-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
Transforming growth factor β (TGF-β) signaling acts through Smad proteins to play fundamental roles in cell proliferation, differentiation, apoptosis, and metabolism. The Receptor associated Smads (R-Smads) interact with DNA and other nuclear proteins to regulate target gene transcription. Here, we demonstrate that the Caenorhabditis elegans R-Smad DAF-8 partners with the nuclear hormone receptor NHR-69, a C. elegans ortholog of mammalian hepatocyte nuclear factor 4α HNF4α), to repress the exp-2 potassium channel gene and increase insulin secretion. We find that NHR-69 associates with DAF-8 both in vivo and in vitro. Functionally, daf-8 nhr-69 double mutants show defects in neuropeptide secretion and phenotypes consistent with reduced insulin signaling such as increased expression of the sod-3 and gst-10 genes and a longer life span. Expression of the exp-2 gene, encoding a voltage-gated potassium channel, is synergistically increased in daf-8 nhr-69 mutants compared to single mutants and wild-type worms. In turn, exp-2 acts selectively in the ASI neurons to repress the secretion of the insulin-like peptide DAF-28. Importantly, exp-2 mutation shortens the long life span of daf-8 nhr-69 double mutants, demonstrating that exp-2 is required downstream of DAF-8 and NHR-69. Finally, animals over-expressing NHR-69 specifically in DAF-28–secreting ASI neurons exhibit a lethargic, hypoglycemic phenotype that is rescued by exogenous glucose. We propose a model whereby DAF-8/R-Smad and NHR-69 negatively regulate the transcription of exp-2 to promote neuronal DAF-28 secretion, thus demonstrating a physiological crosstalk between TGF-β and HNF4α-like signaling in C. elegans. NHR-69 and DAF-8 dependent regulation of exp-2 and DAF-28 also provides a novel molecular mechanism that contributes to the previously recognized link between insulin and TGF-β signaling in C. elegans.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002519
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002519Summary
Transforming growth factor β (TGF-β) signaling acts through Smad proteins to play fundamental roles in cell proliferation, differentiation, apoptosis, and metabolism. The Receptor associated Smads (R-Smads) interact with DNA and other nuclear proteins to regulate target gene transcription. Here, we demonstrate that the Caenorhabditis elegans R-Smad DAF-8 partners with the nuclear hormone receptor NHR-69, a C. elegans ortholog of mammalian hepatocyte nuclear factor 4α HNF4α), to repress the exp-2 potassium channel gene and increase insulin secretion. We find that NHR-69 associates with DAF-8 both in vivo and in vitro. Functionally, daf-8 nhr-69 double mutants show defects in neuropeptide secretion and phenotypes consistent with reduced insulin signaling such as increased expression of the sod-3 and gst-10 genes and a longer life span. Expression of the exp-2 gene, encoding a voltage-gated potassium channel, is synergistically increased in daf-8 nhr-69 mutants compared to single mutants and wild-type worms. In turn, exp-2 acts selectively in the ASI neurons to repress the secretion of the insulin-like peptide DAF-28. Importantly, exp-2 mutation shortens the long life span of daf-8 nhr-69 double mutants, demonstrating that exp-2 is required downstream of DAF-8 and NHR-69. Finally, animals over-expressing NHR-69 specifically in DAF-28–secreting ASI neurons exhibit a lethargic, hypoglycemic phenotype that is rescued by exogenous glucose. We propose a model whereby DAF-8/R-Smad and NHR-69 negatively regulate the transcription of exp-2 to promote neuronal DAF-28 secretion, thus demonstrating a physiological crosstalk between TGF-β and HNF4α-like signaling in C. elegans. NHR-69 and DAF-8 dependent regulation of exp-2 and DAF-28 also provides a novel molecular mechanism that contributes to the previously recognized link between insulin and TGF-β signaling in C. elegans.
Introduction
Transforming growth factor β (TGF-β) signaling plays fundamental roles in cell proliferation, differentiation and apoptosis [1]. TGF-β binding induces the formation of a heterotetrameric type I and type II transmembrane receptor complex, and the type I receptor then phosphorylates the C-terminal Ser-X-Ser motif of receptor-associated Smad (R-Smad) transcription factors [2]. Phosphorylated R-Smads form homodimers and heterotrimers with a Co-Smad and translocate to the nucleus where they regulate the transcription of target genes [1].
TGF-β signaling is evolutionarily conserved; for example, in the lower metazoan Caenorhabditis elegans, two partially overlapping TGF-β signaling pathways have been described. One controls body size and male reproductive development whereas the other controls the formation of a specialized larval stage, the dauer [3]–[5]. The dauer larva is a developmentally arrested, long-lived and stress resistant diapause stage, formation of which is induced by overcrowding, starvation, and high temperatures [5]. Screening for mutations that result either in constitutive (Daf-c) or defective (Daf-d) dauer development [5] revealed multiple signaling circuits that control the dauer developmental switch, including a classical TGF-β signaling pathway. The TGF-β ligand DAF-7 promotes larval growth and inhibits dauer arrest; accordingly, transcription of daf-7, which is expressed in the amphid single ciliated neuron I (ASI) chemosensory neurons, is repressed by the dauer-inducing pheromone [6]. Downstream of daf-7, daf-1 and daf-4 encode the nematode type I and type II TGF-β receptor kinases, respectively, and both genes are required for non-dauer larval development. DAF-1 and DAF-4 phosphorylate DAF-8, an R-Smad that antagonizes the Co-Smad DAF-3 to promote larval growth [7]. In line with these roles, daf-1, daf-4, daf-7, and daf-8 mutants all exhibit Daf-c phenotypes. In an interesting divergence from mammalian TGF-β signaling, the C. elegans Co-Smad DAF-3 represses the transcription of the antagonistic upstream genes daf-7 and daf-8 by directly binding their promoters, thus creating a positive feedback loop [7].
Dauer formation is also influenced by other sensing and signaling pathways, such as cyclic guanosine monophosphate (cGMP) signaling, steroid receptor action and insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS) [5]. Interestingly, several recent reports indicate that IIS and TGF-β signaling interact in C. elegans. In the regulation of fasting/re-feeding induced quiescence, cGMP, insulin/IGF-1, and TGF-β signaling act as parallel activation signals for the protein kinase G (PKG) EGL-4 [8]. All three pathways also affect insulin gene expression, and the gene expression profiles of TGF-β and IIS pathway mutants exhibit a rather broad overlap [9]–[11]. Intriguingly, daf-7 mutants exhibit increased nuclear localization of the transcription factor DAF-16 [12], a key target of IIS that is also nuclear in worms carrying mutations in the insulin receptor gene daf-2. Moreover, the phosphatase PDP-1, which affects TGF-β signaling downstream of daf-7, and likely at the level of daf-8 and daf-14, also promotes DAF-16 nuclear localization and insulin gene expression [13]. Thus, PDP-1 and TGF-β signaling may directly promote the expression of insulin peptides, providing one possible mechanistic link between TGF-β signaling and IIS. Accordingly, both the Daf-c and the longevity phenotypes of TGF-β signaling mutants are suppressed at least partially in a daf-16 mutant [14][9]. These studies support the notion that TGF-β signaling may in part act upstream of IIS to regulate dauer formation and longevity. However, a simple linear relationship between TGF-β and IIS is unlikely even in the regulation of life span, because the longevity of TGF-β pathway mutants is very modest compared to IIS pathway mutants [9]. In any case, the mechanisms that couple IIS and TGF-β signaling remain poorly defined, and an apparent lack of recognizable Smad-binding sites amongst the genes co-regulated in IIS and TGF-β pathway mutants [9] suggests that these signaling pathways integrate upstream of transcriptional regulation.
Another mechanism that acts downstream of TGF-β signaling in the dauer formation cascade centers on the Nuclear Hormone Receptor (NHR) DAF-12 [15]. DAF-12 is one of 284 C. elegans NHRs that are involved in a variety of processes, including embryonic development, dauer formation, regulation of life span, molting, lipid metabolism, xenobiotic metabolism and excretory duct organogenesis [16][17]. Of the 284 NHRs, most are related to a Hepatocyte Nuclear Factor 4 alpha (HNF4α) related ancestor. Most of these remain poorly characterized, although several HNF4α orthologs regulate lipid metabolism and/or life span [18]–[22]. To date however, DAF-12 remains the only NHR with a defined role in the regulation of dauer formation.
To better define the regulatory network and the transcriptional complexes associated with C. elegans DAF-8/R-Smad, we set out to identify its physical interaction partners. We describe a novel physical and functional interaction between DAF-8 and NHR-69 that affects dauer formation, longevity, and the expression of the potassium channel gene exp-2, which in turn affects the secretion of the insulin-like peptide DAF-28. Our data indicate that TGF-β signaling and HNF4α-like signaling exhibit regulatory crosstalk to influence the secretion of an insulin-like peptide in C. elegans, providing a novel mechanistic link between TGF-β signaling and IIS.
Results
NHR-69 and DAF-8 physically interact in vitro and in vivo
To identify proteins that bind DAF-8, we used anti-FLAG immunoprecipitation to purify DAF-8 and associated peptides from a worm strain stably expressing a DAF-8::FLAG transgene [7]. Peptide identities were determined by mass spectrometry and we identified several peptides representing NHR-69 (data not shown). NHR-69 is one of the two closest C. elegans orthologs of human HNF4α with 39% amino acid identity and 60% similarity (as determined by reciprocal Blast analysis; see also [23]). To validate the interaction between these two transcription factors we generated anti-DAF-8 antibodies (Figure S1) and transgenic animals expressing a translational NHR-69::GFP fusion protein (nhr-69p::nhr-69::gfp; see Materials and Methods). Co-immunoprecipitation (Co-IP) showed that NHR-69::GFP associates with DAF-8 in vivo (Figure 1A). We also performed in vitro binding assays using GST-fusions and in vitro transcribed/translated proteins to test for direct interaction between NHR-69 and the functionally distinct SMAD proteins DAF-8, DAF-14, and DAF-3. We found that GST::NHR-69, but not GST alone, associated specifically with DAF-8, but not with DAF-14/R-Smad or DAF-3/Co-Smad (Figure 1B). Together, these data suggest that NHR-69 and DAF-8 specifically interact in vitro and in vivo.
Fig. 1. Interaction of NHR-69 with DAF-8 and expression pattern of nhr-69p::nhr-69::gfp. 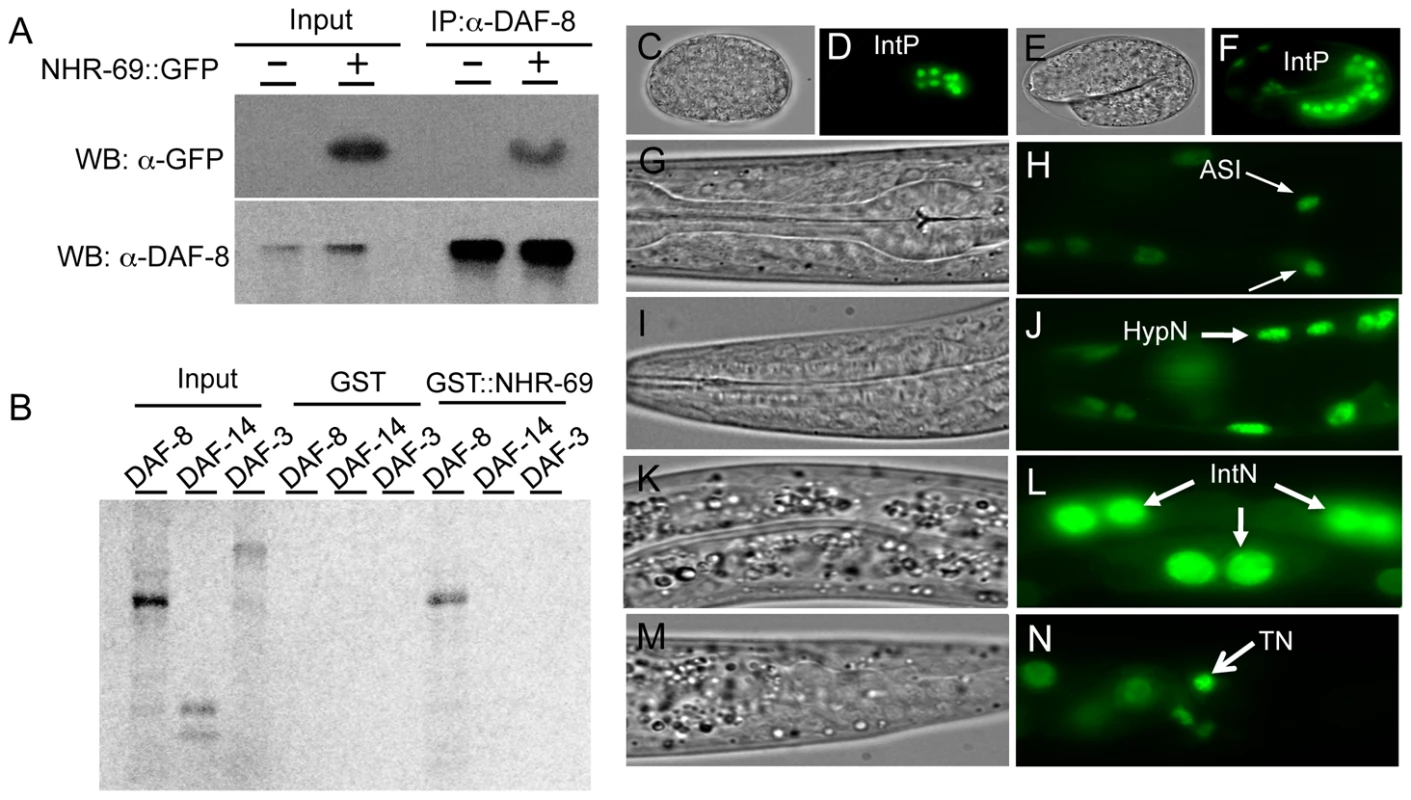
(A) In vivo co-immunoprecipitation was performed with anti-DAF-8 antibody from wild-type worms and nhr-69p::nhr-69::gfp expressing animals, followed by immunoblot with anti-GFP antibody to detect NHR-69::GFP. (B) In vitro GST pull-down assay between NHR-69 and SMAD proteins (DAF-8, DAF-14, or DAF-3). C–N show DIC and fluorescence images of developing embryos (C–F) and of adults (G–N) expressing nhr-69p::nhr-69::gfp. (C and D) Expression at the E16 stage of intestinal precursor cells in embryos. (E and F) Expression at the comma stage. (G and H) Expression in ASI neurons (arrows). (I and J) Hypodermal expression. (K and L) Intestinal expression. (M and N) Expression in tail neurons. IntP; Intestinal precursor cells, HypN; Hypodermal nucleus, IntN; Intestinal nucleus, TN; Tail neurons. As DAF-8 and NHR-69 interact physically, we would expect their in vivo expression patterns to at least partially overlap. Our previous work showed that DAF-8 is expressed in numerous tissues, including the ASI neurons and the intestine [7]. A NHR-69::GFP fusion protein driven by the nhr-69 promoter was detected in the nucleus of the E8 intestinal precursor cells in developing embryos (Figure 1D, 1F), and strong intestinal expression persisted throughout larval development until adulthood (Figure 1L). In adults, we also detected expression in the ASI neurons (Figure 1H, Figure S2), in the hypodermis, and in tail neurons (Figure 1J, 1L and 1N). In all of these tissues, NHR-69::GFP was localized exclusively to the nucleus, as expected for a transcription factor. Taken together with our previous findings, NHR-69 and DAF-8 [7] expression overlaps in the ASI neurons and in the intestine, suggesting that these two factors may interact functionally in these two tissues.
nhr-69 mutation enhances dauer formation and delays dauer recovery of TGF-β Daf-c mutants
Because DAF-8 and NHR-69 interact physically, we next asked whether NHR-69 affects DAF-8 function. As nhr-69 RNAi does not cause any obvious phenotypes in wild-type worms [22], we utilized a sensitized genetic background to test whether nhr-69 contributes to dauer formation. This worm strain, eri-1(mg366); sdf-9(m708), is hypersensitive to RNAi due to the eri-1 mutation [24], and sensitized to dauer formation due a mutation in the tyrosine phosphatase gene sdf-9, which acts parallel to IIS in C. elegans [25]. In this background, nhr-69 RNAi significantly enhanced dauer formation (Figure S3).
To corroborate the RNAi data we obtained a strain carrying a deletion in the nhr-69 gene, nhr-69(ok1926) (see Materials and Methods). The deletion spans the 5′ end of the gene and likely represents a null mutation. In agreement with the RNAi data, the nhr-69(ok1926) mutation enhanced the dauer formation of temperature-sensitive daf-7(e1372), daf-1(m40), daf-8(m85), and daf-14(m77) mutants at the semi-restrictive temperature (22.5°C Table 1; note that even null mutants in the TGF-β pathway like daf-8 are temperature-sensitive because dauer formation is temperature-dependent [26]).
Tab. 1. Loss of nhr-69 enhances the dauer phenotype of TGF-β signaling mutants. 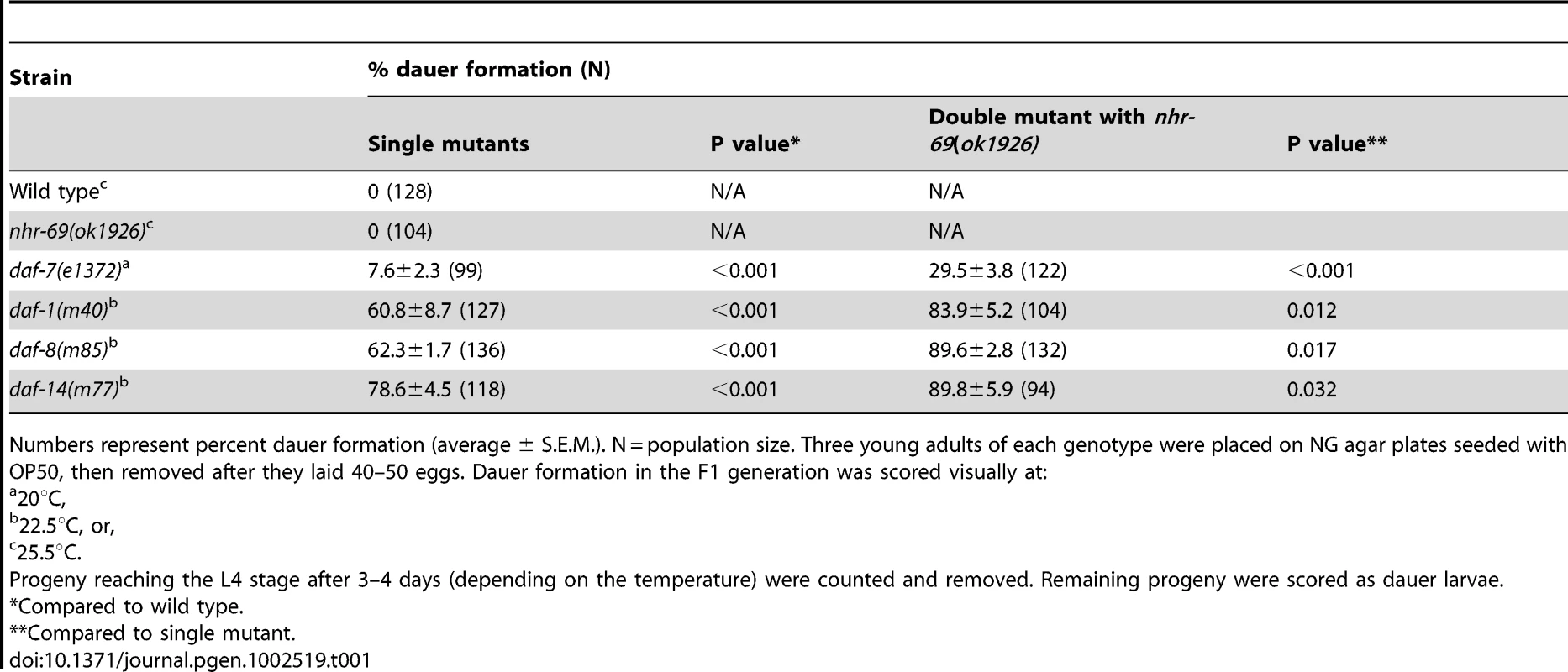
Numbers represent percent dauer formation (average ± S.E.M.). N = population size. Three young adults of each genotype were placed on NG agar plates seeded with OP50, then removed after they laid 40–50 eggs. Dauer formation in the F1 generation was scored visually at: TGF-β pathway mutants that constitutively arrest as dauer larvae at restrictive temperatures (25°C) also exhibit a temperature-dependent dauer exit phenotype. Thus, we tested whether the nhr-69(ok1926) mutation compounded the dauer exit phenotype of TGF-β Daf-c mutants. As expected, TGF-β mutant dauer larvae rapidly recovered from the dauer state, within 24–48 hours after they were transferred to the non-restrictive temperature (15°C; Table 2). In contrast, the nhr-69(ok1926) mutation suppressed the dauer exit capacity of daf-7(e1372), daf-1(m40), daf-8(m85) and daf-14(m77) mutants (Table 2). Hence, loss of nhr-69 function not only enhances dauer formation, but also delays the dauer exit of TGF-β Daf-c mutants.
Tab. 2. Effect of the nhr-69 mutation on dauer recovery of TGF-β mutants. 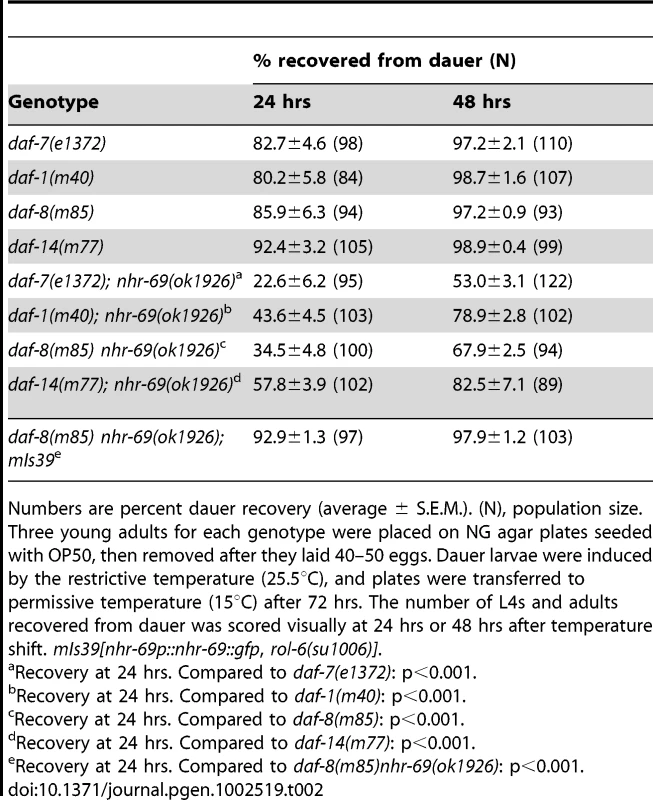
Numbers are percent dauer recovery (average ± S.E.M.). (N), population size. Three young adults for each genotype were placed on NG agar plates seeded with OP50, then removed after they laid 40–50 eggs. Dauer larvae were induced by the restrictive temperature (25.5°C), and plates were transferred to permissive temperature (15°C) after 72 hrs. The number of L4s and adults recovered from dauer was scored visually at 24 hrs or 48 hrs after temperature shift. mIs39[nhr-69p::nhr-69::gfp, rol-6(su1006)]. Insulin signaling, but not TGF-β signaling, regulates NHR-69 expression
Because of the enhanced dauer formation phenotype conveyed by nhr-69 depletion or mutation, and because TGF-β signaling and insulin signaling both regulate dauer entry and exit [27], we next asked whether these pathways regulate nhr-69 expression. We used real-time quantitative PCR (qPCR) to quantify nhr-69 mRNA levels in the TGF-β pathway mutants daf-7(e1372), daf-1(m40), daf-8(m85) and daf-14(m77), and in the insulin signaling mutants daf-2(e1370), daf-16(mgDf47), and daf-2(e1370); daf-16(mgDf47). daf-2 encodes the C. elegans insulin/IGF-1 receptor and daf-16 a downstream transcription factor of the Forkhead Box O family [28][29]. The mRNA levels of nhr-69 were similar in wild-type, daf-1(m40), daf-7(e1372), daf-8(m85), and daf-14(m77) worms (p>0.05), but were downregulated in daf-2(e1370) and upregulated in daf-2(e1370); daf-16(mgDf47) and daf-16(mgDf47) mutants (Figure 2A), suggesting that DAF-2 regulates nhr-69 in DAF-16–dependent fashion.
Fig. 2. Insulin signaling positively regulates nhr-69 expression. 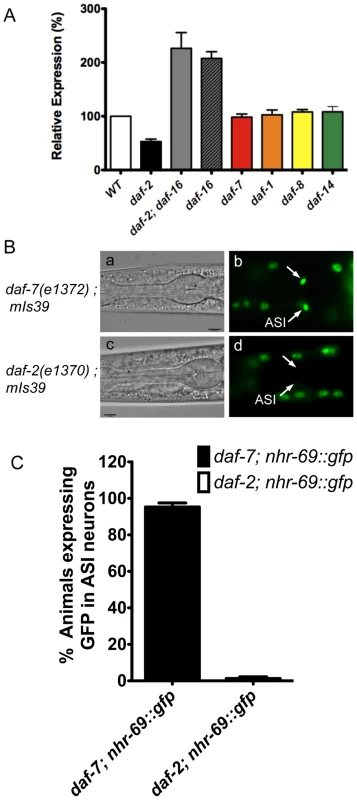
(A) The bars represent relative levels of nhr-69 mRNA as determined by qPCR on RNA isolated from daf-2(e1370), daf-2(e1370); daf-16(mgDf47), daf-16(mgDf47), daf-1(m40), daf-7(e1372), daf-8(m85) and daf-14(m77) mutants (relative to mRNA levels in wild-type worms); mRNA levels were normalized to act-2. Error bars indicate standard errors of the mean (SEM) from four biological replicates. (B) Micrographs show DIC and fluorescence images of NHR-69::GFP in daf-7(e1372) (a and b) and daf-2(e1370) (c and d) mutants. Arrows indicate the ASI neurons. All fluorescence images were taken with identical exposure times. (C) Bar graphs indicate the percentage of animals expressing NHR-69::GFP in ASI neurons in daf-7(e1372) (black bar, N = 78) and in daf-2(e1370) mutant (white bar, N = 83) shown in (B). Error bars indicate SEM. Given this regulation and given that NHR-69::GFP is expressed in multiple tissues, we next addressed whether insulin or TGF-β signaling affect nhr-69 expression in any particular tissue. To this end, we generated strains that express the NHR-69::GFP fusion protein in the daf-2(e1370) and daf-7(e1372) mutant backgrounds. We observed normal NHR-69::GFP expression in the ASI neurons of daf-7(e1372) mutants (96±3% of L2 larvae showed GFP fluorescence in both neurons), but NHR-69::GFP was barely detectable in the ASI neurons of daf-2(e1370) mutants (0.3±0.04% of L2 larvae showed GFP fluorescence in both neurons, p<0.001; Figure 2B c and d). Together, these data suggest that insulin signaling positively regulates NHR-69 expression specifically in the ASI neurons.
Insulin signaling is impaired in a daf-8 nhr-69 double mutant
The daf-8(m85) nhr-69(ok1926) double mutants exhibited defects in dauer entry and exit, processes regulated by insulin signaling [27]; thus, we next asked whether daf-8 and nhr-69 affect insulin signaling in C. elegans. First, we examined the expression of sod-3 and gst-10, two downstream targets of IIS, which are regulated by the transcription factors DAF-16 and SKN-1, respectively [30][31]. SKN-1 is a transcription factor of the bZIP family that acts downstream of daf-2 in C. elegans. qPCR analysis showed that both transcripts were upregulated in the daf-8(m85) single mutant; for sod-3, this is consistent with previous findings that TGF-β signaling regulates daf-16 target genes [9]. Furthermore, sod-3 and gst-10 induction was enhanced in the daf-8(m85) nhr-69(ok1926) double mutant, suggesting that insulin signaling was further reduced in the double mutant (Figure 3A, 3B).
Fig. 3. Reduced insulin signaling and neuropeptide secretion in daf-8 nhr-69 mutants. 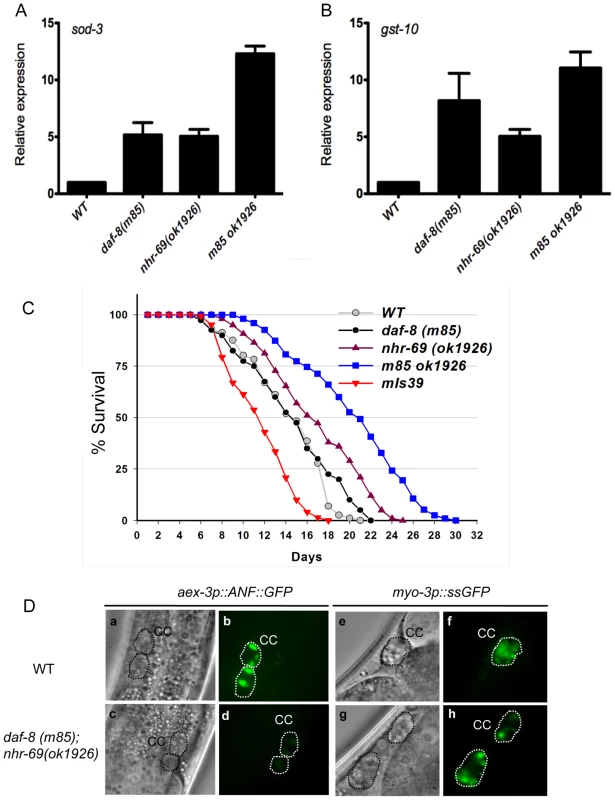
(A and B) Bar graphs show the relative mRNA levels of sod-3 and gst-10 in daf-8(m85), nhr-69(ok1926) and daf-8(m85) nhr-69(ok1926) mutants relative to wild-type worms, as determined by qPCR (normalized to act-2). Age synchronized day-1 adults were used for total RNA extraction. Error bars indicate SEM from three biological replicates. (C) The plots show population survival of wild-type (grey circles), daf-8(m85) (black circles), nhr-69(ok1926) (purple triangles), daf-8(m85) nhr-69(ok1926) (blue squares), and mIs39[nhr-69p::nhr-69::gfp] (red inverted triangles) worms at 25.5°C. (D) DIC and fluorescence micrographs show coelomocyte accumulation (CC, dotted circles) of ANF::GFP (expressed from the oxIs206[aex-3p::ANF::gfp] transgene; b and d) or ssGFP (expressed from the arIs37[myo-3P::ssGFP] transgene; f and h), respectively, in wild-type worms and in daf-8(m85) nhr-69(ok1926) mutants, as indicated. Because adult life span is extended by reduced insulin signaling [28], we next tested whether nhr-69 and daf-8 affect longevity. Figure 3C shows that nhr-69(ok1926) single mutants exhibited a slightly extended life span relative to wild-type worms (mean life span 17.4±1.3 days, p = 0.027), whereas the daf-8(m85) mutant life span was similar to that of wild type (mean life span 14.6±0.8 days, p>0.05; Figure 3C). Notably, the life span of daf-8(m85) nhr-69(ok1926) double mutants was significantly longer than that of wild type worms (mean life span 21.5±1.7 days, p<0.001; Figure 3C), consistent with reduced insulin signaling. By contrast, animals over-expressing NHR-69::GFP exhibited a shortened life span (mean life span 11.8±0.6 days, p<0.001) (Figure 3C and Table S1). Together, these data suggest that daf-8 and nhr-69 cooperatively affect insulin signaling in C. elegans.
Defective neuropeptide secretion in daf-8 nhr-69 mutants
Next, we wished to define the mechanism by which nhr-69 and daf-8 affect insulin signaling. Given that both genes are expressed in the neuropeptide-secreting ASI neurons, we hypothesized that they might control the release of neuropeptides such as the insulin-like peptide DAF-28, which is secreted by the ASI neurons [32]. To examine this possibility, we first attempted to introduce the DAF-28::GFP transgene into the daf-8(m85) mutant background. For unknown reasons, these worms exhibited a non-recoverable Daf-c phenotype, making it impossible to study DAF-28::GFP secretion in this background (data not shown). As an alternative approach, we used worms expressing an aex-3 promoter-driven ANF::GFP (human Atrial Natriuretic Factor-GFP fusion protein), which can be used as a surrogate to study neuropeptide secretion in C. elegans [33]. aex-3p::ANF::GFP is expressed in a pan-neuronal fashion, secreted into the inter-organ space (pseudocoelom), and then endocytosed by the coelomocytes, scavenger-like cells that endocytose secreted material [33]. The accumulation of ANF::GFP in coelomocytes was similar in wild-type worms and daf-8(m85) and nhr-69(ok1926) single mutants, but daf-8(m85) nhr-69(ok1926) double mutants accumulated less ANF::GFP (Figure 3D d, Figure S4A). The overall neuronal ANF::GFP gene expression was comparable in all worm strains (Figure S4C).
The differential ANF::GFP accumulation in coelomocytes could result either from differences in secretion or endocytosis. Thus, we tested whether the daf-8(m85) nhr-69(ok1926) mutants are generally defective for endocytosis. Specifically, we examined a signal sequence GFP (ssGFP) driven by the myo-3 promoter, which is expressed in muscle cells, secreted into the pseudocoelom and endocytosed by coelomocytes [34]. The accumulation of ssGFP in coelomocytes was similar in wild-type worms and in daf-8(m85) nhr-69(ok1926) double mutants (Figure 3D f and h, Figure S4B), suggesting that the daf-8(m85) nhr-69(ok1926) mutants are defective specifically for neuropeptide secretion and not for endocytosis.
NHR-69 directly targets the potassium channel gene exp-2
Although our data suggest that NHR-69 and DAF-8 control neuropeptide secretion, the target genes that might underlie this process remained unknown. Intriguingly, a yeast-one-hybrid (Y1H) study found that NHR-69 binds the promoter of exp-2, which encodes a member of the voltage-gated transmembrane potassium channel (Kv) family [35][36]. Using qPCR, we found that exp-2 expression was slightly upregulated in the daf-8(m85) and nhr-69(ok1926) single mutants and synergistically upregulated in the daf-8(m85) nhr-69(ok1926) double mutant (Figure 4A).
Fig. 4. NHR-69 directly regulates exp-2 transcription. 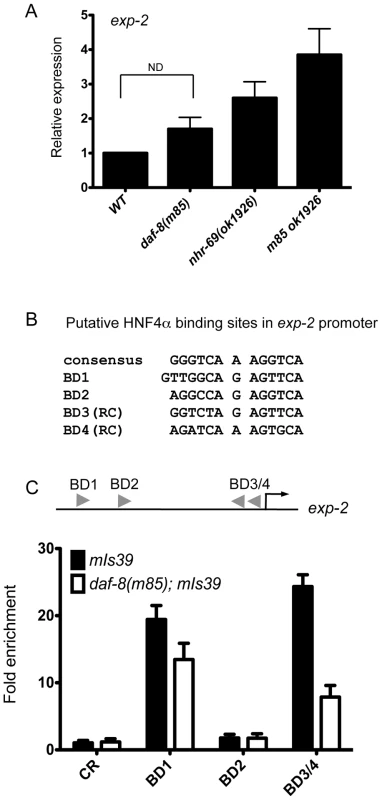
(A) The bar graphs represent the relative levels of exp-2 mRNA in daf-89(m85), nhr-69(ok1926), and daf-8(m85) nhr-69(ok1926) mutants relative to wild type as determined by qPCR (normalized to act-2). Error bars indicate SEM from three biological replicates. Age synchronized day-1 adults were used for total RNA extraction. (B) HNF4α binding consensus sequence and putative NHR-69/HNF4α-like binding sites in the exp-2 promoter region. The positions of possible binding sites 5′ of the ATG of exp-2 are BD1 (−2342 bp), BD2 (−1832 bp), BD3 (−118 bp) and BD4 (−86 bp), respectively. RC, reverse complement. (C) ChIP-qPCR with anti-GFP antibody reveals that NHR-69 is directly associated with at least two sites in the 5′ regulatory region of exp-2. The arrowheads indicate the putative HNF4α binding sites shown in (B). Anti-GFP antibody precipitated sequences from BD1 and BD3 and/or BD4. In a daf-8 background, binding of NHR-69::GFP was reduced 27% for BD1 and 71% for BD3/BD4. Error bars indicate SEM from three biological replicates. The Y1H data suggested that NHR-69 can directly bind the exp-2 promoter. Given the similarity of NHR-69 to mammalian HNF4α, we examined the 2.5 kb region 5′ of the exp-2 start codon for HNF4α-like consensus binding sites [37] and identified four putative elements (Figure 4B). To test whether NHR-69 directly binds any of these sites, we performed chromatin immunoprecipitation (ChIP) on mixed-stage nhr-69p::nhr-69::gfp animals using an anti-GFP antibody. NHR-69 was found to directly associate with at least two of these sites (Figure 4C). Intriguingly, NHR-69 binding was reduced, albeit not entirely lost, in daf-8(m85) mutants, suggesting that DAF-8 is required for optimal NHR-69 binding to the exp-2 promoter (Figure 4C).
To confirm that NHR-69 directly regulates the exp-2 promoter we developed a mammalian reporter assay. We previously showed that the nuclear localization of DAF-8 requires the TGF-β Type 1 receptor DAF-1 [7], and others have reported that mutating two C-terminal serines (Ser465 and Ser467) to glutamates results in phosphomimetic, constitutively active human Smad2 [38]. Generating a phosphomimetic form of DAF-8 (pmDAF-8) by changing two orthologous C-terminal serines (S543 and S544) to glutamate followed by immunohistochemistry showed that pmDAF-8 localizes to the nucleus in transiently transfected HEK293 cells (Figure 5A; resembling the nuclear localization of a DAF-8::GFP fusion protein in C. elegans [7]). NHR-69::GFP was also localized to the nucleus when transiently transfected into HEK293 cells (Figure 5A). Importantly, NHR-69::GFP repressed the activity of a construct in which the wild-type exp-2 promoter is fused to a luciferase reporter (Figure 5B). Moreover, this repression was enhanced by co-transfection of pmDAF-8 (Figure 5B), resembling the results obtained in C. elegans.
Fig. 5. NHR-69 and DAF-8 suppress exp-2 promoter activity in mammalian cells. 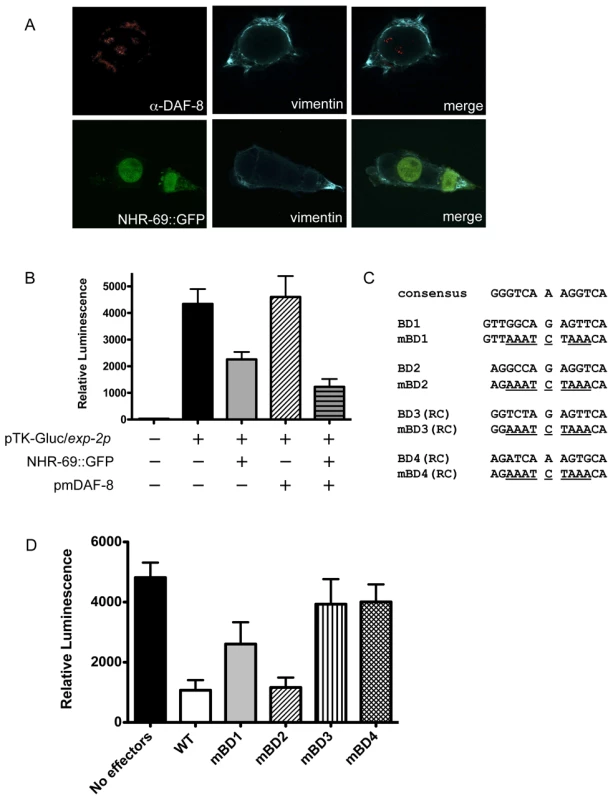
(A) Plasmids expressing NHR-69::GFP and a phosphomimetic variant of DAF-8 (pmDAF-8) were transiently transfected into HEK293 cells and visualized by fluorescence imaging; both proteins show nuclear localization. (B) The bar graphs depict relative luciferase activity resulting from a transiently transfected exp-2-driven luciferase reporter in HEK293 cells. NHR-69, but not pmDAF-8 single transfection represses the exp-2 promoter, and cotransfection of NHR-69 and pmDAF-8 causes an even stronger repression. (C) Details on the site-directed mutagenesis of HNF4α-like binding sites in the exp-2 promoter. (D) The graph depicts the consequence of site directed mutagenesis of HNF4α-like binding sites in the exp-2 promoter on luciferase activity. “No effectors” indicates transfection of the reporter alone, whereas “WT” indicates co-transfection of NHR-69 and pmDAF-8 with the wild-type exp-2 promoter (similar to panel B). Mutating BD1, BD3, or BD4 attenuates the NHR-69 and pmDAF-8-mediated repression of exp-2 activity. The result is an average from three individual trials. To test whether any of the NHR-69 binding sites identified in our ChIP analysis are functionally important, we performed a mutational analysis of the exp-2 promoter (Figure 4B and Figure 5C). Using plasmids carrying exp-2 promoter variants with mutated NHR-69 binding sites (BD1, BD2, BD3 or BD4) resulted in 46%, 76%, 18%, and 17% respective reductions of promoter activity upon NHR-69::GFP and pmDAF-8 co-transfection, whereas the wild-type exp-2 promoter was repressed by approximately 80% (Figure 5D). These data suggest that BD1, BD3, and BD4 contribute to exp-2 repression by NHR-69 whereas BD2 is likely dispensable.
EXP-2 functionally contributes to insulin signaling downstream of NHR-69 and DAF-8
The above data indicate that exp-2 is a direct target of nhr-69 and daf-8; thus, we next wished to test whether exp-2 was functionally required downstream of daf-8 and nhr-69. EXP-2 is a potassium channel that regulates the action potential in pharyngeal muscle [36]. Worms carrying a heterozygous exp-2 gain-of-function allele (exp-2(sa26), encoding an EXP-2 protein with a C480Y point mutation) exhibit defective feeding behavior, hyperactive head movements, and egg-laying defects, whereas homozygous worms arrest terminally after hatching [36][39]. Worms carrying null mutations in exp-2, such as the exp-2(sa26ad1426) allele carrying a T457I mutation, exhibit no defects in defecation or head movement, but have long-lasting pharyngeal muscle contractions [36]. Together, these mutants provide an opportunity to address the functional requirement of exp-2 in insulin signaling.
To study the effect of exp-2 loss, we generated a daf-8(m85) nhr-69(ok1926) exp-2(sa26ad1426) triple mutant and found that the life-span extension due to loss of daf-8 and nhr-69 was abolished by concomitant loss of exp-2 (Figure 6F, Table S1). Taken together, our results suggest that NHR-69 and DAF-8 cooperate to directly regulate exp-2, and that exp-2 is a critical downstream target of these two transcription factors.
Fig. 6. Insulin signaling is impaired in exp-2 mutants. 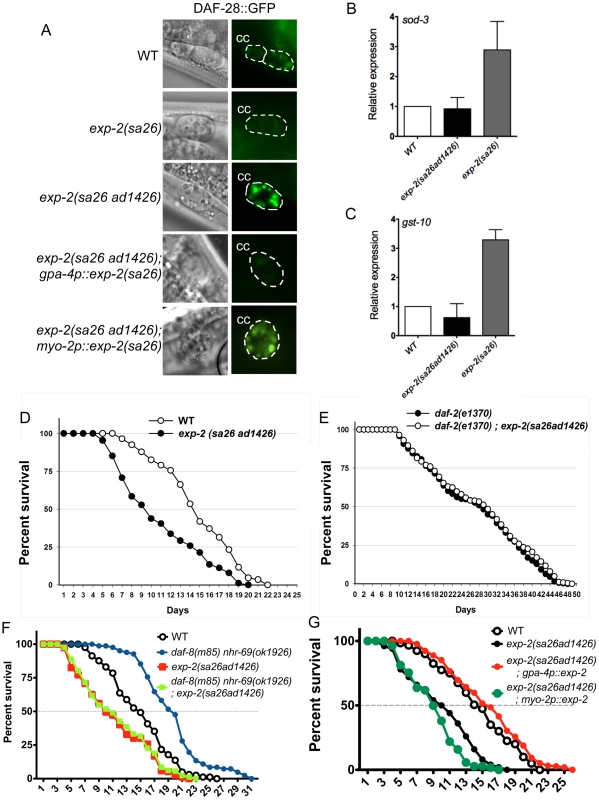
(A) The micrographs show DAF-28::GFP accumulation in coelomocytes (CC) in wild-type worms, in exp-2 mutants, and in exp-2 transgenic strains. exp-2(sa26ad1426) loss-of-function and exp-2(sa26) gain-of-function mutants show increased and decreased DAF-28::GFP intensity, respectively, indicating that EXP-2 represses DAF-28 secretion. ASI-neuron specific expression (driven by the gpa-4 promoter) of the gain-of-function exp-2(sa26) allele in the exp-2(sa26ad1426) background reduces DAF-28::GFP accumulation, resulting in a level similar to that seen in the global exp-2(sa26) gain-of-function mutant; in contrast, pharyngeal-specific exp-2(sa26) expression (driven by the myo-2 promoter) does not alter DAF-28::GFP accumulation in the exp-2(sa26ad1426) background. (B and C) The bar graphs depict average sod-3 and gst-10 mRNA levels in wild-type (white), exp-2(sa26ad1426) (black), and exp-2(sa26) (grey) worms, as determined by qPCR (normalized to act-2). Error bars indicate SEM from four biological replicates. (D) The graph depicts population survival curves for wild type worms (white) and exp-2(sa26ad1426) loss-of-function mutants (black; T = 25.5°C). (E) The graph shows population survival curves of daf-2(e1370) (black) and daf-2(e1370); exp-2(sa26ad1426) mutants (white; T = 25.5°C). (F) The graph depicts population survival curves of wild-type (black circle), daf-8(m85) nhr-69(ok1926) (blue circle), exp-2(sa26ad1426) (red square), and daf-8(m85) nhr-69(ok19260; exp-2(sa26ad1426) (green square) worms (T = 25.5°C). (G) The graph shows population survival curves of wild-type (white), exp-2(sa26ad1426) (black), exp-2(sa26ad1426); gpa-4p::exp-2 (red) and exp-2(sa26ad1426); myo-2p::exp-2 (green) (T = 25.5°C). All strains shown in this panel also harbor the pRF4[rol-6(su1006)] transgene. EXP-2 regulates insulin signaling through its actions in the ASI neurons
Because the exp-2 loss-of-function mutation reduces the longevity of daf-8(m85) nhr-69(ok1926) double mutants, we next tested whether exp-2 directly affects insulin signaling. First, we tested whether loss-of-function and gain-of-function exp-2 mutants [36] affect the accumulation of the agonistic insulin-like peptide DAF-28 in coelomocytes. Using a translational DAF-28::GFP reporter [40], we found that gain-of-function exp-2(sa26) mutants accumulated less DAF-28::GFP in the coelomocytes than wild-type worms, whereas loss-of-function exp-2(sa26ad1426) mutants accumulated more (Figure 6A, Figure S5A).
Because NHR-69 and DAF-8 directly regulate exp-2, and because DAF-28, DAF-8 and NHR-69 are expressed in ASI neurons (Figure S2) [7][32], we reasoned that EXP-2 should also modulate DAF-28::GFP secretion by acting in the ASI neurons. In support of this possibility, exp-2 is strongly expressed in the amphid neurons, as well as in the pharyngeal muscle [36]. To address a putative tissue-specific role for EXP-2, we expressed the exp-2(sa26) mutant gene under the control of the ASI-specific gpa-4 promoter [gpa-4p::exp-2(sa26)] [41] or the pharynx-specific myo-2 promoter [myo-2p::exp-2(sa26)] [42] in exp-2(sa26ad1426); daf-28::gfp animals. We found that, when exp-2(sa26) was expressed selectively in ASI neurons, it caused DAF-28::GFP accumulation at a similar level as seen in exp-2(sa26) worms, whereas pharyngeal expression of exp-2(sa26) resulted in DAF-28::GFP accumulation comparable to that observed in exp-2(sa26ad1426); daf-28::gfp animals (Figure 6A, Figure S5A). Importantly, the mRNA level of daf-28 was comparable in wild-type, exp-2(sa26), exp-2(sa26ad1426), gpa-4p::exp-2(sa26) and myo-2p::exp-2(sa26) worms, as determined by qPCR (Figure S5B). Thus, EXP-2 affects DAF-28 secretion, but not expression, selectively through its function in the ASI neurons.
If EXP-2 regulated DAF-28 secretion, it would be expected to affect downstream insulin target genes and phenotypes such as longevity. Indeed, the mRNA levels of sod-3 and gst-10 were upregulated in gain-of-function exp-2(sa26) mutants (Figure 6B, 6C), and we also observed increased nuclear localization of DAF-16::GFP in the exp-2(sa26) background (Figure S6), consistent with decreased insulin signaling [43]. Moreover, exp-2(sa26ad1426) loss-of-function mutants exhibited a significantly shortened life span compared to that of wild-type worms (mean life span: 9.4±0.6, p = 0.0116; we were unable to assess adult life span of exp-2(sa26) mutants due to lethality). In contrast, the life span of the exp-2(sa26ad1426); daf-2(e1370) double mutants was comparable to that of the daf-2(e1370) mutant itself (Figure 6E, p = 0.7606), suggesting that exp-2 acts upstream of daf-2 to regulate life span, consistent with its role in DAF-28 secretion.
Because exp-2 acts selectively in ASI neurons to control DAF-28 secretion (Figure 6A), we next addressed whether life span regulation by exp-2 also originates from the ASI neurons. Expression of wild-type exp-2 specifically in the ASI neurons (gpa-4p::exp-2) rescued the short life span phenotype of exp-2(sa26ad1426) mutant worms (14.3 days, p = 0.0139), whereas pharyngeal-specific expression did not (8.7 days, p = 0.8326; Figure 6G). This suggests that exp-2 plays an important role in ASI-neurons to regulate adult life span, possibly through its effect on DAF-28 secretion. Taken together, these data provide evidence that life span is inversely related to EXP-2 activity.
Overexpression of NHR-69 in the ASI neurons confers a hypoglycemic phenotype
As NHR-69::GFP is expressed in ASI neurons and DAF-28 is produced and secreted in these cells [32], we asked whether NHR-69 activity in the ASI neurons directly affects insulin signaling. First, we wished to ensure that an nhr-69::gfp translational fusion was biologically functional; thus, we expressed nhr-69::gfp from its own promoter (mIs39[nhr-69p::nhr-69::gfp]). Figure 3C shows that worms expressing this construct in the daf-8(m85) nhr-69(ok1926) mutant background exhibited a shortened life span compared to wild-type and a normal dauer recovery phenotype, suggesting that functional NHR-69 was overexpressed in these worms (Figure 3C and Table 2). Having assured that nhr-69 rescue can work in principle, we generated two transgenic lines that expressed NHR-69::GFP under the control of the ASI-specific gpa-4 promoter (gpa-4p::nhr-69::gfp) [41]. In these worms, we observed a higher accumulation of DAF-28::GFP in coelomocytes (compared to the wild-type background; Figure 7A). Moreover, the ASI specific expression of nhr-69::gfp resulted in a lethargic phenotype with slow, sluggish movement (Figure 7B) reminiscent of hypoglycemia in mammals [44]. Indeed, the endogenous glucose content was reduced in gpa-4p::nhr-69::gfp expressing animals compared to that of wild type (Figure 7C). In agreement with lower glucose levels in gpa-4p::nhr-69::gfp animals, the lethargic phenotype was almost completely rescued by exogenous glucose, but not by non-metabolizable 2-deoxyglucose, suggesting that glucose metabolism, not uptake is impaired in these animals. We conclude that NHR-69 overexpression in the ASI neurons results in hypoglycemia manifested as lethargic behavior, and that NHR-69 acts in the ASI neurons to regulate DAF-28 secretion.
Fig. 7. ASI-specific over-expression of NHR-69 confers hypoglycemia. 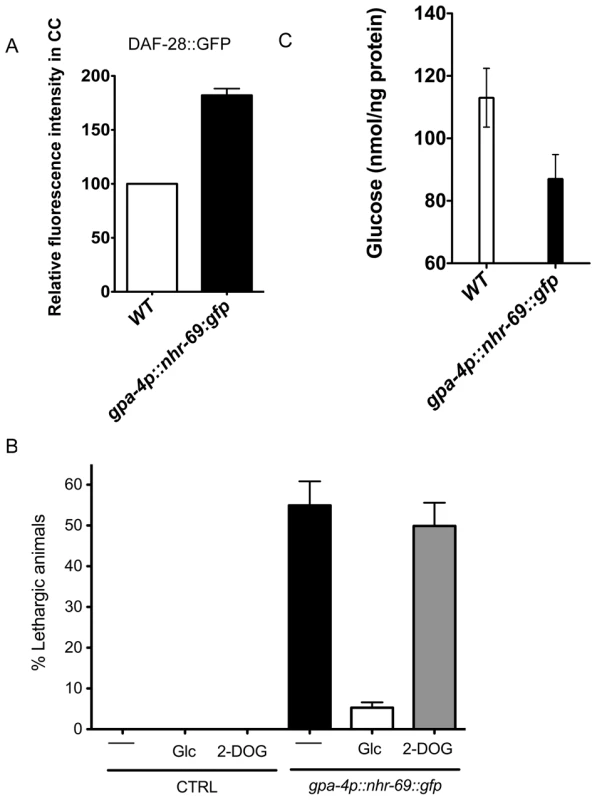
(A) The bar graphs show the relative DAF-28::GFP level in coelomocytes (CC) in wild-type and mIs40[gpa-4p::nhr-69::gfp] day-1 old adult worms. Error bars indicate SEM. N = 62 in wild type, N = 77 in mIs40[gpa-4p::nhr-69::gfp] backgrounds (p<0.001). (B) Bars indicate the percentage of lethargic animals in the presence or absence of exogenous glucose (2 mM Glc) or 2-dexoyglucose (2 mM 2-DOG). Synchronous day-1 adults were video recorded for two minutes individually and worms that stopped and resumed moving more than four times were judged to be lethargic (N = 50). (C) Bars show endogenous glucose content in wild-type and mIs40[gpa-4p::nhr-69::gfp] worms. Synchronous day-1 adults were assayed in four independent biological replicates. Error bars indicate SEM. Discussion
The combinatorial interactions of Smad proteins with other DNA-binding transcription factors provide a critical selectivity required to regulate specific subsets of target genes. In this regard, identifying the transcriptional regulators that cooperate with Smad proteins will help delineate the mechanisms underlying signal integration. Here, we describe a molecular crosstalk in C. elegans whereby the R-Smad DAF-8 partners with NHR-69 to regulate insulin signaling. We find that NHR-69 and DAF-8 regulate the exp-2 potassium channel gene, which in turn modulates the secretion of the insulin-like peptide DAF-28. We further demonstrate that NHR-69 affects DAF-28 secretion and glucose homeostasis through its actions in the neuropeptide-secreting ASI neurons that functionally resemble the mammalian pancreas. These data not only implicate the HNF4α-like NHR-69 in insulin signaling, but also suggest a novel mechanism for the previously suggested link between IIS and TGF-β signaling. Intriguingly, Narasimhan et al. recently showed that the phosphatase PDP-1 modulates IIS by acting at the level of the Smad proteins DAF-8 and DAF-14 [13]. Taken together with our findings, this strengthens the notion that there is significantly more crosstalk between IIS and TGF-β signaling than previously appreciated.
DAF-8 and NHR-69 cooperatively regulate insulin signaling
We set out to find novel transcriptional partners for DAF-8 and identified NHR-69 as a candidate DAF-8 interacting protein. In vivo co-immunoprecipitation and in vitro GST pull-down assays confirmed that NHR-69 and DAF-8 physically associate, whereas the Smads DAF-3 and DAF-14 did not bind to NHR-69, indicating that the interaction is specific for DAF-8. We also found evidence for a functional interaction between DAF-8 and NHR-69, demonstrating that nhr-69 is involved in dauer formation: depletion of nhr-69 by RNAi enhanced dauer formation in sensitized genetic backgrounds, including sdf-9 and several TGF-β Daf-c mutants. Furthermore, resumption of normal larval development of daf-1, daf-7, daf-8 and daf-14 dauer larvae after temperature downshift was delayed approximately two-fold in the nhr-69 mutant background. Because daf-2 signaling is required for temperature-dependent dauer recovery [27], this result suggests that insulin signaling was impaired in nhr-69 mutants.
In line with this notion, we observed an extended life span and upregulation of the insulin-regulated genes sod-3 and gst-10 in the daf-8 nhr-69 double mutant. We note that the observed longevity likely reflects altered expression of genes other than just sod-3 and gst-10, akin to the observations made in long-lived daf-2 mutants, wherein numerous genes are thought to contribute to longevity [9]. Lastly, we observed a reduced accumulation of ANF::GFP in the coelomocytes of daf-8 nhr-69 double mutants, indicating that neuropeptide secretion is impaired in daf-8 nhr-69 double mutants, representing the likely cause for impaired insulin signaling.
The relationship between TGF-β signaling and IIS may be conserved in evolution. Members of the TGF-β superfamily affect the development and physiology of mammalian pancreatic β-cells, although the results are somewhat conflicting. In one study, Smad3 repressed the insulin gene promoter and Smad3 knockdown enhanced glucose stimulated insulin secretion (GSIS) [45], suggesting a repressive role for TGF-β signaling. However, in isolated rat fetus pancreatic β-cells, TGF-β1 stimulates long-term (48 hours) insulin secretion [46], perhaps through transcriptional changes. Similarly, TGF-β1 and Activin A, another TGF-β family member, stimulate short-term (five minutes) insulin secretion in mouse insulinoma cells by increasing cytoplasmic Ca2+ concentration [47][48]. In addition, conditional expression of the inhibitory Smad7 in β-cells resulted in reduced pancreatic insulin levels and in hypoinsulinemia. Lastly, expression of the TGF-β related bone morphogenetic protein 4 (BMP4) and its receptor BMPR1A regulate glucose metabolic genes to enhance GSIS and promote insulin signaling [49]. Our results are more consistent with the latter reports as the daf-8 nhr-69 double mutant exhibited decreased overall insulin signaling.
NHR-69 and DAF-8 control exp-2 expression to regulate DAF-28 secretion
Mechanistically, several lines of evidence suggest that NHR-69 and DAF-8 influence DAF-28 secretion by directly regulating the expression of exp-2, which encodes a voltage-activated (Kv-type) potassium channel. Using ChIP, we found that NHR-69 associated with at least two elements in the exp-2 promoter region that resemble consensus HNF4α binding sites; moreover, this binding was reduced in daf-8 mutants. daf-8 nhr-69 double mutants synergistically induce exp-2 expression, suggesting that exp-2 is repressed by these two factors. Interestingly, using expression profiling, Shaw et al. previously found exp-2 to be induced in several TGF-β Daf-c mutants, including daf-8 mutants [9], although they did not report the extent of the upregulation (we observed a mild induction in daf-8 mutants that was not statistically significant; see Figure 4A). Lastly, we used a heterologous cell culture system to confirm that NHR-69 and DAF-8 collaboratively repress exp-2, with three distinct NHR-69 binding sites being required for full repression.
Together, our results imply that NHR-69 and DAF-8 assemble a complex on the exp-2 promoter that represses exp-2 transcription. Due to the divergent nature of the DNA-binding domain of DAF-8 (only 28% similar to the MH1 domain of human Smad1, and 30% similar to the MH1 domain of Drosophila melanogaster Mad1; Park & Taubert, unpublished), it is difficult to predict whether DAF-8 would bind to the consensus Smad-binding-element (SBE) [50]. Further experiments are required to assess whether DAF-8 directly binds the exp-2 promoter or whether it is tethered to this region through an interaction with NHR-69.
Our data further indicate that exp-2 is critical for insulin signaling downstream of DAF-8 and NHR-69 (but upstream of DAF-2). Firstly, the exp-2(sa26ad1426) mutation, which shortens the life span of wild-type worms, did not shorten the life span of daf-2 mutants. Secondly, exp-2(sa26ad1426) and exp-2(sa26) mutants altered DAF-28 secretion. Thirdly, exp-2(sa26) mutants exhibited increased nuclear localization of DAF-16/FoxO, a typical consequence of reduced DAF-2 activity. Lastly, the exp-2(sa26) mutant showed increased expression of two downstream DAF-2 target genes. Thus, we find evidence for a novel regulatory relationship between insulin secretion and TGF-β signaling. As nhr-69 transcription is itself induced by insulin signaling, a positive regulatory loop to promote larval development and prevent dauer formation is initiated, but only when environmental stimuli activate both TGF-β and insulin signaling.
EXP-2 is a member of the six-transmembrane, voltage-activated potassium channel family, which also controls muscle contraction in the C. elegans pharynx. EXP-2 produces an outward current at the plasma membrane, which terminates the action potential by repolarizing the membrane [36]. As EXP-2 exhibits homology to multiple mammalian Kv-type channels it is difficult to provide a direct parallel to the functions of one mammalian channel. However, several Kv-type channels are known to affect insulin signaling and/or glucose balance [51]. Most prominently, pharmacological inhibition or genetic disruption of Kv2.1 causes increased glucose-stimulated insulin secretion [52][53]. In the future, it will be interesting to test whether NHR action and TGF-β signaling coregulate these mammalian channels.
Neuronal NHR-69 overexpression affects glucose homeostasis and DAF-28 secretion
The expression of nhr-69 overlaps with that of daf-8 in ASI neurons and in the intestine [7], suggesting that the molecular crosstalk between DAF-8 and NHR-69 may occur in these two tissues. Cell ablation analyses demonstrated that the ASI neurons inhibit dauer formation [54], and are required for the expression of the TGF-β ligand DAF-7 and of the insulin-like peptide DAF-28, both of which promote reproductive growth [55][32]. Because of the latter property, the ASI neurons have been proposed to functionally resemble the insulin-producing pancreatic β-cells [40]. To test whether NHR-69 was functionally important in the ASI neurons, we generated a strain that overexpresses nhr-69 specifically in these neurons. These worms exhibited greater accumulation of DAF-28::GFP, a lethargic phenotype, and reduced glucose levels, reminiscent of a hyperinsulinemic/hypoglycemic phenotype [44]. NHR-69 may achieve these effects by regulating other neuropeptides in addition to DAF-28; in any case our results suggest that the regulation of NHR-69 levels and activity in the ASI neurons is critical to achieve glucose homeostasis in C. elegans. They also support the notion that ASI neurons exhibit characteristics of mammalian pancreatic β-cells [40]. We note, however, that NHR-69 and DAF-28 are also co-expressed in the intestine, a tissue that plays a key role in metabolic regulation in C. elegans and may also perform pancreas related functions.
Our overall model (Figure 8) integrates the observations described above: in favorable conditions, DAF-8 and NHR-69 cooperate to repress exp-2 transcription in the ASI neurons, thus causing sustained DAF-28 secretion, which in turn activates IIS and promotes reproductive growth. This mechanism would act in parallel to the one proposed by Narasimhan et al., whereby DAF-8 promotes IIS and reproductive growth by directly inducing the transcription of agonistic insulin peptide genes such as ins-4 [13]. In harsh conditions, exp-2 transcription is de-repressed as a consequence of reduced DAF-8 phosphorylation and lower nhr-69 expression; increased EXP-2 activity would presumably lead to a rapid reduction in the overall levels of agonistic insulins, thus providing a rapid signal for dauer formation (Figure 8).
Fig. 8. Working model for DAF-8 and NHR-69 modulation of insulin secretion in C. elegans. 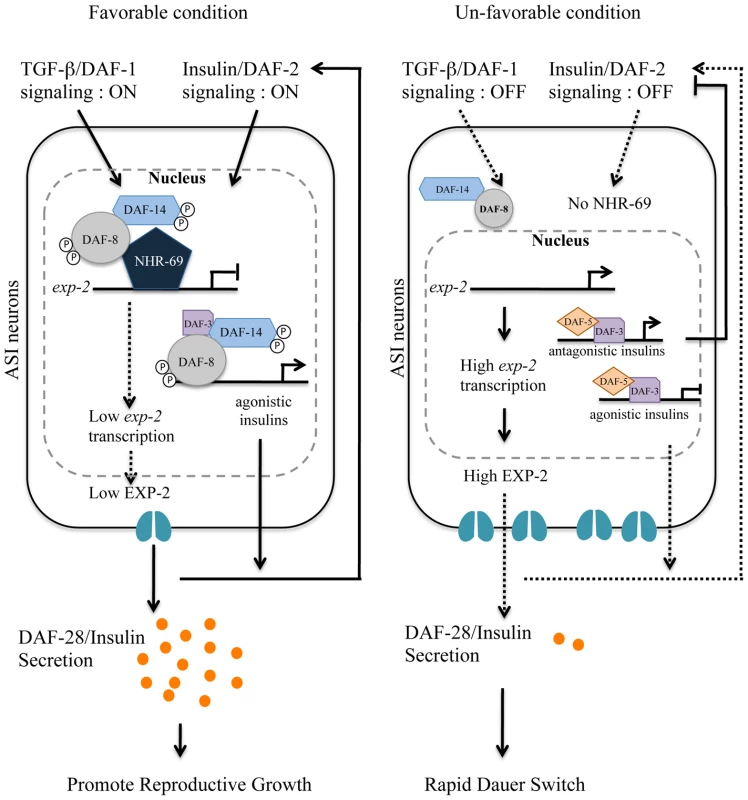
Based on our data, we propose that DAF-2 signaling positively regulates transcription of nhr-69, whereas the TGF-β receptor is known to phosphorylate DAF-8 to activate its function [7]. NHR-69 and DAF-8 cooperatively repress the transcription of the exp-2 Kv channel gene in ASI neurons, which in turn causes sustained secretion of the insulin-like peptide DAF-28, hence activating a feed-forward loop on DAF-2 in favorable conditions. TGF-β and Insulin/IGF-1 signaling crosstalk at the exp-2 promoter through the DAF-8 and NHR-69 transcription factors. Upregulation of exp-2 by the reduction of DAF-8 and NHR-69 activity would attenuate DAF-28 secretion, providing a fast decision for the dauer formation. NHR-69 functionally resembles pancreatic HNF4α
The C. elegans genome encodes 284 NHRs, most of which derive from an HNF4α-like ancestor. The human HNF4A gene encoding HNF4α is mutated in a subtype of MODY (Maturity Onset Diabetes of the Young) and HNF4α is a central regulator of hepatic and pancreatic gene transcription [37]. MODY patients fail to properly secrete insulin in response to glucose challenge [56] and β-cell-specific knockout of HNF4α in mice results in reduced GSIS [57][58]. Intriguingly, akin to our discovery of physical and functional DAF-8:NHR-69 interaction, HNF4α and Smad3 interact physically and functionally in transfected human cells [59], although the physiological impact of this interaction remains unclear. Our study suggests that such HNF4α:Smad complexes may affect glucose homeostasis by regulating potassium channel genes in pancreatic β-cells.
The functions of most C. elegans HNF4α-related NHRs remain unknown and no C. elegans NHR has yet been implicated in glucose and/or insulin metabolism. We provide several lines of evidence that NHR-69 performs these particular functions, somewhat resembling pancreatic HNF4α. Most compellingly, we observe hypoglycemia in animals that overexpress NHR-69 specifically in ASI neurons [32][40]. However, we note that, compared to HNF4α, the defects in nhr-69 single mutants (and in nhr-69 heterozygotes) are relatively mild indicating that possibly other NHRs such as the closely related NHR-64 act redundantly or can compensate for the loss of nhr-69. Given the vast number of HNF4α-related NHRs in C. elegans, other NHRs may also control dauer formation, especially in a sensitized genetic background. As many C. elegans NHRs form heterodimers [60], we speculate that NHR-69 may form heterodimers to implement its regulatory roles.
Human HNF4α targets hundreds of genes in the liver and pancreas to modulate metabolic processes such as lipid biosynthesis, glucose homeostasis and insulin secretion [37]. However, the molecular and physiological functions of individual HNF4α target genes and their combinatorial regulation remains poorly understood. Identifying the functions and the target genes of C. elegans HNF4α-like NHRs can provide insights into the mechanism of related mammalian NHRs. We recently proposed that the functions collectively executed by mammalian HNF4α might be distributed amongst individual C. elegans HNF4α orthologs [17]. For example, NHR-31 regulates the expression of a vacuolar ATPase in the excretory cell in C. elegans [61], possibly hinting at a similar regulatory role of mammalian HNF4α in kidney physiology. Our results suggest that NHR-69 may be a partial functional ortholog of pancreatic HNF4α. Thus, NHR-69 may provide a useful tool to dissect pancreatic HNF4α function, to define in vivo regulators of HNF4α and to delimit additional downstream targets that participate in the regulation of glucose homeostasis.
Materials and Methods
Nematode strains
C. elegans strains were cultured according to standard techniques [62] unless otherwise noted. The E. coli strain OP50 was used as a food source in all assays. Nematode strains and alleles used in this study are LG I: daf-8(sa343), daf-8(m85), nhr-69(ok1926) daf-16(mgDf47); LG III: daf-7(e1372), daf-2(e1370); LG IV: daf-1(m40), eri-1(mg366), daf-14(m77); LG V: exp-2(sa26)/eT1[let-?(n886)], exp-2(ad26ad1426), sdf-9(m708). mIs1[rol-6(su1006)], mIs39[nhr-69p::nhr-69::gfp, rol-6(su1006)], mIs40[gpa-4p::nhr-69::gfp, rol-6(su1006)], mIs4[rol-6(su1006)], mIs27[daf-8p::daf-8::gfp, rol-6(su1006)], nhr-69(ok1926);daf-1(m40), nhr-69(ok1926); daf-7(e1372), daf-8(m85) nhr-69(ok1926), daf-8(m85) nhr-69(ok1926); mIs39, nhr-69(ok1926);daf-14(m77), daf-7(e1372); mIs39, daf-2(e1370);mIs39, daf-8(m85) nhr-69(ok1926); oxIs206[aex-3p::ANF::GFP], daf-8(m85) nhr-69(ok1926); arIs37[myo-3p::ssGFP], exp-2(sa26); svIs69[daf-28p::daf-28::gfp], exp-2(sa26ad1426); svIs69[daf-28p:daf-28::gfp], daf-2(e1370); exp-2(sa26ad1426), daf-8(m85) nhr-69(ok1926); exp-2(sa26ad1426) exp-2(sa26); zIs356[daf-16p::daf-16::gfp], mIs40; svIs69.
In the nhr-69(ok1926) mutant, 1329 bp are deleted, including the 5′ upstream regulatory sequences and the first four exons (www.wormbase.org). The nonsense mutation daf-8(m85) is predicted to truncate DAF-8 in the conserved MH2 domain. It is the strongest daf-8 allele, forming virtually 100% dauer larvae at 25.5°C [7].
Transgene construction and transformation
A genomic fragment containing the entire coding region of nhr-69 plus 3 kb upstream of the ATG was amplified. The gfp gene was amplified from pPD95.75 (kindly provided by A. Fire) and used for PCR fusion (see [63]) to generate nhr-69p::nhr-69::gfp, a translational fusion of gfp at the end of nhr-69 exon 9. The gpa-4p::nhr-69::gfp fusion construct was generated by the same method, with a 2.8 kb 5′ gpa-4 promoter region [41] being used. The fusions were introduced by microinjection into the adult germ line (45 ng/µl) along with the injection marker pRF4 [rol-6 (su1006)] (9 ng/µl).
Life span analysis and measurement of glucose content
Eggs were prepared by alkaline hypochlorite treatment and synchronized by hatching in M9 buffer. After growth on OP50 at 15°C, animals at mid-L4 stage were transferred to NG agar plates containing 40 µM of 5-fluoro-2′-deoxyuridine (FUdR) to prevent progeny production [64]. Life span experiments were performed at 25°C as previously described [65]. We averaged mean life spans from three independent biological replicates. Representative survival curves are shown. We used one-way analysis of variance (ANOVA) for statistical analysis of mean life spans ANOVA [66].
We measured total worm glucose levels with a Glucose Assay Kit (Biovision K606–100). Worms grown on NG agar plates were collected and washed five times in M9 buffer. The worm pellet was resuspended in 500 µl of glucose assay buffer, frozen at −80°C, thawed on ice, and then sonicated (Branson Sonifier 450) five times (output level 3, time 10 seconds) to obtain extracts. After centrifugation at 13,000 rpm at 4°C, the supernatant was used for the glucose assay according to the manufacturer's instructions. We normalized glucose content by protein content in each sample. Statistics were performed using GraphPad Prism 5.
Lethargy assays
Worms were grown synchronously at 20°C from L1 to L4 stage, and then transferred to 25°C. Individual day-1 old adults were video recorded for two minutes, and were judged to be lethargic if they stopped and resumed moving more than four times (N = 50). The plates were tapped manually ten seconds prior to video recording to stimulate movement. For assaying suppression of lethargy by glucose, worms were grown as above and transferred to NG-agar plates supplemented with either 2 mM of D-(+)-Glucose (Sigma, G8270) or 2 mM of 2-deoxy-D-glucose (Sigma D8375).
RNAi and dauer formation and recovery assays
Percent dauer formation was scored visually at 250× magnification for each genotype grown for two days from hatching at 20°C, 22.5°C, and 25.5°C. To assay dauer recovery, the dauer larvae were induced by growth at 25.5°C for each genotype. 50 dauer larvae were put on NG agar plates seeded with OP50 and incubated at 15°C. Dauer recovery was scored visually monitoring pharyngeal pumping and body size 24 and 48 hrs after transfer. RNAi feeding was performed using clones from the Ahringer library [67] on 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) as described [68].
In vitro GST pull down, in vivo co-immunoprecipitation, and immunoblots
For in vitro GST pull down assays, full-length cDNAs encoding NHR-69 or Smad proteins were subcloned into pGEX-4T-1 (GE Healthcare Sciences) and pBluescript SKII vectors, respectively. The GST pull-down assay and in vivo co-IP were performed as previously described [7]. Immunoblots were probed with anti-GFP antibody (1∶2,000, Abcam ab6556) or anti-DAF-8 antibody (1∶1,000; a custom rabbit polyclonal anti-DAF-8 antibody raised against the peptide CQSNRQDDEEPGYYR, generated by GenScript). Total lysates were obtained by sonication (Branson, Sonifier 450). The anti-DAF-8 antibody specifically detected endogenous DAF-8 as well as the exogenous DAF-8::GFP fusion protein, but did not detect DAF-8 in the daf-8(sa343) mutant, in which the epitope is deleted (Figure S1).
Chromatin immunoprecipitation (ChIP) and quantitative PCR
For ChIP, mixed-stage populations were subjected to a 1.5% formaldehyde solution for 30 minutes at room temperature to cross-link DNA and proteins [69]. We performed immunoprecipitation by incubating the lysate with either anti-GFP antibody (1∶250, Abcam) or IgG at 4°C for 16–18 hours. The precipitates were washed, the crosslinks reversed, and the DNA eluted. 5 µl of each eluate were used for quantitative PCR (qPCR).
For qPCR, 1 µg total RNA from each sample was used for reverse transcription with Superscript II reverse transcriptase (Invitrogen) and oligo dT priming. The resulting first-strand cDNA was analyzed by qPCR on an Applied Biosystems 7500 with BioRad iTaq SYBR green supermix using ROX as a normalization dye. The primers used for qPCR are listed in Table S2.
Antibodies, immunohistochemistry, and promoter analysis in HEK293 cells
The phosphomimetic pmDAF-8 construct was amplified by PCR with primers dh_295_L and dh_296_R (Table S2). The resulting cDNA was cloned into the pcDNA3.1 (+) Neo vector using BamHI and EcoRI sites. The cDNA for nhr-69::gfp was amplified from mIs39 animals by using dh_312_L and dh_313_R (Table S2). The resulting PCR product was cloned in the pcDNA3.1 (+) Hygro vector using NheI and ApaI sites. The wild-type 2.5 kb exp-2 promoter was PCR amplified using dh_299_L/dh_300_R (Table S2) and subcloned in pTK-GLuc vector (New England BioLabs N8084) using KpnI and BamHI sites to generate pTK-Gluc/exp-2. The mutant exp-2 promoter constructs were generated using a site directed mutagenesis kit (Invitrogen) with pTK-Gluc/exp-2 as a template. The constructs were delivered to HEK293 cells by transient transfection using Lipofectamine (Invitrogen) according to the manufacturer's manual. The luciferase assays were performed using a BioLux Gaussia Luciferase Flex Assay Kit (New England BioLabs E3308L), and luminescence was measured in a Tecan M200 plate reader.
For immunofluorescent detection of labeled proteins, cells were fixed in 4% paraformaldehyde for 10 minutes, washed in PBS twice for five minutes per wash, permeabilized in 0.1% Triton-X-100 for 30 minutes, washed in PBS, and blocked in 4% Normal Goat Serum for 45 minutes, incubated with primary antibodies overnight in PBS/1% Normal Goat Serum. The following morning, the samples were washed twice with PBS and incubated with secondary antibodies for 1 hour, then washed 3×5 minutes in PBS. The coverslips were mounted using Vectashield (Vector Laboratory). The primary antibodies were: anti-DAF-8 (1/500) custom rabbit polyclonal antibody (Genscript), anti-Vimentin (1/200 Chicken IgY, Millipore #AB5733). The secondary antibodies were Alexa 546-coupled anti-rabbit IgG H+L, highly cross-adsorbed (1/200, Invitrogen/Molecular Probes #A11035), and donkey anti-chicken Cy5 (1/200, Millipore #AP194S).
Microscopy
We used a Zeiss Axioscope equipped with a QImaging camera (RETIGA 2000R) for differential interference contrast (DIC) and fluorescence microscopy. ImageJ software was used to estimate the intensity of the GFP signal in coelomocytes. For DiI staining, cultures were synchronized by hatching purified eggs into M9 buffer, grown on NG agar plates until the L2 stage, washed in M9 buffer, and DiI stained, as described [70]. Fluorescent signals generated by immunostaining were visualized and captured using an Olympus FLUOVIEW FV10i confocal laser-scanning microscope system. Images from different color channels were colorized and merged using ImageJ software.
Supporting Information
Zdroje
1. MassaguéJGomisR 2006 The logic of TGFbeta signaling. FEBS Lett 580 2811 20
2. AttisanoLCarcamoJVenturaFWeisF 1993 Identification of Human Activin and TGF Type I Receptors That Form Heteromeric Kinase Complexes. CELL-CAMBRIDGE MA - 75 671 680
3. SavageCDasPFinelliATownsendSSunC 1996 Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci USA 93 790 4
4. RiddleDLSwansonMMAlbertPS 1981 Interacting genes in nematode dauer larva formation. Nature 290 668 671
5. HuP 2007 Dauer. WormBook: the online review of C elegans biology 1 19
6. SchackwitzWSInoueTThomasJH 1996 Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17 719 728
7. ParkDEstevezARiddleDL 2010 Antagonistic Smad transcription factors control the dauer/non-dauer switch in C. elegans. Development 137 477 485
8. YouY-jaiKimJRaizenDMAveryL 2008 Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metab 7 249 257
9. ShawWMLuoSLandisJAshrafJMurphyCT 2007 The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol 17 1635 1645
10. LiuTZimmermanKKPattersonGI 2004 Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev Biol 4 11
11. HahmJ-HKimSPaikY-K 2009 Endogenous cGMP regulates adult longevity via the insulin signaling pathway in Caenorhabditis elegans. Aging Cell 8 473 483
12. LeeRYHenchJRuvkunG 2001 Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol 11 1950 1957
13. NarasimhanSDYenKBansalAKwonES 2011 PDP-1 Links the TGF-β and IIS Pathways to Regulate Longevity, Development, and Metabolism. PLoS Genet 7 e1001377 doi:10.1371/journal.pgen.1001377
14. VowelsJJThomasJH 1992 Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130 105 123
15. AntebiAYehWTaitDHedgecockERiddleD 2000 daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes & Development 14 1512 27
16. AntebiA 2006 Nuclear hormone receptors in C. elegans. WormBook: the online review of C elegans biology 1 13
17. TaubertSWardJDYamamotoKR 2011 Nuclear hormone receptors in nematodes: Evolution and function. Mol Cell Endocrinol 334 49 55
18. ArdaHETaubertSMacNeilLTConineCCTsudaB 2010 Functional modularity of nuclear hormone receptors in a Caenorhabditis elegans metabolic gene regulatory network. Mol Syst Biol 6 367
19. Van GilstMRHadjivassiliouHJollyAYamamotoKR 2005 Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3 e53 doi:10.1371/journal.pbio.0050053
20. LiangBFergusonKKadykLWattsJL 2010 The role of nuclear receptor NHR-64 in fat storage regulation in Caenorhabditis elegans. PLoS ONE 5 e9869 doi:10.1371/journal.pone.0009869
21. BrockTJBrowseJWattsJL 2006 Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet 2 e108 doi:10.1371/journal.pgen.0020108
22. GissendannerCCrossgroveKKrausKMainaCSluderA 2004 Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol 266 399 416
23. AltschulSFGishWMillerWMyersEWLipmanDJ 1990 Basic local alignment search tool. J Mol Biol 215 403 410
24. KennedySWangDRuvkunG 2004 A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427 645 649
25. JensenVLAlbertPSRiddleDL 2007 Caenorhabditis elegans SDF-9 enhances insulin/insulin-like signaling through interaction with DAF-2. Genetics 177 661 666
26. GoldenJRiddleD 1984 The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Developmental Biology 102 368 78
27. TissenbaumHHawdonJPerregauxMHotezPGuarenteL 2000 A common muscarinic pathway for diapause recovery in the distantly related nematode species Caenorhabditis elegans and Ancylostoma caninum. Proc Natl Acad Sci U S A 97 460 5
28. KimuraKDTissenbaumHALiuYRuvkunG 1997 daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277 942 946
29. OggSParadisSGottliebSPattersonGILeeL 1997 The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389 994 999
30. HondaYHondaS 1999 The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J 13 1385 1393
31. TulletJMAHertweckMAnJHBakerJHwangJY 2008 Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 1025 1038
32. LiWKennedySGRuvkunG 2003 daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev 17 844 858
33. SpeeseSPetrieMSchuskeKAilionMAnnK 2007 UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27 6150 6162
34. FaresHGreenwaldI 2001 Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet 28 64 68
35. DeplanckeBMukhopadhyayAAoWElewaAMGroveCA 2006 A gene-centered C. elegans protein-DNA interaction network. Cell 125 1193 1205
36. DavisMWFleischhauerRDentJAJohoRHAveryL 1999 A mutation in the C. elegans EXP-2 potassium channel that alters feeding behavior. Science 286 2501 2504
37. OdomDZizlspergerNGordonDBellGRinaldiN 2004 Control of pancreas and liver gene expression by HNF transcription factors. Science 303 1378 81
38. FunabaMMathewsLS 2000 Identification and characterization of constitutively active Smad2 mutants: evaluation of formation of Smad complex and subcellular distribution. Mol Endocrinol 14 1583 1591
39. ThomasJH 1990 Genetic analysis of defecation in Caenorhabditis elegans. Genetics 124 855 872
40. KaoGNordensonCStillMRonnlundATuckS 2007 ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell 128 577 87
41. JansenGThijssenKLWernerPvan der HorstMHazendonkE 1999 The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet 21 414 419
42. OkkemaPGHarrisonSWPlungerVAryanaAFireA 1993 Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135 385 404
43. HendersonSTJohnsonTE 2001 daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Current Biology 11 1975 1980
44. LteifANSchwenkWF 1999 HYPOGLYCEMIA IN INFANTS AND CHILDREN. Endocrinology & Metabolism Clinics of North America 28 619 646
45. LinH-MLeeJ-HYadavHKamarajuAKLiuE 2009 Transforming growth factor-beta/Smad3 signaling regulates insulin gene transcription and pancreatic islet beta-cell function. J Biol Chem 284 12246 12257
46. SjoholmAHellerstromC 1991 TGF-beta stimulates insulin secretion and blocks mitogenic response of pancreatic beta-cells to glucose. Am J Physiol 260 C1046 51
47. ShibataHYasudaHSekineNMineTTotsukaY 1993 Activin A increases intracellular free calcium concentrations in rat pancreatic islets. FEBS Lett 329 194 8
48. IshiyamaNShibataHKanzakiMShiozakiSMiyazakiJ 1996 Calcium as a second messenger of the action of transforming growth factor-beta on insulin secretion. Mol Cell Endocrinol 117 1 6
49. GoulleyJDahlUBaezaNMishinaYEdlundH 2007 BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab 5 207 219
50. JonkLJItohSHeldinCHten DijkePKruijerW 1998 Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem 273 21145 21152
51. JacobsonDAPhilipsonLH 2007 Action potentials and insulin secretion: new insights into the role of Kv channels. Diabetes Obes Metab 9 Suppl 2 89 98
52. HerringtonJZhouY-PBugianesiRMDulskiPMFengY 2006 Blockers of the delayed-rectifier potassium current in pancreatic beta-cells enhance glucose-dependent insulin secretion. Diabetes 55 1034 1042
53. JacobsonDAKuznetsovALopezJPKashSÄmmäläCE 2007 Kv2.1 Ablation Alters Glucose-Induced Islet Electrical Activity, Enhancing Insulin Secretion. Cell Metabolism 6 229 235
54. BargmannCHorvitzH 1991 Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251 1243 6
55. RenPLimCSJohnsenRAlbertPSPilgrimD 1996 Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274 1389 1391
56. HermanWFajansSSmithMPolonskyKBellG 1997 Diminished insulin and glucagon secretory responses to arginine in nondiabetic subjects with a mutation in the hepatocyte nuclear factor-4alpha/MODY1 gene. Diabetes 46 1749 1754
57. GuptaRVatamaniukMLeeCFlaschenRFulmerJ 2005 The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. J Clin Invest 115 1006 15
58. MiuraAYamagataKKakeiMHatakeyamaHTakahashiN 2006 Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. J Biol Chem 281 5246 57
59. KardassisDPardaliKZannisVI 2000 SMAD proteins transactivate the human ApoCIII promoter by interacting physically and functionally with hepatocyte nuclear factor 4. J Biol Chem 275 41405 41414
60. LiSArmstrongCMBertinNGeHMilsteinS 2004 A map of the interactome network of the metazoan C. elegans. Science 303 540 543
61. Hahn-WindgassenAVan GilstMR 2009 The Caenorhabditis elegans HNF4alpha Homolog, NHR-31, mediates excretory tube growth and function through coordinate regulation of the vacuolar ATPase. PLoS Genet 5 e1000553 doi:10.1371/journal.pgen.1000553
62. BrennerS 1974 The genetics of Caenorhabditis elegans. Genetics 77 1 71 94
63. HobertO 2002 PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32 728 730
64. GandhiSSantelliJMitchellDHStilesJWSanadiDR 1980 A simple method for maintaining large, aging populations of Caenorhabditis elegans. Mech. Ageing Dev 12 137 150
65. LarsenPLAlbertPSRiddleDL 1995 Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139 1567 1583
66. FisherRA 1970 Statistical Methods for Research Workers. 14th ed Macmillan Pub Co
67. FraserAGKamathRSZipperlenPMartinez-CamposMSohrmannM 2000 Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408 325 330
68. TimmonsLCourtDFireA 2001 Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103 12
69. ErcanSWhittleCLiebJ 2007 Chromatin immunoprecipitation from C. elegans embryos. Nat Protoc http://www.natureprotocols.com/2007/02/15/chromatin_immunoprecipitation.php
70. BlacqueOReardonMLiCMcCarthyJ 2004 Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised. Genes & Dev 18 13 1630 42
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Délka menstruačního cyklu jako marker ženské plodnosti
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



