-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
Plant responses to changes in environmental conditions are mediated by a network of signaling events leading to downstream responses, including changes in gene expression and activation of cell death programs. Arabidopsis thaliana RADICAL-INDUCED CELL DEATH1 (RCD1) has been proposed to regulate plant stress responses by protein-protein interactions with transcription factors. Furthermore, the rcd1 mutant has defective control of cell death in response to apoplastic reactive oxygen species (ROS). Combining transcriptomic and functional genomics approaches we first used microarray analysis in a time series to study changes in gene expression after apoplastic ROS treatment in rcd1. To identify a core set of cell death regulated genes, RCD1-regulated genes were clustered together with other array experiments from plants undergoing cell death or treated with various pathogens, plant hormones or other chemicals. Subsequently, selected rcd1 double mutants were constructed to further define the genetic requirements for the execution of apoplastic ROS induced cell death. Through the genetic analysis we identified WRKY70 and SGT1b as cell death regulators functioning downstream of RCD1 and show that quantitative rather than qualitative differences in gene expression related to cell death appeared to better explain the outcome. Allocation of plant energy to defenses diverts resources from growth. Recently, a plant response termed stress-induced morphogenic response (SIMR) was proposed to regulate the balance between defense and growth. Using a rcd1 double mutant collection we show that SIMR is mostly independent of the classical plant defense signaling pathways and that the redox balance is involved in development of SIMR.
Published in the journal: . PLoS Genet 10(2): e32767. doi:10.1371/journal.pgen.1004112
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004112Summary
Plant responses to changes in environmental conditions are mediated by a network of signaling events leading to downstream responses, including changes in gene expression and activation of cell death programs. Arabidopsis thaliana RADICAL-INDUCED CELL DEATH1 (RCD1) has been proposed to regulate plant stress responses by protein-protein interactions with transcription factors. Furthermore, the rcd1 mutant has defective control of cell death in response to apoplastic reactive oxygen species (ROS). Combining transcriptomic and functional genomics approaches we first used microarray analysis in a time series to study changes in gene expression after apoplastic ROS treatment in rcd1. To identify a core set of cell death regulated genes, RCD1-regulated genes were clustered together with other array experiments from plants undergoing cell death or treated with various pathogens, plant hormones or other chemicals. Subsequently, selected rcd1 double mutants were constructed to further define the genetic requirements for the execution of apoplastic ROS induced cell death. Through the genetic analysis we identified WRKY70 and SGT1b as cell death regulators functioning downstream of RCD1 and show that quantitative rather than qualitative differences in gene expression related to cell death appeared to better explain the outcome. Allocation of plant energy to defenses diverts resources from growth. Recently, a plant response termed stress-induced morphogenic response (SIMR) was proposed to regulate the balance between defense and growth. Using a rcd1 double mutant collection we show that SIMR is mostly independent of the classical plant defense signaling pathways and that the redox balance is involved in development of SIMR.
Introduction
Plants live in a world of change - fluctuating light, temperature, water availability and pathogen attack are among the conditions that require an appropriate response from the plant. Stress responses are energetically costly [1]–[3], hence environmental and growth signals must be integrated and balanced. Reactive oxygen species (ROS), such as superoxide and hydrogen peroxide, which can be generated in different subcellular compartments and fulfill signaling roles during both abiotic/biotic stress and development are among these key signals in plants [4]–[6]. While regulation of ROS production is to some extent understood, the mechanisms of ROS perception and downstream signaling are mostly unknown [7].
Activation of programmed cell death (PCD) is one of the aspects of plant defense responses where ROS play a crucial role [8]–[10]. The understanding of plant cell death execution lags behind that of mammals; however, some key differences have been identified. Apoptosis does not exist in plants; based on morphological criteria plant PCD has been categorized as vacuolar cell death and necrosis, plus several other types that are not easily categorized [11]. Most studies on PCD in the model plant Arabidopsis thaliana have been in the context of immune responses where a localized rapid cell death program termed the hypersensitive response (HR) is a feature of resistance [8], [12]. The air pollutant ozone (O3), when applied in short and high pulses, mimics and activates the plant's own production of an apoplastic ROS burst, similar to that seen in immune responses [13]. Hence, O3 can be used as a tool to study the role of ROS during defense and PCD, where the PCD elicited by O3 shares many similarities with HR [10]. However, many of the regulatory steps governing plant PCD in different contexts remain to be elucidated.
Massive changes in gene expression are observed during plant defense responses, after application of ROS treatments, and during cell death [14]–[19]. The requirement of changes in gene expression for cell death execution is illustrated by the reduction of O3 activated cell death by application of α-amanitin, a transcriptional inhibitor [10]. Furthermore, several lesion mimic mutants, which display spontaneous HR-like cell death in absence of pathogens [20], or mutants with altered pathogen tolerance have been used in various experiments including suppressor mutant screens and protein interaction studies to identify more regulators of cell death. Some of these regulators are involved in epigenetics, transcription or mRNA processing and include the transcription factors MYB30 [21], bZIP10 [22], SR1 [23], WRKY70 [24], mRNA processing proteins MOS2 and MOS4 [25], and chromatin remodeling factors to fine tune the transcriptional status of chromatin (LAZ2, encoding the histone methyltransferase SDG8; [26]). Although numerous abiotic/biotic stress microarray studies have been performed, relatively few experiments available in the public domain directly address the transcriptomics of cell death [27]. Further, the variety of cell death forms, and triggers used to initiate them complicate experimental design and hinder identification of cell death gene expression signatures that would not also simultaneously contain genes regulated during plant defense. Indeed cell death and defense are intimately linked, perhaps even inseparable. Clearly there is need for further analysis of gene expression during cell death in diverse experimental systems to identify potential core regulators of cell death execution.
The O3 sensitive Arabidopsis mutant radical-induced cell death1 (rcd1) is one of the mutants with defects in PCD control; it develops cell death lesions in response to a normally sub-lethal dose of apoplastic superoxide, but not H2O2 [28], [29]. RCD1 belongs to a plant specific SRO (SIMILAR TO RCD-ONE) gene family with five other genes (SRO1-SRO5) [30]–[32], some of which are also involved in stress signaling [33]. RCD1 and its closest homologue SRO1 possess a nuclear localization signal and a WWE domain, which is predicted (but not experimentally shown) to be involved in protein-protein interactions [34]. RCD1 and SRO1 display unequal genetic redundancy: whereas the rcd1 mutant has pleiotropic phenotypes in development and stress responses, the sro1 mutant has only subtle phenotypes. However, the rcd1 sro1 double mutant has severe developmental phenotypes and requires rescue on sugar containing media to generate viable plants [31], [35], [36]. RCD1 has been shown to interact with 21 transcription factors via a novel C-terminal RST (RCD1-SRO-TAF4) domain, and the known target genes of these interaction partners (such as DREB2A, DEHYDRATION-RESPONSIVE ELEMENT BINDING2A) have altered expression in rcd1 [31]. These results suggest that RCD1 may regulate PCD at the level of transcription.
The balancing act of regulating growth while maintaining an appropriate level of defenses includes a response termed stress-induced morphogenic response (SIMR) [14], [36]–[39]. SIMR includes inhibition of shoot elongation and stimulation of auxiliary branching [37], [38]. SIMR is thought to be an adaptive response to stress and is regulated by a complex interplay between ROS, auxin, ethylene and antioxidants [14], [37], [40]. The morphological phenotypes of rcd1 indicate that it could be classified as a mutant with constitutively heightened SIMR response [14], [36], [38]. A screen for modulators of defense responses identified rcd1 as an enhancer of the growth inhibition caused by constitutive activation of defense responses in the mutant snc1 (suppressor of npr1, constituive1), giving further support for a role of RCD1 in balancing between growth and defense [41]. A deeper understanding of SIMR could offer new breeding target(s) for plants with increased tolerance to abiotic/biotic stresses without accompanying growth defects.
Apoplastic ROS, in the form of O3, alter the expression of thousands of genes assigned to biotic and abiotic stress responses [14], [42]; however there are only a few studies comparing both O3 sensitive and tolerant genotypes at gene expression level which is required to dissect signaling pathways involved in PCD regulation [43]. To gain deeper mechanistic understanding of this process we used the rcd1 mutant to perform: (1) microarray analysis of an O3 time course in comparison to Col-0 to explore the role of RCD1 in defense and PCD signaling; (2) analysis of genes differentially regulated in rcd1 gene expression data using public array experiments to find genes regulated during the PCD process; (3) detailed quantitative real time PCR (qPCR) analysis to find marker genes for PCD; (4) a screen of rcd1 double mutants to find regulators of apoplastic ROS induced PCD and SIMR. We identify through the genetic analysis WRKY70 and SGT1b as regulators of cell death in rcd1 and show that quantitative rather than qualitative differences in cell death gene expression appear to better explain outcomes in cell death.
Results and Discussion
Gene expression in Col-0 and O3 sensitive rcd1-1
The O3-induced transcriptional response in rcd1 was studied in order to define cell death signaling events and to address the potential role of RCD1 as a transcriptional co-regulator. Samples of rcd1 and Col-0 were collected 0, 1, 2, 4, 8 and 24 hours after the onset of O3 exposure (6 h 350 nL L−1). All hybridizations were performed with two-color oligonucleotide microarrays against a common reference RNA, facilitating multidirectional comparisons between the genotypes and treatments. Data were analyzed with linear mixed models and genes having two-fold or higher change of expression (log2 ratio ±1, q<0.05) in at least one time point were regarded as differentially regulated in each comparison (Figure 1A). In rcd1, 4102 O3-responsive transcripts were identified over the experimental time course (Figure 1A), slightly more than in the O3-tolerant Col-0 control (3635 transcripts, [14]). In total expression of 475 genes differed between the genotypes; 274 in response to O3 and 114 genes in clean air and 87 under both conditions (Figure 1B). Comparison of the O3 responses of Col-0 and rcd1 revealed remarkable similarity; most O3-responsive transcripts (2897) overlapped between the genotypes (Figure 1B). Transcript levels of altogether 4544 genes responsive to O3-treatment in one or both of the genotypes (Figure 1B) were directly compared to study the quantitative difference in the ROS response between the genotypes. First, lists of O3-induced and –repressed genes were created for each time point and genotype separately. At all time points, there were more O3-regulated genes in rcd1 than in Col-0, and both genotypes possessed uniquely regulated genes, but no transcripts were oppositely regulated by apoplastic ROS in Col-0 and rcd1 backgrounds, i.e., increased in one genotype and decreased in the other at the same time point. Therefore, the genotype-specific lists of regulated genes were combined and the differences in absolute levels between O3-treated rcd1 and O3-treated Col-0 were calculated. The magnitude of O3-induced changes was greater in rcd1 than in Col-0 at all O3 time points for both induced (64–77% of the O3-induced genes) and repressed (58–91% of the O3-repressed genes) transcripts (Figure 1C and 1D, respectively). Therefore, O3-regulated genes had a more pronounced expression change in rcd1, suggesting that the O3 response is quantitatively heightened in rcd1. However, most of the differences between the genotypes were subtle, less than two-fold (Figure 1C and 1D). Four genes were quantified in more detail with qPCR, and the results from this analysis were in agreement with the array data (Figure 1E).
Fig. 1. Gene expression of Col-0 and rcd1 mutant in clean air and O3-treated plants. 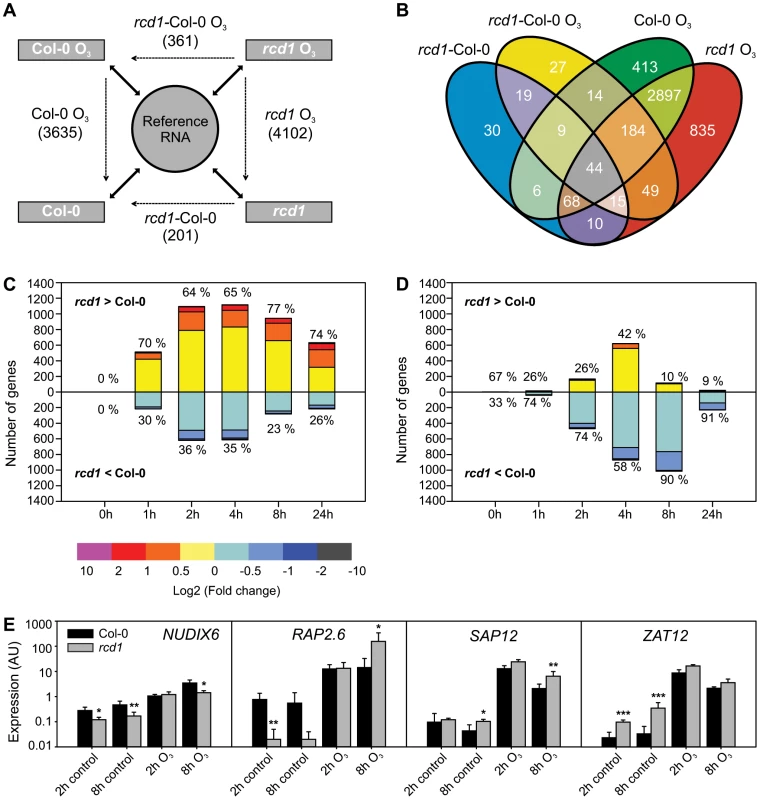
A) Experimental design with each sample hybridized against a reference RNA (solid arrows) enables multidirectional comparisons between genotypes and treatments with a linear mixed model (dotted arrows). The experiment described was repeated at each time point (0, 1, 2, 4, 8 and 24 h) and lists of differentially expressed transcripts (log2 ratio ±1, q<0.05) were made from each comparison. Number of differentially expressed unique genes at all time points is shown. The 0 h comparison of O3-treated rcd1 and O3-treated Col-0 was analyzed together with rcd1 and Col-0 grown in clean air to comprise the list of genes differentially regulated in rcd1 in normal growth conditions (rcd1-Col-0). Simultaneously, this 0 h comparison was omitted from the rcd1-Col-0 O3 data set. B) Venn diagram showing the overlap of gene lists. Transcripts at least two-fold differentially regulated between rcd1 and Col-0 in response to apoplastic ROS are divided into several subcategories discussed in the results. C, D) Transcript levels of 4544 genes responsive to O3-treatment (log2 ratio ±1, q<0.05) in one or both of the genotypes were compared. Genes were divided into O3-induced (C) and O3-repressed (D) according to the O3-response specifically at each time point (0, 1, 2, 4, 8 and 24 h). For each gene the difference between O3-treated rcd1 and Col-0 was calculated (log2 ratio) and the number of genes in each range of differential expression (depicted in color) is shown on the y-axis. The percentage of genes with higher (rcd1>Col-0) and lower (rcd1<Col-0) expression in rcd1 compared to Col-0 was calculated. E) Expression of selected marker genes was studied in Col-0 and rcd1 plants with qPCR. Bars represent means of three biological repeats, error bars show standard deviation. Statistically significant difference between genotypes is depicted with asterisk (P<0.05:*; P<0.01:** and P<0.001:***). Expression of RCD1 and interacting transcription factors
At the whole tissue level, RCD1 transcript levels are slightly responsive to many stresses including O3 (Figure S1, [30], [32]) and show strong induction only in response to high light [44]. To test for local cellular responses in ROS-induced lesions, activity of the RCD1 promoter was monitored by β-glucuronidase (GUS) staining in RCD1-promoter::uidA fusion lines exposed to high O3-concentrations that induced lesions, or treated with the herbicide methyl viologen (MV; paraquat) which induces chloroplastic ROS production. The RCD1 promoter was active specifically in the cells inside O3-induced cell death lesions and also in the cells directly treated with MV (Figure S2). As a comparison, lines expressing uidA under control of the promoter from SRO1, the paralog most similar to RCD1, were also monitored. Although SRO1 and RCD1 share similar developmental expression [31], unlike RCD1, SRO1 expression was not increased in O3-induced lesions or in response to MV treatment (Figure S1). Another member of this gene family, SRO5, regulates salt stress responses [33] and is induced by O3 [32]. The sro1 and sro5 mutants did not display increased O3 sensitivity and the rcd1 sro5 double mutant had unaltered responses compared to rcd1 (Figure S2). The lack of stress regulation of the SRO1 gene or O3 stress phenotypes in the sro1 and sro5 mutants suggests that RCD1 regulates ROS responses and cell death independent of SRO1 and SRO5.
RCD1 interacts with several transcription factors (TFs) and may regulate transcription via protein-protein interactions [31], [45]. Many RCD1 interacting TFs have no established biological functions. Co-expression analysis has been used to suggest the function of previously uncharacterized proteins [46]. Expression profiles of 15 RCD1 interacting TF genes and RCD1 itself were studied in data sets comprising hormone-, abiotic stress-, biotic stress - and O3-treatments (Figure S3; see below for a full discussion of data sets used). DREB2A, ANAC013 and ANAC046 were the only TFs with major expression changes in response to diverse stresses. ANAC013 is localized to both cytosol and nucleus [47] and is a possible membrane anchored protein that after proteolytic cleavage would move to the nucleus [48]. None of the TFs had altered expression in the rcd1 mutant under O3, indicating that RCD1 does not transcriptionally regulate genes encoding its interaction partners. Overall, the RCD1 expression profile was not similar to that of its interaction partners, thus other types of data, for example in vivo protein stability or double mutants will be required to position RCD1 in stress signaling pathways.
Clustering of RCD1 regulated genes
To gain further information on processes downstream of RCD1, the expression profiles of 423 RCD1 regulated genes were clustered together with publicly available data from experiments performed on the Affymetrix ATH1 chip (Figure 2). These experiments were selected to distinguish between genes regulated by abiotic and biotic stresses, stress hormones, ROS and cell death (see Materials and Methods for the complete set of experiments). Several constitutive defense mutants (siz1, sni1 [suppressor of npr1-1, inducible1], lht1 [lysine histidine transporter1], cs26 [cysteine synthase26]) clustered together with the salicylic acid (SA) analog benzothiadiazole (BTH) treatment (Figure 2). The strongest change in gene expression was in cell death associated treatments including Pseudomonas syringae pv. maculicola (Psm) ES4326 infection [49], the acd11 (accelerated cell death11) lesion mimic mutant [26], and ROS challenged rcd1 at 8 and 24 h (Figure 2). These late O3 time points exhibited the largest differences between O3-treated rcd1 and Col-0, whereas early O3 time points of both genotypes clustered together with H2O2 and flagellin 22 (flg22) treatments (Figure 2). The rcd1 mutant in clean air had a unique gene expression profile, as discussed in detail below.
Fig. 2. Cluster analysis of genes differentially regulated in rcd1 compared to Col-0. 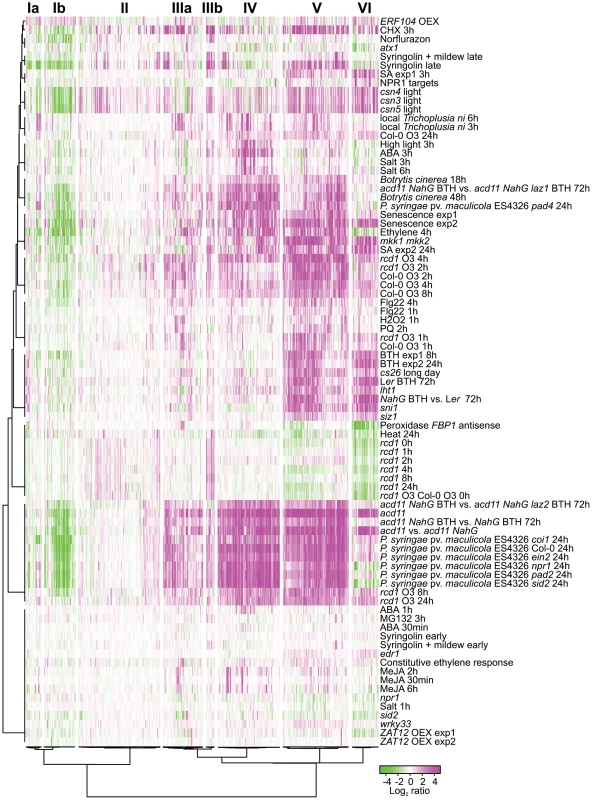
Bootstrapped Bayesian hierarchical clustering of 423 genes with at least two-fold changed expression (log2 ratio ±1, q<0.05) in clean air or O3-treated rcd1 is shown. Data sets used were rcd1 mutant grown in control conditions, O3-treated Col-0, O3-treated rcd1 and several other available experiments related to stress signaling and cell death (see “Materials and Methods” for the complete set of experiments). Six main clusters (I to VI) with subclusters (marked with a or b) were identified. GO and promoter element enrichment results are provided in Table S1. Magenta and green indicate increased and decreased expression as log2 ratio compared with untreated or wild type plants, respectively. RCD1-regulated genes clustered into six different major groups, two of which further divided into subclusters. These were further analyzed for enrichment of gene ontology (GO) classes and promoter elements (Figure 2, see Table S1 for genes belonging to each cluster and statistical results). Clusters Ia and Ib contained genes with reduced transcript accumulation in most experiments analyzed. Cluster Ib was enriched for genes encoding 17 chloroplast located proteins and 5 apoplast proteins (Table S1). Photosynthesis and chloroplast related genes have reduced expression during biotic stress as a defense strategy, likely for energy conservation [50]. However, photosynthesis as a biological process was not enriched in cluster Ib (Table S1).
The heterogeneous cluster II included a few genes with increased expression in rcd1 clean air samples, increased expression in light grown COP9 signalosome mutants (csn3, csn4, csn5; [51] and a late time point after treatment with the proteasome inhibitor Syringolin [52] (Figure 2). The COP9 signalosome regulates the activity and stability of cullin-RING-type E3 ubiquitin ligases (CRL), and Arabidopsis csn mutants arrest growth at the seedling stage, possibly through a DNA damage pathway [51]. Through most other clusters (I, IIIb–VI), expression profiles in csn mutants were very similar to acd11, suggesting that they may undergo cell death during seedling growth arrest. However, in cluster II several genes had increased expression in csn mutants and in clean air rcd1, but were not regulated by other stresses. E3 ligases are involved in targeting specific proteins for degradation by the proteasome, similarly one proposed function of RCD1 is to regulate stability of transcription factors [31]. The specific targets of COP9 and CRLs in Arabidopsis are mostly unknown, but more detailed characterization of genes regulated by both COP9 and RCD1 might reveal new insights into the role of protein degradation in stress responses.
Genes in cluster IIIa had a trend towards increased expression in nearly all treatments studied and included ethylene biosynthesis and signaling genes (ACS6 [1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6], ACO2 [ACC OXIDASE 2], ERF13 [ETHYLENE RESPONSE FACTOR13], ERF104, and the ROS signaling kinase OXI1 (OXIDATIVE SIGNAL-INDUCIBLE1) [53]. Genes in cluster IIIb were characterized by very high expression in clean air rcd1. This cluster of eleven genes contained AOX1a (ALTERNATIVE OXIDASE 1A), UPOX1 (UPREGULATED BY OXIDATIVE STRESS1) and a putative RCD1 interacting transcription factor ANAC013 [31]. The high expression of AOX1a and cluster IIIb genes indicated that rcd1 was under constitutive stress as previously proposed [30], [31]. The major RCD1 interactor DREB2A is a regulator of both drought and heat stress [54]. Of the stress treatments included in this study, rcd1 expression profile in clean air shared the highest similarity to heat stress (Figure 2). Many DREB2A targets are also heat-responsive [54], of which the NFXL1 (NF-X-LIKE1) transcription factor, involved in heat acclimation [55], was found in cluster IIIb. Therefore, the rcd1 mutant may have a heat tolerance phenotype in line with its higher accumulation of heat shock proteins [45]. Transcript levels in cluster IIIb were decreased by MV (Figure 2), which might be connected to the MV tolerance of rcd1 [29], [30]. Further studies with cluster IIIb genes may also reveal pathways contributing to the MV tolerance of the rcd1 mutant. Intriguingly, the cell death regulator LAZ2 involved in chromatin remodeling [26] reversed the expression of cluster IIIb genes hinting that these genes may be involved in cell death and defense responses.
Cluster IV genes generally exhibited increased transcript accumulation under most stress treatments including salt stress, high light and abscisic acid (ABA) (3 h), which suggested that expression of these genes is governed by a “general” stress regulatory circuit. Consistent with a role for ABA, NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3), an early stress-induced ABA biosynthesis gene [56], [57] was found in this cluster, and the GO category “response to water deprivation” was significantly enriched together with cis-elements related to ABA responses and abiotic stress (Table S1). ALD1 (AGD2-LIKE DEFENSE RESPONSE PROTEIN 1), a regulator of biotic stress responses and cell death [58], was found is in this cluster together with transcription factors RAP2.6 (RELATED TO AP2 6), WRKY28 and ANAC019.
Genes in cluster V had reduced expression in clean air rcd1 and were strongly induced by O3, biotic stress, ethylene, SA, BTH, senescence and in constitutive defense mutants. This cluster included multiple TFs, including WRKY18, WRKY25, WRKY48, WRKY75 and bZIP60. Promoters of cluster V genes were enriched in CONSERVED MOTIF2 (CM2), a binding site for CAMTA TFs [59] (Table S1). Furthermore, this cluster contained several regulators of biotic stress responses including PAD4 (PHYTOALEXIN DEFICIENT4), SAG101 (SENESCENCE-ASSOCIATED GENE101), FMO1 (FLAVIN-DEPENDENT MONOOXYGENASE1) and NUDX6 (NUCLEOSIDE DIPHOSPHATES LINKED TO SOME MOIETY X 6). EDS1 (ENHANCED DISEASE SUSCEPTIBILITY1) and its interacting partners PAD4 and SAG101, are regulators of biotic stress, SA responses and ROS signaling [60]. Cluster VI had genes with strongly reduced expression in clean air rcd1 and increased expression by biotic stress, SA, BTH and in constitutive defense and cell death mutants. Expression of these genes was reduced by ethylene and cluster VI was the only cluster where ethylene gave the opposite result to SA/BTH. Reduced expression of these genes in the SA biosynthesis mutant sid2 (salicylic acid induction deficient2) and the SA receptor/transcriptional co-factor npr1 (nonexpressor of pathogenesis-related genes1) strongly suggested that SA and SA signaling were required for expression of these genes. Cluster VI included several direct targets of NPR1, such as WRKY38, WRKY54 and WRKY70 [61] and the SA marker genes PR-2 (PATHOGENESIS-RELATED PROTEIN 2) and PR-5 (Figure 2). GO analysis revealed significant enrichment of “response to salicylic acid stimulus” and “response to bacterium” in cluster VI (Table S1). The SA-responsive cell death regulator ACD6 (ACCELERATED CELL DEATH6) [62] also belonged to cluster VI. The rcd1 mutant in clean air did not display any similarity to the constitutive defense mutants sni1, siz1, cs26, mkk1 mkk2 [mitogen activated protein kinase kinase1/2] or lht1 [63]–[67], which all had high expression of SA and BTH responsive genes (clusters V and VI). In contrast, rcd1 had reduced expression of these genes suggesting that RCD1 is a previously unrecognized positive regulator of SA signaling. In cluster VI, rcd1 in clean air was also very similar to plants with silenced apoplastic peroxidases [68]. Since the rcd1 mutant is specifically sensitive to apoplastic ROS, this expression profile similarity indicates a role for apoplastic signaling events resulting in lowered expression of SA responsive genes. Consistent with this interpretation, signaling activated by flg22 (which is perceived by the FLAGELLIN SENSITIVE2 (FLS2) receptor in the apoplast) leads to reduced expression of SA responsive genes [69]. Importantly, this apoplastic signaling does not involve cluster IIIb genes regulated by heat stress, hence partially dissecting these signaling pathways.
Overall the clustering of experiments indicated that there was no specific cell death profile. Under O3, the early rcd1 time points clustered closely with Col-0. Indeed, many of the genes with decreased expression in rcd1 in clean air, for instance WRKY70, PR-2 and PR-5, exhibited normal O3-induction in rcd1 (Figure 2). At late time points, post O3 treatment, expression levels nearly returned to basal clean air levels in Col-0. In contrast, rcd1 at 8 and 24 hours maintained a highly altered expression profile similar to biotic stress treatments and the acd11 mutant undergoing cell death. Especially genes in cluster IV were strongly induced in these experiments (Figure 2). Cluster IV was enriched for GO classes related to water stress and the ABA response element (ABRE), but not defense (Table S1), and therefore this suggests that cell death and ABA responses coincide at late time points. Spontaneous cell death mutants with associated high expression of various defense genes can arise from various disturbances in cellular homeostasis or signaling [20], which makes it difficult to deconvolute the response to cell death versus activation of defense gene expression. Both basal and effector triggered immunity are qualitatively similar, i.e., the same set of defense genes are induced, but in the latter there is a quantitative difference in that the genes are induced higher and faster [70]. We observed a similar phenomenon in the O3 response of rcd1 where apoplastic ROS induced a gene expression response qualitatively similar to Col-0, but faster and to a higher level. Overall, clustering of genes differentially expressed in rcd1 suggests that the ROS-triggered lesion formation might depend on a fine-tuned threshold and timing, rather than of an on-off regulation of gene expression.
Depending on the particular stress, the hormones ABA, SA, jasmonic acid (JA) and ethylene can show synergistic or antagonistic interactions [71], [72]. SA and ethylene promote ROS-induced cell death, when JA antagonizes cell death [73]. The clustering clearly separated the function of the defense hormones ABA, JA, SA and ethylene in regulating ROS-induced gene expression in rcd1 (Figure 2). ABA and JA mostly had a minor role with only a few genes having strong induction (ABA only regulated genes in cluster IV). In contrast SA/BTH at 24 h and ethylene at 4 h had a very similar and strong effect on many genes in clusters I and IV–V. Timing is clearly important since SA at 3 h did not have strong effect on clusters I and IV. Unfortunately, the only publicly available ethylene Affymetrix experiment using fully grown plants was done only at the 4 h time point [74], and most public SA/BTH experiments only have late time points (8 or 24 h). Given this limitation, and the known kinetics for O3 induced biosynthesis of SA (5 h; [10] and ethylene (1 h or earlier; [28], [75]) it is likely that ethylene is the initial stress hormone, later being augmented with SA in regulation of O3 induced gene expression. One exception to the synergistic role of ethylene and SA were the cluster VI genes which were regulated positively by SA and negatively by ethylene. A similar SA-ethylene antagonism in gene expression was detected for a subset of genes responding to PsmES4326 treatment [76].
Cell death marker genes
To further characterize marker genes and potential cell death regulatory genes, a subset of RCD1 regulated genes with functions related to defense, stress or cell death signaling were studied with qPCR in mutants related to cell death and defense signaling (Figure 2). These genes were NUDX6, SAP12 (STRESS-ASSOCIATED PROTEIN 12) and transcription factors RAP2.6, WRKY38, WRKY62, WRKY70, WRKY75 and ZAT12. In addition, O3-responsive genes ACS6, ALD1, JAZ1 (JASMONATE-ZIM-DOMAIN PROTEIN1) and FMO1 were included in the analysis. Some of these genes have also been shown to be regulators of cell death based on mutant analysis (ald1, fmo1, [58]) or to suppress constitutive defense signaling (wrky70, [24]). Before using these genes in O3 experiments they were validated in an independent cell death experimental system with lesion mimic mutants acd2 (accelerated cell death2), acd5 (accelerated cell death5) and lsd1 (lesion simulating disease resistance1). The three lesion mimic mutants were selected to have contrasting mechanisms for lesion formation. ACD2 encodes a protein with multiple subcellular localizations (chloroplast, mitochondria, cytosol) which likely antagonizes cell death via binding to PCD-inducing metabolic products [77]. ACD5 encodes a ceramide kinase involved in lipid metabolism and signaling [78], [79], and LSD1 has been proposed to act as a cellular hub negatively regulating PCD by interacting with other proteins, such as bZIP10 [22], AtMC1 (METACASPASE1; [80]) and GILP (GSH-INDUCED LITAF DOMAIN PROTEIN; [81]).
The lesion mimic mutants were first grown lesion free in permissive conditions for three weeks and then shifted to lesion-inducing long day (LD) conditions. Three days after the shift to LD, extensive lesion formation was observed in acd5 and lsd1 plants, and to a lesser extent in acd2 plants. Samples were harvested separately from Col-0 and from lesioned leaves (+), lesion-free leaves from lesion-containing plants (−) and lesion free plants (0). This harvesting scheme allowed the separation of gene expression effects before lesions (the 0 samples), cell death (the + samples) and systemic signaling from dying cells (the − samples). Overall, the expression of marker genes before visible lesions (0 samples) was similar to Col-0 and increased with the appearance of lesions, especially in acd2, and to some extent elevated in systemic (−) leaves (Figure 3). However, NUDX6 expression decreased in lesioned leaves (Figure 3). NUDX6 has pyrophosphohydrolase activity towards NADH and regulates redox balance and gene expression in SA signaling [82]. Its close homologue NUDX7 has been extensively characterized in ROS related cell death and defense responses, where knock-out mutants display spontaneous cell death, altered redox balance and constitutive defense gene expression [83]–[85]. SA signaling is dependent on redox changes of NPR1 [86] and NUDX6 is proposed to be involved in regulating this redox balance [82]. Since SA is a positive regulator of cell death [10], [73], and knockout of NUDX6 leads to increased sensitivity to SA [82], the lower expression of NUDX6 in lesion-leaves could be involved in fine-tuning the redox balance and hence downstream events in cell death execution.
Fig. 3. Expression of selected marker genes in lesion mimic mutants. 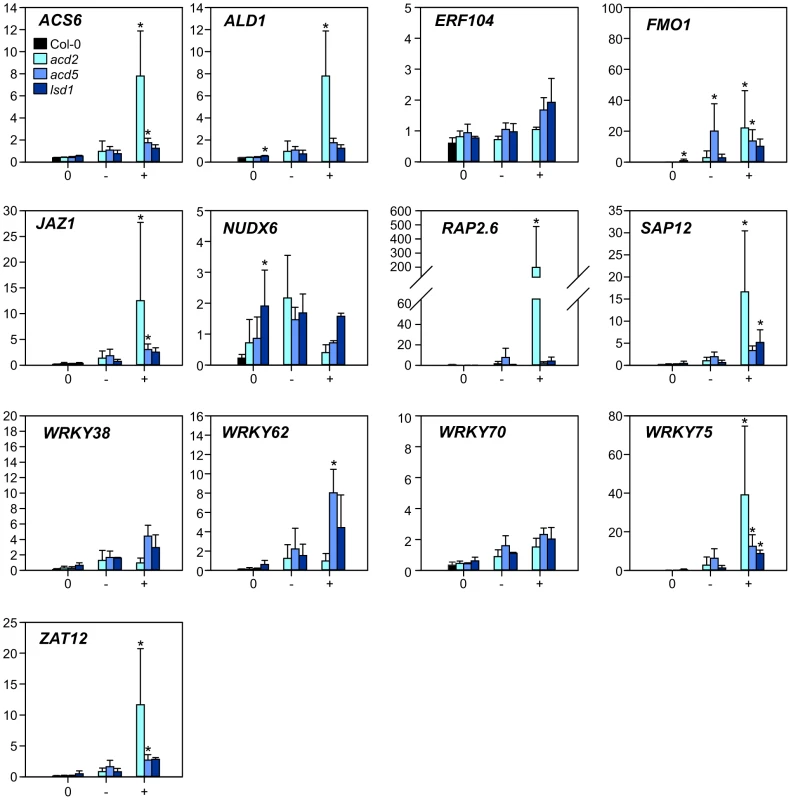
Samples from three lesion mimic genotypes (acd2, acd5 and lsd1) 3 d after the transfer to LD were classified as samples from plants with no lesions (labelled 0); leaves with no lesions from plants with lesions (labelled −); leaves with lesions (labelled +). Averages of qPCR results (arbitrary units) from three biological replicates are shown; error bars depict standard deviation. Asterisks depict statistical significance (P<0.05) between to Col-0 (within 0 samples) or to 0 samples within the respective genotype (− and + samples). Having established that the chosen set of marker genes were regulated during PCD (Figure 3), qPCR analysis of rcd1 and several other single and corresponding double mutants with rcd1 was performed at 2 and 8 h after the start of O3 treatment. The mutants were chosen to disrupt signaling of the major stress hormones JA, ethylene and SA (coi1-16 the receptor for JA, etr1-1 a dominant negative allele of the ethylene receptor, npr1 the SA receptor and transcriptional co-regulator). Furthermore, two other mutants were included: mpk6, a knockout for MAP KINASE6, a regulator of many stress responses including O3 [87], [88] and wrky70, a positive regulator of SA signaling and negative regulator of JA signaling [89]. Previously we have established that removing JA function enhances, and depletion of SA reduces cell death in rcd1 [10], [14].
The qPCR data was clustered with bootstrapped Bayesian hierarchical clustering to find similarities between genes and mutants (Figure 4A) and statistically analyzed with linear mixed models for differences to the respective Col-0 (Figure 4B and 4C). In control conditions many differences were observed in the mutants, in particular ZAT12 expression appeared to be very sensitive to any perturbation since its expression increased in almost all mutants compared to Col-0 (Figure 4C). Increased expression of WRKY38, WRKY62, ALD1, FMO1 and NUDX6 in wrky70 suggests that WRKY70 is a negative regulator of these genes (Figure 4C). NPR1 was a positive regulator of WRKY38, WRKY62 and WRKY70, consistent with previous analysis of NPR1 regulated genes [61]. JAZ1 is a transcriptional repressor of JA responses and is rapidly degraded after binding of the bioactive JA-Ile to the receptor COI1 [90]. The JAZ genes are also regulated at the transcriptional level by JA [91], and in coi1-16 there was low JAZ1 expression (Figure 4C); collectively this indicates that JAZ1 makes a good marker for the output of JA signaling (Figure 4A and 4B).
Fig. 4. Cluster analysis of gene expression in clean air and O3-treated plants. 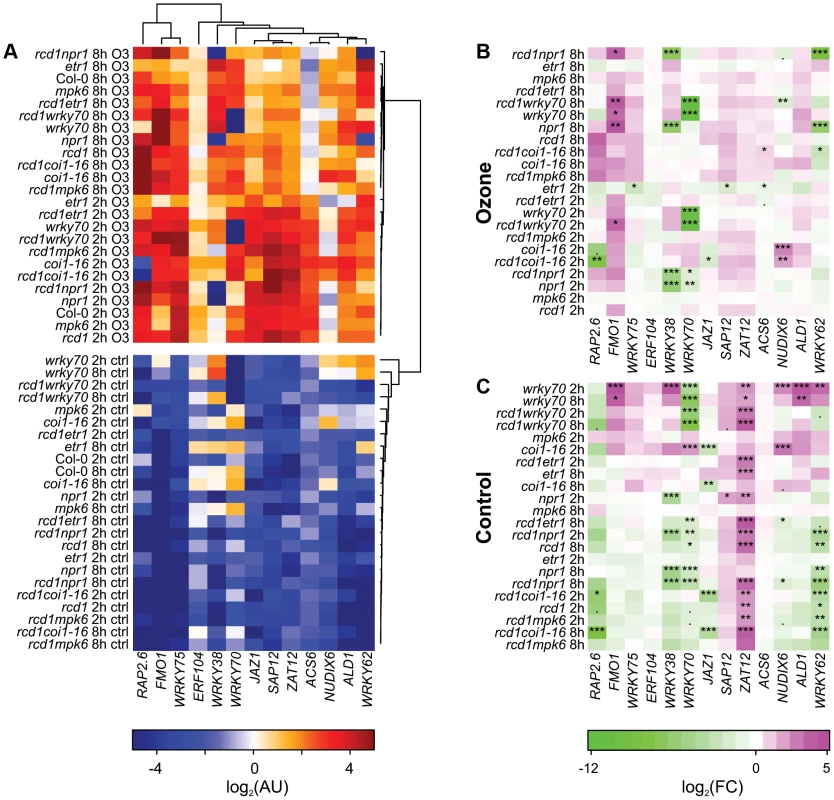
Three-week-old plants were treated with 6 h of O3 (350 nL L−1) and samples harvested at 2 and 8 h after the start of the O3 exposure. Expression of selected marker genes in each genotype was studied with qPCR and bootstrapped Bayesian hierarchical clustering was applied to log2-transformed expression values in arbitrary units (A). Gene expression in mutant genotypes was compared to the respective Col-0 sample at each time point (2 h, 8 h) and log2-transformed fold changes were calculated for O3 treatment (B) and control plants (C). Asterisks mark statistical significance according to the linear model (P<0.10:“.”; P<0.05:“*”; P<0.01:“**”; P<0.001:“***”.). Many of the genes (SAP12, ZAT12, ACS6, ERF104, JAZ1, ALD1 and WRKY75) were induced by apoplastic ROS early at 2 h, after which their expression started to decline 8 h after the start of the O3 treatment (Figure 4A). ACS6 encodes a stress-inducible ethylene biosynthesis gene often used as a marker for ethylene signaling and consistent with this, ACS6 expression was lower after O3 in the etr1 mutant (Figure 4B). The remaining marker genes (FMO1, RAP2.6, NUDX6, WRKY38, WRKY62 and WRKY70) had the highest expression at 8 h (Figure 4 A). The most dramatic change from WT expression pattern was seen for WRKY38, and to a lesser extent WRKY62 in npr1 and rcd1 npr1, where the O3 induction was abolished; hence, NPR1 is an essential positive O3 regulator of these genes (Figure 4A and 4B). SA and Pseudomonas syringae induction of these WRKY TFs also requires NPR1 [92], thus in particular WRKY38 can be used as a convenient reporter to follow signaling via NPR1. In contrast, FMO1 expression was enhanced in npr1, wrky70 and corresponding rcd1 double mutants (Figure 4B), thus NPR1 has both positive and negative signaling roles in apoplastic ROS signaling. The coi1-16 mutant also revealed both positive and negative roles for JA signaling: RAP2.6 expression was reduced and NUDX6 enhanced at 2 h O3 in coi1-16 (Figure 4B). Apparently, JA regulation is most important at early signaling since at 8 h there were no longer any differences compared to WT (Figure 4B).
The marker genes chosen are related to cell death or defense signaling, and were induced during lesion formation (Figure 3). Could they also provide insights to the cell death process in rcd1? The relative severity of cell death at 8 h is in the order rcd1 coi1-16>rcd1, rcd1 mpk6, coi1-16>rcd1 wrky70, rcd1 npr1 ([10], [14]; Figure 5), but there was no indication that rcd1 coi1-16 would be strikingly different from rcd1 at this time point, except for somewhat enhanced ACS6 expression. Instead, it appeared that signaling prior to visible lesion formation (i.e., the 2 h time point) could determine the extent of later cell death. In particular NUDX6 expression was enhanced in the O3 sensitive rcd1 coi1-16 and coi1-16. Although both the cluster analysis (Figure 2) and qPCR results (Figure 3 and 4) revealed some interesting correlations between cell death and gene expression, it appears that gene expression data alone is insufficient to identify “unique” cell death regulators, i.e., genes that would be regulated only during cell death and not by other abiotic or biotic stresses. Identification of genes specifically regulated during cell death could, e.g., be identified with a much higher temporal and spatial resolution, i.e., cells undergoing cell death (as well as neighboring cells) would have to be microdissected out from leaves preferably in a time course [93], [94]. Analysis of whole leaves/rosettes from dying plants is likely to contain a mix of cell death process and neighboring cells with activated defense responses. It is also a distinct possibility that unique cell death regulators might be rare and instead, regulators (such as various transcription factors) are recruited at different times to fulfill signaling roles both during stress and cell death.
Fig. 5. Quantification of O3 -induced cell death in Col-0, rcd1 and various double mutants. 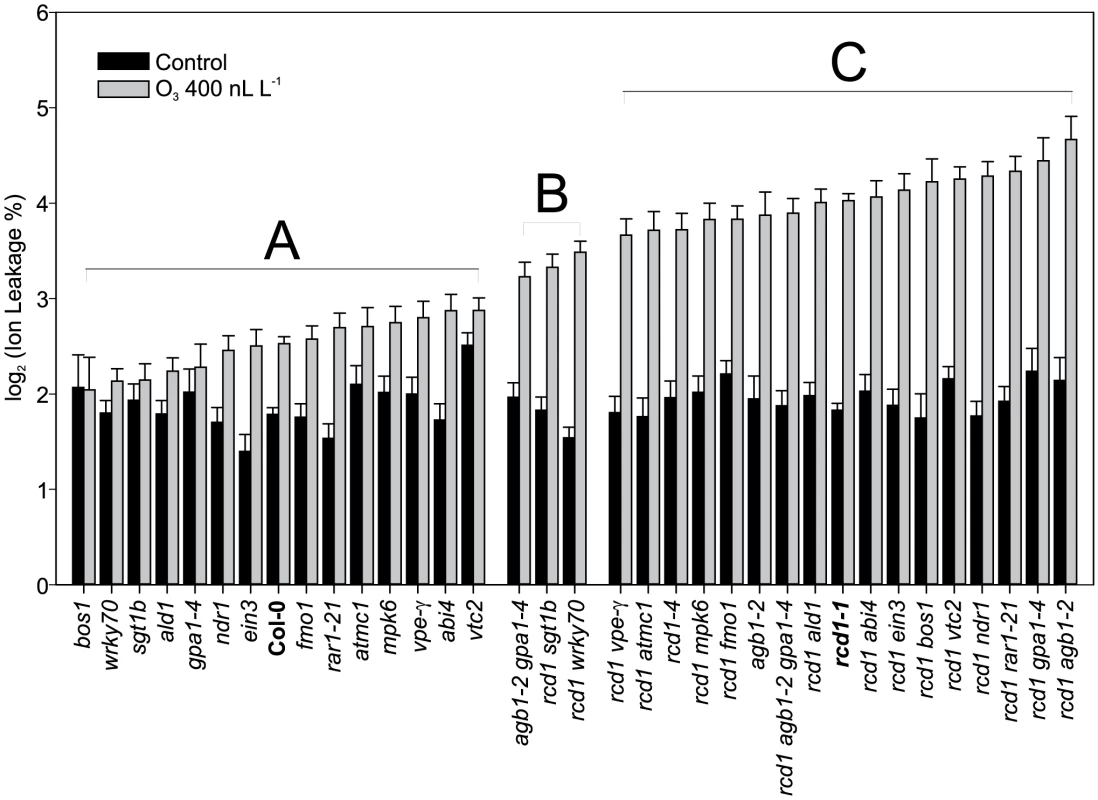
Plants were exposed to 63 (400 nL L−1) and after 2 h recovery in the clean air, they were harvested for ion leakage measurements. Samples of untreated plants grown in clean air were simultaneously collected. Samples are ranked according to their ion leakage in O3. Ion leakage percentages of O3 samples were compared to O3-treated Col-0 and rcd1-1 with linear models. Ion leakages of samples belonging to groups A and C did not differ from Col-0 and rcd1, respectively. Group B and C samples showed elevated O3-damage compared to Col-0 (P<0.01), whereas samples in groups A and B had decreased O3-damage in comparison to rcd1 (P<0.01). In clean air samples, only vtc2 differed from Col-0 (P<0.001). Bars represent means of two to seven biological repeats with standard error. rcd1 cell death execution is distinctly different from pathogen induced cell death processes
Cell death in rcd1 is reduced by application of a transcriptional inhibitor [10]. This suggests that some O3 responsive genes in rcd1 are candidate regulators of cell death. Also, many cell death regulators have been identified through the study of pathogen induced cell death. Mutant analysis was used to directly test the role of these cell death regulatory genes in O3-induced cell death. Based on rcd1 and O3 gene expression data, as well as known regulators of pathogen defenses and suppression of lesion mimic phenotypes, several rcd1 double mutants were constructed and evaluated for O3 induced PCD (Figure 5). Mutants included ald1 and fmo1, suppressors of a SYNTAXIN lesion mimic mutant syp121 syp122 [58]; mpk6, a regulator of stress responses [95]; abi4, a TF regulator of stress signals originating from chloroplasts and mitochondria [96]; ein3, a TF in the ethylene signaling pathway; wrky70 a TF with positive regulation of SA signaling and negative regulator of JA signaling [89]; rar1 (required for MLA12 resistance) and sgt1b (suppressor of the G2 allele of SKP1b) regulators of various aspects of pathogen defenses, SA signaling and PCD [97]; ndr1 (nonrace specific disease resistance1) a regulator of ROS mediated cell death in lsd1, and avirulent Pseudomonas infection [98], [99]; mc1 (metacsapase1) a positive regulator of PCD [80]; vtc2 (vitamin C defective2) a mutant with low concentration of the important antioxidant ascorbic acid [100]; vpe-gamma (vacuolar processing enzyme) [101], gpa1 and agb1, the alpha and beta subunits of heterotrimeric G-protein signaling complex [102].
Despite the well-documented role for many of these genes in regulating cell death during pathogen infection and in lesion mimic mutants, only sgt1b and wrky70 were able to partially suppress O3 induced cell death in rcd1 (Figure 5). WRKY70 acts as an integrator between SA and JA signaling [89]. In addition, wrky70 was isolated from a suppressor screen for mutants that restore normal growth to a dwarfed constitutive defense mutant snc2-1D (suppressor of npr1-1, constitutive 2) [24]. Hence the decreased cell death in rcd1 wrky70 could be the result of reduced expression of a WRKY70-dependent positive regulator of cell death. SGT1b is an accessory factor to SCF (Skp1/Cullin1/F-box) E3 ligases, which are master regulators of ubiquitin targeted protein degradation [103]. Since the SCF E3 ligases have multiple targets in plants, most of which are unknown, we speculate that in rcd1 sgt1b there is negative regulator of cell death that is stabilized when SGT1b function is removed. The lack of influence on O3-induced cell death by other regulators previously shown to alter pathogen-induced cell death or to suppress lesion mimic phenotypes, including ald1 and fmo1, indicate that despite many similarities between pathogen and O3-induced cell death [10], the execution of the O3 cell death program requires different components. Alternatively, RCD1 could function as one of the final downstream steps in cell death execution, hence previously identified cell death regulators might be mostly up-stream of RCD1 and epistatic in double mutant analysis. A suppressor mutant screen of rcd1 has the potential to identify new regulators of PCD execution.
Cluster IIIb (Figure 2) contained genes with very high expression in clean air rcd1 and included two mitochondria localized proteins AOX1a and UPOX1. AOX1a acts to bypass the last step of mitochondrial electron transport and is proposed to reduce ROS production in times of stress and to act as a regulator of PCD [104]. To determine whether constitutively higher expression of these two genes is committing rcd1 for PCD, respective rcd1 double mutants were constructed. Furthermore, plants overexpressing AOX1a (AOX1a OE) or a constitutively active AOX1a (AOX1a OE-CA) were included in the experiments [105]. In contrast to the situation in tobacco where overexpression of AOX1 leads to O3 sensitivity [106], there was no increased cell death in any of the AOX1a OE or AOX1a OE-CA lines, nor in the single aox1a, and no changes in the O3 damage in rcd1-1 aox1a plants in comparison to rcd1 (Figure 6). Increased AOX activity has been proposed to lead to stress tolerance by reducing mitochondrial ROS production and/or maintenance of mitochondrial function during stress [104], [107]. However, the lack of O3 phenotypes in various transgenic lines with altered AOX1a levels suggest that the role of mitochondrial ROS production during cell death is more complicated than anticipated.
Fig. 6. Response to apoplastic ROS in rcd1 is not influenced by AOX1 or UPOX. 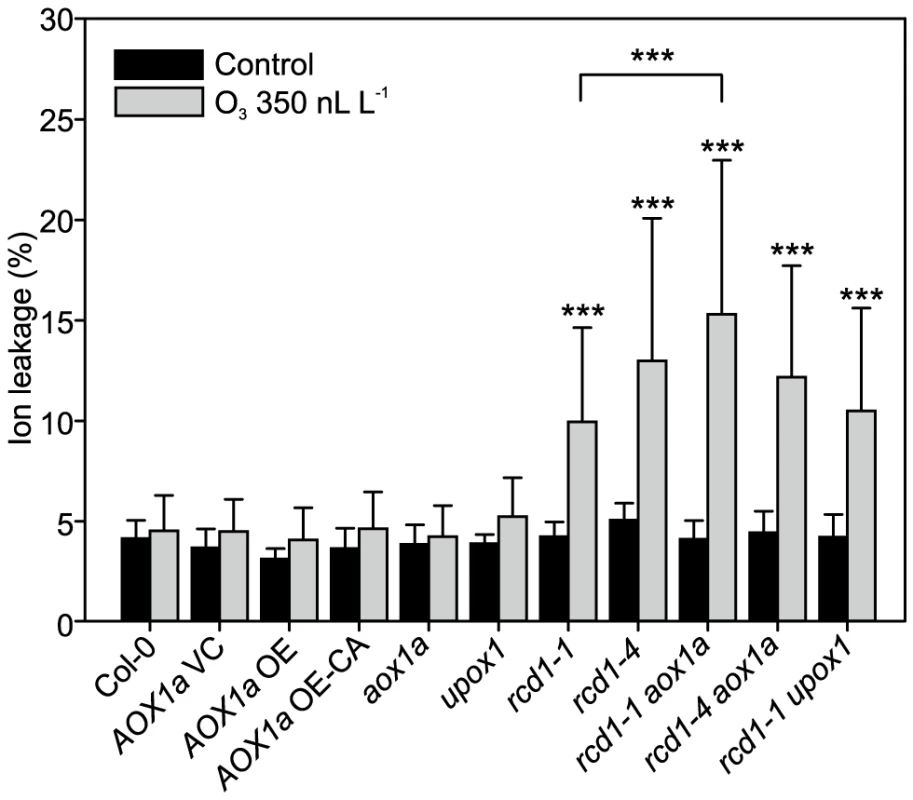
Genotypes studied were Col-0, AOX1a OE (overexpression), AOX1a OE-CA (overexpression of constitutively active AOX1a), the corresponding vector control (VC) and the mutants aox1a, upox1, rcd1-1, rcd1-4, rcd1-1 upox1, rcd1-1 aox1a and rcd1-4 aox1a. Plants were exposed to 6 h of O3 (350 nL L−1) and after 2 hour recovery in the clean air they were harvested for ion leakage measurements. Samples of untreated plants grown in clean air were simultaneously collected. Clean air and O3 samples were compared to wild type Col-0 with linear models (P<0.05:*; P<0.01:** and P<0.001:***). Double mutants (rcd1-1 aox1a, rcd1-4 aox1a and rcd1-1 upox1) were also compared to rcd1-1 and rcd1-4. Bars represent means of four to six biological repeats with standard deviation. O3-induced cell death in Col-0 and rcd1 was also independent of the mitochondrial protein UPOX1 [108] (Figure 6), which is universally induced by oxidative stress [15]. Recently, cytosolic localization of UPOX1 was also demonstrated [47]. It should be noted that the T-DNA insertion in UPOX1 is located at the end of the coding sequence and would only remove the last five amino acids of the protein, and although there was an altered UPOX1 transcript size in upox1 and rcd1 upox1 (data not shown), these plants may still have a functional protein. Apparently, neither AOX1a nor UPOX1 modulate apoplastic ROS cell death of rcd1.
Regulation of AOX1a expression has been extensively studied to reveal components of stress and/or mitochondrial retrograde signaling [109], [110]. A mutant screen for regulators of AOX1a expression identified rao1 (regulator of alternative oxidase1), encoding CYCLIN-DEPENDENT KINASE E1 (CDKE1), as a regulator of stress and growth responses [111]. Since both CDKE1 and RCD1 are regulators of AOX1a expression this prompted us to compare the expression profiles of both mutants. Of 423 genes misregulated in rcd1 (Figure 2), 123 were also misregulated in rao1 [111]. Subsequent clustering of these genes using raw data from [111] and the rcd1/O3 data did not reveal any striking similarities between the two mutants (data not shown). However, the opposite AOX1a expression phenotypes of the two mutants, i.e., higher expression in rcd1 and lower in rao1, suggest that rcd1 may be a valuable tool to further dissect mitochondrial retrograde signaling.
rcd1 growth defects are not suppressed by defense signaling or cell death regulators
The rcd1 mutant displays an altered growth phenotype indicative of constitutive stress-induced morphogenic response (SIMR), a growth response including inhibition of shoot elongation and stimulation of auxiliary branching [37], [38]. SIMR is thought to be an adaptive response to stress and is regulated by a complex interplay between ROS, auxin, ethylene and antioxidants [14], [37]. The effect of ROS may at least partially be mediated by direct oxidation of indole-3-acetic acid to form inactive 2-oxindole-3-acid acid [112]. Since rcd1 displays constitutive SIMR, the rcd1 double mutant collection allows the dissection of signaling pathways contributing to SIMR. Most double mutants exhibited no alterations in rcd1 growth habitus, thus excluding a role for ethylene, SA, JA as well as several other defense regulators in the regulation of SIMR (Table 1). RCD1 has also been shown to regulate growth suppression down-stream from defense activation, a response dependent on ROS production and proper redox balance [41]. The few rcd1 double mutants with altered growth phenotypes include rcd1 axr1, which displayed an additive growth inhibition [14] thus highlighting a role for auxin in regulation of SIMR. Furthermore, both ascorbic acid biosynthesis mutants, vtc1 and vtc2, enhanced growth suppression of rcd1 (Figure 7). In conclusion, the SIMR response was governed by a set of regulators distinct from classical defense signaling and the rcd1 mutant represents a useful tool in dissecting SIMR.
Fig. 7. Constitutive SIMR in the rcd1 mutant is enhanced by ascorbate deficiency. 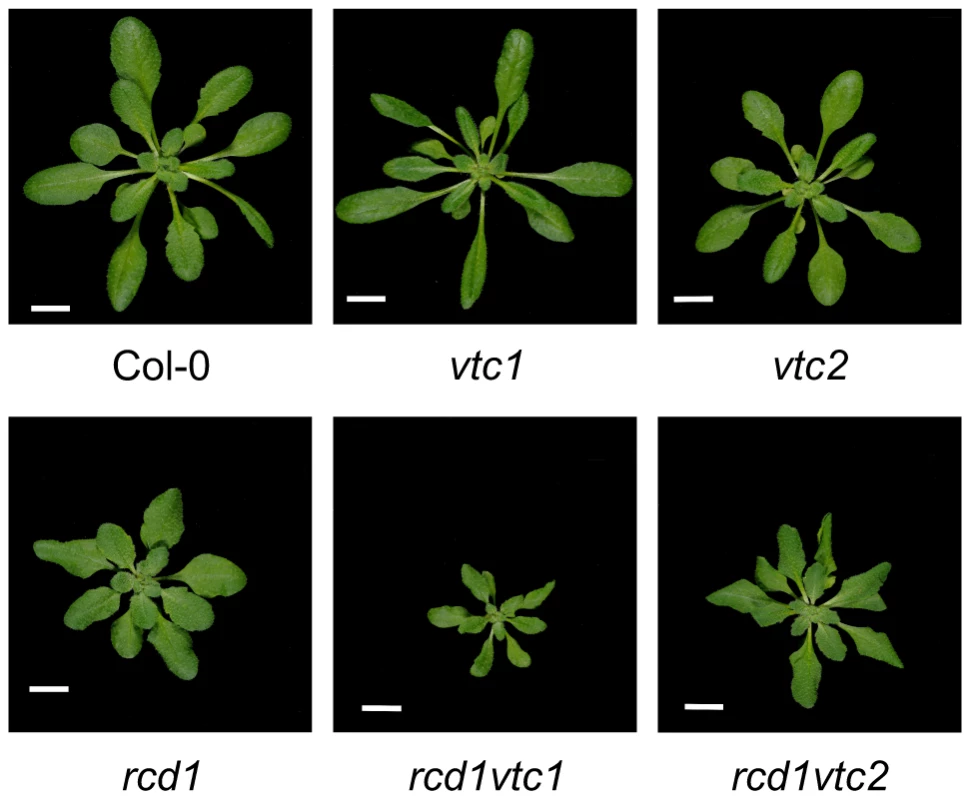
Plants were grown for four weeks and photos taken. Scale bar = 1 cm. Tab. 1. Enhanced SIMR in rcd1 double mutants. 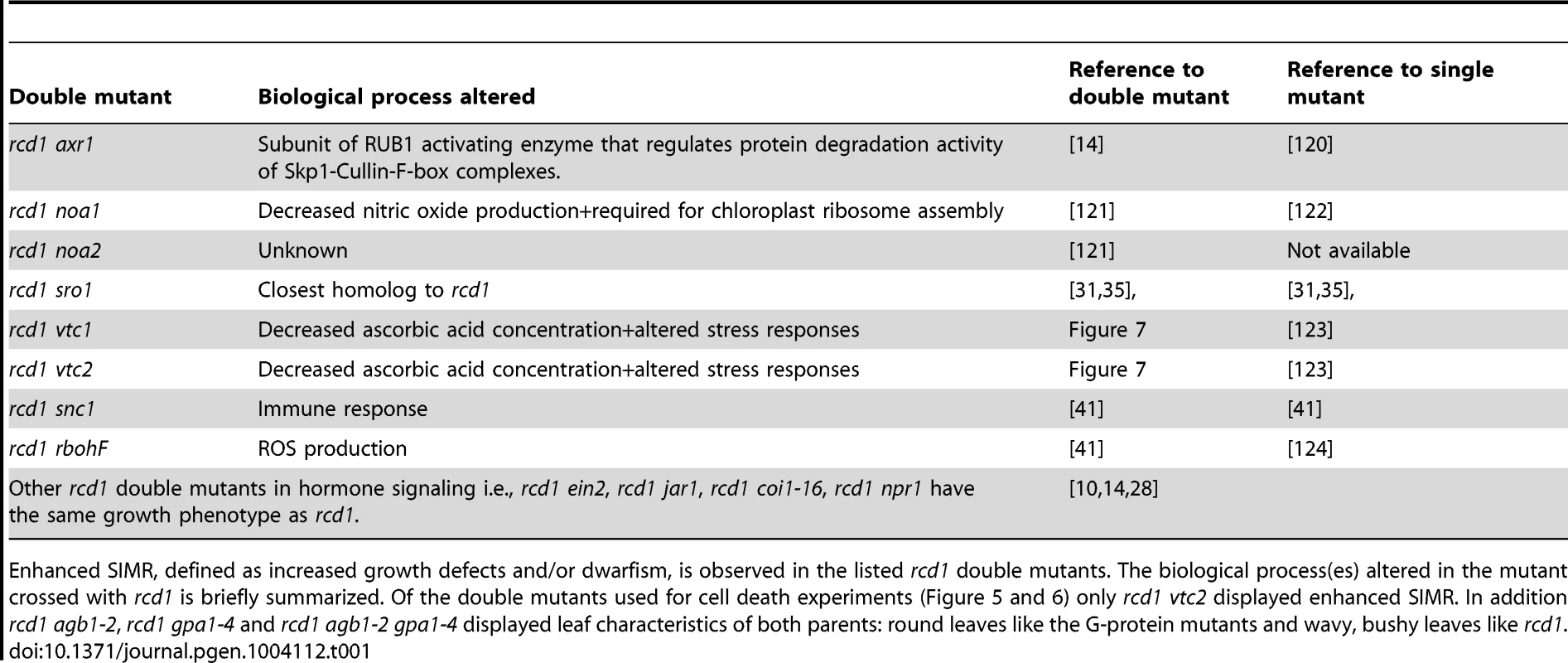
Enhanced SIMR, defined as increased growth defects and/or dwarfism, is observed in the listed rcd1 double mutants. The biological process(es) altered in the mutant crossed with rcd1 is briefly summarized. Of the double mutants used for cell death experiments (Figure 5 and 6) only rcd1 vtc2 displayed enhanced SIMR. In addition rcd1 agb1-2, rcd1 gpa1-4 and rcd1 agb1-2 gpa1-4 displayed leaf characteristics of both parents: round leaves like the G-protein mutants and wavy, bushy leaves like rcd1. Conclusions
Numerous regulators of cell death have been identified through work on plant PCD and lesion mimic mutants. O3-elicited cell death in rcd1 requires a partially distinct set of regulators, indicating further complexity in plant cell death regulation. At the gene expression level quantitatively higher expression of stress related genes was more important than qualitative difference in individual genes. The identification of wrky70 and sgt1b as partial suppressors of rcd1 cell death indicate that there are more cell death regulators to be identified by studying WRKY70 target genes and SGT1B –SCF E3-ligase target proteins. The convergence of stress, growth response and mitochondrial retrograde signaling in RCD1, its nuclear localization and interaction with various TFs, indicate a role for RCD1 in transcriptional regulation or possibly chromatin regulation or other epigenetic changes.
Materials and Methods
Plant material and O3 treatment
The growth conditions used and collection of plant material for microarray experiments are described in [14]. In brief, Arabidopsis thaliana ecotype Col-0 and rcd1-1 were grown in controlled environment chambers (Weiss Bio1300; Weiss Gallenkamp, (http://www.weiss-gallenkamp.com/) with 12-h/12-h (day/night) cycle, temperature 22/19°C, relative humidity 70/90%. O3 experiments (6 hours of 350 nL L−1) were performed with three-week-old plants, which were collected at 0, 1, 2, 4, 8 and 24 h after the start of the O3 treatment. The experiment was repeated three times, in addition to which a fourth identical repeat was used as the common reference RNA. Lesion mimic mutants acd2-2 and lsd1-3 [aka chs4-1 [113]] and T-DNA knockouts were obtained from NASC (http://arabidopsis.info/) and acd5 was a gift from Dr Jean Greenberg. Lesion mimics were grown in growth rooms with the same conditions as above for 22 days (no lesions were visible at this point), and then moved to long day greenhouse to induce lesions (18/6 h day/night). Samples were harvested 72 hours later. Col-0 and lesion mimic genotypes with no visible lesions were marked with 0, individuals with lesion leaves were marked with + and leaves without lesions from the same plant as lesion leaves were marked with -. In this experimental design leaves undergoing cell death (+ leaves) are separated from systemic leaves ( − leaves) which might receive a signal from dying leaves; and leaves/plants which have not yet started the cell death program (0 leaves).
Plants were harvested for cell death quantification with ion leakage after 6 h of 350 (or 400 nL L−1) of O3 and 2 h in clean air into 15 ml MilliQ-water. Ion leakage caused by O3 was measured with a conductivity meter 2 h after harvesting. For total ion content measurements, samples were frozen and thawed to release the content of cells. The rcd1 double mutants were constructed with rcd1-1 or rcd1-4 as pollen acceptor and various other defense related mutants as pollen donors, see Table S2 for full details. Mutants were obtained from NASC (http://arabidopsis.info/) or were gifts from Dr Günter Brader (wrky70), Dr Hans Thordal-Christensen (ald1 and fmo1), Dr Patricia Conklin (vtc1 and vtc2), Dr Heribert Hirt (mpk6), Dr Tesfay Mengiste (bos1), Dr. Jeff Dangl (ndr1, mc1, rar1-21, and edm1), Dr Alan Jones (gpa1, agb1) and Dr. Ikuko Hara-Nishimura (vpe-gamma). Double mutants were initially screened for the visible phenotype of rcd1 (curly leaves and compact rosette), subsequently the mutations were verified with PCR based markers (Table S2). Other rcd1 double mutants have been described previously [10], [14]. GUS staining and RCD1 and SRO1 promoter uidA-lines were described previously [31].
Arabidopsis overexpressing AOX1a or a constitutively active AOX1a and vector controls are described in [105] and [114]. These lines are available from NASC with the stock codes N6589–N6598. Initially all lines were screened for O3 sensitivity, subsequently lines N6589, N6591 and N6595 were characterized in more detail.
Microarray hybridizations and data analysis
RNA extraction, microarray hybridizations, data preprocessing and analysis with scripts in R are reported in [14]. Genes with at least 2-fold change in expression with statistical significance q<0.05 were considered as differentially expressed in each comparison made between treatments, genotypes and time points. Overlap between multiple gene lists was studied with Venn diagrams [115]. Gene expression data is deposited into ArrayExpress, accession number: E-MTAB-662.
Analysis of rcd1 gene expression in comparison with publicly available gene expression data
The raw .cel files were downloaded from public databases, normalized with Robust Multi-array Average (RMA) normalization, and manually annotated to control and treatment conditions. For each experiment the log2-base fold changes of treatment versus control were computed. The preprocessed data was clustered using bootstrapped Bayesian hierarchical clustering as described in [116]. Publicly available experiments using the Affymetrix ATH1-121501 platform were obtained from several data sources: NASC Arrays http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl (ABA - NASCARRAYS-176; CHX - NASCARRAYS-189; MG132 - NASCARRAYS-190; SA experiment 1 - NASCARRAYS-192; BTH experiment 1 - NASCARRAYS-392; ZAT12 OEX experiment 1 - NASCARRAYS-353; Senescence experiment 1 - NASCARRAYS-52; Senescence experiment 2 - NASCARRAYS-150). ArrayExpress http://www.ebi.ac.uk/microarrayas/ae/ (MeJA - EATMX-13; PQ - E-ATMX-28; Syringolin A - E-MEXP-739). Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo/ (H2O2 and ZAT12 OEX experiment 2 - GSE5530; Salt - GSE5623; Heat - GSE19603; High light - GSE7743; Flg22 - GSE5615; sid2 - GSE9955; lht1 - GSE19109; edr1 - GSE26679; Pseudomonas syringae ES4326 - GSE18978; sni1 - GSE6827; csn3, csn4 and csn5 - GSE9728; cs26 long day - GSE19241; Norflurazon - GSE12887; SA experiment 2 - GSE14961; siz1 - GSE6583; BTH experiment 2 and mkk1mkk2 - GSE10646; Ethylene and amiR-ETP1/2 (constitutive ethylene response) - GSE14247; npr1 - GSE13833; ERF104 OEX - GSE11807; Botrytis cinerea infection - GSE5684). The Integrated Microarray Database System http://ausubellab.mgh.harvard.edu/imds (Experiment names: NPR1 direct targets full genome, FBP1-antisense transgenic and Local and systemic responses to Trichoplusia ni feeding). Raw data for wrky33, acd11, laz1 and laz2 [26], [117] were obtained from Dr John Mundy. Raw data for atx1 [118] was obtained from Dr Zoya Avramova.
Gene ontology and promoter element enrichment analysis
Gene ontology enrichment analysis was carried out with GO information downloaded from TAIR (ftp://ftp.arabidopsis.org/) on 30th November 2010. Enrichment was computed with scripts in R implementing Fisher exact test for the set of genes in each cluster in comparison to the clustering gene list (423 genes). Promoter analysis of the genes was carried out with 500 base pair upstream promoter sequences from TAIR 10, available from ftp://ftp.arabidopsis.org/. Matching of 196 known binding motifs was carried out with scripts in R for both plus and minus DNA strands of the promoter areas as described by [14]. Cluster-specific enrichment of motifs was determined with Fisher exact test.
qPCR gene expression analysis
Gene expression analysis of selected marker genes was performed with qRT-PCR (Table S2 has primer sequences and primer amplification efficiencies). RNA was isolated and treated with DNaseI as in [31]. Reverse transcription was performed with 5 µg of RNA with RevertAid Premium RT and Ribolock RNase inhibitor (Thermo Scientific Fermentas) and the reaction diluted to the final volume of 100 µl. PCR was performed in triplicate using iQ SYBR GREEN supermix (Bio-Rad). The cycle conditions with Bio-Rad CFX384 were: 1 cycle initiating with 95°C 10 min, 39 cycles with 95°C 15 s, 60°C 30 s, 72°C 30 s and ending with melting curve analysis. Normalization of the data was performed in qBase 2.0 (Biogazelle), with three reference genes TIP41, At5g08290 and PP2AA3 selected from [119] and validated with geNorm to have stable expression in the samples used in this study. Primer amplification efficiencies were determined in qBase from a cDNA dilution series. Statistical significances in qPCR data were evaluated with scripts in R. In statistical analysis, a 2-base logarithm was first taken from the data to improve the model fit. Then a linear mixed model was fitted in using R package nlme, having fixed effects for Genotype, Treatment and Time and their interactions, plus a random effect for the biological repeat. The model contrasts were then computed with multcomp package, and the subsequent p-values were adjusted for multiple comparisons by Benjamini-Hochberg correction.
Analysis of ion leakage data
Statistical analysis of ion leakage data was carried out with scripts in R. A linear mixed model with fixed effects for Genotype, Treatment and their interaction was fitted to the data, plus a random effect for biological repeat. The model contrasts were estimated with multcomp package, and the estimated p-values were subjected to single-step p-value correction.
Supporting Information
Zdroje
1. HeidelAJ, ClarkeJD, AntonovicsJ, DongX (2004) Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168 : 2197–2206.
2. HeilM, HilpertA, KaiserW, LinsenmairKE (2000) Reduced growth and seed set following chemical induction of pathogen defence: does systemic acquired resistance (SAR) incur allocation costs? J Ecol 88 : 645–654.
3. van HultenM, PelserM, van LoonLC, PieterseCMJ, TonJ (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A 103 : 5602–5607.
4. JaspersP, KangasjärviJ (2010) Reactive oxygen species in abiotic stress signaling. Physiol Plant 138 : 405–413.
5. ShapiguzovA, VainonenJP, WrzaczekM, KangasjärviJ (2012) ROS-talk - how the apoplast, the chloroplast and the nucleus get the message through. Front Plant Sci 3 : 292.
6. SuzukiN, MillerG, MoralesJ, ShulaevV, TorresMA, et al. (2011) Respiratory burst oxidases: the engines of ROS signaling. Curr Opin Plant Biol 14 : 691–699.
7. SierlaM, RahikainenM, SalojärviJ, KangasjärviJ, KangasjärviS (2013) Apoplastic and chloroplastic redox signaling networks in plant stress responses. Antioxid Redox Signal 18 : 2220–2239.
8. CollNS, EppleP, DanglJL (2011) Programmed cell death in the plant immune system. Cell Death Differ 18 : 1247–1256.
9. De PintoMC, LocatoV, De GaraL (2012) Redox regulation in plant programmed cell death. Plant Cell Environ 35 : 234–244.
10. OvermyerK, BroschéM, PellinenR, KuittinenT, TuominenH, et al. (2005) Ozone-induced programmed cell death in the Arabidopsis radical-induced cell death1 mutant. Plant Physiol 137 : 1092–1104.
11. van DoornWG, BeersEP, DanglJL, Franklin-TongVE, GalloisP, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18 : 1241–1246.
12. HofiusD, MunchD, BressendorffS, MundyJ, PetersenM (2011) Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ 18 : 1257–1262.
13. WohlgemuthH, MittelstrassK, KschieschanS, BenderJ, WeigelHJ, et al. (2002) Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ 25 : 717–726.
14. BlomsterT, SalojärviJ, SipariN, BroschéM, AhlforsR, et al. (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol 157 : 1866–1883.
15. GadjevI, VanderauweraS, GechevTS, LaloiC, MinkovIN, et al. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141 : 436–445.
16. GechevTS, FerwerdaMA, MehterovN, LaloiC, QureshiMK, et al. (2008) Arabidopsis AAL-toxin-resistant mutant atr1 shows enhanced tolerance to programmed cell death induced by reactive oxygen species. Biochem Biophys Res Commun 375 : 639–644.
17. MittlerR, VanderauweraS, SuzukiN, MillerG, TognettiVB, et al. (2011) ROS signaling: the new wave? Trends Plant Sci 16 : 300–309.
18. ToufighiK, BradySM, AustinR, LyE, ProvartNJ (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43 : 153–163.
19. WituszyńskaW, ŚlesakI, VanderauweraS, Szechyńska-HebdaM, KornaśA, et al. (2013) LESION SIMULATING DISEASE1, ENHANCED DISEASE SUSCEPTIBILITY1, and PHYTOALEXIN DEFICIENT4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol 161 : 1795–1805.
20. LorrainS, VailleauF, BalaguéC, RobyD (2003) Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci 8 : 263–271.
21. VailleauF, DanielX, TronchetM, MontilletJL, TriantaphylidèsC, et al. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci U S A 99 : 10179–10184.
22. KaminakaH, NäkeC, EppleP, DittgenJ, SchützeK, et al. (2006) bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J 25 : 4400–4411.
23. NieH, ZhaoC, WuG, WuY, ChenY, et al. (2012) SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol 158 : 1847–1859.
24. ZhangY, YangY, FangB, GannonP, DingP, et al. (2010) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22 : 3153–3163.
25. MonaghanJ, GermainH, WeihmannT, LiX (2010) Dissecting plant defence signal transduction: modifiers of snc1 in Arabidopsis. Can J Plant Pathol 32 : 35–42.
26. PalmaK, ThorgrimsenS, MalinovskyFG, FiilBK, NielsenHB, et al. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6: e1001137.
27. HruzT, LauleO, SzaboG, WessendorpF, BleulerS, et al. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008 : 420747.
28. OvermyerK, TuominenH, KettunenR, BetzC, LangebartelsC, et al. (2000) Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12 : 1849–1862.
29. FujibeT, SajiH, ArakawaK, YabeN, TakeuchiY, et al. (2004) A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol 134 : 275–285.
30. AhlforsR, LångS, OvermyerK, JaspersP, BroschéM, et al. (2004) Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction-domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16 : 1925–1937.
31. JaspersP, BlomsterT, BroschéM, SalojärviJ, AhlforsR, et al. (2009) Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 60 : 268–279.
32. JaspersP, OvermyerK, WrzaczekM, VainonenJP, BlomsterT, et al. (2010) The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics 11 : 170.
33. BorsaniO, ZhuJ, VersluesPE, SunkarR, ZhuJK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 : 1279–1291.
34. AravindL (2001) The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci 26 : 273–275.
35. TeotiaS, LambRS (2009) The paralogous genes RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 have partially redundant functions during Arabidopsis development. Plant Physiol 151 : 180–198.
36. TeotiaS, LambRS (2011) RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J Exp Bot 62 : 1271–1284.
37. PottersG, PasternakTP, GuisezY, JansenMAK (2009) Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32 : 158–169.
38. TeotiaS, MuthuswamyS, LambRS (2010) RADICAL-INDUCED CELL DEATH1 and SIMILAR TO RCD ONE1 and the stress-induced morphogenetic response. Plant Signal Behav 5 : 143–145.
39. PottersG, PasternakTP, GuisezY, PalmeKJ, JansenMAK (2007) Stress-induced morphogenic responses: growing out of trouble? Trends Plant Sci 12 : 98–105.
40. TognettiVB, MühlenbockP, Van BreusegemF (2012) Stress homeostasis - the redox and auxin perspective. Plant Cell Environ 35 : 321–333.
41. ZhuY, DuB, QianJ, ZouB, HuaJ (2013) Disease resistance gene-induced growth inhibition is enhanced by rcd1 independent of defense activation in Arabidopsis. Plant Physiol 161 : 2005–2013.
42. LudwikowA, SadowskiJ (2008) Gene networks in plant ozone stress response and tolerance. J Integr Plant Biol 50 : 1256–1267.
43. LiP, ManeSP, SiosonAA, Vasquez RobinetC, HeathLS, et al. (2006) Effects of chronic ozone exposure on gene expression in Arabidopsis thaliana ecotypes and in Thellungiella halophila. Plant Cell Environ 29 : 854–868.
44. BechtoldU, RichardO, ZamboniA, GapperC, GeislerM, et al. (2008) Impact of chloroplastic - and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J Exp Bot 59 : 121–133.
45. VainonenJP, JaspersP, WrzaczekM, LamminmäkiA, ReddyRA, et al. (2012) RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem J 442 : 573–581.
46. AokiK, OgataY, ShibataD (2007) Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol 48 : 381–390.
47. InzéA, VanderauweraS, HoeberichtsFA, VandorpeM, Van GaeverT, et al. (2012) A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ 35 : 308–320.
48. KimSY, KimSG, KimYS, SeoPJ, BaeM, et al. (2007) Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res 35 : 203–213.
49. WangL, MitraRM, HasselmannKD, SatoM, Lenarz-WyattL, et al. (2008) The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol Plant Microbe Interact 21 : 1408–1420.
50. BilginDD, ZavalaJA, ZhuJ, CloughSJ, OrtDR, et al. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33 : 1597–1613.
51. DohmannEMN, LevesqueMP, De VeylderL, ReichardtI, JürgensG, et al. (2008) The Arabidopsis COP9 signalosome is essential for G2 phase progression and genomic stability. Development 135 : 2013–2022.
52. KolodziejekI, Misas-VillamilJC, KaschaniF, ClercJ, GuC, et al. (2011) Proteasome activity imaging and profiling characterizes bacterial effector syringolin A. Plant Physiol 155 : 477–489.
53. RentelMC, LecourieuxD, OuakedF, UsherSL, PetersenL, et al. (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 : 858–861.
54. SakumaY, MaruyamaK, QinF, OsakabeY, ShinozakiK, et al. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci U S A 103 : 18822–18827.
55. LarkindaleJ, VierlingE (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146 : 748–761.
56. IuchiS, KobayashiM, TajiT, NaramotoM, SekiM, et al. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27 : 325–333.
57. TanBC, JosephLM, DengWT, LiuL, LiQB, et al. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35 : 44–56.
58. ZhangZ, LenkA, AnderssonMX, GjettingT, PedersenC, et al. (2008) A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol Plant 1 : 510–527.
59. DohertyCJ, Van BuskirkHA, MyersSJ, ThomashowMF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21 : 972–984.
60. WiermerM, FeysBJ, ParkerJE (2005) Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8 : 383–389.
61. WangD, AmornsiripanitchN, DongX (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2: e123.
62. LuH, RateDN, SongJT, GreenbergJT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15 : 2408–2420.
63. MosherRA, DurrantWE, WangD, SongJ, DongX (2006) A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 : 1750–1765.
64. LeeJ, NamJ, ParkHC, NaG, MiuraK, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49 : 79–90.
65. BermúdezMA, Páez-OchoaMA, GotorC, RomeroLC (2010) Arabidopsis S-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22 : 403–416.
66. QiuJL, ZhouL, YunBW, NielsenHB, FiilBK, et al. (2008) Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol 148 : 212–222.
67. LiuG, JiY, BhuiyanNH, PilotG, SelvarajG, et al. (2010) Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22 : 3845–3863.
68. BindschedlerLV, DewdneyJ, BleeKA, StoneJM, AsaiT, et al. (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47 : 851–863.
69. SatoM, TsudaK, WangL, CollerJ, WatanabeY, et al. (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog 6: e1001011.
70. TaoY, XieZ, ChenW, GlazebrookJ, ChangHS, et al. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15 : 317–330.
71. RaoMV, LeeHI, DavisKR (2002) Ozone-induced ethylene production is dependent on salicylic acid, and both salicylic acid and ethylene act in concert to regulate ozone-induced cell death. Plant J 32 : 447–456.
72. PieterseCMJ, Leon-ReyesA, Van der EntS, Van WeesSCM (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5 : 308–316.
73. OvermyerK, BroschéM, KangasjärviJ (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8 : 335–342.
74. QiaoH, ChangKN, YazakiJ, EckerJR (2009) Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev 23 : 512–521.
75. TuominenH, OvermyerK, KeinänenM, KollistH, KangasjärviJ (2004) Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J 39 : 59–69.
76. GlazebrookJ, ChenW, EstesB, ChangHS, NawrathC, et al. (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34 : 217–228.
77. YaoN, GreenbergJT (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18 : 397–411.
78. LiangH, YaoN, SongJT, LuoS, LuH, et al. (2003) Ceramides modulate programmed cell death in plants. Genes Dev 17 : 2636–2641.
79. GreenbergJT, SilvermanFP, LiangH (2000) Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156 : 341–350.
80. CollNS, VercammenD, SmidlerA, CloverC, Van BreusegemF, et al. (2010) Arabidopsis type I metacaspases control cell death. Science 330 : 1393–1397.
81. HeS, TanG, LiuQ, HuangK, RenJ, et al. (2011) The LSD1-interacting protein GILP is a LITAF domain protein that negatively regulates hypersensitive cell death in Arabidopsis. PLoS One 6: e18750.
82. IshikawaK, YoshimuraK, HaradaK, FukusakiE, OgawaT, et al. (2010) AtNUDX6, an ADP-ribose/NADH pyrophosphohydrolase in Arabidopsis, positively regulates NPR1-dependent salicylic acid signaling. Plant Physiol 152 : 2000–2012.
83. GeX, LiGJ, WangSB, ZhuH, ZhuT, et al. (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145 : 204–215.
84. JambunathanN, PenagantiA, TangY, MahalingamR (2010) Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol 10 : 173.
85. StrausMR, RietzS, Ver Loren van ThemaatE, BartschM, ParkerJE (2010) Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J 62 : 628–640.
86. TadaY, SpoelSH, Pajerowska-MukhtarK, MouZ, SongJ, et al. (2008) Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321 : 952–956.
87. AhlforsR, MacioszekV, RuddJ, BroschéM, SchlichtingR, et al. (2004) Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J 40 : 512–522.
88. MilesGP, SamuelMA, ZhangY, EllisBE (2005) RNA interference-based (RNAi) suppression of AtMPK6, an Arabidopsis mitogen-activated protein kinase, results in hypersensitivity to ozone and misregulation of AtMPK3. Environ Pollut 138 : 230–237.
89. LiJ, BraderG, PalvaET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16 : 319–331.
90. SheardLB, TanX, MaoH, WithersJ, Ben-NissanG, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468 : 400–405.
91. ThinesB, KatsirL, MelottoM, NiuY, MandaokarA, et al. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448 : 661–665.
92. KimKC, LaiZ, FanB, ChenZ (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20 : 2357–2371.
93. LongTA (2011) Many needles in a haystack: cell-type specific abiotic stress responses. Curr Opin Plant Biol 14 : 325–331.
94. Taylor-TeeplesM, RonM, BradySM (2011) Novel biological insights revealed from cell type-specific expression profiling. Curr Opin Plant Biol 14 : 601–607.
95. AndreassonE, EllisB (2010) Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci 15 : 106–113.
96. LeónP, GregorioJ, CordobaE (2013) ABI4 and its role in chloroplast retrograde communication. Front Plant Sci 3 : 304.
97. ZhouF, MosherS, TianM, SassiG, ParkerJ, et al. (2008) The Arabidopsis gain-of-function mutant ssi4 requires RAR1 and SGT1b differentially for defense activation and morphological alterations. Mol Plant Microbe Interact 21 : 40–49.
98. RustérucciC, AvivDH, HoltBFIII, DanglJL, ParkerJE (2001) The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13 : 2211–2224.
99. ShapiroAD, ZhangC (2001) The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol 127 : 1089–1101.
100. ConklinPL, SaraccoSA, NorrisSR, LastRL (2000) Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154 : 847–856.
101. Hara-NishimuraI, HatsugaiN, NakauneS, KuroyanagiM, NishimuraM (2005) Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol 8 : 404–408.
102. UranoD, ChenJG, BotellaJR, JonesAM (2013) Heterotrimeric G protein signalling in the plant kingdom. Open Biol 3 : 120186.
103. SmalleJ, VierstraRD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55 : 555–590.
104. Van AkenO, GiraudE, CliftonR, WhelanJ (2009) Alternative oxidase: a target and regulator of stress responses. Physiol Plant 137 : 354–361.
105. FioraniF, UmbachAL, SiedowJN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature. A study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139 : 1795–1805.
106. PasqualiniS, PaolocciF, BorgogniA, MorettiniR, EderliL (2007) The overexpression of an alternative oxidase gene triggers ozone sensitivity in tobacco plants. Plant Cell Environ 30 : 1545–1556.
107. GiraudE, HoLHM, CliftonR, CarrollA, EstavilloG, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147 : 595–610.
108. SweetloveLJ, HeazlewoodJL, HeraldV, HoltzapffelR, DayDA, et al. (2002) The impact of oxidative stress on Arabidopsis mitochondria. Plant J 32 : 891–904.
109. GiraudE, Van AkenO, HoLHM, WhelanJ (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150 : 1286–1296.
110. Van AkenO, ZhangB, LawS, NarsaiR, WhelanJ (2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162 : 254–271.
111. NgS, GiraudE, DuncanO, LawSR, WangY, et al. (2013) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288 : 3449–3459.
112. PeerWA, ChengY, MurphyAS (2013) Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64 : 2629–2639.
113. HuangX, LiY, ZhangX, ZuoJ, YangS (2010) The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol 187 : 301–312.
114. UmbachAL, FioraniF, SiedowJN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139 : 1806–1820.
115. Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
116. WrzaczekM, BroschéM, SalojärviJ, KangasjärviS, IdänheimoN, et al. (2010) Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol 10 : 95.
117. MalinovskyFG, BrodersenP, FiilBK, McKinneyLV, ThorgrimsenS, et al. (2010) Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS One 5: e12586.
118. Alvarez-VenegasR, SadderM, HlavackaA, BaluskaF, XiaY, et al. (2006) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci U S A 103 : 6049–6054.
119. CzechowskiT, StittM, AltmannT, UdvardiMK, ScheibleWR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139 : 5–17.
120. DharmasiriN, DharmasiriS, WeijersD, KarunarathnaN, JurgensG, et al. (2007) AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52 : 114–123.
121. AhlforsR, BroschéM, KollistH, KangasjärviJ (2009) Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J 58 : 1–12.
122. Flores-PérezÚ, Sauret-GüetoS, GasE, JarvisP, Rodríguez-ConcepciónM (2008) A mutant impaired in the production of plastome-encoded proteins uncovers a mechanism for the homeostasis of isoprenoid biosynthetic enzymes in Arabidopsis plastids. Plant Cell 20 : 1303–1315.
123. PavetV, OlmosE, KiddleG, MowlaS, KumarS, et al. (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139 : 1291–1303.
124. TorresMA, DanglJL, JonesJDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A 99 : 517–522.
Štítky
Genetika Reprodukční medicína
Článek Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the RatČlánek MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease PathogenesisČlánek Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene FunctionČlánek Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Fifteen Years Later: Hard and Soft Selection Sweeps Confirm a Large Population Number for HIV In Vivo
- The Same but Different: Worms Reveal the Pervasiveness of Developmental System Drift
- Serine Carboxypeptidase SCPEP1 and Cathepsin A Play Complementary Roles in Regulation of Vasoconstriction via Inactivation of Endothelin-1
- Coherent Functional Modules Improve Transcription Factor Target Identification, Cooperativity Prediction, and Disease Association
- A Long-Chain Flavodoxin Protects from Oxidative Stress and Host Bacterial Clearance
- Mammalian E-type Cyclins Control Chromosome Pairing, Telomere Stability and CDK2 Localization in Male Meiosis
- Influenza Virus Drug Resistance: A Time-Sampled Population Genetics Perspective
- Transcriptome-Wide Analyses of 5′-Ends in RNase J Mutants of a Gram-Positive Pathogen Reveal a Role in RNA Maturation, Regulation and Degradation
- Selective Disruption of Aurora C Kinase Reveals Distinct Functions from Aurora B Kinase during Meiosis in Mouse Oocytes
- X Chromosome Control of Meiotic Chromosome Synapsis in Mouse Inter-Subspecific Hybrids
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Extreme Population Differences in the Human Zinc Transporter ZIP4 (SLC39A4) Are Explained by Positive Selection in Sub-Saharan Africa
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Genomic Networks of Hybrid Sterility
- Natural Polymorphisms in Influence Negative Selection and CD4∶CD8 Lineage Commitment in the Rat
- Oxidative Stress Is Not a Major Contributor to Somatic Mitochondrial DNA Mutations
- Molecular Identification of Collagen 17a1 as a Major Genetic Modifier of Laminin Gamma 2 Mutation-Induced Junctional Epidermolysis Bullosa in Mice
- Uncoupling of Molecular Maturation from Peripheral Target Innervation in Nociceptors Expressing a Chimeric TrkA/TrkC Receptor
- MicroRNAs Located in the Hox Gene Clusters Are Implicated in Huntington's Disease Pathogenesis
- Loss of Trabid, a New Negative Regulator of the Immune-Deficiency Pathway at the Level of TAK1, Reduces Life Span
- Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response
- Identification of Novel Genetic Loci Associated with Thyroid Peroxidase Antibodies and Clinical Thyroid Disease
- CEP-1, the p53 Homolog, Mediates Opposing Longevity Outcomes in Mitochondrial Electron Transport Chain Mutants
- Transcriptomics and Functional Genomics of ROS-Induced Cell Death Regulation by
- Quantitative Genome-Wide Genetic Interaction Screens Reveal Global Epistatic Relationships of Protein Complexes in
- Cascades of Genetic Instability Resulting from Compromised Break-Induced Replication
- Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging
- Zfp322a Regulates Mouse ES Cell Pluripotency and Enhances Reprogramming Efficiency
- Insertional Mutagenesis and Deep Profiling Reveals Gene Hierarchies and a -Dependent Bottleneck in Lymphomagenesis
- DAAM Is Required for Thin Filament Formation and Sarcomerogenesis during Muscle Development in Drosophila
- Plasma Cholesterol–Induced Lesion Networks Activated before Regression of Early, Mature, and Advanced Atherosclerosis
- High-Resolution Profiling of Stationary-Phase Survival Reveals Yeast Longevity Factors and Their Genetic Interactions
- Comparative RNAi Screens in and Reveal the Impact of Developmental System Drift on Gene Function
- Accurate and Robust Genomic Prediction of Celiac Disease Using Statistical Learning
- Sex-Specific Embryonic Gene Expression in Species with Newly Evolved Sex Chromosomes
- Chromosome X-Wide Association Study Identifies Loci for Fasting Insulin and Height and Evidence for Incomplete Dosage Compensation
- Negative Feedback and Transcriptional Overshooting in a Regulatory Network for Horizontal Gene Transfer
- DNA Sequence Explains Seemingly Disordered Methylation Levels in Partially Methylated Domains of Mammalian Genomes
- Insights into the Genomic Landscape: Comparative Genomics Reveals Variations in Ploidy and Nutrient Utilisation Potential amongst Wine Isolates
- Molecular Evidence for the Inverse Comorbidity between Central Nervous System Disorders and Cancers Detected by Transcriptomic Meta-analyses
- The Centriolar Satellite Protein AZI1 Interacts with BBS4 and Regulates Ciliary Trafficking of the BBSome
- Fine-Mapping the Region Detects Common Variants Tagging a Rare Coding Allele: Evidence for Synthetic Association in Prostate Cancer
- Transmission Distortion Affecting Human Noncrossover but Not Crossover Recombination: A Hidden Source of Meiotic Drive
- A Variant in the Neuropeptide Receptor is a Major Determinant of Growth and Physiology
- Mutation of SLC35D3 Causes Metabolic Syndrome by Impairing Dopamine Signaling in Striatal D1 Neurons
- NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly
- MicroRNA-133 Inhibits Behavioral Aggregation by Controlling Dopamine Synthesis in Locusts
- Convergence of Light and ABA Signaling on the Promoter
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
- Distinct Requirements for Cranial Ectoderm and Mesenchyme-Derived Wnts in Specification and Differentiation of Osteoblast and Dermal Progenitors
- Chk2 and P53 Regulate the Transmission of Healed Chromosomes in the Male Germline
- Ddc2 Mediates Mec1 Activation through a Ddc1- or Dpb11-Independent Mechanism
- Mapping the Fitness Landscape of Gene Expression Uncovers the Cause of Antagonism and Sign Epistasis between Adaptive Mutations
- Euchromatic Transposon Insertions Trigger Production of Novel Pi- and Endo-siRNAs at the Target Sites in the Germline
- miR-100 Induces Epithelial-Mesenchymal Transition but Suppresses Tumorigenesis, Migration and Invasion
- Canine Hereditary Ataxia in Old English Sheepdogs and Gordon Setters Is Associated with a Defect in the Autophagy Gene Encoding
- Within-Host Spatiotemporal Dynamics of Plant Virus Infection at the Cellular Level
- Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function
- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- Mechanistically Distinct Mouse Models for -Associated Retinopathy
- DAF-16/FoxO Directly Regulates an Atypical AMP-Activated Protein Kinase Gamma Isoform to Mediate the Effects of Insulin/IGF-1 Signaling on Aging in
- Chromosome I Controls Chromosome II Replication in
- Integrated Genomic Characterization Reveals Novel, Therapeutically Relevant Drug Targets in FGFR and EGFR Pathways in Sporadic Intrahepatic Cholangiocarcinoma
- The Iodotyrosine Deiodinase Ortholog SUP-18 Functions through a Conserved Channel SC-Box to Regulate the Muscle Two-Pore Domain Potassium Channel SUP-9
- The Genome of Highlights a Fish Pathogen Adapted to Fluctuating Environments
- Distinct DNA Binding Sites Contribute to the TCF Transcriptional Switch in and
- The Streamlined Genome of spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study of Metabolic Traits Reveals Novel Gene-Metabolite-Disease Links
- A Cohesin-Independent Role for NIPBL at Promoters Provides Insights in CdLS
- Classic Selective Sweeps Revealed by Massive Sequencing in Cattle
- Arf4 Is Required for Mammalian Development but Dispensable for Ciliary Assembly
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



