-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
Embryonic stem (ES) cells derived from the inner cell mass (ICM) of blastocysts are characterised by their ability to self-renew and their potential to differentiate into many different cell types. Recent studies have shown that zinc finger proteins are crucial for maintaining pluripotent ES cells. Mouse zinc finger protein 322a (Zfp322a) is expressed in the ICM of early mouse embryos. However, little is known regarding the role of Zfp322a in the pluripotency maintenance of mouse ES cells. Here, we report that Zfp322a is required for mES cell identity since depletion of Zfp322a directs mES cells towards differentiation. Chromatin immunoprecipitation (ChIP) and dual-luciferase reporter assays revealed that Zfp322a binds to Pou5f1 and Nanog promoters and regulates their transcription. These data along with the results obtained from our ChIP-seq experiment showed that Zfp322a is an essential component of mES cell transcription regulatory network. Targets which are directly regulated by Zfp322a were identified by correlating the gene expression profile of Zfp322a RNAi-treated mES cells with the ChIP-seq results. These experiments revealed that Zfp322a inhibits mES cell differentiation by suppressing MAPK pathway. Additionally, Zfp322a is found to be a novel reprogramming factor that can replace Sox2 in the classical Yamanaka's factors (OSKM). It can be even used in combination with Yamanaka's factors and that addition leads to a higher reprogramming efficiency and to acceleration of the onset of the reprogramming process. Together, our results demonstrate that Zfp322a is a novel essential component of the transcription factor network which maintains the identity of mouse ES cells.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004038
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004038Summary
Embryonic stem (ES) cells derived from the inner cell mass (ICM) of blastocysts are characterised by their ability to self-renew and their potential to differentiate into many different cell types. Recent studies have shown that zinc finger proteins are crucial for maintaining pluripotent ES cells. Mouse zinc finger protein 322a (Zfp322a) is expressed in the ICM of early mouse embryos. However, little is known regarding the role of Zfp322a in the pluripotency maintenance of mouse ES cells. Here, we report that Zfp322a is required for mES cell identity since depletion of Zfp322a directs mES cells towards differentiation. Chromatin immunoprecipitation (ChIP) and dual-luciferase reporter assays revealed that Zfp322a binds to Pou5f1 and Nanog promoters and regulates their transcription. These data along with the results obtained from our ChIP-seq experiment showed that Zfp322a is an essential component of mES cell transcription regulatory network. Targets which are directly regulated by Zfp322a were identified by correlating the gene expression profile of Zfp322a RNAi-treated mES cells with the ChIP-seq results. These experiments revealed that Zfp322a inhibits mES cell differentiation by suppressing MAPK pathway. Additionally, Zfp322a is found to be a novel reprogramming factor that can replace Sox2 in the classical Yamanaka's factors (OSKM). It can be even used in combination with Yamanaka's factors and that addition leads to a higher reprogramming efficiency and to acceleration of the onset of the reprogramming process. Together, our results demonstrate that Zfp322a is a novel essential component of the transcription factor network which maintains the identity of mouse ES cells.
Introduction
Embryonic stem (ES) cells, which are derived from the inner cell mass (ICM) of mammalian blastocysts, are characterised by their ability to self-renew and by their potential to differentiate into many different cell types [1], [2]. ES cells provide a prime platform for biomedical research since the investigation of factors and pathways that control pluripotency and differentiation provides us with valuable data that will aid in the advancement of regenerative [3]. The discovery that differentiated cells can be reprogrammed into induced pluripotent stem cells (iPSCs) gave great promises for the progress of regenerative medicine and gene therapy [4]–[7].
It has been discovered that transcription factors play crucial roles in controlling ES cell identity. Genome-wide analyses revealed that in mammalian ES cells, Oct4, Nanog and Sox2 form the core transcriptional circuitry that activate genes involved in self-renewal and pluripotency and repress genes that promote differentiation into different lineages [8], [9]. The importance of this transcription network was subsequently highlighted by the finding that the expression of just four transcription factors, Oct4, Sox2, c-Myc and Klf4 (OSKM) was sufficient to transform mouse embryonic fibroblasts (MEFs) back to pluripotent stem cells, and the expression of OCT4, SOX2, NANOG and LIN28 was sufficient for in human somatic cell reprogramming [6], [7]. Along with these core factors, there are many other transcription factors which closely interact with these factors (i.e. form the gene regulatory network). Hence it is important to unravel the functions of all components in order to fully understand how this regulatory network functions to regulate various target genes. Therefore, it is of great value to expand our knowledge of this transcription regulatory network.
Krüeppel associated box (KRAB) C2H2 zinc finger family can interact directly with specific cis-regulatory DNA elements to regulate genes' activities [10]. Several studies have revealed that proteins from this family, such as ZSCAN4, Zfp296, Zfp206, and Zfp42, are key components of the ES transcriptional network and are crucial for maintaining pluripotent ES cells [11]–[15]. Mouse zinc finger protein 322a (Zfp322a) is another evolutionarily conserved protein that belongs to this family [16]. We proposed that Zfp322a acts as a transcription factor in mouse ES cells for two main reasons: First, ChIP-seq data from a previous study suggested that pluripotency factors Oct4, Zfx, E2F1, Klf4 and Myc bind to the genomic region of Zfp322a [17]. Second, Zfp322a is expressed at a higher level in mES cells compared to trophoblast cells [18]. Additionally, single cell RNA-seq results have demonstrated Zfp322a expression in both mouse ICM and ES cells [19]. These results suggested a potential role for Zfp322a in controlling the identity of mES cells. However, there has been little information regarding the function of Zfp322a.
In this study, we demonstrated that Zfp322a is required for the maintenance of mouse ES cell identity. Depletion of Zfp322a impairs mES cell self-renewal and induces them to differentiate. We found that Zfp322a positively regulates Pou5f1 and Nanog expression, and possibly represses MAPK/ERK pathway, thus preventing mES cells from differentiation. Further genome wide studies also identified the targets of Zfp322a which are involved in a variety of biological processes, including DNA transcription and translation, chromosome organization, development, DNA repair, cell cycle and apoptosis. Through iPSC formation assays we discovered that Zfp322a can be used as a novel reprogramming factor and the reprogramming efficiency was enhanced by the addition of Zfp322a to OKSM. Together, these results established Zfp322a as a novel pluripotency factor that can enhance reprogramming process.
Results
1. Zfp322a is required for the maintenance of mES cell self-renewal and pluripotency
Chen et al. demonstrated that Oct4, Zfx, E2F1, Klf4 and Myc bound to an 800 bp-region in the third intron of Zfp332a [17] (Figure S1A). Our ChIP results confirmed the interaction between Oct4 and Zfp322a intron (Figure S1B). This suggested that Zfp322a is a direct target of regulation by these transcription factors. In previous studies Zfp322a was discovered to be expressed at a higher level in ICM compared to trophectoderm [18]. We further examined the expression of Zfp322a in mES cells by immunostaining assay (Figure S2A), using the antibody specific for Zfp322a protein (Figure S2B). We found that Zfp322a was mainly localized in the nucleus of mES cells, which indicates that as a zinc finger protein, Zfp322a may function as a transcription factor in mES cells.
To determine the change in the expression of Zfp322a upon mES cell differentiation, mouse ES cells were induced to differentiate by culturing in LIF (leukemia inhibitory factor) withdrawal medium. Zfp322a mRNA level was reduced during the differentiation process, dropping to 20% at 7 days after LIF removal (Figure 1A). Similarly, Zfp322a protein also decreased upon mES cell differentiation (Figure 1B). The expression of Zfp322a in undifferentiated mES cells and its repression upon ES cell differentiation further suggested a possible involvement of Zfp322a in pluripotency maintenance in mouse ES cells.
Fig. 1. Zfp322a is required for ES pluripotency. 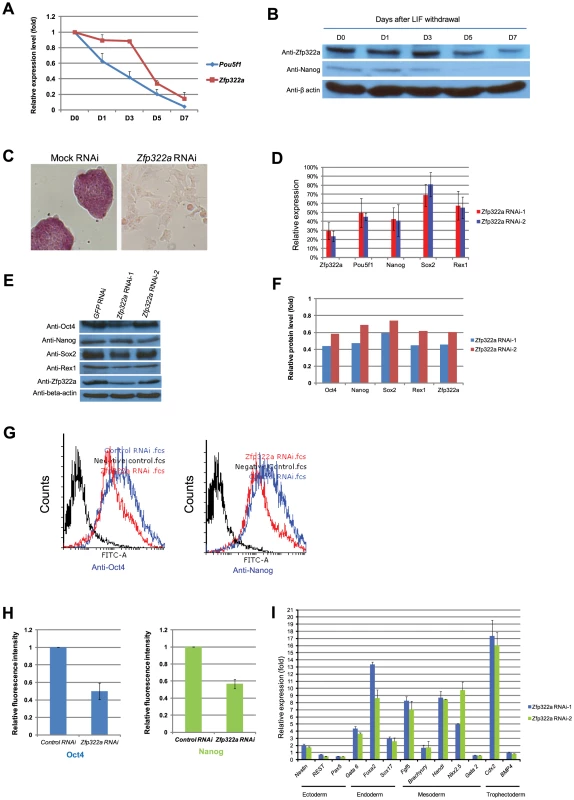
(A) Zfp322a mRNA level was decreased in mES cells cultured in LIF withdrawal ESC medium. The level of the Zfp322a and Pou5f1 mRNA were compared to control cells cultured in normal ESC medium and normalized against β-actin. (B) Zfp322A protein level showed a reduction as mES cells differentiated. β-actin served as loading control. (C) Zfp322a RNAi caused ES cell differentiation. AP staining was conducted on the fourth day of selection after the cells were transfected with Zfp322a shRNAs. Zfp322a RNAi cells displayed a lighter colour compared to the dark red colour of mock RNAi cells. (D) Pluripotency genes were down-regulated upon Zfp322a RNAi. ES cells transfected with empty pSUPER.puro vector were used as a control and gene expression levels were normalized against β-actin. (E) Zfp322a RNAi resulted in decreased Oct4, Nanog, Sox2 and Rex1 protein. β-actin served as loading control. (F) Quantification of the protein level changes. Protein levels of Zfp322a knocked-down cells were normalized against β-actin and compared to control RNAi cells using software ImageJ. (G) Representative flow cytometry results showed that fluorescence intensities of Oct4 and Nanog were repressed in Zfp322a depleted cells as compared to control cells. (H) Zfp322a depletion led to reductions of Oct4 and Nanog expression at cellular levels. The mean of fluorescence intensity was calculated using software flow software 2.5.0. Relative fluorescent intensities of Zfp322a RNAi cells were normalized against control knocked-down cells. Standard deviations were derived from three independent experiments. (I) Zfp322a RNAi caused up-regulation of lineage specific markers for endoderm, mesoderm and trophectoderm. Specific primers were used to check the respective gene expression levels by real-time PCR. In order to investigate the role of Zfp322a in mouse ES cell pluripotency, we examined the effect of Zfp322a depletion in mES cells by RNAi. E14 mES cells were transfected with two independent Zfp322a shRNAs targeting different regions of Zfp322a gene. Both shRNAs were effective in depleting the level of Zfp322a mRNA to 30% of the control (Figure 1D). Upon knock-down of Zfp322a, mES cells lost their characteristics, including the round colony-like morphology and alkaline phosphatise (AP) activity. Instead, RNAi-treated cells exhibited flattened, differentiated cell morphology (Figure 1C). These results indicated that Zfp322a depletion caused mES cell differentiation and impaired self-renewal of mES cells.
2. Depletion of Zfp322a activates developmental genes while repressing pluripotency related genes
We further examined the alteration in gene expression induced by Zfp322a depletion. The mRNA levels of pluripotency genes Pou5f1, Nanog, Sox2 and Zfp42 were significantly reduced in RNAi-treated E14 cells (Figure 1D). Consistently, protein levels of these pluripotency factors were also reduced upon Zfp322a depletion (Figure 1E, 1F). In addition, immunofluorescence (IF) of Oct4 and Nanog were performed to examine their expression at cellular level. The fluorescence intensities of Zfp322a knocked-down cells and control cells were measured with flow cytometry. Upon Zfp322a RNAi, there was a significant reduction of the fluorescence intensities observed both in anti-Oct4 antibody and anti-Nanog antibody stained cells (Figure 1G). Further we examined the population mean value of fluorescence intensities. We found that Zfp322a depletion supressed Oct4 and Nanog IF mean intensities by 50%, 40% respectively as compared to control (Figure 1H). Similar results were obtained in another mES cell line HM1 (Figure S5A). Since Oct4 and Nanog are essential for maintenance of pluripotency, these results are consistent with the observation that Zfp322a RNAi induces differentiation of mES cells. The differentiation of mES cells was further supported by the dramatic increase in various lineage markers after Zfp322a depletion (Figure 1I). Upon knock-down of Zfp322a in mES cells, we observed an up-regulation of endodermal markers: Gata6 (4 fold), Foxa2 (8 fold) and Sox17 (3 fold), which indicated that Zfp322a could maintain mES cell pluripotency by repressing endodermal specification. Trophectoderm maker Cdx2 displayed a 16 fold increase while mesoderm markers, Fgf5, Hand1 and Nkx2.5 increased by 7, 8 and 10 fold respectively (Figure 1I). Thus consistent with the AP staining results, this suggested that Zfp322a is required to suppress lineage specific gene expressions to maintain mES cells in their undifferentiated state.
To further understand how Zfp322a depletion led to mES cell differentiation, we used gene expression microarrays to investigate the global gene expression profile changes induced by Zfp322a depletion (Figure 2A). qPCR experiments were performed to validate the results of the microarray analysis (Figure S3). As an internal control, we examined the level of Zfp322a by the microarray, and consistent with qPCR results (Figure 1D), we found an approximately 3-fold reduction in the mRNA level of Zfp322a. Upon Zfp322a depletion, 1574 genes were up-regulated (increased by >1.5 fold) and 904 genes were down-regulated (reduced by >1.5 fold) (Figure 2A). Importantly, the microarray data analysis revealed that many known pluripotency genes were down-regulated (Figure 2A). This indicated that Zfp322a is a high-level regulator in the mES cell gene regulatory network, which does not only regulate a subset of genes required for pluripotency, but is an essential component of the core network required for the maintenance of mES cell identity.
Fig. 2. Changes of global gene expression upon Zfp322a knock-down in ES cells. 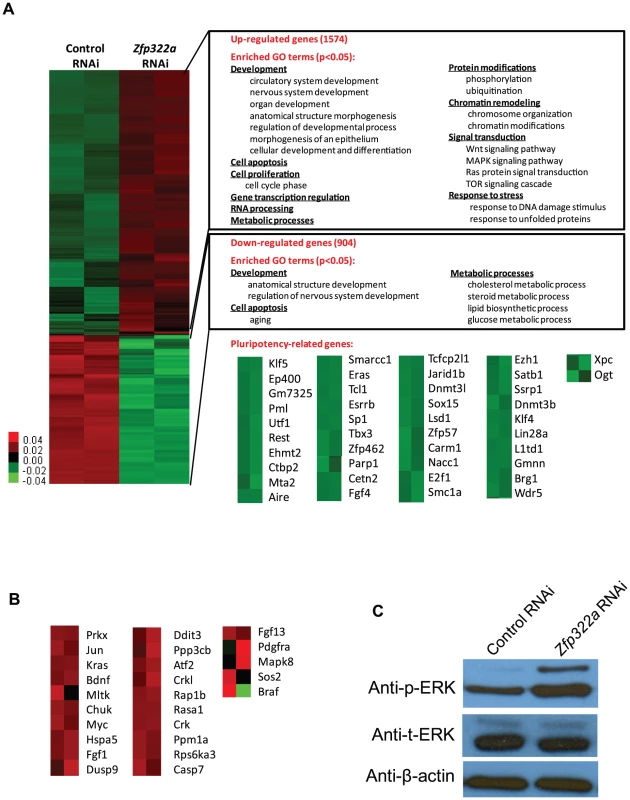
(A) Microarray heat map generated from relative gene expression levels. Zfp322a was knocked down in E14 cells and the cells were selected for 96 hours before whole genome cDNA microarray hybridization was performed. Duplicates were performed to ensure reproducibility of results. Relative highly expressed genes were shown in red and low expressed genes in green. 1574 genes showed an increased expression level of more than 1.5 fold after Zfp322a RNAi. Gene onthology analysis was performed relating to “biological process”. The enriched terms were classified into several function groups and listed in the figure. Many terms related to developmental processes were enriched. 904 genes expression level showed more than 1.5 fold reduction. Examples of down-regulated pluripotency-related genes upon Zfp322a knock-down in ES cells. Genes were selected according to their known functions in pluripotency or ES cells. Each selected gene was taken as individual tiles from the thumbnail-dendogram duplicates. (B) List of up-regulated MAPK pathway related genes upon Zfp322a RNAi. Genes were selected as they fell into the cluster “MAPK signaling pathway” according to gene ontology analysis for enriched KEGG pathways. Each selected gene was taken as individual tiles from the thumbnail-dendogram duplicates. (C) Phosphorylated ERK (p-ERK) level was elevated in Zfp322a depleted cells as compared to control cells, while the total ERK (t-ERK) level was not affected. β-actin served as a control for normalization. To determine whether Zfp322a regulated specific types of gene, we conducted gene ontology (GO) analysis of up-regulated and down-regulated genes. The enriched terms were summarized in Figures 2A (Full list of the enriched terms can be found in Table S1.). For the up-regulated genes, many terms related to development were enriched. This is consistent with the role of Zfp322a as a repressor of differentiation. Furthermore, Zfp322a depletion activated cell apoptosis related genes, also explained the increased apoptosis in Zfp322a RNAi-treated cells. Interestingly, many terms were related to chromosome remodelling, suggesting that Zfp322a may contribute to maintenance of the unique mES chromatin structure. Notably, several signaling pathways implicated in pluripotency such as MAPK pathway, Wnt signaling pathway, Ras signal cascade were also affected after Zfp322a depletion (Table S1, S2, Figure 2A, 2B). Importantly, Zfp322a depletion caused an up-regulation of phosphorylated ERK (p-ERK) level while the total levels of ERK (t-ERK) expression was not affected (Figure 2C). This substantiated that Zfp322a indeed is implicated in the repression of MAPK/ERK cascade without changing ERK expression in mES cells (Figure 2C).
It has been shown that inhibition of MAPK/ERK pathway is important for mES ground state pluripotency [20], [21]. Activation of Ras-MAPK pathway promotes trophectoderm induction and is specifically suppressed by Oct4 in undifferentiated mES cells [22]. ERK pathway has been found to promote the onset of mES cell differentiation and it is involved in multiple developmental processes [23]. Therefore, Zfp322a may maintain mES cell in undifferentiated state via repression of the MAPK/ERK cascade.
To further investigate the role of Zfp322a in MAPK pathway, we next examined whether inhibition of the MAPK pathway could rescue the effects caused by Zfp322a depletion. Zfp322a depleted mES cells were subjected to 50 nM, 250 nM and 1 uM of ERK inhibitors (PD0325901, Sigma). We found that the addition of ERK inhibitors could not rescue the down-regulated Pou5f1, Sox2 and Zfp42 upon Zfp322a depletion, and the ERK inhibitor treated cells exhibited differentiated morphology same as DMSO treated control cells (Figure S3B). However, with the addition of ERK inhibitors, levels of Nanog, Sox2 and Rex1 were higher than that of Zfp322a depleted cells without ERK inhibitor and the endoderm and ectoderm lineage markers were lower (Figure S3B, S3C). This is consistent with previous findings in which the blocking of ERK pathway induces elevated expression of Nanog in ES cells [24], [25]. However, ERK inhibitor did not rescue stem cell differentiation caused by Zfp322a knocked-down cells (Figure S3B, S3C). Interestingly, addition of ERK inhibitors seemed to facilitate Zfp322a knocked-down cells to differentiate into mesoderm lineage, other than endoderm or ectoderm lineages cells (Figure S3B, S3C). This is in consistent with previous finding that Erk2-null mES cells tend to differentiate to mesoderm lineage more efficiently than wild type mES cells [25].
Taken together, it appears that Zfp322a depletion leads to activation of MAPK/ERK pathway, which could drive mES cells towards differentiation. However, the inhibition of MAPK/ERK pathway could not rescue the differentiation phenotype caused by Zfp322a loss.
3. Transcriptional regulation of Pou5f1 and Nanog by Zfp322a
Oct4 and Nanog are master regulators of mES pluripotency [26], [27]. Many pluripotency factors were found to bind to promoters of Pou5f1 and Nanog to regulate their transcriptions [17]. Since Nanog and Pou5f1 were down-regulated upon Zfp322a depletion, we speculated that Zfp322a may bind to Pou5f1 and Nanog promoters to regulate their transcription. To test whether Zfp322a binds to cis-regulatory elements of Pou5f1 and Nanog, ChIP experiments were performed using an anti-Zfp322a antibody to pull-down wild type mES cell chromatin. Real-time PCR was used to determine whether Zfp322a preferentially bound to known enhancer elements upstream of Pou5f1 and Nanog promoters. We found a clear peak in the Pou5f1 distal enhancer, which is also known as CR4 (conserved region 4), showing a 28 fold enrichment (Figure 3A, S5B). CR4 is the main enhancer that drives Pou5f1 expression in mouse ES cells and early embryos, and it is the site bound by many transcription factors, including Nanog, Sox2 and Oct4 itself [8], [28], [29]. Similarly, Zfp322a was also shown to bind to the Nanog proximal promoter. Strong enrichment for amplicon 4 was found in the multiple transcription factor binding locus of Nanog promoter (MTL) (Figure 3B, S5C) [13]. These results showed that Zfp322a could directly bind to Pou5f1 distal enhancer and Nanog proximal promoter in mES cells and may cooperate with other transcription factors in the regulation of Pou5f1 and Nanog transcription.
Fig. 3. Zfp322a positively regulates Oct4 and Nanog transcription. 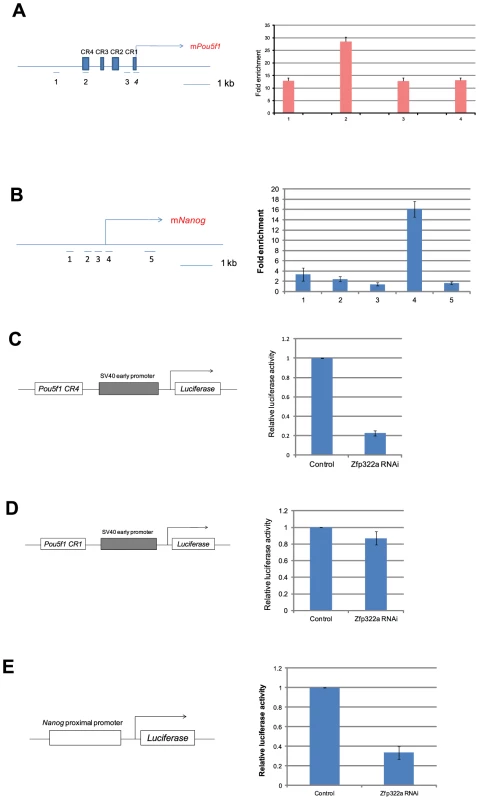
(A) Zfp322a binds to Oct4 distal enhancer regions. Zfp322a ChIP DNA was analyzed by real-time PCR. Locations of primers used in qPCR were mapped to the Pou5f1 genomic region. (B) Zfp322a binds to Nanog proximal promoter, with a highest enrichment fold at TSS starting site. Locations of primers were pictured on mouse Nanog genomic region. Relative luciferase activities were down-regulated upon Zfp322a RNAi using Pou5f1 CR4-pSV40-Luc construct (C) and pNanog PP-Luc construct (E), but not Pou5f1 CR1-pSV40-Luc construct (D). Schematic structures of the constructs were presented. Empty pSUPER.puro vector were transfected in ES cells as a control RNAi. Renilla luciferase vector were transfected simultaneously and relative luciferase activities were normalized against Renilla luciferase activity. To determine whether Zfp322a regulates the transcription of Pou5f1 and Nanog, dual-luciferase assays were performed using two constructs Pou5f1 CR4-pSV40-Luc and pNanog pp-Luc. Interestingly, upon knock-down of Zfp322a, the luciferase activities were strikingly reduced to 20% and 30% respectively in constructs carrying the CR4 or the Nanog proximal promoter (Figure 3C, 3E, S5D). To determine whether this reduction was led by Zfp322a loss directly, Pou5f1 CR1 was chosen as negative control. ChIP experiments showed a relatively lower enrichment fold at this region as compared to CR4. As expected, in the experiment with Pou5f1 CR1-pSV40-Luc construct, the luciferase activity was only reduced by 15% upon Zfp322a depletion, much less than 80% reduction observed in the CR4 experiment (Figure 3D). These strongly suggested that Zfp322a directly regulated Pou5f1 and Nanog through binding to these cis-regulatory elements. Interestingly, it was also observed that compared to single knock-down of Pou5f1, double knock-down of Pou5f1 and Zfp322a further suppressed enhancer activities, (Figure S4A, S4B). Given that Oct4 also binds to CR4 and Nanog MTL to regulate the transcriptions [8], we hypothesized that Zfp322a may cooperate with Oct4 to regulate gene transcriptions.
4. Genome-wide mapping of Zfp322a binding sites in mES cells
To gain more insights into the downstream pathways through which Zfp322a functions, we identified genome-wide binding sites of Zfp322a in mouse ES cells. Following chromatin immunoprecipitation using anti-Zfp322a antibody to enrich the DNA fragments bound by Zfp322a, we used high-throughput sequencing (ChIP-seq) to analyze the ChIP-enriched DNA. Genomic regions defined by multiple overlapping DNA fragments derived from the ChIP enrichments were considered as putative binding sites. To confirm the validity of these putative binding sites, genomic loci with peaks of various fold changes were arbitrarily selected and tested by qPCR. The final threshold value was determined based on enrichment of 2 fold in qPCR validation (Figure S6), which corresponded to 9-fold or higher enrichment in the ChIP-seq experiment. This gave a total of 4382 putative binding sites of Zfp322a that were associated with 4056 genes (Table S3).
The location of the binding site within the gene was mapped as well (Figure 4A). Notably, after putative Zfp322a binding sites were mapped to nearest genes, 62% fall within the TSS of the nearest to gene, showing an obvious preference for TSSs (Figure 4B). 19% of the loci were within gene intronic regions, followed by 5′ UTR, distant promoter (>3 kb from TSS) and promoter (<3 kb from TSS) which occupied 6% each (Figure 4B). Thus, we proposed that Zfp322a is primarily associated with gene promoters.
Fig. 4. Genomic-wide analysis of Zfp322a binding sites. 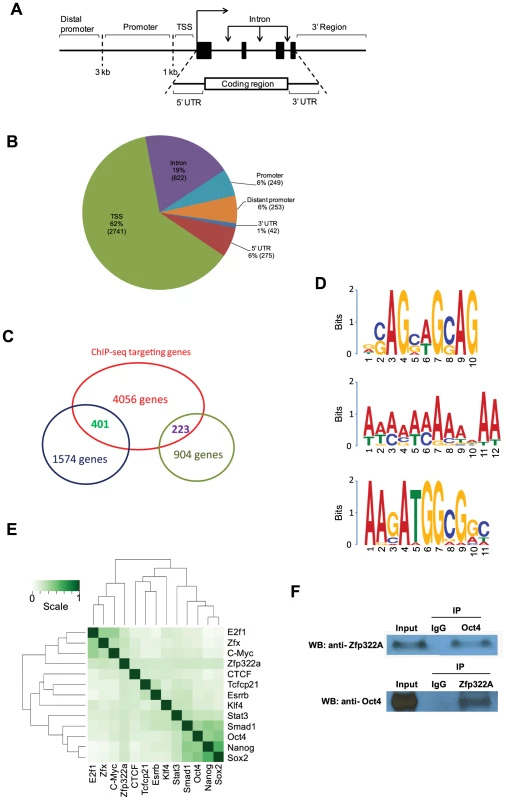
(A) Schematic definitions of locations of the putative Zfp322a binding sites relative to the nearest transcriptional unit. TSS referred to −1000 to +100 bp from 5′-end of annotated RNA. (B) Genomic distributions of Zfp322a binding loci. (C) Identification of genes that were predicted to be directly regulated by Zfp322a. The datasets from microarray analysis and ChIP-Seq targets were calculated for overlapping genes. The results revealed 1574 tentative genes that likely were activated and 223 tentative genes repressed directly by Zfp322a. (D) Predicted binding motifs for Zfp322a. Motifs were computationally determined based on the ChIP-Seq data. Three different motifs were identified, namely motif 1, motif 2, motif 3, with frequencies of 9%, 5%, 4% respectively. (E) Zfp322a can be integrated within ES cell transcription regulatory network. Shown was co-occurrence of transcription factors at the multiple binding loci. Colours in the heat map reflected the co-localization frequency of each pair of transcription factors (the darker the color was, the more frequently colocalized). All the transcription factors were clustered according to the colocalization frequency with other factors, which was calculated based on their co-occurrence at the same binding loci. (F) Zfp322a can interact with Oct4. Cell lysate of wild type ES cells were immunoprecipitated using either anti-ZNF322A antibody or anti-Oct4 antibody. Western blot was subsequently carried out with anti-Oct4 antibody or anti-ZNF322a antibody. Control IP was performed using anti-IgG antibody. Among highly enriched binding sites-associated genes, there were many known key components of ES transcription regulatory network, such as Ino80d, Zfp206, Zfx, Nrf1, Smarce1, implying that Zfp322a could directly regulate transcription of these pluripotency genes (Table S3). To further examine whether Zfp322a targets have preferentially any particular biological functions in mES cells, genes associated with the putative binding sites were subjected to Gene Ontology to search for enriched biological process terms. Large numbers of terms were found to be related to cellular metabolic and biosynthetic processes. Other enriched terms were classified into function groups and summarised in Table S4. Similar to GO analysis of our microarray data, Zfp322a targets were involved in regulation of gene transcription and translation, especially transcription from RNA polymerase II promoter. Notably, Zfp322a binding sites were found near genes encoding core components of RNA polymerase, such asPolr2a, Polr2j, Polr3e. We also found that the targets of Zfp322a were related to developmental processes, implying that Zfp322a may participate in mouse embryo development through regulation of these genes. In addition, many terms surrounding the functions “DNA repair”, “protein modifications”, “cellular component localization”, and “RNA processing”, were enriched.
Besides Pou5f1 and Nanog, we sought to refine our prediction of Zfp322a targets by combining ChIP-seq and microarray data in pluripotent mES cells. We analysed the ChIP-seq in concert with microarray dataset. Overlapping genes between these two sets of data indicated that these genes could be directly regulated by Zfp322a, either positively or negatively. We found that 401 of the 1574 up-regulated genes in the Zfp322a-RNAi microarray data analysis were directly repressed by Zfp322a in mES cells, while 223 genes were activated directly (Figure 4C). Further GO analysis of the directly repressed targets, showed that MAPK pathway related genes was enriched (p value = 0.006, Table S5). This reaffirmed our hypothesis that Zfp322a represses MAPK signalling pathway to maintain mES cell pluripotency.
5. Computational analysis of Zfp322a binding sites
Next, we aimed to identify the Zfp322a binding motif. Through bioinformatic computation, we found three different motifs which repeatedly occurred in Zfp322a binding sites; albeit at low frequencies (Figure 4D). Motif 1 had the highest frequency, presenting in 9% of all the binding sites. Motif 2 was a 12 bp-polyA-sequence with a frequency of 5%. The third motif, which was found in 4% of all binding sites, showed a high similarity to the Oct4/Sox2 binding motif [17]. This suggested that Zfp322a, Oct4 and Sox2 often bind to the same enhancer element, either acting as a complex or interacting in other ways. Notably, the first and the third element are present in the CR4 element of Pou5f1 and the proximal promoter of Nanog. It is expected that the actual Zfp322a binding site was not identified as a consensus binding motif, given that Zfp322a protein harbours 10 zinc fingers, while only 3–5 zinc fingers were needed for specific DNA binding [30]. Therefore, different zinc fingers of Zfp322a possibly recognise distinct sequences, leading to a wide variety of Zfp322a binding motifs.
Given the possibility that Oct4/Sox2 binding sites tended to be present near Zfp322a binding sites, we compared Zfp322a binding sites with target sequences of other transcription factors mapped in previous studies [17]. All the transcription factors were clustered according to the similarity of the co-localization with other factors. The results showed that Zfp322a was closer to the Myc cluster (Figure 4E). But with a closer check of the results, Zfp322a actually had a ubiquitous comparable correlation with all the 12 transcription factors, and did not show any significant preference for either Myc or Oct4/Sox2 centred clusters. Indeed Zfp322a showed a slightly higher co-localization frequency with Oct4 cluster target, from which it was inferred that Zfp322a may facilitate or cooperate with Oct4 in mES cells.
Given the correlation between Zfp322a targets and Oct4 targets, similarities between Oct4/Sox2 and Zfp322a binding motifs, together with our observation that both Zfp322a and Oct4 bound to the CR4 region of Pou5f1 distal promoter and Nanog proximal promoter at the same regions, we sought to determine whether Zfp322a could physically interact with Oct4. Co-IP experiments were performed with either anti-Zfp322a antibody or anti-Oct4 antibody. Western blots were then carried out with anti-Oct4 antibody or anti-Zfp322a antibody. We observed an Oct4 band in Zfp322 IP lane and Zfp322a band in Oct4 IP lane, indicating that Zfp322a physically interacts with Oct4 in mES cells (Figure 4F). This confirmed our hypothesis that Zfp322a functions as a partner of Oct4 in the regulation of gene transcription, though previous studies did not list Zfp322a as an Oct4 partner [31], [32].
6. Zfp322a can enhance OKSM reprogramming of MEFs into iPSCs
Since Zfp322a is involved in mES cell self-renewal and pluripotency regulation, it would be interesting to investigate whether overexpression of Zfp322a can enhance OSKM reprogramming or act as a novel reprogramming factor to replace any of the OSKM factors in generating iPSCs. MEFs transfected with a Pou5f1-GFP reporter were used to identify putative iPSC colonies [33]. It was observed that MEFs infected with OSKM plus Zfp322a showed a more efficient and faster reprogramming process than OSKM alone (Figure 5A). Addition of Zfp322a could enhance the kinetics of OKSM reprogramming as GFP expressing colonies were detected earlier than OKSM control. The number of iPSCs, when counted as GFP+ colonies formed by OKSM plus Zfp322a was higher than OKSM throughout the whole reprogramming process. By day 14 of reprogramming process, the number of GFP+ colonies generated from OKSM plus Zfp322a was 1.4 fold higher than control. Further examination of these iPSC colonies by AP staining also showed more AP positive colonies formed by OKSM plus Zfp322a as compared to OKSM alone (Figure 5B).
Fig. 5. Zfp322a can enhance OSKM reprogramming and replace Sox2. 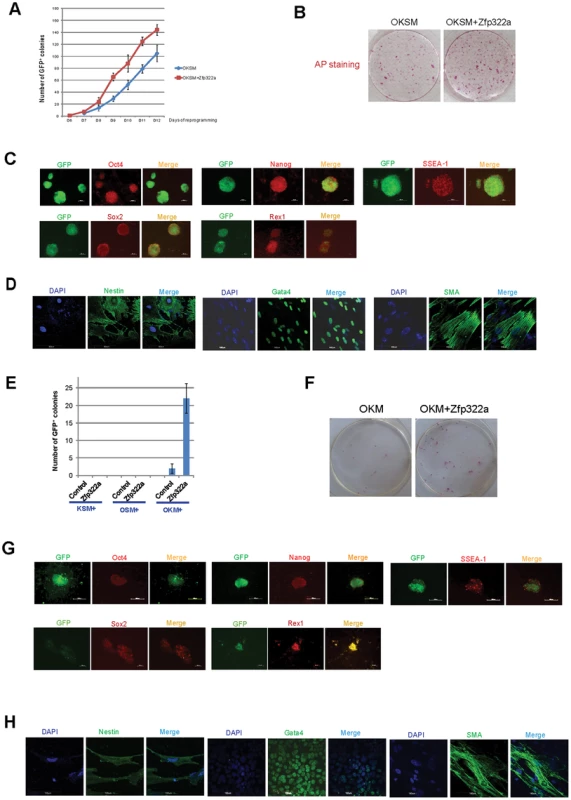
(A) Zfp322a enhanced reprogramming efficiency and accelerated the onset of reprogramming process. OSKM serves as control experiment. (B) The iPSCs generated from OSKM plus Zfp322a presented alkaline phosphatase activity. There were more AP stained colonies generated from OKSM+Zfp322a compare to OKSM. (C) The iPSCs expressed endogenous Oct4, Nanog, Sox2, Rex1 and SSEA-1, indicating that they were ES-cell like. Immunostaining using anti-Oct4, anti-Nanog anti-Sox2, anti-Rex1 and anti-SSEA-1 antibodies were performed with GFP+ iPSCs generated from OKSM+Zfp322a. (D) GFP+ iPSCs generated by OKSM+Zfp322a were able to express ectoderm, mesoderm and endoderm lineage markers in the EB formation assay. iPSCs were stained with anti-Nestin, anti-Gata4 and anti-alpha smooth muscle actin (SMA) antibodies and pictures were taken at 60× magnification. DAPI (blue) served as nucleus marker. (E) Zfp322a was able to replace Sox2, but not Oct4 or Klf4 in OSKM reprogramming process. Results from three independent experiments were presented. (F) iPSCs generated from OKM plus Zfp322a were positive with AP staining and more AP positive colonies were observed in OKM+Zfp322a as compared to OKSM. (G) iPSCs generated by OKM plus Zfp322a expressed pluripotency markers Oct4, Nanog, Sox2, Rex1 and SSEA-1. (H) iPSCs derived from OKM+Zfp322a could differentiate into ectoderm, mesoderm and endoderm lineages, which were showed by anti-Nestin, anti-Gata4, anti-SMA staining respectively. Next, the pluripotency of the iPSCs generated by OKSM plus Zfp322a were evaluated. The expression of GFP suggested that Pou5f1 promoter was strongly reactivated in iPSCs generated from OKSM plus Zfp322a. IF staining results confirmed that these iPSCs expressed pluripotency markers Oct4, Nanog, Sox2 and Rex1. ES cell marker SSEA-1 was also expressed in the iPSCs generated from OKSM plus Zfp322a (Figure 5C). To further characterize the pluripotency of these iPSCs, embryoid body (EB) formation assays were performed to examine whether these reprogrammed cells were able to differentiate into three germ layers. iPSCs were cultured in suspension to form EBs and then transferred to coated plate with EB differentiation media for 14 days before they were stained with lineage markers. We found that EBs derived from those iPSCs were able to express endoderm marker Gata4, mesoderm marker alpha smooth muscle actin (SMA) and ectoderm marker Nestin (Figure 5D). Therefore, it was demonstrated that iPSCs generated from OKSM plus Zfp322a were pluripotent and closely resembled mES cells. Taken together, we concluded that Zfp322a could induce OKSM reprogramming of MEFs into iPSCs more efficiently and faster.
7. Zfp322a can replace Sox2 in the OKSM reprogramming
We then investigated whether Zfp322a can replace the core reprogramming factors in addition to enhancement of reprogramming efficiency. Given that c-Myc is dispensable for reprogramming [34], [35], we only investigated whether Zfp322a can replace any of the OKS factors to generate iPSC colonies from MEFs. Zfp322a was unable to replace Oct4 or Klf4, however, there were GFP+ colonies observed in the MEFs infected with Zfp322a plus OKM (Figure 5E). This indicated that exogenous Zfp322a could replace Sox2 in OKSM reprogramming, albeit at a lower efficiency than OKSM. The first GFP+ colony generated from Zfp322a plus OKM was observed later as compared to OKSM control. In addition, the expression of GFP was weaker and the number of GFP+ colonies was fewer (Figure 5E, 5F, Figure S7A). Similar to iPSCs formed from OKSM plus Zfp322a, these iPSC colonies were positive for AP staining (Figure 5F, Figure S7B). Further examination of the pluripotency profile of these iPSCs showed that these iPS colonies could be stained with anti-SSEA-1, anti-Oct4, anti-Nanog, anti-Sox2 and anti-Rex1 antibodies, and were able to express all three lineage markers when they were induced to differentiate in the EB formation assays (Figure 5G, 5H). These suggested that Zfp322a could replace Sox2, but the combination of OKM plus Zfp322a might have relatively slower kinetics in generating iPSCs than OKSM.
Discussion
The unique properties of mES cells are governed by the master regulators Oct4, Nanog and Sox2, along with a variety of transcription factors (TFs) [17]. These transcription factors form a complex network to regulate mES cell identity. So far, a lot of TFs have been identified to be important for mES cell identity. However, it is still important to identify novel transcription factors since depletion of a single factor will alter mES cell pluripotency [36], [37]. Our research revealed for the first time that Zfp322a is a transcription factor which is important in maintaining mES cells in undifferentiated state.
Our results have demonstrated that depletion of Zfp322a through RNAi induced differentiation of mES cells. The differentiation of mES cells could be due to the suppression of Pou5f1 and Nanog expression after Zfp322a was depleted. Zfp322a was shown to actively regulate Pou5f1 transcription through binding to the CR4 region, and activate Nanog transcription via Nanog MTL, which are the co-binding sites of other pluripotency factors. Our ChIP-seq and microarray analysis further revealed the binding of Zfp322a to the locus of many other key pluripotency genes that were down-regulated upon Zfp322a depletion. It is interesting that Zfp322a can regulate Pou5f1 and Nanog transcription, while Zfp322a itself may also be a target of Oct4 and other pluripotency genes (Figure S1). It appears that these pluripotency factors can form a regulatory loop within the transcriptional network controlling the pluripotency of mES cells. Thus it can be inferred that Zfp322a is a regulator of mES cells by targeting or possibly cooperating with other pluripotency factors.
Moreover, our gene ontology analysis of ChIP-seq and microarray data strongly suggested that Zfp322a targets are involved in embryonic development process and related pathways. Zfp322a is implicated as a pluripotency factor because of its relatively high expression in ICM and undifferentiated mouse ES cells. Since the general reduction trend of Zfp322a expression correlates with a commitment to differentiation and a transient up-regulation of Zfp322a at third day of differentiation possibly indicates a specific role of Zfp322a in regulating early lineage commitments. We speculate that Zfp322a not only simply maintains mES cell in their undifferentiated state but also has certain roles in lineage specifications in embryo development. However, these would warrant further studies.
Notably over-expression of Zfp322a in mES cells did not significantly change cell morphology (Figure S8A). Interestingly, Nanog level was activated via Nanog proximal promoter in Zfp322a over-expressing mES cells, while Pou5f1 level showed no significant change over control (Figure S8B, C). Nanog is a well-known core regulator of ES cells, which can sustain ES cell pluripotency in the absence of LIF [27], . The activation of Nanog was also observed in a lot of other pluripotency factors, such as Zfp296, Nr5a2, Zic3 etc. [12], [39], [40]. All of these factors are required for maintaining ES cells in their undifferentiated state and can induce Nanog expression when over-expressed in ES cells. Moreover, our microarray data and ChIP-seq results showed that Zfp322a can repress MAPK/ERK pathway. It is highly possible that Zfp322a, when over-expressed, may serve as an ERK pathway repressor which results in elevated Nanog expression, mimicking high and homogeneous Nanog expression in “2i+LIF” ES culturing media containing ERK inhibitors [20], [24], [25]. Therefore we hypothesize that Zfp322a can activate Nanog expression either directly or indirectly via MARK/ERK pathway inhibition to maintain the ground pluripotency in mES cells.
In our results, we also discovered the interaction between Oct4 and Zfp322a and presence of a similar binding motif for Zfp322a and Oct4/Sox2. This is in concert with the observed higher co-occurrence frequency between Oct4 and Zfp322a binding loci based on the ChIP-seq analysis. Indeed, when we compared the gene expression profile changes after Zfp322a RNAi with Pou5f1 RNAi, a large number of high overlapping targets were identified (Figure S9) [8]. Gene ontology analysis of these co-targeting genes displayed a large number of terms related to cellular, organic, embryonic development, cell proliferation and apoptosis, chromatin remodelling, DNA transcription etc. (Table S6). In addition, many Oct4-interacting proteins were also affected in the microarray data analysis (Table S7). These implied a close correlation between Zfp322a and Oct4.
Furthermore, the observation of replacement of Zfp322a to Sox2 in OKSM reprogramming verified Zfp322a facilitation of Oct4 functions. Sox2 was discovered as a transcription factor that binds next to Oct4 motif, thus acting synergistically to active gene transcriptions [8], [9]. But subsequent studies have indicated that Sox2 functions redundantly in the activation of Oct-Sox element [41]. It is then suspected that Zfp322a could have roles partially redundant with Sox2 to interact with Oct4 and participate in Oct4/Sox2 gene regulations. Besides, it is noteworthy that Zfp322a knock-down also depleted expression of Sox2, which suggested that Zfp322a is required for Sox2 activation (Figure 1D, 1E). Therefore, Zfp322a may play a similar function of Sox2 or activate Sox2 expression to mediate its functions.
However, as a Krüppel-like zinc finger transcription factor, Zfp322a may have more complex roles in mES cells since these zinc finger transcription factors have evolved to fill roles in many different biological processes [42]. Indeed, our further analysis of genome-wide Zfp322a target sequence revealed that Zfp322a displayed a non-preferential consensus sequence binding and ubiquitously co-localized with other key pluripotency regulators. Therefore, the ubiquitous association with other factors and diverse implicated functions from microarray and ChIP-seq analysis render Zfp322a functions more complicated. Given that Zfp322a protein contains 10 zinc finger motifs, it might associate with a wide variety of co-factors through different fingers, and therefore its function is depending on the co-factors it interacts with. This pattern may be similar to other C2H2 Zfps harbouring multiple zinc fingers [43], [44]. Taken together, we hypothesize that Zfp322a function as a coordinator that fine tunes the association and recruitments of various factors, including Oct4. It would be interesting to further examine the association of Zfp322a with other components of Oct4/Sox2 regulatory cluster and also Myc cluster proteins to examine their interactions.
Considering the relatively low efficiency and considerable time for the OKSM reprogramming process, many studies focused on finding new factors or developing new methods that can accelerate the kinetics of reprogramming process or defining the reprogramming mechanism. Our results from iPSC formation assays identified Zfp322a as a novel reprogramming enhancer that can replace Sox2, thus expanding the current reprogramming code. Addition of Zfp322a was shown to induce OKSM reprogramming more efficiently and faster than OKSM alone, suggesting that Zfp322a, as a partner of Oct4 and a key regulator of mES pluripotency, can accelerate and enhance the efficiency of this process. To our understanding, Zfp322a seems to have a more remarkable role in increasing the portion of GFP+ colonies than increasing the number of AP+ colonies. Since the expression of GFP indicates the reactivation of ES cell marker Pou5f1, this implies that Zfp322a could facilitate the transition of partially reprogrammed AP+ colonies towards fully reprogrammed GFP+ colonies.
There are several possible ways in which Zfp322a can enhance reprogramming. Firstly, overexpression of Zfp322a has been shown to activate Nanog expression. Although Nanog is not one of the canonical quartets of transcription factors used for reprogramming, it is essential for the transition from dedifferentiated intermediates to ground state pluripotency [45]. Thus the enhancement and acceleration of reprogramming brought by Zfp322a could be partially facilitated by Nanog induction. This may also be the same mechanism shared by Nr5a2 and Zfp296 in their enhancement of reprogramming efficiency [12], [40]. Secondly, reprogramming process consists of down-regulation of lineage specific markers, activation of ES cell genes and widespread chromatin remodelling to re-establish the unique chromosomal confirmation of ES cells. As mentioned previously, Zfp322a was shown to repress the lineage specific markers and act as an activator for mES pluripotency genes (Figure 1). ChIP-seq analysis revealed that Zfp322a has many targets involved in chromosome assembly and modifications. Recruitment of epigenetic modifiers, such as histone acetyltransferases, and inhibition of DNA methytransferases and histone deacetylases, can promote reprogramming by loosening the condensed chromatin and thus enabling the exogenous reprogramming factors to access and transcribe pluripotency genes and jumpstart the pluripotency transcriptional network. Therefore, Zfp322a could possibly aid in reprogramming by activating directly or establishing a permissive chromatin state to allow the transcriptions of mES cell-specific genes. Thirdly, Zfp322a may promote the reprogramming via facilitating Oct4/Sox2 functions. Given our observation that Zfp322a is an interacting partner of Oct4 that can replace Sox2 in the OKSM reprogramming, it can be inferred that Zfp322a has similar functions of Sox2. Fourthly, the suppression of MAPK/ERK pathway is implicated in the predicted Zfp322a direct repressed targets. Although the total ERK level was not affected, the elevated p-ERK level upon Zfp322a RNAi indicates that Zfp322a could repress ERK pathway but not ERK expression. ERK pathway has been shown to trigger mES cell differentiation [46]. Inhibition of this pathway is important for maintaining the ground pluripotent state of mES cells and can improve somatic cell reprogramming efficiency as well [21], [47]. Thus Zfp322a could also possibly enhance the reprogramming efficiency via the suppression of MAPK/ERK cascade. Therefore, Zfp322a can maintain mES cell properties and promote reprogramming process in many aspects, yet the underlying mechanisms warrant further investigations.
Although human and mouse ES cells are differed in the signaling networks and epigenetic landscapes, it has been revealed that they share the same core regulators Oct4/Sox2/Nanog and similar transcriptional regulatory network, and the well-known Yamanaka factors OKSM are able to drive reprogramming of both human and mouse somatic cells [6], [48], [49], [50]. Given that Zfp322a is a conserved zinc finger protein in human and mouse [16], we propose that Zfp322a is extremely possible to have similar functions regarding the maintenance and acquaintance of pluripotency in human cells, which is very worthy to be elucidated in future studies.
Materials and Methods
Cell culture
Murine ES cells (E14) were cultured in ES cell medium consisting of Glasgow Minimum Essential Medium (GMEM; Invitrogen), 15% ES cell qualified fetal bovine serum (FBS; Invitrogen), 0.055 mM β-mercaptoethanol (Sigma), 100 mM sodium pyruvate (Invitrogen), 0.1 mM MEM nonessential amino acid (NEAA, Invitrogen) and 1,000 units/ml of LIF (Millipore).
Platinum-E (Plat-E) cells were maintained in Plat-E medium consisting of Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) containing 10% FBS, 1% penicillin and streptomycin. For cells transfected with retroviruses, medium were supplemented with 1 ug/ml puromycin (Sigma-Aldrich) and 10 ug/ml blasticidin (Invitrogen).
SNL feeder cells were maintained in GMEM, 10% FBS and 1% P/S. Medium was changed every 2 days and cells were passaged every 2–3 days. Inactivated SNL feeder cells were prepared by adding Mitomycin C solution (12 µg/ml, Sigma) to cell culture medium and cells were left to incubate for 2.5 h in 37°C with 5% CO2 incubator. The inactivated cells were then passaged and seeded at 80% confluence for culturing iPS cells.
Pou5f1-GFP MEFs were cultured in mESC medium without LIF. For iPSC formation, MEFs that have been infected with retroviruses were maintained in mESC medium without LIF till 5 days post infection and then maintained in Knockout Serum Replacement (KSR) medium. KSR medium contains DMEM, 15% KSR (Invitrogen), 2 mM L - Glutamine (PAA), 1 mM sodium pyruvate, 1000 units/ml of LIF, 1% P/S, 0.055 mM β-mercaptoethanol and 0.1 mM MEM NEAA. All the cells were cultured in 37°C with 5% CO2 incubator.
Plasmid construction
For RNAi design and construction of plasmids for shRNA synthesis, 19 base-pair gene-specific regions were designed. Oligonucleotides were cloned into pSuper.puro (Oligoengine). All sequences were analysed by BLAST to ensure specificity. For overexpression studies, full-length Zfp322a (NM_172586.3) was amplified by PCR and inserted into BamH1 and Xho1 site of pPyCAGIP.
For plasmids used in luciferase assays, Pou5f1 CR4 region and CR1 region was amplified and cloned into the pGL3-Promoter vector (Promega) upstream of the firefly luciferase gene to give the Pou5f1 CR4-pSV40-Luc and Pou5f1 CR1-pSV40-Luc luciferase reporter plasmids respectively; Nanog proximal promoter was amplified and inserted into pGL3-Basic vector to generate the pNanog PP-Luc plasmid.
For retrovirus packaging plasmids, full-length Zfp322a cDNA was amplified by PCR and ligated into MunI and NotI restriction sites of pMX plasmid (Addgene).
The primers being used are available in the Supplementary Table S8.
Transfection, RNA extraction, reverse transcription and quantitative real-time PCR
Transfection was conducted using Lipofectamine 2000 (Invitrogen) according to the manual provided. For shRNA, cells were selected in 6-well culture plate for 3 d using puromycin. ES cells transfected with overexpression vectors were selected using puromycin for 1 week before transferring to 100 mm plate for further selection for another 1 week. Single colonies were picked up and passaged to 6-well dishes. The cells were then harvested for either extraction of protein or RNA. RNA extraction, reverse transcription and qPCR for examine of gene expressions were conducted according to previous studies [51]. For ChIP experiments, relative occupancy values were calculated by determining the apparent IP efficiency (ratios of the amount of ChIP enriched DNA over that of the input sample) and normalized to the level observed at a control region. All the qPCR primers are available in Supplementary Table S8.
Microarray analysis
E14 cells were transfected as described above, with plasmid expressing shRNA targeted against either Zfp322a or control. Cells were harvested after 4 days selection. Total RNA was extracted and purified as described above. Then the RNA was diluted to 200 ug/ul and was analyzed using Affymetrix Mouse Genome MG430 Plus 2.0 Array according the manufacture's instruction.
Microarray data was processed to extract the representative intensities from each probe set using RMA [52]. Fold change of 1.5 was used to identify differential expression between the two sample groups (knockdown versus wild type), and only differentially expressed genes were subjected for further analysis. Prior to hierarchical clustering, log2 transformation was first performed and the transformed data were subtracted from the mean of the means of the two sample groups. To identify the enriched “Gene Ontology” (GO) terms in the differentially expressed genes, the GO TermFinder [53] was applied. For presentation of enriched KEGG pathways in the differentially expressed genes, the GATHER [54] was used. The p value cut-off of 0.05 was employed for both significant enriched GO terms and KEGG pathways. For overlapping genes between ChIP-seq predicted targets and Microarray altered gene targets, two sets of genes were analysed using VLOOKUP functions in Microsoft Excel. The microarray data has been deposited to NCBI Gene Expression Omnibus (GEO) with accession number GSE45139.
ChIP-seq
Chromatin was prepared using E14 cells as previously described [55]. Sonicated chromatin was incubated with Protein G magnetic Dynabeads (Invitrogen) coated with 40 µl of anti-ZNF322 antibody (sc-102205, Santa Cruz) overnight. The beads were then washed and Elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) was added to the beads and incubated for 45 min at 68°C with agitating at 1400 rpm. The eluent was decrosslinked by pronase. Lastly, DNA was precipitated and dissolved in nuclease free water for real-time PCR.
For ChIP-seq, ChIP DNA library was prepared by utilizing the ChIP-seq Sample Prep Kit (Illumina). Sequencing was then performed using the Genome Analyzer IIx (Illumina) and reads were mapped to the M. musculus genome assembly mm9.
ChIP-seq data analysis
ChIP-seq peak detection was performed using Partek software with an average fragment size of 300 bps and 0.05 as the cutoff p-value of Mann-Whitney U test for the separation of forward and reverse reads in a peak. In fact, the Partek software combined several methods of fragment size estimation [56], peak identification [57] and peak filtering using the Mann-Whitney U test. We further enriched the peaks by using the fold change of Zfp322a peak heights to IgG peak heights (fold change 3 as cut-off), and a minimal Zfp322a peak height at 9 reads as a further cut-off criterion (Figure S6). The final list of the inferred peaks was subjected to Zfp322a motif finding. MEME-ChIP in the MEME suite (http://meme.nbcr.net/meme/cgi-bin/meme-chip.cgi) was applied to the inferred peaks. Clustering of Zfp322a with other transcription factors (TFs) was used to evaluate the similarity of the TF targeting. The co-localization between the TFs was first computed and the correlation coefficients between each pair of co-localization vector were then determined. With the completion of all pair-wise correlation, a correlation matrix was obtained. With the matrix, a heatmap reflecting the hierarchical clustering of the correlation coefficients was generated. The Zfp322a ChIP-seq data has been deposited to NCBI Gene Expression Omnibus (GEO) with accession number GSE45138.
Western blot
Western Blot was carried out as previously described [51]. Primary antibodies used were: anti-ZNF322 antibody (sc-102205, Santa Cruz), anti-β-actin (sc-81178, Santa Cruz), anti-Oct4 (sc-8628, Santa Cruz), and anti-Nanog (sc-33760, Santa Cruz), anti-Sox2 (sc-99000, Santa Cruz), anti-Rex1 (sc-377095. Santa Cruz), anti-p-ERK (sc-7383, Santa Cruz), anti-t-ERK (137F5, Cell Signaling).
Immunofluorescence staining
Cells cultured in 24-well dishes were fixed in 4% paraformaldehyde and permeabilized with 0.25% Triton X-100, followed by blocking with 3% BSA in PBS. Then cells were probed with primary antibodyin 3% BSA for 1 h at 4°C and secondary antibodyin 3% BSA for 30 min at room temperature. A drop of Vectashield mounting medium with 4′, 6-diamidino-2-phenylindole (DAPI; Vector Laboratories) was placed on the microscope slide and the cover slip was sealed with nail polish in a way that the ES cells were in contact with the mounting medium. Staining signal was then observed through the Axio Observer A1 inverted light microscope (Zeiss). Primary antibody used were: anti-Oct4 (sc-8628, Santa Cruz), and anti-Nanog (sc-33760, Santa Cruz), anti-Sox2 (sc-99000, Santa Cruz), anti-Rex1 (sc-377095, Santa Cruz), anti-SSEA-1 (mab34301, Millipore), anti-alpha smooth muscle Actin (ab5694, Abcam), anti-Nestin (mab2736, R&D), anti-Gata4 (sc-25310, Santa Cruz).
Flow cytometry
Cells were collected by centrifugation and resuspended in PBS with 4% formaldehyde for 10 min at 37°C. After chilled on ice for 1 min, the cells were resuspended in 90% methanol. The cells were then incubated on ice for 30 min for permeabilization. The cells were rinsed with incubation buffer (0.5% BSA in PBS). The cells were incubated with primary antibody overnight at 4°C. The cells were rinsed twice and then incubated with secondary antibody for 1 h at room temperature in darkness. The cells were rinsed twice before analysed on the flow cytometer (BD FACSCanto). The flow cytometry results were analysed with flow software 2.5.0.
Dual-luciferase assays
Gene-specific shRNA (600 ng) was cotransfected with Pou5f1CR4-Luc reporter (600 ng), Pou5f1 PP-Luc or Nanog pp-Luc reporter (600 ng) and an internal control pRL-TK (30 ng, Promega) encoding Renilla luciferase. Firefly and Renilla luciferase activities were measured with the dual-luciferase reporter system (Promega) 72 h post-transfection by Ultra 384 Microplate Reader (Tecan). The data generated from gene-specific shRNA cells were expressed as relative to non-targeting shRNA control transfection, after normalization to Renilla luciferase readings. Transfections were performed in duplicate and on three independent occasions.
Alkaline phosphatase (AP) staining
AP staining was performed using Alkaline Phosphatase Detection Kit (Millipore) according to manufacturer's instructions and results were obtained using the Axio Observer A1 inverted light microscope (Zeiss).
Retrovirus packaging and infection
Plat-E cells were seeded onto a 10 cm tissue culture plate at 50–70% confluency and transfected with specific retrovirus packaging plasmids 4–6 h later. Transfection was performed as normal RNAi assays but in this experiment, 24 µg of plasmid, 60 µl of Lipofectamine 2000, and 1.5 ml of Optimen (Invitrogen) were used instead. Cells were incubated overnight in a 37°C, 5% CO2 incubator before changing to fresh medium. Virus-containing medium was collected 48 h post transfection, filtered using a 0.22 µm cellulose acetate filter (TPP) and concentrated 100× using Amicon Centrifugal Filter Units-100 kDa (Millipore) by centrifugation at 3800 rpm for 45 min. The concentrated viruses were stored in −80°C for infection use. Pou5f1-GFP MEFs were seeded onto a gelatin-coated 24 well plate at 50–70% confluency 6 h before infection. 10 ul of each concentrated retrovirus, supplemented with 8 µg/ml polybrene (Sigma), were added to the MEF cells. MEFs were then passaged onto the inactivated feeder layer 2 dpi and maintained in mESC medium without LIF before replacing with KSR medium at 5 dpi. KSR medium was replaced every day and appearance of GFP+ colonies was observed.
EB formation assay
iPSCs were treated with 0.05%Trypsin/EDTA and then cultured in ES medium without LIF in Ultra Low Culture Dish (Corning) for 5 days. Then the EB were transferred to gelatin-coated 24-well plate and cultured for 14 days. Immunostaining were performed with antibodies for specific lineage markers. Images were captured under a confocal microscope (Olympus FV1000) at 60× magnification.
Supporting Information
Zdroje
1. EvansMJ, KaufmanMH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292 : 154–156.
2. MartinGR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78 : 7634–7638.
3. KellerG (2005) Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Development 19 : 1129–1155.
4. VitaleAM, WolvetangE, Mackay-SimA (2011) Induced pluripotent stem cells: A new technology to study human diseases. The International Journal of Biochemistry & Cell Biology 43 : 843–846.
5. AmabileG, MeissnerA (2009) Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends in Molecular Medicine 15 : 59–68.
6. TakahashiK, YamanakaS (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 : 663–676.
7. YuJ, VodyanikMA, Smuga-OttoK, Antosiewicz-BourgetJ, FraneJL, et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318 : 1917–1920.
8. LohY-H, WuQ, ChewJ-L, VegaVB, ZhangW, et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genetics 38 : 431–440.
9. BoyerLA, LeeTI, ColeMF, JohnstoneSE, LevineSS, et al. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122 : 947–956.
10. DangDT, PevsnerJ, YangVW (2000) The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol 32 : 1103–1121.
11. HirataT, AmanoT, NakatakeY, AmanoM, PiaoY, et al. (2012) Zscan4 transiently reactivates embryonic genes during the generation of induced pluripotent stem cells. Scientific Reports 2: srep00208.
12. FischedickG, KleinDC, WuG, EschD, HöingS, et al. (2012) Zfp296 is a novel, pluripotent-specific reprogramming factor. PLoS ONE 7: e34645.
13. ScotlandKB, ChenS, SylvesterR, GudasLJ (2009) Analysis of Rex1 (zfp42) function in embryonic stem cell differentiation. Developmental Dynamics 238 : 1863–1877.
14. WangZX, KuehJL, TehCH, RossbachM, LimL, et al. (2007) Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells 25 : 2173–2182.
15. Yu H-B, KunarsoG, HongFH, StantonLW (2009) Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. Journal of Biological Chemistry 284 : 31327–31335.
16. LiY, WangY, ZhangC, YuanW, WangJ, et al. (2004) ZNF322, a novel human C2H2 Krüppel-like zinc-finger protein, regulates transcriptional activation in MAPK signaling pathways. Biochemical and Biophysical Research Communications 325 : 1383–1392.
17. ChenX, XuH, YuanP, FangF, HussM, et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133 : 1106–1117.
18. YoshikawaT, PiaoY, ZhongJ, MatobaR, CarterMG, et al. (2006) High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expression Patterns 6 : 213–224.
19. TangF, BarbacioruC, BaoS, LeeC, NordmanE, et al. (2010) Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-seq analysis. Cell Stem Cell 6 : 468–478.
20. YingQ-L, WrayJ, NicholsJ, Batlle-MoreraL, DobleB, et al. (2008) The ground state of embryonic stem cell self-renewal. Nature 453 : 519–523.
21. NicholsJ, SilvaJ, RoodeM, SmithA (2009) Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 136 : 3215–3222.
22. LiL, SunL, GaoF, JiangJ, YangY, et al. (2010) Stk40 links the pluripotency factor Oct4 to the Erk/MAPK pathway and controls extraembryonic endoderm differentiation. Proceedings of the National Academy of Sciences 107 : 1402–1407.
23. YangSH, KalkanT, MorrisroeC, SmithA, SharrocksAD (2012) A genome-wide RNAi screen reveals MAP kinase phosphatases as key ERK pathway regulators during embryonic stem cell differentiation. PLoS Genet 8: e1003112.
24. LuoY, LimCL, NicholsJ, Martinez-AriasA, WernischL (2012) Cell signalling regulates dynamics of Nanog distribution in embryonic stem cell populations. J R Soc Interface 10 : 20120525.
25. HamiltonWB, KajiK, KunathT (2013) ERK2 suppresses self-renewal capacity of embryonic stem cells, but is not required for multi-lineage commitment. PLoS One 8: e60907.
26. NiwaH, MiyazakiJ, SmithAG (2000) Quantitative expression of Oct4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genetics 24 : 372–376.
27. ChambersI, ColbyD, RobertsonM, NicholsJ, LeeS, et al. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113 : 643–655.
28. ChewJ-L, LohY-H, ZhangW, ChenX, TamW-L, et al. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Molecular and Cellular Biology 25 : 6031–6046.
29. YeomYI, FuhrmannG, OvittCE, BrehmA, OhboK, et al. (1996) Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122 : 881–894.
30. WolfeSA, NekludovaL, PaboCO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29 : 183–212.
31. PardoM, LangB, YuL, ProsserH, BradleyA, et al. (2010) An expanded Oct4 interaction network: implications for stem cell biology, Development, and Disease. Cell Stem Cell 6 : 382–395.
32. Van den BergDLC, SnoekT, MullinNP, YatesA, BezstarostiK, et al. (2010) An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 6 : 369–381.
33. KimJB, ZaehresH, WuG, GentileL, KoK, et al. (2008) Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454 : 646–650.
34. WernigM, MeissnerA, CassadyJP, JaenischR (2008) c-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell 2 : 10–12.
35. NakagawaM, KoyanagiM, TanabeK, TakahashiK, IchisakaT, et al. (2007) Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnology 26 : 101–106.
36. ChambersI, TomlinsonSR (2009) The transcriptional foundation of pluripotency. Development 136 : 2311–2322.
37. YeoJ-C, NgH-H (2012) The transcriptional regulation of pluripotency. Cell Research 23 : 20–32.
38. MitsuiK, TokuzawaY, ItohH, SegawaK, MurakamiM, et al. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113 : 631–642.
39. LimLS, HongFH, KunarsoG, StantonLW (2010) The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. STEM CELLS 28 : 1961–1969.
40. HengJ-CD, FengB, HanJ, JiangJ, KrausP, et al. (2010) The Nuclear Receptor Nr5a2 Can Replace Oct4 in the Reprogramming of Murine Somatic Cells to Pluripotent Cells. Cell Stem Cell 6 : 167–174.
41. MasuiS, NakatakeY, ToyookaY, ShimosatoD, YagiR, et al. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nature Cell Biology 9 : 625–635.
42. HuntleyS, BaggottDM, HamiltonAT, Tran-GyamfiM, YangS, et al. (2006) A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Research 16(5): 669–677.
43. BrayerKJ, SegalDJ (2008) Keep your fingers off my DNA: protein–protein interactions mediated by C2H2 zinc finger domains. Cell Biochemistry and Biophysics 50 : 111–131.
44. NowickK, CarneiroM, FariaR (2013) A prominent role of KRAB-ZNF transcription factors in mammalian speciation? Trends in Genetics 29(3): 130–139.
45. SilvaJ, NicholsJ, TheunissenTW, GuoG, Van OostenAL, et al. (2009) Nanog is the gateway to the pluripotent ground state. Cell 138 : 722–737.
46. LannerF, RossantJ (2010) The role of FGF/Erk signaling in pluripotent cells. Development 137 : 3351–3360.
47. LinT, AmbasudhanR, YuanX, LiW, HilcoveS, et al. (2009) A chemical platform for improved induction of human iPSCs. Nature Methods 6 : 805–808.
48. SchnerchA, CerdanC, BhatiaM (2010) Distinguishing between mouse and human pluripotent stem cell regulation: the best laid plans of mice and men. Stem Cells 28 : 419–430.
49. JaenischR, YoungR (2008) Stem cells, the molecular circuitry of pluripotentcy and nuclear reprogramming. Cell 132 : 567–582.
50. LowryWE, RichterL, YachechkoR, PyleAD, TchieuJ, et al. (2008) Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America 105 : 2883–2888.
51. LeeYH, MaH, TanTZ, NgSS, SoongR, et al. (2012) Protein arginine methyltransferase 6 regulates embryonic stem cell identity. Stem Cells and Development 21 : 2613–2622.
52. IrizarryRA, BolstadBM, CollinF, CopeLM, HobbsB, et al. (2003) Summaries of affymetrix genechip probe level data. Nucleic Acids Research 31: e15.
53. BoyleEI, WengS, GollubJ, JinH, BotsteinD, et al. (2004) GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20(18): 3710–3715.
54. ChangJT, NevinsJR (2006) GATHER: A systems approach to interpreting genomic signatures. Bioinformatics 22(23): 2926–2933.
55. WuQ, BruceAW, JedrusikA, EllisPD, AndrewsRM, et al. (2009) CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells 27 : 2637–2645.
56. KharchenkoPV, TolstorukovMT, ParkPJ (2008) Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnology 26(12): 1351–1359.
57. JiH, JiangH, MaW, JohnsonDS, MyersRM, et al. (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnology 26(11): 1293–1300.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Hypogonadotropní hypogonadismus u žen a vliv na výsledky reprodukce po IVF
- Molekulární vyšetření pro stanovení prognózy pacientů s chronickou lymfocytární leukémií
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



