-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Strong Deletion Bias in Nonallelic Gene Conversion
Gene conversion is the unidirectional transfer of genetic information between orthologous (allelic) or paralogous (nonallelic) genomic segments. Though a number of studies have examined nucleotide replacements, little is known about length difference mutations produced by gene conversion. Here, we investigate insertions and deletions produced by nonallelic gene conversion in 338 Drosophila and 10,149 primate paralogs. Using a direct phylogenetic approach, we identify 179 insertions and 614 deletions in Drosophila paralogs, and 132 insertions and 455 deletions in primate paralogs. Thus, nonallelic gene conversion is strongly deletion-biased in both lineages, with almost 3.5 times as many conversion-induced deletions as insertions. In primates, the deletion bias is considerably stronger for long indels and, in both lineages, the per-site rate of gene conversion is orders of magnitudes higher than that of ordinary mutation. Due to this high rate, deletion-biased nonallelic gene conversion plays a key role in genome size evolution, leading to the cooperative shrinkage and eventual disappearance of selectively neutral paralogs.
Published in the journal: . PLoS Genet 8(2): e32767. doi:10.1371/journal.pgen.1002508
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002508Summary
Gene conversion is the unidirectional transfer of genetic information between orthologous (allelic) or paralogous (nonallelic) genomic segments. Though a number of studies have examined nucleotide replacements, little is known about length difference mutations produced by gene conversion. Here, we investigate insertions and deletions produced by nonallelic gene conversion in 338 Drosophila and 10,149 primate paralogs. Using a direct phylogenetic approach, we identify 179 insertions and 614 deletions in Drosophila paralogs, and 132 insertions and 455 deletions in primate paralogs. Thus, nonallelic gene conversion is strongly deletion-biased in both lineages, with almost 3.5 times as many conversion-induced deletions as insertions. In primates, the deletion bias is considerably stronger for long indels and, in both lineages, the per-site rate of gene conversion is orders of magnitudes higher than that of ordinary mutation. Due to this high rate, deletion-biased nonallelic gene conversion plays a key role in genome size evolution, leading to the cooperative shrinkage and eventual disappearance of selectively neutral paralogs.
Introduction
All genomes contain similar DNA segments. In diploids, such segments can be classified as orthologs or paralogs. Orthologs, or allelic segments, are paired copies located at the same genomic loci on maternal and paternal chromosomes. In contrast, paralogs, or nonallelic segments, are found at different genomic loci and can have any copy number, in which each copy is derived from an ancestral sequence via gene duplication [1].
The sequences of related DNA segments can diverge via ordinary mutation or converge via gene conversion. Ordinary mutation is generally AT-biased for nucleotide replacements [2]–[4] and deletion-biased for length difference mutations [5]. A number of studies have examined nucleotide replacements produced by allelic and nonallelic gene conversion, some of which have uncovered a GC bias [6]–[9]. Here, we explore length difference mutations produced by nonallelic gene conversion.
In contrast to orthologs, paralogs have their own independent long-term phylogenies, making it possible to apply a direct phylogenetic approach to study their coevolution by gene conversion (Figure 1). For this approach, we utilized multiple alignments of pairs of paralogs in two sister species and an outgroup. First, we ascertained all cases in which, at a particular alignment position, there was an ancestral length difference between the paralogs, i.e., the difference was present in one sister and in the outgroup. We then examined orthologous positions in the other sister and identified those cases for which there was no length difference between paralogs. Elimination of a length difference was due to an insertion if one paralog acquired an additional nucleotide(s) at that position, and was due to a deletion if it lost a nucleotide(s) at that position. If the event resulted in the paralogs having identical states at the affected position, it was consistent with gene conversion. A benefit of this approach is that it assumes nothing about the process or biases of ordinary mutation, because an ancestral length difference between paralogs can be caused by either an insertion or a deletion. Moreover, only a small proportion of indels identified using this approach were due to either ordinary mutation or sequencing errors (see Text S1).
Fig. 1. A phylogenetic approach for detecting insertions and deletions produced by nonallelic gene conversion. 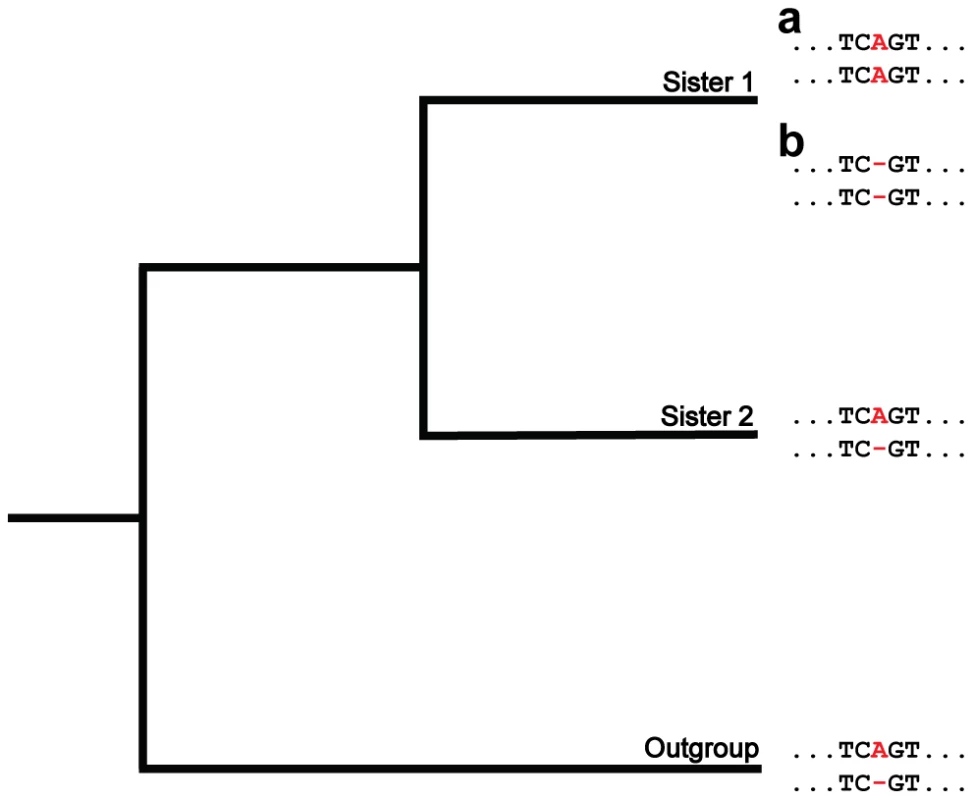
Depicted is a hypothetical multiple alignment for pairs of paralogs in two sisters and an outgroup. The two sequences for each species represent a pair of paralogs, and the position of interest is colored in red. At this position, a length difference (A/−) exists between the paralogs in sister 2 and the outgroup (ancestral state). In the lineage of sister 1, an insertion (a) or deletion (b) of a nucleotide occurs in one paralog. Because these events result in the paralogs having matching states (A/A or −/−), they are consistent with gene conversion. Results/Discussion
Since our approach required that paralogs be present in the genomes of triplets of closely-related species, we chose to study gene conversion in Drosophila and primate lineages, for which whole-genome sequences of multiple close species are available. For Drosophila, we used D. melanogaster and D. simulans as sister species and D. yakuba as an outgroup, and for primates, we used human and chimpanzee as sisters and orangutan as an outgroup. We obtained 338 (199 coding) and 10,149 (1,740 coding) pairs of paralogs that are present in all three species of Drosophila and primates, respectively (Figure 2). Of these, 267 are intra-chromosomal in Drosophila, and 5,997 are intra-chromosomal in primates. Our phylogenetic analysis revealed that 101 Drosophila paralogs and 400 primate paralogs underwent gene conversion during the evolutionary timeframes considered. A general prediction of nonallelic gene conversion is that a pair of paralogs should be more similar in the genome of the species in which they underwent conversion than in the genomes of the other sister or outgroup. As expected, 95 paralogs in Drosophila, and 385 paralogs in primates display this trend.
Fig. 2. Properties of paralogs. 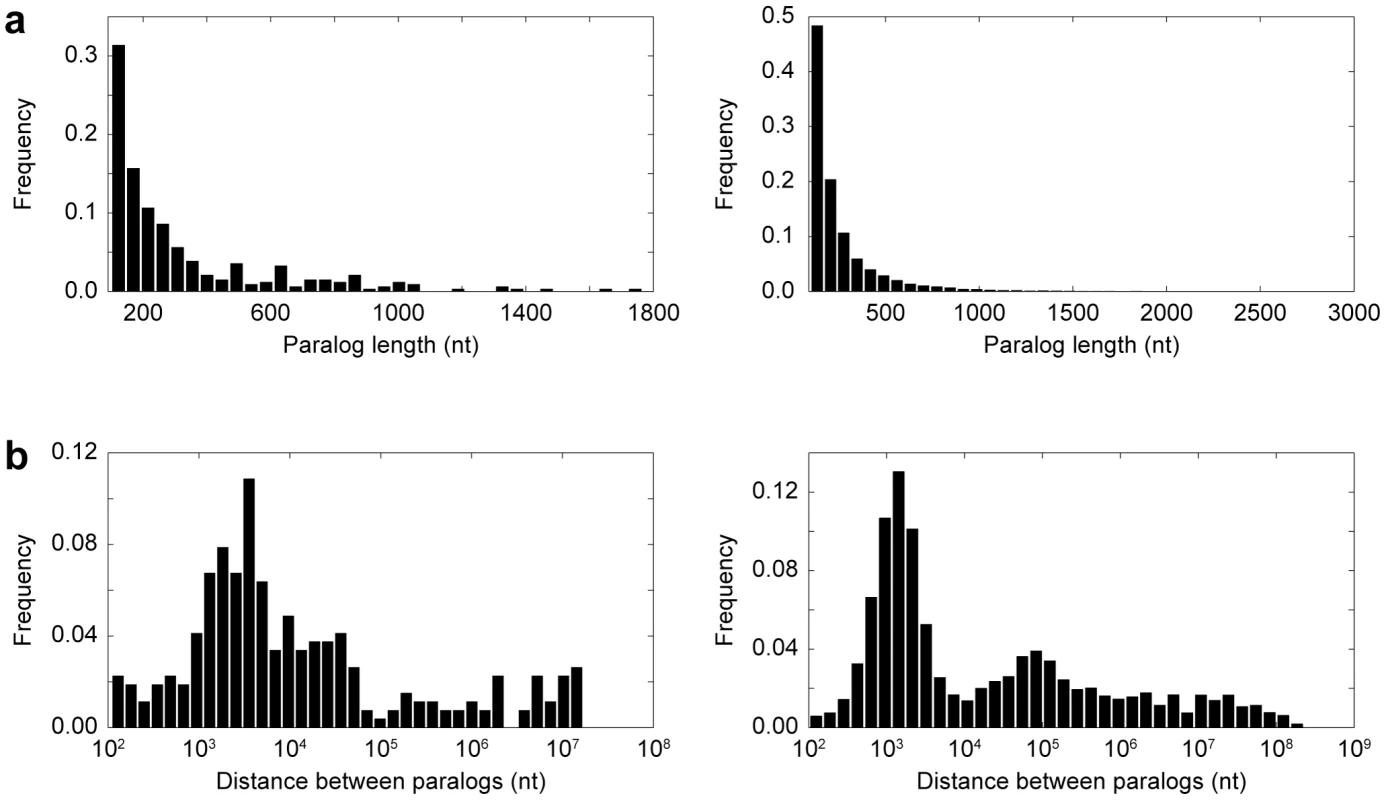
(a) Distribution of paralog sequence lengths in Drosophila (left) and primates (right). (b) Distribution of distances between pairs of paralogs located on the same chromosome in Drosophila (left) and primates (right). Distances are plotted on a log scale. Within our set of paralogs, we identified 179 insertions and 614 deletions consistent with gene conversion in Drosophila, and 132 insertions and 455 deletions consistent with gene conversion in primates (Figure 3a). Thus, there were ∼3.4 times as many deletions as insertions in both lineages, which was highly significant (p<0.0001). In primates, we found that the deletion bias was substantially larger for long than for short indels (Figure 3b). Exclusion of indels that occurred in coding regions, which were rare (45 in Drosophila and 27 in primates), did not alter the deletion bias in either lineage, implying that selection on coding paralogs did not affect the overall deletion/insertion ratios observed.
Fig. 3. Indels consistent with gene conversion. 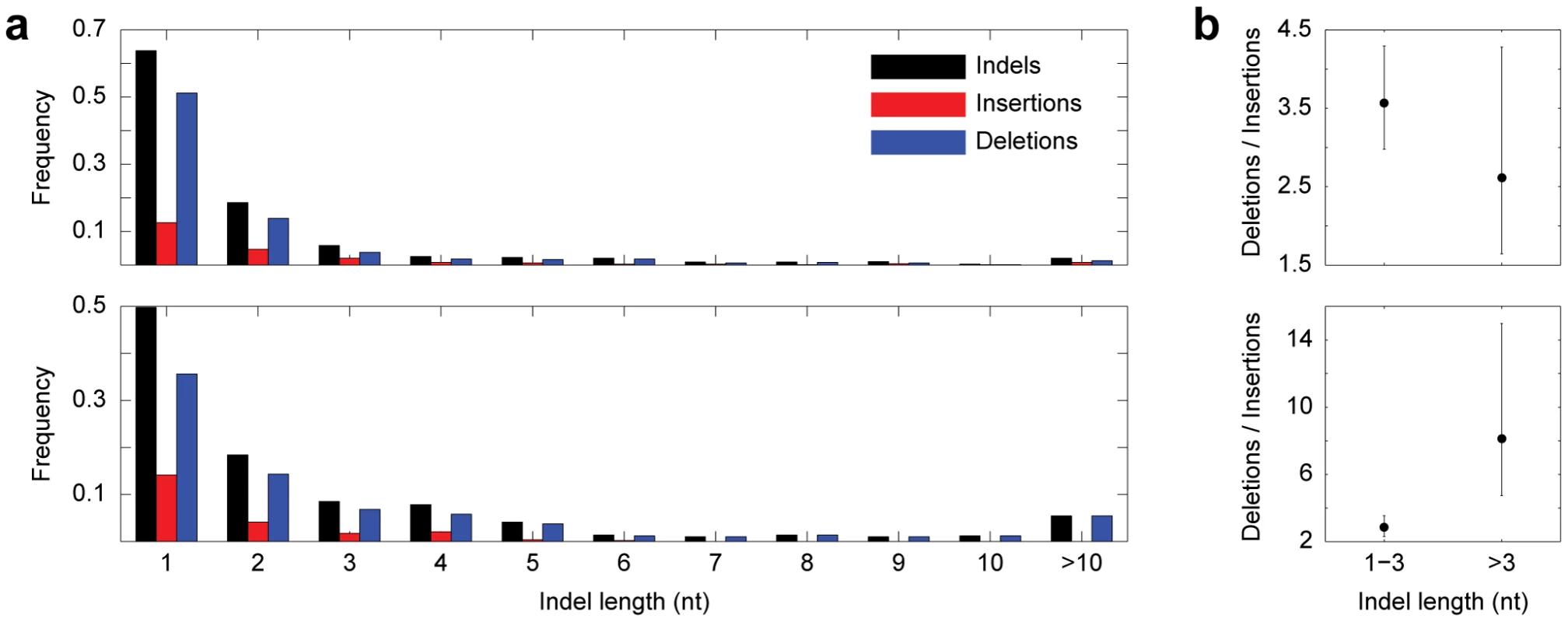
(a) Length distributions of all indels, insertions, and deletions in Drosophila (top) and primates (bottom). (b) Strength of deletion bias as a function of indel length in Drosophila (top) and primates (bottom). Error bars represent confidence limits from binomial sign tests (see Methods). One concern of our parsimony-based approach is homoplasy, which would cause us to misidentify two ordinary mutation-induced indels of the same type, one in sister 2 and one in the outgroup, as one conversion-consistent indel in sister 1 (see Figure 1). However, homoplasy is unlikely in our dataset for several reasons. First, in contrast to nucleotide replacements, identical independent indel mutation events are rare [10], [11]. Though one group did uncover evidence of homoplastic indels [12], their analysis compared orthologs in very distantly-related species, the closest sisters being human and mouse, which have an average synonymous substitution rate, or Ks, of ∼0.77 [13]. In contrast, the Ks between D. melanogaster (D. simulans) and D. yakuba is 0.23 (0.21) [14], and the Ks between human (chimpanzee) and orangutan is 0.03 (0.03) [15]. Thus, there was much less time for multiple independent indel mutations to occur. Second, paralogs in our dataset do not contain any satellite sequences, which are prone to homoplastic mutations [16]–[19], and are minimally repetitive in general (1.69% of Drosophila sequences, and 1.51% of primate sequences). Third, paralogs containing conversion-consistent indels follow the same genomic distribution as the entire set of paralogs (Figure S1), making it unlikely that spatial variation in mutation rate led to the observed patterns. Finally, most conversion events occurred between noncoding paralogs, which are less likely to be under selection for similar function, and also are not as limited as coding paralogs in the types of indels (nucleotide content, size) that can occur.
Even if homoplasy did occur, it would much more likely cause misidentifications of conversion-consistent insertions than deletions, leading to downward biases of deletion/insertion ratios. This is because, for one, homplastic events resembling conversion-consistent deletions require two insertions, which have lower mutation rates than insertions. Additionally, these insertions must be identical in sequence. In the case of single nucleotide insertions, there is a ¼ probability of the second insertion being identical to the first, and this probability rapidly decreases with increasing insertion sequence length.
We next estimated the rate of nonallelic gene conversion in Drosophila and primates. For primates, we performed a simple calculation. There are 28,701 sites at which there was an ancestral length difference between paralogs. Conversion-consistent indels occurred at 587 of these sites, resulting in ∼0.02 indels per site. For Drosophila, a more complex estimate was needed. There were 793 conversion-consistent indels that occurred at 960 possible sites, resulting in 0.83 indels per site. Due to this high proportion, it was necessary to correct for multiple conversion events per site. If we assume that gene conversion is a Poisson process, like ordinary mutation, the mean number of events per site is −ln(1−0.83), or ∼1.8. Because the number of events per site was much smaller in primates than in Drosophila, applying this correction to primate conversion events did not alter the original rate estimate.
Strong sequence similarity of paralogs is associated with high gene conversion rate [20]. To study this phenomenon, we computed Spearman correlation coefficients between paralog similarities and the number of gene conversion indels identified. In Drosophila, similarity was indeed positively correlated with gene conversion rate (ρ = 0.35; p = 2.3×10−11). However, in primates, there was instead a very weak negative relationship between similarity and gene conversion rate (ρ = −0.04; p = 2.45×10−5). Though it is possible that gene conversion rate does not increase with similarity in primates, it is more likely that this result is due to properties of our data. In particular, the primates compared in this study are an order of magnitude more closely related than the Drosophila: the Ks between human and chimpanzee is 0.01 [15], whereas the Ks between D. melanogaster and D. simulans is 0.11 [14]. Thus, similarities between primate paralogs tend to be higher and have a narrower range, making it difficult to assess the relationship between similarity and gene conversion rate in primates. Additionally, our data do not reflect “invisible” gene conversion events, or those that occurred between two identical sequences, which are likely to be more prevalent in primates due to the higher similarities of paralogs. The absence of such cases may have also affected our calculation in Drosophila, producing an underestimate of the correlation between sequence similarity and nonallelic gene conversion rate.
Physical distance between paralogs is also believed to influence gene conversion rate, with paralogs separated by smaller distances hypothesized to undergo faster gene conversion [21]. To study this relationship, we calculated end-to-end distances between pairs of paralogs located on the same chromosomes and computed Spearman coefficients between these distances and numbers of gene conversion indels detected after correcting for divergence between paralogs. In both lineages, there was a very weak positive correlation (ρ = 0.07 for Drosophila, ρ = 0.05 for primates), which was statistically significant only in primates (p = 2.79×10−5). These findings agree with those of McGrath et al. [22], who pointed out that negative correlations between distance and gene conversion rate are likely due to the fact that adjacent paralogs tend to be more similar to each other because of their recent ancestry [23].
Though a higher meiotic recombination rate is associated with elevated rates of allelic gene conversion, the relationship between recombination rate and nonallelic gene conversion is unclear. To investigate the potential relationship between these two parameters, we computed Spearman correlation coefficients between mean recombination rates for pairs of paralogs and numbers of gene conversion indels ascertained. Interestingly, in Drosophila, meiotic recombination rate was negatively correlated with nonallelic gene conversion rate (ρ = −0.22; p = 7.38×10−5). In contrast, we did not detect a correlation between these parameters in primates (ρ = 0.005; p = 0.63).
Next, we calculated the indel mutation rate in Drosophila and primates. To do this, we applied a parsimony-based approach to identify indels produced by ordinary mutation in each lineage. In Drosophila, we observed 202 indels produced by ordinary mutation. Of these indels, 18 were insertions and 184 were deletions, resulting in a deletion bias of ∼10∶1, which is consistent with previous estimates [24]. The target size for such mutations was half the length of all paralogs, which was 104,478 nt. However, as with our conversion analysis, we assumed indel mutations did not occur at the ends of sequences. Subtracting 338 positions, the sequence length along which indels could occur was 104,140 nt, resulting in ∼1.94×10−3 indel mutations per site in Drosophila. In primates, we observed 1,095 indels (533 insertions and 562 deletions) within a total sequence length of 2,448,263 nt, giving a rate of ∼4.47×10−4 indel mutations per site in primates.
Comparison of the rates of indels produced by nonallelic gene conversion and ordinary mutation revealed that nonallelic gene conversion is ∼927.8 times faster in Drosophila and ∼44.7 times faster in primates. This rapid deletion-biased process has a significant effect on genome size evolution. To illustrate this hypothesis, let us consider the life cycle of a length difference mutation within two paralogs. First, ordinary mutation introduces an insertion or deletion in one paralog. Then, deletion-biased gene conversion occurs between the paralogs. If the initial mutation was an insertion, it is removed. Otherwise, the deletion is transmitted to the second paralog, i.e., fixed within the pair of paralogs. In the absence of selection, this process results in the cooperative shrinkage of these paralogous sequence segments.
Cooperative shrinkage of paralogs can be quantified by phylogenetic detection of fixed conversion-induced indels (Figure 4). To perform this analysis, we ascertained all cases for which, ancestrally, two paralogs had identical lengths at a particular site and, in one sister, they acquired matching indels at that position. This condition implies that, in the ancestral lineage of the sister, ordinary mutation produced an indel in one paralog, and that this indel was later copied to the other paralog, or “fixed”, by gene conversion. In Drosophila, we detected 74 fixed insertions, with a total inserted sequence length of 391 nt, and 176 fixed deletions, with a total deleted sequence length of 1,660 nt. In primates, we detected four fixed insertions, with a total inserted sequence length of 4 nt, and 24 fixed deletions, with a total deleted sequence length of 438 nt. Thus, in both lineages, fixed deletions were much longer and more frequent than fixed insertions. Subtracting total insertion lengths from total deletion lengths, we arrived at effective deletion lengths of 1,269 nt in Drosophila and 434 nt in primates. The total sequence length of all paralogs was 208,956 nt in Drosophila and 4,916,824 nt in primates. Therefore, the shrinkage rate of paralogs by gene conversion is ∼0.11 per Ks unit in Drosophila and ∼0.015 per Ks unit in primates. This result implies that, in the absence of selection, these paralogs will exponentially shrink and disappear in ∼138 Ks units, or ∼6,210 million years, in Drosophila and ∼1,021 Ks units, or ∼612,600 million years, in primates.
Fig. 4. A phylogenetic approach for detecting fixed indels. 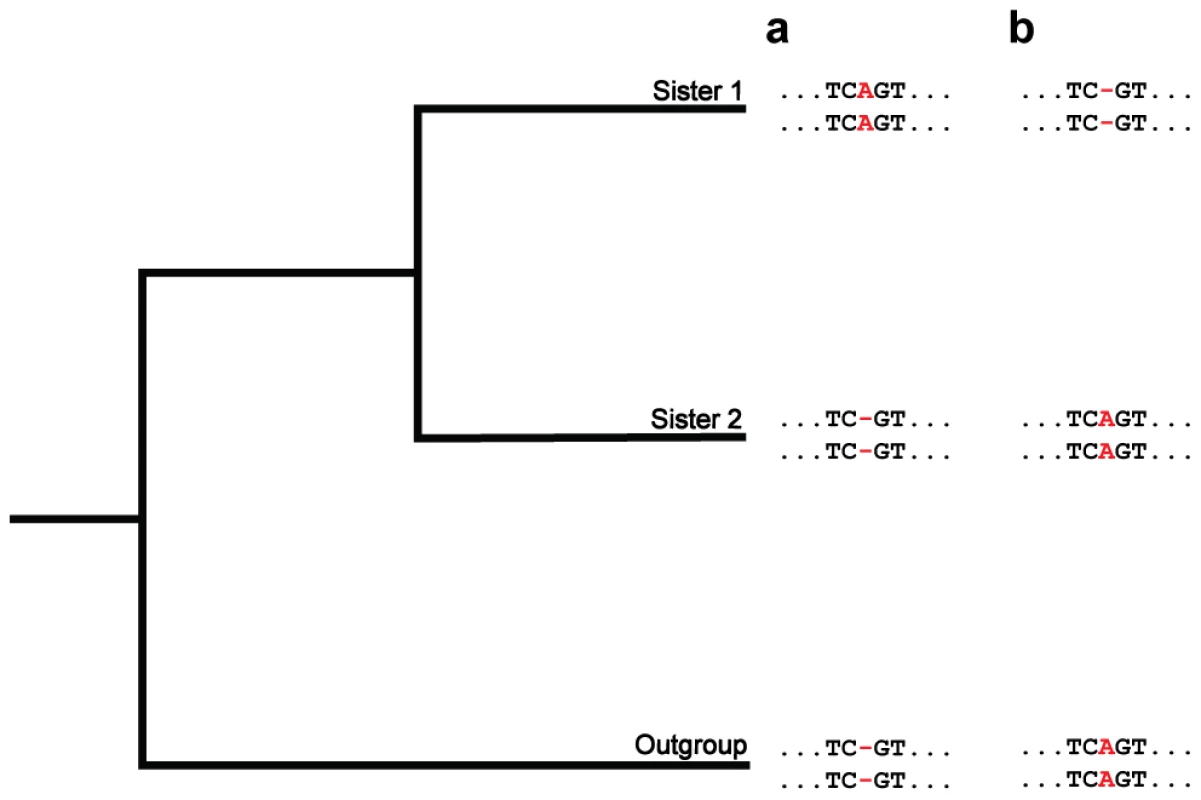
Depicted are hypothetical multiple alignments for pairs of paralogs in two sisters and an outgroup. The two sequences for each species represent a pair of paralogs, and the position of interest is colored in red. (a and b) At this position, both paralogs have identical lengths in sister 2 and the outgroup (ancestral state). In the lineage of sister 1, identical insertions (a) or deletions (b) occur in the paralogs. Each of these situations corresponds to an ordinary mutation producing an indel in one paralog, and this indel subsequently being transferred to the other paralog, or fixed, by gene conversion. Methods
Whole-genome sequences of Drosophila melanogaster, Drosophila simulans, Drosophila yakuba, Homo sapiens (human), Pan troglodytes (chimpanzee), and Pongo pygmaeus (orangutan) were downloaded from the UCSC Genome Bioinformatics site at http://genome.ucsc.edu. We used Mega BLAST [25] (default parameters) and Bridges [26] (KM = 13, FilterDBase = 20, FilterQuery = 20, KS = 12, CoeffMis = 0.01, CoeffGap = 0.05, FlatGap = 10, MaxDist = 50, MinWeight = 100, CoeffMisPost = 0.1, MaxDistPost = 1000) to locate unique pairs of similar sequence segments (both coding and noncoding) in the genomes of D. melanogaster and H. sapiens. To avoid short repeats, we required that each sequence in a pair was greater than 100 nt long. After examining the output from these methods, we set a cutoff of 78% sequence identity between pairs of paralogs. If both paralogs were located on the same chromosome, we required that they were separated by greater than 100 nt to avoid sequencing or genome mapping errors. We used the BLASTN [27] (default parameters) and Mega BLAST (default parameters) algorithms to locate orthologs for each paralog in sister and outgroup species, using conserved synteny of 1,000 nt on either side of each sequence to ensure that orthologs were correctly assigned. Orthologs obtained via this method were verified using multiple species sequence alignments downloaded from the UCSC Genome Bioinformatics website (http://genome.ucsc.edu). To identify repetitive regions within paralogs, we ran RepeatMasker (http://www.repeatmasker.org) with cross_match (http://www.phrap.org) on human paralog sequences with a human-specific repeat library, and on D. melanogaster paralog sequences with a Drosophila-specific repeat library. Pairs of paralogs present in all three species of a lineage were aligned with MUSCLE [28] (default parameters), and alignments, particularly at indel positions, were checked by eye to ensure accuracy. Indels (both conversion-consistent and ordinary mutations) were removed from the analysis if they had different lengths or were located at either end of an alignment. We also excluded cases in which, at a particular position where an indel occurred in one sister, all other orthologous sequences were not identical. Meiotic recombination rates were obtained from the Drosophila melanogaster recombination rate calculator [29] for D. melanogaster and from the HapMap website at http://www.hapmap.org [30] for human. Statistical significance was determined with binomial sign tests for deletion biases, and paired t-tests for Spearman correlation coefficients. For each test, we used α = 0.05 and reported two-tailed probabilities.
Supporting Information
Zdroje
1. KooninEV 2005 Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39 309 338
2. GojoboriTLiWHGraurD 1982 Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol 18 360 369
3. Alvarez-ValinFLamolleaGBernardiG 2002 Isochores, GC3 and mutation biases in the human genome. Gene 300 161 168
4. EcholsNHarrisonPBalasubramanianSLuscombeNMBertoneP 2002 Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res 30 2515 2523
5. PetrovDA 2002 Mutational Equilibrium Model of Genome Size Evolution. Theor Popul Biol 61 531 544
6. MaraisG 2003 Biased gene conversion: implications for genome and sex evolution. Trends Genet 19 330 338
7. ManceraEBourgonRBrozziAHuberWSteinmetzLM 2008 High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454 479 485
8. LiuGLiH 2008 The correlation between recombination rate and dinucleotide bias in Drosophila melanogaster. J Mol Evol 67 358 367
9. BerglundJPollardKSWebsterMT 2009 Hotspots of biased nucleotide substitutions in human genes. PLoS Biol 7 e1000026 doi:10.1371/journal.pbio.1000026
10. RokasAHollandPW 2000 Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15 454 459
11. BaptesteEPhilippeH 2002 The potential value of indels as phylogenetic markers: position of trichomonads as a case study. Mol Biol Evol 19 972 977
12. BelinkyFCohenOHuchonD 2009 Large-scale parsimony analysis of metazoan indels in protein-coding genes. Mol Biol Evol 27 441 451
13. SmithNGCEyre-WalkerA 2003 Human disease genes: patterns and predictions. Gene 318 169 175
14. LazzaroB 2005 Elevated polymorphism and divergence in the class c scavenger receptors of Drosophila melanogaster and D. simulans. Genetics 169 2023 2034
15. ChenFCLiWH 2001 Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet 68 444 456
16. EstoupATailliezCCornuetJMSolignacM 1995 Size homoplasy and mutational processes of interrupted microsatellites in two bee species, Apis mellifera and Bombus terrestris (Apidae). Mol Biol Evol 12 1074 1084
17. GarzaJCFreimerNB 1996 Homoplasy for size at microsatellite loci in humans and chimpanzee. Genome Res 6 211 217
18. AngersBBernatchezL 1997 Complex evolution of a salmonid microsatellite locus and its consequences in inferring allelic divergence from size information. Mol Biol Evol 14 230 238
19. van OppenMJHRicoCTurnerGFHewittGM 2000 Extensive homoplasy, nonstepwise mutations, and shared ancestral polymorphism at a complex microsatellite locus in Lake Malawi cichlids. Mol Biol Evol 17 489 498
20. LukacsovichTWaldmanAS 1999 Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics 151 1559 1568
21. HastingsPJ 2010 Mechanisms of ectopic gene conversion. Gene 1 427 439
22. McGrathCLCasolaCHahnMW 2009 Minimal effect of ectopic gene conversion among recent duplicates in four mammalian genomes. Genetics 182 615 622
23. KatjuVLynchM 2003 The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165 1793 1803
24. PetrovDAHartlDL 1998 High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol 15 293 302
25. ZhangZSchwartzSWagnerLMillerW 2000 A greedy algorithm for aligning DNA sequences. J Comput Biol 7 203 214
26. KondrashovASAssisR 2010 Bridges: a tool for identifying local similarities in long sequences. Bioinformatics 26 2055 2056
27. AltschulSFGishWMillerWMyersEWLipmanDJ 1990 Basic local alignment search tool. J Mol Biol 215 403 410
28. EdgarRC 2004 MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32 1792 1797
29. Fiston-LavierASSinghNDLipatovMPetrovDA 2010 Drosophila melanogaster recombination rate calculator. Gene 463 18 20
30. The International HapMap Consortium 2003 The international HapMap project. Nature 426 789 796
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 2- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Upsetting the Dogma: Germline Selection in Human Males
- A Strong Deletion Bias in Nonallelic Gene Conversion
- Positive Selection for New Disease Mutations in the Human Germline: Evidence from the Heritable Cancer Syndrome Multiple Endocrine Neoplasia Type 2B
- Genome-Wide Association Study in East Asians Identifies Novel Susceptibility Loci for Breast Cancer
- Mixed Effects Modeling of Proliferation Rates in Cell-Based Models: Consequence for Pharmacogenomics and Cancer
- Reduction of NADPH-Oxidase Activity Ameliorates the Cardiovascular Phenotype in a Mouse Model of Williams-Beuren Syndrome
- Genome-Wide Association Study Identifies Chromosome 10q24.32 Variants Associated with Arsenic Metabolism and Toxicity Phenotypes in Bangladesh
- Structural Basis of Transcriptional Gene Silencing Mediated by MOM1
- Genomic Restructuring in the Tasmanian Devil Facial Tumour: Chromosome Painting and Gene Mapping Provide Clues to Evolution of a Transmissible Tumour
- Genome-Wide Association Study Identifies Novel Loci Associated with Circulating Phospho- and Sphingolipid Concentrations
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- The Origin and Nature of Tightly Clustered Deletions in Precursor B-Cell Acute Lymphoblastic Leukemia Support a Model of Multiclonal Evolution
- Ultrafast Evolution and Loss of CRISPRs Following a Host Shift in a Novel Wildlife Pathogen,
- Phosphorylation of Chromosome Core Components May Serve as Axis Marks for the Status of Chromosomal Events during Mammalian Meiosis
- Psoriasis Patients Are Enriched for Genetic Variants That Protect against HIV-1 Disease
- A Pathogenic Mechanism in Huntington's Disease Involves Small CAG-Repeated RNAs with Neurotoxic Activity
- The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Developmental Transcriptional Networks Are Required to Maintain Neuronal Subtype Identity in the Mature Nervous System
- Down-Regulating Sphingolipid Synthesis Increases Yeast Lifespan
- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Loss of Tgif Function Causes Holoprosencephaly by Disrupting the Shh Signaling Pathway
- Sequestration of Highly Expressed mRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability
- Discovery of a Modified Tetrapolar Sexual Cycle in and the Evolution of in the Species Complex
- The Role of Glypicans in Wnt Inhibitory Factor-1 Activity and the Structural Basis of Wif1's Effects on Wnt and Hedgehog Signaling
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
- A Regulatory Network for Coordinated Flower Maturation
- Coexpression Network Analysis in Abdominal and Gluteal Adipose Tissue Reveals Regulatory Genetic Loci for Metabolic Syndrome and Related Phenotypes
- Diced Triplets Expose Neurons to RISC
- The Williams-Beuren Syndrome—A Window into Genetic Variants Leading to the Development of Cardiovascular Disease
- The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals
- Systematic Detection of Epistatic Interactions Based on Allele Pair Frequencies
- Familial Identification: Population Structure and Relationship Distinguishability
- Raf1 Is a DCAF for the Rik1 DDB1-Like Protein and Has Separable Roles in siRNA Generation and Chromatin Modification
- Loss of Function of the Cik1/Kar3 Motor Complex Results in Chromosomes with Syntelic Attachment That Are Sensed by the Tension Checkpoint
- Computational Prediction and Molecular Characterization of an Oomycete Effector and the Cognate Resistance Gene
- The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women's Cancer
- GTPase Activity and Neuronal Toxicity of Parkinson's Disease–Associated LRRK2 Is Regulated by ArfGAP1
- Evaluation of the Role of Functional Constraints on the Integrity of an Ultraconserved Region in the Genus
- Neurophysiological Defects and Neuronal Gene Deregulation in Mutants
- Genetic and Functional Analyses of Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders
- Negative Supercoiling Creates Single-Stranded Patches of DNA That Are Substrates for AID–Mediated Mutagenesis
- Rewiring of PDZ Domain-Ligand Interaction Network Contributed to Eukaryotic Evolution
- The Eph Receptor Activates NCK and N-WASP, and Inhibits Ena/VASP to Regulate Growth Cone Dynamics during Axon Guidance
- Repression of a Potassium Channel by Nuclear Hormone Receptor and TGF-β Signaling Modulates Insulin Signaling in
- The Retrohoming of Linear Group II Intron RNAs in Occurs by Both DNA Ligase 4–Dependent and –Independent Mechanisms
- Cell Lineage Analysis of the Mammalian Female Germline
- Association of a Functional Variant in the Wnt Co-Receptor with Early Onset Ileal Crohn's Disease
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Gene Expression and Stress Response Mediated by the Epigenetic Regulation of a Transposable Element Small RNA
- Contrasting Properties of Gene-Specific Regulatory, Coding, and Copy Number Mutations in : Frequency, Effects, and Dominance
- Homeobox Genes Critically Regulate Embryo Implantation by Controlling Paracrine Signaling between Uterine Stroma and Epithelium
- Nondisjunction of a Single Chromosome Leads to Breakage and Activation of DNA Damage Checkpoint in G2
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



