-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
Genome-wide association studies and follow-up meta-analyses in Crohn's disease (CD) and ulcerative colitis (UC) have recently identified 163 disease-associated loci that meet genome-wide significance for these two inflammatory bowel diseases (IBD). These discoveries have already had a tremendous impact on our understanding of the genetic architecture of these diseases and have directed functional studies that have revealed some of the biological functions that are important to IBD (e.g. autophagy). Nonetheless, these loci can only explain a small proportion of disease variance (∼14% in CD and 7.5% in UC), suggesting that not only are additional loci to be found but that the known loci may contain high effect rare risk variants that have gone undetected by GWAS. To test this, we have used a targeted sequencing approach in 200 UC cases and 150 healthy controls (HC), all of French Canadian descent, to study 55 genes in regions associated with UC. We performed follow-up genotyping of 42 rare non-synonymous variants in independent case-control cohorts (totaling 14,435 UC cases and 20,204 HC). Our results confirmed significant association to rare non-synonymous coding variants in both IL23R and CARD9, previously identified from sequencing of CD loci, as well as identified a novel association in RNF186. With the exception of CARD9 (OR = 0.39), the rare non-synonymous variants identified were of moderate effect (OR = 1.49 for RNF186 and OR = 0.79 for IL23R). RNF186 encodes a protein with a RING domain having predicted E3 ubiquitin-protein ligase activity and two transmembrane domains. Importantly, the disease-coding variant is located in the ubiquitin ligase domain. Finally, our results suggest that rare variants in genes identified by genome-wide association in UC are unlikely to contribute significantly to the overall variance for the disease. Rather, these are expected to help focus functional studies of the corresponding disease loci.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003723
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003723Summary
Genome-wide association studies and follow-up meta-analyses in Crohn's disease (CD) and ulcerative colitis (UC) have recently identified 163 disease-associated loci that meet genome-wide significance for these two inflammatory bowel diseases (IBD). These discoveries have already had a tremendous impact on our understanding of the genetic architecture of these diseases and have directed functional studies that have revealed some of the biological functions that are important to IBD (e.g. autophagy). Nonetheless, these loci can only explain a small proportion of disease variance (∼14% in CD and 7.5% in UC), suggesting that not only are additional loci to be found but that the known loci may contain high effect rare risk variants that have gone undetected by GWAS. To test this, we have used a targeted sequencing approach in 200 UC cases and 150 healthy controls (HC), all of French Canadian descent, to study 55 genes in regions associated with UC. We performed follow-up genotyping of 42 rare non-synonymous variants in independent case-control cohorts (totaling 14,435 UC cases and 20,204 HC). Our results confirmed significant association to rare non-synonymous coding variants in both IL23R and CARD9, previously identified from sequencing of CD loci, as well as identified a novel association in RNF186. With the exception of CARD9 (OR = 0.39), the rare non-synonymous variants identified were of moderate effect (OR = 1.49 for RNF186 and OR = 0.79 for IL23R). RNF186 encodes a protein with a RING domain having predicted E3 ubiquitin-protein ligase activity and two transmembrane domains. Importantly, the disease-coding variant is located in the ubiquitin ligase domain. Finally, our results suggest that rare variants in genes identified by genome-wide association in UC are unlikely to contribute significantly to the overall variance for the disease. Rather, these are expected to help focus functional studies of the corresponding disease loci.
Introduction
Inflammatory bowel diseases (IBDs) are classified as chronic relapsing inflammatory diseases of the gastrointestinal tract. The two major forms of IBDs are Crohn's disease (CD, OMIM 266600) and ulcerative colitis (UC, OMIM 191390). Both genetic and environment factors play a central role in the pathogenesis of the inflammatory response of IBDs [1].
Recent genome-wide association (GWA) studies and meta-analyses in IBD have shown great success, with the identification of 163 independent IBD risk loci. While some loci were shown to be specific to either CD or UC risk, most have been shown to impact on both diseases, supporting earlier claims that these diseases share genetic risk factors [2]. These recent studies have identified important disease pathways but the common SNPs identified, with generally modest effects, explain only 14% and 7.5% of disease variance for CD and UC, respectively [3].
Due to linkage disequilibrium in the genome and limitations of GWAS chip designs to date, genome-wide scans typically identify common variants that tag regions of variable sizes containing multiple candidate genes for disease susceptibility. Although there have been a few notable exceptions, most of the common associated SNPs do not clearly identify causal variants, and further studies are needed to highlight the causal gene in many associated regions [4]–[6]. Sequencing of exons within associated regions in order to identify rare variants with strong effect on disease has been proposed as a means to help identify the causal genes and to help explain a further portion of disease variance. We have recently performed a pooled next-generation sequencing study in Crohn's disease, and identified association to novel low-frequency and rare protein altering variants in NOD2, IL23R, and CARD9, as well as IL18RAP, CUL2, C1orf106, PTPN22 and MUC19 [7]. We opted to use a similar targeted pooled next-generation sequencing approach to study UC-associated regions from our recent meta-analysis of 3 independent genome-wide scans for UC [8]. Using this approach we identified putative causal variants significantly associated to UC in three of the 22 loci examined and identified variants of interest for an additional six loci.
Results
Sequence analyses
We selected 200 ulcerative colitis cases and 150 healthy controls of French Canadian ancestry from among samples collected by the NIDDK IBD Genetics Consortium. Samples were pooled in batches of 50 cases or 50 controls and normalized in order for the DNA pool to reflect sample allele frequencies. We targeted 55 genes from 14 UC-associated regions, as well as 7 regions identified in CD showing nominal replication in our UC GWAS study and an additional candidate gene (ECM1) reported in recent literature [6], [8]–[10]. PCR amplification primers were successfully designed to capture a total of 508 amplicons for a total of 305 Kb or 70% of our original target sequences. Of these 508 PCR reactions, 472 (93%) successfully amplified in each of the 7 sample pools and we used these to construct libraries for high-throughput sequencing on an Illumina Genome Analyzer II. This sequencing yielded large amounts of high-quality data for each pool, that captured 99% of our amplified target regions (283 Kb total; 117 Kb exonic sequences) and achieved 1575× median coverage per pool (corresponding to 31.5× per sample).
We used the previously described variant calling method Syzygy, designed to accommodate pooled study designs, to identify rare and low-frequency single nucleotide variants in our pooled samples [7]. Syzygy detected 1590 high confidence variants in our target regions, including 309 coding region variants (189 missense, 114 synonymous, 2 nonsense and 4 essential splice junction variants) with 56% of these already reported in dbSNP version 132, a non-synonymous/synonymous ratio of 1.7 and a transition/transversion ratio of 2.38 (Table S1). These results are similar to those obtained from our recent re-sequencing study in CD, as well as those reported by the 1000 Genomes Project, and are indicative of a relatively high true-positive rate for our dataset. This was confirmed by genotyping the 350 discovery DNA samples for a random subset of 237 variants from the total of 1590 high quality variants (Table S2).
Follow-up genotyping and association analyses
After removal of variants that did not validate, variants observed only once in our sequencing dataset (singletons) and variants from the MHC region, 84 non-synonymous coding variants (missense, non-sense and splicing variants), were used for subsequent analyses. Following removal of common variants (frequency >5%) and variants that did not design in our genotyping assays, we carried out follow-up genotyping for 42 of these variants. Genotyping was performed in 6 independent case-control cohorts totaling 7,292 UC cases and 8,018 HC (Table S3), and additional data was obtained for 7,143 UC cases and 12,186 HC from the International IBD Genetics Consortium (IIBDGC) Immunochip project for 14 of these variants [3].
Since our study focuses on infrequent and rare variants, we expect few non-reference alleles for these variants in each subcohort studied, which precludes the use of asymptotic statistics utilized in typical association studies of common variants. Also, given the low frequencies of the variants tested, population structure is likely to be a more substantial problem and thus requires a stratified analysis with strict population case-control matching. We used a previously described mega-analysis of rare variants (MARV) approach that provides a permutation-based estimate of significance, within each sub-cohort, and accommodates variable numbers of case-control samples in each independent population for single-marker analysis [7].
With a target set of 42 variants we can define a traditional corrected significance level of P = 0.0012 for our study. Three variants, located in the CARD9, IL23R and RNF186 genes, reach this significance threshold suggesting that these could possibly be the causal genes/variants within these two loci (Table 1). Specifically, our results show that the c.IVS11+1G>C CARD9 splice variant confers significant protection to UC (P = 1.47×10−11; OR = 0.39 [0.30–0.53]). We previously identified this splice variant in a sequencing project of CD loci and demonstrated that it leads to an alternatively spliced transcript that is missing exon 11 [7]. Our results also identify significant association to the valine to isoleucine substitution at position 362 (Val362Ile) in IL23R (P = 1.18×10−03; OR = 0.79 [0.68–0.91]) previously reported by a recent re-sequencing of positional candidates in Crohn's disease [7], [11]. The significantly associated rare variant that we identified in RNF186 (P = 8.69×10−4; OR = 1.49 [1.17–1.90]) encodes an alanine to threonine substitution at position 64 (Ala64Thr). RNF186 encodes a protein with a RING domain and two transmembrane domains. Importantly, the disease-coding variant is located in the RING domain, a domain with a predicted E3 ubiquitin-protein ligase activity (Fig. 1).
Fig. 1. Functional characterization of RNF186. 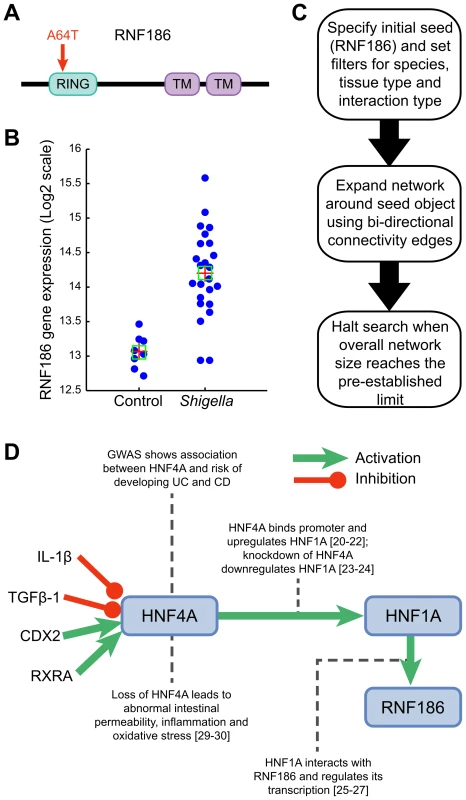
(A) RNF186 encodes a protein with RING domain and two transmembrane domains. E3 ubiquitin-protein ligase activity is intrinsic to the RING domain. This domain contains the disease-coding variant (A64T). (B) RNF186 expression response to S. flexneri in young mice (see also Figure S11). (C) Network building steps. Network is generated by mining multiple sources of interaction databases in Metacore that span human protein-protein, protein-DNA, Protein-RNA and protein-compounds interactions. (D) Transcriptional regulation model for RNF186. IL1-beta and TGF-beta 1 decrease HNF4A mRNA expression [39]–[41]. Knockdown of retinoid X receptor, alpha (RXRA) down-regulates HNF4A gene expression; RXRA interacts with HNF4A gene [24]. HNF4A is a direct target gene of caudal type homeobox 2 (CDX2); CDX2 increases HNF4A mRNA expression in intestinal epithelial cells [42], [43]. HNF4A binds promoter region of HNF1A and up-regulates its expression. HNF1A interacts with RNF186 and regulates its transcription. Tab. 1. Identification of rare variants associated with ulcerative colitis. 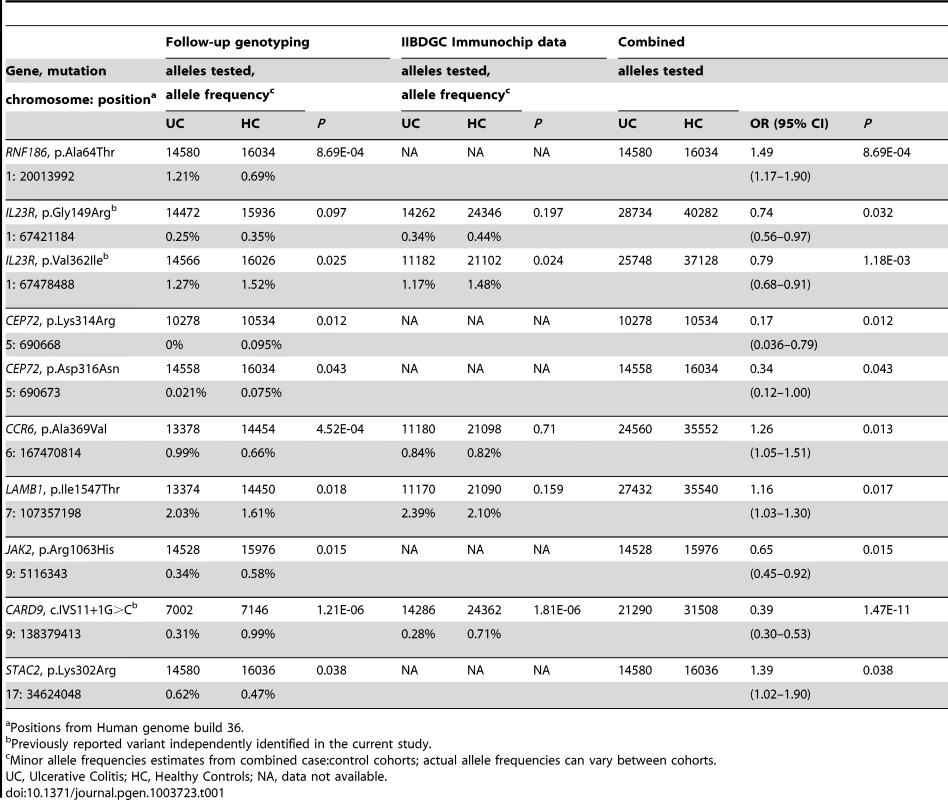
Positions from Human genome build 36. Independence of effect between rare variants in IL23R and CARD9 and the reported common association signals in these genes has previously been shown [7], [11]. For RNF186, the Ala64Thr variant is mostly found on the protective haplotype background from the previously identified common variant, indicating that the reported association is not likely due to partial LD with the common variant. In addition, reciprocal conditional logistic regression analysis, using a subset of samples where both variants were genotyped (3548 UC cases and 3607 healthy controls) shows that these are independent association signals (data not shown).
Given the challenge inherent in achieving corrected significance thresholds for rare variants, even with large cohorts, we expect that some of the other variants that we identified and found to have nominal significance (0.0012<P<0.05) are truly associated with UC. In fact with a target set of 42 variants included in follow-up genotyping, and supposing these are independent and under the null, we would expect <1 SNP to exceed P<0.01 (with a probability of less than 1% to observe 3 or more associations at this level) and ∼2 SNPs to exceed P<0.05 by chance alone (with a probability less than 0.0001 to observe 9 or more association at this level), whereas we observe 3 SNPs with P<0.01 and 9 SNPs with P<0.05, suggesting that there are additional true positives that have not met the more stringent threshold. Indeed, within the group of SNPs that we found to have nominal significance are two non-synonymous coding variants (Gly149Arg and Val362Ile) in IL23R that we and others have shown to be associated with protection from IBD (Table 1) [7], [11]. In addition to these previously-validated variants in IL23R, we have found variants that are nominally associated with UC in the genes encoding CEP72, LAMB1, CCR6, JAK2, and STAC2 (Table 1). Specifically, we identified two nominally associated rare variants in CEP72 (Lys314Arg and Asp316Asn) in perfect LD with each other that appear to protect from UC (Table 1). As we also sequenced the only other gene in this locus (TPPP), but did not find any associated variants in it, this suggests that CEP72 is potentially causal. Similarly, we sequenced both genes in the LAMB1-DLD locus on chromosome 7, with the nominally associated rare variant in LAMB1 (Ile154Thr) suggesting a role for this gene in risk to UC, especially as the associated allele is located in its DUF287 domain and is predicted to have a damaging effect [12]. All genes within the CCR6-FGFR1OP-RNASET2 locus were sequenced, with a single nominally-associated variant (Ala369Val) in CCR6, consistent with this gene's probable role in the migration and recruitment of dendritic and T cells during inflammatory and immunological responses [13]. Within the JAK2-INSL6-LHX3 locus, we only sequenced JAK2 given its key role in signaling from the IL12R/IL23R, a biological pathway proven to be associated with IBD, and identified a nominally associated variant (Arg1063His) within its catalytic domain. STAC2 is within a locus with 16 other genes including ORMDL3, which has been suggested to be the most likely causal gene based previous genetic and functional studies in IBD and asthma [8], [14]. Although we find a nominally associated variant in STAC2 (Lys302Arg) and none in ORMDL3, we have only sequenced 10 of the 17 genes within this locus (Table S4). Studies of each of these variants to determine their functional impact will be essential to prove causality.
Discussion
Genome-wide association studies in IBD have been very successful in identifying genomic regions associated with CD, UC or both. Only infrequently have these GWA studies also directly identified the causal genes/variants, with NOD2, IL23R and ATG16L1 being the few known examples. A recent targeted (exons and exon-intron boundaries) sequencing approach of known CD loci resulted in the identification of potentially causal variants in eight of the 36 loci examined [7]. The primary objective of the current study therefore was to use the same approach to identify likely causal variants within genes that were located in genomic regions associated with UC. While there are over 100 UC loci that have been identified and validated to date, we examined 22 UC loci that were known at the time of the initiation of this project. Of these 22 loci, the current study identified potentially causal variation in three of the loci: two protective alleles in CARD9 and IL23R, and an allele increasing risk in RNF186.
The identification of a rare variant (Ala64Thr) in RNF186 that shows significant association to UC strongly suggests that this is the causal gene within this locus. Importantly, the disease-coding variant is located in the RING domain, a domain with a predicted E3 ubiquitin-protein ligase activity. Ubiquitin ligases have been shown to regulate key adaptors of proinflammatory pathways [15]–[17]. We previously reported that RNF186 expression was higher in human intestinal tissues than in immune tissues [8]. We showed by immunostaining that the RNF186 protein was expressed at the basal pole of epithelial cells and lamina propria within colonic tissues. Using GEO public microarray datasets, we pursued a systematic follow-up analysis of expression profiles of epithelial cells in response to bacterial products, PAMPs/pathogens. We found that RNF186 gene expression was significantly up-regulated in small intestine epithelium and induced by Shigella infection in mice (P = 4.21×10−8) (Figure 1, Panel A) [18], [19]. Both invasive (INV+) and non-invasive (INV−) strains of Shigella induced significant overexpression of RNF186 in intestinal tissues of 4-day - and 7-day-old mice infected for 2 or 4 hours. To further identify putative transcriptional regulators of RNF186 expression, we employed a text-mining and network-generating analysis of human protein-protein, protein-DNA, protein-RNA and protein-compound interactions. Specifically, from our analyses we hypothesize that RNF186 is transcriptionally regulated in a two-step process by the transcription factor Hepatocyte Nuclear Factor 4, alpha (HNF4A) (Figure 1, Panels B,C). Several studies have shown that HNF4A binds to the promoter region and up-regulates the expression of yet another transcription factor HNF1A [20]–[22]. Knockdown of HNF4A has been shown to down-regulate HNF1A gene expression [23], [24]. HNF1A, in turn, regulates RNF186 and this interaction has been confirmed by chromatin immunoprecipitation and chip-on-chip assay [25]–[27]. Our own analysis of transcriptional profiles of HNF4A-Null colons recovered from HNF4AloxP/loxPFoxa3Cre and HNF4AloxP/−Foxa3Cre mice uncovered a significant up-regulation of RNF186 transcript [28]. Expression profiling of human tissues also supports this hypothesis, as HNF4A and RNF186 are clearly co-expressed in the small intestine and the colon (Figure S1). This putative interaction is particularly relevant given that HNF4A has previously been shown to be associated, with genome-wide significance, with risk to developing UC [9]. Our analysis now indicates a direct genetic interaction between two IBD susceptibility genes namely, HNF4A and RNF186. While a singular loss-of-function mutation in HNF4A has already been shown to be associated with susceptibility to abnormal intestinal permeability, inflammation and oxidative stress, we speculate that a dual loss-of-function with additional mutation in RNF186 would further exacerbate one's susceptibility to develop chronic inflammation in the gut [29], [30].
In addition to the variants in IL23R, CARD9, and RNF186, we also identified variants of interest in an additional five loci (specifically within the CEP72, LAMB1, CCR6, JAK2, and STAC2 genes). While these latter six still require confirmation, we estimate that many will validate given that we observed an excess of nominally-associated variants. Examining the data from the current study along with the data derived from prior association and sequencing studies suggests that at a minimum, there currently is strong evidence of association to causal variation in IBD (i.e. missense, nonsense or splice junction variants) in the NOD2, ATG16L1, IL23R, MST1, CARD9, IL18RAP and RNF186 genes, and at least suggestive evidence for causal variation in the CUL2, C1orf106, PTPN22, MUC19, CEP72, LAMB1, CCR6, JAK2, and STAC2 genes (Current study and references [4], [5], [7], [11], [31]). While only a small fraction of the recently identified 163 IBD loci have been sequenced (36 CD, 22 UC for total of 42 independent loci) in IBD patients and controls, this would suggest that from ∼10% (15 of 163 total loci) to ∼35% (15 of 42 loci sequenced) of IBD loci have causal variation affecting the protein-coding or splice junctions. There are an additional 5 loci (ITLN1, GSDMB, YDGL, SLC22A4, and FCGR2A) for which there are non-synonymous coding or splice variants present in public databases (dbSNP, 1KG) that are correlated with the index SNP identified in the GWA studies that have yet been tested directly, thus potentially increasing the estimated number of IBD loci with causal variation within the coding and splice regions [3], [32].
Furthermore, it should be noted that with the exception of a small number of variants with significant effect (e.g. R702W, G908R, fs107insC in NOD2; R381Q in IL23R; IVS11+1G>C in CARD9; V527L in IL18RAP – all of which had 0.5>OR>2) most of the rare variants identified by targeted sequencing of loci from GWAS regions have relatively modest effect sizes that are comparable to those observed for the common variants identified by GWA studies. Consequently, very large sample sizes are required to detect statistically significant association. In the current study, for the majority (93%) of variants with an observed minor allele frequency greater than 0.3%, we had more than 80% power to detect significant association if the OR is 2 or greater with the number of samples typed (up to ∼14,000 cases and ∼20,000 controls) (see Table S5). Moreover, should this observation not be limited to risk loci identified by GWA studies, this has implications with respect to future efforts for discovering risk loci. Specifically, if the occurrence of rare variants with large effects sizes is relatively infrequent, then this may favor the current paradigm of locus discovery by GWA followed by targeted sequencing rather than whole-exome or whole-genome sequencing for locus discovery as this would require even larger sample sizes. Alternatively, given the ever - growing size of public databases of common and rare variants, targeted genotyping of known variants within risk loci identified by GWA may prove to be an efficient approach. For example, all but two of the 22 candidate causal variants identified in the current study or that of Rivas and colleagues are now found in the Exome Sequencing Project database.
Regardless of the study design, these results suggest that a significant proportion of IBD loci contain causal variants within exons or exon-intron boundaries. While these rare/infrequent variants may not account for what has been termed “the missing heritability” of common traits, discovering these variants certainly can provide focus for follow-up functional studies. For example, the current sequencing and follow-up genotyping of the chromosome 1p36 locus, which was first identified in a GWA study of UC, identified significant association to the Ala64Thr variant within RNF186. While further studies will be required, the initial bioinformatics and experimental studies described above suggest that this ring finger protein with an ubiquitin-ligase domain may have an important role in the response to microbes/microbial products. Going forward, systematic evaluation of genes within risk loci via expression-driven functional studies in cellular models (i.e. knock-down or over expression) with sensitive high throughput/high content readouts may very well be a complementary approach given that at least a third of IBD risk loci appear to act via gene expression [3].
Materials and Methods
DNA preparation and pooling
We selected 200 ulcerative colitis patients and 150 healthy control of French-Canadian descent from the NIDDK IBD Genetics Consortium repository samples. The NIDDK IBDGC samples were collected under rigorous clinical phenotyping and control matching for the purpose of genetic studies [33]. Genomic DNA concentrations were measured by Quant-iT PicoGreen dsDNA reagent (Invitrogen) and detected on the Biotek Synergy 2 plate reader. All DNAs were normalized with at least two round of dilution and quantification down to a concentration of 10 ng/µl as described previously [7]. Equimolar amounts of samples were pooled together in batches of 50 cases and 50 controls for a total of 7 pooled groups.
Target selection and design
Target exonic sequences were selected based on the coding exons of 55 genes in 14 UC-associated regions and 7 regions identified in CD with nominal replication in our recent UC GWAS study, as well ECM1 identified from recent candidate-gene study in UC [6], [8]–[10], [34]. Specifically, amplicons were designed from genome build Hg18 using a web-base automated pipeline (Optimus primer: Website (http://op.pgx.ca)) that uses the Primer 3 design software and user defined parameters [35]. Design parameters included amplicon sizes between 400 and 600 base pairs, as well as the inclusion of Not1 tails for subsequent concatenation and shearing steps in library construction. PCR amplification reactions contained 40 ng of pooled genomic DNA, 1× HotStar buffer, 0.8 mM dNTPs, 2 mM MgCl2, 0.4 units of HotStar Enzyme (Qiagen), and 0.25 µM forward and reverse primers in a 10-µl reaction volume. PCR cycling parameters were as follows: one cycle of 95°C for 5 min; 30 or 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; followed by one cycle of 72°C for 5 min. Each DNA pools were amplified for 508 PCR reactions; amplification products were then dosed by Quant-iT PicoGreen dsDNA reagent (Invitrogen) quantification and amplification specificity was validated by agarose gel electrophoresis. In total, 472 PCR amplicons (93% amplification success rate, capturing 283 Kb including 117 Kb of target exonic sequences) (Table S6) for each DNA pool were combined in equimolar amounts to obtain equal representation of all target in library construction.
Sequencing and variant discovery
The combined PCR products from each pooled DNA group were concatenated using the NotI adapters and sheared into fragments as previously described [36]. Libraries were constructed according to Illumina single-end library protocol, with 150–200 bp gel size selection and PCR enrichment using 10 cycles of PCR, and then single-end sequenced with 36 cycles on an Illumina Genome Analyzer II. Each sample pool was sequenced using a single lane of Illumina GAII analyzer flowcell; 36-base pair reads were aligned to the genome using MAQ algorithm [37] and base qualities were recalibrated using GATK (Genome Analysis ToolKit) [38]. Finally, variant discovery was performed using the previously described Syzygy software, designed to analyze sequencing data from pooled DNA sequencing [7].
Genotyping, validation and follow-up genotyping
We randomly selected 237 high quality variants for validation in our 350 discovery DNAs samples using Sequenom MassARRAY iPlex200 chemistry. Genotyping assay designs were obtained from the Assay Designer v.3.1 software, and genotyping oligonucleotides were synthesized at Integrated DNA Technologies. The correlation coefficient between observed minor allele frequencies and frequencies estimated from Syzygy for validated variants was calculated in order to evaluate the overall quality of our dataset (Figure S2). Eighty-four high quality non-synonymous coding variants (missense, nonsense and splicing variants (within 2 bp of a splice site)) remained after the exclusion of singletons from our sequencing results, variants that did not validate and variants within the MHC region. We then evaluated these variants in an independent cohort of North-American individual of European descent from the NIDDK IBD genetics consortium (754 cases and 1008 controls); only variants detected in this independent cohort were kept for follow-up genotyping. Following assay design, 42 SNPs were genotyped using Sequenom MassARRAY iPlex200 chemistry in 6 independent follow-up case-control cohorts (7292 cases and 8018 controls) (Table S3). Because of design constraints and assay failures, not all markers were examined in all follow-up sample sets. For a subset of these variants, further genotyping data was obtained from the International IBD Genetics Consortium Immunochip data (7143 UC, 12186 controls)
Cohort descriptions
For all cohorts, UC was diagnosed according to accepted clinical, endoscopic, radiological and histological findings.
Genotyping of the NIDDK IBDGC cohort, as well as the Italian and Dutch cohorts was performed at the Laboratory for Genetics and Genomic Medicine of Inflammation (www.inflammgen.org) of the Université de Montréal.
NIDDK IBD Genetics Consortium (IBDGC) samples were recruited by the centers included in the NIDDK IBDGC: Cedars Sinai, Johns Hopkins University, University of Chicago and Yale, University of Montreal, University of Pittsburgh and University of Toronto. Additional samples were obtained from the Queensland Institute for Medical Research, Emory University and the University of Utah. Medical history was collected with standardized NIDDK IBDGC phenotype forms. Healthy controls are defined as those with no personal or family history of IBD.
The Italian samples were collected at the S. Giovanni Rotondo “CSS” (SGRC) Hospital in Italy.
The Dutch cohort is composed of ulcerative colitis cases recruited through the Inflammatory Bowel Disease unit of the University Medical Center Groningen (Groningen), the Academic Medical Center (Amsterdam), the Leiden University Medical Center (Leiden) and the Radboud University Medical Center (Nijmegen), and of healthy controls (n = 804) of self-declared European ancestry from volunteers at the University Medical Center (Utrecht).
Genotyping of the German cohort was performed at the Institute for Clinical Molecular Biology
Christian-Albrechts-University in Kiel. German patients were recruited either at the Department of General Internal Medicine of the Christian-Albrechts-University Kiel, the Charité University Hospital Berlin, through local outpatient services, or nationwide with the support of the German Crohn and Colitis Foundation. German healthy control individuals were obtained from the popgen biobank.
Genotyping of Swedish UC cases and controls was performed at Karolinska Institutet's Mutational Analysis core facility (MAF). Swedish ulcerative colitis patients and controls were recruited at the Karolinska University Hospital, Stockholm, and at the Örebro University Hospital, Örebro, Sweden.
Genotyping of the Belgian cohort was performed at the Genomics Core Facility at UZ Leuven, using a MassARRAY iPLEX (Sequenom). Belgian patients were all recruited at the IBD unit of the University Hospital Leuven, Belgium; control samples are all unrelated, and without family history of IBD or other immune related disorders.
Ethics statement
All patients and control subjects provided informed consent. Recruitment protocols and consent forms were approved by Institutional Review Boards at each participating institutions. All DNA samples and data in this study were denominalized.
Association analysis
Association analysis of follow-up genotyping data was performed using the previously described mega-analysis of rare variants (MARV) approach [7]. Briefly, this method evaluates significance of association from stratified sample, using within sub-cohort permutation of individual phenotypes to provide the test statistic. This approach is robust to population stratification and to deviation from Hardy-Weinburg equilibrium.
Network analyses
We downloaded and analyzed several Gene Expression Omnibus (GEO) public microarray datasets including: (a) Expression data from newborn mice infected with Shigella flexneri; GSE9785 (b) Transcription profiles of colon biopsies from UC patients and healthy controls; GSE11223 (c) Steady-state gene expression data of Tuberculosis infected human primary dendritic cells; GSE34151 (d) PBMC transcriptional profiles in healthy subjects, patients with Crohn's Disease, and patients with Ulcerative Colitis; GSE3365, (e) Transcription profiles of colon biopsies from Crohn's patients and healthy controls; GSE20881, (f) Transcription profile of mouse small intestine epithelium vs. mesenchyme; GSE6383, (g) Gene expression in HNF4 null mouse colons compared to control colons; GSE3116, and (h) Microarray profiles of mouse epithelial colon harboring conditional knock out of HFN4A; GSE11759. Each of these datasets was normalized using quantile normalization routine in MATLAB. Genes were tested for significant differences between pairs of control and stimulated/treated samples within each experiment. After selecting genes with nominal P<0.05, estimated using an unpaired T-test, expression of RNF186 was evaluated whether it passed the significance threshold or not. The results of processing all these datasets are shown in Table S7 and Figures S3, S4, S5, S6, S7, S8, S9, S10, S11, S12, S13, S14. For transcriptional network analysis, we used Metacore's suite of network building algorithms to expand the sub-network around RNF186. The algorithm searches through a manually curated knowledgebase of molecular interaction to identify bidirectional connectivity with genes, proteins and small molecules. The search was constrained to expand the overall network size up to 50 components. Given that the bioinformatic analyses suggested that HNF4A controlled the expression of RNF186, we directly tested for their co-expression in a panel of RNAs from a variety of human tissues. Specifically, expression levels of RNF186 and HNF4A were evaluated using a custom expression array from Agilent, which was designed to include an independent probe for each exon of the genes tested (Figure S1). Briefly, total RNA from bone marrow, heart, skeletal muscle, uterus, liver, fetal liver, spleen, thymus, thyroid, prostate, brain, lung, small intestine and colon were purchased from Clontech Laboratories. A reference RNA sample was also included that consisted of an equal mix from 10 different human tissues (adrenal gland, cerebellum, whole brain, heart, liver, prostate, spleen, thymus, colon, bone marrow). With the exception of the small intestine (RIN = 7.6), all RNAs had a RNA Integrity Value (RIN) value ≥8 (range 8.0–9.3) as measured by Agilent 2100 Bioanalyzer using the RNA Nano 6000 kit (Agilent Technologies). Labeled cRNA was then synthesized from 50 ng of each RNA sample using the Low Input Quick Amp WT labeling kit (Agilent Technologies) according to the manufacturer's protocol. Quantity and quality of labeled cRNA samples were assessed by NanoDrop UV-VIS Spectrophotometer. Sample hybridization was performed according to the manufacturer's standard protocol and microarrays were scanned using the Sure Scan Microarray Scanner (Agilent technologies). An expression value was obtained for each gene in each replicate by calculating the geometric mean of all probes within the gene, followed by a median normalization across all genes on the array. A geometric mean and geometric standard deviation was calculated from at least 3 independent measurements for each tissue.
Supporting Information
Zdroje
1. XavierRJ, PodolskyDK (2007) Unravelling the pathogenesis of inflammatory bowel disease. Nature 448 : 427–434.
2. AndersonCA, BoucherG, LeesCW, FrankeA, D'AmatoM, et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43 : 246–252.
3. JostinsL, RipkeS, WeersmaRK, DuerrRH, McGovernDP, et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491 : 119–124.
4. DuerrRH, TaylorKD, BrantSR, RiouxJD, SilverbergMS, et al. (2006) A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314 : 1461–1463.
5. RiouxJD, XavierRJ, TaylorKD, SilverbergMS, GoyetteP, et al. (2007) Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39 : 596–604.
6. HampeJ, FrankeA, RosenstielP, TillA, TeuberM, et al. (2007) A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genetics 39 : 207–211.
7. RivasMA, BeaudoinM, GardetA, StevensC, SharmaY, et al. (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43 : 1066–1073.
8. McGovernDP, GardetA, TorkvistL, GoyetteP, EssersJ, et al. (2010) Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 42 : 332–337.
9. BarrettJC, HansoulS, NicolaeDL, ChoJH, DuerrRH, et al. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 40 : 955–962.
10. FestenEA, GoyetteP, ScottR, AnneseV, ZhernakovaA, et al. (2009) Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut 58 : 799–804.
11. MomozawaY, MniM, NakamuraK, CoppietersW, AlmerS, et al. (2011) Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet 43 : 43–47.
12. AdzhubeiIA, SchmidtS, PeshkinL, RamenskyVE, GerasimovaA, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7 : 248–249.
13. WilliamsIR (2004) Chemokine receptors and leukocyte trafficking in the mucosal immune system. Immunol Res 29 : 283–292.
14. MoffattMF, KabeschM, LiangL, DixonAL, StrachanD, et al. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448 : 470–473.
15. NatoliG, ChioccaS (2008) Nuclear ubiquitin ligases, NF-kappaB degradation, and the control of inflammation. Science signaling 1: pe1.
16. ZhaoW, WangL, ZhangM, YuanC, GaoC (2012) E3 Ubiquitin Ligase Tripartite Motif 38 Negatively Regulates TLR-Mediated Immune Responses by Proteasomal Degradation of TNF Receptor-Associated Factor 6 in Macrophages. J Immunol 188 : 2567–2574.
17. JinW, ChangM, SunSC (2012) Peli: a family of signal-responsive E3 ubiquitin ligases mediating TLR signaling and T-cell tolerance. Cell Mol Immunol 9 : 113–122.
18. FernandezMI, RegnaultB, MuletC, TanguyM, JayP, et al. (2008) Maturation of paneth cells induces the refractory state of newborn mice to Shigella infection. J Immunol 180 : 4924–4930.
19. LiX, MadisonBB, ZachariasW, KolterudA, StatesD, et al. (2007) Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol Genomics 29 : 290–301.
20. BoydM, BressendorffS, MollerJ, OlsenJ, TroelsenJT (2009) Mapping of HNF4alpha target genes in intestinal epithelial cells. BMC Gastroenterol 9 : 68.
21. FangB, Mane-PadrosD, BolotinE, JiangT, SladekFM (2012) Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res 40 : 5343–5356.
22. KtistakiE, TalianidisI (1997) Modulation of hepatic gene expression by hepatocyte nuclear factor 1. Science 277 : 109–112.
23. KyrmiziI, HatzisP, KatrakiliN, TroncheF, GonzalezFJ, et al. (2006) Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20 : 2293–2305.
24. TomaruY, NakanishiM, MiuraH, KimuraY, OhkawaH, et al. (2009) Identification of an inter-transcription factor regulatory network in human hepatoma cells by Matrix RNAi. Nucleic Acids Res 37 : 1049–1060.
25. BandyopadhyayS, ChiangCY, SrivastavaJ, GerstenM, WhiteS, et al. (2010) A human MAP kinase interactome. Nat Methods 7 : 801–805.
26. DasD, NahleZ, ZhangMQ (2006) Adaptively inferring human transcriptional subnetworks. Mol Syst Biol 2 : 2006 0029.
27. OdomDT, ZizlspergerN, GordonDB, BellGW, RinaldiNJ, et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303 : 1378–1381.
28. GarrisonWD, BattleMA, YangC, KaestnerKH, SladekFM, et al. (2006) Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology 130 : 1207–1220.
29. DarsignyM, BabeuJP, DupuisAA, FurthEE, SeidmanEG, et al. (2009) Loss of hepatocyte-nuclear-factor-4alpha affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS One 4: e7609.
30. MarcilV, SeidmanE, SinnettD, BoudreauF, GendronFP, et al. (2010) Modification in oxidative stress, inflammation, and lipoprotein assembly in response to hepatocyte nuclear factor 4alpha knockdown in intestinal epithelial cells. J Biol Chem 285 : 40448–40460.
31. GoyetteP, LefebvreC, NgA, BrantSR, ChoJH, et al. (2008) Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol 1 : 131–138.
32. AsanoK, MatsushitaT, UmenoJ, HosonoN, TakahashiA, et al. (2009) A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet 41 : 1325–1329.
33. DassopoulosT, NguyenGC, BittonA, BromfieldGP, SchummLP, et al. (2007) Assessment of reliability and validity of IBD phenotyping within the National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) IBD Genetics Consortium (IBDGC). Inflamm Bowel Dis 13 : 975–983.
34. FisherSA, TremellingM, AndersonCA, GwilliamR, BumpsteadS, et al. (2008) Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet 40 : 710–712.
35. BrownAM, LoKS, GuelpaP, BeaudoinM, RiouxJD, et al. (2010) Optimus Primer: A PCR enrichment primer design program for next-generation sequencing of human exonic regions. BMC Res Notes 3 : 185.
36. GnirkeA, MelnikovA, MaguireJ, RogovP, LeProustEM, et al. (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27 : 182–189.
37. LiH, RuanJ, DurbinR (2008) Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18 : 1851–1858.
38. McKennaA, HannaM, BanksE, SivachenkoA, CibulskisK, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20 : 1297–1303.
39. CajaL, BertranE, CampbellJ, FaustoN, FabregatI (2011) The transforming growth factor-beta (TGF-beta) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. J Cell Physiol 226 : 1214–1223.
40. HongMH, ChouYC, WuYC, TsaiKN, HuCP, et al. (2012) Transforming growth factor-beta1 suppresses hepatitis B virus replication by the reduction of hepatocyte nuclear factor-4alpha expression. PLoS One 7: e30360.
41. WangB, CaiSR, GaoC, SladekFM, PonderKP (2001) Lipopolysaccharide results in a marked decrease in hepatocyte nuclear factor 4 alpha in rat liver. Hepatology 34 : 979–989.
42. BoydM, HansenM, JensenTG, PerearnauA, OlsenAK, et al. (2010) Genome-wide analysis of CDX2 binding in intestinal epithelial cells (Caco-2). J Biol Chem 285 : 25115–25125.
43. McKinney-FreemanSL, LengerkeC, JangIH, SchmittS, WangY, et al. (2008) Modulation of murine embryonic stem cell-derived CD41+c-kit+ hematopoietic progenitors by ectopic expression of Cdx genes. Blood 111 : 4944–4953.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



