-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
Genetic variation within and between species can be shaped by population-level processes and mutation; however, the relative impact of “survival of the fittest” and “arrival of the fittest” on phenotypic evolution remains unclear. Assessing the influence of mutation on evolution requires understanding the relative rates of different types of mutations and their genetic properties, yet little is known about the functional consequences of new mutations. Here, we examine the spectrum of mutations affecting a focal gene in Saccharomyces cerevisiae by characterizing 231 novel haploid genotypes with altered activity of a fluorescent reporter gene. 7% of these genotypes had a nonsynonymous mutation in the coding sequence for the fluorescent protein and were classified as “coding” mutants; 2% had a change in the S. cerevisiae TDH3 promoter sequence controlling expression of the fluorescent protein and were classified as “cis-regulatory” mutants; 10% contained two copies of the reporter gene and were classified as “copy number” mutants; and the remaining 81% showed altered fluorescence without a change in the reporter gene itself and were classified as “trans-acting” mutants. As a group, coding mutants had the strongest effect on reporter gene activity and always decreased it. By contrast, 50%–95% of the mutants in each of the other three classes increased gene activity, with mutants affecting copy number and cis-regulatory sequences having larger median effects on gene activity than trans-acting mutants. When made heterozygous in diploid cells, coding, cis-regulatory, and copy number mutant genotypes all had significant effects on gene activity, whereas 88% of the trans-acting mutants appeared to be recessive. These differences in the frequency, effects, and dominance among functional classes of mutations might help explain why some types of mutations are found to be segregating within or fixed between species more often than others.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002497
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002497Summary
Genetic variation within and between species can be shaped by population-level processes and mutation; however, the relative impact of “survival of the fittest” and “arrival of the fittest” on phenotypic evolution remains unclear. Assessing the influence of mutation on evolution requires understanding the relative rates of different types of mutations and their genetic properties, yet little is known about the functional consequences of new mutations. Here, we examine the spectrum of mutations affecting a focal gene in Saccharomyces cerevisiae by characterizing 231 novel haploid genotypes with altered activity of a fluorescent reporter gene. 7% of these genotypes had a nonsynonymous mutation in the coding sequence for the fluorescent protein and were classified as “coding” mutants; 2% had a change in the S. cerevisiae TDH3 promoter sequence controlling expression of the fluorescent protein and were classified as “cis-regulatory” mutants; 10% contained two copies of the reporter gene and were classified as “copy number” mutants; and the remaining 81% showed altered fluorescence without a change in the reporter gene itself and were classified as “trans-acting” mutants. As a group, coding mutants had the strongest effect on reporter gene activity and always decreased it. By contrast, 50%–95% of the mutants in each of the other three classes increased gene activity, with mutants affecting copy number and cis-regulatory sequences having larger median effects on gene activity than trans-acting mutants. When made heterozygous in diploid cells, coding, cis-regulatory, and copy number mutant genotypes all had significant effects on gene activity, whereas 88% of the trans-acting mutants appeared to be recessive. These differences in the frequency, effects, and dominance among functional classes of mutations might help explain why some types of mutations are found to be segregating within or fixed between species more often than others.
Introduction
Mutations are the ultimate source of genetic variation, thus understanding the properties of new mutations is important for both medical and evolutionary genetics. Large-scale sequencing surveys have recently measured mutation rates for different types of DNA lesions (e.g., transitions, transversions, indels, rearrangements, duplications) in a variety of organisms [1]–[3], but little remains known about the genetic properties of these mutations or their effects on the activity of individual genes. Although not often incorporated into population genetic models of the evolutionary process, differences in the frequency and properties of different types of mutations can influence evolutionary paths [4]–[6].
From the perspective of a single gene, mutations affecting its activity can be divided into four functional classes: [nonsynonymous] coding mutations that alter the sequence of the encoded RNA or protein gene product, cis-regulatory mutations that alter (typically, non-coding) sequences that regulate the gene's expression in an allele-specific manner, trans-acting mutations that alter coding or cis-regulatory sequences of other genes in the genome and affect activity of the focal gene via a diffusible gene product, and copy number mutations resulting from duplications or deletions that change the number of copies of the focal gene in the genome. As the raw material of evolutionary change, all of these types of mutations have the potential to become polymorphisms segregating at an appreciable frequency within a species and/or substitutions fixed between species, yet studies identifying the genetic basis of trait differences suggest that some types of changes underlie phenotypic differences more often than others (reviewed by [7]–[9]).
The apparent inequality in the contribution of different types of mutations to phenotypic evolution is often explained by invoking differences in pleiotropy (i.e., the number of traits affected by a mutation) among functional classes. Increased pleiotropy is assumed to increase the chance that a mutation has deleterious effects on fitness and will be disfavored by natural selection. One example of this is that coding mutations are commonly expected to be more pleiotropic (and hence have lower average fitness) than cis-regulatory mutations ([7], [9], [10], but see [11]). Although undoubtedly important, pleiotropy is only one factor influencing the probability that a certain type of mutation is fixed. For example, the direction and magnitude of a mutation's effect on gene activity and whether or not the mutation is dominant to the wildtype allele are also expected influence the evolutionary trajectories of new mutations in diploid populations. Of course, these factors matter only after a mutation has occurred, thus mutation rates can influence the evolutionary process as well [7], [12]–[14]. The frequency, effects, and dominance of new mutations have all been predicted to vary among functional classes of mutations [15]–[17], but little data has been available to test these predictions [13], [15].
To directly compare these parameters among functional classes of mutations, we systematically isolated and quantitatively characterized over 200 mutations in Saccharomyces cerevisiae affecting activity of a focal gene. To make this experiment feasible, we used a mutagen to elevate the mutation rate and studied mutations affecting activity of a reporter gene expressing Yellow Fluorescent Protein (YFP) that could be scored quantitatively in thousands of living cells per second using flow cytometry. Expression of this heterologous fluorescent protein was controlled by native S. cerevisiae promoter and terminator sequences (which allowed us to interrogate endogenous S. cerevisiae transcriptional regulatory networks) and the mutagen was expected to cause mutations relatively uniformly across the genome.
Using this experimental system, we measured the proportion of cells with new mutations that altered activity of the reporter gene and used this proportion to estimate the spontaneous mutation rate for this phenotypic change. We then isolated 231 mutants with altered activity of the reporter gene and subjected them to further characterization, including determining the relative frequency of different types of mutations, comparing their effects on reporter gene activity, and assessing their dominance relative to the wildtype allele. These data revealed differences in the frequency, effects, and dominance among coding, cis-regulatory, trans-acting, and copy number mutations that are expected to influence the relative contribution of different types of mutations to phenotypic variation within and between species.
Results
To characterize the spectrum of mutations affecting activity of a focal gene, we screened mutagenized cells containing a fluorescent reporter gene and quantified cellular fluorescence using flow cytometry. Mutagenesis was performed using ethyl methanesulfonate (EMS), and the increase in mutation rate was controlled by titrating exposure of cells to this chemical. The reporter gene was constructed by fusing the coding sequence of the Venus variant [18] of YFP to the S. cerevisiae CYC1 terminator [19], and placing them both under the control of 5′ intergenic sequence of the S. cerevisiae TDH3 gene. This chimeric transgene (PTDH3-YFP) was integrated into a pseudogene on the first chromosome of S. cerevisiae, where integration of fluorescent reporter genes has previously been found to have no measurable effect on fitness (B. Williams, personal communication). For each cell, the activity of PTDH3-YFP was measured as YFP fluorescence per unit of “forward scatter” (FSC); FSC is proportional to cell size [20] and is linearly related to YFP fluorescence (Figure 1A). In the absence of amino acid changes in YFP, cellular fluorescence is expected to be linearly related to YFP protein abundance, as has been shown for the related Green Fluorescent Protein [21].
Fig. 1. EMS treatment increased the frequency of cells with extreme YFP fluorescence. 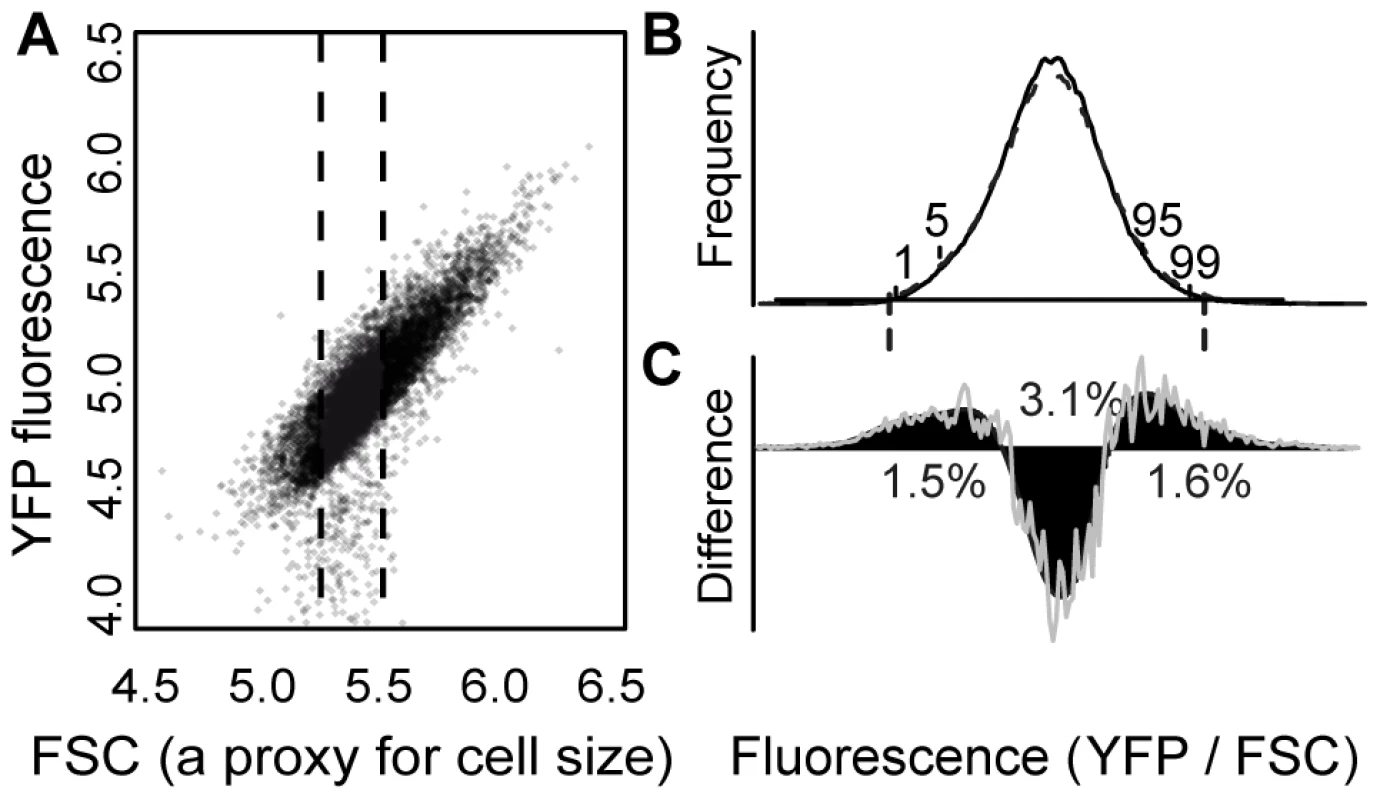
(A) The relationship between “forward scatter” (“FSC”, a proxy for cell size) and YFP fluorescence is shown for a population of control cells. FSC and YFP fluorescence are both reported in arbitrary units on a log scale. A similar linear relationship was observed for other genotypes. Approximately 2% of flow cytometry “events” had YFP fluorescence less than the range plotted and are not shown. The fluorescence phenotype of each sample in the secondary screen was calculated as the median YFP/FSC ratio for FACS events with FSC values between 5.30 and 5.55 (indicated with dotted lines). (B) The distribution of YFP fluorescence phenotypes is shown for EMS-treated (dashed curve) and control cells (solid curve) from one of the nine replicate populations of EMS-treated and control cells analyzed in the primary screen. Locations of the 1st, 5th, 95th, and 99th percentiles of the control sub-population are indicated, and vertical dashed lines show the average thresholds used for cell sorting (see also Table S1). (C) The difference between the number of EMS-treated and control cells in the population is plotted for a range of fluorescence levels (grey line). The black curve shows a spline fit to these data. Positive values indicate fluorescence phenotypes that were more abundant in the EMS-treated sample, whereas negative values indicate fluorescence phenotypes that were more abundant in the control sample. The spline crosses zero at approximately the 17th and 84th percentiles of the control population. The percentage of the EMS-treated population that is either over or under represented is shown for the following percentile ranges: 1–17, 17–84, 84–99. The X-axis representing YFP fluorescence levels has the same scale as in panel B. A spontaneous mutation rate affecting PTDH3-YFP activity was estimated from an EMS-treated population
To determine the frequency of mutations that affected PTDH3-YFP activity, which is expected to reflect the genome-wide mutational target size for this phenotype, we measured YFP fluorescence in each cell of a mixed sample containing both EMS-treated and untreated cells. Cells that were not exposed to EMS were considered control cells and labelled with Cy5 (a fluorescent dye), but otherwise processed identically to the EMS-treated cells. Comparing YFP fluorescence between >25,000 control cells labelled with Cy5 and >20,000 unlabelled control cells showed that Cy5 labeling had no significant effect on the measurement of YFP fluorescence phenotypes (P = 0.9, t-test).
The EMS-treated population displayed an approximately equal increase of cells with YFP fluorescence levels in both tails of the distribution (Figure 1B, 1C), suggesting that mutations increasing and decreasing fluorescence occurred at similar rates. The increase in cells with both high and low fluorescence was taken as the frequency (f = 0.0298) of EMS-induced mutants with altered activity of PTDH3-YFP, whereas the remaining cells were assumed not to carry any mutations affecting PTDH3-YFP activity. Assuming mutations affecting PTDH3-YFP activity were Poisson distributed, the inferred frequency of genotypes without a relevant mutation (P0 = 1−f = 0.9702) suggested an average of 0.0303 mutations affecting PTDH3-YFP activity per genome in the EMS-treated cells. Given this mutation rate, a Poisson process predicts that 2.94% of the EMS-treated cells should have exactly one relevant mutation (P1), whereas just 0.04% of the EMS-treated cells (1% of all mutants) should have more than one mutation affecting PTDH3-YFP activity (P>1).
To estimate a spontaneous mutation rate for PTDH3-YFP activity from this mutagen-treated population, we measured the frequency of canavanine resistance mutants in the same EMS-treated population, and found that it was 5737-fold higher than the spontaneous canavanine resistance mutation rate reported by [22]. Assuming that a similar proportion of sites affecting canavanine susceptibility and PTDH3-YFP activity were targeted by EMS, our data suggest a spontaneous mutation rate for quantitative changes in PTDH3-YFP activity of 0.0303/5737 or 5.3×10−6 per haploid genome per generation (Text S1). An alternative estimate of the spontaneous mutation rate, based on the number of empirically confirmed mutants in each tail of the EMS-treated distribution (described in the next section), was also calculated and is presented in Table S4.
A collection of 231 mutants affecting PTDH3-YFP activity was established using fluorescence activated cell sorting (FACS)
To isolate individual cells with abnormal PTDH3-YFP activity for further characterization, cells exhibiting YFP fluorescence less than a minimum threshold near the 1st percentile of control cells and greater than a maximum threshold near the 99th percentile of control cells (Figure 1B) were collected using FACS. Exact sorting thresholds for the nine replicate sorting experiments are shown in Table S1. On average, cells with YFP fluorescence similar to that of the lowest fluorescing 0.82% and highest fluorescing 0.64% of the control population were sorted. These threshold levels of YFP fluorescence resulted in sorting cells from the lowest fluorescing 1.21% and highest fluorescing 1.04% of the mutagenized population, suggesting that (1.21–0.82)/1.21, or 32.2%, of EMS-treated cells sorted from the low-fluorescence tail and (1.04–0.64)/1.04, or 38.5%, of EMS-treated cells sorted from the high-fluorescence tail were mutants. In all, 864 FACS “events” (i.e., cells or other particles) were sorted from each tail of the EMS-treated subpopulation, and 864 FACS events were sorted from each tail of the control population, for a total of 3456 FACS events arrayed individually on solid media. The percentage of sorted events that formed colonies was similar between the EMS-treated and control populations (68% vs 70%, P = 0.26, Fisher's Exact test), suggesting that EMS-induced mutations severely limiting growth were rare among cells sorted from this population. A slightly larger, and statistically significant, difference was observed, however, between the percentage of sorted events from the high - and low-fluorescing tails that formed colonies for both EMS-treated and control cells (65% vs 70% for mutagenized cells, and 67% vs 72% for control cells; P = 0.03 in each comparison, Fisher's Exact Test). The similar asymmetry observed in the mutagenized and control populations suggests that it was not caused by the EMS treatment.
Each colony was used to inoculate a liquid culture, and YFP fluorescence was measured in at least 5,000 cells from each of these clonal cultures by flow cytometry. The YFP fluorescence phenotype of each culture was calculated as the median YFP/FSC ratio of all cells within a fixed range of FSC values (Figure 1A). To determine the effect on PTDH3-YFP activity of any mutation(s) present in a recovered genotype, we calculated the difference in YFP fluorescence between each genotype and the mean YFP fluorescence of replicate control cultures, and then divided it by the standard deviation of YFP fluorescence phenotypes among the replicate control cultures. This value is a test statistic known as a Z-score (Z), and reflects the magnitude and direction of each genotype's effect on YFP fluorescence relative to the starting (unmutagenized) genotype as well as the likelihood that this effect is significantly different from 0. Given our experimental design, only mutations that prevented colony formation on solid media, slowed growth in liquid culture enough to preclude sampling 5,000 cells, or had effects on YFP fluorescence below our detection limits should have been systematically eliminated from our collection.
Genotypes isolated from the EMS-treated population with |Z|>2.58 were considered mutants and subjected to further analysis. This statistical threshold corresponds to a 99% confidence interval for the mean of the control population, and implies that all genotypes considered mutants showed a change in YFP fluorescence supported by a [two-tailed] p-value<0.01. On the basis of this statistical cut-off, 231 (22%) of the 1064 liquid cultures derived from the EMS-treated colonies were considered mutants (Table S2). By contrast, only 16 (1%) of the 1137 cultures derived from the control colonies exceeded the |Z| = 2.58 significance threshold (Table S2); these 16 isolates are not included in the collection of mutants discussed below. In addition to these changes in median YFP fluorescence, 18.6% of EMS-treated genotypes classified as mutants, 4.4% of EMS-treated genotypes not classified as mutants, and 1.8% of genotypes isolated from the control population showed significant changes in the variance of YFP fluorescence (Figure S1). Although changes in both the median and variance of YFP fluorescence might or might not be caused by the same mutation, the elevated proportion of genotypes with altered variance among EMS-treated genotypes classified as YFP fluorescence mutants suggests that they might often be one and the same.
Frequency of mutants affecting PTDH3-YFP activity differed among mutational classes
Mutants affecting PTDH3-YFP activity were identified solely on the basis of their YFP fluorescence phenotype, thus we expected them to include genotypes with (1) mutations in the coding sequence of PTDH3-YFP, (2) mutations in cis-acting sequences of PTDH3-YFP, (3) mutations outside of the known cis-regulatory and coding sequences of PTDH3-YFP that putatively have trans-acting effects on the cell's YFP fluorescence phenotype, and (4) duplications or deletions changing the copy number of PTDH3-YFP (copy number variants, CNVs).
To identify genotypes with mutations in the coding and cis-regulatory sequences of PTDH3-YFP, we sequenced the entire PTDH3-YFP transgene in each of the 231 mutants. 16 independently isolated genotypes were each found to contain a mutation predicted to change an amino acid or introduce a stop codon in YFP; two of these observed mutations were found in two genotypes each (Figure 2A). Additionally, one mutant was found to contain a synonymous mutation within the YFP coding region (Figure 2A). Four mutants had mutations within the cis-regulatory promoter region of PTDH3-YFP, two of which carried the same mutation (Figure 2A). None of the 231 mutants had a mutation in the CYC1 terminator (Figure 2A), nor did any contain more than one mutation in the entire PTDH3-YFP gene (i.e., TDH3 promoter, YFP coding sequence and CYC1 terminator). Cases where the same mutation was found in two mutants could have resulted from recurrent mutation or common ancestry, although the experiment was designed to minimize the potential for recovering clonally related mutants (see Text S1) and in at least one case (described below) the shared mutation exists on different genetic backgrounds, suggesting independent origins.
Fig. 2. Mutations in the coding and promoter sequences of PTDH3-YFP as well as changes in its copy number were found among the 231 mutant genotypes. 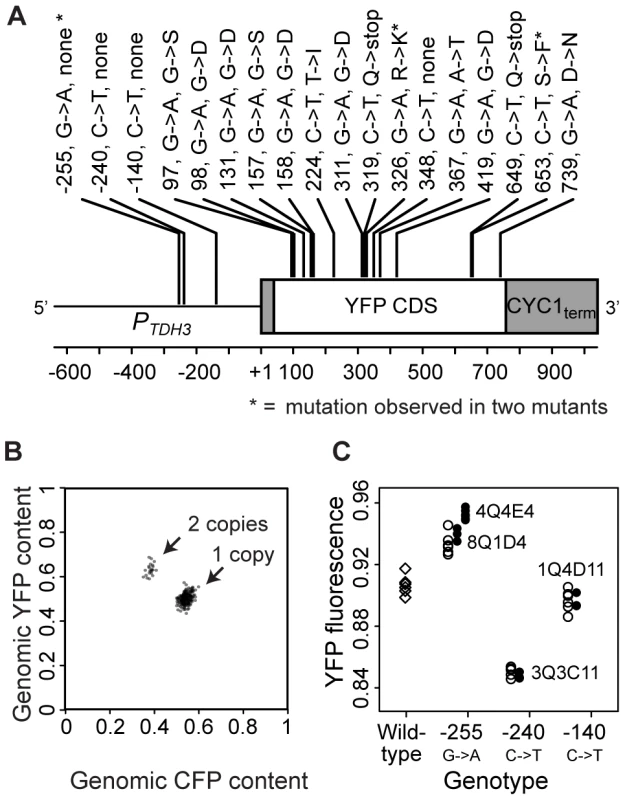
(A) The schematic depicts the PTDH3-YFP reporter gene and is drawn to scale. Annotations indicate the location of each mutation relative to the transcription start site (+1) of TDH3 [68], the nucleotide substitution observed, and the change in amino acid sequence if applicable. Asterisks indicate mutations observed in two mutant genotypes. Mutations at positions −255, −240 and −140 are located within the promoter, and the mutation at position 348 is a synonymous change. (B) In 221 of the 231 mutants, the relative copy number of PTDH3-YFP and PTDH3-CFP was determined by pyrosequencing. Clusters of points representing genotypes with one copy of PTDH3-YFP (CFP and YFP≈1/2) and two copies of PTDH3-YFP (CFP≈1/3 and YFP≈2/3) are indicated with arrows. (C) Median YFP fluorescence is plotted for each of the replicate populations analyzed for eight distinct genotypes: unmutagenized cells containing the “wildtype” PTDH3-YFP sequence (diamonds); each of the four regulatory mutant genotypes (8Q1D4, 4Q4E4, 3Q3C11, and 1Q4D11 in Table S3) that contained a promoter mutation (filled circles); and genotypes in which one of the promoter mutations (−255, −240, or −140) was introduced into same genetic background as the wildtype PTDH3-YFP gene (open circles). Populations of cells containing any one of the promoter mutations showed a significant change in YFP fluorescence relative to the wildtype genotype (P−255 = 0.002, P−240 = 0.002, P−140 = 0.015, MWW test). The mutation at −255 had an effect on YFP fluorescence equivalent to that of mutant genotype 8Q1D4 (P = 0.261, MWW), but not to that of mutant genotype 4Q4E4 (P = 0.0002, MWW). The effects on YFP fluorescence of mutations at −240 and −140 were equivalent to those of the mutant genotypes (3Q3C11 and 1Q4D11, respectively) that harbored them (P−240 = 0.262 and P−140 = 0.262, MWW). EMS is not generally thought to induce changes in copy number [23], [24], but spontaneous duplications are common in S. cerevisiae [1]. Therefore, we tested for changes in the copy number of PTDH3-YFP by mating each haploid mutant to a closely related genotype (of the opposite mating type) in which the YFP coding sequence in PTDH3-YFP was replaced with the coding sequence for a Cyan Fluorescent Protein (CFP, 95% amino acid sequence identity with YFP [25]). Pyrosequencing was then used to compare the relative frequency of YFP and CFP alleles in genomic DNA extracted from each of the resulting diploid genotypes. 22 (10%) of the 221 mutants tested showed evidence of PTDH3-YFP duplications (Figure 2B); 10 mutants were not analyzed because they either failed to produce diploids or showed evidence of contamination. 16 genotypes isolated from the control population with |Z|>2.58 were also tested, and 5 (31%) showed evidence of more than one copy of PTDH3-YFP. This high frequency of copy number variants in the control population is consistent with the idea that copy number variants in the EMS-treated population also resulted primarily from spontaneous duplications. This in turn suggests that duplications are the most common type of spontaneous mutation affecting PTDH3-YFP activity, given that we estimated the frequency of point mutations was elevated ∼5700-fold by EMS in our screen.
On the basis of these data, we divided the 221 mutants tested for copy number variation into four classes (Table S3): the 16 mutants containing a mutation affecting the amino acid sequence of YFP were classified as “coding”; the 22 mutants containing a duplication of PTDH3-YFP were classified as copy number variants, or “CNV”s; the 4 mutants containing a mutation in the TDH3 promoter were classified as “cis-regulatory”; and the 179 mutants that had neither a cis-regulatory or non-synonymous mutation in PTDH3-YFP nor a change in its copy number were classified as “trans-acting”. This large trans-acting class of mutants is expected to include coding and noncoding changes in genes other than PTDH3-YFP that regulate its transcription and post-transcriptional processing as well as mutations that impact elements of the cell that affect fluorescence per unit cell size (i.e., FSC) (e.g., pH [26]). Epigenetic changes are also possible. Of the 10 mutant genotypes that we were unable to test for PTDH3-YFP copy number, none showed any sequence differences in PTDH3-YFP, indicating that they could be either CNVs or trans-acting mutants (Table S3). Because of this ambiguity, these 10 genotypes were excluded from the comparisons among mutant classes described below.
Mutations affecting the amino acid sequence of YFP or the number of copies of PTDH3-YFP were assumed to explain the mutant phenotypes of genotypes in which they occur; however, we were less confident of this assumption for the promoter mutations. Therefore, we empirically tested whether each of the three mutations identified in the promoter region was (1) sufficient to alter YFP fluorescence and (2) sufficient to recreate the YFP fluorescence phenotype of the mutant genotype(s) that harbored it. Site-directed mutagenesis was used to introduce each mutation into the ancestral (unmutagenized) genotype, and YFP fluorescence was analyzed in a haploid population of these genetically modified cells using flow cytometry. In all three cases, populations of cells containing one of these promoter mutations showed in a significant change in YFP fluorescence relative to cells with the ancestral promoter (P<0.05, Mann-Whitney-Wilcoxon (MWW); Figure 2C). For three of the four genotypes containing a promoter mutation, this mutation was sufficient to recapitulate the change in YFP fluorescence (Figure 2C), showing that the promoter mutation was solely responsible for the observed mutant phenotype. The one exception was a genotype that carried the same promoter mutation as another strain; in this case, the promoter mutation only partially recreated the mutant's YFP fluorescence (Figure 2C), indicating that this genotype (mutant 4Q4E4) contained more than one mutation affecting YFP fluorescence.
Effects of mutations on PTDH3-YFP activity differed among mutational classes
As described above, the Z-score calculated for each sorted genotype describes the magnitude and direction of its effects on PTDH3-YFP activity relative to the control (unmutagenized) genotype. To determine the relative frequency of mutations that increased and decreased YFP fluorescence, we examined the sign of the Z-score for each of the 231 mutants with |Z|>2.58 and found that 162 (70.1%) showed increased fluorescence (Z>0). When alternative thresholds of either |Z|>1.96 or |Z|>1.645, corresponding to p<0.05 and p<0.1, respectively, were used to identify mutants, 68% showed increased fluorescence. This excess of mutants with increased YFP fluorescence was surprising given the similar increases in cell number observed in both tails of the full EMS-treated population (Figure 1B, 1C). As described above, differences in colony formation rates were observed between the high - and low-fluorescing tails; however, they are unlikely to explain the apparent excess of high-fluorescing mutants: 65.3% of FACS events sorted from the low-fluorescing tail formed colonies and 38.5% of this group were expected to be mutants, whereas 70.1% of FACS events sorted from the high-fluorescing tail formed colonies and 32.2% of these were expected to be mutants, suggesting that about half (52.7%) of recovered mutants should increase fluorescence. This discrepancy might instead result from a nonuniform distribution of mutants within each tail, such that sorting from a slightly larger tail at the low-fluorescing end of the distribution (1.22% vs 1.04% of cells in the EMS-treated population) resulted in a lower proportion of sorted cells being classified as mutants.
Comparing the distributions of Z-scores among the four mutational classes showed differences in both the magnitude and direction of effects among groups (Figure 3). For example, none of the 16 coding mutants increased fluorescence, compared to 21 (95%) of the 22 CNVs, 2 (50%) of the 4 cis-regulatory mutants, and 131 (73%) of the 179 trans-acting mutants. Considering only the magnitude of the change in YFP fluorescence caused by each mutant (|Z|), we found statistically significant pairwise differences among coding, CNV, and trans-acting mutants (P≤0.02 in all 3 comparisons, MWW test). With only four cis-regulatory mutants recovered from our screen, we had little power to detect differences in comparison with other classes, and all three pairwise tests involving this class failed to reach statistical significance (P≥0.15 in all cases, MWW test). Overall, coding mutants had the largest effect on gene activity (median |Z| = 48), followed by cis-regulatory mutants (median |Z| = 8.1) and CNVs (median |Z| = 8.0), and finally trans-acting mutants (median |Z| = 4.6).
Fig. 3. Effects on YFP fluorescence in haploid cells differ among mutational classes. 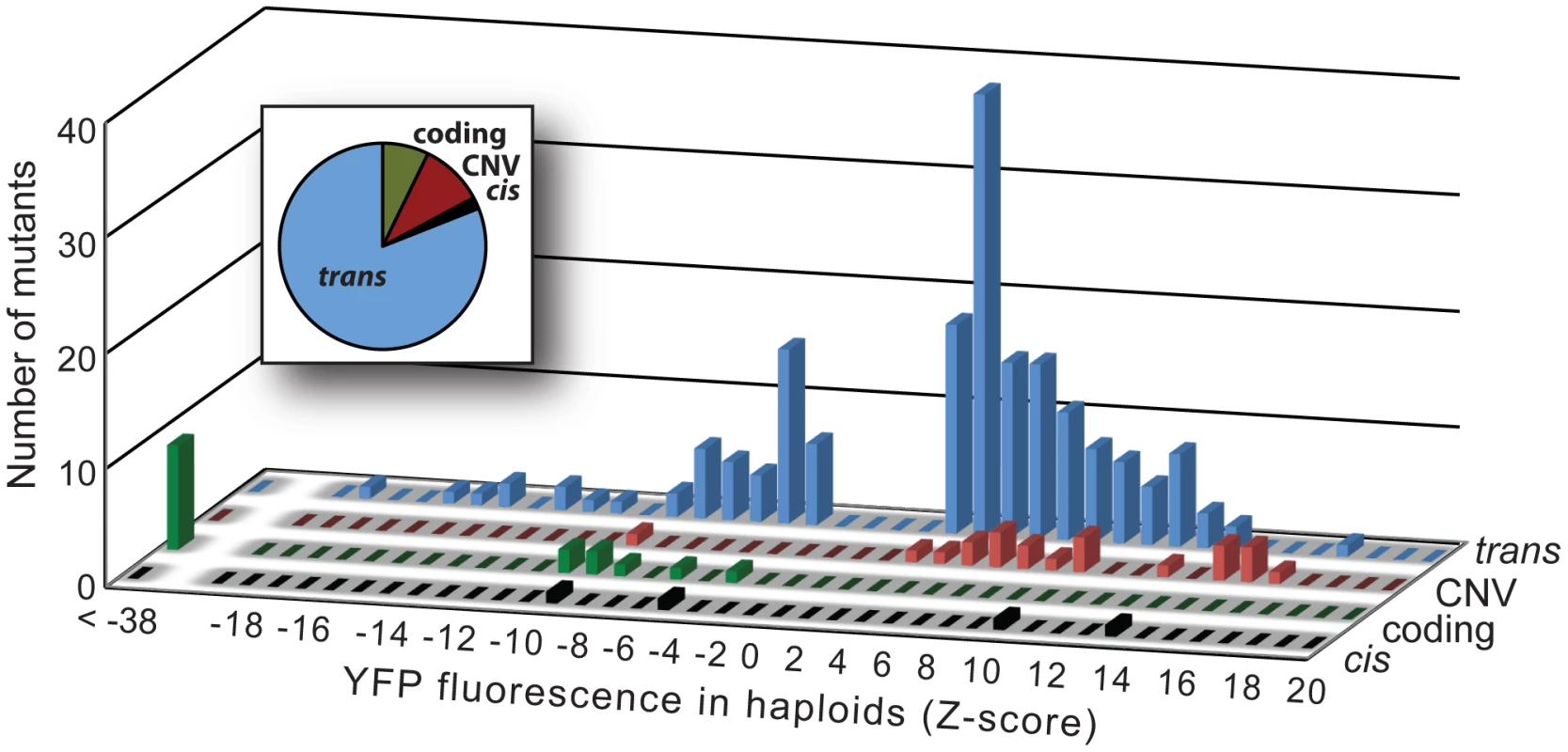
The effects on YFP fluorescence of the 4 cis-regulatory (black), 16 coding (green), 22 CNV (red), and 179 trans-acting (blue) mutants are summarized in histograms. For each mutational class, the height of each bar indicates the number of mutants with the corresponding effect (as measured by Z-score) on YFP fluorescence in haploid cells. Positive Z-scores indicate increases in YFP fluorescence relative to control cells and negative Z-scores indicate decreases in YFP fluorescence relative to control cells. The relative frequency of mutants in each of the four mutational classes is also shown in the inset pie chart. Dominance of mutations affecting PTDH3-YFP activity differed among mutational classes
We isolated mutants in haploid cells so that we could recover recessive mutations; however, wild populations of many eukaryotes, including S. cerevisiae, tend to be diploid. To determine how the haploid mutant genotypes in our collection act in diploid cells, we again crossed each mutant genotype with the reference strain containing PTDH3-CFP that was used to identify CNVs. YFP and CFP fluorescence was measured in at least 9,000 diploid cells for each mutant genotype using flow cytometry. Z-scores describing YFP and CFP fluorescence were calculated for each mutant by comparing their fluorescence to that of replicate populations of control diploid cells resulting from mating the ancestral, unmutagenized PTDH3-YFP haploid genotype to the reference haploid genotype containing PTDH3-CFP. We successfully tested all 16 coding mutants, all 4 cis-regulatory mutants, 21 of the 22 CNVs, and 171 of the 179 trans-acting mutants for their effects on YFP and CFP fluorescence in diploid cells.
To assess the dominance of each mutant relative to the reference strain (i.e., its ability to affect PTDH3-YFP activity in heterozygous, diploid cells), we compared the Z-scores for YFP fluorescence of each mutant from haploid and diploid cells (Figure 4A). We found that the effects of coding, CNV, and cis-regulatory mutants in diploid cells (median |Z| = 96, 15, and 10, respectively) were more similar to their effects in haploid cells (regression coefficients (b) of 1.67, 1.50, and 1.34, respectively, from a model II regression) than were those of trans-acting mutants (median |Z| = 1, b = 0.12). These data show that as a group, the trans-regulatory mutants were much more recessive than mutants from any of the other classes. This was also seen using a threshold of |Z| = 2.58 to classify mutants as recessive (i.e., no significant effect on YFP fluorescence in diploids): none of the coding, CNV, or cis-regulatory mutants were called recessive, whereas 151 (88%) of the 171 trans-acting mutants tested were.
Fig. 4. Effects of heterozygous mutant alleles on YFP and CFP fluorescence differ among mutational classes in diploid cells. 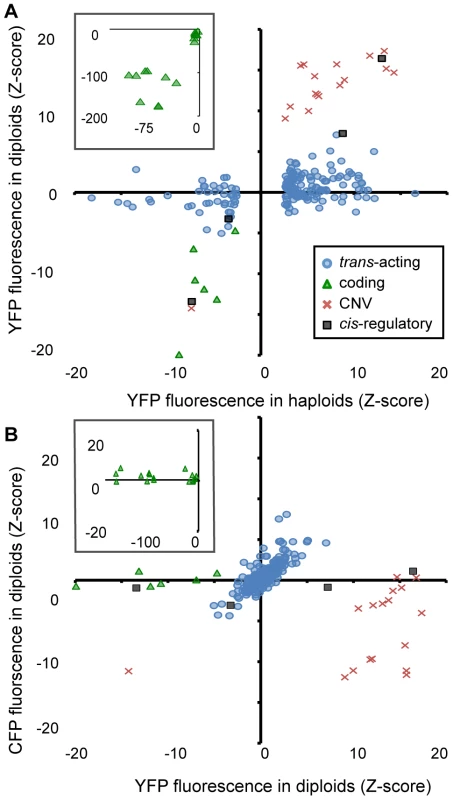
(A) The effect of each mutant genotype on YFP fluorescence in haploid cells (X-axis) is compared to the effect of the heterozygous mutant genotype on YFP fluorescence in diploid cells (Y-axis). These diploid cells were heterozygous for the mutant PTDH3-YFP haploid genome and a reference haploid genome containing PTDH3-CFP. Black squares indicate cis-regulatory mutants, blue circles indicate trans-acting mutants, green triangles indicate coding mutants, and red crosses indicate CNV mutants. (B) Using the same symbols to represent the four mutational classes as in (A), the effect of each mutant on YFP (X-axis) and CFP (Y-axis) fluorescence in diploid cells is shown. Insets in (A) and (B) show only coding mutants and cover the larger ranges of Z-scores needed to plot all of the mutants in this class. To determine whether a mutant genotype had similar effects on both alleles of the reporter gene present in diploid cells, we compared the effects of each genotype on [diploid] YFP and CFP fluorescence (Figure 4B). This analysis showed that CNVs had the largest effect on CFP fluorescence (median |Z| = 4.2 compared to median |Z| = 1 for all other mutant classes). Surprisingly, 15 of the CNVs showed a significant decrease in CFP fluorescence despite a significant increase in YFP fluorescence. Of the coding mutants tested, the majority (14 of 16) showed no significant effect on CFP fluorescence (|Z|<2.58), as expected. The remaining two showed small increases in CFP fluorescence (Z = 2.6 and 2.8, respectively) despite showing decreases in YFP fluorescence (Z = −167 and −28). These genotypes might harbor amino acid changes that alter the emission spectrum of the mutant YFP protein and cause it to overlap that of CFP. Mutants classified as cis-acting were also not expected to alter CFP fluorescence, and three of the four cis-acting mutants did not (|Z| = 0.75, 1.1, and 0.68). The one cis-regulatory mutant that showed an effect on CFP fluorescence (1Q4D11, whose mutant phenotype in haploids appeared to be caused solely by the identified promoter mutation, Figure 2C) decreased both CFP and YFP fluorescence (Z = −2.7, P = 0.006), suggesting transvection [27]. Finally, non-recessive mutants in the trans-acting class were expected to have similar effects on both YFP and CFP fluorescence in diploid cells, and 13 (65%) of the 20 trans-acting mutants with |Z|>2.58 for YFP fluorescence in diploid cells also showed a significant (|Z|>2.58) effect on CFP fluorescence in the same direction. Considering all mutants in all classes, the relationship between CFP and YFP fluorescence in diploid cells was strongest for the trans-acting mutant class (Figure 4B; b = 1 compared with b = 0.00, 0.37, and 0.08 for coding, CNV, and cis-regulatory mutants, respectively).
Discussion
This study provides a systematic survey and functional analysis of mutations affecting activity of a reporter gene (PTDH3-YFP) in S. cerevisiae. By comparing unmutagenized and mutagenized subpopulations of a clonal culture, we estimated a spontaneous mutation rate for activity of PTDH3-YFP of 5.3×10−6 per haploid genome per generation, which, in S. cerevisiae, is intermediate between spontaneous mutation rates reported for single gene loss-of-function phenotypes (10−6–10−8, [22], [28]–[32] and more complex organismal phenotypes such as growth rate (10−3, [33]), and suggests that PTDH3-YFP activity is controlled by a moderate number of genes. Further characterization of 231 mutants with changes in PTDH3-YFP activity revealed differences in the relative frequency, effects, and dominance of different types of mutations (Table 1) that are expected to influence their likelihood of contributing to phenotypic evolution.
Tab. 1. Comparison of properties among mutational classes. 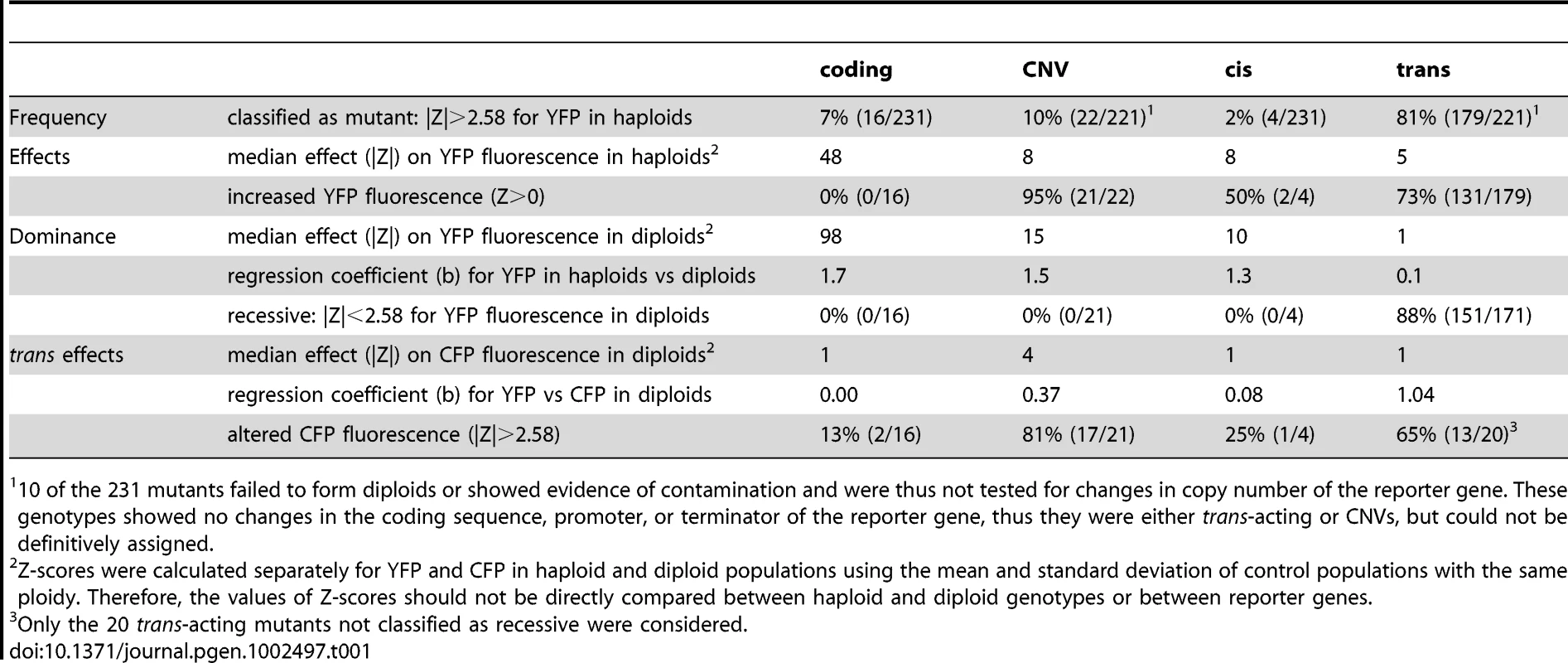
10 of the 231 mutants failed to form diploids or showed evidence of contamination and were thus not tested for changes in copy number of the reporter gene. These genotypes showed no changes in the coding sequence, promoter, or terminator of the reporter gene, thus they were either trans-acting or CNVs, but could not be definitively assigned. EMS-induced mutations affecting PTDH3-YFP are expected to be similar to spontaneous mutations affecting endogenous genes
Before discussing the evolutionary implications of our results, it is important to consider how the use of the fluorescent reporter gene PTDH3-YFP and chemical mutagen EMS might cause our data to differ from a spectrum of spontaneous mutations affecting activity of an endogenous gene.
Activity of the chimeric PTDH3-YFP reporter gene was regulated by the native S. cerevisiae TDH3 promoter and CYC1 terminator sequences. The CYC1 terminator controls proper 3′ end formation of mRNA by binding to factors such as Rat1p and Sen1p that are important for the termination of many genes transcribed by DNA polymerase I and II in S. cerevisiae [34]. The TDH3 gene encodes isozyme 3 of glyceraldehyde-3-phosphate dehydrogenase [35], is not required for viability under normal culture conditions [36], and is transcribed during growth on both fermentable and non-fermentable carbon sources [37] with minimal fluctuations during the cell cycle [38]. The TDH3 promoter exemplifies regulatory principles shared by many eukaryotic genes. For example, it includes both activating [39] and repressing [37] sequences that bind transcription factors such as Gcr1p, Gcr2p, Hsf1p, Pho2p, and Rap1p [40], [41]. TDH3 is one of the ∼19% of genes in the S. cerevisiae genome whose promoter has a TATA box; this is important to note because the expression of such genes appears to be more mutable than genes whose promoters lack this sequence [17]. Promoters with simple repetitive sequences have also been shown to have increased evolvability of gene expression in yeast [42], but the TDH3 promoter appears to lack such sequences. Differences in promoter and terminator sequences among genes are expected to cause differences in gene-specific mutational spectra, but we do not expect the regulatory mutation spectrum recovered for PTDH3-YFP to be fundamentally different from that of an endogenous gene such as TDH3.
Unlike the regulatory sequences of PTDH3-YFP, its coding sequence was not native to yeast: it encoded a Yellow Fluorescent Protein derived from the Green Fluorescent Protein originally isolated from Aequorea victoria [43]. The Venus variant of YFP used in this study was previously optimized to speed maturation, improve stability, and minimize sensitivity to environmental changes such as pH and chloride concentration [18]. This optimization might explain why none of the YFP coding mutants we recovered showed increased fluorescence and suggests that mutants affecting YFP fluorescence by altering the cellular environment might be rare. Native yeast proteins might not always have such optimal activity; however, nonsynonymous mutations are generally thought to decrease a protein's function more often than they increase it, suggesting that the YFP coding sequence is not extremely unrealistic in this respect. The length and GC-content of the YFP coding region are also expected to influence the coding mutation rate measured in this study by affecting the mutational target size: at 238 amino acids, YFP is near the 20th percentile for the length of native S. cerevisiae proteins [44], and its GC-content of 35.56% is similar to the median GC-content for all S. cerevisiae genes of 39.95% (Figure S2).
The use of the chemical mutagen EMS is perhaps the most artificial element of our experimental design, although we are not the first to use it to make inferences about spontaneous mutations [45]–[48]. EMS predominantly causes G/C to A/T transitions [23], [24] and thus generates a subset of possible spontaneous mutations. However, G/C to A/T transitions are tied with G/C to T/A transversions as the most common type of spontaneous point mutation in yeast [1]. We anticipate that many types of point mutations will have similar distributions of genetic and phenotypic effects, although it will be interesting to test this hypothesis in future work. More importantly, we expect the proportion of sites targeted by EMS to be similar for coding, cis-regulatory, and trans-acting mutations, suggesting that comparing EMS-induced mutations among these classes reveals differences that should also be observed for spontaneous point mutations. We do anticipate, however, that spontaneous mutations involving more than one base-pair (e.g., insertions/deletions (indels), segmental duplications, chromosomal rearrangements) will have different distributions of effects than single point mutations, and this was observed when we compared mutants with (presumably spontaneous) duplications of PTDH3-YFP to those with single copies of the PTDH3-YFP gene. Consequently, we believe that the spectrum of mutational effects described in this work provides a reasonable approximation of the spectrum of mutational effects caused by different types of spontaneous point mutations, but might not be representative of other types of DNA lesions.
Differences in the frequency, effects, and dominance among functional classes of mutations may impact their relative contributions to phenotypic evolution
For any mutation, the likelihood of fixation depends upon the probability that the mutational event occurs and the probability that, once it occurs, it becomes fixed within a population. This latter probability depends upon the phenotypic effects of the mutation (specifically, its impact on fitness) and (for diploid organisms) dominance. Mutations that arise more frequently have more opportunities to become fixed; mutations with larger effects on fitness should either be removed from or fixed within a population faster than mutations with smaller effects on fitness [49]; and adaptive mutations that are recessive are less likely to fix in a diploid population than equally adaptive mutations that are not recessive [50]. As described above, we observed differences in the frequency, effects (on YFP fluorescence), and dominance of different types of mutations affecting PTDH3-YFP activity. Below, we discuss how these differences might influence the evolutionary trajectories of (i) coding and regulatory mutations, (ii) cis-regulatory and trans-regulatory mutations, and (iii) copy number variants.
Coding and regulatory mutations
In recent years, the relative contributions of regulatory and coding changes to phenotypic evolution have been discussed extensively; however different authors have defined these categories differently [7], [15]. Some authors contrast only cis-regulatory and coding changes [7]–[10], [13], while others make an additional distinction between coding changes in transcription factors and coding changes in other types of proteins [11]. A broader contrast is also common in which all changes that alter gene expression are compared to changes that alter the coding sequence of the gene of interest [7], [51]. Our data are suitable for contrasting coding mutations with either cis-regulatory or broadly defined regulatory mutations, but we do not currently have enough information to identify which of the regulatory mutants, if any, harbor coding changes in transcription factors.
Comparing only the coding and cis-regulatory mutants in our collection suggests that coding mutations are more common than cis-regulatory mutations (at least for genes similar to PTDH3-YFP), and that cis-regulatory mutations have more moderate effects (Table 1). Both classes of mutations showed similar effects in haploid and diploid cells, suggesting that they are both non-recessive and selectable as soon as they arise in diploid populations. If the large changes in PTDH3-YFP activity we observed in the majority of coding mutations tend to be strongly deleterious, the more moderate effects of cis-regulatory mutations (especially when coupled with the presumed lower degree of pleiotropy of cis-regulatory mutations) might make them more likely to fix than coding mutations despite their lower mutation rate. This might be especially true when a favored phenotype requires an increase in gene activity, as this type of change was observed for 50% of the cis-regulatory mutants recovered, but none of the coding mutants.
The broader definition of “regulatory” mutants described above includes all genotypes classified as cis-regulatory, trans-acting, and copy number mutants in this study because they all change YFP fluorescence without altering the coding sequence of YFP. Considering these three classes together, regulatory mutations were over 10-times more common than coding mutations in PTDH3-YFP. And this is without accounting for the fact that the gene duplication rate was not expected to be increased by EMS. Regulatory mutations (broadly defined) may therefore be a much more abundant source of variation in a gene's activity than mutations changing its protein sequence. This higher frequency might be at least partially offset, however, by the fact that over 75% of the regulatory mutants we characterized (all of which were categorized as trans-acting) appeared to be recessive. Like cis-regulatory mutants, this broader class of regulatory mutants had more moderate effects on YFP fluorescence than coding mutants and the activity of PTDH3-YFP was increased at least half of the time. The frequent recovery of regulatory mutants with elevated YFP fluorescence was unexpected and particularly surprising given that the TDH3 promoter drives high levels of expression in wildtype cells [52] and Tdh3p is in the 98th percentile for protein abundance in S. cerevisiae [53]. It will be interesting to see in future work whether mutational spectra for other genes also show a high rate of mutations that increase the gene's activity.
cis-regulatory and trans-acting changes
The relative contribution of cis - and trans-acting mutations to polymorphic and divergent gene expression has also been examined in a variety of species, either using genetic mapping or allele-specific expression to infer changes in cis - and trans-regulation (reviewed by [14], [54]–[56]). These analyses have revealed differences in the frequency, effects, and dominance of segregating cis - and trans-regulatory variation that our data suggest might result (at least in part) from inherent properties of cis - and trans-regulatory mutations rather than selection for a biased subset of regulatory mutations.
For example, trans-regulatory variation has been observed to be more abundant within a species than cis-acting variation, and we found that the mutational target size for trans-acting mutations affecting activity of PTDH3-YFP was ∼45 times larger than for cis-regulatory mutations affecting activity of this gene. A larger mutational target size for trans-regulatory variation was also inferred from mutation accumulation lines [17], [57]. cis-acting quantitative trait loci affecting gene expression (eQTL) generally have larger average effects on a gene's expression than trans-regulatory eQTL, and we found that cis-regulatory mutations trended toward having larger effects on YFP fluorescence than trans-regulatory mutations in both haploid (P = 0.07, one-sided MWW test) and heterozygous diploid (P = 0.0007, one-sided MWW test) cells. Finally, segregating cis-regulatory alleles have been shown to be recessive less often than trans-regulatory alleles [58], [59], and we found that the effects of cis-regulatory mutants in heterozygous cells were masked less often than the effects of trans-acting mutants.
Taken together, these observations suggest that, trans-acting variation might be more common within a species than cis-regulatory variation for individual genes because of the higher rate of trans-acting mutations, their tendency to have smaller effects on gene activity than cis-acting mutations, and their propensity to be recessive in diploid cells. If the size of the effect on gene activity is correlated with the selection coefficient, cis-regulatory variants might contribute more to expression differences between than within species (as has been observed for Drosophila [60] and Saccharomyces [61]) because the tendency of cis-regulatory mutations to have larger and more non-recessive effects on gene activity than trans-regulatory mutations should cause them to be selected for or against more strongly. This should lower the probability that a cis-regulatory mutation segregates for long periods of time within a species and raise the probability that a cis-regulatory mutation contributes to adaptive regulatory changes between species.
Copy number variants
Gene duplications or deletions can alter the number of copies of a gene and, assuming all copies are expressed, this will affect the abundance of the gene's product. Such copy number variants are known to contribute to phenotypic diversity in humans, yeast and other eukaryotes [62]–[64]. In S. cerevisiae, spontaneous gene duplications are almost 5 times more common than spontaneous point mutations: in one genome replication, 0.019 gene duplications are expected, compared to only 0.004 point mutations [1]. Consistent with this high spontaneous mutation rate, duplications containing PTDH3-YFP were found in 10% of the mutants we examined despite the fact that EMS is not expected to influence their occurrence. Duplications of other genomic regions that affect PTDH3-YFP activity might also be present in our mutant collection as trans-acting mutants, but determining the frequency and location of any such duplications (as well as the extent of the duplications including PTDH3-YFP) requires further analysis. As expected, nearly all mutant genotypes harboring a duplication of PTDH3-YFP increased the YFP fluorescence of both haploid and diploid cells. The one exception was a mutant that showed decreased YFP fluorescence, which could be caused by the duplication of one or more negative regulators of PTDH3-YFP. Interestingly, the majority of duplications that increased YFP fluorescence decreased CFP fluorescence in diploids, which might reflect a mechanism that silences both the original and new copy of a gene following a duplication event [65]. Despite their high mutation rate, copy number variants appear to rarely be fixed between yeast species [66], suggesting that they are often deleterious and eliminated by natural selection or have a high rate of spontaneous reversion.
Unresolved questions: Pleiotropy, fitness, and the genomic locations of mutations affecting PTDH3-YFP activity
This study provides an unprecedented survey of the functional characteristics of new mutations; however, our understanding of mutational properties remains far from complete, even for the PTDH3-YFP reporter gene. For example, the effect of a mutation on fitness is what matters most for evolution, but we measured only the effects of mutations on YFP fluorescence. If fluorescence were an adaptive trait, a relationship between mutational effects on PTDH3-YFP activity and fitness is expected, but the effects of each mutation on other traits (i.e., pleiotropy) will also influence fitness, complicating this relationship. Similarly, we assessed the dominance of mutant effects on YFP fluorescence, but dominance at the level of a single gene's activity might not always translate to dominance at the level of higher-order phenotypes, which are more likely to be the targets of natural selection.
The unique collection of mutants described here provides a rare opportunity to address these issues, however, by mapping each of the mutations affecting PTDH3-YFP activity, engineering them individually into the ancestral genetic background, and directly measuring pleiotropy (quantified as the number of genes in the genome that change expression in response to the mutation) and fitness under different conditions (quantified as the effect of the mutation on relative growth rate). Determining the identity of mutations responsible for these mutant phenotypes will also allow us to assess their distribution within the genome and among factors expected to influence PTDH3-YFP activity and to compare the overall contributions of coding and non-coding changes to the mutational spectrum for PTDH3-YFP. Integrating these data with those presented here, as well as performing similar analyses of reporter genes using promoters from other S. cerevisiae genes, should greatly improve our ability to predict the types of genetic changes most likely to contribute to phenotypic evolution under different conditions.
Materials and Methods
An abbreviated version of the materials and methods follows. Complete materials and methods, including calculation of the spontaneous mutation rate, are included as Supporting Information (Text S1).
Mutagenesis
Chemical mutagenesis with Ethyl Methane Sulfonate (EMS) was performed as previously described [67], except that the volume of the cell suspension was doubled to 2 ml, the cell density was reduced by 50% to 6×107 cells/ml, the concentration of EMS was reduced by 75% to 7.5%, and the time of exposure was reduced by 25% to 45 minutes. Following mutagenesis, control and mutagen-treated cells were cultured at 30–32°C for 42 hours in arginine dropout liquid media (Synthetic Complete media lacking arginine) [67].
Canavanine resistance assay
The canavanine resistance mutation rate in the EMS-treated population was calculated by comparing colony forming units on arginine dropout plates with and without 60 mg/l canavanine sulfate (Sigma-Aldrich, St. Louis, MO) [67].
Flow cytometry and primary screen for prospective mutants
Prior to analysis and sorting, cells from the control culture were stained with Cy5 (GE Healthcare, Piscataway, NJ) so that they could be distinguished from EMS-treated cells when analyzed simultaneously. Aliquots of both populations were mixed together in Phosphate Buffered Saline (PBS) for analysis and sorted in a FACSaria flow cytometer/cell sorter (BD Biosystems, San Jose, CA). The sorting and analysis of the mixed suspensions was restricted to a FSC-defined subset of events to reduce the influence of non-cell particles. In each of nine consecutive sorting runs, 96 events were collected from each tail of the EMS-treated and control populations, for a total of 384 events collected in each sorting run. Thresholds used for sorting during each run are presented in Table S1. Sorted events were arrayed onto YPD agar plates [67], then incubated at 30°C for 28 hours.
Liquid cultures for secondary screen of candidate mutants and diploid testing
High-throughput parallel liquid culturing of genotypes was performed by inoculating from a colony or patch and culturing for 24 hours at 30°C in 96-well deep well plates in YPD. Saturated cultures were diluted 100× into arginine dropout liquid and cultured at 30°C for at least 2 doublings until the density reached 0.5–1.0×107 cells/ml.
Quantification of fluorescence in flow cytometry data
Haploid YFP fluorescence and FSC were evaluated by a C6 flow cytometer (Accuri, Ann Arbor, MI); diploid YFP, CFP, and FSC were evaluated by a FACSaria flow cytometer (BD Biosciences, San Jose, CA). After log-transformation of FSC and fluorescence values, filters were applied to cull events with extreme FSC values; generally, these filters corresponded to approximately the 20th and 80th percentiles of FSC. The fluorescence phenotype of each genotype was defined as its median YFP/FSC or CFP/FSC ratio. This ratio was converted to a Z-score using the mean and standard deviation calculated from at least 10 (and up to 143) replicate control cultures containing cells with the unmutagenized, ancestral genotype.
Pyrosequencing to assay YFP copy number
PCR primers hybridizing to DNA sequences shared by YFP and CFP that flanked a position of dissimilarity were used to amplify a small region of DNA for analysis. Using the PSQ96 pyrosequencer (Qiagen, Valencia, CA), an internal sequencing primer was hybridized to these amplified fragments and extended to a diagnostic position that differed between YFP and CFP, allowing the relative frequency of YFP and CFP alleles to be compared in heterozygous diploid cells.
Testing the sufficiency of promoter mutations
After introducing each promoter mutation into the unmutagenized ancestral genotype using site-directed mutagenesis, we quantified YFP fluorescence using the C6 flow cytometer (Accuri, Ann Arbor, MI). Two 80,000-event samples were collected for genotypes into which the mutations identified at positions −255, −240, and −140 had been introduced into the unmutagenized progenitor, a strain in which a wildtype copy of the promoter had been re-introduced in parallel, and the four mutant genotypes in which the promoter mutations were originally detected (including the two different isolates with the mutation at −255). Taking the median YFP/FSC as the fluorescence phenotype of each culture, we compared the genotypes with site-directed promoter mutations to the reengineered wildtype control, and the genotypes with site-directed promoter mutations to the mutant(s) in which it was originally observed using MWW tests.
Supporting Information
Zdroje
1. LynchMSungWMorrisKCoffeyNLandryCR 2008 A genome-wide view of the spectrum of spontaneous mutations in yeast. Proceedings of the National Academy of Sciences 105 9272 9277
2. NishantKTSinghNDAlaniE 2009 Genomic mutation rates: what high-throughput methods can tell us. BioEssays 31 912 920
3. ConradDFKeeblerJEDePristoMALindsaySJZhangY 2011 Variation in genome-wide mutation rates within and between human families. Nat Genet 43 712 714
4. BraendleCBaerCFFelixMA 2010 Bias and evolution of the mutationally accessible phenotypic space in a developmental system. PLoS Genet 6 e1000877 doi:10.1371/journal.pgen.1000877
5. YampolskyLYStoltzfusA 2001 Bias in the introduction of variation as an orienting factor in evolution. Evolution & development 3 73 83
6. StoltzfusAYampolskyLY 2009 Climbing mount probable: mutation as a cause of nonrandomness in evolution. The Journal of heredity 100 637 647
7. SternDLOrgogozoV 2008 The loci of evolution: How predictable is genetic evolution? Evolution 62 2155 2177
8. HoekstraHCoyneJ 2007 The locus of evolution: Evo-Devo and the genetics of adaptation. Evolution 61 995 1016
9. WrayGA 2007 The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics 8 206 216
10. CarrollSB 2008 Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134 25 36
11. LynchVJWagnerGP 2008 Resurrecting the role of transcription factor change in developmental evolution. Evolution; international journal of organic evolution 62 2131 2154
12. LynchM 1988 The rate of polygenic mutation. Genetical research 51 137 148
13. SternDLOrgogozoV 2009 Is genetic evolution predictable? Science 323 746 751
14. FayJCWittkoppPJ 2008 Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100 191 199
15. StreisfeldMARausherMD 2011 Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution; international journal of organic evolution 65 629 642
16. WittkoppPJ 2005 Genomic sources of regulatory variation in cis and in trans. Cell Mol Life Sci 62 1779 1783
17. LandryCRLemosBRifkinSADickinsonWJHartlDL 2007 Genetic Properties Influencing the Evolvability of Gene Expression. Science 317 118 121
18. NagaiTIbataKParkESKubotaMMikoshibaK 2002 A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotech 20 87 90
19. ZaretKSShermanF 1982 DNA sequence required for efficient transcription termination in yeast. Cell 28 563 573
20. SalzmanHMSinghamSBJohnstonRGBohrenCF 1990 Light scattering and cytometry. MelamedMRLindmoTMendelsohnML Flow cytometry and sorting. 2nd ed New York Wiley 81 107
21. KudlaGMurrayAWTollerveyDPlotkinJB 2009 Coding-sequence determinants of gene expression in Escherichia coli. Science 324 255 258
22. LangGIMurrayAW 2008 Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics 178 67 82
23. CoulondreCMillerJH 1977 Genetic studies of the lac repressor : IV. Mutagenic specificity in the lacI gene of Escherichia coli. Journal of Molecular Biology 117 577 606
24. GreeneEACodomoCATaylorNEHenikoffJGTillBJ 2003 Spectrum of Chemically Induced Mutations From a Large-Scale Reverse-Genetic Screen in Arabidopsis. Genetics 164 731 740
25. RizzoMASpringerGHGranadaBPistonDW 2004 An improved cyan fluorescent protein variant useful for FRET. Nat Biotech 22 445 449
26. KneenMFarinasJLiYVerkmanAS 1998 Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J 74 1591 1599
27. DuncanIW 2002 Transvection effects in Drosophila. Annual Review of Genetics 36 521 556
28. DrakeJW 1991 A constant rate of spontaneous mutation in DNA-based microbes. Proceedings of the National Academy of Sciences 88 7160 7164
29. KunzBAKohalmiLKangXLMagnussonKA 1990 Specificity of the mutator effect caused by disruption of the RAD1 excision repair gene of Saccharomyces cerevisiae. J Bacteriol 172 3009 3014
30. MagniGE 1964 Origin and Nature of Spontaneous Mutations in Meiotic Organisms. J Cell Physiol 64: SUPPL 1 165 171
31. MagniGEVonborstelRCSoraS 1964 Mutagenic Action during Meiosis and Antimutagenic Action during Mitosis by 5-Aminoacridine in Yeast. Mutat Res 106 227 230
32. GottliebDJvon BorstelRC 1976 Mutators in Saccharomyces cerevisiae: MUT1-1, MUT1-2 and MUT2-1. Genetics 83 655 666
33. WlochDMSzafraniecKBortsRHKoronaR 2001 Direct estimate of the mutation rate and the distribution of fitness effects in the yeast Saccharomyces cerevisiae. Genetics 159 441 452
34. KawauchiJMischoHBragliaPRondonAProudfootNJ 2008 Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes & Development 22 1082 1092
35. McAlisterLHollandMJ 1985 Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. The Journal of biological chemistry 260 15019 15027
36. GiaeverGChuAMNiLConnellyCRilesL 2002 Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387 391
37. KurodaSOtakaSFujisawaY 1994 Fermentable and nonfermentable carbon sources sustain constitutive levels of expression of yeast triosephosphate dehydrogenase 3 gene from distinct promoter elements. J Biol Chem 269 6153 6162
38. SpellmanPTSherlockGZhangMQIyerVRAndersK 1998 Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Molecular biology of the cell 9 3273 3297
39. BitterGAChangKKHEganKM 1991 A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Molecular and General Genetics MGG 231 22 32
40. LiebJDLiuXBotsteinDBrownPO 2001 Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28 327 334
41. LeeTIRinaldiNJRobertFOdomDTBar-JosephZ 2002 Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298 799 804
42. VincesMDLegendreMCaldaraMHagiharaMVerstrepenKJ 2009 Unstable Tandem Repeats in Promoters Confer Transcriptional Evolvability. Science 324 1213 1216
43. ShimomuraOJohnsonFHSaigaY 1962 Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. Journal of Cellular and Comparative Physiology 59 223 239
44. WarringerJBlombergA 2006 Evolutionary constraints on yeast protein size. BMC evolutionary biology 6 61
45. KeightleyPDOhnishiO 1998 EMS-induced polygenic mutation rates for nine quantitative characters in Drosophila melanogaster. Genetics 148 753 766
46. YangHPTanikawaAYVan VoorhiesWASilvaJCKondrashovAS 2001 Whole-genome effects of ethyl methanesulfonate-induced mutation on nine quantitative traits in outbred Drosophila melanogaster. Genetics 157 1257 1265
47. OhnishiO 1977 Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. I. Recessive lethal mutations. Genetics 87 519 527
48. KeightleyPDDaviesEKPetersADShawRG 2000 Properties of ethylmethane sulfonate-induced mutations affecting life-history traits in Caenorhabditis elegans and inferences about bivariate distributions of mutation effects. Genetics 156 143 154
49. HartlDLClarkAG 1989 Principles of Population Genetics Sunderland, MA Sinauer Associates, Inc
50. HaldaneJBS 1927 A mathematical theory of natural and artificial selection part V: selection and mutation. Proceedings of the Cambridge Philosophical Society Mathematical and physical sciences 23 838 844
51. StreisfeldMARausherMD 2009 Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. The New phytologist 183 751 763
52. MumbergDMüllerRFunkM 1995 Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156 119 122
53. GhaemmaghamiSHuhWKBowerKHowsonRWBelleA 2003 Global analysis of protein expression in yeast. Nature 425 737 741
54. GiladYRifkinSAPritchardJK 2008 Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet 24 408 415
55. RockmanMVKruglyakL 2006 Genetics of global gene expression. Nat Rev Genet 7 862 872
56. KliebensteinDJ 2009 Quantification of variation in expression networks. Methods in Molecular Biology 553 227 245
57. DenverDRMorrisKStreelmanJTKimSKLynchM 2005 The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet 37 544 548
58. LemosBAraripeLOFontanillasPHartlDL 2008 Dominance and the evolutionary accumulation of cis - and trans-effects on gene expression. Proc Natl Acad Sci U S A 105 14471 14476
59. McManusCJCoolonJDDuffMOEipper-MainsJGraveleyBR 2010 Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20 816 825
60. WittkoppPJHaerumBKClarkAG 2008 Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40 346 350
61. EmersonJJHsiehLCSungHMWangTYHuangCJ 2010 Natural selection on cis and trans regulation in yeasts. Genome Res 20 826 836
62. SchriderDRHahnMW 2010 Gene copy-number polymorphism in nature. Proceedings Biological sciences/The Royal Society 277 3213 3221
63. CarretoLEirizMFGomesACPereiraPMSchullerD 2008 Comparative genomics of wild type yeast strains unveils important genome diversity. BMC genomics 9 524
64. CooperGMNickersonDAEichlerEE 2007 Mutational and selective effects on copy-number variants in the human genome. Nature Genetics 39 S22 29
65. CamblongJBeyrouthyNGuffantiESchlaepferGSteinmetzLM 2009 Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes & Development 23 1534 1545
66. TiroshIReikhavSLevyAABarkaiN 2009 A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324 659 662
67. AmbergDCBurkeDJStrathernJN 2005 Methods in Yeast Genetics
68. ZhangZDietrichFS 2005 Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res 33 2838 2851
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Intrauterinní inseminace a její úspěšnost
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



