-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
Animals have many ways of protecting themselves against stress; for example, they can induce animal-wide, stress-protective pathways and they can kill damaged cells via apoptosis. We have discovered an unexpected regulatory relationship between these two types of stress responses. We find that C. elegans mutations blocking the normal course of programmed cell death and clearance confer animal-wide resistance to a specific set of environmental stressors; namely, ER, heat and osmotic stress. Remarkably, this pattern of stress resistance is induced by mutations that affect cell death in different ways, including ced-3 (cell death defective) mutations, which block programmed cell death, ced-1 and ced-2 mutations, which prevent the engulfment of dying cells, and progranulin (pgrn-1) mutations, which accelerate the clearance of apoptotic cells. Stress resistance conferred by ced and pgrn-1 mutations is not additive and these mutants share altered patterns of gene expression, suggesting that they may act within the same pathway to achieve stress resistance. Together, our findings demonstrate that programmed cell death effectors influence the degree to which C. elegans tolerates environmental stress. While the mechanism is not entirely clear, it is intriguing that animals lacking the ability to efficiently and correctly remove dying cells should switch to a more global animal-wide system of stress resistance.
Published in the journal: . PLoS Genet 9(9): e32767. doi:10.1371/journal.pgen.1003714
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003714Summary
Animals have many ways of protecting themselves against stress; for example, they can induce animal-wide, stress-protective pathways and they can kill damaged cells via apoptosis. We have discovered an unexpected regulatory relationship between these two types of stress responses. We find that C. elegans mutations blocking the normal course of programmed cell death and clearance confer animal-wide resistance to a specific set of environmental stressors; namely, ER, heat and osmotic stress. Remarkably, this pattern of stress resistance is induced by mutations that affect cell death in different ways, including ced-3 (cell death defective) mutations, which block programmed cell death, ced-1 and ced-2 mutations, which prevent the engulfment of dying cells, and progranulin (pgrn-1) mutations, which accelerate the clearance of apoptotic cells. Stress resistance conferred by ced and pgrn-1 mutations is not additive and these mutants share altered patterns of gene expression, suggesting that they may act within the same pathway to achieve stress resistance. Together, our findings demonstrate that programmed cell death effectors influence the degree to which C. elegans tolerates environmental stress. While the mechanism is not entirely clear, it is intriguing that animals lacking the ability to efficiently and correctly remove dying cells should switch to a more global animal-wide system of stress resistance.
Introduction
In nature, animals are constantly exposed to changing environmental conditions. In order to survive, organisms must weather normal and oftentimes extreme variations in temperature, water availability, salt levels, xenobiotics and other environmental factors. To cope, animals have developed mechanisms for stress protection. At the cellular level, DNA damage and other forms of stress can induce apoptosis to remove damaged cells; at the organismal level, stressful conditions can sometimes induce responses that make the entire animal more stress resistant [1], [2].
The nematode C. elegans has evolved several stress-protective responses. These include aversive behaviors, such as avoidance of noxious stimuli, and the activation of alternative developmental programs, such as entry into the dauer state of diapause [3], [4]. The animal can also induce environmental stress resistance by turning on gene transcription to manage the stressor using, for example, the heat-shock transcription factor HSF-1 to combat heat stress [5], SKN-1/Nrf2 to combat xenobiotic stress [6] and the hypoxia-inducible factor HIF-1 to combat hypoxia [7].
In addition to coordinated stress responses at the organismal level, C. elegans, like other organisms, can protect itself against stress at the cellular level. For example, in C. elegans, germ cells undergo apoptosis in response to DNA damage from ionizing radiation [8], [9]. In general, these types of single-cell, live-or-die decisions may be made to sacrifice a part for the betterment of the whole. How these decisions are made and the mechanistic and molecular relationship, if any, between animal-wide stress responses and programmed cell death are, however, poorly understood.
As a fundamental process by which organisms remove unnecessary, abnormal or damaged cells, programmed cell death involves both cell killing via apoptosis and cell corpse removal via phagocytosis and degradation [10]. Although once considered a disinterested second party that simply removes the dead cell, the engulfing cell is now known to be an active participant in the cell death program. For example, in weak C. elegans caspase mutants, in which decisions about whether to complete the cell death program are made stochastically, a second mutation in an engulfment gene further reduces cell death [11], [12]. Likewise, in mammals, mutations affecting either the dying or engulfing cell can disrupt tissue homeostasis and produce developmental disorders, autoimmune disease, cancer and neurodegeneration [13].
Genes responsible for carrying out apoptosis and apoptotic-cell engulfment were first described in C. elegans (Figure S1) [14], [15]. In the apoptotic cell, the canonical programmed cell death pathway involves the Apaf-1 like protein CED-4, which is inhibited by the BCL-2-like protein, CED-9 [16]–[19]. When disinhibited by developmental cues, CED-4 activates the C. elegans executioner caspase CED-3 [20], [21]. In the engulfing cell, several partially redundant pathways govern the membrane and cytoskeletal rearrangements required for phagocytosis of the dying cell (Figure S1) [22]–[25].
C. elegans was instrumental in illuminating the core features of programmed cell death and clearance because it is highly amenable to genetic and experimental manipulation. Recently, we implicated the human disease gene progranulin in the regulation of programmed cell death using a C. elegans mutant [26]. The regulation and function of progranulin are particularly interesting because of its links to disease. Progranulin haploinsufficiency causes the human neurodegenerative disease frontotemporal lobar degeneration while homozygous null carriers develop neuronal ceroid lipofuscinosis [27]–[29]. Allelic variations in the gene have also been linked to Alzheimer Disease, Parkinson Disease and amyotrophic lateral sclerosis (ALS) [30]–[34], and altered progranulin levels have been implicated in autoimmune disease [35], [36], cancer [37]–[42] and ischemic injury [43], [44]. Thus, precise regulation of progranulin levels is important for maintaining health and homeostasis.
Previously, we showed that progranulin normally functions to regulate the rate of apoptotic cell engulfment during the process of programmed cell death [26]. In pgrn-1(-) mutants, apoptotic cells are cleared approximately twice as fast as normal. We also showed that macrophages from progranulin null-mutant mice are able to engulf apoptotic cells more rapidly than are wild-type macrophages. Thus, progranulin, like mtm-1, abl-1 and srgp-1, is a negative regulator of programmed cell death clearance [25], [26], [45]–[47] (Figure S1).
Given the close relationship between environmental stress and age-related disease, we asked whether pgrn-1(-) mutants exhibited an altered response to cellular stressors. We found that they did. However, unexpectedly, they demonstrated increased stress resistance. Even more surprisingly, we found that the same was true of mutations that perturb cell death in other ways, suggesting that a stress response pathway is activated when any part of the programmed cell death pathway does not proceed normally. Our findings reveal an unexpected link between mechanisms that control life-or-death decisions at the level of the individual cell and at the level of the entire animal.
Results
Loss of pgrn-1 Confers Stress Resistance
We tested the resistance of progranulin mutants to several environmental stressors. We found that compared to wild-type controls, pgrn-1(tm985) mutants were resistant to osmotic, heat and endoplasmic reticulum (ER) stress (the latter as measured by resistance to tunicamycin, an inhibitor of N-linked glycosylation) (Figure 1A–C, Tables S1A–C). In contrast, pgrn-1 mutants had normal responses to oxidative stress (paraquat), genotoxic stress (UV light) and pathogen exposure (P. aeruginosa and S. enterica; Figure S2A–C and data not shown). Reintroducing either the C. elegans or human progranulin gene into pgrn-1(-) mutants rescued or partially rescued the mutant stress resistance phenotypes (Figure 1C–D, Tables S1C–D). The partial rescue by human progranulin only at higher doses of tunicamycin could be due to species differences or differences in binding affinities to the progranulin receptor.
Fig. 1. pgrn-1(-) mutants are resistant to osmotic, heat and ER stress. 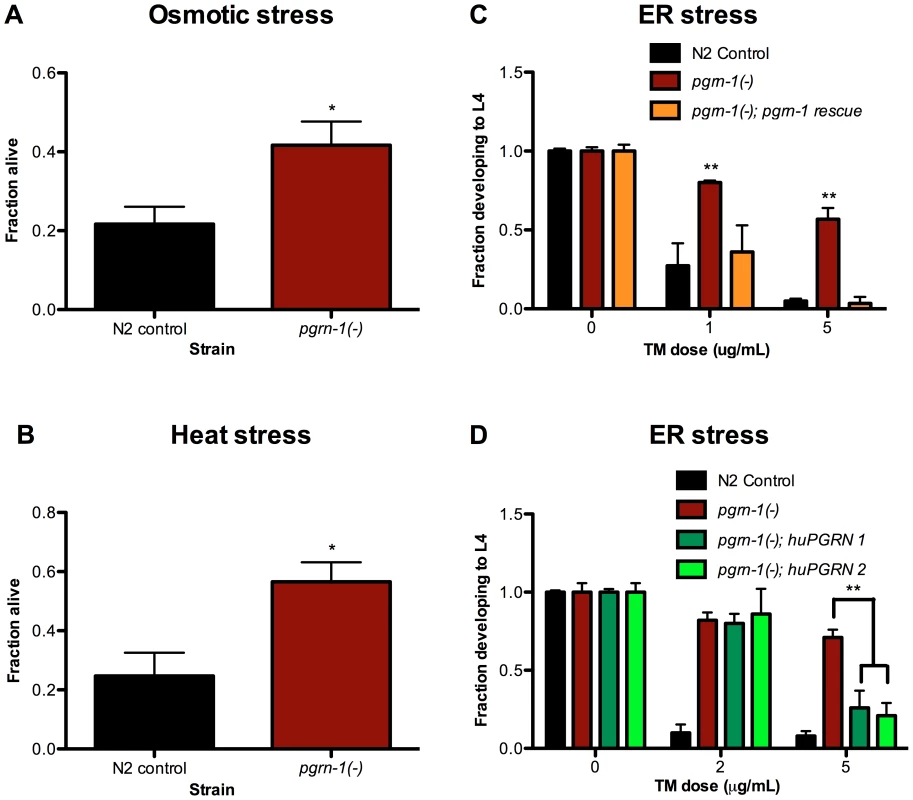
N2 control and pgrn-1(tm985) animals were subjected to various stressors and then scored for survival or ability to develop from egg to L4 stage. (A) Day 1 adult animals were treated with 600 mM NaCl for 24 hours and scored for survival (Student's t test). (B) Day 1 adult animals were incubated at 35°C for 8 hours and scored for survival (Student's t test). (C) Embryos from WT, pgrn-1(-) or pgrn-1(-) expressing a C. elegans progranulin rescue construct were treated with 0, 1 or 5 µg/mL tunicamycin (TM) for 3 days to induce ER stress and scored for ability to develop to the L4 stage (Two-way ANOVA with Bonferroni post-tests). (D) Embryos from WT, pgrn-1(-) or pgrn-1(-) expressing a human progranulin rescue construct (two independently integrated and outcrossed strains) were treated with TM as in (C) (Two-way ANOVA with Bonferroni post-tests). Results shown are representative of at least 2 experiments. Error bars represent standard deviation. Statistical comparisons here are to N2 control. n.s. not significant, *p<0.05, **p<0.01, ***p<0.001. For additional statistical data, please see Table S1A–D. What do heat, osmotic stress and tunicamycin have in common? One possibility is that they all beget unfolded proteins and induce the ER unfolded protein response. To address this idea, we tested the ability of each stressor to increase expression of hsp-4. HSP-4 is the nematode ortholog of mammalian grp78/BiP/HSP70, and is upregulated by heat and ER stress [48], [49]. We confirmed that heat stress and tunicamycin increased Phsp-4::gfp reporter levels (Figure S3A–B), and found that paraquat, UV irradiation and exposure to P. aeruginosa did not (Figure S4A–C). However, under our conditions, osmotic stress did not increase Phsp-4::gfp levels (Figure S3C). Thus, induction of the ER stress-resistance marker Phsp-4::gfp is not a feature that unifies heat, tunicamycin and osmotic stress.
Apoptosis-Defective Mutants Are Stress Resistant
Because loss of function of pgrn-1, a regulator of programmed cell death clearance, caused stress resistance, we asked whether other mutations affecting programmed cell death would also affect the stress response of the whole animal. In contrast to progranulin mutants, loss-of-function mutations in the gene encoding the executioner caspase ced-3 prevent apoptosis [15]. In ced-3 loss-of-function mutants, cells that normally die during development instead persist. Surprisingly, we found that ced-3(n717) mutant animals also exhibited increased resistance to ER stress (Figure 2A and Tables S2A). Moreover, pgrn-1(-); ced-3(n717) double mutants were no more stress-resistant than were either of the single mutants (Figure 2A and Table S2A), suggesting that pgrn-1 and ced-3 mutations may activate the same stress response pathway. ced-3(n717) mutants could also exhibit osmotic and heat stress resistance, albeit not as consistently as ER stress resistance (Figure 2B–C and Table S2B). Like pgrn-1 mutants, ced-3(n717) mutants did not exhibit resistance to paraquat or UV light (Figure S5).
Fig. 2. Mutations in programmed cell death genes ced-3 and ced-9 confer stress resistance. 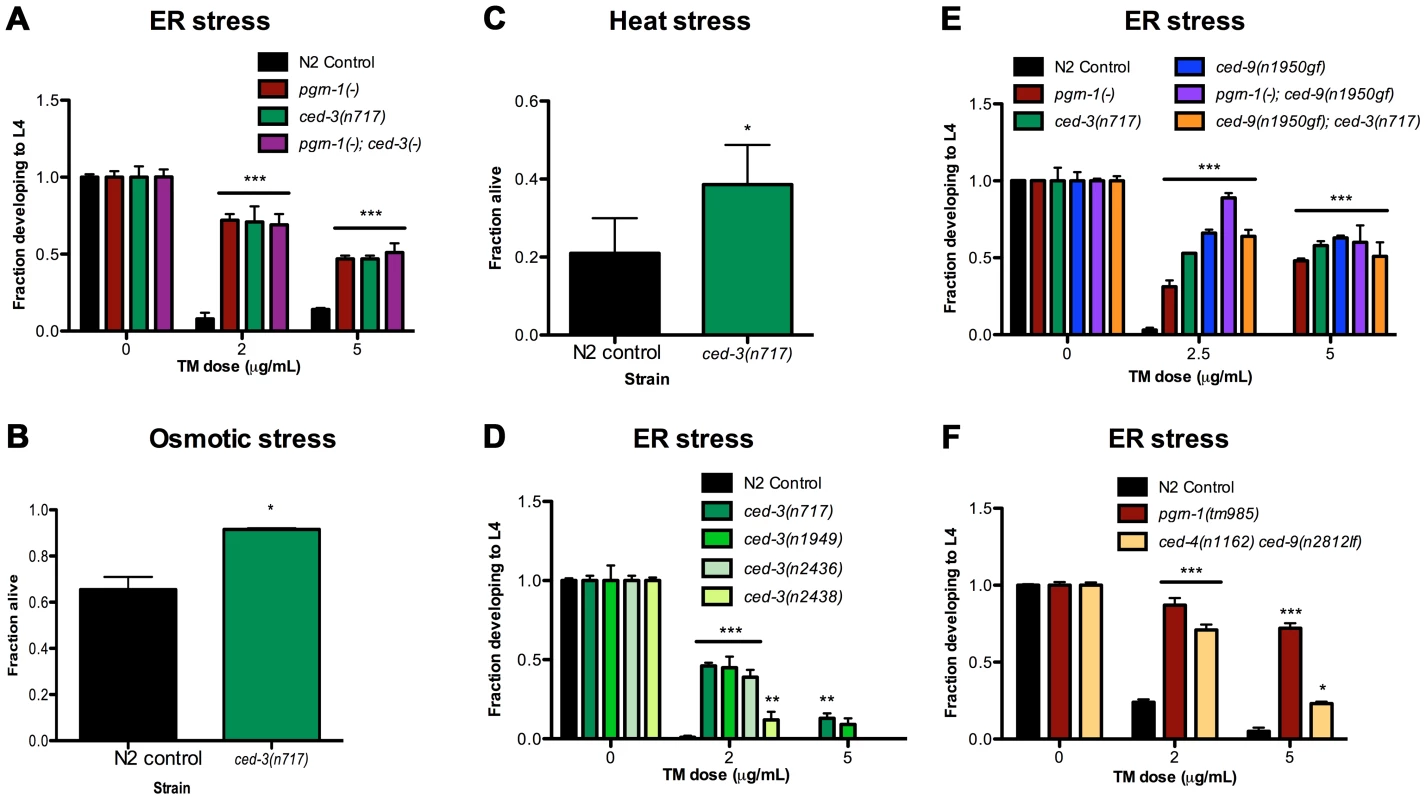
(A) ced-3(n717) animals were subjected to ER stress with indicated doses of tunicamycin (TM) and then scored for the ability to develop from egg to L4 stage. (B–C) Day 1 adult ced-3(n717) animals were exposed to 600 mM NaCl for 24 hours or thermal stress at 35°C for 8 hours and scored for survival. (D) Strong (n717), moderate (n1949, n2436) or weak (n2436) alleles of ced-3 were tested for response to ER stress. (E) A ced-9(n1950) gain-of-function allele was tested for ER stress resistance in wild-type, pgrn-1(-) or ced-3(-) backgrounds. (F) ced-4(n1162) and ced-9(n2812) loss-of-function mutants were treated with TM and scored for the ability to develop from egg to L4 stage. Results shown are representative of at least 2 experiments except for (C), which is an average of 3 experiments. Error bars represent standard deviation. Statistical comparisons are to N2 control (Student's t test or ANOVA with Bonferroni post-tests). n.s. not significant, *p<0.05, **p<0.01 ***p<0.001. For additional statistical data, see Table S2A–D. Many alleles of ced-3 have been isolated, and they form an allelic series based on their ability to inhibit programmed cell death [50]. We found that ced-3 alleles exhibited graded levels of ER stress resistance that were correlated with their ability to block programmed cell death. The strong ced-3 allele n717 was more resistant to ER stress than were two intermediate strength (n1949 and n2436) alleles, and these, in turn, were more resistant than the weak (n2438) allele (Figure 2D and Table S2A). These results are consistent with a model in which the stress resistance conferred by ced-3 mutations is mechanistically related to the apoptotic killing conferred by ced-3.
In C. elegans, ced-9 and ced-4 regulate the ability of ced-3 to activate programmed cell death [17]. The Bcl-2-like protein CED-9 inhibits CED-4/Apaf1 activity, blocking cell death; whereas activated CED-4 cleaves CED-3 and activates its caspase function, leading to cell death [18], [51]. Therefore, ced-9 gain-of-function (gf) and ced-4 loss-of-function (lf or -) mutations are similar to ced-3(-) mutations in the sense that they impair programmed cell death, whereas ced-9(lf) mutations cause excessive cell death (and animal lethality) due to uncontrolled activity of CED-4 and CED-3. We tested ced-9(n1950gf) and ced-4(n1162lf) single mutants, as well as a ced-4(n1162lf) ced-9(n2812lf) double mutant, for their responses to ER stress. We found that ced-9(gf) mutants were resistant to ER stress (Figure 2E and Table S2C). Further, the stress resistance conferred by ced-9(gf) mutations was not additive with that conferred by either pgrn-1(-) or ced-3(-) mutations, once again suggesting that these genes affect the same stress-response pathway (Figure 2E, Table S2C). We also found that ced-4(lf) single mutants were resistant to ER stress at low doses of tunicamycin in one of two experiments (Table S2D), and that these mutants did not require intact ced-9 for this stress resistance (Figure 2F, Table S2D). These findings suggest that ced-4 (and likely ced-3) may be genetically downstream of ced-9 in the stress-resistance pathway, as it is in the cell death pathway. However, since ced-4(lf) animals displayed an incomplete degree of stress resistance compared to ced-3(lf) and ced-9(gf) mutants, it remains possible that ced-4 is dispensable or redundant in this stress response pathway.
Inhibition of Apoptotic Cell Engulfment Also Confers Stress Resistance
In addition to mutations that accelerate the clearance of apoptotic corpses, or prevent apoptosis altogether, we also asked whether mutations in apoptotic cell engulfment pathways affected stress resistance. We found that certain engulfment mutant alleles increased ER stress resistance, although the degree of resistance seen in engulfment mutants was generally less than that seen in animals carrying pgrn-1(-) or strong ced-3(-) mutant alleles. The engulfment mutants ced-1(e1735), ced-6(n1813), ced-7(n1892) and ced-2(e1752), ced-5(n1812), ced-10(n3246) exhibited resistance to ER stress at low doses of tunicamycin (1 µg/mL). However they exhibited variable responses to tunicamycin at higher doses (5 µg/mL), with ced-5, 7 and 10 mutants demonstrating ER stress resistance but not ced-1, 2 or 6 mutants (Figure 3A–B and Table S3A–B). Once again, double mutants containing pgrn-1 and engulfment mutations were not more resistant than pgrn-1(-) alone, suggesting that these mutations could potentially induce a common ER stress-resistance pathway (Figure 3A–B and Table S3A–B).
Fig. 3. Engulfment mutations enhance ER stress resistance. 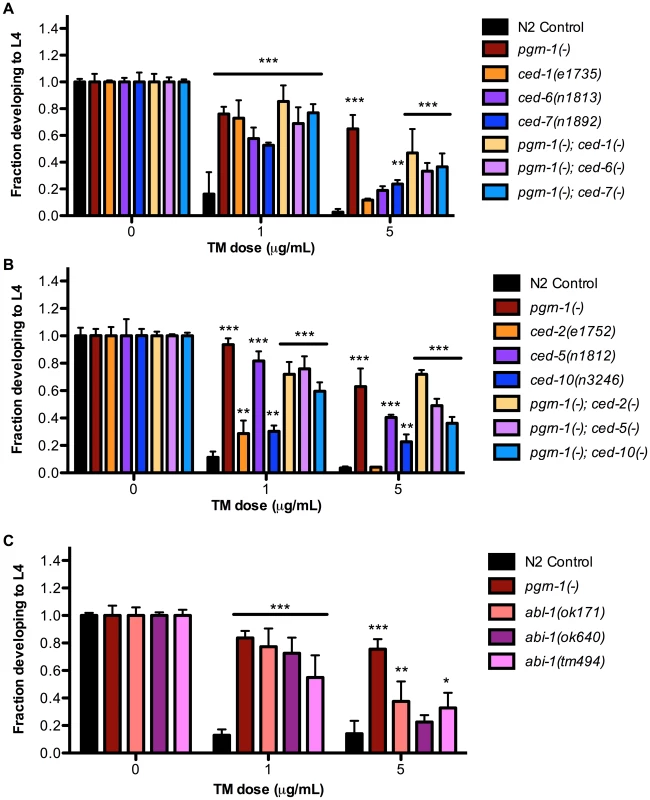
(A) Mutations in ced-1(e1735), ced-6(n1813) and ced-7(n1892) with or without pgrn-1(tm985) in the background were tested for ER stress resistance by tunicamycin (TM) treatment. (B) Mutations in ced-2(e1752), ced-5(n1812) and ced-10(n3246) with or without pgrn-1(tm985) in the background were tested for ER stress resistance by tunicamycin treatment. (C) Mutations in abl-1(ok171), abi-1(ok640) and abi-1(tm494) were tested for ER stress resistance by tunicamycin treatment. Results shown are representative of at least 2 experiments except in the case of (C) which was performed once. Error bars represent standard deviation. Statistical comparisons are to N2 control (ANOVA with Bonferroni post-tests). n.s. not significant, *p<0.05, **p<0.01, ***p<0.001. For additional statistical data, see Table S3A, B, and D. We also tested the response of engulfment mutants to other stressors. We found that ced-1(e1735) and ced-2(e1752) mutants were resistant to osmotic stress (Figure S6A and Table S3C). The ced-2 mutant was also resistant to thermal stress in 1 of 2 trials (Figure S6 and Table S3C). As a group, the engulfment mutants were not as robustly resistant to environmental stressors as pgrn-1 and ced-3 mutants, which may be due to the partial functional redundancy of engulfment pathway genes (See Figure S1).
Like pgrn-1, the tyrosine kinase ABL-1 is a negative regulator of apoptotic corpse engulfment [45]. However, unlike pgrn-1, abl-1 does not act through the canonical engulfment pathways. Instead, abl-1 negatively regulates the engulfment gene abi-1 to inhibit cell death clearance (See Figure S1). We asked whether these two genes might influence ER stress resistance, and found that both abl-1 and abi-1 mutants exhibited resistance to low doses of tunicamycin (Figure 3C, Table S3D). At higher doses of TM, abl-1(-) and one mutant allele of abi-1, ok171, were resistant to tunicamycin stress compared to wild type. Curiously, in some situations, abl-1 mutations actually reduced ER stress resistance. For example, abl-1(n1963) mutations alone have no visible effect on engulfment of apoptotic corpses; however, abl-1 mutations reduce the severity of the engulfment phenotype of ced-1(n2091) and ced-6(n2095) mutants [45]. Likewise, we found that abl-1(n1963) mutations reduced the level of ER stress resistance conferred by ced-1(n2091) and ced-6(n2095) mutations (Figure S7A and Table S3E). We do not have a simple unifying explanation for these findings at this time, but they indicate that pgrn-1 is not the only negative regulator of cell engulfment that can affect ER stress resistance.
We also tested two additional genes that may modulate but are not directly involved in programmed cell death for stress response phenotypes. A mutation in unc-73 enhances the effect of other engulfment mutants but alone has no engulfment defect [52]. A mutation in unc-53 results in defective migration of cells and neuronal processes and the UNC-53 protein interacts with ABI-1 [53]. However, neither unc-73(e936) nor unc-53(e404) mutants exhibited ER stress resistance phenotypes (Figure S7B and Table S3F). Thus, not all genes involved in apoptotic cell engulfment are utilized for stress response.
Stress Resistance in a Non-Apoptotic Programmed Cell Death Pathway
Recently, pqn-41 was identified as a mediator of a type of non-apoptotic cell death [54]. This type of cell death, characterized by crenellation of the nuclear envelope and organelle swelling, occurs independently of the ced-3 caspase and engulfment genes [55], [56]. To determine if pqn-41 affects ER stress resistance, we tested a deletion mutant, ns924. We found that pqn-41(ns924) mutants were resistant to ER stress at a low dose of tunicamycin but not at a high dose, similar to some of the engulfment mutants we tested such as ced-1, ced-2 and ced-6 (Figure S7C and Table S3G). These data suggest that organismal stress resistance may be linked to both apoptotic and non-apoptotic programmed cell death.
Together, these findings indicate that perturbing C. elegans programmed cell death in a variety of ways, either by affecting the initiation of cell death or the engulfment of the dying cell, can confer whole-animal resistance to environmental stress.
The Unfolded Protein Response (UPR) Gene ire-1 Mediates ER Stress Resistance of pgrn-1 Mutants
Because of recent findings connecting the unfolded protein response with neurodegenerative diseases [57], we decided to investigate the mechanism by which a pgrn-1 mutation affected ER stress resistance. The UPR is the cellular program that responds to ER stress. The UPR is mediated by three ER resident proteins encoded by ire-1, pek-1 (mammalian Perk) and atf-6 [58]. Mutations in these genes impair the response to ER stress in C. elegans [59], [60]. Part of this stress response includes alternative splicing of xbp-1 mRNA by activated IRE-1 and the consequent upregulation of XBP-1 target genes, such as hsp-4, the nematode ortholog of mammalian grp78/BiP/HSP70 [48], [49]. To investigate the role of the UPR in the stress resistance of apoptosis mutants, we first tested whether pgrn-1 mutants required ire-1 for ER stress resistance. We tested a pgrn-1; ire-1 double mutant and found that resistance in pgrn-1 mutants was dependent on ire-1 (Figure 4A and Table S4A) suggesting that the mechanism of stress resistance of cell death mutants requires this branch of the UPR pathway. One possibility was that the IRE-1 pathway is constitutively activated in pgrn-1 mutants. To test this, we measured the levels of spliced xbp-1 mRNA. Interestingly, we found no changes in the levels of spliced xbp-1 mRNA in pgrn-1 mutants compared to wild type (Figure S8). We also investigated whether ced-3 or ced-1 mutants exhibited increased levels of spliced xbp-1 mRNA. Similar to pgrn-1 mutants, they did not (Figure S8).
Fig. 4. pgrn-1(-) resistance to ER stress may be partially dependent on the UPR pathway, daf-16 and pmk-1. 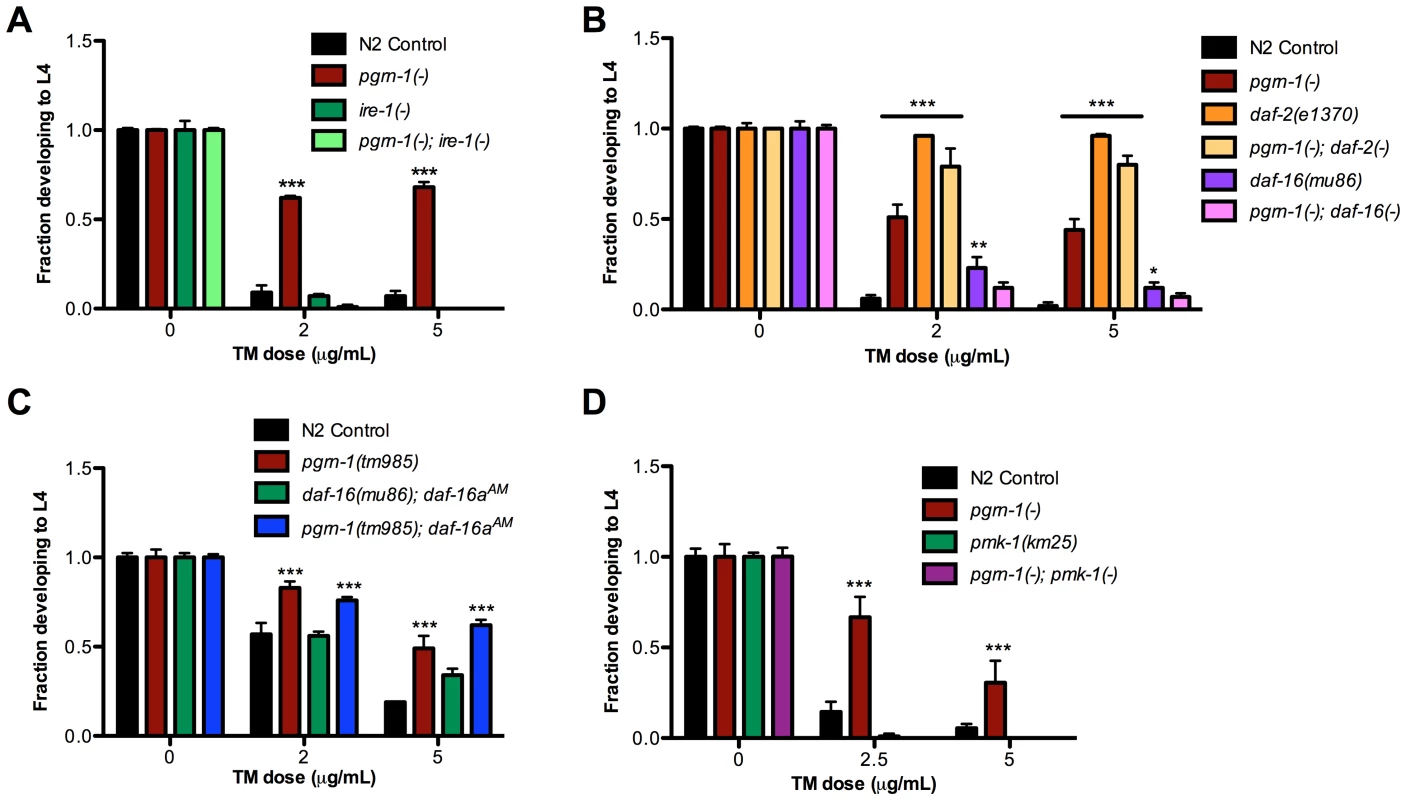
(A) Embryos from the indicated strains were grown on plates with tunicamycin (TM) and assessed for their ability to develop to the L4 stage. (B) daf-2(e1370) and daf-16(mu86) were tested for ER stress resistance with and without pgrn-1(-) in the background. (C) daf-16(mu86); muIs113 and pgrn-1(tm985); muIs113 animals were tested for ER stress resistance. (D) pmk-1(km25) and pgrn-1(-); pmk-1(-) mutants were tested for ER stress resistance. Error bars represent standard deviation. Statistical comparisons are to N2 control (Two-way ANOVA with Bonferroni post-tests). *p<0.05, **p<0.01, ***p<0.001. For additional statistical data, see Table S4A–D. Since the hsp-4 gene is a target of active XBP-1, we measured whether pgrn-1 mutants displayed increased Phsp-4::gfp reporter levels. We found that except for one time point at the L4 stage of larval development, Phsp-4::gfp levels in pgrn-1 mutants were largely unchanged compared to controls. Correspondingly, levels of Phsp-4::gfp in ced-3(n717) mutants were also generally unchanged compared to controls (Figure S9). These data suggest that although pgrn-1 mutations affect the ER stress response through the ire-1 gene, the downstream splicing of xbp-1 mRNA and expression of Phsp-4::gfp is not affected. pgrn-1 mutations may somehow make the IRE-1 branch of the UPR more effective without dramatically changing its activity.
The FOXO Transcription Factor DAF-16 and the MAPK PMK-1 Are Required for ER Stress Resistance of pgrn-1 Mutants
Mammalian progranulin has been demonstrated to activate the insulin/IGF-1 pathway and downstream MAP kinases [37], [61], [62]. Animals carrying mutations in the C. elegans insulin/IGF-1 receptor, daf-2, are long-lived [63], [64] and resistant to many stressors, including heat, osmotic stress and ER stress [63], [64]. The longevity and stress resistance of daf-2 mutants require the FOXO transcription factor daf-16 [60]. Upon inactivation of daf-2, DAF-16 accumulates in the nucleus [65] where it regulates transcription of stress response genes. We found that the degree of ER stress resistance of pgrn-1(tm985); daf-2(e1370) double mutants was similar to that of daf-2(-) single mutants (Figure 4B and Table S4B). pgrn-1 mutants also required intact daf-16 for stress resistance, as daf-16(mu86) pgrn-1(tm985) double mutants were no more stress resistant than were single daf-16(mu86) mutants (Figure 4B and Table S4B). These findings suggest that pgrn-1 may be part of the daf-2 pathway or act with daf-2 to confer stress resistance. However, unlike mutations in daf-2, pgrn-1(-) does not affect nuclear localization of DAF-16::GFP protein (Figure S10). DAF-16::GFP localization is also unaffected in ced-1 and ced-3 mutant animals (Figure S10). To determine if nuclear localization of DAF-16 would further increase stress resistance in pgrn-1 mutants, we crossed the daf-16aAM transgene (which causes DAF-16 nuclear accumulation due to mutation of its AKT-phosphorylation sites) into a pgrn-1(-) background [65]. We found that a pgrn-1(-); daf-16aAM strain was no more stress resistant than was the pgrn-1 mutant alone (Figure 4C and Table S4C).
Others have shown that adult-only ced-3 RNAi extends lifespan without altering DAF-16::GFP localization [66]. Given the correlation between lifespan extension and some forms of stress resistance [67], we tested the lifespan of ced-3, ced-1 and ced-2 mutants. In earlier work, we showed that pgrn-1(-) mutant lifespan is no different than wild type [26]. Whereas a ced-3(-) mutation significantly extended lifespan compared to wild type, ced-1 and ced-2 mutations did not (Figure S11), indicating that longevity and this type of organismal stress resistance can be dissociated.
Several MAP kinases are required for responses to cellular stressors in C. elegans. The PMK-1/p38 MAP kinase encoded by pmk-1 is required for resistance to oxidative stressors [68], pathogenic bacteria [69] and exogenously induced ER stress [70]. We confirmed that pmk-1 mutations increased sensitivity to ER stress and found that pgrn-1(-); pmk-1(km25) double mutants were no more resistant to ER stress than were pmk-1 single mutants. Thus, pmk-1 is required for the ER stress resistance induced by pgrn-1 mutations (Figure 4D and Table S4D).
Progranulin is a secreted protein. In mammals, two progranulin receptors have been identified, the tumor necrosis factor receptor (TNFR) and sortilin [71]. Thus, we tested a downstream TNF receptor associated factor (TRAF) mutant, trf-1(nr2014), for stress resistance and epistasis with pgrn-1(-). trf-1 mutants were not stress resistant compared to wild type and pgrn-1(-) did not require trf-1 for its stress resistance (Figure S12A). We also tested two mutant alleles of trk-1, a C. elegans neurotrophin receptor similar to a co-receptor for sortilin, the other mammalian progranulin receptor. Again, pgrn-1 mutants did not require trk-1 for stress resistance (Figure S12B). Thus, an as yet unidentified receptor(s) appears to be required for progranulin to influence ER stress resistance in C. elegans.
Apoptotic Cell Death Mutants Share Co-regulated Genes
If mutations that perturb cell death in different ways act in the same stress-resistance pathway, then they might share gene expression patterns that differ from wild type. To test this, we performed gene expression profiling by RNA sequencing (RNA-seq), comparing day 1 adult pgrn-1(tm985), ced-3(n717) and ced-1(e1735) mutants to wild-type animals. This allowed us to assess 1) whether these strains have altered gene expression, and 2) whether their differentially expressed genes are shared, suggesting the involvement of a common pathway. In spite of different cell death phenotypes of these mutants, RNA-seq revealed that all three mutants down-regulated the same 95 genes and up-regulated the same 9 genes, a highly significant portion of the total transcriptome (p<10−16) (Figure 5 and Table S5). Of the genes that share differential regulation in our mutants, a significant number are regulated by DAF-16 (Table S5). These findings are consistent with the possibility that the stress resistance phenotypes of these three mutants may be due to the involvement of shared pathways.
Fig. 5. Programmed cell death mutants share differentially regulated genes. 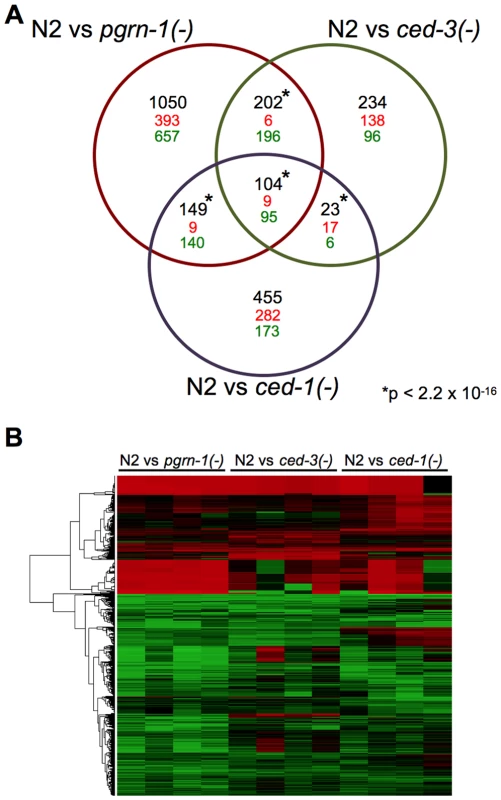
(A) Venn diagram of differentially regulated genes in day 1 adult wild-type (N2) animals versus pgrn-1(tm985), ced-3(n717) or ced-1(e1735) mutants. Significance cut-off was an FDR of <0.05. The numbers in black represent the total number of overlapping genes, with direction of change (up-regulated = red, down-regulated = green) indicated below. The significance of the overlaps between the mutant strains was calculated using Fisher's exact test. See the Excel file (Table S5) for gene list. *p value<2.2×10−16. (B) Heat map depicting the fold changes of gene expression in pgrn-1(tm985), ced-3(n717) and ced-1(e1735) mutants compared to N2 wild-type animals for each of four biological replicates. Discussion
We have shown that mutations that impair C. elegans programmed cell death in any of three ways—by inhibiting apoptosis, by impairing corpse clearance or by accelerating corpse clearance—all enhance resistance to certain environmental stressors. These mutations do not confer resistance to all cellular stressors, as pgrn-1 and ced-3 mutants exhibit normal sensitivity to UV light and oxidative stress.
Several questions naturally follow from these findings. First, why are these mutants resistant to specific stressors? Tunicamycin is an N-linked glycosylation inhibitor that causes retention of translated proteins in the ER and induces the unfolded protein response. Similarly, heat and osmotic stress cause ER and/or cytosolic proteins to unfold and activate signaling programs that induce expression of heat shock proteins and other chaperones. Thus, it seems possible that cell death mutations all trigger a response specific for unfolded proteins, such as the ER unfolded protein response. Consistent with this, we found that the cell-death-related stressors tunicamycin and heat both activated the ER stress-response gene hsp-4/BIP, whereas the cell-death-unrelated stressors Pseudomonas, UV and paraquat did not. However, Phsp-4::gfp was not induced by osmotic stress and pgrn-1(-) mutation did not increase basal Phsp-4::gfp expression levels. Thus, defects in cell death can do more to protect the animal than simply to induce the canonical UPR. They must generate a more multifaceted response that can maintain proteostasis.
The mechanism by which cell death mutations induce animal-wide stress resistance is not known. If different cell-death mutations affected different pathways, or the same pathway to different extents, then one would expect double mutants to be even more stress resistant than the individual single mutants. As this was not the case, it is possible that all of these mutations trigger the same stress response, though other interpretations remain possible. We identified some of the genes required for pgrn-1 mutants to resist the ER stressor tunicamycin. We found that this stress resistance requires an intact ire-1 gene, the MAP kinase PMK-1 and the transcription factor DAF-16/FOXO. We also identified a number of genes that are differentially regulated by all three of our mutants (compared to wild type), suggesting that they may achieve stress resistance by recruiting shared genes and/or pathways. It will be very interesting to explore these shared genes in future studies.
In contrast to the selective stress resistance of pgrn-1 mutants, daf-2 mutants are resistant to most environmental stressors. Thus, decreased DAF-2/insulin/IGF-1 signaling activates a genetic program that more generally elevates organismal resilience. daf-2 mutants appear to increase their resistance to ER stress by making the ire-1/xbp-1 pathway more efficient, possibly by activating stress-response transcription factors like DAF-16 that collaborate with ire-1/xbp-1 to induce new protective genes [60]. In concordance with this, daf-2 mutants actually have reduced levels of expression of ire-1/xbp-1-regulated genes such as hsp-4/BIP. While this was not the case for pgrn-1 or ced-3 mutants, whose Phsp-4::gfp levels were not decreased, there are some unexpected similarities between the ER stress-resistance phenotypes of cell-death mutants and daf-2 mutants. First, the ER stress resistance phenotypes of pgrn-1 and daf-2 mutants require daf-16, either fully (pgrn-1 mutant) or partially (daf-2 mutant). Second, both ER stress responses are completely dependent on ire-1, yet in neither case is xbp-1 splicing increased. Additionally, the cell death mutants exhibited either no increase or only slight increases in Phsp-4::gfp expression, rather than the substantial increase one might expect if this branch of the ER pathway were constitutively active. Finally, the daf-2 and pgrn-1 mutant ER stress-resistance phenotypes are not additive. Thus the ER-stress resistance pathways activated by these mutations likely share at least some mechanistic features.
It was striking that mutations that perturb programmed cell death in such different ways all had similar effects on environmental stress resistance. Why should mutations that lead to undead cells (apoptosis mutations), lingering corpses (engulfment mutations), and prematurely-engulfed corpses (progranulin mutations) all activate what appears to be (from genetic tests) the same stress resistance pathway? From an evolutionary perspective, this linkage may make sense. Presumably apoptosis evolved not only to sculpt tissues during development, but also to remove cells that are damaged and unable to perform their normal functions, or perhaps that are overtly harmful to the animal. Viewed in this way, impairments in programmed cell death could be interpreted by the organism as an inability to respond normally to stress. Perhaps, under these conditions, the animal uses an alternative, back-up system to survive; namely, the system we have described in this study. Specifically, animals could have developed a sensitive surveillance system that can detect abnormalities in cell death, and respond to them by activating another pathway that enhances their overall level of stress resistance. The existence of this type of alternative system could have increased animal fitness during evolution in turbulent or adverse environments.
While this model makes sense from an evolutionary perspective, other models are possible as well. For example, perhaps cell-death proteins, which act in a multi-step pathway to remove unwanted cells, also act together in a different pathway that has the effect of sensitizing the animal to various forms of stress. Non-cell death functions have, in fact, been described for cell-death effectors. In C. elegans, ced-10(-) mutations impair not only cell engulfment but also cell migration [72]. Another engulfment gene, ced-1, has also been implicated in neuronal regulation of innate immunity [73], [74]. In mammals, a defect in the BCL2-family protein BID impairs cytokine production in response to immune activation independently of its cell death signaling function [75]. Further, certain mammalian executioner caspases can activate microglia in response to inflammogens without causing microglial death [76]. Finally, in a mouse model of Alzheimer Disease, caspase activation may be responsible for tau cleavage and aggregate formation, thereby serving a protective function [77]. Thus, programmed cell death effectors could hypothetically act together to sensitize animals to certain stressors or, alternatively, to inhibit a stress-response pathway. In either case, perturbing programmed cell death would increase organismal stress resistance.
A protein in the flowering plant Arabidopsis may support the model that programmed cell death effectors can sensitize an animal to environmental stress. Arabidopsis can express a protein called RD21 that, like CED-3, is a cysteine protease, and, like progranulin, contains a granulin domain. Osmotic stress induces RD21 and, possibly as a consequence, leaf senescence. Interestingly, in response to stress RD21 undergoes a process of maturation in which its caspase domain cleaves and releases the granulin domain [78]–[80]. In RD21, the caspase and granulin domains are contained within the same molecule. However, perhaps in C. elegans, the two domains reside in different proteins but nevertheless act together to influence organismal stress resistance.
In summary, our findings indicate that programmed cell death effectors not only kill and remove individual cells, but also influence environmental stress resistance at the level of the whole animal. To our knowledge this is the first time that cell-death effectors like ced-3, pgrn-1 and ced-1 have been implicated in organismal stress resistance, and these findings raise many interesting new questions about both mechanism and evolution.
Materials and Methods
Strains
Unless otherwise indicated, C. elegans were cultured at 20°C using standard procedures [81]. Strains were kindly provided by the Mitani Laboratory (National Bioresource Project) at the Tokyo Women's Medical University and the Caenorhabditis Genetics Center (CGC) at the University of Minnesota. Strains were outcrossed four times to the laboratory N2 control strain (N2 Bristol). Descriptions of strains can be found at www.wormbase.org. The following strains were used:
AWK2 ced-9(n1950gf) III; ced-3(n717) IV
AWK74 daf-16(mu86) I; ced-3(n717) IV
AWK76 daf-16(mu86) pgrn-1(tm985) I; muIs109[Pdaf-16::daf-16::gfp+Podr-1::RFP]
AWK77 daf-16(mu86) I; ced-3(n717) IV; muIs109[Pdaf-16::daf-16::gfp+Podr-1::RFP]
AWK78 ced-1(e1735) daf-16(mu86) I
AWK80 ced-1(e1735) daf-16(mu86) I; muIs109[Pdaf-16::daf-16::gfp+Podr-1::RFP]
AWK109 pgrn-1(tm985) I; pqn-41(ns294) III
AWK111 pgrn-1(tm985) I; muIs113[Pdaf-16::daf-16AM::gfp+rol-6]
CB404 unc-53(e404) II
CB936 unc-73(e936) I
CF1037 daf-16(mu86) I
CF1041 daf-2(e1370) III
CF1934 daf-16(mu86) I; muIs109[Pdaf-16::daf-16::gfp+Podr-1::RFP]
CF2260 N2; zcIs4[Phsp-4::gfp] V
CF2473 ire-1(ok799) II
CF3050 pgrn-1(tm985) I
CF3165 pgrn-1(tm985) I; zcIs4[Phsp-4-4::gfp] V
CF3170 pgrn-1(tm985) I; ire-1(ok799) II
CF3196 daf-16(mu86) pgrn-1(tm985) I
CF3206 pgrn-1(tm985) I; daf-2(e1370) III
CF3324 ced-3(n717) IV
CF3419 pgrn-1(tm985) I; ced-3(n717) IV
CF3447 pgrn-1(tm985) I; muIs189[Ppgrn-1:: pgrn-1::polycistronic mCherry+Podr-1::CFP]
CF3762 ced-3(n2436) IV
CF3656 ced-2(e1752) IV
CF3660 ced-10(n3246) IV
CF3662 pgrn-1(tm985) I; ced-2(e1752) IV
CF3667 ced-1(e1735) I
CF3672 pgrn-1(tm985) ced-1(e1735) I
CF3675 pgrn-1(tm985) I; ced-10(n3246) IV
CF3680 ced-5(n1812) IV
CF3683 pgrn-1(tm985) I; ced-7(n1892) III
CF3684 pgrn-1(tm985) I; ced-5(n1812) IV
CF3685 pgrn-1(tm985) I; ced-6(n1813) III
CF3687 pgrn-1(tm986) I; muIs211[Pegl-3::huPGRN::polycistronic mCherry+Podr-1::CFP] Line 1
CF3688 pgrn-1(tm986) I; muIs211[Pegl-3::huPGRN::polycistronic mCherry+Podr-1::CFP] Line 2
CF3762 ced-3(n1949) IV
CF3802 ced-3(n717) IV; zcIs4[Phsp-4::gfp] V
CF3808 trk-1(tm3985) X
CF3809 trk-1(tm4054) X
CF3817 pgrn-1(tm985) I; trk-1(tm3985) X
CF3818 pgrn-1(tm985) I; trk-1(tm4054) X
CF3821 trf-1(nr2014) III
CF3833 pgrn-1(tm985) I; trf-1(nr2014) III
CF3879 pgrn-1(tm985) I; pmk-1(km25) IV
CF3881 pgrn-1(tm985) I; ced-9(n1950gf) III
FX494 abi-1(tm494) III
KU25 pmk-1(km25) IV
MT2547 ced-4(n1162) III
MT4433 ced-6(n1813) III
MT4982 ced-7(n1892) III
MT4770 ced-9(n1950gf) III
MT7384 ced-4(n1162) ced-9(n2812lf) III
MT16077 ced-1(n2091) I; abl-1(n1963) X
MT19956 ced-6(n2095) III; abl-1(ok171) X
OS4023 pqn-41(ns294) III
RB829 abi-1(ok640) III
XR1 abl-1(ok171) X
Generation of Transgenic C. elegans Strains
To generate a C. elegans pgrn-1 rescue construct, full-length pgrn-1a+TAA stop codon and its endogenous 0.5 kB upstream promoter were cloned into a Gateway polycistronic mCherry vector (courtesy K. Ashrafi lab, UCSF). The resulting plasmid (Ppgrn-1::pgrn-1+TAA::polycistronic mCherry) expresses both progranulin and mCherry and functions as a full length rescuing construct when expressed in the pgrn-1 mutant. To generate a human progranulin rescue strain, the pan-neuronal egl-3 promoter (courtesy of K. Ashrafi lab) and the human progranulin cDNA sequence were cloned into a Gateway polycistronic mCherry vector (Pegl-1::human PGRN::polycistronic mCherry).
The constructs were microinjected separately into the gonads of day 1 adult C. elegans. Stable monogenic lines were isolated and analyzed using Leica fluorescent, Zeiss Axioplan 2 or Nikon Spectral Confocal microscopes. Extrachromosomal arrays were integrated by UV irradiation by the method of C. Frank et al. [82] and outcrossed at least 5 times to our lab's wild-type N2 control strain.
Stress Assays
For thermal and osmotic stress assays on Day 1 worms, L4-stage animals were picked and grown at 20°C overnight. For thermal stress assays, worms were moved to a 35°C incubator for 12 hours and then scored for survival. Osmotic stress assays were performed by the method of Lamitina et al. [83] with the following modifications: worms were fed OP50 bacteria, worms were cultured at 20°C prior to the assay, and assays were performed on NG-based plates at 20°C with increasing amounts of NaCl added as indicated. For paraquat stress assays, individual animals were placed in 96-well plates with 100 µL of 250 µM methyl viologen (paraquat, Sigma-Aldrich) dissolved in M9 and scored for movement every 1 hour at 25°C. For genotoxic stress assays, day 1 adult animals were transferred to unseeded plates and treated with 1200 J/m2 UV light in a Stratalinker 1800 (Stratagene). Animals were then scored for survival every 24 hours. Pathogen stress was performed by transferring worms to plates seeded with P. aeruginosa or S. enterica starting at day 1 and scoring each subsequent day for survival. In all assays, animals that failed to move in response to a gentle touch with a metal pick were scored as dead.
For ER stress assays, synchronized eggs were transferred to plates containing 0, 1, 2 or 5 µg/mL of tunicamycin (EMD Chemicals). After 3 days, animals that developed to the L4 stage were quantified. Figures show fraction of animals that develop to L4 stage normalized to percent hatching on 0 µg/mL tunicamycin for each strain.
Statistical analyses were performed in GraphPad Prism statistical package with tests as indicated in figure legends.
Lifespan Analysis
Wild-type, ced-1(e1735), ced-2(e1752) and ced-3(n717) strains were grown at 20 degrees Celsius (C), then picked to fresh OP50 at the L4 stage and shifted to 25 degrees C. Subsequent lifespan analysis was done at this temperature. Animals were transferred every day to fresh plates until progeny production ceased. Animals that crawled off the plate, exploded, bagged, or became contaminated were censored. GraphPad Prism was used to calculate mean life spans and perform statistical analyses. P values were determined using log-rank (Mantel-Cox) statistics.
xbp-1 RT-PCR
C. elegans eggs were obtained by bleaching, then plated onto E. coli OP50 and allowed to develop at 20°C to day 1 of adulthood. At this point, positive controls were exposed to 5 mg/ml tunicamycin for 5 hours while all other worms were left untreated. After washing animals off plates, Trizol was added and samples were frozen in liquid nitrogen. Animals were lysed in a Mini-Beadbeater (Biospec products) for 10 minutes at the maximal setting. Total RNA was isolated using a phenol/chloroform extraction and DNA contamination was removed with DNA-free treatment (Ambion). cDNA was synthesized (iScript) using oligo (dT) primers and RT-PCR was performed using primers that amplify an ∼200 bp unspliced transcript and an ∼180 bp spliced transcript. Forward primer sequence: 5′ ctacgaagaagaagtcgtcgg 3′ and reverse primer sequence: 5′ ttcttgttgcgatccatgtg 3′. RT-PCR products were analyzed by running them out on a 3% agarose gel stained with ethidium bromide.
Quantification of Fluorescence
Animals expressing the Phsp-4::gfp transgene were anaesthetized on agarose pads containing 2.5 mM levamisole. Whole worm images were taken using a Retiga EXi Fast1394 CCD digital camera (QImaging, Burnaby, BC, Canada) using the 5× objective on a Zeiss Axioplan 2 compound microscope (Zeiss Corporation, Germany). Each image was taken so that the intestine was in focus and exposure time was calibrated to minimize number of saturated pixels for the set of animals. Images within each experiment were acquired using identical settings and exposure times to allow direct comparisons. Fluorescence intensity was measured by outlining the entire worm. Openlab 4.0.2 software (Improvision, Coventry, UK) was then used to quantify total intensity of each pixel in the selected area. Measurements were obtained by subtracting the minimum intensity from the mean intensity and taking the average of these calculations for 8–10 animals per time-point.
RNA-Seq Analysis
Total RNA was isolated from each of the strains pgrn-1(tm985), ced-3(n717), ced-1(e1735) and wild-type (N2E) using a phenol/chloroform extraction, and DNA contamination was removed with DNA-free treatment (Ambion). Samples were extracted in quadruplicates (four biological replicates for each strain), for a total of 16 samples. Total RNA was quantified using the RiboGreen assay and RNA quality was checked using an Agilent Bioanalyzer (Agilent). RNA Integrity Numbers (RINs) were >8 in all the samples. Libraries for RNA-seq were prepared using the Illumina TruSeq library preparation protocol (Illumina Inc), multiplexed into a single pool and sequenced using an Illumina HiSeq 2500 sequencer across 4 lanes of 2 Rapid Run SR 1×50 flow cells. After demultiplexing, we obtained between 13 and 32 million reads per sample, each one 50 bases long. Quality control was performed on base qualities and nucleotide composition of sequences. Alignment to the C. elegans genome (ce10) was performed using the STAR spliced read aligner (PMID 23104886) with default parameters. Additional QC was performed after the alignment to examine the following: level of mismatch rate, mapping rate to the whole genome, repeats, chromosomes, and key transcriptomic regions (exons, introns, UTRs, genes). Between 92 and 93% of the reads mapped uniquely to the worm genome. Total counts of read-fragments aligned to candidate gene regions within the C. elegans reference gene annotation were derived using HTS-seq program and used as a basis for the quantification of gene expression. Only uniquely mapped reads were used for subsequent analyses. Following alignment and read quantification, we performed quality control using a variety of indices, including sample clustering, consistency of replicates, and average gene coverage. Differential expression analysis was performed using the EdgeR Bioconductor package (19910308), and differentially expressed genes were selected based on False Discovery Rate (FDR Benjamini Hochberg adjusted p - values) estimated at ≤5%. Clustering and overlap analyses were performed using Bioconductor packages within the statistical environment R (www.r-project.org/). Gene Ontology annotation was performed using DAVID (david.abcc.ncifcrf.gov/). DAF-16 dependent genes were curated from published reports [84], [85] and Wormmart annotation (http://caprica.caltech.edu:9002/biomart/martview).
Supporting Information
Zdroje
1. CypserJR, TedescoP, JohnsonTE (2006) Hormesis and aging in Caenorhabditis elegans. Exp Gerontol 41 : 935–939.
2. GartnerA, BoagPR, BlackwellTK (2008) Germline survival and apoptosis. WormBook 1–20.
3. BargmannCI (1993) Genetic and cellular analysis of behavior in C. elegans. Annu Rev Neurosci 16 : 47–71.
4. GoldenJW, RiddleDL (1982) A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218 : 578–580.
5. Hajdu-CroninYM, ChenWJ, SternbergPW (2004) The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168 : 1937–1949.
6. AnJH, BlackwellTK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17 : 1882–1893.
7. JiangH, GuoR, Powell-CoffmanJA (2001) The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A 98 : 7916–7921.
8. GartnerA, MilsteinS, AhmedS, HodgkinJ, HengartnerMO (2000) A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5 : 435–443.
9. GumiennyTL, LambieE, HartwiegE, HorvitzHR, HengartnerMO (1999) Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126 : 1011–1022.
10. KerrJF, WyllieAH, CurrieAR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26 : 239–257.
11. HoeppnerDJ, HengartnerMO, SchnabelR (2001) Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412 : 202–206.
12. ReddienPW, CameronS, HorvitzHR (2001) Phagocytosis promotes programmed cell death in C. elegans. Nature 412 : 198–202.
13. ElliottMR, RavichandranKS (2010) Clearance of apoptotic cells: implications in health and disease. J Cell Biol 189 : 1059–1070.
14. SulstonJE, SchierenbergE, WhiteJG, ThomsonJN (1983) The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100 : 64–119.
15. EllisHM, HorvitzHR (1986) Genetic control of programmed cell death in the nematode C. elegans. Cell 44 : 817–829.
16. HengartnerMO, EllisRE, HorvitzHR (1992) Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 356 : 494–499.
17. SpectorMS, DesnoyersS, HoeppnerDJ, HengartnerMO (1997) Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385 : 653–656.
18. WuD, WallenHD, NunezG (1997) Interaction and regulation of subcellular localization of CED-4 by CED-9. Science 275 : 1126–1129.
19. ConradtB, HorvitzHR (1998) The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93 : 519–529.
20. YuanJY, HorvitzHR (1990) The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol 138 : 33–41.
21. YuanJ, ShahamS, LedouxS, EllisHM, HorvitzHR (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75 : 641–652.
22. HedgecockEM, SulstonJE, ThomsonJN (1983) Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220 : 1277–1279.
23. EllisRE, JacobsonDM, HorvitzHR (1991) Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129 : 79–94.
24. ConradtB, XueD (2005) Programmed cell death. WormBook 1–13.
25. ZouW, LuQ, ZhaoD, LiW, MapesJ, et al. (2009) Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet 5: e1000679.
26. KaoAW, EisenhutRJ, Herl MartensL, NakamuraA, HuangA, et al. (2011) A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proc Natl Acad Sci U S A 108 : 4441–4446.
27. BakerM, MackenzieIR, Pickering-BrownSM, GassJ, RademakersR, et al. (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442 : 916–919.
28. CrutsM, Kumar-SinghS, Van BroeckhovenC (2006) Progranulin mutations in ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Curr Alzheimer Res 3 : 485–491.
29. SmithKR, DamianoJ, FranceschettiS, CarpenterS, CanafogliaL, et al. (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 90 : 1102–1107.
30. BrouwersN, NuytemansK, van der ZeeJ, GijselinckI, EngelborghsS, et al. (2007) Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol 64 : 1436–1446.
31. BrouwersN, SleegersK, EngelborghsS, Maurer-StrohS, GijselinckI, et al. (2008) Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology 71 : 656–664.
32. SleegersK, BrouwersN, Maurer-StrohS, van EsMA, Van DammeP, et al. (2008) Progranulin genetic variability contributes to amyotrophic lateral sclerosis. Neurology 71 : 253–259.
33. ViswanathanJ, MakinenP, HelisalmiS, HaapasaloA, SoininenH, et al. (2009) An association study between granulin gene polymorphisms and Alzheimer's disease in Finnish population. Am J Med Genet B Neuropsychiatr Genet 150B: 747–750.
34. LeeMJ, ChenTF, ChengTW, ChiuMJ (2011) rs5848 variant of progranulin gene is a risk of Alzheimer's disease in the Taiwanese population. Neurodegener Dis 8 : 216–220.
35. TangW, LuY, TianQY, ZhangY, GuoFJ, et al. (2011) The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science 332 : 478–484.
36. MatsubaraT, MitaA, MinamiK, HosookaT, KitazawaS, et al. (2012) PGRN is a key adipokine mediating high fat diet-induced insulin resistance and obesity through IL-6 in adipose tissue. Cell Metab 15 : 38–50.
37. HeZ, IsmailA, KriazhevL, SadvakassovaG, BatemanA (2002) Progranulin (PC-cell-derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res 62 : 5590–5596.
38. HoJC, IpYC, CheungST, LeeYT, ChanKF, et al. (2008) Granulin-epithelin precursor as a therapeutic target for hepatocellular carcinoma. Hepatology 47 : 1524–1532.
39. KamravaM, SimpkinsF, AlejandroE, MichenerC, MeltzerE, et al. (2005) Lysophosphatidic acid and endothelin-induced proliferation of ovarian cancer cell lines is mitigated by neutralization of granulin-epithelin precursor (GEP), a prosurvival factor for ovarian cancer. Oncogene 24 : 7084–7093.
40. MatsumuraN, MandaiM, MiyanishiM, FukuharaK, BabaT, et al. (2006) Oncogenic property of acrogranin in human uterine leiomyosarcoma: direct evidence of genetic contribution in in vivo tumorigenesis. Clin Cancer Res 12 : 1402–1411.
41. PanCX, KinchMS, KienerPA, LangermannS, SerreroG, et al. (2004) PC cell-derived growth factor expression in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. Clin Cancer Res 10 : 1333–1337.
42. WangM, LiG, YinJ, LinT, ZhangJ (2011) Progranulin overexpression predicts overall survival in patients with glioblastoma. Med Oncol 29 ((4)): 2423–31.
43. TaoJ, JiF, WangF, LiuB, ZhuY (2011) Neuroprotective effects of progranulin in ischemic mice. Brain Res 1436 : 130–136.
44. XuJ, XilouriM, BrubanJ, ShioiJ, ShaoZ, et al. (2011) Extracellular progranulin protects cortical neurons from toxic insults by activating survival signaling. Neurobiol Aging 32 : 2326 e2325–2316.
45. HurwitzME, VanderzalmPJ, BloomL, GoldmanJ, GarrigaG, et al. (2009) Abl kinase inhibits the engulfment of apoptotic cells in Caenorhabditis elegans. PLoS Biol 7: e99.
46. NeukommLJ, FreiAP, CabelloJ, KinchenJM, Zaidel-BarR, et al. (2011) Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat Cell Biol 13 : 79–86.
47. NeukommLJ, NicotAS, KinchenJM, AlmendingerJ, PintoSM, et al. (2011) The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in C. elegans. Development 138 : 2003–2014.
48. OlsenA, VantipalliMC, LithgowGJ (2006) Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology 7 : 221–230.
49. UranoF, CalfonM, YonedaT, YunC, KiralyM, et al. (2002) A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol 158 : 639–646.
50. ShahamS, ReddienPW, DaviesB, HorvitzHR (1999) Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153 : 1655–1671.
51. YangX, ChangHY, BaltimoreD (1998) Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281 : 1355–1357.
52. deBakkerCD, HaneyLB, KinchenJM, GrimsleyC, LuM, et al. (2004) Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol 14 : 2208–2216.
53. SchmidtKL, Marcus-GueretN, AdeleyeA, WebberJ, BaillieD, et al. (2009) The cell migration molecule UNC-53/NAV2 is linked to the ARP2/3 complex by ABI-1. Development 136 : 563–574.
54. BlumES, AbrahamMC, YoshimuraS, LuY, ShahamS (2012) Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science 335 : 970–973.
55. AbrahamMC, LuY, ShahamS (2007) A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev Cell 12 : 73–86.
56. BlumES, DriscollM, ShahamS (2008) Noncanonical cell death programs in the nematode Caenorhabditis elegans. Cell Death Differ 15 : 1124–1131.
57. OzcanL, TabasI (2012) Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu Rev Med 63 : 317–328.
58. RonD, WalterP (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8 : 519–529.
59. ShenX, EllisRE, LeeK, LiuCY, YangK, et al. (2001) Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107 : 893–903.
60. Henis-KorenblitS, ZhangP, HansenM, McCormickM, LeeSJ, et al. (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A 107 : 9730–9735.
61. XuSQ, TangD, ChamberlainS, PronkG, MasiarzFR, et al. (1998) The granulin/epithelin precursor abrogates the requirement for the insulin-like growth factor 1 receptor for growth in vitro. J Biol Chem 273 : 20078–20083.
62. Zanocco-MaraniT, BatemanA, RomanoG, ValentinisB, HeZH, et al. (1999) Biological activities and signaling pathways of the granulin/epithelin precursor. Cancer Res 59 : 5331–5340.
63. KenyonC, ChangJ, GenschE, RudnerA, TabtiangR (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464.
64. LithgowGJ, WhiteTM, MelovS, JohnsonTE (1995) Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A 92 : 7540–7544.
65. LinK, HsinH, LibinaN, KenyonC (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 : 139–145.
66. CurranSP, RuvkunG (2007) Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet 3: e56.
67. ShoreDE, CarrCE, RuvkunG (2012) Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet 8: e1002792.
68. InoueH, HisamotoN, AnJH, OliveiraRP, NishidaE, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 : 2278–2283.
69. AballayA, DrenkardE, HilbunLR, AusubelFM (2003) Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol 13 : 47–52.
70. RichardsonCE, KinkelS, KimDH (2011) Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet 7: e1002391.
71. HuF, PadukkavidanaT, VægterCB, BradyOA, ZhengY, et al. (2010) Sortilin-Mediated Endocytosis Determines Levels of the Frontotemporal Dementia Protein, Progranulin. Neuron 68 : 654–667.
72. ReddienPW, HorvitzHR (2000) CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol 2 : 131–136.
73. HaskinsKA, RussellJF, GaddisN, DressmanHK, AballayA (2008) Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell 15 : 87–97.
74. SunJ, SinghV, Kajino-SakamotoR, AballayA (2011) Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332 : 729–732.
75. YeretssianG, CorreaRG, DoironK, FitzgeraldP, DillonCP, et al. (2011) Non-apoptotic role of BID in inflammation and innate immunity. Nature 474 : 96–99.
76. BurguillosMA, DeierborgT, KavanaghE, PerssonA, HajjiN, et al. (2010) Caspase signalling controls microglia activation and neurotoxicity. Nature 472 : 319–324.
77. de CalignonA, FoxLM, PitstickR, CarlsonGA, BacskaiBJ, et al. (2011) Caspase activation precedes and leads to tangles. Nature 464 : 1201–1204.
78. GuC, ShababM, StrasserR, WoltersPJ, ShindoT, et al. (2012) Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS One 7: e32422.
79. YamadaK, MatsushimaR, NishimuraM, Hara-NishimuraI (2001) A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol 127 : 1626–1634.
80. KoizumiM, Yamaguchi-ShinozakiK, TsujiH, ShinozakiK (1993) Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129 : 175–182.
81. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
82. FrankCA, BaumPD, GarrigaG (2003) HLH-14 is a C. elegans achaete-scute protein that promotes neurogenesis through asymmetric cell division. Development 130 : 6507–6518.
83. LamitinaST, MorrisonR, MoeckelGW, StrangeK (2004) Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am J Physiol Cell Physiol 286: C785–791.
84. SchusterE, McElweeJJ, TulletJM, DoonanR, MatthijssensF, et al. (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6 : 399.
85. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 : 277–283.
Štítky
Genetika Reprodukční medicína
Článek Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive SelectionČlánek Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand SkillČlánek Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage EvolvabilityČlánek Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation inČlánek Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 ModelČlánek Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period inČlánek VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival inČlánek Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2AČlánek A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding YeastČlánek Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on PhenotypeČlánek Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding SitesČlánek Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 9- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- The Pathway Gene Functions together with the -Dependent Isoprenoid Biosynthetic Pathway to Orchestrate Germ Cell Migration
- Take Off, Landing, and Fly Anesthesia
- Nucleosome Assembly Proteins Get SET to Defeat the Guardian of Chromosome Cohesion
- Whole-Exome Sequencing Reveals a Rapid Change in the Frequency of Rare Functional Variants in a Founding Population of Humans
- Evidence Is Evidence: An Interview with Mary-Claire King
- Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection
- Convergent Transcription Induces Dynamic DNA Methylation at Loci
- Environmental Stresses Disrupt Telomere Length Homeostasis
- Ultra-Sensitive Sequencing Reveals an Age-Related Increase in Somatic Mitochondrial Mutations That Are Inconsistent with Oxidative Damage
- Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill
- Genetic and Anatomical Basis of the Barrier Separating Wakefulness and Anesthetic-Induced Unresponsiveness
- The Locus, Exclusive to the Ambulacrarians, Encodes a Chromatin Insulator Binding Protein in the Sea Urchin Embryo
- Binding of NF-κB to Nucleosomes: Effect of Translational Positioning, Nucleosome Remodeling and Linker Histone H1
- Manipulating or Superseding Host Recombination Functions: A Dilemma That Shapes Phage Evolvability
- Dynamics of DNA Methylation in Recent Human and Great Ape Evolution
- Functional Dissection of Regulatory Models Using Gene Expression Data of Deletion Mutants
- PAQR-2 Regulates Fatty Acid Desaturation during Cold Adaptation in
- N-alpha-terminal Acetylation of Histone H4 Regulates Arginine Methylation and Ribosomal DNA Silencing
- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in
- miR-1/133a Clusters Cooperatively Specify the Cardiomyogenic Lineage by Adjustment of Myocardin Levels during Embryonic Heart Development
- Hsp104 Suppresses Polyglutamine-Induced Degeneration Post Onset in a Drosophila MJD/SCA3 Model
- Genome-Wide Analysis of Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of L
- Deep Resequencing of GWAS Loci Identifies Rare Variants in , and That Are Associated with Ulcerative Colitis
- Cooperative Interaction between Phosphorylation Sites on PERIOD Maintains Circadian Period in
- VAPB/ALS8 MSP Ligands Regulate Striated Muscle Energy Metabolism Critical for Adult Survival in
- Analysis of Genes Reveals Redundant and Independent Functions in the Inner Ear
- Predicting the Risk of Rheumatoid Arthritis and Its Age of Onset through Modelling Genetic Risk Variants with Smoking
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
- A Shift to Organismal Stress Resistance in Programmed Cell Death Mutants
- Fragile Site Instability in Causes Loss of Heterozygosity by Mitotic Crossovers and Break-Induced Replication
- Tracking of Chromosome and Replisome Dynamics in Reveals a Novel Chromosome Arrangement
- The Condition-Dependent Transcriptional Landscape of
- Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells
- Nebula/DSCR1 Upregulation Delays Neurodegeneration and Protects against APP-Induced Axonal Transport Defects by Restoring Calcineurin and GSK-3β Signaling
- System-Wide Analysis Reveals a Complex Network of Tumor-Fibroblast Interactions Involved in Tumorigenicity
- Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations
- and Are Required for Cellularization and Differentiation during Female Gametogenesis in
- Growth factor independent-1 Maintains Notch1-Dependent Transcriptional Programming of Lymphoid Precursors
- Whole Genome Sequencing Identifies a Deletion in Protein Phosphatase 2A That Affects Its Stability and Localization in
- An Alteration in ELMOD3, an Arl2 GTPase-Activating Protein, Is Associated with Hearing Impairment in Humans
- Genomic Identification of Founding Haplotypes Reveals the History of the Selfing Species
- Plasticity Regulators Modulate Specific Root Traits in Discrete Nitrogen Environments
- The IDD14, IDD15, and IDD16 Cooperatively Regulate Lateral Organ Morphogenesis and Gravitropism by Promoting Auxin Biosynthesis and Transport
- Stochastic Loss of Silencing of the Imprinted Allele, in a Mouse Model and Humans with Prader-Willi Syndrome, Has Functional Consequences
- The Prefoldin Complex Regulates Chromatin Dynamics during Transcription Elongation
- PKA Controls Calcium Influx into Motor Neurons during a Rhythmic Behavior
- A Pre-mRNA-Splicing Factor Is Required for RNA-Directed DNA Methylation in
- Cell-Type Specific Features of Circular RNA Expression
- The Uve1 Endonuclease Is Regulated by the White Collar Complex to Protect from UV Damage
- An Atypical Kinase under Balancing Selection Confers Broad-Spectrum Disease Resistance in Arabidopsis
- Genome-Wide Mutation Avalanches Induced in Diploid Yeast Cells by a Base Analog or an APOBEC Deaminase
- Extensive Divergence of Transcription Factor Binding in Embryos with Highly Conserved Gene Expression
- Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Cell Cycle
- Cell Interactions and Patterned Intercalations Shape and Link Epithelial Tubes in
- A Link between ORC-Origin Binding Mechanisms and Origin Activation Time Revealed in Budding Yeast
- The Genome and Development-Dependent Transcriptomes of : A Window into Fungal Evolution
- SKN-1/Nrf, A New Unfolded Protein Response Factor?
- The Highly Prolific Phenotype of Lacaune Sheep Is Associated with an Ectopic Expression of the Gene within the Ovary
- Fusion of Large-Scale Genomic Knowledge and Frequency Data Computationally Prioritizes Variants in Epilepsy
- IL-17 Attenuates Degradation of ARE-mRNAs by Changing the Cooperation between AU-Binding Proteins and microRNA16
- An Enhancer Element Harboring Variants Associated with Systemic Lupus Erythematosus Engages the Promoter to Influence A20 Expression
- Genome Analysis of a Transmissible Lineage of Reveals Pathoadaptive Mutations and Distinct Evolutionary Paths of Hypermutators
- Type I-E CRISPR-Cas Systems Discriminate Target from Non-Target DNA through Base Pairing-Independent PAM Recognition
- Divergent Transcriptional Regulatory Logic at the Intersection of Tissue Growth and Developmental Patterning
- MEIOB Targets Single-Strand DNA and Is Necessary for Meiotic Recombination
- Transmission of Hypervirulence Traits via Sexual Reproduction within and between Lineages of the Human Fungal Pathogen
- Integration of the Unfolded Protein and Oxidative Stress Responses through SKN-1/Nrf
- Guanine Holes Are Prominent Targets for Mutation in Cancer and Inherited Disease
- Regulation of the Boundaries of Accessible Chromatin
- Natural Genetic Transformation Generates a Population of Merodiploids in
- Ablating Adult Neurogenesis in the Rat Has No Effect on Spatial Processing: Evidence from a Novel Pharmacogenetic Model
- Genotype-Environment Interactions Reveal Causal Pathways That Mediate Genetic Effects on Phenotype
- The Molecular Mechanism of a -Regulatory Adaptation in Yeast
- Phenotypic and Genetic Consequences of Protein Damage
- Recent Acquisition of by Baka Pygmies
- Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons
- A Critical Role for PDGFRα Signaling in Medial Nasal Process Development
- Chromatin-Specific Regulation of Mammalian rDNA Transcription by Clustered TTF-I Binding Sites
- Meiotic Recombination in Arabidopsis Is Catalysed by DMC1, with RAD51 Playing a Supporting Role
- dTULP, the Homolog of Tubby, Regulates Transient Receptor Potential Channel Localization in Cilia
- Widespread Dysregulation of Peptide Hormone Release in Mice Lacking Adaptor Protein AP-3
- , a Direct Transcriptional Target, Modulates T-Box Factor Activity in Orofacial Clefting
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Genome-Wide Systematic Analysis Reveals Different and Predictive Proliferation Expression Signatures of Cancerous vs. Non-Cancerous Cells
- Recent Acquisition of by Baka Pygmies
- The Condition-Dependent Transcriptional Landscape of
- Histone Chaperone NAP1 Mediates Sister Chromatid Resolution by Counteracting Protein Phosphatase 2A
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



