, Hormonal and Epigenetic Regulation
- Functional EF-Hands in Neuronal Calcium Sensor GCAP2 Determine Its Phosphorylation State and Subcellular Distribution , and Are Essential for Photoreceptor Cell Integrity
- Comparison of Methods to Account for Relatedness in Genome-Wide Association Studies with Family-Based Data
- Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age
- Cis and Trans Effects of Human Genomic Variants on Gene Expression
- 8.2% of the Human Genome Is Constrained: Variation in Rates of Turnover across Functional Element Classes in the Human Lineage
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- A Loss of Function Screen of Identified Genome-Wide Association Study Loci Reveals New Genes Controlling Hematopoiesis
- Unraveling Genetic Modifiers in the Mouse Model of Absence Epilepsy
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
- The Coding and Noncoding Architecture of the Genome
- A Novel Locus Is Associated with Large Artery Atherosclerotic Stroke Using a Genome-Wide Age-at-Onset Informed Approach
- Brg1 Loss Attenuates Aberrant Wnt-Signalling and Prevents Wnt-Dependent Tumourigenesis in the Murine Small Intestine
- The PTK7-Related Transmembrane Proteins Off-track and Off-track 2 Are Co-receptors for Wnt2 Required for Male Fertility
- The Co-factor of LIM Domains (CLIM/LDB/NLI) Maintains Basal Mammary Epithelial Stem Cells and Promotes Breast Tumorigenesis
- Essential Genetic Interactors of Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates
- Meiosis-Specific Cohesin Component, Is Essential for Maintaining Centromere Chromatid Cohesion, and Required for DNA Repair and Synapsis between Homologous Chromosomes
- Silencing Is Noisy: Population and Cell Level Noise in Telomere-Adjacent Genes Is Dependent on Telomere Position and Sir2
- The Two Cis-Acting Sites, and , Contribute to the Longitudinal Organisation of Chromosome I
- A Broadly Conserved G-Protein-Coupled Receptor Kinase Phosphorylation Mechanism Controls Smoothened Activity
- Requirements for Acute Burn and Chronic Surgical Wound Infection
- LIN-42, the PERIOD homolog, Negatively Regulates MicroRNA Transcription
- WAPL Is Essential for the Prophase Removal of Cohesin during Meiosis
- Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration
- Sox11 Is Required to Maintain Proper Levels of Hedgehog Signaling during Vertebrate Ocular Morphogenesis
- Accumulation of a Threonine Biosynthetic Intermediate Attenuates General Amino Acid Control by Accelerating Degradation of Gcn4 via Pho85 and Cdk8
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Wnt Signaling Interacts with Bmp and Edn1 to Regulate Dorsal-Ventral Patterning and Growth of the Craniofacial Skeleton
- Novel Approach Identifies SNPs in and with Evidence for Parent-of-Origin Effect on Body Mass Index
- Hypoxia Adaptations in the Grey Wolf () from Qinghai-Tibet Plateau
- DNA Topoisomerase 1α Promotes Transcriptional Silencing of Transposable Elements through DNA Methylation and Histone Lysine 9 Dimethylation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




