-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation
In plants and fungi, small RNAs silence gene expression in the nucleus by establishing repressive chromatin states. The role of endogenous small RNAs in metazoan nuclei is largely unknown. Here we show that endogenous small interfering RNAs (endo-siRNAs) direct Histone H3 Lysine 9 methylation (H3K9me) in Caenorhabditis elegans. In addition, we report the identification and characterization of nuclear RNAi defective (nrde)-1 and nrde-4. Endo-siRNA–driven H3K9me requires the nuclear RNAi pathway including the Argonaute (Ago) NRDE-3, the conserved nuclear RNAi factor NRDE-2, as well as NRDE-1 and NRDE-4. Small RNAs direct NRDE-1 to associate with the pre-mRNA and chromatin of genes, which have been targeted by RNAi. NRDE-3 and NRDE-2 are required for the association of NRDE-1 with pre-mRNA and chromatin. NRDE-4 is required for NRDE-1/chromatin association, but not NRDE-1/pre-mRNA association. These data establish that NRDE-1 is a novel pre-mRNA and chromatin-associating factor that links small RNAs to H3K9 methylation. In addition, these results demonstrate that endo-siRNAs direct chromatin modifications via the Nrde pathway in C. elegans.
Published in the journal: . PLoS Genet 7(8): e32767. doi:10.1371/journal.pgen.1002249
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002249Summary
In plants and fungi, small RNAs silence gene expression in the nucleus by establishing repressive chromatin states. The role of endogenous small RNAs in metazoan nuclei is largely unknown. Here we show that endogenous small interfering RNAs (endo-siRNAs) direct Histone H3 Lysine 9 methylation (H3K9me) in Caenorhabditis elegans. In addition, we report the identification and characterization of nuclear RNAi defective (nrde)-1 and nrde-4. Endo-siRNA–driven H3K9me requires the nuclear RNAi pathway including the Argonaute (Ago) NRDE-3, the conserved nuclear RNAi factor NRDE-2, as well as NRDE-1 and NRDE-4. Small RNAs direct NRDE-1 to associate with the pre-mRNA and chromatin of genes, which have been targeted by RNAi. NRDE-3 and NRDE-2 are required for the association of NRDE-1 with pre-mRNA and chromatin. NRDE-4 is required for NRDE-1/chromatin association, but not NRDE-1/pre-mRNA association. These data establish that NRDE-1 is a novel pre-mRNA and chromatin-associating factor that links small RNAs to H3K9 methylation. In addition, these results demonstrate that endo-siRNAs direct chromatin modifications via the Nrde pathway in C. elegans.
Introduction
Small regulatory RNAs can silence gene expression in the nucleus by establishing repressive chromatin states. This process, termed Transcriptional Gene Silencing (TGS), was first observed in plants, where small RNAs direct DNA methylation and histone modifications (reviewed in [1]). In addition, the fission yeast, Schizosaccharomyces pombe has been an important model in defining the role of small RNAs in heterochromatin formation. In S. pombe, small RNAs direct the formation of heterochromatin primarily at repetitive DNA elements surrounding centromeres [2], [3]. At these repetitive elements, nascent RNAs, transcribed by RNA Polymerase II (RNAP II), serve as platforms for the assembly of RNAi machinery. For instance, the RNA Induced Transcriptional Silencing (RITS) complex, composed of the Argonaute Ago1, the chromodomain protein Chp1, and the glycine and tryptophan (GW)-motif-containing protein Tas3, is guided to nascent transcripts by Argonaute and centromeric siRNAs [4]. The RITS complex recruits chromatin-modifying machinery, such as the histone methyltransferase Clr4, to genomic sites of nuclear RNAi [5], [6]. Clr4 catalyzes the methylation of Histone H3 on Lysine 9 (H3K9me) [7]. H3K9me is a conserved molecular mark of heterochromatin [8]. Thus, in plants and S. pombe, small RNAs play a central role in regulating chromatin dynamics. The role of TGS and heterochromatin formation in metazoan silencing processes is less clear [3].
Experimentally provided small RNAs can elicit transcriptional silencing and induce heterochromatic marks in metazoans. In mammalian cells, experimentally provided siRNAs directed against promoter regions can lead to transcriptional silencing and induce heterochromatic marks [9]–[12]. Paradoxically, experimentally provided small RNAs can also enhance transcription and decrease H3K9me marks [13], [14]. In C. elegans, experimentally provided siRNAs are bound by the Ago NRDE-3 in the cytoplasm, and escorted into the nucleus [15]. NRDE-3/siRNA ribonucleoprotein complexes bind nascent transcripts and recruit the conserved nuclear RNAi factor NRDE-2. The Nrde pathway inhibits RNA Polymerase (RNAP) II during the elongation phase of transcription, and directs the deposition of H3K9me marks at genomic sites that exhibit homology to experimentally introduced siRNAs [16].
How and if endogenously expressed small regulatory RNAs silence gene expression in metazoan nuclei is unclear. Dicer deficient mouse embryonic stem cells express high levels of centromeric repeat RNAs and exhibit altered heterochromatic marks at centromeres [17]. In Drosophila, heterochromatic marks, including H3K9me and HP1, are mislocalized in flies lacking components of the RNAi machinery such as Piwi, Aubergine, and Homeless [18]. In addition, the Drosophila Ago-like protein PIWI binds small RNAs, termed piRNAs, and associates with chromatin [19]. Loss of piwi has variable effects on chromatin states at genomic sites homologous to piRNAs [20]–[24]. Finally, in C. elegans, animals lacking two RNAi-related factors: the RNA-dependent RNA Polymerase EGO-1, or the Ago CSR-1, exhibit large-scale changes in chromosomal H3K9me patterns during germline development [25], [26]. Thus, endogenous small regulatory RNAs have been implicated in chromatin regulation in metazoans. However, a direct link has yet to be established, and the molecular mechanisms by which this might occur are unknown.
Here we show that the endogenous small RNAs, termed endo-siRNAs, direct H3K9me marks at discrete genomic loci in C. elegans. Small RNA-directed H3K9 methylation requires the Nrde pathway and results in the inhibition of transcription from these loci. In addition, we identify two novel nuclear RNAi factors termed NRDE-1 and NRDE-4, and show that these factors are required for small RNA-directed H3K9 methylation. Finally, we show that small RNAs direct NRDE-1 to associate with pre-mRNA and chromatin of genes, which have been targeted by RNAi. Thus, the Nrde pathway links endogenously expressed small regulatory RNAs to the regulation of transcription and chromatin dynamics in C. elegans.
Results
A genetic screen identifies novel nrde genes
We previously reported a forward genetic screen that identified two genes (termed nrde-2 and nrde-3) required for nuclear RNAi [15], [16]. The mechanism(s) by which NRDE-2/3 silence nascent transcripts and inhibit RNAP II transcription are unknown. To understand this mechanism we continued screening for nuclear RNAi factors. >80% of the nrde alleles identified in our original genetic screen were alleles of nrde-3 (Table S1, [15]). To maximize our chances of identifying novel nuclear RNAi factors, we performed our modified screen in animals harboring ectopic copies of nrde-3 (nrde-3::gfp), which was integrated into the genome on chromosome V (Figure S1). eri-1 encodes an exonuclease that negatively regulates RNAi [27]. Our original screen was conducted in an eri-1(−) genetic background. Our modified screen was conducted in eri-1(+) animals (Figure S1). Our modified screen identified twenty-three alleles of nrde-2, nineteen alleles of nrde-1, nine alleles of the RNA-dependent RNA Polymerase (RdRP) rrf-1, four alleles of nrde-4, and one additional nrde allele, which complements the known nrde genes, but has not yet been assigned a nrde gene designation (Figure 1a, and Table S1). Here we report the identification and characterization of nrde-1 and nrde-4.
Fig. 1. nrde-1 encodes a nuclear-localized protein that is required for nuclear RNAi. 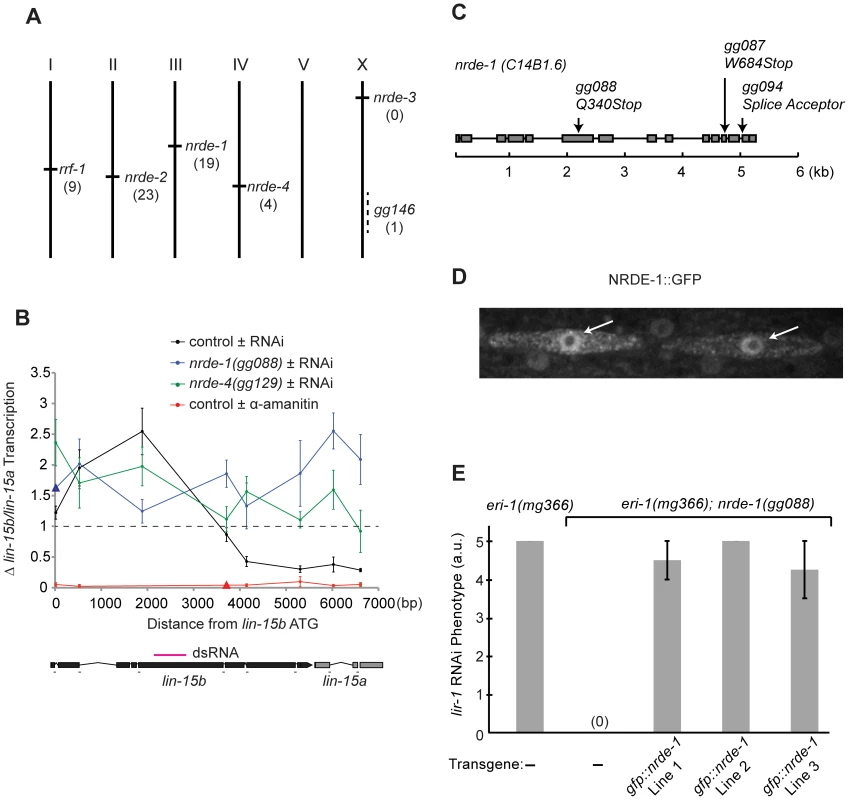
(A) Genetic map positions of genes identified in genetic screen. Number of alleles identified in screen are indicated. (B) RNAi-driven transcriptional inhibition requires nrde-1 and nrde-4. Nuclear Run On (NRO) analysis of transcription from the lin-15b/a gene from nuclei of animals exposed to +/− lin-15b dsRNA or +/− α-amanitin. Data are expressed as a ratio +/− lin-15b dsRNA (normalized to transcription detected from eft-3 gene) or +/− α-amanitin (5 µg/ml). Dotted line indicates a ratio of 1; i.e. no change. The genetic background for this experiment was eri-1(mg366). Control +/− lin-15b dsRNA (n = 3–8, +/− s.e.m.), nrde-1(gg088) (n = 4–6, +/− s.e.m., data point represented by triangle n = 1), nrde-4(gg129)(n = 5, +/− s.e.m.), control +/− α-amanitin (n = 3 +/− s.e.m., data point represented by triangle n = 1). Δ = fold change. Below: diagram of lin-15b/lin-15a gene structure indicating location of primers and trigger dsRNA (magenta). (C) Predicted nrde-1 gene structure. Arrows indicate mutant alleles. (D) NRDE-1 localizes to the nucleus. Fluorescence microscopy of two seam cells in a L4 larval animal expressing gfp::nrde-1. Arrows indicate nuclei. (E) gfp::nrde-1 fusion gene rescues nrde-1 mutant phenotype. Animals of the indicated genotypes were exposed to lir-1 dsRNA. A score of 5 indicates all animals died during larval development and a score of 0 indicates animals did not exhibit any developmental defects. Plates were scored blind and in triplicate for lir-1 RNAi-mediated lethality (a.u. arbitrary units). nrde-1 is a component of the nuclear RNAi pathway
We first focused our attention on characterizing the role of nrde-1 in nuclear RNAi. Three lines of evidence indicate that nrde-1 functions with nrde-2 and nrde-3 to silence nuclear-localized RNAs during nuclear RNAi. First, NRDE-1, like NRDE-2/3, is required for RNAi-based silencing of nuclear-localized RNAs. For instance, the lir-1 and lin-26 genes are expressed in an operon; these genes are co-transcribed as a polycistronic pre-mRNA, which is spliced into distinct mRNAs in the nucleus before export to the cytoplasm [28], [29]. lir-1(−) mutant animals are viable, whereas lin-26(−) mutant animals exhibit a lethal phenotype [30]. RNAi targeting lir-1 induces a lethal phenotype, indicating that lir-1 RNAi silences the nuclear-localized lir-1/lin-26 RNA [15], [30]. nrde-2(−) and nrde-3(−) animals are viable following lir-1 RNAi, indicating that NRDE-2 and NRDE-3 are required for lir-1-mediated silencing of the lir-1/lin-26 RNA [15], [16]. nrde-1 mutant animals were also viable when exposed to lir-1 RNAi, indicating that, like NRDE-2 and NRDE-3, NRDE-1 is required for silencing of the lir-1/lin-26 RNA (Table 1). Similarly, NRDE-1/2/3 are required to silence the nuclear-localized lin-15b/lin-15a RNA. lin-15b and lin-15a genes are encoded in an operon. Mutations in lin-15b or lin-15a alone produce no obvious phenotype, but animals harboring mutations in both lin-15b and lin-15a exhibit a Multi-vulva (Muv) phenotype [31]. RNAi targeting lin-15b induces a Muv phenotype, indicating that lin-15b RNAi silences the nuclear-localized lin-15b/lin-15a RNA [15], [16]. nrde-1/2/3 mutant animals do not exhibit a Muv phenotype in response to lin-15b RNAi, indicating that NRDE-1/2/3 are required for silencing the lin-15b/lin-15a RNA (Table 1). Second, nuclear-localized siRNAs direct a NRDE-2/3 dependent inhibition of RNAP II during the elongation phase of transcription [16]. For instance, lin-15b RNAi inhibits RNAP II transcription 3′ to the site of lin-15b RNAi (Figure 1b). RNAi-mediated inhibition of RNAP II transcription is dependent upon NRDE-2 and NRDE-3 [15], [16]. nrde-1 was also required to link small RNAs to RNAP II inhibition; in nrde-1 mutant animals, lin-15b RNAi did not result in transcription inhibition (Figure 1b). Thus, like nrde-2/3, a wild-type copy of the nrde-1 gene is required for RNAi to inhibit transcription elongation. Third, we conducted a genetic analysis using double mutant combinations of the Nrde factors. This analysis indicated that nrde-1 functions in a genetic pathway with nrde-2 and nrde-3 (Figure S2). Taken together, these data argue that nrde-1 is a component of the Nrde silencing pathway.
Tab. 1. nrde-1 is required for silencing nuclear localized RNAs. 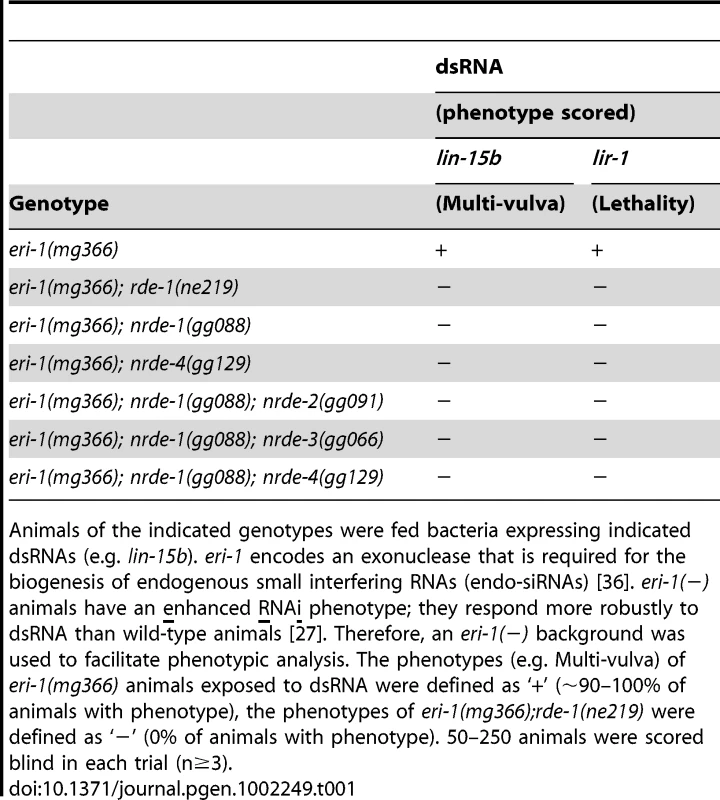
Animals of the indicated genotypes were fed bacteria expressing indicated dsRNAs (e.g. lin-15b). eri-1 encodes an exonuclease that is required for the biogenesis of endogenous small interfering RNAs (endo-siRNAs) [36]. eri-1(−) animals have an enhanced RNAi phenotype; they respond more robustly to dsRNA than wild-type animals [27]. Therefore, an eri-1(−) background was used to facilitate phenotypic analysis. The phenotypes (e.g. Multi-vulva) of eri-1(mg366) animals exposed to dsRNA were defined as ‘+’ (∼90–100% of animals with phenotype), the phenotypes of eri-1(mg366);rde-1(ne219) were defined as ‘−’ (0% of animals with phenotype). 50–250 animals were scored blind in each trial (n≥3). NRDE-1 is a nuclear-localized protein
To determine the molecular identity of nrde-1, we used a single nucleotide polymorphism (SNP)-based mapping approach [32]. We mapped nrde-1 to a 0.86cM interval on Chromosome III that contained 42 genes. The open reading frame (ORF) c14b1.6 lies within this mapping interval. Sequencing of c14b1.6 from three independent nrde-1 alleles revealed three mutations in c14b1.6 (Figure 1c). Two of these alleles encode premature stop codons, and therefore likely reveal the null phenotype of nrde-1. Expression of a wild-type copy of c14b1.6 was sufficient to rescue the Nrde phenotype associated with nrde-1 (see below). We conclude that c14b1.6 corresponds to nrde-1. Analysis of nrde-1 expressed sequence tags (ESTs) indicated that nrde-1 encodes a protein containing 793 amino acids [33]. Database searches revealed that nrde-1 is conserved in other nematode species, but these searches failed to detect any obvious orthologues of nrde-1 outside nematodes. In addition, these database searches did not identify any obvious protein domains within NRDE-1.
We assessed the sub-cellular distribution of NRDE-1. We constructed a NRDE-1 and Green Fluorescent Protein fusion protein (NRDE-1::GFP), which encodes GFP 5′ to a full length copy of nrde-1. We observed fluorescence in nuclei of NRDE-1::GFP expressing animals (Figure 1d). NRDE-1::GFP rescued Nrde phenotypes associated with nrde-1(−) animals (Figure 1e), suggesting that the NRDE-1::GFP expression pattern reflects the expression pattern of endogenous NRDE-1. We conclude that NRDE-1 is a nuclear localized protein.
NRDE-1 functions downstream of NRDE-3
The Ago protein NRDE-3 binds siRNAs in the cytoplasm and transports these siRNAs to the nucleus to facilitate nuclear RNAi [15]. NRDE-3 can bind small RNAs generated from exogenously provided dsRNAs, which are termed exogenous (exo) siRNAs. NRDE-3 also associates with endogenously expressed small RNAs termed endo-siRNAs [15]. NRDE-3 shuttles siRNAs from the cytoplasm to the nucleus; NRDE-3 localizes to the nucleus when bound to either endo or exo-siRNAs, and localizes to the cytoplasm in the absence of these siRNAs [15]. We asked if NRDE-1 was required for NRDE-3/siRNA shuttling. NRDE-3 retained the ability to bind endo-siRNAs in nrde-1(−) animals, indicating that nrde-1 is not required for loading NRDE-3 with siRNAs (Figure 2a). In addition, NRDE-3 remained localized in the nucleus in nrde-1(−) animals, indicating that NRDE-1 activity was not required for NRDE-3 shuttling (Figure 2b). These data suggest that NRDE-1 functions downstream of NRDE-3 siRNAs transport. Following exposure to dsRNA, NRDE-3 associates with un-spliced RNAs (pre-mRNA) that exhibit sequence homology to the trigger dsRNA. The association of NRDE-3 with pre-mRNA is dependent upon the ability of NRDE-3 to localize to the nucleus, the ability of NRDE-3 to bind siRNAs, and is restricted to those pre-mRNAs that have been targeted by RNAi [15]. Thus, siRNAs direct NRDE-3 to associate with pre-mRNAs. To test the idea that NRDE-1 functions downstream of NRDE-3 shuttling, we asked if NRDE-1 was required for the association of NRDE-3 with pre-mRNA in response to RNAi. We performed NRDE-3 RNA Immuno-Precipitation (RIP) and found that, in response to lin-15b RNAi, NRDE-3 retained the ability to bind lin-15b pre-mRNA in nrde-1(−) animals, indicating that NRDE-1 functions downstream of NRDE-3/pre-mRNA association during nuclear silencing events (Figure 2c).
Fig. 2. NRDE-1 is recruited by NRDE-2/-3 to pre-mRNAs that have been targeted by RNAi. 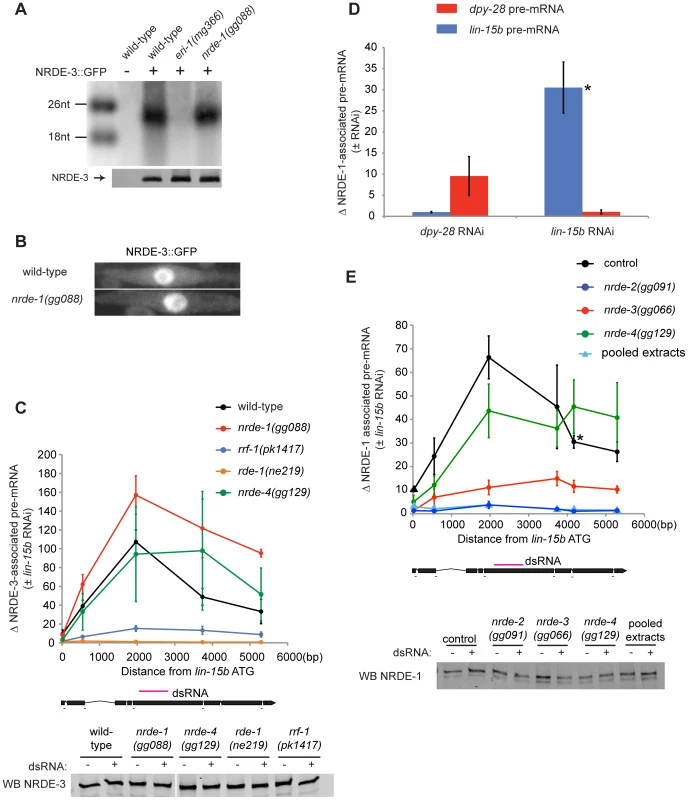
(A) NRDE-3 retains the ability to bind siRNAs in nrde-1(−) animals. FLAG::NRDE-3 co-precipitating RNAs were radiolabeled with 32P and analyzed by polyacrylamide gel electrophoresis. (B) NRDE-3 localizes to the nucleus in nrde-1(−) animals. Fluorescence microscopy of NRDE-3::GFP in seam cells from ∼L3 animals. (C) The recruitment of NRDE-3 to pre-mRNAs by RNAi is unaffected in nrde-1(−) animals. FLAG::NRDE-3 co-precipitating RNAs were converted to cDNA and quantified by qRT-PCR using primers that span exon-intron junctions. Throughout the remainder of this manuscript pre-mRNA levels are measured using exon-intron primer pairs. Data are expressed as a ratio of NRDE-3 precipitating pre-mRNA with or without lin-15b RNAi. For wild-type (n = 7,+/− s.e.m.), nrde-1(−) (n = 5, +/− s.e.m.), rrf-1(−) (n = 5, +/− s.e.m.), nrde-4(−) (n = 5, +/− s.e.m.), rde-1(−) (n = 3, +/− s.e.m.). Below, western blot detecting FLAG::NRDE-3 verified similar levels of NRDE-3 were Immuno-Precipitated (IP'ed) from each strain and for each condition. (D) NRDE-1 associates with pre-mRNAs that have been targeted by RNAi. NRDE-1 co-precipitating RNAs were converted to cDNA and quantified by qRT-PCR. Data are expressed as a ratio of NRDE-1 co-precipitating pre-mRNAs with or without indicated RNAi. Samples exposed to dpy-28 RNAi or lin-15b RNAi were probed with primers targeting either dpy-28 pre-mRNA or lin-15b pre-mRNA. For dpy-28 RNAi (n = 3, +/− s.e.m.), lin-15b RNAi (n = 5, +/− s.e.m.). For lin-15b RNAi, one data point is also shown in panel E and is marked with * in both panels. (E) NRDE-1 association with pre-mRNAs requires NRDE-2 and NRDE-3. FLAG::NRDE-1 co-precipitating pre-mRNAs were converted to cDNA and quantified by qRT-PCR. Data are expressed as a ratio of co-precipitating lin-15b pre-mRNA with or without lin-15b RNAi. Control (n = 5, +/− s.e.m., data point represented by triangle n = 1), nrde-2(−) (n = 3, +/− s.e.m.), nrde-3(−) (n = 6, +/− s.e.m.), nrde-4(−) (n = 5, +/− s.e.m.), n = 1 for pooled extracts. This experiment was performed in an nrde-1(gg088) background. Below, western blot of FLAG::NRDE-1 verified similar amounts of NRDE-1 were IP'ed. Our genetic screen identified nine alleles of the gene rrf-1 (Figure 1a, Table S1). rrf-1 encodes one of four C. elegans RNA-dependent RNA Polymerases (RdRPs) [34]. We sought to position rrf-1 in the nuclear RNAi pathway. In animals lacking RRF-1, NRDE-3 binds fewer small RNAs, suggesting that RRF-1 may generate the small RNAs bound by NRDE-3 [15]. Consistent with this idea, in rrf-1(−) animals, NRDE-3/lin-15b pre-mRNA association was reduced relative to rrf-1(+) animals, indicating that RRF-1 acts upstream of NRDE-3/pre-mRNA association during nuclear silencing (Figure 2c). Taken together, these data indicate that our genetic screen is identifying components of the Nrde pathway that function both upstream and downstream of NRDE-3-mediated siRNA transport.
NRDE-1 is recruited to pre-mRNAs in response to RNAi
We asked if NRDE-1 was recruited to pre-mRNA following RNAi. We performed NRDE-1 RNA Immuno-Precipitation (RIP) experiments in animals exposed to lin-15b dsRNA. lin-15b RNAi induced a ∼30–70× enrichment in un-spliced lin-15b RNA that co-precipitated with FLAG::NRDE-1 (Figure 2d, 2e). The dpy-28 gene encodes a subunit of the C. elegans dosage compensation complex [35]. We tested if dpy-28 dsRNA would induce NRDE-1-dpy-28 pre-mRNA association. Following dpy-28 RNAi, NRDE-1 associated with dpy-28 pre-mRNA (Figure 2d). Finally, dpy-28 or lin-15b RNAi did not result in enrichment of NRDE-1 with lin-15b or dpy-28 pre-mRNA, respectively, indicating that the association of NRDE-1 with pre-mRNA (induced by RNAi) is sequence specific (Figure 2d). We were concerned that NRDE-1 might associate with pre-mRNA targets, in vitro, during sample preparation. To address this issue we pooled extracts from animals exposed to lin-15b dsRNA, and extracts from NRDE-1::GFP expressing animals not exposed to lin-15b, dsRNA and failed to detect an association of NRDE-1 with lin-15b pre-mRNA, indicating that NRDE-1/pre-mRNA interactions likely occurs in vivo (Figure 2e). Taken together, these data show that NRDE-1 associates with pre-mRNAs that have been targeted by RNAi.
NRDE-1 co-precipitating pre-mRNA was enriched for RNA sequences encoded at, or near, the site of RNAi - relative to sequences encoded 5′ or 3′ to the site of RNAi (Figure 2e). We have previously shown that NRDE factors fail to associate with pre-mRNA sequences encoded 3′ to the site of RNAi due to RNAi-mediated inhibition of transcription elongation [16]. We investigated the apparent lack of pre-mRNA sequences encoded 5′ to the site of RNAi and found that, while the NRDE factors fail to associate with un-spliced RNA 5′ to the site of RNAi, the Nrdes do associate with spliced RNA 5′ to the site of RNAi (Figure S3). Splicing is thought to occur co-transcriptionally [29]. Therefore, the apparent lack of NRDE-1/pre-mRNA association 5′ to sites of RNAi may be due to co-transcriptional splicing of nascent transcripts.
We investigated the genetic requirements of NRDE-1/pre-mRNA association. In nrde-2(−) animals, RNAi failed to induce an association of NRDE-1 with pre-mRNA (Figure 2e). In addition, ∼10× less lin-15b pre-mRNA co-precipitated with NRDE-1 in nrde-3(−) animals than in nrde-3(+) animals (Figure 2e). We conclude that the recruitment of NRDE-1 to pre-mRNAs by small RNAs requires NRDE-2 and is largely dependent upon NRDE-3 (see discussion).
NRDE-1 promotes RNAi-directed Histone 3 Lysine 9 methylation
In plants and S. pombe small RNAs direct the methylation of Histone 3 Lysine 9 (H3K9me). Histone methylation results from small RNA-mediated recruitment of histone methyltransferase enzymes to genomic sites exhibiting sequence homology to small RNAs [5]. RNAi also directs H3K9 methylation in C. elegans [16]. nrde-2 is required for RNAi-mediated H3K9 methylation in C. elegans [16]. The mechanism by which the C. elegans Nrde pathway mediates H3K9 methylation is unknown. We conducted H3K9me Chromatin Immuno Precipitation (ChIP) to determine if NRDE-1 was required to link small RNAs to H3K9 methylation. lin-15b RNAi induced a ∼30× increase in H3K9me marks at the lin-15 locus (Figure 3). In nrde-1(−) animals, however, lin-15b RNAi had no effect on the methylation status of chromatin at the lin-15b gene (Figure 3). We conclude that NRDE-1 is required to link small RNAs to H3K9 methylation at a genomic site that has been targeted by RNAi.
Fig. 3. NRDE-1 is required for RNAi-directed H3K9 methylation. 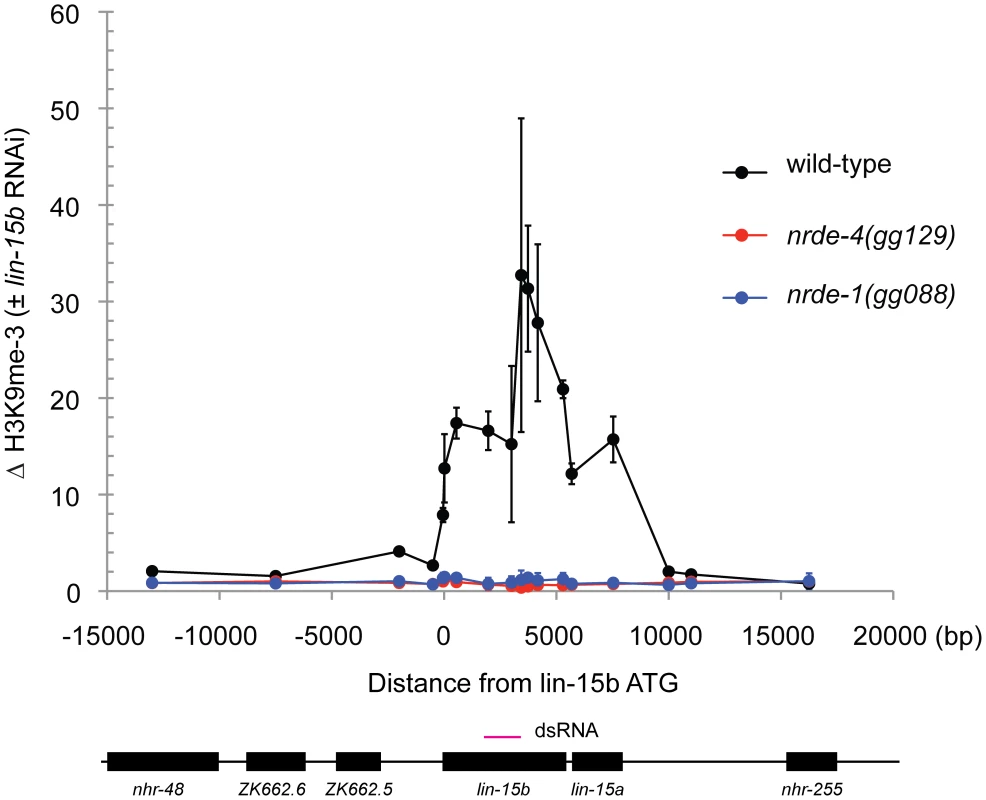
Chromatin Immunoprecipitation (ChIP) with anti-H3K9me3 (Upstate, 07-523) was performed on extracts derived from embryos of animals exposed to +/− lin-15b RNAi. Co-precipitating H3K9me3 DNA was quantified with qRT-PCR and data are expressed as ratios of samples exposed to lin-15b RNAi or no RNAi (n = 3 +/− s.d). RNAi directs NRDE-1 to associate with chromatin
We asked if the NRDE factors themselves might become associated with chromatin in response to RNAi. In order to address this question, we performed NRDE-1/2/3 ChIP experiments before or after exposure of animals to dsRNA. In response to lin-15b RNAi, we did not detect any significant increase in the association of NRDE-2 or NRDE-3 with chromatin at the lin-15b gene (Figure 4a). Interestingly, NRDE-1 precipitated ∼6× more lin-15b DNA following lin-15b RNAi (Figure 4a). In nrde-2(−) and nrde-3(−) animals, lin-15b RNAi failed to trigger an increase in lin-15b DNA that co-precipitated with NRDE-1 (Figure 4b). We conclude that NRDE-1 is able to IP chromatin of a gene that has been targeted by RNAi, and that the association of NRDE-1 with chromatin requires NRDE-2 and NRDE-3. It is possible that the ability of NRDE-1 to co-precipitate with chromatin may occur as an indirect consequence of NRDE-1/pre-mRNA interactions. To address this issue, we turned our attention to nrde-4.
Fig. 4. NRDE-1 associates with chromatin at genomic loci targeted by RNAi. 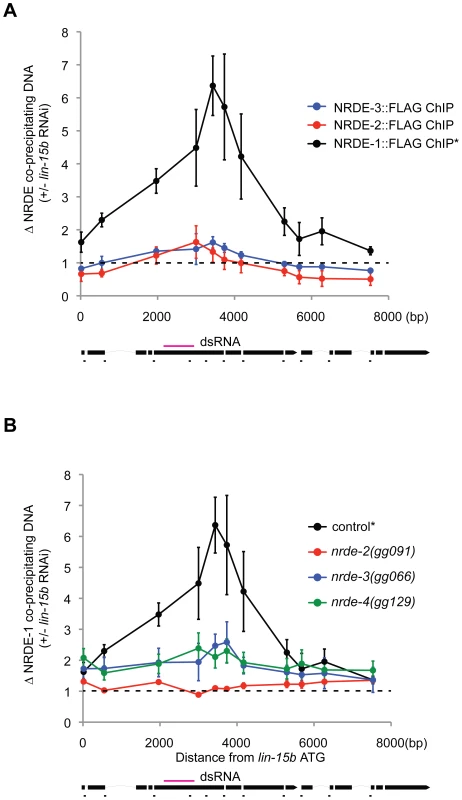
(A) FLAG::NRDE co-precipitating DNA was quantified with qRT-PCR. Data are expressed as ratios of NRDE associated DNA with or without lin-15b RNAi. Data was normalized to input. NRDE-1 (n = 4, +/− s.e.m.), NRDE-2 (n = 3, +/− s.e.m.), NRDE-3 (n = 3 +/− s.e.m.) * indicates the same FLAG::NRDE-1 ChIP data are shown in panel A and B. (B) Genetic requirements for RNAi-driven NRDE-1/chromatin association. FLAG::NRDE-1 co-precipitating DNA from indicated genetic backgrounds was quantified with qRT-PCR. Data are expressed as ratios of NRDE-1 co-IPed DNA with or without lin-15b RNAi. Control (n = 4, +/− s.e.m.), for all other strains (n = 3,+/− s.e.m.). NRDE-4 is required for RNAi-directed recruitment of NRDE-1 to chromatin
We mapped and cloned nrde-4 (Figure S4). nrde-4 is predicted to encode a protein containing 788 amino acids [33]. Database searches revealed that nrde-4 is conserved within other nematode species, but not in other species. nrde-4 encodes a predicted bipartite nuclear localization signal (NLS) and no other obvious protein domains (Figure S4). NRDE-4 is required for silencing nuclear localized RNAs (Table 1), for linking small RNAs to the inhibition of transcription (Figure 1b), and for linking small RNAs to H3K9 methylation (Figure 3). Interestingly, the recruitment of NRDE-1 (and NRDE-2/3) to pre-mRNA was largely unaffected in animals lacking NRDE-4 (Figure 2c, 2e, Figure S5). NRDE-4 was, however, required for recruitment of NRDE-1 to chromatin in response to RNAi (Figure 4b). These data indicate that NRDE-4 functions downstream of NRDE-1/2/3/pre-mRNA interactions during nuclear RNAi. These data also demonstrate that the ability of NRDE-1 to associate with chromatin is dissociable from the ability of NRDE-1 to associate with pre-mRNA, supporting the idea that NRDE-1 associates with chromatin at genomic sites targeted by RNAi.
Endo-siRNAs direct NRDE-dependent H3K9 methylation
C. elegans express at least three types of endogenous small RNAs; the microRNAs, the piRNAs, and the endo-siRNAs. A sub-set of the endo-siRNAs requires ERI-1 for their expression [36], [37]. NRDE-3 associates with the ERI-1-dependent endo-siRNAs, but not the other classes of endogenous small RNAs [15], [37]. Five lines of evidence cumulatively argue that ERI-1 dependent endo-siRNAs are able to direct the deposition of H3K9me marks in C. elegans. First, in animals that fail to express endo-siRNAs H3K9me marks are depleted at genomic regions exhibiting sequence complementarity to endo-siRNAs. For instance, e01g4.5 siRNAs are amongst the most abundant endo-siRNAs expressed in C. elegans [36]. eri-1(−) animals do not express e01g4.5 endo-siRNAs ([36], [37] and Figure 5a). We conducted H3K9me ChIP and detected a ∼6× depletion of H3K9me marks at the e01g4.5 gene in eri-1(−) animals (Figure 5b). The changes in H3K9me marks were restricted to genomic regions exhibiting homology to endo-siRNAs; surrounding genomic regions, which are not homologous to known small regulatory RNAs, did not exhibited altered H3K9me marks (Figure 5b). Second, in nrde-1/2/3/4 mutant animals we observed a similar localized depletion of H3K9me marks at e01g4.5 (Figure 5c and Figure S6). Third, the e01g4.5 pre-mRNA was over-expressed 2–5× in eri-1 and nrde-1/2/3/4 mutant animals ([15], Figure S7, and data not shown). Fourth, we performed NRDE-1 RIP and quantified the amount of e01g4.5 pre-mRNA that co-precipitated with NRDE-1. We conducted this experiment in both nrde-2(+) and nrde-2(−) animals as NRDE-2 is required for NRDE-1 recruitment to pre-mRNAs in response to feeding RNAi. We found that NRDE-1 associated with ∼5× more e01g4.5 pre-mRNA in nrde-2(+) animals than in nrde-2(−) animals (Figure 5d). These data suggest that NRDE-1 can associate with pre-mRNAs that are homologous to endo-siRNAs, and that this process depends upon components of the Nrde pathway. Five, we detected a subtle and complex, yet reproducible, increase in transcription at the e01g4.5 gene in eri-1 and nrde-1/2/4 mutant animals (Figure 5e and Figure S8). Taken together, these data indicate that e01g4.5 endo-siRNAs are able to direct chromatin modification in C. elegans and that this process requires the Nrde pathway.
Fig. 5. Endo-siRNAs promote H3K9 methylation. 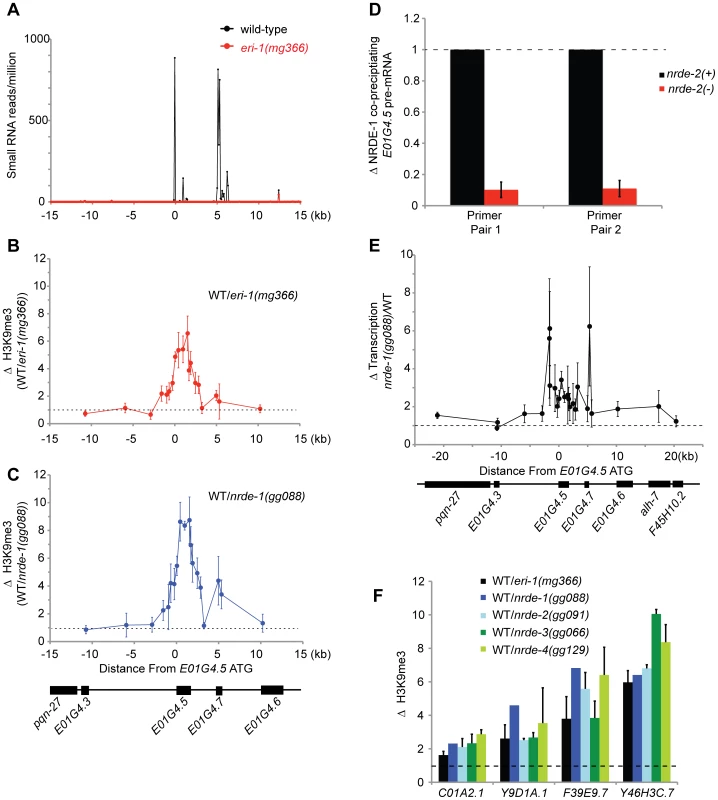
(A) e01g4.5 is an endo-siRNA target. Small RNAs cloned from wild type or eri-1(mg366) L4 larval animals [36] were counted in 100 bp non-overlapping windows across a 30 kb region surrounding e01g4.5. Small RNAs were normalized to total sequenced small RNAs from each sample. (B–C) eri-1(−) and nrde-1(−) animals are depleted for H3K9me3 at e01g4.5. H3K9me3 ChIPs were performed in wild-type (WT), eri-1(mg366), and nrde-1(gg088) animals. Data are represented as ratios of H3K9me3 co-precipitating DNA in WT/eri-1(mg366) (B) or WT/nrde-1(gg088) (C). Data in B and C are normalized to ChIPed eft-3 DNA (n = 3, +/− s.d.). (D) NRDE-1 associates with the pre-mRNA of an endo-siRNA target. Co-precipitating FLAG::NRDE-1 RNAs were isolated from +/− nrde-2 animals, converted to cDNA, and quantified with qRT-PCR. Data are expressed as ratios of NRDE-1 associated e01g4.5 pre-mRNA +/− nrde-2 (n = 2, +/− s.d.). (E) Endo-siRNAs inhibit transcription. NRO transcription analysis from wild-type and nrde-1 mutant animals. Data are represented as a ratio of transcription in nrde-1(−)/WT (n = 4, +/− s.e.m.). The genomic region surrounding e01g4.5 is depicted below the graph. (F) endo-siRNAs direct H3K9me marks at genomic loci homologous to endo-siRNAs. H3K9me3 ChIPs were performed in wild-type (WT) or animals of the indicated genotypes. Data are expressed as ratios of H3K9me co-precipitating DNA in WT/indicated genotype. eri-1(−) (n = 3 +/− s.d.), nrde-1(−) (n = 1) , nrde-2(−) (n = 2, +/− s.d.), nrde-3(−) (n = 2, +/− s.d.), nrde-4(−) (n = 3, +/− s.d.). Lastly, we investigated the generality of small RNA-mediated chromatin regulation in C. elegans. We queried seven additional genomic sites that exhibit sequence homology to eri-1-dependent endo-siRNAs. At four of these loci H3K9me marks were depleted in eri-1 and nrde-1/2/3/4 animals (Figure 5f). At three of these loci, no significant differences in H3K9me marks were observed. We conclude that Nrde-dependent endogenous small RNA-mediated chromatin modification occurs at multiple loci in C. elegans.
Discussion
Here we report that small RNAs are necessary and sufficient to direct chromatin modification in C. elegans. We show that a class of endogenous small RNAs, termed the endo-siRNAs, direct H3K9 methylation at discrete genomic loci, and that this process requires the Nrde pathway. Finally, we identify two novel nuclear RNAi factors including NRDE-1, which we show is recruited to pre-mRNAs and chromatin by RNAi, and is required to link small RNAs to chromatin regulation.
Hierarchical assembly of nrde factors on nascent RNAs
In S. pombe silencing factors assemble upon nascent transcripts during nuclear RNAi [3]. Here, we present evidence that nascent transcripts serve a similar role in C. elegans. The Ago NRDE-3 is guided to nascent transcripts via base pairing between NRDE-3 bound siRNAs and nascent transcripts [15]. In nrde-1 mutant animals, NRDE-3 can still associate with the target pre-mRNA, but nuclear silencing does not occur (Figure 2c). Thus, NRDE-3 bound siRNAs provide the information of where to silence, but additional downstream factors, such as NRDE-1, are required for silencing to occur. NRDE-3 is required for recruitment of NRDE-2 to pre-mRNA in response to RNAi [16]. NRDE-3 and NRDE-2 are required for the recruitment of NRDE-1 to pre-mRNAs in response to RNAi (Figure 2e). Thus, we propose that the NRDE factors assemble in a hierarchical manner on pre-mRNA; NRDE-3 identifies pre-mRNAs, and in association with NRDE-2, recruits NRDE-1 to pre-mRNAs that have been targeted by RNAi (Figure 6).
Fig. 6. Model of NRDE pathway. 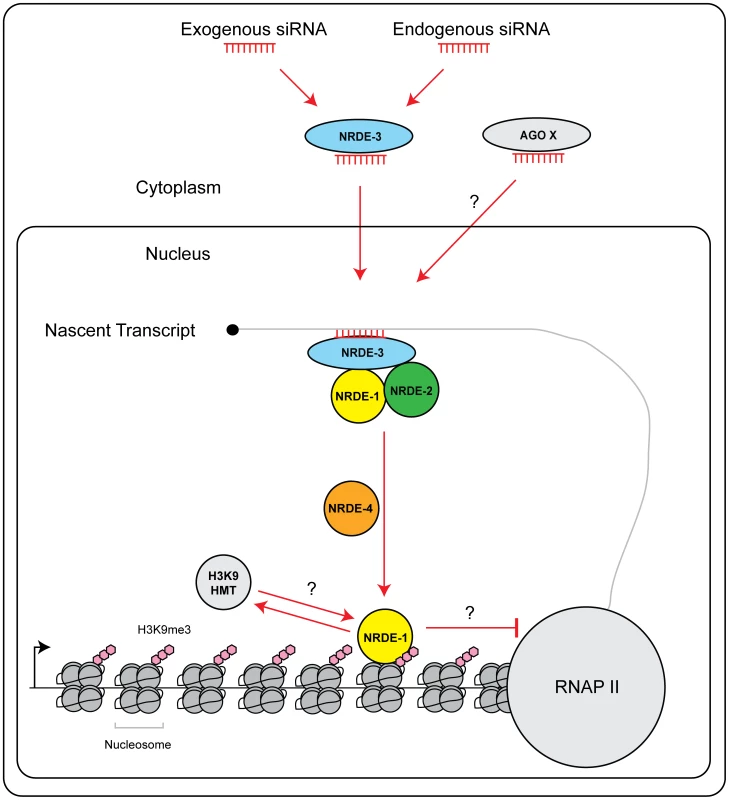
siRNAs (from either an exogenous or endogenous source) are bound by the Ago NRDE-3 in the cytoplasm and escorted into the nucleus. Once in the nucleus, NRDE-3/siRNA complexes bind nascent transcripts synthesized by RNAP II. NRDE-3/siRNA complexes recruit NRDE-2, and NRDE-2/3 recruit NRDE-1 to these nascent transcripts. NRDE-1 is deposited on chromatin in a NRDE-4 dependent manner. An unknown histone methyltransferase (HMT) catalyzes the methylation of histone 3 lysine 9 (H3K9me3). H3K9me marks may facilitate recruitment of NRDE-1 to chromatin. Alternatively, NRDE-1 may recruit an H3K9 methyltransferase to sites of RNAi. Together, H3K9me marks and NRDE-1 inhibit RNAP II elongation. Additional small RNAs and Argonaute proteins (Ago X) may engage NRDE-1/2/4 to direct chromatin modifications. NRDE-1 and NRDE-4 in nuclear RNAi
What is the role of NRDE-4 in nuclear RNAi? Our preliminary investigation has shown that: NRDE-4 is required to link small RNAs to transcription and chromatin regulation. Interestingly, we find that NRDE-4 is not required for small RNA-directed NRDE-1/pre-mRNA association, but is, required for the recruitment of NRDE-1 to chromatin. Therefore, it seems reasonable to speculate that one role of NRDE-4 during nuclear RNAi may be to load/stabilize NRDE-1 on chromatin, following the recruitment of NRDE-1 to pre-mRNAs by NRDE-2/3 (Figure 6).
What is the role of NRDE-1 in nuclear RNAi? In response to RNAi, NRDE-1 co-precipitates with both pre-mRNAs and chromatin. We did not detect an association of NRDE-3 or NRDE-2 with chromatin despite the fact that NRDE-2/3 are able, like NRDE-1, to associate with pre-mRNA in response to RNAi (Figure 4a). These data hint that NRDE-1 may possess a chromatin associating property not exhibited by NRDE-2/3. We considered the possibility that NRDE-1 might IP chromatin indirectly via pre-mRNA/RNAP II intermediates. However, we found that in nrde-4(−) animals, NRDE-1 is recruited to pre-mRNAs by RNAi, but does not become associated with chromatin (Figure 2e and Figure 4b). These data demonstrate that the RNA and chromatin associating properties of NRDE-1 can be separated. Additionally, we find that NRDE-1 association with RNA occurs predominantly 5′ to the site of RNAi, whereas NRDE-1 association with chromatin occurs predominantly 3′ to the site of RNAi (Figure 2e and Figure 4a). Taken together, these data argue that NRDE-1 associates with chromatin in response to RNAi, and that NRDE-1 interacts with pre-mRNAs first and chromatin second during nuclear silencing processes (Figure 6). The question then becomes; what is the role of NRDE-1 at chromatin?
H3K9me and nuclear RNAi
Here we show that small RNAs promote H3K9 methylation in C. elegans. We show that experimentally introduced small RNAs are sufficient to direct H3K9me marks at genomic sites targeted by RNAi. We also show that small RNAs are necessary to establish H3K9me marks; in animals that fail to express endogenous siRNAs, H3K9me marks are depleted at genomic sites homologous to endo-siRNAs. In S. Pombe, the RNAi machinery directs H3K9 methylation at pericentromeric repeats via recruitment of the H3K9 methyltransferase Clr4 to pre-mRNAs exhibiting homology to pericentromeric siRNAs [5]. Interestingly, fungi lacking H3K9me, due to loss of Clr4, fail to express abundant pericentromeric siRNAs [38]. Thus, H3K9me and the RNAi machinery are thought to comprise a self-reinforcing loop that facilitates heterochromatin formation at pericentromeric regions in S. pombe [39], [40]. We find that, in C. elegans, RNAi directs both H3K9 methylation and the association of NRDE-1 with chromatin. These data hint that C. elegans may employ a similar strategy as S. pombe for establishing heterochromatin; e.g. RNAi promotes H3K9 methylation and H3K9 methylation may help recruit components of the RNAi machinery, such as NRDE-1, to chromatin. In order to test this model, the C. elegans methyltransferase(s) responsible for depositing H3K9me marks in response to RNAi will need to be identified.
H3K9me and transcription
We find that H3K9me marks become distributed throughout a gene that has been targeted by RNAi (Figure 3). These data raise several interesting questions. First, how do H3K9me marks spread from the site of RNAi, and how are these marks prevented from spreading into adjacent genes? A simple model posits that the deposition of H3K9me marks (directed by small RNAs) is coupled to transcription in C. elegans. In other words, the act of transcription may alter chromatin in such a way as to permit (and limit) H3K9me spreading. Another question that arises is; what is the connection between H3K9 methylation and RNAP II transcription in C. elegans? We show that both endo-siRNAs and exo-siRNAs direct H3K9 methylation, which correlates with decreases in transcription. These data are consistent with the established repressive role of H3K9 methylation on transcription [8]. We find that RNAi-directed H3K9me marks peak 3′ to sites of RNAi (Figure 3). In addition, we find that NRDE-1 associates with chromatin predominantly 3′ to sites of RNAi (Figure 4a), and RNAP II transcription is inhibited by RNAi predominantly 3′ to the site of RNAi (Figure 1b). Therefore, H3K9me marks and NRDE-1 may contribute to the inhibition of RNAP II elongation by small RNAs (Figure 6). It should be noted, however, that while H3K9me marks peak 3′ to sites of RNAi, we observe H3K9 methylation throughout genes targeted by RNAi, hinting that H3K9me marks alone may not be sufficient to inhibit RNAP II transcription in C. elegans (Figure 3).
Why nuclear RNAi?
In S. pombe small RNAs primarily target repetitive genomic elements. RNAi-directed heterochromatization at pericentromeric repeats permits efficient segregation of chromosomes during meiosis [41]. In plants, small RNAs silence genomic regions enriched in transposons, pericentromeric regions, and rRNA genes [1]. Here we show that ERI-1-dependent endo-siRNAs direct the establishment of heterochromatic marks on chromatin. The biological role(s) of this small RNA-mediated chromatin regulation in C. elegans is unknown. The ERI-1-dependent endo-siRNAs are anti-sense to several hundred cellular mRNAs [36]. In general, these mRNAs appear to be poorly conserved and repetitive, hinting that these mRNAs may represent the products of dead and dying genes [42]. The purpose of nuclear RNAi may be to prevent expression of these dysfunctional genes. Alternatively, these mRNAs may simply serve as templates for the creation of small RNAs, which, in turn, regulate chromatin dynamics.
There are 26 Agos encoded in the worm genome, in addition to nrde-3 [43]. We have detected pleiotropic fertility defects exhibited by nrde-1/2/4(−), but not nrde-3(−), animals, hinting that other Ago proteins and, perhaps, other types of small RNAs, may engage NRDE-1/2/4 to promote H3K9 methylation during development (Figure S9). In support of this idea, we find that the recruitment of NRDE-1 to pre-mRNAs and chromatin, in response to RNAi, is not completely abolished in animals harboring null alleles of nrde-3 (Figure 2e). These data support the idea that other Ago proteins may engage the Nrde pathway to elicit nuclear silencing and chromatin regulation in C. elegans (Figure 2e, Figure 6). The identification of these Ago factors and their small RNA partners will be important for unraveling the cellular connections that exist between endogenous small RNAs and chromatin dynamics in metazoans.
Materials and Methods
Strains
N2, (YY160) nrde-1(gg088), (YY186) nrde-2(gg091), (YY158) nrde-3(gg066), (YY453) nrde-4(gg129), (GR1373) eri-1(mg366), (YY191) eri-1(mg366); nrde-1(gg088), (YY468) eri-1(mg366); nrde-4(gg129), (YY268) nrde-1(gg088); ggIS12[nrde-3p::3xflag::gfp::nrde-1], (YY464) nrde-1(gg088); nrde-2(gg091); ggIS12, (YY459) nrde-1(gg088); nrde-3(gg066); ggIS12, (YY462) nrde-1(gg088); nrde-4(gg129); ggIS12, (YY174) ggIS1[nrde-3p::3xflag::gfp::nrde-3], (YY225) rde-1(ne219); ggIS1, (YY228) nrde-1(gg088); ggIS1, (YY454) nrde-4(gg129); ggIS1, (YY230) rrf-1(pk1417); ggIS1, (YY346) nrde-2(gg091); ggIS28[nrde-3p::3xflag::gfp::nrde-2].
Construction of plasmids and transgenic strains
For FLAG::GFP::NRDE-1 (referred to as GFP::NRDE-1 when assaying NRDE-1 expression or FLAG::NRDE-1 when referring to NRDE-1 immunoprecipitation or western blotting) the nrde-1 coding region and predicted 3′UTR were amplified by PCR from genomic N2 DNA and inserted into the pSG082 plasmid 3′ to the nrde-3p::3xFLAG::GFP. Low copy integrated transgenes were generated by biolistic transformation [44].
RNAi
RNAi experiments were conducted as described previously [45]. The lir-1 and unc-15 bacterial clones were taken from the Ahringer library [46]. The lin-15b clone was described previously [15].
RNA IP (RIP)
RIPs were performed as described previously [15]. Hypochlorite-isolated embryos were used for all RIPs. FLAG::NRDE-1 and FLAG::NRDE-3 proteins were immuno-precipitated with anti-FLAG M2 antibody (Sigma, A2220).
Chromatin IP (ChIP)
ChIP experiments were performed as described previously [16]. Hypochlorite-isolated embryos were used for ChIP experiments. Isolated embryos were snap-frozen in liquid-Nitrogen before performing ChIP. FLAG::NRDE-1, FLAG::NRDE-2, and FLAG::NRDE-3 proteins were immuno-precipitated with anti-FLAG M2 antibody (Sigma, A2220). H3K9me3 antibody was from Upstate (07-523).
Nuclear run on (NRO) assay
NRO was performed as described previously [16]. Hypochlorite-isolated embryos were used for NROs.
cDNA preparation
RNAs were converted to cDNA by the iScript cDNA Synthesis Kit (Bio-Rad, 170–8890) following the vendor's protocol.
Supporting Information
Zdroje
1. SimonSAMeyersBC 2010 Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol
2. GrewalSI 2010 RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20 134 141
3. MoazedD 2009 Small RNAs in transcriptional gene silencing and genome defence. Nature 457 413 420
4. VerdelAJiaSGerberSSugiyamaTGygiS 2004 RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672 676
5. BayneEHWhiteSAKaganskyABijosDASanchez-PulidoL 2010 Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140 666 677
6. ZhangKMoschKFischleWGrewalSI 2008 Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15 381 388
7. NakayamaJRiceJCStrahlBDAllisCDGrewalSI 2001 Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 110 113
8. LachnerMJenuweinT 2002 The many faces of histone lysine methylation. Curr Opin Cell Biol 14 286 298
9. JanowskiBAHuffmanKESchwartzJCRamRNordsellR 2006 Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13 787 792
10. KimDHVilleneuveLMMorrisKVRossiJJ 2006 Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol 13 793 797
11. MorrisKVChanSWJacobsenSELooneyDJ 2004 Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305 1289 1292
12. WeinbergMSVilleneuveLMEhsaniAAmarzguiouiMAagaardL 2006 The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12 256 262
13. JanowskiBAYoungerSTHardyDBRamRHuffmanKE 2007 Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol 3 166 173
14. LiLCOkinoSTZhaoHPookotDPlaceRF 2006 Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 103 17337 17342
15. GuangSBochnerAFPavelecDMBurkhartKBHardingS 2008 An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321 537 541
16. GuangSBochnerAFBurkhartKBBurtonNPavelecDM 2010 Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465 1097 1101
17. KanellopoulouCMuljoSAKungALGanesanSDrapkinR 2005 Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19 489 501
18. Pal-BhadraMLeibovitchBAGandhiSGRaoMBhadraU 2004 Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669 672
19. Brower-TolandBFindleySDJiangLLiuLYinH 2007 Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21 2300 2311
20. LiCVaginVVLeeSXuJMaS 2009 Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137 509 521
21. MaloneCDBrenneckeJDusMStarkAMcCombieWR 2009 Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137 522 535
22. MoshkovichNLeiEP 2010 HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6 e1000880 doi:10.1371/journal.pgen.1000880
23. VaginVVSigovaALiCSeitzHGvozdevV 2006 A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313 320 324
24. YinHLinH 2007 An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450 304 308
25. MaineEMHauthJRatliffTVoughtVESheX 2005 EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired dna during C. elegans meiosis. Curr Biol 15 1972 1978
26. SheXXuXFedotovAKellyWGMaineEM 2009 Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet 5 e1000624 doi:10.1371/journal.pgen.1000624
27. KennedySWangDRuvkunG 2004 A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427 645 649
28. DufourcqPChanalPVicaireSCamutEQuintinS 1999 lir-2, lir-1 and lin-26 encode a new class of zinc-finger proteins and are organized in two overlapping operons both in Caenorhabditis elegans and in Caenorhabditis briggsae. Genetics 152 221 235
29. MooreMJProudfootNJ 2009 Pre-mRNA Processing Reaches Back to Transcription and Ahead to Translation. Cell 136 688 700
30. BosherJMDufourcqPSookhareeaSLabouesseM 1999 RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics 153 1245 1256
31. ClarkSGLuXHorvitzHR 1994 The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137 987 997
32. DavisMWHammarlundMHarrachTHullettPOlsenS 2005 Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6 118
33. Wormbase website. http://www.wormbase.org. Accessed 2011 March 2
34. AokiKMoriguchiHYoshiokaTOkawaKTabaraH 2007 In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26 5007 5019
35. TsaiCJMetsDGAlbrechtMRNixPChanA 2008 Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev 22 194 211
36. GentJILammATPavelecDMManiarJMParameswaranP 2010 Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37 679 689
37. PavelecDMLachowiecJDuchaineTFSmithHEKennedyS 2009 Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics 183 1283 1295
38. HalicMMoazedD 2010 Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140 504 516
39. NomaKSugiyamaTCamHVerdelAZofallM 2004 RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet 36 1174 1180
40. SugiyamaTCamHVerdelAMoazedDGrewalSI 2005 RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A 102 152 157
41. GrewalSIJiaS 2007 Heterochromatin revisited. Nat Rev Genet 8 35 46
42. PavalecD 2010 Identification of Novel ERI Factors and the Role of the ERI/DICER Complex in Endgoenous RNAi Madison University of Wisconsin 202
43. YigitEBatistaPJBeiYPangKMChenCC 2006 Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127 747 757
44. BerezikovEBargmannCIPlasterkRH 2004 Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res 32 e40
45. TimmonsLCourtDLFireA 2001 Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 103 112
46. KamathRSFraserAGDongYPoulinGDurbinR 2003 Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 231 237
Štítky
Genetika Reprodukční medicína
Článek The T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle SpecificationČlánek B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid FishesČlánek Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover RecombinationČlánek Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch SignalingČlánek Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Polo, Greatwall, and Protein Phosphatase PP2A Jostle for Pole Position
- Genome-Wide Association Analysis of Incident Coronary Heart Disease (CHD) in African Americans: A Short Report
- The T-Box Factor MLS-1 Requires Groucho Co-Repressor Interaction for Uterine Muscle Specification
- B Chromosomes Have a Functional Effect on Female Sex Determination in Lake Victoria Cichlid Fishes
- Analysis of DNA Methylation in a Three-Generation Family Reveals Widespread Genetic Influence on Epigenetic Regulation
- PP2A-Twins Is Antagonized by Greatwall and Collaborates with Polo for Cell Cycle Progression and Centrosome Attachment to Nuclei in Drosophila Embryos
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Pervasive Sharing of Genetic Effects in Autoimmune Disease
- DNA Methylation and Histone Modifications Regulate Shoot Regeneration in by Modulating Expression and Auxin Signaling
- Mutations in and Reveal That Cartilage Matrix Controls Timing of Endochondral Ossification by Inhibiting Chondrocyte Maturation
- Variance of Gene Expression Identifies Altered Network Constraints in Neurological Disease
- Frequent Beneficial Mutations during Single-Colony Serial Transfer of
- Increased Gene Dosage Affects Genomic Stability Potentially Contributing to 17p13.3 Duplication Syndrome
- Distinct Cdk1 Requirements during Single-Strand Annealing, Noncrossover, and Crossover Recombination
- Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency
- Suppression of Scant Identifies Endos as a Substrate of Greatwall Kinase and a Negative Regulator of Protein Phosphatase 2A in Mitosis
- Temporal Dynamics of Host Molecular Responses Differentiate Symptomatic and Asymptomatic Influenza A Infection
- MK2-Dependent p38b Signalling Protects Hindgut Enterocytes against JNK-Induced Apoptosis under Chronic Stress
- Specification of Corpora Cardiaca Neuroendocrine Cells from Mesoderm Is Regulated by Notch Signaling
- Genome-Wide Gene-Environment Study Identifies Glutamate Receptor Gene as a Parkinson's Disease Modifier Gene via Interaction with Coffee
- Identification of Functional Toxin/Immunity Genes Linked to Contact-Dependent Growth Inhibition (CDI) and Rearrangement Hotspot (Rhs) Systems
- Genomic Analysis of the Necrotrophic Fungal Pathogens and
- Celsr3 Is Required for Normal Development of GABA Circuits in the Inner Retina
- Genetic Architecture of Aluminum Tolerance in Rice () Determined through Genome-Wide Association Analysis and QTL Mapping
- Predisposition to Cancer Caused by Genetic and Functional Defects of Mammalian
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
- and but Not Interact in Genetic Models of Amyotrophic Lateral Sclerosis
- Gamma-Tubulin Is Required for Bipolar Spindle Assembly and for Proper Kinetochore Microtubule Attachments during Prometaphase I in Oocytes
- Ongoing Phenotypic and Genomic Changes in Experimental Coevolution of RNA Bacteriophage Qβ and
- Genetic Architecture of a Reinforced, Postmating, Reproductive Isolation Barrier between Species Indicates Evolution via Natural Selection
- -eQTLs Reveal That Independent Genetic Variants Associated with a Complex Phenotype Converge on Intermediate Genes, with a Major Role for the HLA
- The GATA Factor ELT-1 Works through the Cell Proliferation Regulator BRO-1 and the Fusogen EFF-1 to Maintain the Seam Stem-Like Fate
- and Control Optic Cup Regeneration in a Prototypic Eye
- A Comprehensive Map of Mobile Element Insertion Polymorphisms in Humans
- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Evidence for Hitchhiking of Deleterious Mutations within the Human Genome
- A Broad Brush, Global Overview of Bacterial Sexuality
- Global Chromosomal Structural Instability in a Subpopulation of Starving Cells
- A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation
- Glutamine Synthetase Is a Genetic Determinant of Cell Type–Specific Glutamine Independence in Breast Epithelia
- The Repertoire of ICE in Prokaryotes Underscores the Unity, Diversity, and Ubiquity of Conjugation
- Genome-Wide Association Analysis of Autoantibody Positivity in Type 1 Diabetes Cases
- Natural Polymorphism in BUL2 Links Cellular Amino Acid Availability with Chronological Aging and Telomere Maintenance in Yeast
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Ku Must Load Directly onto the Chromosome End in Order to Mediate Its Telomeric Functions
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An EMT–Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype
- Chromosome Painting Reveals Asynaptic Full Alignment of Homologs and HIM-8–Dependent Remodeling of Chromosome Territories during Meiosis
- Discovery of Sexual Dimorphisms in Metabolic and Genetic Biomarkers
- Regulation of p53/CEP-1–Dependent Germ Cell Apoptosis by Ras/MAPK Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



