-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
The Unfolded Protein Response (UPR) maintains homeostasis in the endoplasmic reticulum (ER) and defends against ER stress, an underlying factor in various human diseases. During the UPR, numerous genes are activated that sustain and protect the ER. These responses are known to involve the canonical UPR transcription factors XBP1, ATF4, and ATF6. Here, we show in C. elegans that the conserved stress defense factor SKN-1/Nrf plays a central and essential role in the transcriptional UPR. While SKN-1/Nrf has a well-established function in protection against oxidative and xenobiotic stress, we find that it also mobilizes an overlapping but distinct response to ER stress. SKN-1/Nrf is regulated by the UPR, directly controls UPR signaling and transcription factor genes, binds to common downstream targets with XBP-1 and ATF-6, and is present at the ER. SKN-1/Nrf is also essential for resistance to ER stress, including reductive stress. Remarkably, SKN-1/Nrf-mediated responses to oxidative stress depend upon signaling from the ER. We conclude that SKN-1/Nrf plays a critical role in the UPR, but orchestrates a distinct oxidative stress response that is licensed by ER signaling. Regulatory integration through SKN-1/Nrf may coordinate ER and cytoplasmic homeostasis.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003701
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003701Summary
The Unfolded Protein Response (UPR) maintains homeostasis in the endoplasmic reticulum (ER) and defends against ER stress, an underlying factor in various human diseases. During the UPR, numerous genes are activated that sustain and protect the ER. These responses are known to involve the canonical UPR transcription factors XBP1, ATF4, and ATF6. Here, we show in C. elegans that the conserved stress defense factor SKN-1/Nrf plays a central and essential role in the transcriptional UPR. While SKN-1/Nrf has a well-established function in protection against oxidative and xenobiotic stress, we find that it also mobilizes an overlapping but distinct response to ER stress. SKN-1/Nrf is regulated by the UPR, directly controls UPR signaling and transcription factor genes, binds to common downstream targets with XBP-1 and ATF-6, and is present at the ER. SKN-1/Nrf is also essential for resistance to ER stress, including reductive stress. Remarkably, SKN-1/Nrf-mediated responses to oxidative stress depend upon signaling from the ER. We conclude that SKN-1/Nrf plays a critical role in the UPR, but orchestrates a distinct oxidative stress response that is licensed by ER signaling. Regulatory integration through SKN-1/Nrf may coordinate ER and cytoplasmic homeostasis.
Introduction
The endoplasmic reticulum (ER) is responsible for multiple functions in protein synthesis and processing, lipid metabolism, xeno/endobiotic detoxification, and Ca2+ storage (reviewed in [1], [2]). The ER forms a continuous structure with the nuclear envelope and maintains extensive contact with mitochondria [3], [4]. Consequently, the ER is well positioned to sense and respond to changes in the cellular environment.
All secretory and membrane-bound proteins are synthesized in the rough ER, a process that is highly regulated so that only properly folded and modified proteins are released to the Golgi [1], [2], [5], [6]. Maturation and folding of these proteins involves glycosylation and formation of appropriate Cys-Cys crosslinks. When its protein folding capacity is exceeded (ER stress), the ER protects itself through the Unfolded Protein Response (UPR) (Figure S1A) [2], [5], [6]. This signaling and transcription program decreases protein translation, expands ER size and folding capacity, and directs misfolded proteins to be degraded in the cytosol. The UPR functions continuously to maintain ER homeostasis, but is amplified and diversified under ER stress conditions [5], [7]–[10]. In response to severe ER stress, the UPR promotes ER absorption through autophagy and ultimately may induce cell death. ER stress and the UPR have been implicated in many human diseases, including diabetes, inflammatory disease, neurodegenerative disease, secretory cell malignancies, and other cancers [6], [11], [12].
The canonical metazoan UPR is orchestrated by three major ER transmembrane signaling proteins (IRE1, PERK, and ATF6), and three bZIP-family transcription factors (XBP1, ATF4, and cleaved ATF6) (Figure S1A) [2], [5], [6]. The most ancient of these transmembrane proteins, IRE1, is a cytoplasmic endoribonuclease and kinase that senses unfolded proteins in the ER. In response to ER stress, the IRE1 RNAse initiates cytoplasmic splicing of the mRNA encoding XBP1, the transcription factor that is most central to the UPR. The IRE1 kinase contributes to ER homeostasis by regulating the IRE-1 endonuclease activity, and transmits signals through JNK, p38, and other pathways. The kinase PERK phosphorylates the translation initiation factor eIF2α, thereby globally decreasing translation. This reduces the ER protein-folding load, but also favors translation of mRNAs that encode protective proteins, including ATF4. ATF6 resides in the ER membrane but is transported to the Golgi and cleaved in response to ER stress. The activation status of these transmembrane proteins is influenced by their interactions with the ER chaperone BiP (HSP-3/-4 in C. elegans).
The ER lumen maintains an oxidative environment, in contrast to the cytoplasm, because the ER enzyme systems that form disulfide bonds generate reactive oxygen species (ROS) [1], [13], [14]. Accordingly, ER stress may eventually lead to cellular oxidative stress and activation of oxidative stress defense genes [15]. Metazoan oxidative and xenobiotic stress responses are orchestrated mainly by the Nrf bZIP-family transcription factors (Nrf1, 2, 3 in mammals). Nrf-family proteins regulate genes involved in various small molecule detoxification processes, including glutathione biosynthesis and conjugation, and have been implicated in longevity assurance in invertebrates and mammals [16]–[21]. These transcription factors have recently been shown to function in proteasome regulation, stem cell maintenance, and metabolism, suggesting that they may control a wider range of processes than previously realized [22]–[26]. It has been reported that mammalian Nrf1 and Nrf3 associate with the ER membrane and nuclear envelope [27]–[30], and that Nrf2 is phosphorylated by PERK [31], [32]. While these last observations are intriguing, it is unknown whether Nrf-family proteins might actually be involved in ER stress defenses, either through mobilizing an oxidative stress response or participating in the UPR itself.
The nematode C. elegans has been a valuable system for investigating how Nrf proteins function and are regulated in vivo, because of its advantages for employing genetics to elucidate regulatory networks, and performing whole-organism analyses of stress resistance and survival. The C. elegans Nrf ortholog SKN-1 plays a critical role in resistance to oxidative and xenobiotic stress, and in various pathways that extend lifespan [16], [17], [19], [23], [33]. Here we describe a comprehensive analysis of whether SKN-1 might be involved in the UPR. We found that under ER stress conditions SKN-1 directly activates many genes involved in ER function, including canonical ER signaling and transcription factors that in turn induce skn-1 transcription. Importantly, this response is distinct from that which SKN-1 mobilizes under oxidative stress conditions. SKN-1 is required for resistance to ER stress, including reductive stress, a surprising finding given the importance of SKN-1 for oxidative stress defense. Unexpectedly, UPR signaling is needed for SKN-1 to mobilize an oxidative stress response, suggesting that the ER has a licensing and possibly sensing role during oxidative and xenobiotic stress responses.
Results
SKN-1 Directly Regulates ER Stress Genes
Several observations led us to investigate whether SKN-1/Nrf might be involved in ER stress defenses. Expression profiling that we performed in C. elegans under normal and oxidative stress conditions suggested that SKN-1 regulates a number of genes that are involved in UPR or ER functions [21]. These included atf-5 (UPR transcription factor ATF4), ckb-4 (choline kinase), pcp-2 (prolyl carboxypeptidase), and many genes encoding xenobiotic metabolism enzymes that localize to the smooth ER (Table S1). Moreover, a genome-wide Chromatin Immunoprecipitation (ChIP) analysis of C. elegans L1 stage larvae (MOD-ENCODE) [34] detected binding of transgenically expressed SKN-1 at the predicted regulatory regions of numerous genes involved in UPR - or ER processes, including UPR signaling and transcription (ire-1, xbp-1, pek-1, and atf-6), Ca++ signaling, and protein folding and degradation (Table S1).
To investigate whether SKN-1 might be involved in the UPR, we first used quantitative (q) RT-PCR to investigate whether it is needed for expression of representative ER stress-induced or ER maintenance genes, many of which are predicted to be SKN-1 targets (Table S1). In these initial gene expression studies we induced ER stress by treating C. elegans with the N-linked glycosylation inhibitor tunicamycin (TM), at a concentration that readily induces the UPR but does not cause detectable toxicity (5 µg/ml, Figure S1B) [15]. TM treatment resulted in skn-1-dependent upregulation of numerous canonical or predicted UPR - or ER-related genes (Figures 1A and 1B, Table S1). skn-1 was also required for the basal expression of psd-1, R05G6.7, and cnb-1, even though these genes were not activated by TM (Figures 1A and 1B). TM-induced ER stress also upregulated two direct SKN-1 targets that are involved in glutathione metabolism (gcs-1 and gst-4) [19] in a skn-1–dependent manner, and transgenic reporter analysis detected gcs-1 activation in the intestine, the C. elegans counterpart to the gut, liver, and adipose tissue (Figures 1C and 1D). Importantly, however, ER stress did not activate various other genes that are typically induced by SKN-1 under oxidative stress conditions (Figure S1C). Taken together, the data indicate that SKN-1 mediates a response to ER stress, but also that this response does not correspond simply to its oxidative stress defense function.
Fig. 1. SKN-1 regulates diverse functions in response to ER stress. 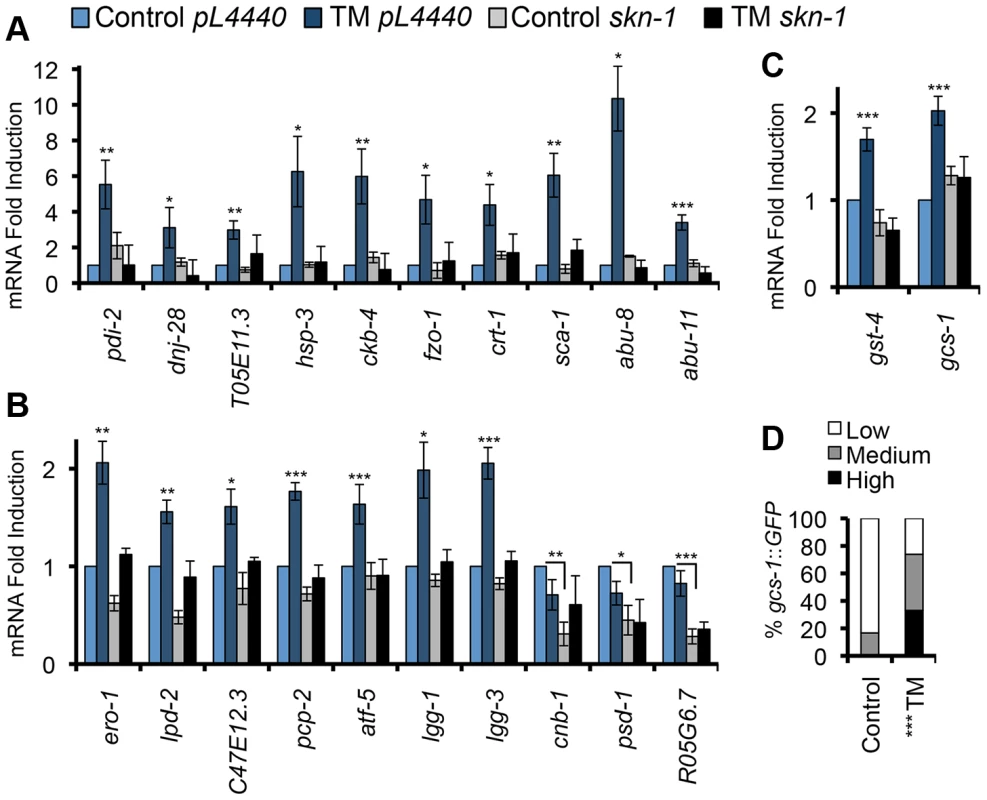
(A, B) ER stress induces skn-1-dependent activation of ER- or UPR-associated genes. qRT-PCR was performed after RNAi Control (pL4440 in all panels) or skn-1 RNAi, and Control or 5 µg/ml TM treatment. Known or predicted functions of these genes are described in Table S1. Genes are grouped in (A) or (B) according to the extent of TM-induced activation, and plotted on different scales. All analyses of TM-regulated gene expression involved a 16 hr TM treatment, based upon a time-course experiment (Figure S1B) and published work in C. elegans [15]. Shorter time courses were chosen for other ER stress treatments (Figure 4, legend). (C) Upregulation of SKN-1-regulated oxidative stress defense genes in response to TM. Error bars represent SEM, * p≤.05, ** p≤.01, *** p≤.001, relative to pL4440 Control. All qRT-PCR p-values were calculated as one or two-sided t-test as appropriate with n≥3. (D) Activation of the gcs-1::GFP transgene in the intestine, with GFP expression scored as High, Medium, or Low. *** p<.0001 chi2 method. See Experimental Procedures for scoring method. See also Figure S1 and Table S1. To investigate whether SKN-1 activates genes directly during ER stress, we used ChIP to detect endogenous SKN-1 and markers of transcription activity at pcp-2, atf-5, and gst-4, each of which is flanked by SKN-1 binding sites and upregulated by oxidative and ER stress in a skn-1-dependent manner [21] (Figures 1B and 1C). SKN-1 was readily recruited to these genes in response to either TM-induced ER stress or Arsenite (AS)-induced oxidative stress (Figures 2A, 2E, 2I, and S2A-S2C). During transcription, RNA Polymerase II (Pol II) is phosphorylated on Ser 2 of its C-terminal domain (CTD) repeat (P-Ser2) [35]. At each gene we examined, ER stress increased Ser 2 phosphorylation levels (Figures 2B, 2F, and 2J). Also consistent with transcriptional activation, at these loci ER stress increased acetylation of Histone H3, another marker of transcription activity [36], but reduced overall Histone H3 occupancy (Figures 2C, 2D, 2G, 2H, 2K, and 2L). Taken together, our findings suggest that SKN-1 directly activates a major transcriptional response to ER stress.
Fig. 2. SKN-1 directly regulates target genes during the UPR. 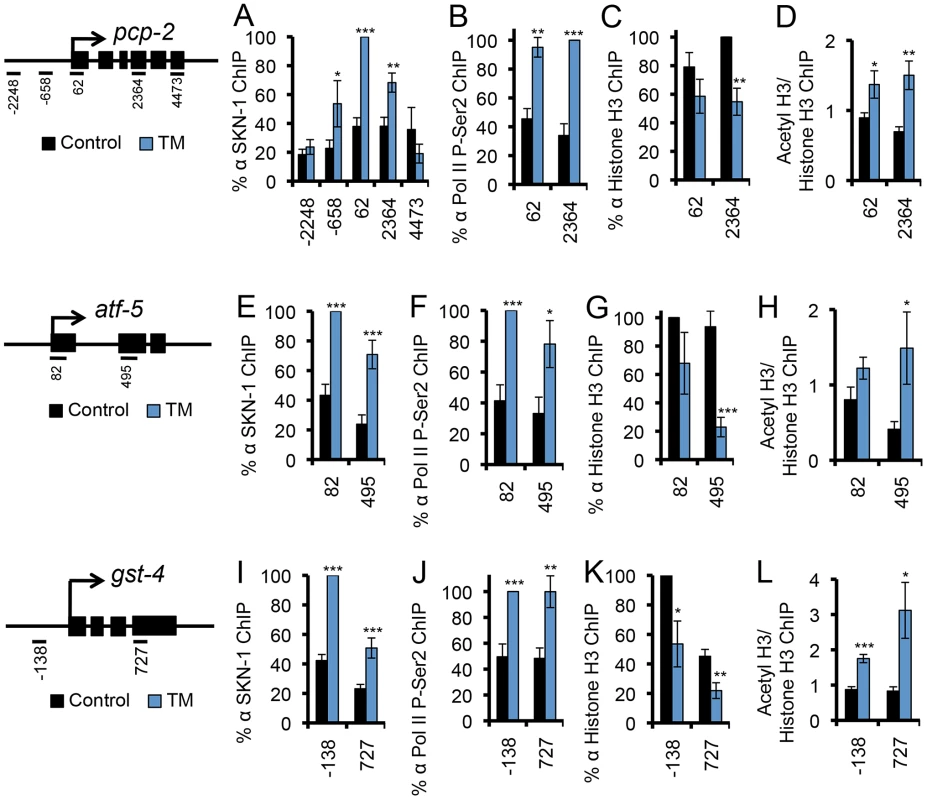
(A–L) ER stress-induced SKN-1 recruitment and transcriptional activation was analyzed at the SKN-1-regulated genes pcp-2 (A–D), atf-5 (E–H), and gst-4 (I–L). TM treatment leads to SKN-1 recruitment (A, E, I), accumulation of Pol II that is phosphorylated at CTD Ser 2 (P-Ser2) (B, F, J), decreased Histone H3 occupancy (C, G, K), and increased H3-AcK56 density (D, H, L) at the site of transcription. Maps mark qPCR amplicons relative to the predicted transcription start site, with exons marked as black boxes. % ChIP signal is relative to input, and normalized to the highest signal for each run [44]. In (D, H, L), a ratio of acetyl histone to histone signal is presented. For ChIP experiments in this study error bars represent SEM, and * p≤.05, ** p≤.01, *** p≤.001, relative to pL4440 Control calculated using one-sided student's t-test. See also Figure S2. Dependence of Core UPR Gene Induction on SKN-1
We next investigated whether SKN-1 might regulate expression of core UPR signaling and transcription factors, as predicted by the MOD-ENCODE data [34]. XBP-1 is central to the UPR, and in mammals it controls transcription of other core UPR genes (atf4/atf-5, and BiP/hsp-4) along with many downstream genes [6], [37]. During the UPR, xbp-1 expression is regulated at the level of transcription, as well as through cytoplasmic splicing of its mRNA by the IRE-1 endoribonuclease (Figure S1A) [5], [6]. The spliced form of the xbp-1 mRNA (xbp-1s) encodes the transcriptionally active form of XBP-1 (XBP-1s). When SKN-1 was lacking, ER stress failed to induce accumulation of each xbp-1 mRNA form and, remarkably, decreased the ratio of xbp-1s to the unspliced xbp-1 form (xbp-1u) (Figures 3A, 3B, and S3A). The xbp-1 locus includes a predicted SKN-1 binding site (not shown), and ChIP results indicated that endogenous SKN-1 accumulates at the xbp-1 site of transcription in response to ER stress (Figure 3C). This evidence that SKN-1 directly regulates xbp-1 could account for the reduction in total xbp-1 mRNA, but not the apparent effect of SKN-1 on xbp-1 splicing. A plausible explanation is that lack of SKN-1 also reduced basal and ER stress-induced expression of ire-1 (Figures 3D and 3E). Moreover, we observed that SKN-1 is recruited to the ire-1 locus in response to ER stress (Figure 3F), consistent with MOD-ENCODE evidence that ire-1 may be a SKN-1 target [34].
Fig. 3. SKN-1 regulates core UPR genes. 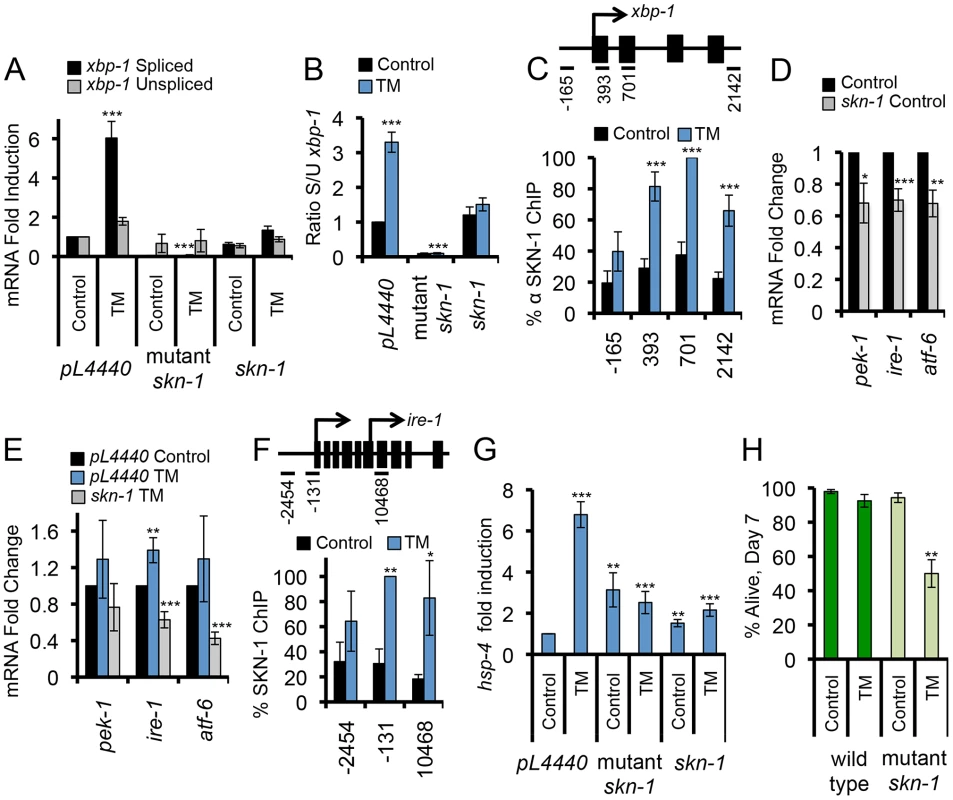
(A) SKN-1 is required for TM-induced accumulation of spliced xbp-1 mRNA. Levels of xbp-1 mRNA forms were analyzed by qRT-PCR with isoform-specific primers, and are presented as the xbp-1s/xbp-1u ratio in (B). skn-1 RNAi and mutant animals were analyzed compared to wild type. skn-1 refers to skn-1 RNAi, and skn-1 mutant refers to the skn-1(zu67) allele in all figures unless otherwise indicated. (C) ER stress induces SKN-1 recruitment along the xbp-1 gene. ChIP analysis is presented as in Figure 2. (D, E) Importance of skn-1 for expression of core UPR genes under basal (D) and TM-treatment (E) conditions, assayed by qRT-PCR. (F) Binding of SKN-1 to the ire-1 locus, analyzed by ChIP. (G) SKN-1-dependence of TM-induced hsp-4/BiP expression, assayed by qRT-PCR. (H) skn-1 mutants are sensitized to TM-induced ER stress. Survival of wild type and skn-1 mutant animals was assayed after 7 days of Control or high-dose TM treatment (35 µg/ml). Error bars represent SEM, and * p≤.05, ** p≤.01, *** p≤.001, relative to pL4440 Control calculated using student's t-test. See also Figure S3 and Table S2. SKN-1 was also required for expression of other core UPR genes. Mutation or RNAi knockdown of skn-1 prevented ER stress-induced expression of the unfolded protein chaperone and sensor HSP-4 (BiP) (Figure S1A)(Figures 3G, S3B, and S3C). Binding of SKN-1 at hsp-4 was not detected in the MOD-ENCODE study of L1 larvae [34], but our ChIP evidence indicated that both SKN-1 and XBP-1 bind directly to the hsp-4 locus (Figures S3D and S3E), which includes predicted SKN-1 binding sites (not shown). SKN-1 similarly contributed to expression of the core UPR factors pek-1 and atf-6 (Figures 3D and 3E). Our evidence that SKN-1 is important for transcriptional induction of core UPR signaling and regulatory factors predicts that it should be important for C. elegans survival under ER stress conditions. Treatment with TM at a 7-fold higher concentration (35 µg/ml) than is sufficient to induce the UPR impaired the survival of skn-1 mutants but not wild type animals (Figure 3H and Table S2). We conclude that SKN-1 plays a critical role in the UPR through its direct transcriptional regulation of core UPR factors, along with many downstream genes.
Activation of SKN-1 by ER Stress Independently of Oxidative Stress
We next examined whether expression of skn-1 itself is increased when the ER becomes stressed, and whether various conditions that cause ER stress affect SKN-1 activity. Treatment with TM increased the levels of multiple mRNA species that encode SKN-1 isoforms (Figure 4A and S4A). In addition, non-lethal treatment with either the Ca++ pump inhibitor thapsigargin (Thap) or the proteasome inhibitor Bortezomib upregulated transcription of skn-1, and various SKN-1-regulated genes (Figures 1, 4A, and S4B–S4C). Finally, knockdown of either the ER chaperone hsp-4 or the UPR transcription factor atf-6 resulted in transcriptional upregulation of skn-1 and many of its ER stress targets in the absence of drug treatment, presumably because of an elevated level of ER stress (Figures 4A, S4D and S4E). We conclude that skn-1 transcription and activity are increased in response to a variety of conditions that are associated with ER stress.
Fig. 4. ER stress activates SKN-1 independently of oxidative stress. 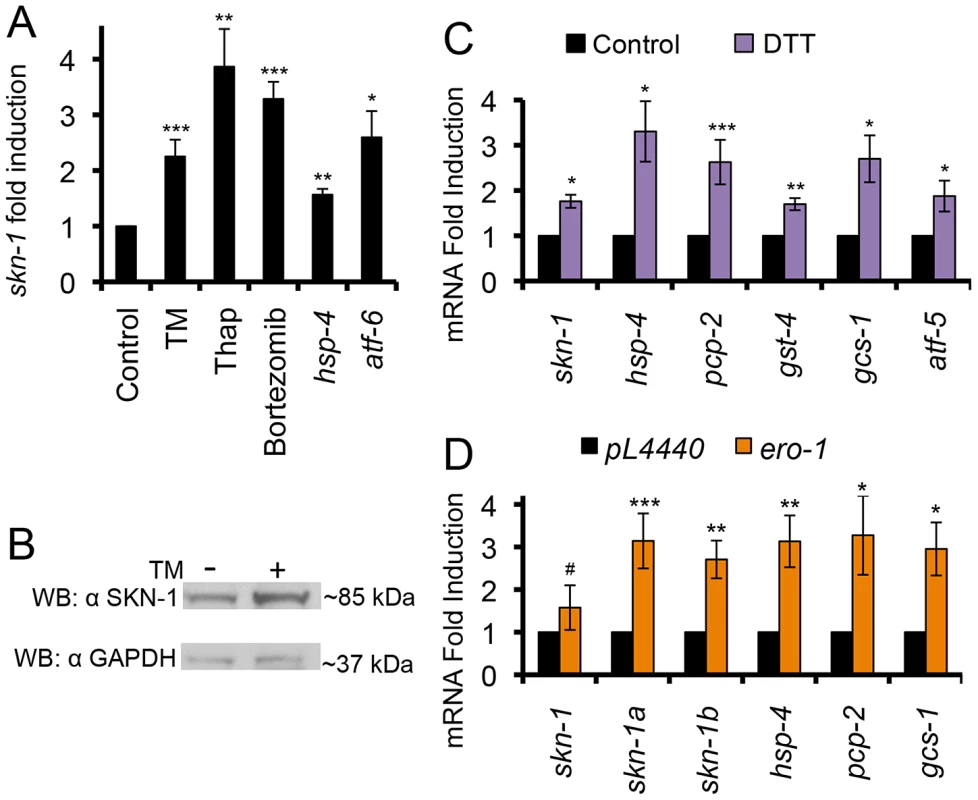
(A) Treatment with TM (16 hrs), thapsigargin (Thap, 2 hrs), or bortezomib (6 hrs) increased skn-1 mRNA levels, as determined by qRT-PCR. RNAi knockdown of hsp-4 or atf-6 also increased skn-1 mRNA levels. (B) Increased endogenous SKN-1 protein levels in response to TM-induced ER stress. SKN-1 was detected by Western blotting with the polyclonal antibody, with GAPDH serving as the loading control. (C) Induction of skn-1 expression and SKN-1-regulated UPR target genes by reductive ER stress (DTT treatment for 2 hrs), assayed by qRT-PCR. (D) Induction of the UPR, skn-1 expression, and SKN-1 target genes by ero-1 RNAi, assayed by qRT-PCR. Different primer sets were used to distinguish among mRNAs that correspond to different skn-1 isoforms. Error bars represent SEM, and * p≤.05, ** p≤.01, *** p≤.001, relative to pL4440 Control calculated using student's t-test. See also Figure S4 and Table S3. An important hallmark of the UPR is a decrease in the overall levels of translation [5], [6]. This relieves stress on the ER, and allows translation of atf4 and other protective genes to be maintained or even increased. We investigated whether SKN-1 translation is similarly “spared” under ER stress conditions. Supporting this idea, TM treatment increased SKN-1 protein levels, a trend that was observed in Western and IP-Western analyses of whole animals with two specific SKN-1 antibodies (Figures 4B and S4F–S4I). Based upon its size, this approximately 85 kD SKN-1 species is likely to represent SKN-1a, the largest SKN-1 isoform. While this size is larger than the expected SKN-1a MW of 70 kD, SKN-1 is phosphorylated and predicted to be glycosylated, as is characteristic of Nrf1 and Nrf3 (not shown) [17], [28], [38]–[40]. Our finding that SKN-1 protein levels are increased by ER stress is consistent with earlier evidence that SKN-1 translation seemed to be preserved when translation initiation was inhibited [41].
Prolonged ER stress leads to accumulation of reactive oxygen species (ROS) and induction of an oxidative stress response [15], [42], making it important to determine whether ER stress treatments might activate SKN-1 simply through a secondary response to oxidative stress. Arguing against this interpretation, even though SKN-1 is well known to defend against oxidative stress, we found that reductive ER stress also induced a SKN-1-dependent response. The reducing agent dithiothreitol (DTT) initiates the UPR through reduction of Cys-Cys bonds in the ER [43]. DTT treatment resulted in transcriptional induction of skn-1 and many of its target genes, and increased SKN-1 protein levels (Figures 4C and S4J). SKN-1 appeared to be required for its downstream targets to be activated by DTT-induced reductive stress (Fig. S4K), and knockdown of either skn-1 or hsp-4 rendered C. elegans comparably sensitive to reductive stress from DTT (Figure S4L and Table S3). Another way to reduce oxidation in the ER is through inhibiting expression of the oxidase ERO-1, which promotes Cys-Cys crosslinking [43]. ero-1 RNAi decreases ROS levels, initiates the UPR, and extends lifespan [15]. As observed with DTT, ero-1 RNAi transcriptionally activated skn-1 and several of its downstream targets (Figure 4D).
Additional lines of evidence support the idea that SKN-1 acts in the UPR independently of its role in oxidative stress defense. Many genes that are activated by SKN-1 under oxidative stress conditions were not upregulated by ER stress (Figures S1C and S4M). Oxidative stress from AS treatment induced the SKN-1::GFP (green fluorescent protein) fusion to accumulate to high levels in intestinal nuclei, as previously described (Inoue, et al., 2005), but this did not occur in response to ER stress (Figure S4N). Finally, we did not observe increased levels of oxidized proteins under conditions of TM-induced ER stress (Figure S4O). Taken together, the data show that ER stress directs SKN-1 to activate a specific set of its target genes independently of any secondary oxidative stress response.
Regulation of SKN-1 by UPR Factors
If ER signaling pathways regulate SKN-1, then key UPR signaling and transcription factors should be required for ER stress to activate SKN-1 and its target genes. Accordingly, RNAi or mutation of ire-1, atf-5, pek-1, or hsp-4 essentially prevented ER stress from inducing transcription of skn-1 and several of its target genes (Figure 5A). Knockdown of xbp-1 under control conditions increased background expression of some SKN-1 isoforms and target genes (skn-1b, pcp-2, gst-4, hsp-4), possibly because ER stress was increased, but also interfered with ER stress-induced activation of several of these genes (skn-1a, pcp-2, gcs-1, hsp-4) (Figure S5A). RNAi against ire-1, which is essential for XBP-1s expression [5], [6], also blocked TM-induced accumulation of SKN-1, Pol II, or P-Ser2 at the gst-4, pcp-2, and atf-5 loci (Figures 5B–5E, S5B and S5C). Knockdown of hsp-4 or pek-1 had a similar effect (Figure S5D–S5G). The evidence indicates that, in general, core UPR factors are required for ER stress to upregulate expression of SKN-1 and its target genes.
Fig. 5. UPR factors required for ER stress-induced SKN-1 activation. 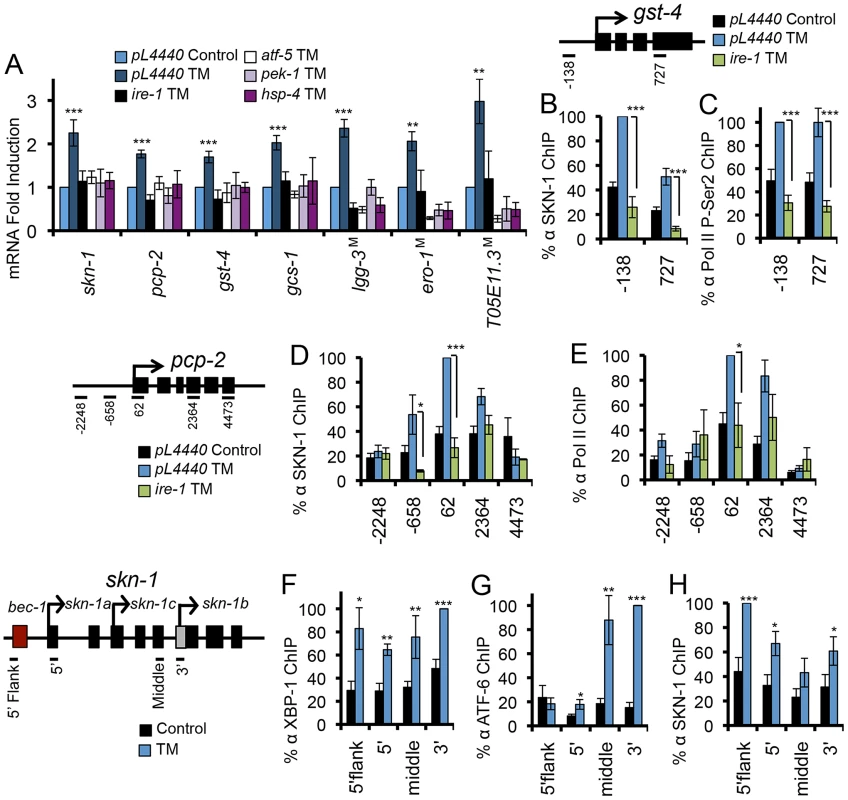
(A) ER stress-induced activation of skn-1 and its target genes requires core UPR factors. RNA levels were assayed by qRT-PCR after RNAi against core UPR genes or in core UPR factor mutants (indicated by M) after TM treatment. (B-E) IRE-1 is required for ER stress-induced SKN-1 accumulation and activity at SKN-1 target genes gst-4 and pcp-2. Presence of SKN-1 and transcription markers was assayed by ChIP as in Figure 2, and ire-1 was knocked down by RNAi. (F–H) Endogenous XBP-1 (F), ATF-6 (G), and SKN-1 (H) bind within the skn-1 gene locus in response to TM-induced ER stress, with binding assayed by ChIP. Multiple start sites are noted within the skn-1 locus. Error bars represent SEM, and * p≤.05, ** p≤.01, *** p≤.001 by student's t-test, relative to pL4440 Control unless otherwise indicated. See also Figure S5. The most straightforward mechanism through which ER stress could increase skn-1 transcription is through the direct regulation of skn-1 by one or more of the canonical UPR transcription factors. During the UPR, downstream gene transcription is controlled largely by XBP1 and ATF4, which may regulate each other directly, with ATF-6 playing a more specialized role [8], [15], [37]. The skn-1 locus contains possible XBP-1 and ATF-6/XBP-1 binding elements (not shown), and genome-wide ChIP studies suggest that mammalian Nrf3 may be a direct XBP1 target [37]. We determined that XBP-1 binds within the skn-1 locus in response to ER stress, suggesting direct regulation (Figure 5F), a remarkable parallel to the direct regulation of xbp-1 by SKN-1 (Figure 3C). Moreover, ATF-6 was also recruited to the skn-1 locus in response to ER stress (Figure 5G). In mammals, XBP-1 may regulate its own expression [37]. Our ChIP analysis indicated that SKN-1 also binds to its own locus with ER stress (Figure 5H), suggesting that SKN-1, XBP-1, and ATF-6 together regulate skn-1 transcription. ER stress also resulted in XBP-1 and ATF-6 recruitment to the direct SKN-1 targets pcp-2 and gst-4 (Figures S5H–S5K). Together, the evidence suggests that SKN-1, XBP-1, and ATF-6 may function together to regulate several downstream genes. We conclude that SKN-1 is transcriptionally integrated into the UPR, in which it functions upstream, downstream, and in parallel to the known core UPR transcription factors.
The mammalian SKN-1 orthologs Nrf1 and Nrf3 have been detected in association with the ER (see Introduction), raising the question of whether this might also be true for a proportion of SKN-1. Consistent with this idea, Nrf1 and the SKN-1a isoform each contain a predicted transmembrane domain [27] (Figure S6A). To investigate whether SKN-1 might be present at the ER, we asked whether it might be detected in association with the ER-resident chaperone BiP (HSP-3/-4)(Figure S1A). We performed co-immunoprecipitation (IP) analyses of intact worms that had been crosslinked with formaldehyde as in our ChIP experiments. These conditions capture direct and indirect in vivo interactions that occur within approximately 2 Å, and allow for high-stringency detergent and salt-based washings that minimize non-specific binding [44], [45]. Under both normal and ER stress conditions, association between HSP-4 and SKN-1 was readily detected by high-stringency IP performed in either direction (Figure 6A and 6B). As in Figure 4B, the size of this SKN-1 species suggested that it may correspond to SKN-1a. The data suggest that some SKN-1 may be produced at the ER and might remain associated with this organelle.
Fig. 6. Association of SKN-1 with the ER. 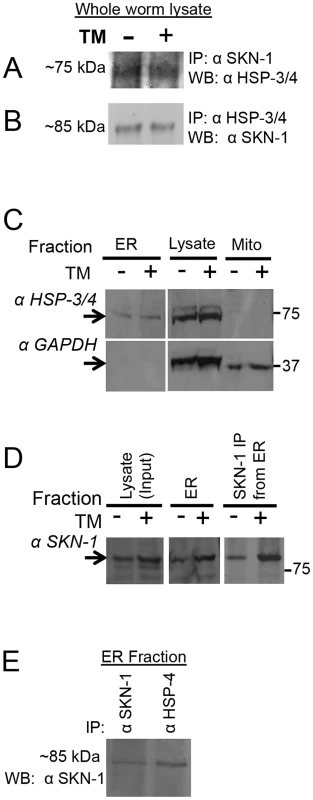
(A, B) Interaction between endogenous SKN-1 and HSP-3/4, detected by IP/Western. Lysates were prepared from animals in which proteins had been crosslinked under ChIP conditions. (A) Monoclonal αSKN-1 IP blotted with αHsc3 (HSP-3/4). (B) αHsc3 (HSP-3/4) IP blotted with monoclonal αSKN-1. (C–E) Analyses of ER fractions prepared from whole worms. The fractionation scheme is described in Fig. S6B. (C) Detection of endogenous HSP-3/4 and the cytoplasmic marker GAPDH in ER and Mitochondrial fractions, and total worm lysate. Note the enrichment of the ER marker HSP-3/4 compared to GAPDH in the ER fraction. TM indicates lysates from animals that had been treated with TM. (D) Presence of endogenous SKN-1 in the ER fraction, detected by western and IP/western blotting. Note that TM treatment increased the levels of SKN-1 protein. (E) Association between endogenous SKN-1 and HSP-3/4 within the ER fraction, detected with polyclonal αSKN-1 and αBiP (HSP-3/4), by IP/Western that was performed without crosslinking. Fractionations and analyses were performed independently twice, with similar results. See also Figure S6. Given that BiP has been found in other cellular locations besides the ER [46], we also investigated whether SKN-1 is present in a cellular fraction that is enriched for the ER (Figure S6B). SKN-1 was readily detectable in an ER fraction that included HSP-4, but not the cytoplasmic protein GAPDH (Figures 6C and 6D). The interaction between endogenous SKN-1 and HSP-4 was confirmed within this ER fraction by a co-IP that was performed without crosslinking (Figure 6E). Together, our findings suggest that the association of SKN-1/Nrf proteins with the ER is evolutionarily conserved.
SKN-1-Mediated Oxidative Stress Responses Depend upon ER Signaling
Our finding that UPR factors are required for SKN-1 activity to be increased under ER stress conditions raised a related question: might UPR-related mechanisms also be involved in SKN-1 responses to oxidative stress? Surprisingly, we found that RNAi or mutation of core UPR signaling and transcription factors (atf-5, pek-1, ire-1, hsp-4 and xbp-1) impaired oxidative stress (AS)-induced activation of several SKN-1 target genes, including skn-1 itself (Figures 7A, 7C, and S7A). Similarly, ire-1 RNAi attenuated activation of the gcs-1::GFP reporter in the intestine (Figure S7B). This impairment of the oxidative stress response is particularly striking because ire-1 RNAi actually increased oxidized protein levels, in contrast to the mild AS treatment conditions used for gene expression analyses (Figure S4O).
Fig. 7. Dependence of oxidative stress responses on UPR components. 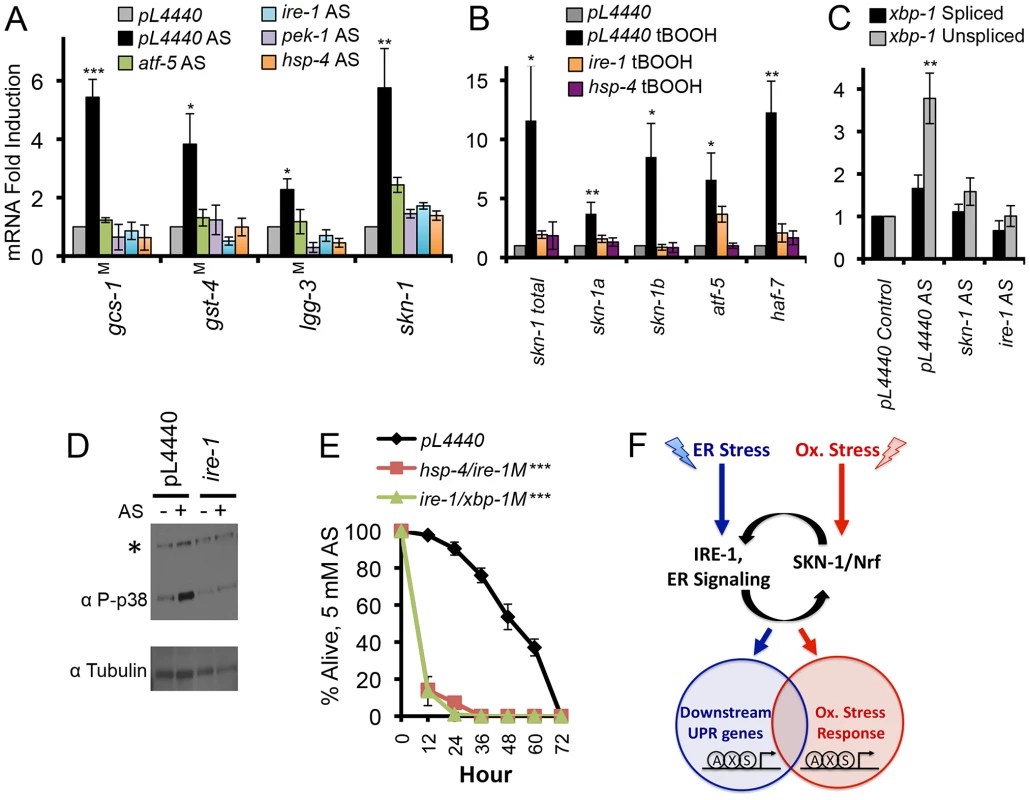
(A, B) Importance of core UPR genes for SKN-1-mediated oxidative stress responses. Induction of skn-1 and skn-1 target gene transcription by AS (A) or t-BOOH (B) was impaired by RNAi against core UPR genes or in core UPR factor mutants (indicated by M). qRT-PCR was performed after treatment with 5 mM AS for 1 hour, or 12 mM t-BOOH for 1 hour. (C) Accumulation of xbp-1 mRNA in response to AS-induced oxidative stress. Note the predominant increase in the unspliced form. (D) Dependence of AS-induced p38 phosphorylation on ire-1. Phosphorylated (active) p38 was assayed by phospho-specific antibody as in [38], and ire-1 expression was knocked down by RNAi. *background signal. (E) UPR factors are required for oxidative stress defense. Survival of AS treatment (5 mM) was scored in RNAi Control, hsp-4(RNAi)/ire-1(zc14), and ire-1(RNAi)/xbp-1(SJ17) animals (M indicates mutant). Error bars represent SEM, and * p≤.05, ** p≤.01, *** p≤.001, relative to pL4440 Control calculated using student's t-test. (F) Functional integration of the ER and oxidative stress responses through SKN-1 and canonical UPR components (see text). SKN-1 is essential for the UPR because it directly controls transcription of most UPR signaling and transcription factors. These UPR factors in turn regulate SKN-1 expression, and function in concert with SKN-1 at downstream targets. This is shown arbitrarily as SKN-1 (S) binding to target promoters together with XBP-1 (X) and ATF- 6 (A). SKN-1 and mammalian Nrf proteins are present in the ER, suggesting a possible signaling role. UPR factors are required not only for SKN-1 to function in the context of the UPR, but also for SKN-1 to mobilize distinct oxidative stress responses. See also Figure S7 and Table S4. Importantly, oxidative stress from AS did not simply activate the canonical UPR. Many SKN-1-regulated genes that were induced by oxidative stress were not upregulated by ER stress, and vice-versa (Figures S1C, S4M, and S7C). This shows that SKN-1 mobilizes distinct transcriptional responses to oxidative and ER stress, even if these responses overlap to an extent. Moreover, AS primarily increased accumulation of the unspliced xbp-1 mRNA form (xbp-1u), in striking contrast to the increase in xbp-1s levels that is characteristic of ER stress (Figures 3A and 7C). Treatment with the oxidative stressor tert-butyl hydrogen peroxide (tBOOH) induces a SKN-1-dependent response that overlaps with the AS response, but includes SKN-1-independent activation of many genes that are otherwise SKN-1-dependent [21]. Knockdown of ire-1 or hsp-4 inhibited tBOOH from upregulating skn-1 and some SKN-1 targets (Figure 7B), but did not eliminate activation of other genes (gcs-1, sdz-8, and gst-10; not shown). The data suggest that core UPR factors are needed for SKN-1 to function properly under oxidative stress conditions, in addition to the setting of ER stress.
The extensive regulatory integration that exists among UPR transcription factors, as described by others and in this study (Figures 7A, 7B, and S7A) [8], [15], [37], could explain why multiple UPR-associated signaling and transcription factors are needed for skn-1 expression to be increased in response to oxidative stress. However, we considered that the UPR might also influence SKN-1 regulation at a post-translational level. In the C. elegans intestine SKN-1 is predominantly cytoplasmic under normal conditions, but accumulates in nuclei in response to oxidative stress from AS treatment [38]. This nuclear accumulation was dramatically reduced in animals that had been exposed to ire-1 RNAi (Figure S7D). The presence of SKN-1 in intestinal nuclei is dependent upon its phosphorylation by the p38 kinase, which is activated by oxidative stress [23], [38], [47]. The IRE-1 kinase activity transmits signals through the JNK and p38 MAPK pathways [6], [48]–[50], and we determined that ire-1 knockdown largely prevented the increase in p38 signaling that occurs in response to oxidative stress (Figures 7D and S7D). Taken together, these data suggest that IRE-1 is required for oxidative stress to activate SKN-1 post-translationally.
If UPR signaling and transcription factors are required for SKN-1 to mobilize appropriate oxidative stress responses, then oxidative stress sensitivity should be increased when these canonical UPR factors are lacking. Accordingly, RNAi or mutation of these genes significantly increased sensitivity to oxidative stress from exposure to AS, paraquat, or t-BOOH (Figures 7E, S7E, and S7F; Table S4). We conclude that signaling from the ER is required for SKN-1 to respond to oxidative stress, and therefore that UPR-mediated regulation of SKN-1 plays a central role in the homeostatic integration of ER and oxidative stress responses.
Discussion
SKN-1 Is a Critical UPR Factor
It is well-established that the canonical UPR transcription factors XBP1, ATF4, and ATF6 control overlapping sets of downstream genes and processes [5], [6], but much less is known about how their responses to ER stress might be integrated with other mechanisms that maintain cellular stress defense and homeostasis. We have determined that the oxidative/xenobiotic stress response regulator SKN-1/Nrf functions as a fourth major UPR transcription factor in C. elegans. Without SKN-1, ER stress failed to increase the expression of core UPR signaling and transcription factors, many of which are regulated directly by SKN-1 (ire-1, xbp-1, atf-5, and hsp-4; Figures 1, 2, 3 and S3). It was particularly striking that SKN-1 was disproportionally required for production of spliced xbp-1 mRNA (xbp-1s), presumably because of its importance for IRE-1 expression (Figures 3D–F). SKN-1 was also needed for ER stress to upregulate numerous genes that are known or predicted to be involved in various ER - or UPR-related processes, including ER homeostasis (ero-1, pdi-2), chaperone-mediated protein folding (hsp-3, hsp-4, dnj-28, T05E11.3 (HSP-90/GRP94)), autophagy (lgg-1, lgg-3), calcium homeostasis (sca-1, crt-1), ER membrane integrity (ckb-4), and a pathway that defends against ER stress when the canonical UPR is blocked (abu-8, abu-11 [51]) (Figure 1, 3G and Table S1). Together, our data indicate that SKN-1 regulates transcription of essentially the entire core UPR apparatus and many downstream ER stress defense genes in vivo.
We were surprised to find that SKN-1 was so broadly important for UPR transcription events. A trivial explanation for our findings would be that skn-1 mutants did not need to induce the UPR robustly because they were resistant to ER stress. This explanation was ruled out, however, by our finding that skn-1 mutants are actually sensitized to ER stress from diverse sources (Figures 3H and S4L). Importantly, our ChIP studies and MOD-ENCODE data [34] indicate that SKN-1 controls many core and downstream UPR genes directly by binding to their promoters (Figures 2, 3, and S3E, Table S1). We also found that ER stress induces SKN-1, XBP-1, and ATF-6 to bind promoters directly to regulate many of the same genes, including skn-1 itself (Figures 5, S3, and S5). In addition, under ER stress conditions, UPR signaling increased levels of skn-1 mRNA and protein (Figures 4 and S4), indicating that SKN-1 is controlled by the UPR and is an active participant in this response. Together, our data reveal that a remarkable degree of regulatory and functional integration exists between SKN-1 and the three canonical UPR transcription factors (Figures 7F and S1A).
Although ER stress increases skn-1-dependent transcription and SKN-1 occupancy at several downstream gene promoters, it did not detectably alter the overall levels of SKN-1 in intestinal nuclei, at least as indicated by levels of a transgenic GFP fusion protein (Figure S4N). While this might seem paradoxical, we observed a similar situation with reduced TORC1 signaling [19]. Under conditions of low TORC1 activity SKN-1 target genes were activated in a skn-1-dependent manner, and this was accompanied by increased SKN-1 binding to their promoters, but not by an obvious increase in the bulk levels of SKN-1 in nuclei. Our finding that SKN-1 binds to downstream UPR genes together with other UPR transcription factors suggests a paradigm that could explain this phenomenon. If SKN-1 binds cooperatively with UPR factors or other co-regulators to some of its targets, this could shift the binding equilibrium to allow those targets to be activated by SKN-1 that is already present in the nucleus, without it being necessary to “flood” the nucleus with higher levels of SKN-1. This scheme might be important for fine-tuning of SKN-1 downstream functions, and for allowing SKN-1 to activate different targets in different situations, as we have observed in this study.
In performing these analyses, we were mindful of the concern that the involvement of SKN-1 in the UPR might derive from its possible role in a secondary oxidative stress response. Several lines of evidence argued against this interpretation. For example, the direct involvement of SKN-1 in regulating multiple core UPR signaling and transcription factors during the UPR (Figures 3 and S3) is not consistent with its UPR functions deriving simply from a secondary oxidative stress response. Moreover, under our ER stress conditions SKN-1 was required for accumulation of the spliced form of the xbp-1 mRNA, whereas oxidative stress increased levels of the unspliced xbp-1 message (Figures 3A, 3B, and 7C). It was particularly striking that SKN-1 defended against reductive ER stresses (Figures 4C, 4D, S4J, S4K, and S4L), given the extensively described role of SKN-1/Nrf proteins in oxidative stress responses. These last observations indicated that SKN-1 defends against ER stress per se, and not only against oxidative conditions. Importantly, ER stress and the UPR directed SKN-1 to activate some of its target genes that are induced by oxidative stress, but not others (Figure S1C and S4M). On the other hand, many genes that SKN-1 activated under ER stress conditions were not induced by oxidative stress (Figure S7C). Taken together, the data show that SKN-1 does not simply activate oxidative stress defenses in the context of ER stress, but orchestrates a specific transcriptional ER stress response that is integrated into the broader UPR.
Our finding that SKN-1 mobilizes overlapping but distinct responses to ER and oxidative stress defines a new function for this surprisingly versatile transcription factor. It also supports our model that SKN-1/Nrf proteins do not control the same genes under all circumstances, but instead induce protective responses that are customized to the challenge at hand [19], [26]. The idea that SKN-1 works together with canonical UPR transcription factors at downstream genes may provide a model for understanding how particular SKN-1 functions can be mobilized under different conditions, if these proteins and other SKN-1 “partners” guide its activities.
Consistent with reports that Nrf1 and Nrf3 are present at the ER [27]–[30], we found that some SKN-1 also localizes to the ER. We detected association between SKN-1 and the ER chaperone HSP-3/4 (BiP) in crosslinking analyses of intact animals, the presence of SKN-1 within an ER fraction, and association between SKN-1 and HSP-3/4 within that fraction (Figure 6 and S6). Each of these experiments involved analysis of endogenous proteins. These strategies would have detected either direct or indirect interactions, so they do not demonstrate that SKN-1 binds directly to HSP-3/4 (BiP), but they do show that these proteins reside very close to each other at the ER. Apparently, association between SKN-1/Nrf proteins and the ER is evolutionarily conserved. The example of ATF-6, which is activated through cleavage in the Golgi (Figure S1A), predicts that ER-associated SKN-1 might have a signaling function in which it is cleaved in response to ER stress. However, the relative instability of SKN-1 and the presence of smaller isoforms have so far confounded the resolution of this question (not shown). We recently determined that some SKN-1 also localizes to mitochondria and that SKN-1 can promote a starvation-like state when overexpressed, a function that also appears to be conserved in Nrf proteins [26]. Given the extensive communication between the ER and mitochondria [4], [52], our results suggest that SKN-1/Nrf might respond directly to the status of each of these organelles. Consistent with this notion, SKN-1 is required for expression of the C. elegans ortholog of mitofusin (fzo-1) (Figure 1A), which mediates mitochondrial fusion and mitochondria-ER interactions [4].
Taken together, our findings show that processes controlled by SKN-1/Nrf proteins are critical for ER stress defense and homeostasis, and that SKN-1 is extensively intertwined with the UPR in vivo. While differences could exist between C. elegans and mammals with respect to regulatory networks, the extent of the functional interactions we have observed predicts that mammalian Nrf proteins are likely to play an important role in the UPR that is distinct from their familiar function in oxidative stress responses.
Regulation of Oxidative Stress Responses by the UPR
Perhaps our most surprising finding was that core UPR signaling and transcription factors were required for SKN-1 to mount a transcriptional response to oxidative stress (Figures 7 and S7). Cooperative interactions between SKN-1 and UPR transcription factors could account for some of these findings, through their effects on SKN-1 expression, but it was striking that ire-1 was needed for AS to induce SKN-1 nuclear accumulation, a phenomenon that does not occur under ER stress conditions (Figures S4N and S7D). Moreover, ire-1 was required for the AS-induced p38 signal that is needed for SKN-1 to be present in nuclei (Figure 7D). These last findings indicate that IRE-1 affects the oxidative stress response at a step upstream of SKN-1. One speculative possibility for further investigation is that the IRE-1 kinase activity might be needed to initiate the oxidative stress-induced p38 signal. Together, our data show that signaling from the ER is required to “license” the oxidative/xenobiotic stress response, and suggest that the ER might function in effect as a stress sensor. This importance of the UPR for SKN-1 activity may have implications for our understanding of aging and longevity assurance. SKN-1/Nrf not only defends against resistance to various stresses, but is also important in pathways that affect longevity, including insulin-like, TORC1, and TORC2 signaling, and dietary restriction [16], [17], [19], [20]. IRE-1 and XBP-1 have each been implicated in longevity [53], [54], making it important to determine the extent to which these UPR-based mechanisms might influence aging through regulation of SKN-1/Nrf and its functions.
Why would such extensive integration have arisen, in which SKN-1/Nrf is essential for the UPR, and signaling from the ER is needed for SKN-1/Nrf activities that are distinct from the UPR (Figure 7F)? SKN-1/Nrf controls cellular processes that profoundly influence the ER. Its target genes drive synthesis of glutathione, the major redox buffer within the ER, and encode many endobiotic and xenobiotic metabolism enzymes that reside on or within the smooth ER (Table S1) [20], [21], [55]. Under some circumstances SKN-1/Nrf also regulates proteasome expression and activity, and numerous chaperone genes [20], [21], [23]–[25]. One possibility is that the influence of SKN-1 could attune the UPR to events taking place in the cytoplasm. It might be advantageous to mount a robust transcriptional UPR if the cytoplasm is under duress, for example, and to moderate the UPR when cytoplasmic stress is low. Under these conditions, SKN-1 activity would be relatively high and low, respectively. SKN-1 activity is also comparatively low when translation rates are high [19], [23]. If the ER becomes stressed under growth conditions it might be useful to limit the transcriptional UPR initially, because a reduction in translation rates might largely suffice to restore homeostasis. Again, under these conditions low SKN-1 activity could act as a brake on the transcriptional UPR. With respect to the oxidative/xenobiotic stress response, it could be important for the ER to have a “vote” on its intensity, given the profound influence of SKN-1/Nrf on cellular redox status and resources devoted to the ER. It seems likely, therefore, that the ER not only manages its own homeostasis, but through SKN-1/Nrf has a broader impact on cellular stress defense networks that is likely to be critical in their normal and pathological functions.
Materials and Methods
Gene Expression Analysis
For each condition studied, RNA was extracted from approximately 100 µl of packed mixed-stage worms that were collected in M9 at the indicated time point. To induce UPR-associated gene expression, at day three of adulthood worms were treated with 5 µg/ml TM (Sigma) for 16 hours [15], or at day four with 5 mM DTT (Sigma) [54] for two hours, 5 µM thapsigargin (Enzo) [56] for two hours, or 5 µM Bortezomib (proteasome inhibitor, LC Labs) for six hours (similar to published C. elegans MG132 proteasome inhibitor treatment [57]). In each case, these treatments were non-lethal. For arsenite (AS) and tBOOH exposure, up to 100 µl of packed worms were collected and nutated in 5 mM AS or 12 mM tBOOH for 1 hour (a non-lethal duration). Each of these treatments was performed in a volume of 1 ml, and was followed by pelleting. RNA was analyzed by qRT-PCR as described, with values normalized to an internal standard curve for each amplicon [19], [44]. The same treatment conditions were used for ChIP experiments.
Transgenic Reporter Scoring
Expression or nuclear accumulation of transgenic GFP proteins was scored as “low,” “medium,” or “high” essentially as published [19], or were quantified using ImageJ 1.45S.
ChIP Lysates and Analysis
ChIP was performed essentially as described [19], [44]. 2 ml of packed mixed-stage worms were crosslinked with formaldehyde at room temperature for 20 minutes. After quenching, lysis, and determination of protein concentration, 1 mg/ml samples were frozen as aliquots at −80°C. The resolution of the assay was approximately 250–500 bp [44]. The monoclonal antibody FC4 [58] was used for SKN-1 ChIP experiments, as in previous ChIP analyses [19]. Other antibodies are described in the Supplemental Experimental Procedures. Analyses of intergenic regions and control genes (not shown) indicated that average signals of 14%, 11%, 26%, 4%, 11%, 7%, and 8% represent thresholds for specific presence of SKN-1, Pol II, PSer2, and H3-AcK56, XBP-1, ATF-6, and Histone H3 respectively.
ER Fractionation
Worms from five confluent 20 cm2 plates were collected in M9 with or without TM treatment (5 µg/ml) for 16 hours, in order to generate 2× 1 ml of packed mixed-stage animals. Worms were sonicated 3× for 20 seconds in homogenization buffer (supplied by IMGENEX kit, supplemented with HDAC inhibitors, protease inhibitors, phosphatase inhibitors, and MG132) with the Branson midiprobe 4900 Sonifer before fractionation with the IMGENEX Endoplasmic Reticulum Enrichment Kit (Cat No. 10088K) [59]. Mitochondrial and ER fractions were washed 3× with 1 ml PBS and resuspended in 400 µl PBS (supplemented with HDAC, protease, and phosphatase inhibitors and MG132). Up to 100 µl of the ER or cytoplasmic fractions were used for each IP.
Immunoprecipitation and Western Blotting
Controls for a polyclonal rabbit antiserum raised against SKN-1c (JDC7, referred to as pSKN-1) are shown in Figures S4F–S4J. HSP-3/4/BiP was detected with either C-terminal Drosophila Hsc3 [60] (Figures 6A and 6B) or N-terminal human BiP antibody (Sigma et21) [61], [62] (Figures 6C and 6E). Note that both BiP antibodies recognized the same 75 kD band. ATF-6 (Abcam ab11909), Tubulin (Sigma #9026), and GAPDH (Santa Cruz sc25778) antibodies were also used. Phosphorylated p38 was detected using an antibody from Cell Signaling T180/Y182 as described previously [23]. For Western blotting, antibodies were used at the following dilution: 1∶200 FC4 monoclonal αSKN-1, 1∶200 polyclonal αSKN-1, 1∶1000 αPol II, and 1∶1000 for αHsc3. All other antibodies were used at manufacturer's recommended concentrations.
For IPs, the indicated antibodies (50 µl FC4 monoclonal αSKN-1 or polyclonal αSKN-1,10 µl Hsc3 (BiP) or 20 µl BiP (Sigma)) and pre-blocked Salmon Sperm DNA/Protein A beads (Zymed) were added to lysates or samples from the fractionation described above. The final volume was brought to 500 µl in 1× PIC, 1× PMSF, and 1∶1000 MG132 diluted in 1× PBS. Samples were nutated overnight at 4°C and washed three times for 5 minutes at 4°C the next day with NP-40 wash buffer. Beads were spun down at 3000 rpm and resuspended in 4× SDS Laemmli Buffer. Samples were boiled for 15 minutes with 20 µl β-mercaptoethanol and 50 µl 4× SDS Laemmli. Samples were loaded (50 µl each) onto NuPAGE Novex Bis-Tris 10% Gels. Pierce ECL or Femto Western Blotting Substrate was used for detection.
Other methods are available in Text S1 (Supplementary Materials and Methods).
Supporting Information
Zdroje
1. GorlachA, KlappaP, KietzmannT (2006) The endoplasmic reticulum: folding, calcium homeostasis, signaling, and redox control. Antioxid Redox Signal 8 : 1391–1418.
2. SchroderM (2008) Endoplasmic reticulum stress responses. Cell Mol Life Sci 65 : 862–894.
3. CsordasG, HajnoczkyG (2009) SR/ER-mitochondrial local communication: calcium and ROS. Biochim Biophys Acta 1787 : 1352–1362.
4. SimmenT, LynesEM, GessonK, ThomasG (2010) Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM). Biochim Biophys Acta 1798 : 1465–1473.
5. WalterP, RonD (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 : 1081–1086.
6. HetzC (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13 : 89–102.
7. RichardsonCE, KinkelS, KimDH (2011) Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet 7: e1002391.
8. ShenX, EllisRE, SakakiK, KaufmanRJ (2005) Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet 1: e37.
9. RutkowskiDT, HegdeRS (2010) Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol 189 : 783–794.
10. MalhotraJD, KaufmanRJ (2007) The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol 18 : 716–731.
11. WangS, KaufmanRJ (2012) The impact of the unfolded protein response on human disease. J Cell Biol 197 : 857–867.
12. FuS, WatkinsSM, HotamisligilGS (2012) The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab 15 : 623–634.
13. TuBP, WeissmanJS (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164 : 341–346.
14. SevierCS, KaiserCA (2008) Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta 1783 : 549–556.
15. HardingHP, ZhangY, ZengH, NovoaI, LuPD, et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11 : 619–633.
16. BishopNA, GuarenteL (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447 : 545–549.
17. TulletJM, HertweckM, AnJH, BakerJ, HwangJY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 : 1025–1038.
18. SteinbaughMJ, SunLY, BartkeA, MillerRA (2012) Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am J Physiol Endocrinol Metab 303: E488–495.
19. Robida-StubbsS, Glover-CutterK, LammingDW, MizunumaM, NarasimhanSD, et al. (2012) TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15 : 713–724.
20. SykiotisGP, BohmannD (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell 14 : 76–85.
21. OliveiraRP, Porter AbateJ, DilksK, LandisJ, AshrafJ, et al. (2009) Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8 : 524–541.
22. HochmuthCE, BiteauB, BohmannD, JasperH (2011) Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8 : 188–199.
23. LiX, MatilainenO, JinC, Glover-CutterKM, HolmbergCI, et al. (2011) Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet 7: e1002119.
24. SteffenJ, SeegerM, KochA, KrugerE (2010) Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell 40 : 147–158.
25. RadhakrishnanSK, LeeCS, YoungP, BeskowA, ChanJY, et al. (2010) Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38 : 17–28.
26. PaekJ, LoJ, NarasimhanSD, NguyenT, Glover-CutterK, et al. (2012) Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab 16 : 526–537.
27. WangW, ChanJY (2006) Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem 281 : 19676–19687.
28. ChevillardG, BlankV (2011) NFE2L3 (NRF3): the Cinderella of the Cap‘n’Collar transcription factors. Cell Mol Life Sci 68 : 3337–3348.
29. ZhangY, KobayashiA, YamamotoM, HayesJD (2009) The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol Chem 284 : 3195–3210.
30. ZhangY, HayesJD (2010) Identification of topological determinants in the N-terminal domain of transcription factor Nrf1 that control its orientation in the endoplasmic reticulum membrane. Biochem J 430 : 497–510.
31. CullinanSB, DiehlJA (2004) PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279 : 20108–20117.
32. CullinanSB, ZhangD, HanninkM, ArvisaisE, KaufmanRJ, et al. (2003) Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol 23 : 7198–7209.
33. AnJH, BlackwellTK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17 : 1882–1893.
34. NiuW, LuZJ, ZhongM, SarovM, MurrayJI, et al. (2011) Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21 : 245–254.
35. BentleyDL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17 : 251–256.
36. LiB, CareyM, WorkmanJL (2007) The role of chromatin during transcription. Cell 128 : 707–719.
37. Acosta-AlvearD, ZhouY, BlaisA, TsikitisM, LentsNH, et al. (2007) XBP1 controls diverse cell type - and condition-specific transcriptional regulatory networks. Mol Cell 27 : 53–66.
38. InoueH, HisamotoN, AnJH, OliveiraRP, NishidaE, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 : 2278–2283.
39. AnJH, VranasK, LuckeM, InoueH, HisamotoN, et al. (2005) Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A 102 : 16275–16280.
40. ZhangY, LucocqJM, HayesJD (2009) The Nrf1 CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem J 418 : 293–310.
41. RogersAN, ChenD, McCollG, CzerwieniecG, FelkeyK, et al. (2011) Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab 14 : 55–66.
42. HigaA, ChevetE (2012) Redox signaling loops in the unfolded protein response. Cell Signal 24 : 1548–1555.
43. PollardMG, TraversKJ, WeissmanJS (1998) Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1 : 171–182.
44. Glover-CutterK, KimS, EspinosaJ, BentleyDL (2008) RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15 : 71–78.
45. NadeauOW, CarlsonGM (2007) Protein interactions captured by chemical cross-linking: one-step cross-linking with formaldehyde. CSH Protoc 2007: pdb prot4634.
46. LynesEM, SimmenT (2011) Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta 1813 : 1893–1905.
47. WangJ, Robida-StubbsS, TulletJM, RualJF, VidalM, et al. (2010) RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet 6: e1001048 doi:10.1371/journal.pgen.1001048
48. UranoF, BertolottiA, RonD (2000) IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci 113 Pt 21 : 3697–3702.
49. NguyenDT, KebacheS, FazelA, WongHN, JennaS, et al. (2004) Nck-dependent activation of extracellular signal-regulated kinase-1 and regulation of cell survival during endoplasmic reticulum stress. Mol Biol Cell 15 : 4248–4260.
50. ChoiCH, JungYK, OhSH (2010) Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol 78 : 114–125.
51. UranoF, CalfonM, YonedaT, YunC, KiralyM, et al. (2002) A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol 158 : 639–646.
52. KornmannB, WalterP (2010) ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci 123 : 1389–1393.
53. ChenD, ThomasEL, KapahiP (2009) HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet 5: e1000486.
54. Henis-KorenblitS, ZhangP, HansenM, McCormickM, LeeSJ, et al. (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A 107 : 9730–9735.
55. CribbAE, PeyrouM, MuruganandanS, SchneiderL (2005) The endoplasmic reticulum in xenobiotic toxicity. Drug Metab Rev 37 : 405–442.
56. SasagawaY, YamanakaK, OguraT (2007) ER E3 ubiquitin ligase HRD-1 and its specific partner chaperone BiP play important roles in ERAD and developmental growth in Caenorhabditis elegans. Genes Cells 12 : 1063–1073.
57. OrsbornAM, LiW, McEwenTJ, MizunoT, KuzminE, et al. (2007) GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development 134 : 3383–3392.
58. BowermanB, DraperBW, MelloC, PriessJ (1993) The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74 : 443–452.
59. OhtaE, ItohT, NemotoT, KumagaiJ, KoSB, et al. (2009) Pancreas-specific aquaporin 12 null mice showed increased susceptibility to caerulein-induced acute pancreatitis. Am J Physiol Cell Physiol 297: C1368–1378.
60. RyooHD, DomingosPM, KangMJ, StellerH (2007) Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J 26 : 242–252.
61. LaiCW, AronsonDE, SnappEL (2010) BiP availability distinguishes states of homeostasis and stress in the endoplasmic reticulum of living cells. Mol Biol Cell 21 : 1909–1921.
62. BuchkovichNJ, MaguireTG, PatonAW, PatonJC, AlwineJC (2009) The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J Virol 83 : 11421–11428.
63. PincusD, ChevalierMW, AragonT, van AnkenE, VidalSE, et al. (2010) BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415.
64. BonillaM, NastaseKK, CunninghamKW (2002) Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J 21 : 2343–2353.
65. BolloM, ParedesRM, HolsteinD, ZheleznovaN, CamachoP, et al. (2010) Calcineurin interacts with PERK and dephosphorylates calnexin to relieve ER stress in mammals and frogs. PLoS One 5: e11925.
66. KapulkinWJ, HiesterBG, LinkCD (2005) Compensatory regulation among ER chaperones in C. elegans. FEBS Lett 579 : 3063–3068.
67. LewisMJ, SweetDJ, PelhamHR (1990) The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell 61 : 1359–1363.
68. NovoaI, ZengH, HardingHP, RonD (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol 153 : 1011–1022.
69. ChetyrkinSV, HuJ, GoughWH, DumaualN, KedishviliNY (2001) Further characterization of human microsomal 3alpha-hydroxysteroid dehydrogenase. Arch Biochem Biophys 386 : 1–10.
70. IchishitaR, TanakaK, SugiuraY, SayanoT, MiharaK, et al. (2008) An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. J Biochem 143 : 449–454.
71. InadaM, GuthrieC (2004) Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc Natl Acad Sci U S A 101 : 434–439.
72. RzymskiT, MilaniM, PikeL, BuffaF, MellorHR, et al. (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29 : 4424–4435.
73. KourokuY, FujitaE, TanidaI, UenoT, IsoaiA, et al. (2007) ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 14 : 230–239.
74. McKayRM, McKayJP, AveryL, GraffJM (2003) C. elegans: a model for exploring the genetics of fat storage. Dev Cell 4 : 131–142.
75. EschenlauerSC, PageAP (2003) The Caenorhabditis elegans ERp60 homolog protein disulfide isomerase-3 has disulfide isomerase and transglutaminase-like cross-linking activity and is involved in the maintenance of body morphology. J Biol Chem 278 : 4227–4237.
76. KornmannB, CurrieE, CollinsSR, SchuldinerM, NunnariJ, et al. (2009) An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325 : 477–481.
77. LeeW, KimKR, SingaraveluG, ParkBJ, KimDH, et al. (2006) Alternative chaperone machinery may compensate for calreticulin/calnexin deficiency in Caenorhabditis elegans. Proteomics 6 : 1329–1339.
78. LynesEM, BuiM, YapMC, BensonMD, SchneiderB, et al. (2012) Palmitoylated TMX and calnexin target to the mitochondria-associated membrane. EMBO J 31 : 457–470.
79. Radominska-PandyaA, CzernikPJ, LittleJM, BattagliaE, MackenziePI (1999) Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev 31 : 817–899.
80. SriburiR, JackowskiS, MoriK, BrewerJW (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol 167 : 35–41.
81. NakajimaS, HiramatsuN, HayakawaK, SaitoY, KatoH, et al. (2011) Selective abrogation of BiP/GRP78 blunts activation of NF-kappaB through the ATF6 branch of the UPR: involvement of C/EBPbeta and mTOR-dependent dephosphorylation of Akt. Mol Cell Biol 31 : 1710–1718.
82. ChetyrkinSV, BelyaevaOV, GoughWH, KedishviliNY (2001) Characterization of a novel type of human microsomal 3alpha -hydroxysteroid dehydrogenase: unique tissue distribution and catalytic properties. J Biol Chem 276 : 22278–22286.
83. HapalaI, MarzaE, FerreiraT (2011) Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol Cell 103 : 271–285.
84. UranoF, WangX, BertolottiA, ZhangY, ChungP, et al. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287 : 664–666.
85. XiaR, WebbJA, GnallLLM, CutlerK, AbramsonJJ (2003) Skeletal muscle sarcoplasmic reticulum contains a NADH-dependent oxidase that generates superoxide. Am J Physiol: Cell Physiol 285: C215–C221.
86. SantosCX, TanakaLY, WosniakJ, LaurindoFR (2009) Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid Redox Signal 11 : 2409–2427.
87. BarbeL, LundbergE, OksvoldP, SteniusA, LewinE, et al. (2007) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7 : 499–508.
88. LochnitG, GeyerR (2003) Evidence for the presence of the Kennedy and Bremer - Greenberg pathways in Caenorhabditis elegans. Acta Biochim Pol 50 : 1239–1243.
89. HardingHP, ZhangY, RonD (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397 : 271–274.
90. CalfonM, ZengH, UranoF, TillJH, HubbardSR, et al. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415 : 92–96.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



